ABSTRACT
In Drosophila, telomere-capping proteins have the remarkable capacity to recognize chromosome ends in a sequence-independent manner. This epigenetic protection is essential to prevent catastrophic ligations of chromosome extremities. Interestingly, capping proteins occupy a large telomere chromatin domain of several kilobases; however, the functional relevance of this to end protection is unknown. Here, we investigate the role of the large capping domain by manipulating HOAP (encoded by caravaggio) capping-protein expression in the male germ cells, where telomere protection can be challenged without compromising viability. We show that the exhaustion of HOAP results in a dramatic reduction of other capping proteins at telomeres, including K81 [encoded by ms(3)K81], which is essential for male fertility. Strikingly however, we demonstrate that, although capping complexes are barely detected in HOAP-depleted male germ cells, telomere protection and male fertility are not dramatically affected. Our study thus demonstrates that efficient protection of Drosophila telomeres can be achieved with surprisingly low amounts of capping complexes. We propose that these complexes prevent fusions by acting at the very extremity of chromosomes, reminiscent of the protection conferred by extremely short telomeric arrays in yeast or mammalian systems.
KEY WORDS: Drosophila melanogaster, Telomere, Capping protein, Male germline, Spermatogenesis
INTRODUCTION
Telomeres are specialized structures that organize the extremities of linear chromosomes in a way that prevents their recognition as DNA double-strand breaks (DSBs) by the DNA repair machinery. Another important function of telomeres is to counteract the gradual erosion of chromosomes that naturally occurs over cell divisions and results from the incomplete replication of DNA ends (reviewed in Fulcher et al., 2014). In the vast majority of eukaryotes, including human and yeasts, telomeres comprise arrays of variable lengths (from several hundred base pairs in yeast to 5–60 kb in mammals) of small G-rich DNA repeats that are added by the enzymatic complex telomerase (reviewed in Jain and Cooper, 2010). These DNA repeats are specifically recognized and bound by capping-protein complexes that inhibit the processing of chromosome extremities through the non-homologous end-joining (NHEJ) repair pathway (reviewed in Oganesian and Karlseder, 2009). In mammals, for instance, TRF1 and TRF2 (also known as TERF1 and TERF2, respectively) recognize the TTAGGG repeats and interact with other capping proteins of the shelterin complex (reviewed in Palm and de Lange, 2008; Stewart et al., 2012; Lu et al., 2013).
Dipterans represent an exception in Metazoa as they lack telomerase (Mason et al., 2008; Villasante et al., 2008). Telomere elongation thus uses different mechanisms, such as homologous recombination or transposition of telomere-specific retrotransposons. In Drosophila melanogaster, terminal sequences contain an array – called the HTT array – of complete and incomplete copies of three non-long-terminal-repeat (non-LTR) retrotransposons (Het-A, TAHRE and TART) (reviewed in Pardue and DeBaryshe, 2011) that has been estimated to vary in length from approximately 20 to 150 kb (Capkova Frydrychova et al., 2008). Drosophila chromosome ends are also bound by a protective protein complex that constitutes a functional analog of the mammalian shelterin complex (Raffa et al., 2011; Raffa et al., 2013). This capping complex includes the fast evolving, non-conserved proteins HP1/ORC-associated protein (HOAP; encoded by caravaggio), HipHop, Verrocchio (Ver) and Modigliani (Moi), which are called terminin proteins, and the conserved Heterochromatin Protein 1a [HP1a; encoded by Su(var)205] (Raffa et al., 2011; Raffa et al., 2013). HP1a localizes not only at telomeres but also throughout heterochromatin and at many euchromatic loci, whereas terminin proteins are specifically enriched at telomeres throughout the cell cycle and are involved in chromosome-end protection. Capping proteins are essential for viability, and loss-of-function mutants usually die during larval stages, displaying chromosome end-to-end fusions at high frequency in rapidly dividing cells (Fanti et al., 1998; Cenci et al., 2003; Raffa et al., 2009; Raffa et al., 2010; Gao et al., 2010). In addition – and despite the fact that HOAP, HipHop, Ver and Moi are not conserved outside Drosophilidae and closely related Diptera – these proteins share some properties with their mammalian or yeast counterparts (Cenci et al., 2005). For instance, Ver and Moi localize exclusively at the tip of the chromosome end, where Ver would bind to the single-stranded DNA 3′-overhang through its oligonucleotide/oligosaccharide-binding (OB)-fold domain, in a manner similar to that of POT1 in mammals (Raffa et al., 2010; Raffa et al., 2013). By contrast, HOAP, HipHop and HP1a form a complex that localizes not only in the direct vicinity of the chromosome extremities, where HOAP interacts directly with Moi (Raffa et al., 2009), but also occupies a relatively large terminal region of at least 10 kb (Gao et al., 2010). Importantly, unlike their mammalian counterparts, Drosophila capping proteins all share the remarkable property of recognizing chromosome ends independently of the underlying DNA sequence (Perrini et al., 2004; Cenci et al., 2005; Pimpinelli, 2006; Rong, 2008; Raffa et al., 2013). Indeed, chromosomes with a terminal deletion that completely removes the HTT array can be generated and maintained through generations (Levis, 1989; Ahmad and Golic, 1998; Gao et al., 2010). Remarkably, such chromosomes accumulate an apparently normal amount of capping proteins at their extremities (Gao et al., 2010). Similarly, no change in the level of capping is observed on chromosomes that have naturally longer HTT arrays (Gao et al., 2010). Hence, although the size of the capping domain in organisms that express telomerase is directly linked to the length of terminal DNA repeats, the establishment of a functional cap in the absence of telomerase remains poorly understood. Several studies have nevertheless established that some effectors of the DNA damage response (DDR), including the conserved Mre11–Rad50–Nbs complex and the ATM and ATR checkpoint kinases, are required for the prevention of telomere fusions and for the initial recruitment of HOAP onto chromosome ends (Ciapponi et al., 2004; Bi et al., 2005; Oikemus et al., 2006; Ciapponi et al., 2006). It has been proposed that the spreading of capping complexes toward the centromere follows this initial recruitment of a protective cap at the extremity (Pimpinelli, 2006; Gao et al., 2010), although the mechanism that regulates the size of the capping domain is currently unknown. Moreover, the relative contribution of this large domain to telomere protection is unclear.
Among the functionally characterized capping proteins, HOAP and HipHop play a pivotal role in establishing a functional telomere. They are interdependent for their stability (Gao et al., 2010), and HOAP is required for the localization of Ver, Moi and HP1a at telomeres. However, the fact that these proteins are essential for viability is a serious limitation when analyzing their function during cell proliferation and differentiation. Spermatogenesis represents a powerful system for functional analysis of capping genes in vivo. Their invalidation in the male germline does not compromise viability, and post-meiotic spermatids are an abundant source of synchronized, differentiated nuclei that are particularly tractable for imaging of telomeres. Furthermore, the existence of a HipHop paralog that is specific to the male germline, K81 [encoded by ms(3)K81], provides a unique opportunity to manipulate capping-complex assembly throughout spermatogenesis (Dubruille et al., 2010; Gao et al., 2011; Dubruille and Loppin, 2011).
By specifically reducing the expression of HOAP in the male germline, we show that this protein plays a central role in determining the size of the capping domain. Surprisingly, we demonstrate that the amount of capping proteins at telomeres can be dramatically decreased without severely disrupting chromosome-end protection and male fertility.
RESULTS
HipHop and K81 successively contribute to telomere protection in male germ cells
We have recently discovered that, in the male germline, HipHop is replaced by K81 in spermatid nuclei (Dubruille et al., 2010). At fertilization, K81 is transmitted to the zygote, where it is required to protect paternal telomeres during the first division. Interestingly, although K81 is absolutely necessary for telomere protection at fertilization, it does not seem to be required during spermatogenesis (Dubruille et al., 2010; Gao et al., 2011). Indeed, no chromosomal defects have been observed in the male germline of K81-mutant males (Fuyama, 1984; Yasuda et al., 1995), suggesting that other types of capping complexes, probably involving HipHop, are required in these cells.
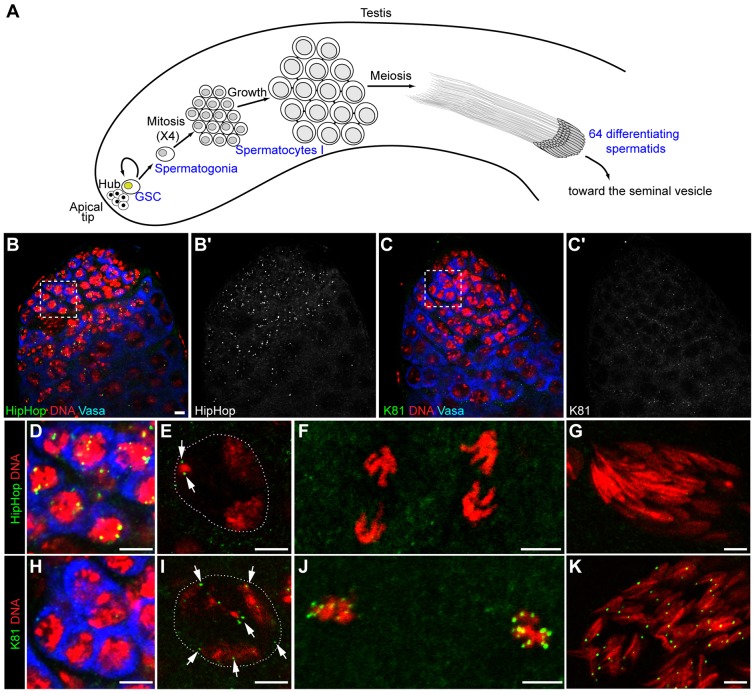
To gain insight into the respective contributions of HipHop and K81 in the protection of telomeres during spermatogenesis, we first analyzed their expression dynamics in the male germline. We used previously characterized transgenic flies expressing green fluorescent protein (GFP)-fusion proteins under the regulation of their own promoter (Dubruille et al., 2010). The 5′hiphop-GFP::hiphop transgene was introduced into an otherwise lethal trans-heterozygous hiphop-mutant background to avoid competition with the endogenous protein. In these rescued animals, GFP–HipHop protein was detected as discrete foci in the nuclei of germline stem cells (GSCs) and their progeny, spermatogonial cells (see Fig. 1A for a schematic description of Drosophila spermatogenesis; Fig. 1B,B′,D). Then, the intensity of GFP–HipHop foci decreased progressively in dividing spermatogonia. In primary spermatocytes, a weak staining often persisted as two foci (Fig. 1E). Then, GFP–HipHop became undetectable throughout meiosis and in spermatid nuclei (Fig. 1F,G).
Fig. 1.
K81 and HipHop show a complementary localization at telomeres in the male germline. (A) Schematic representation of Drosophila spermatogenesis. Spermatogenesis is organized in differentiating units called cysts that progress along the axis of the testis (Fuller, 1993). At the apical end of the testis, a group of somatic cells called the hub forms the niche of germline stem cells (GSCs) and somatic cyst stem cells (not represented on the scheme). The asymmetric division of a GSC produces another GSC and a spermatogonial cell. Spermatogonia undertake four rounds of mitosis without complete cytokinesis leading to a cyst of 16 interconnected spermatocytes. After a growth step, spermatocytes undergo meiotic divisions in synchrony giving rise to 64 haploid spermatids. The maturation of spermatids is notably characterized by the elimination of most histones and their replacement by sperm-specific proteins, such as Mst77F and the protamine-like proteins Mst35Ba/b (not shown on the scheme). At the end of spermatogenesis, sperm cells individualize and are transferred to the seminal vesicle. (B–K) Confocal images of whole-mount testes from 5′hiphop-GFP::hiphop hiphop1/5′hiphop-GFP::hiphop hiphopEY7584 males (B,B′,D–G) and 5′K81-GFP::K81/5′K81-GFP::K81; K812/K812 males (C,C′,H–K) to locate HipHop and K81, respectively. (B–C′) View of the apical tip of testes stained with an antibody against Vasa (blue) to identify germ cells, an antibody against GFP (green) and for DNA (red). GFP–HipHop foci are visible in the nuclei of GSCs and early spermatogonia (B,B′, magnification of the ‘boxed’ islet is shown in D), whereas GFP–K81 is barely detected in these cells (C,C′, magnification of the ‘boxed’ islet is shown in H). In spermatocyte nuclei, only a few GFP–HipHop foci (from 0 to 2) are still visible (E, arrows), whereas GFP–K81 decorates all telomeres (I, arrows). During meiosis (F, two anaphase-II nuclei) and in spermatid nuclei (G), GFP–HipHop is not detected. By contrast, bright GFP–K81 foci are visible throughout meiosis (J, two metaphase-I nuclei) and in spermatid nuclei (K). The dashed white lines in E and I delineate the nucleus. Scale bars: 5 µm.
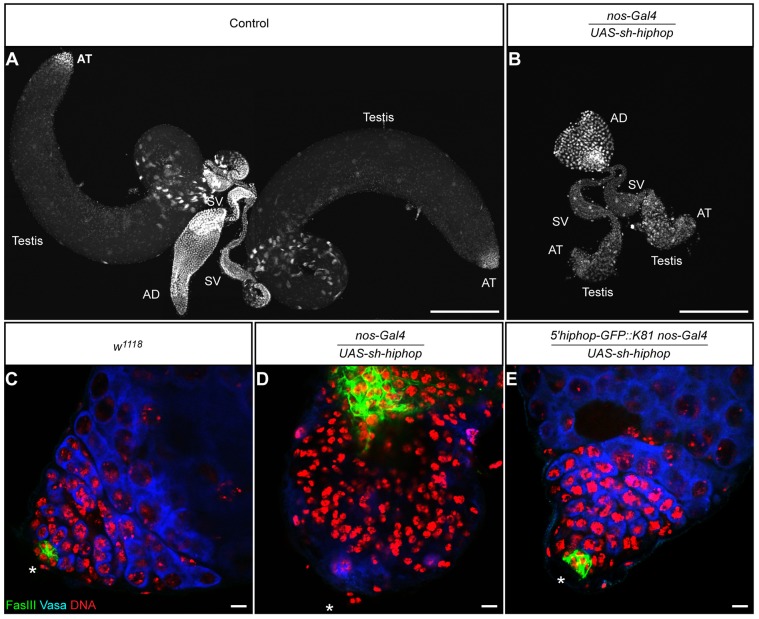
For GFP–K81, very weak signals were occasionally detected in GSCs and the more apical spermatogonia (Fig. 1C,C′,H). Then, the signals increased in late spermatogonia and in primary spermatocytes, forming several GFP foci in each nucleus (Fig. 1I). Finally, GFP–K81 brightly decorated telomeres during meiotic divisions and in spermatid nuclei, as previously reported (Fig. 1J,K) (Dubruille et al., 2010; Dubruille and Loppin, 2011). Thus, these paralogs displayed complementary expression patterns, suggesting that, in contrast to the post-meiotic function of K81, HipHop is required for telomere protection in early male germ cells. To address this question, we chose to knock down HipHop in the male germline by using RNA interference through the UAS-GAL4 system. We generated transgenic flies expressing a small hairpin (sh)RNA targeting hiphop that were then crossed to flies bearing a nos-Gal4 transgene, which drives expression of GAL4 in GSCs and early spermatogonia (White-Cooper, 2012). The hiphop-knockdown males were totally sterile (Table 1). They had severely atrophied testes that were almost systematically devoid of germ cells (97.6%, n=84; Fig. 2A–D). This phenotype, which is characteristic of testes in which GSCs have been genetically ablated (Castrillon et al., 1993; Gönczy and DiNardo, 1996; Kauffman et al., 2003), was also obtained when an shRNA targeting the GFP coding sequence was expressed in the germline of GFP::hiphop-rescued males (Table 1 and data not shown). Our results thus demonstrate that HipHop is necessary for the survival of GSCs.
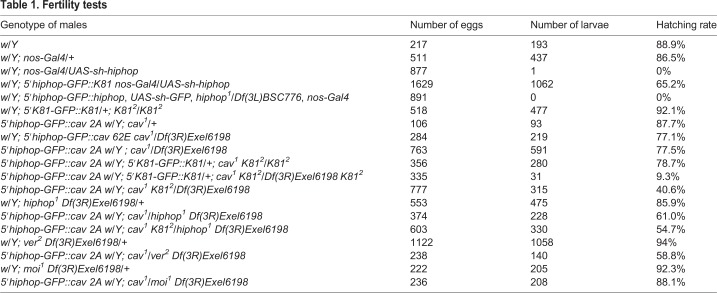
Table 1.
Fertility tests
Fig. 2.
hiphop knockdown in the male germline dramatically impairs early spermatogenesis. (A,B) Confocal images showing a pair of testes from a control male (A) and a hiphop knockdown male (B, nos-Gal4/UAS-sh-hiphop), stained with a pan-histone antibody. The scale is the same for both images. Note the drastic reduction in size of the hiphop knockdown testes. AT, apical tip; AD, anterior ejaculatory duct; SV, seminal vesicle. Almost all of the hiphop knockdown testes were vestigial as shown in B (97.6%, n=84). We observed a few testes with a normal shape but with a reduced size (2.4%, n=84). (C–E) Confocal sections of whole-mount testes stained for the hub [Fas III (green)], stem cells [Vasa (blue)] and DNA (red). Asterisks indicate the apical tip of the testes. The scale is the same for all images. Approximately 5% of the testis is visible on the panels C and E, whereas in D, more than half of the testis is visible. In the testes from hiphop knockdown males (D), the staining of Vasa appears to be severely reduced compared with control testis (C), showing that germ cells are depleted. As previously reported, hub cells expand in agametic testes, as revealed by using the Fas III marker (Gönczy and DiNardo, 1996). This phenotype is partially rescued with a 5′hiphop-GFP::K81 transgene, as about 33% of the males (5′hiphop-GFP::K81 nos-Gal4/UAS-sh-hiphop) have testes of wild-type size, in which the hub and germ cells are clearly detected (E). Scale bars: 250 µm (A,B); 10 µm (C–E).
Interestingly, a single copy of a 5′hiphop-GFP::K81 transgene [which allows expression of GFP–K81 under the control of the hiphop promoter (Dubruille et al., 2010) and is not targeted by hiphop shRNAs] partially rescued the fertility of the hiphop-knockdown flies (Table 1). Accordingly, we observed that 33% of the 5′hiphop-GFP::K81, nos-Gal4/UAS-sh-hiphop flies had testes that appeared normal in shape and size (n=60) and contained germ cells (Fig. 2E). These results demonstrate that K81 can partially replace HipHop during the division of early male germ cells.
Low levels of capping proteins are sufficient to protect telomeres during spermatogenesis
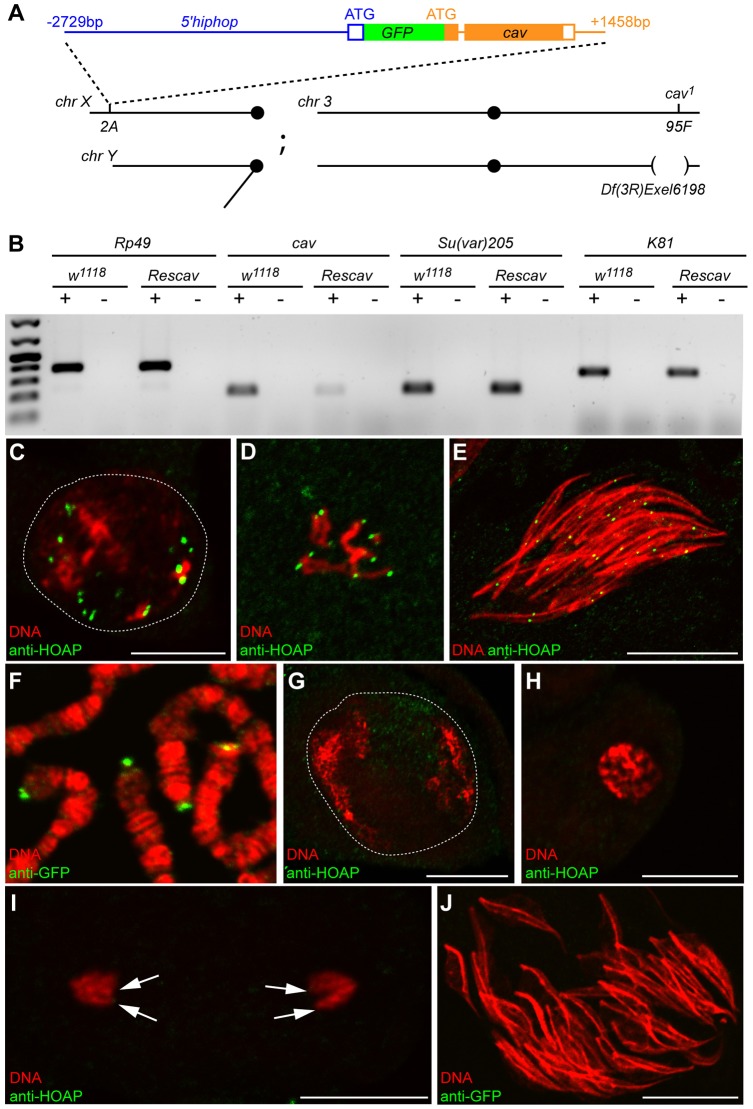
The observed functional redundancy between HipHop and K81 suggests that K81 ensures telomere capping in spermatocytes when HipHop is no longer detected. However, as mentioned above, K81-null mutants do not show any defects during spermatogenesis (Fuyama, 1984; Yasuda et al., 1995), opening the possibility that a low, residual amount of HipHop can protect telomeres during meiotic divisions. To more generally test the adaptability of telomeres to a limiting amount of capping proteins, we turned to HOAP, an interacting protein of HipHop encoded by the caravaggio (cav) gene. In contrast to hiphop, cav has a much stronger expression in testis (Flybase) and does not have a paralog that is specific to the male germline in D. melanogaster (Dubruille et al., 2012). Using an antibody against HOAP, we indeed observed bright HOAP foci in all germ cell nuclei during pre-meiotic and meiotic stages (Fig. 3C,D). We also observed HOAP foci in spermatid nuclei (Fig. 3E), as previously described (Dubruille et al., 2010), suggesting that this capping protein is required to protect telomeres at all stages of spermatogenesis. Because cav is essential for viability, we designed a partial rescue strategy to stably and specifically reduce its expression in the male germline. We generated flies expressing a GFP–HOAP fusion protein under the control of the hiphop promoter (5′hiphop-GFP::cav; Fig. 3A). We reasoned that this transgene, introduced into a cav-null-mutant background, would restore HOAP expression in somatic cells but limit cav expression to GSCs and proliferative spermatogonia in testes.
Fig. 3.
cav is downregulated in male germ cells of Rescav flies. (A) A scheme representing the genotype of Rescav males [5′hiphop-GFP::cav/Y; cav1/Df(3R)Exel6198]. The organization of the 5′hiphop-GFP:cav transgene is represented with filled and empty boxes corresponding to, respectively, coding exons and UTRs; and with lines for introns and intergenic regions. The position relative to the ATG of the corresponding gene is indicated. This transgene was inserted into the 2A platform on the X chromosome (chr) and combined with a cav-null allele (cav1) and a deficiency that uncovers cav [Df(3R)Exel6198]. bp, base pair. (B) RT-PCR analyses were performed on total RNAs from the testes of control (w1118) or Rescav flies. Rp49 was used as a control gene. +, with reverse transcriptase; −, without reverse transcriptase. The expression level of cav mRNA is reduced in Rescav testes. The downregulation of cav does not affect the expression of the capping genes K81 and Su(var)205 (encoding HP1a). (C–E) Confocal images of squashed testes from w1118 males stained with an antibody against HOAP (green) and for DNA (red). (C) A nucleus of a primary spermatocyte (the white dashed line delineates the nucleus), (D) metaphase of meiosis II, (E) a cyst of spermatid nuclei. (F) A confocal image of squashed salivary glands of a 5′hiphop-GFP::cav third instar larvae stained with an antibody against GFP (green) and for DNA (red). The fusion protein GFP–HOAP localizes at telomeres of polytene chromosomes. (G–J) Confocal images of squashed testes from Rescav males stained for HOAP or GFP (green) and for DNA (red). (G) A nucleus of a mature primary spermatocyte (white dashed line), (H) prophase of meiosis, (I) anaphase of male meiosis II and (J) nuclei of a cyst of 64 spermatids. In Rescav males, HOAP is undetectable at telomeres (arrows in I) during meiosis and in spermatid nuclei. Scale bars: 10 µm.
As expected, the 5′hiphop-GFP::cav transgene fully rescued the lethality of cav mutants. We obtained 39.6% of the rescued 5′hiphop-GFP::cav; cav1/Df(3R)Exel6198 genotype (designated as Rescav for simplicity; see Materials and Methods and Fig. 3A) over the 33% expected Mendelian ratio (n=416). GFP–HOAP was normally observed at somatic telomeres of 5′hiphop-GFP::cav animals, such as in salivary gland polytene chromosomes (Fig. 3F). In addition, adult Rescav males had testes of normal size, indicating that telomere protection was not compromised, at least in early male germ cells. However, semi-quantitative reverse-transcription (RT)-PCR analysis confirmed the severe reduction of cav transcripts in Rescav testes compared with that of the wild-type control (Fig. 3B). Accordingly, in testes of adult Rescav males, GFP–HOAP could not be detected above background levels in spermatocytes or spermatid nuclei using antibodies against either GFP or HOAP (Fig. 3G–J).
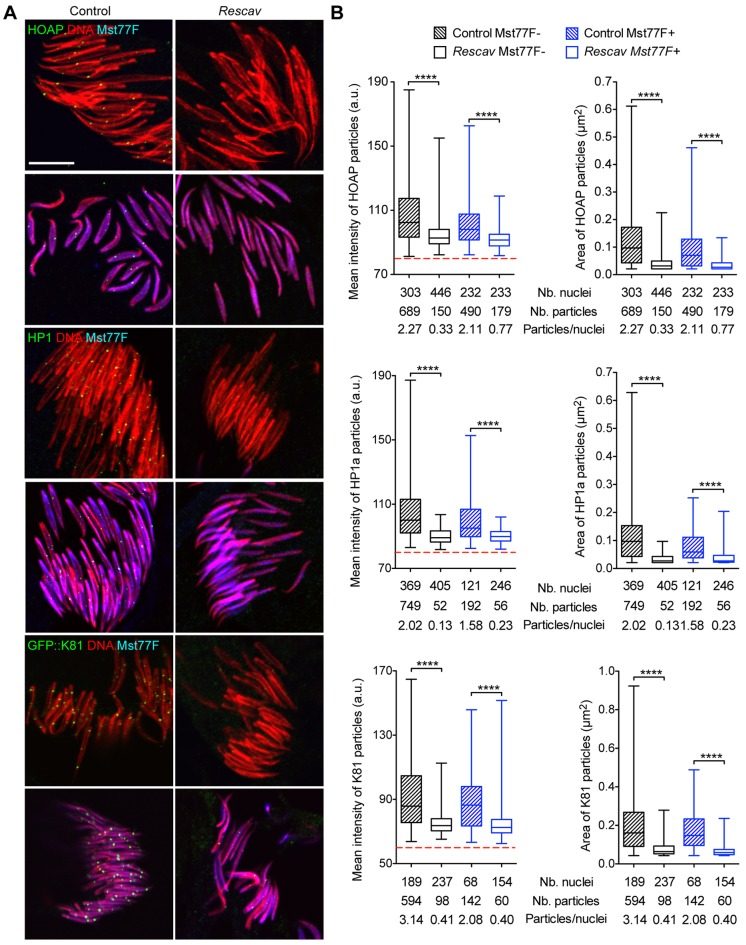
Surprisingly, however, the fertility of Rescav males was only slightly affected by the observed reduction of HOAP levels at telomeres in the male germline (Table 1). In somatic cells, HOAP is crucial for the recruitment or the stabilization of other capping proteins at telomeres (Raffa et al., 2009; Gao et al., 2010; Raffa et al., 2010). We thus wondered whether the accumulation of the capping proteins HP1a and K81, which are present in spermatocytes and early spermatids (Dubruille et al., 2010), are affected in germ cells of Rescav males. We thus systematically analyzed the level of these capping proteins in Rescav spermatid nuclei by quantifying immunofluorescence signals. To exclude any impact of nuclear condensation on antibody accessibility, we only considered spermatid nuclei at the canoe stage and distinguished in the analyses those that were negative or positive for Mst77F, a sperm-specific nuclear protein that is incorporated at the histone-to-protamine transition (Rathke et al., 2010). Using an antibody specific to HOAP, we first confirmed the severe reduction of HOAP at spermatid telomeres in Rescav males. HOAP foci were readily detected in control spermatid nuclei before and after the histone-to-protamine transition, whereas HOAP was barely detectable in Rescav spermatids at both stages (Fig. 4). We then analyzed HP1a and K81 accumulation using, respectively, an antibody against HP1a and the 5′K81-GFP::K81 transgene that was introduced in the Rescav background. Strikingly, both the size and intensity of HP1a and GFP–K81 foci were also drastically reduced in spermatids of Rescav males (Fig. 4).
Fig. 4.
The reduction of HOAP in spermatid nuclei affects the localization of K81 and HP1a at telomeres. (A) Representative confocal images of squashed testes stained for HOAP, HP1a or GFP (green), Mst77F (blue) and DNA (red). For HOAP and HP1a staining, flies were w1118 (Control) or 5′hiphop-GFP::cav/Y; cav1/Df(3R)Exel6198 (Rescav). For GFP–K81, control flies were 5′hiphop-GFP::cav/Y; 5′K81-GFP::K81/+; K812 cav1/K812 and Rescav flies were 5′hiphop-GFP::cav/Y; 5′K81-GFP::K81/+; K812 cav1/K812 Df(3R)Exel6198. For each staining, spermatid nuclei before and after the histone-to-protamine transition are visualized through, respectively, the presence or the absence of the Mst77F staining (blue). (B) Quantification of the mean intensity of fluorescence (graphs on the left) and area (graphs on the right) of particles detected in A are represented as box plots in which whiskers show minimum and maximum values, boxes show the middle 50% of the values, and horizontal lines show the median. The dashed red lines show the threshold under which fluorescence signals were considered as background. Each graph corresponds to the quantification of a single experiment that was repeated two or three times. The number of nuclei (Nb. nuclei) and detected particles (Nb. particles) analyzed are shown under the graph. In all experiments, the mean intensity of fluorescence and the area of HOAP, HP1a and GFP–K81 particles were significantly reduced in the Rescav background compared to in control flies. Mann–Whitney test; ****P<0.0001. a.u., arbitrary unit. Scale bar: 10 µm.
Thus, the reduction of HOAP levels in Rescav males has a general effect on the accumulation of the capping proteins HP1a and K81 at spermatid telomeres. However, we know that at least the capping function of K81 is absolutely required for male fertility, by preventing fusion of paternal telomeres at fertilization (Loppin et al., 2005; Dubruille et al., 2010; Gao et al., 2011). Our analysis thus demonstrates that the very limited quantity of K81 present at telomeres in Rescav post-meiotic male germ cells is sufficient to ensure efficient protection of paternal chromosomes during zygote formation.
Sensitized telomere protection in Rescav males unveils a protective role of K81 in meiosis
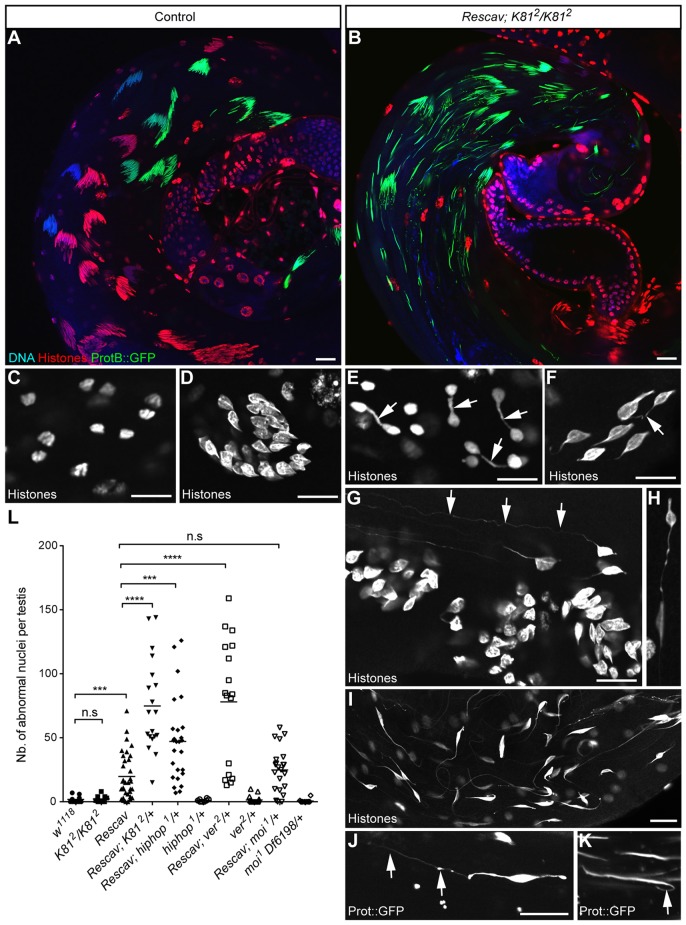
Although the reduction of capping-protein recruitment at telomeres in Rescav testes had no significant impact on spermatogenesis and fertility, we nevertheless occasionally noticed the presence of spermatid nuclei connected by a chromatin thread. This indicated that telomeres occasionally fuse before or during meiotic divisions, thus suggesting that telomere capping is sensitized in the Rescav male germline. To test this hypothesis, we challenged telomere capping by knocking down other capping genes in the Rescav background. Strikingly, in Rescav males that were homozygous for a null allele of K81 (K812), spermatogenesis appeared severely disorganized, with the majority of spermatid nuclei failing to differentiate normally (Fig. 5B). They were notably connected through gigantic chromatin threads of several tens of micrometres (Fig. 5G–J). Moreover, in telophase of meiosis, we could frequently observe chromatin bridges (Fig. 5E), which are likely to result from the fusion of uncapped telomeres. The chromatin threads connecting spermatid nuclei probably result from the stretch of unbroken chromatin bridges that form during male meiosis II, as similarly reported by Titen and colleagues (Titen et al., 2014).
Fig. 5.
Loss of K81 in the Rescav background results in chromatin bridges at high frequency. (A,B) Representative images of a testis from a control fly bearing a protB::GFP transgene in a wild-type (A) or in a Rescav; K812/K812 (B) background stained with a pan-histone antibody (red) and DNA (blue). In Rescav; K812/K812, the cysts of spermatids are disorganized (B). (C,D) Confocal images of nuclei in testes of wild-type flies stained with an antibody against histone. (C) Anaphase of male meiosis II. (D) A group of young spermatids. (E–K) Examples of chromatin bridges observed in the Rescav; K812/K812 flies that have been stained with an antibody against histone (E–I) or expressing a protB::GFP transgene (J,K). (E) Telophase of meiosis. Arrows indicate chromosome bridges between haploid nuclei. (F–H) Young spermatid nuclei presenting chromatin bridges (arrows). Some nuclei present very long chromatin threads (>50 µm; arrows in G). (I) A larger view of a testis with disorganized spermatid nuclei that harbor long chromatin threads. (J,K) Chromatin threads observed in spermatid nuclei that have incorporated protamines (arrows). Scale bars: 10 µm. (L) Quantification of abnormal spermatid nuclei in the indicated genotypes are represented as a scatter plot in which each point shows the number of nuclei with a chromatin thread counted in a single testis. Horizontal lines represent means. The loss of a single copy of K81 or hiphop in the Rescav background results in chromatin bridges at high frequency. Mann–Whitney test. n.s., non significant; ***P<0.001; ****P<0.0001. Note that abnormal spermatid nuclei in Rescav; K812/K812 testes were too numerous for reliable quantification (>300).
We also observed chromatin bridges, albeit at a lower frequency, when Rescav males had a single copy of K81. Quantification of this phenotype confirmed that Rescav; K812/+ testes contained significantly more abnormal nuclei than Rescav testes (Fig. 5L). Accordingly, Rescav; K812/+ males had reduced fertility compared to control flies (Rescav; Table 1). Taken together, these results demonstrate that, in the Rescav background, K81 is required for telomere protection during male meiosis. As we hypothesized above, the fact that no defects are observed in meiosis in K81-mutant males suggests that HipHop efficiently protects telomeres until completion of male meiosis. Interestingly, when we combined Rescav with the hiphop1 null allele, we also observed a significant increase of abnormal spermatid nuclei compared with that Rescav testes (Fig. 5L). This confirms that HipHop is also involved in telomere protection during male meiosis in the Rescav background.
These genetic interactions between Rescav and K81 or hiphop show that a further decrease of capping-protein expression in the Rescav background destabilizes capping complexes and impairs chromosome protection during meiosis. We thus tested, in this context, a partial loss of other capping genes, such as ver and moi, that are essential to prevent telomere fusions in somatic cells and for viability (Raffa et al., 2009; Raffa et al., 2010). We introduced the ver2 mutant allele into the Rescav background and examined whole-mount testes of these flies. Interestingly, we again observed the presence of many spermatids with giant chromatin threads. The quantification of abnormal spermatid nuclei in these flies revealed a phenotype similar to that of Rescav, K812/+ and Rescav, hiphop1/+ males (Fig. 5L). This implies that Ver, like HipHop, is involved in telomere protection in the male germline, at least until completion of meiosis. By contrast, however, combining the moi1 null allele with Rescav did not aggravate the Rescav phenotype (Fig. 5L). This result suggests that either Moi is not required for telomere protection in the male germline or that Moi is not limiting in this context. The fact that, in larval salivary glands, Ver and Moi are interdependent for their localization at telomeres supports the latter hypothesis (Raffa et al., 2010).
Taken together, our results show that during male meiosis, telomere protection relies on capping complexes that contain HipHop or K81, and at least the proteins HOAP and Ver.
Forcing hiphop expression in spermatids increases the recruitment of capping complexes at telomeres
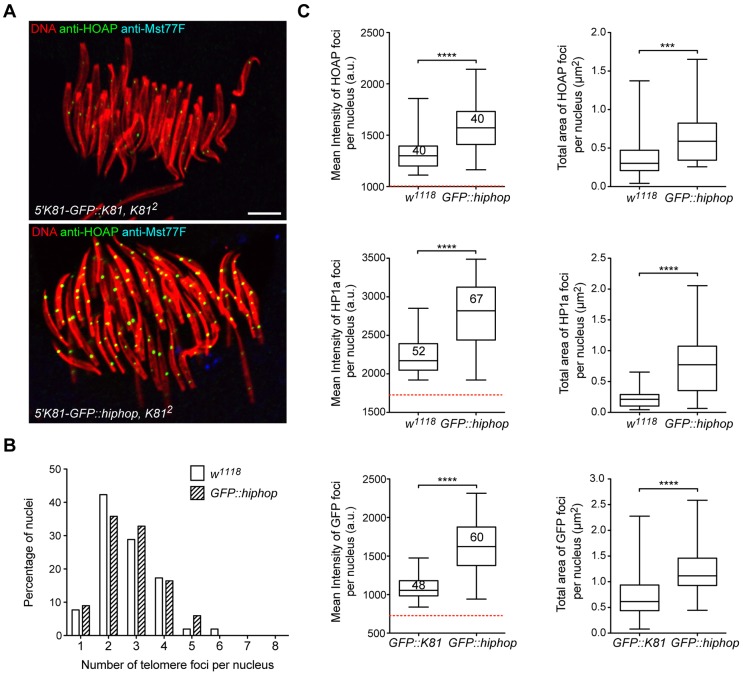
Our analysis of Rescav males showed that reducing HOAP expression level dramatically decreases the level of capping proteins present at telomeres. On the opposing side, we wondered whether it would be possible to increase the amount of capping proteins recruited at telomeres. Interestingly, over the course of our experiments, we noticed in a particular genotype that HOAP foci in young spermatid nuclei appeared brighter and larger than those in control flies (Fig. 6A). These flies bear two copies of a 5′K81-GFP::hiphop transgene, which drives ectopic expression of a GFP–HipHop fusion protein in spermatids (Dubruille et al., 2010). To confirm our observation, we quantified HOAP signals in spermatid nuclei. We and others have previously observed that telomeres cluster in spermatid nuclei and in various somatic cell types (Dubruille et al., 2010; Wesolowska et al., 2013). We thus first evaluated the clustering in spermatid nuclei of 5′K81-GFP::hiphop flies by counting the number of foci in individual nuclei. We did not observe any statistically significant changes of the clustering between 5′K81-GFP::hiphop flies and control flies (Fig. 6B). To nevertheless exclude any clustering effect, we quantified the total area and the mean intensity of all HOAP foci detected in individual nuclei (Fig. 6C, top panels). This analysis confirmed that both the area and intensity of HOAP foci were higher in spermatids of 5′K81-GFP::hiphop males compared with those of control males. We then asked whether the increase of HOAP levels affected the recruitment of other capping proteins. Therefore, we quantified HP1a and GFP–HipHop signals, which appeared, indeed, to be significantly higher than in control flies (GFP–HipHop signals were compared to GFP–K81 expressed with the same promoter; Fig. 6C, middle and bottom panels). These results thus demonstrate that in flies expressing high levels of GFP–HipHop in post-meiotic stages, capping complexes are more abundant at telomeres.
Fig. 6.
The expression of a GFP–HipHop fusion protein in spermatids increases the recruitment of capping proteins at spermatid telomeres. (A) Representative confocal images of spermatid nuclei in squashed testes stained with an antibody against HOAP (green) or Mst77F (blue) and for DNA (red). Flies were 5′K81-GFP::K81/5′K81-GFP::K81; K812/K812 (top panel) and 5′K81-GFP::hiphop K812/5′K81-GFP::hiphop K812 (bottom panel). We show here Mst77F-negative nuclei because GFP–HipHop is lost after the histone-to-protamine transition (Dubruille et al., 2010). The settings for the acquisition of both images were the same. In Mst77F-negative spermatid nuclei, telomere foci appear brighter and larger in 5′K81-GFP::hiphop, K812 flies. Scale bar: 5 µm. (B) A representative experiment to evaluate the clustering in spermatid nuclei. Telomere foci were revealed using an antibody against HP1a. The bar histogram represents the percentage of nuclei observed for each category in control flies (w1118; n=52) or 5′K81-GFP::hiphop K812/5′K81-GFP::hiphop K812 flies (GFP::hiphop; n=67). In all experiments, telomere clustering in spermatid nuclei was not significantly different (Kolmogorov–Smirnov test). (C) Quantification of the total mean intensity of fluorescence (graphs on the left) or total area (graphs on the right) of telomere foci measured in each analyzed nucleus are represented as box plots in which whiskers show the minimum and maximum values, boxes show the middle 50% of the values, and horizontal lines show the median. The dashed red line shows the threshold under which fluorescence signals were considered as background. For HOAP and HP1a, flies were w1118 (control) or 5′K81-GFP::hiphop, K812/5′K81-GFP::hiphop, K812 (GFP::hiphop). 5′K81-GFP::K81/5′K81-GFP::K81; K812/K812 flies (GFP::K81) were used as control for GFP. Each set of graphs (mean intensity and area) corresponds to the quantification of a single experiment that was repeated two or three times. For each staining, the number of individual nuclei analyzed for quantification is written in the box. In all experiments, the mean intensity of fluorescence and total area of telomere foci in 5′K81-GFP::hiphop flies were significantly higher than in control flies. Mann–Whitney test; ***P=0.0002; ****P<0.0001. a.u., arbitrary unit.
DISCUSSION
A capping complex switch during spermatogenesis
We have previously shown that capping protein K81, which is specific to the male germline, localizes at telomeres in spermatids and is essential for the protection of sperm telomeres at fertilization (Dubruille et al., 2010). However, despite the essential role of K81 for male fertility through a paternal effect, spermatogenesis itself, including meiotic divisions, occurs normally in K81-null mutant males (Fuyama, 1984; Yasuda et al., 1995). Here, we show that telomere protection in male germ cells relies on two types of capping complexes, each involving one of the HipHop and K81 paralogs. The onset of spermatogenesis requires HipHop, which is expressed in GSCs and is crucial for their survival, demonstrating that this ubiquitous capping protein is essential in order to protect telomeres in these cells. The requirement of HipHop in GSCs is thus similar to its role in somatic cells. In proliferative spermatogonia and in spermatocytes, our data show that a gradual switch between somatic-like and male-germline-specific complexes occurs, leading to the complete replacement of HipHop by K81 in post-meiotic cells. Although K81 is specialized in the capping of sperm chromosomes, it has been previously reported that K81 is efficiently targeted to somatic telomeres (Dubruille et al., 2010) where it can achieve at least some level of protection (Gao et al., 2011). Our observation that K81 expression in GSCs partially restores testicular development and fertility of nos-Gal4/UAS-sh-hiphop flies further supports the notion that K81 is able to form functional capping complexes outside of the sperm chromatin environment.
The fact that the HipHop-K81 switch occurs gradually in dividing cells suggests that HipHop is progressively diluted as K81 is loaded onto telomeres during each cell cycle. Although the precise dynamics of capping-protein loading onto telomeres is currently unknown, it is tempting to speculate that the recruitment of capping-protein complexes is coupled with DNA replication as new chromosome extremities are produced. Supporting this assumption, we have previously shown that, in fertilized eggs, HipHop is not detected in the decondensing male pronucleus but is subsequently present at paternal telomeres when the apposed pronuclei initiate the first zygotic replication (Dubruille et al., 2010). A coupling of DNA replication with the recruitment of capping proteins would explain why K81, the function of which is essential in post-meiotic nuclei, is loaded during pre-meiotic stages.
Telomere protection with a minimal amount of capping
The most surprising result of our study is the finding that the amount of capping proteins present at telomeres can be severely reduced without dramatically impairing chromosome-end protection. Indeed, K81, HOAP and HP1a are barely detected in spermatid nuclei of Rescav males, which are nevertheless fertile. This implies that the residual amount of K81 in Rescav spermatid nuclei is sufficient to protect paternal chromosomes at fertilization. Moreover, the relatively low number of spermatid nuclei that are connected by a chromatin thread in Rescav testes demonstrates that telomeres are efficiently protected during male meiosis, despite the fact that HOAP is barely detected at this stage. Finally, we also propose that, in absence of K81, residual levels of HipHop can ensure telomere protection in meiosis. Using chromosomes with terminal deletions that removed the HTT array allowing ChIP analyses, Gao and colleagues (Gao et al., 2010) have previously shown that HipHop–HOAP–HP1a capping complexes bind to a large region (≥10 kb) of terminal chromatin. The current model suggests that the recruitment of terminin complexes onto chromosome extremities is mediated by some effectors of the DDR, such as the MRN complex and the ATM and ATR–ATRIP kinases (Raffa et al., 2011). Following this initial recruitment, other HipHop–HOAP–HP1a capping complexes spread over a broader terminal region (Gao et al., 2010). The fact that, in Rescav males, telomere protection is globally not disrupted implies that the presence of a small amount of terminin complexes is sufficient to efficiently inhibit their recognition as DNA DSBs. Although we could not investigate the chromatin occupancy of the HOAP, K81 and HP1a proteins in Rescav males, we hypothesize that the few capping complexes concentrate within the last base pairs of the DNA molecule.
Interestingly, several studies have reported that very short telomeres in human cells and yeast are efficiently protected. Telomeres with extremely short tracts called ‘t-stumps’ have been characterized in transformed and cancer cells (Xu and Blackburn, 2007). Surprisingly however, these ‘t-stumps’ do not compromise cell proliferation. In the yeast Saccharomyces cerevisiae, the characterization of telomere fusion events revealed that a majority of chromosome end-to-end fusions engage with short telomeric tracts of less than 100 bp (Chan and Blackburn, 2003). Similarly, in cultured human cells, telomeres become fusogenic when they contain fewer than ca. 13 repeats, suggesting that 78 bp would be sufficient to recruit shelterin complexes and efficiently protect chromosome ends (Capper et al., 2007). Moreover, in vitro studies have demonstrated that seven TTAGGG repeats are bound by at least one molecule of the human shelterin proteins TRF1 or TRF2 (Xu and Blackburn, 2007) and that a minimum of 12 TTAGGG repeats is required to inhibit the fusion of DNA ends in the presence of TRF2 and RAP1 (also known as TERF2IP in mammals) shelterin proteins (Bae and Baumann, 2007). Thus, in species that rely on telomerase, an extremely small number of DNA repeats is actually sufficient to prevent end-to-end fusions. Remarkably, our study supports that Drosophila telomeres, like their yeast and vertebrate counterparts, can be protected from telomere fusions with a small amount of capping proteins.
What could be the functions of a large telomeric capping domain?
The fact that telomere protection can be achieved with low amounts of terminin complexes raises questions about the role of the relatively large chromatin domain that is occupied by capping proteins at Drosophila telomeres. In a variety of organisms, neo-telomeres can be formed through a mechanism called chromosome healing, which comprises the stabilization of chromosome breaks through the recruitment of capping proteins. In Drosophila, chromosomes with new telomeres have been recovered in the progeny of wild-type males in which chromosome breaks have been induced in the germline or from irradiated females mutant for the mutator-2 gene (Mason et al., 1984; Levis, 1989; Ahmad and Golic, 1998; Beaucher et al., 2012; Titen et al., 2014). De novo telomere formation has also been described in S. cerevisiae and in mammalian cells (Kramer and Haber, 1993; Sprung et al., 1999; Diede and Gottschling, 1999; Lo et al., 2002; Gao et al., 2008; Kulkarni et al., 2010). Interestingly, in all these systems, including Drosophila, it has been observed that chromosome healing is facilitated when the break occurs relatively near to the natural ends of the chromosomes (Levis, 1989; Tower et al., 1993; Sprung et al., 1999; Diede and Gottschling, 1999; Lo et al., 2002; Capkova Frydrychova et al., 2008; Gao et al., 2008; Kulkarni et al., 2010). Kulkarni and colleagues have demonstrated that de novo telomere formation in human tumor cells is tenfold less frequent when the break occurs 100 kb from the extremity compared with 3 kb (Kulkarni et al., 2010). Importantly, they rule out the possibility that this is due to the loss of genetic material essential for cell viability. It has been alternatively proposed that de novo telomere formation is facilitated by the availability of capping proteins (Mason et al., 2008; Beaucher et al., 2012). Hence, a large capping domain might constitute a reserve of capping proteins that could be rapidly mobilized to stabilize a DNA break occurring in the vicinity of the natural telomere. In addition, this ‘reserve’ of capping proteins might also be involved in the stabilization of new chromosomal extremities that are generated when chromosomes replicate or when a telomeric retrotransposon is added to a chromosome end during telomere elongation.
Capping proteins could also be implicated in the formation and the stabilization of DNA structures that further protect telomeres. Indeed, in vertebrates, although very short telomeres are still protected from DNA repair machinery, they are too short to fold into a t-loop (Bae and Baumann, 2007), which is thought to participate in the protection of the chromosome extremity (Griffith et al., 1999; de Lange, 2004; Giraud-Panis et al., 2013). T-loops are formed through the folding and the sequestration of the 3′ single-strand DNA overhang in the double-strand extremity, and their formation is thought to hide the DNA extremity from the repair machinery (Griffith et al., 1999; Doksani et al., 2013). It has been shown that the human shelterin protein TRF2 can induce the formation of t-loops both in vitro and in vivo (Griffith et al., 1999; Stansel et al., 2001; Amiard et al., 2007; Doksani et al., 2013). The ability of Drosophila telomeres to adopt a t-loop structure in vivo remains to be established (Pimpinelli, 2006). However, among the terminin proteins, Verrocchio contains an OB-fold domain known to bind to single-strand DNA, which suggests that Drosophila telomeres terminate with a 3′ single-strand overhang (Raffa et al., 2010). Moreover, the sequence of the telomeric retrotransposon Het-A is prone to form a DNA structure called G-quadruplex, thought to appear in t-loops (Abad and Villasante, 1999). It would thus be interesting to determine whether HOAP or HipHop have biochemical properties similar to those of TRF2.
The telomeric domain occupied by capping complexes is remarkably plastic
Although limiting the expression of HOAP severely reduced the level of HP1a and K81 capping proteins recruited at telomeres, forcing the expression of HipHop in post-meiotic stages increased the abundance of HipHop, HOAP and HP1a at chromosome ends. This observation strengthens the view that HOAP and HipHop and K81 are interdependent for their localization at telomeres. Furthermore, although we cannot exclude that the density of capping complexes is higher within the capping domain of the 5′K81-GFP::hiphop spermatid telomeres, we favor the hypothesis that capping proteins bind to a larger region of terminal chromatin. Indeed, it has been proposed that in the absence of sequence recognition, capping proteins are recruited through a spreading mechanism, from the end towards the centromere (Gao et al., 2010). In this model, the relative availability of key components of the complex, such as HOAP and HipHop, can directly influence the length of the capping domain. This is also supported by the fact that HOAP is limiting for the assembly of capping complexes on telomeres, as shown using Rescav males.
In mammals, very short tracts of telomeric repeats are sufficient to achieve efficient chromosome-end protection through the recruitment of the shelterin proteins TRF2 and RAP1 (Bae and Baumann, 2007; Price, 2007). Because Drosophila telomeres are epigenetically determined structures, the sizes of the HTT array and the capping domain are naturally uncoupled (Gao et al., 2010; Raffa et al., 2013). Therefore, a shorter HTT array has no impact on the amount of capping proteins that are recruited onto chromosome termini. Remarkably, our work shows that limiting the expression of the capping protein HOAP leads to a dramatic decrease of the capping complexes at telomeres without impairing their protection. This study supports that, despite their divergence, Drosophila and telomerase-dependent telomeres share the ability to efficiently inhibit chromosome-end fusions when their capping domain is severely reduced. Further studies should aim to understand how the size of the fly capping domain is regulated.
MATERIALS AND METHODS
Drosophila genetics
w1118 flies were used as a control. The K812, hiphop1 and hiphopEY7584 alleles and the 5′K81-GFP::K81, 5′hiphop-GFP::hiphop, 5′hiphop-GFP::K81 and 5′K81-GFP::hiphop transgenic stocks have been described previously (Dubruille et al., 2010).
The cav1 allele (Cenci et al., 2003) and moi1 and ver2 (l(3)S147910) mutant stocks (Raffa et al., 2009; Raffa et al., 2010) were generous gifts from, respectively, Stéphane Ronsseray (Université Pierre et Marie Curie, Paris, France), Maurizio Gatti and Grazia Raffa (Università di Roma, Roma, Italy). The ProtB-GFP transgenic strain was kindly provided by Renate Renkawitz-Pohl (Philipps-Universität Marburg, Marburg, Germany) (Jayaramaiah-Raja and Renkawitz-Pohl, 2005). Df(3L)BSC776 and Df(3R)Exel6198 deficiencies, UAS-sh-GFP strain (y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=VALIUM20-EGFP}attP2), nos-GAL4 (w[1118]; P{w[+mC]=GAL4::VP16-nos.UTR}CG6325[MVD1]) were obtained at the Bloomington Drosophila Stock Center.
The 5′hiphop-GFP::cav and UAS-sh-hiphop transgenic lines were obtained using the phiC31-mediated integration system (Bischof et al., 2007). The pValium22-sh-hiphop construct (see below) was inserted into the P{y[+t7.7]=CaryP}attP2 platform (68A4) and the pW8-attB-5′hiphop-GFP::cav construct (see below) was inserted into the M{vas-int.Dm}ZH-2A (2A3) and the PBAc{y[+]-attP-3B}VK00031 (62E1) platforms. The 5′hiphop-GFP::cav 2A transgene was combined with the cav1 null allele and Rescav males were obtained by crossing virgin 5′hiphop-GFP::cav/5′hiphop-GFP::cav; cav1/TM6 females with w; Df(3R)Exel6198/TM6 males.
Fertility test
Male fertility tests were performed as described previously (Dubruille et al., 2010).
Plasmid constructions
To obtain the pW8-attB-5′hiphop-GFP::cav construct, the hiphop coding sequence in the previously described pW8-attB-5′hiphop-GFP::hiphop construct (Dubruille et al., 2010) was removed and replaced by a fragment encoding cav that was amplified, by using PCR, from genomic DNA using the 5′-TAGCGGCCGCCATGTCGGGGACGCAAATGTC-3′ and 5′-GAGGATCCAGAACTAGACAGTGGC-3′ primers.
The pValium22-sh-hiphop construction was obtained by cloning the two annealed primers 5′-CTAGCAGTTAAGTTGTGACTAGAGAAC-AATAGTTATATTCAAGCATATTGTTCTCTAGTCACAACTTAGCG-3′ and 5′-AATTCGCTAAGTTGTGACTAGAGAACAATATGCTTGAATA-TAACTATTGTTCTCTAGTCACAACTTAACTG-3′ into a pValium22 vector following instructions of the Transgenic RNAi Project at Harvard Medical School (http://www.flyrnai.org/TRiP-HOME.html) (Ni et al., 2011). The cloned DNA fragment encoded a small hairpin that targets a 21-nucleotide sequence in the 3′UTR of hiphop.
Reverse-transcription and semi-quantitative PCR
Total RNAs were extracted from 50 pairs of testes using the Trizol method (Invitrogen, Thermo Fisher Scientific, Waltham, MA). They were then treated with DNase1 (Ambion, Thermo Fisher Scientific) and reverse transcribed with Superscript II (Invitrogen) and polyA primers. A control reaction was set up without Superscript. cDNAs were PCR amplified using the following primers – 5′-ATGTCGGATTCGCCCCATG-3′ and 5′-TAGTGGTTGATTCTTGCTCCTC-3′ for K81, 5′-AAGATCGTGA-AGAAGCGCAC-3′ and 5′-ACTCGTTCTCTTGAGAACGC-3′ for Rp49, 5′-CAAGCGAAAGTCCGAAGAA-3′ and 5′-ACCATTTCTGCTTGG-TCCAC-3′ for Su(var)205 and 5′-ATCTGATCTTAGCGGCATCG-3′ and 5′-AGTCCGGATCAGACTCCGTA-3′ for cav.
Immunostaining
Polytene chromosomes
Polytene chromosomes were prepared, as described previously (Dubruille et al., 2010), and stained with a mouse monoclonal antibody against GFP, clones 7.1 and 13.1 (Roche, Basel, Switzerland) at a 1:250 dilution and an AlexaFluor-conjugated goat anti-mouse secondary antibody (Molecular Probes, Thermo Fisher Scientific) at a 1:300 dilution.
Squashed and whole-mount testes
Squashed and whole-mount testes were stained as described previously (Dubruille et al., 2010) with the following antibodies – mouse IgG1 anti-GFP (1:100; Roche), mouse anti-HP1a, clone C1A9 [1:25; Developmental Studies Hybridoma Bank (DSHB), University of Iowa], mouse IgG2a anti-FasIII, clone 7G10 (1:50; DSHB), guinea pig anti-HOAP (1:200) (Gao et al., 2010) and rabbit anti-Mst77F (1:1000) (Rathke et al., 2010), rat anti-VASA (1:50; DHSB), mouse monoclonal anti-pan-histone, clone F152 (1:1000; Millipore). Secondary antibodies include AlexaFluor goat anti-mouse, anti-mouse IgG1, anti-mouse IgG2a and anti-rat, anti-rabbit (Molecular Probes or Jackson ImmunoResearch, West Grove, PA) and DyLight donkey anti-guinea pig (Jackson ImmunoResearch). They were used at a 1:300 dilution for squashed or 1:1000 for whole-mount testes. The immunostaining of Mst77F allowed us to distinguish spermatid nuclei that undergo the histone-to-protamine transition. All tissues were directly mounted in mounting medium (Dako, Glostrup, Denmark) containing 5 µg/ml propidium iodide (Sigma-Aldrich, St Louis, MO) or 1 µM DRAQ 5 (BioStatus, Leicestershire, UK).
Imaging and quantification of fluorescent signals in spermatid nuclei
Single-stack images were acquired on an LSM510 confocal microscope (Zeiss, Oberkochen, Germany) in an 8-bit (for analyses in Fig. 4) or 12-bit mode (for analyses in Fig. 6). For quantification, optimal settings (laser power, amplifier offset, etc.) were set using the ‘palette’ command of the LSM software to avoid saturation of fluorescence signals. Then, all images were acquired using the same settings for samples that were compared to each other. Quantification of results was performed using the ImageJ64 software. First, areas corresponding to nuclei were selected in the red channel using the ‘threshold’ and the ‘analyze particle’ commands and the region of interest (ROI) manager. Then, the particles corresponding to HOAP, HP1a or GFP foci in the nucleus areas were selected in the green channel using a threshold above the background level, and particles were analyzed using the ‘analyze particle’ command. Results were plotted and statistical analyses were performed using the Prism software (GraphPad, La Jolla, CA).
Quantification of chromatin bridges in fly testes
Whole-mount testes stained with an antibody against histone (Millipore, Billerica, MA) were observed on a fluorescence microscope (Zeiss). All spermatid nuclei that exhibited a chromatin thread were counted. Note that protamine-positive nuclei were not taken into account. We used the Prism software to plot and analyze data.
Acknowledgements
We thank Renate Renkawitz-Pohl, Christina Rathke (Philipps-Universität Marburg, Marburg, Germany), Yikang Rong (National Cancer Institute NIH, Bethesda, MD, USA), Stéphane Ronsseray (Université Pierre et Marie Curie, Paris, France), Grazia Raffa and Maurizio Gatti (Università di Roma, Roma, Italy) for antibodies and/or fly stocks. We also thank the Transgenic RNAi project (TRiP) at Harvard Medical School [grant NIH/NIGMS R01-GM084947] and the Bloomington Stock Center for providing fly stocks and plasmid vectors used in this study. We are grateful to Julien Falk for his help in using ImageJ for quantification analyses and Laure Sapey-Triomphe for her technical assistance. We also thank all members of B. Loppin's lab for helpful discussions and feedback on this work. Confocal microscopy was performed at the Centre Technologique des Microstructures of the University Claude Bernard Lyon1.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
R.D. and B.L. designed the experiments, R.D. performed the experiments, R.D. and B.L. analyzed the data, R.D. and B.L. wrote the paper.
Funding
This work was supported by the Centre National de Recherche Scientique and a grant from the Fondation ARC pour la Recherche sur le Cancer to B.L. [grant SFI20121205765]. R.D. was supported by a post-doctoral fellowship from the Fondation ARC pour la Recherche sur le Cancer.
References
- Abad J. P. and Villasante A. (1999). The 3′ non-coding region of the Drosophila melanogaster HeT-A telomeric retrotransposon contains sequences with propensity to form G-quadruplex DNA. FEBS Lett. 453, 59-62. 10.1016/S0014-5793(99)00695-X [DOI] [PubMed] [Google Scholar]
- Ahmad K. and Golic K. G. (1998). The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics 148, 775-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard S., Doudeau M., Pinte S., Poulet A., Lenain C., Faivre-Moskalenko C., Angelov D., Hug N., Vindigni A., Bouvet P. et al. (2007). A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 14, 147-154. 10.1038/nsmb1192 [DOI] [PubMed] [Google Scholar]
- Bae N. S. and Baumann P. (2007). A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell 26, 323-334. 10.1016/j.molcel.2007.03.023 [DOI] [PubMed] [Google Scholar]
- Beaucher M., Zheng X. F., Amariei F. and Rong Y. S. (2012). Multiple pathways suppress telomere addition to DNA breaks in the Drosophila germline. Genetics 191, 407-417. 10.1534/genetics.112.138818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Srikanta D., Fanti L., Pimpinelli S., Badugu R., Kellum R. and Rong Y. S. (2005). Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc. Natl. Acad. Sci. USA 102, 15167-15172. 10.1073/pnas.0504981102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F. and Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312-3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capkova Frydrychova R., Biessmann H. and Mason J. M. (2008). Regulation of telomere length in Drosophila. Cytogenet. Genome Res. 122, 356-364. 10.1159/000167823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capper R., Britt-Compton B., Tankimanova M., Rowson J., Letsolo B., Man S., Haughton M. and Baird D. M. (2007). The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 21, 2495-2508. 10.1101/gad.439107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon D. H., Gönczy P., Alexander S., Rawson R., Eberhart C. G., Viswanathan S., DiNardo S. and Wasserman S. A. (1993). Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135, 489-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G., Siriaco G., Raffa G. D., Kellum R. and Gatti M. (2003). The Drosophila HOAP protein is required for telomere capping. Nat. Cell Biol. 5, 82-84. 10.1038/ncb902 [DOI] [PubMed] [Google Scholar]
- Cenci G., Ciapponi L. and Gatti M. (2005). The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma 114, 135-145. 10.1007/s00412-005-0005-9 [DOI] [PubMed] [Google Scholar]
- Chan S. W.-L. and Blackburn E. H. (2003). Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell 11, 1379-1387. 10.1016/S1097-2765(03)00174-6 [DOI] [PubMed] [Google Scholar]
- Ciapponi L., Cenci G., Ducau J., Flores C., Johnson-Schlitz D., Gorski M. M., Engels W. R. and Gatti M. (2004). The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr. Biol. 14, 1360-1366. 10.1016/j.cub.2004.07.019 [DOI] [PubMed] [Google Scholar]
- Ciapponi L., Cenci G. and Gatti M. (2006). The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics 173, 1447-1454. 10.1534/genetics.106.058081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. (2004). T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 5, 323-329. 10.1038/nrm1359 [DOI] [PubMed] [Google Scholar]
- Diede S. J. and Gottschling D. E. (1999). Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99, 723-733. 10.1016/S0092-8674(00)81670-0 [DOI] [PubMed] [Google Scholar]
- Doksani Y., Wu J. Y., de Lange T. and Zhuang X. (2013). Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155, 345-356. 10.1016/j.cell.2013.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R. and Loppin B. (2011). Epigenetic maintenance of telomere identity in Drosophila: buckle up for the sperm ride. Cell Cycle 10, 1037-1042. 10.4161/cc.10.7.15071 [DOI] [PubMed] [Google Scholar]
- Dubruille R., Orsi G. A., Delabaere L., Cortier E., Couble P., Marais G. A. B. and Loppin B. (2010). Specialization of a Drosophila capping protein essential for the protection of sperm telomeres. Curr. Biol. 20, 2090-2099. 10.1016/j.cub.2010.11.013 [DOI] [PubMed] [Google Scholar]
- Dubruille R., Marais G. A. B. and Loppin B. (2012). Repeated evolution of testis-specific new genes: the case of telomere-capping genes in Drosophila. Int. J. Evol. Biol. 2012, 708980 10.1155/2012/708980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L., Giovinazzo G., Berloco M. and Pimpinelli S. (1998). The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2, 527-538. 10.1016/S1097-2765(00)80152-5 [DOI] [PubMed] [Google Scholar]
- Fulcher N., Derboven E., Valuchova S. and Riha K. (2014). If the cap fits, wear it: an overview of telomeric structures over evolution. Cell. Mol. Life Sci. 71, 847-865. 10.1007/s00018-013-1469-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M. T. (1993). Spermatogenesis. In The Development of Drosophila Melanogaster (ed. Bate M. and Martinez-Arias A.), pp. 71-146. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Fuyama Y. (1984). Gynogenesis in Drosophila melanogaster. Jpn. J. Genet. 59, 91-96. 10.1266/jjg.59.91 [DOI] [Google Scholar]
- Gao Q., Reynolds G. E., Wilcox A., Miller D., Cheung P., Artandi S. E. and Murnane J. P. (2008). Telomerase-dependent and -independent chromosome healing in mouse embryonic stem cells. DNA Repair (Amst.) 7, 1233-1249. 10.1016/j.dnarep.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Walser J.-C., Beaucher M. L., Morciano P., Wesolowska N., Chen J. and Rong Y. S. (2010). HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 29, 819-829. 10.1038/emboj.2009.394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Cheng Y., Wesolowska N. and Rong Y. S. (2011). Paternal imprint essential for the inheritance of telomere identity in Drosophila. Proc. Natl. Acad. Sci. USA 108, 4932-4937. 10.1073/pnas.1016792108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Panis M.-J., Pisano S., Benarroch-Popivker D., Pei B., Le Du M.-H. and Gilson E. (2013). One identity or more for telomeres? Front. Oncol. 3, 48 10.3389/fonc.2013.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. and DiNardo S. (1996). The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development 122, 2437-2447. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H. and de Lange T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97, 503-514. 10.1016/S0092-8674(00)80760-6 [DOI] [PubMed] [Google Scholar]
- Jain D. and Cooper J. P. (2010). Telomeric strategies: means to an end. Annu. Rev. Genet. 44, 243-269. 10.1146/annurev-genet-102108-134841 [DOI] [PubMed] [Google Scholar]
- Jayaramaiah Raja S. and Renkawitz-Pohl R. (2005). Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol. Cell. Biol. 25, 6165-6177. 10.1128/MCB.25.14.6165-6177.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman T., Tran J. and DiNardo S. (2003). Mutations in Nop60B, the Drosophila homolog of human dyskeratosis congenita 1, affect the maintenance of the germ-line stem cell lineage during spermatogenesis. Dev. Biol. 253, 189-199. 10.1016/S0012-1606(02)00013-1 [DOI] [PubMed] [Google Scholar]
- Kramer K. M. and Haber J. E. (1993). New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 7 12A, 2345-2356. 10.1101/gad.7.12a.2345 [DOI] [PubMed] [Google Scholar]
- Kulkarni A., Zschenker O., Reynolds G., Miller D. and Murnane J. P. (2010). Effect of telomere proximity on telomere position effect, chromosome healing, and sensitivity to DNA double-strand breaks in a human tumor cell line. Mol. Cell. Biol. 30, 578-589. 10.1128/MCB.01137-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R. W. (1989). Viable deletions of a telomere from a Drosophila chromosome. Cell 58, 791-801. 10.1016/0092-8674(89)90112-8 [DOI] [PubMed] [Google Scholar]
- Lo A. W., Sprung C. N., Fouladi B., Pedram M., Sabatier L., Ricoul M., Reynolds G. E. and Murnane J. P. (2002). Chromosome instability as a result of double-strand breaks near telomeres in mouse embryonic stem cells. Mol. Cell. Biol. 22, 4836-4850. 10.1128/MCB.22.13.4836-4850.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B., Lepetit D., Dorus S., Couble P. and Karr T. L. (2005). Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr. Biol. 15, 87-93. 10.1016/j.cub.2004.12.071 [DOI] [PubMed] [Google Scholar]
- Lu W., Zhang Y., Liu D., Songyang Z. and Wan M. (2013). Telomeres-structure, function, and regulation. Exp. Cell Res. 319, 133-141. 10.1016/j.yexcr.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., Strobel E. and Green M. M. (1984). mu-2: mutator gene in Drosophila that potentiates the induction of terminal deficiencies. Proc. Natl. Acad. Sci. USA 81, 6090-6094. 10.1073/pnas.81.19.6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., Frydrychova R. C. and Biessmann H. (2008). Drosophila telomeres: an exception providing new insights. BioEssays 30, 25-37. 10.1002/bies.20688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., Yang-Zhou D., Shim H. S., Tao R., Handler D., Karpowicz P. et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405-407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L. and Karlseder J. (2009). Telomeric armor: the layers of end protection. J. Cell Sci. 122, 4013-4025. 10.1242/jcs.050567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikemus S. R., Queiroz-Machado J., Lai K., McGinnis N., Sunkel C. and Brodsky M. H. (2006). Epigenetic telomere protection by Drosophila DNA damage response pathways. PLoS Genet. 2, e71 10.1371/journal.pgen.0020071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W. and de Lange T. (2008). How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301-334. 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- Pardue M.-L. and DeBaryshe P. G. (2011). Retrotransposons that maintain chromosome ends. Proc. Natl. Acad. Sci. USA 108, 20317-20324. 10.1073/pnas.1100278108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrini B., Piacentini L., Fanti L., Altieri F., Chichiarelli S., Berloco M., Turano C., Ferraro A. and Pimpinelli S. (2004). HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell 15, 467-476. 10.1016/j.molcel.2004.06.036 [DOI] [PubMed] [Google Scholar]
- Pimpinelli S. (2006). Drosophila Telomeres. In Telomeres (ed. de Lange T., Lundblad V. and Blackburn E. H.), pp. 433-463. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Price C. M. (2007). wRAPing up the end to prevent telomere fusions. Mol. Cell 26, 463-464. 10.1016/j.molcel.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Raffa G. D., Siriaco G., Cugusi S., Ciapponi L., Cenci G., Wojcik E. and Gatti M. (2009). The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc. Natl. Acad. Sci. USA 106, 2271-2276. 10.1073/pnas.0812702106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa G. D., Raimondo D., Sorino C., Cugusi S., Cenci G., Cacchione S., Gatti M. and Ciapponi L. (2010). Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 24, 1596-1601. 10.1101/gad.574810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa G. D., Ciapponi L., Cenci G. and Gatti M. (2011). Terminin: a protein complex that mediates epigenetic maintenance of Drosophila telomeres. Nucleus 2, 383-391. 10.4161/nucl.2.5.17873 [DOI] [PubMed] [Google Scholar]
- Raffa G. D., Cenci G., Ciapponi L. and Gatti M. (2013). Organization and evolution of Drosophila terminin: similarities and differences between Drosophila and human telomeres. Front. Oncol. 3, 112 10.3389/fonc.2013.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C., Barckmann B., Burkhard S., Jayaramaiah-Raja S., Roote J. and Renkawitz-Pohl R. (2010). Distinct functions of Mst77F and protamines in nuclear shaping and chromatin condensation during Drosophila spermiogenesis. Eur. J. Cell Biol. 89, 326-338. 10.1016/j.ejcb.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Rong Y. S. (2008). Telomere capping in Drosophila: dealing with chromosome ends that most resemble DNA breaks. Chromosoma 117, 235-242. 10.1007/s00412-007-0144-2 [DOI] [PubMed] [Google Scholar]
- Sprung C. N., Reynolds G. E., Jasin M. and Murnane J. P. (1999). Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 96, 6781-6786. 10.1073/pnas.96.12.6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel R. M., de Lange T. and Griffith J. D. (2001). T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20, 5532-5540. 10.1093/emboj/20.19.5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J. A., Chaiken M. F., Wang F. and Price C. M. (2012). Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 730, 12-19. 10.1016/j.mrfmmm.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titen S. W. A., Lin H.-C., Bhandari J. and Golic K. G. (2014). Chk2 and p53 regulate the transmission of healed chromosomes in the Drosophila male germline. PLoS Genet. 10, e1004130 10.1371/journal.pgen.1004130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., Karpen G. H., Craig N. and Spradling A. C. (1993). Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics 133, 347-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasante A., de Pablos B., Méndez-Lago M. and Abad J. P. (2008). Telomere maintenance in Drosophila: rapid transposon evolution at chromosome ends. Cell Cycle 7, 2134-2138. 10.4161/cc.7.14.6275 [DOI] [PubMed] [Google Scholar]
- Wesolowska N., Amariei F. L. and Rong Y. S. (2013). Clustering and protein dynamics of Drosophila melanogaster telomeres. Genetics 195, 381-391. 10.1534/genetics.113.155408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H. (2012). Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis 2, 11-22. 10.4161/spmg.19088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. and Blackburn E. H. (2007). Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol. Cell 28, 315-327. 10.1016/j.molcel.2007.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda G. K., Schubiger G. and Wakimoto B. T. (1995). Genetic characterization of ms (3) K81, a paternal effect gene of Drosophila melanogaster. Genetics 140, 219-229. [DOI] [PMC free article] [PubMed] [Google Scholar]