Abstract
Ribonucleotides (rNMPs) mis-incorporated during DNA replication are removed by RNase H2 dependent excision repair or by Topoisomerase I – catalyzed cleavage. Top1 cleavage of rNMPs produces 3’ ends harboring terminal adducts, such as 2’, 3’ cyclic phosphate or Top1 cleavage complex (Top1cc), and leads to frequent mutagenesis and DNA damage checkpoint induction. We surveyed a range of candidate enzymes from Saccharomyces cerevisiae for potential roles in Top1 dependent genomic rNMP removal. Genetic and biochemical analyses reveal that Apn2 resolves phosphotyrosine-DNA conjugates, terminal 2’, 3’ cyclic phosphates and their hydrolyzed products. APN2 also suppresses 2-bp slippage mutagenesis in RNH201-deficient cells. Our results define additional activities of Apn2 in resolving a wide range of 3’- end blocks and identify a role of Apn2 in maintaining genome integrity during rNMP repair.
Introduction
Ribonucleotides (rNMPs) are incorporated into genomic DNA at a frequency of one ribonucleotide per every few thousand deoxyribonucleotides due to errors in DNA polymerase action and to inherent imbalances in deoxyribonucleotide (dNTP): rNTP pools that become more severe upon treatment of certain anti-cancer drugs 1,2. rNMP mis-incorporation can give rise to mutations that pose a threat to genetic integrity 3,4. Accumulation of genomic rNMPs also underlies the recessive human genetic disorders Aicardi-Goutieres syndrome 5 and ataxia oculomotor apraxia 1 6.
At least two enzymatic activities, RNase H2 and topoisomerase I (Top1), have been found to incise rNMPs and trigger multistep-repair events 4,7. While RNase H2 recognizes and produces a nick 5’ to rNMPs, Top1 creates a nick 3’ to rNMPs and introduces an aberrant linkage between the 2’ hydroxyl group and 3’ phosphate group of rNMPs (2’, 3’ cyclic phosphate) via nucleophilic attack of the 2’ hydroxyl group to the phosphotyrosyl bond 8-11. Top1-dependent nucleophilic attack of the 5’ hydroxyl may then reverse the process and regenerate rNMPs in biochemically reconstituted reactions 8,9,12. Top1 can also induce additional cleavage a few bases from the nick and form a Top1 cleavage complex (Top1cc) similar to that found in cells treated with the Top1 poison, camptothecin (CPT)8,9,11. At short repeat sequences, secondary Top1 cleavage and ligation may produce slippage type mutagenesis 4,13,14. Unwinding and degradation of the 5’ nicks by Srs2 and Exo1 suppress Top1-mediated ligation across the gap and initiates an error-free repair 13.
Several enzymatic activities have been implicated in the repair of Top1cc based on their ability to confer cellular resistance to Top1 poisons, such as CPT, that trap Top1cc intermediates 15. These enzymes are candidates to resolve Top1cc formed during two consecutive Top1 cleavages at rNMP loaded genomic DNA; yet their contributions in Top1-dependent ribonucleotide removal have not been elucidated. Alternatively, 2’, 3’ cyclic phosphates may represent stable intermediates in cells, and cells may depend on 3’ end processing enzymes to remove these lesions. Using a combination of genetic and biochemical approaches, we have systematically investigated the role of 3’ end modification enzymes for their contributions to process Top1 cleavage intermediates at genomic rNMPs. Our findings reveal that Apn2 plays critical roles in non-mutagenic Top1-dependent genomic rNMP removal and is biochemically capable of processing 3’-phosphotyrosine (pTyr)-DNA conjugates, 2’, 3’ cyclic phosphates and its hydrolyzed products, viz. terminal ribonucleotide with a monophosphate attached.
Results
Terminal phosphate adducts block the 3’-exonuclease activity from Polδ.
Top1 cleaves 3’ of genomic rNMP and generates a 2’, 3’ cyclic phosphate-terminated nick 8-10. To determine whether the exonuclease activity of Polδ, the major DNA polymerase involved in DNA repair synthesis, removes the 2’, 3’- cyclic phosphate or the degenerated monophosphate terminated end, we expressed and purified Polδ complex (Pol3-Pol31-Pol32) (Supplementary Figure 1A) and tested its exonuclease activity with ssDNA substrates terminated at the 3’ end by either a deoxyribonucleotide, a deoxyribonucleotide bearing a 3’-phosphate, a ribonucleotide, or a nucleotide mixture of 2’, 3’- cyclic phosphate and monophosphate (~40-60% of each). While robust Polδ exonuclease activity was detected on ssDNA or ssDNA bearing a terminal ribonucleotide, a terminal phosphate group (2’, 3’ cyclic phosphate and monophosphate) completely blocked the exonuclease digestion (Figure 1A and 1B). A terminal phosphate also efficiently blocked Polδ -catalyzed extension on a primer/template duplex substrates in either the absence or presence of PCNA (Figure 1C). The priming site with deoxyribonucleotide termini was extended efficiently by Polδ and was stimulated by PCNA. Interestingly, the presence of a ribonucleotide at the 3’-primer terminus activated Polδ 3’-exonuclease activity and led to progressive digestion of the primer, which was partially suppressed by PCNA (Figure 1C). We concluded that the terminal 2’, 3’ cyclic phosphate must be processed prior to Polδ-catalyzed gap filling reaction.
Figure 1. Terminal phosphate adducts block Polδ activities.
A. Polδ (20 nM)-catalyzed digestion of ssDNA substrates terminated at the 3’ end by a deoxyribonucleotide or a ribonucleotide (rU) B. Polδ (20 nM)-catalyzed digestion of ssDNA terminated at the 3’ end by a deoxyribonucleotide bearing a 3’-phosphate (PO4) or a nucleotide harboring a mixture of 2’, 3’- cyclic phosphate (Δ) and monophosphate. C. Polδ-catalyzed primer extension primed by a deoxyribonucleotide, a ribonucleotide, or a nucleotide harboring a mixture of 2’, 3’- cyclic phosphate and monophosphate.
Apn2 processes toxic Top1 cleavage intermediates at rNMP in genomic DNA.
We surmised that a 3’ nuclease may remove the terminal 2’, 3’ cyclic phosphate or other adducts after Top1 cleavage at genomic rNMPs. To identify the 3’ nuclease, we used tetrad analysis to examine the growth effects of deletion of 3’ end processing genes in RNH201 deletion strains that express mutant DNA polymerase (pol2-M644G). In comparison to wild-type, the mutant Polε incorporated about ten-fold more rNMPs during DNA replication due to its reduced ability to discriminate rNTPs from dNTPs 7. Deletion of RNH201 channels rNMP removal towards the Top1-dependent pathway 4,16. Since srs2∆ is synthetic lethal with rnh202∆ pol2-M644G 13, a defect in processing 3’ blocked ends likely impairs cell growth following Top1 cleavage at rNMP, and most importantly, the growth defect should be fully offset by TOP1 deletion. Previously, Apn1, Apn2, Tpp1, and Rad1/Rad10 have been implicated in processing 3’ blocked DNA termini under alkylating or oxidative conditions 17-19. Tdp1, Mre11, Slx4, and Rad1/Rad10 are also implicated in processing Top1cc upon treatment with the Top1 poison, camptothecin (CPT)20-22. One or more of these nucleases are candidates for 3’ end processing in rNMP repair. We also monitored the sensitivity of yeast strains to hydroxyurea (HU) treatment 7, as the inhibition of ribonucleotide reductase by hydroxyurea raises the cellular rNTP/dNTP ratio and increases genomic ribonucleotide incorporation 23.
We found that deletion of APN1, RAD1, SLX4, TDP1, TPP1 caused no apparent growth changes in rnh201Δ pol2-M644G mutant cells nor sensitivity to HU treatment (Supplementary Figure 2A-E, data not shown). We also found that pol3-01, a proof-reading defective mutation, did not affect the viability of rnh201Δ pol2-M644G cells (Supplementary Figure 2F), indicating that the proofreading nuclease activity of Polδ does not process 3’ blocked ends. Deletion of RAD1 in rnh201Δ pol2-M644G mutant showed moderate sensitivity to a low dose HU treatment (Supplementary Figure 2I). However, deletion of TOP1 enhanced rather than reduced the HU sensitivity of pol2-M644G rnh201Δ rad1Δ mutants, indicating that HU sensitivity is not due solely to a defect in repairing blocked 3’ ends during Top1-catalyzed rNMP removal. Previously, deletion of TDP1 or RAD1 alone did not confer sensitivity to CPT treatment due to their functional redundancy in resolving Top1cc 19,20. The deletion of both RAD1 and TDP1, however, did not lead to Top1 dependent growth defects nor HU sensitivity (Supplementary Figure 2G, I). We also found that deletion of MRE11 is lethal in pol2-M644G rnh201Δ mutant in tetrad analysis, but deletion of TOP1 did not restore viability (Supplementary Figure 2H).
Unique among the nucleases examined, deletion of APN2 is synthetic lethal to pol2-M644G rnh201Δ as seen in srs2∆ (Figure 2A)13. Importantly, viability is fully rescued by the concomitant deletion of TOP1 (Figure 2A). Expression of Top1 from a galactose-inducible promoter caused lethality in pol2-M644G rnh201Δ apn2Δ top1Δ cells (Figure 2B). Expression of the nuclease deficient apn2-E59A is lethal in pol2-M644G rnh201Δ apn2Δ (Figure 2C). Deletion of APN2 is also lethal in pol3-L612G rnh201Δ mutant that frequently incorporates rNMPs during lagging strand DNA synthesis 24 (Figure 2D). Importantly, APN2 deletion in rnh201Δ mutants did not increase the level of alkali labile rNMPs in DNA, monitored by denaturing agarose gel electrophoresis (Supplementary Figure 3A) and purified Apn2 protein does not react with duplex DNA with a single embedded rNMP (Supplementary Figure 3B). Apn2, therefore, does not directly remove rNMPs from DNA but likely acts as the major end-healing enzyme to facilitate Top1-catalyzed rNMP repair.
Figure 2. apn2 is synthetic lethal in pol2-M644G rnh201 cells.
A. Tetrad analysis results of pol2-M644G rnh201Δ apn2Δ top1Δ heterozygote diploid cells. Circles and dotted circles mark spores that are pol2-M644G rnh201Δ apn2Δ or pol2-M644G rnh201Δ apn2Δ top1Δ, respectively. B. Five-fold serial dilutions of haploid yeasts containing pGAL-TOP1 were plated on YEPD and YEP-galactose medium to induce Top1 expression. Failed growth on the galactose containing medium of pol2-M644G rnh201Δ apn2Δ cells carrying pGAL-TOP1 indicates synthetic lethality. C. Tetrad analysis results of pol2-M644G rnh201Δ apn2-E59A heterozygote diploid cells. Circles mark spores that are pol2-M644G rnh201Δ apn2-E59A. D. Tetrad analysis results of pol3-L612G rnh201Δ apn2Δ heterozygote diploid cells. Circles mark spores that are pol3-L612G rnh201Δ apn2Δ.
Apn2 processes 2’, 3’-cyclic phosphate terminated ends.
We next examined biochemically the processing of the terminal 2’, 3’-cyclic phosphate by the purified recombinant 3’ cleaning enzymes, Apn1, Apn2 and Tpp1 (Supplementary Figure 1D, E). To create a DNA substrate with a pure 2’, 3’ cyclic phosphate terminated nick, duplex DNA with a single embedded ribonucleotide 12 nt away from the 5’- end was treated with top1-T722A, a ligation defective Top1 mutant that produces nearly 3-fold more cyclic phosphate terminated ends than wild-type Top1 25(Supplementary Figure 3C). PNK, but not CIP, can remove 2’, 3’ cyclic phosphate from a terminal ribonucleotide 8,26, and we confirmed that only PNK removed the terminal 2’, 3’-cyclic phosphate from the substrate (Figure 3A). Strikingly, Apn2, a far less active AP endonuclease compared to Apn1 (Supplementary Figure 3D), removed a dinucleotide from the 2’, 3’-cyclic phosphate terminated end and generated a 10-nt long species (Figure 3A). Since the additional cyclic phosphate causes the 12-nt oligonucleotides to migrate near the 11-nt position, we further treated the Apn2 digestion products with PNK, but did not observe a 11-nt product. Thus, the observed band after Apn2 treatment represents a dinucleotide cleavage product (Figure 3A). In contrast, neither apn2-E59A, Apn1 or Tpp1 reacts with the terminal 2’, 3’ cyclic phosphate (Figure 3A and Supplementary Figure 3E). A similar dinucleotide removal was also observed at the 2’, 3’ cyclic phosphate terminated end generated by RNase If (Figure 3B). In the same reaction, we also detected a 12-nt species, likely derived from removal of the terminal monophosphate by Apn2. To confirm this observation, 5’-overhang DNA substrates harboring various 3’ termini, viz. a clean deoxyribonucleotide, a deoxyribonucleotide with 3’-phosphate, a clean ribonucleotide or a monophosphate attached ribonucleotide (synthesized by IDT) were further tested. The results showed that Apn1, Apn2 and Tpp1 are end modification enzymes with minimal 3’ -exo/endo nuclease activities toward a 3’ end with a clean nucleotide (Figure 3C). Interestingly, Apn2 is able to remove the monophosphate attached to a ribonucleotide (Figure 3C) but not to a deoxyribonucleotide (Figure 3C), while both Apn1 and Tpp1 are capable of removing monophosphate attached to either a deoxyribonucleotide or a ribonucleotide (Figure 3C). Since terminal cyclic-phosphate is a relatively unstable intermediate that may be hydrolyzed to monophosphate in cells, the end-cleaning activities of Apn2 are highly specific to 2’, 3’ cyclic phosphate related adducts. Apn2 could also remove 2’, 3’ cyclic phosphate and monophosphate at terminal ribonucleotide on both 5’-overhang and nicked duplex substrates with similar efficiency, but not on a single stranded substrate (Supplementary Figure 3F). Thus, Apn2 preferentially removes a 3’- adduct from terminal ribonucleotides, consistent with its role in end repair during rNMP cleavage by Top1.
Figure 3. Apn2 clips dinucleotide at the 2’, 3’ cyclic phosphate terminated end.
A. Cleavage assays for Apn2 and apn2-E59A (20 – 100 nM) at a Top1-generated 2’, 3’ cyclic phosphate terminated nick. DNA ends were occluded by biotin-bound streptavidin as diagramed to preserve the 5’ labeling phosphate from treatments with alkaline phosphatase (CIP) and polynucleotide kinase (PNK). B. Apn2-catalyzed cleavage at a recessed 3’ end terminated by a ribonucleotide harboring a mixture of terminal 2’, 3’ cyclic phosphate and monophosphate generated by RNase If. DNA ends were occluded as in panel A to preserve the 5’ labeling phosphate from alkaline phosphatase (CIP) and polynucleotide kinase (PNK) treatment. C. Processing of the terminal monophosphate by Apn2 (5 nM and 50 nM), Apn1 (5 nM and 50 nM) and Tpp1 (0.05 nM and 0.5 nM).
PCNA stimulates Apn2’s exonuclease activity on 2’, 3’- cyclic phosphate terminated ends.
PCNA physically interacts with Apn2 and stimulates both its 3’ – 5’ exonuclease and 3’- phosphodiesterase activities on a deoxyribonucleotide terminated 3’- end 27. We therefore examined the effect of PCNA on terminal 2’, 3’ cyclic phosphate removal by Apn2. To retain PCNA on the substrate DNA molecules, we constructed 5’ overhanging duplex DNA that allow both ends to be occluded by biotin-bound streptavidin (Supplementary Figure 4A). In the presence of streptavidin, the 3’-5’ exonuclease activity of Apn2 was apparent on a clean, recessed 3’-DNA end upon loading of PCNA (Supplementary Figure 4A). End occlusion of the 5’ overhang side can also be achieved by addition of single-stranded DNA binding proteins RPA or E.coli SSB (Supplementary Figure 4A). PCNA also stimulated Apn2 degradation of 3’-termini with a clean ribonucleotide or a ribonucleotide with a monophosphate attached (Supplementary Figure 4B). Importantly, PCNA stimulated Apn2, but not apn2-E59A, in the digestion of the 2’, 3’-cyclic phosphate-terminated nicked duplex (Figure 4A). Apn2, but not apn2-E59A, also enabled primer extension by Polδ from a 2’, 3’ cyclic phosphate-terminated priming site. We note that these reactions contain only dATP, dGTP and dTTP to constrain extension to 6 nucleotides, allowing us to discriminate primer extension products from the full-length substrate molecule. The extension products are resistant to alkaline (NaOH) treatment (Figure 4A), indicating that the terminal ribonucleotide has been removed. The presence of Exo1, while having minimal effect on Apn2-catalyzed cleavage, improved the Polδ extension reaction. While PCNA stimulates Apn2-catalyzed initial clipping at the 2’, 3’-cyclic phosphate-terminated end by 3 to 6-fold at 30°C (Figure 4B), a similar reaction at 16°C clearly revealed early cleavage products, which, following PNK treatment, showed an intermediate corresponding to a dinucleotide removal (Figure 4C). Thus, exonucleolytic digestion by Apn2-PCNA likely occurs after removal of the initial dinucleotide. Apn1 and Tpp1 cannot substitute Apn2 for the observed exonuclease activity on a 2’, 3’-cyclic phosphate terminated end (Supplementary Figure 4C, D), and fail to support Polδ – catalyzed extension (Supplementary Figure 4C, D). PCNA also improves the exonuclease activity of Apn2 (Supplementary Figure 4E) and enables Polδ to extend a terminal ribonucleotide with a monophosphate (Supplementary Figure 4E). In control assays, Polδ-PCNA was able to extend from a clean terminal ribonucleotide (Supplementary Figure 4F) with a minimal effects of addition of Apn2. Thus, the ability of Apn2 to promote Polδ-catalyzed repair synthesis is adduct specific.
Figure 4. Apn2 exonuclease activity on a 2’, 3’ cyclic phosphate terminated end is stimulated by PCNA and permits Polδ-catalyzed primer extension.
A. Polδ–PCNA catalyzed extension from a priming site containing a 2’, 3’ cyclic phosphate in the absence or presence of Apn2 (20 nM) or apn2-E59A (20 nM). For the Polδ catalyzed extension assay, only dATP, dGTP and dTTP were added to trap a 6-nt extension product. B. Apn2-catalyzed digestion of a 2’, 3’ cyclic phosphate terminated end in the absence or presence of DNA bound PCNA loaded by RFC in (i). The data were quantified in (ii) and the error bars represent s.d.from three independent experiments. C. Apn2-catalyzed digestion of a 2’, 3’ cyclic phosphate terminated end in the presence of PCNA. Reactions were maintained under 16°C for 2 min to visualize the early intermediates. DNA ends were occluded in A, B and C by biotin bound streptavidin as diagramed to retain PCNA on DNA. D. Dinucleotide slippage mutation rate at the (AG)4 reporter in rnh201Δ, rnh201Δ apn1∆, rnh201Δ apn2∆, rnh201Δ tdp1∆, and rnh201Δ apn2∆ tdp1∆ mutant strains. Median reversion frequencies were measured by fluctuation tests using n=12 independent cultures per experiments except in the rnh201Δ tdp1∆ mutant strain we used n=15 independent cultures to measure the reversion frequency. Error bars represent 95% confidence intervals.
Role of 3’ end processing in Top1-induced mutagenesis.
Evidence suggests that Top1-dependent cleavage at ribonucleotides induces an increase in 2-5 bp slippage mutation events, which is apparent in RNase H2 deletion mutants 4,13. A secondary Top1 cleavage less than six nucleotides from the initial Top1 cleavage site, followed by ligation across a 2-5 base pair gap, could account for these mutations. Since tdp1∆ and apn2∆ led to a modest increase of slippage mutation rate in rnh202∆ 14, we examined the effect of APN2 gene deletion on Top1-induced mutagenesis in wild type and tdp1 deleted cells using the lys2ΔA746(AG)4 frameshift reversion assay, which detects 2- to 5-bp deletion events 4,13(Figure 4D). The effect of APN1 deletion was also measured as a control. Deletion of APN1, APN2 or TDP1 in strains with functional RNase H2 did not alter lys2 reversion frequency (Figure 4D). Analysis of reversion frequencies indicated that the 2-5 bp slippage events at the Top1 hotspot correspond to 36% in apn2 single gene deletion mutant (Supplementary Table 1). As expected, deletion of rnh201 led to a dramatic increase (75-fold) in the frequency of lys2 reversion that was dependent on TOP1 (Figure 4D). Analysis of the mutation spectrum revealed that 94% of these mutations are 2-bp deletions at the Top1 consensus site (Supplementary Table 1). Deletion of APN2, but not APN1, in a rnh201 mutant strain increased the lys2 reversion frequency 143-fold (Figure 4D). Deletion of TDP1 in rnh201 also (103-fold) increased 2-bp deletions among lys2 revertants. Most importantly, deletion of both APN2 and TDP1 in rnh201 further increased lys2 reversion frequency higher than that observed in apn2 rnh201 or tdp1 rnh201 cells (232-fold). Over 90% of the reversion events in all these mutants were 2-bp deletion at Top1 consensus site (Supplementary Table 1). Taken together, these results suggest that Apn2 and Tdp1 represent two distinct mechanisms that suppress slippage mutagenesis after Top1 cleavage at genomic rNMPs.
Apn2 processes Top1-cc intermediates in vitro.
The synthetic phenotype of apn2∆ and tdp1∆ on slippage mutagenesis prompted us to ask whether Apn2 could process Top1cc. Although deletion of APN2 alone or APN2 and TDP1 does not render cells sensitive to CPT treatment (Figure 5A and data not shown), deletion of APN2, but not APN1, in TDP1- and RAD1-deleted cells severely impaired growth and showed sensitivity to CPT treatment that is far more severe than that of TDP1 RAD1 deleted cells (Figure 5A). Deletion of TOP1 completely restored growth of mutant yeast to levels only slightly lessthan those seen in strains without cognate gene deletions (Figure 5A). Interestingly, while Apn2 does not remove 3’ phosphate attached to DNA end (Figure 3C), purified Apn2, like Tdp1, is capable of removing tyrosine from a duplex DNA harboring a 3’- phosphotyrosine (pTyr) (Figure 5B), an activity that is not observed with either apn2-E59A or Apn1. Unlike Tdp1, which leaves a 3’-phosphate, Apn2 generates a clean 3’-OH end that can be directly modified by terminal transferase to attach a radiolabeled nucleotide (Figure 5C). However, Apn2 is unable to process the 3’-PO4 adduct generated by Tdp1 (Figure 5D) consistent with prior assays (Figure 3C). PCNA also stimulates pTyr removal by Apn2, which enables Polδ – catalyzed extension of the 3’-pTyr blocked primer end (Figure 5E). These results suggest that Apn2 may resolve Top1cc and produce 3’ termini suitable for DNA synthesis.
Figure 5. Top1-cc removal by Apn2.
A. apn2∆, rad1∆ and tdp1∆ show synthetic phenotype in sensitivity to camptothecin (CPT). B. Removal of a 3’- pTyr adduct at a recessed 3’ end by Apn2 (4, 20 and 100 nM), apn2-E59A (20 and 100 nM), Tdp1 (0.05 and 0.5 nM) or Apn1 (5 and 50 nM). C. Labeling of a 3’-OH or a 3’-pTyr end (5 nM) by terminal transferase (Tdt) and cordycepin 5’-triphosphate-[α – P32] in the absence or presence of Apn2 (20 nM). D. Reaction to 3’-PO4 by Apn2 (100 nM) or Tpp1 (10 nM) following the pretreatment of 3’-pTyr end with Tdp1 (5 nM). E. Removal of 3’-pTyr by Apn2 in the absence or presence of RFC-PCNA-RPA in (i). In the extension assays where Polδ was present, only dATP, dGTP and dTTP were added to trap a 6-nt long extension product. DNA ends were occluded by biotin bound streptavidin as diagramed to retain PCNA on DNA. The pTyr removal data were quantified in (ii) and error bars represent s.d. from three independent experiments.
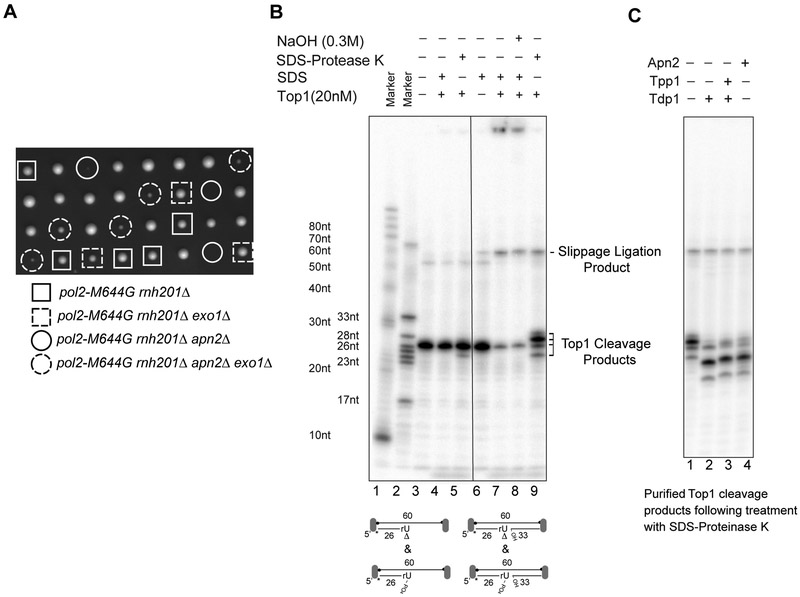
5’ DNA gap limits Top1-dependent mutagenesis.
Cleavage by eukaryotic topoisomerase I relies on duplex regions flanking the cleavage site 28. Hence, gap generation by Srs2/Exo1 could shunt repair to pathways that depend on terminal 2’, 3’ cyclic phosphate processing rather than secondary Top1 cleavage. Thus, inhibition of 5’ gap formation might bypass the importance of Apn2-dependent 2’, 3’ cyclic phosphate processing. Deletion of EXO1 or SRS2 indeed increases lys2 reversion rate 13. Most importantly, deletion of EXO1 partially restored the viability of rnh201∆ pol2-M644G apn2∆ strain (Figure 6A). To directly test whether a 5’ gap determines the type of end repair, we examined the secondary cleavage by Top1 on either a 5’- overhang duplex or a nicked duplex substrate bearing a mixture of terminal 2’, 3’-cyclic phosphate and monophosphate. Consistent with prior reports28, minimal cleavage by Top1 was observed on a 5’ overhang duplex (Figure 6B), while SDS-resistant Top1-cc was detected in the reaction with the nicked duplex substrate. Proteinase K treatment of Top1cc products yielded a series of peptide-DNA conjugates corresponding to secondary cleavage by Top1. An alkaline-resistant ligation product resulting from Top1-catalyzed ligation following secondary cleavage was also observed (Figure 6B)8. Hence the formation of a 5’ DNA gap may dictate the type of 3’ end processing and the level of slippage mutagenesis following Top1-catalyzed cleavage at rNMP sites. Notably, the purified peptide-DNA conjugates derived from Top1cc can be processed by Apn2 to yield clean 3’-OH ends, which reinforces the role of Apn2 in Top1cc removal (Figure 6C).
Figure 6. exo1∆ suppresses the lethality of pol2-M644G rnh201∆ apn2∆.
A. Tetrad analysis results of pol2-M644G rnh201Δ apn2Δ exo1Δ heterozygote diploid cells. Genotypes of spores are indicated. B. Top1-catalyzed cleavage on duplex DNA with either a recessed 3’- end or a nick where the terminal ribonucleotide is attached by a mixture of either a 2’, 3’ –cyclic phosphate or a monophosphate. C. Removal of Top1-cc derived peptide conjugates by Apn2 (100 nM) or Tdp1/Tpp1 (25 nM each).
Discussion
Blocked 3’ ends including protein-DNA conjugates, damaged bases and oxidation of replication forks arise from a wide range of cellular events 20,29,30. The blocked 3’ end, if not processed, could interfere with downstream enzymatic processes and thus be lethal or mutagenic. Herein, we discovered that Apn2 is specialized to remove multiple types of 3’ adducts on DNA terminus, particularly those associated with Top1 cleavage at genomic rNMPs 7,16. We also analyzed the biochemical attributes of Apn2 in processing 3’ ends blocked by phosphate or peptide adducts. Our findings reveal a new role of Apn2 in genome maintenance and mutation avoidance, and shed light on the types of genome stability associated with various blocked 3’ ends produced following Top1 cleavage at rNMPs in yeast.
Apn2 was originally identified as a back-up AP endonuclease to Apn1 31,32. Subsequent studies, however, uncovered 3’ nuclease and phosphodiesterase activities of Apn2, which are stimulated upon its binding to PCNA 27,29,30. Using biochemical approaches, we discovered several new Apn2 substrates. Our results suggest that Apn2 is uniquely capable of removing pTyr-DNA conjugates to generate 3’ hydroxyl ends that are substrates for DNA synthesis and ligation without further modification steps. This activity contrasts to that of Tdp1, whose cleavage produces yet another blocked 3’ phosphate end that requires Tpp1-dependent processing33. Interestingly, the 3’ phosphate generated by Tdp1 is resistant to Apn2. Thus, Apn2 and Tdp1-Tpp1 likely function as independent pathways in Top1cc removal. Furthermore, the presence of a terminal 2’, 3’ cyclic phosphate, but not a monophosphate, triggers Apn2 nuclease to remove two terminal nucleotides from the 3’ end. Taken together, we have demonstrated that Apn2 is a highly versatile nuclease against a diverse set of 3’ blocked ends. Enzymatic activities of Apn2 are likely modulated according to the types of substrates and interacting proteins to optimize their functions under different physiological conditions.
Our results show that apn2∆ is synthetic lethal with pol2-M644G rnh201∆ in a Top1-dependent manner, indicating that Apn2 plays a pivotal role in Top1-dependent rNMP removal. Since purified Apn2 processes both Top1-cc and 2’, 3’ cyclic phosphate termini, the essential function of Apn2 is likely derived from processing one or both types of adducts. Several lines of evidence, depicted below, suggest that the 2’, 3’ cyclic phosphate, not the Top1cc, is the lesion responsible for the lethality observed in pol2-M644G apn2∆ rnh201∆. First, Top1cc can be removed by other enzymes than Apn2, such as the Tdp1/Tpp1 pathway. However, deletion of TDP1 or TPP1 does not impair viability of pol2-M644G rnh201∆ cells. In contrast, Apn2 is uniquely capable of removing the terminal 2’, 3’ cyclic phosphate. Second, the synthetic lethality of apn2∆ or srs2∆ in pol2-M644G rnh201∆ suggests that Srs2 and Apn2 might operate at two different ends of the same intermediate, where the deletion of EXO1, which functions with Srs2 helicase in gap generation, partially offsets the lethality of pol2-M644G rnh201∆ apn2∆ mutant13. Alternatively, we surmise that the terminal 2’, 3’ cyclic phosphate may be removed by Apn2 or by Top1-catalyzed second cleavage 8,9,12. The formation of a gapped duplex by Srs2 and Exo1 suppresses secondary Top1 cleavage, making Apn2 the sole enzyme capable of removing the 2’, 3’ cyclic phosphate from a gapped DNA. Deletion of EXO1 may allow cells to use these other options, rendering the Apn2 pathway partially dispensable for end processing in Top1-mediated rNMP removal. These results suggest that Srs2/Exo1-mediated gap formation may act as the default pathway, and that secondary Top1 cleavage operates only when 5’ gap formation is defective or not yet initiated.
Although Tdp1 doesn’t process the 2’, 3’ cyclic phosphate terminated end, it plays a role in the avoidance of slippage mutations, presumably through the removal of Top1-cc following the second Top1 cleavage (14 and Supplementary Figure 5). apn2∆ also led to an increased slippage mutation rate, which may be attributed to Apn2’s activity in Top1cc removal. Alternatively, Apn2-catalyzed 2’, 3’ cyclic phosphate removal might suppress slippage mutagenesis by inhibiting Top1 cleavage because the presence of a terminal 2’, 3’ cyclic phosphate at a nick stimulates Top1-catalyzed secondary cleavage 8. Taken together, our results suggest that 5’ gap formation could dictate the type of blocked 3’ end processing and thereby modulate repair outcomes and the associated mutagenesis (Supplementary Figure 5).
We discovered that PCNA stimulates Apn2’s ability to remove a terminal 2’, 3’ cyclic phosphate via dinucleotide cleavage. Intriguingly, apn2-∆PIP, a PCNA interaction-deficient variant, fully supports viability of pol2-M644G rnh201∆ cells and does not alter the frequency of Top1-dependent slippage mutagenesis (Supplementary Table 1 and data not shown). These results challenge the requirement for PCNA-dependent stimulation of Apn2 in Top1-mediated rNMP removal. We note, however, that a previous study 27 suggested two different modes of binding between Apn2 and PCNA that depend on whether PCNA is loaded onto DNA. The PIP box motif in Apn2 is largely responsible for the interaction between Apn2 and DNA-free PCNA, which may explain the moderate defect of apn2-PIP mutant in PCNA-stimulated 3’-digestion. Similar to prior reports 27, the stimulation of Apn2 on 2’, 3’ cyclic phosphate end processing observed in our system also requires DNA-bound PCNA (data not shown). Most recently, another PCNA-binding domain of Xenopus Ape2 was identified that promotes exonuclease activity by Ape234. More detailed analyses of PCNA-interacting interfaces and their contributions to Apn2 activities will help determine the nature of Top1-induced blocked 3’ ends and their cellular levels.
Apn2 is an evolutionarily conserved nuclease with homologues found from bacteria to human cells 31,35,36. APE2 (the mammalian Apn2 homolog) knockout mice exhibit growth retardation and mild dyslymphopoiesis 37. APE2 deficiency also leads to defects in class switch recombination 38. In mammals, APE2 activates Chk1 by degrading DNA and generating single stranded DNA at sites of oxidative damage 35,39. However, the basis of this host of symptoms and phenotypes in APE2 deleted cells and organisms is not yet known. It will be important to test whether APE2 has equivalent functions in healing 3’ blocked ends in mammals and whether some of the defects associated with APE2 deficiency could be attributed to elevated genome instability caused by the lack of enzymatic activities we have elucidated here.
Materials and Methods.
Strains.
Strains used for the experiments are listed in Supplementary Table 2. Gene deletions were created by transforming drug-resistant, PCR-derived DNA fragments of the KANMX, CLONAT, HYGROMYCIN B or PCR-derived, yeast metabolic marker genes URA3, TRP1 or HIS3 into the corresponding strains. Gene sequence specific mutation was generated by two-step transformation as described 40. Genomic mutations were verified by PCR and restriction digestion, and DNA sequencing.
lys2 reversion assays.
The Lys+ rate in lys2ΔA746NR(AG)4 reporter cells was determined as described previously4,13. At least 12 independent cultures per isolate (2~3 independent isolates per genotype) were used to determine the rates, and 95% confidence intervals were determined in fluctuation assay. 40~120 Lys+ colonies growing on lysine dropout plate were randomly chosen to verify the types of mutations by sequencing the region flanking the (AG)4 repeat. Primers (5’-AGTTTTTCTAACAAATACGCTGTCG-3’ and 5’-CGCATTATTACTGGTATCCAAATTC-3’) were used to amplify region flanking the inserted (AG)4 repeat.
Drug Sensitivity assay.
Yeast cells with indicated gene deletion or mutation were cultured in YEPD medium for 24~36 h to reach the density of ~1×108 cells/ml, ~2×106 cells were transferred to 200 μl sterilized distilled water, and applied to 1:5 serial dilution. Cells with different density were spotted on to YEPD agar plate with indicated concentration of camptothecin (CPT) and kept at 30°C for 48 ~72 hours.
Expression and Purification FLAG-His6-Top1 and FLAG-His6-top1-T722A from insect cells.
Top1 coding sequence was PCR amplified from yeast genomic DNA (S288C) and cloned into a modified pFASTBac-HTB vector 41 and fused in frame to the FLAG and His6 tags in this vector. Using this construction, a bacmid was generated from the E.coli strain DH10Bac (Invitrogen) to express Top1 in insect cells. For Top1 purification, The insect cell pellet (~15 g from 1 L culture) was resuspended in 100 ml of K buffer (20 mM KH2PO4, pH 7.4, 10% glycerol, 0.5 mM EDTA, 0.01% NP-40, 1 mM DTT) with a cocktail of protease inhibitors (aprotinin, chymostatin, leupeptin, and pepstatin A at 5 μg/ml each, and also 1 mM phenyl-methylsulfonyl fluoride) and 500 mM KCl. Cells were disrupted by sonication for 30 seconds, and the lysate was clarified by centrifugation (20,000 g for 20 min). The supernatant was filtered through Whatman glass microfiber filter (GE Healthcare) and loaded onto a 3 mL Ni-NTA column (Thermo Fisher) using a BioRad Econo pump. The column was washed with 60 mL K buffer containing 500 mM KCl and 15 mM imidazol. Top1 was eluted with 20 mL K buffer containing 500 mM KCl and 200 mM imidazol. The eluate was further incubated with 0.5 mL anti-FLAG M2 agarose resin (Sigma) for 2 hrs. The matrix was washed 40 ml K buffer containing 500 mM KCl and 0.1% NP-40 before Top1 was eluted with 1 ml of K buffer containing 200 μg/ml FLAG peptide. The eluate was filter dialyzed to remove excess FLAG peptide, further concentrated in an Ultracel-30K concentrator (Amicon). The purified Top1 protein (~50 μg) was frozen in liquid nitrogen and stored at −80°C as small aliquots. To express top1-T722A, T722A mutation was introduced via site-directed mutagenesis. The mutant protein was expressed and purified similarly as wild-type Top1 with a slightly better yield (~100 μg).
Expression and Purification of N-terminal His6 tagged Apn1, Apn2, Tdp1 and Tpp1 from E.coli.
To express, purify and systematically characterize 3’ – adduct cleaning enzymes (Apn1, Apn2, Tdp1 and Tpp1), DNA fragment encoding individual enzymes was amplified from yeast genomic DNA (S288C) and fused in frame to the His6 tag in pETDuet1. These four enzymes were expressed in E. coli and purified similarly. For their expression, BL21-codon plus strain was transformed with respective expression vector. An overnight culture was diluted 1:50 into 12 L LB medium containing 100 mg/L Ampicilin and 37 mg/L Chloramphenicol. Expression of the target protein was induced overnight at 16°C with 0.2 mM IPTG added when the culture reached OD600=0.8. Cells were harvested via centrifugation, which yields a 60 g or so cell pellet for each protein expressed. For their purification, the respective cell pellet was resuspended in 240 ml of K buffer containing 150 mM KCl and lysed by sonication (eight times of 15 sec with 15 sec pulse in between). After sample clarification by centrifugation (20,000Xg), the lysate was loaded onto a 5 ml SP sepharose column. The column was washed with 100 ml K buffer plus 150 mM KCl before it was developed with a 100 ml linear gradient from 150 to 575 mM KCl. The fractions containing the target protein were pooled and incubated with 0.5 ml of Ni-NTA resin (Thermo Fisher) for 2 hrs. The matrix was washed with 30 ml K buffer with 500 mM KCl and 15 mM imidizole. The target proteins were then eluted with K buffer containing 500 mM KCl and 200 mM imidizole. The eluate was filter dialyzed to remove excess imidizole using either Ultracel-10K or Ultracel-30K concentrator according to the target protein’s molecular weight. The purified proteins were further concentrated, frozen with liquid nitrogen and stored at 80°C. This procedure yields ~ 100 μg of Apn1, ~ 100 μg of Apn2, ~ 500 μg of Tdp1 and ~ 1 mg of Tpp1. The full length of His6-Apn2 is subjected to proteolysis at its carboxyl-terminus. Thus, the yeast expression system is further employed to express and purify Apn2.
Expression and Purification of Apn2-FLAG from S. cerevisiae.
For yeast expression of Apn2, the coding region of APN2 was cloned between the Not1 and SpeI sites in pESC-URA (Agilent), placing the gene under the control of the GAL10 promotor with a FLAG tag fused to the carboxyl terminus of APN2. Apn2 protein was expressed in the protease-deficient yeast strain 334 (MATα pep4-3 prb1-1122 ura3-52 leu2-3,112 reg1-501 gal1) (kind gift from Dr. Nancy Hollingsworth, Stony Brook University). For Apn2 expression, an overnight culture was diluted 1:50 into 12 L of –ura omission medium with 2% glucose. Cells were cultured at 30°C until the OD660 reached 0.8, at which time galactose was added to reach 2% final concentration to induce Apn2 expression. Cells were harvested by centrifugation after 12 hrs of further incubation, and the pellet (50 g) was stored frozen at −80°C. After grinding the yeast pellet with dry ice, 50 ml of K buffer (20 mM KH2PO4, pH 7.4, 10% glycerol, 0.5 mM EDTA, 0.1% NP-40, 1 mM βME) with a cocktail of protease inhibitors (aprotinin, chymostatin, leupeptin, and pepstatin A at 5 μg/ml each, and also 1 mM phenyl-methylsulfonyl fluoride) and 150 mM KCl was added to the extract. All the subsequent steps were carried out at 0-4°C. The extract was clarified by centrifugation (20,000Xg for 20 min) and then applied onto a 5 ml column of SP agarose (GE). The column was washed with 100 ml K buffer with 150 mM KCl. Apn2 was eluted with 30 ml of K buffer with 500 ml KCl, and the eluate was mixed gently with 0.4 ml anti-FLAG-M2 resin for 2 hrs. After washing the matrix with 30 ml of K buffer with 500 mM KCl and 0.1% NP-40 and 10 ml of K buffer with 500 mM KCl, Apn2 was eluted with 1.5 ml of K buffer plus 500 mM KCl and 200 μg/ml FLAG peptide for 1 hr. The eluate that contained purified Apn2 was filter dialyzed and concentrated using an Ultracel-30K concentrator (Amicon) to 1 mg/ml and stored at −80°C in aliquots. The overall yield of Apn2 was ~100 μg.
The apn2-E59A point mutation was introduced into pESC-URA-APN2 vector by site-directed mutagenesis and the mutant was expressed and purified similarly as wild type. The yield of apn2-E59A was lower than wild type (~ 5 μg from 12 L culture).
Yeast DNA Polymerase δ, RFC, PCNA, RNase H2 and RPA.
The yeast DNA Polymerase δ complex, containing FLAG-Pol3, GST-Pol31 and Pol32, was expressed in yeast protease deficient strain BJ5464 (MATα pep4::HIS3 prb1∆1.6R ura3-52 trp1 leu2∆1 can1 GAL) using pBJ1445 and pBJ1524 (kind gift from Patrick Sung, Yale University). Purification was conducted similarly as described 42. RFC complex (Gst-Rfc1, Rfc2, Rfc3, Rfc4, Rfc5) was also expressed in yeast strain BJ5464 using pBJ1476 (2μ, GAL-PGK-GST-RFC1/RFC4/RFC5, LEU-2d) and pBJ1469 (2μ, GAL-PGK-RFC2/RFC3, TRP1) (kind gift from Patrick Sung, Yale University) and purified similarly as described 42. PCNA was expressed in E. coli using pQE16-Pol30 (kind gift from Patrick Sung, Yale University) and purified similarly as described 42. RNase H2 complex was affinity purified from a yeast strain from the “Yeast GST Fusion Collection” (Horizon/Dharmacon), where the GST tag is fused in-frame at the N-terminus of Rnh201. Single strand DNA binding protein RPA was expressed in yeast and purified similarly as described before 43.
DNA substrates.
Generating 2’, 3’ cyclic phosphate terminated substrates.
Oligos used to construct substrates for biochemical assays were listed in Supplementary Table 3. To generate 2’, 3’ cyclic phosphate terminated nicked duplex, duplex DNA with a single embedded ribonucloetide was treated with top1-T722A, a ligation defective mutant of Top1 that generates about three-fold more cyclic phosphate terminated nicks compared to wild-type Top1 (Supplementary Figure 3C). Using duplex DNA from annealing H1 to H2 and H3 to H4, two nicked duplex substrates were created with DNA fragment harboring 2’, 3’-cyclic phosphate end 12 nt and 27 nt in length respectively. To generate partially 2’, 3’ cyclic phosphate terminated oligos to test exonuclease activity of Polδ and Apn2 nuclease activity on different structured DNA, a 14 nt long oligo (H5) with two consecutive ribonucloetides and a protective deoxyribonucleotide at the 3’ end was partially digested by RNase If (New England Biolabs) for 5 minutes at 37°C, which yields ~ 40-60% of 2’, 3’ cyclic phosphate terminated ends that are resistant to alkaline phosphatase (New England Biolabs).
DNA substrates with other end structures.
Oligos with a terminal deoxyribonucleotide (H6 and H7), a terminal 3’-phosphate (H8 and H9), a terminal ribonucloetide (H10 and H11) were purchased from IDT. Oligos with a 3’-pTyr adduct (H12 and H13) were synthesized by Midland. To create respective 5’-overhanging with a 12 base pair or 27 base pair duplex region, H6, H8, H10 or H12 was annealed to H1 and H7, H9, H11 or H13 was annealed to H3. H14 was added to the annealing reaction respectively to create the corresponding nicked duplex substrates. To block one end of the H3 containing duplex, biotin was engineered to 3’ end of H3 to give the oligo H15.
Apn2 nuclease assays
For Apn2 nuclease assays, unless specified, DNA substrate (10 nM) was incubated with the indicated amount of Apn2 in the reaction buffer (20 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 1 mM DTT, 200 ng/ml BSA, 50 mM KCl) for 10 min at 30°C. Reaction involving PCNA was conducted in a similar reaction buffer except the concentration of KCl was raised to 100 mM. The reaction was stopped by treatment with SDS (0.2% final) and proteinase K (0.5 mg/ml) for 10 min at 37°C. Equal volume of loading dye containing 85% formamide with 25 mM EDTA, and 1 μM unlabeled H1 (ribo-containing strand) was then added to reaction mixtures preventing the re-annealing of the radiolabeled H1 to its complementary strand. The mixture was heated at 95 °C for 5 min before fractionated in a 19% or 12% denaturing polyacrylamide gel in TBE (Tris-Boric Acid-EDTA) buffer for 12 nt or 27 nt substrate respectively, and followed by phosphor imaging analysis. Other nucleases, viz. Apn1, Tdp1, Tpp1, were assayed similarly.
Polδ Extension Assays.
The 2’, 3’ cyclic phosphate terminated nicked duplex was made as described above. In brief, 20 nM duplex DNA with a single ribonucleotide embedded was incubated with 10 nM yeast Top1-T722A for 10 min at 30°C in 10 μl buffer (20 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 100 mM KCl, 1 mM ATP). We note that either 0.1 mM of each of the four dNTPs were added to the reaction to allow full extension by Polδ or only dATP, dGTP and dTTP were added to trap a 6 nucleotide long extension product for quantification purposes. Following the ribonucleotide cleavage reaction, 2 μl protein mix with the indicated components from Apn2, RFC, PCNA, RPA, Exo1, Polδ was added to the reaction and incubated for additional 10 min at 30°C. The reaction was stopped by adding 12 μl stopping buffer containing 85% formamide with 0.4% SDS, 25 mM EDTA, and 1 μM unlabeled H1 (ribonucleotide-containing strand). The reaction mixtures were heated at 75°C for 5 min and resolved in 12% denaturing polyacrylamide gel before subjected to phosphorimaging analysis.
Statistics and Reproducibility
Each experiment was repeated 3-5 times independently with similar results.
Data Availability:
The data that support the findings in this work are available from the corresponding authors upon request.
Supplementary Material
Acknowledgements:
We are grateful to Patrick Sung, Nancy Hollingsworth, Nayun Kim, Hannah Klein, Thomas Kunkel, and Sue Jinks-Robertson for providing plasmids and yeast strains, Hayley Bedwell for technical support, Steve Bell and Brian Calvi for critical reading of the manuscript. This work was supported by William and Ella Owens Medical Research Foundation, Nathan Shock Center Pilot grant, and NIH research grant GM71011 to S.E.L., ThriveWell Foundation to E. Y. S., and GM124765 to H. N.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Jinks-Robertson S & Klein HL Ribonucleotides in DNA: hidden in plain sight. Nat Struct Mol Biol 22, 176–8 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Williams JS & Kunkel TA Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst) 19, 27–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark AB, Lujan SA, Kissling GE & Kunkel TA Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst) 10, 476–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim N et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332, 1561–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigby RE, Leitch A & Jackson AP Nucleic acid-mediated inflammatory diseases. Bioessays 30, 833–42 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Schellenberg MJ, Tumbale PP & Williams RS Molecular underpinnings of Aprataxin RNA/DNA deadenylase function and dysfunction in neurological disease. Prog Biophys Mol Biol 117, 157–165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol 6, 774–81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sparks JL & Burgers PM Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides. EMBO J 34, 1259–69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang SY, Ghosh S & Pommier Y Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J Biol Chem 290, 14068–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekiguchi J & Shuman S Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell 1, 89–97 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Cho JE, Kim N, Li YC & Jinks-Robertson S Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA Repair (Amst) 12, 205–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuman S Polynucleotide ligase activity of eukaryotic topoisomerase I. Mol Cell 1, 741–8 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Potenski CJ, Niu H, Sung P & Klein HL Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature 511, 251–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu H, Potenski CJ, Epshtein A, Sung P & Klein HL Roles of DNA helicases and Exo1 in the avoidance of mutations induced by Top1-mediated cleavage at ribonucleotides in DNA. Cell Cycle 15, 331–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiorani P & Bjornsti MA Mechanisms of DNA topoisomerase I-induced cell killing in the yeast Saccharomyces cerevisiae. Ann N Y Acad Sci 922, 65–75 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Williams JS et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell 49, 1010–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzder SN et al. Requirement of yeast Rad1-Rad10 nuclease for the removal of 3’-blocked termini from DNA strand breaks induced by reactive oxygen species. Genes Dev 18, 2283–91 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillet M & Boiteux S Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J 21, 2833–41 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vance JR & Wilson TE Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci U S A 99, 13669–74 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Pouliot JJ & Nash HA Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc Natl Acad Sci U S A 99, 14970–5 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng C, Brown JA, You D & Brown JM Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170, 591–600 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton NK & Maizels N MRE11 function in response to topoisomerase poisons is independent of its function in double-strand break repair in Saccharomyces cerevisiae. PLoS One 5, e15387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elford HL Effect of hydroxyurea on ribonucleotide reductase. Biochem Biophys Res Commun 33, 129–35 (1968). [DOI] [PubMed] [Google Scholar]
- 24.Nick McElhinny SA, Stith CM, Burgers PM & Kunkel TA Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem 282, 2324–32 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Megonigal MD, Fertala J & Bjornsti MA Alterations in the catalytic activity of yeast DNA topoisomerase I result in cell cycle arrest and cell death. J Biol Chem 272, 12801–8 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Das U & Shuman S Mechanism of RNA 2’,3’-cyclic phosphate end healing by T4 polynucleotide kinase-phosphatase. Nucleic Acids Res 41, 355–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unk I et al. Stimulation of 3’-->5’ exonuclease and 3’-phosphodiesterase activities of yeast Apn2 by proliferating cell nuclear antigen. Mol Cell Biol 22, 6480–6 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christiansen K, Svejstrup AB, Andersen AH & Westergaard O Eukaryotic topoisomerase I-mediated cleavage requires bipartite DNA interaction. Cleavage of DNA substrates containing strand interruptions implicates a role for topoisomerase I in illegitimate recombination. J Biol Chem 268, 9690–701 (1993). [PubMed] [Google Scholar]
- 29.Vance JR & Wilson TE Repair of DNA strand breaks by the overlapping functions of lesion-specific and non-lesion-specific DNA 3’ phosphatases. Mol Cell Biol 21, 7191–8 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unk I, Haracska L, Prakash S & Prakash L 3’-phosphodiesterase and 3’-->5’ exonuclease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Mol Cell Biol 21, 1656–61 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson RE et al. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev 12, 3137–43 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unk I, Haracska L, Johnson RE, Prakash S & Prakash L Apurinic endonuclease activity of yeast Apn2 protein. J Biol Chem 275, 22427–34 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Pouliot JJ, Yao KC, Robertson CA & Nash HA Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286, 552–5 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Lin Y et al. APE2 promotes DNA damage response pathway from a single-strand break. Nucleic Acids Res 46, 2479–2494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willis J, Patel Y, Lentz BL & Yan S APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc Natl Acad Sci U S A 110, 10592–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribar B, Izumi T & Mitra S The major role of human AP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe. Nucleic Acids Res 32, 115–26 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ide Y et al. Growth retardation and dyslymphopoiesis accompanied by G2/M arrest in APEX2-null mice. Blood 104, 4097–103 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Guikema JE et al. Apurinic/apyrimidinic endonuclease 2 is necessary for normal B cell development and recovery of lymphoid progenitors after chemotherapeutic challenge. J Immunol 186, 1943–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallace BD et al. APE2 Zf-GRF facilitates 3’–5’ resection of DNA damage following oxidative stress. Proc Natl Acad Sci U S A 114, 304–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li F et al. Role of Saw1 in Rad1/Rad10 complex assembly at recombination intermediates in budding yeast. EMBO J 32, 461–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niu H et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467, 108–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson MA et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature 502, 393–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Komen S, Macris M, Sehorn MG & Sung P Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol 408, 445–63 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings in this work are available from the corresponding authors upon request.