Abstract
Bisphenol A (BPA) is the raw material of 71% of polycarbonate-based resins and 27% of epoxy-based resins which are used for coating metal-based food and beverage cans. Meanwhile, it is taken into account as a typical environmental pollutant. Hormesis may occur in algae exposed to BPA. In this study, the effects of BPA on Chlorella pyrenoidosa were assessed based on growth inhibition and transcriptome analysis. We have focused on two exposure scenarios as follows: (1) exposure to a low stimulation concentration (0.1 mg.L−1, 19.35% promotion in cell density on the 3rd day); (2) exposure to a high inhibition concentration (10 mg.L−1, 64.71% inhibition in cell density on the 3rd day). Transcriptome analysis showed enrichment in nucleotide transport, single-organism transport, cellular respiration. Among them, adenosine triphosphate (ATP) synthase and Nicotinamide adenine dinucleotide (NADH) dehydrogenase were upregulated under 0.1 mg.L−1 BPA treatment. These changes enhanced the physiological and energy metabolic pathways of C. pyrenoidosa, thereby stimulating cell proliferation. At exposure to the high BPA, severe inhibited changes in the expression levels of several pathways were observed, which were related to tricarboxylic acid (TCA) cycle, glycolysis, fatty acid metabolism, oxidative phosphorylation, and photosynthesis. Therefore, BPA could negatively affect growth inhibition through the multiple energy metabolism processes. These results may result in a deeper insight into BPA-induced biphasic responses in algae, and provide vital information to assess the potential ecological risks of exposure to BPA in an aquatic ecosystem.
Keywords: Bisphenol A (BPA), Chlorella pyrenoidosa, inhibitory effect, stimulation effect, transcriptome analyse
1. Introduction
Bisphenol A (BPA) is an environmental endocrine-disrupting chemical (EDC), associating with the estrogen effect, that is widely utilized in industrial applications [1]. In addition, BPA disrupts the homeostasis of internal environment and affects the growth and development of organisms by interfering with the normal synthesis of hormones in organisms [2]. BPA is considered to be associated with diabetes, obesity, cardiovascular disease, reproductive diseases, breast cancer. [3,4]. Global production of BPA has exceeded 6 million pounds (lbs) since 2003 [5]. Nowadays, pollution by BPA almost exists to varying degrees throughout the world. Moreover, its production rate account for about 43.5% of the gross world production, and the trend tends to increase [6]. In recent years, due to the intensification of industrial activities, a great number of BPA have been discharged into the water body and transmitted through the food chain in an aquatic ecosystem, seriously threatening the safety of drinking water and the balance of the aquatic ecosystem [7].
Algae are the most important primary producers in the aquatic ecosystem, playing a pivotal role in maintaining the ecological balance [8]. In addition, algae are also able to accumulate highly toxic substances (e.g., selenium, zinc, and arsenic) in their cells and/or bodies, thereby eliminating such substances from an aquatic environment [9]. Recently, the impact of BPA on algae has significantly attracted scholars’ attention. Exposure to high concentration of BPA may inhibit the growth of algal cells [10]. However, BPA also acts as a hormetic substance at low concentration. Hormesis refers to a biphasic dose-response relationship characterized by stimulation at low doses and inhibition at high doses [11]. To date, it has been reported that low concentration of toxic or organic substances could stimulate the growth of microalgae, and this phenomenon might contribute to the outbreak of algal blooms to some extent [12,13]. Hence, it is necessary to elucidate the molecular mechanism of hormesis. The majority of previous studies have mainly focused on how BPA might affect the physiological indicators (e.g., growth rate, chlorophyll synthesis) in algae, while no in-depth study on toxicological mechanism has been carried out yet [14,15]. Moreover, a limited number of studies have concentrated on the hormetic mechanism of BPA or other organic pollutants, and the inhibitory effects have been mainly studied. To date, with the development of RNA sequencing (RNA-Seq), the recognition of molecular level of eukaryotic organisms has been continuously improved [6]. Fan et al. [16] conducted a transcriptome-based analysis on gene expression of C. pyrenoidosa under different CO2 concentrations, and found that the CO2-concentrating mechanism was activated at low CO2 environment to compensate for the low activity of RuBisCO in the Calvin cycle. Qian et al. [17] investigated the inhibitory effects of linoleic acid (LA) on C. pyrenoidosa by transcriptome analyses, and reported that genes related to photosynthesis, carbon metabolism, and amino acid metabolism were inhibited, which could be key targets for LA in C. pyrenoidosa. Cheng et al. [18] performed RNA-Seq to justify why astaxanthin yield of the Haematococcus pluvialis mutant was 1.7 times higher than that of a wild strain, owning to the enhancement of pyruvate and fatty acid metabolism for supporting biosynthesis of astaxanthin. Therefore, the hormetic mechanism of BPA needs to be effectively studied.
Chlorella pyrenoidosa is one of the most representative green algae, associating with strong adaptability, rapid reproduction, and sensitivity to pollutants. To date, the hormetic mechanism of BPA on algae (e.g., C. pyrenoidosa) has been rarely studied. In this study, we aimed to apply RNA-Seq to disclose the hormetic mechanism of green algae.
2. Materials and Methods
2.1. Algal Strains and Growth Conditions
Here, C. pyrenoidosa was provided by theResearch Center of Hydrobiology, Jinan University, Guangzhou, China, which was isolated from a freshwater sample. The algae cells were grown in a flask, containing 100 mL of BG11 medium. Algal fluid was grown in an artificial climate box (CC275TL2H; Hangzhou Xutemp Temptech Co., Ltd., Hangzhou, China). The light intensity was 1200 lux, with the temperature of 25 ± 2 °C under 12:12 h light-dark cycle. The flask was shaken three times/day, and the conical flask was changed randomly to ensure that the microalgae are exposed to the uniform light in the conical flask.
2.2. Experimental Process
Here, BPA was dissolved into methanol, and it was essential to ensure that the concentration of methanol did not exceed 0.5%. According to a previous experiment [19], under this threshold, the toxicity of methanol would be negligible. We incubated 100 mL of algal culture in 150-mL flask for subsequent experiments for 5 days. The initial cultivated density of algae in exponential growth phase was 2.8 × 105 cells/L. In order to investigate the effects of BPA on C. pyrenoidosa, different concentrations of BPA (0, 0.1, 1, and 10 mg. L−1) were added into algal culture. The experiments were conducted in triplicate. The flasks were shaken three times/day, and changed randomly to ensure that the microalgae exposed to uniform light in the flask.
To verify changes at transcription level, samples treated for 72 h by BPA at concentrations of 0, 0.1, and 10 mg.L−1 were selected. Furthermore, 100 mL of C. pyrenoidosa was centrifuged at 8000 rpm for 5 min, and then supernatant and extracted cell pellets were removed.
2.3. Growth and Chlorophyll Fluorescence Analysis
The cell density was daily measured by a flow cytometer (BD Accuri C6, Becton Dickinson, Franklin Lakes, NJ, USA). A chlorophyll fluorometer (TD700; Turner Design, Inc., Chicago, IL, USA) determined the content of chlorophyll a (Chla). Maximum quantum efficiency of PSII photochemistry (Fv/Fm) is a common parameter. Under normal conditions, Fv/Fm is extremely stable; when algae or plants are stressed, Fv/Fm significantly decreases. Therefore, Fv/Fm is an important index to study the influences of various stresses on photosynthesis [20]. The Fv/Fm was detected by a portable plant efficiency analyzer (PEA; Hansatech Instruments Ltd., Norfolk, UK). The cultivation of the algae was carried out at dark for 20 min before measurement at room temperature (26 ± 1 °C).
2.4. RNA Extraction and cDNA Library Construction
Total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) on the basis of the manufacturer’s instructions. After that, oligo (dT) magnetic beads were highly enriched with mRNA. Second-strand cDNA was then synthesized for 2 h at 16 °C using the total 20 µL first-strand product plus 80 µL of second strand mix containing 63 µL water, 10 µL second-strand buffer, 4 µL dNTPs, 2 µL DNA Polymerase and 1 µL of RNase H. Then, the cDNA fragments were purified with a QIAquick Polymerase Chain Reaction (PCR) Purification Kit (Qiagen, Hilden, Germany). The ligation products were selected by agarose gel electrophoresis, PCR amplified, as well as sequencing using Illumina HiSeqTM 2000 (Illumina, Inc., San Diego, CA, USA).
2.5. Analysis of Differentially Expressed Genes
To identify differentially expressed genes across samples or groups, the edgeR package (http://www.r-project.org/) was used. Differentially expressed genes (DEGs) were further annotated to Gene Ontology (GO) database and Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
2.6. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) was carried out to confirm the expression of C. pyrenoidosa gene profiles. RNA samples extracted from three biological replicates exposed to 0.1, 1.0, and 10 mg.L−1 BPA were analyzed by RT-qPCR. In addition, RT-qPCR was conducted using a SYBR Green Master kit (Takara, Tokyo, Japan) in accordance with manufacturer’s protocol, and the results were normalized by 18S rRNA. The primers were designed according to the sequence of the transcriptome Denevo. Primers used in RT-qPCR are listed in Table 1.
Table 1.
Primer used in quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis.
| Gene Name | Primer Sequence (5′–3′) |
|---|---|
| 18S (Control) | F:TGGTGCCCTTCCGTCAAT |
| R:CGGCACCTTACGAGAAATCA | |
| Adenosine triphosphate (ATP) Synthase | F:GAAGTCGGCAATGGTGTCC |
| R:CTGGGAGATGAGCACTACGG | |
| Nicotinamide adenine dinucleotide (NADH) dehydrogenase | F:ATCAAGGAAAAGCAGGGGCA |
| R:TGTCCCAAAGCATGAAGGCA | |
| PsbA (photosystem II D1 protein) | F:TGAACGAGAGTTGTTGAAAGAAGC |
| R:TGCTTGGTGTTGCTGGTGTA |
2.7. Statistical Analysis
All experiments were repeated three times independently. Data were analyzed using one-way analysis of variance (ANOVA) with the help of SPSS 13.0 software (IBM, Armonk, NY, USA), and were presented as the mean ± standard deviation (SD). The significance level of differences was set at p < 0.05.
3. Results
3.1. Effects of Bisphenol A (BPA) on Growth and Photosynthetic Efficiency of Algae
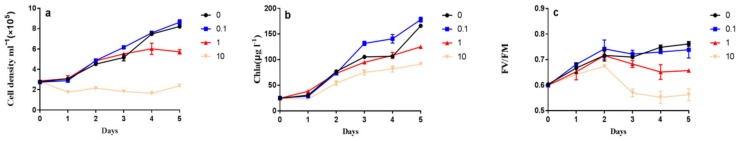
As shown in Figure 1a, on the first two days, no significant effects on growth of algae were observed after exposure to 0.1 and 1 mg.L−1 (p > 0.05). When the BPA concentration reached 10 mg.L−1, cell density was lower than that in other therapeutic groups. On the 3rd day, the cell density in the therapeutic groups of 0.1 and 10 mg.L−1 treatment groups was significantly increased (p < 0.001) by 19.35% and decreased (p < 0.001) by 64.71% compared with the control group, respectively. The results for Chla content (Figure 1b) showed a similar trend compared with the cell density. The Chla content of algae treated with 0.1 mg.L−1 BPA was markedly increased (p < 0.001) from the 3rd day. Obvious promotion occurred on the 3rd day of exposure to BPA, and the Chla content on the 3rd day was increased (p < 0.001) by 24.94% of the control group at concentration of 0.1 mg.L−1. In addition, with respect to the control group, at a high-dose of exposure to BPA (10 mg.L−1), the content of Chla was inhibited (p < 0.001). The change of Fv/Fm value was given in Figure 1c, and C. pyrenoidosa cells treated for 2 days showed a slightly promotion of low concentration and inhibition of high concentration.
Figure 1.
C. pyrenoidosa in response to bisphenol A (BPA). (a) cell density, (b) the content of Chla, (c) photosynthesis activity.
3.2. Global Transcriptional Changes of C. pyrenoidosa
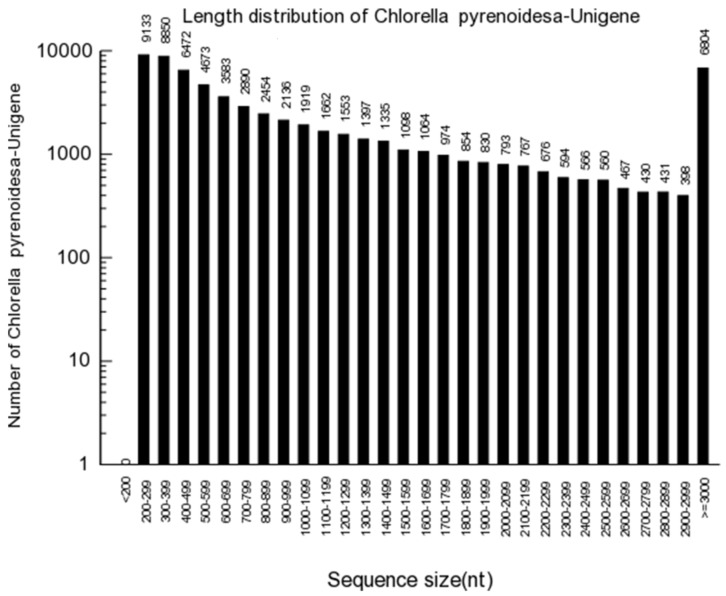
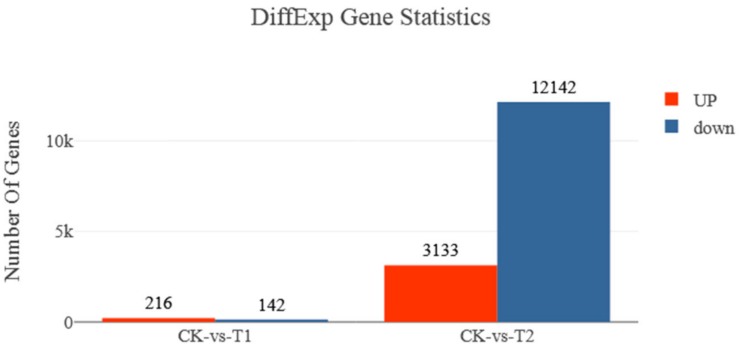
A total of 65,363 unigenes with 2298-bp of unigene N50 were obtained. The distribution of unigenes is illustrated in Figure 2. A false discovery rate (FDR) ≤0.05 and |log2| ≥1 were used as threshold to judge the significance of DEGs, as shown in Figure 3. After exposure to 0.1 mg.L−1 BPA, 216 genes were upregulated, whereas 142 genes were downregulated. At the same time, 10 mg.L−1 treatment group showed that 3133 genes were upregulated, while 12,142 genes were downregulated.
Figure 2.
The length of distribution of unigens. The x-axis shows the scope of the sequence size.
Figure 3.
The number of differentially expressed genes (DEGs) at two groups compared with that at CK group. CK represents Control group, T1 represents 0.1 mg.L−1 BPA treatment group, and T2 shows 10 mg.L−1 BPA treatment group.
3.3. Differentially Expressed Genes (DEGs) Exposed to Low Concentration of BPA
Results of GO enrichment analysis are presented in Table 2; no downregulated GO term was significantly enriched in low concentration of BPA (T1 represents 0.1 mg.L−1 BPA treatment group), while the GO term enrichment of gene was upregulated in T1, the terms “single-organism transport (GO:0044765)” and “single-organism localization (GO:1902578)” were sigificantly overrepsented, as well as the enrichment of terms “nucleoside transport (GO:0006810)” and “cellular respiration (GO:0045333)”. The KEGG database was also used to analyze pathways, in which DEGs were involved as well (Table 3). Besides, T1 affected ribosomes, oxidative phosphorylation, and sulfur metabolism. In oxidative phosphorylation, the DEGs were all upregulated, including ATPF1B, COX1, COX3, and ndhB (Figure 4).
Table 2.
Results of Gene Ontology (GO) enrichment analysis.
| GO ID | Description | p-Value |
|---|---|---|
| T1-UP | ||
| GO:0044765 | single-organism transport | 0.0015563 |
| GO:1902578 | single-organism localization | 0.0017646 |
| GO:0006810 | transport | 0.0020785 |
| GO:0005840 | ribosome | 0.0022349 |
| GO:0016843 | amine-lyase activity | 0.0049691 |
| GO:0015858 | nucleoside transport | 0.0052014 |
| GO:0005198 | structural molecule activity | 0.0071512 |
| GO:0045333 | cellular respiration | 0.007919 |
| GO:0051234 | establishment of localization | 0.0081377 |
| T2-UP | ||
| GO:0042995 | cell projection | 3.81 × 10−6 |
| GO:0033013 | tetrapyrrole metabolic process | 1.18 × 10−5 |
| GO:0051186 | cofactor metabolic process | 2.35 × 10−5 |
| GO:0098796 | membrane protein complex | 2.56 × 10−5 |
| GO:0006778 | porphyrin-containing compound metabolic process | 3.79 × 10−5 |
| GO:0044422 | organelle part | 9.72 × 10−5 |
| T2-DOWN | ||
| GO:0009081 | branched-chain amino acid metabolic process | 0.0059409 |
| GO:0016301 | kinase activity | 9.19 × 10−6 |
| GO:0036094 | small molecule binding | 0.0001015 |
| GO:0016773 | phosphotransferase activity, alcohol group as acceptor | 0.0001521 |
| GO:0016772 | transferase activity, transferring phosphorus-containing groups | 0.0003224 |
| GO:0001883 | purine nucleoside binding | 0.000369 |
| GO:0032549 | ribonucleoside binding | 0.000369 |
| GO:0032550 | purine ribonucleoside binding | 0.000369 |
| GO:0019899 | enzyme binding | 0.0004276 |
| GO:0004672 | protein kinase activity | 0.0004357 |
| GO:0017016 | Ras GTPase binding | 0.0004891 |
| GO:0031267 | small GTPase binding | 0.0004891 |
| GO:0051020 | GTPase binding | 0.0004891 |
| GO:0001882 | nucleoside binding | 0.0005225 |
| GO:0097367 | carbohydrate derivative binding | 0.0006553 |
| GO:0031981 | nuclear lumen | 0.0092762 |
| GO:0043233 | organelle lumen | 0.0092762 |
| GO:0070013 | intracellular organelle lumen | 0.0092762 |
Table 3.
The upregulated and downregulated genes within the Kyoto Encyclopedia of Genes and Genomes (KEGG).
| Pathway | Upregulated | Downregulated | p-Value |
|---|---|---|---|
| T1vsCK | |||
| Ribosome | 11 | 2 | 0.000351118 |
| Oxidative phosphorylation | 8 | / | 0.001435787 |
| Sulfur metabolism | 4 | / | 0.002234359 |
| Propanoate metabolism | 3 | 1 | 0.008392114 |
| Carbon metabolism | 8 | 1 | 0.01697323 |
| Sulfur relay system | 1 | 1 | 0.03425709 |
| Cysteine and methionine metabolism | 4 | / | 0.03562303 |
| Microbial metabolism in diverse environments | 9 | 1 | 0.03764153 |
| Biosynthesis of antibiotics | 10 | 1 | 0.0470081 |
| T2vsCK | |||
| Amino sugar and nucleotide sugar metabolism | 3 | 59 | 9.38 × 10−5 |
| Photosynthesis-antenna proteins | 21 | 22 | 0.000106969 |
| Fatty acid degradation | 13 | 30 | 0.000759285 |
| Galactose metabolism | 10 | 26 | 0.003986388 |
| Microbial metabolism in diverse environments | 58 | 213 | 0.004257043 |
| Biosynthesis of unsaturated fatty acids | 9 | 26 | 0.007765076 |
| Valine, leucine and isoleucine degradation | 12 | 44 | 0.008513731 |
| Ether lipid metabolism | 5 | 15 | 0.008521474 |
| Fructose and mannose metabolism | 13 | 29 | 0.00854504 |
| Biosynthesis of antibiotics | 55 | 259 | 0.009106183 |
| Fatty acid metabolism | 21 | 47 | 0.01303567 |
| Valine, leucine and isoleucine biosynthesis | 1 | 31 | 0.01553805 |
| DNA replication | 5 | 52 | 0.0200779 |
| Metabolic pathways | 204 | 919 | 0.02039731 |
| alpha-Linolenic acid metabolism | 6 | 19 | 0.02238955 |
| Sphingolipid metabolism | 7 | 25 | 0.0240593 |
| Biosynthesis of secondary metabolites | 117 | 416 | 0.03031573 |
| Peroxisome | 18 | 62 | 0.0312795 |
| Carbon metabolism | 46 | 151 | 0.04453267 |
| Nitrogen metabolism | 6 | 19 | 0.04458744 |
| Pyruvate metabolism | 18 | 57 | 0.04607645 |
Figure 4.
Differentially expressed genes related to oxidative decarboxylation in C. pyrenoidosa under CK (control group) and T1 BPA (red color: upregulated genes).
3.4. DEGs Exposed to High Concentration of BPA
At higher BPA concentration, the cell projection (GO:0042995) and tetrapyrrole metabolic process (GO:0033013) were significantly upregulated, while branched-chain amino acid metabolic process (GO:0009081) and kinase activity (GO:0016301) were downregulated. Meanwhile, amino sugar and nucleotide sugar metabolism, photosynthesis-antenna proteins, and fatty acids were enriched in T2 (T2 represents 10 mg.L−1 BPA treatment group). In T1, upregulated genes were more frequent. By contrast, downregulated genes were predominant in T2. In other metabolic pathways, the expression of genes was significantly downregulated (Table 3).
3.5. RT-qPCR for Analysis of Expression of Related Genes
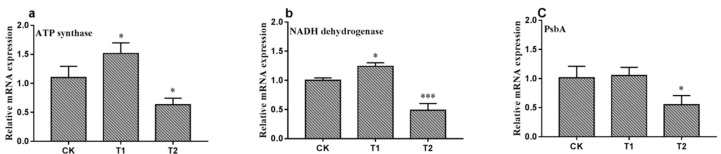
The relative mRNA expression level of several genes after exposure to 0.1 and 10 mg.L−1 BPA were detected by RT-qPCR (Figure 5). The expression levels of ATP synthase and NADH dehydrogenase were significantly upregulated at exposure to 0.1 mg.L−1 BPA. In contrast, at 10 mg.L−1 BPA group, the expression levels of three genes were significantly decreased. This result indicates that synthesis of ATP is stimulated at low concentration level, and BPA at high concentration level is harmful to mitochondria and photosynthesis.
Figure 5.
The mRNA expression of ATP synthase (a), NADH dehydrogenase (b), and PsbA (c) in C. pyrenoidosa exposed to 0.1 mg.L−1 BPA (T1) and 10 mg.L−1 BPA (T2). C. pyrenoidosa without exposure to BPA were used as control group. The data were expressed as mean ± standard deviation (SD) compared with control group * p < 0.05 vs. control group, *** p < 0.001 vs. control group
4. Discussion
4.1. Effects of BPA on Growth Inhibition
In the present study, in the highest concentration, the cell density, Chla content, and Fv/Fm were all decreased, and the growth was inhibited, which was consistent with Zhang et al.’s results [21]. Photosynthesis is sensitive to toxicity [22], thus we speculate that growth inhibition might be partly related to disturbance of Chla, in addition to synthesis of photosythesis. At 0.1 mg.L−1 BPA treatment group, the growth of C. pyrenoidosa and the content of Chla were stimulated. The stimulation response to BPA concentration is similar to observation of another algae, Stephanodiscus hantzschii [23], in which at 3 mg.L−1 BPA treatment, growth of algae was slightly stimulated. In addition, a hormetic effect on algae was also found in other EDCs or organic pollutants [24,25]. In brief, BPA is toxic or hormetic substance depending on its concentration.
4.2. Molecular Response to Low Concentration of BPA
In this study, the low concentration of BPA increased the growth of C. pyrenoidosa, and at the molecular level, the stimulating effect of BPA by promoting energy metabolism was observed. Electron transport chain and oxidative phosphorylation are the most critical reactions in life-sustaining activities, in which the absence of one can affect cellular respiration and even lead to cell death [26]. Mitochondrial electron transport chain, as an important component of mitochondrial structure, plays a exemplary role in regulating cell proliferation and apoptosis [27,28]. The crucial process of oxidative decarboxylation mainly occurs in electron transport chain. The gene ATPF1B encoded ATP synthase subunit beta is important cellular oxidative phosphorylation, regulating the rate of ATP synthesis in eukaryotic cells [29]. ATPase activity has been previously considered to be closely associated with cell proliferation and growth, and the technology of ATP bioluminescence for cell proliferation and determination of cytotoxicity has been developed for a long-time [30]. The upregulation of ATPF1B means the process of the synthesis of ATP from adenosine diphosphate (ADP), in which the content of inorganic phosphate was increased. The increased intracellular ATP promotes the activation of cell-cycle regulators and promotes cell proliferation as well [31]. This finding was similar to Pillai et al.’s result, in which it was revealed that low concentration of Ag+ could stimulate ATP synthesis in Chlamydomonas reinhardtii [32]. NADH dehydrogenase is the largest and most complex enzyme in mitochondrial proton transferase. It catalyzes oxidation of NADH to NAD+ and transfers electron to ubiquinone [33]. The upregulation of NADH dehydrogenase (EC:1.6.5.3) accelerated the rate of electron transport in respiratory chain. The other DEGs in oxidative phosphorylation were upregulated as well. These results showed the increase of ATP synthesis. The ATP synthesis was also affected the level of nucleotide, which were found to be essential for cell proliferation [34]. In GO term related to nucleotide, transport was significantly upregulated. Consistent with these results, the GO term related to ribosome was also upregulated, and it was previously noted that ribosome plays a significant role in proliferation of cells [35]. Briefly, the data showed that the rate of cell proliferation was accelerated and energy and materials were provided for the generation of cell stimulatory effects as well.
4.3. Molecular Response to High Concentration of BPA
Glycolysis, tricarboxylic acid (TCA) cycle, and mitochondrial electron transport chain are essential for energy provision in physiological functions [36]. The gltA encoded citrate synthase, that is a key point to control the speed of metabolism and the rate of turnover of the TCA cycle in some conditions, was downregulated (Table 4) [37]. Meanwhile, IDH1, IDH2, and IDH3 encoded isocitrate dehydrogenase, contributing to oxidative decarboxylation of isocitric acid to produce α-ketoglutarate (α-KG) in TCA, were also downregulated [38]. A downregulation of the genes indicated the production of α-KG and acetyl-CoA was suppressed in C. pyrenoidosa, and the process of the TCA cycle was inhibited as well. The TCA cycle is a metabolic pathway of three major nutrients (carbohydrates, lipids, and amino acids), and intermediate products include raw materials for the synthesis of carbohydrates, lipids, and amino acids [39]. The restriction of the TCA cycle associated with sugars, lipids, and amino acids are not fully oxidized to obtain energy, and their synthesis is also affected. Several genes associating with glycolysis were downregulated, demonstrating similar results for TCA cycle.
Table 4.
Annotation and expression changes of unigenes related to the Tricarboxylic acid (TCA) cycle, glycolysis, oxidative phosphorylation, and fatty acid metabolism.
| K-ID | Gene ID | Annotation | log2 Ratio | p-Value |
|---|---|---|---|---|
| Citrate Cycle (TCA Cycle) | ||||
| K00030 | 0047088 | IDH3; isocitrate dehydrogenase (NAD+) | −1.33141 | 6.5 × 10−7 |
| K00031 | 0059207 | IDH1, IDH2, icd; isocitrate dehydrogenase | −1.31562 | 4.1 × 10−14 |
| K00161 | 0045616 | PDHA, pdhA; pyruvate dehydrogenase E1 component alpha subunit | −8.30485 | 0.00024 |
| K00162 | 0053032 | PDHB, pdhB; pyruvate dehydrogenaseE1 component beta subunit | −1.17676 | 1.4 × 10−8 |
| K01647 | 0010385 | CS, gltA; citrate synthase | −1.81821 | 5.8 × 10−11 |
| Glycolysis | ||||
| K00134 | 0018211 | GAPDH, gapA; glyceraldehyde 3-phosphate dehydrogenase |
−2.95214 | 0.01767 |
| K00850 | 0047002 | pfkA, PFK; 6-phosphofructokinase 1 | −1.51129 | 7.6 × 10−10 |
| Oxidative phosphorylation | ||||
| K00234 | 0003771 | SDHA, SDH1; succinate dehydrogenase (ubiquinone) flavoprotein subunit | −1.06437 | 1.1 × 10−11 |
| K00235 | 0044774 | SDHB, SDH2; succinate dehydrogenase (ubiquinone) iron-sulfur subunit | −1.01169 | 2.3 × 10−8 |
| K02256 | 0064509 | COX1; cytochrome c oxidase subunit 1 |
−5.49004 | 1.2 × 10−10 |
| K02261 | 0028443 | COX2; cytochrome c oxidase subunit 2 |
−2.62114 | 0.01281 |
| K09034 | 0047163 | NDUFB10; NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 10 | −1.32268 | 4.45 × 10−15 |
| K02132 | 0050561 | ATPF1B, atpD; F-type H+-transporting ATPase subunit beta | −1.32455 | 4.6 × 10−11 |
| Fatty acid metabolism | ||||
| K01962 | 0058323 | accA; acetyl-CoA carboxylase carboxyl transferase subunit alpha | −1.15419 | 4.2 × 10−8 |
| K02160 | 0053179 | accB, bccP; acetyl-CoA carboxylase biotin carboxyl carrier protein | −1.16966 | 0.00244 |
| K00645 | 0016765 | fabD; | −9.43886 | 4.2 × 10−45 |
In fatty acids metabolism, acetyl-CoA is catalyzed to form malonyl-CoA [40]. Acetyl-CoA carboxylase is inhibited, thereby reducing the carboxylation of acetyl-CoA to malony-CoA. The fatty acid metabolism is inhibited in some extent as well. The process of energy acquisition by C. pyrenoidosa through oxidative decomposition of sugars and nutrients is inhibited in presence of exposure to high concentration of BPA [41].
In our study, mitochondrial function was also damaged via exposure to high concentrations of BPA. Similarly, Jiang et al. found that the mitochondrial membrane potential and the expression of some oxidative phosphorylation-related genes were downregulated even during exposure to 50 μg/kg/day BPA in rats [42]. The injury to mitochondria indicated that ATP synthesis decreased, and the algae cells could not obtain enough energy to maintain cell survival.
5. Conclusions
In the present study, a novel framework for physiological and molecular responses of C. pyrenoidosa to BPA was presented. Treatment under 0.1 mg.L−1 BPA led to slight growth stimulation. The energy metabolic and nucleotide pathways enhancing provided more energy and material for the promotion of C. pyrenoidosa. At exposure to 10 mg.L−1 BPA, the repression of genes associated with the TCA cycle, glycolysis, fatty acid metabolism, and mitochondrial electron transport could be observed, suggesting that the BPA inhibits growth of algal through multiple pathways. In a word, BPA is a toxic or hormetic agent to C. pyrenoidosa depending on concentration, thereby inhibiting or stimulating their growth through regulating energy metabolism.
Acknowledgments
We are grateful to Ling Xu for providing algae used for our experiments. We also thank the Wencong Zhong and Min Zhao for technical assistances with some of the experiments.
Author Contributions
Conceptualization, L.D.; Data curation, Q.C.; Formal analysis, L.D.; Funding acquisition, S.D.; Methodology, L.D.; Project administration, S.D.; Writing—original draft, L.D.; Writing—review and editing, Q.C.
Funding
This study was founded by the National Natural Science Foundation of China (NSFC; Grant Nos. 41476099 and 41676099), NSFC-Guangdong Joint Fund (Grant No. U1133003) and China Postdoctoral Science Foundation (Grant No. 55350257).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Nomiri S., Hoshyar R., Ambrosino C., Tyler C.R., Mansouri B. A mini review of bisphenol A (BPA) effects on cancer-related cellular signaling pathways. Environ. Sci. Pollut. Res. 2019 doi: 10.1007/s11356-019-04228-9. [DOI] [PubMed] [Google Scholar]
- 2.Amaral Mendes J.J. The endocrine disrupters: A major medical challenge. Food Chem. Toxicol. 2002;40:781–788. doi: 10.1016/S0278-6915(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 3.Vandenberg L.N., Hauser R., Marcus M., Olea N., Welshons W.V. Human exposure to bisphenol A (BPA) Reprod. Toxicol. 2007;24:139–177. doi: 10.1016/j.reprotox.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Eladak S., Grisin T., Moison D., Guerquin M.-J., N’Tumba-Byn T., Pozzi-Gaudin S., Benachi A., Livera G., Rouiller-Fabre V., Habert R. A new chapter in the bisphenol A story: Bisphenol S and bisphenol F are not safe alternatives to this compound. Fertility Sterility. 2015;103:11–21. doi: 10.1016/j.fertnstert.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Burridge E. Bisphenol A: Product profile. Eur. Chem. News. 2003;17:14–20. [Google Scholar]
- 6.Jiao F.R., Sun X.J., Pang Z.T. Production and Market Analysis of Bisphenol A. Chem. Ind. 2008;26:21–33. [Google Scholar]
- 7.Kleywegt S., Pileggi V., Yang P., Hao C., Zhao X., Rocks C., Thach S., Cheung P., Whitehead B. Pharmaceuticals, hormones and bisphenol A in untreated source and finished drinking water in Ontario, Canada—Occurrence and treatment efficiency. Sci. Total Environ. 2011;409:1481–1488. doi: 10.1016/j.scitotenv.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Chapman R.L. Algae: The world’s most important “plants”—An introduction. Mitig. Adapt. Strateg. Glob. Chang. 2013;18:5–12. doi: 10.1007/s11027-010-9255-9. [DOI] [Google Scholar]
- 9.Nelson Y.M., Thampy R.J., Motelin G.K., Raini J.A., Disante C.J., Lion L.W. Model for Trace Metal Exposure in Filter-Feeding Flamingos at Alkaline Rift Valley Lake, Kenya. Environ. Toxicol. Chem. 2010;17:2302–2309. doi: 10.1002/etc.5620171122. [DOI] [Google Scholar]
- 10.Xiang K., Wang J., Si T., Duan S. Interference Effects of Bisphenol A on Platymonas helgolanidica. Asian J. Ecotoxicol. 2015;10:262–267. doi: 10.7524/AJE.1673-5897.20141219003. [DOI] [Google Scholar]
- 11.Calabrese E.J. Hormesis: Changing view of the dose-response, a personal account of the history and current status. Mutat. Res./Rev. Mutat. Res. 2002;511:181–189. doi: 10.1016/S1383-5742(02)00013-3. [DOI] [PubMed] [Google Scholar]
- 12.Dabney B.L., Patiño R. Low-dose stimulation of growth of the harmful alga, Prymnesium parvum, by glyphosate and glyphosate-based herbicides. Harmful Algae. 2018;80:130–139. doi: 10.1016/j.hal.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Bai L., Cao C., Wang C., Zhang H., Deng J., Jiang H. Response of bloom-forming cyanobacterium Microcystis aeruginosa to 17β-estradiol at different nitrogen levels. Chemosphere. 2019;219:174–182. doi: 10.1016/j.chemosphere.2018.11.214. [DOI] [PubMed] [Google Scholar]
- 14.Ji M.K., Kabra A.N., Choi J., Hwang J.H., Kim J.R., Abou-Shanab R.A.I., Oh Y.K., Jeon B.H. Biodegradation of bisphenol A by the freshwater microalgae Chlamydomonas mexicana and Chlorella vulgaris. Ecol. Eng. 2014;73:260–269. doi: 10.1016/j.ecoleng.2014.09.070. [DOI] [Google Scholar]
- 15.Wang Z., Hu F., Song W., Jing G., He W., Feng D. Chronic toxic effect of three estrogens to algae (Scenedesmus obliquus); Proceedings of the International Conference on Human Health & Biomedical Engineering; Jilin, China. 19–22 August 2011. [Google Scholar]
- 16.Fan J., Xu H., Li Y. Transcriptome-based global analysis of gene expression in response to carbon dioxide deprivation in the green algae Chlorella pyrenoidosa. Algal Res. 2016;16:12–19. doi: 10.1016/j.algal.2016.02.032. [DOI] [Google Scholar]
- 17.Qian H., Xu J., Lu T., Zhang Q., Qu Q., Yang Z., Pan X. Responses of Unicellular Alga Chlorella pyrenoidosa to Allelochemical Linoleic Acid. Sci. Total Environ. 2018;625:1415–1422. doi: 10.1016/j.scitotenv.2018.01.053. [DOI] [PubMed] [Google Scholar]
- 18.Cheng J., Li K., Zhu Y., Yang W., Zhou J., Cen K. Transcriptome sequencing and metabolic pathways of astaxanthin accumulated in Haematococcus pluvialis mutant under 15% CO2. Bioresour. Technol. 2017;228:99–105. doi: 10.1016/j.biortech.2016.12.084. [DOI] [PubMed] [Google Scholar]
- 19.Wang R., Diao P., Chen Q., Wu H., Xu N., Duan S. Identification of novel pathways for biodegradation of bisphenol A by the green alga Desmodesmus sp.WR1, combined with mechanistic analysis at the transcriptome level. Chem. Eng. J. 2017;321:424–431. doi: 10.1016/j.cej.2017.03.121. [DOI] [Google Scholar]
- 20.Santabarbara S., Villafiorita Monteleone F., Remelli W., Rizzo F., Menin B., Casazza A.P. Comparative excitation-emission dependence of the FV/FM ratio in model green algae and cyanobacterial strains. Physiol. Plant. 2019 doi: 10.1111/ppl.12931. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W., Xiong B., Sun W.-F., An S., Lin K.-F., Guo M.-J., Cui X.-H. Acute and chronic toxic effects of bisphenol a on Chlorella pyrenoidosa and Scenedesmus obliquus. Environ. Toxicol. 2014;29:714–722. doi: 10.1002/tox.21806. [DOI] [PubMed] [Google Scholar]
- 22.Lu T., Zhu Y., Xu J., Ke M., Zhang M., Tan C., Fu Z., Qian H. Evaluation of the toxic response induced by azoxystrobin in the non-target green alga Chlorella pyrenoidosa. Environ. Pollut. 2018;234:379–388. doi: 10.1016/j.envpol.2017.11.081. [DOI] [PubMed] [Google Scholar]
- 23.Li R., Chen G.-Z., Tam N.F.Y., Luan T.-G., Shin P.K.S., Cheung S.G., Liu Y. Toxicity of bisphenol A and its bioaccumulation and removal by a marine microalga Stephanodiscus hantzschii. Ecotoxicol. Environ. Saf. 2009;72:321–328. doi: 10.1016/j.ecoenv.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Z.-L., Wang S.-C., Zhao F.-F., Wang S.-G., Liu F.-F., Liu G.-Z. Joint toxicity of microplastics with triclosan to marine microalgae Skeletonema costatum. Environ. Pollut. 2019;246:509–517. doi: 10.1016/j.envpol.2018.12.044. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Guo J., Yao T., Zhang Y., Zhou X., Chu H. The influence of four pharmaceuticals on Chlorellapyrenoidosa culture. Sci. Rep. 2019;9:1624. doi: 10.1038/s41598-018-36609-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chance B., Williams G.R. The respiratory chain and oxidative phosphorylation. Adv. Enzymol. Relat. Areas Mol. Biol. 1956;17:65–134. doi: 10.1002/9780470122624.ch2. [DOI] [PubMed] [Google Scholar]
- 27.Yao C.-H., Wang R., Wang Y., Kung C.-P., Weber J.D., Patti G.J. Mitochondrial fusion supports increased oxidative phosphorylation during cell proliferation. eLife. 2019;8:e41351. doi: 10.7554/eLife.41351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Márquez-Jurado S., Díaz-Colunga J., Neves R.P., Martinez-Lorente A., Almazán F., Guantes R., Iborra F.J. Mitochondrial levels determine variability in cell death by modulating apoptotic gene expression. Nat. Commun. 2018;9:389. doi: 10.1038/s41467-017-02787-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo J., Lei M., Wen M., Chen Y., Liu Z. Overexpression of ATP5b promotes cell proliferation in asthma. Mol. Med. Rep. 2017;16:6946. doi: 10.3892/mmr.2017.7413. [DOI] [PubMed] [Google Scholar]
- 30.Crouch S.P.M., Kozlowski R., Slater K.J., Fletcher J. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J. Immunol. Methods. 1993;160:81. doi: 10.1016/0022-1759(93)90011-U. [DOI] [PubMed] [Google Scholar]
- 31.Jones R.G., Thompson C.B. Tumor suppressors and cell metabolism: A recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillai S., Behra R., Nestler H., Suter M.J.-F., Sigg L., Schirmer K. Linking toxicity and adaptive responses across the transcriptome, proteome, and phenotype of Chlamydomonas reinhardtii exposed to silver. Proc. Natl. Acad. Sci. USA. 2014;111:3490–3495. doi: 10.1073/pnas.1319388111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss H., Friedrich T., Hofhaus G., Preis D. The Respiratory-Chain NADH Dehydrogenase (Complex I) of Mitochondria. Eur. J. Biochem. 1991;197:563–576. doi: 10.1111/j.1432-1033.1991.tb15945.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa Y., Tayama S. Metabolism and cytotoxicity of bisphenol A and other bisphenols in isolated rat hepatocytes. Arch. Toxicol. 2000;74:99–105. doi: 10.1007/s002040050659. [DOI] [PubMed] [Google Scholar]
- 35.Volarević S., Stewart M.J., Ledermann B., Zilberman F., Terracciano L., Montini E., Grompe M., Kozma S.C., Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 36.Fernie A.R., Carrari F., Sweetlove L. Respiratory Metabolism: Glycolysis, the TCA Cycle and Mitochondrial Electron Transport. Curr. Opin. Plant Biol. 2004;7:254–261. doi: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Walsh K., Koshland D.E., Jr. Characterization of rate-controlling steps in vivo by use of an adjustable expression vector. Proc. Natl. Acad. Sci. USA. 1985;82:3577–3581. doi: 10.1073/pnas.82.11.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Khallaf H. Isocitrate dehydrogenases in physiology and cancer: Biochemical and molecular insight. Cell Biosci. 2017;7:37. doi: 10.1186/s13578-017-0165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweetlove L.J., Beard K.F.M., Nunes-Nesi A., Fernie A.R., Ratcliffe R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010;15:462–470. doi: 10.1016/j.tplants.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Hansen H.J.M., Carey E.M., Dils R. Fatty acid biosynthesis. VIII. The fate of malonyl-CoA in fatty acid biosynthesis by purified enzymes from lactating-rabbit mammary gland. Biochim. Biophys. Acta. 1971;248:391–405. doi: 10.1016/0005-2760(71)90228-1. [DOI] [Google Scholar]
- 41.Xiang R., Shi J., Zhang H., Dong C., Liu L., Fu J., He X., Yan Y., Wu Z. Chlorophyll a Fluorescence and Transcriptome Reveal the Toxicological Effects of Bisphenol A on an Invasive Cyanobacterium, Cylindrospermopsis Raciborskii. Aquat. Toxicol. 2018;200:188–196. doi: 10.1016/j.aquatox.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Jiang Y., Liu J., Li Y., Chang H., Li G., Xu B., Chen X., Li W., Xia W., Xu S. Prenatal exposure to bisphenol A at the reference dose impairs mitochondria in the heart of neonatal rats. J. Appl. Toxicol. 2014;34:1012–1022. doi: 10.1002/jat.2924. [DOI] [PubMed] [Google Scholar]