Abstract
Microscopic features (i.e., microstructure) of axons affect neural circuit activity through characteristics such as conduction speed. To what extent axonal microstructure in white matter relates to functional connectivity (synchrony) between brain regions is largely unknown. Using magnetic resonance imaging data in 11,354 subjects, we constructed multi-variate models that predict functional connectivity of pairs of brain regions from the microstructural signature of white matter pathways that connect them. Microstructure-derived models provide predictions of functional connectivity that explained 3.5% of cross-subject variance on average (ranging from 1-13%, or r=0.1-0.36) and reached statistical significance in 90% of the brain regions considered. The microstructure-function relationships were associated to genetic variants, co-located with genes DAAM1 and LPAR1, that have previously been linked to neural development. Our results demonstrate that variation in white matter microstructure predicts a fraction of functional connectivity across individuals, and that this relationship is underpinned by genetic variability in some brain areas.
Introduction
Communication between brain regions is achieved by axons grouped in white matter pathways. Properties of these structural connections are highly relevant to brain function, often described as functional connectivity. However, it is not simply the presence of a connection, but also the microscopic tissue architecture (i.e., microstructure) of white matter that influences brain function. For example, axonal diameter, myelination and length all affect the precise timing of neural signals, which is crucial to synchronizing network dynamics1.
Much of our knowledge about structural connectivity in the brain comes from animals2, human lesions3, and post-mortem human dissections4. These approaches have relatively high biological specificity and interpretability but are limited in their ability to characterize inter-individual differences. More recently, diffusion MRI (dMRI) has emerged as a powerful in vivo tool for studying the brain’s structural connections5. Although limited in spatial resolution6, dMRI has the unique ability to estimate the trajectories of white matter bundles (i.e., tractography) as well as some microstructural properties of these bundles, through models linking the within-voxel dMRI signal to tissue architecture. An important benefit of dMRI is that it enables us to characterize inter-individual differences, even in large cohorts (e.g., UK Biobank7). dMRI thus has the potential to relate individual variations in white matter microstructure to differences in brain function, which can also be characterized with MRI.
Diffusion and functional MRI have been used to investigate structure – function relationships, relating the anatomy of a white matter tract to the functional coupling between the regions it connects8–11. Importantly, these studies relate the macroscopic organization of the network to brain function but did not aim to establish whether the microstructural properties of a white matter tract relate to the functional communication it establishes between brain areas.
A few studies have demonstrated the potential for dMRI to establish relationships between microstructure and function. For instance, the commonly-used metric fractional anisotropy (FA) is a measure of diffusion directionality that is biologically non-specific, being sensitive to many properties including axon density, size and myelination12. Mean FA in a given white matter tract has been demonstrated to correlate with strength of functional connectivity13,14. However, these studies focus on the tract connecting a single pair of regions and summarise a tract’s microstructure with a single quantity (e.g. FA averaged over the entire tract).
In this work, we address whether functional connectivity between brain regions is mediated by microstructure of white matter pathways that connect them. We hypothesize that a data-driven model based on dMRI metrics can predict cross-subject variation in functional connectivity, and more specifically that this is a general principle that holds across many brain regions and the pathways connecting them. Unlike previous literature, we generate models that capture rich spatial representation of a tract’s microstructural profile (i.e., a microstructural signature). In addition to diffusion tensor based metrics, we incorporated estimates from a more sophisticated biophysical model that aims to provide greater biological specificity15. We consider interhemispheric connectivity between pairs of homotopic regions (i.e. the homologous region in the two cerebral hemispheres) that are connected by commissural white matter axons that run through the corpus callosum. We build a set of regression models to relate the tract’s microstructural profile to functional connectivity for a large number of paired homotopic regions.
The models described above linking white matter microstructure to functional connectivity were trained (n=7481) and replicated (n=3873) on data from the UK Biobank7. We show that these models can predict up to 13% of the cross-subject variance in functional connectivity, and demonstrate that the microstructure-function link exists for a large number of brain regions and is highly reproducible. We additionally performed genome-wide association studies (GWASs) to identify single-nucleotide polymorphisms (SNPs) that are significantly associated with functionally relevant microstructure in the brain16. The identified SNPs are co-located with genes that have been reported to play an important role in axonal guidance and cortical development.
Results
In our primary analysis, we tested for microstructure-function relationships between homotopic brain regions and the callosal pathways connecting them using dMRI and resting-state functional MRI (fMRI) data from subjects in the UK Biobank project7. All subjects were selected based upon usable resting-state fMRI and dMRI data, in addition to genetic inclusion criteria (seeMethods section). The activity of homotopic region pairs is often synchronized, with high functional connectivity17,18. These pairs are primarily connected through the corpus callosum, the largest commissural pathway in the brain, which is well defined at typical imaging resolutions employed with dMRI.
Functional connectivity
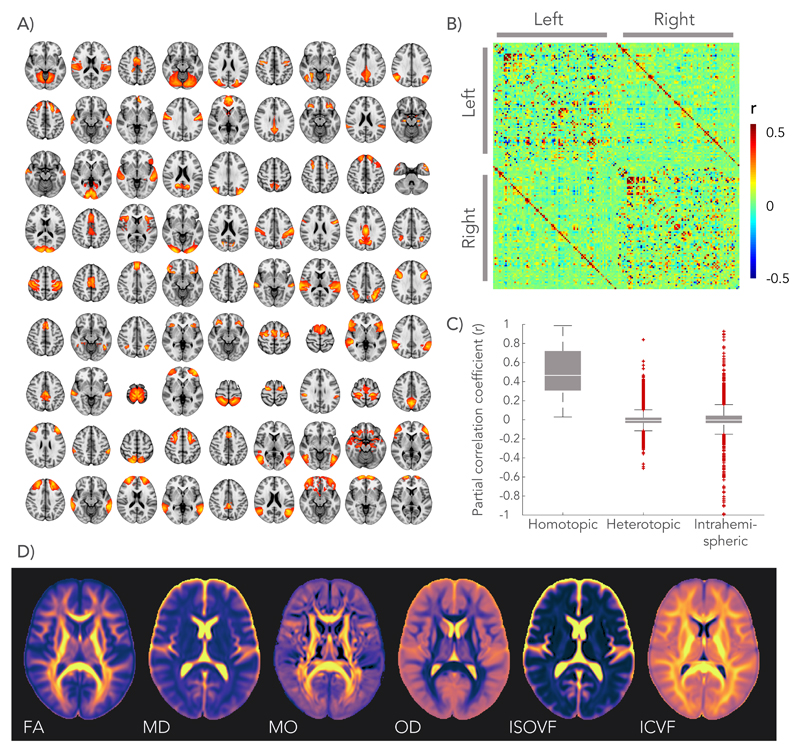
We previously conducted a group-average decomposition of resting-state fMRI data using independent component analysis (ICA), which yielded 55 components corresponding to resting-state networks7,19. For the work here, more finely-grained functional “nodes” were then generated from these components by first splitting each component into its constituent parts for right and left hemispheres, and further splitting if a component still contained non-contiguous brain areas. Homologous regions for the two hemispheres were then identified as nodes with strong similarity, producing 81 homotopic pairs (see Fig. 1.A). Functional connectivity was estimated at the single-subject level by partial correlation of the average BOLD signal time-series (equivalent to regressing out the time-series from all other regions prior to calculating pairwise correlations). This gives a connectivity matrix for each subject, which is summarized in Fig. 1.B as the mean partial correlation across all subjects. Entries in this matrix are ordered first by hemisphere and then by region number, such that inter-hemispheric connections are given in the upper-right and lower-left quadrants. Homotopic connections, shown on the diagonals of these quadrants, were found to express on average the strongest connections in the brain, larger than intra-hemispheric or heterotopic inter-hemispheric connections (see Fig 1.C), in agreement with previous studies17,18.
Figure 1.
Definition of homotopic brain regions and dMRI derived microstructural maps. A) Functional “nodes” were defined by applying independent component analysis to the resting-state fMRI data, splitting between the hemispheres, and isolating contiguous regions. These were then matched between hemispheres into 81 homotopic pairs, most automatically identified from the same independent component but 10 manually identified from different components. B) Connectivity between homotopic pairs was estimated by partial correlation of the average time-series of each node as shown in the connectome (matrix entries are sorted first by hemisphere and then by node number). C) Strength (partial correlation) of different functional connections in the brain, sorted by type. The centre line depicts the median correlation coefficient for a specific type of connection; box limits, the 25th and 75th percentiles of the correlation coefficients; the whiskers extend to the most extreme data points excluding outliers (marked with a + symbol). Group-average estimates from n = 11,354 subjects for n = 3240 connections evaluated. D) Group-averaged microstructure maps derived from the dMRI data.
White matter microstructural signature
A range of microstructural features was derived from the dMRI data for the white matter pathway connecting each pair of homotopic grey matter regions. The diffusion tensor model describes the 3D water displacement profile at each voxel using an ellipsoid20. We extracted estimates of fractional anisotropy (FA), mean diffusivity (MD) and anisotropy mode (MO)21 from this tensor fit. Neurite Orientation Dispersion and Density Imaging (NODDI)15 is a more biologically motivated model that aims to decompose the diffusion signal into an intra-cellular volume fraction (ICVF) and an isotropic volume fraction (ISOVF), the latter representing interstitial and cerebrospinal fluids. In addition, NODDI estimates an Orientation Dispersion (OD) index that quantifies the spread of fibres within the intra-cellular compartment. These dMRI-derived metrics represent an average across thousands of cellular components within each imaging voxel (2x2x2 mm3). Fig. 1.D depicts a brain map of each microstructural metric averaged across all subjects. The white matter pathway that connects a given homotopic region pair was identified using probabilistic tractography22 performed on the dMRI data between the regions.
Predicting functional connectivity with microstructure
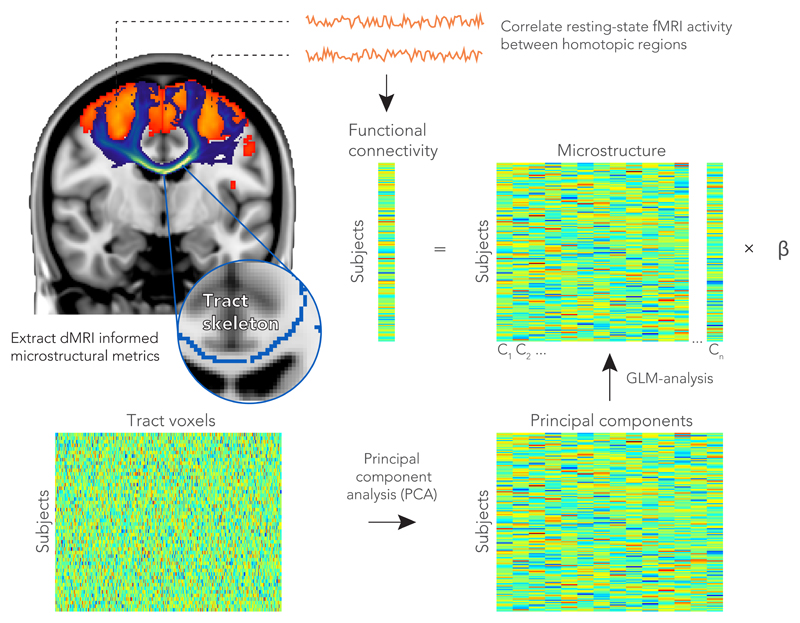
We performed a multiple regression analysis to test whether the microstructural features could predict cross-subject patterns of functional connectivity in the main cohort of 7,481 subjects. For a given homotopic pair of regions, the functional connectivity for all subjects was represented as a vector (Nsubjects x 1). To model the spatial patterns of white matter microstructure in a given tract, we begin by constructing a matrix that contains the dMRI-derived metric of interest for every subject (i.e., a Nsubjects x Nvoxels matrix). The included voxels are restricted to the centre of the tract of interest using a standard “skeletonization” procedure23. Because these microstructure matrices are too large to robustly perform a direct regression (Nvoxels = 5750±4000), we use principal component analysis to reduce the matrix dimensionality. The top 30 principal components (see Supplementary Fig. 1) were extracted to serve as a set of regressors, resulting in an Nsubjects x 30 regression matrix (see Fig. 2 for an overview). Seven linear models were created for each homotopic pair: one for each of the dMRI-derived metrics (FA, MD, MO, OD, ISOVF, ICVF) and a multimodal approach combining all these microstructural metrics in a single matrix. For the multi-modal analysis, the microstructural matrix for each metric was first normalized by its first singular value, and these normalized matrices were concatenated to form a single multimodal matrix (of size Nsubjects x 6Nvoxels) that was again reduced to include only the top 30 principal components.
Figure 2.
Prediction of functional homotopic connectivity from white matter microstructure. Between a pair of functionally defined homotopic areas (shown in orange in the brain), probabilistic tractography was performed to delineate the neuronal tract (shown in blue). The white matter skeleton voxels within a tract were stored as rows in a matrix for each subject. To extract the highest cross-subject variance among the TBSS voxels for a given microstructure metric, we performed a dimensionality reduction on this matrix using a principal components analysis. The top principal components (n = 30) were fed into a linear regression model as explanatory variables for the functional connectivity between a homotopic pair.
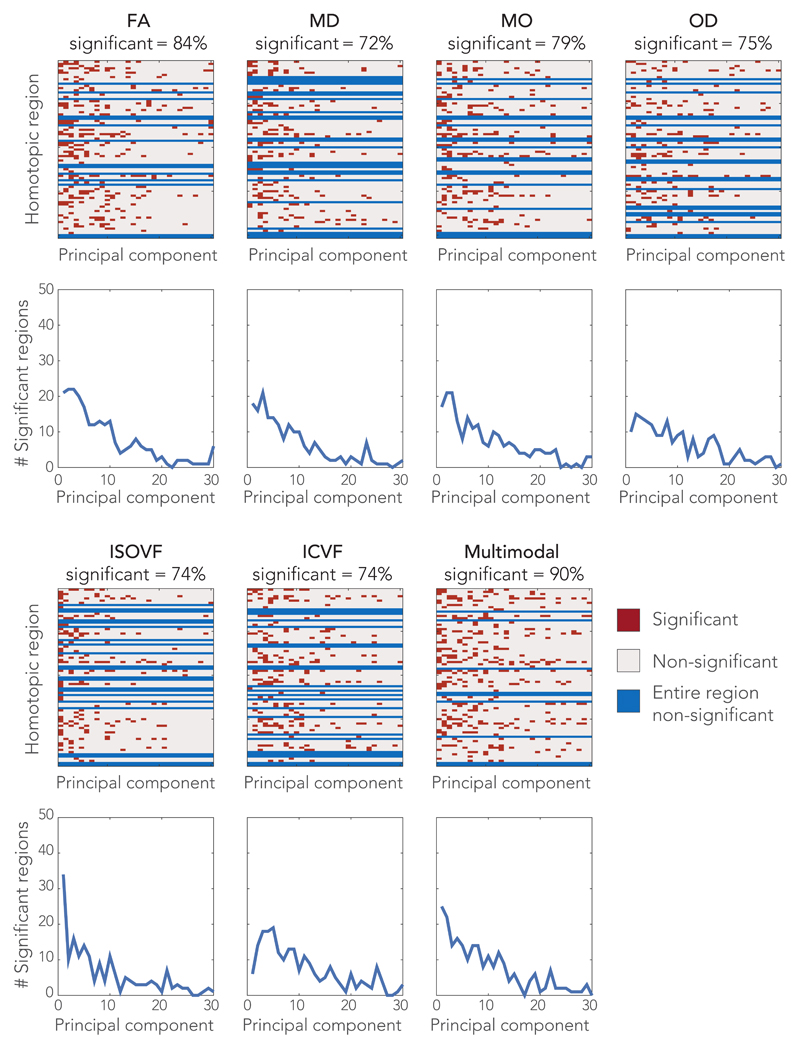
We first test the hypothesis that dMRI-based microstructure can be used to predict cross-subject variation in functional connectivity consistently across many brain regions. We assessed the statistical significance of each model using permutation testing, performed independently across the homotopic pairs and models, and then corrected for multiple comparisons (see Methods section). The significance (p<0.05, corrected) is indicated per microstructural metric in Fig. 3, and in more detail in Supplementary Fig. 2 as Manhattan plots of corrected p-values (family wise error). The overall regression model was able to predict a statistically significant amount of cross-subject variance in 72-90% of the homotopic brain regions (depending on the dMRI metric). The multi-modal microstructure model combining the six dMRI metrics provided a prediction of functional connectivity for the largest number of regions (72, representing 90% of the total brain areas considered). This result is not trivially guaranteed given that this model had the same number of regressors (30) as the other models. These results suggest a general relationship between microstructure and functional connectivity. We can further consider individual regressors (i.e., specific principal components). The statistically significant regressors generally correspond to the top principal components (left-most columns in Fig. 3). This indicates that the highest cross-subject modes of microstructural variation also explain the most cross-subject variation in functional connectivity. As the regressors reflect the primary modes of variation in the dMRI data but are used to model the fMRI data, this property is not trivially guaranteed. For some regions, no significant associations were found between homotopic functional connectivity and a given microstructure metric. The multi-modal microstructure model again resulted in the largest number of significant regressors.
Figure 3.
Significant associations between functional connectivity and microstructure of the connecting white matter tract. Each row in the matrices represents a homotopic region pair with each entry a regressor (on the microstructural principal components) of the linear model. Significance of the regressors is color-coded. The graphs depict the number of regions for which a particular rank order principal component yielded a significant regressor (so for example, in the multimodal models, the first principle component was significant for 28 brain regions). The percentage of homotopic region pairs demonstrating at least one significant regressor is given in the label of each matrix. Statistical significance was determined using permutation testing (two-sided, n = 100,000 permutations, Puncorrected < 2.9 x10-6, Pcorrected < 0.05, corrected for multiple comparisons).
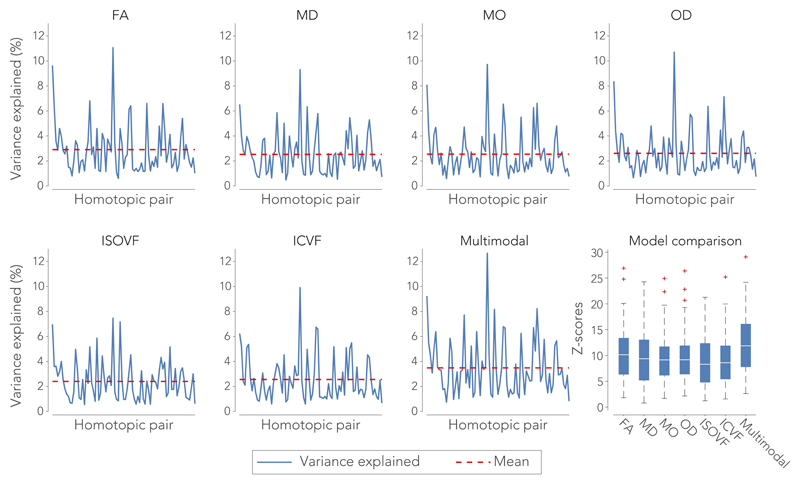
Having established that a microstructure-function link exists in most brain regions, we now consider the apparent strength of this relationship. Effect sizes of the regression models were evaluated in terms of percentage variance explained (equivalent to r2) in functional connectivity by the microstructural metrics. The average variance explained across all significant model fits was 3.5% (r = 0.19) for the multi-modal model that combines all dMRI microstructure metrics. Substantial variation in variance explained was found across the different brain regions investigated (Fig. 4). In the multimodal regression, variance explained was lowest for the middle temporal gyrus (1.1%, r = 0.09) and largest in the posterior cingulate cortex (12.7%, r = 0.36). These effect sizes are mapped back to the 81 homotopic region pairs to visualize how strongly functional connectivity is explained by the underlying microstructure across the cortex (Fig. 5A). In addition, Z-scores were computed to summarize the overall model fits. The multi-modal microstructure regression model yielded on average a higher score than the regressions with any single microstructural metric (Z = 12.0 Fig. 4), suggesting that the different microstructural metrics explain different variance in functional connectivity. The model incorporating FA shows the highest average Z-score of all individual metrics (Z = 10.5), although the different metrics perform overall fairly similar (Fig. 4). A list of all brain areas investigated with their corresponding effect sizes for the multimodal microstructure model is given in Supplementary Table 1.
Figure 4.
Percentage variance explained (r2) in the functional connectivity of each homotopic region pair by the microstructural metrics derived from the connecting white matter tract in the training cohort (n = 7481 subjects). The box-and-whisker plots on the bottom right depicts the model performance of each metric in terms of an F-to-Z transformed score. The centre line depicts the median Z-scores across the homotopic region pairs; box limits, the 25th and 75th percentiles of the Z-scores; the whiskers extend to the most extreme data points excluding outliers (marked with a + symbol).
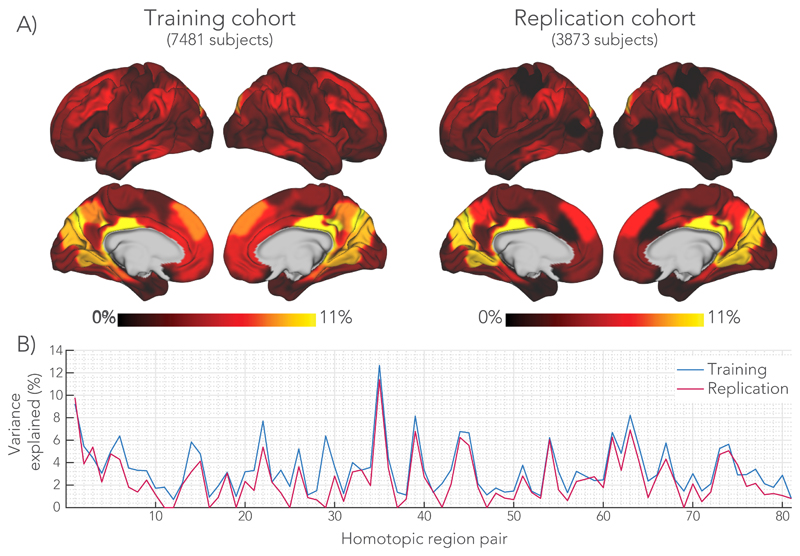
Figure 5.
Total variance explained (TVE) by the multimodal regression model in the training and replication cohorts. A) Variance explained mapped onto the brain surface. The maps were smoothed with 2 mm Gaussian kernel to aid visualization. A similar pattern across the brain was found for the regression models incorporating the individual microstructural metrics. B) Graph reporting percentage variance explained for each homotopic region. The model was trained on the main cohort of 7481 subjects. By applying the regression models trained on the main cohort, we could predict functional connectivity in the replication cohort of 3873 unseen subjects. The homotopic region numbers on the horizontal axis correspond to the brain areas listed in Supplementary Table 1.
Tensor-based features (FA and MD in particular) have been shown to provide sensitive indicators of changes to tissue microstructure in a broad range of contexts. However, these measures can be influenced by multiple aspects of tissue microstructure12, making interpretation difficult. We tested whether functional connectivity relates to a microstructure feature with greater biological specificity. We build on our previous work demonstrating quantitative agreement of OD estimates derived from dMRI data and with myelin stains in the same post-mortem human brain tissue24. The callosal OD profile correlated well between the ex-vivo imaging data (both MRI and microscopy) and the in-vivo dMRI NODDI analyses presented above, with both methods indicating high dispersion on the midline and lower dispersion in the lateral aspects of the callosum (Supplementary Fig. 3C). Furthermore, OD estimates at the midline of the corpus callosum was able to explain significant variance in interhemispheric functional connectivity (Supplementary Fig. 3). While the explained variance, 0.21% on average, was much less than with the spatially-extended microstructure models presented above, the validation against histology demonstrates biological specificity of this particular association.
Model replication
We further tested the validity of the above models by applying them to the replication cohort of 3,873 subjects. Each replication subject’s data was projected onto the 30 regressors and then multiplied by the regression coefficients estimated from the main cohort to predict that subject’s functional connectivity. That is, the models are applied directly and not retrained on the new subjects. This therefore constitutes a direct prediction of functional connectivity from dMRI data in unseen subjects. As shown in Fig. 5, percentage variance explained was quantitatively very similar from region to region (2.5% on average) in the previously unseen subjects as in the main cohort upon which the model was based.
Several medial regions demonstrate notably high effect sizes, with in particular the posterior cingulate cortex and the intra-calcarine cortex having over 10% variance explained. Regions in the temporal lobe, ventral parts of frontal lobe and lateral aspect of the occipital lobe demonstrate the lowest variance explained. In addition to the corpus callosum, temporal lobe regions are connected via the anterior commissure. For these regions, we performed additional analyses in which the microstructural signature from the anterior commissure was used to predict functional connectivity (see Supplementary Fig. 4). While the anterior commissure microstructure was able to predict functional connectivity, it did not explain the data better than callosal microstructure, nor did a model including both tracts.
Negative control analysis
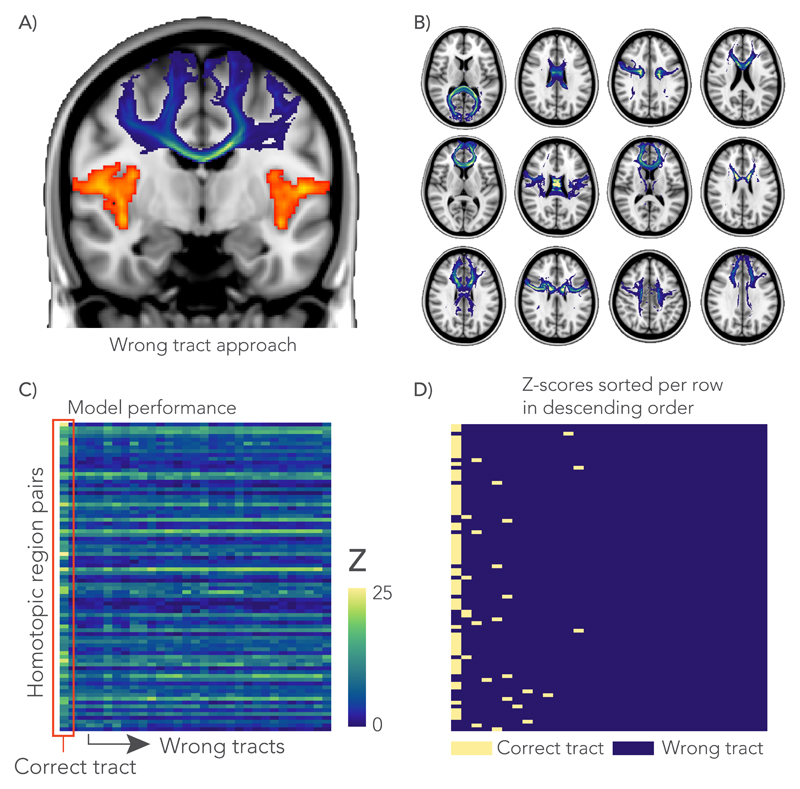
Although the above analyses suggest a general microstructure-function relationship, it is not clear whether these associations are specific to the pathway connecting a given pair of regions, or whether functional connectivity reflects global variance in the microstructural metrics across subjects. A new series of regression analyses were performed similar to those depicted in Fig. 2, but instead of taking microstructure values from a different “wrong” callosal tract (Fig. 6.A). From the 81 callosal sub-regions defined above, we selected a subset of 30 distinct tracts with minimal spatial overlap (Supplementary Fig. 5) for use as control (“wrong”) tracts (Fig. 6.B). We then assessed whether any of the control tract regressions had similar or better performance compared to the correct tract (Fig. 6.C). For 70% (60% in the replication cohort) of the homotopic areas, the highest Z-score was obtained when the model was performed with the anatomically correct tract; overall, for 81% of brain areas the correct tract ranked among the top three models (Fig. 6.D).
Figure 6.
Negative control analysis. A) In the wrong tract approach, the GLM-analysis was performed with a tract (shown in blue) that does not directly connect between a homotopic pair of interest (shown in orange). B) A total of 30 distinct tracts (12 shown here) were chosen based on minimal spatial overlap between them. C) For each GLM, an F-statistic (across the whole model, with the degrees of freedom for model and error, 30 and 7450, respectively) was calculated and transformed to a Z-score to compare between the correct tract and 30 wrong tracts. All GLMs in this analysis were derived from the multimodal microstructural information. D) Rows from the matrix in C were sorted in descending order of Z-score and labelled according to whether they represent the correct pathway or a different tract. The highest Z-scores (left-most column) correspond to the anatomically correct tract in 70% of cases, and overall the correct tract was in the top three models in 81% of cases.
Genome-wide associations
We studied the influence of genetics on the microstructure-function relationships identified above with a series of genome-wide association studies (GWASs). All subjects in this analysis were selected based on recent British ancestry and availability of genotype data that passed the quality control procedures of UK Biobank25. The target phenotypes used in the GWASs were the cross-subject variation in functional connectivity predicted by the microstructure model (i.e., the model fits; see Methods section and Supplementary Fig. 6). For each homotopic region pair, the GWAS consisted of a series of univariate correlations of the model fit with 11,734,353 single-nucleotide polymorphisms (SNPs). These GWASs were fully multiple comparison corrected.
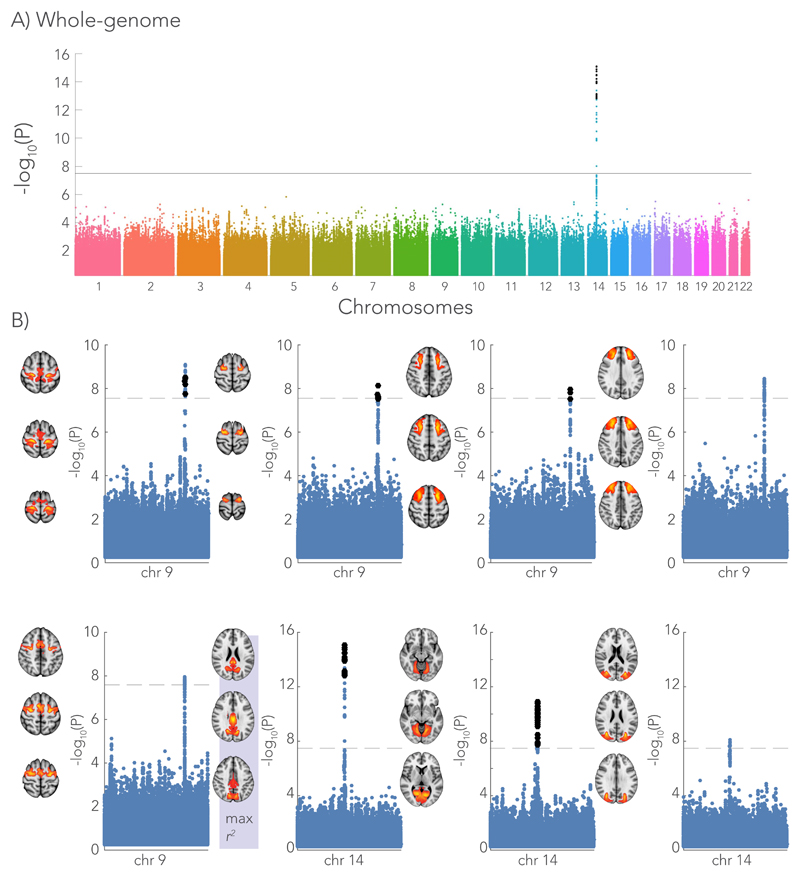
Figure 7 depicts the association across SNPs for the homotopic pair with the largest variance explained in the multi-modal microstructure model (i.e., the posterior cingulate cortex). A group of SNPs in chromosome 14 demonstrated a strong association with the microstructure-function phenotype. These SNPs were co-located with the DAAM1 gene (Dishevelled Associated Activator of Morphogenesis 1), some were also within DAAM1’s promoter region (regulating expression of the gene)26. The DAAM1-protein plays an important role in the Wnt signalling pathway inside the cell, indirectly regulating cell polarity and movement during development. In the central nervous system, this protein has been shown to facilitate the guidance of commissural axons at embryonic stage in mice and drosophila27,28. Expression of the JKAMP gene (Jun N-Terminal Kinase 1-Associated Membrane Protein) was also regulated by these SNPs, as demonstrated by 3D chromatin interaction data29 (Virtual 4C30). Furthermore, the GWAS revealed many SNPs within the LPAR1 gene (Lysophosphatidic Acid Receptor 1) located in chromosome 9. LPAR1 encodes one of the six receptors involved in the lysophosphatidic acid signaling pathway in the cell31. SNPs co-located with both DAAM1 and LPAR1 were found for the microstructure-function association of multiple brain areas (Fig. 7 and Table 1). Detailed Manhattan plots at the location of LPAR1 and DAAM1 are given in Supplementary Figures 7 and 8, respectively. Manhattan plots depicting the GWAS for the microstructure-function model fits of each homotopic region pair in the discovery cohort can be found in Supplementary Figure 9.
Figure 7.
Genome-wide associations with the microstructure-function phenotype (i.e. the pattern of functional connectivity that can be predicted from white matter microstructure). The Manhattan plot depicts the associations with each SNP across all chromosomes expressed as the -log10 p-value. A) As an example, the genome-wide Manhattan plot is given for the homotopic brain region showing the highest variance explained by the microstructure – function model (n = 7481 subjects). The strongest association is with a SNP (rs74826997) in chromosome 14 (linear regression, two-sided). B) Single chromosome Manhattan plots are shown for brain regions that associate with SNPs in either chromosome 9 or 14 that co-located with the genes LPAR1 and DAAM1, respectively (linear regression, two-sided). The -log10 p-value of the SNPs in the discovery GWAS (7481 subjects) are depicted by the blue dots. In an additional cohort of 3873 subjects, we aimed to replicate the significant hits (black dots in single chromosome Manhattan plots). The ICA spatial maps of these brain areas are given for each of Manhattan plot. The brain area (posterior cingulate cortex) highlighted with max r2 corresponds to the genome-wide Manhattan plot in (A). A significance threshold is given for a -log10(p-value) equal to 7.5 corresponding to a p-value of ~3 x 10-8. Significance threshold for the replication GWAS was determined using Bonferroni correction (p < 1.47x10-4).
Table 1.
Genome-wide associations (linear regression, two-sided) with the microstructure-function phenotype (i.e. the pattern of functional connectivity that can be predicted from white matter microstructure). Listed are rsids of the SNPs showing the most significant association that were replicated in the replication cohort. Some SNPs were associated with the microstructure-function model fits of multiple homotopic region pairs (highlighted in gray). The nearest gene of each SNP is reported with its possible function in the human central nervous system. Furthermore, the base-pair position, the SNP alleles, minor allele frequency (maf) and the p-values of the discovery (n = 7481 subjects) and the replication GWAS (n = 3873 subjects) are given. A significance threshold is given for a -log10(p-value) equal to 7.5 corresponding to a p-value of ~3 × 10-8. Significance threshold for the replication GWAS was determined using Bonferroni correction (p < 1.47x10-4)
| chr | node | rsid | nearest gene | function in central nervous system | position | ref allele | minor allele | maf | discovery p-value | replication p-value |
|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 60 | rs10980625 | LPAR1 | Lysophosphatidic Acid Signaling in central nervous system | 113665018 | C | C | 0.11 | 7.24E-09 | 5.56E-06 |
| 62 | rs34860245 | LPAR1 | 113709884 | T | T | 0.14 | 8.29E-10 | 3.76E-05 | ||
| 67 | rs4556147 | LPAR1 | 113651161 | A | A | 0.22 | 1.09E-08 | 7.12E-08 | ||
| 14 | 1 | rs76341705 | DAAM1 | Wnt signaling pathway, axonal growth and guidance | 59628679 | G | G | 0.12 | 1.28E-11 | 7.15E-08 |
| 35 | rs74826997 | DAAM1 | 59628609 | T | T | 0.12 | 8.15E-16 | 1.63E-05 | ||
| 15 | 33 | rs1080066 | C15orf54 | Associated with spinal cord tumor | 39634222 | A | A | 0.09 | 3.48E-13 | 3.18E-11 |
The GWAS was repeated for subjects in the replication cohort. Rather than using the model prediction approach described above, the multi-modal microstructure models were first re-trained to better explain functional connectivity with microstructure for these subjects (see Supplementary Fig. 10 for the effect of re-training). This approach was motivated to make the genetic replication analysis more fully independent of the discovery dataset. Replication GWAS was performed on microstructure-function phenotypes from the homotopic regions showing an association in chromosome 9 and 14 in the original subjects. Following common practice for replication GWA studies32, only SNPs that demonstrated a significant association in the discovery GWAS were tested. For SNPs within the LPAR1 gene in chromosome 9, associations with three out of five brain areas were replicated. The SNPs in chromosome 14 corresponding to the DAAM1 were replicated in two out of three brain areas (Fig. 7).
The GWAS results described above used microstructure-function model fits as the target phenotype. These results could simply reflect correlations of these SNPs with both functional connectivity and microstructure. To test for specificity, two additional GWASs were run using the following target phenotypes for each homotopic pair: (i) the functional connectivity that remains unexplained by white matter microstructure (i.e., the residuals from each multimodal microstructure-function model) and (ii) the first principal component of the multimodal microstructure for the corresponding callosal pathway (i.e. the first regressor in the microstructure model). SNPs that were significantly associated with either of these two phenotyes in the discovery cohort and in the replication cohort are listed in Supplementary Tables 2 and 3. These GWASs did not find any SNPs co-located with either DAAM1 or LPAR1 in any homotopic region. This suggests that the relationship to DAAM1 and LPAR1 is specific to the component of functional connectivity that can be predicted by white matter microstructure. The GWAS associating with the first principal component of multimodal microstructure yielded SNPs within the VCAN gene, which were previously found to associate with ICVF throughout white matter in the brain16.
Discussion
Although basic principles relating axonal properties to neural signalling are well established, the degree to which functional connectivity is mediated by microstructural organization at the level of macroscopic tracts is largely unknown. Several studies have related the “strength” and topology of structural connections to functional activity based on fMRI and dMRI10,33, but these studies are uninformative about microstructure. Here we focused on commissural fibres through the corpus callosum, a set of connections which can be estimated using MRI both structurally and functionally. Our results are consistent with previous work17,18 in that connections between pairs of homotopic areas were the strongest functional connections in the brain. Furthermore, studies have demonstrated that severing the corpus callosum reduces or extinguishes interhemispheric functional connectivity, providing evidence that communication between these regions is primarily facilitated by axons running through the callosum34,35.
In this study, we have demonstrated that white matter microstructure is associated with functional connectivity at the macroscopic level probed by imaging. The majority of brain regions (90%) show statistical evidence for a relationship between white matter microstructure and functional connectivity. Replication in nearly 4000 subjects demonstrates that the regression models fit in the main cohort have predictive power in unseen subjects.
On average these models account for 3-4% of the cross-subject variance in a given brain region, with considerable variation across regions – ranging from 1% to 13% variance explained. It is likely that our results underestimate the true relationships due to methodological limitations. MRI provides indirect estimates of functional connectivity and microstructure. In addition, the model order (linear with 30 regressors), choice of confounds (Supplementary Table 4), and potential for remaining indirect connections (regions not included in the partial correlation) could all lead to unexplained variance. This could be one source of the inter-regional variation, although some true biological variation is also likely. Methodological improvements may well increase the strength of the observed effect sizes. The ability to identify subtle relationships on the order of 1% (r = 0.1) is directly related to our large sample size; indeed, it is common for even smaller effect sizes to be considered valuable in genetic studies, provided replication is demonstrated. The recent advent of population-level imaging requires particular caution in distinguishing between the statistical significance and biological meaningfulness of a given result36. Nevertheless, the identification of small effects can be a first step toward aggregate measures with greater explanatory power; for example, polygenic risk factors for disease combining univariate GWAS outcomes with small effect sizes have been enabled by population-level genetics studies37.
In both the main and replication cohort, functional connectivity was best explained in regions close to the medial aspect of the brain, for example the intra-calcarine and posterior cingulate cortex (see Fig. 5). It should be acknowledged that some of these regions emerged as single contiguous nodes after spatial ICA, where more distal homotopic pairs were separated by other brain structures (see Fig. 1). This could reflect fMRI signal blurring between the hemispheres, driving up the apparent functional connectivity. However, it is unclear why this functional connectivity would be better predicted by a completely independent measure of white matter microstructure estimated from diffusion MRI. To investigate this effect further, we conducted a correlation of the white matter tract length with the variance explained across homotopic region pairs, finding no significant relationship (r = -0.04, p = 0.70).
For the majority of the considered homotopic pairs (70%), the strongest model prediction was derived from microstructure in the anatomically-correct pathway, compared to microstructure obtained from any of the 30 other callosal pathways. This negative control analysis is informative because it establishes that microstructure-function relationships have a degree of regional specificity and do not simply reflect global (brain-wide) inter-individual differences in microstructure and associated function. A similar result has been demonstrated for resting-state functional connectivity between posterior cingulate and medial-frontal cortices, with FA from the correct white matter pathway (cingulum) being more highly correlated than an unrelated tract13. Interestingly, for a minority of the brain areas investigated, functional connectivity was better explained by microstructure from a “wrong” white matter tract. Success of control tracts in predicting a given brain region could be driven by confounded microstructural estimates in the correct tract, partial overlap of tract segmentations, or global (brain-wide) variations in microstructure and functional connectivity.
A general and important confound in our models is “partial volume” (spatial overlap) of tracts. Ideally one would estimate the properties of each axon connecting two brain regions. Instead, dMRI averages within a voxel or region of interest (in our case, a white matter tract). As such, dMRI measurements often mix multiple different white matter bundles; for example, in the centrum semiovale, callosal fibres cross the corticospinal tract and superior longitudinal fasciculus. As a result, our regression models will have included microstructural estimates from other tracts. More advanced modelling to exclude or model these partial volume effects would be valuable to increase specificity without reducing sensitivity.
Frontotemporal regions were particularly prone to being explained by a control tract. These region also tend to have lower functional homotopic connectivity, in agreement with previous literature38. These regions may have fractionally less callosal input and be primarily connected to intra-hemispheric brain areas via associations fibres39. Many temporo-polar regions also have interhemispheric connections via the anterior commissure. We therefore also constructed microstructural models from connections running through the anterior commissure; however, these models did not improve the explained variance in functional connectivity (see Supplementary Fig. 4). The effect sizes for models based on the anterior commissure and the corpus callosum varied similarly across brain regions. This may reflect spatial overlap in the defined tracts close to grey matter, image registration errors, or variation in non-relevant variance in functional connectivity across regions (providing a ceiling on the explainable variance). In addition to brain activity, other sources may also contribute to the resting-state signal (see refs 40–42 for some excellent reviews).
Imaging microstructure with dMRI is a rapidly evolving field, including many models that were only recently developed. The biological interpretation of microstructural metrics is challenging and it is therefore not trivial to decompose the specific contributions of each microstructure parameter in explaining functional connectivity. The microstructural metrics used here each explain some unique aspects of the dMRI signal, but also share some mutual information (e.g., both ICVF and OD correlate with FA). The results presented here demonstrate that combining these metrics yields a more comprehensive characterization of the underlying microstructure. In addition, having the rich representation of microstructure along the entire tract better explained functional connectivity than simply using the mean of the tract (Supplementary Fig. 11).
To gain further insight into the microscopic tissue features driving the dMRI-derived metrics, evaluation against reference measures such as histology is essential. As such, we demonstrated good correspondence between OD profiles derived from the corpus callosum in ex-vivo dMRI and myelin staining24, providing confidence in the biological meaning of this specific measure. In agreement with histology24,43, the dMRI data used in our study indicates that fibres are more dispersed at the centre of the corpus callosum as compared to its lateral aspects. In Supplementary Fig. 3, we use this validated measure of fibre dispersion for a simple (single regressor) model to predict functional connectivity. However, this model provided much lower explanatory power (0.21% on average) than the multivariate regression models described above. This is likely because these more comprehensive models capture the spatial richness of microstructure metrics across the white matter tract, demonstrating how pooling multiple white matter phenotypes can explain more variance in functional connectivity.
Data richness in the UK Biobank project allowed us to associate genetic variants with the imaging derived phenotypes in this study. Meta-analyses in the ENIGMA consortium previously revealed genetic variants that were associated to imaging markers such as hippocampal volume44 and other subcortical structures32 in over 30,000 subjects. ENIGMA pools a vast collection of imaging data from several studies acquired with heterogeneous protocols. It remains to be seen whether the inclusion of a large number of subjects in ENIGMA effectively mitigates this data heterogeneity. In contrast, the UK Biobank project aims to maximise data homogeneity in 100,000 subjects with a common protocol and imaging platform7, which may enable more efficient identification of associations between imaging phenotypes and genetic variants16. Combining datasets from the UK Biobank and ENIGMA can be used to further boost statistical power or can be used separately to replicate discoveries.
We conducted a genome-wide association study (GWAS) for each homotopic region pair to associate SNPs with the fraction of functional connectivity that was predicted by microstructure. In chromosomes 9 and 14, a group of SNPs was found showing a strong association with the cross-subject pattern of functional connectivity predicted by microstructure for multiple brain areas (Fig. 7). As no SNPs associated with these same genes were found in GWASs relating solely to functional connectivity or microstructure, these associations appear to be unique to the microstructure–function relationship (see Supplementary Tables 2 and 3). For the replication cohort, the SNPs in chromosome 9 – co-located with LPAR1 – were replicated for three of the five brain areas showing hits in the discovery GWAS. The SNPs in chromosome 14 were replicated in two out of the three brain areas.
The identified SNPs in chromosomes 9 and 14 are associated with genes that have previously been shown to be important for brain development. The DAAM1 gene is expressed in many tissue of the human body and plays an important role in the Wnt signalling pathway45. In neuronal tissue, the DAAM1 protein is primarily found in the shaft of neuronal dendrites46 and in the developing brain it aids axonal guidance in targeting distal brain regions47. Knock-out studies in mice and drosophila have shown deficits in the central nervous system when DAAM1 is not expressed27. In particular, the formation of commissural fibres at an embryonic stage was disturbed28. Previous work relating cortical thickness to genetic variants also reported SNPs co-located with DAAM1 in the cuneus area16 (http://big.stats.ox.ac.uk). 3D chromatin data revealed that the SNPs in chromosome 14 also regulate expression of the JKAMP gene29. While diseases associated with JKAMP include medulloblastomas48, its exact mechanism in brain development is not well described in literature. For chromosome 9, several SNPs were located in the LPAR1 gene, encoding a receptor involved in the lysophosphatidic acid signalling pathway. These receptors are found on the membranes of most cell types in the central nervous system and have been linked to some neural processes including but not limited to neurogenesis, myelination, microglial activation, and astrocytes responses31,49.
The degree to which functional connectivity between brain regions is mediated by microscopic properties (microstructure) of the white matter pathways is a fundamental question in neuroscience. We demonstrated that a fraction of cross-subject variation in inter-hemispheric functional connectivity can be predicted from white matter tract microstructure connecting two homotopic regions. Our results suggest that microstructure-function relationships are general (across many brain regions), specific (with the correct tract out-predicting control tracts) and reproducible (as a prediction in a replication cohort). Furthermore, the microstructure-function association was underpinned by genetic variants and in particular with SNPs co-located with the genes DAAM1 and LPAR1. Attribution of these relationships to specific biological sources, ideally in a causal manner, cannot be achieved with this kind of observational study but would likely require interventional studies in animals50.
Materials & Methods
Data acquisition and pre-processing
We used resting-state functional MRI and diffusion MRI data provided by the UK Biobank project. An extensive overview of the data acquisition protocols and image processing carried out on behalf of UK Biobank can be found elsewhere7,19. Description of post-processing pipelines and acquisition protocols of MRI data in UK Biobank are available at http://biobank.ctsu.ox.ac.uk/crystal/docs/brain_mri.pdf. Unless stated otherwise, processing of the MR images was performed using FSL51. All imaging data was acquired on a 3T Siemens Skyra MRI scanner (software platform VD13) using a 32-channel receive head coil.
Resting-state fMRI data with 2.4 mm isotropic resolution and whole-brain coverage (field of view, 88x88x64 matrix) was acquired in a six-minute session (multiband acceleration 8, TR=0.735 ms, 490 time-points). The functional data was motion corrected 52 and FIX-cleaned53 to remove physiological noise and image artefacts, before transforming the data to a 2 mm MNI-template.
Diffusion MRI data were acquired at 2 mm isotropic resolution achieving whole brain coverage (field of view, 104x104x72 matrix) with two b-values (b=1000, 2000 s/mm2), with 100 unique gradient directions over the two shells (50 directions/shell). The total acquisition time was 7 minutes (multi-band acceleration 3, TE/TR was 92/3600 ms). After eddy current correction of all images54, tensor metrics (FA, MD, MO) were calculated from the lower shell (b=1000 s/mm2) using DTIFIT. Both shells were used to estimate the NODDI model15 metrics (ICVF, ISOVF, OD) using the AMICO toolbox55.
While not being explicitly used in this study, the UK Biobank imaging protocol includes several structural acquisitions that informed quality control pipeline and served as registration references for the functional and diffusion data7,19. T1-weighted structural scans were acquired using a 3D MPRAGE protocol (1.0x1.0x1.0 mm resolution, matrix 208x256x256, TI/TR = 880/2000 ms, in-plane acceleration 2). T2-weighted imaging using fluid-attenuated inversion recovery (FLAIR) contrast provides estimates of white matter hyperintensities (3D SPACE, 1.05x1.0x1.0 mm resolution, 192x256x56 matrix, TI/TR=1800/5000 ms, in-plane acceleration 2).
Quality control
Quality control (QC) is applied at several stages in this study. First, all raw data is subject to a standard pre-processing pipeline19 that generates several QC measures. The starting point for QC is the T1-weighted structural scan, which is essential for further processing of the other modalities (e.g., the generation of brain masks, tissue segmentations and as a reference for registration). Subjects are excluded if registration to standard space fails, likely due to excessive head motion, atypical structure and/or anatomical abnormalities (e.g., large ventricles). The full list of QC measures relating derived from the T1-weighted images is given elsewhere19. Based on the T1-weighted anatomical, 98% of all subjects were deemed suitable for further analysis. Next, the volume of white matter hyperintensities, used as confound variable, derived from the T2-FLAIR images is characterized with BIANCA56. This feature detects atypical structures and individuals with overt pathology19. Subjects can additionally be excluded from further analysis on the basis of their dMRI and fMRI data due to bad EPI distortions, failed registration to T1, extreme bias fields, unusable fieldmaps and/or severe motion artefacts. 87% of the dMRI datasets and 94% of the rfMRI datasets were considered suitable for further analysis based on these QC measures. The relatively large number of dMRI scans being excluded was caused by a change in processing protocol that deemed some early scans unsuitable.
All subjects selected in this study had both usable dMRI and rfMRI data in addition to suitable genetics data (see section UK Biobank genetics data for more information). This yielded a total of 11,354 subjects; 7481 in the main cohort and 3873 in the replication cohort (randomly assigned). Overall, 5393 females were included, the mean age was 62.8 (SD 7.4) years and all subjects had recent British ancestry. No power calculation was needed in advance and we used all samples available. UK Biobank is an observational prospective epidemiological study, and all analyses in our study use all available subjects that fulfil the criteria described above. Hence there is no equivalent process of randomization that comes into this analysis. For the exact same reason, no blinding step was involved.
Variations in white matter microstructure and/or functional connectivity may be influenced by some of the QC measures (e.g. head motion) in a subtle way that does not require subject exclusion, but which could confound associations. A set of variables of no interest (confounds) are listed in Supplementary Table 4 that are used to deconfound the data prior to modelling (see section Predicting functional connectivity from white matter microstructure).
fMRI processing
The resting-state fMRI data were fed into an Independent Component Analysis (ICA) using the MELODIC tool57 to identify resting-state networks present on average in the whole population. First, data was reduced to 100 dimensions using PCA and then fed into spatial ICA, from which 55 components corresponded to functional regions, and the other 45 judged to reflect physiological noise or image artifacts (“noise”)7,19. A functional component was split if it consisted of non-contiguous brain regions, yielding 81 bilateral (homotopic) regions that were further split between the hemispheres to estimate interhemispheric connectivity (see Supplementary Table 1). Average time-series were generated for all ICA components (i.e., homotopic areas and noise components) by a spatial regression of the subject’s voxelwise resting-state fMRI time-series with the ICA spatial maps. Further analyses were performed using the FSLNets toolbox (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSLNets). The average time-series within a homotopic area was demeaned and “cleaned” by regressing out the time-series from the 45 “noise” component time courses. Functional connectivity was estimated between all pairs of components (2x81) by means of partial correlation of the cleaned time-series using Ridge regression with a regularization factor ρ=1. Partial correlation aims to estimate direct connectivity between two areas by first regressing out all other regions’ time-series before calculating the correlation (i.e., established through inversion of the covariance matrix).
dMRI tractography
White matter tracts between functional regions were delineated using tractography. Up to three fibre orientations were fitted at each dMRI voxel in a Bayesian approach using bedpostX58 modified for multi-shell data59. Probabilistic tractography was then performed with the probtrackx2 algorithm22 by generating streamlines from a seed region (5000/voxel) in one hemisphere and only saving streamlines that passed through the corpus callosum and terminated in the same region in the contralateral hemisphere. This process was repeated by switching the seed and the target area between the hemispheres. The overlap of the identified tracts in this two-step approach were used to generate the mask corresponding to the tract of interest. The tracts were generated for all 81 homotopic pairs (each representing either the seed or the target area) for 10 subjects drawn from the UK Biobank dataset. Tracts between a given homotopic pair were then averaged across these subjects and served as a tract mask for all subjects stored in 1 mm MNI-space.
Tract based spatial statistics
Tract-based spatial statistics23 (TBSS) was used to align white matter tracts between subjects and extract microstructural information from the tract centre (skeleton). The version of TBSS used here employs an optimised non-linear registration (FNIRT) that avoids the need for the projection step in the original version of TBSS60. This avoids misalignment problems in which voxels can be projected onto a different tract that is in close proximity, an issue that has been highlighted from the original method61. The choice of FNIRT-based registration was motivated by its performance compared to other registration algorithms, as described previously19. We also evaluated the use of DTITK registration, which incorporates the full diffusion tensor to further improve the alignment of dMRI scans61, finding equivalent performance between the two algorithms (Supplementary Fig. 12 and 13). The tract reconstructions obtained with probabilistic tracking were used to mask the white matter skeleton voxels for a given homotopic region pair. Microstructural features derived from the diffusion tensor and NODDI fits were extracted from this final tract mask.
Predicting functional connectivity from white matter microstructure
We used a multiple linear regression model to predict homotopic functional connectivity from a set of regressors describing the spatial pattern of microstructure along a white matter tract. A rank-based inverse normal transformation was applied to all data to ensure normality. The regression model was constructed for each pair of homotopic regions separately:
Here Yi (Nsubjects x 1) is a vector that contains the functional connectivity values of all subjects derived from homotopic region i (over n = 81 regions). To build a model using p microstructural regressors, we need to estimate a set of regression coefficients β (p x 1) that describe the relative contribution from the microstructural metrics Xi (Nsubjects x p) along the white matter tract.
The regressors are derived in two stages. First, the microstructural metrics were extracted from the TBSS-voxels (white matter skeleton) corresponding to the tract of interest for every subject, yielding a matrix X°i (Nsubjects x Nvoxels). As the matrix X°i is very large, a direct regression with functional connectivity is ill conditioned. We therefore perform a dimensionality reduction on X†I to derive a set of regressors reflecting the primary modes of variation of a given microstructural metric across space for the cohort of subjects. The microstructural matrices were first demeaned, and then a singular value decomposition was computed from matrix X°i. The top p components were retained, yielding matrix Xi (Nsubjects x p). In practice, p was set to 30 principal components, which approximately corresponded to a transition in the spectrum of singular values in terms of variance explained. This provides a somewhat conservative model order below the point around p=100 at which variance explained roughly tracked noise singular vectors (Supplementary Fig. 1) and linear regression is prone to overfitting.
Matrices Xi were constructed for each of the microstructure metrics separately, yielding six single-metric linear regression models per homotopic region. In addition, a multimodal regression model was created that combined across all microstructure metrics. For the multimodal regression, all raw microstructure matrices (X†i) were demeaned and normalized through division by their first singular value to ensure comparable range of values. The six normalized matrices were then concatenated along the voxel dimension (Nsubjects x 6Nvoxels) and this matrix was reduced to the top 30 principal components as described above.
We defined a set of 64 confound variables of no interest that might bias the estimated regressors by correlating with the estimated microstructural measures (e.g. through artefacts such as partial volume). An overview of all confound variables is given in Supplementary Table 4. The confound variables were regressed out of the functional and microstructural data before fitting the regression models.
Statistical analysis
Statistical significance of the regression models was assessed by means of permutation testing, evaluating each regressor using a t-statistic. A null distribution was constructed for each model t-statistic by randomly permuting the functional connectivity values across subjects (100,000 permutations). Because multiple models were evaluated, correction for the family wise error (FWE) is also essential, where we corrected along three different dimensions of multiple comparison, as follows. First, we test multiple hypotheses in each model, i.e., which of the 30 microstructural principal components explains a significant amount of functional connectivity. Second, the models were applied to each of the 81 homotopic region pairs. Finally, a total of seven models (six individual microstructural models and one multimodal model) were evaluated for each homotopic pair. Following the approach demonstrated by Winkler et al62, a maximum t-statistic null-distribution across all dimensions (regressor, regions and models) was generated from the permuted t-statistics. From this maximum t-statistics null-distribution, a corrected p-value was estimated for each of the non-permuted t-statistics. Furthermore, an F-statistic was computed to judge the overall performance of each regression model (degrees of freedom model and error, 30 and 7450, respectively). The F-statistics were converted to Z-scores. Finally, the effect size of the regression models was expressed in terms of percentage variance explained (equivalent to r2), describing the strength of the relationship between microstructure and functional connectivity.
Negative control analysis
The statistical tests described above test whether there is a relationship between functional connectivity in a given brain region and the microstructure in the white matter pathway that connects them. However, this does not provide any insight into whether these relationships are specific: for example, microstructure and function could correlate at the whole-brain level. In this case, a regression model could indicate a statistically significant relationship even when using a white matter pathway that does not connect a given homotopic pair. Such a relationship could still be biologically meaningful, but the interpretation would change (e.g., demonstrating that individual brains vary globally from hypo- to hyper-connected).
To test this, a negative control analysis was performed to evaluate the uniqueness of the microstructure-function relationships. From the 81 tracts in our study, a subset of 30 tracts with minimal mutual overlap were selected as canonical control (“wrong”) tracts. To identify the set of canonical control tracts, the Dice similarity index was computed among all tracts to quantify spatial overlap. Using k-means clustering (k=3 clusters), a cluster of tracts with the lowest average similarity indices was selected (Supplementary Fig. 5).
The regression models were then re-evaluated for each homotopic area using the control tracts, rather than microstructure from the anatomically correct tract for the homotopic pair of interest. If, for a homotopic area, the anatomically correct tract was among the control tracts, an additional control tract was selected. To summarize, the regression models of the homotopic regions were performed once for microstructure from the correct tract and 30 times for the control tracts. Comparison between the correct and control tract analyses was conducted using the F-statistic converted to Z-scores.
UK Biobank genetics data
The GWASs were performed using the BGENIE software25. Acquisition and processing steps of the genetics dataset for all subjects in the UK Biobank project can be found in25. For the discovery cohort, we began with the set of 12,623 brain imaged UK Biobank subjects for whom data were released in July 2017. As in Elliott et al16, to avoid confounding effects that may arise from population structure or environmental effects, we selected a subset of 11,354 unrelated subjects with recent British ancestry. Ancestry was determined using sample quality control information provided by UK Biobank25. We then filtered the genetic data to remove SNPs with minor allele frequency < 0.01% or a Hardy-Weinberg equilibrium p-value of less than 10-7, yielding a total of 11,734,353 SNPs distributed across the 22 autosomes. Not all of the UK Biobank subjects who underwent brain imaging have usable data with a given MRI modality. All the 11,354 unrelated samples, were subjects which had usable dMRI and fMRI data according to previous quality control19. Subjects were assigned to the discovery and replication cohorts in a similar fashion as for the MRI analyses.
Ex-vivo MRI and histology data
MRI and microscopy data from three ex-vivo corpus callosum specimens were acquired and processed as described previously24. In brief, formalin fixed human brain tissue sections were scanned on a preclinical 9.4 T Varian MRI system. Diffusion MRI was performed with a spin-echo sequence with TE = 29 ms and TR = 2.4 s. Two shells were acquired (b = 2500 s/mm2 and b = 5000 s/mm2), each with 120 gradient directions and 0.4 mm isotropic resolution. Eight images with no diffusion weighting were acquired. A parametric model was fit to the dMRI signals from the b = 5000 s/mm2 dataset to obtain orientation dispersion (OD) estimates63.
Following MR scanning, the specimens were histologically sectioned and immunohistochemically stained for myelin (proteo-lipid-protein). The sections were digitized and we obtained fibre orientation estimates at each pixel using structure tensor analysis64. From a 2D local neighbourhood (0.4 x 0.4 mm) corresponding to the size of an MRI voxel, a fibre orientation distribution was computed from which orientation dispersion (OD) was derived. After registration of dMRI and microscopy data to the same image space65, dispersion estimates were compared against each other in the corpus callosum.
Ethics and informed consent
All participants in the UK Biobank project signed an informed consent which is controlled by a dedicated Ethics and Guidance Council (http://www.ukbiobank.ac.uk/ethics). The Ethics and Governance Framework can be found at http://www.ukbiobank.ac.uk/wp-content/uploads/2011/05/EGF20082.pdf. IRB approval, also from the North West Multi-center Research Ethics Committee, was obtained for the Ethics and Governance Framework.
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
All data in this study were obtained from the UK Biobank project (access number 8107). We are very grateful to all individuals who donated their time to participate in the UK Biobank study. K.L.M., M.K. and J.Mo. are supported by the Wellcome Trust (091509/Z/10/Z, 202788/Z/16/Z, 098369/Z/12/Z). The authors gratefully acknowledge funding from the Wellcome Trust UK Strategic Award [098369/Z/12/Z]. UK Biobank brain imaging and F.A.-A. are funded by the UK Medical Research Council and the Wellcome Trust. J.Ma. acknowledges funding for this work from the European Research Council (ERC; grant 617306) and the Leverhulme Trust. S.J. is supported by the UK Medical Research Council (MR/L009013/1). The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z). Finally, we would like to thank David G. Norris and Jose P. Marques for their valuable input on this work.
Footnotes
Data availability
All source data (including raw and processed brain imaging data and genetics data) is available from UK Biobank via their standard data access procedure (see http://www.ukbiobank.ac.uk/register-apply).
Code availability
The image processing pipelines of the MRI data in the UK Biobank project can be found at http://www.fmrib.ox.ac.uk/ukbiobank. Custom written Matlab code including the microstructure-function modelling is freely available at https://users.fmrib.ox.ac.uk/~jmollink/Biobank/Biobank.html.
Author contributions
J.Mo., S.M.S., S.J and K.L.M. designed the research. J.Mo. performed the research. K.L.M., F.A.-A., and S.M.S, developed acquisition and processing pipelines for the MRI data. L.T.E. and J.Ma. processed genetics data, provided tools for genome-wide associations analysis and gave feedback on genetics results. J.Mo., S.M.S., M.K., M.H., A.M.C.W., S.J and K.L.M. analysed the data and interpreted its outcomes. J.Mo. and K.L.M. wrote the manuscript, which was edited by all authors.
Conflict of interest
J.Ma. is a co-founder and director of GENSCI Ltd. S.M.S. is a co-founder of SBGneuro.
References
- 1.Buzsáki G. Rhythms of the brain. Oxford University Press; 2006. [Google Scholar]
- 2.Kötter R, Sommer FT. Global relationship between anatomical connectivity and activity propagation in the cerebral cortex. Philos Trans R Soc Lond B Biol Sci. 2000;355:127–34. doi: 10.1098/rstb.2000.0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rorden C, Karnath H-O. Opinion: Using human brain lesions to infer function: a relic from a past era in the fMRI age? Nat Rev Neurosci. 2004;5:812–819. doi: 10.1038/nrn1521. [DOI] [PubMed] [Google Scholar]
- 4.Schmahmann JD, Pandya DN. Fiber Pathways of the Brain. 2006. pp. 1–37. [Google Scholar]
- 5.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magn Reson Med. 2000;44:625–32. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 6.Jbabdi S, Johansen-Berg H. Tractography: where do we go from here? Brain Connect. 2011;1:169–83. doi: 10.1089/brain.2011.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller KL, et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci. 2016 doi: 10.1038/nn.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-State Functional Connectivity Reflects Structural Connectivity in the Default Mode Network. Cereb Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Heuvel MP, Mandl RCW, Kahn RS, Hulshoff Pol HE. Functionally linked resting-state networks reflect the underlying structural connectivity architecture of the human brain. Hum Brain Mapp. 2009;30:3127–3141. doi: 10.1002/hbm.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagmann P, et al. Mapping the Structural Core of Human Cerebral Cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koch MA, Norris DG, Hund-Georgiadis M. An Investigation of Functional and Anatomical Connectivity Using Magnetic Resonance Imaging. Neuroimage. 2002;16:241–250. doi: 10.1006/nimg.2001.1052. [DOI] [PubMed] [Google Scholar]
- 12.Beaulieu C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR Biomed. 2002;15:435–55. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- 13.van den Heuvel M, Mandl R, Luigjes J, Hulshoff Pol H. Microstructural organization of the cingulum tract and the level of default mode functional connectivity. J Neurosci. 2008;28:10844–51. doi: 10.1523/JNEUROSCI.2964-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahl M, et al. Human motor corpus callosum: topography, somatotopy, and link between microstructure and function. J Neurosci. 2007;27:12132–8. doi: 10.1523/JNEUROSCI.2320-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61:1000–16. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
- 16.Elliott LT, et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562:210–216. doi: 10.1038/s41586-018-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stark DE, et al. Regional Variation in Interhemispheric Coordination of Intrinsic Hemodynamic Fluctuations. J Neurosci. 2008;28:13754–13764. doi: 10.1523/JNEUROSCI.4544-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen K, et al. Stable long-range interhemispheric coordination is supported by direct anatomical projections. Proc Natl Acad Sci. 2015;112:6473–6478. doi: 10.1073/pnas.1503436112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alfaro-Almagro F, et al. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage. 2018;166:400–424. doi: 10.1016/j.neuroimage.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B. 1994;103:247–54. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 21.Ennis DB, Kindlmann G. Orthogonal tensor invariants and the analysis of diffusion tensor magnetic resonance images. Magn Reson Med. 2006;55:136–146. doi: 10.1002/mrm.20741. [DOI] [PubMed] [Google Scholar]
- 22.Behrens TEJ, Berg HJ, Jbabdi S, Rushworth MFS, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34:144–55. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith SM, et al. Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 24.Mollink J, et al. Evaluating fibre orientation dispersion in white matter: Comparison of diffusion MRI, histology and polarized light imaging. Neuroimage. 2017;157 doi: 10.1016/j.neuroimage.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matusek T, et al. Formin Proteins of the DAAM Subfamily Play a Role during Axon Growth. J Neurosci. 2008;28:13310–13319. doi: 10.1523/JNEUROSCI.2727-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avilés EC, Stoeckli ET. Canonical wnt signaling is required for commissural axon guidance. Dev Neurobiol. 2016;76:190–208. doi: 10.1002/dneu.22307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Won H, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. bioRxiv. 2017 doi: 10.1101/112268. 112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yung YC, Stoddard NC, Mirendil H, Chun J. Lysophosphatidic Acid signaling in the nervous system. Neuron. 2015;85:669–82. doi: 10.1016/j.neuron.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hibar DP, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honey CJ, et al. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A. 2009;106:2035–40. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Reilly JX, et al. Causal effect of disconnection lesions on interhemispheric functional connectivity in rhesus monkeys. Proc Natl Acad Sci. 2013;110:13982–13987. doi: 10.1073/pnas.1305062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roland JL, et al. On the role of the corpus callosum in interhemispheric functional connectivity in humans. Proc Natl Acad Sci U S A. 2017;114:13278–13283. doi: 10.1073/pnas.1707050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith SM, Nichols TE. Statistical Challenges in “Big Data” Human Neuroimaging. Neuron. 2018;97:263–268. doi: 10.1016/j.neuron.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 38.Tobyne SM, et al. A surface-based technique for mapping homotopic interhemispheric connectivity: Development, characterization, and clinical application. Hum Brain Mapp. 2016;37:2849–2868. doi: 10.1002/hbm.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmahmann JD, et al. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain. 2007;130:630–653. doi: 10.1093/brain/awl359. [DOI] [PubMed] [Google Scholar]
- 40.Smith SM. The future of FMRI connectivity. Neuroimage. 2012;62:1257–66. doi: 10.1016/j.neuroimage.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 41.Cordes D, et al. Frequencies contributing to functional connectivity in the cerebral cortex in “resting-state” data. AJNR Am J Neuroradiol. 2001;22:1326–33. [PMC free article] [PubMed] [Google Scholar]
- 42.Friston KJ. Functional and Effective Connectivity: A Review. Brain Connect. 2011;1:13–36. doi: 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- 43.Budde MD, Annese J. Quantification of anisotropy and fiber orientation in human brain histological sections. Front Integr Neurosci. 2013;7:3. doi: 10.3389/fnint.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hibar DP, et al. Novel genetic loci associated with hippocampal volume. Nat Commun. 2017;8 doi: 10.1038/ncomms13624. 13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–54. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 46.Salomon SN, Haber M, Murai KK, Dunn RJ. Localization of the Diaphanous-related formin Daam1 to neuronal dendrites. Neurosci Lett. 2008;447:62–67. doi: 10.1016/j.neulet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 47.Kida Y, Shiraishi T, Ogura T. Identification of chick and mouse Daam1 and Daam2 genes and their expression patterns in the central nervous system. Dev Brain Res. 2004;153:143–150. doi: 10.1016/j.devbrainres.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 48.Behrends U, et al. Novel tumor antigens identified by autologous antibody screening of childhood medulloblastoma cDNA libraries. Int J Cancer. 2003;106:244–251. doi: 10.1002/ijc.11208. [DOI] [PubMed] [Google Scholar]
- 49.González de San Román E, et al. Anatomical location of LPA 1 activation and LPA phospholipid precursors in rodent and human brain. J Neurochem. 2015;134:471–485. doi: 10.1111/jnc.13112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fields RD. A new mechanism of nervous system plasticity: activity-dependent myelination. Nat Rev Neurosci. 2015;16:756–767. doi: 10.1038/nrn4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–90. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 52.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–41. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 53.Salimi-Khorshidi G, et al. Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersson JLR, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–1078. doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daducci A, et al. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage. 2015;105:32–44. doi: 10.1016/j.neuroimage.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 56.Griffanti L, et al. BIANCA (Brain Intensity AbNormality Classification Algorithm): A new tool for automated segmentation of white matter hyperintensities. Neuroimage. 2016;141:191–205. doi: 10.1016/j.neuroimage.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beckmann CF, Smith SM. Probabilistic Independent Component Analysis for Functional Magnetic Resonance Imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- 58.Behrens TEJ, et al. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med. 2003;50:1077–88. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- 59.Jbabdi S, Sotiropoulos SN, Savio AM, Graña M, Behrens TEJ. Model-based analysis of multishell diffusion MR data for tractography: How to get over fitting problems. Magn Reson Med. 2012;68:1846–1855. doi: 10.1002/mrm.24204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Groot M, et al. Improving alignment in Tract-based spatial statistics: Evaluation and optimization of image registration. Neuroimage. 2013;76:400–411. doi: 10.1016/j.neuroimage.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bach M, et al. Methodological considerations on tract-based spatial statistics (TBSS) Neuroimage. 2014;100:358–369. doi: 10.1016/j.neuroimage.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 62.Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE. Permutation inference for the general linear model. Neuroimage. 2014;92:381–397. doi: 10.1016/j.neuroimage.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sotiropoulos SN, Behrens TEJ, Jbabdi S. Ball and rackets: Inferring fiber fanning from diffusion-weighted MRI. Neuroimage. 2012;60:1412–25. doi: 10.1016/j.neuroimage.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Budde MD, Frank JA. Examining brain microstructure using structure tensor analysis of histological sections. Neuroimage. 2012;63:1–10. doi: 10.1016/j.neuroimage.2012.06.042. [DOI] [PubMed] [Google Scholar]
- 65.Heinrich MP, et al. MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal. 2012;16:1423–1435. doi: 10.1016/j.media.2012.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.