Abstract
Despite major strides in reducing cardiovascular disease (CVD) burden with modification of classical CVD risk factors, significant residual risks remain. Recent discoveries linking intestinal microbiota and CVDs have broadened our understanding of how dietary nutrients may impact cardiovascular health and disease. While next-generation sequencing techniques can identify gut microbial community participants and provide insights into microbial composition shifts in response to physiologic responses and dietary exposures, provision of a prebiotic or probiotic has yet to show therapeutic benefit for CVD. Meanwhile, our evolving understanding of intestinal microbiota-derived physiologic modulators (e.g. short-chain fatty acids) and pathogenic mediators (e.g. trimethylamine N-oxide) of host disease susceptibility have created novel potential therapeutic opportunities for improved cardiovascular health. This review will discuss the roles of human intestinal microbiota in normal physiology, their associations with CVD susceptibilities, and the potential of modulating intestinal microbiota composition and metabolism as a novel therapeutic target for CVD.
Condensed Abstract
Recent discoveries reveal a mechanistic link between intestinal microbiota and physiological and pathophysiological processes linked to cardiovascular health and disease. This review discusses the role of human intestinal microbiota in normal physiology, their associations with CVD susceptibilities, and the potential of modulating intestinal microbiota composition and metabolism as a novel therapeutic target for CVD and its prevention.
Introduction
Nutrition is one of the key modifiable risk factors for cardiovascular health. However, the prevalence of ideal levels of diet remained low, reaching 0.6% in children and 1.5% in adults (1). On the other hand, 50% of young adults (ages 20–49 years) and 30.9% older adults (ages 50 years and above) reported poor levels of diet (1). Recent studies have highlighted dietary nutrient intake as a key contributor to global health and disease susceptibility.(2) What we eat also provides nutrients for intestinal microbial metabolism. Thus, a more global view of metabolism is evolving whereby it is the combination of both intestinal microbiota and host metabolic transformations that contribute to our overall metabolism and inter-individual variations in metabolic profiles. Intestinal microbiota serve as a filter of our largest environmental exposure - what we eat. By virtue of the fact that numerous intestinal microbiota generated metabolites are biologically active and impact host phenotypes, the intestinal microbiome thus also functions as a major endocrine organ that is responsive to dietary intake. It communicates with distal organs in the host through complex pathways via intestinal microbiota generated metabolites, and has been shown to impact phenotypes relevant to CVD ranging from inflammation, obesity and insulin resistance, to more direct processes like atherosclerosis and thrombosis susceptibility.(3) This review will discuss the roles of human intestinal microbiota in normal physiology, their associations with disease susceptibilities, and the potential of modulating intestinal microbiota as novel therapeutic targets for CVD.
The Normal Intestinal Microbiome
The human intestine harbors trillions of microbial cells as an essential part of our healthy physiologic ecosystem. Including communities of bacteria, fungi, archaea and viruses, they are often collectively referred to as “microbiota,” and their genome as the “microbiome.” The majority of the known intestinal microbial community is composed of bacteria in the phyla Bacteroidetes, Firmicutes (especially Clostridia species), Actinobacteria, Proteobacteria and Verrucomicrobia (Figure 1) (4). Understanding or defining what constitutes a “normal microbiome” is challenging, and may encompass considerations of the functional core, the healthy community ecology, and the resistance, resilience, and stability perspectives of the microbial ecology and related metabolites.(5) Nevertheless, taxa in these phyla are relatively stable over time within an individual, and relatively consistent among family members while varying widely between unrelated individuals living in different households.(6) However, the microbial communities in a household can be substantially altered, especially in those with physical contacts (7). The full extent of how much the microbiota is altered over time within an individual remains to be determined. Beginning at childbirth, there are substantial environmental influences on an individual’s intestinal microbial composition, function, and metabolism that can directly or indirectly impact host metabolism (8). Under physiological conditions, intestinal microbiota continue to stimulate the immune system especially via intestinal-associated lymphoid tissues. In addition, intestinal microbiota is involved in activating and differentiating a wide range of T- and B-lymphocytes, as well as modulating the mucosal production of immunoglobulins (especially immunoglobulin A) (4).
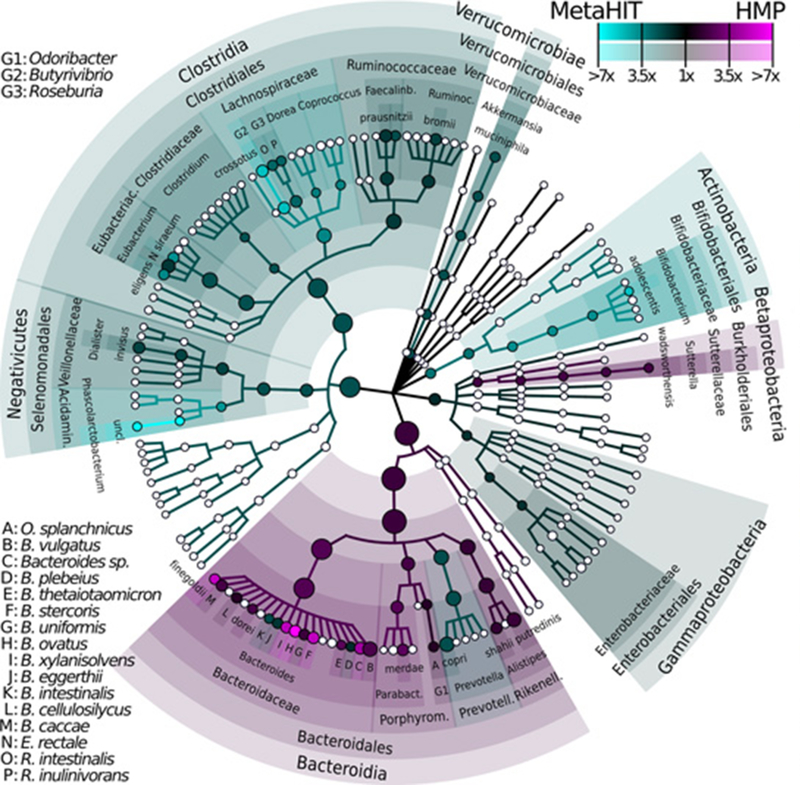
Figure 1: Phytogenetic Tree Representing Human Gut Microbiota.
Taxonomic cladogram reporting all clades present in one or both cohorts of gut microbiota in asymptomatic Western populations as inferred by MetaPhlAn on 224 samples combining Human Microbiome Project (n=139) and MetaHIT (n=85) cohorts. Circle size is proportional to the log of average abundance, color represents relative enrichment of the most abundant taxa (≥1% average in ≥1 cohort). Nodes’ size reflect their relative abundance.(143).
Next-Generation Sequencing Methods to Assess Microbial Diversity
Historically, microbial composition was investigated by traditional culture-based methods, which can be tedious, and only permit sampling of a small proportion of intestinal microbial inhabitants. Culture-based approaches provide a valuable opportunity to directly obtain information regarding bacterial metabolism and growth requirements, and the potential future use of such cultured strains for experimental investigations. Accordingly, significant effort has recently been placed on isolating and culture strains from the human gut.(9,10) More recently, culture-independent “next-generation sequencing” focuses on taxonomic assignments via DNA sequences, allowing previously “unculturable” bacteria to be identified with targeted “culturomics” to gain new mechanistic insights (11). The term “metagenomics” originally refers to a collective genome of microorganisms from an environmental sample that informs the microbial ecosystem. Microbial nucleic sequences are then used as a proxy for estimation of organism identity and relative abundance of complex microbial communities. As shown in Figure 2, two common strategies (12) are used to obtain this sequence information:
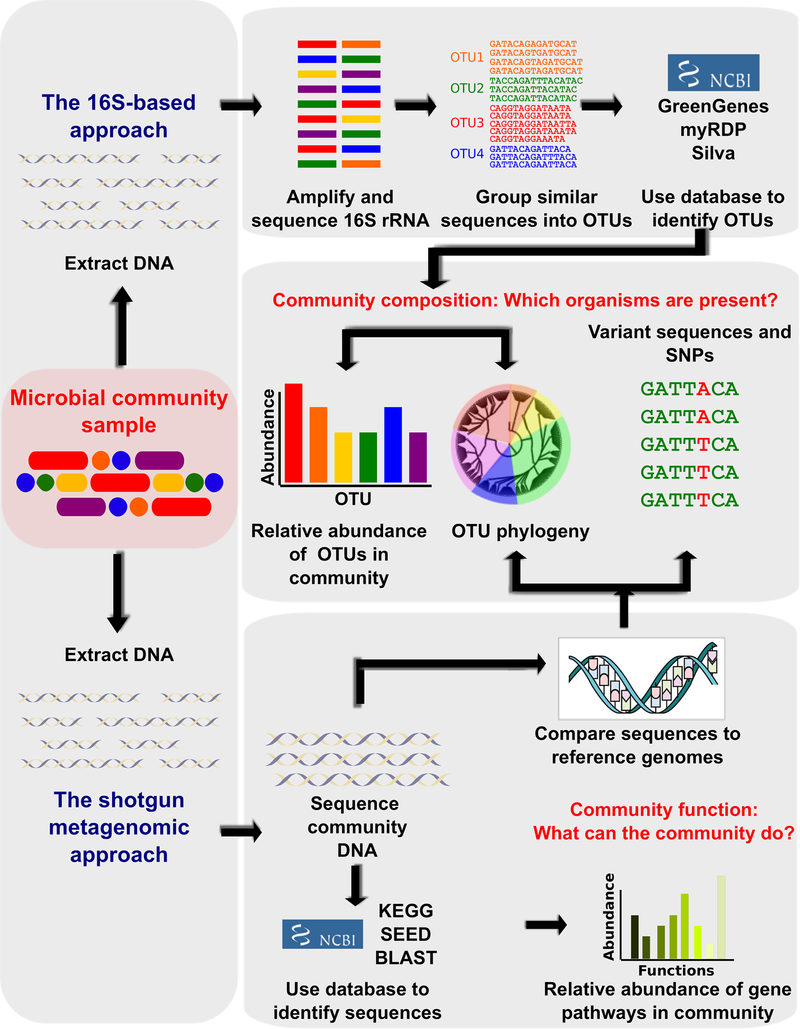
Figure 2: Contemporary Techniques for Metagenomic Sequencing.
Bioinformatic methods for functional metagenomics to define the composition and function of uncultured microbial communities using sequencing-based assays. Community DNA Is extracted from a sample and the bacterial taxa present are most frequently defined by amplifying the 16S rRNA gene and sequencing it with highly similar sequences grouped into Operational Taxonomic Units (OTUs). Direct metagenomic sequencing using a “shotgun” approach to directly compare with reference genomes or gene catalogues improves taxonomic resolution and allow observation of single nucleotide polymorphisms and other variant sequences with functional capabilities determined by comparing the sequences to functional databases (144).
Targeted Amplicon Sequencing. One common sequencing method detects the sequence differences of hypervariable region of the bacterial 16s ribosomal RNA (rRNA) for the taxonomic identification of the bacteria present. While facile and informative, regions of variability with so called “16S” analyses are typically insufficient to provide species-level resolution. Furthermore, different results can be generated based on different regions analyzed.
Whole Genomic Shotgun Sequencing. Using high-throughput genomic sequencing combined with advanced computational bioinformatics, identification of taxonomic and potential functional profiles of both known and unknown microbial communities can be achieved without the need to culture them in the laboratory (many yet to be explored).(13) While theoretically comprehensive and insightful, many studies are still underpowered.
Despite advances in next-generation microbial sequencing, there are some limitations in their clinical and research applications (14). Quality and quantity of recovered nucleic acids may vary depending on time of sample collection (sample types, source location, and collection/processing methods), storage, to processing techniques (from microbial genome extraction to library preparation, sequencing, quality filter, sequence identification steps). Technically, amplification bias, inadequate internal sequencing controls (positive and negative), or contamination may give rise to changes in microbial composition that are not reflecting the true changes. Meanwhile, tissue samples will be dominated by host DNA, and very high sequencing depth or approaches are necessary to enrich the microbial DNA. In addition, results of such analyses typically are presented as a proportion, rather than absolute level, and the presence of specific microbes in a specimen may not equate to pathogenicity.
Spectrum of Microbial Diversity Across Species
Beyond inter-individual variabilities, humans have intestinal microbiota that are distinct from other species. However, the majority of intestinal microbiota mechanistic research have utilized the murine model with well-defined phenotypes and genotypes and carefully controlled environmental variations such as diets and housing conditions in experiments. Mice can vary widely across inbred strains and can be distinctly different from humans in anatomy, genetics and physiology, not to mention dietary intake and environmental exposures. Different mouse models can also give rise to diverged shifts in intestinal microbiota composition and may have different cross-talks between intestinal microbiota and the host. Nevertheless, many proof-of-concept demonstrations have utilized gnotobiotic (all microorganisms are either known or excluded (as in germ-free)) mouse models. Some of the strongest data showing a potential participation of intestinal microbiota in CVD susceptibility have utilized germ-free mice during microbial transplantation studies to demonstrate transmission of a phenotype or disease – fulfilling an essential element of the “Koch’s postulate” for microbial pathogenesis (15,16). Examples where microbial transfer mice studies have shown transmission of phenotypes relevant to CVD include obesity/adiposity (17,18), atherosclerosis,(19,20), hypertension (21–23), thrombosis (24), renal insufficiency (25), and insulin resistance (26,27). More recently, the introduction of human fecal samples to demonstrate transmissibility of disease phenotypes such as features of metabolic syndrome have been reported (28). Discussions regarding the contributory role of intestinal microbiota on obesity and inflammatory diseases have recently been extensively reviewed elsewhere.(29).
Alterations in Composition of Intestinal Microbiota: Dysbiosis
The term “dysbiosis” refers to the condition of having an imbalance in the microbial communities either in or on the body. Intestinal microbial compositional changes associated with the presence of numerous diseases and/or phenotypes have been the focus of a large majority of human microbiome research studies over the past decade. Indeed, the ability to identify specific compositional patterns of microbiota that are associated with enhanced disease susceptibilities over time is an attractive concept. In the healthy intestines, Bacteroidetes and Firmicutes contribute a majority of the total bacterial species, and their ratio is often considered a relative estimate of intestinal microbial health. While intriguing and potentially insightful, Bacteroidetes and Firmicutes represent diverse phyla, and such analyses are thus relatively coarse and also purely associative in nature. Furthermore, host-microbial interactions are often dynamic, depending on local nutrient availability, oxygen tension, pH, gastric motility, and many other parameters. Hence, microbial communities are often unique and distinct across regions, and along the entire alimentary tract. This poses challenges in interpreting microbial composition based on analyses on fecal materials (30). Meanwhile, viruses, fungi and archaea also contribute to the non-host DNA sequence data obtained during deep sequencing analyses beyond bacteria, thereby contributing to further complexity to the analyses and interpretations.
Dysbiosis in Atherosclerosis and Coronary Artery Disease
Distinct microbial compositional changes have been described in the setting of atherosclerotic coronary artery disease (CAD) in various case-controlled studies utilizing fecal samples from patients with different phenotypes (Table 1). Whether these represent microbiota taxa associated with CAD versus medications or risk factors that contribute to CAD development has not been fully characterized. The earliest study exploring microbial composition changes associated with atherosclerotic plaques, oral and intestinal microbiota was reported by Bäckhed and colleagues, and employed pyrosequencing of 16S rRNA genes to survey bacterial taxa whose proportions were associated with CAD (31). More contemporary studies have reported characteristic changes in CAD patients where by a significant increase in Lactobacillales (Firmicutes) coupled with decrease in Bacteroidetes (Bacteroides and Prevotella) that was not observed in a comparative cohort of patients with diabetes.(32) In one of the largest metagenome-wide association studies to date, Jie et al observed an increased abundance of Enterobacteriaceae and oral cavity-asscuated bacteria and relatively depleted butyrate-producing bacteria in patients with atherosclerotic cardiovascular disease vs healthy controls. (33) It is important to highlight that the potential influence of dysbiosis in ACVD pathogenesis should not be confused with earlier investigations that focused on microbial pathogens that were implicated from epidemiologic associations. Mechanistic studies have implicated specific microbial pathogens such as Chlamydia pneumonia, Helicobacter pylori and Porphyromonas gingivalis may directly invade vascular cells and leukocytes and promote inflammation of the gums or lungs. However, randomized controlled trials using antibiotics to target these microbial pathogens have not yielded any clinical benefits in reducing morbidity and mortality in patients with CAD, and the pathogenic contributions of these microbes remain unclear.(34) Indeed, careful examination in early studies revealed that Chryseomonas is identifiable in atherosclerotic plaque samples, and the combined abundances of Veillonella and Streptococcus in atherosclerotic plaques correlated with their abundance in the oral cavity - another indirect link between periodontal disease and atherosclerosis (31). A larger cohort with available endarterectomy samples revealed predominantly taxa belonging to Proteobacteria and Actinobacteria, with no difference between asymptomatic and symptomatic patients, or plaque regions, suggesting less influence on plaque vulnerability (35).
TABLE 1.
Altered Intestinal Microbiota Composition Associated With Cardiovascular Diseases
| Study | Population | Technique | Increase in CVD | Decrease in CVD |
|---|---|---|---|---|
| Karlsson et al, 2012(145) | 12 MI/stroke Swedish patients vs 13 age/sex-matched controls | Gut metagenome | Collinsella |
Eubacterium Roseburia |
| Dinakaran et al, 2014(146) | 80 Indian CVD (valvular, ischemic, congenital heart) patients vs 40 healthy controls | 16s rRNA & metagenome in blood |
Proteobacteria Actinobacteria Propionibacterium phages Pseudomonas phages Rhizobium phages Lymphocystis virus Torque Teno viruses |
|
| Yin et al, 2015(147) | 141 stroke/TIA Chinese patients vs 94 asymptomatic controls | 16S rRNA V4 region |
Enterobacteriaceae Proteobacteria Escherichia/Shigella |
Bacteriodetes Bacteriodales Bacteroidaceae Bacteriodes |
| Emoto et al 2016(32) | 39 CAD Japanese patients vs 30 age/sex-matched controls with risk factors vs 50 healthy controls | Terminal RFLP |
Firmicutes/bateriodetes ratio Lactobacillales |
Bacteroides + Prevotella |
| Feng et al, 2016(148) | 59 CAD Chinese patients vs 43 healthy controls | Gut metagenome |
Streptococcus sp. M334 and M143 Clostridium sp. HGF2 |
|
| Jie et al, 2017(33) | 218 ACVD (stable/unstable angina, acute MI) Chinese patients vs 187 healthy controls | Gut metagenome |
Enterobaceriaceae ■ Escherichia coli ■ Klebsiella spp ■ Enterobacter aerogenes Oral cavity microbiome ■ Streptococcus sp ■ Lactobacillus salivarius ■ Solobacterium moorei ■ Atropobium parvulum Other microbiome members ■ Ruminococcus gnavus ■ Eggerthella lenta |
Butyrate-producing bacteria ■ Roseburia intestinalis ■ Faecalibacterium cf. prausnitzii Common microbiome members ■ Bacteriodes spp ■ Prevotella copri ■ Alistipes shahii |
Abbreviations: RFLP, restriction fragment length polymorphism; CAD, coronary artery disease, ACVD, atherosclerotic cardiovascular disease; TIA, transient ischemic attack; MI, myocardial infarction; CVD, cardiovascular disease.
Dysbiosis in Heart Failure
Alterations in intestinal microbial composition have been well described in patients with HF, especially with reduced diversity and depletion of core intestinal microbiota.(36) As a potential result, patients with heart failure (HF) have long been observed to be more prone to Clostridium difficile infections.(37) Careful characterization of intestinal luminal surfaces have revealed significantly increased bacterial overgrowth with mucosal biofilm and increased bacterial adhesions with HF versus non-HF controls.(38) With bowel wall edema during splanchnic congestion accompanying HF, intestinal barrier function is impaired, and structural components of microbiota may have enhanced interaction with host intestinal mucosa (Central Illustration). When such interactions occur with surface intestinal epithelial cells via pattern recognition receptors, it can stimulate host immune responses, as well as lead to vascular inflammation. This has been observed in the case of HF whereby acute exacerbations were associated with increased circulating lipopolysaccharide detection and downstream inflammator responses (39). Enhanced abundance of pathogenic microbial colonies of fungi (Candida spp) and bacteria (Campylobacter, Shigella, and Yersinia) can be isolated from fecal samples of HF patients, especially in those with elevated right atrial pressure and impaired intestinal barrier function (40). In three independent HF cohorts using sequencing techniques to characterize intestinal microbial compositions, there has been reported a consistent decrease in microbial diversity and a depletion of several butyrate producers (Faecalibacterium prasusnitzii, Lachnospiraceae family, Eubacterium hallii) that were inversely associated with inflammatory biomarkers (36,41–43).
Central Illustration. Intestinal microbiota and its metabolic contributions to cardiovascular health and disease.
Dietary nutrients are filtered by intestinal microbiota by both metabolism-dependent effects (generation of microbial metabolites such as short-chain fatty acids and trimethylamine from dietary carbohydrates and choline/carnitine, respectively) and metabolism-independent effects (lipopolysaccharides and peptidoglycans) leading to downstream metabolic alterations affect cardiovascular and end-organ functions. (145)
Dysbiosis and Hypertension and Diabetes Mellitus
While there have been varying reports over the intestinal microbiota profiles in particular in patients with type-2 diabetes, these patients have distinct intestinal microbiota signatures compared to non-diabetic subjects (44). Specifically, lower concentrations of butyrate-producing microbes such as Roseburia intestinalis and Faecalibacterium prausnitzii, and higher concentrations of Lactobacillus gasseri, Streptococcus mutans, and some Clostridiales, Desulfovibrio, and Proteobacteria species have been observed.(45) Metformin administration increased Akkermansia muciniphila and resulted in a relative increase in abundance of some Escherichia species that may have beneficial effects on glucose homeostasis (46,47). Meanwhile, insulin resistant patients have increased levels of branched chain amino acids (BCAA), especially associated with the presence of Prevotella copri and Bacteroides vulgatus that may drive insulin resistance and increased BCAA levels in mice.(48) Interestingly, human studies revealed that postprandial glucose responses to dietary intake can be modulated by intestinal microbiota (49).
High blood pressure is a leading cause of cardiovascular disease, and high dietary salt intake has been implicated in the pathophysiology of hypertension. Alterations of the intestinal microbiome in response to high salt intake have recently been observed in an experimental study in mice.(50) High salt intake resulted in a depletion of_Lactobacillus murinus. Consequently, treatment of mice with Lactobacillus murinus prevented salt-sensitive hypertension partly by modulating TH17 cells.(50) These results warrant further study in humans.
Probiotic and Dietary Interventions Targeting Intestinal Microbiota
Insights gained from intestinal microbiota and its associated metabolic pathways provided an opportunity to explore the contributory role of intestinal microbiota in generating the variability of physiologic responses to dietary nutrients. Leveraging the ability to monitor continuous glucose levels, an individualized glycemic pattern emerges when carefully curated dietary information with machine learning algorithms based on metagenomic information. (49) Specifically, by measuring microbiome responses in over 800 people using 16S rRNA and shotgun metagenomic profiling to assess taxonomy and function, respectively, tailoring dietary intake to an individual’s intestinal microbiome can potentially minimize post-prandial glucose rise (49). These results underscore how food-advice/interventions may have the potential to be individually tailored to each person (given the huge variation in post-prandial glucose response to traditionally “bad” and “good” food). These observations may allow a greatly advanced accuracy of nutritional advice in the future, as we enter the area of “personalized medicine”. Nevertheless, rigorous prospective studies to determine whether such approaches can impact cardiovascular risk are still warranted.
Theoretically, direct modulation of microbial composition has the potential to restore healthy microbial communities and promote cardiovascular health. In a rat myocardial infarction model, administration of broad-spectrum antibiotics was associated with changes in leptin levels and aromatic amino acid catabolites as well as reduction in infarct sizes.(51) Furthermore, administration of either Lactobacillus plantarum or Lactobacillus rhamnosus GR-1 have been associated with attenuation of post-infarction cardiac remodeling in rats.(51,52) Interestingly, foodborne microbes only transiently colonize the intestines. Unlike antibiotic-associated and Clostridium difficile-associated diarrhea, whether probiotics and prebiotics can directly influence the overall microbial distributions has not been established in human studies (53,54). Even though there are varying reports of lipid and blood pressure lowering effects with probiotics, human intervention studies showing their efficacies are limited, and there are currently no clinical recommendations for their prescription (55).
Microbial Metabolites as Physiologic Modulators: Short Chain Fatty Acids and Bile Acids
One key role for intestinal microbiota is to support every day physiological functions in food digestion via various fermentation processes in response to dietary intake of substrates.(56) Some metabolites can even be directly absorbed into the host circulation and serve as “hormones” to distant organs as sites of action. Other metabolites may be further metabolized by host enzymes similar to “prohormones,” serving as downstream mediators or signaling molecules. It is likely that the majority of microbial-generated metabolites can provide synergistic effects that promote health. However, toxic metabolites can also accumulate, especially when pathogenic species are colonizing or when normal host clearance mechanisms (e.g. renal function) of these metabolites are compromised. Detection of this “food metabolome” provides a unique opportunity to gain insight not only on quality and quantity of food intake, but also the functional consequences as a result of complex microbial-host metabolism (57).
Physiologic Effects of Short-Chain Fatty Acids
Anaerobic fermentation of undigested nutrients such as resistant starch, dietary fiber, and various complex polysaccharides produce fatty acids ranging from one to six carbon chains that are commonly referred to as “short-chain fatty acids” (SCFAs). Examples include acetate, propionate, and butyrate, which are actively as well as passively absorbed at the colonic epithelium into the portal vein.(58) While providing about 5–10% of energy source for the human host, SCFAs serve as signaling molecules, including modulation of autonomic systems and systemic blood pressure, as well as inflammatory responses and other cellular functions. SCFAs exhibit a wide range of physiologic functions, including histone deacetylases (HDACs) inhibition, chemotaxis and phagocytosis modulation, reactive oxygen species induction, cell proliferation and intestinal barrier integrity alteration.(58) It is important to note that patients with type 2 diabetes mellitus have less abundance of butyrate-producing bacteria and more Lactobacillus spp.(44,45,59) Indeed, SCFAs, in particular butyrate, can serve as energy substrates for epithelial cells of the intestines.(60,61) Furthermore, vancomycin treatment reduces the abundance of butyrate-producing bacteria in patients with metabolic syndrome, highlighting their important role in maintaining insulin sensitivity (62).
Recent mechanistic demonstrations also revealed that SCFAs can directly activate specific distinct G-protein-coupled receptors (GPCRs). Some of the GPCR identified to interact with SCFA from genetic and mouse model studies include G-protein receptor 41 (Gpr41), and olfactory receptor 78 (Olfr78, Figure 2) (63). In particular, Olf78 is highly expressed in the renal juxtaglomerular apparatus, where it mediates renin secretion in response to SCFAs. In addition, both Olfr78 and Gpr41 are expressed in smooth muscle cells of small resistance vessels, where they differentially mediate vascular tone. Interestingly, Olfr78 knock-out mice are hypotensive,(64) whereas Gpr41 knock-out mice are hypertensive (65), implying that these pathways may be physiologically important links between SCFAs and host blood pressure control. The three carbon SCFA propionate may stimulate Olfr78 to raise blood pressure, while stimulation of GPR41 can lower blood pressure.(64,66) The obligatory role of intestinal microbiota in generating SCFAs was demonstrated by antibiotic treatment that raised blood pressure in Olfr78 knockout mice, thereby further supporting the involvement of these receptors in blood pressure control. Recent animal studies demonstrated that intestinal microbiota-derived SCFAs are critical for the host immune response and the cardiac repair capacity after myocardial infarction in a mouse model with or without antibiotics (67). However, direct demonstration of such effects in human cardiovascular disease remains limited.
Physiological Effects of Bile Acids and Microbial Modulations
Bile acids facilitate the absorption of dietary fat and fat-soluble molecules. Several bile acids can regulate energy metabolism through activation of nuclear receptors such as G-protein-coupled bile acid receptor 1 (TGR5) and farnesoid X receptor (FXR) (68). Intestinal FXR appears to regulate hepatic cholesterol 7α-hydroxylase (CYP7A1), the rate-limiting enzyme in bile acid synthesis, through a fibroblast growth factor 15 (FGF-15/19)-dependent mechanism.(69) Hence, humans produce a large, conjugated hydrophilic bile acid pool, maintained through positive-feedback antagonism of FXR in intestine and liver. Meanwhile, through bile salt hydrolysis and bile acid 7α-dehydroxylation, intestinal microbiota is capable of producing secondary bile acid hormones that affect host physiology by agonism of FXR in intestine and liver, resulting in a smaller, unconjugated hydrophobic bile acid pool.(68) Interestingly, bile acids such as deoxycholic acid can serve as a direct antimicrobial agent owing to its hydrophobicity and detergent properties on bacterial membranes.(70) Hence, a dynamic equilibrium exists between diet-intestinal microbiome-bile acid pool size and composition. Hydrophilicity of the bile acid pool can be associated with disease states, while reduced bile acid levels in the gut can be associated with bacterial overgrowth and inflammation. In fact, a semi-synthetic bile acid analogue and a potent FXR agonist that is recently approved for treatment of non-alcoholic steatohepatitis, obeticholic acid, may reduce bacterial translocation and intestinal inflammation (71).
Therapeutic Potential of Dietary Interventions to Modulate SCFAs
The many links between the altered intestinal microbial community, metabolites, and susceptibility for CVD and metabolic diseases have placed a spotlight on the intestinal microbiome as a potential novel target for therapeutics. Currently diet modulation is the major therapeutic tool utilized in clinical practice to impact chronic metabolic diseases, and while lifestyle interactions can clearly impact intestinal microbial community structure and function, there are few studies that explore the impact of dietary interventions on the intestinal microbiome in humans. Existing studies of diet on the intestinal microbiota in humans have generally seen modest effects over the short term (72). Nevertheless, extreme shifts from animal-based to plant-based diets can modify regional and systemic productions of SCFAs, thereby potentially contributing to some of the proposed beneficial effects of these diets. In one study, an animal-based diet is associated with increases in the abundance of bile-tolerant microorganisms (Alistipes, Bilophila and Bacteroides) and decreases in the levels of Firmicutes that metabolize dietary plant polysaccharides (Roseburia, Eubacterium rectale andRuminococcus bromii) (53). As a result, there are significant reductions in fecal acetate and butyrate concentrations when switching from plant-based to animal based-diets (53). It is also interesting to note that fecal microbiota transplantation from lean donors to insulin resistant patients with metabolic syndrome leads to improved insulin sensitivity, and was associated with increased abundance of butyrate-producing bacteria such as Roseburia (28). By colonizing germ-free apolipoprotein E-deficient (Apoe−/−) mice with intentional microbial communities with or without Roseburia intestinalis revealed that microbe-diet interactions are crucial to understand the interplay between microbiota and CVD (73). In the presence of plant polysaccharides R. intestinalis could produce butyrate and confer protection against atherosclerosis, whereas on low plant polysaccharide diets no protection was observed (73). Similarly, the microbiota protected Apoe−/− mice against atherosclerosis when fed chow diet, rich in plant polysaccharides, but not when fed Western style diets (74).
Microbial Metabolites as Pathogenic Mediators: Trimethylamine N-oxide (TMAO)
Discoveries of potential pathogenic mediators that directly or indirectly modulate disease susceptibilities have provided a valuable window into microbial-host interactions that can modulate cardio-renal risks. In an initial discovery based untargeted metabolomic analysis, Wang et al identified 18 small-molecule analytes that in subsequent validation case-control cohorts (n~2000 subjects) repeatedly distinguished between patient with versus without future development of major adverse cardiovascular events (death, myocardial infraction [MI] and stroke) (75). Interestingly, some of these metabolites are now identified as known predictors of CVD risk that are not associated with intestinal microbiota (such as L-citrulline).(76) Three of the analytes (m/z 76, 104, and 118) were closely correlated with each other suggesting participation in a common pathway, and one in particular (m/z 76) seemed to be driving the association with incident CVD risks, and was subsequently shown to be trimethylamine N-oxide (TMAO) a intestinal microbiota dependent by-product of dietary choline and phosphatidylcholine (75). Another unknown metabolite whose levels strongly are associated with incident CVD risks has since been identified as the amino acid trimethyllysine (TML), and shown to serve as a nutrient precursor for intestinal microbiota-dependent TMAO generation (77).
Microbial Generation of Trimethylamine and Host Production of Trimethylamine N-oxide
Microbial catabolism of dietary nutrients that possess a trimethylamine (TMA; - N(CH3)3) moiety, such as choline, phosphatidylcholine and L-carnitine, can serve as precursors for TMA generation by specific microbial enzymes (“TMA lyase”) residing in the intestines.(75,78,79) TMA, an odorous gas with rotten fish smell, is then absorbed by the host, and following delivery to the liver via portal circulation, is rapidly converted into trimethylamine N-oxide (TMAO) by hepatic flavin monooxygenase (FMOs, particularly FMO3) (80). While female mice appeared to have greater FMO3 activity than male mice, human genome-wide association studies have yet to identify any sex differences in FMO3 variants (81,82). Meanwhile, patients with genetic polymorphisms of FMO3 experienced a metabolic disorder of excessive TMA called fish malodor syndrome (or trimethylaminuria) (83). Eventually, TMAO is predominantly excreted by the kidneys (Central Illustration) (84).
It is interesting to note that much of the chemistry of TMA as a toxic metabolite was originally studied because of its accumulation as a result of purification in sewage. (85) Choline is an abundant chemical moiety in bile, and is continuously delivered into the intestines in omnivore and vegan alike. Carnitine is an abundant nutrient in meat, especially red meats. Both choline and carnitine present within the gut are absorbed within the small bowel via specific transporters, but absorption is incomplete, particularly with large meals that can saturate the uptake systems. Consequently, both dietary choline and carnitine ingestion can give rise to significant elevations in TMA and TMAO, which has been shown to have many adverse effects on host metabolism, particularly those impacting cardiovascular health (86).
Dietary-Induced TMAO Generation and Atherogenesis
After observing that plasma levels of TMAO are dose dependently associated with CAD in subjects, initial functional studies sought to determine if the associations observed were mechanistically linked to disease causation. To directly demonstrate a proatherogenic contribution of the meta-organismal (i.e., involving both microbe and host) TMAO pathway, initial studies fed mice with a choline-rich or carnitine-rich diet, and demonstrated increases in plasma TMAO levels, cholesterol laden macrophage foam cell formation, and enhanced aortic atherosclerotic plaque development(75,79) Conversely, germ-free mice (lacking intestinal microbes) or short-term broad-spectrum antibiotic suppression of intestinal microbiota eliminated TMAO generating capacity, and suppressed diet (choline or carnitine) dependent atherosclerotic plaque enhancement.(75,79) Of interest, microbial transplantation experiments using cecal microbial communities recovered from a high TMA/TMAO-producer in-bred strain of mice (C57BL/6J) compared to a low TMA/TMAO-producer (NZW/LacJ mice) were shown to transmit choline diet dependent enhancement in atherosclerosis (19). Similarly, Zhu and colleagues demonstrated that microbial transplantation of a high TMA producing community can transmit TMAO generation and enhanced thrombosis potential into recipient germ free mice.(24) In further studies employing germ free mice as recipients and synthetic microbial communities capable of producing TMA versus not, it has been firmly established that the atherogenic metabolite TMAO and choline metabolism are integrally linked to intestinal microbiota and its adverse functional consequences (20,87).
Human Studies of Prognostic Value of Circulating TMAO Levels
Translating to humans, suppression of TMAO production is readily observed in healthy individuals taking a short course of poorly absorbed antibiotics, further illustrating the obligatory role of intestinal microbiota in TMA/TMAO generation.(79,88) In a cohort of >1,800 subjects, plasma TMAO levels were positively associated with CAD, peripheral artery disease, and history of MI independent of traditional risk factors.(75) In a subsequent study combining data from over 4,000 subjects undergoing elective coronary angiography, elevated TMAO predicted major adverse cardiac events (MACE), such as death, myocardial infarction (MI), and stroke over a 3-year period, even in the presence of elevated choline and betaine levels (non-microbial metabolites).(88,89) These prognostic effects were also observed in subsets of patients with history of HF,(90) diabetes mellitus,(91) peripheral artery disease,(92) chronic kidney disease (CKD),(93) high atherosclerotic burden,(94), acute coronary syndrome or MI,(95–97) and even non-CVD patients,(98) all independent of traditional risk factors. In addition, elevated TMAO levels track with degree of atherosclerotic burden as defined by the number of vessels with CAD by angiography, and with SYNTAX scores (99). These findings have now been validated by independent cohorts on multiple different continents, and reviewed recently in several meta-analyses (100–102), suggesting an estimated cut-off value of over 6 μM (approximately the third/fourth quartile cut-point of many cohorts) as predictive of the highest risk of adverse cardiac events (Figure 3). However, in those with excessive circulating TMAO (such as end-stage kidney disease on hemodialysis, with the first quartile in excess of 25–50 pM) the incremental prognostic value of TMAO appeared to be diminished,(103,104) and probiotics had no effects on TMAO levels in this population (105).
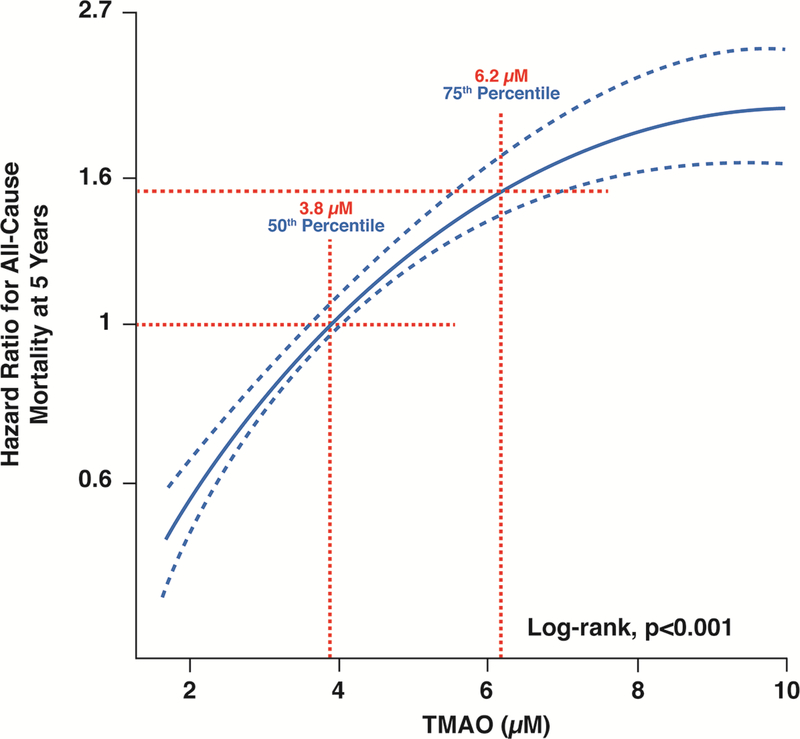
Figure 3: Relationship between plasma levels of TMAO and all-cause mortality in stable cardiac patients.
Increase in proportional hazards with fasting TMAO levels in a cohort of 2,235 patients undergoing elective coronary angiography. The fasting TMAO 4th quartile cut-off of 6.2 μM represented heightened 5-year all-cause mortality risk.
While TMAO in animal models exerts numerous atherosclerosis / thrombosis and inflammation promoting effects, the precise “receptor” or chemical sensor detecting the TMAO remains unknown. Animal studies have indicated that TMA/TMAO production leads to modulation of cholesterol and sterol/bile acid metabolism, as well as alterations of bile acid pool size, composition, and transport.(79) Additional global effects of TMAO include impairment of reverse cholesterol transport and promotion of forward cholesterol transport. There are also in vitro and in vivo experimental evidence of TMAO modulating vascular dysfunction(106) and inflammatory responses,(106) including the NLRP3 inflammasome pathways (107,108). In older mice, antibiotics reversed endothelial dysfunction and arterial stiffening accompanied by lower TMAO levels and oxidative stress as well as greater antioxidant enzyme expression (109).
Recently, TMAO has been shown to have direct effects on platelets, altering stimulus dependent intracellular calcium signaling in response to multiple agonists, and fostering enhanced platelet reactivity and thrombotic risks. For example, either direct injection of TMAO, or dietary choline and intestinal microbiota dependent TMAO elevation, has been shown to heighten platelet responsiveness, and promote faster clot formation in vivo using multiple different thrombosis models in mice (24). Similarly, genetic manipulation of host hepatic FMO3 expression levels in mice (e.g., reduction via targeted antisense oligonucleotides, or over-expression as a transgene) have been shown to alter systemic TMAO levels, and correspondingly, both alter platelet responsiveness and in vivo thrombosis potential (110). Remarkably, enhancement in platelet responsiveness with dietary choline supplementation and TMAO elevation has also been observed in recent interventional studies examining platelet aggregation responses in humans.(111) These latter studies, interestingly, also showed low dose aspirin mildly reduced TMAO levels in subjects, and attenuated the pro-thrombotic (heightened platelet aggregation responses) effect observed with elevated TMAO levels (111).
High levels of TMAO in subjects with normal renal function in the Framingham cohort are reported to herald increased risk for development of CKD (112), and in animal model studies, chronic elevation of TMAO has been shown to foster both renal functional impairment and tubulointerstitial fibrosis, along with activation of the pro-fibrotic TGFβ pathways.(93) Interestingly, in an adenine-induced mice model of CKD, concomitant increase in hepatic FMO3 expression occurs with increased circulating TMAO levels (113). Similarly, human subjects with HF have elevated levels of TMAO and experience worse outcomes (90,114–116). And in murine models, heightened TMAO levels exacerbate cardiac remodeling with pressure overload via transaortic constriction (117).
Other Related Intestinal Microbiota Generated Metabolites
It is important to highlight the fact that the same nutrient may be processed by various metabolic pathways and at different regions of the intestines, and thus the food metabolome detected from plasma samples can only represent the sum total of their balance. In the complex carnitine biosynthesis pathway, precursors of carnitine, gamma-butyrobetaine (γBB) and TML, have also been thoroughly investigated as potential substrates for TMA/TMAO production (77,118). Specifically, dietary l-carnitine is converted into TMAO via 2 sequential intestinal microbiota-dependent transformations in humans—an initial rapid generation of the atherogenic intermediate γBB, followed by transformation into TMA via low-abundance microbiota communities rather than single species in response to omnivorous diet patterns (119). Elevated levels of both metabolites have tracked with adverse long-term outcomes in patient cohorts, yet mechanistic studies have revealed that they may exert somewhat different physiologic/pathogenic effects on cardiovascular health than TMAO (77,79). For example, while dietary carnitine in the Apoe−/− model accelerates atherosclerosis, this effect appears to be mediated via intestinal microbiota generated TMA (and thus TMAO) since the dietary effect of carnitine is no longer observed when intestinal microbiota are suppressed with oral poorly absorbed antibiotics, and human clinical studies show the prognostic value of carnitine in incident CVD risk prediction is attenuated in statistical models were TMAO is included.(79) Studies examining the association between circulating levels of γBB and incident adverse cardiovascular risks have not yet been adequately examined; similarly, examination of the clinical prognostic value of γBB before versus following adjustments for TMAO in future work is needed, since dietary provision of γBB to atherosclerosis prone mice enhanced aortic root atherosclerosis, but only in the presence of intact intestinal microbiota and TMAO generation (118). It is likely that we only observed the tip of the iceberg of microbially produced metabolites modulating cardiovascular diseases. The recent identification of imidazole propionate as microbially produced metabolite that is enriched in type 2 diabetes and that induce insulin resistance when injected in mice suggest that other classes of metabolites should be considered as disease modulators (120).
Dietary and Drug Interventions Targeting TMAO Levels
Like SCFAs, it is clear that TMA/TMAO formation is largely dependent on nutrient sources. Dietary sources of choline/phosphatidylcholine and carnitine can clearly influence systemic levels. Overall, vegans and vegetarians have lower circulating TMAO levels and fecal TMA/TMAO-generating capacities than their omnivores counterparts (79). Specifically, diet rich in red meat is associated with higher levels of circulating TMAO as well as significantly reduced fractional renal excretion of TMAO compared to diet with white meat or no meat (121). Interestingly, chronic exposure to oral L-carnitine supplementations can also induce TMA/TMAO-generating capacities in humans. This same effect of dietary L-carnitine supplementation has been observed in mice(118) and in humans (122,123). Since these are common over-the-counter nutritional supplements and serve as food additives both for human and livestock dietary consumption, their overall long-term impact on cardiovascular health is unknown and needs to be investigated. Plasma TMAO levels can also increase with a high fat content, (124) although the effects are less clear in isocaloric diets.(121) It is also of clinical interest that cardioprotective drugs like statins may also have effects on TMAO.(125) Healthy individuals receiving choline or carnitine supplementation can raise circulating TMAO levels, yet such levels can be partially attenuated by concomitant ingestion of oral aspirin.(111,119,126) In a recent clinical interventional study, fecal microbial transplantation from lean vegan donors to metabolic syndrome recipients did not alter TMAO levels, despite improving indices of glycemic control.(127) It should be noted, however, that the baseline TMAO levels in the metabolic syndrome subjects enrolled in the study were in general low at baseline, making the study more powerful for observing an improvement in abnormalities in glucose metabolism, than reduction in microbiota dependent TMAO generation (127).
A minority of fish have elevated TMAO levels (especially deep sea fish) where the metabolite is used as part of the freeze avoidance mechanism. Thus, fish intake can generate high levels of TMAO in humans, though the specific fish, and even time of year (temperature of water fish are harvested from) can impact TMAO levels in fish.(128,129) It is therefore not surprising that questions have been raised as to whether TMAO is merely a marker of cardiovascular diseases, as decades of epidemiologic research have shown that high fish consumption may lower risk of cardiovascular diseases (130,131). Indeed, there have been reports of variable acute or short-term effects of food groups such as eggs (rich with phosphatidylcholine) and red meat (rich with carnitine) on circulating TMAO levels.(129) Yet one needs to be careful about making causal assumptions that counting food groups equate modulating microbial/host metabolic pathways. From a scientific perspective, TMA concentrations are indeed high in fish compared to equivalent protein quantities from soy or casein (milk). Yet when equivalent amounts of proteins from these sources are fed to atherogenic prone mice, those fed with fish proteins had the highest quantity of atheroma formation.(132) It is also important to highlight that not all fish are equivalent. Most fish don’t harbor omega-3 fatty acids either. And it has not been shown directly that changes in fish consumption is beneficial in all patients (130), nor in those who are vulnerable to TMAO accumulation (e.g., in CKD patients).
With the availability of TMAO as an in vitro clinical diagnostic test to identify those at increased CVD risks, it remains to be seen if one can tailor dietary recommendations monitoring TMAO, much in the same way one can monitor triglycerides, or blood glucose, and provide dietary advice. Whether current dietary recommendations that promote cardiovascular health have potential beneficial impact on the intestinal microbiome in general, or TMAO specifically, remains to be determined. It is important to recognize that a large majority of dietary advice recommends approaches that would be expected to lower TMAO, including lowering of caloric and fat consumption, and focusing on reduction in high fat, high cholesterol foods (animal products in general). In fact, a recent European panel took into account the potential impact of intestinal microbiota-generated TMAO for their dietary reference recommendations for choline (133). Preliminary data supporting the TMAO-lowering effects of hypocaloric diet and exercise (134), as well as intermittent fasting (135) are promising.
Potential of Therapeutic Interventions to Directly Modulate TMAO Production
Approaches to manipulate the intestinal microbiome and its metabolic pathways hold some promise, though they have yet to be realized in clinical practice. Identification of microbial enzymes that work in tandem to convert nutrient components such as choline or carnitine into TMA have spawned the discovery of several small molecule inhibitors that can modulate TMA/TMAO levels and adverse cardiovascular effects in experimental studies.(79,136–138) The prototype of such a “TMA lyase inhibitor” is called 3,3-dimethyl-1-butanol (DMB), which in microbial cell cultures and in vivo mouse models can reduce TMA/TMAO production without compromising microbial cell survival.(138) Mice receiving DMB appeared to reduce atherosclerotic burden, lower macrophage foam cell formation, and attenuate cardio-renal disease progression (138). Surveying common dietary sources have identified DMB in some samples of cold pressed extra-virgin olive oil, a major component of the Mediterranean diet that has been associated with cardiovascular health improvement (138). However, the impact of Mediterranean diet on changes in TMAO have been somewhat mixed (139–141). Next-generation TMA lyase inhibitors have recently been reported that are highly potent, suppressing TMAO levels in animal models. These agents have also shown potential to selectively target and accumulate within intestinal microbiota, permitting sustained inhibition of TMA-producing microbial enzymes with limited systemic exposure within the host (142).
Conclusions and Future Perspectives
Over the past years, accumulating data suggest an important link between the intestinal microbiome and cardiovascular disease. It has become clear that the microbiome plays an important role at the intersection of diet and cardiovascular disease, e.g. by metabolizing dietary components leading to the release of short chain fatty acids, some of which likely promote important beneficial cardiovascular effects. However, a major knowledge gap exists as the majority of studies have focused on characterizing the microbial composition rather than their functional alterations and downstream consequences. We now recognize that microbiome-dependent metabolism may also lead to production of metabolites with potential adverse cardiovascular effects, such as TMAO, that may promote atherosclerosis and heightened thrombosis risks. These observations provide an excellent opportunity for the development and testing of novel therapeutic strategies targeting the intestinal microbiome for prevention and treatment of cardiovascular disease. Approaches may include personalized dietary interventions, probiotics/prebiotics, or non-lethal microbial inhibitors that target specific pathways once they are identified (e.g., TMA production). Agents that target the TMAO pathway would also be expected to have multiple additional potential therapeutic benefits, including reduction in progression of renal functional decline, HF progression, and adverse outcomes in numerous high-risk cohorts (type 2 diabetes, CKD, and HF). However, well-powered prospective intervention studies are needed to validate this novel therapeutic approach. It is also important to stress that cardiometabolic diseases likely result from several metabolites, which may contribute to a variable extent in different individuals with high or low susceptibility, and that TMAO is likely only the “tip of the iceberg”. Future identification of microbially produced metabolites and investigation of whether they are causally linked to cardiometabolic disease will provide exciting potential novel opportunities to improve cardiovascular health and prevention.
HIGHLIGHTS.
Intestinal microbiota are mechanistically linked to physiological processes that impact cardiovascular health
Dietary nutrients serve as key environmental influences to intestinal microbiota and human host metabolism
Modulating intestinal microbiota composition and metabolism may serve as targets for cardiovascular disease prevention.
Acknowledgments
Funding: This work is supported by grants from the National Institutes of Health (NIH) and the Office of Dietary Supplements (R01HL103866, P20HL113452, R01DK106000, R01HL126827) and the Fondation Leducq (17CVD01).
Abbreviations
- BCAA
Branched chain amino acids
- CKD
Chronic kidney disease
- CVD
Cardiovascular disease
- DNA
Deoxyribonucleic acid
- HF
Heart failure
- gBB
Gamma-butyrobetaine
- LPS
lipopolysaccharides
- rRNA
Ribosomal ribonucleic acid
- SCFA
Short-chain fatty acid
- TMA
Trimethylamine
- TMAO
Trimethylamine N-oxide
- TML
Trimethyllysine
Footnotes
Disclosure: Dr. Hazen reports being named as co-inventor on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics and therapeutics. Dr. Hazen also reports being a paid consultant for Proctor and Gamble (P&G), having received research funds from P&G, and Roche Diagnostics, and being eligible to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics or therapeutics from Cleveland HeartLab and P&G. The other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Benjamin EJ, Virani SS, Callaway CW et al. Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation 2018;137:e67–e492. [DOI] [PubMed] [Google Scholar]
- 2.Estruch R, Ros E, Salas-Salvado J et al. Primary prevention of cardiovascular disease with a Mediterranean diet. The New England journal of medicine 2013;368:1279–90. [DOI] [PubMed] [Google Scholar]
- 3.Tang WH, Hazen SL. The Gut Microbiome and Its Role in Cardiovascular Diseases. Circulation 2017;135:1008–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol 2010;10:735–44. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med 2016;8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd-Price J, Mahurkar A, Rahnavard G et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017;550:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lax S, Smith DP, Hampton-Marcell J et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 2014;345:1048–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson RC, Manges AR, Finlay BB, Prendergast AJ. The Human Microbiome and Child Growth - First 1000 Days and Beyond. Trends Microbiol 2019;27:131–147. [DOI] [PubMed] [Google Scholar]
- 9.Forster SC, Kumar N, Anonye BO et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol 2019;37:186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Y, Xue W, Luo G et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol 2019;37:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagier JC, Khelaifia S, Alou MT et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 2016; 1: 16203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Lefterova MI, Suarez CJ, Banaei N, Pinsky BA. Next-Generation Sequencing for Infectious Disease Diagnosis and Management: A Report of the Association for Molecular Pathology. J Mol Diagn 2015;17:623–34. [DOI] [PubMed] [Google Scholar]
- 13.Pasolli E, Asnicar F, Manara S et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019;176:649–662 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y, Fanning S, Proos S, Jordan K, Srikumar S. A Review on the Applications of Next Generation Sequencing Technologies as Applied to Food-Related Microbiome Studies. Front Microbiol 2017;8:1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neville BA, Forster SC, Lawley TD. Commensal Koch’s postulates: establishing causation in human microbiota research. Curr Opin Microbiol 2018;42:47–52. [DOI] [PubMed] [Google Scholar]
- 16.Vonaesch P, Anderson M, Sansonetti PJ. Pathogens, microbiome and the host: emergence of the ecological Koch’s postulates. FEMS Microbiol Rev 2018. [DOI] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, Hamady M, Yatsunenko T et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–31. [DOI] [PubMed] [Google Scholar]
- 19.Gregory JC, Buffa JA, Org E et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem 2015;290:5647–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio 2015;6:e02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Zhao F, Wang Y et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durgan DJ, Ganesh BP, Cope JL et al. Role of the Gut Microbiome in Obstructive Sleep Apnea-Induced Hypertension. Hypertension 2016;67:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mell B, Jala VR, Mathew AV et al. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiological genomics 2015;47:187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu W, Gregory JC, Org E et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity Thrombosis Risk. Cell 2016;165:111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu KY, Xia GH, Lu JQ et al. Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci Rep 2017;7:1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ussar S, Griffin NW, Bezy O et al. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab 2015;22:516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Bahl MI, Roager HM et al. Environmental spread of microbes impacts the development of metabolic phenotypes in mice transplanted with microbial communities from humans. ISME J 2017;11:676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vrieze A, Van Nood E, Holleman F et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012;143:913–6 e7. [DOI] [PubMed] [Google Scholar]
- 29.Kasselman LJ, Vernice NA, DeLeon J, Reiss AB. The gut microbiome and elevated cardiovascular risk in obesity and autoimmunity. Atherosclerosis 2018;271:203–213. [DOI] [PubMed] [Google Scholar]
- 30.Lavelle A, Lennon G, O’Sullivan O et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015;64:1553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koren O, Spor A, Felin J et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proceedings of the National Academy of Sciences of the United States of America 2011;108 Suppl 1:4592–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emoto T, Yamashita T, Sasaki N et al. Analysis of Gut Microbiota in Coronary Artery Disease Patients: a Possible Link between Gut Microbiota and Coronary Artery Disease. J Atheroscler Thromb 2016;23:908–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jie Z, Xia H, Zhong SL et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 2017;8:845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andraws R, Berger JS, Brown DL. Effects of antibiotic therapy on outcomes of patients with coronary artery disease: a meta-analysis of randomized controlled trials. JAMA 2005;293:2641–7. [DOI] [PubMed] [Google Scholar]
- 35.Lindskog Jonsson A, Hallenius FF, Akrami R et al. Bacterial profile in human atherosclerotic plaques. Atherosclerosis 2017;263:177–183. [DOI] [PubMed] [Google Scholar]
- 36.Luedde M, Winkler T, Heinsen FA et al. Heart failure is associated with depletion of core intestinal microbiota. ESC Heart Fail 2017;4:282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mamic P, Heidenreich PA, Hedlin H, Tennakoon L, Staudenmayer KL. Hospitalized Patients with Heart Failure and Common Bacterial Infections: A Nationwide Analysis of Concomitant Clostridium Difficile Infection Rates and In-Hospital Mortality. Journal of cardiac failure 2016;22:891–900. [DOI] [PubMed] [Google Scholar]
- 38.Sandek A, Bauditz J, Swidsinski A et al. Altered intestinal function in patients with chronic heart failure. Journal of the American College of Cardiology 2007;50:1561–9. [DOI] [PubMed] [Google Scholar]
- 39.Sandek A, Bjarnason I, Volk HD et al. Studies on bacterial endotoxin and intestinal absorption function in patients with chronic heart failure. Internation J cardiol. 2012;157:80–5. [DOI] [PubMed] [Google Scholar]
- 40.Pasini E, Aquilani R, Testa C et al. Pathogenic Gut Flora in Patients With Chronic Heart Failure. JACC Heart failure 2016;4:220–7. [DOI] [PubMed] [Google Scholar]
- 41.Cui X, Ye L, Li J et al. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Sci Rep 2018;8:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamo T, Akazawa H, Suda W et al. Dysbiosis and compositional alterations with aging in the gut microbiota of patients with heart failure. PloS one 2017;12:e0174099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kummen M, Mayerhofer CCK, Vestad B et al. Gut Microbiota Signature in Heart Failure Defined From Profiling of 2 Independent Cohorts. Journal of the American College of Cardiology 2018;71:1184–1186. [DOI] [PubMed] [Google Scholar]
- 44.Karlsson FH, Tremaroli V, Nookaew I et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013;498:99–103. [DOI] [PubMed] [Google Scholar]
- 45.Qin J, Li Y, Cai Z et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55–60. [DOI] [PubMed] [Google Scholar]
- 46.Forslund K, Hildebrand F, Nielsen T et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015;528:262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin NR, Lee JC, Lee HY et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014;63:727–35. [DOI] [PubMed] [Google Scholar]
- 48.Pedersen HK, Gudmundsdottir V, Nielsen HB et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016;535:376–81. [DOI] [PubMed] [Google Scholar]
- 49.Zeevi D, Korem T, Zmora N et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015;163:1079–1094. [DOI] [PubMed] [Google Scholar]
- 50.Wilck N, Matus MG, Kearney SM et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017;551:585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam V, Su J, Koprowski S et al. Intestinal microbiota determine severity of myocardial infarction in rats. FASEB J 2012;26:1727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gan XT, Ettinger G, Huang CX et al. Probiotic administration attenuates myocardial hypertrophy and heart failure after myocardial infarction in the rat. Circulation Heart failure 2014;7:491–9. [DOI] [PubMed] [Google Scholar]
- 53.David LA, Maurice CF, Carmody RN et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khalesi S, Bellissimo N, Vandelanotte C, Williams S, Stanley D, Irwin C. A review of probiotic supplementation in healthy adults: helpful or hype? Eur J Clin Nutr 2018. [DOI] [PubMed] [Google Scholar]
- 55.Cicero AFG, Colletti A, Bajraktari G et al. Lipid-lowering nutraceuticals in clinical practice: position paper from an International Lipid Expert Panel. Nutr Rev 2017;75:731–767. [DOI] [PubMed] [Google Scholar]
- 56.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res 2013;69:52–60. [DOI] [PubMed] [Google Scholar]
- 57.Scalbert A, Brennan L, Manach C et al. The food metabolome: a window over dietary exposure. Am J Clin Nutr 2014;99:1286–308. [DOI] [PubMed] [Google Scholar]
- 58.Ohira H, Tsutsui W, Fujioka Y. Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis? J Atheroscler Thromb 2017;24:660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larsen N, Vogensen FK, van den Berg FW et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS one 2010;5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donohoe DR, Garge N, Zhang X et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 2011;13:517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PloS one 2012;7:e46589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vrieze A, Out C, Fuentes S et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol 2014;60:824–31. [DOI] [PubMed] [Google Scholar]
- 63.Pluznick JL. Renal and cardiovascular sensory receptors and blood pressure regulation. American journal of physiology Renal physiology 2013;305:F439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pluznick JL, Protzko RJ, Gevorgyan H et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proceedings of the National Academy of Sciences of the United States of America 2013;110:4410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Natarajan N, Hori D, Flavahan S et al. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiological genomics 2016;48:826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pluznick JL, Zou DJ, Zhang X et al. Functional expression of the olfactory signaling system in the kidney. Proceedings of the National Academy of Sciences of the United States of America 2009;106:2059–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang TWH, Chen HC, Chen CY et al. Loss of Gut Microbiota Alters Immune System Composition and Cripples Postinfarction Cardiac Repair. Circulation 2019;139:647–659. [DOI] [PubMed] [Google Scholar]
- 68.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol 2014;30:332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim I, Ahn SH, Inagaki T et al. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res 2007;48:2664–72. [DOI] [PubMed] [Google Scholar]
- 70.Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev 2005;29:625–51. [DOI] [PubMed] [Google Scholar]
- 71.Ubeda M, Lario M, Munoz L et al. Obeticholic acid reduces bacterial translocation and inhibits intestinal inflammation in cirrhotic rats. J Hepatol 2016;64:1049–1057. [DOI] [PubMed] [Google Scholar]
- 72.Wu GD, Chen J, Hoffmann C et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kasahara K, Krautkramer KA, Org E et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol 2018;3:1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lindskog Jonsson A, Caesar R, Akrami R et al. Impact of Gut Microbiota and Diet on the Development of Atherosclerosis in Apoe(−/−) Mice. Arterioscler Thromb Vasc Biol 2018;38:2318–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Z, Klipfell E, Bennett BJ et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011;472:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang WH, Shrestha K, Wang Z, Troughton RW, Klein AL, Hazen SL. Diminished global arginine bioavailability as a metabolic defect in chronic systolic heart failure. Journal of cardiac failure 2013;19:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li XS, Wang Z, Cajka T et al. Untargeted metabolomics identifies trimethyllysine, a TMAO-producing nutrient precursor, as a predictor of incident cardiovascular disease risk. JCI Insight 2018;3:e99096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rath S, Heidrich B, Pieper DH, Vital M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome 2017;5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koeth RA, Wang Z, Levison BS et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature medicine 2013;19:576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bennett BJ, de Aguiar Vallim TQ, Wang Z et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab 2013;17:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hartiala J, Bennett BJ, Tang WH et al. Comparative genome-wide association studies in mice and humans for trimethylamine N-oxide, a proatherogenic metabolite of choline and L-carnitine. Arterioscler Thromb Vasc Biol 2014;34:1307–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Warrier M, Shih DM, Burrows AC et al. The TMAO-Generating Enzyme Flavin Monooxygenase 3 Is a Central Regulator of Cholesterol Balance. Cell Rep 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Humbert JA, Hammond KB, Hathaway WE. Trimethylaminuria: the fish-odour syndrome. Lancet 1970;2:770–1. [DOI] [PubMed] [Google Scholar]
- 84.Hai X, Landeras V, Dobre MA, DeOreo P, Meyer TW, Hostetter TH. Mechanism of Prominent Trimethylamine Oxide (TMAO) Accumulation in Hemodialysis Patients. PloS one 2015;10:e0143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomas JM, Alexander M. Microbial formation of secondary and tertiary amines in municipal sewage. Appl Environ Microbiol 1981;42:461–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tang WH, Kitai T, Hazen SL. Gut Microbiota in Cardiovascular Health and Disease. Circulation research 2017;120:1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Skye SM, Zhu W, Romano KA et al. Microbial Transplantation With Human Gut Commensals Containing CutC Is Sufficient to Transmit Enhanced Platelet Reactivity and Thrombosis Potential. Circulation research 2018;123:1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang WH, Wang Z, Levison BS et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. The New England journal of medicine 2013;368:1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Z, Tang WH, Buffa JA et al. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. European heart journal 2014;35:904–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang WH, Wang Z, Fan Y et al. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: refining the gut hypothesis. Journal of the American College of Cardiology 2014;64:1908–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang WH, Wang Z, Li XS et al. Increased Trimethylamine N-Oxide Portends High Mortality Risk Independent of Glycemic Control in Patients with Type 2 Diabetes Mellitus. Clin Chem 2017;63:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Senthong V, Wang Z, Fan Y, Wu Y, Hazen SL, Tang WH. Trimethylamine N-Oxide and Mortality Risk in Patients With Peripheral Artery Disease. Journal of the American Heart Association 2016;5:e004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang WH, Wang Z, Kennedy DJ et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circulation research 2015;116:448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Senthong V, Wang Z, Li XS et al. Intestinal Microbiota-Generated Metabolite Trimethylamine-N-Oxide and 5-Year Mortality Risk in Stable Coronary Artery Disease: The Contributory Role of Intestinal Microbiota in a COURAGE-Like Patient Cohort. Journal of the American Heart Association 2016;5:e002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li XS, Obeid S, Klingenberg R et al. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes: a prognostic marker for incident cardiovascular events beyond traditional risk factors. European heart journal 2017;38:814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sheng Z, Tan Y, Liu C et al. Relation of Circulating Trimethylamine N-Oxide With Coronary Atherosclerotic Burden in Patients With ST-segment Elevation Myocardial Infarction. Am J Cardiol 2018. [DOI] [PubMed] [Google Scholar]
- 97.Tan Y, Sheng Z, Zhou P et al. Plasma Trimethylamine N-Oxide as a Novel Biomarker for Plaque Rupture in Patients With ST-Segment-Elevation Myocardial Infarction. Circ Cardiovasc Interv 2019;12:e007281. [DOI] [PubMed] [Google Scholar]
- 98.Yu D, Shu XO, Rivera ES et al. Urinary Levels of Trimethylamine-N-Oxide and Incident Coronary Heart Disease: A Prospective Investigation Among Urban Chinese Adults. Journal of the American Heart Association 2019;8:e010606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Senthong V, Li XS, Hudec T et al. Plasma Trimethylamine N-Oxide, a Gut Microbe-Generated Phosphatidylcholine Metabolite, Is Associated With Atherosclerotic Burden. Journal of the American College of Cardiology 2016;67:2620–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heianza Y, Ma W, Manson JE, Rexrode KM, Qi L. Gut Microbiota Metabolites and Risk of Major Adverse Cardiovascular Disease Events and Death: A Systematic Review and Meta-Analysis of Prospective Studies. Journal of the American Heart Association 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qi J, You T, Li J et al. Circulating trimethylamine N-oxide and the risk of cardiovascular diseases: a systematic review and meta-analysis of 11 prospective cohort studies. J Cell Mol Med 2018;22:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schiattarella GG, Sannino A, Toscano E et al. Gut microbe-generated metabolite trimethylamine-N-oxide as cardiovascular risk biomarker: a systematic review and dose-response meta-analysis. European heart journal 2017;38:2948–2956. [DOI] [PubMed] [Google Scholar]
- 103.Kaysen GA, Johansen KL, Chertow GM et al. Associations of Trimethylamine N-Oxide With Nutritional and Inflammatory Biomarkers and Cardiovascular Outcomes in Patients New to Dialysis. J Ren Nutr 2015;25:351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stubbs JR, Stedman MR, Liu S et al. Trimethylamine N-Oxide and Cardiovascular Outcomes in Patients with End-stage Kidney Disease Receiving Maintenance Hemodialysis. Clinical journal of the American Society of Nephrology : CJASN 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Borges NA, Stenvinkel P, Bergman P et al. Effects of Probiotic Supplementation on Trimethylamine-N-Oxide Plasma Levels in Hemodialysis Patients: a Pilot Study. Probiotics Antimicrob Proteins 2018. [DOI] [PubMed] [Google Scholar]
- 106.Seldin MM, Meng Y, Qi H et al. Trimethylamine N-Oxide Promotes Vascular Inflammation Through Signaling of Mitogen-Activated Protein Kinase and Nuclear Factor-kappaB. Journal of the American Heart Association 2016;5:e002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boini KM, Hussain T, Li PL, Koka S. Trimethylamine-N-Oxide Instigates NLRP3 Inflammasome Activation and Endothelial Dysfunction. Cell Physiol Biochem 2017;44:152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen ML, Zhu XH, Ran L, Lang HD, Yi L, Mi MT. Trimethylamine-N-Oxide Induces Vascular Inflammation by Activating the NLRP3 Inflammasome Through the SIRT3-SOD2-mtROS Signaling Pathway. Journal of the American Heart Association 2017;6:e006347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brunt VE, Gioscia-Ryan RA, Richey JJ et al. Suppression of the gut microbiome ameliorates age-related arterial dysfunction and oxidative stress in mice. The Journal of physiology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhu W, Buffa JA, Wang Z et al. Flavin monooxygenase 3, the host hepatic enzyme in the metaorganismal trimethylamine N-oxide-generating pathway, modulates platelet responsiveness and thrombosis risk. J Thromb Haemost 2018;16:1857–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu W, Wang Z, Tang WHW, Hazen SL. Gut Microbe-Generated Trimethylamine N-Oxide From Dietary Choline Is Prothrombotic in Subjects. Circulation 2017;135:1671–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rhee EP, Clish CB, Ghorbani A et al. A combined epidemiologic and metabolomic approach improves CKD prediction. Journal of the American Society of Nephrology : JASN 2013;24:1330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Johnson C, Prokopienko AJ, West RE 3rd, Nolin TD, Stubbs JR. Decreased Kidney Function Is Associated with Enhanced Hepatic Flavin Monooxygenase Activity and Increased Circulating Trimethylamine N-Oxide Concentrations in Mice. Drug Metab Dispos 2018;46:1304–1309. [DOI] [PubMed] [Google Scholar]
- 114.Suzuki T, Heaney LM, Bhandari SS, Jones DJ, Ng LL. Trimethylamine N-oxide and prognosis in acute heart failure. Heart 2016;102:841–8. [DOI] [PubMed] [Google Scholar]
- 115.Tang WH, Wang Z, Shrestha K et al. Intestinal microbiota-dependent phosphatidylcholine metabolites, diastolic dysfunction, and adverse clinical outcomes in chronic systolic heart failure. Journal of cardiac failure 2015;21:91–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Troseid M, Ueland T, Hov JR et al. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J Intern Med 2015;277:717–26. [DOI] [PubMed] [Google Scholar]
- 117.Organ CL, Otsuka H, Bhushan S et al. Choline Diet and Its Gut Microbe-Derived Metabolite, Trimethylamine N-Oxide, Exacerbate Pressure Overload-Induced Heart Failure. Circulation Heart failure 2016;9:e002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Koeth RA, Levison BS, Culley MK et al. gamma-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab 2014;20:799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Koeth RA, Lam-Galvez BR, Kirsop J et al. l-Carnitine in omnivorous diets induces an atherogenic gut microbial pathway in humans. The Journal of clinical investigation 2019;129:373–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Koh A, Molinaro A, Stahlman M et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018;175:947–961 e17. [DOI] [PubMed] [Google Scholar]
- 121.Wang Z, Bergeron N, Levison BS et al. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. European heart journal 2019;40:583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fukami K, Yamagishi S, Sakai K et al. Oral L-carnitine supplementation increases trimethylamine-N-oxide but reduces markers of vascular injury in hemodialysis patients. J Cardiovasc Pharmacol 2015;65:289–95. [DOI] [PubMed] [Google Scholar]
- 123.Miller MJ, Bostwick BL, Kennedy AD et al. Chronic Oral L-Carnitine Supplementation Drives Marked Plasma TMAO Elevations in Patients with Organic Acidemias Despite Dietary Meat Restrictions. JIMD Rep 2016;30:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Boutagy NE, Neilson AP, Osterberg KL et al. Probiotic supplementation and trimethylamine-N-oxide production following a high-fat diet. Obesity (Silver Spring) 2015;23:2357–63. [DOI] [PubMed] [Google Scholar]
- 125.Li DY, Wang Z, Li XS, Hazen SL, Tang WH. Relationship between Statin Use and Trimethylamine N-oxide in Cardiovascular Risk Assessment. Journal of the American College of Cardiology 2018;71:A115. [Google Scholar]
- 126.Vallance HD, Koochin A, Branov J et al. Marked elevation in plasma trimethylamine-N-oxide (TMAO) in patients with mitochondrial disorders treated with oral l-carnitine. Mol Genet Metab Rep 2018;15:130–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smits LP, Kootte RS, Levin E et al. Effect of Vegan Fecal Microbiota Transplantation on Carnitine- and Choline-Derived Trimethylamine-N-Oxide Production and Vascular Inflammation in Patients With Metabolic Syndrome. J Am Heart Assoc. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kruger R, Merz B, Rist MJ et al. Associations of current diet with plasma and urine TMAO in the KarMeN study: direct and indirect contributions. Molecular nutrition & food research 2017;61. [DOI] [PubMed] [Google Scholar]
- 129.Cho CE, Taesuwan S, Malysheva OV et al. Trimethylamine-N-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition: A randomized controlled trial. Molecular nutrition & food research 2017;61. [DOI] [PubMed] [Google Scholar]
- 130.Lajous M, Willett WC, Robins J et al. Changes in fish consumption in midlife and the risk of coronary heart disease in men and women. Am J Epidemiol 2013;178:382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mozaffarian D, Lemaitre RN, Kuller LH et al. Cardiac benefits of fish consumption may depend on the type of fish meal consumed: the Cardiovascular Health Study. Circulation 2003;107:1372–7. [DOI] [PubMed] [Google Scholar]
- 132.Yazdekhasti N, Brandsch C, Schmidt N et al. Fish protein increases circulating levels of trimethylamine-N-oxide and accelerates aortic lesion formation in apoE null mice. Molecular nutrition & food research 2016;60:358–68. [DOI] [PubMed] [Google Scholar]
- 133.Dietary Reference Values for choline. EFSA Journal 2016;14:e04484. [Google Scholar]
- 134.Erickson ML, Malin SK, Wang Z, Brown JM, Hazen SL, Kirwan JP. Effects of Lifestyle Intervention on Plasma Trimethylamine N-Oxide in Obese Adults. Nutrients 2019; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Washburn RL, Cox JE, Muhlestein JB et al. Pilot Study of Novel Intermittent Fasting Effects on Metabolomic and Trimethylamine N-oxide Changes During 24-hour Water-Only Fasting in the FEELGOOD Trial. Nutrients 2019; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proceed Nat Acad Sciences USA. 2012;109:21307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhu Y, Jameson E, Crosatti M et al. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proceed Nat Acad Sciences USA. 2014;111:4268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang Z, Roberts AB, Buffa JA et al. Non-lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell 2015;163:1585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.De Filippis F, Pellegrini N, Vannini L et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016; 65:1812–1821. [DOI] [PubMed] [Google Scholar]
- 140.Guasch-Ferre M, Hu FB, Ruiz-Canela M et al. Plasma Metabolites From Choline Pathway and Risk of Cardiovascular Disease in the PREDIMED (Prevention With Mediterranean Diet) Study. J Am Heart Association 2017;6:e006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pignanelli M, Just C, Bogiatzi C et al. Mediterranean Diet Score: Associations with Metabolic Products of the Intestinal Microbiome, Carotid Plaque Burden, and Renal Function. Nutrients 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Roberts AB, Gu X, Buffa JA et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nature med. 2018;24:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 2012;9:811–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Morgan XC, Huttenhower C. Chapter 12: Human microbiome analysis. PLoS Comput Biol 2012;8:e1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Karlsson FH, Fak F, Nookaew I et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 2012;3:1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dinakaran V, Rathinavel A, Pushpanathan M, Sivakumar R, Gunasekaran P, Rajendhran J. Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PloS one 2014;9:e105221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yin J, Liao SX, He Y et al. Dysbiosis of Gut Microbiota With Reduced Trimethylamine-N-Oxide Level in Patients With Large-Artery Atherosclerotic Stroke or Transient Ischemic Attack. Journal of the American Heart Association 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Feng Q, Liu Z, Zhong S et al. Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease. Sci Rep 2016;6:22525. [DOI] [PMC free article] [PubMed] [Google Scholar]