Abstract
Magnetic resonance imaging (MRI) volumetric measures have become a standard tool for the detection of incipient Alzheimer's Disease (AD) dementia in mild cognitive impairment (MCI). Focused on providing an earlier and more accurate diagnosis, sophisticated MRI machine learning algorithms have been developed over the recent years, most of them learning their non-disease patterns from MCI that remained stable over 2–3 years. In this work, we analyzed whether these stable MCI over short-term periods are actually appropriate training examples of non-disease patterns. To this aim, we compared the diagnosis of MCI patients at 2 and 5 years of follow-up and investigated its impact on the predictive performance of baseline volumetric MRI measures primarily involved in AD, i.e., hippocampal and entorhinal cortex volumes. Predictive power was evaluated in terms of the area under the ROC curve (AUC), sensitivity, and specificity in a trial sample of 248 MCI patients followed-up over 5 years. We further compared the sensitivity in those MCI that converted before 2 years and those that converted after 2 years. Our results indicate that 23% of the stable MCI at 2 years progressed in the next three years and that MRI volumetric measures are good predictors of conversion to AD dementia even at the mid-term, showing a better specificity and AUC as follow-up time increases. The combination of hippocampus and entorhinal cortex yielded an AUC that was significantly higher for the 5-year follow-up (AUC = 73% at 2 years vs. AUC = 84% at 5 years), as well as for specificity (56% vs. 71%). Sensitivity showed a non-significant slight decrease (81% vs. 78%). Remarkably, the performance of this model was comparable to machine learning models at the same follow-up times. MRI correctly identified most of the patients that converted after 2 years (with sensitivity >60%), and these patients showed a similar degree of abnormalities to those that converted before 2 years. This implies that most of the MCI patients that remained stable over short periods and subsequently progressed to AD dementia had evident atrophies at baseline. Therefore, machine learning models that use these patients to learn non-disease patterns are including an important fraction of patients with evident pathological changes related to the disease, something that might result in reduced performance and lack of biological interpretability.
Keywords: Late MCI, MCI, Alzheimer, Machine learning, MRI
Highlights
-
•
MRI predicted AD dementia significantly better when extending follow-up from 2 to 5 years.
-
•
MRI was similarly able to detect AD in short and mid-term converters.
-
•
Predictive models should not learn non-disease patterns from stable MCI over short periods.
1. Introduction
Alzheimer's disease (AD), the most common form of dementia in the elderly population, is still presenting challenges for an early diagnosis. Since the only definite way to identify AD is by means of a brain biopsy, this disease is predominantly diagnosed clinically (McKhann et al., 2011) and, when possible, diagnosis is supported with biomarkers derived from cerebrospinal fluid or imaging. Although certainty in diagnosis augments by tracking the progression in time of cognitive performance, early symptoms of AD are difficult to distinguish from age-related cognitive impairment and other neuropsychological disorders. These early symptoms are common to a condition known as mild cognitive impairment (MCI) (Petersen et al., 1999) in which a cognitive decline is evident but not sufficiently specific to be considered incipient AD. At this stage, the mean annual conversion rate from MCI to probable AD is about 7%, with most of the MCI patients not progressing to AD within 10 years (Mitchell and Shiri-Feshki, 2009). This result suggests that the origin of the MCI condition is variable and cannot be only attributed to AD, making difficult to filter out those MCI patients with incipient AD.
As pointed out in Greenberg et al. (2013)), a possible reason of the failure of some clinical trials could be the selection of participants with clinical AD at a disease stage that might be too progressed to benefit from the treatment. Restricting the selection to MCI participants seemed the most straightforward solution to this issue. However, the aforementioned heterogeneity of MCI, both in definition and etiology, along with low conversion rates, critically contributed to the impossibility of detecting differences between treatment and placebo groups (Schneider et al., 2014). Furthermore, in order to be approved as a clinical target, both the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) stated that MCI must be defined through robust and validated criteria, being the EMA even more restrictive by requiring positivity in at least one biomarker of amyloidosis or neurodegeneration (Drug Administration Peripheral and Central Nervous System Drugs Advisory Committee, 2001; European medicines agency. pre-authorisation evaluation of medicines for human use, 2008). These requirements resulted in clinicopathological diagnostic definitions, known as prodromal AD or MCI due to AD (Dubois et al., 2007; Albert et al., 2011), incorporating biomarkers. These definitions are widely used nowadays in clinical trials of disease-modifying drugs (Cummings et al., 2017).
Among biomarkers of AD, structural Magnetic Resonance Imaging (MRI) is the most commonly used technique to identify brain atrophies related to AD. Hippocampal atrophy, i.e., volumetric abnormality, assessed visually or quantitatively, is the best established MRI biomarker of AD (Hill et al., 2014), although there is accumulating evidence that atrophies in other parts of the brain such as the entorhinal cortex provide complementary prognostic information as well (Killiany et al., 2002; Du et al., 2001; Dickerson et al., 2001; Devanand et al., 2012; Devanand et al., 2007). Beyond visual assessment and volumetric measurements, several different machine learning approaches have been explored for both feature extraction and classification, reporting an earlier and better detection of AD standard metrics (Rathore et al., 2017). Although the most emphasized strength of these approaches is the power to predict AD dementia earlier, a few models were also designed with the aim of predicting imminent conversion, so that clinical trials could use these approaches for sample enrichment. Despite the good reported results, these techniques have not been used yet either in clinical routine nor trials (Rathore et al., 2017; Arbabshirani et al., 2017), probably due to poor performance when evaluated in new data sets (Arbabshirani et al., 2017). As an example, in (Cuingnet et al., 2011), 10 methods that reported significant prognostic ability using MRI were evaluated using new data. None of them showed to be different from random chance, indicating that the previously reported results were probably biased. Experimenter interventions in the testing process, tuning some of the free parameters of the model, and circular analysis or double-dipping (Kriegeskorte et al., 2009) might explain the lack of generalizability of these models.
Apart from the previously mentioned methodological issues, we also found issues concerning the training sample. The typical approach when training a machine learning classifier is to label as progressive MCI (pMCI) those MCI patients who converted to AD within a fixed follow-up time, typically 1–3 years, and to label as stable MCI (sMCI) those who have not converted within that period. Using this labeling, machine learning algorithms learn the pattern that, theoretically, best discriminates between incipient AD and the rest of MCI. The conceptual validity of this approach relies on the assumption that only a negligible fraction of the sMCI patients are affected by AD or, alternatively, those sMCI actually affected by AD present a too early disease stage in which no major structural changes have occurred yet. Based on the latter assumption, an important number of machine learning classifiers were trained to capture the subtle structural changes that may contribute to an earlier prediction of AD (Rathore et al., 2017). However, neither the hypothesis of the absence of evident structural changes in mid-term pMCI nor the small size of its effect have been confirmed yet, leaving unclear how accurate is the correspondence between sMCI and absence of AD. If an important deviation of this correspondence is finally confirmed, it would imply that this common labeling suffers from poor biological interpretability and, therefore, may result in reduced performance and lack of generalizability.
In this work, we studied the predictive power of MRI measurements of hippocampal and entorhinal cortex volumes for short (2 years) and mid-term (5 years) follow-up times. We also investigated the performance in the subgroups of MCI who converted before 2 years (short-term pMCI), and in those who converted after 2 years and before 5 (mid-term pMCI), as well as in sMCI over 5 years versus sMCI with shorter follow-up times. To this aim, we used all the available data from the ADNI database, including data from (Petersen et al., 1999) MCI patients enrolled in ADNI1 and ADNI2. To our knowledge, this is the first study in which the predictive power of MRI is evaluated at 5-year follow-up using the ADNI1 and ADNI2 MCI cohorts, providing a detailed comparison of performance between the short and mid-term.
2. Materials and methods
2.1. Data
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database http://adni.loni.usc.edu. The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD.
The ADNI project comprises 4 different studies, namely the completed ADNI1, ADNIGO and ADNI2, and the ongoing ADNI3, launched in September 2016. Among the completed studies, ADNI1 and ADNI2 recruited the majority of the participants while ADNIGO recruited only early MCI participants and its main purpose was to serve as a continuation of ADNI1.
2.2. MRI
ADNI2 used only 3 T scanners while ADNI1 employed different protocols and acquired images using 1.5 T scanners, although a small fraction of participants was rescanned with 3 T devices. We restricted to the 1.5 T scan for ADNI1 participants. A detailed MRI acquisition protocol of the different studies of the ADNI project can be found at http://adni.loni.usc.edu/methods/documents/. The ADNI collaborators at the Center for Imaging of Neurodegnerative Diseases at University of California, San Francisco (UCSF) provided Freesurfer (version 4.3 in ADNI1 and 5.1 in ADNI2) (Fischl, 2012) segmentations of MRI T1 images from ADNI participants. Quality of the segmentations was assessed by the researchers at UCSF to ensure a correct parcellation of the brain. A detailed description of the quality control process can be found at http://adni.loni.usc.edu/data-samples/access-data/. The most common failures occur at the cortical level. About 69% of the segmentations in ADNI passed the entire quality control. In this study, only segmentations that fulfilled all the quality criteria were included. Among the available images for each patient, only non-accelerated MPRAGE or IR-SPGR sequences were used. Cortical segmentation was performed using the Desikan-Killiany Atlas (Desikan et al., 2006). Subcortical segmentation scheme was the same as the one described in (Filipek et al., 1994). For simplicity, only total volumes (left plus right) of the hippocampus and entorhinal cortex were considered in this study. We estimated the total intracranial volume (TIV) with SPM12 (Penny et al., 2011) using the utility ‘Tissue Volumes’ (Malone et al., 2015). We provided the Image IDs, as well as the computed TIVs, in the Supplementary Material.
2.3. Subjects
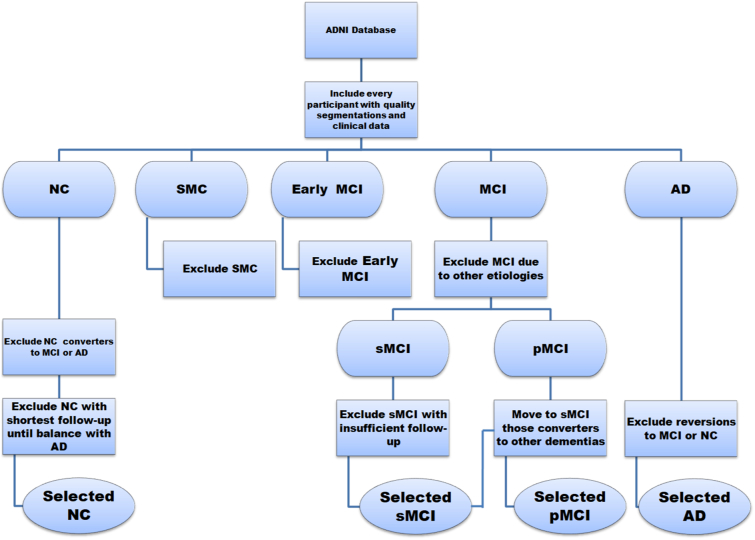
We only included patients from the following diagnostic groups in the ADNI1/2 study: Normal Controls (NC), late MCI (MCI) and AD. Patients in the diagnostic group of Subjective Memory Complaints (SMC) or early MCI were not included. We did not include Early MCI because the aim of this study is to compare the performance of well-known volumetric biomarker metrics with the performance of state-of-the-art machine learning models, which, to the best of our knowledge, were mainly applied to late MCI data (Rathore et al., 2017; Arbabshirani et al., 2017). The inclusion criteria of the diagnostic groups in the ADNI1/GO/2 study can be found at http://adni.loni.usc.edu/methods/documents/. All the participants without clinical evaluation at baseline were excluded. We selected all the participants with available segmentations that passed the quality control. Only MCI patients with a suspected MCI due to Alzheimer's disease at baseline were included. We excluded all the MCI participants that remained stable and withdrew before the minimum follow-up considered in this study, 2 years. MCI patients were considered progressive MCI (pMCI) if they converted at any point within the follow-up time, regardless of the observed outcome in longer follow-up times. For these pMCI patients, conversion time was estimated as the mid-point between the visit in which AD is diagnosed for the first time and the previous visit. MCI patients who progressed to other types of dementia were regarded as stable MCI (sMCI). We also excluded NC participants in which a progression to MCI or AD was observed at any point in their follow-up, as well as AD patients in which a reversion to MCI or NC occurred. After this selection, the number of NC participants was superior to AD in both ADNI1 and ADNI2 cohorts. In order to obtain a balanced sample of NC and AD in both ADNI studies, we excluded those participants with the shortest follow-up times until we balanced the number of NC and AD. We chose a balanced design to exclude informative prior probability (Chawla et al., 2004). Fig. 1 shows a schematic diagram of the whole selection process.
Fig. 1.
Diagram showing the selection criteria used in this study.
It should be noted that the inclusion criteria for all the previously mentioned diagnostic categories are equivalent across ADNI1/2, the only difference being that ADNI2 required a lumbar puncture to participate in the study. Thus, and given the differences in imaging protocols, we decided to analyze ADNI1 and ADNI2 MRI data of common diagnostic cohorts (NC, MCI, and AD) separately. RID identification numbers and diagnosis for the different follow-up times were provided in the Supplementary Material.
2.4. Performance evaluation
In order to obtain an index for the prediction of MCI progression based on baseline MRI data, we fitted logistic regressions on each NC vs. AD sample from ADNI1 (1.5 T) and ADNI2 (3 T). We fitted three models with different inputs, i.e., using hippocampal volume alone, using entorhinal cortex volume alone, and using hippocampal and entorhinal cortex volumes (MRI model). All the inputs were measured at baseline. The models included Age and TIV to account for aging and cranial size (model equations are provided in the Supplementary Material). To derive classification cut points, we followed the recommendations of (Neurobiol. Aging, 1998) and established a cut point in which a sensitivity of 85% in identifying AD patients was obtained (the derived cut-points are reported in the Supplementary Material). Logit values were used as predictive indexes in the trial MCI cohort. Note that this approach is completely bias-free since the MCI cohort did not play any role when training the models.
The first analysis was carried out to compare the predictive performance of MRI as a function of follow-up time from short (2 years) to mid-term (5 years) follow-up times, in annual steps. We computed the area under the Receiver Operating Characteristic Curve (AUC), sensitivity (proportion of pMCI correctly classified) and specificity (proportion of sMCI correctly classified).
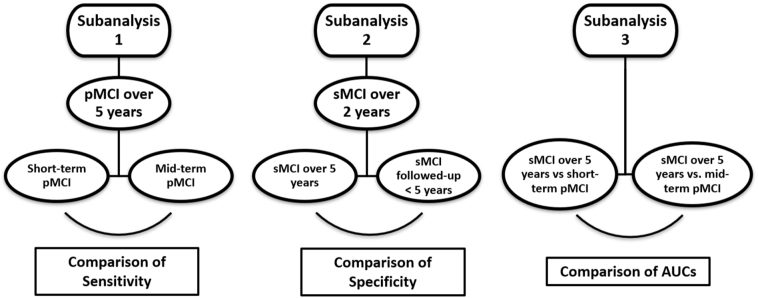
We also performed three different subgroup analyses to evaluate how conversion time and time remaining stable affected classification accuracy of MRI. Specifically, in Subanalysis 1 we compared sensitivities in short-term pMCI (converters within 2 years) and mid-term pMCI (converters after 2 years and before 5) to investigate how NC-like patients at baseline that then progressed within 5 years affect the overall sensitivity. In Subanalysis 2 we stratified the sMCI patients over 2 years into those that remained stable for 5 years and those with shorter follow-up times. We then compared specificities between these two independent samples, so we can assess that our results are not influenced due to the exclusion of sMCI that did not reach the 5 year endpoint. In Subanalysis 3, we separated converters into short-term and mid-term converters, as in Subanalysis 1. We then computed the AUCs of these two types of converters versus sMCI over 5 years, and compared the results. In this way, we can evaluate whether short-term and mid-term converter distributions of atrophies present different degrees of overlapping with the distribution of sMCI over five years. A schematic summary of the subanalyses can ben seen in Fig. 2.
Fig. 2.
Schematic summary of the subanalyses involving different times to conversion and stability.
2.5. Statistical analysis
95% confidence intervals (CI) for AUC, sensitivity, and specificity were calculated using Clopper-Pearson confidence intervals. As previously mentioned, ADNI1 and ADNI2 MCI cohorts followed slightly different inclusion and imaging criteria. Therefore, we pooled AUC, sensitivity, and specificity of these two studies, as well as estimated confidence intervals, following a fixed effects model with a Freeman-Tukey transformation (Freeman and Tukey, 1950). The standard deviations used for pooling AUCs were computed according to the DeLong method (DeLong et al., 1988).
In Subanalyses 1 and 2, we compared sensitivities and specificities using a chi-square test. No separation between the different studies of ADNI was done in these tests. We tested that all the classifiers were significantly different from chance using permutation tests (Ojala and Garriga, 2010). Significance level was set to α = 0.05.
3. Results
3.1. Population
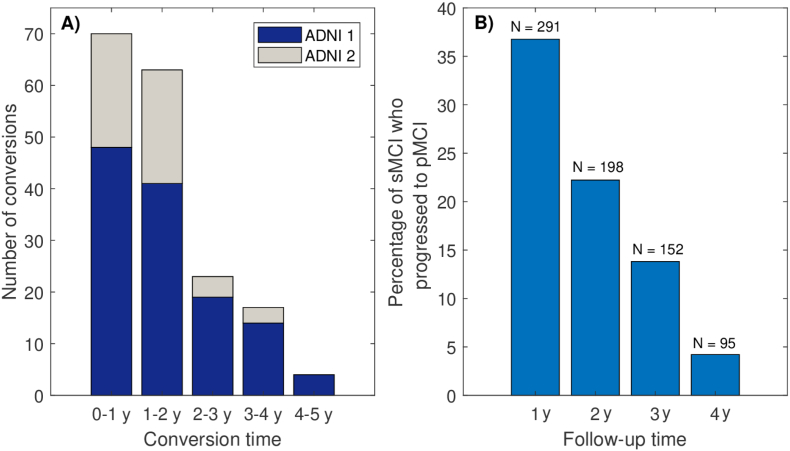
The selection process described in Section 2.3 resulted in a sample of 230 NC and 230 AD (124 from ADNI1 and 106 from ADNI2). Sample sizes MCI varied with follow-up time due to withdrawal. Table 1 shows the number of sMCI and pMCI at 2 and 5 years follow-up. Demographic information for intermediate follow-up times is provided in Supplementary Table 1. The number of conversions per year can be seen in Fig. 3 A). Fig. 3 B) shows that 23% of sMCI at 2 years progressed in the next 3 years. Three MCI converted to other types of dementia and were regarded as sMCI. Descriptive statistics of sex, age, MMSE and APOE status are also provided in Table 1.
Table 1.
Demographic information of the different samples studied in this work. Suffixes -2y and -5y stand for 2 and 5 year follow-up times, respectively. Age and MMSE Score are reported as median [range]. Hippocampal and entorhinal cortex volumes represent the average between left and right volumes, expressed as mean ± standard deviation.
| Study | Diagnostic group | Number | Gender (M/F) | Age (years) | MMSE score | APOE ε4 (heterozygote/homozygote) | Hippocampal volume (cm3) | Entorhinal cortex volume (cm3) |
|---|---|---|---|---|---|---|---|---|
| ADNI1 | NC | 124 | 62/62 | 74 [60–90] | 29 [25–30] | 26/2 | 3.7 ± 0.4 | 1.9 ± 0.3 |
| AD | 124 | 65/59 | 75 [55–90] | 23 [20–27] | 59/25 | 2.8 ± 0.5 | 1.4 ± 0.3 | |
| MCI | ||||||||
| sMCI-2y | 134 | 85/49 | 75 [55–88] | 28 [24–30] | 53/12 | 3.3 ± 0.5 | 1.7 ± 0.4 | |
| pMCI-2y | 89 | 51/38 | 75 [56–88] | 26 [23−30] | 49/15 | 2.9 ± 0.5 | 1.5 ± 0.4 | |
| sMCI-5y | 47 | 33/14 | 75 [60–86] | 28 [24–30] | 17/0 | 3.5 ± 0.4 | 1.9 ± 0.3 | |
| pMCI-5y | 126 | 72/54 | 74 [55–88] | 27 [23–30] | 67/22 | 3.0 ± 0.5 | 1.5 ± 0.4 | |
| ADNIGO/2 | NC | 106 | 50/56 | 72 [56–85] | 30 [24–30] | 28/2 | 3.8 ± 0.5 | 1.9 ± 0.3 |
| AD | 106 | 57/49 | 75 [56–90] | 23 [19–26] | 45/24 | 3.0 ± 0.5 | 1.5 ± 0.3 | |
| MCI | ||||||||
| sMCI-2y | 64 | 34/30 | 71[55–85] | 28 [24–30] | 18/7 | 3.6 ± 0.5 | 1.8 ± 0.3 | |
| pMCI-2y | 44 | 24/20 | 73 [57–85] | 26 [24–30] | 23/9 | 3.0 ± 0.5 | 1.6 ± 0.4 | |
| sMCI-5y | 24 | 13/11 | 68 [55–85] | 29 [25–30] | 7/3 | 3.8 ± 0.5 | 1.9 ± 0.3 | |
| pMCI-5y | 51 | 28/23 | 73 [57–85] | 27 [24–30] | 25/10 | 3.1 ± 0.5 | 1.6 ± 0.3 | |
Fig. 3.
A): Number of conversions per year of MCI patients. B): Proportions of stable MCI that progressed to AD dementia in subsequent years, for each follow-up time. N indicates the number of stable MCI.
3.2. Performance of MRI in the short and mid-term
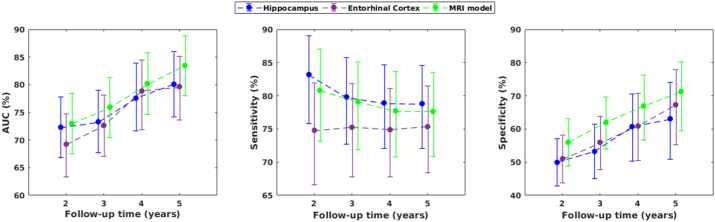
Fig. 4 shows the results for the prediction of AD dementia in the MCI cohort as a function of follow-up time. Each model was significantly different from chance, at every follow-up time and for ADNI1 and ADNI2 (p < 0.001). Discrimination power between sMCI and pMCI was stronger for the 5-year follow-up on each model. The best discrimination at the 5-year follow-up was achieved by the MRI model with an AUC = 84%, CI: [78–89], showing an increase of 11% in AUC compared to the 2-year follow-up (AUC = 73%, CI: [68–78]). There was also a strong rise in specificity, increasing about 14% on each model while sensitivity slightly decreased for the hippocampus and the MRI model but increased for the entorhinal cortex. Fig. 4 shows that this increase in AUC and specificity is the endpoint of a consistent trend as follow-up time increases. All these trends were observed in ADNI1 and ADNI2 for each model (Supplementary Tables 2 and 6).
Fig. 4.
Area under the ROC curve (AUC), sensitivity and specificity of hippocampus, entorhinal cortex, and MRI model for the prediction of AD dementia in MCI as a function of follow-up time. Vertical bars represent 95% confidence intervals.
In order to test whether patients that withdrew potentially biased our results, we compared the performance of each model on the 5-year follow-up set of MCI patients with the performance on that set after relabeling the diagnosis of MCI patients at the 2-year follow-up (Table 2). Results were almost identical to those obtained in the previous analysis.
Table 2.
Area under the ROC curve (AUC), sensitivity and specificity of each model for the prediction of AD dementia, evaluated on the 5-year follow-up set of MCI patients and for the diagnosis at 2 and 5 years of follow-up.
| Model | AUC (%) |
Sensitivity (%) |
Specificity (%) |
|||
|---|---|---|---|---|---|---|
| 2-year | 5-year | 2-year | 5-year | 2-year | 5-year | |
| Hippocampus | 74 [68–80] | 80 [74–86] | 83 [76–89] | 79 [72–84] | 52 [43–61] | 63 [51–74] |
| Entorhinal cortex | 69 [63–76] | 80 [74–86] | 77 [69–84] | 78 [71–84] | 50 [39–58] | 66 [54–77] |
| MRI model | 74 [68–80] | 84 [78–89] | 86 [79–91] | 78 [71–84] | 56 [47–66] | 71 [60–81] |
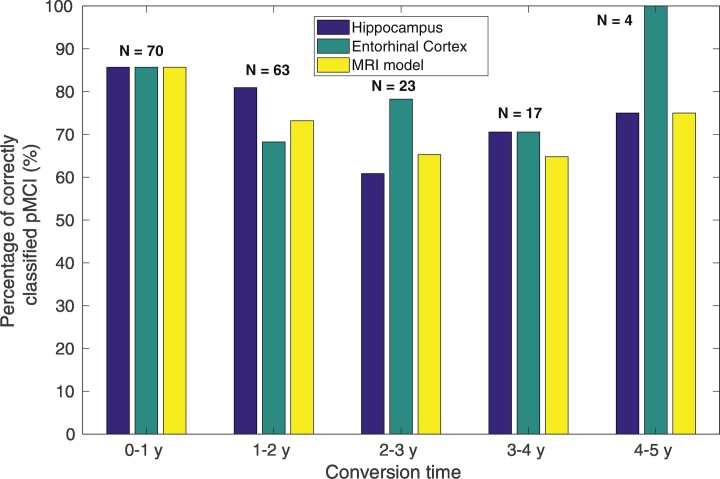
Subanalysis 1 showed only a significant decrease in classification accuracy of mid-term pMCI compared to short-term pMCI in the hippocampus model (65%, CI: [50–79] vs. 83%, CI: [76–89], p = 0.013), while entorhinal cortex model increased (80%, CI: [66–90] vs. 77%, CI: [69–84], p = 0.77) and the MRI model decreased (67%, CI: [52–80] vs. 80%, CI: [73–87], p = 0.072). Despite this sensitivity degradation in hippocampus and MRI model, most of the patients were correctly classified regardless of conversion time, as can be seen in Fig. 5. Subanalysis 2 showed that classification accuracy in the sMCI sample with shorter follow-up times than 5 years was significantly lower than the corresponding classfication accuracy of sMCI over 5 years, for all the models, (43%, CI: [34–52] vs. 63%, CI: [51–74], p < 0.01 for hippocampus, 40%, CI: [32–49] vs. 66%, CI: [54–77], p < 0.001 for entorhinal cortex, and 47%, CI: [38–56] vs. 71%, CI: [60–81], p < 0.001 for the MRI model), and in both ADNI1 and ADNI2 studies (Supplementary Tables 4 and 8). In Subanalysis 3, the AUCs of the MRI model were similar for both short and mid-term pMCI, (AUC = 84%, CI: [78–90] vs. AUC = 82%, CI: [74–90]). AUCs were similarly high for both types of converters in the rest of the models (Supplementary Table 12). Detailed results for ADNI1 and ADNI2 were provided in Supplementary Tables 2–12.
Fig. 5.
Percentage of correctly classified MCI converters for each marker as a function of conversion time. N indicates the number of conversions per year.
4. Discussion
In this work, we investigated the impact of the extension of follow-up time from short (2 years) to mid-term (5 years) on the predictive performance of MRI. Our results were derived using all the available data from ADNI1 and ADNI2, resulting in 248 MCI trial patients with a 5-year follow-up. This work is, to our knowledge, the first that thoroughly investigated the predictive power of MRI beyond the short-term in a relatively large cohort.
We demonstrated that an extension of follow-up time from 2 to 5 years results in a change in diagnosis of 23% sMCI patients and in unexpected increases of about 10% and 15% in AUC and specificity, respectively, in predictive performance. It should be noted that the increase in AUC is independent from cut-point definition, so the improvement cannot be explained due to cut-point selection. This result is closely related to the similar and high AUCs (>82%) found in Subanalysis 3, indicating that mid-term pMCI patients have comparable atrophies to short-term pMCI. The observation of a significant improvement in classification performance when extending follow-up time in MCI patients can be explained by a more accurate ground-truth diagnosis of sMCI. In other words, some patients with incipient AD dementia, presenting measurable pathological features, did not have enough time to progress to AD dementia due to insufficient follow-up time. This is supported by Subanalysis 2, showing that specificity in sMCI at 5 years was significantly higher (71%, CI: [60–81]) than the corresponding for the rest of the sMCI with shorter follow-up (47%, CI: [38–56]), and by the fact that most of the mid-term pMCI presented evident atrophies (Fig. 5). As a result, a significantly lower specificity and an overall decrease in discriminative power were observed at 2 years follow-up. Although this effect was already known in early-stage biomarkers such as brain amyloidosis (Buchhave et al., 2012), we provide first-time evidence that it also matters in a late stage biomarker such as atrophy in MRI. Regarding the behavior of sensitivity, there are two competing factors that determine its trend with follow-up time. On the one hand, atrophy is a late stage pathophysiological change in the course of AD (Jack Jr and Holtzman, 2013) and, thus, by increasing follow-up time we provide more time for the development of atrophies and AD dementia in patients that had no atrophies at baseline. This effect results in an increased number of false negatives, so we expect certain decrease in sensitivity as we increase follow-up time. On the other hand, there is a significant number of patients with atrophies that only progressed in the mid-term, so these false positive cases turned out to be true positive, increasing sensitivity. Looking at Fig. 3, it seems that these two factors compensate each other, resulting in a relatively stable sensitivity for the follow-up times studied here.
From a clinical perspective, the fact that many atrophied patients do not progress in the short-term but in subsequent years indicates that hippocampal and entorhinal atrophy can be ascertained at mid-term time frames prior to the onset of dementia and that this pathological feature is more specific than what was previously thought. This finding can contribute to re-examine the role of hippocampal and entorhinal atrophy as indicators of short-term decline, reinforcing their early prognostic ability, and to re-evaluate their importance at the time of diagnosing early AD dementia. Moreover, the observation of the same levels of atrophy in patients who progressed in the short-term and in the mid-term supports the hypothesis that progression depends on more factors than just biomarker evidence of AD. Among the possible factors, it is likely that cognitive reserve (Stern, 2012) drives most of the variance in conversion times and, thus, comparing clinical symptoms with the expected clinical symptoms for the observed atrophy might help to better predict progression.
Our findings also have implications on the novel machine learning algorithms that learn discriminative patterns from a sample of sMCI and pMCI over a short follow-up time. Table 3 presents a summary of the aforementioned algorithms reviewed in (Rathore et al., 2017) that reported AUC as a performance measure. We selected AUC for being the most robust performance measure against sample imbalance. They used a wide variety of methods, some of high complexity, to detect AD dementia in MCI patients, claiming in some cases to be earlier and more accurate predictors than common volumetric measures. In this context, it is surprising that a simple model based solely on the hippocampus and the entorhinal cortex, such as the one studied in this paper, evaluated in a larger and independent sample, and in a longer time frame with conversions to AD dementia after 4 and 5 years from diagnosis, outperformed in terms of AUC all the complex machine learning methods. Although the reason for this improvement is simply the more refined sample of sMCI patients resulting from a longer follow-up or, equivalently, the good ability to predict mid-term conversion, the performance of our model at 2 and 3 years of follow-up was comparable with most of the machine learning methods (Table 3), casting doubt on the supposed improvement that these methods provide compared to simple volumetric measures. At short follow-up times, machine learning algorithms are forced to learn their complex, subtle patterns using a misleading ground-truth diagnosis in an important proportion of patients while ignoring evident pathological features such as measurable hippocampal or entorhinal atrophies, contributing to spoil any biological interpretation of the results and making the algorithm a black box whose behaviour might be unpredictable in new datasets. For instance, and even if an MRI-only-based ML algorithm is designed with the only purpose of sample enrichment in clinical trials, we believe that training on the basis of discriminating converter and stable patients within a short period might present both generalization and interpretation problems. As mentioned before, inter subject differences in time to progression to AD dementia in patients with the same level of neurodegeneration are probably explained by differences in cognitive reserve (Jack Jr and Holtzman, 2013; Stern, 2012). This implies that an important part of the variance in conversion times is driven by variables that are independent of MRI and, therefore, variations in these variables across different populations (more educated cohorts or cultural differences) will probably affect the performance of the ML model. For instance, consider an ML model which is tested in a cohort that is less educated than the cohort used to train the model. A higher level of education implies a higher cognitive reserve (Meng and DArcy, 2012) and, therefore, we expect that patients progressing within 2 years will present a more advanced pathophysiological stage, i.e., higher baseline atrophies than those progressive patients in the less educated cohort. Thus, the ML model will learn that a very advanced atrophy is necessary to ascertain conversion within 2 years, resulting in an increased number of false negatives in the less educated test cohort, in which the level of atrophy required to progress is expected to be lower. This implies that a fraction of patients with evident atrophies will be labeled as non-progressive, even when these atrophies might be severe and we will actually observe a progression, spoiling the reliability and interpretability of the model.
Table 3.
Summary of MRI machine learning algorithms reviewed in (Rathore et al., 2017). p stands for converter MCI, s for stable MCI, CV indicates that cross-validation was used for evaluation.
| Study | Training sample size | Evaluation method | Follow-up time (years) | AUC |
|---|---|---|---|---|
| Misra et al. (2009) (Misra et al., 2009) | 27p/76 s | CV | Variable (Mean = 2 years) | 77% |
| Liu et al. (2013) (Liu et al., 2013) | 97p/93 s | CV | 3 | 72% |
| Eskildsen et al. (2013) (Eskildsen et al., 2013) | 128p/227 s | CV | Variable (Mean = 1.5 years) | 68% |
| Min et al. (2014) (Min et al., 2014) | 98AD/128NC | CV (117p/117 s) | Not reported | 67% |
| Liu et al. (2015) (Liu et al., 2015) | 117p/117 s | CV | Not reported | 81% |
| Tang et al. (2015) (Tang et al., 2015) | 175AD/210NC | CV (135p/87 s)) | 3 | 74% |
| Chincarini et al. (2011) (Chincarini et al., 2011) | 144AD/189NC | Independent set (136p/166 s) | 2 | 74% |
| Wee et al. (2013) (Wee et al., 2013) | 45p/56 s | Repeated hold-out (44p/55 s) | 3 | 84% |
| Sorensen et al. (2016) (Sorensen et al., 2016) | 101AD/169NC | Independent set (93p/140 s) | 2 | 74% |
| Hippocampus + Entorhinal Cortex | 230AD/230NC | Independent set (133p/198 s) | 2 | 73% |
| Hippocampus + Entorhinal Cortex | 230AD/230NC | Independent set (156p/152 s) | 3 | 76% |
| Hippocampus + Entorhinal Cortex | 230AD/230NC | Independent set (177p/84 s) | 5 | 84% |
Although we consider that the aforementioned issues contribute to the non-generalizability of machine learning methods in incipient AD dementia detection (Arbabshirani et al., 2017), we also consider that they are not the only ones. As stated in Section 1, it is equally important the report of non-biased results and a statistically powerful comparison between novel and standard metrics. As an example, the two best-performing algorithms from Table 3 manually tuned some of the multiple parameters of their complex algorithms, which may result in optimistically biased results. These pitfalls, but also others of a different nature, were also common in the rest of the algorithms and critically contribute, in our view, to poor generalizability.
Looking at Table 3, it seems evident that if MRI machine learning algorithms can actually provide an improvement in AD dementia detection, this improvement has to be relatively small. However, even if this small improvement exists, this result is going to be impossible to ascertain due to the uncertainty generated by 20% of sMCI patients who are misleadingly labeled as non-AD. It is of capital importance to provide an accurate ground-truth diagnosis in training phases, probably including only those stable MCI with the longest follow-up times, in order to exploit the potential of machine learning to improve AD dementia detection in the usually small available cohorts. Although we explored a relatively long 5-year follow-up, it is likely that a nonnegligible proportion of our stable MCI over 5 years is still affected by AD and presented evident atrophies at baseline, as suggested by the fact that we obtained a significant number of false positives. In this context, and given the lack of data with long follow-ups, the usually abandoned strategy of using NC vs. AD for training might be more adequate than what was expected.
This study had some limitations. ADNI1 and ADNI2 used MRI scanners with different field strengths, so combining results from both studies might have influenced our results. However, our models were fitted separately for ADNI1 and ADNI2 data, contributing to better control of bias and generalizability. The heterogeneous protocols in ADNI1 and ADNI2, combining different sequences, are sources of variability whose assessment is beyond the scope of this work but that definitively need to be addressed in future studies. Since we focused on patients with long follow-up, our results might suffer from survivor bias. Nevertheless, Subanalysis 2, performed on the patients that did not reach the follow-up time, supported our conclusions, so we conclude that this source of bias is not significantly affecting our conclusions.
5. Conclusions
In conclusion, we found that MRI was highly predictive in mid-term pMCI, correctly classifying most of them. As a consequence, specificity and discriminative power increased at 5-year follow-up, outperforming complex machine learning approaches of incipient AD detection with shorter follow-up times.
The unexpected good performance of MRI in the mid-term revealed the problem that an insufficient follow-up time may create for machine learning algorithms, given that these algorithms regard as non-diseased an important proportion of patients actually affected by AD and with evident atrophies. This may result in poor performance and lack of generalizability, as well as in non-interpretability of algorithm predictions. If sample enrichment for short-term clinical trials is required, other variables that predict short-term conversion must be included along with MRI.
A short follow-up might be also problematic when deriving biomarker cut-points from MCI samples, especially for those representing the early stages of the disease.
Acknowledgements
Funding: This work was partially supported by the project PI16/01416 (ISCIII co-funded FEDER) and RYC-2015/17430 (Ramón y Cajal, Pablo Aguiar). Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2019.101837.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
References
- Albert M.S., DeKosky S.T., Dickson D., Dubois B. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbabshirani M.R., Plis S., Sui J., Calhoun V.D. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. Neuroimage. 2017;145(Pt B):137–165. doi: 10.1016/j.neuroimage.2016.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchhave P., Minthon L., Zetterberg H., Wallin A.K., Blennow K., Hansson O. Cerebrospinal fluid levels of -amyloid 1-42, but not of tau, are fully changed already 5 to 10 years before the onset of Alzheimer dementia. Arch. Gen. Psychiatry. 2012;69(1):98–106. doi: 10.1001/archgenpsychiatry.2011.155. [DOI] [PubMed] [Google Scholar]
- Chawla N.V., Japkowicz N., Kotcz A. Editorial: Special issue on learning from imbalanced data sets. Newsl. ACM SIGKDD Explor. Newsl. - Special Issue on Learning from Imbalanced Datasets. 2004;6(1):1–6. [Google Scholar]
- Chincarini A., Bosco P., Calvini P., Gemme G. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer's disease. NeuroImage. 2011;58(2):469–480. doi: 10.1016/j.neuroimage.2011.05.083. [DOI] [PubMed] [Google Scholar]
- Cuingnet R., Gerardin E., Tessieras J., Auzias G. Automatic classification of patients with Alzheimer's disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2011;56(2):766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Cummings J., Lee G., Mortsdorf T., Ritter A. Alzheimer's disease drug development pipeline: 2017. Alzheimers Dement. (N Y) 2017;3(3) doi: 10.1016/j.trci.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- Desikan R.S., Sègonne F., Fischl B., Quinn B.T. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Devanand D.P., Pradhaban G., Liu X., Khandji A. Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology. 2007;68(11):828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- Devanand D.P., Bansal R., Liu J., Hao X. MRI hippocampal and entorhinal cortex mapping in predicting conversion to Alzheimer's disease. Neuroimage. 2012;60(3):1622–1629. doi: 10.1016/j.neuroimage.2012.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson B.C., Goncharova I., Sullivan M.P., Forchetti C. Cognitive reserve in ageing and Alzheimer's disease. Neurobiol. Aging. 2001;22(5):747–754. doi: 10.1016/s0197-4580(01)00271-8. [DOI] [PubMed] [Google Scholar]
- Drug Administration Peripheral and Central Nervous System Drugs Advisory Committee Mild Cognitive Impairment. 2001. https://www.fda.gov/ohrms/dockets/ac/01/transcripts/3724t1.pdf
- Du A.T., Schuff N., Amend D., Laakso M.P. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer's disease. J. Neurol. Neurosurg. Psychiatry. 2001;71(4):441–447. doi: 10.1136/jnnp.71.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B., Feldman H.H., Jacova C., DeKosky S.T. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6(8):734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- Eskildsen S.F., Coupé P., Garca-Lorenzo D., Fonov V. Prediction of Alzheimer's disease in subjects with mild cognitive impairment from the ADNI cohort using patterns of cortical thinning. Neuroimage. 2013;65:511–521. doi: 10.1016/j.neuroimage.2012.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Medicines Agency. Pre-authorisation Evaluation of Medicines for Human Use . 2008. Guideline on Medicinal Products for the Treatment of alzheimer's Disease and Other Dementias. [Google Scholar]
- Filipek P.A., Richelme C., Kennedy D.N., Caviness V.S., Jr. The young adult human brain: an MRI-based morphometric analysis. Cereb. Cortex. 1994;4(4):344–360. doi: 10.1093/cercor/4.4.344. [DOI] [PubMed] [Google Scholar]
- Fischl B. Freesurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M.F., Tukey J.W. Transformations related to the angular and the square root. Ann. Math. Statist. 1950;21(4):607–611. [Google Scholar]
- Greenberg B.D., Carrillo M.C., Ryan J.M., Gold M. Improving Alzheimer's disease phase II clinical trials. Alzheimers Dement. 2013;9(1):39–49. doi: 10.1016/j.jalz.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Hill D.L.G., Schwarz A.J., Isaac M., Pani L. Coalition Against Major Diseases/European Medicines Agency biomarker qualification of hippocampal volume for enrichment of clinical trials in predementia stages of Alzheimer's disease. Alzheimers Dement. 2014;10(4):421–429. doi: 10.1016/j.jalz.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Jack C.R., Jr., Holtzman D.M. Biomarker modeling of Alzheimer's disease. Neuron. 2013;80(6):1347–1358. doi: 10.1016/j.neuron.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killiany R.J., Hyman B.T., Gomez-Isla T., Moss M.B. MRI measures of entorhinal cortex vs hippocampus in preclinical AD. Neurology. 2002;58(8):1188–1196. doi: 10.1212/wnl.58.8.1188. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N., Simmons W.K., Bellgowan P.S., Baker C.I. Circular analysis in systems neuroscience: the dangers of double dipping. Nat. Neurosci. 2009;12(5):535–540. doi: 10.1038/nn.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Tosun D., Weiner M.W., Schuff N. Locally linear embedding (LLE) for MRI based Alzheimer's disease classification. Neuroimage. 2013;83:148–157. doi: 10.1016/j.neuroimage.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zhang D., Shen D., for the Alzheimer's Disease Neuroimaging Initiative View-centralized multi-atlas classification for Alzheimer's disease diagnosis. Hum. Brain Mapp. 2015;36(5):1847–1865. doi: 10.1002/hbm.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone I.B., Leung K.K., Clegg S., Barnes J. Accurate automatic estimation of total intracranial volume: a nuisance variable with less nuisance. Neuroimage. 2015;104(1):366–372. doi: 10.1016/j.neuroimage.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., DArcy C. Education and dementia in the context of the cognitive reserve hypothesis: a systematic review with meta-analyses and qualitative analyses. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0038268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min R., Wu G., Cheng J., Wang Q. Multi-atlas based representations for Alzheimer's disease diagnosis. Hum. Brain Mapp. 2014;35(10):5052–5070. doi: 10.1002/hbm.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra C., Fan Y., Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage. 2009;44(4):1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A.J., Shiri-Feshki M. Rate of progression of mild cognitive impairment to dementia–meta-analysis of 41 robust inception cohort studies. Acta Psychiatr. Scand. 2009;119(4):252–265. doi: 10.1111/j.1600-0447.2008.01326.x. [DOI] [PubMed] [Google Scholar]
- Consensus report of the Working Group on: "Molecular and Biochemical Markers of Alzheimer's Disease". The Ronald and Nancy Reagan Research Institute of the Alzheimer's Association and the National Institute on Aging Working GroupNeurobiol. Aging. 1998;19(2):109–116. [PubMed] [Google Scholar]
- Ojala M., Garriga G.C. Permutation tests for studying classifier performance. J. Mach. Learn. Res. 2010;11(6):1833–1863. [Google Scholar]
- Penny W., Friston K., Ashburner J., Kiebel S. Academic Press; 2011. Statistical Parametric Mapping: The Analysis of Functional Brain Images. [Google Scholar]
- Petersen R.C., Smith G.E., Waring S.C., Ivnik R.J. Mild cognitive impairment: clinical characterization and outcome. Arch. Neurol. 1999;56(3):303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- Rathore S., Habes M., Iftikhar M.A., Shacklett A. A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer's disease and its prodromal stages. Neuroimage. 2017;155:530–548. doi: 10.1016/j.neuroimage.2017.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L.S., Mangialasche F., Andreasen N., Feldman H. Clinical trials and late-stage drug development for Alzheimer's disease: an appraisal from 1984 to 2014. J. Intern. Med. 2014;275(3):251–283. doi: 10.1111/joim.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen L., Igel C., Hansen N.L., Osler M. Early detection of Alzheimer's disease using MRI hippocampal texture. Hum. Brain Mapp. 2016;37(3):1148–1161. doi: 10.1002/hbm.23091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y. Cognitive reserve in ageing and Alzheimer's disease. Lancet Neurol. 2012;11(11):1006–1012. doi: 10.1016/S1474-4422(12)70191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Holland D., Dale A.M., Younes L. Baseline shape diffeomorphometry patterns of subcortical and ventricular structures in predicting conversion of mild cognitive impairment to Alzheimer's disease. J. Alzheimers Dis. 2015;44(2):599–611. doi: 10.3233/JAD-141605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee C.-Y., Yap P.-T., Shen D. Prediction of Alzheimer's disease and mild cognitive impairment using cortical morphological patterns. Hum. Brain Mapp. 2013;34(12):3411–3425. doi: 10.1002/hbm.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4