Abstract
Mesenchymal stem cells (MSCs) are widely considered to be an attractive cell source for regenerative therapies, but maintaining multipotency and self-renewal in cultured MSCs is especially challenging. Hence, the development and mechanistic description of strategies that help promote multipotency in MSCs will be vital to future clinical use. Here, using an array of techniques and approaches, including cell biology, RT-quantitative PCR, immunoblotting, immunofluorescence, flow cytometry, and ChIP assays, we show that the extracellular domain of epithelial cell adhesion molecule (EpCAM) (EpEX) significantly increases the levels of pluripotency factors through a signaling cascade that includes epidermal growth factor receptor (EGFR), signal transducer and activator of transcription 3 (STAT3), and Lin-28 homolog A (LIN28) and enhances the proliferation of human bone marrow MSCs. Moreover, we found that EpEX-induced LIN28 expression reduces the expression of the microRNA LET7 and up-regulates that of the transcription factor high-mobility group AT-hook 2 (HMGA2), which activates the transcription of pluripotency factors. Surprisingly, we found that EpEX treatment also enhances osteogenesis of MSCs under differentiation conditions, as evidenced by increases in osteogenic markers, including Runt-related transcription factor 2 (RUNX2). Taken together, our results indicate that EpEX stimulates EGFR signaling and thereby context-dependently controls MSC states and activities, promoting cell proliferation and multipotency under maintenance conditions and osteogenesis under differentiation conditions.
Keywords: mesenchymal stem cells (MSCs), epithelial cell adhesion molecule (EpCAM), epidermal growth factor receptor (EGFR), pluripotency, proliferation, HMGA2, Lin28, multipotency
Introduction
MSCs3 are found in compact bone, tendon, adipose, placenta, and umbilical cord (1), where they have the potential to differentiate into multiple lineages, including bone, fat, cartilage, and muscle (1, 2). These cells are also known to be critical in the bone-healing process, in particular during healing of difficult nonunion fractures resulting from blood insufficiency, trauma, and other conditions. The regeneration of bone tissue is initiated by MSCs with the formation of soft and hard calluses (3). Because of these varied actions, MSCs are considered to be promising therapeutic candidates with wide-ranging clinical applications. The uses for MSCs are not only limited to tissue engineering, but also include utilization of reparative and immunomodulatory properties in wound healing and realignment of dysregulated immune systems (4). Recently, there have been many publications regarding MSC biology and clinical application, and to date, 32 clinical trials have been undertaken using MSCs to treat a variety of adult heart conditions, including acute myocardial infarction, ischemic cardiomyopathy, and nonischemic dilated cardiomyopathy (5). For an adult, 1.8 × 108 cells are typically required for cell therapy, or based on body weight, the number of required cells may be calculated as 6.64 × 106 cells/kg (3). Therefore, the further development of methods for efficient expansion of MSCs is an important undertaking. Aging of human MSCs is known to attenuate their proliferation, while increasing oxidative damage and senescence (6, 7). Therefore, the use of aged MSCs for autologous cell-based therapies is especially challenging (6, 8). Along with reduced proliferation of the MSC pool, aging is also associated with decreased proliferative capacity in MSC-derived osteoprogenitor cells, which leads to decreased osteoblast cell number and eventually hinders bone formation (7, 8).
EpCAM is a type I transmembrane protein with 314 amino acids and a molecular mass of 39–42 kDa (9). It contains an extracellular domain (EpEX, 265 amino acids), a single transmembrane domain, and a short intracellular domain (EpICD, 26 amino acids). EpCAM is a well-known tumor-associated antigen, which is enriched in various carcinomas and is involved in homotypic cell–cell adhesion in normal epithelium (9). Previous research demonstrated that active proliferation is associated with enhanced EpCAM expression in neoplastic tissues. Furthermore, EpCAM is known to be relatively stable within the membrane of normal epithelial tissue, but is prone to cleavage in cancer tissue (10). Maetzel et al. (10) first shed light on the mechanisms of EpCAM activation, showing that it occurs via regulated intramembrane proteolysis (RIP). During this process, EpCAM is cleaved, generating two products (EpEX and EpICD), which then induce EpCAM-mediated proliferative signaling (10). After RIP of EpCAM, EpICD associates with FHL2, β-catenin, and Lef-1 to form a nuclear complex that binds to DNA at Lef-1 consensus sites and regulates gene transcription, potentially contributing to carcinogenesis.
In a recent study, we reported that EpCAM is enriched in human embryonic stem cells (hESCs), where it not only serves as an important surface marker but also regulates the four Yamanaka factors (11). Similarly, EpCAM plays a critical role in regulating self-renewal, cancer-initiating ability, and invasiveness in colon cancer cells (12). It is also notable that overexpression of EpCAM or EpICD decreased the levels of p53 and p21 and increased the promoter activity of Oct4 during induced pluripotent stem cell (iPSC) derivation (13). Based on these findings, we further discovered that EpCAM/EpEX, together with Oct4 or Klf4 expression, can generate iPSCs (14). Despite this growing knowledge about EpCAM function in stem cells, the function of EpCAM/EpEX in human MSCs has not been previously described.
The main purpose for this study was to investigate whether EpCAM signaling can promote multipotency and increase cell proliferation in MSCs. Herein, we not only describe a novel molecular mechanism for the regulation of self-renewal in MSCs through EGFR–STAT3 signaling, but we also provide a new method for maintaining multipotency of MSCs that may be useful to advance research in regenerative medicine.
Results
EpEX enhances cell proliferation and self-renewal in mesenchymal stem cells
A recent study showed that CD49f increases growth of MSCs and sustains multipotency via the regulatory effects on Oct4 and Sox2 (15). We have previously defined EpCAM as a critical stem cell marker, and we showed that EpICD can regulate Oct4 and Sox2 gene expression by binding to their promoters (11). We also recently reported that EpCAM/EpEX cooperates with Oct4 or Klf4 to induce iPSC formation from mouse embryonic fibroblasts and discovered a novel mechanism through which EpCAM/EpEX regulates STAT3–HIF2α signaling (14). Based on these previous reports, we suspected that EpEX may be beneficial for maintenance of pluripotency in MSCs.
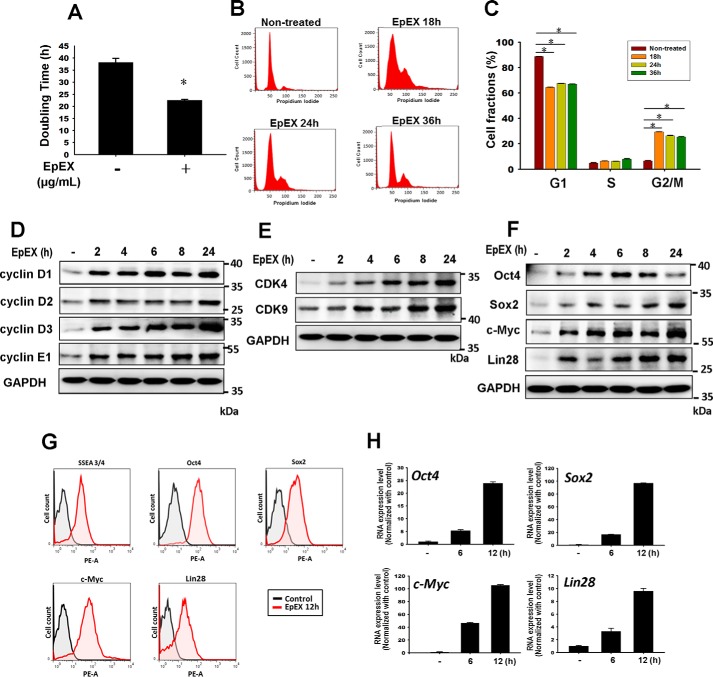
We used human bone marrow-derived MSCs to study the effects of EpEX, first investigating whether EpEX promotes cell proliferation. We assayed MSC doubling time after applying different doses of EpEX and found it was decreased in a dose-dependent manner. The most effective dose of EpEX was 3 μg/ml (Table S1), and EpEX shortened the doubling time of MSCs from 38.2 to 22.5 h (Table 1 and Fig. 1A). Next, we examined the effect of EpEX on cell cycle progression by flow cytometry with propidium iodide (PI) staining. We found that EpEX increased the percentage of cells in the G2/M phase from 6.5 to 29.3% at 18 h (Fig. 1, B and C). EpCAM has been reported to enhance cell cycle progression through up-regulation of the proto-oncogene c-Myc and cyclin A/E (16). Additionally, EpCAM is known to up-regulate cyclin D1 via its direct interaction partner, FHL2, and downstream events such as phosphorylation of the retinoblastoma protein, Rb (17). Therefore, we further asked whether EpEX can up-regulate the expression of cell cycle regulators and pluripotency markers. We first performed Western blotting and found that EpEX significantly increased the protein expression of cell cycle regulators, including cyclin A2, cyclin D1, cyclin D2, cyclin D3, and cyclin E1 (Fig. 1D), as well as CDK4 and CDK9 (Fig. 1E). Surprisingly, EpEX also significantly increased the protein expression of pluripotency markers, including Oct4, Sox2, c-Myc, and Lin28 (Fig. 1F). We then used flow cytometry to confirm that EpEX increased the protein levels of the stemness markers, Oct4, Sox2, c-Myc, and EpCAM, and we used qPCR to probe mRNA expression levels (Fig. 1, G and H). From these experiments, we found that EpEX accelerates MSC proliferation and enhances expression of multipotency markers.
Table 1.
Effect of EpCAM and EpEX on MSC doubling time
| EpEX (μg/ml) | 0 | 3 |
|---|---|---|
| P3 | 17.6 ± 0.3 h | 16.1 ± 0.1 h |
| P9 | 38.2 ± 1.7 h | 22.5 ± 0.4 h |
Figure 1.
EpEX increases cell proliferation and multipotency factors in mesenchymal stem cells. A, proliferation of MSCs was examined by measuring doubling time. MSCs were treated with EpEX (3 μg/ml) for 24 and 48 h. Cell number was counted, and then doubling time was calculated. B and C, MSCs were treated with EpEX for the indicated times. Flow cytometry and PI staining were performed to examine cell cycle progression. Fraction of cells in each phase (G1, S, and G2/M) of the cell cycle was evaluated. D–F, MSCs were treated with EpEX (3 μg/ml) for the indicated times. After treatment, protein expression of cell cycle regulators (cyclin D1, cyclin D2, cyclin D3, cyclin E1, CDK4, and CDK9) and pluripotency factors (Oct4, Sox2, c-Myc, and Lin28) was examined by Western blotting. The results of D and E were from one and the same experiment and are shown separately because of the size of the image; therefore, D and E were with the same GAPDH. G, MSCs were treated with EpEX (3 μg/ml) for the indicated times. After treatment, the protein expression of pluripotency factors (SSEA4, Oct4, Sox2, c-Myc, and Lin28) was examined by flow cytometry. H, MSCs were treated with EpEX (3 μg/ml) for the indicated times. After treatment, the gene expression of pluripotency factors (Oct4, Sox2, c-Myc, and Lin28) was examined by qPCR. Data represent the mean ± S.D. *, p < 0.05.
EpEX induces cell proliferation and self-renewal through EGFR signaling
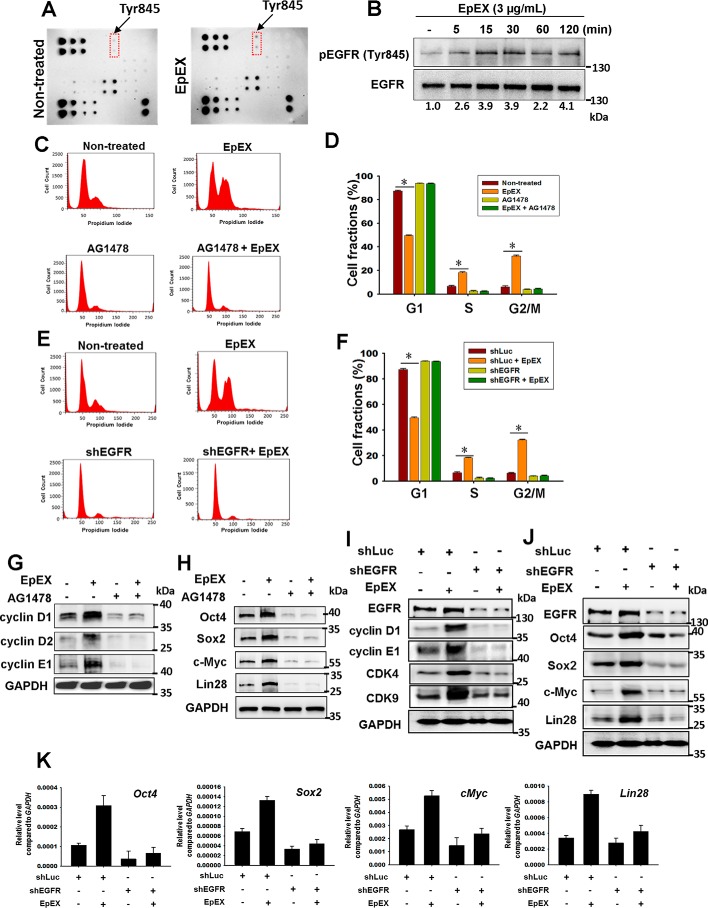
The EGF–EGFR-signaling pathway has been shown to be critical for cell proliferation (18) and self-renewal in MSCs (19). Based on the knowledge that EpEX contains an EGF-like domain and activates EGFR signaling, as measured by a receptor kinase array, we hypothesized that EpCAM/EpEX may serve as a cytokine or a growth factor to activate EGFR signaling and regulate cell growth and multipotency. Hence, we evaluated the phosphorylation state of EGFR by an EGFR membrane antibody array. We found that EpEX induced the phosphorylation of EGFR at Tyr-845 (Fig. 2A). By Western blotting, we confirmed EpEX induced EGFR phosphorylation at Tyr-845 in a time-dependent manner (Fig. 2B).
Figure 2.
EpEX up-regulates cell cycle regulators and stemness markers via EGFR signaling. A, MSCs were treated with EpEX (3 μg/ml) for 15 min, and the phosphorylation of EGF receptors was detected by an EGFR phosphorylation antibody array. B, MSCs were treated with EpEX (3 μg/ml) for the indicated times. Phospho-EGFR (Tyr-845) was detected by Western blotting. C and D, MSCs were pretreated with or without an EGFR inhibitor (AG1478, 25 μm) for 30 min, and then cells were incubated with EpEX for 18 h. After treatment, cell cycle progression was investigated by flow cytometry with PI staining. Fraction of cells in each phase (G1, S, and G2/M) of the cell cycle was evaluated. E and F, cells that expressed EGFR shRNA or shLuc were treated with EpEX for 18 h. After treatment, cell cycle progression was investigated by flow cytometry with PI staining. Fraction of cells in each phase (G1, S, and G2/M) of the cell cycle was evaluated. G and H, MSCs were pretreated with or without AG1478 and then stimulated by EpEX. The protein expression of cell cycle regulators (cyclin D1, cyclin D2, and cyclin E1) and pluripotency factors (Oct4, Sox2, c-Myc, and Lin28) was examined by Western blotting. I and J, MSCs expressing EGFR shRNA were stimulated by EpEX. After treatment, the protein expression of cell cycle regulators (cyclin D1, cyclin E1, CDK4, and CDK9) and pluripotency factors (Oct4, Sox2, c-Myc, and Lin 28) was examined by Western blotting. The results of I and J were from one and the same experiment and are shown separately because of the size of the image; therefore, I and J were with the same EGFR and GAPDH. K, MSCs expressing EGFR shRNA were stimulated with EpEX. After treatment, the gene expression of pluripotency factors (Oct4, Sox2, c-Myc, Lin28, and EpCAM) was examined by qPCR. Data represent the mean ± S.D. *, p < 0.05.
We also showed that both EGFR inhibitor (AG1478) and EGFR shRNA attenuated EpEX-induced cell cycle progression (Fig. 2, C–F). By Western blotting, we showed that inhibition of EGFR by shRNA or inhibitor abolished EpEX-induced protein expression of the cell cycle regulators cyclin D1, cyclin D2, cyclin E1, CDK4, and CDK9 (Fig. 2, G and I), and the pluripotency markers Oct4, Sox2, c-Myc, and Lin28 (Fig. 2, H and J). By qPCR, we found that shEGFR also reversed the EpEX-induced increases in transcript levels of pluripotency markers, including Oct4, Sox2, c-Myc, and Lin28 (Fig. 2K). Taking these results together, we conclude that EpEX may induce cell proliferation and multipotency in MSCs through activation of EGFR.
EpEX induces cell proliferation and self-renewal via STAT3
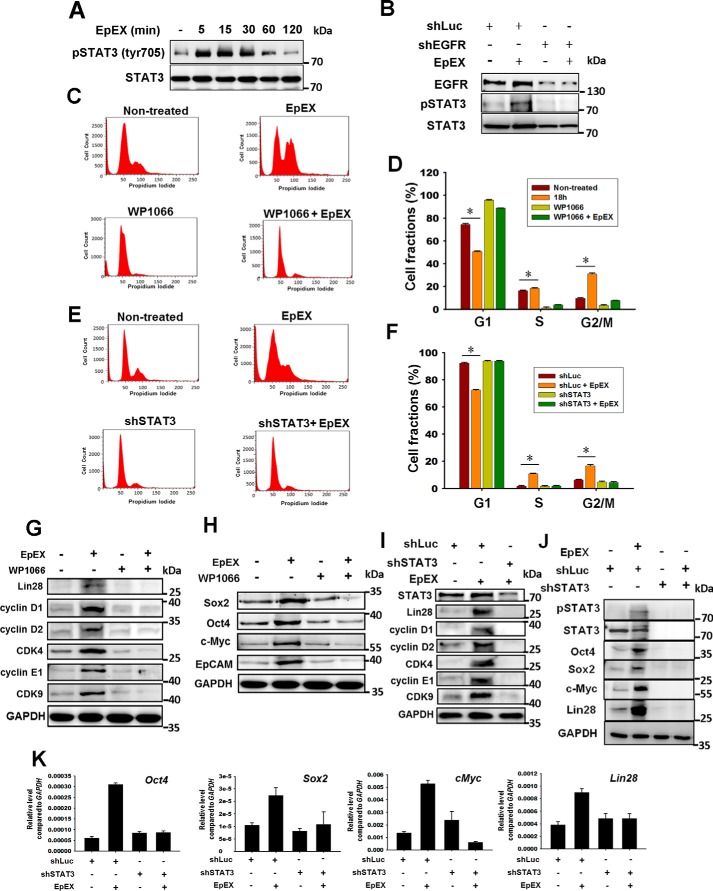
Previous studies have shown that STAT3 is a potent downstream effector of EGFR (20) and also that STAT3 plays a crucial role in pluripotency maintenance (21). Furthermore, we have demonstrated that STAT3 signaling is essential for EpCAM/EpEX promotion of iPSC reprogramming (14). Here, we found that EpEX stimulates STAT3 phosphorylation shortly after treatment (Fig. 3A). Moreover, we found that EpEX-induced phosphorylation of STAT3 was abolished by EGFR knockdown, suggesting that EpEX induces STAT3 signaling through EGFR activation (Fig. 3B).
Figure 3.
EpEX up-regulates cell cycle regulators and stemness markers via EGFR–STAT3 signaling. A, MSCs were treated with EpEX (3 μg/ml) for the indicated times. After treatment, the protein levels of total STAT3 and phospho-STAT3 (Tyr-705) were examined by Western blotting. B, MSCs were treated with EGFR shRNA or shLuc, and total STAT3 and phospho-STAT3 (Tyr-705) was examined by Western blotting with or without EpEX treatment. The results in Figs. 2, I and J, and 3B were from one and the same experiment and are shown separately because of the size of the image; therefore, Figs. 2, I and J, and 3B were with the same EGFR. C and D, cells were pretreated with a STAT3 inhibitor (WP1066, 5 μm), followed by stimulation with EpEX for 18 h. Cell cycle progression was investigated by flow cytometry with PI staining. Fraction of cells in each phase (G1, S, and G2/M) of the cell cycle was evaluated. E and F, cells expressing STAT3 shRNA were treated with EpEX for 18 h, after which cell cycle progression was investigated by flow cytometry with PI staining. Fraction of cells in each phase (G1, S, and G2/M) of the cell cycle was evaluated. G and H, MSCs were pretreated with or without WP1066 and then stimulated by EpEX. The protein levels of cell cycle regulators (cyclin D1, cyclin D2, cyclin E1, CDK4, and CDK9) and pluripotency factors (Oct4, Sox2, c-Myc, and Lin28) were examined by Western blotting. The results of G and H were from one and the same experiment and are shown separately because of the size of the image; therefore, G and H were with the same GAPDH. I and J, MSCs expressing STAT3 shRNA or shLuc were stimulated with EpEX. After treatment, the protein levels of cell cycle regulators (cyclin D1, cyclin D2, cyclin E1, CDK4, and CDK9) and pluripotency factors (Oct4, Sox2, c-Myc, and Lin 28) were examined by Western blotting. K, MSCs expressing STAT3 shRNA or shLuc were stimulated with EpEX. After treatment, the gene expression of pluripotency factors (Oct4, Sox2, c-Myc, Lin28, and EpCAM) was examined by qPCR. Data represent the mean ± S.D. *, p < 0.05.
Because EGF is a cognate ligand of EGFR, we also tested the effects of EGF on EGFR activation and STAT3 phosphorylation. Results showed that EpEX can induce EGFR phosphorylation as well as EGF (Fig. S1A). Moreover, we found that pretreatment of the EGFR inhibitor, AG1478, can attenuate the activation of EGFR by either EGF or EpEX (Fig. S1A). Interestingly, we also confirmed that EGF can induce the phosphorylation of STAT3, similar to EpEX (Fig. S1B), and that AG1478 can attenuate the activation of STAT3 by either EGF or EpEX (Fig. S1B).
We further investigated whether STAT3 signaling is involved in EpEX-induced cell growth and stemness of MSCs. By flow cytometry, we showed that the STAT3 inhibitor (WP1066) and knockdown of STAT3 both attenuated EpEX-induced changes in cell cycle progression (Fig. 3, C–F). By Western blotting, we also found that inhibition of STAT3 blocked EpEX-induced protein expression of the cell cycle regulators cyclin D1, cyclin D2, cyclin D3, cyclin E1, CDK4, and CDK9 (Fig. 3, G and I) and the pluripotency markers Oct4, Sox2, c-Myc, and Lin28 (Fig. 3, H and J). By qPCR, we showed that inhibition of STAT3 prevented EpEX-increased gene expression of the stemness markers Oct4, Sox2, c-Myc, and Lin28 (Fig. 3K). Furthermore, we also showed that knockdown of EpCAM decreased the level of phospho-STAT3 (Fig. S1C), stemness markers (Fig. S1D), and cell cycle regulators (Fig. S1E).
EpEX suppresses let-7 through EGFR–STAT3 signaling
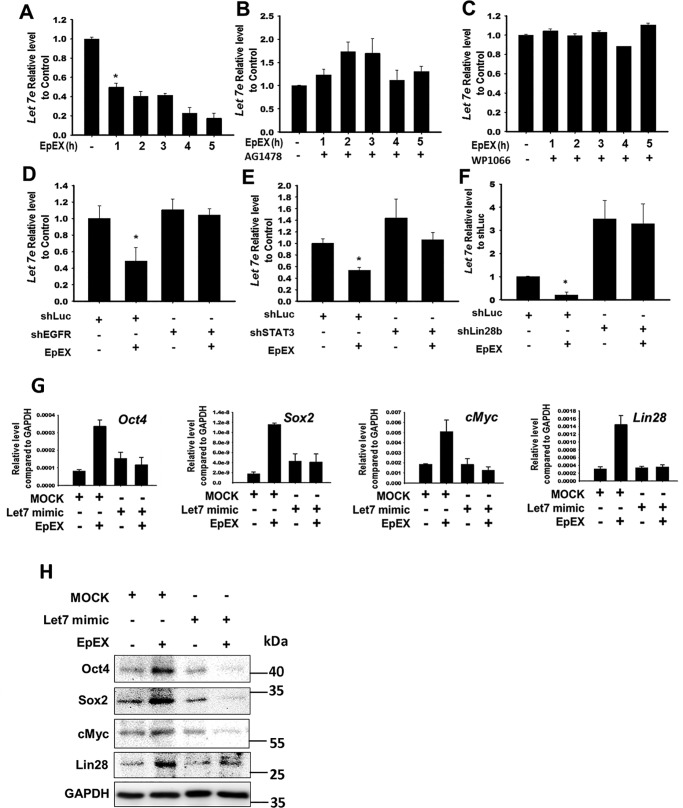
Previous studies have shown that Lin28 inhibits the miRNA, let-7, thereby increasing the levels of pluripotency factors (22–26). Thus, we further examined whether EpEX affects the level of let-7. By qPCR, we showed that EpEX decreased the level of let-7 (Fig. 4A). We next tested whether EpEX-induced inhibition of let-7 expression occurs via STAT3 and EGFR and found that EpEX suppression of let-7 expression was attenuated by shEGFR, shSTAT3, or shLin28 (Fig. 4, B–F). These results indicated that EGFR, STAT3, and Lin28 are necessary in EpEX regulation of let-7 expression.
Figure 4.
EpEX suppresses miRNA, let-7, through EGFR–STAT3–Lin28 signaling. A, cells were treated with EpEX (3 μg/ml), and the expression of let-7 was detected by qPCR. B and C, MSCs were pretreated with or without AG1478, or WP1066, and then stimulated with EpEX. The expression of let-7 was detected by qPCR. D–F, MSCs expressing EGFR shRNA, STAT3 shRNA, or Lin28b shRNA were stimulated with EpEX. The expression of let-7 was detected by qPCR. G, MSCs were transfected with a let-7 inhibitor or a let-7 mimetic, and then stimulated with EpEX. Expression of pluripotency factors (Oct4, Sox2, c-Myc, and Lin28) was examined by qPCR. H, MSCs were transfected with a let-7 mimetic, and then stimulated with EpEX. Protein levels of pluripotency factors (Oct4, Sox2, c-Myc, and Lin28) were examined by Western blotting. Data represent the mean ± S.D. *, p < 0.05.
Next, we used a let-7 mimetic to test whether let-7 suppression is necessary for EpEX-induced increase of pluripotency markers. We found that pretreatment with the let-7 mimetic abolished EpEX-induced gene and protein expression of Oct4, Sox2, c-Myc, and Lin28 (Fig. 4, G and H), confirming the importance of let-7 suppression in this process.
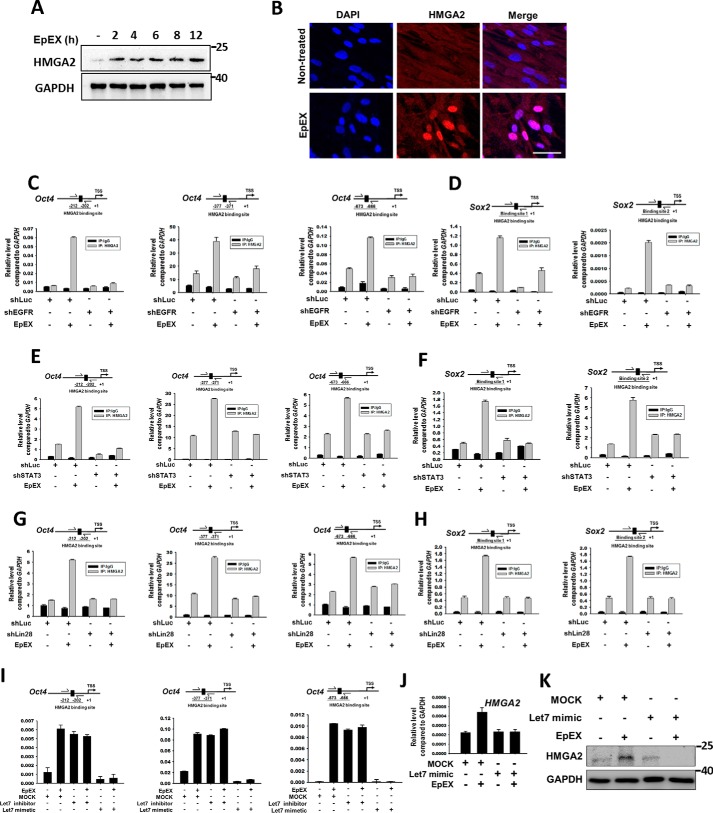
Similar to our findings, Lin28 was previously shown to decrease the level of let-7 (22); let-7 was further shown to suppress transcription of Oct4 and Sox2 through inhibition of transcription cofactors, AT-rich interaction domain molecule 3B (ARID3B), and high-mobility group AT-hook 2 (HMGA2) (27–29). The expression of HMGA2 is ubiquitous and abundant, and it has an important role during embryonic development (30). Moreover, HMGA2 expression has been shown to promote stem cell self-renewal, while decreased expression is associated with stem cell aging (31–34). In normal adult tissues, the level of HMGA2 is very low, but the protein is highly expressed in many types of cancer cells, where it facilitates oncogene expression (35–38). In addition, Lin28, which can suppress let-7 and up-regulate expression of HMGA2, is important for self-renewal (39) and maintenance of an undifferentiated state in cancer cells (40, 41). Based on this information, we hypothesized that EpEX may also regulate HMGA2. Interestingly, we found that EpEX not only induced the level of HMGA2, but also induced its nuclear translocation, as evidenced by Western blotting (Fig. 5A) and immunofluorescent staining (Fig. 5B). Because HMGA2 belongs to the high mobility group with the AT-hook DNA-binding domain family of proteins, it changes DNA conformation by binding to AT-rich regions in the DNA and interacts with other transcription factors, rather than directly activating transcription (34, 42). Therefore, we asked whether EpEX treatment induces HMGA2 to bind to the promoters of pluripotency genes. By ChIP, we showed that EpEX can induce HMGA2 binding to the promoters of Oct4 and Sox2, whereas ablation of EGFR, STAT3, or Lin28 prevented the effect (Fig. 5, C–H). We further tested whether EpEX-induced HMGA2 binding depends on let-7. Pretreatment with the let-7 mimetic abrogated the effect of EpEX, whereas treatment of the let-7 inhibitor was sufficient to induce the binding of HMGA2 to the promoters of Oct4 (Fig. 5I). In addition, we showed that the let-7 mimetic abolished EpEX-induced gene and protein expression of HMGA2 (Fig. 5, J and K).
Figure 5.
EpEX up-regulates HMGA2 and increases its binding to the promoters of Oct4 and Sox2 through EGFR–STAT3–Lin28–let-7 signaling. A, MSCs were treated with EpEX (3 μg/ml) for the indicated times, and expression of HMGA2 was detected by Western blotting. B, MSCs were treated with EpEX (3 μg/ml), and the expression of HMGA2 was detected by immunofluorescence staining. Scale bar, 50 μm. C–H, MSCs expressing EGFR shRNA, STAT3 shRNA, or Lin28b shRNA were treated with EpEX (3 μg/ml) for 12 h. The promoter binding of HMGA2 was examined by ChIP assay. HMGA2 protein was pulled down by a specific anti-HMGA2 antibody. To detect the binding of HMGA2 to the Oct4 and Sox2 promoters, cross-linked DNA was isolated and then amplified with specific primers by qPCR. I, MSCs were treated with let-7 mimetic or let-7 inhibitor and then treated with EpEX (3 μg/ml) for 12 h. To detect the binding of HMGA2 to Oct4 promoters, cross-linked DNA was isolated and then amplified with specific primers by qPCR. J, MSCs were treated with let-7 mimetic and then treated with EpEX (3 μg/ml) for 12 h. The gene expression of HMGA2 was detected by qPCR. K, MSCs were treated with let-7 mimetic and then treated with EpEX (3 μg/ml) for 12 h. The protein abundance of HMGA2 was examined by Western blotting. The results of Figs. 4H and 5K were from one and the same experiment and are shown separately because of the size of the image; therefore, Figs. 4H and 5K were with the same GAPDH.
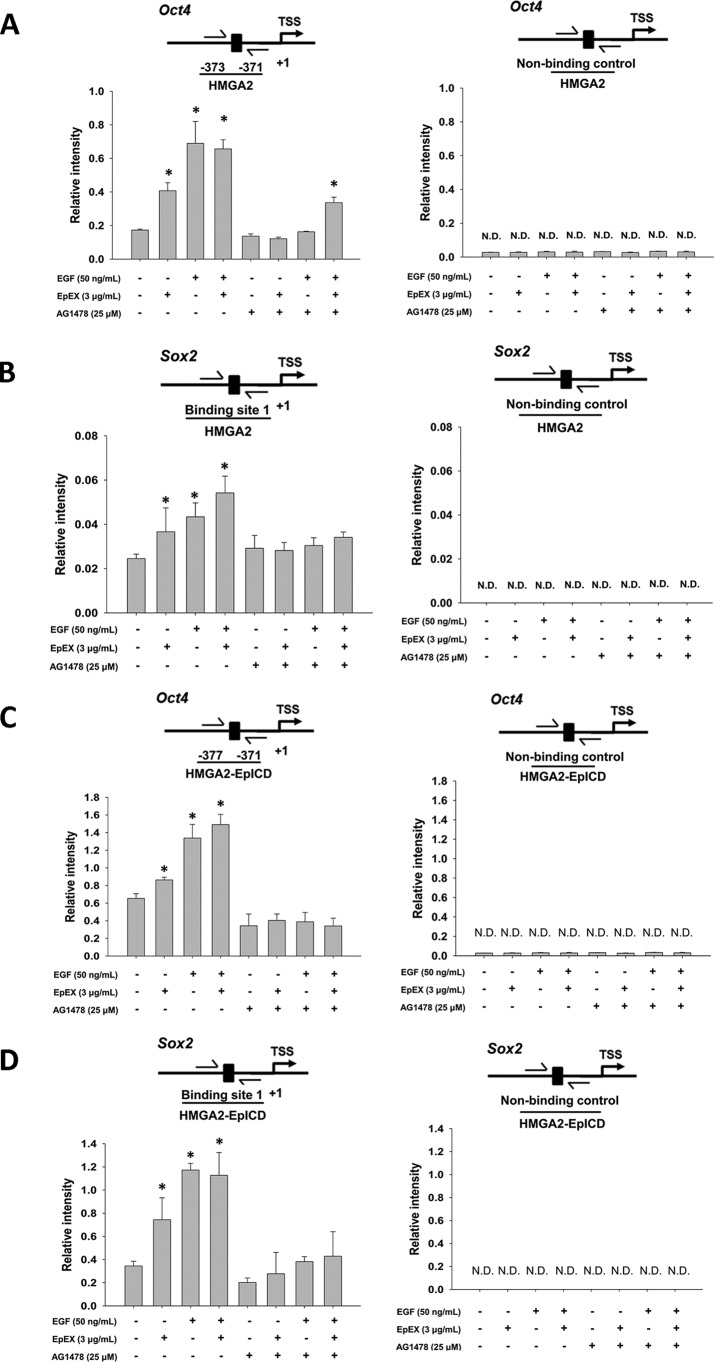
The result showed that EpEX can induce HMGA2 binding to the promoters of Oct4 and Sox2, whereas ablation of EGFR prevented the effect. We further tested whether EpEX- and EGF-induced HMGA2 binding depends on regulation of EGFR. Pretreatment with the EGFR inhibitor AG1478 abrogated the effect of EpEX, and EGF induced the binding of HMGA2 to the promoters of Oct4 and Sox2 (Fig. 6, A and B). Next, we investigated whether EpICD and HMGA2 could form a complex to bind to the promoter of Oct4 and Sox2. We sequentially pulled down EpICD and HMGA2, followed by probing of the binding site within the Oct4 and Sox2 promoter (Fig. 6, C and D). The result showed the EpEX and EGF induction of the EpICD–HMGA2 complex binding to Oct4 and Sox2 promoter depends on EGFR. Pretreatment with the EGFR inhibitor AG1478 abrogated the effect of EpEX, and EGF induced the binding of EpICD–HMGA2 to the Oct4 and Sox2 promoters.
Figure 6.
EpEX increases the binding of HMGA2 and EpICD to the promoter of Oct4 and Sox2. MSCs were treated with the EGF inhibitor AG1478 and then treated with EpEX (3 μg/ml) and EGF (50 nm). Binding of HMGA2 to the Oct4 (A) and Sox2 (B) promoter was examined by ChIP. HMGA2 was pulled down by a specific anti-HMGA2 antibody. The cross-linked DNA was isolated and then probed by qPCR with specific primers for the Oct4 and Sox2 promoter. MSCs were treated with EGF inhibitor AG1478 and then treated with EpEX (3 μg/ml) and EGF (50 nm). Binding of HMGA2–EpICD was examined by sequential ChIP. EpICD was pulled down by a specific anti-EpICD antibody, followed by pulldown with a HMGA2 antibody. To detect bound Oct4 (C) and Sox2 (D) promoter, the cross-linked DNA was isolated and then amplified by qPCR with specific primers. N.D., not detected. Data represent the mean ± S.D. *, p < 0.05, compared with control without EGF, EpEX, and AG1478 group.
Because we showed that EpEX treatment can induce Oct4 gene expression, we examined the binding of EpICD to the Oct4 promoter by single-ChIP and double-ChIP assays. We pulled down EpICD and probed for a specific binding site in the Oct4 promoters, finding that EpICD can indeed associate with the Oct4 promoter after cells are treated with let-7 inhibitor or mimetic (Fig. S2A). We then wanted to investigate whether EpICD and HMGA2 form a complex to bind to the promoter of Oct4. To do so, we sequentially pulled down EpICD and HMGA2 and then probed for the binding site within the Oct4 promoter in the presence of let-7 inhibitor or mimetic (Fig. S2B). Pretreatment with the let-7 inhibitor enhanced the EpEX-induced binding of HMGA2 or EpICD–HMGA2 to the promoter of Oct4, whereas pretreatment with let-7 mimetic abrogated the effect. Indeed, we found that EpEX facilitates EpICD–HMGA2 complex binding to the Oct4 promotor through the let-7 inhibition pathway.
EpEX promotes MSC osteogenesis by up-regulating RUNX2
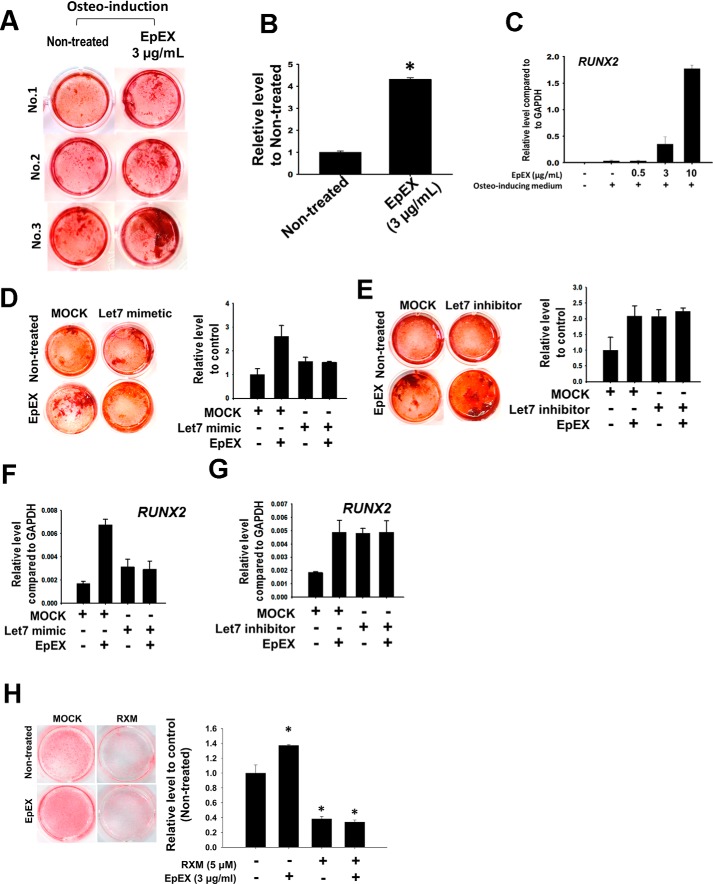
A previous study demonstrated that the up-regulation of Oct4 and Sox2 can promote osteogenesis of MSCs (43). Therefore, we surmised that EpEX may also promote osteogenesis via up-regulation of Oct4 and Sox2. Our results show that EpEX treatment during osteo-induction promoted osteogenesis when compared with cells without EpEX treatment (Fig. 7, A and B). We also measured gene expression of the osteogenetic marker, RUNX2, and found that EpEX increased the transcript level (Fig. 7C). We then analyzed whether RUNX2 participates with let-7 in EpEX-enhanced osteogenesis. First, we examined whether EpEX-enhanced osteogenesis depends on down-regulation of let-7. We found that pretreatment of let-7 mimetic can abolish EpEX enhancements in osteogenesis, whereas the let-7 inhibitor can increase osteogenesis (Fig. 7, D and E). Finally, we showed that let-7 mimetic attenuated EpEX-induced gene expression of RUNX2 (Fig. 7F), whereas the let-7 inhibitor increased RUNX2 gene expression (Fig. 7G). We also showed that treatment of RUNX2 inhibitor roxithromycin (RXM) can abolish the EpEX-enhanced osteogenesis in MSCs (Fig. 7H). In contrast with osteogenesis, EpEX treatment during adipo-induction inhibited adipogenesis (Fig. S3).
Figure 7.
EpEX enhances MSC bone formation by up-regulating RUNX2. A, MSCs were treated with EpEX for 14 days during osteo-induction. Calcium precipitation was measured by ARS staining to probe the efficiency of osteogenesis. This method shows higher calcium precipitation in EpEX (day 14)-treated cells than nontreated controls. B, quantification of osteogenesis, as measured by ARS staining, is shown for each group. C, MSCs were induced by osteogenetic medium and treated with EpEX at indicated doses. The gene expression of RUNX2 was examined by qPCR. D, MSCs were pretreated with let-7 mimetic and then treated with EpEX for 14 days during osteo-induction. ARS staining was performed to check the efficiency of osteogenesis. E, MSCs were pretreated with let-7 inhibitor and then treated with EpEX for 14 days during osteo-induction. ARS staining was performed to check the efficiency of osteogenesis. F, MSCs were pretreated with let-7 mimetic and then induced by EpEX. RUNX2 gene expression was measured by qPCR. G, MSCs were pretreated with let-7 inhibitor and induced by EpEX. RUNX2 gene expression was examined by qPCR. H, MSCs were treated with 0 or 5 μm RUNX2 inhibitor RXM and then treated with EpEX (3 μg/ml) for 21 days during osteo-induction. Calcium precipitation was measured by ARS staining to probe the efficiency of osteogenesis (left panel). The quantification results of the ARS staining are shown in the right panel. Data represent the mean ± S.D. *, p < 0.05, compared with control without RXM and EpEX group.
EpEX induces the phosphorylation and activity of TACE and γ-secretase
Previous studies indicate that EpCAM can be cleaved by the sheddase, TACE, leading to the release of soluble EpEX. This release may then trigger an autocrine cell-signaling response (10). Because EpCAM signaling is processed both by TACE and γ-secretase, we investigated the effect of EpEX on TACE and γ-secretase activities. We detected the phosphorylation and activation of TACE and γ-secretase in EpEX-stimulated MSCs. The results of these assays showed that the activation of TACE and γ-secretase was induced by EpEX treatment in MSCs (Fig. S4, A and B). We also showed the phosphorylation of TACE and γ-secretase was induced by EpEX (Fig. S4, C and D). Next, we used an EGFR inhibitor to examine whether EpEX-induced activation of TACE and γ-secretase requires EGFR signaling. We also investigated the upstream signaling that may result in activation of the TACE enzyme. ERK1/2 has been reported to regulate the activity of TACE, and we showed that EpEX can induce the EGFR-dependent phosphorylation of ERK1/2 (Fig. S4, E and F). We showed that EpEX-induced phosphorylation of TACE and presenilin-2 can be abolished by the addition of the EGFR inhibitor (Fig. S4, G and H). Next, we wanted to examine whether TACE and γ-secretase play roles in maintaining protein levels of cell cycle regulators and pluripotency factors. We found that knockdown of TACE or γ-secretase can inhibit the expression of cell cycle regulators and pluripotency markers (Fig. S5).
Discussion
In this study, we provide evidence for the novel concept that EpEX/EGFR signaling may aid multipotency and self-renewal of MSCs by providing a mitogenic signal. We further elucidated the signaling mechanism that underlies this phenomenon. We found that EpEX induces the phosphorylation of EGFR, which then initiates downstream events, including the sequential up-regulation of STAT3 and Lin28. Lin28 and STAT3 are responsible for inhibiting expression of the miRNA, let-7, which acts to control the expression of cell cycle regulators and pluripotency factors. Expression of these factors culminates in the stimulation of cell proliferation and enhancement of multipotency. These findings are consistent with a previous report that activation of the EGFR axis is important for cell proliferation and self-renewal in MSCs (19). Thus, our study uncovers a novel molecular mechanism in the sophisticated regulation of MSCs' multipotency and self-renewal.
MSCs are characterized by their ability to self-renew and differentiate into tissues of mesodermal origin, including bone, cartilage, adipose, and connective tissues. Thus, they contribute to many types of tissue regeneration (44). MSCs play vital roles in the repair and reconstruction of normal and injured tissue, mainly via interactions with other cell types, such as endothelial cells, vascular SMCs, and leukocytes (45). Therefore, it is of great interest to identify factors that may have a role in MSCs' expansion and maintenance of stem cell plasticity.
The molecular mechanisms that regulate proliferation and multipotency of MSCs are not well understood. In contrast, the molecular basis of proliferation and multipotency of ESCs has been the focus of intense research and is described in detail. In ESCs, the expression of three transcription factors, Oct4, Sox2, and Nanog, are essential for the maintenance of the stemness, self-renewal, and pluripotency (46, 47). Among these transcription factors, Oct4 belongs to the family of Pou-domain transcription factors and is found in developing embryos, developing endoderm, and developing neuroectoderm (48–50). Together with Sox2, Oct4 can up-regulate the expression of Nanog (51), and it is a dose-dependent pluripotency regulator that controls lineage commitment of ESCs (52). In addition to ESCs, the Oct4 gene has been found to be expressed in tumor cells but has not differentiated cells (53, 54). Instead, Oct4 expression has been confirmed in bone marrow (55), dental pulp (56), heart, liver (57), and adipose tissue-derived stem cells (58). Another of these core pluripotency transcription factors is Sox2. This protein is a member of the SRY-related HMG-box (SOX) transcription factor family and plays a diverse role in stem cell potency and maintenance, embryonic development, and cancer (59–62). It is closely co-regulated alongside core pluripotency factors Oct4 and Nanog in ESCs, embryonic carcinoma cells, and induced pluripotent stem cells (62–64). Recently, Sox2 has been implicated in the maintenance and differentiation of adult stem cells, and its expression has been reported in bone marrow, neuronal tissues, and sensory epithelia (55, 65). However, the roles of Oct4, Sox2, and Nanog are not well described in MSCs.
It has been speculated that the core pluripotency transcription factors play an important role in the maintenance of multipotency and self-renewal of adult stem cells, including MSCs. Adult stem cells exist within various tissues, with the purpose of repopulating the tissue after injury or physiological loss. Human MSCs, which are derived from bone marrow, are progenitors that can differentiate into cells of multiple types, including osteogenic, chondrogenic, adipogenic, and myogenic lineages (44, 66, 67). Like human ESCs, human MSCs depend on FGF to maintain self-renewal and pluripotency (68). FGF inhibits differentiation by bone morphogenetic protein signaling and sustains expression of Oct4, Sox2, and Nanog pluripotency-associated genes (69). Upon differentiation, expression of pluripotency transcription factors is expected to be down-regulated (70). However, there is some controversy among different research groups regarding the expression of pluripotency regulators in adult tissues (71).
Oct4 is a transcription factor that is highly expressed in undifferentiated embryonic stem cells and embryonic germ cells (72). A network of key factors, which includes Oct4, Nanog, and Sox2, is necessary for the maintenance of pluripotency, and down-regulation of Oct-4 has been shown to trigger differentiation (50, 73). Recent studies have also demonstrated that Oct4 is a useful germ cell tumor marker (74).
EpCAM was recently found to be highly expressed in ESCs and to promote pluripotency (13). Previously, we found that EpCAM expression in hESCs is regulated by an epigenetic mechanism, and it activates gene expression of c-Myc, Oct4, Nanog, Sox2, and Klf4 via EpICD nuclear activity. These events result in promotion of self-renewal and the maintenance of pluripotency in ES cells (11). However, the underlying molecular mechanisms of EpCAM-mediated effects on reprogramming are unclear.
We previously found that EpCAM plays an important role in regulating cancer-initiating abilities in colon cancer. In tumor-initiating cells, elevation of EpCAM enhanced the formation of tumor spheres in vitro and tumors in vivo and led to increased production of EpICD, which up-regulated expression of reprogramming factors (11). Meanwhile, another study showed that knockdown of EpCAM inhibited the expression of reprogramming factors and epithelial–mesenchymal transition-related genes, thereby suppressing tumor initiation, self-renewal, and invasiveness (12). Moreover, it has been reported that overexpression of EpCAM or EpICD decreased the expression of p53 and p21 and activated the promoter activity of Oct4 in the reprogramming of mouse embryo fibroblasts (13). However, the mechanistic role of EpCAM in pluripotency reprogramming is still unknown. In this study, we found that EpICD can associate with the Oct4 promoter and that this association may occur in a complex with HMGA2. Thus, we suggest that EpCAM not only regulates Oct4 by EpEX signaling, but EpICD also directly regulates Oct4 transcription.
The interaction between EGFR and EpCAM was explored previously (75). Liang et al. (75) showed that the extracellular domain of EpCAM (EpEX) can function as a growth factor and activate ERK and AKT through EGFR signaling. EpEX enhances colon cancer cell proliferation through activating EGFR to induce EpICD shedding and downstream β-catenin and HIF1α-mediated signaling (75). Bidirectional co-immunoprecipitation of endogenous proteins was performed in FaDu, Cal27, and HCT8 cells, revealing co-precipitation of EGFR and EpCAM. The report also used cross-linking experiments to show that EpEX binds directly to the extracellular domain of EGFR (76). Another previous study revealed that the GTP-binding protein, RAS associated with diabetes, can physically associate with EGFR to activate STAT3 and induce expression of the stem cell expression factors Oct4, Nanog, and Sox2, thereby enhancing self-renewal in malignant glioblastoma (77). Our results are consistent with this finding. We showed that EpEX can activate EGFR signaling, induce STAT3, and up-regulate cell cycle regulators and pluripotency factors (Fig. 1). Furthermore, our study shows that the EGFR–STAT3 axis may induce Lin28, which then blocks let-7 and up-regulates cell cycle regulators and stemness markers in MSCs (Fig. 2). Thus, EpEX can serve as a growth factor to stimulate EGFR signaling and trigger the cascade that can help MSCs to cell proliferation and increase the expression of stemness markers.
Several studies have reported that Lin28 inhibits let-7 (78, 79) and that Sox2 plays a fundamental role in regulating Lin28, allowing neural progenitor cell proliferation and preventing neuronal differentiation (79). Moreover, Lin28 can rescue neural progenitor cell proliferation and maintain neurogenic potential in the absence of Sox2 (79). Because Lin28 function is important in these ectoderm-derived progenitor cells, it is consistent that the Lin28/let-7 pathway is also important for MSC proliferation and self-renewal. Interestingly, STAT3 has been shown to down-regulate let-7 family members and miR-200 (80). According to Guo et al. (80), STAT3 enhances Lin28 expression by directly binding to the Lin28 promoter, resulting in the repression of let-7 and concomitant up-regulation of the let-7 target HMGA2. In this way, the STAT3–Lin28B–let-7–HMGA2 circuit can participate with miR-200–ZEB1 to coordinate cytokine-mediated reprogramming of breast cancer cells (80).
HMGA2 is enriched in hESCs (32) and is known to interact with nucleosomes to create a specific type of chromatin domain, which is critical for the establishment of both hESC identity and the regulation of differentiation (33). HMGA2 is critically involved in the regulation of hESC proliferation, where it acts through undifferentiated transcription factor 1 (UTF1) to exert its function (33). Additionally, high-mobility group B protein 2 (HMGB2) has been shown to be expressed in stem cells and to inhibit differentiation in MSCs (81). In this study, we not only showed that EpEX up-regulates EGFR–STAT3–Lin28–let-7, but we also report the novel finding that EpEX can regulate cell proliferation and enhance multipotency via the chromatin-remodeling protein, HMGA2. Furthermore, we show that the EpICD–HMGA2 complex can associate with the promoter of Oct4, suggesting that HMGA2 can associate with EpICD to regulate Oct4 in transcriptional level.
Osteoblasts and adipocytes are known to be derived from multipotential MSCs (82), and increased adipogenesis in bone marrow has been associated with a decrease in osteogenesis. A probable mechanism underlying the commitment of MSC differentiation is the activation of RUNX2, the key transcription factor for osteoblast and adipose differentiation (83, 84). In addition, we found that EpEX can increase the osteogenic markers during osteogenic induction (Fig. S6). A previous report indicated that adiponectin increased osteoblast differentiation by inhibiting osteoclastogenesis and osteoclast activity (85). Moreover, fully differentiated 3T3-L1 adipocytes significantly decreased mRNA and protein expression of RUNX2 in osteoblastic cells. The present results suggest that EpEX promotes MSC osteogenesis by up-regulating RUNX2, while inhibiting adipogenesis. These findings prompted us to further examine the impact of EpEX on adipogenesis, chondrogenesis, and myogenesis by detecting specific markers of these processes (Figs. S7 and S8). Interestingly, we showed that EpEX treatment increased the mRNA and protein levels of chondrogenesis marker, SOX9, in chondrogenesis-induced cultures (Figs. S7 and S8). However, in adipogenic and myogenic-induced cultures, EpEX had no significant impact on the mRNA and protein levels of specific markers, including adipogenesis marker PPARγ and myogenesis marker MyoD (Figs. S7 and S8). We also found that the protein levels of adipogenesis markers, adiponectin, fatty-acid synthase, and acetyl-CoA carboxylase, were not increased by EpEX (Fig. S8). Thus, we conclude that EpEX can promote osteogenesis and may promote chondrogenesis, but does not affect adipogenesis.
Because Notch1 signaling is also regulated by RIP, we further examined whether EpEX is capable of promoting Notch1 cleavage. We found that EpEX has no significant impact on the mRNA level for Notch1 or protein levels of Notch1 and cleaved Notch1 (Fig. S9).
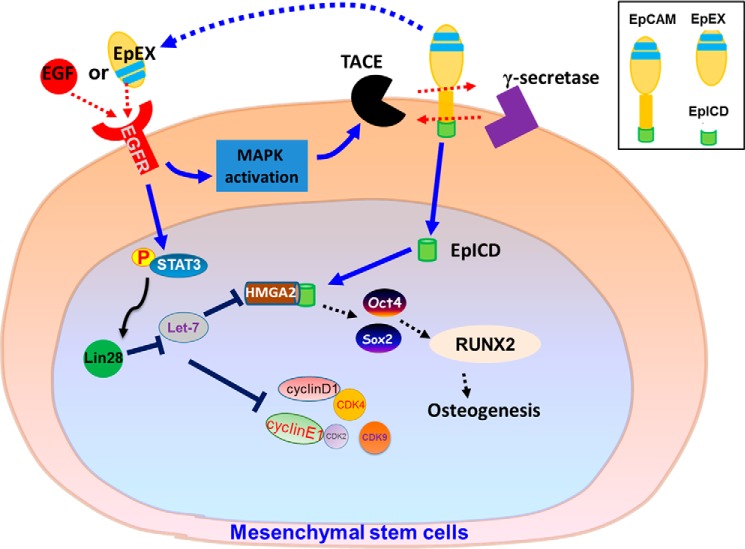
In conclusion, our investigation into the effects of EpEX/EGFR/STAT3 signaling in MSCs has produced fundamental knowledge that elucidates the role of EpCAM in promoting multipotency in this cell type. Our data allow us to propose the possible pathway shown in Fig. 8. These findings not only support the concept that EpEX serves as a cytokine with pleiotropic effects in MSCs, but they also suggest a new strategy for enhancing cell proliferation and multipotency of MSCs.
Figure 8.
Schematic showing the functional roles of EpCAM/EpEX in MSCs. Upon EpEX stimulation, phosphorylation of EGFR–STAT3 signaling is induced and subsequently up-regulates the level of Lin28 which inhibits let-7. When let-7 is inhibited, the transcription factor, HMGA2, is increased and binds to the promoters of Oct4 and Sox2. The EpEX-mediated increases of Oct4 and Sox2 can promote osteogenesis of MSCs during osteo-induction.
Experimental procedures
Cell culture
All experiments with primary human cells were conducted in accordance with relevant guidelines and regulations. Human primary bone marrow mesenchymal stem cells (BMMSCs) were purchased from LONZA and were cultured with Dulbecco's modified Eagle's media–low glucose (DMEM-LG) medium containing 16.6% FBS, 1 mm l-glutamine (Invitrogen), 100 μg/ml penicillin/streptomycin (Gibco). All cells were cultured at 37 °C and 5% CO2. All experiments on primary cells were performed within 10 passages.
Production and purification of EpEX–Fc recombinant protein
The DNA fragment encoding EpEX (amino acid residues 24–262) was amplified by PCR with PfuTurbo DNA polymerase (Stratagene). The PCR product was digested and ligated into pSecTag2 vector (Invitrogen) with C-terminal Fc tag to generate pSecTag2–EpEX–Fc. The EpEX–Fc fusion protein was produced using the Expi293FTM expression system (Thermo Fisher Scientific) and purified with protein G affinity chromatography (GE Healthcare) (75).
Plasmids and lentivirus preparation
For knockdown experiments, human EGFR, EpCAM, STAT3, and Lin28 shRNAs in the pLKO vector were obtained from RNAi core facility (Academia Sinica, Taipei). Lentivirus was produced according to standard protocols with minor modifications. In brief, 293T cells were seeded at a density of 70% in a 100-mm dish and transfected with packaging vectors (pCMV-ΔR8.91, containing gag, pol, and rev genes), envelope vectors (pMD2.G; VSV-G–expressing plasmid), and an individual shRNA vector. The shRNA plasmids were transfected into 293T cells by Polyjet transfection reagent (SignaGen Laboratories). After overnight incubation, the medium was changed to BSA-containing media. MSCs were infected with viral supernatant, containing Polybrene (8 μg/ml), for 24 h. The infection procedure was repeated, and cells were incubated in puromycin (2 μg/ml) for 7 days to select cells with stable shRNA expression.
Osteogenic differentiation
Human primary BMMSCs were cultured in DMEM-LG medium with 10% FBS. Fibroblasts were cultured in DMEM-HG with 10% FBS. To induce differentiation, cells (1 × 104 cells/cm2) were cultured with osteogenic induction medium (90% DMEM-HG, 10% FBS, 0.1 μm dexamethasone, 10 mm β-glycerophosphate, and 0.05 mm l-ascorbic acid phosphate). The media were replaced twice per week during the differentiation period.
Adipogenic differentiation
BMMSCs were cultured in adipogenic induction medium (Biological Industries, Kibbutz Beit-Haemek, Israel). The medium was replaced every 3 days during the differentiation period.
Chondrogenic differentiation
BMMSC pellets were formed by centrifugation at 500 × g for 10 min, and the pellets were incubated with chondrogenic induction medium (Biological Industries) overnight. The next day, the cell pellets formed a spherical aggregate and were treated with chondrogenic induction medium for 14 days.
Myogenic differentiation
BMMSCs were incubated in myogenic induction medium DMEM/Ham's F-12. DM1 (differentiation medium 1) contained 2% donor horse serum (DHS, Biochrom AG) and 1% l-glutamine; DM2 (differentiation medium 2) contained 2% DHS, 1% l-glutamine, 1 ng/ml basic FGF (Sigma), and 0.4 μg/ml dexamethasone (Sigma). The medium was replaced every 3 days during the differentiation period.
Alizarin Red S staining
After 14 days of osteogenic differentiation, cells were fixed with ice-cold 70% ethanol at −20 °C for 1 h and then washed with PBS. The cells were then stained with 40 mm Alizarin Red S (ARS) (pH 4.2) for 10 min and subsequently washed five times with double-distilled H2O before being air-dried. For quantification, the cells were incubated with 1 ml of acetyl pyridinium chloride buffer for 1 h to extract ARS, and the absorbance at 550 nm was recorded.
Quantitative real-time RT-PCR
Total RNA was extracted using TRI Reagent (Invitrogen), and 5 μg of total RNA was reverse-transcribed using oligo(dT) primer (Fermentas, Glen Burnie, MD) with SuperScript III reverse transcriptase (Invitrogen). Quantitative real-time RT-PCR (qPCR) was performed on cDNA using the Light Cycler 480 SYBR Green I Master kit (Roche Applied Science) and the LightCycler480 System (Roche Applied Science). The gene expression levels of each sample were normalized to the expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Western blot analysis and phosphokinase array
Western blotting was performed as described previously (86). Cells were lysed in lysis buffer (150 mm NaCl, 50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40), containing a protease inhibitor mix (Roche Applied Science). Nuclear fractions and cytoplasmic fractions were separated by the nuclear/cytosol fractionation kit according to the manufacturer's instructions (BioVision Inc., Milpitas, CA). Protein samples were separated by SDS-PAGE under denaturing conditions and transferred to a polyvinylidene difluoride membrane (Millipore). To probe for pluripotency markers, membranes were incubated with the indicated antibodies against Oct4 (1:1000, Abcam, Cambridge, UK), Nanog (1:1000, Genetex), Lin28 (1:1000, Genetex), or Sox2 (1:1000, Genetex). The CDK and cyclin antibodies were from the CDK and cyclin antibody sampler kits (Cell Signaling Technology, 9868 and 9869 respectively), including antibodies against cyclin D1, D2, and E1 and CDK4 and CDK9 (1:1000). EpCAM (1:1000, Genetex), phospho-EGFR (1:1000, Cell Signaling), EGFR (1:1000, Cell Signaling), Notch1 (1:1000, Cell Signaling), cleaved Notch (1:1000, Cell Signaling), adiponectin (1:1000, Cell Signaling), PPARγ (1:1000, Cell Signaling), acetyl-CoA carboxylase (1:1000, Cell Signaling), SOX9 (1:1000, Cell Signaling), Osterix/Sp7 (1:1000 R&D Systems), thrombopoietin, (1:1000, R&D Systems), MEPE/OF45 (1:1000 R&D Systems), HMGA2 (1:1000, Cell Signaling), or GAPDH (1:10000, Abcam) were also used. After incubation with primary antibody, the membranes were incubated with horseradish peroxidase–conjugated secondary antibodies, goat anti-mouse IgG (1:3000, Santa Cruz Biotechnology), or goat anti-rabbit IgG (1:3000, Santa Cruz Biotechnology). Finally, membranes were washed three more times and developed using Chemiluminescence Reagent Plus (Thermo Fisher Scientific, Runcorn, UK). The phosphokinase array kit (Proteome Profiler Antibody Array, R&D Systems) was used according to the manufacturer's instructions.
Flow cytometry analysis
Cells were dissociated with 0.25% trypsin/EDTA (1 mm) (Invitrogen) for 3 min, washed with FACS buffer (FACS buffer, PBS containing 1% fetal bovine serum), fixed in 4% PFA, and then permeabilized with 0.1% Triton X-100 in PBS. Subsequently, cells were stained with Oct4, Sox2, or Nanog antibodies (1:100, ab107156, Abcam, UK), washed, suspended in FACS buffer, and incubated with secondary antibody (1:200, Jackson ImmunoResearch) for 60 min at room temperature. Flow cytometry analysis was performed with a FACSCanto II flow cytometer (BD Biosciences).
Immunofluorescence staining
MSCs were seeded onto Millicell EZ slides (Millipore), and then iPSCs or ESCs were seeded. Cells were washed, fixed in 4% PFA for 10 min, and then permeabilized with 0.1% Triton X-100 for 10 min. Cells were stained with HMGA2 antibody (1:1000, Cell Signaling) for 60 min at room temperature and then washed with PBS. Then, the slides were incubated with goat anti-rabbit antibody conjugated with Alexa Fluor 568 (1:250; Invitrogen) for 1 h. After washing, the nuclei were stained with 4′,6-diamidino-2-phenylindole (1:1000) (Invitrogen). Cells were observed by confocal microscopy (TCS SP5; Leica, Wetzlar, Germany).
Chromatin immunoprecipitation
We performed chromatin immunoprecipitation (ChIP) with the PierceTM magnetic ChIP kit (Thermo Fisher Scientific), according to the manufacturer's instructions. In brief, the protein–DNA complexes were cross-linked with 1% formaldehyde and quenched by adding glycine to a final concentration of 200 mm. The chromatin complexes were sonicated to an average size of 250 bp by MISONIX Sonicator 3000. For immunoprecipitation, 4 μg of anti-HIF2α (Novus) was incubated with protein G beads (Invitrogen) for 4 h. The immunocomplexes were further incubated with chromatin for another 4 h. The bound fraction was isolated by protein G beads according to the manufacturer's instructions, and the immunocomplexes were subjected to reverse cross-linking. In double ChIP analysis, sequential (double) immunoprecipitation of two chromatin-binding proteins was performed to detect co-occupancy of proteins on promoter regions of pluripotency genes. We followed a previously described protocol (87). Briefly, we performed the first-round ChIP by using the anti-HMGA2 antibody (Cell Signaling Technologies). The cross-linked DNA–protein complex was washed and eluted with 10 mm dithiothreitol (DTT) at 37 °C for 1 h. The eluents were then diluted 50-fold in a ChIP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl (pH 8.1), 167 mm NaCl). A second-round of ChIP was performed with anti-HIF2α (Novus) or the control IgG antibody (Thermo Fisher Scientific). Chromatin was collected from the protein G-agarose beads after washing by elution with sodium bicarbonate/SDS buffer.
The immunoprecipitated DNA was recovered by a PCR purification kit (Thermo Fisher Scientific), and the purified DNA was subjected to real-time quantitative PCR for further analysis. The primers for the promoter of Oct4 and Sox2 were according to Ref. 29. Immunoprecipitation/input was calculated for each gene, and each gene was further normalized to the level of mouse β-actin promoter.
TACE activity and γ-secretase activity assay
ADAM17 activity was measured using the InnoZyme ADAM17 activity kit (Calbiochem). In brief, cell lysates were harvested and loaded into a TACE antibody-coated plate. After 1 h of incubation, the lysate was removed, and the plate was washed twice. Substrate was added into each well for 5 h at 37 °C. After incubation, the fluorescence signal of the reaction product was detected at excitation of 324 nm and emission of 405 nm. For the detection of γ-secretase activity, cell lysates were extracted, and 500 μg of protein was used. γ-Secretase activity was detected by γ-secretase substrate (35 μm).
Statistical analysis
All data are presented as mean ± S.E. for the indicated number of experiments. Unpaired Student's t test was performed to calculate the statistical significance of the expression percentages versus those of control cultures. A p value of less than 0.05 was considered statistically significant.
Author contributions
I.-I. K. and H.-C. W. conceptualization; I.-I. K., C.-C. L., and C.-H. C. data curation; I.-I. K., C.-C. L., and C.-H. C. formal analysis; I.-I. K., C.-C. L., and C.-H. C. validation; I.-I. K., C.-C. L., and C.-H. C. investigation; I.-I. K., C.-C. L., and C.-H. C. methodology; I.-I. K. and C.-C. L. writing-original draft; I.-I. K., C.-C. L., J. L., Y.-S. K., and H.-C. W. writing-review and editing; J. L. resources; H.-C. W. supervision; H.-C. W. funding acquisition; H.-C. W. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Jean Lu for providing human bone marrow MSCs. We are grateful for the technical assistance of the Imaging Core Facilities at the Institute of Cellular and Organismic Biology and the RNAi Core at the Genomics Research Center, Academia Sinica.
This work was supported by Academia Sinica and Ministry of Science and Technology Grants 106-0210-01-15-02 and 107-0210-01-19-01. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9 and Table S1.
- MSC
- mesenchymal stem cell
- EGFR
- epidermal growth factor receptor
- EGF
- epidermal growth factor
- EpCAM
- epithelial cell adhesion molecule
- EpEX
- extracellular domain of EpCAM
- EpICD
- epithelial intracellular domain of EpCAM
- CDK
- cyclin-dependent kinase
- ARS
- Alizarin Red S
- GAPDH
- of glyceraldehyde-3-phosphate dehydrogenase
- PFA
- paraformaldehyde
- FGF
- fibroblast growth factor
- DHS
- donor horse serum
- DMEM
- Dulbecco's modified Eagle's medium
- DMEM-LG
- DMEM-low glucose
- DMEM-HG
- DMEM-high glucose
- BMMSC
- bone marrow mesenchymal stem cell
- RXM
- roxithromycin
- PPAR
- peroxisome proliferator-activated receptor
- iPSC
- induced pluripotent stem cell
- TACE
- tumor necrosis factor-α–converting enzyme
- ESC
- embryonic stem cell
- hESC
- human ESC
- RIP
- regulated intramembrane proteolysis
- PI
- propidium iodide
- ERK
- extracellular signal-regulated kinase.
References
- 1. Brighton C. T., and Hunt R. M. (1997) Early histologic and ultrastructural changes in microvessels of periosteal callus. J. Orthop. Trauma 11, 244–253 10.1097/00005131-199705000-00002 [DOI] [PubMed] [Google Scholar]
- 2. Valero M. C., Huntsman H. D., Liu J., Zou K., and Boppart M. D. (2012) Eccentric exercise facilitates mesenchymal stem cell appearance in skeletal muscle. PLoS One 7, e29760 10.1371/journal.pone.0029760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oryan A., Kamali A., Moshiri A., and Baghaban Eslaminejad M. (2017) Role of mesenchymal stem cells in bone regenerative medicine: what is the evidence? Cells Tissues Organs 204, 59–83 10.1159/000469704 [DOI] [PubMed] [Google Scholar]
- 4. Le Blanc K., and Davies L. C. (2018) MSCs–cells with many sides. Cytotherapy 20, 273–278 10.1016/j.jcyt.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 5. Bittle G. J., Morales D., Deatrick K. B., Parchment N., Saha P., Mishra R., Sharma S., Pietris N., Vasilenko A., Bor C., Ambastha C., Gunasekaran M., Li D., and Kaushal S. (2018) Stem cell therapy for hypoplastic left heart syndrome: mechanism, clinical application, and future directions. Circ. Res. 123, 288–300 10.1161/CIRCRESAHA.117.311206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stolzing A., Jones E., McGonagle D., and Scutt A. (2008) Age-related changes in human bone marrow-derived mesenchymal stem cells: consequences for cell therapies. Mech. Ageing Dev. 129, 163–173 10.1016/j.mad.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 7. Zhou S., Greenberger J. S., Epperly M. W., Goff J. P., Adler C., Leboff M. S., and Glowacki J. (2008) Age-related intrinsic changes in human bone-marrow–derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell 7, 335–343 10.1111/j.1474-9726.2008.00377.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stenderup K., Justesen J., Clausen C., and Kassem M. (2003) Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone 33, 919–926 10.1016/j.bone.2003.07.005 [DOI] [PubMed] [Google Scholar]
- 9. Litvinov S. V., Velders M. P., Bakker H. A., Fleuren G. J., and Warnaar S. O. (1994) Ep-CAM: a human epithelial antigen is a homophilic cell–cell adhesion molecule. J. Cell Biol. 125, 437–446 10.1083/jcb.125.2.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maetzel D., Denzel S., Mack B., Canis M., Went P., Benk M., Kieu C., Papior P., Baeuerle P. A., Munz M., and Gires O. (2009) Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 11, 162–171 10.1038/ncb1824 [DOI] [PubMed] [Google Scholar]
- 11. Lu T. Y., Lu R. M., Liao M. Y., Yu J., Chung C. H., Kao C. F., and Wu H. C. (2010) Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J. Biol. Chem. 285, 8719–8732 10.1074/jbc.M109.077081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin C. W., Liao M. Y., Lin W. W., Wang Y. P., Lu T. Y., and Wu H. C. (2012) Epithelial cell adhesion molecule regulates tumor initiation and tumorigenesis via activating reprogramming factors and epithelial–mesenchymal transition gene expression in colon cancer. J. Biol. Chem. 287, 39449–39459 10.1074/jbc.M112.386235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang H. P., Chen P. H., Yu C. Y., Chuang C. Y., Stone L., Hsiao W. C., Li C. L., Tsai S. C., Chen K. Y., Chen H. F., Ho H. N., and Kuo H. C. (2011) Epithelial cell adhesion molecule (EpCAM) complex proteins promote transcription factor-mediated pluripotency reprogramming. J. Biol. Chem. 286, 33520–33532 10.1074/jbc.M111.256164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuan I. I., Liang K. H., Wang Y. P., Kuo T. W., Meir Y. J., Wu S. C., Yang S. C., Lu J., and Wu H. C. (2017) EpEX/EpCAM and Oct4 or Klf4 alone are sufficient to generate induced pluripotent stem cells through STAT3 and HIF2α. Sci. Rep. 7, 41852 10.1038/srep41852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu K. R., Yang S. R., Jung J. W., Kim H., Ko K., Han D. W., Park S. B., Choi S. W., Kang S. K., Schöler H., and Kang K. S. (2012) CD49f enhances multipotency and maintains stemness through the direct regulation of OCT4 and SOX2. Stem Cells 30, 876–887 10.1002/stem.1052 [DOI] [PubMed] [Google Scholar]
- 16. Münz M., Kieu C., Mack B., Schmitt B., Zeidler R., and Gires O. (2004) The carcinoma-associated antigen EpCAM up-regulates c-myc and induces cell proliferation. Oncogene 23, 5748–5758 10.1038/sj.onc.1207610 [DOI] [PubMed] [Google Scholar]
- 17. Chaves-Pérez A., Mack B., Maetzel D., Kremling H., Eggert C., Harréus U., and Gires O. (2013) EpCAM regulates cell cycle progression via control of cyclin D1 expression. Oncogene 32, 641–650 10.1038/onc.2012.75 [DOI] [PubMed] [Google Scholar]
- 18. Platt M. O., Roman A. J., Wells A., Lauffenburger D. A., and Griffith L. G. (2009) Sustained epidermal growth factor receptor levels and activation by tethered ligand binding enhances osteogenic differentiation of multi-potent marrow stromal cells. J Cell. Physiol. 221, 306–317 10.1002/jcp.21854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krampera M., Pasini A., Rigo A., Scupoli M. T., Tecchio C., Malpeli G., Scarpa A., Dazzi F., Pizzolo G., and Vinante F. (2005) HB-EGF/HER-1 signaling in bone marrow mesenchymal stem cells: inducing cell expansion and reversibly preventing multilineage differentiation. Blood 106, 59–66 10.1182/blood-2004-09-3645 [DOI] [PubMed] [Google Scholar]
- 20. Markovic A., and Chung C. H. (2012) Current role of EGF receptor monoclonal antibodies and tyrosine kinase inhibitors in the management of head and neck squamous cell carcinoma. Expert Rev. Anticancer Ther. 12, 1149–1159 10.1586/era.12.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raz R., Lee C. K., Cannizzaro L. A., d'Eustachio P., and Levy D. E. (1999) Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. U.S.A. 96, 2846–2851 10.1073/pnas.96.6.2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Piskounova E., Polytarchou C., Thornton J. E., LaPierre R. J., Pothoulakis C., Hagan J. P., Iliopoulos D., and Gregory R. I. (2011) Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147, 1066–1079 10.1016/j.cell.2011.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stefani G., Chen X., Zhao H., and Slack F. J. (2015) A novel mechanism of LIN-28 regulation of let-7 microRNA expression revealed by in vivo HITS-CLIP in C. elegans. RNA 21, 985–996 10.1261/rna.045542.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Triboulet R., Pirouz M., and Gregory R. I. (2015) A single Let-7 microRNA bypasses LIN28-mediated repression. Cell Rep. 13, 260–266 10.1016/j.celrep.2015.08.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee H., Han S., Kwon C. S., and Lee D. (2016) Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 7, 100–113 10.1007/s13238-015-0212-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang T., Wang G., Hao D., Liu X., Wang D., Ning N., and Li X. (2015) Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol. Cancer 14, 125 10.1186/s12943-015-0402-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liao T. T., Hsu W. H., Ho C. H., Hwang W. L., Lan H. Y., Lo T., Chang C. C., Tai S. K., and Yang M. H. (2016) Let-7 modulates chromatin configuration and target gene repression through regulation of the ARID3B complex. Cell Rep. 14, 520–533 10.1016/j.celrep.2015.12.064 [DOI] [PubMed] [Google Scholar]
- 28. Guo Y., Chen Y., Ito H., Watanabe A., Ge X., Kodama T., and Aburatani H. (2006) Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 384, 51–61 10.1016/j.gene.2006.07.011 [DOI] [PubMed] [Google Scholar]
- 29. Chien C. S., Wang M. L., Chu P. Y., Chang Y. L., Liu W. H., Yu C. C., Lan Y. T., Huang P. I., Lee Y. Y., Chen Y. W., Lo W. L., and Chiou S. H. (2015) Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Cancer Res. 75, 2553–2565 10.1158/0008-5472.CAN-14-2215 [DOI] [PubMed] [Google Scholar]
- 30. Monzen K., Ito Y., Naito A. T., Kasai H., Hiroi Y., Hayashi D., Shiojima I., Yamazaki T., Miyazono K., Asashima M., Nagai R., and Komuro I. (2008) A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nat. Cell Biol. 10, 567–574 10.1038/ncb1719 [DOI] [PubMed] [Google Scholar]
- 31. Nishino J., Kim I., Chada K., and Morrison S. J. (2008) Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135, 227–239 10.1016/j.cell.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li O., Vasudevan D., Davey C. A., and Dröge P. (2006) High-level expression of DNA architectural factor HMGA2 and its association with nucleosomes in human embryonic stem cells. Genesis 44, 523–529 10.1002/dvg.20242 [DOI] [PubMed] [Google Scholar]
- 33. Li O., Li J., and Dröge P. (2007) DNA architectural factor and proto-oncogene HMGA2 regulates key developmental genes in pluripotent human embryonic stem cells. FEBS Lett. 581, 3533–3537 10.1016/j.febslet.2007.06.072 [DOI] [PubMed] [Google Scholar]
- 34. Pfannkuche K., Summer H., Li O., Hescheler J., and Dröge P. (2009) The high mobility group protein HMGA2: a co-regulator of chromatin structure and pluripotency in stem cells? Stem Cell Rev. 5, 224–230 10.1007/s12015-009-9078-9 [DOI] [PubMed] [Google Scholar]
- 35. Fusco A., and Fedele M. (2007) Roles of HMGA proteins in cancer. Nat. Rev. Cancer 7, 899–910 10.1038/nrc2271 [DOI] [PubMed] [Google Scholar]
- 36. Rawlinson N. J., West W. W., Nelson M., and Bridge J. A. (2008) Aggressive angiomyxoma with t(12;21) and HMGA2 rearrangement: report of a case and review of the literature. Cancer Genet. Cytogenet. 181, 119–124 10.1016/j.cancergencyto.2007.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei J. J., Wu J., Luan C., Yeldandi A., Lee P., Keh P., and Liu J. (2010) HMGA2: a potential biomarker complement to P53 for detection of early-stage high-grade papillary serous carcinoma in fallopian tubes. Am. J. Surg. Pathol. 34, 18–26 10.1097/PAS.0b013e3181be5d72 [DOI] [PubMed] [Google Scholar]
- 38. Mahajan A., Liu Z., Gellert L., Zou X., Yang G., Lee P., Yang X., and Wei J. J. (2010) HMGA2: a biomarker significantly overexpressed in high-grade ovarian serous carcinoma. Modern Pathol. 23, 673–681 10.1038/modpathol.2010.49 [DOI] [PubMed] [Google Scholar]
- 39. Li Z., Gilbert J. A., Zhang Y., Zhang M., Qiu Q., Ramanujan K., Shavlakadze T., Eash J. K., Scaramozza A., Goddeeris M. M., Kirsch D. G., Campbell K. P., Brack A. S., and Glass D. J. (2012) An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev. Cell 23, 1176–1188 10.1016/j.devcel.2012.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thornton J. E., and Gregory R. I. (2012) How does Lin28 let-7 control development and disease? Trends Cell Biol. 22, 474–482 10.1016/j.tcb.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shell S., Park S. M., Radjabi A. R., Schickel R., Kistner E. O., Jewell D. A., Feig C., Lengyel E., and Peter M. E. (2007) Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 11400–11405 10.1073/pnas.0704372104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cleynen I., and Van de Ven W. J. (2008) The HMGA proteins: a myriad of functions (review). Int. J. Oncol. 32, 289–305 [PubMed] [Google Scholar]
- 43. Matic I., Antunovic M., Brkic S., Josipovic P., Mihalic K. C., Karlak I., Ivkovic A., and Marijanovic I. (2016) Expression of OCT-4 and SOX-2 in bone marrow-derived human mesenchymal stem cells during osteogenic differentiation. Open Access Maced. J. Med. Sci. 4, 9–16 10.3889/oamjms.2016.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., and Marshak D. R. (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147 10.1126/science.284.5411.143 [DOI] [PubMed] [Google Scholar]
- 45. Ge W., Jiang J., Baroja M. L., Arp J., Zassoko R., Liu W., Bartholomew A., Garcia B., and Wang H. (2009) Infusion of mesenchymal stem cells and rapamycin synergize to attenuate alloimmune responses and promote cardiac allograft tolerance. Am. J. Transpl. 9, 1760–1772 10.1111/j.1600-6143.2009.02721.x [DOI] [PubMed] [Google Scholar]
- 46. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., and Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boiani M., and Schöler H. R. (2005) Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 6, 872–884 10.1038/nrm1744 [DOI] [PubMed] [Google Scholar]
- 48. Reim G., and Brand M. (2002) Spiel-ohne-grenzen/pou2 mediates regional competence to respond to Fgf8 during zebrafish early neural development. Development 129, 917–933 [DOI] [PubMed] [Google Scholar]
- 49. Palmieri S. L., Peter W., Hess H., and Schöler H. R. (1994) Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 166, 259–267 10.1006/dbio.1994.1312 [DOI] [PubMed] [Google Scholar]
- 50. Pesce M., and Schöler H. R. (2001) Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells 19, 271–278 10.1634/stemcells.19-4-271 [DOI] [PubMed] [Google Scholar]
- 51. Kim J. H., Jee M. K., Lee S. Y., Han T. H., Kim B. S., Kang K. S., and Kang S. K. (2009) Regulation of adipose tissue stromal cells behaviors by endogenic Oct4 expression control. PLoS One 4, e7166 10.1371/journal.pone.0007166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Niwa H., Miyazaki J., and Smith A. G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 10.1038/74199 [DOI] [PubMed] [Google Scholar]
- 53. Tai M. H., Chang C. C., Kiupel M., Webster J. D., Olson L. K., and Trosko J. E. (2005) Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis 26, 495–502 [DOI] [PubMed] [Google Scholar]
- 54. Monk M., and Holding C. (2001) Human embryonic genes re-expressed in cancer cells. Oncogene 20, 8085–8091 10.1038/sj.onc.1205088 [DOI] [PubMed] [Google Scholar]
- 55. Greco S. J., Liu K., and Rameshwar P. (2007) Functional similarities among genes regulated by OCT4 in human mesenchymal and embryonic stem cells. Stem Cells 25, 3143–3154 10.1634/stemcells.2007-0351 [DOI] [PubMed] [Google Scholar]
- 56. Kerkis I., Kerkis A., Dozortsev D., Stukart-Parsons G. C., Gomes Massironi S. M., Pereira L. V., Caplan A. I., and Cerruti H. F. (2006) Isolation and characterization of a population of immature dental pulp stem cells expressing OCT-4 and other embryonic stem cell markers. Cells Tissues Organs 184, 105–116 10.1159/000099617 [DOI] [PubMed] [Google Scholar]
- 57. Beltrami A. P., Cesselli D., Bergamin N., Marcon P., Rigo S., Puppato E., D'Aurizio F., Verardo R., Piazza S., Pignatelli A., Poz A., Baccarani U., Damiani D., Fanin R., Mariuzzi L., et al. (2007) Multipotent cells can be generated in vitro from several adult human organs (heart, liver, and bone marrow). Blood 110, 3438–3446 10.1182/blood-2006-11-055566 [DOI] [PubMed] [Google Scholar]
- 58. Lin G., Garcia M., Ning H., Banie L., Guo Y. L., Lue T. F., and Lin C. S. (2008) Defining stem and progenitor cells within adipose tissue. Stem Cells Dev. 17, 1053–1063 10.1089/scd.2008.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fang X., Yoon J. G., Li L., Yu W., Shao J., Hua D., Zheng S., Hood L., Goodlett D. R., Foltz G., and Lin B. (2011) The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics 12, 11 10.1186/1471-2164-12-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Girouard S. D., Laga A. C., Mihm M. C., Scolyer R. A., Thompson J. F., Zhan Q., Widlund H. R., Lee C. W., and Murphy G. F. (2012) SOX2 contributes to melanoma cell invasion. Lab. Invest. 92, 362–370 10.1038/labinvest.2011.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu K., Lin B., Zhao M., Yang X., Chen M., Gao A., Liu F., Que J., and Lan X. (2013) The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell. Signal. 25, 1264–1271 10.1016/j.cellsig.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A. A., Ko M. S., and Niwa H. (2007) Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9, 625–635 10.1038/ncb1589 [DOI] [PubMed] [Google Scholar]
- 63. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 64. Sperger J. M., Chen X., Draper J. S., Antosiewicz J. E., Chon C. H., Jones S. B., Brooks J. D., Andrews P. W., Brown P. O., and Thomson J. A. (2003) Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. U.S.A. 100, 13350–13355 10.1073/pnas.2235735100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lengler J., Bittner T., Münster D., Gawad Ael-D, and Graw J. (2005) Agonistic and antagonistic action of AP2, Msx2, Pax6, Prox1 AND Six3 in the regulation of Sox2 expression. Ophthalmic Res. 37, 301–309 10.1159/000087774 [DOI] [PubMed] [Google Scholar]
- 66. Dezawa M., Ishikawa H., Itokazu Y., Yoshihara T., Hoshino M., Takeda S., Ide C., and Nabeshima Y. (2005) Bone marrow stromal cells generate muscle cells and repair muscle degeneration. Science 309, 314–317 10.1126/science.1110364 [DOI] [PubMed] [Google Scholar]
- 67. Pierantozzi E., Gava B., Manini I., Roviello F., Marotta G., Chiavarelli M., and Sorrentino V. (2011) Pluripotency regulators in human mesenchymal stem cells: expression of NANOG but not of OCT-4 and SOX-2. Stem Cells Dev. 20, 915–923 10.1089/scd.2010.0353 [DOI] [PubMed] [Google Scholar]
- 68. Greber B., Lehrach H., and Adjaye J. (2007) Silencing of core transcription factors in human EC cells highlights the importance of autocrine FGF signaling for self-renewal. BMC Dev. Biol. 7, 46 10.1186/1471-213X-7-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xu R. H., Sampsell-Barron T. L., Gu F., Root S., Peck R. M., Pan G., Yu J., Antosiewicz-Bourget J., Tian S., Stewart R., and Thomson J. A. (2008) NANOG is a direct target of TGFβ/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell 3, 196–206 10.1016/j.stem.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chambers I., and Tomlinson S. R. (2009) The transcriptional foundation of pluripotency. Development 136, 2311–2322 10.1242/dev.024398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lengner C. J., Welstead G. G., and Jaenisch R. (2008) The pluripotency regulator Oct4: a role in somatic stem cells? Cell Cycle 7, 725–728 10.4161/cc.7.6.5573 [DOI] [PubMed] [Google Scholar]
- 72. Looijenga L. H., Stoop H., de Leeuw H. P., de Gouveia Brazao C. A., Gillis A. J., van Roozendaal K. E., van Zoelen E. J., Weber R. F., Wolffenbuttel K. P., van Dekken H., Honecker F., Bokemeyer C., Perlman E. J., Schneider D. T., Kononen J., et al. (2003) POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res. 63, 2244–2250 [PubMed] [Google Scholar]
- 73. Pan G., and Thomson J. A. (2007) Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17, 42–49 10.1038/sj.cr.7310125 [DOI] [PubMed] [Google Scholar]
- 74. Cheng L., Sung M. T., Cossu-Rocca P., Jones T. D., MacLennan G. T., De Jong J., Lopez-Beltran A., Montironi R., and Looijenga L. H. (2007) OCT4: biological functions and clinical applications as a marker of germ cell neoplasia. J. Pathol. 211, 1–9 10.1002/path.2105 [DOI] [PubMed] [Google Scholar]
- 75. Liang K. H., Tso H. C., Hung S. H., Kuan I. I., Lai J. K., Fe F. Y., Chuang Y. T., Liu I. J., Wang Y. P., Chen R. H., and Wu H. C. (2018) Extracellular domain of EpCAM enhances tumor progression through EGFR signaling in colon cancer cells. Cancer Lett. 433, 165–175 10.1016/j.canlet.2018.06.040 [DOI] [PubMed] [Google Scholar]
- 76. Pan M., Schinke H., Luxenburger E., Kranz G., Shakhtour J., Libl D., Huang Y., Gaber A., Pavšič M., Lenarčič B., Kitz J., Jakob M., Schwenk-Zieger S., Canis M., Hess J., et al. (2018) EpCAM ectodomain EpEX is a ligand of EGFR that counteracts EGF-mediated epithelial–mesenchymal transition through modulation of phospho- CDK, cyclin-dependent kinase 1/2 in head and neck cancers. PLoS Biol. 16, e2006624 10.1371/journal.pbio.2006624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yeom S. Y., Nam D. H., and Park C. (2014) RRAD promotes EGFR-mediated STAT3 activation and induces temozolomide resistance of malignant glioblastoma. Mol. Cancer Ther. 13, 3049–3061 10.1158/1535-7163.MCT-14-0244 [DOI] [PubMed] [Google Scholar]
- 78. Liu Y., Li H., Feng J., Cui X., Huang W., Li Y., Su F., Liu Q., Zhu J., Lv X., Chen J., Huang D., and Yu F. (2013) Lin28 induces epithelial–to–mesenchymal transition and stemness via downregulation of let-7a in breast cancer cells. PLoS One 8, e83083 10.1371/journal.pone.0083083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cimadamore F., Amador-Arjona A., Chen C., Huang C. T., and Terskikh A. V. (2013) SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc. Natl. Acad. Sci. U.S.A. 110, E3017–E3026 10.1073/pnas.1220176110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Guo L., Chen C., Shi M., Wang F., Chen X., Diao D., Hu M., Yu M., Qian L., and Guo N. (2013) Stat3-coordinated Lin-28–let-7–HMGA2 and miR-200–ZEB1 circuits initiate and maintain oncostatin M-driven epithelial–mesenchymal transition. Oncogene 32, 5272–5282 10.1038/onc.2012.573 [DOI] [PubMed] [Google Scholar]
- 81. Taniguchi N., Caramés B., Hsu E., Cherqui S., Kawakami Y., and Lotz M. (2011) Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation. J. Biol. Chem. 286, 41489–41498 10.1074/jbc.M111.236984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chamberlain G., Fox J., Ashton B., and Middleton J. (2007) Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells 25, 2739–2749 10.1634/stemcells.2007-0197 [DOI] [PubMed] [Google Scholar]
- 83. Duque G. (2008) Bone and fat connection in aging bone. Curr. Opin. Rheumatol. 20, 429–434 10.1097/BOR.0b013e3283025e9c [DOI] [PubMed] [Google Scholar]
- 84. Liu L. F., Shen W. J., Zhang Z. H., Wang L. J., and Kraemer F. B. (2010) Adipocytes decrease Runx2 expression in osteoblastic cells: roles of PPARγ and adiponectin. J. Cell. Physiol. 225, 837–845 10.1002/jcp.22291 [DOI] [PubMed] [Google Scholar]
- 85. Oshima K., Nampei A., Matsuda M., Iwaki M., Fukuhara A., Hashimoto J., Yoshikawa H., and Shimomura I. (2005) Adiponectin increases bone mass by suppressing osteoclast and activating osteoblast. Biochem. Biophys. Res. Commun. 331, 520–526 10.1016/j.bbrc.2005.03.210 [DOI] [PubMed] [Google Scholar]
- 86. Takahashi K., Mitsui K., and Yamanaka S. (2003) Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature 423, 541–545 10.1038/nature01646 [DOI] [PubMed] [Google Scholar]
- 87. Peng G. H., and Chen S. (2013) Double chromatin immunoprecipitation: analysis of target co-occupancy of retinal transcription factors. Methods Mol. Biol. 935, 311–328 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.