Abstract
We present cisTopic, a probabilistic framework to simultaneously discover co-accessible enhancers and stable cell states from sparse single-cell epigenomics data (http://github.com/aertslab/cistopic). On a compendium of single-cell ATAC-seq datasets from differentiating hematopoietic cells, brain, and transcription-factor perturbations, we demonstrate that topic modelling can be exploited for a robust identification of cell types, enhancers, and relevant transcription factors. cisTopic provides insight into the mechanisms underlying regulatory heterogeneity within cell populations.
Genomic regulatory programs are driven by combinations of transcription factors that bind to cis-regulatory control regions, such as enhancers and promoters, thereby regulating the transcription of target genes. Although single-cell transcriptomics allows an unbiased detection of cellular diversity, reverse engineering the genomic regulatory code from the transcriptome remains a challenge. On the other hand, single-cell epigenomic techniques, such as scATAC-seq1, provide a more direct prediction of the genome-wide activity of enhancers and promoters in heterogeneous cell populations. In comparison to single-cell RNA-seq, the computational analysis of scATAC-seq data is more challenging due to the high dimensionality and sparsity of the data (Supplementary Table 1). Current methods to analyze scATAC-seq data can be divided in two classes (Supplementary Table 2), depending on whether they first cluster cells in a lower dimensional space and then infer differentially accessible regions between clusters2–4; or whether they first aggregate regions into cistromes (based on annotations or k-mer/motif enrichment) before cell clustering5–7. The first class is less suitable for the analysis of dynamic processes (where clusters are not clearly defined); and the second class relies on pre-existing annotations. In addition, neither of them is optimized for the unsupervised clustering of regulatory regions.
We reasoned that a co-optimized clustering of cells and regulatory regions can improve the discovery of cell states. To this end, we developed cisTopic, an unsupervised Bayesian framework based on topic modelling to classify regions into regulatory topics and to cluster cells based on their regulatory topic contributions. cisTopic uses Latent Dirichlet Allocation (LDA)8 with a Collapsed Gibbs Sampler9 to iteratively optimize two probability distributions: (1) the probability of a region belonging to a topic (region-topic distribution) and (2) the contribution of a topic within a cell (topic-cell distribution) (Fig. 1a, Supplementary Fig. 1 and Methods). The inferred “cis-regulatory topics” can be directly exploited for motif discovery to predict (combinations of) transcription factors and to explore variations in chromatin state. We evaluated cisTopic on a variety of data sets, including semi-simulated and real scATAC-seq data, as well as other types of single-cell epigenomics data, and found that cisTopic accurately recovers the expected cell types. Particularly at low read depth, topic modelling is more robust compared with previously published approaches. This is illustrated for one case study in Fig. 1b; for additional benchmarking we refer to the supplementary material (Supplementary Fig. 2-7). Importantly, cisTopic yields bona fide regulatory topics that reveal distinct regulatory programs with specific combinations of transcription factors. In addition, we found that topic modelling with Gibbs sampling is very fast, which allows up-scaling to large data sets such as the Mouse Cell Atlas2 (Supplementary Note 1; Supplementary Fig. 7).
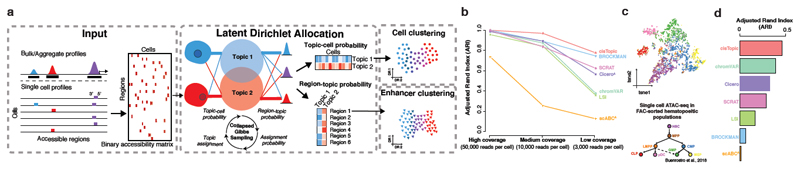
Figure 1. cisTopic workflow and application to hematopoietic differentiation.
a. The input for cisTopic is an accessibility matrix, which can be provided by the user or can be created from single-cell BAM files and candidate regulatory regions. Modelling with LDA is performed using a collapsed Gibbs sampler for the estimation of the region-topic and the topic-cell probability distributions. During this process, each region in each cell is iteratively assigned to a topic, based on the contribution of that topic to the cell and the contribution of that region (across the data set) to that topic. The resulting probability distributions can be used for cell clustering (topic-cell) and region clustering (region-topic). b. Adjusted Rand Index for current scATAC-seq analysis methods using 650 single-cell profiles simulated from bulk ATAC-seq data from hematopoietic populations26. Three data sets were simulated, using different read depth to assess the robustness of the methods. cisTopic has the highest ARI value even at low coverage. c. cisTopic cell-tSNE (based on the topic contributions to each of the 2,755 cells) colored by the FAC-sorted population of origin as annotated by Buenrostro et al.10. d. Adjusted Rand Index for current scATAC-seq analysis methods using 2,755 single-cell profiles from FAC-sorted populations in the hematopoietic system from Buenrostro et al.10. e. Example of 4 of the 17 topics found by the analysis of FAC-sorted populations from the hematopoietic system. Top: t-SNE based on topic-cell distributions colored by the normalized topic contribution in each cell. Middle: tSNE based on the region-topic distributions colored by the topic normalized region score. Bottom: Top enriched motifs in each topic with Normalized Enrichment Score (NES). (A) scABC and Cicero were run with minor adaptations compared to the original workflow, see Methods for details.
To further illustrate the principles of cisTopic, we applied it to a scATAC-seq data set with FAC-sorted differentiating cell types from the human hematopoietic lineage10. On this continuous data set, cisTopic correctly identifies the cell types and the expected developmental trajectory - based on 17 regulatory topics (Fig. 1c, Supplementary Fig. 8a-c)- with higher accuracy than alternative approaches (Fig 1d). Topic contributions per cell are used to reconstruct the developmental trajectory, to reveal differentiation states, and to uncover patient-specific batch effects (Supplementary Fig 8a-d; Supplementary Note 1); while the region-topic likelihood is used to visualize and cluster high confidence co-accessible regions (Fig. 1e). Among the 17 topics, we found one general topic (Topic 3), which contributes to all cells and represents mainly proximal promoters, with higher GC content (Fig. 1e, Supplementary Fig. 8e); while other topics are more specific to differentiation stages. For example, topics 12, 10 and 1 are predominant in the Common Lymphoid Progenitors (CLP), plasmacytoid Dendritic Cells (pDC) and Granulocyte-Macrophage Progenitors (GMP) populations respectively, and motif enrichment of the regions belonging to these topics reveals known master regulators of these cell types, such as EBF1, PU.1 and IRF, and PU.1 and CEBP, respectively11–13.
Interestingly, we can exploit this "enhancer-tSNE" to validate the clustering of regions using independent epigenomic data sets. For example, three different topics (Topics 15, 13 and 5) are enriched for GATA motifs, and genomic regions enriched in these topics in the tSNE are also enriched for GATA2 ChIP-seq peaks (Supplementary Fig. 9a). These topics delineate the differentiation path from Hematopoietic Stem Cells (HSC) to Megakaryocyte-Erythroid Progenitors (MEP). Differential motif enrichment between these regions (see Methods) finds PU.1 and ETS-like motifs in early GATA regions (HSC), RUNX in intermediate GATA regions and E-box motifs in the late GATA cistrome (MEP) (Suplementary Fig. 9b), in agreement with literature13–15. Note that these time-point specific GATA topics are only found by cisTopic; but not by cistrome-based methods such as chromVAR, which can only detect an average GATA cistrome across the time points since the cistrome has to be defined a priori (Supplementary Fig. 9c).
Next, we asked whether cisTopic could provide insights into more biologically complex populations, such as the mammalian brain. We applied cisTopic to a large scTHS-seq data set from the human brain16, including cell populations from the cerebellum, frontal cortex and visual cortex; and to a scATAC-seq data set from the mouse prefrontal cortex17. In both analyses, cisTopic was able to recover the major cell types in the brain, and revealed subpopulations of neuronal types (Fig. 2a,b). On the human brain data set, cisTopic revealed three subpopulations of excitatory neurons, linked to the cortical layer of origin (ExL23, ExL4 and ExL56); and two subpopulations of interneurons, distinguished by their developmental origin from subcortical regions of the medial or lateral/caudal ganglionic eminences (Fig. 2a, Supplementary Fig. 10a-c). In addition, using SCENIC18 on matching scRNA-seq data, we confirm that the inferred epigenomic topics correspond to cell type specific transcriptomes (Supplementary Fig. 10d,e). In the mouse brain data set, cisTopic identified previously unannotated cells as interneurons and four subpopulations of excitatory neurons: one that resembles the dentate gyrus neurons (also found by Preissl et al.17), and three populations linked to the cortical layers (ExL23, ExL4, ExL56) (Fig. 2b, Supplementary Fig. 11a-d). These subpopulations were validated by the enrichment of layer-specific signatures from FAC-sorted bulk ATAC profiles19, cross-species mapping with the human layer-specific topics, and motif enrichment of layer-specific master regulators (Fig. 2c-d, Supplementary Fig. 11d). For example, we find Egr motifs to be significantly enriched in ExL23 enhancers (Egr4), Ror motifs in ExL4 enhancers (Rorb), and Fezf2 motifs in ExL56; each of which are linked to transcription factors specifically expressed in those respective layers (Fig. 2c, Supplementary Fig. 11e) (see Methods).
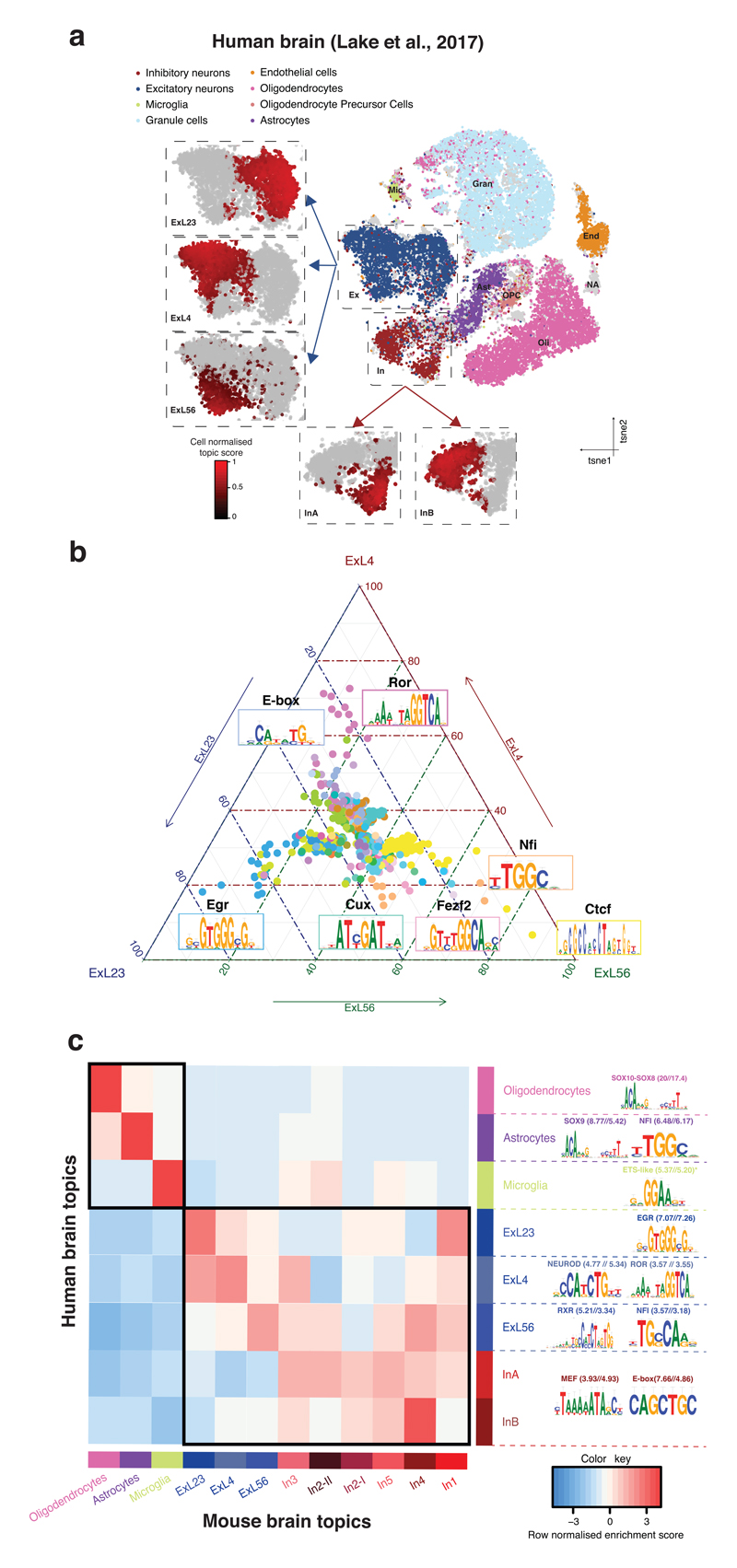
Figure 2. cisTopic unravels the regulatory heterogeneity in the mammalian brain.
a. cisTopic tSNE based on topic-cell contributions from the analysis of the human brain data set (34,520 cells). cisTopic identifies the main cell types and subpopulations of interneurons (InA and InB) and excitatory neurons (ExL23, ExL4 and ExL56). The insets show cell-type specific topic enrichment scores. b. cisTopic tSNE based on topic-cell contributions from the analysis of the mouse brain data set (3,034 cells). cisTopic identifies subpopulations of interneurons previously unannotated (in grey) and excitatory neurons (Dentate Gyrus, ExL23, ExL4 and ExL56; insets). The insets show cell-type specific topic enrichment scores. c. Ternary plot based on the mean Cis-Regulatory Module (CRM) scores per region set (i.e. topic) for differentially enriched motifs between the layer-specific topics. Each corner represents a layer-specific topic, dots represent enriched motifs and axes represent scaled CRM scores for each topic. The colors of the dots are used to indicate which motifs belong to the same transcription factor (based on STAMP clustering). d. Enrichment of human topics ('lifted over' to mm10) in the mouse topics.
We also tested to what extent the neuronal and glial cell type-specific topics in the human and mouse brain are orthologous to each other (Fig. 2d). The strongest conservation is observed for glial cell types, namely oligodendrocytes, astrocytes, and microglia; for which both the topic and the underlying enhancer architectures are strongly conserved (Fig. 2c, Supplementary Fig. 12). These candidate enhancer signatures in oligodendrocytes and astrocytes were further validated by cross-species comparisons, correspondence with single-cell RNA-seq data, and correspondence with independent epigenomic signatures (Supplementary Fig. 12).
Finally, we used cisTopic to investigate dynamic changes in chromatin accessibility during the perturbation of a transcription factor, using SOX10 in melanoma cell lines as a model system, where SOX10 is a key regulator20. We performed scATAC-seq in time series after knockdown (KD) of SOX10 in two short-term patient cultures (MM057 and MM087), sampling at 0, 24, 48 and 72 hours after SOX10 KD20,21 (Fig. 3a, Supplementary Fig. 13). cisTopic recapitulated dynamic cell state changes upon SOX10 KD, finding 15 topics (Fig 3b). The SOX10 gene regulatory network is significantly affected after SOX10 KD, as apparent by the loss of accessibility of known and experimentally validated SOX10 target regions of DCT, TYR and ERBB322–24; while regions that gain accessibility are enriched for AP-1 and TEAD binding sites, as expected20 (Fig. 3c). We identified three topics that represent a decline in accessibility: one including common regions (topic 14) and two cell-line specific topics (topic 11 and 12) (Fig. 3d). The enhancers composing these three “loss of accessibility topics” are highly enriched for the SOX10 motif and represent bona fide SOX10 binding sites, overlapping significantly with SOX10 ChIP-seq peaks25 (Fig. 3d) (p-value < 2.2x10-16). In addition, comparison of the melanoma SOX10 binding sites with oligodendrocyte and astrocyte cell-type specific SOXE binding sites revealed cell-type specific cofactors of the SOXE factors such as TFAP2 and AP-1, OLIG1/2, and NFIA/B in melanoma, oligodendrocytes and astrocytes, respectively (Supplementary Note 2, Supplementary Fig. 14).
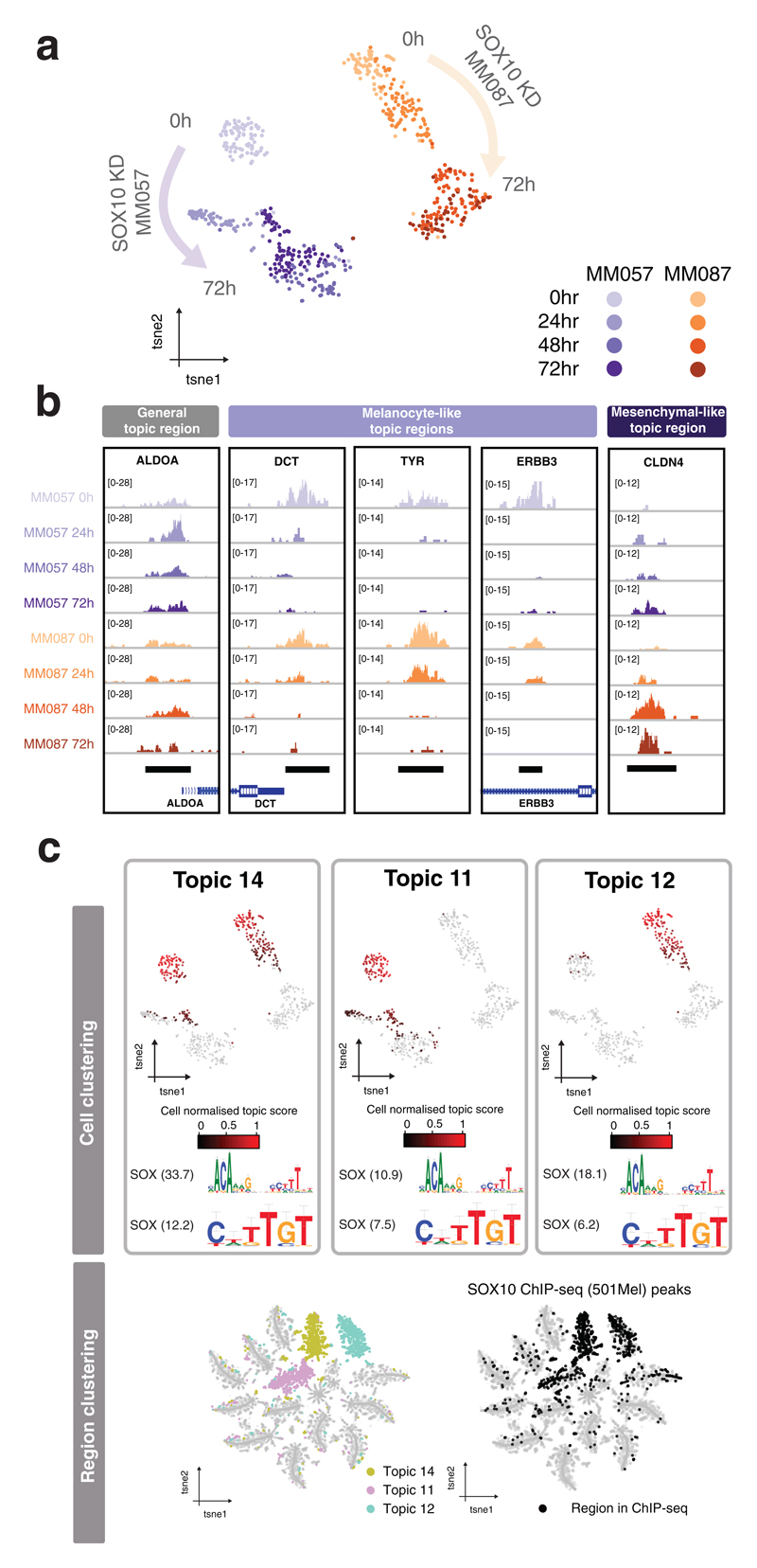
Figure 3. scATAC-seq during SOX10 knockdown in melanoma reveals a core set of melanoma SOX10 enhancers.
a. scATAC-seq was performed with the Fluidigm C1 on two melanocyte-like melanoma lines (MM057 and MM087) during SOX10 KD at four different timepoints (0, 24, 48 and 72 hours post-SOX10 KD). b. tSNE-representation (598 cells) generated by cisTopic using the topic-cell distributions. c. Aggregated scATAC-seq profiles of single cells per condition on a region of a general topic, four melanocyte-like topic regions (topic 14) that are known SOX10 target genes and a mesenchymal-like topic region. d. cisTopic identified three regulatory topics (topic 14, 11 and 12) enriched for SOX10 binding sites that loose accessibility during SOX10 KD. Left: Cell-tSNE colored by normalized topic score, together with motifs enriched in these topic regions (NES scores are mentioned). Right: Region-tSNE colored by topic normalized region scores and overlap with SOX10 ChIP-seq peaks.
Our results show that topic modelling provides a valuable component in the analysis of large-scale single-cell epigenomics data sets. It allows to jointly optimize cell clustering and enhancer categorization, to identify subpopulations of cells and co-accessible enhancers that represent shared epigenomic programs. In summary, cisTopic is a generally applicable method to discover and interpret regulatory topics and cell states from sparse single-cell epigenomics data.
Online methods
cisTopic workflow
cisTopic consists of 4 main steps: (1) generation of a binary accessibility matrix as input for Latent Dirichlet Allocation (LDA); (2) LDA and model selection; (3) cell state identification using the topic-cell distributions from LDA and (4) exploration of the region-topic distributions. cisTopic is available as an R/Bioconductor package at: http://github.com/aertslab/cistopic.
Input and binarization
The input for cisTopic is an accessibility matrix, which can be built from a set of single-cell bam files and a bed file with candidate regulatory regions (e.g. from peak calling on the aggregate or the bulk profile). In the case of single-end reads, we count a fragment if its 5' end falls within the region; in the case of paired end data, if any of its ends falls within the region. By default, we consider a region accessible if at least one read is found, leading to a binarized count matrix. In the case of single-cell methylation data, the matrix can be built from the beta values scores per region per cell, which can also be calculated if the user provides the methylation call files (i.e. tab-delimited files containing chromosome, position, number of methylated reads and total number of reads). By default, we consider a region methylated if the beta value is above 0.5; however, LDA can be run using directly the beta values (see Data Analysis). Note that regions should be blacklisted for potential artefacts prior to the analysis (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/).
Modelling via Latent Dirichlet Allocation
The next step in the cisTopic workflow is to use Latent Dirichlet Allocation (LDA) for the modelling of cis-regulatory topics. LDA allows to derive, from the original high-dimensional and sparse data, (1) the probability distributions over the topics for each cell in the data set (θ) and (2) the probability distributions over the regions for each topic (ϕ)8. These distributions indicate, respectively, how important a regulatory topic is for a cell (θ), and how important regions are for the regulatory topic (ϕ). Here, we use a collapsed Gibbs sampler9, in which we assign each region in each cell to a certain topic by randomly sampling from a distribution where the probability of a region being assigned to a topic is proportional to the contributions of that region to the topic and the contributions of that topic to the cell:
Where:
zi is the current assignment to be made,
z-i are the rest of assignments in the data set,
t is the given topic,
r is the given region,
and P(zi = t | z-i, r) is the probability of assigning the given region r to a regulatory topic t given the rest of the assignments in the data set.
is the number of times the given region r is assigned to topic t across the data set without considering the current assignment to be done,
β is the Dirichlet hyperparameter of the prior distribution for the categorical distribution over regions in a topic . Here, we use symmetric Dirichlet priors for all topics, using 0.1 as value for β.
n−i,t is the total number of assignments to topic t through the data set,
R is the total number of regions in the data set,
and expresses the probability of region r under topic t.
is the total number of region assignments to topic t within the given cell c (without considering the region to be assigned),
α is the Dirichlet hyperparameter of the prior distribution for the categorical distribution over topics in a cell θ(c). Here, we use symmetric Dirichlet priors for all cells, using 50/T as value for α.
is the total number of assignments within the given cell c,
T is the total number of topics in the model. The total number of topics has to be provided (see Model selection),
and is the probability of topic t under cell c.
Collapsed Gibbs sampling allows to reduce the complexity of the model by only sampling the topic assignment of each region per cell without the need of sampling from the region-topic and the topic-cell distributions, thus reducing the exploration space. The topic assignments are recorded through several iterations (after a burn-in), and can be used to estimate the region-topic and the topic-cell distributions (Fig. 1a). The speed of this approach depends on the size of the data set (number of cells and regions), the number of models, the CPU cores used and the number of topics and iterations per model.
In most cases, we used 500 as burn-in and 500 recording iterations (see Model selection and Data analysis). LDA provides two matrices, one containing the total number of assignments per topic in each cell, and another containing the total number of assignments per region to each topic. Models are built using the lda R package27.
Model selection
For performing LDA, values for the Dirichlet priors α and β, the number of topics T and the number of iterations (burn-in and recording iterations) must be provided. We used 50/T and 0.1 for α and β, respectively, as recommended by Griffiths & Steyvers (2004). The log-likelihood per iteration in each model was plotted to confirm that the number of burn-in and recording iterations was correctly chosen (i.e. log-likelihood of the model must be stabilized when the recording of iterations starts). Several models with different number of topics were run (generally, from 5 to 50 topics; see Data analysis), and the optimal number of topics is selected based on the highest log-likelihood in the last iteration. In cases where the highest log-likelihood is stabilized when adding more models, we select the simplest model (i.e. with the lowest number of topics).
Cell state identification
Cell states can be identified based on the normalized topic-cell distributions (i.e. a matrix containing cells as columns, topics as rows, and normalized assignments, as probabilities (dividing the assignments to a topic by the total number of assignments in the cell) or Z-scores per cell as values). These distributions can be used to cluster cells or to visualize the cell states using dimensionality reduction methods such as tSNE (R package Rtsne28), Umap (R package UMAP29), PCA and/or diffusion maps (R package Destiny30). For the topic-cell heatmaps we used ward hierarchical clustering with euclidean distances.
In addition, cisTopic also calculates the predictive distribution, which describes the probability of each region in each cell, by multiplying the topic-cell and the region-topic distributions:
Where:
P(ri | cj) is the probability of region i in cell j,
P(ri | Tk) is the probability of region i in topic k
and P(Tk | cj) is the probability of topic k in cell j.
The predictive distribution is a method for the imputation of drop-outs (Supplementary Fig. 15), and it can be used to analyze the enrichment of epigenomic signatures in the cells. To calculate the enrichment for each signature in each cell, we use a rank-based approach, AUCell18, in which all regions in each cell are ranked based on the predictive distribution. By default, we set a requirement of 40% overlap when the epigenomic signatures are intersected with the regions in the data set. AUCell was used with default settings, using the top 10% regions in the rankings for estimating the AUC.
Topic exploration
Region assignments can be normalized as probabilities of a region in a topic, Z-scores or using the following formula:
Where:
βr,t is a number proportional to the probability of seeing region r in topic t.
T is the total number of topics in the model.
The region-topic distributions can be explored in different ways to understand the biological nature of the regulatory topics.
Enrichment of epigenomic signatures in the topics: Epigenomic signatures are intersected with the regulatory regions in the data set (by default, with at least 40% overlap) and summarised into region sets. All regions are ranked per topic based on the normalized region-topic distribution. The region sets and the topic-specific rankings are used as input for AUCell18. Here, we used 3% of the total number of regions in the data set as a threshold to calculate the AUC.
Region annotation: Regions in the data set are annotated using the R package ChIPseeker31. This annotation includes the region type and the closest gene. Enrichment of region types within the topics is calculated as previously explained (i.e. using as region sets the regions per region class).
Topic binarization: When region-topic distributions cannot be exploited (i.e. for the usage of tools that function with region sets rather than rankings like GREAT32 or cisTarget33), representative regions per topic must be selected. These regions can be selected by rescaling the normalized region-topic assignments to the unit, and fitting a gamma distribution to these values. A threshold is given to select regions above a certain probability (see Data analysis). Note that this threshold must be taken after the density (based on the fitted gamma distribution) is stabilized (i.e. in the tail of the distribution). Alternatively, a set of representative regions can be selected from the top of the distribution (with a user-specified value).
Gene Ontology analysis: GO analyses were performed by using rGREAT32 on the binarized topics.
Motif enrichment: Motif enrichment was performed using RcisTarget18. cisTopic includes functions for performing motif enrichment analysis in sets of regions, rather than sets of genes. Here, we used the region-based hg19 and the mm9 cisTarget feather databases (v8), using liftover between genomes when needed (see Data analysis). The cisTarget motif collection contains more than 20,000 PWMs obtained from JASPAR34, cis-bp35 and Hocomoco36, among others37. We used a minimum fraction overlap of 0.4; a minimum Normalized Enrichment Score (NES) threshold of 3; a ROC threshold for AUC calculation of 0.005 and a threshold for visualization of 20,000. Region-based feather databases are available at: https://resources.aertslab.org/cistarget/. Motif annotation is available within the RcisTarget package.
Topic-specific cistrome formation: Cistromes can be formed based on RcisTarget results (per topic); by selecting the regions that pass the given thresholds. These sets of regions are linked to transcription factors based on motif annotations (direct and inferred). These cistromes are initially formed by Ctx regions33, that are mapped back to the original coordinates in the data set (here, regions are mapped back if there is at least 40% of overlap).
Data analysis
Validation of cisTopic
Comparison of parameter estimation methods for LDA: We compared four different methods for estimating the topic-cell and the region-topic distributions for LDA, namely Collapsed Gibbs Sampling (as implemented by Chang27 and Grün & Hornik38), Variational EM38 and MAP39 (Taddy, 2016). We simulated 650 single-cell epigenomes from 13 FAC-sorted bulk ATAC-seq profiles from the hematopoietic system26 (50 cells per bulk sample, see below) by randomly sampling a given number of reads (50,000; 10,000 and 3,000 reads per cell in each experiment), in order to test their robustness to drop-outs; and tested models between 2 and 100 topics (from 2 to 30, 1 by 1; 50 and 100). Gibbs Sampling was run in each simulation using α=50/T; β=0.1; burn-in iterations=500 and recording iterations=1000 (in both implementations); VEM was run using α=50/T and default parameters; and MAP was run with default parameters (calculating the Bayes Factor in each model). The number of topics selected (from high to low coverage experiment) per method was: Collapsed Gibbs Sampler27: 21, 14 and 12; Collapsed Gibbs Sampler38: 21, 16 and 13; Variational EM: 13, 13 and 7; MAP: 9, 6 and 5. For calculating the Adjusted Rand Index, we used as ground truth the bulk epigenome of each cell and determined the cell labels from each method using euclidean distance and ward clustering (using the tSNE projections).
Simulated epigenomes from FAC-sorted bulk ATAC-seq profiles from the hematopoietic system: After mapping 13 FAC-sorted bulk ATAC-seq profiles from the hematopoietic system26 to the human genome (hg19-Gencode v18) using STAR (v2.5.1) (applying the parameters --alignIntronMax 1, --aslignIntronMin 2 and --alignMatesGapMax 2000) and merging the bam files per cell type using SAMtools (v1.2), we simulated 650 single-cell epigenomes (50 cells per bulk) by randomly sampling a given number of reads (50,000; 10,000 and 3,000 reads per cell in each experiment). Candidate regulatory regions were defined by peak calling with MACS2 in each bulk profile merged per cell type (v.2.0.10, with q < 0.001 and nomodel parameters) and merging of overlapping peaks. For each simulation we ran cisTopic (parameters: α=50/T; β=0.1; burn-in iterations=500; recording iterations=1000) for models with a number of topics between 2 and 100 (from 2 to 30, 1 by 1; 50 and 100). The best model in each simulation was selected based on the highest log-likelihood, resulting in selected models with 21, 14 and 12 topics, from highest to lowest coverage. We binarized the topics using a probability threshold of 0.975 to perform motif enrichment analysis (with default settings). Latent Semantic Indexing (LSI) was performed as described by Cusanovich et al.3. The number of PCs selected was 5, 5 and 7, for the different coverages respectively; and the first principal component was removed in all cases as it was correlated with the read depth. Values of the LSI matrix were rescaled between ±1.5. We ran chromVAR5 with default parameters and including the GC bias correction. We ran scABC4 with default parameters, resulting in models with 7, 2 and 2 landmarks. Since the number of landmarks calculated by scABC differed significantly from the real number of cell types (13), we used the cell-to-landmark correlation matrix for the following steps, rather than directly the scABC cluster assignments (which resulted in lower clustering accuracy). SCRAT was run using default parameters and the cistrome collection with the co-regulated DNAse I hypersensitive sites from ENCODE7. We ran BROCKMAN (v1.0)6 on the fastq files generated from the simulated bam files using bamtofastq (as part of bedtools; v2.23.0), and used default parameters (for single-end reads) for the determination of PCs and tSNE coordinates. We ran Cicero40 with default parameters and chromatin hubs obtained were used as cistromes, and cistrome enrichment per cell was calculated by aggregating hub regions and normalizing each cistrome score using a Z-score. Rtsne was used for visualization in all cases with 50 PCs and 30 as perplexity (after testing other values to ensure the stability of the results)28. For calculating the Adjusted Rand Index, we used as ground truth the bulk epigenome of each cell and determined the cell labels from each method using euclidean distance and ward clustering on the tSNE projections. These tSNE projections are based on the topic-cell distributions matrix from cisTopic, the LSI matrix, the cistrome enrichment matrix from SCRAT, the cistrome enrichment matrix from chromVAR, the cell-to-landmark matrix from scABC, the kmer-PCs from BROCKMAN, and the chromatin hub enrichment matrix from Cicero, respectively.
Simulated epigenomes from melanoma cell lines: We simulated 700 single-cell epigenomes from 14 bulk H3K27Ac ChIP-seq melanoma profiles (50 cells per bulk) by randomly sampling a given number of reads. Eleven of these bulk epigenomes were taken from Verfaille et al.20 (GSE60666); and three have been generated in this work with the same protocol and analysis pipeline. Candidate regulatory regions were defined by peak calling with MACS2 in each bulk profile (v.2.0.10, with q < 0.001 and nomodel parameters and using as control the merged control profiles of five cell lines; namely A375, MM011, MM032, MM047 and MM057) and merging of overlapping peaks. The number of reads per cell was selected randomly from the intervals corresponding to each simulation, namely 26,940-59,580 reads per cell; 8,980-19,860 reads per cell; 5,388-11,916 reads per cell and 2,694-5,958 reads per cell. For each simulation we ran cisTopic (parameters: α=50/T; β=0.1; burn-in iterations=500; recording iterations=1000) for models with a number of topics between 2 to 50 (from 2 to 30, 1 by 1; from 30 to 50, 5 by 5). The best model in each simulation was selected based on the highest log-likelihood, resulting in selected models with 22, 22, 19 and 12 topics, from highest to lowest coverage. We binarized the topics using a probability threshold of 0.975, and performed GO enrichment analysis with rGREAT and motif enrichment analysis with RcisTarget. Latent Semantic Indexing (LSI) was performed as described by Cusanovich et al.3. The number of PCs selected was 7, 5, 5 and 5, for the different coverages respectively; and the first principal component was removed in all cases as it was correlated with the read depth. Values of the LSI matrix were rescaled between ±1.5. SCRAT was run using default parameters and the cistrome collection with the co-regulated DNAse I hypersensitive sites from ENCODE7. We ran chromVAR5 with default parameters and adding the GC bias. We ran scABC with default parameters, resulting in models with 14, 14, 13 and 7 landmarks4. We used the cell-to-landmark correlation matrix for the following steps, rather than directly scABC4 cluster assignments (which resulted in lower clustering accuracy). BROCKMAN6 was run using default parameters for the determination of PCs and tSNE coordinates. Chromatin hubs obtained using default parameters in Cicero40 were used as cistromes, and cistrome enrichment per cell was calculated by aggregating hub regions and normalizing each cistrome score using a Z-score. Rtsne was used for visualization in all cases with 50 PCs and 30 as perplexity (after testing other values to ensure the stability of the results)28. The Adjusted Rand Index was calculated as previously explained for each method. We also tested the robustness of these methods to find rare subpopulations by reducing the number of single-cell epigenomes from 50 to 5 for 3 of these cell lines (A375, MM001 and MM099). Methods were run as previously described, and precision and recall values were calculated by using as ground truth the bulk epigenome of each cell. The cells were labelled for each method using euclidean distances and ward clustering on the using the low dimensional representation of the data. The optimal number of clusters was selected using the dynamicCutTree package41. The clusters with the highest ratio of true positives versus false positives were selected for the calculations.
scATAC-seq from FAC-sorted single-cell populations from the hematopoietic system: We used cisTopic on a publicly available scATAC-seq data set from FAC-sorted populations from the hematopoietic system10, containing 8 different cell types from the hematopoietic lineage. The single-cell reads were first cleaned for adapters using fastq-mcf using fastq-mcf (as part of ea utils; v1.1.2-686). Read quality was then checked using FastQC (v0.11.5). Paired-end reads were mapped to the human genome (hg19-Gencode v18) using STAR (v2.5.1) applying the parameters --alignIntronMax 1, --aslignIntronMin 2 and --alignMatesGapMax 2000. Mapped reads were filtered for quality using SAMtools (v1.2) view with parameter –q4, sorted with SAMtools sort and indexed using SAMtools index. Duplicates were removed using Picard (v1.134) MarkDuplicates using OPTICAL_DUPLICATE_PIXEL_DISTANCE=2500. We used as input for cisTopic the bam files and the regions defined by Buenrostro et al.10, resulting in a count matrix with 2,755 cells and 488,825 regions. We ran cisTopic using α=50/T; β=0.1; burn-in iterations=500; recording iterations=1000 and models with a number of topics between 2 and 500 (from 2 to 30, 1 by 1; 50, 100, 200 and 500). The selected model had 17 topics. We binarized the topics with a probability threshold of 0.985 for motif enrichment analysis. SCRAT was run using default parameters and the cistrome collection with the co-regulated DNAse I hypersensitive sites from ENCODE7. chromVAR was run with default parameters and adding the GC bias. LSI was run as described by Cusanovich et al.3, resulting in 7 PCs selected. Values of the LSI matrix were rescaled between ±1.5. scABC was run with default parameters, resulting in 2 landmarks. As previously, we used the cell-to-landmark correlation matrix for the following steps, rather than directly scABC cluster assignments (which resulted in lower clustering accuracy). BROCKMAN was run using default parameters for the determination of PCs and tSNE coordinates. Cicero was run aggregating the peaks within 5000 bp and using default parameters for the estimation of differentially accessible sites (with p-val < 0.05)40; resulting in 642 ordering sites. Rtsne was used for visualization in all cases with 50 PCs and 30 as perplexity. For cisTopic, the patient-specific topic (Topic 7) was removed from the matrix prior to the use of Rtsne. The Adjusted Rand Index was calculated as previously explained for each method, based on their tsne projections. Differential motif analysis was performed using MAST42. As candidate features we used a combination of known and de novo motifs. Known motifs enriched in each set were retrieved from the cisTarget motif collection33,43 and de novo motifs were obtained by comparing each cistrome versus the others using Homer44 and RSAT peak-motifs45,46 with default parameters. The motifs were scored in the regions using Cluster-Buster47, taking as values the best Cis-Regulatory Module (CRM) score per region to perform MAST (comparing each cistrome versus the others). The top 100 motifs per comparison are shown in the ternary plot, which is done using the average CRM values per motif in each group. The colors in the plot indicate the cluster to which the motif is assigned by STAMP48.
scnmC-seq in human neuronal populations from the frontal cortex: We applied cisTopic on a publicly available scnmC-seq data set from human neurons from the human cortex49. This matrix contains the raw mCH levels for 2,784 cells across 28,342 binned regions. In this case, we ran LDA without binarizing the input matrix. We performed models using α=50/T; β=0.1; burn-in iterations=250; recording iterations=500; and a number of topics between 5 and 100 (from 2 to 30, 1 by 1; 50 and 100), resulting in a model with 21 topics as selected.
sci-ATAC Mouse Cell Atlas: We applied cisTopic on a publicly available sci-ATAC-seq data set containing 80,254 samples (and 436,206 regulatory regions) from several mouse tissues2. We performed models using α=50/T; β=0.1; burn-in iterations=250; recording iterations=500; and a number of topics between 2 and 200 (2, 10 to 100, 10 by 10; and 100 to 200, 20 by 20), resulting in a selected model with 50 topics. We binarized the topics with a probability threshold of 0.99 for performing motif enrichment analysis.
Conservation of regulatory programs in the mouse and the human brain
scTHS-seq and scRNA-seq in the human brain: We analyzed a data set from the human brain16 with 34,520 cells and 287,381 regulatory regions (GSE97942). This data set contains cells from the visual cortex, the frontal cortex and the cerebellum. We ran cisTopic with α=50/T; β=0.1; burn-in iterations=500; recording iterations=1000; and a number of topics between 2 and 50 (from 2 to 30 by 1; from 30 to 50 by 5), resulting in a model with 23 topics to be selected. We binarized the topics with a probability threshold of 0.99 and liftovered the regions from hg38 to hg19 for motif enrichment analysis. Enrichment of epigenomic signatures in cells and topics was performed as previously explained. Enrichment of the transcriptomic signature was performed by taking the regions linked to the signature genes (based on the closest gene).
We filtered the scRNA-seq data from Lake et al.16 (GSE97930) keeping only cells with at least 800 genes expressed, resulting in a data set with 15,884 cells. SCENIC was run using default parameters18, resulting in a matrix with 250 regulons. Next, we mapped the regions to their closest gene, and this dictionary was used to convert the gene-based regulons to region-based regulons. These region sets were used as epigenomic signatures to determine their enrichment within the topics using AUCell as previously explained.
scATAC-seq in the mouse prefrontal cortex: We applied cisTopic on a publicly available scATAC-seq data set of the mouse prefrontal cortex17 with 3,034 cells and 139,504 regions (GSE100033). We ran cisTopic with α=50/T; β=0.1; burn-in iterations=250; recording iterations=500; and a number of topics between 2 and 100 (from 2 to 40, 1 by 1; 50 and 100), resulting in a model with 23 topics to be selected. We binarized the topics with a probability threshold of 0.99 and liftovered the regions from mm10 to mm9 for motif enrichment analysis. Comparison between the layer-specific topics was performed using MAST as previously described, by taking the top 200 motifs per comparison.
Comparison between mouse and human brain: Human binarized topics from the analysis of Lake et al.16 were lifted over from hg38 to mm10. Human-to-mouse lifted over regions were mapped to the coordinates on the mouse brain data set17 given 40% of overlap, and enrichment in the mouse topics was performed using AUCell as previously described. Conserved enhancers were selected based on their presence in the matching topic (e.g. region in the human oligodendrocyte topic that maps to a region in the mouse oligodendrocyte topic). Regions were aligned using MUSCLE (v3.8.31)50. Cluster-Buster47 was used to score the enriched motifs in the sequences and a customised version of TOUCAN51,52 was used for visualizing the motifs in the aligned sequences.
Transcription factor knockdown
scATAC-seq during SOX10 KD in melanoma: We generated scATAC-seq data on different time points (0, 24, 48 and 72h) for two melanoma cell lines (MM057 and MM087) upon SOX10 KD, resulting in a data set with 598 and 78,262 accessible regions (see below). We ran cisTopic with α=50/T; β=0.1; burn-in iterations=500; recording iterations=1000; and a number of topics between 5 and 50 (from 2 to 30, 1 by 1; from 30 to 50, 5 by 5), finding a model with 15 topics to be optimal. Topics were binarized using a probability threshold of 0.975 before RcisTarget and rGREAT analyses. Comparison between the SOX cistromes was performed as previously described, taking the top 200 motifs per comparison for use in the ternary plot. Significance of SOX10 binding sites on topic 14 was calculated using a one-side proportion test (H1: Proportion of SOX10 ChIP-seq peaks in topic 14 > Proportion of SOX10 ChIP-seq peaks in the data set).
Experimental work and data processing
Cell culture and treatment
The two melanoma cultures (MM057 and MM087) are short-term cultures derived from patient biopsies20,21. Cells were kept at 37°C, with 5% CO2 and were maintained in Ham's F10 nutrient mix (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 100 µg ml-1 penicillin/streptomycin (Thermo Fisher Scientific). SOX10 KD was performed using a SMARTpool of four siRNAs against SOX10 (SMARTpool: ON-TARGETplus SOX10 siRNA, number L017192-00-0005, Dharmacon) at a concentration of 20nM using as medium Opti-MEM (Thermo Fisher Scientific) and omitting antibiotics. The cells were incubated for 24, 48 or 72 hours before processing.
OmniATAC-seq
Data generation: OmniATAC-seq was performed as described previously53. Cells were washed, trypsinized, spun down at 1000 RPM for 5 min to remove the medium and resuspended in 1 mL medium. Cells were counted and experiments were only continued when a viability of above 90% was observed. 50,000 cells were pelleted at 500 RCF at 4°C for 5 min, medium was carefully aspirated and the cells were washed and lysed using 50 uL of cold ATAC-Resupension Buffer (RSB) (see Corces et al.53 for composition) containing 0.1% NP40, 0.1% Tween-20 and 0.01% digitonin by pipetting up and down three times and incubating the cells for 3 min on ice. The lysis was washed out by adding 1 mL of cold ATAC-RSB containing 0.1% Tween-20 and inverting the tube three times. Nuclei were pelleted at 500 RCF for 10 min at 4°C, the supernatant was carefully removed and nuclei were resuspended in 50 uL of transposition mixture (25 uL 2x TD buffer (see Corces et al.53 for composition), 2.5 uL transposase (100 nM), 16.5 uL DPBS, 0.5 uL 1% digitonin, 0.5 uL 10% Tween-20, 5 uL H2O) by pipetting six times up and down, followed by 30 minutes incubation at 37°C at 1000 RPM mixing rate. After MinElute clean-up and elution in 21 uL elution buffer, the transposed fragments were pre-amplified with Nextera primers by mixing 20 uL of transposed sample, 2.5 uL of both forward and reverse primers (25 uM) and 25 uL of 2x NEBNext Master Mix (program: 72°C for 5 min, 98°C for 30 sec and 5 cycles of [98°C for 10 sec, 63 °C for 30 sec, 72°C for 1 min] and hold at 4°C). To determine the required number of additional PCR cycles, a qPCR was performed (see Buenrostro et al.54 for the determination of the number of cycles to be added). The final amplification was done with the additional number of cycles, samples were cleaned-up by MinElute and libraries were prepped using the KAPA Library Quantification Kit as previously described53. Samples were sequenced on a NextSeq500 High Output chip, generating between 41 and 70 million reads per sample.
Data processing: Adapter sequences were trimmed from the fastq files using fastq-mcf (as part of ea utils; v1.04.807). Read quality was then checked using FastQC (v0.11.5). Reads were mapped to the human genome (hg19-Gencode v18) using STAR (v2.5.1) applying the parameters --alignIntronMax 1 and --aslignIntronMin 2. Mapped reads were filtered for quality using SAMtools (v1.2) view with parameter –q4, sorted with SAMtools sort and indexed using SAMtools index. Peaks were called using MACS2 (v2.1.1) callpeak using the parameters --nomodel and --call-summits on the 8 conditions separately. A count matrix was generated by using featureCounts (as part of Subread; v1.4.6) of all separate bam files on the merged peak file (after conversion of the merged peak bed file to a gff format using a custom script). Normalized bedGraphs were produced by genomeCoverageBed (as part of bedtools; v2.23.0) using as scaling parameter (-scale) size factors obtained from DEseq2 (v1.18.1). BedGraphs were converted to bigWigs by the bedtools suit functions bedSort to sort the bedGraphs, followed by bedGraphToBigWig to create the bigWigs, which were used in IGV for visualization.
scATAC-seq
Data generation: scATAC-seq was performed using the Fluidigm C1 system as described before55. Briefly, cells were trysinized, spun down (1000 RPM, 5 min), medium was removed and cells were resuspended in fresh medium and passed through a 40 um filter, counted and diluted till 200,000 cells per mL. Cells were loaded (using a 40:60 ratio of RGT:cells) on a primed Open App IFC (10-17 um, the protocol for ATAC-seq from the C1 Script Hub was used). After cell loading, the plate was visually checked under a microscope and the number of cells in each of the capture chambers was noted. Next, the “Sample prep” was performed on the Fluidigm C1 during which the cells underwent lysis and ATAC-seq fragments were prepared. In a 96-well plate, the harvested libraries were amplified in a 25 uL PCR reaction. The PCR products were pooled and purified on a single MinElute PCR purification column for a final library volume of 15 uL. Quality checks were performed using the Bioanalyzer high sensitivity chips. Fragments under 150 bp were removed by bead-cleanup using AMPure XP beads (1.2x bead ratio) (Beckman Coulter). All scATAC-seq libraries were sequenced on a HiSeq4000 paired-end run, generating a median of 170,769 raw reads per single cell.
Data processing: The reads from scATAC-seq samples were first cleaned for adapters using fastq-mcf (as part of ea utils; v1.1.2-686) and read quality was checked using FastQC (v0.11.5). Paired-end reads were mapped to the human genome (hg19-Gencode v18) using STAR (v2.5.1) applying the parameters --alignIntronMax 1, --aslignIntronMin 2 and --alignMatesGapMax 2000. Mapped reads were filtered for quality using SAMtools (v1.2) view with parameter –q4, sorted with SAMtools sort and indexed using SAMtools index. Duplicates were removed using Picard (v1.134) MarkDuplicates using OPTICAL_DUPLICATE_PIXEL_DISTANCE=2500. To filter out cell of bad quality, transcription start site aggregation plots were made using a custom script and cell having a low signal-to-noise profile were removed from further analyses. This led to a final data set of 598 good quality cells over 8 Fluidigm C1 runs. Bam files of good quality single cells were aggregated per condition and peaks were called on these aggregated samples using MACS2 (v2.1.1) callpeak using the parameters --nomodel and --call-summits. The peak files per condition were merged and blacklisted using the blacklisted regions of hg19 listed on http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/hg19-human/ (Anshul Kundaje), leading to a total of 78,262 peaks after blacklisting. This peak file was used, together with the bam files of the good quality single cells, as input for cisTopic. To visualize the aggregated cells per sample, normalized bedGraphs were produced by genomeCoverageBed (as part of bedtools; v2.23.0) using as scaling parameter (-scale) size factors obtained from DEseq2 (v1.18.1). BedGraphs were converted to bigWigs by the bedtools suit functions bedSort to sort the bedGraphs, followed by bedGraphToBigWig to create the bigWigs.
Publicly available data used in this work
For the simulations of single cells from bulk ATAC-seq profiles from the hematopoietic system, data was downloaded from GEO GSE74912. For the simulations of single cells from bulk melanoma cell line epigenomes, we used the H3K27Ac data from Verfaillie et al.20 (GEO GSE60666). Methylation data from Luo et al.49 was obtained from http://brainome.org. sciATAC-seq Mouse Cell Atlas data2 was taken from http://atlas.gs.washington.edu/mouse-atac/. FAC-sorted single-cell ATAC-seq data from the hematopoietic system from Buenrostro et al.10 was retrieved from GEO GSE96772. scTHS-seq and scRNA-seq data from the human brain16 was downloaded from GEO GSE97942 and GEO GSE97930, respectively; and scATAC-seq data from the mouse brain from Preissl et al.17 was retrieved from GEO GSE100033. Layer-specific regions from the mouse brain were taken from Gray et al.19, interneuron signatures56; from GEO GSE63137 and the dentate gyrus signature57 from GEO GSE82010. scRNA-seq oligodendrocyte and astrocyte signatures for the mouse were obtained from Habib et al.58 and methylation signatures for the mouse and the human brain were retrieved from Mo et al.56 and Kozlenkov et al.59, respectively. GATA2 ChIP-seq peaks (GEO GSE32465) were downloaded as bed files from ChIP-Atlas (with q < 1E-20). SOX10 ChIP-seq was downloaded as raw fastq files from GEO GSE61965 and were mapped to the human genome using Bowtie2 (v2.1.0) and peaks were called by MACS2 (v2.1.1).
Supplementary Material
Acknowledgements
This work is funded by an ERC Consolidator Grant to S. Aerts (724226_cis-CONTROL); by the KU Leuven (grant C14/18/092 to S. Aerts), the Harry J. Lloyd Charitable Trust, the Foundation Against Cancer (2016-070; to S. Aerts), PhD fellowships from the F.W.O. (C.B.G-B., 11F1519N; L.M., 1S03317N; D.P., 1S75219N) and a postdoctoral research fellowship from Kom op tegen Kanker (Stand up to Cancer), the Flemish Cancer Society (J.W.). Computing was performed at the Vlaams Supercomputer Center (VSC). Single-cell infrastructure was funded by the Hercules Foundation (AKUL/13/41). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The authors thank Jean-Christophe Marine, Florian Rambow, and Michael Dewaele for helpful discussions; and Blue Lake and Kun Zhang for the information provided regards the human brain data. The authors also thank various groups that make curated position weight matrices publicly available, including T. Hughes (cis-bp), M. Bulyk (Uniprobe), A. Mathelier (Jaspar), V. Makeev (Hocomoco) and many others.
Footnotes
Data availability
The data generated for this study have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE114557.
Code availability
cisTopic is available as an R package at: http://github.com/aertslab/cistopic.
Author contributions
S.Aerts, C.B.G-B. and L.M. conceived the study; C.B.G-B. developed cisTopic and implemented the R package; L.M. performed the experimental work with the help of V.C, K.D. and J.W; C.B.G-B. and L.M. analyzed the data with the help of D.P., S.Aibar, G.H. and K.D.; C.B.G-B., L.M. and S.Aerts wrote the manuscript.
Competing interest
The authors declare that no competing interests exist.
References
- 1.Fiers MWEJ, et al. Mapping gene regulatory networks from single-cell omics data. Brief Funct Genomics. 2018;17:246–254. doi: 10.1093/bfgp/elx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cusanovich DA, et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell. 2018;174:1309–1324.e18. doi: 10.1016/j.cell.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cusanovich DA, et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zamanighomi M, et al. Unsupervised clustering and epigenetic classification of single cells. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14 doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer CG, Regev A. BROCKMAN: deciphering variance in epigenomic regulators by k-mer factorization. BMC Bioinformatics. 2018;19 doi: 10.1186/s12859-018-2255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji Z, Zhou W, Ji H. Single-cell regulome data analysis by SCRAT. Bioinformatics. 2017;33:2930–2932. doi: 10.1093/bioinformatics/btx315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blei DM, Ng AY, Jordan MI. Latent Dirichlet Allocation. J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- 9.Griffiths TL, Steyvers M. Finding scientific topics. Proc Natl Acad Sci U S A. 2004;101(Suppl):5228–35. doi: 10.1073/pnas.0307752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buenrostro JD, et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell. 2018;173:1535–1548.e16. doi: 10.1016/j.cell.2018.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilagos B, et al. Essential role of EBF1 in the generation and function of distinct mature B cell types. J Exp Med. 2012;209:775–792. doi: 10.1084/jem.20112422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cisse B, et al. Transcription Factor E2-2 Is an Essential and Specific Regulator of Plasmacytoid Dendritic Cell Development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta P, Gurudutta GU, Saluja D, Tripathi RP. PU.1 and partners: regulation of haematopoietic stem cell fate in normal and malignant haematopoiesis. J Cell Mol Med. 2009;13:4349–4363. doi: 10.1111/j.1582-4934.2009.00757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elagib KE. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- 15.Nottingham WT, et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lake BB, et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 2017;36:70–80. doi: 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preissl S, et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci. 2018 doi: 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aibar S, et al. SCENIC: Single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray LT, et al. Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex. eLife. 2017;6 doi: 10.7554/eLife.21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verfaillie A, et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun. 2015;6:6683–6683. doi: 10.1038/ncomms7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gembarska A, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med. 2012;18:1239–47. doi: 10.1038/nm.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernd A, et al. Levels of dopachrome tautomerase in human melanocytes cultured in vitro. Melanoma Res. 1994;4:287–291. doi: 10.1097/00008390-199410000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Iozumi K, Hoganson GE, Pennella R, Everett MA, Fuller BB. Role of Tyrosinase as the Determinant of Pigmentation in Cultured Human Melanocytes. J Invest Dermatol. 1993;100:806–811. doi: 10.1111/1523-1747.ep12476630. [DOI] [PubMed] [Google Scholar]
- 24.Buac K, et al. NRG1/ERBB3 signaling in melanocyte development and melanoma: inhibition of differentiation and promotion of proliferation. Pigment Cell Melanoma Res. 2011;22:773–784. doi: 10.1111/j.1755-148X.2009.00616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laurette P, et al. Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. eLife. 2015;2015:1–40. doi: 10.7554/eLife.06857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang J. lda: Collapsed Gibbs Sampling Methods for Topic Models. R package version 1.2.3. 2015 URL http://CRAN.R-project.org/package=lda.
- 28.Krijthe J, van der Maaten L. Package ‘Rtsne’. R package version 0.13. 2017 URL https://github.com/jkrijthe/Rtsne.
- 29.McInnes L, Healy J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv180203426 Cs Stat. 2018 [Google Scholar]
- 30.Angerer P, et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 31.Yu G, Wang L-G, He Q-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 32.Gu Z. rGREAT: Client for GREAT Analysis. R package version 3.7. 2018 URL https://github.com/jokergoo/rGREAT, http://great.stanford.edu/public/html/
- 33.Imrichová H, Hulselmans G, Kalender Atak Z, Potier D, Aerts S. i-cisTarget 2015 update: generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res. 2015;43:W57–W64. doi: 10.1093/nar/gkv395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portales-Casamar E, et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulakovskiy IV, et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018;46:D252–D259. doi: 10.1093/nar/gkx1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janky R, et al. iRegulon: From a Gene List to a Gene Regulatory Network Using Large Motif and Track Collections. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grün B, Hornik K. topicmodels: An R Package for Fitting Topic Models. J Stat Softw. 2011;40 [Google Scholar]
- 39.Taddy M. On Estimation and Selection for Topic Models. In: Lawrence ND, Girolami M, editors. Proceedings of the Fifteenth International Conference on Artificial Intelligence and Statistics; 2012. pp. 1184–1193. PMLR. [Google Scholar]
- 40.Pliner HA, et al. Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol Cell. 2018;71:858–871.e8. doi: 10.1016/j.molcel.2018.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langfelder P, Zhang B. dynamicTreeCut: Methods for Detection of Clusters in Hierarchical Clustering Dendrograms. 2016 [Google Scholar]
- 42.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015:1–13. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrmann C, Van De Sande B, Potier D, Aerts S. i-cisTarget: An integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas-Chollier M, et al. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat Protoc. 2011;6:1860–1869. doi: 10.1038/nprot.2011.409. [DOI] [PubMed] [Google Scholar]
- 46.Thomas-Chollier M, et al. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 2012;40:e31–e31. doi: 10.1093/nar/gkr1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frith MC, Li MC, Weng Z. Cluster-Buster: Finding dense clusters of motifs in DNA sequences. Nucleic Acids Res. 2003;31:3666–3668. doi: 10.1093/nar/gkg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo C, et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357:600–604. doi: 10.1126/science.aan3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aerts S, et al. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aerts S, et al. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 2005;33:W393–396. doi: 10.1093/nar/gki354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corces MR, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14 doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10 doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mo A, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su Y, et al. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat Neurosci. 2017;20:476–483. doi: 10.1038/nn.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Habib N, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozlenkov A, et al. A unique role for DNA (hydroxy)methylation in epigenetic regulation of human inhibitory neurons. Sci Adv. 2018;4 doi: 10.1126/sciadv.aau6190. eaau6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.