Abstract
Objective
To identify novel epigenetic signatures that could provide predictive information that is complementary to promoter methylation status of the O-6-methylguanine-DNA methyltransferase (MGMT) gene for predicting temozolomide (TMZ) response, among glioblastomas (GBMs) without glioma-CpGs island methylator phenotype (G-CIMP)
Methods
Different cohorts of primary non-G-CIMP GBMs with genome-wide DNA methylation microarray data were included for discovery and validation of a multimarker signature, combined using a RISK score model. Different statistical analyses and functional experiments were performed for clinical and biological validation.
Results
By employing discovery cohorts with radiotherapy (RT) and TMZ versus RT alone and a strict multistep selection strategy, we identified seven CpGs, each of which was significantly correlated with overall survival (OS) of non-G-CIMP GBMs with RT/TMZ, independent of age, MGMT promoter methylation status, and other identified CpGs. A RISK score signature of the 7 CpGs was developed and validated to distinguish non-G-CIMP GBMs with differential survival outcomes to RT/TMZ, but not to RT alone. The interaction analyses also showed differential outcomes to RT/TMZ versus RT alone within the RISK score-based subgroups. The signature could also improve the risk classification by age and MGMT promoter methylation status. Functional experiments showed that HSBP2 appeared to be epigenetically regulated by one identified CpG and was associated with TMZ resistance, but it was not associated with cell proliferation or apoptosis in GBM cell lines. The predictive value of the single CpG methylation of HSBP2 by pyrosequencing was observed in a local cohort of isocitrate dehydrogenase 1 (IDH1) R132H wild-type GBMs.
Conclusions
This novel epigenetic signature might be a promising predictive (but not a general prognostic) biomarker and be helpful for refining the MGMT-based guiding approach to TMZ usage in non-G-CIMP GBMs.
Electronic supplementary material
The online version of this article (10.1186/s13148-019-0670-9) contains supplementary material, which is available to authorized users.
Keywords: Glioma-CpGs island methylator phenotype; Glioblastoma; Temozolomide, Predictive biomarker; DNA methylation
Introduction
Glioblastomas ᅟ (GBMs) are a group of clinically refractory disease with apparent intertumor heterogeneity and a generally poor prognosis [1]. Over the past decade, despite extensive explorations on novel therapeutic strategies such as anti-angiogenic therapy [2, 3], immunotherapy [4], and the use of tumor treating fields (TTFs) [5], the combination of radiotherapy (RT) and temozolomide (TMZ) had remained the standard adjuvant treatment for newly diagnosed GBMs [6]. Unfortunately, these tumors often have variable responses to TMZ, and some do not benefit from the combined RT/TMZ treatment. The promoter methylation status of the O-6-methylguanine-DNA methyltransferase (MGMT), encoding a DNA repair enzyme that confers resistance to alkylating drugs, has been by far the most informative predictive biomarker for TMZ outcome in GBMs [7]. However this single-gene methylation status has limitations for clinical utility, especially for guiding the choice of TMZ in unmethylated tumors [8]. Therefore, novel predictive biomarkers with a high predictive value that are independent of MGMT promoter methylation status could be useful. In this study, we investigated the major subgroup of GBMs that do not have the glioma-CpGs island methylator phenotype (G-CIMP), which is exclusively featured by the absence of isocitrate dehydrogenase (IDH) mutations (mostly IDH1R132H) [9]. We developed a novel 7-CpG signature using genome-wide DNA methylation data; the signature may confer predictive information for TMZ usage that is complementary to MGMT promoter methylation status. In addition, we selected the HSBP2 gene for further analysis, the expression of which might be epigenetically regulated by one of the 7 CpGs. Functional studies on this epigenetically regulated gene (HSBP2) provide additional biological and clinical insights to the multimarker signature.
Methods
Molecular datasets from Rennes and Angers University Hospitals
A total of 125 primary non-G-CIMP GBMs were collected between 2004 and 2013 from the Neurosurgery Departments of Rennes and Angers University Hospitals (RAUH), including a new cohort of 77 samples (RAUH-new cohort) and a published cohort of 48 samples (RAUH-GSE22891) [10]. Snap-frozen samples were collected at the time of surgery, following written informed consent, in accordance with the French regulations and the Helsinki Declaration. Initial histological diagnoses were confirmed by a central review panel including at least two neuropathologists. Degree of surgical resection was defined by MRI 72 h after surgery. All patients were treated with RT plus concurrent and adjuvant TMZ. Only samples with > 80% tumor cells were selected for microarray profiling and molecular detection. DNA and RNA were isolated as previously described [10]. DNA methylation and gene expression microarrays were performed according to the manufacturer’s instructions. Specifically, RAUH-new cohort was profiled by the Infinium HumanMethylation450k BeadChip for DNA methylation (deposited in The ArrayExpress at http://www.ebi.ac.uk/arrayexpress/ under the accession number of E-MTAB-4969). Image processing and intensity data extraction were performed within Genome Studio (Illumina Inc.). The novel BMIQ (Beta MIxture Quantile dilation) algorithm was used for intra-array normalization [11]. The methylation data of each CpG is summarized as β value, ranging from 0 (completely unmethylated) to 1 (completely methylated). All but two samples were also profiled by Agilent Whole HumanGenome 8 × 60 K Microarray Kit (Agilent Technologies) for gene expression. Expression intensity was log2 transformed and normalized (scale 50th percentile and baseline transformation) within GeneSpring GX software (Agilent Technologies). DNA methylation and gene expression profiling for RAUH-GSE22891 were reported previously (deposited in Gene Expression Omnibus [GEO] at https://www.ncbi.nlm.nih.gov/geo/ under the accession number of GSE22891) [10]. G-CIMP status was determined by k-means (k = 3) clustering on the 1503 featured probes reported by The Cancer Genome Atlas (TCGA) [9]. MGMT promoter methylation status was determined using a logistic regression model based on two Illumina array probes, i.e., cg12434587 and cg12981137 [12].
Molecular datasets from public databases
Additional DNA methylation or gene expression data of primary non-G-CIMP GBMs were obtained from public databases, including the clinically annotated cohort from TCGA (TCGA-Brennan et al-RT/TMZ [n = 219] or -RT [n = 73]) [13], and two cohorts from GEO (GSE50923-RT/TMZ [n = 49] [14]; GSE60274-RT/TMZ [n = 32] or -RT [n = 27] [15]). Moreover, DNA methylation and gene expression data of lower-grade gliomas (LGGs) and nontumor brains from TCGA [16], gliomas of all grades from Chinese Glioma Genome Atlas (CGGA) [17], and nontumor brains from GSE63347 [18] were obtained for additional validation.
Selection and information of all included samples were summarized in Additional file 1: Figure S1 and Additional file 2: Table S1.
Probe selection and RISK score modeling
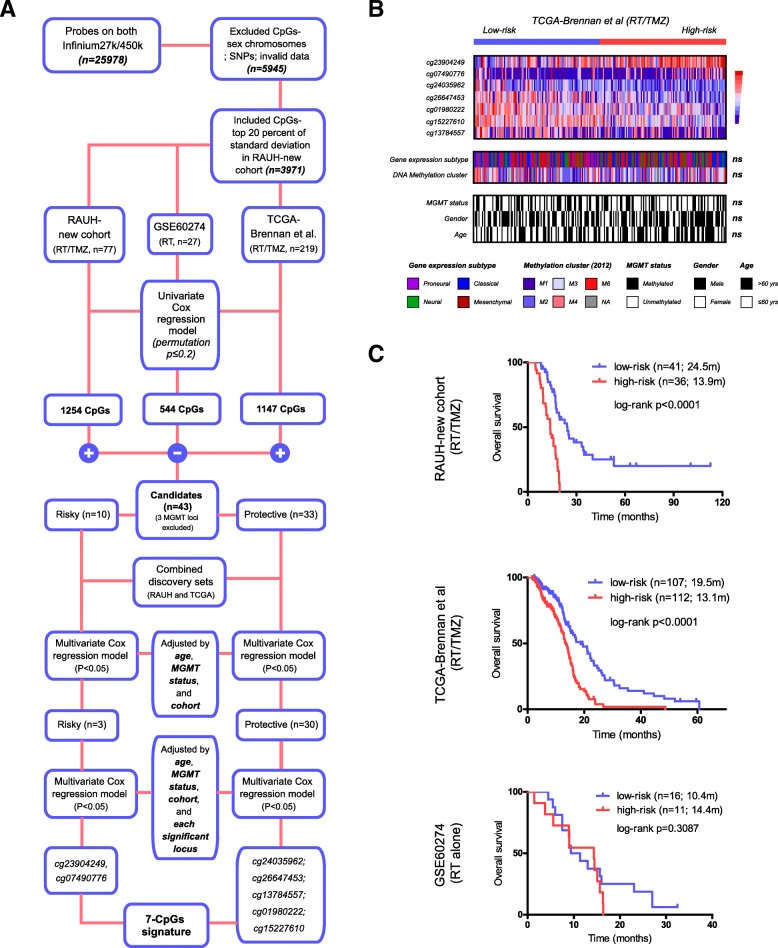
Prior probe selection was performed by removal of those not interrogated on both the Infinium HumanMethylation27k and 450k platforms, those targeting the sex chromosomes, and those associated with single-nucleotide polymorphisms. To make DNA methylation microarray data comparable across each dataset, batch effects between each platform and dataset were adjusted by M-value transformation and the empirical Bayes approach (ber R package) [19, 20]. Missing β values were imputed by impute R package. Discovery-validation approach was employed for predictive model construction. Two cohorts with RT/TMZ (e.g., RAUH-new cohort, TCGA-Brennan et al-RT/TMZ) and one cohort with RT alone (e.g., GSE60274-RT) were used for discovery phase. Selected CpGs with higher variability in methylation levels (top 20 percent of standard deviation of β value) across tumors from RAUH-new cohort were used to correlate with overall survival (OS) using univariate Cox regression model and permutation test (Fig. 1a). After removing inconsistent results, an overlap of 43 candidates (permutation p < 0.2; excluding three MGMT relevant loci) from the discovery cohorts was subjected to multivariate Cox regression models adjusted by different ages, MGMT methylation status, and different cohorts and then to multivariate models incorporating other significant CpGs (Fig. 1a). Finally, a panel of 7 CpGs was identified for constructing a RISK score model (Table 1), which is the sum of β values of each CpG weighted by their multivariate Cox coefficients, adjusted by age, MGMT methylation status, patient cohorts, and other loci. The cutoff for low-risk and high-risk tumors were predefined as the median risk score from the combined discovery cohorts with RT/TMZ.
Fig. 1.
The identification of the 7-CpG signature. a The study workflow regarding the selection of predictive epigenetic panel. b Molecular and clinical correlations of the multi-CpG signature within TCGA-Brennan et al.-RT/TMZ. Heat maps of the methylation patterns of the 7 CpGs are presented, where each row represents a CpG, and each column represents a sample. Clinical and molecular features are indicated for each sample. ns not significant. c Patient classification by the 7-CpG RISK score classifier in two discovery cohorts of non-G-CIMP GBMs with RT/TMZ (RAUH-new cohort and TCGA-Brennan et al.) and one discovery cohort with RT alone (GSE60274);.GBM glioblastoma, G-CIMP glioma-CpGs island methylator phenotype, RT radiotherapy, TMZ temozolomide
Table 1.
Characteristics of the seven-CpG panel
| Probe ID | Chr. | Relevant gene symbol | Relation to gene region | Relation to CpGs islandb | Multivariate Cox coefficientsa |
|---|---|---|---|---|---|
| ᅟcg23904249 | 11 | CCDC86 | TSS1500 | Shore | 1.095 |
| cg07490776 | 8 | AP3M2 | TSS1500 | Island | 1.575 |
| cg24035962 | 10 | NCOA4 | TSS200 | Island | − 1.365 |
| cg26647453 | 4 | C4orf17 | 5′UTR | Open sea | − 1.574 |
| cg01980222 | 6 | TREM2 | 1stExon | Open sea | − 1.248 |
| cg15227610 | 11 | HSPB2 | TSS1500 | Open sea | − 1.120 |
| cg13784557 | 6 | HCP5 | TSS200 | Open sea | − 0.975 |
Chr chromosome, TSS transcriptional start site
aCox coefficients were calculated from multivariate analysis incorporating age, different discovery sets, MGMT methylation status, and the seven CpGs within the combined RT/TMZ discovery cohorts
bOpen sea and shore refers to regions away from relevant CpGs islands more than 4000 bp or less than 2000 bp, respectively
In vitro functional experiments
The human glioma cell lines T98G, U87, U251, U373, and Hs683 were obtained from American Type Culture Collection (ATCC) and were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum at 37 °C in 5% CO2. TMZ (Sigma-Aldrich) was dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich) at a stock concentration of 100 mmol/l at 20 °C. Total RNA was purified using TRIzol reagents (Shanghai Pufei Biotechnology) and reverse transcribed with M-MLV RT kit (Promega) [21]. Polymerase chain reaction (PCR) amplification was performed with SYBR Master Mixture (Takara) using LighterCycler 480 II System (Rcoche). The expression values were normalized to the levels of GAPDH. Total DNA was extracted and bisulfate-modified using EZ DNA Methylation-GoldTM Kit (Beijing Tianmo Biotechnology). Pyrosequencing was performed by Pyromark Q96 ID platform and analyzed by PyroMark CpG software (Qiagen). Cell samples were lysed in RIPA buffer [21]. Equal amounts of proteins were resolved by SDS-PAGE gel electrophoresis and transferred to nitrocellulose membranes. The primary antibodies against HSPB2 (Proteintech Group, 21755-1-AP), MGMT (Proteintech Group, 17195-1-AP), and GAPDH (Santa Cruz Biotechnology, SC-32233) were used according to the manufacturers’ recommendations. Each immunoblot was done at least thrice and the signals were quantified using ImageJ software (Bethesda, MD, USA). For HSBP2 in vitro overexpression, lentivirus/GV358-HSBP2 (Ubi-HSBP2-3FLAG-SV40-EGFP-IRES-puromycin) and lentivirus/GV358 (control) were obtained from Genechem. Viruses were used to infect GBM cells in the presence of 6 μg/ml polybrene. At 48 to 72 hours after virus infection, puromycin selection (1 μg/ml) was applied and cells without subcloning were used for experiments [21]. CCK-8 kit (Yeasen Inc.) was assayed for cell viability analysis. For 5-Aza-2′-deoxycytidine (5-Aza-dC) demethylation treatment, U251, U87, and U373 cells were grown for 4 days in the presence of DMSO control, 5 μM, and 10 μM 5-Aza-dC (Sigma-Aldrich). Fresh 5-Aza-dC was added every 24 h. For cell apoptosis analysis, TUNEL assay was tested using In Situ Cell Death Detection kit (Roche Diagnostics). Annexin V–fluorescein isothiocyanate/propidium iodide double staining (Roche Diagnostics) was used to sort cells in early or late apoptotic phase.
Validation cohort of formalin-fixed paraffin-embedded (FFPE) GBM samples
Surgical samples of 333 primary GBMs (Grade IV, WHO) were totally collected from the Department of Neurosurgery, Xijing Hospital, between 2012 and 2016. Inclusion criteria included (1) adult patients (> 18-years old), (2) no prior therapy before surgery, (3) IDH1R132H wild-type tumors, (4) treatment with standard RT plus (adjuvant or concurrent) TMZ or standard RT alone, (5) available OS or progression-free survival (PFS) data, and (6) available FFPE tissue samples. Finally, 54 samples were included for validation analysis, including 32 samples with RT/TMZ and 22 with RT alone. The treatment choices between RT/TMZ and RT were made according to physician’s suggestions and family or patient’s will. Extent of resection was defined by post-operative MRI or CT within one week. The presence of IDH1R132H mutant protein was assessed by immunohistochemistry (IHC) (obtained from the Department of Pathology, Xijing Hospital). FFPE tissues were also employed for IHC with anti-HSPB2 antibody (Proteintech Group, 21755-1-AP). The intensity and percentage of positive cells were evaluated in at least five separate fields at × 400 magnification. The scores were evaluated by two researchers who were blinded to clinical data. Immunoreactivity was scored as follows: 0, no staining; 1, weak staining in < 50% cells; 2, weak staining in w 50% cells; 3, strong staining in < 50%, cells; and 4, strong staining in a 50% cells [21]. Disputes were resolved through discussion. Seven CpGs (74–81) in the promoter region of MGMT and the single CpG (cg155227610) at the non-CpGs island (CGI) region of HSPB2 were detected on the PyroMark Q96ID platform (Qiagen). The average percentage of CGI methylation of 10% was defined as the threshold for unmethylated and methylated MGMT promoter [22]. The median value of HSPB2 single CpG methylation was used as cutoff for hypermethylation and hypomethylation. The primer sequences for PCR and pyrosequencing were listed in Additional file 3: Table S2. All patients provided written informed consent and this study was approved by the Institutional Review Board.
Statistical analysis
Difference in clinical or molecular features within each risk subgroup was tested by unpaired t test, Fisher’s exact, or Chi-square test. Spearman correlation analysis was performed to correlate DNA methylation and gene expression. OS was the time interval from the date of diagnosis or treatment to the date of death or last follow-up. PFS was the time interval from the date of diagnosis or treatment to the date of progression defined by the Macdonald criteria or Response Assessment in Neuro-Oncology (RANO criteria) [23, 24], or the date of death or last follow-up. Survival data was estimated by the Kaplan-Meier Method and compared by log-rank test. Univariate and multivariate Cox regression models were used to evaluate the correlation and independence of each variable. The discriminating ability for prognosis was also evaluated by time-dependent receiver operating characteristic (ROC) curve (survcomp R package) [25]. All the calculations were done within SPSS statistics (SPSS Software Inc.) and R software, with two-side p values ≤ 0.05 for significance.
Results
Identification of a RISK score signature of seven CpGs with potential linkage to TMZ efficacy
According to a strict selection strategy (Fig. 1a), we identified a panel of seven CpGs from the discovery cohorts (Table 1). Each CpG was significantly associated with OS of non-G-CIMP GBMs treated with RT/TMZ, but not RT alone, which was also independent of age, MGMT promoter methylation status, and other CpG members. These 7 CpGs were not among the reported G-CIMP classifiers [9] but seemed to be among the genomic CpGs affected by this molecular status as the CpG panel showed significant but inconsistent alterations in methylation levels among tumors of each G-CIMP status and nontumor brains (Additional file 4: Figure S2A). In addition, the panel appeared not to be correlated with genome hypomethylation, i.e., LINE-1 methylation (Additional file 4: Figure S2A).
The 7-CpG panel was combined using a RISK score model as follows: risk score = (1.095 × β value of cg23904249) + (1.575 × β value of cg07490776) + (− 1.365 × β value of cg24035962) + (− 1.574 × β value of cg01980222) + (− 1.248 × β value of cg01980222) + (− 1.120 × β value of cg15227610) + (− 0.975 × β value of cg13784557). Using a predefined cutoff (median risk score, − 1.083), all patients were divided into a low-risk group (with lower risk scores) and a high-risk group (with higher risk scores).
Correlation with known clinical or molecular features in TCGA samples showed that the risk subgroups appeared to not be correlated with gene expression subtypes [26], DNA methylation clusters [13], MGMT promoter methylation status, gender, or age subgroup (Fig. 1b).
By applying the RISK score-based classification to the discovery cohorts, we found that, in RAUH-new cohort (RT/TMZ), high-risk patients had shorter OS than low-risk patients (p < 0.0001; Fig. 1c). In TCGA-Brennan et al.-RT/TMZ, high-risk patients also had poorer OS in comparison with low-risk patients (p < 0.0001; Fig. 1c). By contrast, in GSE60274-RT, OS was not significantly different between the risk subgroups (p = 0.3087; Fig. 1c). The results together suggested a potential linkage of the RISK score signature to TMZ efficacy, instead of a treatment-independent impact on prognosis.
The prognostic performance of the RISK score signature in validation cohorts
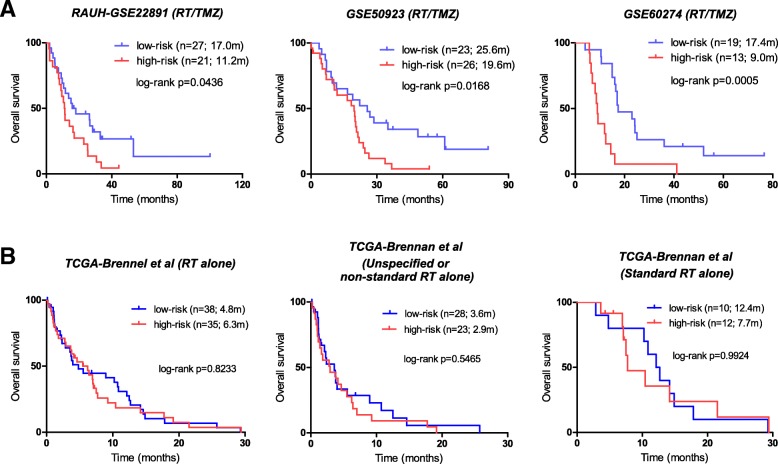
To further investigate the prognostic performance of the RISK score signature, we then applied it to a series of validation cohorts with different treatments. The signature accurately predicted OS in the validation cohorts with RT/TMZ: RAUH-GSE22891 (p = 0.0436), GSE50923-RT/TMZ (p = 0.0168), and GSE60274-RT/TMZ (p = 0.0005; Fig. 2a). The signature also predicted PFS in two available cohorts with RT/TMZ: RAUH-new cohort (p < 0.0001) and TCGA-Brennan et al.-RT/TMZ (p = 0.0151; Additional file 5: Figure S3A). However, the signature was unable to predict OS in a validation cohort with RT alone, i.e., TCGA-Brennan et al.-RT (Fig. 2b). Cox regression analyses of RAUH-new cohort and TCGA-Brennan et al.-RT/TMZ confirmed the RISK score signature as a significant risk factor that is independent of MGMT methylation status, as well as other variables (e.g., age, treatment schedules, treatment at progression), among non-G-CIMP GBMs with RT/TMZ (Additional file 6: Table S3), instead of RT alone (Additional file 7: Table S4). Those findings indicate that the RISK score signature might not be a general prognostic biomarker that is independent of treatment, but rather has a specific linkage to TMZ efficacy in non-G-CIMP GBMs.
Fig. 2.
The prognostic performance of the RISK score signature in different validation cohorts of non-G-CIMP GBMs, a Patient classification in validation cohorts with the combination of RT and TMZ, b Patient classification in validation cohorts with RT alone. GBM glioblastoma, G-CIMP glioma-CpGs island methylator phenotype, RT radiotherapy, TMZ temozolomide
The RISK score signature might be a promising predictive indicator of TMZ response
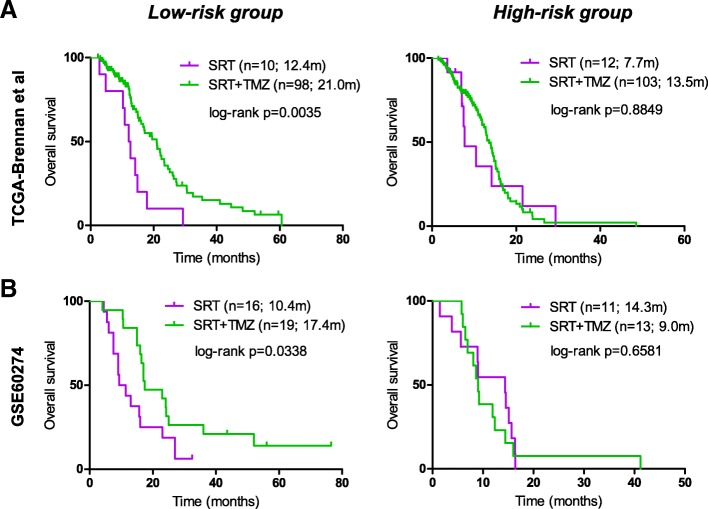
To investigate whether the RISK score signature has a predictive ability for TMZ response, interaction analyses were carried out between the risk subgroups and treatments, incorporating only patients with standard RT alone or combined with (concurrent or adjuvant) TMZ from TCGA-Brennan et al. and GSE60274. No significant difference was observed in baseline information (e.g., age, pre-adjuvant KPS, gender) between patients with different treatments in each risk subgroup (data not shown). The interaction analyses showed that standard RT/TMZ did confer a clear OS benefit to low-risk patients compared to standard RT, and this treatment was associated with a similar OS in high-risk patients (Fig. 3). Similar results were observed in terms of PFS (Additional file 8: Figure S4). Cox regression analyses of TCGA-Brennan et al. and GSE60274 confirmed standard RT/TMZ as a favorable indicator for OS benefit, independent of MGMT methylation status and age, in low-risk patients (Additional file 9: Table S5), but not in high-risk patients (Additional file 10: Table S6). Together, those results indicate that the RISK score signature might be a potential predictive indicator of TMZ outcome and be helpful for identifying subpopulations of patients who are likely to benefit from TMZ treatment. These findings should be conservatively interpreted due to the potential patient bias during treatment assignment in a retrospective series.
Fig. 3.
The predictive performance of the RISK score signature in different validation cohorts. Interaction analysis between treatments (with versus without TMZ) and risk subgroups (low-risk versus high-risk) in a TCGA-Brennan et al. and b GSE60274. Only patients with standard RT regimen were included for analysis in order to reduce potential bias by heterogeneous treatment regimen. RT radiotherapy, TMZ temozolomide
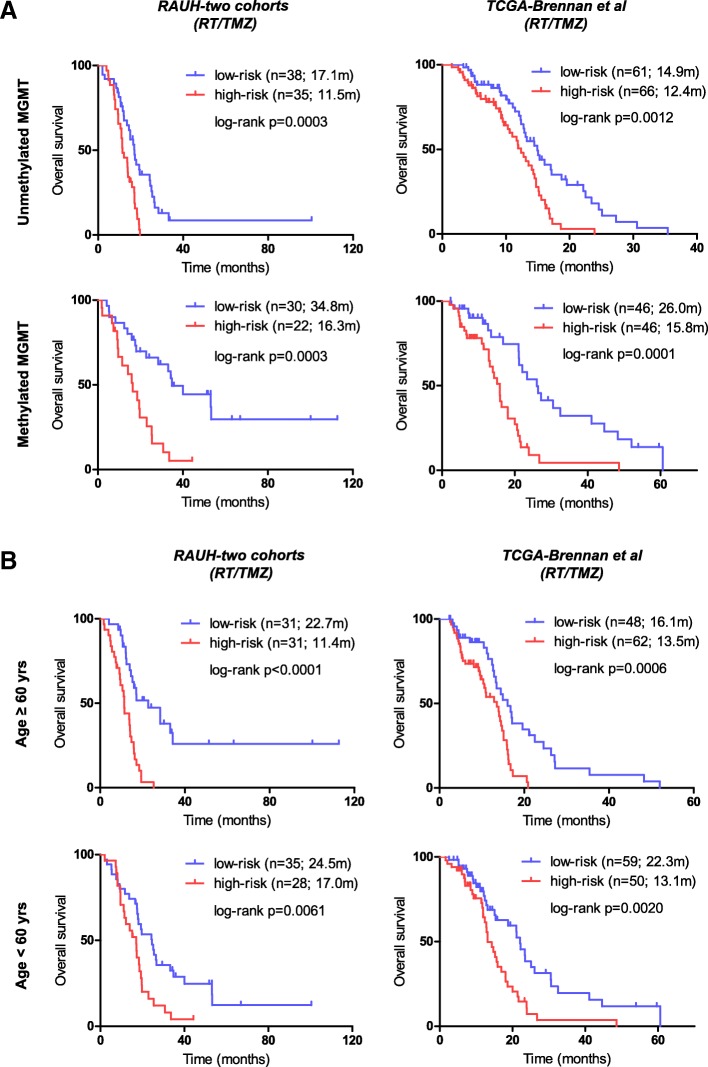
Patient classification in stratified cohorts by MGMT promoter methylation status and age
To further explore the clinical impact of the epigenetic signature, we also evaluated its performance in cohorts stratified by MGMT methylation status and age. We found that the RISK score signature showed significant discriminating value for OS of patients with each MGMT methylation status in the combined RAUH cohorts (RAUH-two cohorts) and in TCGA-Brennan et al.-RT/TMZ (Fig. 4a). Similarly, the signature robustly predicted OS in each age subgroup (age < vs. ≥ 60 years old; Fig. 4b). The results were also observed in two GEO cohorts (Additional file 11: Figure S5A-B). The combination of the RISK score signature with the two conventional risk factors could provide optimized risk classification in non-G-CIMP GBMs (Additional file 12: Figure S6A-B). Moreover, for those who underwent RT/TMZ, the time-dependent ROC showed that the RISK score signature was superior to MGMT methylation status in predicting OS among older patients (≥ 60 years) but had less discriminating value among younger patients (Additional file 12: Figure S6C).
Fig. 4.
The patient classification of the RISK score signature in cohorts stratified by conventional risk factors. a Cohorts stratified by MGMT promoter methylation status. b Cohorts stratified by patient age (≥ 60 vs. < 60 yrs). MGMT O-6-methylguanine-DNA methyltransferase, RT radiotherapy, TMZ temozolomide
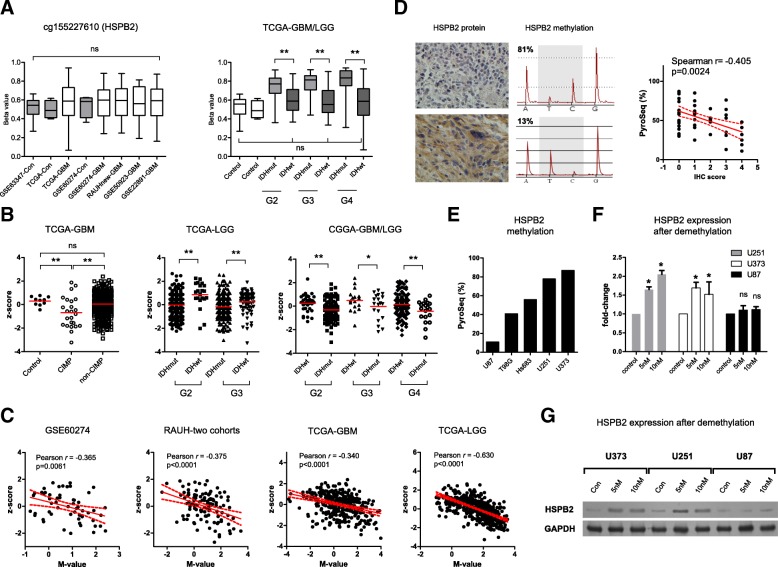
HSPB2 appeared to be epigenetically regulated by non-CGI methylation and was associated with TMZ resistance in GBM cell lines
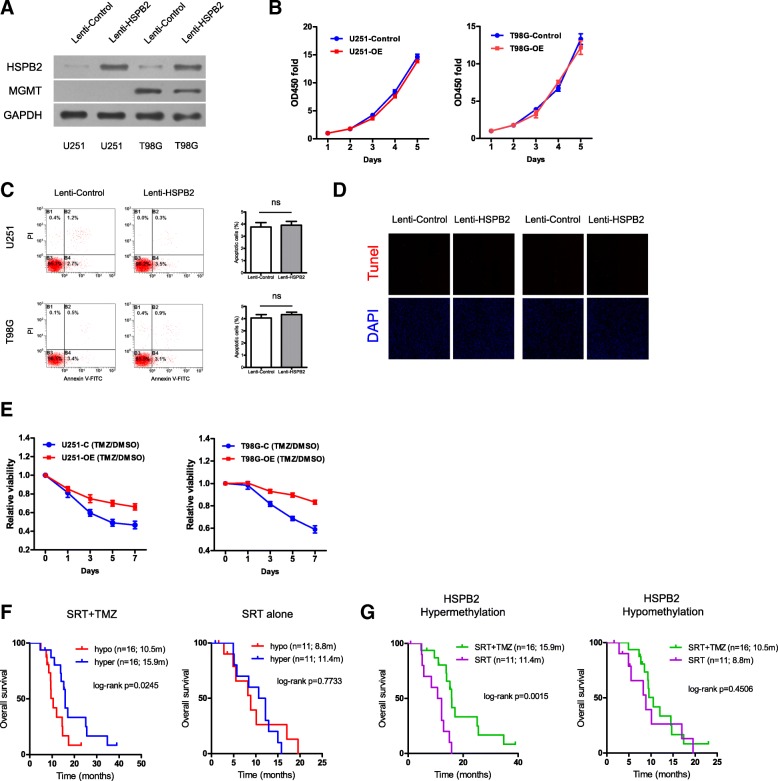
To gain biological insight into the multimarker epigenetic signature, we selected one of the 7 CpGs (cg15227610) for further analysis (Table 1). This single CpG was located at the non-CGI transcriptional regulatory region of HSPB2. The single CpG methylation and HSPB2 expression differed regarding the G-CIMP status (or IDH mutations), but they were not significantly altered in GBMs or in IDH mutant gliomas compared to nontumor brains (Fig. 5a, b). The status of single CpG methylation and HSPB2 expression did not differ across tumors of different grades (Fig. 5b). Notably, the single CpG methylation was consistently and significantly negatively correlated with HSPB2 expression (Fig. 5c). The negative correlation between methylation and protein levels was also observed in a local cohort of FFPE samples (Fig. 5d). Demethylation treatment with 5-Aza-dC showed that HSBP2 expression was increased in GBM cells that had the original hypermethylated CpG, e.g., U373 and U251, but not in cells with hypomethylated CpG, e.g., U87 (Fig. 5e, g). Considering that HSPB2 expression is relatively low in GBM cell lines (data not shown), HSPB2 overexpression by lentivirus infections was conducted and confirmed by western blot (Fig. 6a) and was not associated with significant alterations in proliferation and apoptosis (Fig. 6b–d). However, HSPB2 overexpression did confer resistance to TMZ treatment in GBM cells regardless of MGMT expression (Fig. 6e).
Fig. 5.
DNA methylation and gene expression of HSPB2. a The single CpG methylation of HSPB2 among nontumor brains and GBMs or gliomas of each grade. b gene expression patterns of HSPB2 among nontumor brains and GBMs or gliomas of each grade. c Correlations of the single CpG methylation and gene expression of HSPB2 in each cohort. d Representative HSPB2 IHC staining and pyrosequencing results (left) and the correlation of IHC staining scores and pyrosequencing data (right) in FFPE samples of IDHR132H wild-type GBMs. e HSPB2 single CpG pyrosequencing data in each GBM cell line. f, g Real-time PCR and western blot results of HSPB2 expressions in different GBM cell lines with either hypermethylated CpG (e.g., U251 and U373) or hypomethylated CpG (e.g., U87) after demethylation treatment with 5-Aza-dC at different doses (e.g., control, 5 nM, 10 nM). GBM glioblastoma, G-CIMP glioma-CpGs island methylator phenotype, FFPE formalin-fixed paraffin-embedded, wt wild type
Fig. 6.
The impacts of HSPB2 methylation and expression on TMZ resistance. a HSPB2 overexpression using lentivirus infections was confirmed by western blot in U251 and T98G cells regardless of MGMT expression. b CCK-8 proliferation assays of U251 and T98G cells infected with lentivirus containing human HSPB2 ORF or empty control lentivirus. c, d Flow cytometry and TUNEL assay analysis detecting apoptotic cells in U251 and T98G cells after infections with lentivirus containing human HSPB2 ORF or empty control lentivirus. e CCK-8 assays of U251 and T98G cells with or without HSPB2 overexpression after the treatment of TMZ at 100 μM or an equal volume of DMSO for 7 consecutive days. f The single CpG methylation of HSPB2 by pyrosequencing predicted OS in FFPE samples of IDH1R132H wild-type GBMs treated with the combination of RT and TMZ (left) but not in those with RT alone (right). g Interaction analysis showed that standard RT plus TMZ conferred a clear OS benefit to patients with hypermethylated HSPB2, rather than those with hypomethylated HSPB2, in comparison with standard RT alone. TMZ temozolomide, ORF open reading frame, RT radiotherapy, DMSO dimethyl sulfoxide, OS overall survival, FFPE formalin-fixed paraffin-embedded, GBM glioblastoma
Clinical performance of the single CpG methylation of HSPB2 by pyrosequencing in an IDH1R132H wild-type GBM cohort of FFPE samples
The validation cohort of 54 IDH1R132H wild-type GBMs was associated with similar patient age, gender, and pre-adjuvant therapy KPS compared to all collected IDH1R132H wild-type samples from our center (Additional file 1: Figure S1). Similarly, the clinical characteristics appeared to be similar between those patients who received RT/TMZ versus RT alone (Additional file 2: Table S1). The single CpG methylation of HSPB2 by pyrosequencing predicted OS in patients who received RT/TMZ (log-rank p = 0.0245) but not in those who received RT alone (log-rank p = 0.7733; Fig. 6f). Despite not reaching significance, similar findings were observed for PFS outcome (Additional file 5: Figure S3B). Interaction analyses and Cox analyses both supported an independent predictive potential of the single CpG methylation of HSPB2 to TMZ efficacy in clinically available FFPE samples (Fig. 6g, Additional file 8: Figure S4B and Additional file 9: Table S5 and Additional file 10: Table S6). Moreover, HSPB2 methylation could optimize the risk classification by MGMT promoter methylation status (Additional file 11: Figure S5C).
Discussion
Clinically informative biomarkers predictive of the likely benefit from specific treatments are of great significance in guiding precision medicine in cancer patients [27]. Epigenetic marks and DNA methylation in particular have long been regarded as the leading candidates for biomarker discovery as they have many advantages over genetic or expression-based information such as having reliable DNA samples, altered patterns that have stability, tolerance of nontumor cell contamination, multilevel biological relevance, and drug-induced reversibility [28, 29]. The promoter methylation status of MGMT for predicting TMZ outcome is considered an example of using DNA methylation as a more powerful indicator compared to other molecular information [30]. Unfortunately, this single-gene epigenetic status has critical blind spots regarding guiding treatment decisions for the very heterogeneous groups of GBM patients.[8, 31] Therefore, the development of powerful predictive indicator that could take advantage of multimarker information and provide complementary information to MGMT promoter methylation status would be greatly helpful for improving current clinical decision-making.
In this study, the Illumina 450k array provided a dramatic increase in the genomic coverage of CpGs compared to the 27k array. However, only a limited number of samples were available with Illumina 450k data. To ensure that there were enough samples to employ a discovery-validation approach, we decided to limit CpGs to those that appeared both on the 27k and 450k arrays, at the expense of higher genomic coverage. Regarding the outcome measurements, we choose OS (rather than PFS) as the primary endpoint for observation of the clinical benefit of TMZ. OS had been reported to be a reasonable endpoint to measure the clinical benefit of a given therapy, especially for advanced diseases with poor prognoses [32]. Additional considerations were the inconsistent assessment criteria for tumor progression and the unavailability of PFS data in the included cohorts. We employed a strict selection strategy to search novel CpGs with potential predictive value for TMZ outcome; the approach appeared to be efficient, as it could also identify MGMT-related loci. Following this strategy, we identified a panel of 7 CpGs from the discovery cohorts of RT/TMZ versus RT alone, each of which predicted OS of non-G-CIMP GBM patients, independent of their MGMT methylation status, age, and other identified CpGs. To coordinate these 7 CpGs, a RISKscore algorithm was used to produce a multimarker signature. Applying the signature to different validation cohorts with either RT/TMZ or RT alone showed that it was not a general prognostic biomarker for non-G-CIMP GBMs, as a prognostic factor is commonly defined as a clinical or biological characteristic that provides information on the likely outcome of a disease regardless of treatment [8, 33]. In contrast, a predictive factor is to provide information on the likely response to treatment and is used to identify subpopulations of patients who are most likely to benefit from a given therapy [8, 33]. Accordingly, the interaction analyses between the risk subgroups and treatments found a promising predictive value of the RISK score signature for the efficacy of TMZ, as the signature could identify the subgroups of low-risk patients who appeared to benefit more from RT/TMZ compared to RT alone. Thus, we concluded that the 7-CpG signature might not be a general treatment-independent prognostic biomarker but rather a promising predictive indicator of TMZ response in non-G-CIMP GBMs. These conclusions should be conservatively interpreted due to the following study limitations: (1) the prognostic effect has not been finally validated in patients who received no adjuvant therapies [33], and (2) the predictive effect has not been retrospectively or prospectively verified in a randomized controlled trial (RCT) [33]. Other limitations also confounded our results such as small sample size especially for patients who underwent only RT, potential biases in treatment assignment and patient baseline characteristics, heterogeneous treatment regimens, and insufficient clinical information (e.g., PFS, corticosteroid usage, regimens and salvage treatment). In addition, G-CIMP GBMs were excluded from our analyses because those tumors, exclusively carrying IDH mutations, represented a small subtype (~ 10% of all GBMs) and showed a very distinct molecular background and clinical prognosis compared to non-G-CIMP GBMs [9, 16].
The promoter methylation status of MGMT has been by far the most informative biomarker for GBMs; however, mandatory testing for this is highly controversial due to insufficient evidence showing a direct relationship between MGMT testing and TMZ usage [8]. Previous data has shown that despite with the much reduced benefits, the combination of RT and TMZ could still confer a significant improvement in OS among patients with unmethylated tumors, especially in younger patients [7]. Considering the absence of effective alternative therapies and a generally good tolerance to the aggressive combined treatment, TMZ is unlikely to be withheld from standard care [8]. The clinical value of MGMT testing is thus much compromised. In this study, we found that the RISK score signature might provide complementary information to MGMT for predicting TMZ outcome as it showed good and consistent discriminating value for the prognosis of non-G-CIMP tumors, independent of MGMT methylation status. The RISK score signature might be helpful for refining current TMZ usage practices by identifying patients with unmethylated tumors who might also benefit from TMZ, while sparing those who might not benefit from the high cost and potential toxicity related to this treatment. Though encouraging, the findings of this study should be fully validated in a RCT. The application of the combined RT/TMZ is complicated for elderly GBM patients [34]. Many physicians are still reluctant to treat elderly patients as aggressively as younger patients, citing concerns about overall poor physical condition, the presence of comorbidities, and a decreased tolerance to effective therapies. An increasing number of studies have reported clear benefits of RT/TMZ in elderly patients [34, 35]. However they all have highlighted the importance of appropriate patient selection for the optimal usage of the aggressive therapy in this frail population [34]. In addition to clinical factors (e.g., extent of resection, KPS, and comprehensive geriatric assessment), molecular data may also provide useful information [35]. Two recent phase III randomized trials have demonstrated a direct linkage between MGMT testing and TMZ usage in elderly patients [36, 37]. In our study, the RISK score signature showed good discriminating value for the prognosis of patients with different ages and even had a superior predictive ability compared to MGMT methylation status in older populations. Overall, the RISK score signature could be of potential use for individualized therapy by aiding in the appropriate patient selection and optimizing the current risk classification in GBMs.
The biological implications of the multimarker signature were exemplified by one CpG component and its relevant gene, HSPB2. HSPB2 is a member of the small heat shock protein gene family, encoding a molecular chaperone, and usually plays a protective role for cells from deleterious stresses such oxidative stress, heat shock, radiation and toxic drugs [38]. HSPB2 has been reported to be epigenetically or transcriptionally altered in some human cancers, with potential roles in tumor growth, metastasis, and in particular drug resistance [39]. In this study, the single CpG methylation was found to be negatively correlated with HSPB2 expression, indicating an epigenetic regulatory mechanism for this gene. The DNA methylation status and gene expression of HSPB2 were not significantly altered between nontumor brains and non-G-CIMP/IDHwt gliomas of each grade, indicating that it is not a good candidate for diagnostic use. In vitro experimental data showed that HSPB2 overexpression did not affect cell proliferation and apoptosis in GBM cells but conferred resistance to TMZ treatment regardless of MGMT expression. Together, these data may provide some biological explanations regarding the predictive effects of the multimarker signature—the epigenetic panel might contribute to TMZ resistance via epigenetically regulating the expression levels of drug-resistant genes. Genes (or RNAs) related to other identified CpGs have also been reported to have implications in cancers such as the long non-coding (lnc) RNA HCP5 and the gene TREM2 in gliomas [40, 41], and the genes NCOA4 and C4orf17 in prostate and ovarian cancers[42–44]. Notably, among the 7-CpG panel, some CpGs did not show a significant correlation between DNA methylation and the corresponding gene expression (data not shown). The biological impacts of those epigenetic alterations on TMZ resistance remained largely unclear. Recent studies reported that in addition to a direct alteration in gene expression, cancer-linked DNA methylation abnormalities may have functional impacts via contributing to disrupted heterochromatin, leading to loss of both epigenetic and transcriptional regulatory mechanisms and resulting in hypervariable expression [45]. Future studies will be needed to explore the biological relevance of other identified CpGs and the specific molecular mechanism of HSBP2-mediated TMZ resistance.
Unfortunately, our microarray-based signature is not available for clinical use due to the inconvenience and unavailability of the genome-wide detection technique in real clinical setting. Quantitative pyrosequencing is a well-established and widely used method for single CpG methylation detection. A comparative study has showed excellent congruence of DNA methylation data from the Illumina DNA methylation array with pyrosequencing data [46]. Therefore, the conventional technique of pyrosequencing may represent a good alternative solution for the 7-CpG profiling. Validation of the predictive ability of the single CpG methylation of HSPB2 by pyrosequencing in clinically available FFPE samples was reported not only to provide additional validation of the multi-CpG signature at a clinical level, but also to highlight the promising translational potential of the array-based signature to routine clinical testing. Independent validation studies will, however, be needed for adjusting the microarray-based signature to a pyrosequencing-based signature because there are inconsistencies between the two methods in the DNA methylation data for individual loci [46].
Recent multiplatform molecular profiling analysis had revealed that LGGs with wide-type IDH and intact 1p/19q have remarkable genomic and clinical similarity to primary non-G-CIMP GBMs [16]. Such similarity supports the potential inclusion of this subtype of LGGs within the broad spectrum of GBM-related clinical investigation and biomarker validation [16]. In this study, we have validated that the GBM-derived epigenetic signature could predict OS in an RT/TMZ cohort of GBM-like LGGs without IDH mutations and 1p/19q codeletions from TCGA (unknown doses of RT and TMZ, p = 0.0077; Additional file 13: Figure S7), its predictive performance however should be further investigated in this LGGs subtype without TMZ therapy.
In summary, we proposed a RISK score signature comprising 7 CpGs and the single CpG methylation of HSPB2, both with promising predictive values for the outcome of TMZ therapy in non-G-CIMP (or wild-type IDH) GBMs. These predictors might also provide complementary information to the current best MGMT-based predictor. Future prospective studies are needed to definitively establish these conclusions.
Additional files
Figure S1. The diagram of patient selection in each cohort; (A) selection of non-G-CIMP GBMs with RT/TMZ or RT alone; patient characteristics of included and excluded samples from TCGA-Brennan et al. were compared and presented; (B) selection of IDHwt+1p/19q-intact LGGs with RT/TMZ; patient characteristics of included and excluded samples from TCGA-LGGs were compared and presented; (C) selection of IDH1R132H wild-type GBMs with RT/TMZ or RT alone; patient characteristics of included and excluded FFPE samples from Xijing hospital were compared and presented; RT=radiotherapy; TMZ=temozolomide; G-CIMP=glioma CpG island methylator phenotype; GBM=glioblastoma; LGG=lower-grade gliomas; TCGA=The Cancer Genome Atlas; FFPE= formalin-fixed paraffin-embedded; OS=overall survival; M=male; F=female. (PDF 342 kb)
Table S1. Sample information of discovery and validation cohorts. (XLSX 16 kb)
Table S2. The primer sequences for PCR and pyrosequencing. (XLSX 13 kb)
Figure S2. The molecular correlation of each CpGs with genome DNA methylation status in GBMs; (A) the methylation status of each CpGs and the RISK scores between non-tumor brains, G-CIMP and non-G-CIMP tumors; (B) the correlation of the methylation status of each CpGs and the RISK scores with LINE-1 methylation; non-tumor brains from GSE63347 and TCGA as controls; * and ** indicates P <0.01 and <0.001; GBM=glioblastoma; G-CIMP=glioma CpG island methylator phenotype; TCGA=The Cancer Genome Atlas; ns=non-significance (p >0.05). (PDF 956 kb)
Figure S3. The prognostic performance in terms of PFS outcome; (A) patient classification by the RISK-score signature in RAUH-new cohort (left) and TCGA-Brennan et al. (right); (B) patient classification by HSPB2 methylation pyrosequencing in validation cohort of FFPE samples with RT/TMZ (left) and RT alone (right); PFS=progression-free survival; RT=radiotherapy; TMZ=temozolomide; RAUH=Rennes and Angers University Hospitals; FFPE=formalin-fixed paraffin-embedded. (PDF 424 kb)
Table S3. Univariate and multivariate Cox regression analyses in non-G-CIMP/IDH1R132H wt GBMs with RT/TMZ. (XLSX 27 kb)
Table S4. Univariate and multivariate Cox regression analyses in non-G-CIMP/IDH1R132H wt GBMs with RT alone. (XLSX 25 kb)
Figure S4. The predictive performance in terms of PFS outcome; (A) interaction analysis between treatments (with versus without TMZ) and risk subgroups (low-risk versus high-risk) in TCGA-Brennan et al, and (B) interaction analysis between treatments (with versus without TMZ) and risk subgroups (HSPB2 hypomethylation versus hypermethylation) in Xijing cohort; only patients with standard RT regimen were included for analysis in order to reduce potential bias by heterogeneous treatment regimen; SRT= standard radiotherapy; TMZ=temozolomide. (PDF 399 kb)
Table S5. Univariate and multivariate Cox regression analyses in low-risk group of non-G-CIMP (or HSPB2 hypermethylated IDH1R132H wt) GBMs. (XLSX 23 kb)
Table S6. Univariate and multivariate Cox regression analyses in high-risk group of non-G-CIMP (or HSPB2 hypomethylated IDH1R132H wt) GBMs. (XLSX 23 kb)
Figure S5. The patient classification in cohorts stratified by MGMT promoter methylation status and age; The RISK-score signature in the pooled GEO cohorts with RT/TMZ (GSE50923 and GSE60274 collectively) having (A) MGMT unmethylated (left) and methylated tumors (right) or (B) having older age (left) and younger age (right); HSPB2 methylation in Xijing cohort of FFPE tissues with RT/TMZ with MGMT unmethylated (left) and methylated tumors (right); GEO=Gene Expression Omnibus; FFPE=formalin-fixed paraffin-embedded; RT=radiotherapy; TMZ=temozolomide; MGMT=O-6-methylguanine-DNA methyltransferase. (PDF 508 kb)
Figure S6. The clinical performance of the RISK-score signature in combination with conventional risk factors in the setting of the combination treatment of RT and TMZ; (A) the risk classification of the RISK-score signature in combination with age (≥60 vs. <60 yrs; left) and MGMT promoter methylation status (right); (B) time-dependent ROC values of the RISK-score signature, age, MGMT promoter methylation status, and their combinations at each time point within all patients who underwent RT/TMZ from RAUH and TCGA-Brennan et al; (C) time-dependent ROC values of the RISK-score signature in comparison with MGMT promoter methylation status in subgroups of different ages at each time point; TCGA=The Cancer Genome Atlas; RAUH=Rennes and Angers University Hospitals; RT=radiotherapy; TMZ=temozolomide; MGMT=O-6-methylguanine-DNA methyltransferase; yrs=years. (PDF 749 kb)
Figure S7. The prognostic performance of the GBM-derived epigenetic signature in a RT/TMZ cohort of GBM-like LGGs without IDH mutations and 1p/19q co-deletion from TCGA; TCGA=The Cancer Genome Atlas; GBM=glioblastoma; RT=radiotherapy; TMZ=temozolomide; LGGs=lower-grade glioma; IDH= isocitrate dehydrogenase. (PDF 227 kb)
Acknowledgements
We gratefully acknowledge all the patients who agreed to participate in this study and to those who have provided their medical care. We gratefully acknowledge Pr Menei, Dr Le Reste, Dr. Vauleon, Dr. Quillien, and the Tumor Banks from Angers and Rennes for their constant support in the collection, processing, and histological analysis of tumor samples. We also gratefully thank those who are willing to share their valuable scientific data (Prof. Rose K Lai). I (AAY) thank my wife (Dr. Yu Dong) for her great support. The results published here are in part based upon data generated by The Cancer Genome Atlas, German Cancer Research Center, Chinese Glioma Gemoe Atlas, and the research teams mentioned in our study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81402049, No. 81471266 and No. 81302174) and China Postdoctoral Science Foundation, and by grants from the Brittany Region (France) et the FEDER (Europe).
Availability of data and materials
RAUH: http://www.ebi.ac.uk/arrayexpress/; TCGA: https://tcga-data.nci.nih.gov; CGGA: http://www.cgga.org.cn/; GEO: https://www.ncbi.nlm.nih.gov/geo/
Abbreviation
- ATCC
American Type Culture Collection
- CI
Confidence interval
- DMSO
Dimethyl sulfoxide
- FFPE
Formalin-fixed paraffin-embedded
- GBM
Glioblastoma
- G-CIMP
Glioma-CpGs island methylator phenotype
- HR
Hazard ratios
- IDH
Isocitrate dehydrogenase
- IHC
Immunohistochemistry
- lncRNA
Long non-coding RNA
- KPS
Karnofsky performance score
- LGG
Lower-grade glioma
- MGMT
O-6-methylguanine-DNA methyltransferase
- ORF
Open reading frame
- OS
Overall survival
- PCR
Polymerase Chain Reaction
- RAUH
Rennes and Angers University Hospitals
- RCT
Randomized controlled trial
- ROC
Receiver operating characteristic
- RT
Radiotherapy
- TCGA
The Cancer Genome Atlas
- TMZ
Temozolomide
Authors’ contributions
AAY, AE, BLL, and XZ conceived and designed the study. AAY, AE, YLH, JBS, and JM acquired the data. AAY, YHL, MA, and BLL analyzed and interpreted the data. AAY and YLH wrote and revised the paper. JM, XZ, and ZFL provided administrative, technical, or material support and supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained for all participates from Rennes and Angers University Hospitals, Xijing Hospital, The Cancer Genome Atlas, Chinese Glioma Genome Atlas, and other previously reported cohorts. All procedures performed in studies involving human were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zi-Fan Lu, Email: lzffmmu@163.com.
Xiang Zhang, Phone: +86 29 84771323, Email: xzhangneurosurgery@aliyun.com.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ, Mehta MP. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, Brandes AA, Hilton M, Abrey L, Cloughesy T. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 4.Ampie L, Woolf EC, Dardis C. Immunotherapeutic advancements for glioblastoma. Front Oncol. 2015;5:12. doi: 10.3389/fonc.2015.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Taillibert S, Kanner AA, Kesari S, Steinberg DM, Toms SA, Taylor LP, Lieberman F, Silvani A, Fink KL, Barnett GH, Zhu JJ, Henson JW, Engelhard HH, Chen TC, Tran DD, Sroubek J, Tran ND, Hottinger AF, Landolfi J, Desai R, Caroli M, Kew Y, Honnorat J, Idbaih A, Kirson ED, Weinberg U, Palti Y, Hegi ME, Ram Z. maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma: a randomized clinical trial. JAMA. 2015;314:2535–2543. doi: 10.1001/jama.2015.16669. [DOI] [PubMed] [Google Scholar]
- 6.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 7.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 8.Yin AA, Zhang LH, Cheng JX, Dong Y, Liu BL, Han N, Zhang X. The predictive but not prognostic value of MGMT promoter methylation status in elderly glioblastoma patients: a meta-analysis. PLoS One. 2014;9:e85102. doi: 10.1371/journal.pone.0085102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etcheverry A, Aubry M, de Tayrac M, Vauleon E, Boniface R, Guenot F, Saikali S, Hamlat A, Riffaud L, Menei P, Quillien V, Mosser J. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bady P, Sciuscio D, Diserens AC, Bloch J, van den Bent MJ, Marosi C, Dietrich PY, Weller M, Mariani L, Heppner FL, Mcdonald DR, Lacombe D, Stupp R, Delorenzi M, Hegi ME. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124:547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai RK, Chen Y, Guan X, Nousome D, Sharma C, Canoll P, Bruce J, Sloan AE, Cortes E, Vonsattel JP, Su T, Delgado-Cruzata L, Gurvich I, Santella RM, Ostrom Q, Lee A, Gregersen P, Barnholtz-Sloan J. Genome-wide methylation analyses in glioblastoma multiforme. PLoS One. 2014;9:e89376. doi: 10.1371/journal.pone.0089376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurscheid S, Bady P, Sciuscio D, Samarzija I, Shay T, Vassallo I, Criekinge WV, Daniel RT, van den Bent MJ, Marosi C, Weller M, Mason WP, Domany E, Stupp R, Delorenzi M, Hegi ME. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015;16:16. doi: 10.1186/s13059-015-0583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, Manyam G, Zoppoli P, Ling S, Rao AA, Grifford M, Cherniack AD, Zhang H, Poisson L, Carlotti CJ, Tirapelli DP, Rao A, Mikkelsen T, Lau CC, Yung WK, Rabadan R, Huse J, Brat DJ, Lehman NL, Barnholtz-Sloan JS, Zheng S, Hess K, Rao G, Meyerson M, Beroukhim R, Cooper L, Akbani R, Wrensch M, Haussler D, Aldape KD, Laird PW, Gutmann DH, Noushmehr H, Iavarone A, Verhaak RG. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Zhang W, Chen D, Lv Y, Zheng J, Lilljebjorn H, Ran L, Bao Z, Soneson C, Sjogren HO, Salford LG, Ji J, French PJ, Fioretos T, Jiang T, Fan X. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA. Proc Natl Acad Sci U S A. 2014;111:3538–3543. doi: 10.1073/pnas.1313814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S, Garagnani P, Bacalini MG, Pirazzini C, Salvioli S, Gentilini D, Di Blasio AM, Giuliani C, Tung S, Vinters HV, Franceschi C. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015;14:491–495. doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 20.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang LH, Yin AA, Cheng JX, Huang HY, Li XM, Zhang YQ, Han N, Zhang X. TRIM24 promotes glioma progression and enhances chemoresistance through activation of the PI3K/Akt signaling pathway. Oncogene. 2015;34:600–610. doi: 10.1038/onc.2013.593. [DOI] [PubMed] [Google Scholar]
- 22.Xi YB, Guo F, Xu ZL, Li C, Wei W, Tian P, Liu TT, Liu L, Chen G, Ye J, Cheng G, Cui LB, Zhang HJ, Qin W, Yin H. Radiomics signature: A potential biomarker for the prediction of MGMT promoter methylation in glioblastoma. J Magn Reson Imaging. 2018;47:1380–1387. doi: 10.1002/jmri.25860. [DOI] [PubMed] [Google Scholar]
- 23.Macdonald DR, Cascino TL, Schold SJ, Cairncross JG. Response criteria for phase II studies of supratentorial malignant glioma. J Clin Oncol. 1990;8:1277–1280. doi: 10.1200/JCO.1990.8.7.1277. [DOI] [PubMed] [Google Scholar]
- 24.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, Degroot J, Wick W, Gilbert MR, Lassman AB, Tsien C, Mikkelsen T, Wong ET, Chamberlain MC, Stupp R, Lamborn KR, Vogelbaum MA, van den Bent MJ, Chang SM. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 25.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 26.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roychowdhury S, Chinnaiyan AM. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin. 2016;66:75–88. doi: 10.3322/caac.21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Issa JP. DNA methylation as a clinical marker in oncology. J Clin Oncol. 2012;30:2566–2568. doi: 10.1200/JCO.2012.42.1016. [DOI] [PubMed] [Google Scholar]
- 29.Zhu J, Yao X. Use of DNA methylation for cancer detection: promises and challenges. Int J Biochem Cell Biol. 2009;41:147–154. doi: 10.1016/j.biocel.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 31.Berghoff AS, Preusser M. Clinical neuropathology practice guide 06-2012: MGMT testing in elderly glioblastoma patients--yes, but how? Clin Neuropathol. 2012;31:405–408. doi: 10.5414/NP300576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korn EL, Freidlin B, Abrams JS. Overall survival as the outcome for randomized clinical trials with effective subsequent therapies. J Clin Oncol. 2011;29:2439–2442. doi: 10.1200/JCO.2011.34.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Italiano A. Prognostic or predictive? It’s time to get back to definitions! J Clin Oncol. 2011;29:4718, 4718–4718, 4719. doi: 10.1200/JCO.2011.38.3729. [DOI] [PubMed] [Google Scholar]
- 34.Yin AA, Zhang LH, Cheng JX, Dong Y, Liu BL, Han N, Zhang X. Radiotherapy plus concurrent or sequential temozolomide for glioblastoma in the elderly: a meta-analysis. PLoS One. 2013;8:e74242. doi: 10.1371/journal.pone.0074242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauchet L, Zouaoui S, Darlix A, Menjot DCN, Ferreira E, Fabbro M, Kerr C, Taillandier L. Assessment and treatment relevance in elderly glioblastoma patients. Neuro Oncol. 2014;16:1459–1468. doi: 10.1093/neuonc/nou063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malmstrom A, Gronberg BH, Marosi C, Stupp R, Frappaz D, Schultz H, Abacioglu U, Tavelin B, Lhermitte B, Hegi ME, Rosell J, Henriksson R. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13:916–926. doi: 10.1016/S1470-2045(12)70265-6. [DOI] [PubMed] [Google Scholar]
- 37.Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, Nikkhah G, Papsdorf K, Steinbach JP, Sabel M, Combs SE, Vesper J, Braun C, Meixensberger J, Ketter R, Mayer-Steinacker R, Reifenberger G, Weller M. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13:707–715. doi: 10.1016/S1470-2045(12)70164-X. [DOI] [PubMed] [Google Scholar]
- 38.Chang X, Yamashita K, Sidransky D, Kim MS. Promoter methylation of heat shock protein B2 in human esophageal squamous cell carcinoma. Int J Oncol. 2011;38:1129–1135. doi: 10.3892/ijo.2011.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng G, Zhang Z, Liu H, Xiong Y, Luo L, Jia X, Peng C, Zhang Q, Li N, Gu Y, Lu M, Song Y, Pan H, Liu J, Liu W, He Z. HSP27-mediated extracellular and intracellular signaling pathways synergistically confer chemoresistance in squamous cell carcinoma of tongue. Clin Cancer Res. 2018;24:1163–1175. doi: 10.1158/1078-0432.CCR-17-2619. [DOI] [PubMed] [Google Scholar]
- 40.Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H, Xi Z, Li Z, Liu Y. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of glioma cells. Mol Ther. 2016;24:1806–1822. doi: 10.1038/mt.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Wang XQ, Tao BB, Li B, Wang XH, Zhang WC, Wan L, Hua XM, Li ST. Overexpression of TREM2 enhances glioma cell proliferation and invasion: a therapeutic target in human glioma. Oncotarget. 2016;7:2354–2366. doi: 10.18632/oncotarget.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rockfield S, Flores I, Nanjundan M. Expression and function of nuclear receptor coactivator 4 isoforms in transformed endometriotic and malignant ovarian cells. Oncotarget. 2018;9:5344–5367. doi: 10.18632/oncotarget.23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li P, Yu X, Ge K, Melamed J, Roeder RG, Wang Z. Heterogeneous expression and functions of androgen receptor co-factors in primary prostate cancer. Am J Pathol. 2002;161:1467–1474. doi: 10.1016/S0002-9440(10)64422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margiotti K, Wafa LA, Cheng H, Novelli G, Nelson CC, Rennie PS. Androgen-regulated genes differentially modulated by the androgen receptor coactivator L-dopa decarboxylase in human prostate cancer cells. Mol Cancer. 2007;6:38. doi: 10.1186/1476-4598-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timp W, Bravo HC, McDonald OG, Goggins M, Umbricht C, Zeiger M, Feinberg AP, Irizarry RA. Large hypomethylated blocks as a universal defining epigenetic alteration in human solid tumors. Genome Med. 2014;6:61. doi: 10.1186/s13073-014-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roessler J, Ammerpohl O, Gutwein J, Hasemeier B, Anwar SL, Kreipe H, Lehmann U. Quantitative cross-validation and content analysis of the 450 k DNA methylation array from Illumina, Inc. BMC Res Notes. 2012;5:210. doi: 10.1186/1756-0500-5-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The diagram of patient selection in each cohort; (A) selection of non-G-CIMP GBMs with RT/TMZ or RT alone; patient characteristics of included and excluded samples from TCGA-Brennan et al. were compared and presented; (B) selection of IDHwt+1p/19q-intact LGGs with RT/TMZ; patient characteristics of included and excluded samples from TCGA-LGGs were compared and presented; (C) selection of IDH1R132H wild-type GBMs with RT/TMZ or RT alone; patient characteristics of included and excluded FFPE samples from Xijing hospital were compared and presented; RT=radiotherapy; TMZ=temozolomide; G-CIMP=glioma CpG island methylator phenotype; GBM=glioblastoma; LGG=lower-grade gliomas; TCGA=The Cancer Genome Atlas; FFPE= formalin-fixed paraffin-embedded; OS=overall survival; M=male; F=female. (PDF 342 kb)
Table S1. Sample information of discovery and validation cohorts. (XLSX 16 kb)
Table S2. The primer sequences for PCR and pyrosequencing. (XLSX 13 kb)
Figure S2. The molecular correlation of each CpGs with genome DNA methylation status in GBMs; (A) the methylation status of each CpGs and the RISK scores between non-tumor brains, G-CIMP and non-G-CIMP tumors; (B) the correlation of the methylation status of each CpGs and the RISK scores with LINE-1 methylation; non-tumor brains from GSE63347 and TCGA as controls; * and ** indicates P <0.01 and <0.001; GBM=glioblastoma; G-CIMP=glioma CpG island methylator phenotype; TCGA=The Cancer Genome Atlas; ns=non-significance (p >0.05). (PDF 956 kb)
Figure S3. The prognostic performance in terms of PFS outcome; (A) patient classification by the RISK-score signature in RAUH-new cohort (left) and TCGA-Brennan et al. (right); (B) patient classification by HSPB2 methylation pyrosequencing in validation cohort of FFPE samples with RT/TMZ (left) and RT alone (right); PFS=progression-free survival; RT=radiotherapy; TMZ=temozolomide; RAUH=Rennes and Angers University Hospitals; FFPE=formalin-fixed paraffin-embedded. (PDF 424 kb)
Table S3. Univariate and multivariate Cox regression analyses in non-G-CIMP/IDH1R132H wt GBMs with RT/TMZ. (XLSX 27 kb)
Table S4. Univariate and multivariate Cox regression analyses in non-G-CIMP/IDH1R132H wt GBMs with RT alone. (XLSX 25 kb)
Figure S4. The predictive performance in terms of PFS outcome; (A) interaction analysis between treatments (with versus without TMZ) and risk subgroups (low-risk versus high-risk) in TCGA-Brennan et al, and (B) interaction analysis between treatments (with versus without TMZ) and risk subgroups (HSPB2 hypomethylation versus hypermethylation) in Xijing cohort; only patients with standard RT regimen were included for analysis in order to reduce potential bias by heterogeneous treatment regimen; SRT= standard radiotherapy; TMZ=temozolomide. (PDF 399 kb)
Table S5. Univariate and multivariate Cox regression analyses in low-risk group of non-G-CIMP (or HSPB2 hypermethylated IDH1R132H wt) GBMs. (XLSX 23 kb)
Table S6. Univariate and multivariate Cox regression analyses in high-risk group of non-G-CIMP (or HSPB2 hypomethylated IDH1R132H wt) GBMs. (XLSX 23 kb)
Figure S5. The patient classification in cohorts stratified by MGMT promoter methylation status and age; The RISK-score signature in the pooled GEO cohorts with RT/TMZ (GSE50923 and GSE60274 collectively) having (A) MGMT unmethylated (left) and methylated tumors (right) or (B) having older age (left) and younger age (right); HSPB2 methylation in Xijing cohort of FFPE tissues with RT/TMZ with MGMT unmethylated (left) and methylated tumors (right); GEO=Gene Expression Omnibus; FFPE=formalin-fixed paraffin-embedded; RT=radiotherapy; TMZ=temozolomide; MGMT=O-6-methylguanine-DNA methyltransferase. (PDF 508 kb)
Figure S6. The clinical performance of the RISK-score signature in combination with conventional risk factors in the setting of the combination treatment of RT and TMZ; (A) the risk classification of the RISK-score signature in combination with age (≥60 vs. <60 yrs; left) and MGMT promoter methylation status (right); (B) time-dependent ROC values of the RISK-score signature, age, MGMT promoter methylation status, and their combinations at each time point within all patients who underwent RT/TMZ from RAUH and TCGA-Brennan et al; (C) time-dependent ROC values of the RISK-score signature in comparison with MGMT promoter methylation status in subgroups of different ages at each time point; TCGA=The Cancer Genome Atlas; RAUH=Rennes and Angers University Hospitals; RT=radiotherapy; TMZ=temozolomide; MGMT=O-6-methylguanine-DNA methyltransferase; yrs=years. (PDF 749 kb)
Figure S7. The prognostic performance of the GBM-derived epigenetic signature in a RT/TMZ cohort of GBM-like LGGs without IDH mutations and 1p/19q co-deletion from TCGA; TCGA=The Cancer Genome Atlas; GBM=glioblastoma; RT=radiotherapy; TMZ=temozolomide; LGGs=lower-grade glioma; IDH= isocitrate dehydrogenase. (PDF 227 kb)
Data Availability Statement
RAUH: http://www.ebi.ac.uk/arrayexpress/; TCGA: https://tcga-data.nci.nih.gov; CGGA: http://www.cgga.org.cn/; GEO: https://www.ncbi.nlm.nih.gov/geo/