Abstract
Symptomless grape plants (Vitis vinifera) cultivated in Jind, Punjab, have been found to carry a Grapevine red blotch virus (GRBV). Evaluation of full length DNA sequence (3204 bp) of the virus (KU522121) has revealed similarity with mastrevirus, begomovirus, and other Grapevine red blotch viruses reported in the US and Canada. Similar to naturally growing plants, agroinfiltrated model plants with infectious clone of GRBV do not show any visible disease warning sign. To the best of our knowledge, this is the first report of a symptomless host Vitis vinifera from Indian vineyards harbouring a Grapevine geminivirus.
Electronic supplementary material
The online version of this article (10.1007/s13337-018-0477-x) contains supplementary material, which is available to authorized users.
Keywords: Vitis vinifera, Geminivirus, GRBV, India
Geminiviruses are one of the largest group of plant viruses containing single stranded circular DNA, which is encapsulated in geminate particles. This virus is prevalent in the tropical and subtropical regions of the world and is known to infect crops, ornamentals, and weed plants [15, 22]. A study examining genome wide pairwise sequence identity, genome organization, host range, and insect transmission vector has recently classified the family Geminiviridae into nine genera: Mastrevirus, Curtovirus, Begomovirus, Turncurtovirus, Topocuvirus, Eragrovirus, Becurtovirus, Capulavirus, and Grablovirus [23]. Vitis vinifera (Vitaceae) is a deciduous woody climber with coiled climbing tendrils and large leaves. It contains diverse forms of active compounds such as polyphenols, Melatonin (N-acetyl-5-methoxytryptamine), oils, stilbene resveratrol organic acids, etc. [20]. Vitis vinifera and it’s by-products have remarkable applications in the healthcare systems which include photoprotection properties against UV radiation, antioxidant, anti-aging, depigmenting, anti-inflammatory action, wound healing, antimicrobial, and anti-obesity properties [4, 8]. It is one of the important plant species because of its diverse uses in the production of wine, grape juice, and other food products [10]. In grapevines, graft-transmissible agents (GTAs) that have been recognized so far include several viruses (around 60 known species), viroids, phytoplasmas, and other parasites (insect-transmitted xylematic bacteria) [19]. The present study was aimed to characterize geminiviruses prevailing in the plants latently, without causing any visible disease symptoms. Keeping in mind the growing interest in plant viruses which are unable to reveal infection symptoms [24], the present study was conducted on Vitis vinifera growing in the fields of the state of Punjab, India.
Leaf samples were collected from the grape plantations in Jind, Punjab (Supplementary 1). The samples were then cleaned, cut, rolled in a piece of tissue paper, and stored at − 20 °C until DNA isolation. To investigate the potential geminiviral prevalence, total DNA was extracted from the leaves of grapevine plants using the Cetyl Trimethyl Ammonium Bromide (CTAB) method (kit for viral DNA from serum/plants, HiMedia Laboratories Pvt Ltd), as per the manufacturer’s instructions [9, 12]. During the survey for the identification of symptomless geminiviruses, 5 grapevine plants, each from 9 different vineyards/fields (total collected samples 5 × 9 = 45) of the northern regions of India, were screened; of these plants, 1 from the field number 4 gave non-specific amplification in PCR with begomovirus-specific primers. PCR reaction was carried in a 25 μL volume, containing 100 ng DNA template, Taq 10 × buffers (10 mmol/L Tris–HCl, pH 8.8; 50 mmol/L KCl) 25 mmol/L MgCl2, 200 μmol/L of each dNTPs, 2 units of Taq DNA Polymerase, Nuclease free water and 10 pmol/L of each primer. The reaction profile was pre-PCR denaturation at 94 °C for 120 s followed by 35 cycles of denaturing at 94 °C for 45 s, annealing at 55 °C for 45 s and extension at 72 °C for 45 s, and a final extension at 72 °C for 5 min. After amplification, 4 µL aliquot from each sample was electrophoresed in a 1% agarose gel and visualized by staining with ethidium bromide and UV illumination [14].
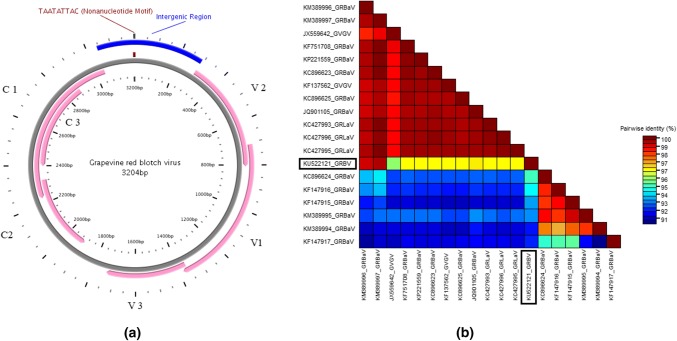
The entire 45 samples were subjected to Rolling Circle Amplification using a TempliPhi DNA amplification kit (GE Healthcare, Little Chalfont, Bukinghamshire, UK), as per the manufacturer’s instructions [18]; remarkably, 1 of the samples was amplified. To identify the possible geminivirus for better assessment and to authenticate the results, entire grapevine plantation from the field number 4 was screened. Eight out of 68 plants showed similar amplification. The whole viral genome (tentative) amplified as a RCA product was successfully digested with Pst I restriction enzyme, and resulted fragments (~ 3200 bp) were cloned in suitable pBluescript SK + vector (Stratagene, La Jolla, USA). The complete nucleotide sequence of all the 8 isolates was determined by sequencing; upon alignment with CLUSTAL-W, all the isolates showed identical sequences. Therefore, only one sequence was submitted to NCBI, USA, under accession number KU522121 (clone RK-1: 3204 bp). The result revealed that DNA contains six predicted open reading frames (ORFs V2, V1, V3, C2, C1 and C3) with three ORFs (V2, V1, V3) in the virion-sense and three ORFs (C2, C1, C3) in the complementary-sense strand (Table 1). The sequence showed a predicted stem-loop structure (between 3196 to 2 bp), which is the origin of replication of the virion-sense strand [1], and an Intergenic Region (between 3045 to 291 bp), as drawn with the help of PlasMapper [6] (Fig. 1a). RK-1 harbouring in Vitis vinifera has a putative conserved domain of the geminivirus coat protein superfamily encoded by V1 (coat protein) and putative conserved domain of the geminivirus replication-associated protein (Rep) encoded by C1, with an E-value of 1.18e−10 and 1.09e−10, respectively, that are conserved among geminiviruses as per the Conserved Domain Search Service (CD Search) tool [13].
Table 1.
Positions and coding capacity of predicted genes for the genome of geminivirus DNA isolated from grape plants, and their highest amino acid sequence identities
| Description | ORFs | Strand | Start codon (nucleotide coordinates) | Stop codon (nucleotide coordinates) | Predicted size (no. of amino acids) | Predicted molecular weight (kDa) | % amino acid identity |
|---|---|---|---|---|---|---|---|
| V2 protein | V2 | Sense strand | 292 | 807 | 171 | 19.20 | 99 with Grapevine red-blotch associated virus (YP_008400113) |
| Coat protein | V1 | Sense strand | 710 | 1384 | 224 | 25.29 | 99 with Grapevine geminivirus (AGV40193) |
| V3 Protein | V3 | Sense strand | 1365 | 1736 | 123 | 14.60 | 99 with Grapevine red-blotch associated virus (AGS42418) |
| Replication associated protein | C2 | Complement strand | 1898 | 2323 | 141 | 16.58 | 94 with Grapevine red-blotch associated virus (AHL69796) |
| Replication associated protein | C1 | Complement strand | 2229 | 3044 | 271 | 31.66 | 92 with Grapevine red-blotch associated virus (YP_008400117) |
| C3 Protein | C3 | Complement strand | 2399 | 2881 | 160 | 17.88 | 100 with Grapevine red-blotch associated virus (AGS42415) |
Fig. 1.
a Genome map of Grapevine virus isolated from Vitis vinifera drawn with the help of PlasMapper. ORFs are marked in the virion sense and complementary strand. b Sequence identity matrix drawn with the help of SDT tool (species demarcation tool) reveals the percentage identity among selected geminiviruses. Grapevine red-blotch associated virus (GRBaV), Grapevine redleaf-associated virus (GRLaV) and Grapevine geminivirus (GVGV)
To assess infectivity of the cloned virus and symptoms it induces in the plants, RK-1 full-length construct was produced. Next, RCA products were partially digested with PstI enzymes to obtain monomer and head-to-tail tandem repeat dimers of full-length DNA, which were later cloned into pCAMBIA2301 vector, as described by Ferreira et al. in 2008. The cloning was confirmed by restriction digestion and sequencing [7]. The clones were agroinfiltrated to 5 plants (newly emerged leaf) each from Chenopodium album, Lycopersicon esculentum and Vitis vinifera (Supplementary 1). Similarly, 5 plants from each species were taken as negative control (not inoculated with the construct). Following agroinfiltration, plants were incubated in a growth chamber at 25 °C, and symptoms were allowed to develop for 2–4 weeks with regular monitoring. Neither Chenopodium album nor Lycopersicon esculentum plants developed any symptoms. Even the green house raised grapevine plants were devoid of any observable symptoms. When no symptoms appeared at the end of 4 weeks, plants were assayed (old as well as young leaves were under investigation) for the presence of virus by PCR investigation. To examine the presence of infectious molecule that could increase the infectivity of the geminivirus, the virus DNA was successfully amplified by PCR with primer pair GVF1 5′-CCACATTCACGTGCAGCATT-3′–GVR1 5′-ATGCAACATTCAAGCCGTGG-3′ developed for RK-1 identification. For dot-blot hybridization, a non-radioactive DIG-DNA labelling kit (Sigma-Aldrich) was used to prepare the probes. Probes for RK-1 isolate were prepared by PCR, which amplified the regions using specific primer pair i.e. GVF2 5′-TACCCGTCTTCGATGGTCCT-3′–GVR2 5′-CCAAGATGGCCAAGATGGCA-3′ and GVF3 5′-GCGGAGAGGTGACAAAGACT-3′–GVR3 5′- AAGGAGGAAGAGCCAAGGAC-3′; the virus DNA was identified as per the kit manufacturer’s instructions. All the sample plants were positive for geminivirus infection, despite lacking the symptoms. The five samples from each plant taken as negative control did not show any viral sequence (Supplementary 2).
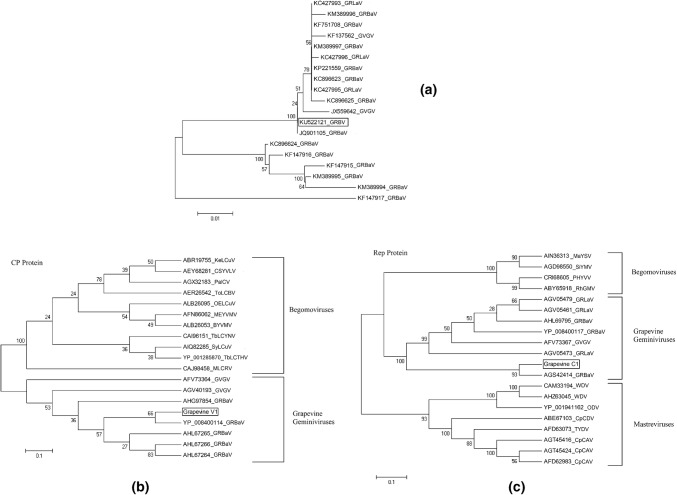
Based on pairwise sequence alignment and identity calculation by SDT tool (species demarcation tool) [17], the complete DNA sequence of the RK-1 isolates showed a high nucleotide sequence identity (93–97%) with other geminiviruses (Fig. 1b) reported from the USA and Canada (Grapevine red-blotch associated virus: GRBaV [JQ901105, KF751708, KC896623, KP221559, KC896625, KC896624, KF147916, KF147915, KF147917], Grapevine redleaf-associated virus: GRLaV [KC427993, KC427996, KC427995] and Grapevine geminivirus: GVGV [KF137562, JX559642]) [11] which are all isolates of geminiviruses infecting grapevine. A nucleotide phylogenetic tree was generated by MEGA 7.0 software, using the neighbor-joining method with 1000 bootstrap replications [21]. Phylogenetic analysis based on the complete DNA sequence of isolate RK-1 geminivirus and other selected complete DNA sequences indicated that isolate RK-1 clusters with the isolates of Grapevine geminiviruses (Fig. 2a). As CP and Rep are the only genes that are homologous to other geminiviruses, we focused on these genes to explore the probable evolutionary relationships between isolate RK-1 and other geminiviruses. We also constructed individual Maximum Likelihood phylogenetic trees for CP (Fig. 2b) and Rep (Fig. 2c) from translated amino acid sequences. While the predicted CP amino acid sequence of RK-1 was clearly most similar to those of Grapevine geminiviruses and other begomoviruses, its Rep amino acid sequence was apparently most closely related to those of mastreviruses and begomoviruses. This indicates a wonderful example of inter-genus geminivirus recombinant. Based on the DNA sequence comparison and presently applicable species demarcation threshold of 91% [5], we concluded that the virus in this study, isolate RK-1 obtained from Vitis vinifera, belongs to the species Grapevine red blotch virus.
Fig. 2.
a Phylogenetic dendrograms based on alignments of geminiviruses genome sequences. Maximum Likelihood trees of the b coat protein (CP) and Rep protein (c) of Grapevine virus isolated from Vitis vinifera with other geminiviruses protein sequences reported in NCBI. The genome sequence accession numbers for coat protein region comparison are as follows: YP_008400114; Grapevine red-blotch associated virus (GRBaV), AGV40193; Grapevine geminivirus (GVGV), AHG97854; Grapevine red-blotch associated virus (GRBaV), AHL67265; Grapevine red-blotch associated virus (GRBaV), AFV73364; Grapevine geminivirus (GVGV), AHL67266; Grapevine red-blotch associated virus (GRBaV), AHL67264; Grapevine red-blotch associated virus (GRBaV), ALB26095; Okra enation leaf curl virus (OELCuV), AFN86062; Mesta yellow vein mosaic virus (MEYVMV), CAJ98458; Malvastrum leaf curl virus (MLCRV), ABR19755; Kenaf leaf curl virus (KeLCuV), AER26542; Tomato leaf curl Bhatinda virus (ToLCBV), CAI96151; Tobacco leaf curl Yunnan virus (TbLCYNV), ALB26053; Bhendi yellow vein mosaic virus (BYVMV), AIQ82285; Synedrella leaf curl virus (SyLCuV), AEY68281; Croton sparsiflorus yellow vein Lakshmangarh virus (CSYVLV), AGX32183; Papaya leaf curl virus (PalCV), YP_001285870; Tobacco leaf curl Thailand virus (TbLCTHV). The genome sequence accession number for rep protein comparison are as follows: AGS42414; Grapevine red-blotch associated virus (GRBaV), YP_008400117; Grapevine red-blotch associated virus (GRBaV), AGV05473; Grapevine redleaf-associated virus (GRLaV), AGV05479; Grapevine redleaf-associated virus (GRLaV), AHL69795; Grapevine red-blotch associated virus (GRBaV), AFV73367; Grapevine geminivirus (GVGV), AGV05461; Grapevine redleaf-associated virus (GRLaV), AIN36313; Macroptilium yellow spot virus (MaYSV), AGD98550; Sida yellow mosaic virus (SiYMV), CRI68605; Pepper huasteco yellow vein virus (PHYVV), ABY65918; Rhynchosia golden mosaic virus (RhGMV), AGT45424; Chickpea chlorosis Australia virus (CpCAV), ABE67103; Chickpea chlorotic dwarf virus (CpCDV), YP_001941162; Oat dwarf virus (ODV), AFD63073; Tobacco yellow dwarf virus (TYDV), AGT45416; Chickpea chlorosis Australia virus (CpCAV), CAM33194; Wheat dwarf virus (WDV), AHZ63045; Wheat dwarf virus (WDV), AFD62983; Chickpea chlorosis Australia virus (CpCAV)
The virus was first reported in 2012 and has been subsequently detected in major grape production regions [3]. The recombination studies on Grapevine red-blotch virus have been done previously by the pioneers who have identified the virus in the US and Canada, and our results are in line with the previous studies [2]. Recently, AL Rwahnih et al. [2] have detected a novel virus in Vitis vinifera from South Korea. They have also discovered a naturally occurring defective subviral DNA. Our study discovered a geminivirus associated with Vitis vinifera in India; moreover, the disease has not been investigated previously in Indian vineyards. Therefore, to the best of our knowledge, this is the first report on molecular characterization of Grapevine red-blotch virus from India. This species of Grapevine red-blotch virus might not be well adapted to Indian Vitis vinifera, suggesting a reason for symptomless temperament. Many plant viruses are recognized on the basis of leaf symptoms, whereas infected grape plant leaves are characterized during the viral infection without visible disease symptoms. The economic loss caused by this disease is difficult to assess, since infected plants are often symptomless. For this reason, appropriate management strategies are needed [16]. As there are no visible disease or infection symptoms, we suggest hosting of Grapevine red-blotch virus in Vitis vinifera. This identification might represent the possibility of a serious threat to the entire wine production system and other economically important plants in India in near future. The outcome of the present investigation is that grape plants are symptomless carriers of Grapevine geminivirus.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1.Vitis vinifera symptomless sample (A, B, C) collected from the grape plantation fields in Punjab. Neither the Chenopodium album (D) nor the Lycopersicon esculentum (E) plants developed any symptoms when induced with infectious construct of isolate RK-1. Even the Green House raised Grapevine plants were devoid of any observable symptoms (F). (PNG 1098 kb)
Supplementary material 2. Dot Blot hybridization of DNA samples extracted from the infectious construct inoculated (1 to 5) and non inoculated/healthy model plants (Abbreviated as Ca = Chenopodium album, Le = Lycopersicon esculentum and Vv = Vitis vinifera) (6 to 10). (PNG 235 kb)
Acknowledgements
The authors are thankful to Science and Engineering Research Board – Department of Science and Technology, New Delhi, India for the financial assistance (File Nos. YSS/2015/000265 and EMR/2016/000579).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Al Rwahnih M, Dave A, Anderson MM, Rowhani A, Uyemoto JK, Sudarshana MR. Association of a DNA virus with grapevines affected by red blotch disease in California. Phytopathology. 2013;103:1069–1076. doi: 10.1094/PHYTO-10-12-0253-R. [DOI] [PubMed] [Google Scholar]
- 2.Al Rwahnih M, Alabi OJ, Westrick NM, Golino D, Rowhani A. Description of a novel monopartite geminivirus and its defective subviral sequence in grapevine (Vitis vinifera L.) Phytopathology. 2016;107:240–251. doi: 10.1094/PHYTO-07-16-0282-R. [DOI] [PubMed] [Google Scholar]
- 3.Bahder BW, Zalom F, Jayanth M, Sudarshana MR. Phylogeny of geminivirus coat protein sequences and digital PCR aid in identifying Spissistilus festinus (Say) as a vector of Grapevine red blotch-associated virus. Phytopathology. 2016;106:1223–1230. doi: 10.1094/PHYTO-03-16-0125-FI. [DOI] [PubMed] [Google Scholar]
- 4.Biasi F, Deiana M, Guina T, Gamba P, Leonarduzzi G, Poli G. Wine consumption and intestinal redox homeostasis. Redox Biol. 2014;2:795–802. doi: 10.1016/j.redox.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JF, Fiallo-Olivé E, Briddon RW, Hernández-Zepeda C, Idris A, Malathi VG, Martin DP, Rivera-Bustamante R, Ueda S, Varsani A. Revision of begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol. 2015;160:1593–1619. doi: 10.1007/s00705-015-2398-y. [DOI] [PubMed] [Google Scholar]
- 6.Dong X, Stothard P, Forsythe IJ, Wishart DS. PlasMapper: a web server for drawing and auto-annotating plasmid maps. Nucleic Acids Res. 2004;32(Web Server issue):W660–W664. doi: 10.1093/nar/gkh410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira PTO, Lemos TO, Nagata T, Inoue-Nagata AK. One-step cloning approach for construction of agroinfectious begomovirus clones. J Virol Methods. 2008;147:351–354. doi: 10.1016/j.jviromet.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Friedman M. Antibacterial, antiviral, and antifungal properties of wines and winery byproducts in relation to their flavonoid content. J Agric Food Chem. 2014;62:6025–6042. doi: 10.1021/jf501266s. [DOI] [PubMed] [Google Scholar]
- 9.Gaur RK, Prajapat R, Marwal A, Sahu A, Rathore MS. First report of a begomovirus infecting Mimosa pudica in India. J Plant Pathol. 2011;93(S4):80. [Google Scholar]
- 10.Georgiev V, Ananga A, Tsolova V. Recent advances and uses of grape flavonoids as nutraceuticals. Nutrients. 2014;6:391–415. doi: 10.3390/nu6010391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krenz B, Thompson JR, Fuchs M, Perry KL. Complete genome sequence of a new circular DNA virus from grapevine. J Virol. 2012;86:7715. doi: 10.1128/JVI.00943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manen JF, Sinitsyna O, Aeschbach L, Markov AV, Sinitsyn A. A fully automatable enzymatic method for DNA extraction from plant tissue. BMC Plant Biol. 2005;5:3–23. doi: 10.1186/1471-2229-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marwal A, Sahu AK, Gaur RK. First report of airborne begomovirus infection in Melia azedarach (Pride of India), an ornamental tree in India. Aerobiologia. 2014;30:211–215. doi: 10.1007/s10453-013-9319-x. [DOI] [Google Scholar]
- 15.Marwal A, Mishra M, Sekhsaria C, Gaur RK. Computational analysis and predicting ligand binding site in the rose leaf curl virus and its betasatellite proteins: a step forward for antiviral agent designing. In: Saxena S, Tiwari AK, editors. Begomoviruses: occurrence and management in Asia and Africa, Chapter 9. Berlin: Springer; 2017. pp. 157–168. [Google Scholar]
- 16.Marwal A, Gaur RK, Khurana SMP. Possible approaches for developing different strategies to prevent transmission of Geminiviruses to important crops. Chapter 18. In: Gaur RK, Paul Khurana SM, Dorokhov Y, editors. Plant viruses: diversity, interaction and management. Boca Raton: CRC Press; 2018. [Google Scholar]
- 17.Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLOS One. 2014;9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nehra C, Marwal A, Verma RK, Gaur RK. Molecular characterization of begomoviruses DNA-A and associated beta satellites with new host Ocimum sanctum in India. Proc Natl Acad Sci India Sect B Biol Sci. 2018 [Google Scholar]
- 19.Oliver JE, Fuchs MF. Tolerance and resistance to viruses and their vectors in Vitis sp.: a virologist’s perspective of the literature. Am J Enol Vitic. 2011;62:438–451. doi: 10.5344/ajev.2011.11036. [DOI] [Google Scholar]
- 20.Pezzuto JM, Kondratyuk TP, Ogas T. Resveratrol derivatives: a patent review (2009–2012) Expert Opin Ther Pat. 2013;23:1529–1546. doi: 10.1517/13543776.2013.834888. [DOI] [PubMed] [Google Scholar]
- 21.Prajapat R, Marwal A, Sahu A, Gaur RK. Molecular In Silico structure and recombination analysis of betasatellite in Calotropis procera associated with begomovirus. Arch Phytopathol Plant Prot. 2012;45:1980–1990. doi: 10.1080/03235408.2012.720460. [DOI] [Google Scholar]
- 22.Prajapat R, Marwal A, Gaur RK. Begomovirus associated with alternative host weeds: a critical appraisal. Arch Phytopathol Plant Prot. 2013;47:157–170. doi: 10.1080/03235408.2013.805497. [DOI] [Google Scholar]
- 23.Varsani A, Roumagnac P, Fuchs M, Navas-Castillo J, Moriones E, Idris A, Briddon RW, Rivera-Bustamante R, Murilo Zerbini F, Martin DP. Capulavirus and grablovirus: two new genera in the family Geminiviridae. Arch Virol. 2017;162:1819–1831. doi: 10.1007/s00705-017-3268-6. [DOI] [PubMed] [Google Scholar]
- 24.Wylie S, Li H, Jones MGK. Donkey orchid symptomless virus: a viral ‘Platypus’ from Australian terrestrial orchids. PLoS ONE. 2013;8:e79587. doi: 10.1371/journal.pone.0079587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1.Vitis vinifera symptomless sample (A, B, C) collected from the grape plantation fields in Punjab. Neither the Chenopodium album (D) nor the Lycopersicon esculentum (E) plants developed any symptoms when induced with infectious construct of isolate RK-1. Even the Green House raised Grapevine plants were devoid of any observable symptoms (F). (PNG 1098 kb)
Supplementary material 2. Dot Blot hybridization of DNA samples extracted from the infectious construct inoculated (1 to 5) and non inoculated/healthy model plants (Abbreviated as Ca = Chenopodium album, Le = Lycopersicon esculentum and Vv = Vitis vinifera) (6 to 10). (PNG 235 kb)