Abstract
Background
Resistance to antiretroviral therapy (ART) among people living with human immunodeficiency virus (HIV) compromises treatment effectiveness, often leading to virological failure and mortality. Antiretroviral drug resistance tests may be used at the time of initiation of therapy, or when treatment failure occurs, to inform the choice of ART regimen. Resistance tests (genotypic or phenotypic) are widely used in high‐income countries, but not in resource‐limited settings. This systematic review summarizes the relative merits of resistance testing in treatment‐naive and treatment‐exposed people living with HIV.
Objectives
To evaluate the effectiveness of antiretroviral resistance testing (genotypic or phenotypic) in reducing mortality and morbidity in HIV‐positive people.
Search methods
We attempted to identify all relevant studies, regardless of language or publication status, through searches of electronic databases and conference proceedings up to 26 January 2018. We searched MEDLINE, Embase, the Cochrane Central Register of Controlled Trials (CENTRAL), in the Cochrane Library, the World Health Organization (WHO) International Clinical Trials Registry Platform (ICTRP), and ClinicalTrials.gov to 26 January 2018. We searched Latin American and Caribbean Health Sciences Literature (LILACS) and the Web of Science for publications from 1996 to 26 January 2018.
Selection criteria
We included all randomized controlled trials (RCTs) and observational studies that compared resistance testing to no resistance testing in people with HIV irrespective of their exposure to ART.
Primary outcomes of interest were mortality and virological failure. Secondary outcomes were change in mean CD4‐T‐lymphocyte count, clinical progression to AIDS, development of a second or new opportunistic infection, change in viral load, and quality of life.
Data collection and analysis
Two review authors independently assessed each reference for prespecified inclusion criteria. Two review authors then independently extracted data from each included study using a standardized data extraction form. We analysed data on an intention‐to‐treat basis using a random‐effects model. We performed subgroup analyses for the type of resistance test used (phenotypic or genotypic), use of expert advice to interpret resistance tests, and age (children and adolescents versus adults). We followed standard Cochrane methodological procedures.
Main results
Eleven RCTs (published between 1999 and 2006), which included 2531 participants, met our inclusion criteria. All of these trials exclusively enrolled patients who had previous exposure to ART. We found no observational studies. Length of follow‐up time, study settings, and types of resistance testing varied greatly. Follow‐up ranged from 12 to 150 weeks. All studies were conducted in Europe, USA, or South America. Seven studies used genotypic testing, two used phenotypic testing, and two used both phenotypic and genotypic testing. Only one study was funded by a manufacturer of resistance tests.
Resistance testing made little or no difference in mortality (odds ratio (OR) 0.89, 95% confidence interval (CI) 0.36 to 2.22; 5 trials, 1140 participants; moderate‐certainty evidence), and may have slightly reduced the number of people with virological failure (OR 0.70, 95% CI 0.56 to 0.87; 10 trials, 1728 participants; low‐certainty evidence); and probably made little or no difference in change in CD4 cell count (mean difference (MD) ‐1.00 cells/mm³, 95% CI ‐12.49 to 10.50; 7 trials, 1349 participants; moderate‐certainty evidence) or progression to AIDS (OR 0.64, 95% CI 0.31 to 1.29; 3 trials, 809 participants; moderate‐certainty evidence). Resistance testing made little or no difference in adverse events (OR 0.89, 95% CI 0.51 to 1.55; 4 trials, 808 participants; low‐certainty evidence) and probably reduced viral load (MD ‐0.23, 95% CI ‐0.35 to ‐0.11; 10 trials, 1837 participants; moderate‐certainty evidence). No studies reported on development of new opportunistic infections or quality of life. We found no statistically significant heterogeneity for any outcomes, and the I² statistic value ranged from 0 to 25%. We found no subgroup effects for types of resistance testing (genotypic versus phenotypic), the addition of expert advice to interpretation of resistance tests, or age. Results for mortality were consistent when we compared studies at high or unclear risk of bias versus studies at low risk of bias.
Authors' conclusions
Resistance testing probably improved virological outcomes in people who have had virological failure in trials conducted 12 or more years ago. We found no evidence in treatment‐naive people. Resistance testing did not demonstrate important patient benefits in terms of risk of death or progression to AIDS. The trials included very few participants from low‐ and middle‐income countries.
11 April 2019
Up to date
All studies incorporated from most recent search
All eligible published studies found in the last search (26 Jan, 2018) were included
Plain language summary
Antiretroviral resistance testing in people living with HIV
What is the aim of this review?
The aim of this review was to find out whether a drug resistance test for people living with HIV (starting antiretroviral therapy (ART) or already on ART but with unsuppressed HIV) would reduce the number of deaths or improve HIV suppression.
Background
Drug resistance to ART implies that specific antiretroviral drugs will be less effective. This happens either because the virus has changed to become resistant, or because an individual was infected with a resistant virus. To determine which drugs will be less effective, healthcare providers may conduct a resistance test. Two kinds of resistance tests are available: the genotypic test, in which the virus is examined to determine which drugs it is resistant to, and the phenotypic test, in which the virus is exposed to antiretroviral drugs to see which one it is resistant to. Use of resistance tests is common only in high‐income countries. Before we prepared this review, we did not know how well the use of resistance tests may reduce the number of deaths and improve HIV suppression.
Main results
Cochrane review authors collected and analysed all relevant studies up to 26 January 2018 to answer this question and included 11 randomized controlled trials (published between 1999 and 2006) with a total of 2531 people. Trials included only people who had detectable HIV despite being on antiretroviral drugs; no trials included patients starting therapy for the first time. Studies were conducted in Europe, USA, or South America. Seven studies used genotypic testing, two used phenotypic testing, and two used both phenotypic and genotypic testing. Only one study was funded by a manufacturer of resistance tests.
Resistance testing probably made little or no difference to the risk of dying (moderate‐certainty evidence) or progression to AIDS (moderate‐certainty evidence). Resistance testing probably increased the chance of successful suppression of HIV replication (low‐certainty evidence) but probably made little or no difference in CD4 cell counts (cells affected by HIV) (moderate‐certainty evidence). Resistance testing made little or no difference in the number of people who experience medication side effects (low‐certainty evidence). No studies examined how many people developed a new opportunistic infection, and no studies examined patient quality of life.
Conclusion
For people for whom treatment no longer works, the use of resistance tests to select new treatments led to suppression of the HIV virus as measured by a blood test, but probably did not reduce the risk of death or progression to AIDS. Whether or not resistance testing provides any benefit for patients who are starting HIV treatment for the first time remains uncertain because no studies have evaluated this. These conclusions are based on studies conducted up to 12 years ago and included very few participants from low‐ and middle‐income countries.
Summary of findings
Summary of findings for the main comparison. Resistance testing versus no resistance testing in HIV‐positive people.
| Resistance testing versus no resistance testing in HIV‐positive people | ||||||
| Patient or population: HIV‐positive people Setting: all settings Intervention: resistance testing Comparison: no resistance testing | ||||||
| Outcomes | Anticipated absolute effects* (95% CI) | Relative effect (95% CI) | Number of participants (trials) | Certainty of the evidence (GRADE) | Comments | |
| Risk with no resistance testing | Risk with resistance testing | |||||
| Mortality | Study population | OR 0.89 (0.36 to 2.22) | 1140 (5 RCTs) | ⊕⊕⊕⊝
MODERATEa,b,c Due to imprecision |
Resistance testing probably has little or no impact on mortality | |
| 22 per 1000 | 20 per 1000 (8 to 47) | |||||
| Virological failure | Study population | OR 0.70 (0.56 to 0.87) | 1728 (10 RCTs) | ⊕⊕⊝⊝
LOWa,d,e,f Due to risk of bias and publication bias |
Resistance testing may reduce the risk of virological failure | |
| 660 per 1000 | 576 per 1000 (521 to 628) | |||||
| Change in CD4 cell count | Mean change in CD4 cell count was 0. | MD 1 lower (12.49 lower to 10.5 higher) | ‐ | 1349 (7 RCTs) | ⊕⊕⊕⊝
MODERATEa,d Due to risk of bias |
Resistance testing probably has little or no effect on change in CD4 cell count |
| Progression to AIDS | Study population | OR 0.64 (0.31 to 1.29) | 809 (3 RCTs) | ⊕⊕⊕⊝
MODERATEg Due to indirectness |
Resistance testing probably has little or no impact on progression to AIDS | |
| 67 per 1000 | 44 per 1000 (22 to 85) | |||||
| Adverse events | Study population | OR 0.89 (0.51 to 1.55) | 808 (4 RCTs) | ⊕⊕⊝⊝
LOWh,i Due to risk of bias and indirectness |
Resistance testing may have little or no effect on adverse effects | |
| 74 per 1000 | 66 per 1000 (39 to 110) | |||||
| Change in viral load | Mean change in viral load was 0. | MD 0.23 lower (0.35 lower to 0.11 lower) | ‐ | 1837 (10 RCTs) | ⊕⊕⊕⊝
MODERATEa,j Due to risk of bias |
Resistance testing probably results in a lower viral load |
| Quality of life ‐ not reported | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| New opportunistic infection ‐ not reported | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| *The risk in the intervention group (and its 95% CI) is based on the assumed risk in the comparison group and the relative effect of the intervention (and its 95% CI). Abbreviations: CI: confidence interval; MD: mean difference; OR: odds ratio; RCT: randomized controlled trial | ||||||
| GRADE Working Group grades of evidence High certainty: we are very confident that the true effect lies close to that of the estimate of the effect Moderate certainty: we are moderately confident in the effect estimate: the true effect is likely to be close to the estimate of the effect, but there is a possibility that it is substantially different Low certainty: our confidence in the effect estimate is limited: the true effect may be substantially different from the estimate of the effect Very low certainty: we have very little confidence in the effect estimate: the true effect is likely to be substantially different from the estimate of effect | ||||||
aRisk of bias: all studies included for this outcome were not blinded, but we did not downgrade for this, as lack of blinding is unlikely to introduce bias. bRisk of bias: one included study was at high risk of bias overall, but it contributed 16.1% of the data (Wegner 2004). We did not downgrade for this. cDowngraded by 1 for imprecision: CIs for the odds ratio include considerable harm and considerable benefit. We downgraded one point for this. dDowngraded by 1 for risk of bias: four of the included studies (˜ 37% of data) were at high or unclear risk of bias (Cingolani 2002;Cohen 2002;Haubrich 2005;Rubini 2002). eIndirectness: the included studies used different cutoffs to define virological failure. We did not downgrade for this. fDowngraded by 1 for publication bias: based on funnel plot asymmetry and a positive Egger's test. gDowngraded by 1 for indirectness: "progression to AIDS" was not defined uniformly across studies. hDowngraded by 1 for risk of bias: all studies included for this outcome were not blinded and adverse events could be interpreted subjectively. iDowngraded by 1 for indirectness: "adverse event" was not defined uniformly across studies. We downgraded one point for this. jDowngraded by 1 for risk of bias: four of the included studies (˜ 34% of data) were at high or unclear risk of bias (Cingolani 2002;Cohen 2002;Haubrich 2005;Rubini 2002).
Background
Description of the condition
Almost 36.7 million people are living with human immunodeficiency virus (HIV) worldwide (UNAIDS 2016). Widespread use of antiretroviral therapy (ART) has reduced the mortality and morbidity associated with HIV infection. Although only 40% of those eligible for ART are currently receiving it, efforts are under way to improve access to ART (WHO 2017). Effective ART inhibits viral replication and reduces viral load. The World Health Organization (WHO) Model List of Essential Medicines contains a list of antiretroviral drugs that are grouped into four classes: nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs), non‐nucleoside reverse transcriptase inhibitors (NNRTIs), protease inhibitors (PIs), and integrase strand transfer inhibitors (INSTIs; WHO 2017b). Recommendations on how they should be used include consideration of cost, availability, ease of administration, toxicity, and efficacy (WHO 2016). The potency of these medications can be severely compromised in the presence of drug resistance. In addition, the combination of drugs prescribed should be chosen with care, given that in some classes of antiretroviral drugs, especially NNRTIs, cross‐resistance is possible: any mutation that confers resistance to one drug will lead to resistance with all other drugs in the class (Sluis‐Cremer 2014). This is a matter of particular concern, given the widespread use of NNRTIs in low‐resource settings (Sluis‐Cremer 2014). The WHO currently recommends initiating ART with two NRTIs in addition to one NNRTI, or one INSTI. In the event of treatment failure, the NNRTI should be switched to a PI (WHO 2016).
Many problems exist in HIV care, including limited access to ART in some parts of the world, suboptimal levels of adherence, drug resistance, and treatment failure (UNAIDS 2014; UNAIDS 2016). International commitment to a unified response to the HIV pandemic is improving access to ART, and numerous research efforts have attempted to pinpoint the best adherence enhancement strategies (Chaiyachati 2014; Kanters 2017; Nachega 2012; Ramjan 2014; Thompson 2012). However, viral resistance to ART can have an important impact on the lives of those affected.
For HIV‐positive people who are taking ART, emergence of drug resistance poses a serious threat to a sustained virological response to treatment; it reduces effective therapeutic options and may increase morbidity, mortality, and infectivity (Cambiano 2013; Gupta 2012). All of the above have led to an increased healthcare burden for individuals and for society. Drug resistance may be transmitted (transmitted drug resistance: TDR) or acquired (drug resistance mutation: DRM; Rojas Sánchez 2014). People infected with highly resistant strains more often need to take ART (in spite of its higher costs, toxicity, and inconvenience) than those infected with HIV that is not antiretroviral‐resistant.
Close to 68% of treatment‐experienced patients who experience failed treatment have resistance to at least one drug (WHO 2017c). Even though recent data suggest that these numbers are dropping (De Luca 2015), treatment‐experienced patients who are failing treatment may still benefit from earlier detection of resistance and an informed selection of a new regimen.
The incidence of primary antiretroviral resistance is increasing (Bakhouch 2009; Barrow 2013; Duwe 2001; Rojas Sánchez 2014; Torti 2004). This drug resistance would most likely involve TDR in newly infected individuals. The prevalence of TDR may be stable in high‐income countries (10% to 17% have resistance to at least one drug; WHO 2017c), and it is on the rise in low‐income countries (Pham 2014). As the number of people on ART is increasing, the frequency of DRMs is also increasing (Boender 2016; Rowley 2016; Villabona‐Arenas 2016). People with pretreatment drug resistance are more likely to experience treatment failure (Hamers 2012; Wittkop 2011), have higher mortality rates (Cambiano 2013; Pinoges 2015), and need more frequent treatment switches during their course of care (Boender 2015). Irrespective of how drug resistance occurs, it represents a serious threat to the potency of ART in both ART‐naive and ART‐experienced patients.
Description of the intervention
Antiretroviral resistance testing is conducted in one of two ways: genotypic testing sequences the viral RNA and compares it against a database of known DRMs to determine which medications it is resistant to; phenotypic testing measures susceptibility of the virus to antiretroviral medications in a controlled environment exposed to specific antiretrovirals. Genotypic testing is less expensive and is more widely available. These tests provide information on resistance to the four main classes of ART (NRTIs, NNRTIs, PIs, and INSTIs; AIDSinfo 2016). Genotypic testing can be completed within one to two weeks, but interpretation of results may be challenging without knowledge of specific gene mutations and the potential for cross‐resistance. Interpretation of genotypic testing is often aided by simultaneous reporting of predicted antiretroviral susceptibilities. Expert advice is often helpful in choosing the optimal ART after genotypic testing (Tural 2002). Phenotypic tests take longer (two to three weeks), and they involve determining the ability of the virus to grow in various concentrations of antiretroviral drugs. Viral replication in the presence of antiretroviral drugs is then compared with replication of a reference HIV strain. Expert assistance may be helpful in guiding interpretation. Genotypic testing is the recommended approach for treatment‐naive and treatment‐experienced patients because it costs less, results can be available faster, and it is more sensitive in detecting mixtures of resistant and wild‐type viruses. Phenotypic testing can be added when complex mutation patterns are known or suspected (AIDSinfo 2016). Overall, both tests are costly, are unlikely to detect resistant viruses that constitute less than 10% to 20% of the circulating virus population, and do not have uniform standards for quality assurance (AIDSinfo 2016).
Additional considerations for the use of resistance testing include timing of the test and viral load. The best results are obtained before or within four weeks of treatment discontinuation, but testing is challenging to perform in patients with low HIV viral loads (AIDSinfo 2016).
How the intervention might work
Currently, the WHO recommends use of population‐based surveys to measure levels of drug resistance. Resistance testing should be considered before ART is initiated when the prevalence of NNRTI TDR is ≥ 10%, and when it is not feasible to provide non‐NNRTI‐based first‐line regimens for all starters owing to costs or availability (WHO 2017a). Data from such population‐based surveys in resource‐limited settings suggest that at 12 months, approximately two‐thirds of those who do not achieve viral suppression on first‐line therapy have drug resistance (Hosseinipour 2013). The prevalence of TDR varies by exposure risk and is probably higher among men who have sex with men (MSM) and people who inject drugs (PWIDs; Burchell 2012; Rocheleau 2017; Sullivan 2013). Therefore population‐based thresholds are not optimal for guiding first‐line ART regimens for many subpopulations. Because viral resistance can compromise virological response and no reliable ways are known to accurately predict primary drug resistance in an individual in the absence of resistance testing, HIV outcomes may be improved if such testing is done before ART is initiated (Barennes 2014). Mathematical models indicate that TDR might have a substantial impact on HIV mortality if not detected (Cambiano 2013).
Why it is important to do this review
Both European HIV Drug Resistance Guidelines and International Antiviral Society‐USA guidelines recommend resistance testing before initiation of ART (EACS 2017; Saag 2018). The WHO does not recommend this approach for resource‐limited settings in which available treatment options are limited, costs of resistance testing are prohibitive, and prevalence of ART resistance may be low. The WHO recommends a survey‐based method to detect population‐level prevalence of resistance and to support adherence and drug supply continuity (WHO 2017a). However, in settings that provide empirical first‐line therapy for most people, the most appropriate thresholds of population‐level resistance chosen for discontinuation of a first‐line ART regimen are unclear. In addition, certain subpopulations (such as pregnant women, MSM, PWIDs, and commercial sex workers (CSWs)) may have higher levels of TDR than are seen in the general population (Bissio 2017; Burchell 2012; Rocheleau 2017; Sullivan 2013).
The US Centers for Disease Control and Prevention (CDC) recommends routine testing of all patients initiating ART based on observational data (AIDSinfo 2016). One economic evaluation, based on a hypothetical cohort, reported that resistance testing is cost‐effective for treatment‐naive individuals (Sax 2005). Cost‐effectiveness results are conflicting for treatment‐experienced patients, depending on which type of resistance testing is used (Corzillius 2004; Phillips 2014). Previous systematic reviews prepared more than 10 years ago have focused solely on patients experiencing treatment failure and on short‐term outcomes (12 months) in high‐income countries (Dunn 2004; Ena 2006; Torre 2002). Torre 2002 found that viral suppression was more likely at three and six months after genotypic resistance testing but not after phenotypic testing. Viral suppression was also more likely when expert advice was provided. Dunn 2004 reported that a higher proportion of participants achieved viral suppression at three to six months among those who underwent resistance testing. Ena 2006 reported similar results at three months and additional benefits if genotypic resistance testing was coupled with expert interpretation. However, given that more options are now available for second‐ and third‐line treatment in resource‐limited settings (WHO 2016), the question of how to choose a new regimen after failure of the first‐line regimen in such settings is important, and more recent and applicable evidence is needed to answer this question.
In this systematic review, we will use data from randomized trials and cohort studies to highlight the benefits or harms of conducting resistance testing in individuals before initiation of ART and after failure of first‐line treatment.
Objectives
To evaluate the effectiveness of antiretroviral resistance testing (genotypic or phenotypic) in reducing mortality and morbidity in HIV‐positive people.
Methods
Criteria for considering studies for this review
Types of studies
We searched for randomized controlled trials (RCTs) and cohort studies conducted to evaluate clinical and biological outcomes in treatment‐naive or treatment‐experienced people with HIV who undergo resistance testing compared to no resistance testing.
Types of participants
We included people of any age who are HIV‐positive (with documented HIV infection as reported by study authors).
Types of interventions
Intervention
Interventions consisted of any type of resistance testing in people with HIV before initiation of therapy or after failure of first‐line therapy. We included genotypic and phenotypic tests. We included studies that compared different methods of resistance testing only if they included a control arm that underwent no testing.
Control
Control groups were given no resistance testing.
Types of outcome measures
Primary outcomes
Mortality (proportion of deaths)
Virological success: the proportion of participants achieving undetectable viral load (using lower limits for detection and time frames defined by study authors)
Secondary outcomes
Change in mean CD4‐T‐lymphocyte count (immunological response) over time
Clinical progression to AIDS: the proportion of participants who develop CDC‐defined AIDS (stages III and IV)
Development of a second or new opportunistic infection
Quality of life (as reported by study authors)
Change in viral load
Adverse events
Any adverse events reported by study authors
Search methods for identification of studies
We performed our literature search with the assistance of an Information Specialist. We adopted a comprehensive and exhaustive search strategy to identify studies reported in all languages irrespective of publication status (published, unpublished, in press, or in progress). We conducted searches of literature from 1989 ‐ the year in which the first case of antiretroviral drug resistance was identified (Larder 1989).
In addition to key antiretroviral terms used in standard searches performed by the Cochrane Infectious Diseases Group, we included all appropriate terms relevant to antiretroviral resistance testing, including Medical Subject Heading (MeSH) terms. We also used Cochrane's Highly Sensitive Strategy for identifying reports of RCTs and additional terms for identifying observational studies.
Electronic searches
We searched the following electronic databases for relevant RCTs and observational studies.
Cochrane Central Register of Controlled Trials (CENTRAL), in the Cochrane Library (1 January 1989 to 26 January 2018)
PubMed (1 January 1989 to 26 January 2018)
Embase (1 January 1989 to 26 January 2018)
Cumulative Index to Nursing and Allied Health Literature (CINAHL) (1 January 1989 to 26 January 2018)
Latin American and Caribbean Health Sciences Literature (LILACS) (1 January 1989 to 26 January 2018)
Web of Science (1 January 1989 to 26 January 2018)
We have outlined our detailed search strategies in Appendix 1, Appendix 2, and Appendix 3.
Conference abstracts
We searched conference abstract archives on the websites of the Conference on Retroviruses and Opportunistic Infections (CROI); the International AIDS Conference (IAC); and the International AIDS Society Conference on HIV Pathogenesis, Treatment, and Prevention (IAS), for all available abstracts presented at all conferences from 1989 to 2017.
Ongoing trials
World Health Organization (WHO) International Clinical Trials Registry Platform (ICTRP)
ClinicalTrials.gov
Searching other resources
We checked the reference lists of pertinent studies for other relevant studies. In addition, we contacted experts in the field. We invited experts who contributed to the WHO 2017 Guidelines Panel on Management of Drug Resistance to provide additional resources that would inform this review (WHO 2017a).
Data collection and analysis
We prepared a PRISMA diagram to present a summary of identification, screening, and inclusion of studies in this review (Moher 2010).
Selection of studies
Two review authors (TA, RS, or JT) independently inspected for relevance the titles and abstracts of all references identified by the search. We obtained full‐text copies of all potentially relevant articles and screened them using a pretested eligibility form. We included only studies that fulfilled our inclusion criteria. We resolved disagreements by consensus. When consensus could not be reached, we consulted a third review author (LM) for adjudication. We examined included manuscripts to ensure that they described unique patients whose data were not reported in another included study. If we had found overlapping studies, we would have included the larger, more comprehensive study. We reported the excluded studies and their reasons for exclusion in a Characteristics of excluded studies table. We illustrated the study selection process using a PRISMA diagram.
Data extraction and management
Two review authors (TA, RS, or JT) used pilot‐tested data extraction forms to independently extract and record data from the included studies. When disagreement arose between the two review authors, a third independent review author adjudicated (LM). When necessary (missing information or unclear reports), we contacted study authors for clarification. For reports not published in English, we invited other scientists with expertise in Cochrane methods to assist with screening and data extraction. We collected bibliometric information and data on participants, interventions, comparisons, outcomes, and study duration.
Bibliometric information
Full reference
Country of study
Participants
Inclusion criteria
Exclusion criteria
Age
Comorbidities
ART exposure (naive versus experienced)
Numbers in intervention and control arms
Interventions
Type of testing used
Use of expert interpretation
Drug regimens
Comparisons
Details on nature of control group (no testing or delayed testing)
Outcomes
Number of participants who experienced an event for dichotomous outcomes
Means and standard deviations for normally distributed continuous outcomes (we standardized continuous data not reported on the same scale)
Study duration
Duration of the study
Timing of outcome measurement (in weeks or months)
Assessment of risk of bias in included studies
We assessed the risk of bias of randomized trials using the Cochrane ‘Risk of bias' assessment tool for the following items (Appendix 4).
Sequence generation: how the allocation sequence was generated and whether this was adequate
Allocation concealment: how the allocation sequence was concealed and whether this was adequate
Blinding of participants, personnel, and outcome assessors
Description of the completeness of outcome data for each main outcome
Selective outcome reporting
Other potential sources of bias (e.g. funding)
We graded included studies as having high, low, or unclear risk of bias, corresponding to assessments of yes, no, or unclear risk of bias. Two review authors independently performed ‘Risk of bias' assessments and prepared ‘Risk of bias' tables.
Regarding the methodological quality of cohort studies, we planned to appraise this using the Newcastle‐Ottawa Scale (NOS; Wells 2009), which consists of assessments for the following items in three domains.
Selection: representativeness of the exposed cohort, selection of the non‐exposed cohort, ascertainment of exposure, demonstration that the outcome of interest was not present at the start of the study
Comparability: comparability of cohorts on the basis of design or analysis
Outcomes: assessment of outcomes, duration of follow‐up, and adequacy of follow‐up
Measures of treatment effect
We analysed the data using Review Manager 5 (RevMan 5) (RevMan 2014). We calculated the odds ratio (OR) for binary data, the weighted mean difference (WMD) for continuous data measured on the same scale, and the standard mean difference (SMD) for continuous data measured on different scales. We presented these results along with 95% confidence interval (CI) values.
Unit of analysis issues
The unit of analysis was the individual. We did not anticipate finding any cluster trials or cross‐over trials.
Dealing with missing data
For missing or unclear data, we contacted the authors of included studies during the eligibility assessment and data extraction stages. We also sought missing data from secondary publications of the same study. In the event that we were unable to obtain the missing data, we conducted a complete‐case analysis.
Assessment of heterogeneity
We first assessed included studies for clinical heterogeneity. If studies were similar enough that we could combine them (with regards to participants, interventions, comparisons, and outcomes), we performed a meta‐analysis and assessed statistical heterogeneity using the Chi² test for homogeneity with a level of significance of alpha = 0.10 and the I² statistic to quantify inconsistency.
Assessment of reporting biases
We assessed reporting bias (selective outcome reporting) using the Cochrane ‘Risk of bias' assessment tool (Appendix 4). We planned to assess publication bias using a funnel plot if 10 or more studies met the inclusion criteria (Higgins 2011).
Data synthesis
We performed a random‐effects meta‐analysis to synthesize sufficiently similar quantitative data. We pooled the results of included studies to determine the odds ratio or the mean difference. We did not intend to pool data from randomized and non‐randomized studies. We planned to conduct analyses per exposure (i.e. data from treatment‐naive and treatment‐experienced people would be analysed separately).
Reporting of change in HIV viral load and CD4 cell count varied across studies; therefore we performed calculations to obtain standard deviations from P values and pooled CIs from group means, as recommended in the Cochrane Handbook for Systematic Reviews of Interventions (Higgins 2011). We converted standard errors and CIs to standard deviations in RevMan 5 (RevMan 2014). We estimated means from medians when sample sizes were greater than 25, which has been shown to be a suitable estimate even in skewed distributions (Hozo 2005). We computed virological failure as the inverse of virological success to permit pooling across studies.
Certainty of the evidence
We assessed the certainty of the body of evidence using the GRADE approach (GRADEpro 2015), which defines the certainty of evidence for each outcome as the extent to which one can be confident that an estimate of effect or association is close to the quantity of specific interest. The certainty rating across studies has four levels: high, moderate, low, or very low. RCTs are categorized as high certainty evidence but can be downgraded; similarly, other types of controlled trials and observational studies are categorized as low certainty but can be upgraded. Factors that decrease the certainty of the evidence include limitations in design, indirectness of evidence, unexplained heterogeneity or inconsistency of results, imprecision of results, or high probability of publication bias. Factors that can increase the certainty level of a body of evidence include having a large magnitude of effect, whether plausible confounding would reduce a demonstrated effect, and if there is a dose‐response gradient (Guyatt 2011). We used the GRADEpro Guideline Development Tool (GDT) software to produce ‘Summary of findings' tables (GRADEpro 2015).
Subgroup analysis and investigation of heterogeneity
We planned to conduct subgroup analyses separately for studies that included treatment‐naive and treatment‐experienced people living with HIV. We prespecified the following subgroups.
Potency of ART used (NNRTI‐ or PI‐based regimens)
Type of resistance testing used (genotype or phenotype)
Level of advancement of disease (Centers for Disease Control and Prevention (CDC) or WHO stage)
Expert interpretation (use of expert advice to guide interpretation of resistance testing results)
Age (children versus adults)
We hypothesized that studies with more potent ART, more sophisticated resistance testing techniques, and patients at early stages of the disease whose choice of regimen is supported by expert advice would have relatively better outcomes with resistance testing compared to no resistance testing. Likewise, we expected that the benefits derived by ART‐naive patients would be better because these patients have a greater variety of potent drugs to choose from after completing resistance testing; patients in settings with a higher population‐level resistance rate would also experience greater benefit from resistance testing before therapy was initiated. We restricted subgroup analyses to those most relevant and those for which we had data. We interpreted subgroup results based on results of the between‐subgroups Chi² interaction test.
Sensitivity analysis
We planned to undertake a sensitivity analysis to evaluate bias introduced by variability in study design (observational versus randomized) and risk of bias.
Results
Description of studies
Results of the search
We conducted literature searches up to 26 January 2018, which yielded 3003 titles. Three review authors (TA, JT, and RS) independently screened the titles, abstracts, and descriptor terms of all material downloaded from the electronic searches to identify potentially relevant studies. After removing duplicates, we screened 2067 titles and abstracts, of which we excluded 2037 studies. We obtained the full‐text articles of 30 potentially relevant or uncertain reports. Eleven of them met our inclusion criteria. We excluded 19 after full‐text review. We have outlined our screening and selection process in Figure 1.
1.
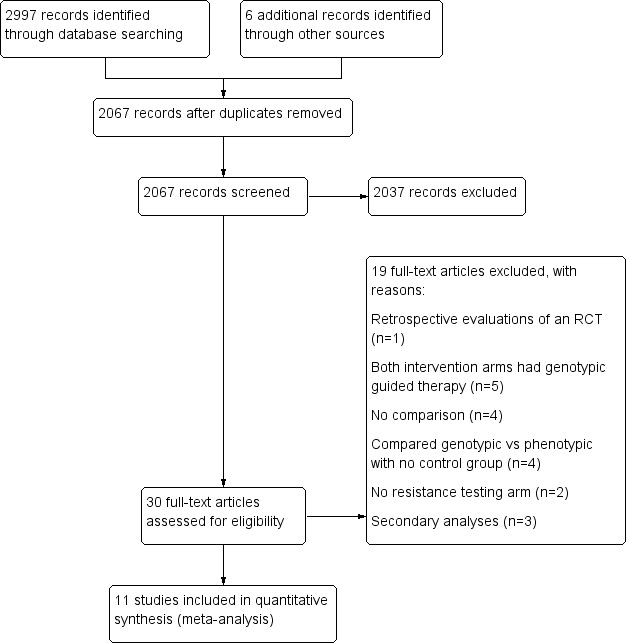
PRISMA study flow diagram.
Included studies
Eleven RCTs met the inclusion criteria of this Cochrane Review (Baxter 2000; Cingolani 2002; Cohen 2002; Dunn 2005; Durant 1999; Green 2006; Haubrich 2005; Meynard 2002; Rubini 2002; Tural 2002; Wegner 2004). The findings reported here are from papers or conference abstracts published between 1999 and 2005. Two study authors provided additional unpublished data (Dunn 2005; Tural 2002). See the Characteristics of included studies table.
Locations of studies
One trial was a multi‐national trial conducted in six countries: Italy, Brazil, UK, Spain, Germany, and Portugal (Green 2006). Four trials were conducted in the USA (Baxter 2000; Cohen 2002; Haubrich 2005; Wegner 2004); two in France (Durant 1999; Meynard 2002); and one each in Italy (Cingolani 2002), Spain (Tural 2002), Brazil (Rubini 2002), and UK (Dunn 2005).
Study participants
All 11 trials exclusively included participants who were treatment‐experienced and whose therapy was failing (Baxter 2000; Cingolani 2002; Cohen 2002; Dunn 2005; Durant 1999; Green 2006; Haubrich 2005; Meynard 2002; Rubini 2002; Tural 2002; Wegner 2004). One study included children exclusively (three months to 18 years of age; Green 2006). Two studies included both adolescents and adults (13 years of age or older; Baxter 2000; Cohen 2002). Another study included children and adolescents (Rubini 2002). One study did not report participant age (Dunn 2005). Remaining studies included adults only (Durant 1999; Haubrich 2005; Meynard 2002; Tural 2002; Wegner 2004).
Interventions provided
Seven trials compared genotypic testing versus usual care (Baxter 2000; Cingolani 2002; Dunn 2005; Durant 1999; Green 2006; Rubini 2002; Tural 2002). Two trials compared phenotypic testing versus usual care (Cohen 2002; Haubrich 2005). Two trials compared genotypic and phenotypic testing versus usual care (Meynard 2002; Wegner 2004). Three trials included expert advice on interpretation of the resistance tests (Baxter 2000; Cingolani 2002; Tural 2002).
Outcomes reported
Six trials reported mortality (Baxter 2000; Durant 1999; Green 2006; Meynard 2002; Tural 2002; Wegner 2004); 10 reported virological success (Baxter 2000; Cingolani 2002; Cohen 2002; Dunn 2005; Durant 1999; Green 2006; Haubrich 2005; Meynard 2002; Rubini 2002; Tural 2002); eight reported change in mean CD4 count (Baxter 2000; Cingolani 2002; Cohen 2002; Dunn 2005; Durant 1999; Green 2006; Haubrich 2005; Meynard 2002); and three reported clinical progression to AIDS (Durant 1999; Green 2006; Meynard 2002). No trials reported on the development of a second or new opportunistic infection or on quality of life. Four trials reported any adverse events (Baxter 2000; Cohen 2002; Durant 1999; Tural 2002). Additional outcomes reported in 10 trials included change in viral load (Baxter 2000; Cingolani 2002; Cohen 2002; Dunn 2005; Durant 1999; Green 2006; Haubrich 2005; Meynard 2002; Rubini 2002; Tural 2002). One study reported time to persistent treatment failure (Wegner 2004).
Length of follow‐up
The shortest length of follow‐up was 12 weeks (Baxter 2000; Cingolani 2002), and the longest was 150 weeks (Wegner 2004). One trial ran for 16 weeks (Cohen 2002); three for 24 weeks (Durant 1999; Rubini 2002; Tural 2002); one for 36 weeks (Meynard 2002); two for 52 weeks (Dunn 2005; Haubrich 2005); and one for 96 weeks (Green 2006). The median follow‐up time was 24 weeks (quartile 1 = 16; quartile 3 = 52).
We have provided further details on the included studies in an additional table (Table 2).
1. Additional characteristics of included studies.
| Trial ID (acronym) | Location | Exposure criteria | Definition of virological failure at entry | Type of resistance testing compared to control | Primary virological success cutoff point (copies/mL) | Expert opinion | Duration of follow‐up (weeks) |
| Baxter 2000 | USA | A combination antiretroviral regimen containing a single PI (indinavir, nelfinavir, saquinavir, or ritonavir) and 2 NRTIs |
Four conditions: (1) patient taking a current triple‐drug regimen for at least 16 weeks; (2) a locally determined screening HIV‐1‐RNA level > 20,000 copies/mL by the Roche Amplicor HIV‐1 assay or > 10,000 copies/mL by the Chiron branched chain (bDNA) assay within 6 weeks before a required baseline visit; (3) documentation that the screening HIV‐1‐RNA level was 3‐fold greater than the nadir HIV‐1‐RNA level while on the triple‐drug regimen, or that the nadir was < 500 copies/mL; and (4) a centrally determined HIV‐1‐RNA level > 5000 copies/mL by the Chiron 2.0 bDNA assay using plasma collected at the baseline visit | Genotype | NA | Yes | 12 |
| Cingolani 2002 (ARGENTA) | Italy | At least 2 months on a highly active antiretroviral regimen |
Two conditions: (1) plasma viral load > 2000 copies/mL in at least two consecutive determinations; or (2) < 1 log reduction in HIV RNA more than 2 months after the start of the last regimen | Genotype | < 500 | Yes | 12 |
| Cohen 2002 | USA | At least 2 NRTIs and only one PI, taken for at least 1 month before screening |
HIV‐1‐RNA plasma levels ≥ 2000 copies/mL | Phenotype | < 400 | No | 16 |
| Dunn 2005 (ERA) | UK | On ART | Most recent HIV‐1‐RNA plasma viral load (VL) exceeding 2000 copies/mL | Genotype | < 50 | No | 52 |
| Durant 1999 (VIRADAPT) | France | At least 6 months of treatment with nucleoside analogues and at least 3 months of treatment with a protease inhibitor |
Plasma HIV‐1‐RNA > 10,000 copies/mL | Genotype | < 200 | No | 24 |
| Green 2006 (PERA (PENTA 8)) | Italy, Brazil, UK, Spain, Germany, Portugal | Greater exposure than 2 or 3 nucleoside reverse transcriptase inhibitors (NRTIs) for < 2 years |
Most recent HIV‐1‐RNA plasma viral load > 2000 copies/mL | Genotype | < 50 | No | 96 |
| Haubrich 2005 (CCTG) | USA | At least 6 months of previous ART, exposure to no more than 2 prior protease inhibitors (PIs) |
HIV RNA > 400 copies/mL | Phenotype | < 400 | No | 52 |
| Meynard 2002 | USA | Previous exposure to at least 1 protease inhibitor (PI) for at least 3 months |
Plasma HIV‐1‐RNA > 1000 copies/mL | Genotype and Phenotype | < 200 | No | 36 |
| Rubini 2002 | Brazil | Previous exposure to at least 2 ART regimens, with failure of their current therapy | Not reported | Genotype | Not reported | No | 24 |
| Tural 2002 (Havana) | Spain | Stable antiretroviral therapy combination for longer than 6 months |
Plasma HIV‐1‐RNA ≥ 1000 copies/mL | Genotype | < 400 | Yes | 24 |
| Wegner 2004 | USA | Receiving a stable ART regimen containing ≥ 2 drugs for at least 8 weeks before randomization |
VL > 3.0 log₁₀ copies/mL concomitant with ≥ 1 of the following conditions: < 1.0 log₁₀ reduction in VL 4 weeks after start of a therapy regimen, failure to suppress VL to < 200 copies/mL 6 weeks after start of therapy, detection of plasma VL > 3.0 log₁₀ copies/mL after initial suppression to < 200 copies/mL, or increase of > 0.5 log₁₀ copies/mL (to > 3.0 log₁₀ copies/mL) from the nadir VL that could not be directly attributed to vaccination or intercurrent illness | Genotype and Phenotype | NA | No | 150 |
Abbreviations: ART: antiretroviral therapy; bDNA: branched DNA assay; NA: not applicable; NRTI: nucleoside reverse transcriptase inhibitor; PI: protease inhibitor; VL: viral load.
Excluded studies
We excluded 19 potentially relevant studies and summarized these in the Characteristics of excluded studies table. Five of these studies used genotypic guided therapy in both study arms (Bossi 2004; Clevenbergh 2000; Dunn 2005a; Gianotti 2006; Hales 2006); and four compared genotypic versus phenotypic testing and included no control arm (Mazzotta 2003; Perez‐Elias 2003; Saracino 2004; Torti 2005). One was a simulation study (Lorenzana 2012). Another compared strategies to interpret HIV genotypic resistance tests (Maggiolo 2007). Two were cross‐sectional studies of drug resistance rates (Pere 2012; Vergne 2003); three were secondary analyses of included studies with no additional outcome information (Clevenbergh 2002; De Luca 2006; Durant 2000); two did not include any relevant intervention arm (Bonjoch 2008; Demeter 2008); and one was a retrospective evaluation of an RCT (Badri 2003).
Risk of bias in included studies
We assessed the risk of bias of each included study using the Cochrane ‘Risk of bias' assessment tool (Appendix 4). We assessed risk of bias in individual trials across seven domains: sequence generation, allocation concealment, blinding of participants and personnel, blinding of outcome assessor, incomplete outcome data, selective outcome reporting, and other potential biases. We summarized these into one overall assessment of risk of bias. See the ‘Risk of bias' summary in Figure 2 and the ‘Risk of bias' graph in Figure 3.
2.
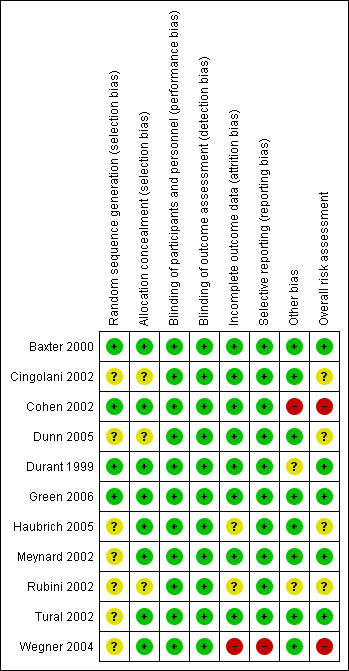
‘Risk of bias' summary: review authors' judgements about each ‘Risk of bias' item for each included study.
3.
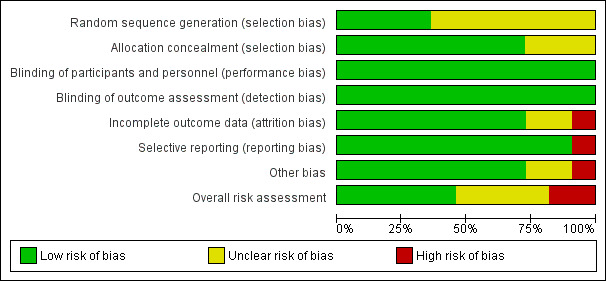
‘Risk of bias' graph: review authors' judgements about each ‘Risk of bias' item presented as percentages across all included studies.
Allocation
Generation of allocation sequence
Only four studies reported how the allocation sequence was generated (Baxter 2000; Cohen 2002; Durant 1999; Green 2006). For all other studies, the method of sequence generation was unclear (Cingolani 2002; Dunn 2005; Haubrich 2005; Meynard 2002; Rubini 2002; Tural 2002; Wegner 2004).
Allocation concealment
Eight studies reported appropriate methods of allocation concealment (Baxter 2000; Cohen 2002; Durant 1999; Green 2006; Haubrich 2005; Meynard 2002; Tural 2002; Wegner 2004). For the remaining three studies, allocation concealment was unclear (Cingolani 2002; Dunn 2005; Rubini 2002).
Blinding of participants and personnel
Nine trials were reported as open‐label or not blinded (Cingolani 2002; Cohen 2002; Dunn 2005; Durant 1999; Green 2006; Haubrich 2005; Meynard 2002; Tural 2002; Wegner 2004), and two studies did not mention blinding (Baxter 2000; Rubini 2002). Given the nature of the intervention, we judged that lack of blinding was unlikely to introduce bias.
Blinding of outcome assessors
No studies reported blinding of outcome assessors. Given the nature of measured outcomes, we judged that lack of blinding was unlikely to introduce bias.
Incomplete outcome data
Eight studies reported levels and causes of attrition. These were balanced between groups, and levels of attrition were low (< 20%; Baxter 2000; Cingolani 2002; Cohen 2002; Dunn 2005; Durant 1999; Green 2006; Meynard 2002; Tural 2002). We judged these studies to be at low risk of bias. One study was at high risk of bias for providing incomplete outcome data (Wegner 2004), and another study was at unclear risk of bias (Haubrich 2005). Rubini 2002 did not report exclusions by group.
Selective reporting
Ten studies reported all outcomes that could reasonably be expected (Baxter 2000; Cingolani 2002; Cohen 2002; Dunn 2005; Durant 1999; Green 2006; Haubrich 2005; Meynard 2002; Rubini 2002; Tural 2002). We judged these studies to be at low risk of bias for selective reporting. One study was at high risk of bias for selective reporting (Wegner 2004).
Other potential sources of bias
Funding
Eight studies reported government or private funding (Baxter 2000; Cingolani 2002; Dunn 2005; Green 2006; Haubrich 2005; Meynard 2002; Tural 2002; Wegner 2004). We judged these studies to be at low risk of bias. One study was funded by a manufacturer of resistance tests (Cohen 2002), and two did not report funding (Green 2006; Rubini 2002). We judged these studies to be at high and unclear risk of bias, respectively.
Publication bias
We assessed publication bias for virological failure using a funnel plot, which appeared asymmetrical on visual inspection (Figure 4). We also ran Egger's regression test and confirmed this asymmetry (P < 0.001) (Egger 1997).
4.
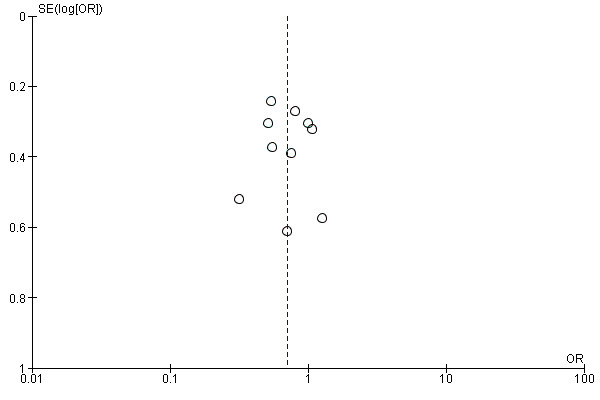
Funnel plot of comparison: 1 Resistance testing versus no resistance testing, outcome: 1.2 Virological failure.
Effects of interventions
See: Table 1
See Table 1.
Resistance testing versus no resistance testing
Mortality
Researchers reported no differences in mortality between the group that received resistance testing and the group that did not (odds ratio (OR) 0.89, 95% CI 0.36 to 2.22; 5 trials, 1140 participants; P = 0.800; Analysis 1.1; Figure 5).
1.1. Analysis.
Comparison 1 Resistance testing (RT) versus no RT, Outcome 1 Mortality.
5.
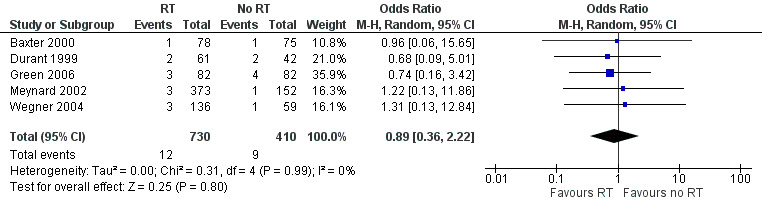
Forest plot of comparison: 1 Resistance testing (RT) versus no RT, outcome: 1.1 Mortality.
Virological failure
Virological failure was less likely in the group that received resistance testing than in the group that did not (OR 0.70, 95% CI 0.56 to 0.87; 10 trials, 1728 participants; P < 0.001; Analysis 1.2; Figure 6).
1.2. Analysis.
Comparison 1 Resistance testing (RT) versus no RT, Outcome 2 Virological failure.
6.
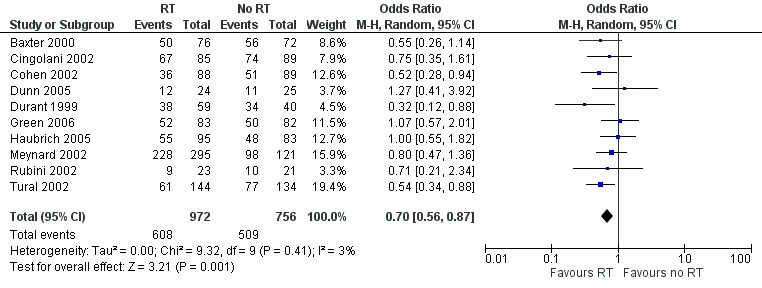
Forest plot of comparison: 1 Resistance testing (RT) versus no RT, outcome: 1.2 Virological failure.
Change in CD4 cell count
The change in CD4 cell count was similar in the group that received resistance testing and in the group that did not (mean difference (MD) ‐1.00 cells/mm³, 95% CI ‐12.49 to 10.50; 7 trials, 1349 participants; P = 0.860; Analysis 1.3; Figure 7).
1.3. Analysis.
Comparison 1 Resistance testing (RT) versus no RT, Outcome 3 Change in CD4 cell count.
7.
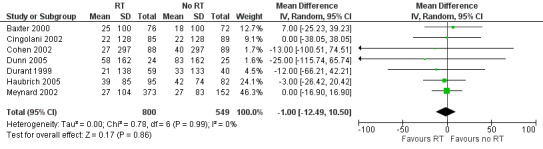
Forest plot of comparison: 1 Resistance testing (RT) versus no RT, outcome: 1.3 Change in CD4 cell count.
Progression to AIDS
Data show no differences in the proportion of participants who progressed to AIDS in the group that received resistance testing compared to the group that did not (OR 0.64, 95% CI 0.31 to 1.29; 3 trials, 809 participants; P = 0.210; Analysis 1.4; Figure 8)
1.4. Analysis.
Comparison 1 Resistance testing (RT) versus no RT, Outcome 4 Progression to AIDS.
8.
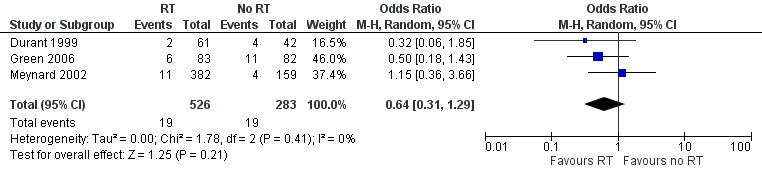
Forest plot of comparison: 1 Resistance testing (RT) versus no RT, outcome: 1.4 Progression to AIDS.
Adverse events
Adverse events were comparable between groups (OR 0.89, 95% CI 0.51 to 1.55; 4 trials, 808 participants; P = 0.670; Analysis 1.5; Figure 9).
1.5. Analysis.
Comparison 1 Resistance testing (RT) versus no RT, Outcome 5 Adverse events.
9.
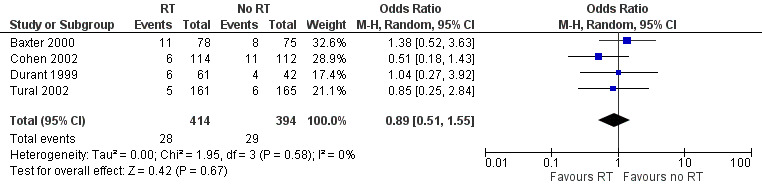
Forest plot of comparison: 1 Resistance testing (RT) versus no RT, outcome: 1.5 Adverse events.
Change in viral load
Change in log₁₀ viral load was greater in the group that received resistance testing (MD ‐0.23, 95% CI ‐0.35 to ‐0.11; 10 trials, 1837 participants; P < 0.001; Analysis 1.6; Figure 10).
1.6. Analysis.
Comparison 1 Resistance testing (RT) versus no RT, Outcome 6 Change in viral load.
10.
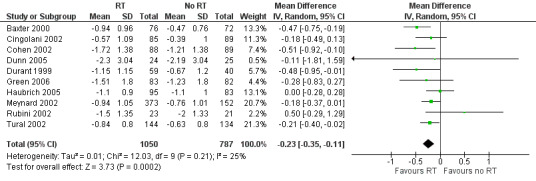
Forest plot of comparison: 1 Resistance testing (RT) versus no RT, outcome: 1.6 Change in viral load.
No studies reported on development of new opportunistic infections or on quality of life.
Subgroup analyses
Statistical heterogeneity was low in all primary analyses, never exceeding I² = 25%. None of the preplanned subgroup analyses revealed any statistically significant subgroup effects for any reported outcomes.
Potency of ART
Data were insufficient to show the potency of ART as a possible effect modifier. Included studies used a wide variety of approaches to describe the potency of ART, including the impact of resistance testing on the choice of ART (Cohen 2002; Durant 1999), the number of new drugs prescribed (Baxter 2000; Dunn 2005), the number of potent drugs given (Meynard 2002), the proportions of participants who received potent drugs (Haubrich 2005), and the prevalence of drug resistance mutations (Cingolani 2002).
Type of resistance test
We noted no subgroup effects when we compared genotypic resistance tests to phenotypic resistance tests for the outcomes of mortality (Analysis 2.1) and virological failure (Analysis 2.2). Contrary to the pooled results, we noted no differences in virological failure in the subgroup of studies that used phenotypic tests. This is likely due to the fact that studies lacked the power needed to detect a difference among the three studies (371 participants) that used phenotypic tests.
2.1. Analysis.
Comparison 2 RT versus no RT: subgroup analyses for type of RT, Outcome 1 Mortality.
2.2. Analysis.
Comparison 2 RT versus no RT: subgroup analyses for type of RT, Outcome 2 Virological failure.
Use of expert advice
We saw no subgroup effects when we compared the addition of expert advice to resistance tests versus resistance testing alone for the outcomes of mortality (Analysis 3.1) and virological failure (Analysis 3.2). However, no difference in virological failure could be seen among studies that did not use expert advice.
3.1. Analysis.
Comparison 3 RT versus no RT: subgroup analyses for expert advice, Outcome 1 Mortality.
3.2. Analysis.
Comparison 3 RT versus no RT: subgroup analyses for expert advice, Outcome 2 Virological failure.
Children versus adults
We observed no subgroup effects when we compared trials that exclusively enrolled children or adolescents versus those that exclusively enrolled adults for the outcomes of mortality (Analysis 4.1) and virological failure (Analysis 4.2). However, studies that included children reported no differences in virological failure.
4.1. Analysis.
Comparison 4 RT versus no RT: subgroup analyses for age, Outcome 1 Mortality.
4.2. Analysis.
Comparison 4 RT versus no RT: subgroup analyses for age, Outcome 2 Virological failure.
Level of advancement of disease
Data were insufficient to show the potency of the level of advancement of disease as a possible effect modifier.
Sensitivity analyses
Risk of bias
Mortality was comparable between studies at high or unclear risk of bias and those at low risk of bias (Analysis 5.1). Results show no significant differences in virological failure between studies at high or unclear risk of bias and those at low risk of bias (Analysis 5.2).
5.1. Analysis.
Comparison 5 RT versus no RT: sensitivity analyses for risk of bias (RoB), Outcome 1 Mortality.
5.2. Analysis.
Comparison 5 RT versus no RT: sensitivity analyses for risk of bias (RoB), Outcome 2 Virological failure.
Discussion
Summary of main results
See Table 1. Eleven RCTs, which included 2531 participants, met the inclusion criteria of this review. All participants were experiencing failing ART (none were starting ART for the first time). We found that resistance testing probably has little or no impact on mortality (moderate‐certainty evidence) nor on progression to AIDS (moderate‐certainty evidence). Resistance testing probably may reduce the risk of virological failure (low‐certainty evidence) and reduces mean HIV viral load (moderate‐certainty evidence). It probably has little or no effect on change in CD4 cell count (moderate‐certainty evidence) nor on adverse events (low‐certainty evidence). Data show no serious inconsistency: we did not find any statistically significant heterogeneity for any outcomes, and the I² statistic ranged from 0 to 25%. Effects of resistance testing were similar by type of resistance test (genotypic versus phenotypic), the addition of expert advice to interpretation of resistance tests, and age. Results for mortality were also consistent in all subgroups.
Overall completeness and applicability of evidence
We identified literature suggesting that resistance testing may be beneficial for people with HIV who are experiencing failing ART. Findings of this Cochrane Review are dominated by data from the USA ‐ Baxter 2000,Cohen 2002,Haubrich 2005,Meynard 2002,Wegner 2004 ‐ and from other high‐income countries such as Italy (Cingolani 2002), UK (Dunn 2005), France (Durant 1999), and Spain (Tural 2002). One study included participants from Italy, Brazil, UK, Spain, Germany, and Portugal (Green 2006). Only two studies included children (Green 2006; Rubini 2002). No studies included treatment‐naive people living with HIV. Therefore, data from low‐income countries and on children and people initiating therapy are lacking. We note that all of these trials were conducted more than 10 years ago, and findings from this review should be interpreted with the understanding that novel antiretroviral drugs lead to more favourable outcomes in patients whose treatment is not informed by a resistance test. Integrase strand inhibitors, especially dolutegravir and bictegravir, are much more potent and have a higher barrier to resistance when compared with most of the non‐nucleoside reverse transcriptase inhibitors (NNRTIs) and early‐generation protease inhibitors (e.g. lopinavir) that were used in the RCTs included in this review (Tsiang 2016). Thus, whether or not resistance testing has a similar impact on viral outcomes among patients starting integrase inhibitor‐based regimens is speculative. Owing to limited data, we were unable to explore the role of the potency of ART in this systematic review. We also acknowledge that newer algorithms for interpreting genotypic tests may be more sensitive (Desai 2007), and some patients with drug resistance included in the older studies may have been missed. These facts would bias results towards the null. Length of follow‐up in many studies was less than one year (median six months). Therefore, we cannot make inferences about the long‐term effects of treatment regimens informed by resistance tests.
Certainty of the evidence
The certainty of evidence for measured outcomes ranged from low to moderate. The main methodological concern of review authors was the lack of blinding noted in included studies. Most study authors declared that studies were open‐label or did not mention blinding. Resistance testing is a complex intervention that includes laboratory technicians, the attending physician(s), and the patient. Blinding, especially of the healthcare provider, may not be possible. Based on this, we did not downgrade for blinding when examining objective outcomes such as mortality and laboratory‐based measures (viral load, CD4 cell count). Many studies did not report how the allocation sequence was generated, and we had concerns about whether the allocation process was truly random. We found evidence of publication bias for the outcome of virological failure and downgraded for this.
Potential biases in the review process
We minimized biases in the review process by considering literature written in any language, by performing a comprehensive search of databases and conference proceedings, and by contacting experts in the field for unpublished and ongoing studies (as part of the consultative process for the development of World Health Organization (WHO) guidelines (WHO 2017a)). We screened articles and extracted data in duplicate. Even though we searched for observational studies, we found none. The absence of validated search strategies for observational studies in HIV precludes our ability to verify whether we missed any studies. Our assessment of publication bias for virological failure through visual inspection of a funnel plot and use of Egger's test suggested the presence of publication bias.
Agreements and disagreements with other studies or reviews
Our findings are comparable with those of three other systematic reviews. Torre 2002 found that undetectable viral load was more likely to be achieved at three months (odds ratio (OR) 1.7, 95% CI 1.3 to 2.2) and at six months (OR 1.6, 95% CI 1.2 to 2.2) if treatment was guided by genotypic or phenotypic resistance testing. This group included six trials (1471 participants) in this systematic review and found that expert guidance conferred additional benefits (OR 2.4, 95% CI 1.5 to 3.7). Ena 2006 included eight trials (1810 participants) and reported similar findings, with better virological success achieved with genotypic or phenotypic resistance testing (risk ratio (RR) 1.23, 95% CI 1.09 to 1.40) and better expert interpretation provided (RR 1.82, 95% CI 1.38 to 2.40). Panidou 2004, which included 10 trials (2258 participants), found that genotypic resistance testing increased the proportions of participants with undetectable viral load at three months by 11% (95% CI 6 to 16) and at six months by 10% (95% CI 5 to 16). All three systematic reviews focused on people who were experiencing failing treatment. We found no subgroup effects for expert advice, even though virological failure appeared to be less likely with expert advice (expert advice: OR 0.59, 95% CI 0.41 to 0.83; no expert advice: OR 0.77, 95% CI 0.57 to 1.04).
Minor differences between the findings of our review and of these three reviews may be explained by the number of included studies and the inclusion criteria applied. We found 11 trials, whereas the largest of the previous reviews included only 10 studies (Panidou 2004). In addition, authors of these three reviews explored head‐to‐head comparisons of different resistance tests (Panidou 2004).
Even though we sought to include data on treatment‐naive patients initiating therapy, we found no relevant studies.
Authors' conclusions
Implications for practice.
Resistance testing probably provides virological benefits (measured up to six months) for people who are experiencing failing therapy and switching to a non‐integrase inhibitor‐based antiretroviral therapy (ART) regimen. American and European guidelines already recommend HIV resistance testing for these patients (EACS 2017; Saag 2018). Even though these guidelines also recommend resistance testing in treatment‐naive people, we found no trials that included treatment‐naive people. However, inferences on how resistance testing will perform in treatment‐naive people can be drawn from these findings and from other systematic reviews demonstrating that the odds of virological failure were higher when HIV‐positive people with transmitted drug resistance (TDR) were started on ART without resistance testing (odds ratio (OR) 2.96, 95% CI 1.89 to 4.65; 12 observational studies; WHO 2017a). Another observational study reported improved survival among treatment‐experienced patients (hazard ratio (HR) 0.70, 95% CI 0.51 to 0.96) but not in treatment‐naive patients (HR 0.25, 95% CI 0.03 to 1.82) when resistance testing was used (Palella 2009). It is unclear if the benefits of testing may be greater for treatment‐naive patients, who would have more therapeutic options. However, the costs of resistance testing and the feasibility of widespread rollout in resource‐limited settings must be considered.
Implications for research.
Although additional trials would provide a more robust body of evidence, they are unlikely to be conducted, given that resistance testing after initiation of treatment and after treatment failure is already performed as part of routine care in some parts of the world. The major downside of this testing appears to involve costs and feasibility, as resistance testing is unlikely to cause harm. In treatment‐naive people, investigators should consider the ethical implications of randomizing participants to a "no resistance testing" arm, as well as the best strategies for incorporating other factors along the cascade of care that influence outcomes such as adherence to medication and potency of ART regimens. Studies using a non‐randomized design and administrative databases may provide answers to these questions.
Based on the findings of this review, we recommend further investigation of the role of resistance testing in treatment‐naive people with HIV and in subpopulations with a greater incidence of TDR, such as men who have sex with men (MSM), commercial sex workers (CSWs), and people who inject drugs (PWIDs). Those conducting randomized investigations should consider use of "delayed resistance testing" instead of "no resistance testing". Mortality is an important outcome that future studies are encouraged to report, in addition to virological outcomes and quality of life. However, resistance testing is unlikely to lead to reduced mortality in the short term ‐ longer‐term large studies would be needed to enhance our understanding of whether improvement in the surrogate measure (viral load) leads to improvements in outcomes that patients are likely to consider important, such as mortality. We also recommend that studies be conducted in low‐resource settings. Given the prohibitive costs of resistance testing, which preclude its widespread use, economic analyses are imperative for low‐income settings before such testing is widely adopted in these places.
Acknowledgements
The Academic Editor of this review was Dr Nathan Ford.
We developed this review as part of the mentoring requirements for the Aubrey Sheiham Evidence‐Based Health Care in Africa Leadership Award, which is administered by Cochrane South Africa at the South African Medical Research Council. LM is mentoring JT, TA, and RS.
The editorial base of the Cochrane Infectious Diseases Group is funded by UK aid from the UK government for the benefit of low‐ and middle‐income countries (project number 300342‐104). The views expressed do not necessarily reflect the UK government’s official policies.
We acknowledge the support of Joy Oliver, Chief Officer at Cochrane South Africa, South African Medical Research Council for developing the search strategy and running the searches.
We are grateful to the study investigators who shared unpublished data.
Appendices
Appendix 1. CENTRAL search strategy
| ID | Search |
| #1 | MeSH descriptor: [HIV Infections] explode all trees |
| #2 | MeSH descriptor: [HIV] explode all trees |
| #3 | hiv or hiv‐1* or hiv‐2* or hiv1 or hiv2 or (hiv near infect*) or (human immunodeficiency virus) or (human immunedeficiency virus) or (human immune‐deficiency virus) or (human immuno‐deficiency virus) or (human immune deficiency virus) or (human immuno deficiency virus) or (acquired immunodeficiency syndrome) or (acquired immunedeficiency syndrome) or (acquired immuno‐deficiency syndrome) or (acquired immune‐deficiency syndrome) or (acquired immun* deficiency syndrome) |
| #4 | MeSH descriptor: [Lymphoma, AIDS‐Related] this term only |
| #5 | MeSH descriptor: [Sexually Transmitted Diseases, Viral] this term only |
| #6 | #1 or #2 or #3 or #4 or #5 |
| #7 | [mh ^genotype] or genotype:ti,ab,kw or [mh "genotypic techniques"] or genotypic:ti,ab,kw or genotyping:ti,ab,kw or genotypical:ti,ab,kw (Word variations have been searched) |
| #8 | [mh ^phenotype] or phenotype:ti,ab,kw or phenotypic:ti,ab,kw or phenotyping:ti,ab,kw or phenotypical:ti,ab,kw (Word variations have been searched) |
| #9 | (resistance or resistant):ti,ab,kw (Word variations have been searched) |
| #10 | #7 or #8 or #9 |
| #11 | (test or tests or tested or testing or assay or assays):ti,ab,kw (Word variations have been searched) |
| #12 | #10 and #11 |
| #13 | [mh "drug resistance"] or resistance:ti,ab,kw or resistant:ti,ab,kw (Word variations have been searched) |
| #14 | #6 and #12 and #13 Publication Year from 1989 to xxx, in Trials |
Appendix 2. PubMed search strategy
| Search | Query |
| #11 | Search (((#1 AND #7 AND #8 AND #9))) AND ("1989/01/01"[Date ‐ Publication] : "xxxx/xx/xx"[Date ‐ Publication]) |
| #10 | Search (#1 AND #7 AND #8 AND #9) |
| #9 | Search (randomized controlled trial [pt] OR controlled clinical trial [pt] OR randomized [tiab] OR placebo [tiab] OR drug therapy [sh] OR randomly [tiab] OR trial [tiab] OR groups [tiab] OR cohort studies[mh:noexp] OR cohort[tiab] OR longitudinal studies[mh:noexp] OR longitudinal[tiab] OR follow‐up studies[mh:noexp] OR follow‐up[tiab] OR followup[tiab] OR prospective studies[mh:noexp] OR prospective[tiab] OR retrospective studies[mh:noexp] OR retrospective[tiab] OR epidemiologic studies[mh:noexp]) NOT (animals [mh] NOT humans [mh]) |
| #8 | Search (drug resistance[mh] OR resistance[tiab] OR resistant[tiab]) |
| #7 | Search (#5 AND #6) |
| #6 | Search (test[tiab] OR tests[tiab] OR testing[tiab] OR tested[tiab] OR assay[tiab] OR assays[tiab]) |
| #5 | Search (#2 OR #3 OR #4) |
| #4 | Search (resistance[tiab] OR resistant[tiab]) |
| #3 | Search (phenotype[mh:noexp] OR phenotype[tiab] OR phenotypic[tiab] OR phenotyping[tiab] OR phenotypical[tiab]) |
| #2 | Search (genotype[mh:noexp] OR genotypic techniques[mh] genotype[tiab] OR genotypic[tiab] OR genotyping[tiab] OR genotypical[tiab]) |
| #1 | Search (HIV Infections[MeSH] OR HIV[MeSH] OR hiv[tiab] OR hiv‐1*[tiab] OR hiv‐2*[tiab] OR hiv1[tiab] OR hiv2[tiab] OR hiv infect*[tiab] OR human immunodeficiency virus[tiab] OR human immunedeficiency virus[tiab] OR human immuno‐deficiency virus[tiab] OR human immune‐deficiency virus[tiab] OR ((human immun*[tiab]) AND (deficiency virus[tiab])) OR acquired immunodeficiency syndrome[tiab] OR acquired immunedeficiency syndrome[tiab] OR acquired immuno‐deficiency syndrome[tiab] OR acquired immune‐deficiency syndrome[tiab] OR ((acquired immun*[tiab]) AND (deficiency syndrome[tiab])) |
Appendix 3. Embase search strategy
| No. | Query |
| #17 | #1 AND #7 AND #8 AND #16 |
| #16 | #11 NOT #15 |
| #15 | #12 NOT #14 |
| #14 | #12 AND #13 |
| #13 | 'human'/de OR 'normal human'/de OR 'human cell'/de |
| #12 | 'animal'/de OR 'animal experiment'/de OR 'invertebrate'/de OR 'animal tissue'/de OR 'animal cell'/de OR 'nonhuman'/de |
| #11 | #9 OR #10 |
| #10 | 'randomized controlled trial'/de OR 'randomized controlled trial' OR random*:ab,ti OR trial:ti OR allocat*:ab,ti OR factorial*:ab,ti OR placebo*:ab,ti OR assign*:ab,ti OR volunteer*:ab,ti OR 'crossover procedure'/de OR 'crossover procedure' OR 'double‐blind procedure'/de OR 'double‐blind procedure' OR 'single‐blind procedure'/de OR 'single‐blind procedure' OR (doubl* NEAR/3 blind*):ab,ti OR (singl*:ab,ti AND blind*:ab,ti) OR crossover*:ab,ti OR cross+over*:ab,ti OR (cross NEXT/1 over*):ab,ti |
| #9 | 'prospective study'/de OR prospective:ab,ti OR 'cohort analysis'/de OR cohort:ab,ti OR 'longitudinal study' OR longitudinal:ab,ti OR 'experimental design'/de OR 'retrospective study'/de OR retrospective:ab,ti OR 'follow up'/de OR 'follow+up':ab,ti OR followup:ab,ti |
| #8 | 'drug resistance'/exp OR resistance:ab,ti OR resistant:ab,ti |
| #7 | #5 AND #6 |
| #6 | test:ab,ti OR tests:ab,ti OR testing:ab,ti OR tested:ab,ti OR assay:ab,ti OR assays:ab,ti |
| #5 | #2 OR #3 OR #4 |
| #4 | resistance:ab,ti OR resistant:ab,ti |
| #3 | 'phenotype'/de OR phenotype:ab,ti OR phenotypic:ab,ti OR phenotyping:ab,ti OR genotypical:ab,ti |
| #2 | 'genotyping technique'/de OR genotype:ab,ti OR genotypic:ab,ti OR genotyping:ab,ti OR genotypical:ab,ti |
| #1 | 'human immunodeficiency virus infection'/exp OR 'human immunodeficiency virus'/exp OR 'human immunodeficiency virus':ab,ti OR 'human immuno+deficiency virus':ab,ti OR 'human immunedeficiency virus':ab,ti OR 'human immune+deficiency virus':ab,ti OR hiv:ab,ti OR 'hiv‐1':ab,ti OR 'hiv‐2':ab,ti OR 'acquired immunodeficiency syndrome':ab,ti OR 'acquired immuno+deficiency syndrome':ab,ti OR 'acquired immunedeficiency syndrome':ab,ti OR 'acquired immune+deficiency syndrome':ab,ti |
Appendix 4. Cochrane ‘Risk of bias' tool
| Domain | Description | Review authors’ judgement |
| Sequence generation | Describe the method used to generate the allocation sequence in sufficient detail to allow an assessment of whether it should produce comparable groups. | Was the allocation sequence adequately generated? |
| Allocation concealment | Describe the method used to conceal the allocation sequence in sufficient detail to determine whether intervention allocations could have been foreseen in advance of, or during, enrolment. | Was allocation adequately concealed? |
| Blinding of participants, personnel, and outcome assessors Assessments should be made for each main outcome (or class of outcomes) | Describe all measures used, if any, to blind study participants and personnel from knowledge of which intervention a participant received. Provide any information related to whether the intended blinding was effective. | Was knowledge of the allocated intervention adequately prevented during the study? |
| Incomplete outcome data Assessments should be made for each main outcome (or class of outcomes) | Describe the completeness of outcome data for each main outcome, including attrition and exclusions from analysis. State whether attrition and exclusions were reported, the numbers in each intervention group (compared with total randomized participants), reasons for attrition/exclusions when reported, and any re‐inclusions in analyses performed by the review authors. | Were incomplete outcome data adequately addressed? |
| Selective outcome reporting | State how the possibility of selective outcome reporting was examined by the review authors and what was found. | Are reports of the study free of the suggestion of selective outcome reporting? |
| Other sources of bias | State any important concerns about bias not addressed in the other domains in the tool. If particular questions/entries were prespecified in the review protocol, responses should be provided for each question/entry. |
Was the study apparently free of other problems that could put it at high risk of bias? |
Data and analyses
Comparison 1. Resistance testing (RT) versus no RT.
| Outcome or subgroup title | No. of studies | No. of participants | Statistical method | Effect size |
|---|---|---|---|---|
| 1 Mortality | 5 | 1140 | Odds Ratio (M‐H, Random, 95% CI) | 0.89 [0.36, 2.22] |
| 2 Virological failure | 10 | 1728 | Odds Ratio (M‐H, Random, 95% CI) | 0.70 [0.56, 0.87] |
| 3 Change in CD4 cell count | 7 | 1349 | Mean Difference (IV, Random, 95% CI) | 1.00 [‐12.49, 10.50] |
| 4 Progression to AIDS | 3 | 809 | Odds Ratio (M‐H, Random, 95% CI) | 0.64 [0.31, 1.29] |
| 5 Adverse events | 4 | 808 | Odds Ratio (M‐H, Random, 95% CI) | 0.89 [0.51, 1.55] |
| 6 Change in viral load | 10 | 1837 | Mean Difference (IV, Random, 95% CI) | ‐0.23 [‐0.35, ‐0.11] |
Comparison 2. RT versus no RT: subgroup analyses for type of RT.
| Outcome or subgroup title | No. of studies | No. of participants | Statistical method | Effect size |
|---|---|---|---|---|
| 1 Mortality | 5 | 1537 | Odds Ratio (M‐H, Random, 95% CI) | 0.86 [0.37, 1.99] |
| 1.1 Genotype | 5 | 890 | Odds Ratio (M‐H, Random, 95% CI) | 0.85 [0.33, 2.21] |
| 1.2 Phenotype | 2 | 647 | Odds Ratio (M‐H, Random, 95% CI) | 0.86 [0.14, 5.35] |
| 2 Virological failure | 10 | 1601 | Odds Ratio (M‐H, Random, 95% CI) | 0.68 [0.55, 0.85] |
| 2.1 Genotype | 8 | 1230 | Odds Ratio (M‐H, Random, 95% CI) | 0.67 [0.52, 0.87] |
| 2.2 Phenotype | 3 | 371 | Odds Ratio (M‐H, Random, 95% CI) | 0.72 [0.45, 1.17] |
Comparison 3. RT versus no RT: subgroup analyses for expert advice.
| Outcome or subgroup title | No. of studies | No. of participants | Statistical method | Effect size |
|---|---|---|---|---|
| 1 Mortality | 5 | 1140 | Odds Ratio (M‐H, Random, 95% CI) | 0.89 [0.36, 2.22] |
| 1.1 Expert advice | 1 | 153 | Odds Ratio (M‐H, Random, 95% CI) | 0.96 [0.06, 15.65] |
| 1.2 No expert advice | 4 | 987 | Odds Ratio (M‐H, Random, 95% CI) | 0.88 [0.33, 2.32] |
| 2 Virological failure | 10 | 1728 | Odds Ratio (M‐H, Random, 95% CI) | 0.70 [0.56, 0.87] |
| 2.1 Expert advice | 3 | 600 | Odds Ratio (M‐H, Random, 95% CI) | 0.59 [0.41, 0.83] |
| 2.2 No expert advice | 7 | 1128 | Odds Ratio (M‐H, Random, 95% CI) | 0.77 [0.57, 1.04] |
Comparison 4. RT versus no RT: subgroup analyses for age.
| Outcome or subgroup title | No. of studies | No. of participants | Statistical method | Effect size |
|---|---|---|---|---|
| 1 Mortality | 5 | 1140 | Odds Ratio (M‐H, Random, 95% CI) | 0.89 [0.36, 2.22] |
| 1.1 Children | 1 | 164 | Odds Ratio (M‐H, Random, 95% CI) | 0.74 [0.16, 3.42] |
| 1.2 Adults | 4 | 976 | Odds Ratio (M‐H, Random, 95% CI) | 0.99 [0.31, 3.09] |
| 2 Virological failure | 10 | 1728 | Odds Ratio (M‐H, Random, 95% CI) | 0.70 [0.56, 0.87] |
| 2.1 Children | 2 | 209 | Odds Ratio (M‐H, Random, 95% CI) | 0.98 [0.56, 1.71] |
| 2.2 Adults | 8 | 1519 | Odds Ratio (M‐H, Random, 95% CI) | 0.66 [0.52, 0.84] |
Comparison 5. RT versus no RT: sensitivity analyses for risk of bias (RoB).
| Outcome or subgroup title | No. of studies | No. of participants | Statistical method | Effect size |
|---|---|---|---|---|
| 1 Mortality | 5 | Odds Ratio (M‐H, Random, 95% CI) | Subtotals only | |
| 1.1 Low RoB | 4 | 945 | Odds Ratio (M‐H, Random, 95% CI) | 0.83 [0.30, 2.24] |
| 1.2 High or unclear RoB | 1 | 195 | Odds Ratio (M‐H, Random, 95% CI) | 1.31 [0.13, 12.84] |
| 2 Virological failure | 10 | Odds Ratio (M‐H, Random, 95% CI) | Subtotals only | |
| 2.1 Low RoB | 5 | 1106 | Odds Ratio (M‐H, Random, 95% CI) | 0.66 [0.47, 0.92] |
| 2.2 High or unclear RoB | 5 | 622 | Odds Ratio (M‐H, Random, 95% CI) | 0.76 [0.55, 1.07] |
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Baxter 2000.
| Methods | A prospective randomized multi‐centre community‐based controlled trial at the Terry Beirn Community Programs for Clinical Research on AIDS (CPCRA) and the Walter Reed Army Medical Center in the USA | |
| Participants | 153 participants Inclusion criteria: at least 13 years old; experiencing virological failure on combination antiretroviral regimen containing a single protease inhibitor and 2 nucleoside reverse transcriptase inhibitors Exclusion criteria: use of an antiretroviral drug other than those in the qualifying regimen in the 16 weeks before the baseline visit; had access to previous genotypic or phenotypic test results |
|
| Interventions | Intervention: genotypic antiretroviral resistance testing (GART) (i.e. treatment choices based on the GART report, which included mutations identified; interpretation of drug susceptibilities, and treatment suggestions) Control: no GART (i.e. treatment choices made without any input) |
|
| Outcomes | Morality, virological success (< 500 copies/mL), change in viral load, change in CD4 cell count, adverse events Follow‐up: 12 weeks. |
|
| Notes | All participants provided informed consent for this study. This study was funded by the National Institute of Allergy and Infectious Diseases (NIAID). |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Participants were allocated via a stratified randomization schedule with permuted blocks. |
| Allocation concealment (selection bias) | Low risk | Central location randomization was conducted and was communicated to the investigators. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Losses to follow‐up were balanced between groups. Trial authors performed ITT analyses. |
| Selective reporting (reporting bias) | Low risk | Trial authors reported all outcomes of interest. |
| Other bias | Low risk | We found no other sources of bias. |
| Overall risk assessment | Low risk | We had no serious risk of bias concerns. |
Cingolani 2002.
| Methods | A randomized open controlled single‐centre study in Italy | |
| Participants | 174 participants Inclusion: at least 2 months on concomitant use of ≥ 3 antiretroviral agents and (1) a plasma viral load > 2000 copies/mL in at least 2 consecutive determinations; or (2) < 1 log reduction of HIV RNA more than 2 months after the start of the last regimen |
|
| Interventions | Intervention: genotypic resistance assay (i.e. treatment decisions based on the standard of care with additional information on a genotypic resistance assay and expert advice) Control: standard of care (SOC) (i.e. treatment decisions based on evaluation of treatment history, clinical picture, and standard immunological and virological parameters) |
|
| Outcomes | Virological success (< 500 copies/mL), change in viral load, change in CD4 cell count Follow‐up: 12 weeks |
|
| Notes | All participants provided informed consent. This study was funded by the National Research Program on AIDS, National Institute of Health, as part of the Research Project on Pathology, Clinics, and Therapeutics, with funds from the Institute for Research and Health Care, Italian Minister of Health, Italy. Trial acronym: ARGENTA (AntiRetroviral Genotypic resistance and patiENT‐reported Adherence) |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | The method of randomization is not reported. |
| Allocation concealment (selection bias) | Unclear risk | No information on allocation concealment is reported. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | This was an open‐label study (knowledge of allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Follow‐up was balanced between groups. Trial authors performed ITT analyses. |
| Selective reporting (reporting bias) | Low risk | Trial authors reported all outcomes of interest. |
| Other bias | Low risk | We found no other sources of bias. |
| Overall risk assessment | Unclear risk | Risk is due to the uncertainties raised above. |
Cohen 2002.
| Methods | A prospective open‐label randomized controlled clinical trial conducted at 25 university and private practice centres in the USA | |
| Participants | 272 participants Inclusion: at least 13 years of age; documented HIV‐1 infection; HIV‐ 1‐RNA plasma levels ≥ 2000 copies/mL; antiretroviral experience; experiencing virological failure on antiretroviral treatment consisting of ≥ 2 NRTIs and only 1 PI, taken for at least 1 month before screening Exclusion: history of alcohol or drug use judged as likely to interfere with therapy; previous phenotypic resistance testing; had participated in an antiretroviral drug trial within 30 days of selection or during the trial; had a life expectancy < 6 months; had disease that could interfere with assessments NB: Patients with previous NNRTI therapy were later allowed to participate. |
|
| Interventions | Intervention: phenotypic resistance testing (PRT) (i.e. antiretroviral regimens chosen with PRT and external expert advice coupled with access to published treatment guidelines and participants' treatment histories) Control: standard of care (SOC) (i.e. antiretroviral regimens chosen on the basis of access to published treatment guidelines and participants' treatment histories) |
|
| Outcomes | Virological success (< 400 copies/mL), change in viral load, change in CD4 cell count Follow‐up: 16 weeks |
|
| Notes | All participants provided informed consent. Study was funded by Glaxo Wellcome, Inc. (primary) and Virco NV. |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Participants were randomly assigned to intervention and control groups. |
| Allocation concealment (selection bias) | Low risk | Centrally located randomization was used with block size of 4. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | Arm assignments were not blinded (knowledge of allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Losses to follow‐up were balanced between groups. Trial authors performed ITT analyses. |
| Selective reporting (reporting bias) | Low risk | Trial authors reported all outcomes of interest. |
| Other bias | High risk | Study was supported by manufacturers of resistance tests. |
| Overall risk assessment | High risk | Risk is due to the uncertainties raised above. |
Dunn 2005.
| Methods | A 2‐part multi‐centre randomized trial (part B included patients for whom the clinician perceived he/she was unable to select a potent regimen with a resistance test; part A included patients for whom the clinician perceived he/she was able to select a potent regimen of 3 or more drugs without a resistance test ‐ only data from part A used here | |
| Participants | Inclusion: decision had been made to switch ART as a consequence of virological failure, provided the most recent HIV‐1 RNA plasma viral load (VL) exceeded 2000 copies/mL Exclusion: not reported |
|
| Interventions | Intervention: centralized genotypic resistance testing (i.e. a report that showed key drug‐associated mutations and classified each drug as “no evidence of resistance”, “resistance possible”, or “evidence of resistance") Control: no genotypic resistance testing |
|
| Outcomes | Change in viral load, virological success (< 50 copies/mL), change in CD4 cell count Follow‐up: 52 weeks |
|
| Notes | All participants provided informed consent. This study was funded by the NHS London Regional Office, Research and Development Programme. VIRCO (Mechelen, Belgium) provided resistance testing and transport of plasma samples to Belgium. Trial acronym: ERA (Evaluation of Resistance Assays) |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Method of randomization was not reported. |
| Allocation concealment (selection bias) | Unclear risk | Trial authors did not report this information. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Follow‐up was balanced between groups. Trial authors performed ITT analyses. |
| Selective reporting (reporting bias) | Low risk | Trial authors reported all outcomes of interest. |
| Other bias | Low risk | We found no other sources of bias. |
| Overall risk assessment | Unclear risk | Risk is due to the uncertainties raised above. |
Durant 1999.
| Methods | A prospective open randomized controlled study in 3 hospitals in southern France | |
| Participants | 108 participants Inclusion: older than 18 years of age, Karnofsky score > 50 and plasma HIV‐1 RNA > 10,000 copies/mL despite at least 6 months of treatment with nucleoside analogues and at least 3 months of treatment with a protease inhibitor Exclusion: foreseeable non‐compliance; haemoglobin concentration < 6 mmol/L; absolute neutrophil count < 0·8 × 10⁹/L; creatinine concentration > 200 μmol/L; liver aminotransferase values > 5 times the normal upper limit |
|
| Interventions | Intervention: genotypic resistance testing (i.e. treatment adapted with knowledge of genotypic test) Control: treatment based on current optimal care, according to published guidelines |
|
| Outcomes | Change in viral load, virological success (< 200 copies/mL), change in CD4 count Follow‐up: 24 weeks |
|
| Notes | All participants provided informed consent. Funding was not reported. Trial acronym: VIRADAPT (antiretroVIRal ADAPTation) |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Participants were randomly allocated to intervention or control by permutation table in blocks of 6. |
| Allocation concealment (selection bias) | Low risk | Allocation was concealed via opaque sealed envelopes. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | This was an open‐label study (knowledge of the allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Follow‐up was balanced between groups. Trial authors performed ITT analyses. |
| Selective reporting (reporting bias) | Low risk | Trial authors reported all outcomes of interest. |
| Other bias | Unclear risk | Funding was not reported. |
| Overall risk assessment | Low risk | We had no serious risk of bias concerns. |
Green 2006.
| Methods | An open randomized 2‐arm parallel‐group multi‐centre trial in UK and France | |
| Participants | 170 participants Inclusion: children 3 months to 18 years of age switching ART owing to virological failure, with their most recent HIV‐1 RNA plasma viral load > 2000 copies/mL Exclusion: exposure to only 2 or 3 nucleoside reverse transcriptase inhibitors (NRTIs) for < 2 years; changed ART in the last month before randomization; resistance test previously performed on the current regimen; paediatricians and primary caregivers unwilling to wait for a resistance test result before switching therapy |
|
| Interventions | Intervention: genotypic resistance test Control: no genotypic resistance test |
|
| Outcomes | Change in viral load, virological success (< 50 copies/mL), change in CD4 count Follow‐up: 96 weeks |
|
| Notes | All primary caregivers gave written consent to participate; additional written assent was obtained from children, when appropriate, according to their age and knowledge of their HIV status Funding/support: European Commission, supported by BIOMED 2; The Medical ResearchCouncil; The Agence Nationale de Recherche sur le Sida (ANRS); Istituto Superiore di Sanità ‐ Progetto Terapia Antivirale; Comunidad Autonoma de Madrid, Spain. PENTA activities are also supported by the PENTA Foundation. VIRCO (Mechelen, Begium) provided resistance testing and transport of plasma samples to Belgium. Trial acronym: PERA (PENTA 8) Paediatric Evaluation of Resistance Assays (Paediatric European Network for the Treatment of AIDS) |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Participants were randomly allocated to intervention or control groups, stratified by previous use of resistance testing and study centre. |
| Allocation concealment (selection bias) | Low risk | Central location randomization was conducted and was communicated by phone or fax. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | This was an open‐label trial (knowledge of the allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Losses to follow‐up were balanced between groups. Trial authors performed ITT analyses. |
| Selective reporting (reporting bias) | Low risk | All relevant outcome were reported. |
| Other bias | Low risk | We found no other sources of bias. |
| Overall risk assessment | Low risk | We had no serious risk of bias concerns. |
Haubrich 2005.
| Methods | A 7‐centre 2‐arm randomized clinical trial in the USA | |
| Participants | 238 participants Inclusion: at least 6 months of previous ART; exposure to no more than 2 prior protease inhibitors (PIs); failure of the current regimen (defined as HIV RNA > 400 copies/mL); a stable regimen (defined as no change for at least 4 weeks before screening) Exclusion: acute infections; active cancers; a resistance assay performed within 6 months; involvement in another investigational study that dictated the regimen |
|
| Interventions | Intervention: phenotypic testing (PHENO) (i.e. a phenotypic test in addition to a structured clinical history; standard of care (SOC) to guide selection of new and subsequent regimens) Control: a structured clinical history (SOC) guiding selection of new and subsequent regimens |
|
| Outcomes | Change in viral load, virological success (< 400 copies/mL) Follow‐up: 52 weeks |
|
| Notes | All participants provided informed consent. Funding: California University‐wide AIDS Research Program (UARP), UCSD Center for AIDS Research (CFAR), ViroLogic Inc., National Institutes of Health, and the Research Center for AIDS and HIV Infection of the San Diego Veterans Affairs Healthcare System Trial acronym: CCTG (California Collaborative Treatment Group) 575 |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Method of randomization was not reported. |
| Allocation concealment (selection bias) | Low risk | Randomization was performed centrally. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | This was an open‐label study (knowledge of allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | Unclear risk | Losses to follow‐up were considerable and were unbalanced between groups (21% versus 31%). Trial authors performed ITT analyses. |
| Selective reporting (reporting bias) | Low risk | Trial authors reported all relevant outcomes. |
| Other bias | Low risk | We found no other sources of bias. |
| Overall risk assessment | Unclear risk | Risk is due to the uncertainties raised above. |
Meynard 2002.
| Methods | A multi‐centre randomized trial in France | |
| Participants | 541 participants Inclusion: plasma HIV‐1 RNA 1000 copies/mL; previous exposure to at least 1 protease inhibitor (PI) for at least 3 months; unchanged antiretroviral regimen for the 2 preceding months; over 18 years of age; Karnofsky score > 70% Exclusion: active opportunistic infection; previous resistance testing; estimated poor adherence; blood haemoglobin < 8 g/dL; blood neutrophils < 750 × 10⁶/L; serum creatinine > 150 µmol/L; serum amylase > 3 times the upper limit of normal; liver aminotransferase values > 5 times the upper limit of normal |
|
| Interventions | Intervention 1: phenotyping (P) arm in which the treatment choice was guided by a phenotypic test Intervention 2: genotyping (G) arm in which the treatment choice was guided by a genotypic test Control: standard of care (SOC) arm in which treatment was chosen by the clinician |
|
| Outcomes | Virological success (< 200 copies and < 20 copies/mL), change in viral load, change in CD4 count Follow‐up: 36 weeks |
|
| Notes | All participants provided informed consent. Funding: Agence Nationale de Recherche sur le Sida, Glaxo Wellcome, Abbott, Dupont‐Pharma, and Visible Genetics |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Method of randomization was not reported. |
| Allocation concealment (selection bias) | Low risk | Arm assignments were revealed only to the investigator and patients at day 0. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | This was an open‐label study (knowledge of allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Losses to follow‐up were balanced across groups but were considerable (> 20%). |
| Selective reporting (reporting bias) | Low risk | Trial authors reported all outcomes of interest. |
| Other bias | Low risk | We found no other sources of bias. |
| Overall risk assessment | Low risk | We had no serious risk of bias concerns. |
Rubini 2002.
| Methods | A prospective randomized study in Brazil | |
| Participants | 49 participants Inclusion criteria: children and adolescents previously exposed to at least 2 ART treatment regimens and to failing therapy Exclusion criteria: not reported |
|
| Interventions | Intervention: therapy switch was based on genotypic resistance testing (genotypic group; GG) Control: therapy switch was based on the physician's clinical experience (control group; CG) |
|
| Outcomes | Treatment success, change in viral load | |
| Notes | Consent procedures were not reported. Funding was not reported. This study was published as an abstract. |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Method of randomization was not reported. |
| Allocation concealment (selection bias) | Unclear risk | No information was provided. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | No information was provided. |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | No information was provided. |
| Incomplete outcome data (attrition bias) All outcomes | Unclear risk | 10.2% were excluded for compliance or toxicity. Data were not reported by group. |
| Selective reporting (reporting bias) | Low risk | Two key outcomes were reported. |
| Other bias | Unclear risk | We found no other sources of bias. |
| Overall risk assessment | Unclear risk | Risk is due to the uncertainties raised above. |
Tural 2002.
| Methods | A randomized open‐label multi‐centre factorial design trial at 13 hospitals in 10 cities in Spain | |
| Participants | 326 participants Inclusion: plasma RNA > 1000 copies/mL, on stable ART combination for longer than 6 months Exclusion: substantial antiretroviral‐related adverse events history, poor adherence or active drug abuse reported by the treating physician |
|
| Interventions | Intervention 1: genotyping without expert advice (G+/EA‐) Intervention 2: genotyping and expert advice (G+/EA+) Intervention 3: no genotyping with expert advice (G‐/EA+) Control: no genotyping and no expert advice (G‐/EA‐) |
|
| Outcomes | Virological success (< 400 copies/mL), change in viral load | |
| Notes | All participants provided informed consent. Funding: Visible Genetics Trial acronym: Havana |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Method of randomization was not reported. |
| Allocation concealment (selection bias) | Low risk | Central location randomization was conducted. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | This was an open‐label study (knowledge of allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Losses to follow‐up were balanced between groups. Trial authors performed ITT analyses. |
| Selective reporting (reporting bias) | Low risk | All relevant outcomes were reported. |
| Other bias | Low risk | We found no other sources of bias. |
| Overall risk assessment | Low risk | We had no serious risk of bias concerns. |
Wegner 2004.
| Methods | A prospective randomized 3‐arm multi‐centre clinical trial at 6 military hospitals in the USA | |
| Participants | 450 participants Inclusion: at least 18 years of age, receiving a stable ART regimen containing ≥ 2 drugs for at least 8 weeks before randomization, able to provide informed consent, willing to attend regular study visits Exclusion: not reported |
|
| Interventions | Intervention 1: routine access to phenotype testing (PT) Intervention 2: routine access to genotype testing (GT) Control: no access to resistance testing (VB) |
|
| Outcomes | Treatment failure, discontinuation | |
| Notes | All participants provided informed consent. Funding: US Army Medical Research and Material Command and the Henry M. Jackson Foundation for the Advancement of Military Medicine |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Method of randomization was not reported. |
| Allocation concealment (selection bias) | Low risk | The randomization procedure was centrally located. |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | This was an open‐label study (knowledge of allocation is unlikely to introduce bias). |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Trial authors did not report this information (knowledge of allocation is unlikely to introduce bias). |
| Incomplete outcome data (attrition bias) All outcomes | High risk | Losses to follow‐up were balanced between groups but were considerable (> 65%). |
| Selective reporting (reporting bias) | High risk | Only treatment failure and discontinuation were reported. |
| Other bias | Low risk | We found no other sources of bias. |
| Overall risk assessment | High risk | Risk is due to the uncertainties raised above. |
Abbreviations: ART: antiretroviral therapy; CG: control group; EA: expert advice; G: genotyping; GART: genotypic antiretroviral resistance testing; GG: genotypic group; GT: genotype testing; ITT: intention‐to‐treat; NNRTI: non‐nucleoside reverse transcriptase inhibitor; NRTI: nucleoside reverse transcriptase inhibitor; P: phenotyping; PI: protease inhibitor; PRT: phenotypic resistance testing; PT: phenotype testing; SOC: standard of care; VB: XXX; VL: viral load.
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Badri 2003 | Retrospective evaluation of a RCT |
| Bonjoch 2008 | No genotypic or phenotypic guided therapy |
| Bossi 2004 | Genotypic guided therapy in both arms |
| Clevenbergh 2000 | Genotypic guided therapy in both arms |
| Clevenbergh 2002 | Secondary analysis |
| De Luca 2006 | Secondary analysis |
| Demeter 2008 | No genotypic or phenotypic guided therapy |
| Dunn 2005a | Genotypic guided therapy in both arms |
| Durant 2000 | Secondary analysis |
| Gianotti 2006 | Genotypic guided therapy in both arms |
| Hales 2006 | Genotypic guided therapy in both arms |
| Lorenzana 2012 | No comparison of resistance testing versus no resistance testing |
| Maggiolo 2007 | No comparison of resistance testing versus no resistance testing |
| Mazzotta 2003 | Compared genotypic and phenotypic tests versus each other with no control group |
| Pere 2012 | No comparison of resistance testing versus no resistance testing |
| Perez‐Elias 2003 | Compared genotypic and phenotypic tests versus each other with no control group |
| Saracino 2004 | Compared genotypic and phenotypic tests versus each other with no control group |
| Torti 2005 | Compared genotypic and phenotypic tests versus each other with no control group |
| Vergne 2003 | No comparison of resistance testing versus no resistance testing |
Abbreviations: RCT: randomized controlled trial.
Differences between protocol and review
We chose the cutoff of 20% for missing data to assess the risk of incomplete outcome data (attrition bias). More than 20% loss to follow‐up poses threats to study validity (Dettori 2011). This threshold was not mentioned in the protocol.
We did not plan to report the outcome ‘Change in viral load'. However, it was reported by almost all of the included studies, so we decided to include it in the review.
TJ and TA are co‐first authors, as they contributed equally to this work.
Contributions of authors
All review authors contributed to the design and conduct of this review, as well as to manuscript drafting and submission. LM, JT, TA, and RS screened articles for inclusion, extracted data, and assessed risk of bias. LM and TA worked on the ‘Summary of findings' tables.
Sources of support
Internal sources
-
Center for the Development of Best Practices in Health, Yaoundé Central Hospital, Yaoundé, Cameroon.
Provided office space and computer and Internet facilities
Liverpool School of Tropical Medicine, UK.
External sources
-
Aubrey Sheiham Evidence‐Based Health Care in Africa Leadership Award, UK.
Financial support for meetings, travels, and capacity building
-
Department for International Development, UK.
Project number 300342‐104
Declarations of interest
JT has no known conflicts of interest. TA has no known conflicts of interest. RS has no known conflicts of interest. LM has no known conflicts of interest.
These authors contributed equally to this work
These authors contributed equally to this work
Unchanged
References
References to studies included in this review
Baxter 2000 {published data only}
- Baxter JD, Mayers DL, Wentworth DN, Neaton JD, Hoover ML, Winters MA, et al. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS (London, England) 2000;14(9):F83‐93. [PUBMED: 10894268] [DOI] [PubMed] [Google Scholar]
Cingolani 2002 {published data only}
- Cingolani A, Antinori A, Rizzo MG, Murri R, Ammassari A, Baldini F, et al. Usefulness of monitoring HIV drug resistance and adherence in individuals failing highly active antiretroviral therapy: a randomized study (ARGENTA). AIDS (London, England) 2002;16(3):369‐79. [PUBMED: 11834948] [DOI] [PubMed] [Google Scholar]
Cohen 2002 {published data only}
- Cohen CJ, Hunt S, Sension M, Farthing C, Conant M, Jacobson S, et al. A randomized trial assessing the impact of phenotypic resistance testing on antiretroviral therapy. AIDS (London, England) 2002;16(4):579‐88. [PUBMED: 11873001] [DOI] [PubMed] [Google Scholar]
Dunn 2005 {published data only}
- Evaluation of Resistance Assays (ERA) Trial Investigators. A randomized controlled trial of the clinical utility of genotypic resistance testing in patients with limited prior exposure to antiretroviral drugs. HIV Clinical Trials 2005;6(4):183‐6. [PUBMED: 16214734] [DOI] [PubMed] [Google Scholar]
Durant 1999 {published data only}
- Durant J, Clevenbergh P, Halfon P, Delgiudice P, Porsin S, Simonet P, et al. Drug‐resistance genotyping in HIV‐1 therapy: the VIRADAPT randomised controlled trial. Lancet (London, England) 1999;353(9171):2195‐9. [PUBMED: 10392984] [DOI] [PubMed] [Google Scholar]
Green 2006 {published data only}
- Green H, Gibb DM, Compagnucci A, Giacomet V, Rossi A, Harper L, et al. A randomized controlled trial of genotypic HIV drug resistance testing in HIV‐1‐infected children: the PERA (PENTA 8) trial. Antiviral Therapy 2006;11(7):857‐67. [PUBMED: 17302248] [PubMed] [Google Scholar]
Haubrich 2005 {published data only}
- Haubrich RH, Kemper CA, Hellmann NS, Keiser PH, Witt MD, Tilles JG, et al. A randomized, prospective study of phenotype susceptibility testing versus standard of care to manage antiretroviral therapy: CCTG 575. AIDS (London, England) 2005;19(3):295‐302. [PUBMED: 15718840] [PubMed] [Google Scholar]
Meynard 2002 {published data only}
- Meynard JL, Vray M, Morand‐Joubert L, Race E, Descamps D, Peytavin G, et al. Phenotypic or genotypic resistance testing for choosing antiretroviral therapy after treatment failure: a randomized trial. AIDS (London, England) 2002;16(5):727‐36. [PUBMED: 11964529] [DOI] [PubMed] [Google Scholar]
Rubini 2002 {unpublished data only}
- Rubini NPM, Brindeiro PA, Brindeiro RM, Llerena CA, Hertogs K, Tanuri A. Clinical value of HIV‐1 genotyping in children and adolescents heavily experiences in antiretroviral therapy. XIV International AIDS Conference. Barcelona, 2002.
Tural 2002 {published data only}
- Tural C, Ruiz L, Holtzer C, Schapiro J, Viciana P, Gonzalez J, et al. Clinical utility of HIV‐1 genotyping and expert advice: the Havana trial. AIDS (London, England) 2002;16(2):209‐18. [PUBMED: 11807305] [DOI] [PubMed] [Google Scholar]
Wegner 2004 {published data only}
- Wegner SA, Wallace MR, Aronson NE, Tasker SA, Blazes DL, Tamminga C, et al. Long‐term efficacy of routine access to antiretroviral‐resistance testing in HIV type 1‐infected patients: results of the clinical efficacy of resistance testing trial. Clinical Infectious Diseases 2004;38(5):723‐30. [PUBMED: 14986258] [DOI] [PubMed] [Google Scholar]
References to studies excluded from this review
Badri 2003 {published data only}
- Badri SM, Adeyemi OM, Max BE, Zagorski BM, Barker DE. How does expert advice impact genotypic resistance testing in clinical practice?. Clinical Infectious Diseases 2003;37:708‐13. [DOI] [PubMed] [Google Scholar]
Bonjoch 2008 {published data only}
- Bonjoch A, Buzon MJ, Llibre JM, Negredo E, Puig J, Perez‐Alvarez N, et al. Transient treatment exclusively containing nucleoside analogue reverse transcriptase inhibitors in highly antiretroviral‐experienced patients preserves viral benefit when a fully active therapy was initiated. HIV Clinical Trials 2008;9:387‐98. [DOI] [PubMed] [Google Scholar]
Bossi 2004 {published data only}
- Bossi P, Peytavin G, Ait‐Mohand H, Delaugerre C, Ktorza N, Paris L, et al. GENOPHAR: a randomized study of plasma drug measurements in association with genotypic resistance testing and expert advice to optimize therapy in patients failing antiretroviral therapy. HIV Medicine 2004;5:352‐9. [DOI] [PubMed] [Google Scholar]
Clevenbergh 2000 {published data only}
- Clevenbergh P, Durant J, Halfon P, Giudice P, Mondain V, Montagne N, et al. Persisting long‐term benefit of genotype‐guided treatment for HIV‐infected patients failing HAART. The Viradapt Study: week 48 follow‐up. Antiviral Therapy 2000;5:65‐70. [PubMed] [Google Scholar]
Clevenbergh 2002 {published data only}
- Clevenbergh P, Garraffo R, Durant J, Dellamonica P. PharmAdapt: a randomized prospective study to evaluate the benefit of therapeutic monitoring of protease inhibitors: 12 week results. AIDS 2002;16:2311‐5. [DOI] [PubMed] [Google Scholar]
De Luca 2006 {published data only}
- Luca A, Giambenedetto S, Cingolani A, Bacarelli A, Ammassari A, Cauda R. Three‐year clinical outcomes of resistance genotyping and expert advice: extended follow‐up of the Argenta trial. Antiviral Therapy 2006;11:321‐7. [PubMed] [Google Scholar]
Demeter 2008 {published data only}
- Demeter LM, Mukherjee AL, DiFrancesco R, Jiang H, DiCenzo R, Bastow B, et al. The design and implementation of A5146, a prospective trial assessing the utility of therapeutic drug monitoring using an inhibitory quotient in antiretroviral‐experienced HIV‐infected patients. HIV Clinical Trials 2008;9:61‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Dunn 2005a {published data only}
- Dunn D. A randomized controlled trial of the value of phenotypic testing in addition to genotypic testing for HIV drug resistance. Journal of Acquired Immune Deficiency Syndromes 2005;38:553‐9. [DOI] [PubMed] [Google Scholar]
Durant 2000 {published data only}
- Durant J, Clevenbergh P, Garraffo R, Halfon P, Icard S, Giudice P, et al. Importance of protease inhibitor plasma levels in HIV‐infected patients treated with genotypic‐guided therapy: pharmacological data from the Viradapt Study. AIDS 2000;14:1333‐9. [DOI] [PubMed] [Google Scholar]
Gianotti 2006 {published data only}
- Gianotti N, Mondino V, Rossi MC, Chiesa E, Mezzaroma I, Ladisa N, et al. Comparison of a rule‐based algorithm with a phenotype‐based algorithm for the interpretation of HIV genotypes in guiding salvage regimens in HIV‐infected patients by a randomized clinical trial: the mutations and salvage study. Clinical Infectious Diseases 2006; Vol. 42:1470‐80. [DOI] [PubMed]
Hales 2006 {published data only}
- Hales G, Birch C, Crowe S, Workman C, Hoy JF, Law MG, et al. A randomised trial comparing genotypic and virtual phenotypic interpretation of HIV drug resistance: the CREST study. PLoS Clinical Trials 2006; Vol. 1:e18. [DOI] [PMC free article] [PubMed]
Lorenzana 2012 {published data only}
- Lorenzana SB, Hughes MD, Grinsztejn B, Collier AC, Luz PM, Freedberg KA, et al. Genotype assays and third‐line ART in resource‐limited settings: a simulation and cost‐effectiveness analysis of a planned clinical trial. AIDS 2012;26:1083‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Maggiolo 2007 {published data only}
- Maggiolo F, Airoldi M, Callegaro A, Ripamonti D, Gregis G, Quinzan G, et al. Prediction of virologic outcome of salvage antiretroviral treatment by different systems for interpreting genotypic HIV drug resistance. Journal of the International Association of Physicians in AIDS Care 2007;6:87‐93. [DOI] [PubMed] [Google Scholar]
Mazzotta 2003 {published data only}
- Mazzotta F, Lo Caputo S, Torti C, Tinelli C, Pierotti P, Castelli F, et al. Real versus virtual phenotype to guide treatment in heavily pretreated patients: 48‐week follow‐up of the Genotipo‐Fenotipo di Resistenza (GenPheRex) trial. Journal of Acquired Immune Deficiency Syndromes 2003;32:268‐80. [DOI] [PubMed] [Google Scholar]
Pere 2012 {published data only}
- Pere H, Charpentier C, Mbelesso P, Dandy M, Matta M, Moussa S, et al. Virological response and resistance profiles after 24 months of first‐line antiretroviral treatment in adults living in Bangui, central African Republic. AIDS Research and Human Retroviruses 2012;28:315‐23. [DOI] [PubMed] [Google Scholar]
Perez‐Elias 2003 {published data only}
- Perez‐Elias MJ, Garcia‐Arata I, Munoz V, Santos I, Sanz J, Abraira V, et al. Phenotype or virtual phenotype for choosing antiretroviral therapy after failure: a prospective, randomized study. Antiviral Therapy 2003;8:577‐84. [PubMed] [Google Scholar]
Saracino 2004 {published data only}
- Saracino A, Monno L, Locaputo S, Torti C, Scudeller L, Ladisa N, et al. Selection of antiretroviral therapy guided by genotypic or phenotypic resistance testing: an open‐label, randomized, multicenter study (PhenGen). Journal of Acquired Immune Deficiency Syndromes 2004;37:1587‐98. [DOI] [PubMed] [Google Scholar]
Torti 2005 {published data only}
- Torti C, Quiros‐Roldan E, Regazzi M, Luca A, Mazzotta F, Antinori A, et al. A randomized controlled trial to evaluate antiretroviral salvage therapy guided by rules‐based or phenotype‐driven HIV‐1 genotypic drug‐resistance interpretation with or without concentration‐controlled intervention: the resistance and dosage adapted regimens (RADAR) study. Clinical Infectious Diseases 2005;40:1828‐36. [DOI] [PubMed] [Google Scholar]
Vergne 2003 {published data only}
- Vergne L, Kane CT, Laurent C, Diakhate N, Ngom Gueye NF, Gueye PM, et al. Low rate of genotypic HIV‐1 drug‐resistant strains in the Senegalese government initiative of access to antiretroviral therapy. AIDS 2003;17:S31‐8. [DOI] [PubMed] [Google Scholar]
Additional references
AIDSinfo 2016
- AIDSinfo. Guidelines for the Use of Antiretroviral Agents in HIV‐1‐Infected Adults and Adolescents. Laboratory Testing. Last Updated: 14 July 2016. http://aidsinfo.nih.gov/guidelines/html/1/adult‐and‐adolescent‐arv‐guidelines/6/drug‐resistance‐testing (accessed 03 June 2017).
Bakhouch 2009
- Bakhouch K, Oulad‐Lahcen A, Bensghir R, Blaghen M, Elfilali KM, Ezzikouri S, et al. The prevalence of resistance‐associated mutations to protease and reverse transcriptase inhibitors in treatment‐naïve (HIV1)‐infected individuals in Casablanca, Morocco. Journal of Infection in Developing Countries 2009;3(5):380‐91. [DOI] [PubMed] [Google Scholar]
Barennes 2014
- Barennes H, Guillet S, Limsreng S, Him S, Nouhin J, Hak C, et al. Virological failure and HIV‐1 drug resistance mutations among naive and antiretroviral pre‐treated patients entering the ESTHER program of Calmette Hospital in Cambodia. PLoS ONE 2014;9(8):e105736. [DOI: 10.1371/journal.pone.0105736] [DOI] [PMC free article] [PubMed] [Google Scholar]
Barrow 2013
- Barrow GJ, Hylton‐Kong T, Rodriguez N, Yamamura Y, Figueroa JP. HIV‐1 drug resistance in treatment‐naive chronically infected patients in Jamaica. Antiviral Therapy 2013;18(7):941‐4. [PUBMED: 23744572] [DOI] [PMC free article] [PubMed] [Google Scholar]
Bissio 2017
- Bissio E, Barbas MG, Bouzas MB, Cudolá A, Salomón H, Espínola L, et al. Pretreatment HIV‐1 drug resistance in Argentina: results from a surveillance study performed according to WHO‐proposed new methodology in 2014‐15. Journal of Antimicrobial Chemotherapy 2017;72(2):504‐10. [PUBMED: 27789684] [DOI] [PubMed] [Google Scholar]
Boender 2015
- Boender TS, Hoenderboom BM, Sigaloff KC, Hamers RL, Wellington M, Shamu T, et al. Pretreatment HIV drug resistance increases regimen switches in sub‐Saharan Africa. Clinical Infectious Diseases 2015;61(11):1749‐58. [PUBMED: 26240203] [DOI] [PubMed] [Google Scholar]
Boender 2016
- Boender TS, Kityo CM, Boerma RS, Hamers RL, Ondoa P, Wellington M, et al. Accumulation of HIV‐1 drug resistance after continued virological failure on first‐line ART in adults and children in sub‐Saharan Africa. Journal of Antimicrobial Chemotherapy 2016;71(10):2918‐27. [PUBMED: 27342546] [DOI] [PubMed] [Google Scholar]
Burchell 2012
- Burchell AN, Bayoumi AM, Rourke SB, Major C, Gardner S, Sandstrom P, et al. Increase in transmitted HIV drug resistance among persons undergoing genotypic resistance testing in Ontario, Canada, 2002‐09. Journal of Antimicrobial Chemotherapy 2012;67:2755‐65. [DOI] [PubMed] [Google Scholar]
Cambiano 2013
- Cambiano V, Bertagnolio S, Jordan MR, Lundgren JD, Phillips A. Transmission of drug resistant HIV and its potential impact on mortality and treatment outcomes in resource‐limited settings. Journal of Infectious Diseases 2013;207(Suppl 2):S57‐62. [PUBMED: 23687290] [DOI] [PubMed] [Google Scholar]
Chaiyachati 2014
- Chaiyachati KH, Ogbuoji O, Price M, Suthar AB, Negussie EK, Bärnighausen T. Interventions to improve adherence to antiretroviral therapy: a rapid systematic review. AIDS (London, England) 2014;28(Suppl 2):S187‐204. [PUBMED: 24849479] [DOI] [PubMed] [Google Scholar]
Corzillius 2004
- Corzillius M, Mühlberger N, Sroczynski G, Jaeger H, Wasem J, Siebert U. Cost effectiveness analysis of routine use of genotypic antiretroviral resistance testing after failure of antiretroviral treatment for HIV. Antiviral Therapy 2004;9(1):27‐36. [PUBMED: 15040534] [PubMed] [Google Scholar]
De Luca 2015
- Luca A, Zazzi M. Interplay between transmitted and acquired HIV Type 1 drug resistance: reasons for a disconnect. Journal of Infectious Diseases 2015;212(1):5‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Desai 2007
- Desai S, Kyriakides T, Holodniy M, Al‐Salman J, Griffith B, Kozal M. Evolution of genotypic resistance algorithms and their impact on the interpretation of clinical trials: an OPTIMA trial substudy. HIV Clinical Trials 2007;8(5):293‐302. [PUBMED: 17956830] [DOI] [PubMed] [Google Scholar]
Dettori 2011
- Dettori JR. Loss to follow‐up. Evidence‐Based Spine Care Journal 2011;2(1):7‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Dunn 2004
- Dunn DT, Gibb DM, Babiker AG, Green H, Darbyshire JH, Weller IV. HIV drug resistance testing: is the evidence really there?. Antiviral Therapy 2004;9(5):641‐8. [PUBMED: 15535402] [PubMed] [Google Scholar]
Duwe 2001
- Duwe S, Brunn M, Altmann D, Hamouda O, Schmidt B, Walter H, et al. Frequency of genotypic and phenotypic drug resistant HIV‐1 among therapy‐naive patients of the German Seroconverter Study. Journal of Acquired Immune Deficiency Syndromes 2001;26(3):266‐73. [PUBMED: 11242200] [DOI] [PubMed] [Google Scholar]
EACS 2017
- European AIDS Clinical Society. Guidelines 2017. www.eacsociety.org/files/guidelines_9.0‐english.pdf (accessed 2 August 2018).
Egger 1997
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta‐analysis detected by a simple, graphical test. BMJ (Clinical research ed.) 1997;315(7109):629‐34. [PUBMED: 9310563] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ena 2006
- Ena J, Ruiz de Apodaca RF, Amador C, Benito C, Pasquau F. Net benefits of resistance testing directed therapy compared with standard of care in HIV‐infected patients with virological failure: a meta‐analysis. Enfermedades Infecciosas y Microbiologia Clinica 2006;24(4):232‐7. [PUBMED: 16725082] [DOI] [PubMed] [Google Scholar]
GRADEpro 2015 [Computer program]
- McMaster University (developed by Evidence Prime). GRADEpro GDT. Version accessed 3 June 2017. Hamilton (ON): McMaster University (developed by Evidence Prime), 2015.
Gupta 2012
- Gupta RK, Jordan MR, Sultan BJ, Hill A, Davis DH, Gregson J, et al. Global trends in antiretroviral resistance in treatment‐naive individuals with HIV after rollout of antiretroviral treatment in resource‐limited settings: a global collaborative study and meta‐regression analysis. Lancet (London, England) 2012;380(9849):1250‐8. [PUBMED: 22828485] [DOI] [PMC free article] [PubMed] [Google Scholar]
Guyatt 2011
- Guyatt G, Oxman AD, Akl EA, Kunz R, Vist G, Brozek J, et al. GRADE guidelines: 1. Introduction‐GRADE evidence profiles and summary of findings tables. Journal of clinical epidemiology 2011;64(4):383‐94. [PUBMED: 21195583] [DOI] [PubMed] [Google Scholar]
Hamers 2012
- Hamers RL, Schuurman R, Sigaloff KC, Wallis CL, Kityo C, Siwale M, et al. Effect of pretreatment HIV‐1 drug resistance on immunological, virological, and drug‐resistance outcomes of first‐line antiretroviral treatment in sub‐Saharan Africa: a multicentre cohort study. Lancet Infectious Diseases 2012;12(4):307‐17. [PUBMED: 22036233] [DOI] [PubMed] [Google Scholar]
Higgins 2011
- Higgins JP, Green S, editor(s). Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org. Version 5.0.2 [updated 2009]. The Cochrane Collaboration.
Hosseinipour 2013
- Hosseinipour MC, Gupta RK, Zyl G, Eron JJ, Nachega JB. Emergence of HIV drug resistance during first‐ and second‐line antiretroviral therapy in resource‐limited settings. Journal of Infectious Diseases 2013;207(Suppl 2):S49‐56. [PUBMED: 23687289] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hozo 2005
- Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Medical Research Methodology 2005;5:13. [PUBMED: 15840177] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kanters 2017
- Kanters S, Park JJ, Chan K, Socias ME, Ford N, Forrest JI, et al. Interventions to improve adherence to antiretroviral therapy: a systematic review and network meta‐analysis. Lancet HIV 2017;4(1):e31‐40. [PUBMED: 27863996] [DOI] [PubMed] [Google Scholar]
Larder 1989
- Larder BA, Richman DD. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science 1989;243(4899):1731‐4. [PUBMED: 2467383] [DOI] [PubMed] [Google Scholar]
Moher 2010
- Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. International Journal of Surgery (London, England) 2010;8(5):336‐41. [PUBMED: 20171303] [DOI] [PubMed] [Google Scholar]
Nachega 2012
- Nachega JB, Uthman OA, Anderson J, Peltzer K, Wampold S, Cotton MF, et al. Adherence to antiretroviral therapy during and after pregnancy in low‐income, middle‐income, and high‐income countries: a systematic review and meta‐analysis. AIDS (London, England) 2012;26(16):2039‐52. [PUBMED: 22951634] [DOI] [PMC free article] [PubMed] [Google Scholar]
Palella 2009
- Palella FJ Jr, Armon C, Buchacz K, Cole SR, Chmiel JS, Novak RM, et al. The association of HIV susceptibility testing with survival among HIV‐infected patients receiving antiretroviral therapy: a cohort study. Annals of Internal Medicine 2009;151(2):73‐84. [PUBMED: 19620160] [DOI] [PubMed] [Google Scholar]
Panidou 2004
- Panidou ET, Trikalinos TA, Ioannidis JP. Limited benefit of antiretroviral resistance testing in treatment‐experienced patients: a meta‐analysis. AIDS (London, England) 2004;18(16):2153‐61. [PUBMED: 15577648] [DOI] [PubMed] [Google Scholar]
Pham 2014
- Pham QD, Wilson DP, Law MG, Kelleher AD, Zhang L. Global burden of transmitted HIV drug resistance and HIV‐exposure categories: a systematic review and meta‐analysis. AIDS 2014;28(18):2751‐62. [DOI: 10.1097/QAD.0000000000000494] [DOI] [PubMed] [Google Scholar]
Phillips 2014
- Phillips A, Cambiano V, Nakagawa F, Mabugu T, Miners A, Ford D, et al. Cost‐effectiveness of HIV drug resistance testing to inform switching to second line antiretroviral therapy in low income settings. PLoS ONE 2014;9(10):e109148. [PUBMED: 25290340] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pinoges 2015
- Pinoges L, Schramm B, Poulet E, Balkan S, Szumilin E, Ferreyra C, et al. Risk factors and mortality associated with resistance to first‐line antiretroviral therapy: multicentric cross‐sectional and longitudinal analyses. Journal of Acquired Immune Deficiency Syndromes 2015;68(5):527‐35. [PUBMED: 25585301] [DOI] [PubMed] [Google Scholar]
Ramjan 2014
- Ramjan R, Calmy A, Vitoria M, Mills EJ, Hill A, Cooke G, et al. Systematic review and meta‐analysis: patient and programme impact of fixed‐dose combination antiretroviral therapy. Tropical Medicine & International Health 2014;19(5):501‐13. [PUBMED: 24628918] [DOI] [PubMed] [Google Scholar]
RevMan 2014 [Computer program]
- Nordic Cochrane Centre, The Cochrane Collaboration. Review Manager 5 (RevMan 5). Version 5.3. Copenhagen: Nordic Cochrane Centre, The Cochrane Collaboration, 2014.
Rocheleau 2017
- Rocheleau G, Franco‐Villalobos C, Oliveira N, Brumme ZL, Rusch M, Shoveller J, et al. Sociodemographic correlates of HIV drug resistance and access to drug resistance testing in British Columbia, Canada. PLoS One 2017;12:e0184848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rojas Sánchez 2014
- Rojas Sánchez P, Holguín A. Drug resistance in the HIV‐1‐infected paediatric population worldwide: a systematic review. Journal of Antimicrobial Chemotherapy 2014;69(8):2032‐42. [PUBMED: 24788658] [DOI] [PubMed] [Google Scholar]
Rowley 2016
- Rowley CF, MacLeod IJ, Maruapula D, Lekoko B, Gaseitsiwe S, Mine M, et al. Sharp increase in rates of HIV transmitted drug resistance at antenatal clinics in Botswana demonstrates the need for routine surveillance. Journal of Antimicrobial Chemotherapy 2016;71(5):1361‐6. [PUBMED: 26929269] [DOI] [PMC free article] [PubMed] [Google Scholar]
Saag 2018
- Saag MS, Benson CA, Gandhi RT, Hoy JF, Landovitz RJ, Mugavero MJ, et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2018 recommendations of the International Antiviral Society‐USA Panel. JAMA 2018;320(4):379‐96. [PUBMED: 30043070] [DOI] [PMC free article] [PubMed] [Google Scholar]
Sax 2005
- Sax PE, Islam R, Walensky RP, Losina E, Weinstein MC, Goldie SJ, et al. Should resistance testing be performed for treatment‐naive HIV‐infected patients? A cost‐effectiveness analysis. Clinical Infectious Diseases 2005;41(9):1316‐23. [PUBMED: 16206108] [DOI] [PubMed] [Google Scholar]
Sluis‐Cremer 2014
- Sluis‐Cremer N. The emerging profile of cross‐resistance among the nonnucleoside HIV‐1 reverse transcriptase inhibitors. Viruses 2014;6(8):2960‐73. [PUBMED: 25089538] [DOI] [PMC free article] [PubMed] [Google Scholar]
Sullivan 2013
- Sullivan A, Sutcliffe P, Sandre R, Harrigan R, Archibald C, Halverson J, et al. Follow‐up investigation of a cluster of treatment‐naive HIV‐infected patients with multi‐drug resistance in Sudbury, Ontario. Canadian Journal of Infectious Diseases and Medical Microbiology 2013;24:38A. [Google Scholar]
Thompson 2012
- Thompson MA, Mugavero MJ, Amico KR, Cargill VA, Chang LW, Gross R, et al. Guidelines for improving entry into and retention in care and antiretroviral adherence for persons with HIV: evidence‐based recommendations from an International Association of Physicians in AIDS Care Panel. Annals of Internal Medicine 2012;156(11):817‐33, W‐284‐94. [PUBMED: 22393036] [DOI] [PMC free article] [PubMed] [Google Scholar]
Torre 2002
- Torre D, Tambini R. Antiretroviral drug resistance testing in patients with HIV‐1 infection: a meta‐analysis study. HIV Clinical Trials 2002;3(1):1‐8. [PUBMED: 11819179] [DOI] [PubMed] [Google Scholar]
Torti 2004
- Torti C, Bono L, Gargiulo F, Uccelli MC, Quiros‐Roldan E, Patroni A, et al. Prevalence of drug resistance and newly recognised treatment‐related substitutions in the HIV‐1 reverse transcriptase and protease genes from HIV‐positive patients naive for antiretrovirals. Clinical Microbiology and Infection 2004;10(9):826‐30. [PUBMED: 15355414] [DOI] [PubMed] [Google Scholar]
Tsiang 2016
- Tsiang M, Jones GS, Goldsmith J, Mulato A, Hansen D, Kan E, et al. Antiviral activity of bictegravir (GS‐9883), a novel potent HIV‐1 integrase strand transfer Inhibitor with an improved resistance profile. Antimicrobial Agents and Chemotherapy 2016;60(12):7086‐97. [PUBMED: 27645238] [DOI] [PMC free article] [PubMed] [Google Scholar]
UNAIDS 2014
- UNAIDS. The Gap Report. 2014. http://files.unaids.org/en/media/unaids/contentassets/documents/unaidspublication/2014/UNAIDS_Gap_report_en.pdf (accessed 3 June 2017).
UNAIDS 2016
- UNAIDS. Get on the Fast‐Track. The Life‐Cycle Approach to HIV. Finding Solutions for Everyone at Every Stage of Life. 2016. www.unaids.org/sites/default/files/media_asset/Get‐on‐the‐Fast‐Track_en.pdf (accessed 3 June 2017).
Villabona‐Arenas 2016
- Villabona‐Arenas CJ, Vidal N, Guichet E, Serrano L, Delaporte E, Gascuel O, et al. In‐depth analysis of HIV‐1 drug resistance mutations in HIV‐infected individuals failing first‐line regimens in West and Central Africa. AIDS (London, England) 2016;30(17):2577‐89. [PUBMED: 27603287] [DOI] [PubMed] [Google Scholar]
Wells 2009
- Wells GA, Shea B, O'Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle‐Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta‐analyses. 2009. www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed 3 June 2017).
WHO 2016
- World Health Organization. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection. Recommendations for a Public Health Approach. Second Edition. June 2016. www.who.int/hiv/pub/arv/arv‐2016/en/ (accessed 3 June 2017).
WHO 2017
- World Health Organization. Antiretroviral Therapy (ART) Coverage Among All Age Groups. www.who.int/gho/hiv/epidemic_response/ART_text/en/ (accessed 3 June 2017).
WHO 2017a
- World Health Organization. Guidelines on the Public Health Response to Pretreatment HIV Drug Resistance. http://apps.who.int/iris/bitstream/handle/10665/255880/9789241550055‐eng.pdf?sequence=1 (accessed 25 July 2018).
WHO 2017b
- World Health Organization. 20th WHO Model List of Essential Medicines. http://www.who.int/medicines/publications/essentialmedicines/20th_EML2017_FINAL_amendedAug2017.pdf?ua=1 (accessed 24 April 2018).
WHO 2017c
- World Health Organization. WHO HIV Drug Resistance Report 2017. http://apps.who.int/iris/bitstream/handle/10665/255896/9789241512831‐eng.pdf?sequence=1 (accessed 24 July 2018).
Wittkop 2011
- Wittkop L, Günthard HF, Wolf F, Dunn D, Cozzi‐Lepri A, Luca A, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord‐CHAIN joint project): a European multicohort study. Lancet Infectious Diseases 2011;11(5):363‐71. [DOI] [PubMed] [Google Scholar]


