Abstract
The endocycle is a modified cell cycle that lacks M phase. Endocycles are well known for enabling continued growth of post-mitotic tissues. By contrast, we discovered pre-mitotic endocycles in precursors of Drosophila rectal papillae (papillar cells). Unlike all known proliferative Drosophila adult precursors, papillar cells endocycle before dividing. Furthermore, unlike diploid mitotic divisions, these polyploid papillar divisions are frequently error prone, suggesting papillar structures may accumulate long-term aneuploidy. Here, we demonstrate an indispensable requirement for pre-mitotic endocycles during papillar development and also demonstrate that such cycles seed papillar aneuploidy. We find blocking pre-mitotic endocycles disrupts papillar morphogenesis and causes organismal lethality under high-salt dietary stress. We further show that pre-mitotic endocycles differ from post-mitotic endocycles, as we find only the M-phase-capable polyploid cells of the papillae and female germline can retain centrioles. In papillae, this centriole retention contributes to aneuploidy, as centrioles amplify during papillar endocycles, causing multipolar anaphase. Such aneuploidy is well tolerated in papillae, as it does not significantly impair cell viability, organ formation or organ function. Together, our results demonstrate that pre-mitotic endocycles can enable specific organ construction and are a mechanism that promotes highly tolerated aneuploidy.
Keywords: Endocycle, Polyploidy, Multipolar, Aneuploidy, Centrosome, Drosophila
INTRODUCTION
In many post-mitotic tissues, continued growth and development is achieved by a cell cycle modification known as the endocycle. In diverse tissues, including the human trophoblast lineage, endocycles alternate DNA synthesis and gap phases to generate polyploid cells (see cell cycle nomenclature in the Materials and Methods; Fox and Duronio, 2013; Pandit et al., 2013). The polyploidy resulting from endocycles usually increases the size of individual cells, thus enabling post-mitotic tissue growth (Edgar et al., 2014; Epstein, 1967). The use of endocycles in post-mitotic tissue growth was recently shown to be crucial for Drosophila blood-brain barrier integrity (Unhavaithaya and Orr-Weaver, 2012).
Although endocycles frequently occur in post-mitotic cells, a return to an M-phase-like state is possible after the endocycle. In the developing Drosophila ovary, 15 germline nurse cells undergo five pre-mitotic endocycles, which are followed by an M-phase-like period where homologous chromosomes condense and locally disperse via a condensin-mediated mechanism (Dej and Spradling, 1999; Hartl et al., 2008). This mitotic state can progress further in mutants that accumulate mitotic cyclins, leading to spindle formation and metaphase chromosomes in these polyploid nuclei (Reed and Orr-Weaver, 1997). However, nurse cells do not normally segregate sister chromosomes into daughter cells after this M-phase-like period but instead return to further endocycles without mitosis-like chromosome separation/compaction.
Recently, we discovered the first example of a complete return to mitosis after endocycles in normally developing Drosophila. In 2nd instar larvae, the rectum consists of a one-cell thick tube of ∼100 cells. These larval cells undergo two endocycles to generate octoploid cells. Subsequently, during metamorphosis, these octoploid cells undergo, on average, two complete cell divisions as the rectal tube splits into four cone-shaped luminal structures termed rectal papillae (papillae or papillar cells). Papillae are thought to perform significant water, ion and metabolite absorption in diverse insects (Phillips, 1981; Wigglesworth, 1942). Taken together, the above-mentioned nurse and papillar cell studies suggest distinct endocycle programs may dictate whether polyploid cells can later enter mitosis. If so, identifying molecular differences between pre- and post-mitotic endocycling cells can uncover key regulation that promotes or suppresses polyploid cell division.
If polyploid cells do successfully enter mitosis, one recurring challenge for such cells is the propensity for mitotic errors. For endocycled cells, this is most extensively documented in cancer cells, in which endocycles are associated with mitotic chromosome aberrations and aneuploidy (Davoli and de Lange, 2012; Levan and Hauschka, 1953). Similarly, inducing ectopic endocycles in Drosophila can lead to error-prone division and to aneuploidy (Hassel et al., 2014; Vidwans et al., 2002). Much like aberrantly endocycling cells, mitosis of Drosophila papillar precursors is highly error prone (Fox et al., 2010). Given its accessible genetics and development, Drosophila papillar formation provides an amenable system in which to study why error-prone polyploid divisions, rather than faithful diploid divisions, would be used for organ construction.
One potential challenge for polyploid mitotic cells is the amplification of centrosomes and subsequent multipolar division (Duncan et al., 2010; Hassel et al., 2014). In many cells with multipolar spindles at metaphase, mechanisms exist to reduce – but not eliminate – the degree of aneuploidy from a multipolar anaphase. In one mechanism, multipolar spindles are transient. Instead of multipolar division, extra centrosomes cluster. Such clustering leads to merotelic microtubule attachments and minor chromosome mis-segregation during a bipolar division, instead of major chromosome mis-segregation from a tripolar division (Duncan et al., 2010; Ganem et al., 2009; Silkworth et al., 2009). A second mechanism is termed reductive division. In reductive divisions, spindles remain tripolar, but DNA is segregated in a near 2:1:1 ratio (Duncan et al., 2010; Mazia et al., 1960). In tetraploid mouse hepatocytes, this mechanism is thought to generate one near tetraploid and two ploidy-reduced near diploid daughters, thus minimizing the often detrimental imbalance between different chromosomes that results from random tripolar DNA segregation (Duncan et al., 2010; Gentric and Desdouets, 2014). Recent evidence also argues that, in the absence of clustering or reductive divisions, even a single tripolar division can be lethal (Ganem et al., 2009; Stewénius et al., 2005). The cell death associated with lethal tripolar divisions was recently attributed to causing microcephaly in mice (Marthiens et al., 2013).
Here, we address the role of endocycles and resultant error-prone divisions in papillar formation. We find that papillar cells require pre-mitotic endocycles for normal development. In the absence of pre-mitotic endocycling, the larval rectum fails to accurately disperse into four adult papillar structures. As a result, papillar structures are malformed, and the resulting flies are intolerant of high-salt dietary conditions. Although pre-mitotic endocycles are thus required for papillar development, we find they are also a mechanism of papillar aneuploidy. In both the M-phase-capable polyploid papillar cells and stage 5 nurse cells, centrioles can be retained. As a result, papillar cells accumulate supernumerary centrosomes during pre-mitotic endocycles, which then lead to tripolar anaphases. These anaphases lack significant centrosome clustering or 2:1:1 reductive divisions, leading to highly variable DNA segregation and aneuploidy. Furthermore, increasing the rate of such variably aneuploid tripolar divisions does not extensively perturb papillar morphogenesis, viability or function. Taken together, our results define a role for pre-mitotic endocycles in organ formation and aneuploidy generation. Our findings suggest centriole retention distinguishes M-phase-competent endocycled cells, and ties this endocycle variant to significant (yet tolerated) aneuploidy.
RESULTS
Pre-mitotic endocycles retain centrioles
Unlike papillar cells, most endocycled cells do not divide. We thus searched for cellular features that might distinguish pre-mitotic and post-mitotic endocycles. One potential distinguishing feature between non-mitotic endocycling cells and mitotic cells is elimination of centrioles. A previous report found that post-mitotic follicle cells of the adult female Drosophila ovary lack centrioles by stage 10 (Mahowald et al., 1979), at which point follicle cells have completed three rounds of endocycles (Lilly and Spradling, 1996, Calvi et al., 1998). We thus examined whether centriole retention correlates with the mitotic potential of endocycled cells. To examine centrioles in diverse endocycling tissues, we examined localization of the centriole-associated protein Pericentrin-like protein (Plp; Martinez-Campos et al., 2004) and the centriolar scaffold Asterless (Asl; Dzhindzhev et al., 2010). We defined a centriole as a discrete focus containing both of these proteins.
First, we re-examined centriole loss in the post-mitotic follicular epithelium using Plp and Asl antibodies. We detected a sharp decline in centriole number (Asl+, Plp+ foci) between stages 8 and 9, and rarely detected any centrioles by stage 10 (Fig. 1A-C). Next, we examined centrioles in additional non-mitotic endocycling tissues. Beginning 8 h into embryonic development, cells of several Drosophila alimentary canal tissues also enter endocycles and, although such endocycling may be discontinuous, these tissues undergo further endocycling/polyploidization during larval development (Smith and Orr-Weaver, 1991). By late larval development, we observed a lack of centrioles in post-mitotic endocycled cells from four 3rd instar larval cell types: midgut enterocytes, salivary gland secretory cells, enterocytes of the hindgut ileum and polyploid Malpighian tubule cells (Fig. 1D,E,I, supplementary material Fig. S1A). Thus, centriole loss is a frequent property of post-mitotic endocycling tissues.
Fig. 1.
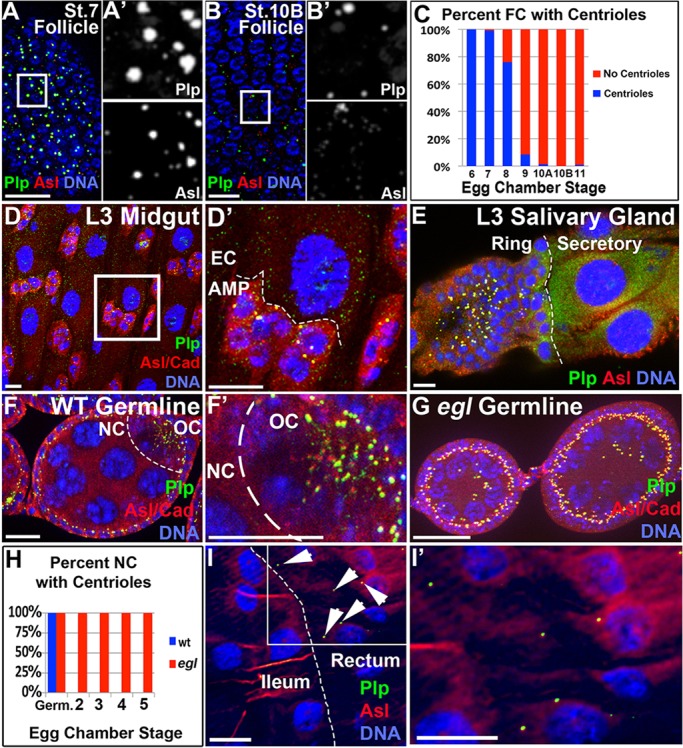
Centriole elimination and retention in polyploid Drosophila tissues. (A) Follicle cell centrioles (co-labeled with Plp and Asl) in a stage 7 egg chamber. (A′) Split Plp and Asl channels, magnified fourfold, from the boxed region in A. (B) Stage 10B follicle cells lacking centrioles, as evidenced by lack of overlapping Asl/Plp foci. (B′) Split Plp and Asl channels, magnified fourfold, from the boxed region in B. (C) Quantitation of centriole loss in follicle cells (FC), where the presence of a centriole is defined by Asl and Plp co-labeling (n=127-2213 cells/stage). (D) Centrioles are absent from polyploid enterocytes (EC) of the 3rd instar larval (L3) midgut, but are present in the diploid adult midgut precursor (AMP) nests. Cadherin (Cad, red) marks cell boundaries in this and in other panels where indicated. (D′) High magnification of boxed region in D. (E) Centrioles are present in the diploid imaginal ring, but not in polyploid secretory L3 salivary gland cells. Dashed line indicates the boundary between diploid and polyploid cells. (F) Stage 5 wild-type egg chamber germ cells with centrioles only in the oocyte (OC), not in the nurse cells (NC). (F′) Higher magnification of F. (G) Stage 4 and 5 egl1 egg chambers germ cells with centrioles retained in the NCs. (H) Quantitation of centriole retention in egl1 mutant and wild-type nurse cells. Germ., germarium. Ten wild-type and five egl chambers were counted per stage. (I) Centrioles are absent in the polyploid ileum of the L3 hindgut but are present in the rectum (arrowheads). (I′) High magnification of boxed region in I. Scale bars: 20 µm.
Our findings in the post-mitotic endocycling cell types we examined contrast our findings in M-phase-prone endocycling cells. Fifteen ovarian nurse cells enter a mitosis-like state after five endocycles (Dej and Spradling, 1999). Although these endocycled nurse cells normally lack centrioles (Fig. 1F), it is not that the centrioles cannot be detected, but rather that they cluster and migrate to the neighboring oocyte through inter-connected ring canals (Fig. 1F,F′; Mahowald and Strassheim, 1970).
We thus blocked oocyte specification to determine whether these M-phase-capable endocycled cells possess a mechanism that actively retains centrioles. To do this, we examined centriole localization in egalitarian (egl1) mutant ovaries. These mutants lack a specified oocyte and instead have 16 polyploid nurse cells, owing to defective dynein-mediated recruitment of oocyte-localized mRNAs mediated by the RNA-binding activity of Egl (Theurkauf et al., 1993; Mach and Lehmann, 1997; Clark et al., 2007; Dienstbier et al., 2009). Indeed, nurse cell centrioles remain in polyploid nurse cells during egl1 mutant endocycles (Fig. 1G,H). Perhaps as a remnant of the migratory process, these centrioles frequently clustered in one area of the egg chamber (Fig. 1G). Importantly, egl1nurse cell centrioles are still present at stage 6, the furthest that egg development progresses in this genotype. At this stage, nurse cells have undergone five endocycles to reach 64C DNA content, and have thus completed more endocycles than wild-type stage 13 follicle cells (16C), which have lost centrioles (Fig. 1C; Dej and Spradling, 1999; Mach and Lehmann, 1997), suggesting that it is not a lesser degree of endocycling that enables egl1 nurse cells to retain centrioles.
Similar to egl1 nurse cells, wild-type papillar precursors of the 3rd instar larval rectum also retain centrioles (Fig. 1I,I′). The ability of rectal cells to retain centrioles for the ∼5 day period after which they have exited the endocycle suggests that cell cycle activity is not required for centriole retention. This observation is further supported by our data that several adult follicle cells begin to lack centrioles by egg chamber stage 8, prior to exit from the endocycle (Fig. 1C; Lilly and Spradling, 1996; Calvi et al., 1998). Taken together, our examination of Drosophila endocycling tissues suggests that centriole retention correlates with future polyploid mitotic potential.
Pre-mitotic endocycles are a mechanism of centriole amplification that seeds aneuploidy generation
We previously characterized programmed cell cycle alterations that accompany Drosophila rectal papillar construction. During the 2nd instar larval stage (L2) of Drosophila development, precursors of the adult papillae endocycle to increase chromosome content from diploid to octoploid (Fig. 2A). Subsequently, during hours 22-48 of pupal development (P1-P2), these precursors undergo canonical mitotic cell cycles as octoploid cells (Fig. 2A; Fox et al., 2010).
Fig. 2.
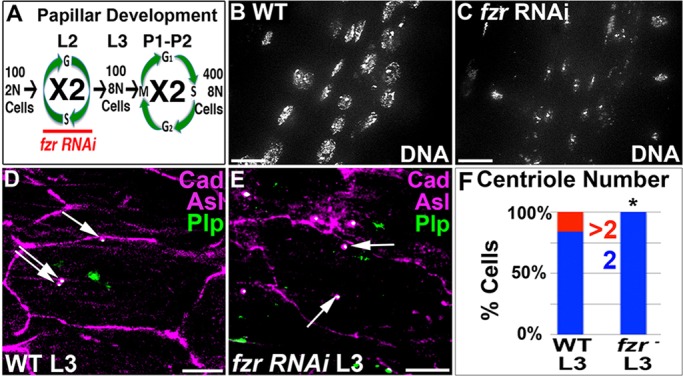
Centrioles amplify during papillar endocycles. (A) Overview of the papillar cell cycle program. L2 and L3 refer to larval instars, P1 and P2 refer to days since pupation. Period of fzr RNAi indicated. (B,C) DAPI (DNA) stain of L3 rectums. (B) Wild type with octoploid nuclei. (C) Transient fzr RNAi, as per the scheme in A, taken at the same exposure settings as B. (D,E) Centrioles in a single representative L3 rectal cell. Cadherin, purple; Plp, green; Asl, purple. (D) Wild-type cell with three centrioles. Anti-Plp occasionally recognizes a structure in the nucleus. (E) fzr RNAi cell with two centrioles. Arrows indicate centrioles. (F) Quantitation of centriole number from D and E. At least 46 cells were counted per experimental condition, each of which included a minimum of two replicates with multiple animals scored per genotype. The distribution of centriole number in fzr RNAi is significantly different from wild type (*P<0.005, χ2 test). Scale bars: 10 µm.
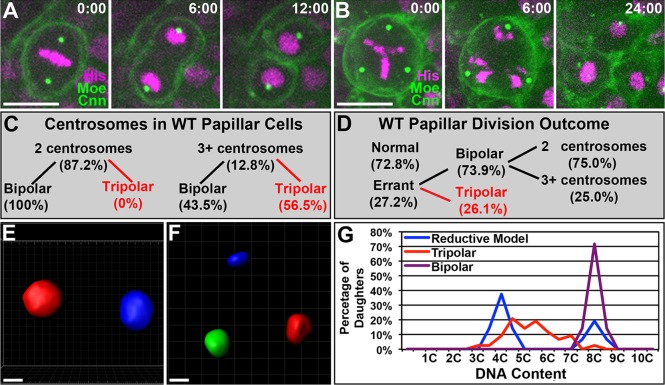
Given that papillar cells retain centrioles in the larval stages, we next examined the consequences of such centriole retention. Plp focus staining is a well-demonstrated readout of centriole pair number (Bettencourt-Dias et al., 2005; Brownlee et al., 2011; Slevin et al., 2012), and Asl co-staining provided us with further verification of centriole pair number. Counting the number of Plp+ Asl+ foci in individual papillar precursors revealed that although most cells have two discernible centriole pairs, 16% of rectal cells contain additional centrioles (Fig. 2D,F).
To determine whether the increased centriole number in a subset of papillar cells depends on endocycle progression, we blocked endocycles throughout the developing hindgut using fizzy-related (fzr) RNAi. fzr is an activator of the anaphase promoting complex/cyclosome (APC/C), which is necessary and sufficient for its oscillations during endocycle progression (Narbonne-Reveau et al., 2008; Sigrist and Lehner, 1997; Zielke et al., 2011). We used the Gal80 temperature-sensitive system to restrict fzr knockdown to the brief period of larval endocycling, thus avoiding any potential effects on pupal mitosis (Fig. 2A, see Materials and Methods). Using byn Gal4, we expressed fzr RNAi in the hindgut during L2 endocycles (Fig. 2A, fzr). In fzr L3 larvae, papillar precursor nuclei are clearly smaller and contain less DNA (compare Fig. 2B with C). As a further test of the efficacy of our fzr RNAi, we examined S phase activity in wild type and fzr. In wild-type L2 animals, EdU frequently accumulated in early and late S-phase patterns in the rectum (supplementary material Fig. S1B,D; Fox et al., 2010). By contrast, fzr animals contained very few cells in S phase (supplementary material Fig. S1C,D). Thus, fzr RNAi effectively blocks rectal endocycles.
Unlike wild type, we could not detect fzr papillar precursors with more than two centriole pairs (Fig. 2E,F). These results suggest that papillar endocycles increase centriole number, albeit inefficiently. Such inefficient centriole amplification could be due to the lack of efficient centrosome licensing that normally takes place as cells pass through mitosis (Nigg and Raff, 2009).
As centriole amplification is tied to aneuploidy, we next examined the contribution of centriole amplification to papillar development. To follow centrosomes during pupal divisions, we expressed Centrosomin GFP (CnnGFP, see Materials and Methods) or used a Cnn antibody (supplementary material Fig. S2A). Thirteen percent of all dividing papillar cells contain three or four Cnn foci (compare Fig. 3A, with B,C). This frequency is very close to the frequency of cells with supernumerary centrioles that we find prior to papillar mitosis (16%, Fig. 2F), suggesting that each Cnn focus is a single centrosome. It remains possible that our centriole/centrosome counting at the light microscope cannot always discern very closely associated centriole pairs, leading to an under-estimation of papillar centrosome number. However, in 10/10 examples, we found only single centriole pairs in mitotic papillar cells using serial section electron microscopy (supplementary material Fig. S2B).
Fig. 3.
Multipolar division generates variable aneuploidy in papillae. (A) Time-lapse of 8N cell undergoing non-errant mitosis. Transgenes: H2AV-RFP (His), Moesin GFP (Moe) and Centrosomin GFP (Cnn). (B) Time-lapse of 8N cell undergoing tripolar mitosis. Transgenes as in A. Time is in minutes relative to anaphase onset. (C) Distribution of bipolar and tripolar divisions in relation to observed centrosome number in wild-type papillae. (D) Distribution of normal, errant bipolar and errant tripolar divisions in wild-type papillar cells. For errant bipolar divisions, the fraction of cells with observed extra centrosomes is also indicated. Data in A-D are from n=379 divisions from among 37 replicate experiments. (E,F) 3D volumetric analysis of daughters from a non-errant bipolar (E) and a tripolar (F) papillar division. (G) Histograms tracking the percentage of volumetrically established DNA content (see Materials and Methods) after a bipolar or tripolar division. These values are plotted relative to the expected segregation pattern for a 2:1:1 reductive division (see Materials and Methods). Data in E-G are from a minimum of n=14 divisions per division outcome. Scale bars: 10 µm.
We next followed papillar divisions in cells with extra centrosomes. Previously, we have reported that many papillar cells contain mitotic bridges or lagging chromosomes (Fox et al., 2010). Given that lagging chromosomes are connected to clustering of extra centrosomes and merotelic spindle attachments (Ganem et al., 2009; Silkworth et al., 2009), we examined whether extra centrosomes correlate with papillar mitosis errors. To monitor chromosomes and cytokinesis, the CnnGFP flies also expressed RFP Histone H2AV (HisRFP) and Moesin GFP (MoeGFP, Fig. 3A; supplementary material Movie 1).
As a result of metaphase centrosome clustering, many studies of cells with extra centrosomes report less than 10% complete multipolar anaphase (Basto et al., 2008; Duncan et al., 2010; Ganem et al., 2009; Marthiens et al., 2013; Silkworth et al., 2009). By contrast, although we could detect efficient centrosome clustering in some papillar cells (defined as two centrosomes moving together prior to a bipolar division; supplementary material Fig. S2C; Fig. 3C,D; supplementary material Movie 2), the majority of papillar cells with extra centrosomes (56.5%) do not efficiently/fully cluster and instead undergo multipolar division (Fig. 3B-D; supplementary material Movie 3). Papillar multipolar divisions are always tripolar and account for approximately one quarter of papillar division errors (Fig. 3D). An additional type of papillar mitotic defect, chromosome bridging/lagging, also occurs. Defects in this class occur not only when we can clearly detect extra centrosomes, but also where we observe only two unclustered centrosomes/spindle poles (Fig. 3D; supplementary material Fig. S2D,F). From these analyses, we conclude that (1) centrosomes do not efficiently cluster during papillar mitosis, leading to frequent multipolar mitosis; and (2) centrosome clustering is not the only cause of papillar division errors.
We next examined the degree of aneuploidy generated by tripolar papillar divisions. In addition to clustering, previous studies (Duncan et al., 2010; Mazia et al., 1960) showed that cells with complete tripolar DNA segregation can decrease the degree of aneuploidy by partitioning the chromosomes in an almost even reductive 2:1:1 ratio. Using 3D rendering of total DNA, we were able to determine whether papillar cells frequently undergo such reductive divisions. Bipolar papillar anaphases evenly distribute half of the DNA into each daughter with high frequency (Fig. 3E,G). By contrast, tripolar papillar anaphases exhibit high variability in DNA segregation (Fig. 3F,G), with some cells undergoing equal (33%) three-way segregation, whereas others have over twofold DNA content differences between daughters (supplementary material Fig. S2E). This resulting DNA content distribution differs significantly from an expected distribution of cells undergoing near 2:1:1 reductive divisions (P<0.05; Fig. 3G, see Materials and Methods). Thus, instead of employing efficient centrosome clustering or reductive division mechanisms, papillar cells with extra centrosomes yield daughters with highly variable aneuploidy.
Papillar development accommodates frequent multipolar aneuploidy
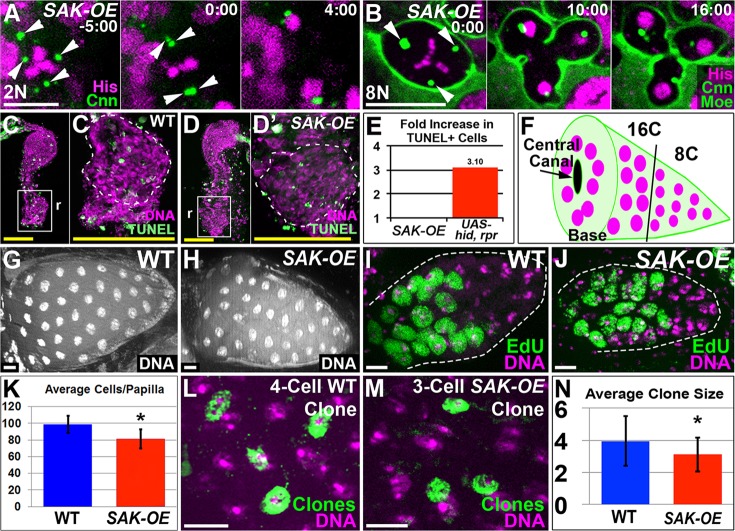
Our above results found tripolar mitosis occurs during wild-type papillar development, and that papillar cells lack efficient clustering or ploidy reduction mechanisms that decrease (but do not eliminate) the ensuing aneuploidy. Although only one round of such divisions can be lethal, the extra chromosome copies in papillar cells might lessen the impact of an aneuploid genome on cell survival. To test this idea, we reasoned that increasing the number of papillar cells with tripolar divisions from the 7% level in wild type would reveal whether such aneuploidy is viable. To increase the frequency of tripolar divisions, we ectopically expressed plk4/SAK overexpression transgenes (SAK-OE, see Materials and Methods), which were previously shown to induce extra centrosomes (Basto et al., 2008; Rodrigues-Martins et al., 2007).
We first expressed SAK-OE in 2N larval neuroblasts. As expected from previous work (Basto et al., 2008), SAK-OE neuroblasts frequently have more than two centrosomes (Fig. 4A; supplementary material Fig. S3A,B). Also consistent with previous results (Basto et al., 2008), we find neuroblasts cluster or inactivate extra centrosomes (Fig. 4A; supplementary material Movie 4) and spindle poles (supplementary material Fig. S3A,C, Movie 5) to undergo bipolar division. Next, we expressed SAK-OE constructs in 8N papillar cells. These constructs increased the percentage of cells with three or more distinct centrosomes from 12.8% in wild type to 89.1%, and the distribution of centrosome number was similar to 2N SAK-OE neuroblasts (supplementary material Fig. S3A,B). However, in contrast to 2N SAK-OE neuroblasts, 8N SAK-OE papillar cells with extra centrosomes maintain a similar rate (57%) of tripolar division as wild type (Fig. 4B; supplementary material Fig. S3A,D,E, Movies 6 and 7). Thus, SAK-OE increases the tissue-wide tripolar division frequency, but does not alter the probability that an individual papillar cell with amplified centrosomes will undergo tripolar division. The lack of papillar centrosome clustering is dependent on fzr activity during endocycles, as expressing fzr RNAi during endocycles (as in Fig. 2) in SAK-OE papillar cells dramatically decreases the rate of multipolar papillar division during pupal development (10% tripolar divisions; supplementary material Fig. S3E-G, Movie 8). Taken together, these data suggest passage through endocycles interferes with the ability of papillar cells to cluster amplified centrosomes.
Fig. 4.
Multipolar aneuploidy is well tolerated and does not perturb papillar development. (A,B) Time-lapse of centrosomes in (A) ubi-SAK neuroblast undergoing centrosome clustering and bipolar division or (B) tripolar division of a ubi-SAK papillar cell. Transgenes are indicated. Arrowheads indicate centrosomes. Time is in minutes relative to anaphase onset. (C-E) TUNEL (green) staining in mitotic stage pupal rectums. DNA, purple; r, rectum. (C) Wild type. (D) ubi-SAK. (C′,D′) High-magnifications of boxed region shown in C and D, respectively. (E) Graph of fold difference in TUNEL staining between wild type versus ubi-SAK or UAS-hid, UAS-rpr (n=10 animals/genotype). (F) Schematic of adult papillar structure, illustrating base with central canal and regional ploidy differences. (G,H) DNA (DAPI) stain of one side of a single wild-type (G) or ubi-SAK (H) adult papilla. (I,J) EdU labeling (green) of post-mitotic S-phase in a 50 h pupal papilla from (I) wild type or (J) ubi-SAK. DNA, purple; EdU, green. (K) Graph of adult papillar cell number in wild type and ubi-SAK from at least 28 papillae/genotype. Data are mean±s.d. The difference in cell number is significantly different where indicated (*P<0.0001 by unpaired t-test). (L) Four-cell clone (green) induced during wild-type papillar development. (M) Three-cell clone (green) induced during ubi-SAK papillar development. (N) Average clone size from lineage experiments. Data are mean±s.d.; n=54 wild-type and 34 ubi-SAK clones; nearly every clone is from a separate animal. A minimum of two experimental replicates/genotype was performed. Clone size in wild type and ubi-SAK are significantly different where indicated (*P<0.01 by two-tailed t-test). Scale bars: 10 µm (white); 250 µm (yellow).
As SAK-OE increases the number of tripolar divisions in developing papillae approximately eightfold, we could determine whether three-way division leads to lethality in this polyploid tissue. By TUNEL staining, we detect some cell death during wild-type rectal development, which probably reflects the extensive cell death known to be associated with remodeling of the hindgut during metamorphosis (Robertson, 1936). However, we could not detect any increase in apoptotic cell death between wild-type and SAK-OE pupal rectums (Fig. 4C-E), whereas a positive pro-apoptotic control (UAS-hid, UAS-rpr) led to an obvious increase in TUNEL labeling (Fig. 4E). Thus, multipolar division in papillar tissue is highly viable throughout development and into adulthood.
Given our observed lack of increased cell death in SAK-OE papillae (up to 75% multipolar anaphase; supplementary material Fig. S3C), and the recent finding that 3.6% multipolar anaphase in neural stem cells causes microcephaly in plk4-OE (SAK-OE) mice (Marthiens et al., 2013), we next examined whether increased tripolar aneuploidy hinders papillar development. Wild-type adult papillae have a cone-like shape (Fig. 4F). Despite the high level of aneuploid divisions in SAK-OE animals, adult papillae appear similar in size and shape to controls (compare Fig. 4G with H).
We further examined whether SAK-OE papillae contained any gross defects in organ morphology. Insect papillae are frequently wider at the base, where cells surround a centrally located canal (Fig. 4F), which is thought to help recycle reabsorbed water and ions back into the fly's circulatory system (Berridge and Gupta, 1967; Wigglesworth, 1942). Although most wild-type papillar cells are close to 8C DNA content after pupal day 2 (P2) divisions, we previously showed that on pupal day 3 (P3) the cells that form the wider bottom region of the papillae go through one post-mitotic S-phase to become 16C (Fox et al., 2010). In addition to these regional ploidy differences, adult papillae also exhibit regional gene expression differences. From a screen of a recently released Gal4 enhancer line collection (Pfeiffer et al., 2008), we identified three separate Gal4-tagged enhancers expressed at the papillar base (supplementary material Fig. S4A,C,E). Thus, developing papillae exhibit localized differences in post-mitotic S-phase and enhancer expression. These localized differences appear grossly normal in SAK-OE animals, as determined by both post-mitotic S-phase pattern (compare Fig. 4I with J) and location of papillar base cell reporter expression (supplementary material Fig. S4B,D,F).
Upon closer examination, we did note a ∼20% cell number decrease in SAK-OE papillae (Fig. 4K). Given the high tripolar division frequency in SAK-OE animals and the production of one extra daughter by these divisions, this number decrease could reflect the lack of additional mitosis following a single tripolar division. If so, the average division number in SAK-OE papillae should decrease relative to wild type. To test this model, we performed clonal analysis of papillar development by sporadically inducing a permanent clonal mark before division (supplementary material Fig. S4G, see Materials and Methods). We then examined the output of each precursor by examining clone size in adults.
By cell counts, we previously estimated that each wild-type papillar precursor produces four cells. This estimate agrees with our wild-type lineage data for both the mean (3.9, Fig. 4L,N) and median (4) clone size. We also detect smaller (two or three cell) and larger (maximum seven cells) clones in wild type, suggesting papillar precursors divide one to three times. Relative to wild type, SAK-OE animals have a smaller mean (3.1, P<0.01, Fig. 4M,N) and median (3) clone size. Furthermore, the number of SAK-OE clones greater than three cells (which must reflect multiple rounds of divisions) decreases in SAK-OE animals (from 67% to 36%; supplementary material Fig. S4H). This latter finding supports our data on lack of cell death in SAK-OE animals, as cell death could prompt a compensatory proliferation response, leading to larger clones from the remaining cells. It remains possible that, over time, some attrition of a minor fraction of tripolar daughters occurs. Overall, our cell death, cell count and lineage results suggest tripolar papillar divisions produce aneuploid daughters, the majority of which remains viable into adulthood.
Rectal papillar development requires pre-mitotic endocycles
Our above results suggest centriole retention distinguishes pre-mitotic endocycles and also contributes to error-prone mitosis and long-term aneuploidy. Thus, this developmentally programmed organ formation event employs a highly error-prone method to generate new cells. Given that all other known mitotic precursors in Drosophila are diploid, we hypothesized that papillar development employs error-prone division because of a requirement for pre-mitotic endocycles. To test this model, we again blocked papillar endocycles by transiently expressing fzr RNAi as in Fig. 2A. We then allowed these animals to proceed through papillar morphogenesis, when the rectal tube splits into four papillae and becomes enveloped by the genital imaginal disc (Fig. 5A,E; Fox et al., 2010). Unlike wild-type pupae, which uniformly exhibit near octoploid papillar karyotypes (Fig. 5B; Fox et al., 2010), fzr pupae frequently exhibit only diploid or only diploid and tetraploid mitotic karyotypes (Fig. 5C,D). Although chromosome number is decreased in fzr mutants, these animals exhibit a similar number of papillar precursors in late larval development, before the mitotic period (Fig. 5G). These results are consistent with efficient knockdown of pre-mitotic endocycles, leading to substantial reduction of polyploidy in papillar precursors following transient fzr RNAi.
Fig. 5.
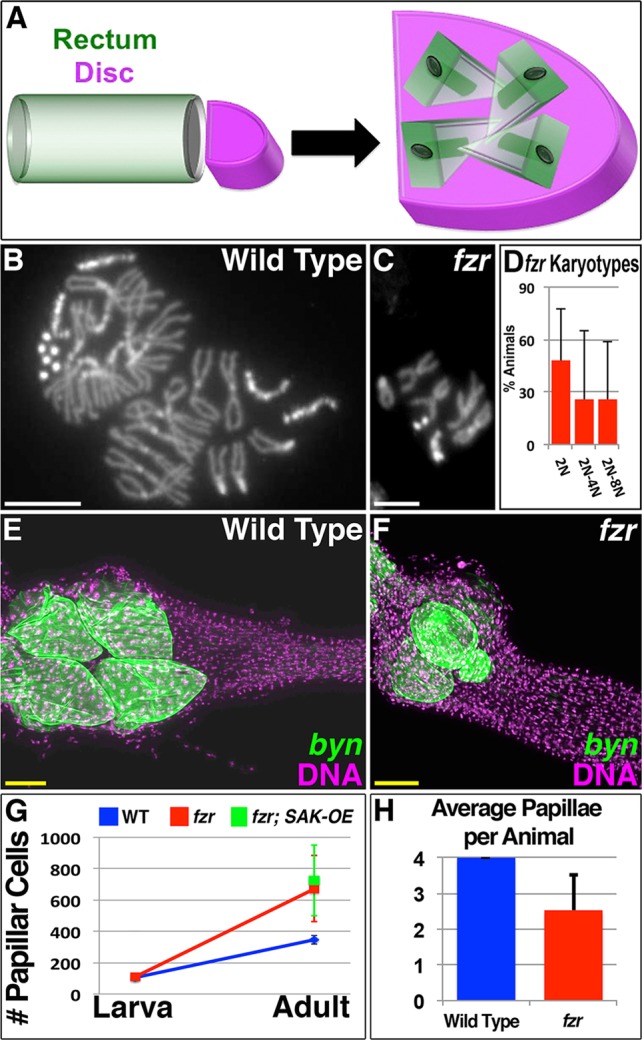
Pre-mitotic endocycles are required for papillar morphogenesis. (A) Schematic of papillar morphogenesis during pupation, with rectal (green) and genital disc (purple) cells indicated. (B) Nearly 8N wild-type male papillar karyotype, with one X- and two 4th-chromosomes missing, likely due to chromosome mis-segregation. (C) fzr RNAi 2N male papillar karyotype. (D) Graph of distribution of karyotypes/animal seen in fzr RNAi experiments. Twenty-seven animals were scored from among 10 experimental replicates. Data are mean±s.d. (E) Wild-type adult rectum with byn Gal4-driven UAS-GFP-labeled papillae in green (DNA is in purple). (F) Transient fzr RNAi adult rectum with byn Gal4-driven UAS-GFP-labeled papillae in green (DNA is in purple). (G) Average papillar cell number in wild-type, fzr RNAi and fzr RNAi; SAK-OE animals, assayed at L3 and/or adult stages. Data are mean±s.d. (H) Average papillar number in wild-type and transient fzr RNAi adults. Data are mean±s.d. For fzr RNAi, 64 animals were scored in total from among six experimental replicates. Scale bars: 5 µm (white); 50 µm (yellow).
After fzr animals progress through papillar morphogenesis, we observed a striking morphological phenotype. Unlike in wild type (four uniform papillae), fzr animals frequently contain three or fewer mis-shaped papillae (Fig. 5E,F,H). This suggests fzr animals have a defect in splitting of the single rectal tube into four papillae. Furthermore, perhaps as a compensatory response to blocking the endocycle in fzr papillae, these animals undergo additional proliferation of the now mostly diploid cells, as overall papillar cell number increases (Fig. 5G), owing to additional S- and M-phase activity during the normal period of pupal number expansion (supplementary material Fig. S5). This cell number increase also occurs in fzr RNAi; SAK-OE animals (Fig. 5G), suggesting the small amount of remaining tripolar divisions in this genotype does not substantially alter the fzr phenotype. Taken together, we find pre-mitotic endocycling promotes tripolar division, but is required for proper formation of Drosophila rectal papillae.
The papillar pre-mitotic endocycle program is required for survival under salt-stress conditions
Finally, we examined the impact of the papillar cell cycle program and its resulting aneuploidy on organismal physiology. Given the papillar-specific defect of fzr animals in the hindgut (supplementary material Fig. S6A versus B), and the lack of significant byn expression in other adult fly tissues (Chintapalli et al., 2007), we were able to use fzr animals to examine the physiological effect of blocking pre-mitotic papillar endocycles. Despite exhibiting severe papillar organogenesis defects, fzr adults are similarly viable relative to controls when fed a control diet (Fig. 6A). As papillae are thought to regulate water and ion balance in dipterans (Berridge and Gupta, 1967; Garrett and Bradley, 1984), we next used a salt-stress assay to examine the physiological function of wild type and various mutant papillae. We subjected wild-type and fzr flies to a high-salt diet (see Materials and Methods). Relative to controls, fzr animals have a marked decrease in survival under high-salt conditions (compare Fig. 6A with B). These results suggest disrupting pre-mitotic endocycles impairs papillar formation to a point that is detrimental to organ function.
Fig. 6.
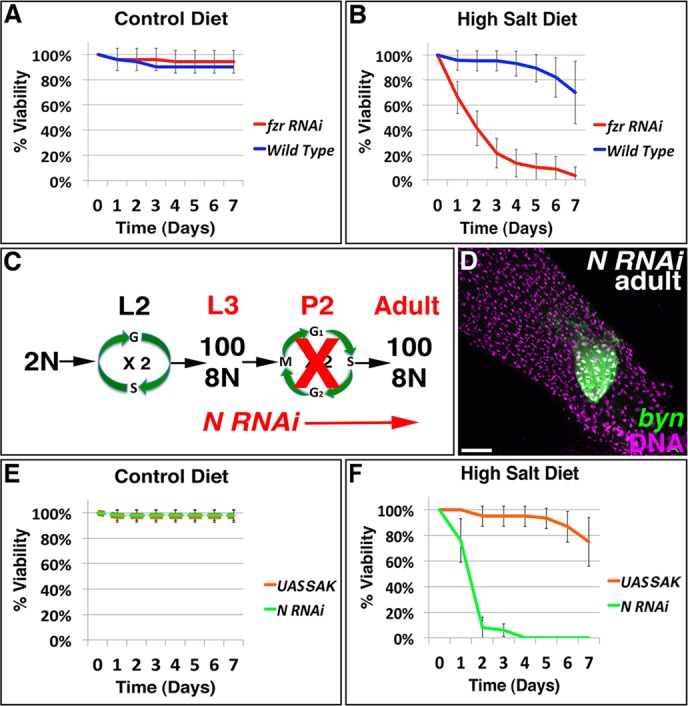
The papillar cell cycle program is required for survival under salt-stress conditions. (A) Graph of percentage survival in wild-type and fzr RNAi animals, as a function of days subjected to sucrose (control) diet. (B) Percentage survival in the same genotypes as A, as a function of days subjected to salt stress. (C) Scheme to block mitosis of 8N papillar cells with Notch (N) RNAi. (D) Rectum of representative N RNAi animal with byn Gal4-driven UAS-GFP-labeled papillae in green (DNA is in purple). (E) Percentage survival in animals expressing either N RNAi or UAS-SAK, as a function of days subjected to sucrose (control) diet. (F) Percentage survival in the same genotypes as E, as a function of days subjected to salt stress diet. Data are from at least five replicates from 15 female flies per genotype (mean±s.d.). Scale bar: 50 µm.
To similarly test whether papillar division after endocycles is required for wild-type papillar function, we exposed mutant flies expressing Notch (N) RNAi to high salt. We have previously shown that N RNAi prevents division and morphogenesis of papillar cells, resulting in one or two papillae with ∼100 total cells, instead of four papillae with ∼400 total cells (Fox et al., 2010). As with fzr RNAi, N RNAi (expressed with byn Gal4 during mitosis, when N acts in the papillae, Fig. 6C) disrupts only rectal papillae and not the rest of the hindgut (Fig. 6D, supplementary material Fig. S6C). Furthermore, as for fzr flies, N RNAi flies die when fed a high-salt diet (Fig. 6E,F). Together, we find pre-mitotic endocycles and subsequent polyploid divisions in Drosophila rectal papillae are key to organ formation and function.
Finally, we examined whether the highly aneuploid papillae formed by SAK-OE retain adult papillar function. Unlike fzr and N animals, SAK-OE animals display high-salt tolerance (compare Fig. 6E with F). Taken together, these results show that the papillar cell cycle program of endocycles followed by mitosis is crucial for adult Drosophila survival under high-salt stress. This essential cell cycle program generates significant aneuploidy through retention and amplification of centrioles, but such aneuploidy does not impair cell survival or function.
DISCUSSION
Recently, we described the contribution of error-prone polyploid mitotic precursors to the generation of Drosophila adult rectal papillar structures. This discovery led to three related questions: (1) why do papillae use a polyploid precursor, when all other known Drosophila precursors are diploid; (2) what distinguishes mitotic papillar cells from non-mitotic polyploid cells; and (3) what is the long-term outcome of errant papillar divisions? In this present study, we explored these inter-related questions. Our data show that: (1) pre-mitotic endocycles are indispensable for normal papillar organ construction, yet (2) they retain and amplify centrioles, unlike non-mitotic polyploid Drosophila cells. However, (3) the resulting centriole-induced aneuploidy is both well tolerated and non-disruptive with regards to tissue formation and function.
Papillar development requires pre-mitotic endocycles
In the absence of pre-mitotic endocycles, papillar formation is severely disrupted. Based on the well-known connection between endocycles/polyploidy and increased cell size, a likely explanation is that fzr RNAi decreases the size of rectal cells, which prevents proper papillar formation. Pre-mitotic endocycles may generate precursors of a large size or specific shape that are needed to form four papillar structures. Related to this potential size requirement, the excess proliferation in fzr pupae may represent a compensatory response to decreased precursor tissue mass. The resulting over-proliferation in fzr RNAi animals could disrupt papillar morphogenesis by keeping papillar cells in a proliferative state when morphogenesis should occur. The potential need for sequential control of endocycles/cell growth followed by mitosis in papillar cells may differ from organs such as the developing Drosophila wing, where endocycling, cell growth and proliferation can be inter-changed without disrupting final organ structure (Neufeld and Edgar, 1998; Weigmann et al., 1997).
Future work is required to see whether a failure in cell size regulation accounts for all aspects of the fzr RNAi papillar phenotype, including the initial failure of the rectal tube to split into four papillae. One alternative, but not mutually exclusive, possibility is the use of the endocycle to promote differentiation and change papillar gene expression. For example, the endocycle may ensure appropriate levels of Notch signaling, which we characterized previously as a key regulator of papillar mitosis and morphogenesis. Similarly, in the adult, it remains possible that the endocycle activates key salt-absorption genes that are crucial for papillar function, in addition to its role in morphogenesis. We also cannot rule out a possible endocycle-independent function of the APC/C in papillar development, given the documented endocycle-independent functions of the APC/C and Fzr/CDH1 proteins in Drosophila and other organisms (van Roessel et al., 2004; Konishi et al., 2004; Herrero-Mendez et al., 2009).
Centriole retention and pre-mitotic endocycles
Based on our data on centriole retention in papillar and nurse cells, we favor the model that distinct endocycle programs in polyploidizing cells determine future mitotic competence. Retaining centrioles (in the absence of secondary centriole elimination mechanisms, such as migration to an oocyte) may thus serve as a useful marker for polyploid cells that retain mitotic potential. However, centriole retention alone is unlikely to be required for future mitotic re-entry of polyploid cells, as morula mutant nurse cells form an acentriolar spindle (Reed and Orr-Weaver, 1997). Rather, centrioles may serve as one easily recognizable marker for retention of a broader polyploid mitotic program. In Drosophila, an additional marker of mitotic competence after an endocycle is likely to be the replication of late-replicating sequences. Both papillar cells and the first five nurse cell endocycles initiate late replication, (Dej and Spradling, 1999; Fox et al., 2010) whereas the other tissues examined in our centriole retention study are known to under-replicate late-replicating sequences (Endow and Gall, 1975; Gall et al., 1971; Sher et al., 2012). Additional study of the differences between pre- and post-mitotic endocycles, such as the mechanism of centriole elimination, will uncover key molecular differences between these distinct classes of endocycles and could reveal the requirements for promoting or suppressing polyploid mitosis.
Aneuploidy tolerance in polyploid cells
Numerous studies link polyploidy in cancer cells to aneuploidy; our work, along with studies in yeast and mouse hepatocytes (Duncan et al., 2010; Storchova et al., 2006), suggests that non-cancerous polyploid cells also generate significant aneuploidy. Our present work shows that papillar aneuploidy generated by tripolar divisions – recognized as a frequently lethal form of aneuploidy after only one division – does not substantially impact survival, development or function of papillar cells. This aneuploidy viability contrasts previous work showing that whole-organism aneuploidy decreases viability in flies (Bridges, 1921a,b; Lindsley et al., 1972). Several possible models might explain this difference. First, changes in the ratio of remaining chromosomes after a tripolar division of an 8N cell may not cause sufficiently detrimental gene/protein dose changes. Second, papillar cells may possess a dose compensation mechanism to neutralize aneuploid gene expression, similar to aneuploid Drosophila S2 cells (Zhang et al., 2010). Third, it remains possible that papillar cells share common cytoplasm, allowing for protein equilibration, as was recently described in polyploid Drosophila follicle cells (McLean and Cooley, 2013). Finally, given the recent finding that polyploid-derived hepatocytes with a chromosome-specific aneuploidy can expand to confer disease resistance (Duncan et al., 2012), it remains possible that papillar aneuploidy generates a diverse pool of aneuploid cells that are poised to respond to future tissue stress. Now that the robust aneuploidy tolerance of papillar cells is established, these possibilities can be explored.
MATERIALS AND METHODS
Fly stocks
Wild-type genotypes: UAS-cnn GFP, his RFP; byn Gal4, UAS-moe GFP for live imaging, and byn Gal4, UAS-moe GFP or w1118 for fixed imaging. Fly stocks have either been described previously (Fox and Spradling, 2009; Fox et al., 2010) or are described on FlyBase (http://flybase.org/). Stocks referred to as SAK-OE refer to ubi-SAK or UAS-SAK. For fzr RNAi, we used UAS-fzr RNAiv25550.
Fly genetics
Except where indicated, all UAS-transgenes were driven by byn Gal4. Lineage analysis was carried out on flies containing hs-FLP and act-FRT-STOP-FRT-lacZ (Struhl and Basler, 1993). Control and SAK-OE flies were subjected to 60 min 37°C heat shock after embryonic rectal divisions, but before pupal polyploid divisions, and then dissected as adults. For temporal UAS expression, flies were kept at 18°C until the time of specified transgene induction, at which time they were shifted to 29°C. For fzr RNAi, embryos/newly hatched UAS-fzr RNAi; Gal80(ts) byn Gal4 larvae were kept at 18°C for 2 days, then shifted to 29°C until wandering 3rd larval instar, then shifted back to 18°C to turn off the RNAi. To visualize adult papillae in these fzr animals (Fig. 5F; supplementary material Fig. S6B), adult flies were shifted back to 29°C again to induce UAS-GFP (papillar cells are quiescent at this time). For salt feeding, newly eclosed adult flies were aged for 1 week on normal food, then placed in empty vials containing a 1.5 ml microcentrifuge cap filled with 5% sucrose only (control) or 250 mM NaCl mixed with 5% sucrose. Flies were transferred to new feeding vials daily and monitored for survival.
Tissue preparation
Tissue was dissected and either fixed or processed for live imaging as previously (Fox and Spradling, 2009; Fox et al., 2010) Antibodies used were: anti-Cnn (1:100; Heuer et al., 1995), Anti-Plp (1:2000; Rogers et al., 2008) and Anti-Asl (1:2000, a gift from Greg Rogers, University of Arizona, Tucson, USA), anti-Phospho-Histone H3 (Cell Signaling, 96705, 1:2000), anti-GFP (Invitrogen, 3E6, 1:2000), anti-DE-Cadherin (1:50, Developmental Studies Hybridoma Bank), anti-γ-Tubulin (Sigma, GTU88, 1:1000) and anti-β-galactosidase (Abcam, Ab9361, 1:1000). TUNEL was performed using the In Situ Cell Death Detection Kit (Roche), as per manufacturer's instructions. EdU labeling has been described by Fox et al. (2010). For supplementary material Fig. S5, 40 ng/ml colchicine (Sigma) was added to the samples for 1 h prior to fixation, to enrich for mitotic cells. For EM, tissue was fixed and processed as described previously (Cox and Spradling, 2003; Ohlstein and Spradling, 2006). Serial sections (80 nm) were cut.
Microscopy
Fixed images were acquired using either a Zeiss AxioImager M.2 with Apotome processing at 20×, 40× or 63×, or a Leica SP5 confocal at 40× or 60×. Live imaging used an Andor XD Spinning Disk Confocal Microscope with a 60× silicon objective. For EM, sections were viewed with a Technai F30 electron microscope at 300 kV.
Image analysis
Z-projections were assembled using ImageJ software. Movies were assembled using MetaMorph software (Molecular Devices). Adobe Photoshop software was used to adjust brightness and contrast. ImageJ was used to count papillar cell number. For volumetric DNA content analysis (Fig. 3), we used IMARIS software to convert His-RFP signal to a 3D object, from which total volume was calculated. The relative per cell distribution of an M-phase 16C octoploid parental cell was then determined by the amount of DNA segregated to each daughter. To generate the model plot of near 2:1:1 reductive divisions, we assumed the small 4th chromosomes contribute negligible DNA and the remaining DNA was evenly distributed between the other 24 chromosomes (48 chromatids) in each octoploid nucleus. One chromatid is thus equivalent to 2.08% total DNA. For each division, the percentage DNA segregated to a daughter was then converted into a chromatid number, and a histogram was generated. To introduce variance caused by mis-segregated chromosomes, we fixed 58% of cells at a perfect 2:1:1 segregation and allowed the other 42% of cells to mis-segregate up to four chromatids. This was based on our observation that 42% wild-type papillar cells with extra centrosomes exhibited lagging/bridging DNA (supplementary material Fig. S2F). Our observed tripolar data was compared with this model data using a Mann-Whitney test.
Cell cycle nomenclature
We use endocycle to refer to any programmed cell cycle in which the genome reduplicates without any entry into mitosis. We recognize the use in the literature of other terms to describe this same event, such as endomitosis, endoreduplication and endoreplication. We use 1N to define the haploid number of chromosomes (four for Drosophila) and use 1C to define the haploid DNA content.
Acknowledgements
The following kindly provided fly stocks: Bloomington Stock Center, Carnegie Protein Trap Collection, Vienna Drosophila Resource Center, Monica Bettencourt-Dias, Dan Kiehart, Ruth Lehmann, Christian Lehner and Jordan Raff. We thank Gerry Rubin, Todd Laverty, and members of Allan Spradling and Ben Ohlstein's Laboratories for assistance with screening Gal4 lines. We thank Allan Spradling, who provided support to D.T.F. while he initiated work related to this manuscript. Greg Rogers kindly provided affinity-purified anti-Plp prior to publication. Sam Johnson, Yasheng Gao and Mike Sepanski provided valuable technical assistance. We thank David MacAlpine, Allan Spradling and members of the Fox and MacAlpine labs for valuable comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
D.T.F. and K.P.S. designed the experiments; all authors carried out the experiments; D.T.F., K.P.S. and R.A.M. wrote the manuscript and assembled the figures.
Funding
D.T.F. is supported by a Pew Scholar award (Pew Charitable Trusts), a Basil O'Connor Scholar Award (The March of Dimes) and a Whitehead Scholar Award (Whitehead Foundation).
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.109850/-/DC1
References
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A. and Raff J. W. (2008). Centrosome amplification can initiate tumorigenesis in flies. Cell 133, 1032-1042. 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M. J. and Gupta B. L. (1967). Fine-structural changes in relation to ion and water transport in the rectal papillae of the blowfly, Calliphora. J. Cell. Sci. 2, 89-112. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G. and Glover D. M. (2005). SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199-2207. 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Bridges C. B. (1921a). Proof of non-disjunction for the fourth chromosome of Drosophila melanogaster. Science 53, 308 10.1126/science.53.1370.308 [DOI] [PubMed] [Google Scholar]
- Bridges C. B. (1921b). Genetical and cytological proof of non-disjunction of the fourth chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 7, 186-192. 10.1073/pnas.7.7.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee C. W., Klebba J. E., Buster D. W. and Rogers G. C. (2011). The Protein Phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 195, 231-243. 10.1083/jcb.201107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi B. R., Lilly M. A. and Spradling A. C. (1998). Cell cycle control of chorion gene amplification. Genes Dev. 12, 734-744. 10.1101/gad.12.5.734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J. and Dow J. A. T. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715-720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Clark A., Meignin C. and Davis I. (2007). A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134, 1955-1965. 10.1242/dev.02832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. T. and Spradling A. C. (2003). A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130, 1579-1590. 10.1242/dev.00365 [DOI] [PubMed] [Google Scholar]
- Davoli T. and de Lange T. (2012). Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell 21, 765-776. 10.1016/j.ccr.2012.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej K. J. and Spradling A. C. (1999). The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development 126, 293-303. [DOI] [PubMed] [Google Scholar]
- Dienstbier M., Boehl F., Li X. and Bullock S. L. (2009). Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23, 1546-1558. 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A. W., Taylor M. H., Hickey R. D., Hanlon Newell A. E., Lenzi M. L., Olson S. B., Finegold M. J. and Grompe M. (2010). The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467, 707-710. 10.1038/nature09414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A. W., Hanlon Newell A. E., Bi W., Finegold M. J., Olson S. B., Beaudet A. L. and Grompe M. (2012). Aneuploidy as a mechanism for stress-induced liver adaptation. J. Clin. Invest. 122, 3307-3315. 10.1172/JCI64026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N. S., Yu Q. D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G. and Glover D. M. (2010). Asterless is a scaffold for the onset of centriole assembly. Nature 467, 714-718. 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Zielke N. and Gutierrez C. (2014). Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15, 197-210. 10.1038/nrm3756 [DOI] [PubMed] [Google Scholar]
- Endow S. A. and Gall J. G. (1975). Differential replication of satellite DNA in polyploid tissues of Drosophila virilis. Chromosoma 50, 175-192. 10.1007/BF00283238 [DOI] [PubMed] [Google Scholar]
- Epstein C. J. (1967). Cell size, nuclear content, and the development of polyploidy in the Mammalian liver. Proc. Natl. Acad. Sci. USA 57, 327-334. 10.1073/pnas.57.2.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D. T. and Duronio R. J. (2013). Endoreplication and polyploidy: insights into development and disease. Development 140, 3-12. 10.1242/dev.080531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D. T. and Spradling A. C. (2009). The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell 5, 290-297. 10.1016/j.stem.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D. T., Gall J. G. and Spradling A. C. (2010). Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 24, 2294-2302. 10.1101/gad.1952710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., Cohen E. H. and Polan M. L. (1971). Repetitive DNA sequences in Drosophila. Chromosoma 33, 319-344. 10.1007/BF00284948 [DOI] [PubMed] [Google Scholar]
- Ganem N. J., Godinho S. A. and Pellman D. (2009). A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278-282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett M. A. and Bradley T. J. (1984). Ultrastructure of osmoregulatory organs in larvae of the brackish-water mosquito, Culiseta inornata (Williston). J. Morphol. 182, 257-277. 10.1002/jmor.1051820303 [DOI] [PubMed] [Google Scholar]
- Gentric G. and Desdouets C. (2014). Polyploidization in liver tissue. Am. J. Pathol. 184, 322-331. 10.1016/j.ajpath.2013.06.035 [DOI] [PubMed] [Google Scholar]
- Hartl T. A., Smith H. F. and Bosco G. (2008). Chromosome alignment and transvection are antagonized by condensin II. Science 322, 1384-1387. 10.1126/science.1164216 [DOI] [PubMed] [Google Scholar]
- Hassel C., Zhang B., Dixon M. and Calvi B. R. (2014). Induction of endocycles represses apoptosis independently of differentiation and predisposes cells to genome instability. Development 141, 112-123. 10.1242/dev.098871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Mendez A., Almeida A., Fernández E., Maestre C., Moncada S. and Bolanos J. P. (2009). The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat. Cell Biol. 11, 747-752. 10.1038/ncb1881 [DOI] [PubMed] [Google Scholar]
- Heuer J. G., Li K. and Kaufman T. C. (1995). The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development 121, 3861-3876. [DOI] [PubMed] [Google Scholar]
- Konishi Y., Stegmüller J., Matsuda T., Bonni S. and Bonni A. (2004). Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science 303, 1026-1030. 10.1126/science.1093712 [DOI] [PubMed] [Google Scholar]
- Levan A. and Hauschka T. S. (1953). Endomitotic reduplication mechanisms in ascites tumors of the mouse. J. Natl. Cancer Inst. 14, 1-43. [PubMed] [Google Scholar]
- Lilly M. A. and Spradling A. C. (1996). The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10, 2514-2526. 10.1101/gad.10.19.2514 [DOI] [PubMed] [Google Scholar]
- Lindsley D. L., Sandler L., Baker B. S., Carpenter A. T., Denell R. E., Hall J. C., Jacobs P. A., Miklos G. L., Davis B. K. and Gethmann R. C. et al. (1972). Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 71, 157-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach J. M. and Lehmann R. (1997). An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 11, 423-435. 10.1101/gad.11.4.423 [DOI] [PubMed] [Google Scholar]
- Mahowald A. P. and Strassheim J. M. (1970). Intercellular migration of centrioles in the germarium of Drosophila melanogaster: an electron microscopic study. J. Cell Biol. 45, 306-320. 10.1083/jcb.45.2.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald A. P., Caulton J. H., Edwards M. K. and Floyd A. D. (1979). Loss of centrioles and polyploidization in follicle cells of Drosophila melanogaster. Exp. Cell Res. 118, 404-410. 10.1016/0014-4827(79)90167-8 [DOI] [PubMed] [Google Scholar]
- Marthiens V., Rujano M. A., Pennetier C., Tessier S., Paul-Gilloteaux P. and Basto R. (2013). Centrosome amplification causes microcephaly. Nat. Cell Biol. 15, 731-740. 10.1038/ncb2746 [DOI] [PubMed] [Google Scholar]
- Martinez-Campos M., Basto R., Baker J., Kernan M. and Raff J. W. (2004). The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165, 673-683. 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazia D., Harris P. J. and Bibring T. (1960). The multiplicity of the mitotic centers and the time-course of their duplication and separation. J. Cell Biol. 7, 1-20. 10.1083/jcb.7.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean P. F. and Cooley L. (2013). Protein equilibration through somatic ring canals in Drosophila. Science 340, 1445-1447. 10.1126/science.1234887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne-Reveau K., Senger S., Pal M., Herr A., Richardson H. E., Asano M., Deak P. and Lilly M. A. (2008). APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development 135, 1451-1461. 10.1242/dev.016295 [DOI] [PubMed] [Google Scholar]
- Neufeld T. P. and Edgar B. A. (1998). Connections between growth and the cell cycle. Curr. Opin. Cell Biol. 10, 784-790. 10.1016/S0955-0674(98)80122-1 [DOI] [PubMed] [Google Scholar]
- Nigg E. A. and Raff J. W. (2009). Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663-678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Ohlstein B. and Spradling A. (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470-474. 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Pandit S. K., Westendorp B. and de Bruin A. (2013). Physiological significance of polyploidization in mammalian cells. Trends Cell Biol. 23, 556-566. 10.1016/j.tcb.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T.-T. B., Misra S., Murphy C., Scully A., Carlson J. W., Wan K. H. and Laverty T. R. et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715-9720. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. (1981). Comparative physiology of insect renal function. Am. J. Physiol. 241, R241-R257. [DOI] [PubMed] [Google Scholar]
- Reed B. H. and Orr-Weaver T. L. (1997). The Drosophila gene morula inhibits mitotic functions in the endo cell cycle and the mitotic cell cycle. Development 124, 3543-3553. [DOI] [PubMed] [Google Scholar]
- Robertson C. W. (1936). The metamorphosis of Drosophila melanogaster, including an accurately timed account of the principal morphological changes. J. Morphol. 59, 351-399. 10.1002/jmor.1050590207 [DOI] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D. M. and Bettencourt-Dias M. (2007). Revisiting the role of the mother centriole in centriole biogenesis. Science 316, 1046-1050. 10.1126/science.1142950 [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rusan N. M., Peifer M. and Rogers S. L. (2008). A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell 19, 3163-3178. 10.1091/mbc.E07-10-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher N., Bell G. W., Li S., Nordman J., Eng T., Eaton M. L., MacAlpine D. M. and Orr-Weaver T. L. (2012). Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 22, 64-75. 10.1101/gr.126003.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist S. J. and Lehner C. F. (1997). Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90, 671-681. 10.1016/S0092-8674(00)80528-0 [DOI] [PubMed] [Google Scholar]
- Silkworth W. T., Nardi I. K., Scholl L. M. and Cimini D. (2009). Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE 4, e6564 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin L. K., Nye J., Pinkerton D. C., Buster D. W., Rogers G. C. and Slep K. C. (2012). The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure 20, 1905-1917. 10.1016/j.str.2012.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. V. and Orr-Weaver T. L. (1991). The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development 112, 997-1008. [DOI] [PubMed] [Google Scholar]
- Stewénius Y., Gorunova L., Jonson T., Larsson N., Höglund M., Mandahl N., Mertens F., Mitelman F. and Gisselsson D. (2005). Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proc. Natl. Acad. Sci. USA 102, 5541-5546. 10.1073/pnas.0408454102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová Z., Breneman A., Cande J., Dunn J., Burbank K., O'Toole E. and Pellman D. (2006). Genome-wide genetic analysis of polyploidy in yeast. Nature 443, 541-547. 10.1038/nature05178 [DOI] [PubMed] [Google Scholar]
- Struhl G. and Basler K. (1993). Organizing activity of wingless protein in Drosophila. Cell 72, 527-540. 10.1016/0092-8674(93)90072-X [DOI] [PubMed] [Google Scholar]
- Theurkauf W. E., Alberts B. M., Jan Y. N. and Jongens T. A. (1993). A central role for microtubules in the differentiation of Drosophila oocytes. Development 118, 1169-1180. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y. and Orr-Weaver T. L. (2012). Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 26, 31-36. 10.1101/gad.177436.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roessel P., Elliott D. A., Robinson I. M., Prokop A. and Brand A. H. (2004). Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell 119, 707-718. 10.1016/j.cell.2004.11.028 [DOI] [PubMed] [Google Scholar]
- Vidwans S. J., DiGregorio P. J., Shermoen A. W., Foat B., Iwasa J., Yakubovich N. and O'Farrell P. H. (2002). Sister chromatids fail to separate during an induced endoreplication cycle in Drosophila embryos. Curr. Biol. 12, 829-833. 10.1016/S0960-9822(02)00845-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigmann K., Cohen S. M. and Lehner C. F. (1997). Cell cycle progression, growth and patterning in imaginal discs despite inhibition of cell division after inactivation of Drosophila Cdc2 kinase. Development 124, 3555-3563. [DOI] [PubMed] [Google Scholar]
- Wigglesworth V. B. (1942). The Principles of Insect Physiology. 2nd edn London: Methuen & Co. Ltd. [Google Scholar]
- Zhang Y., Malone J. H., Powell S. K., Periwal V., Spana E., MacAlpine D. M. and Oliver B. (2010). Expression in Aneuploid Drosophila S2 Cells. PLoS Biol. 8, e1000320 10.1371/journal.pbio.1000320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N., Kim K. J., Tran V., Shibutani S. T., Bravo M.-J., Nagarajan S., van Straaten M., Woods B., von Dassow, G. and Rottig C. et al. (2011). Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 480, 123-127. 10.1038/nature10579 [DOI] [PMC free article] [PubMed] [Google Scholar]