Abstract
Background
Early accurate detection of all skin cancer types is essential to guide appropriate management and to improve morbidity and survival. Melanoma and cutaneous squamous cell carcinoma (cSCC) are high‐risk skin cancers which have the potential to metastasise and ultimately lead to death, whereas basal cell carcinoma (BCC) is usually localised with potential to infiltrate and damage surrounding tissue. Anxiety around missing early curable cases needs to be balanced against inappropriate referral and unnecessary excision of benign lesions. Computer‐assisted diagnosis (CAD) systems use artificial intelligence to analyse lesion data and arrive at a diagnosis of skin cancer. When used in unreferred settings ('primary care'), CAD may assist general practitioners (GPs) or other clinicians to more appropriately triage high‐risk lesions to secondary care. Used alongside clinical and dermoscopic suspicion of malignancy, CAD may reduce unnecessary excisions without missing melanoma cases.
Objectives
To determine the accuracy of CAD systems for diagnosing cutaneous invasive melanoma and atypical intraepidermal melanocytic variants, BCC or cSCC in adults, and to compare its accuracy with that of dermoscopy.
Search methods
We undertook a comprehensive search of the following databases from inception up to August 2016: Cochrane Central Register of Controlled Trials (CENTRAL); MEDLINE; Embase; CINAHL; CPCI; Zetoc; Science Citation Index; US National Institutes of Health Ongoing Trials Register; NIHR Clinical Research Network Portfolio Database; and the World Health Organization International Clinical Trials Registry Platform. We studied reference lists and published systematic review articles.
Selection criteria
Studies of any design that evaluated CAD alone, or in comparison with dermoscopy, in adults with lesions suspicious for melanoma or BCC or cSCC, and compared with a reference standard of either histological confirmation or clinical follow‐up.
Data collection and analysis
Two review authors independently extracted all data using a standardised data extraction and quality assessment form (based on QUADAS‐2). We contacted authors of included studies where information related to the target condition or diagnostic threshold were missing. We estimated summary sensitivities and specificities separately by type of CAD system, using the bivariate hierarchical model. We compared CAD with dermoscopy using (a) all available CAD data (indirect comparisons), and (b) studies providing paired data for both tests (direct comparisons). We tested the contribution of human decision‐making to the accuracy of CAD diagnoses in a sensitivity analysis by removing studies that gave CAD results to clinicians to guide diagnostic decision‐making.
Main results
We included 42 studies, 24 evaluating digital dermoscopy‐based CAD systems (Derm–CAD) in 23 study cohorts with 9602 lesions (1220 melanomas, at least 83 BCCs, 9 cSCCs), providing 32 datasets for Derm–CAD and seven for dermoscopy. Eighteen studies evaluated spectroscopy‐based CAD (Spectro–CAD) in 16 study cohorts with 6336 lesions (934 melanomas, 163 BCC, 49 cSCCs), providing 32 datasets for Spectro–CAD and six for dermoscopy. These consisted of 15 studies using multispectral imaging (MSI), two studies using electrical impedance spectroscopy (EIS) and one study using diffuse‐reflectance spectroscopy. Studies were incompletely reported and at unclear to high risk of bias across all domains. Included studies inadequately address the review question, due to an abundance of low‐quality studies, poor reporting, and recruitment of highly selected groups of participants.
Across all CAD systems, we found considerable variation in the hardware and software technologies used, the types of classification algorithm employed, methods used to train the algorithms, and which lesion morphological features were extracted and analysed across all CAD systems, and even between studies evaluating CAD systems. Meta–analysis found CAD systems had high sensitivity for correct identification of cutaneous invasive melanoma and atypical intraepidermal melanocytic variants in highly selected populations, but with low and very variable specificity, particularly for Spectro–CAD systems. Pooled data from 22 studies estimated the sensitivity of Derm–CAD for the detection of melanoma as 90.1% (95% confidence interval (CI) 84.0% to 94.0%) and specificity as 74.3% (95% CI 63.6% to 82.7%). Pooled data from eight studies estimated the sensitivity of multispectral imaging CAD (MSI–CAD) as 92.9% (95% CI 83.7% to 97.1%) and specificity as 43.6% (95% CI 24.8% to 64.5%). When applied to a hypothetical population of 1000 lesions at the mean observed melanoma prevalence of 20%, Derm–CAD would miss 20 melanomas and would lead to 206 false‐positive results for melanoma. MSI–CAD would miss 14 melanomas and would lead to 451 false diagnoses for melanoma. Preliminary findings suggest CAD systems are at least as sensitive as assessment of dermoscopic images for the diagnosis of invasive melanoma and atypical intraepidermal melanocytic variants. We are unable to make summary statements about the use of CAD in unreferred populations, or its accuracy in detecting keratinocyte cancers, or its use in any setting as a diagnostic aid, because of the paucity of studies.
Authors' conclusions
In highly selected patient populations all CAD types demonstrate high sensitivity, and could prove useful as a back‐up for specialist diagnosis to assist in minimising the risk of missing melanomas. However, the evidence base is currently too poor to understand whether CAD system outputs translate to different clinical decision–making in practice. Insufficient data are available on the use of CAD in community settings, or for the detection of keratinocyte cancers. The evidence base for individual systems is too limited to draw conclusions on which might be preferred for practice. Prospective comparative studies are required that evaluate the use of already evaluated CAD systems as diagnostic aids, by comparison to face–to–face dermoscopy, and in participant populations that are representative of those in which the test would be used in practice.
Keywords: Adult; Humans; Electric Impedance; Algorithms; Carcinoma, Basal Cell; Carcinoma, Basal Cell/diagnosis; Carcinoma, Basal Cell/diagnostic imaging; Carcinoma, Basal Cell/pathology; Carcinoma, Squamous Cell; Carcinoma, Squamous Cell/diagnosis; Carcinoma, Squamous Cell/diagnostic imaging; Carcinoma, Squamous Cell/pathology; Clinical Decision‐Making; Dermoscopy; Dermoscopy/methods; Dermoscopy/standards; Diagnosis, Computer‐Assisted; Diagnosis, Computer‐Assisted/methods; Diagnosis, Computer‐Assisted/standards; False Positive Reactions; Melanoma; Melanoma/diagnosis; Melanoma/diagnostic imaging; Melanoma/pathology; Sensitivity and Specificity; Skin Neoplasms; Skin Neoplasms/diagnosis; Skin Neoplasms/diagnostic imaging; Skin Neoplasms/pathology
Plain language summary
What is the diagnostic accuracy of computer‐assisted diagnosis techniques for the detection of skin cancer in adults?
Why is improving the diagnosis of skin cancer important?
There are a number of different types of skin cancer, including melanoma, squamous cell carcinoma (SCC) and basal cell carcinoma (BCC). Melanoma is one of the most dangerous forms. If it is not recognised early treatment can be delayed and this risks the melanoma spreading to other organs in the body and may eventually lead to death. Cutaneous squamous cell carcinoma (cSCC) and BCC are considered less dangerous, as they are localised (less likely to spread to other parts of the body compared to melanoma). However, cSCC can spread to other parts of the body and BCC can cause disfigurement if not recognised early. Diagnosing a skin cancer when it is not actually present (a false‐positive result) might result in unnecessary surgery and other investigations and can cause stress and anxiety to the patient. Missing a diagnosis of skin cancer may result in the wrong treatment being used or lead to a delay in effective treatment.
What is the aim of the review?
The aim of this Cochrane Review was to find out how accurate computer–assisted diagnosis (CAD) is for diagnosing melanoma, BCC or cSCC. The review also compared the accuracy of two different types of CAD, and compared the accuracy of CAD with diagnosis by a doctor using a handheld illuminated microscope (a dermatoscope or ‘dermoscopy’). We included 42 studies to answer these questions.
What was studied in the review?
A number of tools are available to skin cancer specialists which allow a more detailed examination of the skin compared to examination by the naked eye alone. Currently a dermatoscope which magnifies the skin lesion (a mole or area of skin with an unusual appearance in comparison with the surrounding skin) using a bright light source is used by most skin cancer specialists. CAD tests are computer systems that analyse information about skin lesions obtained from a dermatoscope or other techniques that use light to describe the features of a skin lesion (spectroscopy) to produce a result indicating whether skin cancer is likely to be present. We included CAD systems that get their information from dermoscopic images of lesions (Derm–CAD), or that use data from spectroscopy. Most of the spectroscopy studies used data from multispectral imaging (MSI–CAD) and are the main focus here. When a skin cancer specialist finds a lesion is suspicious using visual examination with or without additional dermoscopy, results from CAD systems can be used alone to make a diagnosis of skin cancer (CAD–based diagnosis), or can be used by doctors in addition to their visual inspection examination of a skin lesion to help them reach a diagnosis (CAD–aided diagnosis). Researchers examined how useful CAD systems are to help diagnose skin cancers in addition to visual inspection and dermoscopy.
What are the main results of the review?
The review included 42 studies looking at CAD systems for the diagnosis of melanoma. There was not enough evidence to determine the accuracy of CAD systems for the diagnosis of BCC (3 studies) or cSCC (1 study).
Derm‐CAD results for diagnosis of melanoma
The main results for Derm‐CAD are based on 22 studies including 8992 lesions.
Applied to a group of 1000 skin lesions, of which 200 (20%) are given a final diagnosis* of melanoma, the results suggest that:
‐ An estimated 386 people will have a Derm–CAD result suggesting that a melanoma is present, and of these 206 (53%) will not actually have a melanoma (false‐positive result)
‐ Of the 614 people with a Derm–CAD result indicating that no melanoma is present, 20 (3%) will in fact actually have a melanoma (false‐negative result)
There was no evidence to suggest that dermoscopy or Derm–CAD was different in its ability to detect or rule out melanoma.
MSI‐CAD results for diagnosis of melanoma
The main results for MSI–CAD are based on eight studies including 2401 lesions. In a group of 1000 people, of whom 200 (20%) actually do have melanoma*, then:
‐ An estimated 637 people will have an MSI–CAD result suggesting that a melanoma is present, and of these 451 (71%) will not actually have a melanoma (false‐positive result)
‐ Of the 363 people with an MSI–CAD result indicating that no melanoma is present, 14 (4%) will in fact have a melanoma (false‐negative result)
MSI–CAD detects more melanomas, but possibly produces more false‐positive results (an increase in unnecessary surgery).
How reliable are the results of the studies of this review?
Incomplete reporting of studies made it difficult for us to judge how reliable they were. Many studies had important limitations. Some studies only included particular types of skin lesions or excluded lesions that were considered difficult to diagnose. Importantly, most of the studies only included skin lesions with a biopsy result, which means that only a sample of lesions that would be seen by a doctor in practice were included. These characteristics may result in CAD systems appearing more or less accurate than they actually are.
Who do the results of this review apply to?
Studies were largely conducted in Europe (29, 69%) and North America (8, 19%). Mean age (reported in 6/42 studies) ranged from 32 to 49 years for melanoma. The percentage of people with a final diagnosis of melanoma ranged from 1% to 52%. It was not always possible to tell whether suspicion of skin cancer in study participants was based on clinical examination alone, or both clinical and dermoscopic examinations. Almost all studies were done in people with skin lesions who were seen at specialist clinics rather than by doctors in primary care.
What are the implications of this review?
CAD systems appear to be accurate for identification of melanomas in skin lesions that have already been selected for excision on the basis of clinical examination (visual inspection and dermoscopy). It is possible that some CAD systems identify more melanomas than doctors using dermoscopy images. However, CAD systems also produced far more false‐positive diagnoses than dermoscopy, and could lead to considerable increases in unnecessary surgery. The performance of CAD systems for detecting BCC and cSCC skin cancers is unclear. More studies are needed to evaluate the use of CAD by doctors for the diagnosis of skin cancer in comparison to face‐to‐face diagnosis using dermoscopy, both in primary care and in specialist skin cancer clinics.
How up‐to‐date is this review?
The review authors searched for and used studies published up to August 2016.
*In these studies, biopsy, clinical follow up, or specialist clinician diagnosis were the reference standards (means of establishing the final diagnosis).
Summary of findings
Summary of findings'. 'Summary of findings table.
| Question: | What is the diagnostic accuracy of computer‐assisted diagnosis for the detection of: i) cutaneous invasive melanoma and atypical intraepidermal melanocyticvariants, ii) BCC, or iii) cSCC in adults? | |||||
| Population: | Adults with lesions suspicious for skin cancer, including:
|
|||||
| Index test: | Computer‐assisted diagnosis (CAD) | |||||
| Comparator test: | Dermoscopy | |||||
| Target condition: | Cutaneous invasive melanoma and atypical intraepidermal melanocytic variants, or basal cell carcinoma (BCC), or cutaneous squamous cell carcinoma (cSCC) | |||||
| Reference standard: | Histology with or without long‐term follow‐up | |||||
| Action: | If accurate, positive results of CAD will identify skin cancers that could otherwise be missed, while negative results will stop patients having unnecessary excision of skin lesions | |||||
| Quantity of evidence | ||||||
| Number of studies | 42a | Total lesions with test results | 13,445 | Total with target condition | 2452 | |
| Limitations | ||||||
| Risk of bias: | Patient selection methods were poorly reported, with some concern (34/42) due to use of case‐control designs, exclusion of difficult‐to‐diagnose types of lesion, and inadequate reporting to assess risks of bias. CAD was generally evaluated in independent populations (35/42). Some concern as it was not clear that the reference standard was interpreted blind to the CAD results in 19/42 studies. Differential verification was used in 6/42 studies, participants were excluded in 10/42, primarily due to technical difficulties with CAD. Timing of tests was not mentioned in 28/42. | |||||
| Applicability of evidence to question: | High concern for poor clinical applicability of included studies. Almost all studies recruited narrowly‐defined populations (41/42) or multiple lesions per participant (14/42) or both, and may not be representative of populations eligible for CAD. Studies provided little information regarding the thresholds used for presence of malignancy (16/42) and often evaluated unestablished thresholds (23/42). Studies performing training of algorithms provided scarce information on the range of conditions included in training sets. Little information was given about the expertise of the histopathologist. | |||||
| FINDINGS: All analyses are undertaken on subgroups of the studies | ||||||
| All included studies considered the detection of melanoma, three of which also looked at the detection of BCC and one at the detection of cSCC. There are therefore insufficient data to make summary statements about the accuracy of CAD for the detection of BCC or cSCC. All results below consider the detection of the primary target condition: cutaneous invasive melanoma and atypical intraepidermal melanocytic variants. | ||||||
| Test: Digital dermoscopy‐based CAD (all systems) | ||||||
| Quantity of evidence | Number of studies | 22 | Total lesions with test results | 8992 | Total with melanoma | 1063 |
|
Sensitivity (95% CI): Specificity (95% CI): |
90.1% (84.0% to 94.0%) 74.3% (63.6% to 82.7%) |
Numbers observed in a cohort of 1000 people being testedb | ||||
| Consistency | Sensitivity estimates consistent. Some heterogeneity in specificity between studies | Consequences | Prevalence | |||
| 7% | 20% | 40% | ||||
| True positives | Receive necessary excision | 63 | 180 | 360 | ||
| False positives | Receive unnecessary excision | 239 | 206 | 154 | ||
| False negatives | Do not receive required excision | 7 | 20 | 40 | ||
| True negatives | Appropriately do not receive excision | 691 | 594 | 446 | ||
| PPV | 21% | 46% | 70% | |||
| NPV | 99% | 97% | 92% | |||
| Test: Multispectral imaging‐based CAD (all systems) | ||||||
| Quantity of evidence | Number of studies | 8 | Total lesions with test results | 2401 | Total with melanoma | 286 |
|
Sensitivity (95% CI): Specificity (95% CI): |
92.9% (83.7% to 97.1%) 43.6% (24.8% to 64.5%) |
Numbers observed in a cohort of 1000 people being tested | ||||
| Consistency | Sensitivity estimates consistent. High heterogeneity in specificity between studies | Consequences | Prevalence | |||
| 7% | 20% | 40% | ||||
| True positives | Receive necessary excision | 65 | 186 | 372 | ||
| False positives | Receive unnecessary excision | 525 | 451 | 338 | ||
| False negatives | Do not receive required excision | 5 | 14 | 28 | ||
| True negatives | Appropriately do not receive excision | 405 | 349 | 262 | ||
| PPV | 12% | 29% | 52% | |||
| NPV | 99% | 96% | 90% | |||
aSix studies with overlapping lesions (Seidenari 1998 and Seidenari 1999; Tomatis 2003 and Bono 2002; Monheit 2011 and Hauschild 2014). bNumbers estimated at 25th, 50th (median) and 75% percentiles of invasive melanoma and atypical intraepidermal melanocytic variants prevalence, observed across 42 studies reporting evaluations of CAD. BCC ‐ Basal cell carcinoma; CAD ‐ computer‐assisted diagnosis; CI ‐ confidence interval; cSCC ‐ cutaneous squamous cell carcinoma; NPV ‐ negative predictive value; PPV ‐ positive predictive value
Background
This review is one of a series of Cochrane Diagnostic Test Accuracy (DTA) Reviews on the diagnosis and staging of melanoma and keratinocyte skin cancers conducted for the National Institute for Health Research (NIHR) Cochrane Systematic Reviews Programme. Appendix 1 shows the content and structure of the programme. Table 2 provides a glossary of terms used, and a table of acronyms used is provided in Appendix 2.
1. Glossary of terms.
| Term | Definition |
| Artificial intelligence | Computer systems undertaking tasks that normally require human intelligence, such as decision–making or visual perception |
| Atypical intraepidermal melanocytic variant | Unusual area of darker pigmentation contained within the epidermis that may progress to an invasive melanoma; includes melanoma in situ and lentigo maligna |
| Atypical naevi | Unusual looking but noncancerous mole or area of darker pigmentation of the skin |
| Basaloid cells | Cells in the skin that look like those in epidermal basal layer |
| BRAF V600 mutation | BRAF is a human gene that makes a protein called B‐Raf which is involved in the control of cell growth. BRAF mutations (damaged DNA) occur in around 40% of melanomas, which can then be treated with particular drugs. |
| BRAF inhibitors | Therapeutic agents which inhibit the serine‐threonine protein kinase BRAF mutated metastatic melanoma. |
| Breslow thickness | A scale for measuring the thickness of melanomas by the pathologist using a microscope, measured in mm from the top layer of skin to the bottom of the tumour. |
| Congenital naevi | A type of mole found on infants at birth |
| Dermoscopy | Whereby a handheld microscope is used to allow more detailed, magnified, examination of the skin compared to examination by the naked eye alone |
| Dermo‐epidermal junction | The area where the lower part of the epidermis and top layer of the dermis meet |
| Dermis | Layer of skin below the epidermis, composed of living tissue and containing blood capillaries, nerve endings, sweat glands, hair follicles and other structures |
| Desmoplastic subtypes of SCC | An aggressive squamous cell carcinoma variant characterised by a proliferation of fibroblasts and formation of fibrous connective tissue |
| Electrodesiccation | The use of high‐frequency electric currents to cut, destroy or cauterise tissue. It is performed with the use of a fine needle‐shaped instrument |
| Electrical impedance spectroscopy | The measurement of electrical current properties as they pass through skin tissues, to retrieve information on cellular structures |
| Epidermis | Outer layer of the skin |
| False negative | An individual who is truly positive for a disease, but whom a diagnostic test classifies them as disease‐free. |
| False positive | An individual who is truly disease‐free, but whom a diagnostic test classifies them as having the disease. |
| Histopathology/Histology | The study of tissue, usually obtained by biopsy or excision, for example under a microscope. |
| Incidence | The number of new cases of a disease in a given time period. |
| Index test | A diagnostic test under evaluation in a primary study |
| Interferometry | The measurement of waves of light or sound after interference in order to extract information |
| Lentigo maligna | Unusual area of darker pigmentation contained within the epidermis which includes malignant cells but with no invasive growth. May progress to an invasive melanoma |
| Lymph node | Lymph nodes filter the lymphatic fluid (clear fluid containing white blood cells) that travels around the body to help fight disease; they are located throughout the body often in clusters (nodal basins). |
| Melanocytic naevus | An area of skin with darker pigmentation (or melanocytes) also referred to as ‘moles’ |
| Meta‐analysis | A form of statistical analysis used to synthesise results from a collection of individual studies. |
| Metastases/metastatic disease | Spread of cancer away from the primary site to somewhere else through the bloodstream or the lymphatic system. |
| Morbidity | Detrimental effects on health. |
| Mortality | Either (1) the condition of being subject to death; or (2) the death rate, which reflects the number of deaths per unit of population in relation to any specific region, age group, disease, treatment or other classification, usually expressed as deaths per 100, 1000, 10,000 or 100,000 people. |
| Multidisciplinary team | A team with members from different healthcare professions and specialties (e.g. urology, oncology, pathology, radiology, and nursing). Cancer care in the National Health Service (NHS) uses this system to ensure that all relevant health professionals are engaged to discuss the best possible care for that patient. |
| Naevus | A mole or collection of pigment cells (plural: naevi or nevi) |
| Optical coherence tomography (OCT) | Based on the same principle as ultrasound, OCT uses a handheld probe to measure the optical scattering of near‐infrared (1310 nm) light waves (rather than sound waves) from under the surface of the skin |
| Prevalence | The proportion of a population found to have a condition. |
| Prognostic factors/indicators | Specific characteristics of a cancer or the person who has it which might affect the patient’s prognosis. |
| Receiver operating characteristic (ROC) plot | A plot of the sensitivity and 1 minus the specificity of a test at the different possible thresholds for test positivity; represents the diagnostic capability of a test with a range of binary test results |
| Receiver operating characteristic (ROC) analysis | The analysis of a ROC plot of a test to select an optimal threshold for test positivity |
| Recurrence | Recurrence is when new cancer cells are detected following treatment. This can occur either at the site of the original tumour or at other sites in the body. |
| Reference Standard | A test or combination of tests used to establish the final or ‘true’ diagnosis of a patient in an evaluation of a diagnostic test |
| Reflectance confocal microscopy (RCM) | A microscopic technique using infrared light (either in a handheld device or a static unit) that can create images of the deeper layers of the skin |
| Resolution | Resolution in an imaging system refers to its ability to distinguish two points in space as being separate points; resolution is measured in two directions: axial and lateral. |
| Sensitivity | In this context the term is used to mean the proportion of individuals with a disease who have that disease correctly identified by the study test |
| Specificity | The proportion of individuals without the disease of interest (in this case with benign skin lesions) who have that absence of disease correctly identified by the study test |
| Spectroscopy | Study of the interaction between matter and electromagnetic radiation |
| Spindle subtypes of SCC | A squamous cell carcinoma variant characterised by poorly differentiated spindle cells surrounded by collagenous stroma |
| Staging | Clinical description of the size and spread of a patient’s tumour, fitting into internationally agreed categories. |
| Stratum corneum | The outermost layer of the epidermis. This layer is the most superficial layer of skin, which is composed of flattened skin cells organised like a brick wall. In normal conditions cells are not nucleated at this layer |
| Subclinical (disease) | Disease that is usually asymptomatic and not easily observable, e.g. by clinical or physical examination. |
Target condition being diagnosed
There are three main forms of skin cancer. Melanoma is the most widely known amongst the general population, yet the commonest skin cancers in white populations are those arising from keratinocytes: basal cell carcinoma (BCC) and cutaneous squamous cell carcinoma (cSCC) (Gordon 2013; Madan 2010). In 2003, the World Health Organization estimated that between two and three million ‘non‐melanoma’ skin cancers (of which BCC and cSCC are estimated to account for around 80% and 16% of cases, respectively) and 132,000 melanoma skin cancers occur globally each year (WHO 2003).
In this DTA review there are three target conditions of interest: (a) melanoma; (b) basal cell carcinoma (BCC); and (c) cutaneous squamous cell carcinoma (cSCC).
Melanoma
Melanoma arises from uncontrolled proliferation of melanocytes, i.e. the epidermal cells that produce pigment or melanin. Cutaneous melanoma refers to any skin lesion with malignant melanocytes present in the dermis, and includes superficial spreading, nodular, acral lentiginous, and lentigo maligna melanoma variants (see Figure 1). Melanoma in situ refers to malignant melanocytes that are contained within the epidermis and have not invaded the dermis, but are at risk of progression to melanoma if left untreated. Lentigo maligna, a subtype of melanoma in situ in chronically sun‐damaged skin, denotes another form of proliferation of abnormal melanocytes. Lentigo maligna can progress to invasive melanoma if its growth breaches the dermo‐epidermal junction during a vertical growth phase (when it is a 'lentigo maligna melanoma'). However its malignant transformation is both lower and slower than for melanoma in situ (Kasprzak 2015). Melanoma in situ and lentigo maligna are both atypical intraepidermal melanocytic variants. Melanoma is one of the most serious forms of skin cancer, with the potential to metastasise to other parts of the body through the lymphatic system and bloodstream. It accounts for only a small proportion of skin cancer cases but is responsible for up to 75% of deaths from this disease (Boring 1994; Cancer Research UK 2017).
1.
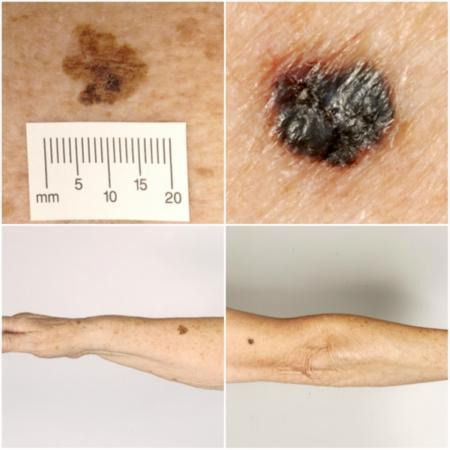
Sample photographs of superficial spreading melanoma (left) and nodular melanoma (right). Copyright © 2010 Dr Rubeta Matin: reproduced with permission.
The incidence of melanoma rose to over 200,000 newly‐diagnosed cases worldwide in 2012 (Erdmann 2013; Ferlay 2015), with an estimated 55,000 deaths (Ferlay 2015). The highest incidence is observed in Australia with 13,134 new cases of melanoma of the skin in 2014 (ACIM 2017) and in New Zealand with 2341 registered cases in 2010 (HPA and MelNet NZ 2014). For 2014 in the USA, the predicted incidence was 73,870 per annum and the predicted number of deaths was 9940 (Siegel 2015). The highest rates in Europe are seen in north‐western Europe and the Scandinavian countries, with a highest incidence reported in Switzerland: 25.8 per 100,000 in 2012. Rates in England have tripled from 4.6 and 6.0 per 100,000 in men and women, respectively, in 1990, to 18.6 and 19.6 per 100,000 in 2012 (EUCAN 2012). Indeed, in the UK, melanoma has one of the fastest‐rising incidence rates of any cancer, and has the biggest projected increase in incidence between 2007 and 2030 (Mistry 2011). In the decade leading up to 2013, age‐standardised incidence increased by 46%, with 14,500 new cases in 2013 and 2459 deaths in 2014 (Cancer Research UK 2017). Rates are higher in women than in men, but the rate of incidence in men is increasing faster than in women (Arnold 2014).The rising incidence of melanoma is thought to be primarily related to an increase in recreational sun exposure and tanning bed use and an increasingly ageing population with higher lifetime recreational ultraviolet (UV) exposure, in conjunction with possible earlier detection (Belbasis 2016; Linos 2009). Putative risk factors are reviewed in detail elsewhere (Belbasis 2016).
A database of over 40,000 US patients from 1998 onwards which assisted the development of the 8th American Joint Committee on Cancer (AJCC) Staging System indicated a five‐year survival of 97% to 99% for stage I melanoma, dropping to between 32% and 93% in stage III disease, depending on tumour thickness, the presence of ulceration and the number of involved nodes (Gershenwald 2017). While these are substantial increases relative to survival in 1975 (Cho 2014), mortality rates have remained static during the same period. This observation, coupled with increasing incidence of localised disease, suggests that improvements in survival may be due to earlier detection and heightened vigilance (Cho 2014). New targeted therapies for advanced (stage IV), melanoma (e.g. BRAF inhibitors), have improved survival, and immunotherapies are evolving such that long‐term survival is being documented (Pasquali 2018; Rozeman 2017). No new data regarding the survival prospects for patients with stage IV disease were analysed for the AJCC 8 staging guidelines due to lack of contemporary data (Gershenwald 2017).
Basal cell carcinoma
BCC can arise from multiple stem cell populations, including from the bulge and interfollicular epidermis (Grachtchouk 2011). BCC growth is usually localised, but it can infiltrate and damage surrounding tissue, sometimes causing considerable destruction and disfigurement, particularly when located on the face (Figure 2). The four main subtypes of BCC are superficial, nodular, morphoeic (infiltrative) and pigmented. Lesions typically present as slow‐growing, asymptomatic papules, plaques, or nodules which bleed or form ulcers that do not heal (Firnhaber 2012). The diagnosis is often made incidentally rather than by people presenting with symptoms (Gordon 2013).
2.
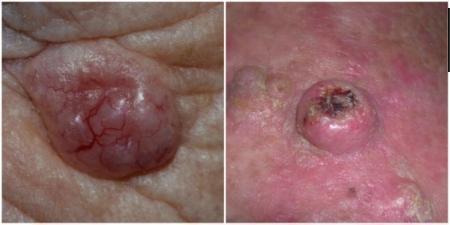
Sample photographs of BCC (left) and cSCC (right). Copyright © 2012 Dr Rubeta Matin: reproduced with permission.
BCC most commonly occurs on sun‐exposed sites on the head and neck (McCormack 1997) and are more common in men and in people over the age of 40. A rising incidence of BCC in younger people has been attributed to increased recreational sun exposure (Bath‐Hextall 2007a; Gordon 2013; Musah 2013). Other risk factors include Fitzpatrick skin types I and II (Fitzpatrick 1975; Lear 1997; Maia 1995), previous skin cancer history, immunosuppression, arsenic exposure, and genetic predisposition such as in basal cell naevus (Gorlin) syndrome (Gorlin 2004; Zak‐Prelich 2004). Annual incidence is increasing worldwide; Europe has experienced an average increase of 5.5% a year over the last four decades, the USA 2% a year, while estimates for the UK show that incidence appears to be increasing more steeply at a rate of an additional 6 per 100,000 persons a year (Lomas 2012). The rising incidence has been attributed to an ageing population, changes in the distribution of known risk factors, particularly ultraviolet radiation, and improved detection due to the increased awareness amongst both practitioners and the general population (Verkouteren 2017). Hoorens 2016 points to evidence for a gradual increase in the size of BCCs over time, with delays in diagnosis ranging from 19 to 25 months.
According to National Institute for Health and Care Excellence (NICE) guidance (NICE 2010), low‐risk BCCs that may be considered for excision are nodular lesions occurring in people older than 24 years who are not immunosuppressed and do not have Gorlin syndrome. Furthermore, low‐risk lesions should be located below the clavicle, should be small (less than 1 cm) with well‐defined margins, not recurrent following incomplete excision and are not difficult to reach surgically or in highly visible locations (NICE 2010). Superficial BCCs are also typically low risk and may be amenable to medical treatments such as photodynamic therapy or topical chemotherapy (Kelleners‐Smeets 2017). Assigning BCCs as low or high risk influences the management options (Batra 2002; Randle 1996).
Advanced locally‐destructive BCC can arise from longstanding untreated lesions or from a recurrence of aggressive basal cell carcinoma after primary treatment (Lear 2012). Very rarely, BCC metastasises to regional and distant sites resulting in death, especially cases of large neglected lesions in those who are immunosuppressed or those with Gorlin syndrome (McCusker 2014). Rates of metastasis are reported at 0.0028% to 0.55% (Lo 1991), with very poor survival rates. It is recognised that baso‐squamous carcinoma (more like a high‐risk cSCC in behaviour and not considered a true BCC) is likely to have accounted for many cases of apparent metastases of BCC, hence the spuriously high reported incidence in some studies of up to 0.55%, which is not seen in clinical practice (Garcia 2009).
Squamous cell carcinoma of the skin
Primary cSCC arises from the keratinocytes in the epidermis or its appendages. People with cSCC often present with an ulcer or firm (indurated) papule, plaque, or nodule (Griffin 2016), often with an adherent crust (Madan 2010). cSCC can arise in the absence of a precursor lesion or it can develop from pre‐existing actinic keratosis (dysplastic epidermis) or Bowen's disease (considered by some to be cSCC in situ). The estimated annual risk of progression is from less than 1% to 20% (Alam 2001), and 5% for lesions developing from pre–existing dysplasia (Kao 1986). It remains locally invasive for a variable length of time, but has the potential to spread to the regional lymph nodes or through the bloodstream to distant sites, especially in immunosuppressed individuals (Lansbury 2010). High‐risk lesions are those arising on the lip or ear, recurrent cSCC, lesions arising on non‐exposed sites, scars or chronic ulcers, tumours larger than 20 mm in diameter or which have a histological depth of invasion greater than 4 mm or poor differentiation status on histopathological examination (Motley 2009).
Chronic ultraviolet light exposure through recreation or occupation is strongly linked to cSCC occurrence (Alam 2001). It is particularly common in people with fair skin and in less common genetic disorders of pigmentation, such as albinism, xeroderma pigmentosum, and recessive dystrophic epidermolysis bullosa (RDEB) (Alam 2001). Other recognised risk factors include immunosuppression; chronic wounds; arsenic or radiation exposure; certain drug treatments, such as voriconazole and BRAF mutation inhibitors; and previous skin cancer history (Baldursson 1993; Chowdri 1996; Dabski 1986; Fasching 1989; Lister 1997; Maloney 1996; O'Gorman 2014). In solid organ transplant recipients, cSCC is the most common form of skin cancer; the risk of developing cSCC has been estimated at 65 to 253 times that of the general population (Hartevelt 1990; Jensen 1999; Lansbury 2010). Overall, local and metastatic recurrence of cSCC at five years is estimated at 8% and 5% respectively. The five‐year survival rate of metastatic cSCC of the head and neck is around 60% (Moeckelmann 2018).
Treatment
For primary melanoma, the mainstay of definitive treatment is wide local surgical excision of the lesion, to remove both the tumour and any malignant cells that might have spread into the surrounding skin (Garbe 2016; Marsden 2010; NICE 2015; SIGN 2017; Sladden 2009). Recommended lateral surgical margins vary according to tumour thickness (Garbe 2016) and stage of disease at presentation (NICE 2015).
Treatment options for BCC and cSCC include surgery, other destructive techniques such as cryotherapy or electrodesiccation and topical chemotherapy. A Cochrane Review of 27 randomised controlled trials (RCTs) of interventions for BCC found very little good‐quality evidence for any of the interventions used (Bath‐Hextall 2007b). Complete surgical excision of primary BCC has a reported five‐year recurrence rate of less than 2% (Griffiths 2005; Walker 2006), leading to significantly fewer recurrences than treatment with radiotherapy (Bath‐Hextall 2007b). After apparent clear histopathological margins (serial vertical sections) after standard excision biopsy with 4 mm surgical peripheral margins taken there is a five‐year reported recurrence rate of around 4% (Drucker 2017). Mohs micrographic surgery, whereby horizontal sections of the tumour are microscopically examined intraoperatively and re‐excision is undertaken until the margins are tumour‐free, can be considered for high‐risk lesions where standard wider excision margins of surrounding healthy skin might lead to considerable functional impairment (Bath‐Hextall 2007b; Lansbury 2010; Motley 2009; Stratigos 2015). Bath‐Hextall and colleagues (Bath‐Hextall 2007b) found a single trial comparing Mohs micrographic surgery with a 3mm standard excision in BCC (Smeets 2004); the update of this study showed non‐significantly lower recurrence at 10 years with Mohs micrographic surgery (4.4% compared to 12.2% after surgical excision, P = 0.10) (Van Loo 2014).
The main treatments for high‐risk BCC are wide local excision, Mohs micrographic surgery and radiotherapy. For low‐risk or superficial subtypes of BCC, or for small and or multiple BCCs at low‐risk sites (Marsden 2010), destructive techniques other than excisional surgery may be used (e.g. electrodesiccation and curettage or cryotherapy (Alam 2001; Bath‐Hextall 2007b)). Alternatively, non‐surgical (or non‐destructive) treatments may be considered (Bath‐Hextall 2007a; Drew 2017; Kim 2014), including topical chemotherapy such as imiquimod (Williams 2017), 5‐fluorouracil (Arits 2013), ingenol mebutate (Nart 2015) and photodynamic therapy (Bath‐Hextall 2007b; Roozeboom 2016). Although non‐surgical techniques are increasingly used, they do not allow histological confirmation of tumour clearance, and their use is dependent on accurate characterisation of the histological subtype and depth of tumour. The 2007 systematic review of BCC interventions found limited evidence from very small RCTs for these approaches (Bath‐Hextall 2007b), which have only partially been addressed by subsequent studies (Bath‐Hextall 2014; Kim 2014; Roozeboom 2012). Most BCC trials have compared interventions within the same treatment class, and few have compared medical versus surgical treatments (Kim 2014).
Vismodegib, a first‐in‐class Hedgehog signalling pathway inhibitor, is now available for the treatment of metastatic or locally‐advanced BCC based on the pivotal study ERIVANCE BCC (Sekulic 2012). It is licensed for use in patients where surgery or radiotherapy is inappropriate, e.g. for treating locally‐advanced periocular and orbital BCCs with orbital salvage of patients who otherwise would have required exenteration (Wong 2017). However, NICE has recently recommended against the use of vismodegib, based on cost effectiveness and uncertainty of evidence (NICE 2017).
A systematic review of interventions for primary cSCC found only one RCT eligible for inclusion (Lansbury 2010). Current practice therefore relies on evidence from observational studies, as reviewed in Lansbury 2013, for example. Surgical excision with predetermined margins is usually the first‐line treatment (Motley 2009; Stratigos 2015). Estimates of recurrence after Mohs micrographic surgery, surgical excision, or radiotherapy, which are likely to have been evaluated in higher‐risk populations, have shown pooled recurrence rates of 3%, 5.4% and 6.4% respectively, with overlapping confidence intervals; the review authors advise caution when comparing results across treatments (Lansbury 2013).
Index test(s)
Computer–assisted diagnosis (CAD) describes a range of artificial intelligence‐based techniques that automate the diagnosis of skin cancer by using a computer to analyse lesion images, and determine the likelihood of malignancy, or the need for excision. Each CAD system has a data collection component, which collects imaging or non‐visual data (e.g. electrical impedance measurements) from the suspicious lesion and feeds it to the data processing component, which then performs a series of analyses to arrive at a diagnostic classification.
Images are acquired using a number of different techniques, although most commonly by digital dermoscopy (Derm–CAD) which creates digital subsurface images of the skin using a computer coupled with a dermatoscope, videocamera and digital television (Rajpara 2009; Esteva 2017). Commercially‐available systems include the DB–MIPS® (DB–Dermo MIPS) (Biomips Engineering SRL, Sienna Italy), MicroDERM (Visiomed AG, Germany), SolarScan (Polartechnics Ltd, Australia) and MoleExpert (DermoScan GmbH, Germany), all of which are hand–held digital or video dermatoscopes that communicate with CAD analysis software (see Figure 3).
3.

Examples of commercially available CAD systems using digital dermoscopy (A), electrical impedance spectroscopy (B) and multispectral imaging (C and D). Reproduced with permission of the manufacturers. Copyright © 2018 MedX Corp, Canada; DermoScanGmbH, Germany; SciBase III, Sweden: reproduced with permission.
Other systems use spectroscopy (Spectro‐CAD), whereby information on cell characteristics (such as cell shape or size) is gathered by measuring how electromagnetic waves pass through skin lesions. This information is most commonly acquired using multispectral imaging (MSI–CAD) that enables computer–generated graphic representations of lesion morphology to be produced from detecting light reflected at several wavelengths across the lesion. By far the most common of these is diffuse reflectance spectrophotometry imaging (DRSi), which uses light that diffusely penetrates the skin to a depth of 2 to 2.5 mm beneath the surface to produce light reflectance images at a number of specific wavelengths across the visible near‐infrared light spectrum (approximately 400 to 1000 nm) to capture variations in light attenuation and scattering from melanin, collagen and blood vessel structures.
DRSi developed from diffuse reflectance spectroscopy, a non‐visual spectroscopic technique which uses optical reflectance to distinguish between lesion types based on spectral shape and calibrated level of reflected light for wavelengths continuously varying from the ultraviolet (320 nm) to the near‐infrared (1100 nm) with a high spectral resolution (4 nm) (e.g. Marchesini 1992; Wallace 2000b). Commercially available DRSi computer‐assisted diagnosis systems include the SIAscope™ (MedX Health Corp, Canada), a hand–held unit that communicates with CAD analysis software (Figure 3). The MelaFind® system (Strata Skin Sciences (formerly Mela Sciences Inc), Horsham, PA, USA) was Food and Drug Administration (FDA)‐approved; however, it no longer appears to be commercially available.
The Nevisense™ system (SciBase III, Sweden; Figure 3) is also commercially available, but is based on electrical impedance spectroscopy (EIS), a non‐optical method which seeks to provide information on cellular features by measuring the feedback from an electrical current once it has passed through the intended tissue. With Nevisense™, an alternating applied voltage (electrical current) is passed by a probe through a skin lesion and the current that is bounced back is measured by the same probe, which measures a combination of tissue resistance and capacitance. At high frequencies, conduction occurs easily through all tissue components, including cells, but at low frequencies current tends to flow only through the extracellular space. The spectral shape is therefore sensitive to cellular components and dimensions, internal structure and cellular arrangements. The Nevisense™ EIS system measures at four multiple depths and at 35 frequencies logarithmically distributed from 1.0 kHz to 2.5 MHz using a 5 x 5 mm area electrode covered in tiny pins that penetrate into the stratum corneum.
Other non‐visual sources of lesion data include Raman spectroscopy, in which a laser is used to excite vibrations in molecules which then impart wavelength shifts to some of the scattered light waves, creating spectral patterns that are related to the molecular structure of lesions (Maglogiannis 2009), and fluorescence spectroscopy which uses a laser to excite electrons, causing molecules to absorb and then re‐emit light in spectral patterns that are also related to the molecular structure of lesions (Rallan 2004).
All CAD systems use machine learning, where a classification algorithm learns features of groups of lesions (i.e. diagnostic types) by exposure to a ‘training set’ of lesions of known histological diagnosis. This process creates a model which is designed to distinguish between these lesion types in future observations. Examples of machine learning algorithms include discriminant analysis, decision trees, neural networks, fuzzy logic, nearest k‐neighbours, logistic regression and support vector machines (SVMs), and all use different mathematical equations to set out how observed features relate to a given diagnosis (Maglogiannis 2009; Masood 2013). Model outputs also vary, in part according to the type of data used to acquire lesion information, and can take the form of binary outputs indicating the presence of malignancy versus benignity (e.g. the Melafind® system), risk scores which can be used at varying thresholds (e.g. the DANAOS system used by MicroDerm), or graphical representations of the CAD pattern analysis which highlight areas of concern within a lesion (e.g. the SIAgraphs produced by SIAscope™). Artificial intelligence systems using continuous learning algorithms, where computer systems continuously develop their classification algorithm as each new case is examined, and do not stop learning at the end of a training period, are not addressed in this review.
Clinical pathway
The diagnosis of skin lesions occurs in primary‐, secondary‐, and tertiary‐care settings by both generalist and specialist healthcare providers. In the UK, people with concerns about a new or changing lesion will present to their general practitioner (GP) rather than directly to a specialist in secondary care. A GP with clinical concerns usually refers a patient to a specialist in secondary care – usually a dermatologist, but sometimes to a surgical specialist such as a plastic surgeon or an ophthalmic surgeon. Suspicious skin lesions may also be identified in a referral setting, for example by a general surgeon, and referred for a consultation with a skin cancer specialist (Figure 4). Skin cancers identified by other specialist surgeons (such as an ear, nose, and throat (ENT) specialist or maxillofacial surgeon) will usually be diagnosed and treated without further referral.
4.
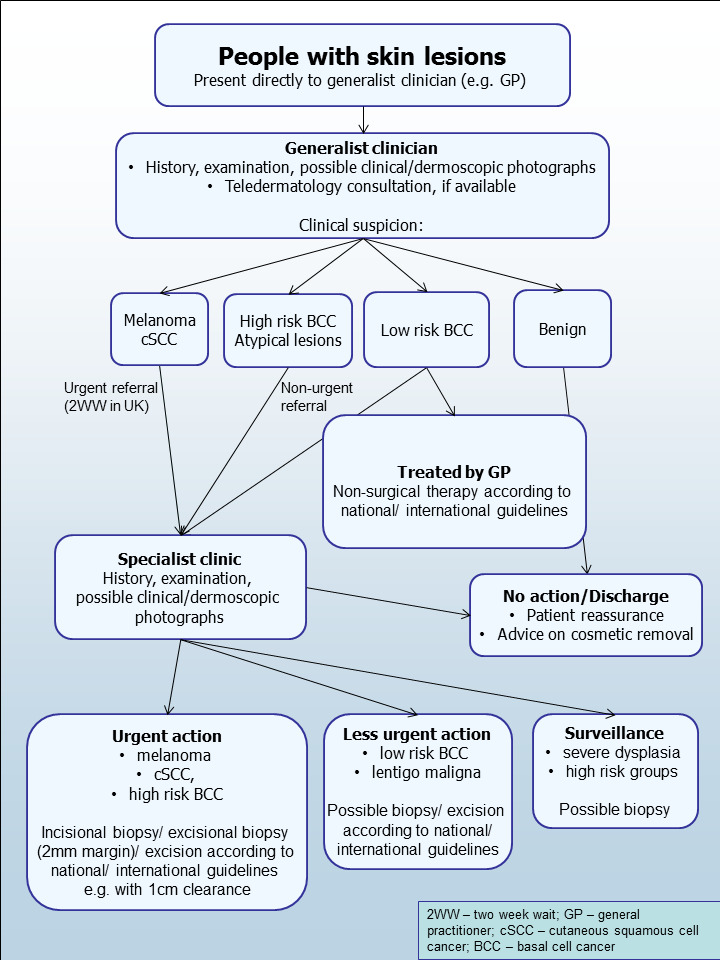
Current clinical pathway for people with skin lesions.
Current UK guidelines recommend that all suspicious pigmented lesions presenting in primary care should be assessed by taking a clinical history and visual inspection (VI) using the seven‐point checklist (MacKie 1990); lesions suspected to be melanoma or cSCC should be referred for appropriate specialist assessment within two weeks (Chao 2013; Marsden 2010; NICE 2015). Evidence is emerging, however, to suggest that excision of melanoma by GPs is not associated with increased risk compared with outcomes in secondary care (Murchie 2017). In the UK, low‐risk BCCs are usually recommended for routine referral, with urgent referral for those in whom a delay could have a significant impact on outcomes, for example due to large lesion size or critical site (NICE 2015). Appropriately‐qualified generalist care providers increasingly undertake management of low‐risk BCC in the UK, such as by excision of low‐risk lesions (NICE 2010). Similar guidance is in place in Australia (CCAAC Network 2008).
For referred lesions, the specialist clinician will use history‐taking, visual inspection of the lesion (in conjunction with other skin lesions), palpation of the lesion and associated lymph nodes in conjunction with dermoscopic examination to inform a clinical decision. If melanoma is suspected, then urgent 2 mm excision biopsy is recommended (Lederman 1985; Lees 1991); for cSCC predetermined surgical margin excision or a diagnostic biopsy may be considered. BCC and pre‐malignant lesions potentially eligible for nonsurgical treatment may undergo a diagnostic biopsy before initiation of therapy if there is diagnostic uncertainty. Equivocal melanocytic lesions for which a definitive clinical diagnosis cannot be reached may undergo surveillance to identify any lesion changes that would indicate excision biopsy or reassurance, and discharge for those lesions that remain stable over a period of time.
Theoretically, teledermatology consultations may aid appropriate triage of lesions into urgent referral; non‐urgent secondary‐care referral (e.g. for suspected basal cell carcinoma); or where available, referral to an intermediate care setting, e.g. clinics run by GPs with a special interest in dermatology. The distinction between setting and examiner qualifications and experience is important, as specialist clinicians might work in primary‐care settings (for example, in the UK, GPs with a special interest in dermatology and skin surgery who have undergone appropriate training), and generalists might practice in secondary‐care settings (for example, plastic surgeons who do not specialise in skin cancer). The level of skill and experience in skin cancer diagnosis will vary for both generalist and specialist care providers and will also impact on test accuracy.
Prior test(s)
Although smartphone applications and community‐based teledermatology services can increasingly be directly accessed by people who have concerns about a skin lesion (Chuchu 2018b), visual inspection of a suspicious lesion by a clinician is usually the first in a series of tests to diagnose skin cancer. In the UK this usually takes place in primary care, but in many countries people with suspicious lesions can present directly to a specialist setting.
A range of technologies has emerged to aid skin cancer diagnosis, both to ensure that malignancies (especially melanoma) are not missed, and at the same time minimising unnecessary surgical procedures. Dermoscopy using a hand‐held microscope has become the most widely used tool for clinicians to improve diagnostic accuracy of pigmented lesions, in particular for melanoma (Argenziano 1998; Argenziano 2012; Haenssle 2010; Kittler 2002), although it is less well established for the diagnosis of BCC or cSCC. Dermoscopy is frequently combined with visual inspection of a lesion in secondary‐care settings, and is also increasingly used in primary care, particularly in countries such as Australia (Youl 2007).The diagnostic accuracy, and comparative accuracy, of visual inspection and dermoscopy have been evaluated in a further three reviews in this series (Dinnes 2018a; Dinnes 2018b; Dinnes 2018c).
Consideration of the degree of prior testing that study participants have undergone is key to interpretation of test accuracy indices, as these are known to vary according to the disease spectrum (or case‐mix) of included participants (Lachs 1992; Leeflang 2013; Moons 1997; Usher‐Smith 2016). Spectrum effects are often observed when tests that are developed further down the referral pathway have lower sensitivity and higher specificity when applied in settings with participants with limited prior testing (Usher‐Smith 2016). Studies of individuals with suspicious lesions at the initial clinical presentation stage ('test‐naïve') are likely to have a wider range of different diagnoses and include a higher proportion of people with benign diagnoses compared with studies of participants who have been referred for a specialist opinion on the basis of visual inspection (with or without dermoscopy) by a generalist practitioner. Furthermore, studies in more specialist settings may focus on equivocal or difficult‐to‐diagnose lesions rather than lesions with a more general level of clinical suspicion. However, this direction of effect is not consistent across tests and diseases, the mechanisms in action often being more complex than prevalence alone, and can be difficult to identify (Leeflang 2013). A simple categorisation of studies according to primary, secondary or specialist setting therefore may not always adequately reflect these key differences in disease spectrum that can affect test performance.
Role of index test(s)
Skin cancer diagnosis, whether by visual inspection alone or with the use of dermoscopy is undertaken iteratively, using both implicit pattern recognition (non‐analytical reasoning) and more explicit ‘rules’ based on conscious analytical reasoning (Norman 2009), the balance of which will vary according to experience and familiarity with the diagnostic question. In the hands of experienced dermatologists, dermoscopy has been shown to enhance the accuracy of skin cancer detection (especially melanoma) when compared to unaided visual examination (Dinnes 2018b; Dinnes 2018c). The subjectivity involved in interpreting lesion morphology is thought to underlie the decrease in accuracy that occurs when the dermatoscope is used by less experienced clinicians (Binder 1995).
The addition of computer–based diagnosis to these investigations has potential to increase the detection of melanomas by reducing the clinicians’ reliance on subjective information, which is necessarily interpreted using their experience of past cases. The additive value of CAD systems is also likely to vary with differences in setting, prior testing and selection of participants, as previously discussed (Prior test(s)). CAD systems could therefore fulfil three different roles in clinical practice: (1) to help GPs, or other clinicians working in unreferred settings, to appropriately triage lesions for referral; (2) as part of a remote diagnostic service; or (3) as an expert–level second opinion to specialists in referral settings. All three roles would rely on CAD being as sensitive for the diagnosis of melanoma as experienced dermatologists. On the other hand, the specificity required for CAD to add value differs for each of these three situations, as discussed below.
If sensitive enough, use of CAD in primary care could allow more appropriate triage of higher‐risk lesions to secondary care by increasing the early detection of potentially malignant lesions. However, although a relatively lower specificity (higher false‐positive rate) may be acceptable in a primary‐care setting, limiting false–positive diagnoses would create health service benefits by avoiding unnecessary referral, and alleviating patient anxiety more promptly. Similarly, the remote use of CAD could inform the need for referral, by sending images or other diagnostic data to specialist clinics, or even to commercial organisations, for remote interpretation, much as teledermatology is already used. In this circumstance, a relatively high specificity would be required in order to avoid unacceptable increases in rates of referral to specialist centres.
Finally, when used in referral settings as a complement to in‐person diagnosis by a specialist, even if CAD could be shown to pick up difficult‐to‐diagnose melanomas that might be missed on visual inspection or dermoscopy, the specificity of the system would need to be very high so as not to inordinately increase the burden of skin surgery. False‐positive diagnoses not only cause unnecessary scarring from a biopsy or excision procedure, but also increase patient anxiety whilst they await the definitive histological results and increase healthcare costs as the number needed to remove to yield one melanoma diagnosis increases. Pigmented lesions are common, so the resource implication for even a small increase in the threshold to excise lesions in populations where melanoma rates are increasing, will avoid a considerable healthcare burden to both patient and healthcare provider, as long as lesions that are not excised turn out to be benign. The use of CAD to detect melanoma in specialist clinics would only be advantageous if it could be shown to detect skin cancers that would otherwise be missed, or to decrease unnecessary surgical intervention (i.e. removal of false–positive lesions) with no loss of sensitivity.
Delay in diagnosis of a BCC as a result of a false‐negative test is not as serious as for melanoma, because BCCs are usually slow‐growing and very unlikely to metastasise. Nevertheless, delayed diagnosis can result in larger and more complex surgical procedures with consequent greater morbidity. Very sensitive diagnostic tests for BCC, however, may compromise on lower specificity, leading to a higher false‐positive rate and an increased burden of skin surgery such that a balance between sensitivity and specificity is needed. The greatest potential advantage of CAD in the management of BCC is likely to lie in its ability to perform rapid, non–invasive assessments of multiple lesions (common in BCC patients (Lear 1997)).
The situation for cSCC is more similar to melanoma, in that the consequences of falsely reassuring a person that they do not have skin cancer can be serious and potentially fatal, given that removal of an early cSCC is usually curative. Thus, a good diagnostic test for cSCC should demonstrate high sensitivity and a corresponding high negative predictive value. A test that can also reduce false‐positive clinical diagnoses without missing true cases of cSCC has patient and resource benefits.
Alternative test(s)
A number of other tests which may have a role in the diagnosis of skin cancer in a specialist setting have been reviewed as part of our series of systematic reviews, including reflectance confocal microscopy (RCM) (Dinnes 2018dDinnes 2018e), optical coherence tomography (OCT) (Ferrante di Ruffano 2018a), high‐frequency ultrasound (Dinnes 2018f) and exfoliative cytology (Ferrante di Ruffano 2018b). Other tests with a role in earlier settings include teledermatology (Chuchu 2018a) and smart–phone applications (Chuchu 2018b). Reviews on the accuracy of gene expression testing and volatile organic compounds could not be performed as planned, due to an absence of relevant studies. Evidence permitting, we will compare the accuracy of available tests in an overview review, exploiting within‐study comparisons of tests and allowing the analysis and comparison of commonly used diagnostic strategies where tests may be used singly or in combination.
We also considered and excluded a number of tests from this review, such as tests used for screening (e.g. total body photography of those with large numbers of typical or atypical naevi) or monitoring (e.g. CAD systems used to monitor the progression of suspicious skin lesions).
Lastly, we did not assess the accuracy of histopathological confirmation following lesion excision, because it is the established reference standard for melanoma diagnosis and will be one of the standards against which the index tests are evaluated in these reviews.
Rationale
Our series of reviews of diagnostic tests used to assist clinical diagnosis of skin cancer aims to identify the most accurate approaches to diagnosis and provide clinical and policy decision‐makers with the highest possible standard of evidence on which to base diagnostic and treatment decisions. With increasing rates of melanoma and basal cell carcinoma and a trend to adopt dermoscopy and other high‐resolution image analysis in primary care, the anxiety around missing early malignant lesions needs to be balanced against the risk of too many unnecessary referrals, and to avoid sending too many people with benign lesions for a specialist opinion. It is questionable whether all skin cancers identified by sophisticated techniques, even in specialist settings, help to reduce morbidity and mortality. It is also a concern that newer technologies incur the risk of increasing false‐positive diagnoses. It is also possible that the use of some technologies, e.g. widespread use of dermoscopy in primary care with little or no training, could actually result in harm by missing melanomas if they are used as replacement technologies for traditional history‐taking and clinical examination of the entire skin. Many branches of medicine have noted the danger of such "gizmo idolatry" amongst doctors (Leff 2008).
The central premise underlying CAD is that it uses quantitative, objective and expert–level assessments of lesion features, which lessen the need for specialist training and lengthy experience in test use. Given the reliance on specialist training and experience to make accurate skin cancer diagnosis using dermoscopy, CAD diagnosis has the potential to improve the health of patients by widening access to specialist diagnostic capabilities in primary and secondary care. If it is sensitive enough, introducing CAD could increase the early detection of skin cancers, which for melanoma and cSCC in particular, is critical to improving outcomes. As with any technology requiring significant investment, a full understanding should be acquired of the benefits including patient acceptability and cost effectiveness compared with usual practice before such an approach can be recommended; establishing the accuracy of diagnosis and referral accuracy is one of the key components.
We identified four published systematic reviews focusing on the accuracy of CAD, two synthesising the performance of Derm–CAD systems (Ali 2012; Rajpara 2009), and two reviewing both Derm–CAD and Spectro–CAD systems (Rosado 2003; Vestergaard 2008). All are limited by out–of–date search periods (Ali 2012 up to 2011, Rajpara 2009 and Vestergaard 2008 up to 2007, Rosado 2003 up to 2002), which is a key concern in the rapidly advancing field of machine learning. Another concern for Ali 2012, Rajpara 2009 and Rosado 2003 is their inclusion of studies which are ineligible for this Cochrane Review due to the absence of an independent validation set, a methodological feature likely to inflate the apparent accuracy of predictive models (Altman 2009). Rosado 2003 also selected datasets on the basis of highest performance, and pooled accuracy estimates for Derm–CAD with Spectro–CAD, which we consider to be two different diagnostic tests. There is therefore a need for an up‐to‐date and rigorous review of the accuracy of dermoscopy–based CAD and of spectroscopy–based CAD which explicitly considers the following key characteristics.
Because CAD models are created by analysing patterns in archived datasets, the degree to which they are likely to make accurate classifications of new observations in real‐life clinical situations relies on the generalisability of the training sets used to develop them (Horsch 2011). Training sets that contain few lesions, or a restricted range of the different diagnoses encountered in clinical practice, are likely to produce models that misclassify new observations due to inadequate learning. Other important attributes thought to influence diagnostic ability are the segmentation process (how the lesion’s border is detected by the computer), which features are selected for analysis (akin to the selection of features for analysis in the algorithms used in epiluminescence microscopy (ELM) dermoscopy, e.g. ABCD or seven–point), the algorithm used, and the type of information produced by the CAD system (e.g. binary outputs indicating presence of malignancy, or visual images of lesions such as macro– or microscopic photographs or graphical representations of highlighting suspect structures).
This review follows a generic protocol which covers the full series of Cochrane DTA Reviews for the diagnosis of melanoma (Dinnes 2015a) and for the diagnosis of keratinocyte cancers (Dinnes 2015b). The Background and Methods sections of this review therefore use some text that was originally published in these protocols (Dinnes 2015a; Dinnes 2015b) and text that overlaps some of our other reviews (Dinnes 2018a; Dinnes 2018b; Ferrante di Ruffano 2018a).
Objectives
To determine the accuracy of CAD systems for diagnosing cutaneous invasive melanoma and atypical intraepidermal melanocytic variants in adults, and to compare the accuracy of CAD systems with that of clinician diagnosis using dermoscopy.
To determine the accuracy of CAD systems for diagnosing BCC in adults, and to compare the accuracy of CAD systems with that of clinician diagnosis using dermoscopy.
To determine the accuracy of CAD systems for diagnosing cSCC in adults, and to compare the accuracy of CAD systems with that of clinician diagnosis using dermoscopy.
Secondary objectives
To determine the accuracy of CAD systems for diagnosing invasive melanoma alone in adults, and to compare the accuracy of CAD systems with that of clinician diagnosis using dermoscopy
To determine the accuracy of CAD systems for identifying any lesion requiring excision (due to any skin cancer or high–grade dysplasia) in adults, and to compare the accuracy of CAD systems with that of clinician diagnosis using dermoscopy
For each of the primary target conditions:
To compare the diagnostic accuracy of CAD systems to clinician diagnosis using dermoscopy, where both tests have been evaluated in the same studies (direct comparisons);
To determine the diagnostic accuracy of individual CAD systems;
To compare the accuracy of CAD–based diagnosis to CAD–assisted diagnosis (CAD results used by clinicians as a diagnostic aid)
Where CAD systems are used as a diagnostic aid, to determine the effect of observer experience on diagnostic accuracy.
Investigation of sources of heterogeneity
We set out to investigate a range of potential sources of heterogeneity across our series of reviews, as outlined in our generic protocols (Dinnes 2015a; Dinnes 2015b) and described in Appendix 3; however, we could not do this because of the available data on each individual test reviewed.
Methods
Criteria for considering studies for this review
Types of studies
We included test accuracy studies that assessed the result of the index test against that of a reference standard, including the following:
Studies where all participants received a single index test and a reference standard;
Studies where all participants received more than one index test and reference standard;
Studies where participants were allocated (by any method) to receive different index tests or combinations of index tests and all receive a reference standard (between‐person comparative studies (BPC));
Studies that recruited series of participants unselected by true disease status (referred to as case series for the purposes of this review);
Diagnostic case‐control studies that separately recruited diseased and non‐diseased groups (see Rutjes 2005); however, we did not include studies that compared results for malignant lesions to those for healthy skin (i.e. with no lesion present);
Both prospective and retrospective studies;
Studies where previously‐acquired clinical or dermoscopic images were retrieved and prospectively interpreted for study purposes.
We excluded studies from which we could not extract or derive 2 x 2 contingency data of the number of true positives, false positives, false negatives and true negatives, or if studies included fewer than five skin cancer cases or fewer than five benign lesions. The size threshold of five is arbitrary, however such small studies are unlikely to add precision to estimate of accuracy.
Studies available only as conference abstracts were excluded; however, attempts were made to identify full papers for potentially relevant conference abstracts (Searching other resources).
Participants
We included studies in adults with pigmented or non–pigmented skin lesions considered to be suspicious for melanoma or an atypical intraepidermal melanocytic variant or a keratinocyte skin cancer (BCC or cSCC). Studies examining adults at high risk of developing skin cancer, including those with a family history or previous history of skin cancer, atypical or dysplastic naevus syndrome, or genetic cancer syndromes were also eligible for inclusion.
We excluded studies that recruited only participants with malignant diagnoses.
We excluded studies conducted in children or which clearly reported inclusion of more than 50% of participants aged 16 and under.
Index tests
Studies reporting accuracy data for tests using automated diagnosis were eligible for inclusion, whether diagnosis was produced independently by the CAD system (system–based diagnosis), or by a clinician using a CAD system as a diagnostic aid (computer–assisted diagnosis). CAD systems using any type of data capture were eligible, including imaging and non–imaging modalities. We included all machine learning algorithms.
We included studies developing new algorithms or methods of diagnosis (i.e. derivation studies) if they evaluated the new approach using a separate 'test set' of participants or images.
We excluded studies if they:
Evaluated a new statistical model or algorithm in the same participants or images as those used to train the model (i.e. absence of an independent test set);
Used cross‐validation approaches such as 'leave‐one‐out' cross‐validation (Efron 1983); or
Evaluated the accuracy of the presence or absence of individual lesion characteristics or morphological features, with no overall diagnosis of malignancy.
Although primary‐care clinicians can in practice be specialists in skin cancer, we considered primary‐care physicians as generalist practitioners and dermatologists as specialists. Within each group, we extracted any reporting of special interest or accreditation in skin cancer.
Target conditions
We defined the primary target conditions as the detection of:
Any form of invasive cutaneous melanoma, or atypical intraepidermal melanocytic variants (i.e. including melanoma in situ, or lentigo maligna, which has a risk of progression to invasive melanoma)
BCC
cSCC
We considered two additional target conditions in secondary analyses, namely the detection of:
Any form of invasive cutaneous melanoma alone
Any skin lesion requiring excision: all forms of skin cancer listed above, as well as melanoma in situ, lentigo maligna, and lesions with severe melanocytic dysplasia.
Reference standards
The ideal reference standard is histopathological diagnosis in all eligible lesions. A qualified pathologist or dermatopathologist should perform histopathology. Ideally, reporting should be standardised, detailing a minimum dataset to include the histopathological features of melanoma to determine the American Joint Committee on Cancer (AJCC) Staging System (e.g. Slater 2014). We did not apply this as a necessary inclusion criterion, but extracted any pertinent information.
Partial verification (applying the reference test only to a subset of those undergoing the index test) was of concern, given that lesion excision or biopsy is unlikely to be carried out for all benign‐appearing lesions within a representative population sample. To reflect what happens in reality, we therefore accepted clinical follow‐up of benign‐appearing lesions as an eligible reference standard, whilst recognising the risk of variable verification bias (as misclassification rates of histopathology and follow‐up will differ).
Additional eligible reference standards included cancer registry follow‐up and 'expert opinion' with no histology or clinical follow‐up. Cancer registry follow‐up is considered less desirable than active clinical follow‐up, as follow‐up is not carried out within the control of the study investigators. Furthermore, if participant‐based analyses as opposed to lesion‐based analyses are presented, it may be difficult to determine whether the detection of a malignant lesion during follow‐up is the same lesion that originally tested negative on the index test.
We considered all of the above to be eligible reference standards, with the following caveats:
All study participants with a final diagnosis of the target skin cancer disorder must have a histological diagnosis, either subsequent to the application of the index test or after a period of clinical follow‐up
At least 50% of all participants with benign lesions must have either a histological diagnosis or clinical follow‐up to confirm benignity.
Search methods for identification of studies
Electronic searches
The Information Specialist (SB) carried out a comprehensive search for published and unpublished studies. A single large literature search was conducted to cover all topics in the programme grant (see Appendix 1 for a summary of reviews included in the programme grant). This allowed for the screening of search results for potentially relevant papers for all reviews at the same time. A search combining disease related terms with terms related to the test names, using both text words and subject headings was formulated. The search strategy was designed to capture studies evaluating tests for the diagnosis or staging of skin cancer. As the majority of records were related to the searches for tests for staging of disease, a filter using terms related to cancer staging and to accuracy indices was applied to the staging test search, to try to eliminate irrelevant studies, for example, those using imaging tests to assess treatment effectiveness. A sample of 300 records that would be missed by applying this filter was screened and the filter adjusted to include potentially relevant studies. When piloted on MEDLINE, inclusion of the filter for the staging tests reduced the overall numbers by around 6000. The final search strategy, incorporating the filter, was subsequently applied to all bibliographic databases as listed below (Appendix 4). The final search result was cross‐checked against the list of studies included in five systematic reviews; our search identified all but one of the studies, and this study was not indexed on MEDLINE. The Information Specialist (SB) devised the search strategy, with input from the Information Specialist from Cochrane Skin. No additional limits were used.
We searched the following bibliographic databases to 29 August 2016 for relevant published studies:
MEDLINE via OVID (from 1946);
MEDLINE In‐Process & Other Non‐Indexed Citations via OVID;
Embase via OVID (from 1980).
We searched the following bibliographic databases to 30 August 2016 for relevant published studies:
Cochrane Central Register of Controlled Trials (CENTRAL) Issue 7, 2016, in the Cochrane Library;
Cochrane Database of Systematic Reviews (CDSR) Issue 8, 2016 in the Cochrane Library;
Cochrane Database of Abstracts of Reviews of Effects (DARE) Issue 2, 2015;
CRD Health Technology Assessment (HTA) database Issue 3, 2016;
Cumulative Index to Nursing and Allied Health Literature (CINAHL) (via EBSCO from 1960).
We searched the following databases for relevant unpublished studies using a strategy based on the MEDLINE search:
CPCI (Conference Proceedings Citation Index), via Web of Science™ (from 1990; searched 28 August 2016);
SCI Science Citation Index Expanded™ via Web of Science™ (from 1900, using the 'Proceedings and Meetings Abstracts' Limit function; searched 29 August 2016).
We searched the following trials registers using search terms 'melanoma', 'squamous cell', 'basal cell' and 'skin cancer' combined with 'diagnosis':
Zetoc (from 1993; searched 28 August 2016)
The US National Institutes of Health Ongoing Trials Register (www.clinicaltrials.gov); searched 29 August 2016
NIHR Clinical Research Network Portfolio Database (www.nihr.ac.uk/research‐and‐impact/nihr‐clinical‐research‐network‐portfolio/); searched 29 August 2016;
The World Health Organization International Clinical Trials Registry Platform (apps.who.int/trialsearch/); searched 29 August 2016.
We aimed to identify all relevant studies, regardless of language or publication status (published, unpublished, in press, or in progress). We applied no date limits.
Searching other resources
We have screened relevant systematic reviews identified by the searches for their included primary studies, and included any missed by our searches. We have checked the reference lists of all included papers, and subject experts within the author team have reviewed the final list of included studies. We did not conduct any electronic citation searching.
Data collection and analysis
Selection of studies
At least one review author (JDi or NC) screened titles and abstracts, with any queries discussed and resolved by consensus. A pilot screen of 539 MEDLINE references showed good agreement (89% with a kappa of 0.77) between screeners. We included primary test accuracy studies and test accuracy reviews (for scanning of reference lists) of any test used to investigate suspected melanoma, BCC, or cSCC at initial screening. Inclusion criteria (Appendix 5) were applied independently by both a clinical reviewer (from one of a team of 12 clinician reviewers) and a methodologist reviewer (JDi, NC or LFR) to all full‐text articles, resolving disagreements by consensus or by a third party (JDe, CD, HW, and RM). We contacted authors of eligible studies when insufficient data were presented to allow for the construction of 2 x 2 contingency tables.
Data extraction and management
One clinical (as detailed above) and one methodologist reviewer (JDi, NC or LFR) independently extracted data for details of the study design, participants, index test(s) or test combinations and criteria for index test positivity, reference standards, and data required to populate a 2 x 2 diagnostic contingency table for each index test, using a piloted data extraction form. We extracted data at all available index test thresholds, resolving disagreements by consensus or by a third party (JDe, CD, HW, and RM).
We contacted authors of included studies where information related to final lesion diagnoses or diagnostic threshold was missing. In particular, invasive cSCC (included as disease‐positive for one of our secondary objectives) is not always differentiated from in situ variants such as Bowens disease (which we did not consider as disease‐positive for any of our definitions of the target condition).
We contacted authors of conference abstracts published from 2013 to 2015, to ask whether full data were available. We marked conference abstracts as 'pending', and will revisit them in a future review update.
Dealing with multiple publications and companion papers
Where we found multiple reports of a primary study, we maximised yield of information by collating all available data. Where there were inconsistencies in reporting or overlapping study populations, we contacted study authors for clarification in the first instance. If this contact with authors was unsuccessful, we used the most complete and up‐to‐date data source where possible.
Assessment of methodological quality
We assessed risks of bias and applicability of included studies using the QUADAS‐2 checklist (Whiting 2011), tailored to the review topic (see Appendix 6). We piloted the modified QUADAS‐2 tool on a small number of included full‐text articles. One clinical (as detailed above) and one methodologist reviewer (JDi, NC or LFR) independently assessed quality for the remaining studies, resolving any disagreement by consensus or by a third party where necessary (JDe, CD, HW, and RM).
Statistical analysis and data synthesis
Our unit of analysis was the lesion rather than the person. This is because (i) in skin cancer initial treatment is directed to the lesion rather than systemically (thus it is important to be able to correctly identify cancerous lesions for each person), and (ii) it is the most common way in which the primary studies reported data. Although there is a theoretical possibility of correlations of test errors when the same person contributes data for multiple lesions, most studies include very few people with multiple lesions and any potential impact on findings is likely to be very small, particularly in comparison with other concerns for risk of bias and applicability. For each analysis, we included only one dataset per study to avoid multiple counting of lesions. Where multiple CAD models or algorithms were assessed in an individual study, we selected one at random, using a random‐number generator. Where studies evaluated CAD as a diagnostic aid by clinicians with varying degrees of experience, we chose the dataset reporting the highest degree of clinical experience. We made these selections without reference to the corresponding accuracy data.
We estimated accuracy of dermoscopy separately according to whether the diagnosis recorded was based on a face–to–face (in‐person) encounter or based on remote (image‐based) assessment. Where multiple algorithms were assessed in an individual study, we selected dermoscopy datasets on the following preferential basis:
‘No algorithm’ reported; data presented for clinician’s overall diagnosis or management decision
Pattern analysis or pattern recognition
ABCD algorithm (or derivatives of)
Seven‐point checklist (also referred to as Glasgow/Mackie checklist)
Menzies algorithm
Three‐point checklist
As for CAD, we preferred dermoscopy datasets reporting the highest degree of clinical experience over studies reporting multiple results using clinicians of varying experience.
We pooled CAD study data for systems using similar methods of data acquisition; thus we considered all studies using digital dermoscopy‐based CAD (Derm‐CAD) to be similar and pooled them. However, spectroscopy‐based CAD (Spectro‐CAD) systems analyse different data types and so were only pooled in these subgroups: multispectral imaging studies (MSI‐CAD), electrical impedance spectroscopy (EIS‐CAD), and diffuse reflectance spectroscopy (DRS‐CAD). For each index test, algorithm or checklist under consideration, we plotted estimates of sensitivity and specificity on coupled forest plots and in receiver operating characteristic (ROC) space. CAD thresholds are created by complex statistical algorithms, and a threshold is difficult to define. We therefore assumed results were binary for the purpose of pooling results across similar CAD systems. We estimated summary operating points (summary sensitivities and specificities) with 95% confidence and prediction regions using the bivariate model (Chu 2006; Reitsma 2005). Where inadequate data were available for the analysis to converge, we simplified the model, first by assuming no correlation between estimates of sensitivity and specificity and secondly by setting variance terms to zero if there was little or no heterogeneity on SROC plots (Takwoingi 2015).
We extracted data on the accuracy of dermoscopy from all included studies that performed both CAD and dermoscopy in the same participants. We performed test comparisons using two analytic strategies. First, we performed indirect comparisons by using all studies of the two tests. Then we made direct comparisons of CAD and dermoscopy by including only comparative studies that assessed the accuracy of both tests in the same study population to enable a robust comparison (Takwoingi 2013). To minimise the risk of bias in the direct comparison, we excluded studies that performed either CAD or dermoscopy on a subsample of the total analysed population. In the comparative meta‐analyses of indirect and direct comparisons, we compared summary points by using a bivariate meta‐regression model that included test type as a covariate. We included covariate terms for sensitivity and specificity. We assessed model fit using likelihood ratio tests to compare nested models. We computed estimates of absolute differences in sensitivity and specificity using the bivariate model parameters, and obtained 95% confidence intervals using the delta method. We obtained P values for the absolute differences using Wald tests. We fitted univariate random‐effects logistic regression models incorporating test type as a covariate when there were few studies or a bivariate meta‐regression analysis did not converge. When the number of studies was insufficient for meta‐analysis, we examined individual study results and calculated 95% CIs using the Newcombe‐Wilson method without continuity correction (Newcombe 1998).
For illustration of the meta‐analytic findings in the 'Summary of findings' table, we computed the numbers of true positives, false positives, false negatives and true negatives, using the summary estimates of sensitivity and specificity together with the lower quartile, median and upper quartile of the prevalence observed in the studies included in the meta‐analysis.
We fitted bivariate models using the xtmelogit command in STATA 15.
Investigations of heterogeneity
We examined heterogeneity between studies by visually inspecting the forest plots of sensitivity and specificity and summary ROC plots. Due to limited data availability, we were unable to formally investigate heterogeneity using meta‐regression.
Sensitivity analyses
The primary analysis included both CAD–based diagnoses and CAD–aided diagnoses. We conducted sensitivity analyses excluding studies of CAD–aided diagnoses.
Assessment of reporting bias
Because of uncertainty about the determinants of publication bias for diagnostic accuracy studies and the inadequacy of tests for detecting funnel plot asymmetry (Deeks 2005), we did not perform tests to detect publication bias.
Results
Results of the search
We identified 34,517 unique references, and screened them for inclusion. Of these, we reviewed 1051 full‐text papers for eligibility for any one of the suite of reviews of tests to assist in the diagnosis of melanoma or keratinocyte skin cancer. Of the 1051 full‐text papers assessed, we excluded 848 from all reviews in our series (see Figure 5 PRISMA flow diagram of search and eligibility results).
5.
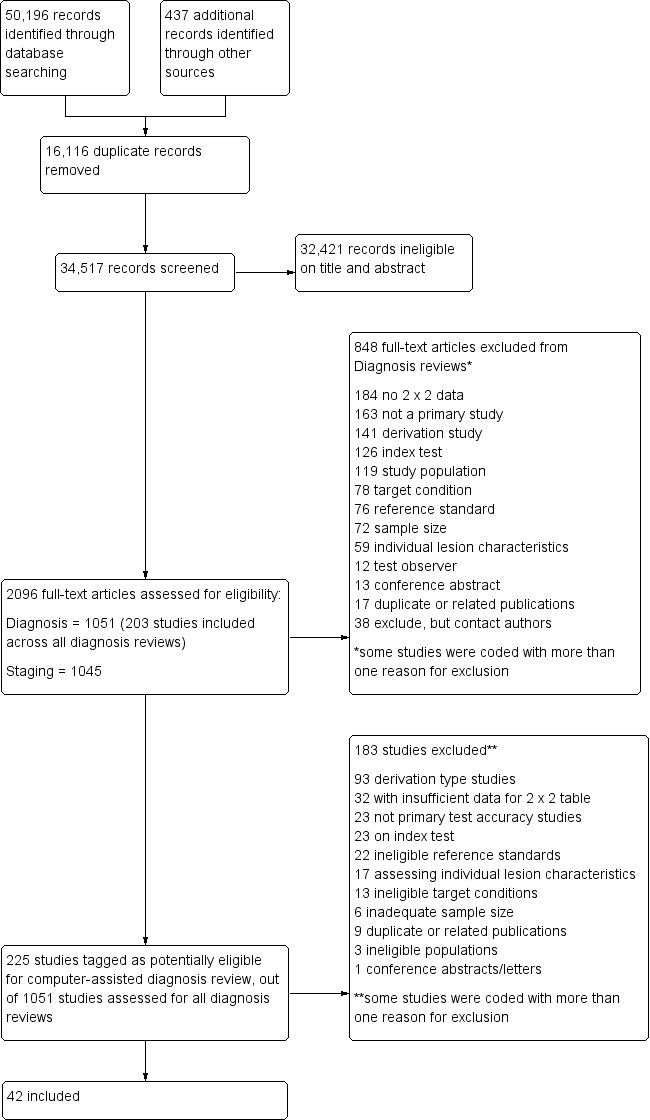
PRISMA flow diagram.
Of the 225 studies tagged as potentially eligible for this review of CAD (164 for Derm–CAD, 61 for Spectro–CAD), we include 42 publications (24 Derm–CAD and 18 Spectro–CAD). Exclusions were mainly due to the absence of a ‘test’ set of lesions used to evaluate CAD's performance independently of the computer algorithm’s development (Derm–CAD n = 76, Spectro‐CAD n = 17); inability to construct a 2 x 2 contingency table based on the data presented (Derm–CAD n = 24, Spectro‐CAD n = 8); the use of ineligible index tests (Derm–CAD n = 18, Spectro‐CAD n = 5) (for example: computers used to measure lesions but not to diagnose them, e.g. Seidenari 2012); or not meeting our requirements for an eligible reference standard (Derm–CAD n = 13, Spectro‐CAD n = 9). Other reasons for exclusion included ineligible definition of the target condition (Derm–CAD n = 10, Spectro‐CAD n = 3) and CAD systems based on evaluating the presence of a single lesion characteristic (Derm–CAD n = 17) (for example, a CAD system analysing the colour balance of a lesion). We provide a list of the 183 publications excluded from this review, with reasons for exclusion in Characteristics of excluded studies, with a list of all studies excluded from the full series of reviews available as a separate pdf (please contact skin.cochrane.org for a copy of the pdf).
We contacted the authors of 10 publications to acquire additional detail on published 2 x 2 data for the accuracy of CAD, with responses for four publications received to date. These did not result in the inclusion of any additional studies, but did permit the inclusion of one additional dataset in an already–included study (Mollersen 2015). One response highlighted an alternative publication that was independently ascertained by the project search, and was included (Serrao 2006); two replies were unable to provide the information requested for two study publications, both of which we subsequently excluded due to incomplete 2 x 2 data. We failed to contact authors of six publications, resulting in the exclusion of those six studies from the review. In addition to these 10 attempted contacts, authors of one other publication (Walter 2012) were contacted as part of another review in this series, i.e. the accuracy of visual inspection for the diagnosis of melanoma (Dinnes 2018a), to provided clarification on methods used; the author response enabled us to include it.
The 42 included studies reported on 39 cohorts of lesions, providing 63 datasets with 13,445 lesions and 2452 malignancies. Most of the studies (n = 24, 57%) contributed data on the diagnostic accuracy of digital dermoscopy–based CAD systems (Derm–CAD), of which seven also compared the diagnostic accuracy of Derm–CAD with dermoscopic diagnosis. The remaining 18 studies contributed data on the diagnostic accuracy of spectroscopy–based CAD (Spectro‐CAD), of which five provided comparative accuracy data with dermoscopy. A cross‐tabulation of studies by CAD type, reported comparisons and target conditions is available in Table 3.
2. A cross‐tabulation of studies by CAD type, reported comparisons and target conditions.
| Study | CAD type | CAD system | CAD diagnosis | Comparison with dermoscopy | Target conditionsa |
| Bauer 2000 | Derm–CAD | DB‐MIPs | CAD diagnostic aid | in–person dermoscopy | 1 |
| Wollina 2007 | Derm–CAD | DB‐MIPs | CAD diagnostic aid | No | 1 |
| Burroni 2004 | Derm–CAD | DB‐MIPs | CAD only | No | 1 |
| Rubegni 2002a | Derm–CAD | DB‐MIPs | CAD only | No | 1 |
| Seidenari 1998 | Derm–CAD | DB‐MIPs | CAD only | image–based assessment | 1 |
| Seidenari 1999 | Derm–CAD | DB‐MIPs | CAD only | No | 1 |
| Stanganelli 2005 | Derm–CAD | DB‐MIPs | CAD only | No | 1 |
| Piccolo 2002 | Derm–CAD | DEM‐MIPS | CAD only | image–based assessment | 1 |
| Binder 1998 | Derm–CAD | IBAS 2000 | CAD only | No | 1 |
| Barzegari 2005 | Derm–CAD | MicroDERM | CAD only | No | 1 |
| Boldrick 2007 | Derm–CAD | MicroDERM | CAD only | No | 1 |
| Serrao 2006 | Derm–CAD | MicroDERM | CAD only | No | 1 |
| Dreiseitl 2009 | Derm–CAD | Image J (MoleMax II) | CAD only | in–person dermoscopy | 1 |
| Maglogiannis 2015 | Derm–CAD | System name NR (MoleMax II) | CAD only | No | 1 |
| Mollersen 2015 | Derm–CAD | Nevus Doctor Mole Expert | CAD only | No | 1,5 |
| Piccolo 2014 | Derm–CAD | Nevuscreen | CAD diagnostic aid | image–based assessment | 1 |
| Cascinelli 1992 | Derm–CAD | Skin View | CAD only | No | 1,5 |
| Cristofolini 1997 | Derm–CAD | Skin View | CAD only | No | 1 |
| Menzies 2005 | Derm–CAD | SolarScan | CAD only | Excluded (different sample sizes) | 1,4 |
| Binder 1994 | Derm–CAD | System name NR | CAD only | image–based assessment | 1 |
| Blum 2004a | Derm–CAD | System name NR | CAD only | Excluded (different sample sizes) | 1 |
| Ferris 2015 | Derm–CAD | System name NR | CAD only | Excluded (different sample sizes) | 1,2,5 |
| Gilmore 2010 | Derm–CAD | System name NR | CAD only | image–based assessment | 1 |
| Menzies 1996 | Derm–CAD | System name NR | CAD only | image–based assessment | 4 |
| Hauschild 2014 | MSI–CAD | MelaFind | CAD diagnostic aid | image–based assessment | 1,4 |
| Winkelmann 2016 | MSI–CAD | MelaFind | CAD diagnostic aid | image–based assessment | 1 |
| Friedman 2008 | MSI–CAD | MelaFind | CAD only | image–based assessment | 1,4 |
| Monheit 2011 | MSI–CAD | MelaFind | CAD only | No | 1 |
| Wells 2012 | MSI–CAD | MelaFind | CAD only | image–based assessment | 1 |
| Sgouros 2014 | MSI–CAD | Siascope (version NR) | CAD only | No | 5 |
| Glud 2009 | MSI–CAD | Siascope II | CAD only | image–based assessment | 1 |
| Terstappen 2013 | MSI–CAD | Siascope V | CAD only | No | 4 |
| Walter 2012 | MSI–CAD | Siascope V (MoleMate) | CAD diagnostic aid | No | 1,4,5 |
| Tomatis 2005 | MSI–CAD | Spectroshade | CAD only | No | 1 |
| Ascierto 2010 | MSI–CAD | Spectroshade | CAD only | in–person dermoscopy | 4 |
| Gutkowicz Krusin 1997 | MSI–CAD | System name NR | CAD only | No | 1 |
| Tomatis 2003 | MSI–CAD | Telespectrophotometric system | CAD only | No | 1 |
| Bono 1996 | MSI–CAD | Telespectrophotometric System | CAD only | No | 4 |
| Bono 2002 | MSI–CAD | Telespectrophotometric System | CAD only | in–person dermoscopy | 1 |
| Garcia Uribe 2012 | DRS–CAD | OIDRS | CAD only | No | 1,5 |
| Malvehy 2014 | EIS–CAD | Nevisense | CAD only | Excluded (different sample sizes) | 1,2,3,4,5 |
| Mohr 2013 | EIS–CAD | Nevisense | CAD only | No | 1,2,4,5 |
Key: Derm‐CAD – Dermoscopy‐based computer–assisted diagnosis; DRS–CAD – diffuse reflectance spectroscopy computer–assisted diagnosis; EIS‐CAD ‐ Electrical impedance‐based computer‐assisted diagnosis; MSI‐CAD ‐ Multispectral imaging‐based computer–assisted diagnosis; NR – Not reported; OIDRS ‐ oblique incidence reflectance spectroscopy
a1 – Invasive melanoma and atypical intraepidermal melanocytic variants; 2 – Basal cell carcinoma; 3 – cutaneous Squamous cell carcinoma; 4 – Invasive melanoma alone; 5 – Any skin cancer or lesion requiring excision
Studies were case series (n = 27, 64%), case‐control (n = 10, 24%), randomised controlled trial (n = 1, 2%), or of unclear design (n = 4, 10%). Lesion selection was most commonly retrospective (n = 22, 52%) or prospective (n = 15, 36%), and was unclear in five studies (12%). Studies included only pigmented (n = 29, 69%) or melanocytic lesions (n = 6, 14%), only suspected melanomas (n = 4, 10%), or any lesions suspected of malignancy (n = 2, 5%). Participant characteristics such as age and gender were reported by 15 of the 42 studies.
Methodological quality of included studies
Most of the included studies were of methodological concern, primarily due to lack of applicability to the current review question, but also due to a high or unclear risk of bias in their design. Since there were no major differences in quality by CAD type, we provide an overview of the quality and applicability of all included studies regardless of CAD type. The methodological quality of studies according to CAD type (Derm–CAD or spectroscopy–based CAD) is summarised in Figure 6 and Figure 7.
6.
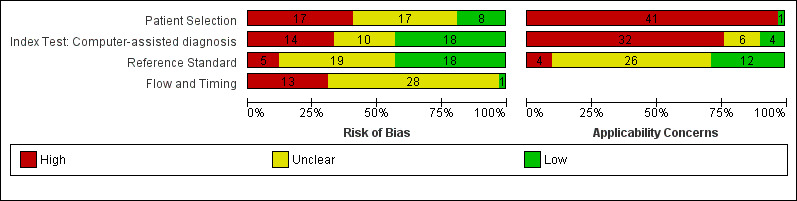
Risk of bias and applicability concerns graph: review authors' judgements about each domain presented as percentages across included studies
7.
Risk of bias and applicability concerns summary: review authors' judgements about each domain for each included study
We judged the risk of participant selection bias to be high in 17 studies (40%), due to the selection of lesions according to their final diagnosis (case–control studies: n = 10, 24%), and/or to the inappropriate exclusion of lesions with particular prognostic characteristics (n = 8, 19%), such as high‐grade dysplastic lesions (Ferris 2015) or small/large lesions (Malvehy 2014; Monheit 2011). Study eligibility criteria and participant exclusions were not reported clearly enough to ascertain the risk of selection bias in 17 studies (40%); this meant that we could not determine whether consecutive or random samples of lesions were recruited (n = 22, 52%), whether participants had been selected according to their final diagnoses (use of 'case–control' selection, n = 5, 12%), or whether participant exclusions were appropriate (n = 22, 52%).
A single study (Sgouros 2014) was of low concern for the applicability of its participant sample to the current review question: it included unexcised lesions and did not recruit participants with multiple lesions. All others (n = 41, 98%) were of high concern due to the use of restricted participant groups and settings (n = 40, 95%), with study populations limited to lesions selected for excision based on the clinical or dermoscopic diagnosis or selected retrospectively from histopathology databases (n = 36, 84%). Six studies did not restrict inclusion to excised lesions, but in three of these the non–excised lesions could not be extracted due to the absence of clinical follow–up in at least 50% of benign cases (Boldrick 2007; Bono 1996; Sgouros 2014). Fourteen studies were also of concern due to their recruitment of participants with multiple lesions (including over 5% more lesions than participants). One of these (Dreiseitl 2009) provided participant–based 2 x 2 data as well as lesion–based data for the accuracy of a Derm–CAD system. These two analyses of the same population highlight the distortion that can occur in study populations that include multiple lesions per participant: lesion–based sensitivity was lower than participant‐based sensitivity (74% versus 89%), but lesion–based specificity was far higher (84% versus 48%) due to the inclusion of many disease‐negative lesions per participant.
Twenty–two (52%) studies did not report the number of participants included (precluding assessment of the inclusion of multiple lesions). Of the 18 studies including model derivation, six (33%) used a wide range of skin conditions to train the classification algorithm. Two (11%) used an inadequately narrow range (absence of non–dysplastic benign conditions), while the remaining 10 (56%) provided inadequate detail of diagnoses included in the training set.
Over half the studies were at high (n = 14, 33%) or unclear (n = 10, 24%) risk of bias due to the methods used to undertake the index test. Most studies (n = 40, 95%) blinded CAD results to the reference standard diagnosis, although almost half (n = 20, 48%) failed to clearly prespecify the diagnostic threshold, of which 13 (30%) were threshold–finding studies that provided accuracy data for the best threshold possible once index test results had been examined. Most studies (n = 35, 83%) evaluated CAD in an independent population from that used to train the classification algorithm, either by external validation (n = 23, 55%) or internal validation (randomised division of a single study group into training and test sets: n = 12, 29%). An additional six studies (14%) used internal validation, but failed to specify whether division of the study group was made randomly (i.e. not selected according to diagnosis), while one study was at risk of bias by selecting which diagnoses to place in the training and test sets (Tomatis 2003).
Of the 18 studies that included CAD model derivation (training of the classification algorithm), eight (44%) accounted for model overfitting by using a Support Vector Machine algorithm (Gilmore 2010; Mohr 2013; Stanganelli 2005), performing a jack–knife calculation (Binder 1994; Burroni 2004), or another method (Rubegni 2002a; Tomatis 2003; Tomatis 2005). One study specified that model optimisation was not incorporated (Garcia Uribe 2012) and nine (50%) did not discuss overfitting. Most studies (n = 32, 76%) were of high concern for the applicability of the index test, due to their evaluation of an unestablished threshold (n = 23), lack of detail about the diagnostic threshold used (n = 16), and/or the use of non–expert clinicians (n = 2) in studies evaluating CAD as a diagnostic aid (n = 7).
Almost all studies reported use of an acceptable reference standard (n = 37, 88%), and around half (n = 19, 45%) clearly reported blinding of the reference standard to the CAD result. For the applicability of the reference standard, four reported using expert diagnosis for some lesions (high concern) and 26 (62%) were unclear as to whether histopathology had been interpreted by an experienced histopathologist or dermatopathologist.
Reporting of study flow and timing was generally poor, with an unclear risk of bias in 28 studies (67%), largely due to ambiguity about the interval between the application of the index test and reference standard (excision for histology or first follow–up visit) (n = 28). Thirteen studies (30%) were at a high risk of bias because they used different reference standards according to diagnosis (differential verification) (n = 6) and/or did not include all participants in the analysis (n = 10), primarily due to technical difficulties with the CAD system (n = 6).
Eleven of the 15 studies comparing CAD with dermoscopy were at high (n = 2) or unclear (n = 9) risk of bias. Six reported blinding between tests, two reported no blinding and seven were unclear. Half (n = 8) did not clearly report the interval between tests.
Findings
The 24 studies evaluating a Derm–CAD system reported 23 cohorts of lesions providing 32 CAD datasets with 9602 lesions including 1313 malignancies of which 1220 were melanomas, at least 83 BCCs (number not specified in one study, Menzies 1996), and nine cSCCs. We cannot estimate the total number of study participants with suspicious lesions, due to lack of reporting in study publications (reported in only 10 studies (with 2400 participants)). Two publications provided data for one cohort of lesions (Seidenari 1998; Seidenari 1999); we included the larger of the two studies (Seidenari 1999) in the primary analysis with data from Seidenari 1998 contributing only to the direct comparison of Derm–CAD with dermoscopy. We evaluated a total of 16 different systems (listed in Table 4), three by multiple independent studies: Microderm (Barzegari 2005; Boldrick 2007; Serrao 2006), DB–MIPS (also called DB–Dermo MIPS) (Bauer 2000; Burroni 2004; Rubegni 2002a; Seidenari 1998; Seidenari 1999; Stanganelli 2005; Wollina 2007), and Skin View (Cascinelli 1992;Cristofolini 1997). The 16 systems differ in the type of dermoscopy used to acquire images, the storage devices used, features analysed and statistical classifier used (summarised in Table 4). The approach to computer–assisted support also differed, with 21 studies (88%) evaluating a stand–alone automated diagnosis (‘system–based diagnosis’), and three studies using Derm–CAD as a diagnostic aid to assist clinical decision–making.
3. Characteristics of included digital dermoscopy‐based CAD (Derm–CAD) systems.
| Derm‐CAD processing machine | Role of machine | Image analysis | Classifier | CAD output | Studies |
| MicroDERM (Visiomed AG, Germany) | Dermoscopy unit with internal camera containing analysis system | DANAOS software combines analytical system based on ABCD with database of 21,000 PSLs. | ANN | DANAOS score indicating risk of malignancy | Barzegari 2005; Serrao 2006; Boldrick 2007 |
| DB–Dermo MIPS (Biomips Engineering, Italy) | Dermoscopy unit, internal stereomicroscope, internal DB, pattern analysis system | DB–MIPS pattern analysis system – integrated database stores the patient's data and the description of the lesion along with the image icons. 38 features analysed (grouped into geometries, colours and Burroni's islands of colours) | ANN | Diagnosis suggested (e.g. melanoma, benign melanocytic naevus) | Bauer 2000; Rubegni 2002a |
| Automatic Data Analysis for Melanoma early detection (ADAM) software which analyses boundary shape, texture and colour distribution | SVM | Low risk, intermediate risk or high risk of melanoma | Stanganelli 2005 | ||
| DB–MIPS pattern analysis system | Multivariate discriminant analysis | Graphical output and numerical output of features provided. Diagnosis suggested (e.g. melanoma, benign melanocytic naevus) |
Seidenari 1998; Seidenari 1999 |
||
| DB–MIPS pattern analysis system | KNN | Diagnosis suggested (e.g. melanoma, benign melanocytic naevus) | Burroni 2004 | ||
| DB–MIPS pattern analysis system | Linear discrimination | Diagnosis suggested (e.g. melanoma, benign melanocytic naevus) | Burroni 2004 | ||
| DDA software analysis – analyses 50 parameters subdivided into 3 categories, i.e. geometries, colours, and textures and islands of colours (Burroni islands) | Euclidian distances | Not specified | Wollina 2007 | ||
| DEM–MIPS (Biomips SRL, Siena, Italy) | Commercially available software coupled to stereomicroscope Wild M‐650 (Leica), video camera, computer, colour monitor. | DEM‐MIPS software (Digital Epi Microscopy Melanoma Image Processing Software; Biomips SRL, Siena, Italy). Evaluates colorimetric and geometric features (not reported). | ANN | Not specified | Piccolo 2002 |
| Skin View | Computerised image analysis system, digital television, videocamera. Connection with the computer is through a digitising board able to process colour images | Features of ABCD system plus clinical data (anatomic site, months of growth, size, shape, colour, ulceration or regression) | NR | ≥ 2 of 8 binary (on/off) indicators indicates malignancy: 1. shape (asymmetry); 2. clinical data (changes through time, regression, ulceration); 3. size (mm); 4. colour (distribution of hue); 5. darkness (percent of black mixed with the hue); 6. saturation (percent of white mixed with the hue); 7. border (sharpness of transition between lesion and healthy skin; 8. texture. |
Cascinelli 1992; Cristofolini 1997 |
| Nevus Doctor | Computerised image analysis system coupled to digital dermatoscope | ND takes a dermoscopic image from the Canon/DermLite device as input and classifies the lesion. Features not reported |
NR | Probability of malignancy | Mollersen 2015 |
| MoleExpert (DermoScan GmbH, Germany) | (micro Version 3.3.30.156). Computerised image analysis with output giving probability of malignancy | Features of ABCD system plus other features (not listed), e.g. colour variation and grey veil | NR | Number between −5.00 and 5.00, where high values indicate suspicion of melanoma | Mollersen 2015 |
|
Image J (NIH, Bethesda, USA) |
Image segmentation, feature extraction, image analysis coupled with dermatoscope MoleMax II | 29 features analysed from 38 extracted features describing shape, form and colour. | ANN | 2 outputs: 1) Visual rendering of analysis showing coloured areas 2) Excision vs no excision decision (system considers the green zone of the scale as benign (0 to 0.1), the yellow zone suspicious (0.1 to 0.4), and the red zone malignant (0.4 to 1)) | Dreiseitl 2009 |
| IBAS 2000 workstation (Zeiss, Oberkochen, Germany) | Digital image analysis workstation attached to Wild binocular stereomicroscope M 650 (Wild Heerbrugg AG, Switzerland) | Analysis of 16 morphometric parameters from the lesion and the border image: lesion area and perimeter: minimum polar distance, maximum polar distance, aspect ratio, circularity shape factor, variances of grey, number different colours, range different colours. Border features and area: maximum and minimum border width, ratio of border area to lesion area, ratio of border perimeter to lesion perimeter | ANN | Dichotomous decision: MM vs Benign (CN or DN) The network was trained to yield a value from 0 to 1 in the output nodes. The node yielding the greatest numerical output was then used as the classification result. |
Binder 1998 |
| Nevuscreen ® (Arkè s.a.s., Avezzano, Italy) | Digital database containing image analysis software, coupled to digital dermatoscope | Nevuscreen software automatically analyses ABCD features | NR | TDS score: < 4.75 benign, 4.75 – 5.45 suspicious, > 5.45 highly suggestive of melanoma | Piccolo 2014 |
| SolarScan (Polartechnics Lts, Australia) | Dermoscopy video unit with internal algorithm for image analysis | 103 automated image analysis variables extracted: consisting of various properties of colour, pattern, and geometry. Number analysed not reported | Linear discriminant analysis | Probability of melanoma, with cut–off (not provided) for benign vs melanoma | Menzies 2005 |
| Name not reported | Computer analysis of stored images captured using different dermoscopy/camera combinations | 54 features analysed, such as border irregularity, eccentricity, length of major and minor axes, and colour histogram properties | Digital forest classifier | Severity score (the fraction of decision trees (n = 1000) in which the path ends in ‘‘malignant’’. Lesion classified as malignant if its image traced a path to a malignant node in at least 40% of the trees) | Ferris 2015 |
| Name not reported | Computer analysis of stored images captured using digital stereomicroscope | Features analysed as present or absent (pattern analysis): Pigment network, brown globules, radial streaming, pseudopods, black dots, margin, pigmentation, depigmentation | ANN | NR | Binder 1994 |
| Name not reported | Computer analysis of digital dermoscopy images (Molemax II) | Features corresponding to the number, size and asymmetry of dots: (a) number of dots, (b) total number of pixels in dots, (c) mean number of pixels in dots, (d) variance of num. pixels in dots (e) fraction of lesion area occupied by dark dots. Asymmetry: radial, angular, primary axis. | Comparison of 5 classifiers: Multilayer perceptron KNN Random forest SVM polykernel c = 5* SVM PUK kernel |
NR | Maglogiannis 2015 |
| Name not reported | Computer analysis of stored images captured using digital microscope | 14 features investigated: 4 asymmetry features, colour variance (red, green, blue), mean colour (red, green, blue, intensity), range colour (red, green, blue) | SVM (principal components analysis) | NR | Gilmore 2010 |
| Name not reported | Computer analysis of stored images captured using digital microscope | Negative features: point and axial symmetry of pigmentation Presence of only a single colour. Positive features: blue‐white, veil, multiple brown dots, pseudopods, radial streaming, scarlike depigmentation, peripheral black dots/globules, multiple (5 ‐ 6) colours, multiple blue/grey dots, broadened network |
Classification and regression tree | Presence of indicative features (Melanoma = 0/2 morphologically negative features AND at least 1/9 positive morphological features) | Menzies 1996 |
| Name not reported | Computer analysis of stored images captured using digital microscope | Analysis of 64 analytical parameters including: a large number of morphological parameters, such as margin, geometric parameters (surface area, extent, largest diameter and largest orthogonal diameter), invariant moments, symmetry, colours (red, green, blue and grey value), texture (energy, entropy, correlation, inverse difference moment and inertia), number of regions, focus and difference of the lesion and its convex cover |
Vision algebra methods | NR | Blum 2004a |
*(classifier selected at random for inclusion in review)
Key: ANN – artificial neural network; CN – common naevus; DN – dysplastic naevus; KNN – K–nearest neighbour; NR – not reported; MM – malignant melanoma; SVM – support vector machine; TDS – total dermoscopic score
The 18 publications evaluating a Spectro–CAD system reported 16 cohorts of lesions contributing 32 datasets with 6336 lesions including 1084 malignancies of which 934 were melanomas, 163 BCCs and 49 cSCCs. We cannot estimate the total number of study participants with suspicious lesions, due to lack of reporting in study publications (reported in only eight studies totaling 4484 participants). Four publications provided data for two patient cohorts (Bono 2002;Tomatis 2003 and Hauschild 2014; Monheit 2011); we included the larger studies (Monheit 2011; Tomatis 2003) in the primary analysis, with data from Bono 2002 (same population as Tomatis 2003) and Hauschild 2014 (same population as Monheit 2011) contributing only to the direct comparison of Spectro–CAD with dermoscopy. These 18 studies reported on five different multispectral systems using DRSi: SpectroShade (Ascierto 2010; Tomatis 2005), Melafind (Friedman 2008; Gutkowicz Krusin 1997; Hauschild 2014; Monheit 2011; Wells 2012; Winkelmann 2016), SIAscope (Glud 2009; Terstappen 2013), and ‘Telespectrophotometric System’ (Bono 1996; Tomatis 2003). One study evaluated a non–imaging diffuse reflectance spectroscopy system: OIDRS (Garcia Uribe 2012), which used analysis of data from incidentally‐reflected diffuse light. Nevisense was the only system to use electrical impedance spectroscopy (EIS), and was evaluated in two large prospective studies; these had overlapping recruitment periods and study centres, so we cannot rule out the possibility of overlap in analysed participants (Malvehy 2014; Mohr 2013). As for the Derm–CAD systems, the Spectro–CAD systems differ in terms of the image acquisition and storage devices used, features analysed, statistical classifier used, and the approach to computer–assisted support (i.e. whether used as a stand–alone automated diagnosis, or as information to assist a clinician’s diagnostic decision) (Table 5).
4. Characteristics of included Spectroscopy‐based CAD (Spectro‐CAD) systems.
| Spectro–CAD system (spectroscopy type) | Role of machine | Image analysis | Classifier | CAD Output | Studies |
|
SpectroShade (MHT, Verona Italy) (MSI‐CAD) |
Illumination assembly located inside a PC and an external detection device placed in a hand‐held probe, with integrated image analysis software | Features analysed: (i) reflectance (R); (ii) variegation (V); (iii) area (A); (iv) dark area ratio (DAR); (v) dark island reflectance (DIR); (vi) dark distribution factor (DDF); (vii) dark permanence (D PER) | ANN (multilayer perceptron) | NR | Tomatis 2005 |
| SpectroShade coupled with a MoleMax II dermatoscope | NR | Diagnostic category: 1 no melanoma, 2 doubtful melanoma, 3 suspected melanoma, 4 probable melanoma | Ascierto 2010 | ||
|
MelaFind (early predecessor) (MSI‐CAD) |
Digital camera and illumination assembly coupled to a computer, with separate image analysis | Features analysed: Lesion asymmetry, border, gradient, centroid, texture, colour | Multiparametric linear classifier | NR | Gutkowicz Krusin 1997 |
|
MelaFind (STRATA Skin Sciences [formerly Mela Sciences Inc], Horsham, PA, USA) (MSI‐CAD) |
Multispectral imaging system with integrated image analysis software; device takes images in vivo | MelaFind image analysis | NR | Binary output: (1) positive, (lesion should be considered for biopsy to rule out melanoma); and (2) negative (lesion should be considered for later evaluation) |
Monheit 2011 |
| 6 constrained linear classifiers | Binary output: excise or follow–up | Friedman 2008 | |||
| NR | Monheit 2011 | Wells 2012 | |||
| Logistic regression | Monheit 2011 | Winkelmann 2016 | |||
| NR | Monheit 2011 | Hauschild 2014 | |||
| Telespectrophotometric system (MSI‐CAD) | Digital camera coupled with an illumination system with interference filters and computer for storage and analysis of multispectral images | From each spectral image, 3 parameters, i.e. mean reflectance, variegation index and lesion area, were derived at the corresponding wavelength | Linear discriminant | NR | Bono 1996 |
| For each spectral image, 5 parameters (lesion descriptors) based on ABCD and related to colour and shape of the imaged lesion were evaluated: mean reflectance, variegation index, compactness, roughness, and area | Linear discriminant vs. ANN* | NR | Tomatis 2003 | ||
| Linear discriminant | NR | Bono 2002 | |||
| SIAscopy ™ (Astron Clinica, UK) (MSI‐CAD) | Spectrophotometric imaging system with hand–held skin probe (SIAscope II) and integrated software | Analysis of dermal melanin, erythematous blush, lesion asymmetry, collagen 'holes', blood commas, or irregularities in the collagen | NR | SIAgraphs and Binary output (based on Australian Scoring System): ‘strong chance of melanoma' or 'low risk of melanoma' | Glud 2009 |
| Spectrophotometric imaging system with hand–held skin probe (SIAscope V) and integrated software (software Dermetrics Version 2.0, Astron Clinica Ltd., Great Britain) | NR | SIAgraphs (no further information) | Terstappen 2013 | ||
|
SIAscope (MedX Health Corp, Canada) (MSI‐CAD) |
Spectrophotometric imaging system with hand–held skin probe (SIAscope, version NR) and integrated software | NR | SIAgraphs and lesion score using Primary Care Scoring Algorithm (6 or more points regarded as suspicious) | Sgouros 2014 | |
|
SIAscope (MedX Health Corp, Canada) (MSI‐CAD) |
SIAscopy with MoleMate (software image management system) viewing platform and integrated Primary Care Scoring Algorithm | NR | SIAgraphs and lesion score using Primary Care Scoring Algorithm (6 or more points regarded as suspicious) | Walter 2012 | |
| Oblique Incidence Diffuse Reflectance Spectroscopy (DRS‐CAD) | Light probe coupled to imaging spectrograph, camera, and computer to store images. | NR | ANN | Diagnostic category (e.g. CN, MM, DN, BCC, cSCC) | Garcia Uribe 2012 |
|
Nevisense (SciBase III, Sweden) (EIS‐CAD) |
Electrical Impedance spectroscopy imaging system with integrated image analysis software | The system measures the overall electrical resistance and reactance at 35 different frequencies | SVM (non‐probabilistic binary linear classifier) | The system computes both a score (0 – 10) and a dichotomous output (EIS negative/positive) at a fixed cut‐off. The fixed threshold is set at 4, i.e. scores < 4 are EIS‐negative and scores of ≥ 4 are EIS positive |
Mohr 2013 Malvehy 2014 |
*Classifier excluded at random
Key: ANN – artificial neural network; BCC – basal cell carcinoma; cSCC – cutaneous squamous cell carcinoma; CAD ‐ computer‐assisted diagnosis; CN – common naevus; DN – dysplastic naevus; EIS ‐ electrical impedance spectroscopy; KNN – K–nearest neighbour; NR – not reported; MM – malignant melanoma; MSI – multispectral imaging; SVM – support vector machine
Study results are summarised below according to target condition, with forest plots of available study data provided in Figures 8 to 23. Results of meta–analysis are provided in Table 6 and Table 7.
5. Summary estimates of sensitivity and specificity for CAD according to target condition.
| Index test, target condition | Studies | Cases/Number of participants | Summary sensitivity (95% CI) % | Summary specificity (95% CI) % |
| Main analyses: Invasive melanoma andatypical intraepidermal melanocytic variants | ||||
| Derm–CAD | 22 | 1063/8992 | 90.1 (84.0 to 94.0) | 74.3 (63.6 to 82.7) |
| MSI–CAD | 8 | 286/2401 | 92.9 (83.7 to 97.1) | 43.6 (24.8 to 64.5) |
| EIS–CAD (Nevisense*) | 2 | 368/2389 | 97.0 (94.7 to 98.3) | 33.6 (31.6 to 35.7) |
| Individual CAD systems: Invasive melanoma andatypical intraepidermal melanocytic variants | ||||
| DB–MIPS (Derm–CAD) | 6 | 502/1903 | 95.2 (89.5 to 97.9) | 89.1 (78.7 to 94.8) |
| Skin View (Derm–CAD) | 2 | 45/220 | 80.0 (65.8 to 89.3) | 47.4 (40.1 to 54.8) |
| MelaFind (MSI–CAD) | 5 | 196/1798 | 97.1 (91.9 to 98.9) | 29.8 (12.3 to 56.3) |
| Main analyses: Basal cell carcinoma | ||||
| EIS–CAD (Nevisense*) | 2 | 69/2389 | 100 (94.7 to 100) | 26.3 (24.5 to 28.1) |
| Secondary target condition: invasive melanoma alone | ||||
| Derm–CAD | 2 | 120/950 | 90.8 (84.2 to 94.9) | 63.5 (60.2 to 66.7) |
| MSI–CAD | 5 | 116/386 | 76.5 (43.0 to 93.3) | 60.7 (38.5 to 79.2) |
| EIS–CAD (Nevisense*) | 2 | 226/2389 | 98.2 (95.4 to 99.3) | 38.0 (36.0 to 40.1) |
| Secondary target condition: any skin cancer or lesions requiring excision | ||||
| EIS–CAD (Nevisense*) | 2 | 644/2389 | 93.5 (91.3 to 95.1) | 32.6 (30.4 to 34.8) |
*For EIS‐CAD the only evidence available was for one system
Key: CAD ‐ computer‐assisted diagnosis; CI ‐ confidence interval; Derm‐CAD ‐ digital dermoscopy‐based computer‐assisted diagnosis; EIS‐CAD ‐ electrical impedance spectroscopy‐based computer‐assisted diagnosis; MSI‐CAD ‐ multispectral imaging‐based computer‐assisted diagnosis.
6. Comparisons of CAD with dermoscopy for detection of the primary target condition: invasive melanoma and atypical intraepidermal melanocytic variants.
| Test | Number of studies | Number of cases | Number of participants | Pooled sensitivity (95% CI) | Pooled specificity (95% CI) |
| Derm‐CAD versus image‐based dermoscopy – indirect comparison | |||||
| Derm‐CAD | 22 | 1063 | 8992 | 90.1 (84.0 to 94.0) | 74.3 (63.6 to 82.7) |
| Image‐based dermoscopy | 5 | 153 | 765 | 93.3 (83.4 to 97.5) | 88.5 (57.3 to 97.8) |
| Difference (95% CI), P value | ‐ | −3.21 (−11.2 to 4.79), P = 0.43 | −14.1 (−34.4 to 6.06), P = 0.17 | ||
| Derm‐CAD versus image‐based dermoscopy – direct comparison | |||||
| Derm‐CAD | 5 | 153 | 765 | 94.1 (89.1 to 96.9) | 80.8 (68.2 to 89.3) |
| Image‐based dermoscopy | 5 | 153 | 765 | 93.9 (85.1 to 97.7) | 88.3 (56.5 to 97.8) |
| Difference (95% CI), P value | ‐ | 0.17 (−6.61 to 6.95), P = 0.96 | −7.44 (−28.4 to 13.6), P = 0.49 | ||
| MSI‐CAD versus image‐based dermoscopy – indirect comparison | |||||
| MSI‐CAD | 8 | 286 | 2401 | 92.9 (83.7 to 97.1) | 43.6 (24.8 to 64.5) |
| Image‐based dermoscopy | 5 | 154 | 371 | 74.0 (66.5 to 80.3) | 58.7 (43.5 to 72.4) |
| Difference (95% CI), P value | ‐ | 18.9 (9.58 to 28.2), P = 0.003 | −15.0 (−40.7 to 10.6), P = 0.26 | ||
| MSI‐CAD versus image‐based dermoscopy – direct comparison | |||||
| MSI‐CAD | 5 | 154 | 371 | 96.8 (92.4 to 98.6) | 29.8 (12.4 to 56.1) |
| Image‐based dermoscopy | 5 | 154 | 371 | 74.0 (66.5 to 80.3) | 58.7 (43.5 to 72.4) |
| Difference (95% CI), P value | ‐ | 22.7 (15.2 to 30.2), P < 0.001 | −28.9 (−56.3 to −1.48), P = 0.039 | ||
| Melafind (MSI–CAD) versus Image‐based dermoscopy – indirect comparison | |||||
| MelaFind | 5 | 196 | 1798 | 97.4 (94.0 to 98.9) | 29.3 (12.1 to 55.6) |
| Image‐based dermoscopy | 4 | 142 | 288 | 72.5 (64.6 to 79.2) | 50.7 (42.6 to 58.7) |
| Difference (95% CI), P value | ‐ | 24.5 (16.5 to 32.4), P < 0.001 | −20.9 (−45.5 to 3.75), P = 0.10 | ||
| Melafind (MSI‐CAD) versus Image‐based dermoscopy – direct comparison | |||||
| MelaFind | 4 | 142 | 288 | 96.5 (91.8 to 98.5) | 22.8 (8.38 to 48.9) |
| Image‐based dermoscopy | 4 | 142 | 288 | 72.5 (64.6 to 79.2) | 50.7 (42.6 to 58.7) |
| Difference (95% CI), P value | ‐ | 23.9 (16.0 to 31.9), P < 0.001 | −27.9 (−50.1 to ‐5.66), P = 0.014 | ||
| Derm‐CAD versus MSI‐CAD ‐ indirect comparison | |||||
| Derm‐CAD | 22 | 1063 | 8992 | 90.1 (84.0 to 94.0) | 74.3 (63.6 to 82.7) |
| MSI‐CAD | 8 | 286 | 2401 | 92.9 (83.7 to 97.1) | 43.6 (24.8 to 64.5) |
| Difference (95% CI), P value | ‐ | 2.83 (−5.04 to 10.7), P = 0.48 |
−30.7 (−53.8 to −7.64), P = 0.009 | ||
Key: CAD ‐ computer‐assisted diagnosis; CI ‐ confidence interval; Derm‐CAD ‐ digital dermoscopy‐ based computer‐assisted diagnosis; MSI‐CAD ‐ multispectral imaging‐based computer‐assisted diagnosis; P ‐ probability.
Target condition: invasive melanoma and atypical intraepidermal melanocytic variants
The diagnostic accuracy of CAD assessment for the detection of invasive cutaneous melanoma or atypical intraepidermal variants was reported by 36 studies with a total of 14,451 lesions including 1889 melanomas. Of these, 23 studies evaluated a Derm–CAD system (total 9082 lesions with 1094 melanomas) and 13 studies evaluated a Spectro‐CAD system (total 5369 lesions with 795 melanomas).
Derm‐CAD
Twenty–two studies were included in a meta–analysis to estimate the accuracy of dermoscopy–based CAD systems in referral settings, regardless of the system manufacturer, algorithm used, or whether CAD was used as a stand–alone automated diagnosis or as a diagnostic aid (Barzegari 2005; Bauer 2000; Binder 1994; Binder 1998; Blum 2004a; Boldrick 2007; Burroni 2004; Cascinelli 1992; Cristofolini 1997; Dreiseitl 2009; Ferris 2015; Gilmore 2010; Maglogiannis 2015; Menzies 2005; Mollersen 2015; Piccolo 2002; Piccolo 2014; Rubegni 2002a; Seidenari 1999; Serrao 2006; Stanganelli 2005; Wollina 2007). We excluded one additional study from this analysis (Seidenari 1998), due to suspected population overlap with an included study (Seidenari 1999). These 22 studies provided 23 datasets for meta–analysis (Mollersen 2015 evaluated two CAD systems, Nevus Doctor and MoleExpert, in the same lesion population). Eleven studies were model derivation studies, eight evaluating the resulting classification algorithm in an independent population (random division of one study group into training and test sets: Binder 1994; Binder 1998; Blum 2004a; Burroni 2004; Ferris 2015; Maglogiannis 2015; Menzies 2005; Stanganelli 2005), and three providing insufficient details to determine the independence of the test population (Cascinelli 1992; Gilmore 2010; Rubegni 2002a). The remaining 11 studies were external validation studies with prospective (Bauer 2000; Cristofolini 1997; Dreiseitl 2009; Wollina 2007), retrospective (Piccolo 2002; Piccolo 2014; Seidenari 1999; Serrao 2006), or unclear (Barzegari 2005; Boldrick 2007; Mollersen 2015) recruitment designs. Only one study did not use excision as an eligibility criterion (Dreiseitl 2009).
Across the 23 datasets, sensitivity ranged from 17% to 100% and specificity from 20% to 98%. We pooled a total of 8992 lesions including 1063 melanomas, giving summary sensitivity of 90.1% (95% confidence interval (CI) 84.0% to 94.0%) and summary specificity of 74.3% (95% CI 63.6% to 82.7%) (Table 6; Figure 8; Figure 9).
8.
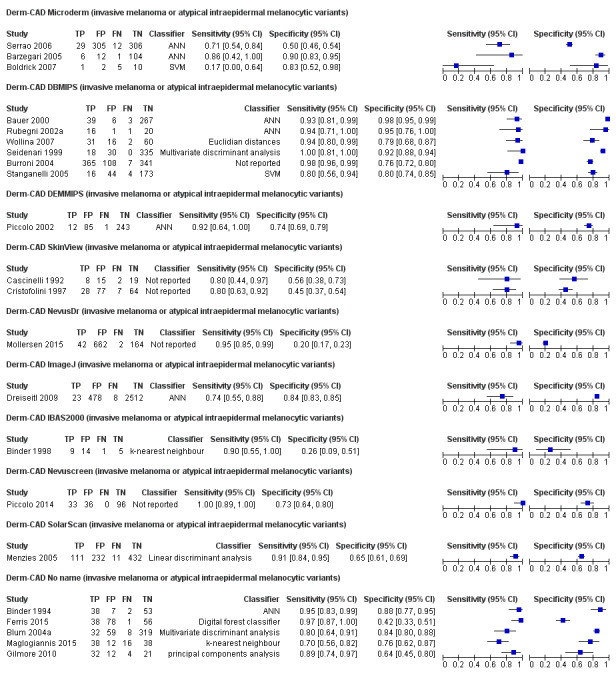
Forest plot of different types of digital dermoscopy CAD systems (DermCAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
9.
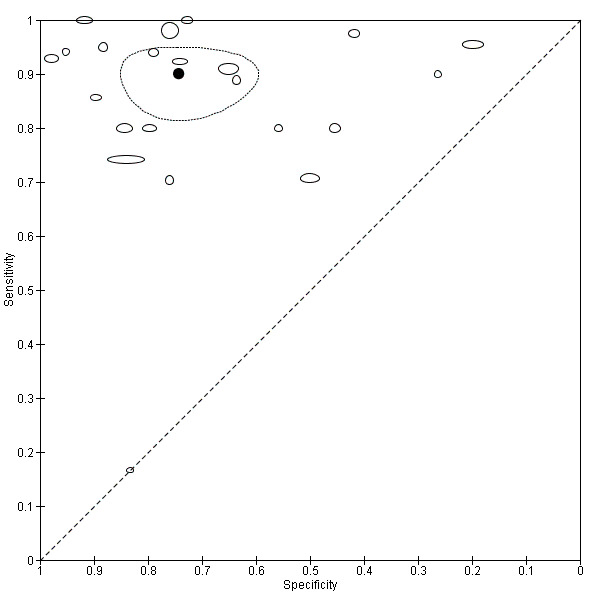
Summary plot of digital dermoscopy CAD systems (Derm‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
The prevalence of melanoma ranged from 1% (Dreiseitl 2009) to 52% (Gilmore 2010; Maglogiannis 2015), and the number of melanomas missed ranged from 0 (Barzegari 2005; Piccolo 2014; Seidenari 1999) to 16 (Maglogiannis 2015) (not reported in four studies). Ten of the 22 studies did not provide clear identification of the target condition (Binder 1994; Blum 2004a; Cascinelli 1992; Cristofolini 1997; Dreiseitl 2009; Gilmore 2010; Maglogiannis 2015; Piccolo 2002; Rubegni 2002a; Stanganelli 2005), and the inclusion of melanoma in situ lesions as disease‐positive was assumed on the basis that the disease‐positive group was described as ‘melanoma’ and not as ‘invasive melanoma’ or ‘malignant melanoma’. Of the 12 studies that clearly reported including in situ lesions (Barzegari 2005; Bauer 2000; Binder 1998; Boldrick 2007; Burroni 2004; Ferris 2015; Menzies 2005; Mollersen 2015; Piccolo 2014; Seidenari 1999; Serrao 2006; Wollina 2007), the percentage of the disease‐positive group (invasive melanoma and atypical intraepidermal melanocytic variants) described as being in situ ranged from 12% to 50%. The number of missed in situ lesions was reported in seven studies, with no missed lesions in three studies (Barzegari 2005; Piccolo 2014; Seidenari 1999) and between 7% (1 of 14, Ferris 2015) and 100% (3 of 3, Boldrick 2007) of in situ lesions misdiagnosed by the remaining studies.
Some studies reported difficulties in excluding a malignancy from clinically benign lesions, particularly non‐melanocytic pigmented lesions such as seborrhoeic keratoses which were reported as an included lesion by five studies (Barzegari 2005; Cascinelli 1992; Ferris 2015; Menzies 2005; Mollersen 2015). Four of these provided lesion diagnoses by CAD result, finding 41 of 61 seborrhoeic keratoses (67%) to have been falsely identified as malignant, while a fifth study highlighted their problematic misclassification of this lesion by both Derm–CAD systems (Mollersen 2015). Other notable false‐positive diagnoses include dysplastic melanocytic naevi (Binder 1994; Binder 1998; Cascinelli 1992; Ferris 2015; Seidenari 1999), actinic keratosis (Barzegari 2005), dermatofibroma (Barzegari 2005; Menzies 2005) and haemangioma (Menzies 2005).
One dataset from one study contributed data for the accuracy of a Derm–CAD system in self–referring patients seeking advice for pigmented naevi (Wollina 2007) (Table 8). This prospective study evaluated the DB–MIPS system, a fully‐integrated dermoscopy unit with internal stereomicroscope, storage database and pattern analysis software, giving it to clinicians to use as a diagnostic aid. Although 3541 lesions (1308 participants) were examined, only the excised lesions (n = 466) were analysed by the authors, of which 357 were recruited in primary‐care clinics. These included 19 melanomas, and 283 dysplastic melanocytic naevi. The authors reported a sensitivity of 89.5% (95% CI 68.6% to 97.1%) and specificity of 84.0% (95% CI 79.7% to 87.5%).
7. Sensitivity and Specificity of CAD systems in Unreferred Populations.
| Derm‐CAD | ||||||||
| CAD System | Study | Target condition | TP | FP | FN | TP | Sensitivity (95% CI) | Specificity (95% CI) |
| DB‐MIPS | Wollina 2007 | MEL | 17 | 54 | 2 | 284 | 89.5 (68.6 to 97.1) | 84.0 (79.7 to 87.5) |
| MSI‐CAD | ||||||||
| CAD System | Study | Target condition | TP | FP | FN | TP | Sensitivity (95% CI) | Specificity (95% CI) |
| MoleMate SIAscope | Walter 2012 | MEL | 18 | 209 | 0 | 539 | 100 (82.4 to 100) | 72.1 (68.7 to 75.2) |
| MoleMate SIAscope | Walter 2012 | MM | 14 | 213 | 0 | 539 | 100 (78.5 to 100) | 71.7 (68.4 to 74.8) |
| MoleMate SIAscope | Walter 2012 | Any | 23 | 204 | 2 | 537 | 92.0 (75.0 to 97.8) | 72.5 (69.1 to 75.6) |
| SIAscope | Sgouros 2014 | Any | 26 | 7 | 5 | 6 | 83.9 (67.4 to 92.9) | 46.2 (23.2 to 70.9) |
Any ‐ any skin cancer or lesion requiring excision (secondary objective); CI ‐ confidence interval; Derm‐CAD ‐ digital dermoscopy‐based computer‐assisted diagnosis; FN ‐ false negative; FP ‐ false positive; MEL ‐ invasive melanoma and atypical intraepidermal melanocytic variants (primary objective); MM ‐ malignant (invasive) melanoma only (secondary objective); MSI‐CAD ‐ multispectral imaging‐based computer‐assisted diagnosis; TN ‐ true negative; TP ‐ true positive
Derm‐CAD versus Dermoscopy
Seven studies (32%) reported accuracy data comparing Derm–CAD with dermoscopy for the detection of invasive melanoma and atypical intraepidermal melanocytic variants, providing seven datasets (4104 lesions including 226 melanomas and no other malignancies). A further four studies reported accuracy data for dermoscopy in a subsample of the total study population, and so we excluded them from analysis (Blum 2004a; Ferris 2015; Menzies 2005; Stanganelli 2005).
Five of the seven studies compared Derm–CAD to diagnosis by expert dermoscopists using dermoscopic images alone (Binder 1994; Gilmore 2010; Piccolo 2014; Seidenari 1998) or alongside clinical images (Piccolo 2002) in 765 lesions (153 melanomas). Two studies compared face‐to‐face dermoscopic diagnosis by an expert dermatologist with Derm–CAD systems DB–MIPS (Bauer 2000) and Image J (Dreiseitl 2009).
The accuracy of Derm‐CAD was compared with the accuracy of dermoscopy in:
(a) all 22 Derm‐CAD studies (8992 lesions and 1063 melanomas) and the five image‐based dermoscopy studies (765 lesions and 153 melanomas) in an indirect comparison (Figure 10; Figure 11), and
10.
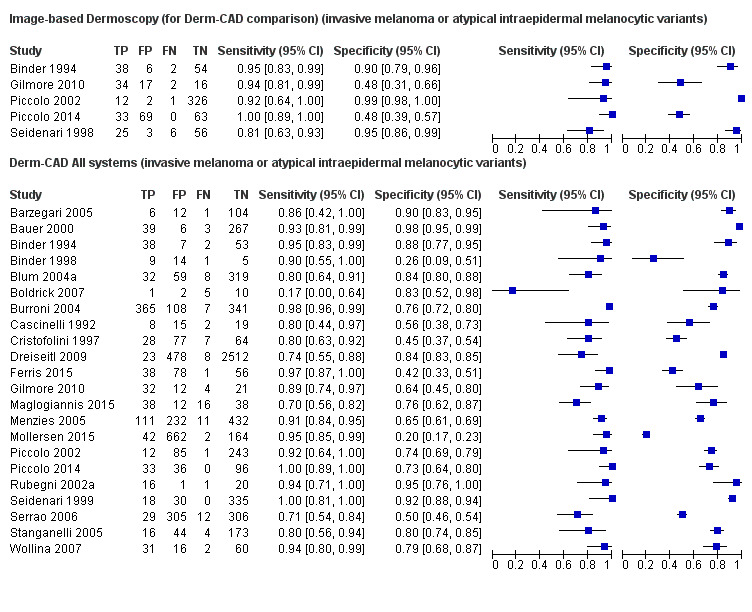
Forest plot of data for image‐based dermoscopy diagnosis and digital dermoscopy CAD systems (Derm‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
11.
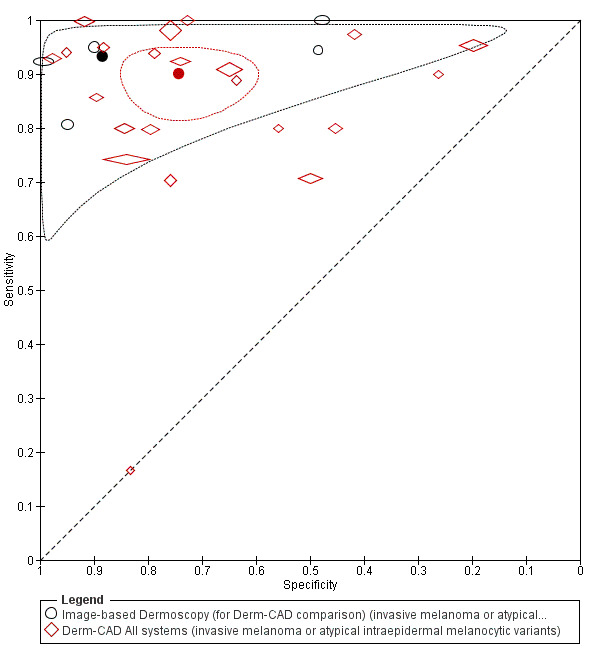
Summary plot of image‐based dermoscopy diagnosis versus digital dermoscopy CAD systems (Derm‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
(b) direct comparisons in the subset of five studies that evaluated both Derm‐CAD and image‐based dermoscopy (765 lesions and 153 melanomas; Figure 12; Appendix 7).
12.
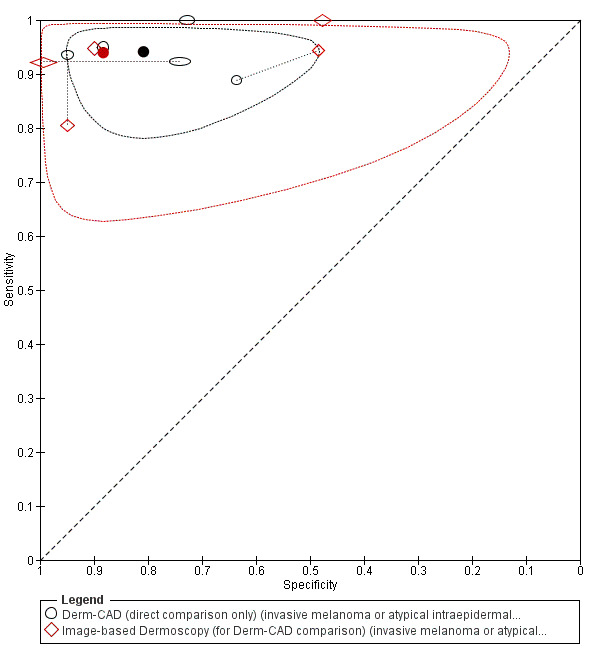
Summary plot of direct comparisons between image‐based dermoscopy diagnosis versus digital dermoscopy CAD systems (Derm‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
In both comparisons similar sensitivities were observed but with lower specificity for CAD compared to dermoscopy, but none of the observed differences were statistically significant (Table 7). For the indirect comparison, the difference (95% CI) in summary sensitivities (Derm‐CAD 90.1% versus dermoscopy 93.3%) was −3.21% (−11.2% to 4.79%), P = 0.43; the difference (95% CI) in summary specificities (Derm CAD 74.3% versus dermoscopy 88.5%) was −14.1% (−34.4% to 6.06%), P = 0.17. For the direct comparison the difference (95% CI) in summary sensitivities (Derm–CAD 94.1% versus dermoscopy 93.9%) was 0.17% (−6.61% to 6.95%), P = 0.96; the difference (95% CI) in summary specificities (CAD‐Derm 80.8% versus dermoscopy 88.3%) was −7.44% (−28.4% to 13.6%), P = 0.49.
Contrasting differences in accuracy were produced by the two studies comparing Derm–CAD with face–to–face dermoscopic diagnosis by an expert (Bauer 2000; Dreiseitl 2009; Table 9), but these differences were imprecise.
8. Direct comparisons of single CAD studies with dermoscopy for diagnosis of melanoma and other types of skin cancer.
| Study | Sensitivity (true positives/cases) % | Difference (95% CI) | Specificity (true negatives/non cases) % | Difference (95% CI) | ||
| MSI–CAD study comparisons: invasive melanoma and atypical intraepidermal melanocytic variants | ||||||
| MSI–CAD | In–person dermoscopy | MSI–CAD | In–person dermoscopy | |||
| Bono 2002 | 80.3 (53/66) | 90.9 (60/66) | −10.6 (−22.8 to 1.58) | 49.0 (121/247) | 74.5 (184/247) | −25.5 (−33.5 to −17.0) |
| MSI–CAD study comparisons: Invasive melanoma | ||||||
| MSI‐CAD | In‐person dermoscopy | MSI‐CAD | In‐person dermoscopy | |||
| Ascierto 2010 | 66.7 (8/12) | 100 (12/12) | −33.3 (−60.9 to −2.20) | 76.2 (32/42) | 45.2 (19/42) | 31.0 (10.1 to 48.4) |
| MelaFind (MSI‐CAD) | Image‐based dermoscopy | Melafind (MSI‐CAD) | Image‐based dermoscopy | |||
| Friedman 2008 | 100 (21/21) | 81.0 (17/21) | 19.1 (−0.15 to 40.0) | 29.5 (23/78) | 44.9 (35/78) | −15.4 (−29.6 to −0.23) |
| Derm–CAD comparisons: invasive melanoma and atypical intraepidermal melanocytic variants | ||||||
| Derm‐CAD | In‐person dermoscopy | Derm‐CAD | In‐person dermoscopy | |||
| Bauer 2000 | 92.9 (39/42) | 78.6 (33/42) | 14.3 (−1.06 to 29.5) | 97.8 (267/273) | 96.3 (263/273) | 1.47 (−1.55 to 4.64) |
| Dreiseitl 2009* | 88.9 (24/27) | 96.3 (26/27) | −7.41 (−24.6 to 8.88) | 48.0 (207/431) | 71.9 (310/431) | −23.9 (−30.1 to −17.4) |
| Unnamed system Derm‐CAD | Image‐based dermoscopy | Unnamed system Derm–CAD | Image‐based dermoscopy | |||
| Menzies 1996 | 88.9 (40/45) | 91.1 (41/45) | −2.22 (−15.7 to 11.2) | 75.6 (90/119) | 70.6 (84/119) | 5.04 (−6.21 to 16.1) |
| DB‐MIPS (Derm‐CAD) | In‐person dermoscopy | DB‐MIPS (Derm‐CAD) | In‐person dermoscopy | |||
| Bauer 2000 | 92.9 (39/42) | 78.6 (33/42) | 14.3 (−1.06 to 29.5) | 97.8 (267/273) | 96.3 (263/273) | 1.47 (−1.55 to 4.64) |
|
DB‐MIPS (Derm–CAD) |
Image‐based dermoscopy |
DB‐MIPS (Derm–CAD) |
Image‐based dermoscopy | |||
| Seidenari 1998 | 93.5 (29/31) | 80.6 (25/31) | 12.9 (−4.62 to 30.5) | 94.9 (56/59) | 94.9 (56/59) | 0.00 (−9.44 to 9.44) |
| CAD‐based diagnosis vs CAD‐aided diagnosis: invasive melanoma and atypical intraepidermal melanocytic variants | ||||||
| CAD‐based diagnosis | CAD‐aided diagnosis | CAD‐based diagnosis | CAD‐aided diagnosis | |||
| Dreiseitl 2009* | 88.9 (24/27) | 74.1 (20/27) | 14.8 (−6.4 to 34.9) | 48.0 (207/431) | 81.9 (353/431) | −33.9 (39.6 to −27.7) |
| Hauschild 2014 | 96.9 (63/65) | 78.5 (51/65) | 18.5 (7.3 to 30.1) | 9.2 (6/65) | 46.2 (30/65) | −36.9 (−49.9 to −22.0) |
*Patient‐based analysis, unlike other studies in the table which were lesion‐based. The Dreiseitl 2009 study did report lesion‐based for the accuracy of CAD‐only Derm‐CAD, used in the meta‐analysis for Derm‐CAD accuracy to detect invasive melanoma or atypical intraepidermal melanocytic variants (data presented in Figure 10).
Key: CAD ‐ computer‐assisted diagnosis; CI ‐ confidence interval; Derm‐CAD ‐ digital dermoscopy‐based computer‐assisted diagnosis; MSI‐CAD ‐ multispectral imaging‐based computer‐assisted diagnosis.
Spectro‐CAD
We meta‐analysed eight datasets to estimate the accuracy of MSI‐CAD systems in referral settings, regardless of the system make, algorithm used, or whether CAD was used as a stand–alone automated diagnosis or as a diagnostic aid (Friedman 2008; Glud 2009; Gutkowicz Krusin 1997; Monheit 2011; Tomatis 2003; Tomatis 2005; Wells 2012; Winkelmann 2016). We excluded one other dataset (Hauschild 2014) from this analysis as it was a subgroup of Monheit 2011. Four were model derivation studies (Friedman 2008; Gutkowicz Krusin 1997; Tomatis 2003; Tomatis 2005), two of which validated the resulting classification algorithm in an independent population (Friedman 2008;Tomatis 2005), and the remaining four were prospective (Glud 2009;Monheit 2011) or retrospective (Wells 2012;Winkelmann 2016) external validation studies. Melanoma prevalence ranged from 7% (Monheit 2011) to 49% (Friedman 2008;Wells 2012). These eight datasets evaluated 2537 excised lesions with 296 melanomas, with sensitivities ranging from 76% (Tomatis 2003) to 100% (Glud 2009;Gutkowicz Krusin 1997), and specificities ranging from 8% (Wells 2012) to 77% (Tomatis 2005). The pooled sensitivity was 92.9% (95% CI 83.7% to 97.1%) and specificity was 43.6% (95% CI 24.8% to 64.5%) (Table 6; Figure 13; Figure 14). The number of melanomas missed ranged from 0 (Glud 2009;Gutkowicz Krusin 1997) to 9 (Tomatis 2003), although the reporting of false‐positive diagnoses was poor so we could not analyse the data.
13.
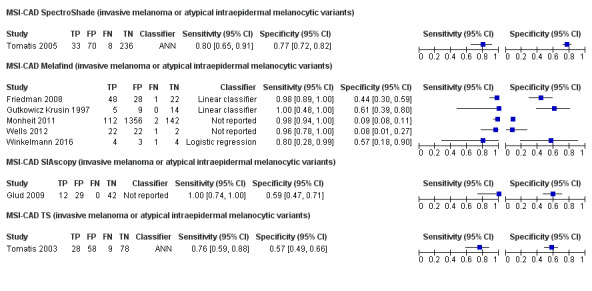
Forest plot of different types of multi‐spectral imaging CAD (MSI‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
14.
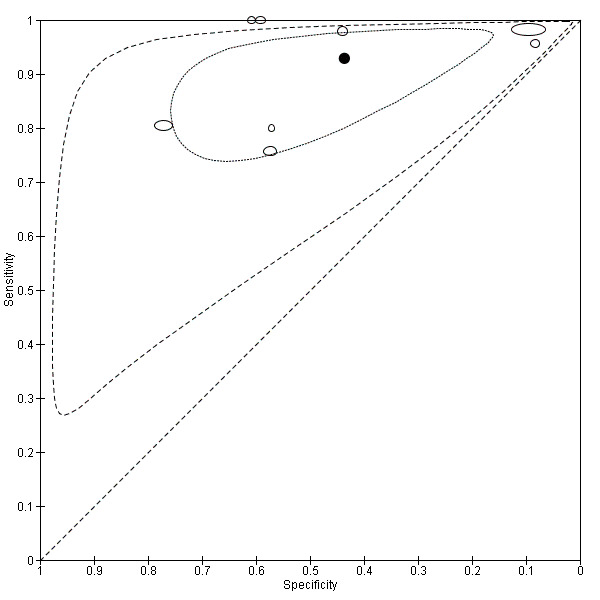
Summary plot of multi‐spectral imaging CAD (MSI‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
One study evaluated a Spectro–CAD system based on the analysis of diffuse reflectance spectroscopy data, a non–imaging CAD system called OIDRS (Garcia Uribe 2012). This model derivation study prospectively recruited 136 pigmented skin lesions with a melanoma prevalence of 7% (final diagnosis determined by histology following biopsy). OIDRS was found to operate with a sensitivity of 90.0% (95% CI 59.6% to 98.2%) and specificity of 89.7% (95% CI 83.2% to 93.9%) (Table 10).
9. Sensitivity and specificity of oblique incidence diffuse reflectance spectrometry CAD (OIDRS–CAD) studies for primary target condition: invasive melanoma and atypical intraepidermal melanocytic variants.
| Index test, target condition | True positives | False positives | False negatives | True negatives | Sensitivity (95% CI) % | Specificity (95% CI) % |
| Garcia Uribe 2012 | 9 | 13 | 1 | 113 | 90.0 (59.6 to 98.2) | 89.7 (83.2 to 93.9) |
The two studies evaluating Nevisense, a CAD system using electrical impedance spectroscopy (EIS), included one internal validation study (Mohr 2013) and one external validation study (Malvehy 2014) (Table 11). Pooling 2389 lesions with 368 melanomas, the summary sensitivity was 97% (95% CI 94.7% to 98.3%) and specificity was 33.6% (95% CI 31.6% to 35.7%) (Table 6). Both studies reported considerable difficulties in the ability of Nevisense to identify seborrhoeic keratoses, with 69/73 (95%) such lesions falsely identified as malignant. Other notable false‐positive diagnoses included dysplastic melanocytic naevi, actinic keratoses and dermatofibroma.
10. Sensitivity and specificity of electrical impedance spectroscopy CAD (EIS–CAD) studies for each primary target condition.
| Index test, target condition | True positives | False positives | False negatives | True negatives | Sensitivity (95% CI) % | Specificity (95% CI) % |
| Invasive melanoma and atypical intraepidermal melanocytic variants | ||||||
| Mohr 2013 | 101 | 246 | 2 | 97 | 98.1 (93.2 to 99.5) | 28.3 (23.8 to 33.3) |
| Malvehy 2014 | 256 | 1095 | 9 | 583 | 96.6 (93.7 to 98.2) | 34.7 (32.5 to 37.1) |
| Basal cell carcinoma | ||||||
| Mohr 2013 | 21 | 351 | 0 | 74 | 100 (84.5 to 100) | 17.4 (14.1 to 21.3) |
| Malvehy 2014 | 48 | 1359 | 0 | 536 | 100 (92.6 to 100) | 28.3 (26.3 to 30.4) |
| Cutaneous Squamous cell carcinoma | ||||||
| Malvehy 2014 | 7 | 1095 | 0 | 841 | 100 (64.6 to 100) | 43.4 (41.3 to 45.7) |
One study evaluated the accuracy of an MSI–CAD system in an unreferred population (Walter 2012). A series of participants with pigmented skin lesions who were presenting for a first clinical assessment were prospectively recruited and imaged by GPs, using a SIAscope™ coupled with MoleMate, a multispectral imaging device with viewing platform and integrated Primary Care Scoring Algorithm (developed by Emery 2010). This was originally an RCT comparing patient referrals by GPs using MSI–CAD (experimental arm) with GP clinical assessment only (control arm), and we extracted the experimental arm data as a prospective case series evaluating MSI–CAD against histology or clinical follow‐up of at least three months for lesions considered benign, as well as expert diagnosis without follow‐up for some benign lesions. Overall 36 melanomas were identified and an additional 209 false diagnoses made amongst 766 included lesions; MSI–CAD sensitivity was 100% (95% CI 82.4% to 100%) and specificity was 72.1% (95% CI 68.7% to 75.2%) (Table 8).
Spectro–CAD versus Dermoscopy
Six MSI–CAD studies (67%) also evaluated the accuracy of dermoscopy, providing six datasets (684 lesions and 229 malignancies comprising 220 melanomas, 8 BCCs and no cSCCs). One of these compared Derm–CAD to in–person dermoscopic diagnosis (Bono 2002), and five studies evaluated dermoscopy using expert dermoscopists (Friedman 2008; Glud 2009; Hauschild 2014) or dermatologists of unreported expertise (Wells 2012;Winkelmann 2016) to interpret stored dermoscopic images of 371 lesions, including 154 melanomas. Four studies also provided additional diagnostic information to clinicians in the form of clinical examination notes (Friedman 2008; Hauschild 2014; Wells 2012) or clinical images (Winkelmann 2016).
The accuracy of MSI‐CAD was compared with the accuracy of dermoscopy in:
(a) all eight MSI‐CAD studies (2401 lesions and 286 melanomas) and the five image‐based dermoscopy studies (371 lesions and 154 melanomas) in an indirect comparison (Figure 15; Figure 16), and
15.
Forest plot of data for image‐based dermoscopy diagnosis and multi‐spectral imaging CAD systems (MSI‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
16.
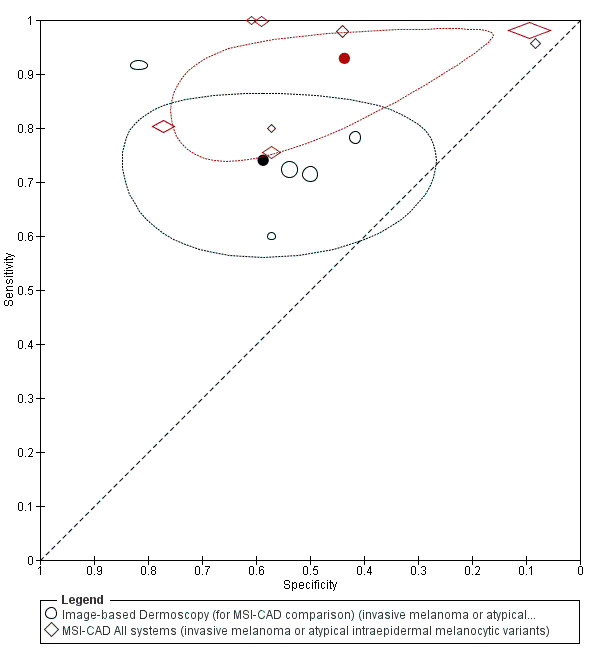
Summary plot of image‐based dermoscopy diagnosis versus multi‐spectral imaging CAD systems (MSI‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
(b) direct comparisons in the subset of five studies that evaluated both MSI‐CAD and image‐based dermoscopy (371 lesions and 154 melanomas; Figure 17; Appendix 7).
17.
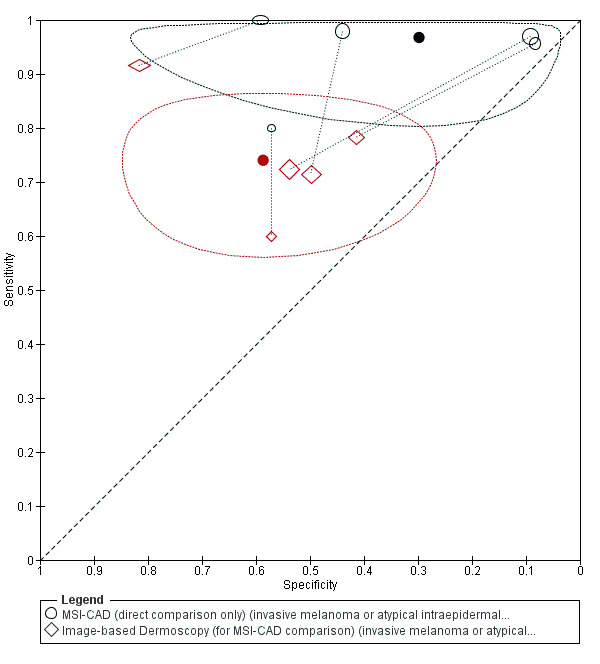
Summary plot of direct comparisons between image‐based dermoscopy diagnosis versus multi‐spectral imaging CAD systems (MSI‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
In both comparisons MSI–CAD was significantly more sensitive with lower specificity, although differences in specificity were only significant for (b), the direct comparison (Table 7). For the indirect comparison (a), the difference (95% CI) in summary sensitivities (MSI–CAD 92.9% versus dermoscopy 74.0%) was 18.9% (9.58% to 28.2%), P = 0.003; the difference (95% CI) in summary specificities (MSI–CAD 43.6% versus dermoscopy 58.7%) was −15.0% (−40.7% to 10.6%), P = 0.26. For the direct comparison (b), the difference (95% CI) in summary sensitivities (MSI–CAD 96.8% versus dermoscopy 74.0%) was 22.7% (15.2% to 30.2%), P < 0.001; the difference (95% CI) in summary specificities (MSI–CAD 29.8% versus dermoscopy 58.7%) was −28.9% (−56.3% to −1.48%), P = 0.039.
Four of the five image–based dermoscopy studies used the MelaFind system (Friedman 2008; Hauschild 2014; Wells 2012; Winkelmann 2016), with the same impact on sensitivity (MelaFind 96.5% versus dermoscopy 72.5%; difference (95% CI) of 23.9% (16.0% to 31.9%), P < 0.001) and specificity (MelaFind 22.8% versus dermoscopy 50.7%; difference (95% CI) −27.9% (−50.1% to −5.66%), P = 0.014) (Table 7).
One study compared the accuracy of MSI–CAD to in–person diagnosis by an expert dermatologist (Bono 2002), and we present the results in Table 9.
Derm–CAD versus Spectro–CAD
None of the studies directly compared the accuracy of Derm‐CAD and MSI‐CAD. An indirect comparison of MSI–CAD (8 studies) and Derm–CAD (22 studies) demonstrated similar sensitivities (Derm–CAD 90.1% versus MSI–CAD 92.9%) with a difference (95% CI) of 2.83% (−5.04% to 10.7%), P = 0.48. However, specificity was lower for MSI–CAD (43.6%) compared to Derm‐CAD (74.3%) with a difference of −30.7% (−53.8% to −7.64%), P = 0.009 (Table 7).
Secondary analyses for the detection of invasive melanoma and atypical intraepidermal melanocytic variants
Accuracy of individual Derm‐CAD systems
Our ability to compare the accuracy of individual CAD systems was limited by lack of data. A sufficient number of datasets to allow separate pooling were available only for the DB‐MIPS system. Six studies evaluated the DB–MIPS system, using varying classification algorithms, in a total of 1903 lesions including 502 melanomas (Bauer 2000; Burroni 2004; Rubegni 2002a; Seidenari 1999; Stanganelli 2005; Wollina 2007; we dropped a seventh study (Seidenari 1998) from this analysis due to population overlap with Seidenari 1999). Three were external validation studies (Bauer 2000; Seidenari 1999; Wollina 2007), and all six included only excised lesions. Summary estimates of sensitivity were 95.2% (95% CI 89.5% to 97.9%) and specificity 89.1% (95% CI 78.7% to 94.8%) (Table 6; Figure 8; Figure 18).
18.
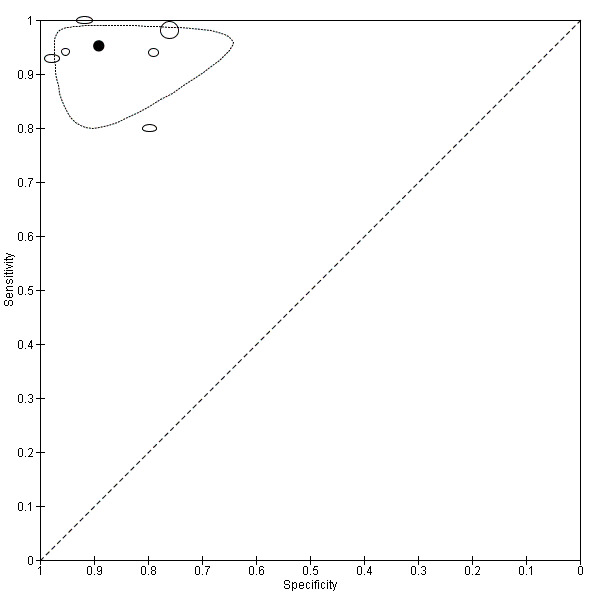
Summary plot for the multi‐spectral imaging CAD system (MSI‐CAD) DBMIPS for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
Three external validation studies evaluated the MicroDERM system in a total of 793 lesions with 54 melanomas. However, due to the limited number of studies and substantial variability between studies, we did not perform meta‐analysis. Sensitivities ranged from 17% (95% CI 0% to 64%) to 86% (95% CI 42% to 100%) and specificities from 50% (95% CI 46% to 54%) to 90% (95% CI 83% to 95%) (Barzegari 2005; Boldrick 2007; Serrao 2006). These represent accuracy for thresholds using DANAOS scores of ≥ 6.5 (Serrao 2006), ≥ 7 (Boldrick 2007) and ≥ 7.34 (Barzegari 2005). The outlying sensitivity of 17% observed in Boldrick 2007 is likely due to a skewed sample of lesions, since of the 1000 pigmented skin lesions assessed in the study, only 18 received an eligible reference standard (histology, clinical follow–up was not reported) and so could be included. The vast majority of the original sample (982/1000) were clinically diagnosed as benign lesions not requiring excision.
Two studies evaluated the Skin View system in 220 excised lesions with 45 melanomas (Cascinelli 1992;Cristofolini 1997). The earlier publication included a model validation phase using internal validation (Cascinelli 1992), while Cristofolini 1997 performed an external validation only. Summary estimates of sensitivity were 80.0% (95% CI 65.8% to 89.3%) and specificity 47.4% (95% CI 40.1% to 54.8%) (Table 6; Figure 8).
Accuracy of individual Spectro‐CAD systems
Five MSI–CAD studies evaluated the MelaFind system in a total of 1798 lesions including 196 melanomas (Friedman 2008; Gutkowicz Krusin 1997; Monheit 2011; Wells 2012; Winkelmann 2016). Summary estimates of sensitivity were 97.1% (95% CI 91.9% to 98.9%) and specificity 29.8% (95% CI 12.3% to 56.3%) (Table 6; Figure 11; Figure 19).
19.
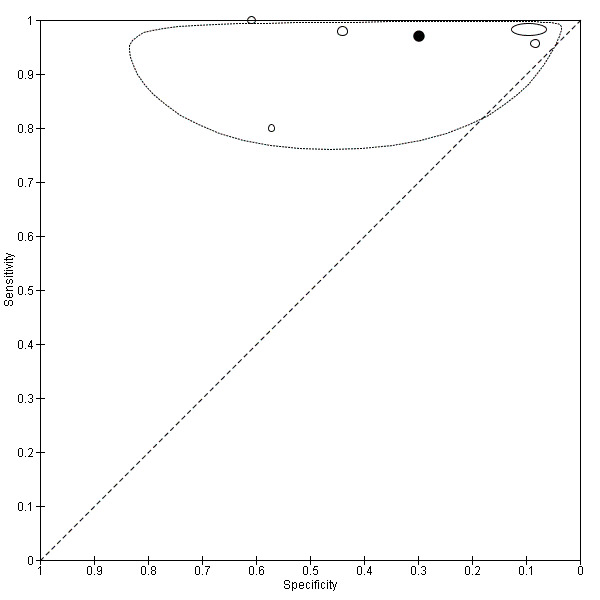
Summary plot for the multi‐spectral imaging CAD system (MSI‐CAD) MelaFind for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
Both EIS–CAD studies reported above evaluated the Nevisense system, in a total of 2389 lesions with 368 melanomas, giving a summary sensitivity of 97.0% (95% CI 94.7% to 98.3%) and specificity of 33.6% (95% CI 31.6% to 35.7%).
CAD‐only performance versus CAD‐aided performance
In sensitivity analyses, we excluded studies that used CAD as a diagnostic aid. Three of the 22 Derm–CAD studies evaluated the Derm–CAD system as a diagnostic aid in referral settings (Bauer 2000; Piccolo 2014; Wollina 2007). Two of the six DB–MIPS studies (both prospective external validation studies) assessed the system as a diagnostic aid. For MSI–CAD, one of the eight studies used CAD as a diagnostic aid in a referral setting (MelaFind, Winkelmann 2016). The results of the sensitivity analyses are shown in Appendix 8. The results indicate very similar findings to the main analyses.
Direct evidence was available from one study (Dreiseitl 2009) which compared Derm–CAD computer diagnoses (CAD only) with diagnoses produced by clinicians using Derm–CAD as a diagnostic aid (CAD–aided). In an external validation study, Dreiseitl 2009 compared CAD–only performance of MoleMax II analysed with the Image J software programme (using a neural network classifier), with CAD–aided diagnosis performed by dermatologists with high experience, low experience, and a third cohort with mixed experience. In this prospective study, 458 consecutive participants were included who were referred to a secondary‐care centre for further investigation of 3021 suspicious pigmented skin lesions. While lesion–based results were reported for the CAD–only diagnosis (pooled for the primary objective Derm‐CAD accuracy estimates in Figure 10), only participant–based results were provided for its comparison to CAD–aided diagnoses; it is notable that the lesion–based sensitivity and specificity for CAD‐only diagnosis differ substantially from the participant–based estimates for CAD‐only diagnosis (74% versus 89% sensitivity, 84% versus 48% specificity), which is to be expected when many more lesions were free of disease (true negatives n = 2512) than were participants (n = 207). The within–study results for the comparison to CAD‐aided diagnosis are reported in Table 9.
A case–control reader study (Hauschild 2014), undertaken in a referred setting, was the only direct comparison of an MSI CAD–only diagnosis with MSI CAD–aided diagnosis. The study included 65 melanomas and 65 benign pigmented skin lesions that had been excised to evaluate the ability of MelaFind to accurately recommend biopsy in pigmented skin lesions. The study sample was a randomly‐selected subset of the consecutively‐recruited, prospective Monheit 2011 population. Differences (95% CI) are reported in Table 9.
Target Condition: Basal Cell Carcinoma (BCC)
Derm‐CAD
Four study populations from referred settings included BCC lesions (Cascinelli 1992; Ferris 2015; Menzies 1996; Mollersen 2015); however two did not provide adequate data to derive 2 x 2 data (Menzies 1996; Mollersen 2015) and one included fewer than the minimum of five lesions to meet our inclusion criteria for this question (Cascinelli 1992). The remaining study evaluated 11 BCCs amongst 173 dermoscopically atypical lesions using the output of an unnamed CAD system (CAD–based diagnosis), in a retrospective case series. Three BCCs were missed, giving a sensitivity of 73% (95% CI 43.4% to 90.3%) with 108 false positives, giving a specificity of 33% (95% CI 26.5% to 40.9%) for the detection of BCC (Ferris 2015). The study used dermoscopic images of skin lesions excised on the basis of clinical suspicion of malignancy, and also included melanomas, cSCCs, seborrhoeic keratosis and benign melanocytic lesions. The very low specificity was as a result of misclassification of seborrhoeic keratoses (7/11), low–grade dysplastic naevi (27/47) and lentigo (8/10) as malignant lesions.
Spectro‐CAD
Two studies evaluated the ability of a Spectro–CAD system to detect BCC lesions, both using the EIS–CAD Nevisense system in participants with suspected melanoma referred for excision (Malvehy 2014; Mohr 2013) (Table 11). Of 2389 analysed lesions, all 69 BCCs were identified, giving a summary sensitivity of 100% (95% CI 94.7% to 100%) and very low specificity of 26.3% (95% CI 24.5% to 28.1%). Since both populations were recruited to rule out melanoma, few benign keratotic lesions were included (including lichenoid keratosis (n = 4), seborrhoeic keratosis (n = 73) and actinic keratosis (n = 8)), a factor which may have contributed to the very low specificity.
Target condition: cutaneous squamous cell carcinoma (cSCC)
The only study to evaluate the performance of a CAD system to detect cSCC used the EIS–based Nevisense system in referred patients, identifying all seven cSCCs amongst 1943 lesions to give a sensitivity of 100% (95% CI 64.6% to 100%) and specificity of 43.4% (95% CI 41.3% to 45.7%) (Malvehy 2014) (Table 11). The high sensitivity could have been influenced by the study's melanoma–focused recruitment selection which resulted in very few benign different diagnoses being included (lichenoid keratosis (n = 4), seborrhoeic keratosis (n = 51) and actinic keratosis (n = 8)).
Secondary target conditions: invasive melanoma alone
The diagnostic accuracy of CAD assessment for the detection of invasive cutaneous melanoma alone was reported by seven studies for a total of 1336 lesions with 236 invasive melanomas.
Of these, two studies evaluated Derm–CAD systems (total 950 lesions with 120 invasive melanomas): SolarScan (Menzies 2005) and an unnamed system (Menzies 1996), both of which included model derivation within the same population (Menzies 1996 did not report the method of dividing lesions into training and test sets). Both also included pigmented skin lesions referred for excision (Menzies 1996; Menzies 2005), of which one may have included BCCs amongst the target disease‐negative group (Menzies 1996 included 18 BCCs in the full study population but did not report how many were included in the independent test set). Melanoma prevalence was 10% in Menzies 2005 and 27% in Menzies 1996. Meta–analysis of these studies provided a summary sensitivity estimate of 90.8% (95% CI 84.2% to 94.9%) and summary specificity of 63.5% (95% CI 60.2% to 66.7%) (Table 6). Menzies 2005 also reported data for the primary target condition, invasive melanoma and atypical intraepidermal melanocytic variants, with little difference in sensitivity (92.0% invasive melanoma only versus 91.0%, 1.0% difference (95% CI −8.7% to 8.2%)) or specificity (61.5% invasive melanoma only versus 65.1%, −3.6% difference (95% CI −8.7% to 1.5%)). Despite in situ lesions making up 39% (47/122) of disease positives, similar proportions of in situ (5/47) and invasive lesions (6/75) were missed by the SolarScan.
One Derm–CAD study, retrospective and of uncertain design, also allowed a comparison with image–based dermoscopy in 164 lesions containing 45 invasive melanomas (Menzies 1996). Observer experience was not reported. Accuracy estimates were similar and are reported in Table 9.
Five studies evaluated an MSI‐CAD system (total 386 lesions with 116 invasive melanomas), regardless of the system make, algorithm used, or whether CAD was used as a stand–alone automate diagnosis or as a diagnostic aid. All were conducted in lesions referred for excision, four solely in pigmented skin lesions (Ascierto 2010; Bono 1996; Friedman 2008; Hauschild 2014) and the fifth in any lesion clinically suspected of being a melanoma (Terstappen 2013). Systems evaluated were MelaFind (Friedman 2008;Hauschild 2014), SpectroShade (Ascierto 2010), Telespectrophotometric System (Bono 1996) and SIAscope, version V (Terstappen 2013).
Sensitivities ranged from 24% (Terstappen 2013) to 100% (Friedman 2008), and specificities from 29% (Friedman 2008) to 84% (Terstappen 2013). The study producing the highest sensitivity and lowest specificity (Friedman 2008) excluded all high‐grade dysplastic lesions and therefore included only melanomas, BCCs (n = 2), low‐grade dysplastic naevi (n = 32) and other benign melanocytic lesions (n = 14). The lowest sensitivity and highest specificity were produced by Terstappen 2013, who further selected their population of clinically suspicious lesions to include only lesions with a positive CAD result (SIAscope). Both datasets that evaluated the MelaFind system (Hauschild 2014;Friedman 2008) generated high sensitivities (81% and 100%) and low specificities (39% and 29%). These five studies (386 lesions including 116 invasive melanomas) gave a summary sensitivity estimate of 76.5% (95% CI 43.0% to 93.3%) and summary specificity of 60.7% (95% CI 38.5% to 79.2%) (Figure 20; Figure 21; Table 6).
20.
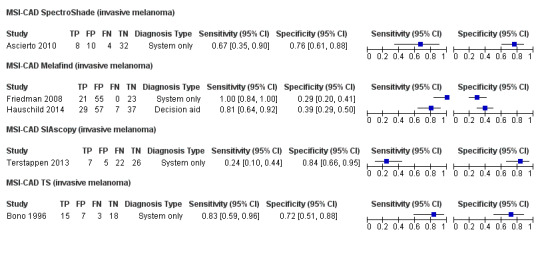
Forest plot of different types of multi‐spectral imaging CAD system (MSI‐CAD) for the detection of invasive melanoma alone (invasive melanoma)
21.
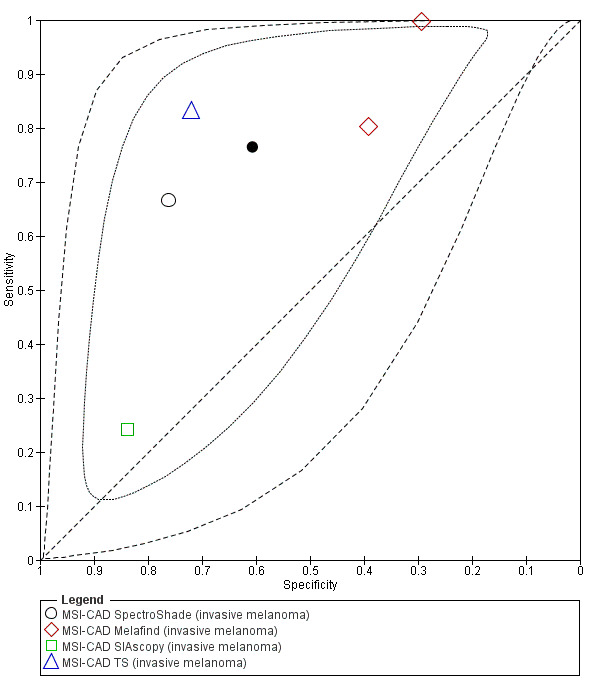
Summary plot of different types of multi‐spectral imaging CAD (MSI‐CAD) for the detection of invasive melanoma alone (invasive melanoma)
In sensitivity analyses, we excluded one study that used CAD as a diagnostic aid, in a referral setting (Hauschild 2014). The results of the sensitivity analysis are shown in Appendix 8. The results indicate very similar findings to the main analysis.
One study compared MSI–CAD (MelaFind) to diagnosis by expert dermoscopists using dermoscopic images in 99 lesions including 21 invasive melanomas, finding Melafind’s sensitivity to be higher (100% versus 81%) and specificity lower (29% versus 45%) than dermoscopy (Friedman 2008) (Table 9).
One other study compared MSI–CAD (SpectroShade) to face–to–face diagnosis by expert dermatologists in 54 lesions with 12 invasive melanomas, finding SpectroShade’s sensitivity to be lower (67% versus 100%) and specificity higher (76% versus 45%) than dermoscopy (Ascierto 2010) (Table 9).
The two studies evaluating the EIS–based Nevisense system produced accuracy data for 2389 lesions with 226 invasive melanomas, giving a summary sensitivity of 98.2% (95% CI 95.4% to 99.3%) and specificity of 38.0% (95% CI 36.0% to 40.1%) (Malvehy 2014; Mohr 2013) (Table 6).
A single study contributed data for the accuracy of detecting invasive melanoma in an unreferred setting, reporting the use of MoleMate with SIAscope by GPs in 766 lesions with 14 invasive melanomas (Walter 2012). No melanomas were missed: sensitivity 100% (95% CI 78.5% to 100%); although false‐positive findings were high, giving a specificity of 71.7% (95% CI 68.4% to 74.8%) (Table 8).
Secondary target conditions: any lesion requiring excision
Derm‐CAD
Four datasets from three studies (Cascinelli 1992; Ferris 2015; Mollersen 2015) provided data to evaluate the accuracy of Derm–CAD to detect any skin cancer or other atypical lesion requiring excision in referred settings. Two were retrospective derivation studies using stored dermoscopic images to train either the Skin View system in clinically suspect pigmented skin lesions referred for excision (Cascinelli 1992), or an unnamed system in lesions suspected of malignancy also referred for excision (Ferris 2015). The third study conducted a head–to–head external validation comparison of two systems, in a case series of pigmented and nonpigmented (if melanoma, BCC, or SCC was a potentially different diagnosis) skin lesions scheduled for excision (Mollersen 2015). The Nevus Doctor system was the subject of evaluation as a system still in development, being compared against the commercially available Mole Expert. The three studies provided a total of 1087 lesions, with the 186 malignancies including 83 BCCs, 9 cSCCs and 1 adnexal carcinoma (Mollersen 2015). Sensitivities were high, ranging from 83% (Cascinelli 1992) to 98% (Mollersen 2015, Nevus Doctor), while specificities were low and more varied ranging from 12% (Mollersen 2015 Nevus Doctor) to 59% (Cascinelli 1992). The lowest specificities (Nevus Doctor 12%, Mole Expert 13%) were produced by the study with the lowest disease prevalence (14%, Mollersen 2015). Mollersen 2015 also produced the highest sensitivity and was the only study to include clinically obvious melanomas amongst its lesions.
These accuracy estimates did not differ substantially from those for the detection of the individual target conditions reported above.
No data were available to compare these results with the use of standard dermoscopy to detect any skin cancer.
Spectro‐CAD
Two datasets evaluated the performance of DRS–CAD systems in referred settings, both from one study (Garcia Uribe 2012) evaluating the performance of the Oblique Incidence Diffuse Reflectance Spectrometry (OIDRS) system in two cohorts of lesions (i.e. CAD algorithms trained separately in each cohort). Amongst 136 pigmented skin lesions including 25 lesions to be excised (10 melanoma, 15 severe dysplastic lesions) OIDRS gave a sensitivity of 92% (95% CI 74.0% to 98.8%) and specificity of 86% (95% CI 78.9% to 91.6%). Amongst 89 non–pigmented skin lesions including 64 lesions to be excised (39 BCC, 25 cSCC) OIDRS gave a sensitivity of 92% (95% CI 83.0% to 96.6%) and specificity of 92% (95% CI 74.0% to 98.8%). However, caution must be used to interpret these estimates, as the spread of disease was different from that intended by our target condition definition of all malignancies: no malignancies other than melanoma were included in the pigmented population, and no melanomas were included in the non–pigmented population.
The two EIS–CAD studies conducted in referred settings produced accuracy data for 2389 lesions with 644 malignancies or highly dysplastic lesions, giving a summary sensitivity of 93.5% (95% CI 91.3% to 95.1%) and specificity of 32.6% (95% CI 30.4% to 34.8%) (Malvehy 2014; Mohr 2013) (Table 6). In addition to the malignancies described above, one Merkel cell carcinoma was included (Malvehy 2014) and detected by Nevisense.
Two datasets provided data for the use of MSI–CAD in unreferred settings (Sgouros 2014; Walter 2012) (Table 8). While both used a SIAscope™ device, Walter 2012 used the MoleMate analysis system incorporating the Primary Care Scoring Algorithm to analyse lesion images and arrive at a diagnosis (Walter 2012), while Sgouros 2014 also used the Primary Care Scoring Algorithm but did not report how images were interpreted. Sensitivity was slightly higher in Walter 2012 (92.0% versus 83.9% in Sgouros 2014) and specificity substantially so (72.5% versus 46.2% Sgouros 2014). In addition to the probable use of different analysis software, this difference is likely explained by two factors: firstly CAD test results were used in different ways, as a diagnostic aid to GPs (after a period of training) by Walter 2012, and as the diagnostic output for a CAD–based diagnosis in Sgouros 2014. Second, our exclusion of 144 benign cases with no reference standard diagnosis from Sgouros 2014 has created a highly selected (excised only) and unrepresentative study population in this study (low sample size, n = 44, and high prevalence of malignancy, 70%). Of the 153 lesions considered benign after clinico‐dermoscopic assessment (of which nine were later excised), 122 were diagnosed as naevi, 23 as seborrhoeic keratoses, seven as dermatofibroma and one as cherry angioma. SIAscopy gave a negative score (< 6) in 100 of these 153 lesions.
Discussion
Summary of main results
Computer–assisted diagnosis has been evaluated using a wide range of computer systems that analyse lesion images obtained by digital dermoscopy, lesion images obtained by multispectral imaging, or that analyse non–visual data from electrical impedance or diffuse reflectance spectroscopy. These computer systems have used a variety of classification algorithms to scrutinise a diverse selection of features. CAD sensitivity estimates were generally high, although with highly variable specificity.
We present six main findings from our review:
1) Included studies inadequately address the review question due to an abundance of low‐quality studies, poor reporting, and recruitment of highly selected groups of participants
This review aimed to assess the accuracy of computer–assisted diagnosis for detecting melanoma, BCC or cSCC in adults. Most included studies focused on its use for detecting or ruling out melanoma in lesions scheduled for excision. These studies therefore do not reveal CAD’s ability to detect clinically missed melanomas, since most have excluded lesions clinically diagnosed as benign. Only three studies, all of Derm–CAD systems, examined lesions not recommended for excision (and thus potentially missed melanomas) using an adequate reference standard in clinically benign lesions.
Studies were poorly reported and generally at unclear to high risk of bias across all domains, particularly for the selection of study participants, the timing of CAD diagnosis in relation to the reference standard diagnosis, and prespecification of CAD thresholds. Most studies used restricted groups of participants and failed to provide details of diagnostic thresholds sufficient for their reproducibility, leading to an almost universally poor clinical applicability of studies.
Poor reporting in the primary studies also hindered attempts to assess sources of heterogeneity, particularly for the lack of reporting CAD results according to the final diagnosis. A substantial number of included studies (half of Derm–CAD studies and a third of Spectro–CAD studies) evaluated experimental versions of CAD systems, in which classification algorithms were trained alongside preliminary assessments of test performance within the same source population. The frequency of these internal validation studies causes high concern for the reliability of accuracy estimates, chiefly because models are likely to give overly optimistic results when training and testing datasets are very similar in the spread of lesion types and severity, as is the case when the same source population is used (Altman 2009). In addition, great caution should be used when considering the applicability of these results to current clinical practice, since the generalisability of a new model can only be estimated in external validation studies that recruit new groups of patients from entirely new source populations.
Most study populations were restricted to excised lesions. Since lesions which are not excised are more likely to be benign, and without an atypical morphological pattern that could be mistaken for a malignancy, their absence from datasets may have reduced CAD specificity estimates from their likely performance in populations where CAD tests would be used in clinical practice, namely those being referred for specialist assessment. Including the appropriate spectrum of benign conditions is key to establishing the accuracy of any test (Lijmer 1999). Spectrum effects are often observed when tests that are developed further down the referral pathway have lower sensitivity and higher specificity when applied in settings with participants with limited prior testing (Usher‐Smith 2016). However, this direction of effect is not consistent across tests and diseases, as Leeflang 2013 clearly demonstrates; the mechanisms in action are often more complex than prevalence alone and can be difficult to identify. It is therefore crucial that tests are evaluated in lesions that are representative of those in which the test will be used in practice. For tests such as CAD that use machine learning, a representative patient population is vital both for training the algorithm and for testing its validity. If a narrow range of benign conditions is used to train a computer algorithm, the resulting CAD system is likely to struggle in discriminating new, previously unseen benign lesions at the validation stage. Unfortunately, very poor reporting of the spectrum of conditions included in studies prevents any assessment of whether mismatches have occurred between training and validation populations.
Limited data about the use of CAD as a diagnostic aid further limits the applicability of study results. Although CAD systems are designed to be used as diagnostic aids, most studies did not evaluate how CAD outputs were interpreted and acted upon by clinicians in their diagnostic decision–making, but instead evaluated the accuracy of the CAD system outputs as stand–alone tests.
2) CAD systems correctly identify melanoma in highly selected populations, but their low and very variable specificity suggest they are unreliable as stand‐alone diagnostic tests, especially in less selected populations
Reflecting the design aims of system developers, almost all the studies sought to evaluate the ability of CAD to identify melanomas. Table 1 presents key results for the primary target condition of cutaneous invasive melanoma or atypical intraepidermal melanocytic variants. For digital dermoscopy CAD systems, pooled results from 22 studies (8992 lesions, 1063 melanomas) provided a sensitivity of 90.1% (95% CI 84.0% to 94.0%) and specificity of 74.3% (95% CI 63.6% to 82.7%). Table 1 illustrates how these estimates would affect diagnoses in a hypothetical cohort of 1000 lesions clinically suspected of being melanomas. At the median melanoma prevalence of 20% that might occur in a highly specialised melanoma referral clinic, digital dermoscopy CAD systems would on average miss 20 out of 200 melanomas and would result in 206 false‐positive diagnoses. At the lower and upper quartile melanoma prevalence of 7% and 40%, 7 and 40 melanomas would be missed, with 239 and 154 false‐positive diagnoses respectively.
For multispectral imaging CAD systems, pooled results from eight studies (2401 lesions, 286 melanomas) provided a sensitivity of 92.9% (95% CI 83.7% to 97.1%) and specificity of 43.6% (95% CI 24.8% to 64.5%). In a hypothetical cohort of 1000 lesions clinically suspected of being melanomas with melanoma prevalence of 20%, MSI–CAD systems would on average miss 14 out of 200 melanomas and would result in 451 false‐positive diagnoses (Table 1). At the lower and upper quartile melanoma prevalence of 7% and 40%, 5 and 28 melanomas would be missed, with 525 and 338 false‐positive diagnoses respectively.
These results demonstrate a consistently high sensitivity for the detection of invasive melanoma and atypical intraepidermal melanocytic variants by CAD, regardless of type (Derm–CAD versus Spectro–CAD). However, specificity tends to be low and varies considerably between studies, particularly for MSI–based systems. The evidence certainly indicates that some benign lesions are more difficult to distinguish from malignancy using both Derm–CAD and Spectro–CAD systems, particularly seborrhoeic keratoses which proved problematic for Derm–CAD and EIS–CAD systems. However the reporting of benign diagnoses by CAD result was very poor, and omitted in 29 of the 42 included studies. As a result, the performance of MSI–CAD systems for these lesions remains uncertain. This difficulty in ruling melanoma out from seborrhoeic keratoses is also encountered when visual examination and dermoscopy are employed, and for CAD systems is equally likely to be due to similarities in the morphological appearance of these non‐melanocytic pigmented lesions and melanomas (Menzies 2005; Mollersen 2015).
Poor reporting of other aspects of study conduct also limit our interpretation of the heterogeneity in specificity, but likely causes include a wide variation in the spread of disease‐negative conditions included in study populations (both for training algorithms and for validating them), as well as considerable variation in CAD system characteristics.
3) There is insufficient evidence to assess the accuracy of CAD systems in primary‐care settings
Insufficient data were available from primary‐care populations to draw firm conclusions, particularly for Derm–CAD, with only one included study which restricted inclusion to excised lesions only. For MSI–CAD, we found some suggestion of high sensitivity, although the evidence base was limited to two studies evaluating differing target conditions and with differing approaches to the use of CAD results (CAD–based versus CAD–aided diagnosis). Limiting study populations to excised lesions is particularly problematic in such settings, since the frequency and distribution of disease is far removed from the range one would expect to see in patients who have received limited prior testing, such as those self–referring to specialist pigmented lesion clinics. Only one study examined all individuals presenting to generalist settings that had lesions which could not immediately be diagnosed as benign (Walter 2012). A second study also included all such lesions (Sgouros 2014), but failed to provide non–excised lesions with any clinical follow–up and so these lesions had to be excluded from our analysis.
4) Preliminary findings suggest CAD systems are at least as sensitive as assessment of dermoscopic images for the diagnosis of invasive melanoma and atypical intraepidermal melanocytic variants
MSI–CAD was significantly more sensitive than image‐based dermoscopy (92.9% versus 74%, P = 0.003), while direct comparisons indicate that it is also significantly less able to rule out truly benign lesions (29.8% versus 58.7%, P = 0.039). However indirect comparisons did not confirm this difference (43.6% versus 58.7%, P = 0.26) (Table 7). Conversely, our evidence suggests Derm–CAD may not differ significantly from dermoscopy in its ability to identify melanomas. However, we caution against drawing firm conclusions from these comparisons in the absence of sufficient data from studies evaluating face–to–face dermoscopy, on the basis that another review in this series has found such studies to demonstrate significantly higher accuracy for dermoscopy than those relying on review of dermoscopic images (Dinnes 2018b).
5) The evidence base for individual systems is too limited to draw conclusions on which might be preferred for practice
Despite the large number of included studies, the evidence base for the accuracy of individual systems to detect the primary target condition of the detection (invasive melanoma and atypical intraepidermal melanocytic variants) remains low, with meta–analysis possible for only two Derm–CAD systems (DB–MIPS and Skin View), one MSI–CAD system (MelaFind) and one EIS–CAD system (Nevisense). Only one of these, DB–MIPS, demonstrated a high specificity (89.1%) alongside high sensitivity (95.2%). Even though they evaluated the same CAD system, the six studies that we pooled for this estimate of DB‐MIPS were very varied in the classification algorithm used (ANN, SVM, K–NN, discriminant analysis, Euclidean distances), disease prevalence (5% to 45%), use of CAD results (two studies evaluated DB–MIPS as a diagnostic aid and four evaluated the DB–MIPS computer output) and range of disease‐negative conditions included (poorly reported, with a mixture of common naevus, dysplastic naevus, and only one study including seborrhoeic keratoses). The clinical settings to which this estimate applies therefore remain unclear.
We observed considerable variation in test characteristics across all CAD systems, such as hardware and software technologies used, the types of classification algorithm employed, methods used to train the algorithms, and which lesion morphological features were extracted and analysed. Wide variations in technology specifications were observed even within the same CAD systems; for example, the four studies evaluating the SIAscope (a commercially available MSI–CAD system) captured between four and eight optical reflectance images at varying wavelengths to inform the final image (not reported in two studies), using different SIAscopes (versions II and V, not reported in two studies) coupled to different software programmes (the MoleMate™ in two studies, and not reported in two studies) using two different thresholds (the Moncrieff 2002 and Emery 2010 methods). We noted this variation in CAD method, alongside limited reporting of important technological variables, in all CAD systems evaluated in this review.
6) Evidence of the ability of CAD to detect keratinocyte cancers is very limited and studies are confined to specialist settings
Only three studies included sufficient numbers of BCC cases for analysis; the one small retrospective study evaluating Derm–CAD is insufficient to draw any conclusions. For EIS–CAD, the two large prospective studies were designed to evaluate Nevisense’s ability to detect melanoma, so that neither recruited populations clinically applicable for keratinocyte cancer detection, casting doubt on the generalisability of pooled estimates to clinical practice. Similarly, evidence for the accuracy of cSCC detection is limited to one Nevisense study, of unlikely clinical applicability to the target condition.
Strengths and weaknesses of the review
The strengths of this review include an in‐depth and comprehensive electronic literature search, systematic review methods including double extraction of papers by both clinicians and methodologists, and contact with authors to allow study inclusion or clarify data. We adopted a clear analysis structure focusing on estimating incremental gains in accuracy, and undertook a detailed and replicable analysis of methodologic quality.
In comparison with the four main existing systematic reviews, our review extends the time period searched for eligible studies (from 2002 in Rosado 2003, from 2007 in Rajpara 2009 and Vestergaard 2008, and from 2011 in Ali 2012), includes all eligible studies regardless of language (Ali 2012; Rosado 2003), the presence of melanocytic lesions (Rajpara 2009), or use of a histological reference standard (Rajpara 2009). Although Vestergaard 2008 reviewed studies evaluating multispectral imaging CAD, electrical impedance spectroscopy CAD, and digital dermoscopy–based CAD systems, ours is the first review to meta‐analyse data and provide pooled accuracy estimates for each of these three types of CAD system. Ours is also the first review to include keratinocyte cancers as a target condition.
Our stringent application of review inclusion criteria meant that we excluded some studies included in previous reviews. For example, we did not include those developing CAD systems and training new algorithms (‘derivation studies’) without assessing their performance in an independent population. Of the 30 studies included in Rosado 2003, we excluded 23, including 17 derivation studies without independent test sets (16 Derm–CAD studies (Andreassi 1999; Binder 2000; Elbaum 2001; Ercal 1994; Ganster 2001; Green 1991; Green 1994; Hintz‐Madsen 2001; Horsch 1997; Kahofer 2002; Pompl 2000; Rubegni 2001a; Sboner 2001; Schindewolf 1994; Schmid‐Saugeon 2003; Smith 2000) and one MSI–CAD study, Farina 2000). We excluded three others for lack of clarity on the 2 x 2 contingency table (Schindewolf 1993), evaluating the diagnostic ability of a single feature (shape, Claridge 1992), and unclear reporting on CAD type and eligibility of the reference standard (Lefevre 2000). Two others were conference abstracts, and so were not eligible for analysis. Of the 12 Derm–CAD studies included in Rajpara 2009, we excluded four derivation studies without independent test sets (Green 1994; Manousaki 2006; Rubegni 2002b; Sboner 2001). Similarly, we excluded four of the nine Derm‐CAD studies in Ali 2012, due to the absence of independent test sets in derivation studies (Iyatomi 2006; Iyatomi 2008a; Iyatomi 2008b; Iyatomi 2010a), and another that did not evaluate diagnosis of the presence of skin cancer (Abbas 2011a). Vestergaard 2008 also required studies to evaluate CAD systems in an independent test set, and we consequently excluded only one of the nine studies included in Vestergaard 2008, due to the absence of a reference standard test in selected participants (Jamora 2003).
Our stringent exclusion of studies without an independent test population has resulted in the exclusion of all studies evaluating non–imaging diffuse reflectance spectroscopy (DRS) systems (e.g. Wallace 2002). This class of technologies used high spectral sampling to achieve a much higher resolution of spectral information from pigmented skin lesions than is found in the included DRSi (multispectral imaging) studies, giving an apparently strong performance for the detection of melanoma (Wallace 2000a; Wallace 2000b). We cannot assess in this review whether these promising results were overoptimistic due to their study design, or are genuine indicators of diagnostic capability. However, as technology improves we are likely to see systems with both imaging capability and high spectral sampling, making further carefully planned evaluation worthwhile.
The main concerns for the review are a result of the poor reporting of primary studies, limiting our assessment of methodological quality, and in particular limiting an understanding of which malignant and benign conditions were correctly and incorrectly identified by the CAD systems. The poor reporting is of particular concern, given the clear heterogeneity in all aspects of study design, and consequently the clinical applicability of results is difficult to determine. Poor reporting also precludes identification of those studies that may have been well designed, but did not document their design and conduct adequately.
Clear identification of the target condition was not provided in nine of the 22 Derm–CAD datasets or in four of the eight MSI–CAD datasets included in our primary analyses for detection of invasive melanoma or atypical intraepidermal melanocytic variants. These studies may or may not have included melanoma in situ lesions. Where studies included other invasive skin cancers in the study population, we tried to class any that were correctly identified as true negative results as opposed to false positives, on the basis that removal of any skin cancer in the attempt to identify melanomas would not be a negative consequence of the test. This relied on studies providing a disaggregation of test results according to final lesion classification and was not always possible.
Our review is limited to studies published up to August 2016, and since computer–assisted diagnosis is a rapidly developing field of technology, the review will have missed some important and more recent developments in the field of machine learning with an application to the detection of skin cancer. Deep learning algorithms (Esteva 2017; Han 2018) are one example where recent advances in computation have been applied to the development of new CAD systems for the detection of skin cancer. In future, it is also probable that CAD systems using continuous machine learning algorithms (where the CAD model continues to refine itself with exposure to clinical cases) will be developed.
Applicability of findings to the review question
Most of the data included in this review are unlikely to be applicable to the current clinical settings of primary care and dermatology clinics that exist in many high‐income countries. The predominance of highly selected lesion groups, scarce documentation of prior testing and frequency of internally validated derivation studies are likely to restrict the applicability of accuracy estimates to clinical practice. Poor documentation of the final diagnoses used to train CAD systems prevents any conclusion about the ideal target patient group, while very limited reporting of CAD results by final diagnosis does not allow us to make clear statements regarding the expected accuracy of different CAD systems, further restricting the transferability of results in practice.
Authors' conclusions
Implications for practice.
The utility of CAD for the primary diagnosis of melanoma in patients referred to specialist care remains largely unknown, since most included studies used CAD to detect malignancy in lesions already scheduled for surgical excision, most commonly with a high clinical suspicion of melanoma. For the detection of melanoma in people with clinically suspicious lesions, the evidence consistently shows all CAD types to have high sensitivity. CAD systems could therefore be useful as a back‐up by specialists to assist in minimising the risk of missing melanomas. However, the evidence base is currently too poor to understand whether CAD system outputs translate to different clinical decision–making in practice, and our sensitivity analysis suggests sensitivity may actually decrease when CAD is used as a diagnostic aid to triage unreferred patients. In addition, any projected gains in the early detection of melanomas must be set against the costs and practicality of implementing new systems.
Our evidence suggests MSI–CAD may be significantly more sensitive than dermoscopy. Given the paucity of data to allow comparison with in–person dermoscopy studies, this finding should be considered as exploratory.
Insufficient data are available to provide conclusive comments on the accuracy of CAD in community settings, or its accuracy to detect BCC and cSCC in any setting.
Implications for research.
Further prospective evaluation of the added value of CAD systems is warranted. Given the technological complexity and variation of CAD systems, it is certainly challenging to evaluate them in a rigorous manner. Nonetheless, studies are needed to evaluate CAD in its intended position in the patient care pathway in comparison to routine clinical examination and dermoscopy. For its use in specialist referral settings, studies should prospectively recruit all consecutive participants that have been referred for investigation of potential malignancy; for melanoma this should include all pigmented skin lesions, and for keratinocyte cancers any lesion suspected of being a BCC or cSCC. These studies should include lesions that are clinically determined to be benign and not excised, using specialist follow–up of at least six months as the accuracy reference standard. Comparisons with dermoscopy should consist of in–person diagnosis by dermatologists with expertise in dermoscopy. In community‐care settings, studies should prospectively recruit all participants presenting to clinic with lesions for which the clinician cannot clearly rule out malignancy, with clinical follow–up of all lesions which are not excised. To understand the clinical validity of CAD systems, studies should further evaluate how CAD system outputs are used to alter clinical decision‐making in real‐world practice settings.
Future studies must also report CAD test results according to their final diagnosis, so that its ability to distinguish between morphologically difficult lesions can be established. Crucially, studies must report the full technological specifications of their CAD systems, including which features were analysed as well as the diagnostic criteria and thresholds used to determine the presence of malignancy. Information on the distribution of lesions used to train the CAD system in previous studies would also be a welcome addition that would enable an interpretation of the system’s accuracy.
In terms of the development of new systems, or refinement of existing ones, cohorts of lesions should be selected so that training sets contain the range of benign and malignant lesions that would be encountered in routine clinical practice. Full descriptions of included lesions should be reported, together with an indication of how diagnostic thresholds have been selected. Validation of preliminary results should be assessed in independent populations, ideally from a different source from that used for model development.
Any future research study needs to be clear about the diagnostic pathway followed by study participants, and should conform to the updated Standards for Reporting of Diagnostic Accuracy (STARD) guideline (Bossuyt 2015).
What's new
| Date | Event | Description |
|---|---|---|
| 18 December 2018 | Amended | Affiliations, Disclaimer and Sources of support updated. |
Acknowledgements
Members of the Cochrane Skin Cancer Diagnostic Test Accuracy Group include:
the full project team (Susan Bayliss, Naomi Chuchu, Clare Davenport, Jonathan Deeks, Jacqueline Dinnes, Lavinia Ferrante di Ruffano, Kathie Godfrey, Rubeta Matin, Colette O'Sullivan, Yemisi Takwoingi, Hywel Williams),
our 12 clinical reviewers ( Rachel Abbott, Ben Aldridge, Oliver Bassett, Sue Ann Chan, Alana Durack, Monica Fawzy, Abha Gulati, Jacqui Moreau, Lopa Patel, Daniel Saleh, David Thompson, Kai Yuen Wong) and methodologist (Louise Johnston) who assisted with full‐text screening, data extraction and quality assessment across the entire suite of reviews of diagnosis and staging and skin cancer,
our expert advisor and co‐author for this review, Jeff Bamber, and on the protocol for keratinocyte skin cancer, Fiona Bath‐Hextall,
and all members of our Advisory Group (Jonathan Bowling, Seau Tak Cheung, Colin Fleming, Matthew Gardiner, Abhilash Jain, Susan O’Connell , Pat Lawton, John Lear, Mariska Leeflang, Richard Motley, Paul Nathan, Julia Newton‐Bishop, Miranda Payne, Rachael Robinson, Simon Rodwell, Julia Schofield, Neil Shroff, Hamid Tehrani, Zoe Traill, Fiona Walter, Angela Webster).
We would also like to thank the manufacturers DermoScan GmbH (Germany), MedX Corp (Canada), and SciBase (Sweden) for providing images of commercially‐available CAD systems.
The Cochrane Skin editorial base wishes to thank Robert Dellavalle, who was the Dermatology Editor for this review; and the clinical referee, Douglas Grossman. We also wish to thank the Cochrane DTA editorial base and colleagues, as well as Kate Cahill, who copy‐edited this review.
Appendices
Appendix 1. Current content and structure of the Programme Grant
| LIST OF REVIEWS | Number of studies | |
| Diagnosis of melanoma | ||
| 1 | Visual inspection | 49 |
| 2 | Dermoscopy +/‐ visual inspection | 104 |
| 3 | Teledermatology | 22 |
| 4 | Smartphone applications | 2 |
| 5a | Computer‐assisted diagnosis – dermoscopy‐based techniques | 42 |
| 5b | Computer‐assisted diagnosis – spectroscopy‐based techniques | Review amalgamated into 5a |
| 6 | Reflectance confocal microscopy | 18 |
| 7 | High‐frequency ultrasound | 5 |
| Diagnosis of keratinocyte skin cancer (BCC and cSCC) | ||
| 8 | Visual inspection +/‐ Dermoscopy | 24 |
| 5c | Computer‐assisted diagnosis – dermoscopy‐based techniques | Review amalgamated into 5a |
| 5d | Computer‐assisted diagnosis – spectroscopy‐based techniques | Review amalgamated into 5a |
| 9 | Optical coherence tomography | 5 |
| 10 | Reflectance confocal microscopy | 10 |
| 11 | Exfoliative cytology | 9 |
| Staging of melanoma | ||
| 12 | Imaging tests (ultrasound, CT, MRI, PET‐CT) | 38 |
| 13 | Sentinel lymph node biopsy | 160 |
| Staging of cSCC | ||
| Imaging tests review | Review dropped; only one study identified | |
| 13 | Sentinel lymph node biopsy | Review amalgamated into 13 above (n = 15 studies) |
Appendix 2. Acronyms
| Acronym | Definition |
| μm | micrometre |
| AK | actinic keratosis |
| ANN | artificial neural network |
| BCC | basal cell carcinoma |
| BD | Bowen’s disease |
| BPC | between person comparison (of tests) |
| CAD | computer assisted diagnosis |
| CCS | case control study |
| CS | case series |
| cSCC | cutaneous squamous cell carcinoma |
| D‐ | disease negative |
| D+ | disease positive |
| Derm–CAD | Digital dermoscopy based computer assisted diagnosis |
| DF | dermatofibroma |
| DRS | diffuse reflectance spectroscopy |
| DRSi | diffuse reflectance spectroscopy imaging |
| Dx | diagnosis |
| EIS | electrical impedance spectroscopy |
| FN | false negative |
| FP | false positive |
| FU | Follow‐ up |
| GP | general practitioner |
| H&E | haematoxylin and eosin stain |
| HFUS | high‐frequency ultrasound |
| Hz | hertz |
| KHz | kilohertz |
| K–NN | k nearest neighbour |
| MHz | megahertz |
| MiS | melanoma in situ (or lentigo maligna) |
| MM | malignant melanoma |
| mm | millimetre |
| MSI | multispectral imaging |
| N/A | not applicable |
| NC | non comparative |
| nm | nanometre |
| NPV | negative predictive value |
| NR | not reported |
| P | prospective |
| PPV | positive predictive value |
| PSL | pigmented skin lesion |
| R | retrospective |
| RCM | reflectance confocal microscopy |
| RCT | randomised controlled trial |
| SCC | squamous cell carcinoma |
| SD | standard deviation |
| se | sensitivity |
| sp | specificity |
| spectro–CAD | spectroscopy based computer–assisted diagnosis |
| SK | seborrhoeic keratosis |
| SSM | superficial spreading melanoma |
| SVM | Support vector machine |
| TN | true negative |
| TS | Telespectrophotometry System |
| VI | visual inspection |
| UNREF | Unreferred population |
| WPC | within person comparison (of tests) |
| WPC‐algs | within person comparison (of algorithms) |
Appendix 3. Proposed sources of heterogeneity
i. Population characteristics
general versus higher risk populations
patient population: Primary /secondary / specialist unit
lesion suspicion: general suspicion/atypical/equivocal/NR
lesion type: any pigmented; melanocytic
inclusion of multiple lesions per participant
ethnicity
ii. Index test characteristics
the nature of and definition of criteria for test positivity
observer experience with the index test
approaches to lesion preparation (e.g., the use of oil or antiseptic gel for dermoscopy)
iii. Reference standard characteristics
reference standard used
whether histology‐reporting meets pathology‐reporting guidelines
use of excisional versus diagnostic biopsy
whether two independent dermatopathologists reviewed histological diagnosis
iv. Study quality
consecutive or random sample of participants recruited
index test interpreted blinded to the reference standard result
index test interpreted blinded to the result of any other index test
presence of partial or differential verification bias (whereby only a sample of those subject to the index test are verified by the reference test or by the same reference test with selection dependent on the index test result)
use of an adequate reference standard
overall risk of bias
Appendix 4. Final search strategies
Melanoma search strategies to August 2016
Database: Ovid MEDLINE(R) 1946 to August week 3 2016
Search strategy:
1 exp melanoma/
2 exp skin cancer/
3 exp basal cell carcinoma/
4 basalioma$1.ti,ab.
5 ((basal cell or skin) adj2 (cancer$1 or carcinoma$1 or mass or masses or tumour$1 or tumor$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or lesion$1 or malignan$ or nodule$1)).ti,ab.
6 (pigmented adj2 (lesion$1 or mole$ or nevus or nevi or naevus or naevi or skin)).ti,ab.
7 (melanom$1 or nonmelanoma$1 or non‐melanoma$1 or melanocyt$ or non‐melanocyt$ or nonmelanocyt$ or keratinocyt$).ti,ab.
8 nmsc.ti,ab.
9 (squamous cell adj2 (cancer$1 or carcinoma$1 or mass or masses or tumor$1 or tumour$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or epithelial or lesion$1 or malignan$ or nodule$1) adj2 (skin or epiderm$ or cutaneous)).ti,ab.
10 (BCC or CSCC or NMSC).ti,ab.
11 keratinocy$.ti,ab.
12 Keratinocytes/
13 or/1‐12
14 dermoscop$.ti,ab.
15 dermatoscop$.ti,ab.
16 photomicrograph$.ti,ab.
17 exp epiluminescence microscopy/
18 (epiluminescence adj2 microscop$).ti,ab.
19 (confocal adj2 microscop$).ti,ab.
20 (incident light adj2 microscop$).ti,ab.
21 (surface adj2 microscop$).ti,ab.
22 (visual adj (inspect$ or examin$)).ti,ab.
23 ((clinical or physical) adj examin$).ti,ab.
24 3 point.ti,ab.
25 three point.ti,ab.
26 pattern analys$.ti,ab.
27 ABCD$.ti,ab.
28 menzies.ti,ab.
29 7 point.ti,ab.
30 seven point.ti,ab.
31 (digital adj2 (dermoscop$ or dermatoscop$)).ti,ab.
32 artificial intelligence.ti,ab.
33 AI.ti,ab.
34 computer assisted.ti,ab.
35 computer‐assisted.ti,ab.
36 neural network$.ti,ab.
37 exp diagnosis, computer‐assisted/
38 MoleMax.ti,ab.
39 image process$.ti,ab.
40 automatic classif$.ti,ab.
41 image analysis.ti,ab.
42 SIAscop$.ti,ab.
43 Aura.ti,ab.
44 (optical adj2 scan$).ti,ab.
45 MelaFind.ti,ab.
46 SIMSYS.ti,ab.
47 MoleMate.ti,ab.
48 SolarScan.ti,ab.
49 VivaScope.ti,ab.
50 (high adj3 ultraso$).ti,ab.
51 (canine adj2 detect$).ti,ab.
52 ((mobile or cell or cellular or smart) adj ((phone$1 adj2 app$1) or application$1)).ti,ab.
53 smartphone$.ti,ab.
54 (DermoScan or SkinVision or DermLink or SpotCheck).ti,ab.
55 Mole Detective.ti,ab.
56 Spot Check.ti,ab.
57 (mole$1 adj2 map$).ti,ab.
58 (total adj2 body).ti,ab.
59 exfoliative cytolog$.ti,ab.
60 digital analys$.ti,ab.
61 (image$1 adj3 software).ti,ab.
62 (teledermatolog$ or tele‐dermatolog$ or telederm or tele‐derm or teledermoscop$ or tele‐dermoscop$ or teledermatoscop$ or tele‐dermatoscop$).ti,ab.
63 (optical coherence adj (technolog$ or tomog$)).ti,ab.
64 (computer adj2 diagnos$).ti,ab.
65 exp sentinel lymph node biopsy/
66 (sentinel adj2 node).ti,ab.
67 nevisense.mp. or HFUS.ti,ab.
68 electrical impedance spectroscopy.ti,ab.
69 history taking.ti,ab.
70 patient history.ti,ab.
71 (naked eye adj (exam$ or assess$)).ti,ab.
72 (skin adj exam$).ti,ab.
73 physical examination/
74 ugly duckling.mp. or UD.ti,ab.
75 ((physician$ or clinical or physical) adj (exam$ or triage or recog$)).ti,ab.
76 ABCDE.mp. or VOC.ti,ab.
77 clinical accuracy.ti,ab.
78 Family Practice/ or Physicians, Family/ or clinical competence/
79 (confocal adj2 microscop$).ti,ab.
80 diagnostic algorithm$1.ti,ab.
81 checklist$.ti,ab.
82 virtual imag$1.ti,ab.
83 volatile organic compound$1.ti,ab.
84 dog$1.ti,ab.
85 gene expression analy$.ti,ab.
86 reflex transmission imag$.ti,ab.
87 thermal imaging.ti,ab.
88 elastography.ti,ab.
89 or/14‐88
90 (CT or PET).ti,ab.
91 PET‐CT.ti,ab.
92 (FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical$).ti,ab.
93 exp Deoxyglucose/
94 deoxy‐glucose.ti,ab.
95 deoxyglucose.ti,ab.
96 CATSCAN.ti,ab.
97 exp Tomography, Emission‐Computed/
98 exp Tomography, X‐ray computed/
99 positron emission tomograph$.ti,ab.
100 exp magnetic resonance imaging/
101 (MRI or fMRI or NMRI or scintigraph$).ti,ab.
102 exp echography/
103 Doppler echography.ti,ab.
104 sonograph$.ti,ab.
105 ultraso$.ti,ab.
106 doppler.ti,ab.
107 magnetic resonance imag$.ti,ab.
108 or/90‐107
109 (stage$ or staging or metasta$ or recurrence or sensitivity or specificity or false negative$ or thickness$).ti,ab.
110 "Sensitivity and Specificity"/
111 exp cancer staging/
112 or/109‐111
113 108 and 112
114 89 or 113
115 13 and 114
Database: Ovid MEDLINE(R) In‐Process & Other Non‐Indexed Citations 29 August 2016
Search strategy:
1 basalioma$1.ti,ab.
2 ((basal cell or skin) adj2 (cancer$1 or carcinoma$1 or mass or masses or tumour$1 or tumor$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or lesion$1 or malignan$ or nodule$1)).ti,ab.
3 (pigmented adj2 (lesion$1 or mole$ or nevus or nevi or naevus or naevi or skin)).ti,ab.
4 (melanom$1 or nonmelanoma$1 or non‐melanoma$1 or melanocyt$ or non‐melanocyt$ or nonmelanocyt$ or keratinocyt$).ti,ab.
5 nmsc.ti,ab.
6 (squamous cell adj2 (cancer$1 or carcinoma$1 or mass or masses or tumor$1 or tumour$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or epithelial or lesion$1 or malignan$ or nodule$1) adj2 (skin or epiderm$ or cutaneous)).ti,ab.
7 (BCC or CSCC or NMSC).ti,ab.
8 keratinocy$.ti,ab.
9 or/1‐8
10 dermoscop$.ti,ab.
11 dermatoscop$.ti,ab.
12 photomicrograph$.ti,ab.
13 (epiluminescence adj2 microscop$).ti,ab.
14 (confocal adj2 microscop$).ti,ab.
15 (incident light adj2 microscop$).ti,ab.
16 (surface adj2 microscop$).ti,ab.
17 (visual adj (inspect$ or examin$)).ti,ab.
18 ((clinical or physical) adj examin$).ti,ab.
19 3 point.ti,ab.
20 three point.ti,ab.
21 pattern analys$.ti,ab.
22 ABCD$.ti,ab.
23 menzies.ti,ab.
24 7 point.ti,ab.
25 seven point.ti,ab.
26 (digital adj2 (dermoscop$ or dermatoscop$)).ti,ab.
27 artificial intelligence.ti,ab.
28 AI.ti,ab.
29 computer assisted.ti,ab.
30 computer‐assisted.ti,ab.
31 neural network$.ti,ab.
32 MoleMax.ti,ab.
33 image process$.ti,ab.
34 automatic classif$.ti,ab.
35 image analysis.ti,ab.
36 SIAscop$.ti,ab.
37 Aura.ti,ab.
38 (optical adj2 scan$).ti,ab.
39 MelaFind.ti,ab.
40 SIMSYS.ti,ab.
41 MoleMate.ti,ab.
42 SolarScan.ti,ab.
43 VivaScope.ti,ab.
44 (high adj3 ultraso$).ti,ab.
45 (canine adj2 detect$).ti,ab.
46 ((mobile or cell or cellular or smart) adj ((phone$1 adj2 app$1) or application$1)).ti,ab.
47 smartphone$.ti,ab.
48 (DermoScan or SkinVision or DermLink or SpotCheck).ti,ab.
49 Mole Detective.ti,ab.
50 Spot Check.ti,ab.
51 (mole$1 adj2 map$).ti,ab.
52 (total adj2 body).ti,ab.
53 exfoliative cytolog$.ti,ab.
54 digital analys$.ti,ab.
55 (image$1 adj3 software).ti,ab.
56 (teledermatolog$ or tele‐dermatolog$ or telederm or tele‐derm or teledermoscop$ or tele‐dermoscop$ or teledermatoscop$ or tele‐dermatoscop$).ti,ab.
57 (optical coherence adj (technolog$ or tomog$)).ti,ab.
58 (computer adj2 diagnos$).ti,ab.
59 (sentinel adj2 node).ti,ab.
60 nevisense.mp. or HFUS.ti,ab.
61 electrical impedance spectroscopy.ti,ab.
62 history taking.ti,ab.
63 patient history.ti,ab.
64 (naked eye adj (exam$ or assess$)).ti,ab.
65 (skin adj exam$).ti,ab.
66 ugly duckling.mp. or UD.ti,ab.
67 ((physician$ or clinical or physical) adj (exam$ or triage or recog$)).ti,ab.
68 ABCDE.mp. or VOC.ti,ab.
69 clinical accuracy.ti,ab.
70 (Family adj (Practice or Physicians)).ti,ab.
71 (confocal adj2 microscop$).ti,ab.
72 clinical competence.ti,ab.
73 diagnostic algorithm$1.ti,ab.
74 checklist$.ti,ab.
75 virtual imag$1.ti,ab.
76 volatile organic compound$1.ti,ab.
77 dog$1.ti,ab.
78 gene expression analy$.ti,ab.
79 reflex transmission imag$.ti,ab.
80 thermal imaging.ti,ab.
81 elastography.ti,ab.
82 or/10‐81
83 (CT or PET).ti,ab.
84 PET‐CT.ti,ab.
85 (FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical$).ti,ab.
86 deoxy‐glucose.ti,ab.
87 deoxyglucose.ti,ab.
88 CATSCAN.ti,ab.
89 positron emission tomograph$.ti,ab.
90 (MRI or fMRI or NMRI or scintigraph$).ti,ab.
91 Doppler echography.ti,ab.
92 sonograph$.ti,ab.
93 ultraso$.ti,ab.
94 doppler.ti,ab.
95 magnetic resonance imag$.ti,ab.
96 or/83‐95
97 (stage$ or staging or metasta$ or recurrence or sensitivity or specificity or false negative$ or thickness$).ti,ab.
98 96 and 97
99 82 or 98
100 9 and 99
Database: Embase 1974 to 29 August 2016
Search strategy:
1 *melanoma/
2 *skin cancer/
3 *basal cell carcinoma/
4 basalioma$.ti,ab.
5 ((basal cell or skin) adj2 (cancer$1 or carcinoma$1 or mass or masses or tumour$1 or tumor$1 or neoplasm$ or adenoma$ or epithelioma$ or lesion$ or malignan$ or nodule$)).ti,ab.
6 (pigmented adj2 (lesion$1 or mole$ or nevus or nevi or naevus or naevi or skin)).ti,ab.
7 (melanom$1 or nonmelanoma$1 or non‐melanoma$1 or melanocyt$ or non‐melanocyt$ or nonmelanocyt$ or keratinocyt$).ti,ab.
8 nmsc.ti,ab.
9 (squamous cell adj2 (cancer$1 or carcinoma$1 or mass or tumor$1 or tumour$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or epithelial or lesion$1 or malignan$ or nodule$1) adj2 (skin or epiderm$ or cutaneous)).ti,ab.
10 (BCC or cscc).mp. or NMSC.ti,ab.
11 keratinocyte.ti,ab.
12 keratinocy$.ti,ab.
13 or/1‐12
14 dermoscop$.ti,ab.
15 dermatoscop$.ti,ab.
16 photomicrograph$.ti,ab.
17 *epiluminescence microscopy/
18 (epiluminescence adj2 microscop$).ti,ab.
19 (confocal adj2 microscop$).ti,ab.
20 (incident light adj2 microscop$).ti,ab.
21 (surface adj2 microscop$).ti,ab.
22 (visual adj (inspect$ or examin$)).ti,ab.
23 ((clinical or physical) adj examin$).ti,ab.
24 3 point.ti,ab.
25 three point.ti,ab.
26 pattern analys$.ti,ab.
27 ABCD$.ti,ab.
28 menzies.ti,ab.
29 7 point.ti,ab.
30 seven point.ti,ab.
31 (digital adj2 (dermoscop$ or dermatoscop$)).ti,ab.
32 artificial intelligence.ti,ab.
33 AI.ti,ab.
34 computer assisted.ti,ab.
35 computer‐assisted.ti,ab.
36 neural network$.ti,ab.
37 MoleMax.ti,ab.
38 exp diagnosis, computer‐assisted/
39 image process$.ti,ab.
40 automatic classif$.ti,ab.
41 image analysis.ti,ab.
42 SIAscop$.ti,ab.
43 (optical adj2 scan$).ti,ab.
44 Aura.ti,ab.
45 MelaFind.ti,ab.
46 SIMSYS.ti,ab.
47 MoleMate.ti,ab.
48 SolarScan.ti,ab.
49 VivaScope.ti,ab.
50 confocal microscop$.ti,ab.
51 (high adj3 ultraso$).ti,ab.
52 (canine adj2 detect$).ti,ab.
53 ((mobile or cell$ or cellular or smart) adj ((phone$1 adj2 app$1) or application$1)).ti,ab.
54 smartphone$.ti,ab.
55 (DermoScan or SkinVision or DermLink or SpotCheck).ti,ab.
56 Spot Check.ti,ab.
57 Mole Detective.ti,ab.
58 (mole$1 adj2 map$).ti,ab.
59 (total adj2 body).ti,ab.
60 exfoliative cytolog$.ti,ab.
61 digital analys$.ti,ab.
62 (image$1 adj3 software).ti,ab.
63 (optical coherence adj (technolog$ or tomog$)).ti,ab.
64 (teledermatolog$ or tele‐dermatolog$ or telederm or tele‐derm or teledermoscop$ or tele‐dermoscop$ or teledermatoscop$).mp. or tele‐dermatoscop$.ti,ab.
65 (computer adj2 diagnos$).ti,ab.
66 *sentinel lymph node biopsy/
67 (sentinel adj2 node).ti,ab.
68 nevisense.ti,ab.
69 HFUS.ti,ab.
70 electrical impedance spectroscopy.ti,ab.
71 history taking.ti,ab.
72 patient history.ti,ab.
73 (naked eye adj (exam$ or assess$)).ti,ab.
74 (skin adj exam$).ti,ab.
75 *physical examination/
76 ugly duckling.ti,ab.
77 UD sign$.ti,ab.
78 ((physician$ or clinical or physical) adj (exam$ or recog$ or triage)).ti,ab.
79 ABCDE.ti,ab.
80 clinical accuracy.ti,ab.
81 *general practice/
82 (confocal adj2 microscop$).ti,ab.
83 clinical competence/
84 diagnostic algorithm$.ti,ab.
85 checklist$1.ti,ab.
86 virtual image$1.ti,ab.
87 volatile organic compound$1.ti,ab.
88 VOC.ti,ab.
89 dog$1.ti,ab.
90 gene expression analys$.ti,ab.
91 reflex transmission imaging.ti,ab.
92 thermal imaging.ti,ab.
93 elastography.ti,ab.
94 dog$1.ti,ab.
95 gene expression analys$.ti,ab.
96 reflex transmission imaging.ti,ab.
97 thermal imaging.ti,ab.
98 elastography.ti,ab.
99 or/14‐93
100 PET‐CT.ti,ab.
101 (CT or PET).ti,ab.
102 (FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical$).ti,ab.
103 exp Deoxyglucose/
104 CATSCAN.ti,ab.
105 deoxyglucose.ti,ab.
106 deoxy‐glucose.ti,ab.
107 *positron emission tomography/
108 *computer assisted tomography/
109 positron emission tomograph$.ti,ab.
110 *nuclear magnetic resonance imaging/
111 (MRI or fMRI or NMRI or scintigraph$).ti,ab.
112 *echography/
113 Doppler.ti,ab.
114 sonograph$.ti,ab.
115 ultraso$.ti,ab.
116 magnetic resonance imag$.ti,ab.
117 or/100‐116
118 (stage$ or staging or metasta$ or recurrence or sensitivity or specificity or false negative$ or thickness$).ti,ab.
119 "Sensitivity and Specificity"/
120 *cancer staging/
121 or/118‐120
122 117 and 121
123 99 or 122
124 13 and 123
Database: Cochrane Library (Wiley) 2016 searched 30 August 2016 CDSR Issue 8 of 12 2016 CENTRAL Issue 7 of 12 2016 HTA Issue 3 of 4 July 2016 DARE Issue 3 of 4 2015
Search strategy:
#1 melanoma* or nonmelanoma* or non‐melanoma* or melanocyt* or non‐melanocyt* or nonmelanocyt* or keratinocyte*
#2 MeSH descriptor: [Melanoma] explode all trees
#3 "skin cancer*"
#4 MeSH descriptor: [Skin Neoplasms] explode all trees
#5 skin near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*)
#6 nmsc
#7 "squamous cell" near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*) near/2 (skin or epiderm* or cutaneous)
#8 "basal cell" near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*)
#9 pigmented near/2 (lesion* or nevus or mole* or naevi or naevus or nevi or skin)
#10 #1 or #2 or #3 or #4 or #5 or #6 or #7 or #8 or #9
#11 dermoscop*
#12 dermatoscop*
#13 Photomicrograph*
#14 MeSH descriptor: [Dermoscopy] explode all trees
#15 confocal near/2 microscop*
#16 epiluminescence near/2 microscop*
#17 incident next light near/2 microscop*
#18 surface near/2 microscop*
#19 "visual inspect*"
#20 "visual exam*"
#21 (clinical or physical) next (exam*)
#22 "3 point"
#23 "three point"
#24 "pattern analys*"
#25 ABDC
#26 menzies
#27 "7 point"
#28 "seven point"
#29 digital near/2 (dermoscop* or dermatoscop*)
#30 "artificial intelligence"
#31 "AI"
#32 "computer assisted"
#33 "computer‐assisted"
#34 AI
#35 "neural network*"
#36 MoleMax
#37 "computer diagnosis"
#38 "image process*"
#39 "automatic classif*"
#40 SIAscope
#41 "image analysis"
#42 "optical near/2 scan*"
#43 Aura
#44 MelaFind
#45 SIMSYS
#46 MoleMate
#47 SolarScan
#48 Vivascope
#49 "confocal microscopy"
#50 high near/3 ultraso*
#51 canine near/2 detect*
#52 Mole* near/2 map*
#53 total near/2 body
#54 mobile* or smart near/2 phone*
#55 cell next phone*
#56 smartphone*
#57 "mitotic index"
#58 DermoScan or SkinVision or DermLink or SpotCheck
#59 "Mole Detective"
#60 "Spot Check"
#61 mole* near/2 map*
#62 total near/2 body
#63 "exfoliative cytolog*"
#64 "digital analys*"
#65 image near/3 software
#66 teledermatolog* or tele‐dermatolog* or telederm or tele‐derm or teledermoscop* or tele‐dermoscop* or teledermatoscop* or tele‐dermatolog*
#67 "optical coherence" next (technolog* or tomog*)
#68 computer near/2 diagnos*
#69 sentinel near/2 node*
#70 #11 or #12 or #13 or #14 or #15 or #16 or #17 or #18 or #19 or #20 or #21 or #22 or #23 or #24 or #25 or #26 or #27 or #28 or #29 or #30 or #31 or #32 or #33 or #34 or #35 or #36 or #37 or #38 or #39 or #40 or #41 or #42 or #43 or #44 or #45 or #46 or #47 or #48 or #49 or #50 or #51 or #52 or #53 or #54 or #55 or #56 or #57 or #58 or #59 or #60 or #61 or #62 or #63 or #64 or #65 or #66 or #67 or #68 or #69
#71 ultraso*
#72 sonograph*
#73 MeSH descriptor: [Ultrasonography] explode all trees
#74 Doppler
#75 CT or PET or PET‐CT
#76 "CAT SCAN" or "CATSCAN"
#77 MeSH descriptor: [Positron‐Emission Tomography] explode all trees
#78 MeSH descriptor: [Tomography, X‐Ray Computed] explode all trees
#79 MRI
#80 MeSH descriptor: [Magnetic Resonance Imaging] explode all trees
#81 MRI or fMRI or NMRI or scintigraph*
#82 "magnetic resonance imag*"
#83 MeSH descriptor: [Deoxyglucose] explode all trees
#84 deoxyglucose or deoxy‐glucose
#85 "positron emission tomograph*"
#86 #71 or #72 or #73 or #74 or #75 or #76 or #77 or #78 or #79 or #80 or #81 or #82 or #83 or #84 or #85
#87 stage* or staging or metasta* or recurrence or sensitivity or specificity or "false negative*" or thickness*
#88 MeSH descriptor: [Neoplasm Staging] explode all trees
#89 #87 or #88
#90 #89 and #86
#91 #70 or #90
#92 #10 and #91
#93 BCC or CSCC or NMCS
#94 keratinocy*
#95 #93 or #94
#96 #10 or #95
#97 nevisense
#98 HFUS
#99 "electrical impedance spectroscopy"
#100 "history taking"
#101 "patient history"
#102 naked next eye near/1 (exam* or assess*)
#103 skin next exam*
#104 "ugly duckling" or (UD sign*)
#105 MeSH descriptor: [Physical Examination] explode all trees
#106 (physician* or clinical or physical) near/1 (exam* or recog* or triage*)
#107 ABCDE
#108 "clinical accuracy"
#109 MeSH descriptor: [General Practice] explode all trees
#110 confocal near microscop*
#111 "diagnostic algorithm*"
#112 MeSH descriptor: [Clinical Competence] explode all trees
#113 checklist*
#114 "virtual image*"
#115 "volatile organic compound*"
#116 dog or dogs
#117 VOC
#118 "gene expression analys*"
#119 "reflex transmission imaging"
#120 "thermal imaging"
#121 elastography
#122 #97 or #98 or #99 or #100 or #101 or #102 or #103 or #104 or #105 or #106 or #107 or #108 or #109 or #110 or #111 or #112 or #113 or #114 or #115 or #116 or #117 or #118 or #119 or #120 or #121
#123 #70 or #122
#124 #96 and #123
#125 #96 and #90
#126 #125 or #124
#127 #10 and #126
Database : CINAHL Plus (EBSCO) 1937 to 30 August 2016
Search strategy:
S1 (MH "Melanoma") OR (MH "Nevi and Melanomas+")
S2 (MH "Skin Neoplasms+")
S3 (MH "Carcinoma, Basal Cell+")
S4 basalioma*
S5 (basal cell) N2 (cancer* or carcinoma* or mass or masses or tumor* or tumour* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*)
S6 (pigmented) N2 (lesion* or mole* or nevus or nevi or naevus or naevi or skin)
S7 melanom* or nonmelanoma* or non‐melanoma* or melanocyt* or non‐melanocyt* or nonmelanocyt*
S8 nmsc
S9 TX BCC or cscc or NMSC
S10 (MH "Keratinocytes")
S11 keratinocyt*
S12 S1 OR S2 OR S3 OR S4 OR S5 OR S6 OR S7 OR S8 OR S9 OR S10 OR S11
S13 dermoscop* or dermatoscop* or photomicrograph* or (3 point) or (three point) or ABCD* or menzies or (7 point) or (seven point) or AI or Molemax or SIASCOP* or Aura or MelaFind or SIMSYS or MoleMate or SolarScan or smartphone* or DermoScan or SkinVision or DermLink or SpotCheck
S14 (epiluminescence or confocal or incident or surface) N2 (microscop*)
S15 visual N1 (inspect* or examin*)
S16 (clinical or physical) N1 (examin*)
S17 pattern analys*
S18 (digital) N2 (dermoscop* or dermatoscop*)
S19 (artificial intelligence)
S20 (computer) N2 (assisted or aided)
S21 (neural network*)
S22 (MH "Diagnosis, Computer Assisted+")
S23 (image process*)
S24 (automatic classif*)
S25 (image analysis)
S26 SIAScop*
S27 (optical) N2 (scan*)
S28 (high) N3 (ultraso*)
S29 elastography
S30 (mobile or cell or cellular or smart) N2 (phone*) N2 (app or application*)
S31 (mole*) N2 (map*)
S32 total N2 body
S33 exfoliative cytolog*
S34 digital analys*
S35 image N3 software
S36 teledermatolog* or tele‐dermatolog* or telederm or tele‐derm or teledermoscop* or tele‐dermoscop* or teledermatoscop* or tele‐dermatoscop* teledermatolog* or tele‐dermatolog* or telederm or tele‐derm or teledermoscop*
S37 (optical coherence) N1 (technolog* or tomog*)
S38 computer N2 diagnos*
S39 sentinel N2 node
S40 (MH "Sentinel Lymph Node Biopsy")
S41 nevisense or HFUS or checklist* or VOC or dog*
S42 electrical impedance spectroscopy
S43 history taking
S44 "Patient history"
S45 naked eye
S46 skin exam*
S47 physical exam*
S48 ugly duckling
S49 UD sign*
S50 (physician* or clinical or physical) N1 (exam*)
S51 clinical accuracy
S52 general practice
S53 (physician* or clinical or physical) N1 (recog* or triage)
S54 confocal microscop*
S55 clinical competence
S56 diagnostic algorithm*
S57 checklist*
S58 virtual image*
S59 volatile organic compound*
S60 gene expression analys*
S61 reflex transmission imag*
S62 thermal imaging
S63 S13 or S14 or S15 OR S16 OR S17 OR S18 OR S19 OR S20 OR S21 OR S22 OR S23 OR S24 OR S25 OR S26 OR S27 OR S28 OR S29 OR S30 OR S31 OR S32 OR S33 OR S34 OR S35 OR S36 OR S37 OR S38 OR S39 OR S40 OR S41 OR S42 OR S43 OR S44 OR S45 OR S46 OR S47 OR S48 OR S49 OR S50 OR S51 OR S52 OR S53 OR S54 OR S55 OR S56 OR S57 OR S58 OR S59 OR S60 OR S61 OR S62
S64 CT or PET
S65 PET‐CT
S66 FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical*
S67 (MH "Deoxyglucose+")
S68 deoxy‐glucose or deoxyglucose
S69 CATSCAN
S70 CAT‐SCAN
S71 (MH "Deoxyglucose+")
S72 (MH "Tomography, Emission‐Computed+")
S73 (MH "Tomography, X‐Ray Computed")
S74 positron emission tomograph*
S75 (MH "Magnetic Resonance Imaging+")
S76 MRI or fMRI or NMRI or scintigraph*
S77 echography
S78 doppler
S79 sonograph*
S80 ultraso*
S81 magnetic resonance imag*
S82 S64 OR S65 OR S66 OR S67 OR S68 OR S69 OR S70 OR S71 OR S72 OR S73 OR S74 OR S75 OR S76 OR S77 OR S78 OR S79 OR S80 OR S81
S83 stage* or staging or metasta* or recurrence or sensitivity or specificity or (false negative*) or thickness
S84 (MH "Neoplasm Staging")
S85 S83 OR S84
S86 S82 AND S85
S87 S63 OR S86
S88 S12 AND S87
Database: Science Citation Index SCI Expanded (Web of Science) 1900 to 30 August 2016
Conference Proceedings Citation Index (Web of Science) 1900 to 1 September 2016
Search strategy:
#1 (melanom* or nonmelanom* or non‐melanoma* or melanocyt* or non‐melanocyt* or nonmelanocyt* or keratinocyt*)
#2 (basalioma*)
#3 ((skin) near/2 (cancer* or carcinoma or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*))
#4 ((basal) near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*))
#5 ((pigmented) near/2 (lesion* or mole* or nevus or nevi or naevus or naevi or skin))
#6 (nmsc or BCC or NMSC or keratinocy*)
#7 ((squamous cell (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*))
#8 (skin or epiderm* or cutaneous)
#9 #8 AND #7
#10 #9 OR #6 OR #5 OR #4 OR #3 OR #2 OR #1
#11 ((dermoscop* or dermatoscop* or photomicrograph* or epiluminescence or confocal or "incident light" or "surface microscop*" or "visual inspect*" or "physical exam*" or 3 point or three point or pattern analy* or ABCDE or menzies or 7 point or seven point or dermoscop* or dermatoscop* or AI or artificial or computer‐assisted or computer assisted or neural network* or Molemax or image process* or automatic classif* or image analysis or siascope or optical scan* or Aura or melafind or simsys or molemate or solarscan or vivascope or confocal microscop* or high ultraso* or canine detect* or cellphone* or mobile* or phone* or smartphone or dermoscan or skinvision or dermlink or spotcheck or spot check or mole detective or mole map* or total body or exfoliative psychology or digital or image software or optical coherence or teledermatology or telederm* or teledermoscop* or teledermatoscop* or computer diagnos* or sentinel))
#12 ((nevisense or HFUS or impedance spectroscopy or history taking or patient history or naked eye or skin exam* or physical exam* or ugly duckling or UD sign* or physician* exam* or physical exam* or ABCDE or clinical accuracy or general practice or confocal microscop* or clinical competence or diagnostic algorithm* or checklist* or virtual image* or volatile organic or VOC or dog* or gene expression or reflex transmission or thermal imag* or elastography))
#13 #11 or #12
#14 ((PET or CT or FDG or deoxyglucose or deoxy‐glucose or fluorodeoxy* or radiopharma* or CATSCAN or positron emission or computer assisted or nuclear magnetic or MRI or FMRI or NMRI or scintigraph* or echograph* or Doppler or sonograph* or ultraso* or magnetic reson*))
#15 ((stage* or staging or metast* or recurrence or sensitivity or specificity or false negative* or thickness*))
#16 #14 AND #15
#17 #16 OR #13
#18 #10 AND #17
Refined by: DOCUMENT TYPES: (MEETING ABSTRACT OR PROCEEDINGS PAPER)
Appendix 5. Full text inclusion criteria
| Criterion | Inclusion | Exclusion |
| Study design |
For diagnostic and staging reviews
|
|
| Target condition |
|
|
| Population |
For diagnostic reviews
For staging reviews
|
|
| Index tests |
For diagnosis
For staging
Any test combination and in any order Any test positivity threshold Any variation in testing procedure (e.g. radioisotope used) |
|
| Reference standard |
For diagnostic studies
For studies of imaging tests for staging
For studies of SLNB accuracy for staging
|
For diagnostic studies
|
| BCC: basal cell carcinoma; cSCC: cutaneous squamous cell carcinoma; CT: computed tomography; FNAC: fine needle aspiration cytology; LND: lymph node dissection; MRI: magnetic resonance imaging; PET: positron emission tomography; PET‐CT: positron emission tomography computed tomography; RCT: randomised controlled trial; SCC: squamous cell carcinoma; SLN+: positive sentinel lymph node; SLN: negative sentinel lymph node; SLNB: sentinel lymph node biopsy. | ||
Appendix 6. Quality assessment (based on QUADAS‐2)
The following tables use text that was originally published in the QUADAS‐2 tool by Whiting and colleagues (Whiting 2011).
| Item | Response (delete as required) |
| PARTICIPANT SELECTION (1) ‐ RISK OF BIAS | |
| 1) Was a consecutive or random sample of participants or images enrolled? |
Yes – if paper states consecutive or random No – if paper describes other method of sampling Unclear – if participant sampling not described |
| 2) Was a case‐control design avoided? |
Yes – if consecutive or random or case‐control design clearly not used No – if study described as case‐control or describes sampling specific numbers of participants with particular diagnoses Unclear – if not described |
3) Did the study avoid inappropriate exclusions, e.g.,
|
Yes ‐ if inappropriate exclusions were avoided No – if lesions were excluded that might affect test accuracy, e.g., 'difficult to diagnose' lesions, or where disagreement between evaluators was observed Unclear – if not clearly reported but there is suspicion that difficult to diagnose lesions may have been excluded |
4) For between‐person comparative studies only (i.e., allocating different tests to different study participants):
|
For A)
For B)
For C)
|
| Could the selection of participants have introduced bias? For non‐comparative and within person‐comparative studies
For between‐person comparative studies
|
For non‐comparative and within person‐comparative studies
For between‐person comparative studies
|
| PARTICIPANT SELECTION (1) ‐ CONCERNS REGARDING APPLICABILITY | |
1) Are the included participants and chosen study setting generalisable to the patient population who will receive the test in practice? (Test set)
|
A) For studies that will contribute to the analysis of participants with a primary presentation of a skin lesion (i.e., test naive) Yes – if participants included in the study appear to be generally representative of those who might present in a usual practice setting No – if study participants appear to be unrepresentative of usual practice, e.g., in terms of severity of disease, demographic features, presence of differential diagnosis or comorbidity, setting of the study, and previous testing protocols Unclear – if insufficient details are provided to determine the generalisability of study participants B) For studies that will contribute to the analysis of referred participants (i.e., who have already undergone some form of testing) Yes – if study participants appear to be representative of those who might be referred for further investigation. If the study focuses only on those with equivocal lesions, for example, we would suggest that this is not representative of the wider referred population No – if study participants appear to be unrepresentative of usual practice, e.g., if a particularly high proportion of participants have been self‐referred or referred for cosmetic reasons. Other factors to consider include severity of disease, demographic features, presence of differential diagnosis or comorbidity, setting of the study, and previous testing protocols Unclear – if insufficient details are provided to determine the generalisability of study participants |
| 2) Did the study avoid including participants with multiple lesions? |
Yes – if the difference between the number of included lesions and number of included participants is less than 5% No – if the difference between the number of included lesions and number of included participants is greater than 5% Unclear – if it is not possible to assess |
| 3) Was an adequate spectrum of cases used to train the algorithm? (training set) |
For melanoma studies: Yes – if all PSLs, main types of melanoma all present (nodular, SSM, Mis), main types of dysplasia present, and a range of benign diagnoses included. For keratinocyte cancer studies: Yes ‐ if the main malignant diagnoses are included (BCC, cSCC), as well as main types of atypia/dysplasia and a range of benign differential diagnoses (AK, SK, BD). No ‐ if study participants appear to be unrepresentative of usual practice, for example with a specific focus on certain lesions groups, e.g. melanoma and common nevus only Unclear – insufficient details to determine generalisability of study participants N/A ‐ algorithm trained in a previous study |
Is there concern that the included participants do not match the review question?
|
|
| INDEX TEST (2) ‐ RISK OF BIAS (to be completed per test evaluated) | |
| 1) Was the index test or testing strategy result interpreted without knowledge of the results of the reference standard? |
Yes – if index test described as interpreted without knowledge of reference standard result or, for prospective studies, if index test is always conducted and interpreted prior to the reference standard No – if index test described as interpreted in knowledge of reference standard result Unclear – if index test blinding is not described |
| 2) Was the diagnostic threshold at which the test was considered positive (i.e., melanoma present) prespecified? |
Yes – if threshold was prespecified (i.e., prior to analysing study results) No – if threshold was not prespecified Unclear – if not possible to tell whether or not diagnostic threshold was prespecified |
| 3) Was the CAD classification algorithm evaluated in an independent patient population? |
Yes – Test set only study (validated in previous study) OR validated within–study in a test set using in a different group of participants recruited from a different source (external validation) OR validated within–study in a test set comprising a randomised subset of one larger participant population also used for model training No – Algorithm developed and evaluated within the same study (internal validation) where training and test sets use the same population which is not divided randomly Unclear – The relationship between training and tests sets is not reported clearly |
| 4) Was model overfitting accounted for? | Yes – a shrinkage method was applied, e.g. bootstrapping for calibration in the large, calibration in the small, overoptimisation, optimisation, use of a verify set to optimise stopping. No – study clearly reports that no method was applied Unclear – overfitting not discussed or corrected N/A ‐ study does not contain a derivation element |
Could the conduct or interpretation of the index test have introduced bias?
|
|
| INDEX TEST (2) ‐ CONCERN ABOUT APPLICABILITY | |
| 1) Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? Study results can only be reproduced if the diagnostic threshold is described in sufficient detail. This item applies equally to studies using pattern recognition and those using checklists or algorithms to aid test interpretation |
Yes – If the criteria for diagnosis of melanoma were reported in sufficient detail to allow replication No – if the criteria for diagnosis of melanoma were not reported in sufficient detail to allow replication Unclear – If some but not sufficient information on criteria for diagnosis to allow replication were provided |
| 2) Was the test interpretation carried out by an experienced examiner? |
Yes – if the test was interpreted by 1 or more speciality‐accredited dermatologists, or by examiners of any clinical background with special interest in dermatology and with any formal training in the use of the test No – if the test was not interpreted by an experienced examiner (see above) Unclear – if the experience of the examiner(s) was not reported in sufficient detail to judge or if examiners were described as 'Expert' with no further detail given N/A – if system‐based diagnosis, i.e., no observer interpretation |
| 3) Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? E.g., previously evaluated/established
|
Yes – if a previously evaluated/established tool to aid diagnosis of melanoma was used or if the diagnostic threshold used was established in a previously published study No – if an unfamiliar/new tool to aid diagnosis of melanoma was used, if no particular algorithm was used, or if the objective threshold reported was chosen based on results in the current study Unclear – if insufficient information was reported |
Is there concern that the index test, its conduct, or interpretation differ from the review question?
|
|
| REFERENCE STANDARD (3) ‐ RISK OF BIAS | |
| 1) Is the reference standard likely to correctly classify the target condition? A) Disease‐positive – 1 or more of the following:
B) Disease‐negative – 1 or more of the following:
|
A) Disease‐positive Yes – if all participants with a final diagnosis of melanoma underwent 1 of the listed reference standards No – If a final diagnosis of melanoma for any participant was reached without histopathology Unclear – if the method of final diagnosis was not reported for any participant with a final diagnosis of melanoma or if the length of clinical follow‐up used was not clear or if a clinical follow‐up reference standard was reported in combination with a participant‐based analysis and it was not possible to determine whether the detection of a malignant lesion during follow‐up is the same lesion that originally tested negative on the index test B) Disease‐negative Yes – If at least 80% of benign diagnoses were reached by histology and up to 20% were reached by clinical follow‐up for a minimum of 3 months following the index test No – if more than 20% of benign diagnoses were reached by clinical follow‐up for a minimum of 3 months following the index test or if clinical follow‐up period was less than 3 months Unclear – if the method of final diagnosis was not reported for any participant with benign or non‐melanoma diagnosis |
| 2) Were the reference standard results interpreted without knowledge of the results of the index test? Please score this item for all studies even though histopathology interpretation is usually conducted with knowledge of the clinical diagnosis (from visual inspection or dermoscopy or both). We will deal with this by not including the response to this item in the 'Risk of bias' assessment for these tests. For reviews of all other tests, this item will be retained |
Yes – if the reference standard diagnosis was reached blinded to the index test result No – if the reference standard diagnosis was reached with knowledge of the index test result Unclear – if blinded reference test interpretation was not clearly reported |
Could the reference standard, its conduct, or its interpretation have introduced bias?
|
|
| REFERENCE STANDARD (3) ‐ CONCERN ABOUT APPLICABILITY | |
| 1) Expert opinion (with no histological confirmation) was not used as a reference standard 'Expert opinion' means diagnosis based on the standard clinical examination, with no histology or lesion follow‐up |
Yes – if expert opinion was not used as a reference standard for any participant No – if expert opinion was used as a reference standard for any participant Unclear – if not clearly reported |
| 2) Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? |
Yes – if histology interpretation was reported to be carried out by an experienced histopathologist or dermatopathologist No – if histology interpretation was reported to be carried out by a less experienced histopathologist Unclear – if the experience/qualifications of the pathologist were not reported |
Is there concern that the target condition as defined by the reference standard does not match the review question?
|
|
| FLOW AND TIMING (4): RISK OF BIAS | |
| 1) Was there an appropriate interval between index test and reference standard? A) For histopathological reference standard, was the interval between index test and reference standard ≤ 1 month? B) If the reference standard includes clinical follow‐up of borderline/benign‐appearing lesions, was there at least 3 months' follow‐up following application of index test(s)? |
A) Yes – if study reports ≤ 1 month between index and reference standard No – if study reports > 1 month between index and reference standard Unclear – if study does not report interval between index and reference standard B) Yes – if study reports ≥ 3 months' follow‐up No – if study reports < 3 months' follow‐up Unclear – if study does not report the length of clinical follow‐up |
| 2) Did all participants receive the same reference standard? |
Yes – if all participants underwent the same reference standard No – if more than 1 reference standard was used Unclear – if not clearly reported |
| 3) Were all participants included in the analysis? |
Yes – if all participants were included in the analysis No – if some participants were excluded from the analysis Unclear– if not clearly reported |
Could the participant flow have introduced bias?
|
|
| BCC = basal cell carcinoma; cSCC = cutaneous squamous cell carcinoma. | |
Appendix 7. Forest plots for the direct comparison of CAD systems vs. image‐based dermoscopy
22.
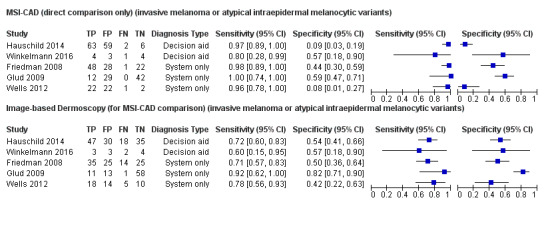
Forest plot of direct comparisons between image‐based dermoscopy diagnosis versus multispectral imaging CAD systems (MSI‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
23.
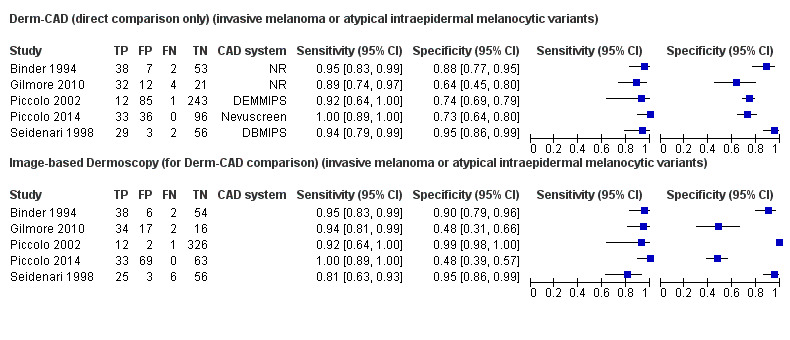
Forest plot of direct comparisons between image‐based dermoscopy diagnosis versus digital dermoscopy CAD systems (Derm‐CAD) for the detection of invasive melanoma or atypical intraepidermal melanocytic variants (invasive melanoma or atypical intraepidermal melanocytic variants)
Appendix 8. Results of sensitivity analysis for CAD systems (excludes diagnostic aid studies)
| Index test | Studies | Cases/Number of participants | Summary sensitivity (95% CI) % | Summary specificity (95% CI) % |
| Primary target condition: Invasive melanoma andatypical intraepidermal melanocytic variants | ||||
| MSI‐CAD | 7 | 281/2389 | 93.7 (83.9 to 97.7) | 41.7 (22.0 to 64.6) |
| Derm‐CAD | 19 | 955/8403 | 88.5 (81.3 to 93.1) | 71.3 (60.0 to 80.4) |
| DB–MIPS | 4 | 427/1479 | 96.4 (84.9 to 99.2) | 85.5 (75.7 to 91.7) |
| MelaFind | 4 | 191/1786 | 97.9 (94.6 to 99.2) | 24.8 (8.82 to 52.9) |
| Secondary target condition: Invasive melanoma alone | ||||
| MSI‐CAD | 4 | 80/256 | 83.1 (26.5 to 98.5) | 66.9 (41.6 to 85.1) |
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
1. Test.
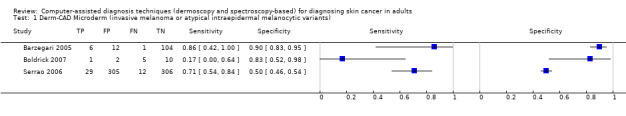
Derm‐CAD Microderm (invasive melanoma or atypical intraepidermal melanocytic variants).
2. Test.
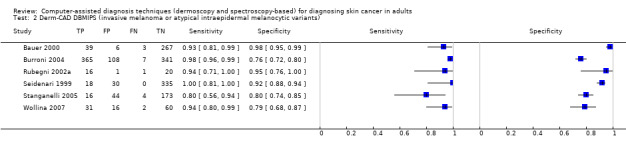
Derm‐CAD DBMIPS (invasive melanoma or atypical intraepidermal melanocytic variants).
3. Test.
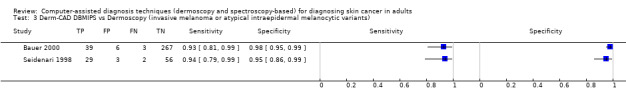
Derm‐CAD DBMIPS vs Dermoscopy (invasive melanoma or atypical intraepidermal melanocytic variants).
4. Test.

Derm‐CAD DEMMIPS (invasive melanoma or atypical intraepidermal melanocytic variants).
6. Test.
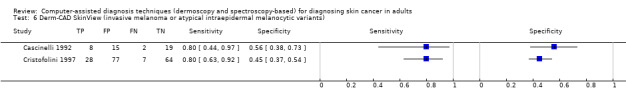
Derm‐CAD SkinView (invasive melanoma or atypical intraepidermal melanocytic variants).
7. Test.

Derm‐CAD SkinView (Any lesion requiring excision).
8. Test.

Derm‐CAD NevusDr (invasive melanoma or atypical intraepidermal melanocytic variants).
9. Test.

Derm‐CAD NevusDr (Any lesion requiring excision).
10. Test.

Derm‐CAD ImageJ (invasive melanoma or atypical intraepidermal melanocytic variants).
11. Test.

Derm‐CAD IBAS2000 (invasive melanoma or atypical intraepidermal melanocytic variants).
12. Test.

Derm‐CAD Nevuscreen (invasive melanoma or atypical intraepidermal melanocytic variants).
13. Test.

Derm‐CAD SolarScan (invasive melanoma).
14. Test.

Derm‐CAD SolarScan (invasive melanoma or atypical intraepidermal melanocytic variants).
15. Test.

Derm‐CAD No name (invasive melanoma).
16. Test.
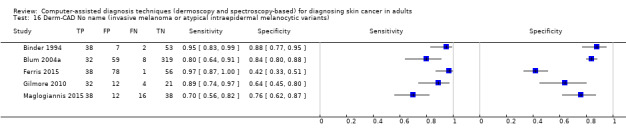
Derm‐CAD No name (invasive melanoma or atypical intraepidermal melanocytic variants).
17. Test.

Derm‐CAD No name (Any lesion requiring excision).
18. Test.

Derm‐CAD No name (BCC).
19. Test.

Derm‐CAD DBMIPS_UNREF (invasive melanoma or atypical intraepidermal melanocytic variants).
20. Test.

MSI‐CAD SIAscope_UNREF (invasive melanoma).
21. Test.

MSI‐CAD SIAscope_UNREF (invasive melanoma or atypical intraepidermal melanocytic variants).
22. Test.

MSI‐CAD SIAscope_UNREF (Any lesion requiring excision).
23. Test.

MSI‐CAD SpectroShade (invasive melanoma).
24. Test.

MSI‐CAD SpectroShade (invasive melanoma or atypical intraepidermal melanocytic variants).
25. Test.
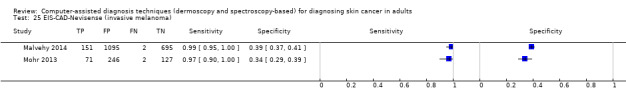
EIS‐CAD‐Nevisense (invasive melanoma).
26. Test.
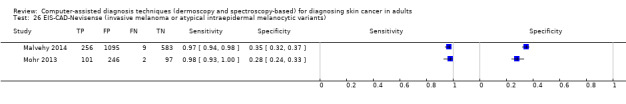
EIS‐CAD‐Nevisense (invasive melanoma or atypical intraepidermal melanocytic variants).
27. Test.
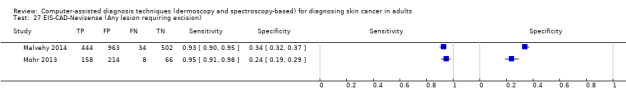
EIS‐CAD‐Nevisense (Any lesion requiring excision).
28. Test.
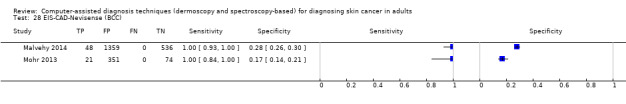
EIS‐CAD‐Nevisense (BCC).
29. Test.

EIS‐CAD‐Nevisense (cSCC).
30. Test.
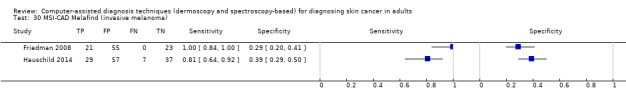
MSI‐CAD Melafind (invasive melanoma).
31. Test.
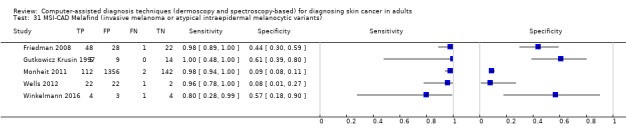
MSI‐CAD Melafind (invasive melanoma or atypical intraepidermal melanocytic variants).
32. Test.
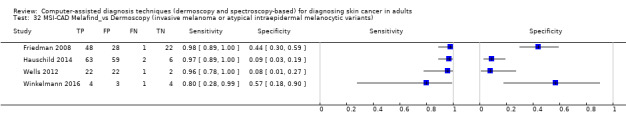
MSI‐CAD Melafind_vs Dermoscopy (invasive melanoma or atypical intraepidermal melanocytic variants).
34. Test.

MSI‐CAD SIAscopy (invasive melanoma).
35. Test.

MSI‐CAD SIAscopy (invasive melanoma or atypical intraepidermal melanocytic variants).
36. Test.

MSI‐CAD SIAscope Only_UNREF (Any lesion requiring excision).
37. Test.

DRS‐CAD OIDRS (invasive melanoma or atypical intraepidermal melanocytic variants).
38. Test.

DRS‐CAD OIDRS (Any + dysplastic).
39. Test.

DRS‐CAD OIDRS (Any lesion requiring excision).
40. Test.

MSI‐CAD TS (invasive melanoma).
41. Test.

MSI‐CAD TS (invasive melanoma or atypical intraepidermal melanocytic variants).
42. Test.

CAD–DRS–TS vs Dermoscopy (invasive melanoma or atypical intraepidermal melanocytic variants).
46. Test.

PersonDERM‐DigidermDBMIPS (invasive melanoma or atypical intraepidermal melanocytic variants).
47. Test.

ImageDERM‐DigidermDBMIPS (invasive melanoma or atypical intraepidermal melanocytic variants).
48. Test.

ImageDERM‐DigidermDEMMIPS (invasive melanoma or atypical intraepidermal melanocytic variants).
49. Test.

PersonDerm‐DigidermImageJ (invasive melanoma or atypical intraepidermal melanocytic variants).
50. Test.

ImageDerm‐DigidermNevuscreen (invasive melanoma or atypical intraepidermal melanocytic variants).
51. Test.

ImageDerm‐DigidermNR (invasive melanoma).
52. Test.
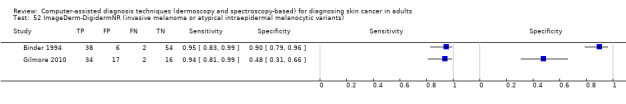
ImageDerm‐DigidermNR (invasive melanoma or atypical intraepidermal melanocytic variants).
53. Test.

PersonDERM‐DRS‐SpectroShade (invasive melanoma).
54. Test.

ImageDERM‐DRSMelafind (invasive melanoma).
55. Test.
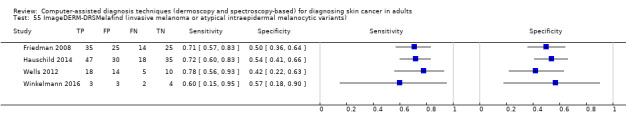
ImageDERM‐DRSMelafind (invasive melanoma or atypical intraepidermal melanocytic variants).
56. Test.

ImageDERM‐DRSSIA (invasive melanoma or atypical intraepidermal melanocytic variants).
57. Test.

PersonDERM‐DRSTS (invasive melanoma or atypical intraepidermal melanocytic variants).
58. Test.
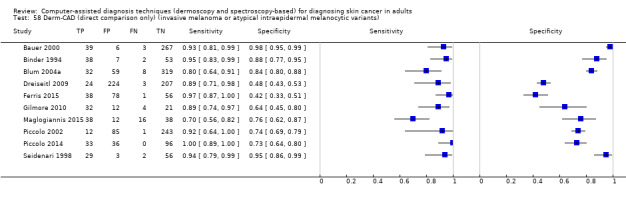
Derm‐CAD (direct comparison only) (invasive melanoma or atypical intraepidermal melanocytic variants).
59. Test.
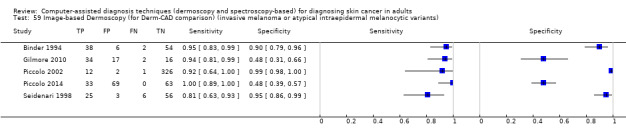
Image‐based Dermoscopy (for Derm‐CAD comparison) (invasive melanoma or atypical intraepidermal melanocytic variants).
60. Test.
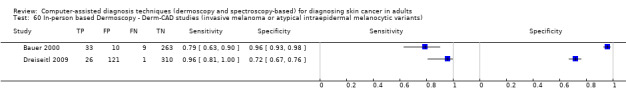
In‐person based Dermoscopy ‐ Derm‐CAD studies (invasive melanoma or atypical intraepidermal melanocytic variants).
61. Test.
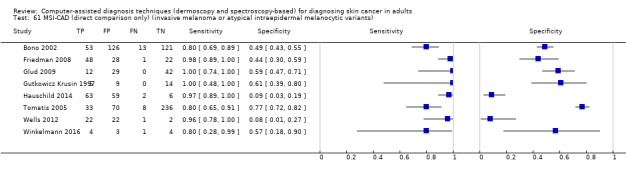
MSI‐CAD (direct comparison only) (invasive melanoma or atypical intraepidermal melanocytic variants).
62. Test.
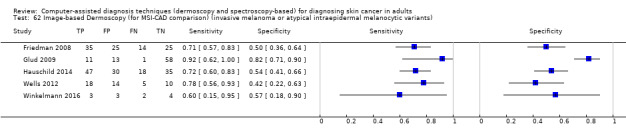
Image‐based Dermoscopy (for MSI‐CAD comparison) (invasive melanoma or atypical intraepidermal melanocytic variants).
63. Test.
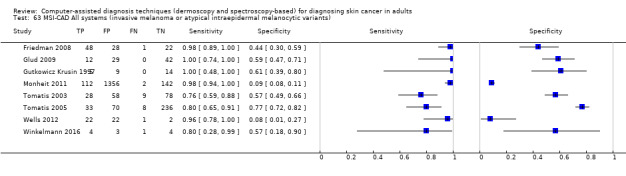
MSI‐CAD All systems (invasive melanoma or atypical intraepidermal melanocytic variants).
64. Test.
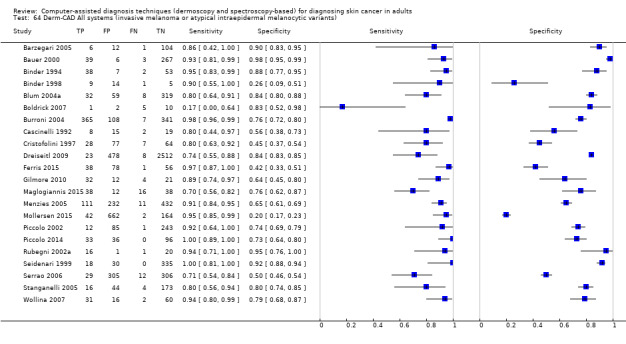
Derm‐CAD All systems (invasive melanoma or atypical intraepidermal melanocytic variants).
65. Test.

Derm‐CAD MoleExpert (Any lesion requiring excision).
66. Test.
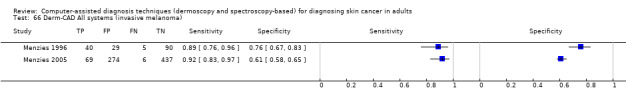
Derm‐CAD All systems (invasive melanoma).
67. Test.

Image‐based Dermoscopy ‐ Derm‐CAD studies (invasive melanoma).
68. Test.
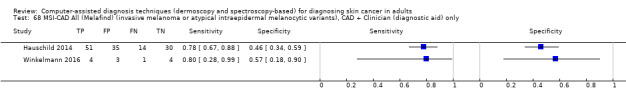
MSI‐CAD All (Melafind) (invasive melanoma or atypical intraepidermal melanocytic variants), CAD + Clinician (diagnostic aid) only.
69. Test.
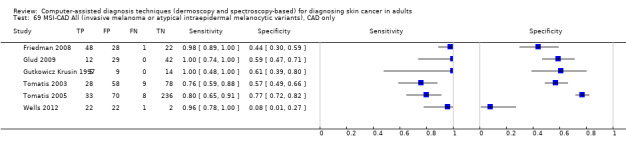
MSI‐CAD All (invasive melanoma or atypical intraepidermal melanocytic variants), CAD only.
70. Test.

MSI‐CAD Melafind (invasive melanoma or atypical intraepidermal melanocytic variants), CAD only.
71. Test.
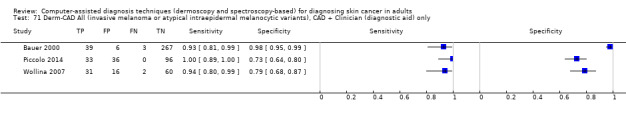
Derm‐CAD All (invasive melanoma or atypical intraepidermal melanocytic variants), CAD + Clinician (diagnostic aid) only.
72. Test.
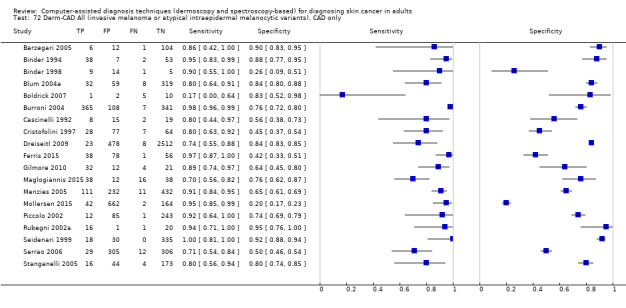
Derm‐CAD All (invasive melanoma or atypical intraepidermal melanocytic variants), CAD only.
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Ascierto 2010.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: Not reported (states in a period of 1 year) Country: Italy Test set derived: The training set consisted of 78 PSL images, comprising 19 MMs and 59 naevi of comparable size. The test set consisted of 383 lesions, including 18 MMs thinner than 0.75 mm (8 in situ). The 59 naevi belonging to the training set were randomly selected from routine material, whereas the 424 naevi of the test set represented consecutive cases. |
||
| Patient characteristics and setting |
Inclusion criteria: Clinically‐relevant cutaneous pigmented lesions, undergoing dermoscopy and excision; only melanocytic lesions meeting at least 2 clinical ABCDE criteria underwent dermoscopy Setting: Specialist unit (skin cancer/pigmented lesions clinic) Prior testing: Clinical suspicion of malignancy without dermatoscopic suspicion Setting for prior testing: Specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: None reported Sample size (participants): N eligible: 54/ N included: 54 Sample size (lesions): 54 Participant characteristics Age (yrs): ‐ Median: 41/ Range: 19 ‐ 73 years Gender: Male: 19 men Lesion characteristics: NR |
||
| Index tests |
Dermoscopy Method of diagnosis: In‐person diagnosis Prior test data: Clinical examination and/or case notes Diagnostic threshold: Qualitative ‐ very high risk ‐ lesion with a pigment network and any of the classical ELM features specific for melanoma (pseudopods, radial streaming, blue‐gray veil, atypical vessel, etc). High risk ‐ lesion with a pigment network and subtle new ELM features that may suggest melanoma but often are also seen in atypical naevi Diagnosis based on: Unclear, NR; evaluations made by expert dermatologists Number of examiners: NR Observer qualifications: Dermatologist Experience in practice: High experience or ‘Expert’ Experience with index test: High experience /‘Expert’ users 'expert dermatologists' Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: SpectroShade (MHT, Verona Italy) (classifier not reported) System details: The system provides information including a series of 15 multispectral images into the near‐infrared bandwidth. 3 spectral areas play a major role in quantification of parameters: 584 nm, 650 ‐ 750 nm, 750 ‐ 950 nm No derivation aspect (external validation study) Lesion characteristics assessed: 7 parameters: mean reflectance (MR); variegation (V); area (A); dark area ratio (DAR); dark island reflectance, DA; dark distribution factor (DDF) Additional predictors included: No further information used Method of diagnosis: In‐person diagnosis CAD‐based diagnosis Prior/other test data: Clinical examination or case notes, or both Dermoscopy CAD output: Diagnostic category: 1 no melanoma, 2 doubtful melanoma, 3 suspected melanoma, 4 probable melanoma Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Disease‐positive: 12 MM; Disease‐negative: 42 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 12 'Benign' diagnoses: 42 |
||
| Flow and timing | No exclusions reported Time interval to reference test: "Before surgery, all patients were investigated by clinical and epiluminescence microscopy (ELM) screenings" Time interval between index test(s): As above |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Was an adequate spectrum of cases used to train the algorithm? | No | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Barzegari 2005.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: NR Period of data collection: NR Country: Iran |
||
| Patient characteristics and setting |
Inclusion criteria: Pigmented skin lesions with a clinical diagnosis of melanocytic lesion ≤ 15 mm diameter referred to dermatology clinic for diagnostic evaluation or cosmetic reasons Setting: Secondary (general dermatology) Prior testing: Clinical suspicion of malignancy without dermatoscopic suspicion Participant request for evaluation/excision Setting for prior testing: NR Exclusion criteria: > 15 mm Sample size (participants): N included: 91 Sample size (lesions): N included: 122 Participant characteristics Age (yrs): Mean: 32.3/ Range: 6 ‐ 94 Gender: Male: 30; 33% Lesion characteristics: NR |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD: microDERM (DANAOS software, ANN classifier) System details: "The system consists of a special camera, which had ability to take images at ×15, ×20, ×30, and ×50 magnifications and contains a 752 × 582 pixel charge coupled device. The image analysis software was Visiomed AG (Ver. 3.50) based on an ANN that was trained using images collected in a Europe‐wide multicentre study (DANAOS)" No derivation aspect (external validation study) Lesion characteristics assessed: Lesion features analysed not described Additional predictors included: No further information used Method of diagnosis: In‐person diagnosis CAD‐based diagnosis Prior/other test data: Clinical examination or case notes, or both CAD Output: The software produces a score per lesion ranging from 0 to 10 Diagnostic threshold: 2 x 2 data for more than one diagnostic threshold Threshold determined based on ROC analysis; threshold chosen on basis of similarity to other microDERM studies: 7.34 |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Disease‐positive: 6; Disease‐negative: 116 Target condition (Final diagnoses) Melanoma (invasive): 3; Melanoma (in situ): 3 Sebhorrheic keratosis: 2; Benign naevus: 104; Dysplastic naevus 7; DF 1 AK |
||
| Flow and timing |
Excluded participants: None Time interval between index test(s): Consecutive |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Low | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Bauer 2000.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: NR. Appears retrospective? But refers to a "campaign for the early diagnosis of cutaneous melanoma (CM) by three dermatologists according to the ABCD system and using ELM evaluation" (Stanganelli 1995) Period of data collection: January 1996 to February 1997 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: "Pigmented skin lesions examined during a campaign for the early diagnosis of cutaneous melanoma (CM)" Setting: Secondary (general dermatology) from authors' institution Prior testing: NR; "campaign for the early diagnosis of cutaneous melanoma (CM)" Setting for prior testing: NR Exclusion criteria: None reported Sample size (participants): N included: 311 Sample size (lesions): N included: 315 Participant characteristics: NR Lesion characteristics NR Thickness/depth: 14 < 0.75 mm, 10 0.75 ‐ 1.5 mm, and 6 > 1.5 mm (n = 42 melanoma) |
||
| Index tests |
Dermoscopy No algorithm, possibly Pattern analysis Method of diagnosis: In‐person diagnosis Prior test data: Clinical examination or case notes or both Diagnostic threshold: Qualitative ‐ Presence of malignancy; threshold not detailed but ELM parameters included irregular and multicomponent pigmentary network pattern, peripheral dark network patches, sharp network margin, pseudopods, radial streaming, blue‐grey areas, pigment dots (blotches, black dots, brown globules), black dots at periphery, whitish veil, depigmentation and hypopigmented areas, erythema, telangiectasia, comedo‐like openings, milia‐like cysts, red‐blue areas. (ABCD appears to related to naked eye exam) Diagnosis based on: Consensus (3 observers) "the evaluation was uniform as the diagnosis was made by consensus amongst the dermatologists (Stanganelli 2005). When they disagreed a fourth dermatologist, an expert in the diagnosis of PSLs, was consulted." Number of examiners: 3 Observer qualifications: Dermatologist Experience in practice: Not described Experience with index test: Not described Any other detail: The dermatologists had all been trained in the recognition of PSLs during a training course on the clinical diagnosis of naevi and melanomas Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: DB–MIPS (Biomips engineering, Italy) (ANN classifier) System details: DBDermoMIPS (Dell'Eva‐Burroni), "which consists of a stereomicroscope (magnification ranging from 36 to 340), a high‐resolution 3CCD RGB video camera and a 486/33 MHz personal computer equipped with a 300 Mb hard disk and 16 Mb of RAM. The digital images of the lesions, shown on a second RGB video monitor, are framed at 768 3 576 true colour pixels and saved onto a 230 Mb magneto‐optic removable disk" Derivation aspect (study type) Lesion characteristics assessed: "Once the borders of the lesion have been automatically detected, the system evaluates 38 variables (grouped into geometries, colours and Burroni's islands of colours). Suspect areas of the lesion are highlighted by means of a proper algorithm called `Burroni's islands filter' based on a local histogram equalisation to produce a new enhanced image in which the darker areas have been enhanced and the shades in the green‐blue dominant areas (when present) are more evident" Additional predictors included: No further information used Method of diagnosis: Dermoscopic images CAD‐aided diagnosis (test operator not reported) Prior/other test data: No further information used CAD output: Diagnosis suggested (e.g. melanoma, benign melanocytic naevus) Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Disease‐positive: 42; Disease‐negative: 273 Target condition (Final diagnoses) Melanoma (invasive): 30; Melanoma (in situ): 12 Severe dysplasia: 25 'atypical' dysplastic; 212 benign naevus; 36 nonmelanocytic (SK, thrombosed angioma) |
||
| Flow and timing |
Excluded participants: None reported After dermoscopy and CAD, all lesions excised and examined histologically Time interval between index test(s): Consecutive Time interval to reference standard: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Binder 1994.
| Study characteristics | |||
| Patient sampling |
Study design: Case‐control Data collection: Retrospective image selection/Prospective interpretation Period of data collection: NR Country: Austria Test set derived: From a sample of 200 PSLs, 2 databases were randomly created for learning and testing purposes. The database was also provided with the histological diagnosis. |
||
| Patient characteristics and setting |
Inclusion criteria: Images of PSLs randomly selected from a PSL image database Setting: Secondary (general dermatology) Prior testing: Selected for excision (no further detail) Setting for prior testing: NR Exclusion criteria: None reported Sample size (participants): NR Sample size (lesions): 200 included (100 test set) Participant characteristics: Lesion characteristics: NR |
||
| Index tests |
Dermoscopy (Modified) pattern analysis Method of diagnosis: Dermoscopic images Prior test data: No further information used Diagnostic threshold: Qualitative ‐ Diagnosis based on: Consensus (2 observers) Number of examiners: 3 Observer qualifications: Dermatologist Experience in practice: High experience or ‘Expert’ Experience with index test: High experience /‘Expert’ users Any other detail: "The images were obtained by photographing the PSL on 24 x 36 mm colour slide film, with oil immersion, using a Wild binocular stereomicroscope M 650 (Wild Heerbrugg AG, Switzerland) at a final magnification of x16 using flashlight illumination." Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: name not reported (ANN classifier) System details: Computer analysis of stored images captured using digital stereomicroscope Derivation study (internal validation) Approach to feature selection: "An input layer of nodes represented external data (ELM characteristics), an output layer represented the class identity (diagnoses). The network processed data by accepting input patterns (the value 0 for "ELM criterion not present") and the value 1 for "ELM criterion present" into the input layer. During the learning process each input pattern (ELM pattern) had a known output pattern (histological diagnosis as the gold standard of truth) the network was expected to produce" Lesion characteristics assessed: Features analysed as present or absent (pattern analysis): Pigment network, brown globules, radial streaming, pseudopods, black dots, margin, pigmentation, depigmentation Additional predictors included: Unclear Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: NR CAD output: NR Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis alone Disease‐positive: 40; Disease‐negative: 60 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 40 Benign naevus: 60 (30 CN, 30 DN) |
||
| Flow and timing |
Excluded participants: None reported Time interval to reference test: NR Time interval between index test(s): NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Binder 1998.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Retrospective image selection/Prospective interpretation Period of data collection: NR Country: Austria Test set derived: Computer‐generated random numbers split data into learning and testing sets; relative proportion of cases in each set was "about 80% and 20%, respectively" |
||
| Patient characteristics and setting |
Inclusion criteria: PSLs with available oil immersion dermoscopic images Setting: Secondary (general dermatology) Prior testing: Selected for excision (no further detail) Setting for prior testing: Unspecified Exclusion criteria: NR Sample size (participants): NR Sample size (lesions): 120 (29 test set); N included: 120 Participant characteristics: NR Thickness/depth: Other: median 0.72 mm (range 0.3 to 1.4 mm) for 39 melanomas |
||
| Index tests |
Computer‐assisted diagnosis ‐ dermoscopy‐based Derm‐CAD system: IBAS 2000 workstation (Zeiss, Oberkochen, Germany) (ANN classifier) System details: Digital image analysis workstation attached to Wild binocular stereomicroscope M 650 (Wild Heerbrugg AG, Switzerland). "The images were obtained by photographing the PSLs on 24 x 36 mm colour slide film using a Wild binocular stereomicroscope M 650 (Wild Heerbrugg AG, Switzerland) at a final magnification of x16 or x25 using flashlight illumination" Derivation study (internal validation) Approach to feature selection: "A 3‐layer, feed‐forward neural‐network with 16 input nodes and 3 hidden nodes was trained with a backpropagation algorithm. Each morphological input feature was assigned a numerical value that was scaled so that each input ranged from 0 to 1. The network was trained to yield a value from 0 to 1 in the output nodes. The node yielding the greatest numerical output was then used as the classification result (the winning node)." Artificial neural networks 2 different ANNs were trained: "the first classified between CN and DN as benign lesions versus MM as a malignant lesion in a dichotomized model, whereas the second classified between the 3 entities of PSL examined, i.e. CN versus DN versus MM. We extracted only data for the first ANN" Lesion characteristics assessed: Analysis of 16 morphometric parameters from the lesion and the border image: lesion area and perimeter: minimum polar distance, maximum polar distance, aspect ratio, circularity shape factor, variances of grey, number of different colours, range of different colours. Border features and area: maximum and minimum border width, ratio of border area to lesion area, ratio of border perimeter to lesion perimeter Additional predictors included: No further information used Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: No further information used CAD output: Dichotomous decision: MM vs. benign (CN or DN) Diagnostic threshold: The network was trained to yield a value from 0 to 1 in the output nodes. The node yielding the greatest numerical output was then used as the classification result |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Histology (not further described): N of participants/lesions: 29/120 lesions included in test set Disease‐positive: 10; Disease‐negative: 19 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 39 melanomas reported for whole dataset including 5 in situ melanoma; the test set included 10 melanomas (number in situ not reported). Other: common naevi and dysplastic naevi: 19 |
||
| Flow and timing |
Exclusion of lesions from analysis: 6 lesions excluded due to incorrect segmentation results Time interval to reference test: NR; "after photography" |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Unclear | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Blum 2004a.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: 11 November 1998 ‐ 2 March 2000 Country: Germany Test set derived For validation of a new CAD procedure the complete collection (837 melanocytic lesions) was divided into 2 equal random subgroups n1 (training set) and n2 (test set) |
||
| Patient characteristics and setting |
Inclusion criteria: Melanocytic skin lesions imaged prospectively at the Pigmented Lesion Clinic of the Department of Dermatology, University of Tuebingen, Germany Setting: Specialist unit (skin cancer/pigmented lesions clinic) Prior testing: NR Setting for prior testing: Specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: images from mucous membrane areas were excluded Sample size (participants): NR Sample size (lesions): N eligible: 837/ N included: 837 (test set 418) Participant characteristics: NR Lesion characteristics Thickness/depth: ≤ 1 mm: Median breslow thickness for all melanomas 0.78 mm (range 0.10 ‐ 3.50) |
||
| Index tests |
Dermoscopy: 7FFM; 7‐point checklist; ABCD; Menzies criteria Method of diagnosis: Dermoscopic images Prior test data: No further information used Diagnostic threshold: NR Diagnosis based on: Single observer (n = 1) Number of examiners: 1 Observer qualifications: Dermatologist Experience in practice: Not described Experience with index test: Not described Any other detail: "The colour video camera MediCam 400 with Y/C 1 signal exit had a ¼‐inch charged‐couple device shooting element with 470,000 pixels (picture elements). The focal area for the dermoscopic pictures was defined from 3.5 cm diameter up to infinity. The focal area for the dermoscopic pictures could be positioned continuously by zoom from 3.2 mm to approx. 1.0 cm, corresponding to a x20–70 magnification on a 17‐inch monitor. Lesions ≤ 12 mm diameter could be imaged completely. The glass plate contacting the skin was always moistened with disinfectant spray." "According to the established dermoscopic classification rules (ABCD rule, Menzies’ score, 7‐point checklist and 7 features for melanoma) the lesions were prospectively classified as benign or malignant melanocytic lesions by the principal investigator (AB)." (referenced to Argenziano 1998; Dal Pozzo 1999; Menzies 1996) Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD: System name NR (Vision algebra classifiers) System details: Computer analysis of stored images captured using digital microscope Derivation study (internal validation) "The analytical parameters of the digital dermoscopy analysis were reduced by means of a factor analysis. In a second step, the impact of the different parameters was examined by logistic regression analysis. The number of parameters included in the multivariate analysis was limited in relation to the number of malignant melanomas: in the sample of large, partially‐imaged lesions it was restricted to 6 parameters and for small, completely‐imaged lesions it was limited to 3 parameters." Lesion characteristics assessed: "Analysis of 64 analytical parameters including: a large number of morphological parameters such as margin, geometric parameters (surface area, extent, largest diameter and largest orthogonal diameter), invariant moments, symmetry, colours (red, green, blue and grey value), texture (energy, entropy, correlation, inverse difference moment and inertia), number of regions, focus and difference of the lesion and its convex cover." Additional predictors included: Unclear Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: Unclear CAD output: NR Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis plus follow‐up Histology Disease‐positive: 84; Disease‐negative: 185 Clinical follow‐up plus histology of suspicious lesions: unexcised lesions were analysed independently by 2 of the investigators 2 ‐ 3 times in 6 months on the basis of dermoscopic criteria. These lesions were classified as benign without any suspicion of malignancy by dermoscopic criteria, and follow‐up records for at least 6 months showed no evidence of malignancy. Disease‐negative: 568 Target consition (Final diagnoses) Melanoma (invasive): 71; Melanoma (in situ): 9; Lentigo maligna 4 'Benign' diagnoses: 766 |
||
| Flow and timing |
Excluded participants: none reported Time interval to reference test: NR |
||
| Comparative | Time interval between index test(s): not reported | ||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| Low | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Unclear | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | No | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | No | ||
| High | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| High | |||
Boldrick 2007.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Retrospective CAD Period of data collection: January 2002 and August 2005 Country: USA |
||
| Patient characteristics and setting |
Inclusion criteria: Patients ≥ 18 years of age Setting: Specialist unit (skin cancer/pigmented lesions clinic) Prior testing: Clinical and/or dermatoscopic suspicion Setting for prior testing: Unspecified Exclusion criteria: None reported Sample size (participants): N eligible: 83; N included: 12 Sample size (lesions): N eligible: 1000; N included: 18 Participant characteristics: |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based CAD‐Derm‐Other MicroDerm/DANAOS Derm‐CAD system: MicroDERM (DANAOS software, ANN classifier) System details: Dermoscopy unit with internal camera containing DANAOS analysis system. "The hand unit contained a miniature charged coupled device (3CCD) with a camera with a resolution of 768 3 576 (440,000) pixels. Digital images were stored and compresed using JPEG format." No derivation aspect (external validation study) Lesion characteristics assessed: Dermoscopic features of PSLs based on the ABCD rule Additional predictors included: None reported Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: No further information used CAD output: DANAOS score indicating risk of malignancy Diagnostic threshold: DANAOS score 2 x 2 data for > 1 diagnostic threshold Per‐patient data reported. Threshold selected on basis of similarity to other microDERM studies: DANAOS score of ≥ 7 |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Histology (biopsy): N patients/lesions: 18 PSL Disease‐positive: 6 MM; Disease‐negative: 12 Target condition (Final diagnoses) Melanoma (invasive): 3; Melanoma (in situ): 2; Lentigo maligna 1 Severe dysplasia: 1; Mild/moderate dysplasia: 6; benign naevus: 5 |
||
| Flow and timing |
Exclusion of lesions from analysis: Review team ‐ only 18 lesions had an adequate reference standard; 982 clinically dx benign lesions without a reference standard were not extracted. Interval between index test and reference standard: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Bono 1996.
| Study characteristics | |||
| Patient sampling |
Study design: Unclear Data collection: NR Period of data collection: Between March 1993 and Oct 1994 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: Pigmented skin lesions at the Instituto Nazionale Tumori of Milan Setting: Specialist unit (skin cancer/pigmented lesions clinic) Instituto Nazionale Tumori of Milan Prior testing: NR Setting for prior testing: NR Exclusion criteria: None reported Sample size (participants): N eligible: 45 Sample size (lesions): N eligible: 54/ N included: 43 Participant characteristics: NR Lesion characteristics: Site ‐ face/ears: 3 (6%); trunk: 39 (72%); limbs: 12 (22%); 10 MM ≤ 1 mm depth; median size: 10 mm (4 to 40 mm) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: Telespectrophotometric System (Linear discriminant classifier) System details: Digital camera coupled with an illumination system with interference filters and computer for storage and analysis of multispectral images No derivation aspect (external validation study) Described in prior study Marchesini 1995 Lesion characteristics assessed: From each spectral image, 3 parameters (i.e. mean reflectance, variegation index and lesion area) were derived at the corresponding wavelength Additional predictors included: None reported Method of diagnosis: Spectroscopic images CAD‐based diagnosis Prior/other test data: Unclear CAD output: NR Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis ‐ Disease‐positive: 18; Disease‐negative: 25 Expert opinion ‐ Disease‐negative: 11 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 18 Mild/moderate dysplasia: 8 dysplactic naevi Benign naevus: 17 common melanocytic naevi |
||
| Flow and timing |
Excluded participants: Only 43 lesions had complete clinical and histological information. 11 lesions not surgically removed had only clinical diagnosis (benign) and were not included in the final accuracy analysis Time interval to reference test: NR Time interval between index test(s): NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | No | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | No | ||
| High | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Bono 2002.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: June 1998 to March 2000 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: "Cutaneous pigmented lesions with clinical and/or dermatoscopic features that suggested a more or less important suspicion of CM" Setting: Specialist unit (skin cancer/pigmented lesions clinic) Prior testing: Clinical and/or dermatoscopic suspicion Setting for prior testing: Specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: Location/site of lesion: Awkwardly situated lesions e.g. interdigital space, ears, nose or eyelids. Lesions on scalp excluded due to hair interference with reflectance Lesion size: obvious large, thick melanomas Sample size (participants): N included: 298 Sample size (lesions): N included: 313 Participant characteristics: Mean age: 40 years (10 ‐ 86); Male: 122 (41%) Lesion characteristics: Lesion site: head/neck: 3%; trunk: 61%; limbs: 36%; Thickness ≤ 1 mm: 70% (46/66); for 55 invasive MM: median thickness 0.64 mm, range 0.17 ‐ 3.24 mm. Median diameter: 11 mm (3 ‐ 31 mm) |
||
| Index tests |
Visual inspection (VI) No algorithm (training in the unit is based on ABCD but subjective experience of the clinician used for diagnosis) Method of diagnosis: In‐person diagnosis Prior test data: N/A; in‐person diagnosis Diagnostic threshold: Clinical diagnostic criteria based on subjective experience; emphasise lesion colour over dimensions. Diagnosis of suspect CM made when the level of suspicion was 'roughly 50% or more'. ABCD criteria have been the basis of training at the unit, but is not implemented in diagnosis; preferred emphasis on colour rather than dimensional character Diagnosis based on: Single observer (n = 1) Observer qualifications: Surgical oncologists Experience in practice: High experience or ‘Expert’; over 5 years Dermoscopy: No algorithm Method of diagnosis: In‐person diagnosis Prior test data: Clinical examination and/or case notes Diagnostic threshold: Presence of at least 1 of the following criteria: radial streaming, pseudopods, grey‐blue veil, regression and erythema, whitish veil, black dots at the periphery (if network present), thick irregular network or milky‐red background with red dots Test observers as described for Visual Inspection (above) Experience with index test: > 5 years Any other detail: Dermatoscopy performed by a hand‐held monocular microscope equipped with an achromatic lens permitting a magnification of x10 (Heine Delta 10) Computer‐assisted diagnosis ‐ Spectroscopy‐based CAD‐Spect‐Other Described as; referenced to MSI‐CAD system: 'telespectrophotometry' (Linear discriminant classifier) System details: The TS consists mainly of a charge‐coupled device camera that is provided with a set of 17 interference filters and a personal computer to allow imaging of cutaneous pigmented lesions at selected wavelengths from 420 to 1040 nm. The acquired 17 spectral images are stored in the personal computer for offline processing. Intensity levels as well as the dimensions of the image picture elements (pixels) were calibrated according to a set of 4 reflectance standards and a geometric reference frame, respectively. Details on the system's feautres have been reported elsewhere (Marchesini 1995). No derivation aspect (external validation study): Derivation described in Marchesini 1995 Lesion characteristics assessed: For each spectral image, 5 parameters (lesion descriptors) based on ABCD and related to colour and shape of the imaged lesion were evaluated: mean reflectance, variegation index, roundness, border irregularity (only four listed in study report). Additional predictors included: No further information used Method of diagnosis: In‐person diagnosis CAD‐based diagnosis Prior/other test data: None CAD output: NR Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis alone Target condition (Final diagnoses) Melanoma (invasive): 55; Melanoma (in situ): 11; BCC: 6 'Benign' diagnoses: 241; 151 compound naevus, 24 junctional naevus, 12 dermal naevus, 12 lentigo simplex, 10 dysplastic naevus, 8 spindle‐cell naevus, 8 sebhorrheic keratosis, 5 blue naevus, 3 spitz naevus, 8 other |
||
| Flow and timing |
Excluded lesions from analysis: None reported Intervals between tests: Appears consecutive but not fully clear |
||
| Comparative | Time interval between index test(s): Appears consecutive but not fully clear | ||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Was an adequate spectrum of cases used to train the algorithm? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Low | High | ||
| DOMAIN 2: Index Test Visual inspection | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Burroni 2004.
| Study characteristics | |||
| Patient sampling |
Study design: Case control Data collection: Retrospective image selection/Prospective interpretation Period of data collection: 1999 ‐ 2003 Country: Italy Test set derived: For the 3 linear classifiers, "lesions from each centre (Rome and Siena) were randomly allocated to training and test sets. Linear classifier 1 was constructed from the Rome training set and tested on all lesions from Siena; Linear classifier 2 was constructed on the Siena training set and tested on all lesions from Rome; Linear classifier 3 was constructed on training sets from both centres and tested on test sets from both centres. For the K‐nearest‐neighbour (K‐nn) classifiers, a separate training set of lesions were selected from the image databases of several institutions that iused the same ELM instrumentation, i.e. IDI‐Rome; Siena University Dermatology Clinic; IDI‐Capranica; and the Italian Cancer League Clinics of Grosseto, Livorno, Arezzo, Trento, and Siena. It was then tested on all lesions from both centres as described above." |
||
| Patient characteristics and setting |
Inclusion criteria: All melanomas undergoing ELM and excision at the 2 centres (1999 ‐ 2003) and random sample of surgically‐removed benign melanocytic lesions, inlcuding 85 histologically atypical naevi Setting: Secondary (general dermatology) Dept Dermatology, University of Siena Specialist unit (skin cancer/pigmented lesions clinic) Instituto Dermatopatico dell'Immacolata (IDI), a research hospital for skin diseases in Rome Prior testing: Selected for excision (no further detail) Setting for prior testing: Secondary (general dermatology) Siena Specialist unit (skin cancer/pigmented lesions clinic) Rome Exclusion criteria: None reported Sample size (participants): NR Sample size (lesions): N included: 821 (475 from Siena; 346 from Rome) Participant characteristics: Thickness/depth: Other: 178 (48% of 372 MM) ≤ 0.75 mm |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: DB‐Mips system (Knn and Linear discrimination classifiers) Classifier selected at random for analysis in review: Knn System details: Dermoscopy unit, internal steromicroscope, internal DB, pattern analysis system Derivation study (internal validation) Discriminant analysis used to identify features for which there was a significant (t test) difference between melanomas and non‐melanomas and, within these diagnostic classes, no significant difference between centres. Details provided. Selected variables were: geometric variables ‐ area, variance of contour symmetry, fractality of borders. Colour variables ‐ mean skin‐lesion gradient, variance of border gradient, and border interruptions. texture variables ‐ mean contrast and entropy of lesion islands of colour variables ‐ dark area, blue‐grey area*, transition region imbalance. Lesion characteristics assessed: 38 parameters belonged to 4 categories (referenced to Soyer 2000): geometries; colors; textures; and islands of colour (i.e. colour clusters inside the lesion). These were all described in detail. Additional predictors included: No further information used Method of diagnosis: In‐person diagnosis CAD‐based diagnosis Prior/other test data: None reported CAD output: Diagnosis suggested (e.g. melanoma, benign melanocytic naevus) Diagnostic threshold: Threshold not reported The method of receiver operating characteristic curves was used to identify the threshold value for a fixed sensitivity of 95%. "The prevalence of melanomas among the first 100 closest neighbours was determined, and the lesion was assigned to the melanoma group if the prevalence was higher than a threshold value T 100. The method of receiver operating characteristic curves was used to identify the T 100 value necessary for a sensitivity of 98%" |
||
| Target condition and reference standard(s) |
Reference standard
‐ Histological diagnosis alone Histology (excision) N participant/lesions: 821 (475 Siena; 346 Rome) Disease‐positive: 372 (217 Siena; 155 Rome); Disease‐negative: 449 (258 Siena; 191 Rome) Target condition (Final diagnoses) Melanoma (invasive): 302; Melanoma (in situ): 70 (higher % at Siena than in Rome) Severe dysplasia: 85 (architectural disorder and melanocytic atypia) Benign naevus: 364 |
||
| Flow and timing |
Participant exclusions: None Intervals between tests: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Cascinelli 1992.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: March ‐ December 1991 Country: Italy Test set derived: Derivation of test set not described.Training set: 169 lesions; 124 benign and 45 malignant lesions, Test series: 44 images, 33 benign lesions and 12 malignant, of which 10 were melanoma |
||
| Patient characteristics and setting |
Inclusion criteria: Study inclusion criteria Training set: Pigmented cutaneous lesions were referred to Institute for a second opinion; Test set: not described Setting: Secondary (general dermatology) Surgical Oncology Prior testing: Training set: Referred for second opinion; basis not reported; Test set: not described Setting for prior testing: Unspecified Exclusion criteria: None reported Sample size (participants): N included: Training set: 165, Test set: not described Sample size (lesions): N included: Training set: 169, Test set: 44 Participant characteristics Age (yrs): Other Training set: 17 aged < 20; 59 aged 21 ‐ 40; 66 aged 41 ‐ 60; 23 aged > 61 Test set: not described Gender: Male: Training set: 70 (42%); Test set: not described Lesion site: Head/neck: Training set: 7.7%; trunk: Training set: 45.5%; limbs: Training set: 46.7% |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: Skin View (classifier NR) System details: Computerised image analysis system, digital television, videocamera. Connection with the computer is through a digitising board able to process colour images Derivation study (internal validation): Lesion characteristics assessed: Features of ABCD system plus clinical data (anatomical site, months of growth, size, shape, colour, ulceration or regression) 8 binary indicators generated: shape, clinical data, size, colour, darkness, saturation, border, and texture (all described) Additional predictors included: Predictors included clinical data, which takes into account anamnestic data provided by the clinician (change in size, change in colour) and an objective evaluation made by the clinician (presence of regression, presence of ulceration) Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: None reported CAD output: CAD–based diagnosis Diagnostic threshold: Malignant lesion ≥ 2 positive indicators |
||
| Target condition and reference standard(s) |
Reference standard
Histological diagnosis alone Histology (excision) N participant/lesions: Test set only: 44 Disease‐positive: 12; Disease‐negative: 32 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 10; BCC: 2 'Benign' diagnoses: 32 |
||
| Flow and timing |
Excluded participants: NR Time interval to reference test: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Unclear | ||
| Was model overfitting accounted for during model development? | Unclear | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Unclear | |||
Cristofolini 1997.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: November 1992 to September 1993 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: "Patients with small and flat common and atypical pigmented skin lesions recruited during a health campaign for the early diagnosis of CM underwent clinical diagnosis, computerised analysis by SVS [Skin View System] and subsequent skin biopsy" Setting: Secondary (general dermatology) Prior testing: No prior testing Setting for prior testing: Secondary (general dermatology) Exclusion criteria: None reported Sample size (participants): 176 included Sample size (lesions): 176 included Participant characteristics: NR Lesion characteristics: NR |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: Skin View (classifier NR) System details: Computerised image analysis system, digital television, videocamera. Connection with the computer is through a digitising board able to process colour images No derivation aspect (external validation) Lesion characteristics assessed: Features of ABCD system plus clinical data (anatomical site, months of growth, size, shape, colour, ulceration or regression) 1. shape (asymmetry); 2. clinical data (changes through time, regression, ulceration); 3. size (mm); 4. colour (distribution of hue); 5. darkness (percent of black mixed with the hue); 6. saturation (percent of white mixed with the hue); 7. border (sharpness of transition between lesion and healthy skin; 8. texture. Additional predictors included: Clinical data Method of diagnosis: Dermoscopy images CAD‐based diagnosis Prior/other test data: None reported CAD output: CAD–based diagnosis Diagnostic threshold: ≥ 2 of 8 binary (on/off) indicators indicates malignancy |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis alone Disease‐positive: 35; Disease‐negative: 141 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 35 Other: 141 melanocytic naevi |
||
| Flow and timing |
Excluded participants: Not reported Time interval to reference test: "subsequent skin biopsy" |
||
| Comparative | Time interval between index test(s): not reported‐appears to be simultaneous | ||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test Visual inspection | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Dreiseitl 2009.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: Test set: February ‐ November 2004 Country: Austria Test set derived: Study focuses on test set but gives detail of separate study in which classifier was trained |
||
| Patient characteristics and setting |
Inclusion criteria: Patients presenting at PSL clinic at Dept Dermatology which serves as a secondary and tertiary referral centre Setting: Specialist unit (skin cancer/pigmented lesions clinic) "The PSL unit of the Department of Dermatology at the Medical University of Vienna serves as a secondary and tertiary referral centre" Prior testing: NR Setting for prior testing: Specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: None reported Sample size (participants): N eligible: 511; N included: 458 with complete information Sample size (lesions): N eligible: 3827; N included: 3021 Participant characteristics: None reported Lesion characteristics: None reported |
||
| Index tests |
Dermoscopy: No algorithm Method of diagnosis: In‐person diagnosis Prior test data: Clinical examination and/or case notes Diagnostic threshold: Not reported Diagnosis based on: Single observer (n = 1) Observer qualifications: Expert dermatologist Experience in practice: High experience or ‘Expert’ Experience with index test: High experience/‘Expert’ users Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: Image J (NIH, Bethesda, USA) (ANN classifier) System details: Image analysis coupled with dermoscope MoleMax II No derivation aspect (external validation) Described in prior study Hable 2004 (PhD thesis) Lesion characteristics assessed: 29 features analysed from 38 extracted features describing shape, form and colour. Approach to feature selection Prior study: A stepwise feature selection method used to identify 29 features relevant for the classification process Additional predictors included: No further information used Method of diagnosis: In‐person diagnosis CAD‐based diagnosis CAD–aided diagnosis Test observers: Single observer (n = 6) Observer qualifications: Other: The educational training of the 6 participating physicians ranged from no training in dermatology to 4 years training in dermatology Experience in practice: Mixed experience (low and high experience combined) Experience with index test: Mixed experience (low and high experience combined) No physician was specifically trained in dermatoscopy Prior/other test data: Clinical examination and/or case notes CAD output: 2 outputs: 1) Visual rendering of analysis showing coloured areas. 2) Excision vs no‐excision decision (system considers the green zone of the scale as benign (0 to 0.1), the yellow zone suspicious (0.1 to 0.4), and the red zone malignant (0.4 to 1)) Diagnostic threshold: Scale 0 – 1: 0 – 0.1 = benign, 0.1 – 4 = suspicious, > 0.4 = malignant |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis plus follow‐up Histology (excision); N participant/lesions: NR Clinical follow‐up plus histology of suspicious lesions: Length of follow‐up: 6 months; N participants: NR Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 27 participants; 31 lesions 'Benign' diagnoses: 431 participants; 2990 lesions |
||
| Flow and timing |
Excluded participants: 806 lesions (53 participants) with inadequate follow‐up Intervals between tests: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | No | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| High | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Ferris 2015.
| Study characteristics | |||
| Patient sampling |
Study design: Unclear. Some dermoscopic images were collected prospectively and some were obtained from a collection of existing images; selection process not described Data collection: Retrospective image selection/Prospective interpretation Period of data collection: NR Country: USA Test set derived: Some dermoscopic images used to train the classifier were obtained from publicly available or purchased image libraries, which were not included in the reader study or used to test the performance of the classifier. The image set was randomly divided into 2 by diagnosis, with half used for training and half used for testing, with the exception that all high‐grade dysplastic naevi were exclusively assigned to the training set to increase the representation of dermoscopic features that could be present in melanoma. Results are presented only for the test set. |
||
| Patient characteristics and setting |
Inclusion criteria: Dermoscopic images of skin lesions excised on the basis of clinical suspicion of malignancy, with available histologic diagnoses Setting: Secondary (general dermatology) Prior testing: Clinical and/or dermatoscopic suspicion; selected for excision (no further detail) Setting for prior testing: Secondary (general dermatology) Exclusion criteria: High‐grade dysplastic naevi were not included in the test set Sample size (participants): N eligible: not reported; N included: not reported Sample size (lesions): N eligible: 473 (includes 273 randomised to training set and 27 non‐biopsied lesions); N included: CAD‐Derm test set 173 lesions; Dermscopy 65 lesions Participant characteristics: None reported Lesion characteristics: Test set: mean lesion thickness 0.76 mm, median 0.5 mm, range 0.2 ‐ 2.98 mm); Reader study: mean 0.93 mm, median 0.74 mm, range 0.2 ‐ 2.98 mm |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: name NR (Digital forest classifier) System details: Computer analysis of stored images captured using different dermoscopy/camera combinations Derivation study (internal validation) Lesion characteristics assessed: 54 features analysed, such as border irregularity, eccentricity, length of major and minor axes, and colour histogram properties. Variations of some features described in Zortea 2014 were included Additional predictors included: Unclear Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: Unclear CAD output: Severity score (the fraction of decision trees (n = 1000) in which the path ends in ‘‘malignant’’) Diagnostic threshold: 0.4; a lesion was classified as malignant if its image traced a path to a malignant node in at least 40% of the decision trees |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone All lesions were biopsied based on clinical suspicion of malignancy. All histologic diagnoses were rendered by at least 1 board‐certified dermatopathologist and were used as the reference standard for diagnosis Disease‐positive: Derm 25 MM; CAD 39 MM; Disease‐negative: Derm 40; CAD 134 Target condition (Final diagnoses) Melanoma (invasive): Derm 15; CAD 25; Melanoma (in situ): Derm 10; CAD 14; BCC: CAD 11; cSCC: CAD 3 Mild/moderate dysplasia: CAD 47; Derm 16. Sebhorrheic keratosis: CAD 11; Derm 4. Benign naevus: CAD 42; Derm 14. Other: CAD 10 lentigines, 5 blue naevi, 2 Spitz naevi, 2 angiomas, and 1 dermatofibroma; Derm 2 blue naevi, 4 lentigines |
||
| Flow and timing |
Excluded participants: None reported Time interval to reference test: "Dermoscopic images of skin lesions were collected before biopsy" |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Unclear | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Friedman 2008.
| Study characteristics | |||
| Patient sampling |
Study design: Case‐control Data collection: Retrospective image selection/Prospective interpretation Period of data collection: NR; lesions selected in July 2005 Country: USA Test set derived: MelaFind data randomly split into training and test sets, but Melafind has previously been evaluated, the only difference here being that only small lesions were included. Would argue that full dataset can reasonably be included here rather than test set only |
||
| Patient characteristics and setting |
!nclusion criteria: A database of images of pigmented skin lesions ≤ 6 mm was used to sample images of melanoma and non‐melanoma lesions; high‐grade dysplastic naevi were excluded Setting: A digital dermoscopic database acquired by Electro‐Optical Sciences Inc for the development and testing of MelaFind; 26 clinical sites have contributed Prior testing: Selected for excision (no further detail). All lesions excised or underwent shave biopsy Setting for prior testing: NR Exclusion criteria: High‐grade dysplastic naevi were excluded. Previously biopsied, ulcerated, or bleeding lesions also excluded, as were those on mucosal surfaces and lesions that contained foreign matter (e.g. tattoos) Sample size (participants): N included: 94 Sample size (lesions): N eligible: 1977; N included: 99 Participant characteristics: None reported Lesion characteristics: 21 invasive MM: median thickness 0.32 mm (0.10 ‐ 1.40 mm). Lesion size: Range: 2 mm to 22 mm |
||
| Index tests |
Dermoscopy: No algorithm Method of diagnosis: Dermoscopic images Prior test data: Clinical examination and/or case notes: sex, age, and lesion location Diagnostic threshold: NR. 2 x 2 reported for: diagnostic sensitivity and specificity, i.e melanoma vs not melanoma and biopsy sensitivity and specificity, i.e excise lesion vs not excise. Each reader had to answer the question: “Is this lesion a melanoma?” and “Would you biopsy/excise this lesion?” with a reason for biopsy. If readers indicated that they would biopsy the lesion because they were sure it was melanoma or to rule out melanoma, then the case was considered true positive (TP) Diagnosis based on: Average; mean and median reported (n = 10) Observer qualifications: 9 dermatologists; 1 nurse practitioner specialising in dermatology Experience in practice: High experience or ‘Expert’ Experience with index test: High experience /‘Expert’ users Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: MelaFind (EO Sciences, USA) (6 constrained linear classifiers) System details: Multispectral imaging system with integrated image analysis software; device takes images in vivo External validation study Derivation described in prior study Gutkowicz Krusin 1997; Elbaum 2001; Gutkowicz‐Krusin 2000. Lesion characteristics assessed: NR Additional predictors included: None reported Method of diagnosis: Spectroscopic images CAD‐based diagnosis Prior/other test data: No further information used CAD output: Binary output: excise or follow–up Diagnostic threshold: A lesion is recommended for biopsy to rule out melanoma only if all scores are above the threshold value Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis alone Disease‐positive: 49; Disease‐negative: 50 Target condition (Final diagnoses) Melanoma (invasive): 21; Melanoma (in situ): 28; BCC: 2 Mild/moderate dysplasia: 32 low‐grade dysplastic; sebhorrheic keratosis: 2; 14 other benign |
||
| Flow and timing |
Excluded participants: None reported Interval between tests: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Garcia Uribe 2012.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: NR Country: USA Test set derived: "Of the 407 PSLs, 271 were used for the training sets of ANN classifiers (Tables 1 and 2) to separate malignant melanoma from varieties of naevi. The remaining 136 data sets were used to test the efficacy of the ANN classifiers." The non‐pigmented lesions consisted of BCCs, SCCs, benign actinic keratoses, and seborrheic keratoses. Among the 266 non‐pigmented lesions, 177 were used to train the ANN classifier and the remaining 89 were used for testing |
||
| Patient characteristics and setting |
Inclusion criteria: NR Setting: Specialist unit (skin cancer/pigmented lesions clinic) Prior testing: NR Setting for prior testing: Unspecified Exclusion criteria: None reported Sample size (participants): NR Sample size (lesions): 136 included Participant characteristics: Pigmented (%): 407 pigmented lesions (60%) Non‐pigmented (%): 266 non‐pigmented (40%) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based DRS‐CAD system: OIDRS (ANN classifier) System details: Light probe coupled to imaging spectrograph, camera, and computer to store images. "The system was built onto a portable cart; it was easily moved to the patient examination rooms. To target both small and large skin lesions, we constructed an optical fiber probe using micromachining technology. The probe consisted of 3 source fibers and 2 linear arrays of 12 collection fibers within an area of 2. The collection fibers were coupled to an imaging spectrograph that generated an optical spectrum from 455 to 765 nm for the collection channel. A charge‐coupled device (CCD) camera collected the spectral images, which were stored on a com‐ puter for data analysis. The data collection took less than 5 minutes, and it did not interfere with the standard health care provided to the patients" Derivation study (internal validation) "A physician identified the lesion(s) to be measured before the scheduled biopsy. To average out the effect of structural anisotropy of the skin tissue, the measurement of each lesion was repeated 4 times to obtain images from different orientations. To provide self‐references, the same measurements were also repeated on the neighboring healthy skin tissues. The anisotropy is defined as the variation of the measurements when conducted in different directions. After the measurements were completed, a biopsy was carried out for each skin lesion and submitted for histopathologic analysis." Lesion characteristics assessed: NR Additional predictors included: Unclear Method of diagnosis: In‐person diagnosis CAD‐based diagnosis Prior/other test data: Unclear CAD output: Diagnostic category (e.g. CN, MM, DN, BCC, cSCC) Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 10 Severe dysplasia: 15; Mild/moderate dysplasia: 83; Benign naevus: common naevi 28 |
||
| Flow and timing |
Excluded participants: NR Time interval to reference test: Biopsy was done after the CAD‐OIDRS measurements were taken |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Yes | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Unclear | ||
| Was model overfitting accounted for during model development? | No | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Unclear | |||
Gilmore 2010.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Retrospective image selection/Prospective interpretation Period of data collection: 2003 ‐ 2008 Country: Austria Test set derived: NR. Training set: 65 melanomas and 65 dysplastic naevi; Test set: 36 melanomas and 33 dysplastic naevi |
||
| Patient characteristics and setting |
Inclusion criteria: Polarised dermoscopic images of atypical melanocytic lesions were obtained from the Department of Dermatology at the Medical University of Graz in Austria; describes database as a "database may be considered a random, but representative, cohort" but does not describe method of selection Setting: Secondary (general dermatology) Prior testing: Clinical and/or dermatoscopic suspicion atypical melanocytic lesions Setting for prior testing: NR Exclusion criteria: None reported Sample size (participants): N included: NR Sample size (lesions): N included: 199: Derivation set N 130 Test set N 69 Participant characteristics: None reported Lesion characteristics: None reported |
||
| Index tests |
Dermoscopy: No algorithm; dermoscopic method of diagnosis NR Method of diagnosis: Dermoscopic images Prior test data: No further information used Diagnostic threshold: NR; subjective impression to excise or not Diagnosis based on: Single observer (n = 1) Observer qualifications: Dermatologist Experience in practice: High experience or ‘Expert’ Experience with index test: High experience/‘Expert’ Images captured using a DermLite FOTO lens (3Gen LLC; Dana Point, CA, USA) coupled to a digital camera (Nikon CoolPix4500; Nikon Corporation, Tokyo, Japan) without flash using the camera’s auto setting Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: name NR (SVM classifier) System details: Computer analysis of stored images captured using digital microscope Derivation study (internal validation) "Feature data from the training set were first normalised to zeromean and unit variance. We then reduced the dimensionality of this set by taking the first three principal components, corresponding to the points to the left and including the infection point of the hyperbolic eigenvalue curve." Training set (p832): "At each step, corresponding to a unique parameter regime, we took 60 random data points (30 of each class) from our 130 training data points to derive a model, and then we tested that model on 30 randomly chosen data points from the same data set. To assess the effectiveness of the model in classification, we performed a tenfold cross‐validation. Because we are using only a subset of the total training set to derive each model Ð this is loosely analogous to the subset selection procedure known as chunking ‐ finding the optimal solution is computationally fast. Each tenfold cross‐validation took approximately 200 s using Mathematica 6.0 on a Macintosh G4 with 4MB of RAM." Lesion characteristics assessed: 14 features investigated: 1. Asymmetry 1 (mean int.) 2. Asymmetry 2 (mean int.) 3. Asymmetry 3 (variance red) 4. Asymmetry 4 (variance red) 5. Variance red 6. Variance green 7. Variance blue 8. Mean red 9. Mean green 10. Mean blue 11. Mean intensity 12. Range red intensit* 13. Range green intensity 14. Range blue intensity Additional predictors included: No further information used Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: Unclear CAD output: NR Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis alone "All lesions were excised and examined microscopically by expert dermatopathologists using standard histopathologic diagnostic criteria" Disease‐positive: 36 test set and 65 derivation set; Disease‐negative: 33 test set and 65 derivation set Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 36 test set and 65 derivation set Dysplastic naevi 33 test set and 65 derivation set |
||
| Flow and timing |
Excluded participants: None reported Intervals between tests: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | No | ||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Unclear | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | No | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Glud 2009.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: January to April 2007 Country: Denmark |
||
| Patient characteristics and setting |
Inclusion criteria: Patients referred for excision biopsy of pigmented lesions where the diagnosis of melanoma could not be excluded on clinical investigation Setting: Secondary (other); Dept Plastic Surgery and Burn Unit Prior testing: Clinical suspicion of malignancy without dermatoscopic suspicion Setting for prior testing: Secondary (not further specified); Department of Plastic Surgery and Burn Unit Exclusion criteria: None reported Sample size (participants): N included: 65 Sample size (lesions): N included: 83 Participant characteristics: Median age 47 years (18 ‐ 90); Male ‐ 29 (45%) Lesion characteristics: Melanoma thickness 0.29 mm to 2.18 mm |
||
| Index tests |
Dermoscopy: No algorithm Method of diagnosis: Dermoscopic images Prior test data: No further information used Diagnostic threshold: NR "dermoscopic images were examined by an experienced dermatologist" Diagnosis based on: Single observer (n = 1) Observer qualifications: Dermatologist Experience in practice: High experience or ‘Expert’ Experience with index test: High experience/‘Expert’ users The dermoscopic and SIAgraphic images were obtained by SIAscope II (Amon Clinica, Cambridge, UK) and stored using the proprietary Dermetrics software (Astron Clinica) Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: SIAscope II (Astron Clinica, UK )(classifier NR) System details: Skin lesion is interrogated with light of different wavelengths and the reflection spectra are analyzed by proprietary algorithms showing distribution, position and quantity of melanin, blood, and collagen within the papillary dermis (the SIAgraphs) No derivation aspect (external validation) Derivation described in prior study – See Moncrieff 2002; Govindan 2007 Lesion characteristics assessed: Analysis of dermal melanin, erythematous blush, lesion asymmetry, collagen 'holes', blood commas, or irregularities in the collagen Additional predictors included: None Method of diagnosis: Spectroscopic images, SIAgraphic images CAD‐based diagnosis Prior/other test data: Unclear CAD output: Binary (based on Australian Scoring System): ‘strong chance of melanoma' or 'low risk of melanoma' Diagnostic threshold: Australian Scoring System; Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis alone (excision biopsy) Breslow thickness and Clark level were determined by standard histopathologic examination. Tumour staging was performed as described by Balch 2001 according to the 2001 melanoma staging system Disease‐positive: 12; Disease‐negative: 71 Target condition (Final diagnoses) Melanoma (invasive): 7; Melanoma (in situ): 5; 1 melanoma metastasis (incl as benign) Sebhorrheic keratosis: 1; Benign naevus: 57; 'Benign' diagnoses: Bowens 1; haemangioma 1; lentigo simplex 2; epidermal naevi 2; DF 6 |
||
| Flow and timing |
Participant excluded from analysis: None reported Interval between tests: NR Interval to reference standard: Images taken prior to biopsy |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Low | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Gutkowicz Krusin 1997.
| Study characteristics | |||
| Patient sampling |
Study design: Unclear Data collection: Retrospective image selection/Prospective interpretation Period of data collection: NR Country: USA (from authors' institution) Test set derived NR. The "classifier was then tested blindly on an independentset of 28 images of melanocytic lesions on slides provided by Dr. A. W. Kopf." |
||
| Patient characteristics and setting |
Inclusion criteria: Lesions suspected of early melanoma or atypical melanocytic naevus Setting: Secondary (general dermatology) From authors' institution Prior testing: Selected for excision (no further detail) No details; all lesions excised Setting for prior testing: Unspecified Exclusion criteria: None reported Sample size (participants): N included: NR Sample size (lesions): N included: 104; 76 training; 28 test set Participant characteristics: NR |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: MelaFind precurser (Multiparametric linear classifier) System details: Digital camera and illumination assembly coupled to a computer, with separate image analysis Derivation study (internal validation) Lesion characteristics assessed: Lesion asymmetry, border, gradient, centroid, texture, colour Additional predictors included: No further information used Method of diagnosis: Spectroscopic images CAD‐based diagnosis Prior/other test data: No further information used CAD output: NR Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Histology (not further described) N participants/lesions: 28 in test set Disease‐positive: 5; Disease‐negative: 23 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 5 Benign naevus: 23 |
||
| Flow and timing |
Excluded participants: NR Time interval to reference test: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | No | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Unclear | ||
| Was model overfitting accounted for during model development? | Unclear | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Hauschild 2014.
| Study characteristics | |||
| Patient sampling |
Study design: Case‐control; all lesions in this study were imaged and analysed by MelaFind in a previous study (Monheit 2011) Data collection: Retrospective image selection/Prospective interpretation Period of data collection: NR Country: USA data from Monheit 2011 study |
||
| Patient characteristics and setting |
Inclusion criteria: Subset of PSLs evaluated in the Monheit trial; melanoma and non‐melanoma randomly selected; none were ulcerated, non‐pigmented, or located on excluded anatomic sites Setting: Lesions sampled from Monheit trial "Seven clinical sites with 23 investigators participated in this trial. Three sites were academic institutions (University of Pittsburgh, Duke University, and Northwestern University), and 4 sites were dermatologic practices highly experienced in managing PLs." Prior testing: Selected for excision (no further detail) Setting for prior testing: NR Exclusion criteria: Ulcerated, non‐pigmented, or located on excluded anatomic sites Sample size (participants): N included: 130 Sample size (lesions): N eligible: 1632 lesions in Monheit trial; N included: 130 Participant characteristics: None reported Lesion characteristics: site Head/neck: 22.3%; trunk: 41.5%; upper limbs/shoulder: 20%; lower limbs/hip: 16.2%. Median thickness (melanomas) 0.39 mm (range 0.12 ‐ 1.2 mm) |
||
| Index tests |
Dermoscopy: No algorithm Method of diagnosis: Clinical photographs and dermoscopic images Prior test data: Clinical examination and/or case notes. Lesion images consisted of a clinical overview at 53 cm/21 inches, a clinical close‐up at 20 cm/8 inches, and a dermatoscopic image. Clinical exam information consisted of 24 items regarding patient demographics and risk factors for melanoma, such as: personal or family history of melanoma, number of atypical naevi, Fitzpatrick skin type, number of severe sunburns before and after age 20, etc. Diagnostic threshold: NR. Responses to the questions about whether or not the dermatologist would biopsy the lesion and reason for biopsy were used to determine dermatologist sensitivity and specificity Diagnosis based on: Average (Arm 1: 101 board‐certified dermatologists; Arm 2 (MelaFind): further 101 board‐certified dermatologists; Arm 3: 9 PSLs) Observer qualifications: Dermatologist (Experts (Arm 3) prospectively identified by the Principal Investigator based on field standing prior to participant recruitment) Experience in practice: High experience or ‘Expert’; > 90% had more than 10 years experience in practice Experience with index test: High experience/‘Expert’ users. All except 6 were trained in dermoscopy use; 155/202 always or almost always used dermoscopy for PSLs Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: MelaFind (classifier NR) System details: Multispectral imaging system with integrated image analysis software; device takes images in vivo No derivation aspect (reader study) Lesion characteristics assessed: NR Additional predictors included: None reported Method of diagnosis: Spectroscopic images CAD‐based diagnosis Prior/other test data: Clinical examination and/or case notes ‐ Study presents system‐based diagnosis plus MelaFind combined with dermatologist decision which was also informed by clinical exam information. CAD output: Binary output: (1) positive, (lesion should be considered for biopsy to rule out melanoma); and (2) negative (lesion should be considered for later evaluation) Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis alone Histology (not further described). Disease‐positive: 65; Disease‐negative: 65 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 65 'Benign' diagnoses: 65 |
||
| Flow and timing |
Participants excluded from analysis: None Time interval between index tests: Unclear Time interval to reference standard: Unclear |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Unclear | ||
| Low | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Maglogiannis 2015.
| Study characteristics | |||
| Patient sampling |
Study design: Case‐control Data collection: Retrospective Period of data collection: NR Country: Greece Random division of 208 lesions into train and test sets (equal numbers) |
||
| Patient characteristics and setting |
Inclusion criteria: Lesions excised (no further details) Setting: Specialist: Department of Plastic Surgery and Dermatology (Athens) Prior testing: Lesions excised (no further details) Setting for prior testing: Not specified Exclusion criteria: None reported Sample size (participants): NR Sample size (lesions): 208 lesions, Participant characteristics: |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: name NR (SVM polykernel c = 5 classifier, selected at random for this review) System details: Computer analysis of digital dermoscopy images captured using the Molemax II dermatoscope Derivation study (internal validation) 5 classifiers trained: Multilayer perceptron, kNN, Random forest, SVM polykernel c = 5, SVM PUK kernel Lesion characteristics assessed: Features corresponding to the number, size and asymmetry of dots: (a) number of dots, (b) total number of pixels in dots, (c) mean number of pixels in dots, (d) variance of num. pixels in dots, (e) fraction of lesion area occupied by dark dots. Asymmetry: radial, angular, primary axis Additional predictors included: None reported Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: No further information used CAD output: NR Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Disease‐positive: 50 MM; Disease‐negative: 54 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 50 'Benign' diagnoses (not further specified): 54 |
||
| Flow and timing | No exclusions reported Time interval to reference test: No details reported |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Unclear | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Malvehy 2014.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective; dermoscopic images assessed remotely from the participant Period of data collection: March 2010 and November 2011 Country: Conducted at 5 American and 17 European investigational sites (Sweden, Germany, Austria, Hungary, U.K. and Spain) |
||
| Patient characteristics and setting |
Inclusion criteria: All patients with skin lesions selected for total excision to rule out melanoma; dermatologists were encouraged to enrol a mix of lesions with an even distribution of low‐, medium‐ and high‐risk lesions Setting: Secondary; authors institutions primarily listed as Dept Dermatology with one "Dermatology Clinical Research Center" Prior testing: Selected for excision Exclusion criteria: Lesions < 2 mm or > 20 mm and those located: on acral skin, e.g. sole or palm; areas of scars, crusts, psoriasis, eczema or similar skin conditions; hair‐covered areas, e.g. scalp, beards, moustaches or whiskers; genitalia; in an area that has been previously biopsied or subjected to any kind of surgical intervention or trauma; mucosal surfaces; with foreign matter, e.g. tattoo or splinter; acute sunburn; or skin surface not measurable, e.g. lesion on a stalk; surface not accessible, e.g. inside ears, under nails or not intact (measurement area) Sample size (participants): N eligible: 1951; N included: 1611 Sample size (lesions): N eligible: 2416; N included: 1943 Participant characteristics: For Nevisense sample: median age: 48 years (range 18 to 91); male 47.5%; 97.5% of white ethnicity. Fitzpatrick skin types: I (7.3%); II (48.6%); III (37%); IV (9.8%); V (1.4%); VI (0.1%) Lesion characteristics: Median Breslow thickness of 0.57 mm (153 invasive melanomas) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based EIS‐CAD system: Nevisense (SciBase III, Sweden) (SVM classifier) System details: Electrical Impedance spectroscopy imaging system with integrated image analysis software. The system measures the overall electrical resistance and reactance at 35 different frequencies Derivation aspect (study type) Lesion characteristics assessed: NR Additional predictors included: None reported Method of diagnosis: Spectroscopic images CAD‐based diagnosis Prior/other test data: None considered by CAD for diagnosis CAD output: The system computes both a score (0 – 10) and a dichotomous output (EIS negative/positive) at a fixed cut‐off Diagnostic threshold: Score: The fixed threshold is set at 4, i.e. scores < 4 are EIS‐negative and scores of ≥ 4 are EIS‐positive. Prior study (Mohr 2013) used dichotomous outcome but recommended score output |
||
| Target condition and reference standard(s) |
Type of reference standard: Histological diagnosis alone Details: Lesions were excised and underwent usual histopathology at investigational site. "A further histopathological evaluation was undertaken for study purposes by a panel of three experienced histopathologists who evaluatedeach lesion independently and were blinded from the investigational site’s original histopathology diagnosis". If they agreed, the diagnosis was considered as the final diagnosis; if there was significant disagreement two additional experts were consulted to establish the final lesion diagnosisif agreement was reached. If disgreement as to lesion diagnosis remained, the lesion was excluded from the efficacy analysis. Disease‐positive: 478; Disease‐negative: 1440 Target condition (Final diagnoses) 153 invasive melanomas, 112 melanoma in situ 48 BCC, 1 invasive cSCC; 1 Merkel cell carcinoma 157 severely dysplastic, 988 mild to moderate dysplasia, 352 benign naevi, 5 spitz naevi, 51 seborrheic keratosis, 6 cSCC in situ; 8 AK; 61 other |
||
| Flow and timing |
Participant exclusions: 473 excluded from Nevisense analysis; all reasons listed; primary reason was investigator oversight or the inability to render a final histopathological diagnosis; 74 exclusions were device‐related (60 with inadequate reference measurement quality and 14 due to device failure) Index test to reference standard interval: Appears consecutive; prospective recruitment with imaging and then "eligible and evaluable lesions were excised and subjected to the investigational site’s histopathology evaluation and managed accordingly." "A postprocedure follow‐up either by a telephone call or at a participant’s visit to the investigational site was conducted at 7 +/‐ 3 days after the Nevisense evaluation, at which time the patient was evaluated for any adverse events." |
||
| Comparative | Interval between index tests Consecutive; "A photograph and dermoscopic image of each included lesion was taken before and after Nevisense measurements to document evaluation according to the protocol.” | ||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Visual inspection | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | |||
| If a threshold was used, was it pre‐specified? | |||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | |||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | |||
| If a threshold was used, was it pre‐specified? | |||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | |||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Menzies 1996.
| Study characteristics | |||
| Patient sampling |
Study design: Unclear Describes including melanomas and randomly‐selected clinically atypical non‐melanoma lesions Data collection: Retrospective image selection/Prospective interpretation Period of data collection NR Country: Australia Test set derived: NR; describes 'division' into a training set and a test set |
||
| Patient characteristics and setting |
Inclusion criteria: PSLs from the Sydney Melanoma Unit with dermoscopic images and histological diagnoses; melanomas and randomly‐selected clinically atypical non‐melanoma lesions were included Setting: Specialist unit (skin cancer/pigmented lesions clinic) From authors' institution Prior testing: Clinical suspicion of malignancy without dermatoscopic suspicion Setting for prior testing: Specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: Unequivocal non‐melanoma excluded Sample size (participants): N included: NR Sample size (lesions): N included: 385 (training set 221, test set 164) Participant characteristics: None reported Lesion characteristics: None reported |
||
| Index tests |
Dermoscopy: Menzies criteria Method of diagnosis: Dermoscopic images Prior test data: No further information used Diagnostic threshold: 2 negative features of melanoma (i.e. cannot be found). Point and axial symmetry of pigmentation. Presence of only a single colour. 9 positive features of melanoma were used (at least 1 feature found). Multiple (5 ‐ 6) colours Blue‐white veil, multiple brown dots, multiple blue/grey, peripheral black dots or globules. A broadened network, Pseudopods, Radial streaming, Scarlike Diagnosis based on: NR Observer qualifications: NR; likely dermatologists Experience in practice: Not described Experience with index test: Not described Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: Name not reported (CART classifier) System details: Computer analysis of stored images captured using digital microscope. PSLs were photographed in vivo by means of immersion oil and a camera (Dermaphot, Heine Ltd). The surface microscopic images were studied on a viewer (Kodak Ektagraphic Viewer, Model 575AF, Eastman Kodak Co, Rochester, NY) Derivation study (Internal validation) Lesion characteristics assessed: Approach to feature selection: A classification and regression tree constructed on the training set produced a 7‐node tree Negative Features: Point and axial symmetry of pigmentation Presence of only a single colour Positive features: blue‐white, veil, multiple brown dots, pseudopods, radial streaming, scarlike depigmentatlon, peripheral black dots/globules, multiple (5 ‐ 6) colours, multiple blue/grey dots, broadened network Additional predictors included: Unclear Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: Unclear CAD output: NR Diagnostic threshold: Presence of indicative features (Melanoma = 0/2 morphologically negative features AND at least 1/9 positive morphological features) |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis alone Histology (not further described); Disease‐positive: 107; Disease‐negative: 278 Target condition (Final diagnoses) Melanoma (invasive): 107; BCC: 18 ?Ephilis lentigo 17; Sebhorrheic keratosis: 23; Benign acquired naevi 58; Dysplastic naevi 105; Blue naevi 11; Spitz naevi 6; spindle cell naevus 2; dermatofibroma 2; haemangioma 13; solar keratosis 9; other 14 |
||
| Flow and timing |
Exclusions from analysis: None reported Time interval to reference test: NR Time interval between index test(s): NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Unclear | ||
| Was model overfitting accounted for during model development? | Unclear | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Menzies 2005.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Retrospective image selection/Prospective interpretation Period of data collection June 1998 to September 2003 Country: Multicentre (Australia, USA, Germany) Test set derived: Study population divided at ratio 2:1 for training: test sets; divison randomised but stratified by diagnostic category and Breslow |
||
| Patient characteristics and setting |
Inclusion criteria: PSLs imaged using SolarScan at 9 different clinical centres including specialist referral centres and private skin cancer clinics; only the 78 lesions from the Sydney Melanoma Unit included in the VI/Dermoscopy evaluation. In all but 1 clinic site, the sole indication for imaging was that the pigmented lesion was to be excised, usually because of a clinical suspicion Setting: Private skin cancer clinics in Australia, staffed by general practitioners Prior testing: In 8/9 clinics, included lesions were to be excised, usually because of a clinical suspicion. "However, clinics were inconsistent in imaging excised lesions from their own practices, with some clinics obtaining images of lesions with a predominately high probability of melanoma." Setting for prior testing: Private care Exclusion criteria: Awkwardly situated lesions (eg, eyelids, some parts of the pinna, some genital sites, and perianal and mucosal surfaces); acral lesions; non‐pigmented pure amelanotic lesions (based on dermoscopy imaging); benign non‐melanocytic lesions excluded from one classifier. Poor‐quality index test image ‐ lesions outside the field of view (24 x 18 mm), contamination of calibration surfaces, or excess artifacts (hair, air bubbles, or movement artifacts). Ulcerated lesions, or diagnosed as pigmented basal cell carcinoma, pigmented Bowen's disease, or squamous cell carcinoma Sample size (participants): N included: NR Sample size (lesions): N eligible: 2430; N included: 1644 training; 786 test set Participant characteristics: None reported Lesion characteristics: None reported |
||
| Index tests |
Visual inspection (VI) Method of diagnosis: Prior test data: Diagnostic threshold: Diagnosis based on: Number of examiners Observer qualifications: Experience in practice: Experience with index test: Dermoscopy No algorithm Method of diagnosis: Dermoscopic images Prior test data: Clinical examination and/or case notes clinical photographs and participant histories Diagnostic threshold: Not reported. No details on lesion characteristics used; data can be extracted at 2 thresholds: correct diagnosis of melanoma (in situ or invasive) ‐ excise decision Diagnosis based on: Average (n = 13) Observer qualifications: GP 3; Dermatology registrar 3; Dermatologist 4 (one local practising dermatologists (Sydney), plus 3 international dermoscopy experts who headed pigmented lesion clinics) Experience in practice: Mixed experience (low and high experience combined) Experience with index test: Mixed experience Had clinical and dermoscopy photographic images (taken with a Heine Dermaphot camera, Heine Ltd, Herrsching, Germany) Computer‐assisted diagnosis – Dermoscopy‐based Derm‐CAD system: SolarScan (Polartechnics Lts, Australia) (Linear discriminant analysis classifier) System details: Dermoscopy video unit with internal algorithm for image analysis. The algorithm model used by SolarScan is an optimised set of fixed discriminant variables with associated weighting factors and relationships features (Australian Patent application No. 20022308395 and Australian Patent No. 2003905998) Derivation study (internal validation) Described in prior study Menzies 2001, referenced. Various properties of colour, pattern, and geometry were extracted from the segmented lesion images.The participant history features (see below) and 103 image analysis variables, in combination with the diagnostic weights (based on a linear representation (range, 0.25 ‐ 20) of correctly classifying the lesion as benign or melanoma), were used in the training set to model 2 diagnostic algorithms Lesion characteristics assessed: 103 automated image analysis variables extracted, consisting of various properties of colour, pattern, and geometry. Number analysed not reported Additional predictors included: Predictors included whether the lesion had, within the previous 2 years, bled without being scratched, changed in colour or pattern, or increased in size Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: None reported CAD output: Probability of melanoma, with cut–off (not provided) for benign vs melanoma Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis plus follow‐up Histology (not further described): 71% of total dataset (n = 1725), presumably including all disease‐positive Clinical follow‐up plus histology of suspicious lesions; Length of follow‐up: 3 months. 26% of full dataset (n = 632) Expert opinion. 3% of image set were diagnosed clinically but not excised Target condition (Final diagnoses). All numbers are for complete dataset Melanoma (invasive): 238; Melanoma (in situ): 144 Benign naevus: 1835, benign melanocytic; Other: 213 benign non‐melanocytic, incl 140 SK |
||
| Flow and timing |
Exclusions from analysis: None Time interval to reference test: NR Time interval between index test(s): NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Yes | ||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Unclear | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Unclear | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | No | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | No | ||
| High | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| High | |||
Mohr 2013.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective; dermoscopic images assessed remotely from the participant Period of data collection: January 2009 to November 2010 Some overlap in study population with Malvehy 2014 possible (no author reply), as ascertained by similar recruitment centres and overlapping periods of data collection Country: Conducted at 19 private and/or academic dermatological centres located in Germany, Hungary, Sweden, Switzerland, and U.K. Test set derived: Data randomised into training set (approximately 40% of data) and test set (approximately 60%) |
||
| Patient characteristics and setting |
Inclusion criteria: Adults of any ethnic group, aged at least 18, with 1 or more primary skin lesion(s), at least 2 mm in diameter, located on normal uninflamed skin and requiring full excision for histopathological analysis Setting: Secondary; authors institutions primarily listed as Dept Dermatology with 1 "Division of Imaging and Technology" Prior testing: Selected for excision Exclusion criteria: > 8 lesions per person; metastatic or recurrent; patients with lesions under finger and toe nails, in sites where the electrode could not reach, e.g. between toes, those lesions with abnormal reference areas (usually inflammatory skin disease like eczema and psoriasis), those with lesion in scars or striae, crusted lesions and those previously subjected to any surgical procedure Sample size (participants): N eligible: NR; N included: 1134 Sample size (lesions): N eligible: NR; N included: 1300 Participant characteristics: NR Lesion characteristics: Median Breslow thickness of 0.43 mm (67 invasive melanomas) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based EIS‐CAD system: Nevisense predecessor (SciBase, Sweden) (SVM classifier) System details: Electrical impedance was measured with the SciBase III electrical impedance spectrometer, equipped with a spring‐loaded probe and a disposable 5‐bar electrode. The system measures bio‐impedance of the skin at 35 different frequencies, logarithmically distributed from 1.0 kHz to 2.5 MHz, at 4 different depths utilising 10 permutations Derivation study (internal validation) Classification algorithm calibration and testing was conducted in 2 stages. In the first stage of development, the data were randomised into 2 cohorts for calibration and verification, utilising 40% and 60% of the available data respectively. In the second stage approximately 55% of the data were used for calibration and the whole data set was used for verification (second algorithm, not extracted) Lesion characteristics assessed: "The electrical impedance data obtained from each measurement represented a very large data set consisting of the complex ratio of voltage to current, composed of the magnitude and phase shift at 35 frequencies for 10 permutations yielding a data set of 700 variables for each measurement. By combining permutations and frequencies, a large EIS feature space could be constructed. The features’ ability to differentiate between melanoma and benign cutaneous lesions was then ranked and, by means of cross‐validation, the optimum number of features was extracted" Additional predictors included: None reported Method of diagnosis: Stored EIS measurements CAD‐based diagnosis Prior/other test data: None reported CAD output: Dichotomous outcome: malignant vs benign Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Type of reference standard: Histological diagnosis alone Details: Lesions were excised and underwent usual histopathology at investigational site. A further histopathological evaluation was undertaken for study purposes by a panel of 3 experienced histopathologists who evaluated each lesion independently using information from clinical diagnosis and histopathology referral reason; blinded from the investigational site’s original histopathology diagnosis. If they agreed, the diagnosis was considered as the histopathological gold standard (HGS) Disease‐positive: 166; Disease‐negative: 280 (Total 446 in test set, after exclusions from analysis) Target condition (Final diagnoses) 67 invasive melanomas, 30 melanoma in situ, 21 BCC, 4 cSCC 38 severely dysplastic, 185 moderate dysplasia, 64 benign naevi, 22 seborrheic keratosis, 9 other |
||
| Flow and timing |
Participants exclusions from analysis: 549/1300 lesions excluded from analysis, mainly due to poor reference measurement quality (n = 290). Other reasons were: screening failure 2, protocol violations 72, no measurements performed 35, unable to map lesions with measurements 9, lesion not excised 18, poor histopathology 11, no consensus diagnosis reached by pathologists 20. Expanded exclusions: bleeding, traumatised or ulcerated lesion 38, lesion located on acral skin 11, surface area not measurable 34, insufficiently covered with measurements 2, no clinical suspicion of melanoma 5, hair‐bearing areas 2 An additional 6 lesions excluded by review team: 6 undefined thickness melanomas excluded from MM1 to give total sample of 446 − 6 = 440 Time interval to reference test: Consecutive, excision within 2 weeks of EIS measurements; prospective recruitment: "After obtaining informed consent from each patient, eligible lesions destined for excision were measured with the SciBase III electrical impedance spectrometer (SciBase AB, Stockholm, Sweden). After a maximum of 14 days the lesions were surgically excised and subjected to histopathological evaluation." |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Yes | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Visual inspection | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | |||
| If a threshold was used, was it pre‐specified? | |||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | |||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | |||
| If a threshold was used, was it pre‐specified? | |||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | |||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Mollersen 2015.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Unclear Period of data collection: March to December 2013 Country: Germany |
||
| Patient characteristics and setting |
Inclusion criteria: Adult patients scheduled for excision of a PSL and those with nonpigmented skin lesions if melanoma, BCC, or SCC was a potential differential diagnosis. The presence of hairs and bubbles, lesion size, inadequate segmentation, etc. were not used as exclusion criteria Setting: Private dermatology practice Prior testing: Scheduled for excision on basis of clinical diagnosis, because of concern about malignancy or when requested by the patient for other reasons (no further details) Setting for prior testing: Unspecified Exclusion criteria: NR Sample size (participants): Eligible NR, Included 516 Sample size (lesions): Eligible NR, Included 877 Participant characteristics: Median age: 53 yrs (range 18 to 93); male 53% Lesion characteristics: Median Breslow thickness of 0.50 mm (23 invasive melanomas); maximum Breslow thickness 2.25 mm |
||
| Index tests |
Computer‐assisted diagnosis – Dermoscopy‐based Derm‐CAD system: Naevus Doctor (ND) (classifier NR) System details: Computerised image analysis system coupled to digital dermoscope. ND takes a dermoscopic image from the Canon/DermLite device as input and classifies the lesion. ND is still in an experimental phase. "All skin lesions were photographed prior to excision with a digital camera (Canon G10, Canon Inc., Tokyo, Japan) with an attached dermoscope (DermLite FOTO, 3Gen LLC, California, USA) and with a videodermoscope (DermoGenius ultra, DermoScan GmbH, Regensburg,Germany)" No derivation aspect (external validation study) Described in previous study Mollersen 2015 in press, reference #37 Lesion characteristics assessed: NR Additional predictors included: None reported Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: Clinical diagnosis CAD output: Probability of malignancy Diagnostic threshold: CAD system tuned to 95% melanoma sensitivity Computer‐assisted diagnosis – Dermoscopy‐based Derm‐CAD system: MoleExpert (ME) (classifier NR) System details: ME (MoleExpert micro Version 3.3.30.156) takes a dermoscopic image from the DermoGenius device as input. ME is intended for use on melanocytic lesions only. "All skin lesions were photographed prior to excision with a digital camera (Canon G10, Canon Inc., Tokyo, Japan) with an attached dermoscope (DermLite FOTO, 3Gen LLC, California, USA) and with a videodermoscope (DermoGenius ultra, DermoScan GmbH, Regensburg,Germany)" No derivation aspect (external validation study) No information provided (ME is a commercial system developed by others and used as a comparator in this study) Lesion characteristics assessed: Features of ABCD system plus other features (not listed), e.g. colour variation and grey veil Additional predictors included: None reported Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: Clinical diagnosis CAD output: Number between −5.00 and 5.00, where high values indicate suspicion of melanoma Diagnostic threshold: CAD system tuned to 95% melanoma sensitivity |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Details: All excised lesions were examined by a dermatopathologist. In the case of a malignant diagnosis, a second dermatopathologist examined the excised lesion and a consensus diagnosis was set Disease‐positive: 107; Disease‐negative: 278 Target condition (Final diagnoses) Melanoma: invasive 25, in situ 19; BCC: 70, cSCC 6, adnexal carcinoma 1 Benign diagnoses: 38 benign non‐melanocytic; 13 collision tumours; 13 AK; 11 Bowen's disease; 79 Sebhorrheic keratosis; 595 benign naevus |
||
| Flow and timing |
Exclusions from analysis: 1 lesion lacking clinical diagnosis; of 875 lesions with histopathological diagnosis, 4 were excluded because ME did not give an output (1 naevus, 1 seborrheic keratosis, 1 BCC, and 1 SCC) and 1 was excluded because the Canon/DermLite image was lost (melanoma in situ) Time interval to reference test: NR Time interval between index test(s): Appears consecutive |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Monheit 2011.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: January 2007 to July 2008 Country: USA |
||
| Patient characteristics and setting |
Inclusion criteria: PSLs scheduled for biopsy in toto, with diameter ≥ 2 mm Setting: Specialist unit (skin cancer/pigmented lesions clinic). 3 sites were academic institutions and 4 sites were dermatologic practices highly experienced in managing PLs Prior testing: Clinical and/or dermatoscopic suspicion Setting for prior testing: Unspecified Exclusion criteria: Difficult‐to‐diagnose lesions; Location/site of lesion anatomic site of pigmented lesion not accessible to the device; within 1 cm of the eye; or on palmar, plantar, or mucosal (eg, lips, genitals) surface or under nails; lesion size diameter < 2 mm or > 22 mm excluded Previous history of skin cancer/prior treatment at site lesion previously biopsied, excised, or traumatised. Other characteristics: known allergy to isopropyl alcohol; skin not intact (e.g. open sores, ulcers, bleeding); in an area of visible scarring; or containing foreign matter (eg, tattoo ink, splinter, marker) Sample size (participants): Eligible: 1383; Included 1257 Sample size (lesions): Eligible: 1831; Included 1632 Participant characteristics: Median age 46 yrs (Range 7 ‐ 97); Male 575 (45.7%); Ethnicity: White 1232 (98%), Black or African‐American 2 (0.2%), Asian 17 (1.4%), Other 6 (0.5%). Thickness/depth: ≤1 mm: 69/70 invasive MM (99%), 1.01 ‐ 2.00 mm: 1/70 (1%) Median Breslow 0.36 mm (70 invasive MM) |
||
| Index tests | MSI‐CAD system: MelaFind (EO Sciences, USA) (classifier NR) System details: Multispectral imaging system with integrated image analysis software; device takes images in vivo. MelaFind takes images at 10 spectral bands, between 430 – 950 nm No derivation aspect (external validation study) The properties of these images as well as image analysis methods have been previously described (Gutkowicz Krusin 1997, Gutkowicz‐Krusin 2000, Gutkowicz‐Krusin 2007 and Elbaum 2001) Lesion characteristics assessed: NR Additional predictors included: None Method of diagnosis: In‐person diagnosis CAD‐based diagnosis Prior/other test data: No further information used CAD output: Binary output: (1) positive, (lesion should be considered for biopsy to rule out melanoma); and (2) negative (lesion should be considered for later evaluation) Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Histology (not further described) N participants/lesions: 1632 Disease‐positive: 175, Disease‐negative: 1457 Target condition (Final diagnoses) Melanoma: invasive 70, in situ 57; BCC: 23; cSCC: 10, Severe dysplasia: 43 Benign diagnoses: Mild/moderate dysplasia 998; Sebhorrheic keratosis 93; Benign naevus 217, atypical melanocytic hyperplasia (AMH) or atypical melanocytic proliferation (AMP) 5, actinic keratosis 16, other keratosis 10, lentigo 16, other 14 |
||
| Flow and timing |
Exclusions from analysis: 20 lesions with the pre‐biopsy dermatologic diagnosis of melanoma were excluded from the primary MelaFind analysis; 1 withdrew, 3 clinician deemed ineligible, 14 dermatopath deemed ineligible, 19 missing/inadequate histology slides, 162 imaging failed (operator errors, too many bubbles, lesion not centred), 36 MelaFind or camera malfunction, 61 operator or MelaFind error (lesion too small to visualise, automatic segmentation falied). Time interval to reference test: Unclear |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test Visual inspection | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | |||
| If a threshold was used, was it pre‐specified? | |||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | |||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Piccolo 2002.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Retrospective image selection/Prospective interpretation Period of data collection: NR; 6‐month period Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: Pigmented lesions excised because of equivocal dermoscopic findings or at the patient’s request Setting: Secondary (general dermatology); from authors' institution Prior testing: Dermatoscopic suspicion; Patient request for evaluation/excision Setting for prior testing: NR Exclusion criteria: None reported Sample size (participants): N included: 289 Sample size (lesions): N included: 341 Participant characteristics: Mean age 33.6 yrs (range 3 – 83); Male: 127 (43.9%); Fitzpatrick phototype I to II (31.4%); Type III (42.2%); Type IV ‐ V (26.4%) Lesion characteristics: None reported |
||
| Index tests |
Dermoscopy No algorithm Method of diagnosis: Clinical photographs and dermoscopic images Cases were clinically and dermoscopically evaluated on a high‐resolution colour monitor, in a random sequence Prior test data: Unclear. Not specifically described but appears to be images only Diagnostic threshold: NR Diagnosis based on: Single observer (n = 2) Observer qualifications: Dermatologist; Resident clinician with minimal training in PSLs Experience in practice: High experience or ‘Expert’ 5 years of experience; Low experience or recently qualified minimal training in PSLs (6 months of experience, comprising 8 hours of specialised training on 3 consecutive days and 2 hours a week in the routine of dermoscopy) Experience with index test: Mixed Any other detail: Stereomicroscope with magnifications varying from x6 to x40 Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: DEM‐MIPS (Digital Epi Microscopy Melanoma Image Processing Software; Biomips SRL, Siena, Italy) (ANN classifier) System details: "DEM‐MIPS is designed to evaluate different colorimetric and geometric parameters of a lesion automatically in real time. All digital images of PSLs were collected in a Truevision Advanced Graphic Array format file with a size of 887 kB for each image." Digital dermoscopic images were framed at x16 magnification before analysis with DEM‐MIPS No derivation aspect (External validation study) Described in prior study "DEM‐MIPS is based on an ANN trained with 100 PSLs (50non‐melanomas and 50 melanomas) and is designed toevaluate different colorimetric and geometric parametersof a lesion automatically in real time." No citation given Lesion characteristics assessed: Evaluates colorimetric and geometric features (NR) Additional predictors included: None reported Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: No further information used CAD output: NR Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Histology (not further described); Disease‐positive: 13; Disease‐negative: 328 Target condition (Final diagnoses) Melanoma (in situ and invasive, or NR): 13 Sebhorrheic keratosis: 3; Benign naevus: 316; Other: 7 dermatofibromas, 2 angiomas |
||
| Flow and timing | Time interval to reference test: NR | ||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Unclear | ||
| Low | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Piccolo 2014.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Retrospective image selection/Prospective interpretation Period of data collection: September 2010 and October 2013 Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: Dermoscopically atypical PSLs selected from the archives of the Dermatology Department at the University of L'Aquila, Italy Setting: Secondary (general dermatology) Prior testing: NR Setting for prior testing: NR Exclusion criteria: Location/site of lesion ‐ acral sites and the face Sample size (participants): N included: 165 Sample size (lesions): N included: 165 Participant characteristics: Mean age 43.5 yrs (range 12 ‐ 84); Male: 59.4% Lesion characteristics: Lesion site: upper extremities 18 (11%); lower extremities 53 (32.1%); 62 (37.5%) on the back; 32 (19.4%) on the chest. Melanoma thickness 87.9% (29/33) < 0.75 mm; 12.1% (4/33) >1 .5 mm |
||
| Index tests |
Visual inspection (VI) Method of diagnosis: Prior test data: Diagnostic threshold: Diagnosis based on: Number of examiners Observer qualifications: Experience in practice: Experience with index test: Dermoscopy ABCD Method of diagnosis: Dermoscopic images Prior test data: No further information used Diagnostic threshold: Semi‐quantitative. Total dermoscopic score is calculated (TDS) ‐ a PSL with a TDS < 4.75 benign, TDS 4.75 to 5.45 suspicious of malignancy, TDS > 5.45 highly suggestive of melanoma Diagnosis based on: Single observer (n = 4) Observer qualifications: 3 dermatologists and 1 GP with different degrees of dermoscopic experience Experience in practice: Mixed Experience with index test: Experience scored using following criteria: number of years specialising in dermoscopy (score: 1. 0 ‐ 1 year; 2. 2 ‐ 5 years; 3. > 5 years); number of PSLs assessed by dermoscopy on a daily basis (1. < 10 lesions/day; 2. 11 ‐ 20 lesions; 3. 21 ‐ 30 lesions; 4. > 30 lesions); number of relevant workshops/seminars attended (1. 0 ‐ 1 workshops/seminars; 2. 2 ‐ 5 workshops/seminars; 3. > 5 workshops/seminars); and the number of authored publications on dermoscopy (1. 0 ‐ 1 publications; 2. 2 ‐ 5 publications; 3. 6 ‐ 10 publications; 4. > 10 publications) Observer 4 considered low experience (underwent dermoscopic training by studying an interactive atlas of dermoscopy between T0 and T1); Observer 1 High experience /‘Expert’; Observers 2 and 3 ); moderately experienced Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: Nevuscreen ® (Arkè s.a.s., Avezzano, Italy) (classifier NR) System details: Digital database containing image analysis software, coupled to digital dermoscope. Nevuscreen software automatically analyses ABCD features No derivation aspect (external validation study) Lesion characteristics assessed: ABCD features. "After image scanning, each pixel is classified in accordance with the main dermoscopic colour to which it is closest. Once the different colour regions are identified, DDA can also calculate asymmetry by considering the overall asymmetry parameter."**Differential dermoscopic structures Pigment network, Globules, Streaks, Black dots and Structureless areas represent notable criteria in dermoscopic evaluation. "Various digital filters (median filters, essentially) are used to obtain a morphological analysis for recognising particles of various dimensions, which are subsequently evaluated for size and shape and compared to numerous sample images. Once the different structures have been recognised, their asymmetry is calculated as a contribution to the overall asymmetry parameter" Additional predictors included: Clinicians use CAD output to assist their diagnosis Method of diagnosis: In‐person diagnosis CAD‐aided diagnosis Prior/other test data: 4 test operators 4 Operator qualifications: GP Dermatologist Experience in practice: Mixed experience (low and high experience combined) Experience with index test: Mixed experience (low and high experience combined) CAD output: TDS Diagnostic threshold: TDS: < 4.75 benign, 4.75 – 5.45 suspicious, > 5.45 highly suggestive of melanoma |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Target condition (Final diagnoses) Melanoma (invasive): 23; Melanoma (in situ): 10 Benign naevus: 105 Clark naevi; 19 Spitz/Reed naevi; 5 blue naevi; 3 dermal naevi |
||
| Flow and timing |
Excluded participants: NR Time interval to reference test: Reference test conducted first not clear what the time interval is between this and the current index test(s) Time interval between index test(s): Not clear ‐ looks like it was simultaneous |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | No | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Unclear | ||
| Low | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Rubegni 2002a.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Retrospective image selection/Prospective interpretation Period of data collection: 1996 to 2001 Country: Italy Test set derived: NR. "To train the SLP‐ANN, 550 of the 588 available cases were used (30 nevi and 200 melanomas in 550 sessions, each with 2 subsets); 549 cases were used for training, and 1 case at a time was used to check overfitting and stepwise feature selection. A third small, independent subset consisting of the other 17 melanomas and 21 nevi was used to test SLP‐ANN diagnostic performance on data not used in the training process." |
||
| Patient characteristics and setting |
Inclusion criteria: Excised PSL, clinically atypical (asymmetrical with variegated colour), flat and impalpable. "All were difficult to diagnose and therefore suitable for morphologic and parametric evaluation of early melanoma" Setting: Secondary (general dermatology) Prior testing: Clinical suspicion of malignancy without dermatoscopic suspicion; selected for excision (no further detail). Described as clinically atypical and difficult to diagnose Setting for prior testing: Secondary (general dermatology) Exclusion criteria: Difficult‐to‐diagnose lesions: Location/site of lesion acral, lesion size only 0.4 – 1 cm in diameter were included; non‐melanocytic appearance pink skin lesions (amelanotic melanoma and classical Spitz naevi); blue naevi were excluded, as were lentigo maligna, lentigo maligna melanoma Sample size (participants): N included: 588 included (1 per participant) Sample size (lesions): N eligible: 4200 PSLs excised; N. included: 588 included (38 in test set) Participant characteristics: Mean age 49 yrs (± 15); Male: 40% (of full sample; n = 588); 100% pigmented. Median Breslow 0.4 mm (157 invasive melanomas) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: DB‐Mips (Biomips Engineering, Italy) (ANN classifier) System details: Dermoscopy unit, internal stereomicroscope, internal DB, pattern analysis system. Lesions were imaged by ELM at a magnification of x16 with the DBDermo‐Mips apparatus Derivation study (internal validation) Described in prior study Andreassi 1999 Lesion characteristics assessed: Approach to feature selection: Computer‐aided stepwise technique to choose the number of discriminant features for optimum generalization. "The parameters, as previously described, belonged to 4 categories: geometries, colors, textures and islands of color. Geometric: area, maximum and minimum* diameters, radius, variance of contour symmetry, circularity*, fractality of borders and ellipsoidality. Color: mean values of red*, green and blue inside the lesion; mean values of red, green* and blue of healthy skin around the lesion; deciles of red*, green and blue inside the lesion; quartiles of red, green and blue* inside the lesion, mean skin‐lesion gradient*, variance of the border gradient, border homogeneity and interruptions of the border. Texture: mean contrast* and entropy of lesion as well as contrast and entropy fractality. Islands of color: peripheral dark regions*; dark area; imbalance of dark region; green area; red area; dominant green region imbalance; blue‐gray area; blue‐gray regions; transition area*; transition region imbalance*; background area*; background region imbalance*; red, green and blue multicomponent; and number of red, green and blue percentiles inside the lesion." (referenced to Andreassi 1999); the asterisks were added by the review team and correspond to the features selected by the model. Additional predictors included: No further information used Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: None reported CAD output: Diagnosis suggested (e.g. melanoma, benign melanocytic naevus) Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Histology (not further described) Histopathologic diagnosis of melanoma and naevi was made according to the criteria of the NIH Consensus Conference (1992). Histopathologic diagnosis discordance was c9%. These were classified as melanoma or naevi when at least 2 of 3 dermopathologists agreed on the diagnosis N participants/lesions: 588, 38 in test set Disease‐positive: 17; Disease‐negative: 21 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 17 Benign diagnoses: 21 (not further specified) |
||
| Flow and timing |
Exclusions from analysis: None reported Time interval to reference test: NR, but appears consecutive |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Was an adequate spectrum of cases used to train the algorithm? | Yes | ||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Unclear | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Seidenari 1998.
| Study characteristics | |||
| Patient sampling |
Study design: Case‐control Data collection: Retrospective image selection/Prospective interpretation Period of data collection NR; 4‐year period Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: Melanomas and benign PSLs from a larger series of PSLs used to develop a new automated classifier; all melanomas with x20 magnification images were included plus a random sample of benign lesions with the same magnification. For the larger series, lesions were referred by dermatologists or general physicians because of 1 or more PSLs that were difficult to interpret on clinical grounds alone, numerous PSLs, or because the patients were at increased risk for melanoma or had had a malignant PSL in the past Setting: Secondary Prior testing: Clinical suspicion of malignancy Setting for prior testing: Primary; secondary (general dermatology) Exclusion criteria: None reported Sample size (participants): NR Sample size (lesions): N eligible: 917; N included: 100 Participant characteristics: None reported Lesion characteristics: Melanoma thickness: ≤ 1 mm : 70.8% (n = 46), < 1 mm 58.5% (n = 38). mean thickness 0.73 ± 0.69 mm |
||
| Index tests |
Dermoscopy No algorithm Method of diagnosis: Dermoscopic images; (obtained via videomicroscopy) Prior test data: No further information used; "Images appeared in a random sequence on the computer screen, and no information about the patient (such as history, skin site, age of the patient, evolution of the lesion) was given to the evaluators" Diagnostic threshold: Clinical diagnosis Diagnosis based on: Single observer (n = 2) Observer qualifications: Dermatologist Experience in practice: Not described Experience with dermoscopy: Low ‐ 1 "untrained" dermatologist; High ‐ 1 routinely used videomicroscopy Any other detail For instrumental examination a 10‐ (39 cases), 20‐ (501 cases), or 50‐fold–magnification (377 cases) was chosen according to the size of the lesion, enabling the whole lesion to be seen on the monitor. For the study, the 31 MMs with x20 magnification were selected plus a random sample of 59 benign Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: DB–MIPS (Biomips Engineering, Italy) (Multivariate discriminant analysis classifier) System details: Digital videomicroscope equipped with a dedicated programme for the diagnosis of melanocytic PSL by evaluating digital features referring to benign and malignant PSL images. For this study an NTSC VMS‐110A videomicroscope (Scalar, Mitsubishi, Tama‐shi, Tokyo, Japan) was used No derivation aspect (external validation study) Lesion characteristics assessed: Radius, area and perimeter of the lesion, symmetry and circularity, fractality (shape), texture analysis, colour expressed as red, green and blue components, skin lesion gradient, 'dark areas' inside the lesion. All described in detail Additional predictors included: Unclear: for each participant personal data and information such as the site of the lesion, the magnification, the clinical and the histological diagnosis were recorded. Unclear how these were used Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: Unclear, NR CAD output: Graphical output and numerical output of features provided. Diagnosis suggested (e.g. melanoma, benign melanocytic naevus) Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Details: describes using "conventional histopathologic criteria" Disease‐positive: 31; Disease‐negative: 59 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 31 'Benign' diagnoses: 59 "nonmelanoma cases consisted of nevi including dysplastic nevi" |
||
| Flow and timing |
Participant exclusions: None reported Index test to reference standard interval: Not described |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Visual inspection | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | |||
| If a threshold was used, was it pre‐specified? | |||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | |||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | |||
| If a threshold was used, was it pre‐specified? | |||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | |||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Seidenari 1999.
| Study characteristics | |||
| Patient sampling |
Study design: Case‐control Data collection: Retrospective image selection/Prospective interpretation Period of data collection: NR Country: Italy Test set derived: Not clearly reported, but appears that the training set was randomly sampled, but the melanomas in the training set were supplemented with images of lesions of comparable size but thicker than 0.75 mm, randomly selected from other melanoma images |
||
| Patient characteristics and setting |
Inclusion criteria: PSLs with x20 magnification images Setting: Secondary (general dermatology) From authors' institution Prior testing: Selected for excision (no further detail) Setting for prior testing: Unspecified Exclusion criteria: None reported Sample size (participants): NR Sample size (lesions): N eligible: 461; N included: 383 in test set, 78 in training set Participant characteristics: Thickness ≤ 1 mm: 18 (100%) < 0.75 mm (8 in situ) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: DB–MIPS (Biomips Engineering, Italy) (Multivariate discriminant analysis classifier) System details: Dermoscopy unit, internal stereomicroscope, internal DB, DB–MIPS pattern analysis system – integrated database stores the patient's data and the description of the lesion along with the image icons. 38 features analysed (grouped into geometries, colours and Burroni's islands of colours) Derivation study (internal validation) Lesion characteristics assessed: The borders of the lesion were automatically identified, plus estimation of radius, area and perimeter of the lesion, symmetry and circularity, fractality (shape), texture analysis, colour expressed as red, green and blue components, skin lesion gradient, 'dark areas' inside the lesion. All described in detail. Approach to feature selection DBDermo‐MIPS software. "Discriminant analysis enables the identification of variables that are important for distinguishing between the groups in the training set in order to develop a procedure for predicting group membership for new cases in which group membership is undetermined (test set). Using the training set data, a threshold score was established that enabled the attribution of each malignant lesion to the right group (100% sensitivity). The same value was employed for discriminating benign and malignant lesions belonging to the test set" Additional predictors included: Unclear; for each participant personal data and information such as the site of the lesion, the magnification, the clinical and the histological diagnosis were recorded. Unclear how these were used Method of diagnosis: In‐person diagnosis CAD‐based diagnosis Prior/other test data: Unclear CAD output: NR Diagnostic threshold: Threshold not reported. Using the training set data, a threshold score was established that enabled the attribution of each malignant lesion to the right group (100% sensitivity). The same value was used for discriminating benign and malignant lesions belonging to the test set |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Histology (not further described) N participants/lesions: 461 (383 in training set) Disease‐positive: 18; Disease‐negative: 365 Target condition (Final diagnoses): Melanoma: invasive 10, in situ 8 'Benign' diagnoses: 365 non‐melanoma cases consisted of benign naevi including common naevi and clinically dysplastic naevi (> 5 mm in diameter, irregular or ill‐defined border, irregular pigmentation) |
||
| Flow and timing |
Exclusions from analysis: None Time interval to reference test: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Serrao 2006.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Retrospective Period of data collection: September 2002 to September 2005 Country: Portugal |
||
| Patient characteristics and setting |
Inclusion criteria: Melanocytic lesions from patients with multiple atypical naevi, personal/familiar melanoma history or doubtful cases on clinical inspection who were referred to a Dermoscopy Unit Setting: Specialist dermatoscopy unit (skin cancer/pigmented lesions clinic) Prior testing: Clinical suspicion of malignancy without dermatoscopic suspicion. Mixed population; high risk or clinically suspicious, or both Setting for prior testing: Secondary (general dermatology) Exclusion criteria: Unequivocal appearance/diagnosis. Clearly benign lesions by clinical examination were not referred Sample size (participants): N eligible: 1186; N included: 344 Sample size (lesions): N included: 652 Participant characteristics: Mean age 40 years (SD ± 14), age range: 11 to 84 years; 49% in the 35 ‐ 64 age group, 33% aged 25 to 34 years old, 3% aged under 18; Male: 33% (114) High‐risk characteristics: history of melanoma/skin cancer (%) 19%, family history of melanoma (%) 3%, 24% history of dysplastic naevi Lesion site: Trunk: back 56% chest 20%, Lower limbs/hip: 13% Thickness/depth: ≤ 1 mm: 29/41 plus 8 in situ; > 1 mm: 3 |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: microDERM (Visiomed AG, Germany) (ANN classifier) System details: Dermoscopy unit with internal camera containing analysis system. DANAOS software combines analytical system based on ABCD with database of 21,000 PSLs. The system has an integrated filter that reduces influence of hairs in the analysis of lesions No derivation aspect (external validation study): Described in prior study Fidalgo 2003 Lesion characteristics assessed: Lesions assessed for about 50 parameters (geometrical, colour and internal pattern) Additional predictors included: None reported Method of diagnosis: Dermoscopic images CAD‐based diagnosis Prior/other test data: Unclear CAD output: DANAOS score indicating risk of malignancy Diagnostic threshold: High‐risk DANAOS score (> 6.5); data also presented for score of > 7.5 |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Criteria used for excision were: Dermoscopic suspicious lesions, irrespective of the DANAOS score All lesions with high‐risk DANAOS score (> 6.5) Significant dermoscopic or clinical architectural change, irrespective of the DANAOS score Disease‐positive 41; Disease‐negative 611 Target condition (Final diagnoses) Melanoma: invasive 32; in situ 9 Benign diagnoses: 472 Benign naevus, 139 dysplastic naevus |
||
| Flow and timing |
Exclusions from analysis: None Time interval to reference test: Unclear; CAD performed in advance of histology |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Unclear | ||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Sgouros 2014.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: 3‐month period; dates not specified Country: Italy |
||
| Patient characteristics and setting |
Inclusion criteria: Pigmented skin tumours, clinically suspicious for the diagnosis of melanoma, BCC or SCC and the inability to establish a definite diagnosis on clinical grounds only Setting: Specialist unit (skin cancer/pigmented lesions clinic) Prior testing: Clinical suspicion of malignancy without dermatoscopic suspicion Setting for prior testing: Specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: None reported Sample size (participants): N eligible: 180 Sample size (lesions): N eligible: 188; N included: 44 excised (authors included remaining 144 non‐excised lesions but these only received expert diagnosis with no follow‐up so not eligible for our review) Participant characteristics: Mean age 43 yrs (range: 2 ‐ 95) (n = 188); Male: 97 (51.6%). |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: SIAscope (MedX Health Corp, Canada) (classifier NR) System details: Spectrophotometric imaging system with hand–held skin probe (SIAscope, version NR) and integrated software No derivation aspect (external validation study) Derivation described in prior study Moncrieff 2002 Lesion characteristics assessed: Analysis of dermal melanin, erythematous blush, lesion asymmetry, collagen 'holes', blood commas, or irregularities in the collagen Additional predictors included: None Method of diagnosis: In‐person diagnosis CAD‐based diagnosis Prior/other test data: Clinical examination and/or case notes Dermoscopy CAD output: SIAgraph and PCSA (see below) Diagnostic threshold: Primary care scoring algorithm (PCSA) (Emery 2010); score ≥ 6 regarded as suspicious |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Histology (excision) N participant/lesions: 44 Disease‐positive: 31; Disease‐negative: 13 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 18 BCC: 10, cSCC: 3 'Benign' diagnoses: 14 |
||
| Flow and timing |
Excluded participants: 144 with only expert final diagnosis (excluded by review team) Time interval between index test(s): Appears consecutive |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | Yes | ||
| Did the study avoid including participants with multiple lesions? | Yes | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Unclear | Low | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Stanganelli 2005.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Retrospective image selection/Prospective interpretation **Dataset previously used in Stanganelli 2000. Period of data collection: NR Country: Italy Test set derived A training set of 22 melanomas and 218 melanocytic nevi was randomised from the dataset. The test set was formed by the complement (the remaining 20 melanomas and 217 naevi). A further subset of images from the original dataset, consisting of 31 melanomas and 103 naevi, was used for the comparison between observers and CAD; derivation of the subset not reported |
||
| Patient characteristics and setting |
Inclusion criteria: Melanocytic lesions from patients referred to the Skin Cancer Unit and undergoing clinical and dermoscopic evaluation Setting: Specialist unit (skin cancer/pigmented lesions clinic) Prior testing: Clinical and/or dermatoscopic suspicion Setting for prior testing: Specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: None reported Sample size (participants): N eligible: 1556 referred; N included: NR Sample size (lesions): N eligible: 3274; N included: 477 melanocytic lesions; 237 in test set and 134 in comparison between CAD and human operators Participant characteristics: None reported Lesion characteristics: Melanoma thickness 61.2% < 0.75 mm |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: DB–MIPS (Biomips Engineering, Italy) (SVM classifier) System details: Dermoscopy unit, internal steromicroscope, internal DB, pattern analysis system. Automatic Data Analysis for Melanoma early detection (ADAM) software which analyses boundary shape, texture and colour distribution Derivation study (internal validation) see also Stanganelli 1995 and Stanganelli 2000 Lesion characteristics assessed: ADAM software is based on a quite recent mathematical technique of shape representation: the Size Functions. "These are very general invariants designed to capture, in a formal and quantitative way, the essential behaviour of some specified aspects (the so called measuring functions) of a signal (27, 28). In the present case, the examined signal is the image of a melanocytic lesion, and the aspects concerned are: boundary shape, texture and color distribution. Size Functions are standardized objects, easy to compute, to store and to compare. So the study is performed on the Size Function instead of the original image. This yields a great simplification and, above all, a greatly focussed analysis. The Size Function obtained from a curve with the distance from point C as measuring function is shown in Figure 1" Additional predictors included: No further information used Method of diagnosis: Dermoscopy images CAD‐based diagnosis Prior/other test data: None reported CAD output: Low risk, intermediate risk or high risk of melanoma Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis plus cancer registry All included lesions underwent histology but some were identified using a cancer‐registry‐based follow‐up of benign diagnoses Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 42 in full sample; 31 in CAD vs human observer interpretation and 20 in test set 'Benign' diagnoses: 435 melanocytic naevi; 103 in CAD‐observer comparison and 217 in test set |
||
| Flow and timing |
Exclusions from analysis: None Time interval to reference test: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | Unclear | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| Low | High | ||
| DOMAIN 2: Index Test Visual inspection | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Unclear | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | Yes | ||
| High | |||
Terstappen 2013.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: NR Period of data collection: NR Country: Sweden |
||
| Patient characteristics and setting |
Inclusion criteria: Lesions clinically suspicious for melanoma and showing positive SIAscopic findings Setting: Secondary (general dermatology) (details from authors' institution) Prior testing: Clinical suspicion of malignancy without dermatoscopic suspicion, showing positive SIAscopic findings Setting for prior testing: Secondary (general dermatology) Exclusion criteria: Poor‐quality index test image: 9 lesions excluded due to technical problems Sample size (participants): N eligible: 69; N included: 60 Sample size (lesions): N eligible: 69; N included: 60 Participant characteristics: ≤ 1 mm thickness: 17/29 melanomas; 8/29 melanomas Breslow thickness < 0.76 mm (Clark II ‐ III) and 9/29 Breslow thickness 0.76 − ≤ 1.0 mm (Clark II ‐ III) and 12 lesions Breslow thickness ≥ 1.1 (Clark III ‐ V) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: SIAscope (Astron Clinica, UK) (classifier NR) System details: Spectrophotometric imaging system with hand–held skin probe (SIAscope V) and integrated software (Dermetrics Version 2.0, Astron Clinica Ltd., Great Britain) No derivation aspect (external validation study) Lesion characteristics assessed: Dermal melanin, blood displacement, collagen holes, erythematous blush Additional predictors included: No further information used Method of diagnosis: In‐person spectroscopic images (SIAgraphs) CAD‐based diagnosis Prior/other test data: None reported CAD output: The instrument generates 4 images depicting the concentration of haemoglobin, melanin, collagen and dermal melanin Diagnostic threshold: "SIAscopic findings indicating melanoma were applied using the method described by Moncrieff (2002)" Results described for: "the combined features (presence of blood displacement with erythematous blush, collagen holes and presence of dermal melanin)". NB Moncrieff 2002 is a derivation study for SIAscope and suggests a number of combinations of features indicative of melanoma, the same features investigated here |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone The excised specimens were routinely processed and the histological sections, 4 μm thick, were stained with haematoxylin and eosin. Before cutting the specimen in slices, the lesion was oriented and the positions of the SIAscopic areas of interest were outlined by comparisons with the overview clinical photo of the lesion N participants/lesions: 60 Disease‐positive: 29; Disease‐negative: 31 Target condition (Final diagnoses) Melanoma: invasive 29, in situ 13 (included as D‐) BCC: 2 Benign diagnoses: 2 sebhorrheic keratosis; 4 melanocytic lesions; 10 dysplastic naevi |
||
| Flow and timing |
Exclusions from analysis: 9/69 lesions (2 invasive melanoma, 2 melanoma in situ, and 5 benign lesions) had to be excluded due to technical problems Time interval to reference test: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | No | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| High | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Tomatis 2003.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Retrospective image selection/Prospective interpretation Period of data collection: January 1995 to March 2000 (test set April 1999 to March 2000) Country: Italy Test set derived: Chronological; acquired in last year of recruitment. Study population not randomised (training set enriched with melanomas) |
||
| Patient characteristics and setting |
Inclusion criteria: Cutaneous pigmented lesions that required a surgical biopsy for diagnosis Setting: Specialist unit (skin cancer/pigmented lesions clinic) Tumour Institute of Milan Prior testing: Selected for excision (no further detail) Setting for prior testing: Unspecified Exclusion criteria: "Difficult to diagnose" lesions, thick and/or large melanomas and awkwardly situated lesions. like those placed at the interdigital spaces, on ears, nose, eyelids, etc.; lesions on the scalp due to hair interference Sample size (participants): N eligible NR; N included: 534 Sample size (lesions): N eligible NR; N included: 573 Participant characteristics: Median age 36 yrs (range 10 ‐ 95); 59.7% Male; thickness ≤ 1 mm: 91/132 MM (68.9%); thickness 0.16 to 3.24 mm. Median Breslow 0.68 mm; Mean diameter: 10 mm (range 3 to 39 mm) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: Telespectrophotometric system (ANN classifier; Linear discriminant classifier also trained and reported, ANN selected at random for review) System details: Digital camera coupled with an illumination system with interference filters and computer for storage and analysis of multispectral images. The Telespectrophotometric System consists of a CCD camera, a set of 17 interference filters. a PC. and an illumination system composed by 2 halogen ( 2 x 100 W) and 2 lamps (2 x 150 W) with emission in the infrared region Derivation study (internal validation) The training set is required for the instruction of the classifiers whose diagnostic performances are evaluated against an independent verify set. The considerable fraction of cases devoted to the classifier training was selected to include an adequate number of positives Derivation described in prior studies Marchesini 1995, Tomatis 1998, Furina 2000 Lesion characteristics assessed: For each spectral image, 5 parameters (lesion descriptors) were evaluated, based on ABCD and related to colour and shape of the imaged lesion: mean reflectance, variegation index, compactness, roughness, and area. Additional predictors included: No further information used Method of diagnosis: Spectroscopic images CAD‐based diagnosis Prior/other test data: None reported CAD output: NR Diagnostic threshold: Threshold selected by authors using ROC analysis. No further information provided |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Histology (not further described) N participants/lesions: 573 (210 in test set) Disease‐positive: 132 (37 in test set); Disease‐negative: 441 (136 in test set) Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 37 (test set) 'Benign' diagnoses: 136 (test set) |
||
| Flow and timing |
Exclusions from analysis: None Time interval to reference test: Appears consecutive: "images acquired in vivo before surgery". Interval not reported |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | |||
| Was the CAD model evaluated in an independent study population? | No | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Tomatis 2005.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: Prospective Period of data collection: September 2002 to April 2004 Country: Italy Test set derived: The study population was split into 3 sets: train, verify and test. The cases were randomly assigned to the above sets among all the 1391 lesions with histology |
||
| Patient characteristics and setting |
Inclusion criteria: Cutaneous pigmented lesions with clinical and/or dermoscopic features that supported a suspicion for cutaneous melanoma and therefore eligible for excision. A further set of lesions all diagnosed as clearly benign common naevi at both clinical and dermoscopic evaluations that did not undergo excision; data for these have not been included as > 50% of benign group must undergo histology or active follow‐up Setting: Specialist unit (skin cancer/pigmented lesions clinic) Prior testing: Clinical and/or dermatoscopic suspicion Setting for prior testing: Specialist unit (skin cancer/pigmented lesions clinic) Exclusion criteria: Difficult‐to‐diagnose lesions, Poor‐quality index test image incorrectly acquired or not correctly segmented by the system Sample size (participants): N eligible: 1359 Sample size (lesions): N eligible: 1485; N included: 1391 excised (94 excluded: lesions out of focus, 46 cases, 3%, or system failure to correctly segment the lesion border (48 cases, 3%)) Participant characteristics: Median age 36 yrs (range 5 to 88); Male: 597 (43.9% of 1359); Pigmented (%): 100%; Located on: head/neck: 2.9%, trunk: 66.2%, upper limbs/shoulder: 9.1%, lower limbs/hip: 17%, limbs: 4.8% Thickness/depth: ≤ 1 mm: 140/164 (85%) invasive MM; median thickness 0.58 mm, (0.1 mm to 2.7 mm), for 164 invasive MM. median size: 9 mm in maximum diameter; ≤ 6 mm in 44 cases (24%) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: SpectroShade (MHT, Verona, Italy) (ANN classifier) System details: Illumination assembly located inside a PC and an external detection device placed in a hand‐held probe, with integrated image analysis software Derivation study (internal validation) "The training group (696 cases, including 90 melanomas) was used to optimise the inner fitting weights of the neural network by means of a training algorithm. The verify set (348 cases, including 53 melanomas) was used to properly stop the training process, preventing the so‐called overlearning, i.e. a drop in the generalisation capabilities of the classifier which would otherwise fit the noise pattern of the data instead of defining a proper boundary between malignant and benign moles. The test set (347 cases, including 41 melanomas) was used to confirm, by independent data, the discrimination performances of the system as obtained from the previous data sets." Also described in prior studies Marchesini 1995, Tomatis 1998, Tomatis 2003 Lesion characteristics assessed: Features analysed: (i) reflectance (R); (ii) variegation (V); (iii) area (A); (iv) dark area ratio (DAR); (v) dark island reflectance (DIR); (vi) dark distribution factor (DDF); (vii) dark permanence (D PER) Additional predictors included: No further information used Method of diagnosis: Spectroscopic images CAD‐based diagnosis Prior/other test data: No further information used CAD output: NR Diagnostic threshold: Threshold set to produce sensitivity of 80% |
||
| Target condition and reference standard(s) |
Reference standard: ‐ Histological diagnosis alone. Data provided for subgroup (test set) that all underwent excision Histology (not further described) N participants/lesions: 1391 (347 in test set) Disease‐positive: 184 (41 in test set); Disease‐negative: 1207 (306 in test set) Target Condition (Final diagnoses) Melanoma (invasive): 164 (full sample, number in test set NR); Melanoma (in situ): 20 (full sample, number in test set NR); ‐ Melanoma (in situ and invasive, or not reported): 41 (test set only) BCC: 7 (full sample, number in test set NR) Sebhorrheic keratosis: 27 (full sample, number in test set NR); 'Benign' diagnoses: 280 dysplastic naevus, rest various benign (893) |
||
| Flow and timing | Time interval to reference test: NR "Before surgery, images of the 1485 pigmented lesions were acquired in vivo." | ||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | Yes | ||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | No | ||
| High | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Walter 2012.
| Study characteristics | |||
| Patient sampling |
Study design: Randomised controlled trial, only experimental group included Data collection: Prospective Period of data collection: March 2008 to May 2010 Country: UK |
||
| Patient characteristics and setting |
Inclusion criteria: Adults with any suspicious PSL, i.e. any lesion presented by a patient, or opportunistically seen by a family doctor or practice nurse, that could not immediately be diagnosed as benign and about which the patient could not be reassured Setting: Primary 15 general practices in eastern England Prior testing: Clinical suspicion of malignancy without dermatoscopic suspicion Setting for prior testing: Primary Exclusion criteria: Those unable to give informed consent or considered inappropriate to include by their family doctor. Sample size (patrticipants): N eligible: 1297; N included: 1293 in RCT, 643 in experimental group Sample size (lesions): N eligible: 1580; N included: 1583 in RCT, 788 in experimental group Participant characteristics (whole population): Mean age: 44.6 yrs (SD 16.8). Male: 465 (36%). Ethnicity: white 1214 (93.9%); mixed 45 (3.5%); missing: 34 (2.6%) Lesion characteristics. Lesion thickness ≤ 1 mm: in 'more than half' of MM |
||
| Index tests |
Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: SIAscope + MoleMate (classifier NR) System details: SIAscopy with MoleMate (software image management system) viewing platform and integrated primary care scoring algorithm No derivation aspect (external validation study) Lesion characteristics assessed: Not described Additional predictors included: None reported Method of diagnosis: In‐person diagnosis, spectroscopic images (SIAgraphs) CAD‐aided diagnosis Prior/other test data: Clinical history and naked‐eye examination Operators: 28 clinicians Operator qualifications: GP 2 nurse practitioners Experience in practice: Mixed experience (low and high experience combined) as previously recorded Experience with index test: Low experience/novice users CAD output: SIAgraphs and lesion score using Primary Care Scoring Algorithm Diagnostic threshold: Primary Care Scoring Algorithm (6 or more points regarded as suspicious) |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis plus follow‐up and expert opinion Histology (not further described) 215 (histology result missing in further 4) Disease‐positive: 35; Disease‐negative: 180 Clinical follow‐up plus histology of suspicious lesions: 22 of the 411 referred participants were monitored (not further described); 566 of the 1162 not referred underwent expert review and were then re‐assessed at 3 ‐ 6 months Disease‐positive: 1; Disease‐negative: 588 Expert opinion. Reviewed by 2 dermatology experts using the recorded clinical history and examination, a digital photograph, and MoleMate image where available Disease‐positive: 0; Disease‐negative: 725 Target condition (Final diagnoses) Melanoma (invasive): 30; Melanoma (in situ): 6; BCC: 10 'Benign' diagnoses: 1306 |
||
| Flow and timing | Excluded participants: 417 withdrew from control group after randomisation ‐ 10 did not attend for dermatology assessment; 19 excluded; 1 died; 4 missing histology (in referred group; included as benign?); plus 12 with unknown outcome (in non‐referred group, assumed benign and included) | ||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Are the included patients and chosen study setting appropriate? | Yes | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Low | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | High | ||
| DOMAIN 2: Index Test Visual inspection | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | No | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | No | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | No | ||
| High | High | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Wells 2012.
| Study characteristics | |||
| Patient sampling |
Study design: Case‐control Data collection: Retrospective ‐ MelaFind diagnoses from acquisition study were used Retrospective image selection/Prospective interpretation ‐ Clinical and dermoscopic images Period of data collection NR Country: USA |
||
| Patient characteristics and setting |
Inclusion criteria: Pigmented lesions cases selected from a repository of lesions amassed during an acquisition study conducted by MELA Sciences Inc for the US Food and Drug Administration Setting: Company database (MELA Sciences Inc) of lesion images amassed during an acquisition study for the FDA Prior testing: Selected for excision (no further detail) Setting for prior testing: NR Exclusion criteria: None reported Sample size (participants): NR Sample size (lesions): N eligible NR; N included: 47 Participant characteristics: None reported Lesion characteristics: None reported |
||
| Index tests |
Dermoscopy No algorithm Method of diagnosis: Dermoscopic images Prior test data: Clinical images and detailed clinical history Diagnostic threshold: NR; Decision to biopsy the lesion/Melanoma or not Diagnosis based on: Average (n = 39) Observer qualifications: Dermatologist Experience in practice: Not described Experience with index test: Not described Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: MelaFind (EO Sciences, USA) (classifier NR) System details: Multispectral imaging system with integrated image analysis software; device takes images in vivo No derivation aspect (external validation study) Described in prior study Monheit 2011 Lesion characteristics assessed: NR Additional predictors included: None reported Method of diagnosis: In‐person diagnosis CAD‐based diagnosis Prior/other test data: Unclear CAD output: Binary output: (1) positive, (lesion should be considered for biopsy to rule out melanoma); and (2) negative (lesion should be considered for later evaluation) Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard: Histological diagnosis alone Disease‐positive: 23; Disease‐negative: 24 Target condition (Final diagnoses) Melanoma (in situ and invasive, or not reported): 23 'Benign' diagnoses: 24 |
||
| Flow and timing |
Exclusions from analysis: None Time interval to reference test: NR; Images taken prior to biopsy |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Unclear | ||
| Unclear | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Unclear | ||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Winkelmann 2016.
| Study characteristics | |||
| Patient sampling |
Study design: Case‐control Data collection: Retrospective image selection/Prospective interpretation Period of data collection: NR Country: NR |
||
| Patient characteristics and setting |
Inclusion criteria: Images of PSLs previously analysed by a digital classifier MSDSLA; method of selection of the 12 not reported Setting: Dermoscopy conference Prior testing: NR Setting for prior testing: Unspecified Exclusion criteria: None reported Sample size (participants): NR Sample size (lesions): N eligible; N included: 12 Participant characteristics: None reported Lesion characteristics: None reported |
||
| Index tests |
Visual inspection (VI) No algorithm Method of diagnosis: Clinical photographs Prior test data: Unclear Diagnostic threshold: NR ‐ biopsy decision Diagnosis based on: Average (n = 70) Observer qualifications: Dermatologist Experience in practice: Not described Experience with index test: Not described Other detail: Practitioners with a particular interest in skin cancer or technology may have chosen to attend this conference and/or self‐selected to take part in the study Dermoscopy No algorithm Method of diagnosis: Dermoscopic images Prior test data: Clinical images Diagnostic threshold: NR ‐ biopsy decision Test observers: As described for Visual Inspection (above) Computer‐assisted diagnosis ‐ Spectroscopy‐based MSI‐CAD system: MelaFind (EO Sciences, USA) (Logistic regression classifier) System details: Multispectral imaging system with integrated image analysis software; device takes images in vivo No derivation aspect (reader study) Lesion characteristics assessed: NR Additional predictors included: None Method of diagnosis: Spectroscopic images CAD‐aided diagnosis Prior/other test data: Clinical and dermoscopic images CAD output: Binary output: (1) positive, (lesion should be considered for biopsy to rule out melanoma); and (2) negative (lesion should be considered for later evaluation) Diagnostic threshold: Threshold not reported |
||
| Target condition and reference standard(s) |
Reference standard Histological diagnosis alone Disease‐positive: 5; Disease‐negative: 7 Target condition (Final diagnoses) Melanoma (invasive): 3; Melanoma (in situ): 2 Mild/moderate dysplasia: 7 low‐grade dysplastic naevi |
||
| Flow and timing |
Exclusions from analysis: None Time interval to reference test: NR |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| High | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Unclear | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | Unclear | ||
| DOMAIN 2: Index Test Visual inspection | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Dermoscopy | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | No | ||
| Was the test interpretation carried out by an experienced examiner? | Yes | ||
| Was the CAD model evaluated in an independent study population? | |||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | |||
| Unclear | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Unclear | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Low | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Wollina 2007.
| Study characteristics | |||
| Patient sampling |
Study design: Case series Data collection: NR Period of data collection: January 2003 to October 2004 Country: Germany (Dresden); Switzerland (Locarno and Lugarno) |
||
| Patient characteristics and setting |
Inclusion criteria: PSLs Setting: Secondary (general dermatology) Dept Dermatology, Dresden; Primary (private clinic), Locarno and Lugano Prior testing: No prior testing Lugano and Locarno (described as representing "a sort of first‐screening check") Clinical and/or dermatoscopic suspicion Dresden (described as having many patients referred for a second‐level control) Setting for prior testing: Unspecified Exclusion criteria: NR Sample size (patients): N eligible: 1308; N included: NR Sample size (lesions): N eligible: 3541; N included: 466 Participant characteristics: Male: 566; 43.2% (full sample) Thickness/depth: ≤ 1 mm: 38 (incl 8 in situ) Dresden: 22 (incl 4 in situ) Locarno: 7 (incl 2 in situ) Lugano 9 (incl 2 in situ) |
||
| Index tests |
Computer‐assisted diagnosis ‐ Dermoscopy‐based Derm‐CAD system: DB‐MIPs (Biomips Engineering, Italy) (Euclidian distances classifier) System details: Dermoscopy unit, internal steromicroscope, internal DB, pattern analysis system. DDA software analysis – analyses 50 parameters subdivided into 3 categories, i.e. geometries, colours, and textures and islands of colours (Burroni islands) No derivation aspect (external validation study) Lesion characteristics assessed: 35 variables including: variance of symmetry, maximum diameter, border’s gradient, skin red average, red average, red tenth, unbalance, dark areas towards periphery, dishomogeneity, blue dominant, transition, unbalance of transition Additional predictors included: No further information used Method of diagnosis: In‐person diagnosis CAD‐aided diagnosis Operator qualifications: Not reported Experience in practice: Not described Experience with index test: Not described. States that test operators do not need special training Prior/other test data: CAD used as part of clinical analysis CAD output: Not specified Diagnostic threshold: The automated diagnosis was run at similprob = 45 and similprob = 75 thresholds, the latter being more sensitive but less specific. Similprob = 75 threshold selected at random for review |
||
| Target condition and reference standard(s) |
Reference standard:
Histological diagnosis alone. Unclear whether data relate to compariaon with histology alone or to whole set of lesions, in which case the reference standard is not reported for most benign lesions, although the 'decision to follow‐up' is mentioned Histology (not further described) N participants/lesions: 466 Disease‐positive: 52 (incl 8 in situ); Disease‐negative: Either 414 or 3489 Target condition (Final diagnoses) Melanoma (invasive): 44; Melanoma (in situ): 8 'Benign' diagnoses: 414 |
||
| Flow and timing |
Exclusions from analysis: None Time interval to reference test: CAD performed during clinical diagnosis, before histopathology. Interval to surgery not reported |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | No | ||
| Was an adequate spectrum of cases used to train the algorithm? | |||
| Unclear | High | ||
| DOMAIN 2: Index Test Computer‐assisted diagnosis | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Was the test interpretation carried out by an experienced examiner? | Unclear | ||
| Was the CAD model evaluated in an independent study population? | Yes | ||
| Was model overfitting accounted for during model development? | |||
| Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? | Yes | ||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Was the use of expert opinion (with no histological confirmation) avoided as the reference standard? | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Were the reference standard results likely to correctly classify the target condition (disease negative)? | Yes | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Included studies: Two quality assessment items could not be evaluated for all studies, and so were left blank:
Patient Selection: Concerns regarding applicability: “Was an adequate spectrum of cases used to train the algorithm? (training set)” (Could only be answered if the study included a derivation aspect). CAD Risk of Bias: "Was model overfitting accounted for during model development?" (Could only be answered if the study included a derivation aspect)
Index test ‐ CAD: Concerns regarding applicability: "Was the test interpretation carried out by an experienced examiner? (Could only be answered if the study gave computer generated CAD results to clinicians to make a diagnosis [CAD–aided diagnosis])"
7FFM: 7 features for melanoma; ABCD: asymmetry, border, colour, dimension; AK: actinic keratosis; BCC: basal cell carcinoma; CM: cutaneous melanoma; CN: common naevus; cSSC: cutaneous squamous cell carcinoma; DF: dermatofibroma; DN: dysplastic naevus; ELM: epiluminescence microscopy; Knn: K nearest neighbour; MM: malignant(invasive) melanoma; NR: not reported; OIDRS: non–imaging diffuse reflectance spectroscopy; PSL: pigmented skin lesion; SK: seborrheic keratosis; SVS: Skin View System; TDS: total dermoscopic score.
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Abbas 2010 | Target condition: border detection not diagnosis |
| Abbas 2011a | Index test: test used to determine border |
| Abbas 2011b | Target condition: border detection not diagnosis |
| Abbas 2012 | No independent test set: Reports training on 20% and testing on 80% of images but results presented only for whole dataset Author contacted, responded but cannot help |
| Abbas 2013a | Target condition: border detection not diagnosis |
| Abbas 2013b | Individual lesion characteristics: focus on border detection Derivation study |
| Abuzaghleh 2015 | Derivation study Authors contacted. In the experiments, 75% of the database images are used for training and 25% are used for testing. No breakdown of D+/D‐ in the test set, cannot back‐calculate from the sensitivity and specificity values given |
| Afonso 2012 | Index test: Not used to differentiate malignant from benign lesions |
| Alfed 2015 | Derivation study: no independent test set used |
| Ali 2012 | Not a primary study |
| Altamura 2008 | Index test: study of optimal surveillance/appropriate follow‐up times not initial diagnosis |
| Andreassi 1999 | Derivation study: Uses jack knife validation |
| Armengol 2011 | Index test; derivation study |
| Arroyo 2011 | Individual lesion characteristics; derivation study |
| Ballerini 2012 | Target condition: D+ includes AK |
| Barata 2012a | Derivation study |
| Barata 2012b | Derivation study |
| Barata 2013 | Derivation study |
| Barata 2015a | Derivation study; uses LOO procedure |
| Barata 2015b | Derivation study: uses 10‐fold classification |
| Barata 2015c | Derivation study; uses 10‐fold classification |
| Binder 2000 | Derivation study; no independent test set |
| Bjerring 2001 | Not a primary study: leaflet |
| Blum 2004b | Overlapping study population (Blum 2004a) |
| Boden 2013 | Derivation study: uses LOO method; no independent test set |
| Bono 1999 | Derivation study; exclude on 2 x 2 data: LFR: not a test accuracy study |
| Borlu 2008 | Not a primary study ‐ not test accuracy; no patient data in this study |
| Brown 2000 | Not a primary study; systematic review |
| Carrara 2007 | Reference standard: < 50% benign lesions had histology or clinical follow‐up |
| Celebi 2008 | Derivation study: no independent test set |
| Chen 2003 | Individual lesion characteristics: investigates colour only; Derivation study; Inadequate 2 x 2 data |
| Cheng 2012 | Individual lesion characteristics: only telengiectasia used to evaluate whether a BCC or not. No other characteristics are evaluated; Derivation study |
| Cheng 2013 | Derivation study; 2 x 2 data |
| Christensen 2010 | Derivation study |
| Claridge 1992 | Individual lesion characteristics; shape of lesion |
| Cukras 2013 | Not a primary study |
| Day 2001 | Reference standard: 16/73 excised; 9 were melanoma |
| Debeir 1999 | Derivation study |
| Di 2010 | Derivation study: primarily derivations study with small paragraph on a validation study with limited details ‐ se/sp given for validation study for individual characteristics but not for overall se/sp of the system; 2 x 2 data |
| Ding 2015 | Derivation study; uses LOO technique |
| Dreiseitl 2005 | Index test: not test accuracy and the CDSS recommendations were simulated |
| Durg 1993 | Target condition: detection of variegated colouring |
| Elbaum 2001 | Derivation study: uses LOO procedure |
| Emery 2010 | Reference standard: 111/1211 lesions excised, remaining lesions did not have any reported follow‐up |
| Engin 2016 | Sample size Reference standard |
| Ercal 1994 | Derivation study |
| Faal 2013 | Derivation study: The 436 images were split into 60% training and 40% test sets. They combined the results of the training and tests sets as an average of the performace of the test, no breakdown of the classifer rate according to the test and training images |
| Farina 2000 | Derivation study ‐ no test set |
| Ferris 2016 | Not a primary study |
| Fidalgo 2003 | 2 x 2 data; Duplicate or related publication: Appears to be superseded by Serrao 2006 |
| Fikrle 2007 | Reference standard: Follow‐up study with < 50% of participants receiving lesion excision |
| Fikrle 2013 | Reference standard: Follow up study <50% of study participants have their final diagnosis reached by histopathology. |
| Fruhauf 2012 | Reference standard: 35/219 underwent histology; 13 followed up; 171 expert clinical diagnosis |
| Fueyo‐Casado 2009 | Reference standard: < 50% of the study population recieved histology as a test. No information given on those who were followed up |
| Ganster 2001 | Derivation study: Uses leave‐one‐out methods for cross‐validation, "test set" for the automated diagnosis but data given in Table 1 incorporates data from the training set |
| Garcia‐Uribe 2004 | Index test: reflectance spectrometry/spectroscopy; Derivation study: results use training and test sets |
| Garcia‐Uribe 2010 | 2 x 2 data: only give the values for sensitivity and specificity but do not give a breakdown of disease positive or negative totals in the test set Contact authors |
| García 2014 | Target condition: test aims to detect presence of pigment network, is not an evaluation of detection of MM |
| Garnavi 2012 | Reference standard: not properly defined, it could be the images were sampled from a database of histopathologically diagnosed lesions but reporting is unclear. |
| Gerger 2003 | Reference standard: no reference standard in 133 naevi, unclear whether these were in the test set; Derivation study: Training sets (n = 2) and test set must overlap as total number of lesions (n = 423) add up to more than total included lesions (n = 136). |
| Glotsos 2015 | Derivation study: uses LOO procedure |
| Gniadecka 2004 | Index test: Raman spectroscopy; Derivation study: methods report use of a 'test set', but this is not further described 2 x 2 data. |
| Govindan 2007 | Reference standard: "Six hundred and twenty seven (71%) lesions were diagnosed as benign and were discharged from the PLC" "As the patients who were clinically diagnosed as having benign lesions did not undergo biopsy of their lesions, the false negative rate for the consultant could not be determined. None of the patients with benign lesions from this study have reported back to the PLC with any concern about their lesions and none have had excision biopsy of their lesions." |
| Green 1991 | Derivation study ‐ no independent test set |
| Green 1994 | Derivation study: no independent test set |
| Guerra‐Rosas 2015 | Derivation study: no test set; 2 x 2 data: no se/sp presented |
| Guillod 1996 | Derivation study |
| Gutkowicz‐Krusin 2000 | Sample size: 2 melanoma; Derivation study. |
| Hacioglu 2013 | Target condition: Does not provide 2 x 2 data for separate keratinocyte cancers |
| Haenssle 2004 | Target condition |
| Haenssle 2010 | 2 x 2 data: Does not report specificity Duplicate or related publication: Same patients as Haenssle 2004 |
| Haniffa 2007 | Reference standard: approximately 20% of patients received a final diagnosis by histology. 179 biopsies were performed. Total sample was 881 lesions. |
| Hintz‐Madsen 2001 | Derivation study: uses LOO technique |
| Hoffmann 2003 | Derivation study: Uses LOO procedure 2 x 2 data: Only reports ROC values, so not able to extract a 2 x 2 table |
| Horsch 1997 | Derivation study |
| Huang 1996 | Individual lesion characteristics: Border irregularity not overall diagnosis 2 x 2 data |
| Ikuma 2013 | Derivation study: uses LOO procedure |
| Isasi 2011 | Derivation study |
| Iyatomi 2006 | Derivation study: uses LOO procedure; 2 x 2 data. |
| Iyatomi 2008a | Derivation study |
| Iyatomi 2008b | Derivation study |
| Iyatomi 2008c | Derivation study: performance evaluated by averaging both combinations (training and test sets). Data not presented separately; uses LOO procedure 2 x 2 data: Not test accuracy; compares automated with manual extraction of tumour area |
| Iyatomi 2010a | Individual lesion characteristics 2 x 2 data: not a test accuracy study |
| Iyatomi 2010b | Target condition: differentiates melanocytic from non‐melanocyitc lesinos and not malignant from benign |
| Iyatomi 2011 | Index test: colour calibration rather than MM detection Individual lesion characteristics 2 x 2 data: not a test accuracy study. |
| Jain 2015 | Not a primary study |
| Jakovels 2013 | Reference standard test not reported |
| Jamora 2003 | Ineligibile reference standard no referene standard for index test negatives |
| Jaworek‐Korjakowska 2016a | Derivation study no separate test set |
| Jaworek‐Korjakowska 2016b | Derivation study: uses cross‐validation with no separate test set; 2 x 2 data: only gives accuracy, not se/sp |
| Jeddi 2016 | Study population: unclear whether data for BCC includes only those with lesions actually suspicious for BCC Ineligible index test: limited details Ineligibile reference standard: appears to be expert Dx |
| Kahofer 2002 | Derivation study. Lesions not split into training/test |
| Kaur 2015 | Ineligibile reference standard: % of benign with histology versus expert dx not reported 2 x 2 data: Only gives AUC and not se/sp Contact authors: 1. reference standard in benign group 2. se/sp for model using all four features needed Dr WV Stoeker e‐mail: wvs@mst.edu |
| Korotkov 2012 | Not a primary study: narrative review |
| Kuzmina 2011 | Derivation study. Ineligibile reference standard: reference standard test not reported |
| Landau 1999 | Derivation study. No indication that system has previously been evaluated |
| LeAnder 2010 | Derivation study. no separate 2 x 2 data for training set and test set |
| Lefevre 2000 | Ineligible index test Limited test detail; cannot tell whether clinical or dermoscopic images used Ineligibile reference standard No details of reference standard |
| Lihacova 2013 | Derivation study. No test set |
| Liu 2012 | Derivation study: asymmetry detection; 10‐fold cross‐validation 2 x 2 data |
| Machado 2015 | Ineligible target condition detection of reticular pattern not MM |
| Maglogiannis 2004 | Derivation study. 2 x 2 data |
| Maglogiannis 2006 | Derivation study. They incorporated the training set population together with the test set as shown below "A training set of 500 cases was randomly selected from the dataset of the total cases. The accuracy of the classification algorithm was examined using a test set consisted of the full set of 1041 cases." |
| Manousaki 2006 | Derivation study. ‐ Training set only |
| Marchesini 1992 | Derivation study. ‐ no independent data set |
| Masood 2013 | Derivation study. |
| Menzies 1999 | Not a primary study |
| Mete 2011 | Derivation study. 2 x 2 data Not test accuracy |
| Mhaske 2013 | Derivation study. 2 x 2 data Not test accuracy |
| Moncrieff 2002 | Derivation study. ‐ no independent test set |
| Morrow 2010 | Not a primary study narrative review |
| Nagaoka 2012 | 2 x 2 data: they did not give the number of lesions in test set, they state "The sensitivity of 90% and specificity of 84% were obtained by applying this threshold value to the validation set." Cannot see how to work out the 2 x 2 data from this information Contact authors: Can you provide us with the number of lesions in the test set, with a breakdown of number of melanoma vs other lesions? We can then use the sensitivity of 90% and specificity of 84% provided to work out the 2 x 2 |
| Nagaoka 2013 | Derivation study. index was developed on nonglabrous skin and is being applied to acral skin for first time, which makes it a derviation study? Ineligibile reference standard No reference standard reported for D‐ |
| Nagaoka 2015 | Ineligibile reference standard Contact authors: reference standard not clearly reported ‐ "Lesions that were clinically judged benign were not biopsied because of ethical reasons." T. Nagaoka e‐mail: nagaoka@aoni.waseda.jp Previous contact for prior studies was unsuccessful |
| Noroozi 2016 | Ineligible index test |
| Oka 2004a | Not a primary study letter |
| Oka 2004b | Derivation study. uses LOO procedure |
| Oka 2006 | Not a primary study letter |
| Pellacani 2004a | Individual lesion characteristics |
| Pellacani 2004b | Assessesindividual lesion characteristics only; looks at colours in melanocytic lesion (ML) images. |
| Pellacani 2006 | Derivation study. looks at detection of asymmetry between clinicians and computer 2 x 2 data: 2 x 2 could be derived for overall asymmetry or border cut‐off but not overall diagnosis |
| Perrinaud 2007 | Inadequate sample size CAD ‐ fewer than 5 melanomas (not including 'typical' melanomas) Ineligible index test Does not provide data for visual inspection alone |
| Pompl 2000 | Derivation study. Training set (60 mel and 60 benign) reincluded for model evaluation |
| Rajpara 2009 | Not a primary study Systematic review |
| Rastgoo 2015 | Derivation study. Describes splitting each set of 180 lesions into 70% for training, 15% for validation and 15% for testing (n=27?). But it's difficult to follow what the results relate to, especially as the classifier is repeated 10 times with different sets of dysplastic lesions but the same melanoma cases |
| Rigel 2012 | Duplicate or related publication CAD ‐ all lesions included in Monheit 2011 |
| Rosado 2003 | Not a primary study Systematic Review |
| Rubegni 2001a | Not a primary study letter |
| Rubegni 2001b | Derivation study. paper states that the accuracy estimates (sensitivity, specificty) are given using the LOO method but they have not given a breakdown of the actually number. It is not possible to tell if they used the whole study sample |
| Rubegni 2002b | Derivation study. the training and test data not given separately |
| Rubegni 2005 | Not a primary study Editorial |
| Rubegni 2010 | Derivation study: uses LOO procedure 2 x 2 data |
| Rubegni 2013 | Derivation study. |
| Sadeghi 2013 | Individual lesion characteristics: irregular streaks 2 x 2 data: Only given the AUC values not possible to work out 2 x 2 from this |
| Safi 2011 | Derivation study. |
| Salerni 2012 | Ineligible index test test used for surveillance |
| Sboner 2001 | Duplicate or related publication same data as Sboner 2004 |
| Sboner 2003 | Derivation study. describes 10‐fold cross‐validation pocess for training/testing classifier. |
| Sboner 2004 | Derivation study. Uses LOO method with no independent test set for validation The whole set of cases is divided in 10 disjoint sets, which are used as test cases. For each test set, the remaining 9 sets are used to train the classifiers (training set). The final results are the average values computed on the 10 test sets |
| Schindewolf 1993 | 2 x 2 data not given enough information to populate 2 x 2 table |
| Schindewolf 1994 | Ineligible index test evaluates CAD not VI Derivation study. uses cross‐validation |
| Schmid‐Saugeon 2003 | Derivation study. no separate test set for validation |
| Schumacher 2016 | Not a primary study comment paper |
| Seidenari 1995 | Ineligible index test comparing 2 ways of attaining videomicroscope images no accuracy data provided 2 x 2 data |
| Seidenari 2005 | Ineligibile reference standard All D+ were excised (n = 95) but only 45% of benign group were excised (76 AN plus 30% of BN (86/288)) and methods of estabishing final diagnosis were not reported for the remainder EXCLUDE but contact authors We would like to include test accuracy results from this study, however in order to do so we would need some further information on how the final diagnosis was reached for thos lesions that were not excised. We have noted that all D+ lesions were excised (n = 95) and 45% of benign lesions were excised (76 atypical naevi plus 30% of benign (86/288)). Can you advise us as to how the final diagnosis was reached for the remaining 126 benign lesions? |
| Seidenari 2007 | Individual lesion characteristics CAD only ‐ CAD system based on single characteristic |
| Seidenari 2012 | Ineligible index test: CAD ONLY ‐ does not evaluate a CAD system Individual lesion characteristics 2 x 2 data Contact authors: Table 3 provides mean ABCD and 7‐point checklist scores; are you able to provide us with a cross‐tabulation of results with each checklist at 'standard' thresholds against final diagnosis? e.g. ABCD > 4.75 and > 5.45 for MIS and benign groups 7‐point checklist: presence ≥ 2 chars and ≥ 3 chars? |
| Shakya 2012 | Ineligible target condition SCC in situ is not included in target condition |
| She 2007 | Derivation study. |
| She 2013 | Derivation study. no separate data for training and test set. used LOO technique |
| Shimizu 2012 | Derivation study. The performance was evaluated under the LOO cross‐validation test |
| Skrovseth 2010 | Not a primary study Statistical paper for developing a new algorithm |
| Smith 2000 | Derivation study. |
| Sober 1994 | Individual lesion characteristics Reports separately for shape, radii and border but not overall classification; note these are results for 1 centre taking part in a larger multicentre study |
| Stanganelli 1995 | Ineligible index test aim of study is to assess the intraobserver agreement |
| Stanley 2007 | Individual lesion characteristics Derivation study. |
| Stanley 2008 | Derivation study. cross‐validation study (check eligibility) |
| Stoecker 2005 | Individual lesion characteristics Derivation study. |
| Swanson 2010 | Ineligible index test reflectance spectroscopy Derivation study. This looks like an include as they mention a pilot study of 47 initial patients. There is an additional 47 patients included, but it seems that they have combined the data so for this reason it would be an excluded if derivation study, as the training data and test data are combined |
| Tehrani 2006 | Derivation study. Borderline exclude ‐ 4 features of NMSC identifed in pilot study and re‐examined in larger sample here; diagnostic model then created based on significaince of characteristics and se/sp data give. No test set presented |
| Terstappen 2007 | Ineligible study population: Includes only BCC ‐ looking for BCC characteristics on Siascope Derivation study. Derivation study; first application of Siascope to pigmented BCC; 21/25 lesions were BCCs |
| Varol 2006 | Not a primary study Link to study with correction "Error in Byline. In the the Study by Menzies et al titled “The Performance of SolarScan: An Automated Dermoscopy Image Analysis Instrument for the Diagnosis of Primary Melanoma,” published in the November 2005 issue of the ARCHIVES (2005;141:1388‐1396), the name of the one of the authors was misspelled. The author’s name is Alexandra Varol, B Med." |
| Vestergaard 2008 | Not a primary study systematic review |
| Wallace 2000a | Derivation study. uses LOO technique |
| Wallace 2000b | 2 x 2 data |
| Wallace 2002 | Derivation study. ‐ Uses LOO cross‐validation |
| Walter 2010 | Not a primary study clinical trial protocol |
| Watson 2009 | 2 x 2 data Not test accuracy; training in MoleMate |
| Wazaefi 2012 | Derivation study. no separate independent test set they used a 20‐fold cross‐validation |
| Wells 2011 | Duplicate or related publication see Wells 2012 |
| Wilson 2013 | 2 x 2 data this study is an economic evaluation of a SIAscopy, which itself was trialled in another paper ‐ not enough data to populate 2 x 2 table |
| Winkelmann 2015a | Duplicate or related publication |
| Winkelmann 2015b | Ineligible target condition D+ includes 15 lesions with moderate dysplasia Inadequate sample size only 1 MM |
| Winkelmann 2015c | Inadequate sample size |
| Winkelmann 2015d | Duplicate or related publication |
| Winkelmann 2016a | Ineligible index test aim of the test to to investigate correlation with clinical histology features (development of new CAD system?) |
| Wood 2008 | 2 x 2 data Not test accuracy ‐ acceptability etc |
| Yoo 2015 | Conference abstract only |
| Zagrouba 2004 | Ineligibile reference standard No reference standard details provided Contacted re reference standard. Author responded ‐ cannot help |
| Zhou 2010a | Derivation study |
| Zhou 2010b | Individual lesion characteristics |
| Zortea 2014 | Derivation study. Although data are divided into training and test sets, the test set data is used more than once over 20 realisations of each model, especially the melanomas, for which the same 10 are used in each realisation |
| Zouridakis 2004 | Ineligible study population Inadequate sample size Derivation study. |
AK: actinic keratosis; AN: atypical naevus; LOO: leave‐one‐out; se: sensitivity; sp: specificity
Differences between protocol and review
We could not conduct reviews on the accuracy of gene expression testing and volatile organic compounds as planned, due to an absence of relevant studies.
For this review, we amended the inclusion criteria to remove inclusion of participants "at high risk of developing melanoma, including those with a family history or previous history of melanoma skin cancer, atypical or dysplastic naevus syndrome, or genetic cancer syndromes" and "at high risk of developing BCC or cSCC, including those with a family history or previous history of skin cancer or genetic cancer syndromes, such as basal cell naevus (Gorlin) syndrome", as these are not target populations for CAD use.
We have changed one of the primary objectives and primary target conditions from diagnosing "cutaneous invasive melanoma alone", to diagnosing "cutaneous invasive melanoma and atypical intraepidermal melanocytic variants", as the latter is more clinically relevant to the practising clinician. The diagnosis of the target condition of invasive melanoma alone has instead been included as a secondary objective.
We amended the text to clarify that studies available only as conference abstracts would be excluded from the review unless full papers could be identified; studies available only as conference abstracts do not allow a comprehensive assessment of study methods or methodological quality.
We excluded rather than included studies using cross‐validation, such as 'leave‐one‐out' cross‐validation, as these methods are not sufficiently robust and are likely to produce unrealistic estimates of test accuracy. To improve clarity of methods, this text from the protocol: "We will include studies developing new algorithms or methods of diagnosis (i.e., derivation studies) if they use a separate independent ’test set’ of participants or images to evaluate the new approach. We will also include studies using other forms of cross validation, such as ‘leave‐one‐out’ cross‐validation (Efron 1983). We will note for future reference (but not extract) any data on the accuracy of lesion characteristics individually, e.g., the presence or absence of a pigment network or detection of asymmetry" has been replaced with:
"Studies developing new algorithms or methods of diagnosis (i.e. derivation studies) wereincluded if they evaluated the new approach using a separate 'test set' of participants or images.
Studies were excluded if they:
evaluated a new statistical model or algorithm in the same participants or images as those used to train the model (i.e. absence of an independent test set)
used cross‐validation approaches such as 'leave‐one‐out' cross‐validation (Efron 1983)"
We proposed to supplement the database searches by searching the annual meetings of appropriate organisations (e.g. British Association of Dermatologists Annual Meeting, American Academy of Dermatology Annual Meeting, European Academy of Dermatology and Venereology Meeting, Society for Melanoma Research Congress, World Congress of Dermatology, European Association of Dermato Oncology); however, due to volume of evidence retrieved from database searches and time restrictions, we were unable to do this.
For quality assessment, we further tailored the QUADAS‐2 tool according to the review topic.
Due to lack of data, we could not perform the following analyses: restriction to analysis of per‐patient data, or comparison of accuracy using diagnosis of stored images (image–based) with in–person diagnosis.
Upon closer review of the topic, but before examination of study data, we planned four secondary analyses in addition to those listed in the protocol: estimation of diagnostic accuracy for individual CAD systems; comparison of the accuracy of CAD to dermoscopy where both tests have been evaluated in the same studies (direct comparisons); the comparison of CAD–based diagnosis to CAD–assisted diagnosis (CAD results used by clinicians as a diagnostic aid); and where CAD systems are used as a diagnostic aid, to determine the effect of observer experience on diagnostic accuracy.
We planned three heterogeneity investigations relating to population characteristics in addition to those listed in the protocol (Patient population: primary/secondary/specialist unit; Lesion type: any pigmented/melanocytic; Inclusion of multiple lesions per participant), but we could not perform these investigations due to insufficient data.
Contributions of authors
JDi was the contact person with the editorial base. LFR co‐ordinated contributions from the co‐authors and wrote the final draft of the review. SB conducted the literature searches. JDi, NC, LFR screened papers against eligibility criteria. LFR, JDi, NC appraised the quality of papers. LFR, JDi, NC, AG, SAC, AD extracted data for the review. LFR sought additional information about papers. LFR, JDi entered data into RevMan. YT, LFR analysed and interpreted data. LFR, JDi, YT worked on the Methods sections. LFR drafted the clinical sections of the Background and responded to the clinical comments of the referees. LFR responded to the methodology and statistics comments of the referees. CO, KG were the consumer co‐authors and checked the review for readability and clarity, as well as ensuring outcomes are relevant to consumers. JDi is the guarantor of the update.
Disclaimer
This project presents independent research supported by the National Institute for Health Research, via Cochrane Infrastructure funding to the Cochrane Skin Group and Cochrane Programme Grant funding, and the NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the Systematic Reviews Programme, NIHR, NHS or the Department of Health and Social Care.
Sources of support
Internal sources
No sources of support supplied
External sources
-
NIHR Systematic Review Programme, UK.
This project was funded by an NIHR Cochrane Systematic Reviews Programme Grant (13/89/15)
-
The National Institute for Health Research (NIHR), UK.
The NIHR, UK, is the largest single funder of the Cochrane Skin Group
NIHR clinical fellowship, UK.
-
NIHR Birmingham Biomedical Research Centre, UK.
JD, JJD and YT receive support from the NIHR Birmingham Biomedical Research Centre
Declarations of interest
Lavinia Ferrante di Ruffano: nothing to declare. Yemisi Takwoingi: nothing to declare. Jac Dinnes: nothing to declare. Naomi Chuchu: nothing to declare. Susan E Bayliss: nothing to declare. Clare Davenport: nothing to declare. Rubeta N Matin: "my institution received a grant for a Barco NV commercially sponsored study to evaluate digital dermoscopy in the skin cancer clinic. My institution also received Oxfordshire Health Services Research Charitable Funds for carrying out a study of feasibility of using the Skin Cancer Quality of Life Impact Tool (SCQOLIT) in non melanoma skin cancer. I have received royalties for the Oxford Handbook of Medical Dermatology (Oxford University Press). I have received payment from Public Health England for the "Be Clear on Cancer" skin cancer report. I have no conflicts of interest to declare that directly relate to the publication of this work." Kathie Godfrey: nothing to declare. Colette O'Sullivan: nothing to declare. Abha Gulati: nothing to declare. Sue Ann Chan: nothing to declare. Alana Durack: nothing to declare. Susan O'Connell: nothing to declare. Matthew D Gardiner: nothing to declare. Jeff Bamber: J Bamber has been a member of the advisory board of Michelson Diagnostics and has received payment from Cancer Research UK and the International Breast Ultrasound School and Queen Mary University of London for lectures given. He has received book royalties from John Wiley and Sons, and acknowledges funding from the Engineering and Physical Sciences Research Council, the NIHR Biomedical Research Centre and the Royal Marsden NHS Foundation Trust and Institute of Cancer Research, and the Cancer Research UK Imaging Centre Grant to the Institute of Cancer Research. There was no involvement of grant funders or other sponsors in the study whatsoever. J Bamber is the first named inventor on a patent jointly held by his institution (The Institute of Cancer Research) and The University of Bern. The priority date was 26 July 2012. The PCT publication date was 30 January 2014. A brief description of the invention is as follows: "A method of ultrasound and photoacoustic imaging in which image clutter is reduced, thus improving image quality and penetration depth, by using a localised vibration to tag true signals so as to discriminate them from false signals which occur from sources outside of the imaged region." Jonathan J Deeks: nothing to declare. Hywel C Williams: I am director of the NIHR HTA Programme. HTA is part of the NIHR which also supports the NIHR systematic reviews programme from which this work is funded.
Edited (no change to conclusions)
References
References to studies included in this review
Ascierto 2010 {published data only}
- Ascierto PA, Palla M, Ayala F, Michele I, Caraco C, Daponte A, et al. The role of spectrophotometry in the diagnosis of melanoma. BMC Dermatology 2010;10:5. [ER4:19728329; PUBMED: 20707921] [DOI] [PMC free article] [PubMed] [Google Scholar]
Barzegari 2005 {published data only}
- Barzegari M, Ghaninezhad H, Mansoori P, Taheri A, Naraghi ZS, Asgari M. Computer‐aided dermoscopy for diagnosis of melanoma. BMC Dermatology 2005;5:8. [ER4:15465860; PUBMED: 16000171] [DOI] [PMC free article] [PubMed] [Google Scholar]
Bauer 2000 {published data only}
- Bauer P, Cristofolini P, Boi S, Burroni M, Dell'Eva G, Micciolo R, et al. Digital epiluminescence microscopy: usefulness in the differential diagnosis of cutaneous pigmentary lesions. A statistical comparison between visual and computer inspection. Melanoma Research 2000;10(4):345‐9. [ER4:15465861; PUBMED: 10985668] [DOI] [PubMed] [Google Scholar]
Binder 1994 {published data only}
- Binder M, Steiner A, Schwarz M, Knollmayer S, Wolff K, Pehamberger H. Application of an artificial neural network in epiluminescence microscopy pattern analysis of pigmented skin lesions: a pilot study. British Journal of Dermatology 1994;130(4):460‐5. [ER4:18375032; PUBMED: 8186110] [DOI] [PubMed] [Google Scholar]
Binder 1998 {published data only}
- Binder M, Kittler H, Seeber A, Steiner A, Pehamberger H, Wolff K. Epiluminescence microscopy‐based classification of pigmented skin lesions using computerized image analysis and an artificial neural network. Melanoma Research 1998;8(3):261‐6. [ER4:15465863; PUBMED: 9664148] [DOI] [PubMed] [Google Scholar]
Blum 2004a {published data only}
- Blum A, Luedtke H, Ellwanger U, Schwabe R, Rassner G, Garbe C. Digital image analysis for diagnosis of cutaneous melanoma. Development of a highly effective computer algorithm based on analysis of 837 melanocytic lesions. British Journal of Dermatology 2004;151(5):1029‐38. [ER4:15465866; PUBMED: 15541081] [DOI] [PubMed] [Google Scholar]
Boldrick 2007 {published data only}
- Boldrick JC, Layton CJ, Nguyen J, Swetter SM. Evaluation of digital dermoscopy in a pigmented lesion clinic: clinician versus computer assessment of malignancy risk. Journal of the American Academy of Dermatology 2007;56(3):417‐21. [ER4:19728338; PUBMED: 17109995] [DOI] [PubMed] [Google Scholar]
Bono 1996 {published data only}
- Bono A, Tomatis S, Bartoli C, Cascinelli N, Clemente C, Cupeta C, et al. The invisible colours of melanoma. A telespectrophotometric diagnostic approach on pigmented skin lesions. European Journal of Cancer 1996;32(4):727‐9. [DOI: ; ER4:20569437] [DOI] [PubMed] [Google Scholar]
Bono 2002 {published data only}
- Bono A, Bartoli C, Cascinelli N, Lualdi M, Maurichi A, Moglia D, et al. Melanoma detection. A prospective study comparing diagnosis with the naked eye, dermatoscopy and telespectrophotometry. Dermatology 2002;205(4):362‐6. [ER4:15465870; PUBMED: 12444332] [DOI] [PubMed] [Google Scholar]
Burroni 2004 {published data only}
- Burroni M, Corona R, Dell'Eva G, Sera F, Bono R, Puddu P, et al. Melanoma computer‐assisted diagnosis: reliability and feasibility study. Clinical Cancer Research 2004;10(6):1881‐6. [ER4:15465878; PUBMED: 15041702] [DOI] [PubMed] [Google Scholar]
Cascinelli 1992 {published data only}
- Cascinelli N, Ferrario M, Bufalino R, Zurrida S, Galimberti V, Mascheroni L, et al. Results obtained by using a computerized image analysis system designed as an aid to diagnosis of cutaneous melanoma. Melanoma Research 1992;2(3):163‐70. [ER4:15465891; PUBMED: 1450670] [DOI] [PubMed] [Google Scholar]
Cristofolini 1997 {published data only}
- Cristofolini M, Bauer P, Boi S, Cristofolini P, Micciolo R, Sicher MC. Diagnosis of cutaneous melanoma: Accuracy of a computerized image analysis system (Skin View). Skin Research and Technology 1997;3(1):23‐7. [ER4:17941039; PUBMED: 27333169] [DOI] [PubMed] [Google Scholar]
Dreiseitl 2009 {published data only}
- Dreiseitl S, Binder M, Hable K, Kittler H. Computer versus human diagnosis of melanoma: evaluation of the feasibility of an automated diagnostic system in a prospective clinical trial. Melanoma Research 2009;19(3):180‐4. [ER4:15465907; PUBMED: 19369900] [DOI] [PubMed] [Google Scholar]
Ferris 2015 {published data only}
- Ferris LK, Harkes JA, Gilbert B, Winger DG, Golubets K, Akilov O, et al. Computer‐aided classification of melanocytic lesions using dermoscopic images. Journal of the American Academy of Dermatology 2015;73(5):769‐76. [ER4:25012337; PUBMED: 26386631] [DOI] [PubMed] [Google Scholar]
Friedman 2008 {published data only}
- Friedman RJ, Gutkowicz‐Krusin D, Farber MJ, Warycha M, Schneider‐Kels L, Papastathis N, et al. The diagnostic performance of expert dermoscopists vs a computer‐vision system on small‐diameter melanomas. Archives of Dermatology 2008;144(4):476‐82. [ER4:15465921; PUBMED: 18427041] [DOI] [PubMed] [Google Scholar]
Garcia Uribe 2012 {published data only}
- Garcia‐Uribe A, Zou J, Duvic M, Cho‐Vega JH, Prieto VG, Wang LV. In vivo diagnosis of melanoma and nonmelanoma skin cancer using oblique incidence diffuse reflectance spectrometry. Cancer Research 2012;72(11):2738‐45. [DOI: 10.1158/0008-5472.can-11-4027; ER4:21450592] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gilmore 2010 {published data only}
- Gilmore S, Hofmann‐Wellenhof R, Soyer HP. A support vector machine for decision support in melanoma recognition. Experimental Dermatology 2010;19(9):830‐5. [ER4:15465935; PUBMED: 20629732] [DOI] [PubMed] [Google Scholar]
Glud 2009 {published data only}
- Glud M, Gniadecki R, Drzewiecki KT. Spectrophotometric intracutaneous analysis versus dermoscopy for the diagnosis of pigmented skin lesions: prospective, double‐blind study in a secondary reference centre. Melanoma Research 2009;19(3):176‐9. [ER4:18375045; PUBMED: 19319002] [DOI] [PubMed] [Google Scholar]
Gutkowicz Krusin 1997 {published data only}
- Gutkowicz‐Krusin D, Elbaum M, Szwaykowski P, Kopf AW. Can early malignant melanoma be differentiated from atypical melanocytic nevus by in vivo techniques? Part II. Automatic machine vision classification. Skin Research and Technology 1997;3(1):15‐22. [ER4:20569466; PUBMED: 27333168] [DOI] [PubMed] [Google Scholar]
Hauschild 2014 {published data only}
- Hauschild A, Chen SC, Weichenthal M, Blum A, King HC, Goldsmith J, et al. To excise or not: impact of MelaFind on German dermatologists' decisions to biopsy atypical lesions. Journal der Deutschen Dermatologischen Gesellschaft 2014;12(7):606‐14. [ER4:17941085; PUBMED: 24944011] [DOI] [PubMed] [Google Scholar]
Maglogiannis 2015 {published data only}
- Maglogiannis I, Delibasis KK. Enhancing classification accuracy utilizing globules and dots features in digital dermoscopy. Computer Methods and Programs in Biomedicine 2015;118(2):124‐33. [ER4:25012309; PUBMED: 25540998] [DOI] [PubMed] [Google Scholar]
Malvehy 2014 {published data only}
- Malvehy J, Hauschild A, Curiel‐Lewandrowski C, Mohr P, Hofmann‐Wellenhof R, Motley R, et al. Clinical performance of the Nevisense system in cutaneous melanoma detection: An international, multicentre, prospective and blinded clinical trial on efficacy and safety. British Journal of Dermatology 2014;171(5):1099‐107. [PUBMED: 24841846] [DOI] [PMC free article] [PubMed] [Google Scholar]
Menzies 1996 {published data only}
- Menzies SW, Ingvar C, Crotty KA, McCarthy WH. Frequency and morphologic characteristics of invasive melanomas lacking specific surface microscopic features. Archives of Dermatology 1996;132(10):1178‐82. [ER4:21450627; PUBMED: 8859028] [PubMed] [Google Scholar]
Menzies 2005 {published data only}
- Menzies SW, Bischof L, Talbot H, Gutenev A, Avramidis M, Wong L, et al. The performance of SolarScan: an automated dermoscopy image analysis instrument for the diagnosis of primary melanoma.[Erratum appears in Arch Dermatol. 2006 May;142(5):558]. Archives of Dermatology 2005;141(11):1388‐96. [ER4:20569478; PUBMED: 16301386] [DOI] [PubMed] [Google Scholar]
Mohr 2013 {published data only}
- Mohr P, Birgersson U, Berking C, Henderson C, Trefzer U, Kemeny L, et al. Electrical impedance spectroscopy as a potential adjunct diagnostic tool for cutaneous melanoma. Skin Research and Technology 2013;19(2):75‐83. [PUBMED: 23350668] [DOI] [PubMed] [Google Scholar]
Mollersen 2015 {published data only}
- Mollersen K, Kirchesch H, Zortea M, Schopf TR, Hindberg K, Godtliebsen F. Computer‐aided decision support for melanoma detection applied on melanocytic and nonmelanocytic skin lesions: A comparison of two systems based on automatic analysis of dermoscopic images. BioMed Research International 2015;2015:579282. [ER4:25012302; PUBMED: 26693486] [DOI] [PMC free article] [PubMed] [Google Scholar]
Monheit 2011 {published data only}
- Monheit G, Cognetta AB, Ferris L, Rabinovitz H, Gross K, Martini M, et al. The performance of MelaFind: a prospective multicenter study. Archives of Dermatology 2011;147(2):188‐94. [ER4:15466015; PUBMED: 20956633] [DOI] [PubMed] [Google Scholar]
Piccolo 2002 {published data only}
- Piccolo D, Ferrari A, Peris K, Diadone R, Ruggeri B, Chimenti S. Dermoscopic diagnosis by a trained clinician vs. a clinician with minimal dermoscopy training vs. computer‐assisted diagnosis of 341 pigmented skin lesions: a comparative study. British Journal of Dermatology 2002;147(3):481‐6. [ER4:15466057; PUBMED: 12207587] [DOI] [PubMed] [Google Scholar]
Piccolo 2014 {published data only}
- Piccolo D, Crisman G, Schoinas S, Altamura D, Peris K. Computer‐automated ABCD versus dermatologists with different degrees of experience in dermoscopy. European Journal of Dermatology 2014;24(4):477‐81. [ER4:17941089; PUBMED: 24721784] [DOI] [PubMed] [Google Scholar]
Rubegni 2002a {published data only}
- Rubegni P, Cevenini G, Burroni M, Perotti R, Dell'Eva G, Sbano P, et al. Automated diagnosis of pigmented skin lesions. International Journal of Cancer 2002;101(6):576‐80. [ER4:20569489; PUBMED: 12237900] [DOI] [PubMed] [Google Scholar]
Seidenari 1998 {published data only}
- Seidenari S, Pellacani G, Pepe P. Digital videomicroscopy improves diagnostic accuracy for melanoma. Journal of the American Academy of Dermatology 1998;39(2 Pt 1):175‐81. [ER4:15466116; PUBMED: 9704824] [DOI] [PubMed] [Google Scholar]
Seidenari 1999 {published data only}
- Seidenari S, Pellacani G, Giannetti A. Digital videomicroscopy and image analysis with automatic classification for detection of thin melanomas. Melanoma Research 1999;9(2):163‐71. [ER4:17940983; PUBMED: 10380939] [DOI] [PubMed] [Google Scholar]
Serrao 2006 {published data only}
- Serrao VV, Baptista J, Paris F, Lopes LC, Fidalgo A, Ferreira A. Digital dermoscopy. Review of 652 lesions analysed by the DANAOS system. Skin Cancer 2006;21(4):185‐98. [ER4:18375096] [Google Scholar]
Sgouros 2014 {published data only}
- Sgouros D, Lallas A, Julian Y, Rigopoulos D, Zalaudek I, Longo C, et al. Assessment of SIAscopy in the triage of suspicious skin tumours. Skin Research and Technology 2014;20(4):440‐4. [ER4:17941094; PUBMED: 24517201] [DOI] [PubMed] [Google Scholar]
Stanganelli 2005 {published data only}
- Stanganelli I, Brucale A, Calori L, Gori R, Lovato A, Magi S, et al. Computer‐aided diagnosis of melanocytic lesions. Anticancer Research 2005;25(6C):4577‐82. [ER4:15466126; PUBMED: 16334145] [PubMed] [Google Scholar]
Terstappen 2013 {published data only}
- Terstappen K, Suurkula M, Hallberg H, Ericson MB, Wennberg AM. Poor correlation between spectrophotometric intracutaneous analysis and histopathology in melanoma and nonmelanoma lesions [Erratum appears in J Biomed Opt. 2013 Jun;18(6):069804]. Journal of Biomedical Optics 2013;18(6):061223. [ER4:15466138; PUBMED: 23296145] [DOI] [PubMed] [Google Scholar]
Tomatis 2003 {published data only}
- Tomatis S, Bono A, Bartoli C, Carrara M, Lualdi M, Tragni G, et al. Automated melanoma detection: multispectral imaging and neural network approach for classification. Medical Physics 2003;30(2):212‐21. [ER4:18375057; PUBMED: 12607839] [DOI] [PubMed] [Google Scholar]
Tomatis 2005 {published data only}
- Tomatis S, Carrara M, Bono A, Bartoli C, Lualdi M, Tragni G, et al. Automated melanoma detection with a novel multispectral imaging system: results of a prospective study. Physics in Medicine and Biology 2005;50(8):1675‐87. [ER4:17941003; PUBMED: 15815089] [DOI] [PubMed] [Google Scholar]
Walter 2012 {published data only}
- Walter FM, Morris HC, Humphrys E, Hall PN, Prevost AT, Burrows N, et al. Effect of adding a diagnostic aid to best practice to manage suspicious pigmented lesions in primary care: randomised controlled trial. BMJ 2012;345:e4110. [ER4:15466154; PUBMED: 22763392] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wells 2012 {published data only}
- Wells R, Gutkowicz‐Krusin D, Veledar E, Toledano A, Chen SC. Comparison of diagnostic and management sensitivity to melanoma between dermatologists and MelaFind: a pilot study. Archives of Dermatology 2012;148(9):1083‐4. [ER4:15466163; PUBMED: 22986873] [DOI] [PubMed] [Google Scholar]
Winkelmann 2016 {published data only}
- Winkelmann RR, Farberg AS, Tucker N, White R, Rigel DS. Enhancement of international dermatologists' pigmented skin lesion biopsy decisions following dermoscopy with subsequent integration of multispectral digital skin lesion analysis. Journal of Clinical and Aesthetic Dermatology 2016;9(7):53‐5. [ER4:25701735; PUBMED: 27672411] [PMC free article] [PubMed] [Google Scholar]
Wollina 2007 {published data only}
- Wollina U, Burroni M, Torricelli R, Gilardi S, Dell'Eva G, Helm C, et al. Digital dermoscopy in clinical practise: a three‐centre analysis. Skin Research and Technology 2007;13(2):133‐42. [ER4:17941010; PUBMED: 17374053] [DOI] [PubMed] [Google Scholar]
References to studies excluded from this review
Abbas 2010 {published data only}
- Abbas Q, Garcia IF, Rashid M. Automatic skin tumour border detection for digital dermoscopy using a new digital image analysis scheme. British Journal of Biomedical Science 2010;67(4):177‐83. [DOI] [PubMed] [Google Scholar]
Abbas 2011a {published data only}
- Abbas Q, Celebi ME, Fondon Garcia I, Rashid M. Lesion border detection in dermoscopy images using dynamic programming. Skin Research and Technology 2011;17(1):91‐100. [PUBMED: 21226876] [DOI] [PubMed] [Google Scholar]
Abbas 2011b {published data only}
- Abbas Q, Fondon I, Rashid M. Unsupervised skin lesions border detection via two‐dimensional image analysis. Computer Methods and Programs in Biomedicine 2011;104(3):e1‐15. [DOI] [PubMed] [Google Scholar]
Abbas 2012 {published data only}
- Abbas Q, Celebi ME, Fondon I. Computer‐aided pattern classification system for dermoscopy images. Skin Research and Technology 2012;18(3):278‐89. [DOI] [PubMed] [Google Scholar]
Abbas 2013a {published data only}
- Abbas Q, Garcia IF, Emre Celebi M, Ahmad W, Mushtaq Q. Unified approach for lesion border detection based on mixture modeling and local entropy thresholding. Skin Research and Technology 2013;19(3):314‐9. [DOI] [PubMed] [Google Scholar]
Abbas 2013b {published data only}
- Abbas Q, Garcia IF, Emre Celebi M, Ahmad W, Mushtaq Q. A perceptually oriented method for contrast enhancement and segmentation of dermoscopy images. Skin Research and Technology 2013;19(1):e490‐7. [DOI] [PubMed] [Google Scholar]
Abuzaghleh 2015 {published data only}
- Abuzaghleh O, Barkana BD, Faezipour M. Noninvasive real‐time automated skin lesion analysis system for melanoma early detection and prevention. IEEE Journal of Translational Engineering in Health and Medicine 2015;3:2900310. [PUBMED: 27170906] [DOI] [PMC free article] [PubMed] [Google Scholar]
Afonso 2012 {published data only}
- Afonso A, Silveira M. Hair detection in dermoscopic images using percolation. IEEE Engineering in Medicine and Biology Magazine 2012;2012:4378‐81. [PUBMED: 23366897] [DOI] [PubMed] [Google Scholar]
Alfed 2015 {published data only}
Ali 2012 {published data only}
- Ali ARA, Deserno TM. A systematic review of automated melanoma detection in dermatoscopic images and its ground truth data. SPIE. Medical Imaging 2012: Image Perception, Observer Performance, and Technology Assessment. 29 February 2012; Vol. 8318. [DOI: 10.1117/12.912389] [DOI]
Altamura 2008 {published data only}
- Altamura D, Avramidis M, Menzies SW. Assessment of the optimal interval for and sensitivity of short‐term sequential digital dermoscopy monitoring for the diagnosis of melanoma. Archives of Dermatology 2008;144(4):502‐6. [DOI] [PubMed] [Google Scholar]
Andreassi 1999 {published data only}
- Andreassi L, Perotti R, Rubegni P, Burroni M, Cevenini G, Biagioli M, et al. Digital dermoscopy analysis for the differentiation of atypical nevi and early melanoma: a new quantitative semiology. Archives of Dermatology 1999;135(12):1459‐65. [DOI] [PubMed] [Google Scholar]
Armengol 2011 {published data only}
- Armengol E. Classification of melanomas in situ using knowledge discovery with explained case‐based reasoning. Artificial Intelligence in Medicine 2011;51(2):93‐105. [DOI] [PubMed] [Google Scholar]
Arroyo 2011 {published data only}
- Arroyo JL, Zapirain BG, Zorrilla AM. Blue‐white veil and dark‐red patch of pigment pattern recognition in dermoscopic images using machine‐learning techniques. IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), Bilbao. 2011:196‐201. [DOI: 10.1109/ISSPIT.2011.6151559] [DOI]
Ballerini 2012 {published data only}
- Ballerini L, Fisher RB, Aldridge B, Rees J. Non‐melanoma skin lesion classification using colour image data in a hierarchical K‐NN classifier. 9th IEEE International Symposium on Biomedical Imaging (ISBI), Barcelona. 2012:358‐61. [DOI: 10.1109/ISBI.2012.6235558] [DOI]
Barata 2012a {published data only}
- Barata C, Marques JS, Rozeira J. A system for the detection of pigment network in dermoscopy images using directional filters. IEEE Transactions on Biomedical Engineering 2012;59(10):2744‐54. [DOI] [PubMed] [Google Scholar]
Barata 2012b {published data only}
- Barata C, Marques JS, Rozeira J. A system for the automatic detection of pigment network. 9th IEEE International Symposium on Biomedical Imaging (ISBI), Barcelona. 2012:1651‐4. [DOI: 10.1109/ISBI.2012.6235894] [DOI]
Barata 2013 {published data only}
- Barata C, Marques JS, Rozeira J. Evaluation of color based keypoints and features for the classification of melanomas using the bag‐of‐features model. Advances in Visual Computing. ISVC. Lecture Notes in Computer Science. Springer, Berlin, Heidelberg, 2013; Vol. 8033:40‐9. [DOI: 10.1007/978-3-642-41914-0_5] [DOI]
Barata 2015a {published data only}
- Barata C, Celebi ME, Marques JS. Improving dermoscopy image classification using color constancy. IEEE Journal of Biomedical and Health Informatics 2015;19(3):1146‐52. [DOI] [PubMed] [Google Scholar]
Barata 2015b {published data only}
Barata 2015c {published data only}
Binder 2000 {published data only}
- Binder M, Kittler H, Dreiseitl S, Ganster H, Wolff K, Pehamberger H. Computer‐aided epiluminescence microscopy of pigmented skin lesions: the value of clinical data for the classification process. Melanoma Research 2000;10(6):556‐61. [DOI] [PubMed] [Google Scholar]
Bjerring 2001 {published data only}
- Bjerring P, Obitz ER, Cotton S. In vivo spectrophotometric evaluation of the skin tumours using a new skin chromophore imaging system SIAscope. Melanoma Research 2001;11:S180. [Google Scholar]
Blum 2004b {published data only}
- Blum A, Hofmann‐Wellenhof R, Luedtke H, Ellwanger U, Steins A, Roehm S, et al. Value of the clinical history for different users of dermoscopy compared with results of digital image analysis. Journal of the European Academy of Dermatology and Venereology : JEADV 2004;18(6):665‐9. [ER4:15465865; PUBMED: 15482291] [DOI] [PubMed] [Google Scholar]
Boden 2013 {published data only}
- Boden I, Nystrom J, Lundskog B, Zazo V, Geladi P, Lindholm‐Sethson B, et al. Non‐invasive identification of melanoma with near‐infrared and skin impedance spectroscopy. Skin Research and Technology 2013;19(1):e473‐8. [DOI] [PubMed] [Google Scholar]
Bono 1999 {published data only}
- Bono A, Tomatis S, Bartoli C, Tragni G, Radaelli G, Maurichi A, et al. The ABCD system of melanoma detection: a spectrophotometric analysis of the Asymmetry, Border, Color, and Dimension. Cancer 1999;85(1):72‐7. [DOI] [PubMed] [Google Scholar]
Borlu 2008 {published data only}
- Borlu M, Yüksel ME. Development of an image processing system for automatic melanoma diagnosis from dermoscopic images: preliminary study. Turkish Journal of Dermatology 2008;2(4):111‐5. [Google Scholar]
Brown 2000 {published data only}
- Brown N. Exploration of diagnostic techniques for malignant melanoma: an integrative review. Clinical Excellence for Nurse Practitioners 2000;4(5):263‐71. [PubMed] [Google Scholar]
Carrara 2007 {published data only}
- Carrara M, Bono A, Bartoli C, Colombo A, Lualdi M, Moglia D, et al. Multispectral imaging and artificial neural network: mimicking the management decision of the clinician facing pigmented skin lesions. Physics in Medicine and Biology 2007;52(9):2599‐613. [ER4:20756747; PUBMED: 17440255] [DOI] [PubMed] [Google Scholar]
Celebi 2008 {published data only}
- Celebi ME, Iyatomi H, Stoecker WV, Moss RH, Rabinovitz HS, Argenziano G, et al. Automatic detection of blue‐white veil and related structures in dermoscopy images. Computerized Medical Imaging and Graphics 2008;32(8):670‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chen 2003 {published data only}
- Chen J, Stanley RJ, Moss RH, Stoecker W. Colour analysis of skin lesion regions for melanoma discrimination in clinical images. Skin Research and Technology 2003;9(2):94‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cheng 2012 {published data only}
- Cheng B, Stanley RJ, Stoecker WV, Hinton K. Automatic telangiectasia analysis in dermoscopy images using adaptive critic design. Skin Research and Technology 2012;18(4):389‐96. [DOI] [PubMed] [Google Scholar]
Cheng 2013 {published data only}
- Cheng B, Stanley RJ, Stoecker WV, Stricklin SM, Hinton KA, Nguyen TK, et al. Analysis of clinical and dermoscopic features for basal cell carcinoma neural network classification. Skin Research and Technology 2013;19(1):e217‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Christensen 2010 {published data only}
- Christensen JH, Soerensen MB, Linghui Z, Chen S, Jensen MO. Pre‐diagnostic digital imaging prediction model to discriminate between malignant melanoma and benign pigmented skin lesion. Skin Research and Technology 2010;16(1):98‐108. [DOI] [PubMed] [Google Scholar]
Claridge 1992 {published data only}
- Claridge E, Hall PN, Keefe M, Allen JP. Shape analysis for classification of malignant melanoma. Journal of Biomedical Engineering 1992;14(3):229‐34. [DOI] [PubMed] [Google Scholar]
Cukras 2013 {published data only}
- Cukras AR. On the comparison of diagnosis and management of melanoma between dermatologists and MelaFind. JAMA Dermatology 2013;149(5):622‐3. [DOI] [PubMed] [Google Scholar]
Day 2001 {published data only}
- Day GR, Barbour RH. Automated skin lesion screening ‐ A new approach. Melanoma Research 2001;11(1):31‐5. [DOI] [PubMed] [Google Scholar]
Debeir 1999 {published data only}
- Debeir O, Decaestecker C, Pasteels JL, Salmon I, Kiss R, Ham P. Computer‐assisted analysis of epiluminescence microscopy images of pigmented skin lesions. Cytometry 1999;37(4):255‐66. [PubMed] [Google Scholar]
Di 2010 {published data only}
- Leo G, Paolillo A, Sommella P, Fabbrocini G. Automatic diagnosis of melanoma: a software system based on the 7‐point check‐list. 43rd Hawaii International Conference on System Sciences (HICSS). IEEE, 2010:2319‐28. [DOI: 10.1109/HICSS.2010.76] [DOI]
Ding 2015 {published data only}
- Ding Y, John NW, Smith L, Sun J, Smith M. Combination of 3D skin surface texture features and 2D ABCD features for improved melanoma diagnosis. Medical & Biological Engineering & Computing 2015;53(10):961‐74. [DOI] [PubMed] [Google Scholar]
Dreiseitl 2005 {published data only}
- Dreiseitl S, Binder M. Do physicians value decision support? A look at the effect of decision support systems on physician opinion. Artificial Intelligence in Medicine 2005;33(1):25‐30. [DOI] [PubMed] [Google Scholar]
Durg 1993 {published data only}
- Durg A, Stoecker WV, Cookson JP, Umbaugh SE, Moss RH. Identification of variegated coloring in skin tumors: Neural network vs. rule‐based induction methods. IEEE Engineering in Medicine and Biology Magazine 1993;12(3):71‐4. [Google Scholar]
Elbaum 2001 {published data only}
- Elbaum M, Kopf AW, Rabinovitz HS, Langley RG, Kamino H, Mihm MC Jr, et al. Automatic differentiation of melanoma from melanocytic nevi with multispectral digital dermoscopy: a feasibility study. Journal of the American Academy of Dermatology 2001;44(2):207‐18. [DOI] [PubMed] [Google Scholar]
Emery 2010 {published data only}
- Emery JD, Hunter J, Hall PN, Watson AJ, Moncrieff M, Walter FM. Accuracy of SIAscopy for pigmented skin lesions encountered in primary care: development and validation of a new diagnostic algorithm. BMC Dermatology 2010;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Engin 2016 {published data only}
- Engin B, Kecici AS, Yilmaz S, Kutlubay Z, Serdaroglu S, Tuzun Y. Infrared imaging in diagnosis of dysplastic nevi and malignant melanoma. Turkiye Klinikleri Journal of Medical Sciences 2016;36(1):14‐21. [Google Scholar]
Ercal 1994 {published data only}
- Ercal F, Chawla A, Stoecker WV, Lee HC, Moss RH. Neural network diagnosis of malignant melanoma from color images. IEEE Transactions on Biomedical Engineering 1994;41(9):837‐45. [DOI] [PubMed] [Google Scholar]
Faal 2013 {published data only}
- Faal M, Miran Baygi MH, Kabir E. Improving the diagnostic accuracy of dysplastic and melanoma lesions using the decision template combination method. Skin Research and Technology 2013;19(1):e113‐22. [DOI] [PubMed] [Google Scholar]
Farina 2000 {published data only}
- Farina B, Bartoli C, Bono A, Colombo A, Lualdi M, Tragni G, et al. Multispectral imaging approach in the diagnosis of cutaneous melanoma: potentiality and limits. Physics in Medicine and Biology 2000;45(5):1243‐54. [DOI] [PubMed] [Google Scholar]
Ferris 2016 {published data only}
- Ferris LK, Satyanarayanan M. Reply to: "Computer‐aided classification of melanocytic lesions using dermoscopic images: Low reported accuracy for reader study on melanomas with low melanoma in situ to invasive melanoma ratio". Journal of the American Academy of Dermatology 2016;75(3):e121. [DOI] [PubMed] [Google Scholar]
Fidalgo 2003 {published data only}
- Fidalgo A, Caldas Lopes L, Macedo Ferreira A. Digital dermatoscopy: One‐year experience with the DANAOS system. Skin Cancer 2003;18(4):211‐8. [Google Scholar]
Fikrle 2007 {published data only}
- Fikrle T, Pizinger K. Digital computer analysis of dermatoscopical images of 260 melanocytic skin lesions; perimeter/area ratio for the differentiation between malignant melanomas and melanocytic nevi. Journal of the European Academy of Dermatology and Venereology 2007;21(1):48‐55. [DOI] [PubMed] [Google Scholar]
Fikrle 2013 {published data only}
- Fikrle T, Pizinger K, Szakos H, Panznerova P, Divisova B, Pavel S. Digital dermatoscopic follow‐up of 1027 melanocytic lesions in 121 patients at risk of malignant melanoma. Journal of the European Academy of Dermatology and Venereology 2013;27(2):180‐6. [DOI] [PubMed] [Google Scholar]
Fruhauf 2012 {published data only}
- Fruhauf J, Leinweber B, Fink‐Puches R, Ahlgrimm‐Siess V, Richtig E, Wolf IH, et al. Patient acceptance and diagnostic utility of automated digital image analysis of pigmented skin lesions. Journal of the European Academy of Dermatology and Venereology 2012;26(3):368‐72. [DOI] [PubMed] [Google Scholar]
Fueyo‐Casado 2009 {published data only}
- Fueyo‐Casado A, Vázquez‐López F, Sanchez‐Martin J, Garcia‐Garcia B, Pérez‐Oliva N. Evaluation of a program for the automatic dermoscopic diagnosis of melanoma in a general dermatology setting. Dermatologic Surgery 2009;35(2):257‐9; discussion 260‐2. [DOI] [PubMed] [Google Scholar]
Ganster 2001 {published data only}
- Ganster H, Pinz P, Rohrer R, Wildling E, Binder M, Kittler H. Automated melanoma recognition. IEEE Transactions on Medical Imaging 2001;20(3):233‐9. [DOI] [PubMed] [Google Scholar]
García 2014 {published data only}
- García Arroyo JL, García Zapirain B. Detection of pigment network in dermoscopy images using supervised machine learning and structural analysis. Computers in Biology and Medicine 2014;44:144‐57. [DOI] [PubMed] [Google Scholar]
Garcia‐Uribe 2004 {published data only}
- Garcia‐Uribe A, Kehtarnavaz N, Marquez G, Prieto V, Duvic M, Wang LV. Skin cancer detection by spectroscopic oblique‐incidence reflectometry: classification and physiological origins. Applied Optics 2004;43(13):2643‐50. [DOI] [PubMed] [Google Scholar]
Garcia‐Uribe 2010 {published data only}
- Garcia‐Uribe A, Zou J, Chang TH, Duvic M, Prieto V, Wang LV. Oblique‐incidence spatially resolved diffuse reflectance spectroscopic diagnosis of skin cancer. SPIE. Optical Diagnostics and Sensing X: Toward Point‐of‐Care Diagnostics. 26 February 2010; Vol. 7572:75720L. [DOI: 10.1117/12.842781] [DOI]
Garnavi 2012 {published data only}
- Garnavi R, Aldeen M, Bailey J. Computer‐aided diagnosis of melanoma using border and wavelet‐based texture analysis. IEEE Transactions on Information Technology in Biomedicine 2012;16(6):1239‐52. [DOI] [PubMed] [Google Scholar]
Gerger 2003 {published data only}
- Gerger A, Pompl R, Smolle J, Stolz W. Automated epiluminescence microscopy‐‐tissue counter analysis using CART and 1‐NN in the diagnosis of melanoma. Skin Research and Technology 2003;9(2):105‐10. [ER4:15465930; PUBMED: 12709127] [DOI] [PubMed] [Google Scholar]
Glotsos 2015 {published data only}
- Glotsos D, Kostopoulos S, Lalissidou S, Sidiropoulos K, Asvestas P, Konstandinou C, et al. Design of a decision support system, trained on GPU, for assisting melanoma diagnosis in dermatoscopy images. 4th International Conference on Mathematical Modeling in Physical Sciences. 2015:012079. [DOI: 10.1088/1742-6596/633/1/012079] [DOI]
Gniadecka 2004 {published data only}
- Gniadecka M, Philipsen PA, Sigurdsson S, Wessel S, Nielsen OF, Christensen DH, et al. Melanoma diagnosis by Raman spectroscopy and neural networks: structure alterations in proteins and lipids in intact cancer tissue. Journal of Investigative Dermatology 2004;122(2):443‐9. [DOI] [PubMed] [Google Scholar]
Govindan 2007 {published data only}
- Govindan K, Smith J, Knowles L, Harvey A, Townsend P, Kenealy J. Assessment of nurse‐led screening of pigmented lesions using SIAscope. Journal of Plastic, Reconstructive & Aesthetic Surgery 2007;60(6):639‐45. [DOI] [PubMed] [Google Scholar]
Green 1991 {published data only}
- Green A, Martin N, McKenzie G, Pfitzner J, Quintarelli F, Thomas BW, et al. Computer image analysis of pigmented skin lesions. Melanoma Research 1991;1(4):231‐236. [PUBMED: 1823631] [DOI] [PubMed] [Google Scholar]
Green 1994 {published data only}
- Green A, Martin N, Pfitzner J, O'Rourke M, Knight N. Computer image analysis in the diagnosis of melanoma. Journal of the American Academy of Dermatology 1994;31(6):958‐64. [DOI] [PubMed] [Google Scholar]
Guerra‐Rosas 2015 {published data only}
- Guerra‐Rosas E, Alvarez‐Borrego J. Methodology for diagnosing of skin cancer on images of dermatologic spots by spectral analysis. Biomedical Optics Express 2015;6(10):3876‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Guillod 1996 {published data only}
- Guillod JF, Schmid Ph, Fischer S, Salomon D, Saurat JH. Detection and classification of pigmented skin lesions by dermatoscopic digital image processing. Dermatology 1996;193(2):169. [Google Scholar]
Gutkowicz‐Krusin 2000 {published data only}
- Gutkowicz‐Krusin D, Elbaum M, Jacobs A, Keem S, Kopf AW, Kamino H, et al. Precision of automatic measurements of pigmented skin lesion parameters with a MelaFind(™) multispectral digital dermatoscope. Melanoma Research 2000;10(6):563‐70. [DOI] [PubMed] [Google Scholar]
Hacioglu 2013 {published data only}
- Hacioglu S, Saricaoglu H, Baskan EB, Uner SI, Aydogan K, Tunali S. The value of spectrophotometric intracutaneous analysis in the noninvasive diagnosis of nonmelanoma skin cancers. Clinical and Experimental Dermatology 2013;38(5):464‐9. [DOI] [PubMed] [Google Scholar]
Haenssle 2004 {published data only}
- Haenssle HA, Vente C, Bertsch HP, Rupprecht R, Abuzahra F, Junghans V, et al. Results of a surveillance programme for patients at high risk of malignant melanoma using digital and conventional dermoscopy. European Journal of Cancer Prevention 2004;13(2):133‐8. [DOI] [PubMed] [Google Scholar]
Haenssle 2010 {published data only}
- Haenssle HA, Korpas B, Hansen‐Hagge C, Buhl T, Kaune KM, Johnsen S, et al. Selection of patients for long‐term surveillance with digital dermoscopy by assessment of melanoma risk factors. Archives of Dermatology 2010;146(3):257‐64. [DOI] [PubMed] [Google Scholar]
Haniffa 2007 {published data only}
- Haniffa MA, Lloyd JJ, Lawrence CM. The use of a spectrophotometric intracutaneous analysis device in the real‐time diagnosis of melanoma in the setting of a melanoma screening clinic. British Journal of Dermatology 2007;156(6):1350‐2. [DOI] [PubMed] [Google Scholar]
Hintz‐Madsen 2001 {published data only}
- Hintz‐Madsen M, Hansen LK, Larsen J, Drzewiecki KT. A probabilistic neural network framework for detection of malignant melanoma. In: Naguib RNG, Sherbet GV editor(s). Artificial Neural Networks in Cancer Diagnosis, Prognosis and Patient Management. CRC Press, 2001:141‐83. [Google Scholar]
Hoffmann 2003 {published data only}
- Hoffmann K, Gambichler T, Rick A, Kreutz M, Anschuetz M, Grunendick T, et al. Diagnostic and neural analysis of skin cancer (DANAOS). A multicentre study for collection and computer‐assisted analysis of data from pigmented skin lesions using digital dermoscopy. British Journal of Dermatology 2003;149(4):801‐9. [DOI] [PubMed] [Google Scholar]
Horsch 1997 {published data only}
- Horsch A, Stolz W, Neiss A, Abmayr W, Pompl R, Bernklau A, et al. Improving early recognition of malignant melanomas by digital image analysis in dermatoscopy. Studies in Health Technology and Informatics 1997;43(Pt B):531‐5. [PubMed] [Google Scholar]
Huang 1996 {published data only}
- Huang CL, Wasti Q, Marghoob AA, Kopf AW, David M, Rao BK, et al. Border irregularity: Atypical moles versus melanoma. European Journal of Dermatology 1996;6(4):270‐3. [Google Scholar]
Ikuma 2013 {published data only}
- Ikuma Y, Iyatomi H. Production of the grounds for melanoma classification using adaptive fuzzy inference neural network. IEEE International Conference on Systems, Man, and Cybernetics (SMC). 2013:2570‐5. [DOI: 10.1109/SMC.2013.439] [DOI]
Isasi 2011 {published data only}
- Isasi AG, Zapirain BG, Zorrilla AM. Melanomas non‐invasive diagnosis application based on the ABCD rule and pattern recognition image processing algorithms. Computers in Biology and Medicine 2011;41(9):742‐55. [DOI] [PubMed] [Google Scholar]
Iyatomi 2006 {published data only}
- Iyatomi H, Oka H, Saito M, Miyake A, Kimoto M, Yamagami J, et al. Quantitative assessment of tumour extraction from dermoscopy images and evaluation of computer‐based extraction methods for an automatic melanoma diagnostic system. Melanoma Research 2006;16(2):183‐90. [DOI] [PubMed] [Google Scholar]
Iyatomi 2008a {published data only}
Iyatomi 2008b {published data only}
- Iyatomi H, Oka H, Celebi ME, Hashimoto M, Hagiwara M, Tanaka M, et al. An improved Internet‐based melanoma screening system with dermatologist‐like tumor area extraction algorithm. Computerized Medical Imaging and Graphics 2008;32(7):566‐79. [DOI] [PubMed] [Google Scholar]
Iyatomi 2008c {published data only}
- Iyatomi H, Oka H, Celebi ME, Ogawa K, Argenziano G, Soyer HP, et al. Computer‐based classification of dermoscopy images of melanocytic lesions on acral volar skin. Journal of Investigative Dermatology 2008;128(8):2049‐54. [DOI] [PubMed] [Google Scholar]
Iyatomi 2010a {published data only}
- Iyatomi H, Celebi ME, Schaefer G, Tanaka M. Automated color normalization for dermoscopy images. 17th IEEE International Conference on Image Processing (ICIP). 2010:4357‐60. [DOI: 10.1109/ICIP.2010.5652370] [DOI]
Iyatomi 2010b {published data only}
Iyatomi 2011 {published data only}
- Iyatomi H, Celebi ME, Schaefer G, Tanaka M. Automated color calibration method for dermoscopy images. Computerized Medical Imaging and Graphics 2011;35(2):89‐98. [DOI] [PubMed] [Google Scholar]
Jain 2015 {published data only}
- Jain S, Jagtap V, Pise N. Computer aided melanoma skin cancer detection using image processing. Procedia Computer Science 2015;48:735‐40. [DOI: 10.1016/j.procs.2015.04.209] [DOI] [Google Scholar]
Jakovels 2013 {published data only}
- Jakovels D, Lihacova I, Kuzmina I, Spigulis J. Application of principal component analysis to multispectral imaging data for evaluation of pigmented skin lesions. SPIE. Biophotonics ‐ Riga 2013. 18 November 2013; Vol. 9032:903204. [DOI: 10.1117/12.2044383; ER4:25233597] [DOI]
Jamora 2003 {published data only}
- Jamora MJ, Wainwright BD, Meehan SA, Bystryn JC. Improved identification of potentially dangerous pigmented skin lesions by computerized image analysis. Archives of Dermatology 2003;139(2):195‐8. [DOI] [PubMed] [Google Scholar]
Jaworek‐Korjakowska 2016a {published data only}
- Jaworek‐Korjakowska J. Computer‐aided diagnosis of micro‐malignant melanoma lesions applying support vector machines. BioMed Research International 2016;2016:4381972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jaworek‐Korjakowska 2016b {published data only}
- Jaworek‐Korjakowska J, Kleczek P. Automatic classification of specific melanocytic lesions using artificial intelligence. BioMed Research International 2016;2016:8934242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jeddi 2016 {published data only}
- Jeddi FR, Arabfard M, Arabkermany Z, Gilasi H. The diagnostic value of skin disease diagnosis expert system. Acta Informatica Medica 2016;24(1):30‐3. [PUBMED: 27046943] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kahofer 2002 {published data only}
- Kahofer P, Hofmann‐Wellenhof R, Smolle J. Tissue counter analysis of dermatoscopic images of melanocytic skin tumours: preliminary findings. Melanoma Research 2002;12(1):71‐5. [DOI] [PubMed] [Google Scholar]
Kaur 2015 {published data only}
- Kaur R, Albano PP, Cole JG, Hagerty J, LeAnder RW, Moss RH, et al. Real‐time supervised detection of pink areas in dermoscopic images of melanoma: importance of color shades, texture and location. Skin Research and Technology 2015;21(4):466‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Korotkov 2012 {published data only}
- Korotkov K, Garcia R. Computerized analysis of pigmented skin lesions: a review. Artificial Intelligence in Medicine 2012;56(2):69‐90. [DOI] [PubMed] [Google Scholar]
Kuzmina 2011 {published data only}
- Kuzmina I, Diebele I, Valeine L, Jakovels D, Kempele A, Kapostinsh J, et al. Multi‐spectral imaging analysis of pigmented and vascular skin lesions: results of a clinical trial. SPIE. Photonic Therapeutics and Diagnostics VII. 3 February 2011; Vol. 7883:788312.
Landau 1999 {published data only}
- Landau M, Matz H, Tur E, Dvir M, Brenner S. Computerized system to enhance the clinical diagnosis of pigmented cutaneous malignancies. International Journal of Dermatology 1999;38(6):443‐6. [DOI] [PubMed] [Google Scholar]
LeAnder 2010 {published data only}
- LeAnder R, Chindam P, Das M, Umbaugh SE. Differentiation of melanoma from benign mimics using the relative‐color method. Skin Research and Technology 2010;16(3):297‐304. [DOI] [PubMed] [Google Scholar]
Lefevre 2000 {published data only}
- Lefevre E, Colot O, Vannoorenberghe P, Brucq D. Knowledge modeling methods in the framework of evidence theory: an experimental comparison for melanoma detection. IEEE International Conference on Systems, Man, and Cybernetics. 2000; Vol. 4:2806‐11. [DOI: 10.1109/ICSMC.2000.884422] [DOI]
Lihacova 2013 {published data only}
- Lihacova I, Derjabo A, Spigulis J. A multispectral imaging approach for diagnostics of skin pathologies. In: Deckert V, Ramanujam N editor(s). SPIE. Clinical and Biomedical Spectroscopy and Imaging III. Vol. 8798, Optical Society of America, 2013:87980X. [Google Scholar]
Liu 2012 {published data only}
- Liu Z, Sun J, Smith L, Smith M, Warr R. Distribution quantification on dermoscopy images for computer‐assisted diagnosis of cutaneous melanomas. Medical & Biological Engineering & Computing 2012;50(5):503‐13. [DOI] [PubMed] [Google Scholar]
Machado 2015 {published data only}
- Machado M, Pereira J, Fonseca‐Pinto R. Classification of reticular pattern and streaks in dermoscopic images based on texture analysis. Journal of Medical Imaging 2015;2(4):044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Maglogiannis 2004 {published data only}
- Maglogiannis IG, Zafiropoulos EP. Characterization of digital medical images utilizing support vector machines. BMC Medical Informatics and Decision Making 2004;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Maglogiannis 2006 {published data only}
- Maglogiannis I, Zafiropoulos E, Kyranoudis C. Intelligent segmentation and classification of pigmented skin lesions in dermatological images. Advances in Artificial Intelligence, 4th Helenic Conference on AI, SETN 2006 May 18‐20; Heraklion, Crete, Greece. Springer, 2006:214‐23.
Manousaki 2006 {published data only}
- Manousaki AG, Manios AG, EI Tsompanaki, Panayiotides JG, Tsiftsis DD, Kostaki AK, et al. A simple digital image processing system to aid in melanoma diagnosis in an everyday melanocytic skin lesion unit. A preliminary report. International Journal of Dermatology 2006;45(4):8. [DOI: 10.1111/j.1365-4632.2006.02726.x] [DOI] [PubMed] [Google Scholar]
Marchesini 1992 {published data only}
- Marchesini R, Cascinelli N, Brambilla M, Clemente C, Mascheroni L, Pignoli E, et al. In vivo spectrophotometric evaluation of neoplastic and non‐neoplastic skin pigmented lesions. II: discriminant analysis between nevus and melanoma. Photochemistry and Photobiology 1992;55(4):515‐22. [DOI] [PubMed] [Google Scholar]
Masood 2013 {published data only}
- Masood A, Al Jumaily AA, Hoshyar AN, Masood O. Automated segmentation of skin lesions: modified Fuzzy C mean thresholding based level set method. 16th International Multi Topic Conference (INMIC). IEEE, 2013:201‐6. [DOI: 10.1109/INMIC.2013.6731350] [DOI]
Menzies 1999 {published data only}
- Menzies SW. Automated epiluminescence microscopy: human vs machine in the diagnosis of melanoma. Archives of Dermatology 1999;135(12):1538‐40. [DOI] [PubMed] [Google Scholar]
Mete 2011 {published data only}
- Mete M, Kockara S, Aydin K. Fast density‐based lesion detection in dermoscopy images. Computerized Medical Imaging and Graphics 2011;35(2):128‐36. [DOI] [PubMed] [Google Scholar]
Mhaske 2013 {published data only}
- Mhaske HR, Phalke DA. Melanoma skin cancer detection and classification based on supervised and unsupervised learning. International Conference on Circuits, Controls and Communications (CCUBE). IEEE, 2013. [DOI: 10.1109/CCUBE.2013.6718539] [DOI]
Moncrieff 2002 {published data only}
- Moncrieff M, Cotton S, Claridge E, Hall P. Spectrophotometric intracutaneous analysis: a new technique for imaging pigmented skin lesions. British Journal of Dermatology 2002;146(3):448‐57. [DOI] [PubMed] [Google Scholar]
Morrow 2010 {published data only}
- Morrow T. MelaFind improves chances for accurate melanoma diagnosis. Managed Care 2010;19(3):54‐5. [PubMed] [Google Scholar]
Nagaoka 2012 {published data only}
- Nagaoka T, Nakamura A, Okutani H, Kiyohara Y, Sota T. A possible melanoma discrimination index based on hyperspectral data: a pilot study. Skin Research and Technology 2012;18(3):301‐10. [DOI] [PubMed] [Google Scholar]
Nagaoka 2013 {published data only}
- Nagaoka T, Nakamura A, Okutani H, Kiyohara Y, Koga H, Saida T, et al. Hyperspectroscopic screening of melanoma on acral volar skin. Skin Research and Technology 2013;19(1):e290‐6. [DOI] [PubMed] [Google Scholar]
Nagaoka 2015 {published data only}
- Nagaoka T, Kiyohara Y, Koga H, Nakamura A, Saida T, Sota T. Modification of a melanoma discrimination index derived from hyperspectral data: a clinical trial conducted in 2 centers between March 2011 and December 2013. Skin Research and Technology 2015;21(3):278‐83. [DOI] [PubMed] [Google Scholar]
Noroozi 2016 {published data only}
- Noroozi N, Zakerolhosseini A. Computer assisted diagnosis of basal cell carcinoma using Z‐transform features. Journal of Visual Communication and Image Representation 2016;40(Part A):128‐48. [DOI: 10.1016/j.jvcir.2016.06.014] [DOI] [Google Scholar]
Oka 2004a {published data only}
- Oka H, Hashimoto M, Iyatomi H, Argenziano G, Soyer HP, Tanaka M. Internet‐based program for automatic discrimination of dermoscopic images between melanomas and Clark naevi. British Journal of Dermatology 2004;150(5):1041. [DOI] [PubMed] [Google Scholar]
Oka 2004b {published data only}
- Oka H, Tanaka M, Kobayashi S, Argenziano G, Soyer HP, Nishikawa T. Linear discriminant analysis of dermoscopic parameters for the differentiation of early melanomas from Clark naevi. Melanoma Research 2004;14(2):131‐4. [DOI] [PubMed] [Google Scholar]
Oka 2006 {published data only}
- Oka H, Iyatomi H, Hashimoto M, Tanaka M. Reply to 'Digital dermoscopy analysis and internet‐based program for discrimination of pigmented skin lesion dermoscopic images'. British Journal of Dermatology 2006;154(3):570‐1; author reply 571‐2. [DOI] [PubMed] [Google Scholar]
Pellacani 2004a {published data only}
- Pellacani G, Grana C, Cucchiara R, Seidenari S. Automated extraction and description of dark areas in surface microscopy melanocytic lesion images. Dermatology 2004;208(1):21‐6. [DOI] [PubMed] [Google Scholar]
Pellacani 2004b {published data only}
- Pellacani G, Grana C, Seidenari S. Automated description of colours in polarized‐light surface microscopy images of melanocytic lesions. Melanoma Research 2004;14(2):125‐30. [DOI] [PubMed] [Google Scholar]
Pellacani 2006 {published data only}
- Pellacani G, Grana C, Seidenari S. Algorithmic reproduction of asymmetry and border cut‐off parameters according to the ABCD rule for dermoscopy. Journal of the European Academy of Dermatology and Venereology 2006;20(10):1214‐9. [DOI] [PubMed] [Google Scholar]
Perrinaud 2007 {published data only}
- Perrinaud A, Gaide O, French LE, Saurat JH, Marghoob AA, Braun RP. Can automated dermoscopy image analysis instruments provide added benefit for the dermatologist? A study comparing the results of three systems. British Journal of Dermatology 2007;157(5):926‐33. [DOI] [PubMed] [Google Scholar]
Pompl 2000 {published data only}
- Pompl R, Bunk W, Horsch A, Stolz W, Abmayr W, Brauer W, et al. MELDOQ: A system to support the early detection of malignant melanoma through digital image processing [MELDOQ: Ein System zur Unterstützung der Früherkennung des malignen Melanoms durch digitale Bildverarbeitung]. In: Horsch A, Lehmann T editor(s). Bildverarbeitung für die Medizin 2000. Springer, Berlin, Heidelberg, 2000:234‐8. [DOI: 10.1007/978-3-642-59757-2_44] [DOI] [Google Scholar]
Rajpara 2009 {published data only}
- Rajpara SM, Botello AP, Townend J, Ormerod AD. Systematic review of dermoscopy and digital dermoscopy/ artificial intelligence for the diagnosis of melanoma. British Journal of Dermatology 2009;161(3):591‐604. [DOI] [PubMed] [Google Scholar]
Rastgoo 2015 {published data only}
- Rastgoo M, Garcia R, Morel O, Marzani F. Automatic differentiation of melanoma from dysplastic nevi. Computerized Medical Imaging and Graphics 2015;43:44‐52. [DOI] [PubMed] [Google Scholar]
Rigel 2012 {published data only}
- Rigel DS, Roy M, Yoo J, Cockerell CJ, Robinson JK, White R. Impact of guidance from a computer‐assisted multispectral digital skin lesion analysis device on decision to biopsy lesions clinically suggestive of melanoma. Archives of Dermatology 2012;148(5):541‐3. [ER4:15466080; PUBMED: 22351788] [DOI] [PubMed] [Google Scholar]
Rosado 2003 {published data only}
- Rosado B, Menzies S, Harbauer A, Pehamberger H, Wolff K, Binder M, et al. Accuracy of computer diagnosis of melanoma: a quantitative meta‐analysis. Archives of Dermatology 2003;139(3):361‐7; discussion 366. [DOI] [PubMed] [Google Scholar]
Rubegni 2001a {published data only}
- Rubegni P, Cevenini G, Burroni M, Perotti R, Dell'Eua G, Andreassi L. Digital dermoscopy analysis of pigmented skin lesions: An important auxiliary for clinical decision and not for automatic diagnosis. Archives of Dermatology 2001;137(3):378. [Google Scholar]
Rubegni 2001b {published data only}
- Rubegni P, Ferrari A, Cevenini G, Piccolo D, Burroni M, Perotti R, et al. Differentiation between pigmented Spitz naevus and melanoma by digital dermoscopy and stepwise logistic discriminant analysis. Melanoma Research 2001;11(1):37‐44. [DOI] [PubMed] [Google Scholar]
Rubegni 2002b {published data only}
- Rubegni P, Burroni M, Cevenini G, Perotti R, Dell'Eva G, Barbini P, et al. Digital dermoscopy analysis and artificial neural network for the differentiation of clinically atypical pigmented skin lesions: a retrospective study. Journal of Investigative Dermatology 2002;119(2):471‐4. [DOI] [PubMed] [Google Scholar]
Rubegni 2005 {published data only}
- Rubegni P, Burroni M, Andreassi A, Fimiani M. The role of dermoscopy and digital dermoscopy analysis in the diagnosis of pigmented skin lesions. Archives of Dermatology 2005;141(11):1444‐6. [DOI] [PubMed] [Google Scholar]
Rubegni 2010 {published data only}
- Rubegni P, Cevenini G, Burroni M, Bono R, Sbano P, Biagioli M, et al. Objective follow‐up of atypical melanocytic skin lesions: a retrospective study. Archives of Dermatological Research 2010;302(7):551‐60. [DOI] [PubMed] [Google Scholar]
Rubegni 2013 {published data only}
- Rubegni P, Cevenini G, Nami N, Argenziano G, Saida T, Burroni M, et al. A simple scoring system for the diagnosis of palmo‐plantar pigmented skin lesions by digital dermoscopy analysis. Journal of the European Academy of Dermatology and Venereology 2013;27(3):e312‐9. [DOI] [PubMed] [Google Scholar]
Sadeghi 2013 {published data only}
- Sadeghi M, Lee TK, McLean D, Lui H, Atkins MS. Detection and analysis of irregular streaks in dermoscopic images of skin lesions. IEEE Transactions on Medical Imaging 2013;32(5):849‐61. [DOI] [PubMed] [Google Scholar]
Safi 2011 {published data only}
- Safi A, Castaneda V, Lasser T, Mateus DC, Navab N. Manifold learning for dimensionality reduction and clustering of skin spectroscopy data. SPIE. Medical Imaging 2011: Computer‐Aided Diagnosis. 2011; Vol. 7963:79631A. [DOI: 10.1117/12.877952] [DOI]
Salerni 2012 {published data only}
- Salerni G, Carrera C, Lovatto L, Marti‐Laborda RM, Isern G, Palou J, et al. Characterization of 1152 lesions excised over 10 years using total‐body photography and digital dermatoscopy in the surveillance of patients at high risk for melanoma. Journal of the American Academy of Dermatology 2012;67(5):836‐45. [DOI] [PubMed] [Google Scholar]
Sboner 2001 {published data only}
- Sboner A, Blanzieri E, Eccher C, Bauer P, Cristofolini M, Zumiani G, et al. A knowledge based system for early melanoma diagnosis support. pdfs.semanticscholar.org/9e12/21c5904bbd310c932414de841d3b126e5579.pdf (accessed prior to 1 May 2018).
Sboner 2003 {published data only}
- Sboner A, Eccher C, Blanzieri E, Bauer P, Cristofolini M, Zumiani G, et al. A multiple classifier system for early melanoma diagnosis. Artificial Intelligence in Medicine 2003;27(1):29‐44. [DOI] [PubMed] [Google Scholar]
Sboner 2004 {published data only}
- Sboner A, Bauer P, Zumiani G, Eccher C, Blanzieri E, Forti S, et al. Clinical validation of an automated system for supporting the early diagnosis of melanoma. Skin Research and Technology 2004;10(3):184‐92. [DOI] [PubMed] [Google Scholar]
Schindewolf 1993 {published data only}
- Schindewolf T, Stolz W, Albert R, Abmayer W, Harms H. Comparison of classification rates for conventional and dermatoscopic images of malignant and benign melanocytic lesions using computerized colour image analysis. European Journal of Dermatology 1993;3(4):299‐303. [Google Scholar]
Schindewolf 1994 {published data only}
- Schindewolf T, Schiffner R, Stolz W, Albert R, Abmayr W, Harms H. Evaluation of different image acquisition techniques for a computer vision system in the diagnosis of malignant melanoma. Journal of the American Academy of Dermatology 1994;31(1):33‐41. [DOI] [PubMed] [Google Scholar]
Schmid‐Saugeon 2003 {published data only}
- Schmid‐Saugeon P, Guillod J, Thiran JP. Towards a computer‐assisted diagnosis system for pigmented skin lesions. Computerized Medical Imaging and Graphics 2003;27(1):65‐78. [DOI] [PubMed] [Google Scholar]
Schumacher 2016 {published data only}
- Schumacher B, Mishra NK, Dusza SW, Halpern AC, Stoecker WV. Computer‐aided classification of melanocytic lesions using dermoscopic images: Low reported accuracy for reader study on melanomas with low melanoma in situ to invasive melanoma ratio. Journal of the American Academy of Dermatology 2016;75(3):e119‐20. [DOI] [PubMed] [Google Scholar]
Seidenari 1995 {published data only}
- Seidenari S, Burroni M, Dell'Eva G, Pepe P, Belletti B. Computerized evaluation of pigmented skin lesion images recorded by a videomicroscope: comparison between polarizing mode observation and oil/slide mode observation. Skin Research and Technology 1995;1(4):187‐91. [DOI] [PubMed] [Google Scholar]
Seidenari 2005 {published data only}
- Seidenari S, Pellacani G, Grana C. Pigment distribution in melanocytic lesion images: a digital parameter to be employed for computer‐assisted diagnosis. Skin Research and Technology 2005;11(4):236‐41. [DOI] [PubMed] [Google Scholar]
Seidenari 2007 {published data only}
- Seidenari S, Grana C, Pellacani G. Colour clusters for computer diagnosis of melanocytic lesions. Dermatology 2007;214(2):137‐43. [DOI] [PubMed] [Google Scholar]
Seidenari 2012 {published data only}
- Seidenari S, Ferrari C, Borsari S, Bassoli S, Cesinaro AM, Giusti F, et al. The dermoscopic variability of pigment network in melanoma in situ. Melanoma Research 2012;22(2):151‐7. [DOI] [PubMed] [Google Scholar]
Shakya 2012 {published data only}
- Shakya NM, LeAnder RW, Hinton KA, Stricklin SM, Rader RK, Hagerty J, et al. Discrimination of squamous cell carcinoma in situ from seborrheic keratosis by color analysis techniques requires information from scale, scale‐crust and surrounding areas in dermoscopy images. Computers in Biology and Medicine 2012;42(12):1165‐9. [DOI] [PubMed] [Google Scholar]
She 2007 {published data only}
- She Z, Liu Y, Damatoa A. Combination of features from skin pattern and ABCD analysis for lesion classification. Skin Research and Technology 2007;13(1):25‐33. [DOI] [PubMed] [Google Scholar]
She 2013 {published data only}
- She Z, Excell PS. Lesion classification using 3D skin surface tilt orientation. Skin Research and Technology 2013;19(1):e305‐11. [DOI] [PubMed] [Google Scholar]
Shimizu 2012 {published data only}
- Shimizu K, Iyatomi H, Norton KA, Celebi ME. Extension of automated melanoma screening for non‐melanocytic skin lesions. Mechatronics and Machine Vision in Practice (M2VIP), 19th International Conference. IEEE, 2012:16‐19. [Google Scholar]
Skrovseth 2010 {published data only}
- Skrovseth SO, Schopf TR, Thon K, Zortea M, Geilhufe M, Mollersen K, et al. A computer‐assisted diagnostic system for malignant melanomas. Applied Sciences in Biomedical and Communication Technologies (ISABEL), 3rd International Symposium 2010. IEEE, 2010. [DOI: ]
Smith 2000 {published data only}
- Smith Y, Weinberg A, Klauss S, Soffer D, Ingber A. Improving screening for melanoma by measuring similarity to pre‐classified images. Melanoma Research 2000;10(3):265‐72. [PubMed] [Google Scholar]
Sober 1994 {published data only}
- Sober AJ, Burstein JM. Computerized digital image analysis: An aid for melanoma diagnosis. Journal of Dermatology 1994;21(11):885‐90. [DOI] [PubMed] [Google Scholar]
Stanganelli 1995 {published data only}
- Stanganelli I, Burroni M, Rafanelli S, Bucchi L. Intraobserver agreement in interpretation of digital epiluminescence microscopy. Journal of the American Academy of Dermatology 1995;33(4):584‐9. [DOI] [PubMed] [Google Scholar]
Stanley 2007 {published data only}
- Stanley RJ, Stoecker WV, Moss RH. A relative color approach to color discrimination for malignant melanoma detection in dermoscopy images. Skin Research and Technology 2007;13(1):62‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Stanley 2008 {published data only}
- Stanley RJ, Stoecker WV, Moss RH, Rabinovitz HS, Cognetta AB Jr, Argenziano G, et al. A basis function feature‐based approach for skin lesion discrimination in dermatology dermoscopy images. Skin Research and Technology 2008;14(4):425‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Stoecker 2005 {published data only}
- Stoecker WV, Gupta K, Stanley RJ, Moss RH, Shrestha B. Detection of asymmetric blotches (asymmetric structureless areas) in dermoscopy images of malignant melanoma using relative color. Skin Research and Technology 2005;11(3):179‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Swanson 2010 {published data only}
- Swanson DL, Laman SD, Biryulina M, Ryzhikov G, Stamnes JJ, Hamre B, et al. Optical transfer diagnosis of pigmented lesions. Dermatologic Surgery 2010;36(12):1979‐86. [DOI] [PubMed] [Google Scholar]
Tehrani 2006 {published data only}
- Tehrani H, Walls J, Price G, Cotton S, Sassoon E, Hall P. A novel imaging technique as an adjunct to the in vivo diagnosis of nonmelanoma skin cancer. British Journal of Dermatology 2006;155(6):1177‐83. [DOI] [PubMed] [Google Scholar]
Terstappen 2007 {published data only}
- Terstappen K, Larko O, Wennberg AM. Pigmented basal cell carcinoma‐‐comparing the diagnostic methods of SIAscopy and dermoscopy. Acta Dermato‐Venereologica 2007;87(3):238‐42. [DOI] [PubMed] [Google Scholar]
Varol 2006 {published data only}
- Varol A. Erratum: The performance of SolarScan: An automated dermoscopy image analysis instrument for the diagnosis of primary melanoma (Archives (November 2005) 141, 1388‐96). Archives of Dermatology 2006;142(5):558. [DOI] [PubMed] [Google Scholar]
Vestergaard 2008 {published data only}
- Vestergaard ME, Menzies SW. Automated diagnostic instruments for cutaneous melanoma. Seminars in Cutaneous Medicine and Surgery 2008;27(1):32‐6. [DOI] [PubMed] [Google Scholar]
Wallace 2000a {published data only}
- Wallace VP, Bamber JC, Crawford DC, Ott RJ, Mortimer PS. Classification of reflectance spectra from pigmented skin lesions, a comparison of multivariate discriminant analysis and artificial neural networks. Physics in Medicine and Biology 2000;45(10):2859‐71. [DOI] [PubMed] [Google Scholar]
Wallace 2000b {published data only}
- Wallace VP, Crawford DC, Mortimer PS, Ott RJ, Bamber JC. Spectrophotometric assessment of pigmented skin lesions: methods and feature selection for evaluation of diagnostic performance. Physics in Medicine and Biology 2000;45(3):735. [DOI] [PubMed] [Google Scholar]
Wallace 2002 {published data only}
- Wallace VP, Bamber JC, Ott RJ, Crawford DC, Mortimer PS. Monitoring pigmented skin lesions. SPIE. International Symposium on Biomedical Optics. Functional Monitoring and Drug‐Tissue Interaction. 2002; Vol. 4623. [DOI: 10.1117/12.469443] [DOI]
Walter 2010 {published data only}
- Walter FM, Morris HC, Humphrys E, Hall PN, Kinmonth AL, Prevost AT, et al. Protocol for the MoleMate UK Trial: a randomised controlled trial of the MoleMate system in the management of pigmented skin lesions in primary care. BMC Family Practice 2010;11:36. [DOI: 10.1186/1471-2296-11-36] [DOI] [PMC free article] [PubMed] [Google Scholar]
Watson 2009 {published data only}
- Watson T, Walter FM, Wood A, Morris H, Hall P, Karner S, et al. Learning a novel technique to identify possible melanomas: are Australian general practitioners better than their U.K. colleagues?. Asia Pacific Family Medicine 2009;8(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Wazaefi 2012 {published data only}
- Wazaefi Y, Paris S, Fertil B. Contribution of a classifier of skin lesions to the dermatologist's decision. 3rd International Conference on Image Processing Theory, Tools and Applications (IPTA). IEEE, 2012. [DOI: 10.1109/IPTA.2012.6469560] [DOI]
Wells 2011 {published data only}
- Wells R. Comparison of diagnostic and biopsy/referral sensitivity to melanoma between dermatologists and MelaFind: A pilot survey study. Journal of Drugs in Dermatology 2011;10(9):1078. [Google Scholar]
Wilson 2013 {published data only}
- Wilson EC, Emery JD, Kinmonth AL, Prevost AT, Morris HC, Humphrys E, et al. The cost‐effectiveness of a novel SIAscopic diagnostic aid for the management of pigmented skin lesions in primary care: a decision‐analytic model. Value in Health 2013;16(2):356‐66. [DOI] [PubMed] [Google Scholar]
Winkelmann 2015a {published data only}
- Winkelmann RR, Hauschild A, Tucker N, White R, Rigel DS. The impact of multispectral digital skin lesion analysis on German dermatologist decisions to biopsy atypical pigmented lesions with clinical characteristics of melanoma. Journal of Clinical and Aesthetic Dermatology 2015;8(10):27‐9. [PMC free article] [PubMed] [Google Scholar]
Winkelmann 2015b {published data only}
- Winkelmann RR, Rigel DS, Kollmann E, Swenson N, Tucker N, Nestor MS. Negative predictive value of pigmented lesion evaluation by multispectral digital skin lesion analysis in a community practice setting. Journal of Clinical and Aesthetic Dermatology 2015;8(3):20‐2. [PMC free article] [PubMed] [Google Scholar]
Winkelmann 2015c {published data only}
- Winkelmann RR, Tucker N, White R, Rigel DS. Pigmented skin lesion biopsies after computer‐assisted multispectral digital skin lesion Analysis. Journal of the American Osteopathic Association 2015;115(11):666‐9. [DOI] [PubMed] [Google Scholar]
Winkelmann 2015d {published data only}
- Winkelmann RR, Yoo J, Tucker N, White R, Rigel DS. Impact of guidance provided by a multispectral digital skin lesion analysis device following dermoscopy on decisions to biopsy atypical melanocytic lesions. Journal of Clinical and Aesthetic Dermatology 2015;8(9):21‐4. [PMC free article] [PubMed] [Google Scholar]
Winkelmann 2016a {published data only}
- Winkelmann RR, Rigel DS, Ferris L, Sober A, Tucker N, Cockerell CJ. Correlation between the evaluation of pigmented lesions by a multi‐spectral digital skin lesion analysis device and the clinical and histological features of melanoma. Journal of Clinical and Aesthetic Dermatology 2016;9(3):36‐8. [PMC free article] [PubMed] [Google Scholar]
Wood 2008 {published data only}
- Wood A, Morris H, Emery J, Hall PN, Cotton S, Prevost AT, et al. Evaluation of the MoleMate training program for assessment of suspicious pigmented lesions in primary care. Informatics in Primary Care 2008;16(1):41‐50. [DOI] [PubMed] [Google Scholar]
Yoo 2015 {published data only}
- Yoo J, Tucker N, White R, Rigel D. The impact of probability of melanoma information provided by a multispectral digital skin lesion analysis device (MSDSLA) on resident dermatologists' decisions to biopsy clinical atypical lesions. Journal of the American Academy of Dermatology 2015;72(5 Suppl 1):AB177. [Google Scholar]
Zagrouba 2004 {published data only}
- Zagrouba E, Barhoumi W. A prelimary approach for the automated recognition of malignant melanoma. Image Analysis and Stereology Journal 2004;23(2):121‐35. [Google Scholar]
Zhou 2010a {published data only}
- Zhou H, Rehg JM, Chen M. Exemplar‐based segmentation of pigmented skin lesions from dermoscopy images. 7th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2010:225‐8. [DOI: 10.1109/ISBI.2010.5490372] [DOI]
Zhou 2010b {published data only}
- Zhou Y, Smith M, Smith L, Warr R. A new method describing border irregularity of pigmented lesions. Skin Research and Technology 2010;16(1):66‐76. [DOI] [PubMed] [Google Scholar]
Zortea 2014 {published data only}
- Zortea M, Schopf TR, Thon K, Geilhufe M, Hindberg K, Kirchesch H, et al. Performance of a dermoscopy‐based computer vision system for the diagnosis of pigmented skin lesions compared with visual evaluation by experienced dermatologists. Artificial Intelligence in Medicine 2014;60(1):13‐26. [DOI] [PubMed] [Google Scholar]
Zouridakis 2004 {published data only}
Additional references
ACIM 2017
- Australian Cancer Database. Melanoma of the skin for Australia (ICD10 C43). Australian Institute of Health and Welfare (AIHW) 2017 Australian Cancer Incidence and Mortality (ACIM) books (www.aihw.gov.au/acim‐books/). Canberra: Australian Institute of Health and Welfare, 2017. [Google Scholar]
Alam 2001
- Alam M, Ratner D. Cutaneous squamous‐cell carcinoma (Review). New England Journal of Medicine 2001;344(13):975‐83. [PUBMED: 11274625] [DOI] [PubMed] [Google Scholar]
Altman 2009
- Altman DG, Vergouwe Y, Royston P, Moons KG. Prognosis and prognostic research: validating a prognostic model. BMJ 2009;338(7708):b605. [doi: 10.1136/bmj.b605; PUBMED: 19477892] [DOI] [PubMed] [Google Scholar]
Argenziano 1998
- Argenziano G, Fabbrocini G, Carli P, Giorgi V, Sammarco E, Delfino M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions. Comparison of the ABCD rule of dermatoscopy and a new 7‐point checklist based on pattern analysis. Archives of Dermatology 1998;134(12):1563‐70. [PUBMED: 9875194] [DOI] [PubMed] [Google Scholar]
Argenziano 2012
- Argenziano G, Albertini G, Castagnetti F, Pace B, Lernia V, Longo C, et al. Early diagnosis of melanoma: what is the impact of dermoscopy?. Dermatologic Therapy 2012;25(5):403‐9. [PUBMED: 23046019] [DOI] [PubMed] [Google Scholar]
Arits 2013
- Arits AH, Mosterd K, Essers BA, Spoorenberg E, Sommer A, Rooij MJ, et al. Photodynamic therapy versus topical imiquimod versus topical fluorouracil for treatment of superficial basal‐cell carcinoma: a single blind, non‐inferiority, randomised controlled trial. Lancet Oncology 2013;14(7):647‐54. [DOI: 10.1016/S1470-2045(13)70143-8] [DOI] [PubMed] [Google Scholar]
Arnold 2014
- Arnold M, Holterhues C, Hollestein LM, Coebergh JW, Nijsten T, Pukkala E, et al. Trends in incidence and predictions of cutaneous melanoma across Europe up to 2015. Journal of the European Academy of Dermatology and Venereology 2014;28(9):1170‐8. [PUBMED: 23962170] [DOI] [PubMed] [Google Scholar]
Balch 2001
- Balch CM, Buzaid AC, Soong SJ, Atkins MB, Cascinelli N, Coit DG, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. Journal of Clinical Oncology 2001;19(16):3635‐48. [DOI] [PubMed] [Google Scholar]
Baldursson 1993
- Baldursson B, Sigurgeirsson B, Lindelof B. Leg ulcers and squamous cell carcinoma. An epidemiological study and a review of the literature. Acta Dermato‐Venereologica 1993;73(3):171‐4. [PUBMED: 8105611] [DOI] [PubMed] [Google Scholar]
Bath‐Hextall 2007a
- Bath‐Hextall F, Leonardi‐Bee J, Smith C, Meal A, Hubbard R. Trends in incidence of skin basal cell carcinoma. Additional evidence from a UK primary care database study. International Journal of Cancer 2007;121(9):2105‐8. [PUBMED: 17640064] [DOI] [PubMed] [Google Scholar]
Bath‐Hextall 2007b
- Bath‐Hextall Fiona J, Perkins W, Bong J, Williams HC. Interventions for basal cell carcinoma of the skin. Cochrane Database of Systematic Reviews 2007, Issue 1. [DOI: 10.1002/14651858.CD003412.pub2] [DOI] [PubMed] [Google Scholar]
Bath‐Hextall 2014
- Bath‐Hextall F, Ozolins M, Armstrong SJ, Colver GB, Perkins W, Miller PS, et al. Surgical excision versus imiquimod 5% cream for nodular and superficial basal‐cell carcinoma (SINS): a multicentre, non‐inferiority, randomised controlled trial. Lancet Oncology 2014;15(1):96‐105. [PUBMED: 24332516] [DOI] [PubMed] [Google Scholar]
Batra 2002
- Batra RS, Kelley LC. A risk scale for predicting extensive subclinical spread of nonmelanoma skin cancer. Dermatologic Surgery 2002;28(2):107‐12; discussion 112. [PUBMED: 11860418] [DOI] [PubMed] [Google Scholar]
Belbasis 2016
- Belbasis L, Stefanaki I, Stratigos AJ, Evangelou E. Non‐genetic risk factors for cutaneous melanoma and keratinocyte skin cancers: An umbrella review of meta‐analyses. Journal of Dermatological Science 2016;84(3):330‐9. [PUBMED: 27663092] [DOI] [PubMed] [Google Scholar]
Binder 1995
- Binder M, Schwarz M, Winkler A, Steiner A, Kaider A, Wolff K, et al. Epiluminescence microscopy. A useful tool for the diagnosis of pigmented skin lesions for formally trained dermatologists. Archives of Dermatology 1995;131(3):286‐91. [PUBMED: 7887657] [DOI] [PubMed] [Google Scholar]
Boring 1994
- Boring CC, Squires TS, Tong T, Montgomery S. Cancer statistics, 1994. CA: a Cancer Journal for Clinicians 1994;44(1):7‐26. [PUBMED: 8281473] [DOI] [PubMed] [Google Scholar]
Bossuyt 2015
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. [DOI: 10.1136/bmj.h5527; PUBMED: 26511519] [DOI] [PMC free article] [PubMed] [Google Scholar]
Cancer Research UK 2017
- Cancer Research UK. Skin cancer statistics. www.cancerresearchuk.org/health‐professional/cancer‐statistics/statistics‐by‐cancer‐type/skin‐cancer#heading‐One (accessed prior to 19 July 2017).
CCAAC Network 2008
- Cancer Council Australia & Australian Cancer Network. Basal Cell Carcinoma, Squamous Cell Carcinoma (and related lesions) ‐ a guide to clinical management in Australia. www.cancer.org.au/content/pdf/HealthProfessionals/ClinicalGuidelines/Basal_cell_carcinoma_Squamous_cell_carcinoma_Guide_Nov_2008‐Final_with_Corrigendums.pdf. Sydney: Cancer Council Australia & Australian Cancer Network, (accessed 19 May 2015).
Chao 2013
- Chao D, London Cancer North and East. London Cancer, Guidelines for Cutaneous Malignant Melanoma Management August 2014. www.londoncancer.org/media/76373/london‐cancer‐melanoma‐guidelines‐2013‐v1.0.pdf. London: London Cancer North and East Alliance, (accessed 12 August 2018).
Cho 2014
- Cho H, Mariotto AB, Schwartz LM, Luo J, Woloshin S. When do changes in cancer survival mean progress? The insight from population incidence and mortality. Journal of the National Cancer Institute. Monographs 2014;2014(49):187‐97. [PUBMED: 25417232] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chowdri 1996
- Chowdri NA, Darzi MA. Postburn scar carcinomas in Kashmiris. Burns 1996;22(6):477‐82. [PUBMED: 8884010] [DOI] [PubMed] [Google Scholar]
Chu 2006
- Chu H, Cole SR. Bivariate meta‐analysis for sensitivity and specificity with sparse data: a generalized linear mixed model approach (comment). Journal of Clinical Epidemiology 2006;59(12):1331‐2. [PUBMED: 17098577] [DOI] [PubMed] [Google Scholar]
Chuchu 2018a
- Chuchu N, Dinnes J, Takwoingi Y, Matin RN, Bayliss SE, Davenport C, et al. Teledermatology for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013193] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chuchu 2018b
- Chuchu N, Takwoingi Y, Dinnes J, Matin RN, Bassett O, Moreau JF, et al. Smartphone applications for triaging adults with skin lesions that are suspicious for melanoma. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013192] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dabski 1986
- Dabski K, Stoll HL Jr, Milgrom H. Squamous cell carcinoma complicating late chronic discoid lupus erythematosus. Journal of Surgical Oncology 1986;32(4):233‐7. [PUBMED: 3736067] [DOI] [PubMed] [Google Scholar]
Dal Pozzo 1999
- Dal Pozzo V, Benelli C, Roscetti E. The seven features for melanoma: a new dermoscopic algorithm for the diagnosis of malignant melanoma. European Journal of Dermatology 1999;9(4):303‐8. [ER4:18375041; PUBMED: 10356410] [PubMed] [Google Scholar]
Deeks 2005
- Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. Journal of Clinical Epidemiology 2005;58(9):882‐93. [PUBMED: 16085191] [DOI] [PubMed] [Google Scholar]
Dinnes 2018a
- Dinnes J, Deeks JJ, Grainge MJ, Chuchu N, Ferrante di Ruffano L, Matin RN, et al. Visual inspection for diagnosing cutaneous melanoma in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013194] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018b
- Dinnes J, Deeks JJ, Chuchu N, Ferrante di Ruffano L, Matin RN, Thomson DR, et al. Dermoscopy, with and without visual inspection, for diagnosing melanoma in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD011902.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018c
- Dinnes J, Deeks JJ, Chuchu N, Matin RN, Wong KY, Aldridge RB, et al. Visual inspection and dermoscopy, alone or in combination, for diagnosing keratinocyte skin cancers in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD011901.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018d
- Dinnes J, Deeks JJ, Saleh D, Chuchu N, Bayliss SE, Patel L, et al. Reflectance confocal microscopy for diagnosing cutaneous melanoma in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013190] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018e
- Dinnes J, Deeks JJ, Chuchu N, Saleh D, Bayliss SE, Takwoingi Y, et al. Reflectance confocal microscopy for diagnosing keratinocyte skin cancers in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013191] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018f
- Dinnes J, Bamber J, Chuchu N, Bayliss SE, Takwoingi Y, Davenport C, et al. High‐frequency ultrasound for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013188] [DOI] [PMC free article] [PubMed] [Google Scholar]
Drew 2017
- Drew BA, Karia PS, Mora AN, Liang CA, Schmults CD. Treatment patterns, outcomes, and patient satisfaction of primary epidermally limited nonmelanoma skin cancer. Dermatologic Surgery 2017;43(12):1423‐30. [DOI: 10.1097/DSS.0000000000001225; PUBMED: 28661992] [DOI] [PubMed] [Google Scholar]
Drucker 2017
- Drucker A, Adam GP, Langberg V, Gazula A, Smith B, Moustafa F, et al. Treatments for Basal Cell and Squamous Cell Carcinoma of the Skin. Comparative Effectiveness Reviews, No. 199. Rockville (MD): Agency for Healthcare Research and Quality (US), 2017. [PubMed] [Google Scholar]
Efron 1983
- Efron B. Estimating the error rate of a prediction rule: improvement on cross‐validation. Journal of the American Statistical Association 1983;78(382):316‐331. [DOI: 10.1080/01621459.1983.10477973] [DOI] [Google Scholar]
Erdmann 2013
- Erdmann F, Lortet‐Tieulent J, Schuz J, Zeeb H, Greinert R, Breitbart EW, et al. International trends in the incidence of malignant melanoma 1953‐2008‐‐are recent generations at higher or lower risk?. International Journal of Cancer 2013;132(2):385‐400. [PUBMED: 22532371] [DOI] [PubMed] [Google Scholar]
Esteva 2017
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature 2017;542(7639):115‐8. [doi: 10.1038/nature21056; PUBMED: 28117445] [DOI] [PMC free article] [PubMed] [Google Scholar]
EUCAN 2012
- EUCAN, International Agency for Research on Cancer. Malignant melanoma of skin: estimated incidence, mortality & prevalence for both sexes, 2012. eco.iarc.fr/eucan/Cancer.aspx?Cancer=20. International Agency for Research on Cancer, (accessed 29 July 2015).
Fasching 1989
- Fasching MC, Meland NB, Woods JE, Wolff BG. Recurrent squamous‐cell carcinoma arising in pilonidal sinus tract‐‐multiple flap reconstructions. Report of a case. Diseases of the Colon and Rectum 1989;32(2):153‐8. [PUBMED: 2914529] [DOI] [PubMed] [Google Scholar]
Ferlay 2015
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer 2015;136(5):E359‐86. [PUBMED: 25220842] [DOI] [PubMed] [Google Scholar]
Ferrante di Ruffano 2018a
- Ferrante di Ruffano L, Dinnes J, Deeks JJ, Chuchu N, Bayliss SE, Davenport C, et al. Optical coherence tomography for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013189] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ferrante di Ruffano 2018b
- Ferrante di Ruffano L, Dinnes J, Chuchu N, Bayliss SE, Takwoingi Y, Davenport C, et al. Exfoliative cytology for diagnosing basal cell carcinoma and other skin cancers in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013187] [DOI] [PMC free article] [PubMed] [Google Scholar]
Firnhaber 2012
- Firnhaber JM. Diagnosis and treatment of basal cell and squamous cell carcinoma. American Family Physician 2012;86(2):161‐8. [PUBMED: 22962928] [PubMed] [Google Scholar]
Fitzpatrick 1975
- Fitzpatrick TB. Soleil et peau. Journal de Médecine Esthétique 1975;2:33‐4. [Google Scholar]
Garbe 2016
- Garbe C, Peris K, Hauschild A, Saiag P, Middleton M, Bastholt L, et al. Diagnosis and treatment of melanoma. European consensus‐based interdisciplinary guideline ‐ Update 2016. European Journal of Cancer 2016;63:201‐17. [PUBMED: 27367293] [DOI] [PubMed] [Google Scholar]
Garcia 2009
- Garcia C, Poletti E, Crowson AN. Basosquamous carcinoma. Journal of the American Academy of Dermatology 2009;60(1):137‐43. [PUBMED: 19103364] [DOI] [PubMed] [Google Scholar]
Gershenwald 2017
- Gershenwald JE, Scolyer RA, Hess KR, Sondak VK, Long GV, Ross MI, et al. Melanoma staging: Evidence‐based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA: A Cancer Journal for Clinicians 2017;67(6):472‐92. [DOI: 10.3322/caac.21409; PUBMED: 29028110] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gordon 2013
- Gordon R. Skin cancer: an overview of epidemiology and risk factors. Seminars in Oncology Nursing 2013;29(3):160‐9. [PUBMED: 23958214] [DOI] [PubMed] [Google Scholar]
Gorlin 2004
- Gorlin RJ. Nevoid basal cell carcinoma (Gorlin) syndrome. Genetics in Medicine 2004;6(6):530‐9. [PUBMED: 15545751] [DOI] [PubMed] [Google Scholar]
Grachtchouk 2011
- Grachtchouk M, Pero J, Yang SH, Ermilov AN, Michael LE, Wang A, et al. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. Journal of Clinical Investigation 2011;121(5):1768‐81. [PUBMED: 21519145] [DOI] [PMC free article] [PubMed] [Google Scholar]
Griffin 2016
- Griffin LL, Ali FR, Lear JT. Non‐melanoma skin cancer. Clinical Medicine 2016;16(1):62‐5. [PUBMED: 26833519] [DOI] [PMC free article] [PubMed] [Google Scholar]
Griffiths 2005
- Griffiths RW, Suvarna SK, Stone J. Do basal cell carcinomas recur after complete conventional surgical excision?. British Journal of Plastic Surgery 2005;58(6):795‐805. [PUBMED: 16086990] [DOI] [PubMed] [Google Scholar]
Gutkowicz‐Krusin 2007
- Gutkowicz‐Krusin D, Elbaum M, Greenebaum M, Jacobs A. Systems and methods for the multispectral imaging and characterization of skin tissue. patents.google.com/patent/US6208749B1/en (accessed prior to 12 November 2018).
Han 2018
- Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. Journal of Investigative Dermatology 2018;138(7):1529‐38. [DOI: 10.1016/j.jid.2018.01.028; PUBMED: 29428356] [DOI] [PubMed] [Google Scholar]
Hartevelt 1990
- Hartevelt MM, Bavinck JN, Kootte AM, Vermeer BJ, Vandenbroucke JP. Incidence of skin cancer after renal transplantation in The Netherlands. Transplantation 1990;49(3):506‐9. [PUBMED: 2316011] [DOI] [PubMed] [Google Scholar]
Hoorens 2016
- Hoorens I, Vossaert K, Pil L, Boone B, Schepper S, Ongenae K, et al. Total‐body examination vs lesion‐directed skin cancer screening. JAMA Dermatology 2016;152(1):27‐34. [PUBMED: 26466155] [DOI] [PubMed] [Google Scholar]
Horsch 2011
- Horsch A. Melanoma diagnosis. In: TM Deserno editor(s). Biomedical Image Processing. Berlin: Springer‐Verlag, 2011:307‐26. [Google Scholar]
HPA and MelNet NZ 2014
- Health Promotion Agency and the Melanoma Network of New Zealand (MelNet). New Zealand Skin Cancer Primary Prevention and Early Detection Strategy 2014 to 2017. www.sunsmart.org.nz//sites/default/files/documents/NZ%20Skin%20Cancer%20PrimaryPrevention%20and%20EarlyDetection%20Strategy%202014%20to%202017%20FINAL%20VERSION%20%23406761.pdf. Cancer Society of New Zealand, (accessed 29 May 2018).
Jensen 1999
- Jensen P, Hansen S, Moller B, Leivestad T, Pfeffer P, Geiran O, et al. Skin cancer in kidney and heart transplant recipients and different long‐term immunosuppressive therapy regimens. Journal of the American Academy of Dermatology 1999;40(2 Pt 1):177‐86. [PUBMED: 10025742] [DOI] [PubMed] [Google Scholar]
Kao 1986
- Kao GF. Carcinoma arising in Bowen's disease. Archives of Dermatology 1986;122(10):1124‐6. [PUBMED: 3767398] [PubMed] [Google Scholar]
Kasprzak 2015
- Kasprzak JM, Xu YG. Diagnosis and management of lentigo maligna: a review. Drugs in Context 2015;4:212281. [PUBMED: 26082796] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kelleners‐Smeets 2017
- Kelleners‐Smeets NW, Mosterd K, Nelemans PJ. Treatment of low‐risk basal cell carcinoma. Journal of Investigative Dermatology 2017;137(3):539‐40. [PUBMED: 28235442] [DOI] [PubMed] [Google Scholar]
Kim 2014
- Kim DD, Tang JY, Ioannidis JP. Network geometry shows evidence sequestration for medical vs. surgical practices: treatments for basal cell carcinoma. Journal of Clinical Epidemiology 2014;67(4):391‐400. [PUBMED: 24491794] [DOI] [PubMed] [Google Scholar]
Kittler 2002
- Kittler H, Pehamberger H, Wolff K, Binder M. Diagnostic accuracy of dermoscopy (Review). Lancet Oncology 2002;3(3):159‐65. [PUBMED: 11902502] [DOI] [PubMed] [Google Scholar]
Lachs 1992
- Lachs MS, Nachamkin I, Edelstein PH, Goldman J, Feinstein AR, Schwartz JS. Spectrum bias in the evaluation of diagnostic tests: lessons from the rapid dipstick test for urinary tract infection. Annals of Internal Medicine 1992;117(2):135‐40. [PUBMED: 1605428] [DOI] [PubMed] [Google Scholar]
Lansbury 2010
- Lansbury L, Leonardi‐Bee J, Perkins W, Goodacre T, Tweed JA, Bath‐Hextall FJ. Interventions for non‐metastatic squamous cell carcinoma of the skin. Cochrane Database of Systematic Reviews 2010, Issue 4. [DOI: 10.1002/14651858.CD007869.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lansbury 2013
- Lansbury L, Bath‐Hextall F, Perkins W, Stanton W, Leonardi‐Bee J. Interventions for non‐metastatic squamous cell carcinoma of the skin: systematic review and pooled analysis of observational studies. BMJ 2013;347:f6153. [PUBMED: 24191270] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lear 1997
- Lear JT, Smith AG, Heagerty AH, Bowers B, Jones PW, Gilford J, et al. Truncal site and detoxifying enzyme polymorphisms significantly reduce time to presentation of further primary cutaneous basal cell carcinoma. Carcinogenesis 1997;18(8):1499‐503. [PUBMED: 9276622] [DOI] [PubMed] [Google Scholar]
Lear 2012
- Lear JT. Oral hedgehog‐pathway inhibitors for basal‐cell carcinoma. New England Journal of Medicine 2012;366(23):2225‐6. [PUBMED: 22670909] [DOI] [PubMed] [Google Scholar]
Lederman 1985
- Lederman JS, Sober AJ. Does biopsy type influence survival in clinical stage I cutaneous melanoma?. Journal of the American Academy of Dermatology 1985;13(6):983‐7. [PUBMED: 4078105] [DOI] [PubMed] [Google Scholar]
Leeflang 2013
- Leeflang MM, Rutjes AW, Reitsma JB, Hooft L, Bossuyt PM. Variation of a test's sensitivity and specificity with disease prevalence. CMAJ 2013;185(11):E537‐44. [PUBMED: 23798453] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lees 1991
- Lees VC, Briggs JC. Effect of initial biopsy procedure on prognosis in stage I invasive cutaneous malignant melanoma: review of1086 patients. British Journal of Surgery 1991;78(9):1108‐10. [PUBMED: 1933198] [DOI] [PubMed] [Google Scholar]
Leff 2008
- Leff B, Finucane TE. Gizmo idolatry. JAMA 2008;299(15):1830‐2. [PUBMED: 18413879] [DOI] [PubMed] [Google Scholar]
Lijmer 1999
- Lijmer JG, Mol BW, Heisterkamp S, Bonsel GJ, Prins MH, Meulen JH, et al. Empirical evidence of design‐related bias in studies of diagnostic tests. JAMA 1999;282(11):1061‐6. [PUBMED: 10493205] [DOI] [PubMed] [Google Scholar]
Linos 2009
- Linos E, Swetter SM, Cockburn MG, Colditz GA, Clarke CA. Increasing burden of melanoma in the United States. Journal of Investigative Dermatology 2009;129(7):1666‐74. [PUBMED: 19131946] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lister 1997
- Lister RK, Black MM, Calonje E, Burnand KG. Squamous cell carcinoma arising in chronic lymphoedema. British Journal of Dermatology 1997;136(3):384‐7. [PUBMED: 9115922] [PubMed] [Google Scholar]
Lo 1991
- Lo JS, Snow SN, Reizner GT, Mohs FE, Larson PO, Hruza GJ. Metastatic basal cell carcinoma: report of twelve cases with a review of the literature. Journal of the American Academy of Dermatology 1991;24(5 Pt 1):715‐9. [PUBMED: 1869642] [DOI] [PubMed] [Google Scholar]
Lomas 2012
- Lomas A, Leonardi‐Bee J, Bath‐Hextall F. A systematic review of worldwide incidence of nonmelanoma skin cancer. British Journal of Dermatology 2012;166(5):1069‐80. [PUBMED: 22251204] [DOI] [PubMed] [Google Scholar]
MacKie 1990
- MacKie RM. Clinical recognition of early invasive malignant melanoma. BMJ 1990;301(6759):1005‐6. [PUBMED: 2249043] [DOI] [PMC free article] [PubMed] [Google Scholar]
Madan 2010
- Madan V, Lear JT, Szeimies RM. Non‐melanoma skin cancer. Lancet 2010;375(9715):673‐85. [PUBMED: 20171403] [DOI] [PubMed] [Google Scholar]
Maglogiannis 2009
- Maglogiannis I, Doukas CN. Overview of advanced computer vision systems for skin lesion characterization. IEEE Transactions on Information Technology in Biomedicine 2009;13(5):721‐33. [DOI: 10.1109/TITB.2009.2017529] [DOI] [PubMed] [Google Scholar]
Maia 1995
- Maia M, Proenca NG, Moraes JC. Risk factors for basal cell carcinoma: a case‐control study. Revista de Saude Publica 1995;29(1):27‐37. [PUBMED: 8525311] [DOI] [PubMed] [Google Scholar]
Maloney 1996
- Maloney ME. Arsenic in dermatology. Dermatologic Surgery 1996;22(3):301‐4. [PUBMED: 8599743] [DOI] [PubMed] [Google Scholar]
Marchesini 1995
- Marchesini R, Tomatis S, Bartoli C, Bono A, Clemente C, Cupeta C, et al. In vivo spectrophotometric evaluation of neoplastic and non‐neoplastic skin pigmented lesions. III. CCD camera‐based reflectance imaging. Photochemistry and Photobiology 1995;62(1):151‐4. [PUBMED: 7638259] [DOI] [PubMed] [Google Scholar]
Marsden 2010
- Marsden JR, Newton‐Bishop JA, Burrows L, Cook M, Corrie PG, Cox NH, et al. BAD Guidelines: Revised UK guidelines for the management of cutaneous melanoma 2010. British Journal of Dermatology 2010;163(2):238‐56. [PUBMED: 20608932] [DOI] [PubMed] [Google Scholar]
McCormack 1997
- McCormack CJ, Kelly JW, Dorevitch AP. Differences in age and body site distribution of the histological subtypes of basal cell carcinoma. A possible indicator of differing causes. Archives of Dermatology 1997;133(5):593‐6. [PUBMED: 9158412] [PubMed] [Google Scholar]
McCusker 2014
- McCusker M, Basset‐Seguin N, Dummer R, Lewis K, Schadendorf D, Sekulic A, et al. Metastatic basal cell carcinoma: Prognosis dependent on anatomic site and spread of disease. European Journal of Cancer 2014;50(4):774‐83. [PUBMED: 24412051] [DOI] [PubMed] [Google Scholar]
Mistry 2011
- Mistry M, Parkin DM, Ahmad AS, Sasieni P. Cancer incidence in the United Kingdom: projections to the year 2030. British Journal of Cancer 2011;105(11):1795‐803. [PUBMED: 22033277] [DOI] [PMC free article] [PubMed] [Google Scholar]
Moeckelmann 2018
- Moeckelmann N, Ebrahimi A, Dirven R, Liu J, Low TH, Gupta R, et al. Analysis and comparison of the 8th edition American Joint Committee on Cancer (AJCC) nodal staging system in cutaneous and oral squamous cell cancer of the head and neck. Annals of Surgical Oncology 2018; Vol. 25, issue 6:1730‐6. [DOI: 10.1245/s10434-018-6340-x] [DOI] [PubMed]
Moons 1997
- Moons KG, Es GA, Deckers JW, Habbema JD, Grobbee DE. Limitations of sensitivity, specificity, likelihood ratio, and Bayes' theorem in assessing diagnostic probabilities: a clinical example. Epidemiology 1997;8(1):12‐7. [PUBMED: 9116087] [DOI] [PubMed] [Google Scholar]
Motley 2009
- Motley RJ, Preston PW, Lawrence CM. Multi‐professional guidelines for the management of the patient with primary cutaneous squamous cell carcinoma. www.bsds.org.uk/uploads/pdfs/SCCguide2009.pdf (accessed 15 November 2017).
Murchie 2017
- Murchie P, Amalraj Raja E, Brewster DH, Iversen L, Lee AJ. Is initial excision of cutaneous melanoma by General Practitioners (GPs) dangerous? Comparing patient outcomes following excision of melanoma by GPs or in hospital using national datasets and meta‐analysis. European Journal of Cancer 2017;86:373‐84. [PUBMED: 29100192] [DOI] [PubMed] [Google Scholar]
Musah 2013
- Musah A, Gibson JE, Leonardi‐Bee J, Cave MR, Ander EL, Bath‐Hextall F. Regional variations of basal cell carcinoma incidence in the UK using The Health Improvement Network database (2004‐10). British Journal of Dermatology 2013;169(5):1093‐9. [PUBMED: 23701520] [DOI] [PubMed] [Google Scholar]
Nart 2015
- Nart IF, Armayones SG, Medina FV, Orti MB, Orpinell XB. Basal cell carcinoma treated with ingenol mebutate. Journal of the American Academy of Dermatology 2015;5(Suppl 1):AB180. [Google Scholar]
Newcombe 1998
- Newcombe RG. Interval estimation for the difference between independent proportions: comparison of eleven methods. Statistics in Medicine 1998;17(8):873‐90. [DOI] [PubMed] [Google Scholar]
NICE 2010
- National Institute for Health and Clinical Excellence. NICE guidance on cancer services. Improving outcomes for people with skin tumours including melanoma (update): The management of low‐risk basal cell carcinomas in the community. www.nice.org.uk/guidance/csg8/resources/improving‐outcomes‐for‐people‐with‐skin‐tumours‐including‐melanoma‐2010‐partial‐update‐773380189. NICE, (accessed 27 November 2017).
NICE 2015
- National Institute for Health and Care Excellence. Melanoma: assessment and management. www.nice.org.uk/guidance/ng14. London: National Institute for Health and Care Excellence, (accessed prior to 2 May 2018).
NICE 2017
- National Institute for Health and Care Excellence. Vismodegib for treating basal cell carcinoma. www.nice.org.uk/guidance/ta489. London: NICE, (accessed prior to 2 May 2018).
Norman 2009
- Norman G, Barraclough K, Dolovich L, Price D. Iterative diagnosis. BMJ 2009;339:b3490. [DOI: 10.1136/bmj.b3490] [DOI] [PubMed] [Google Scholar]
O'Gorman 2014
- O'Gorman SM, Murphy GM. Photosensitizing medications and photocarcinogenesis. Photodermatology, Photoimmunology & Photomedicine 2014;30(1):8‐14. [PUBMED: 24393207] [DOI] [PubMed] [Google Scholar]
Pasquali 2018
- Pasquali S, Hadjinicolaou AV, Chiarion Sileni V, Rossi CR, Mocellin S. Systemic treatments for metastatic cutaneous melanoma. Cochrane Database of Systematic Reviews 2018, Issue 2. [DOI: 10.1002/14651858.CD011123.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Rallan 2004
- Rallan D, Harland CC. Skin imaging: is it clinically useful?. Clinical Dermatology 2004;29(5):453‐59. [PUBMED: 15347322] [DOI] [PubMed] [Google Scholar]
Randle 1996
- Randle HW. Basal cell carcinoma. Identification and treatment of the high‐risk patient. Dermatologic Surgery 1996;22(3):255‐61. [PUBMED: 8599737] [DOI] [PubMed] [Google Scholar]
Reitsma 2005
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. Journal of Clinical Epidemiology 2005;58(10):982‐90. [PUBMED: 16168343] [DOI] [PubMed] [Google Scholar]
Roozeboom 2012
- Roozeboom MH, Arits AH, Nelemans PJ, Kelleners‐Smeets NW. Overall treatment success after treatment of primary superficial basal cell carcinoma: a systematic review and meta‐analysis of randomized and nonrandomized trials. British Journal of Dermatology 2012;167(4):733‐56. [PUBMED: 22612571] [DOI] [PubMed] [Google Scholar]
Roozeboom 2016
- Roozeboom MH, Arits AH, Mosterd K, Sommer A, Essers BA, Rooij MJ, et al. Three‐year follow‐up results of photodynamic therapy vs. imiquimod vs. fluorouracil for treatment of superficial basal cell carcinoma: a single‐blind, noninferiority, randomized controlled trial. Journal of Investigative Dermatology 2016;136(8):1568‐74. [PUBMED: 27113429] [DOI] [PubMed] [Google Scholar]
Rozeman 2017
Rutjes 2005
- Rutjes AW, Reitsma JB, Vandenbroucke JP, Glas AS, Bossuyt PM. Case‐control and two‐gate designs in diagnostic accuracy studies. Clinical Chemistry 2005;51(8):1335‐41. [PUBMED: 15961549] [DOI] [PubMed] [Google Scholar]
Sekulic 2012
- Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, et al. Efficacy and safety of vismodegib in advanced basal‐cell carcinoma. New England Journal of Medicine 2012;366(23):2171‐9. [PUBMED: 22670903] [DOI] [PMC free article] [PubMed] [Google Scholar]
Siegel 2015
- Siegel R, Miller K, Jemal A. Cancer statistics, 2015. CA: a Cancer Journal for Clinicians 2015;65(1):5‐29. [PUBMED: 25559415] [DOI] [PubMed] [Google Scholar]
SIGN 2017
- Scottish Intercollegiate Guidelines Network. Cutaneous Melanoma. www.sign.ac.uk/sign‐146‐melanoma.html. Scotland: SIGN, (accessed prior to 19 July 2017).
Sladden 2009
- Sladden MJ, Balch C, Barzilai DA, Berg D, Freiman A, Handiside T, et al. Surgical excision margins for primary cutaneous melanoma. Cochrane Database of Systematic Reviews 2009, Issue 10. [DOI: 10.1002/14651858.CD004835.pub2] [DOI] [PubMed] [Google Scholar]
Slater 2014
- Slater D, Walsh M. Standards and datasets for reporting cancers: Dataset for the histological reporting of primary cutaneous malignant melanoma and regional lymph nodes, May 2014. www.rcpath.org/Resources/RCPath/Migrated%20Resources/Documents/G/G125_DatasetMaligMelanoma_May14.pdf. London: Royal College of Pathologists, (accessed 29 July 2015).
Smeets 2004
- Smeets NW, Krekels GA, Ostertag JU, Essers BA, Dirksen CD, Nieman FH, et al. Surgical excision vs Mohs' micrographic surgery for basal‐cell carcinoma of the face: randomised controlled trial. Lancet 2004;364(9447):1766‐72. [PUBMED: 15541449] [DOI] [PubMed] [Google Scholar]
Soyer 2000
- Soyer HP, Argenziano G, Chimenti S, Ruocco V. Dermoscopy of pigmented skin lesions. An atlas based on the Consensus Net Meeting on Dermoscopy 2000. Milan, Italy: EDRA Medical Publishing, 2001. [Google Scholar]
Stanganelli 2000
- Stanganelli I, Serafini M, Bucch L. A cancer‐registry‐assisted evaluation of the accuracy of digital epiluminescence microscopy associated with clinical examination of pigmented skin lesions. Dermatology 2000;200(1):11‐6. [PUBMED: 10681607] [DOI] [PubMed] [Google Scholar]
Stratigos 2015
- Stratigos A, Garbe C, Lebbe C, Malvehy J, Marmol V, Pehamberger H, et al. Diagnosis and treatment of invasive squamous cell carcinoma of the skin: European consensus‐based interdisciplinary guideline. European Journal of Cancer 2015;51(14):1989‐2007. [PUBMED: 26219687] [DOI] [PubMed] [Google Scholar]
Takwoingi 2013
- Takwoingi Y, Leeflang MM, Deeks JJ. Empirical evidence of the importance of comparative studies of diagnostic test accuracy. Annals of Internal Medicine 2013;158(7):544‐54. [PUBMED: 23546566] [DOI] [PubMed] [Google Scholar]
Takwoingi 2015
- Takwoingi Y, Guo B, Riley RD, Deeks JJ. Performance of methods for meta‐analysis of diagnostic test accuracy with few studies or sparse data. Statistical Methods in Medical Research 2015;26(4):1896‐911. [DOI: 10.1177/0962280215592269; PUBMED: 26116616] [DOI] [PMC free article] [PubMed] [Google Scholar]
Tomatis 1998
- Tomatis S, Bartoli C, Bono A, Cascinelli N, Clemente C, Marchesini R. Spectrophotometric imaging of cutaneous pigmented lesions: discriminant analysis, optical properties and histological characteristics. Journal of Photochemistry and Photobiology. B, Biology 1998;42(1):32‐9. [DOI] [PubMed] [Google Scholar]
Usher‐Smith 2016
- Usher‐Smith JA, Sharp SJ, Griffin SJ. The spectrum effect in tests for risk prediction, screening, and diagnosis. BMJ 2016;353:i3139. [DOI: 10.1136/bmj.i3139; PUBMED: 27334281] [DOI] [PMC free article] [PubMed] [Google Scholar]
Van Loo 2014
- Loo E, Mosterd K, Krekels GA, Roozeboom MH, Ostertag JU, Dirksen CD, et al. Surgical excision versus Mohs' micrographic surgery for basal cell carcinoma of the face: A randomised clinical trial with 10 year follow‐up. European Journal of Cancer 2014;50(17):3011‐20. [PUBMED: 25262378] [DOI] [PubMed] [Google Scholar]
Verkouteren 2017
- Verkouteren JAC, Ramdas KHR, Wakkee M, Nijsten T. Epidemiology of basal cell carcinoma: scholarly review. British Journal of Dermatology 2017;177(2):359‐72. [DOI: 10.1111/bjd.15321; PUBMED: 28220485] [DOI] [PubMed] [Google Scholar]
Walker 2006
- Walker P, Hill D. Surgical treatment of basal cell carcinomas using standard postoperative histological assessment. Australasian Journal of Dermatology 2006;47(1):1‐12. [PUBMED: 16405477] [DOI] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529‐36. [PUBMED: 22007046] [DOI] [PubMed] [Google Scholar]
WHO 2003
- World Health Organiation. INTERSUN: The Global UV Project. A guide and compendium. www.who.int/uv/publications/en/Intersunguide.pdf 2003 (accessed 20 May 2015).
Williams 2017
- Williams HC, Bath‐Hextall F, Ozolins M, Armstrong SJ, Colver GB, Perkins W, et al. Surgery versus 5% imiquimod for nodular and superficial basal cell carcinoma: 5‐year results of the SINS randomized controlled trial. Journal of Investigative Dermatology 2017;137(3):614‐9. [PUBMED: 27932240] [DOI] [PubMed] [Google Scholar]
Wong 2017
- Wong KY, Fife K, Lear JT, Price RD, Durrani AJ. Vismodegib for locally advanced periocular and orbital basal cell carcinoma: A review of 15 consecutive cases. Plastic and Reconstructive Surgery 2017;5(7):e1424. [PUBMED: 28831360] [DOI] [PMC free article] [PubMed] [Google Scholar]
Youl 2007
- Youl PH, Raasch BA, Janda M, Aitken JF. The effect of an educational programme to improve the skills of general practitioners in diagnosing melanocytic/pigmented lesions. Clinical and Experimental Dermatology 2007;32(4):365‐70. [PUBMED: 17433042] [DOI] [PubMed] [Google Scholar]
Zak‐Prelich 2004
- Zak‐Prelich M, Narbutt J, Sysa‐Jedrzejowska A. Environmental risk factors predisposing to the development of basal cell carcinoma. Dermatologic Surgery 2004;30(2 Pt 2):248–52. [PUBMED: 14871217] [DOI] [PubMed] [Google Scholar]
References to other published versions of this review
Dinnes 2015a
- Dinnes J, Matin RN, Moreau JF, Patel L, Chan SA, Wong KY, et al. Tests to assist in the diagnosis of cutaneous melanoma in adults: a generic protocol. Cochrane Database of Systematic Reviews 2015, Issue 10. [DOI: 10.1002/14651858.CD011902] [DOI] [Google Scholar]
Dinnes 2015b
- Dinnes J, Wong KY, Gulati A, Chuchu N, Leonardi‐Bee J, Bayliss SE, et al. Tests to assist in the diagnosis of keratinocyte skin cancers in adults: a generic protocol. Cochrane Database of Systematic Reviews 2015, Issue 10. [DOI: 10.1002/14651858.CD011901] [DOI] [Google Scholar]


