Abstract
Life builds its molecules from CO2 and breaks them down to CO2 again through the intermediacy of just five metabolites that act as the hubs of biochemistry.1 However, how core biological metabolism initiated and why it uses the intermediates, reactions and pathways that it does remains unclear. Here, we describe a purely chemical reaction network promoted by Fe2+ in which aqueous pyruvate and glyoxylate, two products of abiotic CO2 reduction,2–4 build up nine of the eleven Krebs (tricarboxylic acid, TCA) cycle intermediates, including all five universal metabolic precursors. The intermediates simultaneously break down to CO2 in a life-like regime resembling biological anabolism and catabolism.5 Adding hydroxylamine6–8 and Fe0 into the system produces four biological amino acids in a manner paralleling biosynthesis. The observed network significantly overlaps the Krebs and glyoxylate cycles9,10 and may represent a prebiotic precursor to these core metabolic pathways.
Modern systems views of the origin of life postulate that non-enzymatic reaction networks could have provided an environment from which polymer replicators such as RNA could later have emerged.11 However, the extent to which these primordial networks could have resembled biological ones remains debated.12,13 At the ecosystem level, biochemistry is simultaneously building itself up from CO2 and breaking down to CO2 again. This dynamic process occurs through the intermediacy of just five universal metabolites made of C, H and O: acetate, pyruvate, oxaloacetate, succinate, and α-ketoglutarate.1 These five compounds are found directly on or near life’s core anabolic and catabolic pathways, imparting them with an organizing role within metabolism.1 Theories for the chemical origins of life based on prebiotic analogues of core metabolic pathways therefore hold a high explanatory value for why life uses the compounds, reactions and pathways that it does.14–18 Since molecules must be made before they can be broken down, the question of how C-C bonds could form without relying on energetically uphill ATP-consuming reactions is a major challenge to origins theories rooted in prebiotic analogues of biochemistry.5 Recently, non-enzymatic analogues of the reductive AcCoA pathway have been demonstrated, wherein CO2 can be fixed to acetate and pyruvate, forming C-C bonds.2,19 Beyond this, partial non-enzymatic analogues of core metabolic pathways such as the tricarboxylic acid cycle (TCA cycle or Krebs cycle) and the reductive tricarboxylic acid cycle (rTCA cycle or reverse Krebs cycle) have been reported,9,20,21 but these investigations failed to uncover any of the critical C-C bond forming reactions, nearly all of which are ATP-consuming (e.g. the carboxylation of pyruvate to oxaloacetate; the reductive carboxylation of succinate to α-ketoglutarate).22 A recent theoretical analysis of all known metabolic reactions revealed a robust hypothetical metabolic network, containing all five of the universal metabolic precursors, that does not rely on phosphorus or on phosphorus-containing co-factors such as ATP.23 Of the molecules made up only of C, H and O within this hypothetical phosphorus-free network, the two that represent the biggest branching points are pyruvate and glyoxylate, suggesting that a primitive pre-ATP metabolism, if it existed, would have been critically reliant on these two compounds. Pyruvate and glyoxylate are attractive as starting materials for prebiotic chemistry because they can be accessed through abiotic CO2 fixation2,3,19 as well as by other plausible means,4,24 yet their reactivity with each other has hardly been studied with regards to the origins of metabolism. A lone example used the strong oxidant H2O2 to drive a bicyclic reaction network containing two other TCA cycle intermediates,25 but the influence of transition metal ions as potential naturally occurring catalysts has not yet been explored in this context. Here we show that pyruvate and glyoxylate spontaneously produce a reaction network in warm iron-rich water that recapitulates most of the intermediates and reactions of core biological pathways like the Krebs cycle and glyoxylate cycle, in addition to amino acid biosynthesis.
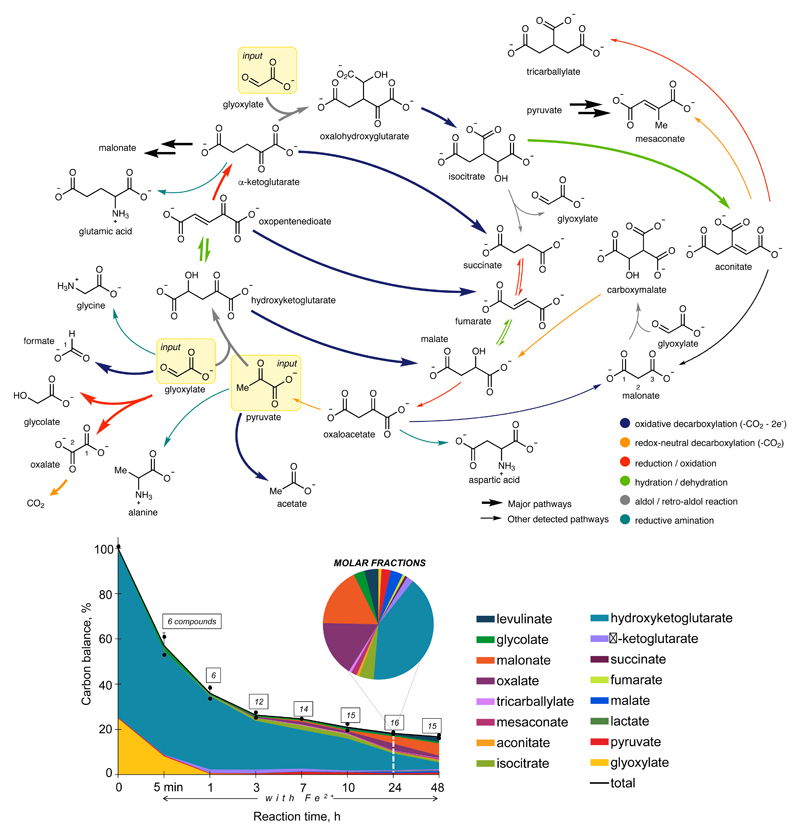
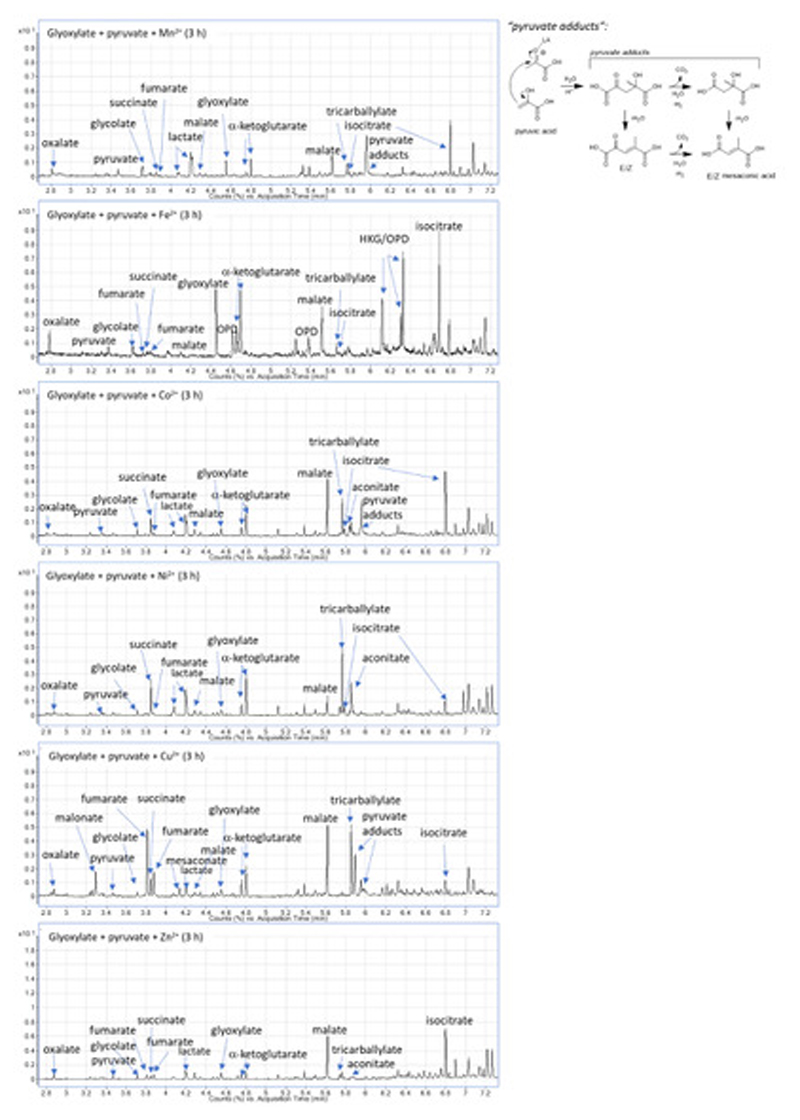
We systematically searched for a reaction network based on pyruvate and glyoxylate by screening a panel of transition metal ions as catalysts (Extended Data Figure 1) and analysing the outcome using GC-MS with comparisons against authentic standards (Extended Data Figure 2, Figure S3, Table S1). A temperature of 70 °C under an inert atmosphere was chosen to simulate a mild hydrothermal environment in accord with previously reported non-enzymatic glycolytic and TCA cycle reactions.20,26 Ferrous iron, thought to be abundant in the Archean crust and waters,27 promoted a reaction network (Figure 1a) whose product distribution evolved over time (Figure 1b). Within 3 h, acetate, pyruvate, malate, fumarate, succinate, α-ketoglutarate, isocitrate and aconitate were detected by GC-MS; acetate was additionally confirmed by NMR (Figures S4-S7, Table S2). Introducing hydroxylamine, an intermediate in biological nitrogen cycles and a prebiotically plausible nitrogen source,6–8 and Fe0 to a typical reaction mixture at t = 1 h produces four amino acids by t = 2 h: glycine, alanine, aspartic acid and glutamic acid (Figure S8). A reductive amination mechanism for the amino acid synthesis was supported by their detection upon exposing four corresponding ketoacids (glyoxylate, pyruvate, oxaloacetate, α-ketoglutarate) to similar conditions (Figure S9, Table S3) and comparing against authentic samples of amino acids (Figures S10-S11). Although oxaloacetate is not detectable by the analytical technique employed, we infer its presence from the detection of aspartic acid and malonate, which we confirmed to arise from reductive amination and oxidative decarboxylation, respectively, of oxaloacetate under the reaction conditions (Figures S8, S12, S13). Thus, the network contains nine of the eleven TCA cycle intermediates, including all five of the universal metabolic precursors.
Figure 1. Synthesis and breakdown of the universal precursors to biological metabolism promoted by ferrous iron.
a The observed Fe2+-promoted reaction network (major pathways are shown using bold reaction arrows). b Time dependence of a reaction network arising from pyruvate and glyoxylate, promoted by Fe2+ at 70 °C. Carbon balance refers to the % of carbon atoms observed in solution relative to 0 h and is reported as the average of two independent runs. Product distribution at 24 h is shown in a pie chart. Error bars correspond to the standard deviation. Values for hydroxyketoglutarate also include oxopentanedioate.
To elucidate mechanisms for the reaction network, authentic samples of most of the observed intermediates were submitted to typical reaction conditions in the presence and absence of glyoxylate and their reactivity studied over time (Figures S13-S14). These experiments, as well as isotopic labelling studies performed using three differentially labelled 13C-pyruvates and doubly 13C-labelled glyoxylate (Figures S15-S17 and S20-S29, Table S4) were consistent with the proposed mechanism described in Figure 1a. The network is primarily composed of four types of reactions: aldol/retro-aldol, hydrations/dehydrations, oxidative decarboxylations, and reductions/oxidations. Notably, both reductive and oxidative reactions occur in the same environment. Some key transformations are discussed here: An aldol reaction between pyruvate and glyoxylate gives hydroxyketoglutarate, followed by dehydration to oxopentenedioate and reduction to α-ketoglutarate. The three intermediates and products of this sequence partially undergo oxidative decarboxylation to give malate, fumarate and succinate, respectively (Figures S4-S5, S13). These oxidative processes were found to be more efficient in the presence of Fe3+ than Fe2+, the former being presumably generated during the reduction of oxopentendioate (Figure S18). α-Ketoglutarate also undergoes an aldol/oxidative decarboxylation sequence to give isocitrate. Reminiscent of the glyoxylate cycle or “glyoxylate shunt”, isocitrate slowly undergoes an Fe2+-catalysed retro-aldol reaction to return small quantities of glyoxylate and succinate but can also slowly dehydrate to give aconitate (Figure S14). Trace amounts of fumarate are observed upon heating succinate under standard conditions, pointing to mechanistic redundancy within the network. Additional evidence for the occurrence of oxidation under the reaction conditions was the observation that malate produced a small amount of acetate, which presumably arises from oxidative decomposition of oxaloacetate (Figure S19). Cannizzaro-type disproportionation of glyoxylate results in the formation of glycolate and oxalate, the latter of which slowly breaks down to CO2 under the reaction conditions (Figures S13-S14). Prolonged reaction time (>24 h) results in a slow build-up of other thermodynamic end-products including malonate, levulinate, succinate, malate and isocitrate (Figure S4). It is plausible that such a network could be kept in a non-equilibrium steady state by continuous input of pyruvate and glyoxylate, both of which can be produced abiotically.2–4,19
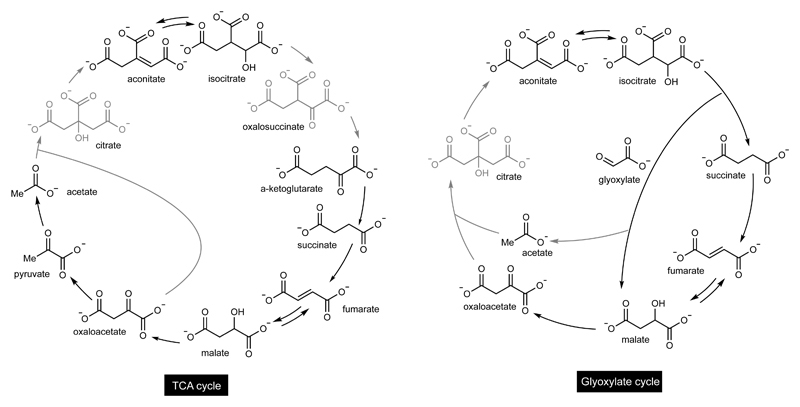
The observed non-enzymatic reaction network bears some remarkable similarities to the TCA cycle (Figure 2), encompassing seven of its eleven reactions and nine of its eleven intermediates (only oxalosuccinate and citrate are missing). It also recapitulates much of the glyoxylate cycle, including eight of its nine intermediates (only citrate is missing) and five of its eight reactions (Figure 2). Therefore, a relatively small amount of catalytic innovation is required for this reaction network to transform into ones used by biology today. Not all of these missing innovations need be biological. For example, the non-enzymatic interconversion of citrate and aconitate is missing in the current network catalysed by Fe2+ but has already been demonstrated to be catalysed by Cr3+, albeit under more extreme conditions (1 M HCl, 140 °C) that are incompatible with the current study.9 A potential evolutionary relationship between the glyoxylate cycle and the TCA cycle has been suggested before.10,28 Since the redox and hydration/dehydration reactions of the TCA and rTCA cycles can occur enzymatically and non-enzymatically in both directions depending on the redox nature of the environment,9,20 the invention of ATP-dependent carboxylation reactions in a reducing environment could also give rise to parts of the rTCA cycle, a possibility in line with the recent discovery of bi-directionality in rTCA cycle-dependent bacteria.33,34
Figure 2. A comparison of the observed reaction network with the TCA cycle (left) and the glyoxylate cycle (right).
Overlapping intermediates and reactions are shown in black. Those not observed in the synthetic reaction cycle are in grey.
In conclusion, an abiotic chemical pathway that resembles core carbon biochemistry is promoted by ferrous iron. Although the ability of simple reaction networks to evolve is limited in the absence of a genetic mechanism,31,32 we speculate that the incorporation of S and P into the current network based on C, H, and O may enable a primitive analogue of bioenergetics that captures decarboxylative processes as high energy thioesters.5,17,33 This, in turn, could drive dehydrative polymerization reactions leading to the eventual emergence of functional polymers such as peptides and RNA. Thereafter, the efficiency of these primitive networks in early life would be improved through enzymatic catalysis.
Methods
General information
GC-MS analysis was performed on a GC System 7820A (G4320) coupled to an MSD block 5977E (G7036A). An Agilent High Resolution Gas Chromatography Column (PN 19091S – 433UI, HP – 5MS UI, 28 m×0.250 mm, 0.25 Micron, SN USD 489634H) was used. Hydrogen (99.999 % purity) was the carrier gas, supplied at a constant flow rate of 1.5 mL min-1. Samples were prepared in ethyl acetate (200 μL sample volume). The analysis was carried out on a 1 μL injection volume (splitless mode). The injection port temperature was 250 °C, and the column oven temperature program was: 60 °C for 1 min, then increased to 310 °C with a 30 °C min-1 ramp, followed by a 3 min hold (total running time 12.33 min). The mass spectrometer was turned on after a 2 min delay and was operated at the electron ionization mode with quadrupole temperature of 150 °C. Data was acquired in the full-scan mode (50-500). Data analysis and integration were performed using Agilent MassHunter Workstation v.B.06.00 software.
1H NMR spectra were recorded on a Bruker Avance400 (400 MHz) spectrometer at ambient temperature in a H2O:D2O mixture (6:1) as solvent, with sodium 3-(trimethylsilyl)-1-propanesulfonate (DSS) as the internal standard (CH3 peak at 0 ppm). Water suppression was achieved using the Bruker ZGESGP pulse program. Relaxation delay D1 was set to 87 s, with time domain size TD = 32768 and sweep width SWH = 4789.27 Hz (11.963 ppm). 32 scans were acquired for each sample. Integration was performed using MestReNova v6.0.2 software.
Materials
Unless otherwise noted, all reagents and solvents were purchased from commercial suppliers and used without further purification. Hydroxyketoglutarate and oxopentenedioate were prepared using a literature procedure.25 Water was obtained from a Milli-Q purification system (18 MΩcm) and was purged with argon before use. All glassware and stir bars were pre-washed with aqua regia, followed by distilled water and acetone, and oven dried to prevent any cross-contamination by metal salts.
Product identification
To facilitate GC-MS analysis, a literature derivatization procedure was applied to the sample to convert carboxy groups to ethyl esters, hydroxy groups to ethyl carbonates, amino groups to ethyl carbamates, ketones to diethyl ketals, and aldehydes to diethyl acetals.2,9 A 700 μL aliquot of the reaction mixture was basified using ~50 mg solid KOH (Merck EMSURE) and centrifuged (6000 rpm, 3 min). To 600 μL of the supernatant was added EtOH (300 μL) and pyridine (40 μL), followed by ethyl chloroformate (ECF, 40 μL). After vortex mixing for 30 s, a second 40 μL portion of ECF was added and vortex mixing was continued for another 30 s. To this, CHCl3 (200 μL) was added, followed by vortex mixing (10 s). Finally, saturated aqueous NaHCO3 (600 μL) was added and the mixture was vortex mixed again for 10 s. The CHCl3 layer was separated and dried over anhydrous Na2SO4. 50 μL of the dry CHCl3 layer was added with 150 μL of ethyl acetate to a vial and subjected to the GC-MS analysis.
Reaction products derivatized to ethyl esters were identified by comparing the mass spectra and retention times to those of analogously derivatized authentic samples, as shown below and also described elsewhere.9
Formate and acetate were determined using an NMR procedure and a GC-MS procedure, both reported in the literature.2
-
a)
NMR procedure: to a 1.5 mL plastic microtube was added ~1 mL of the reaction mixture and ~50 mg solid KOH (Merck EMSURE). The resulting thick suspension was centrifuged at 10 000 rpm for 20 min. To 600 μL of the supernatant was added 100 μL of 0.05 M solution of internal standard (DSS in D2O). The solution was analysed by NMR using the Bruker ZGESGP pulse program, as described above.
-
b)
GC-MS detection of formate and acetate as their amides with N-methylphenylethylamine: To a 120 μL aliquot of a reaction mixture in a 1.5 mL plastic microtube were added: 50 μL of 0.12 M solution of 1-hydroxybenzotriazole in H2O, 75 μL of 0.08 M 1-ethyl-3-(3-dimethyl-aminopropyl)carbodiimide solution (EDC) in acetonitrile, and 75 μL of 0.06 M N-methylphenylethylamine (MPEA) in acetonitrile. The resulting mixture was vortex mixed for 30 s and incubated at 60 °C for 45 min. After cooling to room temperature, 200 μL of CHCl3 was added and the mixture was vortex mixed for 30 s. The CHCl3 layer was removed and dried over anhydrous MgSO4. 50 μL of the CHCl3 supernatant was added to a vial together with 150 μL of EtOAc, and analysed by GC-MS.
Product quantification and error analysis
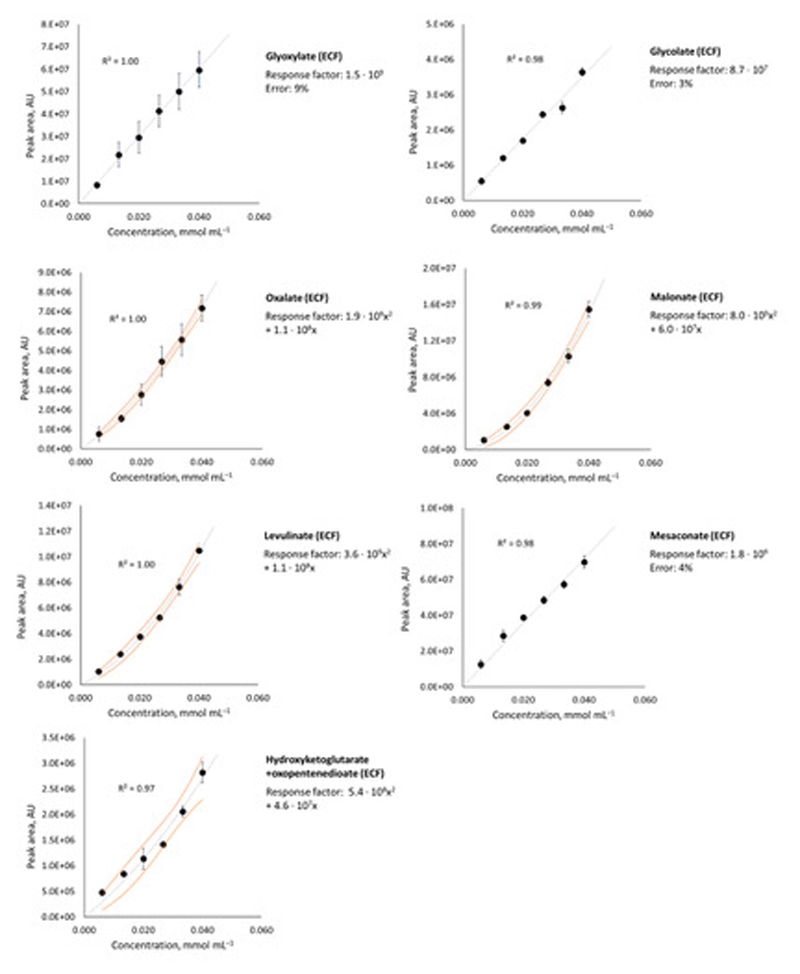
Carboxylic acids and amino acids were quantified according to a literature procedure,9 using 6-point calibration curves prepared from ECF/EtOH-derivatized solutions of authentic samples (0.006 M, 0.013 M, 0.020 M, 0.027 M, 0.033 M and 0.040 M) as described above. Each data point was obtained from three independent measurements and the correlation line was obtained from the least-squares fitting (intercept = 0). Error bars on graphs are shown as ± standard deviation for each data point. For the linear fits, overall percentage error of the response factor corresponds to ± standard deviation for each slope value. For the polynomial fits, 95% confidence envelopes are shown.
Calibration lines for pyruvate, malate, fumarate, succinate, α-ketoglutarate, isocitrate, cis-aconitate, tricarballylate and alanine are identical to those we previously reported, for the same analytical setup.9 Calibration lines for glyoxylate, glycolate, oxalate, malonate, levulinate, mesaconate, hydroxyketoglutarate/oxopentenedioate (HKG/OPD), glycine, aspartate and glutamate are shown in Extended Data Figure 2, and also Figure S2 and S10. Response factors corresponding to calibration lines for all the compounds detected by the GC-MS in this study are listed in the Table S1. Yields were calculated by comparing the GC peak area against the calibration line. Each reaction was performed at least twice, and reported yields are an average of those runs, with an error corresponding to ± standard deviation. Formate and acetate were quantified by NMR with DSS as standard, following a procedure we reported before.2 Equations corresponding to these calibration lines are listed in Table S1.
Hydroxyketoglutarate and oxopentenedioate (HKG/OPD) were quantified together (due to rapid interconversion of the two compounds during derivatization) as obtained in an aqueous reaction mixture (see Synthetic procedures below). The concentration of HKG/OPD in the reaction mixture was determined by NMR (ZGESGP pulse program, 200 μL reaction mixture + 500 μL 0.0360 M solution of sodium fumarate in D2O as internal standard) to be 0.0775 M. From this, six solutions were prepared by dilution, for subsequent ECF/EtOH derivatization and calibration on the GC-MS as described above for other carboxylic acids.
Mass spectra of all the compounds detected in this study through the derivatization with ECF/EtOH are shown in Figure S3.
Synthetic procedures
General procedure (qualitative metal screen): to a 10 mL Pyrex pressure tube were added sodium pyruvate (1.0 equiv, 0.10 mmol, 11 mg), sodium glyoxylate monohydrate (2.0 equiv, 0.20 mmol, 22 mg), transition metal salt (2.0 equiv, 0.20 mmol, 40 mg of MnCl2·4H2O or 40 mg of FeCl2·4H2O, or 48 mg of CoCl2·6H2O, or 48 mg of NiCl2·6H2O, or 34 mg of CuCl2·6H2O, or 27 mg of ZnCl2), and 3 mL of MilliQ water. The contents of the tube were flushed with argon. The tube was then sealed, and the reaction mixture stirred at 70 °C (1000 rpm, external heating block) for 3 h, followed by the KOH workup and ECF/EtOH derivatization, as described above.
General procedure (Fe2+-promoted reactions): to a 10 mL Pyrex pressure tube were added sodium pyruvate (1.0 equiv, 0.20 mmol, 22 mg), sodium glyoxylate monohydrate (2.0 equiv, 0.40 mmol, 44 mg), FeCl2·4H2O (2.0 equiv, 0.40 mmol, 80 mg), and 6 mL of MilliQ water. The contents of the tube were flushed with argon. The tube was then sealed, and the reaction mixture stirred at 70 °C (1000 rpm, external heating block) for up to 48 h, followed by the KOH workup and ECF/EtOH derivatization, as described above.
Reductive amination of glyoxylate/pyruvate network intermediates: to a 10 mL Pyrex pressure tube were added sodium pyruvate (1.0 equiv, 0.10 mmol, 11 mg), sodium glyoxylate monohydrate (2.0 equiv, 0.20 mmol, 22 mg), FeCl2·4H2O (2.0 equiv, 0.20 mmol, 40 mg), and 3 mL of MilliQ water. The contents of the tube were flushed with argon. The tube was then sealed, and the reaction mixture stirred at 70 °C for 1 h (1000 rpm, external heating block). Then, to the reaction mixture were added hydroxylamine hydrochloride (6.0 equiv, 0.60 mmol, 42 mg) and Fe0 powder (10 equiv, 1.0 mmol, 56 mg). The tube was sealed, and stirring was continued at 70 °C for 1 h, followed by the KOH workup and ECF/EtOH derivatization, as described above.
Hydroxyketoglutarate: prepared using a literature procedure23 from oxaloacetic acid (1.00 equiv, 0.902 mmol, 119 mg) in 9.88 mL of 1.0 M potassium phosphate buffer (pH 7.15) and glyoxylic acid (1.20 equiv, 1.08 mmol, added as 119 μL of 50% w/w aq. solution). The reaction mixture was stirred for 3 h at 25 °C, yielding a 0.0775 M solution of hydroxyketoglutarate (86%, NMR yield). 1H NMR (400 MHz, H2O+D2O) δ 4.26 (dd, J = 8.6, 3.4 Hz, 1H), 3.09 (dd, J = 17.8, 3.4 Hz, 1H), 2.95 (dd, J = 17.8, 8.5 Hz, 1H).
Time-point experiment (unlabelled compounds)
The experiment was performed in two replicas, in parallel. 700 μL aliquots of the reaction mixtures were drawn at the following time points: 5 min (substrates added and vortex mixed for 30 s), 1 h, 3 h, 7 h, 10 h, 24 h, 48 h. The results were normalized against a “0 h” mixture of sodium pyruvate (1.0 equiv, 0.20 mmol, 22 mg) and sodium glyoxylate monohydrate (2.0 equiv, 0.40 mmol, 44 mg) in water (3 mL), without Fe2+ added, derivatized with ECF/EtOH using the procedure described above. Reported percentage values are scaled against the number of carbon atoms in each compound, to account for the total carbon mass balance of the system (Table S2). GC chromatograms of the time-point experiment are shown in Figure S4, and mass spectra of all compounds detected at t = 24 h (the highest complexity) are shown in Figure S5.
The reaction mixture pH change over time was measured with an AquaLytic AL10pH handheld pH meter and found to equal ~4.45 at t = “0 h” and ~5.65 at t = 24 h.
Time-point experiment (with pyruvate-2-13C)
The experiment was performed in two replicas, in parallel, using sodium pyruvate-2-13C. 700 μL aliquots of the reaction mixtures were drawn at the following time points: 5 min (substrates added and vortex mixed for 30 s), 1 h, 3 h, 7 h, 10 h, 24 h, 48 h. The results were normalized against a “0 h” mixture of sodium pyruvate-2-13C (1.0 equiv, 0.20 mmol, 22 mg) and sodium glyoxylate monohydrate (2.0 equiv, 0.40 mmol, 44 mg) in water (3 mL), without Fe2+ added, derivatized with ECF/EtOH using the procedure described above. Reported percentage values are scaled against the number of carbon atoms in each compound, to account for the total carbon mass balance of the system (Table S4) and are shown in Figure S15.
The 13C label present in pyruvate was found to be carried over to the following intermediates: levulinate, malonate, malate, fumarate, succinate, α-ketoglutarate, hydroxyketoglutarate/oxopentenedioate, isocitrate, aconitate, tricarballylate, and acetate. Oxalate, glycolate and formate remained label-free, meaning these compounds are derived from glyoxylate.
See Figure S16 for relevant GC chromatograms and mass spectra, and Figure S20 for a graphical depiction of the 13C label propagation in the network.
Additional 13C labelling experiments (with pyruvate-1-13C, pyruvate-3-13C and glyoxylate-1,2-13C2) are described in the SI (see Figures S20-S29).
Detection of formate and acetate
Formate and acetate were detected in the reaction mixtures at the 48-h time-point, using NMR as well as GC-MS (derivatization with ECD/MPEA to amides2). See the Product Identification section for sample preparation details.
Results obtained for unlabelled starting materials are shown in Figure S7 (GC-MS) and Figure S8 (1H NMR), and for 13C-labeled pyruvate – in Figure S18.
Detection of glycine, alanine, aspartic acid and glutamic acid in the Fe2+-promoted reaction mixture
Reductive amination was performed according to the procedure described above. Following KOH workup and ECF/EtOH derivatization, four amino acids were detected as their ethyl esters (glycine, alanine, aspartic acid, glutamic acid). A GC chromatogram of a typical reaction mixture and relevant mass spectra are shown in Figure S8. Mass spectra of authentic amino acids derivatized with ECF/EtOH are shown in Figure S11.
Reductive amination of ketoacids with hydroxylamine
Reductive amination was performed according to the procedure described above on 0.1 mmol of each ketoacid (sodium glyoxylate monohydrate: 11 mg; sodium pyruvate: 11 mg; oxaloacetic acid: 13 mg; α-ketoglutarate: 15 mg), 0.1 mmol of hydroxylamine hydrochloride (1 equiv, 7 mg) and 1 mmol of Fe(0) powder (10 equiv, 56 mg). The reaction was carried out at 100 °C over 16 h to ensure completion. Obtained chromatograms and mass spectra of ECF/EtOH derivatized reaction mixtures are shown in Figure S9. See Table S3 for product yields.
Control experiments with Fe2+ (without glyoxylate)
Control experiments were performed on individual intermediates detected in the reaction network: glyoxylate, glycolate, pyruvate, oxalate, oxaloacetate, malonate, malate, fumarate, succinate, mesaconate, hydroxyketoglutarate/oxopentenedioate (HKG/OPD), α-ketoglutarate, isocitrate and aconitate (Figure S13).
The general procedure was used (except for HKG/OPD): 1.0 equiv (0.10 mmol) of a chosen intermediate, FeCl2·4H2O (2.0 equiv, 0.20 mmol, 40 mg) and 3 mL of MilliQ water. The contents of the tube were flushed with argon. The tube was then sealed, and the reaction mixture stirred at 70 °C (1000 rpm, external heating block) for 16 h, followed by the KOH workup and ECF/EtOH derivatization, as described above.
HKG/OPD was prepared using the procedure described above. From the reaction mixture 3 mL were taken and combined with FeCl3·6H2O (2.0 equiv, 0.20 mmol, 54 mg). The contents of the tube were flushed with argon. The tube was then sealed, and the reaction mixture stirred at 70 °C (1000 rpm, external heating block) for 16 h, followed by the KOH workup and ECF/EtOH derivatization, as described above.
Control experiments with Fe2+ and glyoxylate
Control experiments with Fe2+ and glyoxylate were performed on individual intermediates detected in the reaction network: glycolate, pyruvate, oxalate, oxaloacetate, malonate, malate, fumarate, succinate, mesaconate, hydroxyketoglutarate/oxopentenedioate (HKG-OPD), α-ketoglutarate, isocitrate and aconitate (Figure S14).
The general procedure was used: 1 equiv (0.1 mmol) of a chosen intermediate, sodium glyoxylate monohydrate (2.0 equiv, 0.20 mmol, 22 mg), FeCl2·4H2O (2.0 equiv, 0.20 mmol, 40 mg) and 3 mL of MilliQ water. The contents of the tube were flushed with argon. The tube was then sealed, and the reaction mixture stirred at 70 °C (1000 rpm, external heating block) for 16 h, followed by the KOH workup and ECF/EtOH derivatization, as described above.
Control experiments suggesting oxidation of malate to oxaloacetate
A control experiment was performed to trap oxaloacetate (malate oxidation product undetectable via derivatization with ECF/EtOH) as aspartate, through in situ reductive amination with hydroxylamine and Fe(0).
Two experiments were performed: with Fe2+ and Fe3+. Aspartic acid was detected in both cases, as shown in Figure S12 (see Figure S11 for an MS spectrum of an authentic sample of aspartate).
General reductive amination procedure was used: malic acid (1.0 equiv, 0.10 mmol, 13 mg) and iron salt (2.0 equiv, 0.20 mmol, 40 mg of FeCl2·4H2O or 54 mg of FeCl3·6H2O), and 3 mL of MilliQ water. The contents of the tube were flushed with argon. The tube was then sealed, and the reaction mixture stirred at 70 °C for 1 h (1000 rpm, external heating block). Then, to the reaction mixture were added hydroxylamine hydrochloride (6.0 equiv, 0.60 mmol, 42 mg) and Fe0 powder (10 equiv, 1.0 mmol, 56 mg). The tube was sealed, and stirring was continued at 70 °C for 1 h, followed by the KOH workup and ECF/EtOH derivatization, as described above.
Another indirect proof of malate oxidation is the presence of acetate detected in the reaction mixture comprising of malic acid and iron salts heated at 70 °C over 16 h.
Oxaloacetate is unstable in solution in the presence of transition metal salts, and decarboxylates to pyruvate, which, in turn, decarboxylates to acetate, easily detected by NMR (Figure S19). General procedure was used: malic acid (1.0 equiv, 0.10 mmol, 13 mg) and iron salt (2.0 equiv, 0.20 mmol, 40 mg of FeCl2·4H2O or 54 mg of FeCl3·6H2O), and 3 mL of MilliQ water. The contents of the tube were flushed with argon. The tube was then sealed, and the reaction mixture stirred at 70 °C for 16 h (1000 rpm, external heating block) followed by the KOH workup and NMR sample preparation, as described above.
Control experiment: hydroxyketoglutarate/oxopentenedioate + Fe3+
A control experiment was performed to evidence oxidative decarboxylations of HKG/OPD with Fe3+ species as oxidant. The results, highlighting the presence of fumarate and succinate, are shown in Figure S18.
HKG/OPD was prepared using the procedure described above. From the reaction mixture 3 mL were taken and combined with FeCl3·6H2O (2.0 equiv, 0.20 mmol, 54 mg). The contents of the tube were flushed with argon. The tube was then sealed, and the reaction mixture stirred at 70 °C (1000 rpm, external heating block) for 16 h, followed by the KOH workup and ECF/EtOH derivatization, as described above.
Extended Data
Extended Data Figure 1. Transition metal screen.
GC chromatograms showing a reaction network arising from pyruvate and glyoxylate, promoted by transition metal ions at 70 °C (qualitative screen).
Extended Data Figure 2. Calibration lines for carboxylic acids.
Correlation between the concentration of an aqueous solution of carboxylic acids (glyoxylic, glycolic, oxalic, malonic, levulinic, mesaconic and hydroxyketoglutaric + oxopentenedioic) and the measured gas chromatography peak area. Error bars correspond to the standard deviation (three independent runs). 95% confidence bounds computed for 2nd degree polynomial fits (OriginPro) are shown as orange lines.
Calibration lines for glycine, aspartic acid and glutamic acid are shown in Figure S10.
Calibration lines for the remaining compounds detected in this study (pyruvate, malate, fumarate, succinate, α-ketoglutarate, isocitrate, cis-aconitate, tricarballylate and alanine) are identical to those we previously reported, for the same analytical setup9 (Table S1).
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Acknowledgments
We thank W. F. Martin and D. Segrè for discussions.
Funding: This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement n° 639170) and from ANR LabEx “Chemistry of Complex Systems” (ANR-10-LABX-0026 CSC).
Footnotes
Data availability statement: All data is available in the main text, extended data, or the supplementary information file (Materials and methods, Figures S1 – S29, Tables S1 – S4).
Author contributions: J.M. supervised the research and the other authors performed the experiments. All authors contributed intellectually throughout the study. J.M. and K.B.M wrote the paper and K.B.M. and S.J.V. assembled the Supplementary information.
Author information: Reprints and permissions information is available at www.nature.com/reprints.
Authors declare no competing financial interests.
References
- 1.Braakman R, Smith E. The compositional and evolutionary logic of metabolism. Phys Biol. 2013;10:1478–3975. doi: 10.1088/1478-3975/10/1/011001. [DOI] [PubMed] [Google Scholar]
- 2.Varma SJ, Muchowska KB, Chatelain P, Moran J. Native iron reduces CO2 to intermediates and end-products of the acetyl-CoA pathway. Nature Ecology & Evolution. 2018;2:1019–1024. doi: 10.1038/s41559-018-0542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eggins BR, Brown EM, McNeill EA, Grimshaw J. Carbon dioxide fixation by electrochemical reduction in water to oxalate and glyoxylate. Tetrahedron Letters. 1988;29:945–948. [Google Scholar]
- 4.Marín-Yaseli MR, González-Toril E, Mompeán C, Ruiz-Bermejo M. The Role of Aqueous Aerosols in the “Glyoxylate Scenario”: An Experimental Approach. Chemistry - A European Journal. 2016;22:12785–12799. doi: 10.1002/chem.201602195. [DOI] [PubMed] [Google Scholar]
- 5.Goldford JE, Segrè D. Modern views of ancient metabolic networks. Current Opinion in Systems Biology. 2018;8:117–124. [Google Scholar]
- 6.Canfield DE, Glazer AN, Falkowski PG. The evolution and future of Earth’s nitrogen cycle. Science. 2010;330:192–196. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 7.Kalson N-H, Furman D, Zeiri Y. Cavitation-Induced Synthesis of Biogenic Molecules on Primordial Earth. ACS Central Science. 2017;3:1041–1049. doi: 10.1021/acscentsci.7b00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakurai M, Yanagawa H. Prebiotic synthesis of amino acids from formaldehyde and hydroxylamine in a modified sea medium. Origins of life. 1984;14:171–176. doi: 10.1007/BF00933655. [DOI] [PubMed] [Google Scholar]
- 9.Muchowska KB, et al. Metals promote sequences of the reverse Krebs cycle. Nature Ecology & Evolution. 2017;1:1716–1721. doi: 10.1038/s41559-017-0311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zubay G. The glyoxylate cycle, a possible evolutionary precursor of the TCA cycle. Chemtracts. 2003;16:783–788. [Google Scholar]
- 11.Ruiz-Mirazo K, Briones C, de la Escosura A. Prebiotic Systems Chemistry: New Perspectives for the Origins of Life. Chem Rev. 2014;114:285–366. doi: 10.1021/cr2004844. [DOI] [PubMed] [Google Scholar]
- 12.Sutherland JD. Opinion: Studies on the origin of life — the end of the beginning. Nature Reviews Chemistry. 2017;1 0012. [Google Scholar]
- 13.Harrison S, Lane N. Life as a guide to prebiotic nucleotide synthesis. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07220-y. 5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wächtershäuser G. Evolution of the first metabolic cycles. Proceedings of the National Academy of Sciences. 1990;87:200–204. doi: 10.1073/pnas.87.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith E, Morowitz HJ. Universality in intermediary metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13168–13173. doi: 10.1073/pnas.0404922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin WF, Russell MJ. On the origin of biochemistry at an alkaline hydrothermal vent. Philosophical Transactions of the Royal Society B. 2007;362:1887–1925. doi: 10.1098/rstb.2006.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman H. Speculations on the origin and evolution of metabolism. Journal of Molecular Evolution. 1975;4:359–370. doi: 10.1007/BF01732537. [DOI] [PubMed] [Google Scholar]
- 18.Camprubi E, Jordan S, Vasiliadou R, Lane N. Iron catalysis at the origin of life. IUBMB life. 2017;69:373–381. doi: 10.1002/iub.1632. [DOI] [PubMed] [Google Scholar]
- 19.Roldan A, et al. Bio-inspired CO2 conversion by iron sulfide catalysts under sustainable conditions. Chemical Communications. 2015;51:7501–7504. doi: 10.1039/c5cc02078f. [DOI] [PubMed] [Google Scholar]
- 20.Keller MA, Kampjut D, Harrison SA, Ralser M. Sulfate radicals enable a non-enzymatic Krebs cycle precursor. Nature Ecology & Evolution. 2017;1 doi: 10.1038/s41559-017-0083. 0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang XV, Martin ST. Driving Parts of Krebs Cycle in Reverse through Mineral Photochemistry. Journal of the American Chemical Society. 2007;128:16032–16033. doi: 10.1021/ja066103k. [DOI] [PubMed] [Google Scholar]
- 22.Novikov Y, Copley SD. Reactivity landscape of pyruvate under simulated hydrothermal vent conditions. Proceedings of the National Academy of Sciences. 2013;110:13283–13288. doi: 10.1073/pnas.1304923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldford JE, Hartman H, Smith TF, Segrè D. Remnants of an Ancient Metabolism without Phosphate. Cell. 2017;168:1126–1134. doi: 10.1016/j.cell.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Coggins A, Powner M. Prebiotic synthesis of phosphoenol pyruvate by α-phosphorylation-controlled triose glycolysis. Nat Chem. 2016;9:310–317. doi: 10.1038/nchem.2624. [DOI] [PubMed] [Google Scholar]
- 25.Springsteen G, Yerabolu J, Nelson J, Rhea C, Krishnamurthy R. Linked cycles of oxidative decarboxylation of glyoxylate as protometabolic analogs of the citric acid cycle. Nature Communications. 2013;9 doi: 10.1038/s41467-017-02591-0. 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller M, Turchyn A, Ralser M. Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean. Mol Syst Biol. 2014;10:725–725. doi: 10.1002/msb.20145228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouxel OJ, Bekker A, Edwards KJ. Iron isotope constraints on the Archean and Paleoproterozoic ocean redox state. Science. 2005;307:1088–1091. doi: 10.1126/science.1105692. [DOI] [PubMed] [Google Scholar]
- 28.Eschenmoser A. The search for the chemistry of life’s origin. Tetrahedron. 2007;63:12821–12844. [Google Scholar]
- 29.Mall A, et al. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science. 2018;359:563–567. doi: 10.1126/science.aao2410. [DOI] [PubMed] [Google Scholar]
- 30.Nunoura T, et al. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science. 2018;359:559–563. doi: 10.1126/science.aao3407. [DOI] [PubMed] [Google Scholar]
- 31.Orgel LE. The implausibility of metabolic cycles on the prebiotic Earth. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060018. 0060018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasas V, Szathmáry E, Santos M. Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life. Proceedings of the National Academy of Sciences. 2010;107:1470–1475. doi: 10.1073/pnas.0912628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Duve C. Blueprint for a cell: The nature and origin of life. Neil Patterson Publishers; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.