Abstract
Qiu Ren, Jun-Xing Yang, and Xiao-Yong Chen (2018) Triplophysa stenura is an indigenous and widely distributed loach in the Qinghai-Tibet plateau and adjacent Three Parallel Rivers region of China. Morphological and phylogenetic analyses were performed in order to explore the genetic and morphological variation in T. stenura populations from different geographic regions and infer the divergence time and relationships between populations from the three rivers (Jinsha River, Nu River and Lancang River). Two mitochondrial genes (Cyt b, D-Loop) and 35 morphological characters were selected for genetic and morphological analyses, respectively. Phylogenetic and network analyses reveal that T. stenura is a single lineage with three well supported clades strictly corresponding to specific river systems. Divergence time analysis suggests that the divergence of T. stenura and formation of the Three Parallel Rivers are closely associated with the Kun-Huang Movement (1.1- 0.6 MYA), which lead to the uplift of the Qinghai-Tibet plateau. AMOVA reveals that there is moderate genetic differentiation among populations. Results from ANOVA suggest that several morphological characters show significant variation among populations and drainages. Descriptive morphological variation (e.g., color pattern) in different populations of T. stenura clarifies a set of characters that can be used to accurately identify members of this group in the future. We conclude that T. stenura has moderate population genetic structure and character variation in this study area and the divergence and evolution of T. stenura is associated with the uplift of the Qinghai-Tibet plateau.
Keywords: Triplophysa stenura, Phylogeny, Morphology, Biogeography, Three Parallel Rivers region
BACKGROUND
The Three Parallel Rivers region of China is located in northwest Yunnan Province, and represents the eastern part of area that resulted from the collision between the Indian and Eurasian plates. With the continental collision, three rivers (Jinsha, Lancang and Nu rivers) along with three mountain ranges (Gaoligong, Nu and Yunlong) formed north-southwards (Fig. 1). These ranges demonstrate one of the most unique natura landscapes on Earth and have been recognized as World Heritage locations (Ming and Shi 2006; Ming 2007). Some research has suggested that the formation of the Three Parallel Rivers area was closely correlated with the sharp uplift of the Qinghai-Tibet plateau in the late tertiary (3.6 Million Years Ago, MYA), and this major change in the topography of the Qinghai-Tibet plateau area has had profound impacts on speciation, phylogeographic structure and genetic patterns of species endemic to China (Ren et al. 1959; Li et al. 1995; Li and Fang 1998; Zheng et al. 2006). Therefore, the phylogeographic investigations of endemic fish species can reveal important information as to the origin and relationships between faunas of the Three Parallel Rivers area.
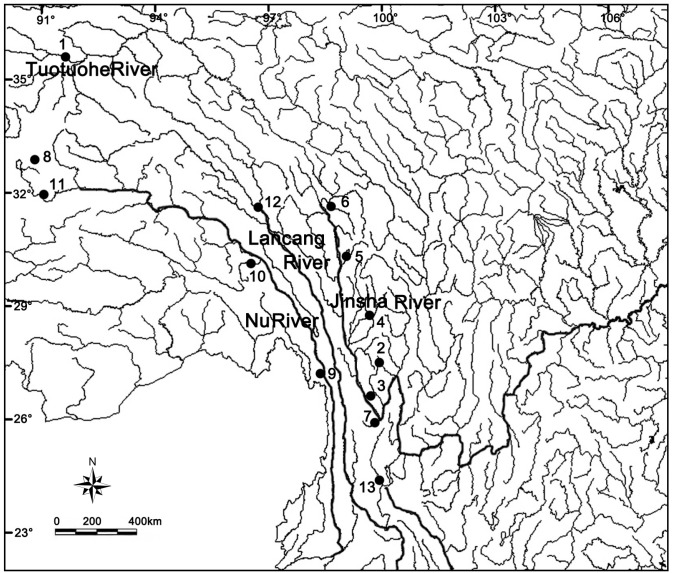
Fig. 1.
Fig. 1. Map showing the collection localities of Triplophysa stenura in this study (numbers correspond to name of localities in Table 1).
The genus Triplophysa Rendahl, 1933 includes approximately 142 species, making it one of the largest loach groups in the family Nemacheilidae, and 85% of this diversity is known from China (He 2008; Ren et al. 2012; Froese and Pauly 2017). The genus is wide spread in the Qinghai-Tibet plateau and adjacent areas (Wu and Wu 1991; Zhu 1989). Due to the similar appearance of Triplophysa species and limited variation between characters currently being examined, it can be difficult to distinguish taxa and geographic variants of species. This is especially true for widespread species whose morphological features (i.e. mouth structure, color pattern, intestine type) vary from different localities or river drainages.
Triplophysa stenura (Herzenstein 1888) is a small to medium sized loach, which is widely distributed in the Qinghai-Tibet plateau and upper reaches of Yangtze, Lancang (upper Mekong River) and Nu (upper Salween River) Rivers of China (Zhu 1989; Chen 1998). This species lives in swift-flowing streams, where it moves amongst rocky crevices by swimming in a saltatory manner. It mainly scrapes surfaces in streams for algae and sometimes feeds on some small aquatic invertebrates (Wu and Wu 1991). T. stenura is diagnosed by a combination of characters: caudal peduncle nearly round, depth reduced sharply towards the caudal-fin base; caudal peduncle length greater than head length; posterior chamber of air bladder degenerate; intestine bending in one to three coils behind stomach; depressed pelvic-fin with tip extending beyond anus.
Mitochondrial DNA is widely used to infer phylogenetic relationships among many species or groups at different taxonomic levels. Cytochrome b (Cyt b) and control region gene (D-Loop) have proven useful in resolving lower level relationships (interspecific and intraspecific) between fishes (Saka et al. 2003; He et al. 2006; Tang et al. 2006; Liu et al. 2007). In this study, these two genes are used as molecular markers to reconstruct the phylogenetics and geographic relationships within T. stenura and explore genetic variation and biogeographic patterns. With a phylogenetic resolution within T. stenura, the estimated age of the clades using a molecular clock inference provides an opportunity to examine the age of the species. Our analysis thus focuses on exploring genetic variation and evolution of T. stenura, and geographic distributions of forms geographically across the Three Parallel Rivers area. We also hypothesize geological and geographical events that occurred in the plateau that may be related to observed divergences. Morphological data and analyses serve as a parallel analysis to explore morphological variation and divergences among different populations of T. stenura. Finally, diagnostic characters typically used in the identification of Triplophysa species are discussed, which provides a reference to the taxonomy and identification of species in the genus for future studies of biodiversity.
MATERIALS AND METHODS
Sampling
Thirty-one tissue samples were collected from 14 localities in the Three Parallel Rivers area for Cyt b and D-Loop analyses. Tissues were from the base of the right pelvic fin of fresh specimens and were preserved in 99.5% ethanol. A total of 91 specimens were collected from eight localities for morphological examination and analyses. These specimens were fixed in 8-10% formalin, rinsed thoroughly and preserved in 75% ethanol for long-term storage. Specimens are deposited in the collection of Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences. Triplophysa stewarti which is thought to be closely related to T. stenura, and two other Triplophysa species - T. orientalis, and T. stoliczkaeas - were chosen as outgroups. Identification of specimens follows Zhu (1989). Detailed information regarding samples is provided in figures 1, 2 and table 1.
Table 1. Information on the sequences and specimens used in this study.
| Species | Location (abbr. and site number) | Drainage | Haplotype NO. | Voucher NO. | Genbank accession number | Specimens examined Catalog & total number | |
| Cytb | D-Loop | ||||||
| Triplophysa stenura | Tuotuohe, Qinghai (TTH) (1) | Jinsha R. | H5 | 20040803001 | KJ650345 | KJ650374 | 2004002175-2004002195 (21) |
| H6 | 20040803002 | KJ650370 | KJ650394 | ||||
| H7 | 20040803003 | KJ650369 | KJ650393 | ||||
| H8 | 20040803007 | KJ650368 | - | ||||
| H4 | 20040803008 | KJ650367 | KJ650392 | ||||
| Xiaozhongdian, Yunnan(XZDH) (2) | Jinsha R. | H1 | 20040620001 | JN837657 | KJ650397 | ||
| H2 | 20040620002 | KJ650353 | KJ650379 | ||||
| H3 | 20040620003 | KJ650371 | KJ650395 | ||||
| H4 | 20040620004 | KJ650352 | KJ650378 | ||||
| Yongsheng, Yunnan (YS) (3) | Jinsha R. | H11 | 041106046 | KJ650365 | KJ650390 | ||
| H12 | 041106047 | KJ650364 | KJ650389 | ||||
| H12 | 041106048 | KJ650363 | KJ650388 | ||||
| H11 | 041106049 | KJ650362 | KJ650387 | ||||
| Xiangcheng, Sichuan (XC) (4) | Jinsha R. | H13 | YJ2009033 | KJ650361 | KJ650386 | ||
| H13 | YJ2009034 | KJ650360 | - | ||||
| H14 | YJ2009035 | KJ650359 | KJ650385 | ||||
| H13 | YJ2009036 | KJ650358 | KJ650384 | ||||
| Batang, Sichuan (BT) (5) | Jinsha R. | H17 | YJ2009079 | KJ650357 | KJ650383 | 2009003584-2009003590 (7) | |
| H16 | YJ2009080 | KJ650356 | KJ650382 | ||||
| H6 | YJ2009081 | KJ650373 | - | ||||
| H15 | YJ2009082 | KJ650350 | KJ650376 | ||||
| Baiyu, Sichuan (BY) (6) | Jinsha R. | H18 | 2006009504 | KJ650344 | - | 2006009504-2006009508 (5) | |
| H16 | 2006009505 | KJ650372 | KJ650396 | ||||
| H19 | 2006009507 | KJ650355 | KJ650381 | ||||
| Shigu, Yunnan (SG) (7) | Jinsha R. | - | - | - | 2011000001-2011000008 (12) | ||
| Anduoqu, Tibet (ADQ) (8) | Nu R. | H9 | 20040804001 | KJ650366 | KJ650391 | 2004002201-2004002213 (13) | |
| H10 | 20040804002 | KJ650351 | KJ650377 | ||||
| Gongshan, Yunnan (GS) (9) | Nu R. | H21 | - | KJ650348 | - | 2000198-2000199 (2) | |
| H20 | CJF1806 | KJ650354 | KJ650380 | ||||
| Baxiu, Tibet (BX) (10) | Nu R. | H22 | - | KJ650347 | - | ||
| Naqu, Tibet (NQ) (11) | Nu R. | H23 | - | KJ650349 | - | ||
| Changdu, Tibet (CD) (12) | Lancang R. | H24 | - | KJ650346 | KJ650375 | 20100042-20100048 (8) | |
| Yangbi, Yunnan (YB) (13) | Lancang R. | - | - | - | 1974000595-1974000617 (23) | ||
| GB1* | H25 | DQ105247 | DQ105319 | ||||
| GB2* | H26 | DQ105246 | DQ105318 | ||||
| T. stewarti | Milin, Tibet | DQ105248 | |||||
| T. orientalis | Xining, Qinghai | DQ105251 | |||||
| T. stoliczkaeas | Gangca, Qinghai | DQ105249 | |||||
*Sequence obtained from Genbank.
Fig. 2.
Fig. 2. Lateral views of Triplophysa stenura individuals from different localities. (A) Changdu, Tibet. KIZ 20100045 138 mm SL; (B) Batang, Sichuan KIZ 2009003584 113 mm SL; (C) Baiyu, Sichuan KIZ 2006009508 121 mm SL; (D) Shigu, Yunnan KIZ 201100003 86.6 mm SL; (E) Gongshan, Yunnan KIZ 20007198 78.4 mm SL; (F) Tuotuohe, Qinghai KIZ 2004002141 68.9 mm SL; (G) Anduoqu, Tibet KIZ 2004002201 79.9 mm SL; (H) Yangbi,Yunnan KIZ 1974000595 60.1 mm SL. Scale bar = 1 cm.
Genetic analyses
Total genomic DNA was extracted from fin tissues following standard methods (Sambrook et al. 1989). The Cyt b and D-Loop genes were amplified by polymerase chain reaction (PCR). Primer sets L14724 (5'-GACTTGAAAAACCACCGTTG-3') and H15915 (5'-CTCCGATCTCCGGATTACAAGAC-3') (Xiao et al. 2001) were used to amplify the Cyt b gene, and DL1 (5'-ACC CCT GGC TCC CAA AGC- 3') and DH2 (5'-ATC TTA GCA TCT TCA GTG- 3') were used to amplify D-Loop gene (Liu et al. 2002). A total volume of 50 μL in each reaction included: 37.75 μL double distilled water, 5 μL of 10 × reaction buffer (Mg2+ Plus), 3 μL of dNTP (each 2.5 mM), 2 μL BSA, 0.25 μL (5U/μL) taq polymerase, 1 μL (10 μm) of each oligonucleotide primer and 1 μL of total genomic DNA template. The PCR amplification was performed at an initial denaturation step at 95°C for 3 mins, followed by denaturing at 94°C for 1 min, annealing at 52°C for 1 min and extending at 72°C for 1 min for a total of 40 cycles; the final cycle was a 10 min extension at 72°C, deposited at 4°C. Primers used for sequencing were the same for PCR amplifications. The unpurified fragments were sequenced by Kunming Shuoyang Technology Co., Ltd. All sequences obtained are deposited in GenBank; accession numbers are listed in table 1.
Nucleotide sequences were initially assembled using SeqMan 6.0 and Editseq 6.0 in the DNA Star package (DNAStar Inc., U.S.), with manual correction if necessary. Sequences were aligned using Clustal W in MEGA 5 (Tamura et al. 2011) and translated into amino acids for confirmation of alignment and assignment of codon positions and detection of stop codons. DnaSP 4.10 (Rozas et al. 2003) and DAMBE 5.1.2 (Xia 2013) were used to identify haplotypes among all sequences. Aligned sequence data were imported into MEGA 5 for nucleotide composition analysis. The Kimura two-parameter model (Kimura 1980) was used to calculate nucleotide divergence (genetic distances). Nucleotide saturation was analyzed using DAMBE 5.1.2, and a plot of absolute number of transitions (Ti) and transversions (Tv) against TN93 distance revealed saturation in substitutions. Hierarchical analysis of molecular variance (AMOVA, Excoffier et al. 1992) was implemented using Arlequin 3.5 (Excoffier et al. 2010) to assess genetic structure and differentiation among populations. A haplotype network was constructed with Network 4.6.0.0 (Bandelt et al. 1999) using median-joining method (Weights = 10; epsilon = 10; Weighting transition/ transversion = 1:3). Maximum Likelihood (ML) was implemented by PAUP*4.0b10 (Swofford 2003), the best-fit model of nucleotide evolution (GTR + I + G) was selected by Akaike Information Criterion (AIC) (Akaike 1974) in Modeltest 3.7. Support for recovered clades was measured using bootstrap analysis with 100 total pseudoreplicates. Bayesian tree resolution and determined posterior probabilities of recovered clades was completed using MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003). The most appropriate model of nucleotide substitution was selected by Modeltest 3.7. Analyses included 10,000,000 generations of Markov chains, which sampled every 1000 generations, to yield 10,000 trees. The first 25% of the trees were discarded as burn-in. A majority rule consensus tree calculated from the remaining 7,501 trees was used to determine posterior probabilities.
Cyt b was used to estimate divergence times in this analysis as we had more data for this particular gene than the D-Loop gene. The average substitution rate of Cyt b for cyprinids, 1.05% per million years (Dowling et al. 2002), was used. Divergence time was estimated using a Bayesian approach with BEAST 2.1.2 (Drummond and Rambaut 2007) and the input file was created using BEAUTi. Analysis was performed using the GTR + I + G model. The coalescent process, which allows for the estimation of population- level parameters, was chosen as the prior to generate patterns of lineage divergences. MCMC process was set as two independent runs of 10,000,000 generations. Trees were sampled every 1,000 generations and the first 10% of trees were discarded as burn-in. The log results were examined in Tracer 1.6 (Rambaut et al. 2013). The maximum credibility tree was summarized using TreeAnnotator in BEAST unit and visualized in Figtree 1.31 (Rambaut 2009).
Morphological analyses
A total of 24 morphometric characters (Table S1) were measured on the left side of specimens using a digital caliper system. Measurements were recorded to the nearest 0.01 mm following methods of Chu and Chen (1989) and He (2008). A total of 11 descriptive characters (Table 2) were carefully examined using a microscope. Preliminary statistical analysis of all morphometric data was performed using Microsoft Excel. To alleviate the influence of allometry and body-size differences and normalize the data, measurements taken on the head were divided by head length and taken on the rest of body were divided by standard length, and all measurements were log10 transformed. To investigate whether there is significant character variation among populations and drainages and where the variation is, one- way analyses of variation (ANOVA) and post- hoc Tukey’s test were conducted using R (R Core Team 2013). Principle component analysis (PCA) was performed using SPSS 20.0 for Windows (SPSS, Chicago, IL, USA).
Table 2. Morphological character comparisons between populations of Triplophysa stenura.
| Gongshan,Yunnan | Anduoqu, Tibet | Shigu, Yunnan | Batang, Sichuan | |
| SL (mm) | 69.3-78.4 | 72.5-92.7 | 61.9-86.6 | 48.6-113.7 |
| Body type | medium, cylindrical | medium, slim | medium, slim | robust, cylindrical |
| Ground color | brownish yellow (ethanol) | pale grey (ethanol) | pale grey (ethanol) | light yellow (ethanol) |
| Mouth | thick with strong furrow | medium thick with slight furrow | medium thick with slight furrow | thick with strong furrow |
| Lower jaw | spade-like | spade-like | spoon-like | spade-like and spoon-like |
| Snout | blunt | pointed | pointed | blunt |
| Color pattern | 6-8 blotches along dorsal midline | 6-8 blotches along dorsal midline | colors mottled, no regular blotches along dorsal midline | 6-10 blotches along dorsal midline |
| Dorsal fin origin | opposite to pelvic-fin origin | opposite to pelvic-fin origin | slightly posterior to pelvic-fin origin | slightly anterior to pelvic-fin origin |
| Intestine type | one coil | three coils | two coils | two coils |
| Caudal fin | slight concave | slight concave | deep concave | slight concave |
| Caudal peduncle | round | round | round | round |
| The tip of pelvic fin | extending beyond anus but not reaching the origin of anal fin | extending beyond anus but not reaching the origin of anal fin | extending beyond anus and reaching the origin of anal fin | extending beyond anus but not reaching the origin of anal fin |
| Baiyu, Sichuan | Tuotuohe, Qinghai | Changdu, Tibet | Yangbi, Yunnan | |
| SL (mm) | 106.5-141.2 | 57.5-78.1 | 91.4-140.6 | 51.8-81.9 |
| Body type | robust, cylindrical | medium, slim | slim, cylindrical | medium, cylindrical |
| Ground color | light yellow (ethanol) | pale grey (ethanol) | light yellow (ethanol) | brownish yellow (formalin) |
| Mouth | thick with strong furrow | medium thick with slight furrow | medium thick with slight furrow | thick with strong furrow |
| Lower jaw | spade-like and spoon-like | spade-like | spade-like | spoon-like |
| Snout | blunt | pointed | blunt | blunt |
| Color pattern | 6-8 blotches along dorsal midline | 6-8 blotches along dorsal midline | colors mottled, no regular blotches along dorsal midline | 6-8 blotches along dorsal midline |
| Dorsal fin origin | slightly posterior to pelvic-fin origin | slightly posterior to pelvic-fin origin | slightly posterior to pelvic-fin origin | slightly anterior to pelvic-fin origin |
| Intestine type | three coils | three coils | two coils | one coil |
| Caudal fin | slight concave | slight concave | slight concave | slight concave |
| Caudal peduncle | round | round | round | slightly depressed |
| The tip of pelvic fin | extending beyond anus but not reaching the origin of anal fin | extending beyond anus and reaching the origin of anal fin | extending beyond anus but not reaching the origin of anal fin | extending beyond anus but not reaching the origin of anal fin |
RESULTS
Sequence variation
In this study, 35 Cyt b sequences (33 from the present study and two from GenBank) yielded 26 unique haplotypes (Table 1). Cyt b gene was 1140bp in length, of which 89 sites were variable. Among the variable sites, 56 were parsimony informative polymorphic sites and 33 were singleton polymorphic sites. The average base composition of the 26 haplotypes was A = 25.5%, T = 29.7%, G = 17.5%, C = 27.3%. The mean base composition of Cyt b sequences have a low G content and are almost equal A, T and C contents, which is consistent with other Cobitid studies (He et al. 2006; Tang et al. 2006). The 26 sequences of D-Loop (plus 2 from GenBank) yielded 18 haplotypes. D-Loop sequences were 928bp, of which 801 sites were constant and 110 were variable. Among the variable sites, 39 were parsimony informative polymorphic sites and 71 were singleton polymorphic sites. The average base compositions of the 18 haplotypes were A=33.7%,T=31.8%,G=13.6%,C=20.9%. Haplotypes were shared by individuals within the same population or among different populations from the same river drainage. Individuals from different drainages, however, did not share haplotypes.
Phylogenetic relationships and divergence times
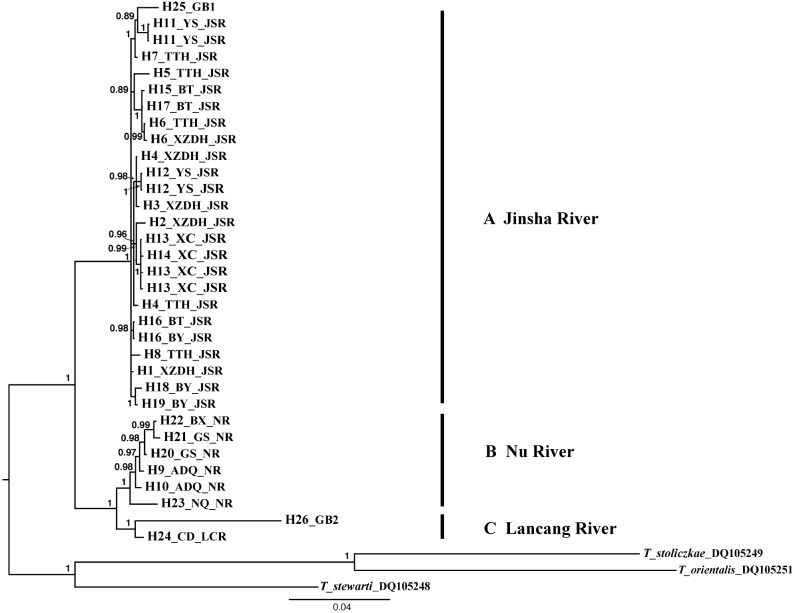
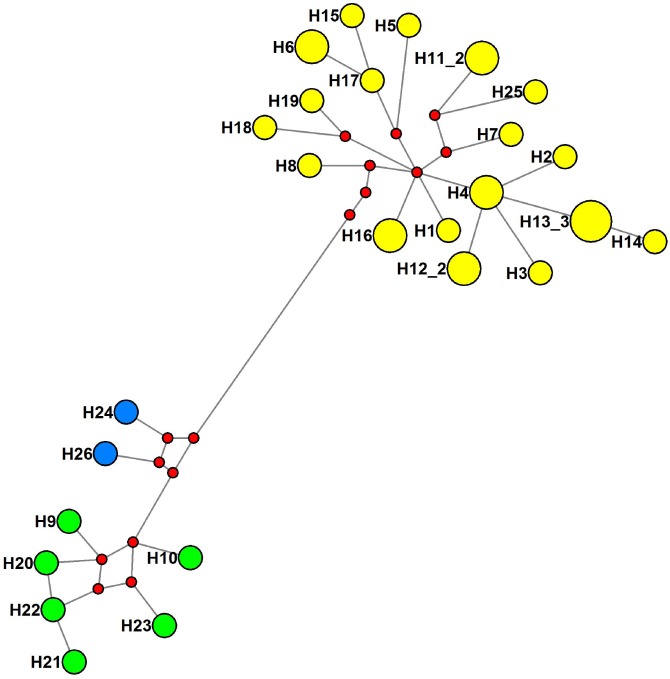
Independent phylogenetic analyses of 33 Cyt b, 26 D-Loop sequences, and the concatenated Cyt b and D-Loop data using ML and BI methods yielded similar topologies. All analyses resolved three strongly supported clades (Fig. 3). The haplotype network analysis yielded a phylogenetic network (Fig. 4) consistent with the phylogenetic trees resolved. The phylogenetic resolution of these genes supported the gene trees of T. stenura as being monophyletic and suggested that individuals within the same drainage but from different populations formed a common lineage. Populations distributed in Jinsha River drainage formed clade A; populations from Nu River drainage formed clade B; and clade C included individuals from the Lancang River drainage. Divergence time analysis estimated that T. stenura diverged about 3.88 MYA. The Jinsha River clade separated from the Nu and Lancang river clade about 1.39 MYA, and Lancang River and Nu River clades diverged about 0.61 MYA.
Fig. 3.
Fig. 3. Phylogenetic relationships within Triplophysa stenura as inferred by Bayesian analysis of combined Cytochrome b and D-Loop sequences. (Posterior probabilities are shown on nodes).
Fig. 4.
Fig. 4. The haplotype network constructed by median-joining method. Haplotypes are named as in table 1. Red dots are median vectors, which are hypothesized sequences to connect existing sequences within the network under maximum parsimony. The yellow circles are haplotypes from the Jinsha River drainage, blue circles are haplotypes from the Lancang River drainage, and green circles represent haplotypes from the Nu River drainage. The size of the circles is proportional to haplotype frequency.
Genetic distances and structures
Genetic distances among individuals (including outgroup individual) ranged from 0.0009 to 0.0450; among populations ranged from 0.0169 to 0.0363; and between ingroups and outgroups ranged from 0.0940 to 0.1488. AMOVA (Table 3) revealed that most of the genetic variation (87.98%) was found within populations (FST = 0.120, P < 0.001); significant genetic differentiation was also found among populations within drainages (FSC = 0.119, P < 0.001); the least genetic variation was present among drainages, which also had no significant differentiation (FCT = 0.001, P > 0.05).
Table 3. Hierarchical analysis of molecular variance (AMOVA) for Triplophysa stenura.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation (%) | P | Fixation indices |
| Among drainages | 2 | 1.08 | 0.00066 | 0.13 | P > 0.05 | FCT = 0.00132 |
| Among populations/ within drainages | 10 | 5.93 | 0.05914 | 11.89 | P < 0.001 | FSC = 0.11908 |
| Within populations | 20 | 8.75 | 0.43750 | 87.98 | P < 0.001 | FST = 0.12025 |
| Total | 32 | 15.76 | 0.49730 | 100.00% |
Morphological variation
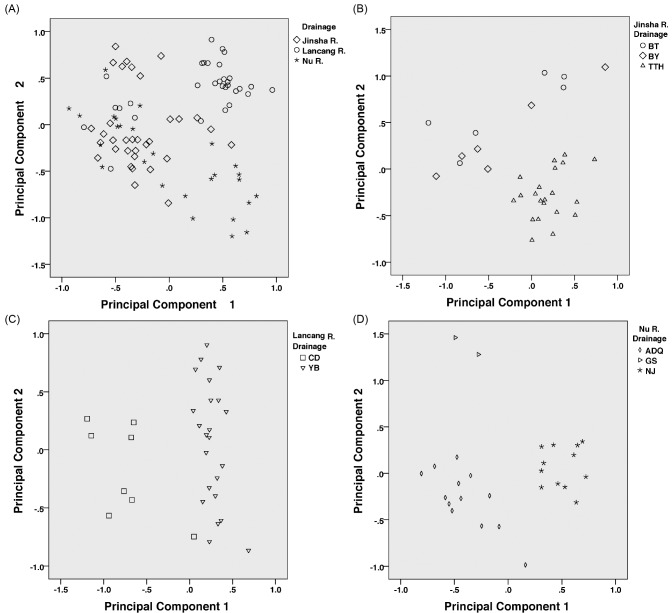
Comparisons of the 11 morphological characters among eight populations are shown in table 3. These results indicate that some characters, such as body shape, color pattern, mouth structure and intestine structure, demonstrated variation among populations of the widespread T. stenura. PCA and ANOVA analyses included 24 morphometric characters from eight geographic populations of the three drainages. Results of the PCA indicate that the first seven principal components (PCs) accounted for 84.6% of total variation. Scatter plots of the first two PCs, which accounted for 53.8%, illustrate that individuals were not clearly distinguishable by drainage (Fig. 5A), but they could be distinguishable by population within drainage separately (Fig. 5B, C and D). ANOVA results indicate that all of characters except for vent-anal distance (VAD) showed significant or extremely significant differences among populations. Sixteen characters demonstrated significant or extremely significant differences among drainages. The post- hoc Tukey’s test identified where the differences are. The detailed ANOVA results are summarized in table S1.
Fig. 5.
Fig. 5. Scatter plots of the first two principal components for Triplophysa stenura individuals from three drainages (A) and populations within three drainages respectively (B: Jinsha R. Drainage; C: Lancang R. Drainage; D: Nu R. Drainage). The locality abbreviations are shown in table 1.
DISCUSSION
Molecular phylogeny and biogeography
Results of the haplotype network and phylogenetic analyses support the hypothesis that T. stenura forms a monophyletic group, including three clades following river drainages. Haplotypes are all unique to river drainages; none are shared among populations from different river drainages. These findings indicate that T. stenura exhibits clear geographical structure in the Three Parallel Rivers region. We assume that geographic barriers are the main factors hindering gene exchange between different drainages and leading to genetic differentiation. Once the independent drainages were formed following the uplift, gene exchange among these drainages must have been impeded, providing the opportunity for divergence in isolation.
The phylogeography of T. stenura reflects relationships between the three rivers, wherein populations from the Nu River were more closely related to those from the Lancang River than to those from the Jinsha River. These findings agree with results from Chu and Chen (1990), which is based on broad research of the ichthyofauna of this region. Chen (1998) and Chu and Chen (1989) proposed that the Nu and Lancang Rivers may have connected during the early-middle Pleistocene. Divergence time results suggest that the Jinsha River clade was first divided from the remaining clades at about 1.39 MYA, and then the Lancang River and Nu River clades split at around 0.61 MYA; both findings are consistent with Ming and Shi (2006) and consistent with the formation of the three rivers in early-middle Pleistocene. Using evidence from quaternary sediment characters, Zhang et al. (1998) suggested the formation of the Jinsha River and its development to its present state dated 1.54 ± 0.178 MYA, and the Nu River dated back to the early-middle Pleistocene (He et al. 1992). Phylogenetic and biogeographic research by Guo et al. (2005) on sisorid catfishes revealed that the Jinsha River was the first formed and isolated, followed by the Lancang River, then the Nu and Irrawardy Rivers; these results are partially consistent with findings supported in the present study.
Some geographic and geological linked research (Li and Fang 1998; Zheng et al. 2006) has shown that the uplift of the Qinghai-Tibetan plateau included four movements: Qinghai-Tibetan Movement (3.6-1.7 Ma), Kun-Huang Movement (1.1-0.6 Ma), Gonghe Movement (0.15 Ma) and Huaxi Movement (30 Ka). This relatively rapid uplift was initiated about 3.4 MYA, and included three phases: A, B and C, at the estimated ages of 3.6 MYA, 2.5 MYA and 1.7 MYA, respectively. Estimates herein identified the time of isolation and origin of the three rivers at approximately the middle period of the Kun-Huang Movement. According to Ming and Shi (2006), the Three Parallel Rivers region located in the northwestern Yunnan Province was formed and influenced by the uplift of the Qinghai-Tibetan and Yunnan Plateaus to the northwest from the Kun-Huang Movement and headwater erosion and incision of river systems and tributaries. The Kun-Huang Movement was the most transformative event of the northwest Lengthways Range and Gorge region as well as the Landform-River development of China, both of which have been proposed to have a major influences on the modern landform formations and ecological and environment changes in the Three Parallel Rivers region. This phenomenon is similar to that hypothesized to have occurred in the highly specialized schizothoracine fishes (He and Chen 2007).
Morphological variation
Examination and comparisons of specimens from different localities revealed several descriptive characters (visible to the eye) in T. stenura with significant variation (Table 2). These features include body size, body color, mouth structure and intestine shape. We assume that the body shape differences are the result of a series of factors relating to population genetic differentiation, food abundance and habitat environment. Color variation might result from environmental conditions, and factors to protect fish from predation. The ability to adjust color patterns of Triplophysa is hypothesized to produce adaptive changes to adjust to the surrounding area in the stream and remain cryptic. Intestinal variability in T. stenura may also be the result of feeding habits and the abundance of different food resources in their environmental settings. If the environmental situation in one population consists of more animal than plant food, then the loach is likely to feed mainly by predation and its intestine would tend to be short; if the habitat is dominated by algae and aquatic plants, then its intestine would tend to be long and form spirals (Zhu 1981).
T. stenura is widely distributed and exists in different environmental conditions, so some of the characters traditionally used to identify the species may display ecophenotypic variation. Thus, we hypothesize that adaptive intraspecific variation exists in T. stenura, and these characters are too unstable to diagnose and differentiate different species of Triplophysa.
For the morphometric data, ANOVA recognized 16 characters that are significantly different among drainages and 23 characters that are significantly different among populations (Table S1). This is consistent with the AMOVA results of moderate genetic variation among populations of T. stenura. PCA analyses (Fig. 5) demonstrated that individuals from different drainages are not clearly distinguishable, but individuals from different populations within drainages can be distinguishable, suggesting that there is some character differentiation among populations within drainages. We hypothesize that characters varying between populations are best explained as the result of genetic and environment influences.
Stable diagnostic characters of Triplophysa
some characters that are stable within species and relatively variable between species can be used to identify and distinguish species of Triplophysa. These include whether the skin smooth or not; whether skin is scaled or scaleless; size and position of eyes; shape of cross-section of base of caudal-peduncle; shape of caudal fin; whether or not tip of depressed pelvic fin reaches anus; possessing or lacking a posterior air bladder and the shape of an existing air bladder; and the shape of intestine.
With the uplift of the Qinghai-Tibet Plateau, the transformation of landforms and water systems, species of Triplophysa gradually dispersed to their present accumulative distributional range and pattern (Zhu 1989). During the long period of geographic isolation and evolution, and the influence of a series of factors such as different climate, elevation, hydrologic conditions, feeding conditions, intraspecific and interspecific competition, different populations of the same species can have degrees of morphological variation. Therefore, most differentiation should be viewed as intraspecific rather than interspecific variation (He 2008). We suggest that the classification and identification of species in Triplophysa should be based on multiple characters, and should not overemphasize some characters with ecophenotypic variation, such as body color, color patterns, beard length and mouth structure.
Supplementary materials
ANOVA depicting the morphometric characters that are significantly different among eight populations and three drainages. Only drainages or populations significantly different from one another are shown. Number of “*” indicates P value1. (download)
Acknowledgments
Acknowledgements: We thank Jian Yang, Jinlu Li, Yuanting Jin and Yapeng Zhao for their help collecting samples. We also thank Lina Du for the help in checking the specimens of T. stenura, and a special thanks to Dr. Richard Mayden and Wansheng Jiang for helping revise the manuscript. This work was supported by the National Natural Science Foundation of China (30730017), Yunnan Provincial Science and Technology Program (2009CC008) and the Southeast Asia Biodiversity Research Institute, CAS (Y4ZK111B01).
Footnotes
Authors’ contributions: RQ, CXY and YJX collected the samples; RQ and CXY analyzed data; RQ and CXY wrote the manuscripts.
Competing interests: The authors declare that they have no conflicts of interest.
Availability of data and materials: The gene sequences are accessible from Genbank by accession numbers. The measurement data are available from the corresponding authors by request.
Consent for publication: All authors agree to the submission of this manuscript.
References
- Akaike H. A new look at the statistical mode identification. IEEE Trans Autom Contr. 19:716–723. [Google Scholar]
- Bandelt H, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Chu X L, Chen Y R. The fishes of Yunnan, China. Part I. Cyprinidae. Beijing: Science Press; 1989. [Google Scholar]
- Chu X L, Chen Y R. The Fishes of Yunnan, China. Part II. Beijing: Cyprinidae. Science Press; 1990. [Google Scholar]
- Chen Y Y. The fishes of the Hengduan Mountains region. Beijing: Science Press; 1998. [Google Scholar]
- Dowling T E, Tibbets C A, Minckley W L, Smith G R. Evolutionary relationships of the Plagopterins (Teleostei: Cyprinidae) from cytochrome b sequences. Copeia. 3:665–678. [Google Scholar]
- Drummond A J, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol; 2007. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse P E, Quattro J M. Analysis of molecular variance inferred from metric distance among DNA restriction data. Genetics. 131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer H E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Froese R, Pauly D. FishBase. World Wide Web electronic publication. Available from: http://www.fish base.org, version August 2017.
- XG Guo, SP He, YG Zhang. Phylogeny and biogeography of Chinese sisorid catfishesre-examined using mitochondrial cytochrome b and 16S rRNA gene sequences. Mol Phylogenet Evol. 2005;35:344–362. doi: 10.1016/j.ympev.2004.12.015. [DOI] [PubMed] [Google Scholar]
- He C L. Taxonomic revision of Triplophysa species in Sichuan Province. 2008.
- He D K, Chen Y F. Molecular phylogeny and biogeography of the highly specialized grade schizothoracine fishes (Teleostei: Cyprinidae) inferred cytochrome b sequences. Chinese Sci Bull. 52:777–788. [Google Scholar]
- He D K, Chen Y X, Chen Y F. Molecular phylogeny and biogeography of the genus Triplophysa. Prog Nat Sci. 16:1395–1404. [Google Scholar]
- He H S, He K Z, Ma Z J. Researches into formed age of Nu River within the boundaries of Yunnan Province. Yunnan Geol. 11:348–355. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Li J J, Fang X M. Uplift of the Tibetan Plateau and environmental changes. Chinese Sci Bull. 43:1569–1574. [Google Scholar]
- Li J J, Shi Y F, Li B Y. Uplift of the Qinghai-Xizang (Tibet) Plateau and globe change. Lanzhou: Lanzhou University Press; 1995. [Google Scholar]
- Liu H, Tzeng C S, Teng H Y. Sequence variations in the mitochondrial DNA control region and their implications for the phylogeny of the Cypriniformes. Can J Zool. 80:569–581. [Google Scholar]
- Liu Z J, Ren B P, Wei F W, Long Y C, Hao Y L, Li M. Phylogeography and population structure of the Yunnan snub-nosed monkey (Rhinopithecus bieti) inferred from mitochondrial control region DNA sequence analysis. Mol Ecol. 16:3334–3349. doi: 10.1111/j.1365-294X.2007.03383.x. [DOI] [PubMed] [Google Scholar]
- Ming Q Z. The landform development and environmental effects of Three Parallel Rivers. Beijing: Science Press; 2007. [Google Scholar]
- Ming Q Z, Shi Z T. The tentative inquiry on the formation time in the region of Three Parallel Rivers. Yunnan Geogr Environ Res. 18:1–4. [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria: 2013. [Google Scholar]
- Rambaut A. Fig Tree, version 1.31. Computer program distributed by the author. 2009.
- Rambaut A, Suchard M A, Xie W, Drummond A J. Tracer v1. 2013. 6
- Ren M E, Bao H S, Han T C. On the geomorphology and river capture of Jinsha River in North Western Yunnan. Acta Geogr Sin. 25:135–155. [Google Scholar]
- Ren Q, Yang J X, Chen X Y. A new species of the genus Triplophysa (Cypriniformes: Nemacheilidae), Triplophysa longliensis sp. Zootaxa. 3586:187–194. [Google Scholar]
- Ronquist F, Huelsenbeck J P. Bioinformatics. 3:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sânchez-Delbarrio J C, Messeguer X, Rozas R. DnaSP, NDA polymorphism analyses by the coalescent and other methods. Bioinformatics. 19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Saka R, Takehana Y, Suguro N, Sakaizumi M. Genetic population structure of Lefua echigonia inferred from allozymic and mitochondrial cytochrome b variations. Ichthyol Res. 50:301–309. [Google Scholar]
- Sambrook J, Fritsch E F, Maniatis T. Molecular cloning A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Swofford D L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10. Sunderland, Massachusetts: Sinaeur Associates; 2003. [Google Scholar]
- Tang Q Y, Liu H Z, Mayden R L, Xiong B X. Comparison of evolutionary rates in the mitochondrial DNA cytochrome b gene and control region and their implications for phylogeny of the Cobitoidea (Teleostei: Cypriniformes). Mol Phylogenet Evol. 2006;39:347–357. doi: 10.1016/j.ympev.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y F, Wu C Z. The fishes of Qinghai-Xizang Plateau. Chengdu: Sichuan Publishing House of Science and Technology; 1991. [Google Scholar]
- Xia X. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 30:1720–1728. doi: 10.1093/molbev/mst064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W H, Zhang Y P, Liu H Z. Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol Phylogenet Evol. 18:63–173. doi: 10.1006/mpev.2000.0879. [DOI] [PubMed] [Google Scholar]
- Zhang Y H, Li J J, Zhu J J, Chen Y, Pan B T, Kuang M S. Studies on development of Jinshajiang river during late Cenozoic. Yunnan Geogr Environ Res. 10:43–48. [Google Scholar]
- Zheng M P, Yuan H R, Zhao X T, Liu X F. The quaternary Pan-Lake (Overflow) period and paleoclimate on the Qinghai-Tibet Plateau. Acta Geol Sin. 80:169–180. [Google Scholar]
- Zhu S Q. Notes on the scaleless loaches (Nemacheilinae, Cobitidae) from Quinghai-Xizang Plateau and adjacent Territories in China. Geol Ecol Stud Quinghai-Xizang Plateau. 2:1061–1070. [Google Scholar]
- Zhu S Q. The Loaches of the subfamily Nemacheilinae in China (Cypriniformes: Cobitidae) Nanjing: Jiangsu Science and Technology Publishing House; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ANOVA depicting the morphometric characters that are significantly different among eight populations and three drainages. Only drainages or populations significantly different from one another are shown. Number of “*” indicates P value1. (download)