ABSTRACT
Many actin filaments in animal cells are co-polymers of actin and tropomyosin. In many cases, non-muscle myosin II associates with these co-polymers to establish a contractile network. However, the temporal relationship of these three proteins in the de novo assembly of actin filaments is not known. Intravital subcellular microscopy of secretory granule exocytosis allows the visualisation and quantification of the formation of an actin scaffold in real time, with the added advantage that it occurs in a living mammal under physiological conditions. We used this model system to investigate the de novo assembly of actin, tropomyosin Tpm3.1 (a short isoform of TPM3) and myosin IIA (the form of non-muscle myosin II with its heavy chain encoded by Myh9) on secretory granules in mouse salivary glands. Blocking actin polymerization with cytochalasin D revealed that Tpm3.1 assembly is dependent on actin assembly. We used time-lapse imaging to determine the timing of the appearance of the actin filament reporter LifeAct–RFP and of Tpm3.1–mNeonGreen on secretory granules in LifeAct–RFP transgenic, Tpm3.1–mNeonGreen and myosin IIA–GFP (GFP-tagged MYH9) knock-in mice. Our findings are consistent with the addition of tropomyosin to actin filaments shortly after the initiation of actin filament nucleation, followed by myosin IIA recruitment.
KEY WORDS: Actin, Tropomyosin, Intravital, Myosin II, Cytoskeleton, Assembly kinetics
Summary: De novo assembly of functional actin–tropomyosin filaments in live rodents is investigated. Unlike non-muscle myosin II, tropomyosin is added to the actin filament shortly after nucleation.
INTRODUCTION
Animal cells contain two types of actin filaments; those containing only actin or, alternatively, those containing co-polymers of actin and tropomyosin (Tpm) (Gunning et al., 2015b; Lazarides, 1975, 1976). Studies in yeast (Johnson et al., 2014), flies (Goins and Mullins, 2015) and mammals (Bryce et al., 2003; Tojkander et al., 2011) have shown that the Tpm isoform determines the functional capacity of the filament (Gunning et al., 2015a). Multiple models, ranging from addition of Tpm after actin filament formation to co-polymerisation, could explain the formation of the actin–Tpm co-polymer (Holmes and Lehman, 2008; Wegner, 1979), but there is no experimental data to discriminate between these models.
Several ‘large-granule’ in vitro model systems have been used to investigate the initial assembly of actin filament structures (Jang et al., 2012; Miklavc et al., 2012, 2015; Nightingale et al., 2011; Sokac et al., 2003; Tran et al., 2015; Yu and Bement, 2007). Actin scaffolds encompassing the granules contain both branched and linear actin filaments (Porat-Shliom et al., 2013; Tran et al., 2015), similar to what is found in the cortical cytoskeleton and podosomes (Bovellan et al., 2014; Panzer et al., 2016). Here, we use the stimulation of secretory granule release in salivary gland acinar cells, where actin scaffold formation can be initiated by β-adrenergic receptor agonist application and then quantified in real time in live rodents using intravital subcellular microscopy (Masedunskas et al., 2011).
RESULTS AND DISCUSSION
Tpm3.1 is associated with actin filaments initiated around fusing secretory granules
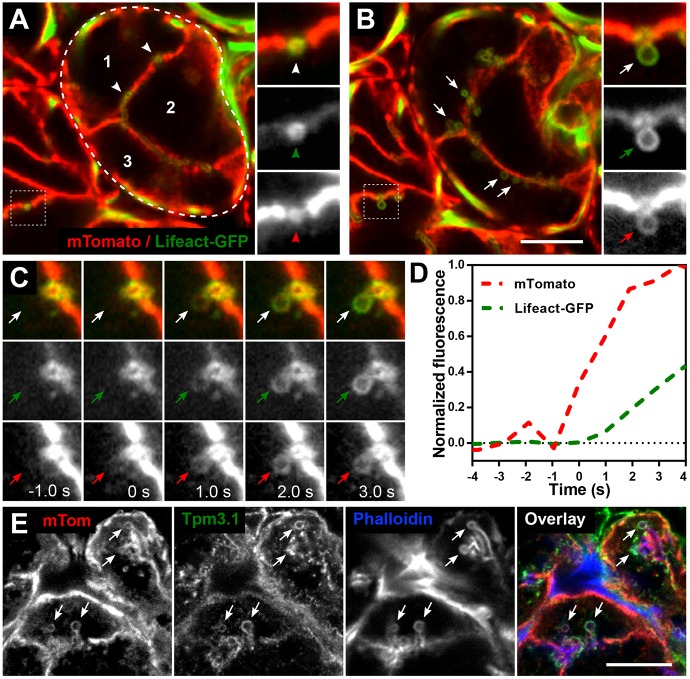
Fig. 1A and Fig. S1A,B show a cross-section of a mouse salivary gland acinus expressing plasma membrane (PM)-localised tdTomato (mTomato) and Lifeact–GFP, which efficiently mark the PM of acinar cells (Muzumdar et al., 2007) and filamentous actin, respectively. Both subcellular markers outline the canaliculi into which secretory granules release their contents (arrowheads). mTomato rapidly transfers to the granule membrane upon fusion with the apical plasma membrane (APM) of the canaliculus (Masedunskas et al., 2011).
Fig. 1.
De novo actin and Tpm 3.1 filament polymerisation forms a scaffold around granules after fusion with the APM. (A–D) Intravital confocal imaging of actin filament assembly during secretory granule exocytosis in submandibular salivary gland acinar cells in the progeny of an mTomato and Lifeact–GFP mouse cross. (A,B) Snapshots of salivary acini in situ showing the membrane marker mTomato (red) and F-actin marker Lifeact–GFP (green). (A) Snapshot was taken at a depth of 12 µm from the surface of the gland. Three individual acinar epithelial cells (numbers 1–3) are part of one acinus (encircled by dashed line). Actin is highly enriched at the APM/canaliculi (white arrowheads). Enlarged split channel images displayed on the right show an APM/canaliculus cross-section (red arrowhead) enriched with F-actin (green arrowhead). (B) The same area as in A was imaged 5 min after subcutaneous injection of isoproterenol. Granules fused to APM are seen (white arrows) after stimulation with isoproterenol. Enlarged split channel images show actin recruitment (green arrow) onto the fused granule as seen by the appearance of the mTomato membrane marker (red arrow). Scale bar: 10 µm; the width of the insets is 5.25 µm. See also Fig. S1 and Movie 1. (C) Enlarged time-course images of a representative granule fusion event. The granule acquires an mTomato signal at t=0 s, marking the fusion event (red arrows), and Lifeact–GFP is first detected around the granule circumference at t=1 s (green arrows). Each panel is 5.25 µm wide and the granule diameter is ∼1.2 µm; temporal sampling is 943 ms per frame. (D) Recruitment profiles of mTomato and Lifeact–GFP during a granule fusion event are shown as normalised fluorescence intensity over time. Increase in mTomato (red line) signal from baseline indicates fusion between the granule membrane and the APM. Increase in Lifeact–GFP (green line) indicates actin filament assembly. The frame before the first detection of actin polymerisation was set at t=0 s for this and all subsequent graphs. (E) Detection of mTomato, Tpm3.1 and actin (phalloidin) on fused granules (arrows) in a fixed salivary gland section from an mTomato mouse at 10 min after isoproterenol injection. The contrast was adjusted for each channel separately to facilitate consistent visualisation of the granules in all figures. Scale bar: 10 µm.
Fig. 1B shows the same region of the gland 5 min after stimulation of secretory granule fusion by subcutaneous injection of 0.01 mg/kg body weight isoproterenol. Arrows identify granules which have fused with the APM. Resting granules are not detectable because they are not associated with either mTomato or the actin polymer prior to fusion (Fig. 1A), as shown previously (Masedunskas et al., 2011). Granules that have fused with the APM display labelling with both mTomato and Lifeact–GFP (expanded views in Fig. 1B; Movie 1). Comparison of the kinetics of mTomato transfer to a granule membrane and Lifeact association with the single granule undergoing fusion shows that actin filament formation closely follows initial granule fusion with the plasma membrane (Fig. 1C; Movie 1). The initial increase in Lifeact–GFP fluorescence lags ∼1 s behind the mTomato signal increase (Fig. 1D; Fig. S1C).
The temporal relationship of actin filament formation and Tpm association with granule membranes was first studied by detecting the endogenous proteins. All granules engaged in APM fusion, as detected by mTomato transfer to the granules, displayed phalloidin staining (Fig. 1E). Co-staining for Tpm3.1 (a short isoform of TPM3) revealed that granules positive for phalloidin were also positive for Tpm3.1 (Fig. 1E). Almost all granules in the four mice displayed coincidence of phalloidin and Tpm3.1 (Table S1). The failure to detect many granules that are both phalloidin-positive and Tpm3.1-negative suggests that Tpm3.1 recruitment to the actin filament occurs at the same time as, or shortly after, actin assembly. Similarly, the failure to detect any granules that are both Tpm3.1-positive and phalloidin-negative is consistent with Tpm3.1 recruitment being dependent on the presence of actin filaments.
Tpm3.1 assembly requires actin, but actin assembly does not require Tpm3.1
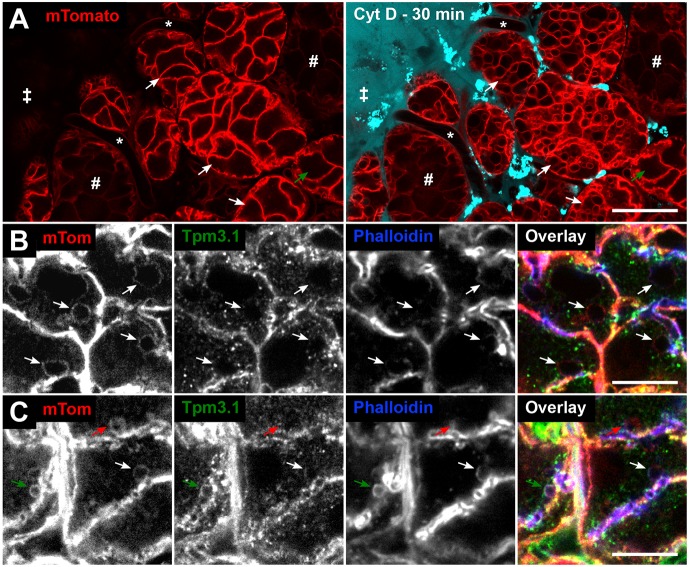
Cytochalasin D, which disrupts actin polymer elongation (Cooper, 1987), was delivered into the salivary glands of mTomato mice via local perfusion. Alexa Fluor 647-labelled dextran, which does not enter the acinar cells, was used to track the presence of the drug solution and the location of cells exposed to it. Prolonged exposure to cytochalasin D results in dramatic swelling of the fused granules and inward blebbing of the APM, thus filling the cells with vacuoles (Masedunskas et al., 2011). Acinar cells further from the edge were less affected by the drug (Fig. 2A, green arrow). Acinar cells that were close to the edge of the organ contained granules with mTomato, indicating membrane fusion; however, the granules displayed a complete lack of phalloidin staining and, in parallel, no detectable Tpm3.1 (Fig. 2B). This indicates that Tpm3.1 is not recruited to granules in the absence of actin filaments and that Tpm3.1 forms a co-polymer with actin.
Fig. 2.
Tpm3.1 recruitment onto fused granules is dependent on actin filaments. (A) Overview of intravital imaging of cytochalasin D (CytD) treatment of salivary glands in mTomato mice [stroma (‡), intact capillaries (*), ducts (#)]. Snapshots of glands before (left) and 30 min after (right) CytD treatment. The drug solution contained 10 kDa Alexa Fluor 647 dextran (cyan). CytD treatment for 30 min results in the formation of large vacuoles in acini (white arrows indicate the same acini pre- and post-CytD). CytD penetrance is not uniform, as shown by an unaffected acinus (green arrow) further from the edge of the organ. Enrichment of bright dextran puncta represent rapid dextran marker uptake by the resident stromal cells, such as dendritic cells and fibroblasts. Scale bar: 20 µm. (B,C) Detection of mTomato, Tpm3.1 and actin (phalloidin) on fused granules (arrows) in a salivary gland section of an mTomato mouse treated with CytD (10 min), followed by isoproterenol injection (10 min). (B) CytD-affected granules are enlarged (arrows) and largely devoid of both Tpm3.1 and F-actin staining. (C) The impact of CytD is variable, with some granules exhibiting full (red arrow) or no (green arrow) impact on the actin scaffold. The white arrow shows a fused granule with partial recruitment (crescent-like patch) of F-actin and Tpm3.1. Scale bars: 10 µm.
Further inside the gland, there was only partial inhibition of actin filament formation (evident in Fig. 2A), resulting in patches of F-actin localising on granule membranes (Fig. 2C). Comparison with Tpm3.1 staining shows co-incidence of phalloidin and Tpm3.1 staining on such partially inhibited granules (white arrow in Fig. 2C). This is consistent with Tpm3.1 depending on actin filament formation to assemble on the granule surface and Tpm3.1 forming a co-polymer with actin in vivo.
Tpm3.1-knockout (KO) mice (Vlahovich et al., 2009) were stimulated with isoproterenol and their glands were stained with phalloidin. Phalloidin staining of granules was not affected by the absence of Tpm3.1 (Fig. S2A,B). This indicates that Tpm3.1 is not required for actin filament assembly on the granules; although, we cannot rule out a role for other Tpm isoforms. Indeed, antibody staining of fixed salivary gland sections revealed the presence of Tpm4.2 (an isoform of TPM4) on the apical plasma membrane and the granules (Fig. S1D).
Generation of a Tpm3.1–mNeonGreen KI mouse
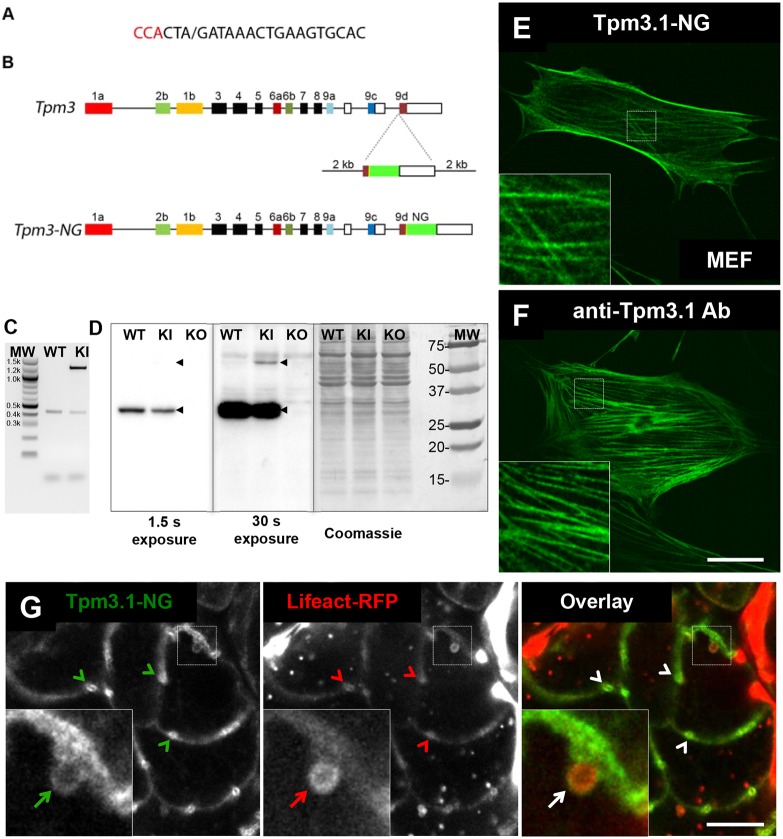
In order to examine the behaviour of the endogenous Tpm3.1 protein in real time, CRISPR-Cas9 was used to introduce the mNeonGreen (NG) tag (Shaner et al., 2013) into the C-terminus of the gene encoding Tpm3.1 (Fig. 3). The mice were fertile and transmitted the tagged allele at a similar frequency to the endogenous allele, and PCR products of mouse DNA confirmed the presence of the wild-type (WT) and knock-in (KI) alleles (Fig. 3C). Western blots of Tpm3.1–NG mouse embryo fibroblasts (MEFs) showed that Tpm3.1–NG accumulates at levels <10% of WT Tpm3.1 (Fig. 3D). Tpm3.1–NG-expressing MEFs displayed a similar pattern of Tpm3.1–NG localization to that seen with anti-Tpm3.1 staining of WT MEFs (Fig. 3E,F; Fig. S2E). Intravital microscopy of the salivary glands of a Tpm3.1–NG KI mouse crossed to a Lifeact–RFP transgenic mouse showed that there was apical enrichment of Tpm3.1–NG (Fig. 3G, arrowheads) and localisation to fused secretory granules (arrows) characteristic of endogenous Tpm3.1 in acinar cells (Fig. 2B and C). Thus, the tagged gene product can incorporate with fidelity into actin filament bundles, but is either synthesised at a lower rate or is more rapidly turned over than the WT product.
Fig. 3.
Generation of Tpm3.1–mNeonGreen KI mouse. (A,B) The design strategy used to create fluorescently tagged KI mice via CRISPR. (A) Guide RNA used with the PAM site in red and the cut site marked with ‘/’. (B) Structure of the Tpm3 gene. (C) Agarose gel showing PCR products using DNA extracted from WT and hemizygous KI mice as a template. (D) Western blots from MEF cells isolated from WT, Tpm3.1–NG KI and Tpm3.1 KO mice. The major product is detected by anti-Tpm3.1 antibody at ∼30 kDa in WT and KI lanes, and not in the KO lane. The KI product of ∼57 kDa is only detected after 30 s exposure. Wild-type and fusion protein Tpm3.1 bands are indicated with black arrowheads. Coomassie staining was used as a loading control (right panel). Molecular mass standards (MW) are shown in kDa. (E,F) MEF cells from Tpm3.1-NG KI and WT mice. (E) Confocal image of Tpm3.1–NG localised to stress fibres (inset) in Tpm3.1–NG KI MEF cells. (F) Confocal image of Tpm3.1 localised to stress fibres (inset) in WT MEF cells detected by anti-Tpm3.1 antibody. Scale bar: 20 µm. (G) Snapshot of a single acinus from the progeny of a Tpm3.1–NG mouse crossed with a Lifeact–RFP mouse, imaged with intravital confocal microscopy after injection of isoproterenol. Cortical distribution and enrichment of Tpm3.1–NG (green) and actin filaments marked by Lifeact (red) at APM/canaliculi is evident (arrowheads). The inset shows a fused granule (white arrow) that is enveloped by an F-actin scaffold (red arrow) containing Tpm3.1 filaments (green arrow). Scale bar: 10 µm; the width of the insets is 6.5 µm.
The Tpm3.1–NG KI mouse shows similar recruitment kinetics for actin and Tpm3.1
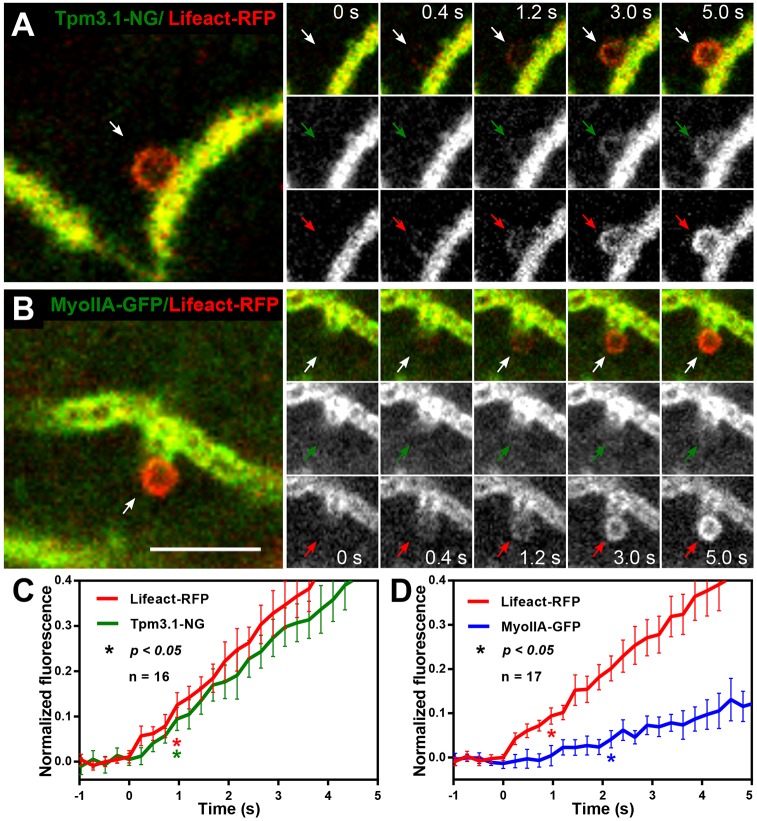
The progeny of the Tpm3.1–NG and Lifeact–RFP mouse cross were used to visualise and quantify de novo assembly of actin and endogenous Tpm3.1. Tpm3.1–NG and Lifeact–RFP exhibit parallel accumulation on the granules during the first few seconds of granule fusion (Fig. 4A; Movie 2). Measurements for 16 granule-fusion events from multiple animals were averaged and the kinetics for the two tags were compared (Fig. 4C). Full kinetics are shown in Fig. S3A,B. The first significant Lifeact signal increase above baseline was detected at 965 ms, the same time point at which the first significant increase in Tpm3.1 signal was registered (Fig. 4C). This further suggests that the initiation of actin polymerisation and the co-assembly of Tpm3.1 are near-simultaneous.
Fig. 4.
The initial actin and Tpm3.1 filament assembly exhibits a close temporal relationship, unlike what is seen with myosin IIA. (A) Live intravital confocal imaging of de novo cytoskeleton assembly after isoproterenol-stimulated secretory granule fusion in the progeny of a Tpm3.1–NG KI and Lifeact-RFP mouse cross. The snapshot (left) shows a fused secretory granule (white arrow) near the APM/canaliculus. The image sequence (right) shows the progression of Tpm3.1–NG (green arrows) and Lifeact–RFP (red arrows) localisation over time. See also Movie 2. (B) Snapshot of intravital confocal imaged salivary acini in situ in the progeny of a myosin IIA–GFP KI and Lifeact–RFP mouse cross (left) after isoproterenol injection. Time-lapse sequence (right) showing the progression of actomyosin scaffold assembly around the granule with F-actin (red arrows) and myosin IIA (green arrows) localisation over time. See also Movie 3. (C) Recruitment kinetics of Lifeact–RFP and Tpm3.1–NG acquired at 241 ms intervals shown as mean normalised fluorescence ±95% CI for each time point. An average of 16 fusion events from four mice were plotted. The first significant (P<0.05) increase in fluorescence intensity versus the zero time point was determined by one-way ANOVA and Dunnett's multiple comparison test for Lifeact–RFP (red asterisk, P=0.0025) and Tpm3.1–NG (green asterisk, P=0.0215). (D) Recruitment kinetics of Lifeact–RFP (red line) and myosin IIA (blue line) acquired at 241 ms intervals are shown as normalised mean fluorescence intensities±95% CI from 17 fusion events in four mice. The first significant (P<0.05) increase in fluorescence intensity versus the zero time point was determined, as in Fig. 4C, for Lifeact–RFP (red asterisk, P=0.0283) and myosin IIA (blue asterisk, P=0.0091). Scale bar: 5 µm; the width of the insets is 6.16 µm.
Myosin IIA recruitment kinetics lag behind those of actin and Tpm3.1
Tpm3.1 has been shown to recruit myosin IIA (the form of non-muscle myosin II with its heavy chain encoded by Myh9) to stress fibres in neuroblastoma cells (Bryce et al., 2003), and myosin IIA and IIB (non-muscle myosin II with its heavy chain encoded by Myh10) isoforms have recently been shown to be recruited to salivary gland granules (Milberg et al., 2017). Interestingly, Tpm3.1 genetic ablation did not affect endogenous myosin IIA localisation on fused granules in mouse salivary gland acinar cells (Fig. S2C,D). We therefore focused on the kinetics of myosin IIA recruitment, and the temporal relationship with actin and Tpm3.1 recruitment in wild-type animals. To compare myosin IIA–GFP (GFP-tagged MYH9) with Tpm3.1–NG kinetics, we first established that at the same imaging parameters, the two tags yield similar signal intensity (Fig. S3C). Myosin IIA is recruited to granules that are positive for Lifeact–RFP (Fig. 4B; Fig. S4A); however, Lifeact was found on all granules before myosin IIA (compare panels in Fig. 4B; Movie 3). Averaged binding curves for Lifeact–RFP and myosin IIA–GFP are shown in Fig. 4D. Full kinetics are shown in Fig. S4B,C. Lifeact–RFP and myosin IIA–GFP show very different kinetics, unlike Lifeact and Tpm3.1, indicating slower recruitment of myosin IIA than of Lifeact (Fig. 4D). Evaluation of the early time points revealed a 1.2 s gap between the first significant increase in Lifeact and myosin IIA signals (Fig. 4D). Differences in kinetics between Lifeact–RFP and myosin IIA–GFP were also supported by line scan analysis of example fusion events (Figs S3C, S4D). A similar lag between initial actin filament formation and myosin II recruitment on fused granules has been shown in the salivary glands of mice (Milberg et al., 2017) and in Drosophila larvae (Tran et al., 2015).
Assembly of actin, Tpm3.1 and myosin IIA on secretory granule membranes
Based on our measurements, we draw the following conclusions: (1) incorporation of Tpm3.1 depends on actin assembly, whereas in contrast actin assembles with the same kinetics regardless of the presence of Tpm3.1, (2) rapid Tpm3.1 incorporation into filaments occurs shortly after actin filaments are first detected (within 240 ms), (3) myosin IIA recruitment lags behind that of Tpm3.1 and (4) actin and Tpm3.1 have very similar initial recruitment kinetics whereas myosin IIA has comparatively slow initial recruitment kinetics. Taken together, these observations point to a process in which, shortly after actin polymerisation commences, Tpm3.1 co-assembles onto these actin filaments, and this is followed later by recruitment of the myosin IIA motor. During the very early stages of the process, new filaments in the structure surrounding the granule are being nucleated and grow in length.
It has been suggested that the actin filament forms first and then Tpm is added to the actin template (Holmes and Lehman, 2008; Wegner, 1979). Several research groups have shown that Tpm will nucleate at multiple sites along a single actin filament (Alioto et al., 2016; Christensen et al., 2017; Nicovich et al., 2016; Schmidt et al., 2015). However, because Tpm3.1 binds to six actin monomers in mammals, there is only a one in six chance that multiple Tpm3.1 nucleation events would join with each other ‘in register’ as they extend along the actin, resulting in gaps in Tpm3.1 coverage. This problem is resolved if Tpm3.1 is added to the pointed end (Hsiao et al., 2015) of growing actin filaments shortly after their nucleation and continues to co-polymerise with actin as the filament is extended. The observation that formin nucleators regulate Tpm incorporation into actin filaments (Alioto et al., 2016; Johnson et al., 2014; Tojkander et al., 2011) is compatible with such a proposal.
MATERIALS AND METHODS
Animals
Animal experiments were performed in accordance with the NSW Animal Research Act (1985) and Australian National Health and Medical Research Council (NHMRC) ‘Code’ 8th edition (2013). All experiments were approved by the UNSW Sydney Animal Care and Ethics Committee under applications 14/92A and 17/100B. Male mice (Mus musculus) aged 8–24 weeks were used in this study. Mice were sourced as described below, and colonies were maintained at the Australian BioResources Facility (Moss Vale, NSW, Australia) and allowed to acclimatise for 1 week prior to experimentation. Animals were housed in specific pathogen-free, humidity- and temperature-controlled facilities, under a 12 h:12 h light:dark cycle, and provided standard laboratory chow and water ad libitum. Tpm3.1 knock-out (KO) mice were previously generated in house (Vlahovich et al., 2009). Tpm3.1 KO mice lack the 9d exon of the Tpm3 gene and are unable to make the Tpm3.1 and Tpm3.2 isoforms. mTomato mice have been described previously (Muzumdar et al., 2007). All tissues in these mice express tandem dimer tomato fluorescent protein (tdTomato) fused to the plasma membrane (PM)-targeting peptide from MARCKS, making this a PM reporter mouse line (Masedunskas et al., 2011). The mTomato mouse strain was purchased from Jackson Laboratory (Bar Harbor, ME, USA) and used alone or in a cross to Lifeact–GFP mice in a homozygous state. Lifeact–GFP and Lifeact–RFP transgenic mice were a gift from Roland Wedlich-Soldner (Riedl et al., 2010) and were used in crosses in a hemizygous state. Myosin IIA–GFP knock-in (KI) mice were generated as described (Zhang et al., 2012) and used in Lifeact–RFP crosses in a homozygous state. The genotypes of the mice were confirmed by PCR, as described in the original publications. Tpm3.1 from the endogenous locus was C-terminally tagged with mNeonGreen (NG) in a mouse line using CRISPR at the South Australia Genome Editing Facility (SAGE, University of Adelaide, Australia). Target sites near Tpm3 exon 9d were identified using the MIT CRISPR Design Tool (Ran et al., 2013). One high-scoring guide RNA (gRNA) sequence was selected that cut at the 5′ intron–exon boundary of exon 9d (CCACTA/GATAAACTGAAGTGCAC; / indicates the cut site). The plasmid repair template was synthesised by GeneArt (Thermo Fisher Scientific, Waltham, MA) and consisted of 2 kb homology arms flanking a mutated PAM site (TCA), exon 9d and a 10 amino acid linker (GGGGSGGGGS), and the NG sequence (a gift from Jiwu Wang, Department of Photobiology and Bioimaging, The Scintillon Institute, San Diego, CA, USA) (Shaner et al., 2013) inserted in frame with exon 9d (Fig. 3). The Tpm3.1–NG construct has been validated by analysing ectopic expression in mouse embryonic fibroblasts (Appaduray et al., 2016). The T in the PAM site was selected as the mutation as it is preferable for splicing efficiency (Dogan et al., 2007). C57Bl/6 mouse zygote pronuclei were injected with Cas9 mRNA (25 ng/μl), CRISPR guide RNA (10 ng/μl) and circular plasmid donor. Surviving zygotes were transferred to pseudopregnant female recipients and the resulting pups were screened for homologous integration of the donor template by PCR using genomic DNA extracted from ear biopsy samples using forward PCR primer, 5′-TCCTTGCAACTGGAGCAAGTC-3′, and reverse PCR primer, 5′-TGCATTGAAGACACTGCGTTG-3′. Hemizygous Tpm3.1–NG KI mice (C57Bl/6 background) were used for experiments and to generate a cross with Lifeact–RFP mice in a hemizygous state. The animals were genotyped by PCR (Fig. 3C) using forward primer, 5′-CAGTAGGCAAGCCAGAGACC-3′, and reverse primer, 5′-CCAACTGGAACTCTGGGAAA-3′, resulting in a 1143 bp band for the KI allele and 403 bp band for the WT allele. New England Biolabs (Ipswich, MA) 100 bp DNA Ladder was used to confirm the size of the bands. The presence of the Tpm3.1–NG protein was confirmed by western blotting and in situ fluorescence microscopy (Fig. 3).
Cell culture and immunofluorescence staining
Mouse embryo fibroblasts (MEFs) were isolated from day 14.5 C57Bl/6 WT, Tpm3.1 KO and Tpm3.1–NG KI mouse embryos and cultured as previously described (Tojkander et al., 2011). Cells were grown in Dulbecco's modified Eagle’s medium (DMEM) with 10% (v/v) fetal bovine serum (FBS) at 37°C and 5% CO2. To visualise Tpm3.1–NG localization, KI MEFs were fixed in 4% paraformaldehyde (PFA) at room temperature (RT) for 15 min and washed in PBS three times. For immunofluorescence staining, WT MEFs at passages 1–3 were seeded into Fluorodish tissue culture dishes (World Precision Instruments Pty Ltd, Hitchin, UK) and grown to 70–90% confluence. Cells were fixed in 4% PFA at RT for 15 min, permeabilised in cold methanol for 15 min, washed in PBS and blocked in 2% BSA in PBS at RT for 60 min. Cells were incubated with CG3 star 635 antibody 1:250 in PBS at RT for 1 h. CG3 Star 635 mouse monoclonal antibody (Novy et al., 1993) conjugated to an Abberior-635 dye recognises the 1b exon from the TPM3 gene. Cells were washed 3 times with PBS, mounted with Immuno-Mount (Thermo Fisher Scientific) and imaged using a Nikon A1 inverted scanning confocal microscope fitted with a Nikon Plan Apochromat lambda series 60×/1.4 NA oil immersion objective. CG3 Star 635 (emission wavelength 654 nm) was excited with a 640 nm laser. mNeonGreen fluorescent protein (emission wavelength 517 nm) was excited with a 488 nm laser.
Western blotting
Cell lysates were made from WT, Tpm3.1 KO and Tm3.1–NG KI MEFs in RIPA buffer and analysed by SDS-PAGE and western blotting, as described previously (Tojkander et al., 2011). Protein concentration was determined by using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of protein (20 µg) were separated on a 12% SDS-PAGE gel and electro-transferred to PVDF membranes. Membranes were incubated with blocking buffer and 5% skimmed milk powder in TBS (100 mM Tris-HCl, pH 7.5, 150 mM NaCl). Tpm 3.1 was recognised by using monoclonal antibody CG3 (1:200 in blocking buffer) (Tojkander et al., 2011) and secondary antibody rabbit anti-mouse IgG antibody-conjugated to horseradish peroxidase (Abcam) (1:10,000 in blocking buffer). The primary antibody was incubated overnight and the secondary antibody was incubated for 2 h, followed by 4×15 min washes in PBS. Blots were developed with the Luminata Crescendo Western HRP Substrate (Millipore), and 1–120 s exposures were taken via the Chemidoc Imaging System (Bio-Rad Laboratories). Protein loading was examined by staining the protein gel blots with 0.1% (w/v) Coomassie Brilliant Blue R350, 20% (v/v) methanol and 10% (v/v) acetic acid.
Tissue sectioning and staining
Fixed sucrose-infused frozen sections (Figs 1 and 2) were made as follows. Adult male C57Bl/6 mice were anaesthetised by intraperitoneal injection of 100 mg/kg body weight ketamine and 15 mg/kg body weight xylazine. Exocytosis of secretory granules was stimulated by subcutaneous injection of 0.01 mg/kg body weight isoproterenol for 10 min. In some experiments, surgically externalised salivary glands (SGs) were locally treated with 10 µM cytochalasin D 10 min prior to exocytosis stimulation. A drug solution was prepared by dissolving in pre-warmed (37°C) saline, which was then added dropwise onto the SGs for the duration of the experiment; 10 min after isoproterenol injection, SGs were excised, immersed in 2% PFA at RT for 1 h and transferred into 15% sucrose overnight. Sucrose-infused tissues were placed in plastic tissue moulds in Tissue-Tek O.C.T. and frozen in isopentane cooled in liquid nitrogen. Then, 10 μm sections were cut (Cryostat, Leica CM1950) and incubated with blocking buffer (5% goat serum, 5% FBS, 1% BSA in PBS) at RT for 1–2 h. Mouse mAb 2G10 (1:2.5 in blocking buffer), which recognises the 9d exon from the Tpm3 gene (Tpm3.1 and 3.2 isoforms) (Tojkander et al., 2011), was added at RT for 12 h, and then secondary antibodies conjugated to Alexa Fluor 488 (mouse; Invitrogen) (1:750 in blocking buffer) with either phalloidin labelled with Atto-647N (ATTO-TEC) or TRITC (Sigma-Aldrich) (1:250 in blocking buffer) were added at RT for 1–2 h. Tpm4.2 and myosin IIA were detected with delta9d rabbit polyclonal (1:100 dilution; Millipore, AB5449) and myosin IIA (1:100 dilution; Covance, PRB-440P), respectively. Images were taken with an Olympus FV1000 laser scanning confocal microscope equipped with an UPLAN 60×/1.35 NA oil immersion objective at 100 nm/pixel spatial sampling density. For granule overlap scoring, a Nikon A1 laser scanning confocal microscope equipped with a CFI Plan Apochromat lambda series 60×/1.4 NA oil immersion objective was used. Seven z-slices were taken every 200 nm sampling over 1.2 µm depth at 120 nm/pixel spatial sampling density. Fused granules were identified in the phalloidin channel as spherical structures with a diameter of 0.5–1.6 µm that were connected to the APM that lines the canaliculi. They were then scored for the presence of Tpm3.1 as detected by 2G10 antibody staining (Table S1).
Intravital microscopy
Mice (18–34 g) were anaesthetised by intraperitoneal injection of 100 mg/kg body weight ketamine and 15 mg/kg body weight xylazine, SGs were surgically externalised and prepared for subcellular intravital microscopy as described (Masedunskas et al., 2013, 2011). The SG was stabilised onto a coverslip on a microscope stage with the vasculature and the innervation still intact. Intravital imaging was performed using a Nikon A1 inverted laser scanning confocal microscope fitted with a CFI Plan Apochromat lambda series 60×/1.27 NA water immersion objective, an Okolab humidified temperature-controlled microscope enclosure, objective heater and a custom-made stage insert. Animal body and SG temperatures were monitored with a MicroTherma T2 thermometer equipped with rectal and thin implantable probes (Braintree Scientific) and heating was adjusted to maintain the temperature at 37–38°C. GFP and NG were excited with a 488 nm laser, and RFP and tdTomato were excited with a 561 nm laser. For time-lapse imaging, frames were acquired in sequential mode at 943 ms/frame (Fig. 1) or 241 ms/frame (all other figures) at a spatial sampling density of 140 nm per pixel. An imaging plane with a visible APM/canaliculus was selected as close to the surface of the organ as possible – at a depth of 8-12 µm. During imaging, any noticeable drift was manually corrected in the x, y or z dimension. For some experiments, 10 µM cytochalasin D was prepared by mixing it with pre-warmed saline [with or without 2 mg/ml Alexa Fluor 647-conjugated 10 kDa dextran (Thermo Fisher Scientific)] and perfused over the surface of the gland at 3 µl/min during the imaging using a PHD Ultra Nanomite programmable syringe pump (Harvard Apparatus). Exocytosis of secretory granules was stimulated by subcutaneous injection of 0.01 mg/kg body weight isoproterenol.
Image quantification and statistics
Images were taken using NIS Elements software. Image processing and data extraction was performed by using ImageJ/Fiji (Schindelin et al., 2012) and Metamorph (Molecular Devices) software. When necessary, drift and motion correction on time-lapse image stacks was carried out with the StackReg ImageJ plugin (Thevenaz et al., 1998). To extract fluorescence trace data from single-granule fusion events, a circular region of interest (ROI) with a diameter of 15 pixels was drawn over the image area capturing the exocytosis event. Only exocytosis events fully captured for the first 20–30 s in the focal plane and devoid of drift, motion artefacts and signal contribution from additional objects, such as plasma membrane, were included in the analysis. Integrated pixel fluorescence intensity values from the ROI were extracted and transferred to Microsoft Office Excel for processing as separate channels. Time t=0 for all events was taken as the time point preceding the first significant increase in Lifeact fluorescence above the baseline value. The baseline or background values for each channel were calculated by averaging ∼5 s worth of data points preceding an increase in fluorescence. The fluorescence intensity values may vary between mice or cells; therefore, the granule fluorescence traces were normalised individually by first subtracting the calculated average baseline value for each trace and then expressing the result as fractional values, with one being the maximum and zero being the minimum/background value. The maximum fluorescence value was picked for each trace and channel by using the MAX function in Excel, which returns the largest value from a sequence of numbers. Separate granule exocytosis events of a given data set were then synchronised to time t=0 with respect to Lifeact fluorescence and plotted from 10 s before the initiation of Lifeact signal increase until 70 s after t=0, when the fluorescence intensity returns to baseline (Figs S3,S4). Mean intensity values±95% CI were calculated and plotted by using Graphpad Prism 7 (GraphPad Software) in Fig. 4. Mean intensity values at each time point were analysed by one-way ANOVA, and the first time point exhibiting a significant increase (P<0.05) in fluorescence was identified with a Dunnett's multiple comparison test of each time point versus the zero time point. A total of 16 or 17 granule fusion events from four animals were quantified. Line scan analysis was performed across fusing granules over time (Figs S3C and S4D). A 3 μm line was drawn across each granule in the raw images and the average (over two pixels) fluorescence intensities in the red and green channels were recorded along the line in Metamorph software. Images for the figures were adjusted for contrast and brightness in Metamorph or ImageJ to have an optimal display range for features of interest, such as secretory granules, and then converted into RGB images. In some cases, the adjustment for secretory granule visualisation may have resulted in unavoidable dynamic range ‘clipping’ of the brightest secondary features such as basolateral PM. The images were then assembled into final figures using Inkscape software (The Inkscape Project). Supplementary figures were assembled in Microsoft PowerPoint software and exported to PDF format.
Acknowledgements
We thank the UNSW BioMedical Imaging Facility (BMIF) under the direction of Renee Whan for help with the microscopy and the UNSW Biological Resource Centre under the direction of Stephen Danon. We thank Michael Carnell for microscopy and image analysis advice.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.M., P.W.G., E.C.H.; Methodology: A.M., R.W., P.W.G., E.C.H.; Validation: A.M., M.A.A.; Investigation: A.M., M.A.A., C.A.L., M.L.C., M. Heydecker, M. Holliday, J.C.M.M.; Resources: J.H., M.W., P.T., Y.Z., R.S.A., N.S.B., P.W.G., E.C.H.; Writing - original draft: A.M., P.W.G.; Writing - review & editing: A.M., T.B., R.W., N.S.B., P.W.G., E.C.H.; Supervision: A.M., A.K., T.M., P.W.G., E.C.H.; Funding acquisition: A.M., P.W.G., E.C.H.
Funding
This work was supported by grants to P.W.G. and E.C.H. from the Australian National Health and Medical Research Council (APP1079866 and APP1100202), to E.C.H., A.M. and R.W. from the Australian Research Council (DP160101623), and to P.W.G., E.C.H., A.M. and M.A.A. from The Kid's Cancer Project.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.212654.supplemental
References
- Alioto S. L., Garabedian M. V., Bellavance D. R. and Goode B. L. (2016). Tropomyosin and profilin cooperate to promote formin-mediated actin nucleation and drive yeast actin cable assembly. Curr. Biol. 26, 3230-3237. 10.1016/j.cub.2016.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appaduray M. A., Masedunskas A., Bryce N. S., Lucas C. A., Warren S. C., Timpson P., Stear J. H., Gunning P. W. and Hardeman E. C. (2016). Recruitment kinetics of tropomyosin Tpm3.1 to actin filament bundles in the cytoskeleton is independent of actin filament kinetics. PLoS ONE 11, e0168203 10.1371/journal.pone.0168203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovellan M., Romeo Y., Biro M., Boden A., Chugh P., Yonis A., Vaghela M., Fritzsche M., Moulding D., Thorogate R. et al. (2014). Cellular control of cortical actin nucleation. Curr. Biol. 24, 1628-1635. 10.1016/j.cub.2014.05.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce N. S., Schevzov G., Ferguson V., Percival J. M., Lin J. J., Matsumura F., Bamburg J. R., Jeffrey P. L., Hardeman E. C., Gunning P. et al. (2003). Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol. Biol. Cell 14, 1002-1016. 10.1091/mbc.E02-04-0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J. R., Hocky G. M., Homa K. E., Morganthaler A. N., Hitchcock-DeGregori S. E., Voth G. A. and Kovar D. R. (2017). Competition between Tropomyosin, Fimbrin, and ADF/Cofilin drives their sorting to distinct actin filament networks. eLife 6, e23152 10.7554/eLife.23152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. A. (1987). Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105, 1473-1478. 10.1083/jcb.105.4.1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan R. I., Getoor L., Wilbur W. J. and Mount S. M. (2007). Features generated for computational splice-site prediction correspond to functional elements. BMC Bioinformatics 8, 410 10.1186/1471-2105-8-410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goins L. M. and Mullins R. D. (2015). A novel tropomyosin isoform functions at the mitotic spindle and Golgi in Drosophila. Mol. Biol. Cell 26, 2491-2504. 10.1091/mbc.E14-12-1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P. W., Ghoshdastider U., Whitaker S., Popp D. and Robinson R. C. (2015a). The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 128, 2009-2019. 10.1242/jcs.165563 [DOI] [PubMed] [Google Scholar]
- Gunning P. W., Hardeman E. C., Lappalainen P. and Mulvihill D. P. (2015b). Tropomyosin - master regulator of actin filament function in the cytoskeleton. J. Cell Sci. 128, 2965-2974. 10.1242/jcs.172502 [DOI] [PubMed] [Google Scholar]
- Holmes K. C. and Lehman W. (2008). Gestalt-binding of tropomyosin to actin filaments. J. Muscle Res. Cell Motil. 29, 213-219. 10.1007/s10974-008-9157-6 [DOI] [PubMed] [Google Scholar]
- Hsiao J. Y., Goins L. M., Petek N. A. and Mullins R. D. (2015). Arp2/3 complex and cofilin modulate binding of tropomyosin to branched actin networks. Curr. Biol. 25, 1573-1582. 10.1016/j.cub.2015.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y., Soekmadji C., Mitchell J. M., Thomas W. G. and Thorn P. (2012). Real-time measurement of F-actin remodelling during exocytosis using Lifeact-EGFP transgenic animals. PLoS ONE 7, e39815 10.1371/journal.pone.0039815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M., East D. A. and Mulvihill D. P. (2014). Formins determine the functional properties of actin filaments in yeast. Curr. Biol. 24, 1525-1530. 10.1016/j.cub.2014.05.034 [DOI] [PubMed] [Google Scholar]
- Lazarides E. (1975). Tropomyosin antibody: the specific localization of tropomyosin in nonmuscle cells. J. Cell Biol. 65, 549-561. 10.1083/jcb.65.3.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarides E. (1976). Actin, alpha-actinin, and tropomyosin interaction in the structural organization of actin filaments in nonmuscle cells. J. Cell Biol. 68, 202-219. 10.1083/jcb.68.2.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masedunskas A., Sramkova M., Parente L., Sales K. U., Amornphimoltham P., Bugge T. H. and Weigert R. (2011). Role for the actomyosin complex in regulated exocytosis revealed by intravital microscopy. Proc. Natl. Acad. Sci. USA 108, 13552-13557. 10.1073/pnas.1016778108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masedunskas A., Appaduray M., Hardeman E. C. and Gunning P. W. (2013). What makes a model system great? A case for intravital microscopy for studying the actin cytoskeleton. IntraVital 2, e26287 10.4161/intv.26287 [DOI] [Google Scholar]
- Miklavc P., Hecht E., Hobi N., Wittekindt O. H., Dietl P., Kranz C. and Frick M. (2012). Actin coating and compression of fused secretory vesicles are essential for surfactant secretion--a role for Rho, formins and myosin II. J. Cell Sci. 125, 2765-2774. 10.1242/jcs.105262 [DOI] [PubMed] [Google Scholar]
- Miklavc P., Ehinger K., Sultan A., Felder T., Paul P., Gottschalk K.-E. and Frick M. (2015). Actin depolymerisation and crosslinking join forces with myosin II to contract actin coats on fused secretory vesicles. J. Cell Sci. 128, 1193-1203. 10.1242/jcs.165571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milberg O., Shitara A., Ebrahim S., Masedunskas A., Tora M., Tran D. T., Chen Y., Conti M. A., Adelstein R. S., Ten Hagen K. G. et al. (2017). Concerted actions of distinct nonmuscle myosin II isoforms drive intracellular membrane remodeling in live animals. J. Cell Biol. 216, 1925-1936. 10.1083/jcb.201612126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L. and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nicovich P. R., Janco M., Sobey T., Gajwani M., Obeidy P., Whan R., Gaus K., Gunning P. W., Coster A. C. F. and Böcking T. (2016). Effect of surface chemistry on tropomyosin binding to actin filaments on surfaces. Cytoskeleton 73, 729-738. 10.1002/cm.21342 [DOI] [PubMed] [Google Scholar]
- Nightingale T. D., White I. J., Doyle E. L., Turmaine M., Harrison-Lavoie K. J., Webb K. F., Cramer L. P. and Cutler D. F. (2011). Actomyosin II contractility expels von Willebrand factor from Weibel-Palade bodies during exocytosis. J. Cell Biol. 194, 613-629. 10.1083/jcb.201011119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novy R. E., Sellers J. R., Liu L.-F. and Lin J. J.-C. (1993). In vitro functional characterization of bacterially expressed human fibroblast tropomyosin isoforms and their chimeric mutants. Cell Motil. Cytoskeleton 26, 248-261. 10.1002/cm.970260308 [DOI] [PubMed] [Google Scholar]
- Panzer L., Trube L., Klose M., Joosten B., Slotman J., Cambi A. and Linder S. (2016). The formins FHOD1 and INF2 regulate inter- and intra-structural contractility of podosomes. J. Cell Sci. 129, 298-313. 10.1242/jcs.177691 [DOI] [PubMed] [Google Scholar]
- Porat-Shliom N., Milberg O., Masedunskas A. and Weigert R. (2013). Multiple roles for the actin cytoskeleton during regulated exocytosis. Cell. Mol. Life Sci. 70, 2099-2121. 10.1007/s00018-012-1156-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A. and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Flynn K. C., Raducanu A., Gärtner F., Beck G., Bösl M., Bradke F., Massberg S., Aszodi A., Sixt M. et al. (2010). Lifeact mice for studying F-actin dynamics. Nat. Methods 7, 168-169. 10.1038/nmeth0310-168 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W. M., Lehman W. and Moore J. R. (2015). Direct observation of tropomyosin binding to actin filaments. Cytoskeleton 72, 292-303. 10.1002/cm.21225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Lambert G. G., Chammas A., Ni Y., Cranfill P. J., Baird M. A., Sell B. R., Allen J. R., Day R. N., Israelsson M. et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407-409. 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokac A. M., Co C., Taunton J. and Bement W. (2003). Cdc42-dependent actin polymerization during compensatory endocytosis in Xenopus eggs. Nat. Cell Biol. 5, 727-732. 10.1038/ncb1025 [DOI] [PubMed] [Google Scholar]
- Thevenaz P., Ruttimann U. E. and Unser M. (1998). A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27-41. 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- Tojkander S., Gateva G., Schevzov G., Hotulainen P., Naumanen P., Martin C., Gunning P. W. and Lappalainen P. (2011). A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 21, 539-550. 10.1016/j.cub.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Tran D. T., Masedunskas A., Weigert R. and Ten Hagen K. G. (2015). Arp2/3-mediated F-actin formation controls regulated exocytosis in vivo. Nat. Commun. 6, 10098 10.1038/ncomms10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahovich N., Kee A. J., Van der Poel C., Kettle E., Hernandez-Deviez D., Lucas C., Lynch G. S., Parton R. G., Gunning P. W. and Hardeman E. C. (2009). Cytoskeletal tropomyosin Tm5NM1 is required for normal excitation-contraction coupling in skeletal muscle. Mol. Biol. Cell 20, 400-409. 10.1091/mbc.E08-06-0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner A. (1979). Equilibrium of the actin-tropomyosin interaction. J. Mol. Biol. 131, 839-853. 10.1016/0022-2836(79)90204-3 [DOI] [PubMed] [Google Scholar]
- Yu H.-Y. E. and Bement W. M. (2007). Multiple myosins are required to coordinate actin assembly with coat compression during compensatory endocytosis. Mol. Biol. Cell 18, 4096-4105. 10.1091/mbc.E06-11-0993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Conti M. A., Malide D., Dong F., Wang A., Shmist Y. A., Liu C., Zerfas P., Daniels M. P., Chan C.-C. et al. (2012). Mouse models of MYH9-related disease: mutations in nonmuscle myosin II-A. Blood 119, 238-250. 10.1182/blood-2011-06-358853 [DOI] [PMC free article] [PubMed] [Google Scholar]