ABSTRACT
Sleep is nearly ubiquitous throughout the animal kingdom, yet little is known about how ecological factors or perturbations to the environment shape the duration and timing of sleep. In diverse animal taxa, poor sleep negatively impacts development, cognitive abilities and longevity. In addition to mammals, sleep has been characterized in genetic model organisms, ranging from the nematode worm to zebrafish, and, more recently, in emergent models with simplified nervous systems such as Aplysia and jellyfish. In addition, evolutionary models ranging from fruit flies to cavefish have leveraged natural genetic variation to investigate the relationship between ecology and sleep. Here, we describe the contributions of classical and emergent genetic model systems to investigate mechanisms underlying sleep regulation. These studies highlight fundamental interactions between sleep and sensory processing, as well as a remarkable plasticity of sleep in response to environmental changes. Understanding how sleep varies throughout the animal kingdom will provide critical insight into fundamental functions and conserved genetic mechanisms underlying sleep regulation. Furthermore, identification of naturally occurring genetic variation regulating sleep may provide novel drug targets and approaches to treat sleep-related diseases.
KEY WORDS: Ecology, zebrafish, Drosophila, Cavefish, Genetic screen, Natural variation
Summary: Although almost all animals sleep, the ecological and evolutionary factors that govern sleep differences throughout the animal kingdom remain unknown. We review sleep literature in non-mammalian models, describing recent advances in zebrafish, C. elegans and Drosophila, as well as emergent genetic model systems.
Introduction
Sleep appears to be fundamental to animal life, yet little is known about how sleep has evolved throughout the animal kingdom. In diverse animal taxa, poor sleep can have detrimental effects on development (Roffwarg et al., 1966; Mirmiran et al., 1983; Kayser et al., 2014), cognitive abilities (Scullin and Bliwise, 2015) and life span (Mazzotti et al., 2014), and it is now appreciated that normal sleep is fundamental to healthy physiology and bodily function (Shaw et al., 2002; Cappuccio et al., 2010; Arble et al., 2015). While the function of sleep remains unknown, studies have identified relationships between sleep and anatomical, physiological or ecological traits (Lesku et al., 2006; Capellini et al., 2008; McNamara et al., 2009). For example, in mammals, parameters such as diet, brain size, social hierarchy and body mass index all affect total sleep times (Campbell and Tobler, 1984; Siegel, 2005; Lesku et al., 2006). A great deal of variation exists in sleep duration and timing among different animal phyla, with animals such as the African Elephant sleeping only 3–4 h a day (Siegel, 2005) while many animals spend the majority of their time sleeping, including the armadillo, which sleeps over 18 h per day (Siegel, 2005; Capellini et al., 2008). Even among humans, sleep times vary widely, ranging from less than 5 h to 10 h or more (Webb and Agnew, 1970; Kronholm et al., 2006). Despite a widespread appreciation for the diversity in sleep duration between and within species, surprisingly little is known about the relationship between sleep and an animal's ecological and evolutionary history.
Large differences in sleep duration and timing among humans suggests that existing genetic variation among individuals potently affects sleep (Hartmann, 1973; Kronholm et al., 2006; He et al., 2009). While many laboratory studies investigating the molecular mechanisms of sleep regulation have relied on highly inbred model systems including mice, zebrafish and fruit flies, the study of sleep in outbred populations has revealed that geographical location, evolutionary history and naturally occurring genetic variation contribute to robust sleep differences within animals of the same species (Duboué et al., 2011; Zimmerman et al., 2012; Banks et al., 2015; Svetec et al., 2015). Even though sleep has been characterized in surprisingly few evolutionary systems, small, non-mammalian model organisms with a well-defined evolutionary history provide opportunities to identify novel mechanisms underlying sleep regulation (Fig. 1). Here, we propose that expanding sleep studies to emergent model systems (see Glossary) is essential to understand how sleep is influenced by evolutionary and ecological history. Identifying how evolution and ecology may shape sleep differences throughout the animal kingdom will provide insight into the fundamental functions of sleep, and the underlying variability in sleep duration throughout the animal kingdom.
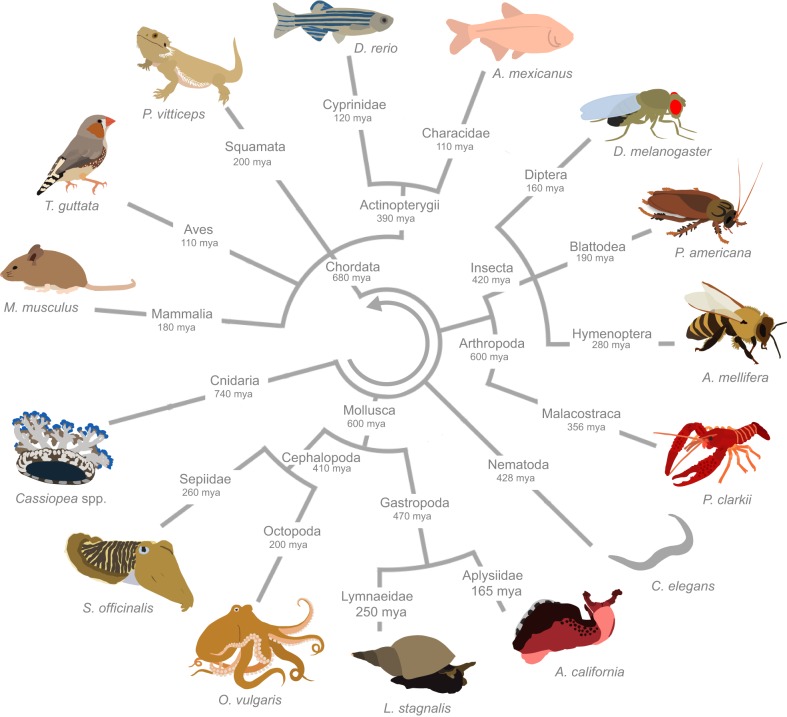
Fig. 1.
Small animal models of sleep research. This unrooted, pruned cladogram is inclusive of animals that have been established as models for studying sleep, demonstrating the diversity of sleep behaviors throughout the animal kingdom. Branches demonstrate broad relationships among animals, but do not represent actual distance measures. Taxonomic information was obtained from the NCBI Taxonomy Database and the tree generated with NCBI Common Tree (Sayers et al., 2012). The circular cladogram was generated using the Interactive Tree of Life (Letunic and Bork, 2016) and estimates of divergence were derived from the TimeTree Database (Hedges et al., 2015). mya, million years ago.
Achieving a full understanding of sleep function requires detailed characterization of the genetic, molecular and neuronal properties associated with sleep and wakefulness. Over the past few decades, non-mammalian genetic model systems including the nematode worm Caenorhabditis elegans (Raizen et al., 2008), the fruit fly Drosophila melanogaster (Hendricks et al., 2000a; Shaw et al., 2000) and the zebrafish Danio rerio (Zhdanova et al., 2001; Prober et al., 2006; Yokogawa et al., 2007) have been particularly advantageous in elucidating genetic and molecular components underlying sleep. Many forward-genetic screens have highlighted the conservation of molecular and neural principles underlying sleep–wake regulation, and initial studies in zebrafish, Drosophila and C. elegans have identified novel and conserved regulators of sleep (Allada and Siegel, 2008; Trojanowski and Raizen, 2016; Levitas-Djerbi and Appelbaum, 2017). Although a mechanistic understanding of sleep regulation is far from complete, these studies provide a framework for interpreting how inter- and intra-species variation may account for naturally occurring differences in sleep.
In this review, we briefly discuss sleep in classic genetic models, as they lay the foundation for work in emergent model systems that focus on the evolution of behavior. We then describe the use of emergent invertebrate and fish model systems to investigate the function of sleep, and the role of evolution in shaping naturally occurring differences. Furthermore, we highlight technology described in other model systems, and discuss how their application in emergent models may be used to investigate highly conserved features of sleep–wake cycles.
Glossary.
Actigraphy
Non-invasive method for measuring levels of activity in humans and other animal subjects.
Arousal threshold
The minimum intensity that an external stimulus needs to elicit a response, typically a locomotor response, in an animal. The threshold to elicit a response is typically higher when animals are in a sleep-like state compared with waking.
Brain local field potential
Electrical potential of an extracellular space of a neuronal area. Recorded through implantation of an extracellular recording electrode.
Central pattern generator
Clusters of neurons that oscillate with a specified rhythmicity. A key feature of central pattern generators is that they produce oscillatory patterns alone, without oscillatory input.
Chronotype
The propensity for an animal to sleep at a given period in a 24 h cycle. Most humans are diurnal organisms, meaning they sleep at night. By contrast, nocturnal organisms sleep during the day.
Circadian rhythm
Fluctuations of locomotor behavior around a 24 h period. Derived from circa (Latin for ‘around’) and dia (Latin for ‘day’).
Connectome
The full repertoire of neural connections in the brain of an organism.
Diurnal
Sleeping during the night and being largely awake during the day.
Emergent model systems
Animal models that are outside those typically considered as genetic model systems (C. elegans, Drosophila, zebrafish and mice).
Locomotor activity
Behavioral movement.
Neuromast
A sensory organ in fish and other aquatic animals that is composed of bundles of neurons. Neuromasts typically lay on the facial structure (superficial neuromasts) or along the body wall (lateral line).
Nocturnal
Sleeping during the day and being largely awake during the night.
Rapid eye movement (REM)
A sleep stage characterized by rapid movement of the eyes. Dreaming in humans occurs in REM sleep.
Rebound
The increase in sleep observed after an individual is sleep deprived.
Sleep latency
The amount of time it takes for an animal to fall asleep when permitted.
Behavioral analysis of sleep in non-mammalian systems
Classical approaches to characterizing sleep
Sleep is characterized in different ways, depending on the available methods in the organism of study. The two hallmarks classically have been (i) electrophysiological or (ii) behavioral changes associated with sleep-like states. Importantly, there is a strong correlation between sleep-like states defined by electrophysiological and behavioral criteria in animals ranging from invertebrates to mammals (Allada and Siegel, 2008). Electrophysiologically, sleep is characterized by changes in brain wave activity, as measured by the electroencephalogram (EEG) in mammals, or local field potential recordings in invertebrates (Berger and Gloor, 1969; Keenan and Hirshkowitz, 2010; van Alphen et al., 2013). The EEG has established its place in sleep research, and provides the ability to compartmentalize sleep into three different stages based on unique patterns of brain wave activity: (i) waking periods, (ii) non-rapid eye movement (NREM), slow-wave sleep, and (iii) REM (see Glossary) or paradoxical sleep (Dement and Kleitman, 1957). Nevertheless, a number of limitations to electrophysiological recordings, including difficulty of recording in small animals and impracticality of recording in a natural setting, highlight the need for behavioral observations that can be used to define sleep.
Animals in a sleep-like state, as measured by an EEG, typically assume stereotypical behaviors, and, by carefully correlating behavior with changes in EEG patterns, behavioral identifiers have been established (Ookawa and Gotoh, 1965; Flanigan et al., 1973; Campbell and Tobler, 1984). Behaviorally, sleep can be characterized by five criteria: (i) prolonged behavioral quiescence, (ii) which is reversible upon stimulation (to differentiate from torpor or coma), (iii) a species-specific posture, (iv) increased arousal threshold (see Glossary) to respond to external stimuli, and (v) rebound (see Glossary) following sleep deprivation. The establishment of behavioral definitions of sleep has opened the door to investigating sleep in small model systems that are not amenable to EEG analysis (Chiu and Prober, 2013; Griffith, 2013; Trojanowski and Raizen, 2016). Although EEG measurements are typically restricted to a limited subset of taxonomic groups, behavioral definitions are largely generalizable to all animals in the animal kingdom, and permit the investigation of sleep from a comparative perspective, as well as in small genetically amenable animal models.
Landmark papers over the past 20 years have defined sleep in classic genetic systems including C. elegans, Drosophila and zebrafish (Hendricks et al., 2000a; Shaw et al., 2000; Zhdanova et al., 2001; Raizen et al., 2008). Each model has unique and shared characteristics that are particularly useful for sleep analysis. In all three model systems, sleep state is associated with prolonged periods of behavioral quiescence, which are readily reversible and correspond to an increase in arousal threshold (Hendricks et al., 2000a; Shaw et al., 2000; Prober et al., 2006; Yokogawa et al., 2007; Raizen et al., 2008; reviewed in Sehgal and Mignot, 2011). Of the classic genetic model systems, worms contain the fewest neurons. The adult hermaphrodite worm has 302 neurons, all of which have been mapped out with sufficient detail, while the larval zebrafish brain and adult fruit fly brain contain 100,000 and 250,000 neurons, respectively (Ward et al., 1975; Ahrens and Engert, 2015). The simple nervous system and powerful genetics of these models has led to the identification of neural circuits, up to single-neuron resolution, that modulate sleep (Appelbaum et al., 2009; Aso et al., 2014a; Singh et al., 2015). The unique application of sophisticated genetic tools and high-throughput genetic screens have paved the way for mechanistic investigation of sleep regulation that has revealed robust conservation across the animal kingdom.
Physiological approaches to characterizing sleep
Although studies in invertebrates and fish have typically relied on behavioral metrics of sleep, there is growing evidence that physiological metrics often used to characterize sleep in mammals are present in small non-mammalian models. Sleep-associated brain activity has been convincingly demonstrated in the fruit fly, where five minutes of inactivity was initially used to define sleep based on the standardized behavioral characteristics, and standardized behavioral systems have been developed to measure sleep (Shaw et al., 2000; Garbe et al., 2015). The detailed behavioral characterization of sleep opened the door to genetic interrogation of sleep in invertebrates and fish models; they lack the precision of EEG or other physiological read-outs of sleep and come with a number of limitations including difficulty of identifying awake animals engaged in torpor.
Later, elegant experimental designs demonstrated that definitions of sleep based on behavior and those based on electrophysiology largely corresponded with one another. A system was devised to record the neural properties of the fly brain in an adult tethered to a tracking ball, providing the opportunity to simultaneously observe neural and behavioral correlates of sleep. Simultaneous recordings of brain local field potentials (see Glossary) and locomotor activity (see Glossary) on a rotating ball revealed reduced neuronal activity (11–40 Hz oscillations) when flies are immobile, which also correlates with elevated arousal threshold (Nitz et al., 2002; van Alphen et al., 2013). In agreement with these findings, dynamic changes in metabolic rate were observed in sleeping flies, supporting the notion that flies, like mammals, suppress metabolic rate when starved of food (Stahl et al., 2017). It is possible that these physiological metrics of sleep will reveal physiological changes akin to sleep stages in animals with more complex brains and can be applied to additional invertebrate and fish models. For example, a study in crayfish identified sleep using all the behavioral correlates, and electrophysiological recordings revealed synchronized activity similar to slow-wave sleep (Ramón et al., 2004). Together, these findings highlight the strength of model organisms in genetic research to identify brain states associated with sleep that are conserved throughout the animal kingdom.
Screen-based identification of sleep regulators using classic model systems
A significant strength of small, genetically amenable model organisms is the ability to screen large numbers of animals for mutations or drugs that affect behavior. In the case of sleep studies, this has been supported by the development of behavioral monitoring systems in C. elegans, Drosophila and zebrafish that allow for high-throughput analysis of locomotor activity. The combination of defined methodology for measuring sleep, and a vast array of genetic mutants and tools provide the capability to identify genetic and pharmacological regulators of sleep.
Classic genetics were first used to investigate circadian rhythms (see Glossary) in fruit flies, and more recently applied to identify novel sleep genes (Hendricks et al., 2000b; Hall, 2003). Large-scale forward-genetic screens in fruit flies have used mutagenesis or transgenic expression of interfering RNA (RNAi) to identify sleep-regulating genes including the K+ channel Shaker, the regulator of cell-cycle modulator Cyclin A, and transmembrane protein Sleepless (Cirelli et al., 2005; Koh et al., 2008; Rogulja and Young, 2012). Similar endeavors in C. elegans and zebrafish have identified additional regulators of sleep, many of which appear to be conserved in mammals (Chiu and Prober, 2013; Singh et al., 2014). For example, a prominent role for the worm homologue of mammalian Neuropeptide Y receptor NPR-1 is required for sleep homeostasis in C. elegans (Nagy et al., 2014), and genome-wide screening identified striking overlap between sleep genes in worms and mammalian systems (Singh et al., 2014). Similarly, screens in zebrafish have implicated the prominent regulators of mammalian sleep Hypocretin/Orexin and melatonin, as well as a number of novel genes required for integration of sleep and sensory systems that detect light (Zhdanova et al., 2001; Prober et al., 2006; Appelbaum et al., 2009; Chen et al., 2016). The diversity of genes identified in these forward-genetic screens highlights the strength of model systems to identify novel genetic architecture contributing to sleep circuits.
The strength of invertebrate and fish models extend beyond genetic applications. Zebrafish, Drosophila and C. elegans are well suited for drug screening, where the quantity of available compounds is often a limiting factor, and pharmacological targets can be validated genetically (Giacomotto and Ségalat, 2010; Rihel and Schier, 2013; Churgin et al., 2017). In a landmark study, over 5000 compounds were assayed in larval zebrafish for their effect on sleep latency (see Glossary), duration and bout number. This study identified a number of sleep- or activity-regulating drugs including inhibitors of ether-a-go-go-related K+ channel, which promote waking activity (Rihel et al., 2010). Similar approaches have been used in fruit flies, where a sleep screen testing 1280 small molecules identified numerous novel regulators of sleep including the vesicular monoamine transporter VMAT (Nall and Sehgal, 2013). Although these approaches have yet to be applied to study sleep in C. elegans, the use of small-molecule screens to study other processes and availability of high-throughput behavioral monitoring systems suggest that this approach is feasible (Schwendeman and Shaham, 2016). These approaches highlight the potential for drug discovery using small-molecule-based screens and sleep itself providing a behavioral read-out of drug efficacy.
The utility of small genetic models extends beyond investigation into the molecular mechanisms of sleep–wake cycles, and studies in these models advanced our understanding of the neuronal circuits that mediate the behavior (Kayser and Biron, 2016). For example, an existing connectome (see Glossary) defining all neural connections within the nervous systems makes C. elegans an extremely tractable model for circuit mapping, and single neurons that either promote or inhibit sleep have been identified (Cho and Sternberg, 2014; Turek et al., 2016). Large collections of transgenic driver lines labelling small subsets of neurons make fruit flies tractable for dissecting neural circuits regulating sleep (Venken et al., 2011; Jenett et al., 2012). Although a number of brain areas have been identified that appear to be crucial regulators of sleep, including the mushroom bodies and fan-shaped body in Drosophila (Joiner et al., 2006; Pitman et al., 2006; Donlea et al., 2011; Aso et al., 2014b) and Orexin/Hypocretin neurons in zebrafish (Prober et al., 2006; Yokogawa et al., 2007; Appelbaum et al., 2009), these findings have not resulted in an integrative model defining how sleep is modulated under different environmental contexts. To date, studies have typically been limited to selectively manipulating or imaging small populations of neurons; however, the recent application of in vivo brain-wide imaging of neural activity in C. elegans, Drosophila and zebrafish provides a novel approach for identifying neural correlates of sleep and wakefulness (Muto et al., 2013; Harris et al., 2015; Nguyen et al., 2015). These studies highlight the complexity of sleep-regulating circuits and the ability to precisely map circuits using genetic tools in these systems. The implementation of transgenic methodology for manipulation of neuronal function or functional imaging into additional animal models will provide novel insights into the biological mechanism and function of sleep regulation.
Use of emergent systems to study how evolution and ecology shape sleep
Investigating the origins of sleep
Genetic variation has been selected against in many laboratory strains of genetic model organisms, often obscuring the ecological relevance of different genetic or neuronal perturbations. Moreover, the small number of genetically amenable models, typically limited to inbred populations of C. elegans, Drosophila, zebrafish and mice, represent a narrow subset of species that sleep. The identification of sleep in C. elegans revealed that sleep exists even in animals with relatively simple nervous systems, suggesting that sleep is an ancient behavior that is probably present throughout the animal kingdom (Zimmerman et al., 2008). The potentially ancient ancestral origins of sleep has led to a prominent question in the field of whether sleep is a property of neural circuits, or rather a property of individual cells (Vyazovskiy and Harris, 2013). Although this is difficult to address in animals with complex brains, organisms with simplified nervous systems provide the ability to ask questions about the fundamental cellular function of sleep.
As stated above, characterizing sleep in novel animal models requires defining sleep based upon behavioral criteria including the period of behavioral quiescence that is associated with changes in arousal threshold. Recently, these metrics have been applied to marine species with simplified nervous systems, called nerve nets, including the Cnidarian jellyfish, as well as multiple different species of molluscs (Brown et al., 2006; Stephenson and Lewis, 2011; Frank et al., 2012; Vorster et al., 2014; Nath et al., 2017). Each of these organisms has nervous systems that may be useful in dissecting the basic neural principles and functions of sleep regulation, although it is noted that the duration, circadian timing and characteristics of sleep vary dramatically between species.
The nerve net in jellyfish consists of rings of neurons along the axial length of the organism (Katsuki and Greenspan, 2013). Sleep has been investigated in different species of the ‘upside-down’ jellyfish, Casseopea, a genus typically found in the shallow coastal waters surrounding Florida and the Caribbean islands (Holland et al., 2004). When active, they display contractions of the nerve net, which in turn causes pulsing behavior that facilitates feeding and flow of nutrients throughout the organism (Jantzen et al., 2010; Santhanakrishnan et al., 2012). Pulsing behavior and their sensitivity to external stimuli is reduced during night-time periods, suggesting that night-time behavioral quiescence is a sleep-like state (Nath et al., 2017). Lastly, when sleep deprived by mechanical stimulation (intermittent water flow during the night), jellyfish exhibit increased immobility the following day that is indicative of a homeostatic rebound in sleep (Nath et al., 2017). Therefore, jellyfish possess many of the behavioral hallmarks of sleep and provide a new model for studying the origins of sleep.
Similarly, sleep has been characterized in several species of molluscs, including the gastropods Aplysia californica, pond snail (Lymnaea stagnalis), the octopus (Octopus vulgaris) and at least one cephalopod, the cuttlefish (Sepia officinalis) (Brown et al., 2006; Stephenson and Lewis, 2011; Frank et al., 2012; Vorster et al., 2014). All of these organisms show periods of behavioral quiescence that correspond to a sleep-like state. Interestingly, circadian modulation of sleep–wake cycles varies among molluscs: Aplysia and cuttlefish display robust diurnal (see Glossary) waking rhythms, whereas the octopus is nocturnal (see Glossary) and the pond snail exhibits infrequent sleep states that occur without influence of time of day (Brown et al., 2006; Stephenson and Lewis, 2011; Vorster et al., 2014). Thus, even between these cephalopod and gastropod species, sleep structure is highly variable, and may provide insight into ecological- and niche-dependent interactions between sleep and circadian rhythms.
Potential applications for gastropod and cephalopod models in sleep research
Mollusc species are well suited for electrophysiological studies, and have been widely used to study the neuronal mechanisms underlying diverse processes including learning and memory, visually guided prey-seeking behavior, and social interactions (Chiao et al., 2015). Characterizing sleep in these mollusc species, therefore, provides a unique opportunity to examine the relationship between sleep and these fundamental processes. For example, recent findings indicate that sleep in Aplysia promotes memory consolidation, while sleep deprivation inhibits the formation of new memories (Krishnan et al., 2016a,b; Levy et al., 2016). Like Aplysia, Lymnaea display robust classical and operant long-term memories, pairing various attractive or aversive stimuli with feeding behavior. Although sleep is sporadic and limited to short bouts in this species, it is possible that loss of sleep will impair memory formation. The ability to form memories varies across strains (Braun et al., 2012), and can thus provide the opportunity to examine the relationship between sleep and memory formation in different strains. The differences in sleep are dependent on the state of a single neuron, RPed1, that is associated with a central pattern generator (see Glossary) (Braun et al., 2012). Given the detailed understanding of the Aplysia and Lymnaea nervous system, and robust assays for measuring sleep and memory, this system has the potential to uncover the functional basis for interactions between sleep and memory formation. Furthermore, the availability of annotated genomes or transcriptomes for Aplysia and Lymnaea (Moroz et al., 2006; Sadamoto et al., 2012) provides the opportunity to investigate the transcriptional basis of sleep-dependent changes in memory formation.
Both cuttlefish and octopus have a highly developed visual system and complex visual behaviors (Young, 1991; Hanlon et al., 2009). The octopus has been used as a model for visual memory formation and forms distinct memories dependent on visually guided or tactile learning (Bradley and Young, 1975; Gutnick et al., 2011). Cuttlefish display remarkably complex visually guided behaviors including changes in body patterning for camouflage and communication, despite a lack of color vision (Wells, 1997). Cuttlefish also have robust visual memories, learning to associate prey with their visual surroundings, but the relationship of these memories to sleep has not been investigated (Jozet-Alves et al., 2013). Interestingly, cuttlefish have stereotyped eye movements that resemble rapid eye movement (REM) sleep, which has not been identified in invertebrates or fish models (Frank et al., 2012). The complex sensory systems of cuttlefish and octopus lend these organisms potential advantages as models for examining interactions between sleep and sensory systems.
While identifying sleep in jellyfish and molluscs opens the door for studies for sleep function, rhythmic circadian behavior or gene expression has been identified in many other systems that have not been analysed for sleep (Joiner, 2016). For example, the starlet sea anemone Nematostella vectensis has robust circadian behavioral and molecular rhythms, a simplified and transparent nervous system, and is amenable to transgenic manipulation (Hendricks et al., 2012; Peres et al., 2014; Oren et al., 2015; Renfer and Technau, 2017). Molecular circadian rhythms have also been identified in the tunicate sea squirt Ciona intestinalis that are accompanied by circadian-dependent regulation of oxygen consumption (Minamoto et al., 2010). Ciona have long been used as a model for embryogenesis, but a recent focus on nervous system structure, and accompanying genetic tools, provide the foundation for behavioral analysis (Takamura et al., 2010; Hozumi et al., 2015). Beyond these simple systems, circadian rhythms have been characterized in many animals with simple body plans including the sea sponge Suberites domuncula (Müller et al., 2012) and the bread mold, Neurospora crassa (Feldman and Hoyle, 1976; Gardner and Feldman, 1980). These studies demonstrate unequivocally that circadian behavior is a cellular property (Müller et al., 2012), and suggest the characterization of sleep in these simple system may shed light on the earliest forms of sleep.
Natural variation in sleep regulation
The naturally occurring variation in human sleep duration or timing appears to be genetically encoded, although little is known about the specific genes that contribute to differences in sleep need between individuals (Parsons et al., 2013; Hu et al., 2016; Lane et al., 2016). Although single genes have been identified that contribute to naturally occurring variation in sleep (Allebrandt et al., 2013; He et al., 2009), most variability undoubtedly comes from a complex genetic architecture. Recent genome-wide association studies (GWAS) have typically relied on self-reported sleep duration, latency and chronotype (see Glossary) (Hu et al., 2016; Jones et al., 2016), while other studies have used actigraphy (see Glossary) (Spada et al., 2016). A significant impediment to these studies has been validating the function of GWAS alleles and investigating their mechanistic role in sleep regulation. One GWAS study that used self-reported sleep data from over 4000 individuals estimated that a locus containing the KATP channel ABCC9 accounted for approximately 5% of variation in human sleep (Allebrandt et al., 2013). Genetic knock-down of the ABCC9 fruit fly ortholog sufonurea receptor 2 (dsur2) in neurons selectively reduced night sleep without affecting daytime sleep (Allebrandt et al., 2013). These findings validate a role for ABCC9/dsur2 and the utility of reverse-genetic approaches to functionally validate genes identified through human GWAS studies.
Naturally occurring variation and its effects on sleep can be leveraged in a laboratory setting, and as such, the contributions of naturally occurring genetic variation to sleep regulation can also be directly investigated in model systems (Fig. 2). While laboratory studies of D. melanogaster typically rely on inbred strains that have been housed in the laboratory for decades, this species is found in diverse climates all over the world (David and Capy, 1988; Stephan and Li, 2007). Drosophila melanogaster from different geographical regions are genetically distinguishable at genetic and behavioral levels (Schmidt et al., 2005; Schmidt and Paaby, 2008; Reinhardt et al., 2014). Multiple studies have found that increased sleep duration is associated with proximity to the equator, suggesting that flies from warmer climates with reduced seasonal variation in temperature sleep longer than flies from northern latitude clines (Fig. 2A; Svetec et al., 2015; Brown et al., 2018). A transcriptome comparison between flies from high and low latitudes revealed enrichment of differentially expressed genes related to circadian clock function (Svetec et al., 2015). These findings, combined with short-sleeping phenotypes of circadian mutants, and a known wake-promoting role for circadian neurons, suggest that selection on latitude-associated changes in the circadian genes may contribute to sleep difference in naturally occurring populations of Drosophila (Parisky et al., 2008; Keene et al., 2010). Investigating the relationship between sleep and circadian function in flies from geographically diverse regions may uncover novel interactions between the circadian and sleep–wake rhythms.
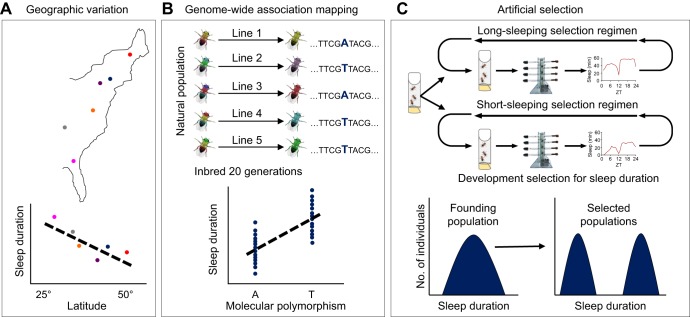
Fig. 2.
Approaches for studying the contributions of natural variation on sleep regulation in fruit flies. (A) Drosophila melanogaster from different geographical regions have been tested for sleep. Increased sleep duration is associated with proximity to the equator (Svetec et al., 2015). (B) Genome-wide association mapping studies have been performed using the Drosophila Genetics Research Panel. Individual inbred fly lines originating from wild-caught populations are tested for sleep and phenotypes are associated with genetic variation (Harbison et al., 2013). A and T represent single nucleotide genomic differences between individual lines. (C) Artificial selection of outbred lines for long- or short-sleep phenotypes, resulting in dramatic shifts in sleep duration. Flies were grown in vials and then selected as long or short sleepers using Drosophila Activity Monitors (DAMs) (Seugnet et al., 2009 and Masek et al., 2014).
Existing natural variation can also be experimentally leveraged to generate genetically variable inbred lines (Fig. 2B). The Drosophila Genome Reference Panel (DGRP) consists of 205 highly inbred strains originating from wild-caught Drosophila (Mackay et al., 2012). These lines have been fully sequenced and an existing whole-body transcriptome allows for genotype–phenotype and expression quantitative trait locus (eQTL) analysis (Huang et al., 2015). Inbred DGRP fly lines have been characterized for numerous behaviors, including feeding, olfaction, alcohol sensitivity, aggression, courtship and sleep (Swarup et al., 2013; Gaertner et al., 2015; Garlapow et al., 2015; Morozova et al., 2015; Shorter et al., 2015). Sleep analysis revealed remarkable diversity between inbred lines, with some exhibiting as much as a threefold difference in day- and night-time sleep, suggesting that the sleep differences caused by naturally occurring variation are as robust as many of the most severe single-gene mutants identified through forward genetic screening (Harbison et al., 2009, 2013). Genotype–phenotype analysis identified 2525 single nucleotide polymorphisms (SNPs) implicated in diverse aspects of sleep regulation, including day- and night-time sleep, bout duration and bout number (Harbison et al., 2013). Although approximately half of SNPs were in intragenic loci, many of the SNPs localized to genes previously associated with fly or mammalian sleep, and a role for five novel genes could be validated using genetic mutants or RNA interference (Harbison et al., 2013). A similar approach investigating the relationship between sleep and whole-body gene expression in these 40 DGRP lines identified numerous sleep-regulating genes including the finding of the polymorphism Catsup, a negative regulator of dopamine synthesis, as well as validation of four additional sleep regulators using genetic mutants (Harbison et al., 2009). Therefore, one can conclude that the study of how naturally occurring variation in sleep associates with transcription or genotype can be used to identify novel sleep regulators.
Studying natural variation in sleep using emergent model systems
Genetic technology in Drosophila may afford the ability to functionally characterize naturally occurring genetic variation identified in non-genetic model systems. A number of additional insect systems provide the opportunity to examine how both genetic variation and life history traits contribute to sleep regulation including honey bees, moths and cockroaches (Anderson, 1968; Tobler, 1983; Sauer et al., 2003). The diversity of insects, each with unique ecological and evolutionary history, may complement the genetic approaches commonly applied in the fruit fly.
Sleep has been studied extensively in the honeybee Apis melifera, a eusocial insect with a complex caste system capable of forming robust memories. Sleep was initially characterized as periods of immobility that were associated with a lack of antennae movement and reduced sensitivity in visual optomotor neurons, providing both behavioral and physiological read-outs of sleep (Kaiser and Steiner-Kaiser, 1983; Sauer et al., 2003). In addition, bees display a rebound following sleep deprivation characterized by increased antennal immobility (Sauer et al., 2004). Electrophysiological approaches have been applied extensively to study memory and odor coding in bees. Recordings of mushroom body neurons, a memory center later found to promote sleep in Drosophila (Joiner et al., 2006; Pitman et al., 2006), revealed episodes of increased activity during sleep, and was proposed to be indicative of slow wave sleep (Schuppe, 1995).
The honeybee, like many eusocial insects, undergoes a series of life transitions. Younger bees tend to be nurse bees that remain in the hive, but at around three weeks of age they shift to foraging traits. This change in task specialization is accompanied by broad changes in gene expression, epigenetic signatures and brain structure (Toth and Robinson, 2007; Patalano et al., 2012). A comparison of sleep between young worker bees and older foraging bees revealed differences in sleep architecture, with young bees spending a greater amount of time in light sleep (Eban-Rothschild et al., 2017). The understanding of variation in gene expression between life stages may be applied to understand how these task transitions are associated with sleep. Furthermore, differences in sleep architecture were observed between different bee colonies, raising the possibility that differences in genetic variation or life history traits contribute to differential sleep regulation. A sequenced genome and the recent implementation of gene-editing or transgenesis approaches in the honeybee may allow for the isolation of sleep genes in the honeybee using similar approaches that have been used in the fruit fly (Weinstock et al., 2006; Schulte et al., 2014).
Like other insects, many species of cockroaches also show robust circadian rhythms of locomotion (Harker, 1956; Roberts, 1960; Page, 1982) mating behavior (Rymer et al., 2007) and neuronal activity (Bult and Mastebroek, 1993). Sleep as a rest-like state has been investigated in at least two species, Leucophea maderae (Tobler, 1983) and the giant cockroach, Blaberus giganteus (Tobler and Neuner-Jehle, 1992). Both species show clear nocturnal rhythms, with peak activity occurring in the first hours after transition to darkness, and prolonged periods of immobility during the lights-on phase. Forced activity by mechanical stimulation in the last hours of the light phases significantly increase behavioral quiescence during the dark period (Tobler, 1983; Tobler and Neuner-Jehle, 1992). Moreover, consistent with immobility as a rest-like state, the threshold for a mechanical stimulus to cause a behavioral response was significantly higher when individuals were immobile (Tobler and Neuner-Jehle, 1992). The large size and ease of access to the central nervous system of cockroaches makes electrophysiological analysis of neural networks easily accessible (Titlow et al., 2013). Moreover, whereas the genomes of B. giganteus or L. maderae have not been published, other cockroach models have had their genomes sequenced (Harrison et al., 2018). Thus, with the advent of genome sequencing, the cockroach may serve as a powerful model for relating genome variation with neural function, and could offer critical insights into how genetic and neural mechanisms underlie variations in sleep-like states.
The characterization of sleep in additional insect species that are studied for circadian or other sleep-related behaviors may provide insight into genetic variation regulating sleep. For example, the marine midge Clunio marinus exhibits circalunar behavior that is dependent on the circadian clock, and they are locally adapted to inhabit rocky patches on the European Atlantic coast (Neumann and Heimbach, 1985). Adult C. marinus synchronously emerge from the sea during low tides, a behavior that requires integration of tidal, lunar and solar cues (Neumann and Heimbach, 1985). QTL analysis of circadian chronotype for adult emergence from the water revealed an association with the circadian gene Calmodulin-dependent protein kinase II (CamKII), splice variants associate with circadian timing, and functional differences in CamKII variants were validated in Drosophila (Kaiser et al., 2016). CamKII has also been implicated in sleep regulation in rodents (Cui et al., 2016), raising the possibility that naturally occurring variants also affect sleep. A similar approach has been used to validate mutations that affect sleep in humans. A variant of the basic helix–loop–helix (bHLH) transcription factor Dec2 resulted in an approximate two-hour reduction in sleep need, and fruit flies carrying the short- sleeping human variant slept less than flies harboring the human wild-type variant (He et al., 2009). Therefore, existing genetic models provide a powerful system for identifying the contributions of naturally occurring genetic variation to sleep regulation.
The adaptive evolution of sleep loss in cavefish
The robust evolution of behavior in response to environmental perturbation provides the opportunity to examine how ecological environment influences sleep (Keene et al., 2015). Fish species trapped in caves across the world have evolved ‘troglomorphic’ traits, including loss of pigmentation and eye loss (Jeffery, 2009; Gross, 2012). In addition, these morphological changes are accompanied by alterations in sleep or activity patterns. Several species of cavefish show altered locomotor rhythms throughout a 24 h period, and thus provide an excellent system to investigate how altered activity rhythms or sleep correlate with different ecological settings (Cavallari et al., 2011; Duboué and Borowsky, 2012; Beale et al., 2013). The convergence on many morphological and behavioral traits, including changes in sleep and activity, in different species of cave animals reveals stereotyped evolutionary responses in the surface-to-cave transition.
Locomotor rhythms have been explored in at least three species of cavefish, the Mexican cavefish Astyanax mexicanus, the Somalian cavefish Phreatichthys andruzzii, and various species of the Hillstream Loach (Balitoridae), found in different regions of Thailand and Laos (Fig. 3). Although it is widely accepted that adaptation to cave life often results in loss of circadian rhythms, as well as altered locomotor activity and sleep, the ecological factors that drive these changes are not well understood. The Somalian cavefish and many populations of the Mexican cavefish have lost light-entrained rhythms (Cavallari et al., 2011; Beale et al., 2013). By contrast, the Hillstream Loach have maintained light-entrained rhythms, although some have diverged from a 24 h period (Duboué and Borowsky, 2012). In the Somalian cavefish, circadian rhythms are entrained by food instead of light (Cavallari et al., 2011). Interestingly, in the Hillstream Loach, all populations show dramatic reduction in sleep despite their maintenance of circadian rhythms (Duboué and Borowsky, 2012). Because both circadian and homeostatic processes contribute to sleep regulation (Borbély, 1982), this work suggests that sleep changes in a cave environment are independent of circadian mechanism, and suggest a reduced sleep need.
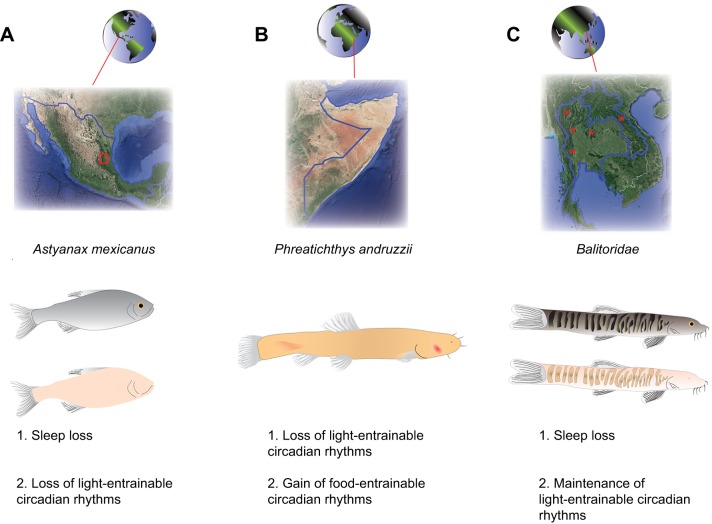
Fig. 3.
Sleep and circadian phenotypes in different cavefish species. (A) Cartoon diagram of A. mexicanus, with eyed surface populations (fish on top) found throughout Mexico and Southern Texas, and eyeless cave populations (fish on bottom) in various caves throughout North East Mexico (red box on map). Both a loss of sleep and loss of circadian rhythms have been described for these fish. (B) The Somalian cavefish, Phreatichthys andruzzii, are blind albino fish that inhabit caves in Somalia (map). A loss of light-entrainable rhythms and a gain of food-entrainable rhythms have been described for these fish. (C) Hillstream loaches, Balitoridae, are found throughout Southeast Asia (map). There are eyed, surface-dwelling species (top cartoon) and various populations of fish that have evolved in caves of Thailand and Laos (red asterisks on map). Cave Balitoridae species are either fully or partially eyeless. For all species of fish listed, average sizes are 5–10 cm in length.
The ability to identify the genetic basis of evolved differences in morphology and evolution has been particularly powerful in the Mexican blind cavefish A. mexicanus, a small freshwater fish species native to Northeast Mexico and southern parts of Texas (Mitchell et al., 1977). The species is found in two different forms: an eyed, surface-dwelling type that inhabits streams and rivers, and at least 29 populations of blind, albino forms found in nearby caves (Gross, 2012). The cavefish have all been derived from a surface-dwelling form. Thus, the model is advantageous to evolutionary biology and ecology as the ancestral form is preserved and extant. Moreover, as many traits such as eye loss and depigmentation are shared between cave and surface fish, yet have evolved in different cave populations through different genetic mechanisms, the model is ideal for investigating mechanisms of convergent evolution.
Like zebrafish, larval and juvenile forms of A. mexicanus display all of the behavioral hallmarks indicative of a sleep-like state (Duboué et al., 2011; Yoshizawa et al., 2015). Surface forms, like zebrafish, are diurnal with high levels of activity during the day and prolonged periods of quiescence at night (Duboué et al., 2011; Yoshizawa et al., 2015). Interestingly, cavefish maintain locomotor rhythms in light–dark conditions, but these are lost in constant darkness, suggesting that they have maintained light-sensing abilities despite the loss of functional eyes, yet do not have a free running clock (Beale et al., 2013). It has been demonstrated that a one-minute period of inactivity corresponds to a sleep-like state in both surface and cavefish; both cave- and surface fish were monitored during the night and, at random periods, mechanical stimulation was delivered to the side of the dish (Duboué et al., 2011). In both surface and cavefish, individuals that were inactive for 1 min or longer were less likely to respond than siblings that had been active in the 1 min period preceding mechanical stimulation. Thus, for both forms, a 1 min period of inactivity is indicative of a sleep-like state. Compared with surface dwelling forms, sleep is reduced by as much as 80% in multiple independently evolved cavefish populations. A central question is how A. mexicanus may have maintained their diurnal rhythms, while losing their functional clock, eyes and much of their sleep, and the ability to disambiguate these processes is a central strength of this model. Nonetheless, the robust differences in sleep between the two different forms of A. mexicanus provide an excellent model to investigate the genetic and neuronal mechanisms underlying sleep variation in naturally occurring animals.
Evolution of sleep loss and circadian disruption in cave animals
The rich repertoire of behaviors in adult fish permit an analysis relating changes in sleep to other biological measures, such as sensory processing. For example, the lateral line – a group of neurons positioned on the body wall of fish that serves an important role in detecting water flow and sensing the external environment – is greatly enhanced in cavefish relative to surface conspecifics (Baker and Montgomery, 1999; Patton et al., 2010). The number of neuromasts (see Glossary) along the lateral line is dramatically increased (Teyke, 1990; Yoshizawa et al., 2010). Interestingly, chemical ablation of the lateral line partially restores sleep levels in cavefish to that of surface fish, suggesting that increased sensitivity to the environment partially underlies changes in sleep (Jaggard et al., 2017). The lateral line has been demonstrated to influence other behaviors, such as the attraction to vibrations (VAB), thought to be important for prey seeking and schooling behavior, yet QTL for genomic regions that underlie VAB appear to be unrelated, suggesting that distinct sensory neuromasts modulate sleep and prey-seeking behavior (Yoshizawa et al., 2010; Kowalko et al., 2013; Yoshizawa et al., 2015). Conversely, the wake-promoting neuropeptide Hypocretin/Orexin is up-regulated in cavefish, and ablation of the lateral line reduced Hypocretin to surface fish levels. These findings suggest that evolved differences in sensory systems contribute to sleep loss by modulating hypothalamic function (Jaggard et al., 2018). The identification of sensory and central-brain mechanisms contributing to sleep loss demonstrate the integral relationship between adaptive changes in sensory processing and sleep regulation.
A unique feature of A. mexicanus is inter-fertility between populations, allowing for the generation of hybrid fish with variable sleep and morphology (Borowsky, 2008; Duboué et al., 2011). Hybrid surface and cavefish have been used for genetic mapping studies and to examine the genetic relationship between traits (Casane and Rétaux, 2016). For example, hybrid fish from two independent eyeless cave populations possess functional eyes, due to distinct genetic architecture modulating each trait (Borowsky, 2008). Therefore, these fish provide a system for testing whether shared or independent factors contribute to sleep loss in each cave population, and relating these mechanisms back to the unique cave ecology. Alternatively, genetic mapping experiments can be applied to surface–cave hybrids. These experiments have been used to identify that sleep traits independently segregate from many morphological traits (Duboué et al., 2011; Yoshizawa et al., 2015). A recent sequencing of the genome of A. mexicanus, and the application of next-generation sequencing approaches, should allow for the application of these approaches to identify genes associated with sleep loss in each cave population (McGaugh et al., 2014).
Many existing fish evolutionary models may provide novel insight into how evolutionary history and ecology affect sleep. The three-spine stickleback, Gasterosteus aculeatus, has rapidly evolved throughout the northern hemisphere, where they uniquely adapt morphology and behavior to their local environments (Bell and Foster, 1994). These fish provide a model for how evolved changes in complex behaviors, including boldness, schooling behavior and aggression, are related to evolutionary history (Ruiz-Gomez and Huntingford, 2012; Jolles et al., 2016; Sanogo and Bell, 2016). Other fish species are evolutionary models for studying ageing (Turquoise Killifish), social behavior (African cichlid), and sexual selection (swordtail) (Basolo, 1990; Alcazar et al., 2016; Harel et al., 2016). Application of approaches currently used to define sleep in zebrafish and Astyanax could define sleep in these models and allow for investigation of the evolutionary relationship between sleep and other complex behavioral and physiological traits. Furthermore, defining sleep in additional fish models would allow for the leveraging of genome mapping approaches and genetic technology currently available in these systems to study the genetic and neural basis of sleep regulation.
Evolved differences in sleep – QTL mapping and artificial selection experiments
The relatively short generation time and robust sleep phenotypes from existing genetic variation make non-mammalian models powerful systems for investigating the evolution of sleep. A number of studies have used experimental evolution or selection experiments to study the evolution of sleep loss. A model of Drosophila insomnia was generated by selecting for short-sleeping individuals through 60 generations, resulting in over 90% sleep loss (Seugnet et al., 2009). A number of biomarkers for sleep deprivation in humans, including the salivary biomarker Amylase and elevated triglyceride levels were present in insomnia-like selected flies, suggesting that selection may provide a model for studying human sleep deprivation. Further insomnia-like flies had similar phenotypes to acutely sleep-deprived flies or short-sleep mutants, including memory deficits, reduced lifespan and reduced stress tolerance, highlighting the conserved functional consequences of sleep loss (Seugnet et al., 2009). These phenotypes are also observed in wild-type laboratory fly populations with mechanically or genetically disrupted sleep, providing multiple lines of evidence that sleep loss has a negative impact on fitness (Li et al., 2009; Bushey et al., 2010; Seugnet et al., 2011).
Evolutionary approaches have also been applied to examine interactions between sleep and other fitness-associated traits. Experimental selection for starvation resistance in outbred flies results in flies with elevated triglyceride levels that survive up to 2 weeks without food, compared with only a few days for controls (Masek et al., 2014). Testing these flies for sleep revealed increased sleep co-evolved with starvation resistance, suggesting that sleep promotes survival during times of starvation, probably through reduced energy expenditure from foraging (Masek et al., 2014; Slocumb et al., 2015). The sleep and metabolic phenotypes were present in three independently selected fly lines originating from a single outbred population, allowing for an investigation of whether shared or distinct genetics underlie the independent generation of these phenotypes. In addition, genomic sequencing and GWAS of these lines have the potential to identify additional genes involved in the regulation of these complex traits.
While the leveraging of experimental evolution or artificial selection to investigate sleep traits has predominantly been studied in Drosophila, these approaches are readily applicable to other model organisms. For example, artificial selection has been used in zebrafish to study complex behaviors (Facchin et al., 2009). In addition, in C. elegans, comparative studies and QTL mapping have been performed between different natural isolate strains (McGrath et al., 2009), although not for sleep traits. If sleep differences are present between these strains, mapping or selection experiments would allow for identification of causal genes contributing to these differences (Li et al., 2006; Elvin et al., 2011). It is possible that examining the genetic contribution to natural variation in sleep will uncover molecular pathways different from those evident from mutagenesis studies, and these natural approaches may have more relevance to the differences in sleep need between humans.
Additional ecologically relevant factors that modulate sleep
While sleep research has predominantly focused on measuring sleep under standardized and stable laboratory conditions, the responses to environmental perturbations are understudied, and are likely to be under strong evolutionary selection (Tourgeron and Abram, 2017). Many environmental factors, including food availability, social interactions and temperature, potently impact sleep in diverse phyla (McNamara et al., 2009; Capellini, 2010). For example, sleep is reduced in flies reared in isolation, and exposure of male flies to females suppresses sleep, revealing robust modulation of sleep by social stimuli (Ganguly-Fitzgerald, 2006; Beckwith et al., 2017; Chen et al., 2017; Machado et al., 2017). These important modulators of sleep are likely to be missed in behavioral assays used in most animal systems that measure each animal in an independent arena, and highlight the need for investigating sleep in an ecologically relevant context.
In addition to social experience, nutrient availability is a crucial modulator of sleep, and animals weigh the cost–benefit of energy savings from sleep against the benefits of wakefulness (Schmidt, 2014). Animals ranging from flies to humans sleep more following a meal (Bernstein, 1974; Stahl et al., 1983; Murphy et al., 2016), and sleep is disrupted during starvation (Macfadyen et al., 1973; Danguir and Nicolaidis, 1979; Keene et al., 2010). Although it has long been presumed that these acute responses to nutrient availability represent a mechanism of maintaining metabolic homeostasis, this hypothesis has been difficult to test experimentally. A number of studies in fruit flies have investigated neural circuits regulating starvation-induced changes in sleep. These studies have found that both sensory perception of taste and signalling molecules that promote food consumption inhibit sleep, supporting the notion that shared pathways regulate both sleep and feeding (Linford et al., 2012; Chung et al., 2017). A genetic screen identified pathways that appear to modulate starvation-induced changes in sleep that do not affect energy stores, metabolic function or feeding behavior (Murakami et al., 2016). While the relationship between sleep and food availability has not been tested in many models, including zebrafish, it is possible that different species have unique adaptations to food availability. For example, prolonged starvation increases sleep in cavefish, similar to what is observed in birds, perhaps representing an energy-saving mechanism to account for long periods with limited food during the dry season (Jaggard et al., 2017). Broader systematic analysis of species-specific responses to changes in food availability and improved understanding of the neural basis for these changes will determine how sleep is modulated by environmental perturbations.
In an ethologically relevant context, many additional factors will have a potent impact on sleep. For example, increased sleep during early development has been documented in flies and humans, and it appears to be crucial for normal brain development (Carskadon, 2011; Kayser et al., 2014). Sleep is affected by many environmental variables, and the small model systems are likely to be amenable to investigation of how sleep is affected by diverse social processes. It is possible that the drive to identify sleep genes and standardization of approaches has obscured many key regulators of sleep that are related to the response to stress, food availability, social behavior or other factors. Investigating the relationship between ecology and these factors is necessary to understand the evolutionary features regulating sleep, and the short lifespan of many models allows for tracing sleep differences throughout development.
Future directions
The development of behavioral assays to measure sleep in fruit flies, C. elegans and zebrafish has led to the rapid discovery of genetic and neural processes regulating sleep. These findings pave the way for investigating the function of sleep, and how it is altered by an animal's ecological environment and evolutionary history. In recent years, progress using behavioral criteria to define sleep in a number of novel model organisms including the jellyfish, Aplysia and cavefish have potential to provide new insight into the biological and functional basis of sleep regulation. Many animals are uniquely suited for studying specific functions of sleep, including the use of Aplysia to study the relationship between sleep and memory formation, and the honeybee to examine interactions between sleep and social experience. We propose that by characterizing sleep in additional animal models of evolution ranging from organisms with simplified nervous systems such as the starlet sea anemone Nematostella to the three-spined stickleback, a model of microevolution, we will gain a better understanding of how ecology and life history traits regulate sleep. The emergence of sleep studies in organisms with simplified nervous systems or defined evolutionary history, combined with the development of gene-editing technology, provide novel avenues to investigate the evolution of function of sleep. Together, these integrative approaches in diverse models will help define the relevance of genetic and neural principles regulating sleep to the broader animal kingdom.
Acknowledgements
The authors would like to thank the reviewers for insightful comments that improved this manuscript. In addition, Bethany Stahl and Elizabeth Brown (FAU) provided support generating figures and critical manuscript feedback.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The authors would like to acknowledge National Institutes of Health R21 NS105071 to E.R.D., R01 NS08152 to A.C.K., and National Science Foundation award 1656575 to A.C.K.
References
- Ahrens M. B. and Engert F. (2015). Large-scale imaging in small brains. Curr. Opin. Neurobiol. 32, 78-86. 10.1016/j.conb.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar R. M., Becker L., Hilliard A. T., Kent K. R. and Fernald R. D. (2016). Two types of dominant male cichlid fish: behavioral and hormonal characteristics. Biol. Open 5, 1061-1071. 10.1242/bio.017640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R. and Siegel J. M. (2008). Unearthing the phylogenetic roots of sleep. Curr. Biol. 18, R670-R679. 10.1016/j.cub.2008.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allebrandt K. V., Amin N., Müller-Myhsok B., Esko T., Teder-Laving M., Azevedo R. V. D. M., Hayward C., van Mill J., Vogelzangs N., Green E. W. et al. (2013). A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol. Psychiatry 18, 122-132. 10.1038/mp.2011.142 [DOI] [PubMed] [Google Scholar]
- Anderson F. (1968). Sleep in moths and its dependence on the frequency of stimulation in Anagasta kuehniella. Opusc Ent 33, 15-24. [Google Scholar]
- Appelbaum L., Wang G. X., Maro G. S., Mori R., Tovin A., Marin W., Yokogawa T., Kawakami K., Smith S. J., Gothilf Y. et al. (2009). Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc. Natl. Acad. Sci. USA 106, 21942-21947. 10.1073/pnas.906637106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arble D. M., Bass J., Behn C. D., Butler M. P., Challet E. and Czeisler C. (2015). Impact of sleep and circadian disruption on energy balance and diabetes: a summary of workshop discussions. Sleep 38, 1849-1860. 10.5665/sleep.5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Sitaraman D., Ichinose T., Kaun K. R., Vogt K., Belliart-Guérin G., Plaçais P.-Y., Robie A. A., Yamagata N., Schnaitmann C. et al. (2014a). Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife 3, e04580 10.7554/eLife.04580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Hattori D., Yu Y., Johnston R. M., Iyer N. A., Ngo T.-T. B., Dionne H., Abbott L. F., Axel R., Tanimoto H. et al. (2014b). The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3, e04577 10.7554/eLife.04580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. F. and Montgomery J. C. (1999). The sensory basis of rheotaxis in the blind Mexican cave fish, Astyanax fasciatus. J. Comp. Physiol. A Neuroethol. Sensory Neural Behav. Physiol. 184, 519-527. 10.1007/s003590050351 [DOI] [Google Scholar]
- Banks G., Heise I., Starbuck B., Osborne T., Wisby L., Potter P., Jackson I. J., Foster R. G., Peirson S. N. and Nolan P. M. (2015). Genetic background influences age-related decline in visual and nonvisual retinal responses, circadian rhythms, and sleep. Neurobiol. Aging 36, 380-393. 10.1016/j.neurobiolaging.2014.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basolo A. L. (1990). Female preference predates the evolution of the sword in swordtail fish. Science 250, 808-810. 10.1126/science.250.4982.808 [DOI] [PubMed] [Google Scholar]
- Beale A., Guibal C., Tamai T. K., Klotz L., Cowen S., Peyric E., Reynoso V. H., Yamamoto Y. and Whitmore D. (2013). Circadian rhythms in Mexican blind cavefish Astyanax mexicanus in the lab and in the field. Nat. Commun. 4, 2769 10.1038/ncomms3769 [DOI] [PubMed] [Google Scholar]
- Beckwith E. J., Geissmann Q., French A. S. and Gilestro G. F. (2017). Regulation of sleep homeostasis by sexual arousal. Elife 6, e27445 10.7554/eLife.27445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M. A. and Foster S. A. (1994). The Evolutionary Biology of the Threespine Stickleback. Oxford: Oxford University Press. [Google Scholar]
- Berger H. and Gloor P. (1969). Hans Berger on the Electroencephalogram of Man: The Fourteen Original Reports on the Human Electroencephalogram (ed. Gloor P.), 350pp Amsterdam: Elsevier. [Google Scholar]
- Bernstein I. L. (1974). Post-prandial EEG synchronization in normal and hypothalamically lesioned rats. Physiol. Behav. 12, 535-545. 10.1016/0031-9384(74)90201-7 [DOI] [PubMed] [Google Scholar]
- Borbély A. A. (1982). A two process model of sleep regulation. Hum. Neurobiol. 1, 195-204. [PubMed] [Google Scholar]
- Borowsky R. (2008). Restoring sight in blind cavefish. Curr. Biol. 18, R23-R24. 10.1016/j.cub.2007.11.023 [DOI] [PubMed] [Google Scholar]
- Bradley E. A. and Young J. Z. (1975). Comparison of visual and tactile learning in octopus after lesions to one of the two memory systems. J. Neurosci. Res. 1, 185-205. 10.1002/jnr.490010302 [DOI] [PubMed] [Google Scholar]
- Braun M. H., Lukowiak K., Karnik V. and Lukowiak K. (2012). Differences in neuronal activity explain differences in memory forming abilities of different populations of Lymnaea stagnalis. Neurobiol. Learn. Mem. 97, 173-182. 10.1016/j.nlm.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Brown E. R., Piscopo S., De Stefano R. and Giuditta A. (2006). Brain and behavioural evidence for rest-activity cycles in Octopus vulgaris. Behav. Brain Res. 172, 355-359. 10.1016/j.bbr.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Brown E. B., Torres J., Bennick R. A., Rozzo V., Kerbs A., DiAngelo J. R. and Keene A. C. (2018). Variation in sleep and metabolic function is associated with latitude and average temperature in Drosophila melanogaster. Ecol. Evol. 8, 4084-4097. 10.1002/ece3.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult R. and Mastebroek H. A. K. (1993). Circadian control of visual information processing in the optic lobe of the giant cockroach Blaberus giganteus. J. Biol. Rhythms 8, 311-323. 10.1177/074873049300800404 [DOI] [PubMed] [Google Scholar]
- Bushey D., Hughes K. A., Tononi G. and Cirelli C. (2010). Sleep, aging, and lifespan in Drosophila. BMC Neurosci. 11, 56 10.1186/1471-2202-11-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S. S. and Tobler I. (1984). Animal sleep: a review of sleep duration across phylogeny. Neurosci. Biobehav. Rev. 8, 269-300. 10.1016/0149-7634(84)90054-X [DOI] [PubMed] [Google Scholar]
- Capellini I. (2010). Ecological constraints on mammalian sleep architecture. In Evolution of Sleep: Phylogenetic and Functional Perspectives (ed. McNamara P., Barton R. A. and Nunn C. L.), pp. 12-33. Cambridge: Cambridge University Press. [Google Scholar]
- Capellini I., Barton R. A., McNamara P., Preston B. T. and Nunn C. L. (2008). Phylogenetic analysis of the ecology and evolution of mammalian sleep. Evolution 62, 1764-1776. 10.1111/j.1558-5646.2008.00392.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuccio F. P., D'Elia L., Strazzullo P. and Miller M. A. (2010). Sleep duration and all-cause mortality: a systematic review and meta-analysis of prospective studies. Sleep 33, 585-592. 10.1093/sleep/33.5.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carskadon M. A. (2011). Sleep's effects on cognition and learning in adolescence. Prog. Brain Res. 190, 137-143. 10.1016/B978-0-444-53817-8.00008-6 [DOI] [PubMed] [Google Scholar]
- Casane D. and Rétaux S. (2016). Evolutionary genetics of the cavefish Astyanax mexicanus. Adv. Genet. 95, 117-159. 10.1016/bs.adgen.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Cavallari N., Frigato E., Vallone D., Fröhlich N., Lopez-Olmeda J. F., Foà A., Berti R., Sánchez-Vázquez F. J., Bertolucci C. and Foulkes N. S. (2011). A blind circadian clock in cavefish reveals that opsins mediate peripheral clock photoreception. PLoS Biol. 9 10.1371/journal.pbio.1001142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Chiu C. N., Mosser E. A., Kahn S., Spence R. and Prober D. A. (2016). QRFP and its receptors regulate locomotor activity and sleep in zebrafish. J. Neurosci. 36, 1823-1840. 10.1523/JNEUROSCI.2579-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Sitaraman D., Chen N., Jin X., Han C., Chen J., Sun M., Baker B. S., Nitabach M. N. and Pan Y. (2017). Genetic and neuronal mechanisms governing the sex-specific interaction between sleep and sexual behaviors in Drosophila. Nat. Commun. 8, 154 10.1038/s41467-017-00087-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiao C.-C., Chubb C. and Hanlon R. T. (2015). A review of visual perception mechanisms that regulate rapid adaptive camouflage in cuttlefish. J. Comp. Physiol. A Neuroethol. Sensory Neural Behav. Physiol. 201, 933-945. 10.1007/s00359-015-0988-5 [DOI] [PubMed] [Google Scholar]
- Chiu C. N. and Prober D. A. (2013). Regulation of zebrafish sleep and arousal states: current and prospective approaches. Front. Neural Circuits 7, 58 10.3389/fncir.2013.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. Y. and Sternberg P. W. (2014). Multilevel modulation of a sensory motor circuit during C. elegans sleep and arousal. Cell 156, 249-260. 10.1016/j.cell.2013.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B. Y., Ro J., Hutter S. A., Miller K. M., Guduguntla L. S., Kondo S. and Pletcher S. D. (2017). Drosophila neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep. 19, 2441-2450. 10.1016/j.celrep.2017.05.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churgin M. A., Jung S.-K., Yu C.-C., Chen X., Raizen D. M. and Fang-Yen C. (2017). Longitudinal imaging of caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging. eLife 6, e26652 10.7554/eLife.26652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Bushey D., Hill S., Huber R., Kreber R., Ganetzky B. and Tononi G. (2005). Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087-1092. 10.1038/nature03486 [DOI] [PubMed] [Google Scholar]
- Cui S.-Y., Li S.-J., Cui X.-Y., Zhang X.-Q., Yu B., Sheng Z.-F., Huang Y.-L., Cao Q., Xu Y.-P., Lin Z.-G. et al. (2016). Phosphorylation of CaMKII in the rat dorsal raphe nucleus plays an important role in sleep-wake regulation. J. Neurochem. 136, 609-619. 10.1111/jnc.13431 [DOI] [PubMed] [Google Scholar]
- Danguir J. and Nicolaidis S. (1979). Dependence of sleep on nutrient's availability. Physiol. Behav. 22, 735-740. 10.1016/0031-9384(79)90240-3 [DOI] [PubMed] [Google Scholar]
- David J. R. and Capy P. (1988). Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 4, 106-111. 10.1016/0168-9525(88)90098-4 [DOI] [PubMed] [Google Scholar]
- Dement W. and Kleitman N. (1957). Cyclic variations in EEG during sleep and their relation to eye movements, body motility, and dreaming. Electroencephalogr. Clin. Neurophysiol. 9, 673-690. 10.1016/0013-4694(57)90088-3 [DOI] [PubMed] [Google Scholar]
- Donlea J. M., Thimgan M. S., Suzuki Y., Gottschalk L. and Shaw P. J. (2011). Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571-1576. 10.1126/science.1202249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboué E. R. and Borowsky R. L. (2012). Altered rest-activity patterns evolve via circadian independent mechanisms in cave adapted balitorid loaches. PLoS ONE 7, e30868 10.1371/journal.pone.0030868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboué E. R., Keene A. C. and Borowsky R. L. (2011). Evolutionary convergence on sleep loss in cavefish populations. Curr. Biol. 21, 671-676. 10.1016/j.cub.2011.03.020 [DOI] [PubMed] [Google Scholar]
- Eban-Rothschild A., Giardino W. J. and de Lecea L. (2017). To sleep or not to sleep: neuronal and ecological insights. Curr. Opin. Neurobiol. 44, 132-138. 10.1016/j.conb.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvin M., Snoek L. B., Frejno M., Klemstein U., Kammenga J. E. and Poulin G. B. (2011). A fitness assay for comparing RNAi effects across multiple C. elegans genotypes. BMC Genomics 12, 510 10.1186/1471-2164-12-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchin L., Argenton F. and Bisazza A. (2009). Lines of Danio rerio selected for opposite behavioural lateralization show differences in anatomical left-right asymmetries. Behav. Brain Res. 197, 157-165. 10.1016/j.bbr.2008.08.033 [DOI] [PubMed] [Google Scholar]
- Feldman J. F. and Hoyle M. N. (1976). Complementation analysis of linked circadian clock mutants of Neurospora crassa. Genetics 82, 9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanigan W. F., Wilcox R. H. and Rechtschaffen A. (1973). The EEG and behavioral continuum of the crocodilian, Caiman sclerops. Electroencephalogr. Clin. Neurophysiol. 34, 521-538. 10.1016/0013-4694(73)90069-2 [DOI] [PubMed] [Google Scholar]
- Frank M. G., Waldrop R. H., Dumoulin M., Aton S. and Boal J. G. (2012). A preliminary analysis of sleep-like states in the cuttlefish Sepia officinalis. PLoS ONE 7, e38125 10.1371/journal.pone.0038125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B. E., Ruedi E. A., McCoy L. J., Moore J. M., Wolfner M. F. and Mackay T. F. C. (2015). Heritable variation in courtship patterns in Drosophila melanogaster. G3 5, 531-539. 10.1534/g3.114.014811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly-Fitzgerald I. (2006). Waking experience affects sleep need in Drosophila. Science 313, 1775-1781. 10.1126/science.1130408 [DOI] [PubMed] [Google Scholar]
- Garbe D. S., Bollinger W. L., Vigderman A., Masek P., Gertowski J., Sehgal A. and Keene A. C. (2015). Context-specific comparison of sleep acquisition systems in Drosophila. Biol. Open 4 10.1242/bio.013011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner G. F. and Feldman J. F. (1980). The frq locus in Neurospora crassa: a key element in circadian clock organization. Genetics 96, 877-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlapow M. E., Huang W., Yarboro M. T., Peterson K. R. and Mackay T. F. C. (2015). Quantitative genetics of food intake in Drosophila melanogaster. PLoS ONE 10, e0138129 10.1371/journal.pone.0138129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomotto J. and Ségalat L. (2010). High-throughput screening and small animal models, where are we? Br. J. Pharmacol. 160, 204-216. 10.1111/j.1476-5381.2010.00725.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith L. C. (2013). Neuromodulatory control of sleep in Drosophila melanogaster: integration of competing and complementary behaviors. Curr. Opin. Neurobiol. 23, 819-823. 10.1016/j.conb.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B. (2012). The complex origin of Astyanax cavefish. BMC Evol. Biol. 12, 105 10.1186/1471-2148-12-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutnick T., Byrne R. A., Hochner B. and Kuba M. (2011). Octopus vulgaris uses visual information to determine the location of its arm. Curr. Biol. 21, 460-462. 10.1016/j.cub.2011.01.052 [DOI] [PubMed] [Google Scholar]
- Hall J. C. (2003). A neurogeneticist's manifesto. J. Neurogenetics 17, 1-90. 10.1080/01677060390230349 [DOI] [PubMed] [Google Scholar]
- Hanlon R., Chiao C.-C., Mathger L., Barbosa A., Buresch K. and Chubb C. (2009). Cephalopod dynamic camouflage: bridging the continuum between background matching and disruptive coloration. Philos. Trans. R. Soc. B Biol. Sci. 364, 429-437. 10.1098/rstb.2008.0270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison S. T., Carbone M. A., Ayroles J. F., Stone E. A., Lyman R. F. and Mackay T. F. C. (2009). Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat. Genet. 41, 371-375. 10.1038/ng.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison S. T., McCoy L. J. and Mackay T. F. C. (2013). Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics 14, 281 10.1186/1471-2164-14-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel I., Valenzano D. R. and Brunet A. (2016). Efficient genome engineering approaches for the short-lived African turquoise killifish. Nat. Protoc. 11, 2010-2028. 10.1038/nprot.2016.103 [DOI] [PubMed] [Google Scholar]
- Harker J. (1956). Factors controlling the diurnal rhythm of activity of Periplaneta americana L. J. Exp. Biol. 33, 224-234. [Google Scholar]
- Harris D. T., Kallman B. R., Mullaney B. C. and Scott K. (2015). Representations of taste modality in the Drosophila brain. Neuron 86, 1449-1460. 10.1016/j.neuron.2015.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. C., Jongepier E., Robertson H. M., Arning N., Bitard-Feildel T., Chao H., Childers C. P., Dinh H., Doddapaneni H., Dugan S. et al. (2018). Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2, 557-566. 10.1038/s41559-017-0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E. L. (1973). The Functions of Sleep. New Haven, CT: Yale University Press. [Google Scholar]
- He Y., Jones C. R., Fujiki N., Xu Y., Guo B., Holder J. L., Rossner M. J., Nishino S. and Fu Y.-H. (2009). The transcriptional repressor DEC2 regulates sleep length in mammals. Science 325, 866-870. 10.1126/science.1174443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S. B., Marin J., Suleski M., Paymer M. and Kumar S. (2015). Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835-845. 10.1093/molbev/msv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks J. C., Finn S. M., Panckeri K. A., Chavkin J., Williams J. A., Sehgal A. and Pack A. I. (2000a). Rest in Drosophila is a sleep-like state. Neuron 25, 129-138. 10.1016/S0896-6273(00)80877-6 [DOI] [PubMed] [Google Scholar]
- Hendricks J. C., Sehgal A. and Pack A. I. (2000b). The need for a simple animal model to understand sleep. Prog. Neurobiol. 61, 339-351. 10.1016/S0301-0082(99)00048-9 [DOI] [PubMed] [Google Scholar]
- Hendricks W. D., Byrum C. A. and Meyer-Bernstein E. L. (2012). Characterization of circadian behavior in the starlet sea anemone, Nematostella vectensis. PLoS ONE 7, e46843 10.1371/journal.pone.0046843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland B. S., Dawson M. N., Crow G. L. and Hofmann D. K. (2004). Global phylogeography of Cassiopea (Scyphozoa: Rhizostomeae): molecular evidence for cryptic species and multiple invasions of the Hawaiian Islands. Mar. Biol. 145, 1119-1128. 10.1007/s00227-004-1409-4 [DOI] [Google Scholar]
- Hozumi A., Horie T. and Sasakura Y. (2015). Neuronal map reveals the highly regionalized pattern of the juvenile central nervous system of the ascidian Ciona intestinalis. Dev. Dyn. 244, 1375-1393. 10.1002/dvdy.24317 [DOI] [PubMed] [Google Scholar]
- Hu Y., Shmygelska A., Tran D., Eriksson N., Tung J. Y. and Hinds D. A. (2016). GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat. Commun. 7, 10448 10.1038/ncomms10448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Carbone M. A., Magwire M. M., Peiffer J. A., Lyman R. F., Stone E. A., Anholt R. R. H. and Mackay T. F. C. (2015). Genetic basis of transcriptome diversity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 112, E6010-E6019. 10.1073/pnas.1519159112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggard J., Robinson B., Stahl B., Oh I., Masek P., Yoshizawa M. and Keene A. (2017). The lateral line confers evolutionarily derived sleep loss in the Mexican cavefish. J. Exp. Biol. (in press) 220, 284-293. 10.1242/jeb.145128 [DOI] [PubMed] [Google Scholar]
- Jaggard J. B., Stahl B. A., Lloyd E., Prober D. A., Duboue E. R. and Keene A. C. (2018). Hypocretin underlies the evolution of sleep loss in the Mexican cavefish. eLife 7, e32637 10.7554/eLife.32637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen C., Wild C., Rasheed M., El-Zibdah M. and Richter C. (2010). Enhanced pore-water nutrient fluxes by the upside-down jellyfish Cassiopea sp. in a Red Sea coral reef. Mar. Ecol. Prog. Ser. 411, 117-125. 10.3354/meps08623 [DOI] [Google Scholar]
- Jeffery W. R. (2009). Regressive evolution in Astyanax cavefish. Annu. Rev. Genet. 43, 25-47. 10.1146/annurev-genet-102108-134216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A., Rubin G. M., Ngo T.-T. B., Shepherd D., Murphy C., Dionne H., Pfeiffer B. D., Cavallaro A., Hall D., Jeter J. et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991-1001. 10.1016/j.celrep.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner W. J. (2016). Unraveling the evolutionary determinants of sleep. Curr. Biol. 26, R1073-R1087. 10.1016/j.cub.2016.08.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner W. J., Crocker A., White B. H. and Sehgal A. (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757-760. 10.1038/nature04811 [DOI] [PubMed] [Google Scholar]
- Jolles J. W., Aaron Taylor B. and Manica A. (2016). Recent social conditions affect boldness repeatability in individual sticklebacks. Anim. Behav. 112, 139-145. 10.1016/j.anbehav.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. E., Tyrrell J., Wood A. R., Beaumont R. N., Ruth K. S., Tuke M. A., Yaghootkar H., Hu Y., Teder-Laving M., Hayward C. et al. (2016). Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 12, e1006125 10.1371/journal.pgen.1006125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozet-Alves C., Bertin M. and Clayton N. S. (2013). Evidence of episodic-like memory in cuttlefish. Curr. Biol. 23 10.1016/j.cub.2013.10.021 [DOI] [PubMed] [Google Scholar]
- Kaiser W. and Steiner-Kaiser J. (1983). Neuronal correlates of sleep, wakefulness and arousal in a diurnal insect. Nature 301, 707-709. 10.1038/301707a0 [DOI] [PubMed] [Google Scholar]
- Kaiser T. S., Poehn B., Szkiba D., Preussner M., Sedlazeck F. J., Zrim A., Neumann T., Nguyen L.-T., Betancourt A. J., Hummel T. et al. (2016). The genomic basis of circadian and circalunar timing adaptations in a midge. Nature 540, 69-73. 10.1038/nature20151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuki T. and Greenspan R. J. (2013). Jellyfish nervous systems. Curr. Biol. 23, R592-R594. 10.1016/j.cub.2013.03.057 [DOI] [PubMed] [Google Scholar]
- Kayser M. S. and Biron D. (2016). Sleep and development in genetically tractable model organisms. Genetics 203, 21-33. 10.1534/genetics.116.189589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M. S., Yue Z. and Sehgal A. (2014). A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science 344, 269-274. 10.1126/science.1250553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan S. and Hirshkowitz M. (2010). Monitoring and staging human sleep. In Principles and Practice of Sleep Medicine, 5th edn (ed. M. Kryger, T. Roth and W. Dement), pp. 1602-1609. Amsterdam: Elsevier. [Google Scholar]
- Keene A. C., Duboué E. R., McDonald D. M., Dus M., Suh G. S. B., Waddell S. and Blau J. (2010). Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr. Biol. 20, 1209-1215. 10.1016/j.cub.2010.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene A., Yoshizawa M. and McGaugh S. (2015). Biology and Evolution of the Mexican Cavefish, 1st edn New York: Academic Press. [Google Scholar]
- Koh K., Joiner W. J., Wu M. N., Yue Z., Smith C. J. and Sehgal A. (2008). Identification of SLEEPLESS, a sleep-promoting factor. Science 321, 372-376. 10.1126/science.1155942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalko J. E., Rohner N., Rompani S. B., Peterson B. K., Linden T. A., Yoshizawa M., Kay E. H., Weber J., Hoekstra H. E., Jeffery W. R. et al. (2013). Loss of schooling behavior in cavefish through sight-dependent and sight-independent mechanisms. Curr. Biol. 23, 1874-1883. 10.1016/j.cub.2013.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan H. C., Gandour C. E., Ramos J. L., Wrinkle M. C., Sanchez-Pacheco J. J. and Lyons L. C. (2016a). Acute sleep deprivation blocks short- and long-term operant memory in Aplysia. Sleep 39, 2161-2171. 10.5665/sleep.6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan H. C., Noakes E. J. and Lyons L. C. (2016b). Chronic sleep deprivation differentially affects short and long-term operant memory in Aplysia. Neurobiol. Learn. Mem. 134, 349-359. 10.1016/j.nlm.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronholm E., Härmä M., Hublin C., Aro A. R. and Partonen T. (2006). Self-reported sleep duration in Finnish general population. J. Sleep Res. 15, 276-290. 10.1111/j.1365-2869.2006.00543.x [DOI] [PubMed] [Google Scholar]
- Lane J. M., Vlasac I., Anderson S. G., Kyle S. D., Dixon W. G., Bechtold D. A., Gill S., Little M. A., Luik A., Loudon A. et al. (2016). Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat. Commun. 7, 10889 10.1038/ncomms10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesku J. A., Roth T. C. II, Amlaner C. J. and Lima S. L. (2006). A phylogenetic analysis of sleep architecture in mammals: the integration of anatomy, physiology, and ecology. Am. Nat. 168, 441-453. 10.1086/506973 [DOI] [PubMed] [Google Scholar]
- Letunic I. and Bork P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242-W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitas-Djerbi T. and Appelbaum L. (2017). Modeling sleep and neuropsychiatric disorders in zebrafish. Curr. Opin. Neurobiol. 44, 89-93. 10.1016/j.conb.2017.02.017 [DOI] [PubMed] [Google Scholar]
- Levy R., Levitan D. and Susswein A. J. (2016). New learning while consolidating memory during sleep is actively blocked by a protein synthesis dependent process. eLife 5, e17769 10.7554/eLife.17769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Álvarez O. A., Gutteling E. W., Tijsterman M., Fu J., Riksen J. A. G., Hazendonk E., Prins P., Plasterk R. H. A., Jansen R. C. et al. (2006). Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2, 2155-2161. 10.1371/journal.pgen.0020222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yu F. and Guo A. (2009). Sleep deprivation specifically impairs short-term olfactory memory in Drosophila. Sleep 32, 1417-1424. 10.1093/sleep/32.11.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linford N. J., Chan T. P. and Pletcher S. D. (2012). Re-patterning sleep architecture in Drosophila through gustatory perception and nutritional quality. PLoS Genet. 8, e1002668 10.1371/journal.pgen.1002668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFadyen U. M., Oswald I. and Lewis S. A. (1973). Starvation and human slow-wave sleep. J. Appl. Physiol. 35, 391-394. 10.1152/jappl.1973.35.3.391 [DOI] [PubMed] [Google Scholar]
- Machado D. R., Afonso D. J. S., Kenny A. R., Öztürk-Çolak A., Moscato E. H., Mainwaring B., Kayser M. and Koh K. (2017). Identification of octopaminergic neurons that modulate sleep suppression by male sex drive. Elife 6, e23130 10.7554/eLife.23130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., Zhu D., Casillas S., Han Y., Magwire M. M., Cridland J. M. et al. (2012). The Drosophila melanogaster genetic reference panel. Nature 482, 173-178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P., Reynolds L. A., Bollinger W. L., Moody C., Mehta A., Murakami K., Yoshizawa M., Gibbs A. G. and Keene A. C. (2014). Altered regulation of sleep and feeding contribute to starvation resistance in Drosophila. J. Exp. Biol. 217, 3122-3132. 10.1242/jeb.103309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzotti D. R., Guindalini C., Moraes W. A. dos S., Andersen M. L., Cendoroglo M. S., Ramos L. R. and Tufik S. (2014). Human longevity is associated with regular sleep patterns, maintenance of slow wave sleep, and favorable lipid profile. Front. Aging Neurosci. 6, 134 10.3389/fnagi.2014.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh S., Gross J. B., Aken B., Blin M., Borowsky R., Chalopin D., Hinaux H., Jeffery W., Keene A., Ma L. et al. (2014). The cavefish genome reveals candidate genes for eye loss. Nat. Commun. 5, 5307 10.1038/ncomms6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P. T., Rockman M. V., Zimmer M., Jang H., Macosko E. Z., Kruglyak L. and Bargmann C. I. (2009). Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron 61, 692-699. 10.1016/j.neuron.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P., Barton R. and Nunn C. (2009). Evolution of Sleep: Phylogenetic and Functional Perspectives (ed. McNamara P., Barton R. A. and Nunn C. L.). Cambridge: Cambridge University Press. [Google Scholar]
- Minamoto T., Hanai S., Kadota K., Oishi K., Matsumae H., Fujie M., Azumi K., Satoh N., Satake M. and Ishida N. (2010). Circadian clock in Ciona intestinalis revealed by microarray analysis and oxygen consumption. J. Biochem. 147, 175-184. 10.1093/jb/mvp160 [DOI] [PubMed] [Google Scholar]
- Mirmiran M., Scholtens J., van de Poll N. E., Uylings H. B. M., van der Gugten J. and Boer G. J. (1983). Effects of experimental suppression of active (REM) sleep during early development upon adult brain and behavior in the rat. Brain Res. 283, 277-286. 10.1016/0165-3806(83)90184-0 [DOI] [PubMed] [Google Scholar]
- Mitchell R. W., Russell W. H. and Elliott W. R. (1977). Mexican Eyeless Characin Fishes, Genus Astyanax: Environment, Distribution, and Evolution (ed. Mackey C. and Barnett G. E.). Texas: Texas Tech Press. [Google Scholar]
- Moroz L. L., Edwards J. R., Puthanveettil S. V., Kohn A. B., Ha T., Heyland A., Knudsen B., Sahni A., Yu F., Liu L. et al. (2006). Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell 127, 1453-1467. 10.1016/j.cell.2006.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova T. V., Huang W., Pray V. A., Whitham T., Anholt R. R. H. and Mackay T. F. C. (2015). Polymorphisms in early neurodevelopmental genes affect natural variation in alcohol sensitivity in adult Drosophila. BMC Genomics 16, 865 10.1186/s12864-015-2064-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E. G., Wang X., Grebenjuk V. A., Korzhev M., Wiens M., Schlossmacher U. and Schröder H. C. (2012). Nocturnin in the demosponge Suberites domuncula: a potential circadian clock protein controlling glycogenin synthesis in sponges. Biochem. J. 448, 233-242. 10.1042/BJ20120357 [DOI] [PubMed] [Google Scholar]
- Murakami K., Yurgel M. E., Stahl B. A., Masek P., Mehta A., Heidker R., Bollinger W., Gingras R. M., Kim Y.-J., Ja W. W. et al. (2016). Translin is required for metabolic regulation of sleep. Curr. Biol. 26, 972-980. 10.1016/j.cub.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. R., Deshpande S. A., Yurgel M. E., Quinn J. P., Weissbach J. L., Keene A. C., Dawson-Scully K., Huber R., Tomchik S. M. and Ja W. W. (2016). Postprandial sleep mechanics in Drosophila. eLife 5, e19334 10.7554/eLife.19334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Ohkura M., Abe G., Nakai J. and Kawakami K. (2013). Real-time visualization of neuronal activity during perception. Curr. Biol. 23, 307-311. 10.1016/j.cub.2012.12.040 [DOI] [PubMed] [Google Scholar]
- Nagy S., Tramm N., Sanders J., Iwanir S., Shirley I. A., Levine E. and Biron D. (2014). Homeostasis in C. elegans sleep is characterized by two behaviorally and genetically distinct mechanisms. eLife 3, e04380 10.7554/eLife.04380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nall A. H. and Sehgal A. (2013). Small-molecule screen in adult Drosophila identifies VMAT as a regulator of sleep. J. Neurosci. 33, 8534-8540. 10.1523/JNEUROSCI.0253-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath R. D., Bedbrook C. N., Abrams M. J., Basinger T., Bois J. S., Prober D. A., Sternberg P. W., Gradinaru V. and Goentoro L. (2017). The jellyfish Cassiopea exhibits a sleep-like state. Curr. Biol. 27, 2984-2990.e3. 10.1016/j.cub.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann D. and Heimbach F. (1985). Circadian range of entrainment in the semilunar eclosion rhythm of the marine insect Clunio marinus. J. Insect Physiol. 31, 549-557. 10.1016/0022-1910(85)90111-8 [DOI] [Google Scholar]
- Nguyen J. P., Shipley F. B., Linder A. N., Plummer G. S., Liu M., Setru S. U., Shaevitz J. W. and Leifer A. M. (2015). Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 113, E1074-E1081. 10.1073/pnas.1507110112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz D. A., van Swinderen B., Tononi G. and Greenspan R. J. (2002). Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr. Biol. 12, 1934-1940. 10.1016/S0960-9822(02)01300-3 [DOI] [PubMed] [Google Scholar]
- Ookawa T. and Gotoh J. (1965). Electroencephalogram of the chicken recorded from the skull under various conditions. J. Comp. Neurol. 124, 1-14. 10.1002/cne.901240102 [DOI] [PubMed] [Google Scholar]
- Oren M., Tarrant A. M., Alon S., Simon-Blecher N., Elbaz I., Appelbaum L. and Levy O. (2015). Profiling molecular and behavioral circadian rhythms in the non-symbiotic sea anemone Nematostella vectensis. Sci. Rep. 5, 11418 10.1038/srep11418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page T. L. (1982). Transplantation of the cockroach circadian pacemaker. Science 216, 73-75. 10.1126/science.216.4541.73 [DOI] [PubMed] [Google Scholar]
- Parisky K. M., Agosto J., Pulver S. R., Shang Y., Kuklin E., Hodge J. J. L., Kang K., Liu X., Garrity P. A., Rosbash M. et al. (2008). PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60, 672-682. 10.1016/j.neuron.2008.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M. J., Lester K. J., Barclay N. L., Nolan P. M., Eley T. C. and Gregory A. M. (2013). Replication of Genome-Wide association studies (GWAS) loci for sleep in the British G1219 cohort. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 162, 431-438. 10.1002/ajmg.b.32106 [DOI] [PubMed] [Google Scholar]
- Patalano S., Hore T. A., Reik W. and Sumner S. (2012). Shifting behaviour: epigenetic reprogramming in eusocial insects. Curr. Opin. Cell Biol. 24, 367-373. 10.1016/j.ceb.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Patton P., Windsor S. and Coombs S. (2010). Active wall following by Mexican blind cavefish (Astyanax mexicanus). J. Comp. Physiol. A Neuroethol. Sensory, Neural, Behav. Physiol. 196, 853-867. 10.1007/s00359-010-0567-8 [DOI] [PubMed] [Google Scholar]
- Peres R., Reitzel A. M., Passamaneck Y., Afeche S., Cipolla-Neto J., Marques A. and Martindale M. Q. (2014). Developmental and light-entrained expression of melatonin and its relationship to the circadian clock in the sea anemone Nematostella vectensis. Evodevo 5, 26 10.1186/2041-9139-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman J. L., McGill J. J., Keegan K. P. and Allada R. (2006). A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753-756. 10.1038/nature04739 [DOI] [PubMed] [Google Scholar]
- Prober D. A., Rihel J., Onah A. A., Sung R.-J. and Schier A. F. (2006). Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 26, 13400-13410. 10.1523/JNEUROSCI.4332-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen D. M., Zimmerman J. E., Maycock M. H., Ta U. D., You Y., Sundaram M. V. and Pack A. I. (2008). Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451, 569-572. 10.1038/nature06535 [DOI] [PubMed] [Google Scholar]
- Ramón F., Hernández-Falcón J., Nguyen B. and Bullock T. H. (2004). Slow wave sleep in crayfish. Proc. Natl. Acad. Sci. USA 101, 11857-11861. 10.1073/pnas.0402015101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt J. A., Kolaczkowski B., Jones C. D., Begun D. J. and Kern A. D. (2014). Parallel geographic variation in Drosophila melanogaster. Genetics 197, 361-373. 10.1534/genetics.114.161463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfer E. and Technau U. (2017). Meganuclease-assisted generation of stable transgenics in the sea anemone Nematostella vectensis. Nat. Protoc. 12, 1844-1854. 10.1038/nprot.2017.075 [DOI] [PubMed] [Google Scholar]
- Rihel J. and Schier A. F. (2013). Sites of action of sleep and wake drugs: insights from model organisms. Curr. Opin. Neurobiol. 23, 831-840. 10.1016/j.conb.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihel J., Prober D. A., Arvanites A., Lam K., Zimmerman S., Jang S., Haggarty S. J., Kokel D., Rubin L. L., Peterson R. T. et al. (2010). Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327, 348-351. 10.1126/science.1183090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. K. de F. (1960). Circadian activity rhythms in cockroaches. I. The free-running rhythm in steady-state. J. Cell. Physiol. 55, 99-110. 10.1002/jcp.1030550112 [DOI] [PubMed] [Google Scholar]
- Roffwarg H. P., Muzio J. N. and Dement W. C. (1966). Ontogenetic development of the human sleep-dream cycle. Science 152, 604-619. 10.1126/science.152.3722.604 [DOI] [PubMed] [Google Scholar]
- Rogulja D. and Young M. W. (2012). Control of sleep by cyclin A and its regulator. Science 335, 1617-1621. 10.1126/science.1212476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gomez M. L. and Huntingford F. A. (2012). Boldness and aggressiveness in early and late hatched three-spined sticklebacks Gasterosteus aculeatus. J. Fish Biol. 81, 966-976. 10.1111/j.1095-8649.2012.03340.x [DOI] [PubMed] [Google Scholar]
- Rymer J., Bauernfeind A. L., Brown S. and Page T. L. (2007). Circadian rhythms in the mating behavior of the cockroach, Leucophaea maderae. J. Biol. Rhythms 22, 43-57. 10.1177/0748730406295462 [DOI] [PubMed] [Google Scholar]
- Sadamoto H., Takahashi H., Okada T., Kenmoku H., Toyota M. and Asakawa Y. (2012). De novo sequencing and transcriptome analysis of the central nervous system of mollusc Lymnaea stagnalis by deep RNA sequencing. PLoS ONE 7, e42546 10.1371/journal.pone.0042546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanogo Y. O. and Bell A. M. (2016). Molecular mechanisms and the conflict between courtship and aggression in three-spined sticklebacks. Mol. Ecol. 25, 4368-4376. 10.1111/mec.13766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanakrishnan A., Dollinger M., Hamlet C. L., Colin S. P. and Miller L. A. (2012). Flow structure and transport characteristics of feeding and exchange currents generated by upside-down Cassiopea jellyfish. J. Exp. Biol. 215, 2369-2381. 10.1242/jeb.053744 [DOI] [PubMed] [Google Scholar]
- Sauer S., Herrmann E. and Kaiser W. (2004). Sleep deprivation in honey bees. J. Sleep Res. 13, 145-152. 10.1111/j.1365-2869.2004.00393.x [DOI] [PubMed] [Google Scholar]
- Sauer S., Kinkelin M., Herrmann E. and Kaiser W. (2003). The dynamics of sleep-like behaviour in honey bees. J. Comp. Physiol. A Neuroethol. Sensory Neural Behav. Physiol. 189, 599-607. 10.1007/s00359-003-0436-9 [DOI] [PubMed] [Google Scholar]
- Sayers E. W., Barrett T., Benson D. A., Bolton E., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Federhen S. et al. (2012). Database resources of the national center for biotechnology information. Nucleic Acids Res. 40, D13-D25. 10.1093/nar/gkr1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. H. (2014). The energy allocation function of sleep: a unifying theory of sleep, torpor, and continuous wakefulness. Neurosci. Biobehav. Rev. 47, 122-153. 10.1016/j.neubiorev.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Schmidt P. S., Matzkin L., Ippolito M. and Eanes W. F. (2005). Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59, 1721-1732. 10.1111/j.0014-3820.2005.tb01821.x [DOI] [PubMed] [Google Scholar]
- Schmidt P. S. and Paaby A. B. (2008). Reproductive diapause and life-history cline in North American populations of Drosophila melanogaster. Evolution 62, 1204-1215. 10.1111/j.1558-5646.2008.00351.x [DOI] [PubMed] [Google Scholar]
- Schulte C., Theilenberg E., Muller-Borg M., Gempe T. and Beye M. (2014). Highly efficient integration and expression of piggyBac-derived cassettes in the honeybee (Apis mellifera). Proc. Natl. Acad. Sci. USA 111, 9003-9008. 10.1073/pnas.1402341111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppe H. (1995). [Rhythmic brain activity in sleeping bees]. Wien. Med. Wochenschr. 145, 463-464. [PubMed] [Google Scholar]
- Schwendeman A. R. and Shaham S. (2016). A high-throughput small molecule screen for C. elegans linker cell death inhibitors. PLoS ONE 11 10.1371/journal.pone.0164595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scullin M. K. and Bliwise D. L. (2015). Is cognitive aging associated with levels of REM sleep or slow wave sleep? Sleep 38, 335-336. 10.5665/sleep.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A. and Mignot E. (2011). Genetics of sleep and sleep disorders. Cell 146, 194-207. 10.1016/j.cell.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Donlea J. M., Gottschalk L. and Shaw P. J. (2011). Sleep deprivation during early-adult development results in long-lasting learning deficits in adult Drosophila. Sleep 34, 137-146. 10.1093/sleep/34.2.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Thimgan M., Donlea J., Gimbel S. I., Gottschalk L., Duntley S. P. and Shaw P. J. (2009). Identifying sleep regulatory genes using a Drosophila model of insomnia. J. Neurosci. 29, 7148-7157. 10.1523/JNEUROSCI.5629-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. J., Cirelli C., Greenspan R. J. and Tononi G. (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834-1837. 10.1126/science.287.5459.1834 [DOI] [PubMed] [Google Scholar]
- Shaw P. J., Tononi G., Greenspan R. J. and Robinson D. F. (2002). Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417, 287-291. 10.1038/417287a [DOI] [PubMed] [Google Scholar]
- Shorter J., Couch C., Huang W., Carbone M. A., Peiffer J., Anholt R. R. H. and Mackay T. F. C. (2015). Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc. Natl. Acad. Sci. USA 112, E3555-E3563. 10.1073/pnas.1510104112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J. M. (2005). Clues to the functions of mammalian sleep. Nature 437, 1264-1271. 10.1038/nature04285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Ju J. Y., Walsh M. B., DiIorio M. A. and Hart A. C. (2014). Deep conservation of genes required for both Drosphila melanogaster and Caenorhabditis elegans sleep includes a role for dopaminergic signaling. Sleep 37, 1439-1451. 10.5665/sleep.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C., Oikonomou G. and Prober D. A. (2015). Norepinephrine is required to promote wakefulness and for hypocretin-induced arousal in zebrafish. eLife 4, e07000 10.7554/eLife.07000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocumb M. E., Regalado J. M., Yoshizawa M., Neely G. G., Masek P., Gibbs A. G. and Keene A. C. (2015). Enhanced sleep is an evolutionarily adaptive response to starvation stress in Drosophila. PLoS ONE 10, e0131275 10.1371/journal.pone.0131275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada J., Scholz M., Kirsten H., Hensch T., Horn K., Jawinski P., Ulke C., Burkhardt R., Wirkner K., Loeffler M. et al. (2016). Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE Adult Study. J. Sleep Res. 25, 690-701. 10.1111/jsr.12421 [DOI] [PubMed] [Google Scholar]
- Stahl M. L., Orr W. C. and Bollinger C. (1983). Postprandial sleepiness: objective documentation via polysomnography. Sleep 6, 29-35. 10.1093/sleep/6.1.29 [DOI] [PubMed] [Google Scholar]
- Stahl B., Slocumb M., Chaitin H., DiAngelo J. and Keene A. (2017). Sleep-dependent modulation of metabolic rate in Drosophila. Sleep ( in press) 40 10.1093/sleep/zsx084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W. and Li H. (2007). The recent demographic and adaptive history of Drosophila melanogaster. Heredity (Edinb) 98, 65-68. 10.1038/sj.hdy.6800901 [DOI] [PubMed] [Google Scholar]
- Stephenson R. and Lewis V. (2011). Behavioural evidence for a sleep-like quiescent state in a pulmonate mollusc, Lymnaea stagnalis (Linnaeus). J. Exp. Biol. 214, 747-756. 10.1242/jeb.050591 [DOI] [PubMed] [Google Scholar]
- Svetec N., Zhao L., Saelao P., Chiu J. C. and Begun D. J. (2015). Evidence that natural selection maintains genetic variation for sleep in Drosophila melanogaster. BMC Evol. Biol. 15, 41 10.1186/s12862-015-0316-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Huang W., Mackay T. F. C. and Anholt R. R. H. (2013). Analysis of natural variation reveals neurogenetic networks for Drosophila olfactory behavior. Proc. Natl. Acad. Sci. U. S. A. 110, 1017-1022. 10.1073/pnas.1220168110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura K., Minamida N. and Okabe S. (2010). Neural map of the larval central nervous system in the ascidian Ciona intestinalis. Zoolog. Sci. 27, 191-203. 10.2108/zsj.27.191 [DOI] [PubMed] [Google Scholar]
- Teyke T. (1990). Morphological differences in neuromasts of the blind cave fish Astyanax hubbsi and the sighted river fish Astyanax mexicanus. Brain. Behav. Evol. 35, 23-30. 10.1159/000115853 [DOI] [PubMed] [Google Scholar]
- Titlow J. S., Majeed Z. R., Hartman H. B., Burns E. and Cooper R. L. (2013). Neural circuit recording from an intact cockroach nervous system. J. Vis. Exp. 81, e50584 10.3791/50584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler I. (1983). Effect of forced locomotion on the rest-activity cycle of the cockroach. Behav. Brain Res. 8, 351-360. 10.1016/0166-4328(83)90180-8 [DOI] [PubMed] [Google Scholar]
- Tobler I. and Neuner Jehle M. (1992). 24h variation of vigilance in the cockroach Blaberus giganteus. J. Sleep Res. 1, 231-239. 10.1111/j.1365-2869.1992.tb00044.x [DOI] [PubMed] [Google Scholar]
- Toth A. L. and Robinson G. E. (2007). Evo-devo and the evolution of social behavior. Trends Genet. 23, 334-341. 10.1016/j.tig.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Tougeron K. and Abram P. K. (2017). An ecological perspective on sleep disruption. Am Nat. 190, E55-E66. 10.1086/692604 [DOI] [PubMed] [Google Scholar]
- Trojanowski N. F. and Raizen D. M. (2016). Call it worm sleep. Trends Neurosci. 39, 54-62. 10.1016/j.tins.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek M., Besseling J., Spies J. P., König S. and Bringmann H. (2016). Sleep-active neuron specification and sleep induction require FLP-11 neuropeptides to systemically induce sleep. Elife 5, e12499 10.7554/eLife.12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Alphen B., Yap M. H. W., Kirszenblat L., Kottler B. and van Swinderen B. (2013). A dynamic deep sleep stage in Drosophila. J. Neurosci. 33, 6917-6927. 10.1523/JNEUROSCI.0061-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., Simpson J. H. and Bellen H. J. (2011). Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 72, 202-230. 10.1016/j.neuron.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorster A. P. A., Krishnan H. C., Cirelli C. and Lyons L. C. (2014). Characterization of sleep in Aplysia californica. Sleep 37, 1453-1463. 10.5665/sleep.3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy V. V. and Harris K. D. (2013). Sleep and the single neuron: the role of global slow oscillations in individual cell rest. Nat. Rev. Neurosci. 14, 443-451. 10.1038/nrn3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Thomson N., White J. G. and Brenner S. (1975). Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J. Comp. Neurol. 160, 313-337. 10.1002/cne.901600305 [DOI] [PubMed] [Google Scholar]
- Webb W. B. and Agnew H. W. (1970). Sleep stage characteristics of long and short sleepers. Science 168, 146-147. 10.1126/science.168.3927.146 [DOI] [PubMed] [Google Scholar]
- Weinstock G. M., Robinson G. E., Gibbs R. A., Worley K. C., Evans J. D., Maleszka R., Robertson H. M., Weaver D. B., Beye M., Bork P. et al. (2006). Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931-949. 10.1038/nature05260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells M. J. (1997). Cephalopod behaviour. Trends Ecol. Evol. 12, 82-83. 10.1016/S0169-5347(97)82690-0 [DOI] [Google Scholar]
- Yokogawa T., Marin W., Faraco J., Pézeron G., Appelbaum L., Zhang J., Rosa F., Mourrain P. and Mignot E. (2007). Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol. 5, 2379-2397. 10.1371/journal.pbio.0050277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M., Gorički Š., Soares D. and Jeffery W. R. (2010). Evolution of a behavioral shift mediated by superficial neuromasts helps cavefish find food in darkness. Curr. Biol. 20, 1631-1636. 10.1016/j.cub.2010.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M., Robinson B. G., Duboué E. R., Masek P., Jaggard J. B., O'Quin K. E., Borowsky R. L., Jeffery W. R. and Keene A. C. (2015). Distinct genetic architecture underlies the emergence of sleep loss and prey-seeking behavior in the Mexican cavefish. BMC Biol. 13, 15 10.1186/s12915-015-0119-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. Z. (1991). Light has many meanings for cephalopods. Vis. Neurosci. 7, 1-12. 10.1017/S0952523800010907 [DOI] [PubMed] [Google Scholar]
- Zhdanova I. V., Wang S. Y., Leclair O. U. and Danilova N. P. (2001). Melatonin promotes sleep-like state in zebrafish. Brain Res. 903, 263-268. 10.1016/S0006-8993(01)02444-1 [DOI] [PubMed] [Google Scholar]
- Zimmerman J. E., Naidoo N., Raizen D. M. and Pack A. I. (2008). Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci. 31, 371-376. 10.1016/j.tins.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman J. E., Chan M. T., Jackson N., Maislin G. and Pack A. I. (2012). Genetic background has a major impact on differences in sleep resulting from environmental influences in Drosophila. Sleep 35, 545-557. 10.5665/sleep.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]