Abstract
Background & Aims
Most targeted therapies against cancer are designed to block growth factor–stimulated oncogenic growth. However, response rates are low, and resistance to therapy is high. One mechanism might relate to the ability of tumor cells to induce growth factor–independent proliferation (GFIP). This project aims to understand how (1) cancer cells preferentially derive a major growth advantage by using critical metabolic products of glucose, such as phosphoenolpyruvate (PEP), to drive proliferation and (2) esophageal squamous cell carcinoma (ESCC) cells, but not esophageal adenocarcinoma cells, can induce GFIP by using glycolysis to activate phosphohistidine (poHis)-mediated signaling through focal adhesion kinase (FAK).
Methods
The hypothesis to be tested is that ESCC GFIP induced by glucose is facilitated by PEP-mediated histidine phosphorylation (poHis) of FAK, leading to the possibility that ESCC progression can be targeted by blocking poHis signaling. Biochemical, molecular biological, and in vivo experiments including bromodeoxyuridine/5-ethynyl-2′-deoxyuridine labeling, radioisotope tracing, CRISPR gene editing, and analysis of signaling gene sets in human cancer tissues and xenograft models were performed to define the mechanisms underlying ESCC GFIP.
Results
Glucose promotes growth factor–independent DNA replication and accumulation of PEP in ESCC cells. PEP is the direct phospho-donor to poHis58-FAK within a known “HG” motif for histidine phosphorylation. Glucose-induced poHis58 promotes growth factor–independent FAK-mediated proliferation. Furthermore, glucose activates phosphatidylinositol-3′-kinase/AKT via poHis58-FAK signaling. Non-phosphorylatable His58A-FAK reduces xenograft growth.
Conclusions
Glucose induces ESCC, but not esophageal adenocarcinoma GFIP via PEP-His58-FAK-AKT signaling. ESCC progression is controlled by actionable growth factor–independent, glucose-induced pathways that regulate proliferation through novel histidine phosphorylation of FAK.
Keywords: Esophageal Cancer, Glucose Metabolism, Growth Factors, Cell Adhesion Molecules, Cell Signaling
Abbreviations used in this paper: Ab, antibody; ANOVA, analysis of variance; ATP, adenosine triphosphate; BrdU, bromodeoxyuridine; DTT, dithiothreitol; EAC, esophageal adenocarcinoma cell; EdU, 5-ethynyl-2′-deoxyuridine; ELISA, enzyme-linked immunosorbent assay; ESCC, esophageal squamous cell carcinoma; FAK, focal adhesion kinase; FBS, fetal bovine serum; GFIP, growth factor–independent proliferation; GSEA, gene set enrichment analysis; IGF-1, insulin-like growth factor-1; LC, liquid chromatography; MS, mass spectrometry; PBS, phosphate-buffered saline; PEP, phosphoenolpyruvate; PI3K, phosphatidylinositol-3′-kinase; poHis, phosphohistidine; SD, standard deviation; TCGA, The Cancer Genome Atlas
Graphical abstract
Summary.
This research addresses how tumor cells utilize glucose as a proliferative hormone to induce growth factor–independent signaling via phosphoenolpyruvate-mediated histidine phosphorylation of focal adhesion kinase. Targeting phosphohistidine signaling may prevent tumor progression in patients who are resistant to growth factor–targeted therapy.
Most anti-neoplastic targeted therapies are designed to block growth factor–stimulated tumor proliferation. Studies from our lab and from others indicate that cancer cells can continue to grow in the presence of glucose under growth factor–deficient conditions.1, 2 It has long been known that glucose availability controls microorganism growth and that cancer cells metabolize large quantities of glucose through enhanced glycolysis as their energy and carbon source (the Warburg effect).3 The role of glucose in directly supporting cancer cell proliferation through growth factor–independent, phosphoenolpyruvate (PEP)-induced histidine signaling pathways is reported here.
Esophageal squamous cell carcinoma (ESCC) has a strong phenotype of aerobic glycolysis, a finding that is used radiologically to identify metastatic disease with glucose-based probes such as 18F-fluorodeoxyglucose.4 In addition, ESCCs exhibit high frequencies of genomic amplifications of the oncogenic drivers, CCND1, SOX2, and TP63, which are also associated with enhanced glycolysis and ESCC progression.5, 6, 7 Tumor cells reprogram glucose metabolism from mitochondrial respiration to glycolysis,1 which is akin to a process often occurring in microorganisms where glucose is used as a less efficient metabolite to supply biosynthetic material and energy. However, in human cancer cells, isotope tracing studies show that amino acids, but not glucose, account for the majority of cancer cell biomass.8 Therefore, the high rates of glucose consumption in human cancer cells support their rapid cell division in excess of supplying carbon sources or energy.
Elevation of glycolytic intermediates such as PEP is a common feature of tumor cells.9 PEP, but not adenosine triphosphate (ATP), can transfer its high-energy phosphoryl group to histidine residues in the bacterial glucose transport system10 and to histidine 11 of the glycolytic enzyme phosphoglycerate mutase in pyruvate kinase 2–expressing tumor cells.9 PEP-mediated phosphorylation of proteins regulates carbohydrate metabolism in response to nutrient availability in bacteria.11 Phosphohistidine (poHis) is a key factor within the signaling cascade that modulates glucose metabolism and proliferation of prokaryotes12 and mammalian cells.9, 13, 14
Focal adhesion kinase (FAK) plays a critical role in glucose-stimulated phosphatidylinositol-3′-kinase (PI3K)-AKT-insulin signaling,15 insulin/insulin-like growth factor 1 (IGF-1) stimulation of glucose metabolism, growth factor–independent survival,1 RTK-induced proliferation, and integrin-stimulated survival.16 On stimulation by growth factors such as insulin and IGF-1, cells activate PI3K/AKT/MTOR signaling via tyrosine kinase signaling. FAK can cross-talk to insulin-independent signaling to promote glucose metabolism.1, 17 FAK activation and up-regulation are associated with uncontrolled proliferation.16, 18 Furthermore, FAK activation has been associated with promotion of ESCC metastases.19
On the basis of data showing that ESCC, but not esophageal adenocarcinoma cancer (EAC), cells exhibited glucose-induced, growth factor–independent proliferation (GFIP), we searched for post-translational modifications/activations stimulated by glucose only. Here, we show that glucose-induced poHis58-FAK mediates GFIP in ESCCs, likely through activation of PI3K/AKT signaling and through a phospho-transfer from the glycolysis intermediate, PEP. Our novel data suggest that ESCCs, but not EAC, have developed this pathway to facilitate GFIP and to escape therapy that targets growth factor signaling. This model identifies potentially new avenues for targeting ESCCs based on antagonizing glucose-mediated pathways.
Results
Glucose Stimulates Esophageal Squamous Cell Carcinoma Growth in the Absence of Growth Factors
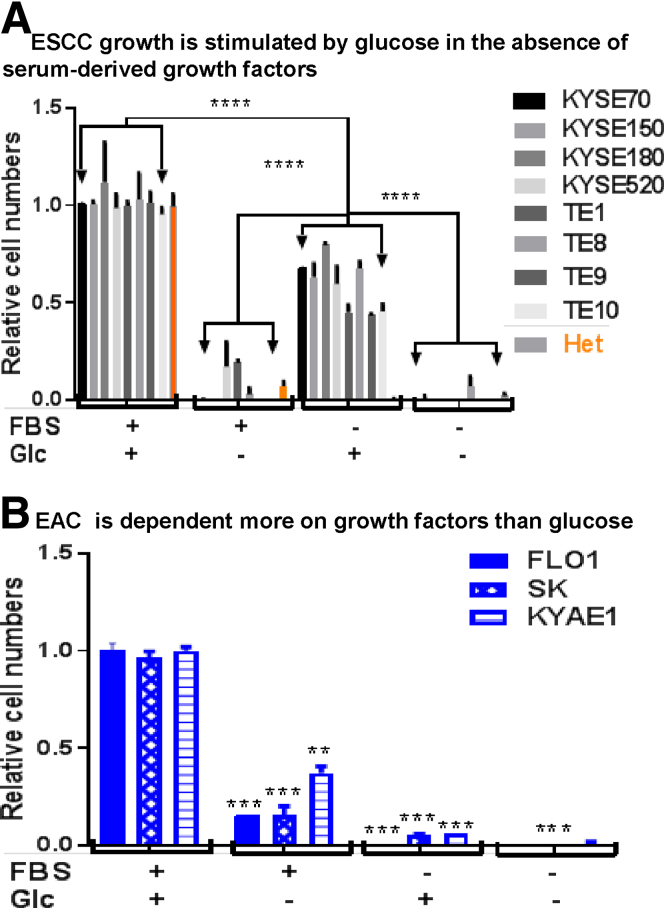
Human ESCCs exhibit one of the highest frequencies of genetic mutations, yet these include few, if any, actionable targets. In fact, no targeted therapy is currently approved for the treatment of ESCC.20 In contrast, clinical radiolabeling of ESCC is facilitated by glucose-based probes, suggesting that tumor metabolism in these tumors might be driven by mediators of the glycolytic pathway. To determine the differential dependence of ESCC on classic growth factor–mediated mitogenic pathways vs those driven by glucose, we cultured cells in serum-free conditions and then induced mitogenesis during a period of 8 days by addition of serum (5% fetal bovine serum [FBS]) and/or glucose (1 mg/mL or 5.56 mmol/L). Compared with [FBS + glucose] conditions, serum alone was a poor mitogen for all ESCC cell lines, whereas glucose alone induced strong proliferation (45%–80% of levels induced by FBS + glucose) (Figure 1A). In contrast, although serum alone could induce proliferation of EAC cell lines, glucose alone was insufficient as a mitogen (Figure 1B). Nonetheless, the finding that EAC proliferation induced by serum was 3- to 5-fold less than under [FBS + glucose] conditions suggests that glucose plays a synergistic role with growth factors.
Figure 1.
Glucose promotes ESCC cell growth in the absence of serum. (A) Cell numbers were counted for untransformed esophageal epithelial (Het) and ESCC cells (KYSE70, KYSE150, KYSE180, KYSE520, TE1, TE8, TE9, and TE10) ± glucose (5.56 mmol/L) stimulation in the presence or absence of FBS (5%) for 8 days. Cell numbers were normalized to the number with glucose and FBS. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, ****P < .0001 vs Glc without FBS. (B) Cell numbers were counted for EAC (FLO1, SK, and KYAE1) ± glucose (5.56 mmol/L) stimulation in the presence or absence of FBS (5%) for 8 days. Relative levels of cell numbers were calculated and shown. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, **P < .01, ***P < .001, ****P < .0001 vs Glc with FBS. Glc, glucose.
Most malignancies consume excessive glucose, and many become addicted to glucose for their uncontrolled growth.21 To determine whether ESCC proliferation is highly glucose-dependent and thus potentially targetable therapeutically, we modified a common protocol for growth factor stimulation studies by depleting glucose for short periods of time (4 hours) in the presence of 5% serum, followed by treatment with glucose (5.56 mmol/L) for 1 hour in the absence of serum. The conditions for assessments of glucose-stimulated cell proliferation are based on the observation that glucose depletion for more than 3 hours followed by glucose addition (5.56 mmol/L) for more than 15 minutes induces new DNA synthesis, as measured by bromodeoxyuridine (BrdU) incorporation (Figure 2A and B). Indeed, ESCC proliferation (BrdU incorporation over background) could be induced by glucose in a dose-dependent manner, whereas EAC or esophageal epithelial cells (Het) were not induced to proliferate under these conditions (Figure 2C). We also showed that short-term (4-hour) glucose treatment in the absence of serum could induce DNA synthesis in a panel of ESCC lines, as measured by 5-ethynyl-2′-deoxyuridine (EdU)–coupled fluorescence flow cytometry (Figure 2D). The ratios of BrdU incorporation induced by glucose over serum-starved conditions were used subsequently as an indicator of glucose-dependent GFIP. For example, glucose alone induced a 2.3-fold increase in the proliferation index of the ESCC line KYSE70, in comparison with no significant increase in Het or the EAC cell line FLO1 (Figure 2E). Furthermore, we examined whether insulin, a known growth factor that stimulates glucose uptake, could supplement glucose-induced proliferation. An insulin responsive breast cancer cell line (MCF7) was used as a positive control. As expected, insulin (5 μg/mL) stimulated MCF7 cell growth in the absence or presence of 5.56 mmol/L glucose (Figure 2F). However, at the concentrations of glucose used (5.56 mmol/L), insulin (5 μg/mL) did not enhance glucose stimulation of ESCC growth (Figure 2F), although it cannot be excluded that it may have effects on more limited glucose concentrations. However, it is important to note that under these conditions, activation of insulin receptor-induced mitogenic pathways was insufficient to augment the ability of glucose to act as an ESCC mitogen, strengthening the notion that glucose can drive ESCC proliferation in a growth factor–independent fashion.
Figure 2.
Glucose stimulates ESCC cell proliferation in the absence of growth factors. (A) Time course of glucose depletion before glucose-induced (5.56 mmol/L) BrdU-DNA. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; one-way ANOVA test, *P < .05, **P < .01, ****P < .0001 vs 0 hour. (B) Glucose increases BrdU-DNA levels in a time-dependent manner. Data are averages with SD from 4 biological replicates; 3 independent experiments that showed similar results; one-way ANOVA test, ***P < .001, ****P < .0001 vs 0 hour. (C) ELISA of BrdU-DNA levels in ESCC (KYSE70, KYSE180, and TE10) and EAC (FLO1, SK, and KYAE1). Cells were preincubated in glucose-free medium containing 5% FBS for 4 hours and then treated with 0–10 mmol/L glucose for 1 hour in the presence of BrdU under serum-deficient conditions. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, ****P < .0001 vs Het. (D) Glucose (5.56 mmol/L) increases EdU-DNA levels in ESCC. EdU coupling fluorescence flow cytometry analysis of EdU-DNA in 5 ESCC cell lines. (E) Proliferative index of glucose. Cell numbers were counted for untransformed esophageal epithelial cells (Het), ESCC (KYSE70), and EAC (FLO1) ± glucose (5.56 mmol/L) stimulation in the absence of serum (FBS)-derived growth factors. Ratios of cell numbers with and without glucose stimulation were calculated and shown. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; one-way ANOVA test, *P < .05 vs Het. (F) WST1 assay of an insulin responsive breast cancer cell line (MCF7) and ESCC cells (KYSE70, KYSE140, TE1, and TE7) ± glucose (5.56 mmol/L) stimulation in the presence or absence of insulin. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; one-way ANOVA test, ****P < .0001 vs Glc alone, Ins alone, or no Glc/Ins. Glc, glucose; Ins, insulin.
Glucose Increases Glycolysis
We sought to determine whether glucose stimulation of ESCC proliferation was mediated through increases in glycolytic pathways. 2-NBDG, a cell-permeable glucose analog that cannot be metabolized via glycolysis, did not induce DNA synthesis in ESCC, whereas glucose did (Figure 3A). Moreover, glucose induced glycolysis in ESCC under serum-free conditions in a dose-dependent manner (Figure 3B) and at 2.5- to 5.2-fold increased levels over those in EAC cell lines (Figure 3C). These results strongly suggest that glucose-increased glycolysis contributes to ESCC proliferation.
Figure 3.
Glucose enhances glycolysis, which is increased in ESCC vs EAC, and glucose-induced proliferation is not mediated through cell viability, redox state, and carbon/energy requirements. (A) EdU-coupled fluorescence flow cytometry of BrdU-DNA in KYSE70 cells stimulated by glucose (Glc) (5.56 mmol/L) or 2-NBDG in the absence of serum. (B) Seahorse analyzer measurement of extracellular acidification rate (ECAR) of KYSE70 cells stimulated by 0–40 mmol/L glucose in the absence of serum. (C) Seahorse assay of glucose-induced (5.56 mmol/L) glycolysis in ESCC and EAC cells in the absence of serum-derived growth factors. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; one-way ANOVA test, ****P < .001 vs ESCC. (D) Glucose repletion promotes DNA synthesis of KYSE70 cells without affecting viability. Simultaneous assessments of viability (WST1) and BrdU-DNA levels in KYSE70 cells that were stimulated by 0–10 mmol/L glucose. (E) WST1 assay of KYSE70, KYSE180, TE10, SK, FLO1, KYAE1, and normal (Het) cells. After glucose depletion, cells were kept in FBS-free medium containing 0–10 mmol/L glucose for 1 hour. (F) Glucose repletion does not alter intracellular ATP levels. Intracellular ATP levels in glucose-stimulated ESCC (KYSE70, TE1) and EAC (FLO1) cells were assessed. (G) Glucose depletion does not alter reactive oxygen species (ROS) levels up to 24 hours and viability up to 12 hours. WST1 and 2′,7′–dichlorofluorescin diacetate (DCFDA) analysis of cell viability and ROS levels in glucose-depleted KYSE70 cells for 0–24 hours in the presence of FBS (5%). Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, **P < .01 vs controls (0 hour). (H) ATP, alternative carbon source, and the effects of an antioxidant do not mimic glucose-induced proliferation. Effects of ATP, alternative carbon (Glutamine, Glut), and an antioxidant (N-acetyl-cysteine, NAC) on proliferation. Data are averages with SD from 3–6 biological replicates; 3 independent experiments that showed similar results; one-way ANOVA test, ****P < .0001 vs vehicle (no FBS, no Glc). Glc, glucose.
Even though we show that glucose increased levels of cell numbers (Figure 1), DNA synthesis (Figure 2), and glycolysis (Figure 3) in ESCC, it is possible that glucose-induced cell numbers are also influenced by increased cell viability. To address this, we simultaneously measured cell viability (WST1) and cell proliferation (BrdU) in KYSE70 cells stimulated with increasing concentrations of glucose. Figure 3D shows that glucose primarily stimulated DNA synthesis and did not serve to rescue cell viability. In addition, glucose repletion did not affect normal or esophageal cancer cell viability (Figure 3E). Alternatively, glucose’s mitogenic role in ESCC might be through its role as a carbon or energy source. However, we noted that the median effective glucose concentrations needed to induce proliferation (27 μmol/L; Figure 3D) are at least ∼150-fold lower than normal blood glucose levels (4.4–10 mmol/L),22 strongly suggesting that glucose functions as a signaling molecule rather than a carbon source under these conditions. Furthermore, increased glucose levels did not alter the levels of intracellular ATP (Figure 3F). Finally, glucose might induce proliferation by promoting a favorable redox status.23 However, reactive oxygen species levels were not altered by glucose depletion for up to 24 hours, although cell viability started to wane between 12 and 24 hours (Figure 3G). Moreover, glucose-promoted proliferation could not be mimicked by the addition of ATP (which is taken up by the cells),24 glutamine as an alternate carbon donor, or the antioxidant N-acetyl-cysteine (Figure 3H). Figure 1A shows total relative cell numbers in the presence of FBS alone (no glucose) for 8 days, whereas Figure 3H shows the relative BrdU-DNA or newly synthesized DNA levels induced by FBS alone (no glucose) for 1 hour. The different effects of FBS alone on ESCC cells suggested that (1) prolonged (8 days) but not brief (1 hour) culture of the cells in media with FBS alone could cause cell death, and therefore, the relative cell numbers could be the combined effects of cell death (loss) and proliferation (gain); and (2) the data suggest that FBS (growth factors) could initiate the entry into S phase (the high BrdU-DNA levels) but could not complete the cell cycle (low relative cell numbers) in the absence of glucose. Taken together, these data demonstrate that glucose-stimulated proliferation is not mediated through effects on cell viability, redox state, or carbon/energy requirements.
Glucose Induces Phosphoenolpyruvate Accumulation and Histidine Phosphorylation of Focal Adhesion Kinase
Metabolic flux studies using 13C-glucose isotope tracing and mass spectrometry (MS) analysis indicate that enhanced glycolysis in tumor cells correlates with the accumulation of glycolytic intermediates including PEP.9 Indeed, glucose treatment shown to induce ESCC proliferation in the absence of serum (Figure 1A) was associated with increased relative levels of intracellular PEP in KYSE70 cells (Figure 4A). Moreover, secreted PEP has been shown to exogenously stimulate proliferation in bacterial and mammalian cells.25, 26 Thus, we treated KYSE70 with increasing concentrations of exogenous PEP in serum-free conditions and showed that PEP induces DNA synthesis (BrdU incorporation over controls) at a median effective concentration of 6 μmol/L but no change in viability (WST1) (Figure 4B).
Figure 4.
Glucose induces PEP accumulation and histidine phosphorylation of FAK. (A) Glucose increases PEP levels. Intracellular PEP levels in KYSE70 cells ± glucose stimulation (5.56 mmol/L). Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; t test, *P < .05 vs cells kept in medium without glucose. (B) PEP stimulates DNA synthesis without reducing viability. BrdU-DNA and WST1 in KYSE70 cells stimulated by 0–1 mmol/L PEP. (C) Glucose induces poHis proteins in KYSE70 cells in a dose-dependent manner. Immunoblot of poHis proteins in glucose-stimulated KYSE70 cells. After glucose depletion for 4 hours, KYSE70 cells were stimulated with 0–1 mmol/L glucose for 1 hour and harvested. A specific anti-poHis Ab was used to detect poHis proteins in the cell lysates. (D) poHis proteins were heat and acid labile. Immunoblot of poHis proteins derived from heat- or acid-treated lysates of KYSE70 cells exposed to 5.56 mmol/L glucose. Lanes 2 and 3 were treated with low pH buffer (acid) or heating to decompose poHis. (E) poHis proteins in FAK KO KYSE70 cells. Immunoblot of poHis proteins in WT and FAK KO KYSE70 cells exposed to 5.56 mmol/L glucose. (F) Immunoprecipitation/Immunoblot of poHis-FAK using anti-FAK Ab to pulldown. The anti-poHis antibody was used to detect poHis-FAK on the polyvinylidene difluoride membrane. The same membrane was stripped and then reprobed with the anti-FAK Ab. (G) Immunoprecipitation/Immunoblot of poHis-FAK using anti-poHis Ab to pulldown. KYSE70 cells were stimulated with glucose (5.56 mmol/L), and lysates were subjected to Immunoprecipitation using the anti-poHis Ab and Immunoblot using the anti-FAK Ab. The membrane was exposed to x-ray film for a short (upper panel) and long (lower panel) period of time. (H) PEP mimics glucose-induced poHis-FAK. KYSE70 and TE9 cells were stimulated with or without glucose (Glc) (5.56 mmol/L) or PEP (5 mmol/L) for 1 hour. IB of poHis-FAK and total FAK was performed. The same membrane was stripped and then reprobed using an anti-actin Ab.
PEP is known to act as the phosphor-donor for the histidine phosphorylation of proteins that up-regulate the expression of proliferative and survival genes in bacteria and rapidly growing cells.9, 12, 14 On the notion that high glucose-induced PEP levels in ESCC might play a similar role, we blotted ESCC lysates with a poHis-specific antibody (Ab), identifying a dose-dependent glucose-induced band at roughly 125 kDa (Figure 4C). This protein was acid- and heat-labile (Figure 4D), a typical property of poHis. FAK has a similar molecular weight and is known to play a critical role in glucose-stimulated PI3K-AKT-insulin signaling,15 GFIP, and survival.1 To address whether the glucose-induced poHis band is FAK, the PTK2 (FAK) gene was disrupted by CRISPR Cas9 in KYSE70 cells, such that its loss correlated with the loss of the poHis–125 kDa band (Figure 4E). The notion that the poHis–125 kDa band was FAK was based on our finding that poHis-Ab reactivity in FAK immunoprecipitates occurred only in glucose-treated cells (Figure 4F, upper panel), even though equal amounts of FAK were found in the immunoprecipitates (Figure 4F, bottom panel). In parallel, poHis immunoprecipitates blotted with FAK Ab only identified the 125 kDa band in glucose-treated cells (Figure 4G). In addition, treatment of ESCC cells, KYSE70 and TE9, with PEP mimicked the ability of glucose to induce the poHis-FAK band (Figure 4H). These results indicate that glucose or PEP likely induces FAK poHis.
Phosphoenolpyruvate Transfers Its Phosphoryl Group to Focal Adhesion Kinase
Several approaches were pursued to confirm that PEP serves as the phospho-donor for FAK in ESCC. First, PEP, but not ATP or pyruvate, induced histidine phosphorylation of recombinant FAK (Figure 5A). We also considered the possibility that the anti-poHis Ab might have weak cross-reactivity with phosphotyrosine, which is known to be induced in FAK on mitogen stimulation.27 To rule out this possibility, phosphotyrosine-linked bovine serum albumin was added to the poHis Ab in our enzyme-linked immunosorbent assay (ELISA), and indeed, addition of poY–bovine serum albumin had no effect on the anti-poHis Ab detection of poHis-FAK levels in varied amounts of ESCC lysate (Figure 5B). In contrast, acidification of ESCC lysates to pH 0.7 ablated detection of the poHis-FAK in ELISA (Figure 5B). Second, addition of active human poHis phosphatase to the PEP/rFAK assay decreased poHis-FAK levels, whereas treatment with heat-inactivated poHis phosphatase did not (Figure 5C). Finally, the ability of 32P-labelled PEP to donate its 32phosphoryl group to purified FAK could be reduced by unlabeled PEP but not by ATP or pyruvate (Figure 5D). In addition, the labeling of FAK by 32P-PEP was dose-dependent (Figure 5E). Taken together, these data strongly suggest that glucose-induced PEP induces a direct histidine phosphorylation of FAK in ESCC.
Figure 5.
PEP donates its phosphoryl group for histidine phosphorylation of FAK. (A) PEP increases poHis-FAK levels. ELISA of poHis-FAK levels after recombinant FAK was incubated with 0–50 mmol/L PEP or controls (ATP, pyruvate). Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, ****P < .0001 vs controls (ATP or pyruvate). (B) The anti-poHis Ab specifically detects poHis but not poTyrosine. ELISA of poHis-FAK in acid-treated lysates (poHis-Low pH). Phospho-tyrosine bovine serum albumin (poY-BSA) was added to the poHis antibody buffer to block its cross-activity to poTyrosine (poY). Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, ****P < .0001 vs control (poHis-Low pH). (C) Human poHis phosphatase treatment. Human poHis phosphatase (PHPT) removal of phosphoryl groups in rFAK and ELISA of poHis on rFAK. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; one-way ANOVA test, **P < .01, and ***P < .001 vs PHPT-treated samples. (D) PEP donates phosphate directly to FAK. Treatment of purified FAK with 32P-PEP, separation of 32P-FAK from unreacted 32P-PEP, and scintillation counting of 32P-poHis-FAK. +PEP/+ATP/+Py: 32P-PEP treatment with excessive cold PEP, ATP, or pyruvate. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; one-way ANOVA test, *P < .05 vs 32P-PEP treated rFAK. (E) Dose response with 32P-PEP. rFAK was incubated with varied concentrations of 32P-PEP. Data are averages with SD from 4 biological replicates; 3 independent experiments that showed similar results; one-way ANOVA test, ***P < .001 vs control (0 mmol/L 32P-PEP).
Phosphohistidine 58–Focal Adhesion Kinase Is Essential for Glucose-Induced Proliferation
We continued to analyze whether poHis-FAK correlated with glucose-induced proliferation in ESCC. Surgical samples from normal human esophagus vs ESCC cases (n = 6) were analyzed for PEP by using PEP Fluorometric Assay Kit. The PEP levels of the human ESCC tumor samples were higher than those of normal esophagus (Figure 6A). The poHis-FAK levels in these human samples were assessed by using ELISA with the poHis Ab for coating/immobilizing poHis proteins and the FAK Ab for detection of poHis-FAK on the well. The tumor samples had statistically significant higher poHis-FAK levels in human ESCC surgical specimens, compared with matched normal esophagus (Figure 6B). This is consistent with the concept that PEP-induced poHis-FAK may play a role in ESCC progression. In ESCC cell lines such as KYSE70, neither serum nor glucose changed total FAK protein levels, yet glucose strongly induced the poHis-FAK band over the level induced by serum (Figure 6C and D). Moreover, the in vitro histidine phosphorylation level of recombinant N-terminal but not C-terminal FAK fragment was similar to that of the full-length FAK (Figure 6E), suggesting that the major poHis sites were located on the N-terminus of FAK.
Figure 6.
poHis-FAK signaling modulates ESCC proliferation. (A) PEP levels in human normal esophagus (ES) or ESCC tumors. Data are averages with SD from 6 biological replicates (2 samples from each subject); 2 independent experiments that showed similar results; unpaired t test, **P < .01 vs ES. (B) ELISA of poHis-FAK levels in human normal esophagus (ES) or ESCC tumors. Data are averages with SD from 6 biological replicates (2 samples from each subject); 2 independent experiments that showed similar results; unpaired t test, **P < .01 vs ES. (C) Immunoblot of poHis-FAK levels in glucose (Glc) (5.56 mmol/L) or FBS (5%)-stimulated KYSE70 cells. (D) ELISA of poHis-FAK levels in KYSE70 cells ± glucose stimulation (5.56 mmol/L). Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; unpaired t test, ***P < .001 vs –Glc. (E) ELISA of poHis-FAK FERM and CD domains. CD, C-terminal domain of FAK; FERM, four-point-one, ezrin, radixin, moesin domain (N-terminus, 1-128 aa); FL, full-length FAK; GST, green florescence protein. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; ordinary one-way ANOVA test, ***P < .001 vs control (FL). (F) Reported poHis proteins that contain the “HG” motif. (G) ELISA of poHis-FAK levels in FAK deficient SCC cells re-expressing WT/H58A FAK. Data are averages with SD from 8 biological replicates; 4 independent experiments that showed similar results; unpaired t test, **P < .01 vs WT. (H) H58A reduces glucose consumption in FAK KO SCC cells. Amounts of glucose taken up by WT/H58A FAK-transfected mouse FAK KO SCC cells. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; unpaired t test, **P < .01 vs WT control. (I) H58A-altered cell morphology. Microscopy examination of WT/H58A FAK-transfected KYSE70 cells. (J) Numbers of WT/H58A FAK-transfected KYSE70 cells with glucose stimulation (5.56 mmol/L). Immunoblot of the transfected cells to assess the levels of WT/H58A-FAK (inset). Data are averages with SD from 9 biological replicates; 3 independent experiments that showed similar results; ordinary one-way ANOVA test, ***P < .001 vs WT. (K) Relative proliferation of WT/H58E FAK-transfected KYSE70 cells ± glucose stimulation (5.56 mmol/L). Data are averages with SD from 6 biological replicates; 3 independent experiments that showed similar results; ordinary one-way ANOVA test, ****P < .0001 vs both H58E lanes.
To define which poHis site or sites on FAK were responsible for controlling glucose-induced proliferation, we scanned FAK for the “HG” motif shared by many poHis proteins,9, 13, 28 ultimately identifying H58 as a candidate site (Figure 6F). This was in agreement with a report on poHis proteomics in Escherichia coli, in which 50% of identified endogenous poHis proteins contained the HG motif.29 We then analyzed recombinant FAK by using liquid chromatography (LC)/MS–MS and identified enrichment of poHis58 (VFHYFESNSEPTTWASIIRpoH58GDATDVRGII). Indeed, the relative glucose-induced poHis level of ectopically expressed FAKH58A was significantly lower than that of WT-FAK in KYSE70 cells (Figure 6G). We noted that KYSE70 cells stably expressing FAKH58A exhibited decreased glucose consumption and altered cell morphology (small and round shape), compared with those expressing WT-FAK (Figure 6H and I). Glucose consumption was decreased in FAK KO SCC cells re-expressing FAKH58A, indicating that H58A inhibition of poHis-FAK signaling decreased utilization of glucose for biomass synthesis (Figure 6H). Furthermore, individual KYSE70-[FAKH58A] clones showed a correlation between FAKH58A expression levels and decreased proliferation (Figure 6J). The stable expression of the phosphomimetic FAKH58E mimicked glucose-increased proliferation (Figure 6K). Taken together, these data suggest that poHis58-FAK is necessary and sufficient to mediate glucose-induced ESCC proliferation and is consistent with our previous finding that dominant negative down-regulation of FAK attenuates glucose-modulated glycolysis and re-sensitizes cancer cells to cell death after serum removal.1
Glucose Modulates Growth Factor–Independent Focal Adhesion Kinase Function and Esophageal Squamous Cell Carcinoma Proliferation
Growth factor/integrin activation of FAK is mediated through autophosphorylation of Y397.16 To determine whether poY397 is required for glucose-induced poHis58, we mutated Y397 to F397 to prevent Y397 phosphorylation. The stable expression of FAKY397F did not attenuate glucose-increased poHis-FAK levels in KYSE70 cells (Figure 7A). Next, we examined whether FAK kinase activity was required for glucose-induced proliferation in ESCC. Whereas treatment with defactinib inhibited serum-induced proliferation, it had no effect on glucose-induced proliferation (Figure 7B). Moreover, FAKY397F attenuated growth factor–induced but not glucose-induced proliferation (Figure 7C). Furthermore, loss of kinase activity (FAK454R, kinase dead30) only slightly attenuated the ability of FAKHis58E-454R to induce proliferation in the absence of glucose (Figure 7D). These data indicate that poHis58-FAK promotes glucose-stimulated proliferation independent of poY397-mediated FAK kinase activity.
Figure 7.
poHis58 mimics poY397-triggered FAK function in glucose-induced proliferation. (A) Immunoblot of poHis-FAK in WT and Y397F FAK-transfected FAK KO mouse embryonic fibroblasts. (B) Effects of FAK kinase inhibitor (defactinib) on FBS- or glucose (5.56 mmol/L)-increased BrdU-DNA levels in KYSE70 cells. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; ordinary one-way ANOVA test, ****P < .0001 vs Glc/FBS. (C) BrdU-DNA levels in WT FAK or FAKY397F-transfected FAK KO mouse embryonic fibroblasts ± glucose and/or FBS stimulation. FAK-WT: FAK KO mouse embryonic fibroblasts re-expressing WT FAK; and FAK-Y397F: FAK KO mouse embryonic fibroblasts re-expressing FAKY397F. Data are averages with SD from 6 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, ****P < .0001 vs Glc/FBS-WT. (D) BrdU-DNA levels in FAKH58E/A-454R-transfected FAK null mouse embryonic fibroblasts ± glucose stimulation. WT: FAK KO mouse embryonic fibroblasts re-expressing WT FAK; H58A-454R: FAK KO mouse embryonic fibroblasts re-expressing FAKH58A-454R; and H58E-454R: FAK KO mouse embryonic fibroblasts re-expressing FAKH58E-454R. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, **P < .01 vs WT +Glc, ****P < .0001 vs WT –Glc. Glc, glucose.
Glucose Activates Phosphatidylinositol-3′-Kinase–AKT Signaling via Phosphohistidine 58–Focal Adhesion Kinase
Growth factors such as insulin and IGF-1 stimulate PI3K/AKT signaling to promote proliferation in normal cells.31 However, it was unclear what mitogenic pathways are induced by glucose in the absence of growth factors. An unbiased screen of major cancer-associated pathways using a Cancer Signaling Phospho Antibody Array (Full Moon Biosystems, Sunnyvale, CA) identified several activated signaling pathways induced by glucose in the absence of growth factors, including those involving ERK1/2, JNK, and p38 mitogen-activated protein kinases, PI3K-AKT, and JAK1/2. In addition, there was also an increase in the relative phospho-levels of FGFR1, EGFR, and FAKpoY925, the latter a known Src phosphorylation site,32 and corresponding to a small increase in relative SrcpoY418 levels (Table 1). Gene set enrichment analysis (GSEA) of transcriptome data from The Cancer Genome Atlas (TCGA) database revealed an enrichment of PI3K/AKT/mTOR signaling gene set expression in ESCC, compared with the less glucose-dependent EAC tumors (Figure 8A). The key components of the PI3K-AKT cascade, including PIK3CD, PIK3CA, PIK3R2, AKT2, and AKT3 expression, were also significantly increased in ESCC (Figure 8B). Indeed, glucose could induce poHis-FAK and poAKT increases in a dose-dependent manner in KYSE70 (ESCC) (Figure 9A) but not in FLO1 (EAC) cells (Figure 9B) under serum-free conditions. The ectopic expression of FAKH58A attenuated glucose-increased relative poAKT levels in KYSE70 cells (Figure 9C), and short hairpin RNA knockdown of AKT expression (using a clone that recognizes AKT1 and 2 isoforms33) ablated glucose-induced proliferation (Figure 9D). In addition, inhibition of PI3K or AKT by wortmannin or AZD5363, respectively, decreased serum- or glucose-stimulated proliferation (Figure 9E). In contrast, the MEK1 inhibitor, PD98059, did not impact FBS- or glucose-induced proliferation (Figure 9E). In addition, a panel of tyrosine kinase inhibitors known to target insulin/IGF-1R, epidermal growth factor receptor, fibroblast growth factor receptor, platelet-derived growth factor receptor, Abl, vascular endothelial growth factor receptor, c-KIT, and Src failed to ablate glucose-induced ESCC proliferation yet significantly inhibited serum-induced proliferation (Figure 9F). Taken together, these data suggest that activation of the PI3K/AKT pathway facilitates glucose-induced proliferation in ESCC via the phosphorylation of FAK on His58.
Table 1.
Ratio Analysis for Paired Antibodies
| Antibody list | Signal ratio |
Fold change |
||
|---|---|---|---|---|
| without glucose | with glucose | with/without glucose | ||
| 14-3-3 zeta (Phospho-Ser58) | 14-3-3 zeta (Ab-58) | 1.92 | 1.88 | 0.98 |
| 4E-BP1 (Phospho-Thr36) | 4E-BP1 (Ab-36) | 1.57 | 1.54 | 0.98 |
| Akt (Phospho-Thr308) | Akt (Ab-308) | 0.77 | 0.84 | 1.09 |
| Akt (Phospho-Ser473) | Akt (Ab-473) | 0.74 | 0.91 | 1.22 |
| Akt2 (Phospho-Ser474) | Akt2 (Ab-474) | 1.17 | 1.18 | 1.01 |
| AMPK1 (Phospho-Thr174) | AMPK1 (Ab-174) | 1.76 | 1.56 | 0.89 |
| BAD (Phospho-Ser112) | BAD (Ab-112) | 0.86 | 0.98 | 1.15 |
| BAD (Phospho-Ser136) | BAD (Ab-136) | 1.00 | 1.10 | 1.10 |
| BAD (Phospho-Ser155) | BAD (Ab-155) | 1.04 | 1.16 | 1.12 |
| BCL-2 (Phospho-Thr56) | BCL-2 (Ab-56) | 0.85 | 0.91 | 1.07 |
| BCL-2 (Phospho-Ser70) | BCL-2 (Ab-70) | 0.66 | 0.72 | 1.09 |
| BCL-XL (Phospho-Ser62) | BCL-XL (Ab-62) | 0.09 | 0.09 | 1.05 |
| Beta-Catenin (Phospho-Ser37) | Beta-Catenin (Ab-37) | 1.12 | 1.21 | 1.08 |
| Beta-Catenin (Phospho-Thr41/Ser45) | Beta-Catenin (Ab-41/45) | 2.10 | 1.99 | 0.95 |
| BRCA1 (Phospho-Ser1423) | BRCA1 (Ab-1423) | 0.14 | 0.15 | 1.05 |
| BRCA1 (Phospho-Ser1524) | BRCA1 (Ab-1524) | 0.88 | 0.91 | 1.04 |
| CaMKII (Phospho-Thr286) | CaMKII (Ab-286) | 0.84 | 0.96 | 1.14 |
| Caspase 3 (Phospho-Ser150) | Caspase 3 (Ab-150) | 1.12 | 1.16 | 1.03 |
| Caspase 9 (Phospho-Thr125) | Caspase 9 (Ab-125) | 0.61 | 0.65 | 1.06 |
| Caspase 9 (Phospho-Ser144) | Caspase 9 (Ab-144) | 0.80 | 0.79 | 0.99 |
| Caspase 9 (Phospho-Tyr153) | Caspase 9 (Ab-153) | 0.88 | 1.02 | 1.17 |
| Caspase 9 (Phospho-Ser196) | Caspase 9 (Ab-196) | 0.40 | 0.37 | 0.93 |
| Caveol in-1 (Phospho-Tyr14) | Caveol in-1 (Ab-14) | 1.81 | 1.91 | 1.06 |
| CDC2 (Phospho-Tyr15) | CDC2 (Ab-15) | 0.72 | 0.96 | 1.32 |
| cdc25A (Phospho-Ser75) | cdc25A (Ab-75) | 1.36 | 1.56 | 1.15 |
| cdc25C (Phospho-Ser216) | cdc25C (Ab-216) | 0.43 | 0.45 | 1.05 |
| CDK2 (Phospho-Thr160) | CDK2 (Ab-160) | 0.87 | 1.00 | 1.16 |
| Chk1 (Phospho-Ser280) | Chk1 (Ab-280) | 1.28 | 1.41 | 1.10 |
| Chk1 (Phospho-Ser317) | Chk1 (Ab-317) | 0.82 | 0.85 | 1.04 |
| Chk1 (Phospho-Ser345) | Chk1 (Ab-345) | 1.19 | 1.19 | 1.00 |
| Chk2 (Phospho-Ser516) | Chk2 (Ab-516) | 0.97 | 1.15 | 1.19 |
| Chk2 (Phospho-Thr68) | Chk2 (Ab-68) | 0.35 | 0.39 | 1.11 |
| c-Jun (Phospho-Thr239) | c-Jun (Ab-239) | 1.04 | 1.10 | 1.05 |
| c-Jun (Phospho-Ser243) | c-Jun (Ab-243) | 0.45 | 0.51 | 1.14 |
| c-Jun (Phospho-Ser73) | c-Jun (Ab-73) | 0.20 | 0.19 | 0.96 |
| c-Kit (Phospho-Tyr721) | c-Kit (Ab-721) | 0.70 | 0.80 | 1.15 |
| CREB (Phospho-Ser133) | CREB (Ab-133) | 0.64 | 0.76 | 1.18 |
| CrkII (Phospho-Tyr221) | CrkII (Ab-221) | 0.61 | 0.62 | 1.01 |
| eEF2K (Phospho-Ser366) | eEF2K (Ab-366) | 0.67 | 0.67 | 1.01 |
| EGFR (Phospho-Tyr1110) | EGFR (Ab-1110) | 0.70 | 0.86 | 1.23 |
| eIF2a (Phospho-Ser51) | eIF2a (Ab-51) | 0.88 | 0.86 | 0.98 |
| elF4E (Phospho-Ser209) | elF4E (Ab-209) | 0.78 | 0.95 | 1.21 |
| Elk-1 (Phospho-Ser383) | Elk-1 (Ab-383) | 1.09 | 1.01 | 0.93 |
| ERK3 (Phospho-Ser189) | ERK3 (Ab-189) | 1.18 | 1.19 | 1.00 |
| ERK8 (Phospho-Thr175/Tyr177) | ERK8 (Ab-175/177) | 0.57 | 0.58 | 1.02 |
| Estrogen Receptor-alpha (Phospho-Ser167) | Estrogen Receptor-alpha (Ab-167) | 1.14 | 1.13 | 0.99 |
| FAK (Phospho-Tyr397) | FAK (Ab-397) | 1.14 | 1.05 | 0.92 |
| FAK (Phospho-Tyr861) | FAK (Ab-861) | 0.75 | 0.79 | 1.06 |
| FAK (Phospho-Tyr925) | FAK (Ab-925) | 1.03 | 1.23 | 1.20 |
| FGF Receptor 1 (Phospho-Tyr154) | FGF Receptor 1 (Ab-154) | 0.72 | 0.89 | 1.23 |
| FKHR (Phospho-Ser256) | FKHR (Ab-256) | 1.23 | 1.23 | 1.00 |
| GSK3 alpha (Phospho-Ser21) | GSK3 alpha (Ab-21) | 0.99 | 0.92 | 0.93 |
| GSK3 beta (Phospho-Ser9) | GSK3 beta (Ab-9) | 0.66 | 0.71 | 1.08 |
| HDAC8 (Phospho-Ser39) | HDAC8 (Ab-39) | 1.06 | 1.10 | 1.04 |
| HER2 (Phospho-Tyr877) | HER2 (Ab-877) | 1.12 | 1.17 | 1.04 |
| Histone H2A.X (Phospho-Ser139) | Histone H2A.X (Ab-139) | 0.84 | 0.88 | 1.05 |
| HSF1 (Phospho-Ser303) | HSF1 (Ab-303) | 0.75 | 1.05 | 1.40 |
| HSP27 (Phospho-Ser15) | HSP27 (Ab-15) | 1.61 | 1.64 | 1.02 |
| HSP27 (Phospho-Ser78) | HSP27 (Ab-78) | 0.96 | 1.23 | 1.28 |
| HSP90B (Phospho-Ser254) | HSP90B (Ab-254) | 1.17 | 1.48 | 1.26 |
| ICAM-1 (Phospho-Tyr512) | ICAM-1 (Ab-512) | 1.89 | 1.86 | 0.99 |
| IGF-1R (Phospho-Tyr1161) | IGF-1R (Ab-1161) | 1.29 | 1.31 | 1.01 |
| IkB-alpha (Phospho-Ser32/Ser36) | IkB-alpha (Ab-32/36) | 5.36 | 6.36 | 1.19 |
| IkB-alpha (Phospho-Tyr42) | IkB-alpha (Ab-42) | 1.76 | 1.88 | 1.06 |
| IkB-epsi lon (Phospho-Ser22) | IkB-epsi lon (Ab-22) | 0.48 | 0.47 | 0.96 |
| IKK alpha (Phospho-Thr23) | IKK alpha (Ab-23) | 0.64 | 0.77 | 1.21 |
| Integrin beta-3 (Phospho-Tyr773) | Integrin beta-3 (Ab-773) | 0.57 | 0.64 | 1.11 |
| Integrin beta-3 (Phospho-Tyr785) | Integrin beta-3 (Ab-785) | 1.11 | 1.10 | 0.99 |
| JAK1 (Phospho-Tyr1022) | JAK1 (Ab-1022) | 0.95 | 1.28 | 1.35 |
| JAK2 (Phospho-Tyr1007) | JAK2 (Ab-1007) | 0.65 | 0.73 | 1.13 |
| JAK2 (Phospho-Tyr221) | JAK2 (Ab-221) | 0.90 | 1.27 | 1.41 |
| JunB (Phospho-Ser259) | JunB (Ab-259) | 1.75 | 1.77 | 1.01 |
| JunB (Phospho-Ser79) | JunB (Ab-79) | 2.08 | 1.87 | 0.90 |
| JunD (Phospho-Ser255) | JunD (Ab-255) | 1.68 | 1.84 | 1.09 |
| Keratin 18 (Phospho-Ser33) | Keratin 18 (Ab-33) | 2.04 | 2.28 | 1.12 |
| MDM2 (Phospho-Ser166) | MDM2 (Ab-166) | 0.06 | 0.07 | 1.06 |
| MEK1 (Phospho-Ser217) | MEK1 (Ab-217) | 1.91 | 1.95 | 1.02 |
| MEK1 (Phospho-Ser221) | MEK1 (Ab-221) | 0.96 | 1.14 | 1.19 |
| MEK1 (Phospho-Thr291) | MEK1 (Ab-291) | 0.61 | 0.68 | 1.10 |
| MEK2 (Phospho-Thr394) | MEK2 (Ab-394) | 0.98 | 0.97 | 0.98 |
| Met (Phospho-Tyr1349) | Met (Ab-1349) | 0.73 | 0.84 | 1.15 |
| MKK3 (Phospho-Ser189) | MKK3 (Ab-189) | 0.84 | 0.98 | 1.16 |
| MSK1 (Phospho-Ser376) | MSK1 (Ab-376) | 1.82 | 1.97 | 1.08 |
| mTOR (Phospho-Ser2448) | mTOR (Ab-2448) | 1.91 | 1.99 | 1.04 |
| Myc (Phospho-Thr358) | Myc (Ab-358) | 0.24 | 0.28 | 1.17 |
| Myc (Phospho-Ser373) | Myc (Ab-373) | 1.57 | 1.58 | 1.00 |
| Myc (Phospho-Thr58) | Myc (Ab-58) | 1.41 | 1.42 | 1.00 |
| Myc (Phospho-Ser62) | Myc (Ab-62) | 1.73 | 1.57 | 0.91 |
| NFkB-p100/p52 (Phospho-Ser865) | NFkB-p100/p52 (Ab-865) | 0.44 | 0.41 | 0.93 |
| NFkB-p100/p52 (Phospho-Ser869) | NFkB-p100/p52 (Ab-869) | 1.02 | 1.04 | 1.02 |
| NFkB-p105/p50 (Phospho-Ser893) | NFkB-p105/p50 (Ab-893) | 0.88 | 0.93 | 1.06 |
| NFkB-p105/p50 (Phospho-Ser907) | NFkB-p105/p50 (Ab-907) | 0.94 | 0.98 | 1.04 |
| NFkB-p65 (Phospho-Thr254) | NFkB-p65 (Ab-254) | 0.90 | 0.83 | 0.92 |
| NFkB-p65 (Phospho-Ser529) | NFkB-p65 (Ab-529) | 1.53 | 1.61 | 1.05 |
| p21Cip1 (Phospho-Thr145) | p21Cip1 (Ab-145) | 1.19 | 1.25 | 1.06 |
| p27Kip1 (Phospho-Ser10) | p27Kip1 (Ab-10) | 1.39 | 1.42 | 1.02 |
| p27Kip1 (Phospho-Thr187) | p27Kip1 (Ab-187) | 1.25 | 1.15 | 0.92 |
| P38 MAPK (Phospho-Tyr182) | P38 MAPK (Ab-182) | 0.36 | 0.45 | 1.26 |
| p44/42 MAP Kinase (Phospho-Thr202) | p44/42 MAP Kinase (Ab-202) | 0.87 | 1.01 | 1.17 |
| p44/42 MAP Kinase (Phospho-Tyr204) | p44/42 MAP Kinase (Ab-204) | 0.96 | 1.21 | 1.26 |
| p53 (Phospho-Ser315) | p53 (Ab-315) | 0.39 | 0.53 | 1.35 |
| p53 (Phospho-Ser6) | p53 (Ab-6) | 1.30 | 1.42 | 1.09 |
| p70 S6 Kinase (Phospho-Ser424) | p70 S6 Kinase (Ab-424) | 1.62 | 1.41 | 0.87 |
| PDGF Receptor beta (Phospho-Tyr751) | PDGF Receptor beta (Ab-751) | 0.52 | 0.59 | 1.14 |
| PDK1 (Phospho-Ser241) | PDK1 (Ab-241) | 0.52 | 0.36 | 0.70 |
| PI3-kinase p85-alpha (Phospho-Tyr607) | PI3-kinase p85-alpha (Ab-607) | 0.68 | 1.01 | 1.47 |
| PI3-kinase p85-subunit alpha/gamma (Phospho-Tyr467/Tyr199) | PI3-kinase p85-subunit alpha/gamma (Ab-467/199) | 0.98 | 0.06 | 1.08 |
| PTEN (Phospho-Ser380/Thr382/Thr383) | PTEN (Ab-380/382/383) | 0.66 | 0.73 | 1.11 |
| Pyk2 (Phospho-Tyr402) | Pyk2 (Ab-402) | 1.48 | 1.58 | 1.07 |
| Rac1/cdc42 (Phospho-Ser71) | Rac1/cdc42 (Ab-71) | 1.83 | 1.79 | 0.98 |
| Raf1 (Phospho-Ser259) | Raf1 (Ab-259) | 1.01 | 1.02 | 1.01 |
| Rb (Phospho-Ser780) | Rb (Ab-780) | 0.93 | 1.07 | 1.15 |
| Rel (Phospho-Ser503) | Rel (Ab-503) | 1.15 | 1.25 | 1.08 |
| SAPK/JNK (Phospho-Thr183) | SAPK/JNK (Ab-183) | 0.77 | 1.08 | 1.40 |
| Shc (Phospho-Tyr349) | Shc (Ab-349) | 0.62 | 0.74 | 1.20 |
| SHP-2 (Phospho-Tyr580) | SHP-2 (Ab-580) | 1.93 | 2.04 | 1.05 |
| Src (Phospho-Tyr418) | Src (Ab-418) | 0.23 | 0.26 | 1.11 |
| Src (Phospho-Tyr529) | Src (Ab-529) | 1.52 | 1.48 | 0.98 |
| STAT1 (Phospho-Tyr701) | STAT1 (Ab-701) | 1.72 | 1.74 | 1.01 |
| STAT1 (Phospho-Ser727) | STAT1 (Ab-727) | 1.78 | 2.11 | 1.19 |
| STAT3 (Phospho-Tyr705) | STAT3 (Ab-705) | 1.18 | 1.16 | 0.98 |
| STAT3 (Phospho-Ser727) | STAT3 (Ab-727) | 1.56 | 1.54 | 0.99 |
| STAT4 (Phospho-Tyr693) | STAT4 (Ab-693) | 1.71 | 1.71 | 1.00 |
| STAT5A (Phospho-Tyr694) | STAT5A (Ab-694) | 1.62 | 1.46 | 0.90 |
| STAT5A (Phospho-Ser780) | STAT5A (Ab-780) | 0.15 | 0.16 | 1.08 |
| STAT6 (Phospho-Tyr641) | STAT6 (Ab-641) | 1.40 | 1.61 | 1.15 |
| STAT6 (Phospho-Thr645) | STAT6 (Ab-645) | 0.22 | 0.24 | 1.13 |
| Tau (Phospho-Ser404) | Tau (Ab-404) | 1.13 | 1.18 | 1.05 |
| Trk B (Phospho-Tyr515) | Trk B (Ab-515) | 0.32 | 0.37 | 1.13 |
| TYK2 (Phospho-Tyr1054) | TYK2 (Ab-1054) | 0.84 | 0.87 | 1.04 |
| VEGFR2 (Phospho-Tyr951) | VEGFR2 (Ab-951) | 0.19 | 0.21 | 1.08 |
Figure 8.
Glucose activates PI3K/AKT signaling via poHis58-FAK. (A) ESCC enriches PI3K/AKT/mTOR signaling genes. Statistical analysis of hallmark PI3K/AKT/MTOR gene sets in ESCC (n = 95) and EAC (n = 88) samples in TCGA database. (B) Levels of PI3K and AKT mRNA are increased in human ESCC tumors. Statistical analysis of key PI3K and AKT expression in ESCC (n = 95) and EAC (n = 88) samples in the TCGA database. ****P < .0001 vs EAC.
Figure 9.
Glucose activates PI3K/AKT signaling via poHis58-FAK. (A) Glucose increases pAKT levels in ESCC (KYSE70) in a dose-dependent manner. Immunoblot of pAKT/AKT levels in KYSE70 exposed to 0–250 μmol/L glucose in the absence of serum (FBS). (B) Glucose does not alter pAKT levels in EAC (FLO1). Immunoblot of pAKT/AKT levels in FLO1 exposed to 0–250 μmol/L glucose in the absence of serum (FBS). (C) H58A attenuates gluocse-increased pAKT levels. Immunoblot of pAKT and total AKT levels in WT/GFP-H58A-FAK-transfected KYSE70 cells exposed to 5.56 mmol/L glucose. Low/Med/High: transfected cells expressing low/medium/high levels of GFP-H58A-FAK. (D) shAKT attenuates glucose-induced proliferation. shRNA inhibition of AKT expression (shAKT) in KYSE70 cells ± glucose (5.56 mmol/L) stimulation in the absence of serum. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; unpaired t test, **P < .01 vs shAKT (with Glc). (E) PI3K/AKT inhibitors reduce glucose-induced proliferation. Effects of PI3K/AKT/ERK inhibition (PI3Ki/AKTi/ERKi) on serum (FBS, 5%) and glucose (Glc) (5.56 mmol/L) stimulation of BrdU-DNA synthesis. Wort, wortmannin; AZD, AZD5363; PD, PD98059. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, ***P < .001, ****P < .0001 vs controls (Vehicle with FBS or Glc). (F) RTKis have minimal effects on glucose-induced proliferation. Effects of receptor tyrosine kinase inhibition (RTKi) on serum-derived growth factor (FBS, 5%) and glucose (Glc, 5.56 mmol/L) stimulation of BrdU-DNA synthesis. BIBW, BIBW2992; BMS, BMS-754807; Das, Dasatinib; Pon, Ponatinib. Data are averages with SD from 3 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, *P < .05, ****P < .0001 vs controls (Vehicle with FBS or Glc).
Blocking Phosphohistidine–Focal Adhesion Kinase Signaling Prevents Glucose-Induced Esophageal Squamous Cell Carcinoma Growth In Vivo
To determine whether poHis58-FAK plays a role in ESCC tumor progression in vivo, under conditions that combine glucose and growth factor stimulation, SCID mice were injected subcutaneously with KYSE70 cells stably expressing either WT- or H58A-FAK. KYSE70-[FAKH58A] tumors grew slower than those expressing WT-FAK (Figure 10A), producing smaller tumors (Figure 10B and C). Importantly, the KYSE70-[FAKH58A] tumors had lower relative poHis-FAK and poAKT levels compared with those in KYSE70-[WT-FAK] tumors (Figure 10D), strongly suggesting that loss of poHis58-FAK inhibits ESCC tumor growth by attenuating PI3K/AKT signaling.
Figure 10.
H58A decreases poHis-FAK signaling in vivo and xenograft growth. (A) H58A reduces tumor growth. Tumor volume of WT- or H58A FAK-transfected xenografts. Data are averages with SD from 6 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, ***P < .001, ****P < .0001 vs WT. (B) H58A reduces tumor weight. Weights of xenografts expressing WT or H58A FAK. Data are averages with SD from 6 biological replicates; 3 independent experiments that showed similar results; unpaired t test, ***P < .001 vs WT. (C) WT-/H58A-FAK xenografts. Images of isolated xenografts derived from WT- or H58A FAK-transfected cells. (D) H58A decreases tumor poHis-FAK and pAKT levels. ELISA of poHis-FAK and pAKT levels in WT/H58A FAK-transfected xenografts. Ratios of pAKT/AKT in H58A xenografts were normalized to those in WT samples. Data are averages with SD from 6 biological replicates; 3 independent experiments that showed similar results; two-way ANOVA test, **P < .01, ****P < .0001 vs WT.
Discussion
Our studies demonstrate a direct onco-proliferative function of glucose (Figure 11). Mammalian cells respond to growth cues such as hormones and/or anchorage by enhancing glucose uptake and glycolysis for biomass synthesis.34, 35 The proliferation of most normal cell types ceases on removal of growth factors, yet many tumors have developed GFIP through the induction of autocrine pathways stimulated by pro-oncogenic mutations.2 However, our data indicate that in the absence of serum-derived growth factors, glucose directly stimulates ESCC cell proliferation, and that this ability is not shared by EAC cells. This can explain resistance to growth factor–targeted therapy that is seen in ESCC as well as other cell types.
Figure 11.
Glucose-induced poHis-FAK signaling and tumor growth.
The pro-proliferative role for glucose in ESCC is not likely through providing energy/carbon sources or by restoring redox balance but more likely as a mitogen. This is because the micromolar levels of glucose capable of inducing ESCC proliferation are several logs lower than serum glucose concentrations involved in providing ATP through the glycolytic pathway. Understanding this glucose-dependent GFIP may allow for novel interventions that target this common abnormality in ESCC.
Our studies demonstrate that glycolytic PEP modification of FAK on His58 contributes to glucose-dependent growth of ESCC cells. This suggests that ESCC cells obtain growth factor independence through poHis-FAK signaling. Common amplification of cyclin D1 gene expression in ESCC has been linked to inhibited mitochondrial activity.6 In addition, cancer cells often express the low activity isoform of pyruvate kinase, PKM2, an enzyme converting PEP to pyruvate, but not the high activity PKM1 isoform. Impaired mitochondrial function and PEP-to-pyruvate conversion can lead to PEP accumulation. Our data demonstrate that PEP elevation in ESCC cells stimulates DNA synthesis and induces poHis58-FAK. By using purified protein preparations of FAK and radiolabeled PEP, we also demonstrate a direct ability of PEP to transfer its phosphoryl group to FAK. This results in increased histidine phosphorylation of FAK. Mutation of the Y397-FAK autophosphorylation site did not alter glucose-induced poHis and did not reduce glucose-induced proliferation but did inhibit growth factor–dependent proliferation. PEP represents a critical source of phosphoryl groups that can drive growth factor–independent histidine signaling. These findings explain why glycolysis is increased in some cancer cells: not to provide nutrients for macromolecular synthesis, but to enable the production of critical intermediates for signaling and proliferation in a growth factor–independent manner.
We have shown that induction of poHis FAK results in PI3K/AKT activation. FAK has been shown to bind to PI3K in 2 major ways: (1) at the poY397 site via the p85-SH2 domain and (2) at the C-terminal PA domain via the p85-SH3 domain. Because poY397-FAK is not required for glucose-induced proliferation, it is likely that poHis58-induced interaction of p85-SH3 to the FAK-PA binding site contributes to AKT activation. Glucose-promoted poHis58-FAK-AKT signaling will allow ESCC cells to avoid their dependence on growth factors such as insulin and/or IGF-1 for proliferation.
Our findings directly link mitogenic glucose to histidine signaling, AKT activation, and tumor proliferation in vitro and in vivo. Thus, whereas other cancer types develop growth factor independence through mutations and/or amplifications of oncogenic mediators that induce autocrine signaling, our data suggest that ESCCs have evolved growth factor independence through a glucose→PEP→poHis58-FAK proliferation pathway. Indeed, this may explain why ESCCs are relatively insensitive to drugs that target the receptor and non-receptor tyrosine kinases activated by growth factors, and why there is no current molecular targeted therapy approved for ESCC. Importantly, this putative glucose dependence pathway represents a therapeutic window that can be exploited.
Epidemiologic studies suggest that there are different factors responsible for the pathogenesis of ESCC compared with those for EAC.36 Despite this, similar treatment regimens are used for these disparate tumor types, resulting in varied response rates. Worldwide, ESCC tumors present a far greater health hazard than EAC.37 Our studies clearly show an increased dependence on glucose and glycolysis for proliferation in ESCC. These results are consistent with the increased uptake of radiolabeled fluorodeoxyglucose on positron emission tomography scans in ESCC compared with EAC, likely reflective of increased glycolysis present in ESCC in vivo.4 In fact, glucose-stimulated ESCC proliferation in eukaryotes resembles nutrient control of prokaryotic growth.12 Finally, patients with metabolic disorders associated with glucose elevation are at increased ESCC cancer risk, supporting the mitogenic role of glucose in this disease.38
Current FAK-targeted or other kinase inhibitors are typically ATP-competitive compounds or inhibitors of scaffolding activity with signaling partners,39 and they are usually assessed for inhibition of growth factor–induced signaling and/or proliferation. Moreover, these inhibitors are likely aimed to patients where the target pathway is activated by mutation and/or gene amplification. However, our studies indicate that glucose-induced poHis58-dependent ESCC proliferation requires neither FAK-Y397 phosphorylation nor FAK kinase activity, both of which are typically induced by growth factor/RTK pathways. Indeed, current FAK inhibitors evaluated in clinical trials in humans have had only modest effects. Although these FAK kinase inhibitors (GSK2256098 or VS-6063 also known as defactinib or PF-045548778) greatly decreased poY397 levels to ∼80% from baseline, minor responses or disease stabilization were observed in only 13%–33% of patients with advanced solid tumors including esophageal cancer.40, 41, 42 It would be of interest to test current growth factor–dependent FAK kinase inhibitors against inhibitors targeting poHis-dependent signaling and glucose-dependent proliferation. In future studies, it would be of interest to develop novel inhibitors of glucose-induced poHis-FAK signaling and compare the effects on ESCC xenograft growth of current FAK tyrosine kinase inhibitors against such novel poHis58-FAK inhibitors. Targeting glucose-induced poHis will be an innovative and specific way to inhibit the proliferation of ESCC, because normal adult tissues are not reliant on aerobic glycolysis-induced proliferation.
Materials and Methods
Cell Models of Glucose-Stimulated Proliferation
In the presence of serum, cells were incubated in glucose-free medium for 3–8 hours to deplete glucose and then in serum-free medium with or without glucose for 1–4 hours. WST1 was used to monitor cell viability/proliferation. BrdU or EdU was used to label glucose-induced DNA synthesis. BrdU/EdU-DNA levels were analyzed by using ELISA or fluorescence flow cytometry. CRISPR-Cas9 gene editing systems were used to interrupt the FAK gene. Standard molecular biological techniques were used to make WT/mutant FAK constructs. All mutations such as FAK His58A/E were verified by sequencing. Cells expressing WT/mutant (His58A/E) FAK were obtained by using flow cytometry sorting, G418 selection, and colony expansion. ESCC cells (KYSE70) expressing WT/His58A FAK were used for xenograft models.
Cell Line and Reagents
To verify that cell lines were not false, misidentified, and were known to be authentic cell lines, we searched the Cellosaurus database ExPASy Bioinformatics Resource Portal for RRID numbers for each verified cell line shown below. Human ESCC KYSE70 (RRID: CVCL_1356), KYSE140 (RRID: CVCL_1347), KYSE150 (RRID: CVCL_1348), KYSE180 (RRID: CVCL_1349), KYSE520 (RRID: CVCL_1355), TE1 (RRID: CVCL_1759), TE7 (RRID: CVCL_9972), TE9 (RRID: CVCL_1767), TE8 (RRID: CVCL_1766), and TE10 (RRID: CVCL_1760) were originally obtained from Takayoshi Tobe lab (KYSE) and the Takashi SASAKI lab (TE). Human esophageal epithelial cell line (Het-1A, RRID: CVCL_3702, ATCC# CRL-2692) and human breast cancer cell line (MCF7, RRID: CVCL_0031, ATCC# HTB-22) were purchased from ATCC. Esophageal adenocarcinoma (FLO1, RRID: CVCL_2045, SK-GT-4, RRID: CVCL_2195, KYSE1, RRID: CVCL_1825, and OACM5.1C, RRID: CVCL_1842) were purchased from Sigma-Aldrich (St Louis, MO). Cancer cell lines were maintained in recommended medium in the presence of 10% FBS and 1% antibiotics: KYSE70/KYSE140//KYSE150/KYSE520/TE1/TE7/TE8/TE9/TE10, RPMI-1640; KYAE1, F-12K + RPMI1640; SK-GT-4, RPMI-1640; and FLO1, DMEM. The human esophageal epithelial cell line was cultured in BEGM.
The antibodies were obtained from the following sources: anti-N3-poHis (Millipore, Burlington, MA; MABS1352), anti-poY397-FAK (Cell Signaling Technology, Danvers, MA; #3283), anti-FAK (Millipore; 05-537), anti-GFP (Abcam, Cambridge, UK; AB290), anti-poS473-AKT (Cell Signaling Technology; #9271), anti-AKT (Cell Signaling Technology; #9272), anti-AKT1 (Santa Cruz Biotechnology, Santa Cruz, CA; sc-5298), anti-AKT2 (Santa Cruz Biotechnology; sc-5270), anti-AKT3 (Santa Cruz Biotechnology; sc-134254), anti-poERK1/2 (Cell Signaling Technology; #4377), anti-ERK1/2 (Cell Signaling Technology; #9102), and anti-β-actin (Sigma-Aldrich; A5441).
AKT and control shRNAs were purchased from Santa Cruz Biotechnology. PI3K/AKT/ERK inhibitors, wortmannin/AZD5363/PD98059, were purchased from Sigma-Aldrich. Receptor tyrosine kinase inhibitors, BMS-754807, BIBW2992, Ponatinib, and Dasatinib, were purchased from Selleck Chemicals (Houston, TX). CRISPR-Cas9 gene editing system (1 × sgRNA/Cas9 all-in-one expression clone targeting PTK2 [NM_001199649.1]) was purchased from GeneCopoeia (Rockville, MD). Lipofectamine 3000 Transfection Reagent was purchased from Life Sciences (Corning, NY). Chemicals such as PEP and pyruvate were purchased from Sigma-Aldrich.
Assessments of Glucose-Induced Proliferation
Three approaches were applied, namely BrdU ELISA, EdU coupling flow cytometry, and trypan blue exclusion cell counting. Briefly, cells were incubated in glucose-free medium containing 5% FBS for 3–8 hours to deplete glucose and then in FBS-free medium with or without varied concentrations of glucose in the presence of BrdU for 1 hour or EdU for 4 hours. For simultaneous assessments of cell viability (WST1) and cell proliferation (BrdU), the treated cells were incubated in RPMI-1640 containing WST1 for 15–60 minutes and measured by using a microplate reader at 450 nm. BrdU-DNA was determined by using the Cell Proliferation ELISA (BrdU, colorimetric; Roche, Basel, Switzerland) by following the manufacturer’s instructions. EdU-DNA was assessed by using Click-iT Plus EdU Pacific Blue Flow Cytometry Assay Kit (Life Science). To count viable cells, the cells were trypsinized, mixed with trypan blue, and counted on a TC-20 cell counter (Bio-Rad Laboratories, Hercules, CA).
Antibody Microarray Analysis
KYSE70 cells (2,000,000/100-mm dish) were incubated in glucose-free RPMI-1640 containing 5% FBS for 8 hours. Next, the cells were incubated in FBS-free medium with or without 1 mmol/L glucose for 1 hour. Cancer Signaling Antibody Arrays (Full Moon Biosystems) were used to detect phosphorylation/activation of key signal proteins by following the manufacturer’s instructions. Briefly, cells were homogenized in lysis buffer, buffer was exchanged, and biotin was labelled. Microarray slides were blocked, coupled with labelled lysates, and detected with C3-streptavidin. The processed antibody array plates were sent to the company for detection and data analysis.
The Cancer Genome Atlas Database Search and Analysis
EC cohort of TCGA consisted of 183 patients (95 ESCC + 88 EAC). The mRNA expression z-scores from RNA-seq were downloaded via cBioPortal Center (http://www.cbioportal.org/).43 Statistical analyses were performed by using R software (http://www.r-project.org/) and Bioconductor (http://bioconductor.org/). GSEA was performed on TCGA cohort by using software provided by the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp), as previously described.43 Patients were classified into 2 groups, ESCC and EAC.
Site-Directed Mutagenesis and Overexpression
GeneArt Site-Directed Mutagenesis System (Invitrogen, Carlsbad, CA) was used for construction of pEGFP-FAK-H58A/E. Mutagenesis primers for H58A were the following: forward, 5′-C AGT ATT ATC AGG GCA GGA GAT GCT ACT GAT G and reverse, 5′-C ATC AGT AGC ATC TCC TGC CCT GAT AAT ACT G; for H58E: forward, 5′-C AGT ATT ATC AGG GAG GGA GAT GCT ACT GAT G and reverse, 5′-C ATC AGT AGC ATC TCC CTC CCT GAT AAT ACT G (the underlined triplets are those that are mutated). The mutation was verified by Sanger sequencing. Cells were transfected with the constructs by using Lipofectamine 3000 Transfection Reagent. G418 selection, fluorescence flow cytometry sorting, and colony expansion were performed as previously described.1
Enzyme-linked Immunosorbent Assay of Phosphohistidine–Focal Adhesion Kinase
The protocol for analysis of poHis-FAK levels was modified because poHis is heat and acid labile. The wells were coated with an anti-poHis-N3 antibody44 (0.98 ng/μL, rabbit, MABS1352; Millipore) in 50 μL coating buffer (phosphate-buffered saline [PBS], pH 7.4) or coating buffer alone at 4°C overnight. After the wells were washed with washing buffer (PBS, pH 7.4, 0.03% Tween-20, 200 μL/well) 3 times, 200 μL of 10% dry milk in washing buffer was added to the wells and kept at room temperature for 2 hours. Then, the wells were rinsed with washing buffer 2 times. To detect PEP-induced poHis-FAK, the reaction mixture containing Na2HPO4/NaH2PO4 buffer (20 mmol/L, pH 7.6), NaCl (50 mmol/L), MgCl2 (5 mmol/L), ethylenediamine tetraacetic acid (1 mmol/L), dithiothreitol (DTT) (2 mmol/L), PEP (20 mmol/L), and purified recombinant FAK (0.26 ng/μL, Cat#: PV3832; Invitrogen) was added to the poHis Ab–coated wells and kept at room temperature for 5 hours. Excessive pyruvate or ATP (100 mmol/L) was used to replace PEP in the parallel reaction mixtures as negative controls. For the assessment of cellular poHis-FAK levels, 50 μL of cell lysates (0.17 ug/μL) was added to the anti-poHis Ab–coated wells. The wells were washed with washing buffer 3 times to remove any unbound materials. The first antibody (FAK4.47, mouse, 1:1000 in 50 μL of antibody buffer [PBS, pH 7.4, 0.03% Tween-20, 1% bovine serum albumin]) was added to the wells and kept at 4°C overnight. After washing 3 times, an anti-mouse immunoglobulin G–horseradish peroxidase (1:1000 in 50 μL antibody buffer) was added to the wells and kept at room temperature for 2 hours. After washing 3 times, the substrate TMB (50 μl/well) was used and detected at 650 nm wavelength on a microplate reader.
Western Blot Analysis and Immunoprecipitation
Preparation of cell lysates in NP-40 lysis buffer (pH 8.0) containing protease and phosphatase inhibitor cocktails (Roche) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis were performed as previously described.1 To detect poHis protein levels, the protocol was modified to retain the acid and heat sensitive poHis on proteins.44 Major changes included the following: (1) cell lysates in Laemmli buffer (pH 8.0) were directly loaded to the gel without heating to 95°–98°C, (2) the membranes were washed in alkaline washing buffer (25 mmol/L Tris, pH 8.5, 137 mmol/L NaCl, 2.7 mmol/L KCl, and 0.04% Tween-20), and (3) dry milk in washing buffer was used to block nonspecific sites on the membranes. For the negative controls, an aliquot of cell lysate was heated at 95°C for 15 minutes or incubated under acidic pH (0.2 mol/L HCl) at 37°C for 15 minutes to reduce histidine phosphorylation.
After glucose depletion, KYSE70 cells were kept in medium with or without glucose (5.56 mmol/L) for 1 hour and harvested. Lysates (1 mg protein) were pre-cleared by using agarose–immunoglobulin G plus for 1 hour and then incubated with agarose-conjugated anti-FAK-AC Ab at 4°C overnight. The beads were washed with 1× PBS (pH 8.0) 3 times. Two times loading buffer (pH 8) was added to the immunoprecipitates. The beads (immunoprecipitates) were directly loaded to a sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel without heating to preserve poHis. The anti-poHis Ab was used to detect poHis-FAK on the polyvinylidene difluoride membrane. The same membrane was stripped and then reprobed with the anti-FAK Ab.
Microscopy Examination
KYSE70 cells were cultured on a 25 cm2 flask. Live cell images were acquired by using a Zeiss (Carl Zeiss GmbH, Oberkochen, Germany) AX10 Observer microscope with a LD APIan 20X/0,30 Ph 1 lens.
Mass Spectrometry
To overcome the labile nature of the poHis phosphoramidite moiety, we used the reported protocols that have been established for poHis detection. Phosphorylated human FAK (PV3832; Invitrogen) was digested with trypsin as previously described.45, 46, 47 The protein samples were processed by using a surfactant-aided precipitation/on-pellet digestion (SOD) procedure, which provides extensive cleanup to remove detergents and non-protein matrix components, deep protein denaturation (by both surfactants and precipitation) for rapid, efficient, and reproducible digestion, and thereby achieves reliable quantification of samples. Briefly, 100 μg protein was reduced with 10 mmol/L DTT with incubation at 37°C for 30 minutes in Eppendorf Thermomixer (Eppendorf, Hauppauge, NY), and cysteine residues were alkylated with 20 mmol/L iodoacetamide at 37°C for 30 minutes in the dark. For protein precipitation, 1 volume of chilled acetone (–20°C) was gently added into each sample and mixed for 1 minute to obtain a cloudy suspension. Then, another 8 volumes of chilled acetone were added to the mixture to precipitate proteins. The solution was vortexed until it became clear and stored at –20°C overnight to allow complete precipitation. Subsequently, samples were centrifuged at 20,000g for 30 minutes at 4°C to obtain a protein pellet. After removing the supernatant, 500 μL chilled acetone/water mixture (85/15, v/v %) was added to wash the pellet. Samples were centrifuged for 3–5 minutes, acetone/water supernatant was discarded, and the sample was allowed to air dry. The system was kept under pH 8.5 all the time to prevent potential acid catalysis of poHis degradation.
For protein digestion, the pellet was dissolved in 100 μL Tris (pH 8.5) buffer and sonicated in a water bath at 37°C. Then, 80 μL Tris buffer was added to 20 μg enzyme powder (Sigma-Aldrich) on ice for activation. The digestion procedure comprised 2 steps: (1) activated trypsin was added to the samples at a ratio of 1:40 (enzyme: substrate) and incubated for 6 hours at 37°C in an Eppendorf Thermomixer, and (2) a second aliquot of trypsin solution with equal volume was added to the samples and incubated overnight. After centrifugation at 20,000g for 20 minutes at 4°C, two-thirds of the digestion solution was carefully transferred into a tube for LC-MS analysis.
The peptide fragments derived from purified human FAK were subjected to HCD MS/MS analysis as previously described.29, 48, 49 The nano-flow reverse phase LC included a Spark Endurance autosampler (Emmen, Holland) and an ultra-high pressure Dionex (Sunnyvale, CA) ultimate Nano-2D Ultra capillary/nano-LC system. Peptide separation used a long nano-LC column (75-μm ID × 100 cm) with Pepmap 3 μm C18 particles. A large-ID trap (300 μm ID × 1 cm) was packed with Zorbax (Agilent Technologies, Santa Clara, CA) 5 μm C18 materials to allow large-capacity loading and removal of hydrophobic and hydrophilic matrix components. Mobile phase A was 10 mmol/L ammonium acetate in 2% acetonitrile (pH 8), and mobile phase B was 10 mmol/L ammonium acetate, pH 8 in 88% acetonitrile. A 4-μg peptide sample was loaded onto the trap with 1% B at 10 μL/min. After the trap was washed for 3 minutes, a 250 nL/min flow rate was used to back-flush the samples onto the nano-LC column for further separation. The column was enclosed in a heating sheath filled with heat-conductive silicone and warmed homogeneously at 40°C, which helps improve the chromatographic resolution and reproducibility. The following was the 2.5 h separation gradient used on the column: 4% B for 15 minutes, 13%–28% B for 110 minutes, 28%–44% B for 5 minutes, 44%–60% B for 5 minutes, 60%–97% B for 1 minute, and 97% B for 17 minutes. The trap was turned offline at 45 minutes to flush hydrophobic components. No perceivable degradation of poHis was observed under this LC condition.
An Orbitrap Fusion Lumos Mass Spectrometer (Thermo Fisher Scientific, San Jose, CA) was used for peptide identification and quantification. Data collection was operated in a 3-second cycle using the data-dependent top-speed mode. The MS1 survey scan (m/z 400–1500) was at a resolution of 120,000, with automated gain control target of 500,000 and a maximum injection time of 50 milliseconds. Precursors were fragmentized in HCD activation mode at normalized collision energy of 35%, and the dynamic exclusion was set with 45 seconds. Precursors were filtered by quadrupole using an isolation window of 1 Th. The MS2 spectra were collected at a resolution of 15,000 in the Orbitrap, with an automated gain control target of 50,000 and a maximum injection time of 50 milliseconds.
The raw files (.raw) generated by LC-MS were matched to database with SEQUEST-HT searching engine embedded in Proteome Discoverer (v2.1; Thermo Scientific). The search parameters were set as follows: (1) precursor ion tolerance, 20 ppm; (2) fragment ion tolerance, 0.6 Da; (3) maximum missed cleavages, 2; (4) static modifications: carbamidomethylation/+57.021 Da (C); (5) dynamic modifications: oxidation/+15.995 Da (M); phosphorylation +79.9663 Da (S, T, Y, H); (6) decoy database search, target FDR 0.01; (7) site probability threshold, 50; and (8) co-isolation interference, 60%.
Extracellular Flux Analysis and Assessments of Glucose, Lactate, and Phosphoenolpyruvate Levels
Seahorse XF243 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA) was used to assess the live cell extracellular acidification rate as previously described.1 Glucose consumption and levels of lactate and PEP were assessed by using glucose/lactate/PEP Colorimetric/Fluorometric Assay Kits (Sigma-Aldrich) as previously described.1
Phosphoenolpyruvate Labelling and 32P-Phosphoenolpyruvate Treatment of rFocal Adhesion Kinase
We adapted a published protocol for labeling PEP with 32P-ATP.9 Briefly, pyruvate was mixed with γ-32P-ATP in the reaction buffer containing rabbit muscle pyruvate kinase and DTT and incubated for 60 minutes. Q column was used to separate unreacted 32P-ATP from 32P-PEP. Thin-layer chromatography was run to verify labelled 32P-PEP. To analyze the phosphoryl group transfer from 32P-PEP to rFAK, rFAK (0.26 ng/μL) was incubated in the phosphate buffer (pH 8) containing 2 mmol/L DTT, 32P-PEP (4 mmol/L) with or without ×10 excessive PEP/ATP/pyruvate at room temperature for 1 hour. FAK was purified by using Pierce Glutathione Spin purification kit (Thermo Fisher Scientific) by following the manufacturer’s instructions. Glutathione-eluted rFAK was subjected to scintillation counting on a scintillation counter (Beckman Coulter, Brea, CA; LS6500).
Xenograft Mouse Models
Animal experimental protocols were approved by the Roswell Park Comprehensive Cancer Center Institutional Animal Care and Use committee. KYSE70 cells (1.5 × 106 cells, 100 μL/mouse) expressing the GFP-FAK-WT or GFP-FAK-H58A gene were mixed with 50% Matrigel Matrix (Fisher Scientific, Corning #356234) and injected into the right (H58A) and left (WT) axilla of 6-week-old female SCID mice (obtained from Roswell Park Comprehensive Cancer Center), respectively.1 Perpendicular diameters of each tumor were measured with calipers. Tumor volume (TV) was calculated by using the following formula: TV = (width)2 × length/2.
Statistical Analyses
GraphPad Prism (GraphPad Software, La Jolla, CA) was used for statistical analysis. Student t tests were used for single comparisons. For comparisons that involve multiple variables and observations, analysis of variance (ANOVA) was used. For in vitro and in vivo studies, the number of biological replicates was calculated by using a statistical analysis for power determination. For all studies, we set an alpha value of 0.05, a power of 0.8, and a standard deviation (SD) of 0.25. For in vivo studies, 6-week-old female SCID mice were randomly assigned to experimental groups.
Footnotes
Author contributions Jianliang Zhang: designed and performed experiments and prepared the manuscript. Irwin Gelman: study design, data interpretation, and critical revision of manuscript. Eriko Katsuta: TCGA database search and analysis. Yuanzi Liang: performed cell viability assays. Xue Wang: performed MS analysis of poHis-FAK. Jun Li: performed MS analysis of poHis-FAK. Jun Qu: designed MS study and interpreted MS results. Li Yan: TCGA database search and analysis. Kazuaki Takabe: designed and guided TCGA database search and analysis. Steven Hochwald: study design, data analysis/interpretation, and manuscript preparation.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the Roswell Park Alliance Foundation Grant, Roswell Park Comprehensive Cancer Center, United States (S.H., J.Z.) and in part through National Cancer Institute Comprehensive Cancer Center funds (P30-CA016056), National Cancer Institute, United States.
References
- 1.Zhang J., Gao Q., Zhou Y., Dier U., Hempel N., Hochwald S.N. Focal adhesion kinase-promoted tumor glucose metabolism is associated with a shift of mitochondrial respiration to glycolysis. Oncogene. 2016;35:1926–1942. doi: 10.1038/onc.2015.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 4.Foley K.G., Fielding P., Lewis W.G., Karran A., Chan D., Blake P., Roberts S.A. Prognostic significance of novel (1)(8)F-FDG PET/CT defined tumour variables in patients with oesophageal cancer. Eur J Radiol. 2014;83:1069–1073. doi: 10.1016/j.ejrad.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Analysis Working Group: Asan University. BC Cancer Agency. Brigham and Women’s Hospital. Broad Institute. Brown University. Case Western Reserve University. Dana-Farber Cancer Institute. Duke University. Greater Poland Cancer Centre. Harvard Medical School. Institute for Systems Biology. KU Leuven. Mayo Clinic. Memorial Sloan Kettering Cancer Center. National Cancer Institute. Nationwide Children’s Hospital. Stanford University. University of Alabama. University of Michigan. University of North Carolina. University of Pittsburgh. University of Rochester. University of Southern California. University of Texas MD Anderson Cancer Center. University of Washington. Van Andel Research Institute. Vanderbilt University. Washington University. Genome Sequencing Center: Broad Institute. Washington University in St Louis. Genome Characterization Centers: BC Cancer Agency. Broad Institute. Harvard Medical School. Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University. University of North Carolina. University of Southern California Epigenome Center. University of Texas MD Anderson Cancer Center. Van Andel Research Institute. Genome Data Analysis Centers: Broad Institute. Brown University. Harvard Medical School. Institute for Systems Biology. Memorial Sloan Kettering Cancer Center. University of California Santa Cruz. University of Texas MD Anderson Cancer Center. Biospecimen Core Resource: International Genomics Consortium. Research Institute at Nationwide Children’s Hospital. Tissue Source Sites: Analytic Biologic Services. Asan Medical Center. Asterand Bioscience. Barretos Cancer Hospital. BioreclamationIVT. Botkin Municipal Clinic. Chonnam National University Medical School. Christiana Care Health System. Cureline. Duke University. Emory University. Erasmus University. Indiana University School of Medicine. Institute of Oncology of Moldova. International Genomics Consortium. Invidumed. Israelitisches Krankenhaus Hamburg. Keimyung University School of Medicine. Memorial Sloan Kettering Cancer Center. National Cancer Center Goyang. Ontario Tumour Bank. Peter MacCallum Cancer Centre. Pusan National University Medical School. Ribeirão Preto Medical School. St Joseph’s Hospital & Medical Center. St Petersburg Academic University. Tayside Tissue Bank. University of Dundee. University of Kansas Medical Center. University of Michigan. University of North Carolina at Chapel Hill. University of Pittsburgh School of Medicine. University of Texas MD Anderson Cancer Center. Disease Working Group: Duke University. Memorial Sloan Kettering Cancer Center. National Cancer Institute. University of Texas MD Anderson Cancer Center. Yonsei University College of Medicine. Data Coordination Center: CSRA Inc. Project Team: National Institutes of Health Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tchakarska G., Roussel M., Troussard X., Sola B. Cyclin D1 inhibits mitochondrial activity in B cells. Cancer Res. 2011;71:1690–1699. doi: 10.1158/0008-5472.CAN-10-2564. [DOI] [PubMed] [Google Scholar]
- 7.D'Aguanno S., Barcaroli D., Rossi C., Zucchelli M., Ciavardelli D., Cortese C., De Cola A., Volpe S., D'Agostino D., Todaro M., Stassi G., Di Ilio C., Urbani A., De Laurenzi V. p63 isoforms regulate metabolism of cancer stem cells. J Proteome Res. 2014;13:2120–2136. doi: 10.1021/pr4012574. [DOI] [PubMed] [Google Scholar]
- 8.Hosios A.M., Hecht V.C., Danai L.V., Johnson M.O., Rathmell J.C., Steinhauser M.L., Manalis S.R., Vander Heiden M.G. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev Cell. 2016;36:540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vander Heiden M.G., Locasale J.W., Swanson K.D., Sharfi H., Heffron G.J., Amador-Noguez D., Christofk H.R., Wagner G., Rabinowitz J.D., Asara J.M., Cantley L.C. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquez J., Reinelt S., Koch B., Engelmann R., Hengstenberg W., Scheffzek K. Structure of the full-length enzyme I of the phosphoenolpyruvate-dependent sugar phosphotransferase system. J Biol Chem. 2006;281:32508–32515. doi: 10.1074/jbc.M513721200. [DOI] [PubMed] [Google Scholar]
- 11.Deutscher J., Francke C., Postma P.W. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puttick J., Baker E.N., Delbaere L.T. Histidine phosphorylation in biological systems. Biochim Biophysica Acta. 2008;1784:100–105. doi: 10.1016/j.bbapap.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Lapek J.D., Jr., Tombline G., Friedman A.E. Mass spectrometry detection of histidine phosphorylation on NM23-H1. J Proteome Res. 2011;10:751–755. doi: 10.1021/pr100905m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besant P.G., Attwood P.V. Histone H4 histidine phosphorylation: kinases, phosphatases, liver regeneration and cancer. Biochem Soc Trans. 2012;40:290–293. doi: 10.1042/BST20110605. [DOI] [PubMed] [Google Scholar]
- 15.Luk C.T., Shi S.Y., Cai E.P., Sivasubramaniyam T., Krishnamurthy M., Brunt J.J., Schroer S.A., Winer D.A., Woo M. FAK signalling controls insulin sensitivity through regulation of adipocyte survival. Nature Communications. 2017;8:14360. doi: 10.1038/ncomms14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frame M.C., Patel H., Serrels B., Lietha D., Eck M.J. The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol. 2010;11:802–814. doi: 10.1038/nrm2996. [DOI] [PubMed] [Google Scholar]
- 17.Muller G., Wied S., Frick W. Cross talk of pp125(FAK) and pp59(Lyn) non-receptor tyrosine kinases to insulin-mimetic signaling in adipocytes. Mol Cell Biol. 2000;20:4708–4723. doi: 10.1128/mcb.20.13.4708-4723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto D., Sonoda Y., Hasegawa M., Funakoshi-Tago M., Aizu-Yokota E., Kasahara T. FAK overexpression upregulates cyclin D3 and enhances cell proliferation via the PKC and PI3-kinase-Akt pathways. Cell Signal. 2003;15:575–583. doi: 10.1016/s0898-6568(02)00142-0. [DOI] [PubMed] [Google Scholar]
- 19.Luo M.L., Zhou Z., Sun L., Yu L., Sun L., Liu J., Yang Z., Ran Y., Yao Y., Hu H. An ADAM12 and FAK positive feedback loop amplifies the interaction signal of tumor cells with extracellular matrix to promote esophageal cancer metastasis. Cancer Lett. 2018;422:118–128. doi: 10.1016/j.canlet.2018.02.031. [DOI] [PubMed] [Google Scholar]
- 20.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L., Boyault S., Burkhardt B., Butler A.P., Caldas C., Davies H.R., Desmedt C., Eils R., Eyfjord J.E., Foekens J.A., Greaves M., Hosoda F., Hutter B., Ilicic T., Imbeaud S., Imielinski M., Jager N., Jones D.T., Jones D., Knappskog S., Kool M., Lakhani S.R., Lopez-Otin C., Martin S., Munshi N.C., Nakamura H., Northcott P.A., Pajic M., Papaemmanuil E., Paradiso A., Pearson J.V., Puente X.S., Raine K., Ramakrishna M., Richardson A.L., Richter J., Rosenstiel P., Schlesner M., Schumacher T.N., Span P.N., Teague J.W., Totoki Y., Tutt A.N., Valdes-Mas R., van Buuren M.M., van 't Veer L., Vincent-Salomon A., Waddell N., Yates L.R., Australian Pancreatic Cancer Genome I, Consortium IBC, Consortium IM-S, PedBrain I. Zucman-Rossi J., Futreal P.A., McDermott U., Lichter P., Meyerson M., Grimmond S.M., Siebert R., Campo E., Shibata T., Pfister S.M., Campbell P.J., Stratton M.R. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birsoy K., Possemato R., Lorbeer F.K., Bayraktar E.C., Thiru P., Yucel B., Wang T., Chen W.W., Clish C.B., Sabatini D.M. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–112. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.American Diabetes Association Glycemic targets: standards of medical care in diabetes—2018. Diabetes Care. 2018;41(Suppl 1):S55–S64. doi: 10.2337/dc18-S006. [DOI] [PubMed] [Google Scholar]
- 23.Cetinbas N.M., Sudderth J., Harris R.C., Cebeci A., Negri G.L., Yilmaz O.H., DeBerardinis R.J., Sorensen P.H. Glucose-dependent anaplerosis in cancer cells is required for cellular redox balance in the absence of glutamine. Scientific Reports. 2016;6:32606. doi: 10.1038/srep32606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudry I.H. Does ATP cross the cell plasma membrane. Yale J Biol Med. 1982;55:1–10. [PMC free article] [PubMed] [Google Scholar]
- 25.Hamasaki N., Wyrwicz A.M., Lubansky J.H., Omachi A. A 31P NMR study of phosphoenolpyruvate transport across the human erythrocyte membrane. Biochem Biophys Res Commun. 1981;100:879–887. doi: 10.1016/s0006-291x(81)80255-0. [DOI] [PubMed] [Google Scholar]
- 26.Saier M.H., Jr., Wentzel D.L., Feucht B.U., Judice J.J. A transport system for phosphoenolpyruvate, 2-phosphoglycerate, and 3-phosphoglycerate in Salmonella typhimurium. J Biol Chem. 1975;250:5089–5096. [PubMed] [Google Scholar]
- 27.Fuhs S.R., Meisenhelder J., Aslanian A., Ma L., Zagorska A., Stankova M., Binnie A., Al-Obeidi F., Mauger J., Lemke G., Yates J.R., 3rd, Hunter T. Monoclonal 1- and 3-phosphohistidine antibodies: new tools to study histidine phosphorylation. Cell. 2015;162:198–210. doi: 10.1016/j.cell.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavalier M.C., Kim S.G., Neau D., Lee Y.H. Molecular basis of the fructose-2,6-bisphosphatase reaction of PFKFB3: transition state and the C-terminal function. Proteins. 2012;80:1143–1153. doi: 10.1002/prot.24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oslund R.C., Kee J.M., Couvillon A.D., Bhatia V.N., Perlman D.H., Muir T.W. A phosphohistidine proteomics strategy based on elucidation of a unique gas-phase phosphopeptide fragmentation mechanism. J Am Chem Soc. 2014;136:12899–12911. doi: 10.1021/ja507614f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X., Peng X., Sun S., Park A.Y., Guan J.L. Role of kinase-independent and -dependent functions of FAK in endothelial cell survival and barrier function during embryonic development. J Cell Biol. 2010;189:955–965. doi: 10.1083/jcb.200912094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitra S.K., Mikolon D., Molina J.E., Hsia D.A., Hanson D.A., Chi A., Lim S.T., Bernard-Trifilo J.A., Ilic D., Stupack D.G., Cheresh D.A., Schlaepfer D.D. Intrinsic FAK activity and Y925 phosphorylation facilitate an angiogenic switch in tumors. Oncogene. 2006;25:5969–5984. doi: 10.1038/sj.onc.1209588. [DOI] [PubMed] [Google Scholar]
- 33.Franke T.F., Yang S.I., Chan T.O., Datta K., Kazlauskas A., Morrison D.K., Kaplan D.R., Tsichlis P.N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 34.Depre C., Rider M.H., Hue L. Mechanisms of control of heart glycolysis. Eur J Biochem. 1998;258:277–290. doi: 10.1046/j.1432-1327.1998.2580277.x. [DOI] [PubMed] [Google Scholar]
- 35.Ata R., Antonescu C.N. Integrins and cell metabolism: an intimate relationship impacting cancer. Int J Mol Sci. 2017;18:189. doi: 10.3390/ijms18010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K., Johnson A., Ali S.M., Klempner S.J., Bekaii-Saab T., Vacirca J.L., Khaira D., Yelensky R., Chmielecki J., Elvin J.A., Lipson D., Miller V.A., Stephens P.J., Ross J.S. Comprehensive genomic profiling of advanced esophageal squamous cell carcinomas and esophageal adenocarcinomas reveals similarities and differences. Oncologist. 2015;20:1132–1139. doi: 10.1634/theoncologist.2015-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnold M., Laversanne M., Brown L.M., Devesa S.S., Bray F. Predicting the future burden of esophageal cancer by histological subtype: international trends in incidence up to 2030. Am J Gastroenterol. 2017;112:1247–1255. doi: 10.1038/ajg.2017.155. [DOI] [PubMed] [Google Scholar]
- 38.Hu D., Peng F., Lin X., Chen G., Liang B., Li C., Zhang H., Liao X., Lin J., Zheng X., Niu W. The elevated preoperative fasting blood glucose predicts a poor prognosis in patients with esophageal squamous cell carcinoma: the fujian prospective investigation of cancer (FIESTA) study. Oncotarget. 2016;7:65247–65256. doi: 10.18632/oncotarget.11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma W.W. Development of focal adhesion kinase inhibitors in cancer therapy. Anti-Cancer Agents in Medicinal Chemistry. 2011;11:638–642. doi: 10.2174/187152011796817628. [DOI] [PubMed] [Google Scholar]
- 40.Soria J.C., Gan H.K., Blagden S.P., Plummer R., Arkenau H.T., Ranson M., Evans T.R., Zalcman G., Bahleda R., Hollebecque A., Lemech C., Dean E., Brown J., Gibson D., Peddareddigari V., Murray S., Nebot N., Mazumdar J., Swartz L., Auger K.R., Fleming R.A., Singh R., Millward M. A phase I, pharmacokinetic and pharmacodynamic study of GSK2256098, a focal adhesion kinase inhibitor, in patients with advanced solid tumors. Ann Oncol. 2016;27:2268–2274. doi: 10.1093/annonc/mdw427. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu T., Fukuoka K., Takeda M., Iwasa T., Yoshida T., Horobin J., Keegan M., Vaickus L., Chavan A., Padval M., Nakagawa K. A first-in-Asian phase 1 study to evaluate safety, pharmacokinetics and clinical activity of VS-6063, a focal adhesion kinase (FAK) inhibitor in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol. 2016;77:997–1003. doi: 10.1007/s00280-016-3010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones S.F., Siu L.L., Bendell J.C., Cleary J.M., Razak A.R., Infante J.R., Pandya S.S., Bedard P.L., Pierce K.J., Houk B., Roberts W.G., Shreeve S.M., Shapiro G.I. A phase I study of VS-6063, a second-generation focal adhesion kinase inhibitor, in patients with advanced solid tumors. Invest New Drugs. 2015;33:1100–1107. doi: 10.1007/s10637-015-0282-y. [DOI] [PubMed] [Google Scholar]
- 43.Kim S.Y., Kawaguchi T., Yan L., Young J., Qi Q., Takabe K. Clinical relevance of microRNA expressions in breast cancer validated using the Cancer Genome Atlas (TCGA) Ann Surg Oncol. 2017;24:2943–2949. doi: 10.1245/s10434-017-5984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kee J.M., Oslund R.C., Perlman D.H., Muir T.W. A pan-specific antibody for direct detection of protein histidine phosphorylation. Nat Chem Biol. 2013;9:416–421. doi: 10.1038/nchembio.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S.C., Chen Y., Mirza S., Xu Y., Lee J., Liu P., Zhao Y. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J Proteome Res. 2006;5:3446–3452. doi: 10.1021/pr0603396. [DOI] [PubMed] [Google Scholar]
- 46.Zu X.L., Besant P.G., Imhof A., Attwood P.V. Mass spectrometric analysis of protein histidine phosphorylation. Amino Acids. 2007;32:347–357. doi: 10.1007/s00726-007-0493-4. [DOI] [PubMed] [Google Scholar]
- 47.An B., Zhang M., Johnson R.W., Qu J. Surfactant-aided precipitation/on-pellet-digestion (SOD) procedure provides robust and rapid sample preparation for reproducible, accurate and sensitive LC/MS quantification of therapeutic protein in plasma and tissues. Anal Chem. 2015;87:4023–4029. doi: 10.1021/acs.analchem.5b00350. [DOI] [PubMed] [Google Scholar]
- 48.Shen X., Shen S., Li J., Hu Q., Nie L., Tu C., Wang X., Orsburn B., Wang J., Qu J. An IonStar experimental strategy for MS1 ion current-based quantification using ultrahigh-field Orbitrap: reproducible, in-depth, and accurate protein measurement in large cohorts. J Proteome Res. 2017;16:2445–2456. doi: 10.1021/acs.jproteome.7b00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Niu J., Li J., Shen X., Shen S., Straubinger R.M., Qu J. Temporal effects of combined birinapant and paclitaxel on pancreatic cancer cells investigated via large-scale, ion-current-based quantitative proteomics (IonStar) Mol Cell Proteomics. 2018;17:655–671. doi: 10.1074/mcp.RA117.000519. [DOI] [PMC free article] [PubMed] [Google Scholar]