ABSTRACT
The GIT proteins, GIT1 and GIT2, are GTPase-activating proteins (inactivators) for the ADP-ribosylation factor (Arf) small GTP-binding proteins, and function to limit the activity of Arf proteins. The PIX proteins, α-PIX and β-PIX (also known as ARHGEF6 and ARHGEF7, respectively), are guanine nucleotide exchange factors (activators) for the Rho family small GTP-binding protein family members Rac1 and Cdc42. Through their multi-domain structures, GIT and PIX proteins can also function as signaling scaffolds by binding to numerous protein partners. Importantly, the constitutive association of GIT and PIX proteins into oligomeric GIT–PIX complexes allows these two proteins to function together as subunits of a larger structure that coordinates two distinct small GTP-binding protein pathways and serves as multivalent scaffold for the partners of both constituent subunits. Studies have revealed the involvement of GIT and PIX proteins, and of the GIT–PIX complex, in numerous fundamental cellular processes through a wide variety of mechanisms, pathways and signaling partners. In this Commentary, we discuss recent findings in key physiological systems that exemplify current understanding of the function of this important regulatory complex. Further, we draw attention to gaps in crucial information that remain to be filled to allow a better understanding of the many roles of the GIT–PIX complex in health and disease.
KEY WORDS: ArfGAP, GIT1, GIT2, RhoGEF, α-PIX, β-PIX
Summary: In this Commentary, we discuss recent findings in key physiological systems that exemplify the current understanding of the functions of GIT, PIX and the GIT–PIX complex.
Introduction
Cell-surface receptors activate intracellular signaling through mediators such as small GTP-binding proteins, heterotrimeric G-proteins and protein kinases. Individual receptors often activate multiple downstream pathways and cells are generally stimulated by more than one kind of signal at once. Cells must integrate these multiple signals in order to perform their specialized functions. The GIT–PIX complex is one prominent mechanism that cells utilize to integrate signals from multiple GTP-binding protein and protein kinase pathways to control cell polarity, adhesion and migration (Hoefen and Berk, 2006; Rosenberger and Kutsche, 2006; Randazzo et al., 2007; Frank and Hansen, 2008). Here, we discuss exciting new data that identified so-far-unknown pathways regulated by this complex, its physiological roles in neurons and immune cells in vivo, and its dysregulation in cancer.
GIT proteins
The GIT proteins, GIT1 and GIT2, are GTPase-activating proteins (GAPs) for ADP-ribosylation factor (Arf) small GTP-binding proteins (Premont et al., 1998; Vitale et al., 2000) and belong to the family of ArfGAP proteins (Kahn et al., 2008). Because Arf proteins have no intrinsic GTPase activity, active GTP-bound Arf proteins require ArfGAP proteins to deactivate them by converting bound GTP to GDP (Randazzo et al., 1994). Mammals express two GIT proteins, GIT1 and GIT2, whereas zebrafish –because they have two GIT2 genes (git2a and git2b) – express three GIT proteins (Yu et al., 2011). Caenorhabditis elegans and Drosophila melanogaster have each only a single GIT gene (git-1 and Git, respectively) (Lucanic and Cheng, 2008; Bahri et al., 2009). Mammalian GIT1 was first identified as a binding partner for G-protein-coupled receptor (GPCR) kinases (GRKs), and named GRK-interacting partner 1 (GIT1) (Premont et al., 1998). GIT1 and GIT2 also were identified soon after by other groups using distinct approaches, and were given different names [e.g. cool-associated tyrosine phosphorylated proteins 1 and 2 (Cat-1 and Cat-2) (Bagrodia et al., 1999); p95 paxillin-kinase linker (p95PKL) (Turner et al., 1999); or p95-ADP ribosylation factor GTPase-activating protein, PIX-, paxillin-interacting proteins 1 and 2’ (p95-APP1 and -2) (Di Cesare et al., 2000). Now, GIT1 and GIT2 are the official protein symbols, and most commonly used (see also Box 1).
Box 1. GIT and PIX gene expression.
GIT and PIX proteins are present in all cells, although individual isoforms exhibit tissue-specific expression. Northern blots of organ tissues derived from human and rat have shown that both GIT1 and GIT2 are ubiquitously expressed, with the GIT2-short variant specifically enriched in immune cells (Premont et al., 1998, 2000). Expression of GIT1 and GIT2 within tissues was compared directly by using gene trap LacZ reporter mice; GIT2 is widely expressed in all tissue, whereas GIT1 is highly expressed in the vasculature and ductular cells of many organs (Schmalzigaug et al., 2007b). GIT1 and GIT2 are widely expressed throughout brain, except for the lack of GIT1 in cerebellar granule cells. Expression of PIX proteins has been mapped by using northern blotting of tissue RNAs and western blotting of tissue lysates, so intra-tissue differences remain mostly unknown. Gene expression of β-PIX is ubiquitous, whereas that of α-PIX is more restricted, with high expression in heart, skeletal muscle and immune cells (Manser et al., 1998).
Three distinct lines of global GIT1-knockout mice have poor post-natal survival (Pang et al., 2009; Schmalzigaug et al., 2009a; Won et al., 2011), whereas two distinct strains of mice lacking GIT2 survive normally (Mazaki et al., 2006; Schmalzigaug et al., 2009b). β-PIX knockout mice exhibit embryonic lethality (Omelchenko et al., 2014), whereas α-PIX-deficient mice survive normally (Missy et al., 2008). In the β-PIX-deficient mouse, lethality results from defective cell migration during early embryogenesis (Omelchenko et al., 2014). Experiments with GIT2 in Xenopus (Köster et al., 2010) and GIT2a in zebrafish (Yu et al., 2011) demonstrated cell migration defects during gastrulation. GIT1-knockout lethality has been reported to result from poor angiogenesis in the lungs (Pang et al., 2009). Interestingly, in zebrafish the mutant bubblehead with a vascular leakage phenotype is a hypomorphic allele of β-PIX (Liu et al., 2007).
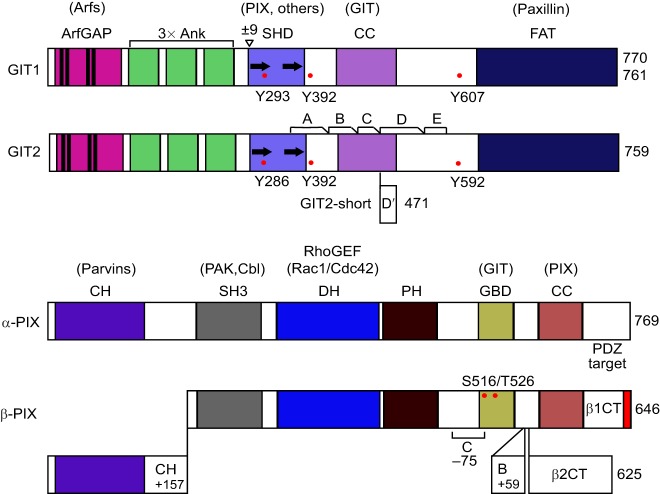
The GIT proteins share a conserved domain architecture, which includes the N-terminal zinc finger ArfGAP domain, three ankyrin repeats, a region containing two near-repeats with homology with the yeast Spa2 protein (Spa2-homology domain, SHD), a coiled-coil, a poorly conserved linker region and a focal adhesion targeting (FAT) domain (Fig. 1). Purified GIT proteins deactivate all Arf proteins (Vitale et al., 2000), but are linked functionally to plasma membrane Arf6 as found in cellular studies (Di Cesare et al., 2000; Claing et al., 2000; Meyer et al., 2006; Miura et al., 2009; Jones et al., 2009). Over 100 GIT-associated proteins and dozens of direct partners have been described (Table SI). The most prominent GIT-binding partners are the p21-activated kinase-interacting exchange factors α-PIX and β-PIX (Bagrodia et al., 1999; Zhao et al., 2000; Premont et al., 2000), which bind to the SHD region. The coiled-coil domain is responsible for dimerization of GIT proteins by forming a parallel coiled-coil (Premont et al., 2004; Schlenker and Rittinger, 2009). The FAT domain is a four-helix bundle and the binding site for the focal adhesion adaptor protein paxillin in a manner that is similar to paxillin binding to the FAT domain of focal adhesion kinase (Schmalzigaug et al., 2007a; Zhang et al., 2008).
Fig. 1.
Domain structure of GIT and PIX proteins, including alternative splicing isoforms. GIT1 and GIT2 share the N-terminal ArfGAP domain, three ankyrin repeats (Ank), the Spa2 homology domain (SHD) – as indicated with two Spa2-like repeats – a coiled-coil (CC) dimerization domain and the C-terminal focal adhesion-targeting (FAT) domain. For GIT1, alternative splicing of a single exon encoding nine amino acids (±9) at the start of the SHD can occur. For GIT2, alternative splicing of five internal in-frame regions (A,B,C,D,E), as well as of a distinct exon (D′) that leads to a truncated C-terminus lacking the FAT domain is possible. α-PIX and β-PIX share the calponin homology (CH) domain, SH3 domain, Dbl homology (DH) domain, pleckstrin homology (PH) domain, a GIT-binding domain (GBD) and a CC domain for trimerization. α-PIX has not been reported to undergo alternative splicing, but β-PIX has alternative N- and C-termini (β1CT and β2CT), and can undergo internal alternative splicing. β1-PIX variants have a PDZ target motif at the extreme C-terminus for binding to PDZ-domain proteins, but β2-PIX variants have a distinct C-terminal sequence that lacks the PDZ target and the coiled-coil domain. Adult β-PIX lacks the N-terminal CH domain that is found in embryonic β-PIX. β-PIX-c lacks the region (denoted as C) between the PH and GBD, whereas β-PIX-b has a sequence inserted (denoted as B) between the GBD and CC domain. Prominent phosphorylation sites are shown in red, and domain partners in parentheses above.
GIT1 has two splice variants in human and mouse due to an alternative exon that encodes nine amino acids at the start of the SHD region (in rat, this sequence is fused to the adjacent exon so that rat GIT1 is always the longer variant). Mammalian GIT2 undergoes extensive alternative splicing (Premont et al., 2000). GIT2-short, a GIT2 variant with a truncated C-terminus that lacks the FAT domain, is abundant in immune cells. GIT2 internal splicing involves five contiguous in-frame blocks of sequence (called A-E) including most of the non-conserved linker region encoded by four exons that are spliced independently. The functional significance of GIT variants remains unclear, beyond GIT2-short being unable to bind paxillin through the (missing) FAT domain – although GIT2-short has been reported to weakly bind to paxillin anyway (Mazaki et al., 2001).
PIX proteins
The two mammalian PIX proteins α-PIX and β-PIX (also known as ARHGEF6 and ARHGEF7, respectively) are guanine nucleotide exchange factors (GEFs, activators) of small GTP-binding proteins that are members of the Rho family (Manser et al., 1998). C. elegans and Drosophila have single PIX genes, PIX-1 and dPIX, respectively (Lucanic and Cheng, 2008; Parnas et al., 2001). PIX proteins were also given alternative names: 85 kDa SH3 domain-containing proline-rich protein (p85SPR) (Oh et al., 1997), and cloned out of library-1 and -2 (cool-1 and cool-2) (Bagrodia et al., 1998), although the most commonly used names are α-PIX and β-PIX (see also Box 1).
The two PIX proteins share a conserved domain structure (Fig. 1). The N-terminus of α-PIX contains a calponin homology (CH) domain that is present in β-PIX forms early in development, whereas the SH3 domain is at the N-terminus of adult β-PIX forms. PIX SH3 domains bind to p21-activated protein kinases 1–3 (PAK1, PAK2, PAK3) (Manser et al., 1998). The Dbl homology (DH) and pleckstrin homology (PH) domains function as a RhoGEF. PIX proteins activate p21 Rac1 and Cdc42, but not RhoA (Manser et al., 1998) and, interestingly, it is Rac1 and Cdc42 that activate PAKs (Manser et al., 1994), making Rac1–(PIX–PAK) and Cdc42–(PIX–PAK) complexes a common functional unit in many reports. The GIT-binding domain (GBD) is an unstructured region that binds to the SHD of GIT proteins (Zhao et al., 2000). The coiled-coil forms a parallel trimer, indicating that PIX-PIX self-association is trimeric (Schlenker and Rittinger, 2009; Im et al., 2010). Finally, one C-terminal variant of β-PIX ends with a PDZ target motif that binds to PDZ proteins, including shank and scribble (Park et al., 2003; Audebert et al., 2004). A large number of α-PIX and β-PIX partners have been identified (Table SI). Whereas α-PIX comprises one isoform containing all domains, β-PIX exhibits complex alternative splicing and multiple bands are found following western blotting of cell extracts. In addition to alternative transcription start sites that include or skip the CH domain, there are two distinct C-termini, with or without the PDZ target motif, as well as variants that lack the coiled-coil region (Kim et al., 2000; Koh et al., 2001). These distinct forms are still mostly uncharacterized.
The GIT–PIX complex
Oligomeric partners
The GIT–PIX complex functions as a scaffold and coordination center that can be recruited for multiple signaling pathways, and moves to several locations in the cell in response to specific signals. Once in place at the membrane, the complex dampens Arf signaling and increases Rac1 and/or Cdc42 activity (Fig. 2), and also serves as a localized scaffold for associated signaling pathways (Fig. 3).
Fig. 2.
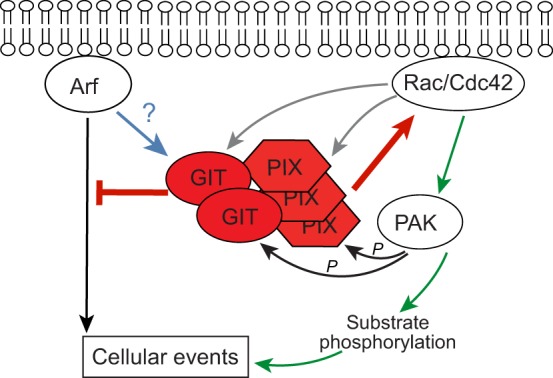
Oligomeric GIT-PIX complexes (GIT2–PIX3) regulate pathways that are mediated by membrane-associated Arf and Rac/Cdc42 small GTPases. GIT proteins hydrolyse GTP bound to membrane-associated active Arf proteins, inhibiting Arf-activated signaling events (red inhibitory line). PIX proteins activate membrane-associated Rac or Cdc42 (Rac/Cdc42) by facilitating both the release of GDP from inactive Rac/Cdc42 and the binding of GTP (red arrow). Active Rac/Cdc42 binds to and activates effectors (green arrow), including PIX-bound PAK, that then phosphorylate substrates to regulate cellular events (black arrow). PAK can also phosphorylate GIT and PIX (black arrows), whereas active Rac/Cdc42 binds to GIT and PIX proteins (grey arrows), suggesting that they are potential Rac/Cdc42 effectors. The net effect of these two types of feedback event remains unclear. Similarly, although ArfGAP proteins have been suggested to act as effectors for activated Arf, it remains unknown whether binding of active Arf to GIT alters other GIT functions (blue arrow). Whether ArfGAP and RhoGEF activities are integrated within the complex is currently unclear.
Fig. 3.
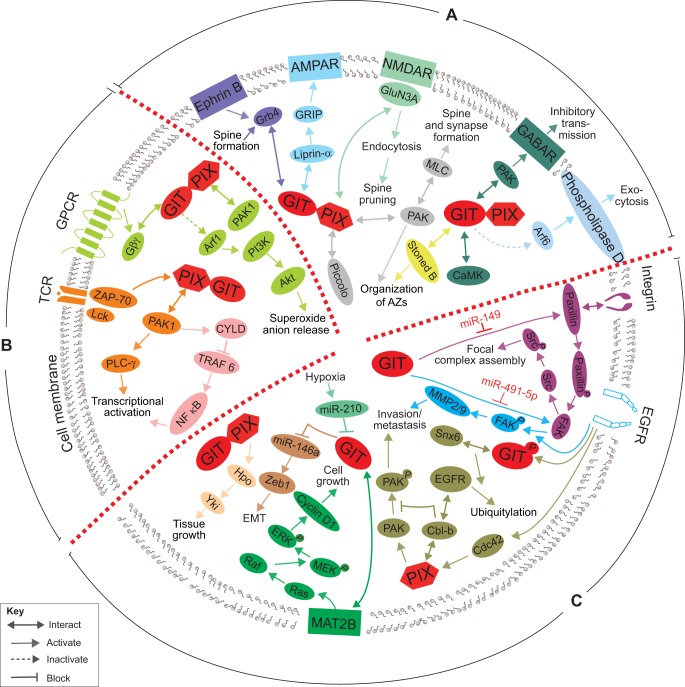
GIT and PIX signaling partners and pathways. Important GIT and PIX partners and pathways are summarized according to their functions in the nervous system (A), immune system (B), and in cancer (C). Arrows of the same colour indicate individual pathways. AMPR, AMPA-type glutamate receptor; Arf, ADP ribosylation factor; AZs, synaptic release active zones; CaMK, Ca2+-dependent protein kinase; CYLD, cylindromatosis lysine 63 deubiquitinase; EGFR, epidermal growth factor receptor; EMT, epithelial–mesenchymal transition; FAK, focal adhesion kinase; GABAR, GABA receptor; GPCR, G-protein-coupled receptor; GRK, GPCR kinase; MLC, myosin II regulatory light chain; NMDAR, NMDA-type glutamate receptor; PAK, p21-activated kinase; PIX, PAK-interacting exchange factor; TCR, T-cell receptor; TKR, tyrosine kinase receptor; TLR, toll-like receptor; TRAF6, TNF receptor-associated factor 6; Cbl-b, Casitas B-lineage lymphoma b; Snx6, sorting nexin 6.
GIT and PIX proteins are unique among the ArfGAPs and RhoGEFs – and, indeed, among signaling regulators in general – in forming a constitutively associated oligomeric complex able to regulate two distinct GTP-binding protein families. Because GIT and PIX proteins each constitutively self-associate through coiled-coils, they form GIT–PIX oligomers (Premont et al., 2004; Schlenker and Rittinger, 2009). In size-exclusion chromatography, native or recombinant GIT–PIX complexes elute together at an apparent mass of ∼1 MDa (Paris et al., 2003; Premont et al., 2004; Totaro et al., 2012). Coiled-coil structures suggest that one GIT dimer binds one PIX trimer to form a five-subunit complex (GIT2–PIX3; Fig. 2) (Schlenker and Rittinger, 2009), although this has not been demonstrated directly and multimers may exist. Pulldown of GIT or PIX quantitatively captures the other partner (Premont et al., 2004). GIT and PIX appear unstable when not in a complex (Premont et al., 2004) because T-cells of α-PIX-deficient mice have reduced levels of GIT2 (Missy et al., 2008), GIT1-deficient mice have reduced levels of PIX in the brain (Won et al., 2011) and GIT2-deficient mice have reduced levels of PIX in immune organs (Hao et al., 2015). Nevertheless, despite clear evidence that GIT and PIX are tightly associated and function together, many publications continue to report on GIT or PIX proteins individually, ignoring potential involvement of the other partner.
The formation of oligomeric GIT–PIX complexes has several implications, some of which remain to be experimentally tested. First, by forming a tight complex, GIT and PIX proteins simultaneously scaffold the partners of either subunit and, potentially multiple copies per complex. For example, the PIX partner PAK can readily be found in association with GITs following co-immunoprecipitation (Zhao et al., 2000; Premont et al., 2000). Therefore, studies reporting the association of a protein with GIT or PIX need to be interpreted with caution when only one of them has been tested, until direct protein–protein interaction is determined (see Table SI).
Next, targeted subcellular localization of one subunit controls the localization of the other. GIT–PIX complexes are found in many locations, including focal adhesion complexes and leading edges in migrating cells, on intracellular vesicles, in neuronal synapses, associated with centrioles, as well as bound to nuclear DNA at sites of damage (Frank and Hansen, 2008; Lu et al., 2015). GIT–PIX complexes are dynamic within cells and exhibit high mobility among compartments, such as when cells adhere or migrate (Manabe et al., 2002) or during activation of chromaffin cells to release stored vesicles (Meyer et al., 2006). GIT and PIX each bind partner proteins that localize the complex to specific cellular locations. These proteins include the GIT partners paxillin (to focal adhesions; Turner et al., 1999), piccolo and stonin-2 (to presynaptic active zones in neurons; Kim et al., 2003; Podufall et al., 2014) and liprin-α (to postsynaptic membranes; Ko et al., 2003), as well as the β-PIX PDZ partners scribble (to epithelial cell–cell contacts and synapses; Audebert et al., 2004; Richier et al., 2010) and shank (to postsynaptic membranes; Park et al., 2003).
Furthermore, the tight linkage of an ArfGAP and RhoGEF suggests a coordination of GAP and GEF functions. In GIT–PIX-dependent systems, elevated Rac1 and/or Cdc42 activity has been found to be associated with reduced Arf6 activity (Albertinazzi et al., 2003; Hajdo-Milasinovic et al., 2009; Jones et al., 2009; Bhanot et al., 2010). However, a coordinated regulation, as opposed to merely a colocalization of GAP and GEF functions, has not been demonstrated.
In addition to the above, GIT–PIX has a complex relationship with small GTP-binding proteins beyond just regulating Arf, Rac and/or Cdc42. Although it has been suggested that ArfGAP proteins act as Arf effectors (Zhang et al., 1998; Kon et al., 2011), it remains unclear whether activated Arf alters the function of GIT or PIX. Similarly, Rac1 and/or Cdc42, and their PAK effectors have been reported to bind to or affect GIT–PIX. But, in addition to PIX binding to inactive Rac1 and/or Cdc42 during activation, active Rac1 and/or Cdc42 (and other Rho family members) have been reported to also bind to GIT–PIX (Brown et al., 2002; Shin et al., 2004; Gu et al., 2006; ten Klooster et al., 2006; Hajdo-Milasinovic et al., 2009; Tay et al., 2010; Wilson et al., 2014). The effects binding of active Rho family proteins has to GIT–PIX remain ill-defined and appear somewhat contradictory because GIT and PIX have each been reported to directly bind Rac1 and/or Cdc42 in cell-based studies where both are present, clouding interpretation. Active Cdc42 was reported to stimulate PIX GEF activity toward Rac1 (Feng et al., 2004; Baird et al., 2005). GIT and PIX can be phosphorylated by PAK (Shin et al., 2002; Webb et al., 2006; Rennefahrt et al., 2007). These multiple feedback loops may amplify local Rac1– and Cdc42–PAK signaling in complex ways (Li et al., 2003; Feng et al., 2006; Mazaki et al., 2006). Furthermore, whereas GIT–PIX has been reported to scaffold many signaling molecules, it remains unclear which partners bind simultaneously and may be able to function together.
Finally, although GIT and PIX are tightly associated, there are reports suggesting that GIT or PIX proteins also act independently (Loo et al., 2004; Feng et al., 2004; Fiuza et al., 2013). As most studies employ the overexpression of GIT or PIX in cells in the presence of the endogenous proteins, accurately interpreting such results can be difficult. For example, the conclusion that GIT1 activates PAK independently of PIX is based on experiments using GIT and PIX mutants that, in protein overlay assays, have been reported to prevent their interaction (Loo et al., 2004); however, these mutants do not inhibit binding to each other in cells (Premont et al., 2004). Another potential explanation for any apparent independent functions is that specific splice variants, such as β2-PIX variants that lack the coiled-coil (Koh et al., 2001), may not form stable GIT–PIX complexes. Further experiments are needed to investigate the structure and dynamics of the GIT–PIX complex, to fully understand the regulation of complex formation and to clarify whether GIT or PIX forms have independent functions.
GIT–PIX complexes: a node for signal integration
The best-known role for GIT–PIX is that in cell adhesion and migration, where binding of GITs to the focal adhesion adaptor protein paxillin localizes GIT–PIX and associated PAKs to integrin-anchored focal adhesions that are forming or dissolving during cell motility (reviewed by Hoefen and Berk, 2006; Frank and Hansen, 2008). In this role, GIT–PIX serves as a dynamic link between cell adhesion and/or motility and cytoskeletal changes.
The GIT–PIX complex is an effector of upstream signaling that is initiated by GPCRs, receptor tyrosine kinases (RTK) and integrins (Frank and Hansen, 2008). These signals are mediated by heterotrimeric G-proteins (Li et al., 2003; Seo et al., 2004; Chahdi et al., 2005; Chahdi and Sorokin, 2010a,b), by tyrosine phosphorylation through RTKs and non-receptor tyrosine kinases including Src and focal ahesion kinase (Brown et al., 2005; Zhang et al., 2005), and by serine/threonine kinases, such as CaM kinase II and protein kinases A, C and D (Chahdi and Sorokin, 2008; Saneyoshi et al., 2008; Huck et al., 2012; Kong et al., 2014; Shirafuji et al., 2014; Nagy et al., 2015). GITs are also prominent substrates for Src family kinases (Haendeler et al., 2000; Brown et al., 2005; Heckel et al., 2009). Src/FAK-mediated phosphorylation of GIT2 at amino acid residues Y286, Y392 and Y592 is required for binding to paxillin at focal adhesions (Brown et al., 2005) but the cognate residues in GIT1 are not necessary for paxillin association (Schmalzigaug et al., 2007a). However, both GIT1 and GIT2 phosphorylated at Y392 bind to the SH2-SH3 adaptor proteins Nck1 and Nck2 (Frese et al., 2006; Segura et al., 2007). Phosphorylated GIT–PIX also binds to 14-3-3 proteins, with GIT2 phosphorylated at S415 being a main 14-3-3-binding site (Angrand et al., 2006; Chan et al., 2011). In β-PIX, amino acid residues S516 and T526 are also main sites for PKA phosphorylation (Chahdi and Sorokin, 2008). How these many events interact remains unknown.
In mammals, GIT1 functions as a scaffold for the Ras–Raf–MEK–ERK pathway after its stimulation by the angiotensin-II–GPCR pathway or the epidermal growth factor (EGF) receptor pathway (Yin et al., 2004, 2005; Zhang et al., 2009). Phosphorylation of GIT1 by Src kinase promotes its interaction with MEK1 (Zhang et al., 2009). GIT2 and PIX involvement remains unexplored, but PAK phosphorylation is known to facilitate MEK–ERK1/2 coupling (Frost et al., 1997; Eblen et al., 2002), suggesting that scaffolding through GIT1 directs PAK-mediated phosphorylation of MEK1.
In Drosophila and mammals, GIT–PIX acts as a scaffold for the Hippo signaling pathway by binding to all three elements of this kinase pathway: Hpo/MST1/2, Wts/LATS1/2 and the transcription coactivator Yki/YAP1 (Heidary Arash et al., 2014; Dent et al., 2015). Active MST1 phosphorylates LATS that, in turn, phosphorylates YAP1, thereby targeting it for degradation. In Drosophila, GIT–PIX scaffolding of the Hippo pathway mediates the activation of Hpo/MST1/2 in response to cadherin cell–cell adhesion signals to reduce tissue growth (Dent et al., 2015).
Compared with the studies addressing the GEF and scaffolding functions, there are fewer reports that examine the ArfGAP activity of GITs or the GIT–PIX complex, possibly owing to the unknowing use of GAP-deficient GIT constructs in many studies. We found that GIT proteins that bear tags at their N-termini, including Flag or 6xHis, lack the ability to act as functional ArfGAPs (Fig. 4). Therefore, in all our studies GIT proteins are tagged at the C-terminus, which does not affect their ArfGAP activity (Premont et al., 1998; Vitale et al., 2000) and does not interfere with their binding to paxillin or association with focal adhesions (Schmalzigaug et al., 2007a).
Fig. 4.
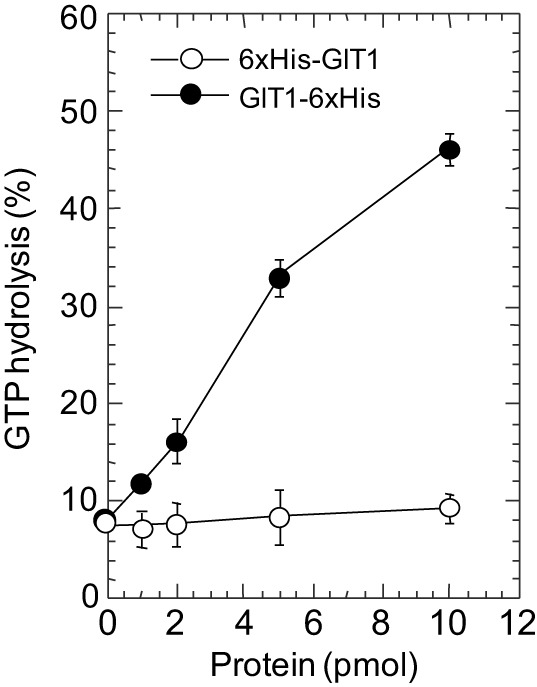
The ArfGAP activity of GIT1 is blocked by an N-terminal tag. Purified bacterial Arf1 preloaded with [α32P]GTP was mixed with the indicated amount of baculoviral-expressed GIT1-6xHis (C-terminal tag) or 6xHis-GIT1 (N-terminal tag) purified from Sf9 cells, and Arf-bound nucleotide (GTP and GDP) was separated by thin-layer chromatography. Whereas GIT1-6xHis catalyzes conversion of GTP to GDP, 6xHis-GIT1 does not, indicating that the N-terminal epitope tag interferes with the ability of GIT1 to recognize or interact with GTP-bound Arf. Similar effects have been noticed by using GIT1 tagged with GST or Flag tag (our unpublished observations). We thank Nicolas Vitale for performing this assay.
Myriad studies have identified new GIT- and PIX-binding partners and functions, expanding the potential roles of these two proteins. However, only few studies have integrated these new findings with prior knowledge, so there is a significant gap in our understanding of how partners and pathways might function together or whether, indeed, they do. However, studies in model organisms that lack GIT or PIX proteins are highlighting the physiological functions regulated by GIT–PIX in particular organs and systems.
Functions in the nervous system
Because ARHGEF6 (α-PIX) (Kutsche et al., 2000) and PAK3 (Allen et al., 1998) are human X-linked intellectual disability genes, GIT proteins have been expected to regulate cognition. Indeed, loss of GIT1 in three distinct knockout mice strains leads to severe defects in learning and memory (Schmalzigaug et al., 2009a; Menon et al., 2010; Won et al., 2011; Hong and Mah, 2015), similar to the cognitive defects in mice that lack α-PIX (Ramakers et al., 2012), or in PAK1/PAK3-double-knockout mice (Huang et al., 2011). Cognitive functions in GIT2- or β-PIX-deficient mice have not been reported but GIT2-knockout mice exhibit anxiety-like behavior (Schmalzigaug et al., 2009b). Loss of GIT1 in mice also leads to microcephaly due to a reduction in neuron size rather than neuron number (Hong and Mah, 2015), which is similar to the lack of PAK1 plus PAK3 in mice (Huang et al., 2011). Drosophila lacking dGIT also have a smaller mushroom body (brain) (Hong and Mah, 2015).
GIT1 has been associated with attention deficit hyperactivity disorder (ADHD) in mice and humans (Lee and Silva, 2011). A specific Git1 gene trap (knockout) mouse strain has been reported to have an ADHD-like phenotype that is characterized by hyperactivity and psychostimulant-induced locomotor calming (Won et al., 2011). However, a different knockout mouse strain with no interfering markers did not display hyperactivity (Schmalzigaug et al., 2009a). Analysis of flies lacking dGIT also failed to demonstrate hyperactivity (Klein et al., 2015). One single-nucleotide polymorphism (SNP) in the human GIT1 gene has been associated with ADHD in a Korean cohort (Won et al., 2011), but studies of GIT1 SNPs found no association with ADHD in a Brazilian cohort (Salatino-Oliveira et al., 2012) or in three distinct cohorts in a large-scale study (Klein et al., 2015). Therefore, at this time, this association remains controversial.
GIT–PIX has also been associated with neurodegenerative diseases. Brain tissue from Huntington's disease patients showed substantial accumulation of a C-terminal proteolytic fragment of GIT1, and GIT1 interacts with huntingtin (and several associated proteins) and enhances the aggregation of mutant huntingtin (Goehler et al., 2004). α-PIX (Eriguchi et al., 2010) and PAK1 (Luo et al., 2008) also interact with huntingtin and enhance its aggregation, suggesting that GIT1, α-PIX and PAK1 function together in the pathology of Huntington's. Mutations in the leucine-rich-repeat kinase 2 (LRRK2) are the most common genetic defect in Parkinson's disease, a motor disability resulting from death of neurons within the striatum (Wallings et al., 2015). LRRK2 has both protein kinase activity and a small GTP-binding protein-like domain. Interestingly, β-PIX is a GEF for the LRRK2 GTPase domain and is phosphorylated by LRRK2 (Haebig et al., 2010; Häbig et al., 2013). Casein-kinase-1α-mediated phosphorylation of LRRK2 reduced β-PIX binding and increased the amount of GTP-bound LRRK2 (Chia et al., 2014). Although GIT proteins have not been directly associated with LRRK2, it is noteworthy that the LRRK2 GTPase domain has been reported to be de-activated by other ArfGAPs (Xiong et al., 2012; Stafa et al., 2012).
GIT–PIX is found in both excitatory and inhibitory neurons, both presynaptically and postsynaptically, consistent with multiple roles in neurotransmission. GIT and PIX regulate migration of neurons, neuritogenesis and branching of neuronal processes (Zhang et al., 2003, 2005; Dyer et al., 2010; Totaro et al., 2012; Häbig et al., 2013; Santiago-Medina et al., 2013). Interestingly, neuronal axon branching in primary rat hippocampal neurons required α-PIX and GIT2 but not β-PIX and GIT1 (Totaro et al., 2012), suggesting a specificity whose molecular basis remains unexplored. The release of neurotransmitter vesicles is restricted to the presynaptic active zone through a macromolecular complex called the presynaptic cytoplasmic matrix or cytomatrix (Gundelfinger and Fejtova, 2012). GIT1, in complex with β-PIX and paxillin, binds the cytomatrix proteins piccolo (Kim et al., 2003) and stonin-2 (Podufall et al., 2014) to regulate localized neurotransmitter vesicle release and reuptake. In Drosophila, dGIT interacts with stoned B at the periphery of the synaptic active zone, and dgit mutant flies have defective synaptic vesicle recycling (Podufall et al., 2014). In the mouse calyx of Held synapse, presynaptic GIT1, or GIT1 together with GIT2, constrains the probability of synaptic vesicle release without altering the readily releasable pool of vesicles; however, GIT2 alone has no significant effect (Montesinos et al., 2015). In adrenal chromaffin cells, neuroendocrine vesicle exocytosis requires both GIT1 and β-PIX, which regulate Arf6 activity to control phospholipase D that promotes vesicle fusion with the membrane (Meyer et al., 2006; Momboisse et al., 2009). β-PIX also contributes to presynaptic function by regulating vesicle targeting (Sun and Bamji, 2011).
Post-synaptic neurotransmitter receptors are localized on dendritic spines, thin projections from neurons containing the post-synaptic density, a specialized network of proteins that enables clustering of neurotransmitter receptors in close proximity to the site of neurotransmitter release from presynaptic neurons (Carlisle and Kennedy, 2005). Dendritic spines undergo rapid actin-based shape changes, and actin regulators are important modulators of spine morphology and synapse formation. GIT and PIX proteins act as key regulators of spine morphology and synapse formation (see Frank and Hansen, 2008). Mice deficient in GIT1 have reduced dendritic spine density in vivo (Menon et al., 2010), whereas α-PIX-deficient mice show increased spine density with an increase in spine length and branching (Ramakers et al., 2012). There are no reports of the effects of GIT2 or β-PIX knockout on dendritic spines in vivo. However, GIT1 and β-PIX were reported to work together with Rac1 and PAK3 to regulate spine morphogenesis and synapse formation in rat primary hippocampal neurons (Zhang et al., 2003, 2005) and in Drosophila (Parnas et al., 2001). GIT1 stabilizes surface expression of post-synaptic glutamatergic AMPA receptors, the primary receptor mediating at excitatory synaptic neurotransmission, through an interaction with a complex formed by liprin-α and GRIP1 bound to the AMPA receptor (Ko et al., 2003). Excitatory synaptic connections can be strengthened by the maturation and stabilization of spines following excitation-induced Ca2+ influx. Ca2+-mediated activation of calmodulin (CaM)-dependent protein kinases (CaMKs) promotes phosphorylation of the GIT1-β-PIX complex at Ser516 of β-PIX; this stimulates GEF activity to increase the level of active Rac1, promoting the formation and stabilization of mature dendritic spines (Saneyoshi et al., 2008). Neuronal activation leads to increased localization of GIT1–β-PIX within spines and, thus, enhanced activity of Rac1 and PAK (Zhang et al., 2003; Webb et al., 2007). In addition to Ca2+ signals, the correct localization of spines involves the regulation of GIT1–β-PIX through integrin-α5 (Webb et al., 2007) and ephrinB (Segura et al., 2007). Signaling through ephrinB promotes Src-mediated phosphorylation of GIT1 or GIT2 at Tyr392, which allows the SH2-SH3 adaptor protein Grb4 to bind to ephrinB, in order to recruit GIT–PIX to regulate spine formation (Segura et al., 2007). During post-natal brain maturation in mammals, maturation of synaptic connections is marked by a transition to adult-type synaptic GluN1/GluN2 NMDA receptors from extrasynaptic NMDA receptors containing GluN3A, a subunit whose expression fades by adulthood. GluN3-containing NMDA receptors repress the formation of stable synaptic spines, in part by direct binding of the tail of GluN3A to GIT1, which prevents localization of GIT1 -β-PIX within forming spines (Fiuza et al., 2013). Intriguingly, binding of GluN3A to GIT1 reduced co-immunoprecipitation of GIT1 with β-PIX, whereas loss of GluN3A increased the association between GIT1 and β-PIX (Fiuza et al., 2013), suggesting that GluN3A binding regulates complex formation.
Functions in the immune system
Cell migration and direction sensing within a chemoattractant gradient are crucial to correct immune function. Leukocytes, such as neutrophils, form the vanguard of the innate immune system and rush to sites of infection to kill invading organisms. The ability of neutrophils to orient themselves in a gradient of chemoattractant requires GIT2, α-PIX and PAK1 (Li et al., 2003; Mazaki et al., 2006). Activation of chemoattractant receptors at the leading edge of the cell releases G-protein βγ-subunits that, together with PIX-activated Cdc42, stimulate PAK. GIT2 and α-PIX function through Arf1 and G-protein βγ-subunits to generate superoxide radicals in order to elicit the immune response (Mazaki et al., 2012). Loss of GIT2 also mislocalizes the direction of superoxide release (Mazaki et al., 2006). In T-cells, IL2 activates Rac1 through PKCθ-mediated phosphorylation of α-PIX at Ser225 and Ser488 (Llavero et al., 2015). In rat basophilic leukemia cells, depletion of GIT1 but not GIT2 alters chemokine-directed cell migration (Gavina et al., 2010). In T lymphocytes, loss of β-PIX reduces their ability to migrate towards the chemokine CXCL12 (Volinsky et al., 2006), whereas knockout of GIT2 increases their migration towards it (Phee et al., 2010).
Adaptive immunity involves the recognition of non-self antigens, which are presented to T- and B-cells by antigen-presenting cells through a specialized cell–cell contact, the immune synapse. In T-cells, activation of the T-cell receptor by antigen leads to activation of PAK1 through a GIT–PIX complex, which together are recruited to the immune synapse (Ku et al., 2001; Phee et al., 2005). The SAP adaptor protein, a T-cell signaling protein that is mutated in X-linked lymphoproliferative disease, binds to the SH3 domain of PIX to couple SLAM receptors to Cdc42 activation during T-cell activation (Gu et al., 2006). Accordingly, mice deficient in α-PIX or GIT2 exhibit immune abnormalities. Loss of α-PIX reduces the number of mature T- and B-cells in blood, and reduces their proliferation in culture by preventing PAK1 activation at immune synapses (Missy et al., 2008). This is in part due to reduced ability of immature CD4+/CD8+ thymocytes to survive selection in the thymus for cells with rearranged T-cell receptors that do not react to self-antigens (Phee et al., 2010).
GIT–PIX has also been implicated in diseases with an autoimmune component. For instance, GIT2-deficient mice exhibit elevated susceptibility to colon damage in two models of colitis and after peritoneal infection with E. coli (Wei et al., 2014). GIT2 interacts with and regulates the NFκB signaling pathway (Wang et al., 2011), and loss of GIT2 augments the signaling of the Toll-like receptor TLR4 to NFκB by facilitating deubiquitination of the E3 ligase TRAF6, which links TLR4 to NFκB (Wei et al., 2014). This leads to a dramatic increase in pro-inflammatory cytokines and amplified tissue damage. By contrast, GIT2-knockout mice exhibit less liver inflammation through T-cells, have lower chemokine levels and reduced liver injury compared to wild-type controls in the concanavalin-A-induced hepatitis model (Hao et al., 2015). Thus, GIT2 has multiple effects on inflammation by affecting the ability of immune cells to mature, to home to sites of injury and to regulate responses to injury. α-PIX has been suggested to be a potential player in Crohn's disease and ulcerative colitis (Chang et al., 2014), whereas β-PIX is thought to facilitate recognition of bacterial muramyl dipeptide through the receptor NOD2 to chemokine production and release (Eitel et al., 2008).
Functions in cancer
GIT and PIX proteins are important regulators of cell adhesion and spreading, cell–cell contact and cell migration (Hoefen and Berk, 2006; Frank and Hansen, 2008), and all these are crucial processes in cancer cells (Hanahan and Weinberg, 2011). Therefore, it is not surprising that GIT and PIX are increasingly found to be associated with cancer. Searches for biomarkers for various tumors have identified both GIT and PIX. GIT1 is elevated in cervical cancer cell lines (Yoo et al., 2012), liver and colon cancer (Peng et al., 2013; Chen et al., 2015), melanoma (Huang et al., 2013), non-small cell lung cancer (Chang et al., 2015) and clear cell renal cancer (Lu et al., 2015), whereas GIT2 is downregulated in breast cancer (Sirirattanakul et al., 2015). α-PIX is overexpressed in glioma (Yokota et al., 2006) and downregulated in melanoma following TNFα treatment (de Lima et al., 2013). β-PIX expression was reported to be elevated in breast cancers (Ahn et al., 2003).
Several mechanisms for GIT–PIX activity in cancer cell proliferation, migration and/or metastasis have been proposed. Among these is the control of epidermal growth factor (EGF) receptor ubiquitylation and degradation by PIX proteins. The SH3 domain of PIX binds to the Casitas B-lineage lymphoma (Cbl) family ubiquitin E3 ligases c-Cbl and Cbl-b (Flanders et al., 2003; Seong et al., 2014). Binding of Cbl or PAK to PIX SH3 is mutually incompatible (Jozic et al., 2005), and overexpression of Cbl displaces PAK from β-PIX (Flanders et al., 2003). Cbl ubiquitin ligases mark EGFR for degradation, limiting cell proliferation; however, Cbl cannot regulate EGFRs when it is bound to PIX (Wu et al., 2003; Feng et al., 2006). Cbl also ubiquitylates PIX proteins for degradation (Schmidt et al., 2006; Seong et al., 2014; Kortüm et al., 2015). Activated Cdc42 promotes binding of Cbl to β-PIX and also promotes PIX GEF activity in a positive feedback loop (Wu et al., 2003). Similarly, stimulation with EGF increases the association of c-Cbl with α-PIX, but α-PIX also strongly promotes EGFR recycling through an undefined mechanism that is independent of its GEF activity or binding to c-Cbl (Kortüm et al., 2015). Knocking down β-PIX (Stevens et al., 2014) or α-PIX (Seong et al., 2014) in glioblastoma cells increased Cbl activity, reduced migration and invasiveness, resulting in reduced tumorigenicity in culture and in vivo. A distinct mechanism for EGFR degradation involves sorting nexin 6 (Snx6) (Cavet et al., 2008). Activation of EGFR promotes GIT1 phosphorylation and binding to Snx6 in endosomes, where Snx6 sorts EGFR for degradation and recycling. Thus, knockdown of GIT1 reduces EGFR degradation rate, whereas overexpression of GIT1 and Snx6 enhances EGFR degradation (Cavet et al., 2008). However, how this mechanism intersects with PIX and/or Cbl in controling EGFR has not yet been examined.
GIT1 also contributes to cell transformation by regulating Arf. NIH3T3 cells that have been transformed by activated mutant Cdc42 lose the ability to form colonies in soft agar when GIT1 is knocked down (Yoo et al., 2012). ArfGAP-defective mutant GIT1 augments growth in soft agar, and coexpression of GIT1 (or GAP-deficient GIT1) with activated mutant Arf6 further increases this (Yoo et al., 2012). This suggests that GIT1 regulates anchorage-independent growth through its paxillin-dependent localization to focal adhesions where it locally regulates Arf proteins, which are required for formation of invadopodia and for anchorage independent growth (Tague et al., 2004; Hashimoto et al., 2004).
GIT1 also interacts with methionine adenosyltransferase 2B (MAT2B) in liver cancer and colon cancer cells. MAT2B binding increases GIT-mediated scaffolding of MEK and Erk proteins, thereby amplifying signaling of the proliferative Ras–MEK–Erk pathway. Knockdown of MAT2B or GIT1 reduces Erk signaling and proliferation of tumor cells, whereas overexpression of GIT1 and of MAT2B increased proliferation and metastasis of lung tumor cells in an orthotopic lung tumor model (Peng et al., 2013; Peng et al., 2015). Intriguingly, MAT2B synthesizes S-adenosyl methionine, the methyl donor required for DNA and histone methylation, suggesting that GIT1 also contributes to the regulation of epigenetic modifications during cancer progression.
MicroRNAs (miRNAs) have received much attention in cancer, both as biomarkers of cancers and as regulators of oncogenes and tumor suppressors (Esquela-Kerscher and Slack, 2006). GIT and PIX genes have been shown to be targets of miRNAs during cancer invasion and metastasis. GIT2 is a target of miR-210 in pancreatic cancer cells, in which hypoxia increases levels of miR-210 to reduce GIT2 (Chen et al., 2012). By contrast, GIT2 regulates levels of miR-146a; knockdown of GIT2 increases the level of miR-146a in an ArfGAP-dependent manner (Zhou and Thiery, 2013), suggesting that Arf has a role in the maturation of this miRNA. Endogenous levels of GIT2 inhibit the processing of miR-146a, which, in turn, inhibits induction of epithelial–mesenchymal transition (EMT) – a key process in metastasis – by reducing the expression of EMT-promoting factors such as Zeb1.
GIT1 is the target of three distinct miRNAs. It is a direct target gene for miR-491-5p; low levels of miR-491-5p in oral squamous carcimona cells have been associated with a poor survival rate of patients, whereas overexpression of miR-491-5p reduces the metastatic potential of tumors (Huang et al., 2013). GIT1 overexpression counteracts miR-491-5p-mediated inhibition of cell migration and in vivo lung metastasis, whereas silencing of GIT1 mimics the ability of miR-491-5p to inhibit migration, invasion and metastasis. In patient samples, low miR-491-5p levels and high expression of GIT1 correlates with the potential to metastasize to lymph nodes (Huang et al., 2013). Huang and colleagues also demonstrated that GIT1 is a direct target of miR-149, which was found to suppress breast cancer metastasis (Chan et al., 2014). There, knockdown of GIT1 reduced migration and invasion of highly metastatic cells in culture, as well as metastasis in vivo, whereas overexpression of GIT1 reversed the miR-149-mediated inhibition of cell migration and invasion, as well as metastasis in a less metastatic cell line. Low miR-149 and high GIT1 expression have been associated with advanced breast cancer and lymph node metastasis (Chan et al., 2014). Finally, GIT1 is a target for miR-138 in non-small cell lung cancer (Li et al., 2015). Expression of miR-138 or knockdown of GIT1 reduces cell growth and EMT, whereas re-expression of GIT1 in miR-138-expressing cells partially reverses these effects. In medulloblastoma cells, ARHGEF6 (α-PIX) has been shown to be a target of miR-135a, and re-expression of miR-135a in highly tumorigenic cells reduces α-PIX expression and tumorigenicity (Hemmesi et al., 2015).
However, to date, no studies have assessed whether miRNA-altered expression level of GIT changes the level of PIX (or vice versa), nor whether overexpression of one member of the GIT–PIX complex in patient tumor samples results in overexpression of other subunits.
Conclusions
The GIT–PIX complex is a crucial integration center for numerous signaling pathways, in addition to its well-known roles in membrane and vesicle trafficking, and cytoskeletal dynamics through regulating Arf and Rac/Cdc42 (Fig. 2). In vivo studies have shown that GIT1 and GIT2, as well as α-PIX and β-PIX, share many functions but that, however, they are not redundant. There is clearly a tight linkage between GIT and PIX proteins and most cells contain the two GIT and PIX isoforms and potentially multiple splice variants of GIT2 and β-PIX. However, the importance of this diversity for cell function is unknown, as are the mechanisms by which complexes are assembled to contain or exclude mixtures of GIT or PIX isoforms. Furthermore, although there are studies that suggest functions for GIT without PIX or for PIX without GIT, it has not yet been unequivocally demonstrated whether GIT and PIX are always associated, or whether they can associate and/or dissociate under particular conditions. The existence of GIT2 and β-PIX variants without coiled-coil domains (Fig. 1) indicates that at least some variants may not function within an oligomeric complex. An intrinsic dynamics of self-association of GIT1 into ‘closed’ and ‘open’ states has also been proposed (Totaro et al., 2007, 2014), but the structure of these states remains to be determined. Structural studies of GIT–PIX are needed to clarify these issues, and to understand how the GAP and GEF activities of GIT and PIX might be coupled. Additionally, research is needed that integrates the knowledge of GIT–PIX regulation, to develop a better understanding of how the many, seemingly distinct, cellular signals that intersect at GIT–PIX complexes are functionally linked.
Acknowledgements
We extend our apologies to investigators whose work was not included due to constraints on space. We thank Nicolas Vitale (Université de Strasbourg, France) for performing the ArfGAP assay.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
W.Z. is supported by the National Natural Science Funds of China (81572879) and Zhejiang Province Natural Science Funds of China [grant number:LY14H160004], X.B.L. is supported by the National Natural Science Foundation of China [grant number: 61373057] and R.T.P. by the National Institutes of Health [grant number: NIH R21 MH090556] and the Department of Defense [grant number: DOD W81XWH-11-2-0112 (DM102564). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.179465/-/DC1
References
- Ahn S.-J., Chung K.-W., Lee R.-A., Park I.-A., Lee S.-H., Park D. E. and Noh D.-Y. (2003). Overexpression of betaPix-a in human breast cancer tissues. Cancer Lett. 193, 99-107. 10.1016/S0304-3835(03)00004-1 [DOI] [PubMed] [Google Scholar]
- Albertinazzi C., Za L., Paris S. and de Curtis I. (2003). ADP-ribosylation factor 6 and a functional PIX/p95-APP1 complex are required for Rac1B-mediated neurite outgrowth. Mol. Biol. Cell 14, 1295-1307. 10.1091/mbc.E02-07-0406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen K. M., Gleeson J. G., Bagrodia S., Partington M. W., MacMillan J. C., Cerione R. A., Mulley J. C. and Walsh C. A. (1998). PAK3 mutation in nonsyndromic X-linked mental retardation. Nat. Genet. 20, 25-30. 10.1038/1675 [DOI] [PubMed] [Google Scholar]
- Angrand P.-O., Segura I., Völkel P., Ghidelli S., Terry R., Brajenovic M., Vintersten K., Klein R., Superti-Furga G., Drewes G. et al. (2006). Transgenic mouse proteomics identifies new 14-3-3-associated proteins involved in cytoskeletal rearrangements and cell signaling. Mol. Cell. Proteomics 5, 2211-2227. 10.1074/mcp.M600147-MCP200 [DOI] [PubMed] [Google Scholar]
- Audebert S., Navarro C., Nourry C., Chasserot-Golaz S., Lécine P., Bellaiche Y., Dupont J.-L., Premont R. T., Sempéré C., Strub J.-M. et al. (2004). Mammalian Scribble forms a tight complex with the betaPIX exchange factor. Curr. Biol. 14, 987-995. 10.1016/j.cub.2004.05.051 [DOI] [PubMed] [Google Scholar]
- Bagrodia S., Taylor S. J., Jordon K. A., Van Aelst L. and Cerione R. A. (1998). A novel regulator of p21-activated kinases. J. Biol. Chem. 273, 23633-23636. 10.1074/jbc.273.37.23633 [DOI] [PubMed] [Google Scholar]
- Bagrodia S., Bailey D., Lenard Z., Hart M., Guan J. L., Premont R. T., Taylor S. J. and Cerione R. A. (1999). A tyrosine-phosphorylated protein that binds to an important regulatory region on the Cool family of p21-activated kinase-binding proteins. J. Biol. Chem. 274, 22393-22400. 10.1074/jbc.274.32.22393 [DOI] [PubMed] [Google Scholar]
- Bahri S. M., Choy J. M., Manser E., Lim L. and Yang X. (2009). The Drosophila homologue of Arf-GAP GIT1, dGIT, is required for proper muscle morphogenesis and guidance during embryogenesis. Dev. Biol. 325, 15-23. 10.1016/j.ydbio.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Baird D., Feng Q. and Cerione R. A. (2005). The Cool-2/alpha-Pix protein mediates a Cdc42-Rac signaling cascade. Curr. Biol. 15, 1-10. 10.1016/j.cub.2004.12.040 [DOI] [PubMed] [Google Scholar]
- Bhanot H., Young A. M., Overmeyer J. H. and Maltese W. A. (2010). Induction of nonapoptotic cell death by activated Ras requires inverse regulation of Rac1 and Arf6. Mol. Cancer Res. 8, 1358-1374. 10.1158/1541-7786.MCR-10-0090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. C., West K. A. and Turner C. E. (2002). Paxillin-dependent paxillin kinase linker and p21-activated kinase localization to focal adhesions involves a multistep activation pathway. Mol. Biol. Cell 13, 1550-1565. 10.1091/mbc.02-02-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. C., Cary L. A., Jamieson J. S., Cooper J. A. and Turner C. E. (2005). Src and FAK kinases cooperate to phosphorylate paxillin kinase linker, stimulate its focal adhesion localization, and regulate cell spreading and protrusiveness. Mol. Biol. Cell 16, 4316-4328. 10.1091/mbc.E05-02-0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle H. J. and Kennedy M. B. (2005). Spine architecture and synaptic plasticity. Trends Neurosci. 28, 182-187. 10.1016/j.tins.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Cavet M. E., Pang J., Yin G. and Berk B. C. (2008). An epidermal growth factor (EGF) -dependent interaction between GIT1 and sorting nexin 6 promotes degradation of the EGF receptor. FASEB J. 22, 3607-3616. 10.1096/fj.07-094086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahdi A. and Sorokin A. (2008). Protein kinase A-dependent phosphorylation modulates beta1Pix guanine nucleotide exchange factor activity through 14-3-3beta binding. Mol. Cell. Biol. 28, 1679-1687. 10.1128/MCB.00898-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahdi A. and Sorokin A. (2010a). The role of beta(1)Pix/caveolin-1 interaction in endothelin signaling through Galpha subunits. Biochem. Biophys. Res. Commun. 391, 1330-1335. 10.1016/j.bbrc.2009.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahdi A. and Sorokin A. (2010b). Endothelin-1 induces p66Shc activation through EGF receptor transactivation: role of beta(1)Pix/Galpha(i3) interaction. Cell. Signal. 22, 325-329. 10.1016/j.cellsig.2009.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahdi A., Miller B. and Sorokin A. (2005). Endothelin 1 induces beta 1Pix translocation and Cdc42 activation via protein kinase A-dependent pathway. J. Biol. Chem. 280, 578-584. 10.1074/jbc.M411130200 [DOI] [PubMed] [Google Scholar]
- Chan P. M., Ng Y.-W. and Manser E. (2011). A robust protocol to map binding sites of the 14–3–3 interactome: Cdc25C requires phosphorylation of both S216 and S263 to bind 14-3-3. Mol. Cell. Proteomics 10, M110.005157 10.1074/mcp.M110.005157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.-H., Huang W.-C., Chang J.-W., Chang K.-J., Kuo W.-H., Wang M.-Y., Lin K.-Y., Uen Y.-H., Hou M.-F., Lin C.-M. et al. (2014). MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene 33, 4496-4507. 10.1038/onc.2014.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D., Gao F., Slavney A., Ma L., Waldman Y. Y., Sams A. J., Billing-Ross P., Madar A., Spritz R. and Keinan A. (2014). Accounting for eXentricities: analysis of the X chromosome in GWAS reveals X-linked genes implicated in autoimmune diseases. PLoS ONE 9, e113684 10.1371/journal.pone.0113684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. S., Su C. Y., Yu W. H., Lee W. J., Liu Y. P., Lai T. C., Jan Y. H., Yang Y. F., Shen C. N., Shew J. Y. et al. (2015). GIT1 promotes lung cancer cell metastasis through modulating Rac1/Cdc42 activity and is associated with poor prognosis. Oncotarget 6, 36278-36291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-Y., Liu W.-J., Zhao Y.-P., Zhou L., Zhang T.-P., Chen G. and Shu H. (2012). Induction, modulation and potential targets of miR-210 in pancreatic cancer cells. Hepatobiliary Pancreat. Dis. Int. 11, 319-324. 10.1016/S1499-3872(12)60168-4 [DOI] [PubMed] [Google Scholar]
- Chen J., Yang P., Yang J., Wen Z., Zhang B. and Zheng X. (2015). GIT1 is a novel prognostic biomarker and facilitates tumor progression via activating ERK/MMP9 signaling in hepatocellular carcinoma. OncoTargets Ther. 8, 3731-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia R., Haddock S., Beilina A., Rudenko I. N., Mamais A., Kaganovich A., Li Y., Kumaran R., Nalls M. A. and Cookson M. R. (2014). Phosphorylation of LRRK2 by casein kinase 1α regulates trans-Golgi clustering via differential interaction with ARHGEF7. Nat. Commun. 5, 5827 10.1038/ncomms6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claing A., Perry S. J., Achiriloaie M., Walker J. K. L., Albanesi J. P., Lefkowitz R. J. and Premont R. T. (2000). Multiple endocytic pathways of G protein-coupled receptors delineated by GIT1 sensitivity. Proc. Natl. Acad. Sci. USA 97, 1119-1124. 10.1073/pnas.97.3.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima V. C. C., de Carvalho A. F., Morato-Marques M., Hashimoto V. L., Spilborghs G. M. G. T., Marques S. M., Landman G., Torres C., Braga Ribeiro K., Brentani H. et al. (2013). TNF-alpha and melphalan modulate a specific group of early expressed genes in a murine melanoma model. Cytokine 62, 217-225. 10.1016/j.cyto.2013.02.022 [DOI] [PubMed] [Google Scholar]
- Dent L. G., Poon C. L. C., Zhang X., Degoutin J. L., Tipping M., Veraksa A. and Harvey K. F. (2015). The GTPase regulatory proteins Pix and Git control tissue growth via the Hippo pathway. Curr. Biol. 25, 124-130. 10.1016/j.cub.2014.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cesare A., Paris S., Albertinazzi C., Dariozzi S., Andersen J., Mann M., Longhi R. and de Curtis I. (2000). p95-APP1 links membrane transport to Rac-mediated reorganization of actin. Nat. Cell Biol. 2, 521-530. 10.1038/35019561 [DOI] [PubMed] [Google Scholar]
- Dyer J. O., Demarco R. S. and Lundquist E. A. (2010). Distinct roles of Rac GTPases and the UNC-73/Trio and PIX-1 Rac GTP exchange factors in neuroblast protrusion and migration in C. elegans. Small GTPases 1, 44-61. 10.4161/sgtp.1.1.12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eblen S. T., Slack J. K., Weber M. J. and Catling A. D. (2002). Rac-PAK signaling stimulates extracellular signal-regulated kinase (ERK) activation by regulating formation of MEK1-ERK complexes. Mol. Cell. Biol. 22, 6023-6033. 10.1128/MCB.22.17.6023-6033.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitel J., Krüll M., Hocke A. C., N'Guessan P. D., Zahlten J., Schmeck B., Slevogt H., Hippenstiel S., Suttorp N. and Opitz B. (2008). Beta-PIX and Rac1 GTPase mediate trafficking and negative regulation of NOD2. J. Immunol. 181, 2664-2671. 10.4049/jimmunol.181.4.2664 [DOI] [PubMed] [Google Scholar]
- Eriguchi M., Mizuta H., Luo S., Kuroda Y., Hara H. and Rubinsztein D. C. (2010). alpha Pix enhances mutant huntingtin aggregation. J. Neurol. Sci. 290, 80-85. 10.1016/j.jns.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A. and Slack F. J. (2006). Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259-269. 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- Feng Q., Baird D. and Cerione R. A. (2004). Novel regulatory mechanisms for the Dbl family guanine nucleotide exchange factor Cool-2/alpha-Pix. EMBO J. 23, 3492-3504. 10.1038/sj.emboj.7600331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Baird D., Peng X., Wang J., Ly T., Guan J.-L. and Cerione R. A. (2006). Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nat. Cell Biol. 8, 945-956. 10.1038/ncb1453 [DOI] [PubMed] [Google Scholar]
- Fiuza M., González-González I. and Pérez-Otaño I. (2013). GluN3A expression restricts spine maturation via inhibition of GIT1/Rac1 signaling. Proc. Natl. Acad. Sci. USA 110, 20807-20812. 10.1073/pnas.1312211110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanders J. A., Feng Q., Bagrodia S., Laux M. T., Singavarapu A. and Cerione R. A. (2003). The Cbl proteins are binding partners for the Cool/Pix family of p21-activated kinase-binding proteins. FEBS Lett. 550, 119-123. 10.1016/S0014-5793(03)00853-6 [DOI] [PubMed] [Google Scholar]
- Frank S. R. and Hansen S. H. (2008). The PIX-GIT complex: a G protein signaling cassette in control of cell shape. Semin. Cell Dev. Biol. 19, 234-244. 10.1016/j.semcdb.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese S., Schubert W.-D., Findeis A. C., Marquardt T., Roske Y. S., Stradal T. E. B. and Heinz D. W. (2006). The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J. Biol. Chem. 281, 18236-18245. 10.1074/jbc.M512917200 [DOI] [PubMed] [Google Scholar]
- Frost J. A., Steen H., Shapiro P., Lewis T., Ahn N., Shaw P. E. and Cobb M. H. (1997). Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. EMBO J. 16, 6426-6438. 10.1093/emboj/16.21.6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavina M., Za L., Molteni R., Pardi R. and de Curtis I. (2010). The GIT–PIX complexes regulate the chemotactic response of rat basophilic leukaemia cells. Biol. Cell 102, 231-244. 10.1042/BC20090074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., Droege A., Lindenberg K. S., Knoblich M., Haenig C. et al. (2004). A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell 15, 853-865. 10.1016/j.molcel.2004.09.016 [DOI] [PubMed] [Google Scholar]
- Gu C., Tangye S. G., Sun X., Luo Y., Lin Z. and Wu J. (2006). The X-linked lymphoproliferative disease gene product SAP associates with PAK-interacting exchange factor and participates in T cell activation. Proc. Natl. Acad. Sci. USA 103, 14447-14452. 10.1073/pnas.0606624103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E. D. and Fejtova A. (2012). Molecular organization and plasticity of the cytomatrix at the active zone. Curr. Opin. Neurobiol. 22, 423-430. 10.1016/j.conb.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Häbig K., Gellhaar S., Heim B., Djuric V., Giesert F., Wurst W., Walter C., Hentrich T., Riess O. and Bonin M. (2013). LRRK2 guides the actin cytoskeleton at growth cones together with ARHGEF7 and Tropomyosin 4. Biochim. Biophys. Acta 1832, 2352-2367. 10.1016/j.bbadis.2013.09.009 [DOI] [PubMed] [Google Scholar]
- Haebig K., Gloeckner C. J., Miralles M. G., Gillardon F., Schulte C., Riess O., Ueffing M., Biskup S. and Bonin M. (2010). ARHGEF7 (Beta-PIX) acts as guanine nucleotide exchange factor for leucine-rich repeat kinase 2. PLoS ONE 5, e13762 10.1371/journal.pone.0013762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendeler J., Hojo Y., Aebersold R., Yan C. and Berk B. C. (2000). Angiotensin II-stimulated phosphorylation of GIT1 determines the relative activation of PLCγ And ERK1/2. Circulation 102, 11-15.10880408 [Google Scholar]
- Hajdo-Milasinovic A., van der Kammen R. A., Moneva Z. and Collard J. G. (2009). Rac3 inhibits adhesion and differentiation of neuronal cells by modifying GIT1 downstream signaling. J. Cell Sci. 122, 2127-2136. 10.1242/jcs.039958 [DOI] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646-674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hao Y.-E., He D.-F., Yin R.-H., Chen H., Wang J., Wang S.-X., Zhan Y.-Q., Ge C.-H., Li C.-Y., Yu M. et al. (2015). GIT2 deficiency attenuates concanavalin A-induced hepatitis in mice. FEBS Open Biol. 5, 688-704. 10.1016/j.fob.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S., Onodera Y., Hashimoto A., Tanaka M., Hamaguchi M., Yamada A. and Sabe H. (2004). Requirement for Arf6 in breast cancer invasive activities. Proc. Natl. Acad. Sci. USA 101, 6647-6652. 10.1073/pnas.0401753101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel T., Czupalla C., Expirto Santo A. I., Anitei M., Arantzazu Sanchez-Fernandez M., Mosch K., Krause E. and Hoflack B. (2009). Src-dependent repression of ARF6 is required to maintain podosome-rich sealing zones in bone-digesting osteoclasts. Proc. Natl. Acad. Sci. USA 106, 1451-1456. 10.1073/pnas.0804464106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidary Arash E., Song K. M., Song S., Shiban A. and Attisano L. (2014). Arhgef7 promotes activation of the Hippo pathway core kinase Lats. EMBO J. 33, 2997-3011. 10.15252/embj.201490230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmesi K., Squadrito M. L., Mestdagh P., Conti V., Cominelli M., Piras I. S., Sergi L. S., Piccinin S., Maestro R., Poliani P. L. et al. (2015). miR-135a inhibits cancer stem cell-driven medulloblastoma development by directly repressing Arhgef6 expression. Stem Cells 33, 1377-1389. 10.1002/stem.1958 [DOI] [PubMed] [Google Scholar]
- Hoefen R. J. and Berk B. C. (2006). The multifunctional GIT family of proteins. J. Cell Sci. 119, 1469-1475. 10.1242/jcs.02925 [DOI] [PubMed] [Google Scholar]
- Hong S.-T. and Mah W. (2015). Critical role of GIT1 in vertebrate and invertebrate brain development. Exp. Neurobiol. 24, 8-16. 10.5607/en.2015.24.1.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Zhou Z., Asrar S., Henkelman M., Xie W. and Jia Z. (2011). p21-Activated kinases 1 and 3 control brain size through coordinating neuronal complexity and synaptic properties. Mol. Cell. Biol. 31, 388-403. 10.1128/MCB.00969-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.-C., Chan S.-H., Jang T.-H., Chang J.-W., Ko Y.-C., Yen T.-C., Chiang S.-L., Chiang W.-F., Shieh T.-Y., Liao C.-T. et al. (2013). miRNA-491–5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 74, 751-764. 10.1158/0008-5472.CAN-13-1297 [DOI] [PubMed] [Google Scholar]
- Huck B., Kemkemer R., Franz-Wachtel M., Macek B., Hausser A. and Olayioye M. A. (2012). GIT1 phosphorylation on serine 46 by PKD3 regulates paxillin trafficking and cellular protrusive activity. J. Biol. Chem. 287, 34604-34613. 10.1074/jbc.M112.374652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Y. J., Kang G. B., Lee J. H., Park K. R., Song H. E., Kim E., Song W. K., Park D. and Eom S. H. (2010). Structural basis for asymmetric association of the betaPIX coiled coil and shank PDZ. J. Mol. Biol. 397, 457-466. 10.1016/j.jmb.2010.01.048 [DOI] [PubMed] [Google Scholar]
- Jones C. A., Nishiya N., London N. R., Zhu W., Sorensen L. K., Chan A. C., Lim C. J., Chen H., Zhang Q., Schultz P. G. et al. (2009). Slit2-Robo4 signalling promotes vascular stability by blocking Arf6 activity. Nat. Cell Biol. 11, 1325-1331. 10.1038/ncb1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozic D., Cárdenes N., Deribe Y. L., Moncalián G., Hoeller D., Groemping Y., Dikic I., Rittinger K. and Bravo J. (2005). Cbl promotes clustering of endocytic adaptor proteins. Nat. Struct. Mol. Biol. 12, 972-979. 10.1038/nsmb1000 [DOI] [PubMed] [Google Scholar]
- Kahn R. A., Bruford E., Inoue H., Logsdon J. M. Jr, Nie Z., Premont R. T., Randazzo P. A., Satake M., Theibert A. B., Zapp M. L. et al. (2008). Consensus nomenclature for the human ArfGAP domain-containing proteins. J. Cell Biol. 182, 1039-1044. 10.1083/jcb.200806041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim T., Lee D., Park S.-H., Kim H. and Park D. (2000). Molecular cloning of neuronally expressed mouse betaPix isoforms. Biochem. Biophys. Res. Commun. 272, 721-725. 10.1006/bbrc.2000.2845 [DOI] [PubMed] [Google Scholar]
- Kim S., Ko J., Shin H., Lee J.-R., Lim C., Han J.-H., Altrock W. D., Garner C. C., Gundelfinger E. D., Premont R. T. et al. (2003). The GIT family of proteins forms multimers and associates with the presynaptic cytomatrix protein Piccolo. J. Biol. Chem. 278, 6291-6300. 10.1074/jbc.M212287200 [DOI] [PubMed] [Google Scholar]
- Klein M., van der Voet M., Harich B., van Hulzen K. J. E., Onnink A. M. H., Hoogman M., Guadalupe T., Zwiers M., Groothuismink J. M., Verberkt A. et al. (2015). Converging evidence does not support GIT1 as an ADHD risk gene. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168, 492-507. 10.1002/ajmg.b.32327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J., Kim S., Valtschanoff J. G., Shin H., Lee J. R., Sheng M., Premont R. T., Weinberg R. J. and Kim E. (2003). Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting . J. Neurosci. 23, 1667-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh C. G., Manser E., Zhao Z. S., Ng C. P. and Lim L. (2001). Beta1PIX, the PAK-interacting exchange factor, requires localization via a coiled-coil region to promote microvillus-like structures and membrane ruffles. J. Cell Sci. 114, 4239-4251. [DOI] [PubMed] [Google Scholar]
- Kon S., Funaki T. and Satake M. (2011). Putative terminator and/or effector functions of Arf GAPs in the trafficking of clathrin-coated vesicles. Cell. Logist. 1, 86-89. 10.4161/cl.1.3.16192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong K.-F., Fu G., Zhang Y., Yokosuka T., Casas J., Canonigo-Balancio A. J., Becart S., Kim G., Yates J. R., Kronenberg M. et al. (2014). Protein kinase C-η controls CTLA-4-mediated regulatory T cell function. Nat. Immunol. 15, 465-472. 10.1038/ni.2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortüm F., Harms F. L., Hennighausen N. and Rosenberger G. (2015). αPIX Is a trafficking regulator that balances recycling and degradation of the epidermal growth factor receptor. PLoS ONE 10, e0132737 10.1371/journal.pone.0132737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster I., Jungwirth M. S. and Steinbeisser H. (2010). xGit2 and xRhoGAP 11A regulate convergent extension and tissue separation in Xenopus gastrulation. Dev. Biol. 344, 26-35. 10.1016/j.ydbio.2010.03.025 [DOI] [PubMed] [Google Scholar]
- Ku G. M., Yablonski D., Manser E., Lim L. and Weiss A. (2001). A PAK1-PIX-PKL complex is activated by the T-cell receptor independent of Nck, Slp-76 and LAT. EMBO J. 20, 457-465. 10.1093/emboj/20.3.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsche K., Yntema H., Brandt A., Jantke I., Nothwang H. G., Orth U., Boavida M. G., David D., Chelly J., Fryns J.-P. et al. (2000). Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat. Genet. 26, 247-250. 10.1038/80002 [DOI] [PubMed] [Google Scholar]
- Lee Y.-S. and Silva A. J. (2011). Modeling hyperactivity: of mice and men. Nat. Med. 17, 541-542. 10.1038/nm0511-541 [DOI] [PubMed] [Google Scholar]
- Li Z., Hannigan M., Mo Z., Liu B., Lu W., Wu Y., Smrcka A. V., Wu G., Li L., Liu M. et al. (2003). Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell 114, 215-227. 10.1016/S0092-8674(03)00559-2 [DOI] [PubMed] [Google Scholar]
- Li J., Wang Q., Wen R., Liang J., Zhong X., Yang W., Su D. and Tang J. (2015). MiR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J. Cell Mol. Med. 19, 2793-2805. 10.1111/jcmm.12666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Fraser S. D., Faloon P. W., Rollins E. L., Vom B. J., Starovic-Subota O., Laliberte A. L., Chen J.-N., Serluca F. C. and Childs S. J. (2007). A betaPix Pak2a signaling pathway regulates cerebral vascular stability in zebrafish. Proc. Natl. Acad. Sci. USA 104, 13990-13995. 10.1073/pnas.0700825104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llavero F., Urzelai B., Osinalde N., Gálvez P., Lacerda H. M., Parada L. A. and Zugaza J. L. (2015). Guanine nucleotide exchange factor αPIX leads to activation of the Rac 1 GTPase/glycogen phosphorylase pathway in interleukin (IL)-2-stimulated T cells. J. Biol. Chem. 290, 9171-9182. 10.1074/jbc.M114.608414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo T.-H., Ng Y.-W., Lim L. and Manser E. (2004). GIT1 activates p21-activated kinase through a mechanism independent of p21 binding. Mol. Cell. Biol. 24, 3849-3859. 10.1128/MCB.24.9.3849-3859.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Cai H., Park S-S., Siddiqui S., Premont R. T., Schmalzigaug R., Paramasivam M., Seidman M., Bodogai I., Biragyn A. et al. (2015). Nuclear GIT2 is an ATM substrate and promotes DNA repair. Mol. Cell. Biol. 35, 1081-1096. 10.1128/MCB.01432-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M. and Cheng H.-J. (2008). A RAC/CDC-42-independent GIT/PIX/PAK signaling pathway mediates cell migration in C. elegans. PLoS Genet. 4, e1000269 10.1371/journal.pgen.1000269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Mizuta H. and Rubinsztein D. C. (2008). p21-activated kinase 1 promotes soluble mutant huntingtin self-interaction and enhances toxicity. Hum. Mol. Genet. 17, 895-905. 10.1093/hmg/ddm362 [DOI] [PubMed] [Google Scholar]
- Manabe R., Kovalenko M., Webb D. J. and Horwitz A. R. (2002). GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J. Cell Sci. 115, 1497-1510. [DOI] [PubMed] [Google Scholar]
- Manser E., Leung T., Salihuddin H., Zhao Z.-S. and Lim L. (1994). A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 367, 40-46. 10.1038/367040a0 [DOI] [PubMed] [Google Scholar]
- Manser E., Loo T.-H., Koh C.-G., Zhao Z.-S., Chen X.-Q., Tan L., Tan I., Leung T. and Lim L. (1998). PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell 1, 183-192. 10.1016/S1097-2765(00)80019-2 [DOI] [PubMed] [Google Scholar]
- Mazaki Y., Hashimoto S., Okawa K., Tsubouchi A., Nakamura K., Yagi R., Yano H., Kondo A., Iwamatsu A., Mizoguchi A. et al. (2001). An ADP-ribosylation factor GTPase-activating protein Git2-short/KIAA0148 is involved in subcellular localization of paxillin and actin cytoskeletal organization. Mol. Biol. Cell 12, 645-662. 10.1091/mbc.12.3.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaki Y., Hashimoto S., Tsujimura T., Morishige M., Hashimoto A., Aritake K., Yamada A., Nam J.-M., Kiyonari H., Nakao K. et al. (2006). Neutrophil direction sensing and superoxide production linked by the GTPase-activating protein GIT2. Nat. Immunol. 7, 724-731. 10.1038/ni1349 [DOI] [PubMed] [Google Scholar]
- Mazaki Y., Nishimura Y. and Sabe H. (2012). GBF1 bears a novel phosphatidylinositol-phosphate binding module, BP3K, to link PI3Kγ activity with Arf1 activation involved in GPCR-mediated neutrophil chemotaxis and superoxide production. Mol. Biol. Cell 23, 2457-2467. 10.1091/mbc.E12-01-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon P., Yin G., Smolock E. M., Zuscik M. J. and Yan C. (2010). GPCR kinase 2 interacting protein 1 (GIT1) regulates osteoclast function and bone mass. J. Cell Physiol. 225, 777-785. 10.1002/jcp.22282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. Z., Déliot N., Chasserot-Golaz S., Premont R. T., Bader M.-F. and Vitale N. (2006). Regulation of neuroendocrine exocytosis by the ARF6 GTPase-activating protein GIT1. J. Biol. Chem. 281, 7919-7926. 10.1074/jbc.M600081200 [DOI] [PubMed] [Google Scholar]
- Missy K., Hu B., Schilling K., Harenberg A., Sakk V., Kuchenbecker K., Kutsche K. and Fischer K.-D. (2008). Alpha PIX Rho GTPase guanine nucleotide exchange factor regulates lymphocyte functions and antigen receptor signaling. Mol. Cell. Biol. 28, 3776-3789. 10.1128/MCB.00507-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Nam J.-M., Kojima C., Mochizuki N. and Sabe H. (2009). EphA2 engages Git1 to suppress Arf6 activity modulating epithelial cell-cell contacts. Mol. Biol. Cell. 20, 1949-1959. 10.1091/mbc.E08-06-0549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momboisse F., Lonchamp E., Calco V., Ceridono M., Vitale N., Bader M.-F. and Gasman S. (2009). betaPIX-activated Rac1 stimulates the activation of phospholipase D, which is associated with exocytosis in neuroendocrine cells. J. Cell Sci. 122, 798-806. 10.1242/jcs.038109 [DOI] [PubMed] [Google Scholar]
- Montesinos M. S., Dong W., Goff K., Das B., Guerrero-Given D., Schmalzigaug R., Premont R. T., Satterfield R., Kamasawa N. and Young S. M. Jr (2015). Presynaptic deletion of GIT proteins results in increased synaptic strength at a mammalian central synapse. Neuron 88, 918-925. 10.1016/j.neuron.2015.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z., Wynne K., von Kriegsheim A., Gambaryan S. and Smolenski A. (2015). Cyclic nucleotide-dependent protein kinases target ARHGAP17 and ARHGEF6 complexes in platelets. J. Biol. Chem. 290, 29974-29983. 10.1074/jbc.M115.678003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh W. K., Yoo J. C., Jo D., Song Y. H., Kim M. G. and Park D. (1997). Cloning of a SH3 domain-containing proline-rich protein, p85SPR, and its localization in focal adhesion. Biochem. Biophys. Res. Commun. 235, 794-798. 10.1006/bbrc.1997.6875 [DOI] [PubMed] [Google Scholar]
- Omelchenko T., Rabadan M. A., Hernández-Martínez R., Grego-Bessa J., Anderson K. V. and Hall A. (2014). β-Pix directs collective migration of anterior visceral endoderm cells in the early mouse embryo. Genes Dev. 28, 2764-2777. 10.1101/gad.251371.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J., Hoefen R., Pryhuber G. S., Wang J., Yin G., White R. J., Xu X., O'Dell M. R., Mohan A., Michaloski H. et al. (2009). G-protein-coupled receptor kinase interacting protein-1 is required for pulmonary vascular development. Circulation 119, 1524-1532. 10.1161/CIRCULATIONAHA.108.823997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris S., Longhi R., Santambrogio P. and de Curtis I. (2003). Leucine-zipper-mediated homo- and hetero-dimerization of GIT family p95-ARF GTPase-activating protein, PIX-, paxillin-interacting proteins 1 and 2. Biochem. J. 372, 391-398. 10.1042/bj20030047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Na M., Choi J., Kim S., Lee J.-R., Yoon J., Park D., Sheng M. and Kim E. (2003). The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J. Biol. Chem. 278, 19220-19229. 10.1074/jbc.M301052200 [DOI] [PubMed] [Google Scholar]
- Parnas D., Haghighi A. P., Fetter R. D., Kim S. W. and Goodman C. S. (2001). Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron 32, 415-424. 10.1016/S0896-6273(01)00485-8 [DOI] [PubMed] [Google Scholar]
- Peng H., Dara L., Li T. W. H., Zheng Y., Yang H., Tomasi M. L., Tomasi I., Giordano P., Mato J. M. and Lu S. C. (2013). MAT2B-GIT1 interplay activates MEK1/ERK 1 and 2 to induce growth in human liver and colon cancer. Hepatology 57, 2299-2313. 10.1002/hep.26258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Li T. W., Yang H., Moyer M. P., Mato J. M. and Lu S. C. (2015). Methionine adenosyltransferase 2B-GIT1 complex serves as a scaffold to regulate Ras/Raf/MEK1/2 activity in human liver and colon cancer cells. Am. J. Pathol. 185, 1135-1144. 10.1016/j.ajpath.2014.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phee H., Abraham R. T. and Weiss A. (2005). Dynamic recruitment of Pak1 to the immunological synapse is mediated by Pix independently of SLP-76 and Vav1. Nat. Immunol. 6, 608-617. 10.1038/ni1199 [DOI] [PubMed] [Google Scholar]
- Phee H., Dzhagalov I., Mollenauer M., Wang Y., Irvine D. J., Robey E. and Weiss A. (2010). Regulation of thymocyte positive selection and motility by GIT2. Nat. Immunol. 11, 503-511. 10.1038/ni.1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podufall J., Tian R., Knoche E., Puchkov D., Walter A. M., Rosa S., Quentin C., Vukoja A., Jung N., Lampe A. et al. (2014). A presynaptic role for the cytomatrix protein GIT in synaptic vesicle recycling. Cell Rep. 7, 1417-1425. 10.1016/j.celrep.2014.04.051 [DOI] [PubMed] [Google Scholar]
- Premont R. T., Claing A., Vitale N., Freeman J. L. R., Pitcher J. A., Patton W. A., Moss J., Vaughan M. and Lefkowitz R. J. (1998). beta2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc. Natl. Acad. Sci. USA 95, 14082-14087. 10.1073/pnas.95.24.14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premont R. T., Claing A., Vitale N., Perry S. J. and Lefkowitz R. J. (2000). The GIT family of ADP-ribosylation factor GTPase-activating proteins: functional diversity of GIT2 through alternative splicing. J. Biol. Chem. 275, 22373-22380. 10.1074/jbc.275.29.22373 [DOI] [PubMed] [Google Scholar]
- Premont R. T., Perry S. J., Schmalzigaug R., Roseman J. T., Xing Y. and Claing A. (2004). The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell. Signal. 16, 1001-1011. 10.1016/S0898-6568(04)00023-3 [DOI] [PubMed] [Google Scholar]
- Ramakers G. J. A., Wolfer D., Rosenberger G., Kuchenbecker K., Kreienkamp H.-J., Prange-Kiel J., Rune G., Richter K., Langnaese K., Masneuf S. et al. (2012). Dysregulation of Rho GTPases in the αPix/Arhgef6 mouse model of X-linked intellectual disability is paralleled by impaired structural and synaptic plasticity and cognitive deficits. Hum. Mol. Genet. 21, 268-286. 10.1093/hmg/ddr457 [DOI] [PubMed] [Google Scholar]
- Randazzo P. A., Terui T., Sturch S. and Kahn R. A. (1994). The amino terminus of ADP-ribosylation factor (ARF) 1 is essential for interaction with Gs and ARF GTPase-activating protein. J. Biol. Chem. 269, 29490-29494. [PubMed] [Google Scholar]
- Randazzo P. A., Inoue H. and Bharti S. (2007). Arf GAPs as regulators of the actin cytoskeleton. Biol. Cell 99, 583-600. 10.1042/BC20070034 [DOI] [PubMed] [Google Scholar]
- Rennefahrt U. E. E., Deacon S. W., Parker S. A., Devarajan K., Beeser A., Chernoff J., Knapp S., Turk B. E. and Peterson J. R. (2007). Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J. Biol. Chem. 282, 15667-15678. 10.1074/jbc.M700253200 [DOI] [PubMed] [Google Scholar]
- Richier L., Williton K., Clattenburg L., Colwill K., O'Brien M., Tsang C., Kolar A., Zinck N., Metalnikov P., Trimble W. S. et al. (2010). NOS1AP associates with Scribble and regulates dendritic spine development. J. Neurosci. 30, 4796-4805. 10.1523/JNEUROSCI.3726-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger G. and Kutsche K. (2006). AlphaPIX and betaPIX and their role in focal adhesion formation. Eur. J. Cell Biol. 85, 265-274. 10.1016/j.ejcb.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Salatino-Oliveira A., Genro J. P., Chazan R., Zeni C., Schmitz M., Polanczyk G., Roman T., Rohde L. A. and Hutz M. H. (2012). Association study of GIT1 gene with attention-deficit hyperactivity disorder in Brazilian children and adolescents. Genes Brain Behav. 11, 864-868. 10.1111/j.1601-183X.2012.00835.x [DOI] [PubMed] [Google Scholar]
- Saneyoshi T., Wayman G., Fortin D., Davare M., Hoshi N., Nozaki N., Natsume T. and Soderling T. R. (2008). Activity-dependent synaptogenesis: regulation by a CaM-kinase kinase/CaM-kinase I/β-Pix signaling complex. Neuron 57, 94-107. 10.1016/j.neuron.2007.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Medina M., Gregus K. A. and Gomez T. M. (2013). PAK-PIX interactions regulate adhesion dynamics and membrane protrusion to control neurite outgrowth. J. Cell Sci. 126, 1122-1133. 10.1242/jcs.112607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenker O. and Rittinger K. (2009). Structures of dimeric GIT1 and trimeric beta-PIX and implications for GIT–PIX complex assembly. J. Mol. Biol. 386, 280-289. 10.1016/j.jmb.2008.12.050 [DOI] [PubMed] [Google Scholar]
- Schmalzigaug R., Garron M. L., Roseman J. T., Xing Y., Davidson C. E., Arold S. T. and Premont R. T. (2007a). GIT1 utilizes a focal adhesion targeting-homology domain to bind paxillin. Cell. Signal. 19, 1733-1744. 10.1016/j.cellsig.2007.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalzigaug R., Phee H., Davidson C. E., Weiss A. and Premont R. T. (2007b). Differential expression of the ARF GAP Genes GIT1 and GIT2 in mouse tissues. J. Histochem. Cytochem. 55, 10393-10348 10.1369/jhc.7A7207.2007 [DOI] [PubMed] [Google Scholar]
- Schmalzigaug R., Rodriguiz R. M., Bonner P. E., Davidson C. E., Wetsel W. C. and Premont R. T. (2009a). Impaired fear response in mice lacking GIT1. Neurosci. Lett. 458, 79-83. 10.1016/j.neulet.2009.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalzigaug R., Rodriguiz R. M., Phillips L. E., Davidson C. E., Wetsel W. C. and Premont R. T. (2009b). Anxiety-like behaviors in mice lacking GIT2. Neurosci. Lett. 451, 156-161. 10.1016/j.neulet.2008.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. H. H., Husnjak K., Szymkiewicz I., Haglund K. and Dikic I. (2006). Cbl escapes Cdc42-mediated inhibition by downregulation of the adaptor molecule betaPix. Oncogene 25, 3071-3078. 10.1038/sj.onc.1209329 [DOI] [PubMed] [Google Scholar]
- Segura I., Essmann C. L., Weinges S. and Acker-Palmer A. (2007). Grb4 and GIT1 transduce ephrinB reverse signals modulating spine morphogenesis and synapse formation. Nat. Neurosci. 10, 301-310. 10.1038/nn1858 [DOI] [PubMed] [Google Scholar]
- Seo M., Cho C.-H., Lee Y.-I., Shin E.-Y., Park D., Bae C.-D., Lee J. W., Lee E.-S. and Juhnn Y.-S. (2004). Cdc42-dependent mediation of UV-induced p38 activation by G protein betagamma subunits. J. Biol. Chem. 279, 17366-17375. 10.1074/jbc.M312442200 [DOI] [PubMed] [Google Scholar]
- Seong M. W., Park J. H., Yoo H. M., Yang S. W., Oh K. H., Ka S. H., Park D. E., Lee S.-T. and Chung C. H. (2014). c-Cbl regulates αPix-mediated cell migration and invasion. Biochem. Biophys. Res. Commun. 455, 153-158. 10.1016/j.bbrc.2014.10.129 [DOI] [PubMed] [Google Scholar]
- Shin E.-Y., Shin K.-S., Lee C.-S., Woo K.-N., Quan S.-H., Soung N.-K., Kim Y. G., Cha C. I., Kim S.-R., Park D. et al. (2002). Phosphorylation of p85 beta PIX, a Rac/Cdc42-specific guanine nucleotide exchange factor, via the Ras/ERK/PAK2 pathway is required for basic fibroblast growth factor-induced neurite outgrowth. J. Biol. Chem. 277, 44417-44430. 10.1074/jbc.M203754200 [DOI] [PubMed] [Google Scholar]
- Shin E.-Y., Woo K.-N., Lee C.-S., Koo S.-H., Kim Y. G., Kim W.-J., Bae C.-D., Chang S.-I. and Kim E.-G. (2004). Basic fibroblast growth factor stimulates activation of Rac1 through a p85 betaPIX phosphorylation-dependent pathway. J. Biol. Chem. 279, 1994-2004. 10.1074/jbc.M307330200 [DOI] [PubMed] [Google Scholar]
- Shirafuji T., Ueyama T., Yoshino K.-I., Takahashi H., Adachi N., Ago Y., Koda K., Nashida T., Hiramatsu N., Matsuda T. et al. (2014). The role of Pak-interacting exchange factor-β phosphorylation at serines 340 and 583 by PKCγ in dopamine release. J. Neurosci. 34, 9268-9280. 10.1523/JNEUROSCI.4278-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirirattanakul S., Wannakrairot P., Tencomnao T. and Santiyanont R. (2015). Gene expression profile in breast cancer comprising predictive markers for metastatic risk. Genet. Mol. Res. 14, 10929-10936. 10.4238/2015.September.21.3 [DOI] [PubMed] [Google Scholar]
- Stafa K., Trancikova A., Webber P. J., Glauser L., West A. B. and Moore D. J. (2012). GTPase activity and neuronal toxicity of Parkinson's disease-associated LRRK2 is regulated by ArfGAP1. PLoS Genet. 8, e1002526 10.1371/journal.pgen.1002526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B. M., Folts C. J., Cui W., Bardin A. L., Walter K., Carson-Walter E., Vescovi A. and Noble M. (2014). Cool-1-mediated inhibition of c-Cbl modulates multiple critical properties of glioblastomas, including the ability to generate tumors in vivo. Stem Cells 32, 1124-1135. 10.1002/stem.1644 [DOI] [PubMed] [Google Scholar]
- Sun Y. and Bamji S. X. (2011). β-Pix modulates actin-mediated recruitment of synaptic vesicles to synapses. J. Neurosci. 31, 17123-17133. 10.1523/JNEUROSCI.2359-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tague S. E., Muralidharan V. and D'Souza-Schorey C. (2004). ADP-ribosylation factor 6 regulates tumor cell invasion through the activation of the MEK/ERK signaling pathway. Proc. Natl. Acad. Sci. USA 101, 9671-9676. 10.1073/pnas.0403531101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay H. G., Ng Y. W. and Manser E. (2010). A vertebrate-specific Chp-PAK-PIX pathway maintains E-cadherin at adherens junctions during zebrafish epiboly. PLoS ONE 5, e10125 10.1371/journal.pone.0010125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Klooster J. P., Jaffer Z. M., Chernoff J. and Hordijk P. L. (2006). Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J. Cell Biol. 172, 759-769. 10.1083/jcb.200509096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A., Paris S., Asperti C. and de Curtis I. (2007). Identification of an intramolecular interaction important for the regulation of GIT1 functions. Mol. Biol. Cell 18, 5124-5138. 10.1091/mbc.E07-06-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A., Tavano S., Filosa G., Gärtner A., Pennucci R., Santambrogio P., Bachi A., Dotti C. G. and de Curtis I. (2012). Biochemical and functional characterisation of αPIX, a specific regulator of axonal and dendritic branching in hippocampal neurons. Biol. Cell 104, 533-552. 10.1111/boc.201100060 [DOI] [PubMed] [Google Scholar]
- Totaro A., Astro V., Tonoli D. and de Curtis I. (2014). Identification of two tyrosine residues required for the intramolecular mechanism implicated in GIT1 activation. PLoS ONE 9, e93199 10.1371/journal.pone.0093199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C. E., Brown M. C., Perrotta J. A., Riedy M. C., Nikolopoulos S. N., McDonald A. R., Bagrodia S., Thomas S. and Leventhal P. S. (1999). Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: a role in cytoskeletal remodeling. J. Cell Biol. 145, 851-863. 10.1083/jcb.145.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale N., Patton W. A., Moss J., Vaughan M., Lefkowitz R. J. and Premont R. T. (2000). GIT proteins, A novel family of phosphatidylinositol 3,4,5-trisphosphate-stimulated GTPase-activating proteins for ARF6. J. Biol. Chem. 275, 13901-13906. 10.1074/jbc.275.18.13901 [DOI] [PubMed] [Google Scholar]
- Volinsky N., Gantman A. and Yablonski D. (2006). A Pak- and Pix-dependent branch of the SDF-1alpha signalling pathway mediates T cell chemotaxis across restrictive barriers. Biochem. J. 397, 213-222. 10.1042/BJ20051655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallings R., Manzoni C. and Bandopadhyay R. (2015). Cellular processes associated with LRRK2 function and dysfunction. FEBS J. 282, 2806-2826. 10.1111/febs.13305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Huo K., Ma L., Tang L., Li D., Huang X., Yuan Y., Li C., Wang W., Guan W. et al. (2011). Toward an understanding of the protein interaction network of the human liver. Mol. Syst. Biol. 7, 536 10.1038/msb.2011.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb D. J., Kovalenko M., Whitmore L. and Horwitz A. F. (2006). Phosphorylation of serine 709 in GIT1 regulates protrusive activity in cells. Biochem. Biophys. Res. Commun. 346, 1284-1288. 10.1016/j.bbrc.2006.06.036 [DOI] [PubMed] [Google Scholar]
- Webb D. J., Zhang H., Majumdar D. and Horwitz A. F. (2007). alpha5 integrin signaling regulates the formation of spines and synapses in hippocampal neurons. J. Biol. Chem. 282, 6929-6935. 10.1074/jbc.M610981200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Wei C., Wang M., Qiu X., Li Y., Yuan Y., Jin C., Leng L., Wang J., Yang X. et al. (2014). The GTPase-activating protein GIT2 protects against colitis by negatively regulating Toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 111, 8883-8888. 10.1073/pnas.1309218111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E., Leszczynska K., Poulter N. S., Edelmann F., Salisbury V. A., Noy P. J., Bacon A., Rappoport J. Z., Heath J. K., Bicknell R. et al. (2014). RhoJ interacts with the GIT–PIX complex and regulates focal adhesion disassembly. J. Cell Sci. 127, 3039-3051. 10.1242/jcs.140434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H., Mah W., Kim E., Kim J.-W., Hahm E.-K., Kim M.-H., Cho S., Kim J., Jang H., Cho S.-C. et al. (2011). GIT1 is associated with ADHD in humans and ADHD-like behaviors in mice. Nat. Med. 17, 566-572. 10.1038/nm.2330 [DOI] [PubMed] [Google Scholar]
- Wu W. J., Tu S. and Cerione R. A. (2003). Activated Cdc42 sequesters c-Cbl and prevents EGF receptor degradation. Cell 114, 715-725. 10.1016/S0092-8674(03)00688-3 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Yuan C., Chen R., Dawson T. M. and Dawson V. L. (2012). ArfGAP1 is a GTPase activating protein for LRRK2: reciprocal regulation of ArfGAP1 by LRRK2. J. Neurosci. 32, 3877-3886. 10.1523/JNEUROSCI.4566-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G., Haendeler J., Yan C. and Berk B. C. (2004). GIT1 functions as a scaffold for MEK1-extracellular signal-regulated kinase 1 and 2 activation by angiotensin II and epidermal growth factor. Mol. Cell. Biol. 24, 875-885. 10.1128/MCB.24.2.875-885.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G., Zheng Q., Yan C. and Berk B. C. (2005). GIT1 is a scaffold for ERK1/2 activation in focal adhesions. J. Biol. Chem. 280, 27705-27712. 10.1074/jbc.M502271200 [DOI] [PubMed] [Google Scholar]
- Yokota T., Kouno J., Adachi K., Takahashi H., Teramoto A., Matsumoto K., Sugisaki Y., Onda M. and Tsunoda T. (2006). Identification of histological markers for malignant glioma by genome-wide expression analysis: dynein, alpha-PIX and sorcin. Acta Neuropathol. 111, 29-38. 10.1007/s00401-005-1085-6 [DOI] [PubMed] [Google Scholar]
- Yoo S. M., Antonyak M. A. and Cerione R. A. (2012). The adaptor protein and Arf GTPase-activating protein Cat-1/Git-1 is required for cellular transformation. J. Biol. Chem. 287, 31462-31470. 10.1074/jbc.M112.353615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. A., Foley F. C., Amack J. D. and Turner C. E. (2011). The cell adhesion-associated protein Git2 regulates morphogenetic movements during zebrafish embryonic development. Dev. Biol. 349, 225-237. 10.1016/j.ydbio.2010.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-J., Cavenagh M. M. and Kahn R. A. (1998). A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273, 19792-19796. 10.1074/jbc.273.31.19792 [DOI] [PubMed] [Google Scholar]
- Zhang H., Webb D. J., Asmussen H. and Horwitz A. F. (2003). Synapse formation is regulated by the signaling adaptor GIT1. J. Cell Biol. 161, 131-142. 10.1083/jcb.200211002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Webb D. J., Asmussen H., Niu S. and Horwitz A. F. (2005). A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J. Neurosci. 25, 3379-3388. 10.1523/JNEUROSCI.3553-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. M., Simmerman J. A., Guibao C. D. and Zheng J. J. (2008). GIT1 paxillin-binding domain is a four-helix bundle, and it binds to both paxillin LD2 and LD4 motifs. J. Biol. Chem. 283, 18685-18693. 10.1074/jbc.M801274200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Cai W., Yin G., Nagel D. J. and Berk B. C. (2009). GIT1 is a novel MEK1-ERK1/2 scaffold that localizes to focal adhesions. Cell Biol. Int. 34, 41-47. 10.1042/cbi20090016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.-S., Manser E., Loo T.-H. and Lim L. (2000). Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol. Cell. Biol. 20, 6354-6363. 10.1128/MCB.20.17.6354-6363.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. and Thiery J. P. (2013). Loss of Git2 induces epithelial–mesenchymal transition by miR146a-Cnot6L-controlled expression of Zeb1. J. Cell Sci. 126, 2740-2746. 10.1242/jcs.126367 [DOI] [PubMed] [Google Scholar]