Abstract
Objective:
Angiogenin is a small protein that exerts potent stimulating effects on angiogenesis. In this study, we aimed to examine the expression of angiogenin in different subtypes of glioblastoma and estimated its independent prognostic value.
Methods:
The genomic and survival data from The Cancer Genome Atlas-glioblastoma were extracted for a secondary study. Results The expression of angiogenin was upregulated in glioblastoma tissues and varied significantly in different subtypes. Although the proneural subtype had the lowest angiogenin expression, high angiogenin expression was associated with significantly worse overall survival. However, this association was not observed in other subtypes. By performing univariate and multivariate analysis using Cox regression model, we observed that high angiogenin expression was an independent indicator of shorter overall survival in proneural glioblastoma (hazard ratio: 1.669, 95% confidence interval: 1.033-2.696, P = .036), after adjustment of age, gender, isocitrate dehydrogenase 1 mutation, temozolomide chemotherapy and radiation therapy. In addition, we also observed a correlation between elevated angiogenin expression and the hypomethylated status of its DNA. The hypermethylation group had significantly better overall survival.
Conclusions:
Angiogenin upregulation might serve as a biomarker for unfavorable overall survival in the proneural subtype of glioblastoma.
Keywords: ANG, overall survival, proneural subtype, glioblastoma
Introduction
Glioblastoma (GBM) is a group of heterogeneous diseases, among which 4 subtypes (proneural, neural, classical, and mesenchymal) were identified by gene expression pattern established by The Cancer Genome Atlas (TCGA) researchers.1 Proneural subtype has a higher frequency of PDGFRA or isocitrate dehydrogenase 1 (IDH1) mutations. Classical subtypes show an abnormally high level of EGFR amplification and homozygous deletions of CDKN2A. 1 Mesenchymal tumors usually contain hemizygous deletions of NF1.1 In comparison, no unique mutations have yet been observed in neural cases.1,2 Each subtype varied significantly in survival length and treatment response. Among the 4 subtypes, the proneural cases have the best survival.1,2 Although several strong prognostic indicators have been identified in GBM, such as isocitrate dehydrogenase (IDH) mutations and O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation,3 the use of these markers in clinical practices still has some limitations due to the heterogeneous properties of GBM.
Angiogenin (encoded by ANG gene) is a 14-kD ribonuclease that exerts potent stimulating effects on angiogenesis during neovascularization process. It shares 33% sequence identity to the pancreatic RNAse A sequence and is also called RNase5.4 Angiogenin interacts with actin of both endothelial and smooth muscle cell surface and forms protein complex that initiates the proteolytic cascades via elevating the expression of proteases and plasmin.5,6 This process leads to the degradation of the basement membrane and extracellular matrix, which enables the endothelial cells to penetrate and migrate into the perivascular tissue.7 Therefore, it does not only play an essential role in normal physiological processes but also involves in the pathological development of tumors.
Previous studies found that ANG expression was elevated in numerous tumors, such as colorectal,8 breast,9 cervical,10 ovarian,11 stomach,12 liver,13 pancreatic14 tumors and gliomas.15 Nuclear translocation of ANG is essential to its angiogenic activity.16 However, not all tumors had nuclear expression of ANG.17 One previous study demonstrated that the glioblastoma (GBM) cells had nuclear expression of ANG.17 Inhibition of its nuclear translocation suppresses its ribonucleolytic activity and 45S ribosomal RNA synthesis.17 Another study observed that ANG promotes GBM cell proliferation via activating the nuclear factor κB signaling pathway and inhibiting the expression of its binding partner Four and a half LIM domains protein 3.18 These findings confirmed that ANG has functional roles in GBM cells.
In this study, using genomic and survival data from the TCGA-GBM, we explored the expression of ANG in different subtypes of GBM and estimated its independent prognostic value in each subtype.
Materials and Methods
In Silico Analysis Using Data From TCGA-GBM
The data in TCGA-GBM were acquired with the access provided by the UCSC Xena Browser (https://xenabrowser.net/heatmap/). Only the primary tumor cases without histological neoadjuvant treatment were included in this study. Gene expression was quantified by Affymetrix Human Genome U133 Array Strip (AffyU133a). The following variables were extracted from the database for subsequent analysis: age at initial pathologic diagnosis, gender, IDH1 mutation status, gene expression subtypes, karnofsky performance score, overall survival (OS) status and time, temozolomide chemotherapy status, radiation therapy status, ANG expression, and ANG DNA methylation (quantified by Infinium HumanMethylation27 BeadChip, which includes cg22723026 and cg19211827 in ANG DNA). O(6)-methylguanine-DNA methyltransferase promoter methylation status was determined by the mean methylation status of the MGMT-STP2 model,19 which includes 2 CpG sites (cg12434587 and cg12981137).
Statistical Analysis
Statistical analyses were conducted using SPSS 25.0 software (SPSS, Chicago, Illinois) and GraphPad Prism 7.04 (GraphPad Inc, La Jolla, California). One-way analysis of variance with Bonferroni post hoc tests and Welch t test (unequal variances t test) were performed to assess the statistical differences. Characteristics of patients (categorical variables) in the 2 groups were analyzed by χ2 test with 2-sided Fisher exact test. Kaplan-Meier OS curves were generated using GraphPad Prism 7.04. Receiver–operating characteristic (ROC) analysis for death detection was performed to identify the best cutoff (Youden index) for ANG expression to separate the patients. Log-rank testing was used to assess the differences between the curves. The independent prognostic value of ANG expression in the proneural subtype was assessed using the univariate and multivariate Cox regression models. P < .05 was considered statistically significant.
Results
ANG Expression Was Upregulated in GBM Tissues and Varied Significantly in Different Subtypes
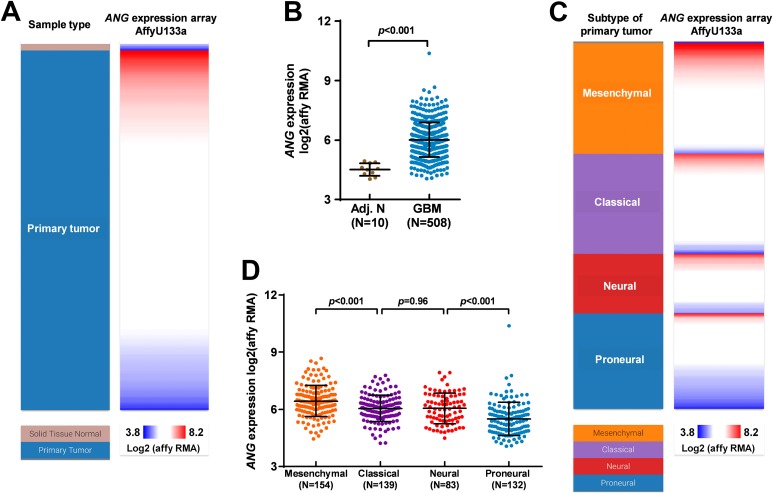
By extracting the microarray data of gene expression in TCGA-GBM, we studied the expression of ANG in GBM tissues and the adjacent normal tissues. Results showed that ANG expression was generally upregulated in GBM tissues (Figure 1A and B). Besides, its expression varied significantly among the 4 subtypes of GBM. Mesenchymal subtype and proneural subtype had the highest and lowest ANG expression, respectively (Figure 1C and D).
Figure 1.
Angiogenin (ANG) expression was upregulated in glioblastoma (GBM) tissues and varied significantly in different subtypes. A-D, Heatmap (A and C) and plots chart (B and D) showing the expression of ANG in GBM tissues and in the adjacent normal tissues (A-B) and among the 4 subtypes of GBM (C-D). Data were extracted from The Cancer Genome Atlas (TCGA)-GBM.
High ANG Expression Was Associated With Significantly Shorter OS in Proneural Subtype But Not Other Subtypes of GBM
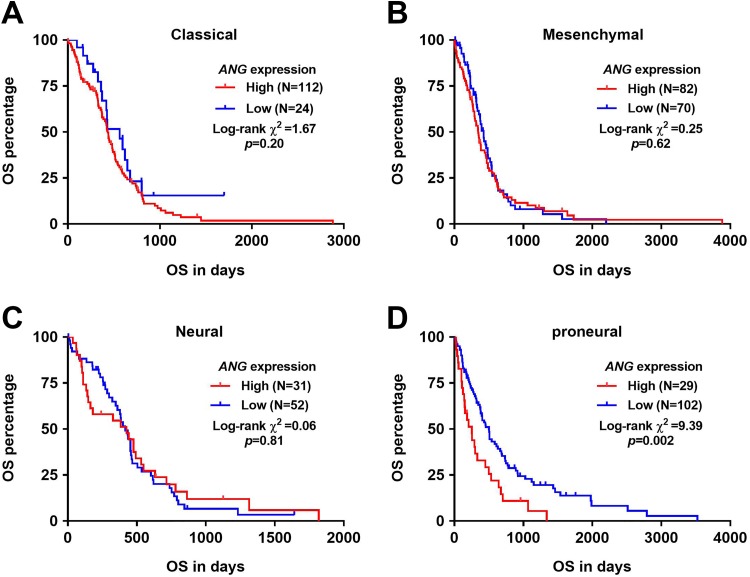
Then, we generated Kaplan-Meier survival curves to investigate the association between ANG expression and OS in each subtype of GBM. By using the best cutoff model, we found that high ANG expression was associated with significantly shorter OS in proneural subtype (P = .002; Figure 2D) but not other subtypes of GBM (Figure 2A–C).
Figure 2.
High angiogenin (ANG) expression was associated with significantly shorter overall survival (OS) in proneural subtype but not other subtypes of GBM. A-D, Kaplan-Meier curves of OS in each subtype of glioblastoma (GBM): classical (A), mesenchymal (B), neural (C), and proneural (D). The Youden Index of ANG expression in the receiver–operating characteristic (ROC) analysis for death detection was applied as the cutoff to separate the patients.
Expression of ANG in Proneural Subtype Might Be Related to its DNA Methylation Status
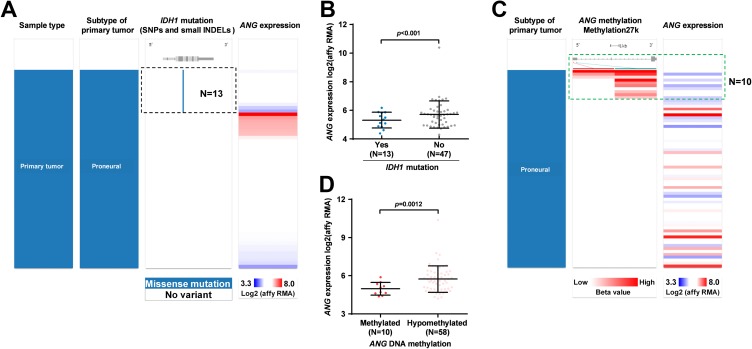
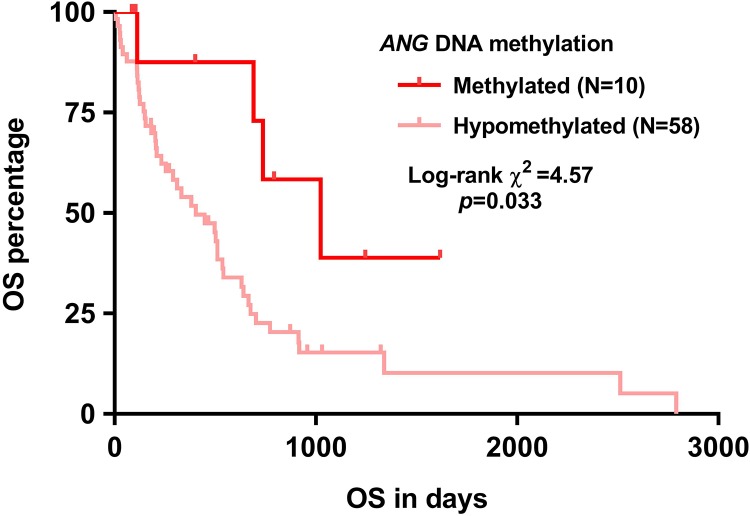
The IDH1 mutation does not only serve as a powerful prognostic marker but also influences the expression of a series of genes via indirectly influencing the activity of DNA demethylases.20 In this study, we examined the correlation between IDH1 mutation and ANG expression in proneural subtype. Results showed that among the 60 cases with proneural subtype having IDH1 mutation identified, 13 cases had mutations. This group of patients also had significantly lower ANG expression, compared to the group without IDH1 mutation (Figure 3A–B). Therefore, we hypothesized that ANG expression might be related to its DNA methylation status. To test this hypothesis, we examined the correlation between ANG expression and the methylation of 2 CpG sites in its DNA. Ten of 68 cases that had at least weak methylation (accumulated methylation ≥0.2) of these 2 CpG sites were defined as the methylated group (Figure 3C). Group comparison showed that the methylated group had significantly lower ANG expression compared to the hypomethylation group (Figure 3D). By generating Kaplan-Meier survival curves, we also examined the association between ANG expression and OS. Results confirmed that the methylated group had significantly longer OS compared to the hypomethylated group (Figure 4).
Figure 3.
Angiogenin (ANG) expression in proneural subtype was related to its DNA methylation status. A-D, Heatmap (A and C) and plots chart (B and D) showing the correlation between isocitrate dehydrogenase 1 (IDH1) mutation and ANG expression (A-B) and between ANG methylation and its expression (C-D) in proneural subtype. Data were extracted from The Cancer Genome Atlas (TCGA)-glioblastoma (GBM).
Figure 4.
Kaplan-Meier overall survival (OS) curves of patients with proneural glioblastoma (GBM) with angiogenin (ANG) DNA methylation measured.
High ANG Expression Might Independently Predict Shorter OS in Patients With Proneural GBM
According to the Youden Index of ANG expression in ROC analysis of death, patients with proneural GBM were separated into ANG high (n = 29) and low (n = 109) expression group. Their clinicopathological features and OS outcome were given and are compared in Table 1. By performing univariate analysis, we found that older age, male patients, without IDH1 mutation, without temozolomide chemotherapy, without radiotherapy, and high ANG expression were risk factors of unfavorable OS (Table 2). Following multivariate analysis confirmed that high ANG expression was an independent indicator of shorter OS in proneural GBM (hazard ratio [HR]: 1.669, 95% confidence interval [CI]: 1.033-2.696, P = .036) after adjustment of other factors (Table 2).
Table 1.
Comparison of the Clinicopathological Parameters Between High and Low ANG Expression Groups in Proneural Subtype.
| Parameters | ANG Expression | P Value | |
|---|---|---|---|
| High, n = 29 | Low, n = 102 | ||
| Age, Mean ± SEM | 63.34 ± 3 | 52.31 ± 1.692 | .002 |
| Gender | |||
| Female | 12 | 40 | .83 |
| Male | 17 | 62 | |
| KPS | |||
| ≤80 | 22 | 67 | .29 |
| >80 | 1 | 11 | |
| No data | 6 | 24 | |
| IDH1 mutations | |||
| No | 11 | 36 | .43 |
| Yes | 1 | 12 | |
| No data | 17 | 54 | |
| MGMT promoter methylation, Mean ± SEM | 0.31 ± 0.06 | 0.30 ± 0.04 | .89 |
| Radiation therapy | |||
| No | 7 | 18 | .43 |
| Yes | 20 | 77 | |
| No data | 2 | 7 | |
| Temozolomide chemotherapy | |||
| No | 12 | 43 | 1.00 |
| Yes | 15 | 52 | |
| No data | 2 | 7 | |
| Living status | |||
| Living | 2 | 26 | .039 |
| Dead | 27 | 76 | |
Abbreviations: ANG, Angiogenin; IDH1, isocitrate dehydrogenase 1; KPS, karnofsky performance score; MGMT, O(6)-methylguanine-DNA methyltransferase; SEM, standard error of mean.
Table 2.
Univariate and Multivariate Analysis of OS in Patients With Proneural GBM.
| Parameters | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| P | HR | 95% CI (Lower/Upper) | P | HR | 95% CI (Lower/Upper) | |||
| Age (Continuous) | <.001 | 1.038 | 1.024 | 1.053 | .018 | 1.020 | 1.003 | 1.036 |
| Gender | ||||||||
| Male (N = 79) | 1.000 | |||||||
| Female (N = 52) | .044 | 0.657 | 0.436 | 0.989 | .001 | 0.460 | 0.287 | 0.737 |
| KPS | ||||||||
| >80 (N = 12) | 1.000 | |||||||
| ≤80 (N = 89) | .062 | 2.105 | 0.964 | 4.595 | ||||
| IDH1 mutations | ||||||||
| Yes (N = 13) | 1.000 | |||||||
| No (N = 47) | .004 | 3.930 | 1.541 | 10.020 | .042 | 2.884 | 1.039 | 8.007 |
| MGMT promoter methylation | .767 | 0.841 | 0.268 | 2.641 | ||||
| Temozolomide chemotherapy | ||||||||
| True (N = 67) | 1.000 | |||||||
| False (N = 55) | .010 | 1.718 | 1.141 | 2.586 | .963 | 1.010 | 0.649 | 1.572 |
| Radiation therapy | ||||||||
| True (N = 97) | 1.000 | |||||||
| False (N = 25) | <.001 | 4.803 | 2.895 | 7.967 | <.001 | 5.326 | 2.920 | 9.715 |
| ANG expression | ||||||||
| Low (N = 102) | 1.000 | |||||||
| High (N = 29) | .003 | 1.989 | 1.270 | 3.116 | .036 | 1.669 | 1.033 | 2.696 |
Abbreviations: ANG, Angiogenin; CI, confidence interval; GBM, glioblastoma; HR, hazard ratio; IDH1, isocitrate dehydrogenase 1; KPS, karnofsky performance score; MGMT, O(6)-methylguanine-DNA methyltransferase; SEM, standard error of mean.
Bold-face values indicates p<0.05.
Discussion
Besides the oncogenic effect of the aberrantly expressed ANG, a series of studies also found that its upregulation might be an unfavorable survival marker in some cancers, such as gastric cancer,12 patients with stage IV melanoma,21 and non-Hodgkin lymphoma.22 One previous study found a trend of increased ANG concentration in the higher grade of malignancy,15 suggesting that ANG might participate in the malignant transformation of GBM. In this study, we confirmed that ANG expression was significantly upregulated in GBM tissues compared to that in adjacent normal tissues. In addition, we found that ANG expression varied significantly in different subtypes of GBM. Therefore, we determined to explore its prognostic value in the subtypes. By generating Kaplan-Meier OS curves in the 4 subtypes of GBM, we found that ANG upregulation might only have prognostic value in the proneural subtype. Since IDH1 mutation and MGMT promoter methylation are 2 well-established prognostic indicators in GBM, we included these 2 factors in the univariate and multivariate analyses to assess the independent prognostic value of ANG expression. Results showed that high ANG expression was an independent indicator of shorter OS in proneural GBM (HR: 1.669, 95%CI: 1.033-2.696, P = .036), after adjustment of age, gender, isocitrate dehydrogenase 1(IDH1) mutation, temozolomide chemotherapy, and radiation therapy. However, we found that MGMT promoter methylation is not associated with OS in proneural subtype of GBM. Although IDH1 mutation has strong predictive value, they are uncommon in primary GBM.20,23 This is a major limitation of its prognostic value. Our study found that ANG expression showed independent prognostic value, even after adjustment of IDH1 mutation. Therefore, it might be used as a biomarker to identify the proneural subtype of GBM with better prognosis.
By checking the association between IDH1 mutation and ANG expression, we found that mutation group had significantly decreased ANG expression. Mechanistically, the mutations result in the loss of isocitrate dehydrogenase activity and following enhanced production of 2-hydroxyglutarate (2-HG),24 which inhibit the enzymic activity of DNA demethylases. Therefore, IDH1 mutation could indirectly increase DNA methylation status of a series of genes. Considering the effect of IDH1 mutation, we examined whether the DNA methylation status of ANG was altered in proneural subtype. As hypothesized, a small proportion of proneural subtype had elevated ANG DNA methylation and decreased ANG expression. In addition, this group also had significantly better OS. These findings suggest that DNA methylation status might play a role in ANG expression. However, since this is an in silico study, no molecular studies were performed to validate these findings. In future, it is meaningful to demonstrate that the causative effect of IDH1 mutation or ANG methylation on ANG expression.
Conclusion
Angiogenin upregulation might serve as an independent predictor of unfavorable OS in proneural subtype of GBM.
Abbreviations
- ANG
Angiogenin
- CI
confidence interval
- GBM
glioblastoma
- HR
hazard ratio
- IDH1
isocitrate dehydrogenase 1
- MGMT
O(6)-methylguanine-DNA methyltransferase
- OS
overall survival
- ROS
receiver–operating characteristic
- TCGA
The Cancer Genome Atlas
Footnotes
Authors’ Note: This is a retrospective study based on online databases. No primary data were collected by any authors in this manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by a grant from the Science and Technology Program of Shenzhen (No: JCYJ20140416122812008).
ORCID iD: Hao Wang, MD  https://orcid.org/0000-0003-4735-7536
https://orcid.org/0000-0003-4735-7536
References
- 1. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ng K, Kim R, Kesari S, Carter B, Chen CC. Genomic profiling of glioblastoma: convergence of fundamental biologic tenets and novel insights. J Neurooncol. 2012;107(1):1–12. [DOI] [PubMed] [Google Scholar]
- 4. Harper JW, Auld DS, Riordan JF, Vallee BL. Enzymatically active angiogenin/ribonuclease A hybrids formed by peptide interchange. Biochemistry. 1988;27(1):219–226. [DOI] [PubMed] [Google Scholar]
- 5. Gao X, Xu Z. Mechanisms of action of angiogenin. Acta Biochim Biophys Sin (Shanghai). 2008;40(7):619–624. [DOI] [PubMed] [Google Scholar]
- 6. Tello-Montoliu A, Patel JV, Lip GY. Angiogenin: a review of the pathophysiology and potential clinical applications. J Thromb Haemost. 2006;4(9):1864–1874. [DOI] [PubMed] [Google Scholar]
- 7. Soncin F. Angiogenin supports endothelial and fibroblast cell adhesion. Proc Natl Acad Sci U S A. 1992;89(6):2232–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Etoh T, Shibuta K, Barnard GF, Kitano S, Mori M. Angiogenin expression in human colorectal cancer: the role of focal macrophage infiltration. Clin Cancer Res. 2000;6(9):3545–3551. [PubMed] [Google Scholar]
- 9. Nilsson UW, Abrahamsson A, Dabrosin C. Angiogenin regulation by estradiol in breast tissue: tamoxifen inhibits angiogenin nuclear translocation and antiangiogenin therapy reduces breast cancer growth in vivo. Clin Cancer Res. 2010;16(14):3659–3669. [DOI] [PubMed] [Google Scholar]
- 10. Landt S, Mordelt K, Schwidde I, et al. Prognostic significance of the angiogenic factors angiogenin, endoglin and endostatin in cervical cancer. Anticancer Res. 2011;31(8):2651–2655. [PubMed] [Google Scholar]
- 11. Chopra V, Dinh TV, Hannigan EV. Angiogenin, interleukins, and growth-factor levels in serum of patients with ovarian cancer: correlation with angiogenesis. Cancer J Sci Am. 1996;2(5):279–285. [PubMed] [Google Scholar]
- 12. Shimoyama S, Kaminishi M. Angiogenin in sera as an independent prognostic factor in gastric cancer. J Cancer Res Clin Oncol. 2003;129(4):239–244. [DOI] [PubMed] [Google Scholar]
- 13. Barcena C, Stefanovic M, Tutusaus A, et al. Angiogenin secretion from hepatoma cells activates hepatic stellate cells to amplify a self-sustained cycle promoting liver cancer. Sci Rep. 2015;5:7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shimoyama S, Gansauge F, Gansauge S, Oohara T, Kaminishi M, Beger HG. Increased angiogenin expression in obstructive chronic pancreatitis surrounding pancreatic cancer but not in pure chronic pancreatitis. Pancreas. 1999;18(3):225–230. [DOI] [PubMed] [Google Scholar]
- 15. Eberle K, Oberpichler A, Trantakis C, et al. The expression of angiogenin in tissue samples of different brain tumours and cultured glioma cells. Anticancer Res. 2000;20(3A):1679–1684. [PubMed] [Google Scholar]
- 16. Moroianu J, Riordan JF. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc Natl Acad Sci U S A. 1994;91(5):1677–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raghu H, Lakka SS, Gondi CS, et al. Suppression of uPA and uPAR attenuates angiogenin mediated angiogenesis in endothelial and glioblastoma cell lines. PLoS One. 2010;5(8):e12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xia W, Fu W, Cai X, et al. Angiogenin promotes U87MG cell proliferation by activating NF-kappaB signaling pathway and downregulating its binding partner FHL3. PLoS One. 2015;10(2):e0116983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van den Bent MJ, Erdem-Eraslan L, Idbaih A, et al. MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic Oligodendrogliomas and Oligoastrocytomas. A report from EORTC study 26951. Clin Cancer Res. 2013;19(19):5513–5522. [DOI] [PubMed] [Google Scholar]
- 20. Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13(5):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vihinen P, Kallioinen M, Vuoristo MS, et al. Serum angiogenin levels predict treatment response in patients with stage IV melanoma. Clin Exp Metastasis. 2007;24(7):567–574. [DOI] [PubMed] [Google Scholar]
- 22. Fang S, Repo H, Joensuu H, Orpana A, Salven P. High serum angiogenin at diagnosis predicts for failure on long-term treatment response and for poor overall survival in non-Hodgkin lymphoma. Eur J Cancer. 2011;47(11):1708–1716. [DOI] [PubMed] [Google Scholar]
- 23. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turkalp Z, Karamchandani J, Das S. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol. 2014;71(10):1319–1325. [DOI] [PubMed] [Google Scholar]