GP78 is an autocrine motility factor (AMF) receptor (AMFR) with E3 ubiquitin ligase activity that plays a significant role in tumor cell proliferation, motility, and metastasis. Aberrant extracellular signal-regulated kinase (ERK) activation via receptor tyrosine kinases promotes tumor proliferation and invasion.
KEYWORDS: DUSP1, EGFR, ERK, GP78, ubiquitination
ABSTRACT
GP78 is an autocrine motility factor (AMF) receptor (AMFR) with E3 ubiquitin ligase activity that plays a significant role in tumor cell proliferation, motility, and metastasis. Aberrant extracellular signal-regulated kinase (ERK) activation via receptor tyrosine kinases promotes tumor proliferation and invasion. The activation of GP78 leads to ERK activation, but its underlying mechanism is not fully understood. Here, we show that GP78 is required for epidermal growth factor receptor (EGFR)-mediated ERK activation. On one hand, GP78 interacts with and promotes the ubiquitination and subsequent degradation of dual-specificity phosphatase 1 (DUSP1), an endogenous negative regulator of mitogen-activated protein kinases (MAPKs), resulting in ERK activation. On the other hand, GP78 maintains the activation status of EGFR, as evidenced by the fact that EGF fails to induce EGFR phosphorylation in GP78-deficient cells. By the regulation of both EGFR and ERK activation, GP78 promotes cell proliferation, motility, and invasion. Therefore, this study identifies a previously unknown signaling pathway by which GP78 stimulates ERK activation via DUSP1 degradation to mediate EGFR-dependent cancer cell proliferation and invasion.
INTRODUCTION
GP78 (also known as AMFR [autocrine motility factor receptor]) is an endoplasmic reticulum (ER)-anchored protein that was initially identified as a receptor for autocrine motility factor (AMF) to promote cell motility and metastasis (1). Subsequent studies showed that GP78 is an E3 ligase that belongs to the RING (really interesting new gene) E3 ubiquitin ligase family (2). GP78 maintains the levels of misfolded proteins by promoting ER-associated degradation (ERAD) activity, which prevents the accumulation of aberrant proteins and subsequent ER stress (3, 4). GP78 can also serve as a negative regulator of cholesterol metabolism by promoting the degradation of key regulators involved in lipid metabolism, including HMG-coenzyme A (CoA) reductase and Insigs (5, 6). A role for GP78 in tumor metastasis has been established, but its underlying mechanism is not fully understood. It has been suggested that tumor metastasis-promoting activity of GP78 depends on the availability of secretory AMF in the tumor microenvironment (7). In addition, GP78 can downregulate the metastatic suppressor CD82/KAI to promote tumor metastasis (8). Our previous study showed that downregulation of GP78 leads to a decrease in extracellular signal-regulated kinase (ERK) activation in response to AMF (9). However, it is not clear how GP78 regulates ERK activation to promote tumor cell proliferation, mobility, and invasion.
Dual-specificity phosphatase 1 (DUSP1) is a member of the mitogen-activated protein kinase (MAPK) phosphatase family. It plays a critical role in regulating the activities of MAPKs, the central regulators that orchestrate many short- and long-term changes in cellular responses to extracellular stimuli (10–13). DUSP1 dephosphorylates both threonine and tyrosine residues on c-Jun N-terminal kinase (JNK), p38, and ERK, thus acting as an endogenous inhibitor of MAPKs (10–14). DUSP1 was originally identified as a mitogen-inducible gene (15), and subsequent studies showed that DUSP1 expression can be induced by various stimuli, including stress, chemotherapeutic agents, and hypoxia (16–18). The deregulation of DUSP1 expression has been implicated in many diseases, including cancer (18–20). For example, DUSP1 is overexpressed in several major types of cancer, including ovarian, lung, and breast cancers (20–23). In addition, DUSP1 can be induced by clinically used chemotherapeutics in cancer cells, which promotes chemoresistance (18, 24).
DUSP1 expression is regulated at multiple mechanistic levels, including transcriptional and posttranslational levels (19). On one hand, the DUSP1 promoter and intron contain several transcription factor-binding sites, including sites for Sp1, AP2, and p53 (25, 26). Binding of transcription factors to those sites can lead to transcriptional induction of DUSP1. On the other hand, DUSP1 can be regulated by posttranscriptional and posttranslational mechanisms, including ubiquitination-mediated degradation (17). It has been shown that DUSP1 protein is ubiquitinated and its level is significantly increased by the proteasome inhibitor MG132 (27). A previous study indicated that the E3 ligase SCFSkp2 mediates exogenous DUSP1 ubiquitination and subsequent degradation (28). However, the mechanism of DUSP1 regulation via E3 ligase-mediated degradation is largely unknown.
In this study, we report that GP78 physically interacts with and promotes DUSP1 ubiquitination and its subsequent degradation. Notably, GP78 mediates downregulation of DUSP1 expression while enhancing epidermal growth factor (EGF)-mediated ERK signaling. In addition, EGF receptor (EGFR) fails to be phosphorylated by EGF when GP78 is downregulated or deleted. Thus, the finding that GP78 mediates DUSP1 degradation provides a novel insight into the mechanism by which GP78 maintains EGFR-mediated ERK signaling.
RESULTS
GP78 mediates DUSP1 ubiquitination and degradation.
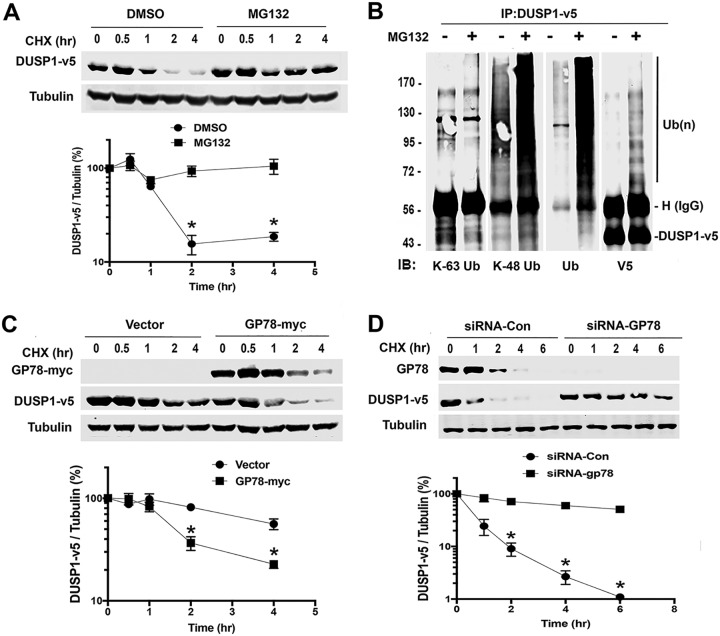
To understand the regulation of DUSP1 protein stability by ubiquitin-mediated degradation, we initially determined the half-life of DUSP1 protein in HEK293T cells transfected with the plasmid pcDNA3.1-DUSP1-v5. When new protein synthesis was inhibited by cycloheximide (CHX), the level of DUSP1-v5 protein started to decrease at 1 h, which was significantly blocked by the proteasome inhibitor MG132 (Fig. 1A), indicating that DUSP1 protein expression is regulated by proteasome-mediated degradation. Using linkage-specific ubiquitin antibodies, we showed that DUSP1 ubiquitination is dependent on K48 but not K63 (Fig. 1B), suggesting that DUSP1 degradation follows a K48 ubiquitin-mediated pathway. Next, we questioned if the E3 ligase GP78 mediates DUSP1 ubiquitination, because GP78 downregulation decreases the level of AMF-stimulated ERK phosphorylation (9) and ERK is a substrate of DUSP1. This prompted us to hypothesize that GP78-mediated DUSP1 degradation results in an increase in ERK phosphorylation. To test this hypothesis, HEK293T cells were cotransfected with DUSP1-v5 and GP78-myc. Figure 1C shows that cotransfection with GP78 decreased the half-life of DUSP1-v5 protein. To confirm the role of GP78 in this downregulation of DUSP1 protein expression, HEK293T cells were transfected with small interfering RNA (siRNA) against GP78 or control siRNA, along with DUSP1-v5, and the cell lysates were immunoblotted with a V5 antibody. Figure 1D shows that a decrease in DUSP1-v5 protein expression was significantly attenuated in cells transfected with siRNA against GP78 compared with cells transfected with control siRNA. We also showed that, relative to cells transfected with control siRNA, knockdown of GP78 significantly increased the half-life of DUSP1 (Fig. 1D). Collectively, these data suggest that DUSP1 protein expression is critically controlled by GP78, possibly via ubiquitination-mediated proteasome degradation.
FIG 1.
GP78 regulates the half-life of DUSP1. (A) CHX chase analysis in HEK293T cells transiently expressing DUSP1-v5. Cells were pretreated with MG132 (10 μM) or dimethyl sulfoxide (DMSO) for 2 h, followed by treatment with CHX (100 μg/ml) for the indicated periods. Western blot analysis of DUSP1-v5 protein (top) and quantification of DUSP1-v5 using ImageJ software (bottom) are shown. HEK293T cells were transfected with DUSP1-v5 for 24 h in the presence or absence of MG132. The data represent means ± SD of the results of three independent experiments. *, P < 0.05. (B) IP with V5 antibody in the lysates of HEK293T cells expressing DUSP1-v5 and Western blotting (immunoblotting [IB]) with K-63-specific, K-48-specific, ubiquitin, or V5 antibody. HEK293T cells were transfected with DUSP1-v5 for 24 h and then treated with MG132 for 2 h. Numbers on the left are kilodaltons. (C) Effect of GP78 overexpression on the half-life of DUSP1-v5 protein in HEK293T cells cotransfected with GP78-myc and DUSP1-v5 (1:1 ratio) for 24 h. The data represent means ± SD of the results of three independent experiments. *, P < 0.05. (D) Effect of GP78 knockdown on the half-life of DUSP1-v5 in HEK293T cells transfected with siRNA-GP78 or control (Con) siRNA for 24 h. The data represent means ± SD of the results of three independent experiments. *, P < 0.05.
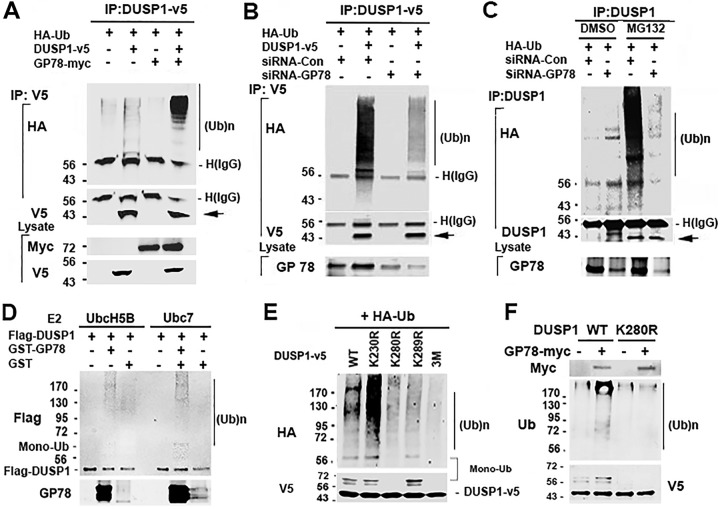
To define the role of GP78 in DUSP1 degradation, we questioned if GP78 causes DUSP1 ubiquitination. First, we tested whether exogenous GP78 affects exogenous DUSP1 ubiquitination. Accordingly, HEK293T cells were cotransfected with hemagglutinin-ubiquitin (HA-Ub) and DUSP1-v5, GP78-myc, or both, and in order to avoid contamination of DUSP1-interacting proteins modified by ubiquitin, cell lysates were prepared, boiled and cooled down, and subjected to immunoprecipitation (IP). Figure 2A shows that exogenous GP78 promoted exogenous DUSP1-v5 ubiquitination. We then asked if endogenous GP78 promotes exogenous DUSP1 ubiquitination. Briefly, HEK293T cells were cotransfected with HA-Ub and DUSP1-v5 or empty vector, along with siRNA against GP78 or control siRNA. Figure 2B shows that DUSP1-v5 ubiquitination was markedly reduced in cells transfected with GP78 siRNA compared to cells transfected with control siRNA. In addition, we tested the ability of endogenous GP78 to promote endogenous DUSP1 ubiquitination by knocking down GP78 with siRNA. As shown in Fig. 2C, knockdown of endogenous GP78 led to a significant reduction of endogenous DUSP1 ubiquitination compared to cells transfected with control siRNA. By demonstrating the effects of both ectopic and endogenous expression of GP78 on DUSP1 ubiquitination, our data suggest that DUSP1 is a novel substrate of GP78.
FIG 2.
GP78 induces DUSP1 ubiquitination. (A) Overexpression of GP78 causes DUSP1-v5 ubiquitination. (Top) Lysates of HEK293T cells transfected with HA-Ub, DUSP1-v5, and GP78-myc constructs (1:2:2 ratio) for 24 h were denatured and reconstituted by boiling and cooling for IP with V5 antibody. (Bottom) The total cell lysates were used to detect the indicated proteins. (B) siRNA-mediated GP78 knockdown reduces DUSP1-v5 ubiquitination. Shown are IP and immunoblotting in cells transfected with siRNA-GP78 and DUSP1-v5 (1:1 ratio) for 48 h. (C) GP78 knockdown decreases endogenous DUSP1 ubiquitination. Cells were transfected with siRNA-GP78 for 48 h and then subjected to IP with anti-DUSP1 antibody, followed by detection of endogenous DUSP1 ubiquitination. (D) In vitro ubiquitination assay. Purified Flag-DUSP1-fused His tag and GST-GP78 proteins were mixed, followed by addition of E1, E2, ATP, and Ub, and then incubated at 30°C for 30 min. Samples were resolved by SDS-PAGE and subjected to immunoblot analysis with anti-Flag and GP78 antibodies. (E) Identification of DUSP1 ubiquitination site. HEK293T cells were transfected with HA-Ub and the wild type (WT) or one of mutant constructs of DUSP1 for 24 h. The K230R, K280R, and K289R constructs have a single mutation, while the 3M construct contains all three mutations (K230R, K280R, and K289R). Monoubiquitinated DUSP1 levels were calculated based on the molecular masses of DUSP1-v5 amino acids plus HA-Ub amino acids. (F) Effect of GP78 on monoubiquitination of the K280R DUSP1 mutant. HEK293T cells were transfected with K280R and GP78-myc for 48 h. The total cell lysates were immunoprecipitated with His antibody and analyzed by Western blotting with ubiquitin antibody. Numbers to the left of the gels are kilodaltons.
To confirm the role of GP78 in mediating DUSP1 ubiquitination in a cell-free system, a glutathione S-transferase (GST)-tagged C terminus of GP78 containing an E3 ligase domain and His-tagged Flag-DUSP1 were expressed in bacteria and purified. An in vitro DUSP1 ubiquitination assay was performed using both purified GST-tagged GP78 C terminus and His-tagged Flag-DUSP1 in the presence of UbcH5B or Ubc7. Of note, UbcH5B and Ubc7 were previously used for in vitro GP78 ubiquitination (2). As shown in Fig. 2D, Flag-DUSP1 was ubiquitinated in the presence of GST-GP78, confirming that GP78 can act as an E3 ligase in vitro to trigger DUSP1 ubiquitination. Next, we asked which specific lysine residue(s) on DUSP1 is the site for its ubiquitination. By analyzing its protein sequence with the algorithm UbPred (www.ubpred.org), we identified 10 lysine (K) residues on DUSP1: K57, K97, K122, K138, K192, K221, K230, K248, K280, and K289. In addition, a quantitative-proteomics approach showed that DUSP1 is frequently modified at K230, K280, and K289 (www.phosphosite.org). On the basis of these findings, we performed site-directed mutagenesis on DUSP1 to replace K230, K280, and K289 with arginine (R) and showed that a mutation in K230R did not affect DUSP1 poly- and monoubiquitination (Fig. 2E). In contrast, a mutation in K280R or K289R led to a significant decrease in DUSP1 polyubiquitination, and DUSP1 monoubiquitination was abolished in K280R but not in K289R. Consistently, DUSP1 mono- and polyubiquitination were completely abolished in the 3 M construct, which contains three mutated lysines (i.e., K230R, K280R, and K289R). Interestingly, GP78 cotransfection slightly induced K280R polyubiquitination without affecting its monoubiquitination (Fig. 2F). These data suggest that K280 and K289 are responsible for DUSP1 polyubiquitination and that K280 is a priming site for DUSP1 ubiquitination, including its monoubiquitination.
DUSP1 physically interacts with GP78.
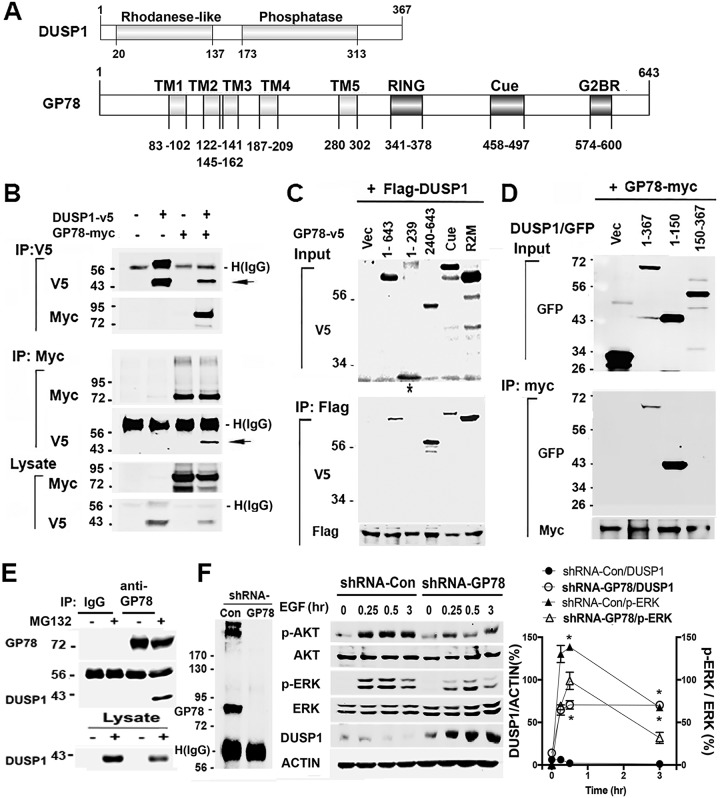
Promoting DUSP1 ubiquitination suggests that GP78 actually interacts with DUSP1. Therefore, we performed coimmunoprecipitation (co-IP) experiments with lysates of HEK293T cells transfected with pcDNA3-GP78-myc and pcDNA3-DUSP1-v5. Figure 3B shows that GP78-myc coimmunoprecipitated with DUSP1-v5 when whole-cell lysates were incubated with V5 antibody. Reciprocally, co-IP with Myc antibody revealed that DUSP1-v5 coimmunoprecipitated with GP78-myc. In addition, we found that GP78 could interact with DUSP4 (data not shown), another member of the DUSP family (18). We then queried the region that was responsible for this observed interaction. GP78 has four main functional domains, i.e., transmembrane, Ring, Cue, and G2BR domains (Fig. 3A). We generated two deletion constructs in pcDNA6-v5 that express either amino acids (aa) 1 to 239 or aa 240 to 643 of GP78. Co-IP experiments with lysates of cells cotransfected with these constructs using anti-Flag antibody showed that aa 240 to 643 were capable of interacting with Flag-DUSP1 while aa 1 to 239 did not, suggesting that DUSP1 interacts with aa 240 to 643 of GP78, the scaffolding region encompassing Ring (E2 binding) and CUE (ubiquitin binding) domains. In addition, a point mutation in either the Cue or R2M domain did not affect their interaction (Fig. 3C). To further define the region that interacts with DUSP1, we generated two additional deletion constructs expressing either aa 1 to 424 or aa 1 to 541 of GP78 and found that aa 1 to 541 were capable of interacting with Flag-DUSP1 while aa 1 to 424 did not (data not shown), suggesting that a region of GP78 containing Cue is required for its interaction with DUSP1. Next, we asked which region of DUSP1 is responsible for its interaction with GP78. DUSP1 contains an N-terminal rhodanese-like domain and a C-terminal phosphatase domain (Fig. 3A). We generated three green fluorescent protein (GFP)-fused constructs that express aa 1 to 367 (full-length), aa 1 to 150, and aa 150 to 367 of DUSP1, respectively. Co-IP with a c-myc antibody revealed that GP78-myc interacts with aa 1 to 150 (Fig. 3D), a region encompassing a rhodanese-like domain that is evolutionarily conserved among members of the DUSP family (29). However, an aa 150-to-367 region of DUSP1 failed to interact with GP78 (Fig. 3D). Importantly, we showed that endogenous DUSP1 is immunoprecipitated by GP78 antibody (Fig. 3E), suggesting that GP78, via a motif containing a G2BR domain, interacts with the rhodanese-like domain of DUSP1.
FIG 3.
DUSP1 physically interacts with GP78. (A) Domain structures of DUSP1 and GP78 proteins. (B) Reciprocal co-IP of HEK293T cells expressing both GP78-myc and DUSP1-v5. Cell lysates were subjected to IP with V5 or myc antibody, respectively. (C) IP with Flag antibody of cells cotransfected with Flag-DUSP1 and one of the GP78 deletion mutant constructs. The Cue mutant construct containing 6 point mutations (M467G, F468G, P469R, V476R, D479V, and L480D) lost its ability to bind to ubiquitin, while the R2M construct, with 2 point mutations (C356G and H362A), lost its ability to bind to E2. (D) Co-IP with GP78-myc in lysates of HEK293T cells expressing DUSP1-fused GFP deletion constructs. (E) Endogenous interaction between GP78 and DUSP1. Shown is co-IP of the lysates of HEK293T cells, pretreated with MG132 or left untreated, with GP78 antibody and immunoblotting with GP78 and DUSP1 antibodies, respectively. (F) (Left and middle) Western blot analyses of GP78 levels using IP with anti-GP78 antibody in vector control HEK293T cells and HEK293T/GP78 knockdown cells (left) and indicated proteins in vector control HEK293T cells and HEK293T/GP78 knockdown cells stimulated with EGF (100 ng/ml) at the indicated time points (middle). (Right) p-ERK and DUSP1 expression after normalization with actin control. *, P < 0.05 (n = 3). The error bars indicate SD. Numbers to the left of the gels are kilodaltons.
DUSP1 negatively regulates GP78-dependent activation of ERK in response to EGF.
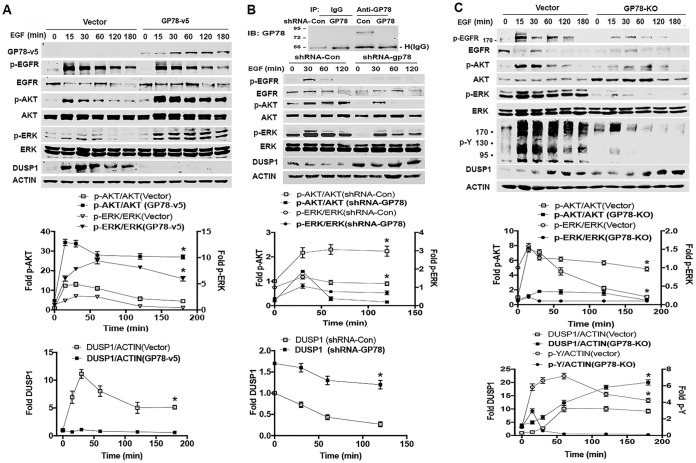
The role of GP78 in DUSP1 degradation led us to test the effect of DUSP1 on ERK activation due to the fact that DUSP1 is able to inactivate ERK (18) while GP78 knockdown leads to a decrease in ERK phosphorylation (9). In this regard, we first generated GP78 stable knockdown HEK293T cells by short hairpin RNA (shRNA) using the lentiviral system (Fig. 3F, left). We treated shRNA-GP78 or shRNA-Con (control) cells with EGF for 15, 30, and 180 min and measured ERK and AKT phosphorylation and DUSP1 protein expression. Figure 3F (middle and right) shows that the basal level of DUSP1 in shRNA-GP78 cells was higher than that in control cells. In response to EGF, DUSP1 was highly induced in a time-dependent manner in shRNA-GP78 cells, but such an increase was not observed in control HEK293T cells (Fig. 3F, middle and right). Consistent with negative regulation of ERK by DUSP1, the level of phosphorylated ERK (p-ERK) was decreased in shRNA-GP78 cells compared with control cells (Fig. 3F). Similar to the effect on ERK activation, AKT phosphorylation was decreased in shRNA-GP78 cells compared with control cells, which is consistent with a role for GP78 in the positive regulation of AKT. These results suggest that GP78 promotes cell survival signals by the DUSP1/ERK axis.
GP78 enhances proliferation and migration in hepatocellular carcinoma cells.
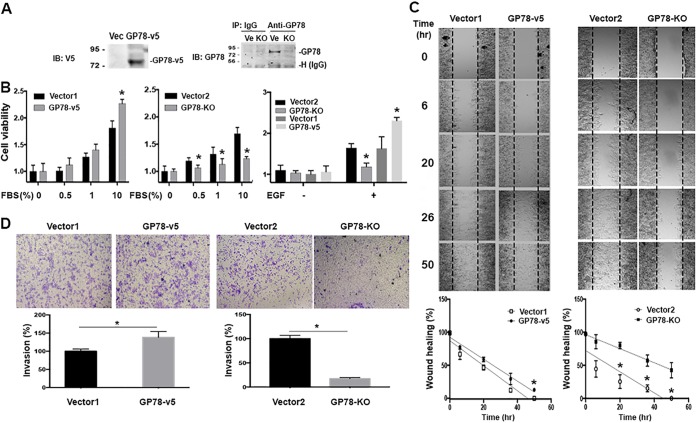
Next, we investigated the possible biological relevance of DUSP1 regulation by GP78 during cancer cell proliferation, motility, and invasion. Since our previous work had indicated an inhibitory role of GP78 in hepatocellular carcinoma (HCC) development (30), we thought to assess the manipulation of GP78 expression in cell proliferation and invasion in human HCC Huh7 cells. First, we generated Huh7 cells in which either GP78 was overexpressed with lenti-GP78-v5 viruses or GP78 was stably knocked out with clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 (Fig. 4A). GP78 knockout (KO) in Huh7 cells was confirmed by DNA sequencing (data not shown). We serum starved both Huh7-GP78 KO and Huh7-GP78-v5 control cells for 24 h; replaced the cell medium with medium containing 0.5%, 1%, or 10% fetal bovine serum (FBS); and then measured cell proliferation. GP78 overexpression promoted FBS- or EGF-stimulated cell proliferation compared to control cells under the same treatment conditions (Fig. 4B, left and right). Conversely, Fig. 4B (middle) shows that knockout of GP78 decreased cell proliferation compared with control cells. A similar result was obtained with EGF stimulation (Fig. 4B, right). Considering a role for GP78 in tumor cell invasion, we examined the effects of GP78 on tumor cell motility and invasion in response to 1% FBS plus EGF stimulation. We used both FBS and EGF because they have higher efficacy than a single stimulus in terms of promoting migration and invasion. As shown in Fig. 4C and D, wound-healing assays and Transwell migration assays revealed that cell migration/invasion was increased in GP78-overexpressing cells compared to vector control cells, while an opposite result was obtained with GP78 KO cells. These results are consistent with the role of GP78 in promoting cancer cell growth, migration, and invasion.
FIG 4.
GP78 promotes cancer cell proliferation, motility, and invasion. (A) Western blot analyses confirmed GP78 overexpression (left) and GP78 knockout (right) in Huh7 cells. Numbers to the left of the gels are kilodaltons. (B) MTT assays of GP78-overexpressing (GP78-v5), overexpressing vector control (Vector1) GP78 knockout (GP78 KO), and knockout control (Vector2) Huh7 cells. Cells were seeded overnight and then cultured in medium containing FBS or EGF (100 ng/ml), and cell proliferation was measured after 24 h. The quantitative data are presented as means and SD from the results of 3 independent experiments (*, P < 0.05). (C) Wound-healing assays of the indicated Huh7 cells cultured in serum-free medium in the presence or absence of EGF for the indicated times. Of note, EGF without FBS was sufficient to promote cell migration. The graphs were generated after normalization with the average distance of the scratched area (n = 3; *, P < 0.05). (D) (Top) Migration and invasion assays of the indicated Huh7 cells incubated with 1% FBS plus 100 ng/ml EGF for 24 h using Transwell chambers. (Bottom) Numbers of cells migrating through the Transwell membrane. The quantitative data are presented as means ± SD from the results of 3 independent experiments (*, P < 0.05).
The GP78/DUSP1/ERK axis regulates EGFR signaling.
To delineate the mechanism by which GP78 promotes cell proliferation and invasion in response to growth factor stimulation, we analyzed the effect of GP78 on DUSP1 levels and ERK phosphorylation in response to EGF. EGF treatment induced DUSP1 expression in vector control cells (Fig. 5A), which is consistent with previous reports that DUSP1 is an immediate-early response gene (31, 32). However, EGF failed to do the same in GP78-overexpressing cells. The basal levels of DUSP1 protein in both vector control and GP78-overexpressing cells were comparable (Fig. 5A). As expected, EGF treatment stimulated ERK phosphorylation within 15 min, which then returned to basal levels at 120 min in vector control cells (Fig. 5A). In contrast, the level of ERK phosphorylation 15 min after EGF stimulation in GP78-overexpressing cells was much higher and remained at the higher levels 120 min after treatment (Fig. 5A). Conversely, we observed an inverse correlation between EGF-stimulated DUSP1 induction and the level of ERK phosphorylation in both GP78 knockdown cells and GP78 KO cells (Fig. 5B and C) compared to corresponding control cells. Similar to ERK activation, the level of AKT phosphorylation was higher in GP78-overexpressing cells and lower in GP78 knockdown or knockout cells compared to the corresponding control cells (Fig. 5), which is consistent with our previous observation (9), thus functionally validating the success of GP78 knockdown/knockout. Interestingly, although GP78 overexpression did not significantly affect levels of total and phosphorylated EGFR (Fig. 5A), the levels of total and phosphorylated p-EGFR were decreased in GP78 knockdown and GP78 KO cells (Fig. 5B and C). These findings indicate that GP78 plays a maintenance role in EGFR activation, as its overexpression does not enhance EGFR phosphorylation but its deficiency impairs EGFR activation. Thus, our results suggest that GP78 interaction with DUSP1 is critical for maintaining ERK signaling in response to EGF in HCC cells.
FIG 5.
Effects of GP78 on EGFR signaling and DUSP1 expression. Huh7 cells were serum starved for 2 h, stimulated with EGF at the indicated time points, and harvested for analyses of the indicated proteins by Western blotting (shown at the top). (A) GP78-v5-overexpressing and vector control Huh7 cells. (B) Huh7 cells infected with shRNA-GP78-containing lentiviruses or control viruses. Downregulation of GP78 by shRNA was confirmed by IP and Western blotting, and the impacts of its downregulation on the indicated proteins are shown. (C) GP78 KO and control cells. p-Y, tyrosine antibody was used to detect EGF-dependent tyrosine phosphorylation. (Top) The impacts of its knockout on the indicated proteins are shown. (A to C, bottom) Fold changes were normalized relative to total forms or actin (n = 3; means ± SD are shown; *, P < 0.05). Numbers to the left of the gels (B and C) are kilodaltons.
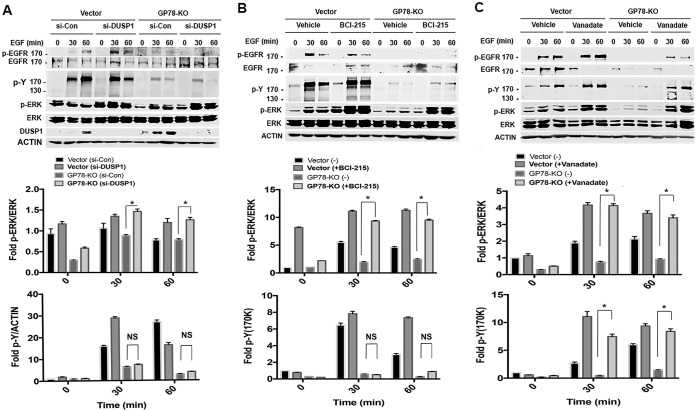
Since it is well established that autophosphorylation on multiple tyrosine residues of EGFR leads to tyrosine phosphorylation of its downstream kinases and adaptor proteins, we assessed the effect of GP78 on EGF-dependent tyrosine phosphorylation. Immunoblotting with phosphotyrosine (p-Y) antibody revealed that tyrosine phosphorylation by EGF was markedly increased at 15 min and then gradually decreased at 60 min in the lysates of vector control cells, whereas such an increase was significantly impaired in GP78 KO cells (Fig. 5C), highlighting the fact that GP78 promotes EGF-dependent tyrosine phosphorylation. Next, using both genetic and pharmacological approaches, we asked whether GP78 affects the phosphorylation of EGFR through DUSP1 and whether DUSP1 impacts GP78-mediated, EGF-stimulated tyrosine phosphorylation. Figure 6A shows that knockdown of DUSP1 increased EGFR phosphorylation upon EGF stimulation in vector control cells but not in GP78 KO cells. However, DUSP1 knockdown relieved the inhibition of ERK phosphorylation in GP78 KO cells. Using BCI-215, a small molecule that inhibits both DUSP1 and DUSP6 (33), we showed that BCI-215 treatment completely restored EGF-induced ERK phosphorylation in GP78 KO cells to the same level as that in control cells with intact GP78. Again, BCI-215 did not affect EGFR phosphorylation. We also found that BCI-215 treatment slightly increased EGF-dependent tyrosine phosphorylation in GP78 KO cells, but the levels of Tyr phosphorylation in BCI-215-treated GP78 KO cells were much lower than those in vector control cells. These results suggest that the GP78-mediated tyrosine phosphorylation upon EGF treatment is likely DUSP1/DUSP6 independent, raising the possibility that other DUSP family members are involved in the process. To test this possibility, we treated control and GP78 KO cells with orthovanadate, a broad-spectrum phosphatase inhibitor, and then examined ERK activation. As shown in Fig. 6C, orthovanadate significantly restored EGF-induced EGFR phosphorylation in GP78 KO cells, although the magnitude of EGFR phosphorylation in GP78 KO cells did not reach the same level as that in control cells. More importantly, EGF-dependent tyrosine phosphorylation and ERK activation were completely restored by orthovanadate in GP78 KO cells. In fact, vector and GP78 KO cells expressed comparable levels of tyrosine phosphorylation and ERK activation (Fig. 6C). On the basis of these findings, we suggest that GP78 positively regulates ERK activation by both maintaining the activation status of upstream EGFR and promoting the degradation of downstream DUSP1.
FIG 6.
Roles of DUSP1 knockdown and pharmacological inhibition of phosphatases in GP78-regulated EGFR signaling. (A to C) (Top) Western blot analyses of the indicated proteins of vector control and GP78 KO cells transfected with siRNA-DUSP1 or control siRNA for 48 h (A) or pretreated with BCI-215 for 2 h (B) and then stimulated with EGF, or Western blot analyses of vector control and GP78 KO cells stimulated with EGF in the presence and absence of orthovanadate (Vanadate) (0.1 mM) (C). (Bottom) Fold induction was calculated by normalization with the indicated proteins (unphosphorylated forms of protein or actin). n = 3; means ± SD are shown; *, P < 0.05; NS, not significant. Numbers to the left of the gels are kilodaltons.
Reexpression of GP78 rescues GP78 KO cells from defects in proliferation and invasion.
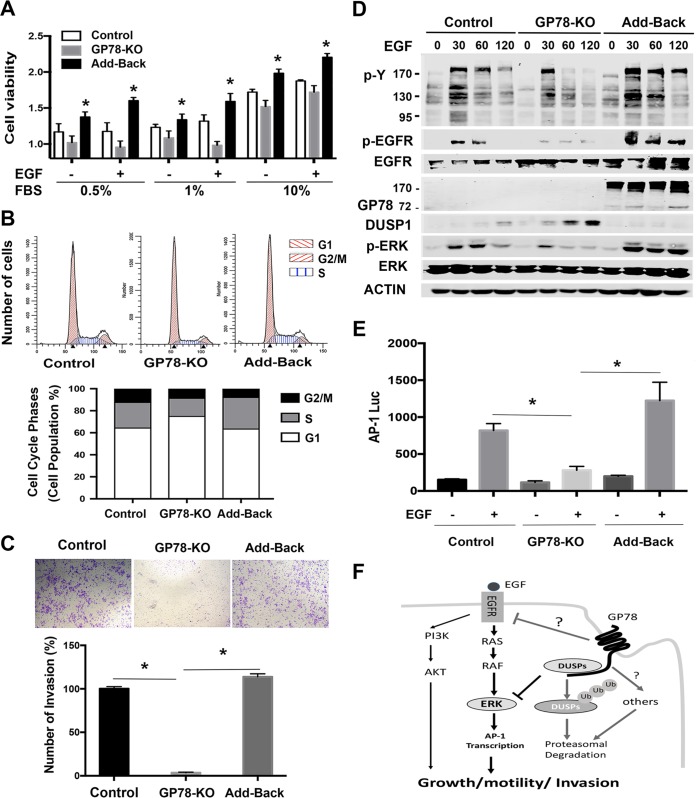
To further determine the impacts of GP78 on the EGFR-ERK/DUSP1 axis, we reintroduced GP78 into GP78 KO cells (Fig. 7D) and then assessed the levels of DUSP1, ERK, and EGFR phosphorylation. We constructed a lenti-GP78-hygro vector and produced GP78-expressing lentiviruses. We infected Huh7-GP78 KO cells with lenti-GP78-hygro or control viruses, followed by selection with hygromycin. Figure 7A shows that GP78 KO/GP78 reexpression (add-back) cells proliferate faster than GP78 KO cells in response to 24-h EGF stimulation. A similar result was obtained with a 48-h treatment (not shown). Cell cycle analysis showed that GP78 KO cells comprised 16% of the cells in S phase of the cell cycle compared to 23% in control cells. This reduction of GP-78 KO cells in S phase recovered to 29% by GP78 add back (Fig. 7B), suggesting that loss of GP78 in GP78 KO cells causes a partial S-phase cell cycle arrest. Furthermore, an invasion assay revealed that the low efficiency of invasion of GP78 KO was restored by GP78 add back to levels comparable to those of control cells (Fig. 7C). Consistent with the role of GP78 in EGFR activation, downregulation of p-EGFR and tyrosine phosphorylation in GP78 KO cells were also rescued by reexpression of GP78, suggesting that GP78 plays a critical role in maintaining EGFR activation status (Fig. 7D). Since loss of GP78 impaired ERK activation while increasing DUSP1 expression in response to EGF stimulation (Fig. 5C), we examined ERK activation and DUSP1 expression by GP78 reexpression and found that downregulation of p-ERK and upregulation of DUSP1 in GP78 KO cells were overcome by reintroduction of GP78 (Fig. 7D). Similar results were obtained with p-AKT in Huh78-GP78 KO cells in which GP78 was reintroduced (not shown). Since the transcription factor AP-1 is one of the downstream targets of ERK, we assessed the effect of GP78 on AP-1 reporter activity. Figure 7E shows that AP-1 luciferase activity was decreased in GP78 KO cells compared to vector control cells and that such a decrease was rescued by GP78 reexpression. These data validate our hypothesis that the GP78-DUSP1 interaction controls ERK signaling in response to EGF.
FIG 7.
Reexpression of GP78 rescues GP78 knockout cells from a defect in proliferation and invasion. (A) MTT assays of vector control, GP78 KO, and GP78 reexpression (Add-Back) cells infected with lenti-GP78-overexpressing viruses. Cells were cultured in medium with FBS in the absence or presence of EGF (100 ng/ml). Representative data from the results of three independent experiments are shown (n = 3; means and SD; *, P < 0.05). (B) FACS analysis of the indicated cells stimulated with EGF for 24 h. (C) Invasion assays of the indicated cells in Transwells coated with Matrigel for 24 h (n = 3; means and SD; *, P < 0.05). (D) Western blot analyses of the indicated proteins. Numbers to the left of the gels are kilodaltons. (E) AP-1 luciferase reporter activity of cells transfected with AP-1 luciferase reporter using a dual-luciferase reporter assay. Fold induction was normalized to Renilla luciferase. Representative data from the results of three independent experiments are shown (means and SD; *, P < 0.05). (F) Proposed role of GP78 in regulating the EGFR-ERK axis in cancer cell growth, motility, and invasion. GP78 plays a double-edged role by targeting DUSP1 for degradation to upregulate ERK signaling while maintaining EGFR activation status. GP78 also positively regulates AKT activation (43). It is unclear how GP78 regulates EGFR activation.
DISCUSSION
The role of GP78 in promoting tumor cell motility, invasion, and metastasis is well documented, but its underlying mechanism is not fully understood (34). Our previous study indicated that knockdown of GP78 decreases AMF-induced ERK activation (9). However, the underlying mechanism by which GP78 regulates ERK activation is not clear. In this study, we presented data showing that knockout of GP78 significantly decreased EGFR activation by EGF stimulation. We believe that GP78-regulated EGFR activation is independent of DUSP1, since DUSP1 knockdown did not impact EGF-mediated EGFR phosphorylation. This suggests that the GP78-EGFR axis is upstream of DUSP1 (Fig. 7F). Interestingly, ERK activation was fully recovered by DUSP1 knockdown, suggesting that DUSP1 is critical for regulating EGFR-mediated ERK activation. This hypothesis was further confirmed by experiments using the DUSP1 inhibitor BCI-215 or the broad-spectrum phosphatase inhibitor orthovanadate, as both inhibitors fully restored ERK activation in GP78 knockout cells in response to EGF stimulation (Fig. 6B and C).
Although DUSP1 knockdown or BCI-215 treatment did not affect GP78 deficiency-mediated suppression of EGFR phosphorylation, orthovanadate was able to partially restore EGFR tyrosine phosphorylation in GP78 knockout cells, suggesting that DUSP1 is not critical for the regulation of EGFR phosphorylation. Since orthovanadate is a broad-spectrum phosphatase inhibitor, incomplete recovery of EGFR phosphorylation in GP78 knockout cells suggests that other tyrosine phosphatases are involved in EGFR dephosphorylation. It has been shown that several tyrosine phosphatases are implicated in the dephosphorylation of EGFR tyrosine, including PTP1B and PTPN2 (35, 36). These phosphatases regulate the amplitude, duration, and amplification of EGFR signaling at the cell plasma membrane and the ER (37). For example, PTPN2 predominantly dephosphorylates EGFR in the ER (36). We found that GP78-proficient and GP78 knockout cells expressed comparable levels of PTP1B and SHP2 (data not shown), which suggests that the GP78 knockout-mediated defect in EGFR phosphorylation is unlikely to be due to the consequences of increased PTP1B and SHP2 expression. Because members of the Cbl family can serve as an E3 ligase to target EGFR degradation (38), we assessed the levels of Cbl-b and Cbl-3 and found that, relative to GP78-proficient cells, GP78 knockout cells expressed a higher level of Cbl-b while the Cbl-3 level remained unchanged (data not shown), suggesting the decrease in EGFR phosphorylation in GP78 knockout or knockdown cells may be in part due to an increase in Cbl-b expression. Since GP78 can be located on either the plasma membrane or the ER, we speculate that the regulation of EGFR phosphorylation by GP78 can occur at the plasma membrane and the ER.
GP78 is known as a tumor metastasis-promoting factor, and increased activation of the GP78-mediated signaling pathway leads to cell proliferation, migration, and invasion, the features that give cancer cells the capacity to metastasize. Consistently, we showed that knockdown of GP78 decreases Huh7 cell proliferation, migration, and invasion (Fig. 4). However, a recent paper indicated that knockout of GP78 increased liver cancer incidence in aged mice (30), thus acting as a tumor suppressor. The discrepancy in terms of being a tumor-promoting factor versus a tumor suppressor could be due to the model systems used. In response to EGF stimulation, GP78 promotes cell proliferation, migration, and invasion. However, in aged mice, GP78 can negatively affect liver cancer growth by impacting the interaction between liver cells and the tumor microenvironment.
The expression levels of DUSP1 protein are tightly regulated by proteasome degradation, but the underlying mechanism is poorly understood. In this study, we provided evidence to show that GP78 acts as an E3 ligase to promote DUSP1 ubiquitination and subsequent degradation. This conclusion was supported by data showing that GP78 expression is positively correlated with ubiquitinated DUSP1 but negatively correlated with total DUSP1 protein at both the exogenous and endogenous levels. Importantly, our data showed that lysine 280 on DUSP1 is the site that can attach to ubiquitin because mutation of this site clearly abolished DUSP1 mono- and polyubiquitination by GP78. In addition, we found that K48, but not K63, on ubiquitin forms polyubiquitin chains to promote DUSP1 degradation. Since a K48-linked polyubiquitin chain involves protein degradation (39), our result suggests that GP78-mediated ubiquitination promotes DUSP1 degradation.
DUSP1 is currently considered a specific MAPK phosphatase, since it was not known to interact with other proteins. Given the fact that GP78 promotes DUSP1 ubiquitination and degradation and that ubiquitination requires a physical interaction between an E3 ligase and its substrates, we reasoned that these two proteins interact with each other. Indeed, we found that exogenous and endogenous GP78 coimmunoprecipitated with DUSP1. In addition, we showed that GP78 interacts with the N terminus of DUSP1, which contains one cdc25 homology domain and a kinase-interacting domain, and that DUSP1 interacts with a region containing the CUE domain of GP78. Therefore, our study suggests that these proteins interact with each other, which was not previously recognized. Since MAPK phosphatases exert their biological functions by interacting only with MAPKs, our data support a previously unrecognized mechanism by which DUSP1 can regulate its biological function through interaction with non-MAPKs.
The upregulation of the ERK signaling pathway is one of the major oncogenic signals in human cancers. ERK can be activated by upstream signaling due to mutations of EGFR/Ras/Raf, the most commonly altered oncogenic pathway in several major types of cancer, including lung cancer and colorectal cancer. The activation of ERK plays a critical role in tumor cell proliferation, invasion, and metastasis. Currently, much effort has been devoted to developing strategies to target the Ras/Raf-EGFR-ERK axis in pursuit of treating cancers with activation of this oncogenic signal. As a result, many tyrosine kinase inhibitors (TKIs) are being used for the treatment of these cancers or are in clinical trials. However, the development of resistance renders TKIs ineffective. Although TKI resistance mechanisms are multifaceted, considering the role of DUSP1 and DUSP family members as a whole in regulating ERK, it is conceivable that the DUSP family members may play important roles in modulating cancer cells’ response to TKIs. Although a recent study indicates that DUSP5, a DUSP family member, affects cancer-causing RAF mutation-mediated ERK activity (40), little is known about the role of DUSPs in modulating cancer cells’ response to TKIs. Thus, understanding the role of DUSPs in regulation of ERK signaling in cancers with EGFR/Ras/Raf mutations would help in developing a better strategy to improve TKI treatment in cancers containing mutations in EGFR/Ras/Raf.
In summary, our data showed that GP78 serves as an E3 ligase to promote DUSP1 ubiquitination and subsequent degradation. In addition, GP78 was found to be required for EGFR activation by EGF. Since GP78 promotes tumor motility/metastasis and ERK activation is an oncogenic signal, further work will study how loss of GP78 impairs EGFR activation and how the GP78-DUSP1 interaction regulates ERK signaling in cancers with EGFR/Ras/Raf mutations.
MATERIALS AND METHODS
Cell culture.
HEK293T and Huh7 cells were obtained from the American Type Culture Collection (ATCC), grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), and maintained at 37°C in a humidified atmosphere of 5% CO2.
Reagents.
The reagents used were as follows: FBS (Sigma-Aldrich; catalog no. F0926), Z-Leu-Leu-Leu-al (MG132) (Sigma-Aldrich; catalog no. C2211), cycloheximide (Sigma-Aldrich; catalog no. C6255), puromycin dihydrochloride (Sigma-Aldrich; catalog no. P9620), Sodium orthovanadate (Sigma-Aldrich; catalog no. S6508), (E)-2-benzylidene-5-bromo-3-(cyclohexylamino)-2,3-dihydro-1H-inden-1-one (BCI-215) (33), penicillin-streptomycin (GE Healthcare; catalog no. SV30010), trypsin-EDTA (1×; Gibco; catalog no. 25300-054), DMEM (1×; Gibco; catalog no. 11995-065), blasticidin S HCl (Thermo Fisher Scientific; catalog no. R21001), hygromycin B (Thermo Fisher Scientific; catalog no. 10687010), Herculase II fusion DNA polymerase (Agilent Technology; catalog no. 600675), glutathione-Sepharose 4B (GE17-0756-01; Sigma), a ubiquitin conjugation initiation kit (BostonBioChem; catalog no. K-995), Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific; 11668027), BD BioCoat Matrigel invasion chambers (BD Biosciences; catalog no. 354480), QuikChange Multi site-directed mutagenesis kit (Agilent Technology; catalog no. 200514), QuikChange II site-directed mutagenesis kit (Agilent Technology; catalog no. 200523), ibidi culture insert (ibidi GmbH, Munich, Germany; catalog no. 81176), Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific; catalog no. 23227), Amicon Ultra-15 centrifugal filter devices (Millipore; Amicon Ultra 100-kDa molecular weight cutoff [MWCO]), EGF, human recombinant (Millipore; catalog no. 01-107).
On-Target plus SMART pool siRNA-DUSP1 contains the following 4 siRNAs: (i) AGGAGGAUACGAAGCGUUU, (Dharmacon; J-003484-21), (ii) UUUGUGAAGCAGAGGCGAA, (Dharmacon; J-003484-22), (iii) GCUUACCUUAUGAGGACUA; (Dharmacon, J-003484-23), and (iv) CCAACCAUUUUGAGGGUCA, (Dharmacon; J-003484-24).
Antibodies.
The antibodies used were as follows: p-AKT (ser473) (Cell Signaling Technology; catalog no. 9271S), AKT (Cell Signaling Technology; catalog no. 9272), p-ERK (42/44)Thr261 (Cell Signaling Technology; catalog no. 9152), p-ERK (42/44) (Thr202/Y204) (Cell Signaling Technology; catalog no. 9101s), ERK (42/44) (Cell Signaling Technology; catalog no. 9102), p-EGFR (Tyr1173/53A3) (Cell Signaling Technology; catalog no. 4407s), p-EGFR (S1046/1047) (Cell Signaling Technology; catalog no. 2238s), p-EGFR (Tyr1045) (Cell Signaling Technology; catalog no. 2237), EGFR (Cell Signaling Technology; catalog no. 2232), EGFR (1005) (Santa Cruz Biotechnology; catalog no. sc-03), GP78 (Cell Signaling Technology; catalog no. 9590S), EGFR (C74B9) (Cell Signaling Technology; catalog no. 2646x), Ub (p4D1) (Cell Signaling Technology; catalog no. 3936), Ub-K48 specific (Cell Signaling Technology; catalog no. 12805S), anti-β-tubulin (Sigma-Aldrich; catalog no. T4026), Ub-K63 specific (EMD Millipore; catalog no. 05-1308), DUSP1 (EMD Millipore; catalog no. 07-535), GP78 (3D9) (Santa Cruz Biotechnology; catalog no. sc-293371), p-Tyr antibody (PY20) (Santa Cruz Biotechnology; catalog no. sc-508), DUSP1 (c-19) (Santa Cruz Biotechnology; catalog no. sc-370), HA (F7) (Santa Cruz Biotechnology; catalog no. sc-7392), V5 (Invitrogen; catalog no. 46-0705), Flag (Sigma-Aldrich; catalog no. F1804), myc (Sigma-Aldrich; catalog no. M4439), and actin (Sigma-Aldrich; catalog no. A1978).
Plasmids and cloning.
pCI-Neo-GP78/JM20 (wild-type GP78) (Addgene; 13303), pCI-Neo-GP78 Cue-m1.2/JM22 (mutations of M467G, F468G, P469R, V476R, D479V, and L480D) (Addgene; 13305), and pCI-Neo-GP78 R2M/JM21 (mutations of C356G and H362A) (Addgene; 13304) were gifts from Allan Weissman and were described previously (2). pcDNA3.1-DUSP1-v5 was generated by subcloning DUSP1 cDNA into pcDNA3.1-V5-His (Invitrogen). DUSP1 cDNA was obtained by double digestion of pIRES2-EGFP-DUSP1 (41) with HindIII/BamHI. pcDNA6-GP78-myc-His was generated by subcloning GP78 cDNA into pcDNA6-myc-His (Invitrogen) at BamHI/XbaI sites. GP78 was obtained from CI-Neo-GP78/JM20. pcDNA6-V5-His-GP78 (aa 1 to 643), pcDNA6-V5-His-GP78 (aa 1 to 239), and pcDNA-myc-His-GP78 (aa 240 to 643) were described previously (42). pcDNA3-DUSP1-Flag was generated by subcloning DUSP1 cDNA into pcDNA3 at BamHI/EcoRI sites. pEGFP-DUSP1 (aa 1 to 367) was generated by subcloning DUSP1 cDNA into pEGFP-N3 (Clontech) at BamHI/XbaI sites. pEGFP-DUSP1 (aa 1 to 150) with the first 450-bp fragment of DUSP1 and pEGFP-DUSP1 (aa 150 to 367) containing a 654-bp fragment of the C terminus of DUSP1 were constructed by PCR amplification and subcloning into pEGFP-N3. The PCR primers used were as follows: 5′-CGCGGATCCACCATGGTCATGGAAGTGGGCACCCTG-3′ and 5′-CGGAATTCCTCAGGGGAAGGCTGAGCCCCAT-3′ for pEGFP-DUSP1 (aa 1 to 150) and 5′-CGGGATCCACTAGCGTCCCTGACAGCGCGGAA-3′ and 5′-CTAGTCTAGATCAGCAGCTGGGAGAGGTCGTAAT-3′ for pEGFP-DUSP1 (aa 150 to 367). pET28a-DUSP1-Flag was generated by subcloning DUSP1 cDNA into pET28a (EMD Bioscience) at BamHI/BglII sites. pGEX6-1-GP7878c (aa 309 to 643) was constructed by subcloning GP78 (aa 309 to 643) into pGEX6P-1 (Amersham) at BamHI/EcoRI sites. LentiGuide-Puro (Addgene plasmid 52963) and lentiCas9-BLAST (Addgene plasmid 52962) were gifts from Feng Zhang. Envelope plasmid pMD2.G (Addgene plasmid 12259) and the packaging plasmid psPAX2 (Addgene plasmid 12260) were gifts from Didier Trono. A Web-based Atum guide RNA (gRNA) design tool (https://www.atum.bio) was used to design gRNA, and CACGGCTGCTTAAGCTGCAACGTGGAGTA was used to target exon 6 of GP78.
Site-directed mutagenesis.
Site-directed mutagenesis was performed on DUSP1 using a QuikChange Lightning Multi site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. Primers were designed using the Agilent Web-based tool by replacing putative lysine (K) with arginine (R), and the mutations were confirmed by DNA sequencing.
Generation of GP78 knockout cells.
HEK293T cells were transfected with GP78 gRNA and lentiCas9-BLAST and then with pMD2.G and psPAX2. Transfection efficacy was monitored by Lenti-GFP and Lenti-RFP (red fluorescent protein) fluorescence. After 96 h, the supernatants were collected and centrifuged to remove debris. Viral particles were concentrated using Amicon Ultra (100K) filter devices (Millipore). To generate GP78 knockout cell lines, HEK293T and Huh7 cells were infected with GP78 gRNA viruses for 96 h, plated in 96-well plates, and selected with blasticidin (Invitrogen) and puromycin (Sigma-Aldrich). Resistant clones were obtained, and the levels of GP78 protein were assessed by IP with anti-GP78 rabbit antibody (30) and immunoblotting with anti-GP78 mouse antibody.
Transfection and CHX chase assay.
HEK293T cells were transfected with GP78, DUSP1, and HA-ubiquitin constructs and then treated with CHX (100 μg/ml). Cells were harvested at various time points, the total cell lysates were subjected to immunoblotting, and the relative ratio of DUSP1 was normalized by actin using NIH (Bethesda, MD) ImageJ software.
Expression and purification of recombinant proteins and in vitro ubiquitination assay.
GST-GP78 fusion protein in pGEX6-1-GP78c (aa 309 to 643) and His-Flag-DUSP1 in pET28a-Flag-DUSP1 were expressed in Escherichia coli (BL21) cells and induced by 0.3 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 3 h at 25°C. The cells were lysed by sonication according to the manufacturer’s protocol. To purify GST fusion proteins, cell extracts were loaded onto a glutathione-agarose column and incubated for 2 h at 4°C, followed by washing with washing buffer, and then eluted with reduced glutathione. To purify His-Flag-DUSP1, the cell extracts were applied to a Ni-nitrilotriacetic acid (Ni-NTA)–agarose column, followed by washing and elution. The eluted proteins were applied to an ultrafiltration filter (100-kDa MWCO; Millipore) for desalting, cleaning, and concentration. To avoid interacting-protein contamination in DUSP1 ubiquitination experiments, cell lysates were suspended in NP-40 buffer containing 2% SDS and 5 mM dithiothreitol (DTT). The samples were heated at 95°C for 5 min, cooled at room temperature, and diluted in buffer containing 0.1% SDS, followed by IP or pulldown.
For the in vitro ubiquitination assay, E1 (catalog no. K-995) and E2 components (UbcH5b/UBE2D2 [catalog no. E2-622]; Ubc7/UBE2G2 [catalog no. E2-680]) were purchased from Boston Biochem. In vitro ubiquitination assays were performed according to the manufacturer’s protocol. Specifically, 0.1 μg of GST-GP78C, 0.1 μg of His-Flag-DUSP1, E1, and E2 were mixed, incubated at 37°C, and then terminated by adding 2× SDS-PAGE sample buffer. Ubiquitinated DUSP1 was analyzed by immunoblotting.
Immunoprecipitation and Western blot analysis.
The preparation of whole-cell lysates, IP, and Western blotting were described previously (43). The levels of protein were quantified using NIH ImageJ software.
MTT assays and FACS analysis.
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays were performed as described previously (44). For EGF and FBS stimulation experiments, cells were stimulated with 100 ng/ml EGF or FBS for 24 or 48 h. For cell cycle analysis, cells were stimulated with EGF or FBS for 24 h and then harvested to perform fluorescence-activated cell sorter (FACS) analysis. Cell cycle distributions were analyzed using Modfit LT 3.0 software (Verity Software House, Topsham, ME, USA).
Wound-healing and Transwell invasion assays.
Wound-healing and Transwell invasion assays were described previously (45).
AP-1 luciferase assays.
Luciferase assays were performed as described previously (45). Specifically, cells were plated at 5 × 105 per well in 6-well plates and then transfected with 100 ng of the reporter (AP-1-luc) and 10 ng of phRL-CMV (Renilla) using lipofectamine 2000. After incubation with EGF for 24 h, luciferase activity was measured with the Dual-Luciferase reporter assay system (Promega, Madison, WI) and normalized to that of Renilla luciferase. Each experiment was performed in triplicate. The fold induction was calculated in comparison with the control vector.
Statistical analysis.
All data were analyzed with Graphpad Prism or Microsoft Office Excel. Data are presented as means and standard deviations (SD). Quantification of Western blots was performed using ImageJ. Comparisons between groups were made using the chi-square test and Student's t test.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant R01 CA174949 to G.S.W.
D.H.K. designed and performed experiments, analyzed and interpreted data, and wrote the manuscript; M.H.U. performed FACS and cell growth analysis; M.C. performed interaction experiments; A.V. provided BCI215; A.R. provided the reagents and contributed to the manuscript; G.S.W. conceived the project, designed experiments, analyzed data, and wrote the manuscript.
We declare that we have no conflict of interest.
REFERENCES
- 1.Watanabe H, Nabi IR, Raz A. 1991. The relationship between motility factor receptor internalization and the lung colonization capacity of murine melanoma cells. Cancer Res 51:2699–2705. [PubMed] [Google Scholar]
- 2.Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. 2001. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A 98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairbank M, St-Pierre P, Nabi IR. 2009. The complex biology of autocrine motility factor/phosphoglucose isomerase (AMF/PGI) and its receptor, the gp78/AMFR E3 ubiquitin ligase. Mol Biosyst 5:793–801. doi: 10.1039/b820820b. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Du S, Fang S. 2012. gp78: a multifaceted ubiquitin ligase that integrates a unique protein degradation pathway from the endoplasmic reticulum. Curr Protein Pept Sci 13:414–424. doi: 10.2174/138920312802430590. [DOI] [PubMed] [Google Scholar]
- 5.Song BL, Sever N, DeBose-Boyd RA. 2005. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell 19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Lee JN, Song B, DeBose-Boyd RA, Ye J. 2006. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J Biol Chem 281:39308–39315. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- 7.Funasaka T, Raz A. 2007. The role of autocrine motility factor in tumor and tumor microenvironment. Cancer Metastasis Rev 26:725–735. doi: 10.1007/s10555-007-9086-7. [DOI] [PubMed] [Google Scholar]
- 8.Tsai YC, Mendoza A, Mariano JM, Zhou M, Kostova Z, Chen B, Veenstra T, Hewitt SM, Helman LJ, Khanna C, Weissman AM. 2007. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat Med 13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
- 9.Kho DH, Nangia-Makker P, Balan V, Hogan V, Tait L, Wang Y, Raz A. 2013. Autocrine motility factor promotes HER2 cleavage and signaling in breast cancer cells. Cancer Res 73:1411–1419. doi: 10.1158/0008-5472.CAN-12-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camps M, Nichols A, Arkinstall S. 2000. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J 14:6–16. doi: 10.1096/fasebj.14.1.6. [DOI] [PubMed] [Google Scholar]
- 11.Franklin CC, Kraft AS. 1997. Conditional expression of the mitogen-activated protein kinase (MAPK) phosphatase MKP-1 preferentially inhibits p38 MAPK and stress-activated protein kinase in U937 cells. J Biol Chem 272:16917–16923. doi: 10.1074/jbc.272.27.16917. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Gorospe M, Yang C, Holbrook NJ. 1995. Role of mitogen-activated protein kinase phosphatase during the cellular response to genotoxic stress. Inhibition of c-Jun N-terminal kinase activity and AP-1-dependent gene activation. J Biol Chem 270:8377–8380. doi: 10.1074/jbc.270.15.8377. [DOI] [PubMed] [Google Scholar]
- 13.Shapiro PS, Ahn NG. 1998. Feedback regulation of Raf-1 and mitogen-activated protein kinase (MAP) kinase kinases 1 and 2 by MAP kinase phosphatase-1 (MKP-1). J Biol Chem 273:1788–1793. doi: 10.1074/jbc.273.3.1788. [DOI] [PubMed] [Google Scholar]
- 14.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R Jr, Martindale JL, Yang X, Gorospe M. 2008. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol 28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau LF, Nathans D. 1985. Identification of a set of genes expressed during the G0/G1 transition of cultured mouse cells. EMBO J 4:3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haagenson KK, Wu GS. 2010. The role of MAP kinases and MAP kinase phosphatase-1 in resistance to breast cancer treatment. Cancer Metastasis Rev 29:143–149. doi: 10.1007/s10555-010-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wancket LM, Frazier WJ, Liu Y. 2012. Mitogen-activated protein kinase phosphatase (MKP)-1 in immunology, physiology, and disease. Life Sci 90:237–248. doi: 10.1016/j.lfs.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keyse SM. 2008. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev 27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Liu Y. 2007. Regulation of innate immune response by MAP kinase phosphatase-1. Cell Signal 19:1372–1382. doi: 10.1016/j.cellsig.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu GS. 2007. Role of mitogen-activated protein kinase phosphatases (MKPs) in cancer. Cancer Metastasis Rev 26:579–585. doi: 10.1007/s10555-007-9079-6. [DOI] [PubMed] [Google Scholar]
- 21.Sun H, Charles CH, Lau LF, Tonks NK. 1993. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell 75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 22.Wang HY, Cheng Z, Malbon CC. 2003. Overexpression of mitogen-activated protein kinase phosphatases MKP1, MKP2 in human breast cancer. Cancer Lett 191:229–237. doi: 10.1016/S0304-3835(02)00612-2. [DOI] [PubMed] [Google Scholar]
- 23.Rojo F, Gonzalez-Navarrete I, Bragado R, Dalmases A, Menendez S, Cortes-Sempere M, Suarez C, Oliva C, Servitja S, Rodriguez-Fanjul V, Sanchez-Perez I, Campas C, Corominas JM, Tusquets I, Bellosillo B, Serrano S, Perona R, Rovira A, Albanell J. 2009. Mitogen-activated protein kinase phosphatase-1 in human breast cancer independently predicts prognosis and is repressed by doxorubicin. Clin Cancer Res 15:3530–3539. doi: 10.1158/1078-0432.CCR-08-2070. [DOI] [PubMed] [Google Scholar]
- 24.Haagenson KK, Wu GS. 2010. Mitogen activated protein kinase phosphatases and cancer. Cancer Biol Ther 9:337–340. doi: 10.4161/cbt.9.5.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak SP, Hakes DJ, Martell KJ, Dixon JE. 1994. Isolation and characterization of a human dual specificity protein-tyrosine phosphatase gene. J Biol Chem 269:3596–3604. [PubMed] [Google Scholar]
- 26.Li M, Zhou JY, Ge Y, Matherly LH, Wu GS. 2003. The phosphatase MKP1 is a transcriptional target of p53 involved in cell cycle regulation. J Biol Chem 278:41059–41068. doi: 10.1074/jbc.M307149200. [DOI] [PubMed] [Google Scholar]
- 27.Brondello JM, Pouyssegur J, McKenzie FR. 1999. Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science 286:2514–2517. doi: 10.1126/science.286.5449.2514. [DOI] [PubMed] [Google Scholar]
- 28.Lin YW, Chuang SM, Yang JL. 2003. ERK1/2 achieves sustained activation by stimulating MAPK phosphatase-1 degradation via the ubiquitin-proteasome pathway. J Biol Chem 278:21534–21541. doi: 10.1074/jbc.M301854200. [DOI] [PubMed] [Google Scholar]
- 29.Dickinson RJ, Keyse SM. 2006. Diverse physiological functions for dual-specificity MAP kinase phosphatases. J Cell Sci 119:4607–4615. doi: 10.1242/jcs.03266. [DOI] [PubMed] [Google Scholar]
- 30.Zhang T, Kho DH, Wang Y, Harazono Y, Nakajima K, Xie Y, Raz A. 2015. gp78, an E3 ubiquitin ligase acts as a gatekeeper suppressing nonalcoholic steatohepatitis (NASH) and liver cancer. PLoS One 10:e0118448. doi: 10.1371/journal.pone.0118448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryser S, Massiha A, Piuz I, Schlegel W. 2004. Stimulated initiation of mitogen-activated protein kinase phosphatase-1 (MKP-1) gene transcription involves the synergistic action of multiple cis-acting elements in the proximal promoter. Biochem J 378:473–484. doi: 10.1042/bj20031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Lau LF, Sturgill TW. 1994. Rapid deactivation of MAP kinase in PC12 cells occurs independently of induction of phosphatase MKP-1. FEBS Lett 353:9–12. doi: 10.1016/0014-5793(94)01000-5. [DOI] [PubMed] [Google Scholar]
- 33.Kaltenmeier CT, Vollmer LL, Vernetti LA, Caprio L, Davis K, Korotchenko VN, Day BW, Tsang M, Hulkower KI, Lotze MT, Vogt A. 2017. A tumor cell-selective inhibitor of mitogen-activated protein kinase phosphatases sensitizes breast cancer cells to lymphokine-activated killer cell activity. J Pharmacol Exp Ther 361:39–50. doi: 10.1124/jpet.116.239756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiu CG, St-Pierre P, Nabi IR, Wiseman SM. 2008. Autocrine motility factor receptor: a clinical review. Expert Rev Anticancer Ther 8:207–217. doi: 10.1586/14737140.8.2.207. [DOI] [PubMed] [Google Scholar]
- 35.Avraham R, Yarden Y. 2011. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol 12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 36.Stanoev A, Mhamane A, Schuermann KC, Grecco HE, Stallaert W, Baumdick M, Brüggemann Y, Joshi MS, Roda-Navarro P, Fengler S, Stockert R, Roßmannek L, Luig J, Koseska A, Bastiaens PIH. 2018. Interdependence between EGFR and phosphatases spatially established by vesicular dynamics generates a growth factor sensing and responding network. Cell Syst 7:295–309. doi: 10.1016/j.cels.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monast CS, Furcht CM, Lazzara MJ. 2012. Computational analysis of the regulation of EGFR by protein tyrosine phosphatases. Biophys J 102:2012–2021. doi: 10.1016/j.bpj.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell 4:1029–1040. doi: 10.1016/S1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 39.Sun L, Chen ZJ. 2004. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol 16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Kidger AM, Rushworth LK, Stellzig J, Davidson J, Bryant CJ, Bayley C, Caddye E, Rogers T, Keyse SM, Caunt CJ. 2017. Dual-specificity phosphatase 5 controls the localized inhibition, propagation, and transforming potential of ERK signaling. Proc Natl Acad Sci U S A 114:E317–E326. doi: 10.1073/pnas.1614684114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou JY, Liu Y, Wu GS. 2006. The role of mitogen-activated protein kinase phosphatase-1 in oxidative damage-induced cell death. Cancer Res 66:4888–4894. doi: 10.1158/0008-5472.CAN-05-4229. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Ha SW, Zhang T, Kho DH, Raz A, Xie Y. 2014. Polyubiquitylation of AMF requires cooperation between the gp78 and TRIM25 ubiquitin ligases. Oncotarget 5:2044–2051. doi: 10.18632/oncotarget.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kho DH, Zhang T, Balan V, Wang Y, Ha SW, Xie Y, Raz A. 2014. Autocrine motility factor modulates EGF-mediated invasion signaling. Cancer Res 74:2229–2237. doi: 10.1158/0008-5472.CAN-13-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Zhou JY, Kho D, Reiners JJ Jr, Wu GS. 2016. Role for DUSP1 (dual-specificity protein phosphatase 1) in the regulation of autophagy. Autophagy 12:1791–1803. doi: 10.1080/15548627.2016.1203483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kho DH, Bae JA, Lee JH, Cho HJ, Cho SH, Lee JH, Seo YW, Ahn KY, Chung IJ, Kim KK. 2009. KITENIN recruits Dishevelled/PKC delta to form a functional complex and controls the migration and invasiveness of colorectal cancer cells. Gut 58:509–519. doi: 10.1136/gut.2008.150938. [DOI] [PubMed] [Google Scholar]