Summary
Cell lines and animal models have provided the foundation of cancer research for many years. However, human pluripotent stem cells (hPSCs) and organoids are increasingly enabling insights into tumor development, progression, and treatment. Here, we review recent studies using hPSCs to elucidate the reciprocal roles played by genetic alterations and cell identity in cancer formation. We also review studies using human organoids as models that recapitulate both intra- and inter-tumoral heterogeneity to gain new insights into tumorigenesis and treatment responses. Finally, we highlight potential opportunities for cancer research using hPSC-derived organoids and genome editing in the future.
Introduction
The study of human cancer has long benefited from the access of scientists to human tumor tissue and from the ability to develop and propagate tumor cell lines. While cell lines are integral to many types of experiments, they tend to drift away from the parent tumor’s genomic features or growth characteristics over time, a widely recognized limitation. Likewise, mouse models of cancer have resulted in great advances in our understanding of tumorigenesis and in the development of therapeutic targets. However, the transition from preclinical mouse modeling and therapeutic discovery to the clinic has been marred with frequent failures. This lack of translational success is often attributed to an array of factors, including heterogeneity of the cancer genome in human tumors and complex tumor-host interactions. Therefore, additional disease modeling strategies are needed to complement existing techniques in cancer research.
The last decade has witnessed enormous leaps in human stem cell technologies. Such advances include greater access to human pluripotent stem cell (hPSC) lines, including embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), as well as the development of diverse hPSC differentiation protocols and increasingly efficient genome editing tools. An ever-growing number of investigators are thus using advanced human cell culture technologies to complement animal models in their studies. Principal among such technologies are methods that use hPSC lines derived from non-malignant cells or tumor cells, as well as techniques using three-dimensional cultures of hPSC progeny or primary samples to derive normal and malignant tissue organoids.
In this review, we will discuss recent studies of cancer using hPSC and human organoid technologies, with a focus on how they highlight tumor formation, progression, and treatment response as developmentally informed, dynamic processes. It is important to mention that these model systems are still maturing and have not yet become as widespread as animal models and conventional cell lines. We will also discuss remaining challenges and considerations for modeling cancer using these strategies, and will highlight opportunities offered by advances in hPSC-derived organoid models and genome editing to cancer researchers.
Developing human cancers outside the patient
It was the physicist Richard Feynman who once famously remarked, “What I cannot create, I do not understand.” Such has been the perspective of cancer researchers who, for the past several decades, have collectively sought to build models of tumorigenesis de novo by initiating genetic changes in otherwise normal cells. Genetic engineering in specific cell types has yielded a wealth of animal models that have contributed enormously to our understanding of cancer. However, efforts to build genetically engineered, human cell-based cancer models have encountered hurdles in many cases, as human cells seem to be more resistant to transformation than their counterparts in other species.
To date, most successful paradigms for ex vivo transformation of human cells have required the introduction of means of escaping replicative senescence, such as overexpression of telomerase, followed by several oncogenes. As an example, the transformation of melanocytes requires the expression of the Simian Virus 40 early region (SV40ER), as well as hTERT followed by the ras oncogene (Gupta et al., 2005), while astrocytes require hTERT, mutant H-ras, and p53/pRB inactivation in order to form high-grade gliomas (Rich et al., 2001; Sonoda et al., 2001). These data are compatible with the concept of cancer formation as a consequence of a series of stepwise oncogenic events that accumulate over time. However, the combinations of oncogenes used in these studies, particularly in the case of melanocyte transformation (Gupta et al., 2005), is not representative of spontaneously occurring human tumors.
Pediatric cancer types, on the other hand, emerge more rapidly than would be predicted by our current understanding of cancer genome evolution (Puisieux et al., 2018; Vogelstein et al., 2013). Moreover, certain types of pediatric cancer can occur or progress due to epigenetic changes with few apparent genetic events, or even none at all (Lee et al., 2012; Mack et al., 2014; Molenaar et al., 2012; Parker et al., 2014; Rausch et al., 2012; Robinson et al., 2012). Taken together, these findings, which are validated by a growing body of data across model systems (Puisieux et al., 2018), would suggest that malignant transformation is highly dependent on cell-of-origin and differentiation state. The identity of a given cell can provide context in the form of transcriptional programs as well as through features of the chromatin landscape, both of which may influence the abilities of certain genetic alterations to drive tumorigenesis.
It has become clear that, if researchers are truly to understand various types of human cancer by creating them, then modeling studies will require precisely defined, physiologically relevant human cell types as well as the genetic lesions that can transform them. Understanding which cell types are most appropriate for cancer modeling studies will more than ever require consideration of the proposed developmental origins of the tumor, so that human cells that share those origins and lineage commitment are properly identified. Studies using human stem cell technologies have revealed that driving cancer cells away from their default states toward other cell identities and lineages can blunt their tumorigenic attributes, even if their oncogenic lesions remain intact – thus highlighting the importance of cell identity in tumorigenesis (Carette et al., 2010; Chao et al., 2017; Funato et al., 2014). These same technologies are now being leveraged to create ever-more-faithful human cancer models, resulting in clear insights into the developmental contexts required for tumorigenesis via certain mutations.
Reprogramming cancer cells to study their origins
It has long been established that tumor cells, through nuclear transfer, can generate embryos with all three germ layers and functional tissues comprised of tumor-derived nuclei (Hochedlinger et al., 2004; McKinnell et al., 1969). That work set the stage for numerous studies demonstrating that, despite their genetic aberrations, cancer cells are capable of recapitulating normal developmental processes. Recent studies have used transcription factor-mediated reprogramming to generate iPSCs from a variety of cancer cell types, paving the way for studies of how cancer genomes affect progression through cellular lineages.
The generation of iPSCs using transcription factor expression in mouse and human somatic cells was first described in 2006 and 2007, respectively (Takahashi et al., 2007; Takahashi and Yamanaka, 2006). Shortly thereafter, it was demonstrated in a murine melanoma cell line that mammalian cancer cells could be reprogrammed to pluripotency using this technique, as had been done previously using nuclear transfer (Utikal et al., 2009). In 2010, successful reprogramming of human BCR-ABL-expressing chronic myeloid leukemia cells into iPSC-like cells was reported, using the classic four Yamanaka factors, OSKM (Carette et al., 2010). The reprogrammed malignant cells acquired pluripotency and differentiated into all three germ layers in a teratoma assay, while retaining expression of the driver oncogene. Additional reports of reprogramming using cell lines representing a range of neoplasms, including solid tumors such as gastrointestinal cancers, glioblastoma and pancreatic cancers, have shown similar findings (Carette et al., 2010; Miyoshi et al., 2010; Nagai et al., 2010; Stricker et al., 2013). While the process of reprogramming cancer cells is often inefficient relative to that of normal cells, such reports suggest that cancer cells do, in fact, retain a broad developmental potential that can be demonstrated by induction of pluripotency.
A corollary to this finding is that induced pluripotency can, at least in some cases, override the cancer phenotype, even if only temporarily. By inducing a widespread remodeling of the epigenetic landscape, reprogramming allows cancer cells to acquire ectoderm, mesoderm and endoderm lineages, irrespective of the cancer cell line’s original identity. In many instances, these induced cancer cell lines show significantly reduced tumorigenesis when implanted in immunosuppressed mice despite retention of the oncogenic genome (Stricker et al., 2013). Furthermore, the alteration in lineage state leads to an increase in chemosensitivity in at least one case, and might lead to a greater response to differentiation therapies as well (Miyoshi et al., 2010). These findings highlight the role that cell identity plays in mediating the oncogenic effects of certain genetic lesions found in cancer.
Recipes for success: directed differentiation of hPSCs to build cancer models
The apparent simplicity with which hPSCs can be coaxed to replicate ontogeny in a lab dish, and thus to give rise to an array of cell types, has led to major efforts in the disease-modeling arena. In addition to the investigation of mechanistic questions, hPSC-based disease modeling has been used for drug discovery, for the development of toxicity assays, and for the derivation of therapeutically relevant cell types on a large scale.
Because hPSC-based “development-in-a-dish” approaches have been so fruitful for diseases of cell type- and/or tissue-specific dysfunction, it is exciting to consider the impact that such approaches could have on modeling specific types of cancer. Indeed, the modeling of cancer initiation in hPSC progeny is a growing field with multiple potential applications: a greater understanding of the early events of cancer initiation, enhancement of animal models of human cancers, and discovery of novel therapeutic approaches.
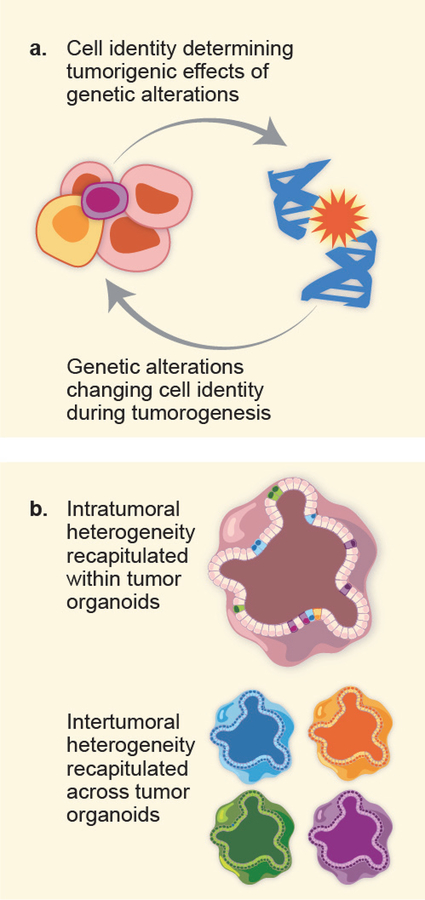
Directed differentiation of hPSCs to certain cell types is frequently achieved with exposure to combinations of morphogens and growth factors applied in specific sequences and precise doses. This ability of hPSCs to progress to defined differentiation states within developmental lineages represents an unprecedented opportunity for cancer researchers. As defective differentiation is a cardinal feature of many, if not all, cancer types, hPSCs can be used to dissect the contributions of oncogenes to these differentiation blockades. Likewise, hPSCs can also be used to dissect the roles played by features of a given cell type – including gene expression, elements of the chromatin landscape, and more – in mediating oncogenic phenotypes associated with certain oncogenic events (Figure 1a).
Figure 1:
(a) Studies using hPSC-based cancer models have revealed a relationship between genetic alterations an cell identity: while a cancer-associated alteration can alter the identity of a given cell, the identity of a cell can dictate its phenotypic response to a given genetic lesion.
(b) Human organoids have proven valuable in understanding the importance of tumor heterogeneity on two levels: intratumoral heterogeneity, in which different populations within a tumor have divergent, clinically relevant phenotypes, and intertumoral heterogeneity, in which tumors of the same type but from different patients have distinct clinical features.
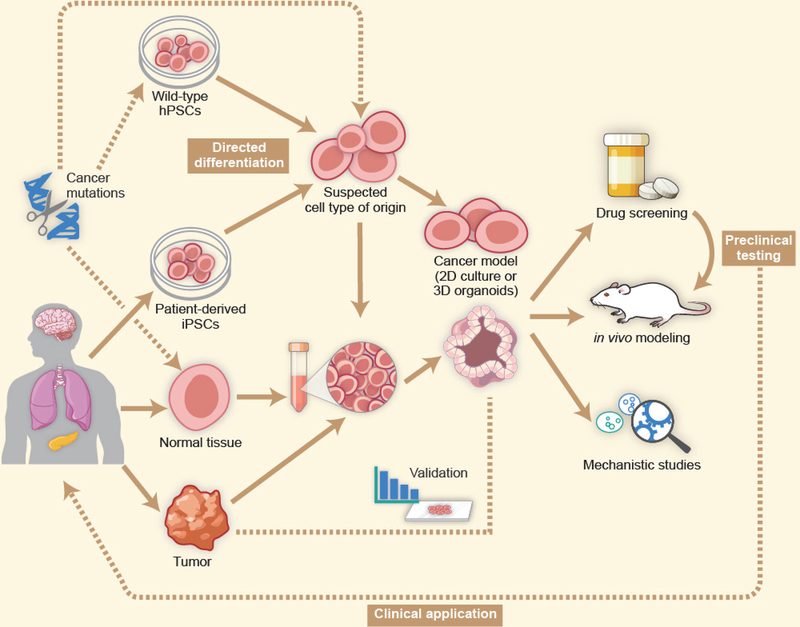
To date, experimental paradigms have consisted broadly of two approaches: those in which one or more human cell types are derived from hPSCs, with oncogenic events induced only after the desired cell type(s) has been obtained (Funato et al., 2014; Huang et al., 2015), and those in which cancer cells are reprogrammed to iPSCs and then re-differentiated along the relevant lineage (from which the putative cell- or tissue-of-origin arose) with the cancer cell’s oncogenic genome intact along the way (Papapetrou, 2016) (Figure 2). While both approaches have proven effective in the laboratory, it should be noted that experiments using the latter approach can, in principle, be affected by the presence of somatic mutations that might alter the lineage progression of iPSCs during directed differentiation.
Figure 2:
HPSCs from established wild-type lines, iPSCs derived from patient cells, and normal and tumor tissues from human patients can be used to establish cancer models. Wild-type or patient-derived hPSCs undergo directed differentiation to a cell type of interest that represents a likely cell-of-origin for a given cancer type. These cells can be maintained as a two-dimensional culture, or can be used to generate three-dimensional organoids. Patient tissues can also be used to generate organoids. Cancer-associated genetic alterations, if not already present in the genome of the source material, are introduced at the hPSC stage, following hPSC directed differentiation or in patient-derived organoids. The resulting cancer model is validated through comparison to patient tumor cells and is used for a variety of in vitro and in vivo studies. Therapeutic strategies that emerge from such work are then validated in primary patient samples and may progress into preclinical studies with an eye towards clinical translation.
When iPSCs are derived from cancer cells, a common strategy consists of reprogramming cancer cells, as well as isogenic normal cells from the same tissue lineage, into iPSCs, then differentiating them along that same lineage. As discussed in the following sections, comparison of these isogenic lines often reveals impaired differentiation of the tumor-derived cells at a specific developmental stage. Functional screens can then define the roles of individual genes in causing or rescuing the oncogenic phenotype, as well as identifying therapeutic means of overcoming the differentiation blockade. This approach is particularly advantageous when addressing complex genomic aberrations that cannot be easily introduced through exogenous means, including copy number alterations and losses of genomic regions containing multiple genes and regulatory sequences (Kotini et al., 2015; Kotini et al., 2017).
Studies using cancer-derived iPSCs or normal hPSCs as a platform to build genetically engineered cancer models have already begun to emerge. These reports provide important insights into how certain cell types and differentiation states are uniquely susceptible to tumorigenesis by a given mutation, as well as into how certain mutations drive shifts in cell identities and differentiation states. Below, we will review what is already revealed by hPSC-based models about the reciprocal relationship between cell identities and genetic events in human cancer.
Cell identity as a determinant of tumorigenic potential in hPSC-based models
There are several longstanding challenges in cancer research that have limited the creation of human cancer models purely through genetic engineering of otherwise normal human cells. Access to the precise cell-of-origin of a given cancer type is often challenging, due to suboptimal knowledge as to the cell-of-origin of some tumors, inaccessibility of a specific lineage or maturation state from which a tumor is thought to have arisen, or difficulty in accessing certain human tissues altogether. HPSC technologies can therefore play a crucial role in facilitating access to a range of lineage and differentiation states to serve as platforms for novel models of human cancer. Such models, in comparison to cell lines or human tumor xenografts, promise to provide valuable results that shed light on the mechanisms through which certain cell types and differentiation states are more or less amenable to transformation by a given mutation. Indeed, such studies of brain tumors and leukemias have already emerged, demonstrating the power of such an approach.
Brain cancer
Diffuse intrinsic pontine glioma (DIPG) is a prime example of a cancer that is well-suited for modeling in hPSC progeny, particularly to address questions of the developmental determinants of cancer formation. It is a pediatric cancer whereby access to primary tumor tissue has been challenging, given the propensity of the tumors to present as an infiltrative mass among sensitive neurological structures in the brainstem. The key genomic aberrations were only recently identified, and they include a previously unknown somatic mutation in the gene coding for the histone variant H3.3 in the majority of cases (Schwartzentruber et al., 2012; Wu et al., 2012). The mutation consists of a single amino acid substitution, whereby the lysine 27 residue is substituted to a methionine (“K27M”), resulting in a dominant-negative effect of a global loss of the H3K27me3 marks on wild-type and mutant histones alike (Bender et al., 2013; Lewis et al., 2013). However, it remains unclear how the histone mutation drives oncogenesis specifically during neural development (Funato and Tabar, 2018), underscoring the need for innovative modeling techniques.
Interestingly, the tumors occur predominantly in young children and only rarely in adults, and are largely confined to the midline of the central nervous system from the thalamus to the pons. They also tend to associate with cooperating mutations, usually involving a receptor tyrosine kinase pathway, in a brain region-specific manner (Jones and Baker, 2014). These findings are highly suggestive of a specific developmental window and cell-of-origin giving rise to the tumors. Indeed, the introduction of relevant mutations into hPSC-derived neural precursor cells (NPCs) leads to in vitro transformation, as well as in vivo brainstem tumors that recapitulate important histological and transcriptomic features of patient tumors (Funato et al., 2014). While the K27M mutation contributed to malignant transformation of NPCs derived from hPSCs, its oncogenic effect was limited to neural precursors within a specific developmental window. It did not have such an effect on any other neural or non-neural cell type tested. Further studies are now underway to understand the mechanisms through which some neural cells, but not others, provide fertile soil for tumorigenesis in the presence of the K27M mutation.
The ability to engineer hPSCs with advanced genetic tools allows the derivation of cancer models with faithful recapitulation of somatic alterations in well-defined genetic backgrounds. This strategy often involves the introduction of oncogenic alterations at the pluripotent cell stage, with differentiation into relevant fates initiated thereafter. A recent example is provided by the generation of neural stem cells (NSCs) from an hPSC line engineered to harbor two null alleles of PTEN (Duan et al., 2015). Alterations leading to PTEN loss-of-function are common in brain tumors (Reifenberger et al., 2017). The PTEN-null NSCs exhibit a more aggressive phenotype than their isogenic wild-type counterparts, but other cell types generated from the PTEN-null hPSCs, such as mesenchymal cells, were phenotypically indistinguishable from wild-type controls. These results suggest that oncogenic events described in adult brain tumors, like those found in pediatric brain tumors, are effective in inducing neoplastic changes only in specific cell contexts.
Hematopoietic cancer
A recent report using iPSCs derived from leukemia cells has shed light on the importance of cell-of-origin to tumorigenesis driven by MLL rearrangements (Chao et al., 2017). When the team carrying out that study generated iPSC lines from patients’ AML cells with chromosomal rearrangements involving the MLL gene, they observed that the resulting iPSCs lose AML-associated gene expression and DNA methylation patterns, as well as the ability to form leukemias in vivo. However, this loss of leukemogenicity is entirely context-dependent – when these AML-iPSCs are directed to differentiate into hematopoietic cells, they regain not only the ability to form leukemias in vivo, but also the gene expression and DNA methylation patterns observed in primary AML cells. These findings highlight the importance of context provided by cell identity to certain cancer-driving mutations: MLL rearrangements are clearly leukemogenic in hematopoietic cells, but they do not have the same power in pluripotent cells that precede hematopoiesis developmentally. Interestingly, the authors of this study report differences in the leukemias that arise from different iPSC clones from the same patient. That observation suggests an ability of hPSC-based cancer modeling to uncover differences among cell subpopulations in heterogeneous tumors as well.
This phenomenon – phenotypes only observed to hematopoietic cells – has also been observed in other studies of iPSCs harboring mutations associated with myelodysplastic syndrome and leukemia. In one case, dependence on the chronic myeloid leukemia (CML)-associated oncogene BCR-ABL was only apparent in CML cells and in cells of the hematopoietic lineage derived from CML-iPSCs, and not in the iPSCs themselves derived from CML cells (Carette et al., 2010; Kotini et al., 2017). In several other cases, iPSCs derived from myelodysplastic syndrome or leukemia cells are reported to have phenotypes that only manifest following the induction of hematopoietic specification (Chang et al., 2018; Kotini et al., 2015; Kotini et al., 2017; Mulero-Navarro et al., 2015).
Pancreatic cancer
In addition to undergoing normal lineage progression, cancer-derived iPSC lines are capable in principle of recapitulating disease progression, upon directed differentiation into their respective tissues-of-origin. This is illustrated by work wherein an “iPSC-like” line was derived from pancreatic ductal adenocarcinoma (PDAC) samples (Kim et al., 2013). It should be noted that the reprogrammed line used in this study was not a true iPSC line, because continuous forced expression of reprogramming factors was required to maintain pluripotency. Despite having been derived from advanced PDAC samples, PDAC “iPSC-like”-derived teratomas formed in mice contained early-stage pancreatic epithelial neoplasia in the first months after injection, with more advanced, invasive PDAC structures only observed at later timepoints. These data suggest that mutations from late-stage disease may not always be sufficient to induce late-stage phenotypes immediately upon reprogramming and redifferentiation, highlighting potential roles for other cell identity characteristics like chromatin features that change or evolve over time in a lineage-specific manner.
While the frequency of this phenomenon in that report was low, and its wide applicability to other epithelial cancer types uncertain, it does provide a window into the evolution of cancer. It also provides an opportunity to study early stages of cancer that are generally not well captured by xenografts or cell lines derived from late-stage disease. Indeed, more recent work using PDAC-derived iPSCs has revealed potential circulating biomarkers that might one day be used for noninvasive screening for that cancer in the clinic (Kim et al., 2017). PDAC has also been studied using hPSC-derived cells of non-tumor origin, with similar observations of early-stage disease features when KRAS and TP53 mutations are introduced into pancreatic progenitors (Huang et al., 2015). As PDAC happens to be a prime example of a disease whose early stages evade both clinicians and scientists, perhaps contributing to poor disease outcomes, early-stage disease insights provided by hPSC-based modeling will be welcome. Insights into the developmental determinants of tumorigenesis in pancreatic lineages may thus be forthcoming.
As more cell types become available from hPSCs, and as more is learned about putative cell types-of-origin of certain cancer types, investigators will gain further abilities to test the effects of cancer-associated genetic lesions in diverse human cell types. Through this work, the role of a cell’s identity and differentiation state in driving or suppressing various cancer types will likely emerge, with implications both for detecting and treating diseases in patients.
Cell identity as a consequence of oncogenic activity in hPSC-based models
In addition to uncovering the roles played by cell type-specific features in mediating tumorigenesis by certain mutations, hPSC-based models can also demonstrate how mutations affect cell identity in turn. An advantage of cancer modeling using specific human cell types is that these cell types are often understood to be part of a well-defined lineage. Such studies can therefore allow investigators to interrogate these cell types both with and without oncogenic events, and to correlate the effects of those events with shifts in cell identity along the relevant lineage. This strategy has been successful in animal models, including a recent breakthrough using a murine lung cancer model that highlights the extraordinary developmental plasticity of cancer cells (Tata et al., 2018). In that study, researchers report that oncogenic events associated with lung cancer are sufficient to impart features of other endoderm-derived tissues, such as those of the gut, on lung epithelial cells. These findings provide further support for the idea that developmental principles underlie plasticity in cell identity driven by cancer mutations (Magnen et al., 2018). Several groups have used hPSC-based models to ask how cancer-associated mutations found in cancer types of the blood, bone, brain, and colon affect a cell’s differentiation status, or capacity therefor, within its lineage of developmental origin.
Hematopoietic cancer
Myelodysplastic syndromes (MDS) are proliferative diseases of the blood that are characterized by the somatic loss of one copy of a genomic region on chromosome 7q. By reprogramming both dysplastic and isogenic normal hematopoietic stem cells, investigators have shown that hematopoietic differentiation is greatly impaired in del7q iPSCs relative to wild-type iPSCs, with a similar phenotype manifesting in hPSCs that have been engineered with hemizygosity of 7q (Kotini et al., 2015). This block of hematopoietic differentiation can be rescued completely by engineered or spontaneous gains of 7q in culture. Similarly, wild-type hPSCs that undergo engineered 7q loss recapitulate the differentiation impairment seen in patient-derived iPSCs. A phenotype-rescue screen identified the roles of candidate haploinsufficient genes that contribute to the differentiation defect. These genes were later validated as necessary for proper hematopoietic differentiation in normal hPSCs and hematopoietic stem cells. More recent work has further confirmed the role of del7q in blocking hematopoietic differentiation, as iPSCs with del7q and cooperating MDS mutations in the splicing factor gene SRSF2 regain competence to differentiate along the hematopoietic lineage when del7q alone is corrected (Chang et al., 2018).
Extending the same approach to a wider range of hematopoietic cancer types, the authors derived iPSCs from different myeloproliferative neoplasm stages – ranging from normal and preleukemic cells to low- and high-risk MDS cells and secondary acute myeloid leukemia (AML) cells (Kotini et al., 2017). They determined that differentiation is impaired to extents that correspond in severity to the stage of the disease from which the iPSC lines have originated. When a genetic lesion, del7q, in advanced-disease iPSCs is corrected, impaired differentiation and other cancer phenotypes are rescued; in contrast, induction of additional genetic lesions in less-advanced-stage disease iPSCs increases the severity of the phenotypes. Additionally, the authors report disease stage-specific responses to a cytotoxic chemotherapy, 5-azacytidine, and a RAS pathway-targeted therapy. These studies provide examples of how hPSCs can be leveraged to interrogate the complex connections between genetic events, effects on differentiation status and cell identity, and disease progression in cancer.
IPSC technology has also been used to establish a novel model of leukemia associated with Noonan Syndrome (NS) (Mulero-Navarro et al., 2015). NS patients have an inherited predisposition to juvenile monomyelocytic leukemia (JMML), which, in many cases, results from germline mutation of the phosphatase-encoding gene PTPN11 (Smpokou et al., 2015). This team generated iPSCs from fibroblasts of NS patients harboring PTPN11 mutations, but with only a fraction of these patients having JMML. Once the iPSCs were directed to a hematopoietic lineage, samples from NS patients with JMML exhibited known phenotypes associated with the disease, including sensitivity to hematopoietic growth factor stimulation and excessive expansion of cells of myeloid fates. Interestingly, samples from NS patients without JMML did not exhibit these phenotypes. Together, these experiments illustrate the potential value of the iPSC platform in establishing a patient-specific risk stratification in familial cancer syndromes, beyond the known risk alleles encoded in the germline. They also demonstrate the value of hPSC-based modeling to uncovering heretofore-unknown mediators of inappropriate expansion of certain subpopulations within a cell lineage.
Bone cancer
Familial cancers associated with Li-Fraumeni Syndrome (LFS) have been modeled with patient-derived iPSCs as well (Lee et al., 2015). Investigators reported the generation of a faithful model of LFS-associated osteosarcoma using osteoblasts differentiated from those iPSCs, finding that LFS osteoblasts are defective in their differentiation capacity and express high levels of genes known to be active in osteosarcoma. Importantly, the LFS osteoblasts and resulting osteosarcoma models lacked many of the cytogenetic aberrations that are often found in primary LFS osteosarcoma samples – highlighting the utility of this iPSC-based system in modeling early disease stages that are inaccessible using patient tissues. In addition to recapitulating key known disease features, this study used LFS osteoblasts to elucidate a novel role for the H19-Decorin pathway in suppressing osteosarcoma formation in LFS patients. H19 gene expression is decreased in LFS osteoblasts relative to wild-type controls, and ectopic expression of H19 in these cells restores osteogenic differentiation while compromising in vivo tumor formation. This study highlights the unique utility of iPSCs from patients with inherited cancer syndromes: these cells can form the basis of human tissue culture models of syndrome-associated cancers from initiation to disease progression, all while incorporating the exact genetic drivers found in patient tissues.
Brain cancer
New details of how mutations found in pediatric and adult brain tumors affect cell identity and differentiation status in the neural cell lineage have been brought forward as well. In addition to providing a glimpse of how cell identity affects oncogenicity of the histone H3 K27M mutation in pediatric gliomas, hPSC-based modeling has also highlighted the effect that this mutation has on neural precursors (NPCs). Transformed NPCs bearing the K27M mutation exhibit impaired glial differentiation relative to isogenic wild-type controls, and their epigenetic and transcriptomic profile are more in line with earlier neurodevelopmental states than are those of the controls (Funato et al., 2014). These results suggest a novel role for the neural stemness-associated gene LIN28B in DIPG tumorigenesis, which is consistent with a mutation-mediated neural lineage differentiation blockade. This work also facilitated the identification of the transcriptional scaffold protein menin as a potential therapeutic target in this disease, with menin inhibition restoring glial differentiation in K27M-bearing NPCs. These findings underscore the integral role that a K27M mutation-mediated differentiation blockade likely plays in mediating the transformation of NPCs into pediatric glioma cells.
Results indicate that mutations found in adult high-grade gliomas affect the differentiation states of neural cells as well. In one study, the authors report that introduction of a combination of mutations into iPSC-derived NPCs drives colony-formation behaviors and metabolic features normally associated with less differentiated cells and tumors in the nervous system through induction of the neural stemness transcription factor SOX2 (Sancho-Martinez et al., 2016). Increased neural stem cell marker expression, including that of SOX2 as well as of OLIG2, has been reported in a more recent hPSC-based modeling study of adult high-grade gliomas as well (Ogawa et al., 2018).
In another study, researchers use hESC-derived NPCs to elucidate the effects of the low-grade glioma-associated IDH1 R132H mutation, in conjunction with losses of p53 and ATRX function, on cell differentiation (Modrek et al., 2017). These authors report marked defects in differentiation capacity in the mutation-bearing NPCs. They further report a mutation-mediated loss of SOX2, with ectopic SOX2 expression rescuing the differentiation defects, potentially via the transcription factor’s role in promoting multipotency in neural stem cells. Together, these studies demonstrate that hPSC-based modeling is a powerful platform upon which to build studies of neural lineage dysregulation in brain tumorigenesis.
Colorectal cancer
Colorectal cancers have been studied using an in vitro model of familial adenomatous polyposis (FAP) that was established using iPSCs derived from patients (Crespo et al., 2017). FAP is an inherited disorder in which patients experience high rates of precancerous polyp formation in the large intestine as a consequence of germline mutations in the WNT pathway regulator APC (Sasikumar et al., 2011). When FAP iPSCs undergo directed differentiation to three-dimensional colonic organoids, the resulting organoids exhibit increased WNT signaling activity as well as higher levels of epithelial cell proliferation relative to APC-wild-type organoids – both hallmarks not only of stem cells in the large intestine, but of FAP-associated polyp formation and progression to colon cancer as well. This work illustrates in a sophisticated human cell-based model that APC mutations are potent mediators of a stem-like cell identity in intestinal cells even before their outright tumorigenesis. Similar findings of APC mutations affecting differentiation in intestinal organoids have been reported by a second group as well (Sommer et al., 2018).
These hPSC-based modeling studies link cell identity with oncogenic alterations in reciprocal ways, and have thus fueled hypotheses regarding developmental principles in cancer biology. They have provided compelling new evidence that cancerous cells may follow some of the same principles of development as healthy tissues, with genetic alterations playing definable roles in the dysregulation of these processes. They have also shown that the potency of genetic alterations is driven to a significant extent by the cell types in which they occur. While some of these experiments may be perceived as products of technical wizardry, they nonetheless converge towards a common theme: oncogenic phenotypes are highly dependent on cell context, even within the same lineage. They also provide further credence to the idea that cancer is a case of “blocked ontogeny,” whereby progression down a specific differentiation path is impaired by genetic alterations, thus creating vulnerability to uncontrolled growth (Potter, 1969, 1978; Sell, 2008). Such a link between the development of normal and cancerous tissues will certainly be the subject of further inquiry using hPSC-based models in the years to come.
Adding a dimension: using human organoids to investigate tumor heterogeneity
While the ability to derive specific cell types on-demand, from hPSCs, has opened exciting new possibilities in cancer research in recent years, so too has the development of techniques to culture entire organ-like structures, known as “organoids,” derived from hPSCs or from human tissues. Using such strategies, organoid models of several human tissues have been described that recapitulate early stages of human organogenesis (Dutta et al., 2017; McCauley and Wells, 2017). Key features of these organoids include the in vitro generation of diverse progenitor cells at various stages of maturation, appropriate 3D organization, histological layers, and migration patterns, among others. These characteristics of organoids have allowed novel studies of early human developmental stages. They have also lent significant relevance to their use in cancer studies, which have traditionally lacked complex human cell-based models.
Over the past few years, organoids derived from hPSCs or human adult stem cell sources, as well as those derived from primary human tumor samples, have become prominent in cancer research laboratories (Drost and Clevers, 2018). Many of the studies discussed here have relied upon patient-derived adult stem cells and/or tumor tissue for organoid generation, while a small but ever-increasing number use hPSC-derived organoids for cancer modeling (Crespo et al., 2017; Huang et al., 2015; Ogawa et al., 2018). The advantages of patient tumor tissue-derived organoids are many: maintenance of the cancer genome driving a patient’s disease, scalability for drug screening and other high-throughput applications, as well as a lack of necessity of complex directed differentiation protocols to generate the tissue or lineage state of interest. Patient adult stem cell-derived organoids also lack a need for complex differentiation schemes, but do not recapitulate the cancer genome and are generally not as scalable as neoplastic organoids. They do, however, permit specific cancer-associated genetic alterations to be introduced at will into samples with isogenic controls available. HPSC-derived organoids, for their part, also have this advantage, along with limitless scalability of hPSCs themselves at the outset of an experiment – but these organoids require optimized differentiation protocols for their generation from hPSCs.
It is very likely that, as methods to generate specific tissue organoids from hPSCs are optimized in the coming years, these techniques will join those using patient-derived tissues in the laboratories of cancer researchers. To date, such techniques have already played a key role in studies of intratumoral heterogeneity in a variety of cancer types, as well as of disease heterogeneity among cancer patients as well (Figure 1b).
Organoids as models of intratumoral heterogeneity
Organoids enable the culture of multiple cell types that comprise a given tumor in a way that facilitates interactions among cells as well as between cells and the surrounding environment. Such a system allows for various groups of cells to behave and respond to stimuli differently from each other – a characteristic of cancer that underlies intratumoral heterogeneity. Because differences in cellular behaviors and responses within tumors can have enormous impact on disease pathogenesis and therapeutic responses, it is important for cancer models to recapitulate such differences among cells.
Brain cancer
A report from a team studying glioblastoma reveals how organoids can be used to study intratumoral heterogeneity (Hubert et al., 2016). The team leverages organoid culture to maintain tumor samples in vitro that give rise to histologically faithful tumor xenografts in vivo, to an extent not seen when cells are maintained in traditional sphere cultures before grafting. The glioblastoma organoids described in this study contain rapidly proliferating cells surrounding a hypoxic core of quiescent cancer stem cells (CSCs) that are more resistant to therapeutic radiation than the rest of the organoid – all structural and microenvironmental features of human glioblastoma that are lost when tumor cells are transitioned to sphere cultures before grafting. This recapitulation of intratumoral heterogeneity in treatment responses lends value to organoids as a model system for glioblastoma. A recent report in fact additionally highlights the roles of CSCs and differentiated cancer cells in glioblastoma progression (Wang et al., 2018), further supporting the hypothesis that maintenance of these subpopulations is key to modeling the tumor.
Bladder cancer
Patient tumor-derived organoids have been characterized in urothelial carcinoma as well (Lee et al., 2018). Both in culture and when transplanted into mice, these bladder cancer organoids evolve much as primary tumors are thought to do in patients. The authors observe significant genetic heterogeneity within a given patient’s organoid model, just as genetic alterations are gained and lost over time in patient tumors. This is consistent with clonal outgrowth of cells bearing certain advantageous genetic lesions as a phenomenon driving tumor, and organoid, evolution. The authors also observe that organoids derived from the most refractory patient tumors display a general resistance to many therapeutic agents tested in the laboratory. This work demonstrates the value of using organoid models to understand intratumoral heterogeneity, along with heterogeneity among patient tumors as well, in bladder cancer.
Gastric cancer
Recent studies have emerged documenting the use of patient-derived organoids to investigate disease mechanisms and therapeutic vulnerabilities in gastric cancer (Nanki et al., 2018; Yan et al., 2018). One report reveals the importance of intratumoral heterogeneity in this disease, with organoids available from multiple tumor regions in single patients (Yan et al., 2018). In one striking case, different organoids derived from the same patient displayed markedly distinct responses to therapeutic PARP inhibition, and in another, a single patient’s organoids with variable APC mutational status had divergent responses to Wnt signaling withdrawal. Organoid models of gastric cancer will likely come to even more widespread use in the near future.
Colorectal cancer
Researchers have used patient tumor-derived organoids to perform several studies of colorectal cancer (CRC). In one study, the authors demonstrated that organoids derived from CRC samples contain diverse histological and morphological features that are found in their respective parent tumors (Fujii et al., 2016). Importantly, they showed that such distinct structural features were driven by underlying differences in the presence or lack thereof of certain genetic alterations, thus confirming a link between genetic and structural heterogeneity that had not previously been shown in human CRC. In the same study, cell-labeling experiments provided data suggesting that certain cell subpopulations in CRC organoids become more or less represented over time. This finding highlights the importance of keeping passaging of organoids, as with conventional cell lines, both limited and carefully monitored. It also raises the eventual possibility of taking advantage of subpopulation outgrowth in organoids to model tumor progression over time. To demonstrate the relevance of organoids to model temporal processes, though, significant optimization and validation of methods will be required.
CRC organoids have also been used to elucidate the biological mechanisms underlying intratumoral heterogeneity. CSCs have been found in human CRC and are believed to be key drivers of tumor regrowth and progression following treatment. In a recent study, researchers found that CRC organoids contain a subpopulation of cells with the characteristic growth potential and aldehyde dehydrogenase (ALDH) expression of colorectal CSCs (Regan et al., 2017). They showed that autocrine non-canonical (GLI-independent) Hedgehog signaling plays a vital role in the maintenance of this subpopulation through reinforcement of WNT signaling, a known driver of CRC tumorigenesis. It has also been shown using human CRC organoids that claudin-2, a tight junction protein, is a key player in the maintenance and propagation of colorectal CSCs, thus contributing to phenotypic heterogeneity and progression in these tumors (Paquet-Fifield et al., 2018).
It has become clear over the past decades that many cancer types are driven by heterogeneous subpopulations of cells with distinct relevance to disease biology and clinical management. It is becoming just as clear now that organoid technologies have the potential to capture these subpopulations for disease modeling and preclinical investigation.
Organoids as a platform to study cancers at the single-patient level
The derivation of organoids from primary tumors of diverse cancer types has enabled their use to study patient-specific attributes. This is an important capability, as significant heterogeneity among patient tumors – so-called “intertumoral” heterogeneity – presents complications to the study of cancer much in the same way that intratumoral heterogeneity does as well. While experimentation using primary tumor samples has long been possible, researchers have traditionally encountered difficulties in acquiring enough material for large-scale studies.
Because organoids preserve many features of primary tumors but are capable of in vitro growth and large-scale expansion, they bridge a key gap between traditional cell line-based and primary sample-based methods. Several groups have now used patient-specific organoids as platforms for drug screening and other methods that require both high quality and high quantity from samples.
Prostate cancer
An early example of organoids as a model system to uncover patient-specific features of cancer came from a collaborative group investigating prostate development and disease. The group developed protocols to form organoids from both normal and neoplastic prostate tissue (Gao et al., 2014; Karthaus et al., 2014) with remarkable structural, histological, and cellular resemblance to human prostate glands and tumors, respectively. The patient-derived tumor organoids represent a diverse array of genetic drivers of prostate cancer, and those genetic underpinnings are shown to correlate with divergent responses to various targeted therapies (Gao et al., 2014).
Results of further studies using human prostate cancer organoids have revealed that the product of SPOP, a gene frequently mutated in prostate cancer, suppresses tumorigenicity and, when intact, confers susceptibility to bromodomain and extraterminal (BET) protein inhibition in tumors by promoting degradation of the BET family member BRD4 (Dai et al., 2017). Organoids with mutated SPOP express high levels of BRD4 and are resistant to BET inhibition, but become sensitive once BRD4 is genetically or pharmacologically targeted.
Other reports have now emerged with more details of mutant SPOP-driven tumorigenesis using human tumor organoids as a model, including a surprising lack of cooperativity between SPOP mutations and another frequent alteration in prostate cancer, ERG overexpression (Shoag et al., 2018). Together, these organoid studies have enabled a deeper understanding of how certain patients’ prostate cancers form, progress, and respond to treatment according to the genetic lesions that drive them.
Gastric cancer
Two recent reports highlight the utility of organoids as a model system for gastric cancer across patients. In one study, investigators showed clear correlations between genotypes and phenotypes in organoids from tumors with a wide array of genetic alterations (Nanki et al., 2018). They demonstrated that certain mutations are responsible for the morphological transition of gastric tissue to malignancy, and that specific genetic alterations and chromatin features are associated with independence from stem cell niche factors like Wnt and R-spondin for survival. In another study, researchers used gastric cancer organoids from a group of patients to model the disease and to screen for therapeutically relevant compounds (Yan et al., 2018).
Colorectal cancer
Extensive work has been done using organoids to study patient-specific disease attributes and drug responses in CRC as well. CRC organoids were first engineered from normal human intestinal stem cells that underwent gene editing to introduce known CRC mutations (Drost et al., 2015; Matano et al., 2015), with tumor-derived organoids reported shortly thereafter (van de Wetering et al., 2015).
Tumor-derived CRC organoids can be expanded and cryopreserved for long-term availability for drug screening, with organoids of diverse CRC genotypes reported. CRC organoids respond to targeted therapies in a mutation-dependent manner, again highlighting the potential role of organoids in tailoring relevant therapeutic regimens to cancer patients with specific mutations. In one report, organoids with mutations in the gene RNF43, but not others, were exquisitely sensitive to inhibition of the Wnt-processing enzyme Porcupine, with divergent responses to other potential therapies uncovered as well (van de Wetering et al., 2015).
Using hPSC-derived organoids, rather than those derived from tumors, a role was defined for Wnt inhibition in tumors harboring inherited APC mutations (Crespo et al., 2017). It has also been shown that gene expression signatures, in addition to mutational statuses, can be derived from organoids and used to predict sensitivity to targeted therapies in a patient-specific manner, as was shown for sensitivity to EGFR inhibition in CRC (Schutte et al., 2017). Recent progress has been made using organoids in establishing patient-specific features of protein abundance in CRC, which represents a level beyond genomic and transcriptomic features on which to interrogate drug responses at the single-patient level (Cristobal et al., 2017).
The culture conditions in which CRC organoids are maintained have also been leveraged to uncover patient-specific aspects of CRC biology. In multiple studies, researchers manipulated the growth conditions of organoids, and, in so doing, revealed what may well be microenvironmental dependencies of human CRC tumors with certain mutations (Drost et al., 2015; Fujii et al., 2016; Matano et al., 2015). The breadth of work done on CRC alone demonstrates that organoids have taken on a starring role in the study of patient-specific disease mechanisms and therapeutic approaches in cancer.
Liver, breast, and pancreas cancer
Organoids have formed the basis of studies describing intertumoral heterogeneity in additional cancer types as well, including pancreatic cancer, liver cancer, and breast cancer. Multiple groups have successfully modeled pancreatic cancer using tumor-derived organoids (Boj et al., 2015; Huang et al., 2015), positioning the field to model patient-specific disease features using this system in the near future. Indeed, one report has already described matching histological and physiological features between pancreatic cancer samples and their organoid progeny, but with heterogeneity of these features among patients (Huang et al., 2015). That group also reported patient-specific therapeutic vulnerabilities, with only a subset of organoids susceptible to inhibition of the histone methyltransferase EZH2.
A similar study emerged more recently using primary liver cancer organoids representing the known diverse subtypes of that entity (Broutier et al., 2017). The investigators in that case demonstrated remarkable phenotypic heterogeneity among the organoids, which corresponded to their parent tumors. They also found a range of responses to many candidate drugs that were screened in organoids, with potentially prognostic genetic alterations found in some cases. An inhibitor of ERK emerged from this work as a widely applicable and promising potential therapy for liver cancers.
Breast cancer organoids have entered the spotlight as well, with recent experiments suggesting their reliability as a model for drug development (Sachs et al., 2018). Not only do breast cancer organoids respond similarly to targeted therapies both in vitro and in vivo, when they are grafted into mice, but they also yield diverse responses to tamoxifen that are predictive of those observed in patients clinically.
It is clear that organoids have become a model system of choice for the investigation of heterogeneous disease features and drug responses among cancer patients. Organoids have now been derived and used to screen drug candidates in even more cancer types than those discussed above, with results shared in a comprehensive report highlighting the pan-cancer type applicability of this approach (Pauli et al., 2017). The importance of organoid technologies to cancer research is poised to grow ever larger in the years to come.
Considerations for future studies using stem cells and organoids to build cancer models
Although the development of organoid platforms with the resolution to capture both intratumoral and intertumoral heterogeneity has progressed substantially, there remain several challenges before human PSC and organoid models can become more integral parts of modern cancer research.
Building cancer models with functional microenvironments
Primary among these challenges is that of recapitulating tumor microenvironments in organoid models, as they contribute greatly to heterogeneity at both levels. Subpopulations of malignant cells with different phenotypes can influence each other’s growth and survival in the tumor microenvironment, as was recently highlighted in the context of glioblastoma (Wang et al., 2018). It is also now recognized that, in addition to cancer cells themselves, solid tumors are often surrounded and infiltrated by an array of cell types and structures that comprise the microenvironment, including immune cells, fibroblasts, neurons and glia, blood vessels, and complex matrix components, among others.
Future improvements to organoid technologies will likely include capabilities to maintain microenvironmental components from patient samples that are lost using current methods, and to integrate primary or stem cell-derived microenvironmental components from non-tumor sources into established organoids. Indeed, recent work using organoid models of PDAC, discussed above, reported the use of cancer-associated fibroblasts as a source of a microenvironmental factor of interest, WNT (Seino et al., 2018). Another recent study features peripheral blood as a source of lymphocytes that are then cultured with organoids to assess tumor-immune interactions (Dijkstra et al., 2018).
A variety of relevant cell types, including hematopoietic cells (Sugimura et al., 2017), lymphocytes (Brauer et al., 2016), microglia (Abud et al., 2017; Douvaras et al., 2017; Muffat et al., 2016; Pandya et al., 2017), central and peripheral neurons and glia (Tao and Zhang, 2016; Tchieu et al., 2017), and endothelial cells (Wilson et al., 2014) are now available from hPSCs and might be used to model tumor microenvironments. Such an approach is compelling and could facilitate further dissection of the role that individual microenvironmental components play in oncogenesis.
Ensuring proper cell maturation for cancer modeling studies
However, the use of hPSC progeny to supplement tumor organoids, as well as the use of hPSC-derived cells and organoids to model oncogenesis, raise the important question of the role of cell and environmental maturity in cancer. It is generally agreed that hPSC-derived cells and organoids represent early stages of development (Studer et al., 2015), and an embryonic or fetal niche may not represent an optimal microenvironment for the study of adult-onset cancer. Nevertheless, organoids may offer distinct advantages over 2D models by supporting cells at different stages of lineage progression, one or more of which may represent the most permissive cell-of-origin state for the development of a given type of neoplastic lesion.
Leveraging further improvements in (epi)genome editing
Genome-engineering and chromatin-modifying techniques in pluripotent stem cells and their progeny in 2D and 3D cultures are continuing to improve in both effectiveness and practicality (Hockemeyer and Jaenisch, 2016; Pulecio et al., 2017; Shi et al., 2017). These improvements are increasingly enabling genetic screens in hPSC-based disease models to identify potential therapeutic strategies. Studies of cancer models arising from these approaches will enable hPSCs and organoids to contribute increasingly to the study of cancer development, as well as to the study of cancer progression and therapy.
Using patient-derived iPSCs to model additional familial cancer syndromes
IPSCs are instrumental in highlighting defects in cell differentiation and tissue development caused by inherited human genetic variants, and thus offer value in understanding inherited cancer syndromes. Many of these syndromes are characterized by predisposition to specific cancer types despite the fact that the inherited variants are of widely expressed genes. In addition, in many syndromes, several variants of the same gene are associated with distinct clinical presentations. Modeling tissue and cancer development using iPSCs from patients with these syndromes promises, in particular, to reveal the molecular underpinnings of these phenomena.
Several iPSC lines have been established from patients with inherited cancer syndromes. These include those from patients harboring germline BRCA1 mutations predisposing to breast and ovarian cancers (Griscelli et al., 2017; Soyombo et al., 2013), from patients with neurofibromatosis type 1 (NF1) (Anastasaki et al., 2015; Wegscheid et al., 2018), and from those with Gorlin syndrome (Hasegawa et al., 2017). Directed differentiation strategies will enable researchers to use these lines to model development of the tissues that give rise to cancer in these syndromes.
Multiple studies have now been reported that include iPSCs generated from patients with inherited bone marrow disorders that predispose to cancer, such as Fanconi anemia (FA) (Liu et al., 2014; Raya et al., 2009), Schwachman-Diamond Syndrome (SBDS) (Tulpule et al., 2013), and Nijmegen Breakage Syndrome (NBS) (Halevy et al., 2016). These studies have provided mechanistic insights into the pathologies associated with these syndromes, with certain phenotypes – such as impaired hematopoietic and pancreatic exocrine differentiation of SBDS iPSCs (Tulpule et al., 2013) – likely relevant to syndrome-associated cancer.
Moving toward widespread use of hPSC-derived organoids in cancer research
An emerging strategy merges organoid cultures with the use of hPSCs derived from established lines or from individual patients with tumors. Several reports have appeared in the literature whereby hES or iPS cells have been used to generate 3D organoid cultures expressing mutations typically found in cancer of the pancreas (Huang et al., 2015), colon (Crespo et al., 2017), and brain (Bian et al., 2018; Ogawa et al., 2018). These studies leverage the unique power of organoid models to study specific cancer-driving mutations or even germline oncogenic alterations in a 3D environment that highlights developmental processes, heterogeneous cell composition, and early tumor-initiating events. The value of these approaches may vary depending on tumor type, the developmental window during which neoplastic events occur, and the specific questions being asked. It can be argued that the value of these systems in some settings, such as drug screens, may be limited since only one or a combination of very few defined mutations are being studied, in comparison to the usually significantly altered genome in tumor-derived organoids. As discussed previously, individually derived iPSCs can also contribute the patient’s own genomic context as a potentially important environmental parameter for the study of cancer-specific mutations.
With expected advances in protocols for the directed differentiation of hPSCs into specific lineages, and in gene-editing technologies, additional cancer models using genetically engineered organoids will likely emerge. Several groups have now reported organoid models of the normal brain (Di Lullo and Kriegstein, 2017; Lancaster et al., 2013), lung (Chen et al., 2017), breast (Qu et al., 2017), stomach (McCracken et al., 2014), intestine (Crespo et al., 2017), liver (Takebe et al., 2013), and kidney (Takasato et al., 2015) tissue derived from pluripotent stem cells. These organoid models are promising tools for the study of cancers arising in these tissues.
Conclusions
In recent years, researchers have gained an increased understanding of the developmental origins of, and relevance of heterogeneity found within and among, human cancers using human stem cell and organoid models. However, these modeling strategies have only begun to enter the spotlight in cancer research. With the continued rise of CRISPR-mediated genome-editing, hPSC differentiation protocols, and chromatin manipulation techniques, these advanced methods to study human cancer in vitro will only grow more sophisticated and more useful. These efforts will surely be informed by an increased understanding of precisely which cell types are represented among the trillions comprising the human body, thanks to the Human Cell Atlas now under development (Regev et al., 2017; Rozenblatt-Rosen et al., 2017) as well as to other single-cell transcriptomic profiling efforts in developmental and disease biology.
These technologies will become valued complements to models created using traditional mouse genetics, with work done in each model system contributing to a concerted effort to understand and to treat cancers as successfully as possible. With each advance in these areas, cancer research as a field stands to benefit.
In this Perspective, Smith and Tabar discuss how models of human pluripotent stem cells (hPSCs) and organoids provide insights into tumor development, progression, and treatment, as well as potential opportunities for cancer research using hPSC-derived organoids and genome editing.
Acknowledgements
We would like to thank members of the Tabar laboratory for regular discussions of advances in the field. We would also like to thank Terry Helms and Wenjing Wu of the MSKCC Department of Communications for their work on the graphics for this article. We gratefully acknowledge funding support from the National Cancer Institute (R01 CA 208405 to V.T.; F31 CA 210408 to R.C.S.), the Keck Foundation (V.T.), the Starr Cancer Consortium (V.T.), and the Louis V. Gerstner, Jr., Graduate School of Biomedical Sciences (R.C.S.). This work was also supported by a core grant from the National Cancer Institute, P30 CA 008748. We apologize to authors whose work we were not able to include due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
Viviane Tabar is a founding investigator of BlueRock Therapeutics.
References:
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, et al. (2017). iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 94, 278–293 e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasaki C, Woo AS, Messiaen LM, and Gutmann DH (2015). Elucidating the impact of neurofibromatosis-1 germline mutations on neurofibromin function and dopamine-based learning. Hum Mol Genet 24, 3518–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, et al. (2013). Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24, 660–672. [DOI] [PubMed] [Google Scholar]
- Bian S, Repic M, Guo Z, Kavirayani A, Burkard T, Bagley JA, Krauditsch C, and Knoblich JA (2018). Genetically engineered cerebral organoids model brain tumor formation. Nature Methods 15, 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, et al. (2015). Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer PM, Singh J, Xhiku S, and Zuniga-Pflucker JC (2016). T Cell Genesis: In Vitro Veritas Est? Trends Immunol 37, 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, et al. (2017). Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med 23, 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, Wernig M, Jaenisch R, and Brummelkamp TR (2010). Generation of iPSCs from cultured human malignant cells. Blood 115, 4039–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-J, Kotini AG, Olszewska M, Georgomanoli M, Teruya-Feldstein J, Sperber H, Sanchez R, DeVita R, Martins TJ, Abdel-Wahab O, et al. (2018). Dissecting the Contributions of Cooperating Gene Mutations to Cancer Phenotypes and Drug Responses with Patient-Derived iPSCs. Stem Cell Reports 10, 1610–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Gentles AJ, Chatterjee S, Lan F, Reinisch A, Corces MR, Xavy S, Shen J, Haag D, Chanda S, et al. (2017). Human AML-iPSCs Reacquire Leukemic Properties after Differentiation and Model Clonal Variation of Disease. Cell Stem Cell 20, 329–344 e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YW, Huang SX, de Carvalho A, Ho SH, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova JL, Bhattacharya J, et al. (2017). A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 19, 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo M, Vilar E, Tsai SY, Chang K, Amin S, Srinivasan T, Zhang T, Pipalia NH, Chen HJ, Witherspoon M, et al. (2017). Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat Med 23, 878–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristobal A, van den Toorn HWP, van de Wetering M, Clevers H, Heck AJR, and Mohammed S (2017). Personalized Proteome Profiles of Healthy and Tumor Human Colon Organoids Reveal Both Individual Diversity and Basic Features of Colorectal Cancer. Cell Rep 18, 263–274. [DOI] [PubMed] [Google Scholar]
- Dai X, Gan W, Li X, Wang S, Zhang W, Huang L, Liu S, Zhong Q, Guo J, Zhang J, et al. (2017). Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med 23, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lullo E, and Kriegstein AR (2017). The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci 18, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, et al. (2018). Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 174, 1586–1598.e1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, Terrenoire C, Zhang B, Gandy S, Schadt E, et al. (2017). Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports 8, 1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, and Clevers H (2018). Organoids in cancer research. Nat Rev Cancer [DOI] [PubMed]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, et al. (2015). Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47. [DOI] [PubMed] [Google Scholar]
- Duan S, Yuan G, Liu X, Ren R, Li J, Zhang W, Wu J, Xu X, Fu L, Li Y, et al. (2015). PTEN deficiency reprogrammes human neural stem cells towards a glioblastoma stem cell-like phenotype. Nat Commun 6, 10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Heo I, and Clevers H (2017). Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol Med 23, 393–410. [DOI] [PubMed] [Google Scholar]
- Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, et al. (2016). A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 18, 827–838. [DOI] [PubMed] [Google Scholar]
- Funato K, Major T, Lewis PW, Allis CD, and Tabar V (2014). Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346, 1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K, and Tabar V (2018). Histone Mutations in Cancer. Annual Review of Cancer Biology 2, 337–351. [Google Scholar]
- Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, et al. (2014). Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griscelli F, Oudrhiri N, Feraud O, Divers D, Portier L, Turhan AG, and Bennaceur Griscelli A (2017). Generation of induced pluripotent stem cell (iPSC) line from a patient with triple negative breast cancer with hereditary exon 17 deletion of BRCA1 gene. Stem Cell Res 24, 135–138. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, and Weinberg RA (2005). The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet 37, 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy T, Akov S, Bohndorf M, Mlody B, Adjaye J, Benvenisty N, and Goldberg M (2016). Chromosomal Instability and Molecular Defects in Induced Pluripotent Stem Cells from Nijmegen Breakage Syndrome Patients. Cell Rep 16, 2499–2511. [DOI] [PubMed] [Google Scholar]
- Hasegawa D, Ochiai-Shino H, Onodera S, Nakamura T, Saito A, Onda T, Watanabe K, Nishimura K, Ohtaka M, Nakanishi M, et al. (2017). Gorlin syndrome-derived induced pluripotent stem cells are hypersensitive to hedgehog-mediated osteogenic induction. PLoS One 12, e0186879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Blelloch R, Brennan C, Yamada Y, Kim M, Chin L, and Jaenisch R (2004). Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev 18, 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, and Jaenisch R (2016). Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell 18, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Holtzinger A, Jagan I, BeGora M, Lohse I, Ngai N, Nostro C, Wang R, Muthuswamy LB, Crawford HC, et al. (2015). Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med 21, 1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, Couce M, McLendon RE, Sloan AE, and Rich JN (2016). A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Res 76, 2465–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, and Baker SJ (2014). Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, et al. (2014). Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bamlet WR, Oberg AL, Chaffee KG, Donahue G, Cao XJ, Chari S, Garcia BA, Petersen GM, and Zaret KS (2017). Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19–9 blood markers. Science translational medicine 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hoffman JP, Alpaugh RK, Rhim AD, Reichert M, Stanger BZ, Furth EE, Sepulveda AR, Yuan CX, Won KJ, et al. (2013). An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep 3, 2088–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotini AG, Chang CJ, Boussaad I, Delrow JJ, Dolezal EK, Nagulapally AB, Perna F, Fishbein GA, Klimek VM, Hawkins RD, et al. (2015). Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat Biotechnol 33, 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotini AG, Chang CJ, Chow A, Yuan H, Ho TC, Wang T, Vora S, Solovyov A, Husser C, Olszewska M, et al. (2017). Stage-Specific Human Induced Pluripotent Stem Cells Map the Progression of Myeloid Transformation to Transplantable Leukemia. Cell Stem Cell 20, 315–328 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, and Knoblich JA (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Su J, Kim HS, Chang B, Papatsenko D, Zhao RY, Yuan Y, Gingold J, Xia WY, Darr H, et al. (2015). Modeling Familial Cancer with Induced Pluripotent Stem Cells. Cell 161, 240–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, Lawrence MS, Auclair D, Mora J, Golub TR, et al. (2012). A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest 122, 2983–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, Williams AB, et al. (2018). Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 173, 515–528.e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, and Allis CD (2013). Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science 340, 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GH, Suzuki K, Li M, Qu J, Montserrat N, Tarantino C, Gu Y, Yi F, Xu X, Zhang W, et al. (2014). Modelling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat Commun 5, 4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM, Wang X, Gallo M, Garzia L, Zayne K, et al. (2014). Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506, 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnen CL, Shen MM, and Abate-Shen C (2018). Lineage Plasticity in Cancer Progression and Treatment. Annual Review of Cancer Biology 2, 271–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, and Sato T (2015). Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 21, 256–262. [DOI] [PubMed] [Google Scholar]
- McCauley HA, and Wells JM (2017). Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 144, 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, et al. (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell RG, Deggins BA, and Labat DD (1969). TRANSPLANTATION OF PLURIPOTENTIAL NUCLEI FROM TRIPLOID FROG TUMORS. Science 165, 394–+. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, Nagano H, Sekimoto M, Doki Y, and Mori M (2010). Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci U S A 107, 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek AS, Golub D, Khan T, Bready D, Prado J, Bowman C, Deng J, Zhang G, Rocha PP, Raviram R, et al. (2017). Low-Grade Astrocytoma Mutations in IDH1, P53, and ATRX Cooperate to Block Differentiation of Human Neural Stem Cells via Repression of SOX2. Cell Reports 21, 1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. (2012). Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483, 589–593. [DOI] [PubMed] [Google Scholar]
- Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai LH, Aubourg P, Ransohoff RM, et al. (2016). Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med 22, 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero-Navarro S, Sevilla A, Roman AC, Lee DF, D’Souza SL, Pardo S, Riess I, Su J, Cohen N, Schaniel C, et al. (2015). Myeloid Dysregulation in a Human Induced Pluripotent Stem Cell Model of PTPN11-Associated Juvenile Myelomonocytic Leukemia. Cell Rep 13, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Ishii H, Miyoshi N, Hoshino H, Saito T, Sato T, Tomimaru Y, Kobayashi S, Nagano H, Sekimoto M, et al. (2010). Long-term culture following ES-like gene-induced reprogramming elicits an aggressive phenotype in mutated cholangiocellular carcinoma cells. Biochem Biophys Res Commun 395, 258–263. [DOI] [PubMed] [Google Scholar]
- Nanki K, Toshimitsu K, Takano A, Fujii M, Shimokawa M, Ohta Y, Matano M, Seino T, Nishikori S, Ishikawa K, et al. (2018). Divergent Routes toward Wnt and R-spondin Niche Independency during Human Gastric Carcinogenesis. Cell 174, 856–869.e817. [DOI] [PubMed] [Google Scholar]
- Ogawa J, Pao GM, Shokhirev MN, and Verma IM (2018). Glioblastoma Model Using Human Cerebral Organoids. Cell Rep 23, 1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, Kim G, Brown MA, Elkahloun AG, Maric D, et al. (2017). Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci 20, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou EP (2016). Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat Med 22, 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet-Fifield S, Koh SL, Cheng L, Beyit LM, Shembrey CE, Moelck C, Behrenbruch C, Papin M, Gironella M, Guelfi S, et al. (2018). Tight junction protein claudin-2 promotes self-renewal of human colorectal cancer stem-like cells. Cancer Res [DOI] [PubMed]
- Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R, et al. (2014). C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature 506, 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, et al. (2017). Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov 7, 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter VR (1969). Recent trends in cancer biochemistry: the importance of studies on fetal tissue. Proceedings Canadian Cancer Conference 8, 9–30. [PubMed] [Google Scholar]
- Potter VR (1978). Phenotypic diversity in experimental hepatomas: the concept of partially blocked ontogeny. The 10th Walter Hubert Lecture. British journal of cancer 38, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puisieux A, Pommier RM, Morel AP, and Lavial F (2018). Cellular Pliancy and the Multistep Process of Tumorigenesis. Cancer Cell 33, 164–172. [DOI] [PubMed] [Google Scholar]
- Pulecio J, Verma N, Mejia-Ramirez E, Huangfu D, and Raya A (2017). CRISPR/Cas9-Based Engineering of the Epigenome. Cell Stem Cell 21, 431–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Han B, Gao B, Bose S, Gong Y, Wawrowsky K, Giuliano AE, Sareen D, and Cui X (2017). Differentiation of Human Induced Pluripotent Stem Cells to Mammary-like Organoids. Stem Cell Reports 8, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. (2012). Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castella M, Rio P, Sleep E, et al. (2009). Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 460, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan JL, Schumacher D, Staudte S, Steffen A, Haybaeck J, Keilholz U, Schweiger C, Golob-Schwarzl N, Mumberg D, Henderson D, et al. (2017). Non-Canonical Hedgehog Signaling Is a Positive Regulator of the WNT Pathway and Is Required for the Survival of Colon Cancer Stem Cells. Cell Rep 21, 2813–2828. [DOI] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. (2017). The Human Cell Atlas. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, and Weller M (2017). Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol 14, 434–452. [DOI] [PubMed] [Google Scholar]
- Rich JN, Guo C, McLendon RE, Bigner DD, Wang X-F, and Counter CM (2001). A Genetically Tractable Model of Human Glioma Formation. Cancer Research 61, 3556–3560. [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. (2012). Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Stubbington MJT, Regev A, and Teichmann SA (2017). The Human Cell Atlas: from vision to reality. Nature 550, 451–453. [DOI] [PubMed] [Google Scholar]
- Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, et al. (2018). A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 172, 373–386 e310. [DOI] [PubMed] [Google Scholar]
- Sancho-Martinez I, Nivet E, Xia Y, Hishida T, Aguirre A, Ocampo A, Ma L, Morey R, Krause MN, Zembrzycki A, et al. (2016). Establishment of human iPSC-based models for the study and targeting of glioma initiating cells. Nature Communications 7, 10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasikumar R, Rejitha JR, Binumon PK, and Manoj M (2011). Role of heterozygous APC mutation in niche succession and initiation of colorectal cancer--a computational study. PLoS One 6, e22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte M, Risch T, Abdavi-Azar N, Boehnke K, Schumacher D, Keil M, Yildiriman R, Jandrasits C, Borodina T, Amstislavskiy V, et al. (2017). Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat Commun 8, 14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. (2012). Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231. [DOI] [PubMed] [Google Scholar]
- Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, Ohta Y, Matano M, Nanki K, Kawasaki K, et al. (2018). Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell [DOI] [PubMed]
- Sell S (2008). Alpha-fetoprotein, stem cells and cancer: how study of the production of alpha-fetoprotein during chemical hepatocarcinogenesis led to reaffirmation of the stem cell theory of cancer. Tumour Biol 29, 161–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Inoue H, Wu JC, and Yamanaka S (2017). Induced pluripotent stem cell technology: a decade of progress. Nature reviews Drug discovery 16, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoag J, Liu D, Blattner M, Sboner A, Park K, Deonarine L, Robinson BD, Mosquera JM, Chen Y, Rubin MA, et al. (2018). SPOP mutation drives prostate neoplasia without stabilizing oncogenic transcription factor ERG. J Clin Invest 128, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smpokou P, Zand DJ, Rosenbaum KN, and Summar ML (2015). Malignancy in Noonan syndrome and related disorders. Clin Genet 88, 516–522. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Capilla A, Molina-Estevez FJ, Gianotti-Sommer A, Skvir N, Caballero I, Chowdhury S, and Mostoslavsky G (2018). Modeling APC mutagenesis and familial adenomatous polyposis using human iPS cells. PLoS One 13, e0200657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, and Pieper RO (2001). Formation of Intracranial Tumors by Genetically Modified Human Astrocytes Defines Four Pathways Critical in the Development of Human Anaplastic Astrocytoma. Cancer Research 61, 4956–4960. [PubMed] [Google Scholar]
- Soyombo AA, Wu Y, Kolski L, Rios JJ, Rakheja D, Chen A, Kehler J, Hampel H, Coughran A, and Ross TS (2013). Analysis of induced pluripotent stem cells from a BRCA1 mutant family. Stem Cell Reports 1, 336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker SH, Feber A, Engstrom PG, Caren H, Kurian KM, Takashima Y, Watts C, Way M, Dirks P, Bertone P, et al. (2013). Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev 27, 654–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer L, Vera E, and Cornacchia D (2015). Programming and Reprogramming Cellular Age in the Era of Induced Pluripotency. Cell Stem Cell 16, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura R, Jha DK, Han A, Soria-Valles C, da Rocha EL, Lu YF, Goettel JA, Serrao E, Rowe RG, Malleshaiah M, et al. (2017). Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, and Yamanaka S (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, et al. (2013). Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. [DOI] [PubMed] [Google Scholar]
- Tao Y, and Zhang SC (2016). Neural Subtype Specification from Human Pluripotent Stem Cells. Cell Stem Cell 19, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Chow RD, Saladi SV, Tata A, Konkimalla A, Bara A, Montoro D, Hariri LP, Shih AR, Mino-Kenudson M, et al. (2018). Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity. Developmental cell 44, 679–693.e675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J, Zimmer B, Fattahi F, Amin S, Zeltner N, Chen S, and Studer L (2017). A Modular Platform for Differentiation of Human PSCs into All Major Ectodermal Lineages. Cell Stem Cell 21, 399–410 e397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulpule A, Kelley JM, Lensch MW, McPherson J, Park IH, Hartung O, Nakamura T, Schlaeger TM, Shimamura A, and Daley GQ (2013). Pluripotent stem cell models of Shwachman-Diamond syndrome reveal a common mechanism for pancreatic and hematopoietic dysfunction. Cell Stem Cell 12, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Maherali N, Kulalert W, and Hochedlinger K (2009). Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci 122, 3502–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. (2015). Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 161, 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, and Kinzler KW (2013). Cancer Genome Landscapes. Science 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Prager BC, Wu Q, Kim LJY, Gimple RC, Shi Y, Yang K, Morton AR, Zhou W, Zhu Z, et al. (2018). Reciprocal Signaling between Glioblastoma Stem Cells and Differentiated Tumor Cells Promotes Malignant Progression. Cell Stem Cell 22, 514–528.e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegscheid ML, Anastasaki C, and Gutmann DH (2018). Human stem cell modeling in neurofibromatosis type 1 (NF1). Exp Neurol 299, 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HK, Canfield SG, Shusta EV, and Palecek SP (2014). Concise review: tissue-specific microvascular endothelial cells derived from human pluripotent stem cells. Stem Cells 32, 3037–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. (2012). Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44, 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HHN, Siu HC, Law S, Ho SL, Yue SSK, Tsui WY, Chan D, Chan AS, Ma S, Lam KO, et al. (2018). A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell [DOI] [PubMed]