Abstract
In plants, RNA silencing provides a major line of defence against viruses. This antiviral immunity involves production of virus-derived small interfering RNAs (vsiRNAs) and results in specific silencing of viruses by vsiRNAs-guided effector complexes. As a counterattack against RNA silencing, many plant viruses encode suppressors of RNA silencing called viral suppressors of RNA silencing (VSRs), which interfere with the silencing pathway by various mechanisms. This review describes various methods that are being used to characterize viral proteins for suppressor function, VSRs found in geminiviruses and associated DNA satellites and their mechanisms of action.
Keywords: Geminivirus, RNA silencing, Viral suppressors of RNA silencing (VSRs)
Introduction
Geminiviruses have circular, single-stranded DNA genome (ssDNA), the size of which can range from ~ 2.5 to 3.0 kb [29]. The family Geminiviridae has been classified recently to comprise nine genera: Capulavirus, Becurtovirus, Begomovirus, Curtovirus, Eragrovirus, Grablovirus, Mastrevirus, Topocuvirus and Turncurtovirus [74]. They are transmitted by insects and infect a wide range of plant species including some of the major food and fibre crops. The genomic DNA is encapsidated within twinned (geminate), incomplete, non-enveloped, icosahedral particles of size ~ 18–30 nm. Geminiviruses can either be bipartite (two DNA molecules, DNA-A and DNA-B (only in genus Begomovirus) or monopartite (one DNA molecule analogous to DNA-A) [30]. Monopartite viruses encode all the proteins essential for successful infection, whereas, in bipartite viruses proteins required for viral DNA encapsidation and replication and a few other functions related to gene expression and RNA silencing suppression are encoded by DNA-A, the functions related to viral movement are encoded by DNA-B. The genes or Open Reading Frames (ORFs) are named according to the coding strand (V-sense or C-sense) and the genomic component.
Plant defence against virus attack: RNA silencing
RNA silencing is a generic term given to diverse yet related pathways, mediated by ~ 21–30 nucleotide double-stranded RNA molecules [small (s) RNAs], leading to regulation of the gene expression in a sequence-specific manner at the transcriptional or translational level. In plants, RNA silencing pathway is a major line of defence against viruses. Upon viral infection, virus-derived siRNAs (vsiRNAs) are formed from dsRNA originating from viral sequences, which then target the viral genome by various mechanisms [22].
Important steps in RNA silencing pathway
The first step in RNA silencing pathway is the formation of partially or perfectly dsRNAs. The latter can arise from various ways, for e.g. from self-complementary fold-back structure [12], imperfect duplexes in the folded regions of genome [41]; convergent, aberrant or RNA induced silencing complex (RISC)-mediated cleaved transcripts, stem-loop structure [46]; conversion of cleaved TAS transcripts [72]; overlapping region of a pair of natural antisense transcripts [6, 77]; transposons or repetitive genomic regions [40, 55, 73]; or gene/pseudogene duplexes [68]. The second step involves the formation of shorter dsRNAs (sRNAs) from the long dsRNA, by the ‘dicing’ activity of an RNaseIII-like enzyme called Dicer-like (DCL) in plants. In the case of viral infection, the siRNAs that are formed during this step are called ‘primary siRNAs’ as they are derived from the target region. Next, the duplex siRNAs are loaded onto a multi protein RISC or RNA-induced transcriptional silencing complex (RITS) [63], containing a protein from the Argonaute family (AGO). One of the strands from duplex sRNA, called passenger strand, is degraded. The strand, called guide strand, binds to complementary region in the homologous mRNA and initiates either post-transcriptional gene silencing (PTGS), leading to degradation of target mRNA or its translational arrest or transcriptional gene silencing (TGS), involving epigenetic changes to the target DNA [18, 22] (Fig. 1).
Fig. 1.
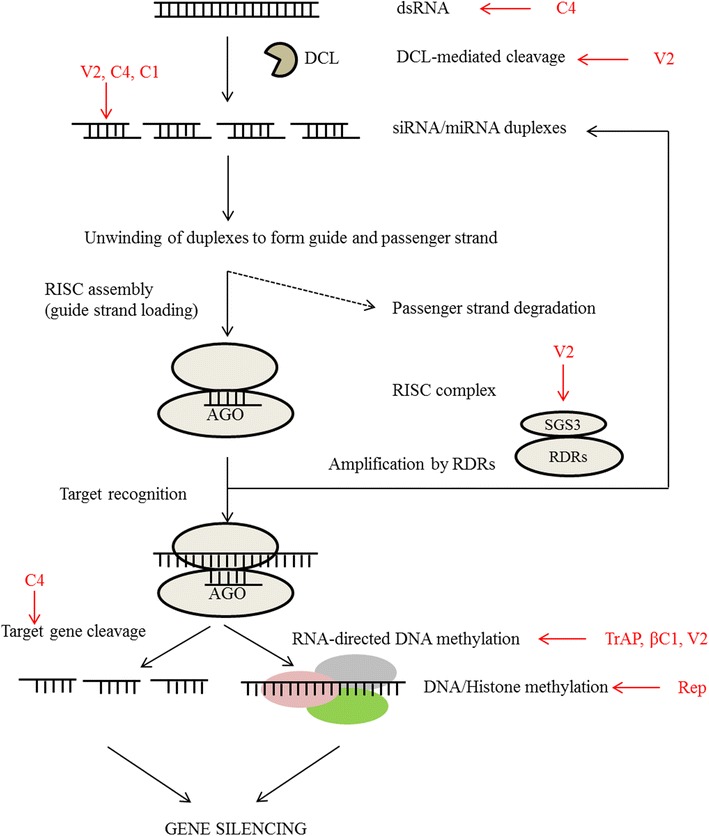
The main steps in RNA silencing pathway where geminiviral suppressors have been shown to act. The dsRNA arising from various sources is cleaved into ~ 21–30 nt small RNA duplexes (siRNA/miRNA). One of the strands in this duplex, called the guide strand is incorporated in the RISC complex containing an AGO protein. This sRNA then guides the AGO protein to the target sequence and mediate the gene silencing by various mechanisms. The RDRs work to amplify/maintain the gene silencing. The red arrow depicts the inhibition of the step shown in the figure (see text for more details) (colour figure online)
Key components of RNA silencing pathway
The key components in RNA silencing pathway include DCL ribonucleases and associated dsRNA binding (DRB) proteins, RNA-dependent RNA polymerases (RDR), and Argonaute (AGO) proteins. The widely studied model plant Arabidopsis thaliana has four DCLs (DCL1 till DCL4), five DRBs (HYL1/DRB1 till DRB5), six RDRs (RDR1 till RDR6) and ten AGO proteins (AGO1 till AGO10) [4, 5, 22, 34, 38, 46]. The functions of the various components of RNAi pathway mentioned above have been recently reviewed [19, 75].
Virus counter-defence against RNA silencing: viral suppressors
As has already been discussed in the preceding section of this review, RNA silencing serves as a major defence mechanism that plants employs to protect themselves against viral infections. Therefore, as a counter-defensive strategy many plant viruses encode proteins, known as viral silencing suppressors (VSRs) that interfere with RNA silencing pathway at one or the other step. VSRs are also found in viruses infecting insects, fish, mammals etc. Most of the plant viruses encode at least one VSR. In addition, several studies showed that mutations giving rise to non-functional VSRs led to increased resistance against viral infection in the host plant. These points emphasize the importance of RNA silencing as a major line of defence against viruses. Apart from acting as suppressors, most of the VSRs are involved in functions that hold importance in viral life cycle like replication, movement, encapsidation, transmission etc. Since the discovery of first viral suppressor, the potyviral P1/Hc-Pro, numerous other VSRs have been discovered [2, 19, 45]. The following sub-sections will describe various methods that have been used to characterize viral proteins for suppressor function, some of the VSRs and their mechanism of action.
Assays for the identification of VSRs which suppress PTGS
Transient expression assays
This method involves the use of transgenic Nicotiana benthamiana plants (16c) constitutively expressing the reporter gene GFP [51]. This assay employs the technique of co-infiltration of two A. tumefaciens strains to the GFP-expressing transgenic plant; one carrying a construct having the GFP gene (inducer of silencing) and the other carrying the candidate viral suppressor gene [37], both the inducer and the candidate suppressor genes under strong constitutive promoters. In the assay for local silencing both the agrobacterium suspensions are co-infiltrated into a leaf and the infiltrated patch is observed for GFP expression over a period of time. The infiltrated patch initially appears green (under ultraviolet illumination) as it expresses high levels of GFP, but turns red in 3–5 days because the inducer triggers silencing of the transgenically-expressed GFP; the red colour resulting due to chlorophyll autofluorescence. If the candidate gene possesses a suppressor activity, the silencing is reversed and the GFP expression does not decrease and the patch retains its green colour.
In the assay for systemic silencing, both the agrobacterium constructs are infiltrated in the leaves of 16c plants and newly-emerging leaves are observed for changes in GFP expression after about 3 weeks. In this particular case, the GFP-inducer construct triggers GFP silencing in the infiltrated leaf and the silencing signal travels systemically, resulting in the emerging leaves appear red. However, if the candidate gene possesses a suppressor activity, such that it prevents the generation and/or spread of the silencing signal, the emerging leaves will appear green.
To assay whether the VSR suppresses the movement of silencing signal, a grafting experiment is generally undertaken. A root-stock, where the silencing of a transgenic reporter gene is initiated by infiltration of the same gene (inducer, as described above), is then used to graft a scion on it, in which the same reporter gene is expressed transgenically. The silencing signal initiates in the root-stock, spreads through the graft to the scion and silences the reporter gene in it. The same effect is seen if the scion is separated from the root-stock by an “intergraft” (middle insert), taken from a wild-type plant, through which the silencing signal can pass and silence the reporter gene in the scion. However, if the intergraft is taken from a plant expressing the VSR capable of blocking the movement of the silencing signal, the reporter gene expressed in the scion is no longer silenced [28, 39].
Stable expression assays
This assay can be used when any of the above assays confirm the suppressor activity of a protein. It exploits two stable transgenic lines: one expressing the candidate suppressor and a line silenced for any well-characterized reporter gene. These lines are then crossed and progeny is tested for the reporter gene expression. The reporter expression will be restored if the putative suppressor eliminates the silencing [2].
Assays for VSRs which suppress TGS
For assaying TGS suppressors, N. benthamiana 16-TGS lines are used. These plants carry a reporter gene (usually GFP) under the control of a cauliflower mosaic virus (CaMV) 35S promoter, which is transcriptionally silenced (by TGS) after infection with a tobacco rattle virus-based vector, which in turn, carries a portion of the CaMV 35S promoter, the inducer of silencing. The silenced promoter cannot drive the expression of GFP and hence the plant appears red under UV illumination. In the presence of TGS suppressor, the promoter becomes active and starts expressing GFP, resulting in the plant showing green fluorescence under UV illumination [11, 64].
Mechanism of action of geminiviral VSRs
VSRs can have diverse mode of action and can interfere with almost any step of silencing pathway. A single VSR can have multiples modes of action as will be evident in following section. Table 1 lists some of the important VSRs and their modes of action.
Table 1.
A List of some of the important viral suppressors of RNA silencing encoded by geminiviruses and associated DNA satellites, and their modes of action
| VSR | Genus | Species | Mode of action | Reference |
|---|---|---|---|---|
| AV2 | Begomovirus | EACMCV | Unknown | [17] |
| V2 | Begomovirus | TYLCV-Is | Acts at a step downstream of dicer-mediated cleavage of dsRNA | [79] |
| ToLCJV-A | HR elicitor | [53] | ||
| AYVV | Unknown | [54] | ||
| TYLCCNV | Sequestration of siRNA molecules | [76] | ||
| CLCuMV | Sequestration of long dsRNA | [1] | ||
| TYLCV | Interaction with SGS3; Binding to CYP1 protein and hindering innate immunity of plant; TGS suppressor | [3, 26, 64] | ||
| Rep | Begomovirus | TYLCSV; TYLCV-Mld, ACMV, TGMV | Decrease the expression of MET1 and CMT3 | [49] |
| Mastrevirus | WDV | siRNA binding | [66] | |
| TrAP | Begomovirus | TYLCV-C | Unknown | [69, 70] |
| EACMCV | Unknown | [62] | ||
| ICMV | Unknown | [62] | ||
| MYMV-Vig | Alteration of host gene expression levels | [61] | ||
| BYVMV | Unknown | [27] | ||
| AYVV | Unknown | [54] | ||
| CLCuMV | Unknown | [1] | ||
| BSCTV | Inhibits proteasomal degradation of SAMDC1 | [78] | ||
| TGMV, CaLCuV | Genome-wide reduction in cytosine methylation; inhibit histone methyl transferases | [11, 13, 65] | ||
| ACMV | Alteration in host gene expression | [57] | ||
| Curtovirus | BCTV | Inhibit SNF1 Kinase and ADK | [31] | |
| AC4/C4 | Begomovirus | ACMV | Binds to ss-sRNA | [16] |
| EACMCV | Unknown (might interfere with spread of silencing) | [25] | ||
| BYVMV | Unknown | [27] | ||
| ToLCV | Unknown (might interact and inhibit activity of shaggy-like kinase and, in turn, modify brassinosteroid signalling) | [23] | ||
| AYVV | Unknown | [54] | ||
| MYMV | Binds to sRNA | [60] | ||
| CLCuMV | Binds to long dsRNA | [1] | ||
| βC1 protein | Betasatellite | TYLCCNB | Inhibit a methyl cycle enzyme, SAHH | [20, 71] |
| BYVMB | Unknown | [27] | ||
| ToLCJB | Unknown | [35] | ||
| CLCuMB | Unknown | [1] | ||
| TYLCCNB | Rgs-CaM mediated suppression of RDR6 expression | [36] |
EACMCV: East African cassava mosaic Cameroon virus; TYLCV-Is: Tomato yellow leaf curl virus- Israel; ToLCJV-A: Tomato leaf curl Java virus-A; AYVV: Ageratum yellow vein virus; TYLCCNV: Tomato yellow leaf curl China virus; CLCuMV: Cotton leaf curl Multan virus; TYLCSV: Tomato yellow leaf curl Sardinia virus; TYLCV-Mld: TYLCV, mild strain; ACMV: African cassava mosaic virus; TGMV: Tomato golden mosaic virus; WDV: Wheat dwarf virus; TYLCV-C: Tomato yellow leaf curl virus-China; ICMV: Indian cassava mosaic virus; MYMV-Vig: Mungbean yellow mosaic virus-Vigna; BYVMV: Bhendi yellow vein mosaic virus; BSCTV: Beet severe curly top virus; CaLCuV: Cabbage leaf curl virus; BCTV: Beet curly top virus; ToLCV: Tomato leaf curl virus; TYLCCNB: Tomato yellow leaf curl China betasatellite; BYVMB: Bhendi yellow vein mosaic betasatellite; ToLCJB: Tomato leaf curl Java betasatellite; CLCuMB: Cotton leaf curl Multan betasatellite
V2 protein
The V2 ORF (AV2 in bipartite begomoviruses) codes for pre-coat protein. V2, along with CP, is required for efficient accumulation of viral DNA forms and therefore, facilitates successful infection in the host [7, 47, 67]. It is also required for efficient viral movement [43, 50]. V2 can suppress both PTGS and TGS. Tomato yellow leaf curl virus (TYLCV) V2 protein interacts with the host SGS3 (SUPPRESSOR OF GENE SILENCING 3) which assists RDR6 in the production of dsRNA. The interaction between V2 and SGS3 affects the amplification step of silencing [26]. V2 protein encoded by Tomato yellow leaf curl China virus possibly sequesters the siRNAs generated against the virus and hinders the silencing pathway [76]. In a similar way it has been speculated that Cotton leaf curl Multan virus (CLCuMV) V2 also sequesters long dsRNA and prevents its dicer-mediated cleavage [1]. The TYLCV V2 also manifests its function as a suppressor by interfering with the innate immunity of the plant by binding to CYP1 protein, a member of the family of papain-like cysteine proteases [3]. V2 has been shown to elicit a hypersensitive response (HR) and acts as a pathogenicity determinant in Tomato leaf curl Java virus-A [53]. In addition to blocking PTGS, V2 can also act as TGS suppressor as has been demonstrated in TYLCV [64].
Replication-associated protein (AC1, C1)
The replication-associated protein (Rep), encoded by C1 or AC1 ORF, plays an indispensable role in viral replication [24]. Rep has been reported to be involved in all the major steps of rolling-circle replication (RCR). Apart from its long recognized role in replication, a recent study implicated Rep as a TGS suppressor, whereby, it was shown to reduce the expression of plant DNA methyl transferases such as METHYLTRANSFERASE 1 (MET1) and CHROMOMETHYLASE 3 (CMT3) [49]. The Rep protein of Wheat dwarf virus (WDV) has been shown to interfere with silencing by binding to 21 and 24 nt siRNA duplexes and ss-siRNA [66].
Transcriptional activator protein (AC2, C2)
The C2 protein [also known as AC2, AL2, L2 or transcriptional activator protein (TrAP)] functions to activate viral gene expression. The C2 has also been shown to act both as PTGS and TGS suppressor. The C2 protein functions as a PTGS suppressor in Tomato yellow leaf curl virus-China [69, 70], East African cassava mosaic Cameroon virus [62], Mungbean yellow mosaic virus-Vigna (MYMV-Vig) [61] and CLCuMV [1]. It has been hypothesized that MYMV-Vig AC2 can alter the expression levels of the host genes involved in gene silencing to bring about its suppression activity. In addition, C2 inhibits the components of methylation machinery such as SUCROSE NONFERMENTING1 (SNF1) Kinase [31], adenosine kinase [11, 65] and certain histone methyltransferases [13]. Therefore, AC2 acts as TGS suppressor. C2 encoded by Beet severe curly top virus also suppresses TGS but with a different mode of action. It attenuates the 26S proteasomal degradation of S-adenosyl-methionine decarboxylase 1, consequently interfering with the DNA-methylation based gene silencing [78].
AC4/C4 protein
The C4 ORF lies entirely within the C1 ORF, however, in a different reading frame. AC4 (bipartite begomoviruses)/C4 (monopartite begomoviruses) ORF is among the least conserved begomoviral ORFs and has been shown to be associated with diverse array of functions, some of them being symptom determination, viral DNA accumulation and virus movement [33, 48, 50]. AC4/C4 has also been proved to act as RNA silencing suppressor in both bipartite and monopartite begomoviruses. The AC4 encoded by ACMV specifically binds to the single stranded form of sRNA [16]. The MYMV-AC4 binds to sRNAs and inhibits cleavage of target mRNA [60]. The CLCuMV-C4, on the other hand, binds both long and short RNAs, with a preference for the dsRNA form [1].
βC1 protein
Betasatellites are circular ssDNAs of ~ 1.35 kb found to be associated with monopartite begomoviruses [14, 15, 21, 32, 52, 58]. However, more recently it has been reported that Cotton leaf curl Multan betasatellite (CLCuMB) contributed to the development of typical leaf curl symptoms when inoculated with DNA-A of a bipartite begomovirus Tomato leaf curl New Delhi virus (ToLCNDV) [56]. DNA β sequence can be demarcated into four different regions namely (1) a conserved stem-loop structure with conserved nonanucleotide sequence, (2) a region of high sequence similarity conserved termed as ‘satellite conserved region’ (SCR), (3) an adenine (A) rich region approximately 370–420 nucleotides (nts) upstream of the SCR, (4) a single gene encoded by the complementary strand, βC1 [8, 10, 59]. The βC1 proteins from several betasatellites have been shown to act as RNA silencing suppressors. Some of the examples include Ageratum yellow vein virus-Indonesia, Bhendi yellow vein mosaic betasatellite (BYVMB), Tomato leaf curl Java betasatellite (ToLCJB), CLCuMB [1, 27, 35, 54]. The βC1 gene of Tomato yellow leaf curl China betasatellite (TYLCCNB) has the ability to suppress TGS by interacting with a methyl cycle enzyme, S-adenosyl homocysteine hydrolase, which is required for methylation [20, 71]. The TYLCCNB encoded βC1 has been reported to enhance the expression of N. benthamiana calmodulin-like protein Nbrgs-CaM. The enhanced expression of Nbrgs-CaM hindered the production of secondary siRNAs, likely through inhibiting the expression of RNA-dependent RNA polymerases 6 (RDR6) [36].
Alphasatellites
Alphasatellites are found to be associated with both old world monopartite begomoviruses and new world bipartite begomoviruses [9, 44]. Similar to betasatellites, they possess circular, ssDNA genome of approximately half the size of their helper begomoviruses. They contain a single gene which encodes replication-associated protein (Alpha-Rep). The Alpha-Rep proteins from two alphasatellites, Gossypium darwinii symptomless alphasatellite and Gossypium mustelinium symptomless alphasatellite have been reported to act PTGS suppressors [42].
Concluding remarks
Geminiviruses are the largest family of plant viruses and are becoming increasingly important as pathogens infecting crop plants in the tropical and sub-tropical regions of the World. A better understanding of the mechanisms of action of geminiviral VSRs can give researchers new insights on the management of geminiviruses, as a means of improving the production of food and fibre crops in many parts of the World. This can also lead to a better understanding of the plant defence mechanisms, especially the small RNA-based defence.
Acknowledgements
RR acknowledges Research Fellowship from University Grants Commission, New Delhi. ID acknowledges financial grant from DU-DST PURSE and R&D Grant from University of Delhi.
References
- 1.Amin I, Hussain K, Akbergenov R, Yadav JS, Qazi J, Mansoor S, Hohn T, Fauquet CM, Briddon RW. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol Plant Microbe Interact. 2011;24:973–983. doi: 10.1094/MPMI-01-11-0001. [DOI] [PubMed] [Google Scholar]
- 2.Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB. A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar-Ziv A, Levy Y, Citovsky V, Gafni Y. The Tomato yellow leaf curl virus (TYLCV) V2 protein inhibits enzymatic activity of the host papain-like cysteine protease CYP1. Biochim Biophys Acta. 2015;460:525–529. doi: 10.1016/j.bbrc.2015.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 5.Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Hohn T, Poogin MM. Four plant dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulton M, Steinkellner H, Donson J, Markham PG, King DI, Davies JW. Mutational analysis of the virion-sense genes of maize streak virus. J Gen Virol. 1989;70:2309–2323. doi: 10.1099/0022-1317-70-9-2309. [DOI] [PubMed] [Google Scholar]
- 8.Briddon RW, Bull SE, Amin I, Idris AM, Mansoor S, Bedford ID, Dhawan P, Rishi N, Siwatch SS, Abdel-Salam AM, Brown JK, Zafar Y, Markham PG. Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology. 2003;312:106–121. doi: 10.1016/S0042-6822(03)00200-9. [DOI] [PubMed] [Google Scholar]
- 9.Briddon RW, Bull SE, Amin I, Mansoor S, Bedford ID, Rishi N, Siwatch SS, Zafar Y, Abdel-Salam AM, Markham PG. Diversity of DNA 1: a satellite-like molecule associated with monopartite begomovirus-DNA beta complexes. Virology. 2004;324:462–474. doi: 10.1016/j.virol.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 10.Briddon RW, Mansoor S, Bedford D, Pinner MS, Saunders K, Stanley J, Zafar Y, Malik KA, Markham PG. Identification of DNA components required for induction of cotton leaf curl disease. Virology. 2001;285:234–243. doi: 10.1006/viro.2001.0949. [DOI] [PubMed] [Google Scholar]
- 11.Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol. 2009;83:5005–5013. doi: 10.1128/JVI.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 13.Castillo-González C, Liu X, Huang C, Zhao C, Ma Z, Hu T, Sun F, Zhou Y, Zhou X, Wang X, Zhang X. Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. Elife. 2015;4:1–31. doi: 10.7554/eLife.06671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee A, Ghosh SK. Association of a satellite DNA β molecule with Mesta yellow vein mosaic disease. Virus Genes. 2007;35:835–844. doi: 10.1007/s11262-007-0160-6. [DOI] [PubMed] [Google Scholar]
- 15.Chattopadhyay B, Singh AK, Yadav T, Fauquet CM, Sarin NB, Chakraborty S. Infectivity of the cloned components of a begomovirus: DNA beta complex causing chilli leaf curl disease in India. Arch Virol. 2008;153:533–539. doi: 10.1007/s00705-007-0017-2. [DOI] [PubMed] [Google Scholar]
- 16.Chellappan P, Vanitharani R, Fauquet CM. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc Natl Acad Sci USA. 2005;102:10381–10386. doi: 10.1073/pnas.0504439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowda-Reddy RV, Achenjang F, Felton C, Etarock MT, Anangfac MT, Nugent P, Fondong VN. Role of a geminivirus AV2 protein putative protein kinase C motif on subcellular localization and pathogenicity. Virus Res. 2008;135:115–124. doi: 10.1016/j.virusres.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Costa AT, Bravo JP, Makiyama RK, Nunes AV, Maia IG. Viral counter defense X antiviral immunity in plants: mechanisms for survival, Current issues in molecular virology. In: Romanowski V editor. Viral Genetics and Biotechnological Applications. InTech; 2013. pp. 251–85. 10.5772/56253
- 19.Csorba T, Kontra L, Burgyán J. Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology. 2015;479–480:85–103. doi: 10.1016/j.virol.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Cui X, Li G, Wang D, Hu D, Zhou X. A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J Gen Virol. 2005;79:10764–10775. doi: 10.1128/JVI.79.16.10764-10775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui X, Tao X, Xie Y, Fauquet CM, Zhou X. A DNA β associated with Tomato yellow leaf curl China virus is required for symptom induction. J Virol. 2004;78:13966–13974. doi: 10.1128/JVI.78.24.13966-13974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dogra SC, Eini O, Rezaian MA, Randles JW. A novel shaggy-like kinase interacts with the tomato leaf curl virus pathogenicity determinant C4 protein. Plant Mol Biol. 2009;71:25–38. doi: 10.1007/s11103-009-9506-x. [DOI] [PubMed] [Google Scholar]
- 24.Etessami P, Saunders K, Watts J, Stanley J. Mutational analysis of complementary-sense genes of African cassava mosaic virus DNA. J Gen Virol. 1991;72:1005–1012. doi: 10.1099/0022-1317-72-5-1005. [DOI] [PubMed] [Google Scholar]
- 25.Fondong VN, Reddy RVC, Lu C, Hankoua B, Felton C, Czymmek K, Achenjang F. The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol Plant Microbe Interact. 2007;20:380–391. doi: 10.1094/MPMI-20-4-0380. [DOI] [PubMed] [Google Scholar]
- 26.Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci USA. 2008;106:157–161. doi: 10.1073/pnas.0709036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gopal P, Kumar PP, Sinilal B, Jose J, Yadunandam AK, Usha R. Differential roles of C4 and βC1 in mediating suppression of post-transcriptional gene silencing: evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 2007;123:9–18. doi: 10.1016/j.virusres.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 28.Guo HS, Ding SW. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 2002;21:398–407. doi: 10.1093/emboj/21.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez C, Ramirez-Parra E, Mar Castellano M, Sanz-Burgos AP, Luque A, Missich R. Geminivirus DNA replication and cell cycle interactions. Vet Microbiol. 1999;98:111–119. doi: 10.1016/j.vetmic.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol. 2013;11:777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- 31.Hao L, Wang H, Sunter G, Bisaro DM. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell. 2003;15:1034–1048. doi: 10.1105/tpc.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jose J, Usha R. Bhendi yellow vein mosaic disease in India is caused by association of a DNA β satellite with a begomovirus. Virology. 2003;305:310–317. doi: 10.1006/viro.2002.1768. [DOI] [PubMed] [Google Scholar]
- 33.Jupin I, De Kouchkovsky F, Jouanneau F, Gronenborn B. Movement of tomato yellow leaf curl geminivirus (TYLCV): involvement of the protein encoded by ORF C4. Virology. 1994;204:82–90. doi: 10.1006/viro.1994.1512. [DOI] [PubMed] [Google Scholar]
- 34.Kalantidis K, Schumacher HT, Alexiadis T, Helm JM. RNA silencing movement in plants. Biol Cell. 2008;58:328–342. doi: 10.1042/BC20070079. [DOI] [PubMed] [Google Scholar]
- 35.Kon T, Sharma P, Ikegami M. Suppressor of RNA silencing encoded by the monopartite tomato leaf curl Java begomovirus. Arch Virol. 2007;152:1273–1282. doi: 10.1007/s00705-007-0957-6. [DOI] [PubMed] [Google Scholar]
- 36.Li F, Huang C, Li Z, Zhou X. Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog. 2014;10:11–14. doi: 10.1371/journal.ppat.1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Llave C, Kasschau KD, Carrington JC. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA. 2000;97:13401–13406. doi: 10.1073/pnas.230334397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22:3879–3889. doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mallory AC, Ely L, Smith TH, Marathe R, Anandalakshmi R, Fagard M, Vaucheret H, Pruss G, Bowman L, Vance VB. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell. 2001;13:571–583. doi: 10.1105/tpc.13.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Molnár A, Csorba T, Lakatos L, Várallyay E, Lacomme C, Burgyán J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nawaz-ul-Rehman MS, Nahid N, Mansoor S, Briddon RW, Fauquet CM. Post-transcriptional gene silencing suppressor activity of two non-pathogenic alphasatellites associated with a begomovirus. Virology. 2010;405:300–308. doi: 10.1016/j.virol.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 43.Padidam M, Beachy RN, Fauquet CM. The role of AV2 (‘precoat’) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology. 1996;224:390–404. doi: 10.1006/viro.1996.0546. [DOI] [PubMed] [Google Scholar]
- 44.Paprotka T, Metzler V, Jeske H. The first DNA 1-like alpha satellites in association with New World begomoviruses in natural infections. Virology. 2010;404:148–157. doi: 10.1016/j.virol.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Pruss G, Ge X, Shi XM, Carrington JC, Bowman Vance V. Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell. 1997;9:859–868. doi: 10.1105/tpc.9.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raja P, Wolf JN, Bisaro DM. RNA silencing directed against geminiviruses: post-transcriptional and epigenetic components. Biochim Biophys Acta. 2010;1799:337–351. doi: 10.1016/j.bbagrm.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 47.Rigden JE, Dry IB, Mullineaux PM, Rezaian MA. Mutagenesis of the virion-sense open reading frames of Tomato leaf curl geminivirus. Virology. 1993;193:1001–1005. doi: 10.1006/viro.1993.1215. [DOI] [PubMed] [Google Scholar]
- 48.Rigden JE, Krake LR, Rezaian MA, Dry IB. ORF C4 of tomato leaf curl geminivirus is a determinant of symptom severity. Virology. 1994;204:847–850. doi: 10.1006/viro.1994.1606. [DOI] [PubMed] [Google Scholar]
- 49.Rodríguez-Negrete E, Lozano-Durán R, Piedra-aguilera A, Cruzado L, Bejarano ER, Castillo AG. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 2013;199:464–475. doi: 10.1111/nph.12286. [DOI] [PubMed] [Google Scholar]
- 50.Rojas MR, Jiang H, Salati R, Xoconostle-Cázares B, Lucas WJ, Gilbertson RL. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology. 2001;125:110–125. doi: 10.1006/viro.2001.1194. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saunders K, Bedford ID, Briddon RW, Markham PG, Wong SM, Stanley J. A unique virus complex causes Ageratum yellow vein disease. Proc Natl Acad Sci USA. 2000;97:6890–6895. doi: 10.1073/pnas.97.12.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma P, Ikegami M. Tomato leaf curl Java virus V2 protein is a determinant of virulence, hypersensitive response and suppression of posttranscriptional gene silencing. Virology. 2010;396:85–93. doi: 10.1016/j.virol.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 53.Sharma P, Ikegami M, Kon T. Identification of the virulence factors and suppressors of posttranscriptional gene silencing encoded by Ageratum yellow vein virus, a monopartite begomovirus. Virus Res. 2010;149:19–27. doi: 10.1016/j.virusres.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 55.Simon SA, Meyers BC. Small RNA-mediated epigenetic modifications in plants. Curr Opin Plant Biol. 2011;14:148–155. doi: 10.1016/j.pbi.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Sivalingam PN, Varma A. Role of betasatellite in the pathogenesis of a bipartite begomovirus affecting tomato in India. Arch Virol. 2012;157:1081–1092. doi: 10.1007/s00705-012-1261-7. [DOI] [PubMed] [Google Scholar]
- 57.Soitamo AJ, Jada B, Lehto K. Expression of geminiviral AC2 RNA silencing suppressor changes sugar and jasmonate responsive gene expression in transgenic tobacco plants. BMC Plant Biol. 2012;12:1–18. doi: 10.1186/1471-2229-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srivastava A, Krishana S, Susheel R, Sunil K, Snehi K. Molecular identification of Ageratum enation virus, betasatellite and alphasatellite molecules isolated from yellow vein diseased Amaranthus cruentus in India. Virus Genes. 2013;47:584–590. doi: 10.1007/s11262-013-0971-6. [DOI] [PubMed] [Google Scholar]
- 59.Stanley J. Subviral DNAs associated with geminivirus disease complexes. Vet Microbiol. 2004;98:121–129. doi: 10.1016/j.vetmic.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Sunitha S, Shanmugapriya G, Balamani V, Veluthambi K. Mungbean yellow mosaic virus (MYMV) AC4 suppresses post-transcriptional gene silencing and an AC4 hairpin RNA gene reduces MYMV DNA accumulation in transgenic tobacco. Virus Genes. 2013;46:496–504. doi: 10.1007/s11262-013-0889-z. [DOI] [PubMed] [Google Scholar]
- 61.Trinks D, Rajeswaran R, Shivaprasad PV, Akbergenov R, Oakeley EJ, Veluthambi K, Hohn T, Pooggin MM. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J Virol. 2005;79:2517–2527. doi: 10.1128/JVI.79.4.2517-2527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanitharani R, Chellappan P, Pita JS, Fauquet CM. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J Virol. 2004;78:9487–9498. doi: 10.1128/JVI.78.17.9487-9498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang B, Li F, Huang C, Yang X, Qian Y, Xie Y, Zhou X. V2 of tomato yellow leaf curl virus can suppress methylation-mediated transcriptional gene silencing in plants. J Gen Virol. 2014;95:225–230. doi: 10.1099/vir.0.055798-0. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Buckley KJ, Yang X, Cody R, Buchmann RC, Bisaro DM. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J Virol. 2005;79:7410–7418. doi: 10.1128/JVI.79.12.7410-7418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Dang M, Hou H, Mei Y, Qian Y, Zhou X. Identification of an RNA silencing suppressor encoded by a mastrevirus. J Gen Virol. 2014;95:2082–2088. doi: 10.1099/vir.0.064246-0. [DOI] [PubMed] [Google Scholar]
- 67.Wartig L, Kheyr-Pour A, Noris E. Genetic analysis of the monopartite tomato yellow leaf curl geminivirus: roles of V1, V2, and C2 ORFs in viral pathogenesis. Virology. 1997;228:132–140. doi: 10.1006/viro.1996.8406. [DOI] [PubMed] [Google Scholar]
- 68.Wen YZ, Zheng LL, Liao JY, Wang MH, Wei Y, Guo XM, Qu LH, Ayala FJ, Lun ZR. Pseudogene-derived small interference RNAs regulate gene expression in African Trypanosoma brucei. Proc Natl Acad Sci USA. 2011;108:8345–8350. doi: 10.1073/pnas.1103894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Wezel R, Dong X, Liu H, Tien P, Stanley J, Hong Y. Mutation of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene—silencing suppression. Mol Plant Microbe Interact. 2002;15:203–208. doi: 10.1094/MPMI.2002.15.3.203. [DOI] [PubMed] [Google Scholar]
- 70.Van Wezel R, Liu H, Tien P, Stanley J, Hong Y. Gene C2 of the monopartite geminivirus tomato yellow leaf curl virus -China encodes a pathogenicity determinant that is localized in the nucleus. Mol Plant Microbe Interact. 2001;14:1125–1128. doi: 10.1094/MPMI.2001.14.9.1125. [DOI] [PubMed] [Google Scholar]
- 71.Yang X, Xie Y, Raja P, Li S, Wolf JN, Shen Q, Bisaro DM, Zhou X. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 2011;7:1–10. doi: 10.1371/journal.ppat.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshikawa M. Biogenesis of trans-acting siRNAs, endogenous secondary siRNAs in plants. Genes Genet Syst. 2013;88:77–84. doi: 10.1266/ggs.88.77. [DOI] [PubMed] [Google Scholar]
- 73.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 74.Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-castillo J, Rivera-Bustamante R, Roumagnac R, Varsani A. ICTV virus taxonomy profile : Geminiviridae. J Gen Virol. 2017;98:131–133. doi: 10.1099/jgv.0.000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang C, Wu Z, Li Y, Wu J. Biogenesis, function, and applications of virus-derived small RNAs in plants. Front Microbiol. 2015;6:1–12. doi: 10.3389/fmicb.2015.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J, Dong J, Xu Y, Wu J. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 2011;163:51–58. doi: 10.1016/j.virusres.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 77.Zhang X, Xia J, Lii Y, Barrera-Figueroa B, Zhou X, Gao S, Lu L, Niu D, Chen Z, Leung C, Wong T, Zhang H, Guo J, Li Y, Liu R, Liang W, Zhu J, Zhang W, Jin H. Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 2012;13:R20. doi: 10.1186/gb-2012-13-3-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z, Chen H, Huang X, Xia R, Zhao Q, Lai J, Teng K, Li Y, Liang L, Du Q, Zhou X, Guo H, Xie Q. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell. 2011;23:273–288. doi: 10.1105/tpc.110.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zrachya A, Glick E, Levy Y, Arazi T, Citovsky V, Gafni Y. Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus—srael. Virology. 2007;358:159–165. doi: 10.1016/j.virol.2006.08.016. [DOI] [PubMed] [Google Scholar]


