ABSTRACT
A full understanding of how ecological factors drive the fixation of genetic changes during speciation is obscured by the lack of appropriate models with clear natural history and powerful genetic toolkits. In a recent study, we described an early stage of ecological speciation in a population of the generalist species Drosophila yakuba (melanogaster subgroup) on the island of Mayotte (Indian Ocean). On this island, flies are strongly associated with the toxic fruits of noni (Morinda citrifolia) and show a partial degree of pre-zygotic reproductive isolation. Here, I mine the nuclear and mitochondrial genomes and provide a full morphological description of this population. Only 29 nuclear sites (< 4 × 10−7 of the genome) are fixed in this population and absent from 3 mainland populations and the closest relative D. santomea, but no mitochondrial or morphological character distinguish Mayotte flies from the mainland. This result indicates that physiological and behavioral traits may evolve faster than morphology at the early stages of speciation. Based on these differences, the Mayotte population is designated as a new subspecies, Drosophila yakuba mayottensis subsp. nov., and its strong potential in understanding the genetics of speciation and plant-insect interactions is discussed.
KEYWORDS: convergent evolution, ecological specialization, genotypic species concept, island speciation, plant-insect interactions, speciation genetics
Introduction
Speciation is a continuing process that culminates in the evolution of complete reproductive isolation and fixed genetic differences between nascent species. Understanding the evolutionary forces driving populations along this continuum (e.g., selection vs. drift) is a major biological problem.1 Modern studies revealed that different forces differently act on various parts of the genome causing a heterogeneous rate of genetic divergence and differentiation.2 This leads to questions about the nature of the earliest genetic differences to become fixed during speciation, the forces driving their fixation, and how they affect reproductive isolation.3
A great deal of our understanding of the genetic basis of speciation and speciation genes draws from studies on Drosophila flies. Most studies have, however, focused on one aspect of the speciation process, i.e. the evolution of reproductive isolation, leading to breakthroughs in the understanding of the genetics of hybrid inviability.4,5 However, the study of speciation should not be equated with reproductive isolation, and hybrid inviability is usually a late step in the speciation process.3,4 In spite of significant progress,6 our knowledge of the natural history of most laboratory model Drosophila species that have been used in speciation genetics research are often quite poor. Indeed, Drosophila research has been criticized for its neglect of the ecological context driving the early stages of speciation, and other models having clear natural history, such as Heliconius butterflies for example, have been proposed to fill this gap.4 However, these models still lack the advanced Drosophila genetic toolkits that can elegantly identify the function of several diverging genes.
A particular case that seemed to be most promising in understanding the genetics of the ecological speciation in Drosophila is the specialization of Drosophila sechellia on the toxic fruits of noni (Morinda citrifolia) on the Seychelles islands in the Indian Ocean.7 D. sechellia is closely-related to Drosophila melanogaster, has its genome fully sequenced and annotated, and can be reared in the laboratory. It has diverged nearly 250,000 y ago from its closest relative, the generalist fly Drosophila simulans, and from which it is partially pre- and post-zygotically isolated.8 In spite of evidence for ancient and recent hybridization between the 2 species in Seychelles,8,9 the 2 species are quite distinct. Orgogozo and Stern10 counted from the literature 5 morphological, 3 physiological and 6 behavioral differences between the 2 species, in addition to the higher attraction, oviposition preference and tolerance to noni toxins in D. sechellia. The capability of crossing D. sechellia with D. simulans and producing fertile hybrid females has led to several quantitative trait loci (QTLs) studies aiming at mapping the genes underlying these differences,11-15 and Drosophila genetics tool elegantly illustrated the function of some genes controlling their phenotypic differences.16-18 However, the old divergence between the 2 species8 and the low level of genetic variation within D. sechellia19 make difficult to finely map these differences or infer the order by which they arose and potentially affected reproductive isolation.
In a recent study, my colleagues and I published the discovery of a parallel case of insular specialization on toxic noni that was also accompanied by partial reproductive isolation.20 It concerns a population of the generalist species, Drosophila yakuba, whose last common ancestor with D. sechellia, D. simulans and D. melanogaster, lived more than 10 million years ago.21 This population lives on Mayotte, an island of the Comoros archipelago, which is also in the Western Indian Ocean like the Seychelles but closer to the African mainland. However, unlike D. sechellia, we inferred the colonization of Mayotte to be relatively recent, ∼29,000 y ago. On Mayotte, the flies were exclusively bred from fallen noni fruits, an unusual habitat for D. yakuba flies that are usually easily collected by fermenting banana traps on the mainland. We confirmed this difference in the laboratory showing that Mayotte flies have higher preference for noni and also higher tolerance to its toxins than mainland flies. Also, genome-wide scan identified high differentiation peaks between Mayotte and the mainland with many peaks falling within identified noni tolerance QTLs of D. sechellia, suggesting some common genetic pathways driving parallel ecological specialization. The differences between D. sechellia and Mayotte D. yakuba on the degrees of specialization and reproductive isolation therefore offer a rare opportunity where early and late steps in speciation may be distinguished. Unlike other non-model organisms for which ecological speciation continuums have also been demonstrated (e.g., sticklebacks, Heliconius butterflies, Rhagoletis flies, etc.),2 most of the advanced genetic and laboratory tools available for D. melanogaster could be applied to both D. sechellia and Mayotte D. yakuba.
Here, I mine the complete genome sequence of mainland and Mayotte D. yakuba and compare their mitochondria and morphology in search of the earliest fixed differences between them. Neither the morphology nor the mitochondrion provides diagnostic characters, whereas among 78,020,004 nuclear genomic sites, only 29 SNPs at 17 genes were specific to Mayotte D. yakuba. I discuss the relevance of these differences and use them, along with the geographical, physiological and behavioral characteristics of this population to define it as a subspecies that will likely become a promising model for ecological speciation genetics.
Materials and methods
Genomic analysis
For the nuclear genomes, I used the alignment to the D. yakuba reference genome r.1.3. (www.flybase.org)22 for a line from Côte d'Ivoire that we generated for D. yakuba from Mayotte (SRA project no. SRP063516),20 20 inbred lines from Cameroon and Kenya (SRP029453)23 and a line of D. santomea, the closest relative of D. yakuba (SRP052875).24 For the mitogenomes, I aligned reads from the pooled 22 lines of D. yakuba to the reference D. yakuba mitogenome (GenBank accession no. NC_001322)25 using the same alignment protocol.20 The consensus sequence was then aligned to complete mitogenomes for 29 and 18 lines of D. yakuba and its closest relative D. santomea, respectively, that were recently published26 using Muscle27 as implemented in MEGA6.28 The D. yakuba lines represent 7 populations from mainland Africa (Côte d'Ivoire, Cameroon, Gabon, Kenya, Tanzania and Zimbabwe) and the Atlantic island of Sao Tomé. To observe polymorphism in the pooled sample of Mayotte, Popoolation229 was used to generate a synchronized mpileup file, to which a column describing sequence variation in the 29 African lines was added using a customized Perl script.
MEGA was used to generate a neighbor-joining (NJ) phylogenetic tree for the mitogenomes under the Kimura 2-parameter model and after 1000 bootstrap iterations. For this analysis, the mitochondrion of D. erecta (GenBank accession number BK006335)22 was used as an outgroup. Character-based approach30 was applied for both mitochondrial and nuclear genome analyses using a customized Perl script which parses the synchronized mpileup files to search sites with nucleotides that are unique to the Mayotte population, i.e., fixed in this population and totally absent in other African genomes including the reference genome. Only sites with at least 44 sequencing depth were evaluated. These sites were then used as molecular diagnostics for Mayotte D. yakuba.
Morphological analysis
Males were obtained from mass cultures of 3 populations of D. yakuba from Mayotte, Kunden (Cameroon) and Andasibe (Madagascar) as well as a culture of D. santomea from Sao Tomé. For each population, the genitalia of 10 males were dissected, mounted on microscopic slides in DMHF mounting medium (Entomopraxis A9001) and observed under a Zeiss light microscope as in Yassin and Orgogozo.31 Male genital structures were also compared to descriptions of D. yakuba from Côte d'Ivoire,32 Kenya33 and Liberia34 covering the whole range of the species.
Results
Nuclear genome
Among 78,020,004 sites, only 36 SNPs belonging to 20 genes contained nucleotides that are fixed in Mayotte D. yakuba and absent from Kenya, Cameroon and the reference genome from Côte d'Ivoire (Table 1). Of these, 21 SNPs (10 genes) were on chromosome X (14,652,942 sites) indicating enrichment for this chromosome at early stages of speciation (Fisher exact test P < 3.1 × 10−7). With the exception of a single SNP, all X-linked fixed SNPs fell within a window of only 0.6 Mb, which constituted the largest peak of differentiation based on our previous FST analysis1 and overlapped with a major QTL conferring larval tolerance to noni toxin in D. sechellia.15
Table 1.
Fixed nucleotidic differences between Mayotte D. yakuba (D. y. mayottensis) and 3 populations of mainland D. yakuba (D. y. yakuba) with their ancestral states from D. santomea genome shown. The three populations of D. y. yakuba represent 21 lines from Côte d'Ivoire, Cameroon and Kenya. Gene names are given according to D. melanogaster ortholog. Sites shared between D. y. mayottensis and D. santomea are underlined.
| Site | Gene | D. y. yakuba | D. y. mayottensis | D. santomea |
|---|---|---|---|---|
| 2L:2216873 | CG4267 (intron) | A | G | A |
| 2L:2217239 | CG4267 (exon) | C | T | C |
| 2L:2220466 | CG4267 (downstream) | A | G | A |
| 2L:2236992 | CG17243 (downstream) | C | A | C |
| 2L:2237074 | CG17243 (downstream) | G | A | G |
| 2L:2237075 | CG17243 (downstream) | G | A | G |
| 2R:11743017 | Egfr (intron) | T | A/C | T |
| 2R:13045177 | CG8908 (exon) | C | G | G |
| 2R:15111406 | CG33958 (intron) | G | A | A |
| 2R:15776372 | Patronin (intron) | T | G | G |
| 2R:19611509 | egl (intron) | C | A | G |
| 2R:19612352 | egl (exon) | A | G | A |
| 2R:19612407 | egl (exon) | T | C | C |
| 2R:19612428 | egl (exon) | A | G | G |
| 2R:19615823 | CG34105/CG12491 (intergenic) | T | C | T |
| X:8980250 | CG9518 (upstream) | G | A | G |
| X:8989321 | CG9514 (downstream) | A | C | A |
| X:8997990 | CG9514 (intron) | C | T | C |
| X:9003564 | CG9514 (upstream) | A | T | T |
| X:9004874 | CG9514/CG9512(intergenic) | A | T | A |
| X:9019266 | CG12398 (exon) | G | A | G |
| X:9019413 | CG12398 (upstream) | C | A | C |
| X:9023586 | CG9503 (exon) | T | A | T |
| X:9024569 | CG9503 (exon) | C | A | C |
| X:9034448 | Flo2 (upstream) | T | A | T |
| X:9040907 | pdgy (intron) | C | G | C |
| X:9060210 | eag (intron) | A | C | A |
| X:9061187 | eag (intron) | C | T | C |
| X:9068430 | eag (intron) | A | G | A |
| X:9069386 | eag (intron) | G | A | G |
| X:9383509 | CG5877 (upstream) | T | C | T |
| X:9387538 | CG5877 (upstream) | T | C | C |
| X:9398249 | gce (intron) | C | G | C |
| X:9407136 | gce (intron) | A | T | A |
| X:9410040 | gce (exon) | A | G | A |
| X:21717401 | stnB (intron) | A | C | A |
For the autosomes, no fixed SNPs distinguishing the island population was found on chromosome 3. The six SNPs on chromosome arm 2L fell within a 20 kb region constituting another peak of differentiation. The SNPs belonged to 2 enzymatic genes, orthologous to the lipase CG4267 and the serine protease CG17243 in D. melanogaster. On chromosome arm 2R, 4 fixed SNPs fell within a 1 kb region within the egalitarian (egl) gene. Although all SNPs consisted of fixed differences between Mayotte and the mainland, one trinucleotide SNP at the Egfr gene consisted of an A/C polymorphism that was specific to Mayotte with both nucleotides absent from the mainland.
Interestingly, 29 out of the 36 sites (17 genes) were also unique to the Mayotte population if compared to D. santomea, suggesting these sites to be likely derived in the Mayotte lineage (Table 1).
Mitochondrial genome
Cytoplasmic introgression is common between D. yakuba and its 2 sibling species, D. santomea that is endemic to the Atlantic island of Sao Tomé and D. teissieri which is confined to the humid rainforests on the African mainland.26 Consequently, mitogenomes should have little taxonomic utility in this species complex. Consistently, the Mayotte D. yakuba mitochondrion branched within the cluster of D. yakuba/D. Santomea mitochondria showing no distinct feature (Fig. 1). Interestingly, the closest mitochondrion to Mayotte D. yakuba belonged to an East African population (Zimbabwe) with a bootstrap value of 84.6%. There is no nuclear genome available at the moment for Zimbabwe D. yakuba. Unfortunately, no mitogenome is available neither from Mozambique, Comoros or Madagascar islands, which may be given their geographical proximity closer to Mayotte D. yakuba and more informative to infer its history.
Figure 1.

Neighbor-joining tree of 29 and 18 mitogenomes of Drosophila yakuba yakuba and D. santomea, respectively, from Llopart et al.26 and the consensus mitogenome of D. y. mayottensis. Bootstrap values are shown above nodes. The tree is rooted using D. erecta mitogenome but the branch leading to the root is not shown because of its long length.
Morphology
No distinct morphological character can distinguish Mayotte D. yakuba from their conspecifics. Considering body pigmentation, which represents the most differentiating trait between D. yakuba and its closest relative D. santomea,35 Mayotte males have the last 2 abdominal tergites darkly pigmented like all D. yakuba flies. For male genitalia, D. yakuba differs from D. santomea in the shape of a 2 accessory articulating structures known as posterior parameres35 (also known as inner paraphyses)36 and the presence of 2 basal protrusions called phallic spines,33 phallic spurs31 or ventral branches.34 Both structures are of the D. yakuba type in the Mayotte population indicating that morphology is not informative at this stage of divergence (Fig. 2).
Figure 2.
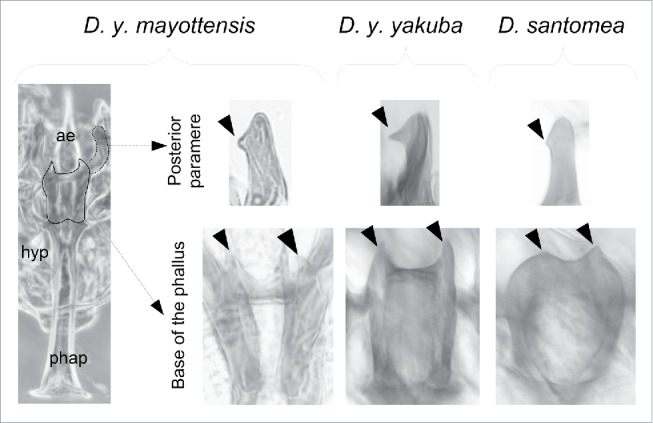
Male phallic apparatus in Drosophila yakuba mayottensis showing the placement of the 2 structures distinguishing D. y. yakuba and D. santomea: posterior parameres (dotted outline, above) and phallic spurs (dashed outline, below). Arrowheads indicate the distinguishing protrusions being longer in both subspecies of D. yakuba. ae = aedeagus; hyp = hypandrium; phap = phallic apodeme.
Taxonomy
In addition to the quantitative physiological differences between Mayotte and mainland D. yakuba regarding the higher preference for and tolerance to noni of the former population that we have previously described,20 I report here some qualitative fixed differences at the nuclear genome level but not at the mitochondrial or morphological levels. Together with the particular geographical locality of Mayotte D. yakuba, I use these differences to split D. yakuba into 2 subspecies, as follows.
Drosophila (Sophophora) yakuba yakuba Burla, 1954:161
Type locality – Côte d'Ivoire.
Diagnosis – Nucleotide differences shown at Table 1.
Description – As in Burla32 and Bock and Wheeler.37
Distribution – Africa: Côte d'Ivoire, Cameroon, Kenya and Sao Tomé, and presumably Benin, Congo, Gabon, Ghana, Madagascar, Malawi, Mozambique, Niger, Nigeria, Sierra Leone, South Africa, Tanzania, Uganda and Zimbabwe.
Drosophila (Sophophora) yakuba mayottensis David, Debat & Yassin, new subspecies
Type locality – Mayotte island (Comoros archipelago). A male holotype and 5 male and 5 female paratypes all drawn from F1 laboratory flies of isofemale lines collected by Jean R. David, Vincent Debat and Nelly Gidazweski in the Bay of Soulou (Grande Terre island, Mayotte) in January 2013 are deposited in the Muséum National d'Histoire Naturelle in Paris.
Diagnosis – Similar to D. y. yakuba, with male and female last abdominal tergites dark, but differs from it with the genomic differences shown in Table 1, its geographical location and its higher ecological specialization on noni fruits reported in Yassin et al.20
Description – (male holotype) Similar to D. y. yakuba. Arista with 5 branches above and 2 below in addition to terminal fork. Pedicel yellow, flagellomere I grayish. Frons narrow and yellow, ocellar triangle brownish and shiny. Face yellow, first and second oral bristles nearly of equal lengths, carina narrow. Cheeks yellow and very narrow. Eyes bright red.
Mesonotum and scutellum yellow. Pleura and legs pale yellow. Eight rows of acrostichal hairs. Anterior scutellars convergent. Anterior to posterior katepisternal bristles (sterno index) about 0.6, with a third, middle bristle, subequal to the anterior one. Sex combs with 7 black, curved bristles on the first tarsomere of the front legs. Wing hyaline to light brown, wing veins light brown.
Abdomen yellow, with dark stripes on the posterior margin of the second to fourth tergite. Fifth and sixth tergite shiny, black.
Epandrium black, with 5 bristles above and a tuff of long bristles on the ventral epandrial lobe, and a vestigial, triangular posterior lobe. Surstylus with 13 stout spines and one long bristle. Cercus boomerang, constricted on the middle, hirsute on its entire length with the bristles on the dorsal portion being shorter and stouter. Hypandrium yellow, rounded, with fine hairs and 2 long bristles on the gonopod, notched on the middle. Outer paraphyses (anterior parameres) with 3 short bristles on the tip. Inner paraphyses (posterior parameres) long, swan-shaped at the tip. Aedeagus membranous, pointed at tip and papillate on its entire length, with 2 sharp protrusions at its base (ventral branches or phallic spurs) (Fig. 2).
Distribution – Africa: Mayotte island (Comoros archipelago).
Discussion
In the genus Drosophila, where the genetics of speciation has been intensively studied in multiple pairs of closely-related species, few taxa were identified as subspecies. This was attributed to the low level of morphological distinction between allopatric populations within Drosophila species compared to other subspecies-rich holometabolous insects such as lepidopterans or coleopterans.38 Of the nearly 1,000 Drosophila species, only 24 have been subdivided into allopatric subspecies showing a relative degree of reproductive isolation (TAXODROS as of 19 November 2015). These subspecies were defined on the basis of morphological differences such as male pigmentation39 or genitalia40, chromosomal41-43 or allozyme traits44,45, since reproductive isolation alone is not enough to designate a subspecies. However, unlike morphological and molecular data, behavioral, karyological and enzymological variation cannot be traced on preserved name-holding type specimens making them of lesser identification utility.46
Drosophila yakuba mayottensis is the first subspecies to be described in the melanogaster subgroup, which consists of 9 African or of African origin species closely related to D. melanogaster.7 This designation is justified in terms of geographical locality, ecological specialization, partial reproductive isolation and the molecular diagnostics given in Table 1. Indeed, the genotypic species concept defines a species by ‘a stable multilocus genetic differentiation in the face of potential gene flow’.47 However, subspecies are reversible since an increase of gene flow may lead to their disappearance and the abortion of the speciation process that has been initiated in their lineage.48,49 Two major endeavors therefore should be undergone in the future. First, we should sequence more nuclear genomes from the African mainland in order to test for the absence of D. y. mayottensis diagnostic nucleotides there. And second, we should functionally test the relevance of these nucleotides to noni adaptation and/or premating reproductive isolation, therefore ecological speciation.
Ecological speciation should not be confounded with sympatric speciation, and indeed natural selection can lead to the rapid divergence of allopatric populations, such as between islands and mainland, due to their different habitats at the early stages of speciation. The fundamental question then becomes, when the 2 populations come in secondary contact, would they continue to be ecologically and reproductively isolated? Our recent population genomics analysis suggested a demographic scenario in which the ancestor of D. y. mayottensis colonized Mayotte long before human introduction of other fruit resources.20 At this time, noni might have been the only predictable source for the flies and natural selection drove the specialization. Further studies on the demographic history of noni on Mayotte and neighboring regions should be conducted to test this hypothesis, i.e. when did noni arrive on the island? Moreover, human agriculture and forest exploitation have drastically changed the flora of Mayotte that only 5% of the original vegetation presently exists.50 Consequently, the drosophilid fauna of Mayotte and neighboring islands is currently dominated by human-commensal species.51,52 This would increase the chances of the introduction of mainland D. y. yakuba to the island, which may outcompete or abort the speciation process. On the other hand, noni itself is currently cultivated and exploited for its pharmaceutical benefits on the African mainland,53 which may ultimately lead to other cases of specialization reminiscent to the ‘parallel speciation’ cases of sticklebacks54 or Timema stick insects.55 Further ecological studies on noni and D. yakuba on Mayotte, neighboring islands and the mainland are thus strongly needed.
The ecological parallelism between D. sechellia and D. yakuba mayottensis is a unique opportunity to distinguish species differences due to natural selection for a similar host from other differences arising during the speciation process. A great endeavor would be to determine the link between noni preference or toxin tolerance loci and the loci underlying mate choice in D. y. mayottensis females. This link should indicate how natural selection could affect reproductive isolation via pleiotropy or linkage, and also inform whether pre-copulatory isolation shares common basis between the 2 cases. Our analysis identified several common noni toxin tolerance genes or genetic pathways between D. sechellia and D. y. mayottensis, with some being implied in specialization on other host plants in some other drosophilids (such as leaf-mining Scaptomyza)56,57 but also in several herbivorous insects, which constitute a majority of animal diversity. This makes this new subspecies a vey promising model to understand the genetics and evolution of insect-plant interactions in Drosophila and beyond.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Coyne JA, Orr HA. Speciation. Sinauer Associates, Incorporated Publishers; 2004 [Google Scholar]
- [2].Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA, Peichel CL, Saetre G-P, Bank C, Brännström Å, et al.. Genomics and the origin of species. Nat Rev Genet 2014; 15:176-92; PMID:24535286; http://dx.doi.org/ 10.1038/nrg3644 [DOI] [PubMed] [Google Scholar]
- [3].Nosil P, Schluter D. The genes underlying the process of speciation. Trends Ecol Evol 2011; 26:160-7; PMID:21310503; http://dx.doi.org/ 10.1016/j.tree.2011.01.001 [DOI] [PubMed] [Google Scholar]
- [4].Mallet J. What does Drosophila genetics tell us about speciation? Trends Ecol Evol 2006; 21:386-93; PMID:16765478; http://dx.doi.org/ 10.1016/j.tree.2006.05.004 [DOI] [PubMed] [Google Scholar]
- [5].Phadnis N, Baker EP, Cooper JC, Frizzell KA, Hsieh E, Cruz AFA, de la Shendure J, Kitzman JO, Malik HS. An essential cell cycle regulation gene causes hybrid inviability in Drosophila. Science 2015; 350:1552-5; PMID:26680200; http://dx.doi.org/ 10.1126/science.aac7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Markow TA, O'Grady PM. Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annu Rev Genet 2005; 39:263-91; PMID:16285861; http://dx.doi.org/ 10.1146/annurev.genet.39.073003.112454 [DOI] [PubMed] [Google Scholar]
- [7].David JR, Lemeunier F, Tsacas L, Yassin A. The historical discovery of the nine species in the Drosophila melanogaster species subgroup. Genetics 2007; 177:1969-73; PMID:18073416; http://dx.doi.org/ 10.1534/genetics.104.84756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, Presgraves DC. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res 2012; 22:1499-511; PMID:22534282; http://dx.doi.org/ 10.1101/gr.130922.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Matute DR, Ayroles JF. Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J Evol Biol 2014; 27:1057-68; PMID:24773151; http://dx.doi.org/ 10.1111/jeb.12391 [DOI] [PubMed] [Google Scholar]
- [10].Orgogozo V, Stern DL. How different are recently diverged species? More than 150 phenotypic differences have been reported for the D. melanogaster species subgroup. Fly (Austin) 2009; 3:117-117; PMID:19440042; http://dx.doi.org/ 10.4161/fly.8836 [DOI] [PubMed] [Google Scholar]
- [11].Orgogozo V, Broman KW, Stern DL. High-resolution quantitative trait locus mapping reveals sign epistasis controlling ovariole number between two Drosophila species. Genetics 2006; 173:197-205; PMID:16489225; http://dx.doi.org/ 10.1534/genetics.105.054098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gleason JM, James RA, Wicker-Thomas C, Ritchie MG. Identification of quantitative trait loci function through analysis of multiple cuticular hydrocarbons differing between Drosophila simulans and Drosophila sechellia females. Heredity 2009; 103:416-24; PMID:19654611; http://dx.doi.org/ 10.1038/hdy.2009.79 [DOI] [PubMed] [Google Scholar]
- [13].Earley EJ, Jones CD. Next-generation mapping of complex traits with phenotype-based selection and introgression. Genetics 2011; 189:1203-1209; PMID:21940681; http://dx.doi.org/ 10.1534/genetics.111.129445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hungate EA, Earley EJ, Boussy IA, Turissini DA, Ting C-T, Moran JR, Wu M-L, Wu C-I, Jones CD. A locus in Drosophila sechellia affecting tolerance of a host plant toxin. Genetics 2013; 195:1063-1075; PMID:24037270; http://dx.doi.org/ 10.1534/genetics.113.154773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Huang Y, Erezyilmaz D. The genetics of resistance to Morinda fruit toxin during the postembryonic stages in Drosophila sechellia. G3 GenesGenomesGenetics 2015; 5:1973-1981; PMID:26224784; http://dx.doi.org/ 10.1534/g3.114.015073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol 2007; 5:e118; PMID:17456006; http://dx.doi.org/ 10.1371/journal.pbio.0050118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 2011; 474:598-603; PMID:21720363; http://dx.doi.org/ 10.1038/nature10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lavista-Llanos S, Svatoš A, Kai M, Riemensperger T, Birman S, Stensmyr MC, Hansson BS. Dopamine drives Drosophila sechellia adaptation to its toxic host. Elife 2014; 3:e03785; PMID:25487989; http://dx.doi.org/ 10.7554/eLife.03785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Legrand D, Tenaillon MI, Matyot P, Gerlach J, Lachaise D, Cariou M-L. Species-wide genetic variation and demographic history of Drosophila sechellia, a species lacking population structure. Genetics 2009; 182:1197-206; PMID:19506309; http://dx.doi.org/ 10.1534/genetics.108.092080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yassin A, Debat V, Bastide H, Gidaszewski N, David JR, Pool JE. Recurrent specialization on a toxic fruit in an island Drosophila population. Proc Natl Acad Sci 2016; 113:4771-6; PMID:27044093; http://dx.doi.org/ 10.1073/pnas.1522559113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Obbard DJ, Maclennan J, Kim K-W, Rambaut A, O'Grady PM, Jiggins FM. Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol Biol Evol 2012; 29:3459-73; PMID:22683811; http://dx.doi.org/ 10.1093/molbev/mss150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al.. Evolution of genes and genomes on the Drosophila phylogeny. Nature 2007; 450:203-18; PMID:17994087; http://dx.doi.org/ 10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- [23].Rogers RL, Cridland JM, Shao L, Hu TT, Andolfatto P, Thornton KR. Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol Biol Evol 2014; 31:1750-66; PMID:24710518; http://dx.doi.org/ 10.1093/molbev/msu124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Turissini DA, Liu G, David JR, Matute DR. The evolution of reproductive isolation in the Drosophila yakuba complex of species. J Evol Biol 2015; 28:557-75; PMID:25611516; http://dx.doi.org/ 10.1111/jeb.12588 [DOI] [PubMed] [Google Scholar]
- [25].Clary DO, Wolstenholme DR. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol 1985; 22:252-71; PMID:3001325; http://dx.doi.org/ 10.1007/BF02099755 [DOI] [PubMed] [Google Scholar]
- [26].Llopart A, Herrig D, Brud E, Stecklein Z. Sequential adaptive introgression of the mitochondrial genome in Drosophila yakuba and Drosophila santomea. Mol Ecol 2014; 23:1124-36; PMID:24460929; http://dx.doi.org/ 10.1111/mec.12678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792-7; PMID:15034147; http://dx.doi.org/ 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/ 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kofler R, Pandey RV, Schlötterer C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 2011; 27:3435-6; PMID:22025480; http://dx.doi.org/ 10.1093/bioinformatics/btr589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yassin A, Markow TA, Narechania A, O'Grady PM, DeSalle R. The genus Drosophila as a model for testing tree- and character-based methods of species identification using DNA barcoding. Mol Phylogenet Evol 2010; 57:509-17; PMID:20800099; http://dx.doi.org/ 10.1016/j.ympev.2010.08.020 [DOI] [PubMed] [Google Scholar]
- [31].Yassin A, Orgogozo V. Coevolution between male and female genitalia in the Drosophila melanogaster species subgroup. PLoS ONE 2013; 8:e57158; PMID:23451172; http://dx.doi.org/ 10.1371/journal.pone.0057158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Burla H. Zur Kenntnis der Drosophiliden der Elfenbeinküste Französisch West-Afrika. Revue Suisse Zool 1954; 61:1-218; http://dx.doi.org/ 10.5962/bhl.part.75413 [DOI] [Google Scholar]
- [33].Kamimura Y, Mitsumoto H. Lock-and-key structural isolation between sibling Drosophila species. Entomol Sci 2012; 15:197-201; http://dx.doi.org/ 10.1111/j.1479-8298.2011.00490.x [DOI] [Google Scholar]
- [34].Peluffo AE, Nuez I, Debat V, Savisaar R, Stern DL, Orgogozo V. A major locus controls a genital shape difference involved in reproductive isolation between Drosophila yakuba and Drosophila santomea. G3 GenesGenomesGenetics 2015; 5:2893-901; PMID:26511499; http://dx.doi.org/ 10.1534/g3.115.023481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lachaise D, Harry M, Solignac M, Lemeunier F, Bénassi V, Cariou ML. Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from São Tomé. Proc Biol Sci 2000; 267:1487-95; PMID:11007323; http://dx.doi.org/ 10.1098/rspb.2000.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yassin A. Phylogenetic classification of the Drosophilidae Rondani (Diptera): the role of morphology in the postgenomic era. Syst Entomol 2013; 38:349-64; http://dx.doi.org/ 10.1111/j.1365-3113.2012.00665.x [DOI] [Google Scholar]
- [37].Bock IR, Wheeler MR. The Drosophila melanogaster species group. The University of Texas Publication 1972; 7213:1-102 [Google Scholar]
- [38].Tsacas L, Bocquet C. L'espèce chez les Drosophilidae. Mém Société Zool Fr 1976; 38:203-47 [Google Scholar]
- [39].Bock IR. Taxonomy of the Drosophila bipectinata species complex. The University of Texas Publication 1971; 7103:273-280 [Google Scholar]
- [40].Pfeiler E, Castrezana S, Reed LK, Markow TA. Genetic, ecological and morphological differences among populations of the cactophilic Drosophila mojavensis from southwestern USA and northwestern Mexico, with descriptions of two new subspecies. J Nat Hist 2009; 43:923-38; http://dx.doi.org/ 10.1080/00222930802610535 [DOI] [Google Scholar]
- [41].Heed WB. Genetic characteristics of island populations. Univ Tex Publ 1962; 6205:173-206 [Google Scholar]
- [42].Wilson FD, Wheeler MR, Harget M, Kambysellis M. Cytogenetic relations in the Drosophila nasuta subgroup of the immigrans group of species. Univ Tex Publ 1969; 6918:207-53 [Google Scholar]
- [43].Matsuda M, Tomimura Y, Tobari YN. Reproductive isolation among geographical populations of Drosophila bipectinata Duda (Diptera, Drosophilidae) with recognition of three subspecies. Genetica 2005; 125:69-78; PMID:16175456; http://dx.doi.org/ 10.1007/s10709-005-0393-z [DOI] [PubMed] [Google Scholar]
- [44].Ayala FJ. Two new subspecies of the Drosophila willistoni group (Diptera: Drosophilidae). Pan-Pac Entomol 1973; 49:273-9 [Google Scholar]
- [45].Ayala FJ, Dobzhansky T. A new subspecies of Drosophila pseudoobscura (Diptera: Drosophilidae). Pan-Pac Entomol 1975; 50:211-9 [Google Scholar]
- [46].DeSalle R, Egan MG, Siddall M. The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philos Trans R Soc B Biol Sci 2005; 360:1905-16; PMID:16214748; http://dx.doi.org/ 10.1098/rstb.2005.1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mallet J. A species definition for the modern synthesis. Trends Ecol Evol 1995; 10:294-9; PMID:21237047; http://dx.doi.org/ 10.1016/0169-5347(95)90031-4 [DOI] [PubMed] [Google Scholar]
- [48].Wilson EO, Brown WL. The subspecies concept and its taxonomic application. Syst Zool 1953; 2:97-111; http://dx.doi.org/ 10.2307/2411818 [DOI] [Google Scholar]
- [49].Mallet J. Subspecies, semispecies, superspecies. Encycl Biodivers 2001; 5:523-6; http://dx.doi.org/ 10.1006/rwbd.1999.0321 [DOI] [Google Scholar]
- [50].Barthelet F, Viscardi G. Flore menacée de l'île de Mayotte: importance patrimoniale et enjeux de conservation. Rev Ecol 2012; 67:15-27 [Google Scholar]
- [51].Yassin A, Gidaszewski N, Albert B, Hivert J, David JR, Orgogozo V, Debat V. The Drosophilidae (Diptera) of the Scattered Islands, with the description of a novel association with Leptadenia madagascariensis Decne. (Apocynaceae). Fly (Austin) 2012; 6:298-302; PMID:23222006; http://dx.doi.org/ 10.4161/fly.21583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].David JR, Yassin A, Gidaszewski N, Debat V. Drosophilids (Diptera) from Mayotte island: an annotated list of species collected in 2013 and comments on the colonisation of Indian Ocean Islands. Ann Soc Entomol Fr 2014; 50:336-342 [Google Scholar]
- [53].Chan-Blanco Y, Vaillant F, Mercedes Perez A, Reynes M, Brillouet J-M, Brat P. The noni fruit (Morinda citrifolia L.): A review of agricultural research, nutritional and therapeutic properties. J Food Compos Anal 2006; 19:645-54; http://dx.doi.org/ 10.1016/j.jfca.2005.10.001 [DOI] [Google Scholar]
- [54].Rundle HD, Nagel L, Boughman JW, Schluter D. Natural selection and parallel speciation in sympatric sticklebacks. Science 2000; 287:306-8; PMID:10634785; http://dx.doi.org/ 10.1126/science.287.5451.306 [DOI] [PubMed] [Google Scholar]
- [55].Soria-Carrasco V, Gompert Z, Comeault AA, Farkas TE, Parchman TL, Johnston JS, Buerkle CA, Feder JL, Bast J, Schwander T, et al.. Stick insect genomes reveal natural selection's role in parallel speciation. Science 2014; 344:738-42; PMID:24833390; http://dx.doi.org/ 10.1126/science.1252136 [DOI] [PubMed] [Google Scholar]
- [56].Whiteman NK, Gloss AD, Sackton TB, Groen SC, Humphrey PT, Lapoint RT, Sønderby IE, Halkier BA, Kocks C, Ausubel FM, et al.. Genes involved in the evolution of herbivory by a leaf-mining, drosophilid fly. Genome Biol Evol 2012; 4:900-16; PMID:22813779; http://dx.doi.org/ 10.1093/gbe/evs063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gloss AD, Vassão DG, Hailey AL, Dittrich ACN, Schramm K, Reichelt M, Rast TJ, Weichsel A, Cravens MG, Gershenzon J, et al.. Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the Drosophilidae. Mol Biol Evol 2014; 31:2441-56; PMID:24974374; http://dx.doi.org/ 10.1093/molbev/msu201 [DOI] [PMC free article] [PubMed] [Google Scholar]


