Abstract
Although cannabinoid receptor 1 (CB1) antagonists have been shown to attenuate diet-induced obesity (DIO) and associated inflammation, the precise molecular mechanisms involved are not clear. In the current study, we investigated the role of microRNA (miR) in the regulation of adipose tissue macrophage (ATM) phenotype following treatment of DIO mice with the CB1 antagonist SR141716A. DIO mice were fed high-fat diet (HFD) for 12 weeks and then treated daily with SR141716A (10 mg/kg) for 4 weeks while continuing HFD. Treated mice experienced weight loss, persistent reduction in fat mass, improvements in metabolic profile, and decreased adipose inflammation. CB1 blockade resulted in down-regulation of several miRs in ATMs, including the miR-466 family and miR-762. Reduced expression of the miR-466 family led to induction of anti-inflammatory M2 transcription factors KLF4 and STAT6, whereas down-regulation of miR-762 promoted induction of AGAP-2, a negative regulator of the neuroimmune retention cues, Netrin-1 and its coreceptor UNC5B. Furthermore, treatment of primary macrophages with SR141716A up-regulated KLF4 and STAT6, reduced secretion of Netrin-1, and increased migration toward the lymph node chemoattractant CCL19. These studies demonstrate for the first time that CB1 receptor blockade attenuates DIO-associated inflammation through alterations in ATM miR expression that promote M2 ATM polarization and macrophage egress from adipose tissue. The current study also identifies additional novel therapeutic targets for diet-induced obesity and metabolic disorder.
Keywords: cannabinoid receptor type 1 (CB1); macrophage; microRNA (miRNA); obesity; adipose tissue; SR141716A; Netrin-1; Unc5b, AGAP-2; inflammation
Introduction
The incidence of obesity has grown significantly in the last 25 years, leading to upward of 1.45 billion overweight adults in the world, of which ∼500 million are obese (1). Chronic low-grade, systemic inflammation associated with obesity plays a major role in the development of various chronic disease states, including type 2 diabetes, metabolic syndrome, and cardiovascular disease, which together contribute to high rates of mortality and morbidity (2). The chronic inflammation observed is primarily driven by the intense migration and accumulation of adipose tissue macrophages (ATMs),2 which drive insulin resistance (3–6). The recruitment of ATMs to adipose tissue positively correlates with the production of proinflammatory molecules, including tumor necrosis factor-α (TNFα), interleukin-1β (IL-1β), and IL-6 that potentiate insulin resistance (7–9).
Macrophages show significant heterogeneity in phenotype and function, as factors in the local milieu determine their activation state and subsequent properties. Macrophages can be activated under various conditions that lead to polarization between two main states, routinely classified as “M1” and “M2.” M1 macrophages are induced during obesity and are said to be “classically activated” cells triggered by lipopolysaccharide and IFNγ that secrete proinflammatory cytokines (TNFα, IL-6, and IL-12) and generate nitric oxide (NO), a reactive oxygen species via iNOS activation (10, 11). However, M2 macrophages are considered “alternatively activated” and populate lean adipose tissue. They can be activated by IL-4 and IL-13, secrete anti-inflammatory cytokines, and up-regulate arginase, which opposes NO production (12). Additionally, studies have shown that the M1 and M2 macrophage phenotypes are not clearly defined because multiple subtypes exist that are activated by various stimuli, and it is possible to dynamically switch their polarization (13). Key signaling molecules such as DNA methyltransferase 3b (DNMT3b) and peroxisome proliferator-activated receptor-γ (PPAR-γ) deregulate ATM polarization, inflammation, and insulin insensitivity (14, 15). In addition, other studies have shown that besides the molecules that trigger macrophage recruitment in adipose tissue, other signaling molecules such as neuronal guidance cues of the Semaphorin, Ephrin, and Netrin families modulate macrophage retention in adipose tissue and regulate immunometabolism (16, 17). Indeed, recent studies have shown Semaphorin 3E and Netrin-1 are involved in chemoattraction and retention of macrophages in the visceral adipose tissue of DIO mice (17, 18). Collectively, studies on neuroimmune guidance cues in macrophages define them as key regulators for ATM accumulation and active participants in the induction of chronic inflammation.
Weight loss is associated with beneficial effects of reducing the underlying inflammation in adipose tissue and subsequent amelioration of insulin resistance (19, 20). The endocannabinoid system plays a major role in food intake and energy balance. Overactivity of the endocannabinoid system in human obesity and in animal models of genetic and diet-induced obesity has been reported (21). Clinical studies on treatment of obesity and metabolic syndrome with the cannabinoid receptor 1 (CB1) antagonist SR141716A (rimonabant) have shown greater weight loss in obese patients compared with placebo (22). However, this drug was removed from the market due to adverse psychiatric effects (23). Nevertheless, the mechanistic effect of CB1 receptor antagonist on inflammation and ATMs has not been well studied. Previous studies have shown that blockade of CB1 receptors suppressed inflammation in adipose tissue of DIO mice (24, 25). However, the signals controlling the beneficial effect of the CB1 receptor antagonists on inflammation in adipose tissue remain poorly understood.
MicroRNAs (miRs) are short noncoding RNAs that can inhibit translation of their mRNA targets (26). Regulatory roles of miRs in many biological processes associated with obesity have been defined. They can synchronize obesity-related pathways such as immune-mediated inflammation, insulin action, fat metabolism, and energy homeostasis (27–29). Recently, we identified miRs involved in regulation of ATM phenotype during DIO (30). However, associations between aberrant miR expression and overactivation of the endocannabinoid system in obesity have not been previously reported to our knowledge. In the present study, we tested the role of miRs in SR141716A-mediated amelioration of obesity-induced inflammation, specifically by focusing on macrophage polarization and retention in visceral adipose tissue. Our study unveiled specific miRs that promote a switch from M1 to M2 phenotype as well as miRs that target neuroimmune guidance cues. Taken together, our findings suggest that CB1 blockade in DIO mice promotes miR dysregulation, which suppresses adipose tissue inflammation and contributes to improved metabolism.
Results
SR141716A attenuates HFD-induced obesity
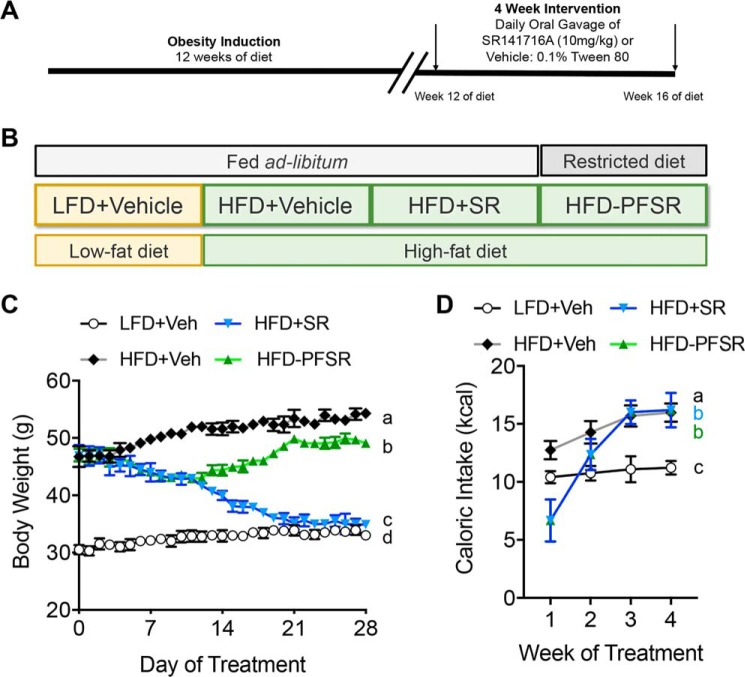
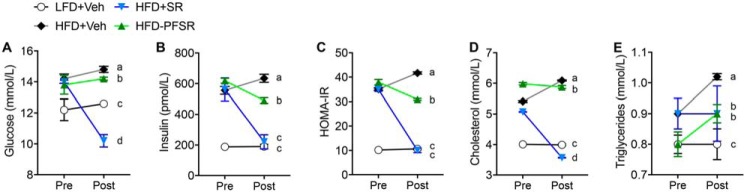
To study the effects of CB1 antagonist on obesity parameters, we treated DIO mice that were fed a 60% HFD for 12 weeks with SR141716A (SR) by daily oral gavage for 4 weeks (Fig. 1A). All other experimental groups were treated with the vehicle (Veh), 0.1% Tween 80. In addition to the HFD+SR group, other experimental groups included mice fed a control diet, 10% low-fat diet (LFD) (LFD+Veh); ad libitum HFD-fed mice (HFD+Veh); and HFD-fed mice that were pair-fed to the HFD+SR group (HFD-PFSR) (Fig. 1B). Blockade of CB1 receptors with SR141716A resulted in acute and persistent weight loss as well as transient reduction of calorie intake in DIO mice (Fig. 1, C and D). At the baseline of the study, the mice were stratified into groups balanced by mean fat mass. After 4 weeks of treatment, reduction in the fat mass and fat percentage was significant in the HFD+SR group when compared with both HFD+Veh and HFD-PFSR groups, whereas the changes in lean mass were not significant (Table S1). Fasting parameters, including blood glucose, insulin, homeostatic model assessment of insulin resistance index (HOMA-IR), total cholesterol, and triglycerides were examined before and after the 4-week intervention with SR compound. Our data demonstrated improvement in metabolic parameters in the HFD+SR group when compared with HFD+Veh and HFD-PFSR (Fig. 2, A–E). Taken together, our data suggested that blocking CB1 receptors in DIO mice results in amelioration of obesity, independent of its effect on calorie intake.
Figure 1.
SR141716A ameliorates DIO phenotype by inducing weight loss. DIO was induced in mice by 12 weeks of purified diet feeding. After week 12, mice were treated orally with either SR (10 mg/kg/day) or Veh (0.1% Tween 80) for 4 weeks. A, experimental timeline. B, experimental groups and description of purified diet. C, daily body weight of each group of mice is shown for the duration of treatment. D, weekly energy intake during the 4-week intervention. For C and D, n = 9–10. Data are shown as mean ± S.D. (error bars). Data were analyzed using two-way ANOVA with a Tukey post hoc test. p < 0.05 if alphabetical characters differ between the groups being compared.
Figure 2.
SR141716A intervention treatment ameliorates metabolic dysfunction in DIO phenotype. Fasted metabolic parameters were assessed before intervention (Pre) and on the day before euthanasia (day 27; Post). A, fasting blood glucose. B, serum insulin concentration. C, HOMA-IR = Fasting insulin (microunits/ml) × Fasting glucose (mmol/liter)/22.5. D, total cholesterol concentration. E, triglyceride concentration. Data are shown as mean ± S.D. (error bars). Data were analyzed using two-way ANOVA with a Tukey post hoc test. p < 0.05 at the Post time point if alphabetical characters differ between the groups being compared. n = 10, except n = 9 for HFD+SR.
SR141716A reduces adipose tissue inflammation and alters ATM miR profile
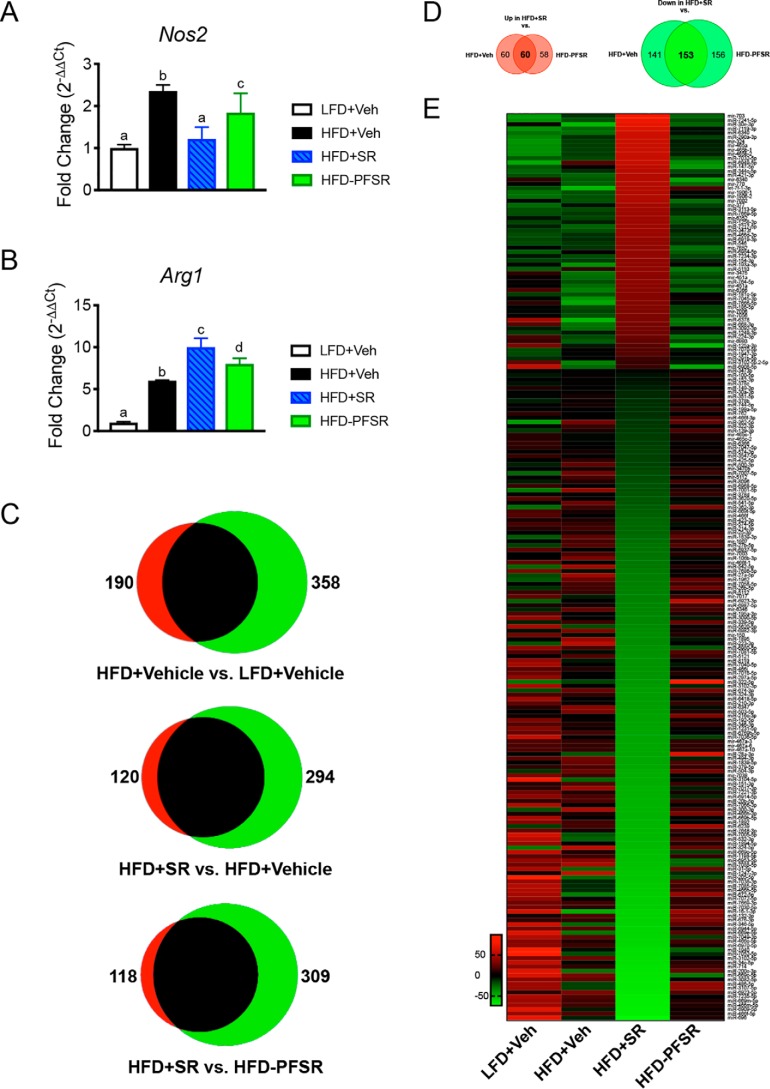
In a previous report, we showed that treatment of DIO mice with SR reduces both ATM infiltration and M1 polarization in epididymal adipose tissue (31). To further confirm the overall inflammatory status in adipose tissue, we quantitated the expression of macrophage polarization genes Nos2 (M1) and Arg1 (M2) in epididymal fat. SR treatment lowered Nos2 expression and increased expression of Arg1 when compared with HFD+Veh and HFD-PFSR, which validated an anti-inflammatory state in the adipose tissue of DIO mice following SR141716A treatment (Fig. 3, A and B). Intermediate expression of Nos2 and Arg1 in the HFD-PFSR group indicated that appetite restriction has beneficial effects; however, SR treatment leads to additional benefits that are independent of appetite restriction.
Figure 3.
SR141716A treatment of DIO mice reduces adipose inflammation and alters miR expression in ATMs. Adipose inflammatory markers and changes in miR expression in ATMs were assessed following SR141716A treatment in DIO mice. A and B, adipose tissue expression of genes for polarization markers iNOS (M1; Nos2) and arginase-1 (M2; Arg1). C, cluster analysis of up- and down-regulated miRs (log2 -fold change of at least ±1.5) in F4/80+ ATMs represented in Venn diagrams. D, Venn diagrams of dysregulated miRs that are independent of appetite. E, heat map of modified miRs due to the effect of SR141716A treatment, independent of its effect on calorie restriction. Data are shown as mean ± S.D. For A, data were analyzed using one-way ANOVA. Groups with different alphabetical characters are significantly different from each other (p < 0.05).
Next, to determine whether SR treatment alters miR expression in ATMs, we performed miR microarray analyses in F4/80+ cells isolated from the stromal vascular fraction of epididymal adipose tissue. Of the more than 3000 miRs tested, 120 were overexpressed and 294 were underexpressed greater than or equal to ±1.5 log2 -fold change in the HFD+SR group when compared with the HFD+Veh group, whereas 118 and 309 of miRs were significantly up- and down-regulated, respectively, in HFD+SR group versus HFD-PFSR group (Fig. 3C). In addition, we noted 190 up- and 358 down-regulated miRs in HFD+Veh versus LFD+Veh (Fig. 3C).
To investigate the effect of SR141716A on the miR profile independent of its effect on calorie restriction, we identified miRs that were similarly dysregulated in the HFD+SR group versus both the HFD+Veh and HFD-PFSR groups (Fig. 3D). A heat map of mean normalized expression of these altered miRs demonstrated that SR treatment alters miR expression independently of food restriction (Fig. 3E).
We next used in silico analyses to identify potential pathways targeted by the dysregulated miRs. First, using Cytoscape analysis modules, we identified the targeted gene ontologies of dysregulated miRs. The main affected pathways following SR treatment included regulation of various components of the immune system (Fig. S1). Next, we performed analysis using Ingenuity Pathway Analysis (IPA; Qiagen). Predicted interactions between miRs and their targeted genes following SR treatment revealed that the altered miR profile might skew the ATM balance to a more anti-inflammatory macrophage phenotype (M2, arginase+) through various cytokine, chemokine, transcription factor, and signaling networks (Fig. S2).
SR141716A-altered miRs promote a shift toward M2 macrophage phenotype
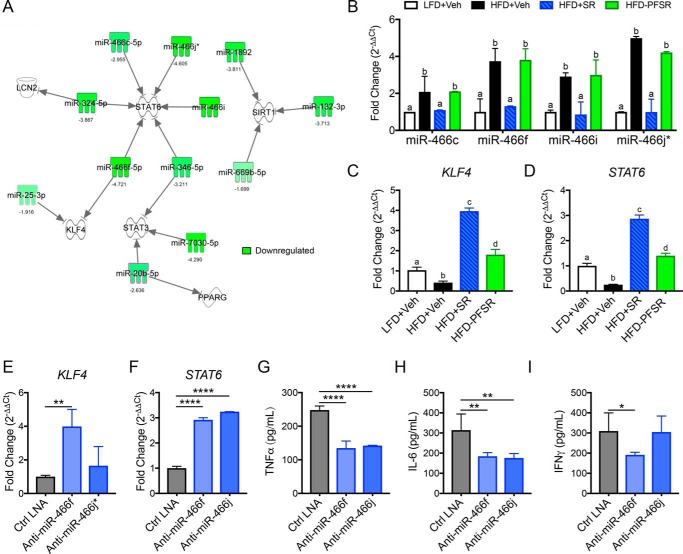
Interestingly, pathway analysis with IPA uncovered SR141716A-mediated alterations in miRs that may induce anti-inflammatory M2 macrophages by targeting the M2-related transcription factors (STAT3, STAT6, LCN2, KLF4, PPAR-γ, and SIRT1) (Fig. 4A). The list of corresponding targets and miRs along with each algorithm is summarized in Table S2. More interestingly, for the first time, we uncovered that most of the members of the miR-466 family were down-regulated following SR141716A treatment regardless of calorie intake (Figs. 3D and 4A). qRT-PCR of the miR-466 family from the ATMs validated that these were down-regulated following SR141716A treatment (Fig. 4B). Furthermore, expression of the M2 transcription factor genes Klf4 and Stat6 was up-regulated in HFD+SR ATMs (Fig. 4, C and D).
Figure 4.
Down-regulation of the miR-466 family following SR141716A treatment promotes M2 polarization in ATMs. Differentially expressed miRs in ATMs due to SR treatment were analyzed for overlap with macrophage polarization genes. A, IPA analysis from filtered miRs revealed that the miR-466 family targets M2 transcription factors. B, qRT-PCR for the miR-466 family in ATMs. C, qRT-PCR for Klf4 in ATMs. D, qRT-PCR for Stat6 in ATMs. For E–I, peritoneal macrophages from naïve mice were transfected with a negative control LNA (Ctrl LNA), miR-466f inhibitor LNA (anti-miR-466f), or miR-466j* inhibitor LNA (anti-miR-466j*) and then cultured in 3T3-L1 adipocyte–conditioned medium. E, qRT-PCR for Klf4. F, qRT-PCR for Stat6. For G–I, cytokines were measured in cell supernatant by ELISAs. G, TNFα ELISA. H, IL-6 ELISA. I, IFNγ ELISA. Data are shown as mean ± S.D. (error bars). For B–D, data with different alphabetical characters are significantly different (p < 0.05) by post hoc one-way ANOVA. For E–I, *, p < 0.05; **, p < 0.01; ****, p < 0.0001 versus Ctrl LNA by post hoc one-way ANOVA.
To validate the impact of the miR-466 family in induction of M2 phenotype in ATMs, peritoneal macrophages were isolated and cultured in conditioned medium from differentiated 3T3-L1 adipocytes following transfection with miR-466j* and miR-466f LNATM Power Inhibitors. The data showed that Klf4 and Stat6 were overexpressed following miR-466 inhibition (Fig. 4, E and F), thereby supporting that miR-466 targeted these transcription factors. Moreover, inflammatory cytokines such as TNFα, IL-6, and IFNγ were suppressed following miR-466 inhibition (Fig. 4, G–I). Taken together, these data suggested that blockade of CB1 in DIO mice results in down-regulation of the miR-466 family in ATMs, which promotes M2 macrophage polarization and thereby induces a switch toward an anti-inflammatory state in adipose tissue.
SR141716A ameliorates ATM retention in adipose tissue
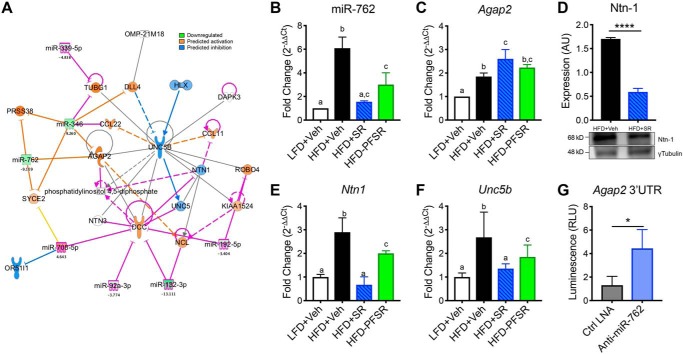
In addition to the effect of SR141716A in promoting M2 macrophages, we previously demonstrated a significant decrease in the number of proinflammatory M1 macrophages in adipose tissue of the HFD+SR-treated group when compared with the HFD+Veh group (31). In the current study, we investigated whether this resulted from altered migration of ATMs due to miR-mediated regulation of neuronal guidance cues. Filtering the IPA modules to the neuroimmune guidance cue canonical pathways demonstrated a novel series of miRs that targeted AGAP-2, which is an upstream negative regulator of the Netrin-1 receptor UNC-5 homology B (UNC5B) (Fig. 5A). This suggested that down-regulation of miR-762 may increase AGAP-2 and thus inhibit Netrin-1–mediated ATM detainment (Fig. 5A). qRT-PCR validated that expression of miR-762 was reduced, whereas Agap2 was increased following SR treatment (Fig. 5, B and C). Interestingly, we noted that the level of expression of Netrin-1 at both protein and transcription levels in ATMs was significantly reduced in HFD+SR mice when compared with HFD+Veh mice (Fig. 5, D and E). Unc5b was also down-regulated in HFD+SR versus HFD+Veh (Fig. 5F). Additionally, we quantified gene expression of other Netrin family receptors (Table S3). To validate the direct interaction of miR-762 and AGAP-2, we used bone marrow–derived macrophages (BMDMs) that were treated with conditioned medium from 3T3-L1 adipocytes. Such BMDMs were transfected with a Renilla luciferase plasmid (transfection efficiency control) containing the 3′-UTR of Agap2 and a firefly luciferase reporter gene. Luciferase expression was suppressed by endogenous miR-762. Subsequently, upon transfection with anti-miR-762 LNA microRNA Inhibitor (5 nm), increased luciferase activity/luminescence was detected, thereby demonstrating that the Agap2 3′-UTR is a direct target of miR-762 (Fig. 5G).
Figure 5.
SR141716A treatment reduces miR-762 in ATMs to promote negative regulation of Netrin-1–mediated retention via induction of AGAP-2. ATMs from different treated groups were examined for changes in the expression of miRs that target neuroimmune guidance cues. A, IPA analysis of differentially expressed miRs following SR treatment overlaid with the neuroimmune guidance cue canonical pathway. The Molecular Activity Predictor (MAP) module was applied for downstream effect prediction. IPA analysis revealed that AGAP-2 is targeted by down-regulated miRs. B, qRT-PCR expression validation of miR-762 in ATMs. C, qRT-PCR of the miR-762 target Agap2 in ATMs. D, Western blotting of Netrin-1 (Ntn-1) in ATMs. E, qRT-PCR of Ntn1 in ATMs. F, qRT-PCR of Unc5b in ATMs. G, luciferase reporter assay of BMDM transfected with the 3′-UTR of Agap2 followed by either Ctrl LNA or anti-miR-762 LNA. Luminescence was measured 24 h after transfection with LNA. Data are shown as mean ± S.D. (error bars). Data with different alphabetical characters are significantly different (p < 0.05) by post hoc one-way ANOVA. ****, p < 0.0001; *, p < 0.05 by two-tailed Student's t test. AU, arbitrary units; RLU, relative luminescence units.
Macrophage-dependent CB1 blockade promotes M2 polarization and decreased retention
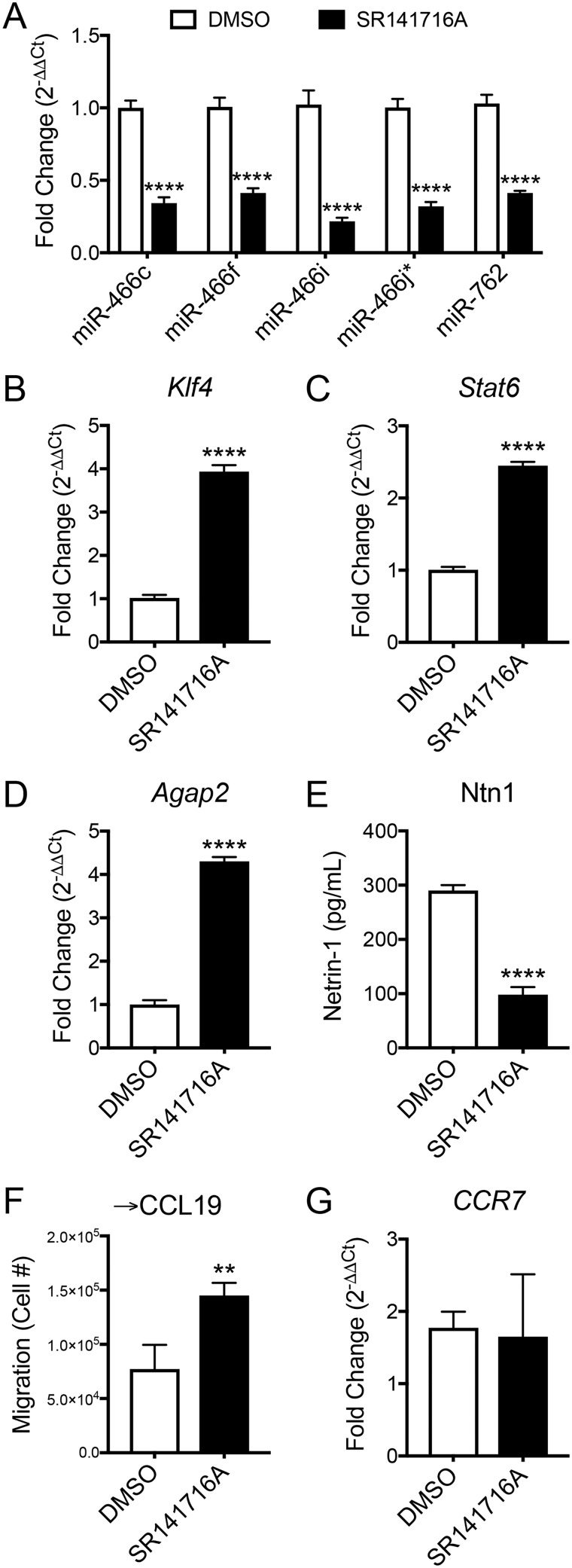
Next, to confirm that the miR-mediated M2 polarization and decreased ATM retention observed in HFD+SR-treated mice were a direct effect of CB1 blockade and not a secondary effect due to weight loss or other factors, we treated primary macrophages in vitro with 3T3-L1 adipocyte–conditioned medium and SR141716A or DMSO vehicle control. Treatment of 3% thioglycollate–elicited peritoneal macrophages with adipocyte-conditioned medium and SR141716A reduced expression of miR-466 family miRs and miR-762, confirming CB1-dependent alterations in macrophage miR expression (Fig. 6A). Consistent with decreased miR-466, SR141716A treatment increased expression of the miR-466 targets Klf4 and Stat6, indicating a shift toward the M2 phenotype (Fig. 6, B and C). Additionally, expression of the miR-762 target Agap2 was increased following SR141716A treatment (Fig. 6D). Concomitantly, Netrin-1 secretion was reduced following SR141716A treatment (Fig. 6E). The macrophage migration rate toward CCL19 was also measured because CCL19 is the primary chemokine implicated in emigration of tissue macrophages toward draining lymph nodes. SR141716A-treated macrophages exhibited higher migration toward CCL19 than DMSO-treated macrophages (Fig. 6F), although the level of expression of the CCL19 receptor CCR7 was similar in both groups (Fig. 6G). These data suggested that SR141716A attenuates the oversecretion of Netrin-1 from ATMs in obese phenotype and potentially promotes their emigration from adipose tissue through miR-762–mediated regulation of AGAP-2. Together, these findings confirmed that blockade of macrophage CB1 signaling has anti-inflammatory and anti-retention effects in macrophages due to altered miR expression.
Figure 6.
Treatment of macrophages with SR141716A in vitro reduces the miR-466 family and miR-762 expression, promotes M2 polarization, and inhibits macrophage retention. Thioglycollate-elicited peritoneal macrophages were isolated from naïve mice and then cultured in 3T3-L1 adipocyte–conditioned medium containing SR141716A (10 μm) or DMSO for 24 h. A, qRT-PCR miRs. B–D, qRT-PCR of Klf4, Stat6, and Agap2. E, Netrin-1 ELISA from cell supernatants. For F and G, macrophages were stained with DilC12(3) and treated with SR141716A or DMSO. The migration rate toward CCL19 (500 ng/ml) was assessed by seeding 2.5 × 105 macrophages in serum-reduced conditioned medium from differentiated 3T3L-1 adipocytes in FluoroBlok plates. 12 h later, cell migration was quantified using Cytation5 imaging, and gene expression analysis was conducted for CCR7. F, the number of cells migrated toward CCL19. G, qRT-PCR expression of Ccr7. Data are shown as mean ± S.D. (error bars). **, p < 0.01; ****, p < 0.0001 versus DMSO vehicle control by two-tailed Student's t test.
Overexpression of miR-466 and miR-762 prevents the effects of CB1 antagonism in macrophages
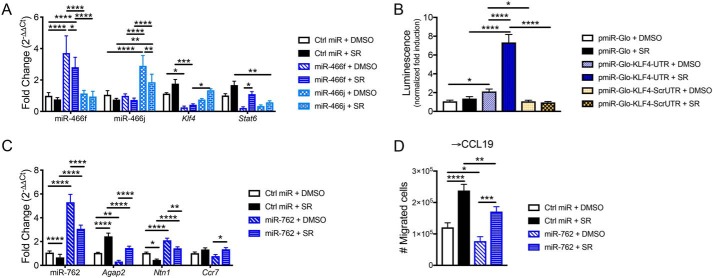
To verify that miRs mediate regulation of macrophage polarization and migration following CB1 antagonist treatment, we performed rescue experiments by overexpressing miR-466 and miR-762 in macrophages treated with SR141716A. Thioglycollate-elicited peritoneal macrophages were harvested from naïve mice and then transfected with miR mimics followed by 24-h incubation with conditioned medium from 3T3-L1 adipocytes and 10 μm SR141716A or DMSO vehicle. Transfection of miR-466f and miR-466j mimics led to up-regulation of the respective miRs, and likewise SR treatment reduced their expressions (Fig. 7A). Consistent with our claim that down-regulation of miR-466 promotes M2 polarization, gene expression of Klf4 and Stat6 showed negative correlation with expression of miR-466f and miR-466j, albeit miR-466f appeared to better regulate Stat6, and miR-466j better regulated Klf4 (Fig. 7A).
Figure 7.
Overexpression of miR-466f, miR-466j, and miR-762 reverses effects of SR141716A treatment in macrophages. Thioglycollate-elicited peritoneal macrophages were harvested from naïve mice; then transfected with a scrambled negative control miR oligonucleotide (Ctrl miR) or miR-466f, miR-466j, or miR-762 mimic; and then cultured for 24 h in conditioned medium from 3T3-L1 adipocytes + 10 μm SR or DMSO vehicle. A, qRT-PCR of miR-466f, miR-466j, Klf4, and Stat6 in macrophages transfected with miR-466f or miR-466j ± SR. For B, luciferase reporter assay was performed in RAW264.7 macrophages transfected with pmirGLO, pmirGLO-KLF4-UTR, or pmirGLO-KLF4-ScrUTR and then incubated for 24 h with SR141716A (10 μm) or DMSO vehicle. B, firefly luciferase activity was measured by luminescence and normalized to pmirGLO + DMSO negative control. For C and D, 5 × 105 peritoneal macrophages were plated in FluoroBlok inserts and then transfected with miR oligonucleotides and cultured for 24 h in conditioned medium from 3T3-L1 adipocytes + 10 μm SR or DMSO vehicle. At 12 h of culture, medium containing 500 ng/ml CCL19 was added to the bottom well, and migrated cells were harvested at 24 h for migration analysis and qRT-PCR. C, qRT-PCR of miR-762, Agap2, Ntn1, and Ccr7 in macrophages transfected with miR-762 ± SR. D, number of cells migrated toward CCL19. Data are mean ± S.D. (error bars). ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05 by one-way ANOVA.
To confirm that miR-466 directly targets the 3′-UTR of Klf4, we performed a luciferase reporter assay in the RAW264.7 macrophage cell line. RAW264.7 cells were transfected with empty vector, vector containing the 3′-UTR–binding site for miR-466, or a scrambled 3′-UTR negative control. The cells were then treated with SR141716A to induce down-regulation of miR-466. Cells transfected with the 3′-UTR of Klf4 + SR had significantly elevated luciferase activity that was not observed in negative controls (Fig. 7B). It should be noted that the luciferase activity was slightly increased in pmirGLO-KLF4-UTR + DMSO versus pmirGLO + DMSO, which could have resulted from experimental variation during transfection of various constructs or differential endogenous expression levels of miR-466 following transfection. Nonetheless, the significant increase in luciferase activity in pmirGLO-KLF4-UTR + SR versus pmirGLO-KLF4-UTR + DMSO confirmed that miR-466 likely directly targets the 3′-UTR of Klf4.
Next, to validate that miR-762 mediates regulation of macrophage chemotaxis following CB1 blockade, we plated peritoneal macrophages in transwell inserts and transfected them with miR-762 mimic followed by culture in conditioned medium from 3T3-L1 adipocytes + SR141716A. Half-way through the 24-h incubation, medium containing CCL19 was added to the bottom well, and the migrated cells were collected for gene expression and migration analysis at the 24-h time point. As expected, miR-762 expression was elevated in macrophages transfected with miR-762 mimic but reduced following SR treatment (Fig. 7C). Expression of the miR-762 target Agap2 inversely correlated with miR-762 expression (Fig. 7C). In accordance with Agap2 being a negative regulator of Netrin-1, Ntn1 expression was negatively correlated with that of Agap2 (Fig. 7C). Overexpression of miR-762 reduced migration toward CCL19 (Fig. 7D). Correspondingly, SR treatment rescued migration toward CCL19 in cells transfected with miR-762 (Fig. 7D). We also noted that expression of Ccr7, which encodes the CCL19 receptor CCR7, was mostly similar between the treatment groups (less than 25% difference from control) (Fig. 7C).
Discussion
The phytocannabinoid Δ9-tetrahydrocannabinol, a CB1 agonist, has been well characterized to stimulate appetite, thereby prompting studies to test whether CB1 antagonists such as SR141716A would exhibit anti-obesity properties (32, 33). SR141716A, also known as rimonabant, was tested in clinical trials, and although it showed great potential as an antiobesity agent, psychiatric side effects prevented it from being approved for use by the Food and Drug Administration (23). Nonetheless, understanding the mechanisms through which SR141716A mediates antiobesity and anti-inflammatory activities may help develop alternative targets against obesity. Consistent with previous studies, the current study demonstrated the beneficial effects of a CB1 receptor antagonist, specifically SR141716A, as an interventional treatment of obese phenotype (24, 34–36). Our data demonstrated that SR141716A suppressed food intake transiently; however, weight loss and reduction in fat mass were persistent.
Interestingly, in the current study, blockade of CB1 receptors in DIO phenotype resulted in altered expression of several miRs in ATMs, specifically down-regulation of the miR-466 family in ATMs, which targeted M2 transcription factors KLF4 and STAT6, thereby promoting M2 macrophage phenotype and accordingly inducing an anti-inflammatory state in adipose tissue. We confirmed that miR-466 directly targets the 3′-UTR of Klf4. SR141716A treatment reduced macrophage miR-466 expression both in vivo and in vitro. Overexpression of miR-466 in macrophages treated with SR141716A reversed SR-mediated induction of Klf4 and Stat6. Together, these data confirm that SR141716A treatment promotes M2 macrophage polarization through regulation of miR-466.
Additionally, we noted that treatment with SR141716A caused down-regulation of miR-762, which was found to target the 3′-UTR of the AGAP-2 gene, a negative regulator of UNC5B and Netrin-1 signaling. Previous studies have shown that Netrin-1, a neuroimmune guidance cue, promotes ATM retention, inflammation, and insulin resistance in obesity (17). Consistent with this observation, our studies demonstrated that treatment with SR141716A led to a decrease in the expression of UNC5B and Netrin-1 and increased macrophage migration toward lymph nodes. Thus, together, our studies demonstrated that treatment with SR141716A leads to alterations in the expression of miRs that promote macrophage differentiation into M2 as well as promote the emigration of ATMs from the adipose tissue, thereby supporting an anti-inflammatory microenvironment, which contributes to attenuation of insulin resistance and obesity.
Various studies have identified miRs involved in the regulation of obesity and macrophage polarization (37–39). More recently, we demonstrated that ATMs from lean and DIO mice display distinct transcript and miR expression profiles, which indicates that miRs are involved in regulation of ATM phenotype (30). Together, these findings suggest that miR-based therapies may be investigated as potential therapeutic interventions for metabolic diseases (40). Our results uncovered a key role for dysregulated miR in ATMs following SR141716A treatment in promoting an anti-inflammatory state in the visceral adipose tissue of DIO mice. Notably, we identified down-regulation of the miR-466 family in ATMs from obese mice following SR141716A treatment. The main target genes of the miR-466 family are Klf4 and Stat6. Previous studies revealed a key role for KLF4 in regulating M2 macrophage polarization (41, 42). Furthermore, a loss of function study of KLF4 in mice bearing myeloid-specific deletion of KLF4 exacerbated DIO severity and metabolic dysfunction (43). Current literature remains limited on the role of the miR-466 family in inflammatory responses. Indeed, a recent report from our laboratory defined a miR-466–TGF-β2 axis in which down-regulation of miR-466 led to induction of the cytokine TGF-β2 and subsequent induction of immunosuppressive Foxp3+ regulatory T cells (44). In addition, miR-466l has been shown to be induced upon acute inflammation and play a role in resolution, suggesting that the miR-466 family plays various roles in inflammatory processes (45, 46). Thus, our studies identified for the first time in ATMs that miR-466 down-regulation may play a critical role in the induction of KLF4 and STAT6 and subsequently M2 polarization following CB1 blockade.
Furthermore, using pathway analysis, we identified the unique interaction between altered miR in the HFD+SR group and molecules involved in the neuroimmune guidance cue pathway. Our data uncovered that therapeutic properties of SR141716A, in attenuation of adipose tissue inflammation, may involve targeting Netrin-1 and its receptor UNC5B in ATMs through miR-762–mediated regulation of AGAP-2. Recent studies demonstrated the novel role of the neuroimmune guidance cue Netrin-1 and its receptor UNC5B in regulating the chemotactic behavior of ATMs (17). Indeed, less accumulation of ATMs in adipose tissue of HFD-fed Netrin-1 knockout mice was identified when compared with HFD-fed WT mice (17). The key regulatory role of Netrin-1 in leukocyte transmigration into tissues (intestine, kidney, and atherosclerosis plaques) has also been identified (47–49). Conversely, abundant expression of Netrin-1 in residual macrophages of atherosclerotic plaques exacerbates the disease state by attracting more macrophages to the site of inflammation (50). Additionally, higher expression of Netrin-1 in endothelial cells of apolipoprotein E knockout and low-density lipoprotein receptor knockout mice ameliorates atherosclerosis by inhibiting leukocyte recruitment (51). Thus, conditional deletion of Netrin-1 and UNC5B in macrophages is required to eliminate the off-target effect of Netrin-1 deletion in other cells such as endothelial and epithelial cells. Our studies suggest that SR141716A may attenuate chronic low-grade inflammation in adipose tissue by targeting Netrin-1 pathways exclusively in macrophages.
Recently, the key functional role of Netrin-1 was established as a monocyte chemoattractant signal, identified by monitoring the migration pattern of macrophages into and out of atherosclerotic plaques as well as adipose tissue of DIO phenotype (52, 53). Utilizing the fluorescent microsphere monocyte labeling technique in HFD-fed Netrin-1 knockout mice, these mice demonstrated less retention of macrophages in adipose tissue and more emigration into draining mesenteric lymph nodes when compared with WT mice (17). We demonstrated herein that cultured peritoneal macrophages in harvested medium from 3T3-L1 adipocytes have reduced expression of miR-762, increased expression of Agap2, and decreased secretion of Netrin-1 and show higher migration to CCL19 in the presence of SR141716A when compared with DMSO, despite equivalent expression of Ccr7. Together, our studies suggested that SR141716A treatment alters miRs that target Netrin-1 signaling in ATMs, leading to lesser retention of macrophages in adipose tissue.
Although the role of Netrin-1 in ATM retention is well documented, the underlying mechanisms remain poorly defined. Herein, for the first time, we identified the key role of miR-762 and AGAP-2 in regulation of UNC5B in ATMs. In an earlier study, the association of AGAP-2 and UNC5B in a p53-dependent manner was identified in a cancer cell line (54). In the current study, we also identified that SR141716A treatment leads to miR-762 down-regulation in vivo in ATMs and in vitro in cultured peritoneal macrophages, leading to induction of AGAP-2, which down-regulates UNC5B expression and Netrin-1 activation. We also demonstrated direct binding of miR-762 to the 3′-UTR of the Agap2 gene. Overexpression of miR-762 in SR-treated macrophages reversed SR-mediated migration toward CCL19. Thus, SR141716A treatment targets both Netrin-1 and UNC5B in ATMs through alterations in miR expression.
This study provides new insights into the role of SR141716A in the induction of activity for Netrin-1 and Netrin-1–related receptors. However, further investigation is required for uncovering the potential effect of SR141716A on other neuronal immune guidance cue families such as Slit, Ephrin, Semaphorin, and their related receptors in ATMs.
Experimental procedures
Animals, diet, and treatment
C57BL/6J male mice (The Jackson Laboratory, Bar Harbor, ME), were fed HFD consisting of 60% kcal from fat (D12492, Research Diets Inc., New Brunswick, NJ) for 12 weeks before starting the intervention treatment with SR141716A. Additionally, age-matched C57BL/6J male mice were fed LFD consisting of 10% kcal from fat (D12450J, Research Diets Inc.) to develop lean controls. Only male mice were used due to their increased susceptibility to DIO development (55, 56). SR141716A was obtained from National Institute on Drug Abuse and administered at a dose of 10 mg/kg daily by oral gavage for 4 weeks, beginning after 12 weeks of diet and ending after 16 weeks. A pair-fed control group was used (HFD-PFSR) in which food intake was restricted to the amount of daily food intake of the HFD+SR group. All mice were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care–accredited animal facility at the University of South Carolina, School of Medicine (Columbia, SC). All procedures were performed according to National Institutes of Health guidelines under protocols approved by the Institutional Animal Care and Use Committee.
Analytical procedures
Body composition in mice was analyzed by dual-energy X-ray absorptiometry (GE Lunar Corp., Madison, WI). The mice were anesthetized and placed in the prone position on the specimen tray to allow scanning of the entire body. For food intake measurements, mice were given a defined amount of large intact food pellet weekly (or daily for the HFD-PFSR group). Food weight was measured using a balance with a precision of 0.01 g. Solid food intake was corrected for any visible spillage.
Assessment of metabolic parameters
Measurements of blood glucose, insulin, total cholesterol, and triglycerides were performed in animals fasted for 5 h. Blood was collected from the tip of the tail. The glucose level in whole blood was measured with a glucometer (Bayer, Leverkusen, Germany). Insulin concentration was determined in isolated plasma using an ELISA kit (Abcam, Cambridge, MA). Total cholesterol (Genzyme, Kent, UK) and triglycerides (Pointe Scientific, Canton, MI) were determined by colorimetric enzymatic reactions according to the manufacturers' instructions (57). To quantify insulin resistance, the HOMA-IR was calculated described as previously (58).
Tissue collection
After 4 weeks of treatment, mice were euthanized for tissue collection. Tissues were removed, weighed, and immediately processed to single-cell suspension, snap frozen in liquid nitrogen, and stored at −80 °C or fixed in 10% formalin.
Adipocyte and adipose tissue macrophage purification
Epididymal fat pads of mice were excised and placed in gentleMACS C Tubes (Miltenyi Biotec, San Diego, CA) containing digestion medium (Hanks' balanced salt solution, 2 mg/ml collagenase (Sigma-Aldrich), and 2% BSA) followed by homogenization utilizing a gentleMACS Dissociator (Miltenyi Biotec). After incubation at 37 °C for 30 min with gentle shaking, the cell suspension was filtered through a 100-μm filter and then spun at 300 × g for 5 min to separate floating adipocytes from the stromal vascular fraction pellet. Isolation of F4/80+ ATMs from the stromal vascular fraction was performed using EasySepTM FITC Positive Selection kit following the manufacturer's protocol (STEMCELL Technologies, Vancouver, British Columbia, Canada). Single-cell suspensions were first incubated with Fc receptor blocker consisting of rat IgG anti-mouse CD16/32 antibody to prevent nonspecific binding of FITC-conjugated antibodies to Fc receptors (BioLegend, catalog number 101301). Next, FITC-anti-F4/80 antibodies (Clone BM8, BioLegend, San Diego, CA) were used to label the cells for positive selection. Isolated cells were lysed for RNA analysis in Qiazol (Qiagen, Valencia, CA) and stored at −80 °C until RNA purification. Flow cytometry of the collected flow-through was used to evaluate selection efficiency, which was routinely >90% (Fig. S3).
RNA purification, cDNA synthesis, and quantitative RT-PCR
ATM RNA was isolated by miRNeasy Mini kit (Qiagen). For RNA extraction from whole adipose tissue, 1 mg of adipose tissue was homogenized in RNA-Solv Reagent with OBI's HiBind® technology reagent (Omega Bio-tek, Norcross, GA), and total RNA was isolated as described previously (57). RNA quality was verified by NanoDrop 2000C (Thermo Scientific, Waltham, MA). Purified RNA (0.5–1 μg) was reverse transcribed using an miScript cDNA Synthesis kit (Qiagen), and qRT-PCR analysis was conducted using a SsoAdvancedTM Universal SYBR® Green Supermix kit (Bio-Rad). Fold change in mRNA expression was calculated using the comparative cycle method (2−ΔΔCt).
miR expression profiling and analysis
The unique expression profile of miRs was assessed in F4/80+ cells isolated from adipose tissue by Affymetrix Gene Chip miRNA 4.0 array platform. The array contains 3100 murine-specific probes from Sanger miRBase. Total RNA was 3′-end labeled using the FlashTag Biotin HSR hybridization technique (Genisphere, Hatfield, PA) and was carried out according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). Microarray CEL files were assessed for quality, normalized, and converted to CHP files using the algorithm RMA-DABG in the software Affymetrix Expression Console version 1.4.1.46. CHP files were further analyzed using Affymetrix Transcriptome Analysis Console version 3.1.0.5 for annotation and differential expression. Expression log2 -fold change was calculated, and mean normalized expression was visualized in the form of a heat map. miRs were considered differentially expressed if the log2 -fold change was greater than ±1.5.
Bioinformatics analysis
The differentially expressed miR target genes were assessed by miR target prediction algorithms miRwalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/)3 (62) and miRmap (http://mirmap.ezlab.org)3 (63). To carry out an enrichment analysis of predicted target genes of miRs in biological pathways, the commercially available analysis tool IPA was used. IPA predicts the top affected canonical pathways, causal connections between differentially altered miRs and their target genes, downstream effectors, and upstream regulators. The Molecular Activity Predictor (MAP) feature of IPA was performed to predict the downstream effect of the differentially expressed miRs that were overlaid to the data set, including miR probes, -fold change, and p value. Gene ontology was assessed in the Cytoscape platform using ClueGO (59).
ELISAs
TNFα, IL-6, and IFNγ ELISA kits were purchased from BioLegend. Netrin-1 ELISA kits were purchased from USCN Life Science (Houston, TX). Concentrations were measured in cell supernatants using mouse standards according to the manufacturers' guidelines.
Migration
The migration of macrophages to CCL19 (500 ng/ml; R&D Systems, Minneapolis, MN) was assessed by FluoroBlok permeable inserts (Corning, 351152) and Cytation5 imaging (BioTek, Winooski, VT). Peritoneal macrophages were harvested from mice primed with 1 ml of 3% (w/v) thioglycolate (i.p.) to elicit peritoneal exudates with macrophage number peaking on day 4. Macrophages were collected in euthanized mice by intraperitoneal wash. For tracking migration toward chemoattractants, macrophages were labeled with DilC12(3) fluorescent dye (Corning). Macrophages were treated with conditioned medium from 3T3-L1 adipocytes as described and treated either with SR141716A (10−6 m) or DMSO as the vehicle control (60, 61).
Western blot analysis
Western blot analyses were carried out according to standard protocols with antibodies raised against Netrin-1 (1:1000; MAB1109, R&D Systems) or γ-tubulin (1:10,000; T5326, Sigma-Aldrich), which was used as a loading control.
Transfection and reporter gene assay
For miR-466 targeting of Klf4, RAW264.7 macrophages were transfected with 5–10 μg of pmirGLO, pmirGLO-KLF4-UTR, or pmirGLO-KLF4-ScrUTR plasmids using Effectene transfection reagent from Qiagen according to the manufacturer's protocol. The reporter construct was generated using mouse KLF4 UTR DNA sequences. To this end, we used pmirGLO reporter vector from Promega (Madison, WI). pmirGLO reporter vector contains two luciferase genes: 1) firefly luciferase reporter gene (luc2) that generates luminescence in the absence of microRNA and 2) Renilla luciferase reporter gene (hRluc-neo fusion protein coding region) that generates luminescence in the presence of microRNA. The mouse KLF4-specific UTR complementary to mouse miR-466F was cloned into pmirGLO vector and was designated as pmirGLO-KLF4-UTR or pmirGLO-KLF4-scrambled UTR (pmirGLO-KLF4-ScrUTR). The details of the KLF4 sequences cloned into pmirGLO are as follows. Both nucleotides of normal and scrambled miR-466F–specific KLF4 UTRs contain PmeI and XbaI restriction sites: miR-466 sense target sequence, 5′-AAACTAGCGGCCGCTAGTCACAACACACACGCACACT-3′; miR-466 antisense target sequence, 5′-CTAGAGTGTGCGTGTGTGTTGTGACTAGCGGCCGCTAGTTT-3′; miR-466 scrambled sense target sequence, 5′-AAACTAGCGGCCGCTAGTCACAACTTCTGTGCACACT-3′; miR-466 scrambled antisense target sequence, 5′-CTAGAGTGTGCTGTCTTGTTGTGACTAGCGGCCGCTAGTTT-3′. Two days post-transfection with plasmids, RAW264.7 cells were replated in triplicate in 96-well plate (75 μl/well), and the cells were treated with vehicle (DMSO) or SR141716A (10 μm) and incubated for 24 h at 37 °C at 5% CO2. Following treatments with vehicle or SR, luciferase assays were performed using the Dual-Glo Luciferase Assay system from Promega according to the manufacturer's protocol. In brief, an equal volume (75 μl/well) of Dual-Glo reagent was added to each well and thoroughly mixed. The cells were incubated for 10–15 min at room temperature to allow cells to lyse. Firefly luciferase activity was measured by reading the sample luminescence using Victor2 (PerkinElmer Life Sciences). After the first reading of the samples, Dual-Glo Stop & Glo reagent (75 μl/well) was added to each well, mixed thoroughly, and incubated for 10–15 min. Renilla luminescence was measured by reading the sample luminescence using Victor2. The ratio of luminescence from experimental samples to luminescence from the control reporter was calculated. The luminescence ratio was then normalized to the ratio of control wells. The relative luminescence ratio was calculated from the normalized ratios, and values were expressed as “normalized -fold induction.”
For miR-762 targeting of Agap2, BMDMs were transfected with a plasmid containing Renilla luciferase (transfection efficiency control) and 3′-UTR AGAP-2 (GenBankTM accession number NM_001301014.1 (UTR length, 808 bp; −383 to −787 bp) in a firefly luciferase reporter gene. Cloning details are as follows: the complete plasmid is 7097 bp; vector, Pezx-MT06; promoter, SV40; antibiotic, ampicillin; 5′ cutting site, AsiSI, EcoRI, BsiWI; 3′ cutting site, XhoI, SpeI; sequencing primers, forward 5′-GATCCGCGAGATCCTGAT-3′ and reverse 5′-CCTATTGGCGTTACTATG-3′ (GeneCopeiaTM, Rockville, MD). Cell cultures were transfected with miR-762 miRCURY LNA microRNA Inhibitor (5 nm) (Exiqon, Woburn, MA). Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher Scientific, Waltham, MA) was used for delivery of oligos into the cell.
Rescue experiments
Rescue experiments were performed by overexpressing miRs and then treating with SR141716A. Thioglycollate-elicited peritoneal macrophages were isolated from naïve mice and then transfected with miR-466f, miR-466j, miR-762, or AllStars negative control (“Ctrl miR”) miScript mimics (Qiagen). Lipofectamine 3000 (Thermo Fisher Scientific) was used as the transfection reagent. The macrophages were then cultured for 24 h in conditioned medium from 3T3-L1 adipocytes + 10 μm SR141716A or DMSO vehicle. For the cells transfected with miR-762, cells were plated in FluoroBlok inserts. Complete Dulbecco's modified Eagle's medium containing 500 ng/ml CCL19 (BioLegend, 587802) was added to the bottom well 12 h prior to harvest to induce migration.
Statistical analysis
For the in vivo mouse experiments, 10 mice were used per experimental group unless otherwise specified. For in vitro assays, all experiments were triplicated. Most experiments were repeated at least twice to see consistency. Statistical analyses were performed using GraphPad Prism version 7.000 (GraphPad Software, La Jolla, CA). Body weight, body composition outcomes, and metabolic outcomes were analyzed using a repeated measures two-way ANOVA. For statistical differences, one-way ANOVA was calculated for each experiment. Tukey's post hoc test was performed to analyze differences between groups. A p value of ≤0.05 was considered statistically significant.
Author contributions
P. M.-B., M. N., and P. S. N. conceptualization; P. M.-B. and K. M. data curation; P. M.-B. and K. M. formal analysis; P. M.-B., K. M., and N. P. S. validation; P. M.-B., K. M., and N. P. S. investigation; P. M.-B. and K. M. visualization; P. M.-B., K. M., N. P. S., E. E. Z., M. N., and P. S. N. methodology; P. M.-B. writing-original draft; P. M.-B., K. M., E. E. Z., M. N., and P. S. N. writing-review and editing; M. N. and P. S. N. resources; M. N. and P. S. N. supervision; M. N. and P. S. N. funding acquisition; M. N. and P. S. N. project administration.
Supplementary Material
This work was supported by National Institutes of Health Grants P01AT003961, P20GM103641, R01AT006888, R01ES019313, and R01MH094755. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Tables S1–S3.
The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE129273.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- ATM
- adipose tissue macrophage
- CB1
- cannabinoid receptor 1
- DIO
- diet-induced obesity
- HFD
- high-fat diet
- LFD
- low-fat diet
- SR
- SR141716A
- HFD+SR
- SR141716A-treated DIO group
- PFSR
- pair-fed to SR141716A group
- miR or miRNA
- microRNA
- TNF
- tumor necrosis factor
- IL
- interleukin
- IFN
- interferon
- iNOS
- inducible nitric oxide synthase
- PPAR
- peroxisome proliferator-activated receptor
- Veh
- vehicle
- HOMA-IR
- homeostatic model assessment of insulin resistance index
- IPA
- Ingenuity Pathway Analysis
- qRT-PCR
- quantitative RT-PCR
- UNC5B
- UNC-5 homology B
- BMDM
- bone marrow–derived macrophage
- TGF
- transforming growth factor
- Scr
- scrambled
- Ctrl
- control
- ANOVA
- analysis of variance
- Ntn
- Netrin
- DilC12(3)
- 1,1′-didodecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate.
References
- 1. Alberti K. G., Eckel R. H., Grundy S. M., Zimmet P. Z., Cleeman J. I., Donato K. A., Fruchart J. C., James W. P., Loria C. M., Smith S. C. Jr., International Diabetes Federation Task Force on Epidemiology and Prevention, National Heart, Lung, and Blood Institute, American Heart Association, World Heart Federation, International Atherosclerosis Society, et al. (2009) Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 120, 1640–1645 10.1161/CIRCULATIONAHA.109.192644 [DOI] [PubMed] [Google Scholar]
- 2. Ogden C. L., Carroll M. D., Kit B. K., and Flegal K. M. (2012) Prevalence of obesity and trends in body mass index among US children and adolescents, 1999–2010. JAMA 307, 483–490 10.1001/jama.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Esser N., L'homme L., De Roover A., Kohnen L., Scheen A. J., Moutschen M., Piette J., Legrand-Poels S., and Paquot N. (2013) Obesity phenotype is related to NLRP3 inflammasome activity and immunological profile of visceral adipose tissue. Diabetologia 56, 2487–2497 10.1007/s00125-013-3023-9 [DOI] [PubMed] [Google Scholar]
- 4. Enos R. T., Velázquez K. T., McClellan J. L., Cranford T. L., Walla M. D., and Murphy E. A. (2014) Reducing the dietary ω-6:ω-3 utilizing α-linolenic acid; not a sufficient therapy for attenuating high-fat-diet-induced obesity development nor related detrimental metabolic and adipose tissue inflammatory outcomes. PLoS One 9, e94897 10.1371/journal.pone.0094897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Rourke R. W., White A. E., Metcalf M. D., Olivas A. S., Mitra P., Larison W. G., Cheang E. C., Varlamov O., Corless C. L., Roberts C. T. Jr., and Marks D. L. (2011) Hypoxia-induced inflammatory cytokine secretion in human adipose tissue stromovascular cells. Diabetologia 54, 1480–1490 10.1007/s00125-011-2103-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weisberg S. P., McCann D., Desai M., Rosenbaum M., Leibel R. L., and Ferrante A. W. (2003) Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 112, 1796–1808 10.1172/JCI200319246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hotamisligil G. S., Shargill N. S., and Spiegelman B. M. (1993) Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science 259, 87–91 10.1126/science.7678183 [DOI] [PubMed] [Google Scholar]
- 8. Vandanmagsar B., Youm Y.-H., Ravussin A., Galgani J. E., Stadler K., Mynatt R. L., Ravussin E., Stephens J. M., and Dixit V. D. (2011) The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 17, 179–188 10.1038/nm.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wen H., Gris D., Lei Y., Jha S., Zhang L., Huang M. T., Brickey W. J., and Ting J. P.-Y. (2011) Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 12, 408–415 10.1038/ni.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gordon S., and Taylor P. R. (2005) Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- 11. Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., and Locati M. (2004) The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25, 677–686 10.1016/j.it.2004.09.015 [DOI] [PubMed] [Google Scholar]
- 12. Gordon S. (2003) Alternative activation of macrophages. Nat. Rev. Immunol. 3, 23–35 10.1038/nri978 [DOI] [PubMed] [Google Scholar]
- 13. Lumeng C. N., Bodzin J. L., and Saltiel A. R. (2007) Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Investig. 117, 175–184 10.1172/JCI29881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Odegaard J. I., Ricardo-Gonzalez R. R., Goforth M. H., Morel C. R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A. W., and Chawla A. (2007) Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447, 1116–1120 10.1038/nature05894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang X., Wang X., Liu D., Yu L., Xue B., and Shi H. (2014) Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Mol. Endocrinol. 28, 565–574 10.1210/me.2013-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takamatsu H., and Kumanogoh A. (2012) Diverse roles for semaphorin-plexin signaling in the immune system. Trends Immunol. 33, 127–135 10.1016/j.it.2012.01.008 [DOI] [PubMed] [Google Scholar]
- 17. Ramkhelawon B., Hennessy E. J., Ménager M., Ray T. D., Sheedy F. J., Hutchison S., Wanschel A., Oldebeken S., Geoffrion M., Spiro W., Miller G., McPherson R., Rayner K. J., and Moore K. J. (2014) Netrin-1 promotes adipose tissue macrophage retention and insulin resistance in obesity. Nat. Med. 20, 377–384 10.1038/nm.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shimizu I., Yoshida Y., Moriya J., Nojima A., Uemura A., Kobayashi Y., and Minamino T. (2013) Semaphorin3E-induced inflammation contributes to insulin resistance in dietary obesity. Cell Metab. 18, 491–504 10.1016/j.cmet.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 19. Ziccardi P., Nappo F., Giugliano G., Esposito K., Marfella R., Cioffi M., D'Andrea F., Molinari A. M., and Giugliano D. (2002) Reduction of inflammatory cytokine concentrations and improvement of endothelial functions in obese women after weight loss over one year. Circulation 105, 804–809 10.1161/hc0702.104279 [DOI] [PubMed] [Google Scholar]
- 20. Yang W. S., Lee W. J., Funahashi T., Tanaka S., Matsuzawa Y., Chao C. L., Chen C. L., Tai T. Y., and Chuang L. M. (2001) Weight reduction increases plasma levels of an adipose-derived anti-inflammatory protein, adiponectin. J. Clin. Endocrinol. Metab. 86, 3815–3819 10.1210/jcem.86.8.7741 [DOI] [PubMed] [Google Scholar]
- 21. Perkins J. M., and Davis S. N. (2008) Endocannabinoid system overactivity and the metabolic syndrome: prospects for treatment. Curr. Diab. Rep. 8, 12–19 10.1007/s11892-008-0004-3 [DOI] [PubMed] [Google Scholar]
- 22. Pi-Sunyer F. X., Aronne L. J., Heshmati H. M., Devin J., Rosenstock J., and RIO-North America Study Group (2006) Effect of rimonabant, a cannabinoid-1 receptor blocker, on weight and cardiometabolic risk factors in overweight or obese patients: RIO-North America: a randomized controlled trial. JAMA 295, 761–775 10.1001/jama.295.7.761 [DOI] [PubMed] [Google Scholar]
- 23. Sam A. H., Salem V., and Ghatei M. A. (2011) Rimonabant: from RIO to ban. J. Obes. 2011, 432607 10.1155/2011/432607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jbilo O., Ravinet-Trillou C., Arnone M., Buisson I., Bribes E., Péleraux A., Pénarier G., Soubrié P., Le Fur G., Galiègue S., and Casellas P. (2005) The CB1 receptor antagonist rimonabant reverses the diet-induced obesity phenotype through the regulation of lipolysis and energy balance. FASEB J. 19, 1567–1569 10.1096/fj.04-3177fje [DOI] [PubMed] [Google Scholar]
- 25. Wang Q., Perrard X. D., Perrard J. L., Mansoori A., Smith C. W., Ballantyne C. M., and Wu H. (2011) Effect of the cannabinoid receptor-1 antagonist rimonabant on inflammation in mice with diet-induced obesity. Obesity 19, 505–513 10.1038/oby.2010.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 27. Gauthier B. R., and Wollheim C. B. (2006) MicroRNAs: “ribo-regulators” of glucose homeostasis. Nat. Med. 12, 36–38 10.1038/nm0106-36 [DOI] [PubMed] [Google Scholar]
- 28. Wang Q., Li Y. C., Wang J., Kong J., Qi Y., Quigg R. J., and Li X. (2008) miR-17–92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl. Acad. Sci. U.S.A. 105, 2889–2894 10.1073/pnas.0800178105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., Subramaniam A., Propp S., Lollo B. A., Freier S., Bennett C. F., et al. (2006) miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98 10.1016/j.cmet.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 30. Miranda K., Yang X., Bam M., Murphy E. A., Nagarkatti P. S., and Nagarkatti M. (2018) MicroRNA-30 modulates metabolic inflammation by regulating Notch signaling in adipose tissue macrophages. Int. J. Obes. 42, 1140–1150 10.1038/s41366-018-0114-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mehrpouya-Bahrami P., Chitrala K. N., Ganewatta M. S., Tang C., Murphy E. A., Enos R. T., Velazquez K. T., McCellan J., Nagarkatti M., and Nagarkatti P. (2017) Blockade of CB1 cannabinoid receptor alters gut microbiota and attenuates inflammation and diet-induced obesity. Sci. Rep. 7, 15645 10.1038/s41598-017-15154-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sofia R. D., and Barry H. 3rd (1974) Acute and chronic effects of Δ9-tetrahydrocannabinol on food intake by rats. Psychopharmacologia 39, 213–222 10.1007/BF00421028 [DOI] [PubMed] [Google Scholar]
- 33. Rinaldi-Carmona M., Barth F., Héaulme M., Shire D., Calandra B., Congy C., Martinez S., Maruani J., Néliat G., and Caput D. (1994) SR141716A, a potent and selective antagonist of the brain cannabinoid receptor. FEBS Lett. 350, 240–244 10.1016/0014-5793(94)00773-X [DOI] [PubMed] [Google Scholar]
- 34. Després J.-P., Golay A., Sjöström L., and Rimonabant in Obesity-Lipids Study Group (2005) Effects of rimonabant on metabolic risk factors in overweight patients with dyslipidemia. N. Engl. J. Med. 353, 2121–2134 [DOI] [PubMed] [Google Scholar]
- 35. Schäfer A., Pfrang J., Neumüller J., Fiedler S., Ertl G., and Bauersachs J. (2008) The cannabinoid receptor-1 antagonist rimonabant inhibits platelet activation and reduces pro-inflammatory chemokines and leukocytes in Zucker rats. Br. J. Pharmacol. 154, 1047–1054 10.1038/bjp.2008.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cota D., Sandoval D. A., Olivieri M., Prodi E., D'Alessio D. A., Woods S. C., Seeley R. J., and Obici S. (2009) Food intake-independent effects of CB1 antagonism on glucose and lipid metabolism. Obesity 17, 1641–1645 10.1038/oby.2009.84 [DOI] [PubMed] [Google Scholar]
- 37. Klöting N., Berthold S., Kovacs P., Schön M. R., Fasshauer M., Ruschke K., Stumvoll M., and Blüher M. (2009) MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One 4, e4699 10.1371/journal.pone.0004699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taganov K. D., Boldin M. P., Chang K.-J., and Baltimore D. (2006) NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y., Zhang M., Zhong M., Suo Q., and Lv K. (2013) Expression profiles of miRNAs in polarized macrophages. Int. J. Mol. Med. 31, 797–802 10.3892/ijmm.2013.1260 [DOI] [PubMed] [Google Scholar]
- 40. Ge Q., Brichard S., Yi X., and Li Q. (2014) microRNAs as a new mechanism regulating adipose tissue inflammation in obesity and as a novel therapeutic strategy in the metabolic syndrome. J. Immunol. Res. 2014, 987285 10.1155/2014/987285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Odegaard J. I., Ricardo-Gonzalez R. R., Red Eagle A., Vats D., Morel C. R., Goforth M. H., Subramanian V., Mukundan L., Ferrante A. W., and Chawla A. (2008) Alternative M2 activation of Kupffer cells by PPARδ ameliorates obesity-induced insulin resistance. Cell Metab. 7, 496–507 10.1016/j.cmet.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eisenstein A., Carroll S. H., Johnston-Cox H., Farb M., Gokce N., and Ravid K. (2014) An adenosine receptor-Krüppel-like factor 4 protein axis inhibits adipogenesis. J. Biol. Chem. 289, 21071–21081 10.1074/jbc.M114.566406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liao X., Sharma N., Kapadia F., Zhou G., Lu Y., Hong H., Paruchuri K., Mahabeleshwar G. H., Dalmas E., Venteclef N., Flask C. A., Kim J., Doreian B. W., Lu K. Q., Kaestner K. H., et al. (2011) Krüppel-like factor 4 regulates macrophage polarization. J. Clin. Investig. 121, 2736–2749 10.1172/JCI45444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Becker W., Nagarkatti M., and Nagarkatti P. S. (2018) miR-466a targeting of TGF-β2 contributes to FoxP3+ regulatory T cell differentiation in a murine model of allogeneic transplantation. Front. Immunol. 9, 688 10.3389/fimmu.2018.00688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma F., Liu X., Li D., Wang P., Li N., Lu L., and Cao X. (2010) MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J. Immunol. 184, 6053–6059 10.4049/jimmunol.0902308 [DOI] [PubMed] [Google Scholar]
- 46. Li Y., Dalli J., Chiang N., Baron R. M., Quintana C., and Serhan C. N. (2013) Plasticity of leukocytic exudates in resolving acute inflammation is regulated by microRNA and proresolving mediators. Immunity 39, 885–898 10.1016/j.immuni.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang W., Reeves W. B., Pays L., Mehlen P., and Ramesh G. (2009) Netrin-1 overexpression protects kidney from ischemia reperfusion injury by suppressing apoptosis. Am. J. Pathol. 175, 1010–1018 10.2353/ajpath.2009.090224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ly N. P., Komatsuzaki K., Fraser I. P., Tseng A. A., Prodhan P., Moore K. J., and Kinane T. B. (2005) Netrin-1 inhibits leukocyte migration in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 14729–14734 10.1073/pnas.0506233102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosenberger P., Schwab J. M., Mirakaj V., Masekowsky E., Mager A., Morote-Garcia J. C., Unertl K., and Eltzschig H. K. (2009) Hypoxia-inducible factor-dependent induction of netrin-1 dampens inflammation caused by hypoxia. Nat. Immunol. 10, 195–202 10.1038/ni.1683 [DOI] [PubMed] [Google Scholar]
- 50. van Gils J. M., Derby M. C., Fernandes L. R., Ramkhelawon B., Ray T. D., Rayner K. J., Parathath S., Distel E., Feig J. L., Alvarez-Leite J. I., Rayner A. J., McDonald T. O., O'Brien K. D., Stuart L. M., Fisher E. A., et al. (2012) The neuroimmune guidance cue netrin-1 promotes atherosclerosis by inhibiting the emigration of macrophages from plaques. Nat. Immunol. 13, 136–143 10.1038/ni.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khan J. A., Cao M., Kang B. Y., Liu Y., Mehta J. L., and Hermonat P. L. (2011) Systemic human Netrin-1 gene delivery by adeno-associated virus type 8 alters leukocyte accumulation and atherogenesis in vivo. Gene Ther. 18, 437–444 10.1038/gt.2010.155 [DOI] [PubMed] [Google Scholar]
- 52. Tacke F., Alvarez D., Kaplan T. J., Jakubzick C., Spanbroek R., Llodra J., Garin A., Liu J., Mack M., van Rooijen N., Lira S. A., Habenicht A. J., and Randolph G. J. (2007) Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J. Clin. Investig. 117, 185–194 10.1172/JCI28549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Feig J. E., Pineda-Torra I., Sanson M., Bradley M. N., Vengrenyuk Y., Bogunovic D., Gautier E. L., Rubinstein D., Hong C., Liu J., Wu C., van Rooijen N., Bhardwaj N., Garabedian M., Tontonoz P., et al. (2010) LXR promotes the maximal egress of monocyte-derived cells from mouse aortic plaques during atherosclerosis regression. J. Clin. Investig. 120, 4415–4424 10.1172/JCI38911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He K., Jang S.-W., Joshi J., Yoo M.-H., and Ye K. (2011) Akt-phosphorylated PIKE-A inhibits UNC5B-induced apoptosis in cancer cell lines in a p53-dependent manner. Mol. Biol. Cell 22, 1943–1954 10.1091/mbc.e10-11-0923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hwang L.-L., Wang C.-H., Li T.-L., Chang S.-D., Lin L.-C., Chen C.-P., Chen C.-T., Liang K.-C., Ho I.-K., Yang W.-S., and Chiou L.-C. (2010) Sex differences in high-fat diet-induced obesity, metabolic alterations and learning, and synaptic plasticity deficits in mice. Obesity 18, 463–469 10.1038/oby.2009.273 [DOI] [PubMed] [Google Scholar]
- 56. Medrikova D., Jilkova Z. M., Bardova K., Janovska P., Rossmeisl M., and Kopecky J. (2012) Sex differences during the course of diet-induced obesity in mice: adipose tissue expandability and glycemic control. Int. J. Obes. 36, 262–272 10.1038/ijo.2011.87 [DOI] [PubMed] [Google Scholar]
- 57. Enos R. T., Davis J. M., Velázquez K. T., McClellan J. L., Day S. D., Carnevale K. A., and Murphy E. A. (2013) Influence of dietary saturated fat content on adiposity, macrophage behavior, inflammation, and metabolism: composition matters. J. Lipid Res. 54, 152–163 10.1194/jlr.M030700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matthews D. R., Hosker J. P., Rudenski A. S., Naylor B. A., Treacher D. F., and Turner R. C. (1985) Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 10.1007/BF00280883 [DOI] [PubMed] [Google Scholar]
- 59. Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.-H., Pagès F., Trajanoski Z., and Galon J. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maianti J. P., McFedries A., Foda Z. H., Kleiner R. E., Du X. Q., Leissring M. A., Tang W.-J., Charron M. J., Seeliger M. A., Saghatelian A., and Liu D. R. (2014) Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature 511, 94–98 10.1038/nature13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sacerdote P., Martucci C., Vaccani A., Bariselli F., Panerai A. E., Colombo A., Parolaro D., and Massi P. (2005) The nonpsychoactive component of marijuana cannabidiol modulates chemotaxis and IL-10 and IL-12 production of murine macrophages both in vivo and in vitro. J. Neuroimmunol. 159, 97–105 10.1016/j.jneuroim.2004.10.003 [DOI] [PubMed] [Google Scholar]
- 62. Dweep H., Sticht C., Pandey P., and Gretz N. (2011) miRWalk - database: prediction of possible miRNA binding sites by “walking” the genes of 3 genomes. J. Biomed. Inform. 44, 839–847 10.1016/j.jbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 63. Vejnar C. E., and Zdobnov E. M. (2012) miRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 40, 11673–11683 10.1093/nar/gks901 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.