Abstract
Long noncoding RNAs (lncRNAs) have been reported to be involved in various human diseases, and increasing studies have revealed that lncRNAs can play a vital role in preeclampsia (PE). In our study, lncRNA hypoxia-inducible factor 1 alpha (HIF1A) antisense RNA 2 (HIF1A-AS2) was found to be significantly downregulated in placenta tissues of PE patients by quantitative real-time PCR analysis. Moreover, Cell Counting Kit-8 (CCK-8) assays showed that downregulation of HIF1A-AS2 can impede cell proliferation of HTR-8/SVneo and JAR trophoblasts cells. Ectopic overexpression of HIF1A-AS2 can increase the function of trophoblasts cell migration and invasion in vitro. RNA-sequencing (RNA-seq), RNA immunoprecipitation (RIP), and chromatin immunoprecipitation (ChIP) experiments showed that HIF1A-AS2 can recruit lysine-specific demethylase 1 (LSD1) and epigenetically repress pleckstrin homology-like domain, family A, member 1 (PHLDA1) transcription in human trophoblasts cells. In summary, our findings suggest that downregulated HIF1A-AS2 may play a role in the pathogenesis and progression of PE, and has potential as a novel prognostic marker in PE.
Keywords: preeclampsia, HIF1A-AS2, LSD1, proliferation, invasion, PHLDA1
Introduction
Preeclampsia (PE) is a significant cause of maternal and perinatal morbidity and mortality worldwide, accounting for approximately one in five maternal deaths and 15%–20% of all premature deliveries.1 According to one estimate, women who had PE in their pregnancies have a significantly increased rate of cardiovascular disease in the future.2, 3 These women who were affected by PE had higher risk for heart failure, stroke, and death as a result of cardiovascular disease compared with women who had uncomplicated pregnancies.3 Furthermore, the rate of long-term cardiovascular morbidity is associated with the severity and the gestational age of onset of PE. Although the exact mechanism of placental-related disorders is yet to be understood, it is believed that inadequate trophoblastic invasion plays a role in the pathogenesis of PE and other disorders. Impaired development of the placenta translates into persistently elevated resistance to blood flow in the uteroplacental circulation.4, 5
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs that are at least 200 nt long.6 Given the biochemical versatility of RNA, lncRNAs play a functional role in various biological processes,7, 8 including post-transcriptional regulation, organization of protein complexes, cell-cell signaling and allosteric regulation of proteins.9, 10 lncRNA hypoxia-inducible factor 1 alpha-antisense RNA 2 (HIF1A-AS2) is a 2,051-bp lncRNA that is located in the chromosome 14q23.2. Recent studies have shown that HIF1A-AS2 may have been involved in the progression of a variety of tumors, including gastric carcinomas,11 bladder cancer,12 colorectal cancer13 and glioblastoma multiforme.14 However, a few studies reported that HIF1A-AS2 may be associated with the development of PE and that its dysregulation may participate in disease progression. The biological functions of HIF1A-AS2 in the control of PE pathogenesis have not been well illustrated. Furthermore, the molecular interactions of HIF1A-AS2 also remain poorly understood. These unanswered questions compelled us to investigate the role of HIF1A-AS2 in the development of PE.
In this study, we explored the potential molecular mechanisms underlying the relationship between HIF1A-AS2 and PE progression. We found that HIF1A-AS2 was significantly downregulated in PE tissues compared with normal pregnant placenta tissues and may be an independent predictor for the development of PE. In addition, HIF1A-AS2 could regulate cell proliferation, invasion, and migration. We demonstrated that HIF1A-AS2 was associated with lysine-specific demethylase 1 (LSD1), and that this association was required for the epigenetic repression of pleckstrin homology-like domain, family A, member 1 (PHLDA1), which plays a significant role in the activation-induced apoptosis.15 In summary, we may offer new insights into the critical role of the lncRNA HIF1A-AS2 in modulating human PE.
Results
HIF1A-AS2 Is Downregulated in Placental Tissue in Patients with PE
We analyzed the clinical characteristics of 104 participants (Table 1). The results indicated that systolic and diastolic blood pressure and proteinuria were higher in PE patients compared with patients with normal pregnancies. The body weight of neonates from PE pregnancies was lower because of the higher rate of early termination in PE pregnancies, which were included in the analysis.
Table 1.
Clinical Characteristics of Patients with PE and Normal Pregnancies
| Variable | PE (N = 52) | Control (N = 52) | p Value (PE versus Control) |
|---|---|---|---|
| Maternal age (year) | 32.154 ± 5.622 | 33.75 ± 3.792 | >0.05 |
| Maternal weight (kg) | 74.760 ± 10.812 | 72.135 ± 8.967 | >0.05 |
| Smoking | 0 | 0 | >0.05 |
| Systolic blood pressure (mm Hg) | 158.712 ± 24.498 | 115.808 ± 7.758 | <0.01 |
| Diastolic blood pressure (mm Hg) | 104.250 ± 11.216 | 74.115 ± 8.375 | <0.01 |
| Proteinuria (g/day) | >0.3 | <0.3 | <0.05 |
| Body weight of infant (g) | 2,366.346 ± 865.652 | 3,411.538 ± 365.100 | <0.05 |
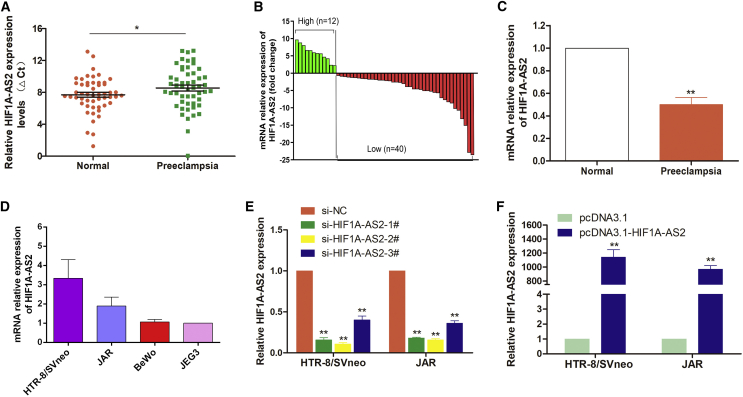
Next, we analyzed the expression levels of HIF1A-AS2 in 52 pairs of PE samples and adjacent normal samples with normalization to GAPDH by quantitative real-time PCR assay. The expression levels of HIF1A-AS2 in PE were significantly lower (p < 0.05) in 76.9% (40/52) of the samples compared with the corresponding normal counterparts (Figure 1A). Furthermore, as shown in Figure 1B, 52 PE patients were divided into two groups according to the median value of their HIF1A-AS2 expression levels: a high-HIF1A-AS2 group (above the median, n = 12) and a low-HIF1A-AS2 group (below the median, n = 40). Also, we found that HIF1A-AS2 was lower in PE with about 2.2-fold downregulation as shown in Figure 1C.
Figure 1.
HIF1A-AS2 Expression Is Decreased in PE
(A–C) The expression of HIF1A-AS2 was analyzed by quantitative real-time PCR and normalized to that of GAPDH. (A) Relative expression levels of HIF1A-AS2 in PE tissues (n = 52) compared with corresponding normal tissues (n = 52). The ΔCt value was determined by subtracting the GAPDH Ct value from the HIF1A-AS2 Ct value (relative to a single reference value). Higher ΔCt values indicate smaller expression. (B) HIF1A-AS2 levels were lower in PE placentas (n = 52) compared with normal placentas (n = 52). (C) Results are presented as the 2.2-fold change in PE placental samples relative to the control, and HIF1A-AS2 expression was classified into two groups. (D) HIF1A-AS2 expression in trophoblast cell lines analyzed by quantitative real-time PCR. The levels of HIF1A-AS2 in HTR-8/SVneo, BeWo, and JAR cell lines were normalized to that in JEG3. (E and F) Relative HIF1A-AS2 expression in HTR-8/SVneo and JAR cells transfected with HIF1A-AS2 siRNAs (E) and pcDNA3.1-HIF1A-AS2 (F). The data are presented as the mean ± SD of three independent experiments; *p < 0.05, **p < 0.01.
Then, we analyzed HIF1A-AS2 expression in four trophoblast cell lines (HTR-8/SVneo, BeWo, JEG3, and JAR) (Figure 1D). We chose cell lines with relatively high HIF1A-AS2 expression (HTR-8/SVneo and JAR) to explore its functional activity.
To assess HIF1A-AS2 effects in trophoblasts, we knocked down or overexpressed HIF1A-AS2 in HTR-8/SVneo and JAR cells by transfecting with siRNA or with overexpression plasmid (pcDNA3.1-HIF1A-AS2), respectively. As shown in Figure 1E, HIF1A-AS2 expression was silenced in HTR/SVneo and JAR cells more efficiently by si-HIF1A-AS2-1# and si-HIF1A-AS2-2# by analyzing quantitative real-time PCR data.
So, we primarily utilized si-HIF1A-AS2-1# and si-HIF1A-AS2-2# to investigate potential mechanisms in subsequent experiments. The upregulation of HIF1A-AS2 expression by transfecting pcDNA3.1-HIF1A-AS2 plasmid was confirmed in two cell lines (Figure 1F).
HIF1A-AS2 Can Promote Proliferation of Trophoblasts and Affect Cell Cycle and Apoptosis in Trophoblasts
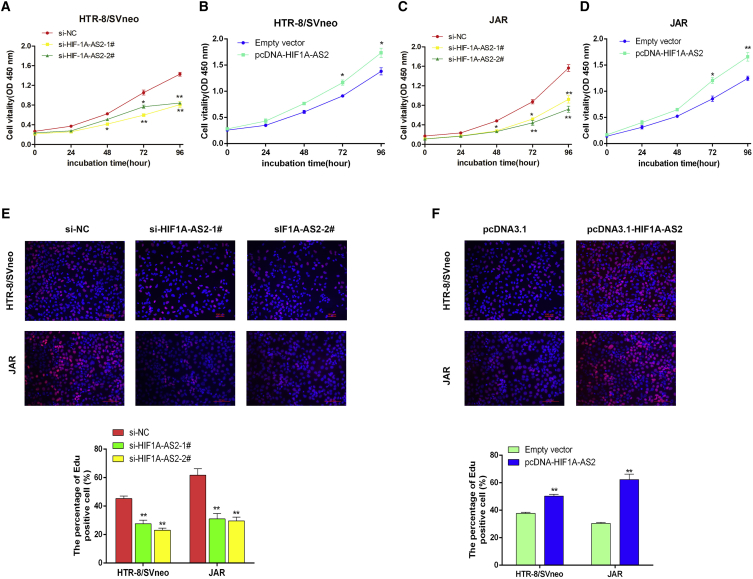
Knockdown of HIF1A-AS2 expression by transfecting with si-HIF1A-AS2 significantly suppressed proliferation activity in HTR-8/SVneo and JAR cells as evidenced by the Cell Counting Kit-8 (CCK-8) assay (Figures 2A and 2C). The effect described above was reversed by HIF1A-AS2 overexpression, which significantly promoted cell proliferation (Figures 2B and 2D). Further, the results of ethynyl deoxyuridine (EdU) immunostaining confirmed the above findings (Figures 2E and 2F). Collectively, these results indicate that HIF1A-AS2 positively regulates the proliferation of HTR/SVneo and JAR trophoblast cells. The results suggest that knockdown of HIF1A-AS2 can play a vital role in the development of PE.
Figure 2.
HIF1A-AS2 Promotes Trophoblast Proliferation In Vitro
(A–D) The viability of si-HIF1A-AS2-transfected HTR-8/SVneo (A) and JAR (C) cells or pCDNA3.1-HIF1A-AS2-transfected HTR-8/SVneo (B) and JAR (D) cells was analyzed by the CCK-8 assay. (E and F) Proliferating HTR-8/SVneo and JAR cells with si-HIF1A-AS2-transfected (E) and pcDNA3.1-HIF1A-AS2-transfected (F) cells are marked with EdU (red) and cell nuclei are stained with DAPI (blue). The data are presented as the mean ± SD of three independent experiments; *p < 0.05, **p < 0.01.
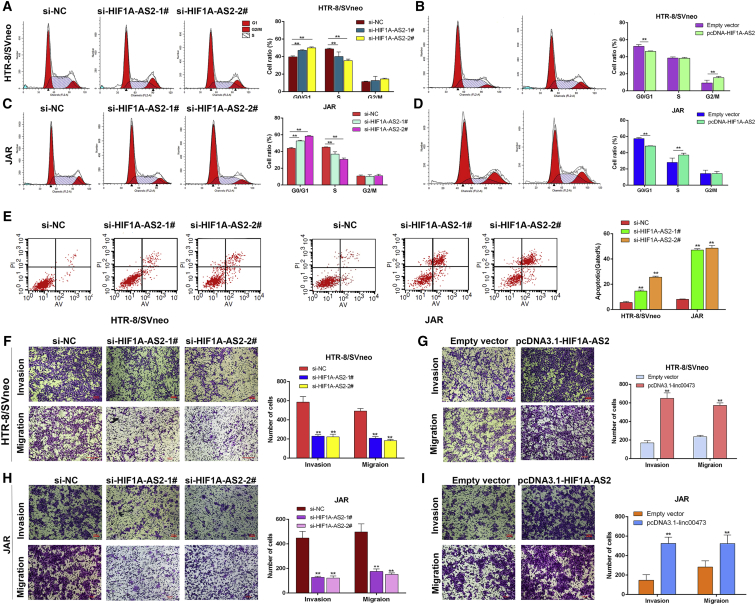
Furthermore, we wanted to explore whether HIF1A-AS2 impacts the proliferation of HTR-8/SVneo and JAR cells through cell-cycle regulation. To investigate this, we tested cell-cycle progression using flow cytometric analysis. The knockdown of HIF1A-AS2 resulted in cell-cycle arrest at the G0/G1 phase (Figures 3A and 3C), whereas HIF1A-AS2 overexpression reversed the effect (Figures 3B and 3D). Consistent with these results, the apoptosis rate in HTR/SVneo and JAR trophoblasts increased after HIF1A-AS2 knockdown by siRNAs (Figure 3E). The phenomena revealed that HIF1A-AS2 was involved in the regulation of cell-cycle progression and apoptosis in trophoblasts.
Figure 3.
Effects of HIF1A-AS2 on Trophoblast Cell Cycle, Apoptosis, Migration, and Invasion In Vitro
HTR-8/SVneo and JAR trophoblasts were transfected with HIF1A-AS2-specific siRNAs (si-HIF1A-AS2-1# and si-HIF1A-AS2-2#) or were transfected with pcDNA3.1-HIF1A-AS2. (A–D) Cell cycle was analyzed by flow cytometry in HTR-8/SVneo (A and B) and JAR (C and D) cells. Representative FACS images and related statistics are shown. (E) Cell apoptosis rates were analyzed by flow cytometry. The data are presented as the mean ± SD of three independent experiments; *p < 0.05, **p < 0.01. (F–I) The migration and invasion capacity of cultured trophoblasts transfected with si-HIF1A-AS2 in HTR-8/SVneo cells (F) and JAR (H) or pcDNA3.1-HIF1A-AS2 in HTR-8/SVneo (G) and JAR (I) cells and analyzed by the Transwell assays was significantly lower or higher, respectively, than that of control cells. The data are presented as the mean ± SD of three independent experiments; **p < 0.01. LR, early apoptotic cells; UR, terminal apoptotic cells.
HIF1A-AS2 Influences the Function of Migration and Invasion Ability of Trophoblasts In Vitro
Next, we evaluated the effects of HIF1A-AS2 on the migration and invasion abilities of HTR-8/SVneo and JAR cells using the Transwell assays. Proper trophoblast migration and invasion are important for the establishment of the blood flow between the mother and embryo, and the development of this placental vasculature is impaired in PE patients. Accordingly, knockdown of HIF1A-AS2 inhibited trophoblast migration and invasion (Figures 3F and 3H), whereas HIF1A-AS2 overexpression exhibited the opposite effect (Figures 3G and 3I). These results suggest that HIF1A-AS2 may influence trophoblast migration and invasion.
HIF1A-AS2 Influences Gene Expression in Trophoblasts
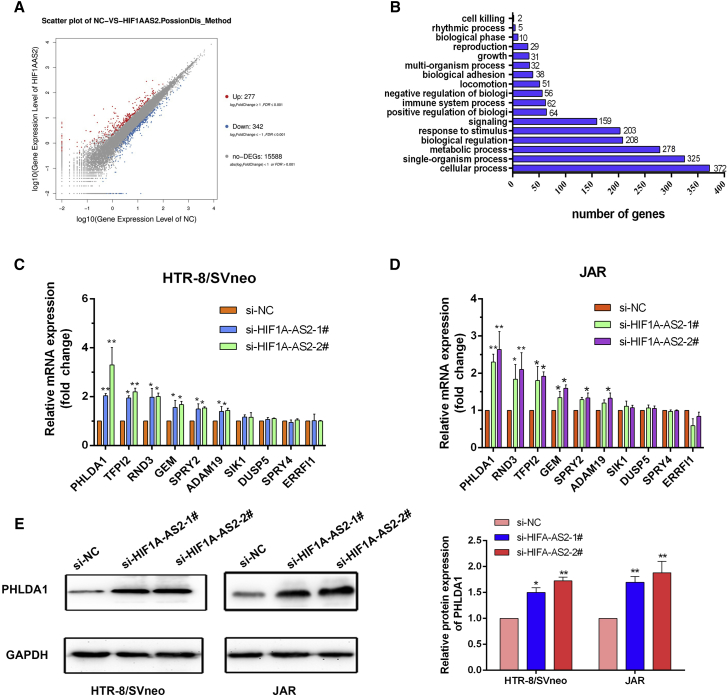
We performed RNA transcriptome sequencing to explore HIF1A-AS2-associated changes in gene expression in negative control siRNA (si-NC) and si-HIF1A-AS2-transfected HTR/SVneo cells. A total of 277 and 342 transcripts were increased or decreased, respectively (Figure 4A; Table S2). Evaluation of the biological pathways activated by HIF1A-AS2 using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases indicated that genes related to cell growth were changed in HIF1A-AS2-knockdown cells (Figure 4B). Expression changes in HTR-8/SVneo and JAR cells were further confirmed by quantitative real-time PCR. Transcripts related to cell growth, apoptosis and migration of the expression of PHLDA1 were significantly upregulated after HIF1A-AS2 knockdown in HTR-8/SVneo and JAR cells (Figures 4C and 4D). Also, western blot analysis discovered the similar effects of HIF1A-AS2 on PHLDA1 protein expression in HTR-8/SVneo and JAR cells (Figure 4E). PHLDA1 encodes an evolutionarily conserved proline-histidine-rich nuclear protein.
Figure 4.
HIF1A-AS2 Knockdown Affected the Expression of Genes Involved in Cell Proliferation and Migration
(A and B) Gene expression profiling was performed by RNA-seq in HTR-8/SVneo cells transfected with si-HIF1A-AS2. Differentially expressed genes are shown (A). GO analysis of genes differentially expressed in si-NC- and si-HIF1A-AS2-transfected cells. Cell growth was among significant biological processes affected by HIF1A-AS2 depletion in trophoblasts as evidenced by the number of genes with altered expression (B). (C and D) Relative mRNA expression of genes involved in cell proliferation and migration in HIF1A-AS2-deficient HTR-8/SVneo cells (C) and JAR cells (D) was analyzed by quantitative real-time PCR. (E) PHLDA1 protein expression in HIF1A-AS2-deficient HTR-8/SVneo cells and JAR cells was assessed by western blotting. The data are presented as the mean ± SD of three independent experiments; *p < 0.05, **p < 0.01.
Although PHLDA1 has been shown to be both pro-apoptotic and antiapoptotic depending on the cell line and the experimental conditions, PHLDA1 is essential for rescuing cells from serum starvation-induced apoptosis in NIH 3T3 cells expressing insulin-like growth factor 1 (IGF1) receptors.16 Conversely, in T cells, neuronal, endothelial, melanoma, cervical carcinoma, and other cell lines, PHLDA1 plays a role in reducing proliferation and inducing cell death.17 Also, reduced PHLDA1 expression has been described in some cancers, and current data indicate that PHLDA1 may act as an apoptotic gene.
HIF1A-AS2 Silences PHLDA1 Epigenetically by Binding to LSD1
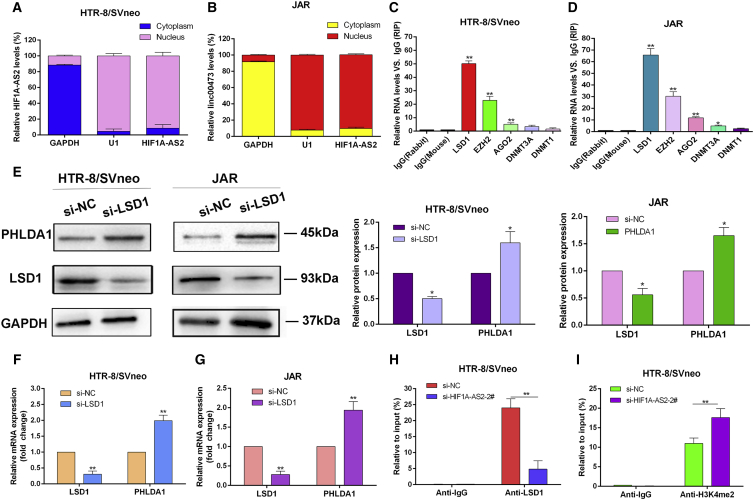
To further examine the effects of HIF1A-AS2 on gene expression, we investigated the subcellular localization of HIF1A-AS2 in HTR/SVneo and JAR cells using GAPDH and small nuclear RNA U1 (RNU1) as cytoplasmic and nuclear markers, respectively, subcellular fractionation, and RNA isolation assays (Figures 5A and 5B). The results indicate that HIF1A-AS2 was mainly localized in the cell nucleus, supporting its role in transcriptional regulation.
Figure 5.
HIF1A-AS2 Binding to LSD1 Inhibits PHLDA1 Expression
(A and B) Cell fractionation assay revealed that HIF1A-AS2 was predominantly localized in the nucleus of HTR-8/SVneo (A) and JAR (B) cells. GAPDH and small nuclear RNA U1 were used as cytoplasmic and nuclear markers, respectively. (C and D) RIP assays showed that HIF1A-AS2 was mainly bound to LSD1 in HTR-8/SVneo cells (C) and JAR (D) cells, respectively. (E–G) LSD1 silencing changed the expression of LSD1 and PHLDA1 at the protein (E) and mRNA (F and G) levels in HTR-8/SVneo and JAR cells. (H and I) Enrichment of LSD1 (H) and H3K4me2 (I) in the promoter region of PHLDA1 detected by the ChIP assay. The data are presented as the mean ± SD of three independent experiments; *p < 0.05, **p < 0.01.
Recently, it was reported that by regulating target gene expression, LSD1 can influence tumorigenesis, embryonic differentiation and the formation of euchromatin.18, 19 Therefore, we hypothesized that HIF1A-AS2 may control PHLDA1 expression by recruiting LSD1 in trophoblasts. Indeed, the RNA immunoprecipitation (RIP) assay showed that HIF1A-AS2 was precipitated with anti-LSD1 antibodies in HTR-8/SVneo and JAR cells (Figures 5C and 5D).
We then investigated the potential functional relationship between HIF1A-AS2 and LSD1. The knockdown of LSD1 by a specific siRNA resulted in PHLDA1 upregulation both at the protein and mRNA levels (Figures 5E–5G), suggesting that HIF1A-AS2 may regulate PHLDA1 expression in trophoblasts through epigenetic mechanisms. The chromatin immunoprecipitation (ChIP) assay with antibodies against LSD1 and H3K4me2 revealed the enrichment of LSD1 and H3K4me2 in the promoter region of the PHLDA1 gene, which was significantly decreased for LSD1 but increased for H3K4me2 after transfection with si-HIF1A-AS2 (Figures 5H and 5I).
PHLDA1 Upregulated in PE Placental Tissues Influences Trophoblast Viability In Vitro
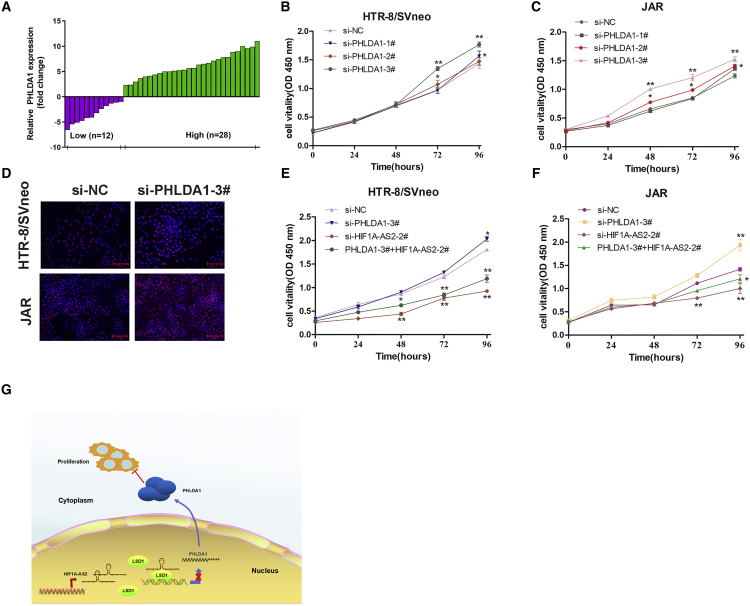
Evaluation of PHLDA1 mRNA expression in 40-pair low-HIF1A-AS2 group placental tissues of PE patients indicated that PHLDA1 transcription was increased in PE compared with normal pregnancy (Figure 6A). In HTR-8/SVneo and JAR trophoblasts, PHLDA1 knockdown promoted cell proliferation by CCK-8 (Figures 6B and 6C) and Edu assays (Figure 6D). Finally, we examined the biological effects of PHLDA1 association with HIF1A-AS2 by co-transfecting HTR-8/SVneo and JAR cells with si-HIF1A-AS2 and PHLDA1 siRNAs, and analyzing cell proliferation using CCK-8 assays (Figures 6E and 6F). The results indicated that si-PHLDA1 could moderately rescue si-HIF1A-AS2-mediated inhibition of proliferation in HTR-8/SVneo and JAR cells.
Figure 6.
PHLDA1 Inhibits Trophoblast Proliferation and Counterbalances the Activity of HIF1A-AS2
(A) Relative expression of PHLDA1 (fold-change) in placental tissues of PE patients compared with those of healthy pregnant women (n = 32). (B and C) CCK-8 assay analysis of cell viability for si-PHLDA1 transfected in HTR-B/SVneo (B) and JAR (C) trophoblasts. (D) Proliferation of HTR-8/SVneo and JAR cells analyzed by the EdU assays. (E and F) Cell proliferation ability was analyzed using CCK-8 assays by co-transfecting HTR-8/SVneo (E) and JAR (F) cells with si-HIF1A-AS2-2# and si-PHLDA1-3#. The data are presented as the mean ± SD of three independent experiments; *p < 0.05, **p < 0.01. (G) A potential schematic pathway illustrating the role of HIF1A-AS2 in the proliferation and migration of trophoblasts in PE.
Discussion
LncRNAs are noncoding RNAs expressed at lower levels compared with protein-coding transcripts and can be either nuclear, nucleolar, or cytoplasmic. LncRNAs in the nucleus have functions in histone modification and block the binding of transcription factors to their promoters or direct transcriptional regulation.10, 20 More than 200 diseases are related to dysregulated or dysfunctional lncRNAs. They can act as structural scaffolds of nuclear domains and influence chromatin organization, as well as transcriptional and post-transcriptional regulation of gene expression.21 Thus, exploration and identification of PE-associated lncRNAs and their functions may provide a new angle in the understanding of PE pathogenesis and define novel therapeutic targets, as well as provide diagnostic and prognostic markers for PE.
The extra-villous trophoblasts (EVTs) play a role in migrating through the endometrium, invade the uterine decidua, meeting only little resistance, and remodel the spiral arteries to establish appropriate nutrient and oxygen supplies for the fetus during the success of pregnancy.22 EVTs are functional cells and may be crucial in the placenta, and the abnormal proliferation, apoptosis, and invasion of EVTs are pivotal contributors to the failure of placentation. EVTs also display a phenotype very similar to that of cancer cells, regarding their capacity for proliferation, apoptosis, migration, and invasion.23 PE is a multi-factorial disease that is caused by the interaction of both genetic and environmental factors.24, 25 However, some of the specific mechanisms underlying the pathogenesis of the disease are unclear. The abnormal invasion of trophoblast cells in the placenta disc and the disturbance of recasting the spiral artery are the important reasons behind the insufficiency of the blood circulation in the placenta typically seen in PE.26 These aspects can partly explain the pathogenesis of PE and help us to further explore it.
In our study, we explored the potential molecular mechanism of HIF1A-AS2 mainly using several different trophoblastic cell lines as a model system. HIF1A-AS2 has been mainly investigated for its role in cancer. In this study, we found that expression of HIF1A-AS2 was downregulated in the placental tissues of women with PE compared with women with normal pregnancies. In vitro, silencing of HIF1A-AS2 expression can effectively suppress the proliferation, cell-cycle progression, invasion, and migration of trophoblast cells, whereas HIF1A-AS2 overexpression caused the opposite results. These findings implicate HIF1A-AS2 as an important regulatory molecule involved in the control of the biological activity of the main players’ PE pathogenesis and suggest that HIF1A-AS2 may be a promising biomarker for the diagnosis of PE.
PHLDA1 was first identified as a potential transcription factor required for Fas expression and activation-induced apoptosis in mouse T cell hybridomas.27 The exact molecular and biological functions of PHLDA1 remain to be elucidated. However, its expression is induced by a variety of external stimuli, and there is evidence that it may function as a transcriptional activator that acts as a mediator of apoptosis,28 proliferation, differentiation,29 and cell migration30 dependent on the cellular type and context. Recently, PHLDA1 has received attention because of its association with cancer.31, 32, 33 Consistent with these findings, PHLDA1 knockdown promoted proliferation of cultured trophoblasts and counterbalanced the inhibitory effects of HIF1A-AS2 deficiency. Our results also indicate that PHLDA1 expression was silenced by LSD1 through epigenetic mechanisms. Based on these findings, we propose that HIF1A-AS2 can inhibit PHLDA1 expression by binding to LSD1 in trophoblasts, thus promoting their invasion and migration, the critical processes for proper uterine spiral artery remodeling in pregnancy, which are deregulated in PE.
In conclusion, this study discovered that the role of HIF1A-AS2 may be as a scaffold and an important indicator of the LSD1-mediated epigenetic regulatory pathway participating in the inhibition of PHLDA1 expression during the development of PE. The present findings show that HIF1A-AS2 can be a novel molecular target for earlier diagnosis and treatment of PE (Figure 6G). Further studies are needed to elucidate other potential mechanisms through which HIF1A-AS2 participates in the biological functions of trophoblasts in the context of PE.
Materials and Methods
Patients and Collection of Tissue Samples
We obtained 52 paired placental samples from women with normal pregnancies and PE who underwent cesarean deliveries in Jiangsu Province Hospital from August 2016 to December 2018. The placenta tissue samples (about 1 cm × 1 cm × 1 cm in size) were taken from the central area of the placenta maternal surface to avoid necrosis and calcification, and were immediately frozen in liquid nitrogen and subsequently used for RNA and protein extraction. Clinicopathological characteristics of the participants are summarized in Table 1. This research was authorized by the Ethnics Board of the First Affiliated Hospital of Nanjing Medical University, China, and all patients provided written informed consent.
Cell Culture
Four human trophoblast cell lines (HTR/SVneo, JAR, JEG3, and BeVo) and human umbilical vein endothelial cells (HUVECs) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). HTR/SVneo and JAR cells were cultured in RPMI 1640, JEG3 cells in MEM, BeVo cells in F12K supplemented with 10% fetal bovine serum (FBS; GIBCO BRL, Invitrogen, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen). All cells were maintained in a humidified atmosphere with 5% CO2 at 37°C.
Cell Transfection
Plasmid vectors (pcDNA, pcDNA3.1-HIF1A-AS2, and pcDNA3.1-PHLDA1) were purified using the DNA Midiprep kit (QIAGEN, Hilden, Germany). Three different HIF1A-AS2-specific siRNAs, LSD1 and PHLDA1 siRNAs and scrambled si-NC, were purchased from Invitrogen; the sequences are shown in Table S1. HTR/SVneo, JAR, and JEG3 cells were cultured in six-well plates until 80% confluence and then transfected with siRNAs using Lipofectamine 2000 (Invitrogen, USA) or with plasmid vectors (4 μg) using X-tremeGENE HP DNA Transfection Reagent (Roche, Pennsburg, Germany) following the manufacturers’ instructions. Cells were harvested 48 h post-transfection and analyzed by real-time qPCR and western blotting.
RNA Preparation and Quantitative Real-Time PCR
Total RNA was isolated from tissues or cultured cells with TRIzol reagent (Invitrogen), and its quality and quantity were assessed using NanoDrop2000c (Thermo Fisher Scientific, Waltham, MA, USA). RNA was reverse-transcribed to cDNA using Primer Script RT Master Mix (Takara, Dalian, China), and quantitative real-time PCR was performed with SYBR Premix Ex Taq (Takara, Dalian, China) and specific primers (Table S1) in an ABI 7500 system following the manufacturer’s protocol. The expression level of HIF1A-AS2 was calculated by the 2−ΔΔCT method, normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and converted to fold changes.
Subcellular Fractionation and RNA Isolation
HTR-8/SVneo and JAR cells were washed with PBS twice. Then, cytoplasmic and nuclear RNA were separated and purified using the PARIS Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Cell Proliferation Assays
Cell proliferation of HTR/SVneo or JAR cells were seeded in 96-well plates (3,000–4,000 cells/well) and transfected with si-HIF1A-AS2 or pcDNA-HIF1A-AS2; five replicates for each point were used. Cell viability was assessed every 24 h using the CCK-8 (Roche) based on the absorbance at 450 nm measured in an ELx-800 University Microplate Reader (Biotech, Winooski, VT, USA). At least three independent experiments were performed.
EdU Assay
The EdU assay was performed for accurate estimation of cell proliferation as a complementary method using the 5-ethynyl-2-deoxyuridine labeling/detection kit (Ribobio, Guangzhou, China). HTR/SVneo and JAR cells were seeded in 24-well plates at ∼2–3 × 104 cells per well and transfected with si-HIF1A-AS2-1#, si-HIF1A-AS2-2#, si-NC, pcDNA-HIF1A-AS2, or pcDNA for ∼24–48 h. Then, 200 μL/well of EdU labeling medium was added, and cells were cultured for another 2 h, fixed in 4% paraformaldehyde (pH 7.4) for 30 min, permeabilized with 0.5% Triton X-100 for 20 min at room temperature, washed in PBS (pH 7.4), and stained with anti-EdU solution for 30 min. After counterstaining with 250 μL DAPI (Invitrogen, Molecular Probes, Eugene, OR, USA) for 25 min, images of EdU-positive cells were captured under a fluorescence microscope (Nikon Corporation, Tokyo, Japan). The experiment was independently repeated three times.
Flow Cytometry Analysis of Cell Cycle and Apoptosis
Cells transfected with si-HIF1A-AS2 or pcDNA-HIF1A-AS2 were harvested about ∼32–48 h post-transfection by trypsinization, washed twice with PBS, stained with propidium iodide (PI) using the Cycle TEST PLUS DNA Reagent Kit (BD Biosciences, San Jose, CA, USA), and analyzed in a FACScan flow cytometer (BD Biosciences) equipped with Cell Quest software (BD Biosciences) to estimate the percentage of cells in G0/G1, S, and G2/M phases. All samples were assayed in triplicate. Apoptosis was assessed by flow cytometry after double staining of the transfected cells with fluorescein isothiocyanate (FITC)-Annexin V and PI using the FITC-Annexin V Apoptosis Detection Kit (BD Biosciences), and the percentages of viable, dead, and early and late apoptotic cells were calculated and compared with those in controls.
Cell Migration and Invasion Assays
Cell migration and invasion assays were performed using 24-well Transwell chambers with 8-μm pore size polycarbonate membranes (Corning, Corning, NY, USA). Cells were plated on the top side of the membrane pre-coated or not with Matrigel (BD, Franklin Lakes, NJ, USA) for invasion or migration assays, respectively. After incubation for ∼36–48 h, cells inside the upper chamber were removed with cottons swabs, whereas those on the lower membrane surface were fixed by methanol and stained with 0.5% crystal violet solution. Five randomly selected fields were analyzed for each well.
Western Blotting Assay
HTR/SVneo and JAR cells cultured in six-well plates were transfected with si-HIF1A-AS2 or pcDNA-HIF1A-AS2. Following transfection, total cellular proteins were extracted with RIPA protein extraction reagent (Beyotime, Beijing, China) supplemented with protease inhibitor cocktail (Roche, Pleasanton, CA, USA) and phenylmethylsulfonyl fluoride (Roche). Proteins were separated by SDS-PAGE in 10% gels and transferred to 0.22-μm polyvinylidene difluoride membranes (Sigma), which were incubated with antibodies against LSD1 (1:1,000; CST, Danvers, MA, USA) or PHLDA1 (1:1,000; Proteintech); anti-GAPDH antibody (1:3,000; Santa Cruz, CA, USA) was used as control. The intensity of protein bands was quantified by densitometry using the Quantity One software (Bio-Rad). At least three independent experiments were performed.
RNA-Sequencing Analysis
The RNA-sequencing (RNA-seq) experiments were conducted in the Huada Genomics Institute (Wuhan, China), and the mRNA-seq library was obtained according to standard protocols (Illumina, San Diego, CA, USA). In brief, total RNA from si-NC- or si-HIF1A-AS2-1#-transfected HTR-8/SVneo cells was isolated as described above, and mRNA was purified using Dynabeads Oligo (dT) (Invitrogen Dynal) and reverse-transcribed into cDNA, which was then fragmented by nebulization to establish the mRNA-seq library.
RIP Assay
RIP assays were performed using the EZ-Magna RIP kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. HTR-8/SVneo and JAR cells were grown in 15-cm plates, collected by centrifugation, and lysed in RIP lysis buffer so that 100 μL contained the lysate of ∼1–2.0 × 107 cells. Cell lysates were used for immunoprecipitation with anti-LSD1, -EZH2, -AGO2, -DNMT1, and -DNMT3A antibodies and normal immunoglobulin G (IgG; Millipore, Billerica, MA, USA), and the immunoprecipitated RNA was amplified by quantitative real-time PCR using the Primer Script RT Master Mix kit and primers specific for HIF1A-AS2 and IgG (Table S1).
ChIP Assay
ChIP assays were performed using the EZ-Magna ChIP A kit (Millipore) according to the manufacturer’s protocol. HTR-8/SVneo and JAR cells were cross-linked by 1% formaldehyde for 10 min at 25°C, lysed, and sonicated to obtain 200- to 500-bp DNA fragments. Primary LSD1 and H3K4me2-specific antibodies (Millipore) or control IgG were added to pre-cleared supernatants, and the mixtures were incubated with rotation for 3 h or overnight at 4°C. Then, Dynabeads protein A/G were added to the mixtures, and the samples were incubated with vortexing for 2 h at 4°C. The beads were then washed sequentially with low-salt and high-salt RIPA, LiCi, and TE, and DNA was isolated by phenol/chloroform extraction. ChIP-qPCR was performed using SYBR Premix Ex Taq and specific primers (Table S1), and the data were normalized to the input. At least three independent experiments were conducted.
Statistical Analysis
The statistical analyses were performed with the SPSS 17.0 statistical software (IBM, Chicago, IL, USA). Each experiment was independently repeated at least three times, and the data were expressed as the mean ± SD. Student’s t test or Mann-Whitney test were used for comparisons between two groups, and ANOVA or Kruskal-Wallis tests were applied for multiple comparisons. p values less than 0.05 indicated statistical significance.
Author Contributions
L.S. and D.W. conceived and designed the study. D.W., N.Y. and Y.X. wrote the article. The experiments were performed by D.W., Y.X. and N.Y. The tissue samples were collected by S.W. and Y.Z. The data were coordinated and analyzed by M.S., B.H. and Z.H. All of the authors contributed to, read, and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The present work was supported by the National Natural Science Foundation of China (grants 81771603 and 81801472), Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant KYCX18_1481), Natural Science Foundation of Jiangsu Province (project numbers: BK20171502, BK20161061, and BK20181080), and National Key Research Plan of China (grant 2018YFC1002205).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.04.009.
Supplemental Information
References
- 1.Pedroso M.A., Palmer K.R., Hodges R.J., Costa F.D.S., Rolnik D.L. Uterine Artery Doppler in Screening for Preeclampsia and Fetal Growth Restriction. Rev. Bras. Ginecol. Obstet. 2018;40:287–293. doi: 10.1055/s-0038-1660777. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pedroso, M.A., Palmer, K.R., Hodges, R.J., Costa, F.D.S., and Rolnik, D.L. (2018). Uterine Artery Doppler in Screening for Preeclampsia and Fetal Growth Restriction. Rev. Bras. Ginecol. Obstet. 40, 287-293. [DOI] [PMC free article] [PubMed]
- 2.Groenhof T.K.J., van Rijn B.B., Franx A., Roeters van Lennep J.E., Bots M.L., Lely A.T. Preventing cardiovascular disease after hypertensive disorders of pregnancy: searching for the how and when. Eur. J. Prev. Cardiol. 2017;24:1735–1745. doi: 10.1177/2047487317730472. [DOI] [PMC free article] [PubMed] [Google Scholar]; Groenhof, T.K.J., van Rijn, B.B., Franx, A., Roeters van Lennep, J.E., Bots, M.L., and Lely, A.T. (2017). Preventing cardiovascular disease after hypertensive disorders of pregnancy: searching for the how and when. Eur. J. Prev. Cardiol. 24, 1735-1745. [DOI] [PMC free article] [PubMed]
- 3.Wu P., Haththotuwa R., Kwok C.S., Babu A., Kotronias R.A., Rushton C., Zaman A., Fryer A.A., Kadam U., Chew-Graham C.A., Mamas M.A. Preeclampsia and Future Cardiovascular Health: A Systematic Review and Meta-Analysis. Circ. Cardiovasc. Qual. Outcomes. 2017;10:e003497. doi: 10.1161/CIRCOUTCOMES.116.003497. [DOI] [PubMed] [Google Scholar]; Wu, P., Haththotuwa, R., Kwok, C.S., Babu, A., Kotronias, R.A., Rushton, C., Zaman, A., Fryer, A.A., Kadam, U., Chew-Graham, C.A., and Mamas, M.A. (2017). Preeclampsia and Future Cardiovascular Health: A Systematic Review and Meta-Analysis. Circ. Cardiovasc. Qual. Outcomes 10, e003497. [DOI] [PubMed]
- 4.Wallace A.E., Fraser R., Gurung S., Goulwara S.S., Whitley G.S., Johnstone A.P., Cartwright J.E. Increased angiogenic factor secretion by decidual natural killer cells from pregnancies with high uterine artery resistance alters trophoblast function. Hum. Reprod. 2014;29:652–660. doi: 10.1093/humrep/deu017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wallace, A.E., Fraser, R., Gurung, S., Goulwara, S.S., Whitley, G.S., Johnstone, A.P., and Cartwright, J.E. (2014). Increased angiogenic factor secretion by decidual natural killer cells from pregnancies with high uterine artery resistance alters trophoblast function. Hum. Reprod. 29, 652-660. [DOI] [PMC free article] [PubMed]
- 5.Wallace A.E., Whitley G.S., Thilaganathan B., Cartwright J.E. Decidual natural killer cell receptor expression is altered in pregnancies with impaired vascular remodeling and a higher risk of pre-eclampsia. J. Leukoc. Biol. 2015;97:79–86. doi: 10.1189/jlb.2A0614-282R. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wallace, A.E., Whitley, G.S., Thilaganathan, B., and Cartwright, J.E. (2015). Decidual natural killer cell receptor expression is altered in pregnancies with impaired vascular remodeling and a higher risk of pre-eclampsia. J. Leukoc. Biol. 97, 79-86. [DOI] [PMC free article] [PubMed]
- 6.Lin P., Wen D.Y., Li Q., He Y., Yang H., Chen G. Genome-Wide Analysis of Prognostic lncRNAs, miRNAs, and mRNAs Forming a Competing Endogenous RNA Network in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2018;48:1953–1967. doi: 10.1159/000492519. [DOI] [PubMed] [Google Scholar]; Lin, P., Wen, D.Y., Li, Q., He, Y., Yang, H., and Chen, G. (2018). Genome-Wide Analysis of Prognostic lncRNAs, miRNAs, and mRNAs Forming a Competing Endogenous RNA Network in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 48, 1953-1967. [DOI] [PubMed]
- 7.Delás M.J., Hannon G.J. lncRNAs in development and disease: from functions to mechanisms. Open Biol. 2017;7:170121. doi: 10.1098/rsob.170121. [DOI] [PMC free article] [PubMed] [Google Scholar]; Delas, M.J., and Hannon, G.J. (2017). lncRNAs in development and disease: from functions to mechanisms. Open Biol. 7, 170121. [DOI] [PMC free article] [PubMed]
- 8.Ernst E.H., Nielsen J., Ipsen M.B., Villesen P., Lykke-Hartmann K. Transcriptome Analysis of Long Non-coding RNAs and Genes Encoding Paraspeckle Proteins During Human Ovarian Follicle Development. Front. Cell Dev. Biol. 2018;6:78. doi: 10.3389/fcell.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ernst, E.H., Nielsen, J., Ipsen, M.B., Villesen, P., and Lykke-Hartmann, K. (2018). Transcriptome Analysis of Long Non-coding RNAs and Genes Encoding Paraspeckle Proteins During Human Ovarian Follicle Development. Front. Cell Dev. Biol. 6, 78. [DOI] [PMC free article] [PubMed]
- 9.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]; Geisler, S., and Coller, J. (2013). RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14, 699-712. [DOI] [PMC free article] [PubMed]
- 10.E, S., Costa M.C., Kurc S., Drozdz A., Cortez-Dias N., Enguita F.J. The circulating non-coding RNA landscape for biomarker research: lessons and prospects from cardiovascular diseases. Acta. Pharmacol. Sin. 2018;39:1085–1099. doi: 10.1038/aps.2018.35. [DOI] [PMC free article] [PubMed] [Google Scholar]; E, S., Costa, M.C., Kurc, S., Drozdz, A., Cortez-Dias, N., and Enguita, F.J. (2018). The circulating non-coding RNA landscape for biomarker research: lessons and prospects from cardiovascular diseases. Acta. Pharmacol. Sin. 39, 1085-1099. [DOI] [PMC free article] [PubMed]
- 11.Chen W.M., Huang M.D., Kong R., Xu T.P., Zhang E.B., Xia R., Sun M., De W., Shu Y.Q. Antisense Long Noncoding RNA HIF1A-AS2 Is Upregulated in Gastric Cancer and Associated with Poor Prognosis. Dig. Dis. Sci. 2015;60:1655–1662. doi: 10.1007/s10620-015-3524-0. [DOI] [PubMed] [Google Scholar]; Chen, W.M., Huang, M.D., Kong, R., Xu, T.P., Zhang, E.B., Xia, R., Sun, M., De, W., and Shu, Y.Q. (2015). Antisense Long Noncoding RNA HIF1A-AS2 Is Upregulated in Gastric Cancer and Associated with Poor Prognosis. Dig. Dis. Sci. 60, 1655-1662. [DOI] [PubMed]
- 12.Chen M., Zhuang C., Liu Y., Li J., Dai F., Xia M., Zhan Y., Lin J., Chen Z., He A. Tetracycline-inducible shRNA targeting antisense long non-coding RNA HIF1A-AS2 represses the malignant phenotypes of bladder cancer. Cancer Lett. 2016;376:155–164. doi: 10.1016/j.canlet.2016.03.037. [DOI] [PubMed] [Google Scholar]; Chen, M., Zhuang, C., Liu, Y., Li, J., Dai, F., Xia, M., Zhan, Y., Lin, J., Chen, Z., He, A., et al. (2016). Tetracycline-inducible shRNA targeting antisense long non-coding RNA HIF1A-AS2 represses the malignant phenotypes of bladder cancer. Cancer Lett. 376, 155-164. [DOI] [PubMed]
- 13.Lin J., Shi Z., Yu Z., He Z. LncRNA HIF1A-AS2 positively affects the progression and EMT formation of colorectal cancer through regulating miR-129-5p and DNMT3A. Biomed. Pharmacother. 2018;98:433–439. doi: 10.1016/j.biopha.2017.12.058. [DOI] [PubMed] [Google Scholar]; Lin, J., Shi, Z., Yu, Z., and He, Z. (2018). LncRNA HIF1A-AS2 positively affects the progression and EMT formation of colorectal cancer through regulating miR-129-5p and DNMT3A. Biomed. Pharmacother. 98, 433-439. [DOI] [PubMed]
- 14.Mineo M., Ricklefs F., Rooj A.K., Lyons S.M., Ivanov P., Ansari K.I., Nakano I., Chiocca E.A., Godlewski J., Bronisz A. The Long Non-coding RNA HIF1A-AS2 Facilitates the Maintenance of Mesenchymal Glioblastoma Stem-like Cells in Hypoxic Niches. Cell Rep. 2016;15:2500–2509. doi: 10.1016/j.celrep.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mineo, M., Ricklefs, F., Rooj, A.K., Lyons, S.M., Ivanov, P., Ansari, K.I., Nakano, I., Chiocca, E.A., Godlewski, J., and Bronisz, A. (2016). The Long Non-coding RNA HIF1A-AS2 Facilitates the Maintenance of Mesenchymal Glioblastoma Stem-like Cells in Hypoxic Niches. Cell Rep. 15, 2500-2509. [DOI] [PMC free article] [PubMed]
- 15.Durbas M., Horwacik I., Boratyn E., Rokita H. Downregulation of the PHLDA1 gene in IMR-32 neuroblastoma cells increases levels of Aurora A, TRKB and affects proteins involved in apoptosis and autophagy pathways. Int. J. Oncol. 2016;49:823–837. doi: 10.3892/ijo.2016.3572. [DOI] [PubMed] [Google Scholar]; Durbas, M., Horwacik, I., Boratyn, E., and Rokita, H. (2016). Downregulation of the PHLDA1 gene in IMR-32 neuroblastoma cells increases levels of Aurora A, TRKB and affects proteins involved in apoptosis and autophagy pathways. Int. J. Oncol. 49, 823-837. [DOI] [PubMed]
- 16.Toyoshima Y., Karas M., Yakar S., Dupont J., Lee Helman, LeRoith D. TDAG51 mediates the effects of insulin-like growth factor I (IGF-I) on cell survival. J. Biol. Chem. 2004;279:25898–25904. doi: 10.1074/jbc.M400661200. [DOI] [PubMed] [Google Scholar]; Toyoshima, Y., Karas, M., Yakar, S., Dupont, J., Lee Helman, and LeRoith, D. (2004). TDAG51 mediates the effects of insulin-like growth factor I (IGF-I) on cell survival. J. Biol. Chem. 279, 25898-25904. [DOI] [PubMed]
- 17.Park C.G., Lee S.Y., Kandala G., Lee S.Y., Choi Y. A novel gene product that couples TCR signaling to Fas(CD95) expression in activation-induced cell death. Immunity. 1996;4:583–591. doi: 10.1016/s1074-7613(00)80484-7. [DOI] [PubMed] [Google Scholar]; Park, C.G., Lee, S.Y., Kandala, G., Lee, S.Y., and Choi, Y. (1996). A novel gene product that couples TCR signaling to Fas(CD95) expression in activation-induced cell death. Immunity 4, 583-591. [DOI] [PubMed]
- 18.Duquette M.L., Kim J., Shi L.Z., Berns M.W. LSD1 mediated changes in the local redox environment during the DNA damage response. PLoS ONE. 2018;13:e0201907. doi: 10.1371/journal.pone.0201907. [DOI] [PMC free article] [PubMed] [Google Scholar]; Duquette, M.L., Kim, J., Shi, L.Z., and Berns, M.W. (2018). LSD1 mediated changes in the local redox environment during the DNA damage response. PLoS ONE 13, e0201907. [DOI] [PMC free article] [PubMed]
- 19.Ambrosio S., Amente S., Saccà C.D., Capasso M., Calogero R.A., Lania L., Majello B. LSD1 mediates MYCN control of epithelial-mesenchymal transition through silencing of metastatic suppressor NDRG1 gene. Oncotarget. 2017;8:3854–3869. doi: 10.18632/oncotarget.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ambrosio, S., Amente, S., Sacca, C.D., Capasso, M., Calogero, R.A., Lania, L., and Majello, B. (2017). LSD1 mediates MYCN control of epithelial-mesenchymal transition through silencing of metastatic suppressor NDRG1 gene. Oncotarget 8, 3854-3869. [DOI] [PMC free article] [PubMed]
- 20.Mansoori Z., Ghaedi H., Sadatamini M., Vahabpour R., Rahimipour A., Shanaki M., Saeidi L., Kazerouni F. Downregulation of long non-coding RNAs LINC00523 and LINC00994 in type 2 diabetes in an Iranian cohort. Mol. Biol. Rep. 2018;45:1227–1233. doi: 10.1007/s11033-018-4276-7. [DOI] [PubMed] [Google Scholar]; Mansoori, Z., Ghaedi, H., Sadatamini, M., Vahabpour, R., Rahimipour, A., Shanaki, M., Saeidi, L., and Kazerouni, F. (2018). Downregulation of long non-coding RNAs LINC00523 and LINC00994 in type 2 diabetes in an Iranian cohort. Mol. Biol. Rep. 45, 1227-1233. [DOI] [PubMed]
- 21.Zampetaki A., Albrecht A., Steinhofel K. Long Non-coding RNA Structure and Function: Is There a Link? Front. Physiol. 2018;9:1201. doi: 10.3389/fphys.2018.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zampetaki, A., Albrecht, A., and Steinhofel, K. (2018). Long Non-coding RNA Structure and Function: Is There a Link? Front. Physiol. 9, 1201. [DOI] [PMC free article] [PubMed]
- 22.Ferretti C., Bruni L., Dangles-Marie V., Pecking A.P., Bellet D. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum. Reprod. Update. 2007;13:121–141. doi: 10.1093/humupd/dml048. [DOI] [PubMed] [Google Scholar]; Ferretti, C., Bruni, L., Dangles-Marie, V., Pecking, A.P., and Bellet, D. (2007). Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum. Reprod. Update 13, 121-141. [DOI] [PubMed]
- 23.Holtan S.G., Creedon D.J., Haluska P., Markovic S.N. Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin. Proc. 2009;84:985–1000. doi: 10.1016/S0025-6196(11)60669-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Holtan, S.G., Creedon, D.J., Haluska, P., and Markovic, S.N. (2009). Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin. Proc. 84, 985-1000. [DOI] [PMC free article] [PubMed]
- 24.Esteve-Valverde E., Ferrer-Oliveras R., Gil-Aliberas N., Baraldès-Farré A., Llurba E., Alijotas-Reig J. Pravastatin for Preventing and Treating Preeclampsia: A Systematic Review. Obstet. Gynecol. Surv. 2018;73:40–55. doi: 10.1097/OGX.0000000000000522. [DOI] [PubMed] [Google Scholar]; Esteve-Valverde, E., Ferrer-Oliveras, R., Gil-Aliberas, N., Baraldes-Farre, A., Llurba, E., and Alijotas-Reig, J. (2018). Pravastatin for Preventing and Treating Preeclampsia: A Systematic Review. Obstet. Gynecol. Surv. 73, 40-55. [DOI] [PubMed]
- 25.Janani F., Changaee F. Seasonal variation in the prevalence of preeclampsia. J. Family Med. Prim. Care. 2017;6:766–769. doi: 10.4103/jfmpc.jfmpc_132_17. [DOI] [PMC free article] [PubMed] [Google Scholar]; Janani, F., and Changaee, F. (2017). Seasonal variation in the prevalence of preeclampsia. J. Family Med. Prim. Care 6, 766-769. [DOI] [PMC free article] [PubMed]
- 26.Lyall F., Robson S.C., Bulmer J.N. Spiral artery remodeling and trophoblast invasion in preeclampsia and fetal growth restriction: relationship to clinical outcome. Hypertension. 2013;62:1046–1054. doi: 10.1161/HYPERTENSIONAHA.113.01892. [DOI] [PubMed] [Google Scholar]; Lyall, F., Robson, S.C., and Bulmer, J.N. (2013). Spiral artery remodeling and trophoblast invasion in preeclampsia and fetal growth restriction: relationship to clinical outcome. Hypertension 62, 1046-1054. [DOI] [PubMed]
- 27.Kuske M.D., Johnson J.P. Assignment of the human PHLDA1 gene to chromosome 12q15 by radiation hybrid mapping. Cytogenet. Cell Genet. 2000;89:1. doi: 10.1159/000015575. [DOI] [PubMed] [Google Scholar]; Kuske, M.D., and Johnson, J.P. (2000). Assignment of the human PHLDA1 gene to chromosome 12q15 by radiation hybrid mapping. Cytogenet. Cell Genet. 89, 1. [DOI] [PubMed]
- 28.Moad A.I., Muhammad T.S., Oon C.E., Tan M.L. Rapamycin induces apoptosis when autophagy is inhibited in T-47D mammary cells and both processes are regulated by Phlda1. Cell Biochem. Biophys. 2013;66:567–587. doi: 10.1007/s12013-012-9504-5. [DOI] [PubMed] [Google Scholar]; Moad, A.I., Muhammad, T.S., Oon, C.E., and Tan, M.L. (2013). Rapamycin induces apoptosis when autophagy is inhibited in T-47D mammary cells and both processes are regulated by Phlda1. Cell Biochem. Biophys. 66, 567-587. [DOI] [PubMed]
- 29.Sellheyer K., Nelson P. Follicular stem cell marker PHLDA1 (TDAG51) is superior to cytokeratin-20 in differentiating between trichoepithelioma and basal cell carcinoma in small biopsy specimens. J. Cutan. Pathol. 2011;38:542–550. doi: 10.1111/j.1600-0560.2011.01693.x. [DOI] [PubMed] [Google Scholar]; Sellheyer, K., and Nelson, P. (2011). Follicular stem cell marker PHLDA1 (TDAG51) is superior to cytokeratin-20 in differentiating between trichoepithelioma and basal cell carcinoma in small biopsy specimens. J. Cutan. Pathol. 38, 542-550. [DOI] [PubMed]
- 30.Johnson E.O., Chang K.H., de Pablo Y., Ghosh S., Mehta R., Badve S., Shah K. PHLDA1 is a crucial negative regulator and effector of Aurora A kinase in breast cancer. J. Cell Sci. 2011;124:2711–2722. doi: 10.1242/jcs.084970. [DOI] [PubMed] [Google Scholar]; Johnson, E.O., Chang, K.H., de Pablo, Y., Ghosh, S., Mehta, R., Badve, S., and Shah, K. (2011). PHLDA1 is a crucial negative regulator and effector of Aurora A kinase in breast cancer. J. Cell Sci. 124, 2711-2722. [DOI] [PubMed]
- 31.Fearon A.E., Carter E.P., Clayton N.S., Wilkes E.H., Baker A.M., Kapitonova E., Bakhouche B.A., Tanner Y., Wang J., Gadaleta E. PHLDA1 Mediates Drug Resistance in Receptor Tyrosine Kinase-Driven Cancer. Cell Rep. 2018;22:2469–2481. doi: 10.1016/j.celrep.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fearon, A.E., Carter, E.P., Clayton, N.S., Wilkes, E.H., Baker, A.M., Kapitonova, E., Bakhouche, B.A., Tanner, Y., Wang, J., Gadaleta, E., et al. (2018). PHLDA1 Mediates Drug Resistance in Receptor Tyrosine Kinase-Driven Cancer. Cell Rep. 22, 2469-2481. [DOI] [PMC free article] [PubMed]
- 32.Marchiori A.C., Casolari D.A., Nagai M.A. Transcriptional up-regulation of PHLDA1 by 17beta-estradiol in MCF-7 breast cancer cells. Braz. J. Med. Biol. Res. 2008;41:579–582. doi: 10.1590/s0100-879x2008005000029. [DOI] [PubMed] [Google Scholar]; Marchiori, A.C., Casolari, D.A., and Nagai, M.A. (2008). Transcriptional up-regulation of PHLDA1 by 17beta-estradiol in MCF-7 breast cancer cells. Braz. J. Med. Biol. Res. 41, 579-582. [DOI] [PubMed]
- 33.Nagai M.A. Pleckstrin homology-like domain, family A, member 1 (PHLDA1) and cancer. Biomed. Rep. 2016;4:275–281. doi: 10.3892/br.2016.580. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nagai, M.A. (2016). Pleckstrin homology-like domain, family A, member 1 (PHLDA1) and cancer. Biomed. Rep. 4, 275-281. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.