Abstract
Idiopathic pulmonary fibrosis (IPF) is a disabling and lethal chronic progressive pulmonary disease. Epigallocatechin gallate (EGCG) is a polyphenol, which is the major biological component of green tea. The anti-oxidative, anti-inflammatory, and anti-fibrotic effects of EGCG have been shown in some studies, whereas its effects in altering gene expression in pulmonary fibroblasts have not been systematically investigated. This study aimed to explore the effect of EGCG on gene expression profiles in fibroblasts of IPF. The pulmonary fibroblasts from an IPF patient were treated with either EGCG or water, and the expression profiles of mRNAs and microRNAs were determined by next-generation sequencing (NGS) and analyzed with the bioinformatics approach. A total of 61 differentially expressed genes and 56 differentially expressed microRNAs were found in EGCG-treated IPF fibroblasts. Gene ontology analyses revealed that the differentially expressed genes were mainly involved in the biosynthetic and metabolic processes of cholesterol. In addition, five potential altered microRNA–mRNA interactions were found, including hsa-miR-939-5p–PLXNA4, hsa-miR-3918–CTIF, hsa-miR-4768-5p–PDE5A, hsa-miR-1273g-3p–VPS53, and hsa-miR-1972–PCSK9. In summary, differentially expressed genes and microRNAs in response to EGCG treatment in IPF fibroblasts were identified in the current study. Our findings provide a scientific basis to evaluate the potential benefits of EGCG in IPF treatment, and warrant future studies to understand the role of molecular pathways underlying cholesterol homeostasis in the pathogenesis of IPF.
Keywords: bioinformatics, fibroblast, next-generation sequencing, epigallocatechin gallate, EGCG, PCSK9
1. Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic progressive pulmonary disease characterized by progressive fibrosing interstitial pneumonitis [1,2,3,4,5]. Patients usually present with non-specific symptoms, such as exertional dyspnea and dry cough, and high-pitched fine inspiratory crackles (the so-called Velcro-like crackles) are usually heard in bilateral basal lung fields on auscultation [2]. A high-resolution computed tomography is a key diagnostic tool, which may reveal the pattern of usual interstitial pneumonia (UIP), characterized by “honey-combing” (subpleural multi-layer cystic lesions), traction bronchiectasis, and peripheral alveolar septal thickening, initially involving the basal and peripheral lungs with gradual progression to involve the whole lungs [3]. With increasing awareness in recent decades, incidence has risen over time, and estimated to be 2–30 cases per 100,000 person-years [2,3,6]. IPF has an increasing global burden, affecting about 3 million people worldwide [3]. Causing progressively disabling dyspnea, IPF has a devastating effect on patients’ quality of life and is a quite lethal disease [4]. The median survival of untreated IPF patients is around 3–5 years, which is worse than the majority of cancers in subjects with similar demographic characters [3,4,7].
Although the precise pathogenic mechanisms of IPF remain largely unclear, it is believed that fibroblasts play a key role in the development and progression of IPF [1,5,8]. Exaggerated proliferation, migration, and activation of fibroblasts, as well as their resistance to apoptosis, differentiation to myofibroblasts, and secretion of extracellular matrix components, have been proposed as a major pathogenic model of IPF [1,3,9,10,11]. Inflammation and oxidative stress are also commonly considered as major factors promoting pulmonary fibrosis [12,13,14].
The treatment modalities for IPF are currently limited. Only two medications, including pirfenidone, an anti-fibrotic agent, and nintedanib, a multi-target tyrosine kinase inhibitor, have been shown to be effective in reducing disease progression and improving quality of life [5,15,16,17,18,19,20,21]. Although these drugs offer hope for IPF patients, not all patients respond to these pharmacotherapies, and no biomarkers are available to predict the treatment response to any drug; therefore, efforts are continuously made to explore potential novel treatment modalities for IPF.
Epigallocatechin gallate (EGCG) is the ester from epigallocatechin and gallic acid [22]. This catechin is the major polyphenol responsible for the biological effects in tea [22,23]. In contrast to black tea, in which the flavan-3-ols are converted to theaflavins and thearubigins during fermentation processing, green tea retains the highest level of EGCG (100 g of dried leaves contain about 7380 mg of EGCG) [22,23]. EGCG is also found in trace amounts in some food sources, such as onions, hazelnuts, plums, and so on [24]. EGCG exhibits a wide range of therapeutic properties, such as anti-oxidative [25], anti-inflammatory [26], and anti-fibrotic effects [27]. It may modulate cell signaling pathways, such as mitogen-activated protein kinase (MAPK), NF-κB, and AMP-activated protein kinase (AMPK) pathways, and may also modulate epigenetic changes, such as DNA methylation and histone acetylation [22,23]. As a well-known mitochondrion-targeting medicinal agent, EGCG may also regulate mitochondrial metabolism, such as mitochondrial biogenesis and mitochondrial bioenergetics, as well as regulate cell cycle and apoptosis via mitochondria-mediated pathways [28].
Some in vitro and in vivo studies have shown the effects of EGCG on fibroblasts, such as attenuating cell proliferation, enhancing antioxidant defense systems, and inhibiting inflammation [27,29,30,31,32]. However, the effects of EGCG in modulating gene expression in pulmonary fibroblasts have not been systematically investigated. We therefore conducted this study to explore the effect of EGCG on gene expression profiles in fibroblasts of IPF using next-generation sequencing (NGS) and bioinformatic analyses.
2. Results
2.1. Gene Expression Profiling and Microrna Changes in IPF Fibroblasts Treated with EGCG
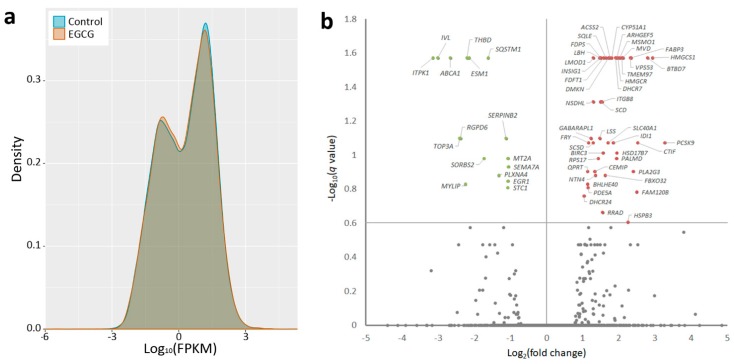
The primary pulmonary fibroblasts from a patient with IPF were treated with EGCG or water, and RNAs were extracted from the cells and sent for NGS followed by bioinformatic analyses. Figure 1a shows the volcano plot of differentially expressed genes in EGCG-treated versus control fibroblasts. Genes with q-value <0.25 and >2-fold changes were selected for further analyses, including 16 significantly downregulated and 45 significantly upregulated genes in EGCG-treated versus control fibroblasts (Figure 1, Table A1). The small RNA-sequencing data generated with NGS were analyzed to identify potentially significant changes in microRNA profiles in EGCG-treated versus control fibroblasts. As shown in Table A2, we identified 56 microRNAs with >2-fold changes (22 upregulated and 34 downregulated).
Figure 1.
Differential gene expression patterns between idiopathic pulmonary fibrosis (IPF) fibroblasts treated with epigallocatechin gallate (EGCG) or water (control). (a) The frequency distribution of fragments per kilobase of transcript per million mapped reads (FPKM) between fibroblasts treated with EGCG or water (control) was compared and are presented in the density plot. (b) The volcano plot of –log10 (q-value) versus log2 (fold change) showed differentially downregulated (left upper quadrant) and upregulated (right upper quadrant) genes expressed in EGCG-treated IPF fibroblasts versus water-treated IPF fibroblasts. The genes with q-values >0.25 and >2-fold changes are plotted in green (downregulated) or red (upregulated).
2.2. Discovering the Altered microRNA–mRNA Interactions in IPF Fibroblasts Treated with EGCG
In order to discover altered microRNA–mRNA interactions in IPF fibroblasts treated with EGCG, we searched the putative targets of the microRNAs with >2-fold changes from NGS results using the miRmap database and selected those with miRmap scores >97.0. We matched the genes showing >2-fold changes with these putative targets. As shown in the intersection Venn diagram (Figure 2), five potential altered microRNA–mRNA interactions were found, including hsa-miR-939-5p–PLXNA4, hsa-miR-3918–CTIF, hsa-miR-4768-5p–PDE5A, hsa-miR-1273g-3p–VPS53, and hsa-miR-1972–PCSK9 (Table 1). For further validation, we also searched these potential microRNA–mRNA interactions in various microRNA target predicting databases via miRWalk 2.0 [33], which included miRWalk, MicroT4, miRanda, miRDB, miRmap, RNA22, RNAhybrid, and TargetScan. Based on the criteria “microRNA target predicted in at least 6 (out of 8) databases”, all five potential altered microRNA–mRNA interactions were validated (Table 1).
Figure 2.
Differentially expressed genes and microRNAs with potential microRNA–target gene interactions identified in idiopathic pulmonary fibrosis (IPF) fibroblasts treated with epigallocatechin gallate (EGCG). A total of (a) 61 differentially expressed genes and (c) 56 differentially expressed microRNAs were identified in the EGCG-treated IPF fibroblasts with next-generation sequencing methods, and the heatmaps according to z-scores are illustrated. (b) Using the miRmap database for microRNA target prediction (selection criteria of miRmap score ≥97.0), 942 putative targets of the 22 upregulated microRNAs and 1334 putative targets of the 34 downregulated microRNAs were identified. Matching to the 16 downregulated genes and 45 upregulated genes identified in the EGCG-treated IPF fibroblasts, the intersection Venn diagram identified five potential microRNA–mRNA interactions (as shown in Table 1).
Table 1.
Potential altered miRNA–mRNA interactions in idiopathic pulmonary fibrosis (IPF) fibroblasts treated with epigallocatechin gallate (EGCG).
| Gene | miRNA | microRNA-Target Gene Prediction in Various Databases * | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Official Symbol | Gene Name | Log2 (ratio) | miRNA Name | Fold Change | mirmap Score | miRWalk | Microt4 | miRanda | miRDB | RNA22 | RNAhybrid | TargetScan |
| PLXNA4 | plexin A4 | −1.32 | hsa-miR-939-5p | 4.85 | 99.97 | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CTIF | cap binding complex dependent translation initiation factor | 2.52 | hsa-miR-3918 | −4.19 | 99.97 | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| PDE5A | phosphodiesterase 5A | 1.14 | hsa-miR-4768-5p | −2.75 | 99.34 | Yes | Yes | Yes | No | No | Yes | Yes |
| VPS53 | VPS53, GARP complex subunit | 2.32 | hsa-miR-1273g-3p | −2.03 | 99.76 | Yes | Yes | Yes | No | No | Yes | Yes |
| PCSK9 | proprotein convertase subtilisin/kexin type 9 | 3.27 | hsa-miR-1972 | −3.11 | 99.58 | Yes | Yes | Yes | No | Yes | Yes | Yes |
2.3. Gene Ontology Annotations of the Differentially Expressed Genes in IPF Fibroblasts Treated with EGCG
Gene ontology analysis of the 61 differentially expressed genes was performed using DAVID, including cellular components, biological processes, and molecular functions. The cellular components significantly associated with these genes included endoplasmic reticulum (15 genes) and endoplasmic reticulum membrane (14 genes) (Table 2). The biological processes significantly associated with these genes included cholesterol biosynthetic process (15 genes), isoprenoid biosynthetic process (6 genes), oxidation-reduction process (11 genes), cholesterol biosynthetic process via lathosterol (3 genes), cholesterol biosynthetic process via desmosterol (3 genes), cholesterol homeostasis (5 genes), and steroid biosynthetic process (4 genes) (Table 2). The analyses failed to identify any molecular functions significantly associated with these genes. In the genes involved in these significantly associated cellular components and biological processes, PCSK9, which is significantly involved in cholesterol homeostasis, was the only gene targeted by a differentially expressed microRNA, hsa-miR-1972 (Table 2).
Table 2.
Gene ontology analysis of the 61 differentially expressed genes in idiopathic pulmonary fibrosis (IPF) fibroblasts treated with epigallocatechin gallate (EGCG) using the Database for Annotation, Visualization and Integrated Discovery (DAVID).
| Category and Term | Gene Count | Genes | Fold Enrichment | p-Value | Adjusted p-Value |
|---|---|---|---|---|---|
| Cellular components | |||||
| Endoplasmic reticulum | 15 | GABARAPL1, MSMO1, HMGCR, CYP51A1, SCD, FDFT1, SQSTM1, SQLE, DHCR7, INSIG1, CEMIP, PCSK9, HSD17B7, NSDHL, DHCR24 | 5.41 | 3.54 × 10−7 | 3.75 × 10−5 |
| Endoplasmic reticulum membrane | 14 | SC5D, MSMO1, CYP51A1, SQLE, HMGCR, SCD, DHCR7, INSIG1, LSS, ABCA1, HSD17B7, NSDHL, FDFT1, DHCR24 | 4.85 | 3.55 × 10−6 | 1.88 × 10−4 |
| Biological processes | |||||
| Cholesterol biosynthetic process | 15 | MSMO1, MVD, HMGCR, CYP51A1, HMGCS1, FDPS, LSS, FDFT1, SQLE, DHCR7, INSIG1, IDI1, HSD17B7, NSDHL, DHCR24 | 108.66 | 9.71 × 10−26 | 4.19 × 10−23 |
| Isoprenoid biosynthetic process | 6 | MVD, HMGCR, FDPS, HMGCS1, IDI1, FDFT1 | 117.98 | 9.59 × 10−10 | 2.07 × 10−7 |
| Oxidation-reduction process | 11 | SC5D, MSMO1, SQLE, HMGCR, CYP51A1, SCD, DHCR7, HSD17B7, NSDHL, FDFT1, DHCR24 | 5.11 | 4.23 × 10−5 | 0.0061 |
| Cholesterol biosynthetic process via lathosterol | 3 | SC5D, DHCR7, DHCR24 | 206.46 | 7.50 × 10−5 | 0.0080 |
| Cholesterol biosynthetic process via desmosterol | 3 | SC5D, DHCR7, DHCR24 | 206.46 | 7.50 × 10−5 | 0.0080 |
| Cholesterol homeostasis | 5 | TMEM97, FABP3, PCSK9, MYLIP, ABCA1 | 21.51 | 7.97 × 10−5 | 0.0068 |
| Steroid biosynthetic process | 4 | CYP51A1, LSS, NSDHL, FDFT1 | 34.41 | 0.0002 | 0.0143 |
* p-values adjusted with false discovery rate using the method by Benjamini et al. The gene predicted as the target of the significantly dysregulated microRNA (Table 1) is highlighted with bold font and underlined.
The KEGG pathways of the differentially expressed genes in EGCG-treated IPF fibroblasts included steroid biosynthesis (fold enrichment of 90.51), biosynthesis of antibiotics (fold enrichment of 11.95), terpenoid backbone biosynthesis (fold enrichment of 41.14), and metabolic pathways (fold enrichment of 2.82) (Table 3).
Table 3.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the significantly differentially expressed genes in idiopathic pulmonary fibrosis (IPF) fibroblasts treated with epigallocatechin gallate (EGCG).
| Description | Count | p-value | Adjusted p-value * |
Genes | Fold Enrichment |
|---|---|---|---|---|---|
| Steroid biosynthesis | 10 | 2.11 × 10−16 | 1.42 × 10−14 | SC5D, MSMO1, SQLE, CYP51A1, DHCR7, LSS, HSD17B7, NSDHL, FDFT1, DHCR24 | 90.51 |
| Biosynthesis of antibiotics | 14 | 2.91 × 10−11 | 9.31 × 10−10 | SC5D, MSMO1, MVD, CYP51A1, SQLE, HMGCR, FDPS, HMGCS1, LSS, IDI1, ACSS2, HSD17B7, NSDHL, FDFT1 | 11.95 |
| Terpenoid backbone biosynthesis | 5 | 4.84 × 10−6 | 1.03 × 10−4 | MVD, HMGCR, FDPS, HMGCS1, IDI1 | 41.14 |
| Metabolic pathways | 19 | 1.53 × 10−5 | 2.45 × 10−4 | SC5D, MSMO1, MVD, CYP51A1, HMGCR, HMGCS1, FDPS, LSS, ACSS2, FDFT1, SQLE, DHCR7, QPRT, PLA2G3, ITPK1, IDI1, HSD17B7, NSDHL, DHCR24 | 2.82 |
* p-values adjusted with false discovery rate using the method by Benjamini et al.
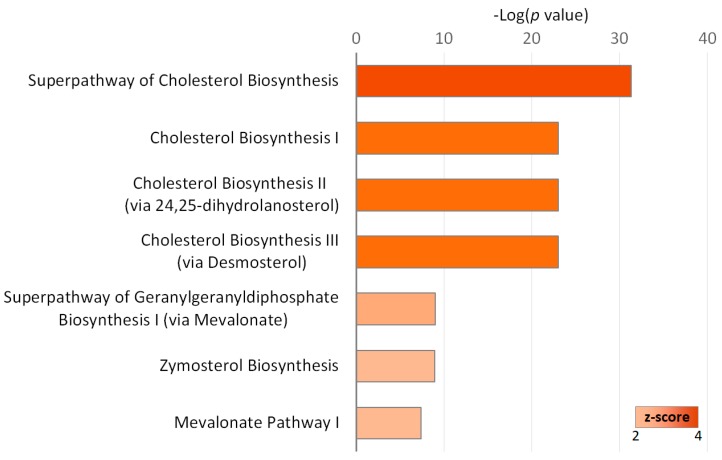
Using IPA, the canonical pathways associated with the 61 differentially expressed genes were investigated. As shown in Figure 3, we found that the associated canonical pathways included superpathway of cholesterol biosynthesis (15 genes), cholesterol biosynthesis I (10 genes), cholesterol biosynthesis II (via 24,25-dihydrolanosterol) (10 genes), cholesterol biosynthesis III (via desmosterol) (10 genes), superpathway of geranylgeranyl-diphosphate biosynthesis I (via mevalonate) (5 genes), zymosterol biosynthesis (4 genes), and mevalonate pathway I (4 genes).
Figure 3.
Canonical pathways significantly associated with the differentially expressed genes in idiopathic pulmonary fibrosis (IPF) fibroblasts treated with epigallocatechin gallate (EGCG) versus water. The z-score represents the magnitude of significant activation.
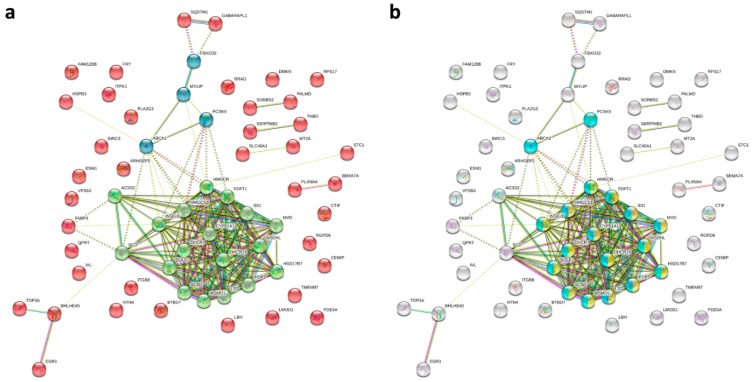
The protein–protein interaction (PPI) network analysis using the STRING database identified a total of 61 nodes and 172 edges, with PPI enrichment p-value <1.0 × 10−16. Using k-means clustering, the network could be further clustered into three clusters (Figure 4a). Within these three clusters, we found a core cluster, in which almost all genes were associated with cholesterol biosynthetic and metabolic processes (Figure 4b).
Figure 4.
Protein–protein interaction (PPI) network analysis of the differentially expressed genes in idiopathic pulmonary fibrosis (IPF) fibroblasts treated with epigallocatechin gallate (EGCG) versus water using the Search Tool for the Retrieval of Interacting Genes (STRING) database. The 61 differentially expressed genes were input into the STRING database for PPI network analysis, and achieved a PPI network of 61 nodes and 172 edges, with PPI enrichment p-value <1.0 × 10−16. (a) Using k-means clustering, the network could be further clustered into three clusters (represented in green, blue, and red colors). (b) The genes associated with cholesterol biosynthetic process and cholesterol metabolic process are highlighted with yellow and light blue colors, respectively.
3. Discussion
The underlying mechanisms of IPF pathogenesis are quite complex, which might include oxidative stress, inflammation, enhanced proliferation and migration of fibroblasts, and so on [2,3,5,7,21]. As EGCG has anti-oxidative, anti-inflammatory, and anti-fibrotic effects, the current study tried to investigate the effects of EGCG on gene expression profiles in IPF fibroblasts. We found 61 differentially expressed genes in EGCG-treated IPF fibroblasts as compared with the control cells. Gene ontology analyses and network analyses revealed that these genes were mainly involved in the biosynthesis and metabolism of cholesterol. In addition, we also identified 56 differentially expressed microRNAs, and further analyses found that five potential altered microRNA–mRNA interactions, including hsa-miR-939-5p–PLXNA4, hsa-miR-3918–CTIF, hsa-miR-4768-5p–PDE5A, hsa-miR-1273g-3p–VPS53, and hsa-miR-1972–PCSK9, might be important changes in response to EGCG treatment in IPF fibroblasts. Among the five genes with potential microRNA–mRNA interactions, PCSK9, which might be upregulated in association with downregulated has-miR-1972, was the only one involved in cholesterol metabolism.
The effects and mechanisms of EGCG have been studied for various diseases. EGCG is considered beneficial for cardiovascular health and may prevent metabolic syndrome due to its effects in preventing or improving atherosclerosis, cardiac hypertrophy, myocardial infarction, diabetes, inflammation, and oxidative stress [34,35]. In type 2 diabetes and obesity, EGCG may modulate muscle homeostasis by increasing the expression of anti-oxidative enzymes, reversing the increased production of reactive oxygen species in skeletal muscle, regulating mitochondria-involved autophagy, stimulating glucose uptake, and increasing lipid oxidation [22]. As EGCG may also block the activity of peroxisome proliferator-activated receptors gamma (PPARγ) via binding to its active site, this polyphenol has been considered a potential anti-obesity compound [36]. Potential neuroprotective effects of EGCG have also been reported [37]. EGCG exhibits multiple immune-modulating effects, regulating intestinal mucosal immune responses, allergic diseases, as well as anticancer immunity [38]. In addition to its effects in inhibiting NF-κB, epithelial-mesenchymal transition, and cell invasion, the anticancer effects of EGCG might also involve the regulation of microRNAs and epigenetic mechanisms responsible for carcinogenesis and cancer progression [39,40].
A few previous studies have investigated the effect of EGCG on pulmonary fibroblasts. EGCG has been shown to inhibit the production of TNF-α, which might play an important role in the pathogenesis of IPF [41]. Sriram et al. have done a series of studies on the effects of EGCG in pulmonary fibrosis using the rat model of pulmonary fibrosis induced by intra-tracheal instillation of bleomycin [27,29,30,31]. EGCG augmented antioxidant activities and alleviated the bleomycin-induced oxidative stress, inflammation (increased levels of NF-κB, TNF-α, and IL-1β), and alveolar damage [30,31]. The increased glycoconjugates, increased activities of matrix degrading lysosomal enzymes, and ultrastructural changes induced by bleomycin were also attenuated by EGCG, suggesting the potential of EGCG as an anti-fibrotic agent [27]. EGCG also reversed the increased expression of gelatinases, including matrix metalloproteinase (MMP)-2 and MMP-9, TGF-β1, SMADs, and α-smooth muscle actin (α-SMA) induced by bleomycin treatment [29]. The in vitro studies using a fibroblast cell line also revealed that EGCG was capable of reversing the TGF-β1-induced proliferation and activation of fibroblasts [29]. Using a rat model of irradiation-induced pulmonary fibrosis, You et al. revealed that EGCG inhibited irradiation-induced alveolitis and pulmonary fibrosis through the effects such as inhibiting fibroblast proliferation, reducing collagen deposition, and regulating inflammatory cytokines [32].
The roles of cholesterol and lipoproteins in pulmonary diseases have been recognized in a few studies, although the detailed mechanisms remain unclear. Dyslipidemia affects innate and adaptive immunities in the lung [42]. Statins, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) inhibitors originally designed as a cholesterol-lowering drug, also have HMGCR-independent properties, such as anti-inflammatory and anti-fibrotic effects. Pitavastatin and simvastatin inhibited TGF-β1-induced production of growth factors and fibrogenic mediators from lung fibroblasts [43,44]. In a mouse study, pravastatin attenuated bleomycin-induced pulmonary fibrosis by reducing the expression of inflammatory and growth factors, such as TGF-β1 and connective tissue growth factor, and the oxidative stress [45]. A large cross-sectional clinical study showed that lower high-density lipoprotein (HDL)-cholesterol was associated with more subclinical interstitial lung disease, shown by more high attenuation areas measured with computed tomography, and more extracellular matrix remodeling, shown by increased serum MMP-7 and surfactant protein-A [46]. Through gene ontology and functional analyses, we found that the EGCG-induced differentially expressed genes mainly involve the biosynthesis and metabolism of cholesterol. Further studies are needed to understand the roles of cholesterol-associated pathways in pulmonary fibrosis.
In this study, five differentially expressed genes were found as the potential targets of corresponding differentially expressed microRNAs, including downregulated PLXNA4 and upregulated CTIF, PDE5A, VPS53, and PCSK9. As a study using the rat model of bleomycin-induced pulmonary fibrosis showed increased expression of PLXNA4 [47], the downregulation of PLXNA4 induced by EGCG might have beneficial effect in treating pulmonary fibrosis. In contrast to our findings that EGCG upregulated PDE5A expression in IPF fibroblasts, PDE5A inhibition by sildenafil improved bleomycin-induced pulmonary fibrosis by reducing oxidative stress [48]. PCSK9 encodes proprotein convertase subtilisin/kexin type 9, which is a regulator of the homeostasis of plasma low-density lipoprotein (LDL)-cholesterol, and is associated with the metabolism of lipid and glucose [49]. Expression of PCSK9 might reverse the abnormal cholesterol accumulation and the development of fibrosis in the liver caused by E2F1 deficiency [50]. Although the roles of these genes in regulating the cell physiology of pulmonary fibroblasts remain largely unknown, these EGCG-induced gene expression alterations might provide potential targets to reverse pulmonary fibrosis and deserve further studies.
Some anti-fibrotic and pro-fibrotic microRNAs have been reported, and some of them might contribute to the pathogenesis of IPF [1,51,52]. The expression of miR-155 in human lung fibroblasts was upregulated by TNF-α and IL-1β and downregulated by TGF-β1; miR-155, which might target keratinocyte growth factor, promoted migration of fibroblasts and enhanced pulmonary fibrosis [53]. In studies using the mice model of bleomycin-induced pulmonary fibrosis, upregulation of miR-155 and downregulation of miR-29 were observed, which correlated with the degree of lung fibrosis [53,54]. The increased expression of miR-155 and decreased expression of miR-29 have been observed in the lungs of IPF patients [51]. In addition, greater expression and localization of miR-34a in pulmonary fibroblasts of IPF have been reported, which might function as an inhibiting mechanism of pulmonary fibrosis via inducing senescence and apoptosis of the fibroblasts [55]. Our findings that EGCG significantly upregulated miR-29b-2-5p and miR-34a-3p and downregulated miR-155-3p in IPF fibroblasts suggested a potential role of EGCG in the treatment of IPF through regulation of these microRNAs.
The dose of EGCG used in this study might be a concern. While most published in vitro studies used 10–100 μM of EGCG [56,57], we chose 25 μM of EGCG. As shown in a few previous studies, this dose of EGCG did not cause significant proliferation inhibition in human fibroblast cell line [29,58] and human colorectal cancer cell lines [59]. In line with these studies, our study showed that 25 μM of EGCG did not significantly alter the expression of genes related to cell proliferation or cell death. In addition, the bioavailability of EGCG is quite poor [56,60]. As shown in a pharmacokinetic study, the peak plasma EGCG level was only about 0.17 μM after drinking two cups of tea [56]. Parenteral routes, such as intravenous injection, might be needed to reach an effective systemic dose. However, a high systemic dose might cause hepatotoxicity [61]. In 2018, the European Food Safety Authority stated that taking ≥800 mg of EGCG daily might increase serum transaminase levels [62]. Further study is needed to determine the optimal systemic dose of EGCG. On the other hand, inhalation of an EGCG aerosol might possibly provide an alternative route of administration to increase EGCG concentration in the lungs while avoiding the potential systemic toxicity [63].
4. Materials and Methods
4.1. Cell Culture and Next-Generation Sequencing (NGS)
Human pulmonary fibroblasts from a patient of IPF (83-year-old Caucasian man, Catalog No. CC-7231), obtained from Lonza (Walkersville, MD, USA) were incubated at 37 °C in a 5% CO2-containing incubator in FGM™-2 Fibroblast Growth Medium-2 (Walkersville, MD, USA, Catalog No. CC-3132) containing 0.5 mL hFGF-B, 0.5 mL insulin, 10 mL FBS and 0.5 mL GA-1000. The medium was changed once every 2 or 3 days and the cells were channeled after distinct cell density. The cells were plated in 6-cm culture plates (1 × 105 cells/well) and, after 24 h of incubation, treated with vehicle alone (ddH2O) or 25 μM of EGCG for 24 h. The dose of EGCG was chosen in reference to previous studies [29,56,57,58,59].
The mRNA and small RNA expression profiles were assessed using NGS as our previous studies [1,64,65,66,67,68]. In brief, total RNAs were extracted using TRIzol® Reagent (Thermo Fisher Scientific, Waltham, MA, USA, Catalog No. 15596018) according to the instruction manual. The purified RNAs were quantified at OD260nm using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and qualitatively analyzed using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) with RNA 6000 LabChip kit (Agilent Technologies, Santa Clara, CA, USA). Library preparation and deep sequencing were carried out at Welgene Biotechnology Company (Taipei, Taiwan) as the official protocol of Illumina (San Diego, CA, USA).
For transcriptome sequencing, the library was constructed with Agilent’s SureSelect Strand Specific RNA Library Preparation Kit followed by AMPure XP Beads size selection. The sequence was directly determined using Illumina’s sequencing-by-synthesis (SBS) technology, and the sequencing data were generated by Welgene’s pipeline based on Illumina’s base-calling program bcl2fastq v2.2.0. For read alignment, HISAT2 [69], a fast and sensitive alignment program for mapping NGS reads to genomes, was used. The new indexing scheme of HISAT2 is based on the hierarchical graph Ferragina–Manzini index (GFM index) [70]. HISAT2 uses the global GFM index and a large set of small GFM indexes providing better performance for splicing junction alignment, which collectively covers the whole genome for rapid and accurate alignment, making HISAT2 an effective tool for transcriptome alignment. Differential expression analysis based on Cuffdiff (Cufflinks 2.2.1) [71] with genome bias detection/correction and Welgene in-house programs was performed. Differential expressed genes of each experiment design were followed by enrichment test for functional assay by clusterProfiler 3.6 [72]. The genes with low expression levels (<0.3 fragment per kilobase of transcript per million mapped reads (FPKM)) in both EGCG-treated and control fibroblasts and the genes with undetected level in either EGCG-treated or control fibroblasts were excluded. The p-values were calculated by Cuffdiff with non-grouped sample using “blind mode”, in which all samples were treated as replicates of a single global “condition” and used to build one model for statistical tests [71,73]. The q-values were p-value adjusted with false discovery rate using the method by Benjamini and Hochberg [74]. Genes with q-value < 0.25 (i.e., −log10 (q-value) > 0.602) and > 2-fold changes were considered significantly differentially expressed.
For small RNA sequencing, samples were prepared using Illumina sample preparation kit according to the TruSeq Small RNA Sample Preparation Guide. After the 3′ and 5′ adaptors were ligated to the RNA, reverse transcription followed by PCR amplification was performed. The enriched cDNA constructs were size-fractionated and purified on a 6% polyacrylamide gel electrophoresis and the bands containing the 18–40 nucleotide RNA fragments (140–155 nucleotides in length with both adapters) were extracted, which were then sequenced on an Illumina instrument (75 bp single-end reads). Sequencing data was processed with the Illumina software. After trimming and removing low-quality data with Trimmomatics v0.36 [75], the qualified data were analyzed with the miRDeep2 [76] to clip the 3′ adapter sequence and discard reads shorter than 18 nucleotides. The reads were then aligned to the human genome from the University of California, Santa Cruz (UCSC). Only reads that mapped perfectly to the genome ≤5 times were used for microRNA detection, because microRNAs usually map to few genomic locations. MiRDeep2 [76] estimates expression levels of known microRNAs, and also identifies novel microRNAs. The microRNAs with low levels (<1 normalized read per million (rpm)) in both EGCG-treated and control fibroblasts were excluded.
4.2. Analyses Using microRNA Target Predicting Databases
We used miRmap, an open-source software library providing comprehensive prediction of microRNA targets (http://mirmap.ezlab.org/) (accessed on 19 September 2018) [77], to predict the potential targets of significantly differentially expressed (>2-fold change) microRNAs. A miRmap score represents the repression strength of a microRNA on a target mRNA. Only the microRNA–mRNA pairs with miRmap score >97.0 were considered in the current study.
For further confirmation, we searched miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) (accessed on 03 February 2019) [33], a comprehensive atlas of predicted and validated microRNA–target interactions, in which 12 computational target prediction databases were available. The information from eight databases, including miRWalk, MicroT4, miRanda, miRDB, miRmap, RNA22, RNAhybrid, and TargetScan, were extracted, because miRBridge, miRNAMap, PICTAR2, and PITA contained relatively little information. Putative microRNA targets suggested by at least six (out of eight) predicting databases were considered meaningful.
4.3. Database for Annotation, Visualization and Integrated Discovery (DAVID) Database Analysis
The DAVID (version 6.8) (https://david.ncifcrf.gov/) (accessed on 06 February 2019) [78] is a powerful tool for gene functional classification, which integrates many functional annotation databases, such as Gene Ontology (GO) biological process, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. By calculating the similarity of global annotation profiles with agglomeration algorithm method, a list of interesting genes can be classified into clusters based on their related biological functions, signaling pathways, or diseases. The differentially expressed genes were analyzed using the methods as in our previous studies [1,64,65,66,67,68]. The p-values (adjusted with false discovery rate using the method by Benjamini et al.) <0.05 were taken as the criteria of significance.
4.4. Ingenuity Pathway Analysis (IPA)
Ingenuity® Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA, USA) integrates many research results and performs multiple analyses providing a comprehensive interpretation of big experimental data. We used IPA (version 2.3) to identify the canonical pathways associated with the candidate genes.
4.5. Search Tool for the Retrieval of Interacting Genes (STRING)
The STRING database (version 11.0) (https://string-db.org/) (accessed on 7 February 2019) covers 5090 organisms, 24.6 million proteins and >2000 million interactions that provides analysis and integration of direct and indirect protein–protein interactions (PPI), and focuses on functional association [79]. The differentially expressed genes identified were uploaded, and interactions with at least medium confidence (interaction score >0.4) were selected. Network was clustered using k-means clustering to a specified number of clusters.
5. Conclusions
In summary, our study demonstrated an innovative model to discover the potential effects of a nature compound on gene expression alterations and microRNA changes using an NGS and bioinformatic approach. We identified differentially expressed genes and microRNAs in response to EGCG treatment in IPF fibroblasts. These gene expression changes were mainly involved in the biosynthesis and metabolism of cholesterol, suggesting that EGCG might have effects on IPF treatment through regulation of the cholesterol-associated genes. Our findings provide a scientific basis to evaluate the potential benefits of EGCG in IPF treatment, and warrant future studies to understand the role of molecular pathways underlying cholesterol homeostasis in the pathogenesis of IPF.
Acknowledgments
The authors wish to thank the staff of the Center for Research Resources and Development of Kaohsiung Medical University for their support.
Appendix A
Table A1.
Differentially expressed genes in idiopathic pulmonary fibrosis (IPF) fibroblasts treated with epigallocatechin gallate (EGCG) versus water (control).
| Official Gene Symbol | FPKM | Ratio (EGCG/Control) |
Log2(ratio) | p-Value | q-Value * | |
|---|---|---|---|---|---|---|
| EGCG | Control | |||||
| PCSK9 | 0.85 | 0.09 | 9.65 | 3.27 | 0.0003 | 0.0846 |
| HMGCS1 | 441.43 | 57.81 | 7.64 | 2.93 | 0.0001 | 0.0268 |
| BTBD7 | 7.42 | 1.07 | 6.96 | 2.80 | 0.0001 | 0.0268 |
| CTIF | 5.12 | 0.89 | 5.75 | 2.52 | 0.0003 | 0.0846 |
| FAM120B | 2.44 | 0.43 | 5.64 | 2.50 | 0.0007 | 0.1650 |
| PLA2G3 | 1.29 | 0.24 | 5.29 | 2.40 | 0.0005 | 0.1250 |
| FABP3 | 117.28 | 23.12 | 5.07 | 2.34 | 0.0001 | 0.0268 |
| VPS53 | 10.80 | 2.16 | 4.99 | 2.32 | 0.0001 | 0.0268 |
| HSPB3 | 5.34 | 1.12 | 4.76 | 2.25 | 0.0011 | 0.2481 |
| MVD | 96.80 | 22.27 | 4.35 | 2.12 | 0.0001 | 0.0268 |
| TMEM97 | 140.13 | 32.98 | 4.25 | 2.09 | 0.0001 | 0.0268 |
| MSMO1 | 224.89 | 55.13 | 4.08 | 2.03 | 0.0001 | 0.0268 |
| HMGCR | 136.04 | 34.55 | 3.94 | 1.98 | 0.0001 | 0.0268 |
| ARHGEF5 | 3.48 | 0.90 | 3.86 | 1.95 | 0.0001 | 0.0268 |
| HSD17B7 | 11.98 | 3.12 | 3.85 | 1.94 | 0.0003 | 0.0973 |
| PALMD | 3.68 | 0.96 | 3.82 | 1.93 | 0.0004 | 0.1047 |
| DHCR7 | 84.31 | 22.39 | 3.76 | 1.91 | 0.0001 | 0.0268 |
| IDI1 | 185.79 | 51.74 | 3.59 | 1.84 | 0.0003 | 0.0846 |
| CYP51A1 | 329.34 | 94.32 | 3.49 | 1.80 | 0.0001 | 0.0268 |
| ACSS2 | 25.91 | 7.69 | 3.37 | 1.75 | 0.0001 | 0.0268 |
| DMKN | 2.32 | 0.71 | 3.28 | 1.71 | 0.0001 | 0.0268 |
| SLC40A1 | 7.87 | 2.43 | 3.24 | 1.70 | 0.0003 | 0.0846 |
| SQLE | 108.28 | 34.77 | 3.11 | 1.64 | 0.0001 | 0.0268 |
| FBXO32 | 5.71 | 1.85 | 3.08 | 1.62 | 0.0005 | 0.1319 |
| FDFT1 | 206.04 | 69.04 | 2.98 | 1.58 | 0.0001 | 0.0268 |
| BIRC3 | 12.03 | 4.07 | 2.95 | 1.56 | 0.0003 | 0.0973 |
| RRAD | 4.19 | 1.43 | 2.94 | 1.55 | 0.0010 | 0.2174 |
| ITGB8 | 6.31 | 2.17 | 2.91 | 1.54 | 0.0001 | 0.0486 |
| FDPS | 260.06 | 91.77 | 2.83 | 1.50 | 0.0001 | 0.0268 |
| SCD | 377.97 | 134.01 | 2.82 | 1.50 | 0.0001 | 0.0486 |
| INSIG1 | 268.58 | 95.40 | 2.82 | 1.49 | 0.0001 | 0.0268 |
| LSS | 129.93 | 46.81 | 2.78 | 1.47 | 0.0002 | 0.0798 |
| LBH | 128.19 | 46.39 | 2.76 | 1.47 | 0.0001 | 0.0268 |
| RPS17 | 60.77 | 22.54 | 2.70 | 1.43 | 0.0004 | 0.1047 |
| NTN4 | 12.06 | 4.72 | 2.56 | 1.35 | 0.0005 | 0.1319 |
| CEMIP | 216.43 | 85.86 | 2.52 | 1.33 | 0.0005 | 0.1250 |
| LMOD1 | 15.53 | 6.33 | 2.46 | 1.30 | 0.0001 | 0.0268 |
| NSDHL | 53.96 | 22.04 | 2.45 | 1.29 | 0.0001 | 0.0486 |
| SC5D | 93.06 | 38.02 | 2.45 | 1.29 | 0.0003 | 0.0846 |
| GABARAPL1 | 20.49 | 8.74 | 2.34 | 1.23 | 0.0002 | 0.0798 |
| FRY | 6.83 | 3.06 | 2.23 | 1.16 | 0.0003 | 0.0846 |
| PDE5A | 278.27 | 125.88 | 2.21 | 1.14 | 0.0007 | 0.1556 |
| BHLHE40 | 16.48 | 7.50 | 2.20 | 1.13 | 0.0006 | 0.1482 |
| QPRT | 12.85 | 5.85 | 2.20 | 1.13 | 0.0005 | 0.1250 |
| DHCR24 | 151.61 | 73.96 | 2.05 | 1.04 | 0.0008 | 0.1742 |
| SEMA7A | 26.51 | 54.97 | 0.48 | -1.05 | 0.0004 | 0.1174 |
| MT2A | 352.79 | 738.65 | 0.48 | -1.07 | 0.0004 | 0.1047 |
| EGR1 | 17.36 | 36.40 | 0.48 | -1.07 | 0.0006 | 0.1426 |
| STC1 | 41.69 | 87.70 | 0.48 | -1.07 | 0.0007 | 0.1556 |
| SERPINB2 | 131.57 | 286.07 | 0.46 | -1.12 | 0.0002 | 0.0798 |
| PLXNA4 | 2.83 | 7.07 | 0.40 | -1.32 | 0.0005 | 0.1319 |
| SQSTM1 | 11.91 | 36.40 | 0.33 | -1.61 | 0.0001 | 0.0268 |
| SORBS2 | 0.12 | 0.41 | 0.30 | -1.73 | 0.0004 | 0.1047 |
| ESM1 | 10.28 | 45.34 | 0.23 | -2.14 | 0.0001 | 0.0268 |
| THBD | 3.13 | 14.37 | 0.22 | -2.20 | 0.0001 | 0.0268 |
| MYLIP | 0.61 | 2.91 | 0.21 | -2.24 | 0.0006 | 0.1482 |
| RGPD6 | 0.57 | 2.99 | 0.19 | -2.38 | 0.0002 | 0.0798 |
| TOP3A | 0.42 | 2.21 | 0.19 | -2.40 | 0.0002 | 0.0798 |
| ABCA1 | 2.09 | 13.21 | 0.16 | -2.66 | 0.0001 | 0.0268 |
| IVL | 0.13 | 1.03 | 0.12 | -3.01 | 0.0001 | 0.0268 |
| ITPK1 | 0.85 | 7.50 | 0.11 | -3.14 | 0.0001 | 0.0268 |
* p-values adjusted with false discovery rate using the method by Benjamini et al. IPF, idiopathic pulmonary fibrosis; EGCG, epigallocatechin gallate; FPKM, fragments per kilobase of transcript per million mapped reads.
Table A2.
MicroRNAs with significant change in idiopathic pulmonary fibrosis (IPF) fibroblasts treated with epigallocatechin gallate (EGCG) versus water (control).
| miRNA | Precursor | Normalized Read Count (rpm) | Fold Change | Up/Down | |
|---|---|---|---|---|---|
| EGCG | Control | ||||
| hsa-miR-491-3p | hsa-mir-491 | 1.39 | 0.08 | 17.38 | Up |
| hsa-miR-4803 | hsa-mir-4803 | 1.21 | 0.08 | 15.13 | Up |
| hsa-miR-1322 | hsa-mir-1322 | 1.04 | 0.17 | 6.12 | Up |
| hsa-miR-939-5p | hsa-mir-939 | 1.65 | 0.34 | 4.85 | Up |
| hsa-miR-101-5p | hsa-mir-101-1 | 1.13 | 0.25 | 4.52 | Up |
| hsa-miR-141-3p | hsa-mir-141 | 1.82 | 0.42 | 4.33 | Up |
| hsa-miR-33a-3p | hsa-mir-33a | 3.03 | 0.84 | 3.61 | Up |
| hsa-miR-34a-3p | hsa-mir-34a | 6.5 | 1.93 | 3.37 | Up |
| hsa-miR-503-3p | hsa-mir-503 | 1.21 | 0.42 | 2.88 | Up |
| hsa-miR-200c-3p | hsa-mir-200c | 1.3 | 0.5 | 2.60 | Up |
| hsa-miR-145-5p | hsa-mir-145 | 93.71 | 37.01 | 2.53 | Up |
| hsa-miR-491-5p | hsa-mir-491 | 1.91 | 0.76 | 2.51 | Up |
| hsa-miR-770-5p | hsa-mir-770 | 2.51 | 1.01 | 2.49 | Up |
| hsa-miR-29b-2-5p | hsa-mir-29b-2 | 2.6 | 1.09 | 2.39 | Up |
| hsa-miR-548at-5p | hsa-mir-548at | 2.17 | 0.93 | 2.33 | Up |
| hsa-miR-4636 | hsa-mir-4636 | 2.51 | 1.09 | 2.30 | Up |
| hsa-miR-3684 | hsa-mir-3684 | 1.65 | 0.76 | 2.17 | Up |
| hsa-miR-6723-5p | hsa-mir-6723 | 1.65 | 0.76 | 2.17 | Up |
| hsa-miR-142-5p | hsa-mir-142 | 1.39 | 0.67 | 2.07 | Up |
| hsa-miR-655-3p | hsa-mir-655 | 4.85 | 2.35 | 2.06 | Up |
| hsa-miR-3130-5p | hsa-mir-3130-2 | 1.56 | 0.76 | 2.05 | Up |
| hsa-miR-3613-3p | hsa-mir-3613 | 1.21 | 0.59 | 2.05 | Up |
| hsa-miR-1273g-3p | hsa-mir-1273g | 0.87 | 1.77 | −2.03 | Down |
| hsa-miR-2116-3p | hsa-mir-2116 | 2.51 | 5.13 | −2.04 | Down |
| hsa-miR-3622a-5p | hsa-mir-3622a | 0.69 | 1.43 | −2.07 | Down |
| hsa-miR-5699-5p | hsa-mir-5699 | 1.65 | 3.45 | −2.09 | Down |
| hsa-miR-138-1-3p | hsa-mir-138-1 | 55.39 | 117.24 | −2.12 | Down |
| hsa-miR-340-3p | hsa-mir-340 | 3.81 | 8.24 | −2.16 | Down |
| hsa-miR-423-5p | hsa-mir-423 | 822.58 | 1779.79 | −2.16 | Down |
| hsa-miR-25-5p | hsa-mir-25 | 27.39 | 59.54 | −2.17 | Down |
| hsa-miR-3605-3p | hsa-mir-3605 | 12.05 | 26.66 | −2.21 | Down |
| hsa-miR-1910-5p | hsa-mir-1910 | 22.71 | 50.63 | −2.23 | Down |
| hsa-miR-4745-5p | hsa-mir-4745 | 0.95 | 2.19 | −2.31 | Down |
| hsa-miR-6840-5p | hsa-mir-6840 | 0.61 | 1.43 | −2.34 | Down |
| hsa-miR-92a-1-5p | hsa-mir-92a-1 | 8.06 | 19.09 | −2.37 | Down |
| hsa-miR-1914-5p | hsa-mir-1914 | 0.52 | 1.26 | −2.42 | Down |
| hsa-miR-937-3p | hsa-mir-937 | 4.33 | 10.85 | −2.51 | Down |
| hsa-miR-323a-5p | hsa-mir-323a | 0.43 | 1.09 | −2.53 | Down |
| hsa-miR-3648 | hsa-mir-3648-1 | 0.78 | 2.02 | −2.59 | Down |
| hsa-miR-3648 | hsa-mir-3648-2 | 0.78 | 2.02 | −2.59 | Down |
| hsa-miR-1228-3p | hsa-mir-1228 | 0.52 | 1.35 | −2.6 | Down |
| hsa-miR-1294 | hsa-mir-1294 | 0.69 | 1.85 | −2.68 | Down |
| hsa-miR-3177-5p | hsa-mir-3177 | 0.69 | 1.85 | −2.68 | Down |
| hsa-miR-4768-5p | hsa-mir-4768 | 0.52 | 1.43 | −2.75 | Down |
| hsa-miR-197-5p | hsa-mir-197 | 0.69 | 2.1 | −3.04 | Down |
| hsa-miR-1972 | hsa-mir-1972-1 | 0.35 | 1.09 | −3.11 | Down |
| hsa-miR-1972 | hsa-mir-1972-2 | 0.35 | 1.09 | −3.11 | Down |
| hsa-miR-3691-5p | hsa-mir-3691 | 1.13 | 3.7 | −3.27 | Down |
| hsa-miR-548al | hsa-mir-548al | 0.35 | 1.18 | −3.37 | Down |
| hsa-miR-4766-3p | hsa-mir-4766 | 0.43 | 1.77 | −4.12 | Down |
| hsa-miR-3918 | hsa-mir-3918 | 0.26 | 1.09 | −4.19 | Down |
| hsa-miR-548t-5p | hsa-mir-548t | 0.26 | 1.09 | −4.19 | Down |
| hsa-miR-6783-3p | hsa-mir-6783 | 0.26 | 1.09 | −4.19 | Down |
| hsa-miR-155-3p | hsa-mir-155 | 0.26 | 1.18 | −4.54 | Down |
| hsa-miR-184 | hsa-mir-184 | 0.61 | 2.78 | −4.56 | Down |
| hsa-miR-6859-5p | hsa-mir-6859-1 | 0.17 | 1.18 | −6.94 | Down |
| hsa-miR-6859-5p | hsa-mir-6859-2 | 0.17 | 1.18 | −6.94 | Down |
| hsa-miR-6859-5p | hsa-mir-6859-3 | 0.17 | 1.18 | −6.94 | Down |
| hsa-miR-6859-5p | hsa-mir-6859-4 | 0.17 | 1.18 | −6.94 | Down |
| hsa-miR-1304-5p | hsa-mir-1304 | 0.09 | 1.43 | −15.89 | Down |
| hsa-miR-891a-5p | hsa-mir-891a | 0.09 | 1.43 | −15.89 | Down |
IPF, idiopathic pulmonary fibrosis; EGCG, epigallocatechin gallate; rpm, reads per million.
Author Contributions
Conceptualization, M.J.T., C.C.S., and P.L.K.; methodology, M.J.T., C.C.S., and P.L.K.; formal analysis, M.J.T., W.A.C., S.H.L., and K.F.C.; investigation, M.J.T., W.A.C., S.H.L., and K.F.C.; resources, M.J.T. and P.L.K.; writing—original draft preparation, M.J.T.; writing—review and editing, M.J.T., W.A.C., S.H.L., K.F.C., C.C.S., and P.L.K.; supervision, C.C.S. and P.L.K.; funding acquisition, M.J.T. and P.L.K.
Funding
This research was funded by grants from the Ministry of Science and Technology (MOST 107-2320-B-037-011-MY3 and MOST 106-2314-B-037-016-MY2), the Kaohsiung Medical University Hospital (KMUHS10701, KMUHS10712, KMUH107-7M06, KMUH107-7M07, KMUH107-7R14, and KMUH106-6T05), and Kaohsiung Medical University (KMU-DK108003 and KMU-Q108005).
Conflicts of Interest
The authors declare no conflict of interest related to this study.
References
- 1.Sheu C.C., Chang W.A., Tsai M.J., Liao S.H., Chong I.W., Kuo P.L. Bioinformatic analysis of nextgeneration sequencing data to identify dysregulated genes in fibroblasts of idiopathic pulmonary fibrosis. Int. J. Mol. Med. 2019;4080:1643–1656. doi: 10.3892/ijmm.2019.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richeldi L., Collard H.R., Jones M.G. Idiopathic pulmonary fibrosis. Lancet. 2017;389:1941–1952. doi: 10.1016/S0140-6736(17)30866-8. [DOI] [PubMed] [Google Scholar]
- 3.Martinez F.J., Collard H.R., Pardo A., Raghu G., Richeldi L., Selman M., Swigris J.J., Taniguchi H., Wells A.U. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Primers. 2017;3:17074. doi: 10.1038/nrdp.2017.74. [DOI] [PubMed] [Google Scholar]
- 4.Raghu G., Collard H.R., Egan J.J., Martinez F.J., Behr J., Brown K.K., Colby T.V., Cordier J.F., Flaherty K.R., Lasky J.A., et al. An official ATS/ERS/JRS/ALAT statement: Idiopathic pulmonary fibrosis: Evidence-based guidelines for diagnosis and management. Am. J. Respir. Crit. Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barratt S.L., Creamer A., Hayton C., Chaudhuri N. Idiopathic Pulmonary Fibrosis (IPF): An Overview. J. Clin. Med. 2018;7:201. doi: 10.3390/jcm7080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutchinson J., Fogarty A., Hubbard R., McKeever T. Global incidence and mortality of idiopathic pulmonary fibrosis: A systematic review. Eur. Respir. J. 2015;46:795–806. doi: 10.1183/09031936.00185114. [DOI] [PubMed] [Google Scholar]
- 7.Vancheri C., Failla M., Crimi N., Raghu G. Idiopathic pulmonary fibrosis: A disease with similarities and links to cancer biology. Eur. Respir. J. 2010;35:496–504. doi: 10.1183/09031936.00077309. [DOI] [PubMed] [Google Scholar]
- 8.Pardo A., Selman M. Lung Fibroblasts, Aging, and Idiopathic Pulmonary Fibrosis. Ann. Am. Thorac. Soc. 2016;13:417–421. doi: 10.1513/AnnalsATS.201605-341AW. [DOI] [PubMed] [Google Scholar]
- 9.Ramos C., Montano M., Garcia-Alvarez J., Ruiz V., Uhal B.D., Selman M., Pardo A. Fibroblasts from idiopathic pulmonary fibrosis and normal lungs differ in growth rate, apoptosis, and tissue inhibitor of metalloproteinases expression. Am. J. Respir. Cell Mol. Biol. 2001;24:591–598. doi: 10.1165/ajrcmb.24.5.4333. [DOI] [PubMed] [Google Scholar]
- 10.Pierce E.M., Carpenter K., Jakubzick C., Kunkel S.L., Evanoff H., Flaherty K.R., Martinez F.J., Toews G.B., Hogaboam C.M. Idiopathic pulmonary fibrosis fibroblasts migrate and proliferate to CC chemokine ligand 21. Eur. Respir. J. 2007;29:1082–1093. doi: 10.1183/09031936.00122806. [DOI] [PubMed] [Google Scholar]
- 11.Vuga L.J., Ben-Yehudah A., Kovkarova-Naumovski E., Oriss T., Gibson K.F., Feghali-Bostwick C., Kaminski N. WNT5A is a regulator of fibroblast proliferation and resistance to apoptosis. Am. J. Respir. Cell Mol. Biol. 2009;41:583–589. doi: 10.1165/rcmb.2008-0201OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veith C., Drent M., Bast A., van Schooten F.J., Boots A.W. The disturbed redox-balance in pulmonary fibrosis is modulated by the plant flavonoid quercetin. Toxicol. Appl. Pharmacol. 2017;336:40–48. doi: 10.1016/j.taap.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu Y., Dobashi K., Sano T., Yamada M. ROCK activation in lung of idiopathic pulmonary fibrosis with oxidative stress. Int. J. Immunopathol. Pharmacol. 2014;27:37–44. doi: 10.1177/039463201402700106. [DOI] [PubMed] [Google Scholar]
- 14.Oldham J.M., Ma S.F., Martinez F.J., Anstrom K.J., Raghu G., Schwartz D.A., Valenzi E., Witt L., Lee C., Vij R., et al. TOLLIP, MUC5B, and the Response to N-Acetylcysteine among Individuals with Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2015;192:1475–1482. doi: 10.1164/rccm.201505-1010OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azuma A., Nukiwa T., Tsuboi E., Suga M., Abe S., Nakata K., Taguchi Y., Nagai S., Itoh H., Ohi M., et al. Double-blind, placebo-controlled trial of pirfenidone in patients with idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2005;171:1040–1047. doi: 10.1164/rccm.200404-571OC. [DOI] [PubMed] [Google Scholar]
- 16.King T.E., Jr., Bradford W.Z., Castro-Bernardini S., Fagan E.A., Glaspole I., Glassberg M.K., Gorina E., Hopkins P.M., Kardatzke D., Lancaster L., et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2014;370:2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 17.Nathan S.D., Costabel U., Glaspole I., Glassberg M.K., Lancaster L.H., Lederer D.J., Pereira C.A., Trzaskoma B., Morgenthien E.A., Limb S.L., et al. Efficacy of Pirfenidone in the Context of Multiple Disease Progression Events in Patients With Idiopathic Pulmonary Fibrosis. Chest. 2018;155:712–719. doi: 10.1016/j.chest.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Richeldi L., Costabel U., Selman M., Kim D.S., Hansell D.M., Nicholson A.G., Brown K.K., Flaherty K.R., Noble P.W., Raghu G., et al. Efficacy of a tyrosine kinase inhibitor in idiopathic pulmonary fibrosis. N. Engl. J. Med. 2011;365:1079–1087. doi: 10.1056/NEJMoa1103690. [DOI] [PubMed] [Google Scholar]
- 19.Richeldi L., du Bois R.M., Raghu G., Azuma A., Brown K.K., Costabel U., Cottin V., Flaherty K.R., Hansell D.M., Inoue Y., et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N. Engl. J. Med. 2014;370:2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 20.Kolb M., Raghu G., Wells A.U., Behr J., Richeldi L., Schinzel B., Quaresma M., Stowasser S., Martinez F.J., Investigators I. Nintedanib plus Sildenafil in Patients with Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 2018;379:1722–1731. doi: 10.1056/NEJMoa1811737. [DOI] [PubMed] [Google Scholar]
- 21.Kropski J.A., Blackwell T.S. Progress in Understanding and Treating Idiopathic Pulmonary Fibrosis. Annu. Rev. Med. 2019;70:211–224. doi: 10.1146/annurev-med-041317-102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casanova E., Salvado J., Crescenti A., Gibert-Ramos A. Epigallocatechin Gallate Modulates Muscle Homeostasis in Type 2 Diabetes and Obesity by Targeting Energetic and Redox Pathways: A Narrative Review. Int. J. Mol. Sci. 2019;20:532. doi: 10.3390/ijms20030532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Negri A., Naponelli V., Rizzi F., Bettuzzi S. Molecular Targets of Epigallocatechin-Gallate (EGCG): A Special Focus on Signal Transduction and Cancer. Nutrients. 2018;10:1936. doi: 10.3390/nu10121936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhagwat S., Haytowitz D.B., Holden J.M. USDA Database for the Flavonoid Content of Selected Foods, Release 3. Agricultural Research Service, U.S. Department of Agriculture; Beltsville, MD, USA: 2011. [Google Scholar]
- 25.Oz H.S. Chronic Inflammatory Diseases and Green Tea Polyphenols. Nutrients. 2017;9:561. doi: 10.3390/nu9060561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minnelli C., Moretti P., Fulgenzi G., Mariani P., Laudadio E., Armeni T., Galeazzi R., Mobbili G. A Poloxamer-407 modified liposome encapsulating epigallocatechin-3-gallate in the presence of magnesium: Characterization and protective effect against oxidative damage. Int. J. Pharm. 2018;552:225–234. doi: 10.1016/j.ijpharm.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Sriram N., Kalayarasan S., Sudhandiran G. Epigallocatechin-3-gallate exhibits anti-fibrotic effect by attenuating bleomycin-induced glycoconjugates, lysosomal hydrolases and ultrastructural changes in rat model pulmonary fibrosis. Chemico-Biol. Interact. 2009;180:271–280. doi: 10.1016/j.cbi.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Shi W., Li L., Ding Y., Yang K., Chen Z., Fan X., Jiang S., Guan Y., Liu Z., Xu D., et al. The critical role of epigallocatechin gallate in regulating mitochondrial metabolism. Future Med. Chem. 2018;10:795–809. doi: 10.4155/fmc-2017-0204. [DOI] [PubMed] [Google Scholar]
- 29.Sriram N., Kalayarasan S., Manikandan R., Arumugam M., Sudhandiran G. Epigallocatechin gallate attenuates fibroblast proliferation and excessive collagen production by effectively intervening TGF-beta1 signalling. Clin. Exp. Pharm. Phys. 2015;42:849–859. doi: 10.1111/1440-1681.12428. [DOI] [PubMed] [Google Scholar]
- 30.Sriram N., Kalayarasan S., Sudhandiran G. Enhancement of antioxidant defense system by epigallocatechin-3-gallate during bleomycin induced experimental pulmonary fibrosis. Biol. Pharm. Bull. 2008;31:1306–1311. doi: 10.1248/bpb.31.1306. [DOI] [PubMed] [Google Scholar]
- 31.Sriram N., Kalayarasan S., Sudhandiran G. Epigallocatechin-3-gallate augments antioxidant activities and inhibits inflammation during bleomycin-induced experimental pulmonary fibrosis through Nrf2-Keap1 signaling. Pulm. Pharm. Ther. 2009;22:221–236. doi: 10.1016/j.pupt.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 32.You H., Wei L., Sun W.L., Wang L., Yang Z.L., Liu Y., Zheng K., Wang Y., Zhang W.J. The green tea extract epigallocatechin-3-gallate inhibits irradiation-induced pulmonary fibrosis in adult rats. Int. J. Mol. Med. 2014;34:92–102. doi: 10.3892/ijmm.2014.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dweep H., Gretz N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 34.Eng Q.Y., Thanikachalam P.V., Ramamurthy S. Molecular understanding of Epigallocatechin gallate (EGCG) in cardiovascular and metabolic diseases. J. Ethnopharmacol. 2018;210:296–310. doi: 10.1016/j.jep.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 35.Legeay S., Rodier M., Fillon L., Faure S., Clere N. Epigallocatechin Gallate: A Review of Its Beneficial Properties to Prevent Metabolic Syndrome. Nutrients. 2015;7:5443–5468. doi: 10.3390/nu7075230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Javaid M.S., Latief N., Ijaz B., Ashfaq U.A. Epigallocatechin Gallate as an anti-obesity therapeutic compound: An in silico approach for structure-based drug designing. Nat. Prod. Res. 2018;32:2121–2125. doi: 10.1080/14786419.2017.1365074. [DOI] [PubMed] [Google Scholar]
- 37.Singh N.A., Mandal A.K., Khan Z.A. Potential neuroprotective properties of epigallocatechin-3-gallate (EGCG) Nutr. J. 2016;15:60. doi: 10.1186/s12937-016-0179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding S., Jiang H., Fang J. Regulation of Immune Function by Polyphenols. J. Immunol. Res. 2018;2018:1264074. doi: 10.1155/2018/1264074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gianfredi V., Nucci D., Vannini S., Villarini M., Moretti M. In vitro Biological Effects of Sulforaphane (SFN), Epigallocatechin-3-gallate (EGCG), and Curcumin on Breast Cancer Cells: A Systematic Review of the Literature. Nutr. Cancer. 2017;69:969–978. doi: 10.1080/01635581.2017.1359322. [DOI] [PubMed] [Google Scholar]
- 40.Pandima Devi K., Rajavel T., Daglia M., Nabavi S.F., Bishayee A., Nabavi S.M. Targeting miRNAs by polyphenols: Novel therapeutic strategy for cancer. Semin. Cancer Biol. 2017;46:146–157. doi: 10.1016/j.semcancer.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Sueoka N., Suganuma M., Sueoka E., Okabe S., Matsuyama S., Imai K., Nakachi K., Fujiki H. A new function of green tea: Prevention of lifestyle-related diseases. Ann. N. Y. Acad. Sci. 2001;928:274–280. doi: 10.1111/j.1749-6632.2001.tb05656.x. [DOI] [PubMed] [Google Scholar]
- 42.Gowdy K.M., Fessler M.B. Emerging roles for cholesterol and lipoproteins in lung disease. Pulm. Pharm. Therap. 2013;26:430–437. doi: 10.1016/j.pupt.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oka H., Ishii H., Iwata A., Kushima H., Toba S., Hashinaga K., Umeki K., Tokimatsu I., Hiramatsu K., Kadota J. Inhibitory effects of pitavastatin on fibrogenic mediator production by human lung fibroblasts. Life Sci. 2013;93:968–974. doi: 10.1016/j.lfs.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 44.Watts K.L., Sampson E.M., Schultz G.S., Spiteri M.A. Simvastatin inhibits growth factor expression and modulates profibrogenic markers in lung fibroblasts. Am. J. Respir. Cell Mol. Biol. 2005;32:290–300. doi: 10.1165/rcmb.2004-0127OC. [DOI] [PubMed] [Google Scholar]
- 45.Kim J.W., Rhee C.K., Kim T.J., Kim Y.H., Lee S.H., Yoon H.K., Kim S.C., Lee S.Y., Kwon S.S., Kim K.H., et al. Effect of pravastatin on bleomycin-induced acute lung injury and pulmonary fibrosis. Clin. Exp. Pharm. Phys. 2010;37:1055–1063. doi: 10.1111/j.1440-1681.2010.05431.x. [DOI] [PubMed] [Google Scholar]
- 46.Podolanczuk A.J., Raghu G., Tsai M.Y., Kawut S.M., Peterson E., Sonti R., Rabinowitz D., Johnson C., Barr R.G., Hinckley Stukovsky K., et al. Cholesterol, lipoproteins and subclinical interstitial lung disease: The MESA study. Thorax. 2017;72:472–474. doi: 10.1136/thoraxjnl-2016-209568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song X., Cao G., Jing L., Lin S., Wang X., Zhang J., Wang M., Liu W., Lv C. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J. Cell Mol. Med. 2014;18:991–1003. doi: 10.1111/jcmm.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemnes A.R., Zaiman A., Champion H.C. PDE5A inhibition attenuates bleomycin-induced pulmonary fibrosis and pulmonary hypertension through inhibition of ROS generation and RhoA/Rho kinase activation. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;294:24–33. doi: 10.1152/ajplung.00245.2007. [DOI] [PubMed] [Google Scholar]
- 49.Coriati A., Arslanian E., Bouvet G.F., Prat A., Seidah N.G., Rabasa-Lhoret R., Berthiaume Y. Proprotein Convertase Subtilisin/Kexin type 9 affects insulin but not lipid metabolism in cystic fibrosis. Clin. Investig. Med. 2017;40:59–65. doi: 10.25011/cim.v40i2.28196. [DOI] [PubMed] [Google Scholar]
- 50.Lai Q., Giralt A., Le May C., Zhang L., Cariou B., Denechaud P.D., Fajas L. E2F1 inhibits circulating cholesterol clearance by regulating Pcsk9 expression in the liver. JCI Insight. 2017;2 doi: 10.1172/jci.insight.89729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pandit K.V., Milosevic J., Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl. Res. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 52.Rajasekaran S., Rajaguru P., Sudhakar Gandhi P.S. MicroRNAs as potential targets for progressive pulmonary fibrosis. Front. Pharm. 2015;6:254. doi: 10.3389/fphar.2015.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pottier N., Maurin T., Chevalier B., Puissegur M.P., Lebrigand K., Robbe-Sermesant K., Bertero T., Lino Cardenas C.L., Courcot E., Rios G., et al. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: Implication in epithelial-mesenchymal interactions. PLoS ONE. 2009;4:e6718. doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cushing L., Kuang P.P., Qian J., Shao F., Wu J., Little F., Thannickal V.J., Cardoso W.V., Lu J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui H., Ge J., Xie N., Banerjee S., Zhou Y., Antony V.B., Thannickal V.J., Liu G. miR-34a Inhibits Lung Fibrosis by Inducing Lung Fibroblast Senescence. Am. J. Respir Cell Mol. Biol. 2017;56:168–178. doi: 10.1165/rcmb.2016-0163OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee M.J., Maliakal P., Chen L., Meng X., Bondoc F.Y., Prabhu S., Lambert G., Mohr S., Yang C.S. Pharmacokinetics of tea catechins after ingestion of green tea and (-)-epigallocatechin-3-gallate by humans: Formation of different metabolites and individual variability. Cancer Epidemiol. Biomark. Prev. 2002;11:1025–1032. [PubMed] [Google Scholar]
- 57.Yang C.S., Maliakal P., Meng X. Inhibition of carcinogenesis by tea. Annu. Rev. Pharm. Toxicol. 2002;42:25–54. doi: 10.1146/annurev.pharmtox.42.082101.154309. [DOI] [PubMed] [Google Scholar]
- 58.Wu Y.R., Choi H.J., Kang Y.G., Kim J.K., Shin J.W. In vitro study on anti-inflammatory effects of epigallocatechin-3-gallate-loaded nano- and microscale particles. Int. J. Nanomed. 2017;12:7007–7013. doi: 10.2147/IJN.S146296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du G.J., Zhang Z., Wen X.D., Yu C., Calway T., Yuan C.S., Wang C.Z. Epigallocatechin Gallate (EGCG) is the most effective cancer chemopreventive polyphenol in green tea. Nutrients. 2012;4:1679–1691. doi: 10.3390/nu4111679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chow H.H., Cai Y., Hakim I.A., Crowell J.A., Shahi F., Brooks C.A., Dorr R.T., Hara Y., Alberts D.S. Pharmacokinetics and safety of green tea polyphenols after multiple-dose administration of epigallocatechin gallate and polyphenon E in healthy individuals. Clin. Cancer Res. 2003;9:3312–3319. [PubMed] [Google Scholar]
- 61.Hu J., Webster D., Cao J., Shao A. The safety of green tea and green tea extract consumption in adults - Results of a systematic review. Regul. Toxicol. Pharm. 2018;95:412–433. doi: 10.1016/j.yrtph.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 62.Younes M., Aggett P., Aguilar F., Crebelli R., Dusemund B., Filipič M., Frutos M.J., Galtier P., Gott D. Scientific opinion on the safety of green tea catechins. EFSA J. 2018;16:e05239. doi: 10.2903/j.efsa.2018.5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Witschi H., Espiritu I., Ly M., Uyeminami D., Morin D., Raabe O.G. Chemoprevention of tobacco smoke-induced lung tumors by inhalation of an epigallocatechin gallate (EGCG) aerosol: A pilot study. Inhal. Toxicol. 2004;16:763–770. doi: 10.1080/08958370490490400. [DOI] [PubMed] [Google Scholar]
- 64.Sheu C.C., Tsai M.J., Chen F.W., Chang K.F., Chang W.A., Chong I.W., Kuo P.L., Hsu Y.L. Identification of novel genetic regulations associated with airway epithelial homeostasis using next-generation sequencing data and bioinformatics approaches. Oncotarget. 2017;8:82674–82688. doi: 10.18632/oncotarget.19752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang W.A., Tsai M.J., Jian S.F., Sheu C.C., Kuo P.L. Systematic analysis of transcriptomic profiles of COPD airway epithelium using next-generation sequencing and bioinformatics. Int. J. Chron. Obstruct. Pulmon. Dis. 2018;13:2387–2398. doi: 10.2147/COPD.S173206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsai M.J., Chang W.A., Jian S.F., Chang K.F., Sheu C.C., Kuo P.L. Possible mechanisms mediating apoptosis of bronchial epithelial cells in chronic obstructive pulmonary disease—A next-generation sequencing approach. Pathol. Res. Pract. 2018;214:1489–1496. doi: 10.1016/j.prp.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 67.Tsai M.J., Tsai Y.C., Chang W.A., Lin Y.S., Tsai P.H., Sheu C.C., Kuo P.L., Hsu Y.L. Deducting MicroRNA-Mediated Changes Common in Bronchial Epithelial Cells of Asthma and Chronic Obstructive Pulmonary Disease-A Next-Generation Sequencing-Guided Bioinformatic Approach. Int. J. Mol. Sci. 2019;20:553. doi: 10.3390/ijms20030553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee W.H., Tsai M.J., Chang W.A., Wu L.Y., Wang H.Y., Chang K.F., Su H.M., Kuo P.L. Deduction of novel genes potentially involved in hypoxic AC16 human cardiomyocytes using next-generation sequencing and bioinformatics approaches. Int. J. Mol. Med. 2018;42:2489–2502. doi: 10.3892/ijmm.2018.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siren J., Valimaki N., Makinen V. Indexing Graphs for Path Queries with Applications in Genome Research. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014;11:375–388. doi: 10.1109/TCBB.2013.2297101. [DOI] [PubMed] [Google Scholar]
- 71.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;1:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Galipon J., Ishii R., Suzuki Y., Tomita M., Ui-Tei K. Differential Binding of Three Major Human ADAR Isoforms to Coding and Long Non-Coding Transcripts. Genes. 2017;8:68. doi: 10.3390/genes8020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 75.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vejnar C.E., Zdobnov E.M. MiRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012;40:11673–11683. doi: 10.1093/nar/gks901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang D.W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:362–368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]