Summary
Epidermal growth factor receptor (EGFR) signaling controls skin development and homeostasis in mice and humans, and its deficiency causes severe skin inflammation, which might affect epidermal stem cell behavior. Here, we describe the inflammation-independent effects of EGFR deficiency during skin morphogenesis and in adult hair follicle stem cells. Expression and alternative splicing analysis of RNA sequencing data from interfollicular epidermis and outer root sheath indicate that EGFR controls genes involved in epidermal differentiation and also in centrosome function, DNA damage, cell cycle, and apoptosis. Genetic experiments employing p53 deletion in EGFR-deficient epidermis reveal that EGFR signaling exhibits p53-dependent functions in proliferative epidermal compartments, as well as p53-independent functions in differentiated hair shaft keratinocytes. Loss of EGFR leads to absence of LEF1 protein specifically in the innermost epithelial hair layers, resulting in disorganization of medulla cells. Thus, our results uncover important spatial and temporal features of cell-autonomous EGFR functions in the epidermis.
Subject Areas: Biological Sciences, Cell Biology, Developmental Biology, Stem Cells Research
Graphical Abstract
Highlights
-
•
EGFR has compartment-specific functions in distinct keratinocyte populations
-
•
EGFR exerts p53-dependent control on DNA integrity in proliferative keratinocytes
-
•
EGFR controls epithelial hair lineage differentiation in a p53-independent manner
-
•
EGFR contributes to onset of anagen induction in adult hair follicle stem cells
Biological Sciences; Cell Biology; Developmental Biology; Stem Cells Research
Introduction
Skin morphogenesis is a complex process requiring the spatial and temporal orchestration of distinct molecular signaling pathways to ensure correct development of epidermal lineages such as interfollicular epidermis (IFE), hair follicles (HF), and sebaceous glands. Initiation of HF development is mainly conducted by β-catenin and SHH signaling (Huelsken et al., 2001, Blanpain and Fuchs, 2009, Hsu et al., 2014), resulting in formation of hair placodes between embryonic day (E)15 and E17 (Tumbar, 2012). After the critical step of placode formation, SHH stimulates proliferation of cells within this structure, which then grow down into the dermis as hair germs and further develop into hair pegs between E17 and E18 (Hsu et al., 2014). Continuous growth of the hair peg leads to the formation of a mature HF, which is constituted of highly proliferative progenitor cells, the so-called matrix, at the bottom. Progeny of the matrix gives rise to the distinct differentiated epithelial cell layers, which comprise the hair or function in hair guidance (Hsu et al., 2014, Adam et al., 2018). The cell layers from the outer to innermost layer are outer root sheath (ORS), companion layer, three layers of inner root sheath (IRS), cuticle, cortex, and medulla. WNT/β-catenin and bone morphogenetic protein (BMP) signaling were shown to orchestrate hair layer differentiation by temporal layering of signaling effectors (Adam et al., 2018). Impairment in the formation of solely one layer is sufficient to induce the collapse of the hair shaft and sustained disruption of hair development (DasGupta and Fuchs, 1999, Kaufman, 2003).
Besides instructive Wnt/β-catenin and BMP signaling pathways, epidermal growth factor receptor (EGFR) signaling strongly contributes to skin development and homeostasis of mice and humans. Mice completely lacking EGFR are born with open eyes and display severe skin defects, including a pronounced delay in HF development (Miettinen et al., 1995, Sibilia and Wagner, 1995, Threadgill et al., 1995). Expression of a dominant-negative mutant of EGFR in keratinocytes (Murillas et al., 1995) and hair transplantation experiments (Hansen et al., 1997) revealed that EGFR-deficient HFs are able to progress through morphogenesis for about 2 weeks after birth, but then fail to enter catagen. Similarly, humanized EGFR mice (hEGFRKI/KI) exhibit impaired HF morphogenesis, alterations in morphology and distribution of HFs, and a failure in anagen to catagen transition owing to inefficient expression of the hEGFR in the skin (Sibilia et al., 2003). At postnatal day (P)90, hEGFRKI/KI mice mostly contained degenerated HFs with thin hair cell layers, which were partly destroyed. Later, studies in conditional knockout mice lacking the EGFR specifically in the epidermal lineage (Nagao et al., 2012, Lichtenberger et al., 2013, Mascia et al., 2013, Bichsel et al., 2016) confirmed that loss of EGFR in keratinocytes is sufficient to induce skin differentiation defects, catagen block, loss of the HF niche, and baldness. Moreover, epidermis-specific EGFR knockout results in severe skin inflammation and confers high susceptibility to S. aureus infections. Similar phenotypes have also been observed in cancer patients receiving anti-EGFR therapies (Lichtenberger et al., 2013, Mascia et al., 2013). In contrast, mice harboring either the naturally occurring EGFR mutation waved2 (Luetteke et al., 1994) or the transforming growth factor (TGF)-α mutation waved1 (Crew, 1933, Luetteke et al., 1993) only show a mild phenotype characterized by wavy hair coat and whiskers but otherwise normal physiology. TGFα shows specific expression in cells of the IRS (Luetteke et al., 1993), suggesting an important role of EGFR signaling in the control of epithelial hair layer development and hair shape, whereas EREG and AREG have been shown to affect sebocyte numbers or sebaceous gland size (Dahlhoff et al., 2014, Li et al., 2016).
So far it has been poorly investigated how much the skin inflammation developing in the absence of epidermal EGFR affects the stem cell compartments thus aggravating the resulting phenotypes. Therefore in this study we employed conditional mouse models allowing us to dissect the cell-autonomous functions of EGFR signaling in distinct keratinocyte and stem cell compartments independently of the non-cell-autonomous contribution of inflammation. Our results indicate that EGFR signaling ensures DNA integrity in proliferative compartments, where accumulation of DNA damage results in TP53-dependent cell death. Moreover, we demonstrate that EGFR controls transcription factor expression in the innermost epithelial hair lineages, which is required for hair shaft differentiation.
Results
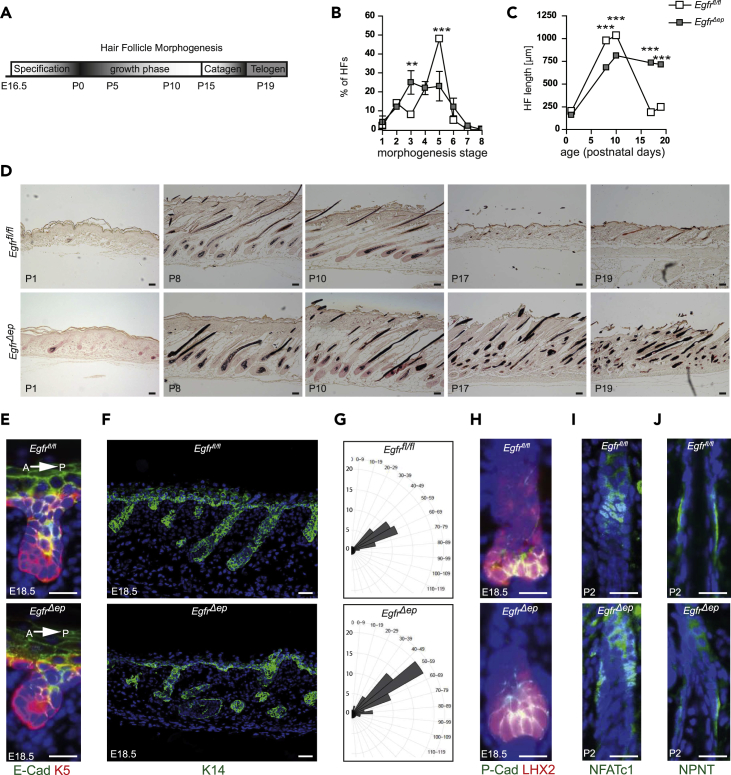
Previous studies have shown that skin-specific genetic ablation of EGFR during murine embryonic development causes profound defects of HF morphogenesis, but so far lacked detailed mechanistic analysis. Developing HFs need to go through several sequential phases to give rise to a mature HF (Figure 1A). To specifically investigate the function of EGFR signaling during the specification and growth phase of HF morphogenesis, Egfrfl/fl mice (Natarajan et al., 2007) were crossed with K5Cre transgenic mice to generate mice lacking the EGFR already during embryonic development in the epidermal lineage (EgfrΔep mice, Figures S1A and S1B; Table S3; Lichtenberger et al., 2013). Time course analysis during the growth, catagen, and telogen phases of HFs showed that EgfrΔep mice present a significant delay in HF growth (Figures 1B–1D), a catagen block (Figures 1C and 1D), and hair shape alterations (Figure S1C).
Figure 1.
EGFR Deficiency Does Not Result in Alterations of HF Specification during Morphogenesis
(A) Schematic time course of HF morphogenesis.
(B) HF morphogenesis staging from control and EgfrΔep mice at P1. Bars show analysis of 15–20 follicles per mouse from n = 3 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with **p < 0.01, ***p < 0.001 as determined by multiple t test using two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli with Q = 1%.
(C) Quantification of HF length from control and EgfrΔep mice at indicated time points. Bars show analysis of 15–20 follicles per mouse from n = 3 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with ***p < 0.001 as determined by multiple t test using two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli with Q = 1%.
(D) Fontana-Masson stainings of back skin sections from control and EgfrΔep mice at indicated time points. Scale bars, 50 μm.
(E) Immunofluorescence staining for E-Cadherin (green), K5 (red), and nuclei (blue) from control, and EgfrΔep mice at E18.5. Arrow indicates anterior-to-posterior orientation of skin section. A, anterior; P, posterior.
(F) Immunofluorescence staining for K14 (green) and nuclei (blue) from E18.5 control and EgfrΔep mice.
(G) Polar plots of angles between IFE and anterior side of HFs from E18.5 control and EgfrΔep mice. Plots show analysis of 8–12 follicles per mouse from n = 3 mice per genotype. Data represent the absolute number of HFs measured at indicated angle ranges.
(H) Immunofluorescence staining for P-Cadherin (green), LHX2 (red), and nuclei (blue) from control and EgfrΔep mice at E18.5.
(I) Immunofluorescence staining for NFATc1 (green) and nuclei (blue) from control and EgfrΔep mice at P2.
(J) Immunofluorescence staining for Nephronectin (NPNT, green) and nuclei (blue) from control and EgfrΔep mice at P2.
Scale bars, 20 μm unless otherwise stated. See also Figure S1.
EGFR Deficiency Does Not Result in Alterations of HF Specification during Morphogenesis
First, we analyzed HF specification at late embryonic time points to investigate whether lack of EGFR may impair early HF development. Staining for E-Cadherin confirmed correct anterior-to-posterior orientation of hair pegs as detected by the absence of E-Cadherin expression at the anterior part of the HF (Muller-Rover et al., 1999, Devenport and Fuchs, 2008) in both control and EgfrΔep mice (Figure 1E). Furthermore, the angle between IFE and HF was mainly similar between control and EgfrΔep mice (Figures 1F and 1G). Despite HF growth differences, we did not find differences in expression of hair follicle stem cell (HFSC) fate factors in newborn mice: histological stainings for the HF progenitor/stem cell markers P-Cadherin and LHX2 (Folgueras et al., 2013) (Figure 1H) and the NFATc1-positive or Nephronectin (NPTN)-positive prospective bulge region in HF (Horsley et al., 2008, Fujiwara et al., 2011) (Figures 1I and 1J) showed robust expression and correct location of those factors in embryonic and newborn skin of EgfrΔep mice. These results demonstrate that EGFR is not required to induce embryonic HFSC specification but exerts its function on proper HF morphogenesis during the growth phase.
EGFR Controls Genes Required for Epidermal Differentiation and DNA Integrity
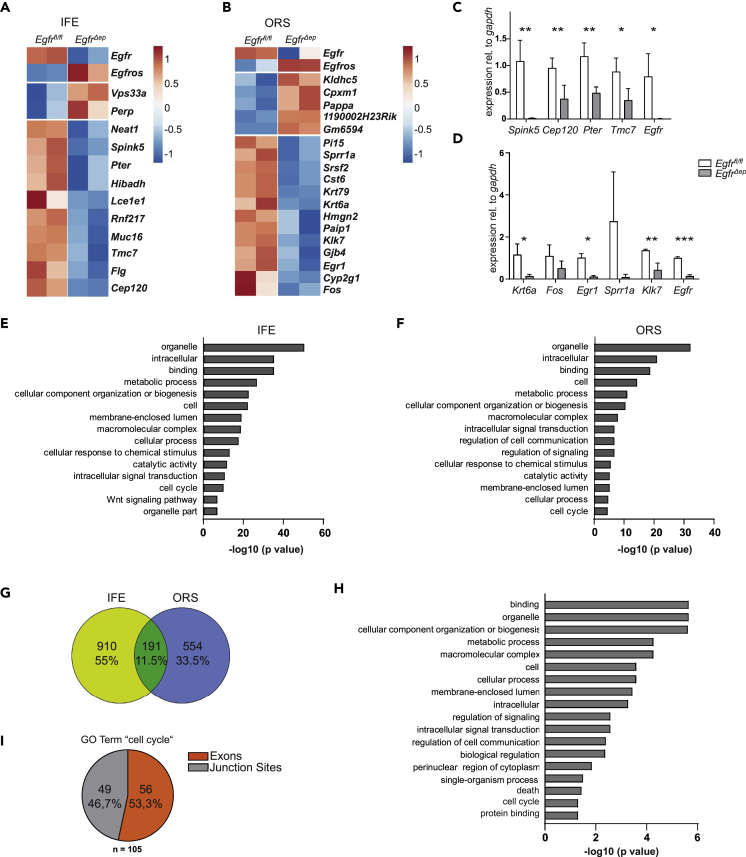
To uncover the genes involved in EGFR-mediated regulation of the growth of epidermal lineages, RNA sequencing (RNA-seq) analysis was performed on sorted IFE and ORS cells from postnatal day (P)2 mice. To label ORS cells we crossed EgfrΔep with LGR5CreERT2GFP mice to make use of LGR5-driven GFP expression as an ORS marker (Jaks et al., 2008), whereas IFE cells were defined as SCA-Ihi ITGA6hi cells (Figures S2A–S2C). The time point P2 was chosen, because at that stage there was still no immune cell infiltration in the skin of EgfrΔep pups (Figure S2D), but a delay in HF growth was already detectable (Figure 1B). We observed reduced GFP expression in ORS cells of EgfrΔep mice at P2, although the total fraction of LGR5-expressing cells was similar to control mice (Figures S2E–S2H). RNA-seq analysis showed strong expression of Egfr, Erbb2, and Erbb3 in IFE and ORS of control mice, whereas there was no expression of Erbb4. Furthermore, we observed no or only low EGFR ligand expression in the different epidermal compartments, like low Epgn expression in the IFE, and low Hb-egf in both IFE and ORS (Figure S2I). Analysis of differentially expressed genes between control and EgfrΔep mice revealed that loss of EGFR significantly affects the expression of structural genes (Spink5, Lce1e1, Flg, Sprr1a, Klk7, Gjb4), as well as of genes regulating cell cycle and apoptosis (Perp, Cep120, Klhdc5, Egr1, Fos) in IFE or ORS cells (Figures 2A–2D; Table S1A).
Figure 2.
EGFR Controls a Variety of Genes Required for Epidermal Differentiation and DNA Integrity
(A) Heatmap of significantly deregulated genes in P2 fluorescence-activated cell sorted IFE from control and EgfrΔep mice. Each column represents one mouse.
(B) Heatmap of significantly deregulated genes in P2 fluorescence-activated cell sorted ORS from control and EgfrΔep mice. Each column represents one mouse.
(C) qRT-PCR from randomly chosen target genes identified by RNA-seq from P2 IFE. Bars show data from n = 3–5 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with *p < 0.05, **p < 0.01 as determined by Student's t test.
(D) qRT-PCR from randomly selected target genes identified by RNA-seq from P2 ORS. Bars show data from n = 3–5 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with *p < 0.05, **p < 0.01, ***p < 0.001 as determined by Student's t test.
(E) Bar graph of the top 15 GO terms obtained from alternative splicing analysis from IFE keratinocytes.
(F) Bar graph of the top 15 GO terms obtained from alternative splicing analysis from ORS keratinocytes.
(G) Venn diagram of alternative splicing targets from P2 IFE and ORS cells.
(H) Bar graph of the significantly enriched GO terms obtained from overlapping genes displaying alternatively spliced exons or junction sites from IFE and ORS keratinocytes.
(I) Pie chart of the ratio of alternatively used exons and junction sites of genes within the GO term “cell cycle.”
EGFR Controls Alternative Splicing, which Regulates Centrosome Function and DNA Integrity
As the list of differentially expressed genes included a variety of targets that were either already shown to be directly linked to EGFR signaling, such as Flg (Franzke et al., 2012), or belonged to an expected biological process, namely, epidermal development, we focused on novel targets not yet reported to regulate epidermal integrity in the context of EGFR signaling. One of those target genes was Srsf2 (Figure 2B). The product of this gene is required for the formation of the earliest ATP-dependent splicing complex and contributes to constitutive as well as alternative splicing (Graveley and Maniatis, 1998). To functionally investigate whether lack of EGFR might affect mRNA splicing, we analyzed our RNA-seq datasets for alternative exon usage employing JunctionSeq (Hartley and Mullikin, 2016). For both compartments, IFE and ORS, we detected a high number of alternative transcripts upon comparison of control and EGFR-deficient keratinocytes. Importantly, these genes group into distinct GO terms like “intracellular signal transduction,” “Wnt pathway,” “cell cycle,” or “organelle part” (Figures 2E and 2F, Table S1B).
To gain more specific information from the large amount of alternatively used exons and junction sites, we focused on the 191 (11.5%) affected overlapping genes between IFE and ORS (Figure 2G, Table S1C). GO term analysis on those overlapping targets elucidated that EGFR is required to control the “perinuclear region of cytoplasm,” “cell cycle,” and “death” in both IFE and ORS (Figure 2H, Table S1D). Deeper investigation of the genes belonging to the term “cell cycle” revealed a high number of genes important in centrosome function and DNA damage repair, e.g., Cntrl, Cep192, Clspn, and Bub1b (Table S1E), and an almost equal number of differentially used exons and junction sites (Figure 2I, Table S1F).
EGFR Regulates Proliferation and Prevents DNA Damage in Epidermal Lineages
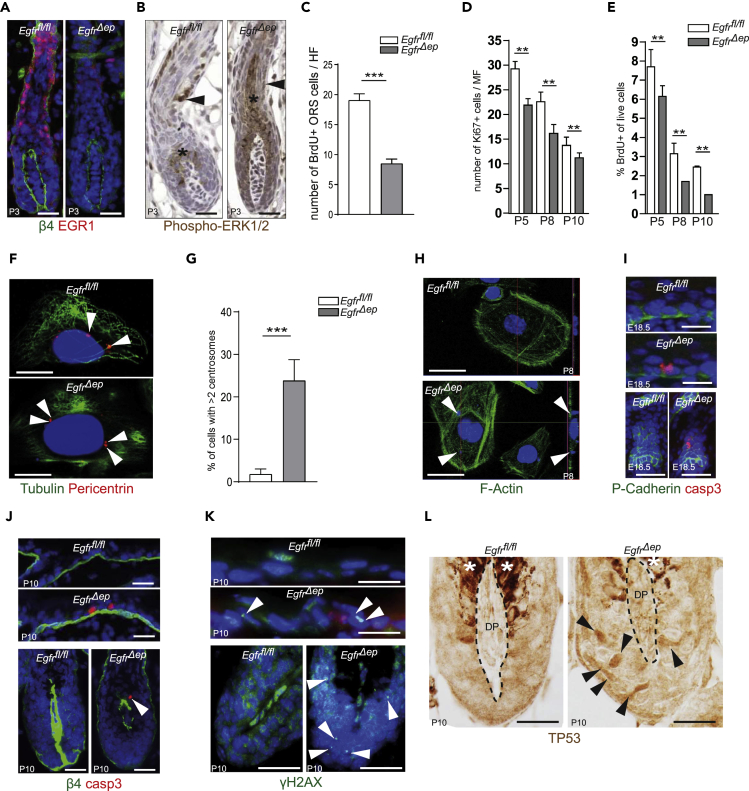
As both global gene expression and alternative splicing analysis identified genes involved in cell cycle, centrosome function, and DNA damage response we hypothesized that a strong disbalance of cell turnover or survival and apoptosis could explain the growth delay of EGFR-deficient HFs. In light of previous reports on an important function of EGFR in promotion of proliferation (Hansen et al., 1997, Bol et al., 1998, Xie et al., 1999, Kiguchi et al., 2000, Lichtenberger et al., 2010), we functionally tested whether EGFR is required for regulating the proliferation rate of distinct keratinocyte populations. RNA-seq analysis showed downregulation of early growth response 1 (Egr1) in the ORS of EgfrΔep mice (Figure 2B). It was previously demonstrated that Egr1 is a downstream target of ERK signaling and involved in cell growth and proliferation (Mayer et al., 2009, Tarcic et al., 2011). We confirmed EGR1 protein absence in the ORS (Figure 3A), as well as strongly reduced phospho-ERK1/2 selectively in ORS cells of EGFR-deficient skin (Figure 3B). In addition, bromodeoxyuridine (BrdU) pulse-chase experiments showed reduced BrdU incorporation into EGFR-deficient ORS cells after 24h (Figures 3C and S3A), providing evidence that EGFR signaling in the ORS is required to induce the ERK signaling cascade, leading to Egr1 induction and cell growth or proliferation. To address whether absence of EGFR also affects proliferation in the IFE, back skin sections from various time points (P5, P8, P10) were stained for Ki67 and additionally analyzed by flow cytometry after a 4h BrdU pulse. Our results show that both numbers of Ki67+ cells and BrdU+ cells are reduced in IFE of EgfrΔep mice when compared with control mice (Figures 3D, 3E, and S3B). These data confirm previous findings (Bol et al., 1998, Xie et al., 1999, Kiguchi et al., 2000, Lichtenberger et al., 2010) and provide more detailed insights into the important role of EGFR in promoting proliferation of the epidermal lineage.
Figure 3.
EGFR Controls Cell Proliferation as well as Centrosome Function and DNA Integrity
(A) Immunofluorescence staining for ITGB4 (green), EGR1 (red), and nuclei (blue) from P3 control and EgfrΔep mice.
(B) Immunohistochemistry staining for phospho-ERK1/2 from P3 control and EgfrΔep mice. Asterisk indicates hair pigmentation.
(C) Quantification of BrdU+ ORS cells of P2 mice 24h after BrdU pulsing. Bars show analysis of 10 follicles per mouse from n = 4 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with ***p < 0.001 as determined by Student's t test.
(D) Quantification of the number of Ki67+ IFE nuclei from frozen back skin sections from control and EgfrΔep mice at indicated time points. MF, microscopic field. Bars show analysis of 6 microscopic fields per mouse from n = 3 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with **p < 0.01 as determined by Student's t test.
(E) Quantification of the number of BrdU+ cells from epidermal cell suspensions analyzed by fluorescence-activated cell sorting from control and EgfrΔep mice at indicated time points. Bars show analysis of n = 3 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with **p < 0.01 as determined by Student's t test.
(F) Immunofluorescence staining for Tubulin (green), Pericentrin (red), and nuclei (blue) from primary keratinocytes derived from control and EgfrΔep mice. Arrows point to centrosomes. Scale bars, 5 μm.
(G) Quantification of the percentage of primary keratinocytes harboring more than two centrosomes. Bars show analysis of 15 cells per mouse from n = 3 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with ***p < 0.001 as determined by Student's t test.
(H) Immunofluorescence staining for F-Actin (green) and nuclei (blue) of primary keratinocytes derived from P8 control and EgfrΔep mice. Arrows highlight nuclear fragments. Scale bars, 5 μm.
(I) Immunofluorescence staining for P-Cadherin (green), activated caspase 3 (red), and nuclei (blue) from control and EgfrΔep mice at E18.5, showing IFE (top) and HF (bottom).
(J) Immunofluorescence staining for ITGB4 (green), activated caspase 3 (red), and nuclei (blue) from control and EgfrΔep mice at P10, showing IFE (top) and HF matrix (bottom). Arrow highlights an apopotic cell.
(K) Immunofluorescence staining for γH2AX (green) and nuclei (blue) from control and EgfrΔep mice at P10, showing IFE (upper panel) and HF matrix (lower panel). Arrows highlight spots of DNA damage.
(L) Immunohistochemical staining of TP53 on sections of P10 control and EgfrΔep mice, showing HF matrix. Asterisk indicates hair pigmentation, delineated area marks dermal papilla (DP).
Scale bars, 20 μm unless otherwise stated. See also Figure S3.
Detailed depiction of exon usage of Centriolin (Cntrl) (Figure S3C) shows that EGFR-deficient keratinocytes preferentially skip exons 11, 15, and 16. Exon 11 encodes an alternative 3′ UTR and leads to a shorter transcript, which is known as a 110-kDa variant of Cntrl termed Cep110. Skipping of the alternative part of exon 11 leads to an increase in full-length transcript, encoding a 270-kDa variant of Cntrl, including a domain required for centrosome localization (Szebenyi et al., 2007). Exons 15 and 16 contribute to the stathmin domain of CNTRL, which is involved in regulation of mitotic spindle assembly. Further investigation of Claspin (Clspn) mRNA shows that EgfrΔep mice skip exon 12 (Figure S3D). Claspin is required for cell-cycle arrest in response to DNA damage via activation of CHK1 (Lee et al., 2003). Interestingly, exon 12 encodes a protein region involved in forming the CHK1-activating domain (CKAD), suggesting that exon skipping leads to reduced binding of CLSPN to CHK1 and subsequently to reduced clearance of DNA damage.
To test this hypothesis, we investigated centrosome numbers, DNA damage, and apoptosis. Pericentrin stainings of primary keratinocytes revealed an increased fraction of EGFR-deficient keratinocytes displaying more than two centrosomes (Figures 3F and 3G). Moreover, we observed nuclear fragmentation in keratinocytes lacking EGFR (Figure 3H). As both, WT and EgfrΔep keratinocyte cultures, were negative for the differentiation marker K1, we exclude that fragmentation of nuclei might be due to accelerated differentiation of EGFR-deficient keratinocytes (Figure S3E). To analyze whether loss of DNA integrity is accompanied by increased cell death, we stained back skin sections for active caspase 3. Apoptotic cells were detected in the IFE and hair pegs of E18.5 epidermis (Figure 3I) as well as in P10 IFE and matrix of EgfrΔep mice (Figure 3J). Furthermore, staining for the DNA damage marker γH2AX uncovered a high number of γH2AX foci in skin of EgfrΔep mice, but not in control mice (Figure 3K). The presence of DNA damage was accompanied by stabilization and accumulation of TP53 in nuclei of matrix cells from EgfrΔep mice (Figure 3L). These data demonstrate that absence of EGFR leads to accumulation of DNA damage, which subsequently induces TP53 stabilization and apoptosis.
Apoptosis in IFE and Matrix Is TP53 Dependent, Whereas Loss of Inner Hair Layers Is TP53 Independent
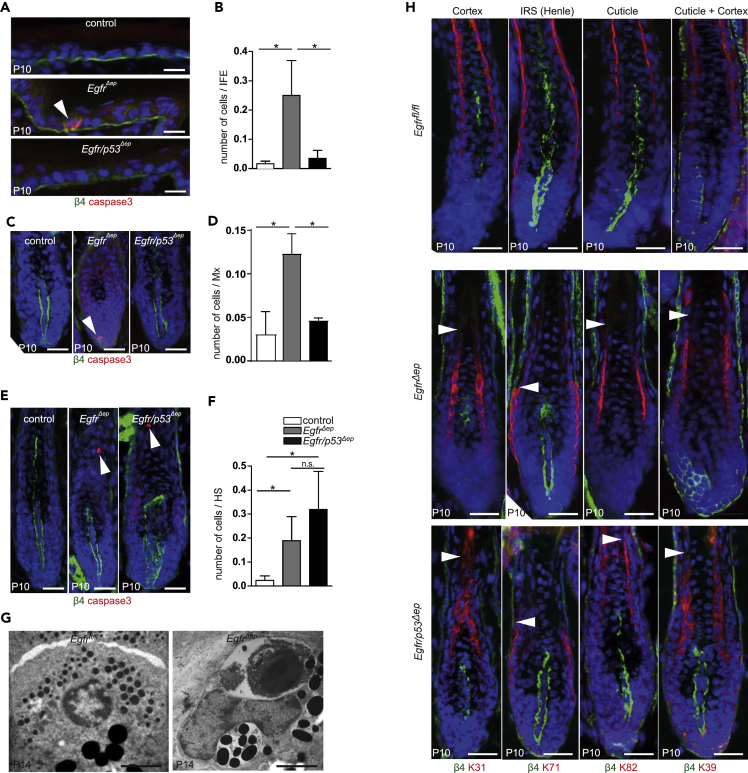
To investigate whether TP53-induced cell death causes impaired HF development in EgfrΔep mice, we crossed EgfrΔep mice with p53fl/fl mice to generate Egfr/p53Δep mice (Figures S4A and S4B). Surprisingly, Egfr/p53Δep mice presented the same phenotype as EgfrΔep mice: skin and hair alterations, reduced survival, and strong skin inflammation (Figures S4C–S4E). However, deletion of TP53 abolished apoptosis in IFE and matrix cells in EgfrΔep mice at P10 (Figures 4A–4D), suggesting a TP53-dependent mechanism for cell death in these highly proliferative epidermal compartments. Upon closer investigation of other epithelial compartments in the epidermal lineage, we also detected apoptotic cells in the hair shaft of EgfrΔep mice, which were, however, not affected by additional TP53 deletion (Figures 4E and 4F), providing evidence for a TP53-independent apoptotic mechanism in postmitotic cells. We could also confirm the presence of apoptosis in innermost hair layers, such as medulla cells, by transmission electron microscopy (Figure 4G). To characterize whether apoptosis in the hair shaft affects the differentiation of epithelial hair layers in mice lacking epidermal EGFR, we stained for hair-layer-specific keratins in control, EgfrΔep mice, and Egfr/p53Δep mice. Both EgfrΔep mice and Egfr/p53Δep mice exhibited onset of hair layer differentiation, but displayed an early loss of those layers (Figure 4H). These data indicate TP53-dependent cell death in proliferative keratinocytes as well as a TP53-independent mechanism of apoptosis in the non-proliferative, differentiated cells of the hair shaft upon lack of EGFR signaling.
Figure 4.
Apoptosis in IFE and Matrix Is TP53 Dependent, whereas Loss of Inner Hair Layers Is TP53 Independent
(A) Immunofluorescence staining for ITGB4 (green), activated caspase 3 (red) and nuclei (blue) from control, EgfrΔep, and Egfr/p53Δep mice at P10, showing IFE. Arrow highlights an apoptotic cell.
(B) Quantification of the number of activated caspase 3+ IFE cells from control, EgfrΔep, and Egfr/p53Δep mice at P10. Bars show analysis of 10 microscopic fields per mouse from n = 3 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with *p < 0.05 as determined by Kruskal-Wallis test.
(C) Immunofluorescence staining for ITGB4 Integrin (green), activated caspase 3 (red), and nuclei (blue) from control, EgfrΔep, and Egfr/p53Δep mice at P10, showing HF matrix (Mx). Arrow highlights an apoptotic cell.
(D) Quantification of the number of activated caspase 3+ matrix cells from control, EgfrΔep, and Egfr/p53Δep mice at P10. Bars show analysis of 20–30 hair follicles per mouse from n = 3 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with *p < 0.05 as determined by Kruskal-Wallis test.
(E) Immunofluorescence staining for ITGB4 (green), activated caspase 3 (red), and nuclei (blue) from control, EgfrΔep, and Egfr/p53Δep mice at P10, showing hair shaft (HS). Arrows highlight apoptotic cells.
(F) Quantification of the number of activated caspase 3+ hair shaft cells from control, EgfrΔep, and Egfr/p53Δep mice at P10. Bars show analysis of 20–30 hair follicles per mouse from n = 3 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with *p < 0.05 as determined by Kruskal-Wallis test.
(G) Transmission electron microscopic image of medulla cells from P14 control and EgfrΔep mice. Scale bars, 2 μm.
(H) Immunofluorescence staining for ITGB4 (green), cortex marker K31 (red), and nuclei (blue) from control, EgfrΔep, and Egfr/p53Δep mice at P10 (far left). Immunofluorescence staining for ITGB4 (green), IRS (Henle layer) marker K71 (red), and nuclei (blue) from control, EgfrΔep, and Egfr/p53Δep mice at P10 (middle left). Immunofluorescence staining for ITGB4 (green), cuticle marker K82 (red), and nuclei (blue) from control, EgfrΔep, and Egfr/p53Δep mice at P10 (middle right). Immunofluorescence staining for ITGB4 (green), cuticle and cortex marker K39 (red), and nuclei (blue) from control, EgfrΔep, and Egfr/p53Δep mice at P10 (far right). Arrows highlight disappearance of layers.
Scale bars, 20 μm unless otherwise stated. See also Figure S4.
EGFR Controls Hair Lineage Transcription Factor Expression
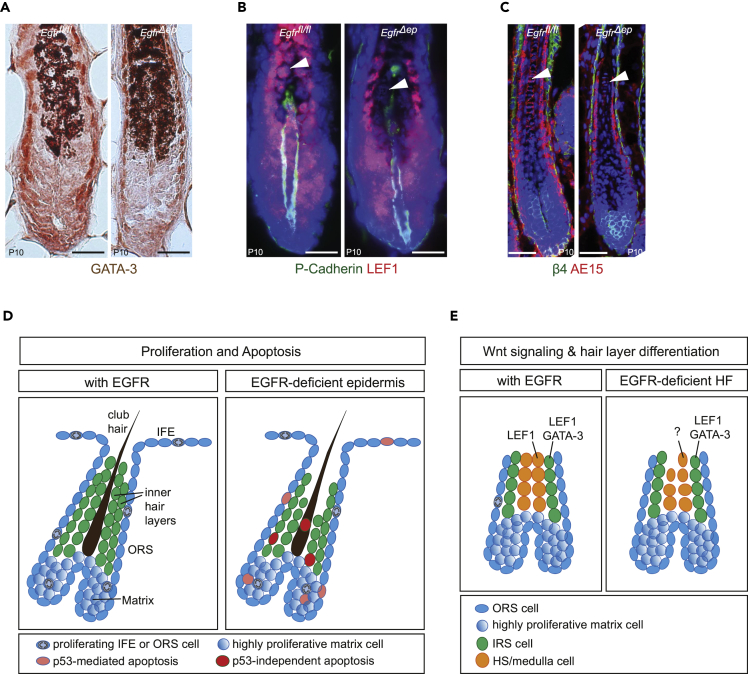
To investigate whether transcription factors involved in hair layer differentiation showed altered expression upon EGFR deletion, we stained for IRS-specific transcription factor GATA-3, as well as for IRS- and HS-specific factor LEF1 (Merrill, 2001, Kaufman, 2003, Adam et al., 2018). GATA-3 expression was not different between control and EgfrΔep mice (Figure 5A). However, LEF1 was only found to be expressed in IRS cells of EgfrΔep mice, but not in hair shaft cells (Figure 5B). Staining for trichohyalin AE15, which is present in differentiated IRS and medulla, revealed lack of properly differentiated medulla cells in EgfrΔep mice (Figure 5C).
Figure 5.
EGFR Controls Hair Lineage Transcription Factor Expression
(A) IHC staining of GATA-3 from P10 control and EgfrΔep mice.
(B) Immunofluorescence staining for P-Cadherin (green), LEF1 (red), and nuclei (blue) from P10 control and EgfrΔep mice. Arrowheads point to hair shaft cells.
(C) Immunofluorescence staining for ITGB4 (green), IRS and medulla marker AE15 (red), and nuclei (blue) from control and EgfrΔep mice at P10. Arrowheads point on medulla cells.
(D) Model summarizing TP53-dependent mechanisms of cell death in IFE, ORS, and medulla during the growth phase of HF morphogenesis in EgfrΔep mice.
(E) Model summarizing TP53-independent effects of EGFR deficiency in epithelial hair lineages.
Scale bars, 20 μm unless otherwise stated.
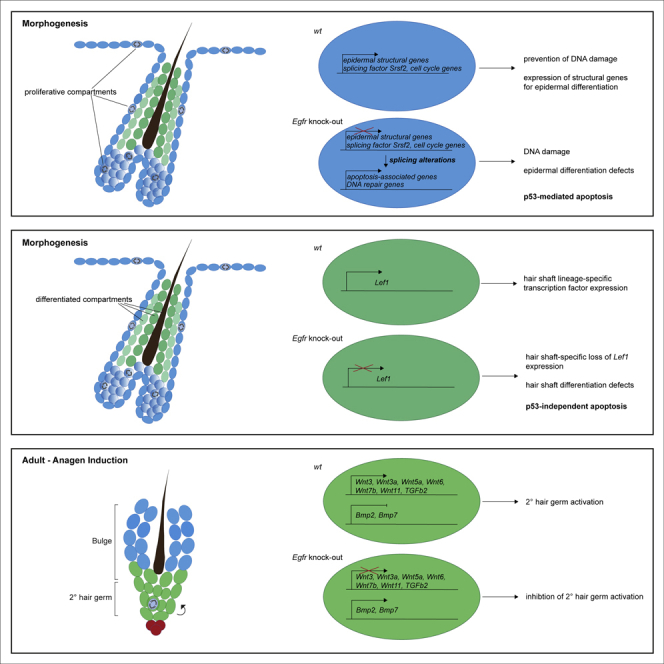
Based on our results, we propose that EGFR signaling affects spatial layering during HF growth and differentiation (Figures 5D and 5E). First, EGFR promotes cell proliferation and ensures DNA integrity in highly proliferative epidermal compartments, such as IFE, ORS, and matrix. Upon loss of EGFR in those compartments, accumulation of DNA damage mediates TP53-dependent apoptosis (Figure 5D). Second, EGFR controls hair-layer-specific transcription factor expression to regulate epithelial hair layer specification and differentiation. EGFR deficiency leads to absence of proper differentiation cues for hair shaft cells, resulting in TP53-independent loss of the medulla layer (Figure 5E).
Loss of EGFR in Adult HFSCs Results in Growth Delay
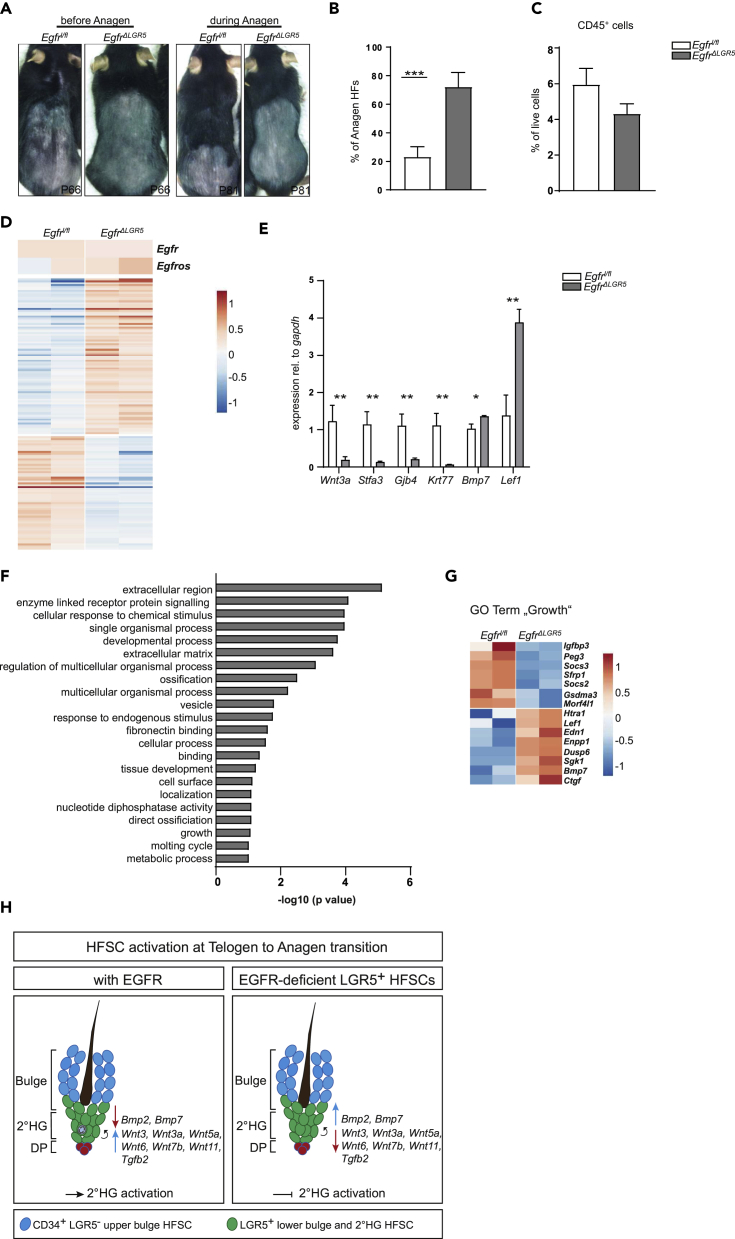
We next investigated whether EGFR also serves a growth-promoting function in adult HFSCs. Therefore, we first tested whether EGFR is expressed in adult HFSCs by applying fluorescently labeled epidermal growth factor to cell suspensions of adult (P45) wild-type mice after completion of the first hair cycle, when hair is in early telogen. We detected high EGFR expression in CD34+ old and new bulge stem cells, intermediate expression in IFE, and no expression in CD45+ hematopoietic cells using flow cytometric analysis (Figure S5A). These results were confirmed by EGFR stainings on back skin sections (Figure S5B). Furthermore, EGFR expression was detected within the matrix, the ORS and the IRS of anagen HFs (Figures S5C and S5D). During catagen, scattered EGFR expression was observed in the retracting HF (Figure S5E). These results suggest that EGFR signaling is required in adult HFSCs. Because it is not possible to investigate activation and growth of adult HFSCs in EgfrΔep mice owing to HF degradation and progressive hair loss (Lichtenberger et al., 2013), we crossed Egfrfl/fl to Lgr5CreERT2GFP mice to delete the EGFR after completion of the first postnatal hair cycle (from P43-P47) in lower bulge and secondary hair germ (2°HG) HFSCs by tamoxifen injection (EGFRΔLGR5−ERT2 mice, Figure S5F). EgfrΔLGR5-ERT2 mice were monitored during progression of the second postnatal hair cycle until P88. EGFR deletion was confirmed by genomic DNA (Figure S5G) and by immunohistochemistry (IHC) analysis (Figure S5H). EgfrΔLGR5-ERT2 mice exhibited delayed entry into the second hair cycle (Figure 6A), demonstrating that EGFR signaling is involved in HFSC activation and proliferation. This was further underlined by the finding that by the time of analysis most follicles of control mice had completed the second hair cycle and already returned to telogen, whereas the majority of HFs of EgfrΔLGR5-ERT2 mice was still in anagen of the second cycle (Figure 6B). Importantly, EgfrΔLGR5-ERT2 mice did not display an immune cell infiltrate (Figure 6C), emphasizing that the growth delay observed in EgfrΔLGR5-ERT2 mice is solely due to cell-autonomous defects in HFSCs.
Figure 6.
Loss of EGFR in Adult HFSCs Results in Growth Delay
(A) Photographs of shaved back skin from control and EgfrΔLGR5 mice before and during anagen of second hair cycle.
(B) Quantification of percentage of anagen HFs in mice of indicated genotypes at P88. Bars show analysis of 10 hair follicles per mouse from n = 7–8 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with ***p < 0.001 as determined by Student's t test.
(C) Quantification of CD45+ cells in P88 control and EgfrΔLGR5 mice. Bars show analysis of n = 3–5 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant. The data sets do not show staticstical significance as determined by Student's t test.
(D) Heatmap of significantly deregulated genes between P88 control and EgfrΔLGR5 mice. Each column represents one mouse.
(E) Graphical representation of qRT-PCR validation of randomly selected target genes from LGR5+ cells of P88 control and EgfrΔLGR5 mice. Bars show analysis of n = 4 mice per genotype. Data are represented as mean ± SD. p values less than 0.05 were considered significant, with *p < 0.05, **p < 0.01 as determined by Student's t test.
(F) Bar diagram of GO terms of significantly deregulated genes.
(G) Heatmap of deregulated genes belonging to GO Term “growth.” Each column represents one mouse.
(H) Model depicting the effects of EGFR in adult HFSCs.
To identify molecules deregulated in adult HFSCs upon loss of EGFR, we performed flow cytometric cell sorting of LGR5+ cells from P88 control and EgfrΔLGR5-ERT2 mice and subjected them to RNA-seq (Figures S5I and S5J). We found 145 genes to be significantly differentially expressed between LGR5+ cells derived from control and EgfrΔLGR5-ERT2 mice (Figures 6D and 6E, Table S2A). Among the differentially expressed genes, we found genes encoding for structural proteins that were deregulated in our dataset obtained from P2 EgfrΔep mice such as Gjb4 and Klk7 and also genes encoding for signaling and growth molecules such as Wnt3a or Bmp7 indicating that lack of EGFR signaling in LGR5+ HFSCs broadly shifts the balance of molecules regulating HFSC behavior. Overall, affected genes show enrichment for GO terms such as “cellular response to chemical stimulus,” “tissue development,” “molting cycle,” or “growth” (Figures 6F and 6G, Tables S2B and S2C). Thus loss of EGFR negatively alters the susceptibility of HFSCs to respond to activating cues (Figure 6H).
Investigation of receptor and ligand expression in control LGR5+ HFSCs revealed that similar to P2 IFE and ORS, these cells highly express Egfr, Erbb2, and Erbb3, but not Erbb4. Moreover, LGR5+ HFSCs display low expression of the ligands Tgfα, Epgn, and Hb-egf, whereas all other ligands are not expressed (Figure S5K).
Furthermore, exons or junction sites of more than 300 genes were affected by alternative splicing alterations in LGR5+ cells of Lgr5+ EgfrΔLGR5-ERT2 mice, enriching for GO terms associated with cell stimulation and intracellular metabolic processes (Figure S5L, Table S2D). Notably, we detected a substantial overlap of alternative transcripts from 213 genes (10.3%) between P2 IFE + ORS and P88 LGR5+ cells (Figure S5M, Table S2E), leading to the assumption that EGFR has some identical targets in distinct epidermal compartments at different time points, as well as a variety of specific targets depending on epidermal cell identity and age. Shared target genes display significant enrichment for GO terms such as “organelle,” “intracellular signal transduction,” and “cell cycle” (Figure S5N, Table S2F), further underlining that EGFR is required for proliferation and proper orchestration of molecules and pathways involved in both HF development and adult HFSC activation and new HF growth.
Discussion
Our study demonstrates that the phenotype of EGFR-deficient skin is multifactorial, displaying alterations in expression of structural proteins, DNA integrity and DNA damage, proliferation, and apoptosis, but not stem cell identity genes. Lack of EGFR in newborn skin reduces proliferation in ORS and also in IFE keratinocytes. Analysis of RNA-seq data and subsequent histological analysis demonstrate reduced ERK activation and loss of expression of the ERK downstream target Egr1 (Mayer et al., 2009, Tarcic et al., 2011) in EGFR-deficient ORS. In various studies on cancer cells and in mouse models of ERBB2 overexpression a prominent function of EGFR signaling in promoting keratinocyte proliferation via ERK phosphorylation has been shown (Lichtenberger et al., 2010) (Bol et al., 1998, Xie et al., 1999, Kiguchi et al., 2000). The proliferation-promoting function of EGFR was mainly assessed in a hyperproliferative context of tumor cells, whereas only little information is available in the framework of epidermal morphogenesis and homeostasis. Albeit a reduction of proliferating keratinocytes was shown for the IFE of Egfr−/− mice (Hansen et al., 1997), this was never formally shown for the ORS. Importantly, previous studies investigating adult EGFR-deficient mice could not show changes in pERK by using in vitro assays (Nagao et al., 2012) or in vivo analysis (Wolf et al., 2016). The absence of effects on pERK levels might be due to assessment of fully developed adult murine skin, and not neonatal mice, undergoing epidermal morphogenesis. In addition, many other pathways have been shown to induce ERK phosphorylation in keratinocytes. Thus our data provide new insight into a growth-promoting function of EGFR in the developing ORS.
We assessed the specification of committed HFSC populations during early HF development by using markers LHX2, NFATc1, and NPNT. All factors were equally expressed and localized in control and EgfrΔep mice. Among the group of epidermal structural genes displaying differential gene expression in our RNA-seq dataset were factors that each alone can result in severe epidermal or hair alterations: mutations in Flg are known to be involved in the development of atopic dermatitis (Tholen et al., 2016), mutations in Spink5 are reported to cause Netherton syndrome (Raghunath et al., 2004), and mutations in Gjb4 are associated with erythrokeratodermia variabilis (Richard et al., 2003). These findings demonstrate that one critical role of EGFR signaling is transcriptional regulation of structural genes directly involved in epidermal differentiation.
We identified centrosome assembly and function as a novel downstream process of EGFR signaling by our expression and alternative splicing analysis. IFE keratinocytes lacking EGFR display centrosome amplifications. Functional consequences of abnormal centrosome numbers due to a disbalance in centrosomal proteins were previously shown to correlate with genome instability (Vitre and Cleveland, 2012, Nam et al., 2015). Importantly, defects in centrosome or mitotic spindle assembly leading to the generation of multiple nuclei are partially causative for the rare disease trichothiodystrophy (Nakabayashi et al., 2005, Zhang et al., 2007), which affects DNA repair and hair structure.
Generally, genome instability and nuclear fragmentation are accompanied by DNA damage. In the epidermis, centrosome amplifications result in impaired mitosis and TP53-mediated cell death (Kulukian et al., 2015). In our datasets, we find transcriptional upregulation of genes associated with cellular response to DNA damage in EgfrΔep mice: TP53 apoptosis effector Perp is known as an apoptosis-associated target of TP53, involved in its stabilization (Attardi et al., 2000, Davies et al., 2011), and Hmgn2 was reported to facilitate chromatin accessibility for DNA repair machinery (Subramanian et al., 2009). So far, cross talk between EGFR and TP53 signaling was only shown in vitro (Kryeziu et al., 2013). Importantly, our data for the first time provide in vivo evidence for a role of EGFR signaling in prevention of DNA damage in proliferating cells and for susceptibility of epidermal cells to TP53-mediated apoptosis upon loss of EGFR. Despite the DNA-damage-induced apoptosis in proliferative keratinocyte compartments, we detect TP53-independent cell loss in differentiated epithelial hair layers comprising the hair shaft. Suprabasal-transit-amplifying cells, which give rise to those innermost hair layers, display a requirement for EGFR signaling to induce correct transcription factor expression of the lineage diversification factor Lef1. During morphogenesis, LEF1 protein is not detectable in hair shaft cells of EGFR-deficient epidermis, thus presumably reducing the expression of hair-shaft-specific genes. These defects ultimately lead to loss of hair shaft cells, hair structure alteration, and hair malformation.
Limitation of the Study
We are formally not able to discriminate between a cell-autonomous function of EGFR in epithelial hair lineage differentiation, or a secondary effect on hair shaft differentiation following absence of EGFR in all epithelial skin lineages. To date, there are no-hair shaft-specific Cre lines available that would allow detailed investigation of the effects of Egfr deletion in these particular hair lineages.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank the Core Facility “Flow Cytometry” of the Medical University of Vienna for help with FACS sorting; and the Core Facility “Genomics” of the Medical University of Vienna for performance of RNA quality control, library preparation, and RNA-seq; as well as Margit Pavelka from the Medical University of Vienna and Daniela Gruber from the University of Vienna for help with Transmission Electron Microscopy (TEM) sample preparation. TEM imaging was performed at the Core Facility “Cell Imaging and Ultrastructure Research” at the University of Vienna. Moreover, we thank Christine Tuppy for assistance in primary keratinocyte culture and are grateful to Martina Hammer and the staff of the Department of Biomedical Research for maintaining our mouse colonies. We thank Khalil Kass Youssef for assistance with histology, Caroline Stremnitzer for critical reading of the manuscript, and Florian Pauler for assistance in R data processing. This work was supported by the Austrian Science Fund (FWF) grants FWF-DK W1212 and the Austrian Federal Government’s GEN-AU program “Austromouse” (GZ 200.147/1-VI/1a/2006 and 820966). M.S.'s research is funded by the European Research Council (ERC) under the Horizon 2020 research and innovation program (grant agreement ERC-2015-AdG 694883), the EU H2020 MSCA ITN-766241, and the Vienna Science and Technology Fund (WWTF, grant agreement LS16-025).
Author Contributions
N.A. designed and performed most of the experiments. P.A.S. provided intellectual and experimental input. G.H. analyzed RNA-seq data. M.H. provided intellectual input. B.M.L. performed hair staging analysis. B.C. performed qRT-PCR. T.B.-G. helped with TEM. C.B. provided intellectual input as well as laboratory space and reagents. M.S. supervised the whole project. N.A., M.H., and M.S. wrote the manuscript.
Declaration of Interests
The authors declare no conflicts of interest.
Published: May 31, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.04.018.
Supplemental Information
A: P2 IFE and ORS significantly differentially expressed genes.
B: Alternative Splicing P2 IFE and ORS Top 15 GO Terms, -log10.
C: Alternative splicing P2 overlapping genes between IFE and ORS.
D: Alternative splicing P2 overlapping GO terms between IFE and ORS.
E: Alternative splicing P2 GO Term “Cell Cycle.”
F: Alternative splicing P2 GO Term “Cell Cycle”—exon usage details.
A: P88 LGR5 significantly differentially expressed genes.
B: P88 LGR5 GO term enrichment of significantly differentially expressed genes.
C: P88 LGR5 significantly differentially expressed genes in GO term “growth.”
D: P88 LGR5 alternative splicing Top 15 GO terms -log10.
E: Alternative splicing overlapping genes between P2 IFE + ORS and P88 LGR5.
F: GO term enrichment of alternative splicing overlapping genes between P2 IFE + ORS and P88 LGR5.
Oligonucleotide Sequences for qRT-PCR primers and for genotyping primers.
Reagents (media, buffers, enzymes, etc.).
Mice and software.
Antibodies used for histology and flow cytometry.
References
- Adam R.C., Yang H., Ge Y., Lien W.H., Wang P., Zhao Y., Polak L., Levorse J., Baksh S.C., Zheng D. Temporal layering of signaling effectors drives chromatin remodeling during hair follicle stem cell lineage progression. Cell Stem Cell. 2018;22:398–413.e7. doi: 10.1016/j.stem.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi L.D., Reczek E.E., Cosmas C., Demicco E.G., McCurrach M.E., Lowe S.W., Jacks T. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–718. [PMC free article] [PubMed] [Google Scholar]
- Bichsel K.J., Hammiller B., Trempus C.S., Li Y., Hansen L.A. The epidermal growth factor receptor decreases Stathmin 1 and triggers catagen entry in the mouse. Exp. Dermatol. 2016;25:275–281. doi: 10.1111/exd.12921. [DOI] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol D., Kiguchi K., Beltran L., Rupp T., Moats S., Gimenez-Conti I., Jorcano J., DiGiovanni J. Severe follicular hyperplasia and spontaneous papilloma formation in transgenic mice expressing the neu oncogene under the control of the bovine keratin 5 promoter. Mol. Carcinog. 1998;21:2–12. [PubMed] [Google Scholar]
- Crew F.A.E. Waved - an autosomal recessive coat form character in the mouse. J. Genet. 1933;27:95–96. [Google Scholar]
- Dahlhoff M., Frances D., Kloepper J.E., Paus R., Schafer M., Niemann C., Schneider M.R. Overexpression of epigen during embryonic development induces reversible, epidermal growth factor receptor-dependent sebaceous gland hyperplasia. Mol. Cell. Biol. 2014;34:3086–3095. doi: 10.1128/MCB.00302-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R., Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- Davies L., Spiller D., White M.R., Grierson I., Paraoan L. PERP expression stabilizes active p53 via modulation of p53-MDM2 interaction in uveal melanoma cells. Cell Death Dis. 2011;2:e136. doi: 10.1038/cddis.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport D., Fuchs E. Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat. Cell Biol. 2008;10:1257–1268. doi: 10.1038/ncb1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueras A.R., Guo X., Pasolli H.A., Stokes N., Polak L., Zheng D., Fuchs E. Architectural niche organization by LHX2 is linked to hair follicle stem cell function. Cell Stem Cell. 2013;13:314–327. doi: 10.1016/j.stem.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke C.W., Cobzaru C., Triantafyllopoulou A., Loffek S., Horiuchi K., Threadgill D.W., Kurz T., van Rooijen N., Bruckner-Tuderman L., Blobel C.P. Epidermal ADAM17 maintains the skin barrier by regulating EGFR ligand-dependent terminal keratinocyte differentiation. J. Exp. Med. 2012;209:1105–1119. doi: 10.1084/jem.20112258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H., Ferreira M., Donati G., Marciano D.K., Linton J.M., Sato Y., Hartner A., Sekiguchi K., Reichardt L.F., Watt F.M. The basement membrane of hair follicle stem cells is a muscle cell niche. Cell. 2011;144:577–589. doi: 10.1016/j.cell.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B.R., Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- Hansen L.A., Alexander N., Hogan M.E., Sundberg J.P., Dlugosz A., Threadgill D.W., Magnuson T., Yuspa S.H. Genetically null mice reveal a central role for epidermal growth factor receptor in the differentiation of the hair follicle and normal hair development. Am. J. Pathol. 1997;150:1959–1975. [PMC free article] [PubMed] [Google Scholar]
- Hartley S.W., Mullikin J.C. Detection and visualization of differential splicing in RNA-Seq data with JunctionSeq. Nucleic Acids Res. 2016;44:e127. doi: 10.1093/nar/gkw501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V., Aliprantis A.O., Polak L., Glimcher L.H., Fuchs E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 2008;132:299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-C., Li L., Fuchs E. Emerging interactions between skin stem cells and their niches. Nat. Med. 2014;20:847–856. doi: 10.1038/nm.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Erdmann B., Cotsarelis G., Birchmeier W. β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Jaks V., Barker N., Kasper M., van Es J.H., Snippert H.J., Clevers H., Toftgård R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Kaufman C.K. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiguchi K., Bol D., Carbajal S., Beltran L., Moats S., Chan K., Jorcano J., DiGiovanni J. Constitutive expression of erbB2 in epidermis of transgenic mice results in epidermal hyperproliferation and spontaneous skin tumor development. Oncogene. 2000;19:4243–4254. doi: 10.1038/sj.onc.1203778. [DOI] [PubMed] [Google Scholar]
- Kryeziu K., Jungwirth U., Hoda M.A., Ferk F., Knasmuller S., Karnthaler-Benbakka C., Kowol C.R., Berger W., Heffeter P. Synergistic anticancer activity of arsenic trioxide with Erlotinib is based on inhibition of EGFR-mediated DNA double-strand break repair. Mol. Cancer Ther. 2013;12:1073–1084. doi: 10.1158/1535-7163.MCT-13-0065. [DOI] [PubMed] [Google Scholar]
- Kulukian A., Holland A.J., Vitre B., Naik S., Cleveland D.W., Fuchs E. Epidermal development, growth control, and homeostasis in the face of centrosome amplification. Proc. Natl. Acad. Sci. U S A. 2015;112:E6311–E6320. doi: 10.1073/pnas.1518376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W.G. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Li Y., Stoll S.W., Sekhon S., Talsma C., Camhi M.I., Jones J.L., Lambert S., Marley H., Rittie L., Grachtchouk M. Transgenic expression of human amphiregulin in mouse skin: inflammatory epidermal hyperplasia and enlarged sebaceous glands. Exp. Dermatol. 2016;25:187–193. doi: 10.1111/exd.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenberger B.M., Gerber P.A., Holcmann M., Buhren B.A., Amberg N., Smolle V., Schrumpf H., Boelke E., Ansari P., Mackenzie C. Epidermal EGFR controls cutaneous host defense and prevents inflammation. Sci. Transl. Med. 2013;5:199ra111. doi: 10.1126/scitranslmed.3005886. [DOI] [PubMed] [Google Scholar]
- Lichtenberger B.M., Tan P.K., Niederleithner H., Ferrara N., Petzelbauer P., Sibilia M. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell. 2010;140:268–279. doi: 10.1016/j.cell.2009.12.046. [DOI] [PubMed] [Google Scholar]
- Luetteke N.C., Phillips H.K., Qiu T.H., Copeland N.G., Earp H.S., Jenkins N.A., Lee D.C. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 1994;8:399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- Luetteke N.C., Qiu T.H., Peiffer R.L., Oliver P., Smithies O., Lee D.C. TGFα deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell. 1993;73:263–278. doi: 10.1016/0092-8674(93)90228-i. [DOI] [PubMed] [Google Scholar]
- Mascia F., Lam G., Keith C., Garber C., Steinberg S.M., Kohn E., Yuspa S.H. Genetic ablation of epidermal EGFR reveals the dynamic origin of adverse effects of anti-EGFR therapy. Sci. Transl. Med. 2013;5:199ra110. doi: 10.1126/scitranslmed.3005773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer S.I., Rossler O.G., Endo T., Charnay P., Thiel G. Epidermal-growth-factor-induced proliferation of astrocytes requires Egr transcription factors. J. Cell Sci. 2009;122:3340–3350. doi: 10.1242/jcs.048272. [DOI] [PubMed] [Google Scholar]
- Merrill B.J. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen P.J., Berger J.E., Meneses J., Phung Y., Pedersen R.A., Werb Z., Derynck R. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- Muller-Rover S., Tokura Y., Welker P., Furukawa F., Wakita H., Takigawa M., Paus R. E- and P-cadherin expression during murine hair follicle morphogenesis and cycling. Exp. Dermatol. 1999;8:237–246. doi: 10.1111/j.1600-0625.1999.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Murillas R., Larcher F., Conti C.J., Santos M., Ullrich A., Jorcano J.L. Expression of a dominant negative mutant of epidermal growth factor receptor in the epidermis of transgenic mice elicits striking alterations in hair follicle development and skin structure. EMBO J. 1995;14:5216–5223. doi: 10.1002/j.1460-2075.1995.tb00206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao K., Kobayashi T., Ohyama M., Akiyama H., Horiuchi K., Amagai M. Brief report: requirement of TACE/ADAM17 for hair follicle bulge niche establishment. Stem Cells. 2012;30:1781–1785. doi: 10.1002/stem.1153. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Amann D., Ren Y., Saarialho-Kere U., Avidan N., Gentles S., MacDonald J.R., Puffenberger E.G., Christiano A.M., Martinez-Mir A. Identification of C7orf11 (TTDN1) gene mutations and genetic heterogeneity in nonphotosensitive trichothiodystrophy. Am. J. Hum. Genet. 2005;76:510–516. doi: 10.1086/428141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H.J., Naylor R.M., van Deursen J.M. Centrosome dynamics as a source of chromosomal instability. Trends Cell Biol. 2015;25:65–73. doi: 10.1016/j.tcb.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A., Wagner B., Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc. Natl. Acad. Sci. U S A. 2007;104:17081–17086. doi: 10.1073/pnas.0704126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunath M., Tontsidou L., Oji V., Aufenvenne K., Schurmeyer-Horst F., Jayakumar A., Stander H., Smolle J., Clayman G.L., Traupe H. SPINK5 and Netherton syndrome: novel mutations, demonstration of missing LEKTI, and differential expression of transglutaminases. J. Invest. Dermatol. 2004;123:474–483. doi: 10.1111/j.0022-202X.2004.23220.x. [DOI] [PubMed] [Google Scholar]
- Richard G., Brown N., Rouan F., Van der Schroeff J.G., Bijlsma E., Eichenfield L.F., Sybert V.P., Greer K.E., Hogan P., Campanelli C. Genetic heterogeneity in erythrokeratodermia variabilis: novel mutations in the connexin gene GJB4 (Cx30.3) and genotype-phenotype correlations. J. Invest. Dermatol. 2003;120:601–609. doi: 10.1046/j.1523-1747.2003.12080.x. [DOI] [PubMed] [Google Scholar]
- Sibilia M., Wagner B., Hoebertz A., Elliott C., Marino S., Jochum W., Wagner E.F. Mice humanised for the EGF receptor display hypomorphic phenotypes in skin, bone and heart. Development. 2003;130:4515–4525. doi: 10.1242/dev.00664. [DOI] [PubMed] [Google Scholar]
- Sibilia M., Wagner E. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- Subramanian M., Gonzalez R.W., Patil H., Ueda T., Lim J.H., Kraemer K.H., Bustin M., Bergel M. The nucleosome-binding protein HMGN2 modulates global genome repair. FEBS J. 2009;276:6646–6657. doi: 10.1111/j.1742-4658.2009.07375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G., Hall B., Yu R., Hashim A.I., Kramer H. Hook2 localizes to the centrosome, binds directly to centriolin/CEP110 and contributes to centrosomal function. Traffic. 2007;8:32–46. doi: 10.1111/j.1600-0854.2006.00511.x. [DOI] [PubMed] [Google Scholar]
- Tarcic G., Avraham R., Pines G., Amit I., Shay T., Lu Y., Zwang Y., Katz M., Ben-Chetrit N., Jacob-Hirsch J. EGR1 and the ERK-ERF axis drive mammary cell migration in response to EGF. FASEB J. 2011;26:1582–1592. doi: 10.1096/fj.11-194654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholen S., Wolf C., Mayer B., Knopf J.D., Loffek S., Qian Y., Kizhakkedathu J.N., Biniossek M.L., Franzke C.W., Schilling O. Skin barrier defects caused by keratinocyte-specific deletion of ADAM17 or EGFR are based on highly similar proteome and degradome alterations. J. Proteome Res. 2016;15:1402–1417. doi: 10.1021/acs.jproteome.5b00691. [DOI] [PubMed] [Google Scholar]
- Threadgill D.W., Dlugosz A.A., Hansen L.A., Tennenbaum T., Lichti U., Yee D., LaMantia C., Mourton T., Herrup K., Harris R.C. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- Tumbar T. Ontogeny and homeostasis of adult epithelial skin stem cells. Stem Cell Rev. 2012;8:561–576. doi: 10.1007/s12015-012-9348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B.D., Cleveland D.W. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr. Opin. Cell Biol. 2012;24:809–815. doi: 10.1016/j.ceb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf C., Qian Y., Brooke M.A., Kelsell D.P., Franzke C.W. ADAM17/EGFR axis promotes transglutaminase-dependent skin barrier formation through phospholipase C gamma1 and protein kinase C pathways. Sci. Rep. 2016;6:39780. doi: 10.1038/srep39780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Chow L.T., Paterson A.J., Chin E., Kudlow J.E. Conditional expression of the ErbB2 oncogene elicits reversible hyperplasia in stratified epithelia and up-regulation of TGFalpha expression in transgenic mice. Oncogene. 1999;18:3593–3607. doi: 10.1038/sj.onc.1202673. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Tian Y., Chen Q., Chen D., Zhai Z., Shu H.B. TTDN1 is a Plk1-interacting protein involved in maintenance of cell cycle integrity. Cell. Mol. Life Sci. 2007;64:632–640. doi: 10.1007/s00018-007-6501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: P2 IFE and ORS significantly differentially expressed genes.
B: Alternative Splicing P2 IFE and ORS Top 15 GO Terms, -log10.
C: Alternative splicing P2 overlapping genes between IFE and ORS.
D: Alternative splicing P2 overlapping GO terms between IFE and ORS.
E: Alternative splicing P2 GO Term “Cell Cycle.”
F: Alternative splicing P2 GO Term “Cell Cycle”—exon usage details.
A: P88 LGR5 significantly differentially expressed genes.
B: P88 LGR5 GO term enrichment of significantly differentially expressed genes.
C: P88 LGR5 significantly differentially expressed genes in GO term “growth.”
D: P88 LGR5 alternative splicing Top 15 GO terms -log10.
E: Alternative splicing overlapping genes between P2 IFE + ORS and P88 LGR5.
F: GO term enrichment of alternative splicing overlapping genes between P2 IFE + ORS and P88 LGR5.
Oligonucleotide Sequences for qRT-PCR primers and for genotyping primers.
Reagents (media, buffers, enzymes, etc.).
Mice and software.
Antibodies used for histology and flow cytometry.