Understanding adipogenesis, the process of adipocyte development, may provide new ways to treat obesity and related metabolic diseases. Adipogenesis is controlled by coordinated actions of lineage-determining transcription factors and epigenomic regulators.
KEYWORDS: adipogenesis, epigenomic regulation, transcriptional regulation
ABSTRACT
Understanding adipogenesis, the process of adipocyte development, may provide new ways to treat obesity and related metabolic diseases. Adipogenesis is controlled by coordinated actions of lineage-determining transcription factors and epigenomic regulators. Peroxisome proliferator-activated receptor gamma (PPARγ) and C/EBPα are master “adipogenic” transcription factors. In recent years, a growing number of studies have reported the identification of novel transcriptional and epigenomic regulators of adipogenesis. However, many of these novel regulators have not been validated in adipocyte development in vivo and their working mechanisms are often far from clear. In this minireview, we discuss recent advances in transcriptional and epigenomic regulation of adipogenesis, with a focus on factors and mechanisms shared by both white adipogenesis and brown adipogenesis. Studies on the transcriptional regulation of adipogenesis highlight the importance of investigating adipocyte differentiation in vivo rather than drawing conclusions based on knockdown experiments in cell culture. Advances in understanding of epigenomic regulation of adipogenesis have revealed critical roles of histone methylation/demethylation, histone acetylation/deacetylation, chromatin remodeling, DNA methylation, and microRNAs in adipocyte differentiation. We also discuss future research directions that may help identify novel factors and mechanisms regulating adipogenesis.
INTRODUCTION
Obesity, characterized by an excessive accumulation of adipose tissue (AT), is a major risk factor for type II diabetes and other metabolic diseases. There are two main types of adipose tissue with distinct functions in human body. White adipose tissue (WAT) stores excess energy in the form of triglycerides, while brown adipose tissue (BAT) burns energy through thermogenesis (1, 2). Promoting BAT development and suppressing WAT development may offer approaches for treating obesity. Therefore, understanding the molecular mechanisms behind adipocyte differentiation (adipogenesis) could provide new insights into a therapeutic strategy for combating obesity.
Adipocytes, which primarily compose adipose tissue, are derived from multipotent mesenchymal stem cells (MSCs). Two phases of adipogenesis are well characterized: commitment of MSCs to preadipocytes and terminal differentiation of preadipocytes toward mature adipocytes (3). Over the last few decades, there has been progress in understanding transcriptional and epigenomic regulation of adipogenesis. The major adipogenic transcription factors (TFs) peroxisome proliferator-activated receptor gamma (PPARγ) and C/EBPα work with other TFs and epigenomic regulators to activate genes required for terminal differentiation of preadipocytes (Fig. 1 and 2).
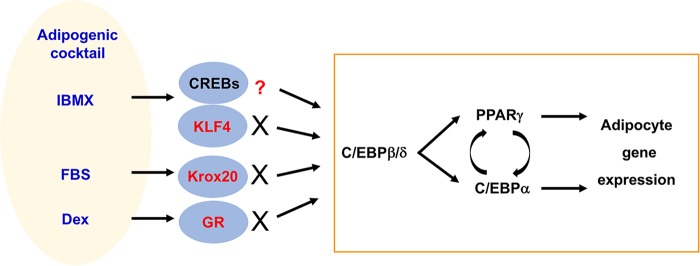
FIG 1.
Activation or induction of TFs during adipogenesis in culture. The chemical cocktail of IBMX (3-isobutyl-1-methylxanthine), fetal bovine serum (FBS), and dexamethasone (Dex) has been routinely used to induce adipogenesis in culture. This cocktail induces expression of TFs KLF4 and Krox20 and activates CREBs and GR, which cooperate to activate C/EBPβ and C/EBPδ. It is generally accepted that C/EBPβ and C/EBPδ induce the expression of master adipogenic TFs PPARγ and C/EBPα, which directly activate adipocyte gene expression (highlighted by a rectangle). Recent studies indicated that induction of KLF4 and Krox20 and activation of GR are artifacts in cell culture. Neither KLF4 nor Krox20 nor GR is required for adipogenesis in culture and in mice. The role of CREBs in adipogenesis in vivo is unclear.
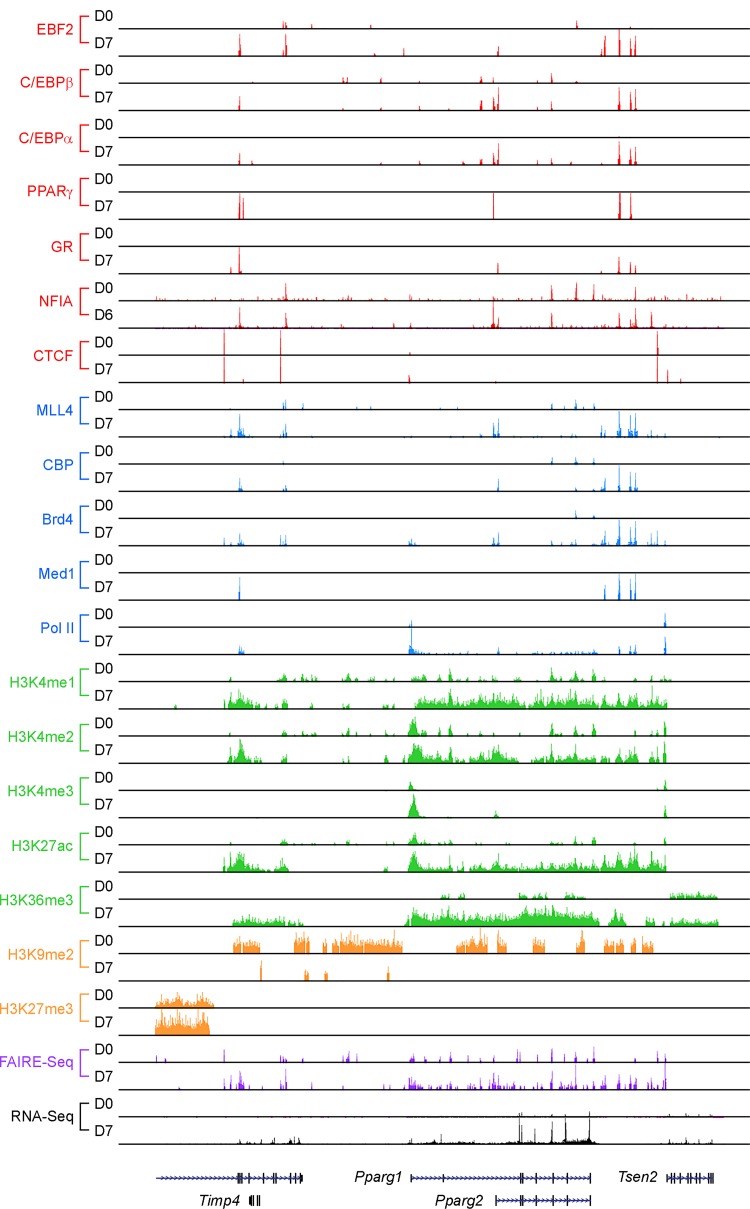
FIG 2.
Transcriptional and epigenomic regulators enriched on Pparg locus in adipogenesis. A genome browser view of the Pparg locus before (day zero [D0]) and after (D7) adipogenesis of brown preadipocytes is shown. Published data were obtained from the GEO database and reanalyzed (NFIA data are from GSE83764, Brd4 data are from GSE99101, and all other data are from GSE74189) (41, 49, 97). CTCF, CCCTC-binding factor; RNA-Seq, transcriptome sequencing.
Many important findings in the adipogenesis field were obtained using immortalized cell lines such as 3T3-L1 preadipocytes for studying terminal differentiation (4) and C3H10T1/2 MSCs for studying adipogenic commitment (5). These cell lines cannot fully represent adipogenesis in vivo, as differentiation in culture is induced by a chemical cocktail that is not present in physiological settings. To overcome this limitation, in vivo studies involving crossing conditional knockout mice with tissue-specific Cre-expressing mice are necessary. These Cre-expressing mouse lines include Myf5-Cre (6), Adipoq-Cre (7, 8), and Fabp4-Cre (9–11). Myf5-Cre deletes floxed genes in progenitor cells of BAT and a subset of skeletal muscles. It is particularly useful for studying adipogenesis during embryonic development. Adipoq-Cre and Fabp4-Cre delete floxed genes in mature adipocytes. However, Fabp4-Cre recombinase activity was also observed in nonadipose tissues such as heart, skeletal muscle, and testis and in multiple cell populations within adipose tissues (12, 13).
In this review, the focus is primarily on TFs and epigenomic regulators implicated in the terminal differentiation process that is common to white adipogenesis and brown adipogenesis (summarized in Tables 1 and 2). The factors that specifically regulate brown adipocyte functions such as Irf4, Zfp516, Jmjd3, and Jmjd1a have been extensively reviewed elsewhere (14–16).
TABLE 1.
Summary of transcription factors and coactivators implicated in adipogenesis
| Factor |
In vitro |
In vivo |
|||||
|---|---|---|---|---|---|---|---|
| Effecta | Cells | Method(s)b | Reference | Mouse model | Phenotype in micec | Reference | |
| DNA binding transcription factors | |||||||
| AP-1 | NAd | NA | NA | NA | A-ZIP/F transgene | No WAT, ↓ BAT | 72 |
| ATF4 | ↑ | hMSCs | KD | 44 | Atf4−/− | Lean, resistant to obesity | 46 |
| ↑ | 3T3-L1 | KD | 45 | ||||
| C/EBPα | ↑ | 3T3-L1 | asRNA | 21 | Cebpa−/−; Albumin-Cebpa | No WAT, delayed BAT development | 17 |
| C/EBPβ/δ | ↑ | MEFs | KO | 35 | Cebpb−/−; Cebpd−/− | ↓ WAT, no lipid in BAT | 35 |
| CREB | ↑ | MEFs, 3T3-L1 | DN, KD | 65 | NA | NA | NA |
| EBF1 | ↑ | NIH 3T3, 3T3-L1 | OE, KD | 37 | Ebf1−/− | ↓ WAT | 42 |
| EBF2 | ↑ | NIH 3T3, 3T3-L1 | OE, KD | 37 | Ebf2−/− | ↓ BAT | 39 |
| ↑ | Brown preadipocytes | OE, KD | 39 | ||||
| GATA2/3 | ↓ | 3T3-F442A, ESe | OE, KO | 69 | NA | NA | NA |
| GR | ↑ | 3T3-L1 | KD | 31 | GRf/f; Myf5-Cre | Normal BAT at E18.5 and 6 wks | 57 |
| ↑ | Preadipocytes | KD, KO | 57 | LSL-luc; Adipoq-Cre; GR−/− | Normal BAT + WAT at E18 | 58 | |
| KLF2 | ↓ | 3T3-L1 | OE | 71 | NA | NA | NA |
| KLF4 | 3T3-L1 | KD | 53 | Klf4f/f; Myf5-Cre | Normal BAT | 54 | |
| — | Preadipocytes | KO | 54 | ||||
| KLF5 | ↑ | MEFs, 3T3-L1 | KD | 66 | Klf5+/− | ↓ WAT at P3, normal at 4 wks | 66 |
| KLF15 | ↑ | NIH 3T3, 3T3-L1 | OE, KD | 67 | NA | NA | NA |
| Krox20 | 3T3-L1 | KD | 52 | Krox20f/f; Myf5-Cre | Normal BAT | 54 | |
| — | Preadipocytes | KO | 54 | ||||
| NFIA/B | ↑ | 3T3-L1 | OE, KD | 48 | Nfia−/− | Normal BAT mass, ↓ Ucp1 | 49 |
| ↑ | Brown preadipocytes | KD | 49 | ||||
| PPARγ | ↑ | NIH 3T3 | OE | 18 | Pparg−/− tetraploid | No BAT + WAT | 19 |
| ↑ | Brown preadipocytes | KO | 54 | Ppargf/f; Myf5-Cre | No BAT | 54 | |
| SREBP1c | ↑ | 3T3-L1 | DN | 74 | Srebp1−/− | Embryonic lethal, normal WAT in surviving mice | 75 |
| STAT5a | ↑ | 3T3-L1 | OE, DN | 68 | NA | NA | NA |
| Zfp423 | ↑ | NIH 3T3, 3T3-L1 | OE, KD | 60 | Zfp423−/− | ↓ WAT and BAT at E18.5 | 60 |
| Zfp423f/f; Adipoq-Cre | ↓ fetal inguinal WAT | 63 | |||||
| Zfp423f/f; Adipoq-rtTa; TRE-Cre | Browning of WAT | 61 | |||||
| Coactivators | |||||||
| Med1 | ↑ | MEFs | KO | 78 | NA | NA | NA |
| Med14 | ↑ | 3T3-L1 | KD | 79 | NA | NA | NA |
| Med23 | ↑ | MEFs, 3T3-L1 | KD, KO | 80 | NA | NA | NA |
| SRC-2 | ↑ | NIH 3T3, 3T3-L1 | OE | 81 | Src-2−/− | ↓ interscapular WAT | 81 |
| SRC-3 | ↑ | MEFs | KO | 82 | Src-3−/− | ↓ WAT | 82 |
| TLE3 | ↑ | 10T1/2, 3T3-L1 | OE, KD | 84 | Fabp4-Tle3 transgene | ↑ WAT, whitening of BAT | 84 |
| ↑ | Brown preadipocytes | OE | 83 | Tle3f/f; Adipoq-Cre | ↑ thermogenic genes in WAT + BAT | 83 | |
↑ and ↓ indicate promoting and inhibitory effects on adipogenesis, respectively; —, no effect on adipogenesis.
KD, knockdown; asRNA, antisense RNA; KO, knockout; OE, overexpression; DN, overexpression of dominant-negative form.
↑, increase; ↓, decrease.
NA, not available.
ES, embryonic stem.
TABLE 2.
Summary of epigenomic regulators implicated in adipogenesis
| Factor |
In vitro |
In vivo |
|||||
|---|---|---|---|---|---|---|---|
| Effectb | Cells | Method(s)a | Reference(s) | Mouse model | Phenotype in micec | Reference | |
| Lysine methyltransferases | |||||||
| Ehmt1 | NAd | NA | NA | NA | Ehmt1f/f; Myf5-Cre | ↓ BAT | 100 |
| Ezh2 | ↑ | Brown preadipocytes | KO | 108 | NA | NA | NA |
| G9a | ↓ | 3T3-L1, brown preadipocytes | OE, KD, KO | 98 | G9af/f; Fabp4-Cre | ↑ WAT + BAT | 98 |
| MLL3/MLL4 | ↑ | 3T3-L1, brown preadipocytes | KD, KO | 91 | Mll4f/f; Myf5-Cre | ↓ BAT | 91 |
| Nsd2 | ↑ | Brown preadipocytes | KD | 109 | LSL-H3.3K36M; Myf5-Cre | ↓ BAT | 109 |
| Fabp4-H3.3K36M transgene | Whitening of BAT | ||||||
| Setdb1 | ↓ | 3T3-L1 | KD | 103 | NA | NA | NA |
| Lysine demethylases | |||||||
| Kdm5 | ↑ | 3T3-L1, brown preadipocytes | KD | 113 | NA | NA | NA |
| Lsd1 | ↑ | 3T3-L1 | KD | 110 | Lsd1f/f; Myf5-Cre | ↓ BAT | 112 |
| Lsd1f/f; Ucp1-Cre | ↑ body fat | ||||||
| Arginine methyltransferases | |||||||
| Carm1 | ↑ | 3T3-L1 | KD | 115 | Carm1−/− | ↓ lipid in BAT | 115 |
| Prmt5 | ↑ | 3T3-L1, C3H10T1/2 | KD, OE, asRNA | 118, 119 | NA | NA | NA |
| Histone acetyltransferases | |||||||
| CBP/p300 | ↑ | 3T3-L1 | Ribozyme | 125 | Crebbp+/− | ↓ WAT | 126 |
| Gcn5/PCAF | ↑ | Brown preadipocytes | KO | 124 | Pcaf−/−; Gcn5f/f; Myf5-Cre | ↓ BAT | 124 |
| Histone deacetylases and sirtuins | |||||||
| Hdac1/2 | ↑ | Fibroblasts | KO | 130 | NA | NA | NA |
| Hdac9 | ↓ | 3T3-L1, preadipocytes | OE, KO | 131 | NA | NA | NA |
| Sirt1 | ↓ | MEFs, C3H10T1/2 | KO, KD | 133 | Sirt1f/f; Fabp4-Cre | ↑ WAT + BAT HFD | 132 |
| Sirt2 | ↓ | 3T3-L1 | KD | 134, 135 | NA | NA | NA |
| Sirt6 | ↑ | MEFs, 3T3-L1 | KD, KO | 137 | NA | NA | NA |
| Sirt7 | ↑ | NA | NA | NA | Sirt7−/− | ↓ WAT | 136 |
| Epigenomic reader Brd4 | ↑ | Brown preadipocytes | KO | 97 | Brd4f/f; Myf5-Cre | ↓ BAT | 97 |
| ↑ | 3T3-L1, C3H10T1/2 | JQ1 | 140 | ||||
| Chromatin remodeling complex SWI/SNF | ↑ | Fibroblasts | DN | 145 | NA | NA | NA |
| DNA methyltransferases/demethylases | |||||||
| DNMT1 | ↓ | 3T3-L1 | KD | 149 | NA | NA | NA |
| ↑ | 5-aza-dC | 150 | |||||
| Tet1/2 | ↑ | MEFs | KO | 153 | NA | NA | NA |
Abbreviations are as defined for Table 1.
↑ and ↓ indicate promoting and inhibitory effects on adipogenesis, respectively.
↑, increase; ↓, decrease.
NA, not available.
TRANSCRIPTIONAL REGULATION OF ADIPOGENESIS
PPARγ and C/EBPs are required for adipogenesis.
PPARγ and C/EBPα are considered master regulators of adipogenesis and have been shown to be essential for adipogenesis both in culture and in vivo (17–21). PPARγ, which has been extensively reviewed, controls terminal differentiation of adipocytes (1, 22–25) and is required for maintaining their differentiated state (26, 27). C/EBPα acts in concert with PPARγ to establish phenotypes of mature adipocytes (28, 29). It is generally accepted that adipogenesis is controlled by a transcriptional cascade (1, 22, 30) initiated through chromatin opening by C/EBPβ (31, 32). C/EBPβ and C/EBPδ are expressed in the early stage of adipogenesis in culture (33) and induce PPARγ and C/EBPα expression (34). However, C/EBPs cannot function efficiently without PPARγ.
C/EBPβ and C/EBPδ are essential for adipose tissue development, but their role in adipogenesis is not fully understood. C/EBPβ and C/EBPδ regulate adipogenesis synergistically as evidenced by lack of lipid droplets in BAT and significantly reduced epididymal WAT levels in double knockout (Cebpb−/− Cebpd−/−) mice (35). Despite the reduced fat mass, Cebpb−/− Cebpd−/− mice express normal levels of PPARγ and C/EBPα, suggesting the presence of factors that enable cells to bypass the requirement for C/EBPβ and C/EBPδ in vivo (1). To clarify the role of C/EBPβ and C/EBPδ in adipogenesis, further studies using tissue-specific knockout mice will be necessary. In 3T3-L1 preadipocytes, opening on Pparg and other adipogenic genes occurs within 4 h after treatment with an adipogenic cocktail. Knockdown of C/EBPβ impairs chromatin opening on these adipogenic gene loci (32). Whether C/EBPβ functions as a pioneering factor working upstream of PPARγ and C/EBPα in vivo needs to be validated.
EBF family TFs.
Early B-cell factor (EBF) family TFs, including EBF1, EBF2, and EBF3, were initially suggested to be early regulators of adipogenesis. Ectopic expression of EBF1/2/3 stimulates adipogenesis in NIH 3T3 fibroblasts, while knockdown of EBF1/2 or expression of dominant-negative EBF1 represses 3T3-L1 adipogenesis (36, 37). EBF2 was recently shown to play a critical role in maintaining brown adipocyte identity (38–40). EBF2 recruits PPARγ to brown adipocyte-selective genes (39). Additionally, EBF2 regulates the expression of Dpf3, a gene encoding a brown adipocyte-specific subunit of chromatin remodeling complex, to activate thermogenic gene expression (40).
Although recent works focused primarily on the role of EBF2 in maintaining brown adipocytes, several lines of evidence suggest that EBF2 regulates adipogenesis and common adipogenic gene expression as well. It has been shown that knockdown of EBF2 in 3T3-L1 preadipocytes and in brown preadipocytes impairs adipogenesis (37, 39) and that Ebf2-null mouse embryos have reduced BAT mass compared with wild-type littermates at embryonic day 18 (E18) (39). In addition, reanalysis of published EBF2 chromatin immunoprecipitation sequencing (ChIP-Seq) data showed that EBF2 binding sites are enriched with motifs of adipogenic TFs C/EBPs and PPARγ (Fig. 3A) (41). Furthermore, EBF2 colocalizes with C/EBPs and PPARγ as well as with enhancer epigenomic regulators MLL4 and CBP on Pparg and Cebpa loci in addition to brown adipocyte marker Ucp1 and Prdm16 loci (Fig. 2 and 3B) (41). Therefore, it is possible that EBF2 cooperates with other adipogenic TFs to activate both general adipogenic gene expression and brown adipocyte-specific gene expression. There might be functional redundancy among EBF family members in regulating adipogenesis in vivo. For instance, Ebf1-null mice have reduced WAT mass but not reduced BAT mass, suggesting that EBF1 and EBF2 may have overlapping function in brown adipogenesis (42). Future studies using tissue-specific Ebf family knockout mice will be helpful to clarify their roles in general adipogenesis and fat depot-specific function. It would be informative to determine genomic localizations of EBF1 and EBF3 during adipogenesis.
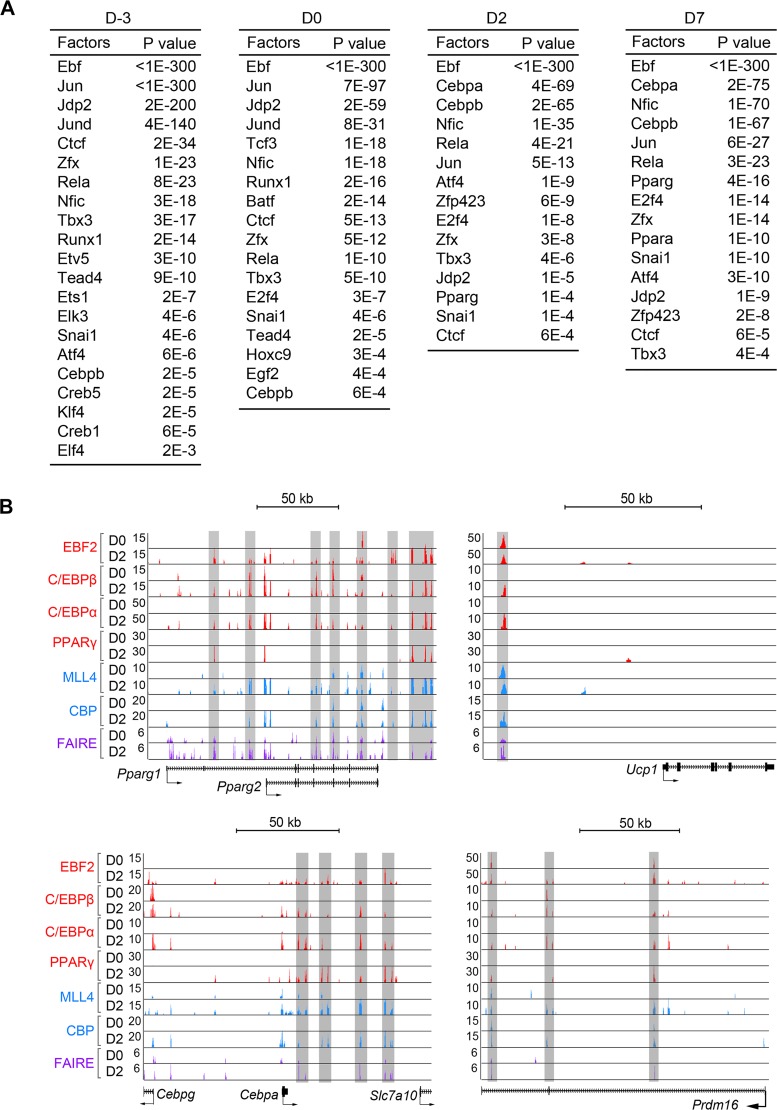
FIG 3.
Genomic binding of EBF2 during brown adipogenesis. (A) Lists of significantly enriched motifs in EBF2 binding sites on enhancers at day −3 (D-3), D0, D2, and D7 of adipogenesis of brown preadipocytes. (B) EBF2 directly regulates expression of both common adipogenic genes Pparg and Cebpa and BAT-specific genes Ucp1 and Prdm16. Highlighted regions indicate active enhancers bound by EBF2 at D2. Published ChIP-Seq data sets of adipogenic TFs EBF2, C/EBPs and PPARγ, and enhancer epigenomic writers MLL4 and CBP, as well as chromatin opening (formaldehyde-assisted isolation of regulatory element sequencing [FAIRE-Seq]) data sets, were obtained from GSE74189 (41) and reanalyzed.
There is some evidence to suggest that EBF family members may function as lineage-specific pioneering TFs. In B cell differentiation, EBF1 was suggested to be a pioneering TF that binds to naive chromatin and induces chromatin opening (43). In preadipocytes, EBF2 premarks a subset of adipogenic superenhancers prior to adipogenesis (41). Future study on whether EBF family members function as lineage-specific pioneering TFs and open chromatin during adipogenesis cooperatively and/or distinctively with C/EBPβ is necessary.
Atf4.
ATF4 has been identified as a positive regulator of adipogenesis in human MSCs (hMSCs) (44). The enhancers with RNA polymerase II (Pol II) binding in confluent hMSCs are enriched with hybrid motifs of C/EBPs and ATF half-sites. While C/EBPβ binds to canonical C/EBP motifs upon induction with adipogenic cocktail, it binds to noncanonical hybrid motifs forming heterodimers with ATF4 in undifferentiated hMSCs. Therefore, ATF4 appears to play a role in the adipogenic commitment stage. ATF4 expression peaks in the confluent hMSCs and gradually decreases upon adipogenic induction. Knockdown of ATF4 reduces C/EBPβ and Pol II binding on enhancers and impairs adipogenesis in hMSCs.
A separate study using 3T3-L1 preadipocytes has shown that levels of ATF4 expression are very low in preadipocytes and peak at a later stage of differentiation and that knockdown of ATF4 inhibits adipogenesis (45). The difference between hMSCs and 3T3-L1 cells in ATF4 expression levels is likely a consequence of the fact that these two cell lines represent distinct stages of adipogenesis. Whole-body Atf4-null mice are lean and resistant to obesity (46). However, it remains unclear whether the leanness of Atf4-null mice is due to failure of adipogenesis or is secondary to growth retardation (47). Lineage-specific knockout of Atf4 will provide insights on the role of ATF4 in adipogenesis in vivo. To understand how ATF4 regulates adipogenesis, genomic localization of ATF4 needs to be determined.
NFI family TFs.
Nuclear factor I (NFI) family TFs were initially identified as positive regulators of adipogenesis by motif analysis of open chromatin regions in differentiated 3T3-L1 cells (48). Knockdown of either NFIA or NFIB inhibits 3T3-L1 adipogenesis, while forced expression of NFIA induces adipogenic genes, including Pparg, Cebpa, and Fabp4, in undifferentiated 3T3-L1 cells. These data suggest that NFIs promote adipogenesis and adipogenic gene expression, although it was not shown whether they are required for adipogenesis in vivo.
A follow-up study using comparative genomic profiling showed that NFI motifs are enriched in BAT-specific open chromatin regions, suggesting that NFIs may play a role in regulating BAT-specific genes (49). Among the four members of NFI family, NFIA expression shows strong induction during adipogenesis of brown preadipocytes, while the expression levels of the others are comparable in brown and 3T3-L1 preadipocytes and remain steady during differentiation. In constrast to previous results in 3T3-L1 cells (48), knockdown of NFIA in brown preadipocytes had no effect on adipogenesis and adipogenic gene expression but decreased the levels of expression of BAT-specific genes. Furthermore, whole-body Nfia-null mouse embryos have normal BAT mass but show decreased expression of Ucp1 and other thermogenic genes (49). In addition, PPARγ binding to the Ucp1 enhancer is impaired in Nfia-null BAT, suggesting a role of NFIA in brown adipocyte-specific gene expression in vivo. Since genomic binding of NFIA is observed with enhancers of adipogenic genes, including Pparg, it is also possible that NFIs redundantly regulate adipogenesis and adipogenic gene expression (Fig. 2).
Krox20, KLF4, and glucocorticoid receptor are dispensable for adipogenesis in vivo.
Our understanding of adipogenesis comes mainly from studies using preadipocyte cells such as 3T3-L1 (50). Adipogenesis is induced by adding chemical cocktail to confluent preadipocytes (Fig. 1). These chemicals activate the TFs CREB and glucocorticoid receptor (GR) and induce expression of Krox20, KLF4, C/EBPβ, and C/EBPδ within hours (51). Activation or ectopic expression of these TFs has been shown to promote adipogenesis in culture (1, 52, 53). However, it was unclear whether endogenous Krox20, KLF4, and GR are essential for adipogenesis in vivo.
A recent study demonstrated that endogenous Krox20 and KLF4 are dispensable for adipogenesis in culture and BAT development in mice (54). Deletion of Krox20 or Klf4 has little effect on either the induction of early adipogenic TFs, including C/EBPβ, C/EBPδ, KLF5, and KLF9, or the expression of adipogenesis markers, including PPARγ, C/EBPα, and Fabp4. Consistent with data determined in vitro, Myf5-Cre-mediated knockout of Krox20 or Klf4 does not affect BAT development in mice. Therefore, the adipogenesis defects observed in the previous knockdown experiments were likely due to off-target effects (52, 53). These recent findings on Krox20 and KLF4 caution us that we cannot solely rely on the knockdown approach and highlight the importance of studying adipogenesis in vivo. Whether Krox20 and KLF4 play specific roles in WAT development remains unclear.
It was believed that GR is critical for adipogenesis, since dexamethasone, a synthetic GR ligand, is commonly used to induce adipogenesis in mouse preadipocytes (Fig. 1), although it is not required for adipogenesis of rat preadipocytes (55, 56). Either knockdown or knockout of GR impairs adipogenesis of 3T3-L1 and primary or immortalized white and brown preadipocytes using a standard 7- to 8-day differentiation protocol (31, 57). Unexpectedly, GR-deficient preadipocytes eventually catch up to wild-type cells and fully differentiate in 3 weeks. In addition, mice that lack GR in Myf5+ progenitors develop normal levels of BAT and are cold tolerant (57). Therefore, even though dexamethasone-bound GR accelerates adipogenesis in culture, endogenous GR is largely dispensable for adipogenesis in culture and BAT development in mice. GR expedites differentiation by recruiting histone acetyltransferase (HAT) CBP to activate C/EBPβ-primed enhancers in culture. An independent study further confirmed that GR is dispensable for adipogenesis in vivo using several approaches (58). First, both wild-type and GR-deficient mouse embryonic fibroblasts (MEFs) can form de novo fat pads in mice. Second, using adipocyte-specific luciferase reporter mice, early inguinal white and brown adipocytes can be detected in GR-deficient embryos, demonstrating that GR is not required for embryonic development of WAT and BAT. Although GR is not required for adipogenesis, it is expressed and involved in lipogenesis in adipocytes (59).
Zfp423.
A quantitative analysis of the transcriptome in clones of Swiss 3T3 fibroblasts identified Zfp423 as a determinant of preadipocytes (60). Compared to nonadipogenic sublines, adipogenic clones highly express Zfp423. Forced expression of Zfp423 in NIH 3T3 fibroblasts induces Pparg2 expression and adipogenesis, while knockdown of Zfp423 in 3T3-L1 inhibits adipogenesis. Whole-body Zfp423 knockout mouse embryos have been shown to have reduced levels of WAT and BAT mass at E18.5, although it is unclear whether this was due to the failure of adipogenesis or secondary to growth defects.
Studies using several tissue-specific Zfp423 knockout mice identified a key role of Zfp423 in maintaining white adipocyte identity by antagonizing EBF2 and repressing thermogenic gene expression (61, 62). Data from a recent study using Adipoq-Cre-mediated Zfp423 knockout mice suggest that Zfp423 is required for the later stage of fetal inguinal WAT development (63). It is worth noting that although Adipoq-Cre is mainly active in mature adipocytes in adult mice, it may target a subset of adipose stromal vascular cells, which likely represent actively differentiating or highly committed preadipocytes, in fetal inguinal WAT (64). It remains unclear whether Zfp423 regulates adipogenesis in general or in a fat depot-specific manner. To understand how Zfp423 regulates adipogenesis, profiling of Zfp423 genomic binding before and after adipogenesis is necessary.
Other TFs implicated in adipogenesis.
Several additional TFs have been implicated in regulating adipogenesis (1, 3, 23). CREB (65), KLF5 (66), KLF15 (67), and STAT5a (68) have been suggested to be positive regulators of adipogenesis based on the finding that expression of dominant-negative form or RNA interference (RNAi)-mediated knockdown of these TFs inhibited adipogenesis in culture. GATA2/3 (69, 70) and KLF2 (71) have been suggested to be negative regulators of adipogenesis, as overexpression of these TFs inhibits 3T3-L1 adipogenesis and Pparg promoter activity in reporter assays. However, most of these TFs have not been validated in physiological settings using animal models and their direct target genes are largely unknown.
Mice with adipose-specific expression of A-ZIP/F, a dominant-negative protein that inhibits C/EBPs and AP-1 family TFs, have no WAT and severely reduced levels of BAT, indicating that either or both of C/EBPs and AP-1 are essential for adipose tissue development in mice (72). Since the AP-1 family includes Jun, Fos, ATF, and JDP, it remains unclear which specific TFs are important for adipogenesis in vivo.
SREBPs (including SREBP-1a, SREBP-1c, and SREBP-2) are key TFs that regulate fatty acid and cholesterol synthesis (73). In culture, SREBP-1c has been shown to promote adipogenesis by providing lipid ligands that mediate PPARγ activation (74). However, whole-body Srebp1-null mice showed no defects in adipose tissue development and adipocyte gene expression (75). Compensatory mechanisms may exist in Srebp1-null mice, as the majority of knockout mice show embryonic lethality. Therefore, phenotypes observed in surviving Srebp1-null mice may not reveal the roles of SREBP-1 in adipose tissues. Future studies using tissue-specific knockout mice are required to better evaluate the roles of SREBPs in adipogenesis.
Transcription coactivators in adipogenesis.
Transcriptional coactivators, such as Mediator proteins and steroid receptor coactivator (SRC) family proteins, function as adaptors that connect TFs to Pol II or histone acetyltransferases to activate gene expression (76, 77). Several components of the Mediator complex, including Med1 (78), Med14 (79), and Med23 (80), and several components of the SRC family, including SRC-2 (81) and SRC-3 (82), have been shown to be required for adipogenesis in culture. However, cell-autonomous roles of these coactivators in adipogenesis have not been demonstrated using tissue-specific knockout mice.
Groucho family member TLE3 has been identified as a white adipocyte-selective PPARγ cofactor. TLE3 inhibits brown adipocyte-enriched gene expression by binding to Prdm16, a brown adipocyte-selective cofactor, and by competing for interactions between Prdm16 and PPARγ, while it promotes white adipocyte-selective gene expression (83). Fabp4 promoter-driven expression of TLE3 in mice leads to an increase in WAT mass (84) and whitening of BAT (83). Adipoq-Cre-mediated knockout of Tle3 induces thermogenic gene expression and improves the thermogenic function of BAT. Genomic binding of TLE3 during white adipogenesis and brown adipogenesis needs to be determined to better understand the fat depot-specific roles of TLE3.
EPIGENOMIC REGULATION OF ADIPOGENESIS
Epigenomic factors play important roles in regulating cell-type-specific gene expression during cell differentiation (85, 86). Key epigenomic regulators include but not limited to the following: histone methyltransferases (MTs)/demethylases, histone acetylases/deacetylases, epigenomic readers, chromatin remodeling factors, DNA methylases/demethylases, and microRNAs (miRNAs) (87, 88).
Lysine methyltransferases.
Lysine residues on histone tails are modified by site-specific methyltransferases (Fig. 4). Histone methylation can be correlated with either gene activation or repression, depending on the methylation sites (89, 90). For example, methylation of histone H3 on lysine 4 (H3K4), lysine 36 (H3K36), or lysine 79 (H3K79) is correlated with gene activation, but methylation on lysine 9 (H3K9) or lysine 27 (H3K27) is correlated with gene repression. Studies have identified distinct roles of H3K4 methyltransferases MLL3 and MLL4 (MLL3/MLL4); H3K9 methyltransferases G9a, Setdb1, and Ehmt1; H3K27 methyltransferase Ezh2; and H3K36 methyltransferase Nsd2 in adipogenesis.
FIG 4.
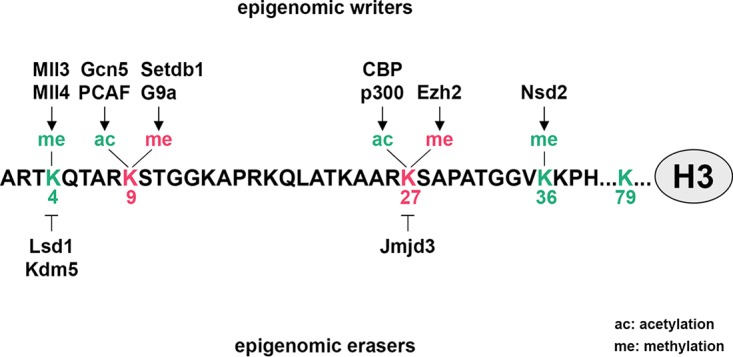
Epigenomic writers and erasers implicated in adipogenesis. A number of site-specific histone lysine methyltransferases, histone demethylases, and histone acetyltransferases play important roles in adipogenesis by adding or removing active (green) or repressive (red) histone marks on lysine residues. The resulting combination of epigenomic marks contributes to activation or repression of downstream target genes regulating adipogenesis. Enzymes that are responsible for methylation/demethylation and acetylation on histone H3 tail are shown.
MLL3/MLL4 are members of the mammalian Set1-like H3K4 methyltransferases, a family which also includes MLL1, MLL2, SET1A, and SET1B (91–93). MLL3/MLL4 are major H3K4 monomethyltransferases enriched on enhancers and required for enhancer activation (91, 94). MLL3/MLL4 and associated PTIP and Ncoa6 have been shown to be required for induction of PPARγ and C/EBPα and subsequently for adipogenesis (91, 95, 96). MLL3/MLL4 are recruited by a pioneering TF C/EBPβ to activate the enhancers of Pparg and Cebpa and induce PPARγ and C/EBPα expression during adipogenesis. Once induced, PPARγ and C/EBPα recruit MLL3/MLL4 to further activate enhancers of downstream target genes (91). Deletion of Mll3 and Mll4 in preadipocytes impairs recruitment of histone acetyltransferases CBP/p300 and the epigenomic reader Brd4 on enhancers, thereby inhibiting induction of adipogenic genes (41, 97). Myf5-Cre-mediated knockout of Mll4 in progenitor cells leads to severe defects in BAT development in mice (91). However, Adipoq-Cre-mediated knockout of Mll4 in differentiated adipocytes does not affect adipose tissue mass or gene expression (94). Therefore, MLL3/MLL4 act as major enhancer writers during adipogenesis, while they are largely dispensable for maintaining differentiated adipocytes. The roles of other H3K4 methyltransferases such as MLL1/MLL2 (MLL1/2) or SET1A/B in adipogenesis have not been reported (51).
H3K9 methyltransferases G9a and Setdb1 have been shown to repress adipogenesis, while Ehmt1 has been shown to promote brown adipogenesis (98, 100, 103). G9a is a major enzyme responsible for the repressive histone mark H3K9 dimethylation (H3K9me2) (101). In preadipocytes, the entire Pparg locus is covered by H3K9me2, where another repressive mark, H3K27me3, is absent (98). During adipogenesis, both H3K9me2 levels and G9a levels decrease, inversely correlating with PPARγ induction. Deletion of G9a in preadipocytes upregulates PPARγ expression by reducing H3K9me2 levels. Fabp4-Cre-mediated G9a knockout mice show increased adipose tissue mass. In addition, G9a promotes expression of Wnt10a, an inhibitor of adipogenesis, independently of its enzymatic activity. Therefore, G9a represses adipogenesis through both enzymatic activity-dependent and enzymatic activity-independent mechanisms. It remains unclear which DNA binding TFs are responsible for G9a recruitment to the Pparg locus. A recent study reported that AP-2α recruits G9a to the Cebpa promoter through sequential chromatin immunoprecipitation (ChIP-reChIP) assays (102). Future studies will be needed to assess whether AP-2α is also responsible for G9a recruitment to Pparg locus. In contrast to G9a, which represses both white and brown adipogenesis, Ehmt1 promotes brown adipogenesis (100). Myf5-Cre-mediated deletion of Ehmt1 impairs BAT development and BAT-selective gene expression in mice. The role of Ehmt1 in white adipogenesis remains unclear. Setdb1, an H3K9 trimethyltransferase, has been shown to poise Pparg and Cebpa expression in preadipocytes by maintaining noncanonical H3K4me3/H3K9me3 bivalent domains (103). Lineage-specific DNA methylation on Pparg and Cebpa recruits the Setdb1-MBD1-MCAF1 complex to establish H3K4me3/H3K9me3 domains. Knockdown of Setdb1 inhibits H3K9me3 deposition on Pparg and Cebpa promoters, allows release of paused Pol II, and induces Pparg and Cebpa expression, which in turn facilitates adipogenesis. However, it remains to be determined whether Setdb1 represses adipogenesis in vivo.
Ezh2 is the catalytic subunit of the dominant H3K27 trimethyltransferase Polycomb repressive complex 2 (PRC2) in mammalian cells (104). PRC2 regulates development by repressing genes that need to be silenced during cell differentiation (105, 106). Secreted Wnt proteins activate the Wnt/β-catenin signaling pathway to inhibit adipogenesis (107). Ezh2 directly represses Wnt genes in a methyltransferase activity-dependent manner to facilitate adipogenesis (108). Deletion of Ezh2 leads to decreases in H3K27me3 levels on Wnt genes and derepression of Wnt expression in preadipocytes and during adipogenesis. This is accompanied by an increase of global acetylation on H3K27 (H3K27ac) (108). With such a dramatic increase in H3K27ac levels, it is possible that other genes in addition to Wnt are activated in Ezh2 knockout cells, contributing to the phenotypes. Determining the genomic binding of Ezh2 or other subunits of PRC2 will be useful to identify genes directly repressed by H3K27me3. Future studies will be needed to identify TFs that recruit the Ezh2-containing PRC2 complex to Wnt genes. In addition, the roles of Ezh2 in adipogenesis in vivo have not been reported.
A recent study has identified Nsd2, an H3K36 dimethyltransferase, as a positive regulator of adipogenesis, based on findings showing that ectopic expression of K36M mutant form of histone H3 (H3K36M) inhibits Nsd2 enzymatic activity, depletes endogenous H3K36 dimethylation, and inhibits adipogenesis by increasing the levels of repressive H3K27me3 on Cebpa and other target genes of PPARγ (109). Knockout or knockdown of Nsd2, but not of other H3K36 methyltransferases (Nsd1 or Setd2), phenocopies the effect of H3K36M expression in adipogenesis. While H3K36M expression in Myf5+ progenitor cells leads to failure of BAT development in mice, Fabp4 promoter-driven H3K36M expression results in whitening of BAT and induces insulin resistance in WAT, indicating that Nsd2-mediated H3K36me2 plays critical roles in adipose tissue development and function. Future work will be needed to validate important roles of Nsd2 in adipogenesis and adipocyte function in vivo.
Lysine demethylases.
Lsd1 is an H3K4 and H3K9 demethylase that has been implicated in both transcriptional activation and repression, depending on the biological context and the associated protein complex. Studies have identified Lsd1 as a positive regulator of adipogenesis, as knockdown or knockout of Lsd1 in preadipocytes inhibits adipogenesis (110, 111). Knockdown of Lsd1 increases the levels of the repressive histone mark H3K9me2 on the Cebpa promoter and impairs the induction of C/EBPα (110). A recent study suggested that Lsd1 is critical for BAT development and function (112). Knockout of Lsd1 in Myf5+ progenitor cells leads to defects in BAT development in mice by increasing H3K9me2 on the Ucp1 promoter and inhibiting Ucp1 expression. Ucp1-Cre-mediated Lsd1 knockout in mice results in increased whole-body adiposity and cold intolerance, suggesting that Lsd1 is required for BAT function. Treating preadipocytes with a chemical inhibitor of Lsd1 prevents adipogenesis, suggesting that Lsd1 enzymatic activity is required for adipogenesis (111). These studies suggested that Lsd1 promotes Cebpa and Ucp1 expression during adipogenesis by demethylating H3K9me2. The role of Lsd1 in WAT development in vivo remains to be determined.
A recent study reported that knockdown of the Kdm5 family of H3K4 demethylases in preadipocytes inhibits adipogenesis (113). Specifically, Kdm5A binds to and activates promoters of genes involved in cell cycle and proliferation. As a result, knockdown of Kdm5 impairs mitotic clonal expansion, which in turn leads to defects in adipogenesis. Reconstitution experiments suggested that the enzymatic activity of Kdm5A is required for the activation of cell cycle genes. Although mitotic clonal expansion is required for adipogenesis in culture, its role in adipose development in vivo is not clear. To better understand the role of Kdm5 family in adipogenesis in vivo, further study using conditional knockout mice will be necessary.
Arginine methyltransferases.
Arginine methylation is mediated by nine protein arginine methyltransferases (PRMTs) in mammals (114). Among these PRMTs, Carm1 and Prmt5 have been implicated in regulating adipogenesis.
Carm1 is a transcriptional coactivator for PPARγ and is required for adipogenesis (115). Transcriptome analysis of whole-body Carm1 knockout mouse embryos suggests that Carm1 is required for adipogenic and lipogenic gene expression downstream of PPARγ. Moreover, E18.5 Carm1 knockout embryos have reduced lipid in BAT compared with wild-type littermates. In vitro, knockdown of Carm1 impairs 3T3-L1 adipogenesis. To prove a cell-autonomous role of Carm1 in adipogenesis, tissue-specific knockout mouse studies are needed. Although reporter assays suggest that Carm1 is a PPARγ coactivator, it needs to be determined whether Carm1 colocalizes with PPARγ on target gene loci. Carm1 has been shown to cooperate with other epigenomic factors, in particular, the enhancer regulators CBP and p300 (116, 117), suggesting the possibility that Carm1 regulates adipogenesis by participating in enhancer activation. Future studies are needed to determine whether the methyltransferase activity of Carm1 is required for adipogenesis.
Prmt5 has also been implicated in regulating adipogenesis (118). Knockdown of Prmt5 inhibits adipogenesis and adipogenic gene expression in 3T3-L1 and C3H10T1/2 cells. A recent study used chromosome conformation capture assay and identified a differentiation-dependent and Prmt5-dependent interaction between Pparg2 promoter and kb −10 enhancer regions in C3H10T1/2 cells after differentiation (119). Prmt5 is also required for Med1 and Brg1 binding at the Pparg2 promoter and the kb −10 enhancer. Given that Prmt5 is essential for cell growth (120), it remains unclear whether Prmt5 directly regulates adipogenic gene expression and whether the methyltransferase activity of Prmt5 is required for adipogenesis.
Histone acetyltransferases.
Acetylation on histone lysine residues generally correlates with gene activation and is catalyzed by site-specific histone acetyltransferases (HATs). For example, Gcn5 and PCAF are major enzymes for acetylation on H3K9 (H3K9ac), while CBP and p300 are responsible for acetylation on H3K18 (H3K18ac) and H3K27 (H3K27ac) in mammalian cells (121–123).
Gcn5 and PCAF function redundantly to regulate adipogenesis (124). Double knockout of Gcn5 and Pcaf by Myf5-Cre impairs BAT development in mice. In vitro, Gcn5/PCAF and their catalytic activities are required for adipogenesis. Interestingly, ectopic PPARγ can rescue adipogenesis but not brown adipocyte-enriched Prdm16 expression in Gcn5 Pcaf double knockout cells, suggesting that Gcn5 and PCAF regulate transcription of general adipogenic genes and brown adipocyte-enriched genes through different mechanisms. Determining genomic localization of Gcn5 and/or PCAF would provide a better understanding of the mechanism by which this functionally redundant pair of HATs regulates adipocyte genes and BAT-enriched genes in brown adipogenesis.
CBP and p300 are required for adipogenesis. Ribozyme-mediated targeting of either CBP or p300 inhibits adipogenesis and PPARγ target gene expression, suggesting that these two enzymes are not mutually compensating each other in regulating adipogenesis (125). Crebbp+/− mice have markedly reduced WAT mass compared to wild-type littermates, suggesting that CBP is essential for adipogenesis in vivo (126). CBP/p300-mediated H3K27ac marks active enhancers (127, 128). Consistently, profiling of CBP/p300 binding sites demonstrated that genomic binding of these HATs drives enhancer activation during adipogenesis (41).
Histone deacetylases and sirtuins.
Histone deacetylation is mediated by classical zinc-dependent histone deacetylases (HDACs) and NAD+-dependent sirtuins (SIRTs) (129). Studies have suggested diverse roles of HDACs and SIRTs in adipogenesis.
Among 11 HDACs, Hdac1, Hdac2, and Hdac9 have been shown to regulate adipogenesis. Hdac1 and Hdac2 positively regulate adipogenesis redundantly, since knockout of both, but not the individual knockout of each, inhibits adipogenesis of fibroblasts (130). In contrast, Hdac9 negatively regulates adipogenesis. Hdac9 expression is downregulated during adipogenesis. Preadipocytes isolated from Hdac9 knockout mice show accelerated adipogenesis, while overexpression of Hdac9 represses 3T3-L1 adipogenesis independently of deacetylase domain (131). The roles of HDACs in adipogenesis in vivo and the genomic localization of HDACs remain to be determined.
Among seven SIRTs, Sirt1, Sirt2, Sirt6, and Sirt7 have been implicated in regulating adipogenesis. Fabp4-Cre-mediated Sirt1 knockout mice show increased adipose tissue mass under conditions of a high-fat diet, suggesting a negative role of Sirt1 in adipose tissue development (132). In culture, Sirt1 knockout enhances adipogenesis in MEFs (133). Sirt2 has also been implicated in negative regulation of adipogenesis. Two studies reported that stable knockdown of Sirt2 promotes adipogenesis in 3T3-L1 cells. Mechanistically, Sirt2 inhibits adipogenesis by deacetylating FOXO1, which represses PPARγ transcriptional activity (134, 135). In contrast, whole-body Sirt7 knockout mice show reduced WAT mass, suggesting that Sirt7 positively regulates adipose tissue development (136). Sirt6 regulates mitotic clonal expansion in cells and is required for adipogenesis (137). Future studies are needed to identify direct target genes of SIRTs in adipogenesis and to demonstrate whether SIRTs play a cell-autonomous role in regulating adipogenesis in vivo.
Epigenomic reader Brd4.
Brd4 is a member of the family of bromodomain and extraterminal domain (BET) proteins, which also includes Brd2 and Brd3 (138). Brd4 binds to acetylated histones and TFs and acts as an epigenomic reader (139). Brd4 is critical for adipogenesis in culture and in mice (97). Deletion of Brd4 in preadipocytes prevents adipogenesis and adipogenic gene induction. During adipogenesis, lineage-determining transcription factors (LDTFs) cooperate with enhancer epigenomic writers MLL3/4 and CBP/p300 to recruit Brd4 to active enhancers, which facilitates the recruitment of Pol II, Mediator, TFIID, and p-TEFb. An independent study also showed that Brd4 is enriched on superenhancers in 3T3-L1 adipogenesis (140). Consistently, BET inhibitors block adipogenesis of 3T3-L1 and C3H10T1/2 cells (140, 141). In mice, Myf5-Cre-mediated knockout of Brd4 impairs BAT development. Although Brd4 is essential for adipogenesis, it is largely dispensable for maintaining differentiated adipocytes, as evidenced by the lack of the phenotype in mice with Adipoq-Cre-mediated Brd4 knockout (97). Interestingly, another BET family protein, Brd2, has been suggested to inhibit adipogenesis (142, 143). Further study is necessary to clarify the shared and distinct functions of BET family members in adipogenesis in vivo.
Chromatin remodeling complex.
SWI/SNF is a multisubunit ATP-dependent chromatin remodeling complex that uses Brg1 or Brm as the ATPase (144). C/EBPs initiate recruitment of SWI/SNF to Pparg2 promoter in adipogenesis (145). Ectopic expression of dominant-negative Brg1 or Brm interferes with PPARγ-, C/EBPα-, or C/EBPβ-mediated adipogenesis in fibroblasts, suggesting that the SWI/SNF complex is required for adipogenesis. Whether the SWI/SNF complex is required for chromatin opening and adipogenesis in vivo and how it regulates adipogenic gene expression remain largely unclear. Recent studies in human cancer cells suggested that the SWI/SNF complex is critical for enhancer activation (146, 147). Whether the SWI/SNF complex is required for activation of enhancers critical for adipogenic gene expression may require further investigation.
DNA methylation.
DNA methylation is catalyzed by DNA methyltransferases (DNMTs) and is removed by TET dioxygenases. There are five DNMTs responsible for establishing or maintaining DNA methylation (148). The role of DNMTs and DNA methylation in adipogenesis has been controversial. An earlier study showed that small interfering RNA (siRNA) knockdown of DNMT1 in 3T3-L1 cells reduces DNA methylation levels on the Pparg gene and accelerates adipogenesis (149). However, a recent study showed that pharmacological inhibition of DNA methylation by 5-aza-dC treatment suppresses adipogenesis and is accompanied by demethylation and upregulation of Wnt10a (150). To clarify the role of DNMTs in adipogenesis in vivo, tissue-specific knockout mouse studies are necessary.
DNA methylation in the form of 5-methylcytosine (5mC) can be reversed by TET1/2/3 (151). A study has shown that 5-hydroxymethylcytosine (5hmC), an intermediate of DNA demethylation process, marks a subset of PPARγ- and C/EBPα-bound enhancers during 3T3-L1 adipogenesis. Addition of 5hmC during adipogenesis coincides with increased levels of enhancer marks H3K4me2 and H3K27ac (152). In line with this finding, double knockout of Tet1 and Tet2 has been shown to block adipogenesis of mouse embryonic fibroblasts (153). The role of TETs in regulating adipogenesis should be investigated in vivo using tissue-specific knockout mice. In addition, whether Tet1 and/or Tet2 localizes on enhancers to regulate adipogenic gene expression needs to be determined.
microRNAs.
MicroRNAs (miRNAs) are noncoding RNAs that posttranscriptionally regulate gene expression by degrading target mRNA or inhibiting protein synthesis (154). Studies have suggested that several miRNAs play a role in adipose tissue development and associated metabolism (155). A study used miRNA microarray analysis to profile PPARγ-regulated miRNAs in human adipocytes (156). Among 27 miRNAs whose expression was altered by treatment of pioglitazone, a synthetic PPARγ ligand, miR-378, has been shown to promote adipogenesis of subcutaneous adipocytes but not of visceral adipocytes. miR-378 is embedded in the first intron of PPARGC1b, which encodes PGC-1β. The target genes of miR-378 and its working mechanisms in adipogenesis have yet to be examined. Another important intronic miRNA is miR-33, which is embedded within the SREBP-2 gene and targets ABCA1 to control cholesterol homeostasis (157, 158). Whole-body miR-33 knockout mice gradually become obese and develop hepatic steatosis through derepression of SREBP-1 in the liver (159). How miR-33 regulates adipogenesis and metabolism in adipocytes remains to be understood.
CONCLUSIONS AND FUTURE DIRECTIONS
In recent years, one major discovery in transcriptional regulation of adipogenesis is the identification of EBF2 as a major regulator of brown adipogenesis. Further study is necessary to clarify the role of EBFs in general versus in brown adipogenesis and their potential redundancy. This can be achieved by generating tissue-specific knockout of single and multiple EBFs in mice. Another important contribution to understanding transcriptional regulation of adipogenesis is the exclusion of Krox20, KLF4, and GR from the list of adipogenic TFs, which underscores the importance of studying adipogenesis in vivo instead of drawing conclusions based on the knockdown approach in cell culture.
During the last decade, many epigenomic factors have been shown to regulate adipogenesis. These factors are involved in regulating histone modification, chromatin remodeling, and DNA methylation. Studies have revealed sequential actions of epigenomic factors on enhancers in adipogenesis. C/EBPβ recruits MLL3/MLL4 to prime enhancers, which facilitates binding of CBP/p300 to activate and label enhancers with H3K27ac. H3K27ac and acetylated TFs are recognized by Brd4, which recruits Mediator and Pol II to activate PPARγ and C/EBPα expression (41, 91, 97). The SWI/SNF complex may participate in these sequential events, leading to lineage-specific enhancer activation in adipogenesis (Fig. 5). The expression of adipogenesis master regulators is also under the control of two mutually exclusive repressive histone marks. G9a-mediated H3K9me2 represses PPARγ expression. Ezh2-mediated H3K27me3 represses Wnt expression (Fig. 5).
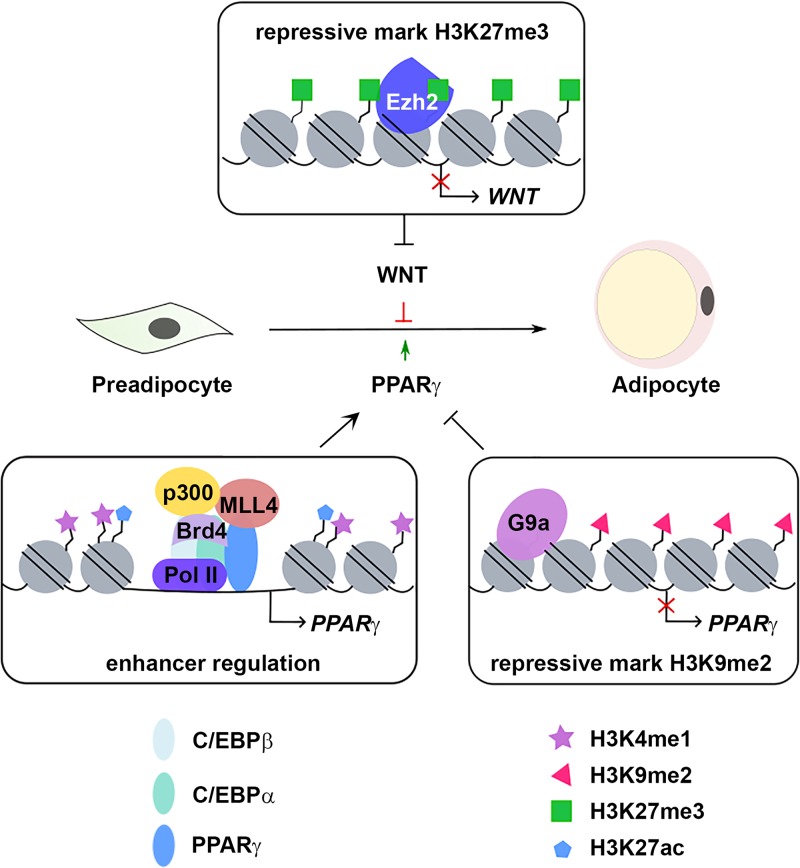
FIG 5.
Epigenomic mechanisms control expression of master regulators of adipogenesis. Epigenomic factors such as MLL4, p300, and Brd4 bind to enhancers to activate expression of adipogenesis genes, including the master adipogenic TF PPARγ gene. Two repressive histone marks oppositely regulate adipogenesis. Ezh2-mediated H3K27me3 represses Wnt gene expression and promotes adipogenesis, whereas G9a-mediated H3K9me2 represses Pparg expression and inhibits adipogenesis.
Despite the rapid progress in the study of transcriptional and epigenomic regulation of adipogenesis, several issues remain to be addressed and new techniques need to be employed. First, although cell culture systems have revealed important mechanisms regulating adipogenesis, they do not fully recapitulate adipogenesis in vivo. Many putative adipogenic TFs identified in cell culture need to be validated using mouse models, and specific and direct target genes of these TFs need to be identified by ChIP-Seq. Second, adipose tissues are heterogeneous and contain multiple cell types (160). Recently developed INTACT, TRAP, and NuTRAP transgenic mice allow isolation of mRNA and nuclei from specific cell types. These techniques will enable transcriptional and epigenomic profiling of adipogenesis in vivo using heterogeneous adipose tissues (161–163). Third, recent advances in single-cell technology provide opportunities for transcriptomic and epigenomic studies at single-cell resolution (164). This will help identify cell lineages and distinct developmental stages during adipogenesis. Fourth, RNAi-mediated knockdown approach often lacks sufficient specificity and tends to create artifacts (165). Artifacts generated by RNAi are also found in the adipogenesis field. The recently developed revolutionary genome editing technique that uses the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system has enabled time- and cost-efficient gene knockout (166). By generating knockout cells by the use of the CRISPR-Cas9 approach, roles of poorly characterized, putative adipogenic factors can be reevaluated. Fifth, studies have implicated noncoding RNAs in regulating adipogenesis in culture (167, 168). Although knockdown of several candidate noncoding RNAs inhibits adipogenesis, more efforts are needed to determine the mechanism by which these noncoding RNAs regulate adipogenesis and to validate their roles with in vivo models. Sixth, higher-order chromatin interactions, in particular, promoter-enhancer looping, are involved in regulating gene expression during differentiation and development (169). It has been shown that promoter-anchored chromatin loops change dynamically during adipogenesis (170). Future efforts are needed to identify the factors that are responsible for dynamic promoter-enhancer looping during adipogenesis.
ACKNOWLEDGMENT
We thank Victoria Noe-Kim for assistance with the figures.
This work was supported by a grant from the Intramural Research Program of NIDDK, NIH, to K.G.
REFERENCES
- 1.Rosen ED, MacDougald OA. 2006. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 2.Cypess AM, Kahn CR. 2010. Brown fat as a therapy for obesity and diabetes. Curr Opin Endocrinol Diabetes Obes 17:143–149. doi: 10.1097/MED.0b013e328337a81f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristancho AG, Lazar MA. 2011. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green H, Meuth M. 1974. An established pre-adipose cell line and its differentiation in culture. Cell 3:127–133. doi: 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- 5.Reznikoff CA, Brankow DW, Heidelberger C. 1973. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res 33:3231–3238. [PubMed] [Google Scholar]
- 6.Tallquist MD, Weismann KE, Hellstrom M, Soriano P. 2000. Early myotome specification regulates PDGFA expression and axial skeleton development. Development 127:5059–5070. [DOI] [PubMed] [Google Scholar]
- 7.Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. 2011. Transcriptional control of adipose lipid handling by IRF4. Cell Metab 13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ZV, Deng Y, Wang QA, Sun K, Scherer PE. 2010. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology 151:2933–2939. doi: 10.1210/en.2010-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barlow C, Schroeder M, Lekstrom-Himes J, Kylefjord H, Deng CX, Wynshaw-Boris A, Spiegelman BM, Xanthopoulos KG. 1997. Targeted expression of Cre recombinase to adipose tissue of transgenic mice directs adipose-specific excision of loxP-flanked gene segments. Nucleic Acids Res 25:2543–2545. doi: 10.1093/nar/25.12.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB. 2001. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 11.He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM. 2003. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A 100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, Smyth G, Rourk M, Cederquist C, Rosen ED, Kahn BB, Kahn CR. 2013. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 62:864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffery E, Berry R, Church CD, Yu S, Shook BA, Horsley V, Rosen ED, Rodeheffer MS. 2014. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte 3:206–211. doi: 10.4161/adip.29674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loft A, Forss I, Mandrup S. 2017. Genome-wide insights into the development and function of thermogenic adipocytes. Trends Endocrinol Metab 28:104–120. doi: 10.1016/j.tem.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Seale P. 2016. Control of brown and beige fat development. Nat Rev Mol Cell Biol 17:691–702. doi: 10.1038/nrm.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inagaki T, Sakai J, Kajimura S. 2016. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat Rev Mol Cell Biol 17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linhart HG, Ishimura-Oka K, DeMayo F, Kibe T, Repka D, Poindexter B, Bick RJ, Darlington GJ. 2001. C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc Natl Acad Sci U S A 98:12532–12537. doi: 10.1073/pnas.211416898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tontonoz P, Hu E, Spiegelman BM. 1994. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79:1147–1156. doi: 10.1016/0092-8674(94)90006-X. [DOI] [PubMed] [Google Scholar]
- 19.Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. 1999. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell 4:585–595. doi: 10.1016/S1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. 1999. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 3:151–158. doi: 10.1016/S1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 21.Lin FT, Lane MD. 1992. Antisense CCAAT/enhancer-binding protein RNA suppresses coordinate gene expression and triglyceride accumulation during differentiation of 3T3-L1 preadipocytes. Genes Dev 6:533–544. doi: 10.1101/gad.6.4.533. [DOI] [PubMed] [Google Scholar]
- 22.Farmer SR. 2006. Transcriptional control of adipocyte formation. Cell Metab 4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JE, Ge K. 2014. Transcriptional and epigenetic regulation of PPARgamma expression during adipogenesis. Cell Biosci 4:29. doi: 10.1186/2045-3701-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefterova MI, Haakonsson AK, Lazar MA, Mandrup S. 2014. PPARgamma and the global map of adipogenesis and beyond. Trends Endocrinol Metab 25:293–302. doi: 10.1016/j.tem.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siersbæk R, Mandrup S. 2011. Transcriptional networks controlling adipocyte differentiation. Cold Spring Harbor Symp Quant Biol 76:247–255. doi: 10.1101/sqb.2011.76.010512. [DOI] [PubMed] [Google Scholar]
- 26.Tamori Y, Masugi J, Nishino N, Kasuga M. 2002. Role of peroxisome proliferator-activated receptor-gamma in maintenance of the characteristics of mature 3T3-L1 adipocytes. Diabetes 51:2045–2055. doi: 10.2337/diabetes.51.7.2045. [DOI] [PubMed] [Google Scholar]
- 27.Imai T, Takakuwa R, Marchand S, Dentz E, Bornert JM, Messaddeq N, Wendling O, Mark M, Desvergne B, Wahli W, Chambon P, Metzger D. 2004. Peroxisome proliferator-activated receptor gamma is required in mature white and brown adipocytes for their survival in the mouse. Proc Natl Acad Sci U S A 101:4543–4547. doi: 10.1073/pnas.0400356101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. 2008. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev 22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ Jr, Liu XS, Lazar MA. 2008. PPAR{gamma} and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. 2000. Transcriptional regulation of adipogenesis. Genes Dev 14:1293–1307. [PubMed] [Google Scholar]
- 31.Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Lazar MA. 2010. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siersbæk R, Nielsen R, John S, Sung M-H, Baek S, Loft A, Hager GL, Mandrup S. 2011. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J 30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao Z, Umek RM, McKnight SL. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 34.Wu Z, Bucher NL, Farmer SR. 1996. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol Cell Biol 16:4128–4136. doi: 10.1128/MCB.16.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka T, Yoshida N, Kishimoto T, Akira S. 1997. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J 16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akerblad P, Lind U, Liberg D, Bamberg K, Sigvardsson M. 2002. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol Cell Biol 22:8015–8025. doi: 10.1128/MCB.22.22.8015-8025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. 2007. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol 27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W, Kissig M, Rajakumari S, Huang L, Lim HW, Won KJ, Seale P. 2014. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci U S A 111:14466–14471. doi: 10.1073/pnas.1412685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. 2013. EBF2 determines and maintains brown adipocyte identity. Cell Metab 17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapira SN, Lim HW, Rajakumari S, Sakers AP, Ishibashi J, Harms MJ, Won KJ, Seale P. 2017. EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev 31:660–673. doi: 10.1101/gad.294405.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai B, Lee JE, Jang Y, Wang L, Peng W, Ge K. 2017. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res 45:6388–6403. doi: 10.1093/nar/gkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fretz JA, Nelson T, Xi Y, Adams DJ, Rosen CJ, Horowitz MC. 2010. Altered metabolism and lipodystrophy in the early B-cell factor 1-deficient mouse. Endocrinology 151:1611–1621. doi: 10.1210/en.2009-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boller S, Li R, Grosschedl R. 2018. Defining B cell chromatin: lessons from EBF1. Trends Genet 34:257–269. doi: 10.1016/j.tig.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 44.Cohen DM, Won KJ, Nguyen N, Lazar MA, Chen CS, Steger DJ. 2015. ATF4 licenses C/EBPbeta activity in human mesenchymal stem cells primed for adipogenesis. Elife 4:e06821. doi: 10.7554/eLife.06821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu K, Mo D, Wu M, Chen H, Chen L, Li M, Chen Y. 2014. Activating transcription factor 4 regulates adipocyte differentiation via altering the coordinate expression of CCATT/enhancer binding protein beta and peroxisome proliferator-activated receptor gamma. FEBS J 281:2399–2409. doi: 10.1111/febs.12792. [DOI] [PubMed] [Google Scholar]
- 46.Seo J, Fortuno ES III, Suh JM, Stenesen D, Tang W, Parks EJ, Adams CM, Townes T, Graff JM. 2009. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes 58:2565–2573. doi: 10.2337/db09-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masuoka HC, Townes TM. 2002. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99:736–745. doi: 10.1182/blood.V99.3.736. [DOI] [PubMed] [Google Scholar]
- 48.Waki H, Nakamura M, Yamauchi T, Wakabayashi K, Yu J, Hirose-Yotsuya L, Take K, Sun W, Iwabu M, Okada-Iwabu M, Fujita T, Aoyama T, Tsutsumi S, Ueki K, Kodama T, Sakai J, Aburatani H, Kadowaki T. 2011. Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet 7:e1002311. doi: 10.1371/journal.pgen.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hiraike Y, Waki H, Yu J, Nakamura M, Miyake K, Nagano G, Nakaki R, Suzuki K, Kobayashi H, Yamamoto S, Sun W, Aoyama T, Hirota Y, Ohno H, Oki K, Yoneda M, White AP, Tseng YH, Cypess AM, Larsen TJ, Jespersen NZ, Scheele C, Tsutsumi S, Aburatani H, Yamauchi T, Kadowaki T. 2017. NFIA co-localizes with PPARgamma and transcriptionally controls the brown fat gene program. Nat Cell Biol 19:1081–1092. doi: 10.1038/ncb3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubin CS, Hirsch A, Fung C, Rosen OM. 1978. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3-L1 cells. J Biol Chem 253:7570–7578. [PubMed] [Google Scholar]
- 51.Ge K. 2012. Epigenetic regulation of adipogenesis by histone methylation. Biochim Biophys Acta 1819:727–732. doi: 10.1016/j.bbagrm.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM. 2005. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab 1:93–106. doi: 10.1016/j.cmet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 53.Birsoy K, Chen Z, Friedman J. 2008. Transcriptional regulation of adipogenesis by KLF4. Cell Metab 7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park YK, Wang L, Giampietro A, Lai B, Lee JE, Ge K. 4 January 2017, posting date Distinct roles of transcription factors KLF4, Krox20, and peroxisome proliferator-activated receptor gamma in adipogenesis. Mol Cell Biol doi: 10.1128/MCB.00554-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deslex S, Negrel R, Ailhaud G. 1987. Development of a chemically defined serum-free medium for differentiation of rat adipose precursor cells. Exp Cell Res 168:15–30. doi: 10.1016/0014-4827(87)90412-5. [DOI] [PubMed] [Google Scholar]
- 56.Gregoire F, Todoroff G, Hauser N, Remacle C. 1990. The stroma-vascular fraction of rat inguinal and epididymal adipose tissue and the adipoconversion of fat cell precursors in primary culture. Biol Cell 69:215–222. doi: 10.1016/0248-4900(90)90348-7. [DOI] [PubMed] [Google Scholar]
- 57.Park YK, Ge K. January 4 2017, posting date Glucocorticoid receptor accelerates, but is dispensable for, adipogenesis. Mol Cell Biol doi: 10.1128/MCB.00260-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bauerle KT, Hutson I, Scheller EL, Harris CA. 2018. Glucocorticoid receptor signaling is not required for in vivo adipogenesis. Endocrinology 159:2050–2061. doi: 10.1210/en.2018-00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang JC, Gray NE, Kuo T, Harris CA. 2012. Regulation of triglyceride metabolism by glucocorticoid receptor. Cell Biosci 2:19. doi: 10.1186/2045-3701-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. 2010. Transcriptional control of preadipocyte determination by Zfp423. Nature 464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shao M, Ishibashi J, Kusminski CM, Wang QA, Hepler C, Vishvanath L, MacPherson KA, Spurgin SB, Sun K, Holland WL, Seale P, Gupta RK. 2016. Zfp423 maintains white adipocyte identity through suppression of the beige cell thermogenic gene program. Cell Metab 23:1167–1184. doi: 10.1016/j.cmet.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hepler C, Shao M, Xia JY, Ghaben AL, Pearson MJ, Vishvanath L, Sharma AX, Morley TS, Holland WL, Gupta RK. 2017. Directing visceral white adipocyte precursors to a thermogenic adipocyte fate improves insulin sensitivity in obese mice. Elife doi: 10.7554/eLife.27669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shao M, Hepler C, Vishvanath L, MacPherson KA, Busbuso NC, Gupta RK. 2017. Fetal development of subcutaneous white adipose tissue is dependent on Zfp423. Mol Metab 6:111–124. doi: 10.1016/j.molmet.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hong KY, Bae H, Park I, Park DY, Kim KH, Kubota Y, Cho ES, Kim H, Adams RH, Yoo OJ, Koh GY. 2015. Perilipin+ embryonic preadipocytes actively proliferate along growing vasculatures for adipose expansion. Development 142:2623–2632. doi: 10.1242/dev.125336. [DOI] [PubMed] [Google Scholar]
- 65.Zhang JW, Klemm DJ, Vinson C, Lane MD. 2004. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J Biol Chem 279:4471–4478. doi: 10.1074/jbc.M311327200. [DOI] [PubMed] [Google Scholar]
- 66.Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R. 2005. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab 1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 67.Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, Kadowaki T, Matsuki Y, Ogawa W, Hiramatsu R, Kasuga M. 2005. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem 280:12867–12875. doi: 10.1074/jbc.M410515200. [DOI] [PubMed] [Google Scholar]
- 68.Nanbu-Wakao R, Morikawa Y, Matsumura I, Masuho Y, Muramatsu MA, Senba E, Wakao H. 2002. Stimulation of 3T3-L1 adipogenesis by signal transducer and activator of transcription 5. Mol Endocrinol 16:1565–1576. doi: 10.1210/mend.16.7.0862. [DOI] [PubMed] [Google Scholar]
- 69.Tong Q, Dalgin G, Xu H, Ting C-N, Leiden JM, Hotamisligil GS. 2000. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 70.Tong Q, Tsai J, Tan G, Dalgin G, Hotamisligil GS. 2005. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol Cell Biol 25:706–715. doi: 10.1128/MCB.25.2.706-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H, Jain MK. 2003. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem 278:2581–2584. doi: 10.1074/jbc.M210859200. [DOI] [PubMed] [Google Scholar]
- 72.Moitra J, Mason MM, Olive M, Krylov D, Gavrilova O, Marcus-Samuels B, Feigenbaum L, Lee E, Aoyama T, Eckhaus M, Reitman ML, Vinson C. 1998. Life without white fat: a transgenic mouse. Genes Dev 12:3168–3181. doi: 10.1101/gad.12.20.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. 2004. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 74.Kim JB, Wright HM, Wright M, Spiegelman BM. 1998. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc Natl Acad Sci U S A 95:4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. 1997. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest 100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.York B, O'Malley BW. 2010. Steroid receptor coactivator (SRC) family: masters of systems biology. J Biol Chem 285:38743–38750. doi: 10.1074/jbc.R110.193367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malik S, Roeder RG. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 78.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. 2002. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 79.Grontved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. 2010. MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis. Mol Cell Biol 30:2155–2169. doi: 10.1128/MCB.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W, Huang L, Huang Y, Yin JW, Berk AJ, Friedman JM, Wang G. 2009. Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev Cell 16:764–771. doi: 10.1016/j.devcel.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O'Malley BW, Chambon P, Auwerx J. 2002. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 111:931–941. doi: 10.1016/S0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 82.Louet JF, Coste A, Amazit L, Tannour-Louet M, Wu RC, Tsai SY, Tsai MJ, Auwerx J, O'Malley BW. 2006. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc Natl Acad Sci U S A 103:17868–17873. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Villanueva CJ, Vergnes L, Wang J, Drew BG, Hong C, Tu Y, Hu Y, Peng X, Xu F, Saez E, Wroblewski K, Hevener AL, Reue K, Fong LG, Young SG, Tontonoz P. 2013. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARgamma specifies lipid storage versus thermogenic gene programs. Cell Metab 17:423–435. doi: 10.1016/j.cmet.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Villanueva CJ, Waki H, Godio C, Nielsen R, Chou WL, Vargas L, Wroblewski K, Schmedt C, Chao LC, Boyadjian R, Mandrup S, Hevener A, Saez E, Tontonoz P. 2011. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab 13:413–427. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ong CT, Corces VG. 2011. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reik W. 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 87.Chen T, Dent SY. 2014. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. 2012. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 90.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 91.Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, Ge K. 2013. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ruthenburg AJ, Allis CD, Wysocka J. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 93.Rao RC, Dou Y. 2015. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer 15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang C, Lee JE, Lai B, Macfarlan TS, Xu S, Zhuang L, Liu C, Peng W, Ge K. 2016. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc Natl Acad Sci U S A 113:11871–11876. doi: 10.1073/pnas.1606857113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cho Y-W, Hong S, Jin Q, Wang L, Lee J-E, Gavrilova O, Ge K. 2009. Histone methylation regulator PTIP is required for PPAR gamma and C/EBP alpha expression and adipogenesis. Cell Metab 10:27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qi C, Surapureddi S, Zhu Y-J, Yu S, Kashireddy P, Rao MS, Reddy JK. 2003. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor {gamma} (PPAR{gamma})-interacting protein, is required for PPAR{gamma}-mediated adipogenesis. J Biol Chem 278:25281–25284. doi: 10.1074/jbc.C300175200. [DOI] [PubMed] [Google Scholar]
- 97.Lee JE, Park YK, Park S, Jang Y, Waring N, Dey A, Ozato K, Lai B, Peng W, Ge K. 2017. Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat Commun 8:2217. doi: 10.1038/s41467-017-02403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L, Xu S, Lee J-E, Baldridge A, Grullon S, Peng W, Ge K. 2013. Histone H3K9 methyltransferase G9a represses PPAR gamma expression and adipogenesis. EMBO J 32:45–59. doi: 10.1038/emboj.2012.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reference deleted.
- 100.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. 2013. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 504:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang ZC, Liu Y, Li SF, Guo L, Zhao Y, Qian SW, Wen B, Tang QQ, Li X. 2014. Suv39h1 mediates AP-2alpha-dependent inhibition of C/EBPalpha expression during adipogenesis. Mol Cell Biol 34:2330–2338. doi: 10.1128/MCB.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsumura Y, Nakaki R, Inagaki T, Yoshida A, Kano Y, Kimura H, Tanaka T, Tsutsumi S, Nakao M, Doi T, Fukami K, Osborne TF, Kodama T, Aburatani H, Sakai J. 2015. H3K4/H3K9me3 bivalent chromatin domains targeted by lineage-specific DNA methylation pauses adipocyte differentiation. Mol Cell 60:584–596. doi: 10.1016/j.molcel.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 104.Simon JA, Kingston RE. 2009. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10:697. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 105.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 107.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 108.Wang L, Jin Q, Lee J-E, Su I, Ge K. 2010. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci U S A 107:7317–7322. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhuang L, Jang Y, Park YK, Lee JE, Jain S, Froimchuk E, Broun A, Liu C, Gavrilova O, Ge K. 2018. Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nat Commun 9:1796. doi: 10.1038/s41467-018-04127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Musri MM, Carmona MC, Hanzu FA, Kaliman P, Gomis R, Parrizas M. 2010. Histone demethylase LSD1 regulates adipogenesis. J Biol Chem 285:30034–30041. doi: 10.1074/jbc.M110.151209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Duteil D, Metzger E, Willmann D, Karagianni P, Friedrichs N, Greschik H, Gunther T, Buettner R, Talianidis I, Metzger D, Schule R. 2014. LSD1 promotes oxidative metabolism of white adipose tissue. Nat Commun 5:4093. doi: 10.1038/ncomms5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sambeat A, Gulyaeva O, Dempersmier J, Tharp KM, Stahl A, Paul SM, Sul HS. 2016. LSD1 interacts with Zfp516 to promote UCP1 transcription and brown fat program. Cell Rep 15:2536–2549. doi: 10.1016/j.celrep.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brier AB, Loft A, Madsen JGS, Rosengren T, Nielsen R, Schmidt SF, Liu Z, Yan Q, Gronemeyer H, Mandrup S. 2017. The KDM5 family is required for activation of pro-proliferative cell cycle genes during adipocyte differentiation. Nucleic Acids Res 45:1743–1759. doi: 10.1093/nar/gkw1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bedford MT, Clarke SG. 2009. Protein arginine methylation in mammals: who, what, and why. Mol Cell 33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, Bedford MT. 2008. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep 9:193–198. doi: 10.1038/sj.embor.7401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. 2002. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol 12:2090–2097. doi: 10.1016/S0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 117.An W, Kim J, Roeder RG. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 118.LeBlanc SE, Konda S, Wu Q, Hu YJ, Oslowski CM, Sif S, Imbalzano AN. 2012. Protein arginine methyltransferase 5 (Prmt5) promotes gene expression of peroxisome proliferator-activated receptor gamma2 (PPARgamma2) and its target genes during adipogenesis. Mol Endocrinol 26:583–597. doi: 10.1210/me.2011-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.LeBlanc SE, Wu Q, Lamba P, Sif S, Imbalzano AN. 2016. Promoter-enhancer looping at the PPARgamma2 locus during adipogenic differentiation requires the Prmt5 methyltransferase. Nucleic Acids Res 44:5133–5147. doi: 10.1093/nar/gkw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. 2011. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci 36:633–641. doi: 10.1016/j.tibs.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jin Q, Yu L-R, Wang L, Zhang Z, Kasper LH, Lee J-E, Wang C, Brindle PK, Dent SYR, Ge K. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kasper LH, Qu C, Obenauer JC, McGoldrick DJ, Brindle PK. 2014. Genome-wide and single-cell analyses reveal a context dependent relationship between CBP recruitment and gene expression. Nucleic Acids Res 42:11363–11382. doi: 10.1093/nar/gku827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, Shaw B, Algire M, Hessler P, Lam LT, Uziel T, Faivre E, Ferguson D, Buchanan FG, Martin RL, Torrent M, Chiang GG, Karukurichi K, Langston JW, Weinert BT, Choudhary C, de Vries P, Van Drie JH, McElligott D, Kesicki E, Marmorstein R, Sun C, Cole PA, Rosenberg SH, Michaelides MR, Lai A, Bromberg KD. 2017. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550:128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jin Q, Wang C, Kuang X, Feng X, Sartorelli V, Ying H, Ge K, Dent SY. 2014. Gcn5 and PCAF regulate PPARgamma and Prdm16 expression to facilitate brown adipogenesis. Mol Cell Biol 34:3746–3753. doi: 10.1128/MCB.00622-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takahashi N, Kawada T, Yamamoto T, Goto T, Taimatsu A, Aoki N, Kawasaki H, Taira K, Yokoyama KK, Kamei Y, Fushiki T. 2002. Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor gamma. J Biol Chem 277:16906–16912. doi: 10.1074/jbc.M200585200. [DOI] [PubMed] [Google Scholar]
- 126.Yamauchi T, Oike Y, Kamon J, Waki H, Komeda K, Tsuchida A, Date Y, Li MX, Miki H, Akanuma Y, Nagai R, Kimura S, Saheki T, Nakazato M, Naitoh T, Yamamura K, Kadowaki T. 2002. Increased insulin sensitivity despite lipodystrophy in Crebbp heterozygous mice. Nat Genet 30:221–226. doi: 10.1038/ng829. [DOI] [PubMed] [Google Scholar]
- 127.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Seto E, Yoshida M. 2014. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Haberland M, Carrer M, Mokalled MH, Montgomery RL, Olson EN. 2010. Redundant control of adipogenesis by histone deacetylases 1 and 2. J Biol Chem 285:14663–14670. doi: 10.1074/jbc.M109.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chatterjee TK, Idelman G, Blanco V, Blomkalns AL, Piegore MG Jr, Weintraub DS, Kumar S, Rajsheker S, Manka D, Rudich SM, Tang Y, Hui DY, Bassel-Duby R, Olson EN, Lingrel JB, Ho SM, Weintraub NL. 2011. Histone deacetylase 9 is a negative regulator of adipogenic differentiation. J Biol Chem 286:27836–27847. doi: 10.1074/jbc.M111.262964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mayoral R, Osborn O, McNelis J, Johnson AM, Oh DY, Izquierdo CL, Chung H, Li P, Traves PG, Bandyopadhyay G, Pessentheiner AR, Ofrecio JM, Cook JR, Qiang L, Accili D, Olefsky JM. 2015. Adipocyte SIRT1 knockout promotes PPARgamma activity, adipogenesis and insulin sensitivity in chronic-HFD and obesity. Mol Metab 4:378–391. doi: 10.1016/j.molmet.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou Y, Song T, Peng J, Zhou Z, Wei H, Zhou R, Jiang S, Peng J. 2016. SIRT1 suppresses adipogenesis by activating Wnt/beta-catenin signaling in vivo and in vitro. Oncotarget 7:77707–77720. doi: 10.18632/oncotarget.12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang F, Tong Q. 2009. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARgamma. Mol Biol Cell 20:801–808. doi: 10.1091/mbc.e08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jing E, Gesta S, Kahn CR. 2007. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab 6:105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fang J, Ianni A, Smolka C, Vakhrusheva O, Nolte H, Kruger M, Wietelmann A, Simonet NG, Adrian-Segarra JM, Vaquero A, Braun T, Bober E. 2017. Sirt7 promotes adipogenesis in the mouse by inhibiting autocatalytic activation of Sirt1. Proc Natl Acad Sci U S A 114:E8352–E8361. doi: 10.1073/pnas.1706945114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen Q, Hao W, Xiao C, Wang R, Xu X, Lu H, Chen W, Deng CX. 2017. SIRT6 is essential for adipocyte differentiation by regulating mitotic clonal expansion. Cell Rep 18:3155–3166. doi: 10.1016/j.celrep.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu SY, Chiang CM. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 139.Shi J, Vakoc CR. 2014. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brown JD, Feldman ZB, Doherty SP, Reyes JM, Rahl PB, Lin CY, Sheng Q, Duan Q, Federation AJ, Kung AL, Haldar SM, Young RA, Plutzky J, Bradner JE. 2018. BET bromodomain proteins regulate enhancer function during adipogenesis. Proc Natl Acad Sci U S A 115:2144–2149. doi: 10.1073/pnas.1711155115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hu YJ, Belaghzal H, Hsiao WY, Qi J, Bradner JE, Guertin DA, Sif S, Imbalzano AN. 2015. Transcriptional and post-transcriptional control of adipocyte differentiation by Jumonji domain-containing protein 6. Nucleic Acids Res 43:7790–7804. doi: 10.1093/nar/gkv645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zang K, Wang J, Dong M, Sun R, Wang Y, Huang Y, Liu X, Li Y, Wang F, Yu M. 2013. Brd2 inhibits adipogenesis via the ERK1/2 signaling pathway in 3T3-L1 adipocytes. PLoS One 8:e78536. doi: 10.1371/journal.pone.0078536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. 2009. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem J 425:71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kadoch C, Crabtree GR. 2015. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv 1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Salma N, Xiao H, Mueller E, Imbalzano AN. 2004. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol Cell Biol 24:4651–4663. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski J, Drier Y, Gillespie SM, Archer TC, Wu JN, Tzvetkov EP, Troisi EC, Pomeroy SL, Biegel JA, Tolstorukov MY, Bernstein BE, Park PJ, Roberts CW. 2017. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 49:289–295. doi: 10.1038/ng.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K, Williams RT, Cassel SH, Qing H, Widmer CJ, Demetri GD, Irizarry RA, Zhao K, Ranish JA, Kadoch C. 2017. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 49:1613–1623. doi: 10.1038/ng.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lyko F. 2018. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet 19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 149.Londono Gentile T, Lu C, Lodato PM, Tse S, Olejniczak SH, Witze ES, Thompson CB, Wellen KE. 2013. DNMT1 is regulated by ATP-citrate lyase and maintains methylation patterns during adipocyte differentiation. Mol Cell Biol 33:3864–3878. doi: 10.1128/MCB.01495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang X, Wu R, Shan W, Yu L, Xue B, Shi H. 2016. DNA methylation biphasically regulates 3T3-L1 preadipocyte differentiation. Mol Endocrinol 30:677–687. doi: 10.1210/me.2015-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wu X, Zhang Y. 2017. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 152.Sérandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, Palierne G, Gheeraert C, Barloy-Hubler F, Péron CL, Madigou T, Durand E, Froguel P, Staels B, Lefebvre P, Métivier R, Eeckhoute J, Salbert G. 2012. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res 40:8255–8265. doi: 10.1093/nar/gks595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wiehle L, Raddatz G, Musch T, Dawlaty MM, Jaenisch R, Lyko F, Breiling A. 2016. Tet1 and Tet2 protect DNA methylation canyons against hypermethylation. Mol Cell Biol 36:452–461. doi: 10.1128/MCB.00587-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 155.Arner P, Kulyte A. 2015. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol 11:276–288. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 156.Yu J, Kong X, Liu J, Lv Y, Sheng Y, Lv S, Di W, Wang C, Zhang F, Ding G. 2014. Expression profiling of PPARgamma-regulated microRNAs in human subcutaneous and visceral adipogenesis in both genders. Endocrinology 155:2155–2165. doi: 10.1210/en.2013-2105. [DOI] [PubMed] [Google Scholar]
- 157.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Näär AM. 2010. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. 2010. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Horie T, Nishino T, Baba O, Kuwabara Y, Nakao T, Nishiga M, Usami S, Izuhara M, Sowa N, Yahagi N, Shimano H, Matsumura S, Inoue K, Marusawa H, Nakamura T, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. 2013. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat Commun 4:2883. doi: 10.1038/ncomms3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee MJ, Wu Y, Fried SK. 2013. Adipose tissue heterogeneity: implication of depot differences in adipose tissue for obesity complications. Mol Aspects Med 34:1–11. doi: 10.1016/j.mam.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Roh HC, Tsai LTY, Lyubetskaya A, Tenen D, Kumari M, Rosen ED. 2017. Simultaneous transcriptional and epigenomic profiling from specific cell types within heterogeneous tissues in vivo. Cell Rep 18:1048–1061. doi: 10.1016/j.celrep.2016.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhou P, Zhang Y, Ma Q, Gu F, Day DS, He A, Zhou B, Li J, Stevens SM, Romo D, Pu WT. 2013. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc Natl Acad Sci U S A 110:15395–15400. doi: 10.1073/pnas.1304124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, Eddy SR, Ecker JR, Nathans J. 2015. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shapiro E, Biezuner T, Linnarsson S. 2013. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 165.Kaelin WG., Jr 2012. Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science 337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shalem O, Sanjana NE, Zhang F. 2015. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, Yuan B, Kellis M, Lodish HF, Rinn JL. 2013. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A 110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yi F, Yang F, Liu X, Chen H, Ji T, Jiang L, Wang X, Yang Z, Zhang LH, Ding X, Liang Z, Du Q. 2013. RNA-seq identified a super-long intergenic transcript functioning in adipogenesis. RNA Biol 10:991–1001. doi: 10.4161/rna.24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pombo A, Dillon N. 2015. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 16:245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 170.Siersbæk R, Madsen JGS, Javierre BM, Nielsen R, Bagge EK, Cairns J, Wingett SW, Traynor S, Spivakov M, Fraser P, Mandrup S. 2017. Dynamic rewiring of promoter-anchored chromatin loops during adipocyte differentiation. Mol Cell 66:420–435.e5. doi: 10.1016/j.molcel.2017.04.010. [DOI] [PubMed] [Google Scholar]