Abstract
Adeno-associated virus (AAV) consists of a simple genome, infects mammalian cells, displays nonpathogenicity in humans, and spans an array of serotypes and variants bearing distinct tissue tropisms. These attributes lend AAV tremendous promise as a gene delivery vector, further substantiated by its extensive testing in human clinical trials. Rational design approaches to capsid engineering leverage current scientific knowledge of AAV to further modulate, enhance and optimize the performance of the vectors. Capsid modification strategies include amino acid point mutations, peptide domain insertions, and chemical biology approaches. Through such efforts, insights regarding AAV capsid sequence-structure-function relationships can be learned. Developments over the last 5 years in rational design-based capsid engineering approaches will be presented and discussed.
Keywords: Adeno-associated virus, AAV, gene therapy, gene delivery, rational design, synthetic virology, viral vector, review
Graphical Abstract
The Basics of AAV
Adeno-associated virus (AAV) is a member of the Parvoviridae family that primarily infects mammalian cells and is purportedly nonpathogenic in humans. First reported in 1965 as a contaminant of adenovirus, it has since been characterized as naturally replication-deficient, requiring helper viruses such as adenovirus for propagation [1].
AAV’s linear single-stranded DNA genome (~4.7kb) encodes two genes, rep and cap, flanked by inverted terminal repeats (ITRs) necessary for packaging the viral genome inside the capsid. The ITRs act as primers for second-strand synthesis and are the only elements in the genome required in cis for viral production [2]. For recombinant AAV (rAAV) vector production, the remainder of the viral genome can be removed, provided in trans on separate plasmids, and replaced with a desired transgene.
The rep gene encodes four overlapping non-structural proteins for replication, integration, and packaging. Rep78 and Rep68 bind to the ITRs and demonstrate helicase and endonuclease activity necessary for AAV genome replication [3]. Rep52 and Rep40 demonstrate 3’ to 5’ helicase activity and package viral genomes into capsids during virus production [4]. The cap gene encodes three structural proteins, VP1, VP2, and VP3, that self-assemble into a 60-mer icosahedral capsid at a ratio of approximately 1:1:10. These three proteins are transcribed from the same open reading frame and share a C-terminal domain but have different N-termini due to alternative start codons and alternative splicing [5]. cap also encodes for a non-structural protein, assembly-activating protein (AAP), in an alternate open reading frame from the VPs initially shown to be required for AAV2 capsid assembly [6]. Capsid assembly dependence on AAP is serotype-specific, as AAV4, −5, and −11 do not require AAP to assemble [7]. Wild-type AAV2 can undergo Rep-mediated site-specific integration into human chromosome 19 without helper virus, and rAAV vectors lacking rep integrate non-specifically at low frequencies [8]. rAAV achieves high levels of long-term gene expression without chromosomal integration and persists episomally in the nucleus in the form of head-to-tail concatemers [9]. rAAV episomes may be able to replicate in proliferating cells, albeit at low frequency [10].
VP1 is the largest (87 kDa), followed by VP2 (72 kDa) and VP3 (62 kDa). They share a common C-terminal domain, while VP1 and VP2 also contain longer N-terminal domains that are packaged inside the capsid but externalize in the endosome during intracellular trafficking [11]. The VP1 N-terminal domain contains a phospholipase A2 domain for endosomal escape and nuclear localization sequences for nuclear trafficking. VP2 is nonessential for capsid formation and viral infection [12]. Techniques including X-ray crystallography and cryo-electron microscopy have been employed to visualize the capsid structures of many serotypes, revealing key domains that can be exploited in rational design strategies to modify functionality [13,14]. All AAV capsids share a core β-barrel motif. The β-strands connected by variable surface loops produce capsid surface topological variations – much of which can be found around the capsid’s three-fold spikes (a region frequently implicated in receptor binding and antibody recognition) [15,16].
Not merely an inert protein shell, the capsid dictates virus-cell receptor interactions and intracellular trafficking. Recent research suggests that VPs may contribute to second-strand synthesis and genomic transcription [17]. Twelve AAV serotypes and numerous variants from human and nonhuman primates have been identified with different serological profiles, cell surface receptor usage, and tissue tropisms [18]. Serotypes can be loosely categorized based on their primary cell surface receptor usage: AAV2, −3, and −6 bind heparan sulfate proteoglycan (HSPG), AAV1, −4, −5 and −6 bind sialic acid, and AAV9 binds galactose [16]. Co-receptors for internalization also vary and include laminin receptor, epidermal growth factor receptor, hepatocyte growth factor receptor, platelet-derived growth factor receptor, and several integrins (reviewed in [19]). A novel receptor AAVR recently identified appears to be required for infection for some variants [20]. Most serotypes depend on AAVR for successful cell internalization, but bind to and interact with it differently [21]. Exceptions include AAV4 and the chimeric variant AAVrh32.33, which use an AAVR-independent pathway [22].
AAV as a Gene Therapy Vector
rAAV was first produced in the early 1980’s; rAAV containing an antibiotic resistance gene in place of cap successfully transduced mammalian cells, establishing AAV’s potential as a gene delivery vector [23,24]. The first FDA approval for gene therapy treatment of a hereditary disease was granted in December 2017 for an AAV2-based product for RPE65-mediated inherited retinal dystrophy.
rAAV possesses several key features that make it highly promising for gene therapy. Its genome and capsid structure are relatively simple, and the ITRs are the only cis-acting elements essential for packaging transgenes into the capsid [2]. Additionally, AAV is nonpathogenic and demonstrates relatively low levels of immunogenicity and genotoxicity [25,26]. Serotypes exhibit a diverse range of tropisms and immune response profiles desirable for different applications. AAV8 is preferential for targeting the liver, whereas cardiac and skeletal muscle gene transfer appears mediated best by AAV1, −6, and −9 [19]. AAV9 and AAVrh.10 have demonstrated the ability to cross the blood-brain barrier (BBB) when injected intravenously [27]. Remarkably, despite AAV predominantly persisting episomally, transgene expression can be detected as long as 10 years post-AAV injection [28].
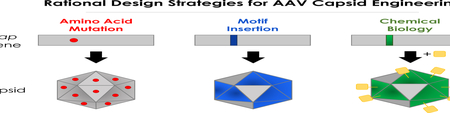
Rational Design Strategies for AAV Capsid Engineering
Despite AAV’s successes as a gene delivery vector, achieving greater control and predictability of function remains a non-trivial task. Fortunately, illuminating studies on AAV structure and biology continue to uncover new insights. Rational design strategies draw from this ever-expanding body of AAV knowledge as a framework for harnessing virus behavior. Three prominent rational capsid engineering strategies employed over the last 5 years are presented below.
Genetic mutation of AAV parts
Several studies have investigated the role of specific capsid amino acid residues in AAV’s functionality. Specifically, the efficiency and specificity of AAV gene delivery can be improved using point mutations on the viral capsid. For example, it has been postulated that undesirable post-translational modification leads to capsid degradation [29]. To address this problem, various serine, threonine and lysine residues in the AAV2 capsid were mutated to alanine or arginine [30]. The majority of these substitutions lead to enhanced transduction efficiency in HeLa cells, as well as greater gene expression in the livers of mice. The triple mutant, Y444F/Y500F/Y730F, is a promising AAV2 vector in the field [31], although similar mutations in other AAV serotype capsids do not enhance gene delivery efficiency. In a different study that addresses gene delivery specificity, an array of naturally occurring AAV variants from non-human primate tissues was isolated and capsid alignment of new isolates to currently available variants identified several residues of interest [32]. Based on this information, a new variant, AAV9.HR, was generated from the parental AAV9 by changing only two residues, H527Y and R533S. This vector transduces cells in the central nervous system (CNS), although not as robustly as AAV9. However, AAV9.HR has increased specificity since its transgene expression in peripheral tissues is reduced. In a murine model of Canavan disease, AAV9.HR-mediated delivery of the human ASPA gene successfully improves motor function. Thus, results demonstrate key point mutations on the AAV capsid can alter gene delivery efficiency and specificity.
Point mutations to the AAV capsid can also be used to mitigate recognition by host antibodies. Using cryo-electron microscopy reconstruction, site-directed mutagenesis of candidate residues, and cellular assays, Bennett et al. identified residue K531 as the contributor to AAV6 recognition by ADK6, a monoclonal antibody [33]. Mutation of K531, therefore, has the potential to impart immune evasion properties to AAV6. In a separate study, an AAV6 mutant, AAV6.2FF, was generated by introducing three point mutations (F129L, Y445F, and Y731F) to the capsid [34]. This mutant exhibits enhanced transduction efficiency in vitro relative to AAV6. Moreover, it is also more resistant to neutralization by intravenous antibodies. Although AAV6.2FF accumulates more rapidly in the lungs and muscles of mice, long-term expression levels do not reveal significant differences with AAV6. Point mutations to the AAV capsid, therefore, can be a useful strategy for preventing vector neutralization by preexisting antibodies in the host.
In addition to point mutations, larger peptide domains from one AAV serotype can be transferred to another serotype to impart new functions. For example, the ‘receptor binding footprint’ of the AAV9 capsid was incorporated into AAV2, which imparted the latter with galactose (Gal) binding properties of AAV9 [35]. The two resulting chimeras, AAV2G9 and AAV2i8G9, effectively bind to both HSPG and Gal receptors for cell entry. Moreover, the latter vector also exhibits liver de-targeting akin to its parental strain AAV2i8. This work revealed that grafting the Gal receptor recognition domain onto the AAV2 capsid does not require substantial sequence alteration and invites further investigation into extending this design approach using receptor binding domains from other AAV serotypes.
More recently, directed evolution was used in combination with rational design to develop AAV variants that could traverse the BBB with greater efficiency and specificity. DNA shuffling was used to generate capsid chimeras between AAV1 and AAVrh.10, which were then selected in vivo for their ability to cross the BBB. Structural analysis of one successful candidate identified three AAVrh.10 domains that may contribute to this property. Further studies reduced the functional domains down to eight key amino acid residues, and this minimal AAVrh.10 ‘BBB traversing footprint’ was grafted onto AAV1. The resulting vector AAV1RX not only transduces cells in the CNS readily, but also demonstrates improved specificity as evidenced by diminished transduction in the liver and vasculature [36].
In order to facilitate the rational design of new AAV capsid chimeras with functional domains of one serotype transplanted into another, the SCHEMA algorithm can be used to calculate the extent of structural disruption during chimeragenesis [37]. Using results from the algorithm as a guide, a small panel of AAV chimeras between AAV2 and AAV4 were generated. Experimental validation revealed that SCHEMA could be a useful tool for AAV capsid design, specifically in assessing capsid intactness and transduction efficiency. In sum, larger peptide domains from one AAV variant can be incorporated into another variant to rationally design new AAV mutants.
Insertion of nonviral parts into AAV capsid
A second rational design approach is to introduce functional domains nonviral in nature into the AAV capsid to elicit desired functions. For example, hexahistidine (His)-tagged designed ankyrin repeat proteins (DARPin) specific for Her2, CD4, and EpCAM have been inserted into the VP2 subunit of AAV2 [38]. Enrichment of DARPin-expressing viral particles by immobilized metal ion affinity chromatography eliminates off-target vector delivery, suggesting that subpopulations of capsids deficient in DARPin moieties lead to off-target transgene expression. The modified vectors demonstrated efficacy in in vitro and in vivo experiments: DARPin (anti-Her2)-AAV carrying a transgene that disrupts DNA replication allows for a temporary halt in breast tumor growth; DARPin (anti-CD4)-AAV selectively transduces target cells both in vitro and in vivo; and DARPin (anti-EpCAM)-AAV can discriminate between tumor cells and blood cells in whole blood samples, transducing only the former.
Nonviral parts can also be inserted into the AAV capsid to render them stimulus-responsive [39,40]. For example, small ‘peptide locks’ consisting of tetra-aspartic acid residues flanked by various protease cleavage sequences have been inserted in close proximity to the HSPG binding domain of AAV2 [41]. The peptide locks prevent the vector from transducing cells until they are cleaved off the capsid by extracellular proteases, such as matrix metalloproteinases (MMPs). Peptide locks with other amino acid compositions have also been tested, and results suggest the locks function primarily via steric obstruction of capsid-receptor binding interactions [42]. The protease-activatable vectors can perform Boolean AND gate logic, requiring detection of two different MMPs to transduce cells. The transduction efficiency of the AAV protease-activatable vectors can be improved by combining different ratios of wild-type and protease-activatable subunits [43]. Transduction efficiency increases with incorporation of more wild-type subunits; however, higher levels of non-specific transduction are also observed.
While endogenous stimuli can prompt viruses to respond accordingly within their microenvironment, external regulation may facilitate more temporal and spatial control of transgene delivery and expression. For instance, AAV transduction may be controlled by an externally applied chemical stimulus [44]. An AAV2 vector was developed with its natural cell receptor binding ability ablated and replaced with human FK-binding protein (FKBP). When supplied with a fusion protein containing an FKBP-rapamycin binding (FRB) domain attached to a DARPin moiety (targeting the human epidermal growth factor receptor - EGFR) as well as the small molecule rapamycin analog, the small molecule induces binding of FKBP to FRB, resulting in the mutant AAV vector able to transduce cells overexpressing EGFR.
In addition to chemical stimuli, externally applied light can be used to control AAV transduction. A light-activatable platform based on the heterodimerization of Phytochrome B (PhyB) and Phytochrome Interacting Factor (PIF) has been developed [45]. Upon delivering AAV2 displaying PIF on its capsid surface, a PhyB-NLS plasmid, and the chromophore phycocyanobilin to mammalian cells, efficiency of virus nuclear translocation can be modulated using different ratios of red to far red light. Moreover, cells exposed to increasing red light intensities through a photomask can exhibit higher gene expression in a spatially controlled fashion [46].
Lastly, nonviral motifs can be inserted into the AAV capsid to bring about new functional outputs. Recently, a panel of mosaic AAV capsids with varying lengths of VP2 truncation mutant subunits with a His tag at the N-terminus have been generated [47]. By harnessing the AAV capsid’s natural mechanism of activatable peptide display, the resulting virus particles exhibit varying degrees of His tag exposure pre- and post- temperature activation. Among the elucidated design principles, capsid mosaicism appears to be a requirement for robust activatable peptide display, with incorporation of fewer mutant subunits improving activatability. The length of the truncation subunits does not impart any significant functional effects. In sum, motifs from nonviral sources can be incorporated into the viral capsid in order to dramatically expand the functionality of AAV vectors.
Chemical biology approaches for AAV capsid modification
The third rational design approach involves using chemical biology strategies to make more precise modifications to the capsid. For example, an aldehyde tag was inserted into all three VP subunits of the AAV2 capsid, to which various types of molecules could be attached [48]. Despite overall low transduction efficiency, conjugating cyclic RGD peptides to the modified AAV improves transduction of HeLa cells compared to controls lacking the functionalized peptides.
In another study, the non-canonical amino acid AzK was genetically incorporated at five different surface-exposed regions on the AAV2 capsid [49]. A synthetic peptide targeting αvβ3 integrin receptors was then chemically conjugated to the AzK residues of two AAV variants, T454AzK and R588AzK. The latter mutant demonstrates effective vector retargeting and transduction of high αvβ3-expressing ovarian cancer cells.
More recently, a small tetracysteine motif was introduced into AAV9 for subsequent chemical attachment of a maleimide dye [50]. Fluorophore labeling does not disrupt the virus’s ability to pass through the BBB in mice, and the fluorescent vectors can be tracked in real-time using intravital microscopy. Insights on the capsid interactome were gleaned from studies employing a maleimide-biotin AAV9 variant in human embryonic kidney (HEK) cells, namely that transduction decreases in the absence of αVβ6 integrin but increases with lower levels of histone deacetylase 4. In sum, chemical biology methods to capsid modification allow for site-specific attachment of moieties, such as targeting ligands and fluorophores, to the vector.
Conclusion
Rational design strategies for AAV capsid engineering have yielded numerous vectors with enhanced functionalities. They rely on fundamental insights derived from the continuous discovery of naturally occurring virus variants, structural characterization, predictive modeling, and mechanistic studies. In consideration of clinical translation, areas that have been explored but still require further progress include improving AAV transduction efficiency, targeting specificity, and minimizing recognition by the host immune system. While mutation of various capsid residues may impart desirable characteristics, such as enhanced transduction or specific tissue de-targeting, the precise mechanisms behind these outcomes often remain poorly understood. Therefore, additional comprehensive experiments are needed to further our understanding of capsid sequence-structure-function relationships. When inserting exogenous motifs into the AAV capsid, it is sometimes difficult to minimize their effects on capsid assembly and hence vector production. If the vectors are to progress towards scale-up and clinical testing, improved designs must be investigated to lessen any adverse impacts of motif insertion on vector titers. As new AAV capsid variants continue to be developed and studied, key design principles will be discovered which will inform future capsid improvement strategies.
Highlights.
Rational design of AAV relies on virus sequence, structure, and function knowledge.
Strategies include point mutations, motif insertions, and chemical biology methods.
Goals are to improve AAV transduction efficiency, specificity, and immune evasion.
Acknowledgments
This work was supported by the National Science Foundation under grant number 1611044, the National Institutes of Health under grant numbers R01CA207497 and R01HL138126, and a Hamill Foundation award to J.S.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
** of outstanding interest
- 1.Atchison RW, Casto BC, Hammon WM: Adenovirus-associated defective virus particles. Science (1965) 149(3685):754–755. [DOI] [PubMed] [Google Scholar]
- 2.McLaughlin SK, Collis P, Hermonat PL, Muzyczka N: Adeno-associated virus general transduction vectors: Analysis of proviral structures. Journal of virology (1988) 62(6):1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Im DS, Muzyczka N: Partial purification of adeno-associated virus rep78, rep52, and rep40 and their biochemical characterization. Journal of virology (1992) 66(2):11191128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King JA, Dubielzig R, Grimm D, Kleinschmidt JA: DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. The EMBO journal (2001) 20(12):3282–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berns KI: Parvovirus replication. Microbiological reviews (1990) 54(3):316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earley LF, Kawano Y, Adachi K, Sun X-X, Dai M-S, Nakai H: Identification and characterization of nuclear and nucleolar localization signals in the adeno-associated virus serotype 2 assembly-activating protein. Journal of virology (2015) 89(6):30383048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earley LF, Powers JM, Adachi K, Baumgart JT, Meyer NL, Xie Q, Chapman MS, Nakai H: Adeno-associated virus (aav) assembly-activating protein is not an essential requirement for capsid assembly of aav serotypes 4, 5, and 11. Journal of virology (2017) 91(3):e01980–01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarty DM, Young SM Jr, Samulski RJ: Integration of adeno-associated virus (aav) and recombinant aav vectors. Annu Rev Genet (2004) 38(819–845. [DOI] [PubMed] [Google Scholar]
- 9.Fisher KJ, Jooss K, Alston J, Yang Y, Haecker SE, High K, Pathak R, Raper SE, Wilson JM: Recombinant adeno-associated virus for muscle directed gene therapy. Nature medicine (1997) 3(3):306–312. [DOI] [PubMed] [Google Scholar]
- 10.Hagedorn C, Schnödt-Fuchs M, Boehme P, Abdelrazik H, Lipps HJ, Büning H: S/mar element facilitates episomal long-term persistence of adeno-associated virus vector genomes in proliferating cells. Human gene therapy (2017) 28(12):1169–1179. [DOI] [PubMed] [Google Scholar]
- 11.Kronenberg S, Böttcher B, Claus W, Bleker S, Kleinschmidt JA: A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden vp1 n termini. Journal of virology (2005) 79(9):5296–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warrington KH, Gorbatyuk OS, Harrison JK, Opie SR, Zolotukhin S, Muzyczka N: Adenoassociated virus type 2 vp2 capsid protein is nonessential and can tolerate large peptide insertions at its n terminus. Journal of virology (2004) 78(12):6595–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS: The atomic structure of adeno-associated virus (aav-2), a vector for human gene therapy.Proceedings of the National Academy of Sciences (2002) 99(16):10405–10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walters RW, Agbandje-McKenna M, Bowman VD, Moninger TO, Olson NH, Seiler M, Chiorini JA, Baker TS, Zabner J: Structure of adeno-associated virus serotype 5. Journal of virology (2004) 78(7):3361–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurda BL, DiMattia MA, Miller EB, Bennett A, McKenna R, Weichert WS, Nelson CD, Chen W-j, Muzyczka N, Olson NH: Capsid antibodies to different adeno-associated virus serotypes bind common regions. Journal of virology (2013) 87(16):9111–9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agbandje-McKenna M, Kleinschmidt J: Aav capsid structure and cell interactions In: Adeno-associated virus. Springer, (2012):47–92. [DOI] [PubMed] [Google Scholar]
- 17.Salganik M, Aydemir F, Nam H-J, McKenna R, Agbandje-McKenna M, Muzyczka N: Adeno-associated virus capsid proteins may play a role in transcription and secondstrand synthesis of recombinant genomes. Journal of virology (2014) 88(2):1071–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z, Asokan A, Samulski RJ: Adeno-associated virus serotypes: Vector toolkit for human gene therapy. Molecular therapy (2006) 14(3):316–327. [DOI] [PubMed] [Google Scholar]
- 19.Asokan A, Schaffer DV, Samulski RJ: The aav vector toolkit: Poised at the clinical crossroads. Molecular Therapy (2012) 20(4):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Ya, Jae LT, Wosen JE, Nagamine CM, Chapman MS: An essential receptor for adeno-associated virus infection. Nature (2016) 530(7588):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pillay S, Zou W, Cheng F, Puschnik AS, Meyer NL, Ganaie SS, Deng X, Wosen JE, Davulcu O, Yan Z, Engelhardt JF et al. : Aav serotypes have distinctive interactions with domains of the cellular receptor aavr. Journal of virology (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudek AM, Pillay S, Puschnik AS, Nagamine CM, Cheng F, Qiu J, Carette JE, Vandenberghe LH: An alternate route for adeno-associated virus entry independent of aavr. Journal of virology (2018) JVI–02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samulski RJ, Berns KI, Tan M, Muzyczka N: Cloning of adeno-associated virus into pbr322: Rescue of intact virus from the recombinant plasmid in human cells. Proceedings of the National Academy of Sciences (1982) 79(6):2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermonat PL, Muzyczka N: Use of adeno-associated virus as a mammalian DNA cloning vector: Transduction of neomycin resistance into mammalian tissue culture cells. Proceedings of the National Academy of Sciences (1984) 81(20):6466–6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calcedo R, Wilson JM: Humoral immune response to aav. Frontiers in immunology (2013) 4(341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Malani N, Hamilton SR, Schlachterman A, Bussadori G, Edmonson SE, Shah R, Arruda VR, Mingozzi F, Wright JF: Assessing the potential for aav vector genotoxicity in a murine model. Blood (2011) 117(12):3311–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Yang B, Mu X, Ahmed SS, Su Q, He R, Wang H, Mueller C, Sena-Esteves M, Brown R: Several raav vectors efficiently cross the blood–brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Molecular therapy (2011) 19(8):1440–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchlis G, Podsakoff GM, Radu A, Hawk SM, Flake AW, Mingozzi F, High KA: Factor ix expression in skeletal muscle of a severe hemophilia b patient 10 years after aavmediated gene transfer. Blood (2012) 119(13):3038–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong LI, Li B, Jayandharan G, Mah CS, Govindasamy L, Agbandje-McKenna M, Herzog RW, Weigel-Van Aken KA, Hobbs JA, Zolotukhin S: Tyrosine-phosphorylation of aav2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology (2008) 381(2):194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabriel N, Hareendran S, Sen D, Gadkari RA, Sudha G, Selot R, Hussain M, Dhaksnamoorthy R, Samuel R, Srinivasan N: Bioengineering of aav2 capsid at specific serine, threonine, or lysine residues improves its transduction efficiency in vitro and in vivo. Human gene therapy methods (2013) 24(2):80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. *.Kanaan NM, Sellnow RC, Boye SL, Coberly B, Bennett A, Agbandje-McKenna M, Sortwell CE, Hauswirth WW, Boye SE, Manfredsson FP: Rationally engineered aav capsids improve transduction and volumetric spread in the cns. Molecular Therapy-Nucleic Acids (2017) 8(184–197. The authors assess the influence of various tyrosine and threonine residue mutations on transduction efficiency in the CNS. Interestingly, outcomes appear to be serotypedependent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Li S, Gessler DJ, Xie J, Zhong L, Li J, Tran K, Van Vliet K, Ren L, Su Q: A rationally engineered capsid variant of aav9 for systemic cns-directed and peripheral tissue-detargeted gene delivery in neonates. Molecular Therapy-Methods & Clinical Development (2018) 9(234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. **.Bennett AD, Wong K, Lewis J, Tseng Y-S, Smith JK, Chipman P, McKenna R, Samulski RJ, Kleinschmidt J, Agbandje-McKenna M: Aav6 k531 serves a dual function in selective receptor and antibody adk6 recognition. Virology (2018) 518(369–376. The authors identify the key amino acid residue responsible for recognition by the ADK6 monoclonal antibody. Mutation of K531 may thus be able to confer AAV6 with immune evasion properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Lieshout LP, Domm JM, Rindler TN, Frost KL, Sorensen DL, Medina SJ, Booth SA, Bridges JP, Wootton SK: A novel triple-mutant aav6 capsid induces rapid and potent transgene expression in the muscle and respiratory tract of mice. Molecular TherapyMethods & Clinical Development (2018) 9(323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen S, Horowitz ED, Troupes AN, Brown SM, Pulicherla N, Samulski RJ, AgbandjeMcKenna M, Asokan A: Engraftment of a galactose receptor footprint onto adenoassociated viral capsids improves transduction efficiency. Journal of Biological Chemistry (2013) 288(40):28814–28823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. **.Albright BH, Storey CM, Murlidharan G, Rivera RMC, Berry GE, Madigan VJ, Asokan A: Mapping the structural determinants required for aavrh. 10 transport across the blood-brain barrier. Molecular Therapy (2017). A minimal AAVrh.10 footprint imparting ability to traverse the BBB is elucidated through combined directed evolution and rational design. Grafting the domain onto AAV1 results in a vector that effectively transduces cells in the CNS, while de-targeting the liver and vasculature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho ML, Adler BA, Torre ML, Silberg JJ, Suh J: Schema computational design of virus capsid chimeras: Calibrating how genome packaging, protection, and transduction correlate with calculated structural disruption. ACS synthetic biology (2013) 2(12):724733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Münch RC, Muth A, Muik A, Friedel T, Schmatz J, Dreier B, Trkola A, Plückthun A, Büning H, Buchholz CJ: Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nature communications (2015) 6(6246. [DOI] [PubMed] [Google Scholar]
- 39.Brun MJ, Gomez EJ, Suh J: Stimulus-responsive viral vectors for controlled delivery of therapeutics. J Control Release (2017) 267(80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans AC, Thadani NN, Suh J: Biocomputing nanoplatforms as therapeutics and diagnostics. J Control Release (2016) 240(387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Judd J, Ho ML, Tiwari A, Gomez EJ, Dempsey C, Van Vliet K, Igoshin OA, Silberg JJ, Agbandje-McKenna M, Suh J: Tunable protease-activatable virus nanonodes. ACS Nano (2014) 8(5):4740–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson TM, Judd J, Ho ML, Suh J: Role of tetra amino acid motif properties on the function of protease-activatable viral vectors. ACS Biomaterials Science & Engineering (2016) in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho ML, Judd J, Kuypers BE, Yamagami M, Wong FF, Suh J: Efficiency of proteaseactivatable virus nanonodes tuned through incorporation of wild-type capsid subunits. Cellular and Molecular Bioengineering (2014) 7(3):334–343. [Google Scholar]
- 44.Hörner M, Kaufmann B, Cotugno G, Wiedtke E, Büning H, Grimm D, Weber W: A chemical switch for controlling viral infectivity. Chemical Communications (2014) 50(71):10319–10322. [DOI] [PubMed] [Google Scholar]
- 45.Gomez EJ, Gerhardt K, Judd J, Tabor JJ, Suh J: Light-activated nuclear translocation of adeno-associated virus nanoparticles using phytochrome b for enhanced, tunable, and spatially programmable gene delivery. ACS Nano (2016) 10(1):225–237. [DOI] [PubMed] [Google Scholar]
- 46.Gerhardt KP, Olson EJ, Castillo-Hair SM, Hartsough LA, Landry BP, Ekness F, Yokoo R, Gomez EJ, Ramakrishnan P, Suh J, Savage DF et al. : An open-hardware platform for optogenetics and photobiology. Scientific reports (2016) 6(35363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. **.Thadani NN, Dempsey C, Zhao J, Vasquez SM, Suh J: Reprogramming the activatable peptide display function of adeno-associated virus nanoparticles. ACS Nano (2018) 12(2):1445–1454. Through the generation of VP truncation mutants, the activatable peptide display behavior of AAV capsids can be altered. Some mutants have a ‘brush-like’ display of peptides, and others exhibit robust activatable display of exogenous peptides. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Fang Y, Zhou Y, Zandi E, Lee CL, Joo KI, Wang P: Site-specific modification of adeno-associated viruses via a genetically engineered aldehyde tag. Small (2013) 9(3):421–429. [DOI] [PubMed] [Google Scholar]
- 49. *.Kelemen RE, Mukherjee R, Cao X, Erickson SB, Zheng Y, Chatterjee A: A precise chemical strategy to alter the receptor specificity of the adeno-associated virus. Angewandte Chemie (2016) 128(36):10803–10807. This study is one of the first to demonstrate the feasibility of incorporating a noncanonical amino acid into the AAV2 capsid. [DOI] [PubMed] [Google Scholar]
- 50. *.Chandran JS, Sharp PS, Karyka E, da Conceição Aves-Cruzeiro JM, Coldicott I, Castelli L, Hautbergue G, Collins MO, Azzouz M: Site specific modification of adeno-associated virus enables both fluorescent imaging of viral particles and characterization of the capsid interactome. Scientific Reports (2017) 7(1):14766 Fluorophore labeling of the AAV9 capsid enables real-time virus particle tracking. It serves as a useful tool for acquiring new insights on capsid interactions in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]


