ABSTRACT
Tether complexes play important roles in endocytic and exocytic trafficking of lipids and proteins. In yeast, the multisubunit transport protein particle (TRAPP) tether regulates endoplasmic reticulum (ER)-to-Golgi and intra-Golgi transport and is also implicated in autophagy. In addition, the TRAPP complex acts as a guanine nucleotide exchange factor (GEF) for Ypt1, which is homologous to human Rab1a and Rab1b. Here, we show that human TRAPPC13 and other TRAPP subunits are critically involved in the survival response to several Golgi-disrupting agents. Loss of TRAPPC13 partially preserves the secretory pathway and viability in response to brefeldin A, in a manner that is dependent on ARF1 and the large GEF GBF1, and concomitant with reduced caspase activation and ER stress marker induction. TRAPPC13 depletion reduces Rab1a and Rab1b activity, impairs autophagy and leads to increased infectivity to the pathogenic bacterium Shigella flexneri in response to brefeldin A. Thus, our results lend support for the existence of a mammalian TRAPPIII complex containing TRAPPC13, which is important for autophagic flux under certain stress conditions.
KEY WORDS: Autophagy, Brefeldin A, TRAPP complex, Golgi apparatus, Shigella flexneri
Highlighted Article: Depletion of the mammalian TRAPPIII complex member TRAPPC13 makes cells resistant to Golgi stressors, diminishes Brefeldin A (BFA)-induced autophagy flux, and reduces Rab1a activity in response to BFA.
INTRODUCTION
Trafficking of proteins and lipids to their correct location is of paramount importance for cellular homeostasis and helps to establish discrete organellar compartments and biochemical entities for maintenance of cell morphology, polarity, synaptic function, motility or secretion. Defects in vesicular transport through the secretory pathway underlie a large number of pathologies including glycosylation defects, lysosomal storage disorders, neurological diseases, diabetes, autoimmune diseases and cancer (Aridor and Hannan, 2000; Munksgaard et al., 2002). As vesicles approach target membranes, initial contacts are made by so called tether factors, which provide the first interaction site between the incoming vesicle and the destination membrane to aid the transition from docking to SNARE [SNAP (soluble NSF attachment protein) receptor]-mediated fusion. Multiple single molecule and multisubunit tether complexes exist in eukaryotes (Bröcker et al., 2010). The multimeric transport protein particle (TRAPP) tether factor was initially discovered in yeast, and comes in three distinct configurations sharing a common core in addition to unique subunits. Yeast TRAPPI acts in endoplasmic reticulum (ER)-to-Golgi transport and mediates COPII tethering via Bet3 (homologous to mammalian TRAPPC3)-Sec23 interaction, whereas TRAPPII functions in COPI-mediated intra-Golgi trafficking. TRAPPIII plays a role in autophagy and is found at the preautophagosomal structure (PAS) (Yu and Liang, 2012). The three yeast TRAPP complexes (TRAPPCs) also serve as guanine nucleotide exchange factors (GEFs) for Ypt1 (mammalian Rab1a and Rab1b orthologs) (Lynch-Day et al., 2010; Wang et al., 2000); in addition TRAPPII has GEF activity toward Ypt31 and Ypt32 (mammalian Rab11a and Rab11b homologs) (Morozova et al., 2006; Zou et al., 2012). The GEF activity of TRAPPCs promotes membrane recruitment of these small GTPases to couple vesicular docking and fusion with the acceptor membrane. The roles of TRAPPCs are best characterized in yeast; in higher eukaryotes its functions are less clear. For instance, little is known about the number and localization of distinct mammalian TRAPPCs or the precise subunit compositions and functions of individual TRAPP subunits (Yu and Liang, 2012). Recent evidence suggests that mammalian TRAPPC composition is more complex and likely distinct from yeast TRAPPI–TRAPPIII (Choi et al., 2011; Zong et al., 2011; Bassik et al., 2013; Scrivens et al., 2011). Although a number of orthologous subunits are shared, additional human TRAPP components including TRAPPC11 (also known as c4orf41) and TRAPPC12 (also known as TTC15) have been identified. This suggests an additional level of regulation or the acquisition of novel roles of this complex extending beyond the characterized functions in yeast. Moreover, several human TRAPPC members are localized in cellular compartments seemingly at odds with TRAPPC tethering functions along the endomembrane system. This raises the possibility that certain mammalian TRAPPC subunits carry out functions independent of their role within the assembled complex. For instance, TRAPPC12 has been implicated in kinetochore assembly and mitosis through association with chromosomes (Milev et al., 2015), and TRAPPC2 can localize to the nucleus, bind to several transcription factors and interact with ion-channel proteins (Jeyabalan et al., 2010; Fan et al., 2003). Importantly, mutations in at least three different human TRAPPC subunits [TRAPPC2 (also known as sedlin), TRAPPC9 and TRAPPC11] are known to lead to a range of phenotypes such as skeletal abnormalities and intellectual disability (Marangi et al., 2013; Bögershausen et al., 2013; Liang et al., 2015). Moreover, a TRAPPC6a mouse mutant displays a hypopigmentation phenotype (Gwynn et al., 2006), and TRAPPC4 was found to interact with and regulate ERK1 and ERK2 (ERK1/2; also known as MAPK3 and MAPK1, respectively) to control tumor formation in mouse xenograft models (Weng et al., 2013). Thus, TRAPPC is implicated in an ever-expanding disease spectrum. The elucidation of the functions of individual mammalian TRAPP units will therefore help to gain insights into disease pathologies associated with TRAPPC misregulation.
We previously identified ADP-ribosylation factor 4 (ARF4) and trafficking protein particle complex 13 (TRAPPC13; also known as C5orf44) loss-of-function mutants in a haploid genetic screen for brefeldin A (BFA) resistance (Reiling et al., 2013). In the present study, we investigated the consequences of loss of TRAPPC13 function in response to Golgi stress and Shigella flexneri infection. Like others, we found TRAPPC13 to be an additional member of human TRAPPC. The effects of TRAPPC13 depletion are dependent on ARF1-GBF1 and mimicked by Rab1 loss-of-function. Aside from TRAPPC13 knockdown, loss of TRAPPC3, TRAPPC8, TRAPPC11 and TRAPPC12, but not TRAPPC9 and TRAPPC10, also caused resistance to several Golgi-disrupting compounds. TRAPPC13-depleted cells show a more preserved secretory pathway, less apoptosis and ER stress induction in response to BFA compared with control cells. Importantly, we found that TRAPPC13 inhibition impairs Rab1 activity and autophagy, the latter process presumably involving ATG9. Moreover, S. flexneri survives substantially better in the presence of BFA in TRAPPC13 knockdown cells compared with controls. These results establish an important role of mammalian TRAPPC13 in regulating autophagy and survival in response to small molecule compound-induced Golgi stress.
RESULTS
TRAPPC13 is part of the TRAPP complex, the loss of which protects against Golgi-disrupting agents
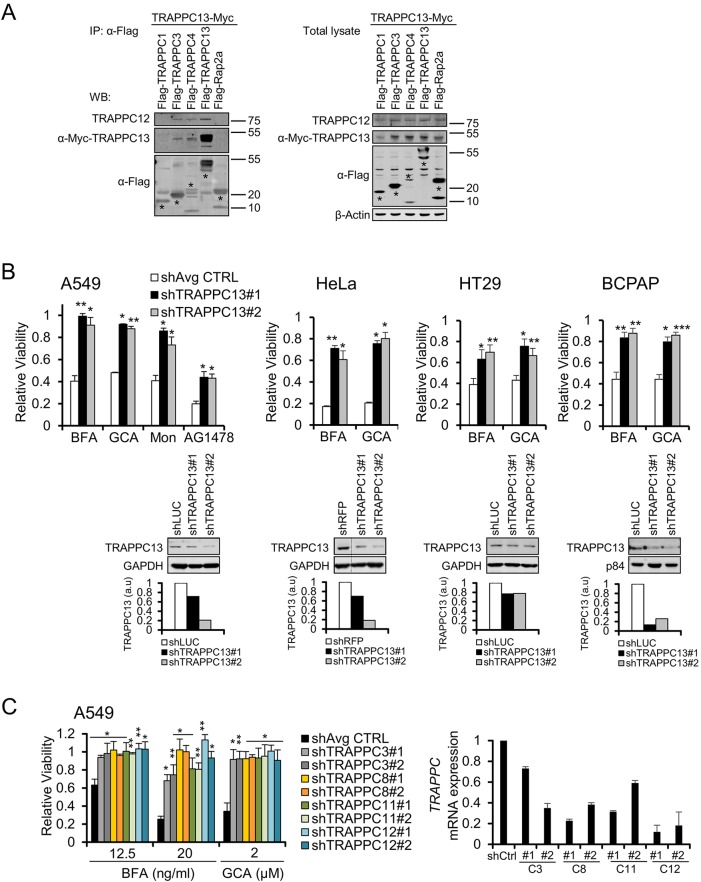
Earlier, we performed an unbiased haploid genetic screen in KBM7 cells for genes mediating the toxic effects of the Golgi disrupting agent and secretion blocker BFA. This screening approach identified ARF4 and TRAPPC13, the loss of function of which rendered cells resistant to BFA (Reiling et al., 2013). To identify potential TRAPPC13-interacting proteins, we immunoprecipitated exogenously expressed epitope-tagged TRAPPC13 in HEK293T and A549 cells, and performed mass spectrometry (MS). The MS analysis revealed the binding of various TRAPPC subunits as some of the most abundant co-purifying peptides suggesting that TRAPPC13 might be an additional component of this multisubunit complex (Fig. S1A). During the course of our studies, TRAPPC13 was reported to bind to TRAPPC2L, TRAPPC3 and TRAPPC8, congruent with our findings (Choi et al., 2011). To verify that TRAPPC13 is incorporated into mammalian TRAPPC, we co-expressed Flag-tagged versions of TRAPPC1, TRAPPC3, TRAPPC4 and TRAPPC13 proteins together with Myc-tagged TRAPPC13 in HEK293T cells. We found that Myc-TRAPPC13 co-purified with Flag-TRAPPC3 and Flag-TRAPPC4. Moreover, endogenous TRAPPC12 was co-precipitated upon TRAPPC13 overexpression. Co-expression of Flag- and Myc-TRAPPC13 revealed that they co-precipitate, indicating that TRAPPC13 is present at least as a dimer within the TRAPPC, consistent with the notion that TRAPPC exists as a multimer (Choi et al., 2011; Scrivens et al., 2011) (Fig. 1A). Next, we investigated whether the knockdown of TRAPPC13 interfered with TRAPPC formation. We thus established A549 control and TRAPPC13 knockdown cells stably expressing Flag-tagged TRAPPC2 or the overexpression control protein Flag-Rap2a. The TRAPPC was precipitated through the Flag tag and blotted for endogenous TRAPPC4 and TRAPPC12 subunits. In the absence of TRAPPC13, TRAPPC2 was still able to bind to TRAPPC4 and TRAPPC12, indicating that the overall architecture of TRAPPC is likely not disrupted upon TRAPPC13 depletion (Fig. S1B). Interestingly, Trs65p, a non-essential yeast protein with homology to TRAPPC13, is required for TRAPPII oligomer stabilization but does not disturb association of other TRAPPII-specific subunits (Choi et al., 2011).
Fig. 1.
Loss of TRAPPC13, a TRAPPC-interacting protein, provides resistance against several Golgi-disrupting agents. (A) TRAPPC13 co-immunoprecipitates TRAPPC3, TRAPPC4, TRAPPC12 and itself. Control and TRAPPC subunits were transiently co-overexpressed in HEK293T cells before Flag IP and western blotting with the indicated antibodies. (B) Viability of several cancer cell lines infected with lentiviral control or TRAPPC13 shRNAs in response to Golgi stress inducers. Relative viability was calculated by dividing fluorescence values (arbitrary units, CTB assay) of BFA-treated cells by their corresponding counterparts under vehicle-treated conditions. The means of TRAPPC13 knockdown and control cells are shown; data are representative of at least three independent experiments. Average controls (shAvg CTRL): the averaged mean survival ratio of one to three control shRNA-infected cell lines (LUC shRNA, RFP shRNA, GFP shRNA); relative viability is shown as mean±s.e.m. Treatment duration, drug concentrations: HeLa: 3 days, 12.5 ng/ml BFA treatment, 2 µM GCA; A549: 3 days 20 ng/ml BFA treatment, 2 µM GCA, 2 µM Mon, 20 µM tyrphostin (AG1478); HT29, BCPAP: 3 days 20 ng/ml BFA treatment, 2 µM GCA; six wells were measured for each genotype and condition. *P<0.05, **P<0.01, ***P<0.001 (two-tailed Student's t-test). Western blot analysis confirmed TRAPPC13 knockdown in the entire cell line panel shown below the viability bar graphs. Quantification of TRAPPC13 levels normalized to GAPDH or p84 are shown; a.u., arbitrary units. (C) Viability of A549 cell lines infected with control or two different hairpins targeting the indicated TRAPPC subunits in response to BFA. Relative viability was calculated as in B. The mean viability values of TRAPPC subunit knockdowns and control cells are shown and are representative of at least three independent experiments. shAvg CTRL: the averaged mean survival ratio of two control shRNA-infected cell lines (LUC shRNA, GFP shRNA); survival ratio is shown as mean±s.e.m. *P<0.05, **P<0.01. Knockdown of TRAPPC3, TRAPPC8, TRAPPC11 and TRAPPC12 transcript levels in A549 cells was confirmed by Q real-time PCR (right graph).
We analyzed the effects of loss of TRAPPC13 function in a panel of additional cancer cell lines including A549, HeLa, HT29 and BCPAP. Several lentiviral vectors targeting TRAPPC13 were produced and used to infect target cells for stable knockdown. Transduced cells were then evaluated for cell viability in the absence or presence of several Golgi-disrupting agents. The BFA and golgicide A (GCA) concentrations used for chronic treatment assays were adjusted for each cell line according to their sensitivities to these compounds. Loss of TRAPPC13 promoted cell survival in response to different Golgi-dispersing agents such as BFA, GCA, monensin (Mon) and tyrphostin (AG1478) (Fig. 1B). Moreover, in vitro colony formation assays showed that TRAPPC13 knockdown cells were able to proliferate after BFA treatment, unlike control cells, which were unable to form colonies under the conditions (Fig. S1C). However, TRAPPC13-depleted cells were not resistant to ER stress inducers, including tunicamycin and thapsigargin, or other small molecule compounds such as DBeQ [ATP-competitive p97 (AAA) ATPase inhibitor] and AZD (SMAC mimetic AZD 5582), pointing to a more specific and localized function of TRAPPC13 at the ER-Golgi network (Fig. S1D).
To determine whether resistance to BFA was unique to TRAPPC13 depletion or also applicable to other TRAPPC components, additional TRAPPC subunits were knocked down in A549 (Fig. 1C) and HeLa cells (Fig. S1E) using several short hairpin RNAs (shRNAs). Strikingly, in comparison with control cells, TRAPPC3, TRAPPC8, TRAPPC11 or TRAPPC12 knockdown cells were largely protected from undergoing cell death when exposed to BFA or GCA, similar to TRAPPC13 knockdown cells. This suggests a conserved role for different mammalian TRAPPC components in mediating BFA and GCA-induced toxicity. Interestingly, depletion of TRAPPC9 and TRAPPC10 had no obvious effect on cell survival when treated with BFA or GCA (Fig. S1F), suggesting that they might not be part of the same complex as TRAPPC13 as can also be inferred from our co-immunoprecipitation (IP)/MS results (Fig. S1A) (see also Discussion). In addition we assessed whether stable TRAPPC13 overexpression influences survival upon BFA treatment. TRAPPC13 gain-of-function did not lead to changes in cell viability in response to BFA exposure, suggesting that it is not sufficient to directly induce cell death (Fig. S1G).
Loss of TRAPPC13 reduces apoptosis and ER stress, and preserves the secretory pathway during BFA treatment
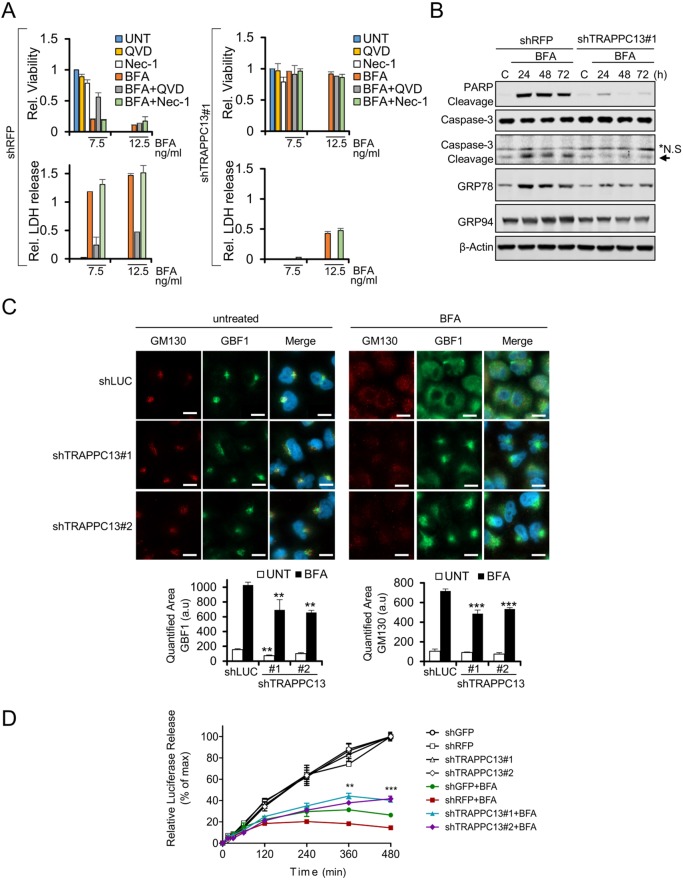
Compromising secretory pathway function can cause the accumulation of proteins in the ER, leading to ER stress and, ultimately, cell death. The results shown in Fig. 2A suggest that BFA induces apoptotic cell death, because QVD-mediated caspase inhibition significantly protected against BFA treatment-induced cell death. The addition of necrostatin-1 (Nec-1), a necroptosis inhibitor, did not rescue cells from BFA-mediated toxicity. The lactate dehydrogenase (LDH) release assay also indicated that secondary necrosis is triggered in response to prolonged exposure to BFA. Cleavage of caspase-3 and the caspase substrate PARP was detected by western blot analysis of BFA-treated control knockdown cells (Fig. 2B). Moreover, BFA led to caspase activity as determined by cleavage of a peptide caspase substrate (Fig. S2A). In contrast to control cells, we observed that loss of TRAPPC13 prevented apoptotic cell death, PARP cleavage and caspase activity when using lower BFA concentrations (Fig. 2B; Fig. S2A). Furthermore, BFA caused the upregulation of several ER stress markers, including GRP94, GRP78 (Fig. 2B) and the proapoptotic CHOP transcription factor (Fig. S2B), in control but not TRAPPC13-depleted cells, suggesting that loss of TRAPPC13 protects against ER stress induction in response to BFA treatment.
Fig. 2.
TRAPPC13 depletion reduces apoptosis and ER stress while partially preserving the secretory pathway in response to BFA. (A) Stable shRFP or shTRAPPC13 HeLa knockdown cells were left untreated or treated with 7.5 or 12.5 ng/ml BFA or vehicle control (0.1% ethanol) in the presence or absence of 20 µM QVD (caspase inhibitor) or 10 µM Nec-1 for 48 h. Viability was determined using the CTB assay (upper graph) and quantification of LDH release (lower graph). Graphs show the mean±s.d. of six replicates from two independent experiments. (B) Stable shRFP or shTRAPPC13 HeLa knockdown cells were cultured for the indicated times in the presence of 12.5 ng/ml BFA or vehicle control (0.1% ethanol). Indicated proteins were resolved on SDS-PAGE gel and detected by immunoblotting. Vehicle control cells (labelled as C) were incubated for 24 h. *N.S., a nonspecific band detected by the anti-caspase-3 antibody. Representative blots from three independent experiments are shown. (C) IF micrographs of control (shLUC) and two stable TRAPPC13 knockdown A549 cell lines reveal that TRAPPC13 depletion leads to a less disrupted Golgi compared to the control cell line when treated with BFA, as assessed by staining for the Golgi markers GM130 and GBF1. Cells were treated with 20 ng/ml BFA for 24 h. Images are representative of three independent experiments. Scale bars: 10 µm. Quantification of Golgi dispersal was calculated using Knime software. a.u., arbitrary units. ***P<0.001, **P<0.01 (one-way ANOVA, Dunnett post test). (D) HeLa cells transfected with Gaussia luciferase were seeded in six-well plates and treated with 40 nM BFA for the indicated duration. Luciferase activity in the medium was assayed at the indicated time points after treatment with BFA. Data are expressed as a percentage of the maximum luciferase signal after 8 h, obtained from the untreated sample of each pair per genotype. Results represent the mean±s.e.m from three independent experiments. **P<0.01, ***P<0.001 (two-way ANOVA with Bonferroni post test for multiple groups).
Next, we investigated the effects of TRAPPC13 knockdown on Golgi structure by immunofluorescence (IF) microscopy. Under untreated conditions, stable lentiviral-mediated knockdown of TRAPPC13 using two different shRNAs did not appreciably alter Golgi morphology compared with control cells infected with an innocuous hairpin, as revealed by staining for the cis-Golgi markers GBF1 or GM130 (also known as GOLGA2) (Fig. 2C, left panel; Fig. S2C). We then examined BFA-treated cells for Golgi complex alterations. In cells infected with control shRNAs and treated with BFA, we found a widespread cytosolic distribution of GBF1- and GM130-positive structures suggestive of Golgi dispersal. By contrast, TRAPPC13 knockdown cells showed somewhat less dispersed GM130 and GBF1 staining after addition of BFA, indicative of a less fragmented Golgi (Fig. 2C, right panel). However, in response to acute short-term treatment with a high BFA concentration, TRAPPC13 knockdown did not provide a consistent protection against compound-induced Golgi dispersal (Fig. S2D). We also assessed whether knockdown of TRAPPC11, TRAPPC12 and TRAPPC13 impaired trafficking through the secretory pathway using a Gaussia luciferase (Gluc) reporter to monitor secretion efficiency (Badr et al., 2007). With the exception of one TRAPPC13 knockdown cell line, which showed a slight increase in Gluc secretion, no significant differences in the amount of secreted Gluc at various time points (0.25-8 h) between knockdown of C11, C12 or C13 TRAPP subunits and control knockdown cells could be detected, suggesting that stable depletion of these TRAPPC components does not diminish secretion of Gluc (Fig. S2E). However, in agreement with our IF results, Gluc secretion following BFA treatment was modestly improved in cells lacking TRAPPC13, albeit not to the levels in untreated cells. The enhanced secretion was not observed at shorter time points (0.25, 0.5, 1, 2 and 4 h), but after 6 h (Fig. 2D). Similar results were obtained with cells depleted of TRAPPC11 or TRAPPC12 (Fig. S2F) (see also Discussion). Thus, the integrity and functionality of the Golgi appears to be sufficiently preserved to warrant protein trafficking and survival in TRAPPC13-depleted cells when challenged with low BFA concentrations.
BFA resistance of TRAPPC13-depleted cells depends on ARF1 activity
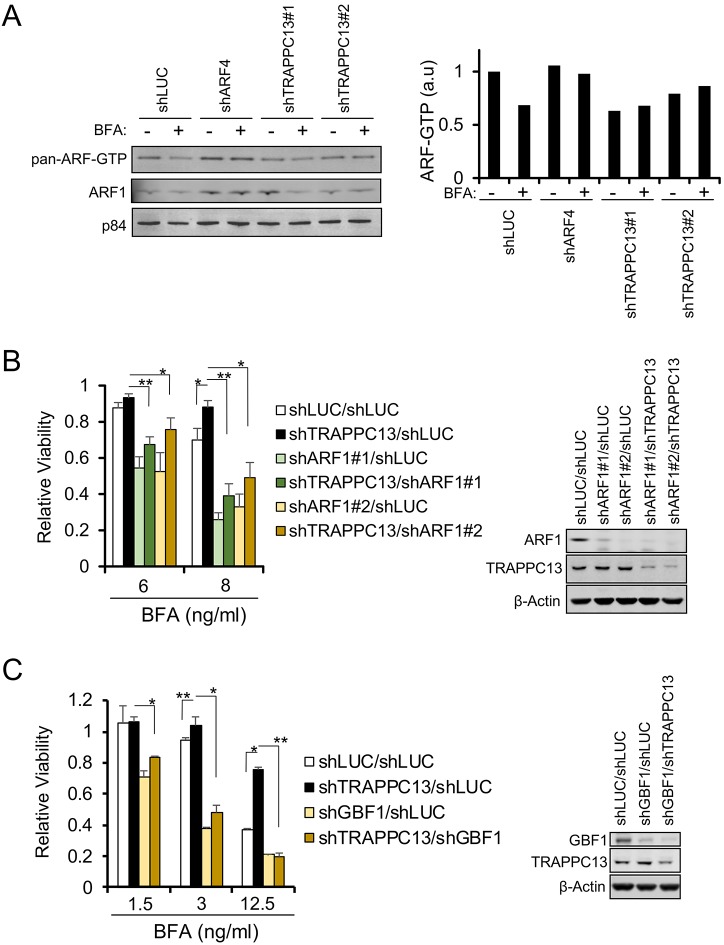
Trafficking through the Golgi apparatus requires members of the ARF family of GTPases, the activation of which is regulated by a number of GEFs. Earlier work by several groups has shown that overexpression of the large ARF GEFs GBF1 and BIG1 (also known as ARFGEF1) and BIG2 (also known as ARFGEF2), or alterations in the expression of ARFs, such as increased ARF1, ARF3, ARF5 levels or loss of ARF4, could bring about BFA resistance in several cancer cells (Donaldson and Jackson, 2011; Reiling et al., 2013; Niu et al., 2005). We did not observe induction of GBF1, BIG1, BIG2 or changes in ARF expression levels in our TRAPPC13 knockdown cells relative to control cells (Fig. S3A). However, these results did not exclude the possibility that changes in ARF activity might occur in the absence of TRAPPC13. We therefore performed ARF GTPase assays to assess the levels of active ARFs (ARF-GTP) with or without BFA treatment in control and TRAPPC13-depleted cells, as well as in ARF4 knockdown cells as a positive control. The ARF-GTP-pulldown experiments showed that although ARF activity was not elevated under untreated conditions in TRAPPC13-depleted cells compared with control cells, it was maintained during BFA treatment, unlike control cells, which displayed a reduction in ARF-GTP levels (Fig. 3A). We thus examined whether reducing ARF1 function in cells without TRAPPC13 could reverse BFA resistance. To test this, we re-infected stable TRAPPC13 knockdown or control cells with lentiviral ARF1 shRNA vectors carrying a different antibiotic marker to select for double-knockdown cells. In comparison to shLUC/shLUC or shLUC/shTRAPPC13 cells, shARF1/shTRAPPC13 double-knockdown cells had a much lower survival ratio after BFA treatment, which was very similar to shARF1/shLUC cells (Fig. 3B). This indicates that ARF1 knockdown is able to overcome the effects of TRAPPC13 depletion. In other words, to manifest BFA resistance, TRAPPC13 knockdown cells require ARF1 function. Similar results were obtained when we created shGBF1/shTRAPPC13 and the respective control double-knockdown cells; upon depletion of GBF1, A549 cells were sensitized to BFA when compared with control or TRAPPC13 knockdown cells (Fig. 3C). However, A549 cells infected with both TRAPPC13 and GBF1 hairpins showed similar survival as single GBF1 knockdown cells. Previous studies have also suggested that altered regulation and activity of other large ARF GEFs, such as BIG1/2, which function at the trans-Golgi network (TGN), may confer resistance to BFA (Flanagan-Steet et al., 2011). To explore this possibility, we first assessed the localization of endogenous BIG1 in A549 cells infected with shLUC or TRAPPC13 hairpins, and co-stained fixed cells with the Golgi marker GM130 (Fig. S3B). Interestingly, after BFA treatment, TRAPPC13-depleted cells had distinct BIG1 punctate structures throughout the cytoplasm instead of the more finely and evenly distributed haze-like appearance observed in control cells. BIG1 localization changes could be an indirect effect of TRAPPC13 knockdown, reflecting perturbations to the cis-Golgi with consequential effects on the late Golgi. Regardless of the exact mechanism underlying BIG localization alterations, shBIG1/shTRAPPC13 double-knockdown cells were less viable than shLUC/shTRAPPC13 double-knockdown cells and slightly sensitized to BFA, compared with cells infected with two innocuous hairpins using lower BFA concentrations (Fig. S3C, left panel). Finally, we checked whether blocking ARF3 function could similarly reverse the BFA resistance phenotype. ARF3 binds to both BIG1 and BIG2 and localizes to the TGN (Manolea and Melancon, 2010). Interestingly, shARF3/shTRAPPC13 double-knockdown cells were almost as resistant to BFA as shLUC/shTRAPPC13 cells, indicating that ARF3 function does not make a major contribution in TRAPPC13-depleted cells to mediate BFA resistance (Fig. S3C, right panel). Altogether, these findings suggest that protection against long-term treatment of BFA upon loss of TRAPPC13 might occur mainly at the early/cis-Golgi and is, at least in part, mediated by ARF1 and its associated large GEF GBF1.
Fig. 3.
ARF1 and GBF1 contribute to BFA resistance in TRAPPC13 knockdown cells. (A) Stable shLUC-transduced control, shARF4 (positive control) or TRAPPC13 knockdown A549 cells were analyzed for total ARF-GTP levels in the absence or presence of 20 ng/ml BFA (24 h treatment) using a GST-VHS-GAT pulldown assay. In this assay, increased ARF binding to VHS-GAT, a truncated GGA3 form and ARF-substrate that only interacts with GTP-bound ARFs, serves as an indicator of ARF activity. Pan-ARF indicates the use of an antibody (1D9), which recognizes all five mammalian ARF isoforms, whereas (1A9/5) specifically detects ARF1 (see also Materials and Methods). A representative quantification from two independent experiments is shown; a.u., arbitrary units. (B,C) Reducing (B) ARF1 or (C) GBF1 function in A549 TRAPPC13 knockdown cells using several independent shRNAs reverses BFA resistance observed in TRAPPC13 single knockdown cells. Cells were treated (B) for 3 days with 6 or 8 ng/ml BFA or (C) for 2 days with 1.5, 3, 12.5 ng/ml BFA; at least five wells per condition were measured. The survival ratio was determined by performing a CTB assay. Data represent at least three independent experiments. **P<0.01, *P<0.05. Western blots of co-depleted cells are shown to confirm knockdown of the indicated proteins.
Loss of Rab1 mimics TRAPPC13 knockdown
Besides acting as a tether factor, TRAPPC also has GEF activity toward Ypt1 and Ypt31/32. In mammalian cells, Rab1 (encoded by Rab1a and Rab1b) and Rab11 (encoded by Rab11a and Rab11b) function as the mammalian Ypt1 and Ypt31/32 homologs, respectively (Barrowman et al., 2010). Overexpression of active (GTP-loaded) Rab1 has been described to protect against BFA-induced Golgi dispersal (Alvarez et al., 2003). Recently, it was shown that depletion of TRAPPC13 in a Rab1a or Rab1b loss-of-function background caused synthetic lethality in HAP1 cells, whereas the single mutants were readily viable (Blomen et al., 2015). This suggests partially overlapping functions between TRAPPC13 and Rab1a/b. On the basis of these findings, we were interested to find out whether loss of TRAPPC13 affects Rab1 activity, and whether the BFA-resistance phenotype observed in TRAPPC13 knockdown cells is related to Rab1a/b function. We were unable to detect consistent changes in expression levels of Rab1a/b under basal and BFA conditions in TRAPPC13 knockdown protein lysates compared with control cells (Fig. S3A). To check for Rab1 activity, we employed a Rab1 effector pulldown assay (Diao et al., 2003; Satoh et al., 2003), in which we immunoprecipitated overexpressed Flag-Golgin-84 from TRAPPC13 knockdown or control cells, and blotted for endogenous Rab1a. Golgin-84 (also known as GOLGA5) binds preferentially to GTP-bound (i.e. active) Rab1, and thus Rab1 signal intensity serves as a readout of Rab1 activity. We found that Rab1a activity in TRAPPC13-depleted cells was lower than in control cells in the presence, but not in the absence, of BFA (Fig. 4A). This indicates that under Golgi stress conditions induced by BFA, TRAPPC13 is a critical regulator of Rab1a-GTP levels. We next examined whether blocking Rab1a/b or Rab11a function would mimic BFA resistance of TRAPPC13-depleted cells. To test this, we infected A549 cells with several lentiviral shRNAs targeting Rab1a or Rab11a. Contrary to TRAPPC13 depletion, Rab11a knockdown caused BFA sensitization in comparison to control cells (Fig. S4A). However, downregulation of Rab1a caused BFA and GCA resistance that was comparable to that seen upon TRAPPC13 knockdown (Fig. 4B). We also tested HAP1 cells harboring a gene trap (GT) insertion in the Rab1a locus, which completely eliminates Rab1a expression, and found these cells to be significantly more resistant to BFA compared to wild-type HAP1 cells (Fig. S4B). To further explore the role of Rab1, we performed viability assays using A549 cells that stably overexpress wild-type Rab1b as well as the respective dominant-negative (Rab1N121I) or active Rab1b-GTP-restricted (Rab1Q67L) mutant. Similar to Rab1a-depleted cells, the dominant-negative, but not the dominant-active, Rab1b mutant conferred increased viability to several Golgi stress inducers when compared with control cells infected with Flag-γTubulin (Fig. 4C). We next tested the effects of blocking Rab1a function in TRAPPC13 knockdown cells. shTRAPPC13/shRab1a double-knockdown, and the respective control, cells were established as described above and grown in the presence or absence of BFA for several days. Interestingly, shTRAPPC13/shRab1a double-knockdown cells exhibited a profound increase in their survival ratio compared to either single mutant (i.e. shTRAPPC13/shLUC or shRab1a/shLUC cells), which was especially apparent using higher BFA concentrations [Fig. 4D (A549), and Fig. S4C (HeLa)]. The synergistic effect of TRAPPC13/Rab1a double-knockdown cells on BFA resistance is indicative of a cooperative interaction, and might be due to incomplete knockdown of one or both factors giving rise to only partial BFA resistance. Strikingly, expression of active Rab1b [Flag-Rab1b(Q67L)] in a TRAPPC13-depleted background reversed BFA resistance and led to diminished viability of those cells compared with TRAPPC13 knockdown cells expressing Flag-γTubulin as an innocuous control protein (Fig. 4E and Fig. S4D). Expression of dominant-negative Rab1b further protected TRAPPC13 knockdown cells against BFA, in agreement with the results shown above (Fig. 4E). Based on these findings, it appears that TRAPPC13 is required for the GEF function of TRAPPC toward Rab1, and that decreased Rab1 function is beneficial for cells to survive in the presence of BFA.
Fig. 4.
TRAPPC13 regulates Rab1-GTP levels, and BFA resistance of TRAPPC13 knockdown cells depends on Rab1. (A) Rab1-effector pulldown assay. Stable shTRAPPC13 or shLUC knockdown A549 cells were stably transduced with Flag-tagged γTubulin or Golgin-84. Cells were treated with 20 ng/ml BFA or left untreated for 24 h. After treatment, Flag IP was performed followed by western blotting with the indicated antibodies. A representative blot of two independent experiments is shown; the bars in the right graph represent Rab1a-IP levels normalized to p84-input levels; a.u., arbitrary units. (B) A549 cells were stably transduced with lentiviral control or Rab1a hairpins. Cell viability was assessed after 3 days of treatment with either 20 or 35 ng/ml BFA or 2 µM GCA using CTB. Graphs display mean±s.e.m. of three independent experiments, each with six wells. A western blot of Rab1a-depleted A549 cells is shown below the survival graph to confirm knockdown. (C) Left panel: A549 cells stably transduced with Flag-tagged γTubulin as a control, Rab1b, Q67L mutant (GTP-bound) or the N121I mutant (GDP-bound) were treated for 3 days as indicated after which cell viability was measured as in B. Overexpression of dominant-negative N121I made cells highly resistant to different Golgi disrupting agents. Cells expressing the dominant-negative Rab1b form are also significantly more protected from Golgi stress than Rab1(wt) or dominant-active (Q67L) overexpressors. Drug concentrations: 12.5, 20, 35 ng/ml BFA, 2 µM GCA, 20 µM AG1478. Results are presented as the mean±s.e.m of three independent experiments (GCA two independent experiments). Right panel: western blot of overexpressed wild-type Flag-Rab1b and Rab1b mutants. Cell lysates were obtained and processed by SDS-PAGE and immunoblotted with anti-Flag epitope antibody. (D) Left panel: reducing Rab1a levels in A549 TRAPPC13 knockdown cells using multiple independent shRNAs induces increased BFA resistance relative to the respective single knockdowns. Cells were treated for 3 days with various BFA concentrations. Six wells were measured per condition and knockdown combination. Graphs display the mean±s.e.m of at least three independent experiments. Right panel: western blot analysis of co-depleted A549 cells to confirm Rab1a knockdown in stable TRAPPC13 knockdown cells. (E) Left panel: viability of A549 cells stably overexpressing wild-type Flag-tagged Rab1b or Rab1b mutants in a TRAPPC13 knockdown background. Cells were treated for 3 days with the indicated BFA concentrations, and six wells were measured per condition and knockdown combination. The graph displays a representative experiment of three, with the mean±s.d. provided. In comparison to TRAPPC13 knockdown cells expressing a control protein (Flag-γTubulin), the same knockdown cells overexpressing Flag-Rab1b(N121I) have a higher survival rate in response to 20 ng/ml BFA. Right panel: western blots of genotypes shown in the survival graph on the left. *P<0.05, **P<0.01, ***P<0.001 (two-tailed Student's t-test).
Loss of TRAPPC13 impairs autophagic flux, the inhibition of which protects cells against BFA-induced toxicity
Autophagy (macroautophagy) is a conserved cellular recycling process in which cytoplasmic compartments are enclosed through the formation of double-membrane vesicles or autophagosomes that subsequently merge with lysosomes to digest the engulfed content. The requirement of Rab1 for autophagy induction has previously been demonstrated (Webster et al., 2016; Winslow et al., 2010; Zoppino et al., 2010). In Saccharomyces cerevisiae, one of the three identified TRAPP complexes (TRAPPIII) has been shown to play a role in the autophagy pathway together with Ypt1 (Sambasivarao, 2014). However, not much is known about the assembly of these complexes in humans, and the existence of only two mammalian TRAPPCs has been inferred so far based on size exclusion chromatography and IP experiments (Bassik et al., 2013). A large-scale study previously showed that knockdown of TRAPPC5, TRAPPC8 or TRAPPC11 reduced autophagosome formation in a human cancer cell line (Behrends et al., 2010). TRAPPC8 as part of a potential mammalian TRAPPIII-like complex was also proposed to be important for autophagy induction by regulating trafficking of ATG9 into/from the Golgi (Lamb et al., 2015). We therefore wanted to further investigate whether autophagy might be altered in TRAPPC13-depleted cells.
First, we determined whether blocking autophagy protects against apoptotic or necrotic cell death induced by Golgi stress. HeLa cells were co-treated with BFA and chloroquine (CQ), a chemical autophagy inhibitor. CQ, although not selective, has been widely employed to analyze autophagic flux in cells, based on its ability to inhibit the last steps of autophagy due to neutralization of the lysosomal pH. Inhibition of lysosomal function with CQ partially protected HeLa and A549 cells against BFA, and curbed LDH release in response to BFA (Fig. S5A). To genetically block autophagy, we infected HeLa cells with a dominant-negative form of ATG4B [ATG4B(C74A)-Flag], which inhibits the lipidation of LC3A and LC3B (also known as MAP1LC3A and MAP1LC3B, respectively) (Fujita et al., 2008). Indeed, ATG4B(C74A) downregulated autophagic flux caused by EBSS or BFA treatment, although inhibition was not complete as judged by the levels of p62 (also known as SQSTM1) and LC3-II (the active phosphatidylethanolamine-conjugated form of LC3) in response to EBSS or BFA (Fig. S5B). Consistent with our CQ/BFA co-treatment results, stable lentiviral-mediated ATG4B(C74A)-expressing cells displayed a significant survival increase and were protected against BFA- and GCA-induced, but not tunicamycin-induced, toxicity compared with control cells (Fig. S5C). Despite the imposed survival benefits, ATG4B(C74A)-overexpression did not cause appreciable preservation of Golgi morphology in response to BFA treatment (Fig. S5D). We conclude from these results that autophagy impairment protects against BFA/GCA-induced cell death, suggesting a deleterious role for autophagy in Golgi stress-mediated apoptosis.
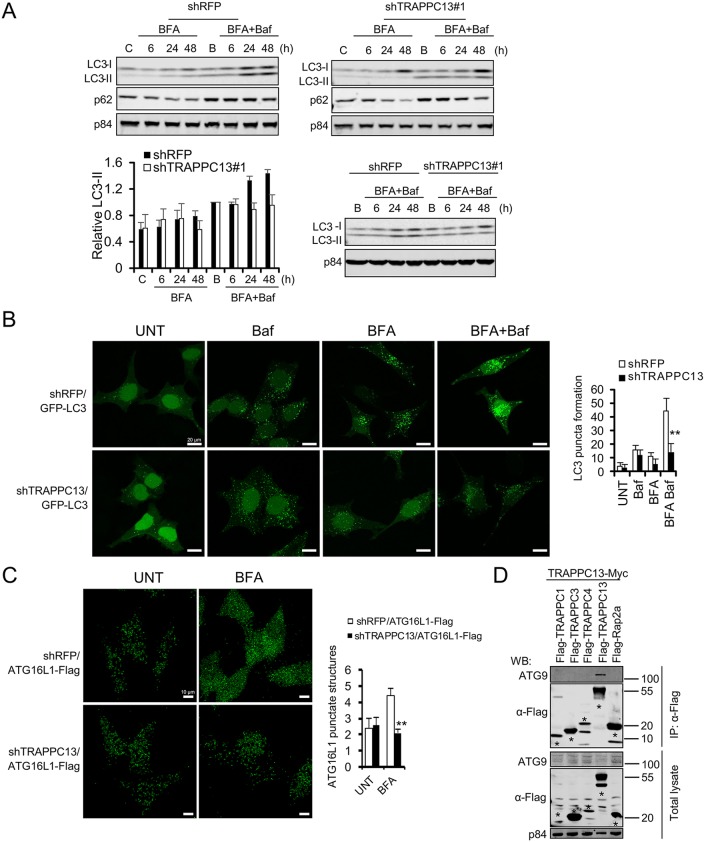
We then checked autophagic flux in TRAPPC13-depleted and in shRFP HeLa control cells by western blot analysis of the levels of LC3-I and its lipidated form, LC3-II, as well as p62, an adaptor protein that plays a critical role in recognizing/loading cargo into autophagosomes for lysosomal degradation. We used bafilomycin A1, a vacuolar H+ ATPase inhibitor, to block lysosomal degradation of autophagosome content. In control cells, LC3-II accumulated after BFA treatment in the presence of bafilomycin A1, indicating that autophagy flux is increased during BFA conditions. Concurrently, p62 levels were reduced. TRAPPC13 knockdown cells had significantly reduced conversion of LC3-I to LC3-II upon BFA treatment, indicating lower levels of autophagic flux (Fig. 5A). Next, stable GFP-LC3 overexpression cells were generated in a TRAPPC13 knockdown context to assess LC3 expression in response to BFA, in the absence or presence of bafilomycin A1, by confocal microscopy. In support of our immunoblotting results, a significantly reduced number of LC3 puncta was present in TRAPPC13 knockdown cells relative to control cells (Fig. 5B). To further substantiate our findings of autophagy interference in TRAPPC13-depleted cells, we used IF to assess the accumulation of ATG16L1, an upstream ATG protein, which is one of the key players in the early steps of autophagosome initiation required to specify the site of LC3 conjugation during autophagy (Zavodszky et al., 2013). HeLa cells stably transduced with epitope-tagged ATG16L1 were depleted of TRAPPC13 by lentiviral shRNA-mediated knockdown. TRAPPC13 downregulation led to reduced accumulation of ATG16L1 compared with control knockdown cells suggestive of diminished autophagosome formation (Fig. 5C). p62 levels were further reduced at longer time points upon TRAPPC13 downregulation compared with shRFP-infected control cells. Since p62 depletion results in inefficient autophagy (Ren et al., 2014; Viiri et al., 2010; Chen et al., 2014), we also tested the survival of HeLa cells infected with several shRNA targeting p62. In agreement with our results presented above, p62 knockdown protected cells against BFA- or GCA-induced apoptosis (Fig. S5E). As p62 is also able to promote aggregation and activation of caspase-8 (Pan et al., 2011; Jin et al., 2009), and autophagosomal membrane can serve as a platform for death-inducing signalling complex (DISC) formation (Young et al., 2012), it could also be that the BFA-protective effects following p62 knockdown might be partially related to decreased caspase-8 activity in these cells.
Fig. 5.
Loss of TRAPPC13 leads to impaired autophagic flux in response to BFA treatment. (A) Stable shTRAPPC13 or shRFP knockdown HeLa cells were treated with 12.5 ng/ml BFA for the indicated times in the presence of 20 nM bafilomycin for the last 3 h of incubation. Untreated control cells were incubated in DMEM for 6 h in the presence (labelled as B) or absence (labelled as C) of bafilomycin A1 (Baf) for the last 3 h of incubation. Protein lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Quantification of relative LC3 II levels was performed as described in the Materials and Methods. Samples shown in the upper two western blot panels were additionally run and evaluated on the same gel as displayed in the lower western blot panel to confirm reduced LC3-II levels in TRAPPC13 knockdown cells compared with control shRFP cells. Data are representative of three independent experiments. (B) HeLa cells stably expressing GFP-LC3 and transduced with shRFP or shTRAPPC13 hairpins were incubated with DMEM in the absence or presence of 20 ng/ml BFA for 24 h with or without Baf for the last 3 h of incubation. The expression of GFP-LC3 was examined by confocal microscopy. LC3 dots in cells were measured as described in the Materials and Methods. Representative quantification results from two independent experiments are shown. **P<0.01 (two-tailed Student's t-test). (C) HeLa cells stably expressing ATG16L1-Flag and transduced with shRFP or shTRAPPC13 hairpins were treated with BFA for 24 h or left untreated. ATG16L1-Flag accumulation was assessed by IF using confocal microscopy. Representative quantification results from two independent experiments are shown. ** P<0.01 (two-tailed Student's t-test). (D) TRAPPC13 interacts with ATG9. The indicated proteins were transiently expressed in HEK293T cells before Flag IP and blotting for endogenous ATG9.
Based on the reduced autophagy flux in TRAPPC13 knockdown cells and on our above results demonstrating that TRAPPC13 cooperates with Rab1 and modulates its activity, we were interested to see whether TRAPPC13 is able to interact with upstream autophagy regulators such as ATG9, a multispanning membrane protein required for autophagosome formation. We therefore immunoprecipitated transfected and epitope-tagged TRAPPC13 or two other TRAPPC core subunits in HEK293T cells, and checked for the presence of endogenous ATG9. ATG9 was found in TRAPPC13-immunoisolates but not in the negative control, indicating that ATG9 and TRAPPC13 are closely associated (Fig. 5D). In summary, these findings suggest that TRAPPC13 depletion leads to attenuation of autophagy through inhibition of Rab1 activity, and presumably through decreased formation and processing of the isolation membrane mediated by ATG9 and ATG16L1, which is necessary for cells to cope with Golgi stress-induced toxicity.
To determine if TRAPPC13 also regulates autophagy flux in response to starvation-induced autophagy, cells were cultured in a medium lacking amino acids (EBSS) for 6 h, and levels of p62 and LC3-II were analyzed by western blotting. Unlike BFA-treated cells, but similar to control knockdown cells, we observed induction of autophagy upon incubation in starvation buffer in TRAPPC13 depleted-cells (Fig. S5F). Similar results were obtained when the rapalog everolimus was added to cells to induce autophagy, as assessed by GFP-LC3 staining (Fig. S5G). These results are consistent with the absence of an effect on LC3-II in siRNA-mediated TRAPPC8 knockdown cells (Lamb et al., 2015). Thus, TRAPPC13 knockdown might become rate-limiting and inhibit autophagy under certain stress conditions, such as BFA treatment, but not under amino acid-deprived conditions or in response to direct pharmacological mTOR inhibition.
Loss of TRAPPC13 increases susceptibility to S. flexneri infection
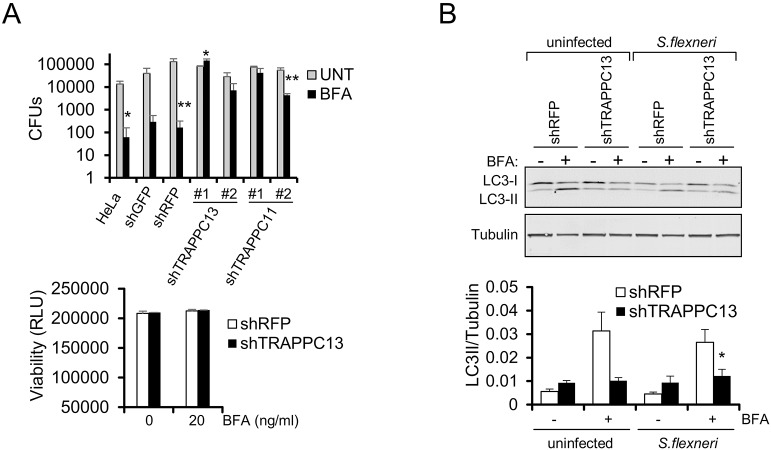
Autophagy can serve as a cellular defense mechanism to disarm pathogenic bacteria after infection. In turn, bacteria have developed strategies to avoid autophagy. In particular, Rab GTPases, which regulate autophagosome maturation, are frequent targets of bacterial pathogens. S. flexneri, for instance, inactivates Rab1 with a GTPase-activating protein (GAP) effector protein called VirA, as one method to resist autophagy-mediated host defense and allow intracellular survival (Dong et al., 2012; Ao et al., 2014). Because TRAPPC13 and Rab1a knockdown cells are both resistant to BFA (Fig. 1B and Fig. 4B), and because loss of TRAPPC13 impairs autophagic flux, we wondered whether knockdown of mammalian TRAPPC subunits impacts intracellular proliferation of S. flexneri. Control, TRAPPC11 and TRAPPC13 knockdown HeLa cells were infected with S. flexneri, and survival of the bacteria in the absence or presence of BFA was assessed by a gentamicin protection assay. In all of our experiments, the cells were infected at a low multiplicity of infection (MOI) of 1. TRAPPC11-depleted cells were included in this experiment because we identified TRAPPC11 as a potential TRAPPC13 interaction partner (Fig. S1A), and because its knockdown made cells resistant to BFA and GCA (Fig. 1C), similar to TRAPPC13 depletion. No differences in colony-forming units (CFUs) between control and TRAPPC knockdown cells were detected 1 h post infection (hpi), with or without BFA addition, indicating that the initial infectivity between the different genotypes is comparable (Fig. S6A). Remarkably, in response to S. flexneri infection, both TRAPPC11 and TRAPPC13 knockdown cells showed a substantial increase in the number of bacteria at 24 hpi compared with control knockdown cells in the presence, but not in the absence, of BFA (Fig. 6A, upper panel). Viability assays showed that at this time point, survival of infected control and shTRAPPC13-depleted HeLa cells is similar, suggesting that the differences in CFUs are not due to unequal cell numbers present (Fig. 6A, lower panel). Interestingly, Rab1 and TRAPPC3 were reported to be essential components for antibacterial autophagy to limit intracellular Salmonella typhimurium growth in response to damage to the Salmonella-containing vacuole (Huang et al., 2011). Furthermore, in line with our p62 knockdown results (Fig. S5E), it was previously shown that the depletion of its ortholog in S. flexneri-infected zebrafish larvae increases bacterial burden, indicating that p62 restricts microbial infection in vivo (Mostowy et al., 2013). We also checked LC3 conversion in TRAPPC13-depleted and shRFP control cells after S. flexneri infection. When infected with S. flexneri, TRAPPC13 knockdown cells displayed reduced LC3-II accumulation (Fig. 6B, lane 8 versus lane 6) compared with shRFP control cells, in response to BFA treatment. This suggests that S. flexneri is able to overcome the lower Rab1 activity in the TRAPPC13 knockdown, but not the control, cells (Fig. 4A) to counteract autophagy for bacterial persistence. It appears that in the absence of BFA, S. flexneri is able to avoid autophagy and survive in both cell lines to a similar extent. However, in response to BFA exposure, there is increased autophagy in control cells that S. flexneri cannot evade, resulting in clearance, but in TRAPPC13 knockdown cells with reduced Rab1 activity and impaired autophagy flux (Fig. 4A and Fig. 5A,B), growth of the bacteria can no longer be efficiently inhibited, resulting in unrestricted S. flexneri survival.
Fig. 6.
Loss of TRAPPC13 increases S. flexneri propagation upon BFA exposure. (A) Increased growth of S. flexneri upon BFA treatment following TRAPPC13 or TRAPPC11 knockdown. Cells were infected with S. flexneri and treated with gentamicin. The cells were then lysed after 24 h growth in medium with or without 20 ng/ml BFA, and dilution plating was used to count the number of CFUs present. Upper panel: TRAPPC13 and TRAPPC11-depleted cells show higher numbers of CFUs compared to control shRNA-infected cells. Data are mean±s.e.m. from three independent experiments performed in triplicate. **P<0.01, *P<0.05. Lower panel: survival of S. flexneri-infected HeLa cells with or without 20 ng/ml BFA for 24 h was determined by a CellTiter-Glo (CTG) assay. Results are representative of three independent experiments; RLU, relative luminescence units. (B) Stable shRFP or shTRAPPC13 HeLa cells were infected with S. flexneri and left untreated or treated with 20 ng/ml BFA for 24 h. Protein lysates were resolved by SDS-PAGE and samples probed with the indicated antibodies. Relative LC3-II levels were calculated. Results are representative of two independent experiments performed in triplicate.
DISCUSSION
In this study, we demonstrate an important role for TRAPPC13 and other TRAPPC subunits in mediating the toxicity of several small molecule compounds that induce Golgi dispersal and inhibit secretion. The observed survival benefit of TRAPPC13 knockdown cells in response to BFA correlates with a more preserved secretory pathway, reduced ER stress and caspase activation, as well as attenuated inhibition of protein secretion, compared with control cells. TRAPPC13 depletion also reduces Rab1 activity and autophagy flux, which makes cells more resistant to BFA and GCA. TRAPPC13 depletion caused attenuated autophagy induced by BFA treatment as evidenced by LC3-II immunoblotting and GFP-LC3 and ATG16L1-flag IF microscopy (Fig. 5A-C). Reassuringly, we find that Rab1a depletion mimics the TRAPPC13 knockdown phenotype displaying protection to several Golgi stress agents. Thus, reducing the rate of autophagy under BFA-induced Golgi stress conditions appears to promote cellular survival. Consistent with diminished autophagic flux in TRAPPC13 knockdown cells, we found much higher S. flexneri survival in TRAPPC13 knockdown cells compared with control cells after treatment with BFA (Fig. 6A). Strengthening the notion of involvement of TRAPPC13 in autophagy, we found that TRAPPC13 co-precipitates ATG9, which is essential for this process. Taken together, these results highlight the importance of autophagy regulation by human TRAPPC13 and suggest that a mammalian TRAPPC exists, which modulates autophagy similarly to yeast TRAPPIII. Interestingly, the downregulation of mammalian TRAPPC function also protects cells against ricin, a plant-derived toxin that enters the cell through endocytosis and traffics in a retrograde manner through the Golgi and ER (Bassik et al., 2013). It is mechanistically unclear, however, whether ricin resistance caused by downregulation of TRAPPIII components is related to its effects on autophagy. Based on co-IP and gel filtration experiments, Bassik et al. identified two distinct mammalian TRAPPCs (designated mTRAPPCII and mTRAPPCIII) containing a core set of proteins in addition to unique subunits (Bassik et al., 2013). Supporting these authors' hypothesis of at least two functionally separate human TRAPPCs, our own Flag-TRAPPC13 co-IP and MS results revealed that TRAPPC13 immunoprecipitates did not recover TRAPPC9 or TRAPPC10, suggesting that TRAPPC13 is part of mTRAPPCIII but not mTRAPPCII. Unlike knockdown of several other mTRAPPCIII subunits, loss of mTRAPPCII-specific TRAPPC9 or TRAPPC10 did not provide resistance to BFA or GCA (Fig. S1F). A large-scale proteomic study has found that TRAPPC5, TRAPPC8, TRAPPC11 and TRAPPC12, components of mTRAPPCIII, play a role in autophagosome formation in U2OS cells (Behrends et al., 2010). Since mTRAPPCIII components can also immunoprecipitate COPII components (Bassik et al., 2013), this suggests that the single mTRAPPCIII fulfils functions similar to both yeast TRAPPI and TRAPPIII, probably depending on the cellular context. Recently, Lamb et al. (2015) showed that TRAPPC8 mediates interaction with TBC1D14 to regulate autophagosome formation, presumably as part of mTRAPPCIII, which also contains TRAPPC12 and TRAPPC4. However, TRAPPC13 was not identified in their study. TRAPPC8 might act through localization and regulation of ATG9 at the Golgi to induce autophagosome structures (Lamb et al., 2015). It is therefore tempting to speculate that TRAPPC13 is similarly involved in retrograde vesicle transport from endosomes to the Golgi, and functions in the recruitment of ATG9 and potentially other autophagy machinery to supply membrane for autophagosome formation. Aside from TRAPPC12 and TRAPPC11, TRAPPC8 is one of the major TRAPPC13-interacting proteins identified in our MS analysis and, thus, both factors might be in close physical association within mTRAPPIII (Fig. S1A).
In addition to TRAPPC13, we originally identified the small GTP-binding protein ARF4 in our gene trap mutagenesis screen for BFA resistance (Reiling et al., 2013). Our results presented here suggest that the observed BFA resistance phenotypes of both TRAPPC13 and ARF4 knockdown cells are only partially related, and are both qualitatively and quantitatively different. For instance, ARF4-depleted cells show upregulation of multiple ARF GTPase family members and are protected from S. flexneri and Chlamydia trachomatis infection (Reiling et al., 2013). In addition to its effect on Rab1 activity, cells with reduced TRAPPC13 function did not display a decline in ARF1 activity in response to BFA treatment, which is normally observed in wild-type cells (Fig. 3A). Moreover, co-depletion of TRAPPC13/ARF1, or TRAPPC13/GBF1, largely suppressed BFA resistance in TRAPPC13 knockdown cells, leading to a survival ratio comparable to GBF1 or ARF1 single knockdown cells. Additionally, another large GEF, BIG1, becomes mislocalized in TRAPPC13 knockdown cells. Thus, our results indicate that TRAPPC13 genetically interacts with ARF1 and its associated large GEFs on multiple levels to cause BFA resistance. In yeast, it was found that ARF1 and the TRAPPC component TRS130 (TRAPPC10 homolog) show a synthetic lethal interaction suggesting compensatory mechanisms between the two factors in the absence of the other (Zhang et al., 2002). Moreover, Gea2, the yeast GBF1 homolog, directly interacts with Trs65, the yeast TRAPPC13 homolog, suggesting that Trs65 might be part of an ARF1-Gea2 effector loop potentially leading to TRAPPII recruitment to membranes. trs65 gea1 gea2 triple mutants showed exacerbated trafficking defects (Chen et al., 2011). Further evidence for a cross talk was shown in yeast, where activated Arf1 recruits TRAPPII to membranes to direct vesicle trafficking (Thomas and Fromme, 2016). The precise functional interaction between human TRAPPC13 and ARF1 signaling awaits further clarification.
It is of interest to note that the conserved oligomeric Golgi (COG) complex, another multisubunit tether factor, has been shown to modulate BFA-induced Golgi morphology changes (Flanagan-Steet et al., 2011). In addition, COG complex mutants exhibit defective autophagy (Yen et al., 2010). Depletion of yet another multisubunit tether factor, Golgi-associated retrograde protein (GARP), which mediates tethering of endosome-derived vesicles to the TGN, also causes autophagy defects (Pérez-Victoria et al., 2010). These findings highlight the importance of TRAPPC and other tethering complexes to control autophagy. Given the multiple roles of autophagy in human disease, the involvement of TRAPPC13/mTRAPPC might offer an additional avenue to manipulate autophagy for therapeutic purposes (Murrow and Debnath, 2012).
Others have reported Golgi fragmentation phenotypes associated with siRNA-mediated TRAPPC2, TRAPPC2L, TRAPPC8, TRAPPC11 or TRAPPC12 knockdown and reduced anterograde or intra Golgi transport of cargo (Lamb et al., 2015; Scrivens et al., 2011; Yamasaki et al., 2009; Loh et al., 2005). Curiously, we neither observed a Golgi fragmentation phenotype upon stable lentiviral TRAPPC13 hairpin transduction, nor a noticeable decrease in Gaussia luciferase secretion, under normal culture conditions (Fig. 2C,D; Fig. S2C,E). TRAPPC2 knockdown cells exhibited impaired procollagen (PC) secretion, though the secretion of other proteins such as VSV-G, CD8α, albumin or α1-antitrypsin was unaffected (Venditti et al., 2012). This could suggest that depletion of individual TRAPPC components might affect trafficking of only a subset of secretory cargoes compatible with cell survival. A protection of the secretory pathway upon TRAPPC13 depletion can also be inferred from the ability of these cells to grow under chronic BFA treatment conditions for days or even weeks. It is also possible that acute versus chronic knockdown in cell culture or differences in remaining expression levels might explain these contrasting findings.
MATERIALS AND METHODS
Plasmids and cloning
TRAPPC13 cDNA was PCR-amplified using a cDNA pool (derived from A549 cells, which was used also for all other cDNA cloning) and oJR397/SalI and oJR398/NotI primers (Table S1). Sequence-verified TRAPPC13 was ligated into SalI/NotI-digested pLJM60 to yield Flag-TRAPPC13. TRAPPC1 (oJR419/SalI and oJR420/NotI), TRAPPC3 (oJR417/SalI and oJR418/NotI) and TRAPPC4 (oJR421/SalI and oJR422/NotI) cDNAs were PCR-amplified from the cDNA pool with the primers indicated in parentheses. After subcloning and sequence verification, the PCR fragments were ligated into pLJM60, thereby generating N-terminally Flag-epitope-tagged proteins. Golgin-84 cDNA was amplified using a cDNA pool and the oligonucleotides Golgin-84/SalI and Golgin-4/NotI; after sequence verification it was ligated into pLJM60 to generate an N-terminal Flag-fusion. ATG4B (wild-type) cDNA was amplified from a cDNA pool using ATG4B/AgeI and ATG4B-flag/EcoRI primers. Through site-directed mutagenesis with ATG4B(C74A)_forw and ATG4B(C74A)_rev primers, ATG4B(C74A)-flag was created, sequence-verified and ligated into AgeI/EcoRI-digested pLJM13 for lentiviral expression. Rab1b WT/Q67L/N121I cDNAs were a gift from Cecilia Alvarez (Universidad Nacional de Córdoba, Argentina) and used as a DNA source for PCR amplification using the oligonucleotides oJR431/SalI and oJR432/NotI. PCR fragments were subsequently inserted into lentiviral vectors for stable expression after sequence verification. The oligonucleotides ATG16L1/AgeI and ATG16L1-flag-EcoRI were used for the PCR reaction in conjunction with an A549 cDNA pool to amplify ATG16L1 cDNA. After sequence verification, ATG16L1-flag was cloned into lentiviral vectors; virus was then produced to generate stable overexpression cell lines. GFP-LC3 (rat) cDNA was PCR-amplified with GFP-LC3/AgeI and GFP-LC3/EcoRI primers. After sequence verification, GFP-LC3 was ligated into the pLJM15 lentiviral vector. Gluc-flag was generated using the pCMV-GLuc 2 plasmid (Promega) as a DNA source for a PCR reaction in combination with GLuc/AgeI and GLuc-flag/EcoRI primers. The sequence-verified PCR product was then ligated into the pLJM13 and pLJM15 vectors. Primer sequences are listed in Table S1.
Cell culture, treatments and reagents
All cell lines described were maintained in high-glucose (25 mM), pyruvate-free DMEM (Invitrogen) supplemented with 2 mM L-glutamine, 200 mg/ml penicillin, 100 mg/ml streptomycin sulfate and 10% heat inactivated fetal serum (IFS) (Invitrogen).
Viability of cells in 96-well assay plates with clear flat bottoms was determined using the CellTiter-Blue (CTB) or CellTiter-Glo (CTG) assay (Promega). HeLa, A549, HT29 and BCPAP cells were seeded at a concentration of 40,000–45,000 cells/ml per well and treated in fresh medium 24 h later at a culture volume of 100 μl per well. The cells were grown in the absence or presence of the tested compound for 48–72 h depending on the experiment; 20 μl CTB reagent was added at the end of the experiment according to the manufacturer's protocol. The fluorescent end product resorufin was measured using a GloMax-Multi Detection System (Promega). Compounds used were as follows: BFA (Sigma-Aldrich), GCA (Santa Cruz Biotechnology), Mon (Enzo Life Sciences), AG1478 (Sigma-Aldrich), tunicamycin (Santa Cruz Biotechnology) and thapsigargin (Santa Cruz Biotechnology).
Western blotting
Cells were washed with ice-cold PBS, lysed by resuspending them in Pierce radioimmune precipitation assay (RIPA) buffer [25 mmol/l Tris-HCl (pH 7.6), 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS; Thermo Fisher Scientific] plus complete protease inhibitor mixture (Roche Applied Science) and PhosSTOP phosphatase inhibitor cocktail tablets (Roche Applied Science), and frozen. Protein concentrations were measured with Pierce BCA protein assay reagent (bicinchoninic acid, Thermo Fisher Scientific). Equal amounts of protein were mixed with Laemmli loading buffer. For western blotting, proteins were resolved on NuPAGE Novex 4–12% Bis-Tris protein gels (Invitrogen) and transferred onto polyvinylidene difluoride membranes (Millipore). Membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences), and secondary antibodies (IRDye 800CW donkey anti-rabbit IgG (1:15.000, LI-COR Biosciences), IRDye 680CW donkey anti-mouse IgG (1:20.000, LI-COR Biosciences) or goat anti-hamster IgG (1:15.000, AbdSerotec) were detected by fluorescence with the Odyssey Fc imaging system.
Primary antibodies used for western blotting were as follows: mouse anti-β-actin (Cell Signaling Technology), mouse anti-FLAG M2 (1:500, F1804/clone M2, Sigma-Aldrich), rabbit anti-FLAG (1:1000, #2368S, Cell Signaling Technology), mouse anti-Myc (1:500, #2276/clone 9B11, Cell Signaling Technology), mouse anti-GBF1 (1:1000, #612116, BD Transduction Laboratories), mouse anti-ARF1 1(1D9)/pan-ARF (1:500, NB300-505/clone 1D9, Novus Biologicals), mouse anti-ARF1 (1:500, sc-53168/clone 1A9/5, Santa Cruz Biotechnology), mouse anti-ARF3 (1:500, sc-135841/clone 41, Santa Cruz Biotechnology), rabbit anti-ARF4 (1:500, 11673-1-AP, Proteintech), mouse anti-ARF5 (1:500, H00000381-M01/clone 1B4, Abnova), hamster anti-ATG9 (1:500, 14F2 8B1, Thermo Fisher Scientific), rabbit anti-cleaved PARP (1:1000, #5625, Cell Signaling Technology), rabbit anti-LC3 A/B (1:1000, #4108S, Cell Signaling Technology), mouse anti-p62 (1:1000, sc-28359, Santa Cruz Biotechnology), rabbit anti-caspase 3 (1:1000, #9662S, Cell Signaling Technology), rabbit anti-cleaved-caspase 3 (1:1000, #9661L, Cell Signaling Technology), rabbit anti-GRP94 (1:1000, GTX103203-25, GeneTex), mouse anti-TTC15/TRAPPC12 (1:1000, ab88751, Abcam), rabbit anti-Rab1a (1:1000, #13075S, Cell Signaling Technology), rabbit anti-Rab1b (1:1000, sc-599, Santa Cruz Biotechnology), rabbit anti-Rab11a (1:1000, #2413, Cell Signaling Technology), rabbit anti-GRP78 (1:1000, sc-13968, Santa Cruz Biotechnology), rabbit anti-BIG1 (1:1000, sc-376866, Santa Cruz Biotechnology), rabbit anti-BIG2 (1:1000, sc-398042, Santa Cruz Biotechnology) and mouse anti-p84 (1:1000, GTX70220, GeneTex).
Quantification of band intensity was performed with Image Studio software 3.1 (LI-COR Biosciences). Fluorescence intensity of bands shown was normalized to β-actin or p84 as control bands from the same membrane.
Quantitative real-time PCR
Cells were grown in 6 cm dishes, and mRNA was isolated using the RNeasy Plus Mini kit (Qiagen). 1 μg total RNA was used for the reverse transcription (RT) reaction using oligo(dT) primers and Superscript III (Invitrogen). cDNA was diluted 1:15 after RT for subsequent use in quantitative real-time PCR (qPCR). QuantiNova SYBR Green PCR Master Mix (Qiagen) was used, and the reaction volume was 25 μl per qPCR reaction (Rotor-Gene Q). Three technical replicates were run per biological replicate for calculating the mean Ct values.
Virus production and generation of stable cell lines
HEK293T cells were seeded at a density of 800×103 cells in 6 cm dishes 24 h before transfection. Plasmids encoding ΔVpr and pCG (VSV-G envelope protein expression vector) and 1 µg shRNA contruct were transfected into HEK293T cells using 6 μl LT1 Transfection Reagent (Mirus). 12 h post-transfection, the medium was changed to a GlutaMAX (Invitrogen) medium plus 30% IFS. Lentiviral supernatants were collected after 48 h. Then, the virus-containing medium was centrifuged to remove cellular debris, and aliquots were frozen.
For lentiviral shRNA, transduction cells (HeLa, A549, HT29, BCPAP) were plated at a density of 150×103 cells in 6 cm dishes and incubated overnight. The culture medium was then replaced by 3 ml DMEM containing 10% IFS, and viral supernatant (200 µl for A549 cells and 400 µl for all other cells) supplemented with 8 μg/ml polybrene (Sigma-Aldrich) was added. After 24 h, infected cells were selected using 2 μg/ml Puromycin (VWR), 350 µg/ml Hygromycin (VWR) or/and 1 mg/ml G418 (Geneticin, VWR). Information about the TRC hairpin clone ID is listed in Table S2.
Immunocytochemistry
Cells were fixed with a fresh solution of 4% paraformaldehyde (Electron Microscopy Sciences) for 20 min at room temperature, washed three times with PBS and permeabilized with 0.05% Triton X-100 in PBS for 20 min. After washing, primary antibodies against GM130 (1:3000, #12480S, Cell Signaling Technology) and GBF1 (1:100, #612116, BD Transduction Laboratories) diluted in 5% normal donkey serum (Jackson ImmunoResearch) were incubated overnight at 4°C. The next day, cells were washed three times with PBS and incubated with the appropriate secondary antibody (1:2000, Life Technologies) and Hoechst (1:2500, Life Technologies) diluted in 5% normal donkey serum for 1 h. Finally, samples were washed five times with PBS. Pictures were acquired with an Olympus Biosystems IX81 inverted microscope at 20× magnification and Olympus ScanR 2.5.0. acquisition software. For image analysis and single cell feature extraction, Knime 3.1.0 software (Stöter et al., 2013) was used. Golgi area is presented as an average derived from at least nine fields per well in triplicate, analyzing ≥1000 cells per condition and genotype; 5000 cells per well were seeded in 96-well plates for imaging (Greiner) with clear flat bottoms, and treated with 5 µg/ml BFA in fresh medium 24 h later (Fig. S2D). The total culture volume per well was 150 μl. Fixation and staining were performed as described above.
Confocal microscopy
HeLa cells were cultured on glass coverslips pretreated with collagen I (Sigma-Aldrich) and treated with the indicated agents. They were then fixed in a fresh solution of 4% paraformaldehyde for 15 min, washed twice with PBS, stained with the indicated antibodies and mounted in Vectashield mounting medium (Vector Laboratories); visualization was performed at room temperature on a Leica TCS SP5 spectral confocal microscope with a HCX PL APO 40×1.3 NA oil objective. The acquisition software used was Leica application suite advanced fluorescence (version 2.6.0.7266). The projections of z-stacks are shown. Vesicles (dots) from z-stacks of whole-field images displaying multiple cells were analyzed with Fiji/ImageJ software followed by the Gaussian Laplacian filter macro. Results are presented as mean dots per cell, and correlate with a measurement of the punctate area in a minimum of five independent images and 40 cells (Fig. 5B,C). A549 cells were plated (200,000 cells/24-well plate). After 24 h, the cells were fixed, permeabilized and stained with primary antibodies. Coverslips were mounted on glass slides with Vectashield mounting medium. Images were acquired at room temperature on a Zeiss LSM780 confocal microscope system with an Objective Plan-Apochromat 40×/1.4 Oil DIC M27 and ZEN acquisition software. The projections of z-stacks are shown (Fig. S2C).
Co-IP assay
HEK293T cells (2×106) were seeded in 10 cm dishes. Transient co-transfection of TRAPPC13 and TRAPPC1, TRAPPC3, TRAPPC4 or Rap2a, was performed using 1000 ng DNA for Flag-TRAPPC13, TRAPPC13-Myc and Flag-Rap2a; 2000 ng DNA for Flag-TRAPPC1 and Flag-TRAPPC4; and 500 ng DNA for Flag-TRAPPC3. TransIT®-LT1 (Mirus) (3 µl) was used as a transfection reagent. 48 h after transfection, the cells were washed with cold PBS and lysed with 500 µl lysis buffer [40 mM Hepes pH 7.5, 120 mM NaCl, 1 mM EDTA, 0.3% CHAPS and protease inhibitor cocktail (Roche)]. Cell lysates were centrifuged for 15 min at 13,000 rpm, and anti-FLAG M2 Affinity Gel (Sigma-Aldrich) that had been washed two times with lysis buffer was added to the HEK293T lysate for 2 h. The immunoprecipitates were washed three times in lysis buffer and eluted in a 2× sample buffer by denaturation at 95°C. Eluates were resolved on 4–12% gradient Bis-Tris gel and probed with anti-Flag, anti-Myc (71D10), and TRAPPC12 or ATG9 antibodies.
For precipitation of different TRAPP subunits in TRAPPC13-depleted cells, 2×106 stably transduced shLUC/Flag-Rap2a, shTRAPPC13/Flag-Rap2a, shLUC/Flag-TRAPPC2 or shTRAPPC13/Flag-TRAPPC2 A549 cells were seeded in 10 cm dishes. After 24 h, the cells were collected and IP was performed as described above. Eluates were resolved on 4–12% gradient Bis-Tris gel and probed with anti-Flag, TRAPPC4 (sc-101311, Santa Cruz Biotechnology) and TRAPPC12 antibodies.
For precipitation of endogenous Rab1a in Flag-Golgin-84 or control protein overexpressing cells, 2×106 stably transduced shLUC/Flag-γTubulin, shLUC/Flag-Golgin-84, shTRAPPC13/Flag-γTubulin and shTRAPPC13/Flag-Golgin-84 A549 cells were seeded in 10 cm dishes. After 24 h, cells were treated with 20 ng/ml BFA. 48 h after seeding, the cells were collected and IP was performed as described above. Eluates were resolved and probed with anti-Flag and Rab1a antibodies.
VHS-GAT pull-down assay
ARF activity was measured using an ARF pull-down assay as described previously (Cohen and Donaldson, 2010). GST-VHS-GAT was bacterially expressed, and the cell pellet spun down at 2831 g. The cell pellet was washed with PBS and lysed by sonication in 20 ml bacterial lysis buffer (40 mM Hepes pH 7.4, 150 mM NaCl, 2 mM EDTA, 1 μM DTT and protease inhibitor cocktail). The bacterial cell lysate was then centrifuged for 30 min at 48,384 g. Glutathione agarose beads (Thermo Fisher Scientific) were washed two times with PBS and once with bacterial lysis buffer, combined with the bacterial supernatant and incubated for 1 h to bind GST-VHS-GAT to the beads. TRAPPC13 knockdown cells were grown in 15 cm dishes overnight, treated with 20 ng/ml BFA for 24 h and lysed in 1 ml 1% NP40 lysis buffer. Total cell protein extracts (2 mg) were incubated with 60 μl glutathione agarose–GST-VHS-GAT slurry for 1 h at 4°C for ARF pull down. Immunoprecipitates were washed three times in 1% NP40 lysis buffer before elution in 50–60 μl 2× sample buffer, and then boiled for 5 min. Eluates were resolved on a 4–12% gradient Bis-Tris gel and probed with anti-ARF1 and pan-ARF antibodies.
Gaussia luciferase assay
Cells were seeded at a density of 500×103 cells in 10 cm dishes, allowed to settle overnight and transfected with 200 ng pCMV-GLuc plasmid (New England Biolabs). After overnight transfection, each transfected cell line was split into two wells of a six-well plate (200×103 cells/well). The following day, one well was treated with 40 nM BFA and the other with an equal volume of vehicle (0.1% ethanol). Samples of culture supernatant (50 µl) were taken at the indicated time points and transferred to a white opaque 96-well plate. Freshly prepared Gaussia luciferase flash assay reagent (Pierce) was added (20 µl), and the luminescent signal was read after a 10 s integration time. Data are expressed as a percentage of the maximum luciferase signal after 8 h, obtained from the untreated sample of each pair per genotype (Fig. 2D; Fig. S2F). Cells stably expressing GLuc-Flag were seeded in a 96-well plate at a density of 5000 cells/well, and treated in fresh medium 24 h later. Samples of culture supernatant (50 µl) were taken at the indicated time points and transferred to a white opaque 96-well plate. Freshly prepared Gaussia luciferase flash assay reagent was added (20 µl), and the luminescent signal was read after a 10 s integration time (Fig. S2E).
Gentamicin protection assay
The bacterial strain used was S. flexneri serovar 2a WT strain 2457T. The day prior to infection, control and knockdown cell lines were seeded in 24-well plates (1×105 cells/well) in triplicate. Cells were infected with S. flexneri by centrifuging exponential phase bacteria diluted in medium onto semi-confluent monolayers of cells at an MOI of 1:1 at 700 g for 10 min. The cells were subsequently incubated at 37°C and 5% CO2 for 20 min, washed three times with PBS, and resuspended in medium with or without 20 ng/ml BFA containing 25 µg/ml gentamicin to kill extracellular bacteria. Cells were then incubated for the indicated amount of time, washed three times with PBS, and lysed in 0.1% sodium deoxycholate/PBS to assess the intracellular bacterial number. Cell lysates were plated on tryptic soy agar (TSA) and colony-forming units were counted after overnight incubation at 37°C.
Acknowledgements
We thank Cornelia Wirth and Rama Kancha for technical assistance and help with cDNA cloning; Eric Spooner for mass spectrometry analysis; and David Sabatini, Andrew Olive, Frank Adolf, Rainer Beck and Felix Wieland for support, insightful discussions and comments on the manuscript. We also thank Thijn Brummelkamp for generously providing the Rab1a GT cells.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.R.-P, T.I.I., R.K.L., J.H.R.; Validation: S.R.-P., T.I.I., E.L.S.; Formal analysis: S.R.-P., J.B.; Investigation: S.R.-P., T.I.I., B.J.v.R., J.B., M.G., J.H.R.; Resources: M.N.S.; Writing - original draft: S.R.-P., J.H.R.; Writing - review & editing: S.R.-P., T.I.I., J.H.R.; Visualization: S.R.-P.; Supervision: J.H.R.; Project administration: J.H.R.
Funding
This study was supported by Merck KGaA.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.199521.supplemental
References
- Alvarez C., Garcia-mata R., Brandon E. and Sztul E. (2003). COPI recruitment is modulated by a Rab1b- dependent mechanism. Mol. Biol. Cell 14, 2116-2127. 10.1091/mbc.E02-09-0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao X., Zou L. and Wu Y. (2014). Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 21, 348-358. 10.1038/cdd.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M. and Hannan L. A. (2000). Traffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic 1, 836-851. 10.1034/j.1600-0854.2000.011104.x [DOI] [PubMed] [Google Scholar]
- Badr C. E., Hewett J. W., Breakefield X. O. and Tannous B. A. (2007). A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS ONE 2, e571 10.1371/journal.pone.0000571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowman J., Bhandari D., Reinisch K. and Ferro-Novick S. (2010). TRAPP complexes in membrane traffic: convergence through a common Rab. Nat. Rev. Mol. Cell Biol. 11, 759-763. 10.1038/nrm2999 [DOI] [PubMed] [Google Scholar]
- Bassik M. C., Kampmann M., Lebbink R. J., Wang S., Hein M. Y., Poser I., Weibezahn J., Horlbeck M., Chen S., Mann M. et al. (2013). A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909-922. 10.1016/j.cell.2013.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C., Sowa M. E., Gygi S. P. and Harper J. W. (2010). Network organization of the human autophagy system. Nature 466, 68-76. 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen V. A., Májek P., Jae L. T., Johannes W., Nieuwenhuis J., Staring J., Van Diemen F. R., Olk N., Stukalov A., Marceau C. et al. (2015). Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092-1096. 10.1126/science.aac7557 [DOI] [PubMed] [Google Scholar]
- Bögershausen N., Shahrzad N., Chong J. X., von Kleist-Retzow J. C., Stanga D., Li Y., Bernier F. P., Loucks C. M., Wirth R., Puffenberger E. G. et al. (2013). Recessive TRAPPC11 mutations cause a disease spectrum of limb girdle muscular dystrophy and myopathy with movement disorder and intellectual disability. Am. J. Hum. Genet 93, 181-190. 10.1016/j.ajhg.2013.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröcker C., Engelbrecht-Vandré S. and Ungermann C. (2010). Multisubunit tethering complexes and their role in membrane fusion. Curr. Biol. 20, R943-R952. 10.1016/j.cub.2010.09.015 [DOI] [PubMed] [Google Scholar]
- Chen S., Cai H., Park S.-K., Menon S., Jackson C. L. and Ferro-Novick S. (2011). Trs65p, a subunit of the Ypt1p GEF TRAPPII, interacts with the Arf1p exchange factor Gea2p to facilitate COPI-mediated vesicle traffic. Mol. Biol. Cell 22, 3634-3644. 10.1091/mbc.E11-03-0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou L., Zhang Y., Leng Y., Pei X.-Y., Lin H., Jones R., Orlowski R. Z., Dai Y. and Grant S. (2014). Targeting SQSTM1/p62 induces cargo-loading failure and converts autophagy to apoptosis via NBK/Bik. Mol. Cell. Biol 34, 3435-3449. 10.1128/MCB.01383-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Davey M., Schluter C., Pandher P., Fang Y., Foster L. J. and Conibear E. (2011). Organization and assembly of the TRAPPII complex. Traffic 12, 715-725. 10.1111/j.1600-0854.2011.01181.x [DOI] [PubMed] [Google Scholar]
- Cohen L. A. and Donaldson J. G. (2010). Analysis of Arf GTP-binding protein function in cells. Curr. Protoc. Cell Biol. Chapter 3, Unit 14, 12.1-17 10.1002/0471143030.cb1412s48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao A., Rahman D., Pappin D. J. C., Lucocq J. and Lowe M. (2003). The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 160, 201-212. 10.1083/jcb.200207045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G. and Jackson C. L. (2011). ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12, 362-375. 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Zhu Y., Lu Q., Hu L., Zheng Y. and Shao F. (2012). Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150, 1029-1041. 10.1016/j.cell.2012.06.050 [DOI] [PubMed] [Google Scholar]
- Fan L., Yu W. and Zhu X. (2003). Interaction of Sedlin with chloride intracellular channel proteins. FEBS Lett. 540, 77-80. 10.1016/S0014-5793(03)00228-X [DOI] [PubMed] [Google Scholar]
- Flanagan-Steet H., Johnson S., Smith R. D., Bangiyeva J., Lupashin V. and Steet R. (2011). Mislocalization of large ARF-GEFs as a potential mechanism for BFA resistance in COG-deficient cells. Exp. Cell Res. 317, 2342-2352. 10.1016/j.yexcr.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T. and Yoshimori T. (2008). An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19, 4651-4659. 10.1091/mbc.E08-03-0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwynn B., Smith R. S., Rowe L. B., Taylor B. A. and Peters L. L. (2006). A mouse TRAPP-related protein is involved in pigmentation. Genomics 88, 196-203. 10.1016/j.ygeno.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Huang J., Birmingham C. L., Shahnazari S., Shiu J., Zheng Y. T., Smith A. C., Campellone K. G., Do Heo W., Gruenheid S., Meyer T. et al. (2011). Antibacterial autophagy occurs at PtdIns(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy 7, 17-26. 10.4161/auto.7.1.13840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyabalan J., Nesbit M. A., Galvanovskis J., Callaghan R., Rorsman P. and Thakker R. V. (2010). SEDLIN forms homodimers: characterisation of SEDLIN mutations and their interactions with transcription factors MBP1, PITX1 and SF1. PLoS ONE 5, e10646 10.1371/journal.pone.0010646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R. and Ashkenazi A. (2009). Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137, 721-735. 10.1016/j.cell.2009.03.015 [DOI] [PubMed] [Google Scholar]
- Lamb C. A., Nühlen S., Judith D., Frith D., Snijders A. P., Behrends C. and Tooze S. A. (2015). TBC 1 D 14 regulates autophagy via the TRAPP complex and ATG 9 traffic. EMBO J. 35, 281-301. 10.15252/embj.201592695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.-C., Zhu W., Mitsuhashi S., Noguchi S., Sacher M., Ogawa M., Shih H.-H., Jong Y.-J. and Nishino I. (2015). Congenital muscular dystrophy with fatty liver and infantile-onset cataract caused by TRAPPC11 mutations: broadening of the phenotype. Skelet. Muscle 5, 29 10.1186/s13395-015-0056-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E., Peter F., Subramaniam V. N. and Hong W. (2005). Mammalian Bet3 functions as a cytosolic factor participating in transport from the ER to the Golgi apparatus. J. Cell Sci. 118, 1209-1222. 10.1242/jcs.01723 [DOI] [PubMed] [Google Scholar]
- Lynch-Day M. A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C. R., Brumell J. H., Ferro-Novick S. and Klionsky D. J. (2010). Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 107, 7811-7816. 10.1073/pnas.1000063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolea F. and Melancon P. (2010). Arf3 is activated uniquely at the trans-Golgi network by brefeldin a-inhibited guanine nucleotide exchange factors. Mol. Biol. Cell 21, 1836-1849. 10.1091/mbc.E10-01-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangi G., Leuzzi V., Manti F., Lattante S., Orteschi D., Pecile V., Neri G. and Zollino M. (2013). TRAPPC9-related autosomal recessive intellectual disability: report of a new mutation and clinical phenotype. Eur. J. Hum. Genet 21, 229-232. 10.1038/ejhg.2012.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev M. P., Hasaj B., Saint-Dic D., Snounou S., Zhao Q. and Sacher M. (2015). TRAMM/TrappC12 plays a role in chromosome congression, kinetochore stability, and CENP-E recruitment. J. Cell Biol. 209, 221-234. 10.1083/jcb.201501090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N., Liang Y., Tokarev A. A., Chen S. H., Cox R., Andrejic J., Lipatova Z., Sciorra V. A., Emr S. D. and Segev N. (2006). TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat. Cell Biol. 8, 1263-1269. 10.1038/ncb1489 [DOI] [PubMed] [Google Scholar]
- Mostowy S., Boucontet L., Mazon Moya M. J., Sirianni A., Boudinot P., Hollinshead M., Cossart P., Herbomel P., Levraud J.-P. and Colucci-Guyon E. (2013). The Zebrafish as a new model for the in vivo study of shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 9, e1003588 10.1371/journal.ppat.1003588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munksgaard B., Aridor M., Hannan L. A. and Hannan L. A. (2002). Traffic jams II: an update of diseases of intracellular transport. Traffic 2, 781-790. 10.1034/j.1600-0854.2002.31103.x [DOI] [PubMed] [Google Scholar]
- Murrow L. and Debnath J. (2013). Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu. Rev. Pathol. 8, 105-137. 10.1146/annurev-pathol-020712-163918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu T.-K., Pfeifer A. C., Lippincott-Schwartz J. and Jackson C. L. (2005). Dynamics of GBF1, a Brefeldin A-sensitive arf1 exchange factor at the golgi. Mol. Biol. Cell 16, 1213-1222. 10.1091/mbc.E04-07-0599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J.-A., Ullman E., Dou Z. and Zong W.-X. (2011). Inhibition of protein degradation induces apoptosis through a microtubule-associated protein 1 light chain 3-mediated activation of caspase-8 at intracellular membranes. Mol. Cell Biol. 31, 3158-3170. 10.1128/MCB.05460-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria F. J., Schindler C., Magadán J. G., Mardones G. A., Delevoye C., Romao M., Raposo G. and Bonifacino J. S. (2010). Ang2/fat-free is a conserved subunit of the Golgi-associated retrograde protein complex. Mol. Biol. Cell 21, 3386-3395. 10.1091/mbc.E10-05-0392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling J. H., Olive A. J., Sanyal S., Carette J. E., Brummelkamp T. R., Ploegh H. L., Starnbach M. N. and Sabatini D. M. (2013). A CREB3-ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat. Cell Biol. 15, 1473-1485. 10.1038/ncb2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Shu G., Liu G., Liu D., Zhou J., Yuan L. and Zhou J. (2014). Knockdown of p62/sequestosome 1 attenuates autophagy and inhibits colorectal cancer cell growth. Mol. Cell. Biochem. 385, 95-102. 10.1007/s11010-013-1818-0 [DOI] [PubMed] [Google Scholar]
- Sambasivarao S. V. (2014). Trs20 is required for TRAPP III complex assembly at the PAS and its function in autophagy. Traffic 15, 327-337. 10.1016/j.micinf.2011.07.011.Innate [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A., Wang Y., Malsam J., Beard M. B. and Warren G. (2003). Golgin-84 is a rab1 Binding Partner Involved in Golgi Structure. Traffic 4, 153-161. 10.1034/j.1600-0854.2003.00103.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrivens P. J., Noueihed B., Shahrzad N., Hul S., Brunet S. and Sacher M. (2011). C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol. Biol. Cell 22, 2083-2093. 10.1091/mbc.E10-11-0873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöter M., Niederlein A., Barsacchi R., Meyenhofer F., Brandl H. and Bickle M. (2013). CellProfiler and KNIME: Open source tools for high content screening. Methods Mol. Biol. 986, 105-122. 10.1007/978-1-62703-311-4_8 [DOI] [PubMed] [Google Scholar]
- Thomas L. L. and Fromme J. C. (2016). GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J. Cell Biol. 215, 499-513. 10.1083/jcb.201608123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti R., Scanu T., Santoro M., Di Tullio G., Spaar A., Gaibisso R., Beznoussenko G. V., Mironov A. A., Mironov A., Zelante L. et al. (2012). Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science 337, 1668-1672. 10.1126/science.1224947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viiri J., Hyttinen J. M. T., Ryhänen T., Rilla K., Paimela T., Kuusisto E., Siitonen A., Urtti A., Salminen A. and Kaarniranta K. (2010). P62/Sequestosome 1 as a regulator of proteasome inhibitor-induced autophagy in human retinal pigment epithelial cells. Mol. Vis. 16, 1399-1414. [PMC free article] [PubMed] [Google Scholar]
- Wang W., Sacher M. and Ferro-Novick S. (2000). Trapp stimulates guanine nucleotide exchange on Ypt1p. J. Cell Biol. 151, 289-296. 10.1083/jcb.151.2.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C. P., Smith E. F., Bauer C. S., Moller A., Guillaume M., Ferraiuolo L., Myszczynska M. A., Higginbottom A., Walsh M. J., Whitworth J. et al. (2016). The C 9 orf 72 protein interacts with Rab 1 a and the ULK 1 complex to regulate initiation of autophagy. EMBO J. 35, 1656-1676. 10.15252/embj [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y.-R., Kong X., Yu Y.-N., Wang Y.-C., Hong J., Zhao S.-L. and Fang J.-Y. (2014). The Role of ERK2 in colorectal carcinogenesis is partly regulated by TRAPPC4. Mol. Carcinog. Suppl 1, E72-84. 10.1002/mc.22031 [DOI] [PubMed] [Google Scholar]
- Winslow A. R., Chen C.-W., Corrochano S., Acevedo-Arozena A., Gordon D. E., Peden A. A., Lichtenberg M., Menzies F. M., Ravikumar B., Imarisio S. et al. (2010). α-Synuclein impairs macroautophagy: Implications for Parkinson's disease. J. Cell Biol. 190, 1023-1037. 10.1083/jcb.201003122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki A., Menon S., Yu S., Barrowman J., Meerloo T., Oorschot V., Klumperman J., Satoh A. and Ferro-novick S. (2009). mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol. Biol. Cell 20, 4205-4215. 10.1091/mbc.E09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.-L., Shintani T., Nair U., Cao Y., Richardson B. C., Li Z., Hughson F. M., Baba M. and Klionsky D. J. (2010). The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 188, 101-114. 10.1083/jcb.200904075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. M., Takahashi Y., Khan O., Park S., Hori T., Yun J., Sharma A. K., Amin S., Hu C.-D., Zhang J. et al. (2012). Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J. Biol. Chem. 287, 12455-12468. 10.1074/jbc.M111.309104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. and Liang Y. (2012). A trapper keeper for TRAPP, its structures and functions. Cell. Mol. Life Sci. 69, 3933-3944. 10.1007/s00018-012-1024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Vicinanza M. and Rubinsztein D. C. (2013). Biology and trafficking of ATG9 and ATG16L1, two proteins that regulate autophagosome formation. FEBS Lett. 587, 1988-1996. 10.1016/j.febslet.2013.04.025 [DOI] [PubMed] [Google Scholar]
- Zhang C.-J., Bowzard J. B., Greene M., Anido A., Stearns K. and Kahn R. A. (2002). Genetic interactions link ARF1, YPT31/32 and TRS130. Yeast 19, 1075-1086. 10.1002/yea.903 [DOI] [PubMed] [Google Scholar]
- Zong M., Wu X. G., Chan C. W. L., Choi M. Y., Chan H. C., Tanner J. A. and Yu S. (2011). The adaptor function of TRAPPC2 in mammalian trapps explains TRAPPC2-associated sedt and TRAPPC9-associated congenital intellectual disability. PLoS ONE 6, e23350 10.1371/journal.pone.0023350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppino F. C. M., Militello R. D., Slavin I., Álvarez C. and Colombo M. I. (2010). Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11, 1246-1261. 10.1111/j.1600-0854.2010.01086.x [DOI] [PubMed] [Google Scholar]
- Zou S., Liu Y., Zhang X. Q., Chen Y., Ye M., Zhu X., Yang S., Lipatova Z., Liang Y. and Segev N. (2012). Modular TRAPP complexes regulate intracellular protein trafficking through multiple Ypt/Rab GTPases in Saccharomyces cerevisiae. Genetics 191, 451-460. 10.1534/genetics.112.139378 [DOI] [PMC free article] [PubMed] [Google Scholar]