Abstract
Probiotics per definition should have beneficial effects on human health, and their consumption has tremendously increased in the last decades. In parallel, the amount of published material and claims for their beneficial efficacy soared continuously. Recently, multiple systemic reviews, meta-analyses, and expert opinions expressed criticism on their claimed effects and safety. The present review describes the dark side of the probiotics, in terms of problematic research design, incomplete reporting, lack of transparency, and under-reported safety. Highlighted are the potential virulent factors and the mode of action in the intestinal lumen, risking the physiological microbiome equilibrium. Finally, regulatory topics are discussed to lighten the heterogeneous guidelines applied worldwide. The shift in the scientific world towards a better understanding of the human microbiome, before consumption of the probiotic cargo, is highly endorsed. It is hoped that better knowledge will extend the probiotic repertoire, re-confirm efficacy or safety, establish their efficacy and substantiate their beneficial effects.
Keywords: probiotics, autoimmune disease, horizontal gene transfer, virulent factor, antibiotic-resistant genes, public health
1. Introduction
For a long time and mainly in the last decades the scientific, medical, industrial, and consumer communities have started to unravel the “superorganism” or “the second brain” presented by the prokaryotes dwelling inside the human enteric lumen [1,2,3]. The gut microbiome is a key player in intestinal eco-events, modulation, homeostasis, and function, dispersing systemically to impact human health [4,5]. Despite the bacterial part in the microbiome, it also contains viruses, archaea, fungi, and protozoa, reaching roughly 1014 cells, equaling the human body’s cell number and outnumbering the number of human genes by 100 fold. On the evolutionary aspect, microorganisms inhabited our globe 3.8 billion years ago, much earlier than the genus Homo emerged in Africa, 2.5 million years ago [6]. Thus, microbes had a much longer time outside us to adjust and adapt, by developing survival mechanisms enabling them to overcome even extreme environments. [7,8]. Notably, human enteric physiological microbiota composition and diversity, beyond infancy, are equilibrated, demonstrating amazing resilience to various perturbations, thus keeping it in the enteric compartment for the benefit of both kingdoms [9].
The situation is completely different with the much later evolutionary introduction of probiotics, most probably during the Neolithic layer of the stone age period, nearly 10,000 years ago [10,11]. Louis Pasteur and E. Metchnikoff should be acknowledged for further defining probiotics toward their actual definition “live microorganisms, which when administered in adequate amounts, confer a health benefit on the host.” Since then numerous publications expanded on the beneficial aspects of probiotic intake on human disease, conditions, mood, behaviors, and performances [12,13,14,15,16]. Just to cite a recent example: “The strongest evidence in favor of probiotics lies in the prevention or treatment of five disorders: necrotizing enterocolitis, acute infectious diarrhea, acute respiratory tract infections, antibiotic-associated diarrhea, and infant colic” [15]. The repertoire of the most consumed probiotics contains lactic acid producing bacteria, mainly lactobacilli, Bifidobacterium, lactococci, and streptococci. Yeast, bacilli, and some non-pathogenic E. coli strains are less often used. The ingested and nutritional, industrially used probiotics and starter cultures were generally considered as safe, for many years, but recently a change in trend has appeared.
The present review will summarize the questionable or unsubstantiated benefits of probiotics in a meta-analysis, highlighting their negative side, side effects, and expanding on their potential harmful capacities, mechanistic pathways, and potential hazards for human health.
1.1. The Probiotic Market is Booming Sky High
According to several reports, probiotics were used, unintentionally, nearly 10,000 years ago, but spread as fermented milk in previous centuries, mainly in Europe, where in the Balkan regions life span longevity and healthy life were attributed to their fermented yogurt. Their food application expanded earlier and on a larger scale compared to the sales in sachets, capsules, or other pharmaceutical preparations [17]. It seems that the trend is changing since direct personal consumption is surging abruptly alongside the adoption of self-care, integrative medicine, social enterprises, and aggressive media advertisement [18,19]. In fact, according to global market analysts, the global probiotic market size is predicted to exceed 3 billion US dollars by 2024 [20]. The market size for lactobacilli strains was valued at 1.2 billion US dollars in 2017, while the Bifidobacterium market size prediction will increase close to 6% until 2024 and the Bacillus strain market size may surpass 180 million US dollars by 2024 [21]. According to the discussion group at the 2017 meeting of the International Scientific Association for Probiotics and Prebiotics, the estimate of product creation and sales will reach 50 billion US dollars within the next five years [19].
1.2. Probiotic: Medical and Non-Medical Uses
The medical and non-medical indications to consume, over the counter, probiotics is constantly increasing. Table 1 summarizes some of those applications.
Table 1.
Medical and non-medical indications for probiotic consumption.
| Medical Applications 1 | Medical Applications 2 | Functional Applications |
|---|---|---|
| Lactose intolerance [20,22,23] | Chronic renal failure [22,24] | Functional digestive complains [22] |
| Hyperlipidemia [18,22] | HIV infection [25] | Mood and behavior changes [26] |
| Nephrolithiasis (oxalate stones) [27] | Cirrhosis, liver encephalopathy, NAFLD [15,23] | Memory improvement [28] |
| Inflammatory bowel disease [22,23,24] | Organ transplant [23] | Anxiety, fatigue, weakness, body or localized pains, nausea [26,29] |
| Irritable bowel syndrome [13,22,23,30] | Metabolic diseases [12,22] | Constipation/loose stools changes [22] |
| Eczema, allergic rhinitis, asthma [12,22,24,30] | Constipation [22] | Day care health [24] |
| Infectious diarrhea [22,23,24,30] | Periodontitis [22] | Working places health [24] |
| Respiratory tract infections [12,22,24] | Depression [26] | Wellbeing [17,24] |
| Traveler’s diarrhea [22,23,24] | Stay in Intensive care unit [31] | Anti-stress [29] |
| Necrotizing enterocolitis [13,24] | Prematurity [32] | Increase longevity [33] |
| Pouchitis [34] | Infant colic [13,15] | Improve sexuality [35] |
| Helicobacter pylori [22,30] | Autoimmune diseases [13,22,23,24,30] | Impaired “intestinal integrity” [22,24] |
| Neurological disorders [21] | Cystic fibrosis, pancreatitis [23,30] | |
| Overweight and obesity [18,21] | Ethanol-induced liver disease [23] | |
| Various cancers [22,23,30] | Small bowel bacterial overgrowth [22] | |
| Along or after antibiotics therapy [22] | Enhancement of oral vaccine administration [30] | |
| Clostridium difficile induced colitis [22,23,30] | Ischemic heart disease [18,22] | |
| Respiratory/urinary tract, rotavirus infections [22,23,24] | Hypertension [36] | |
| Vaginosis [24,30] | Neuropsychiatric/degenerative diseases [37,38] | |
| Dental caries [22,23,24,30] | Enhance growth [22,24,39] | |
| Diabetes type 2 [23] | Enhance weight gain [22,24,39] |
The present review is not intended to cover or update on the various indications for probiotic ingestion but will highlight their usage as a supplement to pharmaceutical therapy of, for example, autoimmune conditions.
1.3. Can Probiotic Intake Change Microbiotic Composition and Restore Eubiosis?
The holistic enteric compartment with its active luminal organ, presented with abundant prokaryotic flora, creates multifunctional defense mechanisms, preventing pathogenic invaders while keeping the microbiome at bay [5,8,9]. However, when failed or transformed to the dysbiome, chronic pathological conditions evolve, resulting in allergic, inflammatory, cancerous, and autoimmune diseases. The hen and chicken dilemma remains an enigma, and the issue of association/causality is far from being resolved. Despite this, taking probiotics to restore the normal, protective microbiome and to “balance” the human body’s flora is booming. The question arises over whether probiotics can modify dysbiosis and reverse the process. According to many recent scientific observations, the ways that the enteric microbiota/dysbiota is influenced by “good bacteria,” or the probiotic, is still unknown, and it seems that probiotic intake does not significantly affect gut flora composition [39,40,41,42,43,44,45,46,47,48,49]. Interestingly, probiotics affect the upper small bowel more than the colon [41,45]. Two major aspects ensue from recent studies: 1. The effects are influenced by the individual microbiome composition and structure, eluding to personal medicine [40,42,43,45]; 2. The probiotics impact is much more related to functional aspects, praising the mobilome, bacterial constituents, metabolomics, as well as the proteinomic effects [40,41,42,43,44,45,46]. After setting the stage of the probiotic clinical usages and impacts, the following is a clinical example of probiotics and the autoimmunity relationship.
1.4. Probiotics as a Supplemental Therapy in Autoimmune Diseases
The etiology, pathomechanisms, initiation, maintenance, progression, and modulation of autoimmunogenesis are continuously evolving. On the other hand, it is not yet understood why 5–8% of the general public is affected by autoimmune conditions. Four mechanisms were suggested for autoimmunity generation: molecular mimicry, self-antigen modification, bystander activation, and immune reactivity modulation [49]. The place of the wide “exposome” and more specifically the dysbiosis associated autoimmune disorders, is far from being clarified. Noteworthy, the relations of the changed composition and diversity in the enteric microbial kingdom to the four pathogenic mechanisms enumerated above is still poorly understood. Despite the lack of the dysbiotic scientific background, the patients and the treating clinical teams are trying to advance the supplemental therapeutic modalities in the direction of restoring the altered microbiome. The application of fecal transplantation, prebiotics, and probiotics are constantly expanding. Multiple autoimmune conditions are treated by probiotics including systemic lupus erythematosus (SLE), rheumatoid arthritis, Celiac and Crohn’s diseases, ulcerative colitis, multiple sclerosis, Sjogren’s syndrome, systemic sclerosis, antiphospholipid syndrome, myasthenia gravis, diabetes type 1 [13,50,51,52,53,54,55,56,57,58,59,60,61,62]. In many local and international scientific conferences, the probiotics trial as a supplement of adjunct therapy is presented. Moreover, the main argument favoring it is: “If it does not help it does not do any harm.”
The purpose of the current review is to protest against that repetitive declaration and to justify the opposite of its dual messages: 1. According to multiple recent meta-analyses, probiotic clinical benefits are questionable or disqualified. 2. Probiotics can present a Trojan horse that works against human health.
2. The Dark Side of Probiotics
Microbes possess an extended arsenal of hostile factors, capable of suppressing or destroying vital eukaryotic host mechanisms, for their advantage. Probiotics are an integral part of the prokaryotic kingdom with evolutionary conserved self-survival systems, in ex vivo and in vivo environments; above all, mainly in the overpopulated, extremely competitive, harsh ecological niche of the human gut. Ingestion of probiotic bacteria or products creates a survival struggle between the well-established inhabitant microbiome. Microbiome in the enteric compartment with the new probiotics.
In addition to the clinically reported multiple side effects in the literature, several pathogenic virulent potential pathways can be expressed in and executed by probiotics, thus affecting human health integrity. The following details several such pathomechanistic avenues.
2.1. Horizontal Gene Transfer (HGT)
HGT is the lateral movement of mobile genetic elements between unicellular or multicellular organisms. It enables the transfer of genes even between distant species mediated usually by transformation, transduction, conjugal transfer, or with specific gene transfer agents [63]. The topic of HGT in the human gut and the transfer of virulent genes to the endogenous microbiome was summarized recently [8]. The human gastrointestinal tract is an ideal environment and represents a hot spot for HGT [8]. As probiotics are extensively used in the processed food and fermented product’s industries and as over the counter additives, the question arises whether they can deliver hostile genetic elements to the microbiome?
Screening the literature, multiple publications describe the existence and transfer of hostile mobile genetic elements in and from probiotics [8]. Taking, for example, the most explored ones, the antibiotic-resistant genes, were found in various dietary supplements [64]. The problem is so widespread that it requires risk assessment measures to be implemented in those nutritional supplements [65]. More so, virulent mobile genetic elements are of a concern when transferred by HGT from probiotics to the enteric commensal communities [66]. More specifically, HGT between probiotic strains was reported for Lactobacillus paracasei [67], Lactobacillus rhamnosus [68], Lactobacillus reuteri [69,70,71], Lactobacillus gasseri [72], Lactobacillus plantarum [71], among other probiotics. Generally, gene flux of antibiotic-resistant genes, from gram-positive cocci to gram-negative microbes has been suggested [73], involving numerous antibiotics [74]. Even if probiotic ingestion does not impact stool microbial composition [48], HGT between ingested probiotic and the endogenous dwellers exist [75]. The cumulative risk of the probiotic double-edged sword effect of lateral genetic transfer of virulent elements is an ongoing enigma [76].
A special compliment should be given to Rosander et al. [77] who wrote a rare publication on the removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730, which is not commonly reported in probiotic research. However, antibiotic gene transfer is only one aspect of virulent genes and was taken just as an example. Gelatinase and hemolytic activities and several enzymes like peptidases, acid phosphatase, phosphohydrolases, α + β- galactosidases, and N-acetyl-β-glucosaminidase were depicted in lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture [78]. Most recently, microbial transglutaminase, a heavily consumed additive by the industrial processed food industry and a prokaryotic survival factor, was recently found to possess virulent factors, with anti-phagocytic being one of them [79,80]. Interestingly, probiotics also secrete the enzyme that was described as a novel potential environmental factor in celiac disease induction [81,82,83,84,85]. Microbial transglutaminase can be considered as a secreted toxin [86], with functional capacities even in pathogenic microbes [87,88,89,90]. Complexed to gliadin, when the microbial transglutaminase is crosslinking gliadin, the complex is immunogenic in celiac patients [84], and multiple deleterious effects on human health were described [91]. Finally, lateral gene transfer might influence the holobiont repertoire in intestinal niches whereby external prokaryotes, including probiotics, can affect genetic stability and evolutionarily conserved processes, threatening human health [92,93,94,95,96,97,98,99].
2.2. Bacteriophages of Probiotics Transfer Mobile Virulent Genes
The success and efficiency of probiotics depends on numerous factors that can be divided into microbiotic-exogenous, host-endogenous, and luminal-environmental [100]. One of the luminal factors are the bacteriophages. They are bacterially infectious small viruses that lyse microbes. The gastrointestinal tract harbors a wide variety of viruses, called the virome, and the phageome constitutes the largest part of this virome [101]. It is estimated that more than 30 billion bacteriophages transcytose human epithelial layers every day. They play a pivotal role shaping the microbiome’s taxonomic and functional compositions. The enteric prophages serve as a mobile repository of genetic elements and are transmitted via our microbiome, thus impacting on the microbiota/dysbiota or symbionts/pathbionts ratios and health and disease [101,102]. The enteric phageome virulence is controlled by the neighboring microbes, fungi, and helminths, thus creating a luminal trans-kingdom relationship [103,104].
They can be regarded as human pathogens, interacting directly or indirectly with prokaryotic, probiotics, as well as eukaryotic cells, including involved in protein misfolding, carrying prion-like domains [102,105]. Probiotics are prokaryotic and as such, are under the influence of the bacteriophages [100,101,102,103,104,105], many of the probiotic strains, orally consumed or used in food industries have their specific phages. A broader view will disclose a global environmental distribution of bacteriophages, carrying their hostile genetic cargo, to most environmental biomes where the bacteria reside [106,107]. Wastewater treatment plants, human fecal samples, food and medical isolates, dairy fermentations, agriculture, and even in the air are where virulent genes were most recently detected in multiple congested metropolitan urban air [108,109,110,111,112,113]. Screening the bacteriophages of commonly used probiotics, Lactobacillus paracasei or gasseri [67,72,114,115], Lactococcus lactic [116,117,118,119,120,121], and many more, were found to have close contact with specific phages.
The probiotic bacteriophages are a potential carrier of hostile genes that by transfecting prokaryote or eukaryote cells, can spread genetic material. Finally, it appears that some bacteriophages contain virulent transglutaminase genes, thus representing additional tranglutaminase activity in the intestinal lumen [86,90], in addition to the microbiome, archaeal, probiotic, and industrially added one [8,82,83,84,85,91]. Finally, the evolution of novel transglutaminase-like peptidase from eukaryotic ciliary compartments was traced back to prokaryotic transglutaminase-like peptidases, thus, deciphering key evolutionary events along the course of the eukaryotic emergence from prokaryotes [122].
2.3. Processed Food and the Probiotic Mobilome
Probiotics are heavily used in the processed food industries, spanning not only dairy fermented products, but also in wider industrial applications including medical, diagnostic, pharmacological, and biotechnological industries [123,124,125,126,127,128]. For many industrial applications, including dairy starter fermentation cultures, they acquired the GRAS (generally regarded as safe) status, which was defined before recent safety concerns were raised, such as the carriage of virulent mobile genetic elements. Notably, genetic transfers in bacteria are more prone to occur in crowded environments, such as the human GI tract, not excluding food reservoirs, manipulations, and products. Probiotics are heavily used, for many years, for processed food manipulations and production. Many of them are used in fermented foods like dairy products, cheese, fermented sausage, fermented vegetables, soy-fermented foods, and fermented cereal products [129]. It is very logical that one of the concerns of their massive usage is the lateral exchange of hostile genes, in-between them, or to the physiological microbiome, to the dysbiota, or even to human cells, as summarized here [8]. Gene acquisition/loss within or between various microbes and probiotic strains were widely described, all across the food chain, be it dairy, meat, or vegetable products and even in the ready-to-go food items [121,130,131,132,133,134]. HGT of antibiotic resistance is wide and was reported for a wide range of probiotics, including Lactobacillus rhamnosus, Lactobacillus gasseri, Lactobacillus paracasei, Lactobacillus reuteri, Lactobacillus plantarum, and many others [8]. In addition to antibiotic-resistant genes, the most extensively explored, numerous additional virulent genes are carried by the probiotic microbial genome. Microbial transglutaminase, mentioned above, is only one of them [79,80,81,82,83,84,85,86,87,88,89,90,91].
2.4. d-lactate, Metabolic Acidosis, and Brain Fogginess
Probiotic consumption is associated with D-lactic academia and acidosis in adults and infants on probiotic-containing formula [41,135,136,137,138,139]. Lactobacillus and Bifidobacterium species are the most used bacteria in probiotic formulations and they produce d-lactate [139,140,141,142] and their consumption was suggested to be avoided in d-lactic acidosis [139]. Intriguingly, d-lactic acidosis and other etiologies for acidosis are associated with neurocognitive symptoms, neurological impairments, and chronic fatigue syndrome [137,143,144], including brain fogginess [41,139]. The syndrome of brain fogginess has, in fact, multiple etiologies, one of which is short bowel syndrome associated with D-lactic acidosis [41,139,144,145]. Despite the critical view on the association with probiotic intake [146,147], the discontinuation of the antibiotics and the resolution of the symptoms on antibiotic therapy, strengthen the causative association [41,139].
2.5. Intestinal Bacterial Overgrowth, Gas, and Bloating
This paragraph is related to the above paragraph and might explain the pathophysiology of the acidosis and brain fogginess described above. Rao et al. described a new syndrome relating post-prandial brain fogginess, gas, and abdominal bloating to small intestinal bacterial overgrowth and probiotic-induced d-lactic acidosis [41,139]. Additional complaints were fatigue, weakness, disorientation, and restlessness. The authors put forward the hypothesis that probiotic fermented carbohydrates in the proximal small bowel induce intestinal bacterial overgrowth, resulting in d-lactic acid production, increased gas output, and abdominal bloating. The d-lactic acidosis is the culprit for the brain fogginess. They suggested that this unique entity is an additional side effect of probiotic consumption.
2.6. Additional Clinical Probiotic Side Effects
The medical literature warns against probiotic consumption in congenital or acquired immune debilitating conditions, heart anomalies, chemo- and radiotherapies, surgical abdomen, HIV-infected, critically ill, post-organ transplantation, post-operation, central venous catheters, autoimmune disease on immune suppression, pregnancy, neutropenia, critically ill patients, including antibiotic-associated diarrhea, active ulcerative colitis, and potential for translocation of probiotic across bowel wall [74,148,149]. Although it is not the main focus of the present review, to wrap up the subject, Table 2 summarizes the reported toxic, unintended, adverse effects following probiotic usage.
Table 2.
Summary of the reported toxic, unintended, adverse effects following probiotic consumption.
| Infectious/Gastrointestinal | Allergic | Genetic | Patho-Toxogenicity |
|---|---|---|---|
| Bacteremia [41,74,150] | Rhinitis [149] | Transfer of virulent factors: | Enhanced adhesion and protein aggregation [74,149] |
| Sepsis [41,74] | Wheezing bronchitis [151] | Antibiotic resistance [74,149,152,153,154] | Mucolysis/hemolysis [74,149] |
| Fungemia [41,155] | Rash [149] | Hemolysin [149,152] | Bile salt hydrolysis [74] |
| Endocarditis, meningitis, endometritis, peritonitis, pneumonia [150,156,157] | Gelatinase [149] | DNA degradation and proteolysis [149] | |
| Liver abscess [150] | Metabolic | DNAse [149] | Innate defense resistance [52,149] |
| Diarrhea, Abdominal cramps [74] | d-lactic acidosis [41,74,149] | Enolase activating plasminogen [149] | Food poisoning [149] |
| Nausea, vomiting, flatulence, taste disturbance [41,74] | Metalloendopeptidase [158] | Immune evasion or over stimulation [74,149] | |
| Low appetite [159] | Cytolysin modification, transport, activation [160] | Facilitated microbial conjugation/translocation [74,149] | |
| Sex pheromones [161] | Macrophage/monocyte chemotactism [162] | ||
| Nanoparticles: Lactomicroselenium [163] | |||
| Gastrointestinal ischemia [41,74] | |||
| Mechanical choking [74] | |||
| Peptide deamidation [164] | |||
| Epigenetic and mobilome manipulation [52] |
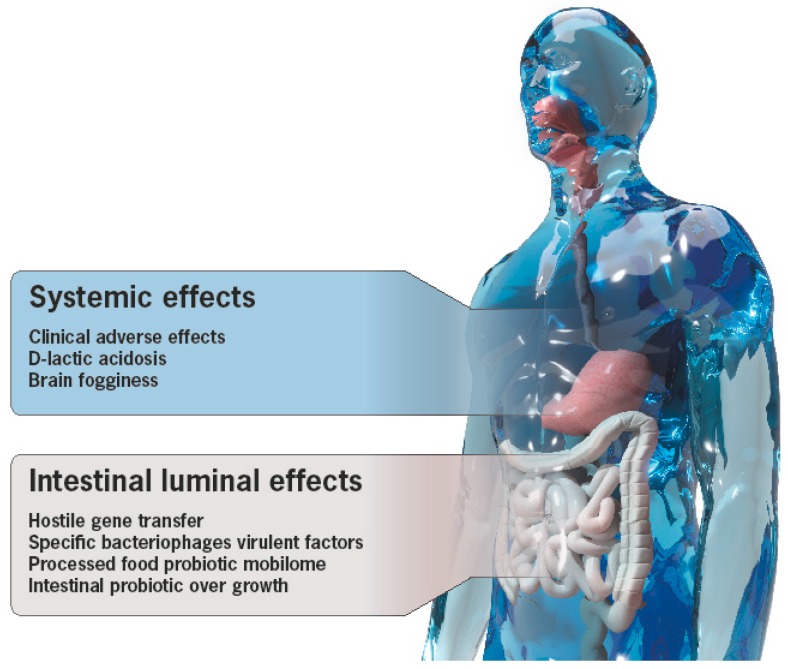
The list of probiotic’s adverse effects is expanding, however, due to a lack of safety and toxicity standardized protocols and regulatory implementations, the list is under-representative. It is clear that more safety and toxicity designed studies are needed to reveal the negative side of probiotic use [74,148,149,154,165]. Figure 1 is a schematic presentation of the local and systemic adverse effects and mechanisms by which the probiotics exert their deleterious effects.
Figure 1.
The local and systemic adverse effects of probiotics.
3. Problematic Inadequate Design, Incomplete Reporting, and Lack of Transparency
The current review aims to highlight the negative side of probiotic consumption. As such, followed herein are the most recent systemic reviews and meta-analyses that criticize multiple aspects of the medical publications on probiotic efficacy and safety (Table 3). Many of them detected a lack of qualified experimental designs, a shortage of standardization, extended data variance, incomplete reporting, high patient withdrawal, and all of which increase the publications’ biases. In a recent review of existing meta-analyses, the authors tried to analyze the contradictory results of the probiotic effectiveness in many frequent conditions [165]. The final results were quite restricted: “Only for antibiotic- and Clostridium difficile-associated diarrhea, and respiratory tract infections the effects of probiotics are considered "evidence-based". Concerning other fields, meta-analyses fail to define the type and biologic effect of probiotic strains, as well as the outcome in a disease state. The authors concluded that: “Further studies are needed, because the available evidence is insufficient to show the efficacy of probiotics themselves. Carefully designed clinical trials are needed to validate the effects of particular strains of probiotics given at specific dosages and for specific treatment durations.”
Table 3.
A summary of recent meta-analyses and systemic reviews criticizing microbiome and probiotic publications.
| Publication | Mal-Designed | Lack of Standardization | High Data Variance | Biased | High Withdrawal | Incomplete Reporting | Reference |
|---|---|---|---|---|---|---|---|
| Review | + | + | + | + | + | [154] | |
| Systemic review | + | + | + | + | + | [148] | |
| Review | + | + | + | + | + | [74] | |
| Systemic review | + | + | + | + | [166] | ||
| Systemic review | + | + | + | + | + | [148] | |
| Systemic review | + | + | + | + | + | [167] | |
| Meta-analysis | + | + | [168] | ||||
| Meta-analysis | + | + | + | [169] | |||
| Meta-analysis | + | + | + | + | [170] | ||
| Meta-analysis | + | + | [171] | ||||
| Meta-analysis | + | + | + | + | [172] | ||
| Meta-analysis | + | + | + | + | [173] | ||
| Systemic review | + | + | [174] | ||||
| Meta-analysis | + | + | + | + | [175] | ||
| Meta-analysis | + | + | + | [62] | |||
| Meta-analysis | + | + | + | [176] | |||
| Meta-analysis | + | + | [177] | ||||
| Meta-analysis | + | + | + | [178] | |||
| Meta-analysis | + | + | + | [179] | |||
| Meta-analysis | + | + | [180] | ||||
| Meta-analysis | + | + | + | + | [181] | ||
| Meta-analysis | + | + | + | [182] | |||
| Systemic review | + | + | + | + | [183] | ||
| Meta-analysis | + | + | + | + | [184] |
+ = exist in the publication.
4. Lack of Effective Regulation of Probiotics
More recent systemic reviews or meta-analyses, from 2018, did not demonstrate differences of outcomes, using probiotics, on the treatment success of: constipation [180], traveler’s diarrhea [179], cancer [173,175], anxiety [171,177], rheumatoid arthritis [62], urinary tract infections [174], decrease in fat mass [172], food allergy [169], childhood asthma [178] and eczema [185], preterm neurodevelopment [182], and adiponectin and leptin levels [181]. Van den Nieuwboer et al. summarized it clearly: “generalizability of conclusions are greatly limited by the inconsistent, imprecise, and potentially incomplete reporting as well as the variation in probiotic strains, dosages, administration regimes, study populations, and reported outcomes” [148].
It seems that scientific and medical societies should “mind the gaps” between published studies praising probiotic therapeutical efficacy and a lack of substantiation when analyzed by more objective, standardized methodologies, such as critical systemic reviews or meta-analyses. More so, when intestinal microbiota composition was assessed on probiotic intake, no significant changes were depicted [44,45,46,47,48,140]. Intriguingly, probiotic effects diminish with time, in mice [186] and their stability and survival markedly decline in frozen capsules [187]. Probiotic colonization is also controversial since studies done on fecal samples alone are insufficient—colonized intestinal biopsies are more indicative [149].
5. Probiotic Safety is Under-Reported
The present review cannot be completed without an update on probiotic safety. The food and drug administration consider some probiotics, as GRAS, when added to food [188], especially when intended to impact taste, aroma, or nutritional value [74,189]. However, most of the reviews, analyzing the safety of probiotics highlight the issue of a lack of structural classification and a wide generalization of conclusions that is limited by imprecise, inconsistent, and incomplete reporting intermingled with variations in strains used, dosages, regimes of administration, experimental designs, and the participating populations [74,148,149,167]. Many contrast the differences that exist between the long history of the large consumption of “safe” probiotics compared to the scarcity of scientific proof for their safety [74,149,167]. Missed microbial identification, misnumbering and mislabeling, and lack of dose-response relationships are additionally reported aspects [149,190,191,192,193,194]. Insufficient and uncontrolled research designs, underpowered studies, and mixed research and outcome results are often encountered [195]. The allergic reaction, or anaphylaxis, is an additional aspect of safety, since probiotic preparations can contain allergens, including cow milk and hen egg proteins [196,197]. Safety is further complicated by the fact that various companies use duplicate cultures of the original strains, by applying fingerprinting techniques, potentially increasing the risk of detrimental effects [149]. To fill the gap in reliability and transparency in probiotics effectiveness and safety the following need to be considered: eliciting side effects data from participants [198], suboptimal adherence to reporting guidelines [199,200,201], over food industry funding mounting to 60% of the screened studies [202], lack of long-term effects in normal and vulnerable populations [203], and occasional lack of viable organisms [204]. Most recently, skepticism was raised concerning the labeled number of bacteria in probiotic preparations, publication bias, the generalizability of findings, and the safety in immune deficient hosts [13]. Finally, it appears that few studies on probiotics are designed to probe safety aspects and much should be improved in this domain [149,205,206].
Quite often patients consume probiotics while physicians encourage probiotic consumption, despite their potentially harmful effects. The notion of “good bacteria” and the manufacturers’ claim of “health promotion” or “balancing” normal gut flora should be taken with a “grain of salt.” In two seminal studies from an Israeli group recently published [42,43] it was shown that “mucosal colonization resistance to empiric probiotics” is host and microbiome dependent and that “post-antibiotic gut mucosal microbiome reconstruction is impaired by probiotics” [40]. Those and multiple reviews, meta-analyses, and studies, mentioned above, reinforce the need for regulation of probiotics for public health protection. It seems that real life habits, hopes, and media-directed information overcome scientific knowledge in real-time. Even the basic categorization of probiotics as drug, food, or dietary supplements is still undetermined and confused [30]. However, there is some light in the regulatory tunnel. The European Food Safety Authority (EFSA) changed their regulatory policy based on the lack of convincing evidence on the claim that probiotics improve human health or wellbeing [195,207,208]. Moreover, when regulation is enforced, consequences are predicted. Within the European Union, all health claims for probiotics were rejected, except for lactose intolerance improvement [209]. Since 2013, no claims concerning the change or improved gut microbiome composition was approved by the EFSA. The American FDA is taking a different approach. Probiotics can be categorized as food, food additive, cosmetics, dietary supplement, or drugs [210], and the responsibility for accuracy and truthfulness of the product is the responsibility of the producer [209,211]. It should be notified that no probiotic was approved for health claims by the FDA in recent years [209]. The tightened regulations impacted the scientific community and the manufacturer’s policies profoundly, as mirrored by the number of publications on the subject in the last two decades. The number of publications or registered studies increased significantly on microbiota while plateauing on probiotics [209]. The shift to explore the microbiota presents an opportunity to uncover new probiotics and understand their mode of action, and explore relationship with their neighboring prokaryotes, eukaryotes, and their secreted mobilomes [212,213].
6. Conclusions
The present review intended to summarize the somber side of probiotics, highlighting the potential detrimental effects embedded in the fact that probiotics are prokaryotes, and as such, contains hostile factors, in order to survive. They are capable of inducing local and systemic adverse effects (Figure 1) thus contradicting their definition as beneficial for human health. More caution, safety exploration, and stringent regulation can prevent these mal-effects. The absence of associated virulence factors should be demonstrated, especially when the probiotic belongs to a bacterial genus with pathogenic capabilities. Consideration of risk-benefit ratio before suggesting probiotics should be highly recommended. In view of potential pathogenic pathways, problematic inadequacy of design, reporting and transparency, and under-reported probiotic safety and non-defined implementable international criteria for regulation, it is encouraging to follow the contemporary back shift to the microbiome. It is hoped that by widening the knowledge of the human intestinal microbiome, that salvation will come from “the ascent of the blessed” probiotics as a preventive/beneficial/therapeutical health promoter. Finally, since intestinal microbiota is a recent new frontier in medicine, further exploration might stage probiotics as a preventive barrier or as a product capable of balancing the dysbiome associated with chronic human morbidity and mortality.
Acknowledgments
Ramesh Ajay for the editing and Neu Alf for designing the figure.
Author Contributions
Conceptualization, Y.S. and T.M.; methodology, Y.S.; formal analysis, A.L.; resources, T.M.; data curation, S.Y and T.M.; writing—original draft preparation, A.L.; writing—review and editing, A.L.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Doré J., Multon M.C., Béhier J.M., Affagard H., Andremont A., Barthélémy P., Batitsa R., Bonneville M., Bonny C., Boyaval G., et al. The human gut microbiome as source of innovation for health: Which physiological and therapeutic outcomes could we expect? Therapie. 2017;72:21–38. doi: 10.1016/j.therap.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Glendinning L., Free A. Supra-organismal interactions in the human intestine. Front. Cell. Infect. Microbiol. 2014;4:47. doi: 10.3389/fcimb.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson J.K., Holmes E., Wilson I.D. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 4.Lerner A., Matthias T. GUT-the Trojan horse in remote organs’ autoimmunity. J. Clin. Cell. Immunol. 2016;7:401. [Google Scholar]
- 5.Lerner A., Neidhöfer S., Matthias T. The gut microbiome feelings of the brain: Perspective for Non-Microbiologists. Microorganisms. 2017;5:66. doi: 10.3390/microorganisms5040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harari Y.N. Sapiens: A Brief History of Humankind. Vintage Books; London, UK: 2011. pp. 1–466. [Google Scholar]
- 7.Lerner A. The last two millennias eco-catastrophes are the driving forces for the potential genetic advantage mechanisms in celiac disease. Med. Hypotheses. 2011;77:773–776. doi: 10.1016/j.mehy.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 8.Lerner A., Aminov R., Matthias T. Potential effects of horizontal gene exchange in the human gut. Front. Microbiol. 2017;8:1630. doi: 10.3389/fimmu.2017.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lerner A., Aminov R., Matthias T. Dysbiosis may trigger autoimmune diseases via inappropriate posttranslational modification of host proteins. Front. Microbiol. 2016;7:84. doi: 10.3389/fmicb.2016.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozen M., Dinleyici E.C. The history of probiotics: The untold story. Benef. Microbes. 2015;6:159–165. doi: 10.3920/BM2014.0103. [DOI] [PubMed] [Google Scholar]
- 11.Gasbarrini G., Bonvicini F., Gramenzi A. Probiotics History; Proceedings of the 8th Probiotics, Prebiotics & New Foods for Microbiota and Human Health Meeting; Rome, Italy. 13–15 September 2015; pp. S116–S119. [Google Scholar]
- 12.Villena J., Kitazawa H. Probiotic Microorganisms: A Closer Look. Microorganisms. 2017;5:17. doi: 10.3390/microorganisms5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Alookaran J.J., Rhoads J.M. Probiotics in Autoimmune and Inflammatory Disorders. Nutrients. 2018;10:1537. doi: 10.3390/nu10101537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathipa M.G., Thantsha M.S. Probiotic engineering: Towards development of robust probiotic strains with enhanced functional properties and for targeted control of enteric pathogens. Gut Pathog. 2017;9:28. doi: 10.1186/s13099-017-0178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., Tran D.Q., Rhoads J.M. Probiotics in Disease Prevention and Treatment. J. Clin. Pharmacol. 2018;58(Suppl. 10):S164–S179. doi: 10.1002/jcph.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pirbaglou M., Katz J., de Souza R.J., Stearns J.C., Motamed M., Ritvo P. Probiotic supplementation can positively affect anxiety and depressive symptoms: A systematic review of randomized controlled trials. Nutr. Res. 2016;36:889–898. doi: 10.1016/j.nutres.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Saxelin M. Probiotic formulations and applications, the current probiotics market, and changes in the marketplace: A European perspective. Clin. Infect. Dis. 2008;46(Suppl. 2):S76–S79; discussion S144–S151. doi: 10.1086/523337. [DOI] [PubMed] [Google Scholar]
- 18.Di Cerbo A., Palmieri B. Review: The market of probiotics. Pak. J. Pharm. Sci. 2015;28:2199–2206. [PubMed] [Google Scholar]
- 19.Reid G., Kort R., Alvarez S., Bourdet-Sicard R., Benoit V., Cunningham M., Saulnier D., Vlieg J.V.H., Verstraelen H., Sybesma W. Expanding the reach of probiotics through social enterprises. Benef. Microbes. 2018;9:707–715. doi: 10.3920/BM2018.0015. [DOI] [PubMed] [Google Scholar]
- 20.Global Market Insights, Inc. [(accessed on 23 November 2018)]; Available online: https://www.gminsights.com/pressrelease/probiotics-market.
- 21.Global Market Insights, Inc. [(accessed on 23 November 2018)]; Available online: https://www.gminsights.com/industry-analysis/probiotics-market.
- 22.de Vrese M., Schrezenmeir J. Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. Biotechnol. 2008;111:1–66. doi: 10.1007/10_2008_097. [DOI] [PubMed] [Google Scholar]
- 23.Goldin B.R., Gorbach S.L. Clinical indications for probiotics: An overview. Clin. Infect. Dis. 2008;46(Suppl. 2):S96–S100; discussion S144–S151. doi: 10.1086/523333. [DOI] [PubMed] [Google Scholar]
- 24.Minocha A. Probiotics for preventive health. Nutr. Clin. Pract. 2009;24:227–241. doi: 10.1177/0884533608331177. [DOI] [PubMed] [Google Scholar]
- 25.D’Angelo C., Reale M., Costantini E. Microbiota and Probiotics in Health and HIV Infection. Nutrients. 2017;9:615. doi: 10.3390/nu9060615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westfall E., Brandenburg D. Probiotics for Symptoms of Depression and Anxiety. Am. Fam. Phys. 2018;97 online. [PubMed] [Google Scholar]
- 27.Lieske J.C. Probiotics for prevention of urinary stones. Ann. Transl. Med. 2017;5:29. doi: 10.21037/atm.2016.11.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misra S., Medhi B. Role of probiotics as memory enhancer. Indian J. Pharmacol. 2013;45:311–312. doi: 10.4103/0253-7613.111917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chong H.X., Yusoff N.A.A., Hor Y.Y., Lew L.C., Jaafar M.H., Choi S.B., Yusoff M., Wahid N., Abdullah M., Zakaria N., et al. Lactobacillus plantarum DR7 alleviates stress and anxiety in adults: A randomised, double-blind, placebo-controlled study. Benef. Microbes. 2019;18:1–20. doi: 10.3920/BM2018.0135. [DOI] [PubMed] [Google Scholar]
- 30.Gorbach S.L. Probiotics in the third millennium. Dig. Liver Dis. 2002;34(Suppl. 2):S2–S7. doi: 10.1016/S1590-8658(02)80155-4. [DOI] [PubMed] [Google Scholar]
- 31.Mahmoodpoor A., Hamishehkar H., Asghari R., Abri R., Shadvar K., Sanaie S. Effect of a Probiotic Preparation on Ventilator-Associated Pneumonia in Critically Ill Patients Admitted to the Intensive Care Unit: A Prospective Double-Blind Randomized Controlled Trial. Nutr. Clin. Pract. 2019;34:156–162. doi: 10.1002/ncp.10191. [DOI] [PubMed] [Google Scholar]
- 32.Sohn K., Underwood M.A. Prenatal and postnatal administration of prebiotics and probiotics. Semin. Fetal Neonatal Med. 2017;22:284–289. doi: 10.1016/j.siny.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayala F.R., Bauman C., Cogliati S., Leñini C., Bartolini M., Grau R. Microbial flora, probiotics, Bacillus subtilis and the search for a long and healthy human longevity. Microb. Cell. 2017;4:133–136. doi: 10.15698/mic2017.04.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guslandi M. Role of Probiotics in Crohn’s Disease and in Pouchitis. J. Clin. Gastroenterol. 2015;49(Suppl. 1):S46–S49. doi: 10.1097/MCG.0000000000000351. [DOI] [PubMed] [Google Scholar]
- 35.Tirandaz H., Ebrahim-Habibi M.B., Moradveisi B., Raoofi S., Salehi-Najafabadi A., Mohammadi E. Microbiota potential for the treatment of sexual dysfunction. Med. Hypotheses. 2018;115:46–49. doi: 10.1016/j.mehy.2018.03.021. [DOI] [PubMed] [Google Scholar]
- 36.Ahtesh F.B., Stojanovska L., Apostolopoulos V. Anti-hypertensive peptides released from milk proteins by probiotics. Maturitas. 2018;115:103–109. doi: 10.1016/j.maturitas.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 37.Santocchi E., Guiducci L., Fulceri F., Billeci L., Buzzigoli E., Apicella F., Calderoni S., Grossi E., Morales M.A., Muratori F. Gut to brain interaction in Autism Spectrum Disorders: A randomized controlled trial on the role of probiotics on clinical, biochemical and neurophysiological parameters. BMC Psychiatry. 2016;16:183. doi: 10.1186/s12888-016-0887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welcome M.O. Current Perspectives and Mechanisms of Relationship between Intestinal Microbiota Dysfunction and Dementia: A Review. Dement. Geriatr. Cogn. Dis. Extra. 2018;8:360–381. doi: 10.1159/000492491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bezirtzoglou E., Stavropoulou E. Immunology and probiotic impact of the newborn and young children intestinal microflora. Anaerobe. 2011;17:369–374. doi: 10.1016/j.anaerobe.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Khoruts A. Targeting the microbiome: From probiotics to fecal microbiota transplantation. Genome Med. 2018;10:80. doi: 10.1186/s13073-018-0592-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao S.S.C., Rehman A., Yu S., Andino N.M. Brain fogginess, gas and bloating: A link between SIBO, probiotics and metabolic acidosis. Clin. Transl. Gastroenterol. 2018;9:162. doi: 10.1038/s41424-018-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zmora N., Zilberman-Schapira G., Suez J., Mor U., Dori-Bachash M., Bashiardes S., Kotler E., Zur M., Regev-Lehavi D., Brik R.B.-Z., et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell. 2018;174:1388–1405. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 43.Suez J., Zmora N., Zilberman-Schapira G., Mor U., Dori-Bachash M., Bashiardes S., Zur M., Regev-Lehavi D., Brik R.B.-Z., Federici S., et al. Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell. 2018;174:1406–1423. doi: 10.1016/j.cell.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 44.Singh A., Sarangi A.N., Goel A., Srivastava R., Bhargava R., Gaur P., Aggarwal A., Aggarwal R. Effect of administration of a probiotic preparation on gut microbiota and immune response in healthy women in India: An open-label, single-arm pilot study. BMC Gastroenterol. 2018;18:85. doi: 10.1186/s12876-018-0819-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Derrien M., van Hylckama Vlieg J.E. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Maldonado-Gómez M.X., Martínez I., Bottacini F., O’Callaghan A., Ventura M., van Sinderen D., Hillmann B., Vangay P., Knights D., Hutkins R.W., et al. Stable Engraftment of Bifidobacterium longum AH1206 in the Human Gut Depends on Individualized Features of the Resident Microbiome. Cell Host Microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Laursen M.F., Laursen R.P., Larnkjær A., Michaelsen K.F., Bahl M.I., Licht T.R. Administration of two probiotic strains during early childhood does not affect the endogenous gut microbiota composition despite probiotic proliferation. BMC Microbiol. 2017;17:175. doi: 10.1186/s12866-017-1090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kristensen N.B., Bryrup T., Allin K.H., Nielsen T., Hansen T.H., Pedersen O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: A systematic review of randomized controlled trials. Genome Med. 2016;8:52. doi: 10.1186/s13073-016-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selmi C., Gao B., Gershwin M.E. The long and latent road to autoimmunity. Cell. Mol. Immunol. 2018;15:543–546. doi: 10.1038/s41423-018-0018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Luca F., Shoenfeld Y. The microbiome in autoimmune diseases. Clin. Exp. Immunol. 2019;195:74–85. doi: 10.1111/cei.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haase S., Haghikia A., Wilck N., Müller D.N., Linker R.A. Impacts of microbiome metabolites on immune regulation and autoimmunity. Immunology. 2018;154:230–238. doi: 10.1111/imm.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen B., Sun L., Zhang X. Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. J. Autoimmun. 2017;83:31–42. doi: 10.1016/j.jaut.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 53.de Oliveira G.L.V., Leite A.Z., Higuchi B.S., Gonzaga M.I., Mariano V.S. Intestinal Dysbiosis and pRobiotic Applications in Autoimmune Diseases. Immunology. 2017;152:1–12. doi: 10.1111/imm.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Oliveira G.L.V. Probiotic applications in autoimmune diseases In Probiotics-Current Knowledge and Future Prospects. IntechOpen; London, UK: 2018. pp. 69–89. [Google Scholar]
- 55.Marietta E., Horwath I., Balakrishnan B., Taneja V. Role of the intestinal microbiome in autoimmune diseases and its use in treatments. Cell Immunol. 2018 doi: 10.1016/j.cellimm.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Balakrishnan B., Taneja V. Microbial modulation of the gut microbiome for treating autoimmune diseases. Expert Rev Gastroenterol. Hepatol. 2018;12:985–996. doi: 10.1080/17474124.2018.1517044. [DOI] [PubMed] [Google Scholar]
- 57.Dwivedi M., Kumar P., Laddha N.C., Kemp E.H. Induction of regulatory T cells: A role for probiotics and prebiotics to suppress autoimmunity. Autoimmun. Rev. 2016;15:379–392. doi: 10.1016/j.autrev.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Tankou S.K., Regev K., Healy B.C., Cox L.M., Tjon E., Kivisakk P., Vanande I.P., Cook S., Gandhi R., Glanz B., et al. Investigation of probiotics in multiple sclerosis. Mult. Scler. 2018;24:58–63. doi: 10.1177/1352458517737390. [DOI] [PubMed] [Google Scholar]
- 59.Kota R.K., Ambati R.R., Yalakurthi A.K., Srirama K., Reddy P.N. Recent advances in probiotics as live biotherapeutics against gastrointestinal diseases. Curr. Pharm. Des. 2018;24:3162–3171. doi: 10.2174/1381612824666180717105128. [DOI] [PubMed] [Google Scholar]
- 60.Eom T., Kim Y.S., Choi C.H., Sadowsky M.J., Unno T. Current understanding of microbiota- and dietary-therapies for treating inflammatory bowel disease. J. Microbiol. 2018;56:189–198. doi: 10.1007/s12275-018-8049-8. [DOI] [PubMed] [Google Scholar]
- 61.Rinaldi E., Consonni A., Guidesi E., Elli M., Mantegazza R., Baggi F. Gut microbiota and probiotics: Novel immune system modulators in myasthenia gravis? Ann. N. Y. Acad. Sci. 2018;1413:49–58. doi: 10.1111/nyas.13567. [DOI] [PubMed] [Google Scholar]
- 62.Aqaeinezhad Rudbane S.M., Rahmdel S., Abdollahzadeh S.M., Zare M., Bazrafshan A., Mazloomi S.M. The efficacy of probiotic supplementation in rheumatoid arthritis: A meta-analysis of randomized, controlled trials. Inflammopharmacology. 2018;26:67–76. doi: 10.1007/s10787-017-0436-y. [DOI] [PubMed] [Google Scholar]
- 63.Aminov R.I. The extent and regulation of lateral gene transfer in natural microbial ecosystems. In: Francino M.P., editor. Horizontal Gene Transfer in Microorganisms. Horizon Scientific Press; Norwich, UK: 2012. pp. 93–100. Chapter 6. [Google Scholar]
- 64.Wong A., Ngu D.Y., Dan L.A., Ooi A., Lim R.L. Detection of antibiotic resistance in probiotics of dietary supplements. Nutr. J. 2015;14:95. doi: 10.1186/s12937-015-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng M., Zhang R., Tian X., Zhou X., Pan X., Wong A. Assessing the Risk of Probiotic Dietary Supplements in the Context of Antibiotic Resistance. Front. Microbiol. 2017;8:908. doi: 10.3389/fmicb.2017.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Reenen C.A., Dicks L.M. Horizontal gene transfer amongst probiotic lactic acid bacteria and other intestinal microbiota: What are the possibilities? A review. Arch. Microbiol. 2011;193:157–168. doi: 10.1007/s00203-010-0668-3. [DOI] [PubMed] [Google Scholar]
- 67.Mercanti D.J., Rousseau G.M., Capra M.L., Quiberoni A., Tremblay D.M., Labrie S.J., Moineau S. Genomic Diversity of Phages Infecting Probiotic Strains of Lactobacillus paracasei. Appl. Environ. Microbiol. 2015;82:95–105. doi: 10.1128/AEM.02723-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ceapa C., Davids M., Ritari J., Lambert J., Wels M., Douillard F.P., Smokvina T., De Vos W.M., Knol J., Kleerebezem M. The Variable Regions of Lactobacillus rhamnosus Genomes Reveal the Dynamic Evolution of Metabolic and Host-Adaptation Repertoires. Genome Biol. Evol. 2016;8:1889–1905. doi: 10.1093/gbe/evw123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tannock G.W., Luchansky J.B., Miller L., Connell H., Thode-Andersen S., Mercer A.A., Klaenhammer T.R. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid. 1994;31:60–71. doi: 10.1006/plas.1994.1007. [DOI] [PubMed] [Google Scholar]
- 70.Lin C.F., Fung Z.F., Wu C.L., Chung T.C. Molecular characterization of a plasmid-borne (pTC82) chloramphenicol resistance determinant (cat-TC) from Lactobacillus reuteri G4. Plasmid. 1996;36:116–124. doi: 10.1006/plas.1996.0039. [DOI] [PubMed] [Google Scholar]
- 71.Egervärn M., Roos S., Lindmark H. Identification and characterization of antibiotic resistance genes in Lactobacillus reuteri and Lactobacillus plantarum. J. Appl. Microbiol. 2009;107:1658–1668. doi: 10.1111/j.1365-2672.2009.04352.x. [DOI] [PubMed] [Google Scholar]
- 72.Baugher J.L., Durmaz E., Klaenhammer T.R. Spontaneously induced prophages in Lactobacillus gasseri contribute to horizontal gene transfer. Appl. Environ. Microbiol. 2014;80:3508–3517. doi: 10.1128/AEM.04092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Courvalin P. Predictable and unpredictable evolution of antibiotic resistance. J. Intern. Med. 2008;264:4–16. doi: 10.1111/j.1365-2796.2008.01940.x. [DOI] [PubMed] [Google Scholar]
- 74.Doron S., Snydman D.R. Risk and safety of probiotics. Clin. Infect. Dis. 2015;60(Suppl. 2):S129–S134. doi: 10.1093/cid/civ085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Egervärn M., Lindmark H., Olsson J., Roos S. Transferability of a tetracycline resistance gene from probiotic Lactobacillus reuteri to bacteria in the gastrointestinal tract of humans. Antonie Van Leeuwenhoek. 2010;97:189–200. doi: 10.1007/s10482-009-9401-0. [DOI] [PubMed] [Google Scholar]
- 76.Imperial I.C., Ibana J.A. Addressing the Antibiotic Resistance Problem with Probiotics: Reducing the Risk of Its Double-Edged Sword Effect. Front. Microbiol. 2016;7:1983. doi: 10.3389/fmicb.2016.01983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosander A., Connolly E., Roos S. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 2008;74:6032–6040. doi: 10.1128/AEM.00991-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muñoz-Atienza E., Gómez-Sala B., Araújo C., Campanero C., del Campo R., Hernández P.E., Herranz C., Cintas L.M. Antimicrobial activity, antibiotic susceptibility and virulence factors of Lactic Acid Bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 2013;13:15. doi: 10.1186/1471-2180-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fittipaldi N., Segura M., Grenier D., Gottschalk M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012;7:259–279. doi: 10.2217/fmb.11.149. [DOI] [PubMed] [Google Scholar]
- 80.Yu J., Pian Y., Ge J., Guo J., Zheng Y., Jiang H., Hao H., Yuan Y., Jiang Y., Yang M. Functional and Structural Characterization of the Antiphagocytic Properties of a Novel Transglutaminase from Streptococcus suis. J. Biol. Chem. 2015;290:19081–19092. doi: 10.1074/jbc.M115.643338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lerner A., Matthias T. Changes in intestinal tight junction permeability associated with industrial food additives explain the rising incidence of autoimmune disease. Autoimmun. Rev. 2015;14:479–489. doi: 10.1016/j.autrev.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 82.Lerner A., Matthias T. Possible association between celiac disease and bacterial transglutaminase in food processing: A hypothesis. Nutr. Rev. 2015;73:544–552. doi: 10.1093/nutrit/nuv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lerner A., Aminov R., Matthias T. Intestinal dysbiotic transglutaminases are potential environmental drivers of systemic autoimmunogenesis. Front. Microbiol. 2017;8:66. doi: 10.3389/fmicb.2017.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matthias T., Jeremias P., Neidhöfer S., Lerner A. The industrial food additive microbial transglutaminase, mimics the tissue transglutaminase and is immunogenic in celiac disease patients. Autoimmun. Rev. 2016;15:1111–1119. doi: 10.1016/j.autrev.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 85.Matthias T., Lerner A. Microbial transglutaminase is immunogenic and potentially pathogenic in pediatric celiac disease. Front. Pediatr. 2018;6:389. doi: 10.3389/fped.2018.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang D., de Souza R.F., Anantharaman V., Lyer L.M., Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Masuda M., Betancourt L., Matsuzawa T., Kashimoto T., Takao T., Shimonishi Y., Horiguchi Y. Activation of Rho through a cross-link with polyamines catalyzed by Bordetella dermonecrotizing toxin. EMBO J. 2000;19:521–530. doi: 10.1093/emboj/19.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jubelin G., Chavez C.V., Taieb F., Banfield M.J., Samba-Louaka A., Nobe R., Nougayrède J.-P., Zumbihl R., Givaudan A., Escoubas J.-M., et al. Cycle Inhibiting Factors (CIFs) Are a Growing Family of Functional Cyclomodulins Present in Invertebrate and Mammal Bacterial Pathogens. PLoS ONE. 2009;4:e4855. doi: 10.1371/journal.pone.0004855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schmidt G., Selzer J., Lerm M., Aktories K. The Rho-deamidating cytotoxic necrotizing factor 1 from Escherichia coli possesses transglutaminase activity. Cysteine 866 and histidine 881 are essential for enzyme activity. J. Biol. Chem. 1998;273:13669–13674. doi: 10.1074/jbc.273.22.13669. [DOI] [PubMed] [Google Scholar]
- 90.Makarova K.S., Aravind L., Koonin E.V. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases. Protein Sci. 1999;8:1714–1719. doi: 10.1110/ps.8.8.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lerner A., Matthias T. Microbial transglutaminase is beneficial to food industries but a caveat to public health. Med One. 2019;4:e190001. doi: 10.20900/mo.20190001. [DOI] [Google Scholar]
- 92.Sitaraman R. Prokaryotic horizontal gene transfer within the human holobiont: Ecological-evolutionary inferences, implications and possibilities. Microbiome. 2018;6:163. doi: 10.1186/s40168-018-0551-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu F., Wang J., Guo Y., Fu P., Zeng H., Li Z., Pei X., Liu X., Wang S. Antibiotic resistance, biochemical typing, and PFGE typing of Bifidobacterium strains commonly used in probiotic health foods. Food Sci. Biotechnol. 2018;27:467–477. doi: 10.1007/s10068-018-0320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gueimonde M., Sánchez B., de Los Reyes-Gavilán C., Margolles A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013;4:202. doi: 10.3389/fmicb.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Comunian R., Daga E., Dupré I., Paba A., Devirgiliis C., Piccioni V., Perozzi G., Zonenschain D., Rebecchi A., Morelli L., et al. Susceptibility to tetracycline and erythromycin of Lactobacillus paracasei strains isolated from traditional Italian fermented foods. Int. J. Food Microbiol. 2010;138:151–156. doi: 10.1016/j.ijfoodmicro.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 96.Guo H., Pan L., Li L., Lu J., Kwok L., Menghe B., Zhang H. Characterization of Antibiotic Resistance Genes from Lactobacillus Isolated from Traditional Dairy Products. J. Food Sci. 2017;82:724–730. doi: 10.1111/1750-3841.13645. [DOI] [PubMed] [Google Scholar]
- 97.Dec M., Nowaczek A., Stępień-Pyśniak D., Wawrzykowski J., Urban-Chmiel R. Identification and antibiotic susceptibility of lactobacilli isolated from turkeys. BMC Microbiol. 2018;18:168. doi: 10.1186/s12866-018-1269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Agersø Y., Stuer-Lauridsen B., Bjerre K., Jensen M.G., Johansen E., Bennedsen M., Brockmann E., Nielsen B. Antimicrobial Susceptibility Testing and Tentative Epidemiological Cutoff Values for Five Bacillus Species Relevant for Use as Animal Feed Additives or for Plant Protection. Appl. Environ. Microbiol. 2018;84:e01108-18. doi: 10.1128/AEM.01108-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abriouel H., Casado Muñoz M.D.C., Lavilla Lerma L., Pérez Montoro B., Bockelmann W., Pichner R., Kabisch J., Cho G.S., Franz C.M.A.P., Gálvez A., et al. New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res. Int. 2015;78:465–481. doi: 10.1016/j.foodres.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 100.Vitetta L., Vitetta G., Hall S. Immunological Tolerance and Function: Associations Between Intestinal Bacteria, Probiotics, Prebiotics, and Phages. Front. Immunol. 2018;9:2240. doi: 10.3389/fimmu.2018.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bakhshinejad B., Ghiasvand S. Bacteriophages in the human gut: Our fellow travelers throughout life and potential biomarkers of heath or disease. Virus Res. 2017;240:47–55. doi: 10.1016/j.virusres.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 102.Tetz G., Tetz V. Bacteriophages as New Human Viral Pathogens. Microorganisms. 2018;6:54. doi: 10.3390/microorganisms6020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pfeiffer J.K., Virgin H.W. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science. 2016;351:aad5872. doi: 10.1126/science.aad5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Proal A., Marshall T. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome in the Era of the Human Microbiome: Persistent Pathogens Drive Chronic Symptoms by Interfering With Host Metabolism, Gene Expression, and Immunity. Front. Pediatr. 2018 doi: 10.3389/fped.2018.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tetz G., Tetz V. Prion-Like Domains in Phagobiota. Front. Microbiol. 2017;8:2239. doi: 10.3389/fmicb.2017.02239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Muniesa M., Colomer-Lluch M., Jofre J. Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Future Microbiol. 2013;8:739–751. doi: 10.2217/fmb.13.32. [DOI] [PubMed] [Google Scholar]
- 107.Muniesa M., Colomer-Lluch M., Jofre J. Could bacteriophages transfer antibiotic resistance genes from environmental bacteria to human-body associated bacterial populations? Mob. Genet. Elem. 2013;3:e25847. doi: 10.4161/mge.25847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lood R., Ertürk G., Mattiasson B. Revisiting Antibiotic Resistance Spreading in Wastewater Treatment Plants—Bacteriophages as a Much Neglected Potential Transmission Vehicle. Front. Microbiol. 2017;8:2298. doi: 10.3389/fmicb.2017.02298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Quirós P., Colomer-Lluch M., Martínez-Castillo A., Miró E., Argente M., Jofre J., Navarro F., Muniesa M. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob. Agents Chemother. 2014;58:606–609. doi: 10.1128/AAC.01684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eaton T.J., Gasson M.J. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marcó M.B., Moineau S., Quiberoni A. Bacteriophages and dairy fermentations. Bacteriophage. 2012;2:149–158. doi: 10.4161/bact.21868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Domingo-Calap P., Delgado-Martínez J. Bacteriophages: Protagonists of a Post-Antibiotic Era. Antibiotics. 2018;7:66. doi: 10.3390/antibiotics7030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li J., Cao J., Zhu Y.G., Chen Q.L., Shen F., Wu Y., Xu S., Fan H., Da G., Huang R.-J., et al. Global Survey of Antibiotic Resistance Genes in Air. Environ. Sci. Technol. 2018;52:10975–10984. doi: 10.1021/acs.est.8b02204. [DOI] [PubMed] [Google Scholar]
- 114.Villion M., Moineau S. Bacteriophages of lactobacillus. Front. Biosci. 2009;14:1661–1683. doi: 10.2741/3332. [DOI] [PubMed] [Google Scholar]
- 115.Capra M.L., Del L Quiberoni A., Ackermann H.W., Moineau S., Reinheimer J.A. Characterization of a new virulent phage (MLC-A) of Lactobacillus paracasei. J. Dairy Sci. 2006;89:2414–2423. doi: 10.3168/jds.S0022-0302(06)72314-1. [DOI] [PubMed] [Google Scholar]
- 116.Aucouturier A., Chain F., Langella P., Bidnenko E. Characterization of a Prophage-Free Derivative Strain of Lactococcus lactis ssp. lactis IL1403 Reveals the Importance of Prophages for Phenotypic Plasticity of the Host. Front. Microbiol. 2018;9:2032. doi: 10.3389/fmicb.2018.02032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Waller A.S., Yamada T., Kristensen D.M., Kultima J.R., Sunagawa S., Koonin E.V., Bork P. Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J. 2014;8:1391–1402. doi: 10.1038/ismej.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Visweswaran G.R., Kurek D., Szeliga M., Pastrana F.R., Kuipers O.P., Kok J., Buist G. Expression of prophage-encoded endolysins contributes to autolysis of Lactococcus lactis. Appl. Microbiol. Biotechnol. 2017;101:1099–1110. doi: 10.1007/s00253-016-7822-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Labrie S.J., Moineau S. Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages. J. Bacteriol. 2007;189:1482–1487. doi: 10.1128/JB.01111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chopin M.C., Chopin A., Bidnenko E. Phage abortive infection in lactococci: Variations on a theme. Curr. Opin. Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 121.Devirgiliis C., Zinno P., Perozzi G. Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front. Microbiol. 2013;4:301. doi: 10.3389/fmicb.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang D., Aravind L. Novel transglutaminase-like peptidase and C2 domains elucidate the structure, biogenesis and evolution of the ciliary compartment. Cell Cycle. 2012;11:3861–3875. doi: 10.4161/cc.22068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khan S.H., Ansari F.A. Probiotics—The friendly bacteria with market potential in global market. Pak. J. Pharm. Sci. 2007;20:76–82. [PubMed] [Google Scholar]
- 124.Kopp-Hoolihan L. Prophylactic and therapeutic uses of probiotics: A review. J. Am. Diet. Assoc. 2001;101:229–238. doi: 10.1016/S0002-8223(01)00060-8. [DOI] [PubMed] [Google Scholar]
- 125.Kaur I.P., Chopra K., Saini A. Probiotics: Potential pharmaceutical applications. Eur. J. Pharm. Sci. 2002;15:1–9. doi: 10.1016/S0928-0987(01)00209-3. [DOI] [PubMed] [Google Scholar]
- 126.O’Toole P.W., Marchesi J.R., Hill C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017;2:17057. doi: 10.1038/nmicrobiol.2017.57. [DOI] [PubMed] [Google Scholar]
- 127.Boesmans L., Valles-Colomer M., Wang J., Eeckhaut V., Falony G., Ducatelle R., van Immerseel F., Raes J., Verbeke K. Butyrate Producers as Potential Next-Generation Probiotics: Safety Assessment of the Administration of Butyricicoccus pullicaecorum to Healthy Volunteers. mSystems. 2018:e00094-18. doi: 10.1128/mSystems.00094-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Roman P., Estévez A.F., Miras A., Sánchez-Labraca N., Cañadas F., Vivas A.B., Cardona D. A Pilot Randomized Controlled Trial to Explore Cognitive and Emotional Effects of Probiotics in Fibromyalgia. Sci. Rep. 2018;8:10965. doi: 10.1038/s41598-018-29388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rezac S., Kok C.R., Heermann M., Hutkins R. Fermented Foods as a Dietary Source of Live Organisms. Front. Microbiol. 2018;9:1785. doi: 10.3389/fmicb.2018.01785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oniciuc E.A., Likotrafiti E., Alvarez-Molina A., Prieto M., Santos J.A., Alvarez-Ordóñez A. The Present and Future of Whole Genome Sequencing (WGS) and Whole Metagenome Sequencing (WMS) for Surveillance of Antimicrobial Resistant Microorganisms and Antimicrobial Resistance Genes across the Food Chain. Genes. 2018;9:268. doi: 10.3390/genes9050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Flórez A.B., Vázquez L., Mayo B. A Functional Metagenomic Analysis of Tetracycline Resistance in Cheese Bacteria. Front. Microbiol. 2017;8:907. doi: 10.3389/fmicb.2017.00907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.de Paula A.C.L., Medeiros J.D., de Azevedo A.C., de Assis Chagas J.M., da Silva V.L., Diniz C.G. Antibiotic Resistance Genetic Markers and Integrons in White Soft Cheese: Aspects of Clinical Resistome and Potentiality of Horizontal Gene Transfer. Genes. 2018;9:106. doi: 10.3390/genes9020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Devirgiliis C., Zinno P., Stirpe M., Barile S., Perozzi G. Functional screening of antibiotic resistance genes from a representative metagenomic library of food fermenting microbiota. Biomed. Res. Int. 2014;2014:290967. doi: 10.1155/2014/290967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cole M.L., Singh O.V. Microbial occurrence and antibiotic resistance in ready-to-go food items. J. Food Sci. Technol. 2018;55:2600–2609. doi: 10.1007/s13197-018-3180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Papagaroufalis K., Fotiou A., Egli D., Tran L.A., Steenhout P. A Randomized Double Blind Controlled Safety Trial Evaluating d-Lactic Acid Production in Healthy Infants Fed a Lactobacillus reuteri-containing Formula. Nutr. Metab. Insights. 2014;7:19–27. doi: 10.4137/NMI.S14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Haschke-Becher E., Brunser O., Cruchet S., Gotteland M., Haschke F., Bachmann C. Urinary D-lactate excretion in infants receiving Lactobacillus johnsonii with formula. Ann. Nutr. Metab. 2008;53:240–244. doi: 10.1159/000185642. [DOI] [PubMed] [Google Scholar]
- 137.Vitetta L., Coulson S., Thomsen M., Nguyen T., Hall S. Probiotics, D-Lactic acidosis, oxidative stress and strain specificity. Gut Microbes. 2017;8:311–322. doi: 10.1080/19490976.2017.1279379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Munakata S., Arakawa C., Kohira R., Fujita Y., Fuchigami T., Mugishima H. A case of D-lactic acid encephalopathy associated with use of probiotics. Brain Dev. 2010;32:691–694. doi: 10.1016/j.braindev.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 139.Rao S.S.C., Yu S., Tetangco E.P., Yan Y. Probiotics can Cause D-Lactic Acidosis and Brain Fogginess: Reply to Quigley et al. Clin. Transl. Gastroenterol. 2018;9:207. doi: 10.1038/s41424-018-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Muñoz J.A., Chenoll E., Casinos B., Bataller E., Ramón D., Genovés S., Montava R., Ribes J.M., Buesa J., Fàbrega J., et al. Novel probiotic Bifidobacterium longum subsp. infantis CECT 7210 strain active against rotavirus infections. Appl. Environ. Microbiol. 2011;77:8775–8783. doi: 10.1128/AEM.05548-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ewaschuk J.B., Naylor J.M., Zello G.A. D-lactate in human and ruminant metabolism. J. Nutr. 2005;135:1619–1625. doi: 10.1093/jn/135.7.1619. [DOI] [PubMed] [Google Scholar]
- 142.Ku W.H., Lau D.C.Y., Huen K.F. Probiotics Provoked D-lactic Acidosis in Short Bowel Syndrome: Case Report and Literature. HK J. Paediatr. 2006;11:246–254. [Google Scholar]
- 143.Petersen C. D-lactic acidosis. Nutr. Clin. Pract. 2005;20:634–645. doi: 10.1177/0115426505020006634. [DOI] [PubMed] [Google Scholar]
- 144.Uribarri J., Oh M.S., Carroll H.J. D-lactic acidosis. A review of clinical presentation, biochemical features, and pathophysiologic mechanisms. Medicine. 1998;77:73–82. doi: 10.1097/00005792-199803000-00001. [DOI] [PubMed] [Google Scholar]
- 145.Oh M.S., Phelps K.R., Traube M., Barbosa-Saldivar J.L., Boxhill C., Carroll H.J. D-lactic acidosis in a man with the short-bowel syndrome. N. Engl. J. Med. 1979;301:249–252. doi: 10.1056/NEJM197908023010505. [DOI] [PubMed] [Google Scholar]
- 146.Quigley E.M.M., Pot B., Sanders M.E. ‘Brain Fogginess’ and D-Lactic Acidosis: Probiotics Are Not the Cause. Clin. Transl. Gastroenterol. 2018;9:187. doi: 10.1038/s41424-018-0057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sachdeva S., Puri A.S., Kumar A., Dalal A., Dahale A.S. Brain Fogginess and SIBO: A Link or Just a Mirage? Clin. Transl. Gastroenterol. 2018;9:184. doi: 10.1038/s41424-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Van den Nieuwboer M., Brummer R.J., Guarner F., Morelli L., Cabana M., Claasen E. The administration of probiotics and synbiotics in immune compromised adults: Is it safe? Benef. Microbes. 2015;6:3–17. doi: 10.3920/BM2014.0079. [DOI] [PubMed] [Google Scholar]
- 149.Sanders M.E., Akkermans L.M., Haller D., Hammerman C., Heimbach J., Hörmannsperger G., Huys G. Safety assessment of probiotics for human use. Gut Microbes. 2010;1:164–185. doi: 10.4161/gmic.1.3.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sherid M., Samo S., Sulaiman S., Husein H., Sifuentes H., Sridhar S. Liver abscess and bacteremia caused by lactobacillus: Role of probiotics? Case report and review of the literature. BMC Gastroenterol. 2016;16:138. doi: 10.1186/s12876-016-0552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kopp M.V., Hennemuth I., Heinzmann A., Urbanek R. Randomized, double-blind, placebo-controlled trial of probiotics for primary prevention: no clinical effects of Lactobacillus GG supplementation. Pediatrics. 2008;121:e850-6. doi: 10.1542/peds.2007-1492. [DOI] [PubMed] [Google Scholar]
- 152.Franz C.M., Muscholl-Silberhorn A.B., Yousif N.M., Vancanneyt M., Swings J., Holzapfel W.H. Incidence of virulence factors and antibiotic resistance among Enterococci isolated from food. Appl. Environ. Microbiol. 2001;67:4385–4389. doi: 10.1128/AEM.67.9.4385-4389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rathnayake I.U., Hargreaves M., Huygens F. Antibiotic resistance and virulence traits in clinical and environmental Enterococcus faecalis and Enterococcus faecium isolates. Syst. Appl. Microbiol. 2012;35:326–333. doi: 10.1016/j.syapm.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 154.Kho Z.Y., Lal S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fatal gastrointestinal mucormycosis in an infant following use of contaminated ABC Dophilus powder from Solgar Inc. [(accessed on 20 February 2015)]; Available online: www.cdc.gov/fungal/outbreaks/rhizopus-investigation.html.
- 156.Naqvi S.S.B., Nagendra V., Hofmeyr A. Probiotic related Lactobacillus rhamnosus endocarditis in a patient with liver cirrhosis. IDCases. 2018;13:e00439. doi: 10.1016/j.idcr.2018.e00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Boumis E., Capone A., Galati V., Venditti C., Petrosillo N. Probiotics and infective endocarditis in patients with hereditary hemorrhagic telangiectasia: A clinical case and a review of the literature. BMC Infect. Dis. 2018;18:65. doi: 10.1186/s12879-018-2956-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jäsberg H., Tervahartiala T., Sorsa T., Söderling E., Haukioja A. Probiotic intervention influences the salivary levels of Matrix Metalloproteinase (MMP)-9 and Tissue Inhibitor of metalloproteinases (TIMP)-1 in healthy adults. Arch. Oral Biol. 2018;85:58–63. doi: 10.1016/j.archoralbio.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 159.Falcinelli S., Rodiles A., Hatef A., Picchietti S., Cossignani L., Merrifield D.L., Unniappan S., Carnevali O. Influence of Probiotics Administration on Gut Microbiota Core: A Review on the Effects on Appetite Control, Glucose, and Lipid Metabolism. J. Clin. Gastroenterol. 2018;52(Suppl. 1):S50–S56. doi: 10.1097/MCG.0000000000001064. [DOI] [PubMed] [Google Scholar]
- 160.Dufour A., Hindré T., Haras D., Le Pennec J.P. The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol. Rev. 2007;31:134–167. doi: 10.1111/j.1574-6976.2006.00045.x. [DOI] [PubMed] [Google Scholar]
- 161.Muthu Selvam R., Vinothini G., Palliyarai Thaiyammal S., Latha S., Chinnathambi A., Dhanasekaran D., Padmanabhan P., Alharbi S.A., Archunan G. The cell aggregating propensity of probiotic actinobacterial isolates: Isolation and characterization of the aggregation inducing peptide pheromone. Biofouling. 2016;32:71–79. doi: 10.1080/08927014.2015.1122759. [DOI] [PubMed] [Google Scholar]
- 162.Kitazawa H., Ino T., Kawai Y., Itoh T., Saito T. A novel immunostimulating aspect of Lactobacillus gasseri: Induction of “Gasserokine” as chemoattractants for macrophages. Int. J. Food Microbiol. 2002;77:29–38. doi: 10.1016/S0168-1605(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 163.Nagy G., Pinczes G., Pinter G., Pocsi I., Prokisch J., Banfalvi G. In Situ Electron Microscopy of Lactomicroselenium Particles in Probiotic Bacteria. Int. J. Mol. Sci. 2016;17:1047. doi: 10.3390/ijms17071047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Washington E.J., Banfield M.J., Dangl J.L. What a difference a Dalton makes: Bacterial virulence factors modulate eukaryotic host cell signaling systems via deamidation. Microbiol. Mol. Biol. Rev. 2013;77:527–539. doi: 10.1128/MMBR.00013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rondanelli M., Faliva M.A., Perna S., Giacosa A., Peroni G., Castellazzi A.M. Using probiotics in clinical practice: Where are we now? A review of existing meta-analyses. Gut Microbes. 2017;8:521–543. doi: 10.1080/19490976.2017.1345414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hempel S., Newberry S., Ruelaz A., Wang Z., Miles J.N., Suttorp M.J., Johnsen B., Shanman R., Slusser W., Fu N., et al. Safety of probiotics used to reduce risk and prevent or treat disease. Evid. Rep. Technol. Assess. 2011;200:1–645. [PMC free article] [PubMed] [Google Scholar]
- 167.van den Nieuwboer M., Brummer R.J., Guarner F., Morelli L., Cabana M., Claassen E. Safety of probiotics and synbiotics in children under 18 years of age. Benef. Microbes. 2015;6:615–630. doi: 10.3920/BM2014.0157. [DOI] [PubMed] [Google Scholar]
- 168.Doherty G.A., Bennett G.C., Cheifetz A.S., Moss A.C. Meta-analysis: Targeting the intestinal microbiota in prophylaxis for post-operative Crohn’s disease. Aliment. Pharmacol. Ther. 2010;31:802–809. doi: 10.1111/j.1365-2036.2010.04231.x. [DOI] [PubMed] [Google Scholar]
- 169.Tan-Lim C.S.C., Esteban-Ipac N.A.R. Probiotics as treatment for food allergies among pediatric patients: A meta-analysis. World Allergy Organ. J. 2018;11:25. doi: 10.1186/s40413-018-0204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Osborn D.A., Sinn J.K. Probiotics in infants for prevention of allergic disease and food hypersensitivity. Cochrane Database Syst. Rev. 2007;4:CD006475. doi: 10.1002/14651858.CD006475.pub2. [DOI] [PubMed] [Google Scholar]
- 171.Reis D.J., Ilardi S.S., Punt S.E.W. The anxiolytic effect of probiotics: A systematic review and meta-analysis of the clinical and preclinical literature. PLoS ONE. 2018;13:e0199041. doi: 10.1371/journal.pone.0199041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Borgeraas H., Johnson L.K., Skattebu J., Hertel J.K., Hjelmesaeth J. Effects of probiotics on body weight, body mass index, fat mass and fat percentage in subjects with overweight or obesity: A systematic review and meta-analysis of randomized controlled trials. Obes. Rev. 2018;19:219–232. doi: 10.1111/obr.12626. [DOI] [PubMed] [Google Scholar]
- 173.Wardill H.R., Van Sebille Y.Z.A., Ciorba M.A., Bowen J.M. Prophylactic probiotics for cancer therapy-induced diarrhoea: A meta-analysis. Curr. Opin. Support Palliat. Care. 2018;12:187–197. doi: 10.1097/SPC.0000000000000338. [DOI] [PubMed] [Google Scholar]
- 174.Canales J., Rada G. Are probiotics effective in preventing urinary tract infection? Medwave. 2018;18:e7186. doi: 10.5867/medwave.2018.02.7185. [DOI] [PubMed] [Google Scholar]
- 175.Hassan H., Rompola M., Glaser A.W., Kinsey S.E., Phillips R.S. Systematic review and meta-analysis investigating the efficacy and safety of probiotics in people with cancer. Support Care Cancer. 2018;26:2503–2509. doi: 10.1007/s00520-018-4216-z. [DOI] [PubMed] [Google Scholar]
- 176.Mohammed A.T., Khattab M., Ahmed A.M., Turk T., Sakr N., Khalil A.M., Abdelhalim M., Sawaf B., Hirayama K., Huy N.T. The therapeutic effect of probiotics on rheumatoid arthritis: A systematic review and meta-analysis of randomized control trials. Clin. Rheumatol. 2017;36:2697–2707. doi: 10.1007/s10067-017-3814-3. [DOI] [PubMed] [Google Scholar]
- 177.Liu B., He Y., Wang M., Liu J., Ju Y., Zhang Y., Liu T., Li L., Li Q. Efficacy of probiotics on anxiety-A meta-analysis of randomized controlled trials. Depress Anxiety. 2018;35:935–945. doi: 10.1002/da.22811. [DOI] [PubMed] [Google Scholar]
- 178.Lin J., Zhang Y., He C., Dai J. Probiotics supplementation in children with asthma: A systematic review and meta-analysis. J. Paediatr. Child Health. 2018;54:953–961. doi: 10.1111/jpc.14126. [DOI] [PubMed] [Google Scholar]
- 179.McFarland L.V., Goh S. Are probiotics and prebiotics effective in the prevention of travellers’ diarrhea: A systematic review and meta-analysis. Travel Med. Infect Dis. 2018 doi: 10.1016/j.tmaid.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 180.Jin L., Deng L., Wu W., Wang Z., Shao W., Liu J. Systematic review and meta-analysis of the effect of probiotic supplementation on functional constipation in children. Medicine. 2018;97:e12174. doi: 10.1097/MD.0000000000012174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rouhani M.H., Hadi A., Ghaedi E., Salehi M., Mahdavi A., Mohammadi H. Do probiotics, prebiotics and synbiotics affect adiponectin and leptin in adults? A systematic review and meta-analysis of clinical trials. Clin. Nutr. 2018 doi: 10.1016/j.clnu.2018.09.033. [DOI] [PubMed] [Google Scholar]
- 182.Upadhyay R.P., Taneja S., Chowdhury R., Strand T.A., Bhandari N. Effect of prebiotic and probiotic supplementation on neurodevelopment in preterm very low birth weight infants: Findings from a meta-analysis. Pediatr. Res. 2018 doi: 10.1038/s41390-018-0211-9. [DOI] [PubMed] [Google Scholar]
- 183.Dalal R., McGee R.G., Riordan S.M., Webster A.C. Probiotics for people with hepatic encephalopathy. Cochrane Database Syst. Rev. 2017;2:CD008716. doi: 10.1002/14651858.CD008716.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Goldenberg J.Z., Lytvyn L., Steurich J., Parkin P., Mahant S., Johnston B.C. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst. Rev. 2015;12:CD004827. doi: 10.1002/14651858.CD004827.pub4. [DOI] [PubMed] [Google Scholar]
- 185.Szajewska H., Horvath A. Lactobacillus rhamnosus GG in the Primary Prevention of Eczema in Children: A Systematic Review and Meta-Analysis. Nutrients. 2018;10:1319. doi: 10.3390/nu10091319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mater D.D., Langella P., Corthier G., Flores M.J. A probiotic Lactobacillus strain can acquire vancomycin resistance during digestive transit in mice. J. Mol. Microbiol. Biotechnol. 2008;14:123–127. doi: 10.1159/000106091. [DOI] [PubMed] [Google Scholar]
- 187.Jalali M., Abedi D., Varshosaz J., Najjarzadeh M., Mirlohi M., Tavakoli N. Stability evaluation of freeze-dried Lactobacillus paracasei subsp. tolerance and Lactobacillus delbrueckii subsp. bulgaricus in oral capsules. Res Pharm Sci. 2012;7:31–36. [PMC free article] [PubMed] [Google Scholar]
- 188.Generally Recognized as Safe (GRAS) [(accessed on 7 February 2019)]; Available online: http://www.fda.gov/food/IngredientspackagingLabeling/GRAS/
- 189.Investigational new drug applications (INDs)-determining whether human research studies can be conducted without an IND. [(accessed on 30 September 2013)]; Available online: www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/UCM229175.pdf.
- 190.Duc L.H., Hong H.A., Barbosa T.M., Henriques A.O., Cutting S.M. Characterization of Bacillus probiotics available for human use. Appl. Environ. Microbiol. 2004;70:2161–2171. doi: 10.1128/AEM.70.4.2161-2171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Green D.H., Wakeley P.R., Page A., Barnes A., Baccigalupi L., Ricca E., Cutting S.M. Characterization of two Bacillus probiotics. Appl. Environ. Microbiol. 1999;65:4288–4291. doi: 10.1128/aem.65.9.4288-4291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Temmerman R., Scheirlinck I., Huys G., Swings J. Culture-independent analysis of probiotic products by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2003;69:220–226. doi: 10.1128/AEM.69.1.220-226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Masco L., Huys G., De Brandt E., Temmerman R., Swings J. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 2005;102:221–230. doi: 10.1016/j.ijfoodmicro.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 194.Ouwehand A.C. A review of dose-responses of probiotics in human studies. Benef. Microbes. 2017;8:143–151. doi: 10.3920/BM2016.0140. [DOI] [PubMed] [Google Scholar]
- 195.Hoffmann D.E. Health claim regulation of probiotics in the USA and the EU: Is there a middle way? Benef. Microbes. 2013;4:109–115. doi: 10.3920/BM2012.0033. [DOI] [PubMed] [Google Scholar]
- 196.Lee T.T., Morisset M., Astier C., Moneret-Vautrin D.A., Cordebar V., Beaudouin E., Codreanu F., Bihain B.E., Kanny G. Contamination of probiotic preparations with milk allergens can cause anaphylaxis in children with cow’s milk allergy. J. Allergy Clin. Immunol. 2007;119:746–747. doi: 10.1016/j.jaci.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 197.Martín-Muñoz M.F., Fortuni M., Caminoa M., Belver T., Quirce S., Caballero T. Anaphylactic reaction to probiotics. Cow’s milk and hen’s egg allergens in probiotic compounds. Pediatr. Allergy Immunol. 2012;23:778–784. doi: 10.1111/j.1399-3038.2012.01338.x. [DOI] [PubMed] [Google Scholar]
- 198.Allen E.N., Chandler C.I., Mandimika N., Leisegang C., Barnes K. Eliciting adverse effects data from participants in clinical trials. Cochrane Database Syst. Rev. 2018;1:MR000039. doi: 10.1002/14651858.MR000039.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jin Y., Sanger N., Shams I., Luo C., Shahid H., Li G., Bhatt M., Zielinski L., Bantoto B., Wang M., et al. Does the medical literature remain inadequately described despite having reporting guidelines for 21 years?—A systematic review of reviews: An update. J. Multidiscip. Healthc. 2018;11:495–510. doi: 10.2147/JMDH.S155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bafeta A., Koh M., Riveros C., Ravaud P. Harms Reporting in Randomized Controlled Trials of Interventions Aimed at Modifying Microbiota: A Systematic Review. Ann. Intern. Med. 2018;169:240–247. doi: 10.7326/M18-0343. [DOI] [PubMed] [Google Scholar]
- 201.van den Nieuwboer M., Claassen E., Morelli L., Guarner F., Brummer R.J. Probiotic and synbiotic safety in infants under two years of age. Benef. Microbes. 2014;5:45–60. doi: 10.3920/BM2013.0046. [DOI] [PubMed] [Google Scholar]
- 202.Mugambi M.N., Musekiwa A., Lombard M., Young T., Blaauw R. Association between funding source, methodological quality and research outcomes in randomized controlled trials of synbiotics, probiotics and prebiotics added to infant formula: A systematic review. BMC Med. Res. Methodol. 2013;13:137. doi: 10.1186/1471-2288-13-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Aceti A., Beghetti I., Maggio L., Martini S., Faldella G., Corvaglia L. Filling the Gaps: Current Research Directions for a Rational Use of Probiotics in Preterm Infants. Nutrients. 2018;10:1472. doi: 10.3390/nu10101472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Huff B.A. Caveat emptor. “Probiotics” might not be what they seem. Can. Fam. Physician. 2004;50:583–587. [PMC free article] [PubMed] [Google Scholar]
- 205.Sanders M. How do we know when something called” probiotic” is really a probiotic? A guideline for consumers and healthcare professionals. Funct. Food Rev. 2009;1:3–12. [Google Scholar]
- 206.Sanders M.E., Klaenhammer T.R., Ouwehand A.C., Pot B., Johansen E., Heimbach J.T., Marco M.L., Tennilä J., Ross R.P., Franz C., et al. Effects of genetic, processing, or product formulation changes on efficacy and safety of probiotics. Ann. N. Y. Acad. Sci. 2014;1309:1–18. doi: 10.1111/nyas.12363. [DOI] [PubMed] [Google Scholar]
- 207.Katan M.B. Why the European Food Safety Authority was right to reject health claims for probiotics. Benef. Microbes. 2012;3:85–89. doi: 10.3920/BM2012.0008. [DOI] [PubMed] [Google Scholar]
- 208.Smug L.N., Salminen S., Sanders M.E., Ebner S. Yoghurt and probiotic bacteria in dietary guidelines of the member states of the European Union. Benef. Microbes. 2014;5:61–66. doi: 10.3920/BM2013.0050. [DOI] [PubMed] [Google Scholar]
- 209.Dronkers T.M.G., Krist L., Van Overveld F.J., Rijkers G.T. The ascent of the blessed: Regulatory issues on health effects and health claims for probiotics in Europe and the rest of the world. Benef. Microbes. 2018;9:717–723. doi: 10.3920/BM2017.0196. [DOI] [PubMed] [Google Scholar]
- 210. [(accessed on 15 November 2012)]; Available online: www.law.umaryland.edu/media/SOL/...Law/FinalWhitePaper.pdf.
- 211.Ebner S., Smug L.N., Kneifel W., Salminen S.J., Sanders M.E. Probiotics in dietary guidelines and clinical recommendations outside the European Union. World J. Gastroenterol. 2014;20:16095–16100. doi: 10.3748/wjg.v20.i43.16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lee E.S., Song E.J., Nam Y.D., Lee S.Y. Probiotics in human health and disease: From nutribiotics to pharmabiotics. J. Microbiol. 2018;56:773–782. doi: 10.1007/s12275-018-8293-y. [DOI] [PubMed] [Google Scholar]
- 213.Azad M.A.K., Sarker M., Li T., Yin J. Probiotic Species in the Modulation of Gut Microbiota: An Overview. Biomed. Res. Int. 2018;2018:9478630. doi: 10.1155/2018/9478630. [DOI] [PMC free article] [PubMed] [Google Scholar]