This article is a retrospective analysis of melanoma cases with BRAF alterations detected using a hybrid capture‐based comprehensive genomic profiling assay, analyzing history of prior BRAF testing and available outcomes data.
Keywords: Melanoma, BRAF testing, V600E, Hybrid capture‐based genomic profiling
Abstract
Background.
BRAF and MEK inhibitors are approved for BRAF V600‐mutated advanced melanoma, with response rates of up to 70%. Responses to targeted therapies have also been observed for diverse non‐V600 BRAF alterations. Thus, sensitive, accurate, and broad detection of BRAF alterations is critical to match patients with available targeted therapies.
Materials and Methods.
Pathology reports were reviewed for 385 consecutive melanoma cases with BRAF mutations or rearrangements identified using a hybrid capture‐based next‐generation sequencing comprehensive genomic profiling (CGP) assay during the course of clinical care.
Results.
Records of prior BRAF molecular testing were available for 79 (21%) cases. Of cases with BRAF V600 mutations, 11/57 (19%) with available data were negative by prior BRAF testing. Prior negative BRAF results were also identified in 16/20 (80%) cases with non‐V600 mutations, 2 of which harbored multiple BRAF alterations, and in 2/2 (100%) cases with activating BRAF fusions. Clinical outcomes for a subset of patients are presented.
Conclusion.
CGP identifies diverse activating BRAF alterations in a significant fraction of cases with prior negative testing. Given the proven clinical benefit of BRAF/MEK inhibitors in BRAF‐mutated melanoma, CGP should be considered for patients with metastatic melanoma, particularly if other testing is negative.
Implications for Practice.
Published guidelines for melanoma treatment recommend BRAF mutational analysis, but little guidance is provided as to selection criteria for testing methodologies, or as to clinical implications for non‐V600 alterations. This study found that hybrid capture‐based next‐generation sequencing can detect BRAF alterations in samples from a significant fraction of patients with advanced melanoma with prior negative BRAF results. This study highlights the need for oncologists and pathologists to be critically aware of coverage and sensitivity limitations of various assays, particularly regarding non‐V600E alterations, of which many are potentially targetable.
Introduction
One of the most compelling examples of clinical utility of targeted therapies is the development of BRAF and MEK inhibitors for the treatment of BRAF V600E/K‐mutated melanoma in stage IV disease [1], [2], [3], [4] and recently in stage III disease in the adjuvant setting [5]. BRAF exon 15 mutations drive proliferation of more than 50% of all cutaneous melanomas [6]. Vemurafenib and dabrafenib [7] have shown remarkable clinical activity in patients with BRAF V600E/K‐mutated melanoma and received U.S. Food and Drug Administration (FDA) approval for the treatment of metastatic melanoma. Subsequently, combinations of BRAF and MEK inhibitors showed improved efficacy when compared with BRAF inhibitor monotherapy, with responses in approximately 70% of cases and median overall survival exceeding 2 years [8], [9], [10].
In addition to V600E/K, other substitutions and indels at V600 and many non‐V600 BRAF mutations have been found, mostly clustered in the activation segment or in the glycine‐rich loop of the kinase domain [11], [12]. BRAF exon 11 mutations have been associated with responses to diverse multikinase inhibitors, such as sorafenib [13] and dasatinib [14]. In addition to BRAF short variant mutations, constitutively activating fusions retaining the BRAF catalytic domain are also found in melanomas, and enriched in Spitzoid melanomas [15], [16]. Because of the rarity and novelty of these fusions, to date no international clinical trials have been initiated for this subgroup, but MEK inhibitors have shown some clinical efficacy in this context and may constitute a crucial therapeutic option for these patients [15], [17].
Current methodology for detecting BRAF alterations in clinical specimens is left to laboratory discretion, and as such multiple assays are used in clinical practice to inform therapy selection [18]. Importantly, limitations and performance characteristics of molecular assays are typically not readily apparent to the treating physician. Given the substantial clinical benefit demonstrated for BRAF and MEK inhibitors in patients with BRAF‐V600E‐mutated melanoma, assessing the limitations of BRAF testing typically used in clinical care is critical. Beyond V600E mutation, other alterations both at V600 and throughout BRAF should be recognized, given early evidence of targetability.
To this end, a comprehensive review of melanoma cases with BRAF alterations detected using a hybrid capture‐based comprehensive genomic profiling (CGP) assay during clinical care was conducted. Both history of prior BRAF testing and available outcomes data were analyzed.
Materials and Methods
A minimum of 50 ng of DNA was extracted from 40 μm of formalin‐fixed paraffin embedded sections of 385 consecutive melanoma cases submitted during the course of clinical care (March 2016 and March 2017), and CGP was performed on hybridization captured, adaptor ligation‐based libraries to a mean coverage depth of >600× for the entire coding sequence of 236 or 315 cancer‐related genes plus 19 to 28 introns from genes frequently rearranged in cancer (including all BRAF exons and introns 7–10) to identify base pair substitutions, insertions/deletions, copy number alterations, and rearrangements [19]. Testing was performed in a Clinical Laboratory Improvement Amendments‐certified/College of American Pathologists‐accredited laboratory (Foundation Medicine Inc., Cambridge, MA). Tumor mutational burden (TMB) was characterized as the number of somatic base substitution or indel alterations per megabase (Mb). Prior test results were extracted from provided pathology reports. Approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817).
Results
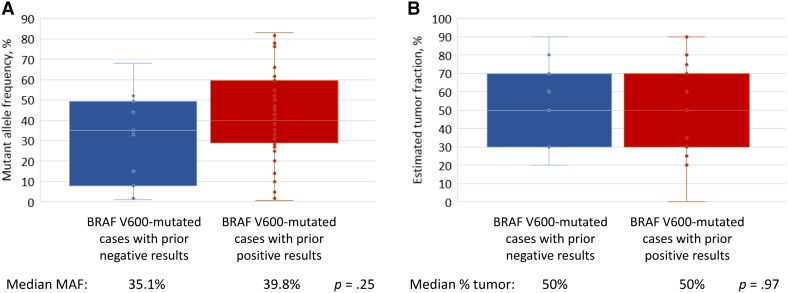
CGP was performed on 385 advanced‐stage melanomas during the course of clinical care. The distribution of cases assessed and results of prior BRAF testing are shown in Figure 1. In this set of 385 advanced melanomas, 38 unique BRAF short variant mutations and 5 unique activating rearrangements (fusions or kinase domain duplications) were represented (Fig. 2). Prior testing records using diverse methodologies were available for 79/385 (21%) cases (Fig. 3). Overall, 29/79 (37%) cases with BRAF alterations detected using CGP were BRAF negative on prior testing. Of BRAF V600‐mutated cases, 11/57 (19%) with available data were negative by prior BRAF testing, including 7/45 (16%) with V600E mutations. In 9/29 cases with prior negative BRAF results, information regarding the prior test methodology was available and suggested that the detected alteration should have been covered (Table 1; Fig. 3C). Biopsies with the same surgical pathology ID were tested in 3/9 cases (including the same block or slide in 2 cases). In the remaining cases, prior testing was done on a confirmed different sample in 5 cases (including one liquid biopsy), and on an unknown sample in 1 case (Table 1; Fig. 3C). Of all cases in which the BRAF mutation was expected to be covered by the prior assay and the same sample was tested, 3/35 (8.6%) appear to have been false negatives (Table 1, patient 5, 6, and 7). For comparison, in cases in which the other test methodology covered the detected BRAF mutation, the same sample was confirmed to have been tested in 3/9 (33%) prior negative cases and 32/50 (64%) prior positive cases (Fig. 3C). Characteristics of cases with BRAF alterations identified by CGP, but not detected by prior BRAF testing, are shown in Figure 2. There was no significant difference in BRAF mutant allele frequencies (median 35% vs. 40%, p = .25) or percentage of tumor nuclei (median 50% for both, p = .97) between BRAF V600‐mutated samples with prior negative and prior positive results (Fig. 4).
Figure 1.
Consort diagram of melanoma patients with BRAF alterations identified using comprehensive genomic profiling.Abbreviation: NGS, next‐generation sequencing.
Figure 2.
Distribution of BRAF alterations detected by comprehensive genomic profiling and associated tumor mutational burden. (A): Distribution of BRAF short variant mutations and rearrangements in our set of 385 advanced melanoma cases. (B): Median tumor mutational burden (mutations per megabase) is shown for each subset of cases.Abbreviations: KDD, kinase domain duplication; SV, short variant; TMB, tumor mutational burden
Figure 3.
Characteristics of cases with BRAF alterations identified using comprehensive genomic profiling (CGP) after prior negative BRAF testing. (A): Characteristics of 29 cases with BRAF alterations identified by hybrid capture‐based next‐generation sequencing (HC‐NGS) after prior negative BRAF result. *Non‐hybrid capture‐based NGS assays. #Two samples with activating K601E mutations also has a second activating or uncharacterized BRAF mutation identified. (B): Thirty‐one BRAF alterations identified in 29 patient samples by HC‐NGS after prior negative result. (C): Fraction of cases in which the detected alteration was expected to be covered by the prior test (100% for prior positive, not shown), as well as the fraction of cases for which the same sample was tested by CGP and the prior BRAF test.Abbreviations: F, female; M, male; MAF, mutant allele frequency; NA, not applicable.
Table 1. Characteristics of 29 melanoma cases positive for BRAF alterations using CGP with prior negative BRAF results.
Prior test details were not provided; however, we make the assumption that any molecular test returning a "BRAF wild‐type" result for melanoma would have covered the canonical V600E mutation.
Both samples were from a single mid‐back specimen, but different slides from the tissue block were tested.
Both samples were from a single left‐thigh specimen.
Liquid biopsy collected 12 months after tissue sample.
Test specifications note probes are designed for V600E but will detect other V600 alterations with limited sensitivity.
Test specifications note primers are designed for V600E/K but may cross react with K601E.
Both samples were from a right‐groin soft tissue mass.
Liquid biopsy collected 4.5 months after tissue sample.
Test methodology says BRAF exon 15 is covered but does not specify individual residues.
Both samples were from a single right‐thigh specimen, but different slides from the tissue block were tested.
Abbreviations: CGP, comprehensive genomic profiling; MAF, mutant allele frequency; N, no; n/a, not applicable; Y, yes.
Figure 4.
BRAF MAF and tumor fraction of 57 cases with BRAF V600 mutation detected by hybrid capture‐based next‐generation sequencing. Comparison of BRAF V600 mutant allele frequency (A), and tumor fraction (B), in 11 cases with prior negative BRAF results (blue) and 46 cases with prior positive BRAF results (red).Abbreviation: MAF, mutant allele frequency.
Prior negative results were identified in 16/20 (80%) cases with BRAF non‐V600 mutations, 2 of which harbored multiple BRAF alterations. Specifically, BRAF non‐V600 mutations not detected by prior testing included nine activating mutations: K601E (n = 4), G464V, G469V, E586K, L597Q, and A589_T599insT; four mutations predicted to result in impaired BRAF kinase activity: D594A/G/N (n = 3) and G466V; and five uncharacterized mutations: S467L (n = 2), L584F (n = 2), and N581I. In addition, 2/2 (100%) cases with activating BRAF fusions (TRIM24‐BRAF and SOX5‐BRAF) also had prior negative BRAF results (Fig. 3A, 3B).
Clinical follow‐up was available for seven patients with BRAF‐activating alterations identified using CGP following prior negative BRAF testing. One patient (Table 1, patient 2), a 27‐year‐old male, directly benefited from CGP testing. Initial BRAF testing using melting curve analysis was negative for V600 mutations. The patient received ipilimumab + nivolumab as first‐line systemic treatment for stage IV melanoma. After 5 months of immunotherapy, CGP of a second biopsy detected BRAF V600E (68% mutant allele frequency) as well as a TMB of 18 mutations/Mb. Immediately following this result, immunotherapy was discontinued because of symptomatic decline and toxicity, and the patient began dabrafenib + trametinib, resulting in symptom improvement and shrinkage of metastatic lung nodules after 3 months of treatment.
Of the remaining patients, four are currently being treated with immunotherapy with ongoing responses, and may pursue BRAF and/or MEK inhibitors as a next line of therapy. In the sixth case, the patient progressed on immunotherapy and then was too ill to pursue a BRAF/MEK inhibitor after the BRAF V600E‐positive result was returned. In the final case, a SOX5 (exons 1–6)‐BRAF (exons 9–18) fusion was identified. The patient received 6 weeks of cobimetinib with no response and is now on an immunotherapy trial. Shortly after our study, an additional patient with an ERC1‐BRAF fusion identified using CGP, with previously negative testing for BRAF, showed a good response to sorafenib after progressing on immunotherapy (Fig. 5), and is still under treatment to date.
Figure 5.
Photographs of a 43‐year‐old female patient with stage IV ERC1‐BRAF fusion melanoma, taken before (A, C) and after (B, D) 2 months of oral sorafenib 400 mg b.i.d. Subcutaneous metastases of the anterior part of the trunk (A, B) and subcutaneous metastases of the back (C, D), responding to sorafenib, after progressing under immunotherapy.
Discussion
Within the last decade, the use of rationally applied targeted therapy has revolutionized the care of metastatic melanoma, beginning with the identification of BRAF V600 mutations, predominantly V600E, that respond to combined BRAF and MEK inhibitors. For such therapies to be optimally delivered, there is an inherent mandate for specific and sensitive clinical testing to detect BRAF mutations. Two recent studies [20], [21] demonstrate that CGP applied in the course of clinical care can identify genomic alterations that guide targeted therapy for patients with advanced non‐small cell lung cancer who have been previously tested "negative" by standard‐of‐care molecular testing.
Despite approved companion diagnostics, significant variability exists in methods used for BRAF testing in the clinical setting. These include BRAF V600E‐specific clone VE1 immunohistochemistry (IHC) [22], [23], polymerase chain reaction (PCR; such as the FDA‐approved COBAS 4800 test), pyrosequencing, high‐resolution melting analysis, Sanger sequencing, and next‐generation sequencing (NGS). Herein, CGP identified BRAF alterations in 37% of cases with prior negative BRAF results returned using a variety of testing methodologies. For cases with BRAF V600 mutations detected by CGP, 19% had prior negative results. For cases with non‐V600 alterations detected by CGP, 80% of cases had prior negative results. No significant differences in mutant allele frequency or tumor fraction were observed in V600‐positive cases with prior positive or prior negative results, suggesting that these variables are unlikely to explain the missed detection in most cases. In approximately two thirds of cases, the prior negative result was likely due to limited coverage of the original BRAF testing method. In two cases, BRAF V600K mutations were not detected when IHC specific for V600E was used, and in one case, V600R (MAF 49%) was not detected by a PCR assay, which indicated that probes were designed for V600E, but that other V600 alterations might be detected with limited sensitivity. Non‐V600 alterations (including 4 K601E and 1 BRAF fusion) were not detected in 14 cases in which the mutation present was known to be excluded from coverage by the assay employed. We also acknowledge that among nine cases with prior negative results in which the original assay should have detected the BRAF alteration, the same sample was only confirmed to have been tested in one third of cases. However, in cases with prior positive results, the same sample was only confirmed to have been tested in 64% of cases, and in 30% of cases a different sample was confirmed to have been tested. Further, studies have shown that driver mutations are strongly conserved between primary and metastatic samples [24].
Although dabrafenib and vemurafenib are specifically developed to inhibit BRAF V600 mutations, responses to RAF and/or MEK inhibitors have been reported for many less common BRAF alterations, including a subset of those observed herein [15], [16], [25], [26], [27]. Currently, multiple clinical trials are enrolling melanoma patients with BRAF non‐V600 alterations (NCT02296112, NCT02465060). This creates an imperative, beyond better BRAF V600 testing, for detection of these diverse kinase‐activating mutations and fusions.
In addition to high sensitivity for detection of diverse BRAF alterations, CGP determines the TMB of a given sample using an algorithm that, based on the genomic alterations detected on 0.83–1.14 Mb of DNA, extrapolates to the genome as a whole [28]. As high TMB may predict responses to immunotherapy in melanoma [29], [30], the option to obtain CGP should be accessible to each patient and doctor prior to treatment decision‐making in the context of metastatic melanoma.
Conclusion
This study highlights the importance of using a sensitive full‐coverage assay, such as the hybrid capture‐based CGP assay employed herein, as opposed to conventional assays that have less sensitivity and/or are limited to small regions of the gene, which may miss less well‐characterized but still actionable alterations. Published guidelines strongly encourage the use of BRAF mutational assays, but without explicit mention of performance characteristics, the necessity of rigorous analytic validation, or the potential value of coverage beyond V600. Previously reported responses to targeted therapies in patients with diverse BRAF alterations, as well as clinical cases described herein, highlight the need for consistent accurate detection of these alterations to allow for selection of matched therapies associated with demonstrated clinical efficacy.
Acknowledgments
We thank the patients who participated in this study and their families.
Author Contributions
Conception/design: Siraj Mahamed Ali, Alexa B. Schrock, Lise Boussemart, Vincent A. Miller
Provision of study material or patients: Michael Wong, Jeffrey Sosman, Janice Mehnert, Gregory Daniels, Kari Kendra
Collection and/or assembly of data: Annie Nelson, Alexa B. Schrock
Data analysis and interpretation: Alexa B. Schrock, Jeffrey S. Ross, Siraj Mahamed Ali
Manuscript writing: Lise Boussemart, Alexa B. Schrock, Siraj Mahamed Ali
Final approval of manuscript: All authors
Disclosures
Lise Boussemart: Novartis, Pierre Fabre (C/A); Annie Nelson: Foundation Medicine, Inc. (E, OI); Jeffrey S. Ross: Foundation Medicine, Inc. (E, OI); Siraj Mahamed Ali: Foundation Medicine, Inc. (E, OI), Incysus Inc. (SAB), Revolution Medicines (C/A); Vincent A. Miller: Foundation Medicine, Inc. (E, OI), Revolution Medicine (Board of Directors); Alexa B. Schrock: Foundation Medicine, Inc. (E, OI). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Chapman PB, Hauschild A, Robert C et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flaherty KT, Puzanov I, Kim KB et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010;363:809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McArthur GA, Chapman PB, Robert C et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation‐positive melanoma (BRIM‐3): Extended follow‐up of a phase 3, randomised, open‐label study. Lancet Oncol 2014;15:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hauschild A, Grob JJ, Demidov LV et al. Dabrafenib in BRAF‐mutated metastatic melanoma: A multicentre, open‐label, phase 3 randomised controlled trial. Lancet 2012;380:358–365. [DOI] [PubMed] [Google Scholar]
- 5.Long GV, Hauschild A, Santinami M et al. Adjuvant dabrafenib plus trametinib in stage III BRAF‐mutated melanoma. N Engl J Med 2017;377:1813–1823. [DOI] [PubMed] [Google Scholar]
- 6.Davies H, Bignell GR, Cox C et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–954. [DOI] [PubMed] [Google Scholar]
- 7.Laquerre S, Arnone M, Moss K et al. A selective RAF kinase inhibitor induces cell death and tumor regression of human cancer cell lines encoding B‐RAFV600E mutation. Mol Cancer Ther 2009;8(suppl 1):B88a. [Google Scholar]
- 8.Bollag G, Hirth P, Tsai J et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF‐mutant melanoma. Nature 2010;467:596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long GV, Stroyakovskiy D, Gogas H et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med 2014;371:1877–1888. [DOI] [PubMed] [Google Scholar]
- 10.Larkin J, Ascierto PA, Dréno B et al. Combined vemurafenib and cobimetinib in BRAF‐mutated melanoma. N Engl J Med 2014;371:1867–1876. [DOI] [PubMed] [Google Scholar]
- 11.Garnett MJ, Rana S, Paterson H et al. Wild‐type and mutant B‐RAF activate C‐RAF through distinct mechanisms involving heterodimerization. Mol Cell 2005;20:963–969. [DOI] [PubMed] [Google Scholar]
- 12.Zheng G, Tseng LH, Chen G et al. Clinical detection and categorization of uncommon and concomitant mutations involving BRAF. BMC Cancer 2015;15:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casadei Gardini A, Chiadini E, Faloppi L et al. Efficacy of sorafenib in BRAF‐mutated non‐small‐cell lung cancer (NSCLC) and no response in synchronous BRAF wild type‐hepatocellular carcinoma: A case report. BMC Cancer 2016;16:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sen B, Peng S, Tang X et al. Kinase‐impaired BRAF mutations in lung cancer confer sensitivity to dasatinib. Sci Transl Med 2012;4:136ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross JS, Wang K, Chmielecki J et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 2016;138:881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchinson KE, Lipson D, Stephens PJ et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res 2013;19:6696–6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menzies AM, Yeh I, Botton T et al. Clinical activity of the MEK inhibitor trametinib in metastatic melanoma containing BRAF kinase fusion. Pigment Cell Melanoma Res 2015;28:607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihle MA, Fassunke J, König K et al. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non‐p.V600E BRAF mutations. BMC Cancer 2014;14:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frampton GM, Fichtenholtz A, Otto GA et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drilon A, Wang L, Arcila ME et al. Broad, hybrid capture‐based next‐generation sequencing identifies actionable genomic alterations in "driver‐negative" lung adenocarcinomas. Clin Cancer Res 2015;21:3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrock AB, Frampton GM, Herndon D et al. Comprehensive genomic profiling identifies frequent drug‐sensitive EGFR exon 19 deletions in NSCLC not identified by prior molecular testing. Clin Cancer Res 2016;22:3281–3285. [DOI] [PubMed] [Google Scholar]
- 22.Capper D, Berghoff AS, Magerle M et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol 2012;123:223–233. [DOI] [PubMed] [Google Scholar]
- 23.Long GV, Wilmott JS, Capper D et al. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. Am J Surg Pathol 2013;37:61–65. [DOI] [PubMed] [Google Scholar]
- 24.Reiter JG, Makohon‐Moore AP, Gerold JM et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 2018;361:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowyer SE, Rao AD, Lyle M et al. Activity of trametinib in K601E and L597Q BRAF mutation‐positive metastatic melanoma. Melanoma Res 2014;24:504–508. [DOI] [PubMed] [Google Scholar]
- 26.Dahlman KB, Xia J, Hutchinson K et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov 2012;2:791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menzies AM, Yeh I, Botton T et al. Clinical activity of the MEK inhibitor trametinib in metastatic melanoma containing BRAF kinase fusion. Pigment Cell Melanoma Res 2015;28:607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalmers ZR, Connelly CF, Fabrizio D et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson DB, Frampton GM, Rioth MJ et al. Targeted next generation sequencing identifies markers of response to PD‐1 blockade. Cancer Immunol Res 2016;4:959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodman AM, Kato S, Bazhenova L et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 2017;16:2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]