Abstract
Aerobic glycolysis and mitochondrial dysfunction are key metabolic features of cancer cells, but their interplay during cancer development remains unclear. We previously reported that human hepatoma cells with mitochondrial defects exhibit down-regulated lactate dehydrogenase subunit B (LDHB) expression. Here, using several molecular and biochemical assays and informatics analyses, we investigated how LDHB suppression regulates mitochondrial respiratory activity and contributes to liver cancer progression. We found that transcriptional LDHB down-regulation is an upstream event during suppressed oxidative phosphorylation. We also observed that LDHB knockdown increases inhibitory phosphorylation of pyruvate dehydrogenase (PDH) via lactate-mediated PDH kinase (PDK) activation and thereby attenuates oxidative phosphorylation activity. Interestingly, monocarboxylate transporter 1 was the major lactate transporter in hepatoma cells, and its expression was essential for PDH phosphorylation by modulating intracellular lactate levels. Finally, bioinformatics analysis of the hepatocellular carcinoma cohort from The Cancer Genome Atlas revealed that a low LDHB/LDHA ratio is statistically significantly associated with poor prognostic outcomes. A low ratio was also associated with a significant enrichment in glycolysis genes and negatively correlated with PDK1 and 2 expression, supporting a close link between LDHB suppression and the PDK–PDH axis. These results suggest that LDHB suppression is a key mechanism that enhances glycolysis and is critically involved in the maintenance and propagation of mitochondrial dysfunction via lactate release in liver cancer progression.
Keywords: hepatocellular carcinoma, lactic acid, mitochondria, pyruvate dehydrogenase complex (PDC), liver cancer, mitochondrial respiratory chain complex
Introduction
One key metabolic feature of cancer cells is a reprogrammed bioenergetic state characterized by impaired mitochondrial respiratory capacity and enhanced glycolysis (1–3). These two alterations can be triggered independently but simultaneously by the same environmental constraint, such as hypoxia. Therefore, it has long been accepted that premalignant cells acquire these hallmarks when they inevitably experience intermittent hypoxia during tumor development (4). Enhanced glycolysis confers a selective advantage for malignant progression by regulating lactic acidosis–mediated tumor invasion, angiogenesis, and immune response (5–7). Mitochondrial dysfunction, despite its association with impaired ATP generation capacity, also promotes malignant tumor progression through retrograde signals, such as reactive oxygen species and deregulated calcium homeostasis (8–14). These circumstances suggest that the bioenergetic hallmarks are not just passive responses to hypoxia but are rather an active strategy to generate advantageous tumoral properties. Moreover, in the absence of hypoxic constraint, these bioenergetic phenotypes can be triggered by several oncogenes or tumor suppressors, including Ras, Myc, and p53 (15–18), further supporting their oncogenic tactical involvement in tumor progression.
Pyruvate dehydrogenase complex (PDC)3 is the priming mitochondrial enzyme that allows pyruvate (the final product of aerobic glycolysis) to enter mitochondrial oxidative processes, the citric acid cycle, and subsequent oxidative phosphorylation (OXPHOS) (19). Therefore, PDC is an important regulatory enzyme in controlling mitochondrial energy metabolism via modulation of metabolite flux. PDC is a large complex containing three core enzymatically active subunits: pyruvate dehydrogenase (PDH), dihydrolipoamide acetyltransferase, and dihydrolipoyl dehydrogenase (20, 21). PDH directly oxidizes pyruvate (22) and harbors regulatory residues for phosphorylation (23). The specific PDH kinase (PDK) phosphorylates residue serine 293 of PDH, blocking its accessibility to the substrate pyruvate (24). Many cancer cells exhibit transcriptional up-regulation of PDK1 expression by hypoxia-inducible factor (HIF-1α), which is stabilized under hypoxia or induced by cMyc (15, 25), implying its pivotal role in hypoxic and oncogenic induction of mitochondrial dysfunction.
Lactate dehydrogenase (LDH) is another key enzyme in determining cellular bioenergetic flow, cellular dependence on glycolytic ATP generation, and oxidative mitochondrial activity. LDH uses the redox power of NADH to catalyze reversible conversion between pyruvate and lactate. Enzymatically functional LDH is a tetramer formed by diverse combinations of two different gene products encoded by LDHA and LDHB genes, which ultimately form five different isozymes: LDH1 (B4), LDH2 (AB3), LDH3 (A2B2), LDH4 (A3B), and LDH5 (A4) (26). LDH5 preferentially catalyzes the conversion of pyruvate to lactate, and its increased expression and activity is linked to the glycolytic phenotype and metastatic activity in many cancers (27, 28). Some studies show that LDH5 activity is mainly enhanced because of increased LDHA expression via the interplay of HIF-1α and cMyc (15, 29). However, we recently reported an LDH isozyme shift toward LDH5 in hepatoma cells, which was unexpectedly achieved by LDHB down-expression rather than LDHA up-expression (30). Moreover, this LDHB suppression restrained mitochondrial respiration, providing a model to elucidate the mechanism by which enhanced glycolysis regulates mitochondrial respiratory activity.
In the present study, we demonstrate that LDHB suppression induced enhanced glycolysis, which lowered mitochondrial respiratory activity via lactate-mediated modulation of the PDK–PDH axis. These findings support an interplay between the tumoral bioenergetics phenomena of aerobic glycolysis and mitochondrial dysfunction in the absence of hypoxic or oncogenic stimulus, emphasizing the glycolytic control of mitochondrial activity.
Results
LDHB suppression occurs upstream of decreased OXPHOS activity in hepatoma cells
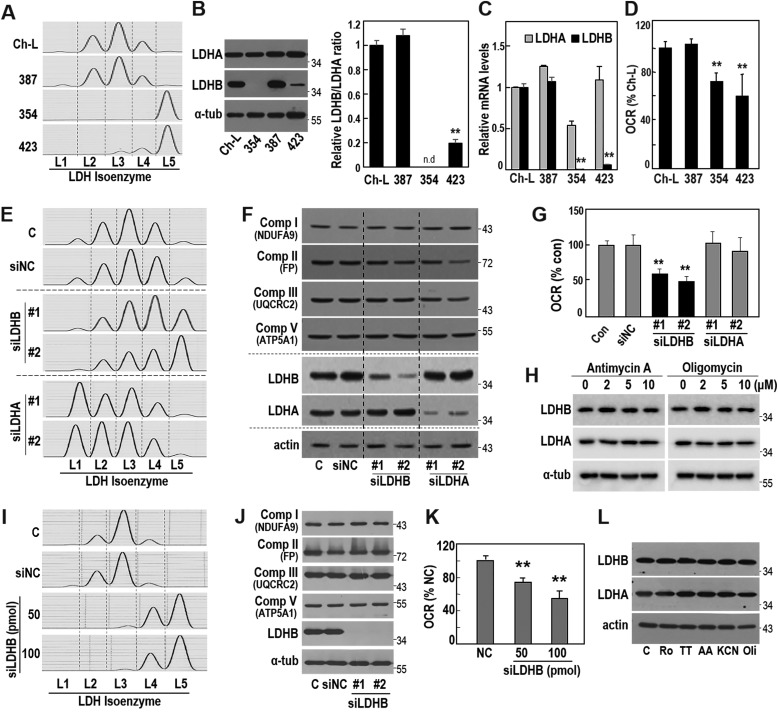
We first examined the metabolic status of three different Seoul National University hepatoma cells (SNU387, SNU354, and SNU423) and of the normal liver cell clone Ch-L (30). Compared with SNU387 and Ch-L, SNU354 and SNU423 showed a clear isoenzyme shift toward LDH5 (Fig. 1A) and harbored decreased LDHB protein expression, showing a low LDHB/LDHA ratio (Fig. 1B). SNU354 and SNU423 cells also exhibited decreased LDHB mRNA levels (Fig. 1C), implying its transcriptional control. Notably, SNU354 exhibited almost complete suppression of LDHB protein, which corresponded well to its LDH5 activity. The low LDHB levels in SNU354 and SNU423 cells were also accompanied by a significantly decreased cellular oxygen consumption rates (OCR) (Fig. 1D), suggesting a possible mechanistic association between the two phenotypes.
Figure 1.
LDHB suppression decreases cellular oxygen consumption rate. A–D, three different SNU hepatoma cell lines (SNU387, SNU354, and SNU423) and a Chang cell clone (Ch-L) were cultured for 2 days to maintain exponentially growing state and subjected to each experiment. A, LDH isozyme assay was performed as described under “Experimental procedures.” B, LDHA and LDHB expressions were assessed by Western blotting analysis using whole-cell lysates (left panel). Quantification of the protein levels is shown as a ratio of LDHB to LDHA (right panel). LDHB protein level of SNU354 was not detected (n.d.). **, <0.01 versus Ch-L. C, mRNA levels of LDHA, LDHB and actin were analyzed by qRT-PCR. mRNA levels of LDHA and LDHB were normalized by mRNA level of actin. **, <0.01 versus Ch-L. D, endogenous cellular oxygen consumption rate (OCR) was measured. **, <0.01 versus Ch-L. E–G, SNU-387 cells were transfected with siRNA for LDHB (siLDHB) and LDHA (siLDHA) for 3 days. E, LDH isozyme assay. F, protein expression levels of the indicated targets were analyzed by Western blotting analysis. G, endogenous cellular oxygen consumption rate. **, <0.01 versus control. H, SNU387 cell was exposed to the indicated concentrations of antimycin A (left panel) and oligomycin (right panel) for 12 h. LDHA and LDHB expressions were examined by Western blotting analysis. I–K, Ch-L cells were transfected with the indicated amounts of LDHB siRNA for 3 days. I, LDH isozyme analysis. J, Western blotting analysis. K, endogenous cellular oxygen consumption rate. **, <0.01 versus negative control (NC). L, Ch-L cells were treated with 10 μm rotenone (Ro, complex I inhibitor), 400 μm TTFA (TT, complex II inhibitor), 10 μm antimycin A (AA, complex III inhibitor), 10 mm KCN (complex IV inhibitor), or 10 μm oligomycin (Oli, complex V inhibitor) for 12 h. Western blotting analysis. C or Con, control.
In SNU387 hepatoma cells (which showed high LDHB expression), LDHB suppression by siRNA-mediated knockdown effectively induced an LDH isozyme shift toward LDH5, indicating dependence on LDHB level (Fig. 1, E and F). Unfortunately, to generate complete isozyme shift toward LDH5 using siRNA-mediated LDHB knockdown was not easy, implying that additional mechanisms, such as LDHA phosphorylation (31, 32) and/or promoter hypermethylation-mediated complete silencing of LDHB (33), might be required. Nevertheless, partial LDH isozyme shift by LDHB knockdown was enough to significantly decrease OCR, without alteration of OXPHOS subunit expressions (Fig. 1, E–G). Although LDHA suppression shifted LDH isozyme toward LDH1, it did not affect OCR (Fig. 1, E–G), showing specificity of the inhibitory effect of LDHB suppression on OCR. To further test the causal relationship, we exposed SNU387 cells to respiratory inhibitors such as antimycin A (complex III inhibitor) and oligomycin (complex V inhibitor) and found that LDHA and LDHB levels were unaltered (Fig. 1H). Similar results were obtained with Ch-L cell (Fig. 1, I–L). These results indicated that LDHB suppression-mediated LDH5 activation occurred upstream of OXPHOS activity suppression.
LDHB suppression increases PDH phosphorylation via PDK activity
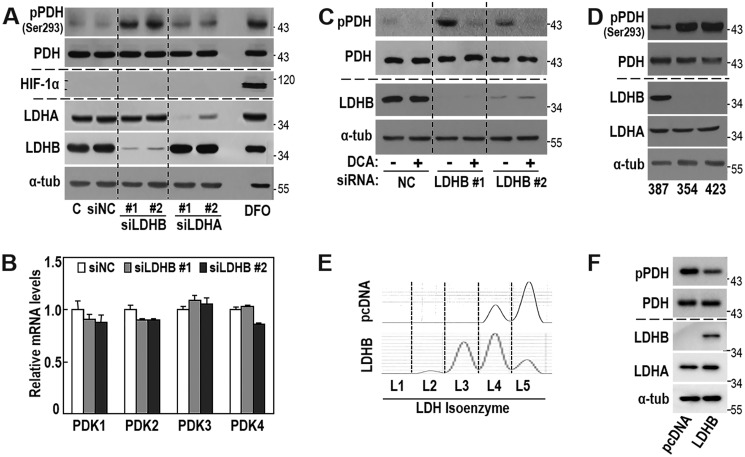
We next investigated the mechanism underlying the regulation of OXPHOS activity via LDHB suppression. LDHB knockdown did not affect protein expressions of OXPHOS subunits (Fig. 1F), and the LDHB-mediated LDH isozyme shift could potentially alter cellular lactate and pyruvate levels; therefore, we examined whether LDHB level controlled the overall OXPHOS flux through PDH activity. LDHB knockdown increased inhibitory phosphorylation of PDH, without stabilizing HIF-1α (Fig. 2A) and induction of mRNA expressions of all four PDK isoforms (Fig. 2B). Moreover, the specific PDK inhibitor dichloroacetate (DCA) effectively abolished the PDH phosphorylation induced by LDHB knockdown (Fig. 2C). This suggested that PDK activity was critically involved in LDHB suppression-mediated PDH phosphorylation. Similarly, SNU354 and SNU423, which harbor low LDHB expression and decreased OXPHOS activity, exhibited enhanced PDH phosphorylation (Fig. 2D). Moreover, overexpression of LDHB in SNU423 cells shifted LDH isozyme toward LDH1 (Fig. 2E) and decreased PDH phosphorylation status (Fig. 2F). These results explained that LDHB suppression played a key role in PDH inactivation and consequent OXPHOS control in hepatoma cells.
Figure 2.
LDHB knockdown induces PDK-mediated PDH phosphorylation without HIF-1 induction. A and B, SNU387 cells were transfected with siRNA for LDHB (siLDHB) and LDHA (siLDHA) for 3 days. Deferoxamine (DFO, hypoxia mimetic agent) was used as a HIF-1α inducer (54). A, Western blotting analysis of the indicated proteins. B, mRNA levels of PDK1∼4 were analyzed by qRT-PCR. C, SNU387 cells were transfected with siRNAs for LDHB for 3 days and exposed to 5 mm DCA for 1 h, followed by Western blotting analysis of the indicated proteins. D, SNU hepatoma cell lines (SNU387, SNU354, and SNU423) were cultured for 2 days to maintain exponentially growing state and subjected to Western blotting analysis. E and F, SNU423 cells were transfected with pcDNA3-LDHB-HA plasmid for 2 days. E, LDH isozyme analysis. F, Western blotting analysis. NC, negative control.
LDHB suppression-mediated lactic acidosis regulates OXPHOS activity via the PDK–PDH axis
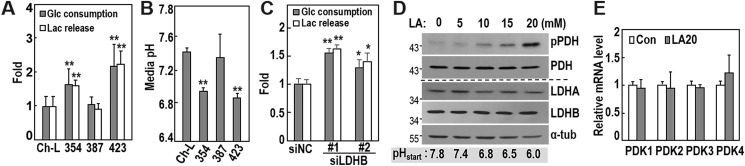
We further confirmed the high glycolytic activity of SNU354 and SNU423 cells, based on cellular glucose consumption, lactate release, and medium acidity (Fig. 3, A and B). Additionally, LDHB knockdown in SNU387 cells (with high LDHB) increased both glucose consumption and lactate release into the medium (Fig. 3C), indicating that LDHB suppression was sufficient to enhance the cellular glycolytic phenotype. These findings suggested that released lactate might control cellular PDH activity, and the subsequent OXPHOS activity. Lactate levels in the glycolytic tumor microenvironment are generally within the range of 5–20 mm (34, 35). We exposed SNU387 cells to lactic acid (LA) levels within this range and observed dose-dependent increases in PDH phosphorylation, without alterations of PDK mRNA (Fig. 3, D and E). These results indicated that lactate released by LDHB suppression augmented PDH phosphorylation in an autocrinal or paracrinal mode.
Figure 3.
Lactate augmented by LDHB suppression increases PDH phosphorylation. A and B, SNU387, SNU354, SNU423, and Ch-L cells were cultured for 2 days to maintain exponentially growing state and subjected to each experiment. A, glucose (Glc) consumption and lactate (Lac) release were estimated by monitoring glucose and lactate concentrations in cultured medium as described under “Experimental procedures.” Actual measurements of consumed Glc and released Lac concentrations of Ch-L were 1.70 ± 0.51 and 3.68 ± 1.11 mm/1 × 105 cells, respectively. **, < 0.01 versus Ch-L. B, medium pH was monitored. **, < 0.01 versus Ch-L. C, SNU387 cells were transfected with siLDHB for 3 days, and then glucose and lactate concentrations of the cultured medium were measured. Actual measurements for consumed Glc and released Lac concentrations of siNC were 2.7 ± 0.28 and 4.48 ± 0.33 mm/5 × 105 cells, respectively. *, < 0.05, **, < 0.01 versus siNC. D, SNU387 cells were treated with the indicated concentrations of LA for 12 h and subjected to Western blotting analysis. Initial pH of LA-treated medium was measured under noncultivating air condition without CO2 supply and indicated in the bottom. E, SNU387 cells were exposed to 20 mm LA for 12 h and subjected to qRT-PCR. Con, control.
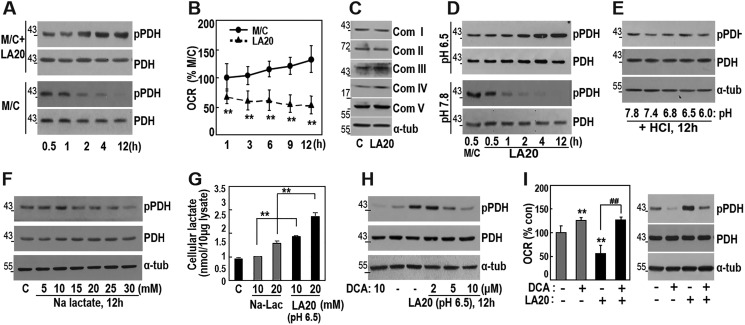
We also examined the time course effect of LA on both PDH phosphorylation and cellular OCR. Exposure of SNU387 cells to 20 mm LA in fresh medium gradually increased PDH phosphorylation over a time course (Fig. 4A, upper panels), whereas medium change alone decreased the phosphorylation (Fig. 4A, lower panels). Consistent with the PDH phosphorylation status, OCR significantly decreased upon LA treatment without alteration of OXPHOS subunit expressions (Fig. 4, B and C), suggesting that LA may regulate OXPHOS activity via PDH phosphorylation (inactivation).
Figure 4.
Lactic acidosis, not lactate alone, suppresses OCR via PDK–PDH axis. A and B, SNU387 cells were replenished with fresh medium alone (M/C, lower panel) or fresh medium containing 20 mm lactic acid (pH 6.5) (M/C + LA20, upper panel) for the indicated period. A, Western blotting analysis. B, cellular oxygen consumption rate (OCR). **, < 0.01 versus M/C. C, SNU387 cells were incubated with 20 mm lactic acid (pH 6.5) for 12 h and subjected to Western blotting analysis. D, SNU387 cells were replenished with fresh medium containing 20 mm lactic acid of which pH was adjusted to 6.5 (upper panel) or 7.8 (lower panel), followed by Western blotting analysis. E, SNU387 cells were replenished with fresh medium of which pH was adjusted to the indicated pH with 100 mm HCl and subjected to Western blotting analysis. F, SNU387 cells were replenished with fresh medium containing the indicated concentrations of sodium lactate (Na-Lac) for 12 h, followed by Western blotting analysis. G, SNU387 cells were replenished with fresh medium containing the indicated concentrations of sodium lactate or lactic acid for 12 h, followed by measurement of intracellular lactate as described under “Experimental procedures.” **, < 0.01 versus sodium lactate. H and I, SNU387 cells were replenished with medium containing 20 mm lactic acid (pH 6.5) for 12 h, followed by treatment with or without DCA for 1 h. H, phosphorylated PDH (pPDH) levels were analyzed by Western blotting. I, oxygen consumption rate (left panel) and Western blotting analysis (right panel). **, < 0.01 versus no drug; ##, < 0.01 versus LA20. Con, control.
Next, we asked whether the PDH phosphorylation is due to lactate per se or the acidic environment. Because extracellular pH values in solid tumors have a pH level of ∼6.5 (36, 37), we adjusted the pH of medium containing 20 mm LA to 6.5 or 7.8 right before medium replenishment, generating the pH of medium to ∼6.4 or 7.6 in cultivating condition with 5% CO2. Medium containing 20 mm LA (pH 6.5) gradually enhanced PDH phosphorylation over a time course, whereas medium containing with 20 mm LA (pH 7.8) showed results similar to those with medium replenishment alone (Fig. 4D). The effect of pH 7.8 medium was confirmed by exposure to different concentrations of sodium lactate up to 30 mm (Fig. 4F). These results clearly indicated that lactate per se without acidity did not affect PDH phosphorylation. Moreover, PDH phosphorylation was not affected by acidification alone of media without lactate (Fig. 4E). Consistently, LA treatment significantly augmented intracellular lactate level, compared with sodium lactate treatment (Fig. 4G), implying that proton concentration facilitated lactate import across cell plasma membrane. These results supported the possibility that lactic acidosis, combined action of lactate level and acidity, by LA treatment was required for intracellular PDH phosphorylation. We further confirmed that lactic acidosis-mediated PDH phosphorylation and OCR inhibition were clearly resolved by pharmacological PDK inhibition (Fig. 4, H and I). Overall, these results indicated that lactic acidosis decreased OXPHOS activity via PDK-mediated PDH inactivation.
PDH phosphorylation involves MCT-mediated lactate transport
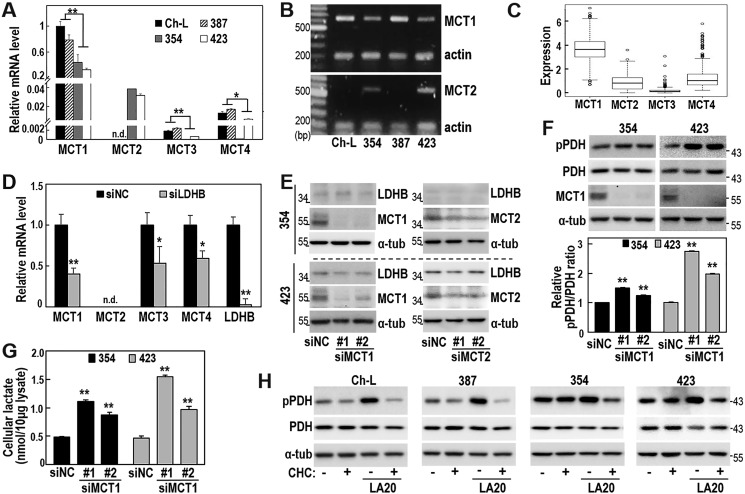
Lactic acid is transported across the plasma membrane by proton-linked monocarboxylate transporters (MCTs), mainly MCT1–4 (38, 39). Interestingly, all four examined cell lines showed very high MCT1 mRNA levels (Fig. 5A), compared with mRNA levels of other MCTs (MCT2–4). Bioinformatic analysis of The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) open database confirmed that LIHC tissues expressed MCT1 at a higher level than MCT2–4 (Fig. 5C), implying a key role of MCT1 in lactate transport in hepatoma cells. SNU354 and SNU423 cells showed markedly high MCT2 expression (Fig. 5A), whereas MCT2 levels were barely detectable in Ch-L and SNU387 cells. Relative mRNA levels of MCT1 and MCT2 in all tested cells were further confirmed by conventional RT-PCR (Fig. 5B). Interestingly, in SNU387 cells, LDHB knockdown attenuated MCT1, MCT3, and MCT4 expression (Fig. 5D), whereas knockdown of MCT1 or MCT2 did not affect LDHB expression (Fig. 5E). These results suggested that LDHB expression is linked to MCT expression and may act as an upstream regulator.
Figure 5.
MCT-mediated lactate transport is importantly linked with the LA-induced PDH phosphorylation. A and B, three different SNU hepatoma cell lines (SNU387, SNU354, and SNU423) and a Chang cell clone (Ch-L) were cultured for 2 days to maintain exponentially growing state and subjected to qRT-PCR (A) and RT-PCR (B). MCT2 mRNA levels were not detected in Ch-L and SNU387 cells (n.d.). *, < 0.05, **, < 0.01 versus Ch-L and SNU387. C, expression analysis of individual MCTs (MCT1∼4) in LIHC tumors (n = 371) from TCGA_LIHC data set. D, SNU387 cells were transfected with siLDHB for 3 days and subjected to qRT-PCR. MCT2 mRNA level of SNU387 cells was not detected (n.d.). *, < 0.05; **, < 0.01 versus siNC. E, SNU354 and SNU423 cells were transfected with siMCT1 or siMCT2 for 3 days and subjected to Western blotting analysis. F and G, SNU354 and SNU423 cells were transfected with siMCT1 for 3 days. F, phosphorylated PDH (pPDH) levels were analyzed by Western blotting (upper panel). The bar graph represents the normalized band intensities of pPDH to PDH using ImageJ software (lower panel). **, < 0.01 versus siNC. G, measurement of intracellular lactate. **, < 0.01 versus siNC. H, Ch-L, SNU387, SNU354, and SNU423 cells were treated with or without 1 mm CHC for 1 h and further incubated with 20 mm lactic acid (pH 6.5) for an additional 12 h. Phosphorylated PDH levels were examined by Western blotting analysis.
We next examined whether lactate transport via MCTs was critical for PDH phosphorylation. Because all four cell lines showed much higher expression of MCT1 compared with MCT2, MCT3, and MCT4, we investigated whether MCT1-mediated lactate transport influenced PDH phosphorylation. In the two glycolytic hepatoma cells (SNU354 and SNU423), which possess high lactate production capacity (Fig. 3A), MCT1 knockdown increased intracellular lactate levels and further augmented PDH phosphorylation status (Fig. 5, F and G). These findings suggested that MCT1 played a key role as a lactate exporter in glycolytic hepatoma cells and that high intracellular lactate regulated PDH phosphorylation. We further investigated whether extracellular lactic acidosis could impact intracellular PDH phosphorylation via MCT-mediated lactate import. As expected, the MCT1/2-specific inhibitor α-cyano-4-hydroxycinnamic acid (CHC) (40, 41) strongly blocked PDH phosphorylation under lactic acidosis in the oxidative SNU387 and Ch-L cells (Fig. 5H). Similar results were also obtained in SNU354 and SNU423 cells (Fig. 5H), implying that lactate could act in an autocrinal mode. Together, our findings suggested that increased intracellular lactate may inhibit mitochondrial OXPHOS activity via PDH phosphorylation, indicating a potential role of lactate as a key player in both autocrinal maintenance and paracrinal propagation of OXPHOS dysfunction.
LDHB/LDHA ratio is a key glycolytic and prognostic marker of LIHC
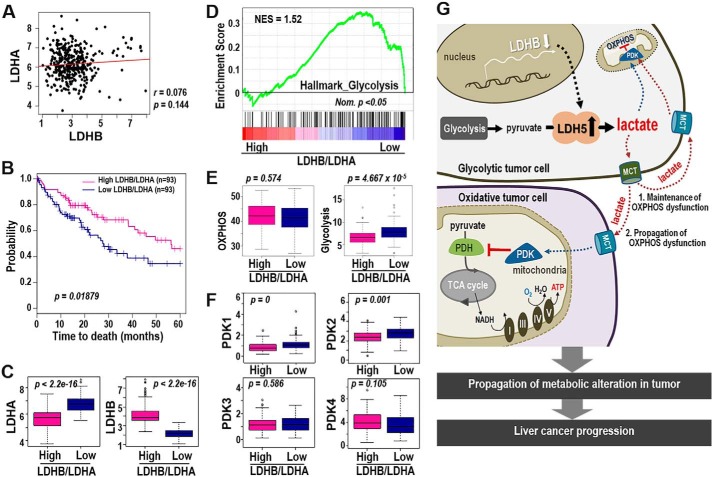
We performed Kaplan–Meier survival analysis to compare clinical outcomes between LDHB-high and LDHB-low expression groups from the TCGA-LIHC cohort. Unexpectedly, we found no significant differences (data not shown). We also could not find any negative association between LDHA and LDHB mRNA levels in the TCGA-LIHC cohort (Fig. 6A). Thus, we hypothesized that individual hepatoma cells may exhibit either LDHB suppression or LDHA induction, such that the LDHB/LDHA ratio may represent the glycolytic status in heterogeneous LIHC tissue context. Interestingly, the low LDHB/LDHA group (n = 93) displayed a poor prognosis with significantly shorter survival compared with the high LDHB/LDHA group (Fig. 6B). Moreover, the low LDHB/LDHA group showed significantly higher LDHA and lower LDHB expression (Fig. 6C). These results suggested that individual LIHC tumor tissue may acquire the LDH5 isozyme via one strategy, either LDHA induction or LDHB suppression.
Figure 6.
LDHB/LDHA ratio is a good prognostic indicator of LIHC harboring mitochondrial dysfunction. A–F, bioinformatics analysis from TCGA_LIHC data set. A, association analysis of LDHA and LDHB expression in LIHC tumors. B, Kaplan–Meier survival analysis of high and low LDHB/LDHA groups was performed (see “Experimental Procedures”). The statistical p value was generated by the Cox–Mantel log-rank test. C, box plot shows expression levels of LDHB (right panel) and LDHA (left panel) of high- and low-LDHB/LDHA group. The p value was calculated based on the Welch two-sample t test. D, enrichment plot from GSEA output based on the gene set of HALLMARK_GLYCOLYSIS (n = 200) is shown. Normalized enrichment score (NES) and nominal p value (Nom. p) are shown. E, single sample GSEA was performed based on the gene sets derived from HALLMARK_OXIDATIVE_PHOSPHORYLATION (n = 200) and HALLMARK_GLYCOLYSIS (n = 200) (MSigDB_V6.1). The p value from the Kolmogorov–Smirnov test was transformed in −log scaled and used in the plot. F, expression levels of PDK1∼4 in high- and low-LDHB/LDHA tissue types were plotted. p value was calculated based on the Welch two-sample t test. G, schematic diagram for our hypothesis explaining the LDHB-mediated glycolytic control of OXPHOS dysfunction in LIHC.
Next, we evaluated whether the LDHB/LDHA ratio was truly associated with an overall metabolic shift from OXPHOS to glycolysis. Gene set enrichment score analysis revealed a significantly more enriched glycolysis hallmark gene signature in the low LDHB/LDHA group than the high LDHB/LDHA group (Fig. 6D). OXPHOS hallmarks were similar between the two groups (Fig. 6E). Finally, we observed that the low LDHB/LDHA group displayed significantly higher PDK1 and PDK2 expressions, whereas PDK3 and PDK4 expressions were similar between groups (Fig. 6F). These results indicated that low LDHB/LDHA ratio was linked to decreased OXPHOS activity, which is possibly mediated by the PDK–PDH axis in LIHC, rather than to OXPHOS gene regulation. Overall, our findings supported that the low LDHB/LDHA ratio is a key marker of both aerobic glycolysis and PDK-mediated mitochondrial dysfunction and is a good prognostic indicator for LIHC with bioenergetic features.
Discussion
The cellular LDH isozyme shift toward LDH5 is a key event in maintaining glycolytic ATP generation by resupplying the NAD+ needed for the early step of glycolysis. This is extremely important for maintaining cellular energy in the absence of mitochondrial oxidation, as occurs in tumor cells with hypoxic conditions and mitochondrial defects. HIF-1α is a key regulator of LDHA gene induction and subsequent LDH5 formation (42), and several oncogenes (e.g. Ras, Src, and Myc) reportedly up-regulate LDHA via HIF-1α–dependent or –independent manner (43, 44). The available data support the importance of oncogenic LDHA up-regulation for gaining aerobic glycolysis and its priming role for bioenergetics features even without hypoxia or mitochondrial dysfunction during tumorigenesis.
In our present study, we clearly demonstrated that LDHB suppression is another effective cellular strategy for driving the LDH isozyme shift toward LDH5 and thus enhancing aerobic glycolysis. Supporting this concept, Cui et al. (45) recently reported similar observations in pancreatic cancer progression. Interestingly, our bioinformatic analysis of the open RNA-Seq database from the TCGA-LIHC cohort revealed that LDHB/LDHA ratio—the best indicator of LDH isozyme shift—was a good prognostic marker for liver cancer, implying the importance of this shift in liver cancer progression. Notably, our findings also suggested that individual tumor tissue may adopt either LDHA induction or LDHB suppression to achieve LDH5-mediated glycolysis in liver cancer. This heterogeneous tactic for LDH5 activation means that clinical outcome cannot be significantly predicted by analysis of LDHA or LDHB alone in liver cancer.
Glycolytic activation in cancer is not only important for maintaining cellular energy but also for producing and releasing lactate. Extracellular lactate promotes chronic acidification of the tumor microenvironment and drives cancer cell metastasis (12, 13). Surprisingly, our present results demonstrated that lactate regulated mitochondrial OXPHOS activity via the PDK–PDH axis. The schematic diagram in Fig. 6G shows that glycolytic hepatoma cells (SNU354 and SNU423), which harbor OXPHOS defect and LDHB depletion-mediated lactic acidosis, utilize lactate to increase PDH phosphorylation, leading to additional OXPHOS dysfunction. Moreover, exposure of oxidative hepatoma cells (Ch-L and SNU387) to lactate decreased OXPHOS activity in a rapid response via PDH phosphorylation. Recently, we reported that lactate also induced mitoribosomal defect in a delayed response via suppressing MRPL13 expression, eventually contributing to the endogenous OXPHOS defect of SNU354 and SNU423 cells (46). Together, these results indicate that lactate may play a key role in the maintenance and propagation of mitochondrial OXPHOS dysfunction in the tumoral context, even under normoxic conditions. High glucose utilization reportedly reduces OXPHOS activity in yeast, termed the Crabtree effect (47). However, to our knowledge, the present study is the first to demonstrate that lactate, the released glycolytic product, inhibits tumoral OXPHOS activity in a rapid response.
Another important question is how lactic acidosis condition regulates PDH phosphorylation. To prevent the intracellular acidification of glycolytic tumor cells, lactate efflux is generally conducted through MCTs, a group of bidirectional proton-linked transporters with varying Km values for lactate (48). Of the four MCTs, MCT1 (high Km for lactate, 3.5–10 mm) was predominantly expressed in our hepatoma model cells and LIHC tissues. Furthermore, compared with the oxidative hepatoma cells (Ch-L and SNU387), the glycolytic hepatoma cells (SNU354 and SNU423) showed decreased but still substantial MCT1 levels, suggesting its major role in liver cancer. MCT2 (low Km for lactate, 0.5–0.75 mm) was detected only in glycolytic hepatoma cells. Further studies are needed to elucidate the role of MCT2 in glycolytic tumor cells, but the absence of MCT2 in oxidative cells implies that these cells may uptake via MCT1 and utilize lactate only when extracellular lactate levels are very high. On the other hand, high lactate-generating glycolytic tumor cells may use MCT1 for lactate efflux and MCT2 for lactate re-entry when necessary. This concept is supported by a previous report that MCT1 acts in both lactate export and import, depending on metabolic state (49, 50).
MCT1 knockdown caused glycolytic tumor cells to accumulate intracellular lactate and exhibit increased PDH phosphorylation by inhibiting lactate efflux. Additionally, in oxidative tumor cells mainly expressing MCT1, pharmacological MCT inhibition almost completely abolished PDH phosphorylation by blocking lactate influx under lactic acidosis condition. These results clearly indicate that intracellular lactate regulates PDH phosphorylation. A previous study reports that lactate accumulation inhibits prolyl hydroxylase-2 (PHD-2) and activates HIF-1α, inducing PDK1 (51) and that lactate stabilizes the NDRG3 protein to mediate hypoxia-induced Raf-ERK pathway activation in a PHD-2–dependent manner (52). However, under our experimental conditions, LDHB depletion did not stabilize HIF-1α and not induce PDK expression. This indicates that LDHB-mediated lactate regulated PDH phosphorylation independently of HIF-1α-mediated PDK expression and possibly via PDK activation by post-translational modification. An ongoing study is investigating this issue in further detail.
In our present study, we present a novel mechanism in which LDHB depletion induces aerobic glycolysis, playing a key role as a priming regulator and a propagator of mitochondrial OXPHOS dysfunction, via lactate-mediated control of the PDK–PDH axis. Our results further emphasize that a low LDHB/LDHA ratio is a useful prognostic indicator for LIHC patients with bioenergetic features. Further studies are needed to determine whether differential therapeutic approaches may be useful for LIHC groups with different metabolic backgrounds.
Experimental procedures
Cell cultures and cell growth rate
Human hepatoma cells (SNU387, SNU354, and SNU423) were purchased from Korean Cell Line Bank (Seoul, Korea) and were cultured in Gibco RPMI 1640 medium (Invitrogen) supplemented with 10% Gibco fetal bovine serum (Invitrogen) and Gibco antibiotics (Invitrogen) at 37 °C in a humidified incubator with 5% CO2. Chang cells were obtained from American Tissue Culture Collection (Manassas, VA), and a Chang cell clone denoted as Chang-L that was isolated by single cell dilution and expansion and had high hepatic characteristics (albumin production and liver-specific carbamoyl-phosphate synthase-1 expression) as described previously (30) was used for this study. Ch-L clone was cultured in Gibco Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% Gibco fetal bovine serum. Cell growth rates were monitored by counting the trypan blue–negative viable cells. The cell lines were tested for mycoplasma contamination (MycoAlertTM mycoplasma detection kit; Lonza, Rockland, ME).
LDH isozyme assay
Activity profiles of the five LDH isozymes are estimated by in gel activity analysis after separating the lysates (5 μg) on agarose native gel electrophoresis according to the protocol provided with an REP LDH isoenzyme assay kit (Helena Laboratories, Beanmont, TX) with Rapid Electrophoresis (Helena Laboratories).
Measurement of lactate and glucose concentrations
Concentrations of lactate and glucose in extracellular medium were measured using a YSI 7100 Bioanalyzer (Yellow Springs, OH). Briefly, the cells were seeded on 12-well plates and cultured for 24 h. After replenishing with fresh medium for 2 days, the conditioned medium was harvested and applied to the chamber assembled with glucose/lactate membrane kit (YSI2365 and YSI2329) to measure simultaneously glucose and lactate concentrations. The concentration values were obtained by comparing with standard calibration curves for lactate and glucose concentrations and normalized by cell numbers. Intracellular lactate levels were measured using a lactate colorimetric/fluorometric assay kit (Biovision, Milpitas, CA) according to the manufacturer's instructions.
Endogenous cellular oxygen consumption rate
Endogenous cellular oxygen consumption rate was measured in situ with cultured cells using the XF-24 extracellular flux analyzer (Seahorse Bioscience) according to the protocol provided. Briefly, the cells were seeded on XF24 cell culture microplates (Seahorse Bioscience) at a density of 10,000 cells/well and preincubated with XF assay medium (Seahorse Bioscience) containing 1 mm pyruvate and 5 mm glucose. Its mitochondrial specificity was confirmed by adding 100 nm antimycin A.
Transfections
All the transfections were performed using OligofectamineTM reagent (Invitrogen), according to the manufacturer's instructions. Target siRNAs were generated from Bioneer (Daejeon, Korea), and single-strand sequences of the siRNAs were as follows: 5′-GGAUAUACCAACUGGGCUA-3′ for LDHB #1, 5′-GCAGAUACCCUGUGGGACA-3′ for LDHB #2, 5′-CGUGCAUUCCCGAUUCCUU-3′ for LDHA #1, 5′-CUGGUUAGUGUGAAAU-3′ for LDHA #2, 5′-GAGUCAGCAUUAUAGU-3′ for MCT1 #1, 5′-CAUGUAUGGAGACUACAAA-3′ for MCT1 #2, 5′-CUCCAUCUGACUACGACUA-3′ for MCT2 #1, 5′-CAGCAAAUAUUCCACACUA-3′ for MCT2 #2, and 5′-CCUACGCCACCAAUUUCGU-3′ for negative control (siNC). The pcDNA-LDHB-HA was generated as previously described (30).
Western blotting analysis
Cultured cells were washed twice with PBS and lysed with lysis buffer (1 m Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% SDS, 1% Nonidet P-40) containing phosphatase inhibitor (200 mm NaF, 200 mm NaVO4) and protease inhibitor (200 mm phenylmethylsulfonyl fluoride, 500 mm leupeptin, 500 mm pepstatin A) at 4 °C for 30 min. The isolated proteins were separated on 8–12% SDS-polyacrylamide gels and then transferred onto nitrocellulose membrane (Amersham Biosciences ProtranTM, GE Healthcare). These membranes were then incubated for 12 h with the following primary antibodies. Next, the membranes were washed with 0.1% PBS-T buffer three times for 15 min each, after which they were incubated with horseradish peroxidase-conjugated anti-mouse, anti-rabbit, anti-sheep, and anti-goat antibody (1:3000–5000, Santa Cruz, Dallas, TX) for 1–2 h. The membranes were then washed three times with 0.1% PBS-T buffer for 15 min. Immunoreactive bands were visualized with the ECL system (West Save, Lab Frontier). Antibody against p-PDH (Ser293) (1:1000, AP1062) was purchased from Calbiochem (San Diego, CA). Antibodies against PDH-E1α (1:2000, ab110330), PDK1 (1:2000, ab90444), and LDHA (1:3000, ab9002) were purchased from Abcam Ltd. (Cambridge, UK). Antibodies against NDUFA9 of complex I (A21344), flavoprotein (A11142) of complex II, UQCRC2 of complex III (A11143), MTCOII of complex IV (A6404), and ATP5A1 of complex V (A21350) were from Molecular Probes Corp. (Eugene, OR). Individual complexes (I to V) are labeled as Comp I, II, III, IV, and V, respectively, in the figures. Antibodies against MCT1 (sc-14916) and MCT2 (sc-22034) were obtained from Santa Cruz Biotechnology.
RT-PCR
Total RNA was isolated using TRIzol (Invitrogen), and cDNA was prepared using avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI). Conventional RT-PCR and subsequent visualization on agarose gel was performed when necessary. Quantitative real-time RT-PCR (qRT-PCR) was performed using THUNDERBIRD® SYBR® quantitative PCR Master Mix (Toyobo, Osaka, Japan), according to the manufacturer's protocol. PCR conditions were different, depending on the primers but generally were within 30–45 cycles of the reaction at 95 °C for 15 s, 55–57 °C for 30 s, and 72 °C for 20–40 s. The PCR primer sets were produced by Bioneer as follows: LDHA, 5′-ATATCTTGACCTACGTGGCTTGGAA-3′ and 5′-ACTCCATACAGGCACACTGGAAT-3′; LDHB, 5′-CTCATTGCACCAGTTGCGGAAG-3′ and 5′-AAGCTCCCATGCTGCAGATCCAT-3′; PDK1, 5′-CTATGAAAATGCTAGGCGTCTGT-3′ and 5′-AACCACTTGTATTGGCTGTCC-3′; PDK2, 5′-AGGACACCTACGGCGATGA-3′ and 5′-TGCCGATGTGTTTGGGATGG-3′; PDK3, 5′-GCCAAAGCGCCAGACAAAC-3′ and 5′-CAACTGTCGCTCTCATTGAGT-3′; PDK4, 5′-TTATACATACTCCACTGCACCA-3′ and 5′-ATAGACTCAGAAGACAAAGCCT-3′; MCT1, 5′-AGTGTCATGGATATCCTCCATAATGT-3′ and 5′-AAGACCTCCAATGACTCCAATACA-3′; MCT2, 5′-GCTGTACCTCACTATGGGATTC-3′ and 5′-CCTACAGAAGGCCTAGCAAAC-3′; MCT3, 5′-TGTCCTCCATCATGCTAGCCAT-3′ and 5′-ATGAGCGACGGCTGGAAGTT-3′; MCT4, 5′-TCTTCGGCTGTTTCGTCATCA-3′ and 5′-ATGCCCAGCGACGCAA-3′; and β-actin, 5′-CCTTCCTGGGCATGGAGTCCTGT-3′ and 5′-GGAGCAATGATCTTGATCTTC-3′. Expression levels of target mRNAs were normalized by β-actin mRNA level.
Chemicals
DCA (347795), l-(+)-lactic acid solution (L1875), sodium lactate (L7022), and CHC (C2020) were purchased from Sigma–Aldrich.
Validation in the open liver cancer cohort
To validate the clinical significance of LDHB, we used liver cancer open database, TCGA-LIHC. Level 3 RNA-SeqV2 data for LIHC were downloaded from TCGA data portal (https://portal.gdc.cancer.gov/). RNASeqV2 expression data of 371 primary tumors was used in the analysis. All expression data were log2 transformed and quantile-normalized using R. In this study, we calculated the ratio of LDHB to LDHA in individual sample, and the top and bottom quartiles of LIHC tissues ranked by LDHB/LDHA ratio were assigned into high and low groups, respectively. To compare clinical significance, we performed Kaplan–Meier (Km) survival analysis, and significance was estimated by Cox–Mantel log-rank test. To elucidate the association between LDHA and LDHB, correlation estimation and p value calculated from Pearson's correlation test were used. We performed gene set enrichment analysis (GSEA) based on the gene set for glycolysis (n = 200) from in Molecular Signatures Database (53) (MSigDB_V6.1;HALLMARK_GLYCOLYSIS) using the GenePattern server (55).
Statistical analysis
Statistical analyses were performed using a two-tailed Student's t test. p < 0.05 was considered statistically significant. All data were confirmed by at least three independent experiments, and all quantitative data were obtained from triplicate samples for each experiment and presented as means ± standard deviation.
Author contributions
S. M. H., Y.-K. L., I. P., and S. M. data curation; S. M. H., Y.-K. L., and I. P. investigation; S. M. H. and G. Y. writing-original draft; Y.-K. L. and I. P. validation; S. M.K. software; S. M.K. formal analysis; G. Y. conceptualization; G. Y. resources; G. Y. supervision; G. Y. funding acquisition; G. Y. writing-review and editing.
This work was supported by Grants 2012R1A5A2048183, 2015R1A2A1A10055038, and 2016R1A6A3A11932420 from the Basic Science Research Program through the National Research Foundation funded by the Ministry of Science, ICT, and Future Planning of Korea. The authors declare that they have no conflicts of interest with the contents of this article.
- PDC
- pyruvate dehydrogenase complex
- LDH
- lactate dehydrogenase
- LDHB
- LDH subunit B
- OXPHOS
- oxidative phosphorylation
- PDH
- pyruvate dehydrogenase
- PDK
- PDH kinase
- MCT
- monocarboxylate transporter
- OCR
- oxygen consumption rates
- TCGA-LIHC
- The Cancer Genome Atlas Liver Hepatocellular Carcinoma
- HIF
- hypoxia-inducible factor
- LA
- lactic acid
- GSEA
- gene set enrichment analysis
- CHC
- α-cyano-4-hydroxycinnamic acid
- qRT-PCR
- quantitative real-time RT-PCR
- DCA
- dichloroacetate
- Tub
- tubulin.
References
- 1. Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 2. Cuezva J. M., Krajewska M., de Heredia M. L., Krajewski S., Santamaría G., Kim H., Zapata J. M., Marusawa H., Chamorro M., and Reed J. C. (2002) The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 62, 6674–6681 [PubMed] [Google Scholar]
- 3. Singh K. K. (2004) Mitochondrial dysfunction is a common phenotype in aging and cancer. Ann. N.Y. Acad. Sci. 1019, 260–264 10.1196/annals.1297.043 [DOI] [PubMed] [Google Scholar]
- 4. Gatenby R. A., and Gillies R. J. (2004) Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 10.1038/nrc1478 [DOI] [PubMed] [Google Scholar]
- 5. Ruan G. X., and Kazlauskas A. (2013) Lactate engages receptor tyrosine kinases Axl, Tie2, and vascular endothelial growth factor receptor 2 to activate phosphoinositide 3-kinase/Akt and promote angiogenesis. J. Biol. Chem. 288, 21161–21172 10.1074/jbc.M113.474619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Romero-Garcia S., Moreno-Altamirano M. M., Prado-Garcia H., and Sánchez-Garcia F. J. (2016) Lactate contribution to the tumor microenvironment: mechanisms, effects on immune cells and therapeutic relevance. Front. Immunol. 7, 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doherty J. R., and Cleveland J. L. (2013) Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 123, 3685–3692 10.1172/JCI69741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen E. I. (2012) Mitochondrial dysfunction and cancer metastasis. J. Bioenerg. Biomembr. 44, 619–622 10.1007/s10863-012-9465-9 [DOI] [PubMed] [Google Scholar]
- 9. Chang C. J., Yin P. H., Yang D. M., Wang C. H., Hung W. Y., Chi C. W., Wei Y. H., and Lee H. C. (2009) Mitochondrial dysfunction-induced amphiregulin upregulation mediates chemo-resistance and cell migration in HepG2 cells. Cell Mol. Life Sci. 66, 1755–1765 10.1007/s00018-009-8767-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hung W. Y., Huang K. H., Wu C. W., Chi C. W., Kao H. L., Li A. F., Yin P. H., and Lee H. C. (2012) Mitochondrial dysfunction promotes cell migration via reactive oxygen species-enhanced β5-integrin expression in human gastric cancer SC-M1 cells. Biochim. Biophys. Acta 1820, 1102–1110 10.1016/j.bbagen.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 11. Ma J., Zhang Q., Chen S., Fang B., Yang Q., Chen C., Miele L., Sarkar F. H., Xia J., and Wang Z. (2013) Mitochondrial dysfunction promotes breast cancer cell migration and invasion through HIF1α accumulation via increased production of reactive oxygen species. PLoS One 8, e69485 10.1371/journal.pone.0069485 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Hsu C. C., Tseng L. M., and Lee H. C. (2016) Role of mitochondrial dysfunction in cancer progression. Exp. Biol. Med. 241, 1281–1295 10.1177/1535370216641787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Srinivasan S., Guha M., and Avadhani N. G. (2016) Mitochondrial respiratory defects promote the Warburg effect and cancer progression. Mol. Cell. Oncol. 3, e1085120 10.1080/23723556.2015.1085120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee Y. K., Jee B. A., Kwon S. M., Yoon Y. S., Xu W. G., Wang H. J., Wang X. W., Thorgeirsson S. S., Lee J. S., Woo H. G., and Yoon G. (2015) Identification of a mitochondrial defect gene signature reveals NUPR1 as a key regulator of liver cancer progression. Hepatology 62, 1174–1189 10.1002/hep.27976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dang C. V., Kim J. W., Gao P., and Yustein J. (2008) The interplay between MYC and HIF in cancer. Nat. Rev. Cancer 8, 51–56 10.1038/nrc2274 [DOI] [PubMed] [Google Scholar]
- 16. Kamp W. M., Wang P. Y., and Hwang P. M. (2016) TP53 mutation, mitochondria and cancer. Curr. Opin. Genet. Dev. 38, 16–22 10.1016/j.gde.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim J. H., Kim H. Y., Lee Y. K., Yoon Y. S., Xu W. G., Yoon J. K., Choi S. E., Ko Y. G., Kim M. J., Lee S. J., Wang H. J., and Yoon G. (2011) Involvement of mitophagy in oncogenic K-Ras-induced transformation: overcoming a cellular energy deficit from glucose deficiency. Autophagy 7, 1187–1198 10.4161/auto.7.10.16643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dang C. V. (1999) c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19, 1–11 10.1128/MCB.19.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris R. A., Bowker-Kinley M. M., Huang B., and Wu P. (2002) Regulation of the activity of the pyruvate dehydrogenase complex. Adv. Enzyme Regul. 42, 249–259 10.1016/S0065-2571(01)00061-9 [DOI] [PubMed] [Google Scholar]
- 20. Hiromasa Y., Fujisawa T., Aso Y., and Roche T. E. (2004) Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J. Biol. Chem. 279, 6921–6933 10.1074/jbc.M308172200 [DOI] [PubMed] [Google Scholar]
- 21. Hitosugi T., Fan J., Chung T. W., Lythgoe K., Wang X., Xie J., Ge Q., Gu T. L., Polakiewicz R. D., Roesel J. L., Chen G. Z., Boggon T. J., Lonial S., Fu H., Khuri F. R., et al. (2011) Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol. Cell 44, 864–877 10.1016/j.molcel.2011.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan J., Kang H. B., Shan C., Elf S., Lin R., Xie J., Gu T. L., Aguiar M., Lonning S., Chung T. W., Arellano M., Khoury H. J., Shin D. M., Khuri F. R., Boggon T. J., et al. (2014) Tyr-301 phosphorylation inhibits pyruvate dehydrogenase by blocking substrate binding and promotes the Warburg effect. J. Biol. Chem. 289, 26533–26541 10.1074/jbc.M114.593970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kolobova E., Tuganova A., Boulatnikov I., and Popov K. M. (2001) Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem. J. 358, 69–77 10.1042/0264-6021:3580069,10.1042/bj3580069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saunier E., Benelli C., and Bortoli S. (2016) The pyruvate dehydrogenase complex in cancer: An old metabolic gatekeeper regulated by new pathways and pharmacological agents. Int. J. Cancer 138, 809–817 10.1002/ijc.29564 [DOI] [PubMed] [Google Scholar]
- 25. Kim J. W., Tchernyshyov I., Semenza G. L., and Dang C. V. (2006) HIF-1–mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 10.1016/j.cmet.2006.02.002 [DOI] [PubMed] [Google Scholar]
- 26. Drent M., Cobben N. A., Henderson R. F., Wouters E. F., and van Dieijen-Visser M. (1996) Usefulness of lactate dehydrogenase and its isoenzymes as indicators of lung damage or inflammation. Eur. Respir. J. 9, 1736–1742 10.1183/09031936.96.09081736 [DOI] [PubMed] [Google Scholar]
- 27. Koukourakis M. I., Giatromanolaki A., Simopoulos C., Polychronidis A., and Sivridis E. (2005) Lactate dehydrogenase 5 (LDH5) relates to up-regulated hypoxia inducible factor pathway and metastasis in colorectal cancer. Clin. Exp. Metastasis 22, 25–30 10.1007/s10585-005-2343-7 [DOI] [PubMed] [Google Scholar]
- 28. Koukourakis M. I., Giatromanolaki A., Sivridis E., Bougioukas G., Didilis V., Gatter K. C., Harris A. L., and Tumour and Angiogenesis Research Group (2003) Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br. J. Cancer 89, 877–885 10.1038/sj.bjc.6601205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lewis B. C., Prescott J. E., Campbell S. E., Shim H., Orlowski R. Z., and Dang C. V. (2000) Tumor induction by the c-Myc target genes rcl and lactate dehydrogenase A. Cancer Res. 60, 6178–6183 [PubMed] [Google Scholar]
- 30. Kim J. H., Kim E. L., Lee Y. K., Park C. B., Kim B. W., Wang H. J., Yoon C. H., Lee S. J., and Yoon G. (2011) Decreased lactate dehydrogenase B expression enhances claudin 1-mediated hepatoma cell invasiveness via mitochondrial defects. Exp. Cell Res. 317, 1108–1118 10.1016/j.yexcr.2011.02.011 [DOI] [PubMed] [Google Scholar]
- 31. Jin L., Chun J., Pan C., Alesi G. N., Li D., Magliocca K. R., Kang Y., Chen Z. G., Shin D. M., Khuri F. R., Fan J., and Kang S. (2017) Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene 36, 3797–3806 10.1038/onc.2017.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ji Y., Yang C., Tang Z., Yang Y., Tian Y., Yao H., Zhu X., Zhang Z., Ji J., and Zheng X. (2017) Adenylate kinase hCINAP determines self-renewal of colorectal cancer stem cells by facilitating LDHA phosphorylation. Nat. Commun. 8, 15308 10.1038/ncomms15308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maekawa M., Taniguchi T., Ishikawa J., Sugimura H., Sugano K., and Kanno T. (2003) Promoter hypermethylation in cancer silences LDHB, eliminating lactate dehydrogenase isoenzymes 1–4. Clin. Chem. 49, 1518–1520 10.1373/49.9.1518 [DOI] [PubMed] [Google Scholar]
- 34. Kato Y., Maeda T., Suzuki A., and Baba Y. (2018) Cancer metabolism: new insights into classic characteristics. Jpn. Dent. Sci. Rev. 54, 8–21 10.1016/j.jdsr.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walenta S., Schroeder T., and Mueller-Klieser W. (2004) Lactate in solid malignant tumors: potential basis of a metabolic classification in clinical oncology. Curr. Med. Chem. 11, 2195–2204 10.2174/0929867043364711 [DOI] [PubMed] [Google Scholar]
- 36. Parks S. K., Chiche J., and Pouysségur J. (2013) Disrupting proton dynamics and energy metabolism for cancer therapy. Nat. Rev. Cancer 13, 611–623 10.1038/nrc3579 [DOI] [PubMed] [Google Scholar]
- 37. Lutz N. W., Le Fur Y., Chiche J., Pouysségur J., and Cozzone P. J. (2013) Quantitative in vivo characterization of intracellular and extracellular pH profiles in heterogeneous tumors: a novel method enabling multiparametric pH analysis. Cancer Res. 73, 4616–4628 10.1158/1538-7445.AM2013-4616,10.1158/0008-5472.CAN-13-0767 [DOI] [PubMed] [Google Scholar]
- 38. Halestrap A. P. (2012) The monocarboxylate transporter family: structure and functional characterization. IUBMB Life 64, 1–9 10.1002/iub.573 [DOI] [PubMed] [Google Scholar]
- 39. Halestrap A. P., and Wilson M. C. (2012) The monocarboxylate transporter family: role and regulation. IUBMB Life 64, 109–119 10.1002/iub.572 [DOI] [PubMed] [Google Scholar]
- 40. Miranda-Gonçalves V., Honavar M., Pinheiro C., Martinho O., Pires M. M., Pinheiro C., Cordeiro M., Bebiano G., Costa P., Palmeirim I., Reis R. M., and Baltazar F. (2013) Monocarboxylate transporters (MCTs) in gliomas: expression and exploitation as therapeutic targets. Neuro. Oncol. 15, 172–188 10.1093/neuonc/nos298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Enerson B. E., and Drewes L. R. (2003) Molecular features, regulation, and function of monocarboxylate transporters: implications for drug delivery. J. Pharm. Sci. 92, 1531–1544 10.1002/jps.10389 [DOI] [PubMed] [Google Scholar]
- 42. Semenza G. L. (2013) HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 123, 3664–3671 10.1172/JCI67230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miao P., Sheng S., Sun X., Liu J., and Huang G. (2013) Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life 65, 904–910 10.1002/iub.1216 [DOI] [PubMed] [Google Scholar]
- 44. Valvona C. J., Fillmore H. L., Nunn P. B., and Pilkington G. J. (2016) The regulation and function of lactate dehydrogenase A: therapeutic potential in brain tumor. Brain Pathol. 26, 3–17 10.1111/bpa.12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cui J., Quan M., Jiang W., Hu H., Jiao F., Li N., Jin Z., Wang L., Wang Y., and Wang L. (2015) Suppressed expression of LDHB promotes pancreatic cancer progression via inducing glycolytic phenotype. Med. Oncol. 32, 143 10.1007/s12032-015-0589-8 [DOI] [PubMed] [Google Scholar]
- 46. Lee Y. K., Lim J. J., Jeoun U. W., Min S., Lee E. B., Kwon S. M., Lee C., and Yoon G. (2017) Lactate-mediated mitoribosomal defects impair mitochondrial oxidative phosphorylation and promote hepatoma cell invasiveness. J. Biol. Chem. 292, 20208–20217 10.1074/jbc.M117.809012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomson J. M., Gaucher E. A., Burgan M. F., De Kee D. W., Li T., Aris J. P., and Benner S. A. (2005) Resurrecting ancestral alcohol dehydrogenases from yeast. Nat. Genet. 37, 630–635 10.1038/ng1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pérez-Escuredo J., Van Hée V. F., Sboarina M., Falces J., Payen V. L., Pellerin L., and Sonveaux P. (2016) Monocarboxylate transporters in the brain and in cancer. Biochim. Biophys. Acta 1863, 2481–2497 10.1016/j.bbamcr.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Doherty J. R., Yang C., Scott K. E., Cameron M. D., Fallahi M., Li W., Hall M. A., Amelio A. L., Mishra J. K., Li F., Tortosa M., Genau H. M., Rounbehler R. J., Lu Y., Dang C. V., et al. (2014) Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis. Cancer Res. 74, 908–920 10.1158/0008-5472.CAN-13-2034,10.1158/1538-7445.AM2014-908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Halestrap A. P., and Meredith D. (2004) The SLC16 gene family: from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 447, 619–628 10.1007/s00424-003-1067-2 [DOI] [PubMed] [Google Scholar]
- 51. San-Millán I., and Brooks G. A. (2017) Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg effect. Carcinogenesis 38, 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee D. C., Sohn H. A., Park Z. Y., Oh S., Kang Y. K., Lee K. M., Kang M., Jang Y. J., Yang S. J., Hong Y. K., Noh H., Kim J. A., Kim D. J., Bae K. H., Kim D. M., et al. (2015) A lactate-induced response to hypoxia. Cell 161, 595–609 10.1016/j.cell.2015.03.011 [DOI] [PubMed] [Google Scholar]
- 53. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guo M., Song L. P., Jiang Y., Liu W., Yu Y., and Chen G. Q. (2006) Hypoxia-mimetic agents desferrioxamine and cobalt chloride induce leukemic cell apoptosis through different hypoxia-inducible factor-1α independent mechanisms. Apoptosis 11, 67–77 10.1007/s10495-005-3085-3 [DOI] [PubMed] [Google Scholar]
- 55. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., and Mesirov J. P. (2006) GenePattern 2.0. Nat. Genet. 38, 500–501 10.1038/ng0506-500 [DOI] [PubMed] [Google Scholar]