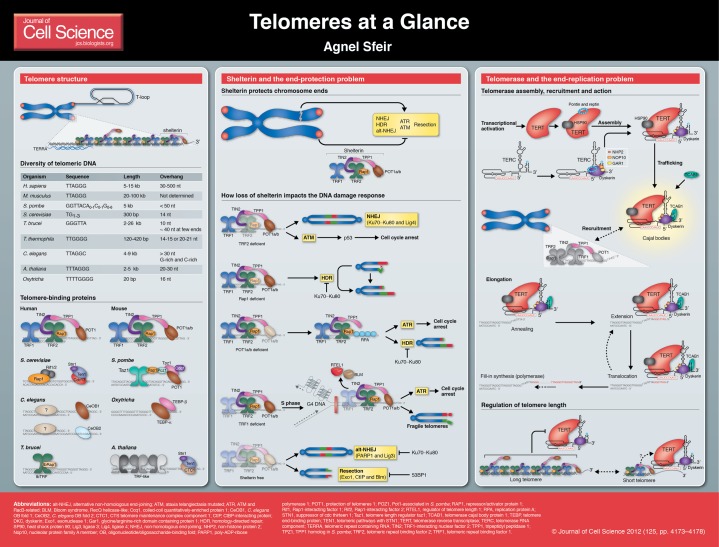
The linearity of chromosomes creates two major problems for eukaryotic cells: the end-replication problem and the end-protection problem. The end-replication problem stems from the inherent inability of the replication machinery to fully duplicate linear templates. The end-protection problem refers to the propensity of linear chromosome ends to be recognized as DNA double-strand breaks (DSBs). Both problems are surmounted by telomeres, the specific nucleoprotein complexes that adorn chromosome ends.
In 2009, the Nobel Prize was awarded to pioneering researchers in the telomere field in recognition of their seminal findings that set the stage for, so far, three decades of vigorous investigation of telomeres and telomerase. In this article, I review our current knowledge and recent findings regarding the function of telomeres, with a focus on the mammalian system. Specifically, I delineate the structure and composition of the telomere and highlight recent discoveries that have elucidated the molecular mechanisms underlying the solutions to both ‘end problems’. Lastly, I describe the perilous consequences of telomere dysfunction during tumorigenesis.
The composition of chromosome ends
Features of telomeric DNA
The telomeric architecture incorporates three known entities: tandem repeats of DNA sequence, a specific set of binding proteins and a non-coding RNA transcript. In mammals, telomeric DNA consists of TTAGGG repeats and their complementary AATCCC sequences and, in humans, its size ranges between 5 and 15 kb. A key feature of the telomere end in all organisms is a 3′ single-stranded G-rich overhang (Makarov et al., 1997; McElligott and Wellinger, 1997). Although, it should also be noted that 5′ single-stranded C-rich overhangs have been observed in worms (Raices et al., 2008) and can occur transiently in some human cancer cells (Oganesian and Karlseder, 2011). Mammalian G-rich overhangs are 30-500 nucleotides long (Chai et al., 2005) and are generated by the removal of the RNA primer from the terminal Okazaki fragment on the lagging strand, as well as by post-replicative processing events that involve nucleases performing 3′ resection on both newly synthesized strands. Recent studies have started to unravel the molecular details of such processing events by identifying the Apollo nuclease that acts on leading end telomeres (Lam et al., 2010; Wu et al., 2010). G-rich overhangs have a central role in sustaining a diverse array of telomeric functions and are thought to invade the preceding duplex region of the telomere to form a lariat-like structure called the t-loop. This higher-order architecture, which was revealed by electron microscopy (Griffith et al., 1999; Nikitina and Woodcock, 2004; Raices et al., 2008), provides one mechanism by which chromosome ends maintain their protective cap.
Telomere-binding proteins
Telomeric DNA is bound by a specialized set of proteins, whose composition, structure and function have diverged across species (de Lange, 2009). In mammalian cells, there are six bona fide telomere-specific proteins: TRF1 (telomeric repeat binding factor 1, also known as TERF1), TRF2 (telomeric repeat binding factor 2, also known as TERF2), RAP1 (TERF2 interacting protein, also known as TERF2IP), TIN2 (TRF1 interacting nuclear factor 2, also known as TINF2), TPP1 (adrenocortical dysplasia protein homolog, also known as ACD) and POT1 (protection of telomeres 1), which together form the shelterin complex (de Lange, 2005). The exquisite specificity with which shelterin binds to the telomeric DNA is conferred by its three DNA-binding modules. TRF1 and TRF2 bind to the duplex region of the DNA (Bianchi et al., 1997; Broccoli et al., 1997; Bilaud et al., 1997), whereas POT1 coats the overhang with its oligonucleotide/oligosaccharide binding (OB) folds (Baumann and Cech, 2001; Lei et al., 2002; Loayza and de Lange, 2003). Rodents express two POT1 paralogs (POT1a and POT1b) that are structurally similar, yet functionally divergent (Hockemeyer et al., 2006; Wu et al., 2006). TIN2 has a bridging function, as it interacts with both TRF1 and TRF2 while simultaneously recruiting TPP1 (Ye et al., 2004a; Ye et al., 2004b; O'Connor et al., 2006; Takai et al., 2011; Houghtaling et al., 2004). In vivo data has suggested that POT1 accumulation at telomeres relies on its interaction with TPP1, as its OB folds are insufficient to tether it to DNA (Kibe et al., 2010). RAP1 is the sixth and most evolutionary conserved member of shelterin (Li and de Lange, 2003). Furthermore, it is the only subunit known to have non-telomeric functions: it also operates as a transcriptional regulator (Martinez et al., 2010) and impinges on nuclear factor kappa B (NF-κB) signaling (Teo et al., 2010).
Biochemical studies have uncovered an increasing number of telomere-associated proteins that are primarily recruited by shelterin and act as accessory factors for the complex (Palm and de Lange, 2008). They include DNA damage factors, nucleases, helicases and DNA replication proteins. Recently, a trimeric complex termed CST, which contains DNA polymerase α, the primase accessory factors CTC1 (for CTS telomere maintenance complex component 1) and STN1 (for suppressor of cdc thirteen 1) in addition to TEN1 (for telomeric pathways with STN1), was found to associate with ∼20% of telomeres (Miyake et al., 2009; Surovtseva et al., 2009). Little is known about the function of CST, but studies on human cells have suggested that STN1 interacts with TPP1 and limits overhang length (Wan et al., 2009).
A long non-coding telomeric RNA
An RNA component, TERRA (for telomeric-repeat-containing RNA), has been identified as the third entity of the telomere nucleoprotein complex (Azzalin et al., 2007). TERRA transcription is mediated by RNA polymerase II and is initiated from the sub-telomeric regions that are found near chromosome ends (Porro et al., 2010). TERRA levels are regulated during the cell cycle, and its localization at telomeres is modulated by the nonsense-mediated decay machinery (Azzalin et al., 2007). It has been postulated that this non-coding RNA is important for telomere maintenance and function (Redon et al., 2010; Flynn et al., 2011), yet the molecular mechanisms underlying this proposed function remain to be uncovered.
Shelterin – the elegant, but not so simple solution to the end-protection problem
The ability of cells to distinguish native chromosome ends from broken DNA – which is unstable and fusogenic – was first documented in the 1940s by McClintock and Muller (Muller, 1938; McClintock, 1941). Linear ends of plasmids are unstable when introduced into eukaryotic cells and often recombine with the genome (Orr-Weaver et al., 1981). Furthermore, when DSBs accumulate in cells, a signaling cascade that leads to cell cycle arrest is initiated (Weinert and Hartwell, 1988). By contrast, natural chromosome ends are inherently stable: they are not targeted by DNA repair pathways, namely non-homologous end-joining (NHEJ) and homology-directed repair (HDR), and do not activate the major DNA damage-induced kinases ataxia telangiectasia mutated (ATM) and ATM and Rad3 related (ATR). It was later recognized that telomeres accomplish chromosome end-protection by means of their protein elements (de Lange, 2005). In recent years, major strides have been made in understanding how shelterin is employed to disguise chromosome ends from DNA damage-sensing and repair pathways. This has been predominantly based on dissecting the phenotypes that are associated with the functional impairment of individual shelterin subunits using conditional knockout mice.
TRF2, the master repressor of ATM signaling and NHEJ
Deleting TRF2 results in the activation of the kinase ATM (Celli and de Lange, 2005) and triggers the accumulation of DNA damage factors, including H2AX (H2A histone family, member X), and 53BP1 (p53 binding protein 1) at telomeres (Dimitrova and de Lange, 2006; Denchi and de Lange, 2007; Dimitrova and de Lange, 2009). ATM activates p53, which induces cell cycle arrest. Furthermore, in the absence of TRF2, ligase 4- and Ku-mediated NHEJ repair is activated at telomeres, which results in chromosome end-to-end fusions (Celli and de Lange, 2005). The mechanism by which TRF2 normally represses ATM and NHEJ is currently under investigation. One model has been proposed, whereby TRF2 is able to sequester the telomere terminus in the t-loop structure. Purified TRF2 binds at the junction of the t-loop (Stansel et al., 2001) and can promote the formation and stabilization of structures resembling t-loops in vitro (Poulet et al., 2009). A t-loop configuration would prevent detection of the telomere end by the MRN (for MRE11, RAD50 and NBS1) complex, which is the DNA damage sensor of the ATM pathway. Similarly, the t-loop structure would exclude the binding of the Ku70–Ku80 heterodimer, thereby blocking NHEJ.
Silencing the kinase ATR by the POT1 proteins
The task of suppressing the second major checkpoint activator, the kinase ATR, is ascribed to POT1 in humans and to POT1a in mice, whereas the main role of mouse POT1b is to regulate the length of the single-stranded overhang (Hockemeyer et al., 2005; Hockemeyer et al., 2006; Wu et al., 2006). The mechanism by which POT1 and POT1a block ATR activation pertains to their ability to prevent replication protein A (RPA), the sensor of the ATR pathway, from binding to single-stranded telomeric DNA. Indeed, RPA foci have been detected at telomeric DNA that has been depleted of POT1 (Gong and de Lange, 2010). Deleting TPP1 or TIN2, which leads to loss of POT1 from telomeres, prompts similar ATR activation (Kibe et al., 2010; Takai et al., 2011).
Cooperative inhibition of HDR by RAP1, POT1 and the Ku70–Ku80 complex
HDR is typically repressed at telomeres, but occurs in a small subset of tumors that do not use telomerase for telomere maintenance (Bryan et al., 1997) as well as during the early cleavage cycles during embryonic development (Liu et al., 2007). Recent work has indicated that recombination is repressed at telomeres in a highly redundant manner. First, it is repressed by the Ku70–Ku80 complex, which inhibits general HDR in the nucleus. Additionally, is it repressed in a telomere-specific manner by RAP1 and POT1 (Palm et al., 2009; Sfeir et al., 2010). The mechanism by which Rap1, POT1 and Ku cooperate to block recombination is not understood.
A distinct protective function for TRF1
The G-rich and highly repetitive nature of telomeric DNA poses major challenges to the DNA replication machinery. The shelterin subunit TRF1 assists the semi-conservative replication machinery during the duplication of bulk telomeric DNA, thereby protecting telomeres from breakage and from acquiring features that are reminiscent of common fragile sites (Martínez et al., 2009; Sfeir et al., 2009). The function of TRF1 is mainly achieved by recruiting helicases, including regulator of telomere length 1 (RTEL1) and Bloom syndrome, RecQ helicase-like (BLM), which unwind spurious secondary structures that can hinder fork progression.
How do shelterin-free telomeres behave?
The studies described above exemplify how the deletion of individual shelterin subunits activates specific DNA damage response pathway(s) without greatly destabilizing the remaining components of the complex. To uncover possible redundancies in the function of shelterin components, we have investigated the consequences of completely depleting telomeres of all shelterin components (Sfeir and de Lange, 2012), and we have identified two additional pathways that can threaten telomere integrity. The first is the alternative NHEJ DNA repair pathway that is mediated by ligase 3 and poly(ADP-ribose) polymerase 1 (PARP1). The second is nucleolytic degradation, which is a marked outcome of telomere deprotection in yeast and has not been previously observed in mammalian cells. Interestingly, both pathways are repressed in a highly redundant manner by shelterin components, as well as by DNA damage factors, whereby alternative NHEJ is inhibited by Ku70–Ku80 and nucleolytic degradation is blocked by 53BP1.
After nearly a decade of investigation, we now have a clear definition of the end-protection problem and are able to better understand the nature of its solution. In summary, with their six-member shelterin complex, mammalian telomeres are properly armed to face the six different challenges (namely ATM, ATR, NHEJ, HDR, alternative NHEJ and resection) that are imposed by the linearity of chromosome ends.
Telomerase – the enzymatic solution to the end-replication problem
In almost all eukaryotes, the task of solving the end-replication problem and counteracting telomere erosion is assigned to the telomerase enzyme complex (Greider and Blackburn, 1985). The active telomerase holoenzyme in mammalian cells exists as a dimer and consists of the telomerase reverse transcriptase (TERT), the telomerase RNA component (TERC) and dyskerin (Cohen et al., 2007). In humans, telomerase is expressed during the early stages of embryogenesis, and its expression is subsequently repressed in most somatic cells, except the male germ line, activated lymphocytes and stem cells found in certain regenerative tissues (Wright et al., 1996). Furthermore, the vast majority of human cancer cells reactivate telomerase, and are thus capable of proliferating indefinitely (Kim et al., 1994). Regulation of telomerase is primarily exerted at the level of TERT transcription. Extensive analysis of the promoter region has uncovered many transcriptional binding sites and regulatory elements, including GC-boxes and E-boxes. In addition, TERT transcription is regulated by oncogenes and tumor suppressor genes, and is influenced by the surrounding chromatin. Post-transcriptional and translational processes are likely to control telomerase activity. Alternative splice variants of human TERT have been reported in some cancer cells. Telomerase is also subject to post-translational modifications including sumoylation, phosphorylation and ubiquitylation, all of which can impact on its activity (Cong et al., 2002).
In vivo, the maturation to a fully active telomerase depends on its assembly into a ribonucleoprotein (RNP) complex and is influenced by several factors, including pontin, reptin and others proteins that bind to TERT and TERC (Venteicher et al., 2008 and see poster). Another important step for telomerase maturation is its trafficking through Cajal bodies, the dynamic subnuclear sites that are involved in RNP biogenesis (Zhu et al., 2004; Tomlinson et al., 2006; Cristofari et al., 2007). Telomerase transit through Cajal bodies is mediated by telomerase Cajal body protein 1 (TCAB1) (Venteicher et al., 2008; Venteicher et al., 2009; Koo et al., 2011).
A key step with regards to telomerase action is its recruitment to chromosome ends in a timely manner. The most coherent picture of telomerase recruitment has emerged from studies in Saccharomyces cerevisiae that have shown that this process is mediated by an interaction between EST1 (ever-shorter telomeres 1), a component of the telomerase complex, and Cdc13, a protein that binds to the single-stranded overhang (Evans and Lundblad, 1999; Qi and Zakian, 2000). Another recruitment pathway involves the Ku complex, which binds to the telomeric DNA, as well as telomerase RNA (Peterson et al., 2001; Stellwagen et al., 2003). Our current understanding of telomerase recruitment in mammalian cells is much more limited. The Ku70–Ku80 complex has been shown to associate with telomerase by interacting with both human TERT and TERC (Chai et al., 2002; Ting et al., 2005). However, Ku70-deficient mice do not show a defect in telomere maintenance (Celli et al., 2006), which possibly argues against a recruitment role for Ku in mammals. In vitro studies have exposed an interaction between TPP1 and TERT (Wang et al., 2007; Xin et al., 2007) that enhances the processivity of telomerase. More recent experiments have suggested that TPP1 might also be involved in recruiting telomerase to the telomere end (Abreu et al., 2010). This has been difficult to prove, because the TERT interaction domain within TPP1 is required to repress the DNA damage response. Therefore, it is difficult to establish a specific function for TPP1 in the context of telomerase recruitment without the confounding activation of the DNA damage response. Another factor that has been suggested to bridge telomerase to the DNA is the heterogenous nuclear RNP protein A1 (hnRNPA1) (LaBranche et al., 1998). Again, this observation has been difficult to confirm, because hnRNPA1 is involved in the biogenesis of a wide spectrum of RNA transcripts. Given the natural low abundance of the telomerase enzyme, more sophisticated methods of telomerase detection are needed to resolve such confounding results and fully characterize the telomerase pathway.
Telomere length regulation
Telomerase research has moved into extra dimensions in recent years, as the enzyme has been found to modulate the Wnt signaling pathways (Park et al., 2009), display an RNA-dependent RNA polymerase activity (Maida et al., 2009) and has been spotted in the mitochondria (Sharma et al., 2012). Whereas the function and impact of telomerase in these pathways is a subject of debate, its indisputable chief role in mammalian cells remains to combat telomere erosion and maintain telomere length. In fact, appropriate telomere length is crucial for cell survival. This leads to the following question: what determines telomere length and how is this process regulated?
The first genes to influence telomere length regulation were identified in S. cerevisiae and, so far, the most comprehensive understanding of telomere length regulation comes from studies in fungi (Shore and Bianchi, 2009). The mechanism underlying telomere length regulation in mammalian cells, whose telomeres are orders of magnitude longer than those of budding yeast, is not fully understood. Nevertheless, some factors are known to influence telomere length; in particular, certain shelterin subunits are known to establish a negative-feedback loop that controls the addition of repeats by telomerase. In effect, overexpressing TRF1 in cancer-derived human cell lines leads to telomere shortening, whereas depleting telomeric DNA of TRF1 results in telomere elongation (van Steensel and de Lange, 1997). Reducing the levels of TIN2 or TRF2 (Ye and de Lange, 2004; Takai et al., 2010) at telomeres leads to a similar extension phenotype. The message from the duplex region of the telomere is relayed to the terminus via POT1, which has been shown in vivo, to act as a negative regulator of telomerase activity (Loayza and de Lange, 2003; Lei et al., 2005). However, as mentioned previously, mechanistic details regarding the regulation of telomerase accessibility and activity at the telomere terminus are still lacking.
Another layer of complexity is added by telomeric chromatin. In contrast with yeast telomeres, which lack histones (Wright et al., 1992), mammalian telomeres are nucleosomal (Makarov et al., 1993; Lejnine et al., 1995) and their chromatin is enriched with repressive histone marks, including H3K9me3 and H4K20me3 (me3 represents trimethylation), and the heterochromatin-specific factors chromobox homolog (CBX) 1, 3 and 5 (García-Cao et al., 2004; Gonzalo et al., 2006). Depletion of these heterochromatic marks correlates with remarkable elongation of telomeres (Marión and Blasco, 2010; Marión et al., 2011; Varela et al., 2011).
Telomere dysfunction and tumorigenesis
Because most somatic cells lack telomerase activity, they are prone to telomere shrinkage. When telomeres become too short and lose the binding of the protective shelterin complex, the cells face two opposing fates. In cells with intact checkpoints, telomere erosion leads to senescence. By contrast, in the absence of p53, telomere dysfunction promotes genomic instability and fuels tumor progression.
Early work on telomerase-knockout mice has provided in vivo evidence in support of telomere dysfunction as an early driver in the evolution of cancer genomes. The short and dysfunctional telomeres in the late-generation telomerase-deficient mice, which usually result in tissue degeneration, are tolerated when p53 is abrogated. The outcome is an increase in the occurrence of epithelial tumors that display hallmarks of genomic instability (Artandi et al., 2000). The progression of these tumors is constrained by the lack of telomerase, which would restore telomere function and allow further growth and more aggressive behavior of tumors. The paradigm has been re-established by recent work that expressed an engineered inducible version of telomerase in mouse models of prostate cancer and T-cell lymphoma (Ding et al., 2012; Hu et al., 2012). Indeed, when telomerase is reactivated in mice that have witnessed a stage of telomere dysfunction, especially aggressive cancers, which possess an increasing propensity of undergoing metastasis, develop.
Telomere dysfunction also drives the progression of human cancers. Many solid tumors, including those of the breast (Chin et al., 2004), prostate (Meeker et al., 1996) and colon (Rudolph et al., 2001), have shorter telomeres when compared with those of the respective normal tissue. Anaphase bridges have been documented at the hyperplasia-to-carcinoma transition, which coincides with telomerase re-activation (Chin et al., 2004). The most compelling evidence supporting a role for telomere dysfunction in tumor progression has emerged from the examination of telomere dynamics in hematological malignancies, particularly in chronic lymphocytic leukemia (CLL). This type of tumor offers a unique opportunity to perform high-resolution telomere length measurements in B cells from patients at different stages of the disease. The results demonstrate that telomere shortening strongly correlates with disease progression, and telomere fusions were noted in the earlier stages of the disease, perhaps setting the stage for the gross genomic rearrangements that drive progression of CLL (Lin et al., 2010).
What lies ahead?
After three decades of research, it is now well established that telomeres, which account for less than 1% of the human genome, have a pivotal role in normal cellular function and that cellular dysfunction ensues if their integrity is compromised. However, despite substantial progress in our understanding of how telomeres protect the integrity of the genetic material, there are many unanswered questions. We have yet to unravel the intricacies by which all the DNA damage pathways are evaded and experimentally validate the function for the t-loop. The interplay between telomere-associated proteins and telomerase, which sets the appropriate telomere length in different cellular stages and cell types, is an area that also requires rigorous investigation. Answering these and other questions will bring us closer to fully understanding the impact of telomere biology on cellular function and might lead to profound improvements in cancer therapy as well as in regenerative medicine.
Acknowledgments
I gratefully acknowledge Titia de Lange for her support during my post-doctoral training. I am thankful to Peng Wu, Eros Lazzerini-Denchi, Nadya Dimitrova and members of the de Lange laboratory for commenting on the manuscript. I apologize to the authors whose publications have not been cited owing to space limitations.
Footnotes
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
A high-resolution version of the poster is available for downloading in the online version of this article at jcs.biologists.org. Individual poster panels are available as JPEG files at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.106831/-/DC1
References
- Abreu E., Aritonovska E., Reichenbach P., Cristofari G., Culp B., Terns R. M., Lingner J., Terns M. P. (2010). TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 30, 2971–2982. 10.1128/MCB.00240-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi S. E., Chang S., Lee S. L., Alson S., Gottlieb G. J., Chin L., DePinho R. A. (2000). Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406, 641–645. 10.1038/35020592 [DOI] [PubMed] [Google Scholar]
- Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801. 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Baumann P., Cech T. R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175. 10.1126/science.1060036 [DOI] [PubMed] [Google Scholar]
- Bianchi A., Smith S., Chong L., Elias P., de Lange T. (1997). TRF1 is a dimer and bends telomeric DNA. EMBO J. 16, 1785–1794. 10.1093/emboj/16.7.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T., Brun C., Ancelin K., Koering C. E., Laroche T., Gilson E. (1997). Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17, 236–239. 10.1038/ng1097-236 [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska A., Chong L., de Lange T. (1997). Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235. 10.1038/ng1097-231 [DOI] [PubMed] [Google Scholar]
- Bryan T. M., Englezou A., Dalla–Pozza L., Dunham M. A., Reddel R. R. (1997). Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3, 1271–1274. 10.1038/nm1197-1271 [DOI] [PubMed] [Google Scholar]
- Celli G. B., de Lange T. (2005). DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718. 10.1038/ncb1275 [DOI] [PubMed] [Google Scholar]
- Celli G. B., Denchi E. L., de Lange T. (2006). Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8, 855–890. 10.1038/ncb1444 [DOI] [PubMed] [Google Scholar]
- Chai W., Ford L. P., Lenertz L., Wright W. E., Shay J. W. (2002). Human Ku70/80 associates physically with telomerase through interaction with hTERT. J Biol. Chem. 277 47242–47247. [DOI] [PubMed] [Google Scholar]
- Chai W., Shay J. W., Wright W. E. (2005). Human telomeres maintain their overhang length at senescence. Mol. Cell. Biol. 25, 2158–2168. 10.1128/MCB.25.6.2158-2168.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K., de Solorzano C. O., Knowles D., Jones A., Chou W., Rodriguez E. G., Kuo W. L., Ljung B. M., Chew K., Myambo K., et al. (2004). In situ analyses of genome instability in breast cancer. Nat. Genet. 36, 984–988. 10.1038/ng1409 [DOI] [PubMed] [Google Scholar]
- Cohen S. B., Graham M. E., Lovrecz G. O., Bache N., Robinson P. J., Reddel R. R. (2007). Protein composition of catalytically active human telomerase from immortal cells. Science 315, 1850–1853. [DOI] [PubMed] [Google Scholar]
- Cong Y. S., Wright W. E., Shay J. W. (2002). Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 66, 407–425. 10.1128/MMBR.66.3.407-425.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G., Adolf E., Reichenbach P., Sikora K., Terns R. M., Terns M. P., Lingner J. (2007). Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell 27, 882–889. 10.1016/j.molcel.2007.07.020 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110. 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2009). How telomeres solve the end-protection problem. Science 326, 948–952. 10.1126/science.1170633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi E. L., de Lange T. (2007). Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071. 10.1038/nature06065 [DOI] [PubMed] [Google Scholar]
- Dimitrova N., de Lange T. (2006). MDC1 accelerates nonhomologous end-joining of dysfunctional telomeres. Genes Dev. 20, 3238–3243. 10.1101/gad.1496606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N., de Lange T. (2009). Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol. Cell. Biol. 29, 5552–5563. 10.1128/MCB.00476-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Wu C. J., Jaskelioff M., Ivanova E., Kost–Alimova M., Protopopov A., Chu G. C., Wang G., Lu X., Labrot E. S., et al. (2012). Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 148, 896–907. 10.1016/j.cell.2012.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S. K., Lundblad V. (1999). Est1 and Cdc13 as comediators of telomerase access. Science 286, 117–120. 10.1126/science.286.5437.117 [DOI] [PubMed] [Google Scholar]
- Flynn R. L., Centore R. C., O'Sullivan R. J., Rai R., Tse A., Songyang Z., Chang S., Karlseder J., Zou L. (2011). TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471, 532–536. 10.1038/nature09772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García–Cao M., O'Sullivan R., Peters A. H., Jenuwein T., Blasco M. A. (2004). Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 36, 94–99. 10.1038/ng1278 [DOI] [PubMed] [Google Scholar]
- Gong Y., de Lange T. (2010). A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol. Cell 40, 377–387. 10.1016/j.molcel.2010.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S., Jaco I., Fraga M. F., Chen T., Li E., Esteller M., Blasco M. A. (2006). DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 8, 416–424. 10.1038/ncb1386 [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H. (1985). Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413. 10.1016/0092-8674(85)90170-9 [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97, 503–514. 10.1016/S0092-8674(00)80760-6 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Sfeir A. J., Shay J. W., Wright W. E., de Lange T. (2005). POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 24, 2667–2678. 10.1038/sj.emboj.7600733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Daniels J. P., Takai H., de Lange T. (2006). Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 126, 63–77. 10.1016/j.cell.2006.04.044 [DOI] [PubMed] [Google Scholar]
- Houghtaling B. R., Cuttonaro L., Chang W., Smith S. (2004). A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14, 1621–1631. 10.1016/j.cub.2004.08.052 [DOI] [PubMed] [Google Scholar]
- Hu J., Hwang S. S., Liesa M., Gan B., Sahin E., Jaskelioff M., Ding Z., Ying H., Boutin A. T., Zhang H., et al. (2012). Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 148, 651–663. 10.1016/j.cell.2011.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibe T., Osawa G. A., Keegan C. E., de Lange T. (2010). Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol. Cell. Biol. 30, 1059–1066. 10.1128/MCB.01498-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015. 10.1126/science.7605428 [DOI] [PubMed] [Google Scholar]
- Koo B. K., Park C. J., Fernandez C. F., Chim N., Ding Y., Chanfreau G., Feigon J. (2011). Structure of H/ACA RNP protein Nhp2p reveals cis/trans isomerization of a conserved proline at the RNA and Nop10 binding interface. J. Mol. Biol. 411, 927–942. 10.1016/j.jmb.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche H., Dupuis S., Ben–David Y., Bani M. R., Wellinger R. J., Chabot B. (1998). Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19, 199–202. 10.1038/575 [DOI] [PubMed] [Google Scholar]
- Lam Y. C., Akhter S., Gu P., Ye J., Poulet A., Giraud–Panis M. J., Bailey S. M., Gilson E., Legerski R. J., Chang S. (2010). SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J. 29, 2230–2241. 10.1038/emboj.2010.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Baumann P., Cech T. R. (2002). Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry 41, 14560–14568. 10.1021/bi026674z [DOI] [PubMed] [Google Scholar]
- Lei M., Zaug A. J., Podell E. R., Cech T. R. (2005). Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem. 280, 20449–20456. 10.1074/jbc.M502212200 [DOI] [PubMed] [Google Scholar]
- Lejnine S., Makarov V. L., Langmore J. P. (1995). Conserved nucleoprotein structure at the ends of vertebrate and invertebrate chromosomes. Proc. Natl. Acad. Sci. USA 92, 2393–2397. 10.1073/pnas.92.6.2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., de Lange T. (2003). Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell 14, 5060–5068. 10.1091/mbc.E03-06-0403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. T., Letsolo B. T., Jones R. E., Rowson J., Pratt G., Hewamana S., Fegan C., Pepper C., Baird D. M. (2010). Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood 116, 1899–1907. 10.1182/blood-2010-02-272104 [DOI] [PubMed] [Google Scholar]
- Liu L., Bailey S. M., Okuka M., Muñoz P., Li C., Zhou L., Wu C., Czerwiec E., Sandler L., Seyfang A., et al. (2007). Telomere lengthening early in development. Nat. Cell Biol. 9, 1436–1441. 10.1038/ncb1664 [DOI] [PubMed] [Google Scholar]
- Loayza D., de Lange T. (2003). POT1 as a terminal transducer of TRF1 telomere length control. Nature 423, 1013–1018. 10.1038/nature01688 [DOI] [PubMed] [Google Scholar]
- Maida Y., Yasukawa M., Furuuchi M., Lassmann T., Possemato R., Okamoto N., Kasim V., Hayashizaki Y., Hahn W. C., Masutomi K. (2009). An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 461, 230–235. 10.1038/nature08283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov V. L., Lejnine S., Bedoyan J., Langmore J. P. (1993). Nucleosomal organization of telomere-specific chromatin in rat. Cell 73, 775–787. 10.1016/0092-8674(93)90256-P [DOI] [PubMed] [Google Scholar]
- Makarov V. L., Hirose Y., Langmore J. P. (1997). Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88, 657–666. 10.1016/S0092-8674(00)81908-X [DOI] [PubMed] [Google Scholar]
- Marión R. M., Blasco M. A. (2010). Telomere rejuvenation during nuclear reprogramming. Curr. Opin. Genet. Dev. 20, 190–196. 10.1016/j.gde.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Marión R. M., Schotta G., Ortega S., Blasco M. A. (2011). Suv4-20h abrogation enhances telomere elongation during reprogramming and confers a higher tumorigenic potential to iPS cells. PLoS ONE 6, e25680 10.1371/journal.pone.0025680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P., Thanasoula M., Muñoz P., Liao C., Tejera A., McNees C., Flores J. M., Fernández–Capetillo O., Tarsounas M., Blasco M. A. (2009). Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 23, 2060–2075. 10.1101/gad.543509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P., Thanasoula M., Carlos A. R., Gómez–López G., Tejera A. M., Schoeftner S., Dominguez O., Pisano D. G., Tarsounas M., Blasco M. A. (2010). Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat. Cell Biol. 12, 768–780. 10.1038/ncb2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. (1941). The stability of broken ends of chromosomes in Zea mays. Genetics 26, 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElligott R., Wellinger R. J. (1997). The terminal DNA structure of mammalian chromosomes. EMBO J. 16, 3705–3714. 10.1093/emboj/16.12.3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker A. K., Sommerfeld H. J., Coffey D. S. (1996). Telomerase is activated in the prostate and seminal vesicles of the castrated rat. Endocrinology 137, 5743–5746. 10.1210/en.137.12.5743 [DOI] [PubMed] [Google Scholar]
- Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., Ishikawa F. (2009). RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206. 10.1016/j.molcel.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Muller H. J. (1938). The remaking of chromosomes. The Collecting Net. Woods Hole 8, 182–195. [Google Scholar]
- Nikitina T., Woodcock C. L. (2004). Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 166, 161–165. 10.1083/jcb.200403118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M. S., Safari A., Xin H., Liu D., Songyang Z. (2006). A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl. Acad. Sci. USA 103, 11874–11879. 10.1073/pnas.0605303103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L., Karlseder J. (2011). Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol. Cell 42, 224–236. 10.1016/j.molcel.2011.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr–Weaver T. L., Szostak J. W., Rothstein R. J. (1981). Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78, 6354–6358. 10.1073/pnas.78.10.6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W., de Lange T. (2008). How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334. 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- Palm W., Hockemeyer D., Kibe T., de Lange T. (2009). Functional dissection of human and mouse POT1 proteins. Mol. Cell. Biol. 29, 471–482. 10.1128/MCB.01352-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. I., Venteicher A. S., Hong J. Y., Choi J., Jun S., Shkreli M., Chang W., Meng Z., Cheung P., Ji H., et al. (2009). Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460, 66–72. 10.1038/nature08137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S. E., Stellwagen A. E., Diede S. J., Singer M. S., Haimberger Z. W., Johnson C. O., Tzoneva M., Gottschling D. E. (2001). The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27, 64–67. 10.1038/83778 [DOI] [PubMed] [Google Scholar]
- Porro A., Feuerhahn S., Reichenbach P., Lingner J. (2010). Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell. Biol. 30, 4808–4817. 10.1128/MCB.00460-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet A., Buisson R., Faivre–Moskalenko C., Koelblen M., Amiard S., Montel F., Cuesta–Lopez S., Bornet O., Guerlesquin F., Godet T., et al. (2009). TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 28, 641–651. 10.1038/emboj.2009.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Zakian V. A. (2000). The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14, 1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Raices M., Verdun R. E., Compton S. A., Haggblom C. I., Griffith J. D., Dillin A., Karlseder J. (2008). C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132, 745–757. 10.1016/j.cell.2007.12.039 [DOI] [PubMed] [Google Scholar]
- Redon S., Reichenbach P., Lingner J. (2010). The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 38, 5797–5806. 10.1093/nar/gkq296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph K. L., Millard M., Bosenberg M. W., DePinho R. A. (2001). Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 28, 155–159. 10.1038/88871 [DOI] [PubMed] [Google Scholar]
- Sfeir A., de Lange T. (2012). Removal of shelterin reveals the telomere end-protection problem. Science 336, 593–597. 10.1126/science.1218498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A., Kosiyatrakul S. T., Hockemeyer D., MacRae S. L., Karlseder J., Schildkraut C. L., de Lange T. (2009). Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103. 10.1016/j.cell.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A., Kabir S., van Overbeek M., Celli G. B., de Lange T. (2010). Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327, 1657–1661. 10.1126/science.1185100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N. K., Reyes A., Green P., Caron M. J., Bonini M. G., Gordon D. M., Holt I. J., Santos J. H. (2012). Human telomerase acts as a hTR-independent reverse transcriptase in mitochondria. Nucleic Acids Res. 40, 712–725. 10.1093/nar/gkr758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Bianchi A. (2009). Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J. 28, 2309–2322. 10.1038/emboj.2009.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel R. M., de Lange T., Griffith J. D. (2001). T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20, 5532–5540. 10.1093/emboj/20.19.5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A. E., Haimberger Z. W., Veatch J. R., Gottschling D. E. (2003). Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17, 2384–2395. 10.1101/gad.1125903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y. V., Churikov D., Boltz K. A., Song X., Lamb J. C., Warrington R., Leehy K., Heacock M., Price C. M., Shippen D. E. (2009). Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36, 207–218. 10.1016/j.molcel.2009.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K. K., Hooper S., Blackwood S., Gandhi R., de Lange T. (2010). In vivo stoichiometry of shelterin components. J. Biol. Chem. 285, 1457–1467. 10.1074/jbc.M109.038026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K. K., Kibe T., Donigian J. R., Frescas D., de Lange T. (2011). Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell 44, 647–659. 10.1016/j.molcel.2011.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo H., Ghosh S., Luesch H., Ghosh A., Wong E. T., Malik N., Orth A., de Jesus P., Perry A. S., Oliver J. D., et al. (2010). Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat. Cell Biol. 12, 758–767. 10.1038/ncb2080 [DOI] [PubMed] [Google Scholar]
- Ting N. S., Yu Y., Pohorelic B., Lees–Miller S. P., Beattie T. L. (2005). Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res. 33, 2090–2098. 10.1093/nar/gki342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson R. L., Ziegler T. D., Supakorndej T., Terns R. M., Terns M. P. (2006). Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 17, 955–965. 10.1091/mbc.E05-09-0903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., de Lange T. (1997). Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743. 10.1038/385740a0 [DOI] [PubMed] [Google Scholar]
- Varela E., Schneider R. P., Ortega S., Blasco M. A. (2011). Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc. Natl. Acad. Sci. USA 108, 15207–15212. 10.1073/pnas.1105414108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A. S., Meng Z., Mason P. J., Veenstra T. D., Artandi S. E. (2008). Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132, 945–957. 10.1016/j.cell.2008.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A. S., Abreu E. B., Meng Z., McCann K. E., Terns R. M., Veenstra T. D., Terns M. P., Artandi S. E. (2009). A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648. 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M., Qin J., Songyang Z., Liu D. (2009). OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 284, 26725–26731. 10.1074/jbc.M109.021105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007). The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510. 10.1038/nature05454 [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. (1988). The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241, 317–322. 10.1126/science.3291120 [DOI] [PubMed] [Google Scholar]
- Wright J. H., Gottschling D. E., Zakian V. A. (1992). Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 6, 197–210. 10.1101/gad.6.2.197 [DOI] [PubMed] [Google Scholar]
- Wright W. E., Piatyszek M. A., Rainey W. E., Byrd W., Shay J. W. (1996). Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18, 173–179. 10.1016/j.cell.2006.05.037 [DOI] [PubMed] [Google Scholar]
- Wu L., Multani A. S., He H., Cosme–Blanco W., Deng Y., Deng J. M., Bachilo O., Pathak S., Tahara H., Bailey S. M., et al. (2006). Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126, 49–62. 10.1016/j.cell.2006.05.037 [DOI] [PubMed] [Google Scholar]
- Wu P., van Overbeek M., Rooney S., de Lange T. (2010). Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol. Cell 39, 606–617. 10.1016/j.molcel.2010.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M. S., Songyang Z. (2007). TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445, 559–562. 10.1038/nature05469 [DOI] [PubMed] [Google Scholar]
- Ye J. Z., de Lange T. (2004). TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat. Genet. 36, 618–623. 10.1038/ng1360 [DOI] [PubMed] [Google Scholar]
- Ye J. Z., Donigian J. R., van Overbeek M., Loayza D., Luo Y., Krutchinsky A. N., Chait B. T., de Lange T. (2004a). TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279, 47264–47271. 10.1074/jbc.M409047200 [DOI] [PubMed] [Google Scholar]
- Ye J. Z., Hockemeyer D., Krutchinsky A. N., Loayza D., Hooper S. M., Chait B. T., de Lange T. (2004b). POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18, 1649–1654. 10.1101/gad.1215404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Tomlinson R. L., Lukowiak A. A., Terns R. M., Terns M. P. (2004). Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell 15, 81–90. 10.1091/mbc.E03-07-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]