Abstract
The ability of amyloid-β peptide (Aβ) to disrupt membrane integrity and cellular homeostasis is believed to be central to Alzheimer's disease pathology. Aβ is reported to have various impacts on the lipid bilayer, but a clearer picture of Aβ influence on membranes is required. Here, we use atomic force and transmission electron microscopies to image the impact of different isolated Aβ assembly types on lipid bilayers. We show that only oligomeric Aβ can profoundly disrupt the bilayer, visualized as widespread lipid extraction and subsequent deposition, which can be likened to an effect expected from the action of a detergent. We further show that Aβ oligomers cause widespread curvature and discontinuities within lipid vesicle membranes. In contrast, this detergent-like effect was not observed for Aβ monomers and fibers, although Aβ fibers did laterally associate and embed into the upper leaflet of the bilayer. The marked impact of Aβ oligomers on membrane integrity identified here reveals a mechanism by which these oligomers may be cytotoxic.
Keywords: Alzheimer disease, amyloid-beta (AB), membrane bilayer, oligomer, atomic force microscopy (AFM), electron microscopy (EM), neurodegeneration, cytotoxicity, large unilamellar vesicles, membrane disruption, mica surface, nanoscale imaging
Introduction
The amyloid cascade is a widely accepted description for the molecular basis of Alzheimer's disease (AD).2 This hypothesis is centered on a small hydrophobic peptide, β-amyloid, which is typically 40 or 42 amino acids in length (Aβ40/42). The self-association of Aβ and subsequent interaction with the lipid bilayer are thought to cause disrupted synaptic function; the resulting loss of cellular homeostasis ultimately leads to hyperphosphorylation of tau, cell death, and dementia (1). Aβ fibrous plaques are abundant in the AD brain and have high lipid content (2, 3).
The interaction of Aβ with lipid membranes has been expertly reviewed (4–6) and draws parallels with the mechanism of toxicity established for antimicrobial peptides (6). Importantly, Aβ42 oligomers are reported to perforate the cellular membrane by forming single, discrete ion channel pores (7). Alternatively, a carpeting effect of Aβ has been proposed, which is thought to result in a general increase in membrane conductance either by membrane thinning or a lateral spreading of lipid headgroups (8–10). Studies have also revealed that Aβ42 can extract and incorporate lipid into growing Aβ42 fibers by an effect likened to the action of a detergent, which has the potential to rupture the lipid bilayer (11). Some of these effects have been observed for other amyloid-forming proteins and include amylin (12–14), α-synuclein (15), and a prion protein fragment (16), which suggests a shared mechanism of membrane disruption. Nevertheless, a clear picture of the impact of Aβ on membranes is lacking, perhaps in part due to insufficient assembly state characterization and separation of Aβ oligomers from monomers and fibers.
Atomic force microscopy (AFM) is particularly well-suited to probe the topography of supported lipid bilayers (17). However, many AFM studies have focused on how the lipid bilayer can influence Aβ fiber assembly (5, 18–21). AFM studies that investigate the impact of Aβ on the bilayer structure are only starting to be explored (22–25).
Here, we report a systematic investigation of the effects of both Aβ40 and Aβ42 isoforms on lipid bilayers. The Aβ isoforms have been studied in three broad assembly states: monomers, prefibrillar heterogeneous oligomeric assemblies, and fibers. We have employed AFM and transmission electron microscopy (TEM) to show that these different Aβ structures have profoundly different impacts on membrane integrity, which can explain the diverse effects reported.
Results and discussion
Lipid bilayers were deposited onto muscovite mica surfaces from an aqueous mixture of phosphatidylcholine (PC), cholesterol, and GM1 ganglioside (68:30:2 by weight, a ratio chosen to mimic the typical composition of the extracellular face of a cellular membrane). A liposomal suspension in aqueous buffer was incubated with the negatively charged mica, allowing the lipid headgroups to adhere and fuse with the mica substrate to form a double-leaflet lipid bilayer. The remaining suspension was then blotted off the mica and air-dried. AFM images indicate this to be an effective method of generating flat, continuous patches of lipid bilayer with a sharp “cliff-edge” boundary between the membrane patch and lipid-free mica, as shown in Fig. S1. A mean bilayer height has been measured at 4.6 ± 0.1 nm, as would be expected for this lipid composition.
To understand the nature of the interactions between monomeric, oligomeric, and fibrillar Aβ40/42 with lipid bilayers, it is important to isolate and characterize the different Aβ assembly types, see “Experimental procedures”; Figs. S2 and S3 for details. We first deposited size-exclusion chromatographically–purified monomeric Aβ40 and Aβ42 preparations (10 μm) onto the lipid bilayer. After 60 min on the bilayer surface, the Aβ solution was blotted off, and the mica was allowed to air-dry for imaging. It is clear that neither monomeric Aβ42 nor Aβ40 has a detectable impact on the integrity of the bilayer, as seen in Fig. 1a and Fig. S4a. There is an even distribution of lipid with minimal disruption both on the lipid surface and at the lipid edge, where a very sharp cliff-edge transition between the lipid-bilayer and the mica is observed. Moreover, the bilayers seen in these images are indistinguishable from the bilayer treated with aqueous buffer.
Figure 1.
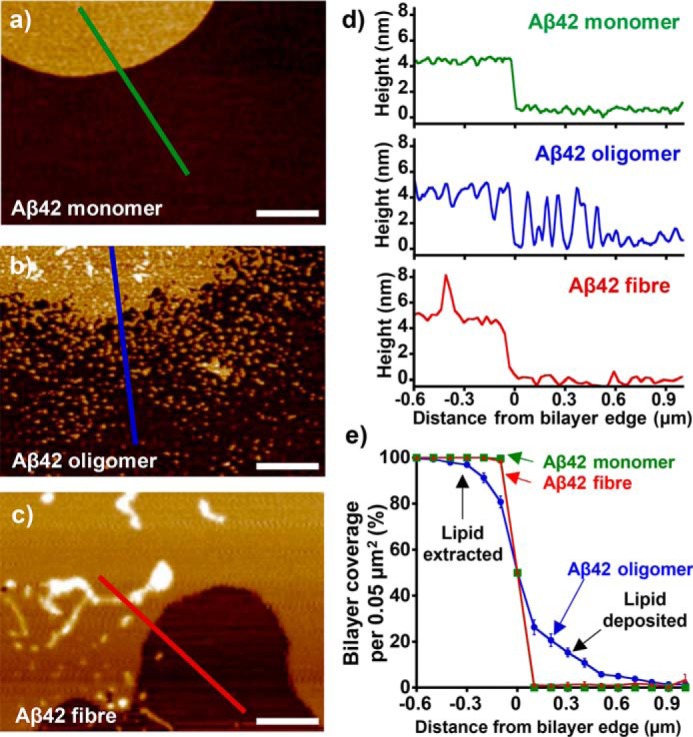
Aβ42 oligomers have a detergent-like effect on the bilayer, which is not seen for Aβ42 monomer nor Aβ42 fibers. AFM topographical images are shown for mica-supported lipid bilayers on exposure to Aβ42 monomer (a), Aβ42 heterogeneous oligomer (b), and Aβ42 fiber (c). Height scale range, 12 nm. d, height cross-sections. e, percentage of bilayer coverage at the edge of a lipid bilayer, exposed to Aβ42 monomer (green), heterogeneous oligomer (blue), and fiber (red). Each data point represents an average percentage of bilayer coverage within a 0.1 × 0.5-μm region. There were 20 measurements per data point, measured across three separate mica-supported lipid bilayer preparations. Error bars, S.E.
In contrast, prefibrillar heterogeneous oligomeric preparations of Aβ42 and Aβ40 (10 μm monomer equivalent) have a very disruptive impact on the integrity of the bilayer (see Fig. 1b and Fig. S4b, respectively). In particular, there is widespread extraction of lipid from bilayer islands, suggesting a very strong detergent effect by Aβ oligomers. Furthermore, small clumps of lipid appear deposited on the surrounding mica in close proximity to the more substantial lipid islands. This is evident in membrane height cross-sections (see Fig. 1d and Fig. S4d). The impact of Aβ42 and Aβ40 mature amyloid fibers was much less pronounced than observed for the oligomeric preparations, with almost no Aβ fiber–induced lipid extraction and deposition (no detergent effect). The lipid bilayers remained predominantly intact in the presence of Aβ fibers, as shown in Fig. 1c and Fig. S4c.
We aimed to quantify the extent of the detergent effect for the three Aβ assembly types. This was achieved by measuring lipid coverage at the lipid-edge interface between regions of bilayer and lipid-free mica for multiple (n = 20) preparations (Fig. 1e and Fig. S4e). These data are presented as percentage of area lipid coverage versus distance from an island of lipid (for details, see Fig. S5). The mean lipid coverage data confirm that neither monomer nor fiber had an impact on the sharp cliff-edge transition between the lipid bilayer and mica substrate. Conversely, a very poorly defined transition between the lipid bilayer and the mica was consistently observed upon the addition of Aβ oligomers. The highly disrupted islands of lipid bilayer were surrounded by small deposits of lipid that become less abundant and dispersed at further distances from the main island of lipid, suggesting an extraction of lipid away from the bilayer origin. We observed this consistently in numerous bilayers and Aβ preparations; mean values from typically 20 measurements are shown for each Aβ type. Further examples are shown for Aβ42 and Aβ40 in Figs. S6 and S7, respectively. Previous studies have utilized Aβ-reconstituted liposomes to form an Aβ-infused mica-supported bilayer, leading to membrane-spanning pore-like annular-oligomer structures, but with this method of bilayer preparation, the extraction and resulting deposition of lipid was not observed (26–28).
In addition to the widespread disruption of lipid at the edges of the bilayer, extraction of lipid by Aβ oligomers also resulted in large holes in the membrane that span the bilayer, as reported previously (22). Quantification of these holes indicates a typical diameter of 50 nm (see Fig. 2). A similar range of hole sizes in the bilayer is observed for Aβ40 (see Fig. S8). These holes, caused by lipid extraction, should not be conflated with the formation of ion channel pores. The diameters of Aβ ion channels have been shown to be between 1.7 and 2.4 nm based on measurements of their ionic conductance (7), imaging of annular Aβ oligomers that resemble channel structures (26–28), and also modeling of Aβ barrel-like structures in the bilayer (29, 30). The holes observed in Fig. 2 are much larger and would be indistinguishable from membrane patch breakdown in ionic conductance measurements (7). These large holes are also very occasionally observed upon the addition of Aβ fibers; however, their occurrence is relatively rare and may be explained by the presence of a small number of oligomers in the Aβ fiber preparation.
Figure 2.
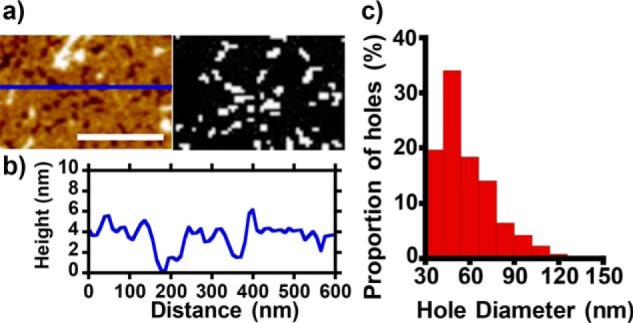
Aβ42 oligomer causes holes within supported lipid bilayers. a (left), AFM topographical image depicting a region of supported lipid bilayer which has holes formed after incubation with Aβ42 heterogeneous oligomer. Height color scale range, 12 nm. Right, binary map of holes, generated by height threshold. b, height cross-section. Typically, both the upper and lower leaflets of bilayer are extracted. Scale bar, 0.3 μm. c, range of hole diameters with a modal value of 50 nm.
We were also interested in quantifying the extent by which the amyloid fibers embed into bilayers. AFM is particularly suited to measuring lipid bilayer heights, measured to be 4.6 ± 0.1 nm, whereas single Aβ fibers are typically 5.3 ± 0.1 nm in height (see Fig. 3 and Figs. S9 and S10). Measurements of the height of fibers laterally associated to the surface of the membrane indicate the extent to which the fibers embed and displace lipids within the bilayer. Fibers that laterally associate onto the surface of the membrane consistently protrude to a height of 2.5 ± 0.05 nm above the lipid bilayer surface. This indicates that the fibers do indeed embed into the membrane to a depth where the upper leaflet of the bilayer is displaced, as shown in Fig. 3c. It seems that fibers do not displace lipid from the lower leaflet of the supported lipid bilayer; nor do fibers rest on the surface of the membrane to leave the bilayer unperturbed. Lateral association of amyloid fibers on the surface of a lipid bilayer has been reported using a range of techniques, including EM and fluorescence microscopy (11, 13, 31–33). However, this is the first clear evidence that laterally associated Aβ fibers are capable of embedding and displacing the upper leaflet of the bilayer rather than simply residing on the charged surface of the bilayer.
Figure 3.
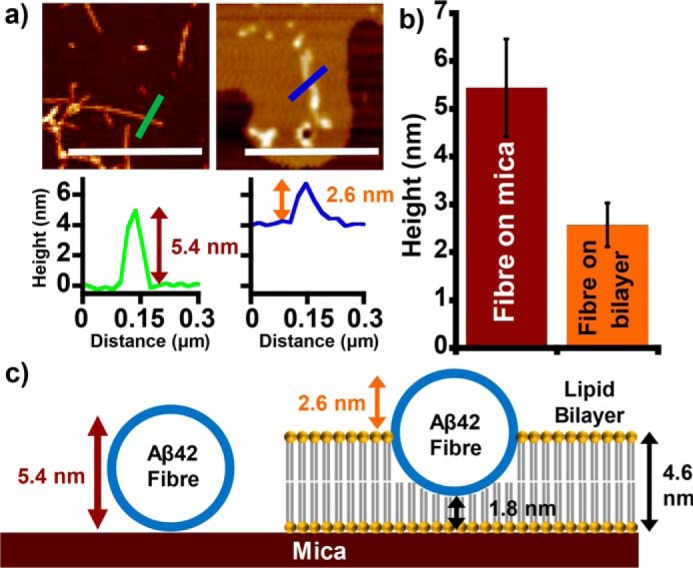
Aβ fibers embed into the lipid bilayer, displacing the upper leaflet. a, AFM topographical images show Aβ42 fibers on mica (left) and on the surface of a lipid bilayer (right). Scale bar, 1 μm. Height scale range, 12 nm. Each image is accompanied by a height cross-section. b, mean Aβ42 fiber height recorded both on mica and above the bilayer surface. c, a scaled schematic showing how Aβ fibers displace the upper leaflet of the membrane as fibers laterally embed into the bilayer. Error bars, S.E.
To investigate the extent to which oligomers and curvy linear protofibrils embed into the lipid bilayer, we used AFM to record both topographical and phase-contrast information. Oligomers and curvy linear protofibrils generated here are typically at least 4 nm in diameter. Phase imaging can be used to distinguish differences in viscoelastic properties that in turn can be explained by different surface properties even when there is little change in height topology. In this way, Aβ oligomers can be observed even when they are fully embedded into the membrane (see Fig. S11). Phase imaging of bilayers exposed to Aβ oligomer revealed dark regions within the bilayer that are indicative of increased probe adhesion to membrane-inserted Aβ; these oligomers were otherwise poorly visualized by height topography. The oligomer and curvy linear protofibrils have a widespread presence in the membrane and are sufficiently buried to be a similar height as the surrounding bilayer, displacing both leaflets of the membrane, which might then facilitate the observed lipid extraction.
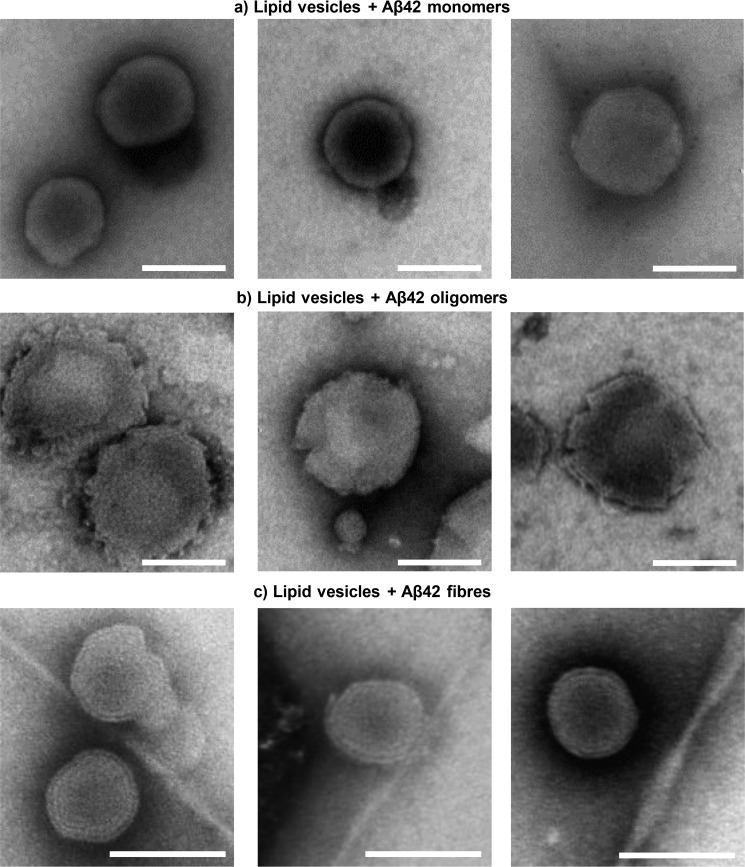
We further investigated the impact of Aβ assemblies on the structure of lipid membranes within large unilamellar vesicles (LUVs) by negative-stain TEM imaging. In agreement with the AFM data, monomers of Aβ have little impact on the membrane, as the vesicles show little difference in appearance from vesicles imaged in the absence of Aβ (Fig. 4a and Fig. S12). Aβ42 oligomers clearly disrupt the shape of the vesicles, causing numerous discontinuities and curvatures within the bilayer (Fig. 4b and Fig. S12). Peptide-induced membrane curvature has also been reported for anti-microbial peptides whose toxic action is linked to a detergent effect dependent on membrane insertion (34, 35). In contrast, Aβ fibers cause less distortion in the vesicle appearance, with only the ends of the fibers causing membrane curvature (Fig. 4c and Fig. S13). This behavior is also apparent for β2-microglobulin and amylin amyloid fibers imaged by TEM (13, 32).
Figure 4.
Aβ oligomer disruption of lipid vesicles. a–c, TEM negative-stain images of LUVs in the presence of Aβ42 (10 μm) monomer (a), Aβ42 (10 μm) heterogeneous oligomers (b), and Aβ42 (10 μm) fibers (c). Scale bars, 100 nm. Aβ42 oligomers cause widespread curvature and discontinuities of the membrane lipid bilayer, whereas vesicles are relatively unaffected by Aβ42 monomers and fibers.
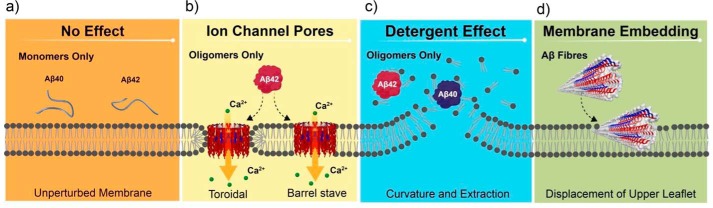
In summary, nanoscale imaging of supported lipid bilayers has revealed very different impacts on the membrane when challenged with Aβ monomers, heterogeneous oligomers, or fibers (Fig. 5). In the presence of isolated monomers of Aβ40 or Aβ42, the lipid membrane remains unperturbed, whereas fibers laterally embed into the bilayer to displace the upper leaflet. Notably, only Aβ oligomers cause extraction of lipid from the membrane, explaining the high levels of lipid deposited within extracellular Aβ plaques in the AD brain. In vivo, this detergent effect is likely to impact cellular homeostasis and integrity and is in line with the relative cytotoxicity of Aβ oligomers compared with fibrillar assembly states (36–38). Cell viability assays report cytotoxicity for both Aβ40 and Aβ42, although Aβ40 is less potent than Aβ42 (38). Here we show there is not a significant difference in membrane disruption caused by Aβ40 oligomers compared with Aβ42, whereas only Aβ42 oligomers form ion channels (7). The differences in ion channel formation and lipid bilayer disruption (Fig. 5) suggest there may be two or more mechanisms by which Aβ can disrupt neuronal homeostasis and trigger the cascade of events that culminate in dementia. Multiple modes of toxic action present numerous therapeutic targets; our study indicates that therapeutic molecules that prevent insertion of the Aβ oligomer into membranes may circumvent both mechanisms of toxicity.
Figure 5.
The mechanisms of β-amyloid mediated membrane disruption depends on Aβ structure. Aβ monomers, oligomers, and fibers have distinct impacts on membrane integrity. a, Aβ monomers have little impact on membrane structure. b, only oligomeric Aβ42 has the ability to form ion channel pores in cellular membranes (7). c, oligomers of both Aβ42 and Aβ40 have a widespread detergent effect causing lipid extraction, curvature, and rupture of the membrane bilayer. d, the ability of Aβ to form ion channels or have a detergent effect is lost on assembly into amyloid fibers; instead, fibers can laterally embed into the membrane and displace the upper leaflet of the bilayer. The fiber structure is taken from Protein Data Bank entry 2MXU (44).
Experimental procedures
Aβ peptides and solubilization
Lyophilized Aβ40 and Aβ42 peptides were purchased from Cambridge Research Biochemicals and EZBiolab Inc. Aβ peptides were synthesized using solid-phase Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry, producing a single elution band in HPLC with correct mass verified by MS. Unless otherwise stated, all other chemicals were purchased from Sigma-Aldrich.
Peptides were solubilized in ultra-high quality (UHQ) water (0.7 mg/ml at pH 10) and left at 4 °C for 12 h. Samples were then centrifuged for 15 min at 16,000 × g at 4 °C to remove any high-molecular weight aggregates. Aβ concentration was determined by absorbance of stock solutions at 280 nm (ϵ = 1280 m−1 cm−1).
Purification of monomeric Aβ by size-exclusion chromatography
Size-exclusion chromatography (SEC) was utilized to separate Aβ monomer from amyloid assemblies of higher molecular weight. Superdex 75 10/300 GL column (GE Healthcare) was used to purify and elute a monomeric fraction of solubilized Aβ preparations using AKTA FPLC. Stock Aβ solution (90 μm) in UHQ water was loaded onto the column, and the monomer peak was eluted with a solution of NaCl (160 mm), HEPES (30 mm) using a 0.5 ml/min flow rate at 4 °C. Eluted monomer had a typical concentration of 30 μm. SEC-purified Aβ was deemed to be seed-free, evidenced by a single elution band and an absence of observable assemblies by negative-stain TEM. Seed-free Aβ, termed here as monomeric, also had no thioflavin T (ThT) fluorescence signal and exhibited a clear lag phase in the nucleation polymerization reaction (see Fig. S2). Monomeric samples were used directly after SEC elution or immediately stored at −80 °C.
Aβ oligomer and fiber preparations
Fiber preparations
Amyloid fiber preparations of Aβ40 and Aβ42 (10 μm monomer-equivalent) were generated in a 96-well plate in aqueous buffer containing NaCl (160 mm) and HEPES (30 mm) at pH 7.4, as described previously (39) (see supporting information for further details). Assembly kinetics were monitored with ThT fluorescence (10 μm). Adjacent sample wells with no ThT added were used to generate assemblies for use in all experiments. To remove any low-molecular weight oligomers, fiber preparations were centrifuged at 16,100 × g for 5 min. The supernatant, which contained small diffusible oligomers, was removed, and the fiber pellet was resuspended. At equilibrium, Aβ assemblies had the typical amyloid fibrous appearance according to AFM and TEM (see Fig. S3). Fiber preparations have a low level of Aβ monomer and oligomer content.
Oligomer preparations and characterization
Similarly, Aβ40 and Aβ42 assemblies with predominantly circular oligomeric structure were obtained from the well plate toward the end of the lag phase, as monitored by ThT fluorescent dye in separate wells. Oligomeric samples were used immediately or stored at −80 °C to halt further assembly. The heterogeneous lag-phase mixture contains a range of oligomer sizes, whereas appreciable monomeric Aβ is still present, as indicated by SEC elusion profiles (Fig. S2d). According to TEM, these oligomeric preparations are heterogeneous in nature but contain a large number of circular oligomeric assemblies that range in size, with typical diameters between 10 and 12 nm, identified by TEM and AFM (see Fig. S3).
TEM micrographs of negatively stained prefibrillar oligomeric assemblies of Aβ42 were further characterized using single-particle analyses. A total data set of 3997 single particles were grouped into 25 class averages (see Fig. S3a). From this data set, 70% were broadly circular in appearance and ranged in diameter between 10 and 12 nm. Using the relationship, volume (nm3) = mass (kDa)/1.27 (nm3/kDa), and assuming spherical protein assemblies, approximate masses of between 400 and 700 kDa are indicated. Whereas SEC suggests smaller oligomers are also present (Fig. S2d), the remaining particles appeared elongated up to 16 nm, with some approaching curvy linear status. Some much longer curvy linear protofibrillar assemblies were indeed present, typically 100 nm long. In agreement with our SEC elution profiles (Fig. S2d), it has been shown much of Aβ remains monomeric in the prefibrillar lag-phase preparations (40). The nature of the heterogeneous Aβ assemblies present in lag-phase preparations has been reviewed previously (40).
Lipid bilayer preparation
Lipid bilayers were prepared using methods similar to those described previously (31). A lipid mixture of egg PC (Avanti Polar Lipids Inc.), GM1 ganglioside (Avanti Polar Lipids Inc.), and cholesterol (Sigma-Aldrich) solubilized to 5 mg/ml at a ratio of 68:2:30 (w/v), in a 2:1 (v/v) mixture of chloroform and methanol. Organic solvent was then gently removed under a stream of nitrogen and left overnight in a fume hood to form a lipid film.
For AFM studies, the dried lipid film of PC/GM1/cholesterol was rehydrated in an aqueous buffer (160 mm NaCl, 30 mm HEPES, pH 7.4) to achieve a 0.6 mg/ml liposome solution. This vesicle suspension was then tip-sonicated at low power for three 30-s periods, until the cloudy lipid solution was clear, ready for deposition onto mica.
For TEM lipid vesicle studies, LUVs were generated by the extrusion method (31). A 1 mg/ml lipid solution was immediately passed through a lipid extruder (Avanti Lipids Ltd.), which contained a 100-nm polycarbonate filter (Avanti Polar Lipids Inc.), seven times to generate a uniform size. LUVs were typically 100–200 nm in diameter.
Preparation of supported lipid bilayers on mica and application of Aβ
Mica discs (Agar Scientific Ltd.) were cleaved five times with sticky tape, and both the mica and the lipid vesicle suspension were heated to 60 °C in a benchtop oven. The heated suspension was then pipetted onto mica discs in a Corning sterile 6-well plate (Sigma-Aldrich). The plate was then sealed in a humid box and incubated at 60 °C for 30 min before decreasing the temperature to 25 °C at a rate of 5 °C every 15 min until reaching room temperature. Slow cooling of the sample enables large micro-islands of bilayer to form on the mica (Fig. S1).
The mica was then washed three times with UHQ water, ready for exposure to Aβ. Typically, 200 μl of Aβ (10 μm) with NaCl (160 mm) and HEPES (30 mm), pH 7.4, was applied by pipette to evenly coat the surface of the mica. The Aβ droplet was incubated in a sealed container for 1 h to minimize evaporation and maintain salt/Aβ concentration. The droplet was then removed using filter paper, and the mica surface was washed again three times with UHQ water. Mica samples were finally left to dry in ambient air conditions under the cover of pierced parafilm. The samples were then imaged by AFM. Aβ was also deposited directly onto the mica in the absence of lipid bilayer using a similar protocol.
AFM and TEM; quantification and characterization
Details of the measurements made using both AFM and TEM can be found in the supporting information. In particular, AFM data quantification of the detergent effect, bilayer hole diameters, (41), lateral fiber embedding, and single particle analysis (42, 43) of TEM micrographs are described.
Author contributions
D. C. B. data curation; D. C. B. software; D. C. B. and J. N. formal analysis; D. C. B. investigation; D. C. B. visualization; D. C. B., M. F., and M. P. methodology; D. C. B. and J. H. V. writing-original draft; D. C. B., M. F., J. N., M. P., and J. H. V. writing-review and editing; M. P. and J. H. V. supervision; J. H. V. conceptualization; J. H. V. funding acquisition; J. H. V. project administration.
Supplementary Material
Acknowledgment
We thank Yao Tian for assistance.
This work was supported by BBSRC Grant BB/M023877/1. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supporting Experimental procedures and Figs. S1–S13.
- AD
- Alzheimer's disease
- GM1
- monosialotetrahexosyl
- Aβ
- amyloid-β peptide
- AFM
- atomic force microscopy
- TEM
- transmission electron microscopy
- PC
- phosphatidylcholine
- LUV
- large unilamellar vesicle
- UHQ
- ultra-high quality
- SEC
- size-exclusion chromatography
- ThT
- thioflavin T.
References
- 1. Selkoe D. J., and Hardy J. (2016) The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 8, 595–608 10.15252/emmm.201606210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kiskis J., Fink H., Nyberg L., Thyr J., Li J. Y., and Enejder A. (2015) Plaque-associated lipids in Alzheimer's diseased brain tissue visualized by nonlinear microscopy. Sci. Rep. 5, 13489 10.1038/srep13489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuzyk A., Kastyak M., Agrawal V., Gallant M., Sivakumar G., Rak M., Del Bigio M. R., Westaway D., Julian R., and Gough K. M. (2010) Association among amyloid plaque, lipid, and creatine in hippocampus of TgCRND8 mouse model for Alzheimer disease. J. Biol. Chem. 285, 31202–31207 10.1074/jbc.M110.142174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams T. L., and Serpell L. C. (2011) Membrane and surface interactions of Alzheimer's Aβ peptide—insights into the mechanism of cytotoxicity. FEBS J. 278, 3905–3917 10.1111/j.1742-4658.2011.08228.x [DOI] [PubMed] [Google Scholar]
- 5. Canale C., Oropesa-Nuñez R., Diaspro A., and Dante S. (2018) Amyloid and membrane complexity: the toxic interplay revealed by AFM. Semin. Cell Dev. Biol. 73, 82–94 10.1016/j.semcdb.2017.08.046 [DOI] [PubMed] [Google Scholar]
- 6. Butterfield S. M., and Lashuel H. A. (2010) Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew. Chem. Int. Ed. Engl. 49, 5628–5654 10.1002/anie.200906670 [DOI] [PubMed] [Google Scholar]
- 7. Bode D. C., Baker M. D., and Viles J. H. (2017) Ion channel formation by amyloid-β42 oligomers but not amyloid-β40 in cellular membranes. J. Biol. Chem. 292, 1404–1413 10.1074/jbc.M116.762526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., and Glabe C. G. (2004) Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 279, 46363–46366 10.1074/jbc.C400260200 [DOI] [PubMed] [Google Scholar]
- 9. Sokolov Y., Kozak J. A., Kayed R., Chanturiya A., Glabe C., and Hall J. E. (2006) Soluble amyloid oligomers increase bilayer conductance by altering dielectric structure. J. Gen. Physiol. 128, 637–647 10.1085/jgp.200609533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Valincius G., Heinrich F., Budvytyte R., Vanderah D. J., McGillivray D. J., Sokolov Y., Hall J. E., and Lösche M. (2008) Soluble amyloid β-oligomers affect dielectric membrane properties by bilayer insertion and domain formation: implications for cell toxicity. Biophys. J. 95, 4845–4861 10.1529/biophysj.108.130997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sasahara K., Morigaki K., and Shinya K. (2013) Effects of membrane interaction and aggregation of amyloid β-peptide on lipid mobility and membrane domain structure. Phys. Chem. Chem. Phys. 15, 8929–8939 10.1039/c3cp44517h [DOI] [PubMed] [Google Scholar]
- 12. Sparr E., Engel M. F., Sakharov D. V., Sprong M., Jacobs J., de Kruijff B., Höppener J. W., and Killian J. A. (2004) Islet amyloid polypeptide-induced membrane leakage involves uptake of lipids by forming amyloid fibers. FEBS Lett. 577, 117–120 10.1016/j.febslet.2004.09.075 [DOI] [PubMed] [Google Scholar]
- 13. Engel M. F., Khemtémourian L., Kleijer C. C., Meeldijk H. J. D., Jacobs J., Verkleij A. J., de Kruijff B., Killian J. A., and Höppener J. W. M. (2008) Membrane damage by human islet amyloid polypeptide through fibril growth at the membrane. Proc. Natl. Acad. Sci. U.S.A. 105, 6033–6038 10.1073/pnas.0708354105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu T., Meng F., Wei Y., Li Y., Wang C., and Li F. (2018) Exploring the relation between the oligomeric structure and membrane damage by a study on rat islet amyloid polypeptide. Phys. Chem. Chem. Phys. 20, 8976–8983 10.1039/C7CP06468C [DOI] [PubMed] [Google Scholar]
- 15. Reynolds N. P., Soragni A., Rabe M., Verdes D., Liverani E., Handschin S., Riek R., and Seeger S. (2011) Mechanism of membrane interaction and disruption by α-synuclein. J. Am. Chem. Soc. 133, 19366–19375 10.1021/ja2029848 [DOI] [PubMed] [Google Scholar]
- 16. Walsh P., Vanderlee G., Yau J., Campeau J., Sim V. L., Yip C. M., and Sharpe S. (2014) The mechanism of membrane disruption by cytotoxic amyloid oligomers formed by prion protein(106–126) is dependent on bilayer composition. J. Biol. Chem. 289, 10419–10430 10.1074/jbc.M113.515866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El Kirat K., Morandat S., and Dufrêne Y. F. (2010) Nanoscale analysis of supported lipid bilayers using atomic force microscopy. Biochim. Biophys. Acta 1798, 750–765 10.1016/j.bbamem.2009.07.026 [DOI] [PubMed] [Google Scholar]
- 18. Yip C. M., Darabie A. A., and McLaurin J. (2002) Aβ42-peptide assembly on lipid bilayers. J. Mol. Biol. 318, 97–107 10.1016/S0022-2836(02)00028-1 [DOI] [PubMed] [Google Scholar]
- 19. Yip C. M., and McLaurin J. (2001) Amyloid-β peptide assembly: a critical step in fibrillogenesis and membrane disruption. Biophys. J. 80, 1359–1371 10.1016/S0006-3495(01)76109-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choucair A., Chakrapani M., Chakravarthy B., Katsaras J., and Johnston L. J. (2007) Preferential accumulation of A beta(1–42) on gel phase domains of lipid bilayers: an AFM and fluorescence study. Biochim. Biophys. Acta 1768, 146–154 10.1016/j.bbamem.2006.09.005 [DOI] [PubMed] [Google Scholar]
- 21. Korshavn K. J., Satriano C., Lin Y., Zhang R., Dulchavsky M., Bhunia A., Ivanova M. I., Lee Y. H., La Rosa C., Lim M. H., and Ramamoorthy A. (2017) Reduced lipid bilayer thickness regulates the aggregation and cytotoxicity of amyloid-β. J. Biol. Chem. 292, 4638–4650 10.1074/jbc.M116.764092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Williams T. L., Johnson B. R., Urbanc B., Jenkins A. T., Connell S. D., and Serpell L. C. (2011) Aβ42 oligomers, but not fibrils, simultaneously bind to and cause damage to ganglioside-containing lipid membranes. Biochem. J. 439, 67–77 10.1042/BJ20110750 [DOI] [PubMed] [Google Scholar]
- 23. Canale C., Seghezza S., Vilasi S., Carrotta R., Bulone D., Diaspro A., San Biagio P. L., and Dante S. (2013) Different effects of Alzheimer's peptide A β(1–40) oligomers and fibrils on supported lipid membranes. Biophys. Chem. 182, 23–29 10.1016/j.bpc.2013.07.010 [DOI] [PubMed] [Google Scholar]
- 24. Lee J., Gillman A. L., Jang H., Ramachandran S., Kagan B. L., Nussinov R., and Teran Arce F. (2014) Role of the fast kinetics of pyroglutamate-modified amyloid-β oligomers in membrane binding and membrane permeability. Biochemistry 53, 4704–4714 10.1021/bi500587p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sheikh K., Giordani C., McManus J. J., Hovgaard M. B., and Jarvis S. P. (2012) Differing modes of interaction between monomeric A β(1–40) peptides and model lipid membranes: an AFM study. Chem. Phys. Lipids 165, 142–150 10.1016/j.chemphyslip.2011.11.011 [DOI] [PubMed] [Google Scholar]
- 26. Lashuel H. A., Hartley D., Petre B. M., Walz T., and Lansbury P. T. Jr. (2002) Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature 418, 291 10.1038/418291a [DOI] [PubMed] [Google Scholar]
- 27. Kayed R., Pensalfini A., Margol L., Sokolov Y., Sarsoza F., Head E., Hall J., and Glabe C. (2009) Annular protofibrils are a structurally and functionally distinct type of amyloid oligomer. J. Biol. Chem. 284, 4230–4237 10.1074/jbc.M808591200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin H., Bhatia R., and Lal R. (2001) Amyloid-β protein forms ion channels: implications for Alzheimer's disease pathophysiology. FASEB J. 15, 2433–2444 10.1096/fj.01-0377com [DOI] [PubMed] [Google Scholar]
- 29. Jang H., Zheng J., Lal R., and Nussinov R. (2008) New structures help the modeling of toxic amyloid β ion channels. Trends Biochem. Sci. 33, 91–100 10.1016/j.tibs.2007.10.007 [DOI] [PubMed] [Google Scholar]
- 30. Serra-Batiste M., Ninot-Pedrosa M., Bayoumi M., Gairí M., Maglia G., and Carulla N. (2016) A β42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments. Proc. Natl. Acad. Sci. U.S.A. 113, 10866–10871 10.1073/pnas.1605104113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matheou C. J., Younan N. D., and Viles J. H. (2015) Cu2+ accentuates distinct misfolding of Aβ1–40 and Aβ1–42 peptides, and potentiates membrane disruption. Biochem. J. 466, 233–242 10.1042/BJ20141168 [DOI] [PubMed] [Google Scholar]
- 32. Milanesi L., Sheynis T., Xue W. F., Orlova E. V., Hellewell A. L., Jelinek R., Hewitt E. W., Radford S. E., and Saibil H. R. (2012) Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 109, 20455–20460 10.1073/pnas.1206325109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han S., Kollmer M., Markx D., Claus S., Walther P., and Fändrich M. (2017) Amyloid plaque structure and cell surface interactions of β-amyloid fibrils revealed by electron tomography. Sci. Rep. 7, 43577 10.1038/srep43577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bechinger B., and Lohner K. (2006) Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta 1758, 1529–1539 10.1016/j.bbamem.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 35. Koller D., and Lohner K. (2014) The role of spontaneous lipid curvature in the interaction of interfacially active peptides with membranes. Biochim. Biophys. Acta 1838, 2250–2259 10.1016/j.bbamem.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 36. Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., and Selkoe D. J. (2002) Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 10.1038/416535a [DOI] [PubMed] [Google Scholar]
- 37. Benilova I., Karran E., and De Strooper B. (2012) The toxic Aβ oligomer and Alzheimer's disease: an emperor in need of clothes. Nat. Neurosci. 15, 349–357 10.1038/nn.3028 [DOI] [PubMed] [Google Scholar]
- 38. Dahlgren K. N., Manelli A. M., Stine W. B. Jr., Baker L. K., Krafft G. A., and LaDu M. J. (2002) Oligomeric and fibrillar species of amyloid-β peptides differentially affect neuronal viability. J. Biol. Chem. 277, 32046–32053 10.1074/jbc.M201750200 [DOI] [PubMed] [Google Scholar]
- 39. Younan N. D., and Viles J. H. (2015) A comparison of three fluorophores for the detection of amyloid fibers and prefibrillar oligomeric assemblies. ThT (thioflavin T); ANS (1-anilinonaphthalene-8-sulfonic acid); and bisANS (4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid). Biochemistry 54, 4297–4306 10.1021/acs.biochem.5b00309 [DOI] [PubMed] [Google Scholar]
- 40. Arosio P., Knowles T. P., and Linse S. (2015) On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 17, 7606–7618 10.1039/C4CP05563B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Samori P., Francke V., Mullen K., and Rabe J. P. (1999) Self-assembly of a conjugated polymer: from molecular rods to a nanoribbon architecture with molecular dimensions. Chem. Eur. J. 5, 2312–2317 [DOI] [Google Scholar]
- 42. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., and Ludtke S. J. (2007) EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 43. van Heel M., Harauz G., Orlova E. V., Schmidt R., and Schatz M. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 10.1006/jsbi.1996.0004 [DOI] [PubMed] [Google Scholar]
- 44. Xiao Y., Ma B., McElheny D., Parthasarathy S., Long F., Hoshi M., Nussinov R., and Ishii Y. (2015) Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease. Nat. Struct. Mol. Biol. 22, 499–505 10.1038/nsmb.2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.