Abstract
Shih-Pin Huang, Feng-Yu Wang, and Tzi-Yuan Wang (2017) The complete mitochondrial genomes of 76 species from 43 genera under Cyprinidae sensu lato were collected to reassess the molecular phylogeny of Opsariichthyinae sensu Liao et al. 2011. The mitogenomes of three species, Candidia barbata, Opsariichthys evolans, and Opsariichthys pachycephalus, were newly sequenced. Phylogenetic trees were reconstructed based on 13 concatenated multiple protein-coding genes with two ribosomal RNA genes. The concatenated dataset provided a new perspective on systematics and relationships. Tree topologies show that a monophyletic group containing Parazacco, Candidia, Nipponocypris, Zacco, and Opsariichthys should belong to the Opsariichthys group. In addition, the present results also strongly support that Candidia and Nipponocypris should be regarded as distinct genera within the Opsariichthys group. Aphyocypris, Yaoshanicus, Nicholsicypris, and Pararasbora form a monophyletic group within Xenocyprididae, distinct from the Opsariichthys group. Furthermore, Hemigrammocypris is nested with four species of Metzia, a genus of ex-Cultrinae in Xenocyprididae. In addition, two major types of distinct stripes - longitudinal and vertical - were observed among species of the Opsariichthys group and were highly correlated with molecular phylogenetic relationships. Such types of vertical and longitudinal stripes presented in the Opsariichthys group might have originated in an ancestor species, after which distinct vertical stripes might have been lost among these cyprinids but retained in the Opsariichthys group.
Keywords: Molecular phylogeny, Mitochondrial genome, Freshwater fish, Cyprinidae, Opsariichthyinae
BACKGROUND
Cyprinidae sensu lato (originally called family Cyprinidae) is the largest family of teleosts in the world, containing 3090 valid species (Eschmeyer et al. 2017). Several recent studies have been carried out to assess the phylogeny and systematics of this group and/or the rest of Cypriniformes based on molecular evidence (Tang et al. 2010 2013; Stout et al. 2016).
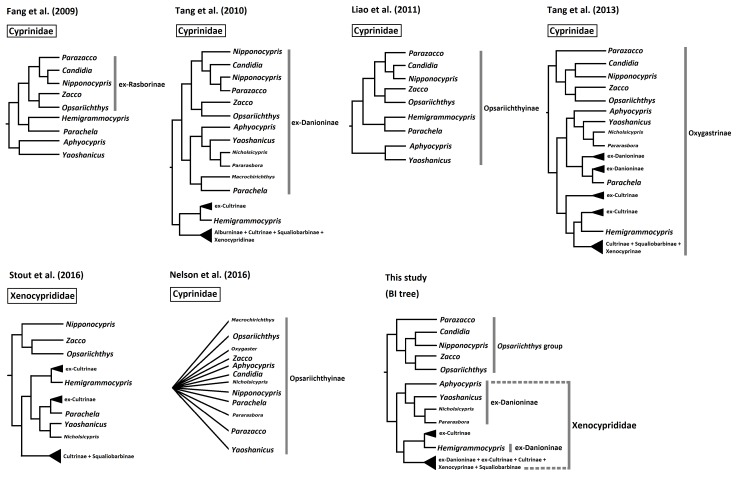
The taxonomic placement of several major subfamilies belonging in Cyprinidae sensu lato has undergone a large change. Several subfamilies, especially Danioninae and Cultrinae, were reported to be paraphyletic or polyphyletic (Tang et al. 2010 2013; Stout et al. 2016). Some were subsequently renamed in an attempt to reflect their new taxonomic placements (Liao et al. 2011c). Among these were Opsariichthyinae, a group of minnows in Cyprinidae sensu lato occurring widely in East Asia that contains the genera Aphyocypris, Candidia, Hemigrammocypris, Nipponocypris, Opsariichthys, Parachela, Parazacco, Yaoshanicus and Zacco (Liao et al. 2011c) (Fig. 1). Most of these genera were previously assigned to Danioninae. However, their taxonomic assignments have been continuously changed over recent years (Mayden et al. 2009; Tang et al. 2010 2013; Liao et al. 2011a; Stout et al. 2016). Among these common minnows, Yaoshanicus, Nicholsicypris, and Pararasbora were considered junior synonyms of Aphyocypris (Liao et al. 2011b), although Huynh and Chen (2013) still considered Nicholsicypris to be a valid genus. Fang et al. (2009) defined ex-Rasborinae, which included Candidia, Nipponocypris, Opsariichthys, Parazacco, and Zacco. Subsequently, Liao et al. (2011c) renamed ex-Rasborinae as Opsariichthyinae to include Aphyocypris, Candidia, Hemigrammocypris, Nipponocypris, Opsariichthys, Parachela, Parazacco, Yaoshanicus and Zacco (Fig. 1). Recently, these genera along with several other subfamilies (Cultrinae, Hypophthalmichthyinae, Squaliobarbinae, Xenocyprindinae, parts of Alburninae, and Danioninae) were reclassified into Oxygastrinae (Tang et al. 2013). Subsequently, Kottelat (2013) proposed that Oxygastrinae was not available, and instead Hypophthalmichthyinae and Xenocypridinae were the earliest available names. Therefore, the genera and subfamilies were assigned to Xenocyprididae (Stout et al. 2016). These taxonomic placements and assignments will be used and discussed in this study.
The subfamilies Cultrinae, Xenocyprinae, Squaliobarbinae, Alburninae, and Opsariichthyinae were formerly in Cyprinidae sensu lato but considered a monophyletic group by Stout et al. (2016) and therefore reassigned to the family Xenocyprididae. However, the taxonomic placements and relatedness of subfamilies under Xenocyprididae remained ambiguous because there was an insufficient number of taxa and none of the taxonomic assignments were included. For example, two species from former subfamily Cultrinae (Chanodichthys erythropterus and Parabramis pekinensis) were nested with Hypophthalmichthys molitrix, Ctenopharyngodon idella, Elopichthys bambusa, and Squaliobarbus curriculus, which were part of the former subfamilies Leuciscinae and Squaliobarbinae (Stout et al. 2016). This classification was inconsistent with another study (Tang et al. 2013). In addition, the taxonomic status of Hemigrammocypris remained controversial. Liao et al. (2011c) proposed that it should be assigned to Opsariichthyinae. However, Tang et al. (2013) and Stout et al. (2016) proposed that it was closest to Metzia, a genus of ex-Cultrinae (Fig. 1). The systematic positions of these genera are summarized in figure 1.
The complete mitochondrial genome could be regarded as an alternative molecular marker for processing at a higher level of phylogenetic analysis (Saitoh et al. 2006; Mayden et al. 2009; Huang et al. 2016). In order to verify the systematic positions of Opsariichthyinae, a reassessment of molecular phylogenetic analysis was performed. We expected that mitogenomes would be useful for resolving these ambiguous relationships in Opsariichthyinae and Xenocyprididae. Fortunately, complete mitochondrial genomes of many species under Opsariichthyinae and Xenocyprididae have been sequenced (Jang-Liaw et al. 2013a b; Chang et al. 2016; Chen et al. 2016a b). However, none of these studies analyzed the complete mitochondrial genome. Recently, mitochondrial DNA has been frequently used for resolving the taxonomic and phylogenetic problems in East Asian cyprinids (Tsao et al. 2016; Huang et al. 2017). In order to verify taxonomic placement and assignment and attempt to provide a new molecular perspective on different genetic marks, this study analyzed more species and genera from Opsariichthyinae and family Xenocyprididae based on complete mitochondrial genomes.
Among East Asian common minnows, there are three major stripe patterns that can be roughly grouped. One is an indistinct stripe or band on the side of the body and the remaining two are a distinct vertical or longitudinal stripe or band on the side of the body. These minnows occur in five genera in East Asia: Candidia, Nipponocypris, Opsariichthys, Parazacco, and Zacco. Among these, Candidia is endemic to Taiwan, Nipponocypris is restricted to Japan and Korea, Opsariichthys is widely distributed in East Asia, Parazacco is restricted to southern China, and Zacco is distributed in northern China, Korea, and Japan. Candidia, Nipponocypris, and Parazacco have visible longitudinal stripes whereas Opsariichthys and Zacco have several vertically aligned stripes or bars (Wu 1977; Chen and Fang 1999; Nakabo 2013). Besides, all these color patterns are also found within Danio but not within single group of Cypriniformes (McCluskey and Postlethwait 2015). In this study, we attempted to understand whether the different stripe patterns were correlated with taxonomic relationships in these common minnows.
Candidia and Nipponocypris have been considered well-separated genera based on molecular evidence (Liao et al. 2011c; Tang et al. 2013; Huynh and Chen 2013), but several contrary findings have been reported in recent years. For example, a phylogenetic tree of twelve Opsariichthyines species was reconstructed based on mitochondrial Cyt b and COI and nuclear RAG1 and Rh1 (Tang et al. 2010). Although they all belonged to a monophyletic group, Candidia, Nipponocypris, and Parazacco were nested together and therefore formed an indeterminate lineage. Yin et al. (2015) revealed the relationship of Candidia, Nipponocypris, Opsariichthys, Parazacco, and Zacco based on complete mitochondrial genomes. Remarkably, Candidia was only nested within Nipponocypris, which is consistent with Tang et al. (2010). In contrast, two Japanese species of Nipponocypris were assigned to Candidia (Nakabo 2013). Thus, this study also used further molecular research to reassess these inconsistencies and the validity of Nipponocypris.
Fig. 1.
Fig. 1. Systematic positions of the Opsariichthys group and related genera from this and other studies.
MATERIALS AND METHODS
Whole mitogenome collection
Seventy-six species from 43 genera of Opsariichthyinae, Xenocyprididae, and related families were used to reassess their molecular phylogenetic analysis (Table 1). Among the East Asian common minnows, three species, Candidia barbata, Opsariichthys evolans, and Opsariichthys pachycephalus, were sequenced for the first time. Mitogenomes for the remaining nine species (Aphyocypris chinensis, Hemigrammocypris rasborella, Nicholsicypris normalis, Nipponocypris temminckii, Opsariichthys uncirostris, Pararasbora moltrechti, Parazacco spilurus, Yaoshanicus arcus, and Zacco platypus), each of which is the type species of its genus, were retrieved from GenBank (Table 1). For the whole phylogenetic analysis, mitogenomes for Cyprinus carpio and five other cyprinids (Danioridae, Acheilognathidae, Gobionidae, Leucisidae, and Tincidae) were also obtained from GenBank (Table 1) and used as outgroups.
Table 1. Species and their GenBank accession numbers used in this study.
| Species | Accession number | Source |
| Opsariichthys group | ||
| Parazacco spilurus | KF971863 | Chang et al. 2016 |
| Candidia barbata | This study | |
| Candidia pingtungensis | KT725246 | Yin et al. 2015 |
| Nipponocypris koreanus | KJ427719 | Chen et al. 2016a |
| Nipponocypris sieboldii | AB218898 | Saitoh et al. 2006 |
| Nipponocypris temminckii | KM213515 | Chen et al. 2016b |
| Zacco acanthogenys | KT290890 | Yin et al. 2015 |
| Zacco platypus | AP012115 | Miya et al. 2015 |
| Opsariichthys acutipinnis | KT725245 | Yin et al. 2015 |
| Opsariichthys bidens | DQ367044 | Wang et al. 2007 |
| Opsariichthys chengtui | KT725244 | Yin et al. 2015 |
| Opsariichthys evolans | This study | |
| Opsariichthys pachycephalus | This study | |
| Opsariichthys uncirostris | AB218897 | Saitoh et al. 2006 |
| Xenocyprididae | ||
| Hemigrammocypris rasborella | AP011422 | Tang et al. 2010 |
| Metzia longinasus | KF955011 | Ma & Luo 2016 |
| Metzia mesembrinum | NC_023797 | Yuan et al. 2016 |
| Metzia formosae | NC_022458 | Lin et al. 2015 |
| Metzia lineata | NC_031541 | GenBank |
| Aphyocypris chinensis | AB218688 | Saitoh et al. 2006 |
| Aphyocypris kikuchii | JX184925 | Jang-Liaw et al. 2013b |
| Yaoshanicus arcus | AP011398 | Tang et al. 2010 |
| Nicholsicypris normalis | AP011396 | Tang et al. 2010 |
| Pararasbora moltrechti | JX311312 | Jang-Liaw et al. 2013a |
| Macrochirichthys macrochirus | NC_015551 | GenBank |
| Paralaubuca typus | AP011211 | Saitoh et al. 2011 |
| Ctenopharyngodon idella | EU391390 | Wang et al. 2008 |
| Elopichthys bambusa | AP011213 | Miya et al. 2015 |
| Hypophthalmichthys molitrix | KJ729094 | Farrington et al. 2015 |
| Hypophthalmichthys nobilis | KJ746959 | Farrington et al. 2015 |
| Squaliobarbus curriculus | KC351187 | Liu et al. 2013 |
| Xenocypris argentea | AP011283 | Mayden et al. 2009 |
| Xenocypris davidi | KF039718 | Liu 2014 |
| Ischikauia steenackeri | NC_008667 | Saitoh et al. 2006 |
| Chanodichthys mongolicus | KF826087 | Wei et al. 2016 |
| Chanodichthys ilishaeformis | NC_029722 | Li et al. 2016 |
| Chanodichthys dabryi | NC_021418 | Zhang et al. 2014 |
| Culter erythropterus | NC_024749 | Chen et al. 2016 |
| Culter recurviceps | NC_024277 | GenBank |
| Culter mongolicus | AP009060 | Saitoh et al. 2006 |
| Parabramis pekinensis | KF857485 | Duan et al. 2016 |
| Megalobrama amblycephala | NC_010341 | GenBank |
| Megalobrama pellegrini | NC_026458 | Liu et al. 2016 |
| Hemiculter bleekeri | NC_029831 | GenBank |
| Hemiculter leucisculus | NC_022929 | GenBank |
| Hemiculter eigenmanni | NC_029388 | GenBank |
| Cprinidae | ||
| Cyprinus carpio | AP017363 | Mabuchi 2016 |
| Danionidae | ||
| Rasbora vaterifloris | NC_015531 | Tang et al. 2010 |
| Rasbora lateristriata | NC_032723 | Kusuma & Kumazawa 2016 |
| Rasbora trilineata | NC_025336 | Ho et al. 2016 |
| Rasbora steineri | NC_020005 | Chang et al. 2013 |
| Danio dangila | NC_015525 | Tang et al. 2010 |
| Danio erythromicron | AP011419 | Tang et al. 2010 |
| Danio rerio | NC_002333 | Broughton et al. 2001 |
| Acheilognathidae | ||
| Acheilognathus macropterus | NC_013711 | Hwang et al. 2014 |
| Acheilognathus typus | NC_008668 | Saitoh et al. 2006 |
| Rhodeus ocellatus | NC_011211 | He et al. 2008 |
| Rhodeus lighti | NC_024885 | Wang et al. 2016 |
| Rhodeus sinensis | NC_022721 | Yang et al. 2015 |
| Rhodeus shitaiensis | NC_022690 | Li et al. 2015 |
| Tanakia limbata | NC_025515 | Luo et al. 2016 |
| Tanakia lanceolata | NC_024566 | Xu et al. 2016 |
| Gobionidae | ||
| Hemibarbus barbus | NC_008644 | Saitoh et al. 2006 |
| Squalidus gracilis | NC_024561 | Liu et al. 2016 |
| Abbottina rivularis | NC_023781 | He et al. 2013 |
| Gobio gobio | NC_008662 | Saitoh et al. 2006 |
| Rhinogobio typus | NC_024423 | Yan et al. 2016 |
| Gnathopogon elongatus | NC_008649 | Saitoh et al. 2006 |
| Sarcocheilichthys variegatus microoculus | NC_004694 | Saitoh et al. 2003 |
| Leucisidae | ||
| Leuciscus burdigalensis | NC_029426 | Hinsinger et al. 2015 |
| Acrocheilus alutaceus | AP012086 | GenBank |
| Cyprinella lutrensis | NC_008643 | Saitoh et al. 2006 |
| Macrhybopsis storeriana | NC_030485 | Gaughan et al. 2016 |
| Tincidae | ||
| Tinca tinca | AB218686 | Saitoh et al. 2006 |
| Tanichthys micagemmae | NC_031631 | GenBank |
| Tanichthys albonubes | NC_015539 | GenBank |
Mitogenomes by illumina shotgun sequencing
Specimens of Candidia barbata, Opsariichthys evolans, and Opsariichthys pachycephalus were collected from an upstream section of the Keelung River located in the Ruifang District of New Taipei City, Taiwan. Genomic DNA was extracted from 100 mg of muscle tissue using a Roche DNA Isolation Kit (Indianapolis, IN, USA) following manufacturer instructions. Whole-genome shotgun sequencing was employed, and a 400- bp insert library was constructed using the Illumina standard protocol (San Diego, CA, USA). Paired- end sequencing was performed using the Illumina NextSeq system to obtain 1-2 Gb of raw reads from the libraries of C. barbata, O. evolans, and O. pachycephalus. The de novo assembly function of CLC Genomics Workbench vers. 7.0 (CLC Bio, Cambridge, MA, USA) was used to construct contigs. For each species, the mitogenome candidate contig was identified by using BLAST on all contigs to the nucleotide database downloaded from NCBI. All reads were mapped onto the candidate contig and the mitogenome consensus sequence was extracted. MitoFish software was used to annotate protein-coding and RNA genes of the mitogenome consensus sequence (Iwasaki et al. 2013).
Phylogenetic analysis
Nucleotide sequence alignment was visually verified using BIOEDIT vers. 5.9 (Hall 2001). Sequence analyses were conducted using Molecular Evolutionary Genetics Analysis (MEGA) vers. 7.0 (Kumar et al. 2016). MEGA 7.0 was also used for aligning sequences of different lengths and then manual modifications were performed before the phylogenetic analysis. All transfer (t) RNA genes were scanned with tRNAscan-SE 1.21 (Lowe and Eddy 1997).
Bayesian inference (BI) and maximum likelihood (ML) methods were employed for phylogenetic analyses in this study. ML analyses were carried out using MEGA 7.0 (Kumar et al. 2016). Branch support for ML trees were established via bootstrap analyses (with 1000 replications). The best-fit model for sequence evolution was selected using jModelTest v.2.1.3 (Darriba et al. 2012) in the BI analyses. The best- fit model of the ML analyses was selected using MEGA 7.0.
All aligned sequences were analyzed and phylogenetic trees were constructed with BI and ML methods. BI analyses were performed using MrBayes 3.0 (Ronquist and Huelsenbeck 2003) over a total of 106 replications. The posterior probabilities of each node were computed from the remaining 75% of all sampled trees.
RESULTS
New mitogenome annotation
The complete mitochondrial genomes of Candidia barbata, Opsariichthys evolans, and Opsariichthys pachycephalus were amplified and sequenced, obtaining respective lengths of 16,608, 16,656, and 16,612 bp. The complete mitochondrial genomes of these three species consisted of 37 genes, including 13 typical vertebrate protein- coding genes, 22 tRNA genes, two ribosomal (r) RNA genes, and one control region. All genes were encoded on the heavy strand except for the ND6 and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer1, tRNAGlu, and tRNAPro). An illustration of the complete mitochondrial genome of Opsariichthys evolans is shown in figure 2.
Molecular phylogeny of the Opsariichthys group
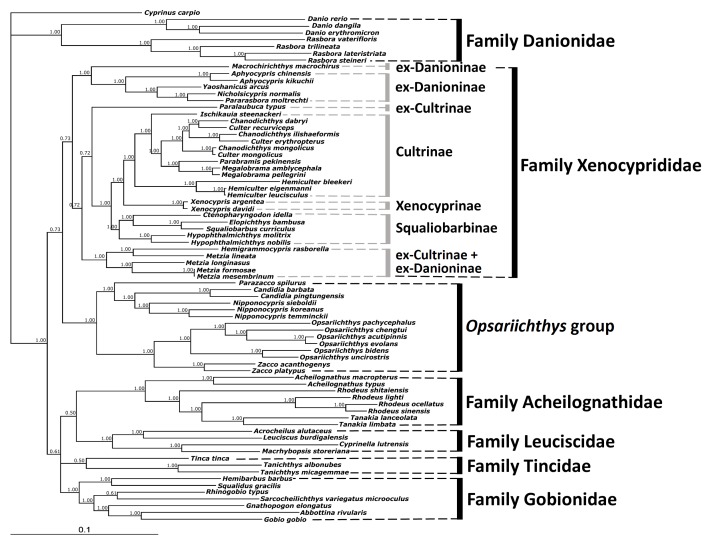
In order to assess the molecular phylogeny of Opsariichthyinae and Xenocyprididae, aligned sequences combined with 13 protein-coding genes and two rRNA genes were used. The lengths of the combined 13 protein-coding genes and two rRNA genes from 76 species were 13,903-14,087 bp in total. This alignment contained 13,287 total mutations and 7,507 polymorphic (segregating) sites, calculated by DNA sequence polymorphisms with DnaSP vers. 5 (Librado and Rozas 2009). The ML tree was reconstructed using concatenated protein-coding genes and rRNA gene sequences based on GTR+G+I models. The HKY+G models were selected as the best-fit models for the BI tree reconstructions based on the concatenated protein-coding genes and rRNA gene sequences. The phylogenetic trees reconstructed using the BI or ML methods based on combined protein- coding and rRNA genes produced slightly different tree topologies (Figs. 3 and 4). BI tree topology revealed that Danionidae is the ancestral group. The remaining OTUs separated into two major lineages. The first one contained the sister pair Xenocyprididae and the Opsariichthys group, which contained five genera of Opsariichthyinae sensu Liao et al. 2011 (Figs. 1 and 3). The second lineage contained the four families Acheilognathidae, Leuciscidae, Tincidae, and Gobionidae. The Xenocyprididae in the rst lineage can be divided into seven clades, three of which followed the traditionally accepted classification (Cultrinae, Xenocyprinae, Squaliobarbinae) while the other four of which were inconsistent (one clade for ex-Cultrinae and ex-Danioninae + ex- Cultrinae, two clades for ex-Danioninae) (Fig. 3). The Opsariichthys group contained two major clades, with Parazacco being sister to Candidia + Nipponocypris in the first clade and Zacco and Opsariichthys a sister pair in the second clade (Fig. 3).
All nodes had high posterior probabilities of 0.72-1.00 in the lineage Xenocyprididae + Opsariichthys group of the BI tree. Relatively low posterior probabilities of 0.50-0.61 occurred at the nodes among the families Acheilognathidae + Leuciscidae + Tincidae + Gobionidae. However, the posterior probability value was as high as 1.00 at the node that separated these two major lineages (Fig. 3).
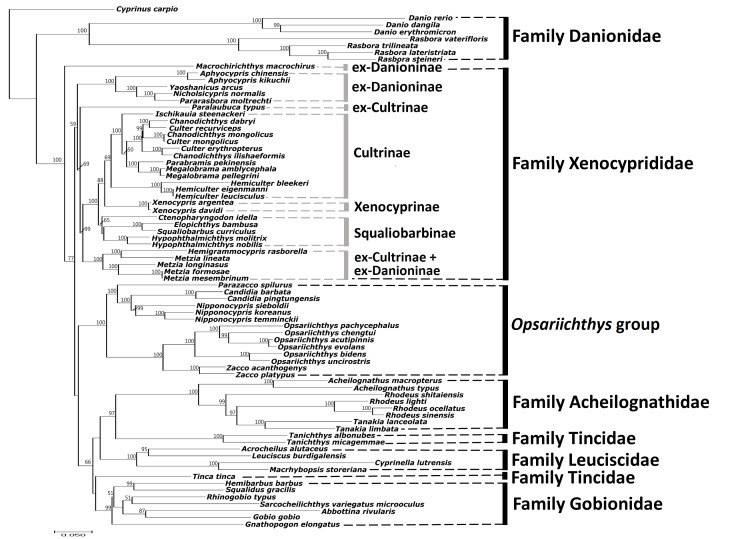
Similar to the BI tree, the ML tree topology revealed that the family Danionidae was the ancestral group but the remainders were divided into three major lineages (Fig. 4). Macrochirichthys macrochirus (ex-Danioninae) formed the first lineage. The second lineage contained the Xenocyprididae lineage, which separated into six clades and three of six clades followed the traditionally accepted classification (Cultrinae, Xenocyprinae, Squaliobarbinae). The third lineage contained the Opsariichthys group, Acheilognathidae, Leuciscidae, Tincidae, and Gobionidae. The ML tree had three different groupings shown in all three lineages when compared to the BI tree in the ML tree, Macrochirichthys macrochirus (ex-Danioninae) formed the first lineage and was outside of the other two lineages. Moreover, the Opsariichthys group was sister to Acheilognathidae + Leuciscidae + Tincidae + Gobionidae. On the other hand, the ML tree revealed that Tincidae was not monophyletic; instead, it was divided into two clades, but with only low bootstrap support (Fig. 4).
In short, the BI tree revealed the Opsariichthys group is sister to Xenocyprididae (Fig. 3). However, the ML tree revealed the inconsistent lineage Opsariichthys group + Acheilognathidae + Leuciscidae + Tincidae + Gobionidae (Fig. 4). These results indicate that higher taxonomic levels might still be unsettled. Nevertheless, ve genera in the Opsariichthys group (Parazacco, Candidia, Nipponocypris, Zacco, and Opsariichthys) indeed formed a monophyletic group distinct to Xenocyprididae (Stout et al. 2016) and other closely-related families in Cypriniformes (Fig. 4).
Fig. 2.
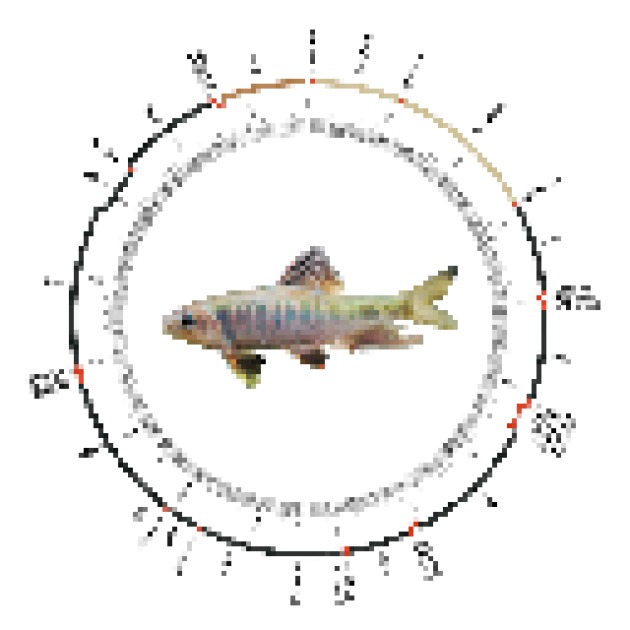
Fig. 2. Mitogenome map of Opsariichthys evolans as a representative species of the Opsariichthys group.
Fig. 3.
Fig. 3. Molecular phylogenetic tree of the Opsariichthys group and related families based on 13 concatenated protein-coding genes and two rRNA gene sequences reconstructed with Bayesian inference (values above the branch are posterior probabilities). Subfamily and family names follow those in Liao et al. (2011c), Nelson et al. (2016), Tang et al. (2013) and Stout et al. (2016).
Fig. 4.
Fig. 4. Molecular phylogenetic tree of the Opsariichthys group and related families based on 13 concatenated protein-coding genes and two rRNA gene sequences reconstructed with the maximum likelihood method (values below the branch are bootstrap numbers, bootstrap values less than 50 not shown). Subfamily and family names follow those in Liao et al. (2011c), Nelson et al. (2016), Tang et al. (2013) and Stout et al. (2016).
DISCUSSION
Molecular evidences (Figs. 3 and 4) reveal a monophyletic Opsariichthys group and further comparative morphological studies might be needed to clarify this taxonomic level in the future.
Opsariichthys group is a monophyletic group
In order to verify taxonomic placements and assignments in the Opsariichthys group, related species from Xenocyprididae (Stout et al. 2016; Tang et al. 2013) were included (Table 1; Fig. 1). Among the two phylogenetic trees reconstructed, the BI tree topology revealed a relatively stable and reliable grouping based on higher posterior probability values and reliable tree topology (Fig. 3). The mitogenomic phylogeny suggests that Opsariichthys is monophyletic and sister to Xenocyprididae. Acheilognathidae, Leuciscidae, Tincidae, and Gobionidae are closely related families as outgroups.
Our results show that Parazacco, Candidia, Nipponocypris, Zacco, and Opsariichthys comprise a stable monophyletic group distinct from Xenocyprididae in both BI and ML trees (Figs. 3, 4). Moreover, all these members were previously confirmed as a monophyletic group (Tang et al. 2013; Stout et al. 2016). Therefore, we propose the Opsariichthys group contains Parazacco, Candidia, Nipponocypris, Zacco, and Opsariichthys.
In addition, Liao et al. (2011b) assigned Aphyocypris and Yaoshanicus to subfamily Opsariichthyinae based on a single mitochondrial cytochrome b gene. Nelson et al. (2016) proposed that several additional genera, including Candidia, Macrochirichthys, Nicholsicypris, Nipponocypris, Oxygaster, Parachela, Pararosbora, and Parazacco, would be assigned to Opsariichthyinae if monophyly of the entire clade is confirmed. Tree topology clearly shows that only Parazacco, Candidia, Nipponocypris, Zacco, and Opsariichthys should be assigned to a monophyletic group, the Opsariichthys group (Figs. 3 and 4). Tang et al. (2013) also showed similar results with different topology (Fig. 1).
Five genera reassigned into Xenocyprididae
Liao et al. (2011c) classified five genera into Opsariichthyinae, which was placed into Xenocyprididae in two later studies (Tang et al. 2013; Stout et al. 2016). Our present results also revealed that Aphyocypris, Yaoshanicus, Nicholsicypris, and Pararasbora are monophyletic with high support at the nodes (1.00 in BI, and 100 in ML), which was confirmed in Tang et al. (2010, 2013). Yaoshanicus, Nicholsicypris, and Pararasbora should be considered to be junior synonyms of Aphyocypris (Liao et al. 2011b). In addition, our study revealed Hemigrammocypris is nested with four species of Metzia and congruent with previous studies (Tang et al. 2013; Stout et al. 2016).
Candidia and Nipponocypris are distinct genera
All valid species of Candidia and Nipponocypris were used in this study to reassess their relatedness. The BI and ML trees both showed that Candidia and Nipponocypris were well separated with high support (1.00 in BI, and 65 in ML); our results were consistent with several previous studies (Huynh and Chen 2013; Liao et al. 2011c; Tang et al. 2013). Morphologically, these two genera can be easily distinguished: Candidia has maxillary barbels, which are absent in Nipponocypris (Chen and Fang 1999; Nakabo 2013). The present study thus strongly suggests that they should be regarded as distinct genera.
Evolutionary implications of the color pattern
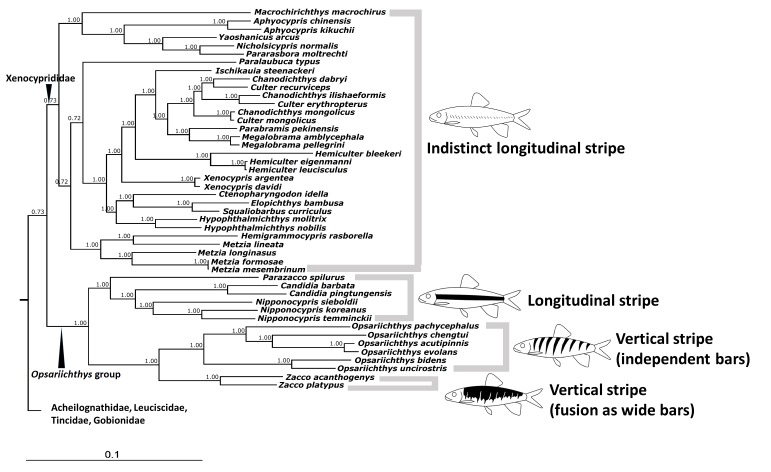
Among all studied species, only those in the Opsariichthys group have distinct longitudinal or vertical stripes. Most of them are known to have color dimorphism, especially Opsariichthys and Zacco (Chen and Chang 2005). This study’s tree topologies revealed that the type of stripe pattern on the sides of the body was highly correlated with molecular phylogeny (Fig. 5). The BI tree showed that Parazacco, Candidia, and Nipponocypris share similar longitudinal stripe patterns. Opsariichthys and Zacco both share similar vertical stripe patterns, although independent bars always appeared in Opsariichthys and the otherwise typically smaller bars are usually fused into a single wide bar in Zacco, but both could be de ned as the same type of color pattern. Otherwise, members of Xenocyprididae have only an indistinct longitudinal stripe.
These representative stripes or bars can also be found in several groups under Cypriniformes, such as Danio, Crossocheilus and Acrosscheilus. The genus Danio, a primitive cyprinids, already shows these stripe and bar patterns of the Opsariichthys group (McCluskey and Postlethwait 2015). Among these, D. erythromicron and D. choprae have distinct vertical stripes and D. nigrofasciatus and D. rerio have distinct longitudinal ones. Furthermore, Danio is the earliest offshoot in tree topologies (Tang et al. 2013; Stout et al. 2016). Therefore, we hypothesize that the types of vertical and longitudinal stripes presented in the Opsariichthys group might have originated from an primitive ancestor, then distinct vertical stripes might have been lost among these cyprinids but retained in the Opsariichthys group.
Fig. 5.
Fig. 5. Correlation between stripe patterns and molecular relationships in the Opsariichthys group (tree reconstructed based on Bayesian inference in this study)
CONCLUSIONS
The phylogenetic trees in this study provide a new perspective on the systematics of the Opsariichthys group and its sister group Xenocyprididae, which includes the related taxa from the following groups, all of which are under Cyprinidae senu lato: Cultrinae, Xenocyprinae, Squaliobarbinae, ex-Danioninae and ex-Cultrinae. The Opsariichthys group formed a stable monophyletic group, which includes five genera: Parazacco, Candidia, Nipponocypris, Zacco, and Opsariichthys. Our results also strongly suggest that Candidia and Nipponocypris be regarded as distinct genera within this family. Lastly, Aphyocypris, Yaoshanicus, Nicholsicypris, Hemigrammocypris and Pararasbora belong to Xenocyprididae.
Acknowledgments
Acknowledgments: This study was funded by the Ministry of Science and Technology, Taiwan (MOST 105-2311-B-001-064, MOST 106-2311-B-001-022).
Footnotes
Authors’ contributions: SPH, FYW and TYW designed the study and prepared the manuscript. SPH and FYW performed the field work and the laboratory experiments. SPH and TYW performed the phylogenetic analyses. All authors participated in revising the manuscript and approved the final manuscript.
Competing interests: SPH, FYW and TYW declare that they have no con ict of interest. FYW and TYW have received research grants from the MOST, Taiwan.
Availability of data and materials: The mito- genomic sequences are deposited to Genbank.
Consent for publication: Not applicable.
Ethics approval consent to participate: All animal experiments in this study were performed in accordance with guidelines of the animal ethics committee and were approved by the Academia Sinica Institutional Animal Care and Use Committee (IAUC 15-12-923).
References
- Broughton R E, Milam J E, Roe B A. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitogenomic. Genome Res. 11:1958–1967. doi: 10.1101/gr.156801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H Y, Yuan L Y, Hu T L, Chien C I, Lin Y C, Tseng S F, Chang T W, Wang W K. Complete mitochondrial DNA genome of Zarazacco spilurus (cypriniformes: Cyprinidae) 2016. [DOI] [PubMed]
- Mitochondrial Dna A Dna. Seq Anal. 27:165–166. [Google Scholar]
- Chang C H, Tsai C L, Jang-Liaw N H. Complete mitochondrial genome of the Chinese rasbora Rasbora steineri (Teleostei, Cyprinidae) Mitochondrial DNA. 24:183–185. doi: 10.3109/19401736.2012.744977. [DOI] [PubMed] [Google Scholar]
- Chen I S, Chang Y C. The photographic guide of inland water fishes of Taiwan. Keelung): I. Cypriniformes. Sheichuan Press; 2005. [Google Scholar]
- Chen I S, Fang L S. The freshwater and estuarine fishes of taiwan. National Museum of Marine Biology and Aquarium. Pingtung): 1999. [Google Scholar]
- Chen I S, Liu Y W, Huang S P, Shen C N. The complete mitochondrial genome of the Korean minnow Nipponocypris koreanus. Cypriniformes, Cyprinidae): 2016. [DOI] [PubMed] [Google Scholar]
- Mitochondrial Dna A Dna. Seq Anal. 27:708–710. doi: 10.3109/19401736.2014.913153. [DOI] [PubMed] [Google Scholar]
- Chen I S, Tsai W L, Huang S P, Chen H M. The complete mitochondrial genome of Temminck's minnow Nipponocypris temminckii. Cypriniformes, Cyprinidae): 2016. [DOI] [PubMed] [Google Scholar]
- Mitochondrial Dna A Dna. Seq Anal. 27:1542–1544. doi: 10.3109/19401736.2014.953129. [DOI] [PubMed] [Google Scholar]
- Chen L, Li B, Zhou L, Zhao G. The complete mitochondrial genome sequence of Predatory carp Chanodichthys erythropterus (Cypriniformes: Cyprinidae) 2016. [DOI] [PubMed]
- Mitochondrial Dna A Dna. Seq Anal. 27:1119–1120. doi: 10.3109/19401736.2014.933328. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada G L, Doallo R, Posada D. Jmodeltest 2: More models, new heuristics and parallel computing. Nat Methods; 2012. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Zhang X, Song W, Wang Y, Gao Z, Wang W. The complete mitochondrial genome sequence of Parabramis pekinensis strenosoma (Cypriniformes: Cyprinidae) 2014. [DOI] [PubMed]
- Mitochondrial Dna A Dna. Seq Anal. 27:86–87. doi: 10.3109/19401736.2013.873915. [DOI] [PubMed] [Google Scholar]
- Eschmeyer W N, Fricke R. CATALOG OF FISHES: GENERA, SPECIES, REFERENCES. http://researcharchive.calacademy.org/ research/ichthyology/catalog/fishcatmain.asp. 2017.
- Fang F, Noren M, Liao T Y, Kallersjo M, Kullander S O. Molecular phylogenetic interrelationships of the south asian cyprinid genera Danio, Devario and Microrasbora (Teleostei, Cyprinidae, Danioninae) Zool Scr. 38:237–256. [Google Scholar]
- Farrington H L, Edwards C E, Guan X, Carr M R, Baerwaldt K, Lance R F. Mitochondrial genome sequencing and development of genetic markers for the detection of DNA of invasive bighead and silver carp (Hypophthalmichthys nobilis and H. molitrix) in environmental water samples from the United States. PLoS One; 2015. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughan S, Johnson R, Wang J, Wachholtz M, Steffensen K, King T, Lu G. The complete mitochondrial genome of the silver chub, Macrhybopsis storeriana. Mitochondrial DNA Part B Resour. 1:789–790. doi: 10.1080/23802359.2016.1197053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. Bioedit: A user-friendly biological sequence alignment editor and analysis, version 5.09. Department of Microbiology. North Carolina): 2001. [Google Scholar]
- He H Y, Wang J, You P. The complete nucleotide sequence of mitochondrial genome of Abbottina rivularis (Cypriniformes: Cyprinidae) Acta Zootaxonomica Sinica. 38:695–704. [Google Scholar]
- He S, Gu X, Mayden R L, Chen W J, Conway K W, Chen Y. Phylogenetic position of the enigmatic genus Psilorhynchus (Ostariophysi: Cypriniformes): evidence from the mitochondrial genome. Mol Phylogenet Evol. 47:419–425. doi: 10.1016/j.ympev.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Hinsinger D D, Debruyne R, Thomas M, Denys Gpj, Menesson M, Utge J, Dettai A. Fishing for barcodes in the Torrent: from COI to complete mitogenomes on NGS platforms. DNA Barcodes. 3:170–186. [Google Scholar]
- Ho C W, Liu M Y, Chen M H. Complete mitochondrial genome of Rasbora trilineata. Cypriniformes, Cyprinidae): 2016. [DOI] [PubMed] [Google Scholar]
- Mitochondrial Dna A Dna. Seq Anal. 27:1755–1757. doi: 10.3109/19401736.2014.963798. [DOI] [PubMed] [Google Scholar]
- Huang S P, Chen I S, Jang-Liaw N H, Shao K T, Yung Mmn. Complete mitochondrial reveals a new phylogenetic perspective on the brackish water goby Mugilogobius group (Teleostei: Gobiidae: Gobionellinae) 33:566–574. doi: 10.2108/zs150154. [DOI] [PubMed] [Google Scholar]
- Huang S P, Zhao Y H, Chen I S, Shao K T. A new species of Microphysogobio (Cypriniformes: Cyprinidae) from Guangxi Province, southern China. Zool Stud; 2017. 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T Q, Chen I S. A new species of cyprinid fish of genus Opsariichthys from Ky Cung-Bang Giang River basin, northern Vietnam with notes on the taxonomic status of the genus from northern Vietnam and southern China. J Mar Sci Tech Suppl. 21:135–145. [Google Scholar]
- Hwang D S, Lee W O, Lee J S. Complete mitochondrial genome of the freshwater fish, Acanthorhodeus macropterus (Cypriniformes, Cyprinidae) Mitochondrial DNA. 25:11–12. doi: 10.3109/19401736.2013.775265. [DOI] [PubMed] [Google Scholar]
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh T P, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. Mitofish and mitoannotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540. doi: 10.1093/molbev/mst141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang-Liaw N H, Tsai C L, Chang C H, Watanabe K. Complete mitochondrial genome of the Moltrecht's minnow, Aphyocypris moltrechti (teleostei, cyprinidae), in comparison with. A. Kikuchii. Mitochondrial DNA. 24:117–119. doi: 10.3109/19401736.2012.731403. [DOI] [PubMed] [Google Scholar]
- Jang-Liaw N H, Tsai C L, Watanabe K. Complete mitochondrial genome of the Kikuchi's minnow Aphyocypris kikuchii (Teleostei, Cyprinidae) Mitochondrial DNA. 24:11–13. doi: 10.3109/19401736.2012.710227. [DOI] [PubMed] [Google Scholar]
- Kottelat M. The fishes of the Inland Waters of Southeast Asia: A catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. Raffles B Zool. 27:1–663. [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Bio Evol. 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuma W E, Kumazawa Y. Complete mitochondrial genome sequences of two Indonesian rasboras (Rasbora aprotaenia and Rasbora lateristriata) Mitochondrial DNA Part A. 27:4222–4223. doi: 10.3109/19401736.2015.1022749. [DOI] [PubMed] [Google Scholar]
- Li F, Shao K T, Lin Y S, Chang C H. The complete mitochondrial genome of the Rhodeus shitaiensis. Teleostei. 26:301–302. doi: 10.3109/19401736.2013.825785. [DOI] [PubMed] [Google Scholar]
- Li S, Liu X, Bai Y, Huang T, Wang J, Qu H, Chen L, Jiang W. The complete mtDNA genome of Erythroculter ilishaeformis: genome characterization and phylogenetic analysis. Mitochondrial DNA Part B. 1:39–40. doi: 10.1080/23802359.2015.1137812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao T Y, Kullander S O, Fang F. Phylogenetic position of rasborin cyprinids and monophyly of major lineages among the danioninae, based on morphological characters (Cypriniformes: Cyprinidae) J Zool Syst Evol Res. 49:224–232. [Google Scholar]
- Liao T Y, Kullander S O, Lin H D. Synonymization of Pararasbora, Yaoshanicus, and Nicholsicypris with Aphyocypris, and description of a new species of Aphyocypris from Taiwan (Teleostei: Cyprinidae) Zool Stud. 50:657–664. [Google Scholar]
- Liao T Y, Unlu E, Kullander S O. Western boundary of the subfamily danioninae in Asia (Teleostei, Cyprinidae): Derived from the systematic position of barilius mesopotamicus based on molecular and morphological data. Zootaxa. 2880:31–40. [Google Scholar]
- Librado P, Rozas J. Dnasp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lin H D, Lin F J, Chiang T Y, Lee T W. The complete mitochondrial genome sequence of Metzia formosae (Cypriniformes, Cyprinidae) Mitochondrial DNA. 26:257–258. doi: 10.3109/19401736.2013.823187. [DOI] [PubMed] [Google Scholar]
- Liu G D, Chen I S, Zhu J Q, Shen C N. The complete mitochondrial genome of small sliver gugeon Squalidus gracilis (Teleostei, Cyprinidae) Mitochondrial DNA A DNA Mapp Seq Anal. 27:603–604. doi: 10.3109/19401736.2014.908366. [DOI] [PubMed] [Google Scholar]
- Liu Q L, Xu B H, Xiao T Y, Su J M, Yao Y B, Liu Y J. Complete mitochondrial genome of the Xiangjiang barbel chub Squaliobarbus curriculus: comparative analysis of the genetic variation associated with geographical population. Mitochondrial DNA. 24:654–656. doi: 10.3109/19401736.2013.772167. [DOI] [PubMed] [Google Scholar]
- Liu X, Xiao K, Zhao X, Liu J, Zhang X, Guo W, Chen L, Du H. The complete mitochondrial genome of the Megalobrama pellegrini (Teleostei: Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal . 2016;27:3069–3070. doi: 10.3109/19401736.2014.1003913. [DOI] [PubMed] [Google Scholar]
- Liu Y. The complete mitochondrial genome sequence of Xenocypris davidi (Bleeker) Mitochondrial DNA. 25:374–376. doi: 10.3109/19401736.2013.809429. [DOI] [PubMed] [Google Scholar]
- Luo Y, Cao X, Zhu Y. The complete mitochondrial genome of Tanakia limbata (Cypriniformes: Cyprinidae) 2016. [DOI] [PubMed]
- Mitochondrial Dna A Dna. Seq Anal. 27:1713–1714. doi: 10.3109/19401736.2014.961135. [DOI] [PubMed] [Google Scholar]
- Lowe T M, Eddy S R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucl Acid Res. 25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Luo W. The complete mitochondrial genome of a cyprinid fish. Teleostei, Cypriniformes): 2016. [DOI] [PubMed] [Google Scholar]
- Mitochondrial Dna A Dna. Seq Anal. 27:185–186. [Google Scholar]
- Mabuchi K. Complete mitochondrial genomes of five introduced strains of common carp (Cyprinus carpio) in Japan with 29 diagnostic SNPs distinguishable by restriction enzyme analysis. Mitochondrial DNA Part B. 1:261–263. doi: 10.1080/23802359.2016.1159931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayden R L, Chen W J, Bart H L, Doosey M H, Simons A M, Tang K L, Wood R M, Agnew M K, Yang L, Hirt M V, Clements M D, Saitoh K, Sado T, Miya M, Nishida M. Reconstructing the phylogenetic relationships of the earth's most diverse clade of freshwater fishes-order Cypriniformes (Actinopterygii: Ostariophysi): A case study using multiple nuclear loci and the mitochondrial genome. Mol Phylogenet Evol. 51:500–514. doi: 10.1016/j.ympev.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Mccluskey B M, Postlethwait J H. Phylogeny of zebrafish, a "model species," within danio, a "model genus. Mol Biol Evol. 32:635–652. doi: 10.1093/molbev/msu325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya M, Sato Y, Fukunaga T, Sado T, Poulsen J Y, Sato K, Minamoto T, Yamamoto S, Yamanaka H, Araki H, Kondoh M, Iwasaki W. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R Soc Open Sci; 2015. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabo T. Fishes of Japan with pictorial keys to the species. Tokyo.): Tokai University Press; 2013. [Google Scholar]
- Nelson J S, Grande T C, Wilson M V. Fishes of the world. John Wiley & Sons; 2016. [Google Scholar]
- Ronquist F, Huelsenbeck J P. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Saitoh K, Miya M, Inoue J G, Ishiguro N B, Nishida M. Mitochondrial genomics of ostariophysan fishes: perspectives on phylogeny and biogeography. J Mol Evol. 56:464–72. doi: 10.1007/s00239-002-2417-y. [DOI] [PubMed] [Google Scholar]
- Saitoh K, Sado T, Doosey M H, Jr Bart, Inoue H L, Nishida J G, Mayden M, Miya R L. Evidence from mitochondrial genomics supports the lower Mesozoic of South Asia as the time and place of basal divergence of cypriniform fishes (Actinopterygii: Ostariophysi) Zool J Linn Soc. 161:633–662. [Google Scholar]
- Saitoh K, Sado T, Mayden R L, Hanzawa N, Nakamura K, Nishida M, Miya M. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): The first evidence toward resolution of higher-level relationships of the world's largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 63:826–841. doi: 10.1007/s00239-005-0293-y. [DOI] [PubMed] [Google Scholar]
- Stout C C, Tan M, Lemmon A R, Lemmon E M, Armbruster J W. Resolving Cypriniformes relationships using an anchored enrichment approach. BMC Evol Biol; 2016. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K L, Agnew M K, Hirt M V, Lumbantobing D N, Sado T, Teoh V H, Yang L, Bart H L, Harris P M, He S, Miya M, Saitoh K, Simons A M, Wood R M, Mayden R L. Limits and phylogenetic relationships of East Asian fishes in the subfamily Oxygastrinae (Teleostei: Cypriniformes: Cyprinidae) 3681:101–135. doi: 10.11646/zootaxa.3681.2.1. [DOI] [PubMed] [Google Scholar]
- Tang K L, Agnew M K, Hirt M V, Sado T, Schneider L M, Freyhof J, Sulaiman Z, Swartz E, Vidthayanon C, Miya M, Saitoh K, Simons A M, Wood R M, Mayden R L. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae) 57:189–214. doi: 10.1016/j.ympev.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Tsao Y F, Lin W W, Chang C H, Ueda T, Jang-Liaw N H, Zhao Y H, Kao H W. Phylogeography, historical demography, and genetic structure of the rose bitterling. Rhodeus ocellatus (Kner, 1866) (Cypriniformes: Acheilognathidae), in East Asia; 2016. 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wang S, Hu M, Wang F. Complete mitochondrial genome of Rhodeus lighti (Cypriniformes: Cyprinidae) 2016. [DOI] [PubMed]
- Mitochondrial Dna A Dna. Seq Anal. 27:1497–1498. doi: 10.3109/19401736.2014.953109. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): insight into its phylogenic position within Cyprinidae. Gene. 424:96–101. doi: 10.1016/j.gene.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang J, He S, Mayden R L. The complete mitochondrial genome of the Chinese hook snout carp Opsariichthys bidens (Actinopterygii: Cypriniformes) and an alternative pattern of mitogenomic evolution in vertebrate. Gene. 399:11–19. doi: 10.1016/j.gene.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Wei M, Yang Y, Yu P, Pan D, Wan Q. The complete mitochondrial genome of Mongolian redfin, Chanodichthys mongolicus (Cypriniformes: Cyprinidae): genome description and related phylogenetic analyses. Mitochondrial DNA A DNA Mapp Seq Anal. 27:20–21. doi: 10.3109/19401736.2013.867440. [DOI] [PubMed] [Google Scholar]
- Wu H W. The cyprinid fishes of China. Shanghai): People's Press; 1977. [Google Scholar]
- Xu X, Cao X, Zhu Y. The complete mitochondrial genome of Tanakia lanceolata (Cypriniformes: Cyprinidae) 2016. [DOI] [PubMed]
- Mitochondrial Dna A Dna. Seq Anal. 27:867–868. doi: 10.3109/19401736.2014.919480. [DOI] [PubMed] [Google Scholar]
- Yan S X, Xiong M H, Zhu B, Tian H, Liao X L, Shao K. The complete mitochondrial genome of Rhinogobio typus (Teleostei, Cyprinidae, Gobioninae) Mitochondrial DNA A DNA Mapp Seq Anal. 27:698–699. doi: 10.3109/19401736.2014.913149. [DOI] [PubMed] [Google Scholar]
- Yang X, Ma Z, Xie L, Yang R, Shen J. Complete mitochondrial genome of the Chinese bitterling Rhodeus sinensis (Cypriniformes: Cyprinidae) Mitochondrial DNA. 26:647–648. doi: 10.3109/19401736.2013.836517. [DOI] [PubMed] [Google Scholar]
- Yin W, Cao K, He H, Fu C. Four complete mitochondrial genomes of the genera Candidia, Opsariichthys, and Zacco (cypriniformes: Cyprinidae) Mitochondrial DNA Part A. 27:4613–4614. doi: 10.3109/19401736.2015.1101582. [DOI] [PubMed] [Google Scholar]
- Yuan L Y, Wang W K, Lee C H, Chiu K H, Li Y M, Kuo P C, Chang H Y. Complete mitochondrial DNA genome of Metzia mesembrinum (Cypriniformes: Cyprinidae) Mitochondrial DNA A DNA Mapp Seq Anal. 27:214–215. doi: 10.3109/19401736.2014.880894. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ma Z, Chen D, Xu W, Yang R. The complete mitochondrial genome of Culter dabryi (Cyprinidae: Cultrinae) Mitochondrial DNA. 25:98–99. doi: 10.3109/19401736.2013.784754. [DOI] [PubMed] [Google Scholar]