Abstract
Objective
To expand the clinical spectrum of lysyl-tRNA synthetase (KARS) gene–related diseases, which so far includes Charcot-Marie-Tooth disease, congenital visual impairment and microcephaly, and nonsyndromic hearing impairment.
Methods
Whole-exome sequencing was performed on index patients from 4 unrelated families with leukoencephalopathy. Candidate pathogenic variants and their cosegregation were confirmed by Sanger sequencing. Effects of mutations on KARS protein function were examined by aminoacylation assays and yeast complementation assays.
Results
Common clinical features of the patients in this study included impaired cognitive ability, seizure, hypotonia, ataxia, and abnormal brain imaging, suggesting that the CNS involvement is the main clinical presentation. Six previously unreported and 1 known KARS mutations were identified and cosegregated in these families. Two patients are compound heterozygous for missense mutations, 1 patient is homozygous for a missense mutation, and 1 patient harbored an insertion mutation and a missense mutation. Functional and structural analyses revealed that these mutations impair aminoacylation activity of lysyl-tRNA synthetase, indicating that defective KARS function is responsible for the phenotypes in these individuals.
Conclusions
Our results demonstrate that patients with loss-of-function KARS mutations can manifest CNS disorders, thus broadening the phenotypic spectrum associated with KARS-related disease.
Aminoacyl-tRNA synthetases (ARSs) play key roles in charging specific tRNAs with cognate amino acids and are critical for enabling protein translational fidelity and cellular integrity. Pathogenic mutations in different ARSs have been reported in patients with a variety of clinical presentations including cardiomyopathy, cancer, autoimmune disorders, and diabetes.1–3 Of interest, a number of mutations in genes encoding ARSs have been linked to neurologic diseases, including inherited peripheral neuropathy, sensorineural deafness, leukodystrophies, or leukoencephalopathies, summarized in table e-1, links.lww.com/NXG/A143.4–27
KARS is one of the 3 bifunctional ARSs and catalyzes the specific attachment of l-lysine to cognate tRNA molecules. Compound heterozygous disease–associated KARS mutations were first identified in a single patient with Charcot-Marie-Tooth disease.28 Later, other KARS mutations were also reported in 3 unrelated families with autosomal recessive nonsyndromic hearing impairment,19 in 1 patient with a suspected mitochondrial disorder,29 in 2 siblings with severe infantile visual loss and progressive microcephaly,15 and in a boy with combined respiratory chain complex deficiencies (I and IV).30 However, dysfunctions of CNS involved in leukoencephalopathy have not been reported or observed as a major clinical presentation in these affected individuals.
In this study, 4 unrelated nonconsanguineous families with leukoencephalopathy were identified by whole-exome sequencing analysis. Functional and structural analyses revealed that each variant was a loss-of-function KARS mutation. Our findings highlight the association of KARS mutations in patients with CNS involvement and broaden the phenotypic spectrum associated with KARS-related disease.
Methods
Standard protocol approval, registrations, and patient consents
Patient blood specimens were submitted to Baylor Genetics, previously Medical Genetics Laboratories at Baylor College of Medicine, Houston, TX, for WES-based analyses. Additional patient specimens were from Fudan University Huashan Hospital, Shanghai, China. The ethical review boards of the participating institutions approved this study.
Exome sequencing in probands and family studies
Library preparation, exome capture, HiSeq next-generation sequencing, and data analyses were conducted as described.31–33 Variants identified in KARS were further validated by Sanger sequencing in these patients. Family members were also tested to evaluate the mode of inheritance and disease segregation.
Functional studies
Aminoacylation assays in vitro were performed as previously described.28,34,35 The initial rate of aminoacylation as a function of tRNA concentration was fit to a hyperbola equation, from which the Michaelis constant (Km) for tRNA and the catalytic turnover (kcat) were derived. Analysis of the catalytic efficiency (kcat/Km) of aminoacylation was presented for each mutant enzyme.
Yeast complementation assays were performed using a haploid Saccharomyces cerevisiae strain with the endogenous KRS1 gene deleted (∆KRS1)28 in both solid and liquid -Leu-Ura media (Teknova, Hollister, CA) containing 0.1% 5-fluoroorotic acid (5-FOA) (Boeke, Trueheart et al. 1987). Each human KARS variant (GenBank accession number AAG30114.1, NM_005548) was modeled in yeast KRS1 (GenBank accession number AAA66916.1) using Gateway technology (Invitrogen). The human KARS residues p.L233, p.E427, p.R505, p.P533, p.T587, and p.L596 correspond to the following yeast residues, respectively: p.L208, p.E403, p.R480, p.P508, p.T562, and p.L571.
Results
Patient history and clinical presentations
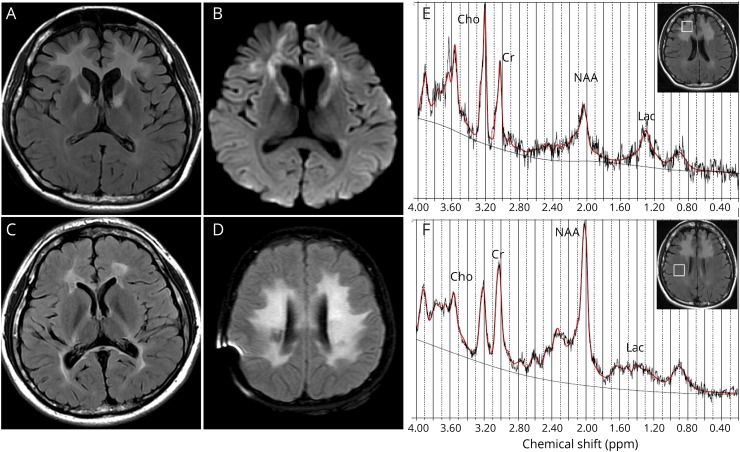
Patient 1 is a 26-year-old woman who developed progressive neurocognitive decline at age 25 years. Her key clinical features included hypotonia, mild intellectual disability, slurred speech, ataxia, and abnormal movement, as well as congenital hearing loss. Two KARS variants c.1514G>A (p.R505H) and c.1597C>T (p.P533S) were identified in the compound heterozygous state. The EMG and histochemical analysis of muscle biopsy did not reveal any myogenic or neurogenic damage. Brain MRI showed symmetric hyperintensity in bilateral frontal white matter, extending along the anterior limb of inner capsule on FLAIR and DWI (figure 1, A and B). Magnetic resonance spectroscopy was performed with Stimulated Echo Acquisition Mode sequence, and data were analyzed with the LC Model (version 6.3). The spectroscopy showed distinctly reduced N-acetylaspartate and slightly elevated lactate peak in the right frontal lesion (figure 1E) compared with that in the ipsilateral normal white matter (figure 1F). The brother of patient 1, who also harbored the same 2 variants, had hearing loss, and his brain MRI performed at age 16 years showed bilateral abnormality in the periventricular white matter (figure 1C). The parents were clinically unaffected and had normal MRI scans, and each was heterozygous for one of these 2 variants. Therefore, variants c.1514G>A (p.R505H) (from the mother) and c.1597C>T (p.P533S) (from the father) segregate with leukoencephalopathy in an autosomal recessive manner in the family of patient 1. She was deceased after 2 years of neurologic features appeared.
Figure 1. Representative cases for novel KARS mutation with leukoencephalopathy.
Characteristics of patients for novel KARS mutations with leukoencephalopathy. (A,B) Brain imaging of Patient 1 showed bilateral FLAIR and DWI signal hyperintensity in the white matter of the frontal lobe. (C) The affacted brother of Patient 1 has also abnormality in the white matter. (D) Brain MRI of Patient 4 showed bilateral T2 signal hyperintensity in the white matter of periventricular area. (E) The MRS showed reduced NAA and elevated lactate peak in the right frontal lesion of Patient 1. (F) The MRS showed the presence of NAA and lactate in the normal white matter of Patient 1.
Patient 2 is a 35-year-old man who developed progressive neurocognitive decline, hypertonia, seizures, ataxia, and abnormal movement over 3 years. Other clinical features included congenital hearing loss, likely secondary hypothyroidism and possible left eye blindness. Brain MRI revealed abnormality in the white matter, and EEG was normal. Family history indicated a sister with congenital deafness and hydrocephaly and 2 second cousins with congenital deafness. He was compound heterozygous for a novel missense variant c.881T>C (p.I294T) and a reported variant c.1760C>T (p.T587M).
Patient 3 is a 3-year-old boy present with developmental delay and regression, failure to thrive, microcephaly, and progressive hypotonia. Other clinical presentations include hearing and vision loss, nystagmus, hyperreflexia, progressive joint contractures, febrile seizures, dysphagia, renal tubular acidosis type I, mild hydronephrosis of the left kidney, and abnormal liver ultrasound. Head CT showed increasing periventricular and cerebellar nuclei calcifications and cerebral atrophy. He was compound heterozygous for a novel insertion variant c.1281_1282insAGA (p.E427_L428insR) and a novel missense variant c.1786C>T (p.L596F).
Patient 4 is an 11-year-old girl whose clinical presentations include global developmental delay, hypotonia, mild intellectual disability, congenital bilateral profound sensorineural hearing loss, history of seizure disorder, mildly elevated lactate, elevated CSF total protein, and developmental regression. Family history was remarkable for a sister with bilateral sensorineural hearing loss. Her brain MRI and CT showed normal at age 1 year. However, the second MRI at age 11 years showed extensive abnormality of the deep white matter of both cerebral hemispheres with increased T2 hyperintensity and involvement of the corticospinal tracts with sparing of the subcortical U fibers (figure 1D). Mild cerebellar volume loss with prominence of the fourth ventricle was also observed. The patient died at age 12 years after a rapid deterioration in her neurologic status over a period of 2 years. Her autopsy showed severe bilateral spongiform leukodystrophy involving the internal capsule, frontal, parietal, and occipital white matter. There was symmetric white matter loss in the descending corticospinal tracts, brainstem, and spinal cord. Dystrophic calcifications were noted in basal ganglia, frontal, and parietal lobes. She harbored a homozygous missense variant c.697C>G (p.L233V). Her mother was found to be heterozygous for this variant. But the father's sample was not enough for further testing.
Computational evaluation of KARS variants
Six novel and 1 known KARS variants were identified by WES in the index patients described here (table). All variants are rare, and none is found in the homozygote state in the ExAC database (table e-2, links.lww.com/NXG/A144). Sequence alignments of KARS proteins from bacteria to human showed that all the affected amino acids are highly conserved (figure 2F). A variety of in silico prediction programs were used to predict the possible effect of each amino acid substitution (table e-2). All 8 novel variants were considered as likely pathogenic, based on predictions by PolyPhen-2, Sorting Intolerant from Tolerant, MutationTaster, MutationAssessor, etc. (table e-2).
Table.
Summary of key clinical manifestations of affected individuals
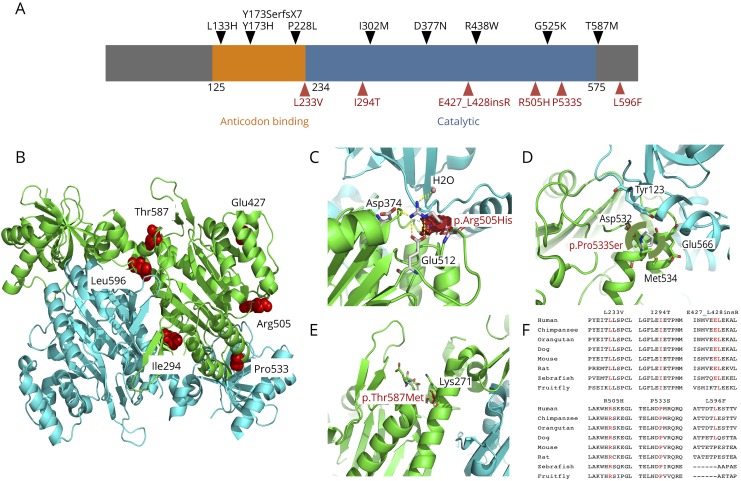
Figure 2. Summary of KARS mutations.
(A) Schematic representation of the KARS gene and the distribution of published mutations (black, above) and mutations found in our patient cohort (red, below). (B) Ribbon diagram of the complex structure model of human lysyl-tRNA synthetase (PDB ID: 3BJU) and mapping the missense mutations onto the structure model. One monomer colored in cyan and the other one in green. The mutation is drawn as a ball-and-stick model and colored in red. (C) Close-up view of the in silico analysis for mutation p.Arg505His. (D) View for mutation p.Pro533Ser. (E) View for mutation p.Thr587Met. (F) Cross species sequence alignment of amino acids. The corresponding positions are indicated in red text.
Functional studies
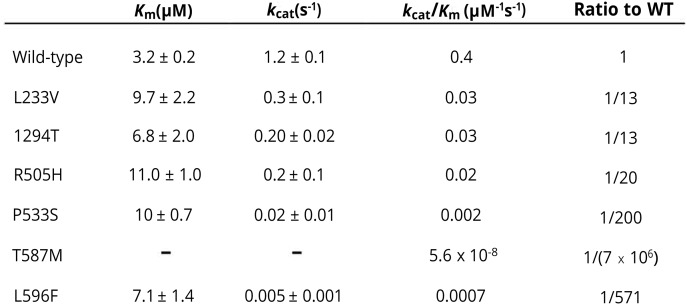
As shown in figure 3, all KARS mutations studied (p.L233V, p.I294T, p.R505H, p.P533S, p.T587M, and p.L596F) reduced enzyme kinetics by at least 13-fold compared with that of wild-type KARS, indicating that these mutations impaired KARS aminoacylation activity.
Figure 3. Summary of the mutations identified in KARS and their effects on tRNA charging.
The column “ratio to WT” indicates the decrease in tRNA charging relative to the WT enzyme for each mutant. All these mutations have a deleterious effect, ranging from 13- to nearly 107-fold.
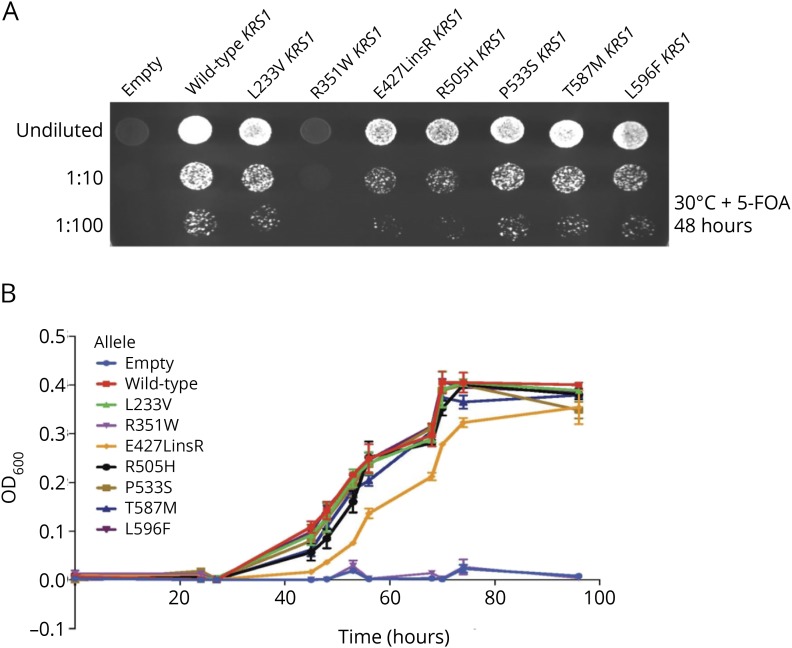
To further evaluate the deleterious effects of KARS mutations, yeast complementation assays were performed by modeling each KARS variant in the S. cerevisiae ortholog KRS1. As shown in figure 4A, yeast expressing p.E427_Lins428R and p.R505H KRS1 demonstrated dramatically reduced, but not ablated, growth. In addition, yeast expressing p.L233V, p.P533S, p.T587M, and p.L596F KRS1 showed a slight but significant reduction of yeast viability compared with wild-type KRS1. These results suggested that p.L233V, p.P533S, p.T587M, p.L596F, p.R505H, and p.E427_Lins428R are hypomorphic alleles. Similar results were obtained from growth curve analyses in liquid media containing 5-FOA (figure 4B).
Figure 4. Yeast complementation analysis of mutant KRS1 alleles.
(A) A haploid yeast strain deleted for endogenous KRS1 was transformed with a LEU2-bearing pRS315 vector containing wild-type KRS1, the indicated mutant form of KRS1, or no insert (“Empty”). Cultures for each strain (labeled along the top) were grown for 2 days in liquid medium and spotted on solid medium containing 5-FOA to determine whether the KRS1 alleles complement loss of KRS1 at 30°C. (B) Cultures for each indicated strain (labeled at right) were grown for 2 days in liquid medium and then diluted in liquid medium containing 5-FOA to determine whether the KRS1 alleles complement loss of KRS1 at 30°C. The optical density (OD600, y-axis) was evaluated for each culture at the indicated time points (x-axis), and error bars indicate SD. 5-FOA = 5-fluoroorotic acid.
Mechanisms of pathogenicity for impaired function of mutated residues
Patient 1 harbors 2 novel KARS missense variants (c.1514G>A [p.R505H] and c.1597C>T [p.P533S]) in the compound heterozygous state. Close examination of the crystal structure revealed that Arg505 forms a hydrogen-bond network with Asp374 and Glu512 of KARS and an adjacent water molecule.36 Replacement with the side chain of His would eliminate the positively charged side chain of Arg and thus impair their hydrogen-bond network (figure 2, B and C). Pro533 is located at the transition point of a helix to a β-strand of KARS protein consisting of amino acids 520–545 to form dimeric interface. The replacement with Ser would not permit the required peptide bond turn of Pro and thus would be expected to have a detrimental effect on the protein secondary structure. Consistent with the predictions from structural analysis, both mutations decreased in tRNA charging activity by 20- and 200-fold relative to the wild-type enzyme, respectively (figure 3). Of interest, although p.P533S mutation caused a 10-fold reduction relative to p.R505H, p.R505H mutation showed a more severe reduction in cellular growth in yeast complementation assays. Thus, these 2 mutations identified in this patient affect critical amino acids and reduce tRNA charging capacity, suggesting a pathogenic role in causing the patient phenotype.
Patient 2 has a novel variant c.881T>C (p.I294T) and a known mutation c.1760C>T (p.T587M). The Ile294 residue forms a hydrophobic interaction with Leu597 located within the same monomer to form the dimeric interface. Replacement of Ile294 with Thr introduces an extra hydroxyl group, which may alter the nature of hydrophobic core. The p.I294T mutant showed a reduction in tRNA charging by 13-fold relative to the wild-type enzyme (figure 3). The p.T587M variant, a previously reported mutation, almost completely eliminated the enzyme activity.
Patient 3 has a novel c.1281_1282insAGA (p.E427_L428insR) insertion mutation and a novel c.1786C>T (p.L596F) missense mutation. Structural analysis indicated that Glu427 and Leu428 are located in a helix tightly packed against another helix consisting of residues 468–484. The insertion of a bulky amino acid Arg into the middle of the first helix will disrupt its secondary structure, affecting the enzyme stability. In addition, Leu596 is embedded in the protein interior and packed tightly with adjacent hydrophobic residues. The replacement with Phe introduced a large bulky side chain creating stereochemical clashes and causing protein instability. Yeast expressing p.E427L_ins428R KRS1 showed a severe reduction of yeast viability compared with wild-type strain, indicating that it is a loss-of-function allele, whereas the p.L596F mutation caused a slight but significant reduction in yeast viability. On the other hand, the tRNA charging activity of p.L596F mutant is reduced by 571-fold, with a 2.2-fold increase in Km, but nearly a 240-fold reduction in kcat (figure 3).
Patient 4 has a single nucleotide variation, c.697C>G (p.L233V), and a copy number loss of KARS at this locus, which were inherited from each parent respectively. The Leu233 is located in the anticodon binding domain. This variant reduced enzyme activity by 13-fold relative to the wild-type protein and slightly decreased yeast viability in yeast complementation studies (figures 3 and 4). Thus, the abnormal kinetic aspect of this mutation affected the catalytic behavior of the enzyme and may likely contribute to disease pathogenesis.
Discussion
KARS mutations have been linked to neurologic disorders with different clinical manifestations. Compound heterozygous mutations p.L133H and p.Y173Sfs*7 were first identified in a patient with Charcot-Marie-Tooth disease.28 Two mutations p.D377N and p.Y173H were found in the homozygous state in patients with autosomal recessive nonsyndromic hearing impairment.19 Compound heterozygous mutations p.P228L and p.T587M were identified in a patient presenting with development delay, hypotonia, and ophthalmoplegia.29 Meanwhile, patients with congenital visual impairment and progressive microcephaly were reported to be compound heterozygous for mutations p.R438W and p.E525K.15 Recently, compound heterozygous mutations (p.V448D and p.I318T) were identified in a boy with combined mitochondrial complex deficiencies.30 In this study, we identified 6 novel KARS mutations in patients with leukoencephalopathy. We also found that the previously reported mutation p.T587M is associated with leukoencephalopathy. Each mutation exerts a loss-of-function effect in at least 1 of the following assays: aminoacylation assay and yeast complementation assay, suggesting that defective KARS charging function is an important component of leukoencephalopathy pathogenesis in our patients. Our results are consistent with the notion that impaired enzyme function is a common characteristic of disease-associated ARSs mutations.
In the patients examined here, CNS involvement is the main clinical presentation. Common clinical features include impaired cognitive ability, seizure, hypotonia, and ataxia. Brain MRI or CT revealed abnormalities in the white matter in all patients. Our results suggest that leukoencephalopathy in these patients is caused by KARS defects and that there are novel clinical features from the previously described KARS-associated neurologic diseases.
Like other bifunctional ARSs, KARS has cytoplasmic and mitochondrial isoforms, which result from alternative splicing of the first 3 exons.36,37 Previously, KARS mutations have been implicated in peripheral neuropathies and sensorineural diseases. In this study, we showed that KARS is also involved in CNS diseases. The mechanisms underlying the tissue specificity of KARS-associated neurologic disorders are unclear. To date, 4 cytoplasmic ARSs (YARS, AARS, HARS, and MARS), but not their mitochondrial counterparts (YARS2, AARS2, HARS2, and MARS2), are associated with peripheral neuropathy, indicating that this disease might be caused by dysfunctions of cytoplasmic protein translation. Mutations in KARS and another bifunctional enzyme (GARS) had also been implicated in peripheral neuropathy. We propose that the dysfunction of cytoplasmic KARS or GARS, but not of mitochondrial KARS or GARS, is involved in peripheral neuropathy. Conversely, most of the mitochondrial ARSs defects have been found to affect the CNS. A functional CNS has an extremely high demand for energy. The oxidative phosphorylation system (OXPHOS) is a key functional unit in mitochondria, and it is the major source of cellular adenosine triphosphate. Defects in mitochondrial ARSs affect mitochondrial protein synthesis and lead to mitochondrial OXPHOS dysfunction38 (tables e-1 and e-2, links.lww.com/NXG/A143 and links.lww.com/NXG/A144). As expected, CNS involvement as the main or sole clinical presentation had been reported in most patients with pathogenic mutations in mitochondrial ARSs, including DARS2, EARS2, MARS2, FARS2, RARS2, VARS2, TARS2, NARS2, and CARS2. Meanwhile, the elevated level of lactate observed in some of our patients is also consistent with mitochondrial dysfunction. Therefore, dysfunction of mitochondrial KARS, but not cytoplasmic KARS, more likely to be contributing to the pathogenesis of leukoencephalopathy reported in this study. This work provides a framework to link the dysfunction of mitochondrial KARS with leukoencephalopathy associated with disorders of the CNS.
Acknowledgment
The authors thank the families and patients for their participation to this study.
Glossary
- ARS
aminoacyl-tRNA synthetase
- 5-FOA
5-fluoroorotic acid
- OXPHOS
oxidative phosphorylation system
Author contributions
C. Sun, J. Song, Y. Jiang, C. Zhao, J. Lin, Y. Li, Y. Wang, M Gao, J. Xi, S. Luo, M. Li, K. Donaldson, S.N. Oprescu, T.P. Slavin, S. Lee, P.L. Magoulas, A.M. Lewis, L. Emrick, S.R. Lalani, Z. Niu, M.L. Landsverk, M. Walkiewicz, R.E. Person, H. Mei, J.A. Rosenfeld, Y. Yang, A. Antonellis, Y-M. Hou, J. Lin, and V.W. Zhang designed the study, performed experiments, and collected and analyzed data. C. Sun, J. Song, Y. Jiang, J. Lin, and V.W. Zhang wrote the manuscript and critically revised the manuscript for important intellectual content. J. Lin and V.W. Zhang supervised the study.
Study funding
This work was supported by the National Natural Science Foundation of China (Nos. 81301203 and 81401035), Shanghai Health and Family Planning Commission key projects (Nos. 201440019 and 15DZ1208002), NIH GM108972 and NIH GM114343 (to YMH), NIH GM118647 (to AA), and a China Scholarship (to ML).
Disclosure
Baylor College of Medicine (BCM) and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of the Baylor Genetics Laboratories (BMGL), which performs clinical exome sequencing. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in the Baylor Genetics Laboratory (BMGL; bmgl.com/BMGL/Default.aspx). V.W. Zhang is employed by and receives a salary from AmCare Genomics Lab. Exome and other panel sequencing are among the commercially available tests available at AmCare Genomics Lab. C. Sun and J. Song report no disclosures. Y. Jiang has been employed by Baylor Genetics Laboratories. C. Zhao has received foundation/society research support from the Shanghai Committee of Science and Technology. J. Lu, Y. Li, Y. Wang, M. Gao, J. Xi, S. Luo, M. Li, K. Donaldson, and S.N. Oprescu report no disclosures. T.P. Slavin has received research support from the NIH/NCI, (1K08CA234394), Oxnard Foundation, Stop Cancer Foundation, and the Israeli Cancer Research Foundation. S. Lee and P.L. Magoulas report no disclosures. A.M. Lewis has received travel funding or speaker honoraria from Texas Parent to Parent Conference. L. Emrick has served on the advisory board of the American Board of Psychiatry and Neurology and has received government research support from the NIH. S.R. Lalani has received publishing royalties from UpToDate. Z. Niu reports no disclosures. M.L. Landsverk has been employed by Ambry Genetics. M. Walkiewicz has provided services for Baylor Miraca Genetics Laboratories. R.E. Person has been employed by GeneDX. H. Mei has been employed by GeneDX and Baylor Genetics. J.A. Rosenfeld has served on the editorial boards of Prenatal Diagnosis and Molecular Syndromology and has received government research support from the NIH. Y. Yang and A. Antonellis report no disclosures. Y.-M. Hou has received government research support from the NIH and has received foundation/society support from the Packard Foundation. J. Lin has received government research support from the National Natural Science Foundation of China and the Shanghai Health and Family Planning Commission. V.W. Zhang has been employed by AmCare Genomic Lab. Disclosures available: Neurology.org/NG.
References
- 1.Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci USA 2008;105:11043–11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genomics Hum Genet 2008;9:87–107. [DOI] [PubMed] [Google Scholar]
- 3.Abbott JA, Francklyn CS, Robey-Bond SM. Transfer RNA and human disease. Front Genet 2014;5:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diodato D, Ghezzi D, Tiranti V. The Mitochondrial aminoacyl tRNA synthetases: Genes and Syndromes. Int J Cell Biol 2014;2014:787956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konovalova S, Tyynismaa H. Mitochondrial aminoacyl-tRNA synthetases in human disease. Mol Genet Metab 2013;108:206–211. [DOI] [PubMed] [Google Scholar]
- 6.Wallen RC, Antonellis A. To charge or not to charge: mechanistic insights into neuropathy-associated tRNA synthetase mutations. Curr Opin Genet Dev 2013;23:302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao P, Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med 2013;5:332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diodato D, Melchionda L, Haack TB, et al. VARS2 and TARS2 mutations in patients with mitochondrial encephalomyopathies. Hum Mutat 2014;35:983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Euro L, Konovalova S, Asin-Cayuela J, et al. Structural modeling of tissue-specific mitochondrial alanyl-tRNA synthetase (AARS2) defects predicts differential effects on aminoacylation. Front Genet 2015;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sofou K, Kollberg G, Holmström M, et al. Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol Genet Genomic Med 2015;3:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanlander AV, Menten B, Smet J, et al. Two siblings with homozygous pathogenic splice-site variant in mitochondrial asparaginyl-tRNA synthetase (NARS2). Hum Mutat 2015;36:222–231. [DOI] [PubMed] [Google Scholar]
- 12.Bansagi B, Antoniadi T, Burton-Jones S, et al. Genotype/phenotype correlations in AARS-related neuropathy in a cohort of patients from the United Kingdom and Ireland. J Neurol 2015;262:1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez M, McLaughlin H, Houlden H, et al. Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J Neurol Neurosurg Psychiatry 2013;84:1247–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin LB, Sakaguchi R, McGuigan D, et al. Impaired function is a common feature of neuropathy-associated glycyl-tRNA synthetase mutations. Hum Mutat 2014;35:1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMillan HJ, Humphreys P, Smith A, et al. Congenital visual impairment and progressive microcephaly due to lysyl-transfer ribonucleic acid (RNA) synthetase (KARS) mutations: the expanding phenotype of aminoacyl-transfer RNA synthetase mutations in human disease. J Child Neurol 2015;30:1037–1043. [DOI] [PubMed] [Google Scholar]
- 16.Safka Brozkova D, Deconinck T, Griffin LB, et al. Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain 2015;138:2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce SB, Chisholm KM, Lynch ED, et al. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc Natl Acad Sci USA 2011;108:6543–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce SB, Gersak K, Michaelson-Cohen R, et al. Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am J Hum Genet 2013;92:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos-Cortez RL, Lee K, Azeem Z, et al. Mutations in KARS, encoding lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am J Hum Genet 2013;93:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puffenberger EG, Jinks RN, Sougnez C, et al. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS One 2012;7:e28936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dallabona C, Diodato D, Kevelam SH, et al. Novel (ovario) leukodystrophy related to AARS2 mutations. Neurology 2014;82:2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahin S, Cansu A, Kalay E, et al. Leukoencephalopathy with thalamus and brainstem involvement and high lactate caused by novel mutations in the EARS2 gene in two siblings. J Neurol Sci 2016;365:54–58. [DOI] [PubMed] [Google Scholar]
- 23.Simons C, Griffin LB, Helman G, et al. Loss-of-function alanyl-tRNA synthetase mutations cause an autosomal-recessive early-onset epileptic encephalopathy with persistent myelination defect. Am J Hum Genet 2015;96:675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steenweg ME, Ghezzi D, Haack T, et al. Leukoencephalopathy with thalamus and brainstem involvement and high lactate “LTBL” caused by EARS2 mutations. Brain 2012;135:1387–1394. [DOI] [PubMed] [Google Scholar]
- 25.van Berge L, Hamilton EM, Linnankivi T, et al. Leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation: clinical and genetic characterization and target for therapy. Brain 2014;137:1019–1029. [DOI] [PubMed] [Google Scholar]
- 26.Wolf NI, Toro C, Kister I, et al. DARS-associated leukoencephalopathy can mimic a steroid-responsive neuroinflammatory disorder. Neurology 2015;84:226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Ling J, Barcia G, et al. Mutations in QARS, encoding glutaminyl-tRNA synthetase, cause progressive microcephaly, cerebral-cerebellar atrophy, and intractable seizures. Am J Hum Genet 2014;94:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLaughlin HM, Sakaguchi R, Liu C, et al. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am J Hum Genet 2010;87:560–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieber DS, Calvo SE, Shanahan K, et al. Targeted exome sequencing of suspected mitochondrial disorders. Neurology 2013;80:1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohda M, Tokuzawa Y, Kishita Y, et al. A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PLoS Genet 2016;12:e1005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Cui H, Lee NC, et al. Clinical application of massively parallel sequencing in the molecular diagnosis of glycogen storage diseases of genetically heterogeneous origin. Genet Med 2013;15:106–114. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 2013;369:1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu H, Zhang VW, Stray-Pedersen A, et al. Rapid molecular diagnostics of severe primary immunodeficiency determined by using targeted next-generation sequencing. J Allergy Clin Immunol 2016;138:1142–1151.e2. [DOI] [PubMed] [Google Scholar]
- 34.Liu C, Gamper H, Liu H, Cooperman BS, Hou YM. Potential for interdependent development of tRNA determinants for aminoacylation and ribosome decoding. Nat Commun 2011;2:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C, Sanders JM, Pascal JM, Hou YM. Adaptation to tRNA acceptor stem structure by flexible adjustment in the catalytic domain of class I tRNA synthetases. RNA 2012;18:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo M, Ignatov M, Musier-Forsyth K, Schimmel P, Yang XL. Crystal structure of tetrameric form of human lysyl-tRNA synthetase: Implications for multisynthetase complex formation. Proc Natl Acad Sci USA 2008;105:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tolkunova E, Park H, Xia J, King MP, Davidson E. The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual alternative splicing of the primary transcript. J Biol Chem 2000;275:35063–35069. [DOI] [PubMed] [Google Scholar]
- 38.Rötig A. Human diseases with impaired mitochondrial protein synthesis. Biochim Biophys Acta 2011;1807:1198–1205. [DOI] [PubMed] [Google Scholar]