Abstract
Transcription factors are the core drivers of gene regulatory networks that control developmental transitions, therefore a more complete understanding of how they access, alter and maintain tissue-specific gene expression patterns remains an important goal. To systematically dissect molecular components that enable or constrain their activity, we investigated the genomic occupancy of FOXA2, GATA4 and OCT4 in several cell types. Despite a classification as pioneer factors, all three factors demonstrate cell type specific enrichment even under super-physiological expression. However, only FOXA2 and GATA4 display, in both endogenous and ectopic conditions, a low enrichment sampling of additional loci that are occupied in alternative cell types. Co-factor expression can lead to increased pioneer factor binding at subsets of previously sampled target sites. Finally, we demonstrate that FOXA2 occupancy and changes to DNA accessibility at silent cis-regulatory elements can occur when the cell cycle is halted in G1, but subsequent loss of DNA methylation requires DNA replication.
Organismal development is orchestrated by selective use and distinctive interpretation of identical genetic material in each cell. During this process, transcription factors (TFs) coordinate protein complexes at associated promoter and distal enhancer elements to modulate gene expression patterns. However, our current understanding of the steps needed for the activation of a silent cis-regulatory element is still incomplete. A generally accepted model assumes that primary access to certain regulatory elements can be restricted by chromatin, which may ensure some spatial and temporal control of gene expression during successive developmental stages1. Thus there is a requirement for a distinct mechanism that transitions repressed cis-regulatory elements towards accessibility to allow coordinated binding of cell type specific TFs. This may be accomplished through so called pioneer TFs, which have the ability to bind to nucleosomal target sites and reorganize such regions leading to increased acessibility2–7. Interestingly, despite the supposed universal targeting and remodeling capabilities of pioneer TFs, they also display a degree of cell type specificity8–10 with recent work suggesting that cell-type specific co-factors11–13, signaling14, and the underlying chromatin landscape5,6 can influence the genomic occupancy of pioneer TFs. Yet it remains a challenge to study the contributions of individual TFs utilizing native developmental systems, where extrinsic signals may induce rapid transitions from an initial TF binding event and stabilization, to local epigenetic remodeling and transcriptional induction without yielding sufficiently stable intermediate states. Moreover, co-factors and partially redundant family members may already be present in these systems, further complicating the isolation and interpretation of their individual roles. To overcome these limitations we compared pioneer TF occupancy at endogenously bound cis-regulatory elements across multiple cell types to individual and combinatoric occupancy in an ectopic environment, thereby allowing us to obtain new insights into the regulatory capabilities of presumed pioneer TFs, FOXA2, GATA4 and OCT4 which are frequently studied in development and utilized in cellular reprogramming.
Results
FOXA2 binds subsets of its motifs in a largely cell type specific manner
The pioneer factor FOXA motif harbors seven core consensus nucleotides with less distinct flanking sequence and is consequently abundant within the human genome (Supplementary Fig. 1a)15. As pioneer factors have the unique ability to access target loci in closed chromatin16,17, one may expect them to extensively occupy genomic sites that contain the core regulatory motif. To investigate this we determined the proportion of its preferred motif sequence that is occupied across a number of human cell types with detectable FOXA2 expression, including HepG2 (hepatocellular liver carcinoma: FPKM 10.9), A549 (lung carcinoma: FPKM 6.2), and dEN (embryonic stem cell (ESC) derived definitive endoderm18; FPKM 20.1). We utilized five position weight matrices (PWM) with varying stringencies, mapped their positions across the human genome, and then only considered motifs that overlapped with region enriched for activating modifications in at least one of the ENCODE/REMC project cell types19 (Supplementary Fig. 1a,b; see Methods). Only 6.3–13.7% of these identified motifs were significantly bound by FOXA2 (Fig. 1a, Supplementary Fig. 1b) and the enrichment was largely cell type specific, consistent with prior studies (Fig. 1b)8–10. It is likely that FOXA2 binding data from additional cell types will confirm more of the FOXA2 motifs to be targets given that we do not observe saturation of the binding spectra with the current selected cell types (Supplementary Fig. 1c).
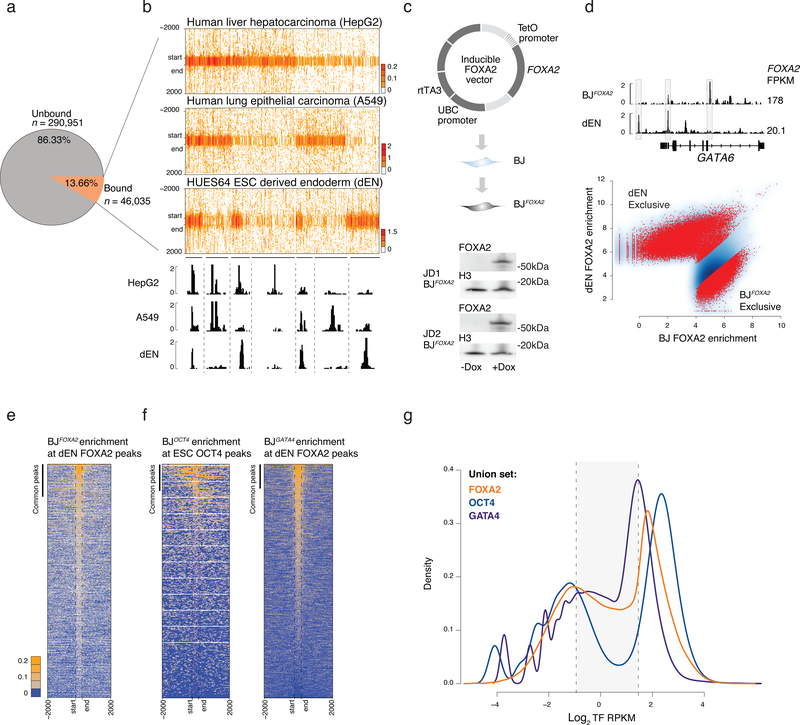
Figure 1 |. Ectopic FOXA2 and GATA4 but not OCT4 display low-level sampling.
a) Pie chart displays the percentage of FOXA motifs (see Supplementary Fig. 1) mapped across the genome that are unbound or bound by FOXA2 at a potentially accessible genomic region in ESC derived endoderm (dEN), HepG2, and A549 cells.
b) Read density heat maps for all IDR called FOXA2 peaks in dEN, A549 and HepG2 cells that overlap a motif instance. Heat maps are clustered by occurrence of binding across the three cell types. IGV tracks highlight shared genomic occupancy across the three cell types (chr18:9,072,728–9,075,158), unique occupancy in HepG2 (chr18:9,202,880–9,225,100), unique in A549 (chr18:9,008,450–9,022,842) shared occupancy in A549 and HepG2 (chr18:8,725,886–8,734,843) shared in HepG2 and dEN (chr4:80,986,601–81,000,201) shared in A549 and dEN chr4:75,017,694–75,029,960 and unique occupancy in dEN (chr4:74,903,404–74,905,306).
c) Schematic of the pTripZ vector used for the generation of a clonal FOXA2 inducible cell line, BJFOXA2. Cropped western blot of FOXA2 and H3 protein levels in two, distinct BJFOXA2 clones (JD1 and JD2).
d) IGV browser shots display differential binding across ectopic BJFOXA2 and dEN (chr18:19,745,852–19,782,939). FOXA2 FPKMs listed on the right. Scatter plot shows output of DiffBind41 differential peak set analysis between dEN and BJFOXA2. Red dots indicate peaks with statistically significant differential enrichment between the two data sets.
e) Read density heat maps of FOXA2 enrichment in BJFOXA2 at dEN FOXA2 regions. Bar indicates peak calls in common between ectopic FOXA2 ChIP-seq data in BJFOXA2 and dEN FOXA2 ChIP-seq. Dashed lines mark the start and end of FOXA2 peaks. Most dEN sites still show low-level enrichment of FOXA2 in BJFOXA2 fibroblasts yet are not called as significantly enriched.
f) Read density heat map of OCT4 signal in the BJs at human ESC OCT4 regions (n=22,477). Bar indicates peak calls in common between ectopic OCT4 ChIP-seq data and ESC OCT4 ChIP-seq (n=1,972). In contrast to FOXA2, very few ESC OCT4 sites show any notable level of OCT4 enrichment in BJOCT4. Read density heat map of GATA4 signal in the BJs at human GATA4 dEN bound genomic regions (n=42,477).
g) Density plot displaying FOXA2, OCT4 and GATA4 log2 RPKM ectopic enrichment in BJs at union sets of ectopic and endogenous sites (FOXA2 – orange, OCT4 – navy, GATA4 - blue). Dashed lines demarcate regions within the background distribution, regions called as sampled sites (shaded) and regions that were called as peaks.
As primary TF engagement cannot be adequately dissected using endogenous systems that already express FOXA2 as part of their regulatory circuitry, we engineered a doxycycline (DOX) inducible system in immortalized foreskin fibroblasts (BJ) that do not normally express FOXA2 or other FOXA family members (Supplementary Fig. 1d). We derived several clonal cell lines (referred to as BJFOXA2) with no detectable FOXA2 in the uninduced state, but rapid, uniform and consistent mRNA/protein induction upon DOX treatment (Fig. 1c, Supplementary Figs. 1e–g). We next performed ChIP-seq for FOXA2 in BJFOXA2 after 1, 4, and 10 days of induction observing a clear increase in FOXA2 binding sites between 1 and 4 days with little change afterwards (Supplementary Figs. 1h,i; Supplementary Table 1) and identified a total of 49,830 consensus IDR FOXA2 peaks for the combined 4 and 10 day time points of which 98% contained a FOX family motif (Supplementary Fig. 1j)20. Despite super-physiological FOXA2 levels, we still primarily observe cell type specific FOXA2 binding with ~70% of FOXA2 peaks showing differential enrichment between dEN and BJFOXA2 (Fig. 1d). Therefore DNA sequence alone is clearly insufficient to direct binding, as the majority of potential FOXA2 targets remain unbound in ectopic conditions.
FOXA2 and GATA4 demonstrate low-level sampling at most of their targets in alternative lineages
While we only observed a partial overlap between significantly called FOXA2 peaks in endogenous and ectopic contexts, we nevertheless noticed consistent, low-level FOXA2 enrichment in BJFOXA2 at the majority of regions occupied by FOXA2 in dEN, HepG2, and A549 (Fig. 1e, Supplementary Fig. 2a). BJ FOXA2 enrichment at the union set of previously defined endogenous (HepG2, A549 and dEN) and ectopic (BJ) FOXA2 peaks (‘FOXA2 union set’) revealed a notable number of regions that were not classified as a highly enriched peak in BJs, but still displayed low to intermediate FOXA2 enrichment (Fig. 1g). To determine whether this low-level enrichment is a general feature of ectopic TF expression, we engineered inducible BJ fibroblasts for two other presumed pioneer factors, OCT4 and GATA4 (BJOCT, BJGATA4; Supplementary Fig. 2b; Supplementary Tables 2,3) and found a comparable low-level enrichment for GATA4 but not for OCT4 (Fig. 1f,g). However, ectopic OCT4 has been shown to display low-level enrichment at ESC OCT4 targets when it is co-expressed with SOX2, KLF4 and cMYC in BJ fibroblasts indicating that this ability is context and co-factor dependent (Supplementary Fig. 2c)5. Additionally, by examining FOXA2 enrichment in HepG2 and dEN cells at all A549 FOXA2 peak regions, we find that this low-level enrichment is also observed in cells endogenously expressing FOXA2 and therefore not just a product of ectopic or super-physiological expression levels (Supplementary Fig. 2d,e).
Differential influence of prior epigenetic state on FOXA2, GATA4 and OCT4 binding
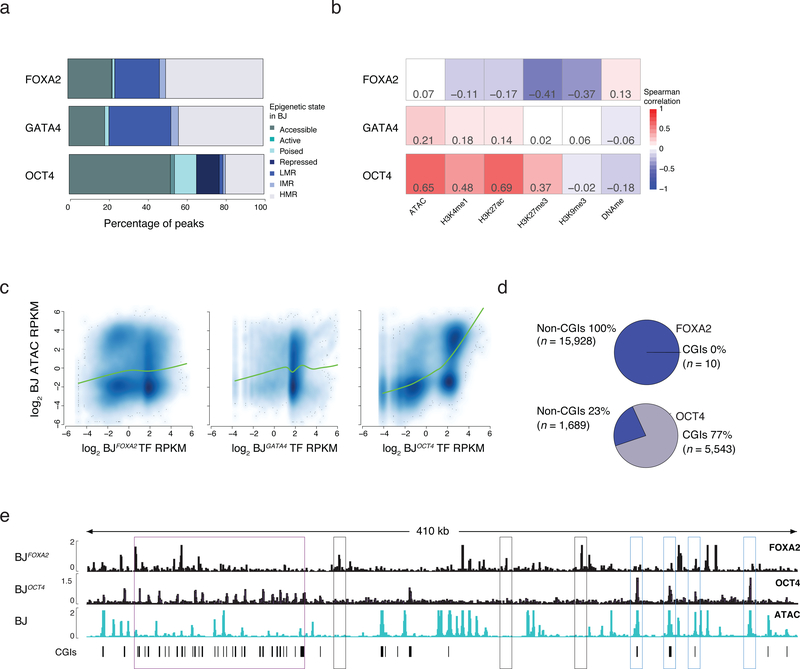
To determine how a cell’s pre-existing epigenome may affect pioneer factor binding, we performed ChIP-seq for select histone modifications associated with active (H3K27ac and H3K4me1) and inactive (H3K27me3) states, measured DNA accessibility by the assay for transposon-accessible chromatin (ATAC-seq)21 and DNA methylation (DNAme) levels by whole genome bisulfite sequencing (WGBS) (Supplementary Table 4). We then defined chromatin states using simple, hierarchical rules that reflect prior knowledge of these modifications and how they interact (Supplementary Fig. 3a). For this analysis we focused on the most highly enriched targets in a given cell type and found that ectopic FOXA2 and GATA4 predominantly engage sites that are devoid of the selected histone modifications and instead contain variable levels of DNAme (Fig. 2a). Endogenous FOXA2 displayed a similar behavior when comparing the epigenome in undifferentiated ESCs at sites that become bound by FOXA2 in dEN (Supplementary Fig. 3b). There is little correlation between FOXA2 or GATA4 enrichment and selected epigenetic features, yet OCT4 binding is positively correlated with pre-existing accessible chromatin (Fig. 2a–c; Supplementary Fig. 3b) and frequently overlaps with CpG islands (CGIs) (Fig. 2d,e, Supplementary Fig. 3c). To also compare these behaviors to the ectopic binding of a presumed non-pioneer factor, we generated another BJ fibroblast line for the hepatocyte nuclear factor, HNF1A and observe no significant HNF1A binding when expressed alone, however in combination with FOXA2, enrichment became readily detectable (Supplementary Fig. 3d–f). Lastly, FOXA2 enrichment was generally depleted in H3K9me3 heterochromatin domains (Supplementary Figs. 3g–i)5. However, few endogenously occupied FOXA2 regions in HepG2, A549 or dEN cells actually fall within BJ K9-domains, and therefore these domains are unlikely the major cause of the cell type specific occupancy observed for FOXA2 (Supplementary Figs. 3j).
Figure 2 |. Influence of prior epigenetic state on pioneer factor occupancy.
a) Percentage of TF bound regions in BJFOXA2, BJGATA4, BJOCT4 falling into assigned chromatin states. State is defined hierarchically using chromatin state in BJ fibroblasts prior to TF induction. First, ‘accessible’ regions were categorized by the occurrence of ATAC-seq enrichment. Then regions highly enrichment for H3K27ac or H3K4me1 were categorized as ‘active’ and ‘poised’, respectively. Regions enriched for H3K27me3 or H3K9me3 were categorized broadly as ‘repressed’ and finally all remaining regions that were not classified into one of the above classes were grouped by their DNAme levels: highly methylated regions (HMRs > 60% mean methylation), intermediate methylated regions (IMR mean methylation: 20–60%) and lowly methylated regions (LMR: < 20% mean methylation). LMRs are equivalent to a ‘low signal’ state that lacks DNA accessibility as well as enrichment of any assessed histone modifications4.
b) Spearman correlations between TF enrichment and enrichment of epigenetic features displayed as heat map.
c) Scatter plots and lowess fit curves (green line) of FOXA2, OCT4 and GATA4 enrichment (Log2 RPKM) versus ATAC-seq enrichment in BJs prior to factor induction (Log2 RPKM).
d) Pie charts summarize the percentage of FOXA2 and OCT4 targets that overlap with defined pre-existing closed chromatin and fall within annotated CpG islands (CGIs).
e) Representative IGV browser tracks displaying FOXA2 and OCT4 enrichment compared to pre-induced BJ ATAC-seq data (chr5:140,657,329–141,085,891). Purple boxes highlight regions of OCT4 binding in pre-existing closed chromatin that overlap with annotated CGIs. Gray boxes highlight FOXA2 binding at pre-existing closed chromatin while blue boxes highlight OCT4 binding in regions of pre-existing open chromatin.
GATA4 co-expression can increase FOXA2 enrichment at a subset of previously sampled targets
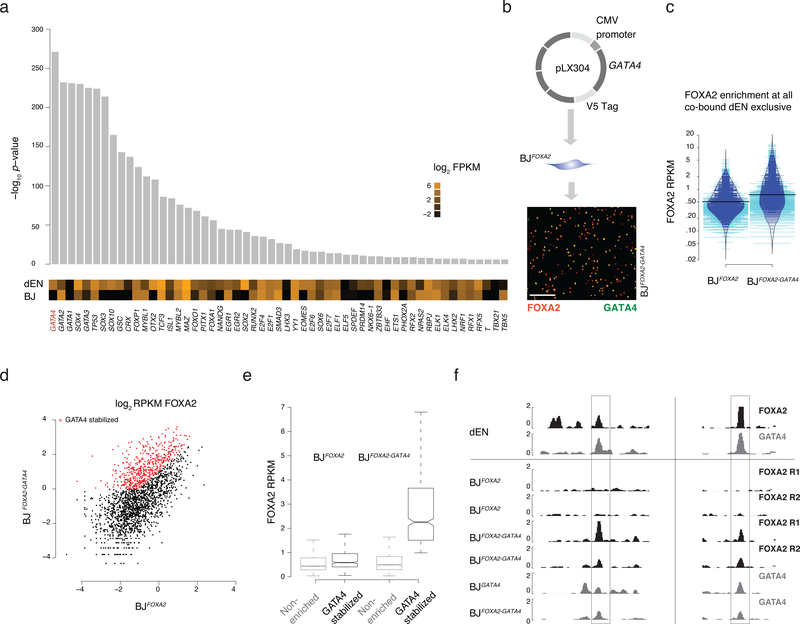
Given the limited ability of the epigenome to determine FOXA2 binding, we speculated that occupancy might instead be mostly directed through cooperativity with cell type specific cofactors, which is common among non-pioneer TFs1,22 and has recently also been suggested for some pioneer TFs12,13. To identify potential co-factors, we searched for differentially enriched motifs between regions bound by FOXA2 exclusively in dEN or BJFOXA2 and cross-referenced these against RNA-seq data for the corresponding TFs (Fig. 3a; Supplementary Fig. 4a). Motif sequences for several known endodermal regulators were enriched at dEN exclusive sites, including GATA4, which is known to bind to the ALB enhancer locus with FOXA2 in early gut endoderm cells prior to ALB expression23–25. Thus we selected GATA4 as a candidate co-factor that might influence FOXA2 binding in the ectopic system. Utilizing our previously published data26 for FOXA2 and GATA4 binding in dEN, we found the two factors co-localized at 2,364 genomic sites, the majority of which overlap with FOXA2 dEN exclusive targets (n=2,093, Supplementary Fig. 4b). We next infected our BJFOXA2 line with a second lentiviral construct containing constitutively expressed, V5-tagged GATA4 and induced simultaneous expression of both factors for four days (Fig. 3b; BJFOXA2/GATA4). Globally FOXA2 enrichment showed only a slight increase (Fig. 3c) while a specific subset of targets displayed a substantial increase (Fig. 3d–f; 504 out of 2,093). Interestingly, these GATA4 stabilized sites showed higher FOXA2 enrichment prior to GATA4 expression indicating they were likely previously sampled by FOXA2 when expressed independently (Fig. 3e; n=318) and in addition, they were occupied by GATA4 alone (Supplementary Fig. 4c). Given that co-expression could only explain a subset of the dEN exclusive co-bound FOXA2 targets, we searched for additional confounding factors and found the GATA motif to be differentially enriched in the GATA4 stabilized subset compared to the non-enriched subset (p-value: 1.0e−5; motif occurring at 76% of regions). In turn, we observed weak differential enrichment of other endodermal factor motifs in the non-enriched subset (T-box; p-value 1.0e−3, Eomes; p-value 1.0e−3; and SOX; p-value 1.0e−3) indicating occupancy of FOXA2 at these regions may be dependent on multiple TFs. Finally, to assess if this cooperativity models dynamic assisted loading12,22, we measured accessibility via ATAC-seq after co-expression and found GATA4 stabilized target sites exhibit little change in DNA accessibility compared to uninduced controls indicating that recruitment of chromatin remodeling machinery has not yet occurred and instead, these two factors cooperatively localize on nucleosomes (Supplementary Fig. 4d). These results support a model where occupancy of a pioneer factor can be partially determined by cofactor engagement at specific subsets of genetically encoded target loci.
Figure 3 |. Co-factor expression modulates FOXA2 enrichment at a subset of target sites.
a) Differential motif analysis displaying –log10 p-value of enriched motifs in dEN exclusive sites versus BJFOXA2 exclusive sites with the most significant motifs on the left. Expression (log2 FPKM) of the TFs associated with the listed motif in both BJFOXA2 and dEN. Of note, while there are many significant differential motifs observed in dEN exclusive sites, not all motifs are associated with factors that display differential expression.
b) Simplified schematic of ectopic expression system used to co-express GATA4 in BJFOXA2. Immunostaining of FOXA2 and V5-GATA4 in co-infected BJFOXA2 fibroblasts. White scale bar is equal to 345nm.
c) Bean plot comparing FOXA2 enrichments at all dEN exclusive co-bound sites when FOXA2 is expressed independently versus when FOXA2 and GATA4 are co-expressed. Thick black bars represent average. Blue lines indicate data points within the distribution while teal bars represent data points outside of the distribution.
d) Scatter plot comparing FOXA2 enrichment of co-bound dEN exclusive sites in BJFOXA2 fibroblasts compared to BJFOXA2-GATA4 fibroblasts. Red dots indicate regions that gain at least 2 fold enrichment and are above RPKM of 1 in BJFOXA2-GATA4 fibroblasts. P-value 2e−16.
e) Box plots displaying the RPKM of FOXA2 enrichment in BJFOXA2 and BJFOXA2-GATA4 at the subset of regions that are GATA4 stabilized compared to the non-enriched subset. Boxes indicate interquartile range and whiskers show maximum and minimum values. Outliers are removed.
f) Representative IGV browser tracks displaying FOXA2 and GATA4 enrichment in dEN and the various fibroblasts as indicated on the left. Gray bar highlights a region that shows the co-factor mediated recruitment of FOXA2 to two of its dEN targets chr13:76,031,782–76,039,815 and chr17:14,352,627–14,360,300.
Transcriptional and epigenetic impact of ectopic FOXA2 binding
To determine the molecular effects of the ectopic TF binding we performed RNA-seq, ATAC-seq and ChIP-seq 48 hours post FOXA2 induction. In line with previous studies on pioneer factors7,18, we find only a small number of genes that were immediately responsive to induction (~299 genes up-regulated and 191 genes down-regulated) and an even smaller number of the activated genes appeared to have a FOXA2 binding site within 1kb of the associated gene promoter (~82 genes; Supplementary Fig. 5a; Supplementary Table 5). Due to the limited trans-activating properties, we focused on chromatin changes upon FOXA2 occupancy and find many regions that either gain de novo or display enhanced enrichment of H3K4me1, H3K4me2 and H3K27ac upon FOXA2 induction (Supplementary Fig. 5b). Such gains in H3K4me histone modifications upon occupancy of FOXA factors have previously been observed when pioneer factors establish competency at cis-regulatory regions7,27–29 however we find that ~40% concomitantly also gain low enrichment of H3K27ac (n=1,937).
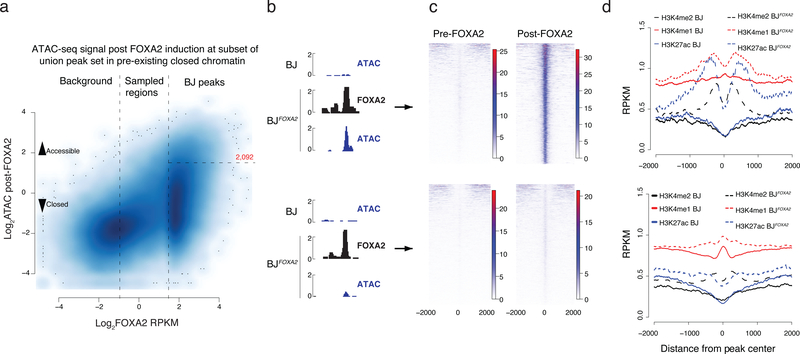
Next we focused on FOXA2 occupied regions that fall in pre-existing closed chromatin and measured induced changes in DNA accessibility and histone modifications. Occupancy alone appears insufficient to affect global accessibility as only a fraction (~13%) of BJ FOXA2 targets demonstrate significant gains in ATAC-seq signal (Fig. 4a,b; n=2,092, p value = < 2.2e-16). Despite the observed infrequent increase in DNA accessibility, 61% of unchanged targets (5,144 out of 8,443) overlap with putative gene regulatory elements that become accessible in at least one other cell type based on all available ENCODE DNase hypersensitive data30. That said, we can detect some low-level increase in ATAC-seq signal even at the target sites that remain inaccessible based on our thresholds (Fig. 4c, Supplementary Fig. 5c). We find a number of distinguishing features that characterize sites that gain significant accessibility. First, FOXA motifs are more highly enriched and widely distributed (Supplementary Fig. 5d–f, top 8 motifs shown). Second, the mean ATAC-seq signal prior to FOXA2 occupancy is slightly higher suggesting some prior, yet minimal, accessibility in these regions (Supplementary Fig. 5g). Third, we observe enrichment of phased nucleosomes modified by H3K4me1/me2 and H3K27ac surrounding the FOXA2 peak summit (Fig. 4d). Scatter plots of binned ATAC-seq signal compared to histone modification enrichment demonstrate a somewhat linear relationship between gain in DNA accessibility and gain in H3K4me1 (along with a weaker enrichment and correlation for H3K4me2 and H3K27ac) (Supplementary Fig. 5h). It is worth stating that the enrichment of activating histone modifications at these regions is quite modest and does not reach similar enrichment levels seen for active promoters (Supplementary Fig. 5i). Furthermore, we observe that the majority of ATAC-seq signal gained after FOXA2 occupancy is already lost within two days of factor withdrawal indicating the transient behavior of this remodeling in agreement with prior work (Supplementary Fig. 5c,–j)3.
Figure 4 |. Epigenetic effect of ectopic FOXA2 binding.
a) Scatter plot displaying FOXA2 enrichment in induced BJFOXA2 compared to post-FOXA2 induction ATAC-seq signal at the union set of FOXA2 binding sites that fall within pre-existing closed chromatin genomic regions (BJ ATAC RPKM <1). Vertical lines separate the union set of FOXA2 peaks into background, sampled and called FOXA2 peaks. Horizontal line indicates boundary to regions that have new BJFOXA2 ATAC enrichment of at least RPKM >3 and are considered newly accessible.
b) Representative IGV browser tracks of regions that become accessible upon FOXA2 binding (top; chr20:36,008,193–36,009,335) versus regions that remain inaccessible as measured by ATAC-seq enrichment post FOXA2 induction (bottom; chr19:1,867,722–1,868,322)
c) Read density heat maps of post- FOXA2 ATAC-seq signal across all regions that become accessible (top) compared to the ones that remain closed upon FOXA2 binding (bottom).
d) Composite plots displaying H3K27ac and H3K4me1 and H3K4me2 enrichment over regions that become accessible and those that remain closed comparing pre- and post- FOXA2 occupancy.
Distance dependent loss of DNAme at selective FOXA2 targets
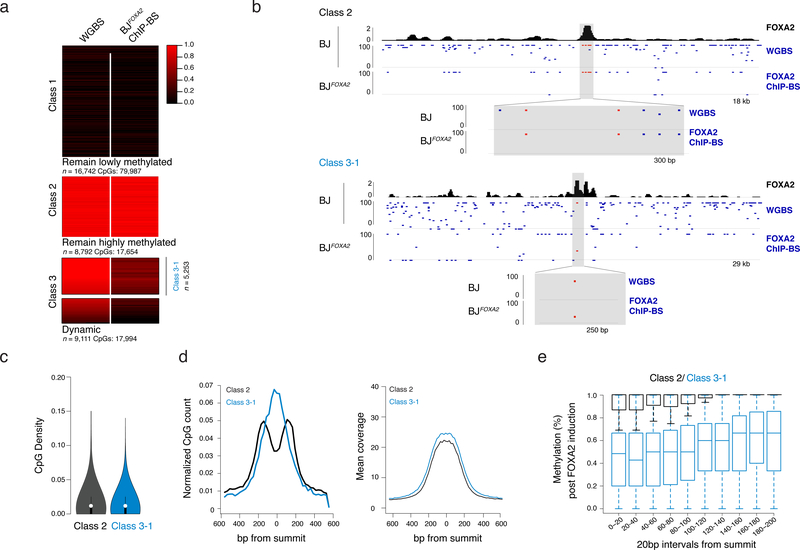
As FOXA2 binding occurs at methylated loci (Fig. 2a) and is associated with loss of methylation at target sites18,28, we investigated FOXA2-mediated demethylation in BJFOXA2. To quantify DNAme levels on fragments that were physically associated with FOXA2 we performed ChIP followed by bisulfite sequencing (ChIP-BS-seq)31 at four days post-induction and compared methylation levels to WGBS data for BJFOXA2 prior to induction (Supplementary Table 6). We found that FOXA2 occupies three distinct sets of genomic regions: those in pre-existing lowly methylated DNA that remained lowly methylated after FOXA2 binding (Fig. 5a, class 1: n=16,742), those that display high DNAme levels before and after FOXA2 binding (Fig. 5a,b, class 2: n=8,794) and a unique class of regions that display a clear loss of DNAme following the binding of FOXA2 (>20% change, Fig. 5a,b, dynamic; class 3: n=9,111). Of note, all three classes were reproducible even after the deletion of either the N- or C-terminal domain of FOXA2 indicating that neither domain was responsible for the local demethylation observed at class 3 targets (Supplementary Fig. 6). As it was unexpected to observe no methylation change upon FOXA2 binding at some targets, we scrutinized the differences between class 2 and 3 target sites in further detail. First, we confirmed the interaction between DNAme and FOXA2 using in vitro electro-mobility shift assays and found no preference for methylated, hemi-methylated or unmethylated DNA (Supplementary Figs. 7a,b). We next selected a stringent subset of class 3 targets with high methylation (≥80%) in uninduced BJFOXA2 (Fig. 5a; Class 3–1, n=5,253) that were comparable to the mean methylation of class 2 targets (>80%). Importantly, FOXA2 enrichment does not correlate with change in DNAme levels at class 3–1 targets (Supplementary Fig. 7c) and both classes 2 and 3–1 targets are largely indistinguishable in their genomic location and CpG density (Fig. 5c; mean CpG count 4.2 and 4.8 for class 2 and 3–1 respectively). However, upon closer inspection, we found that the class 2 targets were comparatively depleted of CpG dinucleotides around the FOXA2 peak summit (Fig. 5d). Additionally, the distance from the peak summit to the nearest CpG was significantly greater for class 2 versus class 3–1 targets (Supplementary Figs. 7d,f; average 74bp and 90bp, respectively p<2.2−16), while the average methylation for these nearest CpGs pre-FOXA2 induction was indistinguishable (Supplementary Figs. 7e,f; p=0.95, average 95% methylated in both). Of note CpGs across the bound regions were increasingly demethylated towards the summit center (Fig. 5e and Supplementary Fig. 7g), which combined with limited observed chromatin dynamics (Supplementary Fig. 7h,i) suggests that loss of DNAme is unlikely the result of recruited histone-modifying enzymes and may be directly linked to FOXA2 occupancy.
Figure 5 |. Distance dependent loss of DNAme at selective FOXA2 targets.
a) Heat map of CpG methylation levels for matched CpGs comparing BJ WGBS (pre-FOXA2 induction) and post-FOXA2 induction ChIP bisulfite sequencing data. Three main classes of FOXA2 binding are displayed. Gray bar in Class 3 indicates the subset of dynamic regions that have a pre-induction methylation level of greater than 80% (regions n=5253) and are further referred to as Class 3–1.
b) Representative IGV browser tracks showing BJFOXA2 FOXA2 enrichment, CpG methylation pre- FOXA2 induction and FOXA2 ChIP-BS data post induction. Top half is an example of a class 2 region (remain hypermethylated) located at chr12:54,002,592–54,021,127. The bottom shot is an example of a class 3 region (dynamic) located at chr18:32,911,411–32,941,267. Zoomed view displays locations of CpG with respect to peak enrichment summit.
c) Violin plot of CpG density of class 2 and class 3 target sites. CpG density is calculated by the number of CpG dinucleotides across 100bp windows divided by total number of base pairs. Dots represent median values while black lines indicate interquartile range.
d) Composite plots showing normalized CpG count of class 2 and class 3 target sites (left). Class 2 is depleted for CpGs toward the center of the peak while class 3 targets are enriched. Mean sequencing coverage between class 2 and class 3 target sites is equivalent (right).
e) Box plot show the percent methylation of CpGs within 20bp windows from the summit of the peak and extended up to 200bp. Methylation measurements were taken from ChIP-BS data after FOXA2 induction. Class 2 black. Class 3–1 blue. Boxes indicate interquartile range and whiskers show maximum and minimum values. Outliers are removed.
Loss of DNAme is dependent on DNA replication
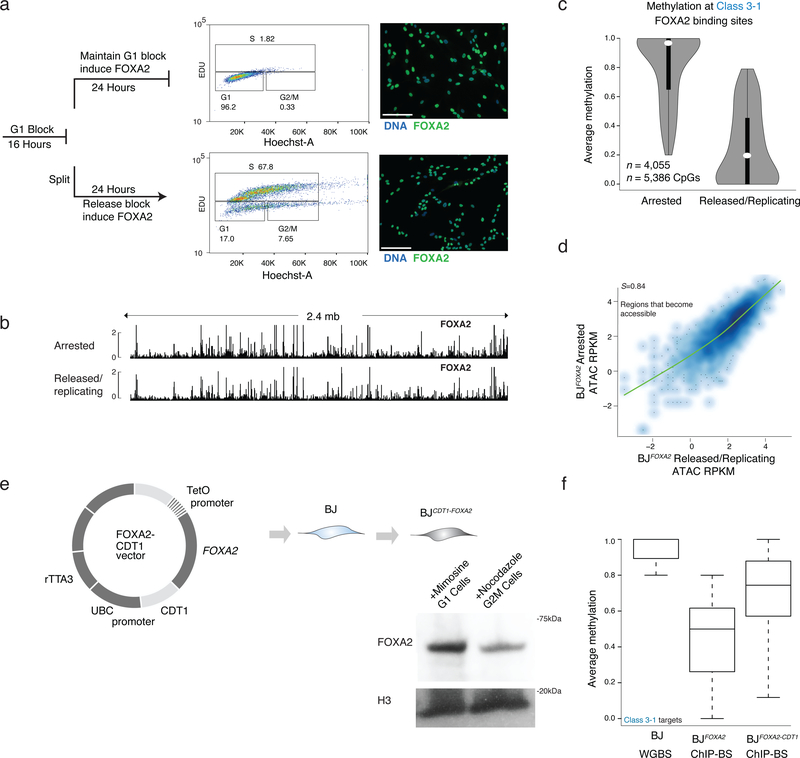
Transitioning a cytosine from methylated to unmethylated requires either an active, enzymatic removal of the methyl group32, a passive, replication dependent loss which would require blocking any maintenance activity following nascent DNA synthesis or a combination of both33,34. To investigate this mechanism at the class 3 FOXA2 targets we reversibly halted BJFOXA2 cells in G1-phase prior to DNA replication with mimosine treatment and induced FOXA2. We subsequently either continued mimosine treatment, or released cells allowing replication (verified by 5-ethynyl-2’-deoxyuridine (EdU) incorporation, approximately 1–2 rounds of cell division Supplementary Fig. 8a), collected cells and performed FOXA2 ChIP-BS-seq (Fig. 6a; Supplementary Tables 7,8). Notably, FOXA2 occupies similar genomic regions in both conditions indicating that even in arrested cells, FOXA2 protein can access similar DNA targets (Fig. 6b; Supplementary Fig. 8b). Quite strikingly however, the dynamic loss of DNAme at the FOXA2 occupied regions was only observed under the replicating conditions (Fig. 6c). Arrested cells displayed no measurable decrease in DNAme levels, despite changes in DNA accessibility (Fig. 6d). Taken together this highlights that FOXA2 binding and its affects on DNA accessibility are replication independent, while loss of DNAme is dependent on it.
Figure 6 |. Loss of DNA methylation but not binding nor nucleosome remodeling is dependent on DNA replication.
a) Cells were halted before DNA replication by blocking the BJFOXA2 cells from progressing in G1 with mimosine, then either released into normal replicating conditions by cell splitting and washing out the chemical treatment or maintaining them halted by persisted treatment with mimosine. FOXA2 was simultaneously induced in both for 24h. FACS analysis using EdU incorporation (488nm) and Hoechst DNA stain shows that halted cells remain in G1 and cells that are removed from chemical block can proliferate at a normal rate. Both populations of cells express FOXA2 highly as shown by immunostaining on the right. White scale bar is equal to 345nm.
b) Representative IGV browser tracks of a 2.4mb window (chr8:57,156,834–59,597,409) showing FOXA2 ChIP-seq data from cells halted in G1 (top track) and cells that were halted and then released back in to normal cycling conditions (bottom track). FOXA2 binding and accumulation is visually similar in both experiments.
c) Violin plots of the average methylation levels for class 3–1 between cells arrested in G1 and cells in normal replicating conditions. Dynamic regions no longer lose DNAme when cells are arrested in G1. Dots represent median values and bar indicates interquartile range.
d) Scatter plot of ATAC-seq signal post FOXA2 induction at regions that become accessible upon FOXA2 binding in G1 arrested cells compared to cells released back into normal cycling conditions. Spearman correlations show the samples are highly correlated.
e) Schematic representation of FOXA2-CDT1 fusion lentiviral construct generated with corresponding cropped western blot for FOXA2 in BJFOXA2-CDT1 cells treated with Mimosine to enhance proportion of cells in G1 and Nocadazole to enhance proportion of cells in G2/M. H3 levels and WT protein levels are shown as loading control.
f) Box plots show average methylation of class 3–1 targets in BJ WGBS, BJFOXA2 ChIP-BS and BJFOXA2-CDT1 ChIP-BS data. Calculated based on regions that had at least 10X coverage. Boxes indicate interquartile range and whiskers show maximum and minimum values. Outliers are removed.
Mechanistically, we hypothesized that the immediate recruitment of FOXA2 to target regions following DNA replication (S-phase) may be sufficient to block maintenance methylation of DNMT1. To explore this, we generated BJ fibroblasts expressing FOXA2 fused to CDT135 (BJFOXA2-CDT1) to specifically deplete FOXA2 expression during S-phase (Fig. 6e). Quantification of FOXA2 protein levels indicates differential expression in G1 compared to G2/M with residual protein observed in S-M that may be the result of our super-physiological expression (Fig. 6e). To ensure similar target site enrichment of FOXA2 in this new system, we performed ChIP-seq for FOXA2 in G1 arrested BJFOXA2-CDT1 and observed high correlation of FOXA2 enrichment in arrested BJFOXA2-CDT1 compared to arrested and/or released BJFOXA2 (Supplementary Fig. 8c, d). We then induced FOXA2 for four days and performed FOXA2 ChIP-BS-seq in normal cycling conditions and observed a reduced loss of DNAme at class 3–1 targets in BJFOXA2-CDT1 cells compared to wildtype BJFOXA2 (Fig. 6f, Supplementary Fig. 8e,f). Taken together the results suggest that FOXA2 occupancy in S-phase may be needed to induce targeted loss of DNA methylation.
Discussion
Here we first compiled a set of cis-regulatory elements that are occupied by FOXA2 in endogenously expressing cell types (HepG2, A549 and dEN cells) and established that ectopic expression of FOXA2 in BJ fibroblasts does not recapitulate the high enrichment occupancy at most of these endogenous targets despite super-physiological expression levels. Instead, we observe a minimal overlap across endogenous and ectopic data sets but observe broad, low-level enrichment (sampling) across most regions occupied by FOXA2 in alternative lineages. Sampling may occur as a result of proposed slow chromatin scanning of pioneer factors36, yet recent evidence from single-molecule tracking studies suggested that pioneer factors may in actuality, have quick DNA residence times that are similar to non-pioneer TFs12. From these experiments, we cannot distinguish if sampled sites are bound with high frequency in a small number of cells or if there is more transient binding at these regions across all cells as standard ChIP-seq signals are averaged across populations. Future application of competition ChIP experiments or similar approaches might yield further insight.
Nevertheless, sampling seems to be a distinctive characteristic of FOXA2 and GATA4 as OCT4 occupancy appeared to predominantly exhibit only highly enriched, cell type specific targets. This is in agreement with a recent study that shows mouse OCT4 also occupies distinct genomic regions when it is expressed alone versus with other reprogramming factors37. Similarly, the authors also observe that enriched OCT4 targets coincide mainly with pre-existing open chromatin regions when expressed independently37. Sampling of alternative target sites may therefore be a defining pioneer TF quality as we observe this for both FOXA2 and GATA4, and that factors like OCT4 may act as a pioneer TF only in specific cellular contexts, but not universally.
TFs uniquely expressed in individual cell types appear to play a partial role in FOXA2 occupancy at genetically encoded targets that FOXA2, itself, can only sample, and not occupy with high enrichment. Modest changes in pioneer factor occupancy due to co-factor expression has also recently been observed by others12,13. The cooperativity we observe between FOXA2 and GATA4 at this specific subset of target sites appears to be distinct from the dynamic assisted loading model of TF binding12,22 as we find little change in DNA accessibility upon co-occupancy of these regions. Given that co-expression of GATA4 only resulted in stabilized binding at a subset of potential sites, it is possible that the precise levels and/or additional factors contribute to FOXA2 occupancy. It will be interesting to investigate how other TFs (including non-pioneer factors), as well as modulations to cofactor motif sequences at particular loci affect pioneer factor occupancy.
It is possible that the known cooperativity of FOXA2 with repressors may explain the limited gain in DNA accessibility observed at the majority of target sites7,38,39. Yet sites that gain significant accessibility concomitantly gain modest enrichment of phased and modified histones potentially indicating the recruitment of chromatin remodeling machinery to this subset of target sites. Additionally, FOXA2 may specifically displace linker histone H1 at this subset of regions leading significant gain in accessibility3.
Mechanistic investigations into the loss of methylation seen at some FOXA2 targets uncovered a dependence on DNA replication as cells arrested in G1 fail to dynamically lose DNAme despite occupancy and change in DNA accessibility. Our study suggests that S-phase binding of FOXA2 may occur rapidly after nascent strand synthesis yet prior to maintenance methylation. The observation that proximity of CpG dinucleotides to the FOXA2 peak summit is an important distinguishing characteristic of targets that become demethylated compared to targets that do not supports a model where occupancy directly interferes with the DNAme machinery. A recent study speculated that loss of DNAme at FOXA1 targets resulted from an active demethylation mechanism involving DNA repair enzymes40, though the authors did not examine DNAme loss in the absence of DNA replication nor report the active demethylating enzyme. Therefore alternative possibilities remain and more work needs to be done to clarify the molecular mechanism. Our data show a clear dependence on DNA replication for the dynamic loss of DNAme, but we cannot rule out that this does not occur via an active mechanism following replication. Our systemic comparison of endogenous and ectopic TF behaviors provides relevant mechanistic details towards a more complete understanding of pioneer factors and their rational application towards cellular reprogramming.
Methods
Cell culture
Clonal FOXA2 doxycycline inducible cells lines were derived from an immortalized BJ foreskin fibroblast cell line from ATCC (BJ-5ta; CRL-4001). Cells were cultured in MEM-Alpha (Life technologies: 32561–037) with 10% FBS, 1% pen-strep, 0.01mg/mL hygromycin B and 5ng/mL bFGF. Derived BJFOXA2 lines were grown in the same conditions plus 0.5ug/mL Puromycin.
BJ cell line generation
Cells were infected with pTRIPZ-FOXA2, pTRIPZ-RFP at an MOI ~1. Following infection, cells were selected with Puromycin (1ug/mL) and replated at a high dilution to ensure separation for clonal expansion and isolation. After two weeks of growth, individual clones were picked, expanded and screened. Criteria for inclusion in the current study included uniform expression of FOXA2 and minimum basal FOXA2 expression in uninduced controls. Clones were maintained in 0.5ug/mL puromycin containing media following expansion. To induce FOXA2, doxycycline was added at 0.5ug/mL.
Cloning and Constructs
To generate pTRIPZ-FOXA2, RFP, FOXA2-CDT1 clones, pTRIPZ inducible lentiviral vector (Thermo Scientific) and full-length FOXA2 were assembled using Gibson Assembly® Master Mix (NEB). pTRIPZ empty vector was digested with XhoI and MluI to remove shRNAmir regulatory sequences, and digested ends were blunted. The linearized pTRIPZ backbone was digested with BsiWI to generate two fragments, each with one sticky end. The fragments were gel extracted, purified, and ligated using the Quick LigationTM Kit (NEB). Primer sequences are listed in Supplementary Table 1.
To generate HaloTagged-FOXA2 construct, full-length FOXA2 was ligated to pFN21A (Promega)
GATA4-V5 and POU5F1-V5 constructs were obtained from the Broad Institutes Genomics Perturbations platform and are available to purchase through Thermo Fisher.
Protein purification
293Ts were transfected with pFN21A-FOXA2. Purification was completed following Promega’s Halotag Protein Purification System. Briefly, 48 hours following transfection, cells were harvested, lysed and gently sonicated four times on Branson Sonifier at 10% amplitude for 15 seconds. Sample was diluted 1:3 with protein purification buffer (1X PBS, 1mM DTT, 0.0005% NP-40) and centrifuged to remove debris. Halo-Resin was washed in purification buffer, added to lysate and incubated at 4C overnight. After incubation resin was washed and FOXA2 protein was cleaved via the addition of TEV-protease during an over night incubation at 4C. Purified protein was assessed via commassie blue gel and western blot.
EMSA
EMSA was performed using LightShift Chemiluminescent EMSA kit (Pierce). Purified halotagged-FOXA2 protein (3–6ug) was mixed with duplexed, biotinlyated probes (20fmol/ul) without competitor DNA. Unlabeled probes (non-biotinlyated) were added at 10X-100X concentrations of biotinlyated probes. Binding reactions were incubated for 20 minutes at room temperature before loading onto a 6% DNA retention gel (Invitrogen). Complexes were transferred to nylon membrane (Invitrogen) and crosslinked via UV radiation in Statalinker. Biotinlyated DNA was detected by chemiluminescence.
Chomatin Immuoprecipitation
Cells were crosslinked with 1% formaldehyde for 10 minutes at room temperature, and quenched with 125mM glycine at room temperature. ChIP was performed as previously described18 by isolating nuclei and shearing DNA to 200–600 basepair fragments using Branson sonicator. Antibody incubation with chromatin was performed overnight. ~10 million cells were used per FOXA2 ChIP with 1ug of antibody/ million cells. ~1 million cells were used for each histone ChIP. Following an overnight incubation, antibody-protein complexes were isolated using Protein G/A beads (Life Technologies) and sequencing libraries were generated. Libraries were generated as previously described18,42 and libraries were sequenced on a HiSeq 2500 at 11pmol.
Chomatin Immuoprecipitation Bisulfite Sequencing
To generate bisulfite converted DNA libraries following ChIP, we used Nugen Ovation UltraLow Methyl-seq Kit (0335–0336). Bisulfite conversion was performed with EpiTect Bisulfite kit (Qiagen) with carrier DNA. Libraries were sequenced on a HiSeq 2500 8pmol with 35% PhiX spike in.
Antibodies
Chips were performed using: FOXA2 (R&D: AF2400), H3K4me1 (Millipore 17–614), H3K4me2 (ActivMotif: 39141), H3K27ac (ActivMotif: 39133), H3K27me3 (ActivMotif: 39155), V5 (MBL: M167–3), OCT4 (ActivMotif: 39811)
Immunostaining performed with the following antibodies: FOXA2 (R&D: AF2400), V5 (MBL: M167–3), OCT4 (ActivMotif: 39811)
Western blots: FOXA2 (R&D: AF2400), V5 (MBL: M167–3), H3 (Abcam: ab1791), OCT4 (ActivMotif: 39811). Complete western blots are shown in Supplementary Figure 9.
IGV Browser shots
All browser shots were created in Illustrator by exporting .svg files from Integrated Genome Viewer (IGV). Data were imported into IGV as normalized TDF files and scaled to the same values (2) unless otherwise specified. Genomic location displayed is listed in figure legend.
Whole genome bisulfite sequencing
WGBS was performed as previously described using Swift Acell-NGS Methyl-Seq DNA kit.
ATAC sequencing
Tagmentation was performed on whole nuclei at 37C for 45 minutes as previously described in Ref 21. DNA was isolated on PCR min-elute columns (Qiagen) and a small amount of the DNA was amplified for 9, 12 and 15 cycles to determine optimal cycling conditions. The rest of the DNA was then amplified using the chosen cycle number and PCR libraries were purified using double-sided Ampure clean up to remove high molecular weight fragments. 0.55x Ampure volume was added to the PCR, mixed and incubated. Supernatant was removed following magnet separation and cleaned-up with a 1X Ampure volume. Libraries were sequenced on a HiSeq 2500 at 8pmol.
RNA sequencing
RNA was isolated with RNeasy columns (Qiagen) and non-stranded libraries were performed using Illumina’s standard Tru-Seq kit. Libraries were sequenced on a HiSeq 2500 at 11pmol.
RTq-PCR
cDNA synthesis was performed with 600–2000 ng of RNA using the RevertAid TM First Strand cDNA Synthesis Kit (Thermo Scientific) with oligo(dT)18 primer. Quantitative PCR (qPCR) primers were designed with Primer-BLAST (NCBI). Primers were designed to span an exon-exon junction, amplify 70–200 bp of cDNA, and amplify all isoforms of a transcript. qPCR was performed with 3–4 technical replicates using a 1:100 or 1:1000 dilution of cDNA, Power SYBR Green Master Mix (Applied Biosystems) and 500 nM of forward and reverse primers on the ViiATM 7 Real-Time PCR System (Applied Biosystems). ACTB and HPRT1 were used as endogenous controls. Relative gene expression was calculated with the comparative CT (ΔΔCT) method using ExpressionSuite Software v1.0.3 (Biosystems).
Western
Nuclear proteins were extracted in standard RIPA buffer supplemented with protease inhibitors (Roche). Equal amounts of extracts were mixed with LDS (Life Technologies) and BME and boiled at 95C for 5 minutes. Samples were loaded onto a NuPage Novex 4–12% Bis-Tris gel (Life technologies) and electrophoresed for 1 hour at 200 volts in 1X MES buffer (Life Technologies). Proteins were transferred to PVDF membrane via iBlot transfer system (Life technologies). Membranes were blocked in 5% Milk/TBST for 1 hour at room temperature and membranes were incubated with primary antibodies in 5% Milk/TBST over night at 4C. FOXA2 primary antibody was diluted at 1:5000 and H3 primary antibody was diluted at 1:10,000. Membranes were washed and incubated in secondary antibodies in TBST at 1:10,000 dilution. Detection was performed with SuperSignalTM West Dura Chemiluminescent Substrate (Thermo Scientific).
Immunostaining
Cells were fixed in 4% formaldehyde for 15 minutes at room temperature. After washing, permeabilization and blocking was performed with 4% FBS/0.4% Triton in PBS for 1 hour at room temperature. Primary antibody staining was performed with 2% FBS/0.2% Triton in PBS overnight at 4°C. Secondary staining was performed with fluorophore-conjugated antibodies in PBS for 1 hour at room temperature.
Cell cycle arrest and FACS analysis
Cells were halted in G1 by the addition of 500mM Mimosine (Sigma) treatment over night.
Cell proliferation was determined using the Click-iT® Plus EdU Flow Cytometry Assay Kit (Life Technologies). 5 μM EdU was added to culture medium, and samples were incubated for 18 hours. Samples were then fixed, permeabilized, and treated with Click-iT® EdU reaction cocktail according to kit instructions. Hoechst and/or Vybrant Dye (Life technologies) were diluted 1:1000 to measure DNA content. FACs analysis was performed on a BD LSR II flow cytometry machine.
ChIP-seq Analysis
All FOXA2 ChIP-seq dataset from different conditions and cell-types were aligned using Bowtie 2 (Ref 43) to hg19 human genome reference assembly using default parameters. Duplicate reads were removed using Picard (http://broadinstitute.github.io/picard/). Genome browser images were created by converting bam files in .tdf files using IGV tools44 by normalizing them to 1 million reads. All data sets were subjected to irreproducible discovery rate (IDR) framework45 with 0.1 as cutoff in combination with using MACS2 (Ref 46) for calling peaks in each replicate separately. For MACS2 peak calling we used corresponding whole cell extract (WCE) as background control and p-value cutoff of 0.01. This initial peak calling using IDR and MACS2 resulted in a set of peaks that are above background for each cell-type. As an additional filtering and also for making peaks from different cell-types and conditions more easily comparable, we developed an in-house computational framework to redefine relative peak positions and to standardize peak width. IDR called peaks from all cell-types were merged together if they were found to be overlapping by at least 20% while keeping track of the summits of the peak that are being merged together. This resulted in a master peak set encompassing all FOXA2 datasets. As several peaks having different peak summits were merged together, we devised a simple weighted framework to define new peak submit. To assign a new peak summit we used the peak height as a measure of weighed distance from peak center. Using this weighted measure of peak height, we calculated a new peak summit, which would be most close to the highest peak that was merged but will also represent contributions from smaller peaks in a distance dependent manner. All peaks were assigned new peak summits using this formula. To define new peak widths we extended each peak by 300bp in both directions from the peak summit to have all peaks of 600bp. Enrichment of different histone marks at these FOXA2 peaks was calculated using standard RPKM formula.
Composite plots
Composite plots to show enrichment of different histone marks at FOXA2 peaks were made by using Homer package47. As described in Homer documentation, we first created tag directories for each sample or histone mark we wish to plot around peak regions. Peaks were extended by 2000bp in each direction and tag directories were then used to create a matrix having tag densities at each nucleotide while normalizing each library for its respective library size. Matrix file having tag density at each position of extended 4000bp window was imported in R to create the plots.
Read Density Heatmaps
Read density heatmaps were created by using EnrichedHeatmap and ComplexHeatmap (https://github.com/jokergoo/EnrichedHeatmap, https://github.com/jokergoo/ComplexHeatmap) package. We first created genome-wide coverage of each sample or histone mark using coverageBed from BEDTools package 48. These coverage files and the peaks regions that are required to be plotted were supplied as input to ComplexHeatmap. Heat on each heatmap was decided based on percentile range by capping the maximum at 99th percentile to remove outliers.
Differential Motif Analysis
Differential motif analysis was performed using Homer47. To calculate differential enrichment between two sets of peaks, we used provided one set as background and vice versa. The motifs were scanned in 200bp region around the peak center in both directions.
Chromatin State Maps
In order to classify FOXA2 bound regions in different chromatin states, we used a hierarchical classification system. All FOXA2 peaks that had either H3K27ac or H3K4me1 were marked as “active”. Excluding these “active” regions, regions having ATAC signal (RPKM) above 3 were marked as “open” and regions having H3K27me3 were classified as “repressed”. After classifying all histone marks, we divided rest of the regions based on their DNAme levels. Regions having DNAme levels below 20% were marked as low methylated regions (LMR), regions having methylation level between 20% and 60% were called intermediate methylated regions (IMR) while those above 60% methylation were termed as hypermethylated regions (HMR). The dynamics of transitions in chromatin state for each peak between different cell types (BJ, hESC and dEN) was visualized using a heat map.
ChIP-BS-seq Analysis
For the analysis of methylation changes associated with FOXA2 binding, we redefined binding sites to maximize overlap with our ChIP-BS-seq data as bisulfite conversion on small amounts of input DNA results in material loss. To do this, we combined FOXA2 ChIP-seq data from BJFOXA2 cells 4days and 10days post induction. The summit of each peak was determined using MACS, then the region 200bp either side was selected, and overlapping regions merged to generate a list of 113,398 sites. We then intersected these regions with BJ fibroblast WGBS and 4day BJFOXA2 ChIP-BS-seq datasets and selected only CpGs covered by at least 3 reads in both samples, giving a total of 42,086 sites and 135,785 CpGs. We used these same regions to select matched CpGs covered at ≥3X in the ChIP-BS-seq data from the BJFOXA2 mimosine treated and released samples (n = 13,494 sites and 18,429 CpGs). We compared individual CpG and mean FOXA2 binding site methylation, generated CpG count and coverage plots and calculated the distances between the summit and nearest CpG using custom R scripts. For comparison to ATAC seq data (described above), we used HOMER 47 to generate enrichment composite plots for 2kb either side of our peaks.
Statistical Methods
All p-value calculations were done using Wilcoxon test unless otherwise stated.
Supplementary Material
Supplementary Table 1: FOXA2 enrichment
Table containing FOXA2 union, IDR peak set with normalized FOXA2 enrichment values listed as RPKM across all union cell types, co-expression experiments and G1 halted ChIP experiments. Corresponding enrichment values of selected histone modifications, ATAC-seq and WGBS data utilized in Figure 2 listed for each region as normalized RPKM values across all union cell types.
Supplementary Table 2: GATA4 enrichment
Table containing OCT4 union, IDR peak set with normalized OCT4 enrichment values listed as RPKM across all union cell types. Corresponding enrichment values of selected histone modifications, ATAC-seq and WGBS data utilized in Figure 2 listed for each region as normalized RPKM values across all union cell types.
Supplementary Table 3: OCT4 enrichment
Table containing GATA4 union, IDR peak set with normalized GATA4 enrichment values listed as RPKM across all union cell types, and co-expression experiments. Corresponding enrichment values of selected histone modifications, ATAC-seq and WGBS data utilized in Figure 2 listed for each region as normalized RPKM values across all union cell types.
Supplementary Table 4: Alignment of data
Table containing number of aligned and unaligned reads per sequencing experiment performed.
Supplementary Table 5: Expression analysis uninduced versus FOXA2 induced
Table containing normalized FPKM expression values at all genes in uninduced BJs versus 4 day FOXA2 induced BJ fibroblasts.
Supplementary Table 6: FOXA2 ChIP-BS-seq
Table containing FOXA2 bound regions, CpGs covered and percent methylation in FOXA2 ChIP-BS-seq experiment from Figure 5.
Supplementary Table 7: FOXA2, G1 block ChIP-BS-seq
Table containing FOXA2 bound regions, CpGs covered and percent methylation in FOXA2 ChIP-BS-seq experiment from G1 blocked condition in Figure 6.
Supplementary Table 8: FOXA2, replicating ChIP-BS-seq
Table containing FOXA2 bound regions, CpGs covered and percent methylation in FOXA2 ChIP-BS-seq experiment from replicating cell condition in Figure 6.
Acknowledgements
We would like to thank all members of the Meissner lab specifically A. Tsankov for helpful discussions and initial data processing and S. Grosswendt for critically reading the manuscript. A.M. is a New York Stem Cell Foundation Robertson Investigator. This work was supported by the New York Stem Cell Foundation, NIH grant 1P50HG006193 and the Max Planck Society.
Footnotes
Data accession: All data have been deposited in GEO under accession number GSE90456.
ReferencesUncategorized References
- 1.Spitz F & Furlong EE Transcription factors: from enhancer binding to developmental control. Nature reviews. Genetics 13, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Cirillo LA et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Molecular cell 9, 279–289 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Iwafuchi-Doi M et al. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Molecular cell 62, 79–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwafuchi-Doi M & Zaret KS Pioneer transcription factors in cell reprogramming. Genes & development 28, 2679–2692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soufi A, Donahue G & Zaret KS Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soufi A et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaret KS & Carroll JS Pioneer transcription factors: establishing competence for gene expression. Genes & development 25, 2227–2241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupien M et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaret KS & Mango SE Pioneer transcription factors, chromatin dynamics, and cell fate control. Current opinion in genetics & development 37, 76–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D & Carroll JS FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nature genetics 43, 27–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swinstead EE et al. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell 165, 593–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z & Kraus WL Catalytic-Independent Functions of PARP-1 Determine Sox2 Pioneer Activity at Intractable Genomic Loci. Molecular cell 65, 589–1704578560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco HL, Nagari A & Kraus WL TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Molecular cell (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuteja G, Jensen ST, White P & Kaestner KH Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic acids research 36, 4149–4157 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Schug J, Tuteja G, White P & Kaestner KH The nucleosome map of the mammalian liver. Nature structural & molecular biology 18, 742–746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirillo LA et al. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. The EMBO journal 17, 244–254 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gifford CA et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149–1163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunham I et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey TL & Machanick P Inferring direct DNA binding from ChIP-seq. Nucleic acids research 40(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voss TC et al. Dynamic Exchange at Regulatory Elements during Chromatin Remodeling Underlies Assisted Loading Mechanism. Cell 146, 544–554 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gualdi R et al. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes & development 10, 1670–1682 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Zaret K Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Developmental biology 209, 1–10 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Bossard P & Zaret KS GATA transcription factors as potentiators of gut endoderm differentiation. Development (Cambridge, England) 125, 4909–4917 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Tsankov AM et al. Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344–349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S & Carroll JS FOXA1 Directs H3K4 Monomethylation at Enhancers via Recruitment of the Methyltransferase MLL3. Cell reports 17, 2715–2723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sérandour AA et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome research 21, 555–565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang A et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell stem cell 16, 386–399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurman RE et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brinkman AB et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome research 22, 1128–1138 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohli RM & Zhang Y TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith ZD & Meissner A DNA methylation: roles in mammalian development. Nature reviews. Genetics 14, 204–220 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Smith ZD & Meissner A The simplest explanation: passive DNA demethylation in PGCs. The EMBO journal 32, 318–321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaue-Sawano A et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Sekiya T, Muthurajan UM, Luger K, Tulin AV & Zaret KS Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes & development 23, 804–809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chronis C et al. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell 168, 442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekiya T & Zaret KS Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Molecular cell 28, 291–303 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J-C et al. Transducin-like enhancer of split proteins, the human homologs of Drosophila groucho, interact with hepatic nuclear factor 3. Journal of Biological Chemistry 275, 18418–18423 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y et al. Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nature genetics 48, 1003–1013 (2016). [DOI] [PubMed] [Google Scholar]
- 41.G, S.R.a.B. DiffBind: differential binding analysis of ChIP-Seq peak data. (2011).
References
- 42.Mikkelsen TS et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell 143, 156–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson JT et al. Integrative genomics viewer. Nature biotechnology 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landt SG et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome research 22, 1813–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 9(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlan AR Current Protocols in Bioinformatics. wiley; (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McPherson CE, Horowitz R, Woodcock CL, Jiang C & Zaret KS Nucleosome positioning properties of the albumin transcriptional enhancer. Nucleic acids research 24, 397–404 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ang SL et al. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development (Cambridge, England) 119, 1301–1315 (1993). [DOI] [PubMed] [Google Scholar]
- 51.Ang SL & Rossant J HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78, 561–574 (1994). [DOI] [PubMed] [Google Scholar]
- 52.Gao N et al. Foxa1 and Foxa2 maintain the metabolic and secretory features of the mature beta-cell. Molecular endocrinology (Baltimore, Md.) 24, 1594–1604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grant CE, Bailey TL & Noble WS FIMO: scanning for occurrences of a given motif. Bioinformatics (Oxford, England) 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: FOXA2 enrichment
Table containing FOXA2 union, IDR peak set with normalized FOXA2 enrichment values listed as RPKM across all union cell types, co-expression experiments and G1 halted ChIP experiments. Corresponding enrichment values of selected histone modifications, ATAC-seq and WGBS data utilized in Figure 2 listed for each region as normalized RPKM values across all union cell types.
Supplementary Table 2: GATA4 enrichment
Table containing OCT4 union, IDR peak set with normalized OCT4 enrichment values listed as RPKM across all union cell types. Corresponding enrichment values of selected histone modifications, ATAC-seq and WGBS data utilized in Figure 2 listed for each region as normalized RPKM values across all union cell types.
Supplementary Table 3: OCT4 enrichment
Table containing GATA4 union, IDR peak set with normalized GATA4 enrichment values listed as RPKM across all union cell types, and co-expression experiments. Corresponding enrichment values of selected histone modifications, ATAC-seq and WGBS data utilized in Figure 2 listed for each region as normalized RPKM values across all union cell types.
Supplementary Table 4: Alignment of data
Table containing number of aligned and unaligned reads per sequencing experiment performed.
Supplementary Table 5: Expression analysis uninduced versus FOXA2 induced
Table containing normalized FPKM expression values at all genes in uninduced BJs versus 4 day FOXA2 induced BJ fibroblasts.
Supplementary Table 6: FOXA2 ChIP-BS-seq
Table containing FOXA2 bound regions, CpGs covered and percent methylation in FOXA2 ChIP-BS-seq experiment from Figure 5.
Supplementary Table 7: FOXA2, G1 block ChIP-BS-seq
Table containing FOXA2 bound regions, CpGs covered and percent methylation in FOXA2 ChIP-BS-seq experiment from G1 blocked condition in Figure 6.
Supplementary Table 8: FOXA2, replicating ChIP-BS-seq
Table containing FOXA2 bound regions, CpGs covered and percent methylation in FOXA2 ChIP-BS-seq experiment from replicating cell condition in Figure 6.