Abstract
Over the past four decades, shrimp aquaculture has turned into a major industry providing jobs for millions of people worldwide especially in countries with large coastal boundaries. While the shrimp industry continues to expand, the sustainability of shrimp aquaculture has been threatened by the emergence of diseases. Diseases caused by single-stranded DNA containing viruses, such as infectious hypodermal and hematopoietic necrosis virus (IHHNV) and hepatopancreatic parvovirus (HPV), have caused immense losses in shrimp aquaculture since the early 1980s. In fact, the disease outbreak in the blue shrimp (Penaeus stylirostris) caused by IHHNV in early 1980s ultimately led to the captive breeding program in shrimp being shifted from P. stylirostris to the white shrimp (Penaeus vannamei), and today P. vannamei is the preferred cultured shrimp species globally. To date, four single-stranded DNA viruses are known to affect shrimp; these include IHHNV, HPV, spawner-isolated mortality virus (SMV) and lymphoidal parvo-like virus (LPV). Due to the economic losses caused by IHHNV and HPV, most studies have focused on these two viruses, and only IHHNV is included in the OIE list of Crustacean Diseases. Hence this review will focus on IHHNV and HPV. IHHNV and HPV virions are icosahedral in morphology measuring 20–22 nm in size and contain a single-stranded DNA (ssDNA) of 4–6 kb in size. Both IHHNV and HPV are classified into the sub-order Brevidensoviruses, family Densovirinae. The genome architecture of both viruses are quite similar as they contain two completely (as in IHHNV) or partially overlapping (as in HPV) non-structural and one structural gene. Histopathology and polymerase chain reaction (PCR)-based methods are available for both viruses. Currently, there is no anti-viral therapy for any viral diseases in shrimp. Therefore, biosecurity and the use of genetically resistant lines remains as the corner stone in the management of viral diseases. In recent years, gene silencing using the RNA interference (RNAi) approach has been reported for both IHHNV and HPV via injection. However, the delivery of RNAi molecules via oral route remains a challenge, and the utility of RNAi-based therapy has yet to be materialized in shrimp aquaculture.
Keywords: Single-stranded DNA viruses, Parvovirus, Shrimp, Infectious hypodermal and hematopoietic necrosis virus, Penaeus stylirostris densovirus, Hepatopancreatic parvovirus, Penaeus monodon densovirus, Spawner-isolated mortality virus, Lymphoidal parvo-like virus, IHHNV, PstDNV, HPV, PmDNV, SMV, LPV
Introduction
During the last four decades, the culture of various marine shrimp species has grown tremendously. Farmed production nowadays contributes over 50% of the world’s shrimp demand (almost 4.5 million metric tons, MMT), and production continues to grow and rapidly replaces capture fisheries as the market demands more providers. In addition to generating billions of dollars from sales and trade, shrimp aquaculture also offers employment for millions of people in developing nations, by incorporating farming into regions of previously unutilized coastal lands; by producing a valuable commodity that is increasingly marketed locally in many countries; and by generating jobs and hard currency through exports.
Shrimp have been cultured in Asia for centuries by farmers who typically cultured them as secondary crops in tidal fish ponds. Modern shrimp culture began in the 1930s, when Dr. Motosaku Fujinaga succeeded in spawning the Kuruma shrimp (Marsupenaeus japonicus). In the early 1970s, entrepreneurs and researchers from several countries in Latin America and Asia became involved in endorsing the development of the industry, which grew steadily. Marine shrimp farming has developed tremendously in the last 25 years, and significant progress has been made in the development of novel technologies and methods to culture shrimp [1, 10].
Marine shrimp farming is currently carried out around the globe in at least 60 countries, however production is mostly concentrated in ~ 15 nations in Asia and Latin America. Since 1992, about a dozen countries have contributed to bulk of shrimp farming, and the top world producers have been, at various times, Ecuador, Taiwan, Indonesia, China and Thailand. Shrimp culture in China has really exploded in recent years, and the country has been the world’s biggest producer since 2003, but other countries like India, Vietnam, Indonesia and Ecuador are not far behind.
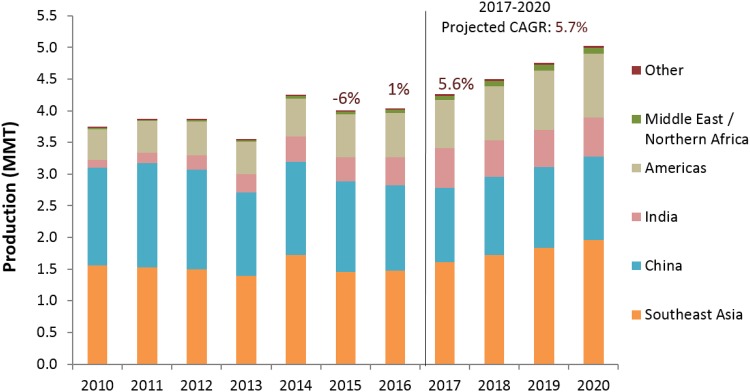
Recently Global Aquaculture Alliance (GAA, www.aquaculturealliance.org) carried out an annual survey of the global production trends in shrimp [2]. The Results for 2018 indicate an annual production of about 4,496,775 metric tons (MT) of farmed shrimp. Figure 1 summarizes the production estimates for global production during 2010–2020, combining data from the United Nations Food and Agriculture Organization (FAO) and the 2011–2018 GAA GOAL surveys.
Fig. 1.
Shrimp farming production by different regions in the world.
(Adapted from Anderson et al. [2])
The 2018 GAA survey reported considerable production declines in China, Thailand, Indonesia and Mexico in 2013 primarily due to outbreaks of Early Mortality Syndrome (EMS) (also known as acute hepatopancreatic necrosis disease, AHPND) that emerged initially in China in 2009. Elevated shrimp prices in the global markets during 2013 were consistent with industry expectations of declining production. Nevertheless, FAO data does not reveal any major impact of diseases in Chinese production during 2009–2013; on the contrary, China is reported by FAO to have increased production from 1.3 to 1.7 million metric tons (MMT) in this period, growing further to 2.0 MMT by 2016. Regarding current trends, production is projected to grow with a 5.7% compound annual growth rate (CAGR) from 2017 to 2020. This will result in growth of 18% over 2017 levels and global shrimp aquaculture harvest of 5.03 MMT (approx. 5.4 MMT including Macrobrachium rosenbergii). GAA forecasts depend on the assumption that major disease crises are averted in the near future.
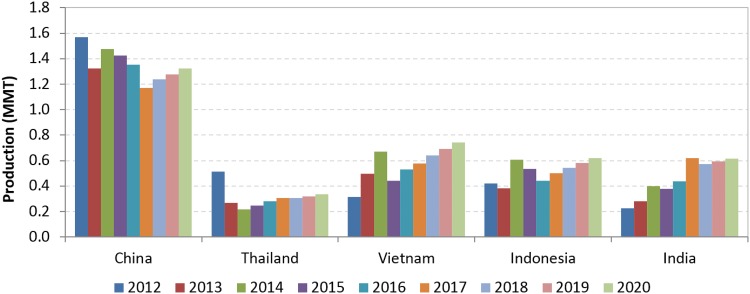
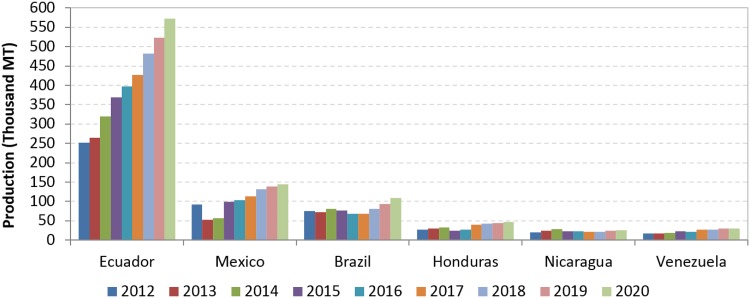
The shrimp farming industry in Asia appears to be recovering, following the substantial production declines during 2012–2015 caused by pervasive disease outbreaks. In 2018, production is expected to exceed pre-EMS levels driven primarily by increase in production in Vietnam, Indonesia and India. China will continue as the main producer, but its growth will be relatively slow (Fig. 2). The most important development in Americas region is the remarkable growth of Ecuadorean shrimp farming in the past few years. As with several Asian countries, Mexico was also able to recover from EMS impact and production is expected to reach 145 thousand MT by 2020. Estimated production for the major producing nations in Latin America is presented in Fig. 3. In addition to Ecuador and Mexico, Brazil, Venezuela, Peru, Honduras and Guatemala reported positive expectations for growth through 2020.
Fig. 2.
Shrimp aquaculture production in major farming nations in Asia.
(Taken from Anderson et al. [2])
Fig. 3.
Shrimp aquaculture production in major farming nations in Latin America.
(Taken from Anderson et al. [2])
Losses in shrimp production due to disease outbreaks have been catastrophic and valued in the billions (US dollars). Estimated economic losses due to different viruses are as follows: White spot syndrome virus (WSSV) 4–6 billion, Taura Syndrome virus (TSV) 1–2 billion, Yellow head virus (YHV) 0.5–1 billion and Infections hypodermal and hematopoietic necrosis virus (IHHNV) 0.5–1 billon [40]. In one of the most recent surveys for Asian producers, the top identified challenges that the shrimp aquaculture industry faces include banned chemicals/antibiotic use, international market prices and cost of feed. However, the top challenge identified by producers was “diseases” as in previous surveys. This is clearly due to the impact that viral diseases have had on the industry throughout the years.
Impact of infectious hypodermal and hematopoietic necrosis virus (IHHNV) in shaping the captive breeding program in shrimp
To our knowledge, the global impact of IHHNV on the captive breeding programs of commercially-important species of penaeid shrimp has not been addressed. In the early 1980s, IHHNV outbreaks occurred in the blue shrimp (Penaeus stylirostris). As a result, the captive breeding program in shrimp was shifted from P. stylirostris to the Pacific white shrimp (Penaeus vannamei), and today P. vannamei is the preferred cultured shrimp species globally. With the shift of the shrimp farming industry towards the culture of P. vannamei, the overall push by the industry has been to exclude broodstock animals that test positive for a variety of viruses, including IHHNV. However, because of the lesser impact of this pathogen on P. vannamei, mostly reduced growth and deformities (Runt Deformity Syndrome), its exclusion has probably been less important than other viruses like TSV and WSSV. This global assessment is overdue.
Recent publications by Sellars et al. [56] are worth noting. According to these authors, IHHNV infection has been typically observed to be relatively benign in aquaculture production of the black tiger shrimp (P. monodon). Despite this general principle, the results of their study show that IHHNV infection in this species needs to be considered more seriously than previously thought. Under circumstances when the shrimp were burdened by a sustained, very high infection load, IHHNV was found to greatly compromising growth, general health, survival and pond harvest yields. With yield losses estimated to be worth AU$67,000 gross for a 1-ha commercial pond. Sellars et al. [56] suggest that in cases where IHHNV is highly prevalent in wild broodstock it is fundamental to quantify the infection severity by quantitative polymerase chain reaction (qPCR), such testing should be used as a means of identifying and removing these high-load broodstock before they are conditioned and spawned. This will ensure that only IHHNV-free or IHHNV-low seedstock are used for production.
Still, in countries like Australia where quarantine regulations severely veto the import of live shrimp for culture purposes, efforts to generate and prolong IHHNV-free breeding lines have not been able to break the cycle of reliance on wild-captured broodstock. Until such Specific Pathogen Free (SPF) lines can be established or acquired, screening-based selection will remain the only means of precluding IHHNV from causing production impacts in Australia.
A list of single-stranded DNA viruses in shrimp
A list of diseases of shrimp caused by single-stranded DNA viruses, the diagnostic tools available to detect these diseases, and the biological and genomic properties of these viruses are summarized in Tables 1 and 2.
Table 1.
Clinical signs, pathology and diagnostic methods of single-stranded DNA viruses infecting marine and freshwater shrimp
| Disease | Etiologic agent | Year of emergence | Geographic distribution | Clinical signs | Pathology | Diagnostic methods | OIE-Listed |
|---|---|---|---|---|---|---|---|
| Infectious hypodermal and hematopoietic necrosis | Infectious hypodermal and haematopoietic necrosis virus (IHHNV) or Penaeus stylirostris densonucleosis virus (PstDNV) | 1981 | America, Asia–Pacific, Africa, Madagascar, Middle-East | Acute infection with very high mortalities occurring in juvenile of P. stylirostris (35-day-old or older) life stages. Bluish with opaque abdominal musculature at terminal-phase IHHNV infections In P. vannamei it causes cuticular deformities, specifically a deformed rostrum bent to the left or right (not always apparent in chronically infected shrimp populations). Nonspecific: reduced growth rates. Reduced food consumption, followed by changes in behavior and appearance |
Prominent intranuclear, Cowdry type A inclusion bodies, eosinophilic and often haloed, in cells of tissues of ectodermal and mesodermal origin | Gross signs, Histology, PCR, Electron Microscopy, DNA Probes, In Situ Hybridization, Bioassay | Yes |
| Hepatopancreatic parvovirus | Hepatopancreatic parvovirus (HPV) or Penaeus monodon densovirus(PmDNV) | 1983 | America, Asia–Pacific, Africa, Madagascar, Middle-East | Nonspecific gross signs: hepatopancreas atrophy, anorexia, poor growth rate, reduced preening activities (and consequential increased tendency for surface and gill fouling epicommensal microorganisms) | Intranuclear basophilic inclusion bodies, mostly in actively dividing E-and F-cells, at the distal end of the hepatopancreatic tubules | Histology, PCR, DNA Probes, In Situ Hybridization, Monoclonal Antibody (Mab) based techniques | NO |
| Spawner-isolated Mortality | Spawner-isolated Mortality Virus (SMV) | 1993 | Australia | Lethargy, failure to feed and redness of carapace and pleiopods, red feces. SMV has been associated to the Mid-Crop Mortality Syndrome (MCMS) epizootics reported in P. monodon |
Hemocytic infiltration in the gut, refractile eosinophilic granular material (REGM) in the hepatopancreas and eyestalk. Hemocytic infiltration around the tubules, pyknotic nuclei, large areas of necrosis. Spheroids in the lymphoidal organ | Histology, Electron Microscopy, Bioassay, DNA Probes, In Situ Hybridization | No |
| Lymphoidal parvo-like virus | Lymphoidal parvo-like virus (LPV) | 1991 | Australia | Nonspecific gross signs | Eosinophilic to basophilic distinctly spherical intranuclear inclusion bodies in cells of the LO, hematopoietic organs, in connective tissues of various organs and tissues, gills,. | Histology, Light and Electron Microscopy | No |
Table 2.
Biophysical and genomic properties of single-stranded DNA viruses infecting marine and freshwater shrimp
| Disease | Virus morphology and size | Genomic properties | Viral encoded protein | Classification | Key references |
|---|---|---|---|---|---|
| Infectious hypodermal and hematopoietic necrosis | Icosahedral, non-enveloped, 20 nm | Single, linear, ssDNA, 4.1 kb | ORF1-NS1 ORF2-NS2 ORF3-VP |
Family: Parvoviridae, Sub-family: Densovirinae, Genus: Brevidensovirus |
[6, 7, 14] |
| Hepadensovirus | Icosahedral, non-enveloped, 22–24 nm | Single, linear, ssDNA, 6.3 kb | ORF1-NS2 ORF2-NS1 ORF3-VP |
Family: Parvoviridae, Sub-family: Densovirinae, Genus: Brevidensovirus | [60, 68] |
| Spawner-isolated Mortality | Icosahedral, non-enveloped, 20 nm | ssDNA | ND | Parvovirus, unclassified | [49] |
| Lymphoidal parvo-like virus | Icosahedral, non-enveloped, 25–30 nm | ssDNA | ND | Parvovirus, unclassified | [48] |
Infectious hypodermal and hematopoietic necrosis virus (IHHNV)
Biology of infectious hypodermal and hematopoietic necrosis disease
Infectious hypodermal and hematopoietic necrosis virus is one of the oldest viruses reported in penaeid shrimp. It is also the smallest of the known penaeid shrimp viruses [8]. Infectious hypodermal and hematopoietic necrosis (IHHN) disease was first reported in the US (Hawaii) in 1981 causing acute mortality in juvenile Penaeus stylirostris in super-intensive raceways [44]. Genetic analysis revealed that IHHNV was introduced to the Americas from the Philippines in the early 1970s most likely through the import of IHHNV-contaminated broodstock of P. monodon imported for experimental aquaculture [65]. See a recent review by Dhar et al. [17]. In Hawaii during the same time period, IHHNV was found in P. vannamei however, it did not cause acute mortalities as reported in P. stylirostris, instead it caused deformities referred to as runt deformity syndrome (RDS, Fig. 4) [29]. In 1987, IHHNV was introduced into Mexico through a batch of IHHNV-infected post-larvae of P. vannamei from Hawaii, reviewed by Dhar et al. [17]. By 1990, IHHNV caused elevated mortalities in shrimp farms rearing P. stylirostris in the coastal north western states in Mexico [17]. The virus spread to the wild populations of penaeid shrimp including P. stylirostris, P. vannamei and P. californiensis in the Gulf of California, Mexico which was later associated with the collapse of wild fishery of P. stylirostris in the Northern Gulf of California [39]. Although the wild P. stylirostris fishery recovered IHHNV is now prevalent in 100% of wild populations suggesting the wild population is tolerant/resistant to the virus [17, 53]. Recently, Escobedo-Bonilla and Rangel [21] found distinct susceptibility in three batches of P. vannamei to IHHNV from the same geographical region (Northern Mexico) and indicated that even within the same species there is considerable difference in IHHNV susceptibility.
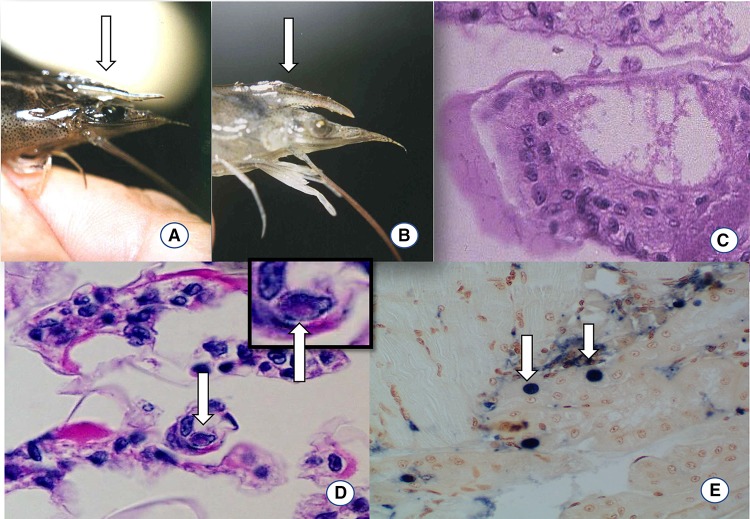
Fig. 4.
Clinical sign and histopathology of IHHNV-infected Penaeus vannamei shrimp. a Example of a juvenile P. vannamei with normal rostrum (arrow). b Example of juvenile P. vannamei, displaying a bent rostrum, an indication of IHHNV infection (arrow). c H&E stained section of a normal gill at ×100. d H&E stained section of a gill showing Cowdry type A inclusion body (arrow) at ×100. In the upper right panel in d, a Cowdry Type A inclusion is shown at a higher magnification (arrow) ×600. e Section of antennal gland of P. stylirostris used in an in situ hybridization using a DIG-labeled IHHNV probe. The blue-purple precipitants are indicative of an IHHNV positive reaction (arrows), ×100
The early detection of IHHNV in South-east Asia including Singapore, Malaysia, Indonesia and the Philippines in shrimp culture facilities that only utilized wild caught P. monodon as broodstock (and previous to the introduction of P. vannamei) indicates that the area is within the natural geographic range of the pathogen, and that P. monodon maybe the natural host [39, 44]. In Thailand, IHHNV appears to cause no problems in the culture of P. monodon.
Although IHHNV could be highly pathogenic to juvenile P. stylirostris [6, 44], shrimp that survive the infection become asymptomatic carriers and can transmit the pathogen by vertical and horizontal transmission [39]. In addition, IHHNV infection causes a decrease in growth rate in infected populations with or without a variety of cuticular deformities of the cephalothorax and abdomen including the rostrum, antennae, thoracic or abdominal segments. These clinical signs together are known as RDS [29]. Recently, Sellers et al. [56], reported reduced growth performance of P. monodon infected with IHHNV indicating that IHHNV still remains as an economically important viral pathogen in shrimp aquaculture.
IHHNV has been reported to infect all life stages including eggs, nauplii, post larvae, juveniles and adults of P. vannamei, the virus is transmitted vertically [46]. Eggs produced by females with high load infections of IHHNV generally fail to develop and hatch. The nauplii produced from these heavily infected females have a high prevalence of IHHNV [46]. Horizontal transmission may occur when healthy shrimp cannibalize on moribund or dead shrimp that are infected or via contaminated water [46].
Diagnosis: histopathology and molecular detection
Histopathology IHHNV produces intranuclear, Cowdry type A (CAIs) inclusions bodies in tissues of ectodermal origin (gills, epidermis, hypodermal epithelium of foregut and hindgut, nerve cord and nerve ganglia) and mesodermal origin (lymphoid organ, connective tissue, antennal gland, gonads and hematopoietic organs). The inclusions occur in the hypertrophied nuclei of infected cells as eosinophilic and surrounded by marginated chromatin (Fig. 4) [22]. Although CAIs are quite representative. The CAIs are comparable to inclusions bodies produced during early stages of WSSV infection. In situ hybridization using a IHHNV-specific probe and/or PCR specific for IHHNV provides a definitive diagnosis [43]. In recent years, the presence of CAIs in P. vannamei is uncommon and the only way to confirm the presence of the pathogen is by in situ hybridization using an IHHNV-specific probe or by PCR specific for IHHNV.
Routine IHHNV detection is conducted by histopathology, in situ hybridization and by PCR [43]. Methods for the detection of IHHNV are described in the Aquatic Animal Health Manual of the World Organization for Animal Health, OIE, Paris, France http://www.oie.int/fileadmin/Home/eng/Health_standards/aahm/current/chapitre_ihhn.pdf. In situ hybridization and dot blot assay using Digoxigenin (DIG)-labeled probes for the detection of IHHNV were the first to be developed for the diagnosis of the disease (Fig. 4) [43, 45]. These methods are more sensitive than detection by histology and could be used to detect the virus in a non-invasive manner using hemolymph and pleopods samples [7] that are particularly useful in screening broodstock [11].
Molecular detection Detection of IHHNV nowadays is one of the major challenges in diagnoses, especially due to the presence of both infectious IHHNV and non-infectious genome integrated forms know as endogenous viral element (EVE) [16, 66]. Several PCR and qPCR methods have been developed to detect IHHNV including [18, 57, 61, 64, 65]. There are many geographical variants of IHHNV that are not detected by a single method. Two sets of primers, 392F/R and 389F/R are most reliable for detecting all the known variants of IHHNV [64, 66]. However, these tests also detect the IHHNV EVE which are integrated into the host genome of certain populations of P. monodon from the western Indo-Pacific, East Africa, Australia and India [63, 66]. In early studies a single PCR (309F/R) was developed to detect the infectious IHHNV that did not amplify IHHNV-EVE present in the P. monodon stocks from Africa, Australia, or Thailand [63, 66]. Primer set MG831F/R can detect only non-infectious genome-integrated Types 3A and 3B [66]. Recently, three new qPCR tests were described using primer sets IHHNV-q309, IHHNV-qEVE and IHHNV 195R/R [13, 16]. While the first two primer sets were developed using a TaqMan assay to detect and quantify infectious and non-infectious forms of IHHNV in shrimp, the later primer set IHHNV 195R/R was developed using a SYBR Green assay for detecting IHHNV in crayfish (Procambarus clarkii).
Since IHHNV is an OIE-listed pathogen, an intensive screening in SPF stocks is conducted by several government agencies in the importation/exportation process. Any PCR/qPCR positive result needs to be confirmed with an alternative diagnostic method such as histology, bioassay or in situ hybridization http://www.oie.int/fileadmin/Home/eng/Health_standards/aahm/current/chapitre_ihhn.pdf.
Molecular biology of IHHNV: biophysical and genomic properties
IHHNV is a small single-stranded DNA (genome ~ 4.1 kb) containing virus with non-enveloped particles, ~ 22 nm in diameter [8]. This is similar to other parvoviruses where positive or negative DNA strands may be encapsidated, but not in the same virus particle [32]. IHHNV is classified as a member of the family Parvoviridae based on morphological, biochemical and genomic properties [8].
Amongst the different members of the family Parvoviridae known to infect marine shrimp, only in IHHNV have the biophysical properties been studied in detail. X-ray crystallography studies reveal that the 20 nm size virus-like particles (VLPs) contain 60 copies of a 37.5 kDa protein that has β–barrel “jelly-roll” motifs analogous to those of other parvoviruses [30].
All members of the family Parvoviridae are single-stranded DNA viruses that contain two large ORFs, located on the same strand of DNA. The first ORF that is located on the 5′-end of the genome encodes two non-structural (NS) proteins, NS1 and NS2 via alternative splicing events [15] and the NS1 protein is involved in viral replication. NS1 is also required for transactivation of viral promoters. The second large ORF, is located on the 3′-end of the genome and encodes capsid protein.
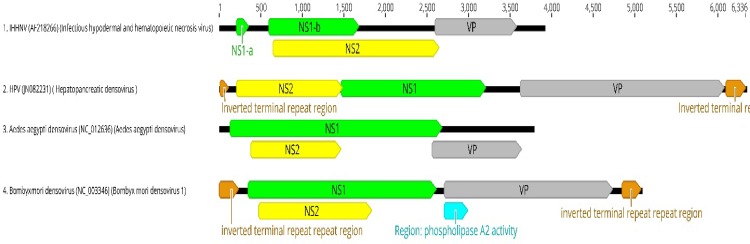
IHHNV was the first shrimp virus for which almost the entire genome was sequenced with the exception of structured regions at the ends of the genome [57]. The IHHNV genome organization is closely related to the densoviruses in the genus Brevidensovirus [57] (Fig. 5). Due to its similarity of the genome organization to other brevidensoviruses. The International Committee for Taxonomy of Viruses (ICTV) has approved the nomenclature for IHHNV as Penaeus stylirostris densovirus (PstDNV).
Fig. 5.
A schematic representation of the genome organization of IHHNV, HPV, Aedes aegypti densovirus and Bombyx mori densovirus. The NS1 ORFs are shown in green, the NS2 ORF are shown in yellow and the viral capsid protein is shown in grey. The inverted terminal repeat regions on HPV and Bombyx mori densovirus genomes are shown in light brown. In Bombyx mori densovirus, the region with phospholipase A2 activity is shown in blue. In IHHNV, the ORF representing NS1-a and the following ORF representing NS1-B are joined upon splicing of the intron located between the two ORFs
The IHHNV genome contains three major open reading frames (ORFs): left, middle and right [57]. The left ORF (referred as NS1-b in Fig. 5) encodes a polypeptide of 666 amino acids with a molecular mass of 75.77 kDa and contains a conserved replication initiation motif, NTP binding and helicase domain. Three putative acceptor sites (A1, A2 and A3) are located at the 5′-end of the left ORF. A small ORF (referred as NS1-a in Fig. 5) is located up stream of the left ORF and contains one proposed 5′ donor site (D1). The NS1 transcript undergoes splicing at D1 and A1 position of NS1-b to generate a mature NS1 transcript [20].
The middle ORF overlays with the left ORF, encodes a protein of 363 amino acids with a molecular mass of 42.11 kDa, this protein shows no similarity with any other proteins reported in online databases. The middle ORF has no assigned function to date. The left ORF also overlaps with the right ORF. The right ORF encodes a protein containing 329 amino acids with a molecular mass of 37.5 kDa and represent the IHHNV capsid protein, VP. In contrast to other Parvoviruses, IHHNV only codes for a single type of capsid protein [57]. A reminiscent of phospholipase A2 (PLA2) catalytic site is present in the N-terminal region of the capsid protein [57].
There are three promoters (P2, P11 and P61) located upstream of the left, middle and right ORFs in the IHHNV genome. The P2 and P61 promoters regulate the expression of NS (1a and 1b) (encoded by left ORF) and VP (encoded by right ORF) genes, respectively, whereas P11 promoter regulates the expression of the middle ORF [20]. The IHHNV promoters are pantropic in nature and have been confirmed to be functional not only in shrimp but also in bacteria, insect and in mammalian cells [19, 20].
Genetic diversity and phylogeny of IHHNV
Initially two different IHHNV genotypes were identified [65]: Type 1 from the Americas and East Asia and the Type 2 from South-East Asia. These genotypes are infectious to P. vannamei and P. monodon. Subsequently, sequences homologous to some parts of the genome of IHHNV were found to be integrated into the chromosomal DNA of P. monodon shrimp as EVE. These insertions were described as Type 3A from East Africa, India and Australia, and Type 3B from the western Indo-Pacific region [63, 66]. Tissues containing the IHHNV-homologous sequences in the P. monodon genome failed to cause infections similar to wild type virus, and hence they are considered as non-infectious [63, 65, 66].
The nucleotide substitution rate for IHHNV is very high (1.39 × 10−4 substitutions/site/year) and is comparable to RNA viruses [53]. These findings differ from what was previously reported by Tang and Lightner [62] based on the capsid gene of 14 IHHNV isolates. In a study by Kim et al. [35] the high rate of evolution for IHHNV was corroborated. To our knowledge, this is the only marine invertebrate parvovirus where high levels of nucleotide substitutions have been estimated. This could play a role in the genetic diversity of IHHNV and has direct implications in PCR- based diagnostics for the detection of all IHHNV genotypes.
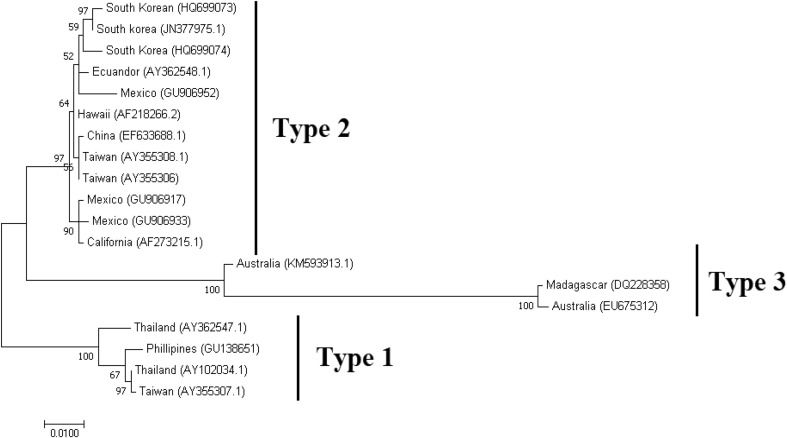
The evolutionary relationships between the distinct IHHNV isolates have been evaluated in numerous studies. The most recent and extensive study was carried out by Robles-Sikisaka et al. [53] where the phylogenetic relationship of 61 IHHNV genotypes were studied. Three clusters were observed (1) one with Mexican southern haplotypes, (2) a second containing western and central Mexican haplotypes, which also contained Asian haplotypes. This illustrates the ancestral relationships of Mexican and Asian IHHNV isolates, (3) finally, a third clade exist that contains a mixture of all geographic isolates [53].
In this review, we analyzed the evolutionary relationships using the IHHNV VP capsid gene from representative geographic isolates from the GenBank database. The multiple alignment of the sequences was made using CLUSTW in Geneious Prime [31] and phylogenetic analysis performed by Maximum Likelihood method using MEGA 7 [36]. The phylogenetic tree shows three defined clades (Fig. 6) that correspond to the suggested genotypes [63, 66]. The first clade closely corresponds to Type 1 and is formed by Southeast Asian isolates including isolates from Thailand, Philippines and Vietnam [63]. The second clade, Type 2 is composed of distinct isolates from the Americas and East Asia. The diversity of this clade shows the effects of the re-introductions due to possible shrimp stocks movements from distinct regions. The third clade, Type 3 contains isolates from the Indo-Pacific region and consists of IHHNV-related non-infectious sequences (Fig. 6).
Fig. 6.
Phylogenetic relationship between distinct geographical isolates of IHHNV determined by Maximum likelihood analysis. The country of origin and the GenBank accession number is given for all the sequences taken for the phylogenetic analysis
Hepatopancreatic parvovirus (HPV)
Biology of disease caused by HPV
Hepatopancreatic parvovirus (HPV) of penaeid shrimp was the original name of a groups of viruses that contained at least three genera including: Penaeus monodon densovirus (PmDNV), Penaeus merguiensis densovirus (PmeDNV) and Penaeus chinensis densovirus (PchDNV) [28]. For the purpose of this review we will call these viruses HPV. Hepatopancreatic parvovirus was first reported in farmed marine prawns P. merguiensis and P. indicus in Singapore [14]. Later on HPV was reported in post-larvae of P. chinensis and P. monodon in the Philippines and P. semisulcatus from Kuwait [41]. HPV is known to infect several wild and cultured penaeid species and is globally distributed [22, 23, 25, 42, 54]. Shrimp affected by HPV usually do not display specific gross signs; however, some signs such as atrophy of the hepatopancreas, anorexia, poor growth rate, presence of epifouling in gills and appendages have been observed [60]. Mortalities associated with HPV during the initial larval stages have been reported from Australia in P. chinensis [58]. A direct correlation between HPV infection and growth stunting has been observed in Thailand in P. monodon [25]. Shrimp positive for HPV were significantly smaller than those from the same pond unaffected by HPV. At a farm level, mortalities due to HPV are difficult to document because HPV is usually found in co-infection with several pathogens where high mortalities have occurred in the early juvenile stages, with cumulative mortalities reaching up to 100% [39]. HPV can cause considerable losses in grow-out ponds without any obvious clinical manifestation.
The transmission of HPV seems to be by both, horizontal and vertical routes. The horizontal transmission of HPV via oral route has been demonstrated in P. monodon PLs [12]. The experimental transmission of HPV to P. vannamei has not been successful at The University of Arizona Aquaculture Pathology Laboratory [39]. Another study using the insect model house cricket (Acheta domesticus) demonstrated horizontal transmission of P. merguiensis densovirus (PmergDNV) [37].
Diagnosis: Histopathology and molecular detection
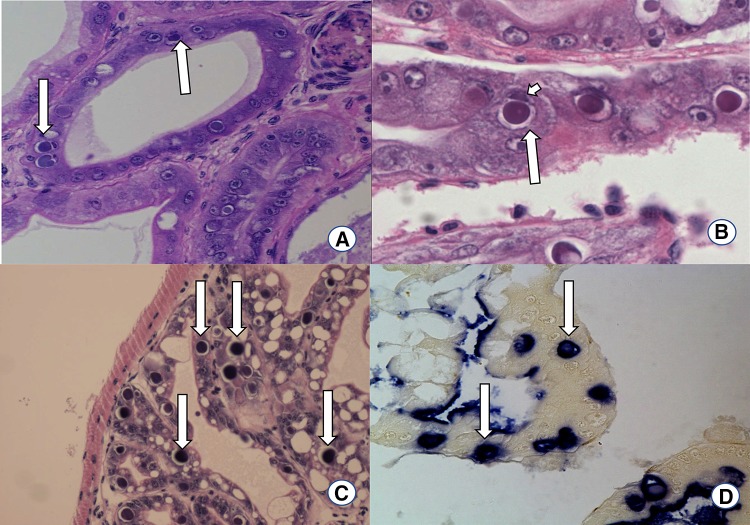
Histopathology Diagnosis of HPV by H&E histology depends of the demonstration of usually single prominent basophilic, Feulgen-positive intranuclear inclusion bodies in the hypertrophied nuclei of hepatopancreatic tubule epithelial cells (Fig. 7). Consequent lateral displacement and compression of the host cell nucleolus, which becomes hypertrophied and remains intimately associated with the developing inclusion body is a key morphological characteristic of an HPV infected nucleus. Chromatin margination is also a typical characteristic of HPV-infected nucleus [39]. The intranuclear inclusion bodies found by H&E, are further confirmed by in situ hybridization using a HPV-specific gene probe [9]. There are some specific probes that are known to react with HPV from Asia, Africa Australia and the Americas but do not present a positive reaction with the HPV present in fresh water Prawn (Macrobrachium rosenbergii) from Malaysia. Recently some HPV-like intranuclear inclusions was recorded in M. rosenbergii at the UA-APL by H&E stain and in situ hybridization that are similar to those reported in penaeids shrimp (Fig. 7).
Fig. 7.
Histopathology of HPV-infected Penaeus vannamei shrimp. a H&E stained section of a hepatopancreas tubule of P. vannamei showing intranuclear basophilic inclusion bodies (arrows), ×100. b H&E stained section of a hepatopancreas tubule of P. vannamei showing intranuclear basophilic inclusion bodies (Large arrow). Displacement and compression of nucleoli are also presented (Small arrow), ×600 c H&E stained section of a hepatopancreas tubules of M. rosenbergii showing intranuclear basophilic inclusion bodies (arrows), ×100. d Section of hepatopancreas tubules cells displaying positive reaction in in situ hybridization using HPV-specific DIG labeled probe. The blue-purple precipitants indicated by arrows are reminiscence of HPV infection (arrows), ×200
Molecular detection Molecular diagnostic tools were developed for a rapid detection of HPV that allows early diagnosis of infection in shrimp larvae, juvenile broodstock and in carrier host. It is worth noting that due to the presence of insertions or deletions in HPV genome in the several strains, for a PCR-based detection of HPV, it is important to either design primers based on the conservative regions in the genome that will enable to detect all isolates or alternatively design PCR test for a specific strain(s). For instances, there is a 30% difference in DNA sequence between HPV in China (HPVchin) and HPV in P. monodon (HPVmon) [51]. A PCR based detection method for HPV in P. monodon (HPVmon) was developed by Sukhumsirichart et al. [59] using the primer sets 121F and 276R that amplifies a 156 bp product and this PCR assay could detect at as low as 1.0 fg of purified HPV DNA. Pantoja et al. [50] designed another primer set (1120F/1120R) based on the sequences of HPV in China (HPVchin) that amplifies a 592 bp product. Another set of primers for HPVmon (H441F and H441R) were published that provides an amplicon of 441 bp and the sensitivity of detection was as low as 1.0 fg of purified HPVmon DNA [51]. The primer set HPV441F/HPV441R, specifically designed for HPVmon has been shown to be detect HPV in P. monodon from India as well.
The wild as well as cultured shrimp are often infected with more than one virus. A multiplex reverse transcription-polymerase chain reaction was developed to detect six major shrimp DNA/RNA viruses including HPV, YHV, WSSV, TSV, IHHNV and MBV [33]. However, the sensitivity of HPV detection by multiplex PCR is greatly reduced [59]. The conventional diagnostic methods involve killing of the organism (broodstock, post larvae, and etc.), however in recent years non-invasive virus detection method by PCR and nested PCR have been developed to detect this enteric pathogen using fecal samples without the need to sacrificing the animals [50, 51, 59].
Molecular biology of HPV
Biophysical and genomic properties
The virus particles are icosahedral in morphology measuring 22–24 nm in length. The viral genome contains a 6321 nucleotide long, negative sense, single-stranded DNA [60]. The 5′ and 3′ ends of the genome contain hairpin-like structures. There are three large ORFs in the HPV genome with the ORFs 1 and 2 partly overlapping (Fig. 5). The left ORF (ORF1), mid ORF (ORF2), and right ORF (ORF3) on the plus (complementary) strand potentially coding for three proteins of 428, 579, and 818 amino acids with molecular masses of 50, 68, and 92 kDa, respectively. ORF1 (NS2) encodes a putative nonstructural protein (Fig. 5). ORF2 (NS1) contains replication initiator motif and NTP-binding and helicase domains. The ORF3 encodes a capsid protein (VP). Apart from the larger genome, the organization of the genome of HPV is very distinct from IHHNV, this highlights the diversity that exists among brevidensoviruses infecting shrimp.
Genetic diversity and phylogeny of HPV
The nucleotide variation calculated using the full-length genomes sequences of HPV isolates from Korea, Thailand, Australia and Tanzania, was found to vary between 12 and 21% [67]. At a protein level, genetic variation is highest in ORF3 with a mean genetic distance of 24% and lowest in ORF2 with a mean genetic distance of 7% [38, 67].
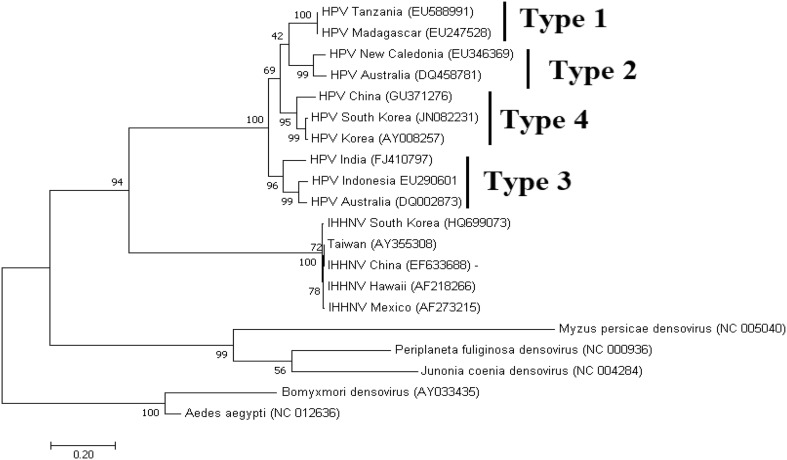
In order to assess phylogenetic relationships among geographically distinct HPV isolates both NS1 and VP1 amino acid sequences were used in several studies. Three distinct HPV genotypes were reported [28, 67]: Type I include isolates from Korea, Madagascar and Tanzania; Type II includes isolates from Thailand and Indonesia, while Type III contains HPV isolates from Australia and New Caledonia [28, 67].
To reevaluate the evolutionary relationships of HPV isolates to other shrimp/insect-infecting parvoviruses, we performed an analysis by Maximum Likelihood method using a multiple alignment of VP1 nucleotide sequences with the MEGA 7 program [36]. The alignment of the sequences was made using CLUSTW in Geneious Prime [31]. The phylogenetic analysis shows all HPV formed a highly supported cluster (Fig. 8). Furthermore, 4 sub clusters are clearly distinguishable that represent the four HPV genotypes. This topology was previously reported by Dhar et al. [17]. The IHHNV isolates formed a distinct separate sister clade indicating a common origin for shrimp parvovirus. A third clade is formed by all insect parvoviruses.
Fig. 8.
Phylogenetic relationship of HPV and IHHNV to insect brevidensoviruses determined by Maximum likelihood analysis. The country of origin and the GenBank accession number is given for all the sequences taken for the phylogenetic analysis
Management of diseases caused by IHHNV and HPV
Disease prevention
The biology of the diseases caused by IHHNV and HPV poses some challenge with respect to their management. IHHNV is a systemic pathogen and listed in the OIE-list of crustacean diseases which implies that once it is detected it must be reported, even in the absence of clinical signs. On the other hand, HPV is an enteric pathogen and does not have much clinical manifestation except stunted growth in the affected populations and it is not included as a OIE-listed pathogen. This leaves no choice but to screen animals regularly for these pathogens irrespective of any clinical manifestation. The application of periodic PCR screening of all life stages and eliminating those that test positive for these viruses is long known to be an effective strategy in minimizing the prevalence of these pathogens.
At farm level, two different approaches have been recently recognized in order to deal with the possible presence of shrimp pathogens. One is the preventative approach that consist of avoidance of pathogens using SPF broodstock and exclusion of potential vectors in culture systems. This strategy is considered as the most effective way to minimize the risk of infection and mitigate the losses caused by pathogens and it is practiced more in south east Asia. On the contrary, in the western hemisphere a commonly used practice is the “inclusion strategy”. This strategy consists of the utilization of All Pathogen Exposed (APE) shrimp where the population has been exposed to multiple pathogens including WSSV, TSV, IHHNV and AHPND for several years (Aranguren et al., unpublished). Animals develop some tolerance/resistance upon continuous exposure to endemic pathogens. The mass selection process carried out to select broodstock to produce the next generations relies in the selection of healthy stocks based on clinical signs including the larger shrimp without any runt or deformity sign. It has been speculated that this selection process has led to the development of shrimp lines that are tolerant to Necrotizing hepatopancreatitis NHP inadvertently [3].
Often, the conditions that the shrimp are cultured such as high stocking density, poor water quality and the inappropriate aquaculture practices contribute to disease introduction and spread. It has been proposed that the supplementation of probiotic bacteria capable of oxidizing wastes (bioaugmentation) could be useful in improving water quality in shrimp ponds. There is a growing interest in the use of prebiotics in shrimp aquaculture. Enhancement of general health conditions in shrimp via the incorporation of compounds such as beta-1, 3-glucans, peptidoglycans, and polysaccharides, into commercial shrimp feed has been shown to stimulate the non-specific immune mechanisms in shrimp. These broad practices may help prevent parvovirus and other viral infections in shrimp aquaculture.
Therapeutic approach: viral inhibition by RNA interference (RNAi)
RNAi therapy in shrimp aquaculture has been successfully tested at experimental levels against a number of viral pathogens like WSSV, YHV, TSV [47, 55, 68], as well as IHHNV [4] although there is no commercially available therapy as of now. It has been demonstrated that RNAi inhibits IHHNV replication both preventatively and therapeutically, within 24 h post infection [26]. Administration of target-specific dsRNA by intramuscular injection showed significant inhibition of viral transcription, as compared with non-specific dsRNA control. There were two distinct RNAi molecules were made to target ORF1-2 and ORF3. Remarkably, the efficiency of gene silencing was higher for the ORF1-2 design as compared to ORF3. The ORF1-2 encodes non-structural proteins whereas ORF3 encodes the viral capsid protein. Since the abundance of transcript representing structural genes are generally more than the non-structural genes, RNAi data suggest that large amounts of viral transcript representing capsid gene may negatively alter the potency of the method. However, it has not been determined if the levels of viral transcripts are the only factor affecting the efficacy of RNAi therapy.
The efficacy of gene silencing in controlling HPV have been observed with the injection of dsRNA into HPV-infected shrimp [4]. The authors showed higher efficacy in eradicating the virus with dsRNA targeting two sites, NS1 and VP genes (ORF2-3), as compared to targeting only NS1 gene [4]. Delivery of RNAi molecules (dsRNA) via injection has been the main routes of administration in laboratory settings. While this approach may be feasible for broodstock, an oral delivery method at farm level still needs to be developed. Methods for oral delivery of the dsRNA are being investigated, with a focus on nanocarriers, including chitosan nanoparticles, cationic liposomes, calcium phosphate, antibodies, cholesterol, aptamers and virus-like particles (VLP) [27]. Recently, IHHNV-VLP carrying plasmid DNA of a marker gene, EGFP was administered via intramuscular injection and was found to successfully deliver the payload into muscle and gills in P. vannamei [34]. This opens a possibility of using VLP as a carrier to deliver RNAi molecules in shrimp.
A study by Attasart et al. [5] was carried out to investigate whether RNAi could be delivered in shrimp by consumption of bacteria expressing dsRNA targeting Rab7 and STAT genes. Target gene suppression was observed in the hepatopancreas and gills implying that the induction of the RNAi was systemic. The results suggest that oral administration of dsRNA using a bacterial expression system could be a feasible method of delivery of dsRNA to shrimp in large scale aquaculture settings. However, oral administration of dsRNA may present some significant challenges, including the off-target activity, innate immune response, and possible toxicity. In addition, the commercial application of the antiviral dsRNA therapy in shrimp for human consumption may raise some safety concerns about introducing synthetic dsRNA molecules and pose a regulatory issue. A recent study by Rao et al. [52] demonstrates that the dsRNA does not transfer to progeny from the broodstock females, indicating that the RNAi used as means of prevention before spawning should not pose a risk to human health.
Viral fragments inserted into the shrimp genome are hypothesized to confer acquired immunity against specific viruses via RNAi pathway [24]. Reverse-transcriptase (RT) present in the shrimp genome, would generate cDNA complementary to mRNA of incoming viruses, and integrase (IN), also present in the genome, would incorporate those fragments into the host genome. Some of the randomly inserted fragments could be then transcribed to viral specific, complimentary pseudogene, that would bind to the viral mRNA transcript and form a dsRNA. This would trigger the RNAi pathway, leading to silencing of cognate viral genes [17]. If this hypothesis is confirmed, it will open the door to creating non-GMO shrimp lines immune to specific viral infections.
Concluding remarks
As shrimp industry grows worldwide and intensive farming is practiced increasingly, the emergence of diseases is inevitable. Often diseases have been propagated well before the identification of the causal agent, the development of diagnostic tools, or understanding the transmission routes and disease epidemiology. The emergence and global distribution of IHHNV in shrimp aquaculture represents an example in this scenario. Among all viral pathogens, parvoviruses especially IHHNV has received much attention for many historical reasons. IHHNV is the first shrimp viral pathogen for which almost the entire genome was sequenced [57] and diagnostic tools such as gene probes [45] and real-time PCR-based assays were developed [18, 61], and the cross protection phenomenon was demonstrated. Despite all the progress IHHNV still remains as a concern in shrimp aquaculture. It remains to be determined how IHHNV and HPV can avoid the host immune response and continue to exist in an infected animal without displaying clinical signs. Understanding the mechanisms that enable shrimp to carry viral infection(s) without a lethal effect may lead to the development of antiviral therapies.
Although there is a lack of availability of any therapeutic measure against parvoviruses, and for that matter for any shrimp viruses, RNAi-based therapy looks promising. The challenge remains in delivering the therapeutic molecules via oral route that can be adapted to develop therapeutic feed for farm level applications. Administering therapeutic molecules using VLP delivery vehicles are opening new opportunities in shrimp disease control. This is a relatively unexplored field and it is likely to receive more attention in near future.
Acknowledgements
Partial funding for this work was provided by College of Agricultural and Life Sciences to Arun K Dhar.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alday-Sanz V. The shrimp book. 1st ed. Alday-Sanz V, editor. Nottingham: Nottingham University Press; 2010.
- 2.Anderson BJL, Valderrama D, Jory DE. Global shrimp production review and forecast: steady growth ahead [Internet]. Glob. Aquac. Alliance. 2018 [cited 2019 Feb 25]. pp. 5–10. https://www.aquaculturealliance.org/advocate/global-shrimp-production-review-and-forecast-steady-growth-ahead/.
- 3.Aranguren LF, Tang KFJ, Lightner DV. Quantification of the bacterial agent of necrotizing hepatopancreatitis (NHP-B) by real-time PCR and comparison of survival and NHP load of two shrimp populations. Aquaculture. 2010;307:187–192. doi: 10.1016/j.aquaculture.2010.07.022. [DOI] [Google Scholar]
- 4.Attasart P, Kaewkhaw R, Chimwai C, Kongphom U, Panyim S. Clearance of Penaeus monodon densovirus in naturally pre-infected shrimp by combined ns1 and vp dsRNAs. Virus Res. 2011;159:79–82. doi: 10.1016/j.virusres.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Attasart P, Namramoon O, Kongphom U, Chimwai C, Panyim S. Ingestion of bacteria expressing dsRNA triggers specific RNA silencing in shrimp. Virus Res. 2013;171:252–256. doi: 10.1016/j.virusres.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Bell TA, Lightner DV. IHHN virus: infectivity and pathogenicity studies in Penaeus stylirostris and Penaeus vannamei. Aquaculture. 1984;38:185–194. doi: 10.1016/0044-8486(84)90142-X. [DOI] [Google Scholar]
- 7.Bell TA, Lightner DV, Brock JA. A biopsy procedure for the nondestructive determination of infectious hypodermal and hematopoietic necrosis virus (IHHNV) infection in Penaeus vannamei. J Aquat Anim Health. 1990;2:151–153. doi: 10.1577/1548-8667(1990)002<0151:ABPFTN>2.3.CO;2. [DOI] [Google Scholar]
- 8.Bonami J-R, Trumper B, Mari J, Brehelin M, Lightner DV. Purification and characterization of the infectious hypodermal and haematopoietic necrosis virus of penaeid shrimps. J Gen Virol. 1990;71:2657–2664. doi: 10.1099/0022-1317-71-11-2657. [DOI] [PubMed] [Google Scholar]
- 9.Bonami JR, Mari J, Poulos BT, Lightner DV. Characterization of hepatopancreatic parvo-like virus, a second unusual parvovirus pathogenic for penaeid shrimps. J Gen Virol. 1995;76:813–817. doi: 10.1099/0022-1317-76-4-813. [DOI] [PubMed] [Google Scholar]
- 10.Browdy CL, Jory DE. The Rising Tide, Proceedings of the Special Session on Sustainable Shrimp Farming. World Aquac 2009. Lousiana, USA; 2009.
- 11.Carr WH, Sweeney JN, Nunan L, Lightner DV, Hirsch HH, Reddington JJ. The use of an infectious hypodermal and hematopoietic necrosis virus gene probe serodiagnostic field kit for the screening of candidate specific pathogen-free Penaeus vannamei broodstock. Aquaculture. 1996;147:1–8. doi: 10.1016/S0044-8486(96)01291-4. [DOI] [Google Scholar]
- 12.Catap ES, Lavilla-Pitogo CR, Maeno Y, Traviña RD. Occurrence, histopathology and experimental transmission of hepatopancreatic parvovirus infection in Penaeus monodon postlarvae. Dis Aquat Organ. 2003;57:11–17. doi: 10.3354/dao057011. [DOI] [PubMed] [Google Scholar]
- 13.Chen B, Dong Z, Pang N, Nian Y, Yan D. A novel real-time PCR approach for detection of infectious hypodermal and haematopoietic necrosis virus (IHHNV) in the freshwater crayfish Procambarus clarkii. J Invertebr Pathol. 2018;157:100-1003. [DOI] [PubMed]
- 14.Chong Y, Loh P. Hepatopancreas chlamydial and parvovirus infections of farmed marine prawns in Singapore. Singapore Vet J. 1984;51–6.
- 15.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, et al. The family Parvoviridae. Arch Virol. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowley JA, Rao M, Coman GJ, Cowley J. Real-time PCR tests to specifically detect infectious hypodermal and haemopoietic necrosis virus (IHHNV) lineages and an IHHNV endogenous viral element (EVE) integrated in the genome of black tiger shrimp (Penaeus monodon). 2018;129:145–58. [DOI] [PubMed]
- 17.Dhar AK, Robles-Sikisaka R, Saksmerprome V, Lakshman DK. Biology, Genome Organization, and Evolution of Parvoviruses in Marine Shrimp. Adv Virus Res. 1st ed. Elsevier Inc.; 2014. p. 85–139. [DOI] [PubMed]
- 18.Dhar AK, Roux MM, Klimpel KR. Detection and quantification of infectious hypodermal and hematopoietic necrosis virus and white spot virus in shrimp using real-time quantitative PCR and SYBR green chemistry. J Clin Microbiol. 2001;39:2835–2845. doi: 10.1128/JCM.39.8.2835-2845.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhar AK, Lakshman DK, Natarajan S, Allnutt FCT, van Beek NAM. Functional characterization of putative promoter elements from infectious hypodermal and hematopoietic necrosis virus (IHHNV) in shrimp and in insect and fish cell lines. Virus Res. 2007;127:1–8. doi: 10.1016/j.virusres.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Dhar AK, Kaizer KN, Lakshman DK. Transcriptional analysis of Penaeus stylirostris densovirus genes. Virology. 2010;402:112–120. doi: 10.1016/j.virol.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Escobedo-Bonilla CM, Rangel JLI. Susceptibility to an inoculum of infectious hypodermal and haematopoietic necrosis virus (IHHNV) in three batches of whiteleg shrimp Litopenaeus vannamei (Boone, 1931) Zookeys. 2014;365:355–365. doi: 10.3897/zookeys.457.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flegel TW. Special topic review: major viral diseases of the black tiger prawn (Penaeus monodon) in Thailand. World J Microbiol Biotechnol. 1997;13:433–442. doi: 10.1023/A:1018580301578. [DOI] [Google Scholar]
- 23.Flegel TW. Detection of major penaeid shrimp viruses in Asia, a historical perspective with emphasis on Thailand. Aquaculture. 2006;258:1–33. doi: 10.1016/j.aquaculture.2006.05.013. [DOI] [Google Scholar]
- 24.Flegel TW. Hypothesis for heritable, anti-viral immunity in crustaceans and insects. Biol Direct. 2009;4:32. doi: 10.1186/1745-6150-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flegel TW, Nielsen L, Thamavit V, Kongtim S, Pasharawipas T. Presence of multiple viruses in non-diseased, cultivated shrimp at harvest. Aquaculture. 2004;240:55–68. doi: 10.1016/j.aquaculture.2004.06.032. [DOI] [Google Scholar]
- 26.Ho T, Yasri P, Panyim S, Udomkit A. Double-stranded RNA confers both preventive and therapeutic effects against Penaeus stylirostris densovirus (PstDNV) in Litopenaeus vannamei. Virus Res. 2011;155:131–136. doi: 10.1016/j.virusres.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Itsathitphaisarn O, Thitamadee S, Weerachatyanukul W, Sritunyalucksana K. Potential of RNAi applications to control viral diseases of farmed shrimp. J Invertebr Pathol. 2017;147:76–85. doi: 10.1016/j.jip.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Jang IK, Jeeva S, Seo HC, Lee Y-S, Choi T-J, Kang S-W. Complete nucleotide sequence analysis of a Korean strain of hepatopancreatic parvovirus (HPV) from Fenneropenaeus chinensis. Virus Genes. 2011;44:89–97. doi: 10.1007/s11262-011-0675-8. [DOI] [PubMed] [Google Scholar]
- 29.Kalagayan H, Godin D, Kanna R, Hagino G, Sweeney J, Wyban J, et al. IHHN virus as an etiological factor in runt-deformity syndrome (RDS) of Juvenile Penaeus vannamei cultured in Hawaii. J World Aquac Soc. 1991;22:235–243. doi: 10.1111/j.1749-7345.1991.tb00740.x. [DOI] [Google Scholar]
- 30.Kaufmann B, Bowman VD, Li Y, Szelei J, Waddell PJ, Tijssen P, et al. Structure of Penaeus stylirostris densovirus, a shrimp pathogen. J Virol. 2010;84:11289–11296. doi: 10.1128/JVI.01240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly DC, Elliott RM. DNA contained by two densonucleosis viruses. JVirol. 1977;21:408–410. doi: 10.1128/jvi.21.1.408-410.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khawsak P, Deesukon W, Chaivisuthangkura P, Sukhumsirichart W. Multiplex RT-PCR assay for simultaneous detection of six viruses of penaeid shrimp. Mol Cell Probes. 2008;22:177–183. doi: 10.1016/j.mcp.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Kiatmetha P, Santimanawong W, Chotwiwatthanakun C, Jariyapong P, Ounjai P, Weerachatyanukul W. Nanocontainer designed from an infectious hypodermal and hematopoietic necrosis virus (IHHNV) has excellent physical stability and ability to deliver shrimp tissues. PeerJ. 2018;6:e6079. doi: 10.7717/peerj.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JH, Choresca CH, Jr, Shin SP, Han JE, Jun JW, Han SY, et al. Detection of infectious hypodermal and hematopoietic necrosis virus (IHHNV) in Litopenaeus vannamei shrimp cultured in South Korea. Aquaculture. 2011;313:161–164. doi: 10.1016/j.aquaculture.2011.01.019. [DOI] [Google Scholar]
- 36.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis Version 7. 0 for Bigger Datasets. Mol B. 2018;33:1870–4. [DOI] [PMC free article] [PubMed]
- 37.La Fauce KA, Owens L. The use of insects as a bioassay for Penaeus merguiensis densovirus (PmergDNV) J Invertebr Pathol. 2008;98:1–6. doi: 10.1016/j.jip.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 38.La Fauce KA, Elliman J, Owens L. Molecular characterisation of hepatopancreatic parvovirus (PmergDNV) from Australian Penaeus merguiensis. Virology. 2007;362:397–403. doi: 10.1016/j.virol.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 39.Lightner DV. Epizootiology, distribution and the impact on international trade of two penaeid shrimp viruses in the Americas. Rev Sci Tech l’OIE. 1996;15:579–601. doi: 10.20506/rst.15.2.944. [DOI] [PubMed] [Google Scholar]
- 40.Lightner V. Biosecurity in shrimp farming : pathogen exclusion through use of SPF stock and routine surveillance. J World Aquac Soc. 2005;36:230–248. [Google Scholar]
- 41.Lightner DV, Redman RM. A parvo-like virus disease of penaeid shrimp. J Invertebr Pathol. 1985;45:47–53. doi: 10.1016/0022-2011(85)90048-5. [DOI] [PubMed] [Google Scholar]
- 42.Lightner D, Redman RM. Penaeid virus diseases of the shrimp culture industry of the Americas. In: Fast A, Lester L, editors. Mar shrimp cult princ pract. Amsterdam: Elsevier; 1992. pp. 569–588. [Google Scholar]
- 43.Lightner DV, Redman RM. Shrimp diseases and current diagnostic methods. Aquaculture. 1998;164:201–220. doi: 10.1016/S0044-8486(98)00187-2. [DOI] [Google Scholar]
- 44.Lightner DV, Redman RM, Bell T, Brock J. Detection of IHHN virus in Penaeys stylirostris and P. vannamei imported into Hawaii. J World Maric Soc. 1983;225:212–225. [Google Scholar]
- 45.Mari J, Bonami JR, Lightner D. Partial cloning of the genome of infectious hypodermal and haematopoietic necrosis virus, an unusual parvovirus pathogenic for penaeid shrimps; diagnosis of the disease using a specific probe. J Gen Virol. 1993;74:2637–2643. doi: 10.1099/0022-1317-74-12-2637. [DOI] [PubMed] [Google Scholar]
- 46.Motte E, Yugcha E, Luzardo J, Castro F, Leclercq G, Rodríguez J, et al. Prevention of IHHNV vertical transmission in the white shrimp Litopenaeus vannamei. Aquaculture. 2003;219:57–70. doi: 10.1016/S0044-8486(02)00631-2. [DOI] [Google Scholar]
- 47.Ongvarrasopone C, Saejia P, Chanasakulniyom M, Panyim S. Inhibition of Taura syndrome virus replication in Litopenaeus vannamei through silencing the LvRab7 gene using double-stranded RNA. Arch Virol. 2011;156:1117–1123. doi: 10.1007/s00705-011-0952-9. [DOI] [PubMed] [Google Scholar]
- 48.Owens L, De Beer S, Smith J. Lymphoidal parvovirus-like particles in Australian penaeid prawns. Dis Aquat Organ. 1991;11:129–34. doi: 10.3354/dao011129. [DOI] [Google Scholar]
- 49.Owens L, Haqshenas G, McElnea C, Coelen R. Putative spawner-isolated mortality virus associated with mid-crop mortality syndrome in farmedPenaeus monodon from northern Australia. Dis Aquat Organ. 1998;34:177–85. doi: 10.3354/dao034177. [DOI] [PubMed] [Google Scholar]
- 50.Pantoja CR, Lightner DV. A non-destructive method based on the polymerase chain reaction for detection of hepatopancreatic parvovirus (HPV) of penaeid shrimp. 2000;39:177–82. [DOI] [PubMed]
- 51.Phromjai J, Boonsaeng V, Withyachumnarnkul B, Flegel TW. Detection of hepatopancreatic parvovirus in Thai shrimp Penaeus monodon by in situ hybridization, dot blot hybridization and PCR amplification. Dis Aquat Organ. 2002;51:227–232. doi: 10.3354/dao051227. [DOI] [PubMed] [Google Scholar]
- 52.Rao M, Cowley JA, Murphy BS, Stratford CN, Sellars MJ. Double-stranded RNA injected into female black tiger shrimp (Penaeus monodon) prior to spawning does not transfer to progeny. Aquaculture. 2019;500:126–134. doi: 10.1016/j.aquaculture.2018.09.059. [DOI] [Google Scholar]
- 53.Robles-Sikisaka R, Bohonak AJ, McClenaghan LR, Dhar AK. Genetic signature of rapid IHHNV (infectious hypodermal and hematopoietic necrosis virus) expansion in wild penaeus shrimp populations. PLoS One. 2010;5:e11799. [DOI] [PMC free article] [PubMed]
- 54.Safeena MP, Rai P. Molecular biology and epidemiology of hepatopancreatic parvovirus of penaeid shrimp. Indian J Virol. 2012;23:191–202. doi: 10.1007/s13337-012-0080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saksmerprome V, Charoonnart P, Gangnonngiw W, Withyachumnarnkul B. A novel and inexpensive application of RNAi technology to protect shrimp from viral disease. J Virol Methods. 2009;162:213–217. doi: 10.1016/j.jviromet.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Sellars MJ, Cowley JA, Musson D, Rao M, Menzies ML, Coman GJ, et al. Reduced growth performance of black tiger shrimp (Penaeus monodon) infected with infectious hypodermal and hematopoietic necrosis virus. Aquaculture. 2019;499:160–166. doi: 10.1016/j.aquaculture.2018.09.032. [DOI] [Google Scholar]
- 57.Shike H, Dhar AK, Burns JC, Shimizu C, Jousset FX, Klimpel KR, et al. Infectious hypodermal and hematopoietic necrosis virus of shrimp is related to mosquito brevidensoviruses. Virology. 2000;277:167–177. doi: 10.1006/viro.2000.0589. [DOI] [PubMed] [Google Scholar]
- 58.Spann K, Adlard R, Hudson D, Pyecroft S, Jones T, Voigt M. Hepatopancreatic parvo-like virus (HPV) of Penaeus japonicus cultured in Australia. Dis Aquat Organ. 1997;31:239–241. doi: 10.3354/dao031239. [DOI] [Google Scholar]
- 59.Sukhumsirichart W, Wongteerasupaya C, Boonsaeng V, Panyim S, Sriurairatana S, Withyachumnarnkul B, et al. Characterization and PCR detection of hepatopancreatic parvovirus (HPV) from Penaeus monodon in Thailand. Dis Aquat Organ. 1999;38:1–10. doi: 10.3354/dao038001. [DOI] [PubMed] [Google Scholar]
- 60.Sukhumsirichart W, Attasart P, Boonsaeng V, Panyim S. Complete nucleotide sequence and genomic organization of hepatopancreatic parvovirus (HPV) of Penaeus monodon. Virology. 2006;346:266–277. doi: 10.1016/j.virol.2005.06.052. [DOI] [PubMed] [Google Scholar]
- 61.Tang KFJ, Lightner DV. Detection and quantification of infectious hypodermal and hematopoietic necrosis virus in penaeid shrimp by real-time PCR. Dis Aquat Organ. 2001;44:79–85. doi: 10.3354/dao044079. [DOI] [PubMed] [Google Scholar]
- 62.Tang K, Lightner D. Low sequence variation among isolates of infectious hypodermal and hematopoietic necrosis virus (IHHNV) originating from Hawaii and the Americas. Dis Aquat Organ. 2002;49:93–97. doi: 10.3354/dao049093. [DOI] [PubMed] [Google Scholar]
- 63.Tang KFJ, Lightner DV. Infectious hypodermal and hematopoietic necrosis virus (IHHNV) -related sequences in the genome of the black tiger prawn Penaeus monodon from Africa and Australia. Virus Res. 2006;118:185–191. doi: 10.1016/j.virusres.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Tang KFJ, Durand SV, White BL, Redman RM, Pantoja CR, Lightner DV. Postlarvae and juveniles of a selected line of Penaeus stylirostris are resistant to infectious hypodermal and hematopoietic necrosis virus infection. Aquaculture. 2000;1:203–210. doi: 10.1016/S0044-8486(00)00407-5. [DOI] [Google Scholar]
- 65.Tang K, Poulos B, Wang J, Redman R, Shih H, Lightner D. Geographic variations among infectious hypodermal and hematopoietic necrosis virus (IHHNV) isolates and characteristics of their infection. Dis Aquat Organ. 2003;53:91–99. doi: 10.3354/dao053091. [DOI] [PubMed] [Google Scholar]
- 66.Tang KFJ, Navarro SA, Lightner DV. PCR assay for discriminating between infectious hypodermal and hematopoietic necrosis virus (IHHNV) and virus-related sequences in the genome of Penaeus monodon. Dis Aquat Organ. 2007;74:165–170. doi: 10.3354/dao074165. [DOI] [PubMed] [Google Scholar]
- 67.Tang K, Pantoja C, Lightner D. Nucleotide sequence of a Madagascar hepatopancreatic parvovirus (HPV) and comparison of genetic variation among geographic isolates. Dis Aquat Organ. 2008;80:105–112. doi: 10.3354/dao01928. [DOI] [PubMed] [Google Scholar]
- 68.Tirasophon W, Yodmuang S, Chinnirunvong W, Plongthongkum N, Panyim S. Therapeutic inhibition of yellow head virus multiplication in infected shrimps by YHV-protease dsRNA. Antiviral Res. 2007;74:150–155. doi: 10.1016/j.antiviral.2006.11.002. [DOI] [PubMed] [Google Scholar]