Abstract
Rationale: Idiopathic pulmonary fibrosis (IPF) causes considerable global morbidity and mortality, and its mechanisms of disease progression are poorly understood. Recent observational studies have reported associations between lung dysbiosis, mortality, and altered host defense gene expression, supporting a role for lung microbiota in IPF. However, the causal significance of altered lung microbiota in disease progression is undetermined.
Objectives: To examine the effect of microbiota on local alveolar inflammation and disease progression using both animal models and human subjects with IPF.
Methods: For human studies, we characterized lung microbiota in BAL fluid from 68 patients with IPF. For animal modeling, we used a murine model of pulmonary fibrosis in conventional and germ-free mice. Lung bacteria were characterized using 16S rRNA gene sequencing with novel techniques optimized for low-biomass sample load. Microbiota were correlated with alveolar inflammation, measures of pulmonary fibrosis, and disease progression.
Measurements and Main Results: Disruption of the lung microbiome predicts disease progression, correlates with local host inflammation, and participates in disease progression. In patients with IPF, lung bacterial burden predicts fibrosis progression, and microbiota diversity and composition correlate with increased alveolar profibrotic cytokines. In murine models of fibrosis, lung dysbiosis precedes peak lung injury and is persistent. In germ-free animals, the absence of a microbiome protects against mortality.
Conclusions: Our results demonstrate that lung microbiota contribute to the progression of IPF. We provide biological plausibility for the hypothesis that lung dysbiosis promotes alveolar inflammation and aberrant repair. Manipulation of lung microbiota may represent a novel target for the treatment of IPF.
Keywords: idiopathic pulmonary fibrosis, inflammation, lung microbiota, bleomycin, germ free
At a Glance Commentary
Scientific Knowledge on the Subject
Mechanisms of disease progression in idiopathic pulmonary fibrosis (IPF) are poorly understood. Recent studies support an association between lung bacterial burden and mortality. Further work has correlated changes in host defense gene expression in peripheral blood with lung microbiota. However, the direct effect of lung microbiota on indices of alveolar inflammation and fibrosis and the causal significance of lung microbiota in IPF remain undetermined.
What This Study Adds to the Field
We report significant correlations between airway microbiota and indices of alveolar inflammation in IPF. We report a significant association between lung bacterial burden and disease progression in human patients. In models of pulmonary fibrosis, dysbiosis precedes peak lung injury and persists in the fibrotic lung. In novel germ-free animal models of pulmonary fibrosis, we found that the absence of microbiota protects against mortality and modulates both humoral and cellular immunity in the fibrotic lung.
Pulmonary fibrosis is a poorly understood, heterogeneous disease with significant global morbidity and mortality. Idiopathic pulmonary fibrosis (IPF), the most common form of pulmonary fibrosis, has poorly understood mechanisms of pathogenesis and disease progression (1, 2). Despite recent progress in the understanding of IPF pathophysiology (3–5), there remains no solution for advanced disease, with the exception of lung transplant (6, 7). Pulmonary fibrosis is characterized by recurrent unknown injury with the loss and abnormal repair of alveolar epithelial cells and deposition of extracellular matrix and collagen (8). This matrix impairs efficient gas exchange within the lung, resulting in respiratory failure and death.
Though IPF has conventionally been considered a sterile lung disorder, the recent revolution in culture-independent microbiology has prompted a reconsideration of IPF pathogenesis (9–11). Pulmonary fibrosis results in irreversible changes to the architecture of the lung, with local alterations in airway and alveolar mucosa, blood flow, and oxygen tension, as well as derangements in host immunity. All of these anatomic, physiologic, and immunologic features of the disease likely exert selective pressure on the bacterial communities of the lower respiratory tract, generating dysbiosis. Recent observational studies in patients with IPF have reported associations between lung dysbiosis, disease progression, and mortality (12–17). Both bacterial burden (12) and bacterial community composition (13) have been correlated with disease outcomes, and the identity of lung bacteria has been correlated with peripheral blood host defense gene expression (14). Impaired host defense and innate immune signaling are contributing factors to IPF disease severity and course (4, 18–21). Systemic immunosuppression is associated with deleterious clinical outcomes in IPF (22), whereas antimicrobials may have a therapeutic role in reduced mortality and limited overt infectious episodes (23). Yet, whether the lung dysbiosis of pulmonary fibrosis is a cause or consequence of disease remains unknown.
In the present study, we used a translational approach to examine the causal role of the lung microbiome in pulmonary fibrosis. In human patients with IPF, increased bacterial burden predicts disease progression, and the diversity and community composition of lung microbiota are correlated with indices of alveolar inflammation. In animal models of pulmonary fibrosis, lung dysbiosis precedes lung injury and persists until the establishment of fibrosis, and germ-free (GF) mice are protected from mortality. Our translational study of lung microbiota demonstrates that the lung microbiome plays a causal role in pulmonary fibrosis progression and may represent a potential target for preventing the aberrant inflammation and dysregulated repair of IPF. Some of the results of these studies were previously reported in the form of an abstract (24).
Methods
Ethics Statement
Human subjects included were patients from a prospective observational study correlating biomarkers with disease progression (COMET [Correlating Outcomes with Biochemical Markers to Estimate Time-Progression in Idiopathic Pulmonary Fibrosis]; www.clinicaltrials.gov identifier NCT01071707) (13). All patients provided written informed consent. All clinical investigations were conducted according to the Declaration of Helsinki. The human study protocol was approved by the institutional review boards of all participating centers. See the online supplement section for further details. The animal studies were approved by the University of Michigan Institutional Animal Care and Use Committee at the University of Michigan under protocol 6821.
Human IPF Study Population
The study population of 68 patients with IPF was a subset of the COMET study who had BAL fluid (BALF) DNA available for analysis. We also included a subset of 39 COMET study patients who had matched BALF microbiota analysis (16S rRNA gene sequencing) and BALF available for cytokine measurements. See the online supplement for details.
Mouse Models
Eight to 10-week-old C57BL/6J mice were purchased from The Jackson Laboratory and housed under specific pathogen–free conditions. GF mice on a C57BL/6 background were bred and maintained by the GF animal core facility at the University of Michigan and were housed in flexible film isolators. Feces of GF mice were assessed for GF status on a regular basis using Gram stain and culture. For further details, see the online supplement.
Preparation of Single-Cell Suspensions from Lung Tissue and Flow Cytometry
Collagenase digestion of mouse lung tissue and flow cytometry were performed as previously described (25). See the online supplement for details.
Bleomycin Model of Pulmonary Fibrosis, Mouse Tissue Collection/Processing, Lung Histologic Sectioning and Staining, and Hydroxyproline Assay
Pulmonary fibrosis was induced with oropharyngeal or intratracheal bleomycin as previously described (26). Control mice received identical volumes of saline instillation. Tissue collection and processing were performed as previously described (27). Histologic sectioning and staining were performed as previously described (28). Collagen deposition was measured using a hydroxyproline assay as described previously (29, 30). See the online supplement for details.
Bacterial DNA Isolation and 16S DNA Sequencing
Genomic DNA extraction from mouse tissue and human BALF was carried out as previously described (13, 31). Droplet digital PCR (ddPCR) for the 16S rRNA gene was performed as previously described (27). See the online supplement for details.
Pulmonary Cytokine Measurements in Lungs of Patients with IPF and Mice
Pulmonary inflammation in vivo was evaluated by cytokine measurements of human BALF and murine lung homogenate using a Luminex platform (MilliporeSigma) as previously described (32). See the online supplement for details.
Statistical Analysis
Principal component analysis (PCA) using BAL cytokine data was performed in comparisons between healthy subjects and subjects with IPF. PCA and redundancy analysis were performed on human microbiota data between tertile groups of patients with IPF and on murine lung microbiota data between murine experimental groups (R vegan package; R Foundation for Statistical Computing). Statistical differences were examined using permutational multivariate ANOVA (PERMANOVA). Univariate and multivariable logistic regression modeling of healthy control and IPF BALF cytokines was also performed using age and sex as variables. Univariate linear regressions between 1) relative abundances of microbiota (at phylum and operational taxonomic unit [OTU] levels), 2) Shannon diversity indices, and 3) bacterial burden by 16S and alveolar cytokine concentrations were performed and adjusted using a multivariable linear regression model with adjustment for the variables age, sex, smoking status, baseline FVC (percent predicted), and baseline DlCO (percent predicted). Multiple comparisons were accounted for using a Benjamini-Hochberg procedure and a false discovery rate (FDR) of 10%. In the OTU regression model, OTUs were examined on the basis of their presence in at least two patients, and relationships with alveolar cytokine concentrations were based on the presence or absence of an OTU in each patient. Univariate and multivariate Cox proportional hazards models were employed to study IPF disease progression with a composite endpoint as previously published (13), adjusted for age, sex, baseline DlCO, baseline FVC, and smoking status. We performed all analyses in R (R: A Language and Environment for Statistical Computing; R Core Team [2017], R Foundation for Statistical Computing) and Prism version 7 software (GraphPad Software).
Identification of Procedural Contaminants and Adequacy of Sequencing
The identification of contaminants in human BAL data and murine lung was carried out as previously described (13, 31). See the online supplement for details.
Results
Lung Bacterial Burden Predicts Disease Progression in IPF
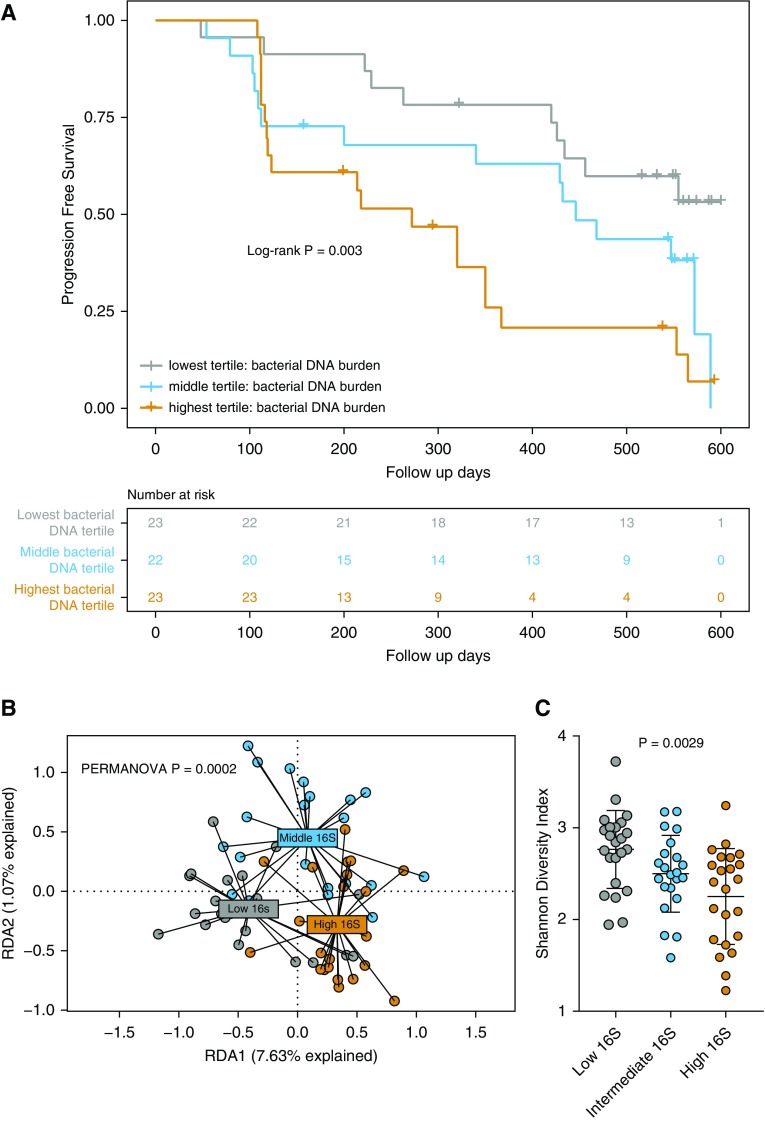
We first tested the hypothesis that increased lung bacterial burden predicts subsequent disease progression, as previously reported (12). To quantify bacterial burden, we studied BALF collected from COMET patients with IPF (33). The cohort clinical characteristics and demographics are reported in Table 1. Our primary outcome was progression-free survival, as measured by a composite endpoint of time until death, acute exacerbation of IPF, lung transplant, or relative decline in FVC greater than 10%, or DlCO greater than 15% (13) (Table E1 in the online supplement). A total of 44 patients had progressive disease based on this composite endpoint. To measure bacterial burden, we used ddPCR, a novel ultrasensitive PCR platform, to quantify copies of the bacterial 16S rRNA gene (27). Patients with IPF with progressive disease had significantly higher bacterial burden than nonprogressors (P = 0.041) (Figure E1). We next stratified the patients with IPF into tertiles based on total bacterial DNA burden (methodology consistent with previous reports [12]) as measured by ddPCR and examined progression-free survival. In an unadjusted Cox proportional hazards model, patients with IPF in the highest tertile (with the highest bacterial burden) had a significantly increased hazard compared with those in the lowest tertile (hazard ratio, 3.58; 95% confidence interval, 1.64–7.81; P = 0.0014) (Figure 1A). This association remained significant after adjustment for age, sex, FVC, DlCO, and smoking status in a multivariable Cox model (hazard ratio, 4.99; 95% confidence interval, 1.96–12.71; P = 0.0007). Results of univariate and multivariable Cox proportional hazards models are reported in Table 2. Endpoint type and frequency are reported in Table E1.
Table 1.
Clinical Characteristics and Demographics of Patients with Idiopathic Pulmonary Fibrosis
| All | Bacterial DNA Tertiles |
P Value* | |||
|---|---|---|---|---|---|
| Low | Intermediate | High | |||
| n | 68 | 23 | 22 | 23 | |
| Age, yr, mean (SD) | 65.0 (7.3) | 63.6 (7.4) | 64.3 (7.6) | 66.9 (6.8) | 0.29 |
| Male, n (%) | 46 (67.6) | 16 (69.6) | 15 (68.2) | 15 (65.2) | 0.94 |
| FVC, mean (SD)† | 70.0 (17.7) | 70.0 (18.2) | 69.0 (14.3) | 71.0 (21.0) | 0.92 |
| DlCO, mean (SD)‡ | 43.5 (14.0) | 44.3 (14.4) | 45.2 (12.6) | 41.0 (15.2) | 0.53 |
| Smoking status, n (%) | 0.96 | ||||
| Nonsmoker | 20 (29) | 7 (30.4) | 6 (27.3) | 7 (30.4) | — |
| Ex-smoker | 46 (68) | 15 (65.2) | 15 (68.2) | 16 (69.6) | — |
| Active smoker | 2 (3) | 1 (4.4) | 1 (4.5) | 0 | — |
P values are calculated by ANOVA for continuous variables and chi-square test/Fisher’s exact test for categorical variables. Smoking status statistical testing by nonsmoker versus ever smoker.
FVC percent predicted at trial enrollment; FVC data missing = 4.
DlCO percent predicted at trial enrollment; DlCO data missing = 7.
Figure 1.
Alveolar bacterial burden predicts disease progression in idiopathic pulmonary fibrosis. (A) BAL was performed in 68 patients with idiopathic pulmonary fibrosis, and bacterial burden was quantified using droplet digital PCR of the 16S rRNA gene. Disease progression was defined by death, acute exacerbation, decline in lung function, or lung transplant. A Kaplan-Meier curve was generated by using a Cox proportional hazards model stratified by bacterial burden (displayed as tertile ranges; lowest tertile range, 9.76e+03–4.64e+04 16S copies; middle tertile range, 4.68E+04–1.31E+05 16S copies; highest tertile range, 1.31E+05–1.14E+07 16S copies). Log-rank P test value is reported. (B and C) Higher bacterial burden is associated with altered lung bacterial community structure or dysbiosis. (B) Redundancy analyses (RDA1, RDA2) of bacterial communities in all three BAL 16S tertile groups, with statistical testing by permutational multivariate ANOVA (PERMANOVA). (C) Increased bacterial burden is associated with a lower α-diversity (Shannon diversity index), as determined by Mann-Whitney U test with Dunn’s multiple comparisons.
Table 2.
Cox Proportional Hazards Model of Disease Progression in Idiopathic Pulmonary Fibrosis
| Variable | HR | 95% CI Lower Limit | 95% CI Upper Limit | P Value |
|---|---|---|---|---|
| Univariate | ||||
| Total bacterial DNA burden: middle tertile* | 2.03 | 0.91 | 4.52 | 0.084 |
| Total bacterial DNA burden: highest tertile* | 3.58 | 1.64 | 7.81 | 0.0014 |
| Shannon diversity index† | 1.19 | 0.46 | 1.56 | 0.59 |
| Age‡ | 1.02 | 0.98 | 1.06 | 0.27 |
| Male§ | 0.94 | 0.50 | 1.76 | 0.85 |
| DlCO|| | 0.98 | 0.96 | 1.01 | 0.36 |
| FVC¶ | 1.00 | 0.98 | 1.02 | 0.65 |
| Smoking status** | 1.19 | 0.67 | 2.11 | 0.55 |
| Multivariate | ||||
| Total bacterial DNA burden: middle tertile* | 2.26 | 0.94 | 5.39 | 0.07 |
| Total bacterial DNA burden: highest tertile* | 4.99 | 1.96 | 12.71 | 0.0007 |
| Age | 1.00 | 0.94 | 1.06 | 0.97 |
| Male | 0.89 | 0.47 | 1.73 | 0.73 |
| DlCO | 0.99 | 0.96 | 1.03 | 0.74 |
| FVC | 1.00 | 0.98 | 1.03 | 0.68 |
| Smoking status | 1.81 | 0.82 | 3.98 | 0.14 |
Definition of abbreviations: CI = confidence interval; HR = hazard ratio.
Versus lowest tertile. N = 68 in multivariate model adjusted for age, sex, DlCO, FVC, and smoking status.
Shannon diversity index, per 1-unit increase.
Age, per 1-year increase.
Male versus female.
DlCO percent predicted at trial enrollment, per 1% increase.
FVC percent predicted at trial enrollment, per 1% increase.
Never smoker versus ever smoker.
We next sought to examine bacterial community structure between these tertile groups by using constrained ordination redundancy analysis based on PCA. These groups, stratified by bacterial burden, demonstrate significantly altered lung bacterial communities (PERMANOVA P = 0.0002) (Figure 1B). Lung bacterial communities in the highest bacterial burden tertile are significantly different from those in the intermediate bacterial burden tertile (PERMANOVA P = 0.007) and the lowest bacterial burden tertile (PERMANOVA P = 0.00009). Given the changes in community structure, we next sought to understand whether there was an associated change in community diversity. We report significant changes in community diversity across all tertile groups (P = 0.0029) (Figure 1C). Diversity in the highest 16S tertile group was significantly reduced compared with the lowest 16S tertile group (P = 0.0020), but it was not significantly different from the intermediate 16S tertile (P = 0.46). We thus concluded that lung bacterial burden predicts disease progression in patients with IPF, a finding now validated across patient cohorts and quantification platforms (12). Increased bacterial burden is also associated with significant differences in community composition and loss of community diversity, suggesting that all three features of the lung microbiome (burden, composition, diversity) may be involved in local inflammation and immune responses.
Alveolar Inflammation Is Dysregulated in Patients with IPF and Correlated with Variation in Lung Microbiota
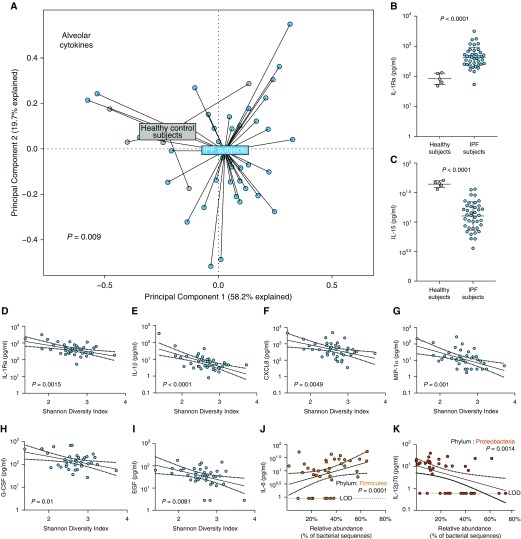
Having confirmed a relationship between the lung microbiome and disease progression in IPF, we then asked if features of the lung microbiome (including burden, composition, and diversity) are associated with alveolar inflammation in patients with IPF. Initially, we examined the degree of dysregulation present in host alveolar inflammation by comparing BALF cytokines in five healthy volunteers with that of patients with IPF. Demographics of the healthy volunteers and this subset of 39 patients with IPF are reported in Table 3. Using PCA and PERMANOVA, we found a significant collective difference in alveolar cytokine concentration in BALF, as measured by multiplex ELISA, between healthy control subjects and patients with IPF (P = 0.009) (Figure 2A). To determine the specific cytokine drivers of this collective variation, we performed logistic regression comparing healthy and IPF BALF, controlling for differences in age and sex. We found significantly increased concentrations of alveolar IL-1Ra in IPF BALF compared with that of healthy volunteers (adjusted P = 0.04) (Table E2, Figure 2B) and significantly decreased concentrations of IL-15 (adjusted P = 0.0035) (Table E1, Figure 2C) and IL-7 (adjusted P = 0.03) (Table E2) in patients with IPF compared with healthy volunteers. Results of the logistic regression model of BALF cytokines in healthy volunteers and patients with IPF are reported in Table E2. We concluded that alveolar inflammation is altered in patients with IPF compared with healthy control subjects.
Table 3.
Clinical Characteristics and Demographics of Patients with Idiopathic Pulmonary Fibrosis and Healthy Control Subjects in Cytokine Lung Microbiota Study
| Healthy Control Subjects | All Patients with IPF | IPF Progressors | IPF Nonprogressors | P Value* | |
|---|---|---|---|---|---|
| n | 5 | 39 | 22 | 17 | |
| Age, yr, mean (SD) | 41.4 (11.6) | 64.3 (8.0) | 65.8 (8.3) | 62.4 (7.3) | 0.19 |
| Male, n (%) | 2 (40) | 26 (66.7) | 13 (59.1) | 13 (76.5) | 0.42 |
| FVC, mean (SD)† | NR | 71.8 (17.9) | 75.0 (18.0) | 67.4 (17.5) | 0.20 |
| DlCO, mean (SD)‡ | NR | 47.6 (12.6) | 48.2 (12.8) | 46.7 (12.7) | 0.74 |
| Smoking status, n (%) | NR | ||||
| Nonsmoker | 13 (33.3) | 7 (31.8) | 6 (35.3) | 0.61 | |
| Ex-smoker | 25 (64.1) | 15 (68.2) | 10 (58.8) | ||
| Active smoker | 1 (2.6) | 0 | 1 (5.9) |
Definition of abbreviations: IPF = idiopathic pulmonary fibrosis; NR = not recorded.
Statistical testing is progressor versus nonprogressor.
P values were calculated by t test for continuous variables and chi-square test/Fisher’s exact test for categorical variables.
FVC percent predicted at trial enrollment.
DlCO percent predicted at trial enrollment.
Figure 2.
Alveolar inflammatory and fibrotic cytokines are disordered in idiopathic pulmonary fibrosis (IPF) and correlated with lung microbiota. Alveolar immunity was characterized using a multiplex ELISA of BAL fluid from 39 patients with IPF and 5 healthy volunteers. Lung bacteria were characterized using 16S rRNA gene sequencing from this subset of 39 patients with IPF. (A) As visualized via principal component analysis and tested using permutational multivariate ANOVA, alveolar cytokine concentrations were altered in patients with IPF compared with healthy control subjects (P = 0.009, permutational multivariate ANOVA). (B and C) These differences in alveolar immunity were driven by increased IL-1Ra in IPF (P < 0.0001; adjusted P = 0.04) and by decreased IL-15 in IPF (P < 0.0001; adjusted P = 0.0035). (D–I) Among patients with IPF, decreased diversity of lung bacteria was correlated with increased alveolar concentration of IL-1Ra (D) (P = 0.0015; adjusted P = 0.0058), IL-1β (E) (P < 0.0001; adjusted P = 0.0004), CXCL8 (C-X-C motif chemokine ligand 8)/IL-8 (F) (P = 0.0049; adjusted P = 0.015), MIP-1α (macrophage inflammatory protein-1α) (G) (P = 0.001; adjusted P = 0.0009), granulocyte colony–stimulating factor (G-CSF) (H) (P = 0.01; adjusted P = 0.02), and EGF (epidermal growth factor) (I) (P = 0.0081; adjusted P = 0.04). (J and K) Alveolar concentrations of IL-6 were positively correlated with relative abundance of the lung Firmicutes phyla (J) (P = 0.02; adjusted P = 0.001), whereas alveolar IL-12p70 was negatively correlated with relative abundance of lung Proteobacteria phylum (K) (P = 0.0056; adjusted P = 0.0014). Statistical significance was determined using univariate and multivariable logistic regression modeling of log-transformed cytokine data, adjusted for age and sex in healthy versus IPF model and univariate/multivariate linear regression models adjusted for age, sex, baseline pulmonary function, and smoking status in the IPF model. Benjamini-Hochberg correction was applied to account for multiple comparisons with a false discovery rate of 0.1 when applicable. Continuous variables were examined using an unpaired t test or Mann-Whitney U test when applicable.
We next asked if variation in alveolar cytokines correlates with diversity of lung microbiota in IPF. We determined community diversity using the Shannon diversity index. We found that decreased lung bacterial diversity was significantly associated with increased alveolar concentrations of proinflammatory profibrotic cytokines and growth factors, including IL-1Ra (adjusted P = 0.0058) (Figure 2D), IL-1β (adjusted P = 0.0004) (Figure 2E), CXCL8 (C-X-C motif chemokine ligand 8) (adjusted P = 0.015) (Figure 2F), MIP-1α (macrophage inflammatory protein-1α) (adjusted P = 0.0009) (Figure 2G), G-CSF (granulocyte colony–stimulating factor) (adjusted P = 0.025) (Figure 2H), VEGF (vascular endothelial growth factor) (adjusted P = 0.015), and epidermal growth factor (EGF) (adjusted P = 0.041) (Figure 2I). These observations remained significant after adjustment for age, sex, FVC, DlCO, and smoking status and multiple comparisons by applying an FDR of 0.1. The results of univariate and multivariable linear regression modeling of diversity and alveolar cytokines are reported in Table E3.
We next asked if variation in alveolar cytokines correlates with variation in the community composition of lung microbiota by prominent lung phyla. After adjusting for age, sex, FVC, DlCO, and smoking status and multiple comparisons by FDR method, we found a positive association between alveolar IL-6 and the relative abundance of the Firmicutes phylum (adjusted P = 0.001) (Figure 2J). IL-6 has known proinflammatory and profibrotic properties (34). We also report a negative association between IL-12p70 and the relative abundance of the Proteobacteria phylum (adjusted P = 0.0014) (Figure 2K). Results of univariate and multivariable linear regression models are reported in Table E4. In an exploratory analysis, we also examined associations between taxa at an OTU (approximately genus/species) level and concentrations of alveolar cytokines using a linear regression model. We report significant associations between varied cytokines and the presence of certain taxa (Table E5). For example, EGF was associated with the presence of OTU0200: Lachnospiraceae (P = 0.0065) and OTU1037: Lachnospiraceae (P < 0.0001). IL-15 was negatively correlated with the presence of OTU1442: Lachnospiraceae (P = 0.0009). IL-1RA was positively correlated with the presence of OTU1463: Veillonella (P = 0.013). IL-1β was positively correlated with the presence of OTU0982: Lactobacillaceae (P = 0.0005) and the presence of OTU1433: Prevotella (P = 0.03). These results are not adjusted for multiple comparisons and are presented for hypothesis generation only. All significant results of linear regression analysis of taxa and alveolar cytokines are reported in Table E5. Given the association between bacterial burden and disease progression, we also sought to identify any association between bacterial burden and alveolar inflammation. We found no significant correlations between lung bacterial burden and alveolar cytokines after adjusting for multiple comparisons, whether analyzing all specimens (Table E6) or restricting our analysis to progressors (Table E7). We concluded that lung bacterial diversity and community composition are correlated with alveolar inflammation in patients with IPF, suggesting a calibrated relationship between lung microbiota and inflammation.
Lung Dysbiosis Precedes Peak Lung Injury and Persists in the Fibrotic Lung
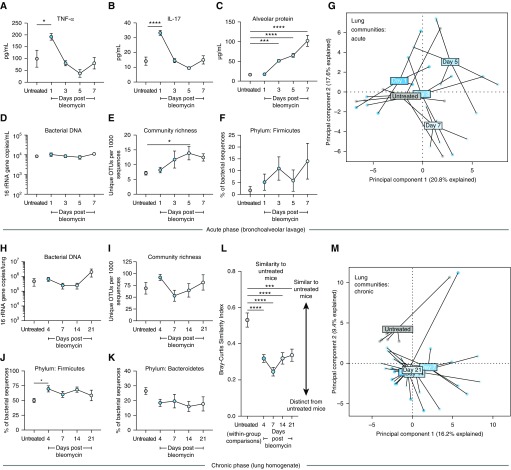
To determine the temporal relationship between lung dysbiosis, inflammation, and fibrogenesis, we administered oropharyngeal bleomycin to mice to elicit acute inflammation (Days 0–7), fibroproliferation (Days 7–14), and fibrosis (Days 14–21). We found peak inflammation in TNF-α (P < 0.05) (bleomycin vs. untreated) (Figure 3A) and IL-17 (P < 0.0001) (bleomycin vs. untreated) (Figure 3B) on Day 1 after bleomycin. Peak lung injury as measured by total BALF protein leak occurred on Day 7 after bleomycin (P < 0.0001) (Figure 3C). Next, we measured bacterial burden and found no significant difference in the burden during the first 7 days in BALF (Figure 3D). We measured community composition by amplicon sequencing of the 16S rRNA gene of BALF and found that BALF bacterial diversity increases after injury and peaks before peak lung injury on Day 7 (P = 0.04) (Figure 3E). We found that community composition was also altered after bleomycin exposure, evidenced by relative abundance of the Firmicutes phylum (Figure 3F) and PCA of BALF microbial communities (Figure 3G), both before peak lung injury.
Figure 3.
After bleomycin exposure, lung dysbiosis precedes peak lung injury and persists until the development of fibrosis. Adult (8–10 wk old) mice (C57BL/6) were challenged with bleomycin, after which their lung microbiota were quantified using droplet digital PCR and characterized using 16S rRNA gene sequencing. (A–C) After bleomycin instillation, acute inflammation peaks within 1 day were reflected in increased alveolar concentrations of inflammatory cytokines TNF-α (A) and IL-17 (B). Peak lung injury does not occur until 7 days after instillation (C). (D) Total bacterial burden in BAL fluid is unchanged in the week after bleomycin exposure. (E) Diversity of lung bacterial communities, as measured by community richness, increases after bleomycin and peaks before peak lung injury. (F and G) The community composition of lung bacteria is altered acutely after bleomycin instillation, reflected in the relative enrichment by the Firmicutes phylum (F) and principal component analysis (G). (H–M) At later time points, alterations in lung microbiota persist until the establishment of pulmonary fibrosis at 21 days. (H and I) Bacterial DNA burden (H) and community richness (I) did not differ significantly at 21 days. (J and K) After bleomycin, lung communities contained increased relative abundance of the Firmicutes phylum (J), with a nonsignificant but consistent decline in Bacteroidetes (K). (L and M) Lung communities of bleomycin-treated mice were significantly distinct from those of untreated mice and remained altered until the establishment of pulmonary fibrosis at 21 days. Significance was determined via ANOVA with the Holm-Sidak multiple comparisons test and the Kruskal-Wallis test with Dunn’s multiple comparisons. OTUs = operational taxonomic units; TNF-α = tumor necrosis factor-α. *P < 0.05, ***P < 0.001, and ****P < 0.0001.
We next examined whether bleomycin-induced lung dysbiosis persists until the establishment of pulmonary fibrosis (Days 14–21) with variables measured in whole lung to capture parenchymal changes. Bacterial burden (Figure 3H) and community richness (Figure 3I) did not significantly differ during fibroproliferation. By contrast, the community composition of lung bacteria was altered after bleomycin exposure and remained altered until Day 21 (Figures 3J–3M). We observed an increase in the relative abundance of the Firmicutes phylum with a nonsignificant but sustained decline in the Bacteroidetes phylum (Figures 3J and 3K). Collectively, lung communities from bleomycin-treated mice differed from those of untreated mice, both via direct comparison of Bray-Curtis similarity (Figure 3L) and via visualization using PCA (Figure 3M). Importantly, this disruption of lung community composition remained over 21 days (Figure 3M), encompassing all stages of the fibroproliferative model and chronic injury. Diversity of fecal bacteria did not change with duration after bleomycin (P > 0.05 at all time points), though community composition of fecal bacteria did differ at all time points (PERMANOVA, P < 0.01 at all time points). We concluded that pulmonary dysbiosis persists through the establishment of fibrosis, both preceding peak injury and in tandem with dysregulated immunity, providing temporal plausibility for the hypothesis that pulmonary microbiota contribute to injury and dysregulated repair in pulmonary fibrosis.
GF Mice Are Protected from Pulmonary Fibrosis–related Mortality
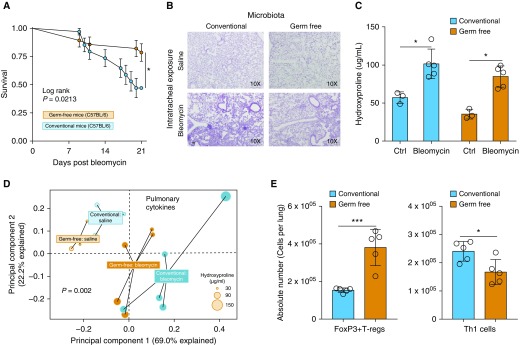
Having shown that dysbiosis of lung microbiota occurs in lung injury and persists throughout the establishment of pulmonary fibrosis, we next sought to test the hypothesis that the absence of lung microbiota would be protective in pulmonary fibrosis. We employed a GF mouse model of bleomycin-induced pulmonary fibrosis and compared outcomes in mice with and without lung microbiota. Compared with genetically identical mice with conventional microbiota, GF mice had a significant reduction in mortality (log-rank P = 0.02, adjusted for batch effect) (Figure 4A) when treated with bleomycin (0.02 U/mouse). Given this protection, we next examined fibrotic outcomes at Day 21 using both histology (Masson’s trichrome stain) and quantification of lung hydroxyproline with a lower dose of bleomycin (0.01 U/mouse). GF and conventional mice did not differ in their histology or in their quantitative amounts of fibrosis at Day 21 (Figures 4B and 4C).
Figure 4.
After bleomycin-induced lung injury, germ-free (GF) mice are protected from mortality and have altered lung immunity. Genetically identical (C57BL/6) GF and conventional mice were challenged with oropharyngeal bleomycin and compared in terms of mortality, fibrosis, and pulmonary inflammation. (A) Compared with conventional mice, GF mice were protected from mortality after bleomycin. The survival difference emerged between Days 10 and 21 after bleomycin exposure. (B and C) Despite this difference in mortality, GF mice had similar severity of pulmonary fibrosis as characterized histologically (B) and by lung concentration of hydroxyproline (C). (D) Pulmonary cytokines measured by multiplex ELISA differed according to microbiome across all four experimental groups. (E) After bleomycin exposure, GF and conventional mice differed in their cellular immunity. Lungs of GF mice contained increased numbers of FOXP3+ regulatory T cells (T-regs) with decreased numbers of T-helper type 1 (Th1) cells. Significance was determined by Kaplan-Meier analysis and batch effect testing, unpaired t test, permutational multivariate ANOVA or Mann-Whitney U test when applicable. *P < 0.05 and ***P < 0.001. Scale bar, 100 μm. Ctrl = control.
Given equivalent fibrosis despite preserved survival in GF mice, we next characterized differences in pulmonary humoral immunity using a multiplex ELISA to characterize concentrations of lung cytokines. We identified significant differences in the concentrations of cytokines (P = 0.002, PERMANOVA) across all experimental groups on Day 21. We visualized specific interactions between microbiome status and bleomycin exposure. As shown in Figure 4D, baseline lung cytokines were similar in GF and conventional mice (P = 0.10, PERMANOVA). Bleomycin exposure significantly altered lung cytokines both in GF mice (P = 0.02, PERMANOVA, saline- vs. bleomycin-treated GF mice) and in conventional mice (P = 0.015, PERMANOVA, saline- vs. bleomycin-treated conventional mice). As seen in Figure 4D, bleomycin exposure increased the within-group dissimilarity among mice, and differences in lung cytokines were associated with differences in severity of fibrosis as measured by lung hydroxyproline concentration (PERMANOVA P = 0.03). Comparing pulmonary cellular immunity (leukocyte profiles on Day 6 after bleomycin exposure) in conventional and GF mice, we found increased absolute numbers of FoxP3+ T-regulatory cells in bleomycin-exposed GF mice compared with conventional mice (P = 0.0008). In contrast, we found reduced absolute numbers of T-helper type 1 (Th1) cells (P = 0.01) (Figure 4E) in bleomycin-exposed GF mice compared with conventional mice. Representative flow plots are shown in Figure E1. Our results suggest a role of lung microbiota in the regulation of Th1/Th2 responses during fibrotic injury and indicate that the injurious effects of lung microbiota in pulmonary fibrosis may occur independently of collagen deposition.
Discussion
The core findings of this study are that lung bacteria 1) predict disease progression in patients with IPF, 2) are correlated with derangements in alveolar immunity, and 3) participate in pathogenesis in animal models of pulmonary fibrosis. In IPF, after adjustment for relevant clinical and physiological variables, lung bacterial burden predicts disease progression. Key features of the lung microbiome correlate with inflammatory and fibrotic mediators and bacterial burden in the lungs of patients with IPF. In animal models of pulmonary fibrosis, lung dysbiosis precedes peak lung injury and persists throughout the periods of fibrogenesis and established fibrosis. The absence of lung microbiota, studied using GF mice, conveys a survival advantage after bleomycin exposure, and GF mice exhibit altered humoral and cellular lung immunity after bleomycin. Our results validate prior observations made using different patient cohorts and different sequencing and quantification platforms (12, 13), and they provide new insights using concurrent analysis with indices of lung immunity as well as complementary animal models. Taken together with the existing literature, our work provides strong corroborating evidence for the hypothesis that lung microbiota contribute to host inflammation and injury in IPF.
Molyneaux and colleagues found an increased risk of mortality with increasing lung bacterial burden (12). They also identified an increased bacterial load as a manifestation of IPF pathobiology. We validate this significant association between the burden of lung microbiota and disease progression in IPF. Provocatively, we found that GF mice are protected from mortality after bleomycin exposure; yet, they exhibit severity of pulmonary fibrosis similar to that of conventional mice. This finding reflects the clinical observation that patients with IPF often die of not merely progressive fibrosis but instead of diverse inflammatory causes of acute-on-chronic respiratory failure such as respiratory infections and poorly understood acute exacerbations of IPF (35). Acute exacerbations of IPF are characterized by diffuse alveolar inflammation and alterations in lung microbiota (16), and their frequency is increased by immunosuppression (22). Our results provide clinical plausibility for the interpretation that the lung microbiome contributes to acute inflammatory dysregulation in IPF, participating in unfavorable outcomes potentially independent of collagen deposition. Supporting this, we found key differences in mice lacking a microbiome, notably a decrease in pulmonary Th1 cellular responses relative to those of conventional mice. We found an enhanced FOXP3+ T-regulatory cell response in bleomycin-treated GF mice, likely skewing the Th1/Th2 balance that has been implicated in IPF pathogenesis (36, 37). Th2 cells have previously been argued to promote tissue repair (38), and although the exact role of FOXP3+ T-regulatory cells in IPF remains undetermined, studies have shown impaired FOXP3+ T-regulatory cell suppressor activity in patients with IPF (39). In contrast, FOXP3+ T cells demonstrated the ability to augment pulmonary fibrosis in silica-induced fibrosis models (40). This finding of higher numbers of T-regulatory cells before peak lung injury would support an early skew toward repair that may contribute to the observed protection. Further studies in GF and monocolonized models will allow for interrogation of the possible mechanism for the reduced mortality observed.
Our findings build on the link between dysregulated systemic immunity and IPF pathogenesis. Previous studies have linked immune gene expression patterns in peripheral blood mononuclear cells and lung microbiota over time in patients with IPF (14, 15). In the present study, we provide evidence demonstrating a calibrated relationship between local alveolar immunity in IPF and the lung microbiome. A disordered lung microbiome has been observed across numerous acute and chronic lung diseases (11). Changes in local mucosal conditions in conjunction with comorbidities may alter the composition and burden of lung microbiota, generating molecular patterns that engage innate immune receptors and promote sustained inflammation. Supporting this interpretation, we report strong correlations between the diversity of lung microbiota in patients with IPF and concentrations of canonical inflammatory and profibrotic cytokines, including IL-1β and IL-8. These cytokines have previously been implicated in pulmonary fibrosis (41, 42). Although most studies to date have focused on the now highly appreciated role of the gut microbiome in shaping systemic immune responses (43–45), the lung microbiome is now recognized to play a pivotal role in directing immune responses in both healthy animals and humans (27, 46). Furthermore, disease results in alterations of the lung microbiome, features of which, in turn, correlate with alveolar inflammation (32, 47, 48). Discriminating between causal links and bystander effect in pulmonary microbiota–host interactions has proven mechanistically difficult. Our translational findings—bacterial burden predicts IPF progression, lung microbiota are correlated with profibrotic alveolar cytokines in patients with IPF, and GF animals are protected from mortality in a model of pulmonary fibrosis—support the hypothesis of a calibrated and causal association between the lung microbiome, lung immunity, and pulmonary fibrosis progression.
There are several limitations to our work. We report correlations between BAL cytokines and microbiota in patients with IPF; however, the number of patients in our study is limited. Our IPF study cohort represents a very well-characterized observational trial cohort through which we validated previous findings between disease progression and pulmonary bacterial load in IPF (12). Procedural and sequencing contamination is always a concern in low-biomass sequencing-based studies. Our protocols are optimized to minimize contamination, including the sequencing of multiple procedural and reagent controls, use of low-biomass protocols for DNA extraction, and randomization of specimen processing to minimize false grouping by kit contamination. We randomized the selection of our animals, recorded cage numbers for multivariable statistical modeling, and controlled diet and environment. However, confounding cannot be completely excluded.
We acknowledge additional limitations to our animal modeling. The bleomycin model does not recapitulate all features of human disease. However, a recent statement supports the use of bleomycin as the best characterized preclinical model available for IPF studies (49). Bleomycin is a glycopeptide antibiotic with antimicrobial efficacy against gram-positive bacteria. However, we do not see significant loss of gram-positive bacteria in exposed mice; in fact, we see progressive enrichment of the gram-positive Firmicutes phylum after exposure. Our control exposure (oropharyngeal saline instillation) likely has an effect on lung bacteria via immigration of pharyngeal microbiota. Future studies should include longitudinal sampling of animals after saline treatment to determine the relative contributions of bleomycin and aspiration to the sustained disruption of lung bacteria in this model. Although our acute phase microbiome comparisons were performed using BALF (to facilitate comparisons with alveolar cytokine and total protein concentrations), our chronic phase microbiome analysis was performed using homogenized lung tissue (to maximize the signal of bacterial DNA and facilitate comparisons with tissue hydroxyproline concentrations). Both tissue types have been used successfully in published murine lung microbiome studies (27, 32, 50), but to our knowledge, there is no published head-to-head comparison using the same mouse-derived specimens. For this reason, we restricted our comparisons of taxonomy to specimens collected using the same tissue type. GF animal models are not representative of human immunology, nor do they replicate microbial conditions that are possible in humans. Preclinical GF models provide a tool for the study of potential mechanisms through which lung microbiota may exert an effect on disease pathobiology in the host. Microbial exposures that contribute to the developing immune response are absent in GF models. Therefore, observations may be related to these immune deficiencies or to developmental differences. Future work will involve exploring the nature of the immune response to injury and fibrosis in monocolonized and conventionalized mouse models.
In conclusion, we provide evidence supporting the link between altered lung microbiota, alveolar inflammation, and disease progression in pulmonary fibrosis, and we provide the first causal evidence that the microbiome participates in the pathogenesis and mortality of fibrotic lung disease. Future work will determine if host–microbiome interactions may be harnessed and leveraged for novel therapeutic strategies in pulmonary fibrosis and other forms of chronic lung disease.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Dr. Jeff Curtis (Veterans Affairs Hospital, Ann Arbor, MI) for the gift of healthy volunteer BAL fluid. The authors also thank COMET study centers, investigators, and patients. In addition, the authors thank Chris Brown (University of Michigan) for assistance with bioinformatics.
Footnotes
Supported by NIH grants K99HL139996 (D.N.O’D.), R01HL121774 (G.B.H.), R01AI117229 (B.B.M.), R01HL127805 (B.B.M.), and K23HL130641 (R.P.D.) and the Host Microbiome Initiative Explorer Program at the University of Michigan.
Sequencing data are available via the National Center for Biotechnology Information Sequence Read Archive (accession numbers PRJNA515255 and PRJNA515279). Operational taxonomic unit, taxonomy, and metadata tables are available from github.com/dicksonlunglab/murine_pulmonary_fibrosis.
Author Contributions: D.N.O’D., J.R.E.-D., B.B.M., and R.P.D.: conceived of the work; acquired, analyzed, and interpreted data; and wrote and revised the manuscript; S.J.G., M.X., and S.M.: analyzed and interpreted data; S.L.A., S.J.G., C.W., N.R.F., K.C.N., K.B.A., M.L.S., M.K.H., K.R.F., E.S.W., and F.J.M.: acquired data; G.B.H.: revised the manuscript. All authors approved the final version of the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201809-1650OC on February 21, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med. 2018;378:1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 2.Macagno F, Varone F, Leone PM, Mari PV, Panico L, Berardini L, et al. New treatment directions for IPF: current status of ongoing and upcoming clinical trials. Expert Rev Respir Med. 2017;11:533–548. doi: 10.1080/17476348.2017.1335601. [DOI] [PubMed] [Google Scholar]
- 3.Yu G, Tzouvelekis A, Wang R, Herazo-Maya JD, Ibarra GH, Srivastava A, et al. Thyroid hormone inhibits lung fibrosis in mice by improving epithelial mitochondrial function. Nat Med. 2018;24:39–49. doi: 10.1038/nm.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rangarajan S, Bone NB, Zmijewska AA, Jiang S, Park DW, Bernard K, et al. Metformin reverses established lung fibrosis in a bleomycin model Nat Med 2018241121–1127.[Published erratum appears in Nat Med 2018;24:1627.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason DP, Brizzio ME, Alster JM, McNeill AM, Murthy SC, Budev MM, et al. Lung transplantation for idiopathic pulmonary fibrosis. Ann Thorac Surg. 2007;84:1121–1128. doi: 10.1016/j.athoracsur.2007.04.096. [DOI] [PubMed] [Google Scholar]
- 7.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Dwyer DN, Ashley SL, Moore BB. Influences of innate immunity, autophagy, and fibroblast activation in the pathogenesis of lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2016;311:L590–L601. doi: 10.1152/ajplung.00221.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Dwyer DN, Habiel D, Hogaboam C. Host–microbial interactions: idiopathic pulmonary fibrosis in technicolor. Am J Respir Crit Care Med. 2017;195:1554–1556. doi: 10.1164/rccm.201701-0092ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384:691–702. doi: 10.1016/S0140-6736(14)61136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190:906–913. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han MK, Zhou Y, Murray S, Tayob N, Noth I, Lama VN, et al. COMET Investigators. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med. 2014;2:548–556. doi: 10.1016/S2213-2600(14)70069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molyneaux PL, Willis-Owen SAG, Cox MJ, James P, Cowman S, Loebinger M, et al. Host–microbial interactions in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;195:1640–1650. doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Ma SF, Espindola MS, Vij R, Oldham JM, Huffnagle GB, et al. COMET-IPF Investigators. Microbes are associated with host innate immune response in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;196:208–219. doi: 10.1164/rccm.201607-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molyneaux PL, Cox MJ, Wells AU, Kim HC, Ji W, Cookson WO, et al. Changes in the respiratory microbiome during acute exacerbations of idiopathic pulmonary fibrosis. Respir Res. 2017;18:29. doi: 10.1186/s12931-017-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris A, Gibson K, Collman RG. The lung microbiome in idiopathic pulmonary fibrosis: what does it mean and what should we do about it? Am J Respir Crit Care Med. 2014;190:850–852. doi: 10.1164/rccm.201409-1626ED. [DOI] [PubMed] [Google Scholar]
- 18.O’Dwyer DN, Norman KC, Xia M, Huang Y, Gurczynski SJ, Ashley SL, et al. The peripheral blood proteome signature of idiopathic pulmonary fibrosis is distinct from normal and is associated with novel immunological processes. Sci Rep. 2017;7:46560. doi: 10.1038/srep46560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, et al. Muc5b is required for airway defence. Nature. 2014;505:412–416. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Dwyer DN, Armstrong ME, Trujillo G, Cooke G, Keane MP, Fallon PG, et al. The Toll-like receptor 3 L412F polymorphism and disease progression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2013;188:1442–1450. doi: 10.1164/rccm.201304-0760OC. [DOI] [PubMed] [Google Scholar]
- 22.Raghu G, Anstrom KJ, King TE, Jr, Lasky JA, Martinez FJ Idiopathic Pulmonary Fibrosis Clinical Research Network. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012;366:1968–1977. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shulgina L, Cahn AP, Chilvers ER, Parfrey H, Clark AB, Wilson EC, et al. Treating idiopathic pulmonary fibrosis with the addition of co-trimoxazole: a randomised controlled trial. Thorax. 2013;68:155–162. doi: 10.1136/thoraxjnl-2012-202403. [DOI] [PubMed] [Google Scholar]
- 24.O’Dwyer DN, Ashley SL, Xia M, Murray S, Norman KC, Salisbury ML, et al. The composition and diversity of pulmonary microbiota correlate with host alveolar inflammation in idiopathic pulmonary fibrosis[abstract]. Am J Respir Crit Care Med 2018197A1060 [Google Scholar]
- 25.Gurczynski SJ, Zhou X, Flaherty M, Wilke CA, Moore BB. Bone marrow transplant-induced alterations in Notch signaling promote pathologic Th17 responses to γ-herpesvirus infection. Mucosal Immunol. 2018;11:881–893. doi: 10.1038/mi.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashley SL, Wilke CA, Kim KK, Moore BB. Periostin regulates fibrocyte function to promote myofibroblast differentiation and lung fibrosis. Mucosal Immunol. 2017;10:341–351. doi: 10.1038/mi.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickson RP, Erb-Downward JR, Falkowski NR, Hunter EM, Ashley SL, Huffnagle GB. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am J Respir Crit Care Med. 2018;198:497–508. doi: 10.1164/rccm.201711-2180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X, Loomis-King H, Gurczynski SJ, Wilke CA, Konopka KE, Ptaschinski C, et al. Bone marrow transplantation alters lung antigen-presenting cells to promote TH17 response and the development of pneumonitis and fibrosis following gammaherpesvirus infection. Mucosal Immunol. 2016;9:610–620. doi: 10.1038/mi.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore BB, Paine R, III, Christensen PJ, Moore TA, Sitterding S, Ngan R, et al. Protection from pulmonary fibrosis in the absence of CCR2 signaling. J Immunol. 2001;167:4368–4377. doi: 10.4049/jimmunol.167.8.4368. [DOI] [PubMed] [Google Scholar]
- 30.Sisson TH, Ajayi IO, Subbotina N, Dodi AE, Rodansky ES, Chibucos LN, et al. Inhibition of myocardin-related transcription factor/serum response factor signaling decreases lung fibrosis and promotes mesenchymal cell apoptosis. Am J Pathol. 2015;185:969–986. doi: 10.1016/j.ajpath.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Dwyer DN, Zhou X, Wilke CA, Xia M, Falkowski NR, Norman KC, et al. Lung dysbiosis, inflammation, and injury in hematopoietic cell transplantation. Am J Respir Crit Care Med. 2018;198:1312–1321. doi: 10.1164/rccm.201712-2456OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naik PK, Bozyk PD, Bentley JK, Popova AP, Birch CM, Wilke CA, et al. COMET Investigators. Periostin promotes fibrosis and predicts progression in patients with idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;303:L1046–L1056. doi: 10.1152/ajplung.00139.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito F, Tasaka S, Inoue K, Miyamoto K, Nakano Y, Ogawa Y, et al. Role of interleukin-6 in bleomycin-induced lung inflammatory changes in mice. Am J Respir Cell Mol Biol. 2008;38:566–571. doi: 10.1165/rcmb.2007-0299OC. [DOI] [PubMed] [Google Scholar]
- 35.Collard HR, Ryerson CJ, Corte TJ, Jenkins G, Kondoh Y, Lederer DJ, et al. Acute exacerbation of idiopathic pulmonary fibrosis: an international working group report. Am J Respir Crit Care Med. 2016;194:265–275. doi: 10.1164/rccm.201604-0801CI. [DOI] [PubMed] [Google Scholar]
- 36.Vasakova M, Striz I, Slavcev A, Jandova S, Kolesar L, Sulc J. Th1/Th2 cytokine gene polymorphisms in patients with idiopathic pulmonary fibrosis. Tissue Antigens. 2006;67:229–232. doi: 10.1111/j.1399-0039.2006.00560.x. [DOI] [PubMed] [Google Scholar]
- 37.Wynn TA. Integrating mechanisms of pulmonary fibrosis. J Exp Med. 2011;208:1339–1350. doi: 10.1084/jem.20110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen JE, Wynn TA. Evolution of Th2 immunity: a rapid repair response to tissue destructive pathogens. PLoS Pathog. 2011;7:e1002003. doi: 10.1371/journal.ppat.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotsianidis I, Nakou E, Bouchliou I, Tzouvelekis A, Spanoudakis E, Steiropoulos P, et al. Global impairment of CD4+CD25+FOXP3+ regulatory T cells in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;179:1121–1130. doi: 10.1164/rccm.200812-1936OC. [DOI] [PubMed] [Google Scholar]
- 40.Lo Re S, Lecocq M, Uwambayinema F, Yakoub Y, Delos M, Demoulin JB, et al. Platelet-derived growth factor-producing CD4+Foxp3+ regulatory T lymphocytes promote lung fibrosis. Am J Respir Crit Care Med. 2011;184:1270–1281. doi: 10.1164/rccm.201103-0516OC. [DOI] [PubMed] [Google Scholar]
- 41.Kolb M, Margetts PJ, Anthony DC, Pitossi F, Gauldie J. Transient expression of IL-1β induces acute lung injury and chronic repair leading to pulmonary fibrosis. J Clin Invest. 2001;107:1529–1536. doi: 10.1172/JCI12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziegenhagen MW, Zabel P, Zissel G, Schlaak M, Müller-Quernheim J. Serum level of interleukin 8 is elevated in idiopathic pulmonary fibrosis and indicates disease activity. Am J Respir Crit Care Med. 1998;157:762–768. doi: 10.1164/ajrccm.157.3.9705014. [DOI] [PubMed] [Google Scholar]
- 43.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 45.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Lesko M, Badri MH, Kapoor BC, Wu BG, Li Y, et al. Lung microbiome and host immune tone in subjects with idiopathic pulmonary fibrosis treated with inhaled interferon-γ. ERJ Open Res. 2017;3:00008-2017. doi: 10.1183/23120541.00008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McVerry BJ, Morris A. The lung microbiome in hematopoietic stem cell transplant: where the money lies. Am J Respir Crit Care Med. 2018;198:1249–1251. doi: 10.1164/rccm.201806-1088ED. [DOI] [PubMed] [Google Scholar]
- 49.Jenkins RG, Moore BB, Chambers RC, Eickelberg O, Königshoff M, Kolb M, et al. ATS Assembly on Respiratory Cell and Molecular Biology. An official American Thoracic Society workshop report: use of animal models for the preclinical assessment of potential therapies for pulmonary fibrosis. Am J Respir Cell Mol Biol. 2017;56:667–679. doi: 10.1165/rcmb.2017-0096ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yadava K, Pattaroni C, Sichelstiel AK, Trompette A, Gollwitzer ES, Salami O, et al. Microbiota promotes chronic pulmonary inflammation by enhancing il-17a and autoantibodies. Am J Respir Crit Care Med. 2016;193:975–987. doi: 10.1164/rccm.201504-0779OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.