SUMMARY
The transduction compartment of inner ear hair cells, the hair bundle, is composed of stereocilia rows of graded height, a property essential for sensory function that remains poorly understood at the molecular level. We previously showed that GPSM2-GNAI is enriched at stereocilia distal tips and required for their postnatal elongation and bundle morphogenesis - two characteristics shared with MYO15A (short isoform), WHRN and EPS8 proteins. Here we first performed a comprehensive genetic analysis of the mouse auditory epithelium to show that GPSM2, GNAI, MYO15A and WHRN operate in series within the same pathway. To understand how these functionally disparate proteins act as an obligate complex, we then systematically analyzed their distribution in normal and mutant bundles over time. We discovered that WHRN-GPSM2-GNAI is an extra module recruited by and added to a pre-existing MYO15A-EPS8 stereocilia tip complex. This extended complex is only present in the first, tallest row, and is required to stabilize larger amounts of MYO15A-EPS8 than in shorter rows, which at tips harbor only MYO15A-EPS8. In absence of GPSM2 or GNAI function, including in the epistatic Myo15a and Whrn mutants, bundles retain an embryonic-like organization that coincides with generic stereocilia at the molecular level. We propose that GPSM2-GNAI confers on the first row its unique tallest identity and participates in generating differential row identity across the hair bundle.
Keywords: hair cell, hair bundle, stereocilia, staircase-like organization, GPSM2, GNAI, MYO15A, WHRN, EPS8
eTOC BLURB
Hair cell stereocilia are precisely arranged in rows of graded height to transduce mechanical stimuli. Tadenev et al. identify GPSM2 and GNAI as new members of the MYO15A stereocilia elongation complex that are limited to the first row, confer its unique tallest identity and help instruct differential identity across rows.
INTRODUCTION
Sound, head motions and gravity are perceived by epithelial sensory hair cells (HCs) in the inner ear via their apical hair bundle, an array of actin-based stereocilia acting as a motion sensor. While stereocilia number, dimensions and organization vary widely among species, sensory organs and positions within an organ [1], stereocilia consistently adopt graded height across rows, probably because a staircase pattern is central to transduction ability. Indeed, tension on oblique tip links connecting stereocilia across rows modulates the opening probability of the mechanoelectrical transducer (MET) channels located in shorter stereocilia [2]. Bundle deflection towards the tallest row increases tension on tip-links and channel conductance, while deflection away from the tallest row has opposite effects, a property known as directional sensitivity [3]. Directional sensitivity is manifested along the epithelial plane, and accordingly, the apical cytoskeleton in individual HCs is planar polarized. Radial asymmetry of the HC apical surface is reflected, for example, by the V- or U-shape of the bundle edge and the off-center position of the kinocilium. Presumably to respond in a coherent manner to shared local stimuli, neighboring HCs usually orient their hair bundles in the same direction, a cell-to-cell communication property known as planar cell polarity (PCP) [4, 5]. Planar polarity mechanisms acting at single cell and intercellular levels thus provide the form behind hair bundle function. It remains unclear however how tissue-, HC- and hair bundle-level mechanisms orchestrate hair bundle shape and orientation. In particular, it remains unknown how stereocilia adopt row-specific morphological and molecular characteristics, like the tallest height for row 1 or MET channel enrichment for subsequent rows.
We and others discovered that G protein signaling modulator 2 (GPSM2, also known as LGN) and inhibitory G proteins alpha (GNAI) become planar polarized at the HC apical membrane during embryogenesis and influence cytoskeleton organization [6-8]. In particular, the GPSM2-GNAI complex (GPSM2-GNAI) labels and defines the “bare zone”, a new apical region conspicuously devoid of protrusions appearing early during bundle development in mouse HCs [7]. Strikingly, GPSM2-GNAI is next found enriched at the distal tip of stereocilia abutting the bare zone, the first tallest row. Inactivating Gpsm2 or impairing GNAI function by expressing Pertussis toxin catalytic subunit (PTXa) result in similar stereocilia stunting and profound deafness [9], possibly explaining and modeling hearing loss in Chudley-McCullough syndrome where GPSM2 is defective [10, 11]. While human and mouse genetics uncovered a range of proteins with distinct contributions to hair bundle morphogenesis [12, 13], Gpsm2 and PTXa mutants exhibit bundle defects reminiscent of a group of proteins previously identified at distal stereocilia tips: the unconventional motor MYO15A, the PDZ-domain scaffolding protein WHRN and the actin-regulator EPS8 [14-20]. In inner hair cells (IHCs) in particular, mutant stereocilia are very short, form an excessive number of rows and only exhibit a very shallow staircase-like architecture even at maturity. Also based on evidence for direct binding between each protein, MYO15A, WHRN, and EPS8 were proposed to work together as a complex at tips to promote F-actin polymerization and stereocilia elongation [14, 19, 21]. Attesting to their crucial importance for inner ear development, all 3 proteins have been associated with congenital human deafness [22-24]. Here, we present a comprehensive genetic and molecular analysis of the interactions between GPSM2-GNAI and MYO15A, WHRN and EPS8 throughout hair bundle development. We confirm and extend recent results reporting a novel interaction between GPSM2 and WHRN and its importance for GPSM2-GNAI trafficking to stereocilia tips and actin dynamics at large [25]. We address in depth the reciprocal influence of GPSM2-GNAI on MYO15A, WHRN and EPS8 stabilization and its role in bundle morphogenesis. We find that the distal stereocilia tip complex is initially and by default composed of MYO15A-EPS8 only, and subsequently extended with an optional WHRN-GPSM2-GNAI module in the first row only. We propose that GPSM2-GNAI specifies the tallest identity of the first row and is required to establish differential row identify across the bundle.
RESULTS
GPSM2, GNAI, MYO15A and WHRN operate in series within the same genetic pathway to shape postnatal hair bundles
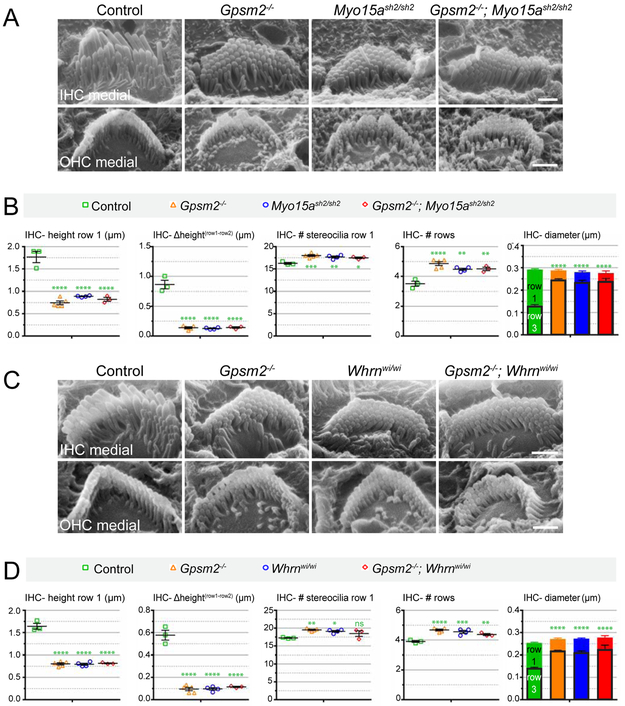
Since GPSM2 and GNAI appeared to share with MYO15A and WHRN related protein localization at stereocilia tips and hair bundle defects in mutants [9], we first sought to define functional relationships between the two groups of proteins. We bred a null allele of Gpsm2 (Gpsm2−) with a motor-defective allele of Myo15a (Myo15ash2) and intercrossed double heterozygotes to assess hair bundles at postnatal day (P)6-7 with SEM (Figure 1A and S1A). We quantified several fine bundle features: stereocilia height in the tallest row (row 1), height differential between row 1 and row 2, number of stereocilia in row 1, number of rows per bundle and stereocilia diameter in row 1 and row 3 (Figure 1B).
Figure 1. Similar hair bundle defects in Gpsm2−/−, Myo15ash2/sh2 and (Gpsm2−/−; Myo15ash2/sh2) double-mutant and in Gpsm2−/−, Whrnwi/wi and (Gpsm2−/−; Whrnwi/wi) double mutants at P6/P7.
(A) Medial views of IHC and OHC hair bundles from Gpsm2−/−, Myo15ash2/sh2, and Gpsm2−/−; Myo15ash2/sh2 double mutant and unaffected littermate control using SEM. (B) Quantification of IHC stereocilia height in row 1, height differential between row 1 and row 2, number of stereocilia in row 1, total number of rows and stereocilia diameter in row 1 (solid-colored bars) and row 3 (black-edged bars) for the genotypes indicated. (C) Medial views of IHC and OHC hair bundles from Gpsm2−/−, Whrnwi/wi, and Gpsm2−/−; Whrnwi/wi double mutant and unaffected littermate control using SEM. (D) Same set of IHC quantification as in (B) for the genotypes indicated. See Figure S1 for OHC quantifications and lateral views of hair bundles in IHCs and OHCs. For all images and quantifications, IHCs were selected at a half-turn position up from the cochlear base. At least 10 IHCs were analyzed for 3 to 5 animals per genotype, and 3 stereocilia were measured per cell for height, height differential and diameter (see Tables S1, S2 for complete dataset). Each point on the graphs represents one animal (average +/− SEM). Statistics were calculated using one-way ANOVA with Tukey's multiple comparisons test, with a single pooled variance (ns, p>0.05; *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001). Green asterisks reflect significance compared to control, whereas orange asterisks reflect significance compared to Gpsm2−/−. Asterisks in the diameter graphs refer to row 3. In all panels and figures, the lateral (abneural) side of the auditory epithelium is up and the medial (neural) side is down. Scale bars are 1 μm. See also Figure S1, S2.
Strikingly, almost identical values were recorded for each feature in single (Gpsm2−/− or Myo15ash2/sh2) and double (Gpsm2−/−; Myo15ash2/sh2) mutant IHCs: row 1 was dramatically shortened, adopting a height close to normal row 2, and contained an excess number of stereocilia (Figure 1B). IHC bundles included extra rows of stereocilia with roughly similar height and diameters, largely eroding the marked difference in diameter normally observed between row 1 and row 3 (Figure 1B). Outer hair cell (OHC) defects were more modest but similar in nature, with two exceptions: row 1 stunting was less severe in Gpsm2−/− than Myo15ash2/sh2, perhaps indicating a lesser role for GPSM2 in OHC development, and the number of stereocilia in row 1 was reduced instead of increased in all mutants (Figure 1A and S1A-B). We next combined Gpsm2− and a mutant allele for Whrn (Whrnwi), performed the same quantitative analysis, and also recorded identical feature-specific defects in single (Gpsm2−/− or Whrnwi/wi) and double (Gpsm2−/−; Whrnwi/wi) mutant IHCs (Figure 1C-D and S1C). As in the Myo15a breeding scheme, mutant OHCs were less severely affected than IHCs, but row 1 shortening was comparable between Gpsm2 and Whrn mutants, and the diameter of row 3 stereocilia appeared to be increased when GPSM2 but not WHRN was missing (Figure 1C and S1C-D). Finally, we confirmed these conclusions qualitatively in mature hair bundles. We analyzed Gpsm2− combined with either Myo15ash2 or Whrnwi, as above, and also generated combined mutants with our Pertussis toxin model where Atoh1-Cre-driven PTXa globally knocks down activity of all GNAIs in HCs [9]. Single and double mutant HCs in all four breeding schemes maintained the bundle defects observed at P6-P7 (Figure S2; Gpsm2− × Myo15ash2, Gpsm2− × Whrnwi, Atoh1-Cre; PTXa × Myo15ash2, Atoh1-Cre; PTXa × Whrnwi).
Altogether, a hallmark of Gpsm2, PTXa, Myo15a and Whrn mutant hair bundles is thus a common erosion of the morphological differences that normally distinguish rows, such as height and diameter, which is particularly dramatic in IHCs. Identical phenotypes among single mutants and absence of new or enhanced defects in double mutants strongly suggest that these functionally disparate proteins are obligate partners acting in the same signaling pathway. We thus set out to analyze their stereocilia distribution in detail, including how they influence each other at different stages of hair bundle maturation.
WHRN-GPSM2-GNAI is a late module only colocalizing with MYO15A-EPS8 in the first stereocilia row
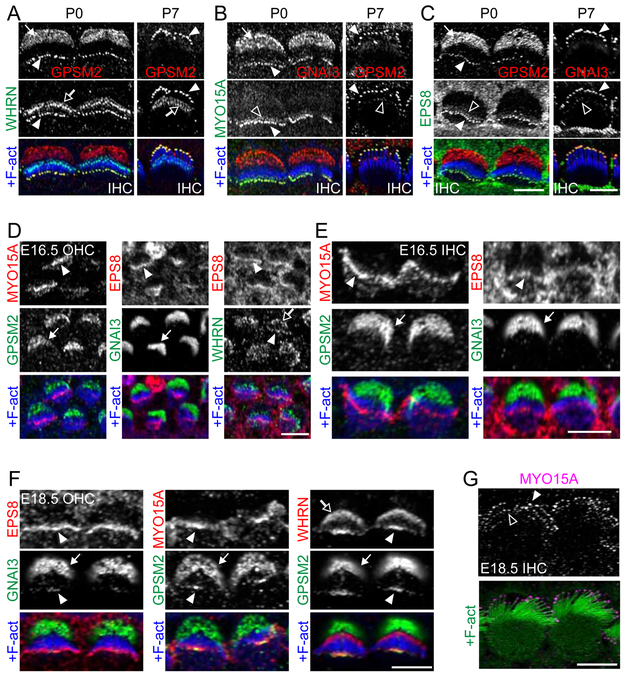
We first analyzed postnatal stages, when hair bundle defects are observed in mutants. In P0 IHCs, GPSM2, GNAI3, WHRN, MYO15A, and EPS8 perfectly colocalized at the tallest stereocilia tips in all pairwise combinations tested using coimmunofluorescence (Figure 2A-C left, solid arrowheads; GPSM2+WHRN, GNAI3+MYO15A, GPSM2+EPS8). Colocalization was confirmed at P7 in IHCs (Figure 2A-C right; GPSM2+WHRN, GPSM2+MYO15A, GNAI3+EPS8) and in OHCs (Figure S3A-C; GPSM2+WHRN, GNAI3+MYO15A, GNAI3+EPS8), and all proteins could be detected until P21 in this compartment (see Figure 5E, 6F and Figure S4B and S6). Although difficult to assess in every cell, MYO15A-EPS8 was also detected in lower amounts at stereocilia tips in row 2, as reported previously (Figure 2B-C, hollow arrowheads)[14, 17, 26]. In contrast, GPSM2-GNAI3 appeared strictly limited to row 1 [9]. The possible enrichment of WHRN at tips in row 2 was challenging to determine due to confusing signal near the stereocilia base (Figure 2A, hollow arrows), and previous studies reached different conclusions [19, 21, 27-30]. We re-examined this question in P4 IHCs using a C-terminal antibody detecting the long and short WHRN isoforms and Airyscan stacks of samples mounted with spacers to avoid bundle compression. We concluded that WHRN is absent at row 2 distal tips and interpreted the signal at stereocilia base as being associated to links interconnecting stereocilia, possibly between row 1 (lower protein amount) and between row 1 and row 2 (higher amount) (Figure S3D-E). This conclusion is compatible with a recent study where a different C-terminal antibody revealed focal accumulation of WHRN (long isoform) near the middle of row 2 stereocilia [29]. Absence at row 2 tips where MYO15A-EPS8 is present suggests that the localization of WHRN shows greater similarity to that of GPSM2-GNAI than that of MYO15A-EPS8.
Figure 2. Distinct stereocilia distribution for WHRN-GPSM2-GNAI3 and MYO15A-EPS8 in postnatal and embryonic hair cells.
(A-C) Coimmunostainings for GPSM2+WHRN (A), GNAI3 or GPSM2+MYO15A (B) and GPSM2 or GNAI3+EPS8 (C) in P0 IHCs at the cochlear base (left) and P7 IHCs at the apical turn (right). All proteins colocalized at row 1 tips (solid arrowheads; see also Figure S3A-C for P7 OHCs). In addition, GPSM2 and GNAI3 localized at the bare zone (solid arrows), WHRN localized at the base of stereocilia (hollow arrows), and MYO15A and EPS8 could be weakly detected at row 2 stereocilia tips (hollow arrowheads). Note that younger bundles tended to fall medially (away from the BZ) during mounting, whereas more mature bundles often fell laterally. (D-E) E16.5 OHCs (D) and IHCs (E) coimmunostainings at the cochlear base. MYO15A and EPS8 were already abundantly enriched at stereocilia tips, whereas WHRN and GPSM2-GNAI3 were low or undetectable there in the same cells (arrowheads). GPSM2-GNAI3 and WHRN already localized to the bare zone (arrow) and the base of stereocilia (hollow arrow), respectively. (F) E18.5 OHCs coimmunostainings at the cochlear base for the proteins indicated. Low amounts of GNAI3 and GPSM2 colocalized with more abundant EPS8 and MYO15A only in central stereocilia, and EPS8 and MYO15A showed a wider distribution encompassing peripheral stereocilia (left and center panels). By contrast, WHRN enrichment matched GPSM2 in central stereocilia (right panels). (G) E18.5 IHCs MYO15A immunostaining. MYO15A was enriched in roughly comparable amounts at stereocilia tips across rows. Two to three rows can be distinguished. In all figures, solid arrows indicate the bare zone, hollow arrows indicate the stereocilia base, solid arrowheads indicate row 1 stereocilia tips, hollow arrowheads indicate row 2 tips and F-act indicates phalloidin staining. (A-C and G) were acquired in Airy mode (single z) (see Methods). Scale bars are 5μm. See also Figure S3.
Looking to support this idea, we next analyzed earlier stages from embryonic day (E)16.5. Remarkably, early immunostainings unambiguously revealed two distinct protein complexes at distal stereocilia. In E16.5 OHCs and IHCs, MYO15A and EPS8 were already consistently present and broadly enriched distally in stereocilia across the whole bundle (Figure 2D-E, solid arrowheads). By contrast, GPSM2-GNAI3 was undetectable or barely detectable in these same hair bundles (Figure 2D-E), but was highly enriched at the bare zone (Figure 2D-E, solid arrow). Interestingly, unlike MYO15A and EPS8, WHRN was barely detectable at tips in E16.5 OHCs, but consistently found at the stereocilia base (Figure 2D, hollow arrow), showing that trafficking to stereocilia tips is a later type of apical enrichment for both WHRN and GPSM2-GNAI proteins. WHRN instead coincided spatially with GPSM2-GNAI3 at tips once these proteins could be detected in the bundle. In E18.5 OHCs, MYO15A and EPS8 were broadly enriched in high amounts across all stereocilia, whereas WHRN, GPSM2, and GNAI3 precisely localized together and were only found in central stereocilia in much lower amounts in the same bundle (Figure 2F; GNAI3+EPS8, GPSM2+MYO15A, GPSM2+WHRN). At E18.5, GPSM2 and GNAI3 levels were lower in IHCs than OHCs, and costainings yielded results comparable to E16.5 (Figure S3F).
Detailed Airyscan analysis of E18.5 IHCs suggested that MYO15A is detected at tips in all rows in roughly comparable amounts, unlike at postnatal stages where row 2 accumulates much less MYO15A than row 1 (Figure 2G, compare with 2B). In summary, temporal and spatial differences in stereocilia localization patterns revealed two distinct protein complexes. While MYO15A-EPS8 formed an early tip complex already broadly enriched at E16.5, WHRN-GPSM2-GNAI was enriched at tips at later stages, initially encompassing central stereocilia only before spanning the whole bundle. We thus propose that WHRN might complex with GPSM2 and GNAI near the medial edge of the bare zone. WHRN-GPSM2-GNAI would then bind MYO15A-EPS8 near the stereocilia base, and the extended complex would traffick to first-row tips only, where it would accumulate along with pre-existing MYO15A-EPS8. To expose the interplay between MYO15A-EPS8 and WHRN-GPSM2-GNAI, we next systematically studied protein localization in the various mutants over time.
MYO15A and WHRN are required for GPSM2 and GNAI3 localization at stereocilia tips
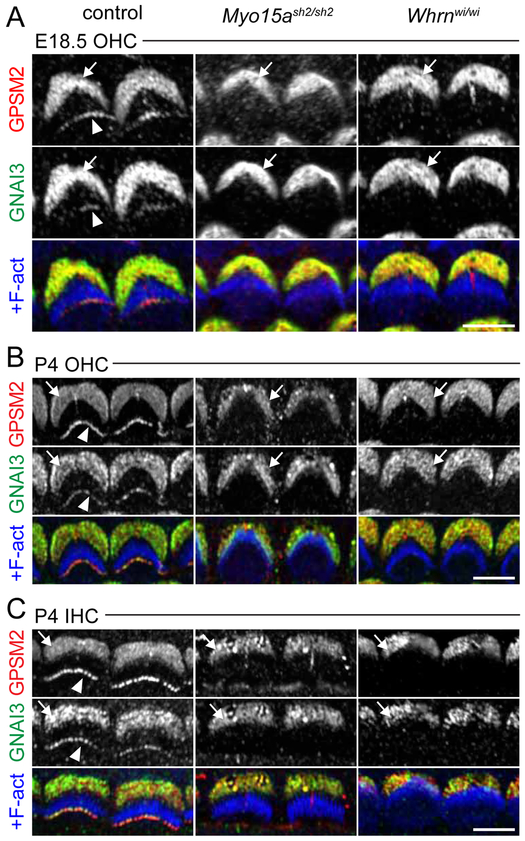
We collected Myo15a mutants at E18.5, when distinct protein complexes were still obvious, and also at P4. GPSM2-GNAI was missing at stereocilia tips in Myo15ash2/sh2 at E18.5 and P4 in both IHCs and OHCs (Figure 3A-C and not shown). MYO15A is thus likely the sole motor bringing GPSM2-GNAI to row 1 tips from their earliest stage of stereocilia trafficking in both IHCs and OHCs. WHRN and EPS8 are well known to require MYO15A for trafficking to tips [14, 21], and were already missing at tips in E18.5 Myo15ash2/sh2 OHCs as well (Figure S4A). Similarly, GPSM2-GNAI3 could not be detected at tips in IHCs or OHCs of Whrnwi/wi mutants lacking both long and short WHRN isoforms at E18.5, P4 or P21 (Figure 3, Figure S4B and not shown). By contrast, GPMS2 and GNAI3 were maintained at P4 stereocilia tips in OHCs and IHCs when only the long WHRN isoform was missing (Figure S4C-D and not shown)[31], showing that WHRN short isoform is sufficient to ensure trafficking. These results support and extend conclusions from Mauriac et al. in P8 IHCs [25]. Interestingly, GPSM2 and GNAI3 enrichment at the bare zone was unaffected at E18.5 and P4 in Myo15ash2 and Whrnwi mutants (Figure 3, arrows), matching the lack of enrichment of MYO15A or WHRN in this region and the absence of stereocilia placement or planar polarity defects in Myo15a or Whrn mutants.
Figure 3. MYO15A and WHRN are required for GPSM2-GNAI3 enrichment at row 1 stereocilia tips.
Coimmunostaining for GPSM2 and GNAI3 in Myo15ash2/sh2 and Whrnwi/wi mutant and a control in E18.5 OHCs at the cochlear base (A), P4 OHCs in the apical turn (B), and P4 IHCs in the basal half-turn (C). GPSM2 and GNAI3 were not detected at stereocilia tips (arrowheads) in either mutant. Note that bare zone localization of GPSM2 and GNAI3 (solid arrow) was unchanged. Control at E18.5 is a Myo15ash2/+ littermate of Myo15ash2/sh2, control for P4 OHCs is a Whrnwi/+ littermate of Whrnwi/wi, and control for P4 IHCs is a Gpsm2+/−; Whrnwi/+ littermate of Whrnwi/wi. Note that kinocilium signal for GPSM2 is not specific, as it is also seen in Gpsm2−/− (not shown). Scale bars are 5μm. See also Figure S4.
GPSM2-GNAI stabilizes large amounts of WHRN-MYO15A-EPS8 at row 1 stereocilia tips
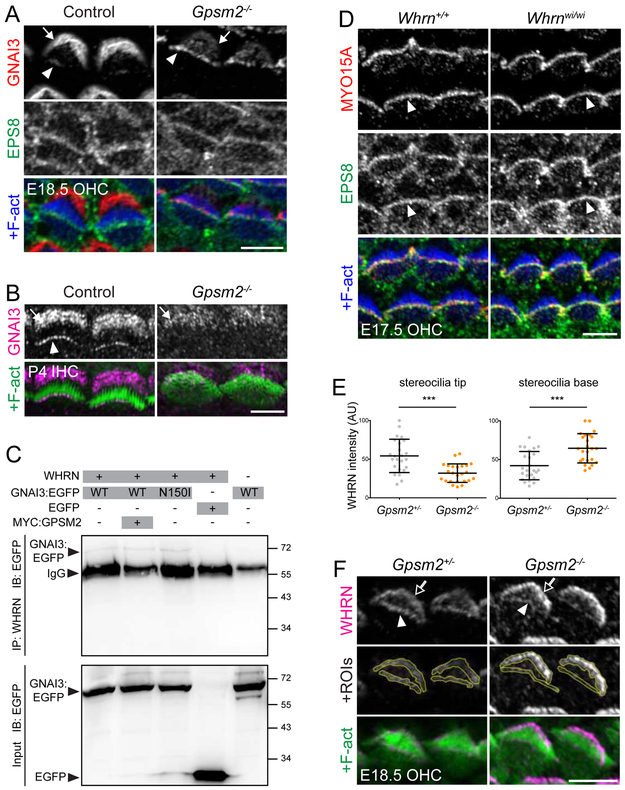
In absence of Gpsm2, GNAI3 was found in excess at stereocilia tips and conversely depleted at the bare zone at E18.5 (Figure 4A and [9]). Although GNAI proteins are well-known to bind GPSM2 Goloco domains [32-34], GNAI3 can thus initially traffic to distal stereocilia without GPSM2. Interestingly, GNAI3 excess appeared to match EPS8 in the same HCs, suggesting that GNAI3 is free to join the early MYO15A-EPS8 complex in absence of GPSM2 (Figure 4A). At postnatal stages, however, reduced GNAI3 enrichment at the bare zone persisted but GNAI3 was no longer detected at tips, showing that GPSM2 is required to maintain GNAI3 there (Figure 4B). In HEK293 cells, WHRN could coimmunoprecipitate GNAI3:EGFP, and overexpressing GPSM2 or transfecting Gnai3:Egfp (N150I) that cannot bind GPSM2 did not alter the amount of GNAI3:EGFP pulled down (Figure 4C; see also Figure S5A for control blots) [35]. We conclude that WHRN can bind GNAI3 directly for trafficking purposes, but that GNAI3 requires GPSM2 to persist at tips.
Figure 4. GPSM2 is not required for early GNAI3 trafficking to stereocilia tips, but is required to stabilize GNAI3 and WHRN at tips.
(A-B) GNAI3 and EPS8 coimmunostaining in E18.5 OHCs (A) and GNAI3 immunostaining in P4 IHCs (B) in Gpsm2−/−. In absence of GPSM2, GNAI3 was found in excess at tips, closely matching EPS8 signal at E18.5, but was missing at tips at P4 (arrowheads). Enrichment at the bare zone was by contrast reduced at both stages (arrows). (C) Immunoprecipitation of GNAI3:EGFP with WHRN in HEK293 cells. GNAI3 constructs were wild-type (WT) or carried a N150I mutation preventing binding to GPSM2, and MYC:GPSM2 was cotransfected in lane 2. Preventing direct binding to GPSM2 (N150I) or adding GPSM2 neither decreased nor increased the amount of GNAI3:EGFP immunoprecipitated, respectively. EGFP alone was not immunoprecipitated by WHRN and GNAI3:EGFP was not immunoprecipitated when WHRN was not transfected (see Figure S5A for extra control blots). (D) MYO15A and EPS8 coimmunostaining in E17.5 Whrnwi/wi OHCs showing normal enrichment at tips (arrowheads). (E-F) WHRN immunostaining in E18.5 Gpsm2−/− OHCs. WHRN signal intensity is reduced at stereocilia tips but increased at stereocilia base compared to littermates (E), as illustrated in (F). Arbitrary unit (AU); n= 4 animals, 6 OHCs per animal; Mann-Whitney U test, ***p=0.0002 for tips and base signal. An example of regions of interest (ROIs) for WHRN signal quantification is shown in (F). Control image in (B) shows the same IHC pair as in Figure 3C because these experiments were performed as GPSM2-GNAI3 coimmunostainings in a Gpsm2− × Whrnwi breeding scheme, and the same Gpsm2+/−; Whrnwi/+ sample was used as littermate control. Solid arrows indicate the bare zone, hollow arrows indicate the stereocilia base, and solid arrowheads indicate row 1 stereocilia tips. Scale bars are 5μm. See also Figure S5.
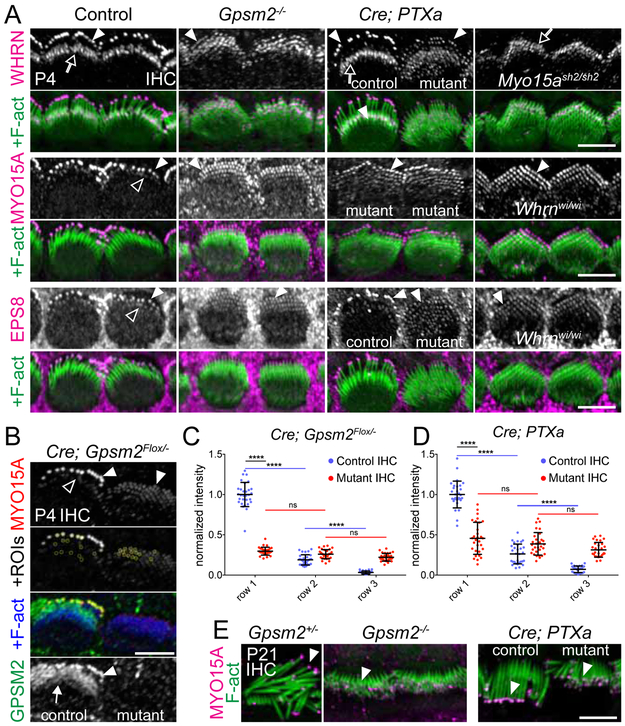
These results suggest that protein trafficking to and stabilization at tips are likely two distinct and successive molecular processes. MYO15A and WHRN are required to bring GPSM2-GNAI to tips, and we hypothesized that the WHRN-GPSM2-GNAI module might in turn be required to build up the large amounts of MYO15A-EPS8 needed to erect the tallest row (Figure 2A-C; Figure 1A-B; Figure S1A-B). To test this hypothesis, we probed MYO15A-EPS8 distribution in Whrn, Gpsm2 and PTXa mutants, and asked whether WHRN might depend on GPSM2-GNAI to accumulate at tips. At E17.5, MYO15A and EPS8 were normally enriched at stereocilia tips in absence of WHRN (and thus GPSM2-GNAI), compatible with the notion that WHRN is an adaptor for the late GPSM2-GNAI module and not part of the early tip complex (Figure 4D). In contrast, members of the late module exhibited variable degrees of codependence. Quantification of signal intensity suggested that WHRN accumulation was slightly reduced at tips in E18.5 Gpsm2 mutant OHCs, and conversely increased at the stereocilia base (Figure 4E-F). PTXa expression largely prevented GPSM2 accumulation at tips at E18.5, showing that GNAI signaling is integral to enriching the GPSM2-GNAI module at row 1 (Figure S5B; see also Figure 6E-F). By P4, WHRN accumulation at tips of row 1 IHC stereocilia was severely reduced in Gpsm2 and Atoh1-Cre; PTXa mutants (Figure 5A). Similarly, MYO15A and EPS8 were severely reduced in row 1 in Gpsm2 and PTXa mutants (Figure 5A), a departure from their intact enrichment in E17.5 Whrnwi/wi OHCs (Figure 4D). In order to quantify defective protein enrichment, we used Atoh1-Cre; Gpsm2Flox/− and Atoh1-Cre; PTXa mice, which both require Cre activity for expression of the mutant phenotype. We selected pairs of IHC neighbors where one “escaper” cell retained apparently normal GPSM2 expression and/or morphology, perhaps through failed or greatly delayed Cre-mediated recombination (Figure 5B). Regardless of their origin, their close similarity to true wild-type cells makes them powerful internal controls to avoid variable antibody labeling across samples. Of note, differences seen between mutant and internal control cells should be, if anything, more modest than the differences between mutant and true wild-type cells. Normalized signal intensity for MYO15A at row 1 tips was reduced ~70% in Gpsm2 mutant and ~50% in PTXa mutant cells (Figure 5C-D). Reduced protein amounts at P4 did not reflect a delayed enrichment, as P21 Gpsm2 and PTXa mutants still exhibited reduced MYO15A (Figure 5E), WHRN and EPS8 (Figure S6) amounts at tips compared to row 1 in controls.
Figure 6. GPSM2 and GNAI are required to establish molecularly distinct stereocilia row identities.
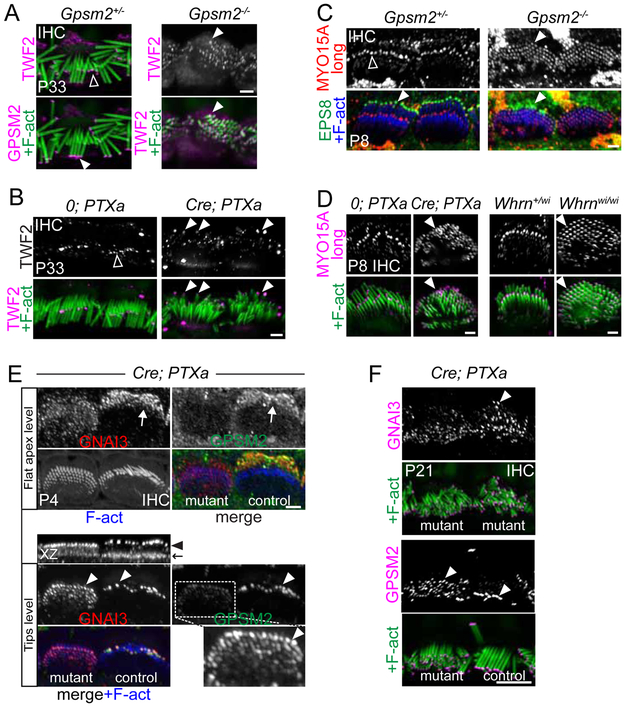
(A-B) TWF2 immunostainings in P33 IHCs. GPSM2 and TWF2 normally adopted complementary localization at row 1 (solid arrowheads) and row 2 (hollow arrowheads) tips, respectively (A), but TWF2 was ectopically found at row 1 in both Gpsm2−/− (A) and Atoh1-Cre; PTXa (B) (arrowheads). Note that the control bundles fell medially upon mounting, whereas the shorter mutant bundles fell laterally. TWF2 levels appeared substantially lower at the stereocilia tips of mutant IHCs compared to control IHCs, particularly in Atoh1-Cre; PTXa samples. (C-D) MYO15A long isoform (MYO15A-long) immunostainings in P8 IHCs. EPS8 and MYO15A-long normally adopted complementary localization at row 1 (solid arrowheads) and row 2 (hollow arrowheads) tips, respectively (C), but MYO15A-long was ectopically found at row 1 in Gpsm2−/− (C), Atoh1-Cre; PTXa and Whrnwi/wi IHCs (D). (E) One representative P4 Isl1-Cre; PTXa IHC pair coimmunostained for GPSM2 and GNAI3 where the apparently normal right IHC served as an internal control (see text and Methods). Top: GNAI3 and GPSM2 were planar polarized at the bare zone (arrow) in the control IHC, but delocalized to the whole apex in the mutant IHC. Bottom: remarkably, GNAI3 and GPSM2 were concomitantly delocalized to all stereocilia rows at tips in the same mutant IHC (arrowheads). Low GPSM2 signal at tips in the mutant IHC was enhanced in the magnified inset. The XZ view shows GNAI3 at tips (black arrowhead) and at the flat IHC surface (black arrow). (F) GNAI3 and GPSM2 immunostainings in P21 IHCs. Delocalization from row 1 to all rows is still clearly observed at maturity in Atoh1-Cre; PTXa IHCs, whereas an internal control IHC only showed GPSM2 at row 1 tips (bottom right IHC). Airy mode was used to acquire images in (A), (B), (D), (F) (single z) and (E) (maximum projection of z-series). In (E), top and bottom panels are maximum intensity projection of flat apical surface and tips level GPSM2-GNAI3 signal, respectively, overlaid with full-depth projection of phalloidin signal. Solid arrows indicate the bare zone, solid arrowheads indicate row 1 stereocilia tips, and hollow arrowheads indicate row 2 tips. Scale bars are 2μm (A-E), 5μm (F).
Figure 5. GPSM2-GNAI is required for graded MYO15A-EPS8 amounts across stereocilia rows and to limit WHRN to row 1 at tips.
(A) WHRN, MYO15A and EPS8 immunostainings in P4 IHCs of Gpsm2−/−, Atoh1-Cre; PTXa and Myo15ash2/sh2 or Whrnwi/wi mutants. WHRN was ectopically located at the tips of row 2 and further stereocilia rows in Gpsm2−/− and Atoh1-Cre; PTXa, but absent at tips in Myo15ash2/sh2. WHRN-MYO15A-EPS8 appeared uniformly enriched at tips across all rows (including supernumerary rows) in lower amounts than row 1 enrichment in control littermates. (B) Example of a P4 IHC pair used to quantify MYO15A accumulation at stereocilia tips in (C) and (D). The left IHC in this Atoh1-Cre; Gpsm2Flox/− sample retained apparently normal GPSM2 distribution and morphology, and was used as an internal control (see text). ROIs for MYO15A signal quantification are indicated as yellow circles at the tips of rows 1, 2, and 3 (control row 3 represents background signal level). (C-D) MYO15A signal per row for Atoh1-Cre; Gpsm2Flox/− (C) and Atoh1-Cre; PTXa (D). MYO15A signal intensity was normalized for each IHC pair to the average signal at row 1 tips in the control IHC. Two IHC pairs were selected for 3 animals and 5 stereocilia quantified per row for each IHC (n=30; two-way ANOVA with Sidak’s multiple comparisons test; ns, p>0.05; ****p≤0.0001). (E) MYO15A immunostaining in P21 Gpsm2−/− and Atoh1-Cre; PTXa IHCs. As at P4, all mutant stereocilia show an approximately equal amount of MYO15A, which is less than in control row 1. Internal control and mutant IHCs are indicated for each Atoh1-Cre; PTXa image. The same cells are shown in (A) for MYO15A and EPS8 (Control, Gpsm2−/− and Whrnwi/wi), and for WHRN and EPS8 (Atoh1-Cre; PTXa) because these experiments were performed as coimmunostainings. Hollow arrows indicate the stereocilia base, solid arrowheads indicate row 1 stereocilia tips, and hollow arrowheads indicate row 2 tips. Scale bars are 5μm. See also Figure S6, S7.
Next, we sought to confirm that GPSM2 can enhance MYO15A enrichment in a different system, as it apparently does at row 1 tips compared to further rows that lack GPSM2. We transfected COS-7 cells with EGFP:MYO15A and mCHERRY:WHRN along with either MYC alone or MYC:GPSM2. MYC:GPSM2 increased the amount of EGFP:MYO15A and mCHERRY:WHRN enriched at filopodia tips and the number of filopodia per cell (Figure S7), complementing previous evidence for increased filopodia length in a similar experiment [25].
Together, these results indicate that GPSM2-GNAI requires WHRN as an adapter to reach stereocilia tips and build a MYO15A-EPS8-WHRN-GPSM2-GNAI complex specific to row 1. In IHCs in particular, the extra WHRN-GPMS2-GNAI module is likely critical to 1) accumulate much higher amounts of the ubiquitous MYO15A-EPS8 stereocilia tip complex compared to further rows, and 2) perhaps as a consequence, greatly elongate row 1 postnatally to give IHCs their steep staircase-like architecture (Figure 1; Figure S2A-B and D-E). In contrast, further rows only hosting low amounts of MYO15A-EPS8 might be unable to elongate to a comparable height.
GPSM2-GNAI confers differential row identity to hair bundle stereocilia
Interestingly, while WHRN and MYO15A-EPS8 accumulation in row 1 was reduced in absence of GPSM2 and GNAI function, their accumulation at tips in row 2 appeared close to normal (Figure 5A-B). Quantified MYO15A accumulation in mutant row 2 was comparable to mutant row 1, and comparable or slightly above control row 2 in both Atoh1-Cre; Gpsm2Flox/− and Atoh1-Cre; PTXa IHCs (Figure 5C-D). MYO15A accumulation in mutant row 3 and further supernumerary rows was well above control row 3 used to define background signal level (Figure 5B-D). Equal enrichment of MYO15A and EPS8 across all rows was also observed in Whrnwi/wi (Figure 5A). This is consistent with the observations that GPSM2-GNAI is not enriched at tips without WHRN (Figure 3; Figure S4B) and that GPSM2-GNAI are required for differential enrichment of MYO15A-EPS8 across rows (Figure 5). WHRN was ectopically enriched at tips in row 2 and detected in similar amounts in all further rows in Gpsm2 and PTXa mutants, demonstrating that GPSM2-GNAI normally ensures restriction of WHRN to row 1 tips (Figure 5A). Equal enrichment across rows was also observed at P21 for MYO15A, EPS8 and WHRN in both Gpsm2 and PTXa mutants (Figure 5E and Figure S6). Importantly, WHRN enriched along with MYO15A-EPS8 at tips was not sufficient to rescue elongation in row 1 (Figure 5A, E; Figure S6; Figure 1A-B), defining GPSM2-GNAI as the essential proteins to confer the tallest row’s identity. These results raise the possibility that stereocilia across rows might be instructed towards distinct molecular identities reflected, for example, by graded MYO15A-EPS8 levels at their tips. Failure to assemble the MYO15A-EPS8-WHRN-GPSM2-GNAI complex in row 1 would abolish molecular identity across rows, result in comparable MYO15A-EPS8-WHRN amounts at all tips across the hair bundle, and explain the erosion of stereocilia identity at the morphological level (Figure 1, Figure S1, S2).
To explore this novel idea, we used as markers proteins known to differentially localize across rows within the normal bundle. TWF2 is an actin capping protein normally found at the tips of shorter stereocilia but excluded from row 1 [36, 37]. In Gpsm2−/− and Atoh1-Cre; PTXa IHCs, however, TWF2 localized to row 1 and all further stereocilia within the P33 bundle (Figure 6A-B). We next examined the localization of MYO15A long isoform (MYO15A-long), which is mostly enriched at the tip of stereocilia in shorter rows from P7 and does not contribute to stereocilia elongation [38]. In both P8 Gpsm2−/− and Atoh1-Cre; PTXa mutants, MYO15A-long was enriched equally at all tips including row 1, and the same was observed in Whrn mutants (Figure 6C-D).
Having established that all Gpsm2 and PTXa mutant stereocilia adopted markers normally restricted to shorter rows, we explored whether they might also all adopt row 1 markers. Besides GPSM2, GNAI and WHRN, proteins specific to row 1 tips have yet to be reported to our knowledge. We thus reasoned that PTXa is expected to affect GNAI function but not its expression. Selecting Isl1-Cre; PTXa IHC pairs at P4 where apparently normal “escapers” served as internal controls, we found that GNAI3 was still enriched at the flat portion of the apical membrane in mutant IHCs. GNAI3 was enriched in reduced amounts, however, and lacked its normal planar polarization at the bare zone (Figure 6E, top). Interestingly, a higher Z capture of the same IHC pairs revealed that GNAI3 concomitantly lost its restriction to the first row, being detectable at tips across all stereocilia including supernumerary rows (Figure 6E, bottom). Coimmunostaining in the same IHC pairs revealed that GPSM2 was similarly depleted but ectopically enriched at all tips (Figure 6E, right panels). Finally, ectopic GPSM2 and GNAI3 localization at tips in all rows was maintained at maturity in Atoh1-Cre; PTXa IHCs (Figure 6F).
We thus propose that in Gpsm2 and PTXa mutants, and by extension in epistatic Myo15a and Whrn mutants, IHC stereocilia lack row identity. Instead, they manifest a non-exclusive, generic molecular profile characterized by the simultaneous presence of markers usually restricted to either row 1 or shorter rows. Interestingly, these mutant bundles strikingly resemble normal immature IHC bundles at E18.5 (see Figure 7): 1) they have about double the number of rows of normal mature bundles (Figure 1, Figure S2), 2) they only have a very shallow staircase architecture (Figure 1, Figure S2), and 3) they broadly enrich comparable levels of MYO15A-EPS8 across all rows (Figure 2G, Figure 5 and Figure S6).
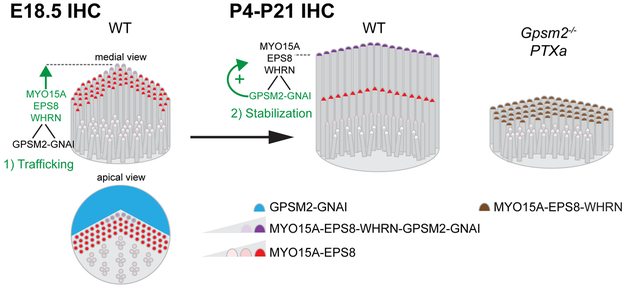
Figure 7. Model describing the contribution of GPSM2-GNAI to IHC hair bundle morphogenesis.
At E18.5 (left), GPSM2-GNAI (blue) is mostly enriched outside the hair bundle at the bare zone, the flat apical sub-region that directly abuts the first stereocilia row (apical view). At this stage, MYO15A-EPS8 (red) broadly localizes at tips throughout the IHC bundle in relatively comparable amounts. 1) Trafficking phase: MYO15A uses WHRN as an adapter to bring low amounts of GPSM2-GNAI (purple) to tips in central stereocilia of the first row only (green arrow). 2) Stabilization phase: by P4 and until maturity (P21) (center), WHRN-GPSM2-GNAI remains restricted to row 1, where it builds up high amounts of MYO15A-EPS8 (green arrow) and specifies the tallest identity of the first row. MYO15A-EPS8 remains enriched at tips in shorter stereocilia but in much lower amounts, as reported previously. In stark contrast, little row-specific differentiation occurs in Gpsm2−/− or PTXa mutant IHCs (right). Stereocilia are uniformly short and maintain an excessive number of rows where MYO15A-EPS8 is enriched in comparable amounts. WHRN is maintained at row 1 tips, but is also ectopically found in all further rows. Although MYO15A-EPS8-WHRN colocalize at tips (brown), they cannot rescue normal elongation in row 1. As a consequence, mature mutant IHC bundles (right) are reminiscent of immature E18.5 embryonic bundles (left).
In conclusion, we find that WHRN-GPSM2-GNAI3 is required in row 1 to modify embryonic bundles where an earlier MYO15A-EPS8 complex is found at all tips, and to create molecular and morphological asymmetry across stereocilia rows. Gpsm2 and PTXa mutants, or epistatic Whrn and Myo15a mutants, do not significantly progress beyond an embryonic bundle organization by the end stage of auditory epithelium development.
DISCUSSION
We previously reported that GPSM2 and GNAI3 are enriched at stereocilia distal tips specifically in the tallest row and that GPSM2-GNAI activity is required for postnatal stereocilia elongation and hearing. We also pointed out similarities with the reported localization and function of MYO15A, WHRN and EPS8 [9]. Recently, Mauriac et al. showed that MYO15A and WHRN are required for GPSM2 and GNAI3 enrichment at stereocilia tips, uncovering a direct interaction between GPSM2 and WHRN and studying its importance for actin regulation in multiple contexts [25]. Here we focused on hair cells (HCs) and the interactions between GPSM2-GNAI and MYO15A, EPS8 and WHRN during hair bundle development. 1) We studied genetic interactions between Gpsm2 or a global GNAI knock-down model (PTXa) and Myo15a or Whrn in a comprehensive four-way breeding scheme. Based on virtually identical hair bundle defects, we formally established that these disparate proteins form an obligate complex and function in a single pathway. 2) We systematically analyzed the distribution of each protein throughout development from E16.5 to maturity. Compiling these results, we discovered that WHRN-GPSM2-GNAI3 forms a protein module added at row 1 only to a prior MYO15A-EPS8 complex that itself lacks row specificity. 3) We systematically studied partner proteins distribution in mutant contexts during development and leveraged mislocalization data to uncover how the accessory WHRN-GPSM2-GNAI module at row 1 impacts stereocilia organization in a row-specific manner.
GPSM2-GNAI is integral to WHRN, MYO15A and EPS8 function in elongating stereocilia
Myo15ash2 is expected to mask any mutation affecting MYO15A cargo proteins, and we and others found that WHRN is strictly required for cargo to include GPSM2 and GNAI (Figure 3 and [25]). While it is thus expected that Myo15ash2/sh2 and Whrnwi/wi would share Gpsm2−/− and PTXa hair bundle defects, it is however remarkable that Gpsm2 and PTXa mutants virtually phenocopy Myo15ash2/sh2 and Whrnwi/wi (Figure 1, Figure S1, S2). We conclude that the classic shaker2 and whirler bundle defects and their associated hearing loss could be largely imputable to a lack of GPSM2-GNAI at row 1 stereocilia tips. In this light, an extended MYO15A-EPS8-WHRN-GPSM2-GNAI complex assembled near the stereocilia base and limited to the tips of row 1, unlike the earlier and ubiquitous MYO15A-EPS8 complex found at tips in all rows, is a departure from the previous notion of a MYO15A-WHRN-EPS8 complex [14, 19, 21]. Eps8 mutants also share similar bundle defects [14, 15], and EPS8 has been proposed to be the effector of the MYO15A complex regulating actin polymerization and/or bundling at tips [14, 15]. It is worth noting however that EPS8 cannot establish normal stereocilia heights without GPSM2-GNAI, even when present along with MYO15A and WHRN at tips (Figure 5A). These results raise the interesting possibility that row 1 elongation and hair bundle morphogenesis largely depend on GNAI signaling through a yet unknown pathway at stereocilia tips.
Cues originating outside the hair bundle could confer distinct stereocilia row identities
Our principal new finding is that GPSM2-GNAI confers on stereocilia in the first embryonic row an exclusive identity distinct from further rows, leading them to adopt the tallest fate postnatally (Figure 7). To our knowledge, this is the first identification of a molecular mechanism establishing asymmetric stereocilia identities across rows during development. Analyzing PTXa mutant IHCs with uniform stereocilia morphology across rows, we established that markers specific to row 1 (GPSM2-GNAI3 themselves) are ectopically found in all rows. Conversely, TWF2 and MYO15A (long isoform), normally preferentially enriched in shorter mechanotransducing rows [36-38] are found in similar amounts in row 1 in Gpsm2 and PTXa mutants (Figure 6). In support of our conclusion that Gpsm2 and PTXa mutant stereocilia lack distinct molecular row identities, previous studies reported ectopic TWF2 at row 1 tips in Myo15ash2 and Whrnwi mutant IHCs [36, 37], which we know now lack GPMS2 and GNAI3 at tips (Figure 3 and [25]). Additional support comes from the distribution of MYO15A-EPS8. Graded enrichment correlating with stereocilia height [14, 17, 26] is lost in Gpsm2 and PTXa mutants, and MYO15A-EPS8 is instead uniformly distributed across rows at reduced levels compared to normal row 1 (Figure 5), reminiscent of normal MYO15A distribution at E18.5 (Figure 2G). Here as well, a similar outcome was noted in Whrnwi mutants [14], where GPSM2-GNAI is missing at tips (Figure 3 and [25]). Interestingly, WHRN was reported missing at row 1 tips in P6 Eps8 mutants [29], consistent with our conclusion that MYO15A-EPS8 represents a prior and prerequisite complex for partner enrichment at tips. We thus speculate that EPS8, like MYO15A, might be required to enrich the entire WHRN-GPSM2-GNAI module at row 1. In Gpsm2 mutants, IHCs are more dysmorphic than OHCs, which harbor less marked differences across rows in stereocilia height and diameter, and OHCs are less affected than in Myo15a mutants (Figure 1, Figure S1, S2). These morphological differences suggest that GPSM2-GNAI could be particularly important where strongly divergent row identity is needed.
We previously proposed that early planar polarization at the bare zone could be the strategy by which GPSM2-GNAI is limited to the adjacent first stereocilia row. We showed that loss of INSC, a protein restricted to the bare zone, or loss of GPSM2 leads to a transient excess of GPSM2-GNAI at stereocilia tips and a concomitant reduction at the bare zone [9]. Since GNAI3 excess in absence of GPSM2 interestingly matches the broader and earlier distribution of EPS8 (Figure 4A), it is tempting to speculate that the GPSM2 adapter is the central protein responsible for the delayed and specific row 1 enrichment of the WHRN-GPSM2-GNAI module. In this scenario, GNAI might simply be unhindered to join the early and ubiquitous MYO15A-EPS8 tip complex in Gpsm2 mutants, a possibility supported by its direct binding to WHRN (Figure 4C). GNAI3 requires GPSM2 to remain durably associated with MYO15A-EPS8 at tips, however (Figure 4B). WHRN is itself not enriched at the bare zone (Figure 2A) and is delocalized to tips in all rows in Gpsm2 and PTXa mutants (Figure 5A, Figure S6), strongly suggesting that GPSM2-GNAI restricts WHRN to row 1 tips. Here our results diverge from Mauriac et al., where WHRN was reported absent from stereocilia tips in Gpsm2 and Gnai3 mutants [25]. How exactly INSC-GPSM2-GNAI planar polarization at the bare zone would restrict WHRN-GPSM2-GNAI trafficking to row 1 remains unclear.
While PTXa prevents GPSM2-GNAI restriction to row 1 (Figure 6E-F) , it is surprising that abolishing GPSM2 normally limited to row 1 indirectly affects morphogenesis of further rows (Figure 1, Figure S1, S2). This result suggests that tallest stereocilia participate in the development of shorter rows, perhaps via tension on interstereocilia links. Several recent studies proposed that MET function in shorter rows plays an important role in stereocilia morphogenesis and/or stability [39-42], and a tighter control over F-actin turnover at tips was reported in row 1 compared to shorter rows [43]. A picture is thus emerging where stereocilia growth in the first row is strictly controlled by developmental mechanisms that could originate in part outside the hair bundle (bare zone), whereas growth in further rows is controlled by the first row and by autonomous mechanisms linked to their mechanosensory function.
In summary, we propose that WHRN is trafficked to stereocilia tips later than MYO15A-EPS8 and is required to bring along GPSM2-GNAI (Figure 7). Reciprocally, the GPSM2-GNAI complex, which is first enriched at the bare zone, ensures that WHRN trafficking to tips is restricted to the first row. Once at tips, the WHRN-GPSM2-GNAI module promotes high abundance of MYO15A-EPS8 compared to further rows and confers upon row 1 its unique tallest identity. Without GPSM2-GNAI, WHRN and MYO15A-EPS8 are present together at row 1 tips but cannot rescue stereocilia elongation to normal levels.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Basile Tarchini (basile.tarchini@jax.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
The Gpsm2 lines are derivatives of the EUCOMM ES cells Gpsm2 tm1a(EUCOMM)Wtsi (MGI:4441912) [7]. Gpsm2− (null allele) was produced via Cre-mediated recombination to permanently remove floxed exon 5, whereas Gpsm2FloX (conditional allele) was produced via FLP-mediated recombination to remove the FRT-ed BetaGeo cassette. References for the other lines are as follows: PTXa (Gt(ROSA)26Sorem1(ptxA)Btar, MGI:6163665) [9], Atoh1-Cre (Tg(Atoh1-cre)1Bfri, MGI:3775845) [44], Isl1-Cre (Isl1tmi(cre)Tmj, MGI;2447758) [45], Myo15ash2 (JAX stock #000109), Whrnwi (JAX stock #000571), Whrnneo (MGI:4462398) [31]. “Cre” notation is Atoh1-Cre except in Figure 6E where it is Isl1-Cre. The inner ear samples analyzed ranged in age from E16.5 to P33 as indicated in each figure and included both males and females. All animals of appropriate age and genotype were included; no additional inclusion or exclusion criteria were applied. Animals were maintained under standard housing and all animal work was reviewed for compliance and approved by the Animal Care and Use Committees of The Jackson Laboratory and the University of Utah.
Cell lines
For immunoprecipitation experiments, HEK293 cells (female, CRL-1573™, ATCC, RRID:CVCL_0045) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37°C. For the filopodia tip assay, COS-7 cells (male, CRL-1651 ™, ATCC, RRID:CVCL_0224) were cultured in DMEM supplemented with 10% fetal bovine serum at 37°C. Neither cell line has been authenticated since our acquisition.
METHOD DETAILS
Scanning electron microscopy
Samples were processed and imaged, and bundle parameters quantified as described [9]. Briefly, for P6/P7 samples, whole inner ears were fixed 1 hour in PFA 4% prior to exposing the sensory epithelium and removing the tectorial membrane. Samples were then fixed for at least one overnight in 2.5% glutaraldehyde and PFA 4% in 1 mM MgCl2 and 0.1 M sodium cacodylate buffer. For P16-P21 samples, inner ears were fixed for at least one overnight in the above glutaraldehyde/PFA fixative before decalcification in 0.11 M EDTA and dissection to expose the sensory epithelium. Following dissection, samples were dehydrated using an ethanol series followed by chemical drying with hexamethyldisilazane (Electron Microscopy Sciences 50-243-18). Dried samples were mounted on aluminum stubs using double-sided carbon tape and sputter coated with gold/palladium before imaging on a Hitachi 3000N VP. Imaging was done at 20kV and 500-10,000x magnification, and tilting of the samples during mounting and imaging was used to minimize parallax to improve the accuracy of stereocilia height measurements.
Immunofluorescence and antibodies
Inner ears were isolated from the skull and either microdissected to expose the cochlear sensory epithelium (embryonic and early postnatal stages) or the cochlea punctured at the apex (stages >P7) prior to fixation with paraformaldehyde (PFA 4%) for 1h at 4°C. Samples >P7 were additionally treated with 0.11 M EDTA overnight at room temperature to soften the bone before dissection to expose the sensory epithelium. After removal of the tectorial membrane, samples were permeabilized and blocked in PBS with 0.5% Triton X-100 and 1% bovine serum albumin for 1h at room temperature before application of the primary antibodies. Following incubation with secondary antibodies and phalloidin, samples were mounted in 10% Mowiol (EMD/Millipore; in 0.1M Tris pH8.5) either flat directly between a slide and a #1.5 coverslip or mounted using spacers to avoid tissue compression (Figure S3D-E). Protein colocalization involved two primary antibodies raised in different species, except in one configuration (GNAI3+MYO15A). In Figure 2B (P0 IHC) and Figure S3B (P7 OHC), rabbit anti-GNAI3 was first incubated alone, detected with an anti-rabbit antibody conjugated to Alexa Fluor (AF) 647, and following washes, the samples were next incubated with rabbit anti-MYO15A alone and then detected with anti-rabbit conjugated to AF555. Since the bare zone where GNAI3 is most abundant was not labeled by AF555, we concluded that AF555 signal was largely specific to MYO15A.
Primary antibodies were goat anti-GPSM2 (PA5-18646, ThermoFisher Scientific, 1:250), rabbit anti-GNAI3 (sc-262, Santa Cruz Biotechnology, 1:400), rabbit anti-MYO15A (2997-3511 aa, NP_034992, 1:500)[46], rabbit anti-WHRN C-terminal (721-800 aa, NP_082916, 1:500)[27], mouse anti-EPS8 (610143, BD Biosciences, 1:250), rabbit anti-TWF1/2 (gift from S. Heller & P. Barr-Gillespie, 1:100) [47], rabbit anti-MYO15A-long (long isoform) (PB888, gift from J. Bird and T. Friedman, 1:500). F-actin was visualized using phalloidin conjugated to AF488 or AF555 (ThermoFisher Scientific A12379, A34055 1:400). Secondary antibodies were raised in donkey and purchased from ThermoFisher Scientific. Most images were captured on a Zeiss LSM800 confocal microscope equipped with an Airyscan detector. Single z or z-series captures in “Airy mode” using the Airyscan detector were deconvoluted using the Zen software (RRID:SCR_013672), and are labeled accordingly in the figure legends. Most other images (unlabeled) were acquired using regular confocal capture. In all panels and figures, the lateral (abneural) side of the auditory epithelium is up and the lateral (neural) side is down. Imaging was typically not performed blind to genotype as we first confirmed that the litter contained pups of the desired genotypes. Each result was confirmed by imaging at least 3 animals per genotype representing at least 2 separate litters.
Immunoprecipitation and western blotting
HEK293 cells were double or triple transfected with WHRN cDNA plasmids [48], EGFP-tagged GNAI3 cDNA plasmids, pEGFP-C1 (Clontech), and MYC-tagged GPSM2 cDNA plasmid using PEI, according to the manufacturer’s protocol (Polysciences, Inc., Warrington, PA). After 24 hours of transfection, cells were homogenized in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% (v/v) Triton X-100, 5 mM EDTA, 1 X protease inhibitor (cOmplete, Sigma/Roche cat# 05056489001) and 1 mM DTT) and centrifuged at 21,000 X g for 10 min. The supernatants were incubated with anti-WHRN (N-terminal, [27])-conjugated protein G sepharose (GE Healthcare cat# 17-0618-01) overnight with gentle agitation. The sepharose beads and their binding proteins were then spun down, washed four times with lysis buffer, and boiled in Laemmli sample buffer for 5 min. Standard immunoblotting procedures were followed using chicken polyclonal anti-WHRN (C-terminal, [27]), mouse monoclonal anti-MYC antibody (631206, Clontech), and rabbit polyclonal anti-EGFP [46]. The immunoprecipitation experiment in Figure 4C was performed twice to confirm the result. An independent experiment using myc-tagged WHRN, EGFP-tagged GNAI3, and an anti-MYC antibody for immunoprecipitation confirmed the WHRN-GNAI3 interaction (data not shown).
Filopodia tip assay
COS-7 cells were transfected with pEgfp-C2-Myo15a(2a) (gift from Jonathan Bird and Thomas Friedman, NIDCD/NIH), pCS2-mCherry:Whrn (long isoform) and either CAGGS-MYC or CAGGS-MYC:Gpsm2 using JETPrime (Polyplus-transfection) as detailed in the manufacturer’s protocol. After transfection, cells were incubated for 36 hours, trypsinized and seeded onto poly-D-Lysine (MP Biomedicals)-coated glass coverslips. Cells were further incubated for 7-8 hours and then fixed in 4% PFA for 15 min at room temperature.
QUANTIFICATION AND STATISTICAL ANALYSIS
The sample size for each experiment was chosen based on previously published work in the field and is detailed below for each experiment. We selected the most appropriate statistical tests available to us based on experimental design and applicability of each test’s inherent assumptions. The choice of test is detailed below for each experiment.
Scanning electron microscopy image analysis
Images for comparison were carefully chosen based on similarity of the angle of the bundle. Images were analyzed blind to genotype in FIJI (RRID:SCR_002285) using the measurement tool and cell counter plugin. 3 to 5 P6 or P7 animals (a1-a5 in Tables S1 and S2) were analyzed per genotype, and for each animal, 10 hair cells were quantified at the half-turn position up from the cochlear base. For stereocilia height in row 1, height differential between row 1 and row 2 and stereocilia diameter, 3 stereocilia were measured per cell. Stereocilia diameter was measured near the distal end, and stereocilia were considered to constitute a separate row if they were regularly spaced and spanned >50% of the length of the bundle. Each point on the Figure 1 and Figure S1 graphs represents one animal (average +/− SEM). Statistics were calculated using one-way ANOVA with Tukey's multiple comparisons test, with a single pooled variance. Statistical analyses and graphical representations of the data were performed using GraphPad Prism (RRID:SCR_002798). We defined significance as follows: not significant (ns) p>0.05; *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001. All stereocilia quantification values are detailed in Tables S1 and S2.
Immunofluorescence image analysis
Orthogonal z-series projections were done using the Zen software (Zeiss). Further image processing was done with Adobe Photoshop CS6 (RRID:SCR_014199), and all images representing different conditions in the same experiment were treated similarly. Note that younger bundles (embryonic and neonatal) tend to fall medially (away from the BZ) during mounting, whereas older postnatal bundles tend to fall laterally, hence the differences in the appearance of the bundles, for example seen in Figure 2A-C between P0 and P7. Mutant bundles with stunted stereocilia behave differently during mounting, often remaining more upright than their control counterparts.
For quantification of WHRN signal in E18.5 Gpsm2 mutant and control OHCs (Figure 4E-F), the left and right cochleae from 4 homozygous null and 4 heterozygous littermates were labeled for WHRN, EPS8 and F-actin. An image was taken of OHCs near the base of each cochlea. Using the F-actin signal, one OHC per row per image was chosen for WHRN quantification based on good physical separation of the signals from the tips and base of the stereocilia bundle. Regions of interest (ROIs) were then drawn for the stereocilia bundle tips and base regions in FIJI [49] using the WHRN channel only. The raw integrated density (RawIntDen) was determined for each ROI, normalized such that the highest value for each region equaled 100, and plotted independently on the graph, yielding a total of 24 data points per region per genotype. A Mann-Whitney U test was performed to examine difference between genotypes.
To quantify MYO15A antibody signal at IHC stereocilia tips at P4 (Figure 5B-D), neighboring IHC pairs were selected in conditional mutants where one cell had apparently normal GPSM2 antibody signal (Figure 5B, bottom) and/or normal hair bundle morphology, including long row 1 stereocilia and absence of supernumerary rows. These “escaper” cells are likely explained by failure or great delay of Cre-mediated Gpsm2 deletion (Figure 5B-C) or Cre-mediated PTXa induction (Figure 5D). Regardless of their origin, their close similarity to true wild-type cells makes them powerful internal controls for variables such as antibody labeling across samples. If anything, differences seen between mutant and internal control cells should be more modest than the differences between mutant and true wild-type cells. This strategy was preferred over selecting control IHCs in littermate animals and using another stereocilia marker like phalloidin as reference, as we observed variation in both MYO15A and phalloidin signal intensity among samples. Images were captured using laser intensity and gain settings that did not saturate signal at row 1 tips in control IHCs. Signal intensity at tips was quantified by centering a circular region of interest of 400nm over the stereocilia tip and using the raw integrated density (RawIntDen) tool provided by FIJI. Regions of interest (ROIs) were placed at tips of rows 1 and 2 in control IHCs and rows 1,2, and 3 in mutant IHCs using the MYO15A channel. Since no MYO15A signal could be detected at row 3 tips in control IHCs, ROIs were selected using the phalloidin channel, and the corresponding intensity values reflected background MYO15A signal in the image. Images were then analyzed by normalizing all density values for each IHC pair to the average signal intensity at row 1 for the internal control IHC, which was set to equal 1. Data were n=30 stereocilia per condition, representing 5 stereocilia per row and 2 IHC pairs from each of 3 different animals. A two-way ANOVA with Sidak’s multiple comparisons test was used to analyze the data.
For detailed localization of GNAI3 and GPSM2 in Figure 6E, subsets of z-series images were selected to isolate signal from either the flat apical surface or the stereocilia tips regions, with separate maximum intensity projections generated for each. In order to show the spatial relation of GNAI3 and GPSM2 domains to the hair bundle, the maximum intensity projection for the entire z-series was used for the phalloidin channel.
For the filopodia tip assay in Figure S7, imaging was done with a ZEISS LSM800 confocal. 488 and 561 nm laser lines were used to sequentially excite the mCHERRY and EGFP proteins. The same digital gain (800V) and laser exposure (0.25%) were selected for all cells imaged. The images were converted to tagged image file format (TIFF). Filopodia were then selected and signals at their tips quantified using the FIJI-based procedure and MATLAB software described by Bird and colleagues (NanoSPD [50]). 33 cells representing 3 independent experiments were selected for each condition, and 126 (MYC) versus 225 (MYC:GPSM2) filopodia that passed the selection process were analyzed. Statistical significance was examined using a Mann-Whitney U test.
Supplementary Material
HIGHLIGHTS.
-
1)
GPSM2, GNAI, WHRN, MYO15A and EPS8 work in the same pathway to shape hair bundles
-
2)
GPSM2-GNAI-WHRN is a late module added to MYO15A-EPS8 at row 1 stereocilia tips only
-
3)
GPSM2-GNAI defines the identity of the tallest, first row stereocilia
-
4)
Mutant bundles comprise generic stereocilia lacking differential row identity
ACKNOWLEDGMENTS
We are grateful to Jonathan Bird and Thomas Friedman (NIDCD/NIH) for sharing the MYO15A-long antibody (PB888) and the pEgfp-C2-Myo15a(2a) construct, and to Stefan Heller (Stanford University) and Peter Barr-Gillespie (OHSU) for sharing the TWF1/2 antibody. This work was supported by a research grant from the National Institute on Deafness and Other Communication Disorders (NIDCD/NIH) to B.T. (R01 DC015242), and NIH grants (EY020853 and EY028130) and department core grants (EY014800 and Research to Prevent Blindness, Inc.) to J.Y.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tilney LG, Tilney MS, and DeRosier DJ (1992). Actin filaments, stereocilia, and hair cells: how cells count and measure. Annu Rev Cell Biol 8, 257–274. [DOI] [PubMed] [Google Scholar]
- 2.Gillespie PG, and Muller U (2009). Mechanotransduction by hair cells: models, molecules, and mechanisms. Cell 139, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shotwell SL, Jacobs R, and Hudspeth AJ (1981). Directional Sensitivity of Individual Vertebrate Hair-Cells to Controlled Deflection of Their Hair Bundles. Annals of the New York Academy of Sciences 374, 1–10. [DOI] [PubMed] [Google Scholar]
- 4.Lu X, and Sipe CW (2016). Developmental regulation of planar cell polarity and hair-bundle morphogenesis in auditory hair cells: lessons from human and mouse genetics. Wiley interdisciplinary reviews. Developmental biology 5, 85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deans MR (2013). A balance of form and function: planar polarity and development of the vestibular maculae. Seminars in cell & developmental biology 24, 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezan J, Lasvaux L, Gezer A, Novakovic A, May-Simera H, Belotti E, Lhoumeau AC, Birnbaumer L, Beer-Hammer S, Borg JP, et al. (2013). Primary cilium migration depends on G-protein signalling control of subapical cytoskeleton. Nature cell biology 15, 1107–1115. [DOI] [PubMed] [Google Scholar]
- 7.Tarchini B, Jolicoeur C, and Cayouette M (2013). A molecular blueprint at the apical surface establishes planar asymmetry in cochlear hair cells. Developmental cell 27, 88–102. [DOI] [PubMed] [Google Scholar]
- 8.Bhonker Y, Abu-Rayyan A, Ushakov K, Amir-Zilberstein L, Shivatzki S, Yizhar-Barnea O, Elkan-Miller T, Tayeb-Fligelman E, Kim SM, Landau M, et al. (2016). The GPSM2/LGN GoLoco motifs are essential for hearing. Mammalian genome : official journal of the International Mammalian Genome Society 27, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarchini B, Tadenev AL, Devanney N, and Cayouette M (2016). A link between planar polarity and staircase-like bundle architecture in hair cells. Development 143, 3926–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty D, Chudley AE, Coghlan G, Ishak GE, Innes AM, Lemire EG, Rogers RC, Mhanni AA, Phelps IG, Jones SJ, et al. (2012). GPSM2 mutations cause the brain malformations and hearing loss in Chudley-McCullough syndrome. American journal of human genetics 90, 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, Abu Rayyan A, Loulus S, Avraham KB, King MC, et al. (2010). Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. American journal of human genetics 87, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barr-Gillespie PG (2015). Assembly of hair bundles, an amazing problem for cell biology. Mol Biol Cell 26, 2727–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGrath J, Roy P, and Perrin BJ (2017). Stereocilia morphogenesis and maintenance through regulation of actin stability. Seminars in cell & developmental biology 65, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manor U, Disanza A, Grati M, Andrade L, Lin H, Di Fiore PP, Scita G, and Kachar B (2011). Regulation of stereocilia length by myosin XVa and whirlin depends on the actin-regulatory protein Eps8. Current biology : CB 21, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zampini V, Ruttiger L, Johnson SL, Franz C, Furness DN, Waldhaus J, Xiong H, Hackney CM, Holley MC, Offenhauser N, et al. (2011). Eps8 regulates hair bundle length and functional maturation of mammalian auditory hair cells. PLoS biology 9, e1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, et al. (1998). Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science 280, 1444–1447. [DOI] [PubMed] [Google Scholar]
- 17.Belyantseva IA, Boger ET, and Friedman TB (2003). Myosin XVa localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. PNAS 100, 13958–13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mburu P, Mustapha M, Varela A, Weil D, El-Amraoui A, Holme RH, Rump A, Hardisty RE, Blanchard S, Coimbra RS, et al. (2003). Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nature genetics 34, 421–428. [DOI] [PubMed] [Google Scholar]
- 19.Delprat B, Michel V, Goodyear R, Yamasaki Y, Michalski N, El-Amraoui A, Perfettini I, Legrain P, Richardson G, Hardelin JP, et al. (2005). Myosin XVa and whirlin, two deafness gene products required for hair bundle growth, are located at the stereocilia tips and interact directly. Human molecular genetics 14, 401–410. [DOI] [PubMed] [Google Scholar]
- 20.Mogensen MM, Rzadzinska A, and Steel KP (2007). The deaf mouse mutant whirler suggests a role for whirlin in actin filament dynamics and stereocilia development. Cell motility and the cytoskeleton 64, 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belyantseva IA, Boger ET, Naz S, Frolenkov GI, Sellers JR, Ahmed ZM, Griffith AJ, and Friedman TB (2005). Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nature cell biology 7, 148–156. [DOI] [PubMed] [Google Scholar]
- 22.Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, et al. (1998). Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 280, 1447–1451. [DOI] [PubMed] [Google Scholar]
- 23.Mustapha M, Chouery E, Chardenoux S, Naboulsi M, Paronnaud J, Lemainque A, Megarbane A, Loiselet J, Weil D, Lathrop M, et al. (2002). DFNB31, a recessive form of sensorineural hearing loss, maps to chromosome 9q32–34. European journal of human genetics : EJHG 10, 210–212. [DOI] [PubMed] [Google Scholar]
- 24.Behlouli A, Bonnet C, Abdi S, Bouaita A, Lelli A, Hardelin JP, Schietroma C, Rous Y, Louha M, Cheknane A, et al. (2014). EPS8, encoding an actin-binding protein of cochlear hair cell stereocilia, is a new causal gene for autosomal recessive profound deafness. Orphanet journal of rare diseases 9, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mauriac SA, Hien YE, Bird JE, Carvalho SD, Peyroutou R, Lee SC, Moreau MM, Blanc JM, Geyser A, Medina C, et al. (2017). Defective Gpsm2/Galphai3 signalling disrupts stereocilia development and growth cone actin dynamics in Chudley-McCullough syndrome. Nature communications 8, 14907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rzadzinska AK, Schneider ME, Davies C, Riordan GP, and Kachar B (2004). An actin molecular treadmill and myosins maintain stereocilia functional architecture and self-renewal. The Journal of cell biology 164, 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathur PD, Zou J, Zheng T, Almishaal A, Wang Y, Chen Q, Wang L, Vashist D, Brown S, Park A, et al. (2015). Distinct expression and function of whirlin isoforms in the inner ear and retina: an insight into pathogenesis of USH2D and DFNB31. Human molecular genetics 24, 6213–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikkawa Y, Mburu P, Morse S, Kominami R, Townsend S, and Brown SD (2005). Mutant analysis reveals whirlin as a dynamic organizer in the growing hair cell stereocilium. Human molecular genetics 14, 391–400. [DOI] [PubMed] [Google Scholar]
- 29.Ebrahim S, Ingham NJ, Lewis MA, Rogers MJC, Cui R, Kachar B, Pass JC, and Steel KP (2016). Alternative Splice Forms Influence Functions of Whirlin in Mechanosensory Hair Cell Stereocilia. Cell reports 15, 935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michalski N, Michel V, Bahloul A, Lefevre G, Barral J, Yagi H, Chardenoux S, Weil D, Martin P, Hardelin JP, et al. (2007). Molecular characterization of the ankle-link complex in cochlear hair cells and its role in the hair bundle functioning. J Neurosci 27, 6478–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Liu X, Zhao Y, Adamian M, Pawlyk B, Sun X, McMillan DR, Liberman MC, and Li T (2010). Ablation of whirlin long isoform disrupts the USH2 protein complex and causes vision and hearing loss. PLoS Genet 6, e1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer M, Petronczki M, Dorner D, Forte M, and Knoblich JA (2001). Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107, 183–194. [DOI] [PubMed] [Google Scholar]
- 33.Schaefer M, Shevchenko A, Shevchenko A, and Knoblich JA (2000). A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Current biology : CB 10, 353–362. [DOI] [PubMed] [Google Scholar]
- 34.Mochizuki N, Cho G, Wen B, and Insel PA (1996). Identification and cDNA cloning of a novel human mosaic protein, LGN, based on interaction with G alpha i2. Gene 181, 39–43. [DOI] [PubMed] [Google Scholar]
- 35.Willard FS, Zheng Z, Guo J, Digby GJ, Kimple AJ, Conley JM, Johnston CA, Bosch D, Willard MD, Watts VJ, et al. (2008). A point mutation to Galphai selectively blocks GoLoco motif binding: direct evidence for Galpha.GoLoco complexes in mitotic spindle dynamics. The Journal of biological chemistry 283, 36698–36710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng AW, Belyantseva IA, Hsu PD, Friedman TB, and Heller S (2009). Twinfilin 2 regulates actin filament lengths in cochlear stereocilia. The Journal of neuroscience : the official journal of the Society for Neuroscience 29, 15083–15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rzadzinska AK, Nevalainen EM, Prosser HM, Lappalainen P, and Steel KP (2009). MyosinVIIa interacts with Twinfilin-2 at the tips of mechanosensory stereocilia in the inner ear. PloS one 4, e7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang Q, Indzhykulian AA, Mustapha M, Riordan GP, Dolan DF, Friedman TB, Belyantseva IA, Frolenkov GI, Camper SA, and Bird JE (2015). The 133-kDa N-terminal domain enables myosin 15 to maintain mechanotransducing stereocilia and is essential for hearing. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caberlotto E, Michel V, de Monvel JB, and Petit C (2011). Coupling of the mechanotransduction machinery and F-actin polymerization in the cochlear hair bundles. Bioarchitecture 1, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velez-Ortega AC, Freeman MJ, Indzhykulian AA, Grossheim JM, and Frolenkov GI (2017). Mechanotransduction current is essential for stability of the transducing stereocilia in mammalian auditory hair cells. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beurg M, Cui R, Goldring AC, Ebrahim S, Fettiplace R, and Kachar B (2018). Variable number of TMC1-dependent mechanotransducer channels underlie tonotopic conductance gradients in the cochlea. Nature communications 9, 2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caberlotto E, Michel V, Foucher I, Bahloul A, Goodyear RJ, Pepermans E, Michalski N, Perfettini I, Alegria-Prevot O, Chardenoux S, et al. (2011). Usher type 1G protein sans is a critical component of the tip-link complex, a structure controlling actin polymerization in stereocilia. Proceedings of the National Academy of Sciences of the United States of America 108, 5825–5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narayanan P, Chatterton P, Ikeda A, Ikeda S, Corey DP, Ervasti JM, and Perrin BJ (2015). Length regulation of mechanosensitive stereocilia depends on very slow actin dynamics and filament-severing proteins. Nature communications 6, 6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, Morris K, Feng F, Jones K, Lee J, and Fritzsch B (2005). Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Developmental dynamics : an official publication of the American Association of Anatomists 234, 633–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou J, Zheng T, Ren C, Askew C, Liu XP, Pan B, Holt JR, Wang Y, and Yang J (2014). Deletion of PDZD7 disrupts the Usher syndrome type 2 protein complex in cochlear hair cells and causes hearing loss in mice. Human molecular genetics 23, 2374–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avenarius MR, Krey JF, Dumont RA, Morgan CP, Benson CB, Vijayakumar S, Cunningham CL, Scheffer DI, Corey DP, Muller U, et al. (2017). Heterodimeric capping protein is required for stereocilia length and width regulation. The Journal of cell biology 216, 3861–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou J, Mathur PD, Zheng T, Wang Y, Almishaal A, Park AH, and Yang J (2015). Individual USH2 proteins make distinct contributions to the ankle link complex during development of the mouse cochlear stereociliary bundle. Human molecular genetics 24, 6944–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bird JE, Barzik M, Drummond MC, Sutton DC, Goodman SM, Morozko EL, Cole SM, Boukhvalova AK, Skidmore J, Syam D, et al. (2017). Harnessing molecular motors for nanoscale pulldown in live cells. Mol Biol Cell 28, 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.