Abstract
In the past two decades, tRNA molecules and their corresponding aminoacyl-tRNA synthetases (aaRS) have been extensively used in synthetic biology to genetically encode post-translationally modified and unnatural amino acids. In this review, we briefly examine one fundamental requirement for the successful application of tRNA/aaRS pairs for expanding the genetic code. This requirement is known as “orthogonality”—the ability of a tRNA and its corresponding aaRS to interact exclusively with each other and avoid cross-reactions with additional types of tRNAs and aaRSs in a given organism.
Keywords: synthetic biology, expanded genetic code, tRNA, aminoacyl-tRNA synthetases, orthogonal translation systems
In the past two decades, over a dozen natural and engineered tRNA molecules have been used to expand or rewrite the genetic code of a living cell. With the help of these tRNAs, numerous strains of bacteria and eukaryotes have been created that are capable of site-specific incorporation of more than 170 non-canonical amino acids into cellular proteins in vivo (reviewed in refs. [1,2,3]). Typically, these engineered organisms possess one or a few additional tRNAs and corresponding tRNA synthetases, which are known as “orthogonal” tRNA/tRNA synthetase pairs. These orthogonal pairs are designed or selected in such a way that they incorporate a corresponding non-canonical amino acid but do not cross-react with other amino acids, tRNAs and tRNA synthetases in a given organism. Therefore, engineering of new tRNA/tRNA synthetase pairs to expand the genetic code requires understanding of what makes these pairs orthogonal.
Early studies revealed that specific tRNA recognition by a cognate tRNA synthetase relies on a small fraction of nucleotides in the tRNA structure (reviewed in refs. [4,5]). These nucleotides were called identity elements and anti-identity elements as they respectively trigger specific tRNA aminoacylation by a cognate tRNA synthetase and prevent tRNA mis-aminoacylation by non-cognate tRNA synthetases. Most typically, both identity and anti-identity elements are located at the two opposite sides of tRNA molecules: the anticodon stem and the acceptor stem. Typical examples of the identity elements include anticodon residues I34, A35, U36 in Saccharomyces cerevisiae tRNAIle or the acceptor stem residue A73 along with anticodon residues U34, U35, U36 in Escherichia coli tRNALys [6,7,8]. The presence of base modification (e.g., thiolation leading to s2U34 in E. coli tRNAGlu) contributes significantly to tRNA identity, as lack of the modification decreases the aminoacylation efficiency drastically [9]. Typical examples of anti-identity elements include base modifications of E. coli tRNAIle residue C34 (lysidine), which prevents tRNAIle mis-aminoacylation by MetRS synthetase, and methylation of G73 base in S. cerevisiae tRNAAsp (m1G), which prevents tRNAAsp mis-aminoacylation by ArgRS synthetase [10,11]. Thus, although tRNA molecules typically comprise ~70–100 nucleotides, only a small fraction of these nucleotides is required for specific recognition of a tRNA by a corresponding tRNA synthetase.
Discovery of the identity elements was empowering as it showed that the correspondence between tRNA molecules and tRNA synthetases can be reprogrammed by transplanting identity elements from one tRNA into another. For example, a transplantation of just three anticodon residues C34, U35, A36 from E. coli tRNAMet to tRNAVal switches the tRNAVal specificity from ValRS to MetRS synthetases, leading to tRNAVal acylation with methionine [12]. Similarly, transplantation of a single G3-U70 base pair from E. coli tRNAAla to tRNAPhe [13,14] or tRNACys [14] turn these tRNAPhe and tRNACys species into specific substrates of alanyl-tRNA synthetase, leading to their aminoacylation with alanine. Thus, discovery of the identity elements allowed to use relatively subtle changes in tRNA structures to redefine the rules of the genetic code.
Before synthetic biologists began to exploit tRNA identity elements to engineer and reprogram the genetic code, many experiments with tRNA identity elements were observed in nature. For example, the fungal pathogen Candida albicans was shown to have an unusual tRNA, tRNASerCGA, with hybrid identity elements that cause tRNASerCGA recognition by two tRNA synthetases, LeuRS and SerRS [15]. Hence, the polysemous tRNASerCGA can be aminoacylated either by leucine or serine, leading to ambiguous translation of CUG codons—a property that helps C. albicans to introduce statistical variation in protein sequences that regulate such virulence attributes as morphogenesis, phenotypic switching, and adhesion [16]. Also, tRNA identity elements were found in a number of tRNA-like molecules. For example, genomic RNA of the turnip yellow mosaic virus carries a stem-loop structure that mimics the tRNAVal anticodon. This mimicry allows the viral RNA to recruit valyl-tRNA synthetase, aminoacylate the 3′-end of the viral RNA with valine, and eventually, initiate translation of viral RNA [17]. Similarly, in bacteria, tmRNA, a tRNA/mRNA hybrid that is used to rescue stalled ribosomes on truncated mRNAs [18], carries an identity element of tRNAAla (the base pair A3-U71) that allows it to aminoacylate tmRNA with alanine and use tmRNA to rescue stalled ribosomes [19]. Overall, these studies showed that specific recruitment of a tRNA synthetase can be achieved even in the absence of canonical tRNA structure, as long as tRNA-like molecules carry a proper set of identity elements.
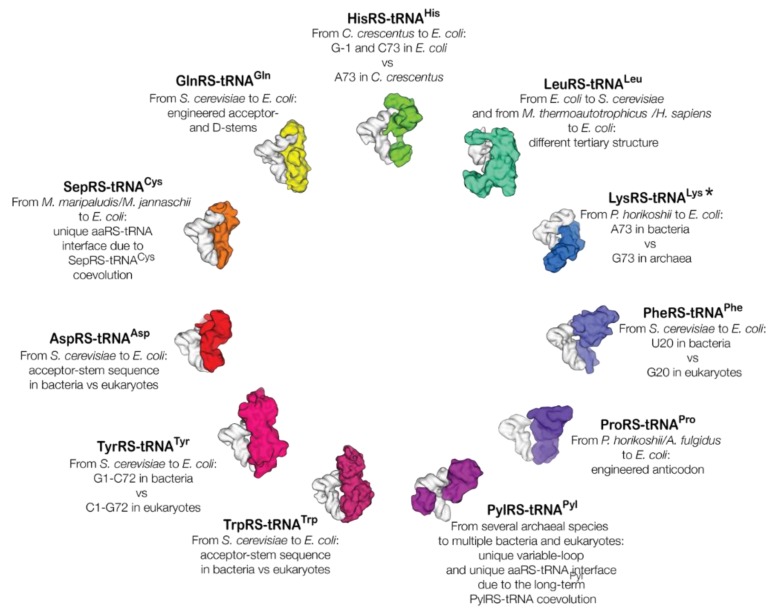
The first successful attempts to use tRNA identity elements to expand the genetic code of live cells exploited variability of identity elements in homologous tRNAs from different species. For example, in bacteria, tRNATyr identity elements include the base pair G1-C72, whereas in archaea and eukaryotes tRNATyr identity elements include the reversed base pair, C1-G72 (reviewed in ref. [20]). This difference allowed the transplant of the tRNATyr/TyrRS pair from the archaeon Methanocaldococcus jannaschii into E. coli (with a few modifications in the tRNATyr) that created a strain carrying an additional tRNATyr/TyrRS pair that does not cross-react with other tRNA synthetases/tRNAs in E. coli, and that allows the genetic encoding of an additional amino acid, O-methyl-l-tyrosine [21]. Similar variations between bacterial, archaeal, and eukaryotic tRNA homologs allowed the creation of over a dozen orthogonal tRNA/synthetase pairs for genetic code expansion (Figure 1), most typically by transplanting archaeal tRNA/synthetase pairs from archaea into bacteria and from bacteria into eukaryotes (reviewed in ref. [1]).
Figure 1.
tRNA molecules and corresponding aminoacyl-tRNAs that have been used to expand the genetic code of live cells. Crystal structures of tRNA molecules (in white) are shown in complex with the corresponding aminoacyl-tRNAs (in the colors of the rainbow). The labels next to each structure indicate the origin of the orthogonal tRNA/tRNA synthetase pair and an organism into which this pair has been transplanted, which is followed by a brief description of tRNA identity or anti-identity elements that make a tRNA/tRNA synthetase pair orthogonal in an organism into which this pair has been transplanted. For example, the label “TyrRS-tRNATyr—From S. cerevisiae to E. coli: G1-C72 in bacteria vs C1-G72 in eukaryotes” means that the TyrRS-tRNATyr pair has been transplanted from S. cerevisiae into E. coli where this pair remains orthogonal due to different identity elements between bacterial and eukaryotic tRNATyr: the G1-C72 pair in bacterial tRNATyr and the C1-G72 pair in eukaryotic tRNATyr. The asterisk next to the LysRS-tRNALys complex indicates that this model was produced by docking the crystal structure of tRNALys into the tRNA-binding pocket of LysRS. The most comprehensive list of orthogonal tRNA/tRNA synthetase pairs and their classes can be found in the Supplemental files of Ref. [1].
More sophisticated orthogonal tRNA/synthetase pairs were engineered by constructing chimeric tRNAs [22] or by focused tRNA mutagenesis and subsequent selection. Two typical examples of these strategies are tRNAUTu [23] and tRNASecUX [24]—engineered tRNAs that were developed for genetic encoding of selenocysteine. tRNAUTu was constructed as a hybrid between tRNASec and tRNASer carrying the determinants for EF-Tu binding [23]. Due to the presence of tRNASec segments, Ser-tRNAUTu is recognized by the protein SelA for conversion to Sec-tRNAUTu. However, due to tRNASer segments, tRNAUtu binds EF-Tu and is delivered to the ribosome—something that natural tRNASec is not capable of doing. Another tRNA for the genetic encoding of selenocysteine, tRNASecUX, was evolved from E. coli wild-type tRNASecUCA and also selectively engineered with the EF-Tu determinants [24]. These examples illustrate that successful engineering of tRNA/synthetase pairs typically require manipulation of both identity and anti-identity elements in the tRNA structure.
Finally, there are a few examples in which orthogonality of tRNA/synthetase pairs stems from a large number of idiosyncratic contacts at the tRNA/synthetase interface, rather than from a small number of identity and anti-identity elements in the tRNA structure. Perhaps the best example of this scenario can be found in the tRNAPyl/pyrrolysyl-tRNA synthetase pair. Phylogenetic analyses indicate that a tRNAPyl/PylRS pair could have originated from a tRNAPhe/PheRS pair about 3 billion years ago [25]. Structural studies show that tRNAPyl is recognized by PylRS in a highly unusual manner: unlike most other tRNA synthetases, PylRS does not bind the tRNA anticodon and instead makes extensive contacts with the acceptor stem and the unusually small variable loop of tRNAPyl [26,27]. In this case, the orthogonality appears to stem from the long-term coevolution of the tRNA and its corresponding tRNA synthetase. This long-term coevolution appears to create a remarkable complementarity of shape and charge between tRNAPyl and PylRS, which makes the tRNAPyl/PylRS pair (e.g., from the archaeon Methanosarcina mazei) exceptionally orthogonal when transplanted into a wide range of bacterial or eukaryotic organisms.
Where do we go from here? As described above, a range of orthogonal tRNA synthetase/tRNA pairs are now available and may be enhanced by additional laboratory evolution. What are the desired characteristics that drive further improvement? The current version of orthogonal tRNA synthetases has moderate catalytic activity and is polyspecific for many non-canonical amino acids [28]. Thus, increasing catalytic power and substrate specificity is of great interest; significant achievements have already been reported for p-azido-PheRS using multiplex automated genome engineering (MAGE) [29] and PylRS using phage-assisted continuous evolution (PACE) [30] or phage-assisted non-continuous evolution (PANCE) [27]. The finding of two classes of PylRS enzymes [31] has led to the development of mutually orthogonal PylRS/tRNAPyl variant pairs that are able to serve different codons with different non-canonical amino acids [32,33]. In addition, the recent finding that three mutually orthogonal tRNA synthetase/tRNA pairs allowed successful site-specific non-canonical amino acid insertion at three distinct sites in a recombinant protein [34] is a sign of the wide range of future applications of genetic code expansion. Also, a recently identified class of tRNA molecules, known as allo-tRNAs [35], and their successful use for selenoprotein synthesis [36], illustrates that there might be many more natural tRNAs and tRNA-type molecules that can serve as templates for the future development of robust and useful orthogonal tRNA synthetase/tRNA pairs for the expanded genetic code.
Acknowledgments
We would like to thank Jeffery Tharp and Ana Crnkovic (Yale University, New Haven, CT, USA) for valuable discussions and critical feedback during preparation of the manuscript.
Author Contributions
S.V.M. and D.S. wrote the manuscript.
Funding
Work in the authors’ laboratory was supported by a grant from the National Institute of General Medical Sciences (R35GM122560 to D.S.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Mukai T., Lajoie M.J., Englert M., Söll D. Rewriting the genetic code. Annu. Rev. Microbiol. 2017;71:557–577. doi: 10.1146/annurev-micro-090816-093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin J.W. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 3.Dumas A., Lercher L., Spicer C.D., Davis B.G. Designing logical codon reassignment—Expanding the chemistry in biology. Chem. Sci. 2015;6:50–69. doi: 10.1039/C4SC01534G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giege R., Sissler M., Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kisselev L.L. The role of the anticodon in recognition of tRNA by aminoacyl-tRNA synthetases. Prog. Nucleic Acid Res. Mol. Biol. 1985;32:237–266. doi: 10.1016/s0079-6603(08)60350-5. [DOI] [PubMed] [Google Scholar]
- 6.Senger B., Auxilien S., Englisch U., Cramer F., Fasiolo F. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry. 1997;36:8269–8275. doi: 10.1021/bi970206l. [DOI] [PubMed] [Google Scholar]
- 7.Normanly J., Kleina L.G., Masson J.M., Abelson J., Miller J.H. Construction of Escherichia coli amber suppressor tRNA genes. III. Determination of tRNA specificity. J. Mol. Biol. 1990;213:719–726. doi: 10.1016/S0022-2836(05)80258-X. [DOI] [PubMed] [Google Scholar]
- 8.McClain W.H., Foss K., Jenkins R.A., Schneider J. Nucleotides that determine Escherichia coli tRNAArg and tRNALys acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc. Natl. Acad. Sci. USA. 1990;87:9260–9264. doi: 10.1073/pnas.87.23.9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sylvers L.A., Rogers K.C., Shimizu M., Ohtsuka E., Söll D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry. 1993;32:3836–3841. doi: 10.1021/bi00066a002. [DOI] [PubMed] [Google Scholar]
- 10.Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 11.Pütz J., Florentz C., Benseler F., Giege R. A single methyl group prevents the mischarging of a tRNA. Nat. Struct. Biol. 1994;1:580–582. doi: 10.1038/nsb0994-580. [DOI] [PubMed] [Google Scholar]
- 12.Schulman L.H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988;242:765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- 13.McClain W.H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 14.Hou Y.M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T., Ueda T., Watanabe K. The ‘polysemous’ codon—A codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 1997;16:1122–1134. doi: 10.1093/emboj/16.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miranda I., Silva-Dias A., Rocha R., Teixeira-Santos R., Coelho C., Goncalves T., Santos M.A., Pina-Vaz C., Solis N.V., Filler S.G., et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. MBio. 2013;4:e00285-13. doi: 10.1128/mBio.00285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinck M., Yot P., Chapeville F., Duranton H.M. Enzymatic binding of valine to the 3′ end of TYMV-RNA. Nature. 1970;226:954–956. doi: 10.1038/226954a0. [DOI] [PubMed] [Google Scholar]
- 18.Karzai A.W., Roche E.D., Sauer R.T. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 19.Komine Y., Kitabatake M., Yokogawa T., Nishikawa K., Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonnefond L., Giege R., Rudinger-Thirion J. Evolution of the tRNATyr/TyrRS aminoacylation systems. Biochimie. 2005;87:873–883. doi: 10.1016/j.biochi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Wang L., Brock A., Herberich B., Schultz P.G. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto K., Hayashi A. Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures. Int. J. Mol. Sci. 2019;20:92. doi: 10.3390/ijms20010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller C., Bröcker M.J., Prat L., Ip K., Chirathivat N., Feiock A., Veszpremi M., Söll D. A synthetic tRNA for EF-Tu mediated selenocysteine incorporation in vivo and in vitro. FEBS Lett. 2015;589:2194–2199. doi: 10.1016/j.febslet.2015.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thyer R., Robotham S.A., Brodbelt J.S., Ellington A.D. Evolving tRNASec for efficient canonical incorporation of selenocysteine. J. Am. Chem. Soc. 2015;137:46–49. doi: 10.1021/ja510695g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavran J.M., Gundllapalli S., O’Donoghue P., Englert M., Söll D., Steitz T.A. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. USA. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nozawa K., O’Donoghue P., Gundllapalli S., Araiso Y., Ishitani R., Umehara T., Söll D., Nureki O. Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature. 2009;457:1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T., Miller C., Guo L.T., Ho J.M.L., Bryson D.I., Wang Y.S., Liu D.R., Söll D. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 2017;13:1261–1266. doi: 10.1038/nchembio.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo L.T., Wang Y.S., Nakamura A., Eiler D., Kavran J.M., Wong M., Kiessling L.L., Steitz T.A., O’Donoghue P., Söll D. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. USA. 2014;111:16724–16729. doi: 10.1073/pnas.1419737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amiram M., Haimovich A.D., Fan C., Wang Y.S., Aerni H.R., Ntai I., Moonan D.W., Ma N.J., Rovner A.J., Hong S.H., et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat. Biotechnol. 2015;33:1272–1279. doi: 10.1038/nbt.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryson D.I., Fan C., Guo L.T., Miller C., Söll D., Liu D.R. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 2017;13:1253–1260. doi: 10.1038/nchembio.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borrel G., Parisot N., Harris H.M., Peyretaillade E., Gaci N., Tottey W., Bardot O., Raymann K., Gribaldo S., Peyret P., et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genom. 2014;15:679. doi: 10.1186/1471-2164-15-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meineke B., Heimgärtner J., Lafranchi L., Elsässer S.J. Methanomethylophilus alvus Mx1201 provides basis for mutual orthogonal pyrrolysyl tRNA/aminoacyl-tRNA synthetase pairs in mammalian cells. ACS Chem. Biol. 2018;13:3087–3096. doi: 10.1021/acschembio.8b00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willis J.C.W., Chin J.W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat. Chem. 2018;10:831–837. doi: 10.1038/s41557-018-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Italia J.S., Addy P.S., Erickson S.B., Peeler J.C., Weerapana E., Chatterjee A. Mutually orthogonal nonsense-suppression systems and conjugation chemistries for precise protein labeling at up to three distinct sites. J Am Chem Soc. 2019;141:6204–6212. doi: 10.1021/jacs.8b12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukai T., Vargas-Rodriguez O., Englert M., Tripp H.J., Ivanova N.N., Rubin E.M., Kyrpides N.C., Söll D. Transfer RNAs with novel cloverleaf structures. Nucleic Acids Res. 2017;45:2776–2785. doi: 10.1093/nar/gkw898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukai T., Sevostyanova A., Suzuki T., Fu X., Söll D. A facile method for producing selenocysteine-containing proteins. Angew. Chem. Int. Ed. Engl. 2018;57:7215–7219. doi: 10.1002/anie.201713215. [DOI] [PMC free article] [PubMed] [Google Scholar]