Abstract
Background
Melanoma accounts for a small proportion of all skin cancer cases but is responsible for most skin cancer‐related deaths. Early detection and treatment can improve survival. Smartphone applications are readily accessible and potentially offer an instant risk assessment of the likelihood of malignancy so that the right people seek further medical attention from a clinician for more detailed assessment of the lesion. There is, however, a risk that melanomas will be missed and treatment delayed if the application reassures the user that their lesion is low risk.
Objectives
To assess the diagnostic accuracy of smartphone applications to rule out cutaneous invasive melanoma and atypical intraepidermal melanocytic variants in adults with concerns about suspicious skin lesions.
Search methods
We undertook a comprehensive search of the following databases from inception to August 2016: Cochrane Central Register of Controlled Trials; MEDLINE; Embase; CINAHL; CPCI; Zetoc; Science Citation Index; US National Institutes of Health Ongoing Trials Register; NIHR Clinical Research Network Portfolio Database; and the World Health Organization International Clinical Trials Registry Platform. We studied reference lists and published systematic review articles.
Selection criteria
Studies of any design evaluating smartphone applications intended for use by individuals in a community setting who have lesions that might be suspicious for melanoma or atypical intraepidermal melanocytic variants versus a reference standard of histological confirmation or clinical follow‐up and expert opinion.
Data collection and analysis
Two review authors independently extracted all data using a standardised data extraction and quality assessment form (based on QUADAS‐2). Due to scarcity of data and poor quality of studies, we did not perform a meta‐analysis for this review. For illustrative purposes, we plotted estimates of sensitivity and specificity on coupled forest plots for each application under consideration.
Main results
This review reports on two cohorts of lesions published in two studies. Both studies were at high risk of bias from selective participant recruitment and high rates of non‐evaluable images. Concerns about applicability of findings were high due to inclusion only of lesions already selected for excision in a dermatology clinic setting, and image acquisition by clinicians rather than by smartphone app users.
We report data for five mobile phone applications and 332 suspicious skin lesions with 86 melanomas across the two studies. Across the four artificial intelligence‐based applications that classified lesion images (photographs) as melanomas (one application) or as high risk or 'problematic' lesions (three applications) using a pre‐programmed algorithm, sensitivities ranged from 7% (95% CI 2% to 16%) to 73% (95% CI 52% to 88%) and specificities from 37% (95% CI 29% to 46%) to 94% (95% CI 87% to 97%). The single application using store‐and‐forward review of lesion images by a dermatologist had a sensitivity of 98% (95% CI 90% to 100%) and specificity of 30% (95% CI 22% to 40%).
The number of test failures (lesion images analysed by the applications but classed as 'unevaluable' and excluded by the study authors) ranged from 3 to 31 (or 2% to 18% of lesions analysed). The store‐and‐forward application had one of the highest rates of test failure (15%). At least one melanoma was classed as unevaluable in three of the four application evaluations.
Authors' conclusions
Smartphone applications using artificial intelligence‐based analysis have not yet demonstrated sufficient promise in terms of accuracy, and they are associated with a high likelihood of missing melanomas. Applications based on store‐and‐forward images could have a potential role in the timely presentation of people with potentially malignant lesions by facilitating active self‐management health practices and early engagement of those with suspicious skin lesions; however, they may incur a significant increase in resource and workload. Given the paucity of evidence and low methodological quality of existing studies, it is not possible to draw any implications for practice. Nevertheless, this is a rapidly advancing field, and new and better applications with robust reporting of studies could change these conclusions substantially.
Keywords: Adult, Humans, Algorithms, Diagnostic Errors, Diagnostic Errors/statistics & numerical data, Early Detection of Cancer, Early Detection of Cancer/instrumentation, Early Detection of Cancer/methods, Melanoma, Melanoma/diagnostic imaging, Mobile Applications, Sensitivity and Specificity, Skin Neoplasms, Skin Neoplasms/diagnostic imaging, Smartphone, Triage, Triage/methods
Plain language summary
How accurate are smartphone applications ('apps') for detecting melanoma in adults?
What is the aim of the review?
We wanted to find out how well smartphone applications can help the general public understand whether their skin lesions might be melanoma.
Why is improving the diagnosis of malignant melanoma skin cancer important?
Melanoma is one of the most dangerous forms of skin cancer. Not recognising a melanoma (a false negative test result) could delay seeking appropriate advice and surgery to remove it. This increases the risk of the cancer spreading to other organs in the body and possibly causing death. Diagnosing a skin lesion as a melanoma when it is not present (a false positive result) may cause anxiety and lead to unnecessary surgery and further investigations.
What was studied in the review?
Specialised applications ('apps') that provide advice on skin lesions or moles that might cause people concern are widely available for smartphones. Some apps allow people to photograph any skin lesion they might be worried about and then receive guidance on whether to get medical advice. Apps may automatically classify lesions as high or low risk, while others can act as store‐and‐forward devices where images are sent to an experienced professional, such as a dermatologist, who then makes a risk assessment based on the photo. Cochrane researchers found two studies, evaluating five apps that used automated analysis of images and one that used a store‐and‐forward approach, to evaluate suspicious skin lesions.
What are the main results of the review?
The review included two studies with 332 lesions, including 86 melanomas, analysed by at least one smartphone application. Both studies used photographs of moles or skin lesions that were about to be removed because doctors had already decided they could be melanomas. The photographs were taken by doctors instead of people taking pictures of their lesions with their own smartphones. For these reasons, we are not able to make a reliable estimate about how well the apps actually work.
Four apps that produce an immediate (automated) assessment of a skin lesion or mole that has been photographed by the smartphone missed between 7 and 55 melanomas.
One app that sends the photograph of a mole or skin lesion to a dermatologist for assessment missed only one melanoma. Another 6 melanomas examined by the dermatologist via the application were not classified as high risk; instead the dermatologist was not able to classify the lesion as either 'atypical' (possibly a melanoma) or 'typical' (definitely not a melanoma).
How reliable are the results of the studies of this review?
The small number and poor quality of included studies reduces the reliability of findings. The people included were not typical of those who would use the applications in real life. The final diagnosis of melanoma was made by histology, which is likely to have been a reliable method for deciding whether patients really had melanoma*. However, the studies excluded between 2% and 18% of images because the applications failed to produce a recommendation.
Who do the results of this review apply to?
Studies took place in the USA and Germany. They did not report key patient information such as age and gender. The percentage of people with a final diagnosis of melanoma was 18% and 35%, much higher than that observed in community settings. The definition of eligible patients was narrow in comparison to likely users of the applications. The photographs used were taken by doctors rather than by smartphone users, which seriously impacts the applicability of results.
What are the implications of this review?
Current smartphone applications using automated analysis are observed to have a high chance of missing melanomas (false negatives). Store‐and‐forward image applications could have a potential role in the timely identification of people with potentially malignant lesions by facilitating early engagement of those with suspicious skin lesions, but they have resource and workload implications.
The development of applications to help identify people who might have melanoma is a fast‐moving field. The emergence of new applications, higher quality and better reported studies could change the conclusions of this review substantially.
How up‐to‐date is this review?
The review authors searched for and used studies published up to August 2016.
*In these studies biopsy was the reference standard (means of establishing final diagnoses).
Summary of findings
Summary of findings'. '.
| Question | What is the diagnostic accuracy of smartphone applications for detecting cutaneous melanoma in adults? | |||||
| Participants | Adults with suspicious skin lesions | |||||
| Prior testing and prevalence | Studies did not report the basis for participant selection. One selected a sample of lesions previously imaged during routine care just before excision of the lesion. The second study evaluated the test on patients who had been referred for further screening of the lesion by a specialist. Prevalence of melanoma was 18% and 35%. | |||||
| Settings | Secondary care | |||||
| Target condition(s) | Invasive melanoma and atypical intraepidermal melanocytic variants | |||||
| Index test | Smartphone applications intended for use by the general public. Lesions not visualised by applications excluded | |||||
| Reference standard | Histology | |||||
| Action | If accurate, positive results of smartphone applications will help to highlight lesions of concern to the lay public, promoting earlier diagnosis of melanoma and reducing consultations for benign lesions. | |||||
| Limitations | ||||||
| Risk of bias | Patient selection methods at high risk of bias due to selective inclusion of lesion types (2/2) and use of a case‐control design (1/2). Test interpretation was blinded to reference standard and pre‐specified for artificial intelligence‐based diagnosis (2/2). Reference standard blinding not described. Timing of index and reference standards not reported. Exclusions due to test failures were not reported (1/2) or their final diagnoses were not described (2/2) | |||||
| Applicability of evidence to question | High concerns about applicability due to unrepresentative participant samples with high disease prevalences (2/2). Test not applied and interpreted by the intended user of the application (2/2). Reference standard interpretation by experienced histopathologists was not described (1/2). | |||||
| Total number of studies:2 | ||||||
| Detection of melanoma | ||||||
| Quantity of evidence | Number of studies | 2 | Total participants with test results | 332 | Total with target condition | 86a |
| Findings | Across the four artificial intelligence‐based applications that classified lesion images (photographs) as either melanomas (one application) or as high risk or 'problematic' lesions (three applications), sensitivities ranged from 7% (95% CI 2% to 16%) to 73% (95% CI 52% to 88%) and specificities from 37% (95% CI 29% to 46%) to 94% (95% CI 87% to 97%). This means that between 27% and 93% of invasive melanoma or atypical intraepidermal melanocytic variants were not picked up as high risk by the automated applications (or as melanomas by one of the four applications). With a prevalence of melanoma ranging between 18% and 37% for these evaluations, the number of melanomas missed was 7 to 55. The single application using store‐and‐forward review of lesion images by a dermatologist had a sensitivity of 98% (95% CI 90% to 100%) and specificity of 30% (95% CI 22% to 40%); the dermatologist missed one melanoma. The number of test failures (lesion images analysed by the applications but classed as unevaluable and excluded by the study authors) ranged from 3 to 31 (or 2% to 18% of lesions analysed). The store‐and‐forward application had one of the highest rates of test failure (15%). At least one melanoma was classed as unevaluable in three of the four application evaluations, the highest number of melanomas excluded by the dermatologist evaluating the store‐and‐forward images (6/60 melanomas assessed). |
|||||
aOf the 60 melanomas included in one study, the four applications successfully analysed 54 to 60.
Background
This is one of a series of Cochrane Diagnostic Test Accuracy (DTA) Reviews on the diagnosis and staging of melanoma and keratinocyte skin cancers as part of the National Institute for Health Research (NIHR) Cochrane Systematic Reviews Programme. Appendix 1 shows the content and structure of the programme. Appendix 2 provides a glossary of terms used.
Target condition being diagnosed
Melanoma arises from uncontrolled proliferation of melanocytes – the epidermal cells that produce pigment or melanin (Thompson 2003). Melanoma can occur in any organ that contains melanocytes, including mucosal surfaces, the back of the eye, and lining around the spinal cord and brain (McLaughlin 2005), but it most commonly arises in the skin (Erdmann 2013; Ferlay 2015). Cutaneous melanoma refers to any skin lesion with malignant melanocytes present in the dermis and includes superficial spreading, nodular, acral lentiginous, and lentigo maligna melanoma variants (see Figure 1). Melanoma in situ refers to malignant melanocytes that are contained within the epidermis and have not yet invaded the dermis, but which are at risk of progression to melanoma if left untreated (Thompson 2003; SEER 2007). Lentigo maligna, a subtype of melanoma in situ in chronically sun‐damaged skin, denotes another form of proliferation of abnormal melanocytes. Lentigo maligna can progress to invasive melanoma if its growth breaches the dermo‐epidermal junction during a vertical growth phase (when it becomes known as 'lentigo maligna melanoma'); however, its malignant transformation is both lower and slower than for melanoma in situ (Kasprzak 2015). Melanoma in situ and lentigo maligna are both atypical intraepidermal melanocytic variants (Thompson 2003; SEER 2007). Melanoma is one of the most dangerous forms of skin cancer, with the potential to metastasise to other parts of the body via the lymphatic system and blood stream; it accounts for only a small percentage of all skin cancer cases but is responsible for up to 75% of skin cancer deaths (Boring 1994; Cancer Research UK 2017a).
1.
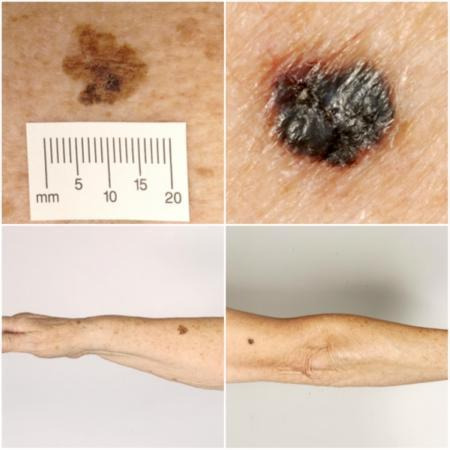
Sample photographs of superficial spreading melanoma (left) and nodular melanoma (right). Copyright © 2010 Dr Rubeta Matin: reproduced with permission.
The annual incidence of melanoma exceeded 200,000 newly diagnosed cases worldwide in 2012 (Erdmann 2013; Ferlay 2015), with an estimated 55,000 deaths (Ferlay 2015). The highest incidence is observed in Australia with 13,134 new cases of melanoma of the skin in 2014 (ACIM 2017) and in New Zealand with 2341 registered cases in 2010 (HPA and MelNet NZ 2014). For 2014 in the USA, the predicted incidence was 73,870 per annum and the predicted number of deaths was 9940 (Siegel 2015). The highest rates in Europe are seen in north‐western Europe and the Scandinavian countries, with a highest incidence reported in Switzerland: 25.8 per 100,000 in 2012. Rates in England have tripled from 4.6 and 6.0 per 100,000 in men and women, respectively, in 1990, to 18.6 and 19.6 per 100,000 in 2012 (EUCAN 2012). Indeed, in the UK, melanoma has one of the fastest rising incidence rates of any cancer and has had the biggest projected increase in incidence between 2007 and 2030 (Mistry 2011). In the decade leading up to 2013, age standardised incidence increased by 46%, with 14,500 new cases in 2013 and 2459 deaths in 2014 (Cancer Research UK 2017b). Rates are higher in women than in men; however, the rate of incidence in males is increasing faster than in females (Arnold 2014).
The rising incidence in melanoma is thought to be primarily related to an increase in recreational sun exposure, tanning bed use and an increasingly ageing population with higher lifetime recreational ultraviolet (UV) exposure, in conjunction with possible earlier detection (Linos 2009; Belbasis 2016). Putative risk factors are reviewed in detail elsewhere (Belbasis 2016); however, risk factors can be broadly divided into host or environmental factors. Host factors include pale skin and light hair or eye colour; older age (Geller 2002); male sex (Geller 2002); previous skin cancer (Tucker 1985); genetically inherited skin disorders, for example xeroderma pigmentosum (Lehmann 2011); a family history of melanoma (Gandini 2005a); and predisposing skin lesions, such as high melanocytic naevus counts (Gandini 2005a), clinically atypical naevi (Gandini 2005a), or large congenital naevi (Swerdlow 1995). Environmental factors include recreational, occupational, and work‐related exposure to sunlight (both cumulative and episodic burning) (Gandini 2005b; Armstrong 2017); artificial tanning (Boniol 2012); and immunosuppression, such as that seen in organ transplant recipients or HIV‐positive individuals (DePry 2011). Lower socioeconomic class may be associated with delayed presentation and thus more advanced disease at diagnosis (Reyes‐Ortiz 2006).
Five‐year survival for stage I melanoma is reported to be 91% to 95%, falling to 27% to 69% in stage III disease (Balch 2009). Tumour thickness, the presence of tumour ulceration and age are the main determinants of melanoma prognosis, and prognostic tools have been developed that include such features (Mahar 2016). Before the advent of targeted and immunotherapies, metastatic melanoma (involving distant sites and visceral organs) resulted in median survival of six to nine months with a three‐year survival of 15% (Balch 2009; Korn 2008). Despite rising incidence, melanoma mortality appears to be stable (Apalla 2017). Between 1975 and 2010, five‐year relative survival for melanoma in the US increased from 80% to 94% but mortality rates showed little change, at 2.1 per 100,000 deaths in 1975 and 2.7 per 100,000 in 2010 (Cho 2014). Increasing incidence in localised disease over the same period (from 5.7 to 21 per 100,000) suggests that the observed survival benefits may be due to earlier detection and heightened vigilance (Cho 2014); however, targeted therapies for stage IV melanoma (e.g. BRAF inhibitors) have improved survival expectation, and immunotherapies are demonstrating potential for long‐term survival (Pasquali 2018).
Treatment of melanoma
For primary melanoma, the mainstay of definitive treatment is wide local excision of the lesion, to remove both the tumour and any malignant cells that might have spread into the surrounding skin (Sladden 2009; Marsden 2010; NICE 2015; Garbe 2016; SIGN 2017). Recommended surgical margins vary according to tumour thickness, as described in Garbe 2016, and stage of disease at presentation, as in NICE 2015 guidelines. The role of narrower (e.g. 1 cm healthy tissue) excision margins for thinner lesions is still debated (Sladden 2009; Wheatley 2016). Following histological confirmation of diagnosis, the lesion is staged according to the American Joint Committee on Cancer (AJCC) Staging System to guide treatment (Balch 2009). Stage 0 refers to melanoma in situ; stages I to II, localised melanoma; stage III, regional metastasis; and stage IV, distant metastasis (Balch 2009). The main prognostic indicators can be divided into histological and clinical factors. Histologically, Breslow thickness is the single most important predictor of survival, as it is a quantitative measure of tumour invasion that correlates with the propensity for metastatic spread (Balch 2001). Microscopic ulceration, mitotic rate, microscopic satellites, regression, lymphovascular invasion, and nodular (rapidly growing) or amelanotic (lacking in melanin pigment) subtypes are also associated with worse prognosis (Shaikh 2012; Moreau 2013).
Independent of tumour thickness, prognosis is worse in older people (Geller 2002); males (Geller 2002); those with recurrent lesions (Dong 2000); and in those with distant lymph node involvement (microscopic or macroscopic), metastatic disease, or both, at the time of primary presentation (Balch 2009). There is debate regarding the prognostic effect from primary lesion site, with some evidence suggesting a worse prognosis for truncal lesions or those on the scalp or neck (Zemelman 2014).
Index test(s)
Smartphones are rapidly evolving from communication and entertainment devices to tools with specialised applications ('apps') that are intimately involved in many aspects of daily life (Kassianos 2015). The processing powers of modern smartphones allow their use in more demanding tasks such as image analysis (Massone 2007). Melanoma risk assessment tools are recent additions and include applications such as Mel App and Skin Scan (Robson 2012).
Once downloaded to a user's mobile phone (both Android and Apple iOS platforms), the applications can act as an information resource about melanoma or other skin cancer, provide guidance on whether people should seek medical advice for a particular lesion that they have photographed with the mobile phone, or be used to monitor skin lesions to identify any changes over time (Kassianos 2015).
Some applications that provide guidance on particular skin lesions can use internally programmed algorithms (or 'artificial intelligence') to catalogue and classify the lesion images. Others are store‐and‐forward applications that forward the photograph of the lesion to an experienced professional such as a dermatologist for review and then communicate a recommendation regarding the nature of the lesion to the user (essentially allowing members of the public direct access to a teledermatology‐type service) (Kassianos 2015).
The artificial intelligence‐based applications use algorithms to compare the acquired image against a bank of exemplar images of malignant and benign lesions or compare the image against a host of benign and malignant lesion characteristics learned from analysing thousands of other images to assess the likelihood of melanoma. These algorithms are generally based on fractal analysis. A fractal, in biology, is a natural phenomenon that exhibits a repeating pattern at every scale (Landini 2011). Fractal analysis can provide a quantitative measure of irregularity where regularity is expected. With regard to melanoma, this includes irregularities in a lesion's physical characteristics, such as those used in established algorithms to assist in the diagnosis of melanoma (e.g. the 'ABCs' of melanoma (Friedman 1985)), as well as texture, patterns, and other geometric features. Fractal analysis has been used for diagnosis of other cancers, for example, mammography for breast cancer (Rangayyan 2007; Raguso 2010), but it has not historically been made available to consumers for assessment of their own malignancy risk. A major benefit of fractal analysis is that it is automated and thus observer‐independent.
A recent review by Kassianos 2015 identified 39 available smartphone applications related to melanoma; most were multifunctional in that they provided information about melanoma in addition to lesion classification or a means of monitoring a given lesion. Just under half of the applications (46%; 18/39) provided some form of image analysis, and a quarter (23%; 9/39) used 'store‐and‐forward' lesion image review by a dermatologist. Those providing image analysis often did not describe how the photographic images were processed and analysed to provide advice on the likelihood of melanoma (Kassianos 2015). Authors described four applications as providing an assessment of the likelihood of melanoma: two used an artificial intelligence‐based algorithm based on the ABCDE method (assessing asymmetry, borders, colour, diameter and evolution), one provided a risk approximation based on the completion of a visual analogue scale by the application user, and one provided insufficient information regarding the method involved (Kassianos 2015). Between 2014 and 2018, 235 new dermatology smartphone applications became available (an increase of 80.8%), including dozens of teledermatology applications, which rose from 32 to 106 (Flaten 2018).
Clinical pathway
Individuals or their relatives are often best placed to recognise suspicious or changing skin lesions and may use a range of resources to become better informed about their concerns. Smartphone applications could have a role very early on in the clinical pathway, as they are readily accessible and potentially offer an instant risk assessment of the likelihood of malignancy, reassuring those with benign appearing lesions and effectively triaging those who need to seek further medical attention from a clinician for more detailed assessment of the lesion (Figure 2).
2.
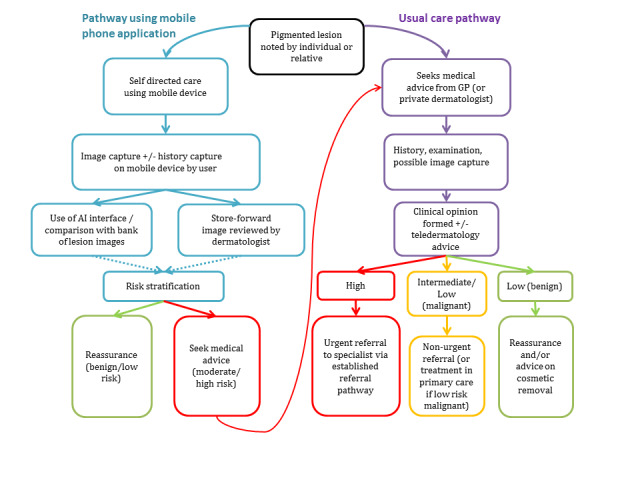
Example pathway for an individual using a smartphone application to examine a suspicious mole in resource settings with smartphones
In the UK, people with concerns about a new or changing lesion (either based on skin self‐examination alone or with the aid of a mobile phone application) will then present to their general practitioner (GP) rather than directly to a specialist in secondary care (Figure 3). If the GP has concerns, they may refer the patient to a specialist in secondary care – usually a dermatologist but sometimes a plastic surgeon or an ophthalmologist. Other systems may be in place in other countries, with the possibility of presenting directly to a skin specialist. Other specialists may also identify suspicious skin lesions, for example, a general surgeon or other specialist surgeon (including ear, nose, and throat (ENT) specialist (Figure 3) and refer patients to a specialist consultation with a dermatologist or plastic surgeon. Current UK guidelines recommend that GPs assess all suspicious pigmented lesions presenting in primary care by taking a clinical history and visually inspecting them using the revised seven‐point checklist (MacKie 1990); GPs should urgently refer suspicious pigmented skin lesions for specialist assessment within two weeks (Marsden 2010; Chao 2013; NICE 2015). Evidence is emerging, however, to suggest that excision of melanoma by GPs is not associated with increased risk compared with outcomes in secondary care (Murchie 2017). The specialist clinician will use history‐taking, visual inspection of the lesion (in comparison with other lesions on the skin), and usually dermoscopy to inform a clinical decision. If clinicians suspect melanoma, then urgent excision is advised. Other lesions such as suspected dysplastic naevi or pre‐malignant lesions such as lentigo maligna may also be referred for a diagnostic biopsy, further surveillance, or reassurance and discharge.
3.
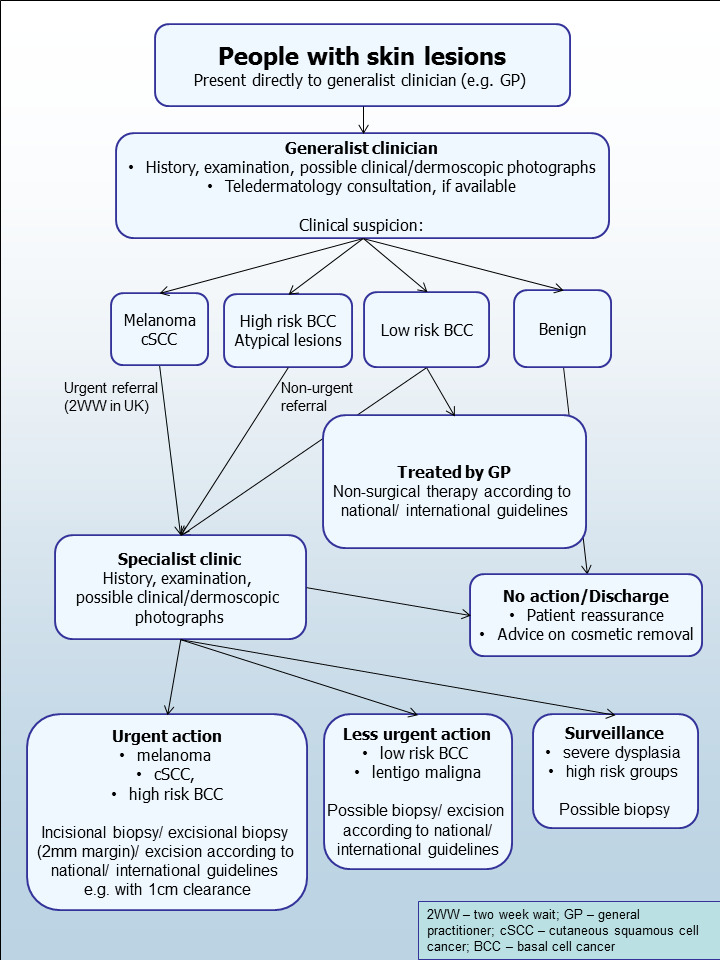
Current clinical pathway for people with skin lesions.
Role of index test(s)
Advances in smartphone technology have provided innovative platforms, where people can become more educated about their medical conditions, leading to better engagement with healthcare professionals (Robertson 2014). The use of smartphone technology can facilitate active self‐management health practices and provide patients information related to their condition (Tyagi 2012). As they are self‐initiated, psychological barriers to seeking medical advice can diminish, as assessments take place outside clinical settings and are often interactive and personalised (Tyagi 2012). The advances in smartphone technology provide new strategies for engaging patients in the management of potentially suspicious skin lesions, increasing the likelihood of detecting melanoma earlier in the progression of the disease. Early detection of melanoma is crucial for patients, dramatically improving survival and reducing morbidity (Balch 2009). There is increased value to the users and healthcare professionals alike, as more educated patients can better engage with their doctors, making consultations more effective and efficient (Robertson 2014).
The greatest concern about mobile phone applications in this context relates to their ability to accurately stratify lesions by level of risk of development of melanoma, particularly given the potential for falsely reassuring people that their lesion is benign. There is real concern that people could be dissuaded from accessing healthcare advice if the app deems their lesion to be low risk (Robson 2012). The most useful applications will therefore be those that maximise sensitivity over specificity for the detection of melanoma. Howeve, this feeds a concern that those who use such applications may be the 'worried well' rather than those who might actually have melanoma, which could flood limited healthcare resources with unnecessary referrals or simply generate profits for private providers who may take advantage of public cancer anxiety.
Alternative test(s)
For the purposes of our series of reviews, we consider each component of the diagnostic process, including visual inspection or clinical examination (whether delivered in‐person or remotely via teledermatology), as a diagnostic or index test, the accuracy of which can be established in comparison with a reference standard of diagnosis, either alone or in combination with other available technologies.
Once an individual or their relatives have identified a suspicious lesion, the only alternative to the use of a mobile phone application is to immediately seek medical advice from their GP or specialist clinician. The clinician will then use history‐taking, visual inspection of the lesion (in comparison with other lesions on the skin), and usually dermoscopy to inform a clinical decision (Figure 2). Our series of systematic reviews has also assessed the accuracy of visual inspection alone and dermoscopy plus visual inspection (Dinnes 2018a; Dinnes 2018b).
Chuchu 2018 has also conducted a review of the accuracy of teledermatology, whereby dermatologists receive clinical photographs or dermoscopic images of a skin lesion, traditionally from non‐specialist clinicians, and give a specialist opinion on a suspicious lesion. This can be done on a store‐and‐forward basis, using digital cameras or mobile phones to acquire photographs or dermoscopic images of a lesion, or via a live video link. According to UK guidelines, 'full dermatology' services (i.e. a replacement for a face‐to‐face consultation) require both clinical and dermoscopic images, whereas 'triage teledermatology' services process only dermoscopic images where facilities permit (BAD 2013).
Teledermatology not only allows clinicians rapid access to expert opinion but may lead to a reduction in waiting times and limit unnecessary referrals (Ndegwa 2010; Warshaw 2010; Bashshur 2015). In rural areas, where people's access to speciality services can have significant and potentially off‐putting travel and time implications, teledermatology has the potential to increase access to specialist opinion.
Teledermatology is also becoming available in a community setting, especially within community or 'high street' pharmacies (for example, the Boots 'Mole Scanning Service', www.boots.com/health‐pharmacy‐advice/skin‐services/mole‐scanning‐service), and is therefore a potential alternative to smartphone applications. Due to their extended opening hours, ease of access, presence of healthcare professionals and availability of consultation rooms, community pharmacies are increasingly providing early detection services (Kjome 2016), such as mole scanning by trained pharmacy staff. In theory, using pharmacies as the first‐line identifier to separate those with skin lesions requiring follow‐up from those who do not, gives general practitioners and specialists more time and resources for those who require intervention (Kjome 2016).
A number of other tests that may have a role in the diagnosis of melanoma in a specialist setting have been reviewed as part of our series of systematic reviews, including reflectance confocal microscopy, optical coherence tomography, computer‐assisted diagnosis or artificial intelligence‐based techniques, and high‐frequency ultrasound (Dinnes 2018c; Ferrante di Ruffano 2018a; Ferrante di Ruffano 2018b; Dinnes 2018d).
Evidence permitting, we will compare the accuracy of available tests in an overview of review, exploiting within‐study comparisons of tests and allowing the analysis and comparison of commonly used diagnostic strategies where tests may be used alone or in combination.
Rationale
Our series of reviews of diagnostic tests used to assist clinical diagnosis of melanoma aims to identify the most accurate approaches to diagnosis and provide clinical and policy decision‐makers with the highest possible standard of evidence on which to base decisions. With increasing rates of melanoma incidence and the push towards the use of dermoscopy and other high‐resolution image analysis in primary care without adequate evidence of effectiveness or safety, the anxiety around missing early cases needs to be balanced against the risk of over referrals, to avoid sending too many people with benign lesions for a specialist opinion. It is questionable whether all skin cancers picked up by sophisticated techniques, even in specialist settings, help to reduce morbidity and mortality or whether newer technologies run the risk of increasing false positive results. It is also possible that use of some technologies (e.g. widespread use of dermoscopy in primary care with no training), could actually result in harm by missing melanomas if they are used as replacement technologies for traditional history‐taking and clinical examination of the entire skin; many branches of medicine have noted the danger of such 'gizmo idolatry' amongst doctors (Leff 2008).
Smartphone applications in general are already widely available and used by consumers, and the popularity of such platforms to offer clinical and diagnostic advice is steadily increasing. Given the rapidly changing evidence base and lack of available systematic reviews on the topic, there is a need for an up‐to‐date analysis of the accuracy of smartphone applications.
As several reviews for each topic area followed the same methodology, we prepared generic protocols in order to avoid duplication of effort: one for diagnosis of melanoma, Dinnes 2015a, and one for diagnosis of keratinocyte skin cancers, Dinnes 2015b. The Background and Methods sections of this review therefore use some text that was originally published in the protocol concerning the evaluation of tests for the diagnosis of melanoma (Dinnes 2015a) and some text that overlaps some of our other reviews (Dinnes 2018b).
Objectives
To assess the diagnostic accuracy of smartphone applications to rule out cutaneous invasive melanoma and atypical intraepidermal melanocytic variants in adults with concerns about suspicious skin lesions.
Methods
Criteria for considering studies for this review
Types of studies
We included test accuracy studies that allow comparison of the result of the index test with that of a reference standard, including the following.
Studies where all participants receive a single index test and a reference standard.
Studies where all participants receive more than one index test(s) and reference standard.
Studies where participants are allocated (by any method) to receive different index tests or combinations of index tests, and all receive a reference standard (between‐person comparative studies (BPC)).
Studies that recruit series of participants unselected by true disease status (referred to as case series for the purposes of this review).
Diagnostic case‐control studies that separately recruit diseased and non‐diseased groups (see Rutjes 2005).
Both prospective and retrospective studies.
Studies where previously acquired clinical or dermoscopic images were retrieved and prospectively interpreted for study purposes.
We excluded studies from which we could not extract 2 × 2 contingency data or small studies with fewer than five disease‐positive or disease‐negative participants. Although the size threshold of five is arbitrary, such small studies are likely to give unreliable estimates of sensitivity or specificity and may be biased, like small randomised controlled trials of treatment effects.
Studies available only as conference abstracts were excluded; however, attempts were made to identify full papers for potentially relevant conference abstracts (Searching other resources).
Participants
We included studies in adults with pigmented skin lesions or lesions suspicious for melanoma. These could include those at high risk of developing melanoma, including those with a family history or previous history of melanoma skin cancer, atypical or dysplastic naevus syndrome, or genetic cancer syndromes. Ideally, participants should be recruited from community settings; however, due to an anticipated paucity of data, we considered participants recruited from any setting as eligible. We excluded studies that recruited only participants with malignant diagnoses and studies that compared test results in participants with malignancy compared with test results based on 'normal' skin as controls, due to the bias inherent in such comparisons (Rutjes 2006). We excluded studies with more than 50% of participants aged 16 and under.
Index tests
We included studies evaluating smartphone applications intended for use by any individual (or member of the public) with a smartphone in a community setting who has a skin lesion that concerns them. Applications intended for use by smartphone users were considered to be those using standard photographs acquired by the mobile phone. We considered applications to be intended for clinician use (e.g. GPs) as a way to access specialist dermatologist opinion (i.e. for store‐and‐forward teledermatology assessments) when the applications required dermoscopic or other microscopic attachments for the acquisition of magnified images.
We included studies developing new mobile phone applications (i.e. derivation studies) if they used a separate independent 'test set' of participants or images to evaluate the new approach.
We excluded studies if they:
used a statistical model to produce a data‐driven equation or algorithm based on multiple diagnostic features, with no separate test set;
used cross‐validation approaches such as 'leave‐one‐out' cross‐validation (Efron 1983); or
evaluated the accuracy of the presence or absence of individual lesion characteristics or morphological features, with no overall diagnosis of malignancy.
Target conditions
The target condition was cutaneous melanoma and atypical intraepidermal melanocytic variants (i.e. including melanoma in situ or lentigo maligna, which has a risk of progression to invasive melanoma).
Reference standards
The preferred reference standard for establishing the final diagnosis of a skin lesion is histopathological diagnosis of the excised lesion or biopsy sample in all eligible lesions. Histopathlogical assessment is not a perfect reference standard because it only samples lesions for examination and may therefore miss tumour cells in non‐sampled portions. As it is a subjective assessment, there is also some degree of inter‐observer variation, especially for borderline lesions. A qualified pathologist or dermatopathologist should perform histopathology. Ideally, reporting should be standardised, detailing a minimum dataset including the histopathological features of melanoma needed to determine staging according to the American Joint Committee on Cancer (AJCC) Staging System (e.g. Slater 2014). We did not apply the reporting standard as a necessary inclusion criterion but extracted any pertinent information.
Due to the potential for partial verification (with lesion excision or biopsy unlikely to be carried out for all benign‐appearing lesions within a representative population sample), we also accepted clinical follow‐up of benign‐appearing lesions, cancer registry follow‐up and 'expert opinion' with no histology or clinical follow‐up as eligible reference standards. We considered the risk of differential verification bias (as misclassification rates of histopathology and follow‐up will differ) in our quality assessment of studies.
We considered all of the above reference standards for establishing final diagnoses of the lesion, with the following caveats:
all study participants with a final diagnosis of the target disorder must have a histological diagnosis, either subsequent to the application of the index test or after a period of clinical follow‐up; and
at least 50% of all participants with benign lesions must have either a histological diagnosis or clinical follow‐up to confirm benignity.
The ability of a smartphone application to correctly triage those who need further assessment of suspicious skin lesions is not the only outcome of interest for this type of test, however. It is possible to estimate referral accuracy (or ability of the smartphone application to approximate an in‐person lesion assessment) by comparing the action recommended by the smartphone application with the management recommendation from face‐to‐face assessment by an appropriately qualified clinician. To this end, 'expert opinion' as the sole reference standard is an eligible reference standard for our reviews of both mobile phone applications and for teledermatology.
Search methods for identification of studies
Electronic searches
The Information Specialist (SB) carried out a comprehensive search for published and unpublished studies. A single large literature search was conducted to cover all topics in the programme grant (see Appendix 1 for a summary of reviews included in the programme grant). This allowed for the screening of search results for potentially relevant papers for all reviews at the same time. A search combining disease related terms with terms related to the test names, using both text words and subject headings was formulated. The search strategy was designed to capture studies evaluating tests for the diagnosis or staging of skin cancer. As the majority of records were related to the searches for tests for staging of disease, a filter using terms related to cancer staging and to accuracy indices was applied to the staging test search, to try to eliminate irrelevant studies, for example, those using imaging tests to assess treatment effectiveness. A sample of 300 records that would be missed by applying this filter was screened and the filter adjusted to include potentially relevant studies. When piloted on MEDLINE, inclusion of the filter for the staging tests reduced the overall numbers by around 6000. The final search strategy, incorporating the filter, was subsequently applied to all bibliographic databases as listed below (Appendix 3). The final search result was cross‐checked against the list of studies included in five systematic reviews; our search identified all but one of the studies, and this study was not indexed on MEDLINE. The Information Specialist devised the search strategy, with input from the Information Specialist from Cochrane Skin. No additional limits were used.
We searched the following bibliographic databases to 29 August 2016 for relevant published studies.
MEDLINE via OVID (from 1946);
MEDLINE In‐Process & Other Non‐Indexed Citations via OVID; and
Embase via OVID (from 1980).
We searched the following bibliographic databases to 30 August 2016 for relevant published studies.
The Cochrane Central Register of Controlled Trials (CENTRAL; 2016, Issue 7), in the Cochrane Library.
The Cochrane Database of Systematic Reviews (CDSR; 2016, Issue 8), in the Cochrane Library.
Cochrane Database of Abstracts of Reviews of Effects (DARE; 2015, Issue 2).
CRD HTA (Health Technology Assessment) database (2016; Issue 3).
CINAHL (Cumulative Index to Nursing and Allied Health Literature via EBSCO from 1960).
We searched the following databases for relevant unpublished studies using a strategy based on the MEDLINE search:
CPCI (Conference Proceedings Citation Index), via Web of Science™ (from 1990; searched 28 August 2016); and
SCI Science Citation Index Expanded™ via Web of Science™ (from 1900, using the 'Proceedings and Meetings Abstracts' Limit function; searched 29 August 2016).
We searched the following trials registers using the search terms 'melanoma', 'squamous cell', 'basal cell' and 'skin cancer' combined with 'diagnosis':
Zetoc (from 1993; searched 28 August 2016).
The US National Institutes of Health Ongoing Trials Register (www.clinicaltrials.gov); searched 29 August 2016.
NIHR Clinical Research Network Portfolio Database (www.nihr.ac.uk/research‐and‐impact/nihr‐clinical‐research‐network‐portfolio/); searched 29 August 2016.
The World Health Organization International Clinical Trials Registry Platform (apps.who.int/trialsearch/); searched 29 August 2016.
We aimed to identify all relevant studies regardless of language or publication status (published, unpublished, in press, or in progress). We applied no date limits.
Searching other resources
We have screened relevant systematic reviews identified by the searches for their included primary studies, and we included any missed by our searches. We have checked the reference lists of all included papers, and subject experts within the author team reviewed the final list of included studies. We did not perform electronic citation searching.
Data collection and analysis
Selection of studies
At least one author (JDi or NC) screened titles and abstracts, discussing and resolving any queries by consensus. A pilot screen of 539 MEDLINE references showed good agreement (89% with a kappa of 0.77) between screeners. At initial screening, we included primary test accuracy studies and test accuracy reviews (for scanning of reference lists) of any test used to investigate suspected melanoma, basal cell carcinoma (BCC), or cutaneous squamous cell carcinoma (cSCC). Both a clinical reviewer (from a team of 12 clinician reviewers) and a methodologist reviewer (JDi or NC) applied inclusion criteria (Appendix 4) to all full text articles, resolving disagreements by consensus or by consultation with a third party (JDe, CD, HW or RM). We contacted authors of eligible studies when they presented insufficient data to allow for the construction of 2 × 2 contingency tables.
Data extraction and management
One clinical (as detailed above) and one methodological reviewer (JDi, NC or LFR) independently extracted data concerning details of the study design, participants, index test(s) or test combinations and criteria for index test positivity, reference standards, and data required to populate a 2 × 2 diagnostic contingency table for each index test using a piloted data extraction form. Data were extracted at all available index test thresholds. We resolved disagreements by consensus or in consultation with a third party (JDe, CD, HW or RM).
We contacted authors of included studies where there was missing information related to the target condition (in particular to allow the differentiation of invasive cancers from in situ variants) or diagnostic threshold. We contacted authors of conference abstracts published from 2013 to 2015 to ask whether full data were available, marking them as 'pending' when we could not obtain a full paper. We will revisit these in future review updates.
Where we identified multiple reports of a primary study, we maximised yield of information by collating all available data. Where there were inconsistencies in reporting or overlapping study populations, we contacted study authors for clarification in the first instance. If contact with authors was unsuccessful, we used the most complete and up‐to‐date data source where possible.
Assessment of methodological quality
We assessed risk of bias and applicability of included studies using the QUADAS‐2 checklist (Whiting 2011), tailored to the topic of skin cancer diagnosis (see Appendix 5). We piloted the modified QUADAS‐2 tool on five full‐text articles. One clinical and one methodological reviewer (JDi, NC or LFR) independently assessed quality for the remaining studies, resolving any disagreement by consensus or in consultation with a third party where necessary (JDe, CD, HW or RM).
Statistical analysis and data synthesis
Due to scarcity of data and the poor quality of studies, we did not undertake a meta‐analysis for this review. For illustrative purposes, we plotted estimates of sensitivity and specificity on coupled forest plots for each application under consideration. Our unit of analysis was the lesion rather than the patient, as initial treatment in skin cancer is directed to the lesion and not systemically to a patient (thus it is important to be able to correctly identify cancerous lesions within each patient). Moreover, this is the most common way in which the primary studies reported data. Although there is a theoretical possibility of correlations of test errors when multiple lesions are included from the same patients, most studies include very few patients with multiple lesions, and any potential impact on findings is likely to be very small, particularly in comparison with other concerns regarding risk of bias and applicability. For each analysis, we included only one dataset per study to avoid multiple counting of lesions.
Investigations of heterogeneity
We examined heterogeneity between studies by visually inspecting the forest plots of sensitivity and specificity and summary receiver operating characteristics (ROC) plots. we did not identify enough studies to allow meta‐regression to investigate potential sources of heterogeneity.
Sensitivity analyses
We did not perform any sensitivity analyses.
Assessment of reporting bias
Due to uncertainty about the determinants of publication bias for diagnostic accuracy studies and the inadequacy of tests for detecting funnel plot asymmetry (Deeks 2005), we did not perform any tests to detect publication bias.
Results
Results of the search
We screened a total of 34,517 unique references for inclusion. Of these, we reviewed 1051 full‐text papers for eligibility and included 203 publications in at least one of the suite of reviews of tests to assist in the diagnosis of melanoma or keratinocyte skin cancer. Figure 4 provides a PRISMA flow diagram of search and eligibility results. We considered 16 studies to be potentially eligible for this review of smartphone applications and ultimately included two publications. Figure 4 lists the reasons for exclusion, while the Characteristics of excluded studies tables list both the studies and the reasons we excluded them. Two studies included fewer than five melanoma cases (Massone 2007; Robson 2012); three studies used an inappropriate index test (including two studies where mobile phones were used to capture dermoscopic (Massone 2007) or otherwise magnified (Diniz 2016) images in a specialist clinic setting); two studies were derivation studies that did not separate data for training and test sets (Ramlakhan 2011; Wadhawan 2011), and one study was a duplicate or related publication (Von Braunmühl 2015 reported data for the same patients as Maier 2015). A list of all studies excluded from the full series of reviews is available as a supplementary file (please contact skin.cochrane.org for a copy).
4.
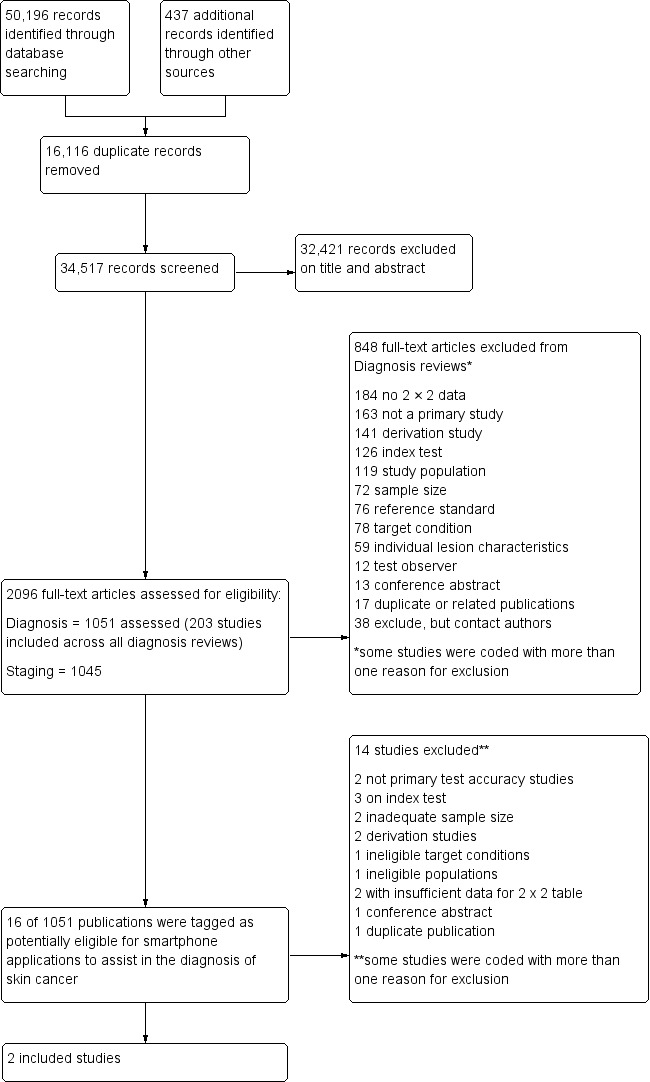
PRISMA flow diagram.
Across all of our reviews, we contacted the corresponding authors of 84 studies to ask them to supply further information needed to allow study inclusion (37 studies), to clarify diagnostic thresholds (18 studies) or to define the target condition (29 studies). We received responses from 39 authors, allowing the inclusion of 4 studies across various reviews (and 1 study for this review, Wolf 2013), and providing data clarifications for 23 others.
This review reports on two cohorts of lesions published in two studies that provide six datasets (Wolf 2013; Maier 2015). The applications successfully analysed a total of 332 lesions, including 86 melanomas. The studies did not report the number of participants with lesions included in the studies.
Wolf 2013 retrospectively evaluated photographs of lesions selected from their dermatology database of lesions that had been scheduled for excision using a case‐control type design. Health professionals routinely captured these lesion images from participants who had presented with suspicious skin lesions in a dermatological setting rather than the community setting where these applications are intended to be used, and the study included only lesions with a final diagnosis of melanoma, melanoma in situ, lentigo, benign naevi (including compound, junctional and low grade dysplastic naevi), dermatofibroma, sebhorrhoeic keratosis and haemangioma. The study included lesions with good quality photographs (as assessed by one or two dermatologists) and with a clear histological diagnosis. The study excluded lesions that were uncommon or had an equivocal diagnosis (such as 'melanoma cannot be ruled out' or 'atypical melanocytic proliferation') and lesions with moderate or high grade atypia. Investigators excluded more than half the images reviewed for inclusion in the study (52%; 202/390) due to poor image quality, the presence of identifiable patient features or insufficient clinical or histological information. The applications analysed 3 to 29 additional lesions, but trialists considered these to be unevaluable or test failures (see Findings).
Maier 2015 conducted a prospective case series of patients with melanocytic skin lesions seen routinely at the department of dermatology for skin cancer screening. It is unclear whether participants were referred or could access the dermatology clinic directly. Up to three smartphone images (photographs) per lesion were taken, presumably by the dermatologist, before excising the lesion; however, authors did not clearly describe the image acquisition process. The study excluded 20/195 lesions (10%) due to poor image quality or incomplete imaging. Study authors excluded an additional 31 lesions they considered as test failures for the purposes of this review (see Findings), including 13 due to "two‐point differences" (explained as non‐consecutive risk classes, presumably for different images of the same lesion) and 18 "tie‐cases" (defined as having an equal number of results in two consecutive risk classes, e.g. 1 high risk, 1 medium risk and 1 low risk result).
Wolf 2013 took place in the USA and Maier 2015 in Germany. Neither study reported information on the number of patients recruited or their characteristics (e.g. age and sex); total numbers of included lesions were 188 in Wolf 2013 and 144 in Maier 2015. Both studies reported on the accuracy of smartphone applications for detecting melanoma and its atypical intraepidermal melanocytic variants. The prevalence of melanoma was 18% in Maier 2015 and 35% in Wolf 2013.
Wolf 2013 evaluated four different smartphone applications, providing no names to avoid consumer bias; they numbered the applications one to four to allow an assessment of accuracy. Three applications were artificial intelligence‐based classifications of lesions as 'problematic' versus 'okay' (App 1), 'melanoma' versus 'not melanoma' (App 2) and 'high risk' versus 'medium/low‐risk' (App 3) as part of the assessment of the images. It was also possible to dichotomise data for App 3 as 'high/medium risk' (test positive) versus 'low risk' (test negative). App 4 used a store‐and‐forward approach with remote lesion assessment by a qualified dermatologist. Users could run this application on either a smartphone or a website, with lesion images uploaded and transmitted remotely to a dermatologist to make an assessment and return it to the user within 24 hours. The output given was 'atypical' versus 'typical'.
Maier 2015 evaluated an automated risk assessment algorithm using the SkinVision App; this relied on fractal image analysis of three images per lesion. The application classified lesions as 'high risk' versus 'medium or low risk'. The study also reported the diagnostic accuracy of face‐to‐face clinical diagnosis by a dermatologist for the same lesions.
In both studies the reference standard diagnosis was made by histology alone (all lesions were either biopsied or excised).
Methodological quality of included studies
Figure 5 and Figure 6 summarise the overall methodological quality of included studies.
5.
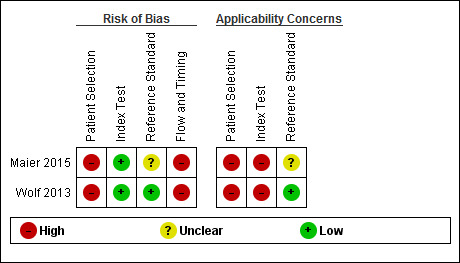
Risk of bias and applicability concerns summary: review authors' judgements about each domain for each included study
6.
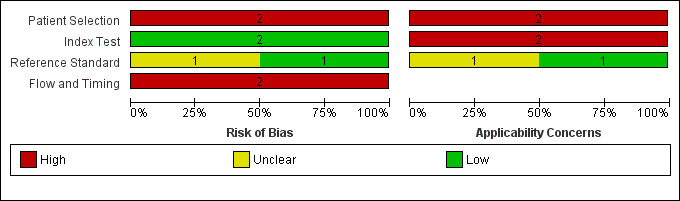
Risk of bias and applicability concerns graph: review authors' judgements about each domain presented as percentages across included studies
We assessed both studies as being at high risk of bias for participant selection due to the inappropriate exclusion of lesions that would have otherwise been eligible for assessment with the applications. Wolf 2013 also used a case‐control design, including only lesions with particular final diagnoses. Similarly, both studies caused high concern regarding included participants and setting, due to unclear reporting of patient samples and whether they included multiple lesions per patient. Wolf 2013 excluded lesions that were common or with equivocal diagnoses. All studies included only lesions selected for excision. This is not a representative spectrum of lesions that would be observed in daily life but rather a highly selected sample of participants. These participants would have already presented to a doctor with concerns about a particular lesion and therefore are likely represent a more severe spectrum of abnormality, which will artificially increase the sensitivity of the test in comparison to use by smartphone users in general.
We assessed both studies as being at low risk of bias in the index test domain, with the artificial intelligence‐based assessment made without knowledge of the histological diagnosis. All had a pre‐specified threshold. However, both studies caused high concern about applicability of the index tests, which were not used as intended in practice. Wolf 2013 used archived photographs of lesions rather than images taken using a phone and did not report the real names of the applications used. Maier 2015 did use smartphone images; however, the details around the imaging process were unclear.
All studies reported the use of an acceptable reference standard; however, Maier 2015 did not report blinding of histology to the index test result. Only Wolf 2013 reported histopathological interpretation by an experienced dermatopathologist.
Both studies were at high risk of bias for flow and timing due to exclusion of unevaluable images from further analysis (authors do state the numbers excluded, allowing computation of test failure rates). Both were unclear on the interval between image capture and performance of the reference standard.
Findings
Detection of invasive melanoma or atypical intraepidermal melanocytic variants
Across the five different applications that the two studies assessed, sensitivities for the detection of invasive melanoma or atypical intraepidermal melanocytic variants ranged from 7% (95% confidence interval (CI) 2% to 16%) to 98% (95% CI 90% to 100%) and specificities from 30% (95% CI 22% to 40%) to 94% (95% CI 87% to 97%; Figure 7).
7.

Forest plot of tests: showing sensitivity and specificity of all the applications for the detection of cutaneous melanoma and atypical intraepidermal variants
1 App 1[problematic vs okay], 2 App 2 [mel vs not mel], 3 App 3(a) [high risk vs medium+low risk], 4 App 3(b) [high+medium risk vs low risk], 5 App 4 (remote diagnosis) [atypical vs typical], 6 SkinVision [high risk vs medium/low risk].
One of the four artificial intelligence‐based applications attempted to correctly identify lesions as melanomas or not. For App 2 in Wolf 2013 (185 lesions and 58 melanoma cases), the resulting sensitivity was 69% (95% CI 55% to 80%), and specificity was 37% (95% CI 29% to 46%).
The remaining three artificial intelligence‐based algorithms attempted to categorise lesions as high risk or 'problematic' or not (Figure 7). Sensitivities were around 70% for App 1 in Wolf 2013 (70%, 95% CI 57% to 81%) and for the SkinVision app in Maier 2015 (73%, 95% CI 52% to 88%) but only 7% (95% CI 2% to 16%) for App 3 in Wolf 2013. The corresponding specificities for the three applications were 39% (95% CI 31% to 49%; Wolf 2013; 182 lesions and 60 melanomas), 83% (95% CI 75% to 89%; Maier 2015; 144 lesions and 26 melanomas), and 94% (95% CI 87% to 97%; Wolf 2013; 170 lesions and 59 melanomas). Decreasing the threshold for considering lesions as test positive (i.e. including both high and medium risk lesions) for App 3 in Wolf 2013 (denoted App 3b in Figure 7) increased sensitivity from 7% to 54% (95% CI 41% to 67%), with a fall in specificity from 94% to 61% (95% CI 52% to 70%).
The final application, App 4 in Wolf 2013, was a store‐and‐forward system, with a dermatologist classifying lesion images as atypical or typical. The sensitivity for this application was 98% (95% CI 90% to 100%) and specificity 30% (95% CI 22% to 40%; 159 lesions and 54 melanoma cases).
This application however, recorded the highest percentage of 'test failures' in the Wolf 2013 study (i.e. eligible lesions analysed by the applications but recorded as unevaluable). The test failure rates were: 3% (App 1), 2% (App 2), 6% (App 3) and 15% (App 4; designated by the dermatologist as 'send another photograph' or 'unable to categorise') (Table 2). Three of the four applications classed at least one melanoma as unevaluable, with the dermatologist conducting the assessment for the store‐and‐forward application (App 4) missing 6 (10%) melanomas for this reason.
1. Index test failures – lesions not evaluable by smartphone application.
| Study | Total number of lesions (melanomas) successfully assessed by the each 'app' | Unevaluable lesions (%) | Number (%) of unevaluable melanomas | |
|
Wolf 2013 188 lesions (60 melanomas)a |
Application 1 | 182 (60) | 6 (3%) | 0 (0%) |
| Application 2 | 185 (58) | 3 (2%) | 2 (3%) | |
| Application 3 | 170 (59) | 12 (6%) | 1 (2%) | |
| Application 4 | 159 (54) | 29 (15%) | 6 (10%) | |
|
Maier 2015 175 lesions (number of melanomas: not reported; between 26 and 40)b |
SkinVision | 144 (26) | 31 (18%) | Not reported (≤ 14) |
a52% (202/390) of all images reviewed for inclusion in the study were excluded prior to analysis by the applications. b10% (20/195) of all images reviewed for inclusion in the study were excluded due to poor image quality or incomplete imaging. The original sample of 195 lesions included 60 melanomas. The number of melanomas excluded on the basis of image quality and the number analysed by the application but not included by the study authors (considered as test failures for the purposes of this review) were not separately reported.
The SkinVision application described in Maier 2015 analysed a total of 31 lesions (18%; 31/175) as unevaluable (Table 2). Although the studies did not report the number of melanomas classed as unevaluable; however, the study authors excluded 35% of all melanomas originally eligible for the study (14/40) due to poor image quality or because the images were classed as unevaluable.
Maier 2015 also reported the accuracy of face‐to‐face clinical diagnosis of the same lesions by a dermatologist. These data are not directly comparable to those that the application generated, as the in‐person assessment relates to the diagnosis of melanoma whereas the SkinVision application was developed to identify lesions at high risk of melanoma. The sensitivity of the face‐to‐face assessment was 85% (95% CI 65% to 96%) and specificity, 97% (95% CI 93% to 99%; Figure 8; 144 lesions and 26 melanomas).
8.

Forest plot of tests: SkinVision automated diagnosis compared to face to face clinical diagnosis by a dermatologist
6 SkinVision [high risk vs medium/low risk], 7 Face‐to‐face clinical diagnosis [high risk vs medium/low risk].
Investigations of heterogeneity
We were unable to undertake investigations of heterogeneity listed in the protocol due to an insufficient number of studies.
Discussion
Summary of main results
This review aimed to assess the accuracy of smartphone applications for detecting invasive melanoma or atypical intraepidermal melanocytic variants. We included two studies with a total of 332 lesions, 86 of which were melanomas (Table 1).
Studies were generally of poor methodological quality. Risk of bias was low for both studies only for the index test domain. Poor reporting did not always allow adequate judgement of the quality of the reference standard. Study participants were highly selected in comparison to those who might choose to use a smartphone application to check a skin lesion that was causing them concern. Both of the studies used photographs of skin lesions that were scheduled for excision in a dermatology clinic setting, and clinicians were the ones who took the photographs instead of people using their own smartphones, potentially leading to the acquisition of higher quality images. Studies were blinded for index test interpretation and used pre‐specified test thresholds and adequate reference standards. One study did not report blinding of the reference standard to the lesion images, and one did not mention interpretation by an experienced histopathologist. We are therefore unable to make a reliable estimate of the accuracy of smartphone applications for detecting melanoma or intra‐epidermal melanocytic variants.
Across the four artificial intelligence‐based applications that classified lesion images (photographs) as melanomas (one application) or as high risk or 'problematic' lesions (three applications), sensitivities ranged from 7% (95% CI 2% to 16%) to 73% (95% CI 52% to 88%) and specificities from 37% (95% CI 29% to 46%) to 94% (95% CI 87% to 97%). This means that between 27% and 93% of invasive melanoma or atypical intraepidermal melanocytic variants were not picked up as requiring further assessment by a clinician by the automated applications (or as melanomas by one of the four applications). With a prevalence of melanoma ranging between 18% and 37% for these evaluations, the number of melanomas missed was between 7 and 55.
The single application using store‐and‐forward review of lesion images by a dermatologist had a sensitivity of 98% (95% CI 90% to100%) and specificity of 30% (95% CI 22% to 40%); the dermatologist missed one melanoma.
The number of test failures (lesion images that the applications analysed but that the study authors classed as unevaluable and excluded) ranged from 3 to 31 (or 2% to 18% of lesions analysed). The store‐and‐forward application had one of the highest rates of test failure (15%). Three of the four applications classed at least one melanoma as unevaluable, with the highest number of excluded melanomas (6/60 melanomas assessed) resulting from dermatologist evaluation of the store‐and‐forward images.
Strengths and weaknesses of the review
The strengths of this review include an in‐depth and comprehensive electronic literature search, systematic review methods including double extraction of papers by both clinicians and methodologists, and contact with authors to allow study inclusion or clarify data. We planned a clear analysis structure to allow estimation of test accuracy in different study populations and undertook a detailed and replicable analysis of methodologic quality.
We did not identify any other systematic reviews of smartphone applications during the preparation of this review. However, Kassianos 2015 systematically attempted to identify all available smartphone applications as of July 2014 by searching the online stores of smartphone providers (Apple and Android) and then systemically extracting data about the applications from their online descriptions. Authors made no attempt to identify any diagnostic test accuracy research underlying the applications. It is notable that Kassianos 2015 identified 39 applications, and we were only able to identify test accuracy evaluations for five. We did not contact developers of commercially available smartphone applications for any further accuracy data; however, a future review update could do this.
The main concerns for the review are the clinical applicability of the findings and exclusion of unevaluable test results, with likely overestimation of sensitivity.
Applicability of findings to the review question
The data included in this review are unlikely to be generally applicable to the intended setting. Study participants were people who had skin lesions already scheduled for excision in a dermatology clinic setting rather than smartphone users with concerns about a new or changing mole or skin lesion, and dermatologists were likely to have taken the photographs used with the applications in the clinic setting in both studies. One study also excluded equivocal lesions and those with moderate or high‐grade atypia, both of which could potentially be more likely to produce unevaluable results or be misclassified by the application.
Authors' conclusions
Implications for practice.
We could not produce any summary estimates of test accuracy to answer the research question for this review. Smartphone applications using artificial intelligence‐based analysis have not yet demonstrated sufficient promise in terms of accuracy, and they are associated with a high likelihood of missing melanomas. Available data have limited applicability in practice due to selective participant recruitment from secondary referral settings and the use of images not acquired by the intended users of the smartphone applications (i.e. members of the public). Applications based on store‐and‐forward images could have a potential role in the timely presentation of people with potentially malignant lesions by facilitating active self‐management health practices and early engagement of those with suspicious skin lesions; however, there are resource and workload implications with a store‐and‐forward approach.
Given the paucity of evidence and low methodological quality, we cannot draw any implications for practice. Nevertheless, this is a fast‐moving field, and new and better apps and better reported studies could change these conclusions substantially.
Implications for research.
Prospective evaluation of smartphone applications for identifying people with suspicious skin lesions who should seek further medical advice from a suitably qualified clinician is required to fully understand the accuracy of these tools. Studies should take place in a clinically relevant community or primary care setting, recruiting smartphone users who may have concerns about their risk of developing melanoma or about a new or changing skin lesion. Studies might compare the recommendation from the smartphone with that of a GP following a face‐to‐face clinical diagnosis of the same lesion. In such a study it is important that the GP assesses all lesions examined using the smartphone in the same way, with blinding to the smartphone recommendation. Although histological confirmation of melanoma versus not melanoma is the ideal reference standard, it is not a practical or ethical one for study participants with lesions at low risk of malignancy. Systematic follow‐up of non‐excised lesions over a five‐year period would avoid over‐reliance on a histological reference standard and would further allow results to be more generalisable to routine practice. Although a pragmatic evaluation amongst the general population of smartphone users would be challenging, studies could include those most at risk of developing melanoma, in whom the prevalence of disease would be higher. Use of the test by smartphone users themselves rather than healthcare professionals or equipment experts is also key to ensuring the clinical applicability of study findings and to determine the true test failure rate, which could seriously inhibit the use of smartphone applications in practice. Any future research study should conform to the updated Standards for Reporting of Diagnostic Accuracy (STARD) guideline (Bossuyt 2015).
What's new
| Date | Event | Description |
|---|---|---|
| 19 December 2018 | Amended | Affiliations, Disclaimer and Sources of support updated |
Acknowledgements
Members of the Cochrane Skin Cancer Diagnostic Test Accuracy Group include:
the full project team (Susan Bayliss, Naomi Chuchu, Clare Davenport, Jonathan Deeks, Jacqueline Dinnes, Lavinia Ferrante di Ruffano, Kathie Godfrey, Rubeta Matin, Colette O'Sullivan, Yemisi Takwoingi, Hywel Williams);
our 12 clinical reviewers (Rachel Abbott, Ben Aldridge, Oliver Bassett, Sue Ann Chan, Alana Durack, Monica Fawzy, Abha Gulati, Jacqui Moreau, Lopa Patel, Daniel Saleh, David Thompson, Kai Yuen Wong) and 2 methodologists (Lavinia Ferrante di Ruffano and Louise Johnston), who assisted with full text screening, data extraction and quality assessment across the entire suite of reviews of diagnosis and staging and skin cancer;
our expert advisors and co‐authors Abhilash Jain and Fiona Walter; and
all members of our Advisory Group (Jonathan Bowling, Seau Tak Cheung, Colin Fleming, Matthew Gardiner, Abhilash Jain, Susan O'Connell, Pat Lawton, John Lear, Mariska Leeflang, Richard Motley, Paul Nathan, Julia Newton‐Bishop, Miranda Payne, Rachael Robinson, Simon Rodwell, Julia Schofield, Neil Shroff, Hamid Tehrani, Zoe Traill, Fiona Walter, Angela Webster).
The Cochrane Skin editorial base wishes to thank Urbà González, who was the Dermatology Editor for this review; and the clinical referees, Saul Halpern and David de Berker. We also wish to thank the Cochrane DTA editorial base and colleagues, as well as Meggan Harris, who copy‐edited this review.
Appendices
Appendix 1. Current content and structure of the Programme Grant
| LIST OF REVIEWS | Number of studies | |
| Diagnosis of melanoma | ||
| 1 | Visual inspection | 49 |
| 2 | Dermoscopy +/‐ visual inspection | 104 |
| 3 | Teledermatology | 22 |
| 4 | Smartphone applications | 2 |
| 5a | Computer‐assisted diagnosis – dermoscopy‐based techniques | 42 |
| 5b | Computer‐assisted diagnosis – spectroscopy‐based techniques | Review amalgamated into 5a |
| 6 | Reflectance confocal microscopy | 18 |
| 7 | High‐frequency ultrasound | 5 |
| Diagnosis of keratinocyte skin cancer (BCC and cSCC) | ||
| 8 | Visual inspection +/‐ Dermoscopy | 24 |
| 5c | Computer‐assisted diagnosis – dermoscopy‐based techniques | Review amalgamated into 5a |
| 5d | Computer‐assisted diagnosis – spectroscopy‐based techniques | Review amalgamated into 5a |
| 9 | Optical coherence tomography | 5 |
| 10 | Reflectance confocal microscopy | 10 |
| 11 | Exfoliative cytology | 9 |
| Staging of melanoma | ||
| 12 | Imaging tests (ultrasound, CT, MRI, PET‐CT) | 38 |
| 13 | Sentinel lymph node biopsy | 160 |
| Staging of cSCC | ||
| Imaging tests review | Review dropped; only one study identified | |
| 13 | Sentinel lymph node biopsy | Review amalgamated into 13 above (n = 15 studies) |
Appendix 2. Glossary of terms
| Term | Definition |
| Atypical intraepidermal melanocytic variant | Unusual area of darker pigmentation contained within the epidermis that may progress to an invasive melanoma; includes melanoma in situ and lentigo maligna |
| Atypical naevi | Unusual looking but non‐cancerous mole or area of darker pigmentation of the skin |
| BRAF V600 mutation | BRAF is a human gene that makes a protein called B‐Raf which is involved in the control of cell growth. BRAF mutations (damaged DNA) occur in around 40% of melanomas, which can then be treated with particular drugs. |
| BRAF inhibitors | Therapeutic agents that inhibit the serine‐threonine protein kinase BRAF mutated metastatic melanoma. |
| Breslow thickness | A scale for measuring the thickness of melanomas by the pathologist using a microscope, measured in mm from the top layer of skin to the bottom of the tumour |
| Congenital naevi | A type of mole found on infants at birth |
| Dermoscopy | Whereby a handheld microscope is used to allow more detailed, magnified, examination of the skin compared to examination by the naked eye alone |
| False negative | An individual who is truly positive for a disease, but whom a diagnostic test classifies them as disease‐free |
| False positive | An individual who is truly disease‐free, but whom a diagnostic test classifies them as having the disease |
| Histopathology/Hhistology | The study of tissue, usually obtained by biopsy or excision, for example under a microscope |
| Incidence | The number of new cases of a disease in a given time period |
| Index test | A diagnostic test under evaluation in a primary study |
| Lentigo maligna | Unusual area of darker pigmentation contained within the epidermis which includes malignant cells but with no invasive growth. May progress to an invasive melanoma |
| Lymph node | Lymph nodes filter the lymphatic fluid (clear fluid containing white blood cells) that travels around the body to help fight disease; they are located throughout the body often in clusters (nodal basins). |
| Melanocytic naevus | An area of skin with darker pigmentation (or melanocytes), also referred to as moles |
| Meta‐analysis | A form of statistical analysis used to synthesise results from a collection of individual studies |
| Metastases/metastatic disease | Spread of cancer away from the primary site to somewhere else through the bloodstream or the lymphatic system |
| Micrometastases | Micrometastases are metastases so small that they can only be seen under a microscope |
| Mitotic rate | Microscopic evaluation of number of cells actively dividing in a tumour |
| Morbidity | Detrimental effects on health |
| Mortality | Either the condition of being subject to death; or the death rate, which reflects the number of deaths per unit of population in relation to any specific region, age group, disease, treatment or other classification, usually expressed as deaths per 100, 1000, 10,000 or 100,000 people |
| Multidisciplinary team | A team with members from different healthcare professions and specialties (e.g. urology, oncology, pathology, radiology, and nursing). Cancer care in the National Health Service (NHS) uses this system to ensure that all relevant health professionals are engaged to discuss the best possible care for that patient. |
| Prevalence | The proportion of a population found to have a condition |
| Prognostic factors/indicators | Specific characteristics of a cancer or the person who has it which might affect the patient's prognosis |
| Receiver operating characteristic (ROC) plot | A plot of the sensitivity and 1 minus the specificity of a test at the different possible thresholds for test positivity; represents the diagnostic capability of a test with a range of binary test results |
| Receiver operating characteristic (ROC) analysis | The analysis of a ROC plot of a test to select an optimal threshold for test positivity |
| Recurrence | Recurrence is when new cancer cells are detected following treatment. This can occur either at the site of the original tumour or at other sites in the body. |
| Reference standard | A test or combination of tests used to establish the final or 'true' diagnosis of a patient in an evaluation of a diagnostic test |
| Reflectance confocal microscopy (RCM) | A microscopic technique using infrared light (either in a handheld device or a static unit) that can create images of the deeper layers of the skin |
| Sensitivity | In this context the term is used to mean the proportion of individuals with a disease who have that disease correctly identified by the study test |
| Specificity | The proportion of individuals without the disease of interest (in this case with benign skin lesions) who have that absence of disease correctly identified by the study test |
| Staging | Clinical description of the size and spread of a patient's tumour, fitting into internationally agreed categories |
| Subclinical (disease) | Disease that is usually asymptomatic and not easily observable, e.g. by clinical or physical examination |
| Systemic treatment | Treatment, usually given by mouth or by injection, that reaches and affects cancer cells throughout the body rather than targeting one specific area |
Appendix 3. Final search strategies
Melanoma search strategies to August 2016
Database: Ovid MEDLINE(R) 1946 to August week 3 2016
Search strategy:
1 exp melanoma/
2 exp skin cancer/
3 exp basal cell carcinoma/
4 basalioma$1.ti,ab.
5 ((basal cell or skin) adj2 (cancer$1 or carcinoma$1 or mass or masses or tumour$1 or tumor$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or lesion$1 or malignan$ or nodule$1)).ti,ab.
6 (pigmented adj2 (lesion$1 or mole$ or nevus or nevi or naevus or naevi or skin)).ti,ab.
7 (melanom$1 or nonmelanoma$1 or non‐melanoma$1 or melanocyt$ or non‐melanocyt$ or nonmelanocyt$ or keratinocyt$).ti,ab.
8 nmsc.ti,ab.
9 (squamous cell adj2 (cancer$1 or carcinoma$1 or mass or masses or tumor$1 or tumour$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or epithelial or lesion$1 or malignan$ or nodule$1) adj2 (skin or epiderm$ or cutaneous)).ti,ab.
10 (BCC or CSCC or NMSC).ti,ab.
11 keratinocy$.ti,ab.
12 Keratinocytes/
13 or/1‐12
14 dermoscop$.ti,ab.
15 dermatoscop$.ti,ab.
16 photomicrograph$.ti,ab.
17 exp epiluminescence microscopy/
18 (epiluminescence adj2 microscop$).ti,ab.
19 (confocal adj2 microscop$).ti,ab.
20 (incident light adj2 microscop$).ti,ab.
21 (surface adj2 microscop$).ti,ab.
22 (visual adj (inspect$ or examin$)).ti,ab.
23 ((clinical or physical) adj examin$).ti,ab.
24 3 point.ti,ab.
25 three point.ti,ab.
26 pattern analys$.ti,ab.
27 ABCD$.ti,ab.
28 menzies.ti,ab.
29 7 point.ti,ab.
30 seven point.ti,ab.
31 (digital adj2 (dermoscop$ or dermatoscop$)).ti,ab.
32 artificial intelligence.ti,ab.
33 AI.ti,ab.
34 computer assisted.ti,ab.
35 computer aided.ti,ab.
36 neural network$.ti,ab.
37 exp diagnosis, computer‐assisted/
38 MoleMax.ti,ab.
39 image process$.ti,ab.
40 automatic classif$.ti,ab.
41 image analysis.ti,ab.
42 SIAscop$.ti,ab.
43 Aura.ti,ab.
44 (optical adj2 scan$).ti,ab.
45 MelaFind.ti,ab.
46 SIMSYS.ti,ab.
47 MoleMate.ti,ab.
48 SolarScan.ti,ab.
49 VivaScope.ti,ab.
50 (high adj3 ultraso$).ti,ab.
51 (canine adj2 detect$).ti,ab.
52 ((mobile or cell or cellular or smart) adj ((phone$1 adj2 app$1) or application$1)).ti,ab.
53 smartphone$.ti,ab.
54 (DermoScan or SkinVision or DermLink or SpotCheck).ti,ab.
55 Mole Detective.ti,ab.
56 Spot Check.ti,ab.
57 (mole$1 adj2 map$).ti,ab.
58 (total adj2 body).ti,ab.
59 exfoliative cytolog$.ti,ab.
60 digital analys$.ti,ab.
61 (image$1 adj3 software).ti,ab.
62 (teledermatolog$ or tele‐dermatolog$ or telederm or tele‐derm or teledermoscop$ or tele‐dermoscop$ or teledermatoscop$ or tele‐dermatoscop$).ti,ab.
63 (optical coherence adj (technolog$ or tomog$)).ti,ab.
64 (computer adj2 diagnos$).ti,ab.
65 exp sentinel lymph node biopsy/
66 (sentinel adj2 node).ti,ab.
67 nevisense.mp. or HFUS.ti,ab.
68 electrical impedance spectroscopy.ti,ab.
69 history taking.ti,ab.
70 patient history.ti,ab.
71 (naked eye adj (exam$ or assess$)).ti,ab.
72 (skin adj exam$).ti,ab.
73 physical examination/
74 ugly duckling.mp. or UD.ti,ab.
75 ((physician$ or clinical or physical) adj (exam$ or triage or recog$)).ti,ab.
76 ABCDE.mp. or VOC.ti,ab.
77 clinical accuracy.ti,ab.
78 Family Practice/ or Physicians, Family/ or clinical competence/
79 (confocal adj2 microscop$).ti,ab.
80 diagnostic algorithm$1.ti,ab.
81 checklist$.ti,ab.
82 virtual imag$1.ti,ab.
83 volatile organic compound$1.ti,ab.
84 dog$1.ti,ab.
85 gene expression analy$.ti,ab.
86 reflex transmission imag$.ti,ab.
87 thermal imaging.ti,ab.
88 elastography.ti,ab.
89 or/14‐88
90 (CT or PET).ti,ab.
91 PET‐CT.ti,ab.
92 (FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical$).ti,ab.
93 exp Deoxyglucose/
94 deoxy‐glucose.ti,ab.
95 deoxyglucose.ti,ab.
96 CATSCAN.ti,ab.
97 exp Tomography, Emission‐Computed/
98 exp Tomography, X‐ray computed/
99 positron emission tomograph$.ti,ab.
100 exp magnetic resonance imaging/
101 (MRI or fMRI or NMRI or scintigraph$).ti,ab.
102 exp echography/
103 Doppler echography.ti,ab.
104 sonograph$.ti,ab.
105 ultraso$.ti,ab.
106 doppler.ti,ab.
107 magnetic resonance imag$.ti,ab.
108 or/90‐107
109 (stage$ or staging or metasta$ or recurrence or sensitivity or specificity or false negative$ or thickness$).ti,ab.
110 "Sensitivity and Specificity"/
111 exp cancer staging/
112 or/109‐111
113 108 and 112
114 89 or 113
115 13 and 114
Database: Ovid MEDLINE(R) In‐Process & Other Non‐Indexed Citations 29 August 2016
Search strategy:
1 basalioma$1.ti,ab.
2 ((basal cell or skin) adj2 (cancer$1 or carcinoma$1 or mass or masses or tumour$1 or tumor$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or lesion$1 or malignan$ or nodule$1)).ti,ab.
3 (pigmented adj2 (lesion$1 or mole$ or nevus or nevi or naevus or naevi or skin)).ti,ab.
4 (melanom$1 or nonmelanoma$1 or non‐melanoma$1 or melanocyt$ or non‐melanocyt$ or nonmelanocyt$ or keratinocyt$).ti,ab.
5 nmsc.ti,ab.
6 (squamous cell adj2 (cancer$1 or carcinoma$1 or mass or masses or tumor$1 or tumour$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or epithelial or lesion$1 or malignan$ or nodule$1) adj2 (skin or epiderm$ or cutaneous)).ti,ab.
7 (BCC or CSCC or NMSC).ti,ab.
8 keratinocy$.ti,ab.
9 or/1‐8
10 dermoscop$.ti,ab.
11 dermatoscop$.ti,ab.
12 photomicrograph$.ti,ab.
13 (epiluminescence adj2 microscop$).ti,ab.
14 (confocal adj2 microscop$).ti,ab.
15 (incident light adj2 microscop$).ti,ab.
16 (surface adj2 microscop$).ti,ab.
17 (visual adj (inspect$ or examin$)).ti,ab.
18 ((clinical or physical) adj examin$).ti,ab.
19 3 point.ti,ab.
20 three point.ti,ab.
21 pattern analys$.ti,ab.
22 ABCD$.ti,ab.
23 menzies.ti,ab.
24 7 point.ti,ab.
25 seven point.ti,ab.
26 (digital adj2 (dermoscop$ or dermatoscop$)).ti,ab.
27 artificial intelligence.ti,ab.
28 AI.ti,ab.
29 computer assisted.ti,ab.
30 computer aided.ti,ab.
31 neural network$.ti,ab.
32 MoleMax.ti,ab.
33 image process$.ti,ab.
34 automatic classif$.ti,ab.
35 image analysis.ti,ab.
36 SIAscop$.ti,ab.
37 Aura.ti,ab.
38 (optical adj2 scan$).ti,ab.
39 MelaFind.ti,ab.
40 SIMSYS.ti,ab.
41 MoleMate.ti,ab.
42 SolarScan.ti,ab.
43 VivaScope.ti,ab.
44 (high adj3 ultraso$).ti,ab.
45 (canine adj2 detect$).ti,ab.
46 ((mobile or cell or cellular or smart) adj ((phone$1 adj2 app$1) or application$1)).ti,ab.
47 smartphone$.ti,ab.
48 (DermoScan or SkinVision or DermLink or SpotCheck).ti,ab.
49 Mole Detective.ti,ab.
50 Spot Check.ti,ab.
51 (mole$1 adj2 map$).ti,ab.
52 (total adj2 body).ti,ab.
53 exfoliative cytolog$.ti,ab.
54 digital analys$.ti,ab.
55 (image$1 adj3 software).ti,ab.
56 (teledermatolog$ or tele‐dermatolog$ or telederm or tele‐derm or teledermoscop$ or tele‐dermoscop$ or teledermatoscop$ or tele‐dermatoscop$).ti,ab.
57 (optical coherence adj (technolog$ or tomog$)).ti,ab.
58 (computer adj2 diagnos$).ti,ab.
59 (sentinel adj2 node).ti,ab.
60 nevisense.mp. or HFUS.ti,ab.
61 electrical impedance spectroscopy.ti,ab.
62 history taking.ti,ab.
63 patient history.ti,ab.
64 (naked eye adj (exam$ or assess$)).ti,ab.
65 (skin adj exam$).ti,ab.
66 ugly duckling.mp. or UD.ti,ab.
67 ((physician$ or clinical or physical) adj (exam$ or triage or recog$)).ti,ab.
68 ABCDE.mp. or VOC.ti,ab.
69 clinical accuracy.ti,ab.
70 (Family adj (Practice or Physicians)).ti,ab.
71 (confocal adj2 microscop$).ti,ab.
72 clinical competence.ti,ab.
73 diagnostic algorithm$1.ti,ab.
74 checklist$.ti,ab.
75 virtual imag$1.ti,ab.
76 volatile organic compound$1.ti,ab.
77 dog$1.ti,ab.
78 gene expression analy$.ti,ab.
79 reflex transmission imag$.ti,ab.
80 thermal imaging.ti,ab.
81 elastography.ti,ab.
82 or/10‐81
83 (CT or PET).ti,ab.
84 PET‐CT.ti,ab.
85 (FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical$).ti,ab.
86 deoxy‐glucose.ti,ab.
87 deoxyglucose.ti,ab.
88 CATSCAN.ti,ab.
89 positron emission tomograph$.ti,ab.
90 (MRI or fMRI or NMRI or scintigraph$).ti,ab.
91 Doppler echography.ti,ab.
92 sonograph$.ti,ab.
93 ultraso$.ti,ab.
94 doppler.ti,ab.
95 magnetic resonance imag$.ti,ab.
96 or/83‐95
97 (stage$ or staging or metasta$ or recurrence or sensitivity or specificity or false negative$ or thickness$).ti,ab.
98 96 and 97
99 82 or 98
100 9 and 99
Database: Embase 1974 to 29 August 2016
Search strategy:
1 *melanoma/
2 *skin cancer/
3 *basal cell carcinoma/
4 basalioma$.ti,ab.
5 ((basal cell or skin) adj2 (cancer$1 or carcinoma$1 or mass or masses or tumour$1 or tumor$1 or neoplasm$ or adenoma$ or epithelioma$ or lesion$ or malignan$ or nodule$)).ti,ab.
6 (pigmented adj2 (lesion$1 or mole$ or nevus or nevi or naevus or naevi or skin)).ti,ab.
7 (melanom$1 or nonmelanoma$1 or non‐melanoma$1 or melanocyt$ or non‐melanocyt$ or nonmelanocyt$ or keratinocyt$).ti,ab.
8 nmsc.ti,ab.
9 (squamous cell adj2 (cancer$1 or carcinoma$1 or mass or tumor$1 or tumour$1 or neoplasm$1 or adenoma$1 or epithelioma$1 or epithelial or lesion$1 or malignan$ or nodule$1) adj2 (skin or epiderm$ or cutaneous)).ti,ab.
10 (BCC or cscc).mp. or NMSC.ti,ab.
11 keratinocyte.ti,ab.
12 keratinocy$.ti,ab.
13 or/1‐12
14 dermoscop$.ti,ab.
15 dermatoscop$.ti,ab.
16 photomicrograph$.ti,ab.
17 *epiluminescence microscopy/
18 (epiluminescence adj2 microscop$).ti,ab.
19 (confocal adj2 microscop$).ti,ab.
20 (incident light adj2 microscop$).ti,ab.
21 (surface adj2 microscop$).ti,ab.
22 (visual adj (inspect$ or examin$)).ti,ab.
23 ((clinical or physical) adj examin$).ti,ab.
24 3 point.ti,ab.
25 three point.ti,ab.
26 pattern analys$.ti,ab.
27 ABCD$.ti,ab.
28 menzies.ti,ab.
29 7 point.ti,ab.
30 seven point.ti,ab.
31 (digital adj2 (dermoscop$ or dermatoscop$)).ti,ab.
32 artificial intelligence.ti,ab.
33 AI.ti,ab.
34 computer assisted.ti,ab.
35 computer aided.ti,ab.
36 neural network$.ti,ab.
37 MoleMax.ti,ab.
38 exp diagnosis, computer‐assisted/
39 image process$.ti,ab.
40 automatic classif$.ti,ab.
41 image analysis.ti,ab.
42 SIAscop$.ti,ab.
43 (optical adj2 scan$).ti,ab.
44 Aura.ti,ab.
45 MelaFind.ti,ab.
46 SIMSYS.ti,ab.
47 MoleMate.ti,ab.
48 SolarScan.ti,ab.
49 VivaScope.ti,ab.
50 confocal microscop$.ti,ab.
51 (high adj3 ultraso$).ti,ab.
52 (canine adj2 detect$).ti,ab.
53 ((mobile or cell$ or cellular or smart) adj ((phone$1 adj2 app$1) or application$1)).ti,ab.
54 smartphone$.ti,ab.
55 (DermoScan or SkinVision or DermLink or SpotCheck).ti,ab.
56 Spot Check.ti,ab.
57 Mole Detective.ti,ab.
58 (mole$1 adj2 map$).ti,ab.
59 (total adj2 body).ti,ab.
60 exfoliative cytolog$.ti,ab.
61 digital analys$.ti,ab.
62 (image$1 adj3 software).ti,ab.
63 (optical coherence adj (technolog$ or tomog$)).ti,ab.
64 (teledermatolog$ or tele‐dermatolog$ or telederm or tele‐derm or teledermoscop$ or tele‐dermoscop$ or teledermatoscop$).mp. or tele‐dermatoscop$.ti,ab.
65 (computer adj2 diagnos$).ti,ab.
66 *sentinel lymph node biopsy/
67 (sentinel adj2 node).ti,ab.
68 nevisense.ti,ab.
69 HFUS.ti,ab.
70 electrical impedance spectroscopy.ti,ab.
71 history taking.ti,ab.
72 patient history.ti,ab.
73 (naked eye adj (exam$ or assess$)).ti,ab.
74 (skin adj exam$).ti,ab.
75 *physical examination/
76 ugly duckling.ti,ab.
77 UD sign$.ti,ab.
78 ((physician$ or clinical or physical) adj (exam$ or recog$ or triage)).ti,ab.
79 ABCDE.ti,ab.
80 clinical accuracy.ti,ab.
81 *general practice/
82 (confocal adj2 microscop$).ti,ab.
83 clinical competence/
84 diagnostic algorithm$.ti,ab.
85 checklist$1.ti,ab.
86 virtual image$1.ti,ab.
87 volatile organic compound$1.ti,ab.
88 VOC.ti,ab.
89 dog$1.ti,ab.
90 gene expression analys$.ti,ab.
91 reflex transmission imaging.ti,ab.
92 thermal imaging.ti,ab.
93 elastography.ti,ab.
94 dog$1.ti,ab.
95 gene expression analys$.ti,ab.
96 reflex transmission imaging.ti,ab.
97 thermal imaging.ti,ab.
98 elastography.ti,ab.
99 or/14‐93
100 PET‐CT.ti,ab.
101 (CT or PET).ti,ab.
102 (FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical$).ti,ab.
103 exp Deoxyglucose/
104 CATSCAN.ti,ab.
105 deoxyglucose.ti,ab.
106 deoxy‐glucose.ti,ab.
107 *positron emission tomography/
108 *computer assisted tomography/
109 positron emission tomograph$.ti,ab.
110 *nuclear magnetic resonance imaging/
111 (MRI or fMRI or NMRI or scintigraph$).ti,ab.
112 *echography/
113 Doppler.ti,ab.
114 sonograph$.ti,ab.
115 ultraso$.ti,ab.
116 magnetic resonance imag$.ti,ab.
117 or/100‐116
118 (stage$ or staging or metasta$ or recurrence or sensitivity or specificity or false negative$ or thickness$).ti,ab.
119 "Sensitivity and Specificity"/
120 *cancer staging/
121 or/118‐120
122 117 and 121
123 99 or 122
124 13 and 123
Database: Cochrane Library (Wiley) 2016 searched 30 August 2016 CDSR Issue 8 of 12 2016 CENTRAL Issue 7 of 12 2016 HTA Issue 3 of 4 July 2016 DARE Issue 3 of 4 2015
Search strategy:
#1 melanoma* or nonmelanoma* or non‐melanoma* or melanocyt* or non‐melanocyt* or nonmelanocyt* or keratinocyte*
#2 MeSH descriptor: [Melanoma] explode all trees
#3 "skin cancer*"
#4 MeSH descriptor: [Skin Neoplasms] explode all trees
#5 skin near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*)
#6 nmsc
#7 "squamous cell" near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*) near/2 (skin or epiderm* or cutaneous)
#8 "basal cell" near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*)
#9 pigmented near/2 (lesion* or nevus or mole* or naevi or naevus or nevi or skin)
#10 #1 or #2 or #3 or #4 or #5 or #6 or #7 or #8 or #9
#11 dermoscop*
#12 dermatoscop*
#13 Photomicrograph*
#14 MeSH descriptor: [Dermoscopy] explode all trees
#15 confocal near/2 microscop*
#16 epiluminescence near/2 microscop*
#17 incident next light near/2 microscop*
#18 surface near/2 microscop*
#19 "visual inspect*"
#20 "visual exam*"
#21 (clinical or physical) next (exam*)
#22 "3 point"
#23 "three point"
#24 "pattern analys*"
#25 ABDC
#26 menzies
#27 "7 point"
#28 "seven point"
#29 digital near/2 (dermoscop* or dermatoscop*)
#30 "artificial intelligence"
#31 "AI"
#32 "computer assisted"
#33 "computer aided"
#34 AI
#35 "neural network*"
#36 MoleMax
#37 "computer diagnosis"
#38 "image process*"
#39 "automatic classif*"
#40 SIAscope
#41 "image analysis"
#42 "optical near/2 scan*"
#43 Aura
#44 MelaFind
#45 SIMSYS
#46 MoleMate
#47 SolarScan
#48 Vivascope
#49 "confocal microscopy"
#50 high near/3 ultraso*
#51 canine near/2 detect*
#52 Mole* near/2 map*
#53 total near/2 body
#54 mobile* or smart near/2 phone*
#55 cell next phone*
#56 smartphone*
#57 "mitotic index"
#58 DermoScan or SkinVision or DermLink or SpotCheck
#59 "Mole Detective"
#60 "Spot Check"
#61 mole* near/2 map*
#62 total near/2 body
#63 "exfoliative cytolog*"
#64 "digital analys*"
#65 image near/3 software
#66 teledermatolog* or tele‐dermatolog* or telederm or tele‐derm or teledermoscop* or tele‐dermoscop* or teledermatoscop* or tele‐dermatolog*
#67 "optical coherence" next (technolog* or tomog*)
#68 computer near/2 diagnos*
#69 sentinel near/2 node*
#70 #11 or #12 or #13 or #14 or #15 or #16 or #17 or #18 or #19 or #20 or #21 or #22 or #23 or #24 or #25 or #26 or #27 or #28 or #29 or #30 or #31 or #32 or #33 or #34 or #35 or #36 or #37 or #38 or #39 or #40 or #41 or #42 or #43 or #44 or #45 or #46 or #47 or #48 or #49 or #50 or #51 or #52 or #53 or #54 or #55 or #56 or #57 or #58 or #59 or #60 or #61 or #62 or #63 or #64 or #65 or #66 or #67 or #68 or #69
#71 ultraso*
#72 sonograph*
#73 MeSH descriptor: [Ultrasonography] explode all trees
#74 Doppler
#75 CT or PET or PET‐CT
#76 "CAT SCAN" or "CATSCAN"
#77 MeSH descriptor: [Positron‐Emission Tomography] explode all trees
#78 MeSH descriptor: [Tomography, X‐Ray Computed] explode all trees
#79 MRI
#80 MeSH descriptor: [Magnetic Resonance Imaging] explode all trees
#81 MRI or fMRI or NMRI or scintigraph*
#82 "magnetic resonance imag*"
#83 MeSH descriptor: [Deoxyglucose] explode all trees
#84 deoxyglucose or deoxy‐glucose
#85 "positron emission tomograph*"
#86 #71 or #72 or #73 or #74 or #75 or #76 or #77 or #78 or #79 or #80 or #81 or #82 or #83 or #84 or #85
#87 stage* or staging or metasta* or recurrence or sensitivity or specificity or "false negative*" or thickness*
#88 MeSH descriptor: [Neoplasm Staging] explode all trees
#89 #87 or #88
#90 #89 and #86
#91 #70 or #90
#92 #10 and #91
#93 BCC or CSCC or NMCS
#94 keratinocy*
#95 #93 or #94
#96 #10 or #95
#97 nevisense
#98 HFUS
#99 "electrical impedance spectroscopy"
#100 "history taking"
#101 "patient history"
#102 naked next eye near/1 (exam* or assess*)
#103 skin next exam*
#104 "ugly duckling" or (UD sign*)
#105 MeSH descriptor: [Physical Examination] explode all trees
#106 (physician* or clinical or physical) near/1 (exam* or recog* or triage*)
#107 ABCDE
#108 "clinical accuracy"
#109 MeSH descriptor: [General Practice] explode all trees
#110 confocal near microscop*
#111 "diagnostic algorithm*"
#112 MeSH descriptor: [Clinical Competence] explode all trees
#113 checklist*
#114 "virtual image*"
#115 "volatile organic compound*"
#116 dog or dogs
#117 VOC
#118 "gene expression analys*"
#119 "reflex transmission imaging"
#120 "thermal imaging"
#121 elastography
#122 #97 or #98 or #99 or #100 or #101 or #102 or #103 or #104 or #105 or #106 or #107 or #108 or #109 or #110 or #111 or #112 or #113 or #114 or #115 or #116 or #117 or #118 or #119 or #120 or #121
#123 #70 or #122
#124 #96 and #123
#125 #96 and #90
#126 #125 or #124
#127 #10 and #126
Database: CINAHL Plus (EBSCO) 1937 to 30 August 2016
Search strategy:
S1 (MH "Melanoma") OR (MH "Nevi and Melanomas+")
S2 (MH "Skin Neoplasms+")
S3 (MH "Carcinoma, Basal Cell+")
S4 basalioma*
S5 (basal cell) N2 (cancer* or carcinoma* or mass or masses or tumor* or tumour* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*)
S6 (pigmented) N2 (lesion* or mole* or nevus or nevi or naevus or naevi or skin)
S7 melanom* or nonmelanoma* or non‐melanoma* or melanocyt* or non‐melanocyt* or nonmelanocyt*
S8 nmsc
S9 TX BCC or cscc or NMSC
S10 (MH "Keratinocytes")
S11 keratinocyt*
S12 S1 OR S2 OR S3 OR S4 OR S5 OR S6 OR S7 OR S8 OR S9 OR S10 OR S11
S13 dermoscop* or dermatoscop* or photomicrograph* or (3 point) or (three point) or ABCD* or menzies or (7 point) or (seven point) or AI or Molemax or SIASCOP* or Aura or MelaFind or SIMSYS or MoleMate or SolarScan or smartphone* or DermoScan or SkinVision or DermLink or SpotCheck
S14 (epiluminescence or confocal or incident or surface) N2 (microscop*)
S15 visual N1 (inspect* or examin*)
S16 (clinical or physical) N1 (examin*)
S17 pattern analys*
S18 (digital) N2 (dermoscop* or dermatoscop*)
S19 (artificial intelligence)
S20 (computer) N2 (assisted or aided)
S21 (neural network*)
S22 (MH "Diagnosis, Computer Assisted+")
S23 (image process*)
S24 (automatic classif*)
S25 (image analysis)
S26 SIAScop*
S27 (optical) N2 (scan*)
S28 (high) N3 (ultraso*)
S29 elastography
S30 (mobile or cell or cellular or smart) N2 (phone*) N2 (app or application*)
S31 (mole*) N2 (map*)
S32 total N2 body
S33 exfoliative cytolog*
S34 digital analys*
S35 image N3 software
S36 teledermatolog* or tele‐dermatolog* or telederm or tele‐derm or teledermoscop* or tele‐dermoscop* or teledermatoscop* or tele‐dermatoscop* teledermatolog* or tele‐dermatolog* or telederm or tele‐derm or teledermoscop*
S37 (optical coherence) N1 (technolog* or tomog*)
S38 computer N2 diagnos*
S39 sentinel N2 node
S40 (MH "Sentinel Lymph Node Biopsy")
S41 nevisense or HFUS or checklist* or VOC or dog*
S42 electrical impedance spectroscopy
S43 history taking
S44 "Patient history"
S45 naked eye
S46 skin exam*
S47 physical exam*
S48 ugly duckling
S49 UD sign*
S50 (physician* or clinical or physical) N1 (exam*)
S51 clinical accuracy
S52 general practice
S53 (physician* or clinical or physical) N1 (recog* or triage)
S54 confocal microscop*
S55 clinical competence
S56 diagnostic algorithm*
S57 checklist*
S58 virtual image*
S59 volatile organic compound*
S60 gene expression analys*
S61 reflex transmission imag*
S62 thermal imaging
S63 S13 or S14 or S15 OR S16 OR S17 OR S18 OR S19 OR S20 OR S21 OR S22 OR S23 OR S24 OR S25 OR S26 OR S27 OR S28 OR S29 OR S30 OR S31 OR S32 OR S33 OR S34 OR S35 OR S36 OR S37 OR S38 OR S39 OR S40 OR S41 OR S42 OR S43 OR S44 OR S45 OR S46 OR S47 OR S48 OR S49 OR S50 OR S51 OR S52 OR S53 OR S54 OR S55 OR S56 OR S57 OR S58 OR S59 OR S60 OR S61 OR S62
S64 CT or PET
S65 PET‐CT
S66 FDG or F18 or Fluorodeoxyglucose or radiopharmaceutical*
S67 (MH "Deoxyglucose+")
S68 deoxy‐glucose or deoxyglucose
S69 CATSCAN
S70 CAT‐SCAN
S71 (MH "Deoxyglucose+")
S72 (MH "Tomography, Emission‐Computed+")
S73 (MH "Tomography, X‐Ray Computed")
S74 positron emission tomograph*
S75 (MH "Magnetic Resonance Imaging+")
S76 MRI or fMRI or NMRI or scintigraph*
S77 echography
S78 doppler
S79 sonograph*
S80 ultraso*
S81 magnetic resonance imag*
S82 S64 OR S65 OR S66 OR S67 OR S68 OR S69 OR S70 OR S71 OR S72 OR S73 OR S74 OR S75 OR S76 OR S77 OR S78 OR S79 OR S80 OR S81
S83 stage* or staging or metasta* or recurrence or sensitivity or specificity or (false negative*) or thickness
S84 (MH "Neoplasm Staging")
S85 S83 OR S84
S86 S82 AND S85
S87 S63 OR S86
S88 S12 AND S87
Database: Science Citation Index SCI Expanded (Web of Science) 1900 to 30 August 2016
Conference Proceedings Citation Index (Web of Science) 1900 to 1 September 2016
Search strategy:
#1 (melanom* or nonmelanom* or non‐melanoma* or melanocyt* or non‐melanocyt* or nonmelanocyt* or keratinocyt*)
#2 (basalioma*)
#3 ((skin) near/2 (cancer* or carcinoma or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*))
#4 ((basal) near/2 (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*))
#5 ((pigmented) near/2 (lesion* or mole* or nevus or nevi or naevus or naevi or skin))
#6 (nmsc or BCC or NMSC or keratinocy*)
#7 ((squamous cell (cancer* or carcinoma* or mass or masses or tumour* or tumor* or neoplasm* or adenoma* or epithelioma* or lesion* or malignan* or nodule*))
#8 (skin or epiderm* or cutaneous)
#9 #8 AND #7
#10 #9 OR #6 OR #5 OR #4 OR #3 OR #2 OR #1
#11 ((dermoscop* or dermatoscop* or photomicrograph* or epiluminescence or confocal or "incident light" or "surface microscop*" or "visual inspect*" or "physical exam*" or 3 point or three point or pattern analy* or ABCDE or menzies or 7 point or seven point or dermoscop* or dermatoscop* or AI or artificial or computer aided or computer assisted or neural network* or Molemax or image process* or automatic classif* or image analysis or siascope or optical scan* or Aura or melafind or simsys or molemate or solarscan or vivascope or confocal microscop* or high ultraso* or canine detect* or cellphone* or mobile* or phone* or smartphone or dermoscan or skinvision or dermlink or spotcheck or spot check or mole detective or mole map* or total body or exfoliative psychology or digital or image software or optical coherence or teledermatology or telederm* or teledermoscop* or teledermatoscop* or computer diagnos* or sentinel))
#12 ((nevisense or HFUS or impedance spectroscopy or history taking or patient history or naked eye or skin exam* or physical exam* or ugly duckling or UD sign* or physician* exam* or physical exam* or ABCDE or clinical accuracy or general practice or confocal microscop* or clinical competence or diagnostic algorithm* or checklist* or virtual image* or volatile organic or VOC or dog* or gene expression or reflex transmission or thermal imag* or elastography))
#13 #11 or #12
#14 ((PET or CT or FDG or deoxyglucose or deoxy‐glucose or fluorodeoxy* or radiopharma* or CATSCAN or positron emission or computer assisted or nuclear magnetic or MRI or FMRI or NMRI or scintigraph* or echograph* or Doppler or sonograph* or ultraso* or magnetic reson*))
#15 ((stage* or staging or metast* or recurrence or sensitivity or specificity or false negative* or thickness*))
#16 #14 AND #15
#17 #16 OR #13
#18 #10 AND #17
Refined by: DOCUMENT TYPES: (MEETING ABSTRACT OR PROCEEDINGS PAPER)
Appendix 4. Full text inclusion criteria
| Criterion | Inclusion | Exclusion |
| Study design |
For diagnostic and staging reviews
|
|
| Target condition |
|
|
| Population |
For diagnostic reviews
For staging reviews
|
|
| Index tests |
For diagnosis
For staging
Any test combination and in any order Any test positivity threshold Any variation in testing procedure (e.g. radioisotope used) |
|
| Reference standard |
For diagnostic studies
For studies of imaging tests for staging
For studies of SLNB accuracy for staging
|
For diagnostic studies
|
| BCC: basal cell carcinoma; cSCC: cutaneous squamous cell carcinoma; CT: computed tomography; FNAC: fine needle aspiration cytology; LND: lymph node dissection; MRI: magnetic resonance imaging; PET: positron emission tomography; PET‐CT: positron emission tomography computed tomography; RCT: randomised controlled trial; SCC: squamous cell carcinoma; SLN+: positive sentinel lymph node; SLn: negative sentinel lymph node; SLNB: sentinel lymph node biopsy. | ||
Appendix 5. Quality assessment (based on QUADAS‐2)
The QUADAS‐2 checklist (Whiting 2011) was tailored to the review topic as follows below.
Participant selection domain (1)
Selective recruitment of study participants can be a key influence on test accuracy. In general terms, all participants eligible to undergo a test should be included in a study, allowing for the intended use of that test within the context of the study. We considered studies that separately sampled malignant and benign lesions to have used a case‐control design; and those that supplemented a series of suspicious lesions with additional malignant or benign lesions to be at unclear risk of bias.
In terms of exclusions, we considered studies that excluded particular lesion types (e.g. lentigo maligna), particular lesion sites, or other lesions on the basis of image quality or lack of observer agreement (e.g. on histopathology) to be at high risk of bias.
In judging the applicability of patient populations to the review question, we considered restriction to particular lesion populations, such as melanocytic, nodular, high risk or restrictions by size to be of high concern for applicability.
Given that diagnosis of skin cancer is primarily lesion‐based, there is the potential for study participants with multiple lesions to contribute disproportionately to estimates of test accuracy, especially if they are at particular risk of having skin cancer. We considered studies that include a high number of lesions in relation to the number of participants in the study to be less representative than studies conducted in a more general population of participants (i.e. if the difference between the number of included lesions and number of included participants is greater than 5%).
Index test domain (2)
Given the potential for subjective differences in test interpretation for melanoma, the interpretation of the index test blinded to the result of the reference standard is a key means of reducing bias. For prospective studies and retrospective studies that used the original index test interpretation, the diagnosis will by nature be interpreted and recorded before the result of the reference standard is known; however, studies using previously acquired images could be particularly susceptible to information bias. For these studies to be at low risk of bias, we required a clear indication that observers were unaware of the reference standard diagnosis at time of test interpretation. We also added an item to assess the presence of blinding between interpretations of different algorithms; however, we did not include this item in the overall assessment of risk of bias.
Pre‐specification of the index test threshold was considered present if the study clearly reported that the threshold used was not data driven, i.e. was not based on study results. We considered studies that did not clearly describe the threshold used but that required clinicians to record a diagnosis or management decision for a lesion to be unclear on this criterion. We considered studies reporting accuracy for multiple numeric thresholds, where ROC analysis was used to select the threshold, or that reported accuracy for the presence of independently significant lesion characteristics with no separate test set of lesions, to be at high risk of bias.
In terms of applicability of the index test to the review question, we required the test to be applied and interpreted as it would be in real life setting, i.e. tests used and interpreted by the intended users: the general public.
Despite the often subjective nature of test interpretation, it is also important for study authors to outline the particular lesion characteristics that were considered to be indicative for melanoma, particularly where established algorithms or checklists were not used. Studies were considered of low concern if they used a threshold established in a prior study or presented sufficient threshold detailsto allow replication.
Reference standard domain (3)
In an ideal study, consecutively recruited participants should all undergo incisional or excisional biopsy of the skin lesion regardless of level of clinical suspicion of melanoma. In reality, both partial and differential verification bias are likely. Partial verification bias may occur where histology is the only reference standard used, and only those participants with a certain degree of suspicion of malignancy based on the result of the index test undergo verification, the others either being excluded from the study or defined as being disease‐negative without further assessment or follow‐up, as discussed above.
Differential verification bias will be present where other reference standards are used in addition to histological verification of suspicious lesions. A typical example of verification bias in skin cancer occurs when investigators do not biopsy people with benign‐appearing lesions but instead follow them up for a period of time to determine whether any malignancy subsequently develops (these would be false‐negatives on the index test). We defined an 'adequate' reference standard as: all disease‐positive individuals having a histological reference standard either at the time of application of the index test or after a period of clinical follow‐up; and at least 80% of disease‐negative participants have received a histological diagnosis, with up to 20% undergoing at least three months' follow‐up of benign‐appearing lesions.
A further challenge is the potential for incorporation bias, i.e. where the result of the index test is used to help determine the reference standard diagnosis. It is normal practice for the clinical diagnosis (usually by visual inspection or dermoscopy) to be included on pathology request forms and for the histopathologist to use this diagnosis to help with the pathology interpretation. Although inclusion of such clinical information on the histopathology request form is theoretically a form of incorporation bias, blinded interpretation of the histopathology reference standard is not normal practice, and enforcement of such conditions would significantly limit the generalisability of the study results. For studies evaluating reflectance confocal microscopy (RCM), we divided this item into two questions, firstly whether the reference standard was blinded to the index test result (RCM), and secondly whether it was blinded to the clinical diagnosis. We included only the response to the first part (i.e. blinding to RCM) in our overall assessment of risk of bias for the reference standard domain.
In judging the applicability of the reference standard to our review question, we scored studies as causing high concern around applicability if they used expert diagnosis (with no follow‐up) as a reference standard in any patient, or did not report histology interpretation by a dermatopathologist.
Flow and timing domain (4)
In the ideal study, the diagnosis based on the index test and reference standard should be made consecutively or as near to each other in time as possible to avoid changes in lesion over time. For lesions with a histological reference standard, we have defined a one‐month period as an appropriate interval between application of the index test and the reference standard. For studies using clinical follow‐up, we defined a minimum three‐month follow‐up period as at low risk of bias for detecting false‐negatives. This interval was chosen based on a study showing that most false‐negative melanomas will be diagnosed within three months of the initial negative index test, although a small number will be diagnosed up to 12 months subsequently (Altamura 2008).
In assessing whether all patients were included in the analysis, we considered studies at high risk of bias if participants were excluded following recruitment.
The following tables use text that was originally published in the QUADAS‐2 tool by Whiting and colleagues (Whiting 2011).
| Item | Response (delete as required) |
| PARTICIPANT SELECTION (1) ‐ RISK OF BIAS | |
| 1) Was a consecutive or random sample of participants or images enrolled? |
Yes – if paper states consecutive or random No – if paper describes other method of sampling Unclear – if participant sampling not described |
| 2) Was a case‐control design avoided? |
Yes – if consecutive or random or case‐control design clearly not used No – if study described as case‐control or describes sampling specific numbers of participants with particular diagnoses Unclear – if not described |
3) Did the study avoid inappropriate exclusions, e.g.
|
Yes ‐ if inappropriate exclusions were avoided No – if lesions were excluded that might affect test accuracy, e.g. 'difficult to diagnose' lesions, or where disagreement between evaluators was observed Unclear – if not clearly reported but there is suspicion that difficult to diagnose lesions may have been excluded |
4) For between‐person comparative studies only (i.e. allocating different tests to different study participants):
|
For A)
For B)
For C)
|
| Could the selection of participants have introduced bias? For non‐comparative and within‐person‐comparative studies
For between‐person comparative studies
|
For non‐comparative and within‐person‐comparative studies
For between‐person comparative studies
|
| PARTICIPANT SELECTION (1) ‐ CONCERNS REGARDING APPLICABILITY | |
1) Are the included participants and chosen study setting appropriate to answer the review question, i.e. are the study results generalisable?
|
A) For studies that will contribute to the analysis of participants with a primary presentation of a skin lesion (i.e. test naive) Yes – if participants included in the study appear to be generally representative of those who might present in a usual practice setting No – if study participants appear to be unrepresentative of usual practice, e.g. in terms of severity of disease, demographic features, presence of differential diagnosis or comorbidity, setting of the study, and previous testing protocols Unclear – if insufficient details are provided to determine the generalisability of study participants B) For studies that will contribute to the analysis of referred participants (i.e. who have already undergone some form of testing) Yes – if study participants appear to be representative of those who might be referred for further investigation. If the study focuses only on those with equivocal lesions, for example, we would suggest that this is not representative of the wider referred population No – if study participants appear to be unrepresentative of usual practice, e.g. if a particularly high proportion of participants have been self‐referred or referred for cosmetic reasons. Other factors to consider include severity of disease, demographic features, presence of differential diagnosis or comorbidity, setting of the study, and previous testing protocols Unclear – if insufficient details are provided to determine the generalisability of study participants |
| 2) Did the study avoid including participants with multiple lesions? |
Yes – if the difference between the number of included lesions and number of included participants is less than 5% No – if the difference between the number of included lesions and number of included participants is greater than 5% Unclear – if it is not possible to assess |
Is there concern that the included participants do not match the review question?
|
|
| INDEX TEST (2) ‐ RISK OF BIAS (to be completed per test evaluated) | |
| 1) Was the index test or testing strategy result interpreted without knowledge of the results of the reference standard? |
Yes – if index test described as interpreted without knowledge of reference standard result or, for prospective studies, if index test is always conducted and interpreted prior to the reference standard No – if index test described as interpreted in knowledge of reference standard result Unclear – if index test blinding is not described |
| 2) Was the diagnostic threshold at which the test was considered positive (i.e. melanoma present) prespecified? |
Yes – if threshold was prespecified (i.e. prior to analysing study results) No – if threshold was not prespecified Unclear – if not possible to tell whether or not diagnostic threshold was prespecified |
| 3) For within‐person comparisons of index tests or testing strategies (i.e. > 1 index test applied per participant): was each index test result interpreted without knowledge of the results of other index tests or testing strategies? |
Yes – if all index tests were described as interpreted without knowledge of the results of the others No – if the index tests were described as interpreted in the knowledge of the results of the others Unclear – if it is not possible to tell whether knowledge of other index tests could have influenced test interpretation NA – if only 1 index test was evaluated |
| Could the conduct or interpretation of the index test have introduced bias? For non‐comparative and between‐person comparison studies
For within‐person comparative studies
|
For non‐comparative and between‐person comparison studies
For within‐person comparative studies
|
| INDEX TEST (2) ‐ CONCERN ABOUT APPLICABILITY | |
| 1) Was the diagnostic threshold to determine presence or absence of disease established in a previously published study? e.g. previously evaluated/established
|
Yes – if a previously evaluated/established tool to aid diagnosis of melanoma was used or if the diagnostic threshold used was established in a previously published study No – if an unfamiliar/new tool to aid diagnosis of melanoma was used, if no particular algorithm was used, or if the objective threshold reported was chosen based on results in the current study Unclear – if insufficient information was reported |
| 2) Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? Study results can only be reproduced if the diagnostic threshold is described in sufficient detail. This item applies equally to studies using pattern recognition and those using checklists or algorithms to aid test interpretation |
Yes – if the criteria for diagnosis of melanoma were reported in sufficient detail to allow replication No – if the criteria for diagnosis of melanoma were not reported in sufficient detail to allow replication Unclear – if some but not sufficient information on criteria for diagnosis to allow replication were provided |
| 3) Was the test interpretation carried out by an experienced examiner? |
Yes – if the test was interpreted by 1 or more speciality‐accredited dermatologists, or by examiners of any clinical background with special interest in dermatology and with any formal training in the use of the test No – if the test was not interpreted by an experienced examiner (see above) Unclear – if the experience of the examiner(s) was not reported in sufficient detail to judge or if examiners were described as 'Expert' with no further detail given NA – if artificial intelligence‐based diagnosis, i.e. no observer interpretation |
Is there concern that the index test, its conduct, or interpretation differ from the review question?
|
|
| REFERENCE STANDARD (3) ‐ RISK OF BIAS | |
| 1) Is the reference standard likely to correctly classify the target condition? A) Disease‐positive – 1 or more of the following:
B) Disease‐negative – 1 or more of the following:
|
A) Disease‐positive Yes – if all participants with a final diagnosis of melanoma underwent 1 of the listed reference standards No – If a final diagnosis of melanoma for any participant was reached without histopathology Unclear – if the method of final diagnosis was not reported for any participant with a final diagnosis of melanoma or if the length of clinical follow‐up used was not clear or if a clinical follow‐up reference standard was reported in combination with a participant‐based analysis and it was not possible to determine whether the detection of a malignant lesion during follow‐up is the same lesion that originally tested negative on the index test B) Disease‐negative Yes – If at least 80% of benign diagnoses were reached by histology and up to 20% were reached by clinical follow‐up for a minimum of 3 months following the index test No – if more than 20% of benign diagnoses were reached by clinical follow‐up for a minimum of 3 months following the index test or if clinical follow‐up period was less than 3 months Unclear – if the method of final diagnosis was not reported for any participant with benign or non‐melanoma diagnosis |
| 2) Were the reference standard results interpreted without knowledge of the results of the index test? Please score this item for all studies even though histopathology interpretation is usually conducted with knowledge of the clinical diagnosis (from visual inspection or dermoscopy or both). We will deal with this by not including the response to this item in the 'Risk of bias' assessment for these tests. For reviews of all other tests, this item will be retained |
Yes – if the reference standard diagnosis was reached blinded to the index test result No – if the reference standard diagnosis was reached with knowledge of the index test result Unclear – if blinded reference test interpretation was not clearly reported |
| Could the reference standard, its conduct, or its interpretation have introduced bias? For visual inspection/dermoscopy evaluations
For all other tests
|
For visual inspection/dermoscopy evaluations
For all other tests
|
| REFERENCE STANDARD (3) ‐ CONCERN ABOUT APPLICABILITY | |
| 1) Are index test results presented separately for each component of the target condition (i.e. separate results presented for those with invasive melanoma, melanoma in situ, lentigo maligna, severe dysplasia, BCC, and cSCC)? |
Yes – if index test results for each component of the target condition can be disaggregated No – if index test results for the different components of the target condition cannot be disaggregated Unclear – if not clearly reported |
| 2) Expert opinion (with no histological confirmation) was not used as a reference standard 'Expert opinion' means diagnosis based on the standard clinical examination, with no histology or lesion follow‐up ***do not complete this item for teledermatology studies |
Yes – if expert opinion was not used as a reference standard for any participant No – if expert opinion was used as a reference standard for any participant Unclear – if not clearly reported |
| 3) Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? |
Yes – if histology interpretation was reported to be carried out by an experienced histopathologist or dermatopathologist No – if histology interpretation was reported to be carried out by a less experienced histopathologist Unclear – if the experience/qualifications of the pathologist were not reported |
Is there concern that the target condition as defined by the reference standard does not match the review question?
***For teledermatology studies only
|
***For teledermatology studies only
|
| FLOW AND TIMING (4):RISK OF BIAS | |
| 1) Was there an appropriate interval between index test and reference standard? A) For histopathological reference standard, was the interval between index test and reference standard ≤ 1 month? B) If the reference standard includes clinical follow‐up of borderline/benign‐appearing lesions, was there at least 3 months' follow‐up following application of index test(s)? |
A) Yes – if study reports ≤ 1 month between index and reference standard No – if study reports > 1 month between index and reference standard Unclear – if study does not report interval between index and reference standard B) Yes – if study reports ≥ 3 months' follow‐up No – if study reports < 3 months' follow‐up Unclear – if study does not report the length of clinical follow‐up |
| 2) Did all participants receive the same reference standard? |
Yes – if all participants underwent the same reference standard No – if more than 1 reference standard was used Unclear – if not clearly reported |
| 3) Were all participants included in the analysis? |
Yes – if all participants were included in the analysis No – if some participants were excluded from the analysis Unclear– if not clearly reported |
| 4) For within‐person comparisons of index tests Was the interval between application of index tests ≤ 1 month? |
Yes – if study reports ≤ 1 month between index tests No – if study reports > 1 month between index tests Unclear – if study does not report the interval between index tests |
| Could the participant flow have introduced bias? For non‐comparative and between‐person comparison studies
For within‐person comparative studies
|
For non‐comparative and between‐person comparison studies
For within‐person comparative studies
|
| BCC = basal cell carcinoma; cSCC = cutaneous squamous cell carcinoma. | |
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
1. Test.

App 1 [decision: problematic vs okay].
2. Test.

App 2 [decision: melanoma vs not melanoma].
3. Test.
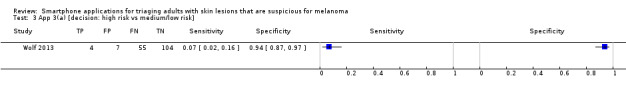
App 3(a) [decision: high risk vs medium/low risk].
4. Test.

App 3(b) [decision: high/medium risk vs low risk].
5. Test.

App 4 (remote diagnosis) [decision: atypical vs typical].
6. Test.

SkinVision [decision: high risk vs medium/low risk].
7. Test.

Face‐to‐face dermatologist diagnosis [decision: melanoma vs not melanoma].
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Maier 2015.
| Study characteristics | |||
| Patient sampling |
Study design: case series Data collection: prospective Period of data collection: not reported Country: Germany |
||
| Patient characteristics and setting |
Inclusion criteria: patients seen routinely for skin cancer screening at the Department of Dermatology Setting: secondary (general dermatology) Prior testing: selected for excision (no further detail) Setting for prior testing: not reported Exclusion criteria: poor quality index test image; (elements in the image not belonging to the lesion e.g. hair, images containing more than one lesion, incomplete imaged lesions); non‐melanocytic lesions. Also excluded "two‐point differences cases" mainly due to inappropriate imaging angle or distance (we assume this to mean lesions with results in non‐consecutive risk classes, e.g. 1 high risk and 2 low risk); and tie cases (described as cases with an equal number of results in two consecutive risk classes, e.g. 1 high risk, 1 medium risk and 1 low risk result). Sample size (patients): not reported Sample size (lesions): no. eligible: 195; no. included: 175 (at least 3 images included per lesion) Participant characteristics: not reported Lesion characteristics: not reported |
||
| Index tests |
Mobile phone application Acquisition and transmission of images: secondary care Nature of images used: clinical photographs Any additional patient information provided: unclear if the clinical and dermoscopic diagnosis was independently documented Diagnostic threshold: the SkinVision application evaluates lesions to be of high risk (red), medium risk (yellow) and low risk (green). We classified histologically proven naevi (benign and dysplastic) as being at low or medium risk Diagnosis based on: artificial intelligence‐based diagnosis # In person assessment Method of diagnosis: visual inspection and dermosocpy Prior test data: not reported Diagnostic threshold: diagnosis of melanoma Diagnosis based on: single observer Number of examiners: two Observer qualifications: dermatologist Experience in practice: unclear – not specified Experience with index test: unclear – not specified |
||
| Target condition and reference standard(s) |
Reference standard: histological diagnosis alone Details: Histology (excision) – 195 eligible lesions including 40 melanomas; 20 lesions excluded due to image quality (lesion types not reported), leaving 175 lesions analysed by the application (number of melanomas remaining not reported) Target condition (final diagnoses) For the sample of 195: melanoma (in situ or invasive): 40; dysplastic naevi (mild/moderate) 42; benign naevi 113 For the analysed sample of 175: lesion diagnoses not reported For the final sample of 144: melanoma (in situ or invasive): 26; dysplastic naevi (mild/moderate) 34; benign naevus: 84 |
||
| Flow and timing |
1. Excluded participants: 20 lesions (10%) excluded due to poor image quality (significant amount of hair, lesion out of focus or multiple lesions in the focus). An additional 31 were excluded as unevaluable (13 lesions (6%) based on two‐point‐differences and 18 (9%) tie cases with an equal number of results in different risk classes) 2. Time interval to reference test: not reported – assume it is < 1 month as images of the lesions were taken prior to excision |
||
| Comparative | |||
| Notes | — | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test Index test | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| If the reference standard includes clinical follow‐up of borderline/benign appearing lesions, was there a minimum follow‐up following application of index test(s) of at least: 3 months for melanoma or cSCC or 6 months for BCC? | |||
| High | |||
Wolf 2013.
| Study characteristics | |||
| Patient sampling |
Study design: case control study Data collection: retrospective image selection/prospective interpretation Period of data collection: not reported Country: USA |
||
| Patient characteristics and setting |
Inclusion criteria: lesion images selected from the institution image database with specific and clear histologic diagnoses including: melanoma, melanoma in situ, lentigo, benign naevi (including compound, junctional and low grade dysplastic naevi), dermatofibroma, sebhorrhoeic keratosis and haemangioma Setting: unspecified Prior testing: selected for excision (no further detail) Setting for prior testing: unspecified Exclusion criteria: Poor quality index test image Images that contained any identifiable features, such as facial features, tattoos, or labels with patient information, were excluded or cropped to remove the identifiable features or information Lesions with specific diagnoses including: Spitz naevi, Reed nevus, uncommon or equivocal lesions; and lesions with moderate or high‐grade atypia Sample size (patients): not reported Sample size (lesions): no. eligible 390; no. included 188 Participant characteristics: not reported Lesion characteristics: not reported Other: (note: Von Braunmühl 2015 extrapolates sensitivity and specificity to the whole dataset of 195 lesions by including the poor quality index test images ‐ excluded as overlapping populations with Maier 2015) |
||
| Index tests |
1. Mobile phone application Acquisition and transmission of images: secondary care Nature of images used: not reported Any additional patient information provided: no further information used Diagnostic threshold: Application 1. The application analyses the image and gives an assessment of 'problematic' (positive test result) or 'okay' (negative test result) Application 2. The output given is 'melanoma' (positive test result) or 'looks good' (negative test result). Application 3. The output given is 'high risk' (positive test result) or 'medium risk' or 'low risk,' both of which we considered to be a negative test result. Application 4. The dermatologist assigns an output of 'atypical' (positive test result) or 'typical' (negative test result); images classified as 'send another photograph' or 'unable to categorise' were considered test failures and excluded by study authors. Observer qualifications (remote diagnosis): application 4 only: images interpreted by a board‐certified dermatologist (n = not reported) |
||
| Target condition and reference standard(s) |
Reference standard: histological diagnosis alone Details: reference standard details Histology (not further described) – No. patients/lesions: a total of 188 lesions – disease positive: 60 melanomas – disease negative: 128 benign Target condition (final diagnoses) Malignant – melanoma (in situ and invasive): 60 Benign – 'Benign' diagnoses: 128 |
||
| Flow and timing | 1. Excluded participants: 202/390 lesion images excluded due to poor image quality, containing identifiable patient information or features, or lacking sufficient clinical or histological information. Between 3 and 29 additional lesions were analysed by the applications but considered unevaluable or test failures. The test failure rates were: 3% (n = 6; App 1), 2% (n = 3; App 2), 6% (n = 12; App 3) and 15% (n = 29; App 4). 2. Time interval to reference test: NA |
||
| Comparative | |||
| Notes | ‐ | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Are the included patients and chosen study setting appropriate? | No | ||
| Did the study avoid including participants with multiple lesions? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test Index test | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Was the test applied and interpreted in a clinically applicable manner? | No | ||
| Were thresholds or criteria for diagnosis reported in sufficient detail to allow replication? | Yes | ||
| Low | High | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Were the reference standard results interpreted without knowledge of the referral diagnosis? | Unclear | ||
| Expert opinion (with no histological confirmation) was not used as a reference standard | Yes | ||
| Was histology interpretation carried out by an experienced histopathologist or by a dermatopathologist? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| If the reference standard includes clinical follow‐up of borderline/benign appearing lesions, was there a minimum follow‐up following application of index test(s) of at least: 3 months for melanoma or cSCC or 6 months for BCC? | |||
| High | |||
NA: not applicable.
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Braun 2015 | Case report |
| Burki 2013 | Not a primary study |
| Diniz 2016 | Inappropriate index test (mobile phone used along with amplifying microscopic lens) |
| Jahan‐Tigh 2016 | Inappropriate index test (telediagnosis of ex vivo pathology specimens) |
| Karargyris 2012 | Inappropriate study population Inappropriate target condition |
| Lai 2015 | Conference abstract No 2 × 2 data |
| Massone 2007 | Small sample size (< 5 cases of melanoma as final diagnosis) Inappropriate index test (mobile phone used to capture dermoscopic images) |
| Ramlakhan 2011 | Derivation study (results of the training and test sets not differentiated Table II) |
| Robson 2012 | Inappropriate sample size |
| Varma 2011 | Not a primary study (Editorial) |
| Von Braunmühl 2015 | Duplicate or related publication (Maier 2015) |
| Wadhawan 2011 | Derivation study |
| Yu 2011 | Inappropriate study population No 2 × 2 data |
| Zouridakis 2015 | Not a primary study (book chapter) |
Differences between protocol and review
We changed the primary objectives and primary target condition from detection of cutaneous invasive melanoma alone, to the detection of cutaneous invasive melanoma and atypical intraepidermal melanocytic variants, as the latter is more clinically relevant in practice.
We also amended the primary objective from "To determine the diagnostic accuracy of smartphone applications for the detection of cutaneous invasive melanoma and atypical intraepidermal melanocytic variants in adults when used by consumers" to "To assess the diagnostic accuracy of smartphone applications to rule out cutaneous invasive melanoma and atypical intraepidermal melanocytic variants in adults with concerns about suspicious skin lesions" in order to better reflect the intended role of smartphone applications.
Due to lack of data, we did not investigate secondary objectives related to the detection of any skin cancer or skin lesion with a high risk of progression to melanoma or the original primary objective related to detection of invasive melanoma alone.
We amended the text to clarify that studies available only as conference abstracts would be excluded from the review unless full papers could be identified; studies available only as conference abstracts do not allow a comprehensive assessment of study methods or methodological quality.
To improve clarity of methods, we replaced the following text from the protocol: "We will include studies developing new algorithms or methods of diagnosis (i.e. derivation studies) if they use a separate independent 'test set' of participants or images to evaluate the new approach. We will also include studies using other forms of cross validation, such as 'leave‐one‐out' cross‐validation (Efron 1983). We will note for future reference (but not extract) any data on the accuracy of lesion characteristics individually, e.g. the presence or absence of a pigment network or detection of asymmetry."
This section now reads as follows.
"We included studies developing new mobile phone applications (i.e. derivation studies) if they used a separate independent 'test set' of participants or images to evaluate the new approach.
"We excluded studies if they:
used a statistical model to produce a data‐driven equation or algorithm based on multiple diagnostic features, with no separate test set;
used cross‐validation approaches such as 'leave‐one‐out' cross‐validation (Efron 1983); or
evaluated the accuracy of the presence or absence of individual lesion characteristics or morphological features, with no overall diagnosis of malignancy."
As per the secondary objectives above, we have removed the target conditions of invasive melanoma alone and of any skin cancer or skin lesion with a high risk of progression to melanoma from the review due to lack of data.
We added a clarification to the Index tests section that smartphone app use by clinicians for a second opinion by specialists is covered in teledermatology, whereas this review covers applications intended for use by the general public.
We planned to supplement the database searches by searching the annual meetings of appropriate organisations (e.g. British Association of Dermatologists Annual Meeting, American Academy of Dermatology Annual Meeting, European Academy of Dermatology and Venereology Meeting, Society for Melanoma Research Congress, World Congress of Dermatology, European Association of Dermato Oncology); however, due to the volume of evidence retrieved from database searches and time restrictions we were unable to do this.
For quality assessment, we further tailored the QUADAS‐2 tool according to the review topic.
In terms of analysis, we could not restrict analysis to per patient data due to lack of data. For the same reason, we could not investigate heterogeneity or perform sensitivity analyses.
Contributions of authors
NC was the contact person with the editorial base. NC co‐ordinated contributions from the co‐authors and wrote the final draft of the review. SB conducted the literature searches. NC, JD, OB and JM screened papers against eligibility criteria. NC obtained data on ongoing and unpublished studies. NC, JD and OB appraised the quality of papers. NC and OB extracted data for the review and sought additional information about papers. NC entered data into RevMan. NC analysed and interpreted data. NC, JD, SB, CD, YT, SO and JJD worked on the Methods sections. NC, HW, RM, OB, JM, SO, AJ and FW drafted the clinical sections of the Background and responded to the clinical comments of the referees. JD responded to the methodology and statistics comments of the referees. KG was the consumer co‐author and checked the review for readability and clarity, as well as ensuring outcomes are relevant to consumers. JD is the guarantor of the update.
Disclaimer
This project presents independent research supported by the National Institute for Health Research, via Cochrane Infrastructure funding to the Cochrane Skin Group and Cochrane Programme Grant funding, and the NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the Systematic Reviews Programme, NIHR, NHS or the Department of Health and Social Care.
Sources of support
Internal sources
No sources of support supplied
External sources
-
NIHR Systematic Review Programme, UK.
This project was funded by an NIHR Cochrane Systematic Reviews Programme Grant (13/89/15)
-
The National Institute for Health Research (NIHR), UK.
The NIHR, UK, is the largest single funder of the Cochrane Skin Group
-
NIHR Birmingham Biomedical Research Centre, UK.
JD, JJD and YT receive support from the NIHR Birmingham Biomedical Research Centre
Declarations of interest
Naomi Chuchu: none known. Yemisi Takwoingi: none known. Jac Dinnes: none known. Rubeta N Matin: my institution received a grant for a Barco NV commercially sponsored study to evaluate digital dermoscopy in the skin cancer clinic. My institution also received Oxfordshire Health Services Research Charitable Funds for carrying out a study of feasibility of using the Skin Cancer Quality of Life Impact Tool (SCQOLIT) in non melanoma skin cancer. I have received royalties for the Oxford Handbook of Medical Dermatology (Oxford University Press) and payment from the UK Photopheresis Society for a lecture on cutaneous graft versus host disease (October 2017). I have no conflicts of interest to declare that directly relate to the publication of this work. Oliver Bassett: none known. Jacqueline F Moreau: I helped draft the paper and performed data analysis for an included study (Wolf 2013). Susan E Bayliss: none known. Clare Davenport: none known. Kathie Godfrey: none known. Susan O'Connell: none known. Abhilash Jain: none known. Fiona M Walter: none known. Jonathan J Deeks: none known. Hywel C Williams: I am director of the NIHR health technology assessment (HTA) Programme. HTA is part of the NIHR which also supports the NIHR systematic reviews programme from which this work is funded.
Clinical referee David de Berker: I am Principal investigator for a single site in a multicentre study for assessment of images in pigmented lesions. The sponsor is Skin Analytics. I receive no payment for this from Skin Analytics, although they pay the hospital for participation of our site.
Edited (no change to conclusions)
References
References to studies included in this review
Maier 2015 {published data only}
- Maier T, Kulichova D, Schotten K, Astrid R, Ruzicka T, Berking C, et al. Accuracy of a smartphone application using fractal image analysis of pigmented moles compared to clinical diagnosis and histological result. Journal of the European Academy of Dermatology and Venereology : JEADV 2015;29(4):663‐7. [ER4:25012308; PUBMED: 25087492] [DOI] [PubMed] [Google Scholar]
Wolf 2013 {published data only}
- Wolf JA, Moreau JF, Akilov O, Patton T, English JC 3rd, Ho J, et al. Diagnostic inaccuracy of smartphone applications for melanoma detection. JAMA Dermatology 2013;149(4):422‐6. [ER4:15466167; PUBMED: 23325302] [DOI] [PMC free article] [PubMed] [Google Scholar]
References to studies excluded from this review
Braun 2015 {published data only}
- Braun RP, Marghoob A. High‐dynamic‐range dermoscopy imaging and diagnosis of hypopigmented skin cancers. JAMA Dermatology 2015;151(4):456‐7. [PUBMED: 25535875] [DOI] [PubMed] [Google Scholar]
Burki 2013 {published data only}
- Burki TK. Diagnostic accuracy of smartphone applications. Lancet Oncology 2013;14(3):e90. [PUBMED: 23580957] [DOI] [PubMed] [Google Scholar]
Diniz 2016 {published data only}
- Diniz LE, Ennser K. Melanoma detection using a mobile phone app. Proceedings of SPIE. March 7, 2016; Vol. 9699. [DOI: 10.1117/12.2212446] [DOI]
Jahan‐Tigh 2016 {published data only}
- Jahan‐Tigh RR, Chinn GM, Rapini RP. A comparative study between smartphone‐based microscopy and conventional light microscopy in 1021 dermatopathology specimens. Archives of Pathology & Laboratory Medicine 2016;140(1):86‐90. [PUBMED: 26717060] [DOI] [PubMed] [Google Scholar]
Karargyris 2012 {published data only}
- Karargyris A, Karargyris O, Pantelopoulos A. DERMA/care: an advanced image‐processing mobile application for monitoring skin cancer. IEEE 24th International Conference on Tools with Artificial Intelligence; 2012 Nov 7‐9; Athens, Greece. 2012; Vol. 2:1‐7. [DOI: 10.1109/ICTAI.2012.180] [DOI]
Lai 2015 {published data only}
- Lai I, Ko J, Pathipati A. DermLens: device for mobile teledermatology. Journal of the American Academy of Dermatology 2015;72(5, Suppl 1):AB88. [EMBASE: 71895108] [Google Scholar]
Massone 2007 {published data only}
- Massone C, Hofmann‐Wellenhof R, Ahlgrimm‐Siess V, Gabler G, Ebner C, Soyer HP. Melanoma screening with cellular phones. PLOS ONE 2007;2(5):e483. [PUBMED: 17534433] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ramlakhan 2011 {published data only}
- Ramlakhan K, Shang Y. A mobile automated skin lesion classification system. IEEE 23rd International Conference on Tools with Artificial Intelligence; 2011 Nov 7‐9; Boca Raton, FL, USA. 2011:138‐41. [DOI: 10.1109/ICTAI.2011.29] [DOI]
Robson 2012 {published data only}
- Robson Y, Blackford S, Roberts D. Caution in melanoma risk analysis with smartphone application technology. British Journal of Dermatology 2012;167(3):703‐4. [PUBMED: 22762381] [DOI] [PubMed] [Google Scholar]
Varma 2011 {published data only}
- Varma S. Mobile teledermatology for skin tumour screening. British Journal of Dermatology 2011;164(5):939‐40. [PUBMED: 21518326] [DOI] [PubMed] [Google Scholar]
Von Braunmühl 2015 {published data only}
- Braunmühl T. Smartphone apps for skin cancer diagnosis? The Munich study [Smartphone Apps für die Hautkrebs‐Diagnose? – die Münchner Studie]. Kosmetische Medizin 2015;36(4):152‐7. [Google Scholar]
Wadhawan 2011 {published data only}
- Wadhawan T, Situ N, Rui H, Lancaster K, Yuan X, Zouridakis G. Implementation of the 7‐point checklist for melanoma detection on smart handheld devices. IEEE Engineering in Medicine and Biology Magazine ‐ Conference Proceedings 2011;2011:3180‐3. [PUBMED: 22255015] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yu 2011 {published data only}
- Yu LS, Joseph AONR, Lindsley EH, Farkas DL. Polarization‐sensitive digital dermoscopy for image processing‐assisted evaluation of atypical nevi: towards step‐wise detection of melanoma. Proceedings of SPIE; 2011 Feb 28; San Francisco, California, United States. 2011; Vol. 7902. [DOI: 10.1117/12.891083] [DOI]
Zouridakis 2015 {published data only}
- Zouridakis G, Wadhawan T, Situ N, Hu R, Yuan X, Lancaster K, et al. Melanoma and other skin lesion detection using smart handheld devices. Methods in Molecular Biology 2015;1256:459‐96. [PUBMED: 25626557] [DOI] [PubMed] [Google Scholar]
Additional references
ACIM 2017
- Australian Cancer Database. Melanoma of the skin for Australia (ICD10 C43). Australian Institute of Health and Welfare (AIHW) 2017 Australian Cancer Incidence and Mortality (ACIM) books (www.aihw.gov.au/acim‐books/). Canberra: Australian Institute of Health and Welfare, 2017. [Google Scholar]
Altamura 2008
- Altamura D, Avramidis M, Menzies SW. Assessment of the optimal interval for and sensitivity of short‐term sequential digital dermoscopy monitoring for the diagnosis of melanoma. Archives of Dermatology 2008;144(4):502‐6. [PUBMED: 18427044] [DOI] [PubMed] [Google Scholar]
Apalla 2017
- Apalla Z, Lallas A, Sotiriou E, Lazaridou E, Ioannides D. Epidemiological trends in skin cancer. Dermatology Practical & Conceptual 2017;7(2):1. [DOI: 10.5826/dpc.0702a01] [DOI] [PMC free article] [PubMed] [Google Scholar]
Armstrong 2017
- Armstrong BK, Cust AE. Sun exposure and skin cancer, and the puzzle of cutaneous melanoma: a perspective on Fears et al. Mathematical models of age and ultraviolet effects on the incidence of skin cancer among whites in the United States. American Journal of Epidemiology 1977; 105: 420‐7. Cancer Epidemiology 2017;48:147‐56. [PUBMED: 28478931] [DOI] [PubMed] [Google Scholar]
Arnold 2014
- Arnold M, Holterhues C, Hollestein LM, Coebergh JW, Nijsten T, Pukkala E, et al. Trends in incidence and predictions of cutaneous melanoma across Europe up to 2015. Journal of the European Academy of Dermatology and Venereology: JEADV 2014;28(9):1170‐8. [PUBMED: 23962170] [DOI] [PubMed] [Google Scholar]
BAD 2013
- British Association of Dermatology. Quality standards for Teledermatology using 'store and forward' images. www.bad.org.uk/shared/get‐file.ashx?itemtype=document&id=794. London: British Association of Dermatology, (accessed prior to 16 May 2018).
Balch 2001
- Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, Cascinelli N, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. Journal of Clinical Oncology 2001;19(16):3622‐34. [PUBMED: 11504744] [DOI] [PubMed] [Google Scholar]
Balch 2009
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final Version of 2009 AJCC Melanoma Staging and Classification. Journal of Clinical Oncology 2009;27(36):6199‐206. [PUBMED: 19917835] [DOI] [PMC free article] [PubMed] [Google Scholar]
Bashshur 2015
- Bashshur RL, Shannon GW, Tejasvi T, Kvedar JC, Gates M. The empirical foundations of teledermatology: a review of the research evidence. Telemedicine Journal and E‐Health 2015;21(12):953‐79. [PUBMED: 26394022] [DOI] [PMC free article] [PubMed] [Google Scholar]
Belbasis 2016
- Belbasis L, Stefanaki I, Stratigos AJ, Evangelou E. Non‐genetic risk factors for cutaneous melanoma and keratinocyte skin cancers: an umbrella review of meta‐analyses. Journal of Dermatological Science 2016;84(3):330‐339. [PUBMED: 27663092] [DOI] [PubMed] [Google Scholar]
Boniol 2012
- Boniol M, Autier P, Boyle P, Gandini S. Cutaneous melanoma attributable to sunbed use: systematic review and meta‐analysis. BMJ 2012;345:e4757. [PUBMED: 22833605] [DOI] [PMC free article] [PubMed] [Google Scholar]
Boring 1994
- Boring CC, Squires TS, Tong T, Montgomery S. Cancer statistics, 1994. CA: a Cancer Journal for Clinicians 1994;44(1):7‐26. [PUBMED: 8281473] [DOI] [PubMed] [Google Scholar]
Bossuyt 2015
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. [DOI: 10.1136/bmj.h5527; PUBMED: 26511519] [DOI] [PMC free article] [PubMed] [Google Scholar]
Cancer Research UK 2017a
- Cancer Research UK. Skin cancer statistics. www.cancerresearchuk.org/health‐professional/cancer‐statistics/statistics‐by‐cancer‐type/skin‐cancer#heading‐One. (accessed prior to 21 July 2017).
Cancer Research UK 2017b
- Skin cancer incidence statistics. www.cancerresearchuk.org/health‐professional/cancer‐statistics/statistics‐by‐cancer‐type/skin‐cancer/incidence (accessed prior to 30 May 2018).
Chao 2013
- Chao D, London Cancer (North and East). Guidelines for cutaneous malignant melanoma management August 2013. www.londoncancer.org/media/76373/london‐cancer‐melanoma‐guidelines‐2013‐v1.0.pdf. London: London Cancer North and East Alliance, (accessed 25 February 2015).
Cho 2014
- Cho H, Mariotto AB, Schwartz LM, Luo J, Woloshin S. When do changes in cancer survival mean progress? The insight from population incidence and mortality. Journal of the National Cancer Institute. Monographs 2014;2014(49):187‐97. [PUBMED: 25417232] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chuchu 2018
- Chuchu N, Dinnes J, Takwoingi Y, Matin RN, Bayliss SE, Davenport C, et al. Teledermatology for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013193] [DOI] [PMC free article] [PubMed] [Google Scholar]
Deeks 2005
- Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. Journal of Clinical Epidemiology 2005;58(9):882‐93. [PUBMED: 16085191] [DOI] [PubMed] [Google Scholar]
DePry 2011
- DePry JL, Reed KB, Cook‐Norris RH, Brewer JD. Iatrogenic immunosuppression and cutaneous malignancy [Review]. Clinics in Dermatology 2011;29(6):602‐13. [PUBMED: 22014982] [DOI] [PubMed] [Google Scholar]
Dinnes 2018a
- Dinnes J, Deeks JJ, Grainge MJ, Chuchu N, Ferrante di Ruffano L, Matin RN, et al. Visual inspection for diagnosing cutaneous melanoma in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013194] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018b
- Dinnes J, Deeks JJ, Chuchu N, Ferrante di Ruffano L, Matin RN, Thomson DR, et al. Dermoscopy, with and without visual inspection, for diagnosing melanoma in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD011902.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018c
- Dinnes J, Deeks JJ, Saleh D, Chuchu N, Bayliss SE, Patel L, et al. Reflectance confocal microscopy for diagnosing cutaneous melanoma in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013190] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dinnes 2018d
- Dinnes J, Bamber J, Chuchu N, Bayliss SE, Takwoingi Y, Davenport C, et al. High‐frequency ultrasound for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013188] [DOI] [PMC free article] [PubMed] [Google Scholar]
Dong 2000
- Dong XD, Tyler D, Johnson JL, DeMatos P, Seigler JF. Analysis of prognosis and disease progression after local recurrence of melanoma. Cancer 2000;88(5):1063‐71. [DOI: 10.1002/(SICI)1097-0142(20000301)88:5%3C1063::AID-CNCR17%3E3.0.CO;2-E; PUBMED: 10699896] [DOI] [PubMed] [Google Scholar]
Efron 1983
- Efron B. Estimating the error rate of a prediction rule: improvement on cross‐validation. Journal of the American Statistical Association 1983;78(382):316‐31. [DOI: 10.1080/01621459.1983.10477973] [DOI] [Google Scholar]
Erdmann 2013
- Erdmann F, Lortet‐Tieulent J, Schuz J, Zeeb H, Greinert R, Breitbart EW, et al. International trends in the incidence of malignant melanoma 1953‐2008‐‐are recent generations at higher or lower risk?. International Journal of Cancer 2013;132(2):385‐400. [PUBMED: 22532371] [DOI] [PubMed] [Google Scholar]
EUCAN 2012
- EUCAN, International Agency for Research on Cancer. Malignant melanoma of skin: estimated incidence, mortality & prevalence for both sexes, 2012. eco.iarc.fr/eucan/Cancer.aspx?Cancer=20. International Agency for Research on Cancer, (accessed 29 July 2015).
Ferlay 2015
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer 2015;136(5):E359‐86. [PUBMED: 25220842] [DOI] [PubMed] [Google Scholar]
Ferrante di Ruffano 2018a
- Ferrante di Ruffano L, Dinnes J, Deeks JJ, Chuchu N, Bayliss SE, Davenport C, et al. Optical coherence tomography for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013189] [DOI] [PMC free article] [PubMed] [Google Scholar]
Ferrante di Ruffano 2018b
- Ferrante di Ruffano L, Takwoingi Y, Dinnes J, Chuchu N, Bayliss SE, Davenport C, et al. Computer‐assisted diagnosis techniques (dermoscopy and spectroscopy‐based) for diagnosing skin cancer in adults. Cochrane Database of Systematic Reviews 2018, Issue 12. [DOI: 10.1002/14651858.CD013186] [DOI] [PMC free article] [PubMed] [Google Scholar]
Flaten 2018
- Flaten HK, Claire C, Schlager E, Dunnick CA, Dellavalle RP. Growth of mobile applications in dermatology ‐ 2017 update. escholarship.org/uc/item/3hs7n9z6 (accessed prior to 15 November 2018). [PUBMED: 29630159] [PubMed]
Friedman 1985
- Friedman RJ, Rigel DS, Kopf AW. Early detection of malignant melanoma: the role of physician examination and self‐examination of the skin. CA: a Cancer Journal for Clinicians 1985;35(3):130‐51. [PUBMED: 3921200] [DOI] [PubMed] [Google Scholar]
Gandini 2005a
- Gandini S, Sera F, Cattaruzza MS, Pasquini P, Abeni D, Boyle P, et al. Meta‐analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. European Journal of Cancer 2005;41(1):28‐44. [PUBMED: 15617989] [DOI] [PubMed] [Google Scholar]
Gandini 2005b
- Gandini S, Sera F, Cattaruzza MS, Pasquini P, Picconi O, Boyle P, et al. Meta‐analysis of risk factors for cutaneous melanoma: II. Sun exposure. European Journal of Cancer 2005;41(1):45‐60. [PUBMED: 15617990] [DOI] [PubMed] [Google Scholar]
Garbe 2016
- Garbe C, Peris K, Hauschild A, Saiag P, Middleton M, Bastholt L, et al. Diagnosis and treatment of melanoma. European consensus‐based interdisciplinary guideline ‐ Update 2016. European Journal of Cancer 2016;63:201‐17. [PUBMED: 27367293] [DOI] [PubMed] [Google Scholar]
Geller 2002
- Geller AC, Miller DR, Annas GD, Demierre MF, Gilchrest BA, Koh HK. Melanoma incidence and mortality among US whites, 1969‐1999. JAMA 2002;288(14):1719‐20. [PUBMED: 12365954] [DOI] [PubMed] [Google Scholar]
HPA and MelNet NZ 2014
- Health Promotion Agency and the Melanoma Network of New Zealand (MelNet). New Zealand Skin Cancer Primary Prevention and Early Detection Strategy 2014 to 2017. www.sunsmart.org.nz//sites/default/files/documents/NZ%20Skin%20Cancer%20PrimaryPrevention%20and%20EarlyDetection%20Strategy%202014%20to%202017%20FINAL%20VERSION%20%23406761.pdf. Cancer Society of New Zealand, (accessed 29 May 2018).
Kasprzak 2015
- Kasprzak JM, Xu YG. Diagnosis and management of lentigo maligna: a review. Drugs in Context 2015;4:212281. [PUBMED: 26082796] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kassianos 2015
- Kassianos AP, Emery JD, Murchie P, Walter FM. Smartphone applications for melanoma detection by community, patient and generalist clinician users: a review. British Journal of Dermatology 2015;172(6):1507–18. [PUBMED: 25600815] [DOI] [PubMed] [Google Scholar]
Kjome 2016
- Kjome RL, Wright DJ, Bjaaen AB, Garstad KW, Valeur M. Dermatological cancer screening: Evaluation of a new community pharmacy service. Research in Social and Administrative Pharmacy 2016;16:30581‐2. [DOI: 10.1016/j.sapharm.2016.12.001; PUBMED: 27964893] [DOI] [PubMed] [Google Scholar]
Korn 2008
- Korn EL, Liu PY, Lee SJ, Chapman JA, Niedzwiecki D, Suman VJ, et al. Meta‐analysis of phase II cooperative group trials in metastatic stage IV melanoma to determine progression‐free and overall survival benchmarks for future phase II trials. Journal of Clinical Oncology 2008;26(4):527‐34. [PUBMED: 18235113] [DOI] [PubMed] [Google Scholar]
Landini 2011
- Landini G. Fractals in microscopy. Journal of Microscopy 2011;241(1):1‐8. [PUBMED: 21118245] [DOI] [PubMed] [Google Scholar]
Leff 2008
- Leff B, Finucane TE. Gizmo idolatry. JAMA 2008;299(15):1830‐2. [PUBMED: 18413879] [DOI] [PubMed] [Google Scholar]
Lehmann 2011
- Lehmann AR, McGibbon D, Stefanini M. Xeroderma pigmentosum. Orphanet Journal Of Rare Diseases 2011;6:70. [PUBMED: 22044607] [DOI] [PMC free article] [PubMed] [Google Scholar]
Linos 2009
- Linos E, Swetter SM, Cockburn MG, Colditz GA, Clarke CA. Increasing burden of melanoma in the United States. Journal of Investigative Dermatology 2009;129(7):1666‐74. [PUBMED: 19131946] [DOI] [PMC free article] [PubMed] [Google Scholar]
MacKie 1990
- MacKie RM. Clinical recognition of early invasive malignant melanoma. BMJ 1990;301(6759):1005‐6. [PUBMED: 2249043] [DOI] [PMC free article] [PubMed] [Google Scholar]
Mahar 2016
- Mahar AL, Compton C, Halabi S, Hess KR, Gershenwald JE, Scolyer RA, et al. Critical assessment of clinical prognostic tools in melanoma. Annals of Surgical Oncology 2016;23(9):2753‐61. [PUBMED: 27052645] [DOI] [PubMed] [Google Scholar]
Marsden 2010
- Marsden JR, Newton‐Bishop JA, Burrows L, Cook M, Corrie PG, Cox NH, et al. BAD Guidelines: revised UK guidelines for the management of cutaneous melanoma 2010. British Journal of Dermatology 2010;163(2):238‐56. [PUBMED: 20608932] [DOI] [PubMed] [Google Scholar]
McLaughlin 2005
- McLaughlin CC, Wu XC, Jemal A, Martin HJ, Roche LM, Chen VW. Incidence of noncutaneous melanomas in the U.S. Cancer 2005;103(5):1000‐7. [PUBMED: 15651058] [DOI] [PubMed] [Google Scholar]
Mistry 2011
- Mistry M, Parkin DM, Ahmad AS, Sasieni P. Cancer incidence in the United Kingdom: projections to the year 2030. British Journal of Cancer 2011;105(11):1795‐803. [DOI: 10.1038/bjc.2011.430; PUBMED: 22033277] [DOI] [PMC free article] [PubMed] [Google Scholar]
Moreau 2013
Murchie 2017
- Murchie P, Amalraj Raja E, Brewster DH, Iversen L, Lee AJ. Is initial excision of cutaneous melanoma by General Practitioners (GPs) dangerous? Comparing patient outcomes following excision of melanoma by GPs or in hospital using national datasets and meta‐analysis. European Journal of Cancer 2017;86:373‐84. [PUBMED: 29100192] [DOI] [PubMed] [Google Scholar]
Ndegwa 2010
- Ndegwa S, Prichett‐Pejic W, McGill S, Murphy G, Severn M. Teledermatology services: rapid review of diagnostic, clinical management, and economic outcomes. Ottawa: Canadian Agency for Drugs and Technologies in Health (CADTH), 2010. [Google Scholar]
NICE 2015
- National Institute for Health and Care Excellence. Melanoma: assessment and management. www.nice.org.uk/guidance/ng14 (accessed prior to 21 July 2017).
Pasquali 2018
- Pasquali S, Hadjinicolaou AV, Chiarion Sileni V, Rossi CR, Mocellin S. Systemic treatments for metastatic cutaneous melanoma. Cochrane Database of Systematic Reviews 2018, Issue 2. [DOI: 10.1002/14651858.CD011123.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Raguso 2010
Rangayyan 2007
- Rangayyan RM, Nguyen TM. Fractal analysis of contours of breast masses in mammograms. Journal of Digital Imaging 2007;20(3):223‐37. [PUBMED: 17021926] [DOI] [PMC free article] [PubMed] [Google Scholar]
Reyes‐Ortiz 2006
- Reyes‐Ortiz CA, Goodwin JS, Freeman JL, Kuo YF. Socioeconomic status and survival in older patients with melanoma. Journal of the American Geriatrics Society 2006;54(11):1758‐64. [PUBMED: 17087705] [DOI] [PMC free article] [PubMed] [Google Scholar]
Robertson 2014
- Robertson N, Polonsky M, McQuilken L. Are my symptoms serious Dr Google? A resource‐based typology of value co‐destruction in online self‐diagnosis. Australasian Marketing Journal 2014;22(3):246‐56. [DOI: 10.1016/j.ausmj.2014.08.009] [DOI] [Google Scholar]
Rutjes 2005
- Rutjes AW, Reitsma JB, Vandenbroucke JP, Glas AS, Bossuyt PM. Case‐control and two‐gate designs in diagnostic accuracy studies. Clinical Chemistry 2005;51(8):1335‐41. [PUBMED: 15961549] [DOI] [PubMed] [Google Scholar]
Rutjes 2006
- Rutjes AW, Reitsma JB, Nisio M, Smidt N, Rijn JC, Bossuyt PM. Evidence of bias and variation in diagnostic accuracy studies. CMAJ 2006;174(4):469‐76. [PUBMED: 16477057] [DOI] [PMC free article] [PubMed] [Google Scholar]
SEER 2007
- SEER. Cutaneous melanoma equivalent terms, definitions and illustrations. C440‐C449 with histology 8720‐8780. seer.cancer.gov/tools/mphrules/2007/melanoma/terms_defs.pdf (accessed 28 February 2018).
Shaikh 2012
- Shaikh WR, Xiong M, Weinstock MA. The contribution of nodular subtype to melanoma mortality in the United States, 1978 to 2007. Archives of Dermatology 2012;148(1):30‐6. [PUBMED: 21931016] [DOI] [PubMed] [Google Scholar]
Siegel 2015
- Siegel R, Miller K, Jemal A. Cancer statistics, 2015. CA: a Cancer Journal for Clinicians 2015;65(1):5‐29. [PUBMED: 25559415] [DOI] [PubMed] [Google Scholar]
SIGN 2017
- Scottish Intercollegiate Guidelines Network. Cutaneous Melanoma. www.sign.ac.uk/sign‐146‐melanoma.html (accessed prior to 21 July 2017).
Sladden 2009
- Sladden MJ, Balch C, Barzilai DA, Berg D, Freiman A, Handiside T, et al. Surgical excision margins for primary cutaneous melanoma. Cochrane Database of Systematic Reviews 2009, Issue 10. [DOI: 10.1002/14651858.CD004835.pub2] [DOI] [PubMed] [Google Scholar]
Slater 2014
- Slater D, Walsh M. Standards and datasets for reporting cancers: Dataset for the histological reporting of primary cutaneous malignant melanoma and regional lymph nodes, May 2014. www.rcpath.org/Resources/RCPath/Migrated%20Resources/Documents/G/G125_DatasetMaligMelanoma_May14.pdf. London: Royal College of Pathologists, (accessed 29 July 2015).
Swerdlow 1995
- Swerdlow AJ, English JS, Qiao Z. The risk of melanoma in patients with congenital nevi: a cohort study. Journal of the American Academy of Dermatology 1995;32(4):595‐9. [PUBMED: 7896948] [DOI] [PubMed] [Google Scholar]
Thompson 2003
- Thompson JF, Morton DL, Kroon BBR. Textbook of Melanoma: Pathology, Diagnosis and Management. CRC Press, 2003. [ISBN 9781901865653] [Google Scholar]
Tucker 1985
- Tucker MA, Boice JD Jr, Hoffman DA. Second cancer following cutaneous melanoma and cancers of the brain, thyroid, connective tissue, bone, and eye in Connecticut, 1935‐82. National Cancer Institute Monographs 1985;68:161‐89. [PUBMED: 4088297] [PubMed] [Google Scholar]
Tyagi 2012
- Tyagi A, Miller K, Cockburn M. e‐Health tools for targeting and improving melanoma screening: a review. Journal of Skin Cancer 2012;2012:437502. [DOI: 10.1155/2012/437502; PUBMED: 23304515] [DOI] [PMC free article] [PubMed] [Google Scholar]
Warshaw 2010
- Warshaw EM, Gravely AA, Nelson DB. Accuracy of teledermatology/teledermoscopy and clinic‐based dermatology for specific categories of skin neoplasms. Journal of the American Academy of Dermatology 2010;63(2):348‐52. [PUBMED: 20633809] [DOI] [PubMed] [Google Scholar]
Wheatley 2016
- Wheatley K, Wilson JS, Gaunt P, Marsden JR. Surgical excision margins in primary cutaneous melanoma: A meta‐analysis and Bayesian probability evaluation. Cancer Treatment Reviews 2016;42:73‐81. [PUBMED: 26563920] [DOI] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529‐36. [PUBMED: 22007046] [DOI] [PubMed] [Google Scholar]
Zemelman 2014
- Zemelman VB, Valenzuela CY, Sazunic I, Araya I. Malignant melanoma in Chile: different site distribution between private and state patients. Biological Research 2014;47(1):34. [PUBMED: 25204018] [DOI] [PMC free article] [PubMed] [Google Scholar]
References to other published versions of this review
Dinnes 2015a
- Dinnes J, Matin RN, Moreau JF, Patel L, Chan SA, Wong KY, et al. Tests to assist in the diagnosis of cutaneous melanoma in adults: a generic protocol. Cochrane Database of Systematic Reviews 2015, Issue 10. [DOI: 10.1002/14651858.CD011902] [DOI] [Google Scholar]
Dinnes 2015b
- Dinnes J, Wong KY, Gulati A, Chuchu N, Leonardi‐Bee J, Bayliss SE, Takwoingi Y, Davenport C, Matin RN, Bath‐Hextall FJ, Jain A, Lear JT, Motley R, O'Sullivan C, Deeks JJ, Williams HC. Tests to assist in the diagnosis of keratinocyte skin cancers in adults: a generic protocol. Cochrane Database of Systematic Reviews 2015, Issue 10. [DOI: 10.1002/14651858.CD011901] [DOI] [Google Scholar]


