Abstract
Single-stranded (ss)DNA viruses are extremely widespread, infect diverse hosts from all three domains of life and include important pathogens. Most ssDNA viruses possess small genomes that replicate by the rolling-circle-like mechanism initiated by a distinct virus-encoded endonuclease. High throughput genome sequencing and improved bioinformatics tools have yielded vast information on presence of ssDNA viruses in diverse habitats. The simple genome of ssDNA viruses have high propensity to undergo mutation and recombination often emerging as threat to human civilization. Interestingly their genome is found embedded in fossils dating back to million years. The unusual evolutionary history of ssDNA viruses reveal evidences of horizontal gene transfer, sometimes between different species and genera.
Keywords: ssDNA, CRESS viruses, Rolling circle replication
Introduction
Recent advances in metagenomic sequencing involving large population of viruses in environmental samples have revealed an astonishing volume of virome in every habitat in the biosphere. It is estimated [56, 58] that viral biomass is about 200 million tonnes which if placed end to end would span 65 galaxies. Untargeted de novo metagenomics and bioinformatics tools have radically changed our understanding of viruses; their diverse nature in genome organization, regulation of gene expression, replication mechanism and their interaction with cellular hosts are keenly analyzed to improve our perception. Metagenomics analysis of thousands of samples had led to approximately 16-fold increase in the number of viral genes identified and above 1,25,000 new viral genomes have been identified. Analysis of viral distribution across diverse ecosystem such as marine or freshwater, atmospheric air, soil, hot springs and habitats like insects, plants, human tissue have revealed that the majority of viral sequences identified are ssDNA viruses with either circular or linear DNA genome. Excellent reviews by Krupovic et al. [31], Kryukov et al. [32], Rosario et al. [50], Liu et al. [36], Zhao et al. [63] on various aspects of ssDNA viruses may kindly be referred. The current issue of virus diseases is dedicated to discuss the discovery, diversity, importance and evolutionary trends in this group of viruses.
The ssDNA viruses constitute an economically, medically and ecologically important group infecting hosts from all three domain of life viz., bacteria, archaea and eukarya. The viruses infecting human beings and domesticated animals do not cause major diseases; however those viruses which are arthropod vector borne and infect plants cause major diseases affecting agricultural productivity. The ssDNA viruses consist of a very minimal genetic blue print of a structural coat protein (CP) gene and replication-associated protein (Rep). Most of the ssDNA viruses have circular DNA as genome and together this group is referred to as circular Rep encodings DNA viruses (CRESS DNA viruses) and is suggested to share common ancestor as they all exhibit rolling circle replication [50].
Discovery
The ancient origin of the ssDNA viruses is borne out by molecular analysis of more than ~ 4000 eukaryotic genome which have endogenized ssDNA virus sequences [32]. These genome fossils give good indications of ssDNA viruses occurrence earlier than what is being hypothesized. For example integrated Rep sequence of the begomovirus in tobacco in South America, puts the virus origin to be nearly 1.8 million year ago [34]. The earliest historical record of the disease caused by ssDNA virus is the yellowing or vein clearing disease in Eupatorium species; the yellowing of the weed plant was an inspiration for an Japanese Empress to pen a poem as early as 752 AD [54]. In animals circovirus infection was observed in Australia in 1888 and was reported to cause bald birds [3]. Despite its ancient origin and ubiquitous presence, the particle morphology and circular ssDNA genome was first delineated only in 1977 for bean golden mosaic virus [15, 19]. The first animal infecting circular DNA virus characterized was porcine circovirus [60, 61].
Application of phi29 DNA polymerase and random hexamer mediated rolling circle amplification (RCA), various types of PCR such as direct PCR, inverse PCR, single primer based PCR, etc., have resulted in explosion of CRESS DNA viral discovery [35, 49].
Taxonomy
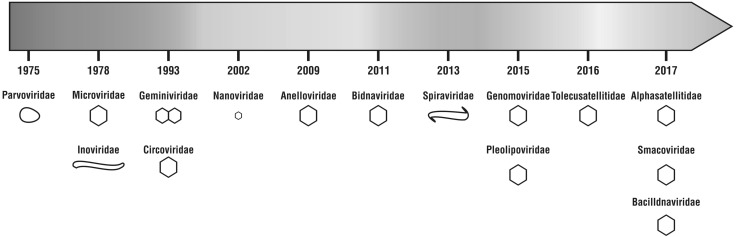
At present nearly 70,000 nucleotide sequence are available in NCBI database belonging to ssDNA virus group. The taxonomy of ssDNA viruses has undergone lot of revisions since 2015, as more and more sequences are available by metagenomics mining. The bacterial ssDNA viruses have been assembled into families fairly early (1978) compared to viruses infecting Eukaryons, in which Parvoviridae member infecting humans are the earliest recognized group. The years between 2015 and 2017 resolved the grouping of CRESS-DNA viruses (Fig. 1). Currently thirteen families have been established (ICTV, http://ictv.global/report) of which eleven families contain circular genome, two families, Parvoviridae and Bidnaviridae have a linear single stranded DNA genome. Seven of this eleven families infect eukaryotes they are Anelloviridae, Bacilladnaviridae, Circoviridae, Geminiviridae, Genomoviridae, Nanoviridae and Smacoviridae; It is interesting to note here that the new family Genomoviridae comprises only a single fungal infecting virus; but more than 120 viral members detected in the insect tissue have also been included in it. The striking feature of members of Anelloviridae is they have negative sense ssDNA genome unlike other ssDNA viruses. It may also be noted that the family Smacoviridae has no cultured representation though they have been detected in faecal matter of mammals and dragonflies. The single stranded DNA viruses infecting bacteria belong to families Microviridae, Inoviridae and those which are harboured by archaea are grouped under two families, Pleolipoviridae and Spiraviridae. Besides viruses, the satellite DNA molecules associated with Nanoviruses and geminiviruses have been reclassified into two families, Alphasatellitidae [6] and Tolecusatellitidae. The details on genome size, capsid morphology, host species infected and number of the genera in each family are given in Table 1.
Fig. 1.
Classification of ssDNA viruses into families with progress of time
Table 1.
Overall properties of ssDNA viruses
| Host | Virus taxon | Sub family | Genera | Virion morphology Size in nms |
Genome morphology | Genome size | Replication |
|---|---|---|---|---|---|---|---|
| Bacteria | Microviridae | 2 | 3 | Icosahedral 30 | Circular | 4.4–6.1 | RC-Rep |
| Inoviridae | – | 7 | Filamentous/rod shaped, 7 × 700–2000 | Circular | 4.5–12.4 | RC-Rep | |
| Archaea | Pleolipoviridae (haloarchaea) | – | 3 | Pleomorphic 40 | Circular ss or ds | 7–10.6 | RC-Rep |
| Spiraviridae (crenarchaea) | – | 1 | Coil shaped antenna like projection 28 × 200 | Circular | 24.9 | Unknown | |
| Eukarya | Anelloviridae (human) | – | 12 | Icosahedral 30–32 | Circular | 2–4 | RC-Rep |
| Bidnaviridae (silk worm) | – | 1 | Icosahedral 25 | Linear segmented with terminal inserted repeats | 6–6.5 per segment | DNA pol B | |
| Circoviridae (birds and animals) | – | 2 | Icosahedral 20 | Circular | 1.7–2.3 | RC-Rep | |
| Geminiviridae (plants) | – | 9 | Icosahedral twinned 22 × 38 | Circular | 2.5–3 | RC-Rep | |
| Nanoviridae (plants) | – | 2 | Icosahedral 18–19 | Circular | 0.9–1.1 per segment | RC-Rep | |
| Smacoviridae (fecal matter of mammals, dragonflies) | – | 6 | Icosahedral unknown Most probably icosahedral |
Circular | 2.2 | RC-Rep | |
| Parvoviridae (vertebrates and arthropods) | 2 | 5 and 8 | Icosahedral 18–26 | Linear TIR | 4–6.3 | RHR-NS1, NS2 | |
| Genomoviridae (one definite mycovirus several in mammals birds insert, plants, sewage and sediments) | – | 9 | Icosahedral 20 | Circular | 2.2 | RC-Rep | |
| Bacilladnaviridae (diatoms) | – | 3 | Icosahedral 33–38 | Circular, partially ds | 5.8–6 | RC-Rep | |
| Alphasatellitidae (plants) | 2 | 4 and 7 | Icosahedral 18–19 | Circular | 0.9–1.1 | RC-Rep | |
| Tolecusatellitidae (plants) | 0 | 2 | Icosahedral 18–19 | Circular | 1.3 | Not coding |
Unifying features
The basic anchoring feature that unite most of the ssDNA viruses is the replication initiation protein/Rep, which shows great degree of conservation; contrastingly CP is highly variable and in the case of circular genome the arrangement of Rep/CP ORFs either divulge or converge towards the origin of replication. In this respect, ssDNA viruses resemble prokaryotic rolling circle plasmids probably indicating evolutionary relationship. All the eukaryotic ssDNA viruses (established members and CRESS virus groups) express Rep and replicate via rolling circle replication (RCR) [17].
The Reps of ssDNA viruses consist of well organised two functional domains. They are HUH (His hydrophobic His) endonuclease domain in the N terminus and super family 3 (SF3) helicase domain at the C terminus [22, 25]. The HUH endonuclease domain is characterised by RCR motifs I, II and III which are essential for RCR initiation and termination. The SF3 helicase domain contains Walker A, Walker B and Walker C motifs which lead to replicative helicase activity during RCR elongation [16]. Kazlauskas et al. [23] identified another catalytic arginine finger in Rep encoded by members of Bacilladnaviridae, Circoviridae, Nanoviridae and Smacocviridae, but not in the Geminiviridae and Genomoviridae [23]. In majority of CRESS DNA viruses both spliced (Rep) and non spliced forms of Rep (Rep A) are expressed, they have multifunctional role in virus genome replication, transcription and gene expression regulation.
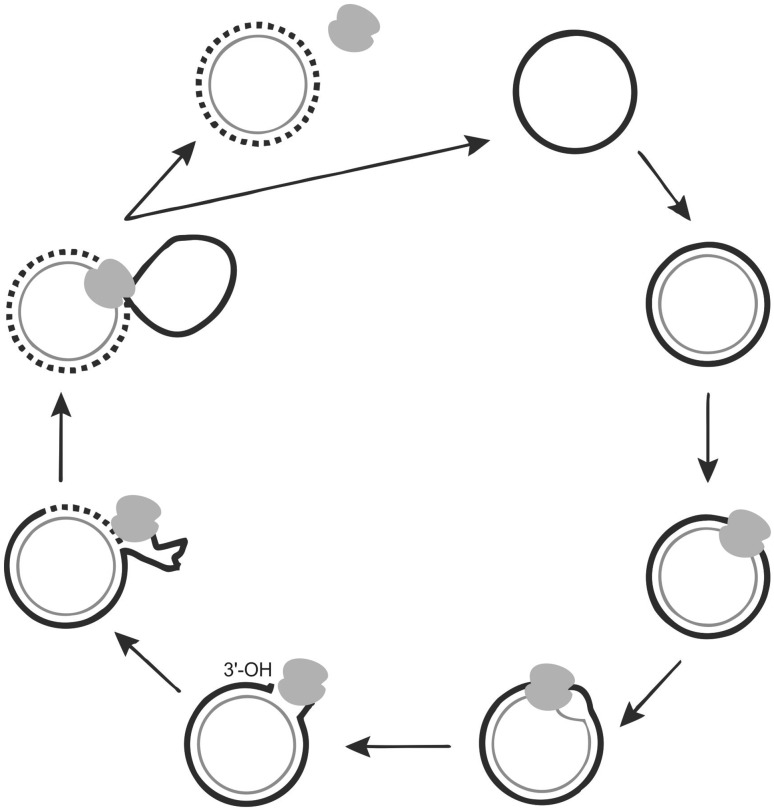
In the circular DNA viruses, the ssDNA virus is converted to transcriptionally active, covalently closed circular DNA by host DNA polymerase (Fig. 2). The dsDNA molecules associate with histone proteins and get assembled into minichromoses suitable for replication. The Rep protein expressed from the viral genome cleaves at specific sites (origin of replication v-ori) at the virion strand to produce replicative form of DNA. The free 3′ OH end is used by host DNA polymerase to synthesize the new strand using the circular strand as template. As the new strands are synthesized, old strands are progressively displaced, after one or two circles, the old displaced strands are ligated to yield circular monomeric or concatameric ssDNA virion strands. The circularized new strands are either encapsidated or converted into replicative forms to enter another round of replication [18].
Fig. 2.
Schematic representation of rolling circle replication (RCR) in circular ssDNA viruses. The encapsidated ssDNA released into host cell enters nucleus and is converted to dsDNA by host enzymes. Viral encoded Rep binds to dsDNA at iteron sequences and creates nick at nonanucleotide sequences to generate 3′ OH end. Rep is covalently bound to 5′ PO4 end. Host DNA polymerase synthesizes the virion sense strand using negative strand as template. As new strand is synthesized, old strand is displaced. When replication is completed Rep joins the nicks and releases initial strand from the replication complex
The v-ori (viral strand, origin of replication) has been identified in all the circular DNA viruses and defined by 10–30 nt long inverted repeat sequence capable of forming a hairpin structure and an highly conserved nonanucleotide sequence [38, 53]. The high level of homology observed in Rep gene among the divergent eukaryotic CRESS viruses is used extensively for a phylogenetic analysis and taxonomical classification [56].
It is not yet known whether anelloviruses replicate via rolling circle mechanism. Detection of dsDNA in liver bone marrow cells of people infected with anelloviruses, is suggestive of similar type of replication. The torque teno virus (TTV) has somewhat similar origin of replication as observed in geminivirus, nanovirus and circovirus replication [42, 43]. On the basis of analysis of high molecular weight double stranded DNA in the host cells recombination dependent replication (RDR) is suggested to be a major mechanism of replication in geminivirus, nanovirus, circovirus and even prokaryotic infecting microviruses [40].
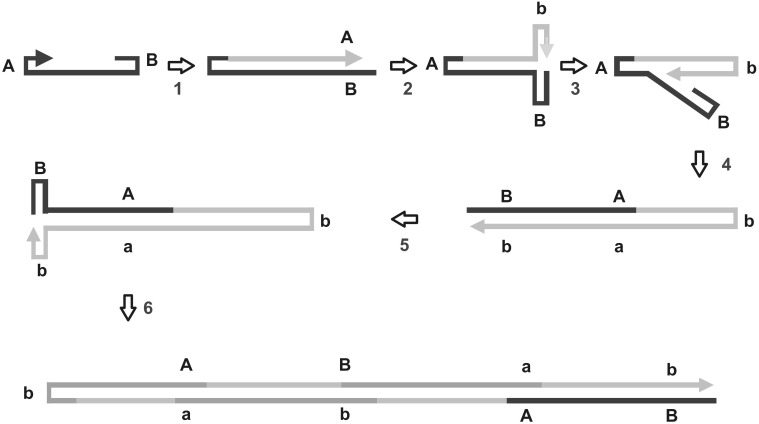
Members of the family Parvoviridae are small non enveloped viruses and DNA genome is single stranded and linear the coding region is bracketed by short (121–421) palindromic terminal sequences. They exhibit rolling hairpin replication (RHR); parvovirus members have only two genes, one for the capsid protein (VP1 and VP2) and the other for nonstructural proteins (NS1 and NS2). The replication is dependent on NS1, a multifunctional nuclear phosphoprotein with helicase, ATPase, transcriptional regulation sequence specific DNA recognition site and strand specific nicking function. In parvoviruses rolling hairpin replication (Fig. 3) is initially primed by 3′ hydroxyl end of the 3′ terminal hairpin which initiates the production of a linear dsDNA molecule. At this point, replication complex switches from the parental strand to identical sequence in new strand and replication continues to produce ds DNA molecules with paired hairpin at one end. This process continues back and forth producing head to head and tail to tail, concatamers which are cleaved by Rep homologue NS1. The displaced ssDNA strand are then packaged [8, 59].
Fig. 3.
Schematic representation of rolling hairpin replication (RHR). Upon infection of host cell, 3′ end of the linear ssDNA serves as primer and host enzymes convert ssDNA to ds DNA. Replication is initiated by viral endonuclease NS1 which creates nicks between coding region and hairpin at 3′ region. New strand is synthesized by host DNA polymerase. Instead of simply ending at this point, the replication complex switches from the parental strand to the identical sequence on the newly synthesized strand and replication continues to produce a dsDNA molecule with paired hairpins at one end. This process then continues back and forth producing head to head and tail to tail, genomic concatemers within which the palindromic genome ends are replicated half as frequently as the coding regions. New single stranded breaks are introduced at the replication origins of these dsDNA molecules (by NS1), which result in the formation of new replication forks that, starting with the replication of the palindromic ends, displace ssDNA strands that are ready for packaging.
Adapted from Martin et al. [40]
Virus members of the family Bidnaviridae which include small isometric viruses that infect the silkworm Bombyx mori have entirely different mode of replication. The genome of bidnavirus consists of two linear ssDNA fragments 6.5 kb (VD1) and 6 kb (VD2) which are packaged into separate virions. Bombyx mori bidensovirus (BxBDV) encodes a type B DNA polymerase and is predicted to replicate its genome via the protein primed mechanism reminiscent of that characterized in adenoviruses. The superhelicase S3H is encoded by VD1 and the endonuclease domain of NS1 is not observed in BxBDV. Pol B proteins of bidnaviruses with other protein, primed replicating viruses such as adenoviruses, tectiviruses contain a N terminal 400 aminoacid domain which is also present in virus like transposon elements Polinto Pol B. They may be involved in priming the initiation of replication. Krupovic and Koonin [29] suggest that bidnaviruses which were initially grouped in Parvoviridae have evolved from a parvoviruses ancestor (capsid protein organization similarity) but acquired Pol B gene to have a different mode of replication. Besides, bidnaviruses code for receptor binding protein and a novel antiviral modulator derived from dsRNA viruses (Reoviridae) and dsDNA viruses, (Baculoviridae) [29].
Capsid protein
The virion particles of the most of the ssDNA viruses have icosahedral morphology, except Inoviridae and Spiraviridae with helical morphology: the pleomorphic 40 nm size enveloped virions are met with only in the case of Pleolipoviridae infecting archaea. In the case of the family Geminiviridae twinned icosahedral capsid particles package a single genome component [19].
The coat protein is assembled either with one protein (e.g.: Geminiviridae) or several coat protein molecules (e.g. Parvoviridae). In all cases wherever high resolution structural information is available, CP of ssDNA viruses were found to display a jelly-roll (antiparallel eight stranded β barrel) fold which is found in majority of positive sense ssRNA viruses infecting eukaryotes. However, the families do not share any similarity at sequence level [26, 27]. The CP genes of many single stranded DNA viruses have been analysed to show evolutionary linkage with many RNA viruses. The capsid proteins of geminiviruses are organised similar to ssRNA plant viruses satellite tobacco necrosis virus [30].
The most unexpected observations are on a unique group of CRESS DNA virus group isolated from boiling spring lake by Diemer and Stedman [11]. The group is given the name as Cruciviruses [46] and more members are being characterized [9, 51, 57]. They represent a RNA–DNA hybrid virus encoding a CRESS DNA viral Rep and CP from unclassified ssRNA viruses similar to Tombusviridae.
Ecology and distribution of ssDNA viruses
ssDNA viral genome sequences have been detected in diverse environments, associated with diverse life forms which need to be analyzed to speculate the global impact such distribution will have. For example CRESS DNA viruses have been isolated from faecal matter of bats, chimpanzies, rodents. It was interesting to see that 700 years old frozen faecal matter of caribou contained ssDNA virus sequences. Interestingly the caribou virus was also shown to be infectious on Nicotiana benthamiana though the plants were symptom free [41]. Viruses have been recorded in the abdomen tissue of insects such as dragonflies, cockroach, damselfly, mosquitoes; nepatopancreas tissue of shrimps. Abundance of bacilladnaviruses in marine sediment is suggestive of its role in marine algal bloom and concomitant modulation of zooplankton.
Three of eukaryotic CRESS viruses are well established pathogens. Geminiviruses and nanoviruses infect economically important crop plants and cause damaging diseases affecting the agricultural productivity. The circoviruses infect both vertebrates and invertebrates. In animals, PCV1 contributes to viral diseases of pigs (post weaning multisystemic wasting syndrome), later it was found to be causative agent for a collection of syndrome known as PCAD including respiratory disease, enteric diseases and reproductive problems [44]. Other circoviruses are known to infect livestock or human companion animals.
In the case of newly identified CRESS DNA viruses identified by metagenomic survey detectable disease symptoms are not yet observed. Where the viruses were detected in faecal matter, cerebrospinal fluid of humans like CyCV, it is speculated that they will emerge as potential pathogen. Abundance of bacilladnaviruses infecting diatoms could be used to curb toxic algal growth. Since many CRESS DNA viruses infect invertebrates, they may play important role in maintenance of food chain and biogeochemistry of global environment.
Endogenisation and horizontal gene transfer of ssDNA viruses
Deep sequencing of eukaryotic genome increasingly has thrown out abundant and more diverse viral sequences. Endogenisation of retroviruses are very common, which comprises of ~ 8% of human genome. Data on genome fossils provide information on host alteration and host virus interactions [20]; such virus integration might also affect the evolutionary history of the host. One of the typical examples of CRESS DNA viruses endogenisation is that of tobacco geminiviruses Rep homolog found in tobacco species [4], they suggest that integration event might have occurred in common hosts a million year ago.
Circovirus like sequences have been observed in animal host genome and integration events of parvoviruses and circoviruses are estimated to be before 40–50 million years [5, 10, 36, 36]. Recently Kryukov et al. [32] analyzed about 4000 eukaryotic genome and found homologous ssDNA viruses endogenized, more than 50% of these sequences hits were seen with plant genome whether these sequences will become active, and whether whole viral genome can be rescued is one major avenue of research to be carried out. Filloux et al. [12] reported that endogenized geminivirus sequence are active in yam and small RNA transcripts were observed.
Interestingly endogenous sequences in host genome cluster with exogenous viruses currently infecting similar hosts in a phylogenetic analysis. For example cyclovirus elements found in insects are similar to those associated with arthropods. Viral genome endogenized data will also inform when, where and which hosts were infected by the ancestors of the currently circulating related virus.
ssDNA viruses adopt different mechanism to integrate into cellular organism and it differs between viruses infecting, bacteria/archaea/eukaryotes. For example the filamentous bacteriophages (Inoviridae), exhibit three mechanisms; they may encode integrase of serine/tyrosine superfamilies; may use DDE transposases type; or as observed in some members of Microviridae and Inoviridae the host XerCD recombination machinery may be exploited. Contrastingly eukaryotic viruses depend on endonucleases activity of RCR similar to transposon like integration [28].
Horizontal gene transfer
Living organism acquire genes not only by vertical transmission but also from other distantly related species through horizontal gene transfer. In the case of bacterial genome integrated phages (prophage) contribute to such transfer. In eukaryotes retroviruses which integrate into host genome play a critical role in gene transfer.
Recent genome studies clearly established the presence of ssDNA virus, Rep protein sequence in vast array of organisms. Rep like genes were also formed in genomes of parasitic protozoan Entamoeba histolytica and Giardia instinalis [14]. Since Rep proteins do not have a cellular homologues a search was made among cellular genomes for the presence of Rep like sequences [36]. They performed phylogenetic analysis to determine the relationship between endogenous viruses and known established CRESS DNA viruses. The endogenous sequences separated into three, geminivirus like, nanovirus like and circovirus like clades. In each clade endogenous virus sequence clustered within known virus; however is did not fall into established viral families. Suggesting that these endogenous viruses might have originated from ancestor of different ssDNA virus lineage.
An exception is the sequence detected in opossum (Monodelphis domestica), which clustered within Circoviridae and was closely related to pig circoviruses. The interesting finding was with Rep like sequences recorded in the tree species Populus. The sequence were degenerate with lot of modifications and these sequences occupied position at the base of the geminiviridae clade indicating that insertion into this species genome happened million years ago.
The copy numbers of integrated viral sequences generally were less than 10 copies species; however more than sixty copies were identified in genome of louse and honeybee mite. Liu et al. [36] have identified geminivirus like transposons is an ectomycorrhizal fungus (Tuber melanosporum) when sequences were reconstructed, it revealed Rep like ORF, two transposons ORF and terminal inserted repeats. Similarly, parvovirus like repetitive element was also recorded in acorn worm. All these findings suggest that eukaryotic transposons could have originated from ssDNA viruses.
By analysis of endogenous virus sequences with EST database it was clear that at least some endogenous viral sequences are expressed as observed in the case of Magnaporthe grisea. It is possible that these viral genes are co-opted to perform cellular function in eukaryotic genome.
Truncated Rep protein gene sequence was detected in mitochondrion of Oomyctes Phytophthora sojae which contained geminivirus AL1 domain. Two circovirus Rep sequences were found in the genome of canarypox virus but not in other pox viruses. Most of the ssDNA virus like sequences were related to Rep gene, only three sequences were related to CP gene. Liu et al. [36] discovered endogenous virus like sequences in at least 35 species among nuclear genomes of plants fungi, animals and protists. Surprisingly no anellovirus like sequences were recorded in any eukaryotic genome. All the viruses like sequences in fungi were related to Sclerotinia sclerotiarum hypovirulence associated DNA virus 1 (SsHADV-1).
Irrespective of sequence diversity, the conserved motif Rep proteins were found in all endogenous virus like sequences. The endogenous sequences are generally interspersed within non coding region of the host genome. In rare cases where they are found within host genes or transposon they have influenced host genome evolution through gene perturbation.
Circoviruses are known to infect only birds and pigs. But recently sequence of these viruses have been detected in dragonflies, fish and humans. Likewise gemini and nanovirus sequences are also found in various environmental samples. Endogenisation of parvo and densovirusesin human and chimpanzee faecal matter was also recorded The endogenous viral sequences identified in recent studies are molecular fossils of invasion of these hosts by viruses, which means the host range of ssDNA virus is much more extended than we have understood.
Evolutionary trends
The eukaryotic ssDNA viruses infecting plants and pet animals emerge as threatening pathogens due to high rate of substitution in genome. This has seen well established by analyzing dataset where statistically detectable recombinational events have been removed. tomato yellow leaf curl virus, maize streak virus and sugarcane streak Reunion virus in Geminiviridae, faba bean yellow necrotic virus in Nanoviridae are such examples. High substitution rate of CRESS DNA viruses often do not result in change in the protein coding genes over a long period of evolution. This time dependent substitution rates is more an adaptation to retain function of essential genes required for biological fitness [1].
It is all the more puzzling how high substitution rates compared to RNA viruses occur in ssDNA viruses, as the replication is mainly mediated by host polymerase. In bacterial viruses such as Phage PhiX14 and M13, they were shown to mutate more rapidly than their host E. coli. Ritchie et al. [47] analyzed circovirus genome and suggested that unpaired single stranded DNA bases may be oxidatively damaged leading to high rates of transition. In begomoviruses substitution are strand specific, which indicate the possibility of oxidative damage while the genome is encapsidated. Presently the accurate measurement of substitution rates in eukaryotic ssDNA viruses and mechanism behind are yet to be understood clearly.
Recombination
Much more than the mutation rates CRESS DNA viruses are known to be capable of frequent recombination. Successful recombinants often emerge as more virulent pathogen. Recombination bring in genetic change more than one point mutation resulting in observable phenotypic change. Recombination between two genera, begomoviruses Rep and mastrevirus CP resulted in emergence of member of a new genus Curtovirus [52]; emergence of severe Ug strain of ACMV/EACMV [45], cotton and tomato leaf curl virus [37], are well known in the family Geminiviridae. Recently emergence of porcine circoviruses 3 which has PCV Rep, an avian circovirus CP [13] was observed. The recombination may occur when a gene is simultaneously replicated and transcribed, the paused polymerases may switch the template resulting in recombination [40]. There are more recombination breakpoints located in the antisense gene in genome, the orientation where replication and transcription meet from opposite direction. Recombination is the key process for generation of variability and the origin of replication is considered to represent the hot spot. High rates of homologous and non-homologous recombination and component exchange are recorded to occur within and between different ssDNA virus species. Lefeuvre et al. [33] observed that there is a significant tendency for recombination to occur either outside or on the peripheries of genes, than within genes. From which it is inferred that there is natural selection operating against expression of recombinant protein. The recombination events occur between strain of same species and also between species sharing < 90% sequence identity. Fewer recombination breakpoints occur within CP gene.
In multipartite viruses (nanoviruses, begomoviruses) sequence similarity in the origin of replication will facilitate reassociation of genomic components, which is also referred to as component capture. The recombinants are known to spread faster with increased virulence, biologically fitter than wild ones. Recombination is recorded to occur in parvoviruses [55], microviruses [48] anelloviruses [39] circoviruses nano and geminiviruses components including DNA B and satellites [2, 40].
Origin and evolution of ssDNA viruses
In the context of ever-growing presence of diverse ssDNA viruses, it is interesting to speculate their origin and how different lineages might have separated from each other. Krupovic et al. proposed that geminivirus may have originated from a phytoplasmal plasmid, which would have acquired capsid protein gene from ssRNA plant virus. However, recent reports on geminivirus like mycovirus and numerous such related sequences suggest that geminiviruses and fungal ssDNA virus are more closely related, than phytoplasma plasmid.
A phylogenetic analysis [36] based on rep like protein from plants, fungi phytoplasma and algae clearly revealed that the plant geminal Reps clustered together with fungal reps, however SsHADV-1 is very distinct from geminiviruses in capsid gene and genome organisation. Therefore it can be inferred that from a common ancestor SsHADV-1 like viruses would have separated long time ago, followed an independent unique path to evolution in their host.
Zhao et al. [63] analysed recombination free data set of 32 Rep sequences of 6 recognized families and CRESS viruses and showed that (a) Genomoviridae Rep is closely related to Geminiviridae, could have a monophyletic grouping (b) Nanoviridae sequences form a clade with Reps from satellite DNA alphasatellite and are closely related to smacoviruses. The bacilladnaviruses are distant from all other eukaryotic viruses.
Unlike capsid proteins, Reps show significant similarity between different groups of CRESS DNA viruses, so it is widely used as marker to estimate the diversity and relationship. Surprisingly, Kazlauskas et al. [24] found that Rep nuclease and helicase domain exhibit incongruent evolutionary histories. The analyses indicated that Reps encoded by members of Bacilladnaviridae, Circoviridae, Geminiviridae, Genomoviridae, Nanoviridae and Smacoviridae display largely congruent evolutionary pattern. But the unclassified CRESS DNA viruses 71% seem to have chimeric Rep. They represent a dynamic population in which exchange of gene fragment of the nuclease and helical domain is frequent. Each one of the group may represent a monophyletic cluster.
Transboundary movement
With increasing trade and movement of agricultural commodities it is inevitable that ssDNA viruses move across continents. Emergence of ToLCNDV in Europe and spread of TYLCV spreading through Caribbean island into central and North America, the new world squash leaf curl virus affecting cucurbit production in Asia Minor are some examples to cite how plant infecting geminiviruses are dispersed across boundaries of nation. By doing phylogenetic analysis it is possible to trace the pathway of the long distance movement of virus. Livestock trading has been shown to be significant in PCV2 emergence in many countries [62]. Like BFDV spread out of Australia may also have resulted due to pet trade. Analysis of large number of sampling over a long period is required to work out the pathway of these viruses which are emerging as serious threat to human welfare.
Concluding remarks
The ssDNA viruses constitute the widespread, diverse and important group of viruses affecting all three domains of life. Metagenomic mining and characterization of Rep like sequences have led to explosion of CRESS DNA virus data which reveal the ubiquitous presence of these viruses in diverse habitat. The high substitution rates and recombination along with capturing of genomic components have resulted in emergence of new strains/species/genera with increase in their biological fitness. Endogenizations of viral genome and horizontal gene transfer happening across different hosts have revealed the past history of many of the viruses. Whatever information gathered is only tip of the iceberg. Detection of ssDNA virus like sequences frequently in large number of eukaryotes, suggest that their origin and invasion of hosts might have occurred long time ago and genome fossils embedded in eukaryotic genome only reveal the past lineages of viruses. Considering its presence in characteristic ecological niches, it may be speculated that ssDNA viruses may have a bigger role in modulation of global ecology and environment.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aiewsakun P, Katzourakis A. Endogenous viruses: connecting recent and ancient viral evolution. Virology. 2015;479–480:26–37. doi: 10.1016/j.virol.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Amin I, Mansoor S, Amrao L, Hussain M, Irum S, Zafar Y, Bull SE, Briddon RW. Mobilisation into cotton and spread of a recombinant cotton leaf curl disease satellite—brief report. Arch Virol. 2006;151:2055–2065. doi: 10.1007/s00705-006-0773-4. [DOI] [PubMed] [Google Scholar]
- 3.Ashby E. Notes on Psephotus haematonotus, the red-rumped grass Parrakeet. Avic Mag. 1921;12:131–133. [Google Scholar]
- 4.Bejarano ER, Khashoggi A, Witty M, Lichtenstein C. Integration of multiple repeats of geminiviral DNA into the nuclear genome of tobacco during evolution. Proc Natl Acad Sci USA. 1996;93:759–764. doi: 10.1073/pnas.93.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belyi VA, Levine AJ, Skalka AM. Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the Parvoviridae and Circoviridae are more than 40 to 50 million years old. J Virol. 2010;84:12458–12462. doi: 10.1128/JVI.01789-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briddon RW, Martin DP, Roumagnac P, Navas-Castillo J, Fiallo-Olive E, Moriones E, Lett J-M, Zerbini FM, Varsani A. Alphasatellitidae: a new family with two subfamilies for the classification of geminivirus- and nanovirus-associated alphasatellites. Arch Virol. 2018 doi: 10.1007/s00705-018-3854-2. [DOI] [PubMed] [Google Scholar]
- 7.Cotmore SF, Tattersall P. Resolution of parvovirus dimer junctions proceeds through a novel heterocruciform intermediate. J Virol. 2003;77:6245–6254. doi: 10.1128/JVI.77.11.6245-6254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotmore SF, Tattersall P. Parvoviruses: small does not mean simple. Annu Rev Virol. 2014;1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- 9.Dayaram A, Galatowitsch ML, Arguello-Astorga GR, van Bysterveldt K, Kraberger S, Stainton D, Harding JS, Roumagnac P, Martin DP, Lefeuvre P, Varsani A. Diverse circular replication-associated protein encoding viruses circulating in invertebrates within a lake ecosystem. Infect Genet Evol. 2016;39:304–316. doi: 10.1016/j.meegid.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Dennis TPW, Flynn PJ, Marciel de Souza W, Singer JB, Moreau CS, Wilson SJ, Gifford RJ. Insights into circovirus host range from the genomic fossil record. J Virol. 2018;9:2e00145-18. doi: 10.1128/JVI.00145-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diemer GS, Stedman KM. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol Direct. 2012;7:13. doi: 10.1186/1745-6150-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filloux D, Murrell S, Koohapitagtam M, Golden M, Julian C, Galzi S, Uzest M, Rodier-Goud M, D’Hont A, Vernerey MS, Wilkin P, Peterschmitt M, Winter S, Murrell B, Martin DP, Roumagnac P. The genomes of many yam species contain transcriptionally active endogenous geminiviral sequences that may be functionally expressed. Virus Evol. 2015;1:vev002. doi: 10.1093/ve/vev002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzo G, Segales J, Tucciarone CM, Cecchinato M, Drigo M. The analysis of genome composition and codon bias reveals distinctive patterns between avian and mammalian circoviruses which suggest a potential recombinant origin for porcine circovirus 3. PLoS ONE. 2018;13:e0199950. doi: 10.1371/journal.pone.0199950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs MJ, Smeianov VV, Steele JL, Upcroft P, Efimov BA. Two families of rep-like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol Biol Evol. 2006;23:1097–1100. doi: 10.1093/molbev/msj122. [DOI] [PubMed] [Google Scholar]
- 15.Goodman RM. Infectious DNA from a whitefly-transmitted virus of Phaseolus vulgaris. Nature. 1977;266:54. doi: 10.1038/266054a0. [DOI] [Google Scholar]
- 16.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure–function relationships. Curr Opin Struct Biol. 1993;3:419–429. doi: 10.1016/S0959-440X(05)80116-2. [DOI] [Google Scholar]
- 17.Gutierrez C, Ramirez-Parra E, Castellano MM, Sanz-Burgos AP, Luque A, Missich R. Geminivirus DNA replication and cell cycle interactions. Vet Microbiol. 2004;98:111–119. doi: 10.1016/j.vetmic.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol. 2013;11:777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- 19.Harrison BD, Barker H, Bock KR, Guthrie EJ, Meredith G, Atkinson M. Plant viruses with circular single-stranded DNA. Nature. 1977;270:760. doi: 10.1038/270760a0. [DOI] [Google Scholar]
- 20.Hayward A, Katzourakis A. Endogenous retroviruses. Curr Biol. 2015;25:R644–R646. doi: 10.1016/j.cub.2015.05.041. [DOI] [PubMed] [Google Scholar]
- 21.Hefferon KL, Moon YS, Fan Y. Multi-tasking of nonstructural gene products is required for bean yellow dwarf geminivirus transcriptional regulation. FEBS J. 2006;273:4482–4494. doi: 10.1111/j.1742-4658.2006.05454.x. [DOI] [PubMed] [Google Scholar]
- 22.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazlauskas D, Dayaram A, Kraberger S, Goldstien S, Varsani A, Krupovic M. Evolutionary history of ssDNA bacilladnaviruses features horizontal acquisition of the capsid gene from ssRNA nodaviruses. Virology. 2017;504:114–121. doi: 10.1016/j.virol.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Kazlauskas D, Varsani A, Krupovic M. Pervasive chimerism in the replication-associated proteins of uncultured single-stranded DNA viruses. Viruses. 2018;10:187. doi: 10.3390/v10040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin EV, Ilyina TV. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA-replication initiator proteins. J Gen Virol. 1992;73:2763–2766. doi: 10.1099/0022-1317-73-10-2763. [DOI] [PubMed] [Google Scholar]
- 26.Krupovic M. Recombination between RNA viruses and plasmids might have played a central role in the origin and evolution of small DNA viruses. BioEssays. 2012;34:867–870. doi: 10.1002/bies.201200083. [DOI] [PubMed] [Google Scholar]
- 27.Krupovic M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr Opin Virol. 2013;3:578–586. doi: 10.1016/j.coviro.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Krupovic M, Forterre P. Single-stranded DNA viruses employ a variety of mechanisms for integration into host genomes. Ann N Y Acad Sci. 2015;1341:41–53. doi: 10.1111/nyas.12675. [DOI] [PubMed] [Google Scholar]
- 29.Krupovic M, Koonin EV. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep. 2014;4:5347. doi: 10.1038/srep05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krupovic M, Ravantti JJ, Bamford DH. Geminiviruses: a tale of a plasmid becoming a virus. BMC Evol Biol. 2009;9:112. doi: 10.1186/1471-2148-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krupovic M, Zhi N, Li J, Hu G, Koonin EV, Wong S, Shevchenko S, Zhao K, Young NS. Multiple layers of chimerism in a single-stranded DNA virus discovered by deep sequencing. Genome Biol Evol. 2015;7:993–1001. doi: 10.1093/gbe/evv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kryukov K, Ueda MT, Imanishi T, Nakagawa S. Systematic survey of non-retroviral virus-like elements in eukaryotic genomes. Virus Res. 2018;262:30–36. doi: 10.1016/j.virusres.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Lefeuvre P, Lett JM, Varsani A, Martin DP. Widely conserved recombination patterns among single-stranded DNA viruses. J Virol. 2009;83:2697–2707. doi: 10.1128/JVI.02152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefeuvre P, Harkins GW, Lett J-M, Briddon RW, Chase MW, Moury B, Martin DP. Evolutionary time-scale of the begomoviruses: evidence from integrated sequences in the Nicotiana genome. PLoS ONE. 2011;6:e19193. doi: 10.1371/journal.pone.0019193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Masroor MA, Wilson ML, Ndjango JB, Peeters M, Gross-Camp ND, Muller MN, Hahn BH, Wolfe ND, Triki H, Bartkus J, Zaidi SZ, Delwart E. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol. 2010;84:1674–1682. doi: 10.1128/JVI.02109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Peng Y, Yi X, Jiang D. Widespread endogenization of densoviruses and parvoviruses in animal and human genomes. J Virol. 2011 doi: 10.1128/JVI.00828-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malathi VG, Renukadevi P, Rageshwari S. Molecular dynamics of geminivirus–host interactome. In: Gaur RK, Khurana SMP, Dorokhov Y, editors. Plant viruses, diversity, interaction and management. Boca Raton: CRC Press; 2017. pp. 173–189. [Google Scholar]
- 38.Mankertz A, Persson F, Mankertz J, Blaess G, Buhk HJ. Mapping and characterization of the origin of DNA replication of porcine circovirus. J Virol. 1997;71:2562–2566. doi: 10.1128/jvi.71.3.2562-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manni F, Rotola A, Caselli E, Bertorelle G, Di Luca D. Detecting recombination in tt virus: a phylogenetic approach. J Mol Evol. 2002;55:563–572. doi: 10.1007/s00239-002-2352-y. [DOI] [PubMed] [Google Scholar]
- 40.Martin DP, Biagini P, Lefeuvre P, Golden M, Roumagnac P, Varsani A. Recombination in eukaryotic single stranded DNA viruses. Viruses. 2011;3(9):1699–1738. doi: 10.3390/v3091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng TF, Chen LF, Zhou Y, Shapiro B, Stiller M, Heintzman PD, Varsani A, Kondov NO, Wong W, Deng X, Andrews TD, Moorman BJ, Meulendyk T, MacKay G, Gilbertson RL, Delwart E. Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc Natl Acad Sci USA. 2014;111:16842–16847. doi: 10.1073/pnas.1410429111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto H, Takahashi M, Nishizawa T, Tawara A, Sugai Y, Sai T, Tanaka T, Tsuda F. Replicative forms of tt virus DNA in bone marrow cells. Biochem Biophys Res Commun. 2000;270:657–662. doi: 10.1006/bbrc.2000.2481. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto H, Ukita M, Nishizawa T, Kishimoto J, Hoshi Y, Mizuo H, Tanaka T, Miyakawa Y, Mayumi M. Circular double-stranded forms of tt virus DNA in the liver. J Virol. 2000;74:5161–5167. doi: 10.1128/JVI.74.11.5161-5167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opriessnig T, Meng X-J, Halbur PG. Porcine circovirus type 2-associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest. 2007;19:591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- 45.Pita JS, Fondong VN, Sangare A, Kokora RNN, Fauquet CM. Genomic and biological diversity of the african cassava geminiviruses. Euphytica. 2001;120:115–125. doi: 10.1023/A:1017536512488. [DOI] [Google Scholar]
- 46.Quaiser A, Krupovic M, Dufresne A, Francez AJ, Roux S. Diversity and comparative genomics of chimeric viruses in sphagnum-dominated peatlands. Virus Evol. 2016;2:vew025. doi: 10.1093/ve/vew025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie PA, Anderson IL, Lambert DM. Evidence for specificity of psittacine beak and feather disease viruses among avian hosts. Virology. 2003;306:109–115. doi: 10.1016/S0042-6822(02)00048-X. [DOI] [PubMed] [Google Scholar]
- 48.Rokyta DR, Wichman HA. Genic incompatibilities in two hybrid bacteriophages. Mol Biol Evol. 2009;26:2831–2839. doi: 10.1093/molbev/msp199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosario K, Duffy S, Breitbart M. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J Gen Virol. 2009;90:2418–2424. doi: 10.1099/vir.0.012955-0. [DOI] [PubMed] [Google Scholar]
- 50.Rosario K, Duffy S, Breitbart M. A field guide to eukaryotic circular single stranded DNA viruses: insights gained from metagenomics. Arch Virol. 2012;157:1851–1871. doi: 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- 51.Roux S, Enault F, Bronner G, Vaulot D, Forterre P, Krupovic M. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat Commun. 2013;4:2700. doi: 10.1038/ncomms3700. [DOI] [PubMed] [Google Scholar]
- 52.Rybicki EP. A phylogenetic and evolutionary justification for three genera of Geminiviridae. Arch Virol. 1994;139:49–77. doi: 10.1007/BF01309454. [DOI] [PubMed] [Google Scholar]
- 53.Saunders K, Lucy A, Stanley J. DNA forms of the geminivirus african cassava mosaic-virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res. 1991;19:2325–2330. doi: 10.1093/nar/19.9.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saunders K, Bedford ID, Yahara T, Stanley J. The earliest recorded plant virus disease. Nature. 2003;422:831. doi: 10.1038/422831a. [DOI] [PubMed] [Google Scholar]
- 55.Shackelton LA, Holmes EC. Phylogenetic evidence for the rapid evolution of human b19 erythrovirus. J Virol. 2006;80:3666–3669. doi: 10.1128/JVI.80.7.3666-3669.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simmonds P, Adams MJ, Benko M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AM, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini FM. Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 57.Steel O, Kraberger S, Sikorski A, Young LM, Catchpole RJ, Stevens AJ, Ladley JJ, Coray DS, Stainton D, Dayarama A, Julian L, van Bysterveldt K, Varsani A. Circular replication-associated protein encoding DNA viruses identified in the faecal matter of various animals in New Zealand. Infect Genet Evol. 2016;43:151–164. doi: 10.1016/j.meegid.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 58.Suttle CA. Viruses: unlocking the greatest biodiversity on Earth. Genome. 2013;56:542–544. doi: 10.1139/gen-2013-0152. [DOI] [PubMed] [Google Scholar]
- 59.Tattersall P, Ward DC. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature. 1976;263:106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- 60.Tischer I, Rasch R, Tochtermann G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralblatt fur€ Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. Erste Abteilung Originale. Reihe A: Medizinische Mikrobiologie und Parasitologie. 1974;226:153–167. [PubMed] [Google Scholar]
- 61.Tischer I, Gelderblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature. 1982;295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 62.Vidigal PMP, Mafra CL, Silva FMF, Fietto JLR, Silva Júnior A, Almeida MR. Tripping over emerging pathogens around the world: a phylogeographical approach for determining the epidemiology of porcine circovirus-2 (PCV-2), considering global trading. Virus Res. 2012;163:320–327. doi: 10.1016/j.virusres.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 63.Zhao L, Rosario K, Breitbart M, Duffy S. Eukaryotic circular Rep encoding single stranded DNA (CRESS DNA) viruses: ubiquitous viruses with small genomes and a diverse host range. Adv Virus Res. 2018 doi: 10.1016/bs.aivir.2018.10.001. [DOI] [PubMed] [Google Scholar]