Abstract
Diarrhoeagenic Escherichia coli (DEC) is a leading cause of infectious diarrhoea worldwide. In recent years, Escherichia albertii has also been implicated as a cause of human enteric diseases. This study describes the occurrence of E. coli pathotypes and serotypes associated with enteric illness and haemolytic uremic syndrome (HUS) isolated in Brazil from 2011 to 2016. Pathotypes isolated included enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC) and Shiga toxin-producing E. coli (STEC). PCR of stool enrichments for DEC pathotypes was employed, and E. albertii was also sought. O:H serotyping was performed on all DEC isolates. A total of 683 DEC and 10 E. albertii strains were isolated from 5047 clinical samples. The frequencies of DEC pathotypes were 52.6% (359/683) for EPEC, 32.5% for EAEC, 6.3% for ETEC, 4.4% for EIEC and 4.2% for STEC. DEC strains occurred in patients from 3 months to 96 years old, but EPEC, EAEC and STEC were most prevalent among children. Both typical and atypical isolates of EPEC and EAEC were recovered and presented great serotype heterogeneity. HUS cases were only associated with STEC serotype O157:H7. Two E. albertii isolates belonged to serogroup O113 and one had the stx2f gene. The higher prevalence of atypical EPEC in relation to EAEC in community-acquired diarrhoea in Brazil suggests a shift in the trend of DEC pathotypes circulation as previously EAEC predominated. This is the first report of E. albertii isolation from active surveillance. These results highlight the need of continuing DEC and E. albertii surveillance, as a mean to detect changes in the pattern of pathotypes and serotypes circulation and provide useful information for intervention and control strategies.
Key words: Bacterial infections, diarrhoea, Escherichia coli (E. coli), molecular epidemiology, surveillance
Introduction
Escherichia coli is as one of the most important enteric human pathogens worldwide [1]. Strains of E. coli causing enteric diseases are collectively designated diarrhoeagenic (DEC) and are currently divided into six main categories or pathotypes based on defined virulence attributes. The known DEC pathotypes are named enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), Shiga toxin-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC) [2].
EPEC and EAEC induce diarrhoea through their ability to adhere to host intestinal mucosa, leading to the formation of attaching and effacing (A/E) lesions in the case of EPEC and the aggregative adhesion (AA) pattern in the case of EAEC [2, 3]. Genes such as eae for A/E lesion and aaf (AA fimbriae) for AA, among others, are responsible for the production of these featured adhesion phenotypes [3, 4]. As pathogenic groups both EPEC and EAEC are subdivided in typical and atypical strains. For EPEC, this division is based upon the presence of EAF plasmid (pEAF) in typical (tEPEC) strains and its absence in atypical (aEPEC) ones [4]. The pEAF contains in its structure an operon termed bfp, which is responsible for the production of a type IV pilus named bundle-forming pilus (BFP). BFP is thought to be involved in bacteria to bacteria interactions during the host colonization by EPEC [4]. The occurrence of gene aggR defines typical EAEC strains while atypical EAEC are devoid of this marker [2]. Gene aggR is regarded as a major transcriptional regulator of many of the genes responsible for EAEC virulence factors production [3]. STEC and ETEC damage the host mainly by elaborating and secreting toxins [2]. STEC produces Shiga toxins (Stx). There are two distinct Stx types, Stx1 and Stx2 [4] with 10 subtypes, 1a, 1c and 1d for Stx1, and 2a, 2b, 2c, 2d, 2e, 2f and 2g for Stx2 [5]. ETEC produces thermolabile (LT) and thermostable (ST) enterotoxins. Both LT and ST toxins can also be divided into the distinct antigenic types LT-I and LT-II and ST-I and STII. Furthermore, ST-I may present human (STh) and porcine (STp) variant forms [2]. EIEC phenotypically resemble the genus Shigella. They are capable of invading the host intestinal mucosa and this invasive behaviour relies on a complex array of effector molecules which are employed by the bacteria in order to penetrate, evade immune response and replicate within intestinal cells [5]. The consequent inflammatory response triggered against the invasion process leads to the damage of the intestinal epithelia, characteristic of the bacillary dysentery [2].
In addition to E. coli, another species within the genus Escherichia, E. albertii, can also be a human pathogen. E. albertii was isolated for the first time from a diarrhoeic child in Bangladeshi and misidentified as Hafnia alvei [6]. Currently, E. albertii is considered an ‘emerging’ human enteric pathogen. Similarly to EPEC, E. albertii also harbours the eae gene and thus may produce A/E lesions. Some isolates may possess additional virulence determinants like cytolethal distending and Stx toxins [7].
EPEC, EAEC and ETEC are leading bacterial causes of acute childhood diarrhoea worldwide [8]. EPEC and EAEC have also been implicated in prolonged diarrhoeal diseases and ETEC along with EAEC are agents of the so-called ‘traveller diarrhoea’. On the other hand, STEC strains have been linked with large outbreaks of diarrhoea, and with the occurrence of haemorrhagic colitis and haemolytic uremic syndrome (HUS) [9].
In Brazil, the presence of DEC strains has been investigated in young children, in studies conducted at specific geographic locations [10]. However, there are no reports assessing the occurrence of DEC pathotypes from official surveillance programmes and involving patients from all age groups. Given the heterogeneous nature of DEC strains and their ability to emerge in new pathogenic forms through the gain or loss of genetic material [11], monitoring their virulence traits is of great utility as it can inform on outbreak detection. In order to provide useful epidemiologic data on the occurrence of DEC in Brazil, the present study aimed to describe the pathotypes and serotypes of E. coli and E. albertii strains associated with human infections.
Material and methods
Bacterial strains
The Brazilian Reference Laboratory for E. coli enteric infections Adolfo Lutz Institute (IAL) receives clinical isolates biochemically characterized as E. coli from several regional and local public health laboratories for molecular pathotype identification and serotyping. From January of 2011 to December of 2016, E. coli isolates representing 5047 cases of human infection, including two cases of HUS, were sent to our laboratory for this purpose. Of these, 82 cases had been previously analysed during the investigation of outbreaks of diarrhoea in the years of 2012 and 2013 [12]. These cases were also included in this study as they contribute to the total cases recorded in the period of the present study. The cases encompassed subjects of all age groups. Due to the fact that commensal E. coli is the predominant facultative anaerobe in the human gut, for the identification of diarrhoeagenic strains, it is necessary to evaluate more than one E. coli-like colony from the same patient. In our laboratory, five to 10 E. coli colonies from each patient are routinely received for the characterization of DEC-specific virulence markers. If more than one colony of the same pathotype is found to be positive, only one colony is considered in each case. In the present study, cases of mixed infection (two distinct DEC pathotypes occurring in the same patient) were not considered. This study also employed reference strains serving as positive controls for each of the following DEC pathotypes: EPEC (E2369/48), EAEC (17-2), ETEC (H10407), STEC/EHEC (EDL933), EIEC (Shigella flexneri, CDC2a). The commensal E. coli K12:H5 served as a negative control for molecular and phenotypic procedures.
DEC pathotypes investigation
Screening for specific virulence genes (Table 1) defining the five most relevant DEC pathotypes (EPEC, EAEC, STEC, ETEC and EIEC) was performed by a multiplex PCR assay. For EPEC, the eae gene which is located in the pathogenicity island locus of enterocyte effacement (LEE) and is responsible for the production of the adhesin intimin was employed. For EAEC, the aatA gene encoding a protein related to an ATP-binding cassette transport system was used. For STEC, genes stx1 and stx2, which are bacteriophage-borne and related to the production of the Stx1 and Stx2 toxins respectively, were chosen. For ETEC, we used ltA and stA related to LT and ST toxins production, and for EIEC, the detection target was ipaH gene, which is associated with the invasion plasmid antigen H. Primers sequences and amplification parameters employed in the assays are described in Table 1. Template DNA for PCR reactions was produced by boiling bacterial suspensions from individual E. coli colonies cultivated on Tryptic Soy agar. After bacterial lysates preparation, five to 10 colonies from each patient were pooled and tested. If a given pool was positive, individual colonies forming this pool were retested with primers for the corresponding amplified gene. If a positive result was achieved, the corresponding colony was confirmed as positive.
Table 1.
Primer sequences, target genes and amplification conditions employed in multiplex and individual PCR assays for characterizing DEC strains analysed in this study
| Target gene (product/related DEC pathotype) | Primers sequences (5′–3′) | Amplification conditions | Amplicon size | Reference |
|---|---|---|---|---|
| stx1 (Shiga toxin type I/STEC) | ATAAATCGCCATTCGTTGACTAC AGAACGCCCACTGAGATCATC |
95 °C 5′, 95 °C 40″, 58 °C 1′, 72 °C 2′ (40 cycles) | 188 | [13] |
| stx2 (Shiga toxin type II/STEC) | GGCACTGTCTGAAACTGCTCC TCGCCAGTTATCTGACATTCTG |
255 | ||
| eae (intimin/STEC and EPEC) | GACCCGGCACAAGCATAAGC CCACCTGCAGCAACAAGAGG |
384 | ||
| ipaH (protein associated with pINV plasmid/EIEC) | CTCGGCACGTTTTAATAGTCTGG GTGGAGAGCTGAAGTTTCTCTGC |
917 | ||
| ltA (thermo-labile toxin/ETEC) | GGCGACAGATTATACCGTGC CGGTCTCTATATTCCCTGTT |
450 | ||
| stA (thermostable toxin/ETEC) | ATTTTTMTTTCTGTATTRTCTT CACCCGGTACARGCAGGATT |
190 | ||
| aatA (protein associated with an ATP-binding cassette transporter system/EAEC) | CTGGCGAAAGACTGTATCAT CAATGTATAGAAATCCGCTGTT |
630 | ||
| bfpA (Bfp fimbriae/EPEC) | CAATGGTGCTTGCGCTTGCT GCCGCTTTATCCAACCTGGT |
95 °C 5′, 94 °C 1′, 56 °C 2′, 72 °C 1′ (30 cycles) | 324 | |
| aggR (transcriptional virulence regulator/EAEC) | CTAATTGTACAATCGATGTA ATGAAGTAATTCTTGAAT |
95 °C 5′, 94 °C 1′, 40 °C 1′, 72 °C 1′ (30 cycles) | 308 | [14] |
| sth (human variant of ETEC thermostable toxin/ETEC) | TTCACCTTTCCCTCAGGATG CTATTCATGCTTTCAGGACCA |
95 °C 5′, 94 °C 30″, 52 °C 30″, 72 °C 1′ (35 cycles) | 120 | [15] |
| stp (porcine variant of ETEC thermostable toxin/ETEC) | TCTTTCCCCTCTTTTAGTCAG ACAGGCAGGATTACAACAAAG |
166 |
E. albertii investigation
E. albertii was investigated by a triplex PCR assay recently described by Lindsey et al. This PCR targets cyclic di-GMP regulator gene (cdgR), DNA-binding transcriptional activator of cysteine biosynthesis gene (EAKF1_ch4033) and palmitoleoyl-acyl carrier protein-dependent acyltransferase gene (EFER_0790) allowing discrimination among E. coli, E. albertii and E. fergusonii [16].
Shiga toxin genes (stx) subtyping
Strains presenting stx1 and/or stx2 genes were subjected to stx subtyping by PCR employing the primers and amplification conditions proposed by Scheutz et al. [17].
Identification of typical and atypical EPEC/EAEC strains and ETEC ST toxin gene (st) variants
Strains presenting eae and aatA genetic markers were further investigated for bfp and aggR genes (Table 1) defining typical EPEC and EAEC, respectively. Strains negative for these genes were classified as atypical EPEC/EAEC. ST toxin gene (st)-positive ETEC strains were submitted to an additional duplex PCR (Table 1) in order to investigate the presence of human and porcine variants.
Phenotypic differentiation between EIEC and Shigella strains
Given that ipaH genetic marker can be present in both EIEC and Shigella, and the possible occurrence of cross-reactivity among serogroups of these two bacteria in serological tests, all the strains positive for ipaH gene in PCR assays were submitted to an extended biochemical profiling [18]. Only strains positive for citrate, mucate and sodium acetate utilization were considered as EIEC.
Serotyping
Strains classified in any of the DEC pathotypes investigated by PCR were O:H serotyped by tube agglutination assays [18] employing absorbed somatic (O1-O188) and flagellar antisera (H1-H56) produced at IAL. Non-motile ETEC strains of serogroup O6 were subjected to PCR-RFLP in order to identify the allelic forms of their fliC genes [19].
Cytotoxicity assays
Strains harbouring stx genes were confirmed as STEC in cytotoxic assays employing cultured Vero cells [20].
Statistical analyses
The χ2 test was employed to test the hypothesis that the distribution of each pathotype was not homogeneous among the distinct age groups of patients. The analysis was performed with SAS 9.3 (SAS Institute, Cary, NC). P-value of <0.05 was considered to indicate statistically significant differences.
Results and discussion
DEC strains are considered major aetiological agents of diarrhoeal diseases in Brazil, and worldwide [1, 10, 21]. Nevertheless, updated information on DEC circulation in Brazilian settings is not currently available. Patterns in the circulation of diarrhoeagenic pathotypes and serotypes tend to change over time and may vary between different countries. Therefore, the primary aim of this study was to describe the occurrence of pathotypes and serotypes of DEC isolated from sporadic and outbreak cases of acute diarrhoea and HUS, during a period of 6 years of active epidemiological surveillance, performed in different Brazilian states. However, among diarrhoeagenic eae-harbouring E. coli-like colonies, we identified 10 E. albertii isolates, and the objective of this study was extended to encompass the analysis of such isolates.
A total of 693 (13.7%) cases were positive for DEC or E. albertii. DEC strains representing one of the five major pathotypes were detected as the sole enteric pathogen in 683 (13.5%) cases. E. albertii could be found in 10 (0.2%) of the total cases. The frequency of DEC strains reported in the present study is similar to previously reported data for China and Nigeria [22, 23], but lower than that reported in Mexico [24]. However, the real prevalence of DEC in Brazil could be greater, since in a large number of diarrhoeal cases reported, including outbreaks, the aetiologic agents are not identified due to insufficient epidemiological investigation or technical limitations. The reliable classification of DEC into distinct pathotypes requires the use of molecular tools. Since many local public health laboratories in Brazil are not adequately equipped to perform molecular techniques, most DEC infections are probably missed.
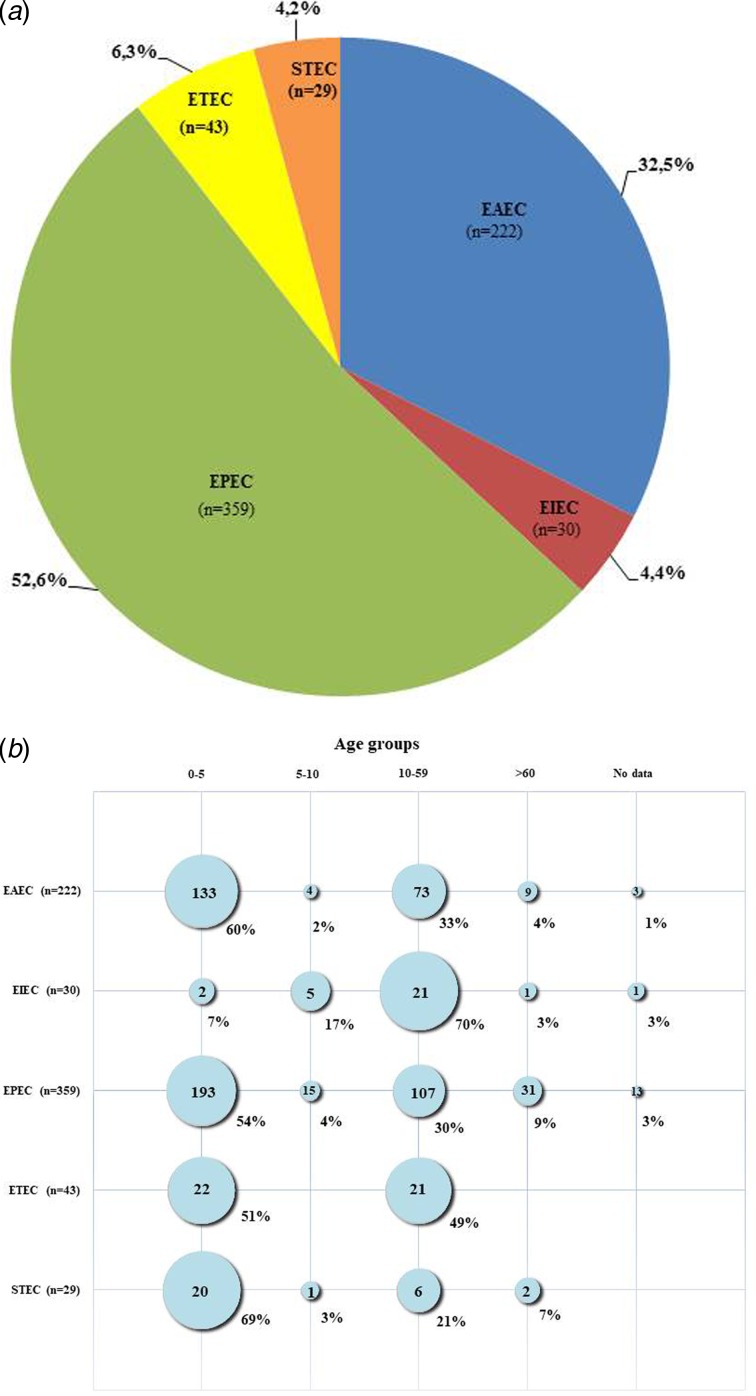
Figure 1a shows the distribution of the different DEC pathotypes among positive DEC strains in this study. The most frequent pathotype was EPEC, found in 359 (52.6%) of the positive DEC cases, followed by EAEC present in 222 (32.5%) of the cases. ETEC, EIEC and STEC were identified in 43 (6.3%), 30 (4.4%) and 29 (4.2%) of the positive cases, respectively. By comparing current results with studies conducted earlier in Brazil, two important differences were noticed: in prior years, EAEC strains were found to be more frequent than EPEC [10, 21], but presently, the occurrence of EPEC was higher than EAEC. In addition, according to previous reports, STEC and EIEC pathotypes were not found or were rarely diagnosed in cases of diarrhoea [10]. In this study, however, both pathotypes were found, albeit at lower frequencies compared with EPEC and EAEC. Our findings support the suggestion that there was a shift in the pattern of circulation of EAEC and EPEC strains in Brazil in recent years. Previously, EAEC were most common but EPEC have become predominant. However, the differences between the results of this study and earlier Brazilian studies may be due to the focus of earlier studies on specific regions and on children under 5 years of age [10]. So, the data they provided regarding the circulation of DEC pathotypes although useful could have been biased by local factors.
Fig. 1.
(a) Pathotypes among 683 cases of enteric infection caused by diarrhoeagenic Escherichia coli (DEC) in Brazil during the years of 2011–2016. (b) Occurrence of pathotypes in different age groups of patients affected by DEC strains in Brazil during the years of 2011–2016.
Figure 1b shows the age distribution for all DEC-positive samples collected from Brazilian patients ranging from 3 months to 96 years old. Most (370; 54%) of the DEC strains were isolated from individuals aged up to 5 years old. However, an analysis of the occurrence of the different pathotypes by age groups showed some differences regarding individual pathotypes. Most EPEC, EAEC and STEC strains were isolated from patients aged <5 years old, whereas most EIEC strains occurred among those aged >10 years old (22; 73%). ETEC infections occurred almost equally in children and adults, being found in 22 (51%) of cases involving children younger than 5 years old and in 21 (49%) cases of individuals older than 10. This pathotype did not occur in subjects older than 60. Statistical analyses demonstrated that DEC pathotypes were not equally distributed among the distinct age categories (P < 0.001), with the exception of ETEC, which was equally distributed between the two age groups from which this pathotype was isolated (P > 0.05). Enteric infections affecting young children may have serious negative consequences, so in most studies [1], including studies performed in Brazil [10, 21], this population is preferentially targeted. There is evidence that frequent and persistent infections due to DEC can lead to impairments in physical and cognitive development [25]. Moreover, age is a risk factor for HUS development after STEC infections, and children <5 years old are considered to be at greater risk [26]. The two laboratory-confirmed HUS cases in this study involved patients aged <5 years. Therefore, considering that infectious diarrhoea more often affects young children, and can also be more detrimental to them, we advise that this group of patients must receive priority for diagnosis and intervention measures.
All the identified EPEC strains of this study, except one, were classified as atypical (aEPEC), as they lacked bfp gene. The only typical (tEPEC) we found was a strain belonging to serotype O157:H39, isolated in 2011 from a child. Since the original description of EPEC in the middle of 1940s [27], tEPEC was the leading cause of childhood diarrhoea. However, in the 1990s, for undetermined reasons, a decline in the incidence of tEPEC was observed worldwide with concomitant rise in the incidence of aEPEC [28, 29], which is nowadays by far more prevalent than tEPEC in many locations. This trend has also been observed in Brazil [29], however, care should be taken in analysing this phenomena, as previously the identification of EPEC was based solely in serogroup determination and the presence of bfp gene was not routinely sought. Atypical EPEC infections affect both children and adults, and have been linked to acute, persistent and outbreaks of diarrhoeal diseases in several countries, including Brazil [12].
Among the aEPEC strains of this study, 86 serogroups were identified and their association with distinct flagellar antigens resulted in 96 different serotypes. The diversity of serogroups and serotypes found among aEPEC strains in our study is shown in Table 2. As it can be noted, no predominant serotype was identified in the period analysed, although some specific ones like O126:H19 and O33:H34 were found more often than the others. Moreover, we also observed the presence of serotypes as O145:HNM, O55:H7, O63:H6 and O26:H11/HNM that are frequently associated with STEC pathotype, raising the speculation that these aEPEC could actually represent strains that were originally STEC before loss of stx genes, which are bacteriophage-borne [30]. There were serotypes such as O39:HNM that have already been linked with EPEC diarrhoeal outbreaks [31]. Serotype O2:H16 in particular has been reported as an agent of aEPEC outbreaks in Brazil [12]. The finding of a great diversity of serotypes among aEPEC in this study is in agreement with other studies [32], demonstrating the heterogeneous nature of aEPEC in terms of antigenic and virulence features. It has been suggested that not all aEPEC strains are in fact pathogenic and many human subjects can be asymptomatic carriers of the bacteria [4]. Currently, we cannot determine whether all the aEPEC serotypes circulating in our settings are indeed relevant in clinical and epidemiological terms. However, it is important to continue monitoring aEPEC strains and to employ whole genome sequencing approaches to uncover the most virulent and potentially epidemic clones in order to clarify questions regarding aEPEC virulence potential.
Table 2.
O:H antigenic types (serotypes) found among 683 DEC strains isolated between 2011 and 2016 in Brazil
| DEC pathotype (total) | Serotypes (no. of isolates) | |
|---|---|---|
| EPEC (359) | O2:H16 (6); O2:H40 (1); O2:H49 (1); O2:HNM (1); O3:H38 (2); O4:HNM (1); O7:HNM (1); O8:H10 (1); O8:H19 (1); O11:H2 (1); O11:H49 (1); O21:H21 (5); O22:H2 (1); O23:HNM (1); O25:HNM (1); O26:H11 (4); O26:H6 (1); O26:H8 (1); O26:HNM (4); O33:H34 (11); O34:H4 (2); O34:HNM (1); O35:H19 (5); O37:H45 (1); O37:HNM (3); O39:H9 (2); O43:H2 (1); O45:HNM (3); O49:H10 (6); O49:H16 (1); O49:H9 (1); O49:HNM (2); O51:H40 (4); O51:HNM (2); O55:H7 (7); O56:H6 (2); O63:H40 (1); O63:H6 (6); O63:HNM (2); O66:H21 (1); O70:H40 (2); O70:H8 (1); O71:H19 (2); O71:H49 (3); O71:HNM (2); O73:H18 (1); O76:H10 (1); O76:HNM (1); O79:H2 (1); O80:H2 (2); O81:H31 (1); O85:H4 (2); O86:H18 (1); O86:H21 (1); O86:HNM (1); O87:H10 (1); O88:H25 (5); O88:H8 (3); O88:HNM (5); O88:HNT (1); O91:H23 (3); O96:H7 (2); O98:HNM (1); O101:H33 (1); O101:HNM (1); O102:H19 (1); O103:H4 (2); O106:HNM (1); O107:H40 (1); O108:H21 (2); O108:H9 (2); O108:HNM (2); O109:H21 (6); O109:H23 (2); O109:H25 (1); O109:HNT (1); O111:H38 (1); O111:H8 (2); O112:H19 (2); O113:H19 (1); O114:HNM (3); O115:H38 (1); O117:HNM (1); O118:HNT (2); O118:HR (3); O119:H21 (1); O119:HNM (2); O121:HNM (1); O123:H19 (6); O123:HNM (2); O124:H4 (2); O125:H5 (1); O125:H6 (1); O126:H19 (12); O127:H21 (2); O127:H40 (3); O127:H45 (1); O128:H2 (2); O129:H11 (1); O131:H46 (2); O132:H34 (3); O132:H6 (1); O132:H8 (1); O133:HNM (1); O136:HNM (1); O137:H6 (3); O142:H34 (2); O144:HNM (1); O145:H34 (5); O145:HNM (7); O145:HNT (1); O145:HR (1); O146:H21 (1); O15:H1 (1); O153:H12 (1); O153:H21 (1); O153:H31 (1); O153:H7 (2); O156:H1 (3); O157:H16 (4); O157:H39a (1); O157:HNM (1); O160:H14 (1); O160:H19 (3); O161:H19 (3); O162:H33 (1); O165:H9 (1); O170:H49 (1); O177:H9 (2); O177:HNM (1); O177:HNM (1); O179:H31 (1); O180:HNM (3); O181:H18 (1); O181:HNM (1); ONT:H10 (2); ONT:H11 (1); ONT:H19 (5); ONT:H2 (1); ONT:H21 (7); ONT:H23 (1); ONT:H30 (1); ONT:H4 (3); ONT:H40 (4); ONT:H43 (1); ONT:H49 (2); ONT:H5 (1); ONT:H6 (5); ONT:H8 (1); ONT:H9 (1); ONT:HNM (7); ONT:HNT (5); ONT:HR (1); OR:H1 (1); OR:H10 (2); OR:H16 (2); OR:H19 (5); OR:H21 (4); OR:H23 (1); OR:H25 (1); OR:H31 (1); OR:H33 (3); OR:H4 (2); OR:H40 (1); OR:H49 (1); OR:HNM (2); OR:HR (1) | |
| EAEC | Typical (187) | O3:H2 (3); O5:H10 (2); O9:H10 (3); O11:H18 (1); O15:H18 (4); O15:H2 (9); O20:H30 (1); O21:H2 (4); O25:H4 (1); O38:H25 (2); O39:H9 (1); O44:H18 (2); O55:H21 (1); O59:H19 (2); O59:HNM (2); O68:H1 (1); O73:H1 (4); O73:H18 (4); O73:H33 (1); O73:HNM (1); O82:H10 (1); O84:HNM (1); O86:H2 (4); O99:H4 (1); O99:H6 (3); O104:H4 (3); O106:H18 (1); O106:HNM (1); O114:H10 (1); O131:H25 (1); O138:H48 (1); O153:H2 (9); O153:H28 (1); O153:HNM (2); O155:H19 (2); O168:HNM (4); O175:H23 (1); O175:H28 (11); O176:H33 (5); O176:H34 (1); O179:HNM (1); O181:H28 (2); ONT:H1 (2); ONT:H10 (16); ONT:H18 (3); ONT:H2 (2); ONT:H21 (2); ONT:H33 (1); ONT:H4 (2); ONT:H9 (1); ONT:HNM (9); OR:H1 (1); OR:H10 (2); OR:H17 (1); OR:H18 (7); OR:H2 (10); OR:H21 (1); OR:H25 (2); OR:H33 (1); OR:H4 (2); OR:H6 (1); OR:HNM (15) |
| Atypical (35) | O11:H18 (1); O15:H2 (1); O38:H25 (1); O43:H2 (1); O55:H25 (2); O70:H8 (1); O73:H18 (1); O80:H10 (3); O81:H27 (1); O139:H19 (1); O170:H49 (1); O175:H28 (1); ONT:H32 (2); ONT:H33 (1); ONT:HNM (2); OR:H10 (3); OR:H2 (1); OR:H21 (1); OR:H25 (2); OR:H33 (3); OR:H35 (2); OR:H45 (1); OR:HNM (2) | |
| STEC (29) | O8:H19 (1); O24:H4 (1); O26:H11 (1); O71:H8 (1); O91:H14 (1); O100:HNM (1); O103:HNM (1); O111:H11 (1); O111:H8 (2); O111:HNM (4); O118:H16 (1); O123:H2 (1); O123:HNM (3); O145:HNM (1); O153:H21 (1); O153:H7 (1); O157:H7 (2); O177:HNM (1); O178:H19 (1);ONT:H19 (1); ONT:H46 (1); OR:HNM (1) | |
| ETEC (43) | O6:H16 (22); O6:HNM (3); O25:HNM (6); O64:H21 (1);O109:H19 (1); O159:H17 (1); O159:H21 (1); O165:H15 (1); O166:H15 (2); ONT:H10 (1); ONT:H2 (1); ONT:H8 (1); ONT:HNM (1); OR:H31 (1) | |
| EIEC (30) | O2:HNM (1); O18:H31 (1); O121:HNM (7); O124:H30 (4); O124:HNM (2); O132:H21 (8); O132:HNM (1); O135:HNM (1); O144:HNM (3); ONT:HNM (2) | |
OR, O rough; ONT, O non-typeable; HNM, H non-motile; HNT, H non-typeable.
Typical EPEC (bfp+).
In our analysis, strains carrying aggR gene, thus classified as typical EAEC, were 84% (187/222) of the total of EAEC-positive strains, while 16% (35/222) of the EAEC in this study were atypical and lacked the gene. These findings are similar to previous reports that the majority of the EAEC strains linked to enteric illness harbour aggR [33]. As the aggR gene product regulates the expression of most of the currently identified virulence factors of EAEC, in some studies, it is suggested that only typical EAEC are pathogenic for humans [3]. However, there has been a report implicating atypical EAEC with diarrhoeal cases [34]. The fact that in this study we did not find any other bacterial or viral enteropathogens in samples positive for atypical EAEC corroborates this previous report and is evidence for a role for atypical EAEC in enteric illness.
EAEC strains in this study included 42 distinct O:H serotypes, 35 of them associated with typical isolates and 12 associated with atypical EAEC (Table 2). Serotypes O11:H18, O15:H2, O175:H28, O38:H25, O73:H18 were common to both typical and atypical strains, and one can speculate that these aEAEC belonging to these serotypes of typical EAEC could have lost aggR-bearing plasmid. Serotypes O175:H28, O15:H2 and O153:H2 were more commonly found among typical EAEC. However, the majority of the EAEC strains of this study, irrespective of the fact they were typical or atypical, could not have their O antigens determined due to roughness or absence of antisera reactivity. The self-agglutinating nature of EAEC strains together with serotype diversity [35, 36] limits the use of serological techniques in outbreaks tracing and laboratory surveillance. Nevertheless, some interesting findings regarding antigenic features of EAEC of this study could be observed. The H2 antigen was frequently associated with typical EAEC. Some of the serotypes found in this study have been reported in other studies conducted in Brazil, but serotypes O175:H28 and O15:H2 were not previously reported, though they were the most common serotypes found in the period of this study [37, 38]. Strains of serogroup O15 with H18 antigen had been reported previously in Brazil as atypical EAEC [37]. In this study, O15 serogroup was found in combination with H2 flagellar type and mostly among typical EAEC.
ETEC is a major cause of moderate-to-severe diarrhoea in developing countries especially in Asia [39].This pathotype is also an important enteric pathogen in South America and earlier studies in Brazil have reported ETEC infections, including outbreaks, in different regions [10, 21, 40]. In the present study, ETEC was isolated in 43 (6.3%) of the total of diarrhoeal cases analysed. Our results confirm that although less frequent than other DEC pathotypes such as EPEC and EAEC, ETEC are still responsible for causing enteric illness in our country, and must therefore continue to be considered in the list of enteric pathogens to be sought for the diagnosis of diarrhoeal diseases. In this study, we found the profile lt+/st+ as the most common toxigenic genotype among ETEC-positive isolates, being present in 56% (24/43) of these strains, while 44% (19/43) of the strains harboured only LT enterotoxin-related gene lt. None of the strains studied carried st gene alone. It has been reported that st or st/lt carrying ETEC strains, rather than lt only harbouring ETEC, are more often associated with moderate-to-severe diarrhoea and a higher risk of death in young children [1]. In the st+ isolates, sth gene variant was carried by 21 strains, while only one strain possessed the stp variant. ST enterotoxin variants STh and STp can both induce diarrhoea in humans; however, the human variant is considered to be more relevant in clinical terms due to its higher prevalence when compared with STp [41]. Three ETEC strains did not give any result in relation to the st gene variants searched. No information regarding the toxigenic profiles of ETEC isolated previously in Brazil is available; therefore, our findings although derived from a small number of strains are the only data available on this topic. Examination of the 43 ETEC isolates for O:H antigens demonstrated the occurrence of 14 distinct serotypes (Table 2). However, 25 (58%) of these strains belonged to the single serotype O6:H16, including 22 motile strains (H16 antigens expressed) and three non-motile strains (H16 type was determined by PCR-RFPL analysis of fliC genes). ETEC of serotype O6:H16 is of worldwide occurrence, being one of the most common serotypes associated with ETEC infections in humans [2].
All the strains positive for stx genes in this study were phenotypically confirmed as STEC in Vero cell cytotoxic assays. We had 19 strains (65.5%) carrying stx1, while nine (31%) carried only stx2 and one strain carried both stx1 and stx2 (Table 3). Previous studies characterizing STEC of clinical origin in Brazil have also reported that most of the isolates harboured only stx1 and were from cases of acute non-complicated diarrhoea [42]. Subtyping of stx genes revealed that stx1a allele was carried by all the stx1-positive strains, except one that had stx1d (Table 3). In relation to stx2 subtypes, we encountered the allele 2a in association with 2c, 2d or 2e in five of the stx2-positive STEC, while subtypes 2c and 2e were found alone in four strains (Table 3). Subtypes 2b and 2g were not present. Stx2 and subtypes 2a, 2c and 2d are more often linked with complicated STEC infections and their association with some specific O serogroups and adherence factors can be a predictor of greater probability of HUS [43–45]. In this study, we were able to demonstrate that these three most problematic stx2 subtypes were found in most of stx2-positive strains. However, STEC strains producing Stx1 can also cause HUS [45], so the possibility that stx1a-bearing Brazilian isolates can be responsible for more complicated infections exist and for this reason they must be carefully monitored.
Table 3.
Serotypes and stx genotypes among 29 STEC strains recovered from human infections in Brazil from 2011 to 2016
| Strain | Year of isolation | Serotype | stx genotype | Presence of eae | Clinical condition |
|---|---|---|---|---|---|
| 179/11 | 2011 | OR:HNM | 2c | + | AD |
| 61/12 | 2012 | O100:HNM | 2e | − | AD |
| 75/12 | O111:HNM | 1a,2a | + | AD | |
| 169/12 | O177:HNM | 2c | + | AD | |
| 343/12 | O153:H21 | 1a | − | AD | |
| 359/12 | O111:H8 | 1a | + | AD | |
| 611/12 | O111:HNM | 1a | + | AD | |
| 340/13 | 2013 | O157:H7 | 2a,2c | + | HUS |
| 377/13 | O24:H4 | 1a | − | AD | |
| 423/13 | O118:H16 | 1a | + | AD | |
| 444/13 | O103:HNM | 1a | + | AD | |
| 502/13 | O111:H8 | 1a | + | AD | |
| 605/13 | O71:H8 | 1a | + | AD | |
| 438/14 | 2014 | O111:H11 | 1a | + | AD |
| 504/14 | O145:HNM | 1a | + | AD | |
| 544/14 | O153:H7 | 1d | − | AD | |
| 579/14 | O123:HNM | 1a | + | AD | |
| 589/14 | O91:H14 | 1a | − | AD | |
| 672/14 | O8:H19 | 2a,2d | − | AD | |
| P059-9/14 | 026:H11 | 1a | + | BD | |
| 302/15 | 2015 | ONT:H46 | 2a,2d | − | AD |
| 516/15 | O123:H2 | 1a | + | AD | |
| 768/15 | O111:HNM | 1a | + | AD | |
| 831/15 | O157:H7 | 2a,2c | + | HUS | |
| 927/15 | O123:HNM | 1a | + | AD | |
| P001/15 | ONT:H19 | 2a, 2e | − | AD | |
| 254/16 | 2016 | O123:HNM | 1a | + | AD |
| 583/16 | O178:H19 | 2c | − | AD | |
| 811/16 | O111:HNM | 1a | + | AD |
AD, acute diarrhoea; BD, bloody diarrhoea; HUS, haemolytic uremic syndrome.
Twenty (69%) of the STEC strains herein analysed possessed eae gene, while nine strains (31%) lacked this marker (Table 3). This indicates that the majority of the human STEC infections in Brazil, in the period covering this study, were caused by strains dotted with the potential ability to express both Shiga toxin and A/E phenotypes. The potential to form A/E lesions by STEC isolates is regarded as an additional risk factor in the clinical outcome of STEC diseases, as there is a higher risk of HUS development [9]. In fact, most of the HUS cases registered in Brazil [46] including the two cases analysed in this study were caused by strains carrying stx2 and eae. In this study, STEC strains fell into 15 distinct serotypes (Table 2), and included serotypes of major epidemiological importance such as O157:H7, O111:H8/NM, O26:H11, O145:HNM, O103:HNM, as well as serotypes which have been implicated in human disease, but isolated less frequently [47]. The most frequent serotype presently observed was O111:H8. The same situation was observed in prior years in Brazil where STEC O111 was the most frequent serogroup encountered in human diseases [42]. By comparing the present results with data about STEC serotypes in Brazil spanning the period of 1979–2004, we could note that the diversity of serotypes detected in this study was greater than that observed before. This may be indicative of the efforts that have been made in Brazil to increase the detection of STEC pathogens by employing molecular approaches targeting stx genes, instead of serogroup-based screening by immunological methods, which were largely performed in the past. This change in methodology must continue and should be implemented in the largest possible number of laboratories in Brazil, for the benefit of future surveys addressing STEC infection epidemiology.
Studies describing the occurrence and markers for EIEC circulation are scarce. This is due to the fact that EIEC differentiation from Shigella is often problematic as these two bacteria are closely related and almost identical in terms of genetic contents. Additionally, surface antigens of EIEC and Shigella cross-react, so serological tests may not be a suitable option. In fact, there is evidence from phylogenetic studies demonstrating that EIEC strains represent intermediate forms in the evolution from commensal E. coli to Shigella [48]. Differentiation among EIEC and Shigella is possible only through extensive biochemical profiling which can be performed solely in reference laboratories. As a consequence, EIEC strains are under-represented in most epidemiological surveys. Therefore, EIEC contribution to the burden of DEC infections is largely overlooked. In this study, phenotypically confirmed EIEC corresponded to 4.4% of DEC strains, showing that this pathotype has a role in community-acquired diarrhoea in Brazil. Serogroups O132, O121 and O124 were the most common, being present in 9/30 (30%), 7/30 (23%) and 6/30 (20%) of strains, respectively, and serotype O132:H21 was the most frequent. Serogroups O121 and O124 are among the most commonly reported among EIEC strains [2], and in this sense, our results only partially agree with previous reports, as in this study the O132 serogroup was the most prevalent. EIEC outbreaks have been reported in other countries [49] and we are currently performing PFGE typing to assess the genetic relatedness among strains of the same serotype isolated in this study.
Ten of the eae-harbouring strains, which had been previously identified in our routine laboratory testing as EPEC, were actually found to be E. albertii. The recognition of E. albertii is challenging in that with few exceptions their biochemical profile and most of the virulence markers resemble DEC pathotypes EPEC and STEC. Only recently genomic approaches have allowed accurate discrimination, reallocating these strains to another taxonomic position [50]. This is the first report on the occurrence of E. albertii from active surveillance of foodborne diseases in Brazil. The majority of the E. albertii were untypeable or rough regarding their somatic antigens, and were non-motile, so their O:H serotypes could not be identified. This is in agreement with previous reports of the antigenic untypeability of E. albertii strains [51]. It is worth mentioning that there is no specific typing scheme for E. albertii and attempts to determine their somatic and flagellar antigens usually employ antisera produced against E. coli strains. This suggests that E. albertii O and H antigens may have distinct characteristics in relation to E. coli antigens. However, there were two exceptions in this study, as two strains reacted with O113 E. coli antisera, but were non-motile, rendering serotype O113:HNM. One of the analysed E. albertii strains in this study was positive for stx2f gene. Production of Stx2f by E. albertii strains has been reported [7] and one can speculate about the potential of these strains to cause more serious diseases. So far, E. albertii appears to represent a small proportion of the diarrhoeagenic strains circulating in Brazilian settings, but even so they must receive attention in surveillance programmes in Brazil and elsewhere, so that it will be possible to determine how their circulation trends will evolve.
We attempted to draw a scenario for DEC strains occurrence in comparison to prior years in Brazil, and with data from other countries. Although several problems were faced, especially related to logistical difficulties in sending bacterial isolates to reference laboratories for analysis, we believe the present study contributes a useful ‘snapshot’ on the aetiology of diarrhoeal diseases caused by DEC strains in our country. Certainly this will be very important for future studies and considering intervention measures. The continuous epidemiological surveillance of food and water transmissible diseases and characterization of DEC strains associated with human infections is essential for the recognition of new patterns of pathogen virulence and circulation. In this regard, it is of paramount importance that public health and clinical laboratories involved in infectious diseases diagnosis and surveillance are capable of correctly recognizing DEC strains.
Acknowledgements
The authors are grateful to Professor J. C. F. Pantoja for assistance with statistical analysis. The authors also acknowledge the work of all the professionals of the State Public Health Laboratories in Brazil involved in the laboratory surveillance of enteric diseases.
Conflict of interest
None.
References
- 1.Kotloff KL et al. (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (The Global Enteric Multicenter Study, GEMS): a prospective, case-control study. The Lancet 382, 209–222. [DOI] [PubMed] [Google Scholar]
- 2.Croxen MA et al. (2013) Recent advances in understanding enteric pathogenic Escherichia coli. Clinical Microbiology Reviews 26, 822–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estrada-Garcia T and Navarro-Garcia F (2012) Enteroaggregative Escherichia coli pathotype: a genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunology and Medical Microbiology 66, 281–298. [DOI] [PubMed] [Google Scholar]
- 4.Hernandes RT et al. (2009) An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiology Letters 297, 137–149. [DOI] [PubMed] [Google Scholar]
- 5.Parsot C (2005) Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiology Letters 252, 11–18. [DOI] [PubMed] [Google Scholar]
- 6.Hyma KE et al. (2005) Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. Journal of Bacteriology 187, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooka T et al. (2012) Clinical significance of Escherichia albertii. Emerging Infectious Diseases 18, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platts-Mills JA et al. (2015) Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). The Lancet Global Health 3, 564–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paton JC and Paton AW (1998) Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clinical Microbiology Reviews 11, 450–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreno ACR et al. (2010) Etiology of childhood diarrhea in the northeast of Brazil: significant emergent diarrheal pathogens. Diagnostic Microbiology and Infectious Diseases 66, 50–57. [DOI] [PubMed] [Google Scholar]
- 11.Bielaszewska M et al. (2007) Aspects of genome plasticity in pathogenic Escherichia coli. International Journal of Medical Microbiology 297, 625–639. [DOI] [PubMed] [Google Scholar]
- 12.Vieira MA et al. (2016) Atypical enteropathogenic Escherichia coli as etiologic agents of sporadic and outbreak-associated diarrhea in Brazil. Journal of Medical Microbiology 65, 998–1006. [DOI] [PubMed] [Google Scholar]
- 13.Peresi JTM et al. (2016) Search for diarrheagenic Escherichia coli in raw kibbe samples reveals the presence of Shiga toxin-producing strains. Food Control 63, 165–170. [Google Scholar]
- 14.Elias WP et al. (2002) Combinations of putative virulence markers in typical and variant enteroaggregative Escherichia coli strains from children with and without diarrhoea. Epidemiology and Infection 129, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodas C et al. (2009) Development of multiplex PCR assays for detection of enterotoxigenic Escherichia coli colonization factors and toxins. Journal of Clinical Microbiology 47, 1218–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindsey RL et al. (2017) Multiplex polymerase chain reaction for identification of Escherichia coli, Escherichia albertii and Escherichia fergusonii. Journal of Microbiological Methods 140, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheutz F et al. (2012) Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. Journal of Clinical Microbiology 50, 2951–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewing WH (1986) Edwards and Ewing's Identification of Enterobacteriaceae, 4th Edn. New York: Elsevier Science Publishing Co., Inc., 536 pp. [Google Scholar]
- 19.Machado J, Grimont F and Grimont PAD (2000) Identification of Escherichia coli flagellar types by restriction of the amplified fliC gene. Research in Microbiology 151, 535–546. [DOI] [PubMed] [Google Scholar]
- 20.Konowalchuk J, Speirs JI and Stavric S (1977) Vero response to a cytotoxin of Escherichia coli. Infection and Immunity 18, 775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bueris V et al. (2007) Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador, Bahia, Brazil. Memorias do Instituto Oswaldo Cruz 102, 839–844. [DOI] [PubMed] [Google Scholar]
- 22.Huang Z et al. (2016) Prevalence and antimicrobial resistance patterns of diarrheagenic Escherichia coli in Shanghai, China. The Pediatric Infectious Disease Journal 35, 1. [DOI] [PubMed] [Google Scholar]
- 23.Ifeanyi CIC et al. (2015) Diarrheagenic Escherichia coli pathotypes isolated from children with diarrhea in the Federal Capital Territory Abuja, Nigeria. Journal of Infection in Developing Countries 9, 165–174. [DOI] [PubMed] [Google Scholar]
- 24.Canizalez-Roman A et al. (2016) Surveillance of diarrheagenic Escherichia coli strains isolated from diarrhea cases from children, adults and elderly at northwest of Mexico. Frontiers in Microbiology 7, 1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrant RL et al. (2008) Malnutrition as an enteric infectious disease with long-term effects on child development. Nutrition Reviews 66, 487–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proulx F, Seidman EG and Karpman D (2001) Pathogenesis of Shiga toxin-associated hemolytic uremic syndrome. Pediatric Research 50, 163–171. [DOI] [PubMed] [Google Scholar]
- 27.Bray J (1945) Isolation of antigenically homogeneous strains of Bacterium coli neopolitanum from summer diarrhea of infantis. The Journal of Pathology and Bacteriology 57, 239–247. [Google Scholar]
- 28.Ochoa TJ et al. (2008) New insights into the epidemiology of enteropathogenic Escherichia coli infections. Transactions of the Royal Society of Tropical Medicine and Hygiene 102, 852–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues J et al. (2004) Reduced etiological role for enteropathogenic Escherichia coli in cases of diarrhea in Brazilian infants. Journal of Clinical Microbiology 42, 398–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bielaszewska M et al. (2007) Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS ONE 2, e1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedberg CW et al. (1997) An outbreak of foodborne illness caused by Escherichia coli O39:NM, an agent not fitting into the existing scheme for classifying diarrheogenic E. coli. The Journal of Infectious Diseases 176, 1625–1628. [DOI] [PubMed] [Google Scholar]
- 32.Tennant SM et al. (2009) Characterisation of atypical enteropathogenic E. coli strains of clinical origin. BMC Microbiology 9, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cennimo D et al. (2009) The prevalence and virulence characteristics of enteroaggregative Escherichia coli at an urgentcare clinic in the USA: a case-control study. Journal of Medical Microbiology 58, 403–407. [DOI] [PubMed] [Google Scholar]
- 34.Itoh Y et al. (1997) Laboratory investigation of enteroaggregative Escherichia coli O Untypeable:H10 associated with a massive outbreak of gastrointestinal illness. Journal of Clinical Microbiology 35, 2546–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weintraub A (2007) Enteroaggregative Escherichia coli: epidemiology, virulence and detection. Journal of Medical Microbiology 56, 4–8. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins C et al. (2006) Genotyping of enteroaggregative Escherichia coli and identification of target genes for the detection of both typical and atypical strains. Diagnostic Microbiology and Infectious Disease 55, 13–19. [DOI] [PubMed] [Google Scholar]
- 37.Regua-Mangia AH et al. (2009) Molecular typing and virulence of enteroaggregative Escherichia coli strains isolated from children with and without diarrhoea in Rio de Janeiro city, Brazil. Journal of Medical Microbiology 58, 414–422. [DOI] [PubMed] [Google Scholar]
- 38.Uber AP et al. (2006) Enteroaggregative Escherichia coli from humans and animals differ in major phenotypical traits and virulence genes. FEMS Microbiology Letters 256, 251–257. [DOI] [PubMed] [Google Scholar]
- 39.Qadri F et al. (2005) Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clinical Microbiology Reviews 18, 465–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vicente ACP et al. (2005) Outbreaks of cholera-like diarrhoea caused by enterotoxigenic Escherichia coli in the Brazilian Amazon rainforest. Transactions of the Royal Society of Tropical Medicine and Hygiene 99, 669–674. [DOI] [PubMed] [Google Scholar]
- 41.Fleckenstein JM et al. (2010) Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes and Infection 12, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaz TMI et al. (2004) Virulence properties and characteristics of Shiga toxin-producing Escherichia coli in São Paulo, Brazil, from 1976 through 1999. Journal of Clinical Microbiology 42, 903–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Usein C-R et al. (2017) Molecular characterisation of human Shiga toxin-producing Escherichia coli O26 strains: results of an outbreak investigation, Romania, February to August 2016. Euro Survaillance 22, 00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bielaszewska M et al. (2006) Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clinical Infectious Diseases 43, 1160–1167. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W et al. (2007) Structural and functional differences between disease-associated genes of enterohaemorrhagic Escherichia coli O111. International Journal of Medical Microbiology 297, 17–26. [DOI] [PubMed] [Google Scholar]
- 46.Souza RL et al. (2011) Hemolytic uremic syndrome in pediatric intensive care units in São Paulo, Brazil. The Open Microbiology Journal 5, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saupe A et al. (2017) Acute diarrhoea due to a Shiga toxin 2e-producing Escherichia coli O8:H19. Journal of Medical Microbiology Case Reports 4, 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng J, Yang J and Jin Q (2009) The molecular evolutionary history of Shigella spp. and enteroinvasive Escherichia coli. Infection, Genetics and Evolution 9, 147–152. [DOI] [PubMed] [Google Scholar]
- 49.Newitt S et al. (2016) Two linked enteroinvasive Escherichia coli outbreaks, Nottingham, UK, June 2014. Emerging Infectious Diseases 22, 1178–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ooka T et al. (2015) Defining the genome features of Escherichia albertii, an emerging enteropathogen closely related to Escherichia coli. Genome Biology and Evolution 7, 3170–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nimri LF (2013) Escherichia albertii, a newly emerging enteric pathogen with poorly defined properties. Diagnostic Microbiology and Infectious Disease 77, 91–95. [DOI] [PubMed] [Google Scholar]