SUMMARY
Ion channels establish and regulate membrane potentials in excitable and non-excitable cells. How functional diversification of ion channels contributed to the evolution of nervous systems may be understood by studying organisms at key positions in the evolution of animal multicellularity. We have carried out the first analysis of ion channels cloned from a marine sponge, Amphimedon queenslandica. Phylogenetic comparison of sequences encoding for poriferan inward-rectifier K+ (Kir) channels suggests that Kir channels from sponges, cnidarians and triploblastic metazoans each arose from a single channel and that duplications arose independently in the different groups. In Xenopus oocytes, AmqKirA and AmqKirB produced K+ currents with strong inward rectification, as seen in the mammalian Kir2 channels, which are found in excitable cells. The pore properties of AmqKir channels demonstrated strong K+ selectivity and block by Cs+ and Ba2+. We present an original analysis of sponge ion channel physiology and an examination of the phylogenetic relationships of this channel with other cloned Kir channels.
KEY WORDS: Kir, evolution, inwardly rectifying, ion channel, metazoa, sponge
INTRODUCTION
Potassium (K+) channels are found in cell membranes throughout all domains of life. They are important for essential cellular functions including the regulation of cell volume, growth, membrane excitability and K+ secretion and recycling. Among the functionally diverse K+ channels, inwardly rectifying (Kir) channels are unusual in that they preferentially conduct K+ from the extracellular compartment into the cell. Channel subunits are comprised of two transmembrane domains (M1 and M2) flanking a central pore domain (P), which is highly conserved among the great variety of K+ channels. Their presence in prokaryotes and a variety of eukaroytic cell types emphasizes their importance to cells. The mechanisms of permeation, selectivity and rectification are known from the structural analysis of K+ channels in prokaryotes (Doyle et al., 1998; Jiang et al., 2002; Kuo et al., 2003; Nishida et al., 2007) and in mice (Nishida and MacKinnon, 2002). Knowledge of how membrane conduction properties changed during the evolution of multicellularity would be aided by the comparative analysis of K+ channels from phyla occupying basal positions.
Relationships among the earliest diverging animal groups are still not clearly understood, with recent molecular evidence supporting one of placozoans, ctenophores or sponges as the sister group to all other metazoans (Dellaporta et al., 2006; Dunn et al., 2008; Srivastava et al., 2008). Most phylogenies, however, place sponges (suspension-feeding animals lacking nerves and muscle) as the most ancient multicellular metazoans. Analyses based on 18S rDNA suggest that sponges arose first but are paraphyletic. This suggests that a sponge-like animal gave rise to other metazoans (Borchiellini et al., 2001; Collins, 1998; Sperling et al., 2007), and that critical metazoan innovations might, therefore, be reflected in modern sponges. Evidence for elements of the postsynaptic scaffold in sponge larvae suggests this might be true (Sakarya et al., 2007). Although sponges lack true nerves, they show many types of non-nervous behavior. Aneural coordination in sponges involves regulation of their feeding current by immediate flagella arrest (Hexactinellida) (Leys and Mackie, 1997) or contraction of canals (Demospongiae, Calcarea) (Elliott and Leys, 2007; Nickel, 2004). In the former, the mechanism is a calcium action potential (Leys et al., 1999; Leys and Eerkes-Medrano, 2006); in the latter it is thought to be by calcium waves triggered by local release of signaling molecules (Ellwanger and Nickel, 2006).
Intracellular recordings have not yet been obtained from any sponge due to the difficulty of penetrating the complex glycocalyx surface coat and the thin nature of cells and tissues lining the surface (Litchfield and Morales, 1976; Muller et al., 1998). Patch-clamp recordings of cation channels have been made from the demosponge Axinella polypoides (Carpaneto et al., 2003; Zocchi et al., 2001) but trivalent ions were required to obtain a seal and no electrical conduction was found. Similar difficulty has been encountered in patching glass sponge tissue and, given the extreme difficulty of collecting and maintaining these deep-water animals, an ex situ approach is considered to offer a better insight into ion channel function in sponges and the evolution of electrical signaling in this phylum.
In this study, we report the cloning and functional characterization of the first ion channels isolated from a sponge (Amphimedon queenslandica, Demospongiae, Porifera). The newly available genome project of the tropical demosponge A. queenslandica (formerly Reniera sp.) (http://www.jgi.doe.gov/sequencing/why/3161.html) has facilitated progress in understanding the collection of metazoan homologues present in the earliest animals. We have prioritized the study of inward-rectifier potassium (Kir) channels for their basic structure and their important role both in stabilizing the resting potential and regulating cellular excitability. We present functional data on two clones, AmqKirA and AmqKirB, whose properties were studied in the Xenopus oocyte expression system. Sponge Kir channels share the phenotype and critical residues that regulate strong inward rectification with vertebrate Kir2 channels, which are typically expressed in excitable cells including neurons and skeletal and cardiac muscle cells (Doupnik et al., 1995). Phylogenetic comparison with other metazoan Kir channels suggests that Kir channels in sponges, cnidaria and triploblastic metazoans each arose from a single channel and that duplications arose independently in the different groups. Diversification into the channel subfamilies from Kir1 to Kir7 occurred independently in the chordates.
MATERIALS AND METHODS
Isolation of cDNAs and DNA sequencing
Amphimedon queenslandica larvae were procured from Heron Island Reef, Queensland, Australia and preserved in RNA later (Qiagen, Valencia, CA, USA). Total RNA was extracted with TRIZOL® according to Invitrogen specifications. Full-length cDNA was prepared in two ways: with the GeneRacer kit (Invitrogen, Carlsbad, CA, USA) and by rolling circle amplification (RCA) (Polidoros et al., 2006). For RCA, cDNA was synthesized from 5 μg total RNA using Superscript III Reverse Transcriptase Module (Invitrogen) with a 5′-phosphorylated OligodT primer (Ellwanger and Nickel, 2006). Following purification with the QIAquick PCR purification kit (Qiagen), full-length transcripts circularized with 150 units CircLigase enzyme (Epicentre, Madison, WI, USA) in 1× Circligase Buffer with 50μ mol l–1 ATP. The reaction was incubated for 1 h at 60°C, inactivated at 80°C for 10 min and purified as above. Circularized templates were amplified using 10 units of phi29 DNA polymerase in 1× phi29 DNA Polymerase Reaction Buffer, supplemented with 200μ gml–1 bovine serum albumin, 200 μmol l–1 dNTPs and 10 μmol l–1 random heptamers, which were modified by 3′-phosphothiote linkages to make the primers resistant to phi29 exonuclease activity. Amplification proceeded for 20 h at 30°C and was inactivated by incubation at 70°C for 10 min.
Putative ion channel sequences were assembled from unannotated trace files of the A. queenslandica (formerly Reniera sp.) genome as previously described (Sakarya et al., 2007). Initial fragments were amplified using specific primers designed against these sequences. The complete cDNA sequences were determined using a combination of inverse PCR on RCA cDNA template (Polidoros et al., 2006) and RACE-PCR on GeneRACE-ready cDNA (Invitrogen GeneRacer kit). Products were cloned into the PCR-4-TOPO (Invitrogen) vector and sequenced. Open reading frames were amplified using primers modified with restriction sites to allow ligation into pXT7 expression plasmid (Dominguez et al., 1995). The full sequence was confirmed from uncloned PCR products from at least two independent PCR reactions, and clones free of PCR errors were selected for expression.
Amino acid sequence and phylogenetic analysis
Sponge inward-rectifier sequences were aligned and compared with amino acid sequences of both invertebrate and vertebrate inward-rectifier sequences compiled from NCBI, Joint Genome Institute (http://www.jgi.doe.gov/sequencing/why/3161.html) and Ensembl databases. To determine pairwise identity values, AmqKirA and AmqKirB amino acid sequences were each in turn aligned using LALIGN online software (http://www.ch.embnet.org/software/LALIGN_form.html) and default parameters for global alignments. Multiple alignments were generated using MUSCLE (Edgar, 2004). Alignments used for phylogenetic analysis were manually adjusted in MacClade (Maddison and Maddison, 2003) and a PERL script was used to remove characters with more than 2% gaps to produce a trimmed alignment of 122 sequences and 280 positions. All data sets are available on request. The optimal tree topology and Bayesian posterior values were obtained using MrBayes v.3.1.2 (Ronquist and Huelsenbeck, 2003) with the default parameters for amino acid sequences. The same data set was also analyzed with RaxML (Stamatakis, 2006), as implemented on the CIPRES server (http://www.phylo.org/) to obtain maximum likelihood bootstrap values.
Functional expression in oocytes
RNA preparation, oocyte isolation and injection were done using standard methods (Boland et al., 2003). In brief, plasmids containing sponge channel cDNAs were linearized and capped RNAs synthesized in vitro using Ambion (Austin, TX, USA) mMessage Machine RNA polymerase kits. RNA was purified using the RNAid kit (Bio 101, Vista, CA, USA) and concentrations were determined by spectrophotometry. Oocytes were surgically harvested from female Xenopus laevis (Xenopus I, Dexter, MI, USA) frogs after anesthesia by immersion in buffered 0.1% 3-aminobenzoic acid ethyl ester (Sigma Chemical Co., St Louis, MO, USA). Oocytes were released and defolliculated by gentle agitation for 1 h or less in 0.5 mg ml–1 collagenase A (Sigma Chemical Co.) dissolved in a Ca2+-free solution containing (in mmol l–1): 96 NaCl, 2 KCl, 1 MgCl2, 5 Hepes, pH 7.4 with NaOH. Oocytes were then washed and stage V/VI oocytes were injected with 50 nl of cRNA dissolved in DEPC-treated water (5–45 ng cRNA/oocyte). Oocytes were maintained at 19°C in a frog Ringer solution of (in mmol l–1): 96 NaCl, 1 KCl, 1 CaCl2, 2 MgCl2, 10 Hepes, 5 sucrose and 2 Na pyruvate, pH 7.4 with NaOH with 50 μ ml–1 penicillin G and 50 μgml–1 streptomycin. Electrophysiological recordings were done 1–6 days post-injection.
Electrophysiology
K+ currents were recorded from oocytes using standard methods of two-electrode voltage clamp (Boland et al., 2003). We used a Geneclamp 500B amplifier (Axon Instruments, Foster City, CA, USA) and an OC-725C amplifier (Warner Instruments, Hamden, CT, USA). Voltage-measuring and current-passing electrodes were backfilled with 3 mol l–1 KCl and had resistances between 0.3–1.0 MΩ. Currents were sampled at 5–10 kHz and filtered at 1–2 kHz. Recordings were done at room temperature (about 23°C) and oocytes were perfused continuously during recordings. Solution composition varied by experiment. The standard, low-chloride, external solution contained (in mmol l–1): 5 K+ methanesulfonate (MES), 95 NMDG–MES, 2 CaCl2, 2 MgCl2, 10 Hepes, pH 7.3 with methanesulfonic acid. K+ concentrations were adjusted by changing the ratios of K+ and NMDG-containing solutions using either the MES or chloride salt of K+. Other solutions are noted in figure legends. Data were recorded on Pentium computers equipped with Digidata 1320A (Axon Instruments) A/D hardware. Axon's Clampex aquisition and Clampfit analysis software (v. 9) were used. Data were also transferred to Microsoft Excel and Microcal Origin (Northampton, MA, USA) for additional analysis, curve-fitting and the production of figures. Results were calculated from cells with negligible background currents and only for stable cells with membrane potentials not more depolarized than –50 mV in 5 mmol l–1 external K+.
RESULTS AND DISCUSSION
Phylogenetic analysis and primary structure
Two inward-rectifier K+ channels, AmqKirA and AmqKirB, were cloned from the demosponge A. queenslandica. Analysis of the genome assembly indicated that these are single exon, tandemly duplicated genes. The cDNAs had open reading frames of 1047 and 1071 bp, respectively, corresponding to predicted amino acid sequences of 349 and 357 residues with 68% shared identity (Fig. 1). As with other Kir channels, AmqKirA and AmqKirB each contain two transmembrane helices M1 and M2 flanking a conserved pore structure containing the TXGYG signature sequence for K+ channels. We identified five and four consensus protein kinase A (PKA) phosphorylation sites in the cytoplasmic domains of AmqKirA and AmqKirB, respectively (Fig. 1). Additionally, in AmqKirB, there are two consensus protein kinase C (PKC) phosphorylation sites. Sequence identity for the full-length clones was 29–35% with Nematostella vectensis (Cnidaria) and 23–30% with mammalian Kir channels; well below the identity levels found across known Kir subfamilies (50–60% with the exception of Kir7, which shares only 38% identity with other mammalian subfamilies) (Doupnik et al., 1995; Krapivinsky et al., 1998). The channel core (M1, P and M2) had higher amino acid identity with mammalian (38–51%) and cnidarian (48–54%) inward-rectifiers (Table 1; supplementary material Fig. S1).
Fig. 1.
LALIGN global alignment of predicted amino acid sequences of two sponge inward-rectifier potassium (Kir) channels, AmqKirA and AmqKirB, cloned from Amphimedon queenslandica. Membrane-spanning regions deduced from hydropathy analysis and comparison with vertebrate Kir channels are noted by bars (M1, M2). Putative protein kinase A (PKA) (•) and protein kinase C (PKC) (▴) phosphorylation sites were predicted by two out of three prediction programs used (KinasePhos, PredPhospho and Group-based Phosphorylation Scoring).
Table 1.
Percentage amino acid identity of AmqKirA and AmqKirB with Nematostella vectensis (Nv) and vertebrate Kir channels calculated from global alignments with LALIGN online software (http://www.ch.embnet.org/software/LALIGN_form.html) for the overall channel sequence and the channel core (transmembrane regions M1 and M2 and pore region)
|
% Identity with AmqKirA |
% Identity with AmqKirB |
|||||
|---|---|---|---|---|---|---|
| Sequence | Overall | Core | Overall | Core | ||
| AmqKirA | – | – | 67.8 | 69.2 | ||
| AmqKirB | 67.8 | 69.2 | – | – | ||
| Nv gi156375411 | 35.4 | 46.8 | 34.6 | 46.8 | ||
| Nv gi156372465 | 29.2 | 54.4 | 29.6 | 50.6 | ||
| Nv gi156395183 | 32.7 | 50.6 | 33.5 | 50.6 | ||
| Nv gi156387460 | 32.7 | 51.9 | 31.7 | 48.1 | ||
| rKir1.1 | 28.4 | 44.3 | 28.7 | 48.1 | ||
| mKir2.1 | 29.5 | 46.8 | 28.2 | 38.0 | ||
| hKir2.2 | 30.1 | 51.3 | 29.5 | 46.2 | ||
| hKir2.3 | 27.9 | 46.2 | 28.1 | 44.9 | ||
| hKir2.4 | 26.9 | 44.9 | 25.7 | 38.5 | ||
| hKir3.1 | 23.3 | 39.7 | 24.5 | 42.3 | ||
| hKir3.2 | 30.2 | 47.4 | 28.9 | 43.6 | ||
| hKir3.3 | 29.1 | 47.4 | 27.8 | 41.0 | ||
| hKir3.4 | 29.3 | 44.9 | 28.6 | 44.9 | ||
| hKir4.1 | 25.9 | 38.0 | 27.1 | 41.8 | ||
| hKir4.2 | 25.3 | 42.3 | 26.6 | 42.3 | ||
| hKir5.1 | 24.8 | 41.0 | 24.3 | 37.2 | ||
| hKir6.1 | 25.1 | 43.0 | 26.6 | 44.3 | ||
| hKir6.2 | 28.7 | 49.4 | 28.6 | 43.0 | ||
| hKir7.1 | 25.0 | 43.6 | 23.2 | 39.7 | ||
Vertebrate Kir sequences are given for human (hKirx.x), rat (rKir1.1) and mouse (mKir2.1) channels, which were used as controls in our electrophysiology experiments
The phylogenetic relationships between sponge and other metazoan inward-rectifiers are shown in Fig. 2. The overall tree topology shows that Kir diversification occurred independently in sponges, cnidarians, invertebrates and chordates and indicates that a single ancestral gene gave rise to inward-rectifiers in the metazoa.
Fig. 2.
Phylogenetic relationships between AmqKir channels and other metazoan inward-rectifier potassium (Kir) channels. The phylogenetic relationship was derived as described in Materials and methods. The best Maximum Likelihood tree is shown with Bayesian support values above and RaxML maximum likelihood bootstrap support values below each node. Relationships not recovered in the RaxML analysis are indicated by nf. The tree was simplified by collapsing vertebrate Kir subfamilies from one through to seven and urochordate, arthropod and nematode clades of inward-rectifiers (represented by open triangles). Complete datasets used in analysis are available upon request.
We rooted the tree between poriferan and cnidarian sequences based on current knowledge of relationships among metazoan phyla (Borchiellini et al., 2001; Collins, 1998; Sperling et al., 2007). Computer-predicted inward-rectifier sequences from the choanoflagellate Monosiga brevicolis (gi167515450 and gi167522960) were highly divergent and their inclusion in the analysis destabilized all branches, suggesting that protozoan and metazoan Kir sequences are distinct. Lack of available sequences for ctenophores prevented a test of the recent hypothesis that this group is the sister to other metazoans (Dunn et al., 2008).
The order of branching of vertebrate Kir subfamilies is consistent with the analysis by Tanaka-Kunishima and colleagues (Tanaka-Kunishima et al., 2007). Branching of vertebrate Kir6 after the triploblastic invertebrates (Fig. 2) was also not supported in their analysis. As shown previously for other ion channels (Okamura et al., 2005), we find that diversification of Kir channel subfamilies occurred in vertebrates and that urochortate Kirs lie at the base of this diversification. (A compressed archive of files used to analyze the phylogeny is available from the authors.)
Electrophysiological properties
To study the functional characteristics of AmqKirA and AmqKirB channels, we cloned full-length cDNAs into the pXT7 expression plasmid, prepared cRNAs by in vitro transcription and injected purified cRNAs into frog oocytes. From a holding potential of –50 or 0 mV, voltage steps or a ramp protocol were used to change the membrane potential. Oocytes injected with either AmqKirA or AmqKirB cRNAs demonstrated prominent inwardly rectifying currents, which did not inactivate during 2 s voltage pulses. Because we injected oocytes with only a single cRNA species, it is clear that AmqKir channels can exist as homo-multimers. Whether they naturally co-assemble with other Kir subunits is not yet known.
The current–voltage relationships for AmqKirA and AmqKirB are strongly rectifying with large inward currents at membrane potentials more hyperpolarized to the equilibrium potential for K+ (EK) and very small currents positive to EK (Fig. 3A,B). In mammalian cells, this phenotype is determined by pore-lining residues, which facilitate voltage-dependent block by internal Mg2+ and polyamines. Mutagenesis of the strongly rectifying mammalian Kir2.1 channels has identified residues that control block by Mg2+ and polyamines, including S165 and D172 in the M2 region and E224, D255, D259 and E299 in the C-terminus (for a review, see Lu, 2004). These are largely conserved in AmqKirA and AmqKirB (supplementary material Fig. S1).
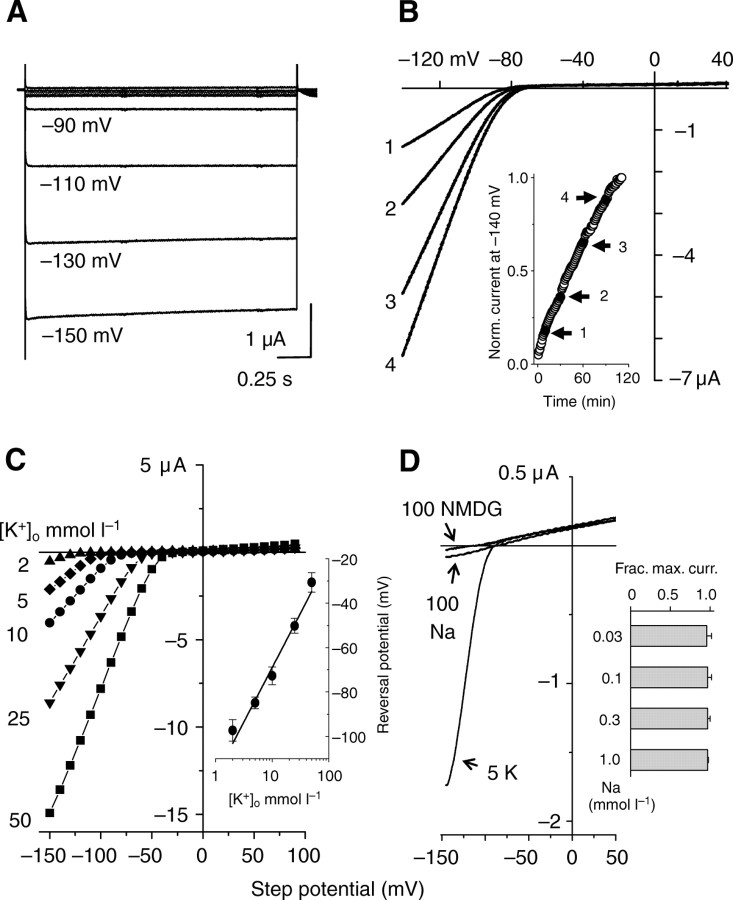
Fig. 3.
Heterologous expression of sponge inward-rectifier potassium (Kir) channels. Strongly rectifying currents in 5 mmol l–1 external K+ (KMES) recorded from (A) AmqKirA using 2 s voltage steps from– 150 to +100 mV in +20 mV increments from a holding potential –50 mV and (B) AmqKirB using a voltage ramp from a holding potential of –50 mV. Current–voltage plots for AmqKirB taken at 10 (#1), 30 (#2), 60 (#3) and 90 (#4) mins. The inset shows, in the same cell, the time-dependent change in the normalized current at –140 mV (until the recording ended). (C) Effect of increasing external K+ on AmqKirA currents. Solutions of 2, 5, 10, 25 and 50 mmol l–1 KMES were used. The inset is a semi-log plot of the reversal potential versus external K+ concentration for AmqKirA (N=6–28 per concentration, means± s.e.m.). The fitted line has a slope of 49 mV. (D) Representative AmqKirA current–voltage relationships recorded in 5 mmol l–1 K+, 100 mmol l–1 NMDG, and 100 mmol l–1 Na (all used as MES salts) showing no Na or NMDG permeability. The inset shows results of the test of Na (0.03–1 mmol l–1 NaCl) block of 5 mmol l–1 KMES currents (N=5, means ± s.e.m.).
Sponge channels were also constitutively active without the addition of activators such as G-proteins or ATP, although we cannot rule out a requirement for an activator that is endogenous to the oocytes. A suggestion for an as yet unknown modulatory mechanism was found for AmqKirB currents, which demonstrated an unusual phenomenon of run-up of current amplitude during recordings. The example (Fig. 3B) shows 5-fold growth of the inward current without a significant change in the reversal potential and without an increase in the outward current over nearly 2 h. Similar results were obtained from all cells injected with AmqKirB although the rate of increase of inward current varied from cell-to-cell. Run-up has been reported for vertebrate Kir2.1 and Kir2.2 currents and may be explained by PKA activity whereas these currents were downregulated by activation of PKC (Fakler et al., 1994; Scherer et al., 2007; Zitron et al., 2004).
Selectivity for K+
Increasing external K+ concentration, [K+]o, caused the reversal potentials for AmqKir currents to shift in accordance with the EK. The current amplitude also increased in a concentration-dependent manner (Fig. 3C). The relationship between reversal potential and [K+]o was linear; the fitted slope was 49 mV, close to the theoretical value of 58 for a perfectly K+ selective channel (Fig. 3C, inset). Selectivity was further tested by perfusing with 5 mmol l–1 K+ or solutions containing 100 mmol l–1 Na+ or 100 mmol l–1 NMDG in the absence of K+ (Fig. 3D). Currents were negligible in the Na+ or NMDG-containing solutions. This high K+ selectivity is typical of most K+ channels (Hille, 2001). In addition, Na+ (0.03–1 mmol l–1) was ineffective as a blocker of K+ (5 mmol l–1) currents (Fig. 3D, inset).
Block by external Cs+ and Ba2+
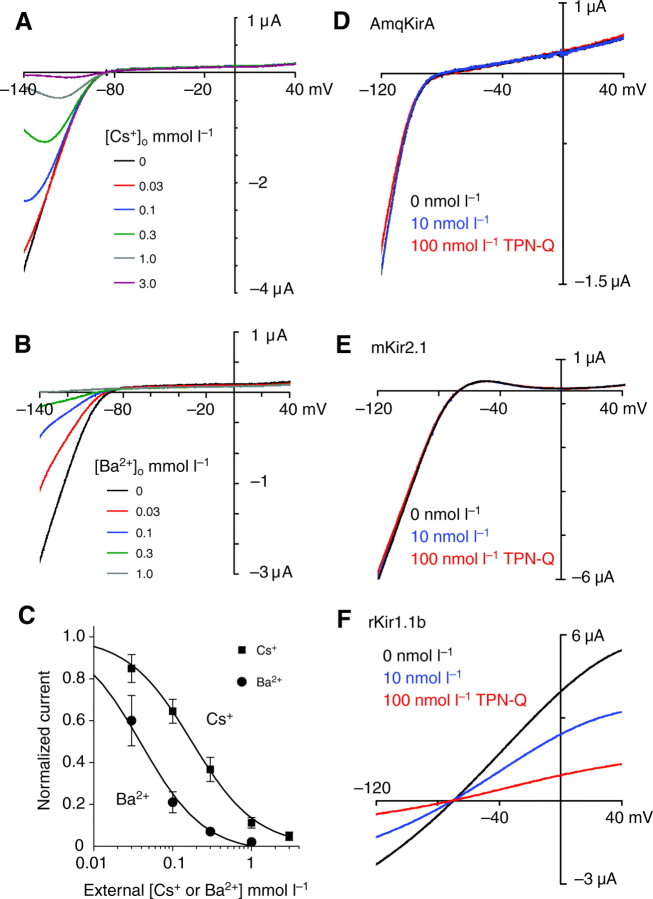
To further characterize the electrophysiology of AmqKir channels, we tested the effect of Cs+ and Ba2+ (monovalent and divalent cations that block K+ current in other inward-rectifiers). Both AmqKirA and AmqKirB currents were blocked by Cs+ and Ba2+ (Fig. 4) but we analyzed data for AmqKirA only because continual current run-up (Fig. 3B) precluded accurate measurement of the percentage block in AmqKirB. Oocytes expressing AmqKirA were perfused with solution containing 0.03–3 mmol l–1 Cs+ (Fig. 4A) and 0.01–1 mmol l–1 Ba2+ (Fig. 4B), and current inhibition was assessed using voltage step and ramp protocols. Block by Cs+ showed prominent voltage-dependence in that hyperpolarization intensified the block and depolarization relieved it. Block of AmqKirA currents in 5 mmol l–1 K+ was concentration-dependent with an IC50 of 37μmoll–1 for Ba2+ and 173μmoll–1 for Cs+ when measured at –140 mV (Fig. 4C).
Fig. 4.
Test of channel block of AmqKirA. Representative current–voltage relationships in 5 mmol l–1 KMES with increasing concentrations of (A) CsCl or (B) BaCl2. (C) The normalized Kir current at –140 mV is plotted versus blocking ion concentration. The data were fit by a logistic equation with an IC50 of 37 μmol l–1 for Ba2+ (N=5–8) and 173 μmol l–1 for Cs+ (N=8, means ± s.e.m.). Test for tertiapin-Q (TPN-Q) toxin block of (D) AmqKirA, (E) mKir2.1 (mouse Kir) and (F) rKir1.1b (rat Kir) in the absence (0 nmol l–1) or presence of 10 and 100 nmol l–1 TPN-Q. Recordings were done in 5 mmol l–1 KMES, pH 7.6. Currents were measured using 500 ms voltage ramps from a holding potential of –50 mV. The block of rKir1.1b was reversible.
Test of block by tertiapin-Q
Tertiapin-Q (TPN-Q), a non-oxidizable form of honeybee toxin, has been shown to selectively inhibit a subset of Kir channels and is one of only a few blockers available to discriminate between the contributions of different Kir channels in situ (Ramu et al., 2004). We tested the ability of TPN-Q to block AmqKirA, relative to the toxin-sensitive rat Kir1.1b and toxin-insensitive mouse Kir2.1. Because the block of Kir1.1b by TPN-Q is pH-sensitive (Ramu et al., 2004), we performed these experiments at an external pH of 7.6. In our experiments, AmqKirA was insensitive to 100 nmol l–1 TPN-Q (Fig. 4D) (N=3), similar to mouse Kir2.1 (Fig. 4E) (N=1). Rat Kir1.1b currents, however, were blocked in a concentration-dependent manner, with 80% block at 100 nmol l–1 (Fig. 4F) (N=2). The structural basis of TPN-Q selectivity is a variable region between M1 and M2. It has been suggested that the variable region in Kir1.1 (11 residues) and Kir3.4 (10 residues) forms an alpha helix that interacts with the helical portion of TPN-Q. Insensitivity of Kir2.1 channels may be explained by a shorter variable region (eight residues) as a complete helical turn may not form (Jin and Lu, 1999; Ramu et al., 2004). The corresponding region in AmqKir channels has only four residues, suggesting that is too short to confer sensitivity.
Concluding remarks
The electrophysiological properties of sponge Kir channels including the absence of substantial outward current at membrane potentials positive to EK, the high selectivity for K+ and the voltage-dependent block by Ba2+ and Cs+ indicate that these key features of strong inwardly rectifying K+ channels are conserved throughout evolution. In mammals, strongly rectifying K+ channels such as Kir2 channels are typically found in skeletal muscle, cardiac muscle and neurons (Kubo et al., 2005; Oliver et al., 2000). Kir2 channels function in setting the excitation threshold by permitting membrane depolarization without much K+ efflux. Cells expressing channels such as the AmqKir channels should be able to maintain a stable resting potential and be able to sustain prolonged depolarizations without massive loss of internal K+. The strong rectification of AmqKir channels is consistent with the idea that some sponge cell membranes may be specialized for active signaling.
The essential structural regions that control K+ selectivity have been conserved throughout the metazoa for more than 600 million years of evolution. Differences in the structures of the AmqKir channel and other metazoan Kir channels may impart functional differences not yet identified but clearly do not impact the fundamental properties of ion selectivity, block and rectification. Our results provide insight into the functional properties of the first ion channels cloned from the oldest lineage of metazoans and they shed light on properties that must have been present in the early evolution of multicellular organisms.
LIST OF ABBREVIATIONS
- AmqKirA
Amphimedon queenslandica inward-rectifier K+ channel A
- AmqKirB
Amphimedon queenslandica inward-rectifier K+ channel B
- EK
equilibrium potential for K+
- Kir
inward-rectifier K+
- [K+]o
external K+ concentration
- M1
transmembrane domain 1
- M2
transmembrane domain 2
- P
pore domain
- PKA
cAMP-dependent protein kinase
- PKC
protein kinase C
- TPN-Q
tertiapin-Q
FOOTNOTES
Supplementary material available online at http://jeb.biologists.org/cgi/content/full/212/6/761/DC1
The authors thank Kenneth Kosik (University of California-Santa Barbara) for sharing sponge ion channel sequences at an early stage in this project. We are also grateful to Henry Sackin (The Chicago Medical School, Rosalind Franklin University) for the rKir1.1b cDNA, and Paulo Kofuji (University of Minnesota) for the mKir2.1 cDNA. We thank Zhe Lu (University of Pennsylvania) for a gift of recombinant tertiapin-Q. We thank Lori Spicer, Michelle Drzewiecki, and Malcolm Hill (University of Richmond) for their respective help with RNA preparation and injection, oocyte preparation and interpretation of the phylogenetic analysis. We acknowledge the contribution of The US Department of Energy Joint Genome Institute in the production of Amphimedon genomic and EST sequences. Supported by NSERC Discovery grants to W.J.G., A. N. Spencer and S.P.L., Australian Research Council grants to B.D. and by funds from the University of Richmond School of Arts and Sciences to L.M.B. Data deposition footnote: sequence data for AmqKirA and AmqKirB has been deposited to GENBANK (Accession Numbers FJ375323 and FJ375324). Conflict of Interest Statement: No conflicts declared.
Present address: Department of Research Informatics, Amgen, Thousand Oaks, CA 91320, USA
References
- Boland, L. M., Jiang, M., Lee, S. Y., Fahrenkrug, S. C., Harnett, M. T. and O'Grady, S. M. (2003). Functional properties of a brain-specific NH2-terminally spliced modulator of kV4 channels. Am. J. Physiol. Cell Physiol. 285,C161 -C170. [DOI] [PubMed] [Google Scholar]
- Borchiellini, C., Manuel, M., Alivon, E., Boury-Esnault, N., Vacelet, J. and Le Parco, Y. (2001). Sponge paraphyly and the origin of Metazoa. J. Evol. Biol. 14,171 -179. [DOI] [PubMed] [Google Scholar]
- Carpaneto, A., Magrassi, R., Zocchi, E., Cerrano, C. and Usai, C. (2003). Patch-clamp recordings in isolated sponge cells (Axinella Polypoides). J. Biochem. Biophys. Methods 55,179 -189. [DOI] [PubMed] [Google Scholar]
- Collins, A. G. (1998). Evaluating multiple alternative hypotheses for the origin of bilateria: an analysis of 18S rRNA molecular evidence. Proc. Natl. Acad. Sci. USA 95,15458 -15463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S. L., Xu, A., Sagasser, S., Jakob, W., Moreno, M. A., Buss, L. W. and Schierwater, B. (2006). Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum. Proc. Natl. Acad. Sci. USA 103,8751 -8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, I., Itoh, K. and Sokol, S. Y. (1995). Role of glycogen synthase kinase 3ss as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc. Natl. Acad. Sci. USA 92,8498 -8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupnik, C. A., Davidson, N. and Lester, H. A. (1995). The inward rectifier potassium channel family. Curr. Opin. Neurobiol. 5, 268-277. [DOI] [PubMed] [Google Scholar]
- Doyle, D. A., Morais Cabral, J., Pfuetzner, R. A., Kuo, A., Gulbis, J. M., Cohen, S. L., Chait, B. T. and MacKinnon, R. (1998). The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69-77. [DOI] [PubMed] [Google Scholar]
- Dunn, C. W., Hejnol, A., Matus, D. Q., Pang, K., Browne, W. E., Smith, S. A., Seaver, E., Rouse, G. W., Obst, M. and Edgecombe, G. D. (2008). Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5,113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, G. R. and Leys, S. P. (2007). Coordinated contractions effectively expel water from the aquiferous system of a freshwater sponge. J. Exp. Biol. 210,3736 -3748. [DOI] [PubMed] [Google Scholar]
- Ellwanger, K. and Nickel, M. (2006). Neuroactive substances specifically modulate rhythmic body contractions in the nerveless metazoon Tethya wilhelma (Demospongiae, Porifera). Front. Zool. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakler, B., Brandle, U., Glowatzki, E., Zenner, H. P. and Ruppersberg, J. P. (1994). Kir2.1 inward rectifier K+ channels are regulated independently by protein kinases and ATP hydrolysis. Neuron 13,1413 -1420. [DOI] [PubMed] [Google Scholar]
- Hille, B. (2001). Potassium channels and chloride channels. In Ion Channels of Excitable Membranes, pp. 143. Sunderland, MA: Sinauer Associates.
- Jiang, Y., Lee, A., Chen, J., Cadene, M., Chait, B. T. and MacKinnon, R. (2002). The open pore conformation of potassium channels. Nature 417,523 -526. [DOI] [PubMed] [Google Scholar]
- Jin, W. and Lu, Z. (1999). Synthesis of a stable form of tertiapin: a high-affinity inhibitor for inward-rectifier K+ channels. Biochemistry 38,14286 -14293. [DOI] [PubMed] [Google Scholar]
- Krapivinsky, G., Medina, I., Eng, L., Krapivinsky, L., Yang, Y. and Clapham, D. E. (1998). A novel inward rectifier K+ channel with unique pore properties. Neuron 20,995 -1005. [DOI] [PubMed] [Google Scholar]
- Kubo, Y., Adelman, J. P., Clapham, D. E., Jan, L. Y., Karschin, A., Kurachi, Y., Lazdunski, M., Nichols, C. G., Seino, S. and Vandenberg, C. A. (2005). International Union of Pharmacology. LIV. Nomenclature and molecular relationships of inwardly rectifying potassium channels. Pharmacol. Rev. 57,509 -526. [DOI] [PubMed] [Google Scholar]
- Kuo, A., Gulbis, J. M., Antcliff, J. F., Rahman, T., Lowe, E. D., Zimmer, J., Cuthbertson, J., Ashcroft, F. M., Ezaki, T. and Doyle, D. A. (2003). Crystal structure of the potassium channel KirBac1.1 in the closed state. Science 300,1922 -1926. [DOI] [PubMed] [Google Scholar]
- Leys, S. P. and Mackie, G. O. (1997). Electrical recording from a glass sponge. Nature 387, 29-30. [Google Scholar]
- Leys, S. P. and Eerkes-Medrano, D. I. (2006). Feeding in a calcareous sponge: particle uptake by pseudopodia. Biol. Bull. 211,157 -171. [DOI] [PubMed] [Google Scholar]
- Leys, S. P., Mackie, G. O. and Meech, R. W. (1999). Impulse conduction in a sponge. J. Exp. Biol. 202,1139 -1150. [DOI] [PubMed] [Google Scholar]
- Litchfield, C. and Morales, R. W. (1976). Are Demospongiae membranes unique among living organisms? In Aspects of Sponge Biology (ed. W. Harrison and C. Cowden), pp.183 -200. New York: Academic Press.
- Lu, Z. (2004). Mechanism of rectification in inward-rectifier K+ channels. Annu. Rev. Physiol. 66,103 -129. [DOI] [PubMed] [Google Scholar]
- Maddison, D. R. and Maddison, W. P. (2003).MacClade 4: analysis of phylogeny and character evolution , version 4.06. Sunderland, MA: Sinauer Associates.
- Muller, W. E., Kruse, M., Koziol, C., Muller, J. M. and Leys, S. P. (1998). Evolution of early Metazoa: phylogenetic status of the Hexactinellida within the Phylum of Porifera (Sponges). Prog. Mol. Subcell. Biol. 21,141 -156. [DOI] [PubMed] [Google Scholar]
- Nickel, M. (2004). Kinetics and rhythm of body contractions in the sponge Tethya wilhelma (Porifera: Demospongiae). J. Exp. Biol. 207,4515 -4524. [DOI] [PubMed] [Google Scholar]
- Nishida, M., Cadene, M., Chait, B. T. and MacKinnon, R. (2007). Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 26,4005 -4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, M. and MacKinnon, R. (2002). Structural basis of inward rectification: cytoplasmic pore of the G protein-gated inward rectifier GIRK1 at 1.8 A resolution. Cell 111,957 -965. [DOI] [PubMed] [Google Scholar]
- Okamura, Y., Nishino, A., Murata, Y., Nakajo, K., Iwasaki, H., Ohtsuka, Y., Tanaka-Kunishima, M., Takahashi, N., Hara, Y., Yoshida, T. et al. (2005). Comprehensive analysis of the Ascidian genome reveals novel insights into the molecular evolution of ion channel genes. Physiol. Genomics 22,269 -282. [DOI] [PubMed] [Google Scholar]
- Oliver, D., Baukrowitz, T. and Fakler, B. (2000). Polyamines as gating molecules of inward-rectifier K+ channels. Eur. J. Biochem. 267,5824 -5829. [DOI] [PubMed] [Google Scholar]
- Polidoros, A. N., Pasentsis, K. and Tsaftaris, A. S. (2006). Rolling circle amplification-RACE: a method for simultaneous isolation of 5′ and 3′ cDNA ends from amplified cDNA templates. BioTechniques 41, 35-36; 38; 40 passim. [DOI] [PubMed] [Google Scholar]
- Ramu, Y., Klem, A. M. and Lu, Z. (2004). Short variable sequence acquired in evolution enables selective inhibition of various inward-rectifier K+ channels. Biochemistry 43,10701 -10709. [DOI] [PubMed] [Google Scholar]
- Ronquist, F. and Huelsenbeck, J. P. (2003). MrBayes 3, Bayesian phylogenetic inference under mixed models. Bioinformatics 19,1572 -1574. [DOI] [PubMed] [Google Scholar]
- Sakarya, O., Armstrong, K. A., Adamska, M., Adamski, M., Wang, I. F., Tidor, B., Degnan, B. M., Oakley, T. H. and Kosik, K. S. (2007). A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE 2,e506 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, D., Kiesecker, C., Kulzer, M., Gunth, M., Scholz, E. P., Kathofer, S., Thomas, D., Maurer, M., Kreuzer, J., Bauer, A. et al. (2007). Activation of inwardly rectifying Kir2.x potassium channels by β3-adrenoceptors is mediated via different signaling pathways with a predominant role of PKC for Kir2.1 and of PKA for Kir2.2. Naunyn Schmiedebergs Arch. Pharmacol. 375,311 -322. [DOI] [PubMed] [Google Scholar]
- Sperling, E. A., Pisani, D. and Peterson, K. J. (2007). Poriferan paraphyly and its implications for precambrian palaeobiology. Geol. Soc. London 286, 355. [Google Scholar]
- Srivastava, M., Begovic, E., Chapman, J., Putnam, N. H., Hellsten, U., Kawashima, T., Kuo, A., Mitros, T., Salamov, A. and Carpenter, M. L. (2008). The Trichoplax genome and the nature of Placozoans. Nature 454, 955. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688. [DOI] [PubMed] [Google Scholar]
- Tanaka-Kunishima, M., Ishida, Y., Takahashi, K., Honda, M. and Oonuma, T. (2007). Ancient intron insertion sites and palindromic genomic duplication evolutionally shapes an elementally functioning membrane protein family. BMC Evol. Biol. 7, 1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitron, E., Kiesecker, C., Luck, S., Kathofer, S., Thomas, D., Kreye, V. A., Kiehn, J., Katus, H. A., Schoels, W. and Karle, C. A. (2004). Human cardiac inwardly rectifying current IKir2.2 is upregulated by activation of protein kinase A. Cardiovasc. Res. 63,520 -527. [DOI] [PubMed] [Google Scholar]
- Zocchi, E., Carpaneto, A., Cerrano, C., Bavestrello, G., Giovine, M., Bruzzone, S., Guida, L., Franco, L. and Usai, C. (2001). The temperature-signaling cascade in sponges involves a heat-gated cation channel, abscisic acid, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 98, 14859. [DOI] [PMC free article] [PubMed] [Google Scholar]