Abstract
Variegated plants are valuable materials for investigating leaf color regulated mechanisms. To unveil the role of posttranslational modification in the variegated phenotype, we conducted global quantitative phosphoproteomic analysis on different leaf color sectors of Maiyuanjinqiu and the corresponding of Catalpa fargesii using Ti4+-IMAC phosphopeptide enrichment. A total of 3778 phosphorylated sites assigned to 1646 phosphoproteins were identified, and 3221 in 1434 proteins were quantified. Differential phosphoproteins (above 1.5 or below 1/1.5) in various leaf color sectors were selected for functional enrichment analyses. Gene ontology (GO) enrichment revealed that processes of photosynthesis, regulation of the generation of precursor metabolites, response to stress, homeostasis, amino acid metabolism, transport–related processes, and most of the energy metabolisms might contribute to leaf color. KEGG pathway enrichment analysis was performed based on differential phosphoproteins (DPs) in different organelles. The result showed that most enriched pathways were located in the chloroplasts and cytosol. The phosphorylation levels of glycometabolism enzymes might greatly affect leaf variegation. Measurements of fluorescence parameters and enzyme activities confirmed that protein phosphorylation could affect plant physiology by regulating enzyme activity. These results provide new clues for further study the formation mechanisms of naturally variegated phenotype.
Keywords: Catalpa fargesii, phosphoproteomics, variegated leaf, regulated mechanism
1. Introduction
Variegated leaves usually consist of green and yellow/white sectors. Variegated species have outstanding application value in landscaping and landscape design. Moreover, they are ideal materials for studying the mechanism of leaf color formation. With advances in high-throughput technology, genomics, transcriptomics, and proteomics approaches have emerged in the study of variegated leaves [1,2,3,4]. It has been reported that the yellow sectors generally had decreased photosynthesis and increased oxidative stress to the green sectors by transcriptome and proteome profiling [1,5]. However, the correlation between mRNA and protein expression was low in the previous researches, and scholars postulated that posttranslational modifications (PTMs) might also play a pivotal role in variegation [6,7]. Protein phosphorylation is the most basic and most common mechanism for regulating and controlling protein activity and function, and it is closely related to many biological processes such as photosynthesis and oxidative stress [8]. We inferred that protein phosphorylation might have a great relationship with the phenotype. However, this speculation lacks detailed data.
Protein phosphorylation could modulate the function of a particular protein, rather than achieving an overall effect by the overexpression/low expression or complete deletion of the gene encoding specific protein. Currently, the regulatory mechanism of photosynthesis proteins by phosphorylation modification is well established [9,10,11]. The well-known kinases STN7 and STN8 play important roles in optimizing photosystems under changing light and regulating PSII photo-damage repair [12,13]. In addition to phosphorylation studies of proteins localized in chloroplasts, an increasing number of phosphoproteins in other organelles have been researched [14]. Previous studies concluded that variegated plants were associated with the physiological phenomenon of state transition [15]. This phenomenon is often accompanied by phosphorylation of the PSII core subunits and the LHCII polypeptides [16]. It would be of interest to study the global levels of phosphorylation in the variegated leaves.
In recent years, the development of quantitative phosphoproteomics made it possible to detect phosphorylation sites on a global scale, which provided useful information for subsequent functional studies of the identified modification sites. At present, comparative phosphoproteomic analysis has been applied for the territory of plant growth, development and stress resistance [17,18,19]. However, the effect of phosphorylation on the natural variegated phenotype is still poorly understood. Comparative phosphoproteomic analysis will deepen our understanding of the effects of posttranslational modifications on leaf variegation.
Catalpa fargesii is a widely distributed native species in China. Maiyuanjinqiu is a novel variety of C. fargesii with natural variability, whose leaves display a typical variegated (yellow edge) phenotype (Identification code: 20150150). In our study, the green and yellow leaf sectors were sampled separately. The corresponding sectors of C. fargesii were used as controls. We performed comparative phosphoproteomic analyses using Ti4+-IMAC phosphopeptide enrichment technology based on TMT labeling. This enrichment technology has higher protein phosphorylation enrichment ability than TiO2 enrichment, conventional immobilized metal ion affinity chromatography (Fe3+-IMAC), and other approaches [20]. Integrative bioinformatics analyses were performed through screening differential phosphoproteins (DPs) among different color sectors. We also compared the fluorescence parameters and key enzyme activities in different leaf color sectors. This study not only helps to understand the role of protein phosphorylation in the variegated phenotype of Maiyuanjinqiu, but also provides a reliable reference for research on natural leaf color variation in other plants.
2. Results
2.1. Phenotypic Character and Physiological Parameters in Maiyuanjinqiu and C. fargesii
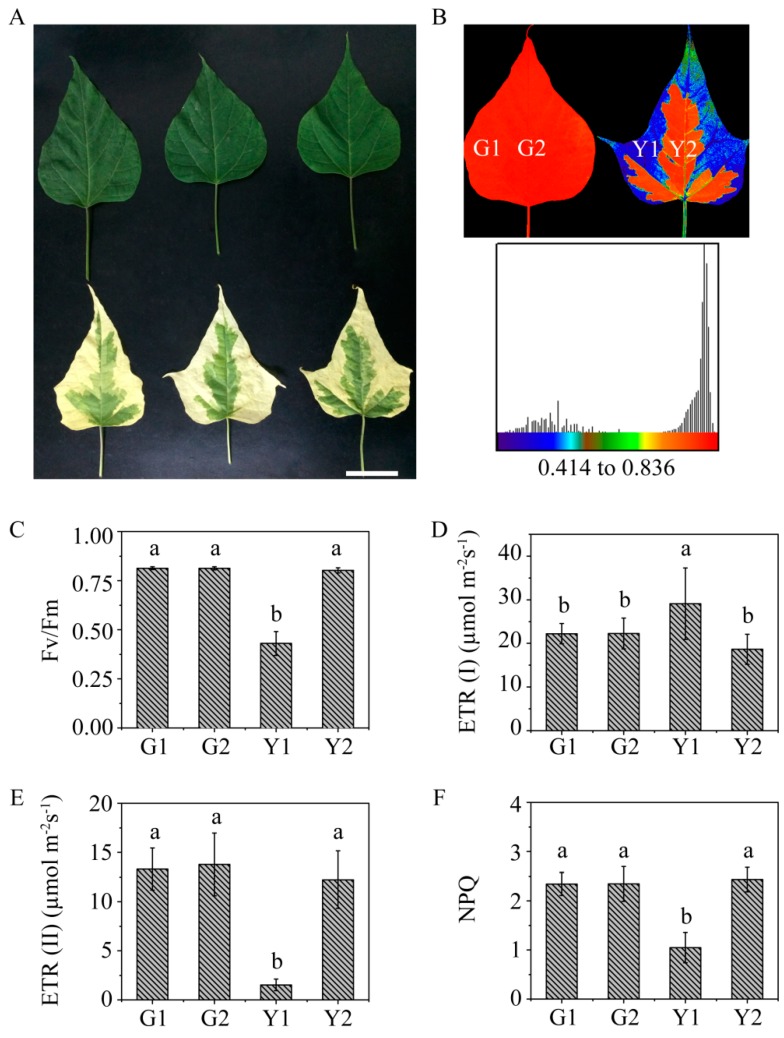
The leaves of C. fargesii were green throughout the whole growth period. Maiyuanjinqiu displayed a variegated leaf phenotype (Figure 1A). Photosynthetic fluorescence parameters were measured in Maiyuanjingqiu (Y1 and Y2) and the corresponding positions of C. fargesii (G1 and G2), respectively (Figure 1B). Fv/Fm represents the maximum photochemical quantum yield of PSII. The values of Fv/Fm were considerably lower in the yellow sectors than in the green sectors, whereas the values were similar in the Y2 and G2 sectors (Figure 1C). Compared with that in the green tissue, the electron transport rate of PSII (ETR(II)) was weakened in the yellow tissue, whereas the electron transport rate of PSI (ETR(I)) was enhanced under 129 µmol·m−2·s−1 in the yellow sectors (Figure 1D,E). These suggested that the energy balance of PSII and PSI was reequilibrated in the yellow sectors. The nonphotochemical quenching (NPQ) in Y1 was also significantly lower than that in the green sectors, which implied a deficiency in photoprotective ability in the yellow sectors (Figure 1F).
Figure 1.
Phenotypic character and determination of photosynthetic fluorescence parameters. (A) Leaf phenotypes of C. fargesii (top) and Maiyuanjinqiu (down); the length of white bar represents 5 cm. (B) Fluorescence imaging of Fv/Fm; (C) The value of Fv/Fm; (D) The value of electron transport rate (ETR(I)); (E) The value of ETR(II); (F) The value of nonphotochemical quenching (NPQ) in G1, G2, Y1, and Y2. The photosynthetically active radiation was 129 µmol·m−2·s−1. Bars express the means ± SD (n = 18), and different letters indicate significant differences (p < 0.05, one-way ANOVA).
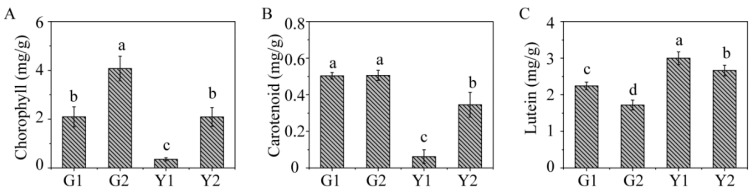
The chlorophyll, carotenoid and lutein were examined in G1, G2, Y1, and Y2, respectively. The results showed that the total chlorophyll and carotenoids were much lower in Maiyuanjinqiu than in C. fargesii (Figure 2A,B). However, the level of lutein was higher in both the green and yellow sectors of Maiyuanjinqiu compared to the corresponding sectors of C. fargesii (Figure 2C). In addition, the chlorophyll and carotenoid contents were higher in Y2 than in Y1, but the level of lutein was lower in Y2.
Figure 2.
The measurement of pigment contents. (A) The level of chlorophyll; (B) The level of carotenoid; (C) The level of lutein. The values are shown as the means ± SD (n ≥ 4), a–c indicate that the differences are significant at p < 0.05 (one-way ANOVA).
2.2. Analyses of Global Phosphorylated Proteins and Sites in Maiyuanjinqiu and C. fargesii
To assess the changes in protein phosphorylation on the variegated phenotype, we carried out a quantitative phosphoproteomics analysis in different leaf color sectors with three biological replicates. In total, 3778 phosphorylation sites in 1646 proteins were identified, among which 3221 sites in 1434 proteins were quantified. The number of total phosphopeptides was 2804 and the enrichment efficiency was 55.4%, respectively. First, the error rate of enriched phosphorylated peptides was counted. All peptide mass error rates were between −5 and 5 ppm, which suggested the high accuracy of the mass spectrometry data (Figure S1A). The number of modification sites for each phosphorylated protein in the phosphorylome was counted. Among which, approximately 54.9% proteins were singly phosphorylated. Phosphoproteins with two, three, four and greater than or equal to five sites comprised 21.4%, 9.19%, 6.34%, and 8.15% of the total (Figure S1B). The distribution of phosphorylated sites in our study was consistent with that in other plant studies [21]. In addition, the length distribution of more than 90% of peptides was between 7 and 18 (Figure S1C), which was consistent with the properties of tryptic peptides. Statistics on the phosphorylated amino acids revealed that the percentages of phosphorylation at serine, threonine and tyrosine residues were 78.3%, 15.3%, and 6.4%, respectively (Figure S1D).
2.3. Secondary Structure and Motif Analysis of Phosphoproteins
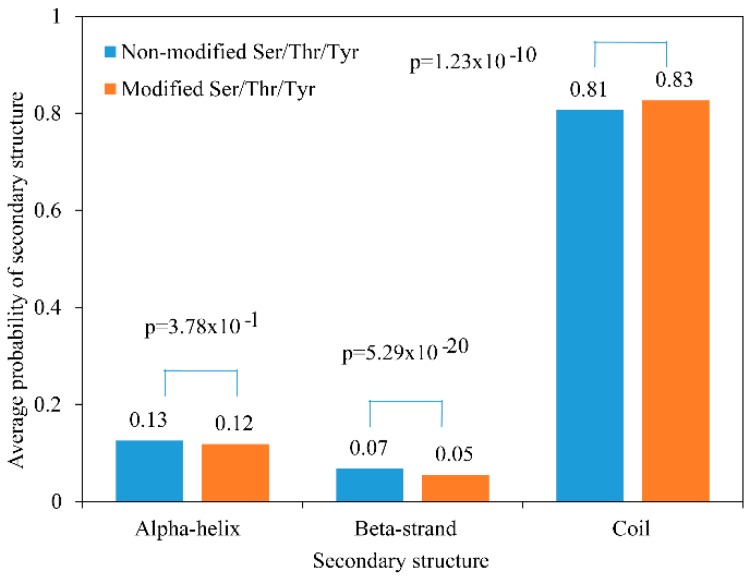
To explore the relationship between phosphorylation and protein secondary structure, we performed a secondary structure analysis of all identified phosphorylated proteins. Approximately 83% of the phosphorylated proteins were in the disordered secondary structure region, while only 17% of the phosphorylated proteins were in the ordered secondary structure region (Figure 3). The modified and non-modified proteins were significantly different in the proportions of beta-strand and coil regions, but not of alpha-helix regions (Figure 3).
Figure 3.
Probabilities of localization to different protein secondary structures.
A motif analysis of the modified amino acid residues from −6 to 6 positions was performed. We identified 28 motifs at serine residues (Figure 4A), 9 motifs at threonine residues (Figure 4B) and 3 motifs at tyrosine residues (Figure 4C). The results showed that arginine (R) and proline (P) near the serine residue (S) were the most conserved amino acids upstream and downstream of modified serine sites (Figure 4D). R, P, and S were enriched around the threonine phosphorylated sites at the −3 to +3 positions (Figure 4E). The conserved amino acid R around the tyrosine modified site was mainly located at the −6, −2, and +4 positions (Figure 4F). These conserved amino acids near serine, threonine, and tyrosine might be functionally important for the occurrence of phosphorylation events.
Figure 4.
The motif analyses of modified sites. (A) Visualized motif analysis logos of serine (the top ten); (B) Visualized motif analysis logos of threonine; (C) visualized motif analysis logos of tyrosine; (D) conserved amino acid around the modified serine; (E) conserved amino acid around the modified threonine; (F) conserved amino acid around the modified tyrosine residues from −6 to +6 positions.
2.4. Identification of Differential Phosphoproteins (DPs)
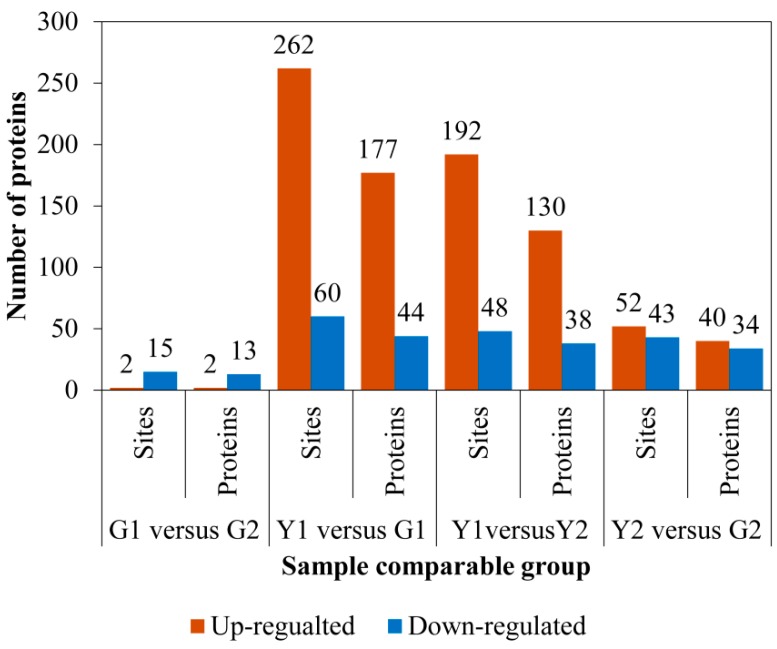
As shown in Figure 5, 221 DPs and 322 differential phosphorylation sites were identified in the Y1 versus G1 comparison, while 74 DPs and 95 differentially phosphorylated sites were identified in the Y2 versus G2 comparison. In the Y1 versus Y2 comparison, 168 differentially phosphorylated proteins and 204 differentially phosphorylated sites were identified. G1 versus G2 has only 15 differentially phosphorylated proteins and 17 phosphorylation sites, which are used to eliminate the positional effect between the Y1 and Y2 sectors. The upregulated DPs and differentially phosphorylated sites were significantly more frequent than the downregulated DPs and sites in the Y1 versus G1 and Y1 versus Y2 comparisons.
Figure 5.
Numbers of phosphorylated proteins and sites in the G1 versus G2, Y1 versus G1, Y1 versus Y2, and Y2 versus G2 comparisons.
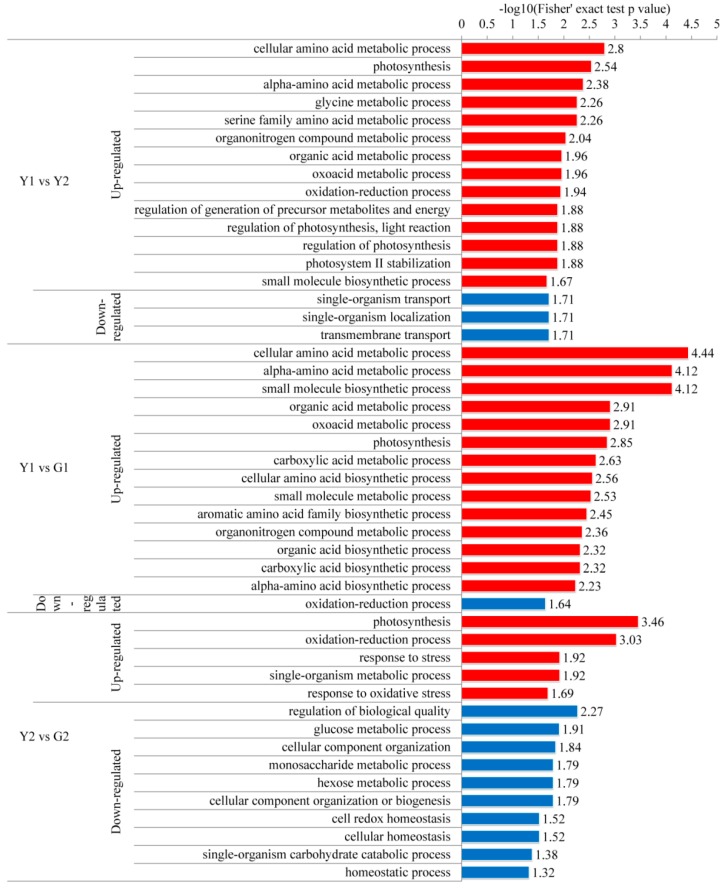
2.5. Gene Ontology (GO) Functional Classification, Subcellular Localization, and GO Enrichment Analysis
GO functional classifications were performed in the Y1 versus G1, Y2 versus G2, and Y1 versus Y2 comparisons. In terms of biological processes, the DPs of three comparisons were all enriched in the metabolic processes, cell processes, single organism processes, localization, biological regulation, stress response, etc with a slightly different scale (Figure S2). For the cellular component category, the DPs were mainly distributed among membranes, cells, macromolecular complexes and organelles (Figure S3). For the molecular function category, DPs were mainly associated with binding, catalysis, transport and structural molecular activity (Figure S4). In terms of subcellular localization, the DPs were mostly located in the chloroplast, nuclear, cytosol, mitochondria and plasma membrane (Figure S5).
GO enrichment analysis were performed based on the upregulated and downregulated levels of DPs in different comparisons. The GO terms of the Y1 versus G1 comparison were very similar to those in the Y1 versus Y2 comparison. In terms of biological processes, the processes of photosynthesis, amino acid metabolism, organic nitrogen complex metabolism, organic acid metabolism, oxoacid metabolism, redox process, and regulation of generation of precursor metabolites were upregulated in the yellow sectors, while processes associated with transport and localization were downregulated (Figure 6). In the Y2 versus G2 comparison, DPs involved in photosynthesis, oxidation–reduction process and response to stress were upregulated, while monosaccharides metabolic processes, glucose and hexose metabolic processes, single-organism carbohydrate catabolic processes and homeostatic processes were downregulated (Figure 6). For the cellular component category, the DPs in Y1 versus G1, Y1 versus Y2 and Y2 versus G2 shared a similar distribution, and were mostly located in the photosynthetic membrane, photosystem, thylakoid and thylakoid membrane (Figure S6). The differences are that non-membrane bounded organelles were downregulated in the Y1 versus G1 comparison, and cell and ribonucleoprotein complexes were downregulated in the Y2 versus G2 comparison. With respect to molecular function category, oxidoreductase activity and binding activity were upregulated, while transmembrane transport activity was downregulated in Y1 versus G1 and Y1 versus Y2 comparisons (Figure S7). In the Y2 versus G2 comparison, oxidoreductase activity, heme and tetrapyrrole binding activity, dioxygenase activity, and peroxidase activity were upregulated while carbon-carbon lyase activity and isomerase activity were downregulated (Figure S7). Taken together, the results showed that phosphorylation of proteins associated with photosynthesis, regulation of the generation of precursor metabolites, response to stress, homeostasis, amino acid metabolism, transport–related processes and most of the energy metabolic processes might contribute to leaf color.
Figure 6.
Gene ontology (GO) enrichment analysis of biological processes in the Y1 versus G1, Y1 versus Y2, and Y2 versus G2 comparisons. GO enrichment analysis of upregulated and downregulated proteins were separately.
2.6. Domain Enrichment Analysis
Domain enrichment analysis showed that the phosphorylation levels of the ubiquitin-related domain, 1,3-beta-glucan synthase subunit, ClpP/crotonase-like, and glycoside hydrolase superfamily domain were significantly upregulated, while the chlorophyll a/b binding protein domain was downregulated in the Y1 versus G1 and Y1 versus Y2 comparisons (Figure S8). In addition, argonaute, linker domains that are required for RNA-mediated gene silencing, were upregulated in the yellow sectors. Similarly, the stress-related domain, including the thioredoxin-like fold and GroES-like domain, were also significantly enriched in the yellow sectors. Furthermore, the tetratricopeptide repeat-containing domain, nucleotide-diphospho-sugar transferases, ATPase, AAA-type, core and aldolase-type TIM barrel domain were enriched in the Y2 versus G2 comparison (Figure S8). The domain enrichment analysis further helps us to link functional domains and phenotypes.
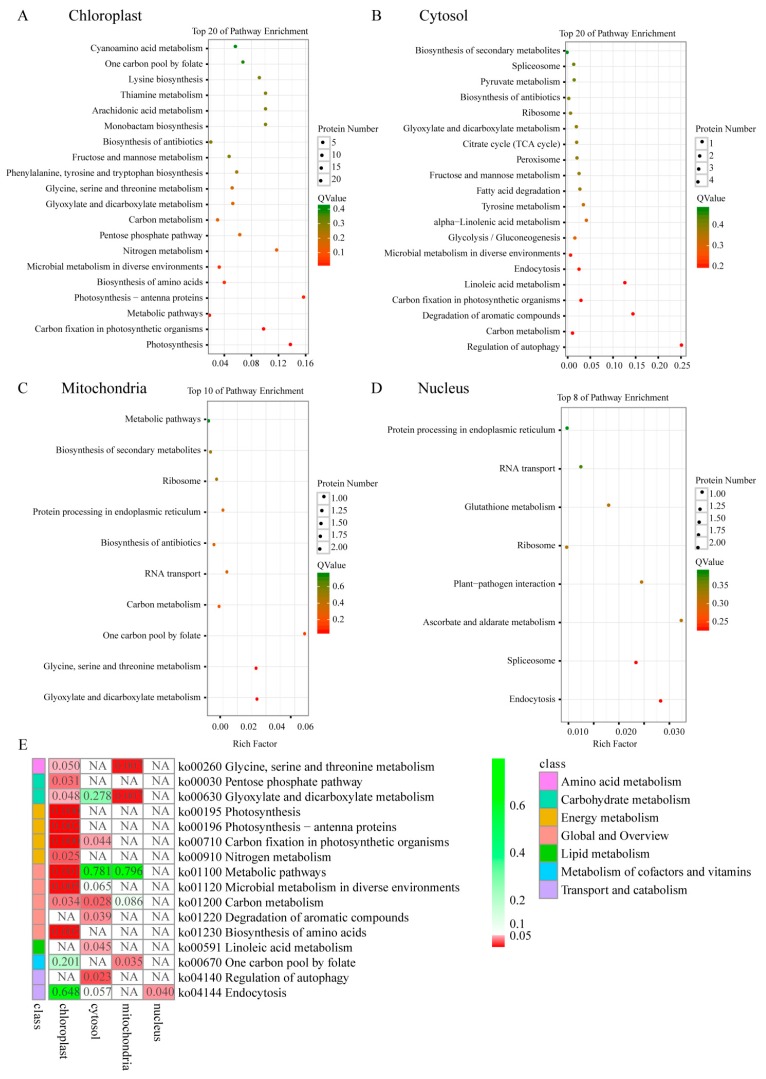
2.7. KEGG Pathway of Differential Phosphoproteins in Different Sectors of Maiyuanjinqiu
The specific phosphoproteins in the Y1 versus Y2 comparison were extracted. The common phosphoproteins identified in the G1 versus G2 comparison were removed. Further detailed KEGG pathway analysis of these specific DPs was performed based on the chloroplast, cytosol, nucleus and mitochondria. The DPs in chloroplasts were enriched in photosynthesis, photosynthetic antenna proteins, carbon fixation in photosynthetic organisms, nitrogen metabolism, carbon metabolism, pentose phosphate pathway, amino acid synthesis and microbial metabolism in diverse environments (Figure 7A). The DPs in the cytosol, DPs were mainly associated with carbon fixation in photosynthetic organisms, carbon metabolism, degradation of aromatic compounds, lipid metabolism and regulation of autophagy (Figure 7B). DPs on the mitochondria were mainly involved in amino acid metabolism, glyoxylate dicarboxylate metabolism and one-carbon pathway (Figure 7C). Although a considerable proportion of DPs were localized in the nucleus, only the endocytosis and spliceosome were significantly enriched in this subcellular compartment (Figure 7D). The DPs in the other components were mainly associated with transport and hydrolase activities. The results of KEGG pathway enrichment were consistent with those of the GO terms. By comparing the phosphorylation levels of different subcellular locations in the green and yellow sectors, we found that the central metabolic pathways were mainly enriched in the chloroplast and cytosol (Figure 7E). This result suggested that the phosphoproteins distributed in the chloroplasts and cytosol might play a key regulatory role in the formation of leaf variegation.
Figure 7.
KEGG pathway enrichment of the specific differential phosphoproteins (DPs) between the green and yellow sectors based on the chloroplasts (A), cytosol (B), mitochondria (C), and nucleus (D), the common sites from the G1 versus G2 comparison were excluded. (E), the heat map of the qv value.
2.8. Protein–Protein Interaction (PPI) Network of Phosphoproteins
Proteins in living organisms often do not function as single entities, but form a complex regulatory network to function together. The differentially phosphorylated proteins in the Y1 versus G1, Y1 versus Y2, and Y2 versus G2 comparisons were used to construct a PPI network. PPI network analysis of Y1 versus G1 and Y1 versus Y2 revealed that the differentially phosphorylated proteins between green and yellow sectors were mainly enriched in photosynthesis, ribosomes and a large number of metabolic pathways (Figure S9A,B). Phosphorylated proteins in the Y2 versus G2 comparison were mainly associated with photosynthesis, ribosomes and glycometabolism (Figure S9C). These phosphoproteins involved in photosynthesis, ribosome and energy metabolism might play an important part in the variegated phenotype.
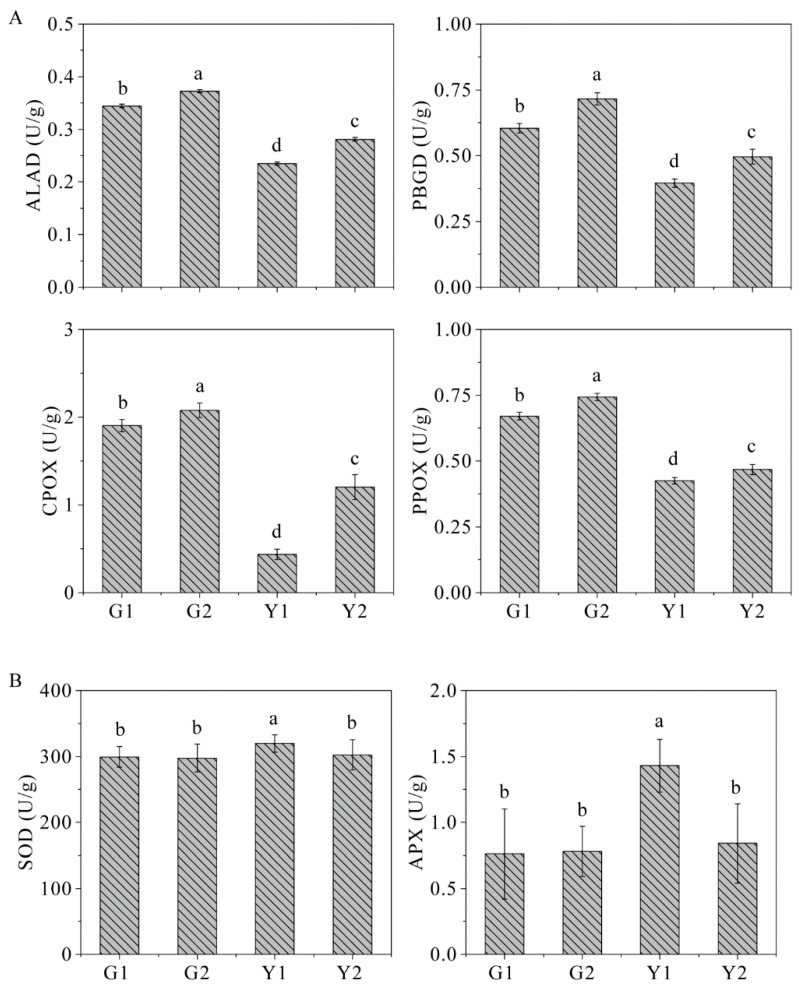
2.9. The Key Enzyme Activities
The activity of enzymes involved in chlorophyll synthesis, including ALA dehydrogenase (ALAD), porphobilinogen deaminase (PBGD), coproporphyrinogen III oxidase (CPOX), and protoporphyrinogen IX oxidase (PPOX), were further determined. The activities of these four enzymes were significantly lower in the yellow sectors than in the green sectors (Figure 8A). The activities of the antioxidant enzymes superoxide dismutase (SOD) and ascorbate peroxidase (APX) were also measured. The activities of SOD and APX were higher in the yellow leaf sectors than in the green sectors, which suggested that the ability to scavenge reactive oxygen was enhanced in the yellow sectors (Figure 8B). In addition, we enriched the phosphorylation of PBGD and APX in the yellow sectors, which suggested the phosphorylation of these enzymes might affect the processes of pigment biosynthesis and stress resistance by regulating the enzyme activities.
Figure 8.
The key enzyme activities in chlorophyll synthesis and stress resistance. (A) The enzyme activities of ALA dehydrogenase (ALAD), porphobilinogen deaminase (PBGD), coproporphyrinogen III oxidase (CPOX), and protoporphyrinogen IX oxidase (PPOX); (B) the enzyme activities of superoxide dismutase (SOD) and ascorbate peroxidase (APX).
3. Discussion
Previous researches have speculated that posttranslation modifications might play important role in the formation of variegation. However, the study of posttranslation modification is limited due to the scarcity of modified antibodies in plant kingdom. High- throughput phosphoproteomic can provide more effective information for us to understand the biological processes. In our study, we observed that the values of ETR (II) in the yellow leaves sectors were lower than those in the green sectors, while the ETR (I) values showed the opposite behavior (Figure 1D,E). These results suggested that the energy balance of PSI and PSII was redistributed in the yellow sectors, which was consistent with the fluorescence measurement results of the yellow cotyledons in the past [22]. This phenomenon attracted our attention to the differences in overall phosphorylation levels between the yellow and green sectors of Maiyuanjinqiu.
3.1. Global Analysis of Phosphorylome Profiling Associated with Leaf Color
In the Y1 versus G1 and Y1 versus Y2 comparisons, photosynthesis, carbohydrate/energy metabolism, protein metabolism, amino acid metabolic process, organonitrogen compound metabolic process, and the oxidation–reduction process were enriched based on GO enrichment (Figure 6). Our enrichment results shared similar pathways to those in the previous proteome of leaves from the chimera Hosta “gold standard” [6]. This result implied that protein phosphorylation might have a significant regulatory effect on protein expression. In the Y2 versus G2 comparison, the decreased phosphorylation levels of monosaccharides might contribute to the accumulation of increased levels of monosaccharides to respond and adapt to oxidative stress, achieving relative homeostasis in the green sectors of Maiyuanjingqiu (Figure 6). Combining domain enrichment analysis and PPI regulation network analysis, we concluded that photosynthesis and energy metabolism, protein homeostasis, and stress response were the key pathways associated with variegation. In the Y1 versus Y2 comparison (not include the common modified sites in the G1 versus G2 comparison), further KEGG pathway analysis was analyzed based on different organelle. The result showed that pigment biosynthesis, photosynthesis, energy metabolisms, stress response and defense, protein homeostasis, transcriptional regulation, and transport were most relevant to leaf color (Table A1).
3.2. DPs Involved in Pigment Biosynthesis and Photosynthesis
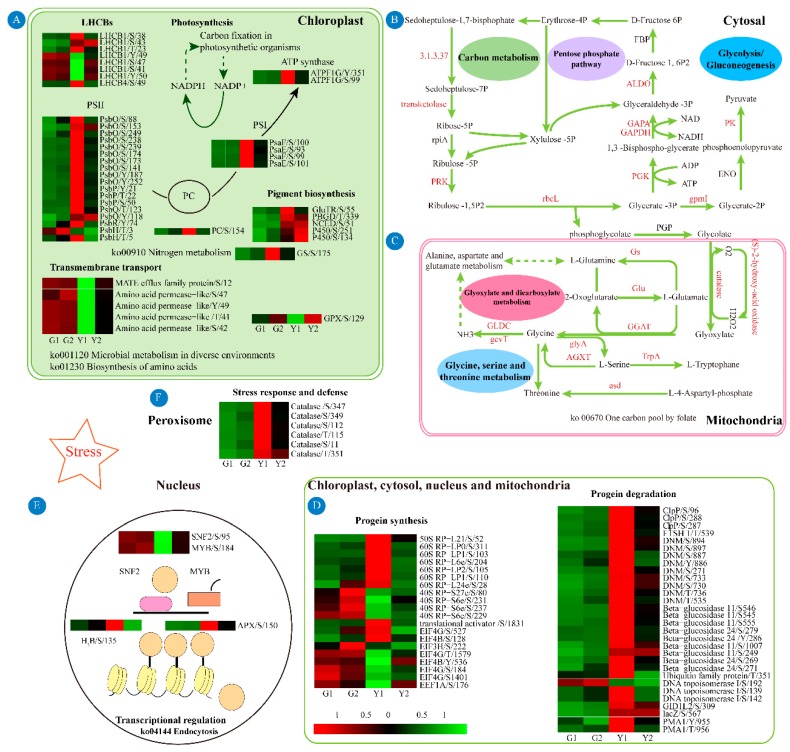
The phosphorylation levels of glutamyl-tRNA reductase (GluTR) and PBGD involved in chlorophyll biosynthesis were significantly higher in the yellow sectors than in the green sectors (Figure 9A). In contrast, the enzyme activities of ALAD, PBGD, CPOX, and PPOX involved in chlorophyll synthesis were lower in the yellow sectors (Figure 8A). These results implied that the phosphorylation of these two proteins might negatively regulate the enzyme activities, thereby affecting chlorophyll biosynthesis. Compared with the yellow sectors, the green sectors had higher phosphorylation levels of P450 family proteins (Figure 9A). P450 family proteins are involved in tetrapyrrole synthesis and catalyze the carotenoid synthesis process [23]. The globally higher phosphorylation levels of NCED in Maiyuanjinqiu might contribute to the biosynthesis of abscisic acid from carotenoids [24]. Taken together, the results indicated that phosphorylation levels of these DPs involved in pigment synthesis might be important targets for regulating leaf color.
Figure 9.
Schematic presentations of proteins involved in central metabolisms at phosphorylation level in green and yellow sectors. Differentially phosphorylated proteins are integrated according to subcellular organelles. (A) DPs localized on chloroplasts, including pathways such as photosynthesis, light-harvesting antennas and pigment biosynthesis. (B) DPs localized on the cytosal, including pathways such as glycolysis/gluconeogenesis, pentose phosphate pathway and carbon metabolism. The phosphorylated enzymes (upper right) are shown in red. The arrows represent chemical reactions. (C) DPs located on mitochondria, including glyoxylate and dicarboxylate metabolism, glycine, serine, and threonine metabolism. (D) DPs involved in protein synthesis and degradation is present in chloroplasts, cytosal, nucleus and mitochondria. (E) DPs localized on nucleus, including transcriptional regulators, such as transcription factors and histones. (F) DPs located on peroxisome, mainly stress- and defense-related proteins were presented. The expression pattern of phosphorylation levels are shown by heat maps in the G1, G2, Y1 and Y2 sectors. The phosphorylated proteins are upregulated or downregulated in red and green, respectively.
Light-harvesting antenna proteins are composed of pigment protein complexes. In our study, in addition to Thr 23, other phosphorylation sites of LHCB1 were all downregulated in the yellow sectors (Figure 9A). Studies have shown that the phosphorylation of LHCB1 is essential for optimizing the photosystem balance under changing environments [25,26]. The different phosphorylation sites of LHCB1 might play different roles in energy balance (Figure 9A). PsbO, PsbP, PsbQ, and PsbR are related to PSII stability [27]. The phosphorylation levels of these four proteins were upregulated in the yellow sectors, which might affect the distribution and rearrangement of protein complexes on the thylakoid membrane [28]. In addition, the phosphorylation levels of PSI proteins (PsaF and PsaE) were also upregulated in the yellow sectors compared to the levels in the green sectors (Figure 9A). The phosphorylation of these two proteins could affect their binding to plastocyanin (PC) and Fd and alter the distribution of LHCII to PSI [29]. The upregulated phosphorylation levels of photosynthesis-related proteins might be a regulatory adaptation mechanism for photosynthetic pigment deficiency in yellow leaves.
3.3. DPs Involved in Energy Metabolism
In addition to photosynthesis, energy metabolic processes were also significantly enriched based on GO enrichment and KEGG pathway analyses. The enriched pathways, such as glycolysis/gluconeogenesis, pentose phosphate pathway, carbon metabolism and nitrogen metabolism, were mainly located in the chloroplast and cytosol. In our study, a large portion of the enzymes involved in glycolysis/gluconeogenesis showed significant differences in phosphorylation levels between the green and yellow sectors (Figure 9B). For instance, fructose-bisphosphate aldolase (ALDO), EST C74302 (E30840) corresponds to a region of the predicted gene (GAPA), glyceraldehyde-3-phosphate dehydrogenase 2, cytosolic (GAPDH), putative 2, 3-bisphosphoglycerate-independent phosphoglycerate mutase (gpmI), 2-oxoglutarate dehydrogenase (OGDH), glucose-1-phosphate adenylyltransferase (glgC) and phosphoenolpyruvate carboxykinase (PGK) (Figure 9B). In addition, the pentose phosphate pathway is crucial to the maintenance of the redox state and plays a protective role in oxidative stress [30]. UDP-glucose 6-dehydrogenase (UGDH) catalyzes the first step of the pentose phosphate pathway. Transketolase (TK) synthesizes UDP-alpha-D-glucuronate from UDP-alpha-D-glucose, and acts as a stress sensor involved in the adaption process [31]. The expression of TK is related to the activity of the pentose phosphate pathway [32]. It has been reported that the balance between chloroplast and cytosol translation regulates the extent of var2 variegation [33]. These phosphoproteins involved in glycometabolism on chloroplast and cytosol might be important for the formation of leaf variegation. Although studies have shown that posttranslational modifications of enzymes involved in glycometabolism can respond to plant growth, development, and stress tolerance [34,35], the regulation of leaf color by the phosphorylation of these enzymes was first reported recently.
In addition to the large proportion of phosphorylated enzymes in glycometabolism, several enzymes involved in carbon fixation and carbon metabolism were also differential phosphorylated between the yellow and green sectors. For instance, phosphoribulokinase (PRK) is involved in the Calvin cycle, which is part of carbohydrate biosynthesis. Previous studies have suggested that posttranslational modifications of this protein might play a key function in the variegated leaf phenotype [6]. We supported this notion by quantifying the differential phosphorylation levels of PRK in different leaf color sectors. Similarly, the phosphorylation levels of proteins involved in nitrogen metabolism differed significantly between the green and yellow sectors. Compared with green sectors, the phosphorylation levels of glutamine synthases at Ser 175 was higher in the yellow sectors. Glutamine syntheses is a central component of nitrogen metabolism and the phosphorylation of this enzyme is related to nitrogen remobilization [36]. Therefore, we inferred that the phosphorylation of these enzymes was a prerequisite for regulating energy metabolism in different leaf color sectors.
Protein post-modification generally requires upstream enzymes to recognize substrate-specific motifs. Therefore, the recognition and study on the conserved motifs of the modified proteins are important to the prediction of post-translational modifications [37]. We found that the kinase PRK phosphorylated around the motif ...R..S......, phosphoenolpyruvate carboxykinase (PEPCK) could regulate the motif ....P.TP....., 6-phosphofructo-2-kinase/fructose-2,6-bisphosphate 2-phosphatase (PFKFB3) targeted the motif ....S.S...... and the motif ......SP..... could be recognized by putative SNF1-related protein kinase family proteins. These kinases are responsive to glucose levels, and their phosphorylation levels might modulate glycometabolism in different leaf sectors of Maiyuanjinqiu [38]. The effect of the phosphorylation sites of these kinases on leaf color remains to be further demonstrated.
3.4. DPs Involved in Stress Response and Defense
The phosphorylation levels of proteins associated with stress or stimulus were upregulated in the yellow sectors. The phosphorylation level of L-ascorbate peroxidase 3 was consistent with the increased APX enzyme activity in the yellow sectors (Figure 6 and Figure 8B). This suggested that phosphorylation at this site might positively regulate APX enzyme activity. However, the phosphorylation level of glutathione peroxidase (GPX) related to antioxidant stress was lower in the yellow sectors than in the green sectors (Figure 6). We speculated that the phosphorylation of different antioxidant enzymes might exhibit different regulatory patterns for enzyme activity. The phosphorylation levels of these proteins relative to stress or stimulus might be an adaptation mechanism in the yellow sectors [39]. Furthermore, the enriched pathways of glyoxylate and dicarboxylate metabolism and glycine, serine and threonine metabolism were upregulated in the yellow sectors. These two pathways also could participate in the response to biotic or abiotic stress (Figure 9C) [40,41]. Taken together, the results showing differential phosphorylation levels of stress-related proteins between the yellow and green sectors were remarkable. Our data supported the notion that the yellow/white sectors suffered from photooxidative stress or other stresses [1,42].
3.5. DPs Involved in Protein Homeostasis
Previous studies have shown that the homeostasis of protein synthesis and degradation is critical for the formation of variegation [43]. In our study, the phosphorylation levels of the translation initiation factors EIF4G-1, EIF4G and EIF4B were not uniformly upregulated or downregulated in the yellow leaves, and these different phosphorylation sites might have different regulatory effects on protein function (Figure 9D). EIF4G acts as a hub protein together with eIF3 and additional initiation factors, recruiting 40S ribosomal subunits to mRNA [44,45]. The chloroplast translation initiation factors (translation initiation factor 3) could regulate the var2-mediated leaf variegation [46,47]. The phosphorylation levels of the 40S ribosomal protein subunits were downregulated in the yellow leaves, while those of the 60S ribosome were upregulated (Figure 9D). This result was contrary to the differential phosphorylation observed in sugarcane mosaic virus infection [48].
Protein hydrolysis or proteolysis is essential for protein homeostasis [49]. The Clp protease system and metalloprotease FTSH are the most important stromal proteases in chloroplast because they are responsible for normal protein turnover and chloroplast development [50]. In the yellow sectors, the phosphorylation levels of ATP -dependent protease Clp protease hydrolysis subunit (Ser 287 and 288) and FTSH1 (Thr 539) were upregulated (Figure 9D). The Clp protease system could control the ALA synthesis through posttranslational regulation of GluTR [51]. The higher phosphorylation level of Clp protease subunits might play a direct or indirect role in chlorophyll biosynthesis. The increased phosphorylation levels of autophagy in the yellow sectors might be related to the phosphorylation of Clp protease [52]. In Arabidopsis thaliana, the absence of FTSH1 presents a typical variegated leaf phenotype [53]. The phosphorylation of FTSH1 can change its hydrolytic activity and affect the formation of the photosynthetic complex [54,55]. In the yellow leaves, the phosphorylation levels of other proteins involved in protein degradation or hydrolysis were also upregulated, including putative dynamin homolog, β-glucosidase family protein, glycoside hydrolase family and ubiquitin family proteins (Figure 9D). These differences in protein synthesis and degradation might affect protein homeostasis and leaf color.
3.6. DPs Involved in Transcriptional Regulation and Transport
Compared with those in the green sectors, the phosphorylation levels of proteins involved in transcriptional regulation, such as SNF2 and MYB, were globally downregulated in the yellow sectors. In contrast, the phosphorylation levels of zinc finger domains were upregulated in the yellow sectors (Figure 9E). A recent study showed that the SNF2 chromatin-remodeling ATPase enzyme BRM could modulate chlorophyll biosynthesis, and that BRM activity could be controlled by phosphorylation /dephosphorylation [56,57]. The phosphorylated MYB transcription factors could optimize the flavonoid metabolism in soybean under salt stress and could regulate anthocyanin biosynthesis by interacting with bHLH and WD40 [58,59]. We also identified upregulated (1.684-fold) phosphorylation at the Ser 135 site of histone H2B in the yellow sectors (Figure 9E). Previous research indicated that the phosphorylation of H2B at Ser6 contributed to chromosomal stability [60]. These phosphoproteins involved in transcriptional regulation might play an important role in leaf color. Furthermore, we found that the phosphorylation levels of enriched pathways were all upregulated except for single-organism transport, localization, and transmembrane transport in yellow leaves (Figure 6 and Figure 9A). The differential phosphorylation of proteins in these processes might mediate the direction of sugar transport in Maiyuanjinqiu, which was of great significance for the variegated phenotype.
4. Materials and Methods
4.1. Plant Material
Maiyuanjinqiu is a variety derived from C. fargesii seedlings. The leaves of Maiyuanjinqiu display a variegated phenotype. Maiyuanjinqiu and C. fargesii were both cultivated in the experimental field (Luoyang, China). The leaves were separately sampled according to the marked positions shown in Figure 1B. The samples were wrapped in tin foil paper, immediately frozen in liquid nitrogen, and then stored at −80 °C until use. The frozen samples were used for the protein extraction and quantitative phosphorylomic analysis.
4.2. Determination of Photosynthetic Fluorescence Parameters and Pigment Contents
Potted plants were developed in the greenhouse of Chinese Academy of Forestry. A chlorophyll fluorescence imager (CFI) (Technologica, Essex, UK) was used to obtain the imaging photo of Fv/Fm, following the method of Baker and Oxborough [61] (n ≥ 10). ETR(I), ETR(II), and NPQ were detected by a Dual-PAM-100 fluorometer (Walz, Effeltrich, Germany) and the photosynthetically active radiation was set as 129 µmol·m−2·s−1, which was suitable for the determination of different leaf color sectors (n = 18). The third leaves from top to bottom were selected for determination. After the determination, samples were taken for the measurement of pigment contents. The levels of chlorophyll and carotenoid were determined according to the method described by Lichtenthaler [62] (n ≥ 4). The lutein content was detected with ELISA Kits (Mlbio, Shanghai, China) (n ≥ 8).
4.3. Protein Extraction, Trypsin Digestion, and TMT Labeling
The protein extraction was performed by the method of Tong, et al. [63] with some modification. The frozen leaves were ground into a fine powder and sonicated on ice 3 times in lysis buffer. After centrifugation, the obtained supernatant was precipitated with precooled TCA, followed by washing 3 times with cold acetone. The acetone was evaporated at 4 °C, and the powder was redissolved in buffer (8 M urea, 100 mM TEAB, pH = 8). Before the next step, the protein concentration was quantified using a 2-D Quant Kit (GE Healthcare, Piscataway, NJ, USA).
Protein reduction, alkylation, and dilution were performed by the method of Xu, et al. [64]. The protein solution was reduced at 37 °C and alkylated at room temperature. We modified the urea concentration to less than 2 M. For subsequent trypsin digestion and TMT labeling, the method refers to the previously described [63].
4.4. HPLC Fractionation and Affinity Enrichment
HPLC fractionation was performed by using an acetonitrile gradient; the 80 fractions obtained by high pH reverse-phase HPLC were combined into 14 components. The phosphopeptides were enriched using IMAC combined with Ti4+ enrichment. The Ti4+-IMAC material was prepared as described in the previous research [20]. The non-specific phosphopeptides were removed and the obtained phosphopeptides were eluted with 10% NH4OH with vibration. The collected phosphopeptides were lyophilized for LC-MS/MS analysis.
4.5. LC-MS/MS Analysis
The peptides were dissolved with 0.1% formic acid (Fluka, Buchs, Switzerland) in 2% acetonitrile (Thermo Scientific, San Jose, CA, USA), and then loaded directly onto the reversed-phase analysis column. Intact peptides were detected by tandem mass spectrometry (MS/MS) in an Orbitrap FusionTM TribridTM (Thermo Scientific, San Jose, CA, USA). For a full range mass scan, the resolution was set to 60,000, while the ion fragments were detected at a resolution of 15,000. For MS/MS, 35% normalized collision energy (NCE) was applied. The threshold of precursor ions was greater than 5E3, and the dynamic exclusion was set to 15.0 s. One MS scan was alternated sequentially with 20 scans. Accumulated 5E4 ions were used for the whole MS/MS spectra of the phosphorylome. The MS scan range was 350 to 1550 m/z, and the fixed first mass was 100 m/z.
4.6. Database Search and Data Analysis
The spectrometric data were searched by MaxQuant software with the Andromeda search engine (v.1.5.2.8, Martinsried, Germany). Tandem mass spectra were matched against the Catalpa bungei protein database) [65]. The false discovery rate (FDR) was specified as 1%. The site location probability was greater than 0.5. The significant fold-change cutoff was 1.5-fold for differential proteins in the Y1 versus G1, Y1 versus Y2 and Y2 versus G2 comparisons. The fold changes of phosphorylated proteins were normalized to the protein fold change in proteome [66]. The raw phosphoproteomics data was deposited in the ProteomeXchange with identifier PXD013065.
4.7. Bioinformatics Methods
Soft motif-x was used to analyze the upstream and downstream amino acids of the modified site in all the phosphoproteins. Functional enrichments were tested using a two tailed Fisher’s exact test, and a corrected p value < 0.05 was considered significant. Subcellular localization prediction was conducted by the updated version of WOLFPSORT (http://www.genscript.com/wolf-psort.html). GO annotation of the phosphoproteins was derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). InterProScan was used to annotate the unmapped proteins using the protein sequence. The identified protein domain descriptions were annotated by the InterPro database (http://www.ebi.ac.uk/interpro/). To better understand the mechanism of why the leaves had two colors with the same genetic background, we focused on the DPs in the Y1 versus Y2 comparison for KEGG pathway analysis, removing the DPs in the G1 versus G2 comparison. To understand which organelles play a key role in the formation of variegation, the analysis was based on the chloroplast, cytosol, nuclear and mitochondria. The KEGG database (http:// www.genome.jp/kegg/) was used to annotate the protein pathways. PPIs were predicted by STRING database version 10.5 (http://string-db.org/). The filtering scores was set as ≥0.7 (high confidence), and the mapping organism was Populus trichocarpa. The PPIs of the DPs in different comparisons were visualized by the Cytoscape software.
4.8. Key Enzyme Activities
The enzyme activities of ALAD, PBGD, CPOX, PPOX, SOD, and APX were measured following the protocol described in the instructions of ELISA Kits (Mlbio, Shanghai, China).
5. Conclusions
This study is the first to dissect the differences in global phosphorylation levels in different leaf color sectors in woody plants. As a result, 1434 phosphoproteins were quantified, among which, 165 phosphoproteins were specifically related to the color sectors in Maiyuanjinqiu. Integrative bioinformatics analysis revealed that pigment biosynthesis, photosynthesis, and energy metabolism, protein homeostasis, stress response, transcriptional regulation, and transport were the key pathways contributing to variegation. KEGG pathway enrichment suggested that the enzymes located in the chloroplasts and cytosol might play a central role in the variegated phenotype. The phosphorylation function of kinases, such as PRK, PEPCK, PFKFB3, and SNF1-related protein kinase family proteins, needs to be further researched. Similarly, further functional studies of the role of FTSH1 phosphorylation in leaf development would be of great significance. In addition, physiological and biochemical measurements confirmed that protein phosphorylation could affect enzyme activities and plant physiology.
Acknowledgments
We thank Jianwei Ma, Qiuling Zhao and Huiling Yun for providing the original material. We also acknowledge Kun Zhao and Yunde Jiao for their assistance on sampling.
Abbreviations
| Fv/Fm | Maximum quantum yield of PSII |
| ETR | Electron transport rate |
| NPQ | Non photochemical quenching |
| PS | Photosystem |
| LC-MS/MS | Liquid chromatography–mass spectrometry/mass spectrometry |
| GO | Gene ontology |
| DPs | Differential phosphoproteins |
| LHCB | Chlorophyll a-b binding protein |
| ATPF 1G | ATP synthase subunit gamma |
| PsbO | Oxygen-evolving enhancer protein 1 |
| PsbP | Oxygen-evolving enhancer protein 2 |
| PsbQ | Oxygen-evolving enhancer protein 3 |
| PsbR | Photosystem II reaction center protein H |
| PsbH | Photosystem II reaction center protein H |
| LHCB | Chlorophyll a-b binding protein |
| PsaE | Putative Photosystem I reaction center subunit IV |
| PsaF | Putative photosystem I reaction center subunit III |
| PC | Plastocyanin |
| GluTR | Glutamyl-tRNA reductase |
| NCED | 9-cis-epoxycarotenoid dioxygenase 1 |
| P450 | Cytochrome P450 family protein, expressed |
| 3.1.3.37 | Putative fructose-1,6-bisphosphatase |
| PRK | Phosphoribulokinase |
| rbcL | Ribulose bisphosphate carboxylase large chain |
| ALDO | Fructose-bisphosphate aldolase |
| GAPA | EST C74302 (E30840) corresponds to a region of the predicted gene |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase 2, cytosolic |
| PGK | Phosphoenolpyruvate carboxykinase |
| PK | Pyruvate kinase |
| gpml | Putative 2, 3-bisphosphoglycerate-independent phosphoglycerate mutase |
| glgC | Glucose-1-phosphate adenylyltransferase |
| Glu | Glutamine syntheses |
| TrpA | Tryptophan synthase alpha chain |
| GLDC | Putative glycine dehydrogenase |
| GGAT | Alanine aminotransferase |
| gcvT | Aminomethyltransferase |
| AGXT | Serine-glyoxylate aminotransferase |
| asd | Aspartate-semialdehyde dehydrogenase family protein, expressed |
| SNF2 | SNF2 transcription factor |
| MYB | MYB transcription factor |
| H2B | Histone H2B |
| APX | Ascorbate peroxidase |
| RP | Ribosomal protein |
| EIF | Eukaryotic translation initiation factor |
| Clp | ATP-dependent Clp protease proteolytic subunit |
| FTSH1 | ATP-dependent zinc metalloprotease FTSH 1, chloroplastic |
| DNM | Putative dynamin homolog |
| GIDIL | Gibberellin receptor GID1L2 |
| lacZ | Glycoside hydrolase family 2 family protein |
| PMA | H-ATPase |
| ALAD | ALA dehydrogenase |
| PBGD | Porphobilinogen deaminase |
| CPOX | Coproporphyrinogen III oxidase |
| PPOX | Protoporphyrinogen IX oxidase |
| SOD | Superoxide dismutase |
| ELISA | Enzyme-linked immunosorbent assay |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/8/1895/s1.
Appendix A
Table A1.
The phosphorylation modification levels of the key proteins on the enriched pathways based on functional enrichment analysis in G1, G2, Y1 and Y2.
| Protein Description (Abbreviation/Phosphorylated Amino Acid/Site) | G1 | G2 | Y1 | Y2 |
|---|---|---|---|---|
| Pigment biosynthesis | ||||
| Glutamyl-tRNA reductase (GluTR/S/55) | 0.53 | 0.69 | 2.37 | 0.92 |
| Porphobilinogen deaminase, chloroplastic (hemC/T/339) | 0.72 | 0.78 | 1.34 | 1.17 |
| 9-cis-epoxycarotenoid dioxygenase 1 (NCED/S/51) | 0.42 | 0.44 | 1.66 | 1.84 |
| Cytochrome P450 family protein, expressed (P450/S/251) | 0.53 | 0.74 | 1.17 | 1.52 |
| Cytochrome P450 family protein, expressed (P450/S/134) | 0.46 | 0.69 | 1.08 | 1.62 |
| Photosynthesis | ||||
| Oxygen-evolving enhancer protein 1 (PsbO/Y/252) | 0.26 | 0.34 | 3.29 | 0.95 |
| Oxygen-evolving enhancer protein 1 (PsbO/S/239) | 0.35 | 0.48 | 3.12 | 0.84 |
| Oxygen-evolving enhancer protein 1 (PsbO/S/174) | 0.20 | 0.27 | 3.02 | 0.88 |
| Oxygen-evolving enhancer protein 1 (PsbO/S/173) | 0.40 | 0.46 | 2.73 | 0.76 |
| Oxygen-evolving enhancer protein 1 (PsbO/S/141) | 0.50 | 0.67 | 2.65 | 0.80 |
| Oxygen-evolving enhancer protein 1 (PsbO/Y/187) | 0.37 | 0.48 | 2.64 | 0.83 |
| Oxygen-evolving enhancer protein 1 (PsbO/S/88) | 0.77 | 0.99 | 1.46 | 0.90 |
| Oxygen-evolving enhancer protein 1 (PsbO/S/153) | 0.77 | 0.82 | 1.35 | 1.18 |
| Oxygen-evolving enhancer protein 1 (PsbO/S/249) | 0.86 | 0.97 | 1.34 | 0.95 |
| Oxygen-evolving enhancer protein 1 (PsbO/S/238) | 0.16 | 0.21 | 3.65 | 0.96 |
| Oxygen-evolving enhancer protein 2 (PsbP/S/50) | 0.26 | 0.30 | 5.13 | 0.94 |
| Oxygen-evolving enhancer 2-1, chloroplastic (PsbP/Y/21) | 0.89 | 0.95 | 1.71 | 1.04 |
| Oxygen-evolving enhancer 2-1, chloroplastic (PsbP/T/22) | 0.89 | 0.94 | 1.64 | 1.07 |
| Oxygen-evolving enhancer protein 3-1 (PsbQ/T/123) | 0.84 | 0.81 | 2.54 | 1.28 |
| Oxygen-evolving enhancer protein 3-1 (PsbQ/Y/118) | 0.82 | 0.92 | 1.30 | 1.39 |
| Photosystem II reaction center protein H (PsbH/T/3) | 1.05 | 1.18 | 0.85 | 0.76 |
| Photosystem II reaction center protein H (PsbH/T/5) | 0.95 | 1.19 | 2.08 | 0.68 |
| Photosystem II 10 kDa polypeptide (PsbR/Y/74) | 0.79 | 1.13 | 1.64 | 0.93 |
| Plastocyanin (PetE/S/154) | 0.79 | 0.88 | 1.32 | 0.85 |
| Putative photosystem I reaction center subunit III (PsaF/S/100) | 0.69 | 0.78 | 3.53 | 1.38 |
| Putative Photosystem I reaction center subunit IV (PsaE/S/93) | 0.42 | 0.50 | 3.46 | 1.37 |
| Putative Photosystem I reaction center subunit IV (PsaE/S/101) | 0.74 | 0.77 | 2.31 | 1.05 |
| Putative photosystem I reaction center subunit III (PsaF/S/99) | 0.69 | 0.78 | 3.39 | 1.35 |
| ATP synthase subunit gamma (ATPF1G/S/348) | 0.53 | 0.65 | 3.33 | 1.13 |
| ATP synthase subunit gamma (ATPF1G/Y/351) | 0.55 | 0.66 | 3.28 | 1.11 |
| ATP synthase subunit gamma (ATPF1G/S/99) | 0.55 | 0.66 | 3.28 | 1.11 |
| Photosynthesis –antenna proteins | ||||
| Chlorophyll a/b-binding protein (LHCB4/S/49) | 0.94 | 0.97 | 2.15 | 0.91 |
| Chlorophyll a/b-binding protein (LHCB4/S/49) | 0.94 | 0.97 | 2.15 | 0.91 |
| Chlorophyll a-b binding protein 2, chloroplastic (LHCB1/S/38) | 0.76 | 0.89 | 3.90 | 0.90 |
| Chlorophyll a-b binding protein 1(LHCB1/T/23) | 0.76 | 0.89 | 1.73 | 1.01 |
| Chlorophyll a-b binding protein 1 (LHCB1/S/43) | 0.48 | 0.59 | 1.66 | 1.97 |
| Chlorophyll a-b binding protein 1 (LHCB1/Y/49) | 1.14 | 1.09 | 0.59 | 0.84 |
| Chlorophyll a-b binding protein 2, chloroplastic (LHCB1/S/47) | 1.01 | 1.00 | 0.45 | 1.01 |
| Chlorophyll a-b binding protein 1 (LHCB1/S/41) | 1.11 | 1.08 | 0.42 | 1.00 |
| Chlorophyll a-b binding protein 2, chloroplastic (LHCB1/Y/50) | 1.05 | 0.97 | 0.27 | 1.01 |
| Stress and stimulus response proteins | ||||
| Catalase (katE/S/347) | 0.52 | 0.59 | 2.23 | 1.12 |
| Catalase (katE/S/349) | 0.49 | 0.70 | 2.16 | 1.03 |
| Catalase (katE/S/112) | 0.55 | 0.66 | 2.00 | 1.15 |
| Catalase (katE/T/115) | 0.59 | 0.65 | 1.95 | 1.14 |
| Catalase (katE/S/11) | 0.78 | 0.85 | 1.35 | 0.98 |
| Catalase (katE/T/351) | 0.47 | 0.59 | 1.85 | 1.41 |
| Probable L-ascorbate peroxidase 3 (E1.11.1.11/S/150) | 0.70 | 0.74 | 2.19 | 1.25 |
| OSJNBb0048E02.12 protein (OSJNBb0048E02.12/S/8) | 0.62 | 0.78 | 1.44 | 0.89 |
| Glutaredoxin domain containing protein (S/154) | 0.66 | 1.02 | 1.43 | 0.94 |
| OSJNBb0048E02.12 protein (OSJNBb0048E02.12/S/50) | 0.80 | 0.77 | 1.40 | 0.81 |
| OSJNBb0048E02.12 protein (OSJNBb0048E02.12/Y/3) | 0.66 | 0.78 | 1.39 | 0.96 |
| OSJNBb0048E02.12 protein (OSJNBb0048E02.12/S/5) | 0.68 | 0.8 | 1.36 | 0.98 |
| Putative cinnamyl-alcohol dehydrogenase family protein (E1.1.1.195/T/7) | 1.00 | 1.08 | 1.39 | 0.90 |
| Glutaredoxin domain containing protein (S/99) | 0.87 | 1.00 | 1.38 | 0.79 |
| Glutaredoxin domain containing protein (S/46) | 0.87 | 1.06 | 0.79 | 1.27 |
| Probable cinnamyl alcohol dehydrogenase 3 (S/20) | 0.90 | 1.03 | 1.38 | 0.83 |
| Glutaredoxin domain containing protein (S/103) | 0.86 | 1.00 | 1.34 | 0.84 |
| Putative nuclear snf4-like (S/48) | 0.91 | 0.98 | 1.32 | 0.87 |
| Homeobox-leucine zipper protein ROC1 (S/46) | 1.01 | 1.08 | 4.2 | 0.40 |
| Lipoxygenase (LOX2S/S/76) | 0.46 | 0.59 | 2.71 | 1.31 |
| Lipoxygenase (LOX2S/S/74) | 0.44 | 0.51 | 2.7 | 1.5 |
| Putative ENTH/ANTH/VHS superfamily protein isoform 1 (S/180) | 0.78 | 0.81 | 1.78 | 0.71 |
| Integral membrane single C2 domain protein (T/475) | 0.83 | 0.90 | 1.66 | 0.73 |
| Integral membrane single C2 domain protein (T/473) | 0.82 | 0.93 | 1.64 | 0.73 |
| Clathrin assembly protein AP180 short form-like (S/306) | 0.84 | 0.91 | 1.62 | 0.78 |
| Clathrin assembly protein AP180 short form-like (T/307) | 0.93 | 0.94 | 1.42 | 0.77 |
| Putative ENTH/ANTH/VHS superfamily protein isoform 1 (T/221) | 0.91 | 0.93 | 1.26 | 0.78 |
| Peroxisomal (S)-2-hydroxy-acid oxidase GLO5 (HAO/T/159) | 0.97 | 1.14 | 1.22 | 0.79 |
| Monodehydroascorbate reductase (E1.6.5.4/S/344) | 0.74 | 1.33 | 1.17 | 0.74 |
| Monodehydroascorbate reductase (E1.6.5.4/T/346) | 0.69 | 1.30 | 1.16 | 0.86 |
| Chaperonin (Chaperonin/T/125) | 0.73 | 0.81 | 1.09 | 1.25 |
| Glutathione peroxidase (gpx/S/129) | 0.98 | 1.19 | 0.75 | 1.33 |
| Putative C2 domain-containing protein (S/406) | 0.87 | 1.05 | 0.71 | 1.32 |
| L-lactate dehydrogenase (LDH/S/26) | 1.22 | 1.24 | 0.75 | 0.81 |
| Energy and metabolism | ||||
| Fructose-bisphosphate aldolase (ALDO/T/88) | 0.67 | 0.67 | 3.48 | 0.97 |
| Fructose-bisphosphate aldolase (ALDO/S/42) | 0.56 | 0.70 | 2.72 | 0.78 |
| Fructose-bisphosphate aldolase (ALDO/S/82) | 0.67 | 0.88 | 1.68 | 0.91 |
| Fructose-bisphosphate aldolase (ALDO/S/73) | 0.76 | 1.06 | 1.44 | 0.93 |
| Fructose-bisphosphate aldolase (ALDO/S/83) | 0.79 | 0.91 | 1.44 | 0.82 |
| Fructose-bisphosphate aldolase (ALDO/S/349) | 0.92 | 0.99 | 1.42 | 0.76 |
| Fructose-bisphosphate aldolase (ALDO/T/80) | 0.83 | 0.96 | 1.36 | 1.13 |
| Transketolase isoform 1 (transketolase [EC:2.2.1.1]/Y/426) | 0.66 | 0.67 | 3.35 | 0.77 |
| Transketolase isoform 1(transketolase ([EC:2.2.1.1]/S/434) | 0.77 | 0.89 | 1.76 | 0.78 |
| Transketolase isoform 1 ( transketolase [EC:2.2.1.1]/S/430) | 0.97 | 1.14 | 1.22 | 0.83 |
| EST C74302(E30840) corresponds to a region of the predicted gene (GAPA/S/26) | 0.40 | 0.57 | 2.35 | 0.79 |
| EST C74302(E30840) corresponds to a region of the predicted gene (GAPA/S/27) | 0.48 | 0.59 | 2.27 | 0.82 |
| EST C74302(E30840) corresponds to a region of the predicted gene (GAPA/S/25) | 0.75 | 0.82 | 1.52 | 0.81 |
| EST C74302(E30840) corresponds to a region of the predicted gene (GAPA/S/278) | 0.81 | 0.90 | 1.37 | 1.14 |
| EST C74302(E30840) corresponds to a region of the predicted gene (GAPA/T/295) | 0.90 | 1.29 | 1.12 | 0.99 |
| putative 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (gpmI/S/81) | 0.84 | 0.88 | 1.53 | 0.77 |
| Phosphoenolpyruvate carboxykinase (ATP), conserved site-containing protein (E4.1.1.49/T/71) | 0.76 | 1.06 | 1.44 | 0.66 |
| Phosphoglucomutase/phosphomannomutase, C-terminal domain, putative (pgn/T/5) | 0.81 | 0.86 | 1.27 | 0.75 |
| Carbonic anhydrase (cynT/S/111) | 0.51 | 0.66 | 2.14 | 0.75 |
| Carbonic anhydrase (cynT/S/109) | 0.80 | 0.86 | 1.19 | 0.78 |
| Carbonic anhydrase (cynT/S/83) | 0.84 | 1.01 | 1.52 | 1.26 |
| Phosphoribulokinase (PRK/T/78) | 0.60 | 0.60 | 2.03 | 0.85 |
| Phosphoribulokinase (PRK/S/219) | 0.64 | 0.80 | 1.45 | 0.94 |
| Phosphoribulokinase (PRK/S/79) | 0.88 | 1.03 | 1.45 | 0.85 |
| Putative fructose-1,6-bisphosphatase (E3.1.3.37/S/320) | 0.72 | 0.82 | 1.80 | 0.83 |
| Serine-glyoxylate aminotransferase (AGXT/Y/35) | 0.85 | 0.87 | 1.76 | 1.02 |
| Serine-glyoxylate aminotransferase (AGXT/S/37) | 0.69 | 0.73 | 1.46 | 1.00 |
| Ribulose bisphosphate carboxylase small chain (rbcS/S/114) | 0.7 | 1.24 | 1.59 | 1.16 |
| Ribulose bisphosphate carboxylase small chain (rbcS/Y/118) | 1.01 | 1.00 | 1.37 | 1.21 |
| Ribulose bisphosphate carboxylase small chain (rbcS/Y/117) | 0.95 | 1.26 | 1.21 | 1.20 |
| GLO5 Peroxisomal (S)-2-hydroxy-acid oxidase GLO5 (HAO/T/159) | 0.79 | 0.97 | 1.42 | 0.79 |
| 2-oxoglutarate dehydrogenase, putative (OGDH/S/344) | 0.94 | 1.01 | 1.33 | 0.91 |
| Putative fructose-1,6-bisphosphatase (E3.1.3.37/S/318) | 0.88 | 0.89 | 1.31 | 1.10 |
| Phosphoglycerate kinase (PGK/S/276) | 0.83 | 0.94 | 1.26 | 1.05 |
| Aminomethyltransferase (gcvT/S/268) | 0.75 | 0.94 | 2.09 | 0.92 |
| Aminomethyltransferase (gcvT/Y/390) | 1.02 | 1.22 | 1.10 | 1.10 |
| Aminomethyltransferase (gcvT/S/339) | 0.74 | 1.02 | 0.83 | 0.82 |
| Glucose-1-phosphate adenylyltransferase (glgC/S/263) | 0.39 | 0.41 | 2.13 | 1.10 |
| Glucose-1-phosphate adenylyltransferase (glgC/S/424) | 0.48 | 0.47 | 2.05 | 1.05 |
| UDP-glucose 6-dehydrogenase (UGDH/S/393) | 1.15 | 1.12 | 0.69 | 0.96 |
| 1,3-beta-glucan synthase component-like (S/1019) | 0.80 | 1.04 | 1.43 | 0.79 |
| Putative beta 1,3 glucan synthase (T/27) | 0.76 | 1.05 | 1.41 | 0.91 |
| Serine hydroxymethyltransferase (glyA/S/384) | 0.72 | 0.82 | 1.8 | 1.06 |
| Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplastic (E2.5.1.54/S/67) | 0.43 | 0.47 | 1.72 | 1.10 |
| Aspartate-semialdehyde dehydrogenase family protein, expressed (ASD/S/137) | 0.72 | 0.79 | 1.65 | 0.78 |
| Orange pericarp1 (trpB/S/76) | 0.79 | 0.97 | 1.48 | 0.91 |
| Cellulose synthase-like protein E6 (IPR029044/S/393) | 0.5 | 0.63 | 1.8 | 1.36 |
| Glyceraldehyde-3-phosphate dehydrogenase 2, cytosolic (GAPDH/Y/257) | 0.75 | 0.82 | 1.52 | 0.74 |
| Putative beta 1,3 glucan synthase (S/29) | 0.81 | 1.08 | 1.29 | 0.90 |
| putative 6-phosphofructo-2-kinase / fructose-2, 6-bisphosphate 2-phosphatase(S/276) | 1.04 | 1.16 | 0.56 | 1.28 |
| Glutamine synthetase (Gs/S/175) | 0.76 | 0.87 | 1.61 | 0.98 |
| Putative glycine dehydrogenase (GLDC/S/51) | 0.77 | 0.89 | 1.76 | 0.95 |
| Thiamine thiazole synthase, chloroplastic (THI4/S/27) | 0.37 | 0.37 | 2.41 | 0.62 |
| Nitrogen regulatory protein P-II homolog (/T114) | 0.51 | 0.62 | 1.71 | 0.63 |
| Nitrogen regulatory protein P-II homolog (/T113) | 0.57 | 0.7 | 1.61 | 0.68 |
| Nitrate-induced NOI protein, expressed (RIN4/T/161) | 0.61 | 0.96 | 1.53 | 0.70 |
| Nitrate-induced NOI protein, expressed (RIN4/T/206) | 0.77 | 1.13 | 1.34 | 0.64 |
| Nitrate-induced NOI protein, expressed (RIN4/T/216) | 0.74 | 1.16 | 1.32 | 0.68 |
| Nitrate-induced NOI protein, expressed (RIN4/T/227) | 0.79 | 1.07 | 1.22 | 0.88 |
| Nitrate-induced NOI protein, expressed (RIN4/T/168) | 0.77 | 0.78 | 1.54 | 0.63 |
| Nitrate-induced NOI protein, expressed (RIN4/S/170) | 0.82 | 0.74 | 1.39 | 0.68 |
| Nitrate-induced NOI protein, expressed (RIN4/S/96) | 0.96 | 0.95 | 1.31 | 0.66 |
| Nitrate-induced NOI protein, expressed (RIN4/T/165) | 0.79 | 0.88 | 1.4 | 0.75 |
| Nitrate-induced NOI protein, expressed (RIN4/T/228) | 0.86 | 0.87 | 1.32 | 0.82 |
| Nitrate-induced NOI protein, expressed (RIN4/S/154) | 0.99 | 1.03 | 1.19 | 0.77 |
| Nitrate-induced NOI protein, expressed (RIN4/S/183) | 0.87 | 0.91 | 1.28 | 0.84 |
| Nitrate-induced NOI protein, expressed (RIN4/S/127) | 0.93 | 0.96 | 1.23 | 0.81 |
| Nitrate-induced NOI protein, expressed (RIN4/S/164) | 0.83 | 0.92 | 1.28 | 0.85 |
| Nitrate-induced NOI protein, expressed (RIN4/S/225) | 0.72 | 1.31 | 1.09 | 0.92 |
| Nitrate-induced NOI protein, expressed (RIN4/S/133) | 1.22 | 1.16 | 0.97 | 0.77 |
| Nitrate-induced NOI protein, expressed (RIN4/S/185) | 1.59 | 1.84 | 0.4 | 0.43 |
| Nitrate-induced NOI protein, expressed (RIN4/S/95) | 1.17 | 1.39 | 0.75 | 0.84 |
| Protein synthesis | ||||
| 50S ribosomal protein L21 (RP-L21/S/52) | 0.32 | 0.38 | 2.4 | 1.18 |
| 60S acidic ribosomal protein P0 (RP-LP0/S/311) | 0.25 | 0.35 | 1.53 | 0.25 |
| 60S acidic ribosomal protein P1 (RP-LP1/S/103) | 0.23 | 0.21 | 1.51 | 0.28 |
| 60S ribosomal protein L6 (RP-L6e/S/204) | 0.9 | 0.92 | 1.36 | 0.86 |
| 60S acidic ribosomal protein P2A (RP-LP2/S/105) | 0.15 | 0.34 | 1.33 | 0.15 |
| 60S acidic ribosomal protein P3 (RP-LP1/S/110) | 0.35 | 0.47 | 1.31 | 0.37 |
| 60S ribosomal protein L24 (RP-L24e/S/28) | 0.82 | 1.09 | 1.24 | 0.86 |
| 40S ribosomal protein S27 (RP-S27e/S/80) | 1.15 | 1.70 | 0.65 | 0.80 |
| 40S ribosomal protein S6 (RP-S6e/S/231) | 1.00 | 1.17 | 0.65 | 0.96 |
| 40S ribosomal protein S6 (RP-S6e/S/237) | 1.02 | 1.3 | 0.51 | 0.76 |
| 40S ribosomal protein S6 (RP-S6e/S/229) | 1.21 | 1.32 | 0.29 | 0.56 |
| Putative translational activator (S/1831) | 0.78 | 0.83 | 1.88 | 0.78 |
| 4G-1 Eukaryotic translation initiation factor isoform 4G-1 (EIF4G/S/527) | 0.86 | 1.11 | 1.34 | 0.74 |
| 4B Eukaryotic translation initiation factor 4B (EIF4B/S/128) | 0.72 | 0.84 | 1.29 | 0.86 |
| 3H Eukaryotic translation initiation factor 3 subunit H (EIF3H/S/222) | 0.96 | 1.27 | 0.91 | 0.79 |
| 4G Eukaryotic translation initiation factor 4G (EIF4G/T/1579) | 1.49 | 1.52 | 0.8 | 1.15 |
| 4B Eukaryotic translation initiation factor 4B (EIF4B/Y/536) | 1.15 | 1.12 | 0.73 | 1.17 |
| 4G Eukaryotic translation initiation factor 4G (EIF4G/S/184) | 1.74 | 1.53 | 0.73 | 1.13 |
| 4G Eukaryotic translation initiation factor 4G (EIF4G/S1401) | 2.24 | 1.76 | 0.59 | 1.00 |
| Elongation factor 1-alpha (EEF1A/S/176) | 1.22 | 1.07 | 0.54 | 1.18 |
| Protein Degradation | G1 | G2 | Y1 | Y2 |
| Putative dynamin homolog (DNM/S/894) | 0.47 | 0.57 | 2.53 | 1.44 |
| Putative dynamin homolog (DNM/S/897) | 0.52 | 0.60 | 2.49 | 1.38 |
| Putative dynamin homolog (DNM/S/887) | 0.77 | 0.84 | 1.67 | 0.80 |
| Putative dynamin homolog (DNM/Y/886) | 0.69 | 0.88 | 1.67 | 0.75 |
| Putative dynamin homolog (DNM/T/736) | 0.79 | 0.88 | 1.56 | 1.11 |
| Putative dynamin homolog (DNM/S/733) | 0.80 | 0.86 | 1.49 | 1.20 |
| Putative dynamin homolog (DNM/S/730) | 0.80 | 0.87 | 1.48 | 1.19 |
| Putative dynamin homolog (DNM/T/535) | 0.88 | 0.95 | 1.32 | 1.00 |
| Dynamin-related protein 1C, putative, expressed (DNM/S/271) | 0.76 | 0.82 | 1.66 | 1.06 |
| ATP-dependent Clp protease proteolytic subunit (ClpP/S/288) | 0.31 | 0.37 | 2.46 | 0.89 |
| ATP-dependent Clp protease proteolytic subunit (ClpP/S/287) | 0.30 | 0.46 | 2.23 | 0.90 |
| ATP-dependent zinc metalloprotease FTSH 1, chloroplastic (FTSH1/T/539) | 0.39 | 0.53 | 2.42 | 0.90 |
| ATP-dependent Clp protease proteolytic subunit (ClpP/S/287) | 0.30 | 0.46 | 2.23 | 0.90 |
| ATP-dependent Clp protease proteolytic subunit (ClpP/S/96) | 0.49 | 0.60 | 1.70 | 1.00 |
| H-ATPase (PMA1/Y/955) | 0.83 | 0.40 | 2.02 | 1.07 |
| H-ATPase (PMA1/T/956) | 0.67 | 0.80 | 2.01 | 0.79 |
| Pleiotropic drug resistance protein 5 (S/26) | 0.56 | 0.77 | 1.83 | 0.98 |
| Pleiotropic drug resistance protein 6 (S/14) | 0.94 | 1.05 | 1.28 | 0.73 |
| Pleiotropic drug resistance protein 6 (S/12) | 0.93 | 1.08 | 1.21 | 0.79 |
| Chloroplast thylakoidal processing peptidase-like protein (S/392) | 0.54 | 0.62 | 1.65 | 1.11 |
| Neutral/alkaline invertase (Neutral/alkaline invertase/S/48) | 0.81 | 0.99 | 1.62 | 0.85 |
| Beta-glucosidase 11 (Beta-glucosidase 11/S546) | 0.57 | 0.60 | 1.62 | 0.99 |
| Endonuclease III homologue (NTH/S/346) | 0.80 | 0.82 | 1.61 | 0.83 |
| Gibberellin receptor GID1L2 (GID1L2/S/309) | 0.62 | 0.71 | 1.60 | 1.25 |
| Beta-glucosidase 11 (Beta-glucosidase 11/S545) | 0.60 | 0.62 | 1.58 | 0.98 |
| Beta-glucosidase 11 (Beta-glucosidase 11/S555) | 0.67 | 0.72 | 1.50 | 0.93 |
| Beta-glucosidase 11 (Beta-glucosidase 11/S/1007) | 0.69 | 0.77 | 1.45 | 1.13 |
| Beta-glucosidase 11 (Beta-glucosidase 11/S/249) | 0.70 | 0.78 | 1.25 | 1.21 |
| Beta-glucosidase 24 (Beta-glucosidase 24/S/279) | 0.66 | 0.64 | 1.47 | 0.8 |
| Beta-glucosidase 24 (Beta-glucosidase 24 /Y/286) | 0.64 | 0.63 | 1.47 | 0.81 |
| Beta-glucosidase 24 (Beta-glucosidase 24/S/271) | 0.66 | 0.83 | 1.35 | 0.87 |
| Beta-glucosidase 24 (Beta-glucosidase 24/S/269) | 0.66 | 0.73 | 1.33 | 0.94 |
| DNA topoisomerase I (DNA topoisomerase I/S/139) | 0.90 | 0.98 | 1.41 | 0.89 |
| DNA topoisomerase I (DNA topoisomerase I/S/142) | 0.90 | 0.98 | 1.41 | 0.89 |
| Glycoside hydrolase family 2 family protein (lacZ/S/567) | 0.68 | 0.74 | 1.35 | 1.35 |
| Ubiquitin family protein (Ubiquitin family protein/T/351) | 0.93 | 0.92 | 1.33 | 0.74 |
| H2B Filamentation temperature-sensitive H2B (S/198) | 0.81 | 1.02 | 1.24 | 0.95 |
| H2B Filamentation temperature-sensitive H2B (S/199) | 0.81 | 1.02 | 1.24 | 0.95 |
| putative endo-1,3;1,4-beta-D-glucanase (S/209) | 0.93 | 1.06 | 1.23 | 0.81 |
| Beta-glucosidase-like SFR2, chloroplastic (S/132) | 0.75 | 1.11 | 0.85 | 1.30 |
| Soluble inorganic pyrophosphatase (ppa/S/33) | 0.82 | 1.17 | 0.77 | 1.25 |
| Soluble inorganic pyrophosphatase (ppa/S/34) | 0.80 | 1.10 | 0.77 | 1.33 |
| DNA topoisomerase I (DNA topoisomerase I/S/192) | 1.11 | 1.21 | 0.71 | 0.63 |
| CER5 (CER5/S/330) | 1.01 | 1.24 | 0.64 | 1.09 |
| Transcriptional regulation | ||||
| Histone H2B (H2B/S/135) | 0.92 | 1.02 | 1.30 | 0.77 |
| Histone H2B (H2B/S/19) | 1.25 | 1.12 | 0.73 | 0.9 |
| SNF2 transcription factor (MSI/S/95) | 1.28 | 1.25 | 0.72 | 1.16 |
| MYB transcription factor (S/184) | 1.15 | 1.19 | 0.66 | 1.06 |
| Putative MYB DNA-binding domain superfamily protein (S/171) | 0.75 | 1.02 | 1.25 | 1.01 |
| Zinc finger CCCH domain-containing protein 8 (S/436) | 1.36 | 1.2 | 0.82 | 0.78 |
| Zinc finger CCCH domain-containing protein 8 (S/238) | 1.27 | 1.34 | 0.59 | 1.01 |
| Zinc-finger homeodomain protein 1 (S/199) | 0.70 | 0.86 | 1.30 | 1.24 |
| Zinc-finger homeodomain protein 1 (T/37) | 0.78 | 1.04 | 1.22 | 1.00 |
| Zinc-finger homeodomain protein 1 (T/17) | 1.01 | 0.88 | 1.40 | 0.73 |
| Zinc-finger homeodomain protein 3 (Y/151) | 0.76 | 1.00 | 1.33 | 1.03 |
| Zinc finger CCCH domain-containing protein 8 (S/436) | 1.36 | 1.20 | 0.82 | 0.78 |
| Zinc finger CCCH domain-containing protein 8 (S/238) | 1.27 | 1.34 | 0.59 | 1.01 |
| Zinc finger CCCH domain-containing protein 63 (S/408) | 0.69 | 0.97 | 1.84 | 0.85 |
| Zinc finger CCCH domain-containing protein 63 (S/410) | 0.72 | 1.01 | 1.80 | 0.81 |
| Zinc finger CCCH domain-containing protein 63 (S/183) | 0.76 | 1.04 | 1.76 | 0.73 |
| Zinc finger CCCH domain-containing protein 63 (T/185) | 0.89 | 1.08 | 1.48 | 0.72 |
| Zinc finger CCCH domain-containing protein 63 (S/424) | 0.90 | 0.93 | 1.38 | 0.93 |
| ROC1 Homeobox-leucine zipper protein ROC1 (ROC1/S/46) | 1.01 | 1.08 | 4.20 | 0.40 |
| DNA binding protein, mRNA (S/176) | 0.94 | 0.92 | 0.75 | 1.28 |
| DNA binding protein, mRNA (S/204) | 0.8 | 0.92 | 1.33 | 1.07 |
| DNA binding protein, mRNA (S/64) | 1.11 | 1.09 | 0.69 | 1.02 |
| Transport | ||||
| Hexose transporter (Hexose transporter/S/643) | 0.55 | 0.64 | 2.11 | 0.86 |
| Hexose transporter (Hexose transporter/S/70) | 0.61 | 0.75 | 1.78 | 0.97 |
| Putative sulfate transporter (S/643) | 0.68 | 0.96 | 1.90 | 0.68 |
| Putative sulfate transporter (Putative sulfate transporter/S/651) | 0.73 | 0.9 | 1.69 | 0.82 |
| Sugar transport1 isoform 1 (Sugar transport1 isoform 1/S/33) | 0.66 | 0.77 | 1.56 | 0.66 |
| Sugar transport1 isoform 1 (Sugar transport1 isoform 1/S/32) | 0.84 | 0.98 | 1.31 | 0.75 |
| Sugar transport1 isoform 1 (Sugar transport1 isoform 1/S/485) | 1.24 | 1.32 | 0.65 | 1.16 |
| Amino acid permease-like (Amino acid permease-like/S/20) | 0.22 | 0.47 | 0.98 | 2.45 |
| Amino acid permease-like (Amino acid permease-like/S/47) | 1.10 | 1.25 | 0.61 | 1.03 |
| Amino acid permease-like (Amino acid permease-like/Y/49) | 1.25 | 1.22 | 0.52 | 1.02 |
| Amino acid permease-like (Amino acid permease-like /T/41) | 1.26 | 1.23 | 0.47 | 1.01 |
| Amino acid permease-like (Amino acid permease-like/S/42) | 1.28 | 1.22 | 0.45 | 1.01 |
| Calcium-transporting ATPase 1, plasma membrane-type (S/45) | 0.87 | 1.17 | 0.73 | 1.15 |
| MATE efflux family protein (MATE efflux family protein/S/12) | 1.10 | 1.07 | 0.65 | 1.01 |
| ABC transporter family protein, putative, expressed (S/99) | 0.69 | 0.89 | 2.43 | 0.61 |
| ABC transporter family protein, putative, expressed (S/7) | 0.70 | 0.92 | 2.36 | 0.61 |
| ABC transporter family protein, putative, expressed (S/63) | 0.81 | 0.72 | 2.30 | 0.72 |
| ABC-2 type transporter family protein (S/617) | 0.75 | 0.80 | 1.53 | 1.13 |
Author Contributions
Conceptualization and Methodology, J.W., W.M. and N.W.; data curation and formal analysis, N.W.; writing—original draft preparation, N.W.; writing—review and editing, T.Z.; validation, N.L., Z.W. and G.Y.; visualization, G.Q. and L.K.; project administration, J.W., W.M. and S.Z.; Supervision, J.W. and W.M.; funding acquisition, J.W.
Funding
This research was supported by Fundamental Research Funds of Chinese Academy of Forestry (CAFYBB2017ZY002).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Aluru M.R., Zola J., Foudree A., Rodermel S.R. Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiol. 2009;150:904–923. doi: 10.1104/pp.109.135780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu W., Zhu Y., Ma Z., Sun Y., Quan Q., Li P., Hu P., Shi T., Lo C., Chu I.K., et al. Proteomic evidence for genetic epistasis: ClpR4 mutations switch leaf variegation to virescence in Arabidopsis. Plant J. 2013;76:943–956. doi: 10.1111/tpj.12344. [DOI] [PubMed] [Google Scholar]
- 3.Miura E., Kato Y., Sakamoto W. Comparative transcriptome analysis of green/white variegated sectors in Arabidopsis yellow variegated2: Responses to oxidative and other stresses in white sectors. J. Exp. Bot. 2010;61:2433–2445. doi: 10.1093/jxb/erq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashe A., Morgan D.K., Whitelaw N.C., Bruxner T.J., Vickaryous N.K., Cox L.L., Butterfield N.C., Wicking C., Blewitt M.E., Wilkins S.J., et al. A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol. 2008;9:R182. doi: 10.1186/gb-2008-9-12-r182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motohashi R., Rodiger A., Agne B., Baerenfaller K., Baginsky S. Common and specific protein accumulation patterns in different albino/pale-green mutants reveals regulon organization at the proteome level. Plant Physiol. 2012;160:2189–2201. doi: 10.1104/pp.112.204032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J., Zhang J., Zhao Q., Liu Y., Chen S., Guo H., Shi L., Dai S. Proteomic analysis reveals the leaf color regulation mechanism in Chimera Hosta “gold standard” leaves. Int. J. Mol. Sci. 2016;17:346. doi: 10.3390/ijms17030346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y., Wu X., Zhang Z., Gao Z. Comparative proteomic analysis of floral color variegation in peach. Biochem. Biophys. Res. Commun. 2015;464:1101–1106. doi: 10.1016/j.bbrc.2015.07.084. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y., Hoehenwarter W. Changes in the phosphoproteome and metabolome link early signaling events to rearrangement of photosynthesis and central metabolism in salinity and oxidative stress response in Arabidopsis. Plant Physiol. 2015;169:3021–3033. doi: 10.1104/pp.15.01486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steiner S., Dietzel L., Schroter Y., Fey V., Wagner R., Pfannschmidt T. The role of phosphorylation in redox regulation of photosynthesis genes psaA and psbA during photosynthetic acclimation of mustard. Mol. Plant. 2009;2:416–429. doi: 10.1093/mp/ssp007. [DOI] [PubMed] [Google Scholar]
- 10.Lundin B., Hansson M., Schoefs B., Vener A.V., Spetea C. The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein. Plant J. 2007;49:528–539. doi: 10.1111/j.1365-313X.2006.02976.x. [DOI] [PubMed] [Google Scholar]
- 11.Longoni P., Douchi D., Cariti F., Fucile G., Goldschmidt-Clermont M. Phosphorylation of the light-harvesting complex II isoform Lhcb2 is central to state transitions. Plant Physiol. 2015;169:2874–2883. doi: 10.1104/pp.15.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trotta A., Suorsa M., Rantala M., Lundin B., Aro E.M. Serine and threonine residues of plant STN7 kinase are differentially phosphorylated upon changing light conditions and specifically influence the activity and stability of the kinase. Plant J. 2016;87:484–494. doi: 10.1111/tpj.13213. [DOI] [PubMed] [Google Scholar]
- 13.Bonardi V., Pesaresi P., Becker T., Schleiff E., Wagner R., Pfannschmidt T., Jahns P., Leister D. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437:1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- 14.Wang J.P., Chuang L., Loziuk P.L., Chen H., Lin Y.C., Shi R., Qu G.Z., Muddiman D.C., Sederoff R.R., Chiang V.L. Phosphorylation is an on/off switch for 5-hydroxyconiferaldehyde O-methyltransferase activity in poplar monolignol biosynthesis. Proc. Natl. Acad. Sci. USA. 2015;112:8481–8486. doi: 10.1073/pnas.1510473112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zagari N., Sandoval-Ibanez O., Sandal N., Su J., Rodriguez-Concepcion M., Stougaard J., Pribil M., Leister D., Pulido P. Snowy cotyledon 2 promotes chloroplast development and has a role in leaf variegation in both Lotus japonicus and Arabidopsis thaliana. Mol. Plant. 2017;10:721–734. doi: 10.1016/j.molp.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Swiatek M., Kuras R., Sokolenko A., Higgs D., Olive J., Cinque G., Muller B., Eichacker L.A., Stern D.B., Bassi R., et al. The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. Plant Cell. 2001;13:1347–1367. doi: 10.1105/tpc.13.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baginsky S. Protein phosphorylation in chloroplasts—a survey of phosphorylation targets. J. Exp. Bot. 2016;67:3873–3882. doi: 10.1093/jxb/erw098. [DOI] [PubMed] [Google Scholar]
- 18.Wioleta W., Anna D., Ilona B., Kamila K., Elżbieta R. Lead induced changes in phosphorylation of PSII proteins in low light grown pea plants. Biometals. 2015;28:151–162. doi: 10.1007/s10534-014-9811-y. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Q., Chen W., Bian J., Xie H., Li Y., Xu C., Ma J., Guo S., Chen J., Cai X., et al. Proteomics and phosphoproteomics of heat stress-responsive mechanisms in spinach. Front Plant Sci. 2018;9:800. doi: 10.3389/fpls.2018.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H., Ye M., Dong J., Corradini E., Cristobal A., Heck A.J.R., Zou H., Mohammed S. Robust phosphoproteome enrichment using monodisperse microsphere–based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 2013;8:461–480. doi: 10.1038/nprot.2013.010. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C., Guo X., Xie H., Li J., Liu X., Zhu B., Liu S., Li H., Li M., He M., et al. Quantitative phosphoproteomics of lectin receptor-like kinase VI.4 dependent abscisic acid response in Arabidopsis thaliana. Physiol. Plant. 2018 doi: 10.1111/ppl.12763. [DOI] [PubMed] [Google Scholar]
- 22.Xue X., Wang Q., Qu Y., Wu H., Dong F., Cao H., Wang H.L., Xiao J., Shen Y., Wan Y. Development of the photosynthetic apparatus of Cunninghamia lanceolata in light and darkness. New Phytol. 2017;213:300–313. doi: 10.1111/nph.14096. [DOI] [PubMed] [Google Scholar]
- 23.Su V., Hsu B.D. Cloning and expression of a putative cytochrome P450 gene that influences the colour of Phalaenopsis flowers. Biotechnol. Lett. 2003;25:1933–1939. doi: 10.1023/B:BILE.0000003989.19657.53. [DOI] [PubMed] [Google Scholar]
- 24.Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., Tabata S., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2010;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 25.Saito A., Shimizu M., Nakamura H., Maeno S., Katase R., Miwa E., Higuchi K., Sonoike K. Fe deficiency induces phosphorylation and translocation of Lhcb1 in barley thylakoid membranes. FEBS Lett. 2014;588:2042–2048. doi: 10.1016/j.febslet.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 26.Grieco M., Tikkanen M., Paakkarinen V., Kangasjarvi S., Aro E.M. Steady-state phosphorylation of light-harvesting complex II proteins preserves photosystem I under fluctuating white light. Plant Physiol. 2012;160:1896–1910. doi: 10.1104/pp.112.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allahverdiyeva Y., Suorsa M., Rossi F., Pavesi A., Kater M.M., Antonacci A., Tadini L., Pribil M., Schneider A., Wanner G., et al. Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 2013;75:671–684. doi: 10.1111/tpj.12230. [DOI] [PubMed] [Google Scholar]
- 28.Gao S., Gu W., Xiong Q., Ge F., Xie X., Li J., Chen W., Pan G., Wang G. Desiccation enhances phosphorylation of PSII and affects the distribution of protein complexes in the thylakoid membrane. Physiol. Plant. 2015;153:492–502. doi: 10.1111/ppl.12258. [DOI] [PubMed] [Google Scholar]
- 29.Grieco M., Jain A., Ebersberger I., Teige M. An evolutionary view on thylakoid protein phosphorylation uncovers novel phosphorylation hotspots with potential functional implications. J. Exp. Bot. 2016;67:3883–3896. doi: 10.1093/jxb/erw164. [DOI] [PubMed] [Google Scholar]
- 30.Slekar K.H., Kosman D.J., Culotta V.C. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- 31.Pandolfi P.P., Sonati F., Rivi R., Mason P., Grosveld F., Luzzatto L. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J. 1995;14:5209–5215. doi: 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapala-Kozik M., Wolak N., Kujda M., Banas A.K. The upregulation of thiamine (vitamin B1) biosynthesis in Arabidopsis thaliana seedlings under salt and osmotic stress conditions is mediated by abscisic acid at the early stages of this stress response. BMC Plant Biol. 2012;12:2. doi: 10.1186/1471-2229-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R., Zhao J., Jia M., Xu N., Liang S., Shao J., Qi Y., Liu X., An L., Yu F. Balance between cytosolic and chloroplast translation affects leaf variegation. Plant Physiol. 2017 doi: 10.1104/pp.17.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Hou Y., Qiu J., Li Z., Zhao J., Tong X., Zhang J. A quantitative acetylomic analysis of early seed development in rice (Oryza sativa L.) Int. J. Mol. Sci. 2017;18:1376. doi: 10.3390/ijms18071376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia Y., Jing D., Kong L., Zhang J., OuYang F., Zhang H., Wang J., Zhang S. Global lysine acetylome analysis of desiccated somatic embryos of Picea asperata. Front Plant Sci. 2016;7:1927. doi: 10.3389/fpls.2016.01927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finnemann J., Schjoerring J.K. Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14-3-3 protein interaction. Plant J. 2010;24:171–181. doi: 10.1046/j.1365-313x.2000.00863.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Bian Y., Cheng K., Gu L.F., Ye M., Zou H., Sun S.S., He J.X. A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J. Proteomics. 2013;78:486–498. doi: 10.1016/j.jprot.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 38.Axel T., Katrin P., Anja B., Natalia P., Rowan M.K., Halford N.G., Peter G. Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 2003;35:490–500. doi: 10.1046/j.1365-313x.2003.01823.x. [DOI] [PubMed] [Google Scholar]
- 39.Sun H., Xia B., Wang X., Gao F., Zhou Y. Quantitative phosphoproteomic analysis provides insight into the response to short-term drought stress in Ammopiptanthus mongolicus roots. Int. J. Mol. Sci. 2017;18:2158. doi: 10.3390/ijms18102158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X., Xu Y., Li J., Ren Y., Wang Z., Xin Z., Lin T. Identification of two novel wheat drought tolerance-related proteins by comparative proteomic analysis combined with virus-induced gene silencing. Int. J. Mol. Sci. 2018;19:4020. doi: 10.3390/ijms19124020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Li D., Zhou R., Wang X., Dossa K., Wang L., Zhang Y., Yu J., Gong H., Zhang X., et al. Transcriptome and metabolome analyses of two contrasting sesame genotypes reveal the crucial biological pathways involved in rapid adaptive response to salt stress. BMC Plant Biol. 2019;19:66. doi: 10.1186/s12870-019-1665-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodermel S. Pathways of plastid-to-nucleus signaling. Trends Plant Sci. 2001;6:471–478. doi: 10.1016/S1360-1385(01)02085-4. [DOI] [PubMed] [Google Scholar]
- 43.Miura E., Kato Y., Matsushima R., Albrecht V., Laalami S., Sakamoto W. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell. 2007;19:1313–1328. doi: 10.1105/tpc.106.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonenberg N., Morgan M.A., Merrick W.C., Shatkin A.J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5’-terminal cap in mRNA. Proc. Natl. Acad. Sci. USA. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamphear B.J., Kirchweger R., Skern T., Rhoads R.E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 46.Zheng M., Liu X., Liang S., Fu S., Qi Y., Zhao J., Shao J., An L., Yu F. Chloroplast translation initiation factors regulate leaf variegation and development. Plant Physiol. 2016;172:1117–1130. doi: 10.1104/pp.15.02040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu F., Liu X., Alsheikh M., Park S., Rodermel S. Mutations in suppressor of variegation 1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell. 2008;20:1786–1804. doi: 10.1105/tpc.107.054965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu L., Wang S., Wu J., Han Z., Wang R., Wu L., Zhang H., Chen Y., Hu X. Phosphoproteomic analysis of the resistant and susceptible genotypes of maize infected with sugarcane mosaic virus. Amino Acids. 2015;47:483–496. doi: 10.1007/s00726-014-1880-2. [DOI] [PubMed] [Google Scholar]
- 49.Kim J., Olinares P.D., Oh S.H., Ghisaura S., Poliakov A., Ponnala L., van Wijk K.J. Modified Clp protease complex in the ClpP3 null mutant and consequences for chloroplast development and function in Arabidopsis. Plant Physiol. 2013;162:157–179. doi: 10.1104/pp.113.215699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adam Z., Adamska I., Zheng B., Vallon O., Rodermel S.R., Shinozaki K., Clarke A.K. Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 2001;125:1912–1918. doi: 10.1104/pp.125.4.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Apitz J., Nishimura K., Schmied J., Wolf A., Hedtke B., van Wijk K.J., Grimm B. Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiol. 2016;170:2040–2051. doi: 10.1104/pp.15.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramundo S., Casero D., Muhlhaus T., Hemme D., Sommer F., Crevecoeur M., Rahire M., Schroda M., Rusch J., Goodenough U., et al. Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell. 2014;26:2201–2222. doi: 10.1105/tpc.114.124842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato Y., Miura E., Ido K., Ifuku K., Sakamoto W. The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 2009;151:1790–1801. doi: 10.1104/pp.109.146589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurokawa M., Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138:838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tikkanen M., Aro E.M. Thylakoid protein phosphorylation in dynamic regulation of photosystem II in higher plants. Biochim. Biophys. Acta. 2012;1817:232–238. doi: 10.1016/j.bbabio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Zhang D., Li Y., Zhang X., Zha P., Lin R. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA regulates chlorophyll biosynthesis in Arabidopsis. Mol. Plant. 2017;10:155–167. doi: 10.1016/j.molp.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Peirats-Llobet M., Han S.K., Gonzalez-Guzman M., Jeong C.W., Rodriguez L., Belda-Palazon B., Wagner D., Rodriguez P.L. A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol. Plant. 2016;9:136–147. doi: 10.1016/j.molp.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Pi E., Zhu C., Fan W., Huang Y., Qu L., Li Y., Zhao Q., Ding F., Qiu L., Wang H. Quantitative phosphoproteomic and metabonomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress. Mol. Cell. Proteomics. 2018;17:1209–1224. doi: 10.1074/mcp.RA117.000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu Z., Zhu J., Hao Q., Yuan Y.W., Duan Y., Men S., Wang Q., Hou Q., An Liu Z., Shu Q., et al. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa) Plant Cell Physiol. 2018;60:599–611. doi: 10.1093/pcp/pcy232. [DOI] [PubMed] [Google Scholar]
- 60.Seibert M., Kruger M., Watson N.A., Sen O., Daum J.R., Slotman J.A., Braun T., Houtsmuller A.B., Gorbsky G.J., Jacob R., et al. CDK1-mediated phosphorylation at H2B serine 6 is required for mitotic chromosome segregation. J. Cell Biol. 2019;218:1164. doi: 10.1083/jcb.201806057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baker N.R., Oxborough K. Chlorophyll fluorescence as a probe of photosynthetic productivity. In: Papageorgiou G.C., Govindjee, editors. Chlorophyll a Fluorescence. Springer; Dordrecht, the Netherlands: 2004. pp. 65–82. [Google Scholar]
- 62.Lichtenthaler H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148C:350–382. [Google Scholar]
- 63.Tong J., Helmy M., Cavalli F.M., Jin L., St-Germain J., Karisch R., Taylor P., Minden M.D., Taylor M.D., Neel B.G., et al. Integrated analysis of proteome, phosphotyrosine-proteome, tyrosine-kinome, and tyrosine-phosphatome in acute myeloid leukemia. Proteomics. 2017;17:1600361. doi: 10.1002/pmic.201600361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu X., Liu T., Yang J., Chen L., Liu B., Wang L., Jin Q. The first whole cell proteome and lysine acetylome based comparison between Trichophyton rubrum conidial and mycelial stages. J. Proteome Res. 2018;17:1436–1451. doi: 10.1021/acs.jproteome.7b00793. [DOI] [PubMed] [Google Scholar]
- 65.Wang J. The Genome of Catalpa bungei Provides Insights into the Disease Resistance and the Wood Formation. 2019 Unpublished work. [Google Scholar]
- 66.Wang N., Zhang S., Wang J. Comprehensive Analyses Reveal the Mechanism of Natural Variegated Leaf in Catalpa fargesii by Transcriptome and Proteome Profiling. 2019 Unpublished work. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.