Abstract
Innovation follows discovery. If the 20th century was a golden age of discovery in the biomolecular biosciences, the current century may be remembered by the explosion of beneficial devices and therapies conceived by the bioengineers of the era. Much as the development of solid-state electronic components made possible the information revolution, the rational combining of millions of basic molecular control modules will enable the development of highly sophisticated biomachines that will make today's smartphones appear rudimentary. The molecular toolbox is already well-stocked, particularly in our ability to manipulate DNA, control transcription, generate functionally novel hybrid proteins, and expand the genetic code to include unnatural amino acids. This review focuses on how RNA-based regulatory modules that direct alternative readings of the genetic code can be employed as basic circuit components to expand our ability to control gene expression.
Keywords: translation, RNA, ribosome, synthetic biology, bioengineering, circuitry, frameshift, readthrough, recoding
Introduction
Since the elucidation of the genetic code in the early 1960s (1), the common perception has been that 1) all codons encode identical information in all organisms, and 2) coding sequences are “fixed” (i.e. ribosomes must maintain translational reading frame to faithfully decode all of the information contained in mRNAs). Shortly thereafter, however, bacteriophage geneticists identified bacterial mutants capable of suppressing stop codons in viral mRNAs (2), which led to the discovery of tRNAs (called suppressor tRNAs) in both bacteria (3) and eukaryotes (4) that are capable of reassigning the meaning of “stop codons” to encode specific amino acids. In the next decade, there were additional discoveries of cis-acting mRNA elements that direct ribosomes to reassign the meanings of codons or induce ribosomes to shift into alternative reading frames. This general phenomenon is collectively referred to as “translational recoding,” which is defined as instances in which “the rules for decoding are temporarily altered through the action of specific signals built into the mRNA sequences” (5).
In a unique and thought-provoking research article, Anzalone et al. (6) exploited RNA recoding elements to create a gene expression control circuit. They employed a directed evolutionary approach in vitro to identify RNA aptamers that stimulated the activity of a translational recoding element. These aptamers were then exploited to construct single-mRNA logic gates that were applied to control a new module governing cell death. This work marked a seminal milestone in the effort to integrate translational reprogramming modules into the synthetic biology toolbox. Importantly, these synthetic elements enabled manipulation of gene expression during the elongation phase of translation, thus addressing a critical gap in the synthetic biology toolbox for regulating gene expression in synthetic biology.
Translational recoding elements
Translational recoding elements direct information encoded in mRNAs to be translated in variance to the canonical rules of the genetic code. Their modular nature enables individual genes to encode multiple proteins, a recurrent theme in mammalian genomes. The processes governing recoding are dynamic, yielding products from both canonical and recoded reading. The relative proportions of the products from each are dependent on the gene configuration. The two most common types of recoding elements program ribosomes to either reassign the meaning of a termination codon (termination codon reassignment (TCR)2) or slip or shift the reading frame of an mRNA (designated programmed ribosomal frameshifting (PRF)). PRF signals typically direct ribosomes to slip by one base in either the 5′ (−1) or the 3′ (+1) direction. Less common are recoding elements that cause ribosomes to bypass defined segments of mRNAs (ribosome shunting) or those that reassign the meaning of a codon to encode an alternative amino acid (e.g. selenocysteine incorporation signals).
This review will focus on how TCR and PRF modules can be employed akin to electronic circuit components (see Box) to differentially engineer gene expression during the process of protein synthesis. In general, these mRNA regulatory elements are relatively small, with lengths ranging from ∼30 to 100 nucleotides. These regulatory elements function in cis and are bipartite, composed of 1) a special sequence at which the recoding event occurs and 2) a downstream (3′) RNA structural element that directs translating ribosomes to pause over the recoding sequence. An exception to this general rule is the Ty1 +1 PRF element, in which only a 7-nucleotide sequence is sufficient to direct recoding.
Recoding elements attenuate translation and thereby function as resistors
Many viral mRNAs (7), and an increasing number of cellular transcripts (8), harbor TCR elements. It is important to emphasize that TCR events are almost never 100% efficient; only a fraction of the ribosomes that encounter a TCR signal misread the stop codon, whereas the remaining decode it “correctly.” I suggest that the analogous electrical circuit component is a resistor, because by decreasing the fraction of ribosomes that are able to translate through them, TCR elements effectively decrease the amplitude of the signal output. For example, by Ohm's law (V = IR), if a 100-ohm resistor (R) is attached across a 12-volt battery (V), then a current (I) of 12/100 = 0.12 amperes flows through that resistor (i.e. the amount of current/signal strength exiting the resistor is attenuated 100-fold). By analogy, if a TCR with 10% efficiency is placed within an ORF, then only 90% of initiating ribosomes terminate at the stop codon, allowing 10% of the ribosomes to continue to translate the remaining ORF (i.e. a 10-fold signal attenuation is achieved) (Fig. 1A). PRF signals can be configured within an mRNA, such that either the majority of (Fig. 1B) or only a few (Fig. 1C) translating ribosomes are able to synthesize the complete, functional protein. As a consequence, TCRs can be engineered to generate a wide range of “resistance values.” Unlike electronics, where the relationship between resistance and signal strength is linear, in biological circuits controlled by recoding signals, an exponential correlation applies. The reasons underlying this difference in scaling are discussed in greater detail below.
Figure 1.
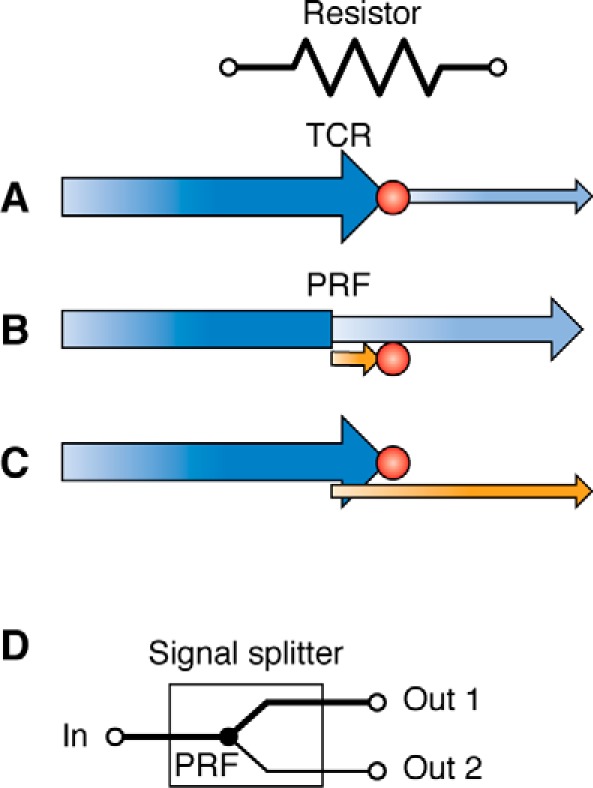
Translational recoding signals are analogous to electrical resistors. The symbol for resistor is shown at the top. Incoming information (5′ coding information) is shown in dark blue. Outgoing information in the original reading frame (3′ coding information) is shown in light blue. Outgoing information in alternative reading frames is shown in orange. The amplitude of each signal is indicated by arrow height. A, a TCR signal functions as a resistor by decreasing the fraction of ribosomes that are able to decode 3′ genetic information, thus decreasing the amount (amplitude) of the total protein synthesized from the gene. B, by directing a fraction of ribosomes to a “dead end” alternative reading frame, a PRF signal decreases the fraction of ribosomes that decode the original ORF, decreasing the amount (signal amplitude) of protein synthesized. This is most often utilized to control cellular gene expression. C, by directing only a small number of ribosomes to the 3′ information required to synthesize the full-length protein, the synthesis of this protein relative to a truncated protein encoded by the 5′ 0-frame ORF can be limited. This is most often used to control viral gene expression. D, a PRF signal can also function as a signal splitter.
In addition to attenuating signal strengths, PRF elements can also function as signal splitters by bifurcating the pathways that ribosomes can take to produce functional proteins (Fig. 1D). This is particularly important when the nonshifted and shifted protein products have different functions. For example, in most retroviruses, the first half of the genome is organized as shown in Fig. 1C. Here, the 5′ portion encodes structural proteins, whereas the frameshifted 3′ product encodes proteins with enzymatic functions. Viruses require relatively large quantities of structural elements to make new viral particles, whereas only a few enzymes are needed for viral replication. Splitting the signal to optimize the ratios of these two products enables controlling two outputs from a single input.
Converting simple resistors into sophisticated circuit elements
Recoding elements can also be configured to function akin to other passive and active circuitry components, including rheostats, transistors, and diodes. To help illuminate this, a brief discussion of translation elongation is first required. This cyclical process can be broken down into an ordered series of biochemical steps, each of which has been kinetically optimized to ensure that the ribosome efficiently utilizes 1) diverse sets of trans-acting factors (elongation factors, tRNAs, and release factors) and 2) intrinsic biochemical activities, to ensure accurate translation of a diverse array of mRNAs (9). Like all synthetic programs, altering the kinetics (speed or rate) of the main reactions can change the probability that side reactions, such as translational recoding, may occur. Importantly, the RNA elements that drive most cases of translational recoding alter the processivity kinetics of elongating ribosomes by directing ribosomes to pause at specific coding sites in mRNAs. Directed pausing is critical, as the mRNA sequence over which a ribosome is stopped helps to lower the energy barrier to the side reaction.
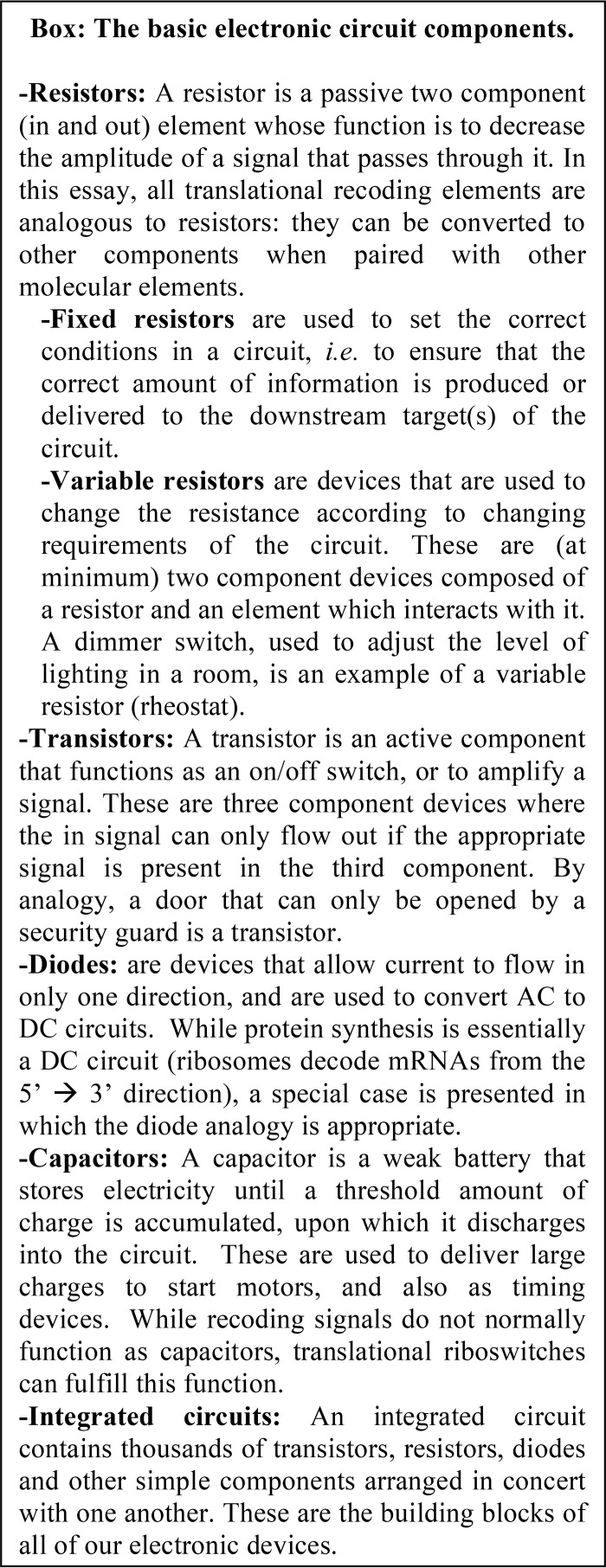
Ribosomes can be directed to pause, or become “kinetically trapped,” on mRNAs by cis-acting sequences, trans-acting factors, or combinations of both. The simplest example of a cis-acting kinetic trap is a termination codon. Because these are poorly recognized by tRNAs, these codons normally cause ribosomes to stop and await recognition by a release factor complex (RF), which then promotes termination of translation. Whereas the normal rate of termination codon misreading is <1%, if RF is depleted, ribosomes are paused at stop codons for longer times, providing the opportunity for them to utilize a tRNA that inefficiently recognizes the termination codon. This process allows TCR rates to be increased by an order of magnitude. Similarly, ribosomes can be kinetically trapped by limiting the concentration of a tRNA that is required to decode a particular codon. This is the basis for +1 PRF promoted by the yeast Ty1 retrotransposable element (10–12). In this system, the kinetic trap is supplied by the rare in-frame (0-frame) A-site AGG codon that is embedded in the heptameric sequence CUU AGG C (where the incoming, or 0-frame, is indicated by spaces). This codon is decoded by a very low-abundance tRNA, which is encoded by the HSX1 gene. As ribosomes pause at this codon awaiting this tRNA, they have time for the tRNA that occupies the ribosomal P-site to slip from the 0-frame CUU to +1-frame UUA, placing the GGC codon in the ribosomal A-site (C UUA GGC). The GGC codon is recognized by a highly abundant tRNA encoding glycine. The resulting kinetic competition between the low-abundance Arg-tRNA for the 0-frame codon and the high-abundance Gly-tRNA for the +1-frame codon results in a kinetic partitioning rate of 0.6 (i.e. ∼40% of the ribosomes shift into the new reading frame). In its simplest form, the codon and adjacent mRNA sequences could be employed as a resistor, reducing the amplitude of the input signal by 40% (if in-frame translation is required for synthesis of the functional protein) (Fig. 2A).
Figure 2.
The Ty1 +1 PRF signal as an example of how a resistor can be converted into a transistor. A, left, the yeast Ty1 +1 PRF signal promotes an efficiency of ∼40%. Thus, the amplitude of the signal leaving this element is 0.4 relative to that of the incoming signal. This is indicated by lighter blue coloring. Right, the Arg-tRNA that recognizes the AGG codon is encoded by the HSX1 gene. When HSX1 is on, the abundance of this tRNA is high, and the AGG codon is decoded normally. When HSX1 is off, the Arg-tRNA is absent, enabling the ribosome to slip by one base in the 3′ (+1) direction. This positions the GGC codon in the A-site, which is decoded by a high abundance Gly-tRNA, establishing the frameshift. B, +1 PRF depends on the concentration of a low-abundance Arg-tRNA, encoded by the yeast HSX1 gene. When HSX1 is turned off, this tRNA is not synthesized, and +1 PRF is 100%. Conversely, when it is turned on, the tRNA is present in high abundance, and +1 PRF is 0% (indicated by white fill). Depending on the orientation of the outgoing ORF, this system can be configured as OR or AND logic gates.
Importantly, ribosome pausing at codons is malleable; +1 PRF can be reduced to zero by overexpression of HSX1, and conversely, it can be increased to 100% efficiency by turning off HSX1 expression (12). This example of “overlaid control” can be exploited to construct logic gates (i.e. transistors) (Fig. 2). For example, controlling HSX1 transcription with an inducible/repressible gene promoter could be used as the basis for turning Ty1 +1 PRF on and off (Fig. 2B). Specifically, repression of HSX1 transcriptional expression could be used to switch the +1 PRF signal into the “on” position, whereas induction of this tRNA would switch this mechanism “off.” This is the essence of a logic gate where one or more inputs are processed to produce a single (binary) output. Depending on whether expression of the functional proteins require continued 0-frame expression or a frameshift event, this system could be configured as an OR gate or an AND gate, respectively.
As discussed above, cis-acting mRNA structural elements located proximally downstream of specific “slippery” sequences are commonly employed to provide kinetic traps that drive recoding. The best-studied systems that employ slippery sequences followed by mRNA structural elements are those that direct programmed −1 ribosomal frameshifting (−1 PRF). The cis-acting mRNA structural elements can be either a simple stem-loop or a more complex mRNA pseudoknot (ΨK; see example in Fig. 3A). Because these mRNA secondary structures are difficult to unwind during translation, they can cause ribosomes to pause, creating a trapping activity. When these mRNA structures are placed at an appropriate distance 3′ of a special heptameric slippery sequences (X XXY YYZ, where the incoming reading frame is denoted by spaces), tRNAs that are base-paired to the XXY YYZ codons are afforded the time to slip backward by one base, repairing their nonwobble bases to the XXX YYY codons (13). Whereas these secondary structures are very effective drivers of −1 PRF, it should be noted that having to build appropriate mRNA structures into amino acid coding sequences presents an added complication to genetic engineering applications. In other words, the desired coding sequence of the mRNA can be restricted by the engineered regulatory secondary structure that differentially drives frameshifts.
Figure 3.
Creating a wide range of variable −1 PRF signals/resistors (rheostats and potentiometers) using trans-acting factors. A, trans-acting nucleic acid–derived elements. A variety of possible −1 PRF elements are shown, graphed relative to ranges of −1 PRF that they can promote. In general, more stable downstream stimulatory elements promote higher levels of −1 PRF and thus serve to increase resistor strength. For example, simple stem-loops (SL) are less thermodynamically stable and thus typically promote low levels of −1 PRF (in the range of 1%); these can be used as weak resistors. In contrast, mRNA pseudoknots (ΨK) are more stable, promote higher rates of −1 PRF (in the range of 10%), and can thus be used as stronger resistors. Trans-acting RNAs, DNAs, or synthetic nucleic acid-derived oligomers can be variously employed to create pseudo-stem-loops (ΨSL) (19, 20), pseudo-pseudoknots (ΨΨK) (21), pseudoknots in combination with micro-RNAs (ΨK+ miR) (22), and potentially even pseudo-pseudo-pseudoknots (ΨΨΨK), thus generating a wide range of resistors. B, −1 PRF stimulated by the encephalomyocarditis virus protein 2A as an example of −1 PRF stimulated by a trans-acting protein (25).
Generating a broad range of resistors
Regardless of whether a −1 PRF signal directs translating ribosomes to produce a functional C-terminally extended protein or a premature termination codon (see Fig. 1), they function as resistors because fewer ribosomes will continue to translate beyond the frameshift signal. As a general rule, −1 PRF efficiency is determined by a combination of the “slipperiness” of the slippery heptamer and the thermodynamic stability of the downstream stimulatory element. Although no precise quantitative formulation has been determined, in general, −1 PRF rates tend to be maximized according to the A/U content of slippery sites and by the thermodynamic stability of downstream stimulatory elements (14–18). Focusing on the downstream elements, because simple stem-loops are relatively easy for ribosomes to unwind, they are thought to make ribosomes pause for short periods of time and generally have weak −1 PRF activity (in the range of a few percent). In contrast, mRNA pseudoknots present a greater challenge because their downstream stems must be fully denatured before an elongating ribosome can completely unwind the upstream stems. As a consequence, mRNA pseudoknots tend to promote higher rates of −1 PRF (Fig. 3A, compare SL with ΨK). Given the large number of −1 PRF signals that have been characterized to date, a wide range of these resistors are immediately available for synthetic biology applications.
Employing trans-acting modulators to make variable resistors
The recoding toolbox need not be limited to naturally occurring −1 PRF signals: trans-acting oligonucleotides that interact with specific mRNA sequences can be used to create new recoding elements in mRNAs. For example, a trans-acting oligonucleotide can anneal to a specific “flat” sequence to form a pseudo-stem-loop (ΨSL) downstream of a slippery site, thus creating a synthetic −1 PRF module (19, 20) (Fig. 3A). Similarly, trans-acting oligonucleotides can be used to bridge the loop of a stem-loop with downstream sequence to form a “pseudo-pseudoknot” (ΨΨK), which can promote highly efficient −1 PRF (21). MicroRNAs (miRNAs) that stabilize a pseudoknot (ΨK + miR) have been shown to further stimulate −1 PRF activity (22). In theory, miRNAs that destabilize −1 PRF-stimulating pseudoknots may be used to inhibit −1 PRF (13). It is also theoretically possible to create entirely novel −1 PRF signals on an otherwise unstructured mRNA by combining a stem-loop–forming oligonucleotide with a downstream pseudoknot-forming oligonucleotide; we can call this a pseudo-pseudo-pseudoknot (ΨΨΨK). By combining different slippery sites with various cis-acting mRNA structural elements and complementary trans-acting oligonucleotides, it is possible to generate a collection of −1 PRF signals with a broad range of frameshifting efficiencies (i.e. a wide selection of resistors). Finally, by placing the expression of the −1 PRF-stimulating/inhibiting RNAs under control of inducible/repressible promoters, one can create variable resistors: rheostats and potentiometers.
Trans-acting modulators of recoding need not be limited to oligonucleotides. Some RNA viruses naturally employ viral and cellular encoded proteins to regulate PRF during viral replication cycles. For example, a novel −1/−2 PRF mechanism is stimulated in the absence of any apparent downstream RNA structural element in porcine reproductive and respiratory syndrome virus (23, 24). Both −1 and −2 PRF events are stimulated by the binding of a trans-acting protein complex composed of the virus-encoded nsP1β replicase subunit and the cellular poly(C)-binding protein. This complex binds the mRNA sequence CCCANCUCC located 11 nucleotides 3′ of the GGGUUUUU shift site. Akin to PRF-stimulating downstream mRNA structural elements, binding of this complex to the target sequence induces ribosome pausing over this slippery site, resulting in slips of 1 or 2 bases in the 5′ direction (−1/−2 PRF). Similarly, the encephalomyocarditis virus protein 2A trans-activates ribosomal frameshifting by binding to (and theoretically stabilizing) a stem-loop structure located 3′ of a GGUUUUU slippery site (Fig. 3B) (25). In conclusion, the synthetic biologist need not be limited to “naturally occurring” recoding elements. Rather, they can employ a wide range of trans-acting nucleic acids and/or proteins to create “designer” translational recoding elements.
Recoding attenuation and diodes
Cis- and trans-acting elements can also be employed to attenuate −1 PRF activity. For example, naturally occurring cis-acting −1 PRF attenuator modules, first identified in coronaviruses (26), consist of stem-loop structures located immediately 5′ of the slippery site sequences. These hairpins are first unwound by elongating ribosomes as they approach the frameshift signal. As the ribosome enters the slippery site, however, it clears the hairpin-forming sequence, enabling the stem-loop to re-form. This structure can then resist the backward slippage of the ribosome caused by the −1 PRF signal. This activity can be artificially created by using trans-acting oligonucleotides that hybridize with slippery site 5′ proximal sequences to similarly generate −1 PRF-inhibiting dsRNA structures (27). Thus, this general approach can be used to further expand the variety of recoding element rheostats. Additionally, whereas these examples attenuate −1 PRF and thus function as variable resistors, they also can be thought of as insuring the unidirectionality of translation; thus, one can view these regulatory these elements as analogous to diodes that can be used to attenuate recoding.
Translational riboswitches as capacitors and variable resistors
Capacitors are weak batteries; they store charge until a critical amount of voltage has been accumulated, at which point they discharge and open the circuit. As such, capacitors are used as on/off switches as a function of voltage. Riboswitches are structured RNA elements that can change their shapes, and thus their functions, when they interact with specific small molecules (28). Cells naturally use riboswitches to turn transcription or translation on or off when their interacting small molecules reach critical concentrations. The application of translational riboswitches as capacitors is rather straightforward in that they can function as on/off switches in response to changing concentrations of the small molecules with which they interact.
Riboswitches can also be exploited to create PRF signals/variable resistors. For example, in the presence of S-adenosylhomocysteine (SAH), the SAH riboswitch folds into a pseudoknotted structure. If placed appropriately downstream of a slippery sequence, this module can induce −1 PRF in an SAH-dependent manner (29). Similarly, the preQ(1) riboswitch aptamer has been exploited to design inducible −1 PRF signals (30). PreQ(1) riboswitches derived from a variety of bacterial species were found to induce between 7 and 20% −1 PRF in response to increasing preQ(1) levels. Importantly, the amenability of translational riboswitches to molecular engineering approaches suggests that they can be “tuned” by varying ligand concentrations. For example, combining elements derived from the severe acute respiratory syndrome–associated coronavirus (SARS-CoV) −1 PRF signal with a theophylline-responsive element was used to generate a range of ligand-responsive −1 PRF modules (31). Critically, the ability to ramp −1 PRF (resistance) up and down in a continuous manner is the fundamental hallmark of a rheostat.
Signal output options and scaling
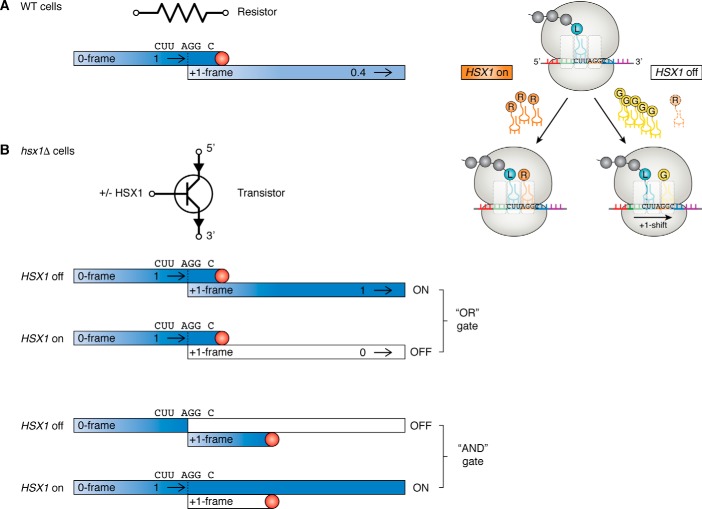
In the case of PRF, two output options are possible; a PRF event can either shift ribosomes into an extended ORF or to a new, proximal termination codon. The former tends to be favored by viruses, which use this arrangement to encode multiple gene products from a small genome and to maximize regulation of gene expression (13). The latter appears to be favored by cellular mRNAs as a mechanism to dampen gene expression (8). The “cellular” orientation (b in Fig. 4A) only slightly limits the amplitude of the downstream signal, whereas the “viral” (c in Fig. 4A) strongly affects it. As a consequence, b is a weak resistor, whereas c is a strong one. However, in eukaryotes, the relationship between PRF efficiency and signal strength is not linear. This is because directing ribosomes to premature termination codons in either orientation channels mRNAs to be rapidly degraded by the nonsense-mediated mRNA decay (NMD) pathway. NMD is a quality control system that evolved to remove mutant mRNAs that may encode truncated proteins that are potentially toxic, which has also evolved to post-transcriptionally control gene expression (32, 33). In orientation b, because only a small fraction of ribosomes encounter a 0-frame termination codon, the effect of NMD is not as strong as in orientation c (22, 34). The relationship between −1 PRF efficiency and NMD on mRNA abundance has been empirically determined to follow an exponential decay profile in which mRNA abundance ≈ e−0.05x where x = percentage of PRF (Fig. 4B) (22, 35). Additionally, the ability of −1 PRF-stimulatory elements (and likely also TCR modules) to stop ribosomes for long periods of time (36, 37) also partially renders these mRNAs substrates for degradation through a second pathway called no-go mRNA decay (NGD) (34). The relative contributions of NMD and NGD to mRNA stability in response to −1 PRF signals are not well understood; a thorough quantitative analysis would fill this important gap in the literature. Finally, because limiting mRNA abundance would negatively impact virus replication, viruses have evolved molecular mechanisms to circumvent NMD (38–40). Theoretically, these NMD-inhibitory elements might be recruited to at least partially linearize the relationship between PRF efficiency and gene expression (Fig. 4A, d).
Figure 4.
The scaling relationship between recoding efficiency and signal output is not linear in eukaryotic cells. A, examples showing the relationship between PRF efficiency and mRNA abundance. a, in the absence of PRF, the input is equal to output, with an assigned value of 1. b, if a −1 PRF event occurs within an ORF with a frequency of 10%, the amount of functional protein product synthesized is ∼0.6. c, in the case where production of the functional protein requires a −1 PRF event, if this occurs with a frequency of 10%, the total output is not 0.1, but rather ∼0.01. d, inactivation of nonsense-mediated mRNA decay (orange triangle) can boost signal output to ∼0.9 in orientation b. B, empirically determined relationship between −1 PRF efficiency and mRNA abundance (35).
Integrated circuits
Given these basic components, it is possible to imagine combining multiple recoding modules to construct complex circuits to control gene expression pathways in response to various inputs. Nature has provided numerous examples, three of which are discussed here. The first is release factor 2 (RF2), encoded by the pfrB gene. Its protein product is used to decode UGA codons in many bacteria. The prfB mRNAs in most bacterial species harbor an in-frame UGA codon located ∼26 codons from their AUG start codons, with the remainder of the protein-coding sequence in the +1-frame (41). When RF2 levels are high, termination is efficient (no PRF), preventing synthesis of excess RF2. However, when RF2 levels drop below a critical threshold, the UGA codon is inefficiently recognized, allowing translating ribosomes to proceed to downstream coding sequences, thus leading to high levels of PRF and to RF2 production (42). This is a classic autoregulatory feedback loop.
A second example is seen in the feedback loop controlling polyamine synthesis in eukaryotes (43). Ornithine decarboxylase (ODC), a key enzyme in this process, is degraded by ornithine decarboxylase antizyme (OAZ). Synthesis of OAZ requires a +1 PRF event that is stimulated by polyamines. Thus, when polyamine levels are low, OAZ is not made, and ODC is available to synthesize polyamines. However, when polyamine levels reach a critical threshold, +1 PRF occurs on the OAZ mRNA, OAZ is synthesized, and it degrades ODC, halting polyamine biosynthesis. Both the RF2 and OAZ frameshifting systems can be viewed as oscillators, toggling between PRF on and off states, depending on the concentrations of downstream products. Importantly, oscillators enable counting of on and off states (i.e. ones and zeros). Oscillators such as these can be used to transform biological systems into digital devices.
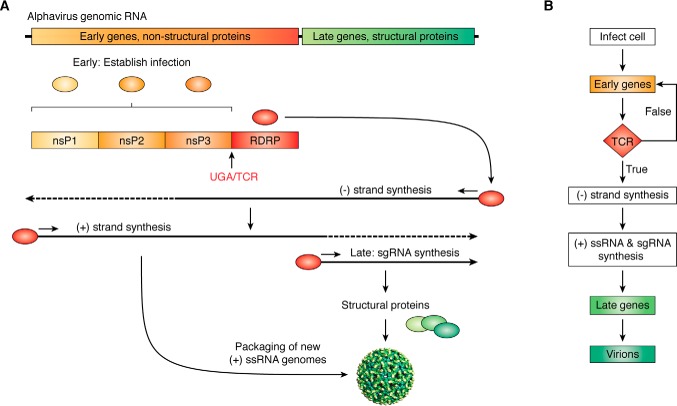
Recoding signals can also be used to control more complex developmental programs. This is most clearly observed in viruses, which by the very nature of having small genomes, have simple life cycles. As a third example, the alphavirus replication cycle can be construed as a simple two-stage program. The viral genome is split into two large ORFs encoding early (nonstructural) and late (structural) proteins (Fig. 5A). Initially, only the 5′ ORF is accessible to ribosomes. Encoded in this ORF are three nonstructural proteins (nsPs), whose functions are to establish infection. A fourth nsP can only be synthesized consequent to a TCR event. This encodes the RNA-dependent RNA polymerase (RDRP in Fig. 5). Upon its synthesis, it directs transcription of the (−)-RNA replicative intermediate strand, which then serves as the template for synthesis of new (+)-strand genomic RNAs and transcription of the 3′ ORF as a subgenomic RNA. The subgenomic RNA encodes the viral structural proteins that, along with new genomic RNA, enable production of progeny virions. This process can be diagrammed as a simple integrated circuit flowchart, where the TCR event provides the conditional operation that serves as the decision point between early and late gene expression (Fig. 5B).
Figure 5.
Viruses use translational recoding to create integrated circuits and developmental programs. A, alphaviruses have (+)-ssRNA genomes harboring two open reading frames. Expression of early genes (nsP1–nsP3) occurs first; their function is to establish infection and hijack the cell. A TCR event produces the RNA-dependent RNA polymerase. This initiates the second phase of the viral program, enabling synthesis of the (-)-ssRNA intermediate, new (+)-ssRNA genomes, and synthesis of the subgenomic RNA (sgRNA), which encodes the structural proteins to produce new virions. B, the alphavirus life cycle depicted as a programming flowchart.
Final thoughts
Science is the application of a set of intellectual tools that enables us to understand and manipulate the material world. Foundational to our understanding of this world is that it is built from the bottom up using functional modules. The intent of this review is to prod a new generation of molecular and cell biologists to build on core concepts used by electrical engineers, to initiate an interdisciplinary train of thought that applies the modular nature of the biological architectonic with the ongoing revolution of molecular and cellular engineering. Our ability to manipulate gene expression in living cells has made tremendous progress over the past half-century. The addition of programmable translational recoding modules to the bioengineering toolbox provides a crucial new level of control, further enhancing our ability to create increasingly complex biological systems and build the amazing biomachines of the 21st century.
Acknowledgments
I thank Rachel Green for encouraging me to explore and expand on this idea, John Woolford and Antony Jose for insightful critiques, and Ron Wek for commissioning and editorially shepherding this review.
This work was supported in part by NIGMS, National Institutes of Health, Grants R01 GM117177 and R01 HL119439. The author declares that he has no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TCR
- termination codon reassignment
- PRF
- programmed ribosomal frameshifting
- RF
- release factor complex
- miRNA
- microRNA
- SAH
- S-adenosylhomocysteine
- NMD
- nonsense-mediated mRNA decay
- NGD
- no-go mRNA decay
- ODC
- ornithine decarboxylase
- OAZ
- ornithine decarboxylase antizyme
- nsP
- nonstructural protein.
References
- 1. Nirenberg M., Caskey T., Marshall R., Brimacombe R., Kellogg D., Doctor B., Hatfield D., Levin J., Rottman F., Pestka S., Wilcox M., and Anderson F. (1966) The RNA code and protein synthesis. Cold Spring Harb. Symp. Quant. Biol. 31, 11–24 10.1101/SQB.1966.031.01.008 [DOI] [PubMed] [Google Scholar]
- 2. Weigert M. G., and Garen A. (1965) Amino acid substitutions resulting from suppression of nonsense mutations. I. Serine insertion by the SU-1 suppressor gene. J. Mol. Biol. 12, 448–455 10.1016/S0022-2836(65)80267-4 [DOI] [PubMed] [Google Scholar]
- 3. Atkins J. F., and Gesteland R. F. (1975) The synthetase gene of the RNA phages R17, MS2 and f2 has a single UAG terminator codon. Mol. Gen. Genet. 139, 19–31 10.1007/BF00267992 [DOI] [PubMed] [Google Scholar]
- 4. Gesteland R. F., Wolfner M., Grisafi P., Fink G., Botstein D., and Roth J. R. (1976) Yeast suppressors of UAA and UAG nonsense codons work efficiently in vitro via tRNA. Cell 7, 381–390 10.1016/0092-8674(76)90167-7 [DOI] [PubMed] [Google Scholar]
- 5. Gesteland R. F., and Atkins J. F. (1996) Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 65, 741–768 10.1146/annurev.bi.65.070196.003521 [DOI] [PubMed] [Google Scholar]
- 6. Anzalone A. V., Lin A. J., Zairis S., Rabadan R., and Cornish V. W. (2016) Reprogramming eukaryotic translation with ligand-responsive synthetic RNA switches. Nat. Methods 13, 453–458 10.1038/nmeth.3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baranov P. V., Gesteland R. F., and Atkins J. F. (2002) Recoding: translational bifurcations in gene expression. Gene 286, 187–201 10.1016/S0378-1119(02)00423-7 [DOI] [PubMed] [Google Scholar]
- 8. Advani V. M., and Dinman J. D. (2016) Reprogramming the genetic code: the emerging role of ribosomal frameshifting in regulating cellular gene expression. Bioessays 38, 21–26 10.1002/bies.201500131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodnina M. V., Fischer N., Maracci C., and Stark H. (2017) Ribosome dynamics during decoding. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160182 10.1098/rstb.2016.0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clare J. J., Belcourt M., and Farabaugh P. J. (1988) Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc. Natl. Acad. Sci. U.S.A. 85, 6816–6820 10.1073/pnas.85.18.6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belcourt M. F., and Farabaugh P. J. (1990) Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62, 339–352 10.1016/0092-8674(90)90371-K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawakami K., Pande S., Faiola B., Moore D. P., Boeke J. D., Farabaugh P. J., Strathern J. N., Nakamura Y., and Garfinkel D. J. (1993) A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dinman J. D. (2012) Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip. Rev. RNA 3, 661–673 10.1002/wrna.1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dinman J. D., Icho T., and Wickner R. B. (1991) A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. U.S.A. 88, 174–178 10.1073/pnas.88.1.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dinman J. D., and Wickner R. B. (1992) Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 66, 3669–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marczinke B., Fisher R., Vidakovic M., Bloys A. J., and Brierley I. (1998) Secondary structure and mutational analysis of the ribosomal frameshift signal of Rous sarcoma virus. J. Mol. Biol. 284, 205–225 10.1006/jmbi.1998.2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brierley I., Jenner A. J., and Inglis S. C. (1992) Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 227, 463–479 10.1016/0022-2836(92)90901-U [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brierley I., Rolley N. J., Jenner A. J., and Inglis S. C. (1991) Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 220, 889–902 10.1016/0022-2836(91)90361-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olsthoorn R. C., Laurs M., Sohet F., Hilbers C. W., Heus H. A., and Pleij C. W. (2004) Novel application of sRNA: stimulation of ribosomal frameshifting. RNA 10, 1702–1703 10.1261/rna.7139704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu C. H., Noteborn M. H., and Olsthoorn R. C. (2010) Stimulation of ribosomal frameshifting by antisense LNA. Nucleic Acids Res. 38, 8277–8283 10.1093/nar/gkq650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plant E. P., and Dinman J. D. (2005) Torsional restraint: a new twist on frameshifting pseudoknots. Nucleic Acids Res. 33, 1825–1833 10.1093/nar/gki329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Belew A. T., Meskauskas A., Musalgaonkar S., Advani V. M., Sulima S. O., Kasprzak W. K., Shapiro B. A., and Dinman J. D. (2014) Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature 512, 265–269 10.1038/nature13429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Y., Treffers E. E., Napthine S., Tas A., Zhu L., Sun Z., Bell S., Mark B. L., van Veelen P. A., van Hemert M. J., Firth A. E., Brierley I., Snijder E. J., and Fang Y. (2014) Transactivation of programmed ribosomal frameshifting by a viral protein. Proc. Natl. Acad. Sci. U.S.A. 111, E2172–E2181 10.1073/pnas.1321930111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Napthine S., Treffers E. E., Bell S., Goodfellow I., Fang Y., Firth A. E., Snijder E. J., and Brierley I. (2016) A novel role for poly(C) binding proteins in programmed ribosomal frameshifting. Nucleic Acids Res. 44, 5491–5503 10.1093/nar/gkw480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Napthine S., Ling R., Finch L. K., Jones J. D., Bell S., Brierley I., and Firth A. E. (2017) Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat. Commun. 8, 15582 10.1038/ncomms15582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su M. C., Chang C. T., Chu C. H., Tsai C. H., and Chang K. Y. (2005) An atypical RNA pseudoknot stimulator and an upstream attenuation signal for −1 ribosomal frameshifting of SARS coronavirus. Nucleic Acids Res. 33, 4265–4275 10.1093/nar/gki731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu H.-T., Cho C.-P., Lin Y.-H., and Chang K.-Y. (2016) A general strategy to inhibiting viral −1 frameshifting based on upstream attenuation duplex formation. Nucleic Acids Res. 44, 256–266 10.1093/nar/gkv1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dambach M. D., and Winkler W. C. (2009) Expanding roles for metabolite-sensing regulatory RNAs. Curr. Opin. Microbiol. 12, 161–169 10.1016/j.mib.2009.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chou M. Y., Lin S. C., and Chang K. Y. (2010) Stimulation of −1 programmed ribosomal frameshifting by a metabolite-responsive RNA pseudoknot. RNA 16, 1236–1244 10.1261/rna.1922410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu C.-H., Luo J., Iwata-Reuyl D., and Olsthoorn R. C. L. (2013) Exploiting preQ(1) riboswitches to regulate ribosomal frameshifting. ACS Chem. Biol. 8, 733–740 10.1021/cb300629b [DOI] [PubMed] [Google Scholar]
- 31. Lin Y.-H., and Chang K.-Y. (2016) Rational design of a synthetic mammalian riboswitch as a ligand-responsive −1 ribosomal frame-shifting stimulator. Nucleic Acids Res. 44, 9005–9015 10.1093/nar/gkw718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Celik A., He F., and Jacobson A. (2017) NMD monitors translational fidelity 24/7. Curr. Genet. 63, 1007–1010 10.1007/s00294-017-0709-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nasif S., Contu L., and Mühlemann O. (2018) Beyond quality control: the role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 75, 78–87 10.1016/j.semcdb.2017.08.053 [DOI] [PubMed] [Google Scholar]
- 34. Belew A. T., Advani V. M., and Dinman J. D. (2011) Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 39, 2799–2808 10.1093/nar/gkq1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Advani V. M., Belew A. T., and Dinman J. D. (2013) Yeast telomere maintenance is globally controlled by programmed ribosomal frameshifting and the nonsense-mediated mRNA decay pathway. Translation (Austin) 1, e24418 10.4161/trla.24418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lopinski J. D., Dinman J. D., and Bruenn J. A. (2000) Kinetics of ribosomal pausing during programmed −1 translational frameshifting. Mol. Cell Biol. 20, 1095–1103 10.1128/MCB.20.4.1095-1103.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Caliskan N., Katunin V. I., Belardinelli R., Peske F., and Rodnina M. V. (2014) Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell 157, 1619–1631 10.1016/j.cell.2014.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang X., Zhu Y., Baker S. L., Bowler M. W., Chen B. J., Chen C., Hogg J. R., Goff S. P., and Song H. (2016) Structural basis of suppression of host translation termination by Moloney murine leukemia virus. Nat. Commun. 7, 12070 10.1038/ncomms12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ge Z., Quek B. L., Beemon K. L., and Hogg J. R. (2016) Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife 5, e11155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. May J. P., Yuan X., Sawicki E., and Simon A. E. (2018) RNA virus evasion of nonsense-mediated decay. PLOS Pathog. 14, e1007459 10.1371/journal.ppat.1007459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larsen B., Peden J., Matsufuji S., Matsufuji T., Brady K., Maldonado R., Wills N. M., Fayet O., Atkins J. F., and Gesteland R. F. (1995) Upstream stimulators for recoding. Biochem. Cell Biol. 73, 1123–1129 10.1139/o95-121 [DOI] [PubMed] [Google Scholar]
- 42. Craigen W. J., and Caskey C. T. (1986) Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 322, 273–275 10.1038/322273a0 [DOI] [PubMed] [Google Scholar]
- 43. Ivanov I. P., Gesteland R. F., and Atkins J. F. (2000) Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 28, 3185–3196 10.1093/nar/28.17.3185 [DOI] [PMC free article] [PubMed] [Google Scholar]