Abstract
Alphasatellites are non-essential satellite-like components associated with geminiviruses. The precise selective advantage to a geminivirus infection of an alphasatellite remains unclear. The ability of the cotton leaf curl Multan alphasatellite (CLCuMuA)-encoded replication-associated protein (Rep) to suppress TGS was investigated by using Nicotiana benthamiana line 16-TGS (16-TGS) harbouring a transcriptionally silenced green fluorescent protein (GFP) transgene. Inoculation of 16-TGS plants with a recombinant Potato virus X (PVX) vector carrying CLCuMuA Rep resulted in restoration of GFP expression. Northern blot analysis confirmed that the observed GFP fluorescence was associated with GFP mRNA accumulation. Inoculation with PVX vectors harbouring a further six Rep proteins, encoded by genetically distinct alphasatellites, were similarly shown to result in 16-TGS plants with restored GFP expression. These results indicate that the alphasatellite-encoded Rep can restore the expression of a transcriptionally silenced GFP transgene in N. benthamiana, indicating that alphasatellites are involved in overcoming host defence.
Keywords: Alphasatellite, Geminivirus, RNA interference, Transcriptional gene silencing
RNA interference (RNAi) is involved in numerous fundamental biological processes that include cellular defence against pathogens and mobile genetic elements such as transposons, regulation of gene expression, de novo DNA methylation and modification of chromatin [5]. A characteristic of RNAi is the production of 21–24 nucleotide long small interfering RNAs (siRNAs) that are derived from processing of large double-stranded RNAs. RNAi (also known as gene silencing) occurs at both the transcriptional and post-transcriptional levels. Post-transcriptional gene silencing (PTGS) acts on target RNAs, including the genomes of RNA viruses, resulting in their degradation. Transcriptional gene silencing (TGS) results from the de novo methylation within promoter sequences that may induce chromatin modifications. TGS is involved in maintaining genome integrity by preventing rearrangement in centromeric and telomeric repeats and by suppressing the activity of transposons and other invasive DNAs [13].
Geminiviruses are single-stranded DNA (ssDNA) viruses that infect a wide range of economically important plants. The family Geminiviridae consists nine genera; Begomovirus, Mastrevirus, Topocuvirus, Curtovirus, Turncurtovirus, Eragrovirus, Becurtovirus, and the recently established Grablovirus and Capulavirus [22]. The genus Begomovirus encompasses the largest number, and economically the most devastating viruses. Begomoviruses are exclusively transmitted by the whitefly Bemisia tabaci, infect dicotyledonous plants and occur in the warmer parts of the world. In the New World (NW) begomoviruses are generally bipartite, with a genome consisting of two ssDNA components known as DNA A and DNA B. Only a single monopartite begomovirus, with a genome consisting of a homolog of the DNA A component of bipartite begomoviruses, has been identified in the NW [11, 20]. In contrast, in the Old World, most begomoviruses are monopartite and associate with small ssDNA satellites known as betasatellites and alphasatellites [26].
In plants, geminivirus infection is countered by repressive histone methylation and TGS. To overcome these geminiviruses have evolved proteins which can suppress TGS; these include the transcriptional-activator protein [3], the replication-associated protein (Rep; a rolling-circle replication initiator protein) [15], the V2 protein [24] and the βC1 protein encoded by betasatellites [19, 25]. These proteins act at distinct steps in the TGS pathway.
Alphasatellites (previously known as DNA1) are small (~ 1380 nt) circular ssDNA, satellite-like molecules that associate with viruses of the family Geminiviridae [2]. Although most commonly identified in association with geminiviruses of the genus Begomovirus, recently an alphasatellite has been shown in association with a geminivirus of the genus Mastrevirus [9]. In the OW alphasatellites are almost exclusively identified in association with monopartite begomoviruses that are also associated with betasatellites. The mastrevirus shown to associate with an alphasatellite was also associated with a betasatellite [9]. Although only few alphasatellites have been identified in the NW, these have been in association with bipartite begomoviruses [8, 16].
Alphasatellites autonomously replicate in permissive host cells but are dependent on their helper viruses for inter-as well as intracellular movement, encapsidation and insect transmission [2]. However, the helper virus is not dependent upon the alphasatellite for infection; the component is dispensable [21, 23]. A single gene is encoded by alphasatellites which codes for a Rep. Alphasatellite DNA sequences also comprise a region rich in adenine residues and a predicted hairpin structure, with the nonanucleotide sequence TAGTATTAC forming part of the loop, with similarity to the origin of replication of viruses of the family Nanoviridae (another family of ssDNA viruses with multipartite genomes) [6]. The similarity in organisation (encoding a single gene in the virion-sense) and the same nonanucleotide sequence led to the suggestion that alphasatellites may have originated as components of nanoviruses which were captured by geminiviruses following a co-infection [10].
The Rep-encoding genes of Cotton leaf curl Multan alphasatellite (CLCuMuA; Acc. No. FR877532), Ageratum yellow vein India alphasatellite (AYVIA; HG515070), Tomato leaf curl alphasatellite (ToLCA; HG515065), Tomato leaf curl Pakistan alphasatellite (ToLCuPKA; HE966422), Cotton leaf curl Gezira alphasatellite (CLCuGeA; KY652676), Gossypium darwinii symptomless alphasatellite (GDarSLA; HG530128) and the Dragonfly associated alphasatellite (DfasA; JX458742; [17]) were PCR amplified with specific primers (Table 1) and cloned into the Potato virus X (PVX) expression vector pGR107 [4]. The production of a pGR107 construct for the expression of Cotton leaf curl Multan betasatellite (CLCuMuB) βC1 has been described previously [14]. The pGR107 constructs were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Nicotiana benthamiana plants were inoculated with A. tumefaciens cultures harbouring PVX constructs as previously described [1].
Table 1.
List of primers used in the study
| Primer | Sequence | Restriction sitea |
|---|---|---|
| Rep09(MuAOld) F | TAAAAGCTTATGCCTACTATTCAGTCACAATG | HindIII |
| Rep09(MuAOld) R | TTAGGATCCTTATTCCATATATTCGCCACAC | BamHI |
| Rep702-AYVIA-F | TCTGGATCCTGTATTGTTAATCGAAGATGC | BamHI |
| RepX-AYVIA-R | AGAGTCGACATTAATCCAAATACTCATCAC | SalI |
| Rep702-ToLCuA-F | CTCGGATCCTTTGCCAAGTAAAAATGCC | BamHI |
| RepX-ToLCuA-R | ACTGTCGACTTTATTCCATATATTCACCAC | SalI |
| Rep702-CLCuGeA-F | CATGAATTCTAACTATAAGATGCCTGC | EcoRI |
| RepX-CLCuGeA-R | TTGGTCGACAATGTTTAATCCAAATATTCC | SalI |
| Rep702-DfasA-F | GCTGGATCCATTTCTGGCTTCCTTTCATG | BamHI |
| RepX-DfasA-R | TCTGTCGACTTCAACATCCAATACAAAGAC | SalI |
| RepX-GDarSLA-F | GAAATCGATTGAGTTCTCGCTCTTTCGC | ClaI |
| RepX-GDarSLA-R | AGAGTCGACAGTAATTAATCCACATAATC | SalI |
| Rep702-ToLCuPKA-F | TCATCTAGATCGTCAAGTAAGAATGCCC | XbaI |
| RepX-ToLCuPKA-R | ACAGTCGACATTATTCCAAATATTCCTC | SalI |
aRestriction enzyme used to clone the gene fragment. The recognition sequence of each endonuclease is underlined in the primer sequence
N. benthamiana line 16-TGS plants harbour a transcriptionally silenced green fluorescence protein (GFP) transgene [18]. 16-TGS plants fluoresce red under UV illumination, due to chlorophyll autofluorescence (Fig. 1, panel A). Inoculation of 16-TGS plants with the PVX vector harbouring the CLCuMuB βC1 gene results in plants that fluoresce green under UV illumination. The βC1 protein encoded by betasatellites has been shown to interfere with TGS activity [19, 25]; in this case restoring expression of the transcriptionally silenced GFP gene. Similarly inoculation of 16-TGS plants with PVX harbouring the Rep gene of CLCuMuA resulted in restoration of GFP expression. Northern blot analysis of leaves developing subsequent to inoculation on 16-TGS plants inoculated with PVX expressing CLCuMuA Rep have higher levels of GFP expression than either non-inoculated or mock inoculated (plants inoculated with an empty PVX vector) but this is significantly lower than GFP expression in N. benthamiana line 16C (16C) plants (a line in which GFP expression is not transcriptionally silenced) (Fig. 2). Overall these results indicate that the Rep protein of CLCuMuA has the ability to interfere with TGS.
Fig. 1.
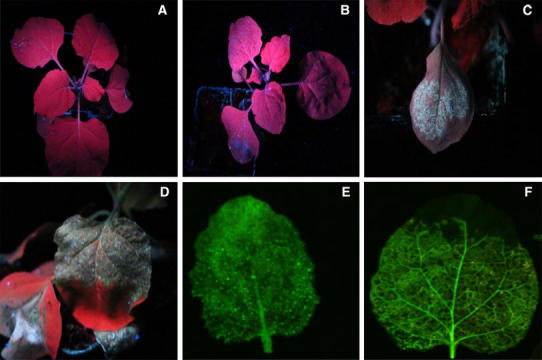
Images of N. benthamiana 16-TGS plants and leaves under UV illumination. The plants were either not inoculated (a), mock inoculated with the pGR107 vector (b), or inoculated with the pGR107 vector harbouring the βC1 gene of CLCuMuB (c) or inoculated with the pGR107 vector harbouring the Rep gene of CLCuMuA (d). The images in panels E and F are of 16-TGS plants inoculated with the pGR107 vector harbouring the βC1 gene of CLCuMuB and the pGR107 vector harbouring the Rep gene of CLCuMuA, respectively, that have been scanned with a PharosFX Molecular Imager (Biorad, www.bio-rad-com) to remove chlorophyll autofluorescence and highlight GFP fluorescence. The photographs were taken at 9 dpi and of leaves developing subsequent to inoculation
Fig. 2.
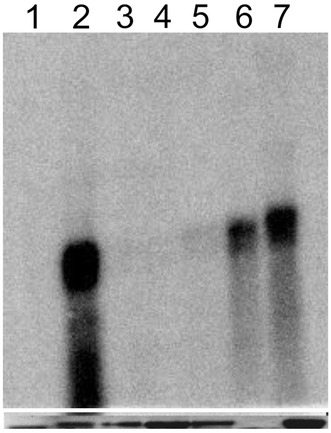
Northern blot analysis of total RNA extracted from N. benthamiana plants hybridised with a probe specific for GFP. Not inoculated wild-type N. benthamiana plant (lane 1), 16C plant (lane 2), 16-TGS plant not inoculated (lane 3), 16-TGS plant mock inoculated with pGR107 (lane 4), and 16-TGS plants inoculated with pGR107 harbouring the Rep gene of CLCuMuA (lanes 5–7). The samples were taken at 9 dpi. The panel below the blot shows a photograph of the ethidium stained rRNA band on the gel to confirm equal loading
To assess how widespread the ability to interfere with TGS is amongst distinct alphasatellites, PVX vectors harbouring the Rep genes encoded by six further alphasatellites were inoculated to 16-TGS plants. For all plants inoculated with PVX vectors harbouring alphasatellite Rep genes green fluorescence was evident at 15 day post inoculation (dpi; Fig. 3). This indicates that interfering with TGS is a common, possibly universal, property of Rep proteins encoded by alphasatellites. The Rep proteins of the alphasatellites investigated, with the exception of the Rep of DfasA, show greater than 86.5% amino acid sequence identity. The Rep of DfasA shows less than 51.2% amino acid sequence identity to the other alphasatellites investigated. This shows that suppression of TGS is conserved between evolutionarily quite distantly related alphasatellites.
Fig. 3.

Images of N. benthamiana leaves under UV illumination. Wild-type N. benthamiana (a), 16C plant (b), 16-TGS plants that were not inoculated (c) and mock inoculated (d), respectively, 16-TGS plants inoculated with the pGR107 vector harbouring the Rep genes of AYVIA (e), ToLCuA (f), ToLCuPKA (g), CLCuGeV (h), GDarSLA (i) and DfasA (j). The photographs were taken at 15 dpi and of leaves developing subsequent to inoculation
Few studies have investigated the contribution of alphasatellites to geminivirus infections. Studies have indicated that some alphasatellites may act to attenuate the symptoms induced by the helper virus by reducing betasatellite accumulation [7]. A similar study by Kumar et al. [23] noted the attenuation of symptoms by alphasatellites but did not show a reduction in betasatellite accumulation. Nawaz-ul-Rehman et al. [12] showed that at least some alphasatellites encode Rep proteins that can suppress PTGS, indicating that alphasatellites are involved in overcoming plant host defence. The results presented here, together with the results of previous studies, show that geminivirus-betasatellite-alphasatellite complexes may encode as many as four proteins with TGS suppressor activity [3, 15, 19, 24, 25]. The geminivirus Rep protein was shown to interfere with two methyltransferases involved in the maintenance methylation pathways [15].
The dispensable nature of alphasatellites for geminivirus infections indicates that the benefit of the presence of an alphasatellite is a subtle one. Nevertheless, the frequency with which alphasatellites are identified in association with, in particular, begomovirus-betasatellite complexes indicates that the selective advantage of the presence of alphasatellites is significant. Future work will centre on investigating the role of each of the identified TGS suppressors play in overcoming host defences, as well as elucidating the process(es) that are mediated by alphasatellite-encoded Rep proteins to interfere with the TGS pathway.
Acknowledgements
The authors are grateful to Arvind Varsani for providing the CLCuGeA and DfasA clones. QA and MS were supported by indigenous Ph.D. scholarships from the Higher Education Commission (HEC), Government of Pakistan. RWB was supported by the HEC under the “Foreign Faculty Hiring Program”.
References
- 1.Amin I, Patil BL, Briddon RW, Mansoor S, Fauquet CM. Comparison of phenotypes produced in response to transient expression of genes encoded by four distinct begomoviruses in Nicotiana benthamiana and their correlation with the levels of developmental miRNAs. Virol J. 2011;8:238. doi: 10.1186/1743-422X-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briddon RW, Bull SE, Amin I, Mansoor S, Bedford ID, Rishi N, et al. Diversity of DNA 1: a satellite-like molecule associated with monopartite begomovirus-DNA β complexes. Virology. 2004;324(2):462–474. doi: 10.1016/j.virol.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 3.Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol. 2009;83(10):5005–5013. doi: 10.1128/JVI.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman S, Kavanagh T, Baulcombe D. Potato virus X as a vector for gene expression in plants. Plant J. 1992;2:549–557. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- 5.Dalakouras A, Wassenegger M. Revisiting RNA-directed DNA methylation. RNA Biol. 2013;10(3):453–455. doi: 10.4161/rna.23542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gronenborn B. Nanoviruses: genome organisation and protein function. Vet Microbiol. 2004;98(2):103–109. doi: 10.1016/j.vetmic.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Idris AM, Shahid MS, Briddon RW, Khan AJ, Zhu J-K, Brown JK. An unusual alphasatellite associated with monopartite begomoviruses attenuates symptoms and reduces betasatellite accumulation. J Gen Virol. 2011;92(3):706–717. doi: 10.1099/vir.0.025288-0. [DOI] [PubMed] [Google Scholar]
- 8.Jeske H, Kober S, Schäfer B, Strohmeier S. Circomics of Cuban geminiviruses reveals the first alpha-satellite DNA in the Caribbean. Virus Genes. 2014;49(2):312–324. doi: 10.1007/s11262-014-1090-8. [DOI] [PubMed] [Google Scholar]
- 9.Kumar J, Kumar J, Singh SP, Tuli R. Association of satellites with a mastrevirus in natural infection: complexity of wheat dwarf India virus disease. J Virol. 2014;88(12):7093–7104. doi: 10.1128/JVI.02911-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansoor S, Khan SH, Bashir A, Saeed M, Zafar Y, Malik KA, et al. Identification of a novel circular single-stranded DNA associated with cotton leaf curl disease in Pakistan. Virology. 1999;259(1):190–199. doi: 10.1006/viro.1999.9766. [DOI] [PubMed] [Google Scholar]
- 11.Melgarejo TA, Kon T, Rojas MR, Paz-Carrasco L, Zerbini FM, Gilbertson RL. Characterization of a new world monopartite begomovirus causing leaf curl disease of tomato in Ecuador and Peru reveals a new direction in geminivirus evolution. J Virol. 2013;87(10):5397–5413. doi: 10.1128/JVI.00234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nawaz-ul-Rehman MS, Nahid N, Mansoor S, Briddon RW, Fauquet CM. Post-transcriptional gene silencing suppressor activity of two non-pathogenic alphasatellites associated with a begomovirus. Virology. 2010;405(2):300–308. doi: 10.1016/j.virol.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 13.Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Micro. 2013;11(11):745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- 14.Qazi J, Amin I, Mansoor S, Iqbal J, Briddon RW. Contribution of the satellite encoded gene βC1 to cotton leaf curl disease symptoms. Virus Res. 2007;128(1–2):135–139. doi: 10.1016/j.virusres.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Rodríguez-Negrete E, Lozano-Durán R, Piedra-Aguilera A, Cruzado L, Bejarano ER, Castillo AG. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 2013;199(2):464–475. doi: 10.1111/nph.12286. [DOI] [PubMed] [Google Scholar]
- 16.Romay G, Chirinos D, Geraud-Pouey F, Desbiez C. Association of an atypical alphasatellite with a bipartite New World begomovirus. Arch Virol. 2010;155(11):1843–1847. doi: 10.1007/s00705-010-0760-7. [DOI] [PubMed] [Google Scholar]
- 17.Rosario K, Padilla-Rodriguez M, Kraberger S, Stainton D, Martin DP, Breitbart M, et al. Discovery of a novel mastrevirus and alphasatellite-like circular DNA in dragonflies (Epiprocta) from Puerto Rico. Virus Res. 2012;171(1):231–237. doi: 10.1016/j.virusres.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10(6):937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saeed M, Krczal G, Wassenegger M. Three gene products of a begomovirus–betasatellite complex restore expression of a transcriptionally silenced green fluorescent protein transgene in Nicotiana benthamiana. Virus Genes. 2015;50(2):340–344. doi: 10.1007/s11262-014-1155-8. [DOI] [PubMed] [Google Scholar]
- 20.Sánchez-Campos S, Martínez-Ayala A, Márquez-Martín B, Aragón-Caballero L, Navas-Castillo J, Moriones E. Fulfilling Koch’s postulates confirms the monopartite nature of tomato leaf deformation virus: a begomovirus native to the New World. Virus Res. 2013;173(2):286–293. doi: 10.1016/j.virusres.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Saunders K, Stanley J. A nanovirus-like component associated with yellow vein disease of Ageratum conyzoides: evidence for interfamilial recombination between plant DNA viruses. Virology. 1999;264(1):142–152. doi: 10.1006/viro.1999.9948. [DOI] [PubMed] [Google Scholar]
- 22.Varsani A, Roumagnac P, Marc F, Navas-Castillo J, Moriones E, Idris A, et al. Capulavirus and Grablovirus: two new genera in the family Geminiviridae. Arch Virol. 2017;162(6):1819–1831. doi: 10.1007/s00705-017-3268-6. [DOI] [PubMed] [Google Scholar]
- 23.Vinoth Kumar R, Singh D, Singh AK, Chakraborty S. Molecular diversity, recombination and population structure of alphasatellites associated with begomovirus disease complexes. Infect Genet Evol. 2017;49:39–47. doi: 10.1016/j.meegid.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Wang B, Li F, Huang C, Yang X, Qian Y, Xie Y, et al. V2 of Tomato yellow leaf curl virus can suppress methylation-mediated plant transcriptional gene silencing. J Gen Virol. 2014;95(1):225–230. doi: 10.1099/vir.0.055798-0. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Xie Y, Raja P, Li S, Wolf JN, Shen Q, et al. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 2011;7(10):e1002329. doi: 10.1371/journal.ppat.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X. Advances in understanding begomovirus satellites. Ann Rev Phytopathol. 2013;51(1):357–381. doi: 10.1146/annurev-phyto-082712-102234. [DOI] [PubMed] [Google Scholar]


